
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 08 November 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.986678
This article is part of the Research Topic Regulation of Alternative Splicing in Plant Stress Responses View all 7 articles
 Tingting Xu†
Tingting Xu† Jingjing Zhang†
Jingjing Zhang† Yiran Liu
Yiran Liu Qiuxin Zhang
Qiuxin Zhang Weiyan Li
Weiyan Li Yueling Zhang
Yueling Zhang Meifeng Wu
Meifeng Wu Tong Chen
Tong Chen Daochun Ding
Daochun Ding Wenyi Wang*
Wenyi Wang* Zemin Zhang*
Zemin Zhang*The formation of leaf color largely depends on the components of pigment accumulation in plastids, which are involved in chloroplast development and division. Here, we isolated and characterized the rice albino leaf 4 (al4) mutant, which exhibited an albino phenotype and eventually died at the three-leaf stage. The chloroplasts in al4 mutant were severely damaged and unable to form intact thylakoid structure. Further analysis revealed that the candidate gene encodes 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE), which participates in the methylerythritol phosphate (MEP) pathway of isoprenoid biosynthesis. We further demonstrated that the mutation at the exon-intron junction site cause alternative splicing factors fail to distinguish the origin of the GT-AG intron, leading to exon skipping and producing a truncated OsIspE in the al4 mutant. Notably, disruption of OsIspE led to the reduced expression of chloroplast-associated genes, including chloroplast biosynthetic and translation related genes and photosynthetic associated nuclear genes (PhANGs). In summary, these findings reveal that OsIspE plays a crucial role in chloroplast biogenesis and provides novel insights into the function of CMK during chloroplast development in rice.
The chloroplast as semi-autonomous organelle, plays an important role in photosynthesis in higher plants (Liu et al., 2021). Defects in chloroplast development can cause abnormal chlorophyll accumulation, such as albino, light green, etiolation, green-revertible yellow, and striped leaves. Leaf color mutations generally affect photosynthetic efficiency in plants, leading to stunted growth even death. Leaf color is a common, easily identifiable phenotype that is considered ideal materials for studying the mechanisms of photosynthesis, chloroplast synthesis and degradation in plants.
Accumulating evidences has revealed that chlorophyll accumulation is regulated by many factors, such as temperature, RNA editing, and splicing. In rice, several pentatricopeptide repeat proteins (PPR) protein have been reported to modulate chloroplast development by regulating RNA editing and splicing, such as OsPPR6 and CDE4 (Tang et al., 2017; Liu et al., 2021). The OsPPR6 gene participates in the editing of ndhB and splicing of ycf3 transcripts, thereby regulating early chloroplast biogenesis in rice (Tang et al., 2017). The cde4 mutant exhibits abnormal chloroplast development and albino seedlings under low temperature conditions, owing to a defect in transcript splicing of the chloroplast genes rpl2, ndhA, and ndhB (Liu et al., 2021). Other genes have also been shown to regulate chloroplast development in different plant species. For example, the AtDPG1 gene encodes a putative chloroplast localized protein, and the dep1 mutant shows a delayed pale-green phenotype in Arabidopsis (Liu et al., 2016a). OsAL1 encodes a sole octotricopeptide repeat protein (OPR) protein, and regulates chloroplast development by coordinating the expression of chloroplast related genes in rice (Zhang et al., 2016). OsAL2 encodes a chloroplast group IIA intron splicing facilitator, and the al2 mutant shows an albino phenotype at the early developmental stage in rice (Liu et al., 2016b). OsCSL1 encodes a MAPK kinase kinase22 (MKKK22), and interacts with OsMKK4. The mutation of OsCSL1 resulted in a chlorosis seedling lethality phenotype (Liang et al., 2022).
Isoprenoids such as chlorophylls, carotenoids, cytokinins, and gibberellins, play an important role in membrane structure, growth, pathogen defense, and photosynthesis in plants (Rodrı´guez-Concepcio´n and Boronat, 2002; Bohlmann and Keeling, 2008). Its precursors are two five-carbon compounds, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). Two pathways are involved in the biosynthesis of IPP and DMAPP, including mevalonic acid (MVA) pathway in the cytoplasm and the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway in plastids. IPP and DMAPP generated by the MVA pathway are used for the synthesis of ubiquinone, sterols, and brassinosteroids (Pulido et al., 2012; Henry et al., 2018). The MEP pathway provides precursor molecules for phylloquinones, gibberellins, and other compounds. Seven enzymatic steps are involved in the MEP pathway (Liu et al., 2018). Genes that participate in the MEP pathway responsible for the leaf color phenotype have been identified in rice (Chen et al., 2018). For example, IspF encodes a 2-C-methyl-D-erythritol-2,4-cyclodiphosphate synthase, which is involved in the MEP pathway. A single nucleotide mutation in IspF caused a yellow-green leaf phenotype in rice (Huang et al., 2018). IspE encodes 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK), and catalyzes 4-diphosphocytidyl-2-C-methyl-D-erythritol (CDP-ME) into 4-diphosphocytidyl-2-C-methyl-D-erythritol-2-phosphate (CDP-MEP). A missense mutation of IspE caused green-revertible yellow leaf phenotype in rice (Chen et al., 2018). Although few studies have revealed the roles of the MEP pathway during chloroplast growth and development, knowledge of the underlying molecular mechanisms remains largely limited.
In the present study, we isolated and characterized a rice mutant albino leaf 4 (al4). The MutMap+ method revealed that the candidate gene encodes IspE, a key enzyme in the MEP pathway. Furthermore, a mutation at the exon-intron junction site in OsIspE led to exon skipping and produced truncated OsIspE in the mutant. Several genes related to chlorophyll synthesis and chloroplast development were significantly down-regulated in the al4 mutant. Our findings suggest that OsIspE is essential for chloroplast biogenesis and development in rice.
The rice (Oryza sativa L.) albino leaf 4 (al4) mutant identified from EMS (Ethyl methyl sulfonate) mutant lines with Xian/Indica rice cultivar Ruanhua2B (RH2B) background. Wild-type (RH2B) and al4 mutant plants were cultivated in the experimental field, located in South China Agricultural University (Guangzhou), during the natural growing season. To examine the albino phenotype of the mutant clearly, the rice plants were grown in the greenhouse under a 12-h-light/12-h-dark cycle at 30°C for laboratory work. No significant differences were observed between al4 mutants grown in the greenhouse and those grown in the experimental field.
Pigment contents of seedlings were measured based on the method of Zhang et al. (2016) with slight modifications. Fresh leaves (approximately 0.1g) were collected and placed in a 10 mL centrifuge tube with 2 mL extraction buffer (ethanol: propanol: H2O=4.5:4.5:1) for 48 h in the dark at 4°C. After centrifuging at 12000 rpm for 5 min, the samples were analyzed measuring the absorbance at 470, 645, and 663 nm. Pigment contents were calculated using the following equations:
The second leaves of the wild-type and al4 mutant plants were cut into small pieces for scanning electron microscopy observation. The pieces were fixed with 2.5% glutaraldehyde in a phosphate buffer (0.1 M, pH 7.4) for 24 h at 4°C. Washed with 0.1 M phosphate buffer for four times, the samples were incubated in 1% (v/v) OsO4 for 4 h. After staining with 1% uranyl acetate overnight, the samples were dehydrated using an ethanol series (30, 50, 70, 85, 95, and 100% ethanol solution) at room temperature, and subjected to incubation of the leaves at each concentration for 15 min. Following the gradient series of epoxy resin (acetone: resin=3:1, 1:1, 1:3, v/v), the samples were infiltrated at each concentration for 3 h, and then the mixtures were incubated in pure Epon812 for 12 h. Subsequently, the samples were embedded in paraffin and cut into thin 60-80nm sections using a microtome. The sections were observed using a transmission electron microscope (Leica EM UC6).
MutMap+ method was used to map OsIspE gene according to Fekih et al. (2013). Briefly, the bulk DNA used for MutMap+ analysis was prepared by mixing DNA of 50 green seedlings (Pool WT) and 50 albino seedlings (Pool MT) from the M4 generation. Subsequently, both bulk DNA samples were subjected to whole-genome sequencing. High-quality clean sequence reads of the mutant pool and WT pool were aligned to the reference genome RHB to enable the identification of SNPs, and the SNP-index and ΔSNP-index were calculated based on the SNP information.
Total RNA was extracted with the RNA extraction kit (TRIgol reagent) (DingGuo, Beijing) based on the manufacturer’s protocol. First strand of cDNA was synthesized using One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing). Quantitative real-time PCR (RT-qPCR) was conducted using the CFX ConnectTM real-time PCR system (Bio-Rad) with SYBR Green Pro Taq HS (AG, GuangZhou) according to the manufacturer’s instructions. The procedure was as follows: 95°C for 2 min, followed by 40 cycles at 95°C for 15 s, 60°C for 30 s and 72°C 30 s, finally extension for 5 min at 72°C. The rice OsUBQ gene was used as an internal control. All primers used in RT−qPCR are listed in Table S1.
To generate OsIspE-cas9 lines, two target sites in the first and sixth exons were designed by CRISPR-GE website online. First target of a 20-bp nucleotide sequence in the first exon was “TGGCTTGCTCCACCCACCTC”, and second target sequence in the sixth exon was “AAGAAGGTCCCTACTGGTGC” (Figure S1). Two sgRNA expression cassette driven by the Zea mays U6a and U3 promoters respectively. Using Golden Gate ligation, two sgRNAs with different target sequences of OsIspE were constructed into a pYLCRISPR/Cas9Pubi-H vector according to the previous methods (Ma et al., 2016), and then transferred into ZH11 by Agrobacterium tumefaciens-mediated transformation. To examine the subcellular localization of the OsIspE protein, the coding region without the stop codon of OsIspE was obtained by PCR from the cDNA of the wild-type (RH2B). The amplified sequences were inserted into the C-terminus of GFP under the control of the Ubi and 35S promoter to generate the pRTV-OsIspE-cGFP and p1305-OsIspE-cGFP vectors. The primers used for vector construction are listed in Table S2.
Rice seeds (ZH11) were treated with 75% ethanol for 1 min, washed with sterile H2O and 5% NaClO for 30 min, and then washed with sterile H2O for four times. The sterilized seeds were placed on 1/2×MS medium and grown in a greenhouse under a 16-h-light/8-h-dark cycle at 30°C for 12~14 days. Rice protoplast isolation was based on the method described by Zhang et al. (2011) with minor modifications. The leaves the seedlings were cut into 0.5 mm strips and submerged in 0.6 M mannitol for 20 min. Afterward, the mannitol solution was replaced with a 10-20 mL enzyme solution and then incubated for 4 h in the dark with gentle shaking at ambient temperature. The enzyme solution was carefully removed and 20 mL W5 medium was added. This was gently shaken for 30 min to release the protoplasts. The protoplasts were filtered through 40 μm nylon meshes into a 50 mL centrifuge tube and centrifuged at 1000 rpm for 10 min at an ambient temperature to collect the protoplast pellet. Thereafter, the protoplasts were washed twice with W5 medium, transferred to several 2 mL centrifugal tubes, and centrifuged at 1000 rpm for 5 min. Finally, 2-5 mL W5 was added to collect the protoplasts and prepared for DNA transfection. The pRTV-OsIspE-cGFP was transferred into rice protoplasts according to the method described by Liang et al. (2022). The samples were examined using a confocal microscopy (Leica TCS SP8).
To investigate the underlying molecular mechanisms that participate in chloroplast development, an albino leaf 4 (al4) mutant was isolated by genetic screening of EMS (Ethyl methyl sulfonate) mutant lines with Xian/Indica rice cultivar Ruanhua2B (RH2B) background. Phenotypic analysis showed that the buds and seedling in al4 exhibited an albino phenotype compared to those in RH2B (wildtype, WT) (Figures 1A–C). Similar to other chlorotic or albino leaf mutants identified in our laboratory, such as al1, al2, and csl1, the seedlings gradually withered and eventually died (Zhang et al., 2016; Liu et al., 2016b; Liang et al., 2022). The albino phenotype in al4 mutant also could not recover at later developmental stages and eventually died at the three-leaf stage.
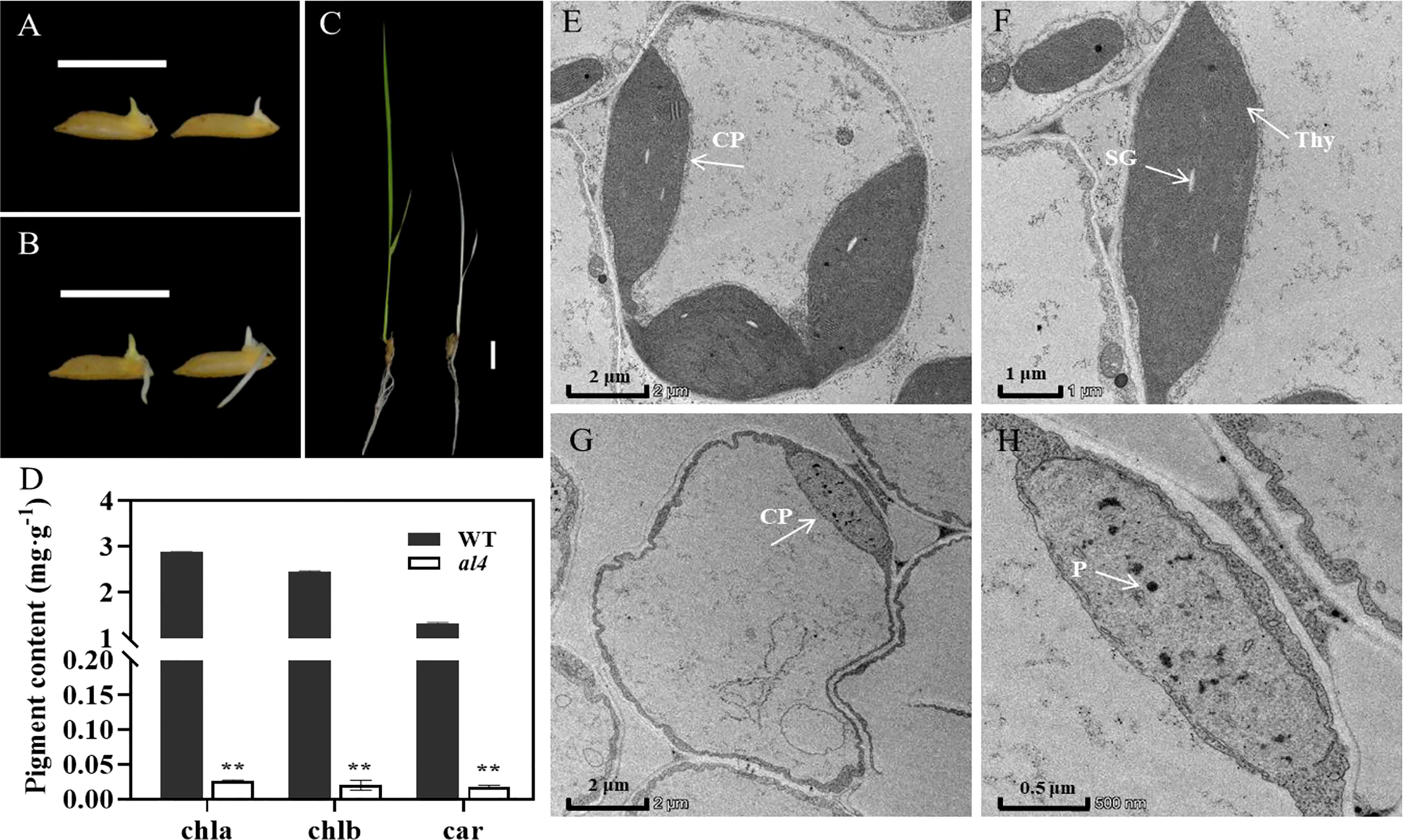
Figure 1 Phenotypic analysis of the albino leaf 4 (al4) mutant. (A–C) Phenotypic comparison of WT (RH2B, left) and al4 mutant (right) at 2 day (A), 3 day (B) and 10 day (C) after germination (DAP). Bars=1 cm. (D) Chlorophyll (Chla, Chlb) and carotenoid (Car) levels between WT and al4 mutant at 18 DAP. Data are presented as mean ± SD from three biological replicates. Two asterisks indicate statistically significant differences compared with the wild type at P<0.01. (E–H) Transmission electron microscopy analysis of the leaves in WT (E, F) and al4 mutant (G, H) at the three-leaf stage. CP, chloroplast; SG, starch granule; Thy, thylakoid lamellar; P, plastoglobuli. Bar=2 μm (E, G), 1 μm (F) and 0.5 μm (H).
Since the photosynthetic pigments chlorophylls and carotenoids are closely associated with capturing light energy in plants, we evaluated the chlorophyll (Chla, Chlb) and carotenoid (Car) content in al4 mutant and WT plants. Consistent with the albino phenotype, al4 had significantly lower Chla, Chlb and Car levels than WT (Figure 1D). These results suggest that the early-stage lethality phenotype in al4 mutant might be caused by abnormal chloroplast development.
Chlorophyll and carotenoid levels are generally modulated through chloroplast biogenesis in plants. Thus, transmission electron microscopy (TEM) was used to observe the chloroplast structure in al4 and WT plants at the three-leaf stage. The results showed that the chloroplasts in WT plants exhibited abundant distinct thylakoids packed with grana and many starch granules (Figures 1E, F), whereas those of al4 contained few undeveloped chloroplasts (Figure 1G). Plastoglobuli (PGs) were increased in al4 mutant compared to WT (Figure 1H). The chloroplasts in al4 were severely damaged and unable to form an intact thylakoid structure (Figure 1H). Collectively, these results suggest that the albino phenotype of al4 is due to defects in chloroplast development.
The segregation ratio of the green and albino seedlings in the heterozygous M3 plants was 284: 94 (χ2 = 0.0035, P>0.05, Table S3, Figure S2), indicating that the al4 phenotype was controlled by a single recessive gene. The MutMap+ method was applied for rapid identification of the gene controlling the albino phenotype in the al4 mutant, a bulk of 50 green seedlings (Pool WT) and 50 albino seedlings (Pool MT) from the M4 generation were separately aligned to the reference sequence. The SNP-index and ΔSNP-index were calculated with each SNP. The ΔSNP-index and chromosome positions are shown in Figure 2A. ΔSNP-index value over 0.35 were further analyzed, and a total of 41 SNPs were identified (Table S4). On the long arm of chromosome 1, a peak of the ΔSNP-index closes to 0.7 was observed (Figure 2A). In this candidate region, a G to A transition at the exon-intron junction site in Os01g0802100 causes abnormal splicing of OsIspE (Figures 2B, C)
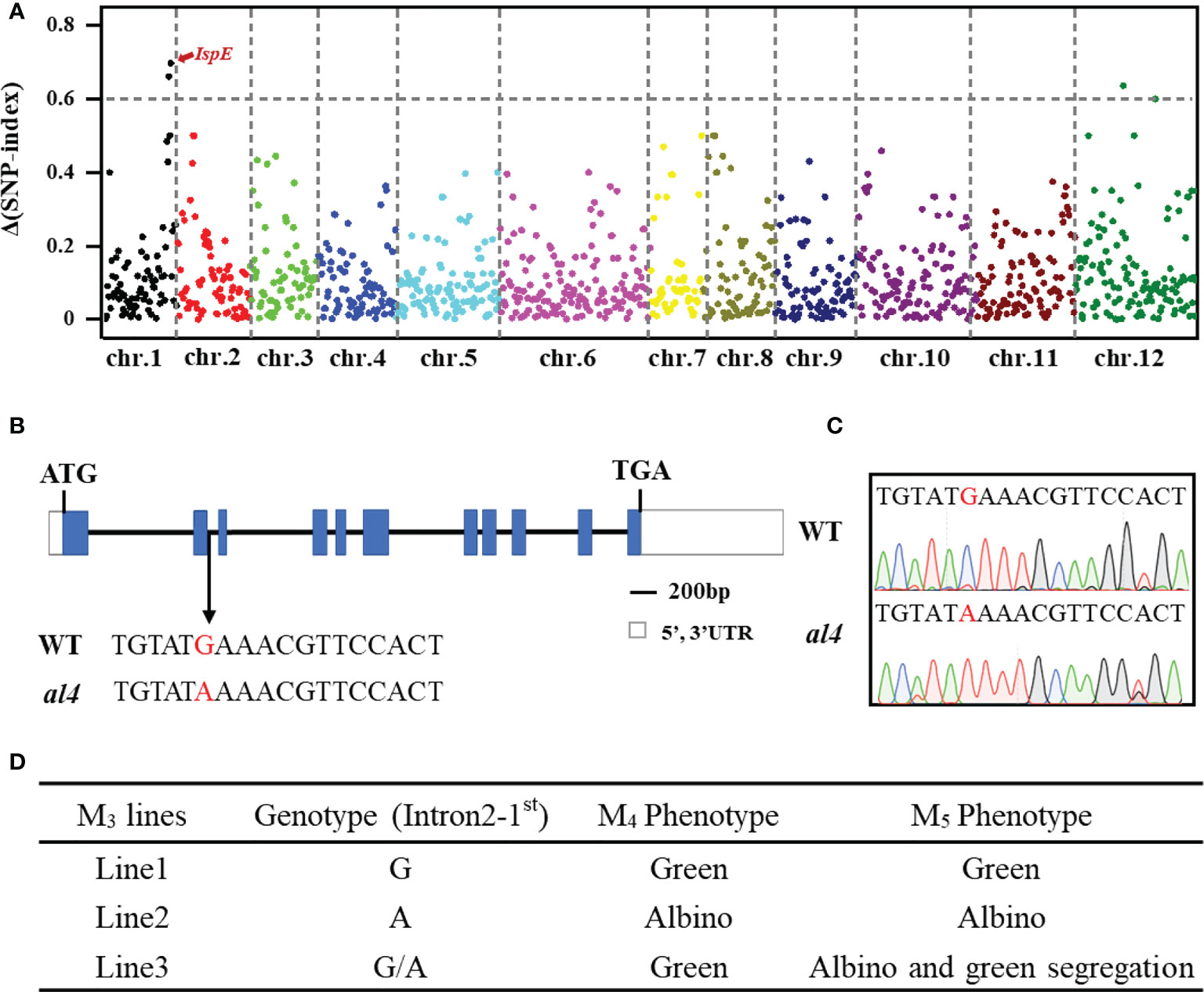
Figure 2 MutMap+ identifies the causal mutation of the al4 early-stage lethality phenotype. (A) ΔSNP-index plot of the whole genome generated by MutMap+ analysis. (B) Schematic diagram of al4 and point mutations between the WT and al4 mutant. Blue boxes represent exons and black lines represent introns. (C) Genomic regions spanning the mutation site of OsIspE were amplified and sequenced. (D) Segregation verification in different EMS generations.
Os01g0802100 encodes 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK), which belongs to the IspE superfamily (Chen et al., 2018). We subsequently examined whether the albino leaf phenotype was consistent with the single deletion mutation in OsIspE, and the point mutation was verified in different EMS generations using genomic DNA sequencing. Leaf color phenotype segregation was observed in the M5 population seedling stage only the M4 plant genotype was heterozygous (Figure 2D; Figure S3), and the green/albino seedling segregation ratio in the M5 population was 94:28. Taken together, these results suggested that Os01g0802100 is a strong candidate gene for OsIspE.
Alternative splicing is a common process in post-transcriptional mRNA regulation, that produces various mature mRNAs with different structures and functions. The full-length OsIspE gene consists of 1206 bp and including 11 exons (Figure 3A). To examine splicing alternation of OsIspE in the mutant, RT-PCR was performed with a forward and a reverse primer in exon1 and 11, respectively (Figure 3C). WT generated a 1206 bp fragment as predicted, whereas PCR product in al4 mutant was smaller than that in WT (Figure 3D). To determine whether exon2 was missing in the al4 mutant, a forward and a reverse primer in exon1 and 2 were designed. RT-PCR results showed that WT produced a 184 bp fragment as expected, but the al4 mutant did not produce a fragment (Figure S4). Sanger sequencing revealed that 96 bp deletion and the entire exon2 was absent in the al4 mutant (Figures 3B, E). This mutation resulted in the deletion of 32 amino acids in al4 mutant (Figures S5, S6). RT-qPCR analysis of OsIspE expression in WT and al4 revealed that OsIspE expression was significantly reduced in the al4 (Figure S7). Protein structure prediction indicated that α-helix was include in the exon2 of OsIspE (Figure S8). Taken together, these results suggest that mutation at the exon-intron junction site cause alternative splicing factors fail to distinguish the origin of the GT-AG intron, leading to exon skipping and producing a truncated OsIspE in the al4 mutant.
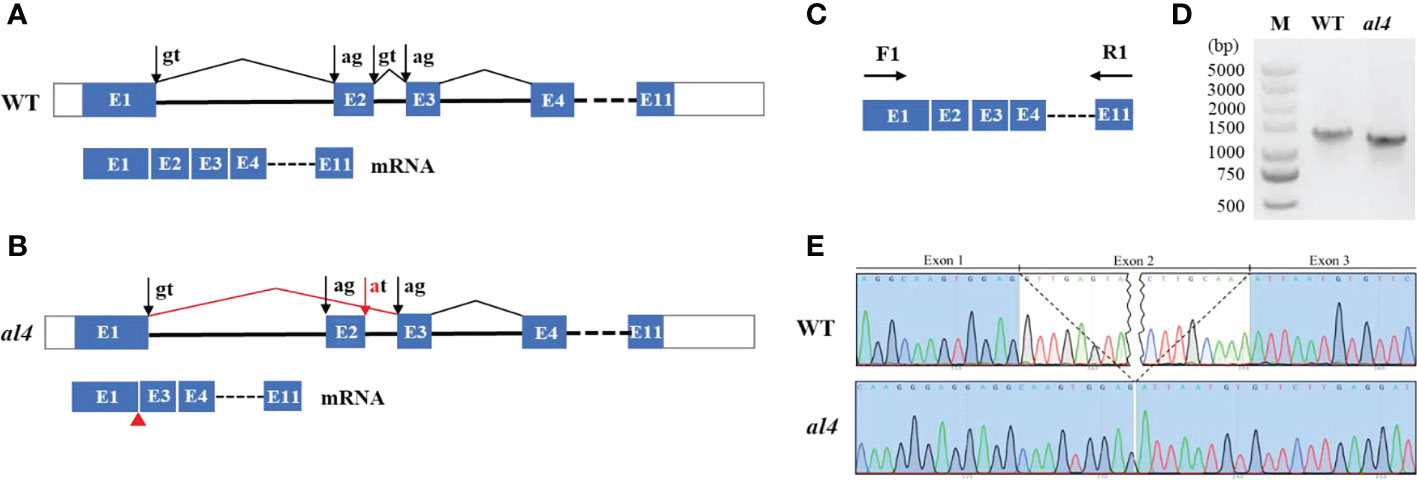
Figure 3 Intron mutation of OsIspE results in exon skipping. (A) Schematic representation of the structure of the WT genomic sequence of OsIspE. The positions of ‘‘canonical’’ splicing sites are shown with black arrows. (B) Exon-intron junction site mutation (red arrow), leading to exon skipping and production of truncated OsIspE in the al4 mutant. (C, D) RT-PCR bands of OsIspE in the wild type and al4 mutant with the specific primers indicated in panel C. (E) Chromatograms of cDNAs regions in the WT and al4 mutant. The entire exon2 is absent in the al4 mutant.
To verify whether the mutation of OsIspE corresponded to the albino phenotype, we generated OsIspE-Cas9 transgenic plants by CRISPR/Cas9 genome-editing approach. Two target sites located on the first and sixth exon were selected and a total of 20 independent transgenic lines were obtained. Deletion mutations at target sites were characterized by DNA sequencing, and three OsIspE-Cas9 transgenic plants resulting in frameshift mutations were selected for further analysis (Figures 4A, B). In OsIspE-Cas9-1(KO-1) plants, 1bp and 16bp deletions were detected in target site 1 and 2, respectively (Figure 4A). In addition, 3bp and 2bp deletions in target site 1 were identified in KO-3 and KO-4 transgenic plants (Figure 4A). OsIspE expression levels were significantly decreased in the OsIspE-Cas9 lines compared to WT (Figure 4C). Phenotypic observation showed that OsIspE-Cas9 lines (KO-1, KO-3 and KO-4) displayed an albino leaf phenotype compared to WT (Figure 4D).
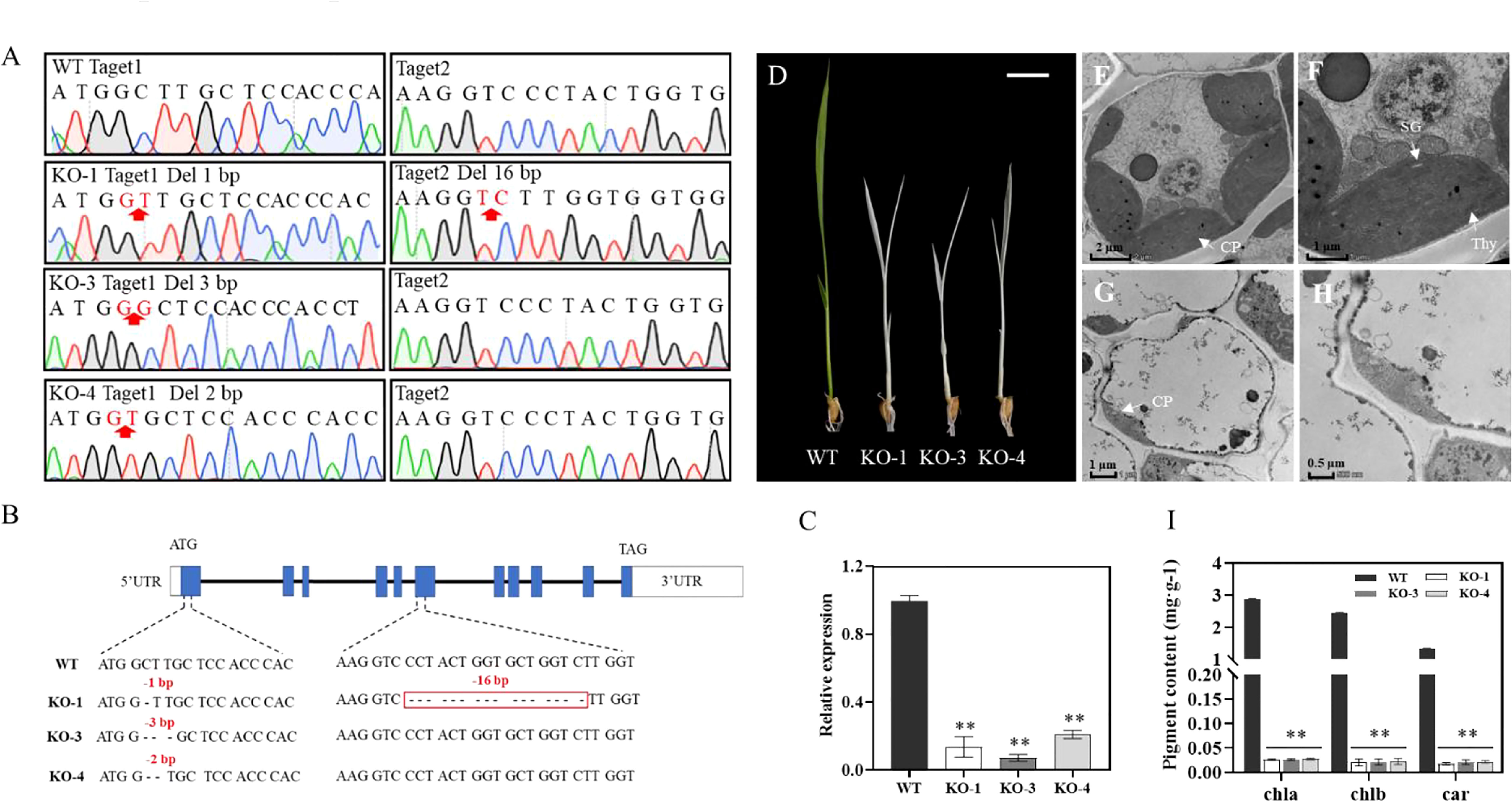
Figure 4 Transgenic confirmation of the role of OsIspE in regulating leaf color. (A, B) OsIspE-Cas9 lines (KO-1, KO-3, KO-4) containing deletion mutations were confirmed via sequencing. (C) RT-qPCR analysis of OsIspE in ZH11 and OsIspE-Cas9 lines. (D) Phenotypic analysis of OsIspE-Cas9 lines at 10 DAP. Bars=1cm. (F–H) Transmission electron microscopy analysis of the leaves of ZH11 (E, F) and KO-1 (G, H) at the three-leaf stage. CP, chloroplast; SG, starch granule; Thy, thylakoid lamellar. Bar=2 μm (E), 1 μm (F, G) and 0.5 μm (H, I) Chlorophyll (Chla, Chlb) and carotenoid (Car) levels of ZH11 and OsIspE-Cas9 lines at 10 DAP. **P < 0.01.
We further applied TEM to investigate the chloroplast structures of KO-1 and WT seedlings at the three-leaf stage. As expected, WT had well-developed chloroplasts, containing normally structured thylakoid lamellar and starch granule (Figures 4E, F), whereas chloroplasts in OsIspE-Cas9 plants displayed abnormal structures, without intact thylakoid lamellae (Figures 4G, H). We next measured the contents of Chla, Chlb and Car in WT and OsIspE-Cas9 lines, and the results showed that chlorophyll and carotenoid levels were significantly decreased in the OsIspE-Cas9 lines (Figure 4I). Taken together, these results demonstrated that OsIspE is essential for chloroplast development in rice.
RT-qPCR was applied to detected transcript profiles of OsIspE in different tissues, including roots (R), pulvinus (P), leaf sheaths (LS), flag leaves (FL), pour two leaves (PTL), young panicles (YP). The results showed that OsIspE was expressed in different tissues and organs, and particularly abundant in flag leaf (Figure 5A). To further examined whether the expression of OsIspE corresponds to the period of early leaf development, we prepared the cDNA samples come from wild-type plants of 3‐week‐old seedlings (Figure 5B). And the results showed that OsIspE was expressed in all leaves, and preferentially expressed in the latest growing leaves, such as the L4 (Figure 5C). Notably, the OsIspE was highly expressed in stem.
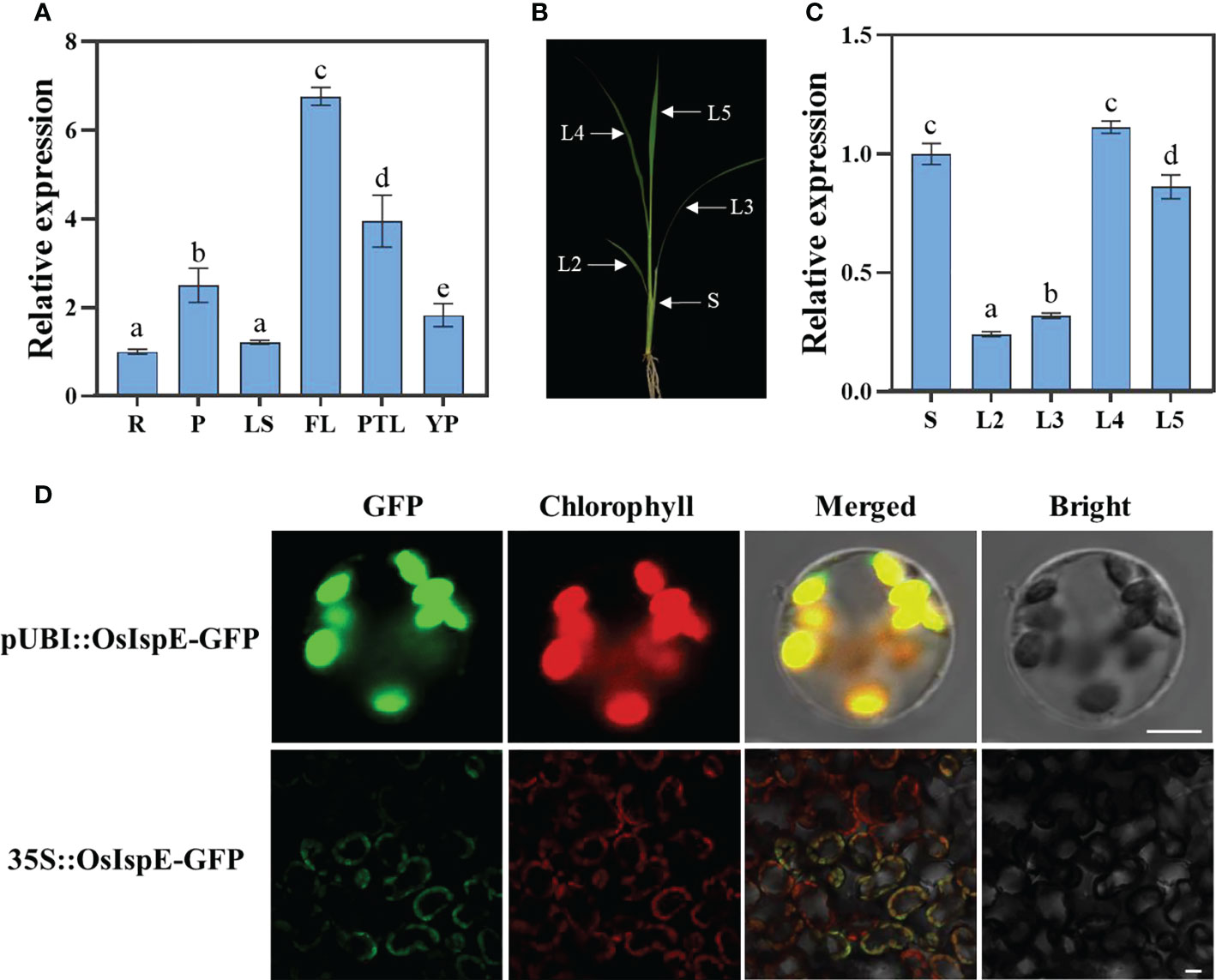
Figure 5 Expression pattern and subcellular localization of OsIspE. (A) OsIspE expression levels in different tissues. R, roots; P, pulvinus; LS, leaf sheaths; FL, flag leaves; PTL, pour two leaves; YP, young panicles. Values are means ± SD from three biological replicates. (B) Schematic of seedling plant used for RT-qPCR. S represent stem; L2-L5 represent the second to the fifth leaves. (C) OsIspE expression levels in different tissues of young seedlings. Values are means ± SD from three biological replicates. Data were analyzed using two-way ANOVA with post hoc Tukey tests (letters indicate significant differences between samples at P< 0.05). (D) Subcellular localization of OsIspE. pUBI::OsIspE-GFP and 35S::OsIspE-GFP were transiently transformed into rice protoplasts and tobacco leaves respectively. Fluorescence signals were detected using confocal microscopy, and representative images are shown. Bars=10 μm (rice protoplast) and 20 μm (tobacco leaves).
To examine the subcellular localization of OsIspE, a transient expression vector harboring the OsIspE-GFP fusion protein was transiently transformed in rice protoplasts. The green fluorescent signals of OsIspE-GFP were co-localized with chlorophyll autofluorescence signals (Figure 5D), which consistent with the previous study (Chen et al., 2018). We further confirm the subcellular localization in tobacco leaves, and the fluorescence signals of OsIspE-GFP was also colocalized with chloroplast, suggesting that OsIspE is a chloroplast-localized protein.
Since the OsIspE gene is involved in the MEP pathway, the mutation of OsIspE would cause an abnormal chloroplast structure. We assumed that the loss of OsIspE would influence the expression of chloroplast-associated genes. To examine this hypothesis, we investigated the expression of chloroplast-associated genes using RT-qPCR, including chloroplast biosynthetic and translation related genes, Photosynthetic associated nuclear genes (PhANGs) and plastid-encoded polymerases (PEPs)-dependent chloroplast genes. Compared to WT, the expression levels of several chloroplast biosynthetic and translation related genes were significantly reduced, such as HAP3A, Cab1R, Cao1, and HEMAI (Figure 6A). Chloroplast biosynthetic and translation related genes and PEP-dependent chloroplast genes displayed diverse expression patterns between WT and al4 mutant, expression levels of rps2 and psaB were not affected, petA and psbA were significantly reduced, and the atpA and psaA were upregulated in the al4 compared to WT (Figures 6A, C). The expression levels of the PhANG genes, including psbO, psbP, lhcb2, psaE, psaD, and rbcS were dramatically reduced in the al4 (Figure 6B). We also investigated the expression pattern of mitochondrial genes between WT and al4 mutant, results showed several genes, such as nad2, nad4, and nad5 were upregulated in the al4 mutant (Fig S9). These results indicate that the mutation of OsIspE affects the expression of chloroplast-associated genes that regulate chloroplast development in rice.
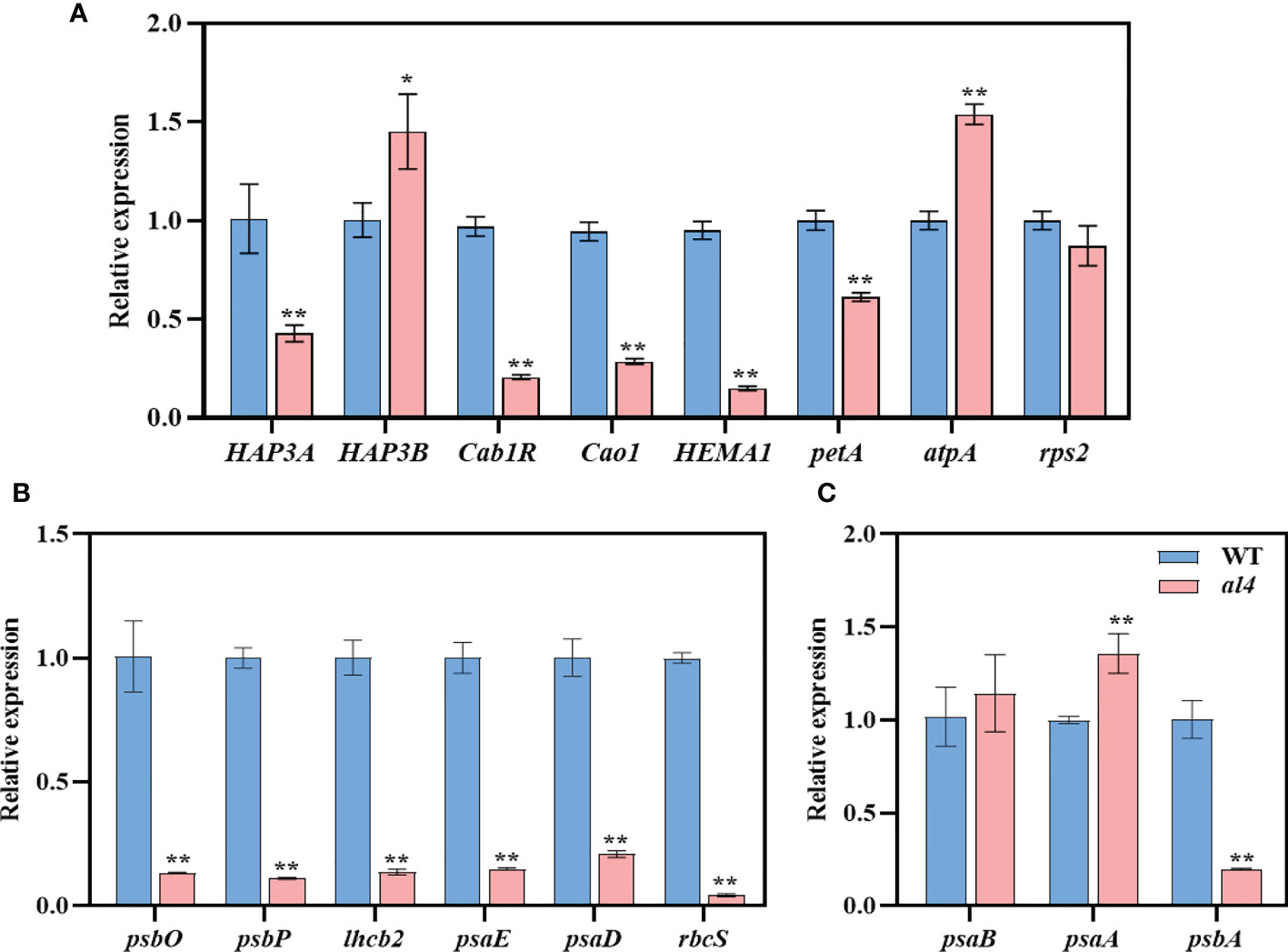
Figure 6 Expression of chloroplast-associated genes in WT and al4. Relative expression of chloroplast biosynthetic and translation related genes (A), Photosynthetic associated nuclear genes (PhANGs) (B), Plastid-encoded polymerases (PEPs)-dependent chloroplast genes (C). Data are presented as mean ± S.D. *P < 0.05; **P < 0.01.
Leaf color mutants are ideal materials for studying chlorophyll biosynthesis and chloroplast development in plants. Here, we isolated and characterized a leaf albino 4 mutant using Ethyl methyl sulfonate (EMS) mutagenesis with Xian/Indica rice cultivar Ruanhua2B (RH2B) background. Further analysis revealed that the early-stage lethality phenotype in the al4 mutant was caused by defective chloroplast development (Figures 1G, H). We used MutMap+ to identified that the mutation in Os01g0802100 gene encoding 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK), which belongs to the IspE superfamily, was responsible for the al4 mutant phenotype (Figure 2). IspE encodes an intermediate enzyme in the methylerythritol phosphate (MEP) pathway of isoprenoid biosynthesis.
Isoprenoids are the largest group of natural products and are derived from the basic building block isopentenyl diphosphate (IPP) and its allylic isomer dimethylallyl diphosphate (DMAPP) (Hoshino and Gaucher, 2018). IPP and DMAPP are mainly synthesized via two pathways: the mevalonate (MVA) pathway and the non-mevalonate pathway, which is also known as the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway (Rohmer, 1999). The MEP-pathway comprises seven nuclear-encoded plastid-localized enzymes (Zeng and Dehesh, 2021). Previous studies have shown that defects in MEP pathway genes may result in leaf color change phenotype in different plant species.
1-Deoxy-D-xylulose-5-phosphate synthase (DXS) catalyzes the first committed step in the MEP pathway, Arabidopsis DXS1 knock-out cla1-1 mutant displayed albino phenotype and can be rescued by the addition of 1-Deoxy-D-Xylulose (DX) (Mandel et al., 1996; Estévez et al., 2000). In tomato, the DXS1 T-DNA mutant wls-2297 has a severe deficiency in chlorophylls and carotenoids, leading to an albino phenotype (García-Alcázar et al., 2017). The second step is catalyzed by 1-deoxy-d-xylulose-5-phosphate reductoisomerase (DXR), which transforms 1-deoxy-D-xylulose 5-phosphate (DOXP) to MEP, notably, disruption of DXR genes also results albino phenotype in plants. For example, the Arabidopsis T-DNA insertion mutant dxr exhibited an albino phenotype and grew purple cotyledons. Further analysis revealed that the dxr mutant only developed proplastids, without normal thylakoids (Xing et al., 2010). Taken together, emerging evidence has demonstrated that the MEP pathway is extensively involved in chloroplast development, leading to changes in leaf color.
IspE catalyzes the ATP-dependent conversion of 4-diphosphocytidyl-2-C-methyl-D-erythritol (CDP-ME) to 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate (CDP-MEP) in the MEP pathway (Rohdich et al., 2000). A defect in IspE activity might decrease the level of cellular metabolites produced by the MEP pathway, such as chlorophylls and carotenoids (Figure S10) (Ahn and Pai, 2008). Virus-induced gene silencing of IspE in Nicotiana benthamiana results in defective biogenesis of both chloroplasts and mitochondria, leading to severe leaf yellowing phenotypes (Ahn and Pai, 2008). The Arabidopsis homozygous T-DNA insertion mutant ispE-1 is albino lethal, and thylakoid development is completely abolished in the ispE-1 mutant (Hsieh et al., 2008). The IspE gene was further characterized in the monocotyledonous model plant rice, and map-based cloning revealed that a missense mutation in OsIspE in gry340 mutant, resulted in a green-revertible yellow leaf phenotype (Chen et al., 2018). The OsIspE protein is localized in the chloroplast (Chen et al., 2018). Further analysis revealed that OsIspE altered the expression of other MEP and MVA pathway genes (Chen et al., 2018).
Notably, in the present study, the ispE (al4) mutant displayed an albino phenotype, which was not completely consistent with gry340. A single nucleotide C to T substitution at position 2,554 of the IspE coding region was detected in the gry340 mutant, whereas deletion of 96 bp and the entire exon2 was absent of IspE in the al4 mutant, suggesting that variable expression might lead to different phenotypic effects in gry340 and al4 mutants. Defects in leaf color could be attributable to the suppression of expression of chloroplast biosynthetic genes, thus we analyzed the expression patterns of a series of chloroplast-related genes. Results showed that most of genes were significantly reduced in the al4 compared to WT, such as photosynthetic associated nuclear genes (PhANGs), suggesting that defective chloroplast development in al4 mutant might be associated with the repression of chloroplast-associated genes (Figure 6). We also investigated the expression pattern of mitochondrial genes between WT and al4 mutant, including cob, atp8, cox1, and nad2 ect, results showed that few genes, such as cox1 have different expression pattern between al4 and gry340 mutants.
Alternative splicing (AS) is an important posttranscriptional process that produces different mRNA isoforms through the selection and utilization of alternative splice sites in the same pre-mRNA (Simpson et al., 2008; Reddy et al., 2013). Several types of AS were identified and classified, including intron retention (IR), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), mutually exclusive exons (MXE), and exon skipping (ES). The excision of introns from a pre-mRNA is directed by special sequences at the intron-exon junctions (Black, 2003), and splicing of pre-mRNA is controlled by a multi-megadalton ribonucleoprotein (RNP) complex, called the spliceosome, which leads to the removal of introns from pre-mRNA and assembly of a mature transcript (Brown and Simpson, 1998; Will and Lührmann, 2011).
The first step in pre-mRNA splicing is the recognition and selection of specific splicing sites. Previous studies have shown that disruption of splice sites commonly results in missing an exon and both flanking introns during the splicing process. For example, a SNP on the splicing site of CsSEP2, resulted in the skipping of exon6 and abolishment of the transcriptional activity in CsSEP2, leading to perturbed floral and fruit development in cucumber (Wang et al., 2016). A single mutation(G-to-A) at the splice site of intron 5 in CRS1, results in exon skipping and perturbed chloroplast development in maize (Wang et al., 2021). In the present study, mutation at the exon-intron junction site caused the spliceosome complex fail to distinguish of the original splice site, leading to 96 bp deletion and entire exon2 was absent in the al4 mutant, which displayed an albino phenotype (Figure 3), therefore the exon skipping occurred during splicing due to the mutation in the splice site.
In conclusion, the present study showed that OsIspE is extensively involved in chloroplast biogenesis and development. Furthermore, mutation at the exon-intron junction site in IspE resulted in exon skipping and produced truncated IspE, eventually exhibiting an albino phenotype. Moreover, the findings of this study suggest that OsIspE regulates chloroplast biogenesis and development through coordinated transcription of chloroplast-associated genes, particularly PhANGs.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
TX and ZZ conceived and designed the study. TX and JZ conduct the most of experiments and analyzed the data. YL performed the Mutmap+ analysis. QZ, WL, YZ, MW, TC, and DD helped with the data interpretation. WW and ZZ analyzed the data and drafted the manuscript. ZZ supervised the study. All authors contributed to the article and approved the submitted version.
This study was supported by the Key-Area Research and Development Program of Guangdong Province (2018B020202012), and the Natural Science Foundation of China (31671645).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.986678/full#supplementary-material
MEP, Methylerythritol phosphate; MVA, Mevalonic acid; al4, albino leaf 4; PPR: Pentatricopeptide repeat protein; IPP: Isopentenyl diphosphate; DMAPP, Dimethylallyl diphosphate; CMK, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; EMS, Ethyl methyl sulfonate; Chla/Chlb, Chlorophyll a/b; Car, Carotenoid; DAP, day after germination; TEM, Transmission electron microscopy; PhANGs, photosynthetic associated nuclear genes; GFP, Green fluorescent protein.
Ahn, C. S., Pai, H. S. (2008). Physiological function of IspE, a plastid MEP pathway gene for isoprenoid biosynthesis, in organelle biogenesis and cell morphogenesis in nicotiana benthamiana. Plant Mol. Biol. 66 (5), 503–517. doi: 10.1007/s11103-007-9286-0
Black, D. L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72 (1), 291–336. doi: 10.1146/annurev.biochem.72.121801.161720
Bohlmann, J., Keeling, C. I. (2008). Terpenoid biomaterials. Plant J. 54 (4), 656–669. doi: 10.1111/j.1365-313X.2008.03449.x
Brown, J. W. S., Simpson, C. G. (1998). Splice site selection in plant pre-mRNA splicing. Annu. Rev. Plant Biol. 49 (1), 77–95. doi: 10.1146/annurev.arplant.49.1.77
Chen, N. G., Wang, P. R., Li, C. M., Wang, Q., Pan, J. H., Xiao, F. L., et al. (2018). A single nucleotide mutation of the IspE gene participating in the MEP pathway for isoprenoid biosynthesis causes a green-revertible yellow leaf phenotype in rice. Plant Cell Physiol. 59 (9), 1905–1917. doi: 10.1093/pcp/pcy108
Estévez, J. M., Cantero, A., Romero, C., Kawaide, H., Jiménez, L. F., Kuzuyama, T., et al. (2000). Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in arabidopsis. Plant Physiol. 124 (1), 95–104. doi: 10.1104/pp.124.1.95
Fekih, R., Takagi, H., Tamiru, M., Abe, A., Natsume, S., Yaegashi, H., et al. (2013). MutMap+: genetic mapping and mutant identification without crossing in rice. PloS One 8 (7), e68529. doi: 10.1371/journal.pone.0068529
García-Alcázar, M., Giménez, E., Pineda, B., Capel, C., García-Sogo, B., Sánchez, S., et al. (2017). Albino T-DNA tomato mutant reveals a key function of 1-deoxy-D-xylulose-5-phosphate synthase (DXS1) in plant development and survival. Sci. Rep. 7 (1), 1–12. doi: 10.1038/srep45333
Henry, L., Thomas, S., Widhalm, J., Lynch, J., Davis, C., Kessler, S., et al. (2018). Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Plants 4 (9), 721–729. doi: 10.1038/s41477-018-0220-z
Hoshino, Y., Gaucher, E. A. (2018). On the origin of isoprenoid biosynthesis. Mol. Biol. Evol. 35 (9), 2185–2197. doi: 10.1093/molbev/msy120
Hsieh, M. H., Chang, C. Y., Hsu, S. J., Chen, J. J. (2008). Chloroplast localization of methylerythritol 4-phosphate pathway enzymes and regulation of mitochondrial genes in ispD and ispE albino mutants in arabidopsis. Plant Mol. Biol. 66 (6), 663–673. doi: 10.1007/s11103-008-9297-5
Huang, R., Wang, Y., Wang, P. R., Li, C. M., Xiao, F. L., Chen, N. G., et al. (2018). A single nucleotide mutation of IspF gene involved in the MEP pathway for isoprenoid biosynthesis causes yellow-green leaf phenotype in rice. Plant Mol. Biol. 96 (1-2), 5–16. doi: 10.1007/s11103-017-0668-7
Liang, J. Y., Zhang, Q. X., Liu, Y. R., Zhang, J. J., Wang, W. Y., Zhang, Z. Z. (2022). Chlorosis seedling lethality 1 encoding a MAP3K protein is essential for chloroplast development in rice. BMC Plant Biol. 22 (1), 20. doi: 10.1186/s12870-021-03404-9
Liu, Z. C., Jin, Y., Liu, W. F., Tao, Y., Wang, G. G. (2018). Crystal structure of IspF from bacillus subtilis and absence of protein complex assembly amongst IspD/IspE/IspF enzymes in the MEP pathway. Bioscience Rep. 38 (1), BSR20171370. doi: 10.1042/BSR20171370
Liu, D., Li, W. C., Cheng, J. F. (2016b). The novel protein delayed pale-greening1 is required for early chloroplast biogenesis in arabidopsis thaliana. Sci. Rep. 6, 25742. doi: 10.1038/srep25742
Liu, X. Y., Zhang, X. C., Cao, R. J., Hu, S. K., Shao, G. N., Sheng, Z. H., et al. (2021). CDE4 encodes a pentatricopeptide repeat protein involved in chloroplast RNA splicing and affects chloroplast development under low-temperature conditions in rice. J. Integr. Plant Biol. 63 (10), 1724–1739. doi: 10.1111/jipb.13147
Liu, C. H., Zhu, H. T., Xing, Y., Tan, J. J., Chen, X. H., Zhang, J. J., et al. (2016a). Albino leaf 2 is involved in the splicing of chloroplast group I and II introns in rice. J. Exp. Bot. 67 (18), 5339–5347. doi: 10.1093/jxb/erw296
Ma, X. L., Liu, Y. G. (2016). CRISPR/Cas9-Based multiplex genome editing in monocot and dicot plants. Curr. Protoc. Mol. Biol. 115, 31.6.1–31.6.21. doi: 10.1002/cpmb.10
Mandel, M. A., Feldmann, K. A., Herrera-Estrella, L., Rocha-Sosa, M., León, P. (1996). CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J. 9 (5), 649–658. doi: 10.1046/j.1365-313x.1996.9050649.x
Pulido, P., Perello, C., Rodríguez-Concepción, M. (2012). New insights into plant isoprenoid metabolism. Mol. Plant 5 (5), 964–967. doi: 10.1093/mp/sss088
Reddy, A. S., Marquez, Y., Kalyna, M., Barta, A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25, 3657–3683. doi: 10.1105/tpc.113.117523
Rodrı´guez-Concepcio´n, M., Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A Metab. milestone achieved through Genomics Plant Physiol. 130 (3), 1079–1089. doi: 10.1104/pp.007138
Rohdich, F., Wungsintaweekul, J., Lüttgen, H., Fischer, M., Eisenreich, W., CA, S., et al. (2000). Biosynthesis of terpenoids: 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase from tomato. Proc. Natl. Acad. Sci. USA 97 (15), 8251–8256. doi: 10.1073/pnas.140209197
Rohmer, M. (1999). The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Natural Product Rep. 16 (5), 565–574. doi: 10.1039/A709175C
Simpson, C. G., Lewandowska, D., Fuller, J., Maronova, M., Kalyna, M., Davidson, D., et al. (2008). Alternative splicing in plants. Biochem. Soc. Trans. 36 (3), 508–510. doi: 10.1042/BST0360508
Tang, J. P., Zhang, W. W., Wen, K., Chen, G. M., Sun, J., Tian, Y. L., et al. (2017). OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Mol. Biol. 295 (4-5), 345–357. doi: 10.1007/s11103-017-0654-0
Wang, X., Gao, D., Sun, J., Liu, M., Lun, Y., Zheng, J., et al. (2016). An exon skipping in a sepallata-like gene is associated with perturbed floral and fruits development in cucumber. J. Integr. Plant Biol. 58 (9), 766–771. doi: 10.1111/jipb.12472
Wang, M., Li, K., Li, Y., Mi, L., Hu, Z., Guo, S., et al. (2021). An exon skipping in CRS1 is associated with perturbed chloroplast development in maize. Int. J. Mol. Sci. 22 (19), 10668. doi: 10.3390/ijms221910668
Will, C. L., Lührmann, R. (2011). Spliceosome structure and function. Cold Spring Harbor Perspect. Biol. 3 (7), a003707. doi: 10.1101/cshperspect.a003707
Xing, S., Miao, J., Li, S., Qin, G., Tang, S., Li, H., et al. (2010). Disruption of the 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) gene results in albino, dwarf and defects in trichome initiation and stomata closure in arabidopsis. Cell Res. 20 (6), 688–700. doi: 10.1038/cr.2010.54
Zeng, L., Dehesh, K. (2021). The eukaryotic MEP-pathway genes are evolutionarily conserved and originated from chlaymidia and cyanobacteria. BMC Genomics 22 (1), 1–12. doi: 10.1186/s12864-021-07448-x
Zhang, Y., Su, J., Duan, S., Ao, Y., Dai, J., Liu, J., et al. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7 (1), 30. doi: 10.1186/1746-4811-7-30
Keywords: rice, albino leaf 4 (al4), methylerythritol phosphate (MEP), exon skipping, OsIspE
Citation: Xu T, Zhang J, Liu Y, Zhang Q, Li W, Zhang Y, Wu M, Chen T, Ding D, Wang W and Zhang Z (2022) Exon skipping in IspE Gene is associated with abnormal chloroplast development in rice albino leaf 4 mutant. Front. Plant Sci. 13:986678. doi: 10.3389/fpls.2022.986678
Received: 13 July 2022; Accepted: 24 October 2022;
Published: 08 November 2022.
Edited by:
Ezequiel Petrillo, Molecular Biology and Neurosciences (IFIBYNE), ArgentinaReviewed by:
Minoru Ueda, RIKEN Center for Sustainable Resource Science (CSRS), JapanCopyright © 2022 Xu, Zhang, Liu, Zhang, Li, Zhang, Wu, Chen, Ding, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zemin Zhang, em16aGFuZ0BzY2F1LmVkdS5jbg==; Wenyi Wang, d3l3YW5nQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.