
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 September 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.986673
This article is part of the Research TopicImproving crop nutritional security for sustainable agriculture in the era of climate changeView all 12 articles
As one of the most important and largest transcription factors, WRKY plays a critical role in plant disease resistance. However, little is known regarding the functions of the WRKY family in cultivated peanuts (Arachis hypogaea L.). In this study, a total of 174 WRKY genes (AhWRKY) were identified from the genome of cultivated peanuts. Phylogenetic analysis revealed that AhWRKY proteins could be divided into four groups, including 35 (20.12%) in group I, 107 (61.49%) in group II, 31 (17.82%) in group III, and 1 (0.57%) in group IV. This division is further supported by the conserved motif compositions and intron/exon structures. All AhWRKY genes were unevenly located on all 20 chromosomes, among which 132 pairs of fragment duplication and seven pairs of tandem duplications existed. Eighteen miRNAs were found to be targeting 50 AhWRKY genes. Most AhWRKY genes from some groups showed tissue-specific expression. AhWRKY46, AhWRKY94, AhWRKY156, AhWRKY68, AhWRKY41, AhWRKY128, AhWRKY104, AhWRKY19, AhWRKY62, AhWRKY155, AhWRKY170, AhWRKY78, AhWRKY34, AhWRKY12, AhWRKY95, and AhWRKY76 were upregulated in ganhua18 and kainong313 genotypes after Ralstonia solanacearum infection. Ten AhWRKY genes (AhWRKY34, AhWRKY76, AhWRKY78, AhWRKY120, AhWRKY153, AhWRKY155, AhWRKY159, AhWRKY160, AhWRKY161, and AhWRKY162) from group III displayed different expression patterns in R. solanacearum sensitive and resistant peanut genotypes infected with the R. solanacearum. Two AhWRKY genes (AhWRKY76 and AhWRKY77) from group III obtained the LRR domain. AhWRKY77 downregulated in both genotypes; AhWRKY76 showed lower-higher expression in ganhua18 and higher expression in kainong313. Both AhWRKY76 and AhWRKY77 are targeted by ahy-miR3512, which may have an important function in peanut disease resistance. This study identified candidate WRKY genes with possible roles in peanut resistance against R. solanacearum infection. These findings not only contribute to our understanding of the novel role of WRKY family genes but also provide valuable information for disease resistance in A. hypogaea.
Transcription factors (TFs) activate diverse signal transduction cascades and regulate the transcriptional mode of targeted genes, which helps crop plants to adapt to diverse environmental stresses (Javed et al., 2020, 2022; Zhu et al., 2021; Haider et al., 2022). Among various TFs, WRKY TFs are one of the largest TF families in plants and regulate various biological processes, including growth, development, stress, and disease resistance (Rushton et al., 2010; Zhu et al., 2021; Javed et al., 2022). WRKY proteins contain a highly conserved WRKYGQK motif at N-terminus and a zinc finger motif (C2H2: CX4CX22−23HX1H, C2HC: CX7CX23HX1C) at C-terminus (Rushton et al., 2010). WRKY proteins can be classified into four groups (I, II, III, and IV). Group I members have two WRKY domains and the C2H2 type zinc finger. Group II and Group III members have one WRKY domain; Group II members have a C2H2 type zinc finger and Group III members have a C2HC-type. Group I can be classified into two subgroups Ia and Ib based on two WRKY domains at the N and C terminus (Chen et al., 2019a; Li Z et al., 2020). Group II WRKY can be further classified into five subgroups (IIa, IIb, IIc, IId, and IIe) based on the sequence of the DNA-binding domain (Zhang and Wang, 2005). Group III is separated into subgroups IIIa and IIIb based on the zinc-finger motif structure (Eulgem et al., 2000; Chen et al., 2019a). Group IV has an incomplete or partial WRKY domain and lacks a zinc-finger motif, suggesting that members of this group may have lost their function as WRKY TFs (Li Z et al., 2020; Javed et al., 2022).
Ralstonia solanacearum is a soil-borne bacterium; it usually infects plants through roots and then spreads through the vascular system to the whole plant, eventually causing mechanical blockage of the water transport system. In response to bacterial wilt (BW) disease, plants become wilt and die when the leaves are still green (Hikichi et al., 2017). A previous study indicated that AtWRKY52 (group III member) in Arabidopsis thaliana confers resistance to the bacterial soil-borne pathogen R. solanacearum (Deslandes et al., 1998, 2002). CaWRKY28, CaWRKY30 (group III member), CaWRKY40, CaWRKY41 (group III member), and CaWRKY58 in pepper regulated the defense response to R. solanacearum in plants (Wang et al., 2012; Muhammad et al., 2018; Dang et al., 2019; Hussain et al., 2021; Liu et al., 2021; Yang et al., 2021). Overexpression of sugarcane class III WRKY5 increased bacterial wilt resistance (Wang et al., 2020). Group III SiWRKY53 is a source of resistance to BW in Solanum incanum L. (Mishra et al., 2021). WRKY, especially group III members, play a vital part in the BW resistance of plants.
So far, WRKY TF families have been identified in many plants. For instance, there are 74 WRKY genes in Arabidopsis (Arabidopsis thaliana L.) (Dong et al., 2003), 153 WRKY genes in rapeseed (Brassica napus L.) (He et al., 2016), 287 WRKY genes in banana (Musa acuminata L.) (Kaliyappan et al., 2016), 88 WRKY genes in common bean (Phaseolus vulgaris L.) (Wu et al., 2017), 54 WRKY genes in pineapple (Ananas comosus L.) (Xie et al., 2018), 97 WRKY genes in Actinidia (Caragana intermedia L.) (Wan et al., 2018), 94 WRKY genes in sorghum (Sorghum bicolor L.) (Baillo et al., 2020), and 224 WRKY genes in Camelina (Camelina sativa L.) (Song et al., 2020). Previously, 77 and 75 WRKY proteins have been identified from the two wild ancestral diploid genomes of cultivated tetraploid peanuts, Arachis duranensis and Arachis ipaënsis (Song et al., 2016). Recently, a study identified 158 WRKY proteins from cultivated tetraploid peanuts using an older version of A. hypogea and gene expression of AhWRKY family members in response to drought stress (Zhao et al., 2020).
Climate change and crop production are directly correlated with each other, and current climate changes significantly impact agricultural productivity (Sharma et al., 2021, 2022; Farooq et al., 2022). Mainly, different biotic diseases and abiotic environmental factors greatly affect the productivity and overall quality of various crop plants around the world (Irfan et al., 2016, 2021; Kumar et al., 2016; Jiang et al., 2017; Ansari et al., 2022; Raza et al., 2022a,b). Peanut (Arachis hypogaea L.) is one of the most important oil crops in the world, but its yield is severely limited by various biotic and abiotic stresses (Mishra et al., 2015; Gangurde et al., 2019). Among these, BW is the major biotic factor that significantly hampers peanut production in the field environment (Jiang et al., 2017). The BW is one of the most important diseases in peanuts, especially destructive in China. When infected with the R. solanacearum, peanut plants wilt and die. It usually causes a 400,000 hm2 incidence area and 10–30% yield loss in 1 year in China (Jiang et al., 2017). This study identified 174 WRKY proteins, comprehensive analysis of gene structures, chromosome location, evolution relationship, cis-acting element, putative miRNAs, and expression analysis of gene response to R. solanacearum. These results provide insights into the evolution of the peanut WRKYs and their novel functions in peanut BW resistance. The identification and characterization of these WRKY genes may provide opportunities for peanut disease improvement and breeding programs in the era of climate change.
The protein data of cultivated peanuts (cv. Tifrunner) were obtained from the peanut genome database (PeanutBase, https://www.peanutbase.org/). The HMM file of the WRKY domain (PF03106) was retrieved from the Pfam database (http://pfam.sanger.ac.uk/) and was used to search the WRKY family proteins in the peanut protein database with an E-value threshold of 1 × 10−5 by simple HMM search TBtools (Toolbox for Biologists) v1.098 (Chen et al., 2020). Arabidopsis WRKY protein sequences were downloaded from the database of The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/). The protein sequences of AhWRKY and AtWRKY were subjected to BLAST alignment by TBtools (Toolbox for Biologists) v1.098 (E value <1 × 10−5) (Chen et al., 2020). Subsequently, all non-redundant peanut WRKY protein sequences were validated for the presence of the WRKY domain by submitting them as search queries to the NCBI CDD (http://www.ncbi.nlm.nih.gov/cdd/) and SMART databases (http://smart.embl.de/). Finally, 174 AhWRKY sequences were identified and named AhWRKY1 to AhWRKY174 according to their physical locations on the chromosome.
Multiple sequence alignment of AhWRKY proteins was conducted by MUSCLE with default parameters. A neighbor-joining (NJ) tree was constructed in MEGA 7.0 with the following criteria: Poisson model, pairwise deletion, and 1,000 bootstrap replications. Maximum likelihood (ML) analysis of the AtWRKY and AhWRKY gene family was conducted to draw a phylogenetic tree.
The MEME online program (Multiple Expectation Maximization for Motif Elicitation: http://meme-suite.org/tools/meme/) was used to identify conserved motifs in the AhWRKY proteins with a maximum motif number of 10. Information on the intron–exon structure was obtained from the peanut genome database (A. hypogaea cv. Tifrunner) and the TBtools software was used for visualization (Chen et al., 2020). TBtools was also used to determine the chromosomal location of the AhWRKY genes and draw the image. The duplication types and collinear blocks were discovered and were analyzed using One Step MCScanX and Simple Ka/Ks calculator (NJ) of Tbtools, and a diagram of gene duplication events was drawn using the Tbtools. Duplication and divergence time was calculated by the following formula as described by Bertioli et al. (2016):
The molecular weight (Mw) and isoelectric point (pI) of the full-length proteins were predicted using the pI/Mw tool (https://web.expasy.org/compute_pi/) in ExPASy (Gasteiger et al., 2003). The 2,000-base-pair (bp) sequences upstream of the start codon ATG of AhWRKY genes were retrieved from the peanut genome using TBtools (Chen et al., 2020) and were submitted to the online software PlantCARE to identify the cis-acting elements. The draft of the element distribution on each promoter was sketched using TBtools.
The CDS of AhWRKY genes were employed to distinguish target miRNAs in the psRNATarget database (https://www.zhaolab.org/psRNATarget/home) with default parameters. The interaction network figure among the miRNAs and target genes was developed with the Cytoscape software (V3.8.2; https://cytoscape.org/download.html).
The expression data of the AhWRKY genes in different tissues were identified using RNA-seq (Tifrunner variety) data obtained from the PeanutBase database (Clevenger et al., 2016). The read counts were transformed to TPM (Transcripts Per Kilobase of exon model per Million mapped reads) by GenomicFeatures in R, and the heatmap diagram was constructed with lg (TPM+1) using TBtools.
Two peanut cultivars (ganhua 18, resistant; and kainong 313, susceptible) were used for expression analysis against BW disease. Plant growth and R. solanacearum inoculation were carried out according to Chen et al. (2014). Plants were grown in 10 cm × 10 cm pots, and every pot contained three seedlings. After 21 days of sowing seeds, they were watered with 20 mL of R. solanacearum strain race 1 suspension (107 cfu ml−1) in one pot. Roots of three individual seedlings were sampled at 0, 6, 12, 24, 48, 72, and 96 h post-inoculation. The samples were frozen in liquid nitrogen immediately and stored at −80°C for further analysis.
The TianGen kit (TianGen, Beijing, China) was used to extract total RNA from roots, and the experimental operations were carried out according to the developer's instructions. Approximately, 5 μg of total RNA was reverse-transcribed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA). The primers were designed using the Primer 5.0 software (Supplementary Table 4), and the quantitative real-time qRT-PCR was performed on LightCycler 96 (Roche) using the Aidlab SYBR Green Mix kit (Aidlab, Beijing, China). The PCR protocol was conducted with a 10 μL volume, which contained 0.5 μL of upstream and downstream primers (10 μmol L−1), 2 μL of cDNA template, 2 μL of double-distilled water (ddH2O), and 5 μL of 2 × SYBR (SYBR Green qPCR Mix). The PCR program was 95°C for 120 s, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. qRT-PCR was performed with three technical repetitions, and the relative expression level was calculated using the 2−ΔΔCT method.
In this study, a total of 174 WRKY sequences were identified in cultivated peanuts (cv. Tifrunner) and named AhWRKY1 to AhWRKY174 according to their physical locations on the chromosome (Supplementary Table 1). The AhWRKY genes vary in length from 498 bp (AhWRKY22 and AhWRKY31) to 10,044 bp (AhWRKY59), and their coding proteins range from 91 (AhWRKY22) to 1,345 (AhWRKY59) amino acids. The MWs of AhWRKYs were between 10.4 (AhWRKY22) and 148.6 kDa (AhWRKY132), with the majority of them ranging from 20 to 85 kDa in peanuts. The predicted pI ranged from 4.88 (AhWRKY55) to 10.08 (AhWRKY132), with an average of 7.06, among which 67 AhWRKYs had pI > 7, and 107 AhWRKYs had pI <7.
To explore the evolutionary relationship between AhWRKY TFs, a phylogenetic tree was constructed using 174 AhWRKY proteins in peanuts and 74 AtWRKYs from Arabidopsis (Figure 1). In brief, 174 WRKY TFs were divided into four groups, group I (35), group II (107), group III (31), and group IV (1); group II was further divided into five subgroups, subgroup II-a (6), subgroup II-b (22), subgroup II-c (43), subgroup II-d (15), and subgroup II-e (21) (Figure 1; Supplementary Table 1).
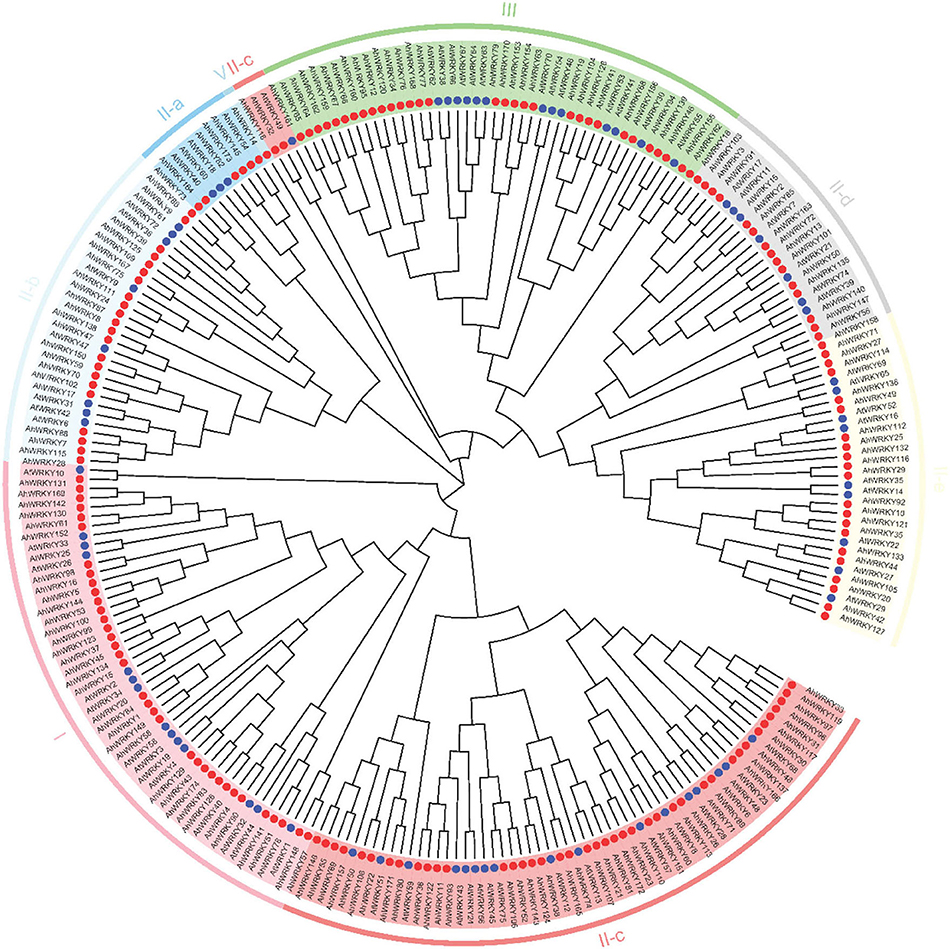
Figure 1. Phylogenetic tree of WRKY in A. hypogaea cv. Tifrunner and Arabidopsis. All full-length protein sequences were aligned by MUSCLE, and the tree was generated using the neighbor-joining (NJ) method in MEGA7 software. Different colors indicated groups I, II-a, II-b, II-c, II-d, II-e, III, and IV subfamily WRKY proteins from peanuts. The blue and red circles indicate the Arabidopsis thaliana and A. hypogea protein.
The mapping results showed that 174 AhWRKYs were found to be unevenly distributed on 20 chromosomes using genome chromosomal location analysis (Figure 2). Most genes were present on chromosomes 4 and 13, whereas 16 genes belonging to the A and B genomes, respectively, accounted for 7.47% (13 genes) and 8.05% (14 genes), 7.47% (13 genes) of the total gene numbers, and the fewest genes were scattered on chromosome 9 from the A genome and chromosome 19 of the B genome, accounting for 1.15% (2 genes) and 1.72% (3 genes), respectively.
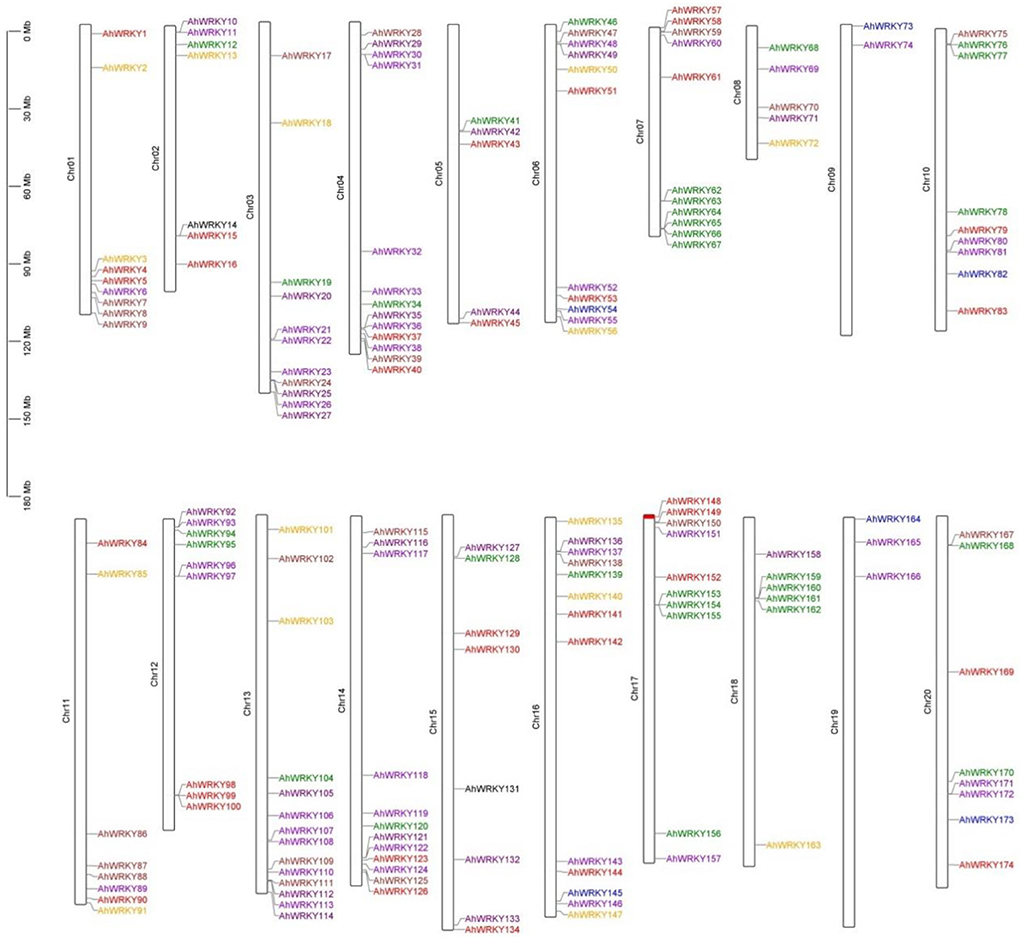
Figure 2. Chromosome mapping of AhWRKY genes. The scale represents a 30-Mb chromosomal distance. Red, sky blue, brown, DarkViolet, orange, purple, green, and blue indicated groups I, II-a, II-b, II-c, II-d, II-e, III, and IV, respectively.
Chromosomal mapping showed there are many gene clusters of AhWRKY in different chromosomes. For example, AhWRKY3-9 on chr1, AhWRKY23-27 on chr3, AhWRKY35-40 on chr4, AhWRKY47-49 on chr6, AhWRKY62-67 on chr7, AhWRKY75-77 on chr10, AhWRKY98-100 on chr12, AhWRKY136-139 on chr16, AhWRKY153-155 on chr17, and AhWRKY159-162 on chr18.
Collinearity analysis showed that among 174 AhWRKYs, 132 pairs of segmental duplication genes were identified; these were divided into seven groups according to the phylogenetic tree (Figure 3). Maximum segmental duplication events occurred in group II-c (34 pairs), followed by group III (22 pairs), group II-e (19 pairs), group II-b (18 pairs), group I (18 pairs), group II-d (13 pairs) and group II-a (8 pairs). Seven pairs of tandem duplication genes were detected, such as AhWRKY99 and AhWRKY100 in group I (Figure 3), AhWRKY30 and AhWRKY31, AhWRKY96 and AhWRKY97 in group II-c, AhWRKY64 and AhWRKY65, AhWRKY66 and AhWRKY67, AhWRKY76 and AhWRKY77, and AhWRKY153 and AhWRKY154 in group III. The Ka/Ks for segmental duplication was 0.05–1.21 with an average of 0.31, while the ratio of tandem duplication ranged from 0.42 to 0.94 with an average of 0.63. Only the Ka/Ks ratio of one pair (AhWRKY119 and AhWRKY33) was 1.21, which indicated the gene pair was subjected to positive selection, and others were strongly subjected to pure selection. These segmental and tandem duplications may occur in 0.34–186.58 and 6.11–35.81 Mya, respectively (Figure 3; Supplementary Table 2).
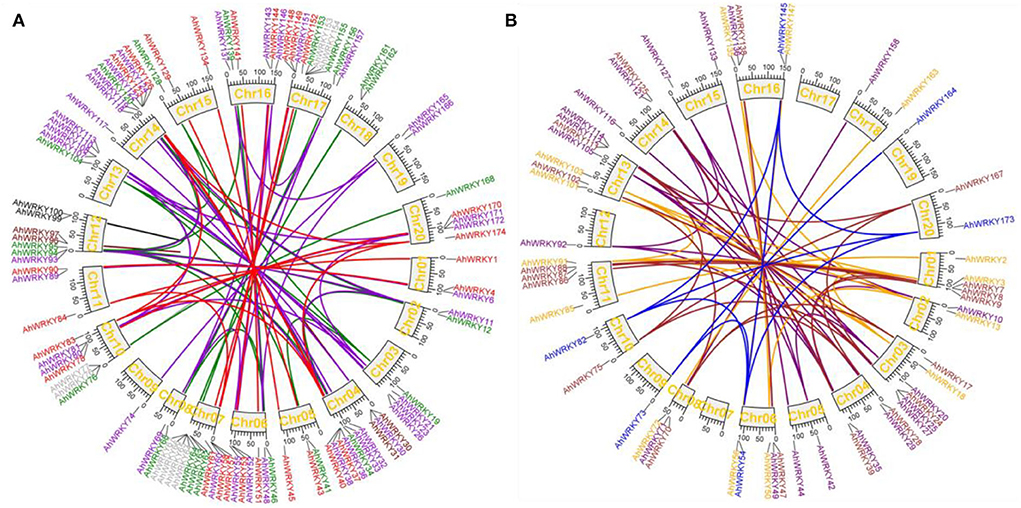
Figure 3. Circos figures for chromosome locations with AhWRKY duplication links. (A) Groups I, II-c, and III subfamily duplication links. Red, dark violet, and green lines indicated groups I, II-c, and III subfamily segmented, and black, maroon, and dark gray lines indicated groups I, II-c, and III subfamily tandem duplicated gene pairs, respectively. (B) Groups II-a, II-b, II-d, and II-e subfamily duplication links. Blue, brown, orange, and purple lines indicated group II-a, II-b, II-d, and II-e subfamily segmented duplicated gene pairs, respectively.
The numbers of introns varied among 174 AhWRKY genes. All the AhWRKY family genes contain one to eight introns. Most members of group I and subgroup II-b have four introns, whereas most members of subgroup II-c, subgroup II-d, subgroup II-e, and group III have two introns. Four members of subgroup II-a have three introns, and two members have four introns. The member of group IV has two introns (Figures 4A,B; Supplementary Table 2). There was some subgroup specificity, possibly attributed to the number of changes of introns during evolution. In contrast, the number and position of the introns were relatively conserved in the same group of plant species (Figures 4A,C).
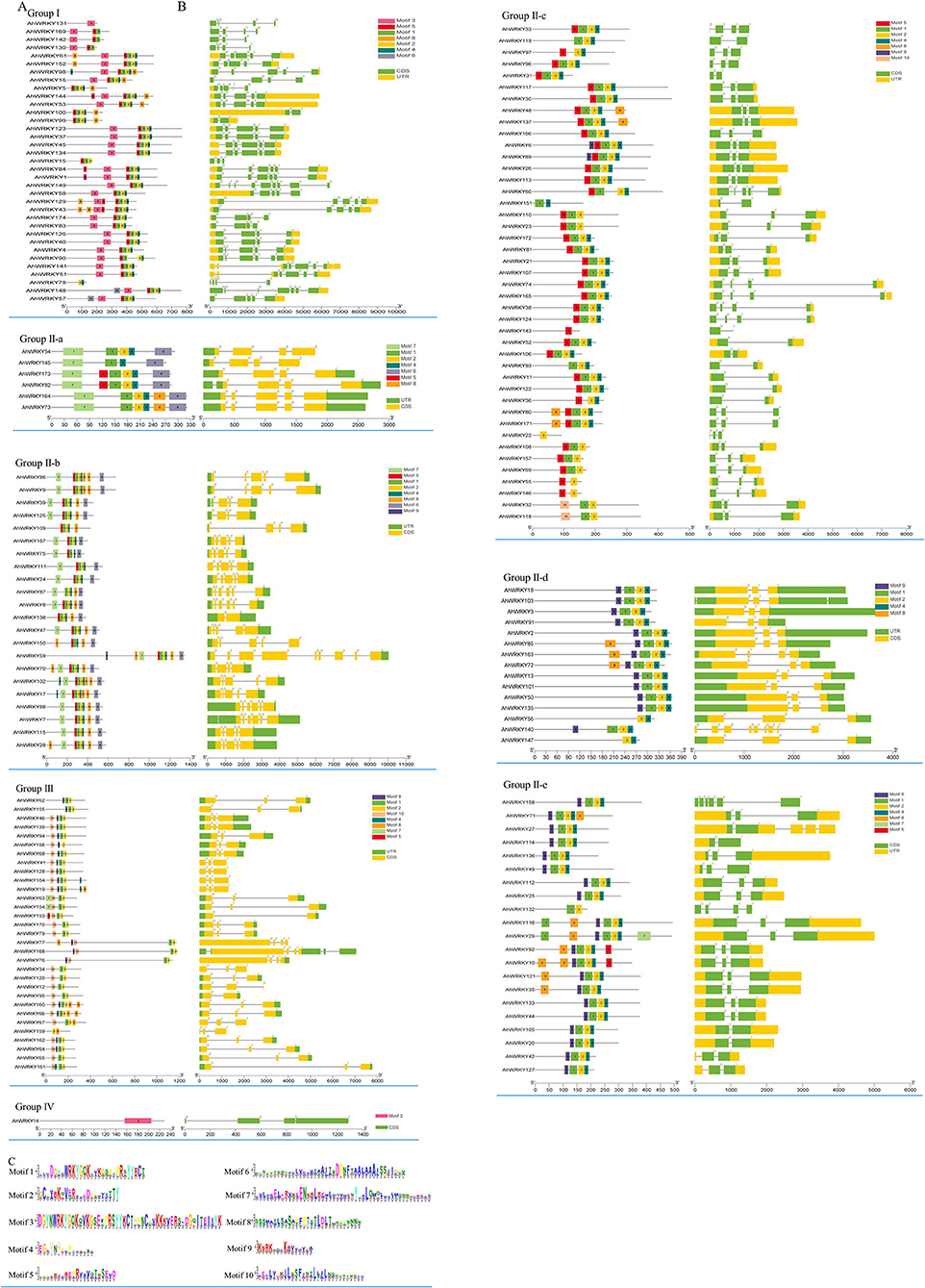
Figure 4. Gene structures and conserved motif analysis of AhWRKY genes. (A) The conserved motif of AhWRKY. (B) The exon–intron structure of AhWRKY. (C) The amino acid composition of each motif. Each conserved motif is represented by various-colored rectangles. Box length corresponds to motif length. Different colors represent different group members. Due to the large image size, we have presented the structures and motifs in various group-wise.
Ten conserved motifs of 174 AhWRKY genes were analyzed using the MEME software (Figures 4A–C). Motif 1 was dominantly present in the WRKY domain regions of most family members. The proteins of the same group showed identical numbers and arrangements of motifs, five motifs were detected in group I (motifs 3, 5, 1, 2, and 4), motif 3 in the N terminal, and motif 1 in the C terminal contained WRKY. Five and seven motifs were detected in subgroup II-a (motifs 7, 1, 2, 4, and 6) and II-b (motifs 7, 5, 1, 2, 4, 8, and 6), four motifs in subgroup II-c (motifs 5, 2, 1, and 4), II-d (motifs 9, 2, 1, and 4) and II-e (motifs 9, 2, 1, and 4), three motifs in group III (motifs 9/10, 1, and 2), and one motif in group IV (motif 3). Motifs in the same group showed great similarity, indicating the functional conservation in different groups. Comparing the intron–exon structure and conserved motif analysis, members of the same group showed great similarity in characteristics, indicating that most AhWRKY genes were highly conserved among groups.
To further study the potential regulatory mechanism of the AhWRKY genes, the 2 kb upstream sequence of the AhWRKY genes translation start site was used to detect the cis-elements. A total of 55 known cis-elements (31 light-related elements, 11 hormone-related elements, seven tissue-specific elements, and six stress-related elements) were detected (Figure 5). MBS (drought inducibility), TC-rich repeats (defense and stress responsiveness), WUN motif (wound responsiveness), LTR (low-temperature responsiveness), ARE (essential for the anaerobic induction), and GC motif (anoxic specific inducibility) involved in stress responses are found in 52.5, 2.9, 14.9, 2.6, 12.1, and 15% of AhWRKY promoters, respectively. Meanwhile, there are a large number of hormonal response elements, including ABRE, AuxRR, and TGA-element, CGTCA motifs and TGACG-motif, GARE motifs, P-box, and TATC-box, O2 site, TCA element, and SARE. Promoters of AhWRKY possess a relatively large number of ARE elements, 83, 100, 82, 78, 73, 76, 81, and 100% family members of the group I, II-a, II-b, II-c, II-d, II-e, III, and IV possessed ARE elements. Notably, 20% of family members of group I possessed AuxRE, 91% of family members of subgroup II-b possessed ABRE elements, 54 and 48% of family members of subgroup II-c and II-e possessed TCA-element, 47% of family members of subgroup II-d possessed P-box, 84% family members of group III possessed CGTCA-motif. There was a divergence in cis-acting elements in promoter regions of different groups.
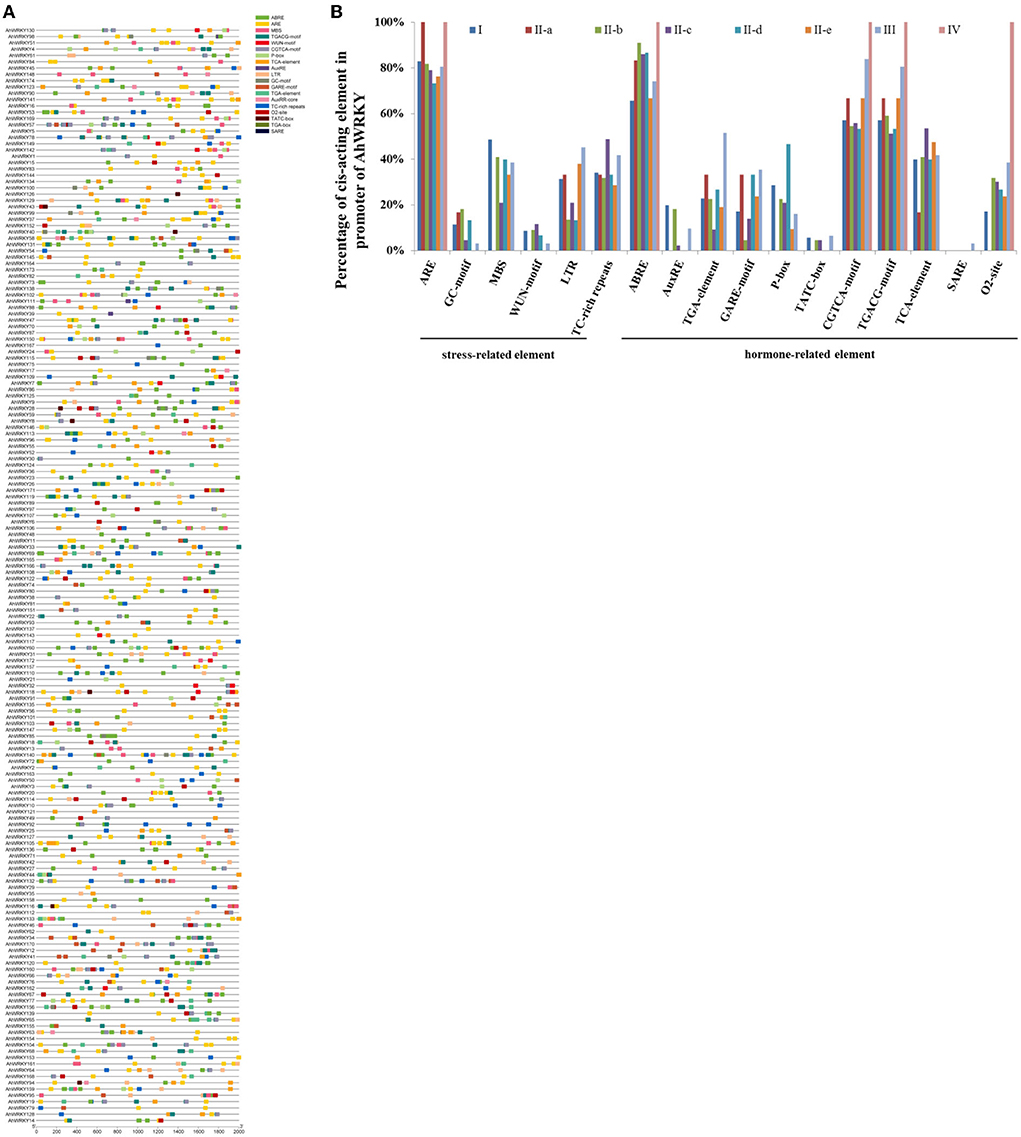
Figure 5. Identification of the cis-acting element in the promoter of AhWRKY genes. (A) Each type of element is represented by a number of colored rectangles. Box length corresponds to element length. Different colors represent different group members. (B) Percentage of each cis-acting element in the promoter of the AhWRKY.
Generally, the gene expression had a similar pattern within the same phylogenetic classes. Members of group I showed high gene expression levels in peg tip Pat, fruit Pat. 1, fruit Pat. 3, pericarp Pat. 5, and pericarp Pat. 6, and two members (AhWRKY61 and AhWRKY152) were also highly expressed in root and nodule. The expression pattern of members of subgroup II-a was similar to group I, and two members (AhWRKY163 and AhWRKY74) showed the same trend as AhWRKY61 and AhWRKY152. Genes of subgroup II-b showed low expression levels in all tissues, but AhWRKY115 and AhWRKY28 had the same expression trend as AhWRKY61 and AhWRKY152. Members of subgroup II-c were highly expressed in fruit Pat. 3, pericarp Pat. 5, and pericarp Pat. 6, most members of subgroup II-d were highly expressed in all tissues, and most members of subgroup II-e and group III were less expressed in all tissues (Figure 6).
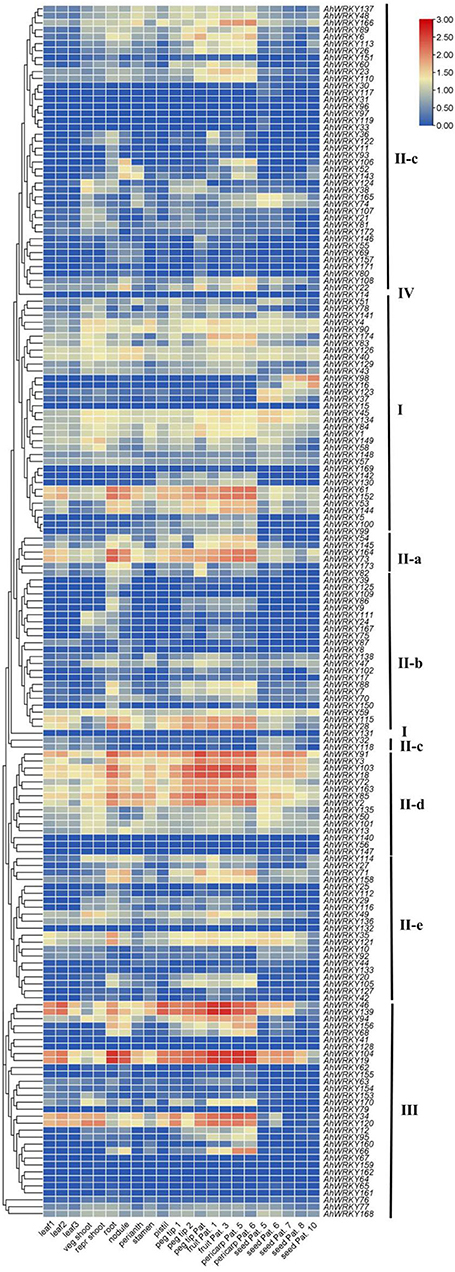
Figure 6. Expression profiles of AhWRKY genes across the different tissues. Gene expression is expressed in l g (TPM+1). Seedling leaf 10 days post-emergence (leaf 1), main stem leaf (leaf 2), lateral stem leaf (leaf 3), vegetative shoot tip from the main stem (veg shoot), reproductive shoot tip from first lateral (repr shoot), 10-day roots (root), 25-day nodules (nodule), aerial gynophore tip (peg tip 1), subterranean peg tip (peg tip 2), Pattee 1 stalk (peg tip Pat. 1), Pattee 1 pod (fruit Pat. 1), Pattee 3 pod (fruit Pat. 3), Pattee 5 pericarp (pericarp Pat. 5), Pattee 6 pericarp (pericarp Pat. 6), Pattee 5 seed (seed Pat. 5), Pattee 6 seed (seed Pat. 6), Pattee 7 seed (seed Pat. 7), Pattee 8 seed (fruit Pat. 8), and Pattee 10 seed (seed Pat. 10).
The expression patterns of AhWRKY genes in group III differed between the two cultivars in response to R. solanacearum infection (Figure 7). One and six members of group III showed the highest and lowest expression in 6 h, eight and five in 12 h, and 13 highest in 24 h after infection with R. solanacearum in ganhua18. Four and eighth members of group III showed the highest and lowest expression in 6 h, seven and one in 12 h, six and three in 24 h, one highest expressed in 48 and 72 h, and five highest expressed in 96 h after infection with R. solanacearum in kanong313. AhWRKY139, AhWRKY63, AhWRKY154, AhWRKY120, AhWRKY160, AhWRKY159, AhWRKY162, AhWRKY64, AhWRKY65, and AhWRKY161 were downregulated in ganhua18 after R. solanacearum infection. Whereas, AhWRKY63, AhWRKY154, AhWRKY153, AhWRKY66, AhWRKY67, AhWRKY159, AhWRKY162, AhWRKY65, AhWRKY77, and AhWRKY161 were downregulated in kainong313 after R. solanacearum infection, these genes may negatively regulate bacterial wilt resistance in peanuts. Moreover, AhWRKY46, AhWRKY94, AhWRKY156, AhWRKY68, AhWRKY41, AhWRKY128, AhWRKY104, AhWRKY19, AhWRKY62, AhWRKY155, AhWRKY153, AhWRKY170, AhWRKY78, AhWRKY34, AhWRKY12, AhWRKY95, AhWRKY66, AhWRKY67, and AhWRKY76 were upregulated in ganhua18 after R. solanacearum infection. On the other hand, AhWRKY46, AhWRKY94, AhWRKY156, AhWRKY68, AhWRKY41, AhWRKY128, AhWRKY104, AhWRKY19, AhWRKY62, AhWRKY155, AhWRKY170, AhWRKY78, AhWRKY34, AhWRKY120, AhWRKY12, AhWRKY95, AhWRKY160, AhWRKY64, AhWRKY161, and AhWRKY76 were upregulated in kainong313 after R. solanacearum infection, indicating that these genes may positively regulate bacterial wilt resistance in peanuts (Figure 7).
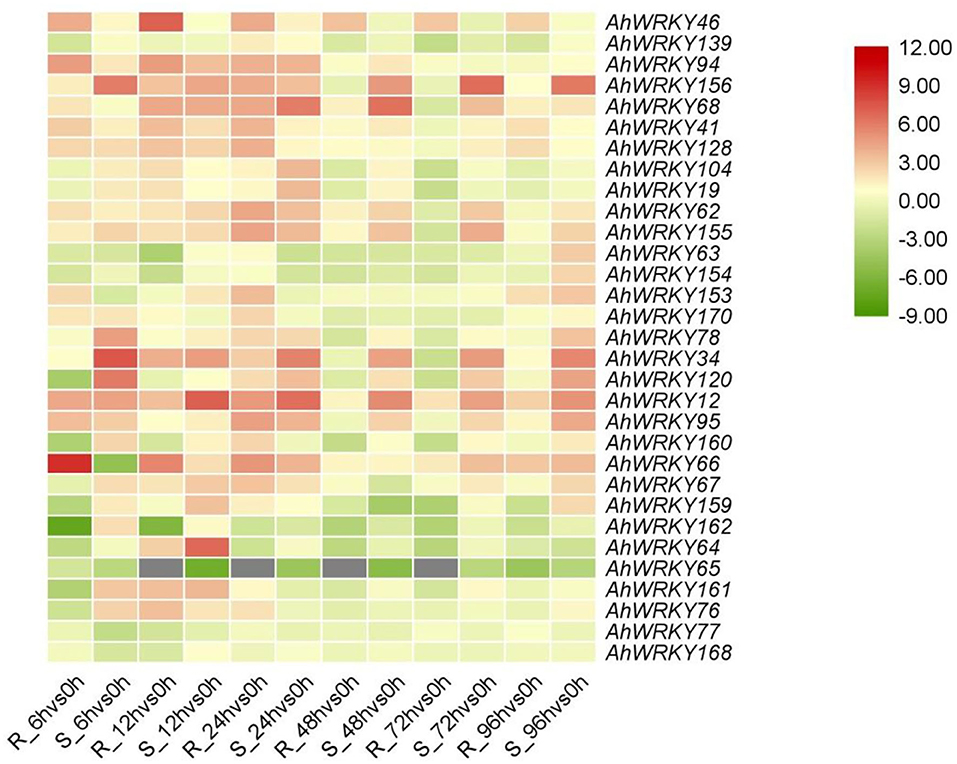
Figure 7. Expression of AhWRKYs in the two cultivars infected with Ralstonia solanacearum. The qRT-PCR of each gene in peanut roots infected with Ralstonia solanacearum in the two cultivars is shown. Heatmap showed the log2-transformed ratio values. R, Resistance; S, Susceptibility.
After 6 h of R. solanacearum infection, the expressions of AhWRKY120, AhWRKY159, AhWRKY161, and AhWRKY76 were downregulated by 0.07, 0.13, 0.1, and 0.28 than control in ganhua18, while that in kainong313 were increased by 66.91, 3.12, 8.19 and 6.05 than control. After 72 h of infection with the R. solanacearum, the expressions of AhWRKY155, AhWRKY78, and AhWRKY34 were downregulated by 0.34, 0.45, and 0.24 than control in ganhua18, while that in kainong313 were increased by 17.46, 2.04, and 27.58 than control. After 6 h and 12 h infection with the R. solanacearum, the expression of AhWRKY162 was downregulated by 0.01 and 0.02 than control in ganhua18, while that in kainong313 was increased by 4.35 and 2.11 than control. After 6 h, 12 h, and 72 h infection with the R. solanacearum, the expression of AhWRKY160 was downregulated by 0.09, 0.39, and 0.21 than control in ganhua18, while that in kainong313 was increased by 5.64, 2.56, and 2.17 than control. After 6 h infection with the R. solanacearum, the expression of AhWRKY153 was upregulated by 5.03 than control in ganhua18, while that in kainong313 was reduced by 0.41 than control (Figure 7). The differences in bacterial wilt resistance of two cultivars may be attributed to the expression patterns of these genes.
This study forecasted 18 miRNAs targeting 50 AhWRKY genes (Figure 8; Supplementary Table 3). The comprehensive data of all miRNAs targeted genes/sites is given in Supplementary Table 3. Overall, the results established that ahy-miR3520-5p target a maximum number of genes (13), followed by ahy-miR159 and ahy-miR156b-3p which targets six AhWRKY genes (Figure 8). Whereas, some genes are only targeted by one miRNA, such as AhWRKY47, AhWRKY16, AhWRKY142, and AhWRKY140 (Figure 8). In further investigations, the transcript levels of these miRNAs and their targeted genes need confirmation to govern their biological acts in peanut breeding.
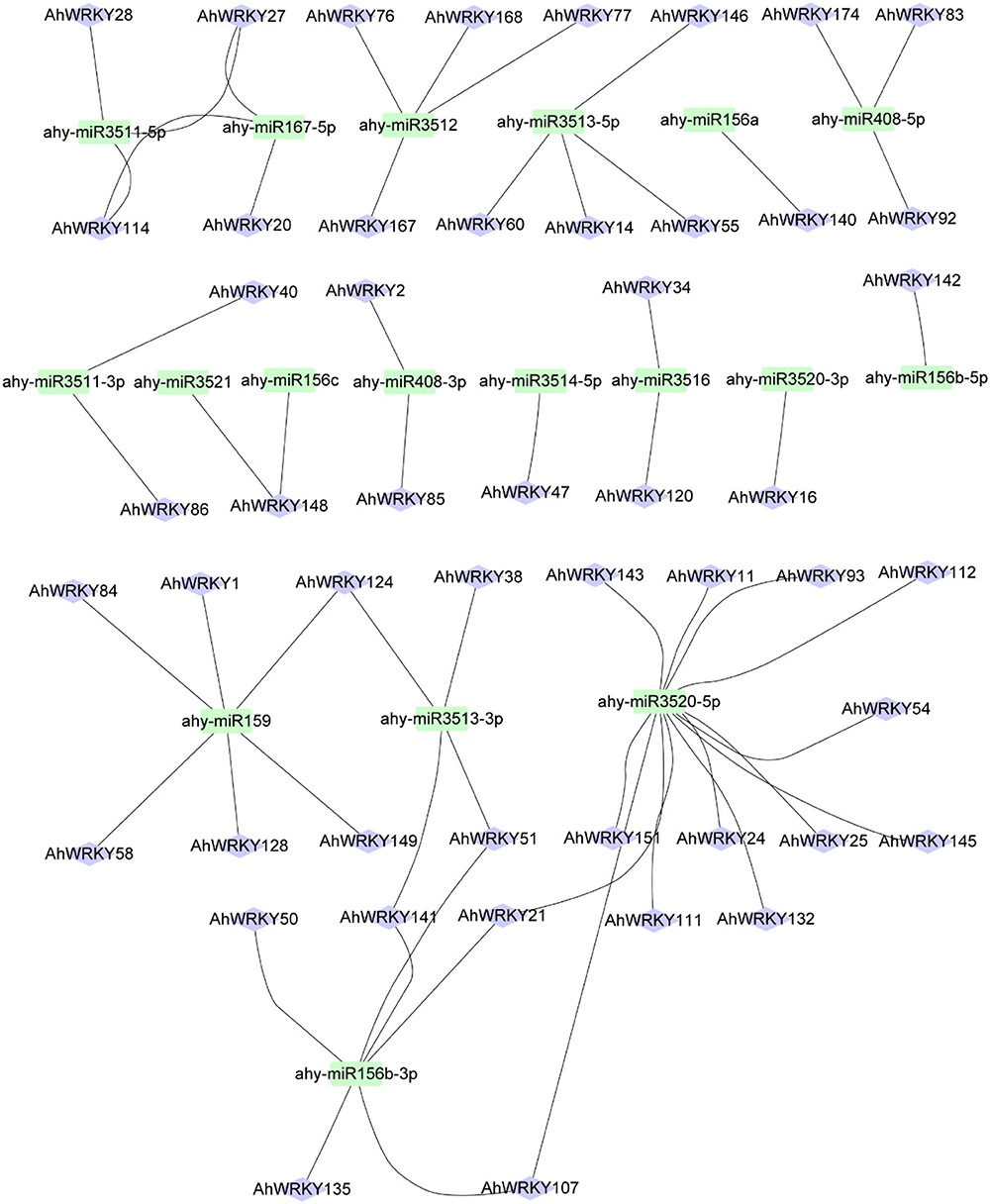
Figure 8. Network figure of identified miRNA targeting AhWRKY genes. Green box colors correspond to miRNAs, and bluish ellipse shapes represent target WRKY genes.
As one of the most important and largest TF, WRKY TFs play a critical role in response to biotic stresses in plants (Rushton et al., 2010; Javed et al., 2020, 2022). WRKY families have been reported in many plants, but little is known regarding the functions of the WRKY family in A. hypogaea. There were 77, 75, and 158 WRKY proteins in Arachis duranensis, Arachis ipaënsis, and Arachis hypogaea with reference to the first version of the A. hypogaea genome, respectively (Song et al., 2016; Zhao et al., 2020). Based on the first and second versions of the A. hypogaea genome released on June 4, 2021, this study identified 174 AhWRKY genes in peanuts, 83 WRKY genes in the A genome, and 91 in the B genome. In our study, there were 17 more genes in A. hypogea genome and one fewer genes in A and B genomes than previous reports. Previous studies analyzed the expressions of AhWRKY under drought stress in peanuts (Zhao et al., 2020); however, this study analyzed expressions of AhWRKY genes in response to R. solanacearum infection in peanuts. The high multiplicity/unpredictability in the quantity of WRKY TFs among diverse plant species could be due to unalike evolutionary events or duplication of whole genomes during the evolutionary period. In particular, the distinctive reports of replicated (tandem/segmental) genes succeeding in the repetition of entire genomes could also be accountable for continuing evolutionary changeovers and crop domestication (Leister, 2004; Bertioli et al., 2016, 2019; Van De Peer et al., 2017; Bohra et al., 2022). Nevertheless, previous reports also validated that segmental and tandem repetitions, particularly for the previous events, could be important driving factors in the evolution and enlargement of WRKY TFs in diverse crop plants (Song et al., 2016; Xie et al., 2018; Zhao et al., 2020; Javed et al., 2022).
Mutations and deletions prevail in the plant WRKY domain (Wei et al., 2012). Five WRKY genes deleted one WRKY domain in Helianthus annuus L. (Li J et al., 2020). Nine WRKY genes deleted one WRKY domain, seven WRKY genes deleted zinc finger, and three WRKY genes deleted one WRKY motif in maize (Hu et al., 2021). Previous reports suggest that mutations also occurred in the WRKY motif of most WRKY genes in peanuts (Song et al., 2016; Zhao et al., 2020). In this study, there were 17 AhWRKYs mutated and 13 AhWRKYs deleted in the WRKY domain. One group I member AhWRKY motif changed to WRKYGEK. Seven members of subgroup II-c AhWRKY motif changed to WRKYGKK, and two members changed to WRKYGEK, two members changed to WRKYGRK. WRKY motif of AhWRKY36 (subgroup II-c), AhWRKY131 (I), AhWRKY97 (subgroup II-e), and AhWRKY14 (IV) changed to WRK, WRMYGQK, WHKYGKK, and GRKYGQK, respectively. Moreover, 12 AhWRKYs mutated and deleted in zinc fingers. Zinc finger deleted in C terminal of AhWRKY130 (group I) and N terminal of AhWRKY169 (group I). Zinc finger deleted in AhWRKY145 (subgroup II-a), AhWRKY75 (subgroup II-b), AhWRKY150 (subgroup II-b), AhWRKY9 (subgroup II-c), AhWRKY143 (subgroup II-c), and AhWRKY151 (subgroup II-c). Zinc finger changed to C-X4-C-X25-H-X-S in AhWRKY22 (subgroup II-c), C-X5-C in AhWRKY132 (subgroup II-e), C-X4-C-X22-H-X-Y in AhWRKY14 (IV), and C-X4-C in AhWRKY131 (I). There are many mutations in the zinc finger of group I and subgroup II-e and the WRKY motif of subgroup II-c presumed that the variation in members of group I, subgroup II-c, and II-e generate new functions.
Gene duplication plays an important role in the amplification and evolution of plant gene families (Cannon et al., 2004; Leister, 2004; Bertioli et al., 2016; Van De Peer et al., 2017). Among gene clusters, five pairs of tandem duplication and nine pairs of segmental duplication genes were identified; seven pairs of tandem duplication and 132 pairs of segmental duplication genes were identified in 14 and 144 AhWRKYs (Figure 3), which were one more tandem duplication and nine segmental duplications than the previous study (Zhao et al., 2020). The divergence of two wild ancestral diploids of cultivated tetraploid peanuts may occur in 2.16 MYA (Chen et al., 2019b). Ninety-nine pairs of segmental duplication genes occurred before the divergence of two wild ancestral diploids, 31 pairs of tandem duplication, and six pairs of segmental duplication genes occurred after the divergence of two wild ancestral diploids. The ka/ks ratios of 99.4% (173/174) of AhWRKYs were less than one, which suggested that most AhWRKYs were selected for purification. The ka/ks ratios of one gene pair, including AhWRKY119 and AhWRKY33, was 1.21 (more than 1), which indicated that these genes were in a state of positive selection in peanuts, evolving rapidly, and might be very important for the evolution of peanuts.
Prediction analysis of cis-acting elements with AhWRKYs indicated many light-related, hormone-related, tissue-specific, and stress-related elements in the promoters of AhWRKY genes (Figure 5), which was in accordance with the previous study (Zhao et al., 2020) and speculated that AhWRKY is involved in various biological processes.
Tissue-specific expression suggested that genes from the same group indicated similar expression patterns (Figure 6). Tissue-specific expression was in agreement with the tissue-specific elements in the promoters of AhWRKY genes. For instance, AhWRKY16 was highly expressed in seed, and an RY-element, which is a cis-acting regulatory element involved in seed-specific regulation in the promoter, and AhWRKY158 was highly expressed in nodule and a nodule-site1 element, which is a nodule-specific factor binding site in its promoter (Figures 5, 6).
LRR is closely related to the disease resistance of plants (Jones and Dangl, 2006; McHale et al., 2006). It is known that some WRKY proteins contain the NBS-LRR domain (Deslandes et al., 2003; Shen et al., 2007; Liu et al., 2016), and WRKY proteins from group III are mainly involved in plant disease resistance (Kalde et al., 2003; Eulgem and Somssich, 2007). AtWRKY52 possessed the NBS-LRR domain and WRKY domain, which is a critical gene in regulating bacterial wilt resistance (Deslandes et al., 1998, 2002). CaWRKY58 and CaWRKY40b (II-a) negatively regulate bacterial wilt resistance, while CaWRKY22 (II-e) and CaWRKY27 (II-e), CaWRKY28 (II-c), CaWRKY30 (III), and CaWRKY41 (III) positively regulate bacterial wilt resistance in pepper (Wang et al., 2012; Dang et al., 2014, 2019; Hussain et al., 2018, 2021; Muhammad et al., 2018; Yang et al., 2021). SiWRKY53 (III) responded to the bacterial wilt resistance (Mishra et al., 2021). WRKY members of group III played a vital role in plant bacterial wilt resistance. In this study, AhWRKY76 and AhWRKY77 of group III had LRR domain. qRT-PCR results showed that AhWRKY77 was downregulated by 0.34 than control in 12 h of ganhua18 and 0.19 than control 6 h of kainong313 after infection with the R. solanacearum, which may negatively regulate bacterial wilt resistance in peanuts. After R. solanacearum infection, AhWRKY76 displayed lower-higher expressions, i.e., 0.28, 10.35, and 3.97 than control in 6 h, 12 h, and 24 h of ganhua18, respectively, and higher expressions by 6.05 and 3.53 than control in 6 h and 12 h in kainong313, respectively, which may contribute to explain the difference in BW resistance between the two cultivars. These outcomes suggest that AhWRKY76 and AhWRKY77 may play a critical role in bacterial wilt resistance in peanuts. AhWRKY76 and AhWRKY77 targeted by ahy-miR3512 showed that ahy-miR3512 might have a vital role in BW resistance (Figure 7).
In conclusion, a total of 174 AhWRKY genes were identified in A. hypogaea. The classification, chromosomal location, collinearity, protein structure, conserved motif composition, cis-acting elements, putative miRNAs, and phylogenetic relationship of AhWRKYs were systematically analyzed, which provide a basis for further study of the molecular and functional structure of AhWRKYs. Furthermore, the expression patterns of tissues-specific and pathogen-responsive AhWRKYs were obtained from RNA-seq data and qRT-PCR results, providing useful information for further functional investigations in response to biotic stresses. Ten AhWRKY genes (AhWRKY34, AhWRKY76, AhWRKY78, AhWRKY120, AhWRKY153, AhWRKY155, AhWRKY159, AhWRKY160, AhWRKY161, and AhWRKY162) from group III displayed different expression patterns in sensitive and resistant peanut genotypes infected with the R. solanacearum. Two AhWRKY proteins (AhWRKY76 and AhWRKY77) from group III obtained the LRR domain; AhWRKY77 was downregulated in two cultivars, and AhWRKY76 showed lower-higher expression ganhua18 and higher expression in kainong313. Both AhWRKY76 and AhWRKY77 were targeted by ahy-miR3512, which may have an important function in peanut disease resistance. The positively upregulated genes could be used for further characterization in peanuts to develop disease-smart peanut cultivars. In short, our study could help researchers better understand the function and regulatory mechanism of AhWRKYs genes in A. hypogaea during pathogen response.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
LY carried out the bioinformatic analysis and drafted the manuscript. LY and HJ collected the plant materials and performed the experiments. AR helped with some analysis and improved the manuscript. LY, AR, and YH participated in handling figures and tables. LY, AR, DG, and XZ edited and revised the manuscript. All authors read and approved the final version of the manuscript.
This work was supported by the Agricultural Collaborative Innovation Project of Jiangxi Province, China (JXXTCXBSJJ2022007 and JXXTCX202112), and the Third Census and Collection of Crop Germplasm Resources in China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.986673/full#supplementary-material
Ansari, S., Kumar, V., Bhatt, D. N., Irfan, M., and Datta, A. (2022). N-acetylglucosamine sensing and metabolic engineering for attenuating human and plant pathogens. Bioengineering 9, 64. doi: 10.3390/bioengineering9020064
Baillo, E. H., Hanif, M. S., Guo, Y., Zhang, Z., Xu, P., and Algam, S. A. (2020). Genome-wide Identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L.) moench). PLoS ONE 15, e0236651. doi: 10.1371/journal.pone.0236651
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2016). The genome sequences of arachis duranensis and arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. doi: 10.1038/ng.3517
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884. doi: 10.1038/s41588-019-0405-z
Bohra, A., Tiwari, A., Kaur, P., Ganie, S. A., Raza, A., Roorkiwal, M., et al. (2022). The key to the future lies in the past: insights from grain legume domestication and improvement should inform future breeding strategies. Plant Cell Physiol. pcac086. doi: 10.1093/pcp/pcac086. [Epub ahead of print].
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., and Xia, R. (2020). TB tools: An integrative toolkit developed for in-teractive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Li, C., Wang, H., and Guo, Z. (2019a). WRKY transcription factors: evolution, binding, and action. Phytopathol. Res. 1, 13. doi: 10.1186/s42483-019-0022-x
Chen, X., Lu, Q., Liu, H., Zhang, J., Hong, Y., Lan, H., et al. (2019b). Sequencing of cultivated peanut, Arachishypogaea, yields insights into genome evolution and oil improvement. Mol. Plant. 12, 920–934. doi: 10.1016/j.molp.2019.03.005
Chen, Y., Ren, X., Zhou, X., Huang, L., Yan, L., Lei, Y., et al. (2014). Dynamics in the resistant and susceptible peanut (Arachis hypogaea L.) root transcriptome on infection with the Ralstonia solanacearum. BMC Genomics 15, 1078. doi: 10.1186/1471-2164-15-1078
Clevenger, J., Chu, Y., Scheffler, B., and Ozias-Akins, P. (2016). A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 7, 1446. doi: 10.3389/fpls.2016.01446
Dang, F., Lin, J., Chen, Y., Li, G. X., and He, S. (2019). A feedback loop between CaWRKY41 and H2O2 coordinates the response to Ralstonia solanacearum and excess cadmium in pepper. J. Exp. Bot. 70, 1581–1595. doi: 10.1093/jxb/erz006
Dang, F., Wang, Y., She, J., Lei, Y., Liu, Z., Eulgem, T., et al. (2014). Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant. 150, 397–411. doi: 10.1111/ppl.12093
Deslandes, L., Olivier, J., Peeters, N., Feng, D. X., Khounlotham, M., Boucher, C., et al. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III ffector targeted to the plant nucleus. Proc Natl Acad Sci USA. 100, 8024–8029. doi: 10.1073/pnas.1230660100
Deslandes, L., Olivier, J., Theulieres, F., Hirsch, J., et al. (2002). Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. U. S. A. 99, 2404–2409. doi: 10.1073/pnas.032485099
Deslandes, L., Pileur, F., Liaubet, L., Camut, S., Can, C., Williams, K., et al. (1998). Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum. Mol. Plant Microbe Interact. 11, 659–667. doi: 10.1094/MPMI.1998.11.7.659
Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. doi: 10.1023/A:1020780022549
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Farooq, M. S., Uzair, M., Raza, A., Habib, M., Xu, Y., Yousuf, M., et al. (2022). Uncovering the research gaps to alleviate the negative impacts of climate change on food security: a review. Front. Plant Sci. 13, 927535. doi: 10.3389/fpls.2022.927535
Gangurde, S. S., Kumar, R., Pandey, A. K., Burow, M., and Pandey, M. K. (2019). “Climate-smart groundnuts for achieving high productivity and improved quality: current status, challenges, and opportunities”, in Genomic Designing of Climate-Smart Oilseed Crops, ed C. Kole (Cham: Springer), 133–172. doi: 10.1007/978-3-319-93536-2_3
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Re. 31, 3784–3788. doi: 10.1093/nar/gkg563
Haider, S., Raza, A., Iqbal, J., Shaukat, M., and Mahmood, T. (2022). Analyzing the regulatory role of heat shock transcription factors in plant heat stress tolerance: a brief appraisal. Mol. Biol. Rep. 49, 5771–5785. doi: 10.1007/s11033-022-07190-x
He, Y., Mao, S., Gao, Y., Zhu, L., and Qian, W. (2016). Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in Brassica napus. PLoS ONE 11, e0157558. doi: 10.1371/journal.pone.0157558
Hikichi, Y., Mori, Y., Ishikawa, S., Hayashi, K., Ohnish, I. K., Kiba, A., et al. (2017). Regulation involved in colonization of intercellular spaces of host plants in Ralstonia solanacearum. Front. Plant Sci. 8, 967. doi: 10.3389/fpls.2017.00967
Hu, W., Ren, Q., Chen, Y., Xu, G., and Qian, Y. (2021). Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 21, 427. doi: 10.1186/s12870-021-03206-z
Hussain, A., Khan, M. I., Albaqami, M., Mahpara, S., Noorka, I. R., Ahmed, M. A. A., et al. (2021). CaWRKY30 positively regulates pepper immunity by targeting CaWRKY40 against Ralstonia solanacearum inoculation through modulating defense-related genes. Int. J. Mol. Sci. 22, 12091. doi: 10.3390/ijms222112091
Hussain, A., Li, X., Weng, Y., Liu, Z., Ashraf, M. F., Noman, A., et al. (2018). CaWRKY22 Acts as a Positive Regulator in Pepper Response to Ralstonia Solanacearum by Constituting Networks with CaWRKY6, CaWRKY27, CaWRKY40, and CaWRKY58. Int J Mol Sci. 19, 1426. doi: 10.3390/ijms19051426
Irfan, M., Ghosh, S., Meli, V. S., Kumar, A., Kumar, V., Chakraborty, N., et al. (2016). Fruit ripening regulation of α-Mannosidase expression by the MADS box transcription factor RIPENING INHIBITOR and ethylene. Front. Plant Sci. 7, 10. doi: 10.3389/fpls.2016.00010
Irfan, M., Kumar, P., Ahmad, I., and Datta, A. (2021). Unraveling the role of tomato Bcl-2-associated athanogene (BAG) proteins during abiotic stress response and fruit ripening. Sci. Rep. 12, 3503. doi: 10.1038/s41598-021-01185-7
Javed, T., Shabbir, R., Ali, A., Afzal, I., Zaheer, U., and Gao, S. J. (2020). Transcription factors in plant stress responses: challenges and potential for sugarcane improvement. Plants 9, 491. doi: 10.3390/plants9040491
Javed, T., Zhou, J. R., Li, J., Hu, Z. T., Wang, Q. N., and Gao, S. J. (2022). Identification and expression profiling of WRKY family genes in sugarcane in response to bacterial pathogen infection and nitrogen implantation dosage. Front. Plant Sci. 13, 917953. doi: 10.3389/fpls.2022.917953
Jiang, G., Wei, Z., Xu, J., Chen, H., Zhang, Y., She, X., et al. (2017). Bacterial wilt in China: history, current status, and future perspectives. Front. Plant Sci. 8, 1549. doi: 10.3389/fpls.2017.01549
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature. 444, 323–329. doi: 10.1038/nature05286
Kalde, M., Barth, M., Somssich, I. E., and Lippok, B. (2003). Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol Plant Microbe Interact. 16, 295–305. doi: 10.1094/MPMI.2003.16.4.295
Kaliyappan, R., Viswanathan, S., Suthanthiram, B., Subbaraya, U., Marimuthu Somasundram, S., and Muthu, M. (2016). Evolutionary expansion of WRKY gene family in banana and its expression profile during the infection of root lesion nematode, Pratylenchus coffeae. PLoS ONE 11, e0162013. doi: 10.1371/journal.pone.0162013
Kumar, V., Chattopadhyay, A., Ghosh, S., Irfan, M., Chakraborty, N., et al. (2016). Improving nutritional quality and fungal tolerance in soya bean and grass pea by expressing an oxalate decarboxylase. Plant Biotechnol. J. 14, 1394–1405. doi: 10.1111/pbi.12503
Leister, D. (2004). Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20, 116–122. doi: 10.1016/j.tig.2004.01.007
Li, J., Islam, F., Huang, Q., Wang, J., Zhou, W., Xu, L., et al. (2020). Genome-wide characterization of WRKYgene family in Helianthus annuus L. and their expression profiles under biotic and abiotic stresses. PLoS ONE 15, e0241965. doi: 10.1371/journal.pone.0241965
Li, Z., Hua, X., Zhong, W., Yuan, Y., Wang, Y., Wang, Z., et al. (2020). Genomewide identification and expression profile analysis of WRKY family genes in the autopolyploid Saccharum spontaneum. Plant Cell Physiol. 61, 616–630. doi: 10.1093/pcp/pcz227
Liu, X., Inoue, H., Hayashi, N., Jiang, C. J., and Takatsuji, H. (2016). CC-NBS-LRR-type R proteins for rice blast commonly interact with specific WRKY transcription factors. Plant Molecular Biology Reporter. 34, 533–537. doi: 10.1007/s11105-015-0932-4
Liu, Z. Q., Shi, L. P., Yang, S., Qiu, S. S., Ma, X. L., Cai, J. S., et al. (2021). A conserved double-W box in the promoter of CaWRKY40 mediates autoregulation during response to pathogen attack and heat stress in pepper. Mol. Plant Pathol. 22, 3–18. doi: 10.1111/mpp.13004
McHale, L., Tan, X., Koehl, P., and Michelmore, R. W. (2006). Plant NBS-LRR proteins: adaptable guards. Genome Biol. 7, 212. doi: 10.1186/gb-2006-7-4-212
Mishra, G. P., Radhakrishnan, T., Kumar, A., Thirumalaisamy, P. P., Kumar, N., Bosamia, T. C., et al (2015). Advancements in molecular marker development and their applications in the management of biotic stresses in peanuts. Crop Protect. 77, 74–86. doi: 10.1016/j.cropro.2015.07.019
Mishra, P., Tripathi, A. N., Kashyap, S. P., Aamir, M., and Tiwari, S. K. (2021). In silico mining of WRKY TFs through Solanum melongena L. and Solanum incanum L. transcriptomes and identification of SiWRKY53 as a source of resistance to bacterial wilt. Plant Gene 26, 100278. doi: 10.1016/j.plgene.2021.100278
Muhammad, I. K., Zhang, Y., Liu, Z., Hu, J., Liu, C., Yang, S., et al. (2018). CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int. J. Mol. Sci. 19, 1403. doi: 10.3390/ijms19051403
Raza, A., Charagh, S., García-Caparrós, P., Rahman, M. A., Ogwugwa, V. H., Saeed, F., and Jin, W. (2022b). Melatonin-mediated temperature stress tolerance in plants. GM Crops & Food 13, 196–217. doi: 10.1080/21645698.2022.2106111
Raza, A., Tabassum, J., Mubarik, M. S., Anwar, S., Zahra, N., Sharif, Y., et al. (2022a). Hydrogen sulfide: an emerging component against abiotic stress in plants. Plant Biol. 24, 540–558. doi: 10.1111/plb.13368
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Sharma, M., Irfan, M., Kumar, A., Kumar, P., and Datta, A. (2021). Recent insights into plant circadian clock response against abiotic stress. J. Plant Growth Regul. 1–14. doi: 10.1007/s00344-021-10531-y. [Epub ahead of print].
Sharma, M., Kumar, P., Verma, V., Sharma, R., Bhargava, B., and Irfan, M. (2022). Understanding plant stress memory response for abiotic stress resilience: molecular insights and prospects. Plant Physiol. Biochem. 179, 10–24. doi: 10.1016/j.plaphy.2022.03.004
Shen, Q. H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103. doi: 10.1126/science.1136372
Song, H., Wang, P., Lin, J. Y., Zhao, C., Bi, Y., and Wang, X. (2016). Genome-wide identification and characterization oF WRKY gene family in peanut. Front. Plant Sci. 7, 534. doi: 10.3389/fpls.2016.00534
Song, Y., Cui, H., Shi, Y., Xue, J., Ji, C., Zhang, C., et al. (2020). Genome-wide identification and functional characterization of the Camelina sativa WRKY gene family in response to abiotic stress. BMC Genomics 21, 786. doi: 10.1186/s12864-020-07189-3
Van De Peer, Y., Mizrachi, E., and Marchal, K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. doi: 10.1038/nrg.2017.26
Wan, Y., Mao, M., Wan, D., Yang, Q., Yang, F., Mandlaa, et al. (2018). Identification of the WRKY gene family and functional analysis of two genes in Caragana intermedia. BMC Plant Biol. 18, 31. doi: 10.1186/s12870-018-1235-3
Wang, D., Wang, L., Su, W., Ren, Y., and Su, Y. (2020). A class III WRKY transcription factor in sugarcane was involved in biotic and abiotic stress responses. Sci. Rep. 10, 20964. doi: 10.1038/s41598-020-78007-9
Wang, Y., Dang, F., Liu, Z., Wang, X., Eulgem, T., Lai, Y., et al. (2012). Cawrky58, encoding a group i wrky transcription factor ofcapsicum annuum, negatively regulates resistance toralstonia solanacearuminfection. Mol. Plant Pathol. 14, 131–144. doi: 10.1111/j.1364-3703.2012.00836.x
Wei, K. F., Chen, J., Chen, Y. F., Wu, L. J., and Xie, D. X. (2012). Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 19, 153–164. doi: 10.1093/dnares/dsr048
Wu, J., Chen, J., Wang, L., and Wang, S. (2017). Genome-wide investigation of WRKY transcription factors involved in terminal drought stress response in common bean. Front. Plant Sci. 8, 380. doi: 10.3389/fpls.2017.00380
Xie, T., Chen, C., Li, C., Liu, J., Liu, C., and He, Y. (2018). Genome-wide investigation of WRKY gene family in pineapple: evolution and expression profiles during development and stress. BMC Genomics 19, 490. doi: 10.1186/s12864-018-4880-x
Yang, S., Zhang, Y., Cai, W., Liu, C., and Shuilin, H. E. (2021). CaWRKY28 Cys249 is required for interaction with CaWRKY40 in the regulation of pepper immunity to Ralstonia solanacearum. Mol. Plant Microbe Interact. 34, 733–745. doi: 10.1094/MPMI-12-20-0361-R
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5, 1. doi: 10.1186/1471-2148-5-1
Zhao, N., He, M., Li, L., Cui, S., Hou, M., Wang, L., et al. (2020). Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea L.). PLoS ONE 15, e0231396. doi: 10.1371/journal.pone.0231396
Keywords: biotic stress, gene structure, oilseed crop, phylogenetic analysis, expression analysis, bacterial wilt
Citation: Yan L, Jin H, Raza A, Huang Y, Gu D and Zou X (2022) WRKY genes provide novel insights into their role against Ralstonia solanacearum infection in cultivated peanut (Arachis hypogaea L.). Front. Plant Sci. 13:986673. doi: 10.3389/fpls.2022.986673
Received: 05 July 2022; Accepted: 16 August 2022;
Published: 20 September 2022.
Edited by:
Mohammad Irfan, Cornell University, United StatesReviewed by:
Vinay Kumar, The Ohio State University, United StatesCopyright © 2022 Yan, Jin, Raza, Huang, Gu and Zou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoyun Zou, anhhdXp4eUAxNjMuY29t; Deping Gu, Z3VkcDE5OTlAc2luYS5jb20=
†ORCID: Lei Yan orcid.org/0000-0003-0226-4862
Ali Raza orcid.org/0000-0002-5120-2791
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.