- 1National Engineering Research Center for Sugarcane, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Institute of Nanfan and Seed Industry, Guangdong Academy of Sciences, Guangzhou, China
Cys-tathionine-β-synthase (CBS) domain-containing proteins (CDCPs) are essential for regulating plant responses to various biotic and abiotic stressors. This study describes the systematic identification and characterization of CDCP family genes in Saccharum spontaneum. A total of 95 SsCDCP genes and eight phylogenetic groups were identified that were distributed over 29 chromosomes of the AP85-441 genome. Most (78/95) SsCDCPs underwent fragment duplication events, and 64 gene pairs were located in synteny blocks. Expression profiling of nine ShCDCPs was also carried out in the Saccharum spp. cultivars ROC22 and MT11-611 that are resistant and susceptible to red stripe, respectively, in response to: (i) Infection by the bacterial pathogen Acidovorax avenue subsp. avenae (Aaa); (ii) abiotic stressors (drought and salinity); and (iii) exogenous salicylic acid (SA) treatment. Members of one gene pair (ShCBSD-PB1-5A and ShCBSD-PB1-7A-1) with a fragment duplication event acted as negative regulators in sugarcane under four stresses, as supported by the significantly decreased expression levels of ShCBSD-PB1-5A (23–83%) and ShCBSD-PB1-7A-1 (15–75%) at all-time points, suggesting that they have functional redundancy. Genes in another pair, ShCBS-4C and ShCBS-4D-1, which have a fragment duplication event, play opposing regulatory roles in sugarcane exposed to multiple stresses, particularly Aaa and NaCl treatments. ShCBS-4C expression was significantly decreased by 32–77%, but ShCBS-4D-1 expression was dramatically upregulated by 1.2–6.2-fold in response to Aaa treatment of both cultivars across all-time points. This result suggested that both genes exhibited functional divergence. Meanwhile, the expression of SsCBSDCBS-5A was significantly upregulated in ROC22 by 1.4–4.6-fold in response to the four stressors. These findings provide important clues for further elucidating the function of ShCDCP genes in sugarcane responding to a diverse range of stresses.
Introduction
Climate change is leading to more frequent extreme environmental events, such as abiotic (drought, low and high temperature, high salinity, and soil salinization) and biotic (invasive arthropod pests and diseases) stressors (González Guzmán et al., 2022). Furthermore, climate change that produces various abiotic stresses has affected the incidence and geographic distribution of plant diseases and pathogens (Burdon and Zhan, 2020). These environmental stressors have major impacts on food production worldwide (González Guzmán et al., 2022; Karthika and Govintharaj, 2022).
Sugarcane (Saccharum spp.) is an important global sugar and biofuels crop that is distributed in tropical and sub-tropical areas, where it is subject to various biotic and abiotic stressors (Javed et al., 2020). Among biotic stressors, the bacterial pathogen Acidovorax avenae subsp. avenae (Aaa) that causes red stripe in sugarcane can lead to serious yield reduction and even plant death (Li et al., 2018). Red stripe disease commonly occurs in main sugarcane-planting areas in China, with varying incidences ranging from 4 to 23% in cultivar FN38 (Fu et al., 2017) and 8–80% in cultivar YZ03-194 (Shan et al., 2017). In Argentina, red stripe affected 30% of the milling stems causing serious economic losses (Fontana et al., 2019). In general, plants respond to pathogen infection via two layers of the immune system, pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) (Zhai et al., 2022). Our previous studies using transcriptome (Chu et al., 2020), proteomic (Zhou et al., 2021), and genome-wide analyses (Chu et al., 2022) revealed that multiple genes/proteins and their related metabolites and signal pathways are involved in defense responses in sugarcane.
Abiotic stresses such as drought, salinity, extreme temperature, and low soil fertility also affect sugarcane growth and yield worldwide (Lakshmanan and Robinson, 2014; Budeguer et al., 2021). Drought is the most important abiotic factor and can reduce sugarcane yield by up to 50–60% (Ferreira et al., 2017; Flack-Prain et al., 2021). Generally, plants or organs responding to water stress exhibit physiological and metabolic changes to minimize water loss under moderate-to-severe short-term stress (Lakshmanan and Robinson, 2014; Ferreira et al., 2017). Salinity is another important abiotic factor that negatively affects sugarcane production in many areas (Budeguer et al., 2021). Sugarcane is more sensitive to soil salinity during germination and early growth stages compared to later stages of plant growth (Lakshmanan and Robinson, 2014). Sugarcane plants that are often reported to be moderately salt-sensitive and grow in saline soil have adverse physiological and developmental disruptions (Lakshmanan and Robinson, 2014; Brindha et al., 2019). Many phytohormones, plant growth regulators, and signaling molecules participate in abiotic stress responses in sugarcane (Ferreira et al., 2017; Budeguer et al., 2021).
Cys-tathionine-β-synthase (CBS) domain-containing proteins (CDCPs) are an evolutionarily conserved superfamily of proteins that contain varying numbers of CBS domains (Kushwaha et al., 2009; Hao et al., 2016; Tomar et al., 2022). The CBS domain was originally discovered in the archaebacterium Methanococcus jannaschii (Bateman, 1997). The CBS domain contains about 60 amino acid residues that form two α-helices and three β-strands and generally exists as tandem repeats, particularly in pairs or quads, in the polypeptide (Anashkin et al., 2017; Tomar et al., 2022). In addition to the CBS domain, CDCP family genes encode other domains such as CNNM (or DUF21), inosine-5′-monophosphate dehydrogenase (IMPDH), Phox and Bem1 (PB1), voltage chloride channel (Voltage CLC) (Kushwaha et al., 2009; Tomar et al., 2022). Phylogenetic groupings of CDCP family genes differ among plant species. There are eight groups in Arabidopsis thaliana (Kushwaha et al., 2009) and Triticum aestivum (Guo et al., 2020), and nine in Oryza sativa (Kushwaha et al., 2009) and Glycine max (Hao et al., 2016). Recently, 14 major clades of CDCP family genes were identified in 11 genomes from ten Oryza species (Tomar et al., 2022).
The CBS domain was found to be widely associated with several proteins that have distinct functions such as AMP-activated protein kinase (AMPK), IMPDH, and CLC (Jeong et al., 2013; Labesse et al., 2015; Subba et al., 2021). The activity of related enzymes and transporter domains was shown to be regulated by the CBS domain that mediates binding of adenosine-based molecules such as AMP, ATP or ASM (S-adenosylmethionine) (Kemp, 2004; Baykov et al., 2011; Anashkin et al., 2017). The CBS domain is an efficient regulatory element and is integrated into proteins with different functions to enhance or weaken protein activity depending on the binding of different ligands (Anashkin et al., 2017). Additionally, signals are transmitted remotely between different subunits of AMPK through the CBS domain and allosteric regulation resulting from the CBS domain can be integrated into more complex regulatory mechanism as in AMPK (Anashkin et al., 2017).
Previous studies have showed that CDCP family genes participate in regulation of plant growth and development, environmental stress, and pathogen infection. In Arabidopsis, some CDCPs in root and shoot tissues are expressed in response to drought, salinity, and wounding stresses (Kushwaha et al., 2009); AtCBSX1 was found to modulate development by regulating the thioredoxin system in chloroplasts (Yoo et al., 2011), whereas AtCBSX3 is involved in plant development and the redox system through regulation of the generation of reactive oxygen species (ROS) in mitochondria (Shin et al., 2020). In Oryza sativa, OsBi1 overexpression can enhance plant resistance against the herbivore brown planthopper (Nilaparvata lugens Stal.) (Wang et al., 2004). Some genes encoding CDCPs from rice were found to be involved in responses to multiple stresses such as drought, salinity, and wounding (Kushwaha et al., 2009). OsCBSX4 overexpression in tobacco plants confers strong resistance to salinity stress (Singh et al., 2012). OsCBSX9 and OsCBSCBS4 displayed significantly higher expression levels under both salinity and drought stress conditions in rice plants (Tomar et al., 2022). For Glycine max, GmCBS21 and GmCBSDUF3 overexpression in Arabidopsis showed that the transgenic plants had enhanced tolerance to low nitrogen stress (Hao et al., 2016) and to drought and salt stress (Hao et al., 2021). A recent iTRAQ proteomic analysis revealed that expression of a CBS domain-containing protein was upregulated in wheat (T. aestivum) in response to waterlogging (Yang et al., 2022).
Modern sugarcane cultivars with an allo-autopolyploid genome contain chromosomes from S. officinarum (80%) and S. spontaneum (10%) (D’Hont et al., 1996), but the characteristics of stress response, disease resistance, and regeneration ability of these cultivars are derived from S. spontaneum (Garsmeur et al., 2018). To better understand features of the sugarcane genome, two reference genome sequences from the S. spontaneum clones AP85-441 (Zhang et al., 2018) and Np-X (Zhang et al., 2022) were assembled at the chromosome level, while three draft genome sequences from hybrid genotypes R570 (Garsmeur et al., 2018), SP80-3280 (Souza et al., 2019), and CC01-1940 (Trujillo-Montenegro et al., 2021) were assembled at a non-chromosome level. These sugarcane genome sequences, particularly in AP85-441 and Np-X, are convenient for exploring how resistance genes are related to various stress responses in sugarcane. It may be hypothesized that some members of the CDCP gene family are involved in alleviating biotic and abiotic stresses in sugarcane. However, systematic identification and analysis of the CDCP gene family in sugarcane remains incomplete. Thus, the objectives of this study were: (i) Identification and characterization of CDCP family genes in S. spontaneum AP85-441; (ii) investigation of expression profiles in ROC22 and MT11-610 cultivars after Aaa inoculation, and drought, salinity as well as exogenous salicylic acid (SA) treatment; and (iii) comparison of functional redundance and divergence of CDCP family genes in stress responses. Our results provide important information about several CDCP genes and how they are involved in response to different stressors.
Materials and methods
Plant growth and experimental treatments
The sugarcane cultivars ROC22 (resistant to red stripe) and MT11-610 (susceptible to red stripe) were provided by the National Engineering Research Center for Sugarcane, Fujian Agriculture and Forestry University, Fuzhou, China (26.0849°N, 119.2397°E). Single buds cut from the two varieties were immersed in flowing water (24 h) and then dipped in hot water at 50°C for 2 h. Sugarcane plants were maintained for 28 days in a growth chamber at 28°C and 60% relative humidity under a 16/8 h photoperiodic cycle. Four stress experiments were performed following the procedure described by Chu et al. (2022). Briefly, the Aaa strain SC-026 (108 CFU/ml) was used for sugarcane seedling inoculation with a leaf-cutting method (Chu et al., 2020). The plants were inoculated with liquid NB medium as a control (CK). Leaf samples were collected at 0, 24, 48, and 72 h post-inoculation (hpi) to examine responses to Aaa stress. Sampling points for seedlings treated with 25% PEG6000 were 0, 3, 6, and 12 h; sampling points for 250 mM NaCl or 0.1 mmol/L SA (containing 0.01% Tween-20) treatments were 0, 6, 12, and 24 h after treatment. Three biologic replicates of six plants were used at each sampling time.
Identification of cys-tathionine-β-synthase domain-containing proteins in Saccharum spontaneum
A. thaliana and T. aestivum CDCP protein sequences were obtained from TAIR1 and UniProt,2 respectively. These sequences were used as bait to search for genes encoding CDCP proteins in the AP85-441 S. spontaneum genome3 using NCBI BLAST-P4 with an e-value < 1e–5 (default parameters were used for other settings). Sequences remaining after manual removal of redundant sequences were used for further analysis. PF00571 (CBS domain) and HMMER software with default parameters were used to search for candidate CDCP genes in S. spontaneum. Furthermore, the Conserved Domain Search,5 Pfam online tools6 and SMART online tools7 with default settings were used to verify each gene containing a CBS domain. The CDCP family genes identified in S. spontaneum were termed SsCDCPs. The nomenclature of all CDCP family genes identified in S. spontaneum and two sugarcane cultivars was identical to Arabidopsis (Kushwaha et al., 2009) and Triticum aestivum (Guo et al., 2020). For example, in the SsCBS-3C gene “Ss” is an abbreviation for S. spontaneum, “CBS” stands for the typical conserved domain, and “3C” refers to the chromosomal location.
Analysis of physico-chemical properties
The ExPASy Proteome Server8 was used to compute physio-chemical traits such as the number of amino acids (aa), molecular weight (MW), and theoretical isoelectric points (pI). The Plant-mPLoc9 online tool was used to predict the sub-cellular localization of SsCDCP members.
Multiple sequence alignment and phylogenetic analysis
Alignment of 256 CDCP protein sequences (A. thaliana = 34, T. aestivum = 127, and S. spontaneum = 95) was performed using the CLUSTALW program implemented in the MEGA version 11 (Tamura et al., 2021). A phylogenetic tree was constructed using MEGA version 11 with the maximum likelihood (ML) method and bootstraps of 1,000 replicates. Visualizations were generated using the EvoIView server.10
Gene structure, cis-regulatory elements, and gene duplication analysis
Conserved motifs of SsCDCP sequences were analyzed with the MEME tool11 with default parameters. Conserved domains were checked with NCBI-CDD (Use default parameters) (see text footnote 5) (Lu et al., 2020). The gene structure of SsCDCPs was visualized using TBtools v0.6655 (Chen et al., 2020). Cis-acting elements in the promoter sequences (2,000 bp) of each SsCDCP gene were analyzed using PlantCARE online software12 with default parameters. TBtools v0.6655 was used to analyze and visualize the gene duplication events among SsCDCP genes (Wang et al., 2012).
Expression profiling using ribonucleic acid-seq data
To determine the expression profiling of CDCP genes in sugarcane cultivars following Aaa infection, a previously published ribonucleic acid (RNA)-seq dataset (PRJNA579959) was used (Chu et al., 2020). The CDCP genes identified in the Saccharum spp. hybrid were termed ShCDCPs. The expression abundance of the ShCDCPs was calculated using the fragments per kilobase of transcript per million fragments mapped (FPKM) value and the relative expression level is shown as log2 (Fold Change) values.
ShCDCP gene expression analysis by RT-qPCR
Transcript expression of nine ShCDCP genes was investigated by RT-qPCR assay. Total RNA was extracted from leaf samples and cDNA was synthesized by reverse transcription as previously described (Chu et al., 2020). Primers for nine ShCDCP genes were designed with Primer 5.0 software (Supplementary Table 1). The SYBR green dye method was used for RT-qPCR amplification (Javed et al., 2022). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a reference gene. The 2–ΔΔCt quantification method was used to determine the expression of each ShCDCP gene. Each sample was assayed with three biological and three technical replicates.
Statistical analysis
The relative expression levels at each time point were analyzed by one-way ANOVA and the test of significance among means was carried out with LSD (least significance difference) at 5% probability level (p ≤ 0.05). All statistical analyses were carried out using SPSS software (IBM SPSS Statistics 25).
Results
Identification and physico-chemical properties of cys-tathionine-β-synthase domain containing proteins family genes in Saccharum spontaneum
A total of 37 SsCDCP genes with varying numbers of alleles were identified in S. spontaneum clone AP85-441. Among these SsCDCPs, 21.6% (8/37) lacked alleles, and the remainder had 1–6 alleles (Supplementary Table 2). Thus, a total of 95 SsCDCP genes were identified in AP85-441. The physico-chemical properties showed that the 95 SsCDCP proteins had: between 169 (SsCBS-3A-1) and 1,367 (SsCBS-SIS-4C) amino acids; molecular weights ranging from 17,138.29 (SsCBS-3A-1) to 147,232.08 (SsCBS-SIS-4C) Da; and isoelectric points between 4.54 (SsTlyc-1C-1) and 10.68 (SsCBS-5A) (Supplementary Table 3). Most (37.9%) SsCDCPs were predicted to localize to the plasma membrane, and the remainder were predicted to localize to the chloroplast (30.5%), cytoplasm (16.8%), and mitochondria (10.5%). Only a few SsCDCPs (4.2%) localized to the nucleus.
Phylogeny of cys-tathionine-β-synthase domain containing proteins family genes in three plant species
A phylogenetic analysis to describe evolutionary relationships among S. spontaneum (95 SsCDCPs), A. thaliana (34 AtCDCPs), and T. aestivum (127 TaCDCPs) showed that all 256 CDCPs could be classified into nine different groups (Supplementary Figure 1). Most (69) CDCPs were in group C, followed by group A (48), and group J had the fewest CDCPs (3). Another phylogenetic tree revealed that all 95 SsCDCPs could be divided into eight groups (lack of group J) as compared to all CDCPs identified in the three species. Notably, group C contained 27 CDCPs and could be sub-divided into two groups (C1 and C2) having 8 and 19 genes, respectively (Figure 1).
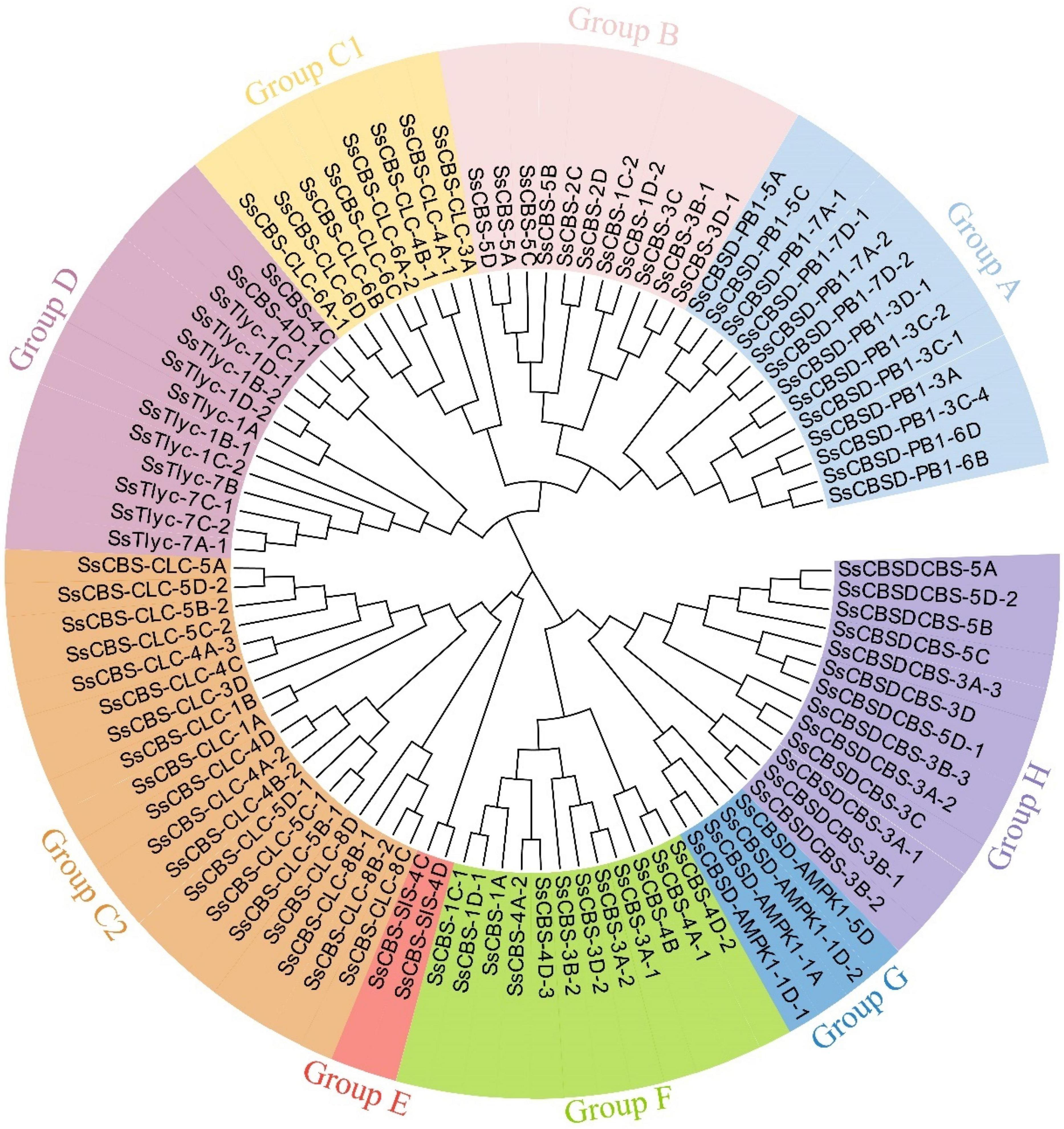
Figure 1. Unrooted phylogenetic tree of SsCDCPs identified in S. spontaneum AP85-441. The tree was conducted using the maximum likelihood (ML) method with 1,000 bootstrap replicates.
Gene structure analysis among SsCDCP genes
Among 95 SsCDCPs, 18 had only 5′-UTR, 21 had only 3′-UTR, and 21 SsCDCPs had both 5′-UTR and 3′-UTR. The other remaining genes had no 5′-UTR or 3′-UTR (Figure 2 and Supplementary Table 3). The number of introns ranged from 0 (SsCBS-3A-1) to 22 (SsCBS-CLC-1A), while the number of exons ranged from 1 (SsCBS-3A-1) to 23 (SsCBS-CLC-1A). SsCBS-CLC-8B-2 had the longest intron, followed by SsCBS-SIS-4C and SsCBS-CLC-5B-2. The longest exon structure was observed in SsCBSD-AMPK1-1A, followed by SsCBS-SIS-4C and SsCBSD-AMPK1-1D-2. Among eight SsCDCP groups, members of group B (11 SsCDCPs), group F (12 SsCDCPs) and group H (13 SsCDCPs) contained 1–3 CBS domains. The members of other groups had additional domains other than the CBS domain. For example, all group A (13 SsCDCPs) members, except for SsCBSD-PB1-7D-1, had an additional Phox/Bem1 (PB1) domain. Genes in groups C1 (8 SsCDCPs) and C2 (19 SsCDCPs) had an additional CLC domain (except for SsCBS-CLC-6A-2). Those in Group D (13 SsCDCPs) had an additional Tlyc domain (except for SsCBS-1D-2 and SsCBS-5D). SsCBS-4C clustered in group D had a unique COG2905 domain. Group E (2 SsCDCPs) contained an additional gutQ domain. Group G (4 SsCDCPs) had an additional AMPK1_CBM domain (Figure 2A). Conserved motif numbers ranged from 1 (SsCBSD-PB1-7A-2) to 10 (SsCBS-CLC-4B-2). Motifs 6 and 7 were found in 96 and 65% of SsCDCPs, respectively (Figure 2B).
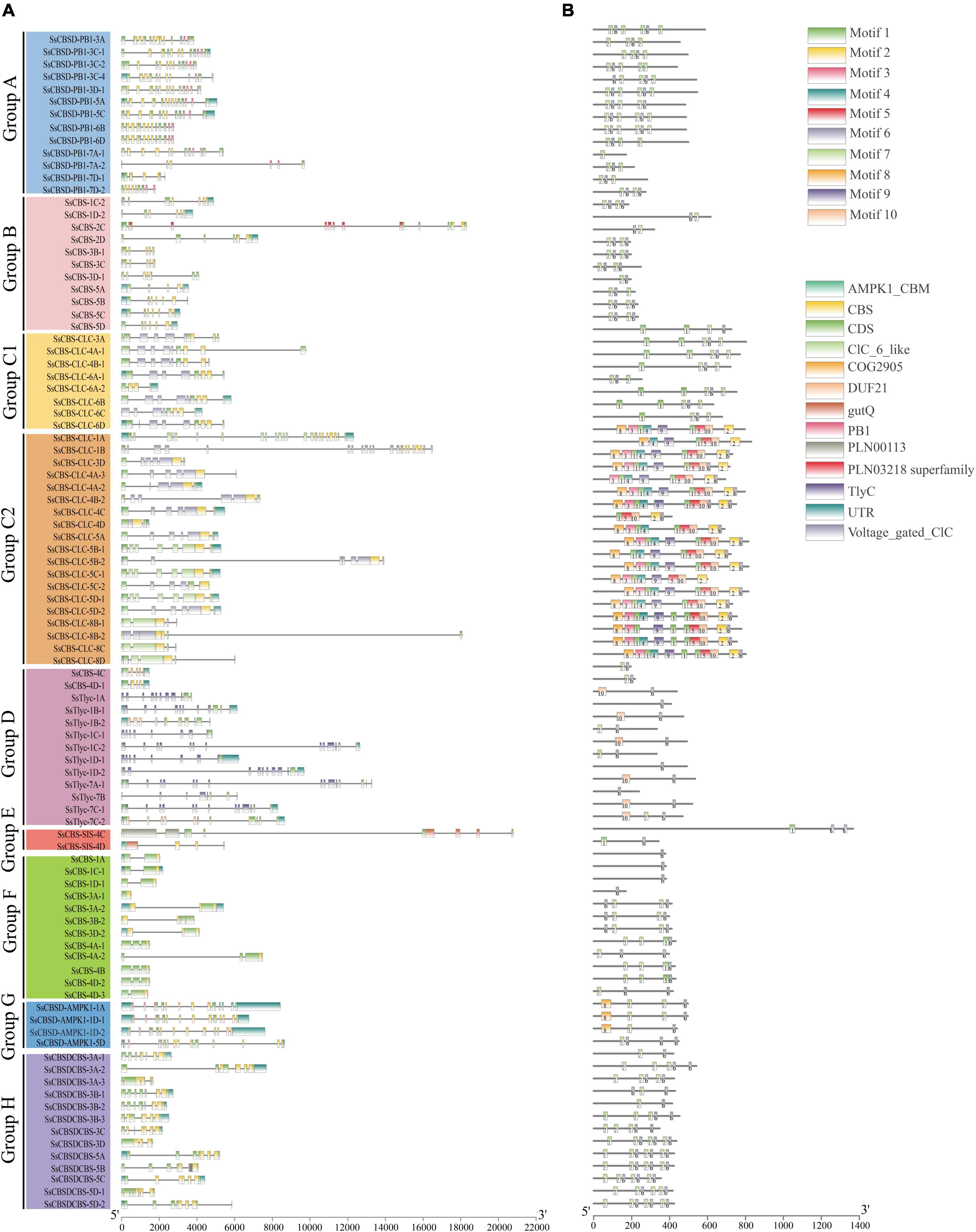
Figure 2. Gene structure (A) and conserved motif (B) of SsCDCPs in S. spontaneum. (A) Untranslated 5′- and 3′-regions, CDS and conserved domains are displayed with different colored boxes. (B) Motifs 1–10 are displayed as different colored boxes. Gene and protein lengths can be estimated using the scales at the bottom.
In silico promoter analysis of SsCDCP genes
A total of 24 cis-acting regulatory elements related to phytohormones, stress and MYB transcription factors were predicted (Figure 3). Potential functions of cis-acting regulatory elements are annotated in Supplementary Table 4. Promoter sequences (2 kb) of SsCDCPs contained different numbers of cis-elements, ranging from 0 (SsCBS-5B) to 56 (SsCBS-4A-2). The promoters of the two genes SsCBS-5A/5C had only 1–2 cis-elements. Most (94%) SsCDCP genes had MYB, and ABRE, STRE, CGTCA, and TGACG motifs were present in 85–88% of SsCDCP genes.
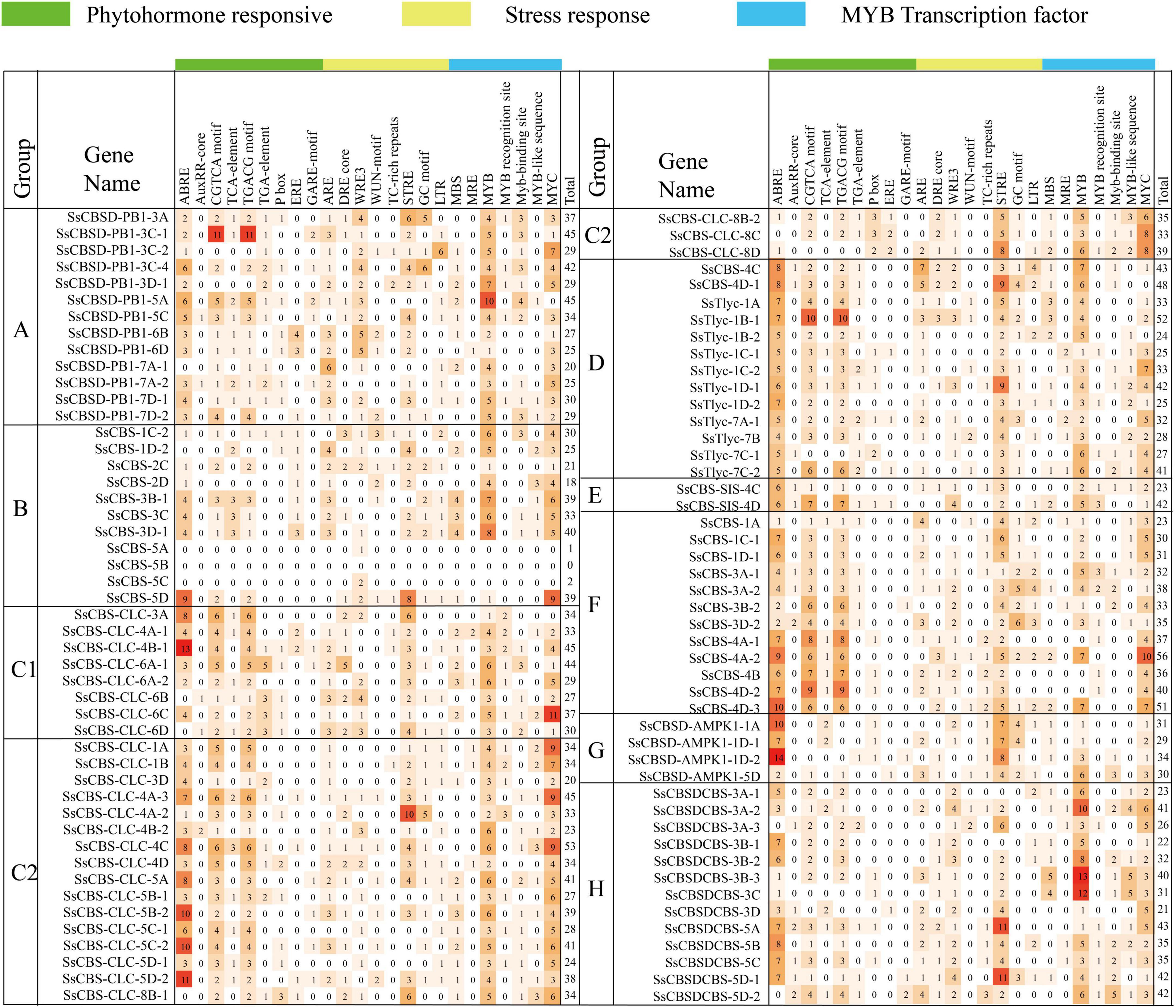
Figure 3. Heatmap of cis-regulatory elements in promoter sequences (2 kb) of SsCDCPs from S. spontaneum.
Chromosomal distribution and gene duplication analysis of SsCDCP genes
All 95 SsCDCPs including 87 alleles were distributed on 29 chromosomes at different densities (Figure 4). Chromosome 3A (Chr3A) had the most SsCDCPs (7), followed by Chr1D and Chr5D, which contained six SsCDCPs, and then chromosomes 3B, 3C, 3D, 4A, 4D, and 5C that each had five SsCDCPs. The other 20 chromosomes had 1–4 SsCDCPs. Most SsCDCPs were located at the proximal end of each chromosome. Gene duplication and collinear correlation analysis showed that fragment duplication occurred in 82% (78/95) SsCDCPs and 64 gene pairs existed in synteny blocks, such as SsCBSD-PB1-3A and SsCBSD-PB1-3C-4, SsCBS-4C and SsCBS-4D-1, and SsCBSD-PB1-5A and SsCBSD-PB1-7A-1. Notably, gene duplication events mainly occurred on chromosomes Chr3A/3B/3C/3D, Chr4A/4B/4C/4D, and Chr5A/5B/5C/5D. The Ka/Ks ratios of all SsCDCP gene pairs were < 1, suggesting that they were under purifying selection (Supplementary Table 5).
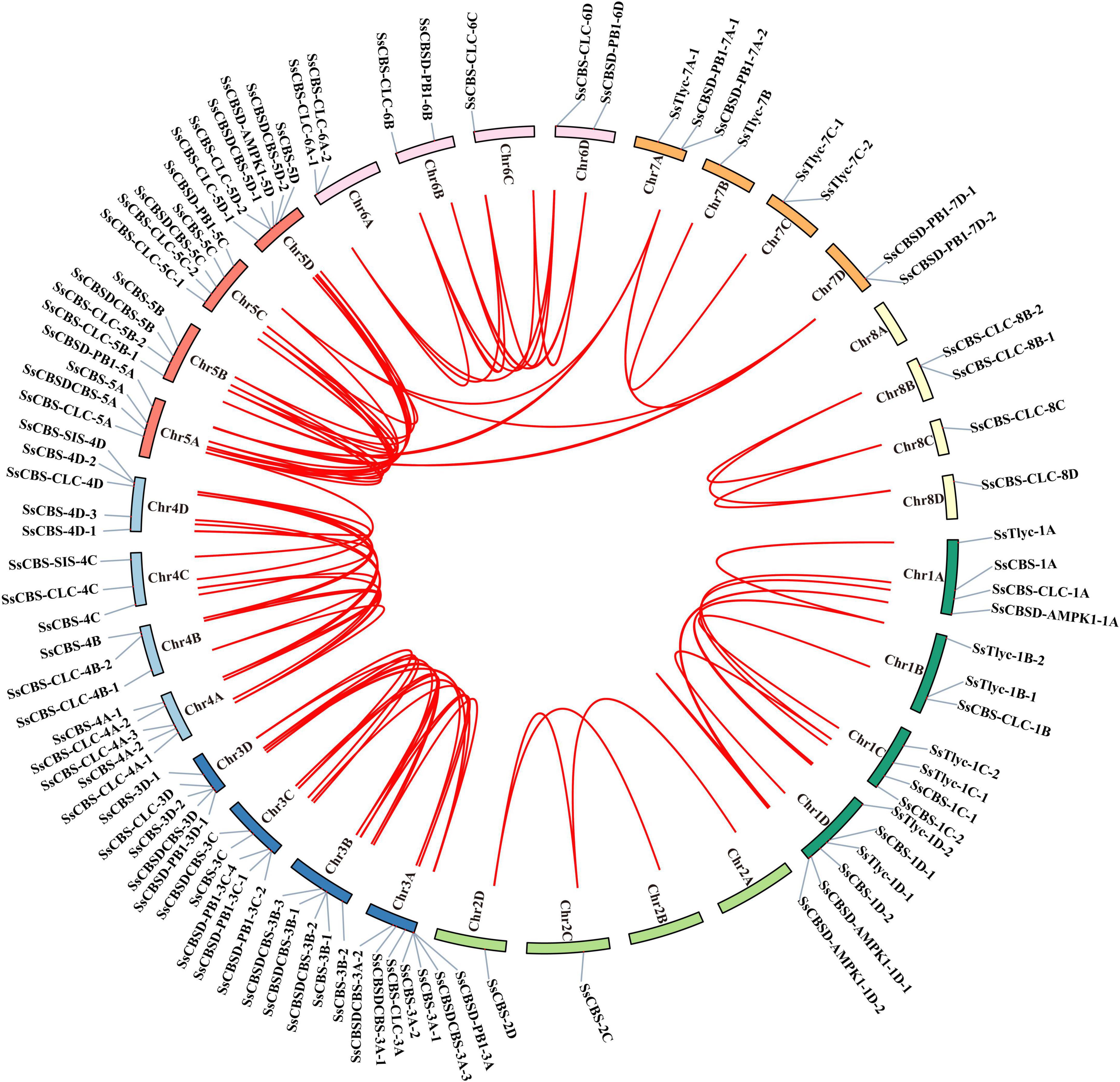
Figure 4. Chromosomal localization and gene replication relationship of SsCDCPs in S. spontaneum. Ribbon links (Red lines) indicate segmental duplication events between genes. Chromosome numbers are indicated inside the outer circle. The gene names on each chromosome are indicated on the outer circle.
Expression analysis of ShCDCP gene responses to Acidovorax avenue subsp. avenae infection
A total of 83 ShCDCPs were identified on the transcriptome database (PRJNA579959). RNA-seq data revealed that these genes showed different expression patterns under Aaa infection. Expression levels of 10 ShCDCPs were upregulated with an increase of 2% to 2.2-fold, but 16 ShCDCPs were downregulated across all-time points in both sugarcane cultivars. Twelve ShCDCPs were significantly upregulated in ROC22 (resistant to red stripe) but were downregulated or unchanged in MT11-610 (susceptible to red stripe) across all-time points (Figure 5 and Supplementary Table 6). For example, the transcript level (log2FC) of the ShCBS-4D-1 gene was increased by between 41% and 2.1-fold in ROC22, but this gene was significantly decreased in MT11-610 relative to the control (0 hpi).
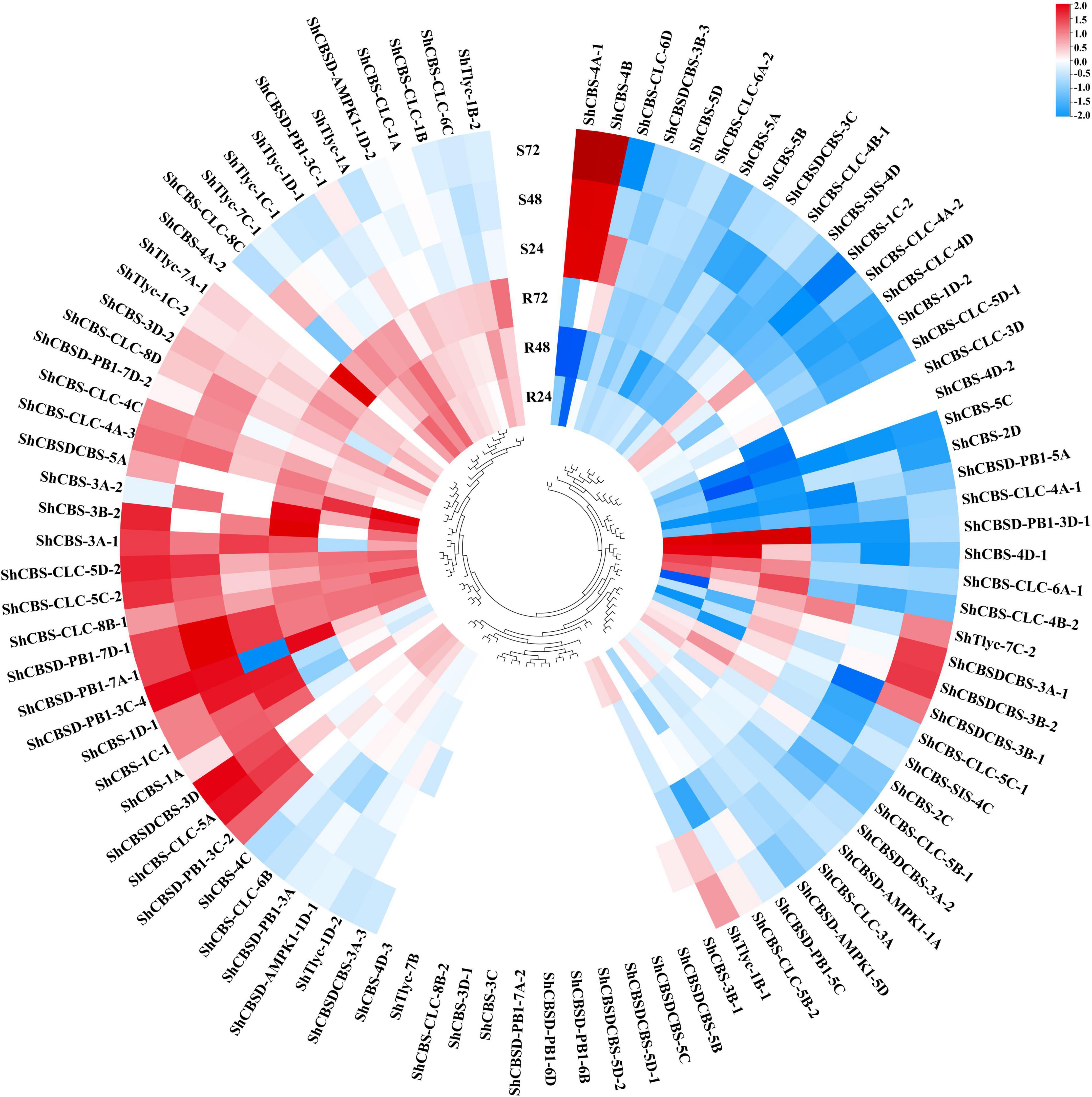
Figure 5. Heatmap of transcript expression profiles of ShCDCPs in two cultivars, ROC22 (resistant to red stripe, R) and MT11-610 (susceptible to red stripe, S) based on RNA-Seq data. The gene names are indicated in the outer circle and the expression values mapped to a color gradient from low (blue) to high (red) expression level are shown inside the outer circle.
Expression patterns of nine ShCDCPs were further investigated by RT-qPCR assay in two cultivars after Aaa infection (Figure 6 and Supplementary Table 7). In ROC22, expression of ShCBS-4D-1 and ShCBSDCBS-5A was upregulated, while ShCBSD-PB1-5A, ShCBSD-PB1-7A-1, and ShCBS-4C expression was downregulated across all-time points compared to the control (0 hpi). Expression of ShCBS-4D-1 and ShCBSDCBS-5A was increased by more than 4.2-fold and 1.5-fold in ROC22 after Aaa infection. In MT11-610, five genes (ShCBS-1D-2, ShCBSD-PB1-3A, ShCBSD-PB1-3C-4, ShCBS-4D-1, and ShCBSDCBS-5A) were significantly upregulated across all or some timepoints under Aaa infection. Three genes (ShCBSD-PB1-5A, ShCBS-4C, and ShCBS-5D) were significantly downregulated or unchanged across all-time points. The ShCBSD-PB1-7A-1 gene showed a fluctuating expression profile after Aaa infection.
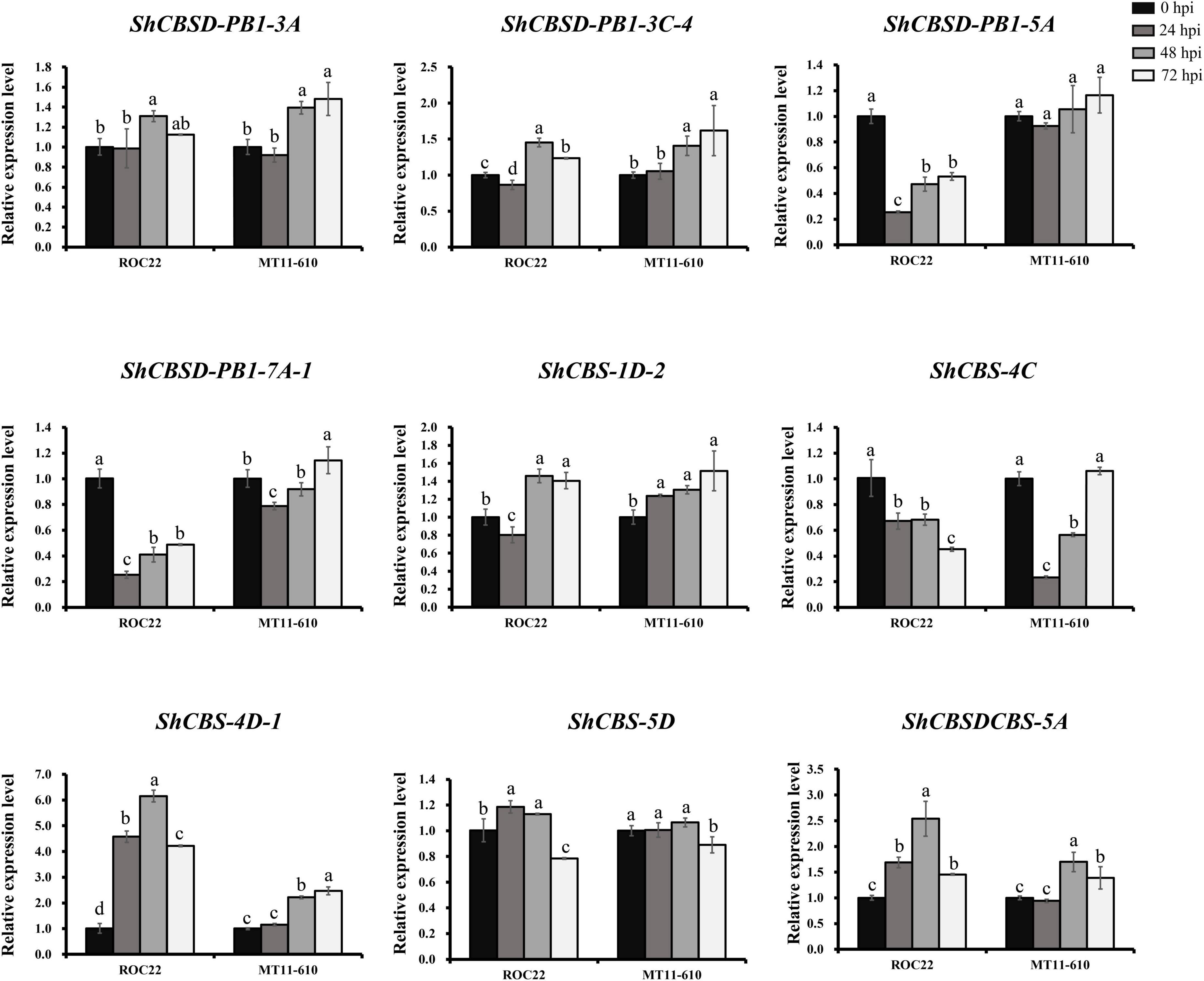
Figure 6. Expression levels of nine ShCDCPs in the cultivars ROC22 and MT11-610 with Aaa infection based on RT-qPCR data. Means ± standard errors are shown. The same letters atop the bars indicate no significant difference at the 5% level.
Expression analysis of ShCDCP genes under diverse abiotic stresses
Under NaCl stress, 7/9 ShCDCPs shared similar expression profiles in two cultivars, while two genes (ShCBS-1D-2 and ShCBSDCBS-5A) showed cultivar-specific behavior. Three ShCDCPs (ShCBSD-PB1-3A, ShCBSD-PB1-3C-4 and ShCBS-4C) were significantly upregulated with an increase of 1.2–3.8-fold, while two (ShCBSD-PB1-5A and ShCBSD-PB1-7A-1) were significantly downregulated in both cultivars across all-time points. ShCBS-1D-2 and ShCBSDCBS-5A were upregulated in ROC22, but downregulated in MT11-610 across all-time points. The expression level of ShCBS-5D was increased for the two cultivars except for 12 h post-NaCl treatment in MT11-610. Additionally, in the two cultivars ShCBS-4D-1 displayed upregulation at early time points (particularly at 6 h) but later was downregulated (Figure 7 and Supplementary Table 7).
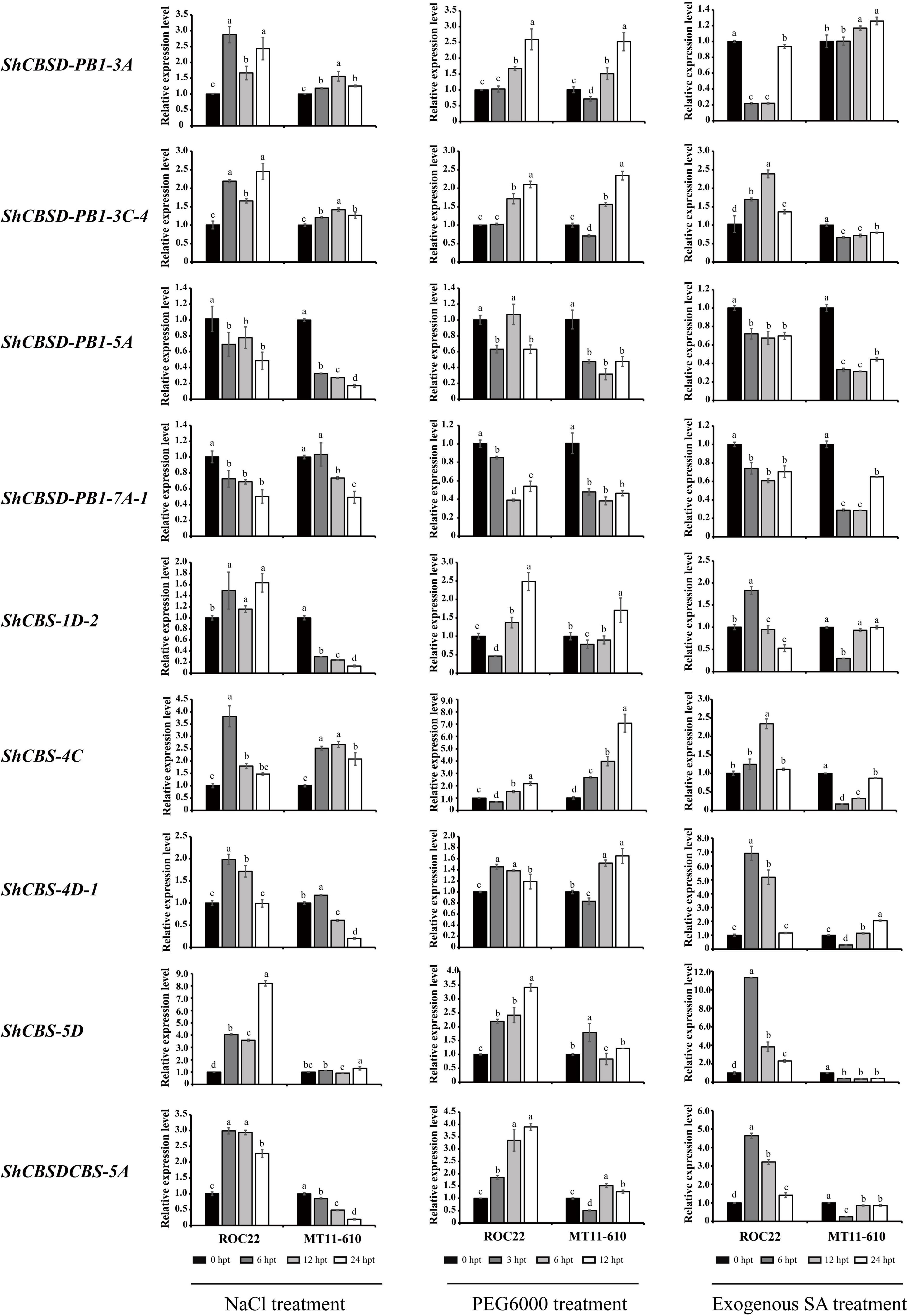
Figure 7. Expression levels of nine ShCDCPs in ROC22 and MT11-610 cultivars treated with NaCl, PEG6000 or exogenous SA treatment based on RT-qPCR data. Means ± standard errors are shown. The same letters atop the bars indicate no significant difference at the 5% level. Rows show the same gene and columns correspond to stressor. hpt, h post treatment.
Under PEG6000 treatment, two ShCDCP genes (ShCBSD-PB1-5A and ShCBSD-PB1-7A-1) were downregulated in both cultivars across all-time points. However, the other seven genes (except for ShCBS-5D) showed downregulation or lower expression levels at early timepoints with PEG6000 treatment (3 h), but thereafter the expression levels progressively increased (Figure 7 and Supplementary Table 7).
Under SA stress, 7/9 ShCDCPs shared cultivar-specific expression profiles. Two ShCDCP genes (ShCBSD-PB1-5A and ShCBSD-PB1-7A-1) were downregulated in both cultivars across all-time points, but ShCBS-1D-2 was also downregulated across all-time points except for 6 h after SA treatment in ROC22. Four genes (ShCBSD-PB1-3C-4, ShCBS-4C, ShCBS-5D, and ShCBSDCBS-5A) were dramatically upregulated by 1.1–11.3-fold in ROC22, but were significantly downregulated in MT11-610 at all-time points. The ShCBSD-PB1-5A gene was upregulated in ROC22, but downregulated in MT11-610. ShCBS-4D-1 was upregulated in both cultivars except for 6 h post-SA treatment in MT11-610 (Figure 7 and Supplementary Table 7).
Functional redundancy and divergence of ShCDCP genes responding to various stressors
To investigate the expression profiles of duplicated gene pairs in sugarcane responses to various stressors, three gene pairs with fragment duplication events were examined (Figures 6, 7). Similar transcript profiles and functions existed for the ShCBSD-PB1-5A and ShCBSD-PB1-7A-1 gene pair in the two cultivars under Aaa infection and three abiotic stresses, suggesting that the two genes displayed functional redundancy in response to multiple stressors. ShCBS-4C and ShCBS-4D-1 as part of a gene pair displayed different expression patterns in sugarcane under biotic and abiotic stresses, particularly Aaa infection, but also with salt and SA treatments, indicating that this gene pair exhibited functional divergence in response to multiple stressors. Two ShCBSD genes, ShCBSD-PB1-3A and ShCBSD-PB1-3C-4, were significantly upregulated in both cultivars under Aaa, salt, and drought stresses, but the two genes had opposite roles in individual cultivars following SA treatment.
Discussion
CDCPs are a large family of proteins that are involved in a variety of biological functions in plants (Hao et al., 2021). Various numbers of CDCP family genes at a genome-wide level were found in previous studies. The number of SsCDCPs (95) identified in S. spontaneum in this study was lower than that of T. aestivum (136) (Guo et al., 2020), but more than that for A. thaliana (34), O. sativa (59) (Kushwaha et al., 2009), and Glycine max (71) (Hao et al., 2016). A large proportion of duplication events (particularly fragment replication) occurred in SsCDCPs, which is similar to the Oryza species (Tomar et al., 2022) and suggests that duplication events contributed to the expansion of the CDCP gene family in plants. Tandem and fragment duplication events are important driving forces for the evolution of gene families in plants, particular in polyploidy crops (Panchy et al., 2016; Van De Peer et al., 2017).
Various expression patterns of ShCDCPs were observed in sugarcane in response to Aaa infection revealed by RNA-seq data and RT-qPCR assay. Of these, two genes, ShCBS-4D-1 and ShCBSDCBS-5A, may be associated with resistance to Aaa in sugarcane since increased expression levels of both genes were present in ROC22, which is resistant to red stripe, compared to MT11-610, which is susceptible to red stripe. Previous studies showed that proteins containing the CBS domain mostly play a positive regulatory role in resistance to stress and disease in other crops. For example, maximal expression levels of the TaCDCP1 (ShCBS-4D-1 homolog) gene were induced in T. aestivum inoculated with Puccinia srtiformis f. sp. tritici at 18 and 96 hpi (Wang et al., 2010). Transcripts of OsCBSX3 (ShCBS-4D-1 homolog) were significantly upregulated by inoculation of M. oryzae and over-expressing OsCBSX3 plants exhibited significantly enhanced resistance to M. oryzae infection (Mou et al., 2015). Additionally, a CsCBS gene could respond positively to cucumber downy mildew and cucumber target leaf spots, as indicated by the continued increase in expression of the CsCBS gene in the resistant cultivar D9320 during the early stages and there was no significant change in the susceptible cultivar D0401 (Qin et al., 2018). Our recent studies revealed that the gene encoding the CBS domain containing protein Cluster-13677.239347, which is identical to ShCBS-4D-1, plays a role as a positive regulator in sugarcane after infection by Aaa based on transcriptome and proteome databases, as well as results of RT-qPCR (Chu et al., 2020; Zhou et al., 2021).
Under three abiotic stresses, nine tested ShCDCPs showed two transcription patterns that were cultivar-dependent and -independent. For instance, ShCBSD-PB1-5A and ShCBSD-PB1-7A-1 were negatively induced by each abiotic stress tested in both cultivars. Rice OsCBSCBSPB4 (homolog of ShCBSD-PB1-5A) can respond to high and low temperature, oxidative and salt stress, whereas overexpression of OsCBSCBSPB4 in Escherichia coli resulted in higher survival rate under salt, oxidative, PEG and high temperature stress, indicating that this gene is likely involved in abiotic stress response and is a potential candidate for producing plants that have enhanced tolerance to multiple abiotic stresses (Kumar et al., 2018). ShCBS-1D-2 was upregulated in ROC22 but downregulated in MT11-610 under NaCl treatment. However, this gene acted as a negative regulator in both cultivars under PEG6000 and exogenous SA treatments. Overexpression of OsCBSX4 (a homolog of ShCBS-1D-2) in transgenic tobacco was associated with better tolerance to salt, oxidation, and heavy metals, as well as a higher survival rate of plants under stress via reduction of H2O2 content (Singh et al., 2012). The Arabidopsis genome contains six CBSXs, which may directly regulate activation of thioredoxin to control levels of intracellular H2O2 and affect plant growth and development (Ok et al., 2012). A recent study showed that CBSX3-Trxo-2 (homolog of ShCBS-1D-2) regulates ROS generation and plays an important role in regulating plant development and the redox system in Arabidopsis (Shin et al., 2020). Other CDCP family genes are also involved in stress responses. For example, overexpression of GmCBS21 (homolog of ShTlyc-1D-2) or GmCBSDUF3 (homolog of ShTlyc-1D-2) enhanced tolerance to low nitrogen levels, drought, and salt stresses in Arabidopsis (Hao et al., 2021). OsCBSX9 (homolog of ShCBS-4D-2) and OsCBSCBS4 (homolog of ShCBSDCBS-3B-1) displayed higher expression under drought as well as salinity stress conditions in rice (Tomar et al., 2022).
In general, two expression profiles of duplicated gene pairs in sugarcane responding to various stressors were observed in this study. Three gene pairs with fragment duplication events exhibited functional redundancy or divergent stress responses. Interestingly, the gene pairs ShCBSD-PB1-5A and ShCBSD-PB1-7A-1 and ShCBSD-PB1-3A and ShCBSD-PB1-3C-4 contained similar domains (two CBS domains and one PB1 domain) that shared functional redundancy of two gene pairs responding to various stressors. On the other hand, the gene pair ShCBS-4C and ShCBS-4D-1 contained different domains, i.e., ShCBS-4C included two domains (CBS and COG2905) while ShCBS-4D-1 contained one CBS domain. The two genes ShCBS-4C and ShCBS-4D-1 exhibited functional divergence in response to various stressors and require further investigation. Hughes et al. (2014) demonstrated that 13% of all duplicated genes (homologous gene pairs) underwent enhanced purifying selection to exhibit regulatory neofunctionalization (functional divergence) in Zea mays. A similar observation by Tomar et al. (2022) reported that functional divergence of two duplicate gene pairs OsCBSCBS2 (homolog of SsCBSDCBS-3D) and OsCBSCBS3 (homolog of SsCBSDCBS-5B) and OsCBSCBSPB2 (homolog of ShCBSD-PB1-5A) and OsCBSCBSPB4 (homolog of ShCBSD-PB1-5A) are expressed in Oryza species under drought stress.
Conclusion
A total of 95 SsCDCP genes with 87 alleles were systematically identified in the S. spontaneum genome (AP85-441) and were classified into eight phylogenetic groups. Gene duplication played an important driving force in the expansion and evolution of SsCDCP genes. The RNA-seq dataset and/or RT-qPCR analysis revealed that ShCDCP genes displayed different expression patterns in sugarcane under biotic (Aaa) and abiotic (drought, salinity, and SA) stresses. ShCBSD-PB1-5A and ShCBSD-PB1-7A-1 served as negative regulators in sugarcane under multiple stress conditions, while ShCBS-4D-1 and ShCBSDCBS-5A played a positive role in sugarcane under Aaa infection and in a specific cultivar (ROC22) under abiotic stress conditions (NaCl, PEG6000, and SA). Although functional redundancy and divergence among ShCDCPs were present in this study, detailed molecular mechanisms must be further explored in sugarcane in response to multiple stressors using forward (overexpression) and reverse (knockdown) genetics.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
J-RZ and S-JG: conceptualization and writing—review and editing. J-RZ and JL: writing—original draft preparation. J-RZ, JL, and J-XL: bioinformatics analysis. J-XL, H-MX, and NC: data reduction. Q-NW and S-JG: supervision, funding acquisition, and project administration. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the China Agriculture Research System (grant nos. CARS-170302 and CARS-170112) and the GDAS Project of Science and Technology Development (grant no. 2022GDASZH-2022010102).
Acknowledgments
We thank Hui-Li Zhang, Hua-Ying Fu, and Mei-Ting Huang for their assistance with this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.985653/full#supplementary-material
Footnotes
- ^ https://www.arabidopsis.org/
- ^ https://www.uniprot.org/
- ^ http://www.life.illinois.edu/ming/downloads/Spontaneum_genome/
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://www.ncbi.nlm.nih.gov/cdd/
- ^ http://pfam.xfam.org/
- ^ http://smart.embl.de/
- ^ https://web.expasy.org/protparam/
- ^ http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/
- ^ https://www.evolgenius.info/evolview/
- ^ http://meme-suite.org/tools/meme
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Anashkin, V. A., Baykov, A. A., and Lahti, R. (2017). Enzymes regulated via cystathionine β-Synthase domains. Biochemistry 82, 1079–1087. doi: 10.1134/S0006297917100017
Bateman, A. (1997). The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 22, 12–13. doi: 10.1016/s0968-0004(96)30046-7
Baykov, A. A., Tuominen, H. K., and Lahti, R. (2011). The CBS domain: A protein module with an emerging prominent role in regulation. ACS Chem. Biol. 6, 1156–1163. doi: 10.1021/cb200231c
Brindha, C., Vasantha, S., and Arunkumar, R. (2019). The response of sugarcane genotypes subjected to salinity stress at different growth phases. J. Plant Physiol. 5, 28–33. doi: 10.25081/jpsp.2019.v5.5643
Budeguer, F., Enrique, R., Perera, M. F., Racedo, J., Castagnaro, A. P., Noguera, A. S., et al. (2021). Genetic transformation of sugarcane, current status and future prospects. Front. Plant Sci. 12:768609. doi: 10.3389/fpls.2021.768609
Burdon, J. J., and Zhan, J. (2020). Climate change and disease in plant communities. PLoS Biol. 18:e3000949. doi: 10.1371/journal.pbio.3000949
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chu, N., Zhou, J. R., Fu, H. Y., Huang, M. T., Zhang, H. L., and Gao, S. J. (2020). Global gene responses of resistant and susceptible sugarcane cultivars to Acidovorax avenae subsp. avenae identified using comparative transcriptome analysis. Microorganisms 8:10. doi: 10.3390/microorganisms8010010
Chu, N., Zhou, J. R., Rott, P. C., Li, J., Fu, H. Y., Huang, M. T., et al. (2022). ScPR1 plays a positive role in the regulation of resistance to diverse stresses in sugarcane (Saccharum spp.) and Arabidopsis thaliana. Ind. Crops Prod. 180:114736. doi: 10.1016/j.indcrop.2022.114736
D’Hont, A., Grivet, L., Feldmann, P., Rao, S., Berding, N., and Glaszmann, J. C. (1996). Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol. Gen. Genet. 250, 405–413. doi: 10.1007/BF02174028
Ferreira, T. H. S., Tsunada, M. S., Bassi, D., Araújo, P., Mattiello, L., Guidelli, G. V., et al. (2017). Sugarcane water stress tolerance mechanisms and its implications on developing biotechnology solutions. Front. Plant Sci. 8:1077. doi: 10.3389/fpls.2017.01077
Flack-Prain, S., Shi, L., Zhu, P., da Rocha, H. R., Cabral, O., Hu, S., et al. (2021). The impact of climate change and climate extremes on sugarcane production. GCB Bioenergy 13, 408–424. doi: 10.1111/gcbb.12797
Fontana, P. D., Tomasini, N., Fontana, C. A., Di Pauli, V., Cocconcelli, P. S., Vignolo, G. M., et al. (2019). MLST reveals a separate and novel clonal group for Acidovorax avenae Strains causing red stripe in sugarcane from Argentina. Phytopathology 109, 358–365. doi: 10.1094/PHYTO-08-18-0303-R
Fu, H. Y., Ge, D. F., Li, X. Y., Wu, X. B., Chen, R. K., and Gao, S. J. (2017). Nested-PCR detection of Acidovorax avenae subsp. avenae, the pathogen of red stripe on sugarcane. J. Plant Prot. Res. 44, 276–282. doi: 10.13802/j.cnki.zwbhxb.2017.2015156
Garsmeur, O., Droc, G., Antonise, R., Grimwood, J., Potier, B., Aitken, K., et al. (2018). A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 9:2638. doi: 10.1038/s41467-018-05051-5
González Guzmán, M., Cellini, F., Fotopoulos, V., Balestrini, R., and Arbona, V. (2022). New approaches to improve crop tolerance to biotic and abiotic stresses. Physiol. Plant 174:e13547. doi: 10.1111/ppl.13547
Guo, F. Y., Cang, J., Lu, Q. W., Tian, Y., Song, C. H., Ren, Z. P., et al. (2020). Genome-wide analysis of CBS gene family in Hexaploid Wheat. J. Triticeae Crops 40, 425–433.
Hao, Q., Shang, W., Zhang, C., Chen, H., Chen, L., Yuan, S., et al. (2016). Identification and comparative analysis of CBS domain-containing proteins in Soybean (Glycine max) and the primary function of GmCBS21 in enhanced tolerance to low nitrogen stress. Int. J. Mol. Sci. 17:620. doi: 10.3390/ijms17050620
Hao, Q., Yang, Y., Shan, Z., Chen, H., Zhang, C., Chen, L., et al. (2021). Genome-wide investigation and expression profiling under abiotic stresses of a Soybean unknown function (DUF21) and cystathionine-β-synthase (CBS) domain-containing protein family. Biochem. Genet. 59, 83–113. doi: 10.1007/s10528-020-09991-w
Hughes, T. E., Langdale, J. A., and Kelly, S. (2014). The impact of widespread regulatory neofunctionalization on homeolog gene evolution following whole-genome duplication in maize. Genome Res. 24, 1348–1355. doi: 10.1101/gr.172684.114
Javed, T., Shabbir, R., Ali, A., Afzal, I., Zaheer, U., and Gao, S. J. (2020). Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 9:491. doi: 10.3390/plants9040491
Javed, T., Zhou, J. R., Li, J., Hu, Z. T., Wang, Q. N., and Gao, S. J. (2022). Identification and expression profiling of WRKY family genes in sugarcane in response to bacterial pathogen infection and nitrogen implantation dosage. Front. Plant Sci. 13:917953. doi: 10.3389/fpls.2022.917953
Jeong, B. C., Park, S. H., Yoo, K. S., Shin, J. S., and Song, H. K. (2013). Change in single cystathionine β-synthase domain-containing protein from a bent to flat conformation upon adenosine monophosphate binding. J. Struct. Biol. 183, 40–46. doi: 10.1016/j.jsb.2013.04.013
Karthika, G., and Govintharaj, P. (2022). “Breeding climate-resilience crops for future agriculture,” in Climate Change and Crop Stress, eds A. K. Shanker, C. Shanker, A. Anand, and M. Maheswari (Cambridge, MA: Academic Press), 1–32. doi: 10.1016/B978-0-12-816091-6.00009-2
Kemp, B. E. (2004). Bateman domains and adenosine derivatives form a binding contract. J. Clin. Invest. 113, 182–184. doi: 10.1172/JCI20846
Kumar, R., Subba, A., Kaur, C., Ariyadasa, T., Sharan, A., Pareek, A., et al. (2018). OsCBSCBSPB4 is a two cystathionine-β-synthase domain-containing protein from rice that functions in abiotic stress tolerance. Curr. Genom. 19, 50–59. doi: 10.2174/1389202918666170228141706
Kushwaha, H. R., Singh, A. K., Sopory, S. K., Singla-Pareek, S. L., and Pareek, A. (2009). Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genom. 10:200. doi: 10.1186/1471-2164-10-200
Labesse, G., Alexandre, T., Gelin, M., Haouz, A., and Munier-Lehmann, H. (2015). Crystallographic studies of two variants of Pseudomonas aeruginosa IMPDH with impaired allosteric regulation. Acta Crystallogr. D Biol. Crystallogr. 71, 1890–1899. doi: 10.1107/S1399004715013115
Lakshmanan, P., and Robinson, N. (2014). “Stress physiology: Abiotic stresses,” in Sugarcane: Physiology, Biochemistry, and Functional Biology, eds P. H. Moore and F. C. Botha (Chichester: John Wiley & Sons Ltd), 411–434. doi: 10.1002/9781118771280.ch16
Li, X. Y., Sun, H. D., Rott, P. C., Wang, J. D., Huang, M. T., Zhang, Q. Q., et al. (2018). Molecular identification and prevalence of Acidovorax avenae subsp. avenae causing red stripe of sugarcane in China. Plant Pathol. 67, 929–937. doi: 10.1111/ppa.12811
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Mou, S., Shi, L., Lin, W., Liu, Y., Shen, L., Guan, D., et al. (2015). Over-expression of rice CBS domain containing protein, OsCBSX3, confers rice resistance to Magnaporthe oryzae inoculation. Int. J. Mol. Sci. 16, 15903–15917. doi: 10.3390/ijms160715903
Ok, S. H., Yoo, K. S., and Shin, J. S. (2012). CBSXs are sensor relay proteins sensing adenosine-containing ligands in Arabidopsis. Plant Signal. Behav. 7, 664–667. doi: 10.4161/psb.19945
Panchy, N., Lehti-Shiu, M., and Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Qin, Z., Wang, Y., Liu, D., Xin, M., and Zhou, X. (2018). CsCBS cloning of cucumber and preliminary verification of resistance to downy mildew and leaf spot diseases. J. Northeast Agric. Univ. 49, 39–47. doi: 10.19720/j.cnki.issn.1005-9369.2018.02.005
Shan, H., Li, W., Huang, Y., Wang, X., Zhang, R., Luo, Z., et al. (2017). First detection of sugarcane red stripe caused by Acidovorax avenae subsp. avenae in Yuanjiang, Yunnan, China. Trop. Plant Pathol. 42, 137–141. doi: 10.1007/s40858-017-0132-x
Shin, J. S., So, W. M., Kim, S. Y., Noh, M., Hyoung, S., Yoo, K. S., et al. (2020). CBSX3-Trxo-2 regulates ROS generation of mitochondrial complex II (succinate dehydrogenase) in Arabidopsis. Plant Sci. 294:110458. doi: 10.1016/j.plantsci.2020.110458
Singh, A. K., Kumar, R., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2012). Overexpression of rice CBS domain containing protein improves salinity, oxidative, and heavy metal tolerance in transgenic tobacco. Mol. Biotechnol. 52, 205–216. doi: 10.1007/s12033-011-9487-2
Souza, G. M., Van Sluys, M. A., Lembke, C. G., Lee, H., Margarido, G. R. A., Hotta, C. T., et al. (2019). Assembly of the 373k gene space of the polyploid sugarcane genome reveals reservoirs of functional diversity in the world’s leading biomass crop. Gigascience 8:giz129. doi: 10.1093/gigascience/giz129
Subba, A., Tomar, S., Pareek, A., and Singla-Pareek, S. L. (2021). The chloride channels: Silently serving the plants. Physiol. Plant 171, 688–702. doi: 10.1111/ppl.13240
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tomar, S., Subba, A., Bala, M., Singh, A. K., Pareek, A., and Singla-Pareek, S. L. (2022). Genetic conservation of CBS domain containing protein family in Oryza species and their association with abiotic stress responses. Int. J. Mol. Sci. 23:1687. doi: 10.3390/ijms23031687
Trujillo-Montenegro, J. H., Rodríguez Cubillos, M. J., Loaiza, C. D., Quintero, M., Espitia-Navarro, H. F., Salazar Villareal, F. A., et al. (2021). Unraveling the genome of a high yielding colombian sugarcane hybrid. Front. Plant Sci. 12:694859. doi: 10.3389/fpls.2021.694859
Van De Peer, Y., Mizrachi, E., and Marchal, K. (2017). The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424. doi: 10.1038/nrg.2017.26
Wang, X., Ren, X., Zhu, L., and He, G. (2004). OsBi1, a rice gene, encodes a novel protein with a CBS-like domain and its expression is induced in responses to herbivore feeding. Plant Sci. 166, 1581–1588. doi: 10.1016/j.plantsci.2004.02.011
Wang, X. M., Feng, H., Sun, Y. F., Liu, B., Wang, X. J., Xu, L. S., et al. (2010). Cloning and expression analysis of a CBS domain containing protein gene TaCDCP1 from Wheat. Zuo Wu Xue Bao 36, 2091–2098.
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Yang, R., Li, M., Harrison, M. T., Fahad, S., Wei, M., Li, X., et al. (2022). iTRAQ proteomic analysis of Wheat (Triticum aestivum L.) genotypes differing in waterlogging tolerance. Front. Plant Sci. 13:890083. doi: 10.3389/fpls.2022.890083
Yoo, K. S., Ok, S. H., Jeong, B. C., Jung, K. W., Cui, M. H., Hyoung, S., et al. (2011). Single cystathionine β-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis. Plant Cell 23, 3577–3594. doi: 10.1105/tpc.111.089847
Zhai, K., Liang, D., Li, H., Jiao, F., Yan, B., Liu, J., et al. (2022). NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature 601, 245–251. doi: 10.1038/s41586-021-04219-2
Zhang, J., Zhang, X., Tang, H., Zhang, Q., Hua, X., Ma, X., et al. (2018). Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 50, 1565–1573. doi: 10.1038/s41588-018-0237-2
Zhang, Q., Qi, Y., Pan, H., Tang, H., Wang, G., Hua, X., et al. (2022). Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat. Genet. 54, 885–896. doi: 10.1038/s41588-022-01084-1
Keywords: CBS domain containing proteins (CDCPs), sugarcane, gene expression, Acidovorax avenue subsp. avenae, abiotic stress, defense response
Citation: Zhou J-R, Li J, Lin J-X, Xu H-M, Chu N, Wang Q-N and Gao S-J (2022) Genome-wide characterization of cys-tathionine-β-synthase domain-containing proteins in sugarcane reveals their role in defense responses under multiple stressors. Front. Plant Sci. 13:985653. doi: 10.3389/fpls.2022.985653
Received: 04 July 2022; Accepted: 02 August 2022;
Published: 25 August 2022.
Edited by:
Shah Fahad, The University of Haripur, PakistanReviewed by:
Nader R. Abdelsalam, Alexandria University, EgyptSunny Ahmar, University of Silesia in Katowice, Poland
Copyright © 2022 Zhou, Li, Lin, Xu, Chu, Wang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qin-Nan Wang, d2FuZ3Fpbm5hbjY2QDE2My5jb20=; San-Ji Gao, Z2Fvc2FuamlAZmFmdS5lZHUuY24=
 Jing-Ru Zhou
Jing-Ru Zhou Juan Li1
Juan Li1 San-Ji Gao
San-Ji Gao