
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 07 September 2022
Sec. Plant Nutrition
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.984700
This article is part of the Research Topic Dissecting Antinutrient Traits Using Omics Approaches View all 6 articles
 Rintu Jha1,2
Rintu Jha1,2 Hemant Kumar Yadav1
Hemant Kumar Yadav1 Rahul Raiya1
Rahul Raiya1 Rajesh Kumar Singh1
Rajesh Kumar Singh1 Uday Chand Jha3
Uday Chand Jha3 Lekshmy Sathee4
Lekshmy Sathee4 Prashant Singh2
Prashant Singh2 Mahendar Thudi5,6,7
Mahendar Thudi5,6,7 Anshuman Singh8
Anshuman Singh8 Sushil Kumar Chaturvedi8
Sushil Kumar Chaturvedi8 Shailesh Tripathi1*
Shailesh Tripathi1*Global food security, both in terms of quantity and quality remains as a challenge with the increasing population. In parallel, micronutrient deficiency in the human diet leads to malnutrition and several health-related problems collectively known as “hidden hunger” more prominent in developing countries around the globe. Biofortification is a potential tool to fortify grain legumes with micronutrients to mitigate the food and nutritional security of the ever-increasing population. Anti-nutritional factors like phytates, raffinose (RFO’s), oxalates, tannin, etc. have adverse effects on human health upon consumption. Reduction of the anti-nutritional factors or preventing their accumulation offers opportunity for enhancing the intake of legumes in diet besides increasing the bioavailability of micronutrients. Integrated breeding methods are routinely being used to exploit the available genetic variability for micronutrients through modern “omic” technologies such as genomics, transcriptomics, ionomics, and metabolomics for developing biofortified grain legumes. Molecular mechanism of Fe/Zn uptake, phytate, and raffinose family oligosaccharides (RFOs) biosynthesis pathways have been elucidated. Transgenic, microRNAs and genome editing tools hold great promise for designing nutrient-dense and anti-nutrient-free grain legumes. In this review, we present the recent efforts toward manipulation of genes/QTLs regulating biofortification and Anti-nutrient accumulation in legumes using genetics-, genomics-, microRNA-, and genome editing-based approaches. We also discuss the success stories in legumes enrichment and recent advances in development of low Anti-nutrient lines. We hope that these emerging tools and techniques will expedite the efforts to develop micronutrient dense legume crop varieties devoid of Anti-nutritional factors that will serve to address the challenges like malnutrition and hidden hunger.
Malnutrition is one of the major challenging issues with the ever-increasing human population globally (Harding et al., 2018). It is widely prevalent in underprivileged population residing in the African and South Asian nations. The diets of almost one-third of the people across the globe is deficient in the required concentration of micronutrients in their food source (Stein, 2010). Malnutrition is gaining serious attention globally as it causes multiple health ailments due to insufficient daily intake of micronutrients including dietary minerals and vitamins in diet (Khoury et al., 2014). About 20 million infants are born annually with below average birth weight owing to micronutrient-related problems leading to neonatal death (Kumar and Pandey, 2020). High consumption of calorie-rich staple insufficient in micronutrition leads to “hidden hunger” especially in human population residing across low-income countries (Dawson et al., 2019). Among the various micronutrient deficiencies, iron deficiency is the most commonly found micronutrient deficiency causing anemia especially, in women and children human population globally (Abbaspour et al., 2014). About 2 billion people are afflicted with iron deficiency across the globe (Viteri, 1998; Miller, 2013). Zinc deficiency is associated with impaired growth and development, compromised immune response-related issues, cardiovascular disease, and chronic kidney disease in human population (Nakatani et al., 2021). Similarly, inadequate folate consumption affects millions of people and enhances women’s risk during the pregnancy (Sen Gupta et al., 2013). Grain legumes are well known for their high protein and vitamin content and are affordable alternatives to animal protein. The United Nations (UN) declared the year 2016 as “International Year of Pulses (IYOP, 2016)” to create awareness about pulse crops as vital components for global food and nutritional security and to promote pulse production. Food legumes such as chickpea (Cicer arietinum L.), common bean (Phaseolus vulgaris L.), lentil (Lens culinaris), mung bean (Vigna radiata L.), pea (Pisum sativum), and pigeon pea (Cajanus cajan [L.] Millsp.) are rich reservoir of dietary micronutrients and essential amino acids and thus could potentially play a pivotal role in overcoming the increasing challenges of malnutrition-related problems.
Understanding the process of mineral acquisition, transport, and accumulation in legume seeds is critical for optimizing nutrient bioavailability (Roorkiwal et al., 2021). Each of these processes is likely governed by a set of genes, many of which are still unknown. Several studies have discovered genes involved in uptake and mobilization of Fe (Palmer and Guerinot, 2009; Xiong et al., 2012; Sen Gupta et al., 2017) and Zn (Palmgren et al., 2008; Urwat et al., 2021) from different parts of plant to the seeds.
Improving grain micronutrient contents through plant breeding approach is an economically viable options to overcome the malnutrition problem. Crop biofortification requires a multidisciplinary effort involving the plant breeders, nutritionists, and agronomists. Thorough assessment of global legume germplasm including cultivated and wild species could greatly assist us in identifying high micronutrient containing genotypes. Pre-breeding and breeding efforts are needed to develop the micronutrient dense lines and test them at multilocations to assess the role of environment on micronutrient assimilation. Likewise, increasing legume genomic repertoire could enable us uncovering the various QTLs/genomic segments controlling micronutrient content through biparental QTL mapping and genome-wide association studies (GWAS). Likewise, unprecedented advancements in functional genomics shed light on the various underlying candidate gene(s) regulating various micronutrient biosynthesis and accumulation with precise function. Similarly, agronomic interventions like foliar application of fertilizers enriched with micronutrients could also aid in the biofortification of grain legumes are mentioned (Poblaciones and Rengel, 2016; Silva et al., 2019). Transgenics for improving micronutrient content with suitable examples are illustrated, and scope of genomic selection, speed breeding and emerging genome editing and microRNA technologies for designing biofortified grain legumes are discussed. An integrated breeding approach embracing all these technologies can surely help us in alleviating the rising malnutrition-related problems and securing “zero hunger” world. Additionally, despite the fact that grain legumes are a rich source of minerals, several Anti-nutritional elements or factors, including as phytic acids (phytate or PA), oxalates, and tannins, chelate the minerals and reduce their bioavailability by limiting their absorption in our dietary system, thus numerous health issues occur. Several studies demonstrated the presence of phytic acids (Urbano et al., 2000; Shi et al., 2018; Pramitha et al., 2021), RFOs (Zhawar et al., 2011; Roorkiwal et al., 2021), oxalates (Noonan and Savage, 1999; Shi et al., 2018) and tannins (Arts et al., 2000; Petroski and Minich, 2020) in legumes and their Anti-nutritional properties.
Metal micronutrients play critical biochemical and structural roles in plants. Terrestrial vascular plants, as sessile organisms, are exposed to a wide range of environmental circumstances and the stresses that come with them, including soil nutrient deficiency signals, which have a detrimental impact on growth and development (Fan et al., 2021). Understanding the process of mineral uptake from the soil and their accumulation in legume seeds is essential for nutrient bioavailability and optimization. Several studies have attempted to identify and establish gene families involved in mineral transport from different vegetative tissues of plant to seed. In recent years, much has been learned about the genes and proteins necessary for primary Fe and Zn uptake from the soil but here we attempt to illustrate the Fe and Zn transport mechanisms in legumes.
In legumes, the proteins HA2 (H+-ATPase), FRO2 (Ferric reductase oxidase2), and IRT1 (iron-regulated transporter) are primarily responsible for Fe absorption and delivery to the roots (Figure 1; Roorkiwal et al., 2021). A plasma membrane-localized transporter protein, YS1, is a crucial protein for acquiring Fe via uptake of Fe(III)-phytosiderophores (Curie et al., 2001). Expression analysis has revealed several genes that encode Fe transporter protein families in several legume species such as in Arachis hypogaea (AhIRT1, Xiong et al., 2012), Medicago truncatula- natural resistance-associated macrophage protein 1 (MtNRAMP1, Tejada-Jiménez et al., 2015; MtZIP3, MtZIP5, and MtZIP6, Lopez-Millan et al., 2004), Glycine max (NRAMP, Qin et al., 2017), Lens culinaris [Ferritin-1, basic helix–loop–helix-1 (bHLH-1), and FER-like transcription factor protein and IRT1, Sen Gupta et al., 2017] and in Cicer arietinum (CaFer1, Parveen et al., 2016). Fe sensing mechanisms are still unknown, despite the discovery of distinct transcriptional networks that regulate absorption and intracellular distribution (Thomine and Vert, 2013). The high-affinity transporter IRT1 is principally responsible for Fe absorption (Palmer and Guerinot, 2009). Transporters like ZIP (zinc inducible protein) and YSL (yellow stripe like) are implicated in metal unloading from the xylem (Roorkiwal et al., 2021). In Arabidopsis, the oligopeptide transporter (OPT3) has been proposed to play a key role in accurate long-distance Fe transmission from shoots to roots, as well as in importing Fe into phloem companion cells (Kumar et al., 2017). The NRAMP (NRAMP3 and NRAMP4) family of genes is known to play a role in Fe homeostasis, while YSL and OPTs are involved in the loading and unloading of Fe2+ NA (nicotianamine) complexes into and out of phloem (Palmer and Guerinot, 2009).
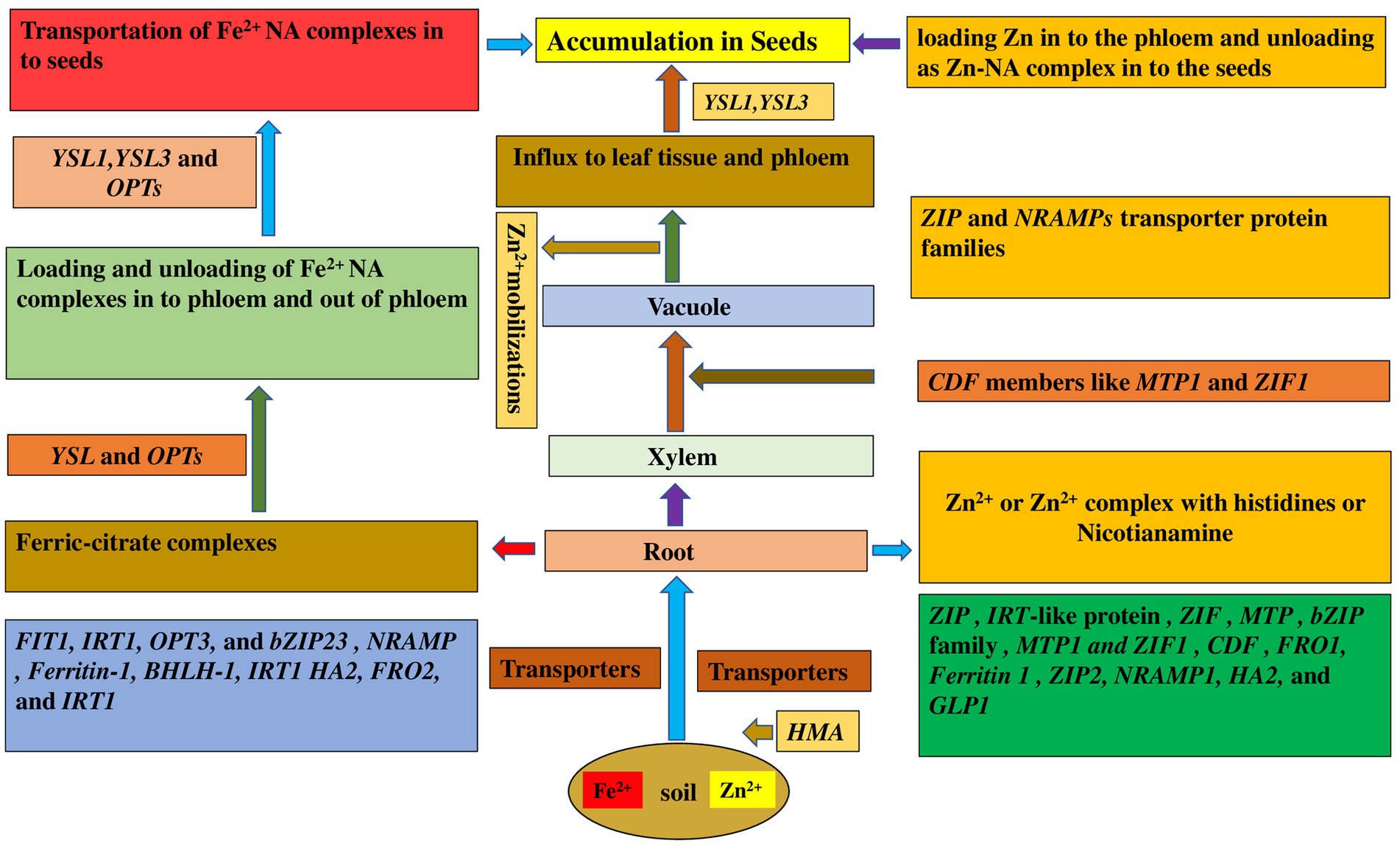
Figure 1. Molecular mechanism involved in uptake and acquisition of Fe and Zn in legume seeds. YSL, Yellow Stripe Like; OPT, Oligopeptide transporter; NRAMP, Natural resistance-associated macrophage protein; IRT, Iron regulated transporter; FER, Ferritin; FRO Ferric reduction oxidase; ZIP, Zinc induced protein; HA, H+ transporting ATPase; GLP, Germin like protein; CDF, Cation diffusion facilitator; MTP, Metal tolerance protein; ZIF, Zinc-induced facilitator; BHLH, basic helix–loop–helix; HMA, heavy metal ATPase.
Mutants with defects in homeostatic regulation of Fe and other metals have been studied for several years. More recently, it has been shown that the Ferric reductase defective3 (FRD3) protein controls Fe localization within the plant (Green and Rogers, 2004). It was observed in pea (Pisum sativum) mutant bronze (brz) that there is overaccumulation of Fe, Cu, Mn, Zn, and other elements in shoots (Cary et al., 1994), and the pea mutant degenerative leaves (dgl) overaccumulates Fe in leaves and seeds (Grusak et al., 1996). Given evidences suggest for the presence of nicotianamine (NA) in shoot tissues and its affinity for different ions. NA is a non-protein amino acid that is ubiquitous in plants. NA has been implicated as important for translocation of Cu, Fe, Mn, and Zn (Pich et al., 1994; Pich and Scholz, 1996). Based on studies of the chln mutant, NA is vital for homeostasis of Fe and other metal micronutrients. The tomato mutant chloronerva (chln) exhibits increased ferric reductase activity and overaccumulates Fe in mature leaves (Stephan and Grun, 1989), yet the younger leaves are chlorotic (Becker et al., 1992). In Arabidopsis, the combination of null mutations in two AtYSLs, YSL1 and YSL3, resulted in a severe phenotype including interveinal chlorosis; altered metal concentrations in leaves, roots, and seeds; and greatly decreased fertility. YSL promoter-β-glucuronidase (GUS) reporter constructs reveal a localization pattern that was consistent with a role for YSL1 and YSL3 in providing metal-NA compounds to leaves, pollen, and developing seeds (Waters et al., 2006).
Metal remobilization from senescent leaves is a major role of YSL genes, which also serve as transporters in seed development, reproductive organ growth, and long-distance transport of metal complexes with NA (Di Donato et al., 2004). Iron and Cu regulate AtYSL2 expression, and YSL2 is abundantly expressed in the root endodermis, pericycle, and xylem cells. AtYSL2 is thought to play a function in transporting metals to and from the vasculature based on its location. Downregulation of the genes YSL1 and YSL3 in Fe-deficiency situations would decrease Fe removal from the xylem into adjacent tissues. Hence, the physiological role of YSL1 and YSL3 is to translocate metals into vascular parenchyma cells for distribution away from veins into interveinal regions in maturing leaves, away from interveinal regions toward phloem tissue in senescing leaves, and out of the vasculature in fruits to supply seeds.
Zinc is primarily taken up as Zn2+ across the plasma membrane of root cells in legumes. Zn absorption and transport from root to seed have been aided by ZIP (Zinc Iron Permease) transporters (Palmgren et al., 2008). The key transport protein families involved in Zn transport are the ZIP family/ZRT, IRT-like proteins, the HMA (Heavy Metal ATPases), and the MTP (Metal Tolerance Protein) family (Gupta et al., 2016). ZIP genes have been identified in various plant species like Arabidopsis (bZIP19, bZIP23, and bZIP24, Assunçao et al., 2010), Soybean (GmZIP1; Moreau et al., 2002), M. truncatula (MtZIP1, MtZIP3, MtZIP4, MtZIP5, MtZIP6, and MtZIP7, Lopez-Millan et al., 2004) and in Phaseolus vulgaris (PvZIP12, PvZIP13, PvZIP16, and PvbZIP1, Astudillo et al., 2013).
Net Zn fluxes in the shoot are influenced by both symplastic and apoplastic fluxes. Apoplastic Zn transport includes Zn entering the cytosol via the cell wall plasma membrane contact, making it less selective than symplastic transport. Symplastic transport, on the other hand, regulates the selectivity and amount of nutrient delivery (Gupta et al., 2016). MTP1 and ZIF1 transporters, members of the cation diffusion facilitator (CDF) family, are involved in Zn sequestration in vacuole, while NRAMPs are involved in Zn mobilization from the vacuole (Haydon and Cobbett, 2007). Zn is loaded into the xylem by HMA (Heavy metal ATPases), and is transported as Zn2+ or in complex with histidines or Nicotianamine within the xylem (Palmgren et al., 2008). YSL is involved in loading Zn into the phloem and discharging it to the seeds as the Zn-NA complex, whereas members of the ZIP family are involved in mediating Zn2+ influx to leaf tissue and the phloem (Waters and Grusak, 2008). The molecular mechanism of uptake and mobilization of Zn2+ from root to seeds have been reviewed in several articles (Waters et al., 2006; Palmgren et al., 2008).
More recently, in common bean, Urwat et al. (2021) demonstrated, the expression of IRT1, FRO1, and Ferritin 1 genes showed an important role in Fe and Zn uptake/transport and signaling under Fe/Zn stress whereas the expression of ZIP2, NRAMP1, HA2, and GLP1 (Germin-like protein) genes in Phaseolus vulgaris was highly responsive to Zn uptake (Figure 2).
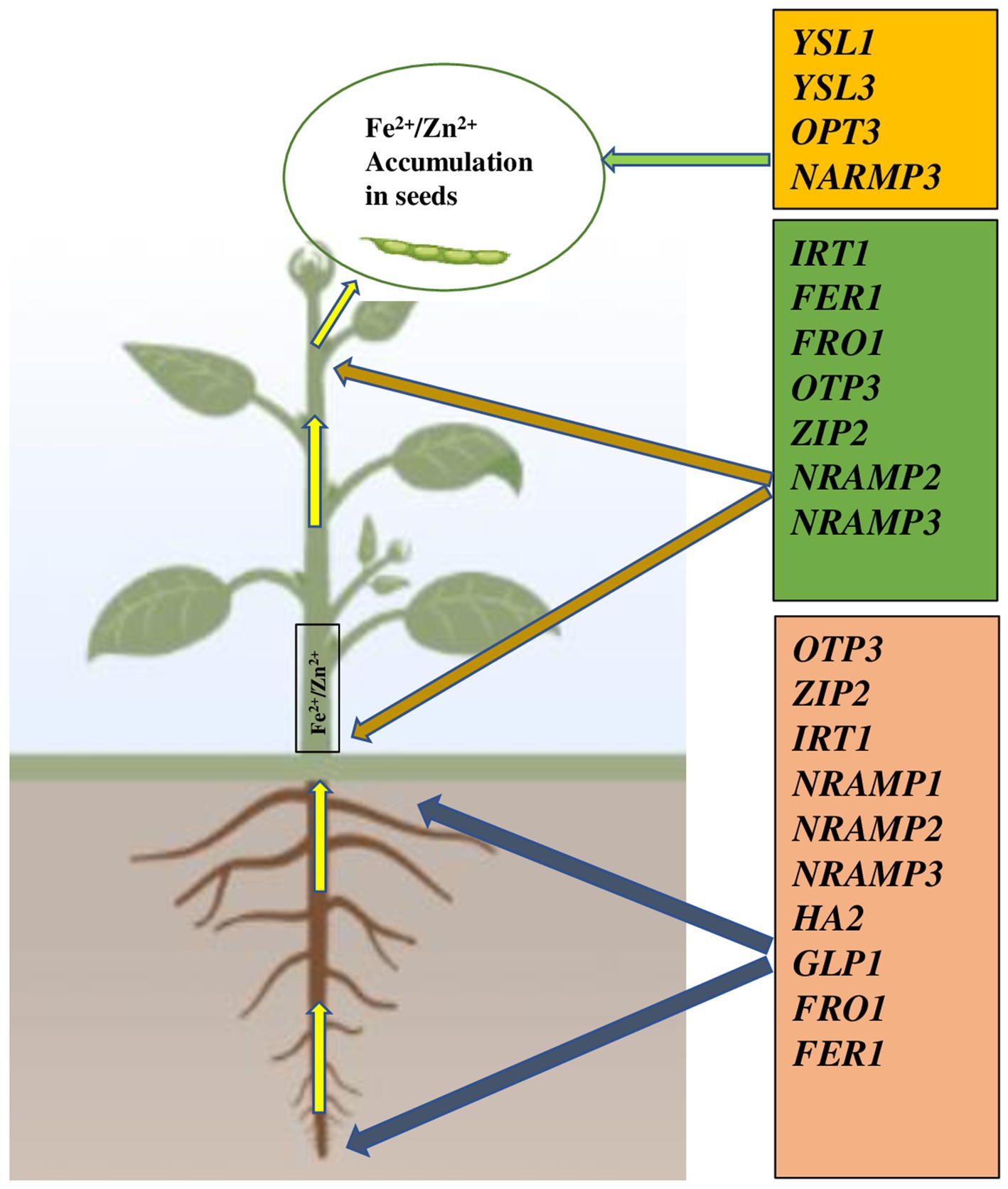
Figure 2. Gene families involved in translocation and accumulation of Fe2+/Zn2+ in seed of common bean (Urwat et al., 2021). YSL1 and YSL3 genes are involved in unloading minerals to seeds. YSL, Yellow Stripe Like; OPT, Oligopeptide transporter; NRAMP, Natural resistance-associated macrophage protein; IRT, Iron regulated transporter; FER, Ferritin; FRO Ferric reduction oxidase; ZIP, Zinc induced protein; HA, H+ transporting ATPase; GLP, Germin like protein. Plant images made with Biorender (https://biorender.com/).
These identified candidate gene families can be potentially deployed in crop plants to enhance the Fe/Zn uptake and its bioavailability.
Conventional breeding programs require screening of a genetically diverse panel and identification of potential donor parents fortified with micronutrient(s) of our interest in order to transfer the gene to the elite genotype to obtain a biofortified crop variety (Guindon et al., 2021). Although biofortification through the use of plant breeding strategies offers the most reliable, sustainable, and cost-effective strategies, developing a bio-fortified product takes a long time. The use of molecular markers could reduce time by allowing rapid identification of the superior donor with the gene of interest.
Genetic diversity for micronutrient concentration in the gene pool is a prerequisite for successful biofortification. Genetic variation for micronutrient concentration has been well studied in legumes (Table 1). Research on legume crops have revealed significant genetic variations for the essential micronutrients in chickpea (Ray et al., 2014; Jha et al., 2015; Rezaei et al., 2016; Vandemark et al., 2018; Karaca et al., 2020), common bean (Beebe et al., 2000; Ray et al., 2014; Murube et al., 2021), cowpea (Gerrano et al., 2022; Sodedji et al., 2022), lentil (Ray et al., 2014; Kumar et al., 2016; Vandemark et al., 2018), mung bean (Dahiya et al., 2013; Kumar and Pandey, 2020), Pea (Ray et al., 2014; Jha et al., 2015; Dissanayaka, 2019; Klein et al., 2020) and soybean (Kumar and Pandey, 2020). The promising genotypes identified can be utilized in biofortification programs for mapping studies and development of breeding material rich in micronutrients. Molecular markers linked to micronutrient content can aid in marker-assisted selection.
Genetic dissection of grain micronutrient content remains complex as it is governed by multiple quantitative trait loci (QTLs). During the last decade, a paradigm shift in developing genomic resources in various legumes allowed unraveling the genetic control of important traits, including grain micronutrient content. Initially, the bi-parental mapping approach has been mainly used to investigate genomic regions controlling micronutrient content in various grain legumes (Cichy et al., 2009; Blair et al., 2010; Park et al., 2019; Diaz et al., 2020; Sab et al., 2020; Table 2).
Availability of high throughput novel single-nucleotide polymorphism (SNP) markers allows implementations of GWAS approach for identifying genomics regions/haplotype assembly rich in micronutrient density in various grain legumes (Diapari et al., 2014; Khazaei et al., 2017; Diaz et al., 2020; Wu et al., 2020). Employing SNP markers derived from genotyping by sequencing technique in a chickpea mapping population derived from the cross MNK1 x Annigeri1 a total of eight QTL contributing to grain Fe content on CaLG03, CaLG04, and CaLG05 and a total of 11 QTLs controlling grain Zn content on CaLG04, CaLG05, and CaLG08 were identified (Sab et al., 2020). Notably, they also found three QTLs viz., CaqFe4.4, CaqFe4.5, and CaqZn4.1 were colocalized on the “QTL-hotspot” region for drought tolerance on CaLG04 (Sab et al., 2020). In a study on a diverse set of chickpea genotypes, GWAS analysis recruiting 1,536 SNPs markers revealed one significant MTA for grain Zn on LG1, three significant MTAs for grain Zn on LG4, two important MTA for Fe content on LG4, and two significant MTA for grain Fe and grain Zn content on LG6 and LG7 (Diapari et al., 2014).
Phenotypic and genotypic evaluation of a mapping population developed from the cross AND969 x G19833 in common bean revealed a significant QTL controlling grain Zn and Fe content colocalized on B6 chromosome and contributing 36% of total phenotypic variation (Cichy et al., 2009). Subsequently, a total of 12 meta-QTLs controlling grain Zn and Fe content were identified by assessing seven mapping populations tested across multi-locations. Among these meta-QTLs, 2 QTLs were found to be specific to grain Fe and two QTLs specific to grain Zn and eight QTLs remained colocalized controlling both Fe and Zn content (Izquierdo et al., 2018). QTL analysis in a multiparent advanced generation intercross (MAGIC) population derived from eight founder parents allowed pinpointing two QTLs (SdFe6.1 andSdFe6.2) controlling grain Fe content in common bean (Diaz et al., 2020). Genetic dissection of micronutrients in common bean unraveled 21 QTLs in an F9:11 RIL (DOR364 × G19833) population for 13 nutrient elements (Blair et al., 2016). In common bean, a genome-wide survey through SNP markers in a set of 192 accessions resulted in one significant MTA for grain Zn on the PV01 chromosome (Caproni et al., 2020). Further, the underlying candidate gene controlling grain Zn content was Phvul001G233500.
In lentils, a total of six QTLs controlling grain Mn content explaining phenotypic variation up to 24% were elucidated on LG1, LG3, and LG7 by evaluating mapping population derived from CDC Redberry × ILL7502 cross assessed for 2 years at three locations in Turkey (Ates et al., 2018). Mapping for Se content in lentil recombinant inbred lines (RILs) developed from PI 320937 × Eston cross identified a total of 4 QTLs on LG2 and LG5 explaining 6.3%–16.9% phenotypic variation (Ates et al., 2016). In a marker-trait-association (MTA) study in lentil, mapping in a panel comprising 96 diverse germplasm for Fe and Zn concentration identified three markers PBALC 13, PBALC 206, and GLLC 563 associated with grain Fe concentration that explained 9%–11% of phenotypic variation and four SSR markers PBALC 353, SSR 317–1, PLC 62, and PBALC 217 associated with grain Zn concentration which explained 14%–21% of phenotypic variation (Singh et al., 2017). Subsequently, association mapping for grain zinc and iron content by deploying 80 SSR markers in a panel of 96 lentil genotypes allowed deciphering two significant marker-trait associations for grain Fe and three significant marker-trait associations for grain Zn content (Kumar et al., 2019). Two significant MTAs for grain Fe content and one important MTA for grain Zn were identified through deploying 1,150 SNP markers in a panel of 138 lentil accession collected from diverse countries (Khazaei et al., 2017). GWAS in a set of 1,150 individuals derived from MAGIC population, evaluated in 2014 and 2016 revealed two QTLs (SdFe6.1, SdFe6.2) controlling grain Fe content (Diaz et al., 2020).
In a GWAS study performed in a set of 96 mung bean genotypes collected from 13 countries five significant MTAs for Fe content, five for Mn content, eight for P content, nine for S content, seven for Zn content, and nine for K content were identified.
MicroRNAs (miRNAs) are potential candidates for increasing grain micronutrients, in addition to protein-coding genes (Sunkar and Zhu, 2004). miRNAs are involved in almost every biological and metabolic process in plants, including development, cell wall production, and stress responses (Chen, 2009). miRNA are one of the potential tools to enrich grain particles with micronutrients by regulating gene expression in food crops. To date, several miRNAs have been discovered with regulatory activity in differentially expressed genes that governs various functions such as uptake, mobilization, and homeostasis of nutrients, ensuring adequate supply without any toxicity and also reported their specific roles in homeostasis of micronutrients like B, Mn, Zn, Cu, Mo, and Ni (Patel et al., 2017).
miRNAs have emerged as important regulators of plant stress responses and for maintaining nutrient homeostasis by regulating the expression of transporters involved in nutrient absorption and mobilization (Paul et al., 2015). miRNAs also have a role in nutritional homeostasis by acting as an endogenous signal for micro and macronutrient availability (Marín-González and Suárez-López, 2012; Kehr, 2013; Jagadhesan et al., 2022). The reports on miRNAs regulating micronutrient homeostasis in legumes are meager, barring the information pertaining to Phaseolus vulgaris (common bean) and Medicago truncatula (Valdes-Lopez et al., 2010; Naya et al., 2014).
During Copper (Cu) deficiency, several miRNA families, such as miR397, miR408, and miR857 are upregulated, suppressing the production of laccase and plastocyanin genes (Abdel-Ghany and Pilon, 2008). The induction of miR398 suppresses Cu/Zn superoxide dismutase CSD1, CSD2, and Cu chaperones for SOD1 (CCS1) gene expression under Cu deficit (Sunkar et al., 2006). When exposed to Cu deficiency, common bean plants with transgenic roots over-expressing miR398 showed a 20-fold increase in mature miR398b and almost no target transcript levels, as well as increased anthocyanin content and expression of Cu-stress responsive genes (Naya et al., 2014). Fe deficit also regulates miR398, one of the primary activators of CSD gene expression during Cu deficiency, albeit in the opposite direction. Cu shortage increases the expression of miR397, miR398a, miR398b, miR398c, miR398s, miR399, miR408, and miR2111, whereas Fe deficiency decreases their expression and hence regulates CSD1 and CSD2 expression (Pant et al., 2008; Waters et al., 2012). As a result, the Cu-Fe interaction is yet another novel finding in the field of Fe homeostasis gene expression research. In addition, eight miRNAs from five families (miR159, miR164, miR172, miR173, and miR394) were previously identified as Fe-responsive families in Arabidopsis, The Fe deficiency responsive cis-acting elements1 and 2 (IDE1/IDE2) were discovered in the promoters of 24 miRNA genes in Arabidopsis, and they mirror the Fe-responsive gene families that are controlled during Fe deficiency (Kong and Yang, 2010).
Valdes-Lopez et al. (2010) found miRNAs in the leaves, roots, and nodules of control and nutrient-stressed common bean plants (deficiency of phosphorus (P), nitrogen (N), or iron (Fe); acidic pH; and manganese (Mn) toxicity). Cu content was shown to be higher in common bean plants grown under N and Fe deficit conditions, where miR398 and miR408 were downregulated, but lower in acidic pH and high Mn, where miR398 was upregulated. These findings are consistent with those of Yamasaki et al. (2009), who discovered that SQUAMOSA promoter binding protein-like 7 (SPL7) increased the transcription of miR397, miR398, and miR408 in Arabidopsis under Low-Cu circumstances. For example, pvu-miR1511, gma-miR1513, gma-miR1515, and gma-miRNA1516 were all found to be significantly enhanced in Fe deficient leaves, suggesting that they may be involved in the Fe signal transduction pathway (Valdes-Lopez et al., 2010). Several miRNAs have been discovered to be upregulated during Mn deficiency which are linked to other mineral deficiencies also. miR319, miR169, miR396, miR170, miR164, miR390, miR395, miR166, miR172, miR157, miR156, and miR167 are upregulated during Mn toxicity in Phaseolus vulgaris and suppresses the expression of a wide group of genes, including various transcription factors such as TEOSINTE-LIKE1, CYCLOIDEA, PROLIFERATING CELL FACTOR1 (TCP), HAPLESS (HAP2), SCARECROW-LIKE, NO APICAL MERISTEM (NAC; Valdes-Lopez et al., 2010).
High concentration of Al, Cd, Hg, and Cd, have also been shown to cause upregulation or downregulation of certain miRNAs, which are thought to play a role in plant adaptation processes by regulating the expression of numerous stress-related genes (Zhou et al., 2012; Li et al., 2013; Zeng et al., 2014). In Arabidopsis sulfate, P, and Cu deprivation stimulate miR395, miR399, and miR398 expression (Sunkar et al., 2007). In Arabidopsis and Brassica, Cu deficiency has been found to enhance the production of miR397 and miR408 genes (Sunkar et al., 2006; Naya et al., 2014). When three-week-old Medicago truncatula seedlings were subjected to deficiency of sulfate, P, or Cu. In response to sulfate deficiency, the abundance of miR395 was increased in shoots and roots. When grown on phosphate-free media, miR399 was activated, while miR398 was upregulated in both shoots and roots of M. truncatula grown on medium without Cu. A similar pattern of induction was seen for miR397 in M. truncatula seedlings grown on Cu-deficient media (Jagadeeswaran et al., 2009; Saini et al., 2012). Mineral ion absorption, transport, and remobilization are all closely related to mineral bioavailability in grains (Aung et al., 2013). The discoveries on nutrient-related miRNAs and their gene regulation mechanism could pave the way for a new platform for boosting legume bio fortification initiatives in the future (Blair et al., 2013).
Agronomic intervention optimizes the application of mineral fertilizers and improves the solubilization and mobilization of mineral elements in the soil (White and Broadley, 2009). Application of fertilizers can improve the micronutrient status in soil to overcome micronutrient deficiencies in humans (Cakmak, 2008). The use of mineral fertilizers is feasible in the developed world, as demonstrated by the success of selenium (Se) fertilization of plants in Finland (Aro et al., 1995), zinc (Zn) fertilization in Turkey (Cakmak et al., 1999) and iodine (I) fertilization in irrigation water in China (Jiang et al., 1997). Biofortification with agronomic approaches has been successfully demonstrated in some legumes. For example; the application of Se as selenate provides more phyto-availability than selenite in cowpea (Silva et al., 2019). A recent study found that combined foliar treatment of ZnSO4.H2O (0.5%) and FeSO4H2O (0.5%) at the pre-flowering and pod formation stages in lentil were most effective in enhancing grain and straw yield, Zn and Fe content, and uptake (Dhaliwal et al., 2021). Likewise, the foliar application of a mixture containing 8 mg ZnSO4·7H2O/kg soil and 0.25% (w/v) ZnSO4·7H2O could serve a better option for fortifying field peas (Poblaciones and Rengel, 2016). Similarly, Zn-fortified soybean sprouts have been successfully achieved by application of ZnSO4 solution, containing, Zn 10 to 20 μg/ml (Zou et al., 2014).
Use of agronomic practices like intercropping, accelerate the P, Fe and Zn uptake efficiency, leading to increased yield and improved grain quality (Xue et al., 2015). Biofortification through agronomic approaches is simple and the least expensive but has some disadvantages like, regular application of mineral fertilizers, could impede the accessibility of other micronutrients and also cause environmental toxicity (Jha and Warkentin, 2020). In addition, soil factors such as higher pH, calcite, organic matter, and high concentration of other ions hinder the accessibility of micronutrients to the crop plants (Alloway, 2009).
In crops, in which the desired variation for a nutrient is not available in cultivated and wild relatives or in which biofortified crop varieties are difficult to produce with the help of conventional breeding approaches, transgenic breeding and genome editing are alternative approaches to produce bio-enriched crops with nutritional traits of interest. In addition to enriching the micronutrient concentration, this approach can also target the removal of Anti-nutrients or the ingestion of promoters that can increase the bioavailability of micronutrients (Carvalho and Vasconcelos, 2013; Garg et al., 2018). Genome editing is a powerful tool for knock out, knock down, or alteration of gene expression without disturbing the genetic makeup of the genotype. CRISPR/Cas9 genome editing can be utilized to enhance the seed micronutrient content as well as to reduce anti-nutritional factors in plants.
CRISPR-edited plants are free from any foreign DNA; thus, they may have better acceptability compared to traditional GM crops. Targeted genome editing through the use of artificial nucleases such as zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs) has the potential to accelerate basic as well as applied research by offering the possibility to quickly and precisely create modified genomes in a predictable way (Bortesi and Fischer, 2014; Jaganathan et al., 2018). Unlike first-generation genome editing tools (ZFNs and TALENs), clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR associated protein Cas9 genome editing represents simple designing and cloning techniques, Cas9 protein with guide RNAs potentially targeting multiple sites in the genome (Jaganathan et al., 2018). Applying these modern genome editing techniques will aid in the development of non-GMO plants with the desired trait that can contribute to increased yield potential under stressful conditions and also improve the nutritional quality. In recent years, genome editing techniques have been applied on several crops like rice (Li et al., 2012; Zhang et al., 2014) wheat (Wang et al., 2014), and tomatoes (Brooks et al., 2014). In addition, transgenic rice with 30% of the estimated average requirement (EAR) for micronutrients Fe and Zn has been developed and tested in flooded field conditions (Trijatmiko et al., 2016). In rice, for Fe and Zn biofortification, the nutritional goals were achieved in field conditions in the Philippines and Colombia (Trijatmiko et al., 2016). Transgenic corn containing multivitamins was made by modifying three different metabolic pathways at the same time to increase levels of three vitamins, namely beta-carotene (169-fold), ascorbate (6-fold), and folate (2-fold), in the endosperm (Naqvi et al., 2009). Metabolic engineering has increased the folate concentration in tomatoes and rice (Blancquaert et al., 2013, 2014). A recent study demonstrated the synergistic use of CRISPR/Cas9 and TALENs technologies to generate a collection of functional mutant plant lines in Glycine max and Medicago truncatula (Curtin et al., 2018). In wheat, the CRISPR/Cas9 editing of TaIPK1 effectively reduced the phytate content and thereby increased the grain Fe and Zinc (Zn) content (Ibrahim et al., 2022). Nucleotide substitutions of inositol trisphosphate six kinase gene (OsITPK6) could significantly reduce rice grain phytic acid content. Targeted editing of the first exon of OsITPK6 using the CRISPR/Cas9 method generated mutant lines OsIPTK6_1 and OsIPTK6_2 with significantly low phytic acid content and higher inorganic P levels than the wild type (Jiang et al., 2019).
The success in developing transgenic fortified pulses has not yet been reported, due to the absence of effective and reproducible plant regeneration methods and biosafety regulations. Recently, genetically engineered chickpea with high Fe content using chickpea nicotianamine synthase 2 (CaNAS2) gene in combination to soybean ferritin (GmFER) genes was developed (Tan et al., 2018). Transgenic chickpea lines with root-specific expression of the cytokinin oxidase/dehydrogenase 6 (CaCKX6) gene had increased concentration of minerals, namely Zn (27%–62%), Cu (26%–61%), Fe (22%–48%), Mg (13%–22%), K (11%–27%), and P (5%–19%) (Khandal et al., 2020). The NAS genes isolated from chickpea (CaNas2) and rice (OsNas2) catalyze the biosynthesis of nicotianamine (NA) and are involved in Fe uptake and translocation in plants. Stable transgenic chickpea lines containing GUS (uidA) as well as Fe-biofortification genes (OsNAS2 and CaNAS2) have successfully been produced (Das Bhowmik et al., 2019). Transgenic common bean expressing methionine-rich storage albumin gene from Brazil nut showed increased methionine and cysteine content (Aragao et al., 1999). Enhanced expression of sunflower seed albumin gene increased methionine content (by up to 94%) in lupin (Molvig et al., 1997).
In cowpea (V. unguiculata), symbiotic nitrogen fixation was successfully disrupted by targeting a symbiosis receptor-like kinase gene using CRISPR/Cas9-mediated gene editing (Ji et al., 2019). In soybean, instance of increasing pro-vitamin A, oleic acid, and seed protein content in soybean by expressing bacterial PSY (phytoene synthase) gene is available (Schmidt et al., 2015). Likewise, improvement of beta-carotene through overexpression of PSY and carotene desaturase in soybean has been reported (Kim et al., 2012). Soybean protein is deficient in sulfur-containing essential amino acids viz., cysteine, and methionine. Kim et al. (2012) demonstrated increased cysteine content through over expression of O-acetylserine sulfhydrylase gene in soybean. Overexpression of maize zein protein enabled in enhancing methionine and cysteine content in soybean (Dinkins et al., 2001). Enhanced methionine content in soybean through overexpression of cystathionine γ-synthase has been reported (Hanafy et al., 2013; Song et al., 2013). Manipulation of maize C1 and R transcription factor-driven gene enhanced isoflavone content in soybean (Yu et al., 2003).
The development of RNA-guided genome editing (CRISPR-Cas9 technology) has been established in some legume crops, like Medicago truncatula, soybean, cowpea, and chickpea for which transformation protocols are available. Multiple GmU6 promoters were tested for their suitability in driving gRNA expression in soybean hairy roots (Di et al., 2019). It was found that the GmU6-8 and GmU6-10 promoters had higher activity in enhancing editing efficiency (20.3% and 20.6%, respectively), compared with the other nine promoters. Gene editing has been successfully carried out by CRISPR/Cas9 technology in soybean for seed-related traits, such as seed oil profile (Do et al., 2019), unpleasant beany flavor of seed product (Wang et al., 2020), and isoflavone content and resistance to soybean mosaic virus (Zhang et al., 2020).
Gene-editing through CRISPR/Cas9 technology has paved way for novel possibilities for functional genomics in grain legume crops. The basic requirement for genetic transformation, including CRISPR/Cas9 gene editing, is the ability to deliver the DNA/RNA components, with the regeneration of an entire plant. Most of the food legumes are resistant to the uptake and integration of introduced DNA (Yadav et al., 2017) and quite a few of them are also recalcitrant (Ochatt et al., 2018). Hence, the availability of effective methods for plant transformation and whole-plant regeneration along with a supportive regulatory environment, and consumer acceptance of gene-edited crops are necessary for the success of genome editing for legume enhancement.
Biofortification strategies have contributed to food security by producing legume varieties fortified with essential micronutrients (Table 3).
Biofortified varieties of lentils have been developed as part of the CGIAR’s Harvest Plus program. Nepal has released Khajurah Masuro-4 with an Fe content of more than 75 ppm.1 Lentil varieties enriched with Fe and Zn content from Bangladesh are Barimasur-9 (78 ppm Fe), Barimasur-4 (86.2 ppm, Fe), Barimasur-5 (86 ppm Fe and 59 ppm Zn), Barimasur-6 (86 ppm Fe and 63 ppm Zn), Barimasur-7 (81 ppm Fe) and Barimasur-8 (75 ppm Fe and 60 ppm Zn). In India biofortified lentil varieties L4704 (125 ppm and 74 ppm) and Pusa Vaibhav (102 ppm Fe) have been released for commercial cultivation while IPL 220 (113 ppm Fe) has been recently identified. Idlib-2 (73 ppm Fe) and Idlib-3 (72 ppm Fe) from Syria while Alemaya (82 ppm Fe and 66 ppm Zn) have been released from Ethiopia.
Cowpea enriched with Fe (Pant Lobia-1, Pant Lobia-2, Pant Lobia-3, and Pant Lobia-4) has been developed from G.B. Pant University of Agriculture and Technology, Pantnagar (India).
Similarly, agronomic practices have supported some important achievements in legumes (Table 3). In chickpeas, Fe and Zn have been fortified by use of arbuscular mycorrhizal fungi (AMF; Pellegrino and Bedini, 2014). Biofortification of chickpeas with Zn and Se by foliar application has also been reported (Poblaciones et al., 2014; Shivay et al., 2015). Zinc biofortification by foliar application of Zn fertilizer has been described in common bean (Ibrahim and Ramadan, 2015; Ram et al., 2016). Soybean fortified with Se was obtained by foliar application of fertilizer complexed with selenium (Yang et al., 2003). Recently, foliar application of Na2SeO4 as a source of Se in cowpea for biofortification has been reported (Ramos et al., 2019).
Phytic acid (PA) is one of the most important anti-micronutrient component in grain which reduces the bioavailability of micronutrients. Ingestion of PA-rich foods leads to deficiencies of minerals such as Fe and Zn in the human population (Pramitha et al., 2021). In legume seeds, phytic acid is commonly present as myoinositol 1,2,3,4,S,6-hexakis (dihydrogen phosphate; Urbano et al., 2000). Phytate (IP6) is a compound that comprising of six phosphate groups connected to an inositol ring that can bind up to 12 protons. The Anti-nutritional effect is attributed to these six phosphate groups which are powerful chelators, binding rapidly to mineral cations mostly Cu2+, Ca2+, Zn2+, and Fe3+ (Castro-Alba et al., 2019). Phytic acid limits the utilization of phosphate as it is present mainly in the organic form of inositol hexakisphosphate (IP6). Hence, low IP6 content can improve crops and grains’ phosphate and mineral bioavailability. Majority of studies on impact of phytate on mineral nutrition suggest that insoluble phytate-metal complexes are formed in the digestive track, preventing metal absorption. The reduced availability of essential minerals in legumes and other protein foods depends on several factors, such as the ability of endogenous carriers in the intestinal mucosa to absorb the essential minerals bound to phytate and other dietary substances, phytic acid, mineral concentrations in foods, phytate hydrolysis by the phytase enzyme in the intestine, phytate inhibition, and food processing methods (Rackis and Anderson, 1977).
Variation in phytic acid content has been detected in legumes. The concentration of phytic acid in legumes grown in Canadian environment ranged from 11.33–14 mg/g in chickpea,15.64–18.82 mg/g in common beans,19.65–22.85 mg/g in faba bean, 8.56–17.1 mg/g in lentil and 9.93–12.40 mg/g in peas (Shi et al., 2018). Recent study demonstrated that cotyledon of common bean is rich in minerals Mg, Ca, Fe, and Zn but it also contain a significant amount of phytic acid that limits mineral absorption during digestion (Rousseau et al., 2020). Chickpea and pigeon pea have low phytic acid contents. Breeding programs should use low-phytic acid genotypes of pulses in order to increase their nutritional value and uptake (Chitra et al., 1995). Significant role of polyamines have been observed in breakdown of phytate. Polyamine spermidine (Spd), could hasten phytic acid breakdown in mung bean sprouts (Zhou et al., 2017). In faba bean, it was observed that phytic acid create very persistent compounds with proteins and making them inaccessible to gastrointestinal digestion and absorption (Rosa-Sibakov et al., 2018). Therefore phytase treatment may be advantageous in plant-based foods where high phytic acid levels may prevent proper protein digestion. Processing techniques such as soaking, fermentation, sprouting, germinating, and cooking procedures can drastically change the phytate content of grains in legumes, allowing improved mineral bioavailability (Petroski and Minich, 2020). For example, cooking legumes for 1 h at 95°C resulted in phytate reduction up to 23% in yellow split peas, 20%–80% in lentils, and 11% in chickpeas. In black beans, just a 0.29% reduction was observed (Shi et al., 2018). In legumes, breeding strategies have been attempted to reduce the PA content. For example, in common beans, low phytic acid (lpa) mutant lines were derived from a mutagenized population with up to 90% reduced PA levels (Campion et al., 2009). A low phytic acid line (lpa1) of common bean has been identified and used to map the Mrp1 gene, which is involved in the metabolism of phytic acid (Panzeri et al., 2011). Similarly, low-phytate pea (Pisum sativum L.) lines have been developed using chemical mutagenesis in cultivar CDC Bronco (Warkentin et al., 2012).
Raffinose series oligosaccharides (RFO’s) are another major anti-nutrients in legumes that shows significant Anti-nutritional effects in monogastric animals and human beings. In contrast to phytic acid which chelates minerals and decreases their bioavailability to humans and other animals, RFOs are responsible for flatulence (Zhawar et al., 2011). In chickpea, considerable amount of raffinose sugars were found (0.38–0.99 g/100 g; Roorkiwal et al., 2021). Efforts are being made to reduce phytic acid and RFO content without compromising on other quality parameters in chickpea.
Oxalates can form insoluble salts with sodium, potassium, calcium, iron, and magnesium. Both water soluble and insoluble oxalates are derived from plant. Water-soluble oxalate are potassium oxalate, sodium oxalate, and ammonium oxalates while insoluble oxalate is calcium oxalate (Noonan and Savage, 1999). According to Savage et al. (2000), oxalates that are soluble can chelate minerals and reduce absorption, or can lead to formation of calcium oxalate kidney stones. Due to negative effect in mineral absorption and health ailments oxalates are considered as Anti-nutritional factors. According to Shi et al. (2018), soybeans had the highest concentration of oxalates (370 mg/100 g dry weight (DW)), followed by lentils and peas (168–293 mg/100 g DW), chickpeas (192 mg/100 g DW), and common beans (98–117 mg/100 g DW). However, studies have shown that cooking can effectively reduce the amount of soluble oxalate that is present in the diet (Petroski and Minich, 2020).
Tannins, in the form of proanthocyanidins or catechins are considered one of the most prevalent secondary plant metabolites that are found in cocoa beans, tea, wines, fruits, juices, nuts, seeds, legumes, and cereal grains (Petroski and Minich, 2020). Tannins serve as antioxidants by scavenging free radicals, and has also ability to chelate dietary elements like Fe2+,Cu2+, and Zn2+ (Karamac, 2009). It has been demonstrated that kidney bean (Phaseolus vulgaris L.) contained a significant amount of catechins (20.1 mg/kg; Arts et al., 2000). Processing and cooking could reduce the total catechin content in food crops.
Saponins are a group of amphipathic compounds composed of lipophilic triterpene and hydrophilic sugar moieties. Pea seeds biosynthesize saponin B (also referred to as soyasaponin I) and DDMP saponin (soyasaponin βg; Reim and Rohn, 2015). The presence of saponin B and DDMP saponin in pea flour causes astringent, and bitter “off-flavor” (Heng et al., 2006). As the biosynthetic genes for these compounds have been reported in legumes (Sundaramoorthy et al., 2019), CRISPR/Cas9 can be applied to either remove or reduce the levels of saponin B and DDMP saponin in pea seeds.
Evaluation of genetic resources in legume crops has revealed significant variation for protein content as well as micronutrients. This review highlights the efforts being made toward screening of germplasm, mapping of QTLs for seed protein and micronutrient content and future thrust areas for enhancing the nutritional quality of major pulses. Micronutrient content in seed appears to be a complex trait under control of number of genes/QTLs and the concentration of these nutrients is highly influenced by multiple factors which include edaphic as well as environmental components. An integrated breeding approach involving QTL mapping, genome-wide association analysis, and differential gene expression profiling is currently the most efficient strategy for genetic dissection of complex traits like nutritional quality.
Ample scope exists for further improving the nutritional quality of pulse crops and development of biofortified varieties. The variability for Fe and Zn content and other micronutrients in food legumes can be exploited for enhancing the micronutrient level and their bioavailability. Minimizing the anti-nutritional factors viz., phytate, trypsin, and chymotrypsin inhibitors can further enhance the bioavailability and consumption of legumes. Search for genotypes with low anti-nutrient content in the germplasm or through induced mutagenesis or genome editing is an important objective in legume improvement program for achieving nutritional security. Crop wild relatives of various legumes could be used in pre-breeding program as they lack these Anti-nutrients (Singh and Jauhar, 2005).
The whole genome information coupled with phenotypic evaluation can identify rare variants for nutritional quality traits. The identification of haplotypes governing high protein and micronutrient content, low anti-nutritional factor in legume crops will be useful for translation genomics to breed biofortified cultivars with superior nutritional quality. Recent findings represent the kudos for future application of transgenic and gene editing approaches in legumes for enhancement of nutritional quality traits.
RJ and ST conceived the idea. RJ, UJ, HY, and RR prepared the manuscript. LS, PS, RS, AS, MT, UJ, SC, and ST edited and finalized the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer KT declared a shared affiliation with the authors ST, RJ, HY, RR, RS, UJ, and LS at the time of the review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbaspour, N., Hurrell, R., and Kelishadi, R. (2014). Review on iron and its importance for human health. J. Res. Med. Sci. 19, 164–174.
Abdel-Ghany, S. E., and Pilon, M. (2008). MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 283, 15932–15945. doi: 10.1074/jbc.M801406200
Alloway, B. J. (2009). Soil factors associated with zinc deficiency in crops and humans. Environ. Geochem. Health 31, 537–548. doi: 10.1007/s10653-009-9255-4
Aragao, F. J. L., Barros, L. M. G., De Sousa, M. V., Grossi de Sa, M. F., Almeida, E. R. P., Gander, E. S., et al. (1999). Expression of a methionine-rich storage albumin from the Brazil nut (Bertholletiaexcelsa H.B.K., Lecythidaceae) in transgenic bean plants (Phaseolus vulgaris L., Fabaceae). Genet. Mol. Biol. 22, 445–449. doi: 10.1590/S1415-47571999000300026
Aro, A., Alfthan, G., and Varo, P. (1995). Effects of supplementation of fertilizers on human selenium status in Finland. Analyst 120, 841–843. doi: 10.1039/an9952000841
Arts, I. C. W., van de Putte, B., and Hollman, P. C. H. (2000). Catechin contents of foods commonly consumed in The Netherlands. 1. Fruits, vegetables, staple foods, and processed foods. J. Agric. Food Chem. 48, 1746–1751. doi: 10.1021/jf000025h
Assunçao, A. G. L., Herrero, E., Lin, Y. F., Huettel, B., Talukdar, S., Smaczniak, C., et al. (2010). Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc. Natl. Acad. Sci. U. S. A. 107, 10296–10301. doi: 10.1073/pnas.1004788107
Astudillo, C., Fernandez, A. C., Blair, M. W., and Cichy, K. A. (2013). The Phaseolus vulgaris ZIP gene family: identification, characterization, mapping, and gene expression. Front. Plant Sci. 4:286. doi: 10.3389/fpls.2013.00286
Ates, D., Aldemir, S., Yagmur, B., Kahraman, A., Ozkan, H., Vandenberg, A., et al. (2018). QTL mapping of genome regions controlling manganese uptake in lentil Seed.G3: genes, genomes. Genetics 8, 1409–1416. doi: 10.1534/g3.118.200259
Ates, D., Sever, T., Aldemir, S., Yagmur, B., Temel, H. Y., Kaya, H. B., et al. (2016). Identification QTLs controlling genes for se uptake in lentil seeds. PLoS One 15:e0149210. doi: 10.1371/journal.pone.0149210
Aung, M. S., Masuda, H., Kobayashi, T., Nakanishi, H., Yamakawa, T., and Nishizawa, N. K. (2013). Iron biofortification of Myanmar rice. Front. Plant Sci. 4. doi: 10.3389/FPLS.2013.00158
Becker, R., Grun, M., and Scholz, G. (1992). Nicotianamine and the distribution of iron into the apoplasm and symplasm of tomato (Lycopersicon esculentum mill.). Planta 187, 48–52. doi: 10.1007/BF00201622
Beebe, S., Gonzalez, A. V., and Rengifo, J. (2000). Research on trace minerals in the common bean. Food Nutr. Bull. 21, 387–391. doi: 10.1177/156482650002100408
Blair, M. W., Izquierdo, P., Astudillo, C., and Grusak, M. A. (2013). A legume biofortification quandary: variability and genetic control of seed coat micronutrient accumulation in common beans. Front. Plant Sci. 4:275. doi: 10.3389/fpls.2013.00275
Blair, M. W., Medina, J. I., Astudillo, C., Rengifo, J., Beebe, S. E., Machado, G., et al. (2010). QTL for seed iron and zinc concentration and content in a Mesoamerican common bean (Phaseolus vulgaris L.) population. Theor. Appl. Genet. 121, 1059–1070. doi: 10.1007/s00122-010-1371-0
Blair, M. W., Wu, X., Bhandari, D., and Astudillo, C. (2016). Genetic dissection of ICP-detected nutrient accumulation in the whole seed of common bean (Phaseolus vulgaris L.). Front. Plant Sci. 7:219. doi: 10.3389/fpls.2016.00219
Blancquaert, D., De Steur, H., Gellynck, X., and Van Der Straeten, D. (2014). Present and future of folate biofortification of crop plants. J. Exp. Bot. 65, 895–906. doi: 10.1093/jxb/ert483
Blancquaert, D., Van Daele, J., Storozhenko, S., Stove, C., Lambert, W., and Van Der Straeten, D. (2013). Rice folate enhancement through metabolic engineering has an impact on rice seed metabolism, but does not affect the expression of the endogenous folate biosynthesis genes. Plant Mol. Biol. 83, 329–349. doi: 10.1007/s11103-013-0091-7
Bohra, A., Sahrawat, K. L., Kumar, S., Joshi, R., Parihar, A. K., Singh, U., et al. (2015). Genetics- and genomics-based interventions for nutritional enhancement of grain legume crops: status and outlook. J. Appl. Genet. 56, 151–161. doi: 10.1007/s13353-014-0268-z
Bortesi, L., and Fischer, R. (2014). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Brooks, C., Nekrasov, V., Lippman, Z. B., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Cakmak, I. (2008). Enrichment of cereal grains with zinc: agronomic or genetic biofortification. Plant and Soil 302, 1–17. doi: 10.1007/s11104-008-9584-6
Cakmak, I., Kalayci, M., Ekiz, H., Braun, H. J., Kilinç, Y., and Yilmaz, A. (1999). Zinc deficiency as a practical problem in plant and human nutrition in Turkey: a NATO- science for stability project. Field Crops Res. 60, 175–188. doi: 10.1016/S0378-4290(98)00139-7
Campion, B., Sparvoli, F., Doria, E., Tagliabue, G., Galasso, I., Fileppi, M., et al. (2009). Isolationand characterisation of an lpa (low phytic acid) mutant in common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 118, 1211–1221. doi: 10.1007/s00122-009-0975-8
Caproni, L., Raggi, L., Talsma, E. F., Wenzl, P., and Negri, V. (2020). European landrace diversity for common bean biofortification: a genome-wide association study. Sci. Rep. 10, 1–3. doi: 10.1038/s41598-020-76417-3
Carvalho, S. M. P., and Vasconcelos, M. W. (2013). Producing more with less: strategies and novel technologies for plant-based food biofortification. Food Res. Int. 54, 961–971. doi: 10.1016/j.foodres.2012.12.021
Cary, E. E., Norvell, W. A., Grunes, D. L., Welch, R. M., and Reid, W. S. (1994). Iron and manganese accumulation by the brz pea mutant grown in soils. Agron. J. 86, 938–941. doi: 10.2134/agronj1994.00021962008600060003x
Castro-Alba, V., Lazarte, C. E., Bergenståhl, B., and Granfeldt, Y. (2019). Phytate, iron, zinc, and calcium content of common Bolivian foods and their estimated mineral bioavailability. Food Sci. Nutr. 7, 2854–2865. doi: 10.1002/fsn3.1127
Chen, X. (2009). Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 25, 21–44. doi: 10.1146/ANNUREV.CELLBIO.042308.113417
Chitra, U., Vimala, V., Singh, U., and Geervani, P. (1995). Variability in phytic acid content and protein digestibility of grain legumes. Plant Foods Hum. Nutr. 47, 163–172. doi: 10.1007/bf01089266
Cichy, K. A., Caldas, G. V., Snapp, S. S., and Blair, M. W. (2009). QTL analysis of seed iron, zinc, and phosphorus levels in an Andean bean population. Crop. Sci. 49, 1742–1750. doi: 10.2135/cropsci2008.10.0605
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J. F., and Walker, E. L. (2001). Maize yellow stripe 1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349. doi: 10.1038/35053080
Curtin, S. J., Xiong, Y., Michno, J. M., Campbell, B. W., Stec, A. O., Cermak, T., et al. (2018). CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 16, 1125–1137. doi: 10.1111/pbi.12857
Dahiya, P. K., Linnemann, A. R., Nout, M. J. R., van Boekel, M. A. J. S., and Grewal, R. B. (2013). Nutrient composition of selected newly bred and established mung bean varieties. LWT 54, 249–256. doi: 10.1016/j.lwt.2013.05.017
Das Bhowmik, S. S., Cheng, A. Y., Long, H., Tan, G. Z. H., Hoang, T. M. L., Karbaschi, M. R., et al. (2019). Robust genetic transformation system to obtain non-chimeric transgenic chickpea. Front. Plant Sci. 10:524. doi: 10.3389/fpls.2019.00524
Dawson, I. K., Powell, W., Hendre, P., Bancic, J., Hickey, J. M., Kindt, R., et al. (2019). The role of genetics in mainstreaming the production of new and orphan crops to diversify food systems and support human nutrition. New Phytol. 224, 37–54. doi: 10.1111/nph.15895
Dhaliwal, S. S., Sharma, V., Shukla, A. K., Kaur, J., Verma, V., Singh, P., et al. (2021). Enrichment of zinc and iron micronutrients in lentil (Lens culinaris Medik.) through biofortification. Molecules 26:7671. doi: 10.3390/molecules26247671
Di Donato, R. J., Roberts, L. A., Sanderson, T., Eisley, R. B., and Walker, E. L. (2004). Arabidopsis yellow stripe-Like 2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J. 39, 403–414. doi: 10.1111/j.1365-313X.2004.02128.x
Di, Y.-H., Sun, X.-J., Hu, Z., Jiang, Q.-Y., Song, G.-H., Zhang, B., et al. (2019). Enhancing the CRISPR/Cas9 system based on multiple GmU6 promoters in soybean. Biochem. Biophys. Res. Commun. 519, 819–823. doi: 10.1016/j.bbrc.2019.09.074
Diapari, M., Sindhu, A., Bett, K., Deokar, A., Warkentin, T. D., and Tar’an, B. (2014). Genetic diversity and association mapping of iron and zinc concentrations in chickpea (Cicer arietinum L.). Genome 57, 459–468. doi: 10.1139/gen-2014-0108
Diaz, S., Ariza Suarez, D., Izquierdo, P., Lobaton, J. D., de la Hoz, J. F., Acevedo, F., et al. (2020). Genetic mapping for agronomic traits in a MAGIC population of common bean (Phaseolus vulgaris L.) under drought conditions. BMC Genomics 21, 1–20. doi: 10.1186/s12864-020-07213-6
Dinkins, R. D., Reddy, M. S. S., Meurer, C. A., Yan, B., Trick, H., Thibaud-Nissen, F., et al. (2001). Increased Sulphur amino acids in soybean plants overexpressing the maize 15 kDazein protein. In vitro cell Dev. Biol. Plant 37, 742–747. doi: 10.1007/s11627-001-0123-x
Dissanayaka, D. (2019). Genome wide association study to identify single nucleotide polymorphism markers for Fe, Zn, and se concentration in field pea seeds. master’s thesis. Saskatoon, SK, Canada: University of Saskatchewan.
Do, P. T., Nguyen, C. X., Bui, H. T., Tran, L. T., Stacey, G., Gillman, J. D., et al. (2019). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2–1A and GmFAD2–1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 19:311. doi: 10.1186/s12870-019-1906-8
Eckert, H., LaVallee, B., Schweiger, B. J., Kinney, A. J., Cahoon, E. B., and Clemente, T. (2006). Co-expression of the borage Δ6 desaturase and the Arabidopsis Δ15 desaturase results in high accumulation of stearidonic acid in the seeds of transgenic soybean. Planta 224, 1050–1057. doi: 10.1007/s00425-006-0291-3
Fan, X., Zhou, X., Chen, H., Tang, M., and Xie, X. (2021). Cross-talks Between macro- and micronutrient uptake and signaling in plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.663477
Garg, M., Sharma, N., Sharma, S., Kapoor, P., Kumar, A., Chunduri, V., et al. (2018). Biofortified crops generated by breeding, agronomy, and transgenic approaches are improving lives of millions of people around the world. Front. Nutr. 5. doi: 10.3389/fnut.2018.00012
Gerrano, A. S., Mbuma, N. W., and Mumm, R. H. (2022). Expression of nutritional traits in vegetable cowpea grown under various south African agro-ecological conditions. Plan. Theory 11:1422. doi: 10.3390/plants11111422
Gerrano, A. S., Thungo, Z. G., Shimelis, H., Mashilo, J., and Mathew, I. (2022). Genotype-by-environment interaction for the contents of micro-nutrients and protein in the Green pods of cowpea (Vigna unguiculata L. Walp). Agriculture 12:531. doi: 10.3390/agriculture12040531
Green, L. S., and Rogers, E. E. (2004). FRD3 controls iron localization in Arabidopsis. Plant Physiol. 136, 2523–2531. doi: 10.1104/pp.104.045633
Grusak, M. A., Pezeshgi, S., O’Brien, K. O., and Abrams, S. A. (1996). Intrinsic 42Ca-Labelling of Green bean pods for use in human bioavailability studies. J. Sci. Food Agric. 70, 11–15. doi: 10.1002/(SICI)1097-0010(199601)70:1<11::AID-JSFA453>3.0.CO;2-8
Guindon, M. F., Cazzola, F., Palacios, T., Gatti, I., Bermejo, C., and Cointry, E. (2021). Biofortification of pea (Pisum sativum L): a review. J. Sci. Food Agric. 101, 3551–3563. doi: 10.1002/jsfa.11059
Gupta, N., Ram, H., and Kumar, B. (2016). Mechanism of zinc absorption in plants: uptake, transport, translocation and accumulation. Rev. Environ. Sci. Biotechnol. 15, 89–109. doi: 10.1007/s11157-016-9390-1
Hanafy, M. S., Rahman, S. M., Nakamoto, Y., Fujiwara, T., Naito, S., Wakasa, K., et al. (2013). Differential response of methionine metabolism in two grain legumes, soybean and azuki bean, expressing a mutated form of Arabidopsis cysta-thionine γ-synthase. J. Plant Physiol. 170, 338–345. doi: 10.1016/j.jplph.2012.10.018
Harding, K. L., Aguayo, V. M., and Webb, P. (2018). Hidden hunger in South Asia: a review of recent trends and persistent challenges. Public Health Nutr. 21, 785–795. doi: 10.1017/S1368980017003202
Haydon, M. J., and Cobbett, C. S. (2007). Transporters of ligands for essential metal ions in plants. New Phytol. 174, 499–506. doi: 10.1111/j.1469-8137.2007.02051.x
Heng, L., Vincken, J. P., van Koningsveld, G., Legger, A., Gruppen, H., van Boekel, T., et al. (2006). Bitterness of saponins and their content in dry peas. J. Sci. Food Agric. 86, 1225–1231. doi: 10.1002/jsfa.2473
Ibrahim, E. A., and Ramadan, W. A. (2015). Effect of zinc foliar spray alone and combined with humic acid or/and chitosan on growth, nutrient elements content and yield of dry bean (Phaseolus vulgaris L.) plants sown at different dates. Sci. Hortic. 184, 101–105. doi: 10.1016/j.scienta.2014.11.010
Ibrahim, S., Saleem, B., Rehman, N., Zafar, S. A., Naeem, M. K., and Khan, M. R. (2022). CRISPR/Cas9 mediated disruption of inositol Pentakisphosphate 2-kinase 1 (TaIPK1) reduces phytic acid and improves iron and zinc accumulation in wheat grains. J. Adv. Res. 37, 33–41. doi: 10.1016/j.jare.2021.07.006
IYOP. (2016). FAO. Available at: https://news.un.org/en/story/2015/11/515012-un-launches-2016-international-year-pulses-celebrating-benefits-legumes
Izquierdo, P., Astudillo, C., Blair, M. W., Iqbal, A. M., Raatz, B., and Cichy, K. A. (2018). Meta-QTL analysis of seed iron and zinc concentration and content in common bean (Phaseolus vulgaris L.). Theo. Appl. Genet. 131, 1645–1658. doi: 10.1007/s00122-018-3104-8
Jagadeeswaran, G., Zheng, Y., Li, Y. F., Shukla, L. I., Matts, J., Hoyt, P., et al. (2009). Cloning and characterization of small RNAs from Medicago truncatula reveals four novel legume-specific microRNA families. New Phytol. 184, 85–98. doi: 10.1111/J.1469-8137.2009.02915.X
Jagadhesan, B., Das, S., Singh, D., Jha, S. K., Durgesh, K., and Sathee, L. (2022). Micro RNA mediated regulation of nutrient response in plants: the case of nitrogen. Plant Physiol. Rep. 1–13. doi: 10.1007/s40502-022-00653-0
Jaganathan, D., Ramasamy, K., Sellamuthu, G., Jayabalan, S., and Venkataraman, G. (2018). CRISPR for crop improvement: an update review. Front. Plant Sci. 9:985. doi: 10.3389/fpls.2018.00985
Jha, A. B., Ashokkuma, K., Diapari, M., Ambrose, S. J., Zhang, H., Taran, B., et al. (2015). Genetic diversity of folate profiles in seeds of common bean, lentil, chickpea and pea. J. Food Comp. Anal. 42, 134–140. doi: 10.1016/j.jfca.2015.03.006
Jha, A. B., and Warkentin, T. D. (2020). Biofortification of pulse crops: status and future perspectives. Plan. Theory 9:73. doi: 10.3390/plants9010073
Ji, J., Zhang, C., Sun, Z., Wang, L., Duanmu, D., and Fan, Q. (2019). Genome editing in cowpea Vignaunguiculata using CRISPR-Cas9. Int. J. Mol. Sci. 20:2471. doi: 10.3390/ijms20102471
Jiang, X. M., Cao, X. Y., Jiang, J. Y., Ma, T., James, D. W., Rakeman, M. A., et al. (1997). Dynamics of environmental supplementation of iodine: four years’ experience in iodin- ation of irrigation water in Hotien, Xinjiang, China. Arch. Environ. Health 52, 399–408. doi: 10.1080/00039899709602218
Jiang, M., Liu, Y., Liu, Y., Tan, Y., Huang, J., and Shu, Q. (2019). Mutation of inositol 1,3,4-trisphosphate 5/6-kinase6 impairs plant growth and Phytic acid synthesis in Rice. Plan. Theory 8:114. doi: 10.3390/plants8050114
Karaca, N., Ates, D., Nemli, S., Ozkuru, E., Yilmaz, H., Yagmur, B., et al. (2020). Identification of SNP markers associated with iron and zinc concentrations in CicerSeeds. Curr. Genomics 21, 212–223. doi: 10.1016/j.ygeno.2019.09.012
Karamac, M. (2009). Chelation of cu(II), Zn(II), and Fe(II) by tannin constituents of selected edible nuts. Int. J. Mol. Sci. 10, 5485–5497. doi: 10.3390/ijms10125485
Kehr, J. (2013). Systemic regulation of mineral homeostasis by micro RNAs. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00145
Khandal, H., Gupta, S. K., Dwivedi, V., Mandal, D., Sharma, N. K., Vishwakarma, N. K., et al. (2020). Root specific expression of chickpea cytokinin oxidase/dehydrogenase 6 leads to enhanced root growth, drought tolerance and yield without compromising nodulation. Plant Biotechnol. J. 18, 2225–2240. doi: 10.1111/pbi.13378
Khazaei, H., Podder, R., Caron, C. T., Kundu, S. S., Diapari, M., Vandenberg, A., et al. (2017). Marker–trait association analysis of iron and zinc concentration in lentil (Medik.) seeds. Plant Genome 10. doi: 10.3835/plantgenome2017.02.0007
Khoury, C. K., Bjorkman, A. D., Dempewolf, H., Ramirez-Villegas, J., Guarino, L., Jarvis, A., et al. (2014). Increasing homogeneity in global food supplies and the implications for food security. Proc. Natl. Acad. Sci. U. S. A. 111, 4001–4006. doi: 10.1073/pnas.1313490111
Kim, M. J., Kim, J. K., Kim, H. J., Pak, J. H., Lee, J. H., Kim, D. H., et al. (2012). Genetic modifica-tion of the soybean to enhance the β-carotene content through seed-specific expression. PLoS One 7:e48287. doi: 10.1371/journal.pone.004828
King, K. E., Peiffer, G. A., Reddy, M., Lauter, N., Lin, S. F., Cianzio, S., et al. (2013). Mapping of iron and zinc quantitative trait loci in soybean for association to iron deficiency chlorosis resistance. J. Plant Nutr. 36, 2132–2153. doi: 10.1080/01904167.2013.766804
Klein, A., Houtin, H., Rond-Coissieux, C., Naudet-Huart, M., Touratier, M., Marget, P., et al. (2020). Meta-analysis of QTL reveals the genetic control of yield-related traits and seed protein content in pea. Sci. Rep. 10:15925. doi: 10.1038/s41598-020-72548-9
Kong, W. W., and Yang, Z. M. (2010). Identification of iron-deficiency responsive microRNA genes and cis-elements in Arabidopsis. Plant Physiol. Biochem. 48, 153–159. doi: 10.1016/j.plaphy.2009.12.008
Kumar, R. K., Chu, H. H., Abundis, C., Vasques, K., Rodriguez, D. C., Chia, J. C., et al. (2017). Iron-nicotianamine transporters are required for proper long distance iron signaling. Plant Physiol. 175, 1254–1268. doi: 10.1104/pp.17.00821
Kumar, J., Gupta, D. S., Kumar, S., Gupta, S., and Singh, N. P. (2016). Current knowledge on genetic biofortification in lentil. J. Agric. Food Chem. 64, 6383–6396. doi: 10.1021/acs.jafc.6b02171
Kumar, S., and Pandey, G. (2020). Biofortification of pulses and legumes to enhance nutrition. Heliyon 6:e03682. doi: 10.1016/j.heliyon.2020.e03682
Kumar, H., Singh, A., Dikshit, H. K., Mishra, G. P., Aski, M., Meena, M. C., et al. (2019). Genetic dissection of grain iron and zinc concentrations in lentil (lens culinarisMedik.). J. Genet. 98, 1–4. doi: 10.1007/s12041-019-1112-3
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Li, Y., Zhang, Y., Shi, D., Liu, X., Qin, J., Ge, Q., et al. (2013). Spatial-temporal analysis of zinc homeostasis reveals the response mechanisms to acute zinc deficiency in Sorghum bicolor. New Phytol. 200, 1102–1115. doi: 10.1111/nph.12434
Lopez-Millan, A. F., Ellis, D. R., and Grusak, M. A. (2004). Identification and characterization of several new members of the zip family of metal ion transporters in Medicago truncatula. Plant Mol. Biol. 54, 583–596. doi: 10.1023/B:PLAN.0000038271.96019.aa
Ma, Y., Coyne, C. J., Grusak, M. A., Mazourek, M., Cheng, P., Main, D., et al. (2017). Genome-wide SNP identification, linkage map construction and QTL mapping for seed mineral concentrations and contents in pea (Pisumsativum L.). BMC Plant Biol. 17:43. doi: 10.1186/s12870-016-0956-4
Marín-González, E., and Suárez-López, P. (2012). And yet it moves: cell-to-cell and long-distance signaling by plant microRNAs. Plant Sci. 196, 18–30. doi: 10.1016/j.plantsci.2012.07.009
Miller, J. L. (2013). Iron deficiency anemia: a common and curable disease. Cold Spring Harb. Perspect. Med. 3:a011866. doi: 10.1101/cshperspect.a011866
Molvig, L., Tabe, L. M., Eggum, B. O., Moore, A. E., Craig, S., Spencer, D., et al. (1997). Enhanced methionine levels and increased nutritive value of seeds of trans-genic lupins (Lupinusangustifolius L.) expressing a sunflower seed albumin gene. Proc. Natl. Acad. Sci. U. S. A. 94, 8393–8398. doi: 10.1073/pnas.94.16.8393
Moreau, S., Thomson, R. M., Kaiser, B. N., Trevaskis, B., Guerinot, M. L., Udvardi, M. K., et al. (2002). Gmzip 1 encodes a symbiosis-specific zinc transporter in soybean. J. Biol. Chem. 277, 4738–4746. doi: 10.1074/jbc.M106754200
Murube, E., Beleggia, R., Pacetti, D., Nartea, A., Frascarelli, G., Lanzavecchia, G., et al. (2021). Characterization of nutritional quality traits of a common bean Germplasm collection. Foods 10:1572. doi: 10.3390/foods10071572
Nakatani, S., Mori, K., Shoji, T., and Emoto, M. (2021). Association of zinc deficiency with development of CVD events in patients with CKD. Nutrients 13:1680. doi: 10.3390/nu13051680
Naqvi, S., Zhu, C., Farre, G., Ramessar, K., Bassie, L., Breitenbach, J., et al. (2009). Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc. Natl. Acad. Sci. U. S. A. 106, 7762–7767. doi: 10.1073/pnas.0901412106
Naya, L., Paul, S., Valdés-López, O., Mendoza-Soto, A. B., Nova-Franco, B., Sosa-Valencia, G., et al. (2014). Regulation of copper homeostasis and biotic interactions by MicroRNA 398b in common bean. PLoS One 9:e84416. doi: 10.1371/JOURNAL.PONE.0084416
Noonan, S. C., and Savage, G. P. (1999). Oxalate content of foods and its effect on humans. Asia Pac. J. Clin. Nutr. 8, 64–74. doi: 10.1046/j.1440-6047.1999.00038.x
Ochatt, S., Conreux, C., Moussa Mcolo, R., Despierre, G., Magnin-Robert, J.-B., and Raffiot, B. (2018). Phytosulfokine-alpha, an enhancer of in vitro regeneration competence in recalcitrant legumes. Plant Cell Tiss. Org. Cult. 135, 189–201. doi: 10.1007/s11240-018-1455-0
Palmer, C. M., and Guerinot, M. L. (2009). Facing the challenges of CuFe and Zn homeostasis in plants. Nat. Chem. Biol. 5, 333–340. doi: 10.1038/nchembio.166
Palmgren, M. G., Clemens, S., Williams, L. E., Krämer, U., Borg, S., Schjørring, J. K., et al. (2008). Zinc biofortification of cereals: problems and solutions. Trends Plant Sci. 13, 464–473. doi: 10.1016/j.tplants.2008.06.005
Pant, B. D., Buhtz, A., Kehr, J., and Scheible, W. R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738. doi: 10.1111/j.1365-313X.2007.03363.x
Panzeri, D., Cassani, E., Doria, E., Tagliabue, G., Forti, L., Campion, B., et al. (2011). A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo−inositol and alters ABA sensitivity. New Phytol. 191, 70–83. doi: 10.1111/j.1469-8137.2011.03666.x
Park, C., Dwiyanti, M. S., Nagano, A. J., Liu, B., Yamada, T., and Abe, J. (2019). Identification of quantitative trait loci for increased α-tocopherol biosynthesis in wild soybean using a high-density genetic map. BMC Plant Biol. 19, 1–5. doi: 10.1186/s12870-019-2117-z
Parveen, S., Gupta, D. B., Dass, S., Kumar, A., Pandey, A., Chakraborty, S., et al. (2016). Chickpea ferritin cafer1 participates in oxidative stress response, and promotes growth and development. Sci. Rep. 6:31218. doi: 10.1038/srep31218
Patel, P., Yadav, K., and Ganapathi, T. R. (2017). Small and hungry: MicroRNAs in micronutrient homeostasis of plants. Microrna 6, 22–41. doi: 10.2174/2211536606666170117160338
Paul, S., Datta, S. K., and Datta, K. (2015). miRNA regulation of nutrient homeostasis in plants. Front. Plant Sci. 6:232. doi: 10.3389/fpls.2015.00232
Pellegrino, E., and Bedini, S. (2014). Enhancing ecosystem services in sustainable agriculture: biofertilization and biofortification of chickpea (Cicer arietinum L.) by arbuscular mycorrhizal fungi. Soil Biol. Biochem. 68, 429–439. doi: 10.1016/j.soilbio.2013.09.030
Petroski, W., and Minich, D. M. (2020). Is there such a thing as “anti-nutrients”? A narrative review of perceived problematic plant compounds. Nutrients 12:2929. doi: 10.3390/nu12102929
Pich, A., and Scholz, G. (1996). Translocation of copper and other micronutrients in tomato plants (Lycopersicon esculentum mill.): nicotianaminestimulated copper transport in the xylem. J. Exp. Bot. 47, 41–47. doi: 10.1093/jxb/47.1.41
Pich, A., Scholz, G., and Stephan, U. W. (1994). Iron-dependent changes of heavy metals,nicotianamine, and citrate in different plant organs and in the xylem exudate of two tomato genotypes. Nicotianamine as possible copper translocator. Plant and Soil 165, 189–196. doi: 10.1007/BF00008061
Poblaciones, M. J., and Rengel, Z. (2016). Soil and foliar zinc biofortification in field pea (Pisum sativum L.). grain accumulation and bioavailability in raw and cooked grains. Food Chem. 212, 427–433. doi: 10.1016/j.foodchem.2016.05.189
Poblaciones, M. J., Rodrigo, S., Santamaria, O., Chen, Y., and McGrath, S. P. (2014). Selenium accumulation and speciation in biofortified chickpea (Cicer arietinum L.) under Mediterranean conditions. J. Sci. Food Agric. 94, 1101–1106. doi: 10.1002/jsfa.6372
Pramitha, J. L., Rana, S., Aggarwal, P. R., Ravikesavan, R., Joel, A. J., and Muthamilarasan, M. (2021). in Chapter Three-Diverse role of phytic acid in Plants and Approaches to Develop low-Phytate Grains to Enhance Bioavailability of Micronutrients. ed. G. Kumar, vol. 107 (Elsevier, United States: Academic Press), 89–120.
Qin, L., Han, P., Chen, L., Walk, T. C., Li, Y., Hu, X., et al. (2017). Genome-wide identification and expression analysis of NRAMP family genes in soybean (glycine max L.). Front. Plant Sci. 8:1436. doi: 10.3389/fpls.2017.01436
Ram, H., Rashid, A., Zhang, W., Duarte, A. P., Phattarakul, N., Simunji, S., et al. (2016). Biofortification of wheat, rice and common bean by applying foliar zinc fertilizer along with pesticides in seven countries. Plant and Soil 403, 389–401. doi: 10.1007/s11104-016-2815-3
Ramos, D. P., Tavares, T. C. O., Sousa, S. A., Nascimento, V. L., Martinez, R. A. S., Junior, A. F. C., et al. (2019). Agronomic biofortification of cowpea with selenium by foliar fertilization: effect of doses in three cultivars. J. Plant Nutr. 43, 538–547. doi: 10.1080/01904167.2019.1685096
Ray, H., Bett, K., Tar’an, B., Vandenberg, A., Thavarajah, D., and Warkentin, T. (2014). Mineral micronutrient content of cultivars of field pea, chickpea, common bean, and lentil grown in Saskatchewan, Canada. Crop. Sci. 54, 1698–1708. doi: 10.2135/cropsci2013.08.0568
Reim, V., and Rohn, S. (2015). Characterization of saponins in peas (Pisum sativum L.) by HPTLC coupled to mass spectrometry and a hemolysis assay. Food Res. Int. 76, 3–10. doi: 10.1016/j.foodres.2014.06.043
Rezaei, M. K., Deokar, A., and Tar’an, B., (2016). Identification and expression analysis of candidate genes involved in carotenoid biosynthesis in chickpea seeds. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01867
Roorkiwal, M., Pandey, S., Thavarajah, D., Hemalatha, R., and Varshney, R. K. (2021). Molecular mechanisms and biochemical pathways for micronutrient acquisition and storage in legumes to support biofortification for nutritional security. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.682842
Rosa-Sibakov, N., Re, M., Karsma, A., Laitila, A., and Nordlund, E. (2018). Phytic acid reduction by bioprocessing as a tool to improve the In vitro digestibility of Faba bean protein. J. Agric. Food Chem. 66, 10394–10399. doi: 10.1021/acs.jafc.8b02948
Rousseau, S., Pallares Pallares, A., Vancoillie, F., Hendrickx, M., and Grauwet, T. (2020). Pectin and phytic acid reduce mineral bioaccessibility in cooked common bean cotyledons regardless of cell wall integrity. Food Res. Int. 137:109685. doi: 10.1016/j.foodres.2020.109685
Sab, S., Lokesha, R., Mannur, D. M., Mallikarjuna, B. P., and Thudi, M. (2020). Genome-wide SNP discovery and mapping QTLs for seed iron and zinc concentrations in chickpea (Cicer arietinum L.). Front. Nutr. 7. doi: 10.3389/fnut.2020.559120
Saini, A., Li, Y., Jagadeeswaran, G., and Sunkar, R. (2012). Role of microRNAs in Plant Adaptation to Environmental Stresses. (Springer, Berlin, Heidelberg), 219–232.
Sathya, A., Vijayabharathi, R., Srinivas, V., and Gopalakrishnan, S. (2016). Plant growth-promoting actinobacteria on chickpea seed mineral density: an upcoming complementary tool for sustainable biofortification strategy. 3 Biotech 6:138. doi: 10.1007/s13205-016-0458-y
Savage, G., Vanhanen, L. P., Mason, S., and Ross, A. B. (2000). Effect of cooking on the soluble and insoluble oxalate content of Some New Zealand foods. J. Food Compos. Anal. 13, 201–206. doi: 10.1006/jfca.2000.0879
Schmidt, M. A., Parrott, W. A., Hildebrand, D. F., Berg, R. H., Cooksey, A., Pendarvis, K., et al. (2015). Transgenic soya bean seeds accumulating β-carotene exhibit the col-lateral enhancements of oleate and protein content traits. Plant Biotechnol. J. 13, 590–600. doi: 10.1111/pbi.12286
Sen Gupta, D., McPhee, K., and Kumar, S. (2017). Development of molecular markers for iron metabolism related genes in lentil and their expression analysis under excess iron stress. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00579
Sen Gupta, D., Thavarajah, D., Knutson, P., Thavarajah, P., McGee, R. J., Coyne, C. J., et al. (2013). Lentils (Lens culinaris L.), a rich source of Folates. J. Agric. Food Chem. 61, 7794–7799. doi: 10.1021/jf401891p
Shi, L., Arntfield, S. D., and Nickerson, M. (2018). Changes in levels of phytic acid, lectins and oxalates during soaking and cooking of Canadian pulses. Food Res. Int. 107, 660–668. doi: 10.1016/j.foodres.2018.02.056
Shivay, Y. S., Prasad, R., and Pal, M. (2015). Effects of source and method of zinc application on yield, zinc biofortification of grain, and Zn uptake and use efficiency in chickpea (Cicer arietinum L.). Commun. Soil Sci. Plant Anal. 46, 2191–2200. doi: 10.1080/00103624.2015.1069320
Silva, V. M., Boleta, E. H. M., Martins, J. T., dos Santos, F. L. M., Silva, A. C. d. R., Alcock, T. D., et al. (2019). Agronomic biofortification of cowpea with selenium: effects of selenate and selenite applications on selenium and phytate concentrations in seeds. J. Sci. Food Agric. 99, 5969–5983. doi: 10.1002/jsfa.9872
Singh, R., and Jauhar, P. (2005). Genetic Resources, Chromosome Engineering, and crop Improvement: Grain Legumes. CRC Press, Boca Raton.
Singh, A., Sharma, V., Dikshit, H. K., Aski, M., Kumar, H., Thirunavukkarasu, N., et al. (2017). Association mapping unveils favorable alleles for grain iron and zinc concentrations in lentil (Lens culinaris subsp. culinaris). PLoS One 21:e0188296. doi: 10.1371/journal.pone.0188296
Sodedji, F. A. K., Ryu, D., Choi, J., Agbahoungba, S., Assogbadjo, A. E., N’Guetta, S.-P. A., et al. (2022). Genetic diversity and association analysis for carotenoid content among sprouts of cowpea (Vigna unguiculata L. Walp). Int. J. Mol. Sci. 23:3696. doi: 10.3390/ijms23073696
Song, S., Hou, W., Godo, I., Wu, C., Yu, Y., Matityahu, I., et al. (2013). Soybean seeds expressing feedback-insensitive cystathionine -synthase exhibit a higher content of methionine. J. Exp. Bot. 64, 1917–1926. doi: 10.1093/jxb/ert053
Stein, A. J. (2010). Global impacts of human mineral nutrition. Plant and Soil 335, 133–154. doi: 10.1007/s11104-009-0228-2
Stephan, U. W., and Grun, M. (1989). Physiological disorders of the nicotianamineauxotroph tomato mutant chloronerva at different levels of iron nutrition. II. Iron deficiency response and heavy metal metabolism. Biochem. Physiol. Pflanz. 185, 189–200. doi: 10.1016/S0015-3796(89)80080-0
Sundaramoorthy, J., Park, G. T., Komagamine, K., Tsukamoto, C., Chang, J. H., Lee, J. D., et al. (2019). Bio- synthesis of DDMP saponins in soybean is regulated by a distinct UDP-glycosyltransferase. New Phytol. 222, 261–274. doi: 10.1111/nph.15588
Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309. doi: 10.1016/j.tplants.2007.05.001
Sunkar, R., Kapoor, A., and Zhu, J. K. (2006). Posttranscriptional induction of two cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065. doi: 10.1105/tpc.106.041673
Sunkar, R., and Zhu, J. K. (2004). Novel and stress regulated microRNAs and other small RNAs from Arabidopsis w inside box sign. Plant Cell 16, 2001–2019. doi: 10.1105/TPC.104.022830
Tan, G. Z. H., Das Bhowmik, S. S., Hoang, T. M. L., Karbaschi, M. R., Long, H., Cheng, A., et al. (2018). Investigation of baseline iron levels in Australian chickpea and evaluation of a transgenic biofortification approach. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00788
Tejada-Jiménez, M., Castro-Rodríguez, R., Kryvoruchko, I., Lucas, M. M., Udvardi, M., Imperial, J., et al. (2015). Medicago truncatula natural resistance-associated macrophage protein 1 is required for iron uptake by rhizobia-infected nodule cells. Plant Physiol. 168, 258–272. doi: 10.1104/pp.114.254672
Thomine, S., and Vert, G. (2013). Iron transport in plants: better be safe than sorry. Curr. Opin. Plant Biol. 16, 322–327. doi: 10.1016/j.pbi.2013.01.003
Trijatmiko, K. R., Dueñas, C., Tsakirpaloglou, N., Torrizo, L., Arines, F. M., Adeva, C., et al. (2016). Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci. Rep. 6:19792. doi: 10.1038/srep19792
Upadhyaya, H. D., Bajaj, D., Das, S., Kumar, V., Gowda, C. L., Sharma, S., et al. (2016). Genetic dissection of seed-iron and zinc concentrations in chickpea. Sci. Rep. 6:24050. doi: 10.1038/srep24050
Urbano, G., Lopez-jurado, M., Aranda, P., Vidal-Valverde, C., Tenorio, E., and Porres, J. (2000). The role of phytic acid in legumes: antinutrient or beneficial function? J. Physiol. Biochem. 56, 283–294. doi: 10.1007/BF03179796
Urwat, U., Ahmad, S. M., Masi, A., Ganai, N. A., Murtaza, I., Khan, I., et al. (2021). Fe and Zn stress induced gene expression analysis unraveled mechanisms of mineral homeostasis in common bean (Phaseolus vulgaris L.). Sci. Rep. 11:24026. doi: 10.1038/s41598-021-03506-2
Valdes-Lopez, O., Yang, S. S., Aparicio-Fabre, R., Graham, P. H., Reyes, J. L., Vance, C. P., et al. (2010). MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytol. 187, 805–818. doi: 10.1111/j.1469-8137.2010.03320.x
Vandemark, G. J., Grusak, M. A., and McGee, R. J. (2018). Mineral concentrations of chickpea and lentil cultivars and breeding lines grown in the US Pacific northwest. Crop J. 6, 253–262. doi: 10.1016/j.cj.2017.12.003
Viteri, F. E. (1998). A new concept in the control of iron deficiency: community-based preventive supplementation of at-risk groups by the weekly intake of iron supplements. Biomed. Environ. Sci. 11, 46–60.
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, J., Kuang, H., Zhang, Z., Yang, Y., Yan, L., Zhang, M., et al. (2020). Generation of seed lipoxygenase-free soybean using CRISPR-Cas9. Crop J. 8, 432–439. doi: 10.1016/j.cj.2019.08.008
Warkentin, T. D., Delgerjav, O., Arganosa, G., Rehman, A. U., Bett, K. E., Anbessa, Y., et al. (2012). Development and characterization of low-Phytate pea. Crop. Sci. 52, 74–78. doi: 10.2135/cropsci2011.05.0285
Waters, B. M., Chu, H. H., Didonato, R. J., Roberts, L. A., Eisley, R. B., Lahner, B., et al. (2006). Mutations in Arabidopsis yellow stripe-like 1 and yellow stripe-like 3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141, 1446–1458. doi: 10.1104/pp.106.082586
Waters, B. M., and Grusak, M. A. (2008). Whole-plant mineral partitioning throughout the life cycle in Arabidopsis thaliana ecotypes Columbia Landsberg erecta, Cape Verde Islands, and the mutant line ysl 1ysl3. New Phytol. 177, 389–405. doi: 10.1111/j.1469-8137.2007.02288.x
Waters, B. M., McInturf, S. A., and Stein, R. J. (2012). Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 63, 5903–5918. doi: 10.1093/JXB/ERS239
Westermann, D. T., Teran, H., Munoz-Perea, C. G., and Singh, S. P. (2011). Plant and seed nutri-ent uptake in common bean in seven organic and conventional production systems. Can. J. Plant Sci. 91, 1089–1099. doi: 10.4141/cjps10114
White, P. J., and Broadley, M. R. (2009). Biofortification of crops with seven mineral elements often lacking in human diets-iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 182, 49–84. doi: 10.1111/j.1469-8137.2008.02738.x
Wu, X., Islam, A. S., Limpot, N., Mackasmiel, L., Mierzwa, J., Cortés, A. J., et al. (2020). Genome-wide Snp identification and association mapping for seed mineral concentration in mung bean (Vigna radiata L.). Front. Genet. 11, 11–656. doi: 10.3389/fgene.2020.00656
Xiong, H., Kobayashi, T., Kakei, Y., Senoura, T., Nakazono, M., Takahashi, H., et al. (2012). AhNRAMP1 iron transporter is involved in iron acquisition in peanut. J. Exp. Bot. 63, 4437–4446. doi: 10.1093/jxb/ers117
Xue, Y., Xia, H., Christie, P., Zhang, Z., Li, L., and Tang, C. (2015). Crop acquisition of phosphorus, iron and zinc from soil in cereal/legume intercropping systems: a critical review. Ann. Bot. 117, 363–377. doi: 10.1093/aob/mcv182
Yadav, R., Mehrotra, M., Singh, A. K., Niranjan, A., Singh, R., Sanyal, I., et al. (2017). Improvement in agrobacterium-mediated transformation of chickpea (Cicer arietinum L.) by the inhibi-tion of polyphenolics released during wounding of cotyledonary node explants. Protoplasma 254, 253–269. doi: 10.1007/s00709-015-0940-0
Yamasaki, H., Hayashi, M., Fukazawa, M., Kobayashi, Y., and Shikanai, T. (2009). SQUAMOSA promoter binding protein--like 7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 21, 374–361. doi: 10.1007/S11104-013-1907-6/FIGURES/2
Yang, F., Chen, L., Hu, Q., and Pan, G. (2003). Effect of the application of selenium on selenium content of soybean and its products. Biol. Trace Elem. Res. 93, 249–256. doi: 10.1385/BTER:93:1-3:249
Yu, O., Shi, J., Hession, A. O., Maxwell, C. A., McGonigle, B., and Odell, J. T. (2003). Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63, 753–763. doi: 10.1016/S0031-9422(03)00345-5
Zeng, H., Wang, G., Hu, X., Wang, H., Du, L., and Zhu, Y. (2014). Role of microRNAs in plant responses to nutrient stress. Plant and Soil 374, 1005–1021. doi: 10.1007/s11104-013-1907-6
Zhang, P., Du, H., Wang, J., Pu, Y., Yang, C., Yan, R., et al. (2020). Multiplex CRISPR/Cas9-mediated metabolic engineer-ing increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol. J. 18, 1384–1395. doi: 10.1111/pbi.13302
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. doi: 10.1111/pbi.12200
Zhawar, V. K., Kaur, N., and Gupta, A. K. (2011). Phytic acid and raffinose series oligosaccharides metabolism in developing chickpea seeds. Physiol. Mol. Biol. Plants 17, 355–362. doi: 10.1007/s12298-011-0080-8
Zhou, T., Wang, P., Yang, R., and Gu, Z. (2017). Polyamines regulating phytic acid degradation in mung bean sprouts. J. Sci. Food Agric. 98, 3299–3308. doi: 10.1002/jsfa.8833
Zhou, Z. S., Zeng, H. Q., Liu, Z. P., and Yang, Z. M. (2012). Genome-wide identification of Medicago truncatula microRNAs and their targets reveals their differential regulation by heavy metal. Plant Cell Environ. 35, 86–99. doi: 10.1111/j.1365-3040.2011.02418.x
Keywords: legumes, micronutrients, hidden hunger, anti-nutritional factors, biofortification
Citation: Jha R, Yadav HK, Raiya R, Singh RK, Jha UC, Sathee L, Singh P, Thudi M, Singh A, Chaturvedi SK and Tripathi S (2022) Integrated breeding approaches to enhance the nutritional quality of food legumes. Front. Plant Sci. 13:984700. doi: 10.3389/fpls.2022.984700
Received: 02 July 2022; Accepted: 26 July 2022;
Published: 07 September 2022.
Edited by:
Manoj Prasad, National Institute of Plant Genome Research (NIPGR), IndiaReviewed by:
Kuldeep Tripathi, National Bureau of Plant Genetic Resources (ICAR), IndiaCopyright © 2022 Jha, Yadav, Raiya, Singh, Jha, Sathee, Singh, Thudi, Singh, Chaturvedi and Tripathi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shailesh Tripathi, c2hhaXRyaUByZWRpZmZtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.