- 1The Key Laboratory of Biology and Genetic Improvement of Oil Crops, The Ministry of Agriculture and Rural Affairs of the PRC, Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences, Wuhan, China
- 2State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan, China
- 3Hubei Collaborative Innovation Center for Green Transformation of Bio-Resources, School of Life Sciences, Hubei University, Wuhan, China
Cupin_1 domain-containing proteins (CDPs) are ubiquitously present in higher plants, which are known to play essential roles in various biological processes. In this study, we carried out genome-wide characterization and systematic investigation of the CDP genes in Brassica napus. A total of 96 BnCDPs, including 71 germin-like proteins (GLPs; proteins with a single cupin_1 domain) and 25 CDP bicupins (proteins with two cupin_1 domains), were identified and clustered into six distinct subfamilies (I–VI) based on the phylogenic analysis, gene structure and motif distribution. Further analysis indicated that whole-genome duplication (WGD) and segmental duplication are main contributors to the species-specific expansion of the BnCDP gene family, and all the duplicated genes subsequently underwent strong purification selection. The promoter region of BnCDPs showed enrichment of cis-regulatory elements associated with development, hormone and stress, as well as transcription factor binding sites, which validates the prediction that BnCDPs are widely involved in plant growth and biotic and abiotic stress responses. The BnCDPs in different subfamilies exhibited obvious differences in expression among 30 developmental tissues/stages of B. napus, implying that BnCDPs may be involved in tissue- and stage-specific developmental processes. Similar trends in expression of most BnCDPs were observed under Sclerotinia sclerotiorum inoculation and four abiotic stresses (dehydration, cold, ABA and salinity), particularly the BnGLPs in subfamily I and III with single cupin_1 domain, revealing that BnCDPs are of great importance in the environmental adaption of B. napus. We then performed a genome-wide association study (GWAS) of 274 B. napus core germplasms on S. sclerotiorum resistance and identified four significantly associated loci harboring five BnGLPs. The expression levels of two candidate genes, BnGLP1.A08 and BnGLP1.C08, were significantly correlated with S. sclerotiorum resistance. Their functional responses to multiple stages of S. sclerotiorum inoculation and four abiotic stresses were further examined through qPCR. Overall, this study provides rich resources for research on the function and evolutionary playground of CDP genes.
Introduction
Adverse environmental conditions including biotic and abiotic stresses pose serious threats to crop productivity in agriculture and food security (Zhu, 2016; Zhao et al., 2022). How plants adapt to adverse environments is a critical issue of biological studies and global agricultural production. It is critical to tune the expression of stress-responsive genes for resistance and adaptation to various biotic and abiotic stresses. The cupin_1 domain-containing protein (CDP) coding genes such as GLP members tend to be induced by pathogen attack and abiotic stress, and play important roles in response to a number of biotic and abiotic stresses to improve the development and environmental adaption of plants (Dunwell et al., 2004). Cupin superfamily proteins, which were named based on a conserved β-barrel fold, were first discovered using a conserved motif found within germin and germin-like proteins from higher plants (Dunwell et al., 2000, 2004). To date, the cupin superfamily has been considered as one of the most functionally diverse super-gene families in plants (Dunwell et al., 2004; Brunetti et al., 2022). This superfamily contains 69 gene families according to the Pfam database (accessed on 1st, June 2022), including the cupin_1 domain-containing family (Mistry et al., 2021). The cupin superfamily can be divided to monocupin (one single cupin domain), bicupin (a duplicated cupin structure) and multicupin (>two cupin domains; Dunwell et al., 2004). Germin and germin-like protein (GLP), which contain a single cupin_1 domain and belong to monocupin (Dunwell et al., 2000, 2004), have been widely deciphered in various plants, while the duplicated cupin_1 domain protein (CDP bicupin) has been rarely characterized. GLPs are defined by their sequence homology to germin, which was initially identified as a germination-specific marker in wheat embryos (Thompson and Lane, 1980; Dunwell et al., 2008; Rietz et al., 2012). Both germin and GLP display extremely high resistance to proteasome activity, heating, extreme pH, and detergents (Woo et al., 2000). Due to the conserved sequences and similarity in structural characteristics, it is difficult to classify GLPs and germins (Agarwal et al., 2009). In general, germins belong to a well-conserved homogeneous group and can be uniquely found within cereal plant species, including barley (Hordeum), maize (Zea), oat (Avena), rice (Oryza), rye (Secale) and wheat (Triticum) (Lane, 2002). In contrast, the GLP proteins have a wider taxonomic distribution and are generally present in other land plants besides cereals (Dunwell et al., 2008). Despite similarities in sequence among the members of GLPs, they have undergone significant functional diversification, and this family comprises numerous classes of important enzymes such as superoxide dismutase (SOD) that converts superoxide to H2O2 and O2 (Gucciardo et al., 2007; Guevara-Olvera et al., 2012), oxalate oxidase (OXO) that degrades oxalic acid to H2O2 and CO2 (Sakamoto et al., 2015), and polyphenol oxidase, dioxygenases, isomerases, epimerases, synthases and decarboxylases (Davidson et al., 2010; Cheng et al., 2014).
The CDP genes have different spatial and temporal expression characteristics during development in a variety of plants, and their expression also varies greatly among different tissues such as roots, stems, leaves, flowers, seeds and embryos and various developmental processes of the same plant (Lu et al., 2010; Wang et al., 2013; Li L. et al., 2016). Among the 69 identified cupin genes in soybean, 35 were found to be expressed in at least one tissue, and most of them displayed distinct tissue-specific expression patterns (Wang et al., 2014). Expression profiling of GLPs in rice and Arabidopsis thaliana has demonstrated that many of the members are only expressed in certain tissues or developmental stages (Li L. et al., 2016). For example, OsGLP3-3 and OsGLP8-2 are specifically expressed in developing seeds and OsGLP8-14 is preferentially expressed in developing panicles in rice (Li L. et al., 2016). These tissue specifically expressed GLPs may have essential functions in plant growth and development. For example, OsGLP2-1, which is specifically expressed in seed scutellum, positively regulates the dormancy of developing seeds through the abscisic acid and gibberellic acid signaling pathways (Wang et al., 2020); while OsGLP1 is predominantly expressed in green vegetative tissues and down-regulation of its expression in transgenic rice resulted in a semi-dwarfism phenotype (Banerjee and Maiti, 2010). GbGLP2 is mainly expressed in elongating fiber at 10 days post anthesis and negatively regulates fiber elongation in cotton (Sun et al., 2020). All these studies have demonstrated that GLPs play various roles in many tissues and organs or certain vital developmental stages of plants.
The CDP genes have long been considered be associated with responses to various biotic and abiotic stresses in different plant species (Thompson et al., 1995; Liu et al., 2004; Zou et al., 2007; Dong et al., 2008). Some GLP members with inherent OXO or SOD enzymatic activity can confer tolerance to biotic stress by hyper-accumulation of H2O2 and enhancement of cross-link between cell wall components during pathogen infection (Banerjee et al., 2010; Gangadhar et al., 2021). Riezt et al. identified 14 BnGLP genes in Brassica napus and demonstrated that both BnGLP3 and BnGLP12 have SOD activity, whose early induction is involved in the oxidative burst, and play a pivotal role in defense against Sclerotinia sclerotiorum (Rietz et al., 2012). GmGLP10 positively regulates the resistance to S. sclerotiorum, and transgenic tobacco overexpressing GmGLP10 from soybean showed significantly enhanced tolerance to oxalate acid and S. sclerotiorum infection (Zhang et al., 2018). Similarly, A. thaliana plants expressing sunflower HaGLP1 exhibited higher reactive oxygen species accumulation and resistance against S. sclerotiorum and Rhizoctonia solani (Beracochea et al., 2015). Overexpression in A. thaliana of a novel GLP gene GhABP19 from Gossypium hirsutum resulted in enhanced resistance to Verticillium dahliae and Fusarium oxysporum infection through its SOD activity and activation of the Jasmonic acid (JA) pathway (Pei et al., 2019). Moreover, GLPs also widely participate in defense against some other fungal pathogens such as Blumeria graminis (Yuan et al., 2021), Verticillium longisporum (Knecht et al., 2010), Magnaporthe oryzae (Liu et al., 2016) and Aspergillus flavus (Wang et al., 2013), as well as responses to viruses, bacteria, and even insect herbivores (Dunwell et al., 2008). Apart from biotic stress, the CDP genes are also widely involved in defense against abiotic stress in plants. For instance, A. thaliana with ectopic overexpression of soybean GmGLP7 exhibited obviously enhanced tolerance to drought, salt and oxidative stress, and was hypersensitive to exogenous ABA treatment (Li Y. et al., 2016). Similarly, A. thaliana overexpressing AhGLP2 or AhGLP3 from peanut showed higher tolerance to salt stress (Wang et al., 2013). Knockout of OsGLP1 by CRISPR/Cas9 resulted in higher sensitivity of rice plants to UV-B, suggesting that OsGLP1 is involved in the acclimation to UV-B radiation (He et al., 2021). In addition, overexpression of StGLP in potato plants increased the H2O2 level, triggered the scavenging signaling pathways of reactive oxygen species and induced the expression of heat stress-responsive genes to enhance the tolerance to heat stress (Gangadhar et al., 2021). Besides, the GLP genes are also responsive to other abiotic stresses such as drought (Anum et al., 2022), heavy metal (Cheng et al., 2018) and wound (Wang et al., 2013).
Brassica napus (2n = 4x = 38, AACC) is an important source of vegetable oil and stock feed in the world, which is cultivated in an area of more than 36 million hectares with an annual seed yield of 72 million tons (FAO STAT1). In actual production, the yield of B. napus is threatened by various biotic (insect pest and disease) and environmental/abiotic (salinity, acidity, alkalinity, drought, heat and water-logging) stresses (Liu et al., 2022). Therefore, it is urgent to explore genes with durable disease resistance and tolerance to diverse abiotic stresses in crop breeding. Considering the important roles of CDP genes in resistance and adaptation of plants to various biotic and abiotic stresses, it is reasonable to speculate that CDPs may also function in the environmental adaptation of B. napus. However, there has been no genome-wide identification and systematic investigation of the CDP family represented by monocupion GLPs in B. napus to date. In this study, we identified a total of 96 CDP genes (including 71 BnGLPs and 25 CDP bicupins) in the B. napus genome through genome-wide analysis. In addition, we comprehensively analyzed their evolutionary relationships, gene structures, conserved motifs, cis-regulatory elements and transcription factor binding sites (TFBSs). Expression profiling in 30 B. napus tissues/stages demonstrated that BnCDPs are involved in tissue- and stage-specific developmental processes. Expression analysis under biotic (S. sclerotiorum infection) and abiotic (dehydration, cold, ABA and salinity) stress treatments together with GWAS on S. sclerotiorum resistance demonstrated that two BnGLPs are commonly responsive to multiple biotic and abiotic stresses. The findings provide important insights into the role of BnCDPs in resistance to biotic and abiotic stresses and lay a foundation for future functional study of the CDP genes.
Results
Identification of BnCDP gene family members in Brassica napus
A total of 96 BnCDP genes with the cupin_1 domain were identified in B. napus through HMMsearch by using PF00190 as the query and subsequent domain verification. Among these genes, 50 genes were located in the An subgenome, while the remaining 46 genes were found in the Cn subgenome. The overall distribution of BnCDP genes was uneven across chromosomes (Supplementary Figure 1). Chromosome A02 (seven genes), A06 (eight genes), A07 (eight genes), A09 (six genes) and C08 (11 genes) had the most BnCDP genes; while A04 and C02 only contained two and one BnCDP gene, respectively; and A05 chromosome even had no BnCDP. The length of BnCDP proteins ranged from 175 aa (BnaC08g43590D) to 652 aa (BnaC03g48460D), with an average length of 286 aa. The exon number of each BnCDP gene ranged from 1 to 7 (only BnaA06g25170D), with most of the members (74%) having no more than three exons. The predicted theoretical pI values varied from 4.77 to 9.77 and the MW values were between 19.45 and 78.20 kDa. Moreover, the GRAVY (grand average of hydrophobicity) index values ranged from –1.300 to 0.545. According to the predicted subcellular location, the BnCDP proteins showed a wide subcellular distribution pattern and were mainly located in extracellular (41/96) and plasma membrane (18/96), and the remaining proteins were specifically located in the chloroplast (11), cytoplasmic region (11), nuclear (7), endoplasmic reticulum (5) and mitochondrion (3) (Supplementary Table 1).
Phylogenetic analysis of BnCDPs
To further characterize and classify the BnCDP family members, we constructed a phylogenetic tree using all the 96 BnCDP proteins from B. napus and 42 AtCDP proteins from A. thaliana. Previous studies have reported 32 AtGLP genes in A. thaliana (Li L. et al., 2016). In this research, we identified ten new AtCDP genes including two monocupin coding genes (AT4G36700 and AT5G44120) and eight bicupin coding genes (AT2G28490, AT3G22640, AT1G03880, AT1G03890, AT4G28520, AT1G07750, AT2G28680 and AT2G18540) in A. thaliana (Supplementary Table 2). Finally, these CDP proteins were assigned to I–VI subfamilies based on the topology of the phylogenetic tree, in which the members in subfamily I–III were all monocupins; the members in subfamily V (except for AT5G44120 and BnaC08g43590D) and VI (except for BnaCnng38930D) are almost all bicupins; while subfamily IV was a mixture of monocupins (six) and bicupins (seven; Figure 1 and Supplementary Table 2). The number of BnCDP genes varied significantly among the six subfamilies, with subfamily I including approximately half of all the BnCDP genes (40), while the subfamily II–VI only comprised 8, 16, 9, 11 and 12 BnCDP members, respectively. Except for four genes (AtGLP1-4, AtGLP1-5, AtGLP1-7 and AtGLP3-8) clustered in subfamily II and three genes (AtGLP5-1, AtGLP1-6 and AtGLP5-15) clustered in subfamily III, the remaining previously reported AtGLP genes (Li L. et al., 2016) all fell into subfamily I, and all the ten newly identified AtCDP members were scattered in subfamily IV (four members), V (four members) and VI (two members). These results indicated that the CDP genes were present in the common ancestors of A. thaliana and B. napus, and some of them might have undergone species-specific expansion and subsequently significant divergence.
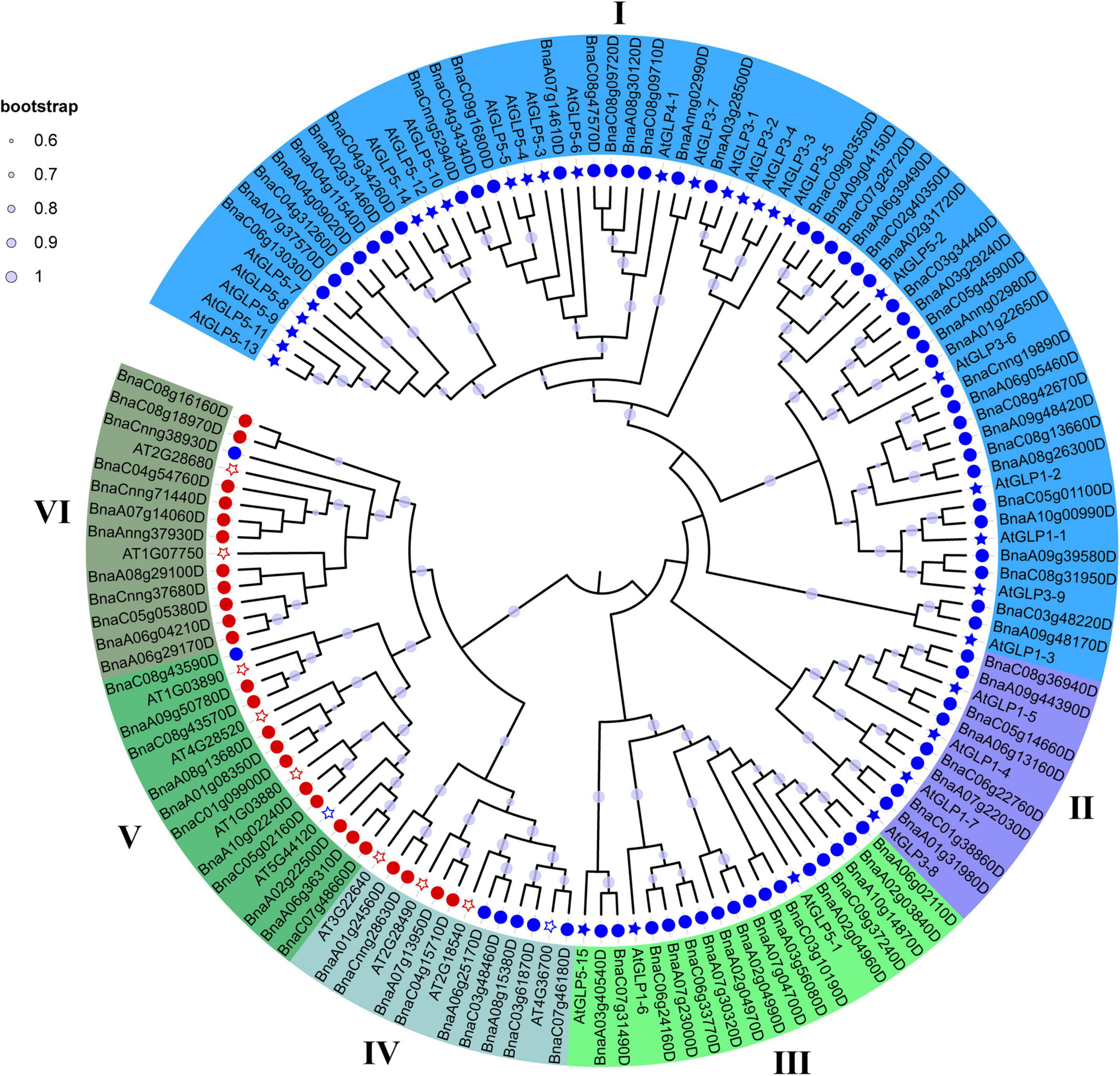
Figure 1. A neighbor-joining (NJ) phylogenetic tree of CDP proteins in Arabidopsis thaliana (At) and Brassica napus (Bn). All CDP proteins were clustered into six subfamilies, and each subfamily was represented by a different color. Light purple dot on a branch in the figure indicates that the bootstrap support is greater than 60%. Circles refer to BnCDPs, while filled stars refer to AtGLPs identified previously and empty stars refer to AtCDPs identified in this research; Blue and Red refer to CDPs with single or duplicated cupin_1 domain, respectively.
Duplication events of the BnCDP genes were detected based on BLAST and MCScan X. Briefly, all 96 BnCDPs were derived from duplication, among which 72 genes (75.0%) were generated from whole-genome duplication (WGD) or segmental duplication, and 18 genes (18.75%) resulted from dispersed duplication (Supplementary Table 1). Moreover, three tandem and three proximal gene duplication types were detected. There were 191 paralogous gene pairs with high identities (identity > 75%, and alignment length > 75%) in B. napus, with 56 gene pairs in the An subgenome, 32 gene pairs in the Cn subgenome, and the remaining 103 duplication events occurring between the two subgenomes (Figure 2 and Supplementary Table 3). To estimate the selection pressure on BnCDP genes in B. napus, the ratio of non-synonymous substitution to synonymous substitution (Ka/Ks) for the 191 paralogous gene pairs was calculated. The results showed that the Ka/Ks ratio for all paralogous gene pairs varied from 0 to 0.52 and lower than 1, suggesting that the BnCDP genes have undergone purification selection during evolution (Supplementary Table 3).
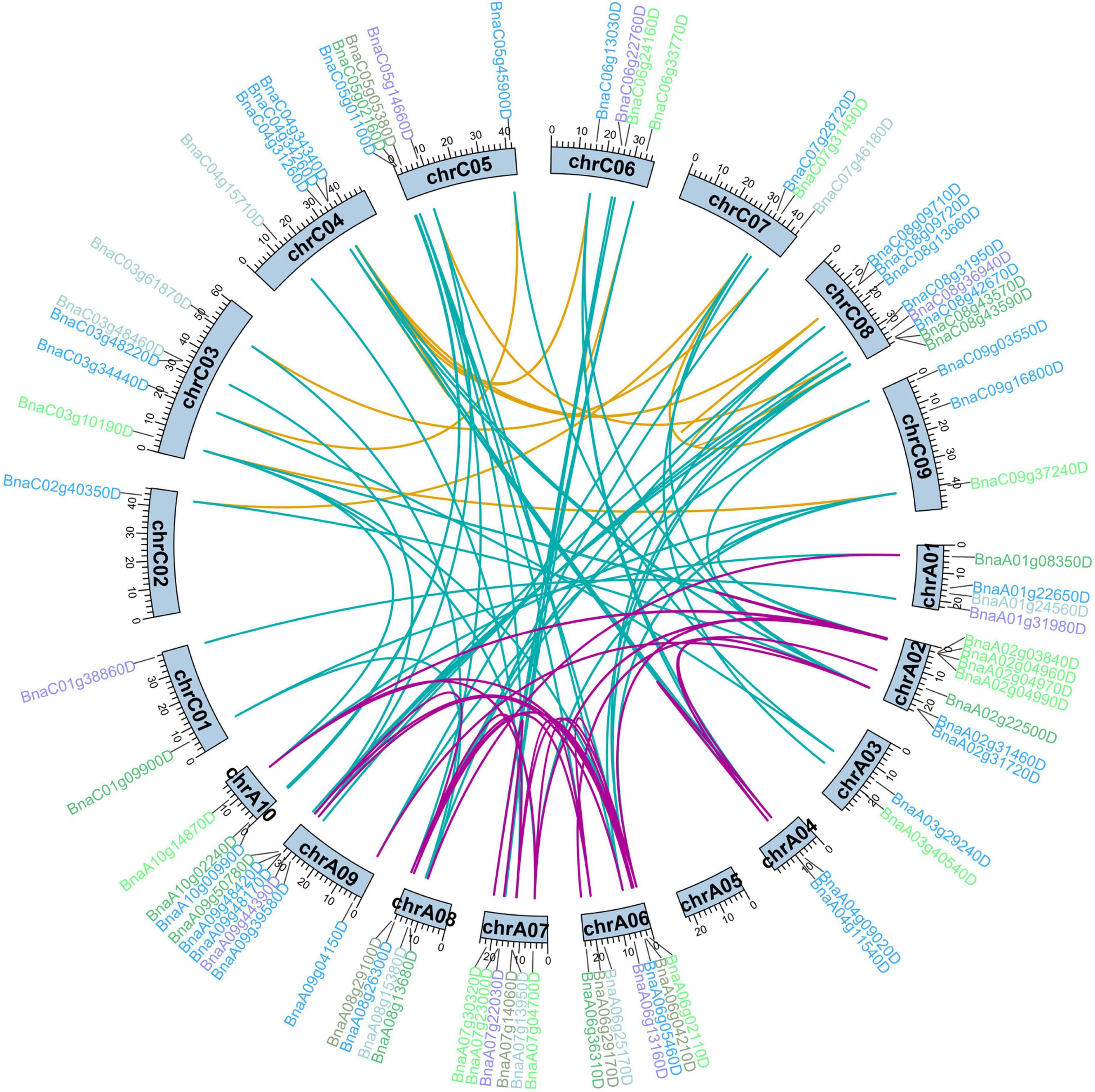
Figure 2. Duplication analysis of BnCDP genes in Brassica napus. The gene names are marked outward of the corresponding chromosomes. The different colors indicate different subfamilies of the BnCDP genes. The duplicated gene pairs are highlighted with connecting lines colored according to the subgenomes, purple indicates that both genes in the gene pair are from the An subgenome, yellow represents that both genes in the gene pair are from the Cn subgenome, while the blue shows that two genes in the gene pair come from different subgenomes.
Gene structure and conserved motif analysis of the BnCDP family in Brassica napus
The exon-intron structure of all the BnCDP genes in six subfamilies was displayed based on their phylogenetic relationships (Figures 3A,B). As shown in Figure 3B, 36 members contained both 5′ and 3′ UTRs; 33 members exhibited no UTR; while the remaining 27 members possessed either a 5′ or 3′ UTR. Furthermore, the gene structure seemed to vary remarkably among different subfamilies, but were relatively conserved within the subfamily. For example, most BnCDPs in subfamily I had two exons, and all members in subfamily II and nearly all members in subfamily III were intronless. Most of members in subfamily VI had three exons, while members in subfamily IV had the maximum number of exons and introns.
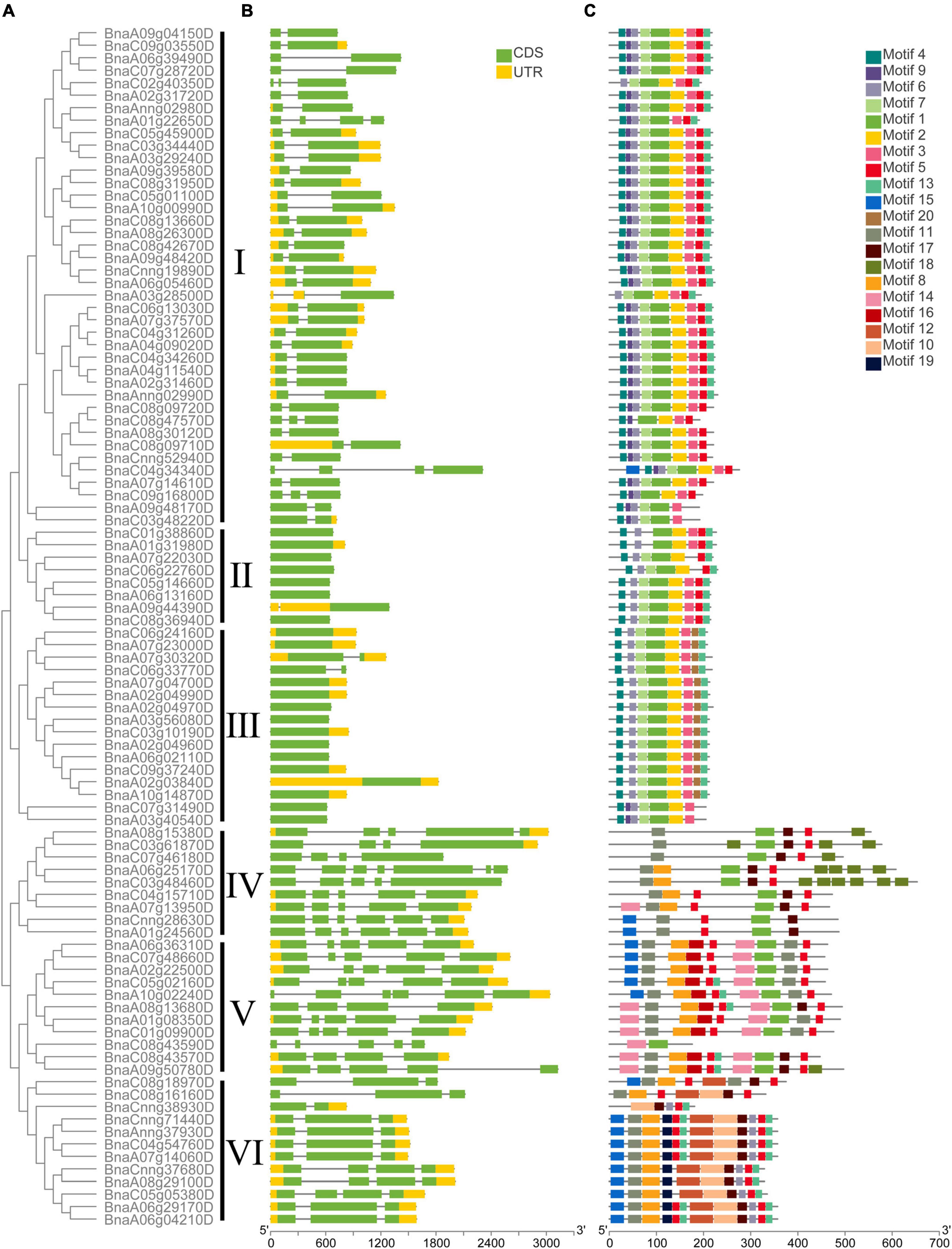
Figure 3. The phylogenetic relationship, exon-intron architecture, and conserved motifs of 96 BnCDP in Brassica napus. (A) The phylogenetic relationships of BnCDP proteins based on the NJ method. (B) Gene structures of BnCDP genes. Yellow boxes represent the untranslated region (UTR), green boxes represent exons and the gray lines represent introns. (C) The conserved motif composition of BnCDP proteins. Scale bars represent gene length (bp) and protein sequence length (aa).
We also analyzed the distribution of conserved motifs in the BnCDP family. In total, 20 distinct conserved motifs were identified (Figure 3C and Supplementary Table 4). In subfamily I–III, the motifs were well conserved. Most genes in these subfamilies simultaneously contained motif 1, 2, 3, 4, 7. However, motif 9 was specifically identified in members in subfamily I except for two genes in subfamily III (BnaC07g31490D and BnaA03g40540D). Members in subfamily III specifically contained motif 20 but were lack of motif 5 prevalently present in other five subfamilies. Motif 10, 12 and 19 were only found in subfamily VI, and motif 11 was present in nearly all members in subfamily IV–VI. The specificity of motifs was conformed to the observed evolutionary characteristics of BnCDP genes, implying that the specifically conserved motifs in different BnCDP subfamilies are associated with their functions.
Analysis of cis-regulatory elements in the promoter region of BnCDPs
The regulatory elements in the promoter region are often relatively conserved in sequence and function throughout evolution, particularly in tolerance to biotic and abiotic stresses (Oudelaar and Higgs, 2021). To explore the potential roles of BnCDP genes, we analyzed the cis-regulatory elements in the 2-kb promoter regions of BnCDPs based on the PlantCARE database. As a result, the cis-acting regulatory elements associated with development, hormone and stress were enriched in these promoters (Figure 4, Supplementary Figure 2, and Supplementary Tables 5, 6). These cis-regulatory elements included development related elements such as the circadian element (involved in circadian control), the GT1-motif, Sp1, ACE, G-box and GT1 motif (involved in light responsiveness); hormone responsive elements such as ABRE (involved in the abscisic acid response), AuxRR-core and TGA-element (involved in auxin response), GARE-motif and TATC-box (involved in gibberellin response), CGTCA-motif and TGACG-motif (involved in MeJA response) and the TCA-element (involved in salicylic acid response); stress-responsive elements such as ARE elements (essential for anaerobic induction), TC-rich repeats (involved in defense and stress responsiveness), AT-rich sequence (element for maximal elicitor-mediated activation), LTR (involved in low-temperature responsiveness), MBS (MYB binding site involved in drought-inducibility), MBSI (involved in flavonoid biosynthesis) and WUN-motif (involved in wound response). There were many MeJA-related cis-acting regulatory elements in the promoters of BnCDP genes: the CGTCA-motif was identified in 71 of the 96 BnCDP gene promoters (2.13 on average for each promoter), and the ABRE elements were present in 75 BnCDP promoters (with 3.39 on average for each promoter). Besides, 65 TC-rich repeats associated with defense and stress response were unevenly scattered in 45 BnCDP gene promoters, which were the most enriched in cluster III (10/16) and VI (8/12), but were rare in cluster V (2/11), while 83 low-temperature response (LTR) elements showed relatively unbiased distribution in subfamily I–VI. Moreover, ARE (84/96, 87.50%) and MBS (49/96, 51.04%) elements were also common in the promoters of BnCDP genes. These results implied that members of the BnCDP family are potentially involved in biotic and abiotic stress response during plant growth and development.
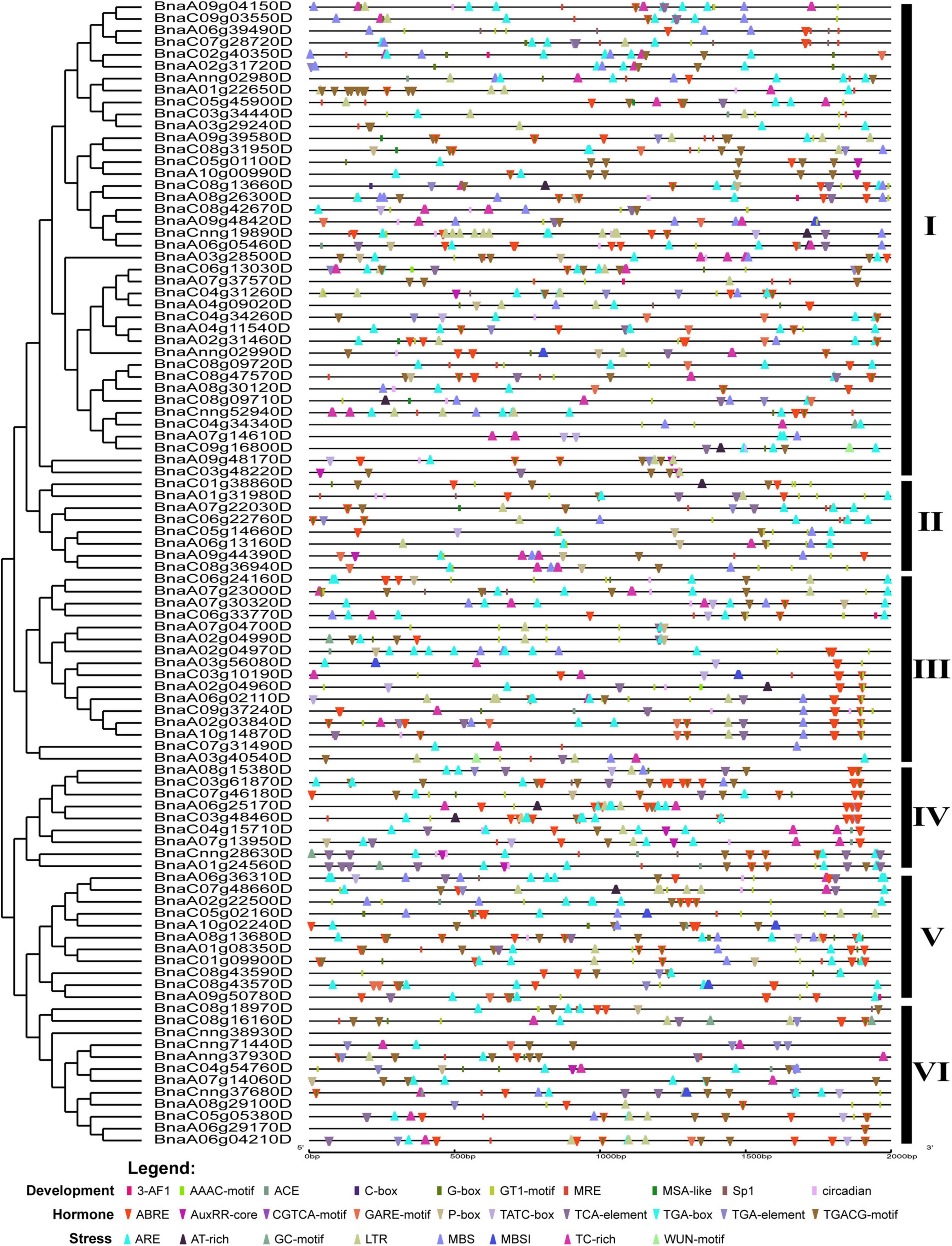
Figure 4. Cis-acting regulatory elements identified in promoters of BnCDP genes in Brassica napus. Boxes indicate development-related elements, down-wedges indicate hormone-related elements, and up-wedges indicate stress-related elements. Different colors indicate different elements.
The spatiotemporal expression patterns of the genes playing essential roles in plant development and stress responses tend to be regulated by their corresponding transcription factors (TFs) (Jin et al., 2014). Hence, we analyzed the transcription factor binding sites (TFBSs) in the promoter regions of all 96 BnCDP genes, and found that 64 of the promoters contain TFBSs, which correspond to 21 TF families (Supplementary Table 7). These TFs included Dof, B3, AP2, MIKC_MADS, MYB, MYB_related, GATA, ERF, C2H2, LBD, GRAS, Nin-like, BBR-BPC, SRS, NAC, E2F/DP, bZIP, ARF, bHLH, SBP, and Trihelix. Among the 64 promoters, 22 and 42 potentially bind to single and multiple TFs, respectively (Supplementary Table 7).
Expression patterns of BnCDP genes in multiple tissues of the whole growth period
To comprehensively explore the potential function of BnCDP genes, we investigated the expression patterns of all 96 BnCDP members in 30 tissues/stages, including the leaf, root, stem, bud, stamen, new pistil, blossomy pistil, wilted pistil, sepal, ovule and ten time-course seeds and silique walls [from 4 to 48 days after pollination (DAP)] based on our previously published transcriptome data (Figure 5 and Supplementary Table 8; He et al., 2022). Overall, 89 genes were expressed (FPKM > 1) in at least one tissue or stage, with 48 genes showing high expression (FPKM > 50), 24 genes exhibiting intermediate expression (FPKM > 10) and three genes being lowly expressed (FPKM < 10). Generally, most of the BnCDP genes showed highly tissue-specific expression patterns, and only three genes (BnaC08g13660D, BnaCnng19890D and BnaC05g05380D) were expressed in all the 30 tissues or stages (Figure 5). Members in subfamily IV and subfamily V displayed nearly identical expression patterns, which were highly expressed in the ovule and silique wall (28 and 40 DAP), and had extremely high expression in the seed at the intermediate to late stages, implying their essential roles in the successful reproduction of B. napus. The genes in subfamily I and subfamily II showed great differences in expression and some of them were highly expressed in the root, bud and pistil. Overall, members in subfamily III showed high expression levels, such as in the leaf, stem and early stages of silique wall development, particularly BnaA02g04960D, BnaA07g23000D, BnaA07g30320D, BnaC06g24160D and BnaC06g33770D; while the members in subfamily VI exhibited relatively low expression levels. These differences in expression pattern among subfamilies implied that the BnCDP genes may be involved in tissue- or stage-specific developmental processes.
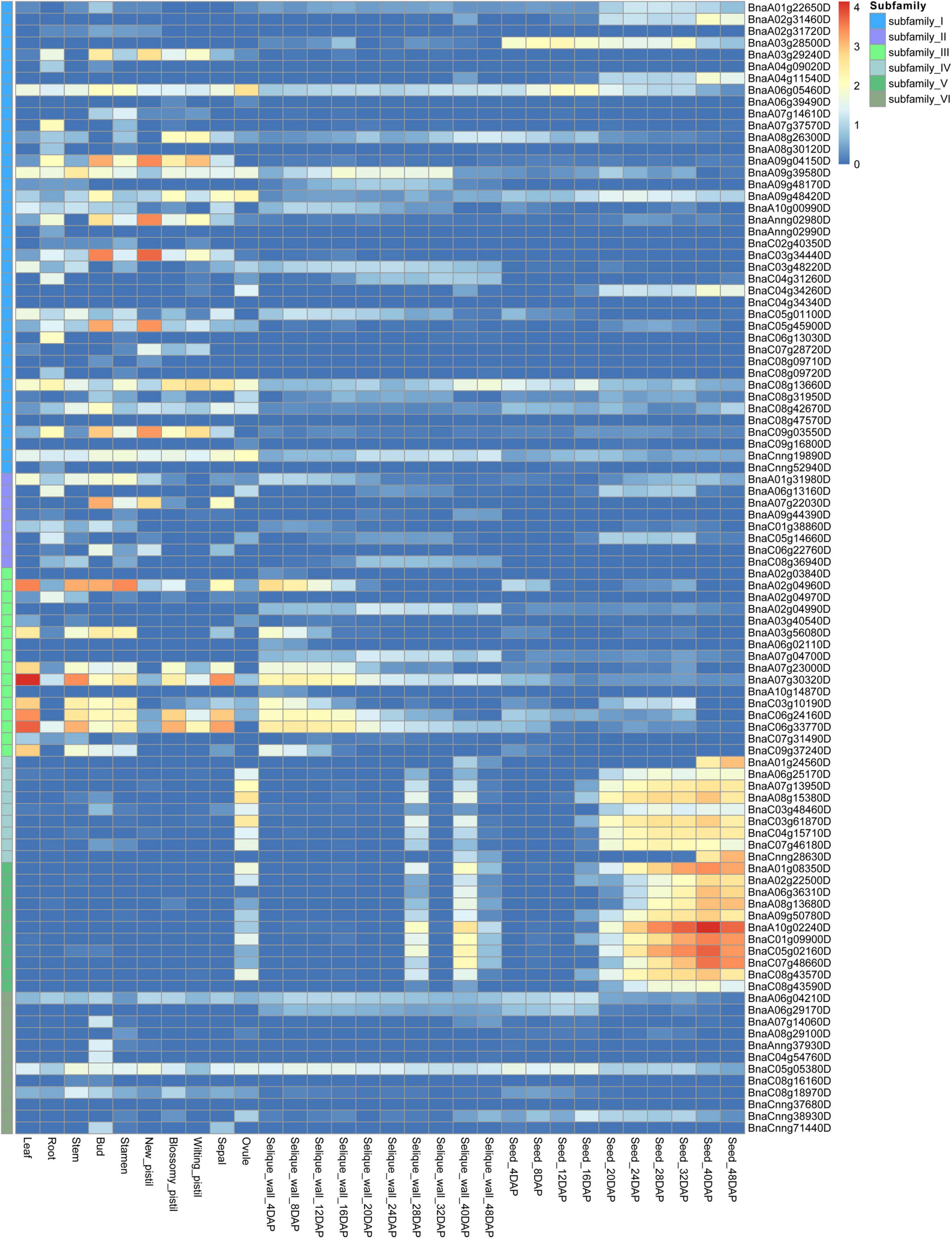
Figure 5. Expression patterns of 96 BnCDP genes in 30 tissues of Brassica napus ZS11 variety. The expression data were processed with the log10 normalization of fragments per kilobase million (FPKM). The color scale represents relative expression levels from low (blue color) to high (red color).
Expression patterns of BnCDP genes under biotic and abiotic stresses
As sessile organisms, the growth and development of plants are constantly challenged by various stresses. S. sclerotiorum is an ascomycete plant pathogen causing Sclerotinia stem rot in B. napus and severely affecting its seed yield and quality. To analyze the role of BnCDPs in the response to biotic stress, we investigated the expression profiles of 96 BnCDPs in the leaves of tolerant variety ZY821 of B. napus at 0 and 24 h after inoculation with S. sclerotiorum (Girard et al., 2017). Interestingly, just a part of BnCDP members were responsive to S. sclerotiorum inoculation, as many of them were inactive in the leaves of ZY821, such as all members in subfamily IV and V, which had no transcript accumulation at all (Figure 6). Among the 18 expressed BnCDP genes (FPKM > 1), 13 exhibited remarkable expression changes at 24 h post-inoculation (hpi), with eight genes (BnaA08g26300D, BnaA09g48420D, BnaC06g13030D, BnaC08g13660D from subfamily I; BnaA09g44390D from subfamily II; and BnaA07g30320D, BnaC06g24160D, BnaC06g33770D from subfamily III) being up-regulated and five genes (BnaA02g31720D, BnaA07g37570D, BnaAnng02990D from subfamily I; and BnaA06g04210D, BnaC05g05380D from subfamily VI) being down-regulated. Particularly, BnaA08g26300D and BnaC08g13660D in subfamily I exhibited high expression (FPKM > 500) and heavily induced by S. sclerotiorum inoculation in ZY821 (Figure 6 and Supplementary Table 9).
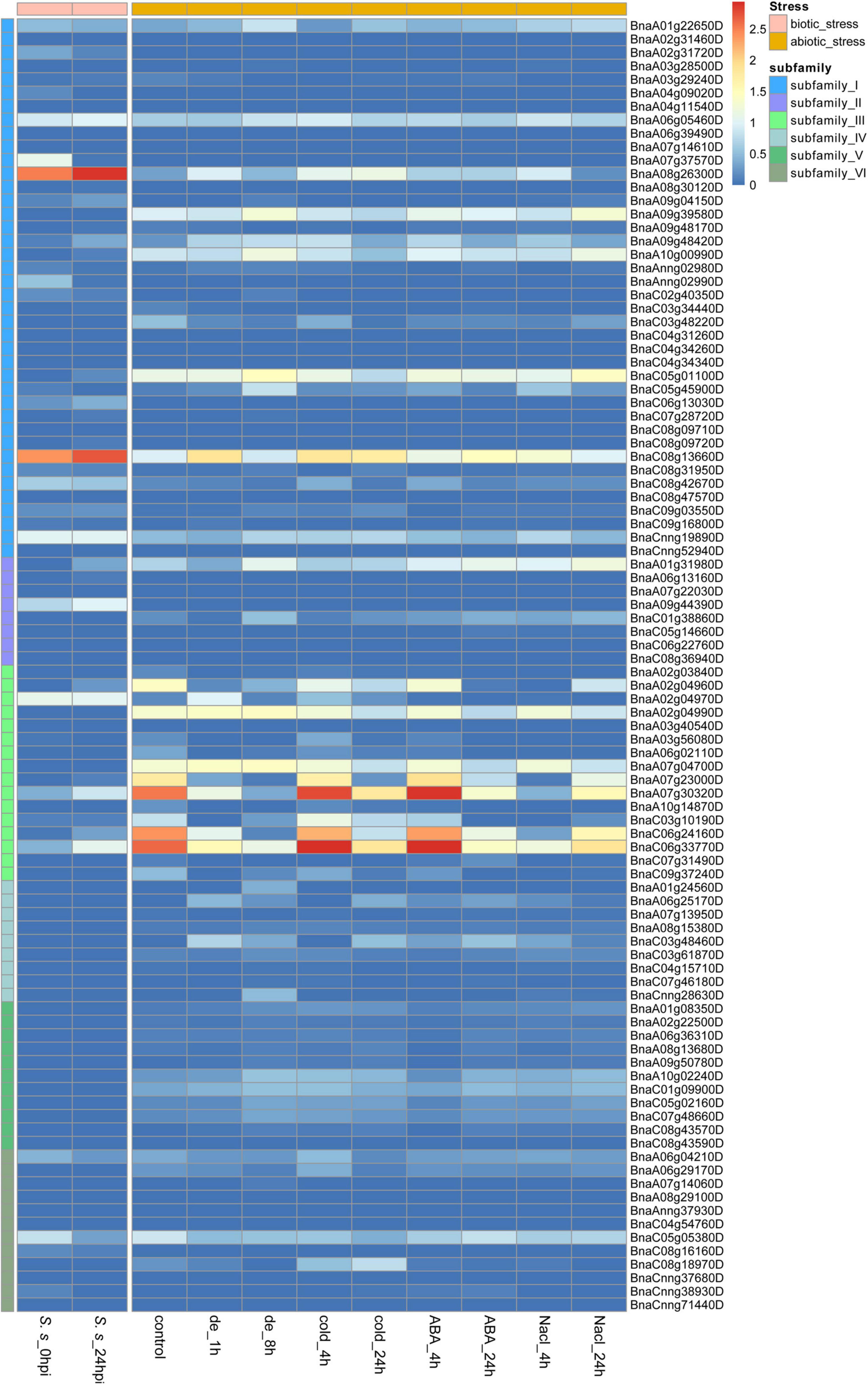
Figure 6. Expression profiles of 96 BnCDP genes under biotic (left) and abiotic (right) stress conditions. The left panel shows the expression level of BnCDP genes in ZY821 at 0 and 24 h after Sclerotinia sclerotiorum inoculation; The left panel shows the expression level of BnCDP genes under different abiotic stress conditions (dehydration, cold, ABA and salinity). The expression data were processed with the log10 normalization of fragments per kilobase million (FPKM). The color scale represents relative expression levels from low (blue color) to high (red color).
To identify the potential functions of BnCDP genes in response to different abiotic stresses, we analyzed the RNA-seq data from samples under dehydration, cold, ABA and salinity treatment, respectively (Zhang et al., 2019). By taking two-fold change as the threshold value, we identified and compared the differentially expressed genes under each of the above-mentioned stresses (Figure 6 and Supplementary Table 10). Under dehydration treatment, 25 out of the 29 expressed BnCDP genes (FPKM > 1) were significantly responsive at 1 h and/or 8 h of treatment, and most of upregulated genes (9/12) were from subfamily I, while the downregulated genes (8/12) were mainly from subfamily III. Besides, one gene (BnaA01g31980D) in subfamily II displayed an opposite pattern, which was downregulated at 1 h of treatment, but subsequently upregulated at 8 h. Under cold treatment, 29 BnCDPs showed differential expression, and more than two-thirds (21/29) of them belong to subfamily I and III. A total of 20 BnCDP genes were significantly responsive to ABA treatment and almost all down-regulated genes (8/9) belonged to subfamily III. Twenty-four BnCDP genes showed significant fold changes in response to NaCl treatment, and 90% of the responsive genes (9/10) in subfamily I were upregulated except for BnaC03g48220D, while all ten responsive members in subfamily III were downregulated. The transcript levels of genes from subfamily VI almost showed no fluctuation during ABA and NaCl treatment. Overall, the BnCDP family showed highly similar expression patterns in response to the above four abiotic stresses, and mainly the members in subfamily I and subfamily III were responsive to the treatments: The former tended to be induced while the latter was usually down-regulated in response to these stresses. Besides, we found that 13 BnCDPs were significantly responsive to all the four abiotic stresses, among which six members (BnaA07g23000D, BnaA07g30320D, BnaA08g26300D, BnaC06g24160D, BnaC06g33770D and BnaC08g13660D) were involved in response to S. sclerotiorum inoculation (Supplementary Figure 3). These genes are common stress-responsive genes shared by multiple biotic and abiotic stresses and may be used for the breeding of varieties with multiple stress resistance in B. napus.
Genome wide association study on Sclerotinia sclerotiorum resistance and functional candidate gene BnGLPs analysis
To further examine the potential effects of BnCDP genes in S. sclerotiorum resistance, we used more than 2.38 million SNPs with MAF > 0.05 across 274 worldwide collected accessions to perform a GWAS based on disease index using the FarmCPU model. As shown in Figure 7A, the frequency distribution of disease index approximates to normal distribution, indicating that this population was suitable for association analysis. Then, the SNP GWAS was conducted and the significant SNPs associated with S. sclerotiorum resistance were displayed on Manhattan plot (Figure 7B) and QQ plot (Figure 7C). A total of 24 significant SNP loci (P < 4.199 × 10–7; Bonferroni-adjusted significance threshold 1/n, n = 2,381,566; Figure 7B) associated with S. sclerotiorum resistance were detected, which constituted 10 QTL distributed on ten chromosomes including A01, A02, A03, A04, A08, A10, C03, C03, C04 and C08 of B. napus. Among them, four QTL containing five BnGLPs (BnaA04g11540D, BnaA08g26300D, BnaC04g34260D, BnaC04g34340D and BnaC08g13660D) were identified. Furthermore, BnaA08g26300D and BnaC08g13660D are WGD genes (ortholog of AT1G09560/AtGLP1-2), while BnaA04g11540D and BnaC04g34260D are WGD genes (ortholog of AT5G39130/AtGLP5-10). Considering that BnaA08g26300D and BnaC08g13660D (named as BnGLP1.A08 and BnGLP1.C08, respectively) gene pairs are both common responsive genes for biotic and abiotic (dehydration, cold, ABA and salinity treatment) stresses, we took the two genes as candidates for further haplotype analysis. Our results revealed that each of BnGLP1.A08 and BnGLP1.C08 had two major haplotypes associated with S. sclerotiorum resistance (Figures 7D,E).
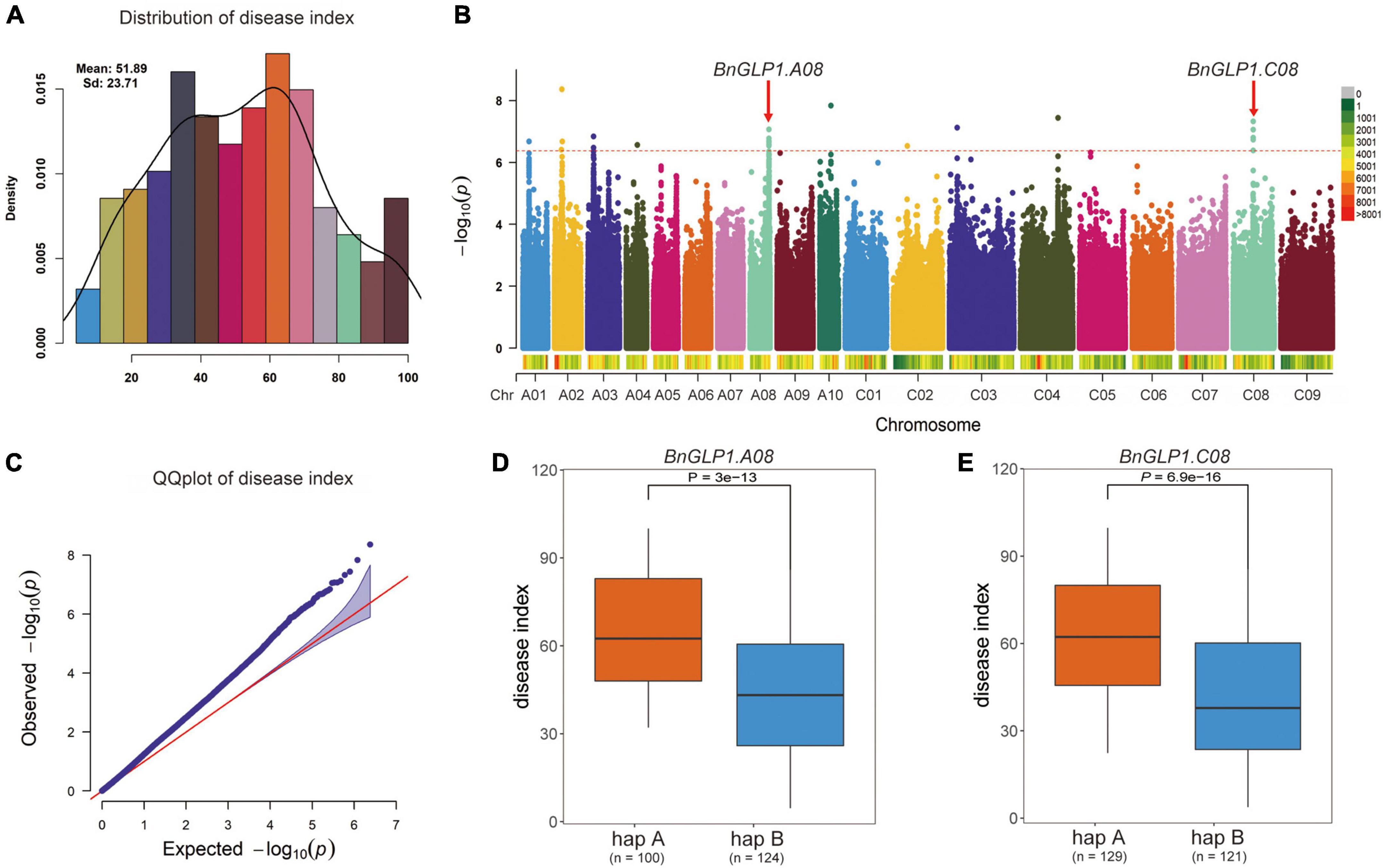
Figure 7. Both BnGLP1.A08 and BnGLP1.C08 are associated with Sclerotinia sclerotiorum resistance in Brassica napus. (A) Frequency distribution of disease index of 274 B. napus accessions. (B) Manhattan plot of the disease index from association analyses by FarmCPU model. Each point represents a SNP, and the SNP that exceeds the threshold (red dotted line) –log10 (1/n) = 6.377 is significant. (C) QQ plot for the disease index from association analyses. (D,E) Boxplots for disease index based on the two haplotypes of BnaGLP1.A08 (D) and BnaGLP1.C08 (E). The phenotypic differences between groups were tested using a two-tailed t-test (P < 0.01).
To confirm that BnGLP1.A08 and BnGLP1.C08 are involved in adaptive stress response, we determined their expression levels of B. napus seedlings under S. sclerotiorum infection and different abiotic stress treatments (dehydration, cold, ABA and salinity) by quantitative real-time PCR (qRT-PCR). The leaves at four time points (12 h, 24 h, 36 h and 48 h) after S. sclerotiorum inoculation were collected to explore the dynamic gene expression changes (Figure 8). The results demonstrated that the expression pattern of BnGLP1.A08 and BnGLP1.C08 after S. sclerotiorum inoculation was very similar, which stayed relatively consistent during the first 24 h and was then upregulated to a peak (Figure 8). For abiotic stress responses, the expression level of both BnGLP1.A08 and BnGLP1.C08 genes showed significantly increased expression under dehydration, cold, ABA or salinity treatment, and displayed more sensitive to ABA treatment, leading to more accumulation in transcripts (Figure 8). Taking together, the qRT-PCR analysis verified the reliability of the RNA-seq data and revealed that both BnGLP1.A08 and BnGLP1.C08 gene pairs are common responsive genes shared by multiple stresses.
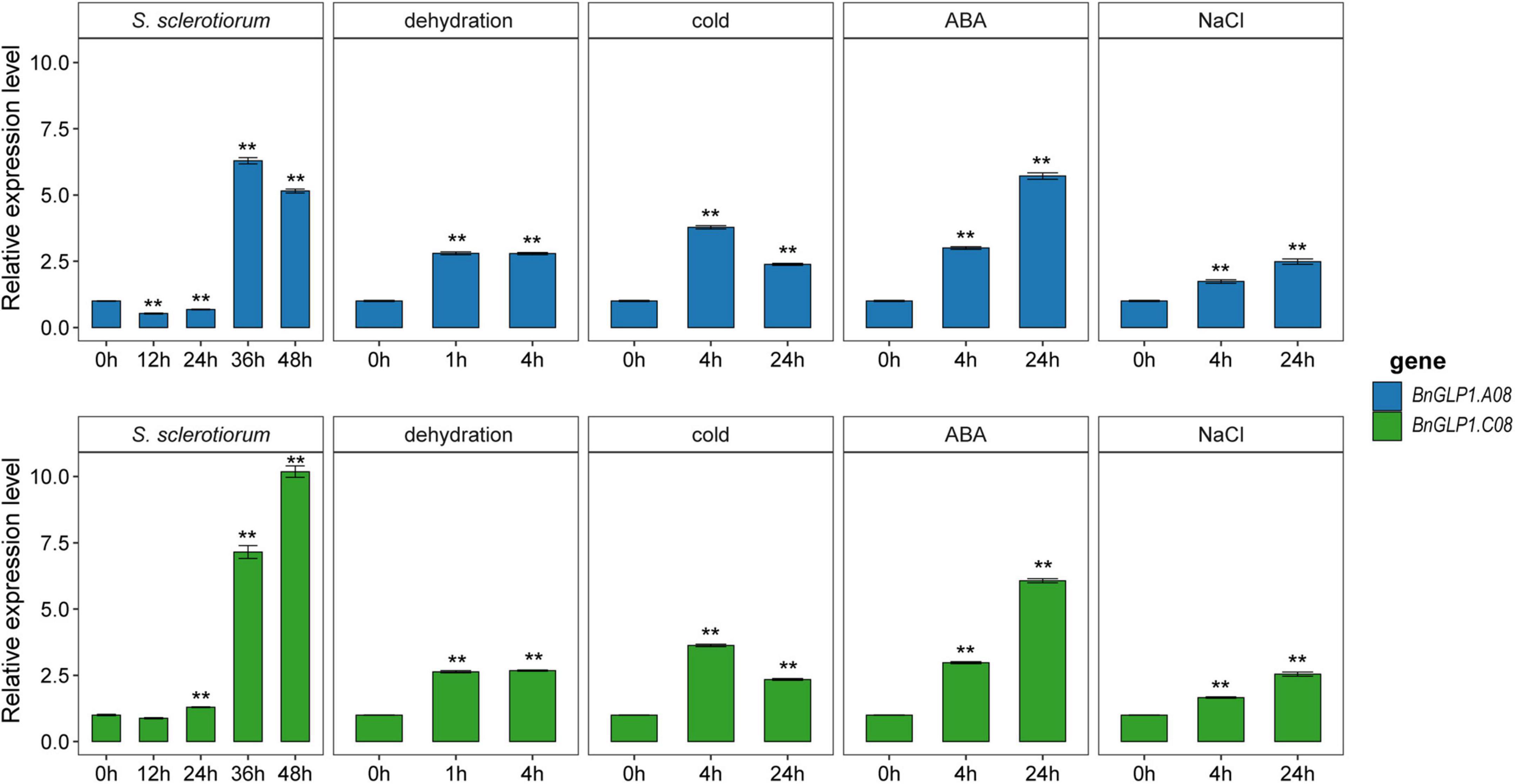
Figure 8. The expression validation of candidate BnGLP1.A08 and BnGLP.C08 genes in response to Sclerotinia sclerotiorum infection and four abiotic stress treatments (dehydration, cold, ABA and salinity) by qRT-PCR. The time points under the x-axis represent hours (h) after corresponding biotic and abiotic treatments. The error bars show the standard error of three replicates. Student’s t-test was used for statistical analysis, ** indicates significant differences at P < 0.01, all compared to the treatment at 0 h.
Discussion
Environmental stresses such as pathogen infection or drought, salinity, heat and cold cause devastating impacts on plant growth and extensive losses in the crop yield (Suzuki et al., 2014). Sclerotinia stem rot caused by S. sclerotiorum is a devastating disease leading to significant yield and economic losses in many crop and vegetable plants, particularly Brassica crops (Liu et al., 2021). Moreover, drought and salt stresses are important limiting factors affecting about 26% and 20% of the agricultural land, respectively (Raza et al., 2021). Therefore, it is urgent to identify more effective loci with durable disease resistance or diverse abiotic stress tolerance in plants, especially in important agricultural crops, to develop biotic and abiotic stress-tolerant genotypes. Single cupin_1 domain GLPs play essential roles in regulating plant development and biotic/abiotic stress resistance (Dunwell et al., 2004). However, the prevalence and functional diversity of the CDP gene family in B. napus have not been thoroughly investigated. In this study, we performed a comprehensive analysis of the BnCDP family in B. napus. The features of BnCDP genes, including their chromosomal distribution, phylogenetic classification, gene structures, conserved motifs, cis-regulatory elements, expression profiles, and responses to various stresses were explored. The results will provide insights into this gene family and offer valid information for predicting their potential functions in plant growth and stress response.
To date, the GLP genes (encoding single cupin_1 domain protein) have been genome-wide identified in numerous plant species, such as A. thaliana (Li L. et al., 2016), rice (Li L. et al., 2016), soybean (Lu et al., 2010), Vitis vinifera (Ilyas et al., 2022), cucumber (Liao et al., 2021), potato (Zaynab et al., 2021), peanut (Wang et al., 2013) and Physcomitrella patens (Nakata et al., 2004). However, the genes encoding duplicated cupin_1 domain protein were till rarely characterized. In this study, 96 BnCDPs in B. napus and 42 AtCDPs (including 32 previously identified AtGLPs) were systematically identified. Gene duplication might account for the differences in the number of CDP family members between B. napus and A. thaliana. In our results, 75.0% of BnCDPs (72 BnCDP genes) were originated from WGD or segmental duplication, which is consistent with the conclusion that WGD and segmental duplication are the main contributors to the expansion of gene families in other researches (Ma et al., 2017; Wu et al., 2018; Xie et al., 2022). Since B. napus was formed by the interspecific hybridization between B. rapa and B. oleracea about 7,500 years ago, both of which had undergone a genome triplication event after divergence from A. thaliana lineage, six homologs for each A. thaliana gene are expected to be present in B. napus (Allender and King, 2010). However, the number of identified BnCDPs was much smaller than expected (less than threefold of AtCDPs), which may be ascribed to the occurrence of gene loss during the diploidization process (Albalat and Canestro, 2016). Despite the uneven distribution of BnCDP genes at the chromosome level, the total number of genes was roughly similar in the An (50 members) and Cn subgenome (46 members) (χ2 = 0.167 < 3.84). Based on the Ka/Ks ratio of paralogous gene pairs (Supplementary Table 3), it can be speculated that purification selection plays a significant role in the evolution of BnCDP genes in B. napus.
A phylogenetic analysis of the CDP members from B. napus and A. thaliana revealed that these CDPs could be divided into six subfamilies (I–VI). All members in subfamily I–III are GLPs, while those in subfamily IV–VI tend to be bicupins, and their monocupin members showed closer evolutionary relationship with bicupins than with other GLPs (Figure 1). Dunwell et al. predicted that bicupins probably evolved from the duplication and then fusion of a single domain ancestor (Dunwell et al., 2004). Our phylogenetic analysis indicated that the fused duplicated domain protein (bicupin) might also lose a domain during the subsequent evolution to produce a new monocupin with higher similarities in sequence and gene structure to its bicupin ancestor. Furthermore, BnCDPs within the same subfamily have high similarities in gene structure and motif distribution (Figure 3), implying that the members in the same subfamily have similar functions.
Spatio-temporal expression pattern can reflect the potential function of a gene to a certain extent. In the present study, we analyzed the expression patterns of all 96 BnCDP members in 30 tissues/stages (He et al., 2022). As shown in Figure 5, most of the BnCDP genes showed preferential expression in specific tissues/stages, which is consistent with the previous findings in cucumber, rice and A. thaliana (Li L. et al., 2016; Liao et al., 2021). For example, almost all members in subfamily IV and subfamily V were predominantly expressed in seeds at the intermediate to the late stage; BnaA09g04150D, BnaAnng02980D and BnaC03g34440D from subfamily I were predominantly expressed in the bud and pistil; BnaA07g22030D from subfamily II was highly expressed in the pistil; BnaA07g30320D and BnaC06g33770D from subfamily III showed high accumulation of transcripts in the leaf. These results indicate that they play essential roles in these tissues/stages. Besides, three genes (BnaC08g13660D, BnaCnng19890D, BnaC05g05380D) were expressed in all the 30 tissues/stages (Figure 5), suggesting their possible essential roles in the entire growth and development stages.
Plant GLP genes also play vital roles in regulating biotic and abiotic stress responses. Numerous studies have demonstrated that the GLPs are widely involved in resistance to diverse pathogens such as S. sclerotiorum (Rietz et al., 2012; Zhang et al., 2018), Rhizoctonia solani (Beracochea et al., 2015), Blumeria graminis (Yuan et al., 2021), Magnaporthe oryzae (Liu et al., 2016), Aspergillus flavus (Wang et al., 2013), and response to UV-B radiation (He et al., 2021), heat (Gangadhar et al., 2021; Zaynab et al., 2021), drought (Wang et al., 2013; Anum et al., 2022), heavy metal (Cheng et al., 2018) and wound (Wang et al., 2013). To further explore the possible function of BnCDPs in stress resistance, the analysis of cis-acting regulatory elements in the promoter regions was conducted in this study. The results revealed the enrichment of elements associated with development, hormone and stress (Figure 4, Supplementary Figure 2 and Supplementary Table 5), implying that the members of the BnCDP family are potentially involved in biotic and abiotic stress responses during plant growth and development. Consistently, many members in the BnCDP family were significantly responsive to one or more stresses according to the transcriptome data under one biotic stress (S. sclerotiorum infection) and four abiotic stresses (dehydration, cold, ABA and salinity). Furthermore, six BnCDP genes were commonly regulated by S. sclerotiorum infection and all the four abiotic stresses, which belong to multiple biotic and abiotic stress-responsive genes. Similar results were previously obtained that several members in both barley HvGER family (Zimmermann et al., 2006) and Peanut AhGLP family (Wang et al., 2013) appeared to participate in multiple biotic and abiotic stress responses. Notably, the bicupins from subfamily IV–VI are also responsive to multiple stresses, such as BnaCnng28630D in subfamily IV responsive to dehydration treatment, BnaA10g02240D in subfamily V responsive to dehydration, cold and ABA treatments, BnaC05g05380D in subfamily VI responsive to dehydration, cold treatments and S. sclerotiorum infection. These results suggested that some other BnCDP genes apart from BnGLPs may also have important functions in environmental adaption, and some of them were promising broad-spectrum stress resistance candidates with tremendous potential in improving crop resistance to different stresses. To further examine the potential roles of BnCDP genes in S. sclerotiorum resistance, we performed a GWAS analysis. The results showed that five BnGLPs are located in the significant associated regions, including a duplicate gene pair BnGLP1.A08 and BnGLP1.C08, whose response patterns to different biotic and abiotic stresses were validated by qPCR experiment. The results support the conclusion that the BnCDP family members are widely involved in environmental adaption of B. napus. This study provides a useful resource for future research on the biological function and evolutionary history of the BnCDP gene family.
Conclusion
In this study, the cupin_1 domain protein (CDP) gene family in B. napus was genome widely characterized and systematically investigated. In total, 96 BnCDP genes were identified and clustered into six distinct subfamilies (I–VI) based on their evolutionary relationships. Genes from the same subfamily have similar gene structure and motif distribution, which are more conserved in subfamily I–III (BnGLP) than in subfamily IV–VI. To better understand their potential functional roles, we analyzed the cis-regulatory elements and TFBSs in the promoters of BnCDPs, as well as their expression patterns in diverse tissues/stages and under various biotic and abiotic stresses. The results demonstrated that the BnCDP family members play important roles in plant development and stress tolerance, particularly the six genes commonly regulated by S. sclerotiorum infection and four abiotic stresses, which may serve as promising broad-spectrum stress tolerance candidates. GWAS on S. sclerotiorum resistance revealed that two (BnGLP1.A08 and BnGLP.C08) of the six common stress response candidate genes were located in significant associated regions, and their expression patterns under different biotic and abiotic stress treatments were further validated by qPCR analysis. In summary, this study provides detailed information about BnCDPs in B. napus, and will facilitate the functional studies and genetic improvement to deal with different stresses.
Materials and methods
Identification of BnCDP gene family in Brassica napus
The genome (v4.1) and annotation (v5) information of the B. napus cultivar “Darmor-bzh” (Chalhoub et al., 2014) was obtained from the Brassicaceae Database (BRAD2). To identify BnCDPs in B. napus, PF00190 from the Pfam database3 (Mistry et al., 2021) was used as a query to search in the entire protein database of B. napus using HMMER 3.3.24 (Mistry et al., 2013; the e-value was set to 1 × 10–5). Then, all putative BnCDPs identified were subjected to the NCBI Conserved Domain Database5 (Lu et al., 2020) and the SMART database6 (Letunic et al., 2021) to verify the presence of cupin_1 domain. Moreover, the peptide length, molecular weight (MW), isoelectric point (pI), instability index, gravy of each BnCDP proteins were calculated using ProtParam7 (Wilkins et al., 1999), an online software of SWISS-PROT. Subcellular location prediction for these BnCDP proteins was conducted using CELLO v2.58 (Yu et al., 2006). The physical locations on the chromosomes and the exon number of the BnCDP genes were obtained from the GFF3 annotation file of the B. napus genome using a custom shell script.
Phylogenetic analysis
To gain insights into the evolutionary relationships of BnCDP genes in B. napus, the amino acid sequences of all 96 BnCDPs identified in this study, combined with 32 previously identified A. thaliana GLPs (Li L. et al., 2016) and 10 newly identified cupins, were subjected to multiple sequence alignment using ClustalW2 program (Larkin et al., 2007) with default parameters. Then, MEGA 11 (Tamura et al., 2021) software was used to generate the phylogenetic tree using the neighbor-joining (NJ) method with 1,000 bootstrap replicates. The final phylogenetic tree was visualized using iTOL v6.9 The BnCDP genes were further categorized into different subfamilies based on the topology of the phylogenetic tree.
Chromosomal distribution and gene duplication analysis of BnCDP genes
To identify gene duplication events, BLASTP with the e-value of 1e–10 was used to align the sequence, and MCScanX (Wang et al., 2012) was used to detect the duplication patterns including segmental and tandem duplication. Chromosomal locations and duplication events were visualized using the TBtools software (Chen et al., 2020). To determine the evolutionary pressure on duplicated genes, the ratio of non-synonymous substitution to synonymous substitution (Ka/Ks) of duplicate gene pairs was calculated using TBtools (Chen et al., 2020).
Gene structure, conserved motif, and cis-regulatory element analysis
The conserved motif analysis of BnCDPs was conducted using the online motif finding tool, MEME (Multiple Expectation Maximization for Motif Elicitation, v5.4.110; Bailey et al., 2015) with 20 motif numbers, and the remaining parameters were set to default values. The identified motifs were annotated by using the Interpro database.11 The TBtools (Chen et al., 2020) software was used to display the gene structures and conserved motifs in BnCDP proteins. To identify the cis-regulatory elements of BnCDP genes, the promoters (2-kb upstream sequences from initiation codon) of BnCDPs were extracted and predicted by PlantCARE12 (Lescot et al., 2002). The location and type of each selected cis-regulatory element were displayed by Gene Structure Display Server (GSDS 2.0; Hu et al., 2015). Besides, the transcription factor binding sites (TFBSs) in the promoter region of BnCDP genes were predicted using PlantRegMap/PlantTFDB v5.013 and the threshold p-value was set to 1e-7.
Expression analysis of BnCDP genes in Brassica napus
To explore the spatial-temporal expression patterns of BnCDPs, transcriptome data from 30 tissues/stages which include leaf, root, stem, bud, stamen, new pistil, blossomy pistil, wilted pistil, sepal, ovule and ten time-course seeds and silique walls (4, 8, 12, 16, 20, 24, 28, 32, 40, 48 days after pollination) of ZS11 were used in this study (He et al., 2022). Furthermore, in order to detect the expression patterns of BnCDP genes under biotic and abiotic stresses, RNA-seq data from tolerant B. napus cultivar ZY821 under the induction of S. sclerotiorum fungi and different abiotic stress conditions (dehydration, cold, ABA and salt) of B. napus cultivar ZS11 were also used in this study (Girard et al., 2017; Zhang et al., 2019). Then, the RNA-seq reads from each sample were aligned to the reference genome of Darmor-bzh (v4.1) using Hisat2 (Kim et al., 2015). Subsequently, the expression levels of BnCDP genes were calculated with Stringtie (Pertea et al., 2015) and displayed by Pheatmap in R.
Ribonucleic acid isolation and quantitative real-time PCR analysis of BnCDP genes
For S. sclerotiorum inoculation, the seedlings of ZY821 were kept growing in greenhouse at 22°C with a 16-h light and 8-h dark photoperiod for 6 weeks. S. sclerotiorum isolate obtained from Wuhan field was cultured on potato dextrose agar (PDA) medium and sub-cultured twice before inoculation at 22°C in darkness. Mycelial agar plug (7 mm in diameter) punched from the margin of a 2-day-old culture of S. sclerotiorum grown on PDA was carefully upended onto the adaxial surface of the latest or penultimate fully extended leaves with similar size. The inoculated plants were placed in a humidification chamber with high relative humidity (>85%) and samples were taken every 12 h. For abiotic stress treatments, the seeds of ZS11 were sterilized in 75% ethanol for 1 min, in 3.1% NaOCl for 10 min, and then rinsed six times with sterile water. Next, the seeds were sowed on Murashige and Skoog (MS) medium (MS, 1% sucrose, 0.7% agar, pH 5.8) in plates. The plates were placed vertically in the growth chamber with the temperature of 22°C and photoperiod of 16 h/8 h day/night. Two-week-old uniform plants/seedlings were removed from MS medium and subjected to dehydration, low temperature (4°C), ABA (25 uM), and salt (200 mM) stress treatment according to previously described methods (Zhang et al., 2019). Whole seedlings were collected at 1 h and 4 h after dehydration while 4 h and 24 h of low temperature, ABA, and salt treatment. All the samples mentioned above were flash frozen in liquid nitrogen and store at –80°C. Total RNA was extracted using the TRIZOL reagent (Invitrogen) according to the manufacturer’s protocol and subjected to reverse transcription with the PrimeScript RT Reagent Kit with genomic DNA Eraser (Takara). Quantitative Real-time PCR (qRT-PCR) was performed by using SYBR Green Real-time PCR Master Mix (Bio-Rad) in 20 ml reaction mixture and run on CFX96 Real-time PCR system (Bio-Rad). β-actin gene was used as internal control. All the results were obtained with three biological replications, and each with three technical replications. The results were analyzed using the 2–ΔΔCT method as described previously (Livak and Schmittgen, 2001). The list of all the primers used in this study is included in Supplementary Table 11.
Genome wide association study on Sclerotinia sclerotiorum resistance
We selected 274 B. napus core germplasm accessions from all over the world to form a natural population. The genotypic data were obtained by 7 × re-sequencing and referring to the genome of ‘Darmor-bzh’ (Ding et al., 2020). SNPs were tested using the Broad Institute’s opensource Genome Analysis Toolkit.14 Then, the sites with SNP deletion of more than 0.9 or with minor allele frequency (MAF) less than 0.05 were filtered using VCFtools (Danecek et al., 2011), and finally 2,381,566 SNPs were obtained for GWAS. The phenotypic data were collected by investigating the disease index of mature rapeseeds grown at the Yangluo test base (Wuhan, China) from 2015 to 2018 (Ding et al., 2020). GWAS was performed for S. sclerotiorum resistance using Fixed and random model Circulating Probability Unification (FarmCPU) model (Yin et al., 2021), and the significance threshold was set to p < 4.199 × 10–7.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YH, YL, and YZ designed the research. SL supervised the research. YH, YL, RZ, and JL performed the experiments. YH, YL, ZB, and MX analyzed the data. CT collected the data. XC and YYL provided the plant materials. YH and YL wrote the manuscript. YZ revised the manuscript. All authors have read and approved the current version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (2018YFE0108000), the Central Public-interest Scientific Institution Basal Research Fund (No. Y2020YJ03), the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ZDRW202105 and CAAS-ASTIP-2013-OCRI), the Young Top-notch Talent Cultivation Program of Hubei Province, and the National Natural Science Foundation of China (31770250).
Acknowledgments
We thank all the laboratory members for investigating the phenotype of Brassica napus population. We also thank Minqiang Tang at the college of forestry of Hainan university for analyzing the SNP data. The numerical calculations in this manuscript have been done on the supercomputing system in the Supercomputing Center of Oil Crops Research Institute, Chinese Academy of Agricultural Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.983786/full#supplementary-material
Supplementary Figure 1. Chromosomal (chr) localization of 96 BnCDP genes on Brassica napus chromosomes and contigs. The different colors in chromosomes represent gene density, and BnCDP genes from different subfamilies are represented in different colors.
Supplementary Figure 2. Number of cis-acting regulatory elements in promoters of BnCDP genes in Brassica napus. The color scale represents amounts from low (blue color) to high (red color).
Supplementary Figure 3. Venn diagrams analysis of the stress-responsive BnCDP genes under different treatments. (A) Venn diagram shows overlapping responsive genes in BnCDP family among different abiotic stresses. (B) Venn diagram shows overlapping responsive genes in BnCDP family under S. sclerotiorum inoculation and four abiotic stress treatments.
Footnotes
- ^ https://www.fao.org/faostat/
- ^ http://brassicadb.cn/
- ^ http://pfam.xfam.org
- ^ http://www.hmmer.org/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://smart.embl.de/
- ^ https://web.expasy.org/protparam/
- ^ http://cello.life.nctu.edu.tw/
- ^ https://itol.embl.de/
- ^ https://meme-suite.org/meme/
- ^ https://www.ebi.ac.uk/interpro/search/sequence/
- ^ https://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://plantregmap.gao-lab.org/binding_site_prediction.php
- ^ https://software.broadinstitute.org/gatk/
References
Agarwal, G., Rajavel, M., Gopal, B., and Srinivasan, N. (2009). Structure-based phylogeny as a diagnostic for functional characterization of proteins with a cupin fold. PLoS One 4:e5736. doi: 10.1371/journal.pone.0005736
Albalat, R., and Canestro, C. (2016). Evolution by gene loss. Nat. Rev. Genet. 17, 379–391. doi: 10.1038/nrg.2016.39
Allender, C. J., and King, G. J. (2010). Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10:54. doi: 10.1186/1471-2229-10-54
Anum, J., O’Shea, C., Zeeshan Hyder, M., Farrukh, S., Skriver, K., Malik, S. I., et al. (2022). Germin like protein genes exhibit modular expression during salt and drought stress in elite rice cultivars. Mol. Biol. Rep. 49, 293–302. doi: 10.1007/s11033-021-06871-3
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Banerjee, J., and Maiti, M. K. (2010). Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem. Biophys. Res. Commun. 394, 178–183. doi: 10.1016/j.bbrc.2010.02.142
Banerjee, J., Das, N., Dey, P., and Maiti, M. K. (2010). Transgenically expressed rice germin-like protein1 in tobacco causes hyper-accumulation of H2O2 and reinforcement of the cell wall components. Biochem. Biophys. Res. Commun. 402, 637–643. doi: 10.1016/j.bbrc.2010.10.073
Beracochea, V. C., Almasia, N. I., Peluffo, L., Nahirñak, V., Hopp, E. H., Paniego, N., et al. (2015). Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell Rep. 34, 1717–1733. doi: 10.1007/s00299-015-1819-4
Brunetti, S. C., Arseneault, M., and Gulick, P. J. (2022). Characterization and expression of the Pirin gene family in Triticum aestivum. Genome doi: 10.1139/gen-2021-0094 [Epub ahead of print].
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, D., Tan, M., Yu, H., Li, L., Zhu, D., Chen, Y., et al. (2018). Comparative analysis of Cd-responsive maize and rice transcriptomes highlights Cd co-modulated orthologs. BMC Genomics 19:709. doi: 10.1186/s12864-018-5109-8
Cheng, X., Huang, X., Liu, S., Tang, M., Hu, W., and Pan, S. (2014). Characterization of germin-like protein with polyphenol oxidase activity from Satsuma mandarine. Biochem. Biophys. Res. Commun. 449, 313–318. doi: 10.1016/j.bbrc.2014.05.027
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Davidson, R. M., Manosalva, P. M., Snelling, J., Bruce, M., Leung, H., and Leach, J. E. (2010). Rice germin-like proteins: allelic diversity and relationships to early stress responses. Rice 3, 43–55. doi: 10.1007/s12284-010-9038-7
Ding, L. N., Li, M., Guo, X. J., Tang, M. Q., Cao, J., Wang, Z., et al. (2020). Arabidopsis GDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus. Plant Biotechnol. J. 18, 1255–1270. doi: 10.1111/pbi.13289
Dong, X., Ji, R., Guo, X., Foster, S. J., Chen, H., Dong, C., et al. (2008). Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 228, 331–340. doi: 10.1007/s00425-008-0740-2
Dunwell, J. M., Gibbings, J. G., Mahmood, T., and Saqlan Naqvi, S. M. (2008). Germin and germin-like proteins: evolution, structure, and function. Crit. Rev. Plant Sci. 27, 342–375. doi: 10.1080/07352680802333938
Dunwell, J. M., Khuri, S., and Gane, P. J. (2000). Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153–179. doi: 10.1128/MMBR.64.1.153-179.2000
Dunwell, J. M., Purvis, A., and Khuri, S. (2004). Cupins: the most functionally diverse protein superfamily? Phytochemistry 65, 7–17. doi: 10.1016/j.phytochem.2003.08.016
Gangadhar, B. H., Mishra, R. K., Kappachery, S., Baskar, V., Venkatesh, J., Nookaraju, A., et al. (2021). Enhanced thermo-tolerance in transgenic potato (Solanum tuberosum L.) overexpressing hydrogen peroxide-producing germin-like protein (GLP). Genomics 113, 3224–3234. doi: 10.1016/j.ygeno.2021.07.013
Girard, I. J., Tong, C., Becker, M. G., Mao, X., Huang, J., de Kievit, T., et al. (2017). RNA sequencing of Brassica napus reveals cellular redox control of Sclerotinia infection. J. Exp. Bot. 68, 5079–5091. doi: 10.1093/jxb/erx338
Gucciardo, S., Wisniewski, J. P., Brewin, N. J., and Bornemann, S. (2007). A germin-like protein with superoxide dismutase activity in pea nodules with high protein sequence identity to a putative rhicadhesin receptor. J. Exp. Bot. 58, 1161–1171. doi: 10.1093/jxb/erl282
Guevara-Olvera, L., Ruíz-Nito, M. L., Rangel-Cano, R. M., Torres-Pacheco, I., Rivera-Bustamante, R. F., Muñoz-Sánchez, C. I., et al. (2012). Expression of a germin-like protein gene (CchGLP) from a geminivirus-resistant pepper (Capsicum chinense Jacq.) enhances tolerance to geminivirus infection in transgenic tobacco. Physiol. Mol. Plant P. 78, 45–50. doi: 10.1016/j.pmpp.2012.01.005
He, Y., Yang, Z., Tang, M., Yang, Q. Y., Zhang, Y., and Liu, S. (2022). Enhancing canola breeding by editing a glucosinolate transporter gene lacking natural variation. Plant Physiol. 188, 1848–1851. doi: 10.1093/plphys/kiac021
He, Z. D., Tao, M. L., Leung, D., Yan, X. Y., Chen, L., Peng, X. X., et al. (2021). The rice germin-like protein OsGLP1 participates in acclimation to UV-B radiation. Plant Physiol. 186, 1254–1268. doi: 10.1093/plphys/kiab125
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Ilyas, M., Rahman, A., Khan, N. H., Haroon, M., Hussain, H., Rehman, L., et al. (2022). Analysis of germin-like protein genes family in Vitis vinifera (VvGLPs) using various in silico approaches. Braz. J. Biol. 84:e256732. doi: 10.1590/1519-6984.256732
Jin, J., Zhang, H., Kong, L., Gao, G., and Luo, J. (2014). PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42, D1182–D1187. doi: 10.1093/nar/gkt1016
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Knecht, K., Seyffarth, M., Desel, C., Thurau, T., Sherameti, I., Lou, B., et al. (2010). Expression of BvGLP-1 encoding a germin-like protein from sugar beet in Arabidopsis thaliana leads to resistance against phytopathogenic fungi. Mol. Plant Microbe Interact. 23, 446–457. doi: 10.1094/MPMI-23-4-0446
Lane, B. G. (2002). Oxalate, germins, and higher-plant pathogens. IUBMB Life 53, 67–75. doi: 10.1080/15216540211474
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, L., Xu, X., Chen, C., and Shen, Z. (2016). Genome-Wide characterization and expression analysis of the germin-like protein family in rice and Arabidopsis. Int. J. Mol. Sci. 17:1622. doi: 10.3390/ijms17101622
Li, Y., Zhang, D., Li, W., Mallano, A. I., Zhang, Y., Wang, T., et al. (2016). Expression study of soybean germin-like gene family reveals a role of GLP7 gene in various abiotic stress tolerances. Can. J. Plant Sci. 96, 296–304. doi: 10.1139/cjps-2015-0213
Liao, L., Hu, Z., Liu, S., Yang, Y., and Zhou, Y. (2021). Characterization of germin-like proteins (GLPs) and their expression in response to abiotic and biotic stresses in cucumber. Horticulturae 7:412. doi: 10.3390/horticulturae7100412
Liu, D., Wu, J., Lin, L., Li, P., Li, S., Wang, Y., et al. (2021). Overexpression of Cinnamoyl-CoA Reductase 2 in Brassica napus increases resistance to Sclerotinia sclerotiorum by affecting lignin biosynthesis. Front. Plant Sci. 12:732733. doi: 10.3389/fpls.2021.732733
Liu, Q., Yang, J., Yan, S., Zhang, S., Zhao, J., Wang, W., et al. (2016). The germin-like protein OsGLP2-1 enhances resistance to fungal blast and bacterial blight in rice. Plant Mol. Biol. 92, 411–423. doi: 10.1007/s11103-016-0521-4
Liu, S., Fitt, B. L., Liu, R., Evans, N., Dong, C., and Huang, Y. (2004). Cloning of plant defensin and oxalic acid oxidase genes and expressions of these genes induced by pathogens and chemicals in Brassica napus. Chin. J. Oil Crop Sci. 26:3.
Liu, S., Raman, H., Xiang, Y., Zhao, C., Huang, J., and Zhang, Y. (2022). De novo design of future rapeseed crops: challenges and opportunities. Crop J. 10, 587–596. doi: 10.1016/j.cj.2022.05.003
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, M., Han, Y. P., Gao, J. G., Wang, X. J., and Li, W. B. (2010). Identification and analysis of the germin-like gene family in soybean. BMC Genomics 11:620. doi: 10.1186/1471-2164-11-620
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Ma, J. Q., Jian, H. J., Yang, B., Lu, K., Zhang, A. X., Liu, P., et al. (2017). Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus L.). Gene 620, 36–45. doi: 10.1016/j.gene.2017.03.030
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., and Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41:e121. doi: 10.1093/nar/gkt263
Nakata, M., Watanabe, Y., Sakurai, Y., Hashimoto, Y., Matsuzaki, M., Takahashi, Y., et al. (2004). Germin-like protein gene family of a moss, Physcomitrella patens, phylogenetically falls into two characteristic new clades. Plant Mol. Biol. 56, 381–395. doi: 10.1007/s11103-004-3475-x
Oudelaar, A. M., and Higgs, D. R. (2021). The relationship between genome structure and function. Nat. Rev. Genet. 22, 154–168. doi: 10.1038/s41576-020-00303-x
Pei, Y., Li, X., Zhu, Y., Ge, X., Sun, Y., Liu, N., et al. (2019). GhABP19, a novel germin-like protein from Gossypium hirsutum, plays an important role in the regulation of resistance to Verticillium and Fusarium wilt pathogens. Front. Plant Sci. 10:583. doi: 10.3389/fpls.2019.00583
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Raza, A., Razzaq, A., Mehmood, S. S., Hussain, M. A., Wei, S., He, H., et al. (2021). Omics: the way forward to enhance abiotic stress tolerance in Brassica napus L. GM Crops Food 12, 251–281. doi: 10.1080/21645698.2020.1859898
Rietz, S., Bernsdorff, F. E., and Cai, D. (2012). Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 63, 5507–5519. doi: 10.1093/jxb/ers203
Sakamoto, A., Nishimura, T., Miyaki, Y. I., Watanabe, S., Takagi, H., Izumi, S., et al. (2015). In vitro and in vivo evidence for oxalate oxidase activity of a germin-like protein from azalea. Biochem. Biophys. Res. Commun. 458, 536–542. doi: 10.1016/j.bbrc.2015.02.002
Sun, M., Ye, Z., Tan, J., Chen, S., Zhang, X., and Tu, L. (2020). A cotton germin-like protein GbGLP2 controls fiber length via regulating genes involved in secondary cell wall synthesis. Mol. Breed. 40:98. doi: 10.1007/s11032-020-01177-x
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., and Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. doi: 10.1111/nph.12797
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thompson, C., Dunwell, J. M., Johnstone, C. E., Lay, V., Ray, J., Schmitt, M., et al. (1995). Degradation of oxalic acid by transgenic oilseed rape plants expressing oxalate oxidase. Euphytica 85, 169–172. doi: 10.1007/BF00023945
Thompson, E. W., and Lane, B. G. (1980). Relation of protein synthesis in imbibing wheat embryos to the cell-free translational capacities of bulk mRNA from dry and imbibing embryos. J. Biol. Chem. 255, 5965–5970. doi: 10.1016/s0021-9258(19)70725-x
Wang, H., Zhang, Y., Xiao, N., Zhang, G., Wang, F., Chen, X., et al. (2020). Rice GERMIN-LIKE PROTEIN 2-1 functions in seed dormancy under the control of abscisic acid and gibberellic acid signaling pathways. Plant Physiol. 183, 1157–1170. doi: 10.1104/pp.20.00253
Wang, T., Chen, X., Zhu, F., Li, H., Li, L., Yang, Q., et al. (2013). Characterization of peanut germin-like proteins, AhGLPs in plant development and defense. PLoS One 8:e61722. doi: 10.1371/journal.pone.0061722
Wang, X., Zhang, H., Gao, Y., Sun, G., Zhang, W., and Qiu, L. (2014). A comprehensive analysis of the Cupin gene family in soybean (Glycine max). PLoS One 9:e110092. doi: 10.1371/journal.pone.0110092
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7
Woo, E. J., Dunwell, J. M., Goodenough, P. W., Marvier, A. C., and Pickersgill, R. W. (2000). Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7, 1036–1040. doi: 10.1038/80954
Wu, Y., Ke, Y., Wen, J., Guo, P., Ran, F., Wang, M., et al. (2018). Evolution and expression analyses of the MADS-box gene family in Brassica napus. PLoS One 13:e0200762. doi: 10.1371/journal.pone.0200762
Xie, M., Zuo, R., Bai, Z., Yang, L., Zhao, C., Gao, F., et al. (2022). Genome-wide characterization of serine/arginine-rich gene family and its genetic effects on agronomic traits of Brassica napus. Front. Plant Sci. 13:829668. doi: 10.3389/fpls.2022.829668
Yin, L., Zhang, H., Tang, Z., Xu, J., Yin, D., Zhang, Z., et al. (2021). RMVP: a memory-efficient, visualization-enhanced, and parallel-accelerated tool for genome-wide association study. Genom. Proteom. Bioinf. 19, 619–628. doi: 10.1016/j.gpb.2020.10.007
Yu, C. S., Chen, Y. C., Lu, C. H., and Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins 64, 643–651. doi: 10.1002/prot.21018
Yuan, B., Yang, Y., Fan, P., Liu, J., Xing, H., Liu, Y., et al. (2021). Genome-wide identification and characterization of germin and germin-like proteins (GLPs) and their response under powdery mildew stress in wheat (Triticum aestivum L.). Plant Mol. Biol. Rep. 39, 821–832. doi: 10.1007/s11105-021-01291-w
Zaynab, M., Peng, J., Sharif, Y., Fatima, M., Albaqami, M., Al-Yahyai, R., et al. (2021). Genome-wide identification and expression profiling of germin-like proteins reveal their role in regulating abiotic stress response in potato. Front. Plant Sci. 12:831140. doi: 10.3389/fpls.2021.831140
Zhang, Y., Ali, U., Zhang, G., Yu, L., Fang, S., Iqbal, S., et al. (2019). Transcriptome analysis reveals genes commonly responding to multiple abiotic stresses in rapeseed. Mol. Breed. 39:158. doi: 10.1007/s11032-019-1052-x
Zhang, Y., Wang, X., Chang, X., Sun, M., Zhang, Y., Li, W., et al. (2018). Overexpression of germin-like protein GmGLP10 enhances resistance to Sclerotinia sclerotiorum in transgenic tobacco. Biochem. Biophys. Res. Commun. 497, 160–166. doi: 10.1016/j.bbrc.2018.02.046
Zhao, Y., Zhu, X., Chen, X., and Zhou, J. M. (2022). From plant immunity to crop disease resistance. J. Genet. Genomics doi: 10.1016/j.jgg.2022.06.003 [Epub ahead of print].
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zimmermann, G., Baumlein, H., Mock, H. P., Himmelbach, A., and Schweizer, P. (2006). The multigene family encoding germin-like proteins of barley. Regulation and function in Basal host resistance. Plant Physiol. 142, 181–192. doi: 10.1104/pp.106.083824
Keywords: cupin_1 domain, germin-like protein, Sclerotinia sclerotiorum resistance, abiotic stress, GWAS, Brassica napus
Citation: He Y, Li Y, Bai Z, Xie M, Zuo R, Liu J, Xia J, Cheng X, Liu Y, Tong C, Zhang Y and Liu S (2022) Genome-wide identification and functional analysis of cupin_1 domain-containing members involved in the responses to Sclerotinia sclerotiorum and abiotic stress in Brassica napus. Front. Plant Sci. 13:983786. doi: 10.3389/fpls.2022.983786
Received: 01 July 2022; Accepted: 11 July 2022;
Published: 01 August 2022.
Edited by:
Cunmin Qu, Southwest University, ChinaReviewed by:
Jinpeng Wang, Institute of Botany (CAS), ChinaZongxiang Zhan, Shenyang Agricultural University, China
Xiaodong Wang, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2022 He, Li, Bai, Xie, Zuo, Liu, Xia, Cheng, Liu, Tong, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyuan Zhang, emhhbmd5eUBjYWFzLmNu
†These authors have contributed equally to this work and share first authorship
 Yizhou He
Yizhou He Yan Li2,3†
Yan Li2,3† Meili Xie
Meili Xie Rong Zuo
Rong Zuo Jie Liu
Jie Liu Chaobo Tong
Chaobo Tong Yuanyuan Zhang
Yuanyuan Zhang Shengyi Liu
Shengyi Liu