
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 September 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.983650
This article is part of the Research Topic Dissection of the Genetic and Molecular Basis of the Physiological Processes Crucial for Improving Yield, Quality, and Resistance in Oil Crops View all 5 articles
 Zhiyong Xu1,2
Zhiyong Xu1,2 Ruikai Wang1
Ruikai Wang1 Keke Kong1
Keke Kong1 Naheeda Begum1
Naheeda Begum1 Aisha Almakas1
Aisha Almakas1 Jun Liu2
Jun Liu2 Hongyu Li2
Hongyu Li2 Bin Liu2
Bin Liu2 Tuanjie Zhao1*
Tuanjie Zhao1* Tao Zhao2*
Tao Zhao2*Plant height is one of the key agronomic traits affecting soybean yield. The cytokinin response factors (CRFs), as a branch of the APETALA2/ethylene responsive factor (AP2/ERF) super gene family, have been reported to play important roles in regulating plant growth and development. However, their functions in soybean remain unknown. This study characterized a soybean CRF gene named GmCRF4a by comparing the performance of the homozygous Gmcrf4a-1 mutant, GmCRF4a overexpression (OX) and co-silencing (CS) lines. Phenotypic analysis showed that overexpression of GmCRF4a resulted in taller hypocotyls and epicotyls, more main stem nodes, and higher plant height. While down-regulation of GmCRF4a conferred shorter hypocotyls and epicotyls, as well as a reduction in plant height. The histological analysis results demonstrated that GmCRF4a promotes epicotyl elongation primarily by increasing cell length. Furthermore, GmCRF4a is required for the expression of GmYUCs genes to elevate endogenous auxin levels, which may subsequently enhance stem elongation. Taken together, these observations describe a novel regulatory mechanism in soybean, and provide the basis for elucidating the function of GmCRF4a in auxin biosynthesis pathway and plant heigh regulation in plants.
Soybean [Glycine max (L.) Merr.] is an economically important crop cultivated globally, providing a valuable source of cooking oil and plant protein for human consumption (Lee et al., 2011). Although, the global soybean production has gradually increased, the yield per unit of soybean is relatively low compared to other cereal crops including wheat, maize, and rice, which have achieved great success in developing ideal plant types (Liu et al., 2020; Vogel et al., 2021). It has been noted that genetic dissection and the use of specific plant-type traits, including plant height, can improve the efficiency of soybean high-yield breeding (Liu et al., 2020). Plant height, as a complex quantitative trait, is governed by abundant genes and a variety of hormones, such as gibberellin, auxin, and cytokinin (Wang et al., 2017, 2021; Zhang et al., 2017; Dong et al., 2019; Liao et al., 2019; Castorina and Consonni, 2020; Bai et al., 2021). During the last century’s “Green Revolution,” the Rht1 (Reduced height 1) gene of wheat (Rockwell and Hongay, 2019), the Sd1 (Semi-dwarf 1) gene of rice (Ren et al., 2019), and the D8 (Dwarf-8) gene of maize (Huang et al., 2019) all regulated plant height by adjusting gibberellic acid (GA) contents. In addition to GA pathway, genes affecting plant height via the auxin pathway have also been identified in several crops. For example, overexpression of the ZmPIN1a (PIN-FORMED1a) gene in maize reduces plant height, internode length, and ear height (Li et al., 2018a). Knocking out the members of the TRANSPORT INHIBITOR RESISTANT1/AUXIN SIGNALING F-box (OsTIR1/AFB) gene family alters plant height and yield in rice (Guo et al., 2021). Growth habit is an agronomically important trait associated with the domestication in soybean. According to the termination time of apical stem growth, most soybeans can be divided into two growth habits: indeterminate and determinate, which brings about changes in plant height and yield. Subsequently, the key genes Determinate 1 (Dt1) and Dt2 genes regulating plant height and growth habits of soybean were found (Tian et al., 2010; Ping et al., 2014). Deletion of Dwarf Mutant 1 (GmDW1) (Li et al., 2018b), GmAP1s (Apetala1) (Chen et al., 2020), and GmLHY (Late elongated hypocotyl) (Cheng et al., 2019) genes in soybean can also regulate plant height by changing endogenous GA content. Overexpression of both an APETALA2-like gene GmTOE4a and an ABI3/VP1 gene GmRAV can cause reduced plant height and internode length with lower GA levels in soybean plants (Zhao et al., 2015; Xue et al., 2022). Nevertheless, few auxin related genes have been reported to affect plant height in soybean.
The AP2/ERF superfamily, as one of the largest transcription factor families in plants, can be divided into five subfamilies: APETALA2 (AP2), RAV (ABI3/VP1), Dehydration Responsive Element Binding Protein, ethylene responsive factor (ERF) and Soloist according to the number and binding sequence of AP2/ERF domains (Nakano et al., 2006; Wang, 2019). The AP2/ERF family genes play multiple roles in regulating plant defense responses to stress as well as growth and development (Licausi et al., 2013; Chen et al., 2015; Gan et al., 2019; Ma et al., 2020; Zhang et al., 2020; Zhao et al., 2020). For example, the cytokinin response factors (CRFs) proteins, as a branch of the ERF subfamily, play essential roles in plant growth and development, nitrogen uptake, biological and abiotic stress, and cytokinin and auxin response (Cutcliffe et al., 2011; Jeon et al., 2016; Kim, 2016; Zwack et al., 2016; Hallmark and Rashotte, 2019; Zong et al., 2021). In Arabidopsis, there are 12 CRF genes, with CRF4 being one of the few whose transcription is not induced by cytokinin (Rashotte et al., 2006; Cutcliffe et al., 2011; Zwack et al., 2012). The crf4 mutant was initially found to be sensitive to low temperature treatment (Zwack et al., 2016). While under normal conditions, crf4 mutants and CRF4 overexpression lines (CRF4OX) showed no obvious phenotypic alterations, except for a slight increase in primary root length of CRF4OX. In addition, under low nitrogen conditions, the CRF4 overexpression significantly reduced 15NO3− uptake and plant biomass, including reduced dry weight, primary root length, and lateral root number (Varala et al., 2018).
In this study, we tested the function of GmCRF4a gene in more detail by characterizing its expression patterns, subcellular localization and by GmCRF4a mutant, overexpressing, and co-silencing lines to dissect the roles of this transcription factor in regulation of plant height. We observed that the plant height of Gmcrf4a-1 mutant and co-silencing lines were significantly decreased than that of the WT. We also observed the relative expression levels of auxin biosynthesis genes and the auxin content in the Gmcrf4a-1 mutant and co-silencing lines were significantly lower than in WT. These results suggested that GmCRF4a may directly regulate plant height by mediating key components of the auxin pathway. Moreover, the phenotype of GmCRF4a overexpression lines further confirmed the above results. Our findings suggest that the manipulation of GmCRF4a gene should facilitate improvement in plant height in soybean.
The soybean cultivar Tianlong 1 (TL1) is an elite material for genetic transformation. For the seedling phenotypes, the wild type (TL1), GmCRF4a knockout mutants (Gmcrf4a-1), GmCRF4a co-silencing line (CS), and GmCRF4a overexpression lines (OX1 and OX2) were grown under 16 h light/8 h dark conditions or 12 h light/12 h dark conditions with 250–300 μmol·m−2 s−1 light at 25°C for 11 days. For the field phenotypes, the materials were grown under natural conditions (about 14.5 h-light conditions) in a field in Beijing (40.1° N, 116.7° E) with two plot replications. The seeds were planted in a 3.0 m row, with 0.6 m separating rows and a space of 0.3 m between adjacent plants. The plant height and node number of main stem were investigated and photographed during the period of co-silencing line (CS) R8, and the growth period and grain status of each material were counted after harvest.
The peptide sequences of AP2/ERF in soybean and Arabidopsis thaliana were selected with the Pfam name PF00847 (ap2 domain) in a threshold e-value < 10−5 from Phytozome v13.0.1 All the soybean AP2/ERF protein sequences were aligned with Multiple Sequence Alignment v7.273 (Katoh and Standley, 2013), and then back-translated into coding sequence by ParaAT (Zhang et al., 2012). At last, Maximum-Likelihood (ML) phylogenetic tree was generated using Fasttree for a larger number of sequences (Price et al., 2009). The detailed list is presented in Supplementary Table S1. For the CRF4 phylogenetic tree the GmCRF4a homologous genes in Arabidopsis, soybean, maize, and rice were used to constructed Maximum Likelihood tree by MEGA 7.0 software. For the protein sequence alignment and critical domain analysis were conducted by DNAMAN software.
Tissue-specific expression patterns data came from the soybean Expression Atlas.2 The median TPM per tissue-stir condition was selected, and the graphic was created by GraphPad Prism 9 software. Tissue-specific expression data is given in Supplementary Table S2.
The full-length cDNA of GmCRF4a was cloned into the pA7-YFP vector. GmCRF4a-YFP was transiently expressed in soybean mesophyll protoplasts as described in our previous work (Xiong et al., 2019). The pA7-GmMYB29-RFP plasmid was used as a nuclear marker (Chu et al., 2017). The fluorescence signal was observed under a confocal microscope (Zeiss LSM700) after 16 h of transformation at room temperature in the dark.
To generate the overexpression vectors, the full-length GmCRF4a was cloned into the Gateway entry vector pDONRZeo, and then fused into the destination binary vector pEarleyGate-101-eYFP vector using the Gateway recombination system (Invitrogen). To generate the CRISPR-Cas9 knockout soybean mutants, gRNAs were designed using the web tool CRISPR-P,3 and constructed into the p0645 vector. According to the previously described method (Lyu et al., 2021), the Agrobacterium-cotyledonary node transformation system was used to generate overexpression lines and gene editing mutants. To screen for homozygous mutants, transgenic lines were propagated by self-pollination for at least two generations. Homozygous mutations were identified by DNA sequencing (Supplementary Figure S4A). For CS lines, it was obtained by screening for co-suppression lines with the opposite phenotype to the overexpressed lines. GmCRF4a expression was detected by qPCR experiments.
For epicotyl longitudinal sections, seedlings were grown under 12 h light/12 h dark with 250–300 μmol·m−2 s−1 light at 25°C for 11 days and then stained with safranin-O-fast green by paraffin sections (De Micco and Aronne, 2007). The images were observed using a ZEISS Imager M2 (Jena, Germany) after staining. The cell size was calculated by ImageJ software.
The seedlings were grown under 12 h light/12 h dark conditions with 250–300 μmol·m−2 s−1 light at 25°C for 11 days. For auxin concentration measurement, five single plants’ epicotyls were measured. The IAA enzyme-linked immunoassay (ELISA) kit (Meimian, MM-0953O1, Yancheng, China) was used for the determination of auxin. Please see kit description for details.
Total RNA was extracted from 13-day old soybean leaves under 12 h light/12 h dark conditions using Trizol (Invitrogen, 15,596–026, California, United States). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed in 384-well optical plates on the QuantStudio 7 Flex (Massachusetts, United States) using SYBR Green RT-PCR kit (Vazyme, Q221-01, Nanjing, China). The GmActin11 was used as an internal control. Three independent biological replicates and two mechanical replicates were evaluated. Related genes and primer lists are presented in Supplementary Table S3.
Two-sided t-test and ANOVA were performed on the data using GraphPad Prism 9 software.
In our previous research we identified an AP2/ERF family gene GmCRF4a (Glyma.14G205600), which originally discovered as one of the fine-mapping candidate genes and turned out to be a non-target gene, involved in the regulation of important traits such as plant height and flowering in soybean. Therefore, we conducted further studies on this gene. GmCRF4a encodes a protein of 282 amino acid residues with a predicted molecular mass of 31.7 kDa. BLASP search of the Arabidopsis database revealed that GmCRF4a shares the highest identity with AT4G27950, Cytokinin Response Factor 4 (CRF4), which led us to designate Glyma.14G205600 as GmCRF4a, the first identified in Arabidopsis CRF4 homologue. Protein analysis of the deduced amino acid sequence revealed that GmCRF4a contains a typical AP2/ ERF domain. Further amino acid alignment analysis showed that GmCRF4a shares high similarity at AP2/ERF domain with the homologues in soybean (Glyma.04G238700, Glyma.13G040400, Glyma.02G236800, Glyma.19G253100, Glyma.03G255500, Glyma.03G094700, Glyma.20G215700, Glyma.05G214400, Glyma.11G199300, Glyma.11G019000, Glyma.14G205600, Glyma.14G123900, Glyma.08G020900, Glyma.01G224100, Glyma.06G125100 and Glyma.16G079600), maize (Zm00001d034605, Zm00001d036251, Zm00001d044004, Zm00001d039324, Zm00001d008968, Zm00001d011499, Zm00001d045262), rice (LOC_Os01g46870, LOC_Os01g12440, LOC_Os06g06540) and Arabidopsis (CRF1, CRF2, CRF3, CRF4, CRF5 and CRF6), (Supplementary Figure S1). Phylogenetic analysis showed that GmCRF4a shares 50–85% high identity with soybean homologues, and about 41% identity with the Arabidopsis genes (Supplementary Figure S2). Homologous genes in rice and maize clustered together independently, with lower homology to GmCRF4a. To determine subgroup of GmCRF4a in AP2/ERF gene family, 354 soybean AP2/ERF genes from cultivated soybean Williams 82 and 146 Arabidopsis AP2/ERF genes were selected for homology comparison, and the phylogenetic tree was constructed by referring to previous AP2 family classification methods (Wang, 2019). Cluster analysis revealed that the gene GmCRF4a (Figure 1A; Red Asterisk) belonged to the ERF-VI subfamily of AP2/ERF.
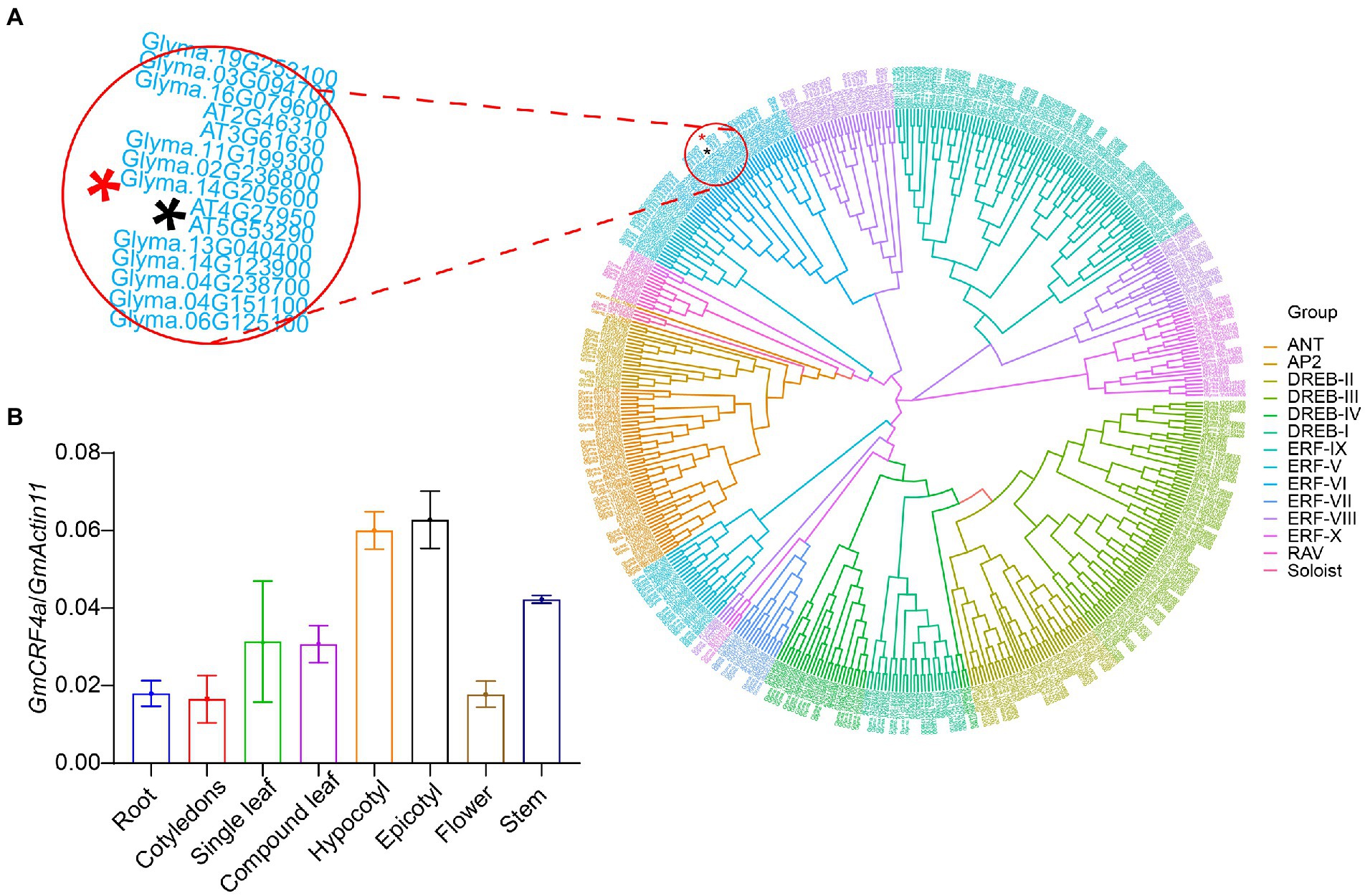
Figure 1. A phylogenetic tree and tissue-specific analysis of (A) homology analysis of AP2/ERF genes in soybean and Arabidopsis thaliana. These include 354 soybean genes and 146 Arabidopsis thaliana genes. Different colors are marked according to different subfamilies, where ANT represents AINTEGUMENTA. The black asterisk represents CRF4 in Arabidopsis thaliana; the red asterisk represents GmCRF4a in soybean. And the upper left corner is a partial enlargement. (B) Transcriptional levels of GmCRF4a in various tissues of TL1. GmActin11 as an internal control. Data are displayed as means ± s.d. (n = 3).
To get insight into the function of the GmCRF4a gene, we detected the expression level of GmCRF4a in different tissues under the background of TL1. The results showed that the GmCRF4a gene was evenly expressed in all tissues (Figure 1B), suggesting that GmCRF4a may be function as a transcriptional factor to regulate soybean growth and development of entire life cycle. Then we compared the GmCRF4a gene expression level among different tissues using the data accessed from the soybean Expression Atlas.4 The results were similar to the data provided by the qPCR, that is, GmCRF4a was expressed in all tissues (Supplementary Figure S3). To determine the subcellular localization of GmCRF4a, we transiently expressed the GmCRF4a-YFP fusion protein in soybean mesophyll protoplasts. The fluorescent images demonstrated that GmCRF4a-YFP was colocalized with nuclear markers (Figure 2), while the control protein YFP was only detectable in the intracellular region.
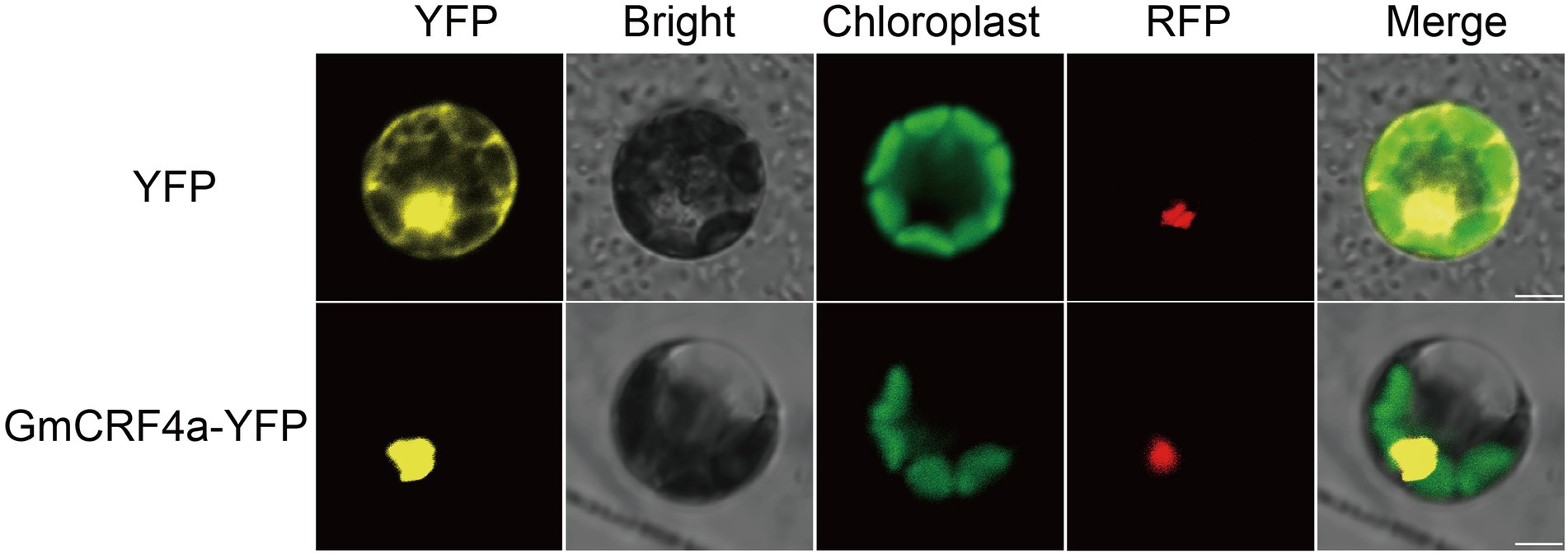
Figure 2. Subcellular localization of GmCRF4a-YFP fusion protein in soybean mesophyll protoplasts. The GmCRF4a-YFP fusion proteins were located in the nucleus. YFP was used as a control. The GmMYB29-RFP fusion protein was used as a nuclear marker Scale bars = 5 μm.
To dissect the role of GmCRF4a in soybean development, we knocked out the GmCRF4a gene by CRISPR/Cas9 genome editing technology. We generated a homozygous mutant line, Gmcrf4a-1, which carries two base deletions (ΔAC) in the exon resulting in premature termination of protein translation (Figures 3A,B; Supplementary Figure S4A). Furthermore, we made the 35S:GmCRF4a-YFP construct and obtained two overexpression lines (OX1 and OX2) and one co-silencing line (CS) using the Agrobacterium-cotyledonary node transformation system (Figure 3C; Supplementary Figure S4B). Subsequently, these lines were planted under 16 h light/8 h dark growth conditions and 12 light/12 dark growth conditions for phenotypic analysis. The results showed that, compared with the wild-type TL1, the OX1 and OX2 lines showed an increased plant height phenotype, with longer hypocotyls and epicotyls, under both 16 h light/8 h dark growth conditions or 12 light/12 dark growth conditions (Figures 3C,D). In sharp contrast, the plant height of the Gmcrf4a-1 mutant and CS line were significantly decreased in comparison to that of TL1 (Figures 3C,D). Under natural field conditions, OX1 line produced more main stem nodes (MSNs) and was taller than TL1, while the CS line was dwarf with less MSN (Supplementary Figures S5A–C). In addition, the growth period of the OX1 line was prolonged, while that of the CS line was shortened in field conditions (Supplementary Figure S5D). Notably, the overall phenotypes of the CS line are relatively stronger than that of the Gmcrf4a-1 mutant. This is possibly because not only the GmCRF4a gene but also its homologous genes Glyma.02G236800 and Glyma.14G123900 were co-silenced in the CS line (Supplementary Figures S6A,B). Taken together, these results suggested that the GmCRF4a gene positively regulates plant height by increasing stem elongation, node number, and growth period.
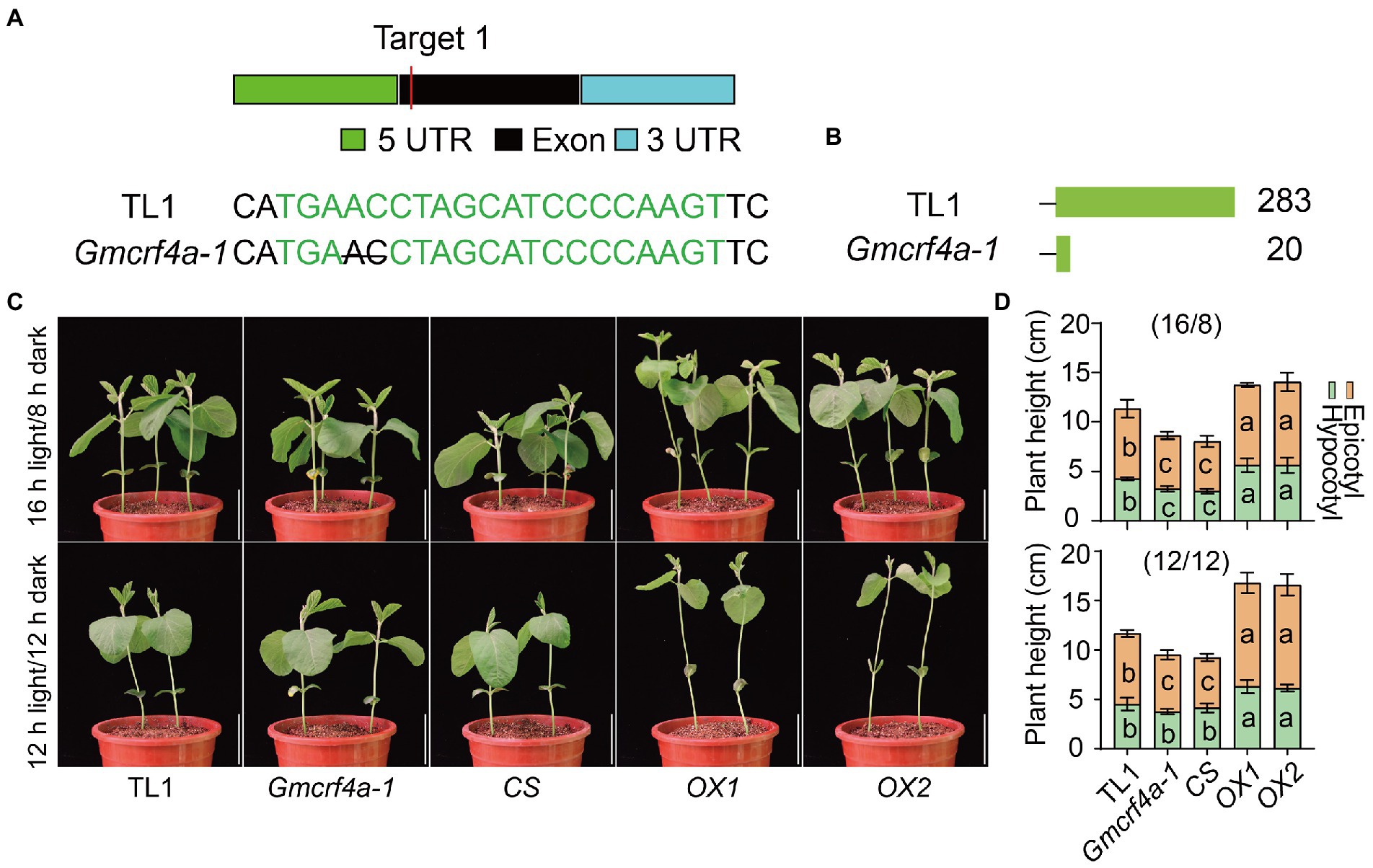
Figure 3. The phenotypes of wild type (TL1), GmCRF4a knockout mutants (Gmcrf4a-1), GmCRF4a co-silencing line (CS), and GmCRF4a overexpression lines (OX1 and OX2) were compared at the seedling stage. (A) The sgRNA designed to position and base change Gmcrf4a-1, the black line within the target site, denotes nucleotide deletion. (B) Schematic diagram comparing intact and mutated GmCRF4a proteins. The number of amino acids behind each protein. (C) Plants height of wild type (TL1), GmCRF4a knockout mutant (Gmcrf4a-1), GmCRF4a co-silencing line (CS), and GmCRF4a overexpression lines (OX1 and OX2) at 11 days under 16 h light/8 h dark or 12 h light/12 h dark conditions. Plant height statistics (D): data from C. The letters in the yellow areas indicate significant differences between epicotyls, and the letters in the green areas indicate significant differences between hypocotyls (p < 0.05). It was determined by one-way ANOVA (Tukey’s multiple comparisons test). Data are means ± s.d. (n > 4).
To investigate how GmCRF4 regulates stem elongation at cellular level, we used the epicotyls of 11-day-old plants grown under 12 light/12 dark conditions for longitudinal section (Figure 4A). The histological results showed that the pith cell length of the OX lines increased, whereas that of the Gmcrf4a-1 mutant and CS line decreased in comparison to the wild-type TL1 (Figure 4A). The statistical results demonstrated the pith cell length of the OX1 and OX2 lines increased by 60 and 70%, the mutant Gmcrf4a-1 decreased by 20%, and the CS line decreased by 60%, respectively (Figure 4B). The number of pith cells in each line was estimated by the ratio of the epicotyl mean length to the pith cell mean length. The results showed that, although the number of pith cells in the CS line increased, the cell number of the OX lines and Gmcrf4a-1 mutant did not change significantly in comparison to TL1 (Figure 4C). These results suggested that GmCRF4a promoted epicotyl elongation mainly by increasing cell length.
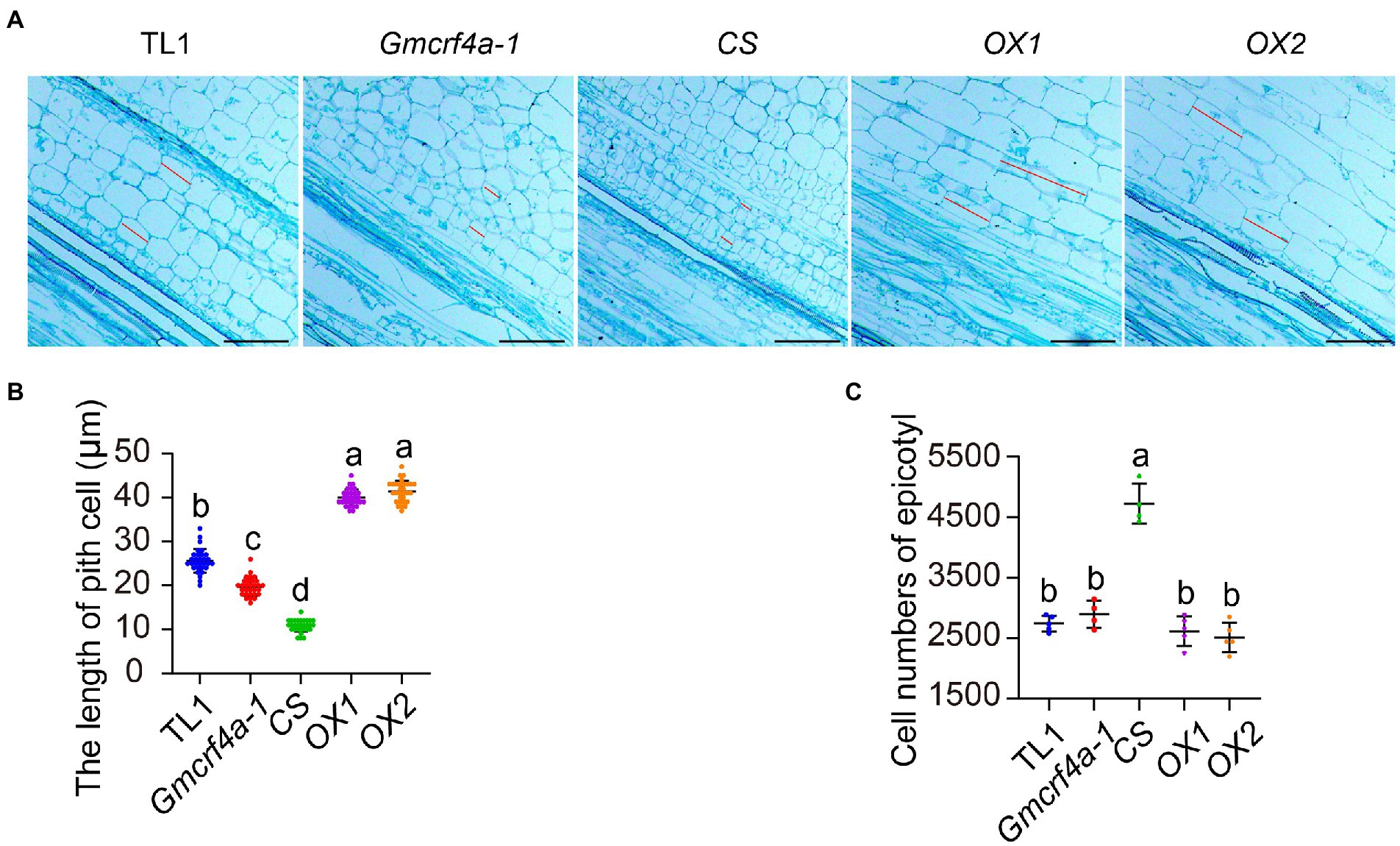
Figure 4. The organization structure and statistical analysis of epicotyls in five individuals. (A) Structure of pith cells in the epicotyls of 5 individuals at 11 days under 12 light/12 dark conditions. The cells between these two red lines represent the measurement part, scale bars = 30 μm. (B) The length of single pith cell in five individuals, data from A, each material measured 30 cells and 3 biological replicates. (C) Cell numbers of epicotyls in five individuals, data from the average length of epicotyl/the average of single cell length. Lowercase letters indicate significant differences (p < 0.05).
To test if GmCRF4a enhances stem elongation through the auxin pathway, we analyzed the endogenous 3-indoleacetic acid (IAA) content in the epicotyl of 11-day-old plants grown under 12 light/12 dark conditions. The results indicated that the total IAA content substantially declined in the Gmcrf4a-1 mutant and CS line, but significantly increased in the OX1 and OX2 lines, respectively (Figure 5A). Next, we test the expression of YUCs (YUCA) genes which play a key role in the auxin synthesis pathway (Chen et al., 2014; Chao et al., 2020). The qRT-PCR experiments were performed using the indicated lines. The results revealed that auxin synthesis genes GmYUC4a, GmYUC4b, and GmYUC10a were down-regulated in the Gmcrf4a-1 mutant and CS line, but up-regulated in the OX1 and OX2 lines (Figures 5B–D). To be noted, the expressions of auxin transport carriers GmPIN1a and GmPIN1b were not changed significantly in the GmCRF4a mutant and overexpression lines (Supplementary Figure S7). These results indicated that GmCRF4a may increase auxin content by up-regulating the expression of GmYUC genes.
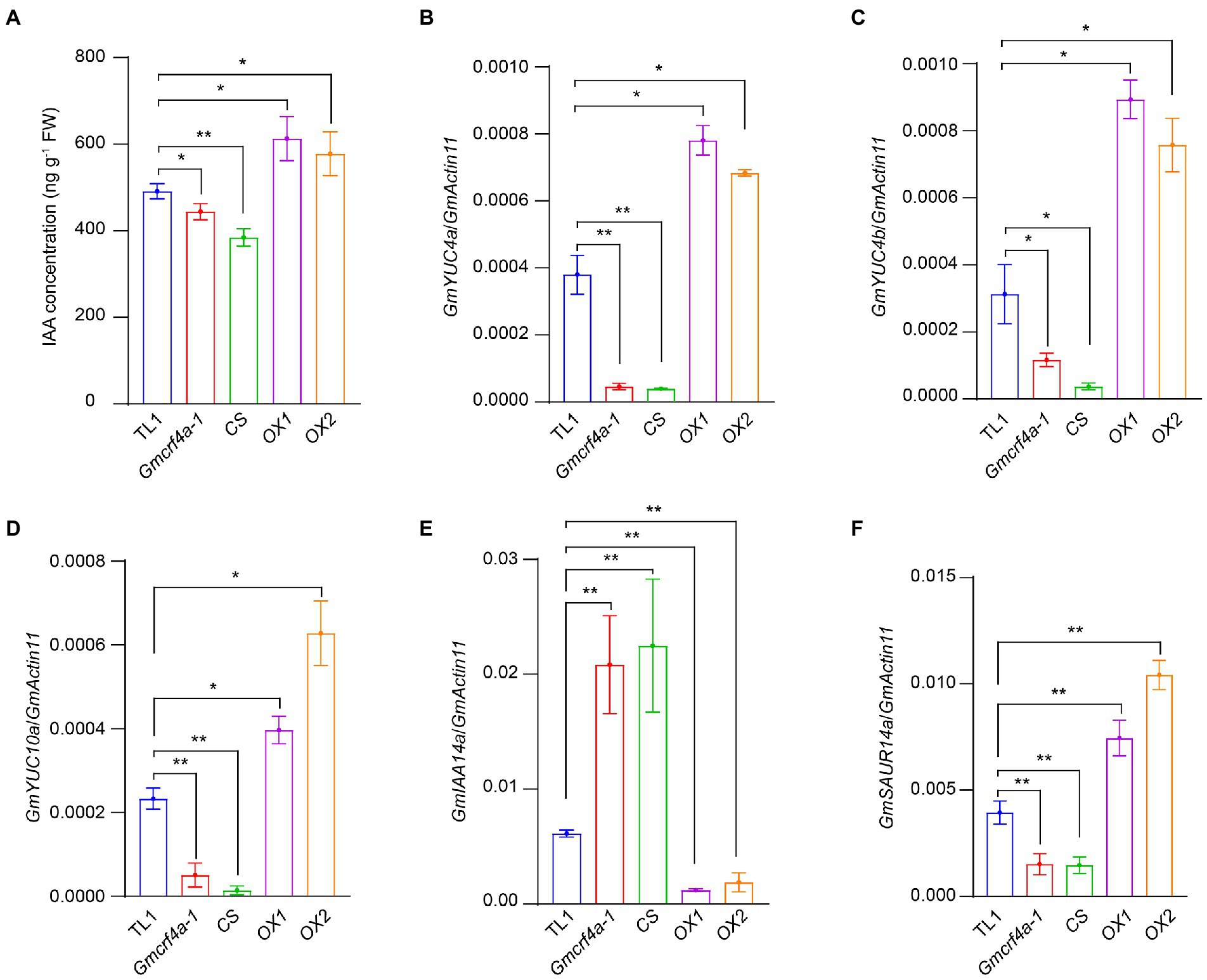
Figure 5. IAA concentration and IAA related gene expression levels in five individuals. (A) The concentration of IAA in five individuals for 11 days under SD. (B–F) Expression levels of IAA-related genes GmYUC4a (B), GmYUC4b (C), GmYUC10a (D), GmSAUR14a (E) and GmIAA14a (F) in the indicated lines. GmActin11 as an internal control. The asterisk indicates a statistically significant difference from the wild type by a two-sided t-test (*p < 0.05, **p < 0.01). Data are displayed as means ± s.d. (n = 3).
We further tested the expression of AUX/IAA (Auxin/indole-3-acetic acid) genes, the negative regulator of auxin signal transduction pathway (Sauer et al., 2006; Trenner et al., 2017; Xu et al., 2019). The results demonstrated that the GmIAA14a gene, a soybean IAA14 member, was up-regulated in the Gmcrf4a-1 mutant and CS line, but down-regulated in the OX1 and OX2 lines (Figure 5E). In addition, a soybean auxin-responsive gene GmSAUR14a (Small auxin up RNA) was down-regulated in the Gmcrf4a-1 mutant and CS line, but up-regulated in the OX1 and OX2 lines (Figure 5F). Taking together, the above results reinforce the hypothesis that GmCRF4a enhances auxin content and further regulates plant height by promoting auxin synthesis gene expression.
In the past few decades, the green revolution has promoted the significant yield increase of gramineous crops such as rice and wheat resulting from the development of semi-dwarf plant architecture. However, it has been difficult to be applied in dicotyledonous crops such as soybeans. Actually, reducing plant height often leads to the reduction of biomass and nodes number, which directly affects the ultimate yield of soybean (Liu et al., 2020). The ideal architecture for soybeans could include strong stems, shorter internode lengths, and higher node numbers. To achieve this goal, the identification and functional characterization of genes that contribute to specific aspects of soybean architecture, such as the plant height of soybeans, will be critical to fully exploiting soybean genomes for crop improvement. In this study, we characterized a soybean CRF gene GmCRF4a, and function analysis results suggested GmCRF4a may positively regulate the plant height through affecting auxin pathway.
CRF genes belong to a subset of AP2/ERF transcription factors superfamily and originally function as CRFs in Arabidopsis, with one exception that CRF4 does not transcriptionally respond to the cytokinin treatment (Rashotte et al., 2006; Cutcliffe et al., 2011; Zwack et al., 2012). Our amino acid alignment and phylogenetic analysis showed that the GmCRF4a gene had the highest identity to the CRF4 gene in Arabidopsis. Supplementary Figures S1, S2, suggesting that GmCRF4a maybe have a similar function to CRF4. Further expression pattern analysis results indicated the soybean GmCRF4a was ubiquitously expressed in all detected tissues, especially highly expressed in hypocotyl, embryo, and suspensor (Figure 1B). In contrast, Arabidopsis CRF4 is also widely expressed in the vascular system of plant root tip, stem, leaf, and flower, but not in stem apex (Zwack et al., 2012). In addition, the homologues of Arabidopsis CRF4 in other species also exhibit various expression patterns. For example, tomato SlCRF4 is also expressed in roots, stems, leaves, and flowers, but more in roots (Shi et al., 2012). Chinese cabbage BrCRF4 is highly expressed in stamens, while BolCRF4c in Brassica oleracea and BnaCRF4e in Brassica napus are preferentially expressed in pistils (Liu et al., 2013; Kong et al., 2018). PtERF85, the homologous gene of CRF4 in aspen trees, is highly expressed in phloem and vascular cambium, the highest in xylem expansion area and less in mature xylem (Seyfferth et al., 2021). The significant differences of expression pattern among the homologues of CRF4 might imply their functional differences in different species.
In Arabidopsis, CRFs play an important role in embryo development, morphological structure, biological and abiotic stress. For example, crf2, crf3, crf5, crf3crf6, crf2,5,6 and crf1,3,5,6 single and multiple mutants led to various morphological changes in hypocotyls, main and lateral roots, leaf size, and leaf senescence (Rashotte et al., 2006; Jeon et al., 2016; Kim, 2016; Raines et al., 2016; Hallmark and Rashotte, 2019; Wang et al., 2020; Park et al., 2021). However, the altered CRF4 expression did not exhibit visible phenotypic changes, except that a slight increase in primary root length of CRF4OX, and it responded to low temperature and low nitrogen levels (Zwack et al., 2016; Varala et al., 2018). Inconsistent with function of Arabidopsis CRF4, altering expression of GmCRF4a in soybean resulted in a variety of phenotypic changes. Knock out GmCRF4a led to the decreased pith cell length, lower hypocotyls and epicotyls, fewer MSNs, lower plant height and a shorter growth period, while GmCRF4a overexpressing plants exhibited opposite phenotype (Figure 3; Supplementary Figure S5), indicating a significant functional difference of GmCRF4a with its Arabidopsis homologue CRF4. In addition, it is reported that Arabidopsis CRF4 homologous genes in other species are related to abiotic stress response. Under salt treatment, the expression of SlCRF4 in tomato, BnaCRF4b and BnaCRF4e in Brassica napus were up-regulated, while the expression of BolCRF4b in Brassica oleracea and BniCRF4c in Brassica nigra were down regulated (Kong et al., 2018). After ethylene and salicylic acid treatment, the expression of SlCRF4 increased significantly, but there was no significant change under jasmonic acid treatment (Shi et al., 2012). After ABA treatment, the expression of BrCRF4, BolCRF4b, BniCRF4a, BniCRF4b and BniCRF4c increased (Liu et al., 2013; Kong et al., 2018). After 6-BA treatment, the expression of BnaCRF4e was up-regulated, while the expression of BolCRF4a,BolCRF4b,BniCRF4a,BniCRF4b and BnaCRF4c were down-regulated (Kong et al., 2018). These results, together with the expression pattern results, indicated homologues of CRF4 in different species tend to have different expression patterns and diverge functionally.
Recent studies in Arabidopsis have shown that CRFs are not only responsive to the cytokinin, but also participate in the regulation of auxin transport (Schlereth et al., 2010; Ckurshumova et al., 2014; Simaskova et al., 2015; Wu et al., 2015; Cucinotta et al., 2016; Waidmann et al., 2019). CRF2 and CRF6 could bind to the promoters of auxin efflux carriers PIN1 and PIN7 and suppress their expression level (Simaskova et al., 2015). PIN7 was upregulated in crf6 mutants, and auxin content in the root tips of crf3crf6 double mutants was increased compared with wild type (Simaskova et al., 2015). In addition, the loss of CRF2 and CRF3 led to changes in lateral root gravity (Waidmann et al., 2019). In this study, GmCRF4a may also be involved in the auxin pathway. The auxin content in mutant Gmcrf4a-1 and co-silencing line (CS) was significantly reduced, while significantly increased in the OX lines (Figure 5A). Auxin governs the form and shape of the plant body by stimulating cell elongation, which might explain the cell elongation and plant height phenotype of Gmcrf4a-1, CS and OX lines (Velasquez et al., 2016). In order to clarify the source of auxin, expression of the key genes in auxin transport and synthesis pathways were detected. The results indicated that the auxin transporter GmPINs exhibited no difference in Gmcrf4a-1, CS and OX lines, while the auxin synthesis gene GmYUCs were significantly upregulated in OX lines and downregulated in Gmcrf4a-1 and CS lines (Supplementary Figure S7; Figures 5B–D). GmIAA14a and GmSAUR14a, the downstream genes of auxin signaling transduction pathway, were also examined and the results showed opposite expression pattern in GmCRF4a mutants and overexpression lines. Overall, although the protein structure of GmCRF4a is more homologous to Arabidopsis CRF4, its expression profile is different, and its function is closer to CRF2, CRF3 and CRF6. Different from CRF2, CRF3 and CRF6, GmCRF4a did not affect auxin transport, but may affected auxin content by increasing auxin key synthase. Therefore, the present findings provide a new possible pathway for CRF to regulate plant height and enrich the functional cognition of CRFs.
In addition, the diffident phenotypes of CS and OX1 in plant height and growth period under natural field condition provides two possible scenarios for their breeding applications. CS plants had a dwarf compact plant architecture and short growth period which make it possible to be introduced from low latitude to high latitude, and the yield may be increased through intercropping and reasonable dense planting. OX1 displayed higher plant height, increased pod setting height, more nodes, and a longer growth period, so it was recommended to be introduced from high latitude to low latitude to achieve the possibly purpose of improving late-maturing yield. Taken together, our study provides an excellent candidate gene GmCRF4a and corresponding plant materials for genetic breeding to improve soybean architecture, which would facilitate future molecular breeding practice.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
TaZ, TuZ, and BL designed the studies and wrote the manuscript. ZX conducted the bioinformatic analyses. ZX, RW, KK, NB, AA, JL, and HL performed the experiments. All authors contributed to the article and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (32072091), the National Natural Science Foundation of China (32171965), Science and Technology Innovation Team of Soybean Modern Seed Industry in Hebei (21326313D), the Jiangsu Seed Industry Revitalization Project [JBGS(2021)014], and the Jiangsu Collaborative Innovation Centre for Modern Crop Production (JCIC-MCP) Program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.983650/full#supplementary-material
1. ^https://phytozome-next.jgi.doe.gov/, accessed on January 7, 2022.
2. ^https://venanciogroup.uenf.br/cgi-bin/gmax_atlas/index.cgi, accessed on January 5, 2022.
3. ^http://crispr.hzau.edu.cn/CRISPR2/, accessed on January 5, 2022.
4. ^https://venanciogroup.uenf.br/cgi-bin/gmax_atlas/index.cgi
Bai, Y., Shen, Y., Zhang, Z., Jia, Q., Xu, M., Zhang, T., et al. (2021). A GPAT1 mutation in Arabidopsis enhances plant height but impairs seed oil biosynthesis. Int. J. Mol. Sci. 22:785. doi: 10.3390/ijms22020785
Castorina, G., and Consonni, G. (2020). The role of brassinosteroids in controlling plant height in Poaceae: A genetic perspective. Int. J. Mol. Sci. 21:1191. doi: 10.3390/ijms21041191
Chao, Y., Shang, J., and Ji, W. (2020). ALKBH5-m(6)A-FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia. Biochem. Biophys. Res. Commun. 521, 499–506. doi: 10.1016/j.bbrc.2019.10.145
Chen, Q., Dai, X., De-Paoli, H., Cheng, Y., Takebayashi, Y., Kasahara, H., et al. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55, 1072–1079. doi: 10.1093/pcp/pcu039
Chen, W. H., Li, P. F., Chen, M. K., Lee, Y. I., and Yang, C. H. (2015). FOREVER YOUNG FLOWER negatively regulates ethylene response DNA-binding factors by activating an ethylene-responsive factor to control Arabidopsis floral organ senescence and abscission. Plant Physiol. 168, 1666–1683. doi: 10.1104/pp.15.00433
Chen, L., Nan, H., Kong, L., Yue, L., Yang, H., Zhao, Q., et al. (2020). Soybean AP1 homologs control flowering time and plant height. J. Integr. Plant Biol. 62, 1868–1879. doi: 10.1111/jipb.12988
Cheng, Q., Dong, L., Su, T., Li, T., Gan, Z., Nan, H., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 19:562. doi: 10.1186/s12870-019-2145-8
Chu, S., Wang, J., Zhu, Y., Liu, S., Zhou, X., Zhang, H., et al. (2017). An R2R3-type MYB transcription factor, GmMYB29, regulates isoflavone biosynthesis in soybean. PLoS Genet. 13:e1006770. doi: 10.1371/journal.pgen.1006770
Ckurshumova, W., Smirnova, T., Marcos, D., Zayed, Y., and Berleth, T. (2014). Irrepressible MONOPTEROS/ARF5 promotes de novo shoot formation. New Phytol. 204, 556–566. doi: 10.1111/nph.13014
Cucinotta, M., Manrique, S., Guazzotti, A., Quadrelli, N. E., Mendes, M. A., Benkova, E., et al. (2016). Cytokinin response factors integrate auxin and cytokinin pathways for female reproductive organ development. Development 143, 4419–4424. doi: 10.1242/dev.143545
Cutcliffe, J. W., Hellmann, E., Heyl, A., and Rashotte, A. M. (2011). CRFs form protein-protein interactions with each other and with members of the cytokinin signalling pathway in Arabidopsis via the CRF domain. J. Exp. Bot. 62, 4995–5002. doi: 10.1093/jxb/err199
De Micco, V., and Aronne, G. (2007). Combined histochemistry and autofluorescence for identifying lignin distribution in cell walls. Biotech. Histochem. 82, 209–216. doi: 10.1080/10520290701713981
Dong, H., Yan, S., Liu, J., Liu, P., and Sun, J. (2019). TaCOLD1 defines a new regulator of plant height in bread wheat. Plant Biotechnol. J. 17, 687–699. doi: 10.1111/pbi.13008
Gan, Z., Wei, W., Wu, J., Zhao, Y., Zhang, L., Wang, T., et al. (2019). Resveratrol and Curcumin improve intestinal mucosal integrity and decrease m(6)A RNA methylation in the intestine of weaning piglets. ACS Omega 4, 17438–17446. doi: 10.1021/acsomega.9b02236
Guo, F., Huang, Y., Qi, P., Lian, G., Hu, X., Han, N., et al. (2021). Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance. New Phytol. 229, 2676–2692. doi: 10.1111/nph.17061
Hallmark, H. T., and Rashotte, A. M. (2019). Review-Cytokinin response factors: responding to more than cytokinin. Plant Sci. 289:110251. doi: 10.1016/j.plantsci.2019.110251
Huang, T., Guo, J., Lv, Y., Zheng, Y., Feng, T., Gao, Q., et al. (2019). Meclofenamic acid represses spermatogonial proliferation through modulating m(6)A RNA modification. J. Anim. Sci. Biotechnol. 10, 63. doi: 10.1186/s40104-019-0361-6
Jeon, J., Cho, C., Lee, M. R., Van Binh, N., and Kim, J. (2016). CYTOKININ RESPONSE FACTOR2 (CRF2) and CRF3 regulate lateral root development in response to cold stress in Arabidopsis. Plant Cell 28, 1828–1843. doi: 10.1105/tpc.15.00909
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, J. (2016). CYTOKININ RESPONSE FACTORs gating environmental signals and hormones. Trends Plant Sci. 21, 993–996. doi: 10.1016/j.tplants.2016.10.004
Kong, L., Zhao, K., Gao, Y., Miao, L., Chen, C., Deng, H., et al. (2018). Comparative analysis of cytokinin response factors in brassica diploids and amphidiploids and insights into the evolution of brassica species. BMC Genomics 19, 728. doi: 10.1186/s12864-018-5114-y
Lee, G. A., Crawford, G. W., Liu, L., Sasaki, Y., and Chen, X. (2011). Archaeological soybean (Glycine max) in East Asia: does size matter? PLoS One 6:e26720. doi: 10.1371/journal.pone.0026720
Li, Z. F., Guo, Y., Ou, L., Hong, H., Wang, J., Liu, Z. X., et al. (2018b). Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor. Appl. Genet. 131, 1001–1016. doi: 10.1007/s00122-017-3044-8
Li, Z., Zhang, X., Zhao, Y., Li, Y., Zhang, G., Peng, Z., et al. (2018a). Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol. J. 16, 86–99. doi: 10.1111/pbi.12751
Liao, Z., Yu, H., Duan, J., Yuan, K., Yu, C., Meng, X., et al. (2019). SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 10:2738. doi: 10.1038/s41467-019-10667-2
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, Z., Kong, L., Zhang, M., Lv, Y., Liu, Y., Zou, M., et al. (2013). Genome-wide identification, phylogeny, evolution and expression patterns of AP2/ERF genes and cytokinin response factors in Brassica rapa ssp. pekinensis. PLoS One 8:e83444. doi: 10.1371/journal.pone.0083444
Liu, S. L., Zhang, M., Feng, F., and Tian, Z. X. (2020). Toward a "green revolution" for soybean. Mol. Plant 13, 688–697. doi: 10.1016/j.molp.2020.03.002
Lyu, X., Cheng, Q., Qin, C., Li, Y., Xu, X., Ji, R., et al. (2021). GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 14, 298–314. doi: 10.1016/j.molp.2020.11.016
Ma, Z., Wu, T., Huang, K., Jin, Y. M., Li, Z., Chen, M., et al. (2020). A novel AP2/ERF transcription factor, OsRPH1, negatively regulates plant height in rice. Front. Plant Sci. 11:709. doi: 10.3389/fpls.2020.00709
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Park, J., Lee, S., Park, G., Cho, H., Choi, D., Umeda, M., et al. (2021). CYTOKININ-RESPONSIVE GROWTH REGULATOR regulates cell expansion and cytokinin-mediated cell cycle progression. Plant Physiol. 186, 1734–1746. doi: 10.1093/plphys/kiab180
Ping, J., Liu, Y., Sun, L., Zhao, M., Li, Y., She, M., et al. (2014). Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26, 2831–2842. doi: 10.1105/tpc.114.126938
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Raines, T., Shanks, C., Cheng, C. Y., McPherson, D., Argueso, C. T., Kim, H. J., et al. (2016). The cytokinin response factors modulate root and shoot growth and promote leaf senescence in Arabidopsis. Plant J. 85, 134–147. doi: 10.1111/tpj.13097
Rashotte, A. M., Mason, M. G., Hutchison, C. E., Ferreira, F. J., Schaller, G. E., and Kieber, J. J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. U. S. A. 103, 11081–11085. doi: 10.1073/pnas.0602038103
Ren, W., Lu, J., Huang, M., Gao, L., Li, D., Wang, G. G., et al. (2019). Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat. Commun. 10, 5042. doi: 10.1038/s41467-019-12923-x
Rockwell, A. L., and Hongay, C. F. (2019). The m(6)A dynamics of profilin in neurogenesis. Front. Genet. 10:987. doi: 10.3389/fgene.2019.00987
Sauer, M., Balla, J., Luschnig, C., Wisniewska, J., Reinohl, V., Friml, J., et al. (2006). Canalization of auxin flow by aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20, 2902–2911. doi: 10.1101/gad.390806
Schlereth, A., Moller, B., Liu, W., Kientz, M., Flipse, J., Rademacher, E. H., et al. (2010). MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464, 913–916. doi: 10.1038/nature08836
Seyfferth, C., Wessels, B. A., Vahala, J., Kangasjarvi, J., Delhomme, N., Hvidsten, T. R., et al. (2021). Populus PtERF85 balances xylem cell expansion and secondary cell wall formation in hybrid aspen. Cell 10:1971. doi: 10.3390/cells10081971
Shi, X., Gupta, S., and Rashotte, A. M. (2012). Solanum lycopersicum cytokinin response factor (SlCRF) genes: characterization of CRF domain-containing ERF genes in tomato. J. Exp. Bot. 63, 973–982. doi: 10.1093/jxb/err325
Simaskova, M., O’Brien, J. A., Khan, M., Van Noorden, G., Otvos, K., Vieten, A., et al. (2015). Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat. Commun. 6:8717. doi: 10.1038/ncomms9717
Tian, Z., Wang, X., Lee, R., Li, Y., Specht, J. E., Nelson, R. L., et al. (2010). Artificial selection for determinate growth habit in soybean. Proc. Natl. Acad. Sci. U. S. A. 107, 8563–8568. doi: 10.1073/pnas.1000088107
Trenner, J., Poeschl, Y., Grau, J., Gogol-Doring, A., Quint, M., and Delker, C. (2017). Auxin-induced expression divergence between Arabidopsis species may originate within the TIR1/AFB-AUX/IAA-ARF module. J. Exp. Bot. 68, 539–552. doi: 10.1093/jxb/erw457
Varala, K., Marshall-Colon, A., Cirrone, J., Brooks, M. D., Pasquino, A. V., Leran, S., et al. (2018). Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. U. S. A. 115, 6494–6499. doi: 10.1073/pnas.1721487115
Velasquez, S. M., Barbez, E., Kleine-Vehn, J., and Estevez, J. M. (2016). Auxin and cellular elongation. Plant Physiol. 170, 1206–1215. doi: 10.1104/pp.15.01863
Vogel, J. T., Liu, W. D., Olhoft, P., Crafts-Brandner, S. J., Pennycooke, J. C., and Christiansen, N. (2021). Soybean yield formation physiology - a foundation for precision breeding based improvement. Front. Plant Sci. 12:719706. doi: 10.3389/fpls.2021.719706
Waidmann, S., Ruiz Rosquete, M., Scholler, M., Sarkel, E., Lindner, H., LaRue, T., et al. (2019). Cytokinin functions as an asymmetric and anti-gravitropic signal in lateral roots. Nat. Commun. 10, 3540. doi: 10.1038/s41467-019-11483-4
Wang, D. O. (2019). Mapping m(6)A and m(1)A with mutation signatures. Nat. Methods 16, 1213–1214. doi: 10.1038/s41592-019-0636-z
Wang, J., Hu, B., Jing, Y., Hu, X., Guo, Y., Chen, J., et al. (2021). Detecting QTL and candidate genes for plant height in soybean via linkage analysis and GWAS. Front. Plant Sci. 12:803820. doi: 10.3389/fpls.2021.803820
Wang, S., Zhang, H., Shi, L., Xu, F., and Ding, G. (2020). Genome-wide dissection of the CRF gene family in Brassica napus indicates that BnaCRF8s specifically regulate root architecture and phosphate homeostasis against phosphate fluctuation in plants. Int. J. Mol. Sci. 21:3660. doi: 10.3390/ijms21103660
Wang, Y., Zhao, J., Lu, W., and Deng, D. (2017). Gibberellin in plant height control: old player, new story. Plant Cell Rep. 36, 391–398. doi: 10.1007/s00299-017-2104-5
Wu, M. F., Yamaguchi, N., Xiao, J., Bargmann, B., Estelle, M., Sang, Y., et al. (2015). Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife 4:e09269. doi: 10.7554/eLife.09269
Xiong, L., Li, C., Li, H., Lyu, X., Zhao, T., Liu, J., et al. (2019). A transient expression system in soybean mesophyll protoplasts reveals the formation of cytoplasmic GmCRY1 photobody-like structures. Sci. China Life Sci. 62, 1070–1077. doi: 10.1007/s11427-018-9496-5
Xu, C., Shen, Y., He, F., Fu, X., Yu, H., Lu, W., et al. (2019). Auxin-mediated aux/IAA-ARF-HB signaling cascade regulates secondary xylem development in Populus. New Phytol. 222, 752–767. doi: 10.1111/nph.15658
Xue, Y., Zhang, Y., Shan, J., Ji, Y., Zhang, X., Li, W., et al. (2022). Growth repressor GmRAV binds to the GmGA3ox promoter to negatively regulate plant height development in soybean. Int. J. Mol. Sci. 23, 1721. doi: 10.3390/ijms23031721
Zhang, L., Liu, P., Wu, J., Qiao, L., Zhao, G., Jia, J., et al. (2020). Identification of a novel ERF gene, TaERF8, associated with plant height and yield in wheat. BMC Plant Biol. 20, 263. doi: 10.1186/s12870-020-02473-6
Zhang, Z., Xiao, J., Wu, J., Zhang, H., Liu, G., Wang, X., et al. (2012). ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 419, 779–781. doi: 10.1016/j.bbrc.2012.02.101
Zhang, Y., Yu, C., Lin, J., Liu, J., Liu, B., Wang, J., et al. (2017). OsMPH1 regulates plant height and improves grain yield in rice. PLoS One 12:e0180825. doi: 10.1371/journal.pone.0180825
Zhao, X., Cao, D., Huang, Z., Wang, J., Lu, S., Xu, Y., et al. (2015). Dual functions of GmTOE4a in the regulation of photoperiod-mediated flowering and plant morphology in soybean. Plant Mol. Biol. 88, 343–355. doi: 10.1007/s11103-015-0322-1
Zhao, Q., Hu, R. S., Liu, D., Liu, X., Wang, J., Xiang, X. H., et al. (2020). The AP2 transcription factor NtERF172 confers drought resistance by modifying NtCAT. Plant Biotechnol. J. 18, 2444–2455. doi: 10.1111/pbi.13419
Zong, Y., Hao, Z., Tu, Z., Shen, Y., Zhang, C., Wen, S., et al. (2021). Genome-wide survey and identification of AP2/ERF genes involved in shoot and leaf development in Liriodendron chinense. BMC Genomics 22, 807. doi: 10.1186/s12864-021-08119-7
Zwack, P. J., Compton, M. A., Adams, C. I., and Rashotte, A. M. (2016). Cytokinin response factor 4 (CRF4) is induced by cold and involved in freezing tolerance. Plant Cell Rep. 35, 573–584. doi: 10.1007/s00299-015-1904-8
Keywords: soybean, plant height, auxin, cytokinin response factor, GmCRF4a
Citation: Xu Z, Wang R, Kong K, Begum N, Almakas A, Liu J, Li H, Liu B, Zhao T and Zhao T (2022) An APETALA2/ethylene responsive factor transcription factor GmCRF4a regulates plant height and auxin biosynthesis in soybean. Front. Plant Sci. 13:983650. doi: 10.3389/fpls.2022.983650
Received: 01 July 2022; Accepted: 17 August 2022;
Published: 06 September 2022.
Edited by:
Sateesh Kagale, National Research Council Canada (NRC-CNRC), CanadaReviewed by:
Liangyu Liu, Capital Normal University, ChinaCopyright © 2022 Xu, Wang, Kong, Begum, Almakas, Liu, Li, Liu, Zhao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tuanjie Zhao, dGp6aGFvQG5qYXUuZWR1LmNu; Tao Zhao, emhhb3RhbzAyQGNhYXMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.