- 1Turfgrass Research Institute, College of Grassland Science, Beijing Forestry University, Beijing, China
- 2Chongqing Key Laboratory of Germplasm Innovation and Utilization of Native Plants, Chongqing Landscape and Gardening Research Institute, Chongqing, China
- 3Beijing Research and Development Center for Grass and Environment, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 4Inner Mongolia Mengcao Ecological Environment (Group) Co., Ltd., Hohhot, China
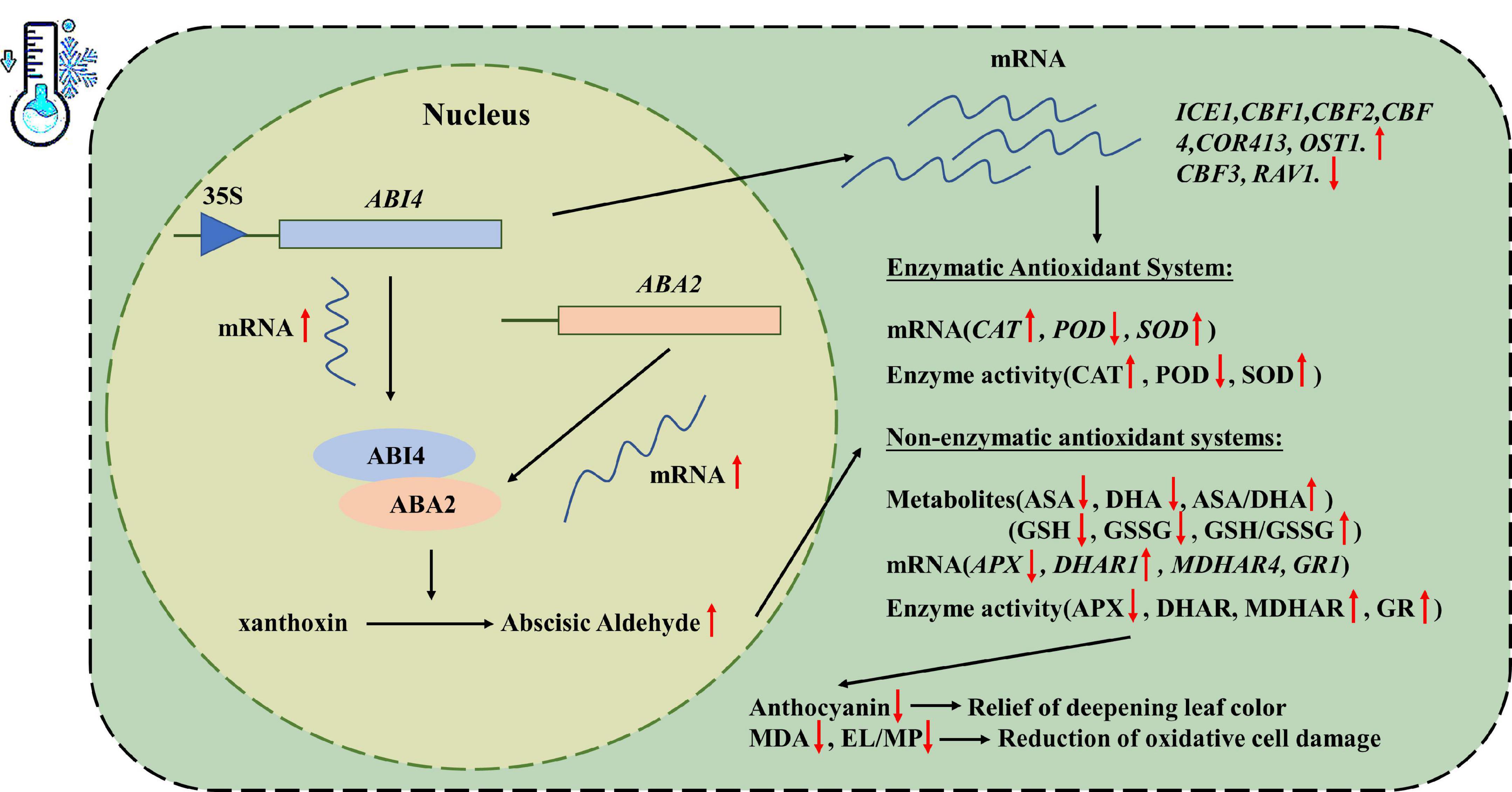
ABI4 is considered an important transcription factor with multiple regulatory functions involved in many biological events. However, its role in abiotic stresses, especially low-temperature-induced stress, is poorly understood. In this study, the MtABI4 gene was derived from M. truncatula, a widely used forage grass. Analysis of subcellular localization indicated that ABI4 was localized in the nucleus. Identification of expression characteristics showed that ABI4 was involved in the regulatory mechanisms of multiple hormones and could be induced by the low temperature. IP-MS assay revealed that MtABI4 protein could interact with xanthoxin dehydrogenase protein (ABA2). The two-hybrid yeast assay and the biomolecular fluorescence complementarity assay further supported this finding. Expression analysis demonstrated that overexpression of MtABI4 induced an increase in ABA2 gene expression both in M. truncatula and Arabidopsis, which in turn increased the ABA level in transgenic plants. In addition, the transgenic lines with the overexpression of MtABI4 exhibited enhanced tolerance to low temperature, including lower malondialdehyde content, electrical conductivity, and cell membrane permeability, compared with the wide-type lines after being cultivated for 5 days in 4°C. Gene expression and enzyme activities of the antioxidant system assay revealed the increased activities of SOD, CAT, MDHAR, and GR, and higher ASA/DHA ratio and GSH/GSSG ratio in transgenic lines. Additionally, overexpression of ABI4 also induced the expression of members of the Inducer of CBF expression genes (ICEs)-C-repeat binding transcription factor genes(CBFs)-Cold regulated genes (CORs) low-temperature response module. In summary, under low-temperature conditions, overexpression of ABI4 could enhance the content of endogenous ABA in plants through interactions with ABA2, which in turn reduced low-temperature damage in plants. This provides a new perspective for further understanding the molecular regulatory mechanism of plant response to low temperature and the improvement of plant cold tolerance.
Introduction
abi4 abscisic acid-insensitive 4 was originally named after a series of ABA-insensitive mutants identified through a forward genetic screen (Finkelstein, 1994). In recent years, ABI4 has been recognized as a core transcription factor with multiple regulatory functions, which is involved in many important biological events. For example, ABI4 was associated with seed germination and seedling morphogenesis. The expression of ABI4 gradually decreased with seed germination and seedling morphology (Shkolnik-Inbar and Bar-Zvi, 2011). Accordingly, overexpression of ABI4 exacerbated the ABA-induced inhibition of seed germination in Arabidopsis (Finkelstein et al., 2011). ABI4 also played some significant function in plastid/mitochondrial reverse signaling, especially acting as a downstream target of the GUN1 in mediating the PGE pathway regulating plastid (Sun et al., 2011). Moreover, ABI4 was also involved in sugar signaling. Some studies showed that ABI4 regulates the expression of the sugar-responsive gene by binding to CE1-like motif elements in the promoter region, such as the ADP-glucose pyrophosphorylase large subunit ApL3 in the starch synthesis pathway and precipitate branching enzyme SBE2.2 in the starch synthesis pathway (Bossi et al., 2009). ABI4 affects lateral root production. abi4 mutants increase lateral root density and length compared to the wild type, while their overexpression reduced lateral root production (Shkolnik-Inbar and Bar-Zvi, 2010).
Additionally, reactive oxygen species (ROS) are also involved in the ABI4-mediated reverse signaling pathway (León et al., 2012). Overall, ABI4 plays an integral role in plant growth, development, and response to adversity. Although in recent years studies have led to a certain understanding of the function of ABI4, its role remains incomplete, such as its regulatory mechanism in the low-temperature response.
Plant cold tolerance is a complex trait controlled by multiple genes, such as COR, ICE, CBF, and GPAT, which all play an important role in plant response to low-temperature stress and in improving cold tolerance (Zhu et al., 2019; Tang et al., 2020). In plants, the most widely studied low-temperature response signaling pathway is the CBF/DREB1 signaling pathway with the main upstream and downstream modules Inducer of CBF expression genes (ICEs)-C-repeat binding transcription factor genes (CBF)/DREB1s-Cold regulated genes (CORs). ICE1 binds to the promoter region of the CBF gene to activate the expression, thereby mediating cold tolerance in plants (Chinnusamy et al., 2007). Low-temperature signals do not directly regulate ICE1 gene transcription, but the E3 ligase HOS1 (high osmotic expression1) is involved in ICE1 ubiquitination and degradation via the 26S protease pathway, thereby affecting ICE1 protein stability (Shi et al., 2015). The activity of OST1 kinase (open stomata1) was found to be rapidly induced by low temperature, and ICE1 and activated OST1 interacted to phosphorylate ICE1 protein and improve its binding capacity to the CBF3 promoter region and ICE1 stability (Ding et al., 2019), and it was also found that OST1 phosphorylated BTF3L and BTF3 and promoted their interaction with CBF (Ding et al., 2018). The Jas domains of JAZ1 and JAZ4 interacted with the C-terminal of the ICE1 protein and inhibited ICE1 activity in A. mimosa. When exposed to low-temperature stress, JAZ proteins were degraded by increased JA content activating JA signaling, thereby relieving the inhibitory effect of ICE1 by JAZ and improving low-temperature tolerance (Hu et al., 2013).
Low temperature triggers the change in the activity and content of hormones in plants, which in turn influence the metabolic processes of plant growth. Abscisic acid (ABA) as a natural hormone defending against the abiotic stresses, plays an important role in stresses such as low temperatures (Jiang and Zhang, 2002). There is a significant increase in ABA content in leaves of Arabidopsis and rice at low temperatures (Eremina et al., 2016). In addition, ABA reduces the water content of plants and promotes the accumulation of soluble sugars and total phenolic content, thus improving the cold resistance of plants (Yang et al., 2016). The first hormone found to be associated with cold resistance was gibberellin (GA), and studies have found that the expression of CRT/DRE transcription-related genes is associated with GAs. For example, studies on the relationship between CBF1 and gibberellin have revealed that constitutive expression of CBF1 reduces gibberellin activity (Chen et al., 2007). Salicylic acid (SA) content increases significantly under low-temperature conditions and is detrimental to plant growth if the xconcentration of SA is too high, while low concentrations of SA promote cell expansion and growth (Klessig et al., 2018). Jasmonic acid (JA), a very important plant signaling molecule, can mediate the expression of those resistance genes, and JA interacts with CBF to regulate downstream cold-responsive genes, thereby enhancing cold tolerance in plants (Hu et al., 2017). Additionally, the increased JA content improves CAT, SOD, and APX activities, thereby enhancing plant cold tolerance (Campos et al., 2019).
There are fewer reports on the function of ABI4 in the low-temperature response. Only one recent study was reviewed, showing that MdABI4 crosstalked jasmonic and abscisic acid signaling to regulate cold tolerance in apples (An et al., 2022). On the other hand, as a model plant of the legume family (Stacey et al., 2006), it is of great importance to investigate the low-temperature stress response genes of M. truncatula for exploring its cold tolerance mechanism and improving the cold tolerance ability, in agricultural production. It also has important implications for the study of other legumes. Therefore, in this study, the function and regulatory mechanisms of MtABI4 in response to low temperature was characterized, aiming to lay the foundation for rapid breeding of cold-tolerance forage varieties of M. truncatula by means of genetic engineering.
Results
Molecular characterization and subcellular localization of MtABI4
Sequence analysis indicated that MtABI4 possessed a complete coding region of 1,011 bp, encoding a polypeptide chain containing 336 amino acids with a predicted molecular weight of 37.8 kD. MtABI4 protein contained an AP2 DNA-binding domain, belonging to the AP2 superfamily (Supplementary Figures 1A,B). Homology and phylogenetic analysis showed that MtABI4 was more closely related to ABI4 from Trifolium pratense, and ABI4 with high homology from ten species all had three motifs and an AP2 domain (Figure 1A). SignalP predicted that MtABI4 had no signal peptide, and subcellular localization prediction showed that MtABI4 was an extracellular protein (Supplementary Figures 1C,D).
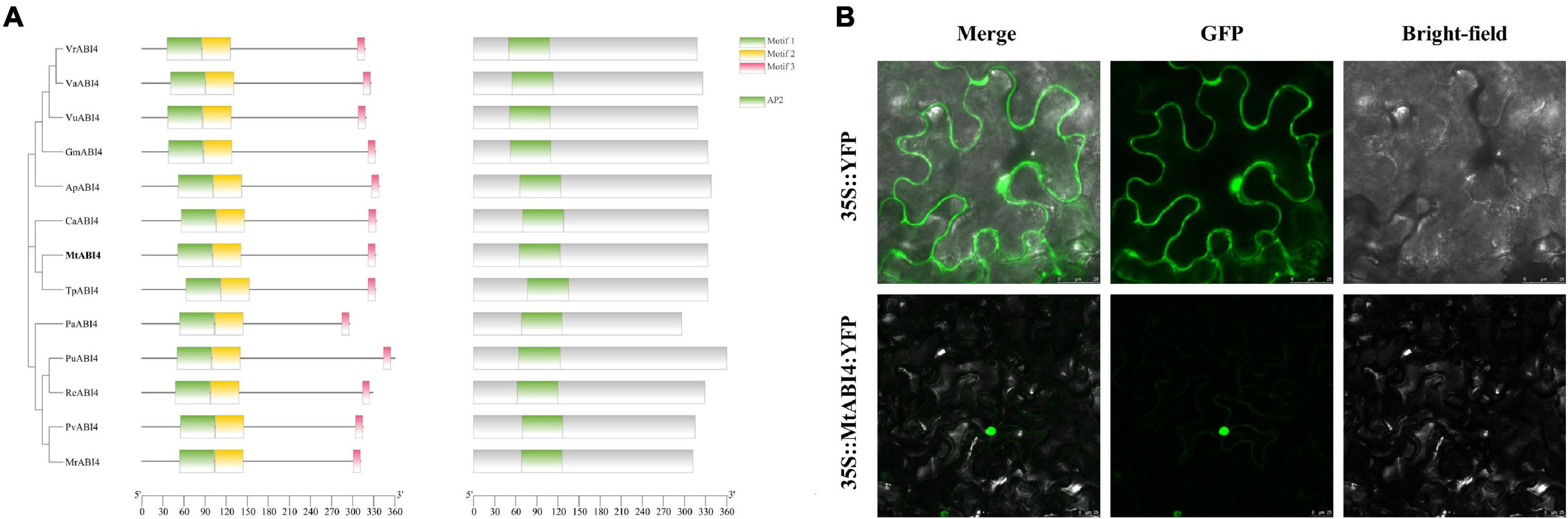
Figure 1. Phylogenetic tree, motif analysis, protein structural domains, and the subcellular localization of ABI4. In (A) different color blocks represent different motifs and structural domains. The scale bar represents the length of the amino acid sequence. (B) Represents the subcellular localization of ABI4.
By observing the expression of an ABI4-GFP fusion protein in tobacco leaves, we found that ABI4 was localized in the nucleus, which was inconsistent with our previous prediction (Figure 1B).
MtABI4 gene responds to multiple hormones
To analyze the regulation mechanism of MtABI4, the upstream sequence of the gene, which was 2,000 bp in length, was obtained. Cis-acting elements analysis showed that the motifs enriched in the promoter region of MtABI4 were widely involved in the abscisic acid responsiveness, anaerobic induction, auxin responsiveness, the MeJA-responsiveness, endosperm expression, low-temperature responsiveness, zein metabolism regulation, gibberellin-responsiveness, defense and stress responsiveness, salicylic acid responsiveness, and light response (Figure 2A), indicating that this gene may play a multifunctional role in response to multiple hormones and stress.
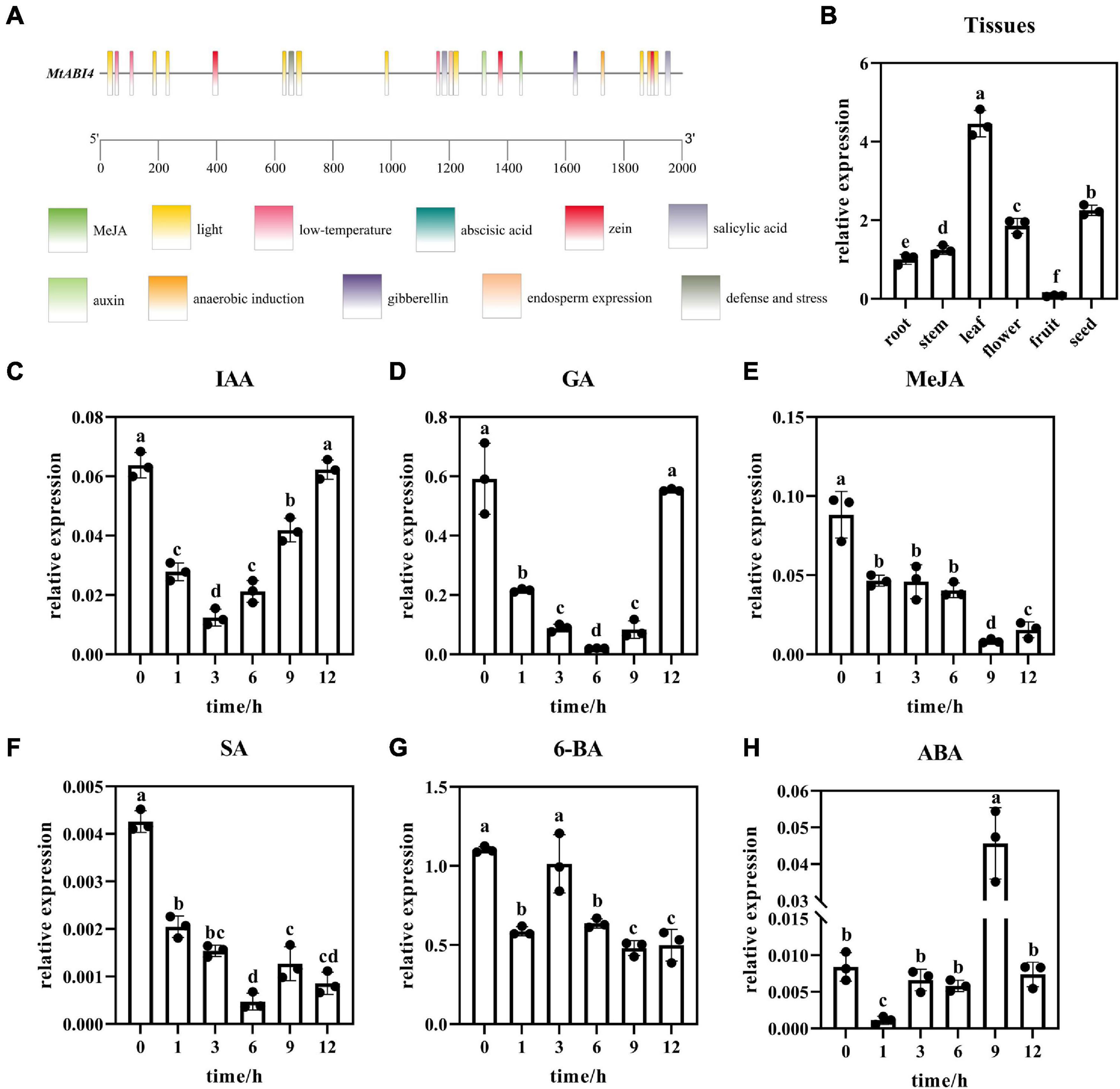
Figure 2. Cis-acting elements analysis and the expression patterns of MtABI4 by qRT-PCR analysis. In graph panel (A), different color blocks represent different cis-acting elements. The scale bar represents the length of the base sequence. Panel (B) represents the expression of ABI4 in different tissues. Different letters in graphs indicate significant differences at p ≤ 0.05. Panels (C–H) represents the expression of ABI4 at different time points in response to IAA, GA, MeJA, SA, 6-BA, and ABA, respectively. The values are means ± SD (n = 3).
Based on this, wild-type plants were sprayed with the corresponding hormones and plant tissues were collected at different time points for analysis of the MtABI4 gene response to these hormones. Quantitative fluorescence analysis indicated that the expression of ABI4 was down-regulated under the treatment of IAA, 6-BA, GA, MEJA, and SA (Figures 2C–G). The difference was that ABI4 had a rebound at 6 h under the treatment of 6-BA, and returned to the initial level at 12 h under the treatment of GA and IAA. In contrast to them, the expression level of ABI4 showed an increasing trend under ABA treatment, with a down-regulation at 1 h and a peak at 9 h (Figure 2H). This would suggest that ABI4 is indeed involved in the regulatory mechanisms of these hormones. In addition, tissue-specific expression analysis showed that the ABI4 gene possessed the highest expression in leaves, followed by fruits, and the lowest expression in seeds (Figure 2B).
MtABI4 improved the cold tolerance of the over-expressed transgenic plants
Considering that the promoter region of the ABI4 gene contains a low-temperature element, the transgenic lines, and the wild type was incubated at 4°C for 5 days to investigate the role of the ABI4 gene in the low-temperature response. Phenotypic observations showed that the plants all exhibited some degree of wilting, with the wild-type plants showing darker leaf color after low-temperature treatment (Figure 3A). The results of chloroplast pigment content measurements showed that chlorophyll a, chlorophyll b, chlorophyll, and carotenoids were slightly lower in the transgenic lines than in WT, but there were no significant changes in chloroplast pigment content in the individual lines, before and after the low-temperature treatment (Figure 3B). WT exhibited higher anthocyanin content than the transgenic strain after low-temperature treatment (Figure 3C). The wild-type and transgenic lines did not show significant differences in electrolytic leakage, membrane permeability, and malondialdehyde content before the low-temperature treatment, however, these indicators in WT showed a significant increase and were much greater than in the transgenic lines, after the low-temperature treatment (Figures 3D,E). These results suggested that the transgenic plants accumulated less anthocyanin and suffered less cell membrane damage under low-temperature conditions.
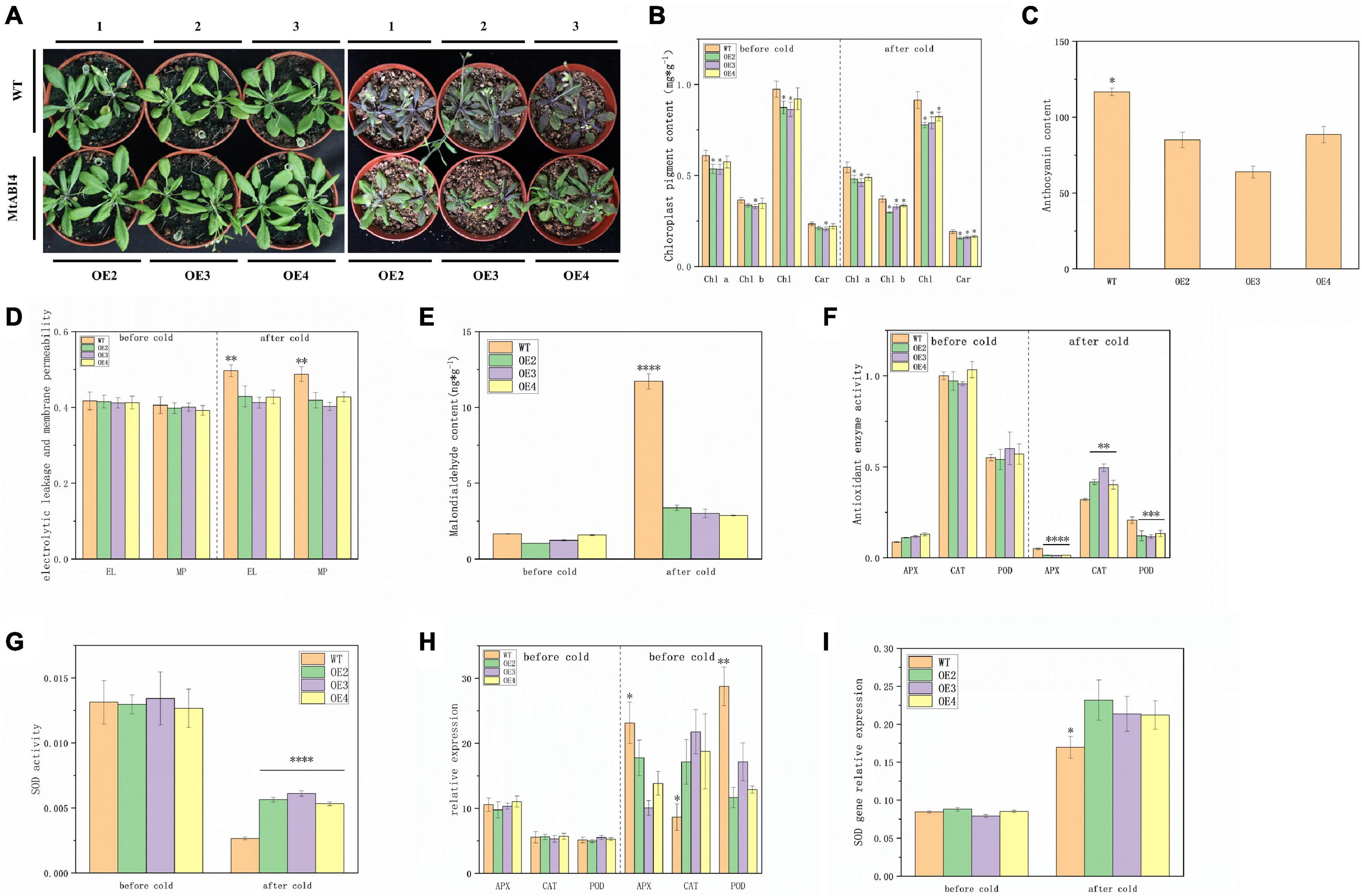
Figure 3. Plant growth status, physiological characteristics, and the expression of antioxidant enzyme genes at low temperatures. Panel (A) represents the growth condition of wild-type and transgenic plants before the low-temperature treatment and after the 5-day low-temperature treatment. Panels (B–E) represents the content of chloroplast pigments, anthocyanin, electrolytic leakage, membrane permeability, and malondialdehyde. Panels (F,G) represent the activity of APX, CAT, POD, and SOD. Panels (H,I) represent the expression of these antioxidant enzyme genes. The values are means ± SD (n = 3). *, in graphs, indicates significant differences in the means at p < 0.05 between each level in transgenic lines and WT plants, and **, ***, and ****, in graphs, indicate significant differences in the means at p < 0.01, p < 0.001, and p < 0.0001, respectively.
Analysis of antioxidant enzyme activities showed that there were no significant differences in APX, CAT, POD, and SOD contents between wild-type and transgenic lines before low-temperature treatment. These antioxidant enzyme activities decreased after low-temperature treatment, in both WT and transgenic lines. Among them, APX and POD activities were significantly lower in the transgenic lines than in the wild type, but the opposite was true for CAT and SOD (Figures 3F,G). The expression of the antioxidant enzyme genes also showed a similar profile to the enzyme activity, the only difference being that the expression of these genes showed an overall upregulation after low-temperature treatment (Figures 3H,I). These results suggested that overexpression of the ABI4 gene under low-temperature conditions affected gene expression and enzyme activity of these antioxidant enzymes.
As a major part of the non-enzymatic antioxidant system, the ASA-GSH cycle played an important role in the scavenging of reactive oxygen species and resistance to stresses in plants (Nahar et al., 2015). Corresponding compound content analysis indicated that the transgenic plants contained lower ASA and DHA, but higher ASA/DHA (Figures 4A–C). GSH, GSSG, and GSH/GSSG also exhibited similar trends (Figures 4D–F). This would suggest that there are lower levels of oxidation in transgenic plants. Meanwhile, the activities of MDHAR and GR in transgenic lines demonstrated a significant enhancement over those in WT, while the activity of DHAR showed no significant difference (Figures 4G–I). The expression of the DHAR1 in transgenic plants was significantly upregulated compared to the WT, but there were no significant differences in MDHAR4 and GR1 (Figures 4J–L).
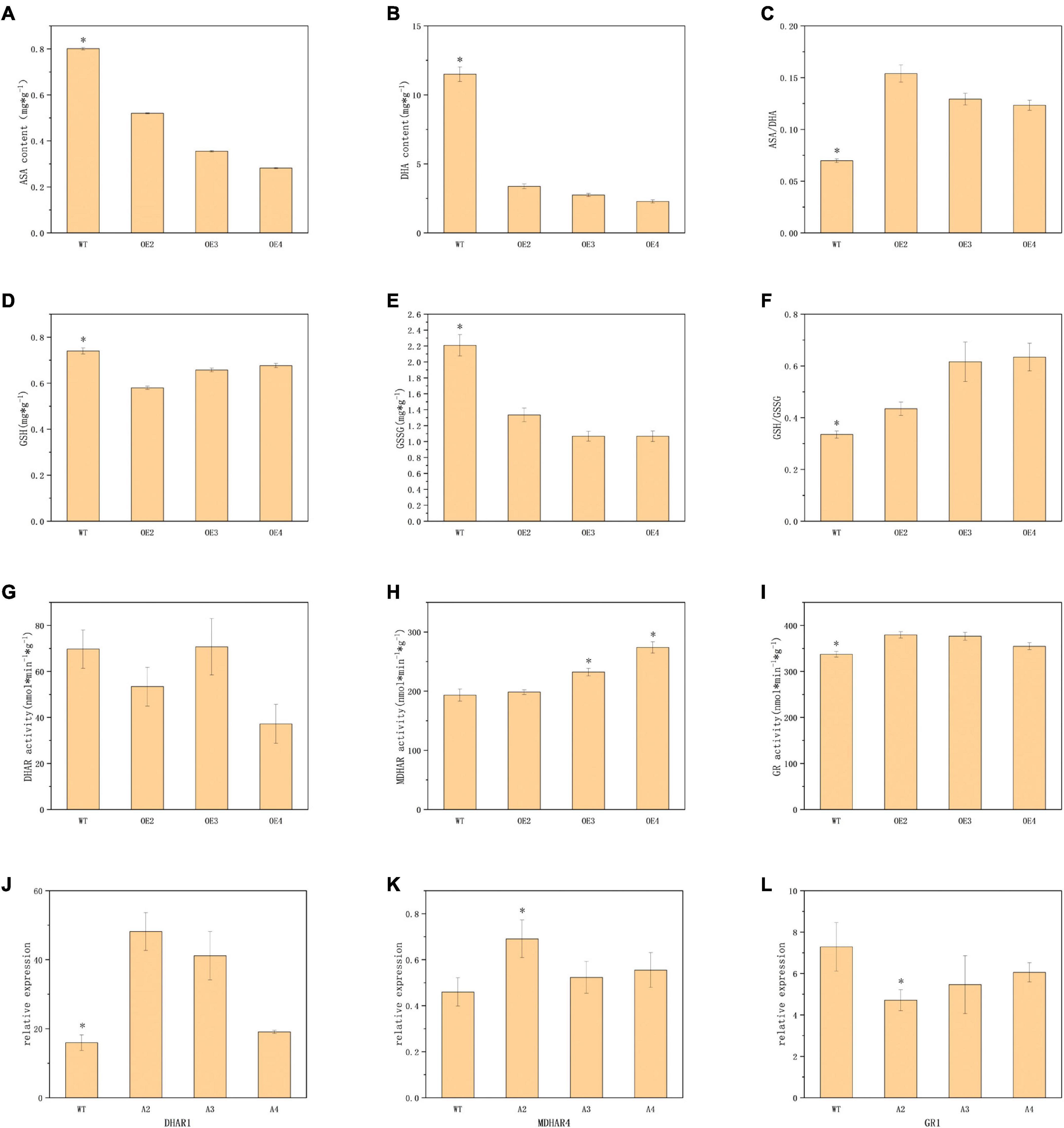
Figure 4. Determination of indicators related to non-enzymatic antioxidant systems. Panels (A,B,D,E) represents the content of ASA, DHA, GSH, and GSSG in turn. Panel (C,F) represents the ASA/DHA and GSH/GSSG ratio, respectively. Panels (G–I) represents the activities of DHAR, MDHAR, and GR. Panels (J–L) represents the relative expression of DHAR1, MDHAR4, and GR1. The values are means ± SD (n = 3). *, in graphs, indicates significant differences in the means at p < 0.05 between each level in transgenic lines and WT plants.
MtABI4 can affect the expression of the Inducer of CBF expression genes-C-repeat binding transcription factor genes-cold regulated genes module and some other low-temperature responsive genes
To further explore the regulation pattern of the ABI4 response pathway to low temperature, the expression pattern of related genes was identified. The results showed that under room temperature conditions, the expression of COR413, ICE1, and OST1 was significantly lower in transgenic plants than in WT, while the opposite was true for CBF1, CBF2, CBF3, and CBF4 (Figures 5A–G). The expression of RAV1 did not exhibit significant differences between the transgenic plants and WT (Figure 5H). This suggested that MtABI4 can affect the expression of the COR-CBF-ICE module and OST1, however, the effect on RAV1 was not significant.
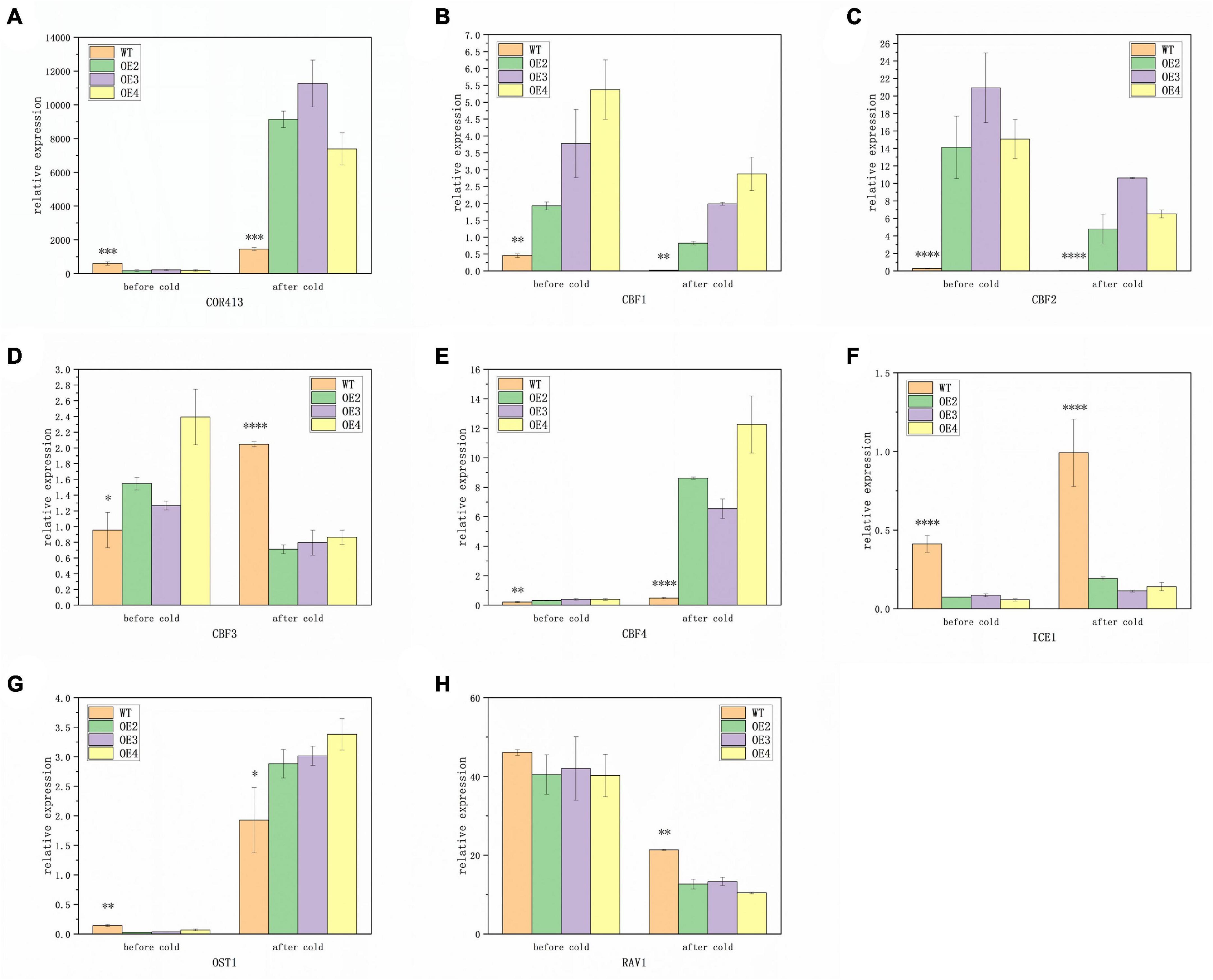
Figure 5. Expression patterns of genes responsive to low temperatures. (A–H) Represent the expression levels of COR413, CBF1-4, ICE1, OST1, and RAV1, in turn, under low temperatures. The values are means ± SD (n = 3). *, in graphs, indicates significant differences in the means at p < 0.05 between each level in transgenic lines and WT plants, and **, ***, and ****, in graphs, indicate significant differences in the means at p < 0.01, p < 0.001, and p < 0.0001, respectively.
After low-temperature treatment, there was a significant increase in the expression of COR413, CBF4, ICE1, and OST1 in all lines, compared to those before low-temperature treatment (Figures 5A,E,F,G). Among them, the expression of COR413 and OST1 was significantly higher in transgenic plants than in WT, in contrast to before treatment. In addition, the expression of ICE1 remained higher in WT, and the expression of CBF4 remained higher in transgenic plants. The expression of the remaining genes, including CBF1, CBF2, and RAV1 was down-regulated both in the transgenic plants and WT after low-temperature treatment (Figures 5B,C,H). Among them, the transgenic plants exhibited a significantly higher expression of CBF1 and CBF2 than WT, while the expression of RAV1 was higher in WT. This indicated that the roles of the COR-CBF-ICE module and OST1 and RAV1 in the molecular response mechanisms induced by low temperature were somewhat altered under the influence of overexpressed ABI4 genes.
MtABI4 directly interacts with MtABA2 protein and promotes the expression of the ABA2 gene, which in turn increases abscisic acid content in transgenic plants
To screen for candidate proteins that might interact with MtABI4 protein, we purified ABI4-3flag fusion proteins transiently expressed in the wild-type callus of Medicago using flag magnetic beads. SDS-page electrophoretic analysis of the purified products showed that some candidate target proteins were obtained (Figure 6A). Mass spectrometry analysis showed that 145 target proteins were collected (Figure 6B), including a xanthoxin dehydrogenase gene (ABA2), considered a key role in abscisic acid biosynthesis (González-Guzmán et al., 2002). Further, the yeast two-hybrid assay revealed that only the mixture of yeast, containing pGADT7-ABA2 and pGBKT7-ABI4, respectively, could be capable of normal growth and turn blue in SD medium lacking the four amino acids, while the other controls could not (Figure 6C). Bimolecular fluorescence complementation (BIFC) assay indicated that GFP fluorescence in nuclear was observed only in Tobacco cells co-transformed with ABI4-NYFP and ABA2-CYFP (Figure 6D). Together, these results demonstrated that MtABI4 could directly interact with MtABA2 protein in vivo. In addition, the ABA2 protein was also located in the nucleus (Figure 6E).
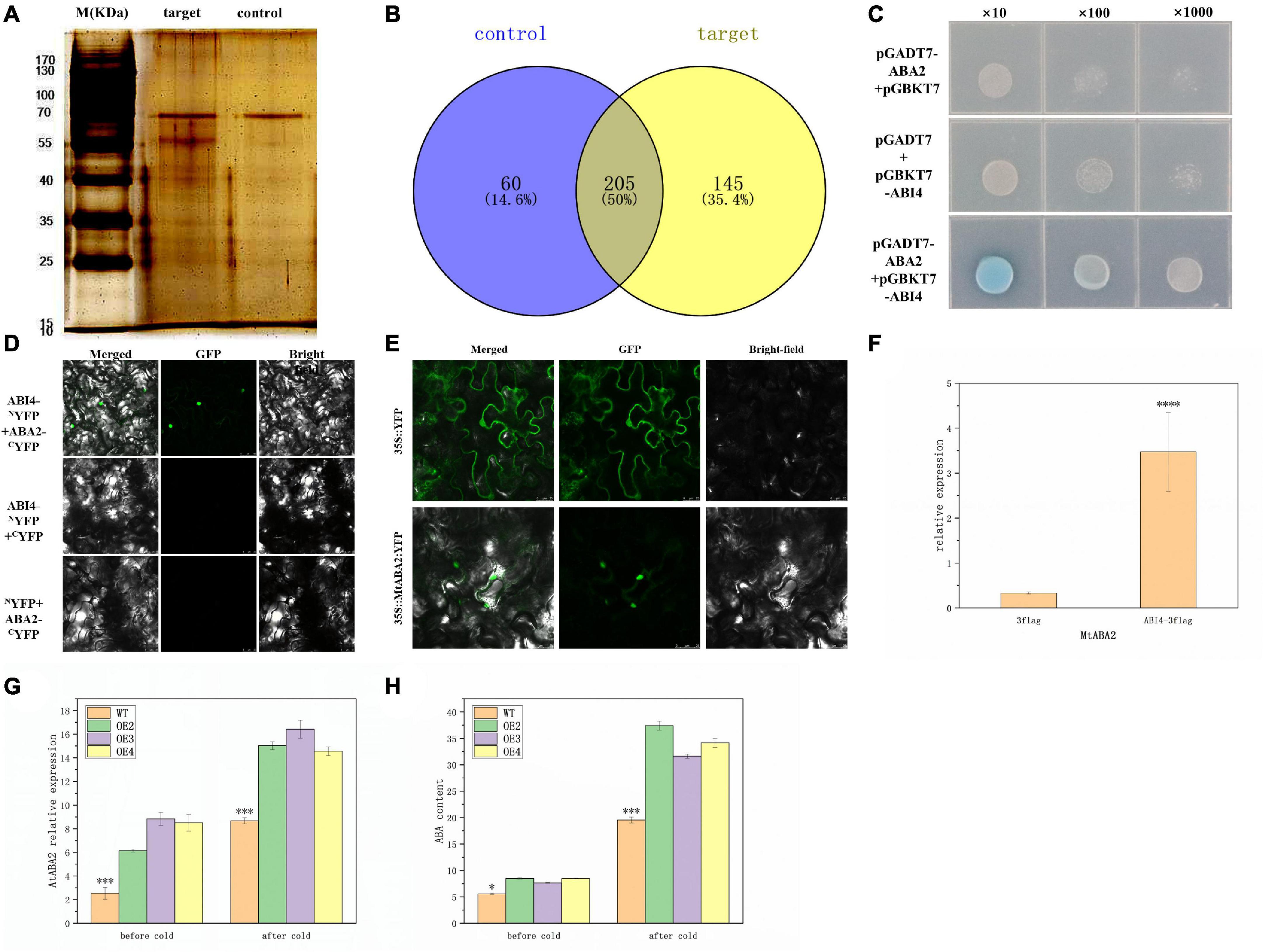
Figure 6. IP-MS, Y2H, BIFC, the expression level of ABA2, and ABA content. (A) Represents the results of the SDS-page electrophoretic. (B) Represents the intersection of the target and control mass spectrometry data. (C) Represents the growth of yeast in the Y2H analysis. (D) Represents the fluorescence observed in the BIFC analysis. (E) Represents the subcellular location of ABA2. (F,G) Represent ABA2 expression levels in M. truncatula callus, and in transgenic Arabidopsis before and after low-temperature treatment. (H) Represents the ABA level in the plant before and after the low-temperature treatment. *, in graphs, indicates significant differences in the means at p < 0.05 between each level in transgenic lines and WT plants, and ***, ****, in graphs, indicates significant differences in the means at p < 0.001 and p < 0.0001, respectively.
To further explore the regulatory pattern of ABI4 on ABA2, the expression of ABA2 in previously transfected callus and transgenic Arabidopsis was characterized by qRT-PCR. The results showed that the expression of MtABA2 and AtABA2 was up-regulated under the induction of MtABI4 (Figures 6F,G). It suggested that over-expression of MtABI4 could promote the expression of the ABA2 gene both in M. truncatula callus and transgenic Arabidopsis. Meanwhile, the ABA content also increased in the transgenic Arabidopsis (Figure 6H). Previous studies showed that the main function of ABA2 is to catalyze the conversion of xanthoxin to Abscisic Aldehyde which was confirmed as one of the last steps of the major ABA biosynthetic pathway (González-Guzmán et al., 2002). In conclusion, this was evidence that MtABI4 could increase the ABA content in the transgenic Arabidopsis through the upregulated expression of ABA2. Furthermore, the overexpression of MtABI4 also enhanced the expression of ABA2 and improved the ABA biosynthesis of the plants, under low-temperature conditions (Figures 6G,H).
Discussion
ABI4 gene plays an integral role in plant growth and development and response to adversity. In this study, we found that the protein sequence of MtABI4 contains three elements and an AP2 structural domain, which is highly conserved with homologous proteins in other closely related legumes. This demonstrated that it was representative of investigating the function of the MtABI4 gene from Medicago truncatula, a kind of model plant (Stacey et al., 2006), which could provide a reference for studying the function of homologous genes in other legumes. The MtABI4 protein was localized in the nucleus, and although this was inconsistent with our previous predictions, it was consistent with the localization of homologous proteins in Arabidopsis (Gregorio et al., 2014). This also confirms the accuracy of our results. It is important to note that YFP and GFP fluorescence have very close excitation light bands. To better avoid background interference, we chose the excitation band of GFP to observe the fluorescence of the YFP protein.
Analysis of the promoter sequence of the ABI4 gene and hormone-induced expression analysis showed that the ABI4 gene was able to respond to a variety of hormones, including IAA, 6-BA, GA, MeJA, SA, and ABA, which are thought to be associated with plant growth and development and stress response (Kazan, 2013; Colebrook et al., 2014). Several previous studies have suggested that ABI4, as a downstream of DES1, is involved in the ABA response in Arabidopsis (Zhou et al., 2021). ABA stabilizes the ABI4 protein, while GA promotes its degradation (Shu et al., 2016). This also coincides with our results that MtABI4 was down-regulated under the treatment of GA, but up-regulated under the treatment of ABA. In addition, ABI4 expression is induced by cytokinin (CTK) but inhibited by IAA (Shkolnik-Inbar and Bar-Zvi, 2010). Notably, our results indicated that although the expression of ABI4 showed a similar decrease in the IAA treatment, it showed a decrease followed by an increase under the 6-BA treatment rather than a simple increase. This may be because the response of ABI4 to CTK is phased in a temporal gradient. Moreover, SA and MeJA induced the expression of PtrWRKY18 and PtrWRKY35, and overexpression of PtrWRKY18 and PtrWRKY35 down-regulated the expression of ABI4 (Guo et al., 2020). It is reasonable to speculate that the inhibition of ABI4 by SA and MeJA in this study may also be mediated by WRKY18 and WRKY35. In summary, ABI4 is widely involved in the molecular response mechanisms of manyp lant hormones and may play a pivotal role in hormone synergy.
In addition, we also discovered a low-temperature response element in the promoter region of ABI4. After the low-temperature treatment, the transgenic plants accumulated less anthocyanin and suffered less cell membrane damage. Synthesis and accumulation of anthocyanins are employed to contribute to plant resistance to low-temperature stress. For example, as previously reported, the cold tolerance of the Hedera helix increases as the total sugar and anthocyanin content of the leaves increases (Parker, 1962). Wild-type Arabidopsis plants are significantly more freeze-resistant than the four cold-sensitive mutants that do not synthesize anthocyanins (McKown et al., 1996). The northern ecotype Populus trichocarpa is more susceptible to surviving the winter than the southern ecotype due to its ability to accumulate more anthocyanins during the progressively shorter photoperiod (Chalker-Scott, 1999). This means that the transgenic plants in this study are insensitive to low temperatures and do not require the accumulation of large amounts of anthocyanin to respond dramatically to low temperatures, as the wild type does. Further, no significant differences in gene expression and enzyme activities of the four antioxidant enzymes appeared in transgenic plants and WT before the low-temperature treatment, different from the situation in low-temperature conditions. This suggests that ABI4 may not act directly on these antioxidant enzyme genes, but indirectly affects their performance at low temperatures. After low-temperature treatment, overexpression of ABI4 promoted the gene expression and enzyme activity of CAT and SOD but inhibited those of APX and POD. Numerous studies have suggested that gene expression and enzyme activity of SOD and CAT are positively correlated with plant cold tolerance (Gao et al., 2009; Zhang et al., 2018, 2021). This is also consistent with our results. In contrast, the significantly lower APX and POD enzyme activities in transgenic plants than in WT may be due to the insensitivity of the transgenic plants to low temperatures. This is worth exploring further in the future. In addition, the activities of antioxidant enzymes in plants are in chronological order, with a trend of increasing and then decreasing with the duration of low-temperature treatment (Ahmad et al., 2010). This may account for the significant decrease in antioxidant enzyme activity in both transgenic plants and WT after up to 5 days of cold treatment.
ICE1 is the initial step in the ICE-CBF-COR signaling pathway, which plays a critical regulatory role in the low-temperature signaling pathway that involves multiple transcription factors (Stockinger et al., 1997). TaICE1a and TaICE1d were induced by cold treatment in wheat (Guo et al., 2019). Our results are also consistent with this, but the regulatory role of MtABI4 on the expression of ICE1 is more likely to be a repressor. And overexpression of MdABI4 is capable of upregulating the MdCBF1, MdCBF2, MdCBF3, and MdCOR47 in apple calli (An et al., 2022). Cold acclimation induces freezing tolerance in two chrysanthemums species by increasing COR413 expression (Chen et al., 2014). Interestingly, except for those genes with the expression pattern consistent with previous results, ABI4 acted as an inhibitor of COR413 expression at room temperature but induced it at low temperature, whereas CBF3 expression displayed the opposite trend. This “switch-like” regulation of CBF3 and COR413 by ABI4 under low-temperature induction deserves further investigation. It has been shown that COR can only be activated to produce cold-regulated proteins only under certain conditions, such as low temperature and short sunlight, thus improving cold tolerance in plants (Jia et al., 2016). Under normal temperature, the HOS15-HD2C complex binds to the CORs promoter and represses expression by inducing CORs chromatin deacetylation. HOS15 enhances the binding of CBFs to the CORs promoter by activating the E3 ligase CUL4 (CULLIN4) for ubiquitinated degradation of HD2C under low-temperature stress (Park et al., 2015). It is reasonable to assume that ABI4 may also play a similar role to that of HOS15 in the regulation of COR413 and CBF3. Apart from this, several studies have suggested that RAV1 and OST1 are also actively involved in adaption to cold stress in plants. For instance, VaRAV1 may promote plant cold tolerance in grapevine cells via improving cell membrane stability and inducing the expression of genes involved in plant cell wall composition (Ren et al., 2021). At low temperatures, OST1 can be activated to phosphorylate ICE1, leading to its higher stability and transcriptional activity (Ding et al., 2015). In our results, their regulatory effects by ABI4 also displayed a similar pattern to that of CBF3 and COR413, which may be a new perspective to explore how ABI4 and these cold-inducible genes are mutually regulated and act together in the process of plant low-temperature response.
ABA2 involved in the final step of the ABA biosynthetic pathway plays a vital role in the regulation of endogenous ABA levels in plants (González-Guzmán et al., 2002). It has been shown that the Aba2-2 mutant has lower levels of endogenous ABA than the wild type (Nambara et al., 1998). This exactly complements our results that the elevation of ABA2 expression levels caused by ABI4 overexpression was accompanied by an increase in ABA content. ABA is an important signaling factor for stressful low temperatures and has a protective function for the microtubule structure of cells, and plants under low-temperature stress accumulate ABA in large quantities and rapidly to perform a protective function (Lang et al., 1994). The foliar application of exogenous ABA increased the endogenous ABA content of the leaves and improved the tolerance of the crop to low temperatures (Hongtao et al., 2017). Under low-temperature conditions, exogenous ABA can increase the content of proline, soluble sugar, and soluble protein in rice leaves, effectively increase SOD and CAT activities, reduce Malondialdehyde (MDA) accumulation and enhance cold tolerance of rice. Another more direct strategy adopted in our study was to promote the expression of ABA2 and enhance the accumulation of endogenous ABA, through the overexpression of ABI4, thus improving the cold tolerance of plants.
Conclusion
MtABI4 protein was localized in the cell nucleus and could be induced by multiple hormones, including IAA, GA, MeJA, SA, 6-BA, and ABA. Moreover, MtABI4 could directly interact with MtABA2 protein and promotes the expression of the ABA2 gene, which in turn increases abscisic acid content in transgenic plants. At low temperatures, the high accumulation of ABA in transgenic plants led to increased SOD, CAT, MDHAR and GR activity, a lower MDA level, a higher ASA/DHA ratio and GSH/GSSG ratio, and reduced anthocyanin content, thus providing some protection against low-temperature damage and leaf color deepening in plants. In addition, overexpression of ABI4 also induced the expression of members of the ICEs-CBFs-CORs low-temperature response module. These factors combined to improve the cold tolerance of ABI4 overexpression plants (Figure 7). This study provides a new insight into the mechanisms of ABA-mediated regulation of plant cold tolerance and offers a molecular theoretical basis for plant cold tolerance improvement.
Materials and methods
Plant materials, growth conditions, and treatments
The seeds of M. truncatula (R108), Arabidopsis thaliana, and Nicotiana benthamiana, and plant expression vector 3302Y, 3302-3flag, and Agrobacterium tumefaciens EHA105 strain used in this study were preserved in the laboratory of turfgrass science and management of Beijing forestry university. These seeds were vernalized for three days at 4°C and then germinated on the sterile filter paper for about one week. The seedlings were transferred to a vegetative soil matrix (peat: vermiculite = 1:1). Medicago and Nicotiana were cultivated in the greenhouse under the condition of a 16 h light/8 h dark cycle at 26°C/24°C, and Arabidopsis were under a 16 h light/8 h dark cycle at 23°C/21°C. After two months of cultivation, Medicago with similar growth was selected and sprayed with 10 μmol/L IAA, 10 μmol/L 6-BA, 10 μmol/L GA3, 10 μmol/L ABA, 10 μmol/L MeJA, and 0.5 mmol/L SA (Guo et al., 2020), respectively, and the plant tissues were collected at 0, 1, 3, 6, 9, and 12 h for temporal expression analysis.
Gene cloning and bioinformatics analysis
Total RNA was extracted from well-grown leaves of M. truncatula, and then reverse transcribed into cDNA by using the PrimeScript RT reagent Kit with gDNA Eraser from TAKARA. Using cDNA as a template and ABI4-CLONE-F, and ABI4-CLONE-R (Supplementary Table 1) as primers, the CDS of this gene was obtained by RT-PCR. And then it was attached to the pMD19-T clone vector from TAKARA, named as ABI4-pMD19-T vector, and was preserved in DH5α E. coli from TAKARA for further analysis.
DNAMAN 6.0 was used to analyze the sequence of MtABI4 and predict the encoded amino acid sequence. Blast homology comparison was performed on the NCBI website. ABI4 homologous proteins in different species were mapped into evolutionary trees by MEGA 5.0. MEME website1 was used for motif analysis. The results above were visualized via TBtools (Chen Chengjie, Guangzhou, Guangdong, China). SignalP 4.1 Server website2 and Softberry website3 were used to predict signal peptide cleavage sites and subcellular localization, respectively.
Subcellular localization and spatio-temporal expression analysis
Using the ABI4-pMD19-T vector as a template and ABI4-3302y-F, ABI4-3302y-R (Supplementary Table 1) as primers, the 35S:MtABI4:YFP expression vector was constructed by PCR and was turned into EHA105 A. tumefaciens for analysis of subcellular localization. The qualified bacterial solution above was injected into the young tender leaves of Nicotiana benthamiana. After 48 h of dark culture, the infected leaves were observed with LEICA SP-8 confocal microscope to determine the localization of the ABI4 protein. The localization of ABA2 protein was analyzed by the same methods.
The root, stem, leaf, flower, seeds, and pods of wild-type M. truncatula were collected and stored at −80°C after quick freezing with liquid nitrogen for further analysis of spatial differential expression. Total RNA of these tissues was extracted and then transcribed into cDNA as a template for quantitative fluorescence detection. Using ABI4-RT-F, ABI4-RT-R (Supplementary Table 1) as primers and the MtActin gene as the internal reference, real-time fluorescence quantitative analysis of the MtABI4 gene was completed by the CFX Connect RT-PCR system. The reaction conditions were as follows: 95°C for 10 min;95°C 15 s, 60°C 1 min, 40 cycles. And the relative expression of ABI4 was determined by the 2–ΔΔCT method (Guo et al., 2020). All samples were processed with three biological replicates. Temporal expression analysis of MtABI4 adopted the same method mentioned above. The primers used were shown in the Supplementary Table 1.
Generation and identification of transgenic plants
The 35S:MtABI4:YFP expression vector was transformed into Arabidopsis plants by the floral dip method (Guo et al., 2020). Positive transgenic plants were screened by spraying 60 mg⋅L–1 glyphosate and then identified by PCR and RT-PCR. T3 generation transgenic lines were cultivated for further treatment and analysis. See Supplementary Table 1 for the primers used.
Chlorophyll content
The chlorophyll content was determined using 95% ethanol as described in the previous report (Teng et al., 2021). Total chlorophyll, Chl a, Chl b, and carotenoid contents were calculated based on the absorbance recorded. All the samples were replicated three times.
Anthocyanin content
The anthocyanin content was determined using 10 mL 1% methanol hydrochloride solution (pH = 0–2). After shaking at 30°C for 5 h, the absorbance at 530 nm was measured.
Electrolyte leakage and cell membrane permeability
0.1 g plant sample in 25 mL distilled water was shaken for 24 h at room temperature. The electrolyte leakage of supernatant (EL1) was measured. After boiling water bath for 30 min, the electrolyte leakage of supernatant (EL2) was measured. The electrolyte leakage of distilled water (EL0) with the same shaking and boiling was also measured as a control. The electrolyte leakage of samples was calculated as EL1/EL2, and the cell membrane permeability was calculated as (EL1- EL0)/(EL2- EL0).
Antioxidant enzyme activity and malondialdehyde content
SOD activity was measured by photoreduction with nitrogen blue tetrazolium (NBT). 3.4 mL of the reaction mix and 0.3 mL of 20 μmol L–1 riboflavin solution were added to 100 μL of the enzyme solution and then mix well, with no enzyme solution shaded with tin foil as control. The tubes were placed under a 4,000 l× lamp for 20–30 min and the OD values were measured at 560 nm.
POD activity was measured by guaiacol colorimetric assay. The reaction solution, containing 1 mL of phosphate buffer with 50 mmol L–1 pH 7.0, 0.95 mL of 0.2%guaiacol, 1 mL of 0.3% H2O2, and 50 μL of enzyme solution was added to a spectrophotometer cup (1 cm radius) and then mix well with no enzyme solution as a control. The kinetic OD values were measured at 470 nm in one minute. The Time interval was 5 s.
CAT activity was measured by UV spectrophotometry. 2.9 mL of the reaction solution, containing 20 mmol L–1 H2O2 solutions was added to 100 μL of enzyme solution and mix well. Distilled water was used as a blank control, the kinetic OD values were measured at 240 nm in one minute. The time interval was 5 s.
APX activity was measured by ascorbic acid colorimetric assay. The reaction solution, containing 2.82 mL of phosphate buffer with 50 mmol L–1 pH 7.0, 30 mL of 50 mmol L–1 ascorbic acid, 30 μL of 10 mmol L–1 EDTA, and 100 μL of enzyme solution was added to a spectrophotometer cup (1 cm radius) and then mix well with no enzyme solution as a control. The kinetic OD values were measured at 290 nm in one minute. The time interval was 5 s.
MDA content was measured by the thiobarbituric acid (TBA) reduction method. 1 mL of the enzyme solution, mixed with 2 mL TBA in a 5 mL centrifugal tube was boiled for 30 min. The tubes were then rapidly transferred to an ice bath for cooling to abort the reaction. After centrifugation at 10,000 g for 10 min, the supernatant was taken and the absorbance values at 600 and 532 nm were measured. All the samples above were replicated three times.
Determination of indicators related to non-enzymatic antioxidant systems
The content of ASA, DHA, GSH, and GSSG and the activities of DHAR, MDHAR, and GR was measured by the corresponding kits (No.G0201F, G0202F, G0206F, G0207F, G0212F, G0213F, and G0209F in turn) from GERUISI-BIO (Suzhou, China).
Abscisic acid content
ABA content were determined by high-performance liquid chromatography (Lu et al., 2010).
Immunoprecipitation, coupled with mass spectrometry
The entire experimental procedure for the Immunoprecipitation-mass spectrometry (IP-MS) analysis was referenced from the previous method with some modifications (Cao and Yan, 2016). Wild-type callus of M. truncatula was obtained via plant tissue culture. The ABI4-3302-3FLAG expression vector, constructed in a similar way to the aforementioned vectors, was transformed into callus by an Agrobacterium-mediated method for transient expression of ABI4 protein. In addition, the 3302-3FLAG expression vector was also transformed into the callus as a control.
After three days of dark incubation, the total protein of the callus was extracted by grinding in PBS buffer (pH = 7.4). The protein samples were purified using BeyoMag™ Anti-Flag Magnetic Beads (Beyotime, China). The end products obtained by elution were subjected to electrophoretic analysis using 10% SDS-PAGE gel. When bands were observed on the silver-stained gel treated with the Fast Silver Stain Kit (Beyotime, China), the gel block was sent to Bio for mass spectrometry analysis (Biomarker, China). The candidate proteins that interacted with the target protein were obtained by subtracting the protein in the control group from the experimental group results.
Yeast two-hybrid
pGADT7-ABA2 and pGBKT7-ABI4 expression vectors were constructed by a similar method above and transformed into Y187 and yeast two-hybrid (Y2H) yeast strains in turn. The mixture of these two kinds of yeast was cultivated in SD solid medium lacking Leu, Trp, His, and Ade, supplemented with 40 μg ml–1 X-α-gal and 125 ng ml–1 Aureobasidin A, for 3–5 days at 29.5°C. Moreover, they matched with the yeast strains containing pGADT7or pGBKT7 empty vector and were cultivated under the same conditions as controls.
Bimolecular fluorescence complementation
ABI4-NYFP and ABA2-CYFP expression vectors were constructed by a method similar to those described above and transformed into Agrobacterium tumefaciens EHA105 strain. These fluids were injected into tobacco leaves by a method similar to subcellular localization. The mixture of strains containing ABI4-NYFP and ABA2-CYFP was the treatment group and these two strains were mixed with those containing the NYFP or CYFP empty vectors, separately, as controls. GFP fluorescence in tobacco cells was observed with LEICA SP-8 confocal microscope, after 2 days of dark incubation.
Statistical analysis
Data were analyzed by SPSS 15.0, Excel 2020, GraphPad Prism 9.0, and Origin 2022. All data were presented as the mean ± SD (n = 3).
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YL proposed ideas and designed the experiments and wrote the manuscript. YL, SL, and MW completed the experiments. YL, TG, and KT analyzed the experimental data. DD, ZL, and YC proposed key opinions that cannot be ignored. LH and CJ played a guiding role in the whole work process. All authors read and reached an agreement with the final manuscript.
Funding
This work is supported by the Fundamental Research Funds for the Central Universities (Grant No. 2021ZY84), the National Natural Science Foundation of China (Grant No. 31971770), the Performance Incentive and Guidance Special Project of Scientific Research Institution, Chongqing Science and Technology Committee, and the Special Support for Chongqing Postdoctoral Research Project. All of them provide funding for the entire research effort, which is used for material maintenance, reagents, data collection, labor costs and so on.
Conflict of interest
CJ was employed by Inner Mongolia Mengcao Ecological Environment (Group) Co., Ltd., Hohhot, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.982715/full#supplementary-material
Supplementary Figure 1 | Bioinformatic analysis of MtABI4.
Footnotes
- ^ https://meme-suite.org/meme/tools/meme
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://www.softberry.com/
References
Ahmad, P., Jaleel, C. A., Salem, M. A., Nabi, G., and Sharma, S. (2010). Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 30, 161–175. doi: 10.3109/07388550903524243
An, J. P., Xu, R. R., Liu, X., Su, L., Yang, K., Wang, X. F., et al. (2022). Abscisic acid insensitive 4 interacts with ice1 and jaz proteins to regulate aba signaling-mediated cold tolerance in apple. J. Exp. Bot. 73, 980–997. doi: 10.1093/jxb/erab433
Bossi, F., Cordoba, E., Dupré, P., Mendoza, M. S., Román, C. S., and León, P. (2009). The Arabidopsis aba-insensitive (abi) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of abi5 and sbe2.2 genes during sugar signaling. Plant J. 59, 359–374. doi: 10.1111/j.1365-313X.2009.03877.x
Campos, C. N., Ávila, R. G., de Souza, K. R. D., Azevedo, L. M., and Alves, J. D. (2019). Melatonin reduces oxidative stress and promotes drought tolerance in young Coffea arabica l. Plants Agric. Water Manag. 211, 37–47. doi: 10.1016/j.agwat.2018.09.025
Cao, X., and Yan, J. (2016). Identification of associated proteins by immunoprecipitation and mass spectrometry analysis. Methods Mol. Biol. 1407, 131–139. doi: 10.1007/978-1-4939-3480-5_10
Chalker-Scott, L. (1999). Environmental significance of anthocyanins in plant stress responses. Photochem. Photobiol. 70, 1–9.
Chen, I., Lee, S., Pan, S., and Hsieh, H. (2007). Gasa4, a ga-stimulated gene, participates in light signaling in Arabidopsis. Plant Sci. 172, 1062–1071. doi: 10.1016/j.plantsci.2007.03.012
Chen, Y., Jiang, J., Chang, Q., Gu, C., Song, A., Chen, S., et al. (2014). Cold acclimation induces freezing tolerance via antioxidative enzymes, proline metabolism and gene expression changes in two Chrysanthemum species. Mol. Biol. Rep. 41, 815–822. doi: 10.1007/s11033-013-2921-8
Chinnusamy, V., Zhu, J., and Zhu, J. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Colebrook, E. H., Thomas, S. G., Phillips, A. L., and Hedden, P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75. doi: 10.1242/jeb.089938
Ding, Y., Jia, Y., Shi, Y., Zhang, X., Song, C., Gong, Z., et al. (2018). Ost1-mediated btf3l phosphorylation positively regulates cbfs during plant cold responses. Embo J. 37:e98228. doi: 10.15252/embj.201798228
Ding, Y., Li, H., Zhang, X., Xie, Q., Gong, Z., and Yang, S. (2015). Ost1 kinase modulates freezing tolerance by enhancing ice1 stability in Arabidopsis. Dev. Cell 32, 278–289. doi: 10.1016/j.devcel.2014.12.023
Ding, Y., Shi, Y., and Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Eremina, M., Rozhon, W., and Poppenberger, B. (2016). Hormonal control of cold stress responses in plants. Cell. Mol. Life Sci. 73, 797–810. doi: 10.1007/s00018-015-2089-6
Finkelstein, R. R. (1994). Mutations at two new Arabidopsis aba response loci are similar to the abi3 mutations. Plant J. 5, 765–771. doi: 10.1046/j.1365-313X.1994.5060765.x
Finkelstein, R., Lynch, T., Reeves, W., Petitfils, M., and Mostachetti, M. (2011). Accumulation of the transcription factor aba-insensitive (abi)4 is tightly regulated post-transcriptionally. J. Exp. Bot. 62, 3971–3979. doi: 10.1093/jxb/err093
Gao, J. J., Qin, A. G., and Yu, X. C. (2009). Effects of grafting on cucumber leaf sod and cat gene expression and activities under low temperature stress. Chin. J. Appl. Ecol. 20, 213–217.
González-Guzmán, M., Apostolova, N., Bellés, J. M., Barrero, J. M., Piqueras, P., Ponce, M. R., et al. (2002). The short-chain alcohol dehydrogenase aba2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14, 1833–1846. doi: 10.1105/tpc.002477
Gregorio, J., Hernández-Bernal, A. F., Cordoba, E., and León, P. (2014). Characterization of evolutionarily conserved motifs involved in activity and regulation of the aba-insensitive (abi) 4 transcription factor. Mol. Plant 7, 422–436. doi: 10.1093/mp/sst132
Guo, J., Ren, Y., Tang, Z., Shi, W., and Zhou, M. (2019). Characterization and expression profiling of the ice-cbf-cor genes in wheat. Peerj 7:e8190. doi: 10.7717/peerj.8190
Guo, T., Wang, S., Li, Y., Yuan, J., Xu, L., Zhang, T., et al. (2020). Expression of a ngatha1 gene from Medicago truncatula delays flowering time and enhances stress tolerance. Int. J. Mol. Sci. 21:2384. doi: 10.3390/ijms21072384
Hongtao, X., Tongtong, W., Dianfeng, Z., Lizhi, W., Yanjiang, F., Yu, L., et al. (2017). Aba pretreatment enhances the chilling tolerance of a chilling-sensitive rice cultivar. Braz. J. Bot. 40, 853–860. doi: 10.1007/s40415-017-0409-9
Hu, Y., Jiang, L., Wang, F., and Yu, D. (2013). Jasmonate regulates the inducer of cbf expression–c-repeat binding factor/dre binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J., and Yu, D. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 68, 1361–1369. doi: 10.1093/jxb/erx004
Jia, Y., Ding, Y., Shi, Y., Zhang, X., Gong, Z., and Yang, S. (2016). The cbfs triple mutants reveal the essential functions of cbfs in cold acclimation and allow the definition of cbf regulons in Arabidopsis. New Phytol. 212, 345–353. doi: 10.1111/nph.14088
Jiang, M., and Zhang, J. (2002). Involvement of plasma-membrane nadph oxidase in abscisic acid- and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 215, 1022–1030. doi: 10.1007/s00425-002-0829-y
Kazan, K. (2013). Auxin and the integration of environmental signals into plant root development. Ann. Bot. 112, 1655–1665. doi: 10.1093/aob/mct229
Klessig, D. F., Choi, H. W., and Dempsey, D. A. (2018). Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant Microbe Interact. 31, 871–888. doi: 10.1094/MPMI-03-18-0067-CR
Lang, V., Mantyla, E., Welin, B., Sundberg, B., and Palva, E. T. (1994). Alterations in water status, endogenous abscisic acid content, and expression of rab18 gene during the development of freezing tolerance in Arabidopsis thaliana. Plant Physiol. 104, 1341–1349. doi: 10.1104/pp.104.4.1341
León, P., Gregorio, J., and Cordoba, E. (2012). Abi4 and its role in chloroplast retrograde communication. Front. Plant Sci. 3:304. doi: 10.3389/fpls.2012.00304
Lu, Q., Chen, L., Lu, M., Chen, G., and Zhang, L. (2010). Extraction and analysis of auxins in plants using dispersive liquid-liquid microextraction followed by high-performance liquid chromatography with fluorescence detection. J. Agric. Food Chem. 58, 2763–2770. doi: 10.1021/jf903274z
McKown, R., Kuroki, G., and Warren, G. (1996). Cold responses of Arabidopsis mutants impaired in freezing tolerance. J. Exp. Bot. 47, 1919–1925. doi: 10.1093/jxb/47.12.1919
Nahar, K., Hasanuzzaman, M., Alam, M., and Fujita, M. (2015). Exogenous spermidine alleviates low temperature injury in mung bean (Vigna radiata l.) Seedlings by modulating ascorbate-glutathione and glyoxalase pathway. Int. J. Mol. Sci. 16, 30117–30132. doi: 10.3390/ijms161226220
Nambara, E. H. U. S., Kawaide, H., Kamiya, Y., and Naito, S. (1998). Characterization of an Arabidopsis thaliana mutant that has a defect in aba accumulation: Aba-dependent and aba-independent accumulation of free amino acids during dehydration. Plant Cell Physiol. 39, 853–858. doi: 10.1093/oxfordjournals.pcp.a029444
Park, S., Lee, C. M., Doherty, C. J., Gilmour, S. J., Kim, Y., and Thomashow, M. F. (2015). Regulation of the Arabidopsis cbf regulon by a complex low-temperature regulatory network. Plant J. 82, 193–207. doi: 10.1111/tpj.12796
Parker, J. (1962). Relationships among cold hardiness, water-soluble protein, anthocyanins, & free sugars in hedera helix l. Plant Physiol. 37, 809–813. doi: 10.1104/pp.37.6.809
Ren, C., Li, H., Wang, Z., Dai, Z., Lecourieux, F., Kuang, Y., et al. (2021). Characterization of chromatin accessibility and gene expression upon cold stress reveals that the rav1 transcription factor functions in cold response in Vitis amurensis. Plant Cell Physiol. 62, 1615–1629. doi: 10.1093/pcp/pcab115
Shi, Y., Ding, Y., and Yang, S. (2015). Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 56, 7–15. doi: 10.1093/pcp/pcu115
Shkolnik-Inbar, D., and Bar-Zvi, D. (2010). Abi4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell 22, 3560–3573. doi: 10.1105/tpc.110.074641
Shkolnik-Inbar, D., and Bar-Zvi, D. (2011). Expression of abscisic acid insensitive 4 (abi4) in developing arabidopsis seedlings. Plant Signal Behav. 6, 694–696. doi: 10.4161/psb.6.5.14978
Shu, K., Chen, Q., Wu, Y., Liu, R., Zhang, H., Wang, P., et al. (2016). Abi4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 85, 348–361. doi: 10.1111/tpj.13109
Stacey, G., Libault, M., Brechenmacher, L., Wan, J., and May, G. D. (2006). Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 9, 110–121. doi: 10.1016/j.pbi.2006.01.005
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana cbf1 encodes an ap2 domain-containing transcriptional activator that binds to the c-repeat/dre, a cis-acting dna regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U. S. A. 3:1035. doi: 10.1073/pnas.94.3.1035
Sun, X., Feng, P., Xu, X., Guo, H., Ma, J., Chi, W., et al. (2011). A chloroplast envelope-bound phd transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2:477. doi: 10.1038/ncomms1486
Tang, K., Zhao, L., Ren, Y., Yang, S., Zhu, J. K., and Zhao, C. (2020). The transcription factor ice1 functions in cold stress response by binding to the promoters of cbf and cor genes. J. Integr. Plant Biol. 62, 258–263. doi: 10.1111/jipb.12918
Teng, K., Tan, P., Guan, J., Dong, D., Liu, L., Guo, Y., et al. (2021). Functional characterization of the chlorophyll b reductase gene nyc1 associated with chlorophyll degradation and photosynthesis in Zoysia japonica. Environ. Exp. Bot. 191:104607. doi: 10.1016/j.envexpbot.2021.104607
Yang, Y., Yao, N., Jia, Z. K., Duan, J., Chen, F. J., Sang, Z. Y., et al. (2016). Effect of exogenous abscisic acid on cold acclimation in two magnolia species. Biol. Plant. 60, 555–562. doi: 10.1007/s10535-016-0623-5
Zhang, G., Ding, Q., and Wei, B. (2021). Genome-wide identification of superoxide dismutase gene families and their expression patterns under low-temperature, salt and osmotic stresses in watermelon and melon. 3 Biotech 11:194. doi: 10.1007/s13205-021-02726-7
Zhang, Y., Ji, H., Yu, J., and Zhang, Z. (2018). Effect of cold and heat shock treatment on the color development of mature green tomatoes and the roles of their antioxidant enzymes. Food Bioprocess Technol. 11, 705–709. doi: 10.1007/s11947-017-2053-6
Zhou, M., Zhang, J., Shen, J., Zhou, H., Zhao, D., Gotor, C., et al. (2021). Hydrogen sulfide-linked persulfidation of abi4 controls aba responses through the transactivation of mapkkk18 in Arabidopsis. Mol. Plant 14, 921–936. doi: 10.1016/j.molp.2021.03.007
Keywords: ABI4, Medicago truncatula, cold tolerance, xanthoxin dehydrogenase, ABA
Citation: Li Y, Wang M, Guo T, Li S, Teng K, Dong D, Liu Z, Jia C, Chao Y and Han L (2022) Overexpression of abscisic acid-insensitive gene ABI4 from Medicago truncatula, which could interact with ABA2, improved plant cold tolerance mediated by ABA signaling. Front. Plant Sci. 13:982715. doi: 10.3389/fpls.2022.982715
Received: 30 June 2022; Accepted: 29 August 2022;
Published: 23 September 2022.
Edited by:
Kehua Wang, China Agricultural University, ChinaCopyright © 2022 Li, Wang, Guo, Li, Teng, Dong, Liu, Jia, Chao and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liebao Han, aGFubGllYmFvQDE2My5jb20=
 Yinruizhi Li
Yinruizhi Li Mengdi Wang1
Mengdi Wang1 Tao Guo
Tao Guo Ke Teng
Ke Teng Yuehui Chao
Yuehui Chao Liebao Han
Liebao Han