
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 31 August 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.982317
This article is part of the Research Topic Artemisinin - From Traditional Chinese Medicine to Artemisinin Combination Therapies; Four Decades of Research on the Biochemistry, Physiology, and Breeding of Artemisia annua, Volume II View all 5 articles
 Tian-Tian Chen
Tian-Tian Chen Xing-Hao Yao
Xing-Hao Yao Hang Liu
Hang Liu Yong-Peng Li
Yong-Peng Li Wei Qin
Wei Qin Xin Yan
Xin Yan Xiu-Yun Wang
Xiu-Yun Wang Bo-Wen Peng
Bo-Wen Peng Yao-Jie Zhang
Yao-Jie Zhang Jin Shao
Jin Shao Xin-Yi Hu
Xin-Yi Hu Qing Miao
Qing Miao Xue-Qing Fu
Xue-Qing Fu Yu-Liang Wang
Yu-Liang Wang Ling Li
Ling Li Ke-Xuan Tang*
Ke-Xuan Tang*The plant Artemisia annua is well known for its production of artemisinin, a sesquiterpene lactone that is an effective antimalarial compound. Although remarkable progress has been made toward understanding artemisinin biosynthesis, the effect of MADS-box family transcription factors on artemisinin biosynthesis is still poorly understood. In this study, we identified a MADS transcription factor, AaSEP4, that was predominantly expressed in trichome. AaSEP4 acts as a nuclear-localized transcriptional activator activating the expression of AaGSW1 (GLANDULAR TRICHOME-SPECIFIC WRKY1). Dual-luciferase and Yeast one-hybrid assays revealed that AaSEP4 directly bound to the CArG motif in the promoter region of AaGSW1. Overexpression of AaSEP4 in A. annua significantly induced the expression of AaGSW1 and four artemisinin biosynthesis genes, including amorpha-4,11-diene synthase (ADS), cytochrome P450 monooxygenase (CYP71AV1), double-bond reductase 2 (DBR2) and aldehyde dehydrogenase 1 (ALDH1). Furthermore, the results of high-performance liquid chromatography (HPLC) showed that the artemisinin content was significantly increased in the AaSEP4-overexpressed plants. In addition, RT-qPCR results showed that AaSEP4 was induced by methyl jasmonic acid (MeJA) treatment. Taken together, these results explicitly demonstrate that AaSEP4 is a positive regulator of artemisinin biosynthesis, which can be used in the development of high-artemisinin yielding A. annua varieties.
Malaria is a mosquito-borne infectious disease that became a life-threatening problem with more than three billion people, especially in South-East Asia and Africa (Garcia, 2010; Ma et al., 2020). Artemisinin, a sesquiterpene lactone endoperoxide, is isolated from the traditional Chinese medicinal plant Artemisia annua (Zhang et al., 2014; Xiong and Huang, 2021). According to the World Health Organization (WHO), Artemisinin-based combination therapy (ACT) is considered the most recommended treatment to Plasmodium falciparum malaria (White, 2008). In addition, recent studies have reported that Artemisinin is also effective in the treatment of several cancers (Efferth, 2017). In yeast, semi-synthetic high production of artemisinin has been successfully developed (Paddon et al., 2013). Although production of artemisinin is quite low (0.1%–1.0% DW of A. annua), A. annua is the only plant source for artemisinin production (Shen et al., 2016). Therefore, improving the content of artemisinin in A. annua is necessary and urgent. The artemisinin biosynthesis pathway has been studied extensively and genes underlying most of the biosynthetic steps have been identified in A. annua. ADS catalyzes farnesyl diphosphate (FPP) converses to amorpha-4, 11-diene is the first step of artemisinin production, then amorpha-4, 11-diene is converted to dihy-droartemisinic acid (DHAA) through the function of CYP71AV1, DBR2, and ALDH1. Finally, DHAA is transformed to artemisinin in the glandular trichome of A. annua. (Mercke et al., 2000; Teoh et al., 2006, 2009; Zhang et al., 2008). Previous studies have shown that analyzing the artemisinin biosynthesis regulatory mechanism may reveal strategies for generating large yields and high-quality artemisinin (Zhang et al., 2013; Lv et al., 2016). A number of transcription factors from various families have been found to enhance the production of artemisinin via up-regulating the expression of ADS, CYP71AV1, DBR2, and ALDH1 (Chen et al., 2017, 2021; Ma et al., 2018; Hao et al., 2019). For instance, the WRKY transcription factor AaGSW1 (GLANDULAR TRICHOME-SPECIFIC WRKY1) was reported to enhance artemisinin biosynthesis by directly binding to the promoter of CYP71AV1 (Chen et al., 2017). It also has been demonstrated that overexpression of the AP2/ERF transcription factors (TFs) such as AaERF1 and AaERF2 increases the artemisinin content through increasing the transcript levels of ADS and CYP71AV1 (Yu et al., 2012). However, knowledge of the transcriptional regulatory mechanisms that control the expression of four enzyme genes in artemisinin biosynthesis is rather limited.
The MADS-box TFs that share conserved DNA-binding domain have been extensively studied in plant, animal, and fungi (Shore and Sharrocks, 1995; Smaczniak et al., 2012; Schilling et al., 2018). Recently, an increasing number of studies have shown that MADS-box TFs participate in the regulation of secondary metabolism in various plants (Vrebalov et al., 2009; Martel et al., 2011). In tomato, MADS-box TF RIN (Ripening Inhibitor) through directly regulating the expression of PSY (phytoene synthase) to promote the lycopene accumulation (Martel et al., 2011). Similarly, silencing of the MADS-box genes TAGL1, and FUL1/2 (FRUITFULL 1/2) significantly decreased carotenoid accumulation (Vrebalov et al., 2009; Bemer et al., 2012; Wang et al., 2014). The citrus transcription factor CsMADS6 positively modulates carotenoid metabolism by directly regulating the transcript level of LCYb1 (Lycopene β-cyclases) and other carotenogenic genes (Lu et al., 2018). However, few researchers have been able to identify any MADS-box TFs that are involved in the regulation of artemisinin biosynthesis in A. annua.
In this study, we identified a MADS-box transcription factor AaSEP4 that directly binding to the promoter of AaGSW1. Overexpression of AaSEP4 obviously increased the transcript levels of AaGSW1 and all four key enzymes (ADS, CYP71AV1, DBR2, ALDH1), thus enhancing the artemisinin biosynthesis in A. annua. In conclusion, our research reveals a novel MADS-box TF that regulates artemisinin biosynthesis, which advances our understanding of the complex transcriptional regulation of artemisinin metabolism in A. annua.
High artemisinin content A. annua cultivar ‘Huhao 1’ was used in this study, which originated in Chongqing and has been developed several years in Shanghai. Artemisia annua and Nicotiana benthamiana seeds were grown in pots at 24 ± 2°C and under a 16 h light photoperiod. For MeJA treatment, 2-week-old A. annua plants were sprayed with 100 μM methyl jasmonate (MeJA; Sigma-Aldrich), 0.1% ethanol as a mock control treatment. Leaf samples were collected at 0, 0.5, 1.5, 3, 6, 12, and 24 h after treatment.
RNA of A. annua tissues and leaves was extracted using a plant RNA isolation reagent (Tiangen Biotech, Beijing, China). Trichomes were isolated from buds as previously described (Wang et al., 2009). Glass beads and a commercial cell disrupter (BioSpec Products) were used to separate trichome cells from the surface of flower buds. Then cells and tissue mixture sequentially pass through a 40 μm and a 30 μm nylon sieves and finally collected glandular trichome cells in 30 μm meshes. RNA samples were extracted using a plant RNA isolation reagent (Tiangen Biotech, Beijing, China), and the reverse transcription of complementary DNA (cDNA) was performed by using a PrimeScript RT Master Mix (Takara, Japan). The expression level of all relative genes was performed on a Roche lightercycler96 real-time PCR machine (Roche, Basel, Switzerland) and using the SuperReal PreMix Plus SYBR-Green (Tiangen Biotech, China). Each sample has three biological replicates. All the primers used are listed in Supplementary Table S1.
The ORF of AaSEP4 was amplified using KOD plus DNA polymerase and then cloned into the plant expression vector pHB-YFP to generate a pHB-AaSEP4–YFP fusion protein. Then the plasmid was transferred into A. tumefaciens strain GV3101 for N. benthamiana leaf transient expression. After 48 h low light condition, the fluorescent signals of N. benthamiana leaves were observed by confocal laser microscopy (Leica TCS SP5-II). 4′, 6-diamidino-2-phenylindole (DAPI) was used for nuclei stain, pHB-YFP was used as negative control.
The 738 bp full-length cDNA of AaSEP4 was amplified by using KOD plus DNA polymerase (Toyobo, Osaka, Japan) and then cloned into the pHB vector. The construct pHB-ANAaSEP4 was introduced into the Agrobacterium tumefaciens strain EHA105 and genetically transformed into A. annua for further analysis as described previously (Hao et al., 2019). Firstly, A. annua seeds were placed on germination medium MS0 and then cultured at 24°C–26°C with 16 h light and 8 h dark treatment (8,000 lux). After 2 weeks, the leaves of the germinated seedlings were collected and cut into 0.5-cm-diameter discs, then these cut leaves were co-cultivated with A. tumefaciens strain EHA105 at 25°C for 3 days. Then the leaves were transferred to the selective medium MS1 (MS0 + 2.5 mg/L N6-benzoyladenine +0.3 mg/L naphthalene-1-acetic acid +50 mg/L hygromycin +250 mg/L carbenicillin), we selected the antibiotic-resistant plantlets sub-cultured three times and then transferred them into rooting medium MS2 (½ MS0 + 250 mg/L carbenicillin). Finally, the rooted plantlets were transferred to soil pots in the growth chamber after 1 month.
To construct 1391Z-proAaSEP4-GUS, the 1,386-bp promoter region upstream of the start codon of AaSEP4 was amplified from the A. annua genomic DNA library and inserted into the pCambia1391Z vector. The plasmids 1391Z- proAaSEP4-GUS were introduced into A. annua plants using Agrobacterium-mediated genetic transformation, as described previously (Hao et al., 2019). Four-week-old 1391Z-proAaSEP4-GUS transgenic A. annua plants were stained to observe the tissue distribution. GUS assay was performed as previously described (Xie et al., 2021). GUS staining solution [1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid, 100 mM Na2HPO4, 50 mM KH2PO4, 10 mM Na2EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, and 0.1% (v/v) Triton X-100] was used to stain leaves, then leaves were incubated at 37°C for 12 h in the dark. After staining, 70% ethanol was used to remove chlorophyll for three times.
The fragment containing AaSEP4 binding site (GArG-box) was amplified from the promoter of AaGSW1 and cloned into the pLacZ vector. The ORF of AaSEP4 was amplified and ligated into the pB42AD vector. Various combinations of pB42AD-AaSEP4/pB42AD and pLacz-3 x CArG-box/pLacz-3 x mCArG-box were co-transformed into the yeast strain EGY48a. An empty pB42AD vector was used as a negative control. The transformed yeast cells were grown on SD/−Trp/-Ura plates at 30°C for 2–4 days. SD/−Trp/-Ura plates with X-gal were used as test media. Yeast one-hybrid assays were conducted as previously described (Zhong et al., 2018).
The promoter of AaGSW1 was cloned into pGREEN II 0800 vector as reporter and transformed into A. tumefaciens strain GV3101 with the helper plasmid pSoup 19. PHB-AaSEP4 was transformed into A. tumefaciens strain GV3101 to act as an effector and pHB empty vector was used as a negative control. The effectors and reporter were mixed in a 9:1 volume ratio to transform 4-week-old tobacco leaves (Hellens et al., 2005). The infiltrated leaves of N. benthamiana were detected after 48 h low light incubation by using the Dual-Luciferase Reporter Assay System (Promega, United States). The activity of LUC was normalized to the activity of REN, and the relative LUC/ REN ratios were used to represent the activity of the promoter. Four biological repeats were performed for each sample.
Leaves of 4-month-old A. annua were gathered and dried at 50°C in an oven. Subsequently, leaves were ground into powder and 0.1 g powder was extracted twice with 2 ml methanol under ultrasound for 30 min (55 W, 30°C). After centrifuging 12,000 g, 10 min, the supernatants were filtered through nitrocellulose (0.22 μm). High-performance liquid chromatography (HPLC) was used to analyze the contents of artemisinin (Qin et al., 2021). Three repeats were measured in all samples.
AaGSW1, a glandular trichome-specific WRKY transcription factor, which is a key positive regulator of artemisinin biosynthesis in A. annua. To identify TFs that regulate artemisinin metabolism, we performed a yeast one-hybrid (Y1H) screen. In this study, the promoter sequences of AaGSW1 were used as bait to screen a cDNA library derived from young leaves of A. annua. Several TFs were identified, one of which encoding a protein belonging to the MADS-box TF superfamily. This MADS-box TF was orthologous gene AtSEP4 from Arabidopsis thaliana by a BLAST search of the TAIR database (Figure 1A). Thus, we named this MADS-box TF in A. annua as AaSEP4. The full-length coding sequence of AaSEP4 encoded a protein of 245 amino acids with a calculated molecular mass of 28.36 kDa and a predicted pI of 8.15.1 To further understand the relationship of AaSEP4 to other MADS proteins, a neighbor-joining tree of AaSEP4 and other MADS-box family members in different plant species was constructed (Figure 1B).
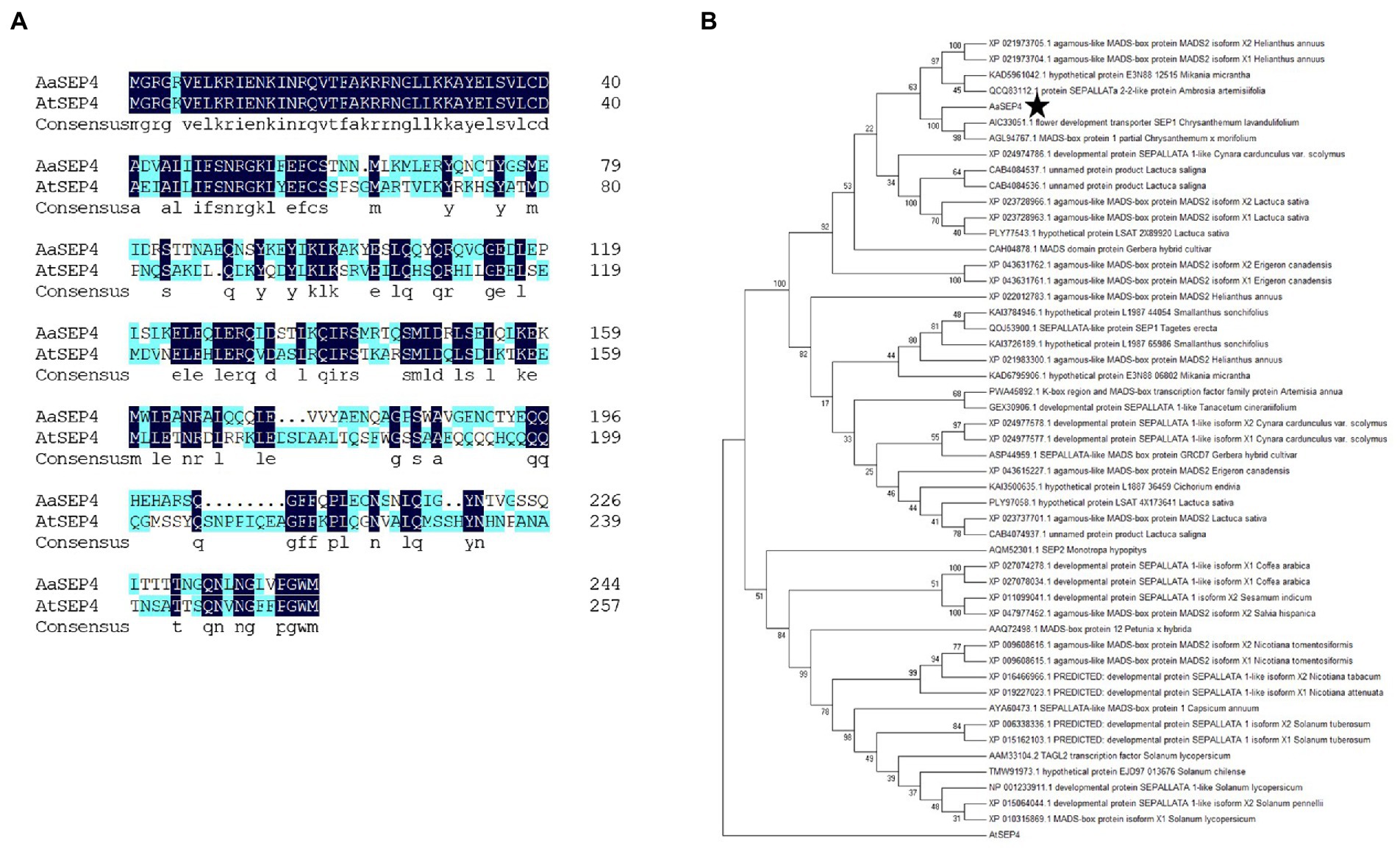
Figure 1. Phylogenetic analysis of AaSEP4. (A) The protein sequence alignment of AaSEP4 and AtSEP4. (B) Phylogenetic analysis was performed using MADS family proteins from various other plant species. The tree presented here is a neighbor-joining tree based on amino acid sequence alignment and constructed using the program MEGA.
To understand the spatial and temporal expression patterns of AaSEP4, we determined its relative transcript levels in different tissues and during the different stages of leaf development in A. annua by RT-qPCR. As Figure 2A shown, AaSEP4 was predominantly expressed in the trichome, flower and bud. During leaf development, the transcript levels of AaSEP4 showed no obvious difference (Figure 2B). Furthermore, 2,186 bp sequence of the AaSEP4 promoter was amplified and generated the construct 1391Z-proAaSEP4-GUS, then transformed it into A. annua. The GUS staining was strongly detected in the glandular secretary trichome (GST) of the transgenic plants (Figure 2C). Previous reports showed that artemisinin biosynthesis is promoted by JA and the expression of AaGSW1 was significantly increased after JA treatment (Zhang et al., 2015; Chen et al., 2017). We therefore investigated whether JA regulates AaSEP4 expression. The results of RT-qPCR experiments revealed that the expression of AaSEP4 was induced drastically increased after 1.5 h of MeJA treatment compared to that in the mock-treated leaves (Figure 2D). These results indicated that AaSEP4 has potentially function in the GST of A. annua and was induced by MeJA treatment.
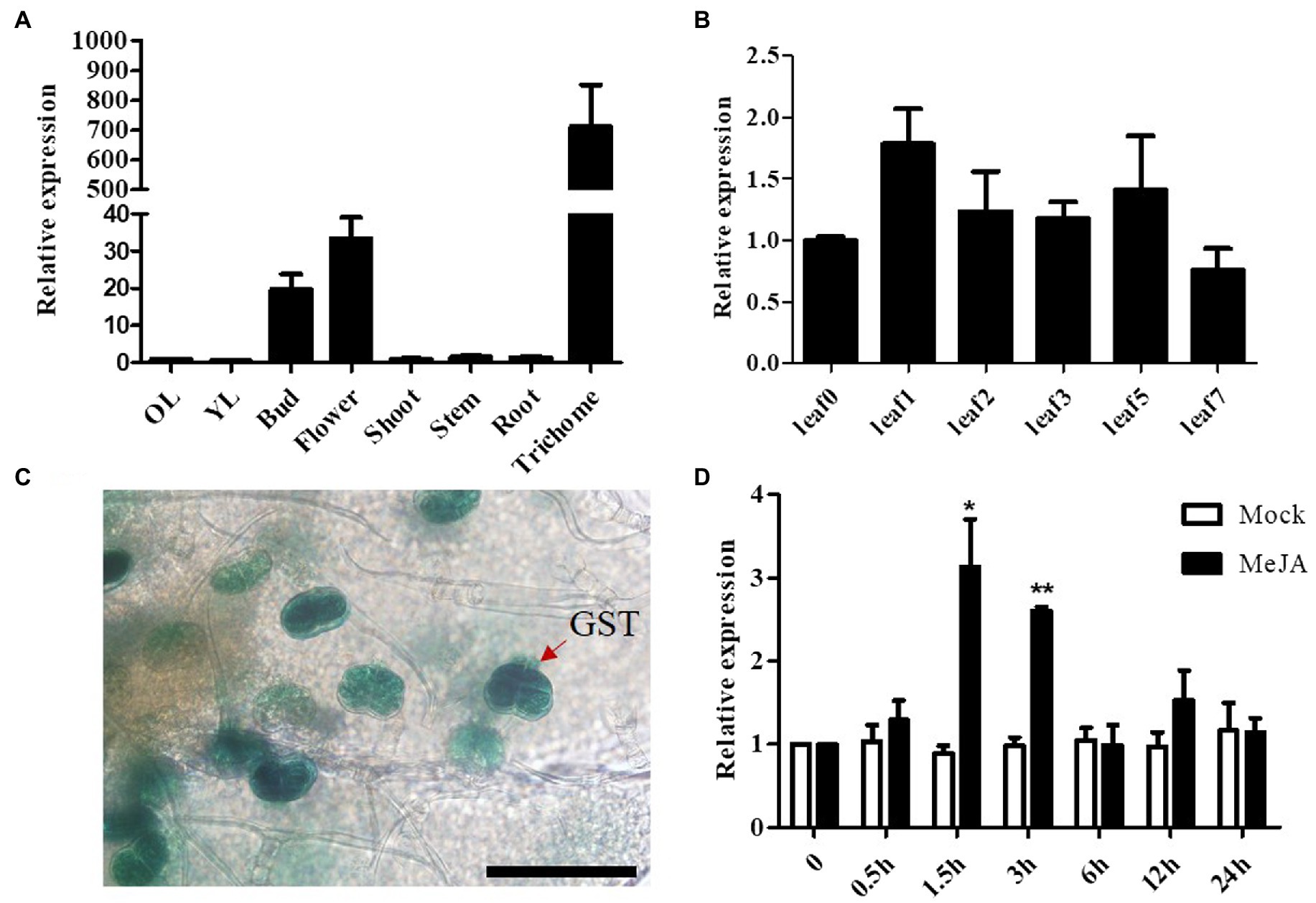
Figure 2. Transcript levels of AaSEP4 in Artemisia annua. (A, B) Relative expression levels of AaSEP4 in different tissues (A) and at different stages of leaves (B). OL, old leaves; YL, young leaves. Data values are means ± SD (n = 3). (C) β-Glucuronidase expression of 1391-proAaSEP4-GUS transgenic A. annua plants. Bars: 100 μm. (D) Relative expression of AaSEP4 in response to methyl jasmonate (MeJA, 100 μM) by RT-qPCR. Plants were treated with ddH2O as mock. All data are given as means ± SD (n = 3) *p < 0.05; **p < 0.01; Student’s t-test.
To further explore the subcellular localization of AaSEP4, we generated a pHB-AaSEP4-YFP (yellow fluorescent protein) fusion construct and transiently expressed in N. benthamiana leaves (Figure 3). Using fluorescence microscopy, we found that the YFP signals exceptionally in the nucleus and overlapped extensively with the DAPI signals. These data indicated that AaSEP4 protein localized to the nucleus, which is consistent with its role as a TF.
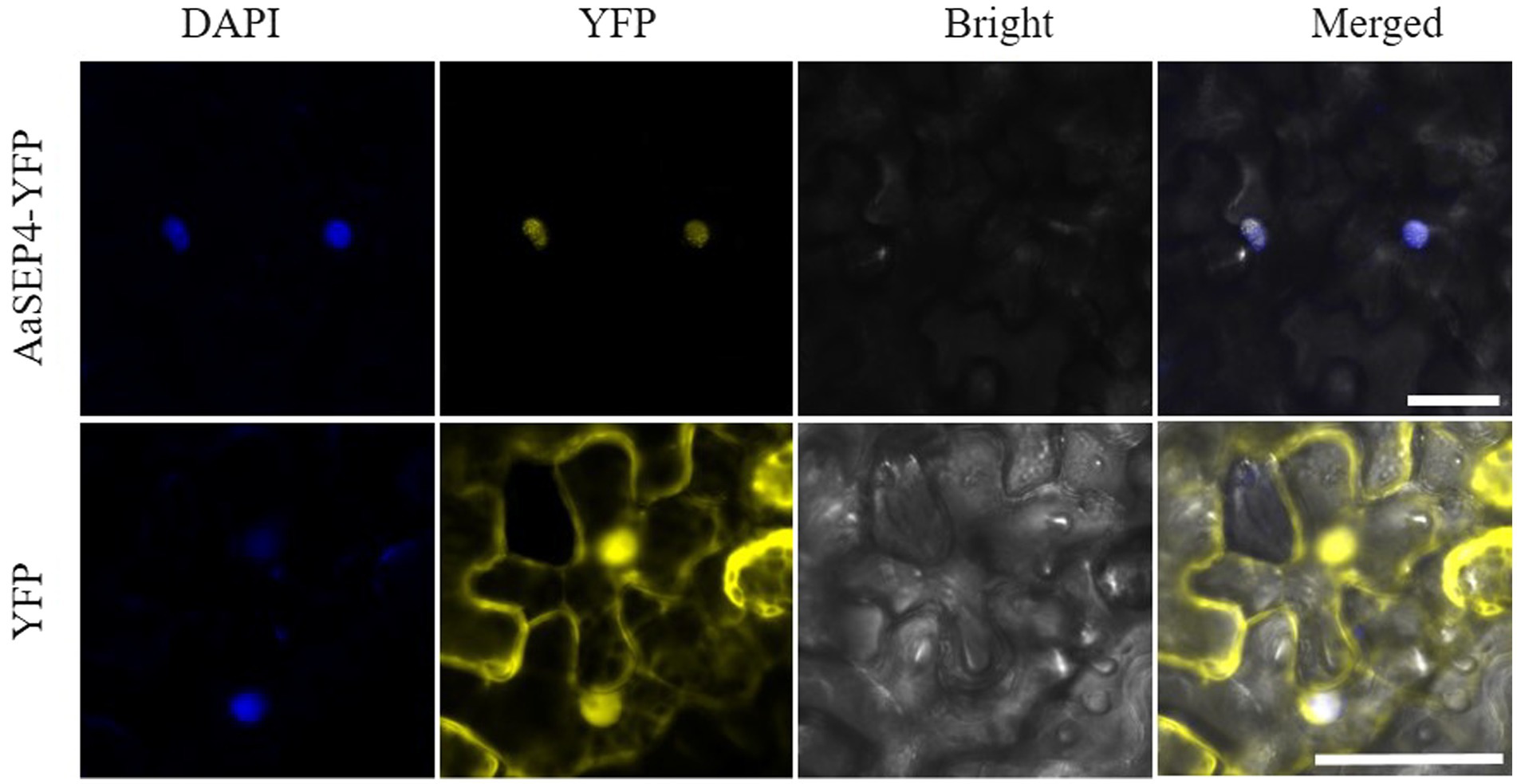
Figure 3. The subcellular localization of AaSEP4 in leaves of N. benthamiana. Yellow, yellow fluorescent protein (YFP). Blue, 4′, 6-diamidino-2-phenylindole staining (DAPI). Bars, 50 μm.
To test the interaction between the AaSEP4 protein and the AaGSW1 promoter as previously described, we first performed a dual-luciferase assay. As shown in Figure 4A, compared with the control (empty PHB vector), the relative luciferase expression driven by the promoter of AaGSW1 was significantly higher in the presence of AaSEP4. This result suggests that AaSEP4 activated the promoter activity of AaGSW1. Plant MADS-box proteins bind to specific DNA sequences known as CArG element sequence 5′-CC(A/T)6GG-3′ (Smaczniak et al., 2012; Käppel et al., 2018; Li et al., 2019), we found one CArG-box by analyzing the AaGSW1 promoter (Supplementary Figure S1). To further confirm the binding activity of AaSEP4 on AaGSW1 promoter, Y1H assay was performed to test if AaSEP4 could bind to this motif. As Figure 4B shown, AaSEP4 bound to CArG-box motif in the promoter region of AaGSW1. Taken together, these results indicated that AaSEP4 protein activated the promoter activity of AaGSW1 by interacting with the CArG element in the promoter region of AaGSW1.
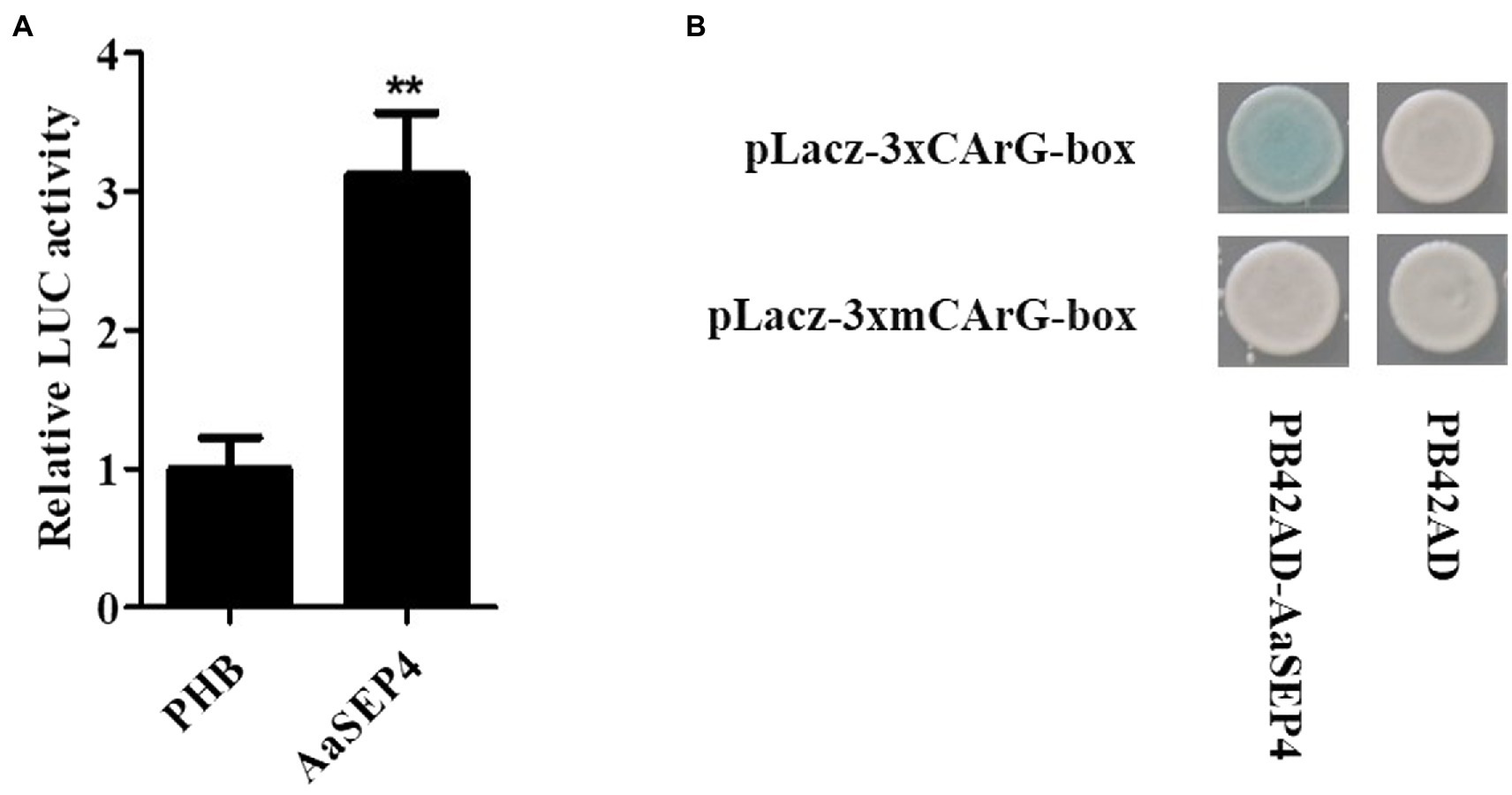
Figure 4. AaSEP4 directly binds and activates the promoter of AaGSW1. (A) Transient dual-LUC detected in tobacco leaves. Effects of AaSEP4 on AaGSW1 promoter activation. The relative LUC activity was normalized to the reference Renilla (REN) luciferase. Error bars indicate SD (n = 3). Student’s t-test: **p < 0.01. (B) Yeast one-hybrid assay of AaSEP4 and GArG-box motif in promoter of AaGSW1. Empty vector pB42AD was used as a negative control.
Since AaSEP4 activated AaGSW1 directly, we further explored the role of AaSEP4 in the artemisinin biosynthesis. According to the results of RT-qRCR, we selected three representative transgenic lines (AaSEP4-OE-1, AaSEP4-OE-2, AaSEP4-OE-3) that accumulated high levels of AaSEP4 transcript for further study (Figure 5A). In AaSEP4-overexpressing lines, the transcript level of AaGSW1 was significantly increased by 2–3 times (Figure 5B), as well as the transcript levels of ADS, CYP71AV1, DBR2 and ALDH1 (Figure 5C). In addition, HPLC was used to measure the artemisinin content of 5-month-old AaSEP4-overexpressing transgenic A. annua. It was found that the artemisinin content of AaSEP4-OE lines was 19%–72% higher than that in the WT A. annua (Figure 5D). These results demonstrated that AaSEP4 positively regulates the artemisinin biosynthesis by up-regulating the transcription level of AaGSW1 and four enzyme genes of the artemisinin biosynthesis.
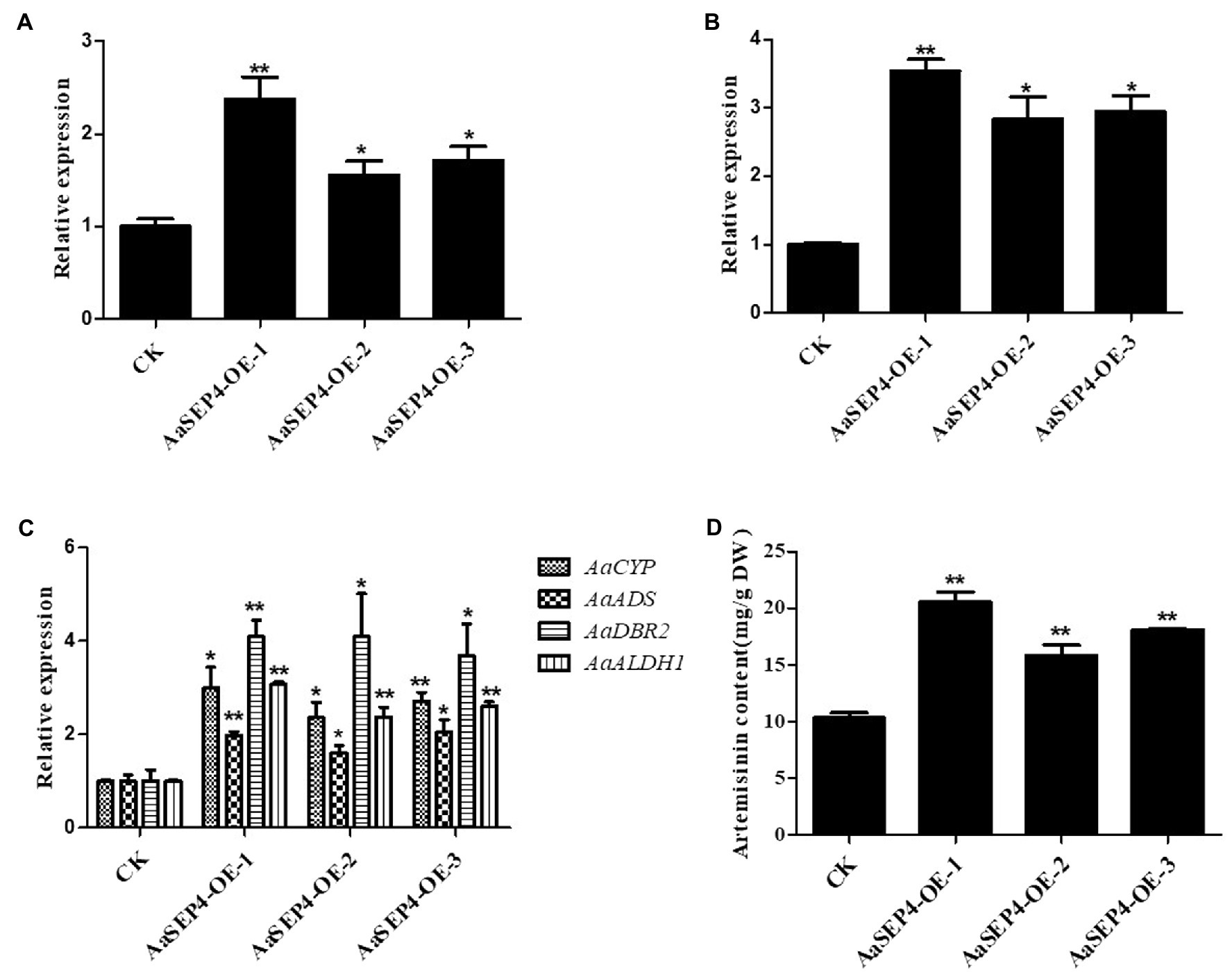
Figure 5. AaSEP4 is a positive regulator of artemisinin biosynthesis. Expression levels of AaSEP4 (A), AaGSW1 (B) and four enzyme genes (C) in AaSEP4 overexpression transgenic plants. Actin was used as internal reference. (D) Artemisinin content in AaSEP4 overexpression lines measured by high-performance liquid chromatography (HPLC). All data are given as means ± SD (n = 3) *p < 0.05; **p < 0.01; Student’s t-test.
Artemisinin is the key component of artemisinin-based combination therapies (ACTs) for malaria (Talman et al., 2019). Although the production of artemisinin is quite low (0.1%–1.0% DW), Chinese traditional herb A. annua is the main source to extract artemisinin currently (Hao et al., 2019). The biosynthetic pathway of artemisinin has been elucidated in depth. Dissection of the regulatory mechanism of artemisinin in A. annua is an effective strategy to improve the artemisinin production. Several transcription factors families such as TCP, AP2/ERF, bHLH and WRKY have been reported to regulate artemisinin biosynthesis by directly or indirectly increasing the transcript levels of four key enzyme genes in A. annua (Xiang et al., 2019; Fu et al., 2021; Ma et al., 2021; Wu et al., 2021). For better understanding of the mechanisms regulating artemisinin metabolism, we identified a MADS-box TF AaSEP4 that with potential roles in regulating the expression of AaGSW1 and accumulation artemisinin in this study. AaSEP4 belongs to the AGAMOUS-like subfamily and is homologous to the AtSEP4 protein from Arabidopsis (Figure 1). In addition, we found that AaSEP4 was strongly expressed in glandular secretary trichomes where the artemisinin is mainly synthesized and stored in A. annua (Figure 2). Using Y1H and dual-luciferase assays, we firstly demonstrated that AaSEP4 directly bound to the promoter of AaGSW1 and activated its promoter activity (Figure 4). In addition, the transcript levels of AaGSW1 were higher in AaSEP4-overexpressing A. annua plants than those in the control (Figures 5A,B). The expression levels of ADS, CYP71AV1, DBR2, and ALDH1 were also strongly induced in AaSEP4-overexpressed plants when compared with control plant (Figure 5C). These results are consistent with previous results that AaGSW1 directly activates CYP71AV1 promoter in vivo and promotes ADS, DBR2 and ALDH1 expression indirectly (Chen et al., 2017). There is no doubt that the artemisinin content was significantly enhanced in AaSEP4-overexpressed plant as Figure 5D shown.
Previous studies have reported that several MADS-box TFs bind to promoters and directly regulate the transcription of their target genes, then affect the related metabolites accumulation (flavonoid carotenoid, lycopene; Wang et al., 2014; Lu et al., 2018; Li et al., 2019). In this study, we identified for the first time a MADS-box TF AaSEP4 that are involved in the regulation of artemisinin metabolism. Although AaSEP4 can only activate the promoter of AaGSW1, the expression of ADS, CYP71AV1, ALDH1 and DBR2 were also altered by the overexpression of AaSEP4. In addition, AaSEP4 was significantly induced by MeJA treatment (Figure 2D). The transcriptional regulation of artemisinin metabolism in A. annua is complex and how AaSEP4 regulates the artemisinin metabolism through JA signaling need to be further explored. This study demonstrates that the transcription factor AaSEP4 functions positively in the artemisinin promotion and provides insight into the engineering of artemisinin biosynthesis in the future.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
T-TC and K-XT designed the project. T-TC, Y-PL, WQ, X-QF, Q-M, X-Y, X-YW, and Y-JZ performed most of the experiments. B-WP, HL, LL, X-HY, JS, X-YH, Y-LW, and K-XT analyzed the data and discussed the article. T-TC wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key R&D Program of China (2018YFA0900600), the Shanghai Science and Technology Innovation Action Plan (19431901700), the Bill & Melinda Gates Foundation (OPP1199872 and INV- 027291), SJTU Trans-med Awards Research (20190104) and the SJTU Global Strategic Partnership Fund (2020 SJTU-CORNELL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.982317/full#supplementary-material
Bemer, M., Karlova, R., Ballester, A. R., Tikunov, Y. M., Bovy, A. G., Wolters-Arts, M., et al. (2012). The tomato fruitfull homologs tdr 4/ful 1 and mbp7/ful2 regulate ethylene-independent aspects of fruit ripeningW. Plant Cell 24, 4437–4451. doi: 10.1105/tpc.112.103283
Chen, T., Li, Y., Xie, L., Hao, X., Liu, H., Qin, W., et al. (2021). AaWRKY17, a positive regulator of artemisinin biosynthesis, is involved in resistance to pseudomonas syringae in Artemisia annua. Hortic. Res. 8, 217. doi: 10.1038/s41438-021-00652-6
Chen, M., Yan, T., Shen, Q., Lu, X., Pan, Q., Huang, Y., et al. (2017). GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 214, 304–316. doi: 10.1111/nph.14373
Efferth, T. (2017). From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Semin. Cancer Biol. 46, 65–83. doi: 10.1016/j.semcancer.2017.02.009
Fu, X., Peng, B., Hassani, D., Xie, L., Liu, H., Li, Y., et al. (2021). AaWRKY9 contributes to light- and jasmonate-mediated to regulate the biosynthesis of artemisinin in Artemisia annua. New Phytol. 231, 1858–1874. doi: 10.1111/nph.17453
Hao, X., Zhong, Y., Nützmann, H. W., Fu, X., Yan, T., Shen, Q., et al. (2019). Light-induced Artemisinin biosynthesis is regulated by the bZIP transcription factor AaHY5 in Artemisia annua. Plant Cell Physiol. 60, 1747–1760. doi: 10.1093/pcp/pcz084
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 1–14. doi: 10.1186/1746-4811-1-13
Käppel, S., Melzer, R., Rümpler, F., Gafert, C., and Theißen, G. (2018). The floral homeotic protein SEPALLATA3 recognizes target DNA sequences by shape readout involving a conserved arginine residue in the MADS-domain. Plant J. 95, 341–357. doi: 10.1111/tpj.13954
Li, S., Chen, K., and Grierson, D. (2019). A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 221, 1724–1741. doi: 10.1111/nph.15545
Lu, S., Zhang, Y., Zhu, K., Yang, W., Ye, J., Chai, L., et al. (2018). The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 176, 2657–2676. doi: 10.1104/pp.17.01830
Lv, Z., Zhang, F., Pan, Q., Fu, X., Jiang, W., Shen, Q., et al. (2016). Branch pathway blocking in Artemisia annua is a useful method for obtaining high yield artemisinin. Plant Cell Physiol. 57, 588–602. doi: 10.1093/pcp/pcw014
Ma, Y. N., Xu, D. B., Li, L., Zhang, F., Fu, X. Q., Shen, Q., et al. (2018). Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua. Sci. Adv. 4, 1–19. doi: 10.1126/sciadv.aas9357
Ma, Y. N., Xu, D. B., Yan, X., Wu, Z. K., Kayani, S. I., Shen, Q., et al. (2021). Jasmonate- and abscisic acid-activated AaGSW1-AaTCP15/AaORA transcriptional cascade promotes artemisinin biosynthesis in Artemisia annua. Plant Biotechnol. J. 19, 1412–1428. doi: 10.1111/pbi.13561
Ma, N., Zhang, Z., Liao, F., Jiang, T., and Tu, Y. (2020). The birth of artemisinin. Pharmacol. Ther. 216, 107658. doi: 10.1016/j.pharmthera.2020.107658
Martel, C., Vrebalov, J., Tafelmeyer, P., and Giovannoni, J. J. (2011). The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157, 1568–1579. doi: 10.1104/pp.111.181107
Mercke, P., Bengtsson, M., Bouwmeester, H. J., Posthumus, M. A., and Brodelius, P. E. (2000). Molecular cloning, expression, and characterization of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L. Arch. Biochem. Biophys. 381, 173–180. doi: 10.1006/abbi.2000.1962
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., McPhee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi: 10.1038/nature12051
Qin, W., Xie, L., Li, Y., Liu, H., Fu, X., Chen, T., et al. (2021). An R2R3-MYB transcription factor positively regulates the glandular secretory Trichome initiation in Artemisia annua L. Front. Plant Sci. 12, 1–10. doi: 10.3389/fpls.2021.657156
Schilling, S., Pan, S., Kennedy, A., and Melzer, R. (2018). MADS-box genes and crop domestication: the jack of all traits. J. Exp. Bot. 69, 1447–1469. doi: 10.1093/jxb/erx479
Shen, Q., Yan, T., Fu, X., and Tang, K. (2016). Transcriptional regulation of artemisinin biosynthesis in Artemisia annua L. Sci. Bull. 61, 18–25. doi: 10.1007/s11434-015-0983-9
Shore, P., and Sharrocks, A. D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229, 1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x
Smaczniak, C., Immink, R. G. H., Angenent, G. C., and Kaufmann, K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098. doi: 10.1242/dev.074674
Talman, A. M., Clain, J., Duval, R., Ménard, R., and Ariey, F. (2019). Artemisinin bioactivity and resistance in malaria parasites. Trends Parasitol. 35, 953–963. doi: 10.1016/j.pt.2019.09.005
Teoh, K. H., Polichuk, D. R., Reed, D. W., and Covello, P. S. (2009). Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. Botany 87, 635–642. doi: 10.1139/B09-032
Teoh, K. H., Polichuk, D. R., Reed, D. W., Nowak, G., and Covello, P. S. (2006). Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580, 1411–1416. doi: 10.1016/j.febslet.2006.01.065
Vrebalov, J., Pan, I. L., Arroyo, A. J. M., McQuinn, R., Chung, M., Poole, M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–3062. doi: 10.1105/tpc.109.066936
Wang, S., Lu, G., Hou, Z., Luo, Z., Wang, T., Li, H., et al. (2014). Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J. Exp. Bot. 65, 3005–3014. doi: 10.1093/jxb/eru137
Wang, W., Wang, Y., Zhang, Q., Qi, Y., and Guo, D. (2009). Global characterization of Artemisia annua glandular trichome transcriptome using 454 pyrosequencing. BMC Genomics 10, 465. doi: 10.1186/1471-2164-10-465
White, N. J. (2008). Qinghaosu (artemisinin): the price of success. Science 320, 330–334. doi: 10.1126/science.1155165
Wu, Z., Li, L., Liu, H., Yan, X., Ma, Y., Li, Y., et al. (2021). AaMYB15, an R2R3-MYB TF in Artemisia annua, acts as a negative regulator of artemisinin biosynthesis. Plant Sci. 308, 110920. doi: 10.1016/j.plantsci.2021.110920
Xiang, L., Jian, D., Zhang, F., Yang, C., Bai, G., Lan, X., et al. (2019). The cold-induced transcription factor bHLH112 promotes artemisinin biosynthesis indirectly via ERF1 in Artemisia annua. J. Exp. Bot. 70, 4835–4848. doi: 10.1093/jxb/erz220
Xie, L., Yan, T., Li, L., Chen, M., Hassani, D., Li, Y., et al. (2021). An HD-ZIP-MYB complex regulates glandular secretory trichome initiation in Artemisia annua. New Phytol. 231, 2050–2064. doi: 10.1111/nph.17514
Xiong, Y., and Huang, J. (2021). Anti-malarial drug: the emerging role of artemisinin and its derivatives in liver disease treatment. Chinese Med. (United Kingdom) 16, 80–19. doi: 10.1186/s13020-021-00489-0
Yu, Z. X., Li, J. X., Yang, C. Q., Hu, W. L., Wang, L. J., and Chen, X. Y. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5, 353–365. doi: 10.1093/mp/ssr087
Zhang, F., Fu, X., Lv, Z., Lu, X., Shen, Q., Zhang, L., et al. (2015). A basic leucine zipper transcription factor, aabzip1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua. Mol. Plant 8, 163–175. doi: 10.1016/j.molp.2014.12.004
Zhang, X. G., Li, G. X., Zhao, S. S., Xu, F. L., Wang, Y. H., and Wang, W. (2014). A review of dihydroartemisinin as another gift from traditional Chinese medicine not only for malaria control but also for schistosomiasis control. Parasitol. Res. 113, 1769–1773. doi: 10.1007/s00436-014-3822-z
Zhang, F., Lu, X., Lv, Z., Zhang, L., Zhu, M., Jiang, W., et al. (2013). Overexpression of the Artemisia Orthologue of ABA receptor, AaPYL9, enhances ABA sensitivity and improves Artemisinin content in Artemisia annua L. PLoS One 8, e56697. doi: 10.1371/journal.pone.0056697
Zhang, Y., Teoh, K. H., Reed, D. W., Maes, L., Goossens, A., Olson, D. J. H., et al. (2008). The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J. Biol. Chem. 283, 21501–21508. doi: 10.1074/jbc.M803090200
Keywords: AaSEP4, MADS-box, CArG-box, artemisinin, AaGSW1
Citation: Chen T-T, Yao X-H, Liu H, Li Y-P, Qin W, Yan X, Wang X-Y, Peng B-W, Zhang Y-J, Shao J, Hu X-Y, Miao Q, Fu X-Q, Wang Y-L, Li L and Tang K-X (2022) MADS-box gene AaSEP4 promotes artemisinin biosynthesis in Artemisia annua. Front. Plant Sci. 13:982317. doi: 10.3389/fpls.2022.982317
Received: 30 June 2022; Accepted: 11 August 2022;
Published: 31 August 2022.
Edited by:
Bin Xu, Nanjing Agricultural University, ChinaReviewed by:
Xiaolong Hao, Zhejiang Chinese Medical University, ChinaCopyright © 2022 Chen, Yao, Liu, Li, Qin, Yan, Wang, Peng, Zhang, Shao, Hu, Miao, Fu, Wang Y-L, Li L and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ke-Xuan Tang, a3h0YW5nQHNqdHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.