- 1State Key Laboratory of Cotton Biology and State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng, China
- 2Department of Molecular Biology and Plant Biology, University of Geneva, Geneva, Switzerland
The chloroplast is a complex cellular organelle that not only performs photosynthesis but also synthesizes amino acids, lipids, and phytohormones. Nuclear and chloroplast genetic activity are closely coordinated through signaling chains from the nucleus to chloroplast, referred to as anterograde signaling, and from chloroplast to the nucleus, named retrograde signaling. The chloroplast can act as an environmental sensor and communicates with other cell compartments during its biogenesis and in response to stress, notably with the nucleus through retrograde signaling to regulate nuclear gene expression in response to developmental cues and stresses that affect photosynthesis and growth. Although several components involved in the generation and transmission of plastid-derived retrograde signals and in the regulation of the responsive nuclear genes have been identified, the plastid retrograde signaling network is still poorly understood. Here, we review the current knowledge on multiple plastid retrograde signaling pathways, and on potential plastid signaling molecules. We also discuss the retrograde signaling–dependent regulation of nuclear gene expression within the frame of a multilayered network of transcription factors.
Introduction
Photosynthesis, the process through which plants and algae capture light energy and convert it into chemical energy, is one of the most important energy sources to sustain life on earth. In plants and algae, the photosynthetic process occurs in chloroplasts, intracellular organelles with a size of a few microns (Abdallah et al., 2000; Ankele et al., 2007; Yin et al., 2022). Plant viability, seed set, development, and crop yield depend to a large extent on chloroplasts. Photosynthesis has a significant impact on our environment and climate by acting as a major carbon sink and by providing carbohydrates and oxygen for sustaining life (Waters and Langdale, 2009; Cottage et al., 2010). The majority of chloroplasts are localized in plant leaves with only a small portion in plant stems where light absorption is considerably less efficient. Chloroplasts contain their own DNA and protein-synthesizing apparatus. Many key components of the photosynthetic machinery are encoded by the chloroplast genome (Sakamoto et al., 2008; Waters and Langdale, 2009; Yin et al., 2022). Chloroplasts divide by binary fission in a similar way as bacteria (Abdallah et al., 2000; Ankele et al., 2007). They are bounded by the chloroplast envelope consisting of two phospholipid bilayers, the outer and inner membranes. In addition, chloroplasts contain an internal thylakoid membrane system consisting of appressed membrane disks, called grana, connected by stromal lamellae (Sakamoto et al., 2008; Tano and Woodson, 2022). These membranes enclose the inner lumen space and contain the photosynthetic complexes that catalyze the primary light reactions of photosynthesis. The chloroplast stroma includes the space between the chloroplast inner envelope membrane and the thylakoid membrane system, and it contains starch granules, ribosomes, soluble enzymes and nucleoids consisting of chloroplast DNA and associated proteins (Figure 1; Sakamoto et al., 2008).
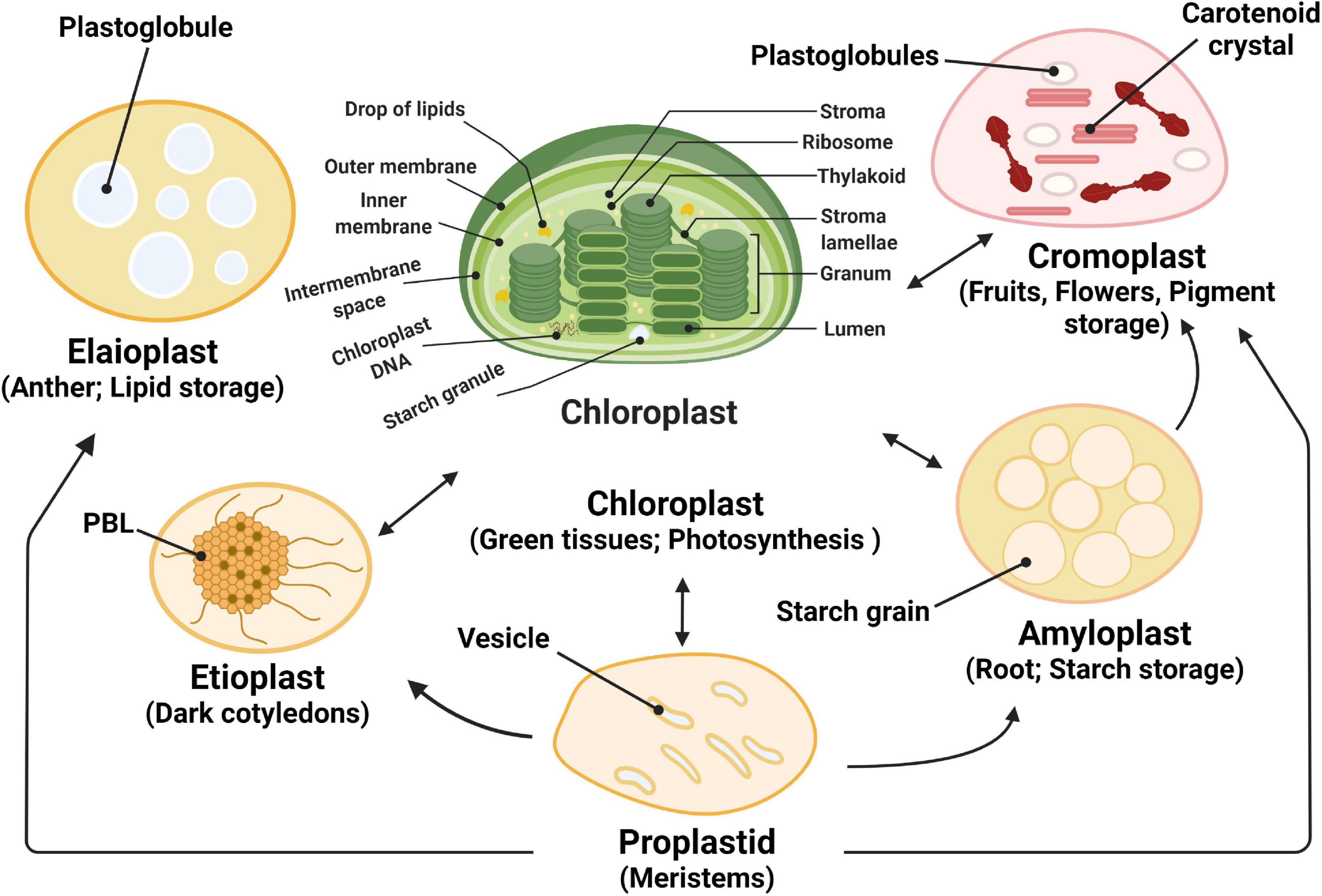
Figure 1. Plastid functions and plastid interconversions. Scheme of the chloroplast with its different compartments. The internal thylakoid membranes fold into stacked grana disks that interconnect via stroma lamellae. Thylakoid membranes enclose the lumen compartment. Plastids exist in different forms, and their identity and abundance are controlled by developmental and environmental cues. Different types interconvert (see the arrows) following a reorganization of the organellar proteome, a process that is controlled by the differentially regulated import of nucleus-encoded proteins. Chloroplasts are photosynthetic plastids, amyloplasts are starch-storing plastids, chromoplasts are carotenoid pigment-accumulating plastids, and proplastids are undifferentiated plastids that can differentiate into the different types of plastids. Etioplasts are chloroplast progenitors that form in darkness and accumulate chlorophyll precursors [in paracrystalline structures called prolamellar bodies (PLB)] that rapidly differentiate upon illumination. Elaioplasts store lipids in lipid droplets known as plastoglobules and exist, for example, in tapetal cells during pollen development.
Evolution and development of chloroplasts
Origin of chloroplasts
It is well accepted that chloroplasts originated more than a billion years ago from cyanobacteria which invaded a primitive eukaryotic cell through endosymbiosis (Abdallah et al., 2000; Ali et al., 2022). This event was followed by a massive gene transfer from the endosymbiont to the host genome (Reyes-Prieto et al., 2007; Arsova et al., 2010). The majority of the 1000–8000 genes of the ancient cyanobacterial genome (Barczak-Brzyżek et al., 2022), were either lost or relocated to the host nuclear genome with close to 100 genes remaining in the chloroplast genome (Barkan, 2011). It contains genes that code for tRNAs, rRNAs, and proteins involved in transcription, translation, RNA splicing, proteolysis, photosynthesis, and other metabolic activities (Barnes et al., 2005; Prabha et al., 2016).
The components of the photosynthetic complexes are encoded by both the chloroplast and nuclear genomes. As a result, nucleus-encoded components must be imported into chloroplasts after translation in the cytosol (Barnes and Mayfield, 2003; Baruah et al., 2009a), and assembled with their chloroplast-encoded partners into functional protein complexes through a carefully orchestrated crosstalk between these two genetic systems. The number of proteins in the chloroplast has been estimated between 2,100 and 4,500 based on the presence of predicted chloroplast transit peptides (cTP) (Leister and Kleine, 2008; Breeze and Mullineaux, 2022). In addition, a study combining nine separate datasets found that 1808 proteins were trustworthy chloroplast proteins, with 1720 encoded by the nuclear genome (Chehab et al., 2006; Llamas et al., 2022). A close coordination of chloroplast and nuclear gene expression is essential for chloroplast function, growth and adaptation to stress (Yu et al., 2008; Tano and Woodson, 2022).
Chloroplast development in land plants
Plants and algae harbor many double-membrane organelles, called plastids, which are derived from undifferentiated non-photosynthetic organelles, called proplastids. Plastids are classified based on their biological functions, in amyloplasts, involved in starch storage, etioplasts, formed in dark-grown leaves that accumulate chlorophyll precursors; chloroplasts, responsible for photosynthesis and biosynthesis of metabolites; chromoplasts that produce carotenoid pigments; elaioplasts with high amounts of plastoglobuli that are used for lipid storage (Figure 1; Chehab et al., 2006; Chen et al., 2008; Sakamoto et al., 2008).
The chloroplast development process is extremely complicated, differing not only between monocotyledonous and dicotyledonous species, but also between developmental stages, organs, and plant tissues, with many underlying mechanisms still unknown (Pogson et al., 2015). Overall, chloroplast biogenesis necessitates the coordination of several biological processes, such as protein and metabolite synthesis and formation of the thylakoid membrane system. The identity and origin of signals that initiate and govern these processes are still largely unknown. Chloroplast development has been shown to have a wide range of effects on plant vitality, seed set, and growth (Abdallah et al., 2000; Pogson et al., 2015). Even when chloroplast formation occurs normally in true leaves, poor chloroplast development at the very earliest cotyledon stages has a detrimental impact on plant growth and yield. Understanding the regulatory mechanisms that govern chloroplast growth is therefore critical (Sakamoto et al., 2008; Cottage et al., 2010; Pogson et al., 2015).
When the leaf primordia develop from the shoot apical meristem (SAM) in true leaves, chloroplasts develop directly from proplastids (Charuvi et al., 2012). The number of proplastids in meristem cells is normally between 10 and 20. These proplastids have a diameter of 0.2–1.0 μm and contain only a few internal membrane vesicles (Sakamoto et al., 2008) formed from either the inner envelope or derived from parental proplastids (Sakamoto et al., 2008). These vesicles differentiate into thylakoids after the perception of light. Chloroplasts are fully developed in mesophyll cells with a significant increase both in their number (20–100) (Lopez-Juez and Pyke, 2005; Schuhmann and Adamska, 2012) and size (5–10 μm) compared to proplastids. This increase in size is likely caused by the newly developed thylakoid membranes and the accumulation of photosynthetic proteins and lipids (Mullet, 2003).
Chloroplasts can also differentiate from etioplasts, an intermediate plastid form in the cotyledons of dark-grown plants. Etioplasts are also present in newly emerged leaves when the plants are placed in the darkness or in the seedlings when they emerge from the earth (Pogson et al., 2015). De-etiolation is the process of differentiation from etioplast to chloroplast upon exposure to light. In contrast to proplastids, which contain vesicles and no differentiated structures, etioplasts feature a prolamellar body (PLB), which has a lattice-like membranous structure. Several essential photosynthetic proteins, such as non-chlorophyll binding photosystem II (PSII) subunits, cytochrome b6f complex, ATP synthase and ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) have been detected in etioplasts although generally in much lower amounts compared to mature chloroplasts (Escoubas et al., 1995; Dietz et al., 2010; Enami et al., 2011; Estavillo et al., 2011; Dogra et al., 2022). After the disassembly of PLB and the onset of chlorophyll production in response to light exposure, thylakoid membranes develop. During the de-etiolation process, photosystem I (PSI) activity can be detected after only 15 min of light exposure, and PSII and ATP synthase activity after 2–3 h (Finkelstein et al., 1998; Estavillo et al., 2011; Dogra et al., 2022). Biogenesis of chloroplasts is completed in 6–24 h (Fischer et al., 2012; Fisher et al., 2022). In rare circumstances, such as in regreening of Valencia oranges, chloroplasts can differentiate from chromoplasts. This is accompanied by a reduction in the size and quantity of osmiophilic globules, as well as the appearance of well-developed grana (Abdallah et al., 2000; Fisher et al., 2022). Chloroplast development is highly complex and involves many different biological processes. This organelle must be able to sense and act on signals from many different light and metabolic pathways.
Light signaling pathways control chloroplast development
One of the most important triggers for initiating chloroplast biogenesis is light. In this part, we will cover light signaling mechanisms that underpin chloroplast development.
Repressors of chloroplast development in darkness
In the dark, germinated seedlings undergo skotomorphogenesis, in which the hypocotyl is extended at the expense of leaf development, an effective strategy for accessing light. The tightly folded apical hook, which helps the seedling to break through the earth crust and protects the unfolded cotyledons as well as the meristem before their emergence from the soil, is another hallmark of this stage. This process ensures that the few nutrients stored in the seeds are used efficiently during the search for light which is essential for photoautotrophic life (Chen et al., 2010; Kami et al., 2010). PHYTOCHROME-INTERACTING FACTORS (PIFs) are a family of basic helix-loop-helix (bHLH) transcription factors that serve as important modulators of the skotomorphogenesis pathway (Leivar and Quail, 2011). Altogether, eight PIFs have been identified, including PIF1, PIF3 to PIF8, and PIF3-LIKE1 (PIL1) which have overlapping and antagonistic roles in skotomorphogenesis (Leivar et al., 2008; Luo et al., 2014). PIF1, PIF3, PIF4, PIF5, and PIF7, for example, stimulate hypocotyl development by activating genes involved in the biosynthetic pathway of the growth hormone auxin (Shin et al., 2009; Stephenson et al., 2009; Li et al., 2012). Moreover, PIF1, PIF3, PIF4, and PIF5 impede chloroplast development by repressing expression of nucleus-encoded photosynthetic proteins (Stephenson et al., 2009; Kim et al., 2011).
Besides preparing for light perception, dark-grown seedlings must deal with soil-induced mechanical stress as the hypocotyl lengthens to emerge from the earth (Liu et al., 2017). ETHYLENE-INSENSITIVE3 (EIN3) which is stabilized by inhibiting the activity of EIN3 BINDING F-BOX 1 and 2 (EBF1/2), is induced by mechanical stress (An et al., 2010; Merchante et al., 2015). EIN3 reacts to mechanical stress by stimulating the expression of a number of downstream genes, including ETHYLENE RESPONSE FACTOR (ERF1), which is important for reinforcing the hypocotyl cell wall and decreasing the pace of hypocotyl elongation (Maier et al., 2011; Zhong et al., 2014). Furthermore, the EBF1/2-EIN3 pathway activates HOOKLESS1 (HLS1) to regulate seedling apical hook development (Woodson et al., 2013; Llamas et al., 2022). It was found that EIN3 has a similar function as PIF3 and that the two proteins interact to suppress chloroplast growth in the dark. EIN3-PIF3 is able to repress protochlorophyllide (Pchlide) production while also upregulating PROTOCHLOROPHYLLIDE OXIDOREDUCTASE (PORA and PORB) (Masuda, 2008; Melonek et al., 2010; Maier et al., 2011; Maruta et al., 2012). As a result, EIN3-PIF3 is crucial for regulating Pchlide and PORA/PORB levels during de-etiolation to protect the seedling from photooxidative damage. EIN3 and PIF3 have been shown to bind to the promoters of LIGHT-HARVESTING CHLOROPHYLL A/B-BINDING (LHC) genes, and to repress numerous LHCA and LHCB genes (Mochizuki et al., 2010; Mittler et al., 2011). Overall, EIN3-PIF3 may be able to control chloroplast development by tying the mechanical and light signaling pathways together. There are a few repressors, including the CONSTITUTIVE PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA (COP/DET/FUS) proteins, which form three distinct protein complexes and modulate the stability of positive transcription factors throughout the greening process (Mochizuki et al., 2008; Schwechheimer et al., 2022). In Arabidopsis thaliana (Arabidopsis), cop/det/fus mutants have very clear phenotypes. Their seedlings display light-grown phenotypes in darkness, and they accumulate high levels of anthocyanin (Palmer, 1985; Op den Camp et al., 2003; Mullineaux et al., 2006; Nott et al., 2006; Noordally Zeenat et al., 2013). In the dark, COP1, an E3 ubiquitin ligase, uses the ubiquitin-26S proteasome system to degrade several light-responsive transcription factors such as ELONGATED HYPOCOTYL 5 (HY5), HY5 HOMOLOGUE (HYH), LONG HYPOCOTYL IN FAR RED (HFR1), LONG AFTER FAR-RED LIGHT 1 (LAF1), and EBF1/2 (Paul and Foyer, 2001; Pesaresi et al., 2006, 2009, 2011; Patel and Berry, 2008; Barczak-Brzyżek et al., 2022). Other COP/DET/FUS proteins, COP9, DET1 as well as COP10, are also required for the proteasomal degradation of light-responsive regulators in the dark (Pesaresi et al., 2007, 2011).
Photoreceptors transmit light information to signaling pathways that control chloroplast development
When a seedling emerges from the earth, light triggers a variety of physiological responses, including hypocotyl growth inhibition, chlorophyll accumulation, and chloroplast development. Massive transcriptional reprogramming of both the nuclear and plastid genomes, as well as the onset of a complex signaling interconnecting network, accompany these morphological changes in response to light. This process is called photomorphogenesis and it includes numerous and poorly understood light-sensing mechanisms (Kusuma and Bugbee, 2021).
Plants possess a large number of photoreceptors to sense light intensity, direction, and wavelength. According to their absorption spectrum, these photoreceptors may be divided into different groups: one UV-B light receptor (UVR8) in Arabidopsis, three cryptochromes (CRY1-CRY3), two phototropins (PHOT1 and PHOT2), and three Zeitlupe family (ZTL) proteins detecting UV-A/blue light, and five phytochromes (PHYA-PHYE) absorbing red (R)/far-red (FR) light (Kleine et al., 2003; Chory, 2010). These light receptors interact in synergistic, antagonistic, and redundant ways with each other (Mazzella, 2001; Pfannschmidt and Liere, 2005; Pfannschmidt, 2010; Kusuma and Bugbee, 2021).
Light is primarily sensed by phytochromes and cryptochromes during photomorphogenesis. Phytochromes contain the linear tetrapyrrole phytochromobilin as their chromophore. PHYs are found in the cytosol in their inactive Pr form in the dark. They are photoconverted into the active Pfr form when exposed to light. This light-induced conversion is followed by translocation of PHYs from the cytosol to the nucleus, where they are directed to subnuclear sites known as the photobodies (Pfannschmidt et al., 2001; Piippo et al., 2006; Pesaresi et al., 2007; Van Buskirk et al., 2012, 2014). Photobodies are sites for protein degradation and/or transcription factors, as they colocalize with a variety of transcriptional regulators and their E3 ubiquitin ligases as well as photoreceptors (Possingham, 1980; Pogson et al., 2015). Photobodies change in size and quantity in response to changes in light, developmental phases, and daily rhythms (Possingham, 1980; Pesaresi et al., 2007; Van Buskirk et al., 2014; Kusuma and Bugbee, 2021).
Numerous essential light-signaling components, including many transcription factors such as PIFs, EIN3, and COP1, are regulated by PHYs. All PIFs interact physically with the PHYs in their Pfr forms and are subsequently phosphorylated, ubiquitinated, and degraded. PIF phosphorylation has been linked to casein kinase II (CK2), brassinosteroid insensitive 2 (BIN2), photoregulatory protein kinases (PPK1–PPK4), and phytochromes. The full intricacies of light-induced phosphorylation of PIFs, however, have yet to be uncovered (Pfalz et al., 2006; Pham et al., 2018). PIFs are ubiquitinated by distinct E3 ligases after phosphorylation; for example, cullin4 (CUL4) and CUL3-based E3 ubiquitin ligase ubiquitinate PIF1 and PIF3, respectively. The 26S proteasome system then degrades ubiquitinated PIFs (Wostrikoff et al., 2004; Pham et al., 2017). PHYB may facilitate chloroplast growth by promoting connections between EBF1/2 and EIN3, resulting in EIN3 degradation by the 26S proteasome (Schwenkert et al., 2022). Furthermore, by reducing the interaction between COP1 and SUPPRESSOR OF PHYA-105 (SPA) proteins, PHYA and PHYB are able to deactivate COP1 and prevent its proteasome-mediated repression of photomorphogenesis (Pham et al., 2018; Schwenkert et al., 2022).
Cryptochromes are photolyase-like flavoproteins with a flavin adenine dinucleotide (FAD) chromophore (Nott et al., 2006; Pogson et al., 2015; Liang et al., 2022). A CRY photoexcitation model has been proposed in which FAD is postulated to have three redox states: oxidized FAD, semi-reduced FAD or FADH, and completely reduced FADH or FADH2. The photoexcitation cycle of FAD in Arabidopsis begins with the ground state, oxidized FAD in darkness, and ends with the semi-reduced FADH upon perception of blue light (Liu et al., 2017). Although this model has been questioned, it is generally agreed that photoexcited CRY adopts an open shape, which is accompanied by metabolic processes such phosphorylation. Various CRY-interacting proteins bind to the altered CRY, establishing a CRY-partner complex, which causes changes in gene expression and developmental programs (Possingham, 1980; Raghavendra and Padmasree, 2003; Pogson et al., 2015). CRY2 also undergoes photodimerization to become active, a process blocked by the BLUE- LIGHT INHIBITOR OF CRYPTOCHROMES 1 (BIC1) protein (Xu et al., 2022). CRY1 and CRY2 control the expression of 5–25% of genes in Arabidopsis in response to blue light at the transcriptional or post-translational level (Rodermel and Park, 2003; Ramel et al., 2012; Llamas et al., 2022). CRY2 binds directly to the DNA region shared by PIF4 and PIF5, enhancing low blue light (LBL) induced hypocotyl development at the transcriptional level (Pogson et al., 2015). CRYs may also exert an indirect effect at the post-translational level (Rodermel and Park, 2003; Pedmale et al., 2016; Lee et al., 2022).
Moreover, expression of numerous nuclear genes involved in plastid transcription and photosynthesis, such as SIGMA FACTOR 5 (SIG5), CHLOROPHYLL A/B-BINDING PROTEIN (CAB), and RIBULOSE BISPHOSPHATE CARBOXYLASE SMALL SUBUNIT (RBCS), is controlled by CRYs in response to blue light (Rodermel and Park, 2003; Rook et al., 2006; Ruckle et al., 2012). Apart from PIFs, SPA1, and COP1 mentioned above, there may be additional proteins that interact with CRYs to mediate the light signaling pathway. It will be interesting to identify these proteins and to elucidate their function in the CRY signaling pathway that controls chloroplast biogenesis (Schröter et al., 2010; Ruckle et al., 2012; Pedmale et al., 2016; Pham et al., 2018; Liebers et al., 2020).
Nuclear components involved in the regulation of chloroplast development
When seedlings are exposed to light, the expression of approximately 35% of the nuclear genes encoding chloroplast proteins are changed (Schwechheimer et al., 2022; Schwenkert et al., 2022). Expression of several chloroplast-targeted proteins which are critical for chloroplast development, is considerably increased. These proteins are subsequently transported into chloroplasts where they participate in several chloroplast activities. Anterograde signaling from nucleus to chloroplasts is a process that involves both positive and negative transcriptional regulators from a variety of TF families, including basic leucine zipper (bZIP), Zinc-finger, GARP, and others. In the next part, we will discuss the involvement of these transcription factors in chloroplast biogenesis. A leucine zipper dimerization motif and a DNA binding domain are found in the bZIP transcription factors. According to their DNA-binding region and other conserved motifs, they are divided into 10 groups A-I and S in Arabidopsis. Members of the same family have comparable DNA-binding motifs, implying that they bind to similar cis regions and, as a result, are functionally redundant, a feature which prevents the appearance of distinctive phenotypes for single mutants (Jakoby et al., 2002; Sherameti et al., 2002; Seo et al., 2008; Pedmale et al., 2016).
HY5 is a well-known factor with a bZIP motif that belongs to the H group. It is able to bind to the promoter region of over 3000 genes and to control a wide range of developmental processes (Jakoby et al., 2002; Lee Keun et al., 2007; Zhang et al., 2011). HY5 was shown to repress the cell elongation genes by directly binding to the G-box (CACGTG) present in their promoter regions (Oyama et al., 1997). HY5 also controls the expression of genes involved in chlorophyll biosynthesis and photosynthesis-associated nuclear genes (PhANGs), such as GUN5, PORC, LHCA4, LHCB1.1, and LHCB1.3 (Toledo-Ortiz et al., 2014). The roots of cop1 and det1 mutants, in which HY5 inhibition is loosened, become green due to a large accumulation of chlorophyll (Nott et al., 2006; Lee Keun et al., 2007; Stern et al., 2010; Pedmale et al., 2016; Pham et al., 2018).
Furthermore, HY5 promotes thylakoid biogenesis by activating the expression of DIGALACTOSYLDIACYLGLYCEROL SYNTHASE1 (DGD1), and it also plays a role in plastid transcription by activating the expression of PURPLE ACID PHOSPHATASE 8 (PAP8), an essential subunit of the plastid-encoded plastid RNA polymerase (Steiner et al., 2009; Sun et al., 2011). DGD1 is involved in chloroplast lipid metabolism and the DGD proteins can be divided into two types (Lyu et al., 2021). In Arabidopsis, DGD1 and DGD2 contain two and one galactosyltransferase domain, respectively (Liu et al., 2004). The function and location of monogalactosyldiacylglycerol (MGDG), and DGDG are determined by the structure of the polar head and the fatty acid tails. Moreover, MGDG is present in both photosystem I (PSI), photosystem II (PSII), and the cytochrome b6f complex whereas DGDG is only present in PSII and light-harvesting complex II (Liu et al., 2004; Liang et al., 2019). These two galactosyldiacylglycerols provide biochemical connections between photosynthesis, chloroplast shape maintenance, jasmonate synthesis, phosphate starvation responses, freezing tolerance, and probably other processes (Liu et al., 2004; Lyu et al., 2021).
The G-group members of nuclear bZIP TF family, bZIP16, bZIP68, and GBF1-3, have been linked to various aspects of photomorphogenesis, including photosynthesis, inhibition of hypocotyl elongation, and phytohormone signaling (Surpin and Chory, 1997; Surpin et al., 2002; Sun et al., 2011). LHCB2.4 has been identified as a target gene of bZIP16, bZIP68, and GBF1 (Surpin et al., 2002; Tano and Woodson, 2022). However, nothing is known regarding the particular role of these genes in the formation of chloroplasts. Functional redundancy and significant physical interactions between these proteins have been discovered previously. Using the yeast two hybrid system, bZIP16, bZIP68, and GBF1, were found to form heterodimers (Tano and Woodson, 2022; Trösch et al., 2022). Such heterodimerizations have also been observed between the GBF1, GBF2, and GBF3 proteins (Pesaresi et al., 2007; Toyokuni et al., 2022; Trösch et al., 2022). GBF1 was also found to interact with other proteins in a variety of ways, including acting antagonistically with HY5 for the expression of RBCS-1A, acting additively with HY5 to regulate the expression of LHCB1.3 (Pesaresi et al., 2007), and acting antagonistically with MYC2, a bHLH TF, in blue light signaling and regulation of hypocotyl elongation (Tyagi, 2001; Tyagi and Gaur, 2003; Pesaresi et al., 2007). Taken together, these findings show that their functional redundancy and diversity as well as their heterodimerization are important for ensuring flexibility to the regulation of this system. Aside from bZIPs, many B-box Zinc Finger proteins (BBXs), including BBX21-BBX25, BBX28, and BBX30-32, play a role in photomorphogenesis. BBX21-BBX23 enhance HY5 activity, with BBX21 directly binding to the HY5 promoter and regulating its expression (Tyagi and Gaur, 2003; Jung and Chory, 2010). BBX24, BBX25, BBX28, and BBX32, on the other hand, are negative regulators of HY5 activity, and HY5 might limit the expression of BBX30 and BBX31 which also operate as repressors of photomorphogenesis (Holtan et al., 2011; Lin et al., 2018). As a result, BBXs-HY5 act as a critical regulatory network for ensuring optimal photomorphogenesis (Holtan et al., 2011; Gangappa et al., 2013; Lin et al., 2018; Xu, 2020).
Golden 2-like (GLK) proteins, which are part of the GARP superfamily of transcription factors, have also been identified as regulators of chloroplast development. GLKs modulate the expression of LHCBs, and genes involved in chlorophyll production directly (Yin et al., 2022). GLK1 and GLK2 control the expression of DGD1 (see above) (Weissmann and Brutnell, 2012; Yin et al., 2022). In Arabidopsis, glk1glk2 double mutants have a pale green phenotype associated with decreased thylakoid grana (Yasumura et al., 2005; Waters et al., 2009). This phenotype can be compensated by overexpression of either of the two GLK genes, suggesting that these two genes have functional redundancy (Waters et al., 2009). Thus, GLK1 and GLK2 appear to play an important role in chloroplast formation. These findings show that the transcriptional response to changes in light quality and quantity and to a shift from darkness to light is governed by a highly coordinated regulatory network (Figure 2).
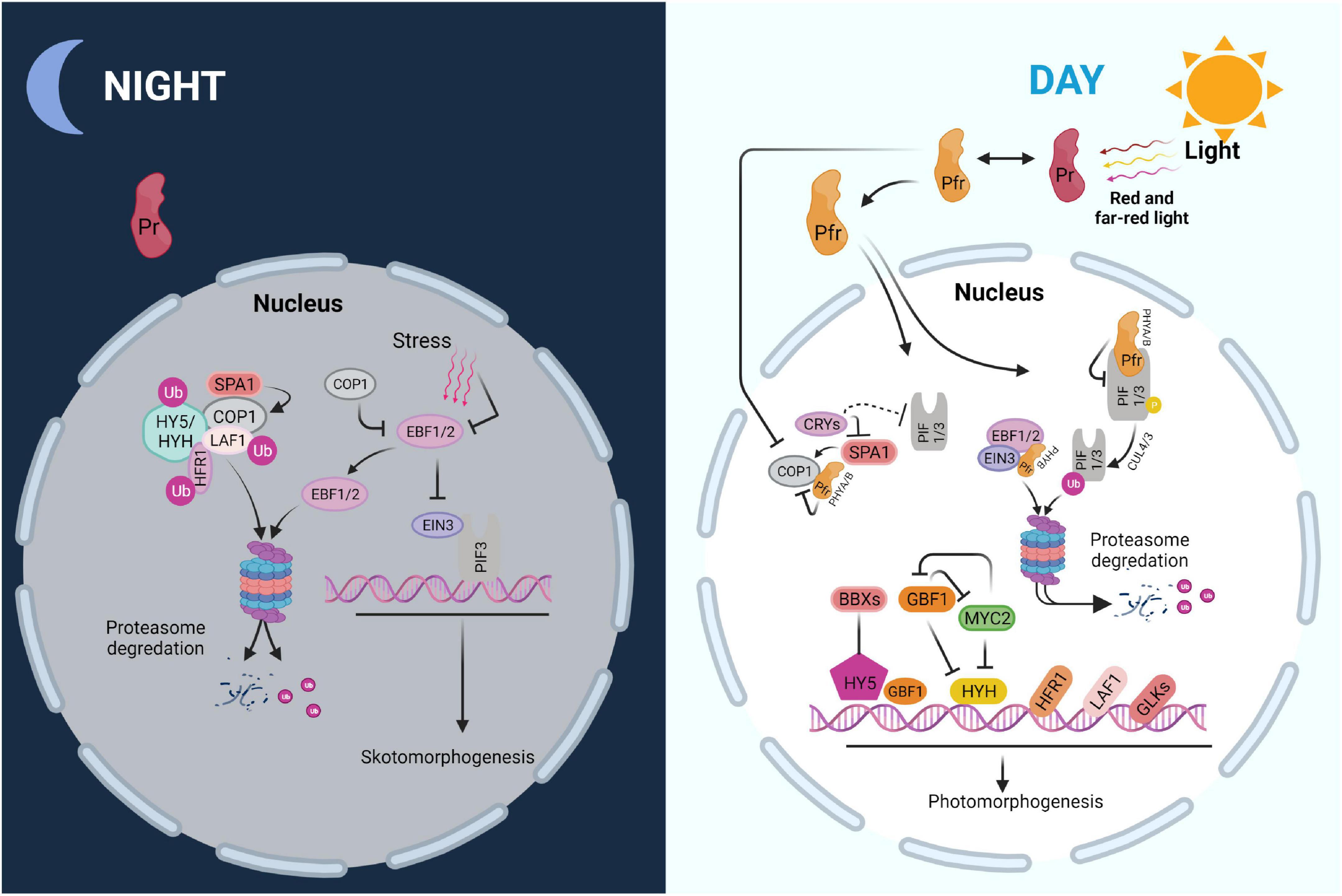
Figure 2. Scheme of major regulatory networks in skoto- and photomorphogenesis. PIF-EIN3 increases skotomorphogenesis and suppresses chloroplast biogenesis in the dark, when combined with soil-induced mechanical stress. COP1, an E3 ubiquitin ligase, degrades EBF1/2, which impedes EIN3 function. Moreover, COP1 degrades numerous light-responsive transcription factors including HY5, HYH, HFR1, and LAF1 via the ubiquitin-26S proteasome system. Photoactivated PHYs in the Pfr form interact with PIFs upon the onset of light, causing their phosphorylation, ubiquitination, and destruction. PHYB may increase EBF1/2-EIN3 interaction, resulting in EIN3 destruction by the 26S proteasome system. PHYs and CRYs deactivate COP1 at the same time, preventing the degradation of positive regulators including HY5, HYH, HFR1, and LAF1. Furthermore, GBF1 and BBX proteins, as well as other regulators such as GLKs, may interact with HY5 to enhance photomorphogenic growth.
Chloroplast transcription systems
The chloroplast DNA (plastome) is around ∼120–160 kb in size and packaged into nucleoids. The nucleoids move from the inner envelope to the thylakoids during chloroplast maturation (Pogson et al., 2015). The plastid genome encodes tRNAs, and rRNAs and key photosynthetic proteins as well as proteins involved in protein synthesis and metabolism. At least two kinds of RNA polymerases transcribe the chloroplast genome. One is nucleus-encoded and consists of a single T3-T7 bacteriophage type subunit (NEP). The other is the plastid-encoded plastid RNA polymerase (PEP), a multi-subunit enzyme of eubacterial type consisting of the core subunits RpoA, RpoB, RpoC1, and RpoC2 that are encoded by plastid genes transcribed by NEP. The PEP complex includes additional bacterial-type nucleus-encoded sigma factors (SIGs) that are imported into the organelle post-translationally. These sigma factors are necessary for targeting the PEP core to the promoter region of defined genes and initiating their transcription. Apart from the sigma factors, PEP is associated to a group of other proteins known as PEP-associated proteins (PAPs), which are likewise nucleus-encoded and required for PEP activity in response to various developmental and environmental signals. In contrast to PEP, NEP can recognize promoters, initiate and terminate transcription on its own as a single catalytic subunit polymerase (Tyagi and Gaur, 2003; Xiao et al., 2012).
Unlike PEP, which can be traced back to cyanobacteria, the evolutionary origin of NEP is unknown. According to current research, the genes for plastid NEP enzymes may have arisen from a duplication of the RPOTm gene, which encodes the mitochondrial RNA polymerase (Chan et al., 2014). In monocot plastids, RPOTp encodes a single NEP, but in dicot plastids, two NEPs have been described, one encoded by RPOTp targeted to plastids and the other by RPOTmp targeted to both mitochondria and plastids (Baier and Dietz, 2005; Chan et al., 2014).
In Arabidopsis, knocking out one of the NEPs, RPOTmp or RPOTp, results in reduced pigmentation, altered leaf morphology, and stunted growth whereas the rpoTmprpoTp double mutant is seedling lethal (von Gromoff et al., 2006; Xu et al., 2022). NEP and PEP recognize distinct promoter components. The –10 and –35 cis-elements, which are comparable to the E. coli σ70 promoters, are used by PEP. NEP promoters are divided into three types based on the conserved sequence motifs they recognize: type Ia, type Ib, and type II. The YRTa motif is characteristic for the Ia motif and is conserved in the type Ib motif but with an extra GAA-box. The type II motif, on the other hand, bears little resemblance to the type I motifs (Xiao et al., 2012; Fortunato et al., 2022; Yin et al., 2022).
The involvement of NEP and PEP in the transcription of chloroplast genes has been intensively investigated. Based on their transcription, chloroplast genes may be divided into three categories: genes transcribed exclusively by NEP or PEP, and genes transcribed by both polymerases. NEP transcribes only a few genes, including some housekeeping genes and rpo genes required mostly at the beginning of chloroplast biogenesis, while PEP transcribes the vast majority of plastid genes (∼80%) in mature chloroplasts (Fortunato et al., 2022). This is consistent with the finding that in the absence of functioning PEP, accumulation of transcripts from photosynthesis-related genes is dramatically decreased (Jung and Chory, 2010; Breeze and Mullineaux, 2022; Fortunato et al., 2022). However, differential RNA sequencing (dRNA-seq) has revealed that NEP transcribes numerous photosynthetic proteins formerly assumed to be transcribed uniquely by PEP (Jung and Chory, 2010).
Furthermore, examination of run-on transcription activities in PEP-deficient plastids revealed that most plastid genes, regardless of gene class, are transcribed in the absence of PEP (Fortunato et al., 2022), suggesting that NEP alone can transcribe the majority of the chloroplast genome. However, NEP was unable to restore the albino phenotype of PEP-deficient mutants suggesting that the number of RNAs produced by NEP alone is inadequate for normal photosynthetic activity (Jung and Chory, 2010; Fortunato et al., 2022). Chloroplast biogenesis is thus associated with a complex transcription apparatus with some overlap between two RNA polymerases. Both polymerases are required for the establishment of a functional photosynthetic apparatus. Further research is needed to determine the specific role of each polymerase and to elucidate how they interact with other factors for efficient plastid transcription.
The specific function of the different plastid sigma factors
In Arabidopsis, sequence alignment of the six sigma factors (SIG1–SIG6) with their prokaryotic counterparts reveals that all of them belong to the σ70 family of transcription factors (Fischer et al., 2012). SIG2 and SIG6 have been shown to play important roles in chloroplast development, while SIG5 is involved in the blue light-stimulated transcription of psbD encoding PHOTOSYSTEM II REACTION CENTER PROTEIN D (Heiber et al., 2007; Larkin and Ruckle, 2008; Stern et al., 2010). The other σ factors are less important because corresponding knock-out plants lack visible phenotypes. SIG2 mediates the transcriptional transition from NEP to PEP through transcription of tRNAGlu, which binds directly to NEP in vitro and could limit NEP transcriptional activity (Surpin and Chory, 1997; Stern et al., 2010). The sig6 mutant, like the sig2 mutant, is deficient in chlorophyll, especially in the cotyledons (Stern et al., 2010). The levels of many PEP-dependent transcripts, including mRNAs of thylakoid proteins, certain tRNAs, and rRNAs, are decreased in sig6 seedlings throughout the early stages of development (Pfannschmidt, 2010; Stern et al., 2010; Toyokuni et al., 2022). These findings suggest that SIG6 is especially important during these early phases. The sig2sig6 double mutant, as predicted, has a more severe pale phenotype and is seedling-lethal, demonstrating critical roles for SIG2 and SIG6 in seedling development (Toyokuni et al., 2022).
The roles of plastid RNA polymerase-associated proteins during chloroplast biogenesis
Gel filtration and mass spectrometry studies in Arabidopsis revealed a total of 35 proteins in transcriptionally active chloroplast DNA-protein complexes (TAC) (Pfalz et al., 2006). Mutants deficient in ten of these TAC proteins exhibit an albino or light green phenotype. T-DNA insertions in the genes of pTAC7 and Mur ligase E domain-containing protein-like (MurE-like) caused a comparable phenotype although they are not physically associated with PEP. They were designated as PAPs based on their phenotype (Pfalz et al., 2006; Grabowski et al., 2008; Fischer et al., 2012). These 12 TAC proteins are known as PAPs because they are thought to be necessary for active PEP transcriptional activity. PAPs were split into three categories based on our present understanding and predictions of their structural domain (Pfannschmidt et al., 2001; Pfannschmidt and Liere, 2005; Piippo et al., 2006; Pfannschmidt, 2010). PAP1/pTAC3, PAP2/pTAC2, PAP3/pTAC10, PAP5/pTAC12, PAP7/pTAC14, PAP8/pTAC6, and PAP12/pTAC7 belong to the first group. This family of proteins binds DNA, RNA, or both, and is thought to play a role in promoter recognition, transcription initiation, and elongation (Nott et al., 2006; Estavillo et al., 2011; Mittler et al., 2011).
Proteins of the second group, PAP4/Fe SUPEROXIDE DISMUTASE 3 (FSD3), PAP6/FRUCTOKINASE-LIKE PROTEIN 1 (FLN1), PAP9/FSD2, and PAP10/THIOREDOXINz (TRXz) are responsible for redox-dependent gene regulation and PEP protection against reactive oxygen species (ROS) (Estavillo et al., 2011; Li and Kim, 2022).
The third category includes the remaining PAPs. The function of PAP11/MurE-like is unknown. PAP3 may interact through its carboxyl-terminal region downstream of the S1 domain with several other PAPs including PAP4, PAP7, PAP9, PAP10, and PAP12. PAP3, on the other hand, does not interact directly with any of the PEP core components, such as RpoA and RpoB (Chen et al., 2008; Estavillo et al., 2011). PAP5 and PAP8, two other members of this family, have a dual location in the nucleus and chloroplast where they have distinct roles. PAP5 functions in gene transcription in chloroplasts (Pfalz et al., 2006) and mediates degradation of PIF1 and PIF3 through phytochrome in the nucleus (Galvão et al., 2012; Qiu et al., 2015). PAP5 interacts directly with PAP7 and PAP12 (Yu et al., 2013) although its biological role is unknown. PAP8, like PAP5, plays a crucial role in the creation of photobodies. PAP8 is implicated in the degradation of PIF1 and PIF3 by phytochromes, as well as the stability of HY5 and the GLKs (Figure 3; Liebers et al., 2020). Because of these multiple roles, PAP5 and PAP8 have been proposed to be crucial coordinators of nuclear and chloroplast gene expression during chloroplast biogenesis (Stern et al., 2010; Galvão et al., 2012; Liebers et al., 2020).
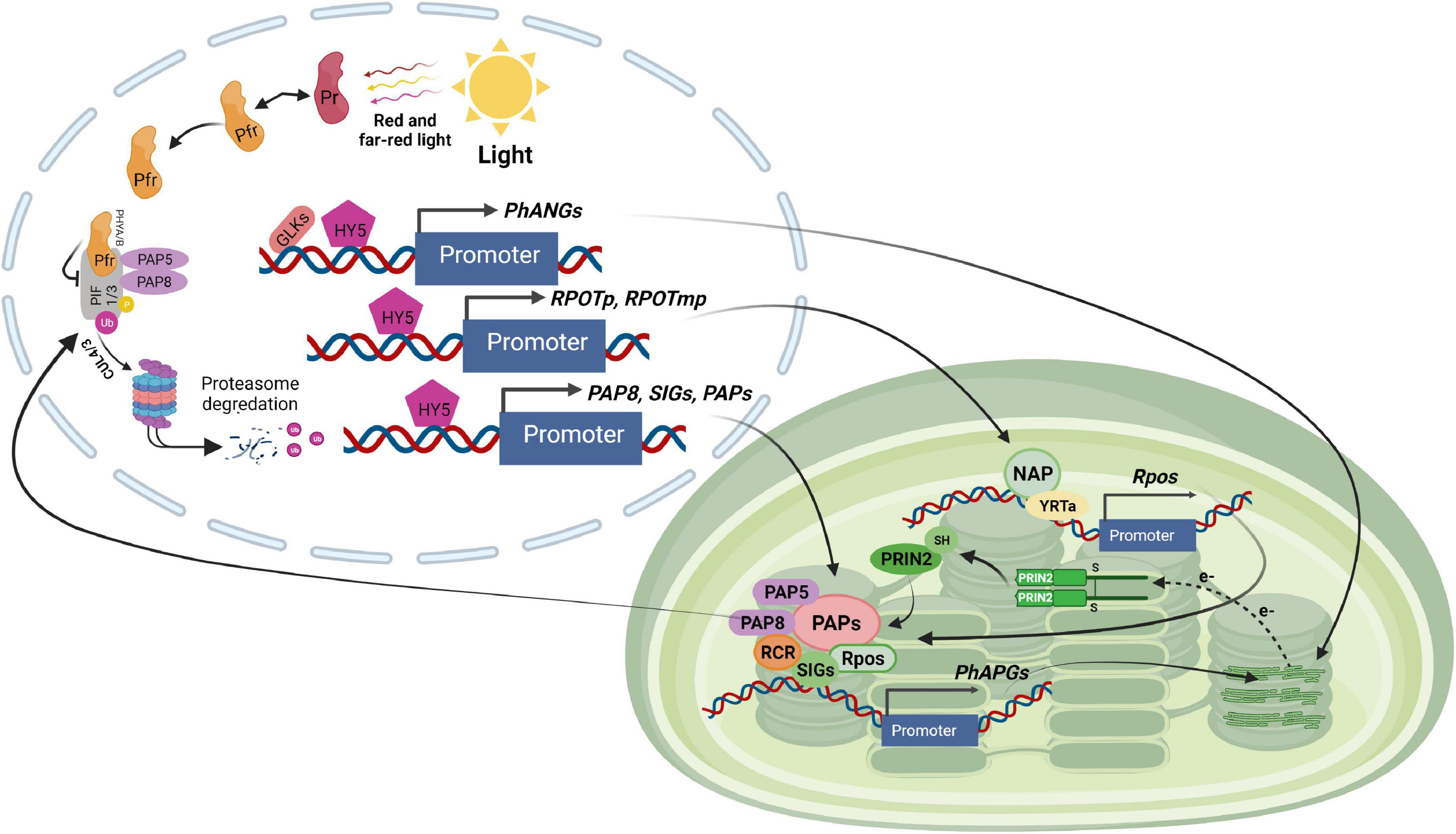
Figure 3. Light signaling pathway controlling plastid transcription. GLKs, HY5 and unknown regulators stimulate the expression of chloroplast proteins encoded by nuclear genes such as PhANGs, RPOTp, and RPOTmp (encoding NEP), SIGs and PAPs (encoding PEP components) in response to light exposure. NEP transcribes the rpo genes of the PEP core proteins. The PEP core is associated with PAPs, which include PAP5 and PAP8, two dual-localized components implicated in PHY-mediated PIF1 and PIF3 degradation in the nucleus. PEP activity is regulated by other proteins such as PRIN2 in a redox-dependent way. The PEP complex transcribes the photosynthesis-associated plastid-encoded genes (PhAPGs). Their products aid in the establishment of photosynthesis when combined with the proteins encoded by PhANGs.
Chloroplast gene expression is a complex and highly regulated process, and the molecular relationship between photosynthetic electron transport activity and regulation of plastid gene expression (PGE) has been studied for many years (Rochaix, 2013). A major target of photosynthetic redox signals is the PEP complex. It fine-tunes plastid transcription by sensing redox signals resulting from unbalanced stimulation of both photosystems, in addition to its direct function in plastid transcription (Galvão et al., 2012; Liebers et al., 2020; Fortunato et al., 2022). However the molecular mechanisms underlying this redox control are still unclear. A previous study demonstrated that the redox modulation of PRIN2 activity is responsible for the thiol-mediated regulation of PEP-dependent transcription (Steiner et al., 2011). The photosynthetic electron transport chain is assembled during the greening phase, and after its components are reduced by light, the FTR/TRX system is activated. Through the reduction of the Cys68–Cys68 disulfide link, TRX transforms the PRIN2 dimer to the active monomeric form (Díaz et al., 2018). The monomeric form increases the light-activated transcription of photosynthetic chloroplast genes. As a result, activation of the PEP complex creates a positive retrograde signal that controls expression of the nuclear-encoded photosynthesis genes, therefore synchronizing the activities of the two genomes in response to light (Díaz et al., 2018). The formation of functioning chloroplasts and the initiation of photosynthesis is a complex process that involves several cellular compartments. First, the growth of chloroplasts is light-dependent, and the initial light signal activates photoreceptors, triggering substantial changes in nuclear transcription (Chen et al., 2004; Waters et al., 2009). Second, the expression of the plastid-encoded photosynthesis genes is dependent on the expression and assembly of the nuclear-encoded components necessary for PEP action (Kindgren and Strand, 2015). Using gel filtration and mass spectrometry, the transcriptionally active complex in chloroplasts was shown to comprise 43 nuclear encoded proteins (Kindgren and Strand, 2015). Consequently, given its complex structure, activation of PEP is most likely a rate-limiting step for the onset of photosynthetic gene expression in the plastids. It was proposed that it represents a developmental bottleneck in the establishment of photosynthesis (Pfalz et al., 2015). In addition, the activation of PEP involves a redox regulatory system that modulates PGE (Pfannschmidt and Liere, 2005).
The nature of the retrograde signal is unknown, but it has been suggested that blocked chloroplast development would either disrupt a positive signal emitted by the plastid, which acts in a GUN1-regulated manner, or induce a negative plastid-emitted signal, which represses nuclear transcription in a GUN1-mediated manner (Vinti et al., 2000; Martín et al., 2016). Recent data describing the establishment of photosynthesis during chloroplast development suggest that full PhANG induction is dependent on a positive signal from healthy developing plastids (Dubreuil et al., 2018). In addition, it is known that LHCB expression is directly correlated with the recovery of PEP activity in the different prin2-2 complemented lines, suggesting that a positive signal generated by PEP activity in the plastids stimulates LHCB expression in the nucleus. Taken together, the results suggest that the monomeric form of PRIN2 is the active form of the protein, and contributes to the activation of the PEP complex during the establishment of functional chloroplasts in response to light (Díaz et al., 2018). Once PEP is activated, a positive retrograde signal is triggered and thus, the status of the PEP complex links the functional state of the chloroplast to the nucleus, enabling the plant to coordinate expression of photosynthetic genes of the nuclear and chloroplast genomes during seedling development (Díaz et al., 2018).
PAP10/TRX z features a thioredoxin domain and has been shown to interact with PAP6, FLN2, and PLASTID REDOX INSENSITIVE (PRIN2) to mediate the redox control of PEP activity (Figure 3; Waters et al., 2009; Wimmelbacher and Börnke, 2014). It is therefore possible that PAP10 is involved in redox-controlled plastid transcription. PAP4 and PAP9, two members of the second group, both include iron superoxide dismutase domains that protect against oxidative stress during the onset of photosynthetic activity in early development (Myouga et al., 2008; Qing-Bo, 2013). Although the function of the remaining PAPs is unknown, loss of function mutants of all PAPs have a PEP-deficient phenotype characterized by the inability to grow autotrophically and an altered plastid transcription profile (Pfalz et al., 2006; Pfannschmidt, 2010), implying that all PAPs are required for PEP-mediated gene expression.
A surprising finding was that redox-inactive versions of TRXz and FLN1 are able to entirely restore PEP function in the corresponding loss of function mutants during early chloroplast development (Weissmann and Brutnell, 2012; Wimmelbacher and Börnke, 2014; Yin et al., 2022). A possible explanation is that some, if not all, of the PAPs have a structural rather than a regulatory role (Steiner et al., 2011; Liebers et al., 2020; Toyokuni et al., 2022). This suggests that the absence of some PAPs has a negative impact on the structural integrity of the PEP complex during chloroplast maturation. PEP evolved from its cyanobacterial ancestor to associate with numerous PAPs in order to optimize its function in the photosynthetically active chloroplast. The precise functions of the various subunits are currently unknown. The PEP complex is thought to undergo a structural rearrangement during chloroplast development, in which PEP only consists of the core subunits in darkness and is assembled with PAPs upon light exposure (Pfannschmidt et al., 2001; Pfannschmidt and Liere, 2005; Pfannschmidt, 2010). The role of each component and the assembly process will need to be investigated further.
The roles of (p)ppGpp in chloroplast stress responses
The hyperphosphorylated nucleotides guanosine pentaphosphate and tetraphosphate (together referred to as ppGpp) have emerged as likely candidates for regulating chloroplast stress signaling. They are synthesized from ATP and GDP/GTP by enzymes of the RelA SpoT Homolog (RSH) family in chloroplasts (Boniecka et al., 2017; Field, 2018). In plants ppGpp was shown to be required for the regulation of plant growth and development (Sugliani et al., 2016; Ono et al., 2021), plant immunity (Abdelkefi et al., 2018), and photosynthesis (Sugliani et al., 2016). Many years ago ppGpp was shown to be involved in the stringent response in bacteria which is triggered by nitrogen limitation and results in growth arrest (Cashel and Gallant, 1969). In plants subjected to nitrogen limitation the chloroplast with its photosynthetic machinery represents a major potential nitrogen source for metabolic remobilization. Under these conditions ppGpp was indeed induced and shown to be important for remodeling the photosynthetic electron transport chain, thereby down-regulating photosynthetic activity and providing protection against oxidative stress and cell damage (Romand et al., 2022). Moreover ppGpp was shown to promote the coordinated down-regulation of both chloroplast and nuclear genes of chloroplast proteins and thus to couple chloroplast and nuclear gene expression during nitrogen starvation (Romand et al., 2022). Thus ppGpp appears to be a key regulator of chloroplast activity with a photoprotective role for the adaptation of plants to environmental stress conditions.
Chloroplast intracellular signaling
In plants, both the plastid and nuclear genomes encode proteins that are essential for photosynthetic function. Most of the chloroplast proteins (∼90%) are encoded by the nucleus (Surpin and Chory, 1997; Surpin et al., 2002; Rodermel and Park, 2003; Barkan, 2011; Ramel et al., 2012; Wu et al., 2019). Many nucleus-encoded gene products interact with plastid-encoded proteins within the chloroplasts to assemble into functional complexes (such as the ATP synthase, PSI, and PSII); others perform regulatory functions within the chloroplasts (such as PPR proteins) (Barkan, 2011). Rubisco, the major CO2 fixation enzyme, represents a classic example of gene coordination between nucleus and chloroplast (Wimmelbacher and Börnke, 2014). Each plastid chromosome contains one rbcL gene, but each plastome can include up to 100 chloroplast DNA copies, and each photosynthetic plant cell contains up to 100 chloroplasts (Possingham, 1980; Palmer, 1985). The Small subunit (SSU), on the other hand, is encoded by a small group of 5–15 rbcS genes in the nucleus (Patel and Berry, 2008). As a result, the number of rbcL genes greatly exceeds the number of rbcS genes within each plant cell. Despite this large difference in gene copy number, the plastid-encoded rbcL gene is coordinated with the nucleus-encoded rbcS genes to produce stoichiometric amounts of Large subunit (LSU) and SSU proteins in the chloroplasts, allowing the L8S8 holoenzyme to be assembled. Multiple steps of post-transcriptional regulation are required for the coordinated synthesis of LSU and SSU synthesis, including mRNA stability, RNA processing, translational initiation, translational elongation, protein stability, and final assembly of the L8S8 holoenzyme (Patel and Berry, 2008; Woodson et al., 2013).
Folding of newly synthesized LSU is mediated by the Cpn60/Cpn20 chaperonin complex. This is followed by formation of LSU oligomers consisting of four dimers and by the assembly of the holoenzyme L8S8, a process which is assisted by the chaperones RbcX, Raf1, and Raf2 (Aigner et al., 2017; Wietrzynski et al., 2021). In both land plants and Chlamydomonas, the stoichiometric expression of the two Rubisco subunits is regulated by the CES process (control by epistasy of synthesis) that represses the translation of LSU in the absence of SSU (Wostrikoff and Stern, 2007). The CES process is also operational during the assembly of the other photosynthetic complexes PSII, PSI, Cytb6f, and ATP synthase in the thylakoid membranes (Choquet and Vallon, 2000), and involves the assembly dependent synthesis of some key subunits of these complexes; it has been mostly studied in Chlamydomonas.
When amaranth seedlings are exposed to different light regimes, translational coordination occurs (Patel and Berry, 2008). In dark-grown seedlings transferred to light (light-shift), the LSU and SSU polypeptides are induced coordinately extremely rapidly with no corresponding increase in their transcript levels. Both rbcL and rbcS mRNAs are not associated with polysomes in dark-grown plants (when there is no synthesis), but bind to polysomes when these seedlings are exposed to light, allowing both Rubisco proteins to be synthesized coordinately. Moreover, when light-grown plants are shifted to darkness (dark-shift), the synthesis of both Rubisco subunits is promptly arrested. Although Rubisco production is drastically decreased in dark-shifted amaranth seedlings, rbcL and rbcS transcripts remain bound to polysomes indicating that translation elongation is also regulated in a coordinated manner. These results show that light-induced synthesis of both Rubisco subunits is well synchronized between the two cellular compartments, owing to control at the translational level (Jung and Chory, 2009; Stern et al., 2010).
There is thus an interactive communication between the two cellular compartments, allowing for the coordinated expression of plastid- and nucleus-encoded photosynthetic proteins (Figure 4). A large number of nuclear encoded factors are involved in this coordination and act at several steps of chloroplast gene expression including transcription, RNA editing, RNA processing and degradation, splicing and translation (Barkan and Goldschmidt-Clermont, 2000). Although nucleus-encoded factors govern most aspects of chloroplast growth and function, communication between the two compartments occurs in both direction, from nucleus to plastids (anterograde signaling) and from plastids to nucleus (retrograde signaling) (Surpin and Chory, 1997; Surpin et al., 2002; Jung and Chory, 2009).
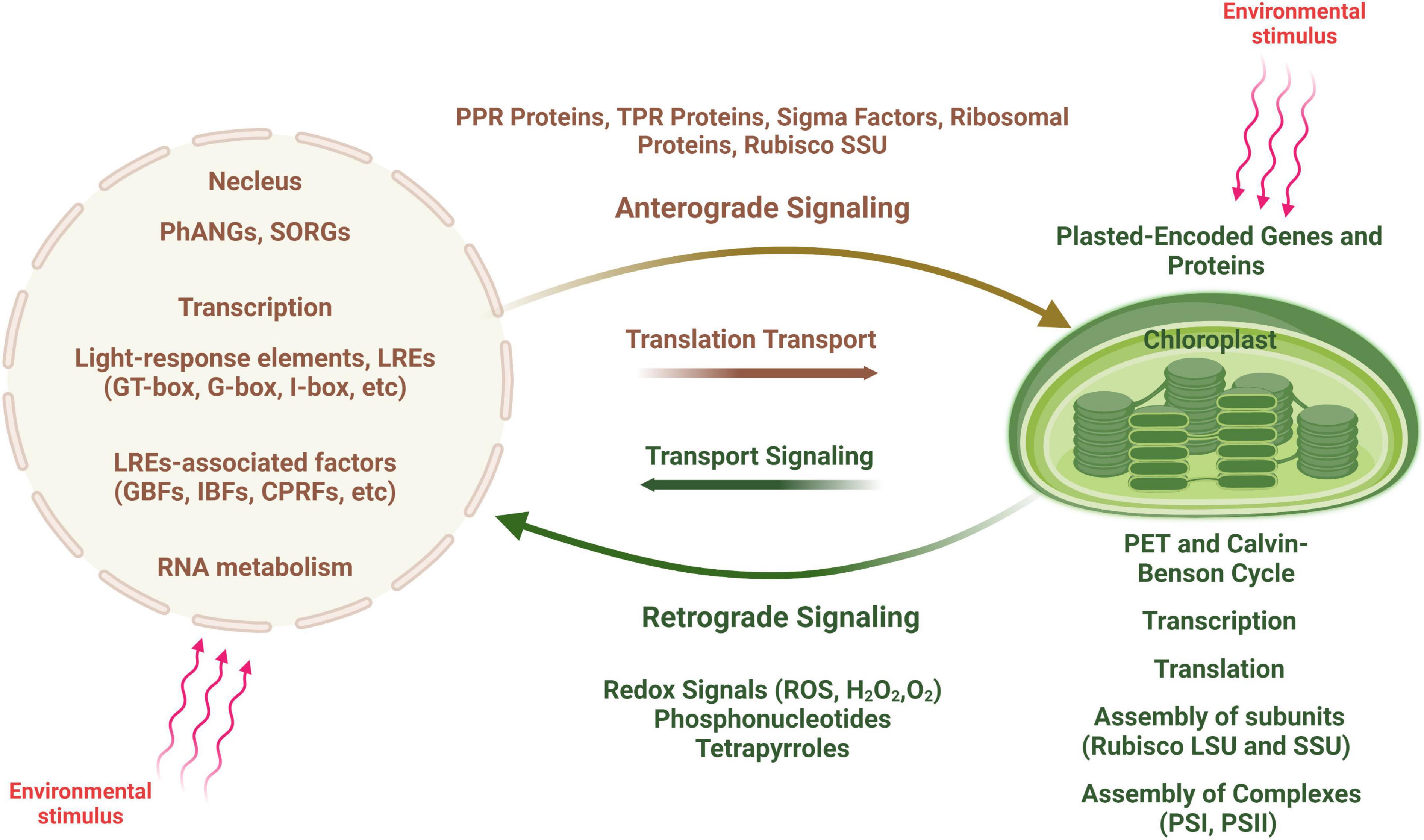
Figure 4. Scheme of anterograde and retrograde signaling in plant cells. An environmental stimulus is perceived by the nucleus, communicated to the chloroplast (shown by red arrows) and leads to changes in chloroplast transcription (anterograde signaling). In contrast, signals generated by the chloroplast are transduced to the nucleus causing changes in nuclear gene expression (retrograde signaling), in particular to regulate the expression of nucleus-encoded photosynthesis associated nuclear genes (PhANGs).
Developmental processes such as leaf maturation and senescence, light irradiance, photosynthetic energy production, redox state, and other factors all affect these signaling networks. Intracellular photosynthetic signaling processes between chloroplasts and nucleus are highly complex, involving the functional integration of components from both inside and outside of these two compartments. Signals from the mitochondria (such as energy and redox state) and the cytosol influence the photosynthetic crosstalk within plant cells (such as transport) (Paul and Foyer, 2001; Barnes and Mayfield, 2003; Strand et al., 2003; Barnes et al., 2005; Stern et al., 2010; Maier et al., 2011; Steiner et al., 2011; Maruta et al., 2012). Protein complexes that control chloroplast transcription, RNA processing and splicing, translation, and assembly/stabilization, as well as pigment-binding complexes participate in these intracellular interactions (Weissmann and Brutnell, 2012; Breeze and Mullineaux, 2022).
Multiple plastid retrograde signaling pathways
In the plastids of higher vascular plants, four distinct plastid retrograde signaling pathways have been traditionally recognized based on the sources of the signals: Accumulation of tetrapyrrole intermediates, Inhibition of PGE, Changes in plastid redox status, and Production of ROS (Nott et al., 2006; Kleine et al., 2009; Pesaresi et al., 2009; Woodson et al., 2013; Pogson et al., 2015; Garmash, 2022). Several additional plastid retrograde signals have been proposed that include SAL1-PAP (3′-phosphoadenosine 5′- phosphate) and methylerythritol cyclodiphosphate (MEcPP) (Estavillo et al., 2011; Xiao et al., 2012; Liebers et al., 2020).
Accumulation of tetrapyrrole intermediates and connection with retrograde signaling
Retrograde signaling from the plastid to the nucleus is important for two reasons. First, functional multiprotein complexes in plastids, such as photosystems, are composed of subunits encoded by both the nuclear and plastid genomes whose expression needs to be coordinated. Second, metabolic activities and the functional state of plastids are affected by the external environment, such as light conditions. To adapt plastids must sense the environmental changes and adjust expression of nuclear genes of chloroplast proteins and the corresponding protein flow (Jung and Chory, 2010; Kindgren et al., 2011). Here we focus on the tetrapyrrole pathway and the mechanisms by which changes in the functional or metabolic state of the plastids elicit signals that affect nuclear gene expression. Analysis of perturbations of the tetrapyrrole biosynthetic pathway in the gun mutants have provided clues for potential plastid signals (Figures 5, 6) (Susek et al., 1993; Rodermel and Park, 2003; Xiao et al., 2012). Chlorophyll, heme, siroheme, and phytochromobilin are generated by the tetrapyrrole biosynthetic pathway in plastids (Tanaka and Tanaka, 2007; Mochizuki et al., 2008). In land plants the chlorophyll biosynthetic pathway begins with glutamyl tRNA and can be separated into three parts: from ALA to protoporphyrin IX in the chloroplast matrix; from magnesium protoporphyrin to chlorophyllide on the chloroplast envelope membrane; and ultimately Chl a and Chl b are synthesized on the thylakoid membrane (Figure 5; Matringe et al., 1992).
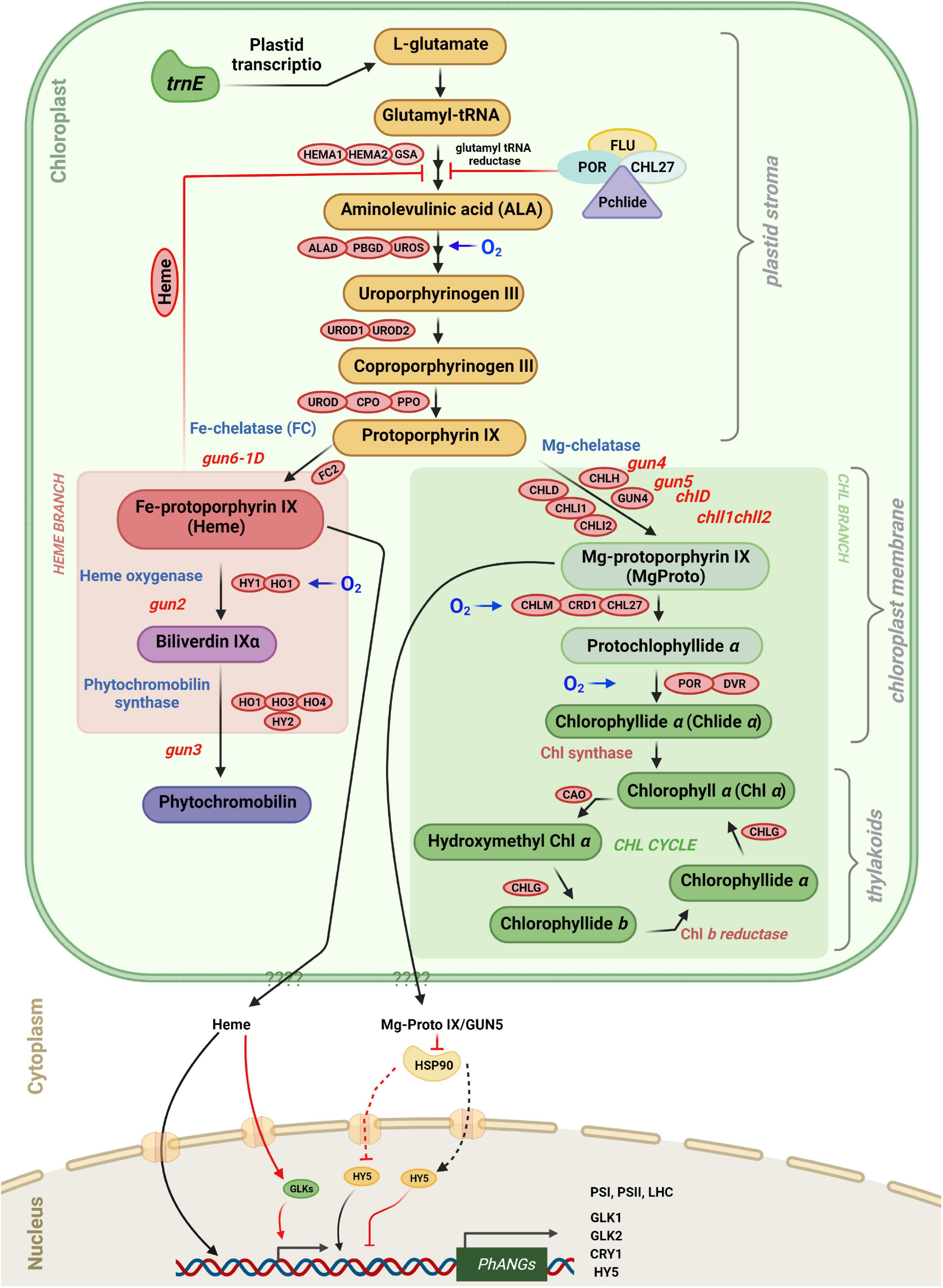
Figure 5. Tetrapyrrole biosynthesis and plastid gene expression with key enzymes. Retrograde signaling is mediated in part by tetrapyrrole intermediates. The different steps in tetrapyrrole biosynthesis are indicated by arrows. FLU as well as the amount of “free” heme control the rate of ALA Synthesis. Protoporphyrin IX can be converted either to Fe-protoporphyrin IX (heme), leading to phytochromobilin, or to Mg-protoporphyrin IX leading to chlorophyll a and chlorophyll b. The heme branch is mediated by GUN6 (Fe-chelatase), GUN2 (heme oxygenase), and GUN3 (phytochromobilin synthase). Heme acts as a positive regulator of photosynthesis-associated nuclear genes (PhANGs), but the nuclear components involved in heme signaling, as well as the precise process by which heme enters the nucleus, remain unclear. GUN5/CHLH and GUN4 (which binds the substrate and product of the Mg-chelatase-catalyzed process and activates the Mg-chelatase) are engaged in the chlorophyll branch. It is proposed that Mg-proto IX is exported from plastids via an unknown route and subsequently binds to HSP90, which in turn represses HY5 and prevents activation of PhANG expression. Enzymes involved in the tetrapyrrole biosynthetic pathway are indicated in red boxes. They are ALA, 5-aminolevulinic acid; HEMA, glutamyl-tRNA reductase; GSA, glutamate-1-semialdehyde 2,1-aminomutase; ALAD, 5-aminolevulinate dehydratase; PBGD, porphobilinogen deaminase; UROS, uroporphyrinogen III synthase; UROD, uroporphyrinogen III decarboxylase; CPO, coproporphyrinogen III oxidase; PPO, protoporphyrinogen IX oxidase; CHLH, Mg-chelatase H subunit; CHLI, Mg-chelatase I subunit; CHLD, Mg-chelatase D subunit; GUN4, regulator of Mg-chelatase; CHLM, Mg-proto IX methyltransferase; CRD1, Mg-proto IX monomethylester cyclase; POR, NADPH:protochlorophyllide oxidoreductase A, B and C; DVR, divinyl-protochlorophyllide reductase; CHLG, chlorophyll synthase; CAO, chlorophyllide a oxygenase; FC, ferrochelatase; HO/HY1, heme oxygenase; and phytochromobilin synthase (HY2).
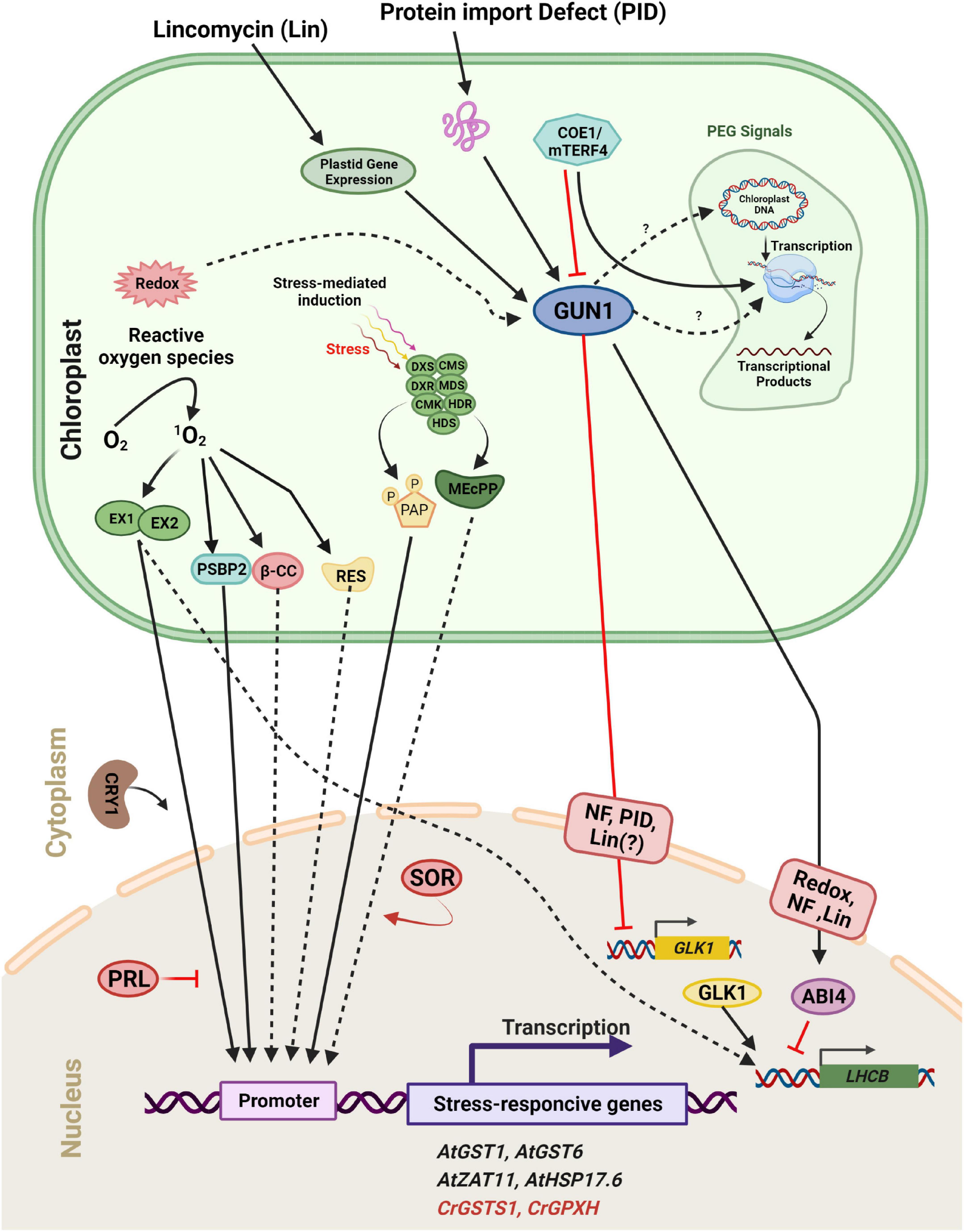
Figure 6. Scheme of plastid retrograde signaling (PGE) in a plant/algal cell. Components are considered to be involved in 1O2-mediated induction of either Arabidopsis thaliana (At, black) or Chlamydomonas reinhardtii (Cr, Red) genes. 1O2 accumulates during exposure to excess light and activates distinct signaling pathways. The 1O2 accumulated in the plastid is sensed and/or transmitted to the nucleus via the concerted action of two chloroplast proteins, EXECUTER 1 and EXECUTER 2 (EX1 and EX2). In the cytosol/nucleus, the blue-light photoreceptor CRY1 is involved in 1O2-mediated stress responses, but PRL may act as a general negative regulator of 1O2 signaling. One product of the 1O2 -caused oxidation of carotenoids, β-cyclocitral (β-CC), may act as a second messenger involved in the 1O2 signaling pathway in plants. A second stress response involves the induction of methylerythritolcyclo diphosphate (MEcPP) and phosphoadenosine phosphate (PAP), two important metabolites that participate in retrograde signaling through chromatin remodeling and changes in nuclear gene expression. Full arrows indicate activation or conversion, and dashed arrows indicate diffusion or transport of molecules. Furthermore, distinct plastid processes [redox change, plastid gene repression by Lincomycin (Lin), Norflurazon (NF), and Protein Import Defect (PID)] generate plastid signals. These signals converge at GUN1 within the plastid. Subsequently, the signal is sent to two transcription factors, ABI4 and GLK1. GUN1 activates ABI4, a negative regulator of LHCB expression and also down-regulates the expression of GLK1, leading to a subsequent decrease in LHCB expression.
The first committed step of tetrapyrrole synthesis is the conversion of glutamyl-tRNA to 5-aminolevulinic acid (ALA) by glutamyl-tRNA reductase (GluTR) and glutamate 1-semialdehyde aminotransferase (GSAT) (Figure 5; Tanaka and Tanaka, 2007). In the dark Pchlide cannot be converted to chlorophyllide because the NADPH:Pchlide oxidoreductase (POR) requires light to convert Pchlide a to chlorophyllide a. Pchlide forms a complex with GluTR and CHL27 (a component of Mg-protoporphyrin monomethylester cyclase3) and the FLUORESCENT (FLU) protein which inhibits GluTR thereby preventing accumulation of free Pchlide which is phototoxic (Kauss et al., 2012; Hou et al., 2019; Abbas et al., 2022). The steady-state level of Pchlide in the flu mutant is unrestrained under moderate hypoxia compared to normoxia and hyperoxia, demonstrating that accumulation of Pchlide in various species is also regulated through oxygen sensing. Furthermore, GluTR acts at the rate-limiting step in the tetrapyrrole pathway, and its level is regulated at the transcriptional and post-transcriptional level (Tanaka and Tanaka, 2007; Tanaka et al., 2011). The biosynthetic pathways of chlorophyll and heme follow the same pathway up to protoporphyrin IX (proto IX) where they split in two branches, the chl branch and the heme branch (Mochizuki et al., 2008).
In green algae and higher plants disruption of the tetrapyrrole pathway impacts the expression of photosynthesis-associated nuclear genes (PhANGs) (Strand et al., 2003; von Gromoff et al., 2008; Zhang et al., 2011). Furthermore, several studies were conducted with Arabidopsis gun mutants (genomes uncoupled) that partially recovered LHCB1 expression following norflurazon (NF) treatment (Susek et al., 1993). When exposed to oxidative stress, the gun1-6 mutant expressed PhANGs, whereas these genes were repressed in the WT supporting the evidence that some tetrapyrrole intermediates are involved in the communication network between chloroplasts and nucleus. Heme and Mg-ProtoIX affect PhANGs, in particular those of proteins involved in the primary reactions of photosynthesis, Calvin cycle enzymes, and enzymes of the tetrapyrrole biosynthetic pathway.
At the branchpoint Mg2+ or Fe2+ are inserted into Proto IX by Mg-chelatase (MgCh) or ferrochelatase (FC) to produce Mg-protoporphyrin IX (MgProto) and heme, respectively (Figure 5). In plants, MgCh consists of three subunits: CHLI, CHLD, and CHLH. CHLD and CHLH are encoded by a single gene in Arabidopsis, while CHLI exists in two isoforms, CHLI1 and CHLI2. CHLI1 is required for photosynthesis (Koncz et al., 1990; Rissler et al., 2002), whereas CHLI2 only plays a minor role in the assembly of MgCh (Kobayashi et al., 2008). Furthermore, GUN4 increases MgCh activity by regulating substrate or product channeling (Larkin et al., 2003; Shimizu and Masuda, 2021).
Analysis of gun mutants of Arabidopsis and mutants affected at different steps of the tetrapyrrole pathway in Chlamydomonas suggests that tetrapyrroles have a regulatory role in plastid signaling in plants and algae and are involved in retrograde signaling. The chlorophyll biosynthesis inhibitor a, a-dipyridyl was reported to limit light-induced LHCB mRNA increase in Chlamydomonas. This compound blocks the chlorophyll biosynthetic pathway at a late step which leads to the buildup of Mg-protoIX methyl ester (Mg-ProtoIX-ME) (Johanningmeier and Howell, 1984; Kleine et al., 2009). Mg-protoIX methyl ester is a porphyrin intermediate that is used as a marker for environmental changes that affect PhANG expression (Kindgren et al., 2011). When the chlorophyll biosynthetic pathway was disrupted upstream of Mg-ProtoIX, the above-mentioned inhibition of chlorophyll biosynthesis did not occur. This suggests that Mg-ProtoIX and Mg-ProtoIX-ME may act as negative factors in chlorophyll biosynthesis and are needed for the repression of LHCB transcription (Jasper et al., 1991). In addition, Mg-ProtoIX and Mg-ProtoIX-ME are required for the induction of HEAT SHOCK PROTEIN 70A/B (HSP70A and HSP70B), the cytosolic and plastid-localized proteins, respectively, in Chlamydomonas (Johanningmeier and Howell, 1984; Fortunato et al., 2022; Li et al., 2022). According to Vasileuskaya et al. (2004), Mg-ProtoIX also stimulates HEMA, which encodes GluTR in Chlamydomonas (Figure 5). Light induction of HSP70 is prevented in the chlorophyll-deficient brs-1 mutant affected at a step upstream of Mg-ProtoIX (Vasileuskaya et al., 2004; Toyokuni et al., 2022). Also Mg-ProtoIX buildup is needed for HSP70 light induction in the Chlamydomonas mutant PC-1/Y-7 which is unable to convert protochlorophyllide to chlorophyllide in the dark (Hess et al., 1994).
Recently, a T-DNA insertion was found 8 kb upstream of the gene ferrochelatase I (FC1) encoding chloroplast heme synthase/plastid ferrochelatase in a gun mutant called gun6-1D. It has the same phenotype as the gun2-gun5 mutants with higher expression of PhANGs than in WT under photooxidative conditions (Woodson et al., 2013). In this mutant FC1 is over-expressed. It was proposed that heme accumulates in gun6-1D and appears to be a positive retrograde signal, as evidenced by increased flux through the heme branch of the tetrapyrrole biosynthesis pathway and increased expression of PhANGs in the mutant (Voss et al., 2011; Woodson et al., 2013; Figure 5).
Ferrochelatase inserts Fe2+ into Proto IX in the heme branch to form protoheme (heme b), the prosthetic group of b-type cytochromes and of proteins like catalase and peroxidase. In Arabidopsis and cucumber, there are two isoforms of FC (FC1 and FC2) which have different tissue-specific and developmental expression profiles: FC2 is light-dependent and mostly expressed in photosynthetic tissues, whereas FC1 is stress-responsive and found throughout the plant (Chow et al., 1998; Nagai et al., 2007). Other hemes, such as heme a and heme c, are synthesized from protoheme. Bilins use protoheme as a substrate. Heme oxygenase oxidizes protoheme to produce biliverdin IX. Phytochromobilin synthase then transforms biliverdin IX into 3Z-phytochromobilin. Finally, 3Z-phytochromobilin is isomerized to 3E-phytochromobilin, which serves as the chromophore of phytochromes (PHY) (Terry et al., 2002; Shimizu and Masuda, 2021).
Recent studies with Chlamydomonas have revealed novel essential functions of bilins in retrograde signaling, notably in the regulation of the expression of genes involved in the detoxification of ROS (Duanmu et al., 2013, 2017). Moreover bilins also function in the maintenance of a functional photosynthetic apparatus as well as in the regulation of chlorophyll synthesis by binding GUN4, an important regulator of Mg chelatase (Zhang et al., 2021).
Plastid gene expression pathway and retrograde signaling
Retrograde signaling is triggered when PGE is disrupted at the transcriptional or translational levels (Woodson and Chory, 2008). PGE is responsible for changing PhANG expression (Zhang et al., 2011). PGE is also closely regulated by redox signals from the thylakoid membrane, which are transmitted through an intricate network of phosphorylation processes (Pfannschmidt et al., 2001; Pfannschmidt and Liere, 2005; Pfannschmidt, 2010). Use of the PGE inhibitors chloramphenicol and lincomycin (LIN) revealed the role of PGE in retrograde signaling (Stern et al., 2010; Pogson et al., 2015; Figure 6). When chloramphenicol or LIN is applied to immature seedlings, PhANGs are repressed. The retrograde signal from PGE was found to be light-independent in two photomorphogenic mutants, cop1-4 in Arabidopsis and lip1 in pea which partially restore LHCB1.2 expression in the dark compared to wild type (WT) upon LIN treatment (Sullivan and Gray, 1999; Tanaka and Tanaka, 2007). Sigma factors have also been shown to influence PGE in vivo through reversible phosphorylation. In addition, the redox state of chloroplasts controls SIG1 activity, which impacts the expression of the psaA gene, implying a link between photosynthetic activity and SIG1 phosphorylation, as well as the expression of photosynthesis genes possibly via the Sensor Kinase (CSK) in chloroplasts (Shimizu et al., 2010; Liebers et al., 2020). A nucleus-encoded circadian oscillator regulates PGE cycles. SIG5 which regulates the transcription of a number of chloroplast genes in Arabidopsis (Shimizu et al., 2010; Noordally Zeenat et al., 2013) is also involved in this circadian gating of light input to chloroplast genes (Noordally Zeenat et al., 2013).
Several components involved in the signaling pathway have been identified (Figure 6). GUN1, a plastid-localized pentatricopeptide repeat protein, is one of the key regulators in plastid-to-nucleus signaling (Koussevitzky et al., 2007). Signals from at least four distinct plastid processes (tetrapyrrole biosynthesis, PGE, redox, and plastid protein import) have been found to be mediated by GUN1 (Kozaki et al., 2000; Koussevitzky et al., 2007), although the links between GUN1 and other components within plastids such as STN7 remain unclear. Two nuclear transcription factors, ABI4 and GLK1, may regulate the expression of photosynthetic genes downstream of GUN1 (Figure 6). ABI4 is a negatively acting transcription factor in response to plastid signals. According to the current model, ABI4 competitively binds to the G-box cis-element in response to plastid signals and inhibits the induction of LHCB expression (Koussevitzky et al., 2007). GUN1 activates ABI4 through an as yet uncharacterized mechanism which is also used by signals arising from tetrapyrrole biosynthesis, PGE, and redox status (Figure 6). However, the signal arising from protein import defects does not use this pathway. Instead, it involves GLK1 (Kozaki et al., 2000). Unlike ABI4, GLK1 is a positive regulator of LHCB expression. GLK1 coordinates the expression of photosynthesis related genes in the nucleus (Nakamura et al., 2009; Waters et al., 2009). When plastids are damaged by stresses such as NF treatment or a protein import defect, the expression of GLK1 is significantly repressed, leading to subsequent down-regulation of photosynthetic genes. Furthermore, GUN1 activity is necessary for the repression of GLK1 (Kozaki et al., 2000). The level of GLK1 also appears to be regulated at the protein level (Waters et al., 2008). Hence unraveling the mechanism by which GLK1 expression/accumulation is regulated may be one of the key tasks for the near future.
In mustard, the retrograde signaling system is active exclusively at an early developmental stage of seedlings (Nott et al., 2006). Analysis of the Arabidopsis prors1 mutant, on the other hand, suggests that the PGE pathway is maintained in mature leaf tissue (Pesaresi et al., 2006, 2009). The Arabidopsis prors1 mutant is deficient in prolyl-tRNA synthetase leading to reduced protein synthesis in both the plastids and mitochondria. PhANGs are specifically down-regulated in mature prors1 plants (Pesaresi et al., 2006). This decreased expression, however, was not observed in the Arabidopsis mrpl11 or prpl11 mutants, which are deficient in the mitochondrial and plastid ribosomal L11 proteins, respectively (Pesaresi et al., 2007, 2011), but was observed in the mrpl11 prpl11 double mutant. This finding indicates that plastid and mitochondrial translation act cooperatively to control PhANG expression (Pesaresi et al., 2011). The analysis of transcriptionally active plastid chromosomes (TAC) has recently shown that additional proteins are necessary for chloroplast transcription besides the main components of PEP (Pfalz et al., 2006). TRXz and its intrinsic partners fructokinase-like protein 1 (FLN1) and 2 (FLN2) are associated with the plastid-encoded RNA polymerase (PEP), suggesting that these proteins mediate the redox regulation of PGE (Steiner et al., 2009, 2011; Stern et al., 2010; Wimmelbacher and Börnke, 2014). Studies of the trx-z Arabidopsis mutant revealed alterations in PGE that are diagnostic for PEP deficiency, suggesting that TRX-z is required for PEP function. Furthermore, Wimmelbacher and Börnke (2014), demonstrated that TRX-z and FLN1 are structurally important components of the PEP complex, and that the fine-tuning of PEP function may be controlled by TRX-z and FLN1 redox activity (Chan et al., 2014; Wimmelbacher and Börnke, 2014).
Role of singlet oxygen in signaling from plastids to nucleus
Chloroplasts are very important sensors of environmental factors such as high light, low or high temperature, and drought that perturb photosynthetic electron transport (Baruah et al., 2009a,b). Under these stress conditions, light energy absorbed by the photosynthetic apparatus may exceed its capacity and lead to an over-reduction of the electron transport chain (Kramarenko et al., 2006). Photosynthetic organisms have developed several ways to dissipate excess light excitation energy to avoid this over-reduction which can produce damaging ROS (Blazek et al., 1989; Baruah et al., 2009a). Oxygen is chemically inactive in its ground state, but it can be turned into ROS through light-induced electron transfer reactions. Different forms of ROS can be formed such as superoxide, hydrogen peroxide (H2O2), hydroxyl radicals and singlet oxygen (1O2) (Carloni et al., 1993). Depending on the amounts of these ROS produced, they can cause oxidative damage or act as signaling molecules.
Conversion of protochlorophyllide to chlorophyllide is a key step in chlorophyll synthesis and requires light in land plants. Protochlorophyllide is highly photodynamic and its accumulation in the dark causes photo-oxidative damage when plants are shifted from the dark to the light. This is prevented by the FLUORESCENT (FLU) protein which inhibits glu-tRNA reductase, an enzyme which catalyzes one of the early steps of chlorophyll synthesis (Kauss et al., 2012; Figure 5). After a shift form darkness to light, mutants deficient in FLU produce 1O2 and initiate a programmed cell death (PCD) response and photobleach (Meskauskiene et al., 2001). However, these mutants survive in constant dark or constant light. A suppressor screen of the flu mutant in Arabidopsis led to the identification of the chloroplast proteins Executer 1 (EX1) and its homolog EX2. In the ex1 suppressor the 1O2-induced programmed cell death response no longer occurs (Wagner et al., 2004; Lee et al., 2007; Pruzinská et al., 2007). EX1 and EX2 play a critical role in this cell response to a burst in 1O2 which elicits changes in gene expression (Figure 6; Brzezowski et al., 2012; Prabha et al., 2016). Further screening was performed with a transgenic flu line containing a reporter driven by the promoter of an 1O2-responsive AAA-ATPase gene for constitutive high-level expression. One of the mutant isolated is affected in the Pleiotropic Response Locus 1 (PRL1) gene. This gene appears to suppress the expression of 1O2-responsive genes and to play an important role in the response of plants to environmental stress. Additional genetic studies also suggest that 1O2-mediated signals are transmitted to the nucleus through a signaling network and that 1O2-mediated retrograde signaling interconnects to other signaling pathways (Baruah et al., 2009a,b).
Other 1O2-response mutants (caa33, soldat8, and soldat10) are similarly deficient in mechanisms that impact plastid homeostasis, resulting in an increased 1O2 response (Lee et al., 2007; Simková et al., 2012). All of these examples show that the response to 1O2 in plants is part of a complex signaling network that is influenced by other signaling pathways (Baruah et al., 2009b). 1O2 signaling affects gene expression not only at the transcriptional level, but also at the translational level (Simková et al., 2012). In response to 1O2, the barley flu-orthologous mutant tigrine-d.12 had decreased expression of a number of chloroplast proteins (Khandal et al., 2009). This inhibition of protein synthesis was due to a halt in translation initiation on 80S cytoplasmic ribosomes which correlated with a decrease in phosphorylation of the ribosomal protein S6 (Liebers et al., 2020).
The HSP70A gene was shown to be induced by 1O2 and several HSP70A-based reporter constructs reacted specifically to either 1O2 or H2O2 in Chlamydomonas. The HSP70A promoter region is composed of two promoters named PA1 and PA2, two separate cis-acting regulatory regions, that confer inducibility by hydrogen peroxide and singlet oxygen (Shao et al., 2007). The GPXH/GPX5 gene is significantly induced by 1O2, but only weakly increased by other ROS, and it is selectively induced by 1O2 under high light (Fischer et al., 2009, 2010). In addition, the transcriptional activation of GPXH is dependent on several regulatory factors different from reactive electrophile species (RES). RES signaling, another 1O2–specific response, is likely to be involved in Rose Bengal (RB) acclimation in C. reinhardtii. This 1O2 specific response includes the strong induction of GPXH and maybe other genes at low RB concentrations and during the early high light response (Fischer et al., 2010). A reporter construct containing a portion of the 1O2-specific GPXH promoter was utilized to test over 5,000 UV-mutagenized clones for their responsiveness to 1O2. The analysis of these mutants revealed that, similar to plants, the 1O2 signal is not conveyed along a single linear chain in C. reinhardtii, as no mutation defining signal transmission could be isolated (Fischer et al., 2006). PSBP2 is a paralog of the oxygen-evolving enhancer gene PSBP1, and it has been proposed that it functions as an 1O2 responsive factor to activate GPXH expression. However activation of the wild-type GPXH gene and adaptation to 1O2 are not completely abolished in psbP2. This suggests that different signaling pathways are involved in the response to 1O2 in C. reinhardtii (Figure 6; Fischer et al., 2010; Brzezowski et al., 2012).
Additional possible plastid signaling molecules
Although several retrograde signaling chains have been identified, the nature of the plastid retrograde signaling molecules is still largely unknown. Norflurazon (NF), an inhibitor of carotenoid synthesis has been used extensively for studying retrograde signaling. Upon treatment of seedlings with NF and exposure to light, they bleach because photooxidation of chlorophyll in the absence of carotenoids leads to the accumulation of tetrapyrrole intermediates that are highly photodynamic. Under these conditions expression of LHCB genes is strongly decreased in wild-type plants. Mg-proto IX levels are 15 times higher in NF treated compared to untreated Arabidopsis seedlings (Strand et al., 2003; Chan et al., 2014; Garmash, 2022). When seedlings were fed tetrapyrrole intermediates, such as heme, porphobilinogen, or proto IX, they did repress LHCB promoter-driven expression of luciferase (Strand et al., 2003). Expression of LHCB was repressed by feeding Mg-proto IX directly to leave protoplasts implying a specific control of expression of PhANGs by Mg-proto IX (Strand et al., 2003). Mg-proto IX increased in both plastids and cytosol under stress (Strand et al., 2003; Tyagi and Gaur, 2003). However these claims have been questioned by other studies which showed no correlation between tetrapyrrole intermediates, including Mg ProtoIX, and LHCB expression (Mochizuki et al., 2008; Moulin et al., 2008). It is thus unlikely that Mg-proto IX is exported from the plastid to the cytosol as a retrograde signal. The signal created by tetrapyrrole intermediates might also be produced from ROS (Ankele et al., 2007), either directly or indirectly, or derived from redox signals, because perturbation of the tetrapyrrole biosynthetic pathway could result in ROS formation or changes in the plastid redox state (Surpin et al., 2002; Mochizuki et al., 2008, 2010; Monaghan et al., 2009).
Another tetrapyrrole intermediate, heme, affects nuclear gene expression and appears to be involved in retrograde control. In the Arabidopsis gun6 gain-of-function mutant, heme was found to be a retrograde signaling molecule (Estavillo et al., 2011). MEcPP (methylerythritolcyclodiphosphate) and PAP (3’-phosphoadenosine 5’-phosphate) two key metabolites generated in plastids that have been proposed to act as signaling molecules in the nucleus, in addition to the tetrapyrrole intermediates. PAP accumulates in the plastid under stress conditions, although it has no known physiological role in plant cells (Estavillo et al., 2011; Xiao et al., 2012; Wu et al., 2019). Although PAP is generated in the plastids, expression of the phosphatase SAL1 in the nucleus lowers overall PAP levels (Estavillo et al., 2011). PAP appears to regulate gene expression by altering RNA metabolism mediated by 5’-3’ exonucleases. MEcPP, like PAP, plays a vital function in retrograde signaling, although it is unknown how it moves from the plastid to the nucleus. The concept of a metabolite signal makes sense since the plastid constantly conveys its metabolic state to the cytosol through the interchange of various metabolites, which ultimately leads to changes in nuclear gene expression (Pfannschmidt, 2010; Estavillo et al., 2011; Leister and Kleine, 2011; Xiao et al., 2012). As a result, a number of metabolites, including carbohydrates and redox valves, have been proposed to be retrograde signals (Kleine et al., 2009; Leister, 2012). It should be noticed that these new signals were discovered during physiologically relevant stress conditions like drought or high light unlike the previously identified signals induced through artificial stress conditions.
Reactive oxygen species molecules have also been proposed to function as retrograde signaling molecules as they have the ability to move across the subcellular compartments, which is one of the main characteristics of the signaling molecules. H2O2 may be the best candidate whereas 1O2 is thought to have a less important role because of its short half-life, high reactivity and limited diffusion (Nott et al., 2006; Larkin and Ruckle, 2008; Leister, 2012; Maruta et al., 2012; Li and Kim, 2022). β-Cyclocitral (β-CC), a 1O2 oxidation product of carotenoids was identified in Arabidopsis as a messenger in the 1O2 signaling pathway and induces gene expression in response to 1O2 (Ramel et al., 2012; Figures 5, 6). As it is volatile, β-CC has the ability to travel through the cytosol by diffusion (Leister and Kleine, 2011; Leister, 2012; Liang et al., 2022), to regulate nuclear gene expression and to act as a downstream messenger for rising levels of 1O2 (Ramel et al., 2012).
Conclusion and future perspectives
Plastids are essential compartments of eukaryotic cells. They are involved in the synthesis of many metabolites and are the sites of important metabolic and regulatory pathways. Here we have discussed several retrograde-signaling pathways that are highly complex and diverse. They are not operating in every cell type but restricted to specific cell types and developmental stages. As an example, the singlet-oxygen–mediated pathway occurs mostly in differentiated photosynthetically active mesophyll cells. In the last few decades, significant advances have been achieved in our understanding of plastid-to-nucleus signaling. However, in spite of this progress, there are still a lot of question marks concerning the identity of the authentic retrograde signals and how they work. Possibly, signals could only accumulate transiently and locally and be therefore difficult to detect. It is also not clear how perturbations of chloroplast processes elicit retrograde signals and how these signals are transmitted from the plastids to the nucleus and how they change the expression of nuclear genes. For a long time, the redox state of the plastids and PGE were thought to be the main determinants of retrograde signaling. Yet we still do not know how changes in redox state or impairment of PGE are transduced into signals and their identity remains elusive. At this time, it is not clear whether the signals are proteins, proteolytic products, oxidized lipids, metabolites or ionic fluxes.
Most of the cellular biosynthetic pathways occur, at least partly, in the plastids, and specific carriers move different metabolites into and out of them. Different retrograde signaling pathways have been proposed based on different approaches and different experimental models. But it is not yet clear how these signaling networks interact with each other and how some key transcriptional factors like PEND, GLK, WHIRLY1, AP2/EREBP operate within these networks. Clearly, these challenging questions will need to be addressed in the coming years.
Author contributions
XS designed the review outline and wrote the “Conclusion and future perspectives” section of the manuscript. MJ, ZL, and J-DR wrote the other sections and designed all figures. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (31670233).
Acknowledgments
We apologize to colleagues whose work could not be cited due to limited space.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbas, M., Sharma, G., Dambire, C., Marquez, J., Alonso-Blanco, C., Proaño, K., et al. (2022). An oxygen-sensing mechanism for angiosperm adaptation to altitude. Nature 606, 565–569. doi: 10.1038/s41586-022-04740-y.
Abdallah, F., Salamini, F., and Leister, D. (2000). A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 5, 141–142. doi: 10.1016/S1360-1385(00)01574-0
Abdelkefi, H., Sugliani, M., Ke, H., Harchouni, S., Soubigou-Taconnat, L., Citerne, S., et al. (2018). Guanosine tetraphosphate modulates salicylic acid signalling and the resistance of Arabidopsis thaliana to turnip mosaic virus. Mol. Plant Pathol. 19, 634–646. doi: 10.1111/mpp.12548
Aigner, H., Wilson, R. H., Bracher, A., Calisse, L., Bhat, J. Y., Hartl, F. U., et al. (2017). Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 358, 1272–1278. doi: 10.1126/science.aap9221
Ali, E., Hussain, S., Hussain, N., Kakar, K. U., Shah, J. M., Zaidi, S. H. R., et al. (2022). Tocopherol as plant protector: An overview of tocopherol biosynthesis enzymes and their role as antioxidant and signaling molecules. Acta Physiol. Plant. 44:20. doi: 10.1007/s11738-021-03350-x
An, F., Zhao, Q., Ji, Y., Li, W., Jiang, Z., Yu, X., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22, 2384–2401. doi: 10.1105/tpc.110.076588
Ankele, E., Kindgren, P., Pesquet, E., and Strand, A. (2007). In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast. Plant Cell 19, 1964–1979. doi: 10.1105/tpc.106.048744
Arsova, B., Hoja, U., Wimmelbacher, M., Greiner, E., Üstün, Ş, Melzer, M., et al. (2010). Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22, 1498–1515. doi: 10.1105/tpc.109.071001
Baier, M., and Dietz, K.-J. (2005). Chloroplasts as source and target of cellular redox regulation: A discussion on chloroplast redox signals in the context of plant physiology. J. Exp. Bot. 56, 1449–1462. doi: 10.1093/jxb/eri161
Barczak-Brzyżek, A., Brzyżek, G., Koter, M., Siedlecka, E., Gawroński, P., and Filipecki, M. (2022). Plastid retrograde regulation of miRNA expression in response to light stress. BMC Plant Biol. 22:150. doi: 10.1186/s12870-022-03525-9
Barkan, A. (2011). Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155, 1520–1532. doi: 10.1104/pp.110.171231
Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. doi: 10.1016/s0300-9084(00)00602-7
Barnes, D., Franklin, S., Schultz, J., Henry, R., Brown, E., Coragliotti, A., et al. (2005). Contribution of 5’- and 3’-untranslated regions of plastid mRNAs to the expression of Chlamydomonas reinhardtii chloroplast genes. Mol. Genet. Geno. 274, 625–636. doi: 10.1007/s00438-005-0055-y
Barnes, D., and Mayfield, S. P. (2003). Redox control of posttranscriptional processes in the chloroplast. Antioxid Redox Signal 5, 89–94. doi: 10.1089/152308603321223577
Baruah, A., Simková, K., Apel, K., and Laloi, C. (2009a). Arabidopsis mutants reveal multiple singlet oxygen signaling pathways involved in stress response and development. Plant Mol. Biol. 70, 547–563. doi: 10.1007/s11103-009-9491-0
Baruah, A., Simková, K., Hincha, D. K., Apel, K., and Laloi, C. (2009b). Modulation of O-mediated retrograde signaling by the PLEIOTROPIC RESPONSE LOCUS 1 (PRL1) protein, a central integrator of stress and energy signaling. Plant J. 60, 22–32. doi: 10.1111/j.1365-313X.2009.03935.x
Blazek, E. R., Peak, J. G., and Peak, M. J. (1989). Singlet oxygen induces frank strand breaks as well as alkali- and piperidine-labile sites in supercoiled plasmid DNA. Photochem. Photobiol. 49, 607–613. doi: 10.1111/j.1751-1097.1989.tb08431.x
Boniecka, J., Prusińska, J., Da̧browska, G. B., and Goc, A. (2017). Within and beyond the stringent response-RSH and (p)ppGpp in plants. Planta 246, 817–842. doi: 10.1007/s00425-017-2780-y
Breeze, E., and Mullineaux, P. M. (2022). The passage of H2O2 from chloroplasts to their associated nucleus during retrograde signalling: Reflections on the role of the nuclear envelope. Plants 11, 552. doi: 10.3390/plants11040552
Brzezowski, P., Wilson, K. E., and Gray, G. R. (2012). The PSBP2 protein of Chlamydomonas reinhardtii is required for singlet oxygen-dependent signaling. Planta 236, 1289–1303. doi: 10.1007/s00425-012-1683-1
Carloni, P., Damiani, E., Greci, L., Stipa, P., Tanfani, F., Tartaglini, E., et al. (1993). On the use of 1,3-diphenylisobenzofuran (DPBF). React. Carbon Oxygen Centered Radi. Model Nat. Syst. 19, 395–405.
Cashel, M., and Gallant, J. (1969). Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841. doi: 10.1038/221838a0
Chan, K. X., Crisp, P. A., Estavillo, G. M., and Pogson, B. J. (2014). Chloroplast-to-nucleus communication. Plant Sign. Behav. 5, 1575–1582. doi: 10.4161/psb.5.12.13758
Charuvi, D., Kiss, V., Nevo, R., Shimoni, E., Adam, Z., and Reich, Z. (2012). Gain and loss of photosynthetic membranes during plastid differentiation in the shoot apex of Arabidopsis. Plant Cell 24, 1143–1157. doi: 10.1105/tpc.111.094458
Chehab, E. W., Raman, G., Walley, J. W., Perea, J. V., Banu, G., Theg, S., et al. (2006). Rice HYDROPEROXIDE LYASES with unique expression patterns generate distinct aldehyde signatures in Arabidopsis. Plant Physiol. 141, 121–134. doi: 10.1104/pp.106.078592
Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Ann. Rev. Genet. 38, 87–117. doi: 10.1146/annurev.genet.38.072902.092259
Chen, M., Galvão, R. M., Li, M., Burger, B., Bugea, J., Bolado, J., et al. (2010). Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141, 1230–1240. doi: 10.1016/j.cell.2010.05.007
Chen, Y.-N., Slabaugh, E., and Brandizzi, F. (2008). Membrane-tethered transcription factors in Arabidopsis thaliana: Novel regulators in stress response and development. Curr. Opin. Plant Biol. 11, 695–701. doi: 10.1016/j.pbi.2008.10.005
Choquet, Y., and Vallon, O. (2000). Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82, 615–634. doi: 10.1016/S0300-9084(00)00609-X
Chory, J. (2010). Light signal transduction: an infinite spectrum of possibilities. Plant J. 61, 982–991. doi: 10.1111/j.1365-313X.2009.04105.x
Chow, K. S., Singh, D. P., Walker, A. R., and Smith, A. G. (1998). Two different genes encode ferrochelatase in Arabidopsis: Mapping, expression and subcellular targeting of the precursor proteins. Plant J. 15, 531–541. doi: 10.1046/j.1365-313x.1998.00235.x
Cottage, A., Mott, E. K., Kempster, J. A., and Gray, J. C. (2010). The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J. Exp. Bot. 61, 3773–3786. doi: 10.1093/jxb/erq186
Díaz, M. G., Hernández-Verdeja, T., Kremnev, D., Crawford, T., Dubreuil, C., and Strand, Å (2018). Redox regulation of PEP activity during seedling establishment in Arabidopsis thaliana. Nat. Commun. 9:50. doi: 10.1038/s41467-017-02468-2
Dietz, K.-J., Vogel, M. O., and Viehhauser, A. (2010). AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245, 3–14. doi: 10.1007/s00709-010-0142-8
Dogra, V., Singh, R. M., Li, M., Li, M., Singh, S., and Kim, C. (2022). EXECUTER2 modulates the EXECUTER1 signalosome through its singlet oxygen-dependent oxidation. Mol. Plant 15, 438–453. doi: 10.1016/j.molp.2021.12.016
Duanmu, D., Casero, D., Dent, R. M., Gallaher, S., Yang, W., Rockwell, N. C., et al. (2013). Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Biol. Sci. 110, 3621–3626. doi: 10.1073/pnas.1222375110
Duanmu, D., Rockwell, N. C., and Lagarias, J. C. (2017). Algal light sensing and photoacclimation in aquatic environments. Plant Cell Environ. 40, 2558–2570. doi: 10.1111/pce.12943
Dubreuil, C., Jin, X., Barajas-López, J. D., Hewitt, T. C., Tanz, S. K., Dobrenel, T., et al. (2018). Establishment of photosynthesis through chloroplast development is controlled by two distinct regulatory phases. Plant Physiol. 176, 1199–1214. doi: 10.1104/pp.17.00435
Enami, K., Ozawa, T., Motohashi, N., Nakamura, M., Tanaka, K., and Hanaoka, M. (2011). Plastid-to-nucleus retrograde signals are essential for the expression of nuclear starch biosynthesis genes during amyloplast differentiation in tobacco BY-2 cultured cells. Plant Physiol. 157, 518–530. doi: 10.1104/pp.111.178897
Escoubas, J. M., Lomas, M., LaRoche, J., and Falkowski, P. G. (1995). Light intensity regulation of cab gene transcription is signaled by the redox state of the plastoquinone pool. Proc. Natl. Acad. Sci. U.S.A. 92, 10237–10241. doi: 10.1073/pnas.92.22.10237
Estavillo, G. M., Crisp, P. A., Pornsiriwong, W., Wirtz, M., Collinge, D., Carrie, C., et al. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012. doi: 10.1105/tpc.111.091033
Field, B. (2018). Green magic: Regulation of the chloroplast stress response by (p)ppGpp in plants and algae. J. Exp. Bot. 69, 2797–2807. doi: 10.1093/jxb/erx485
Finkelstein, R. R., Wang, M. L., Lynch, T. J., Rao, S., and Goodman, H. M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10, 1043–1054. doi: 10.2307/3870689
Fischer, B. B., Dayer, R., Schwarzenbach, Y., Lemaire, S. D., Behra, R., Liedtke, A., et al. (2009). Function and regulation of the glutathione peroxidase homologous gene GPXH/GPX5 in Chlamydomonas reinhardtii. Plant Mol. Biol. 71, 569–583. doi: 10.1007/s11103-009-9540-8
Fischer, B. B., Eggen, R. I. L., and Niyogi, K. K. (2010). Characterization of singlet oxygen-accumulating mutants isolated in a screen for altered oxidative stress response in Chlamydomonas reinhardtii. BMC Plant Biol. 10:279. doi: 10.1186/1471-2229-10-279
Fischer, B. B., Ledford, H. K., Wakao, S., Huang, S. G., Casero, D., Pellegrini, M., et al. (2012). SINGLET OXYGEN RESISTANT 1 links reactive electrophile signaling to singlet oxygen acclimation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 109, E1302–E1311. doi: 10.1073/pnas.1116843109
Fischer, B. B., Wiesendanger, M., and Eggen, R. I. (2006). Growth condition-dependent sensitivity, photodamage and stress response of Chlamydomonas reinhardtii exposed to high light conditions. Plant Cell Physiol. 47, 1135–1145. doi: 10.1093/pcp/pcj085
Fisher, K. E., Krishnamoorthy, P., Joens, M. S., Chory, J., Fitzpatrick, J. A. J., and Woodson, J. D. (2022). Singlet oxygen leads to structural changes to chloroplasts during their degradation in the Arabidopsis thaliana plastid ferrochelatase two mutant. Plant Cell Physiol. 63, 248–264. doi: 10.1093/pcp/pcab167
Fortunato, S., Lasorella, C., Tadini, L., Jeran, N., Vita, F., Pesaresi, P., et al. (2022). GUN1 involvement in the redox changes occurring during biogenic retrograde signaling. Plant Sci. 320:111265. doi: 10.1016/j.plantsci.2022.111265
Galvão, R. M., Li, M., Kothadia, S. M., Haskel, J. D., Decker, P. V., Van Buskirk, E. K., et al. (2012). Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev. 26, 1851–1863. doi: 10.1101/gad.193219.112
Gangappa, S. N., Crocco, C. D., Johansson, H., Datta, S., Hettiarachchi, C., Holm, M., et al. (2013). The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25, 1243–1257. doi: 10.1105/tpc.113.109751
Garmash, E. V. (2022). Signal pathways for regulation of plant alternative oxidase genes’ expression. Russ. J. Plant Physiol. 69:1. doi: 10.1134/S1021443722010058
Grabowski, E., Miao, Y., Mulisch, M., and Krupinska, K. (2008). Single-stranded DNA-binding protein whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol. 147, 1800–1804. doi: 10.1104/pp.108.122796
Heiber, I., Ströher, E., Raatz, B., Busse, I., Kahmann, U., Bevan, M. W., et al. (2007). The redox imbalanced mutants of Arabidopsis differentiate signaling pathways for redox regulation of chloroplast antioxidant enzymes. Plant Physiol. 143, 1774–1788. doi: 10.1104/pp.106.093328
Hess, W. R., Müller, A., Nagy, F., and Börner, T. (1994). Ribosome-deficient plastids affect transcription of light-induced nuclear genes: Genetic evidence for a plastid-derived signal. Mol. Gen. Genet. 242, 305–312. doi: 10.1007/bf00280420
Holtan, H. E., Bandong, S., Marion, C. M., Adam, L., Tiwari, S., Shen, Y., et al. (2011). BBX32, an Arabidopsis B-box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 156, 2109–2123. doi: 10.1104/pp.111.177139
Hou, Z., Yang, Y., Hedtke, B., and Grimm, B. (2019). Fluorescence in blue light (FLU) is involved in inactivation and localization of glutamyl-tRNA reductase during light exposure. Plant J. Cell Mol. Biol. 97, 517–529. doi: 10.1111/tpj.14138
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/s1360-1385(01)02223-3
Jasper, F., Quednau, B., Kortenjann, M., and Johanningmeier, U. (1991). Control of cab gene expression in synchronized Chlamydomonas reinhardtii cells. J. Photochem. Photobiol. Biol. 11, 139–150. doi: 10.1016/1011-1344(91)80256-H
Johanningmeier, U., and Howell, S. H. (1984). Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardi. possible involvement of chlorophyll synthesis precursors. J. Biol. Chem. 259, 13541–13549. doi: 10.1016/S0021-9258(18)90727-1
Jung, H.-S., and Chory, J. (2009). Signaling between chloroplasts and the nucleus: Can a systems biology approach bring clarity to a complex and highly regulated pathway? Plant Physiol. 152, 453–459.
Jung, H.-S., and Chory, J. (2010). Signaling between chloroplasts and the nucleus: Can a systems biology approach bring clarity to a complex and highly regulated pathway. Plant Physiol. 152, 453–459. doi: 10.1104/pp.109.149070
Kami, C., Lorrain, S., Hornitschek, P., and Fankhauser, C. (2010). Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66. doi: 10.1016/s0070-2153(10)91002-8
Kauss, D., Bischof, S., Steiner, S., Apel, K., and Meskauskiene, R. (2012). FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg++-branch of this pathway. FEBS Lett. 586, 211–216. doi: 10.1016/j.febslet.2011.12.029
Khandal, D., Samol, I., Buhr, F., Pollmann, S., Schmidt, H., Clemens, S., et al. (2009). Singlet oxygen-dependent translational control in the tigrina-d.12 mutant of barley. Proc. Natl. Acad. Sci. U.S.A. 106, 13112–13117. doi: 10.1073/pnas.0903522106
Kim, K., Shin, J., Lee, S. H., Kweon, H. S., Maloof, J. N., and Choi, G. (2011). Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 108, 1729–1734. doi: 10.1073/pnas.1011066108
Kindgren, P., Eriksson, M.-J., Benedict, C., Mohapatra, A., Gough, S. P., Hansson, M., et al. (2011). A novel proteomic approach reveals a role for Mg-protoporphyrin IX in response to oxidative stress. Physiol. Plant. 141, 310–320. doi: 10.1111/j.1399-3054.2010.01440.x
Kindgren, P., and Strand, Å (2015). Chloroplast transcription, untangling the gordian knot. New Phytol. 206, 889–891. doi: 10.1111/nph.13388
Kleine, T., Lockhart, P., and Batschauer, A. (2003). An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J. 35, 93–103. doi: 10.1046/j.1365-313x.2003.01787.x
Kleine, T., Voigt, C., and Leister, D. (2009). Plastid signalling to the nucleus: Messengers still lost in the mists? Trends Genet. 25, 185–192. doi: 10.1016/j.tig.2009.02.004
Kobayashi, K., Mochizuki, N., Yoshimura, N., Motohashi, K., Hisabori, T., and Masuda, T. (2008). Functional analysis of Arabidopsis thaliana isoforms of the Mg-chelatase CHLI subunit. Photochem. Photobiol. Sci. 7, 1188–1195. doi: 10.1039/b802604c
Koncz, C., Mayerhofer, R., Koncz-Kalman, Z., Nawrath, C., Reiss, B., Redei, G. P., et al. (1990). Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 9, 1337–1346. doi: 10.1002/j.1460-2075.1990.tb08248.x
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719.
Kozaki, A., Kamada, K., Nagano, Y., Iguchi, H., and Sasaki, Y. (2000). Recombinant carboxyltransferase responsive to redox of pea plastidic acetyl-CoA carboxylase. J. Biol. Chem. 275, 10702–10708. doi: 10.1074/jbc.275.14.10702
Kramarenko, G. G., Hummel, S. G., Martin, S. M., and Buettner, G. R. (2006). Ascorbate reacts with singlet oxygen to produce hydrogen peroxide. Photochem. Photobiol. 82, 1634–1637. doi: 10.1562/2006-01-12-RN-774
Kusuma, P., and Bugbee, B. (2021). Improving the predictive value of phytochrome photoequilibrium: Consideration of spectral distortion within a leaf. Front. Plant Sci. 12:596943. doi: 10.3389/fpls.2021.596943
Larkin, R. M., Alonso, J. M., Ecker, J. R., and Chory, J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. doi: 10.1126/science.1079978
Larkin, R. M., and Ruckle, M. E. (2008). Integration of light and plastid signals. Curr. Opin. Plant Biol. 11, 593–599. doi: 10.1016/j.pbi.2008.10.004
Lee, H. Y., Hwang, O. J., and Back, K. (2022). Phytomelatonin as a signaling molecule for protein quality control via chaperone, autophagy, and ubiquitin–proteasome systems in plants. J. Exp. Bot. 2022:erac002. doi: 10.1093/jxb/erac002
Lee Keun, P., Kim, C., Landgraf, F., and Apel, K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 10270–10275.
Lee, K. P., Kim, C., Landgraf, F., and Apel, K. (2007). EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 10270–10275. doi: 10.1073/pnas.0702061104
Leister, D. (2012). Retrograde signaling in plants: From simple to complex scenarios. Front. Plant Sci. 3:135. doi: 10.3389/fpls.2012.00135
Leister, D., and Kleine, T. (2008). Towards a comprehensive catalog of chloroplast proteins and their interactions. Cell Res. 18, 1081–1083. doi: 10.1038/cr.2008.297
Leister, D., and Kleine, T. (2011). “Chapter three - role of intercompartmental DNA transfer in producing genetic diversity,” in International Review of Cell and Molecular Biology, ed. K. W. Jeon (Academic Press), 73–114. doi: 10.1016/B978-0-12-386035-4.00003-3
Leivar, P., Monte, E., Al-Sady, B., Carle, C., Storer, A., Alonso, J. M., et al. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20, 337–352. doi: 10.1105/tpc.107.052142
Leivar, P., and Quail, P. H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16, 19–28. doi: 10.1016/j.tplants.2010.08.003
Li, J.-Y., Yang, C., Tian, Y.-Y., and Liu, J.-X. (2022). Regulation of chloroplast development and function at adverse temperatures in plants. Plant Cell Physiol. 63, 580–591. doi: 10.1093/pcp/pcac022
Li, L., Ljung, K., Breton, G., Schmitz, R. J., Pruneda-Paz, J., Cowing-Zitron, C., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26, 785–790. doi: 10.1101/gad.187849.112
Li, M., and Kim, C. (2022). Chloroplast ROS and stress signaling. Plant Commun. 3:100264. doi: 10.1016/j.xplc.2021.100264
Liang, J., Wen, F., and Liu, J. (2019). Transcriptomic and lipidomic analysis of an EPA-containing nannochloropsis sp. PJ12 in response to nitrogen deprivation. Sci. Rep. 9:4540. doi: 10.1038/s41598-019-41169-2
Liang, J., Zhang, Q., Liu, Y., Zhang, J., Wang, W., and Zhang, Z. (2022). Chlorosis seedling lethality 1 encoding a MAP3K protein is essential for chloroplast development in rice. BMC Plant Biol. 22:20. doi: 10.1186/s12870-021-03404-9
Liebers, M., Gillet, F. X., Israel, A., Pounot, K., Chambon, L., Chieb, M., et al. (2020). Nucleo-plastidic PAP8/pTAC6 couples chloroplast formation with photomorphogenesis. EMBO J. 39:e104941. doi: 10.15252/embj.2020104941
Lin, F., Jiang, Y., Li, J., Yan, T., Fan, L., Liang, J., et al. (2018). B-BOX DOMAIN PROTEIN28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 30, 2006–2019. doi: 10.1105/tpc.18.00226
Liu, X., Liu, R., Li, Y., Shen, X., Zhong, S., and Shi, H. (2017). EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell 29, 3051–3067. doi: 10.1105/tpc.17.00508
Liu, Z., Yan, H., Wang, K., Kuang, T., Zhang, J., Gui, L., et al. (2004). Crystal structure of spinach major light-harvesting complex at 2.72Å resolution. Nature 428, 287–292. doi: 10.1038/nature02373
Llamas, E., Koyuncu, S., Lee, H. J., Gutierrez-Garcia, R., Dunken, N., Charura, N., et al. (2022). Chloroplast protein import determines plant proteostasis and retrograde signaling. bioRxiv [preprint]. doi: 10.1101/2022.03.19.484971
Lopez-Juez, E., and Pyke, K. A. (2005). Plastids unleashed: their development and their integration in plant development. Int. J. Dev. Biol. 49, 557–577. doi: 10.1387/ijdb.051997el
Luo, Q., Lian, H. L., He, S. B., Li, L., Jia, K. P., and Yang, H. Q. (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26, 2441–2456. doi: 10.1105/tpc.113.121657
Lyu, J., Gao, R., Guo, Z. J. J., and Chemistry, F. (2021). Galactosyldiacylglycerols: From a photosynthesis-associated apparatus to structure-defined in vitro assembling. J. Agric. Food Chem. 69, 8910–8928.
Maier, A., Zell, M. B., and Maurino, V. G. (2011). Malate decarboxylases: evolution and roles of NAD(P)-ME isoforms in species performing C(4) and C(3) photosynthesis. J. Exp. Bot. 62, 3061–3069. doi: 10.1093/jxb/err024
Martín, G., Leivar, P., Ludevid, D., Tepperman, J. M., Quail, P. H., and Monte, E. (2016). Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat. Commun. 7:11431. doi: 10.1038/ncomms11431
Maruta, T., Noshi, M., Tanouchi, A., Tamoi, M., Yabuta, Y., Yoshimura, K., et al. (2012). H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress*. J. Biol. Chem. 287, 11717–11729. doi: 10.1074/jbc.M111.292847
Masuda, T. (2008). Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth. Res. 96, 121–143. doi: 10.1007/s11120-008-9291-4
Matringe, M., Camadro, J. M., Block, M. A., Joyard, J., Scalla, R., Labbe, P., et al. (1992). Localization within chloroplasts of protoporphyrinogen oxidase, the target enzyme for diphenylether-like herbicides. J. Biol. Chem. 267, 4646–4651.
Mazzella, M. A. (2001). Interactive signalling by phytochromes and cryptochromes generates de-etiolation homeostasis in Arabidopsis thaliana. Plant Cell Environ. 24, 155–161. doi: 10.1111/j.1365-3040.2001.00653.x
Melonek, J., Mulisch, M., Schmitz-Linneweber, C., Grabowski, E., Hensel, G., and Krupinska, K. (2010). Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 232, 471–481. doi: 10.1007/s00425-010-1183-0
Merchante, C., Brumos, J., Yun, J., Hu, Q., Spencer, K. R., Enríquez, P., et al. (2015). Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163, 684–697. doi: 10.1016/j.cell.2015.09.036
Meskauskiene, R., Nater, M., Goslings, D., Kessler, F., den Camp, R., and Apel, K. (2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 12826–12831. doi: 10.1073/pnas.221252798
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: The new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Mochizuki, N., Tanaka, R., Grimm, B., Masuda, T., Moulin, M., Smith, A. G., et al. (2010). The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 15, 488–498. doi: 10.1016/j.tplants.2010.05.012
Mochizuki, N., Tanaka, R., Tanaka, A., Masuda, T., and Nagatani, A. (2008). The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 15184–15189. doi: 10.1073/pnas.0803245105
Monaghan, J., Xu, F., Gao, M., Zhao, Q., Palma, K., Long, C., et al. (2009). Two Prp19-like U-box proteins in the MOS4-associated complex play redundant roles in plant innate immunity. PLoS Pathog. 5:e1000526. doi: 10.1371/journal.ppat.1000526
Moulin, M., McCormac, A. C., Terry, M. J., and Smith, A. G. (2008). Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. U.S.A. 105, 15178–15183. doi: 10.1073/pnas.0803054105
Mullet, J. (2003). Chloroplast development and gene expression. Ann. Rev. Plant Physiol. Plant Mol. Biol. 39, 475–502. doi: 10.1146/annurev.pp.39.060188.002355
Mullineaux, P. M., Karpinski, S., and Baker, N. R. (2006). Spatial dependence for hydrogen peroxide-directed signaling in light-stressed plants. Plant Physiol. 141, 346–350. doi: 10.1104/pp.106.078162
Myouga, F., Hosoda, C., Umezawa, T., Iizumi, H., Kuromori, T., Motohashi, R., et al. (2008). A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20, 3148–3162. doi: 10.1105/tpc.108.061341
Nagai, S., Koide, M., Takahashi, S., Kikuta, A., Aono, M., Sasaki-Sekimoto, Y., et al. (2007). Induction of isoforms of tetrapyrrole biosynthetic enzymes, AtHEMA2 and AtFC1, under stress conditions and their physiological functions in Arabidopsis. Plant Physiol. 144, 1039–1051. doi: 10.1104/pp.107.100065
Nakamura, H., Muramatsu, M., Hakata, M., Ueno, O., Nagamura, Y., Hirochika, H., et al. (2009). Ectopic overexpression of the transcription factor OsGLK1 induces chloroplast development in non-green rice cells. Plant Cell Physiol. 50, 1933–1949. doi: 10.1093/pcp/pcp138
Noordally Zeenat, B., Ishii, K., Atkins Kelly, A., Wetherill Sarah, J., Kusakina, J., Walton Eleanor, J., et al. (2013). Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science 339, 1316–1319. doi: 10.1126/science.1230397
Nott, A., Jung, H. S., Koussevitzky, S., and Chory, J. (2006). Plastid-to-nucleus retrograde signaling. Ann. Rev. Plant Biol. 57, 739–759. doi: 10.1146/annurev.arplant.57.032905.105310
Ono, S., Suzuki, S., Ito, D., Tagawa, S., Shiina, T., and Masuda, S. (2021). Plastidial (p)ppGpp synthesis by the Ca2+-dependent RelA-SpoT homolog regulates the adaptation of chloroplast gene expression to darkness in Arabidopsis. Plant Cell Physiol. 61, 2077–2086. doi: 10.1093/pcp/pcaa124
Op den Camp, R. G. L., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., et al. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis[W]. Plant Cell 15, 2320–2332. doi: 10.1105/tpc.014662
Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. doi: 10.1101/gad.11.22.2983
Palmer, J. D. (1985). Comparative organization of chloroplast genomes. Ann. Rev. Genet. 19, 325–354. doi: 10.1146/annurev.ge.19.120185.001545
Patel, M., and Berry, J. O. (2008). Rubisco gene expression in C4 plants. J. Exp. Bot. 59, 1625–1634. doi: 10.1093/jxb/erm368
Paul, M. J., and Foyer, C. H. (2001). Sink regulation of photosynthesis. J. Exp. Bot. 52, 1383–1400. doi: 10.1093/jexbot/52.360.1383
Pedmale, U. V., Huang, S. C., Zander, M., Benjamin, J., Hetzel, J., Ljung, K., et al. (2016). Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell 164, 233–245. doi: 10.1016/j.cell.2015.12.018
Pesaresi, P., Hertle, A., Pribil, M., Kleine, T., Wagner, R., Strissel, H., et al. (2009). Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell 21, 2402–2423. doi: 10.1105/tpc.108.064964
Pesaresi, P., Masiero, S., Eubel, H., Braun, H.-P., Bhushan, S., Glaser, E., et al. (2006). Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell 18, 970–991. doi: 10.1105/tpc.105.039073
Pesaresi, P., Pribil, M., Wunder, T., and Leister, D. (2011). Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: The roles of STN7, STN8 and TAP38. Biochim. Biophys. Acta (BBA) Bioenerg. 1807, 887–896. doi: 10.1016/j.bbabio.2010.08.002
Pesaresi, P., Schneider, A., Kleine, T., and Leister, D. (2007). Interorganellar communication. Curr. Opin. Plant Biol. 10, 600–606. doi: 10.1016/j.pbi.2007.07.007
Pfalz, J., Holtzegel, U., Barkan, A., Weisheit, W., Mittag, M., and Pfannschmidt, T. (2015). ZmpTAC12 binds single-stranded nucleic acids and is essential for accumulation of the plastid-encoded polymerase complex in maize. New Phytol. 206, 1024–1037. doi: 10.1111/nph.13248
Pfalz, J., Liere, K., Kandlbinder, A., Dietz, K.-J., and Oelmüller, R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18, 176–197. doi: 10.1105/tpc.105.036392
Pfannschmidt, T. (2010). Plastidial retrograde signalling – a true “plastid factor” or just metabolite signatures? Trends Plant Sci. 15, 427–435. doi: 10.1016/j.tplants.2010.05.009
Pfannschmidt, T., and Liere, K. (2005). Redox regulation and modification of proteins controlling chloroplast gene expression. Antiox. Redox Sign. 7, 607–618. doi: 10.1089/ars.2005.7.607
Pfannschmidt, T., Schütze, K., Brost, M., and Oelmüller, R. (2001). A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment*. J. Biol. Chem. 276, 36125–36130. doi: 10.1074/jbc.M105701200
Pham, V. N., Kathare, P. K., and Huq, E. (2017). Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038. doi: 10.1104/pp.17.01384
Pham, V. N., Kathare, P. K., and Huq, E. (2018). Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038.
Piippo, M., Allahverdiyeva, Y., Paakkarinen, V., Suoranta, U.-M., Battchikova, N., and Aro, E.-M. (2006). Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol. Genom. 25, 142–152. doi: 10.1152/physiolgenomics.00256.2005
Pogson, B. J., Ganguly, D., and Albrecht-Borth, V. (2015). Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 1847, 1017–1024. doi: 10.1016/j.bbabio.2015.02.003
Possingham, J. V. (1980). Plastid replication and development in the life cycle of higher plants. Ann. Rev. Plant Physiol. 31, 113–129. doi: 10.1146/annurev.pp.31.060180.000553
Prabha, R., Singh, D. P., Somvanshi, P., and Rai, A. (2016). Functional profiling of cyanobacterial genomes and its role in ecological adaptations. Geno. Data 9, 89–94. doi: 10.1016/j.gdata.2016.06.005
Pruzinská, A., Anders, I., Aubry, S., Schenk, N., Tapernoux-Lüthi, E., Müller, T., et al. (2007). In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19, 369–387. doi: 10.1105/tpc.106.044404
Qing-Bo, Y. (2013). TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana. Physiol. Plant. 148, 408–421.
Qiu, Y., Li, M., Pasoreck, E. K., Long, L., Shi, Y., Galvão, R. M., et al. (2015). Hemera couples the proteolysis and transcriptional activity of phytochrome interacting factors in Arabidopsis photomorphogenesis. Plant Cell 27, 1409–1427. doi: 10.1105/tpc.114.136093
Raghavendra, A. S., and Padmasree, K. (2003). Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8, 546–553. doi: 10.1016/j.tplants.2003.09.015
Ramel, F., Birtic, S., Ginies, C., Soubigou-Taconnat, L., Triantaphylidès, C., and Havaux, M. (2012). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5535–5540. doi: 10.1073/pnas.1115982109
Reyes-Prieto, A., Weber, A. P., and Bhattacharya, D. (2007). The origin and establishment of the plastid in algae and plants. Ann. Rev. Genet. 41, 147–168. doi: 10.1146/annurev.genet.41.110306.130134
Rissler, H. M., Collakova, E., DellaPenna, D., Whelan, J., and Pogson, B. J. (2002). Chlorophyll biosynthesis. expression of a second chl I gene of magnesium chelatase in Arabidopsis supports only limited chlorophyll synthesis. Plant Physiol. 128, 770–779. doi: 10.1104/pp.010625
Rochaix, J. D. (2013). Redox regulation of thylakoid protein kinases and photosynthetic gene expression. Antiox. Redox Sign. 18, 2184–2201. doi: 10.1089/ars.2012.5110
Rodermel, S., and Park, S. (2003). Pathways of intracellular communication: tetrapyrroles and plastid-to-nucleus signaling. Bioessays 25, 631–636. doi: 10.1002/bies.10308.
Romand, S., Abdelkefi, H., Lecampion, C., Belaroussi, M., Dussenne, M., Ksas, B., et al. (2022). A guanosine tetraphosphate (ppGpp) mediated brake on photosynthesis is required for acclimation to nitrogen limitation in Arabidopsis. Elife 11:75041. doi: 10.7554/eLife.75041
Rook, F., Hadingham, S. A., Li, Y., and Bevan, M. W. (2006). Sugar and ABA response pathways and the control of gene expression. Plant Cell Environ. 29, 426–434. doi: 10.1111/j.1365-3040.2005.01477.x
Ruckle, M. E., Burgoon, L. D., Lawrence, L. A., Sinkler, C. A., and Larkin, R. M. (2012). Plastids are major regulators of light signaling in Arabidopsis. Plant Physiol. 159, 366–390. doi: 10.1104/pp.112.193599
Sakamoto, W., Miyagishima, S. Y., and Jarvis, P. (2008). Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. Arabid. Book 6:e0110. doi: 10.1199/tab.0110
Schröter, Y., Steiner, S., Matthäi, K., and Pfannschmidt, T. (2010). Analysis of oligomeric protein complexes in the chloroplast sub-proteome of nucleic acid-binding proteins from mustard reveals potential redox regulators of plastid gene expression. Proteomics 10, 2191–2204. doi: 10.1002/pmic.200900678
Schuhmann, H., and Adamska, I. (2012). Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiol. Plant 145, 224–234. doi: 10.1111/j.1399-3054.2011.01533.x
Schwechheimer, C., Schröder, P. M., and Blaby-Haas, C. E. (2022). Plant GATA factors: their biology, phylogeny, and phylogenomics. Ann. Rev. Plant Biol. 73, 123–148. doi: 10.1146/annurev-arplant-072221-092913.
Schwenkert, S., Fernie, A. R., Geigenberger, P., Leister, D., Möhlmann, T., Naranjo, B., et al. (2022). Chloroplasts are key players to cope with light and temperature stress. Trends Plant Sci. 27, 577–587. doi: 10.1016/j.tplants.2021.12.004
Seo, P. J., Kim, S.-G., and Park, C.-M. (2008). Membrane-bound transcription factors in plants. Trends Plant Sci. 13, 550–556. doi: 10.1016/j.tplants.2008.06.008
Shao, N., Krieger-Liszkay, A., Schroda, M., and Beck, C. F. (2007). A reporter system for the individual detection of hydrogen peroxide and singlet oxygen: its use for the assay of reactive oxygen species produced in vivo. Plant J. 50, 475–487. doi: 10.1111/j.1365-313X.2007.03065.x
Sherameti, I., Sopory, S. K., Trebicka, A., Pfannschmidt, T., and Oelmüller, R. (2002). Photosynthetic electron transport determines nitrate reductase gene expression and activity in higher plants*. J. Biol. Chem. 277, 46594–46600. doi: 10.1074/jbc.M202924200
Shimizu, M., Kato, H., Ogawa, T., Kurachi, A., Nakagawa, Y., and Kobayashi, H. (2010). Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc. Natl. Acad. Sci. U.S.A. 107, 10760–10764. doi: 10.1073/pnas.0911692107
Shimizu, T., and Masuda, T. (2021). The role of tetrapyrrole- and GUN1-dependent signaling on chloroplast biogenesis. Plants 10:196. doi: 10.3390/plants10020196
Shin, J., Kim, K., Kang, H., Zulfugarov, I. S., Bae, G., Lee, C. H., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665. doi: 10.1073/pnas.0812219106
Simková, K., Moreau, F., Pawlak, P., Vriet, C., Baruah, A., Alexandre, C., et al. (2012). Integration of stress-related and reactive oxygen species-mediated signals by topoisomerase VI in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109, 16360–16365. doi: 10.1073/pnas.1202041109
Steiner, S., Dietzel, L., Schröter, Y., Fey, V., Wagner, R., and Pfannschmidt, T. (2009). The role of phosphorylation in redox regulation of photosynthesis genes psaA and psbA during photosynthetic acclimation of mustard. Mol. Plant 2, 416–429. doi: 10.1093/mp/ssp007
Steiner, S., Schröter, Y., Pfalz, J., and Pfannschmidt, T. (2011). Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 157, 1043–1055. doi: 10.1104/pp.111.184515
Stephenson, P. G., Fankhauser, C., and Terry, M. J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. U.S.A. 106, 7654–7659.
Stern, D. B., Goldschmidt-Clermont, M., and Hanson, M. R. (2010). Chloroplast RNA metabolism. Ann. Rev. Plant Biol. 61, 125–155. doi: 10.1146/annurev-arplant-042809-112242
Strand, Å, Asami, T., Alonso, J., Ecker, J. R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83. doi: 10.1038/nature01204
Sugliani, M., Abdelkefi, H., Ke, H., Bouveret, E., Robaglia, C., Caffarri, S., et al. (2016). An ancient bacterial signaling pathway regulates chloroplast function to influence growth and development in Arabidopsis. Plant Cell 28, 661–679. doi: 10.1105/tpc.16.00045
Sullivan, J. A., and Gray, J. C. (1999). Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11, 901–910. doi: 10.1105/tpc.11.5.901
Sun, X., Jia, H.-L., Xiao, C.-L., Yin, X.-F., Yang, X.-Y., Lu, J., et al. (2011). Bacterial proteome of Streptococcus pneumoniae through multidimensional separations coupled with LC-MS/MS. OMICS J. Int. Biol. 15, 477–482. doi: 10.1089/omi.2010.0113
Surpin, M., and Chory, J. (1997). The co-ordination of nuclear and organellar genome expression in eukaryotic cells. Essays Biochem. 32, 113–125.
Surpin, M., Larkin, R. M., and Chory, J. (2002). Signal transduction between the chloroplast and the nucleus. Plant Cell 14, S327–S338. doi: 10.1105/tpc.010446
Susek, R. E., Ausubel, F. M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. doi: 10.1016/0092-8674(93)90459-4.
Tanaka, R., Kobayashi, K., and Masuda, T. (2011). Tetrapyrrole metabolism in Arabidopsis thaliana. Arabid. Book 9:e0145. doi: 10.1199/tab.0145
Tanaka, R., and Tanaka, A. (2007). Tetrapyrrole biosynthesis in higher plants. Ann. Rev. Plant Biol. 58, 321–346. doi: 10.1146/annurev.arplant.57.032905.105448.
Tano, D. W., and Woodson, J. D. (2022). Putting the brakes on chloroplast stress signaling. Mol. Plant 15, 388–390. doi: 10.1016/j.molp.2022.02.009
Terry, M. J., Linley, P. J., and Kohchi, T. (2002). Making light of it: the role of plant haem oxygenases in phytochrome chromophore synthesis. Biochem. Soc. Trans. 30, 604–609.
Toledo-Ortiz, G., Johansson, H., Lee, K. P., Bou-Torrent, J., Stewart, K., Steel, G., et al. (2014). The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 10:e1004416. doi: 10.1371/journal.pgen.1004416
Toyokuni, S., Kong, Y., and Mi, D. (2022). Commentary for an article on photooxidation in isolated chloroplasts. Arch. Biochem. Biophys. 2022:109133. doi: 10.1016/j.abb.2022.109133
Trösch, R., Ries, F., Westrich, L. D., Gao, Y., Herkt, C., Hoppstädter, J., et al. (2022). Fast and global reorganization of the chloroplast protein biogenesis network during heat acclimation. Plant Cell 34, 1075–1099. doi: 10.1093/plcell/koab317
Tyagi, A. K., and Gaur, T. (2003). Light regulation of nuclear photosynthetic genes in higher plants. Crit. Rev. Plant Sci. 22, 417–452. doi: 10.1080/07352680390243503
Van Buskirk, E. K., Decker, P. V., and Chen, M. (2012). Photobodies in light signaling. Plant Physiol. 158, 52–60. doi: 10.1104/pp.111.186411
Van Buskirk, E. K., Reddy, A. K., Nagatani, A., and Chen, M. (2014). Photobody localization of phytochrome B is tightly correlated with prolonged and light-dependent inhibition of hypocotyl elongation in the dark. Plant Physiol. 165, 595–607. doi: 10.1104/pp.114.236661
Vasileuskaya, Z., Oster, U., and Beck, C. F. (2004). Involvement of tetrapyrroles in inter-organellar signaling in plants and algae. Photosynth. Res. 82, 289–299. doi: 10.1007/s11120-004-2160-x
Vinti, G., Hills, A., Campbell, S., Bowyer, J. R., Mochizuki, N., Chory, J., et al. (2000). Interactions between hy1 and gun mutants of Arabidopsis, and their implications for plastid/nuclear signalling. Plant J. 24, 883–894. doi: 10.1111/j.1365-313X.2000.00936.x
von Gromoff, E. D., Alawady, A., Meinecke, L., Grimm, B., and Beck, C. F. (2008). Heme, a plastid-derived regulator of nuclear gene expression in chlamydomonas. Plant Cell 20, 552–567. doi: 10.1105/tpc.107.054650
von Gromoff, E. D., Schroda, M., Oster, U., and Beck, C. F. (2006). Identification of a plastid response element that acts as an enhancer within the chlamydomonas HSP70A promoter. Nucleic Acids Res. 34, 4767–4779. doi: 10.1093/nar/gkl602
Voss, I., Goss, T., Murozuka, E., Altmann, B., McLean, K. J., Rigby, S. E. J., et al. (2011). FdC1, a novel ferredoxin protein capable of alternative electron partitioning, increases in conditions of acceptor limitation at photosystem I*. J. Biol. Chem. 286, 50–59. doi: 10.1074/jbc.M110.161562
Wagner, D., Przybyla, D., Op den Camp, R., Kim, C., Landgraf, F., Lee, K. P., et al. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306, 1183–1185. doi: 10.1126/science.1103178
Waters, M. T., and Langdale, J. A. (2009). The making of a chloroplast. EMBO J. 28, 2861–2873. doi: 10.1038/emboj.2009.264
Waters, M. T., Moylan, E. C., Langdale, J. A. J., and Biology, M. (2008). GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 56, 432–444. doi: 10.1111/j.1365-313X.2008.03616.x
Waters, M. T., Wang, P., Korkaric, M., Capper, R. G., Saunders, N. J., and Langdale, J. A. (2009). GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128. doi: 10.1105/tpc.108.065250
Weissmann, S., and Brutnell, T. P. (2012). Engineering C4 photosynthetic regulatory networks. Curr. Opin. Biotechnol. 23, 298–304. doi: 10.1016/j.copbio.2011.12.018
Wietrzynski, W., Traverso, E., Wollman, F.-A., and Wostrikoff, K. (2021). The state of oligomerization of rubisco controls the rate of synthesis of the raaubisco large subunit in Chlamydomonas reinhardtii. Plant Cell 33, 1706–1727. doi: 10.1093/plcell/koab061
Wimmelbacher, M., and Börnke, F. (2014). Redox activity of thioredoxin z and fructokinase-like protein 1 is dispensable for autotrophic growth of Arabidopsis thaliana. J. Exp. Bot. 65, 2405–2413. doi: 10.1093/jxb/eru122
Woodson, J. D., and Chory, J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395. doi: 10.1038/nrg2348
Woodson, J. D., Perez-Ruiz, J. M., Schmitz, R. J., Ecker, J. R., and Chory, J. (2013). Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 73, 1–13. doi: 10.1111/tpj.12011
Wostrikoff, K., Girard-Bascou, J., Wollman, F.-A., and Choquet, Y. (2004). Biogenesis of PSI involves a cascade of translational autoregulation in the chloroplast of Chlamydomonas. EMBO J. 23, 2696–2705. doi: 10.1038/sj.emboj.7600266
Wostrikoff, K., and Stern, D. (2007). Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 104, 6466–6471. doi: 10.1073/pnas.0610586104
Wu, G.-Z., Meyer, E. H., Richter, A. S., Schuster, M., Ling, Q., Schöttler, M. A., et al. (2019). Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants 5, 525–538. doi: 10.1038/s41477-019-0415-y
Xiao, Y., Savchenko, T., Baidoo, E. E., Chehab, W. E., Hayden, D. M., Tolstikov, V., et al. (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149, 1525–1535. doi: 10.1016/j.cell.2012.04.038
Xu, D. (2020). COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol. 228, 1748–1753. doi: 10.1111/nph.16296
Xu, X., Wang, Q., Li, W., Hu, T., Wang, Q., Yin, Y., et al. (2022). Overexpression of SlBBX17 affects plant growth and enhances heat tolerance in tomato. Int. J. Biol. Macromol. 206, 799–811. doi: 10.1016/j.ijbiomac.2022.03.080
Yasumura, Y., Moylan, E. C., and Langdale, J. A. (2005). A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell 17, 1894–1907. doi: 10.1105/tpc.105.033191
Yin, G., Wang, Y., Xiao, Y., Yang, J., Wang, R., Jiang, Y., et al. (2022). Relationships between leaf color changes, pigment levels, enzyme activity, photosynthetic fluorescence characteristics and chloroplast ultrastructure of Liquidambar formosana hance. J. Forest. Res. doi: 10.1007/s11676-021-01441-6.
Yu, Q.-B., Li, G., Wang, G., Sun, J.-C., Wang, P.-C., Wang, C., et al. (2008). Construction of a chloroplast protein interaction network and functional mining of photosynthetic proteins in Arabidopsis thaliana. Cell Res. 18, 1007–1019. doi: 10.1038/cr.2008.286
Yu, Q. B., Lu, Y., Ma, Q., Zhao, T. T., Huang, C., Zhao, H. F., et al. (2013). TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana. Physiol. Plant 148, 408–421. doi: 10.1111/j.1399-3054.2012.01718.x
Zhang, W., Willows, R. D., Deng, R., Li, Z., Li, M., Wang, Y., et al. (2021). Bilin-dependent regulation of chlorophyll biosynthesis by GUN4. Proc. Natl. Acad. Sci. U.S.A. 118:20. doi: 10.1073/pnas.2104443118
Zhang, Z.-W., Yuan, S., Feng, H., Xu, F., Cheng, J., Shang, J., et al. (2011). Transient accumulation of Mg-protoporphyrin IX regulates expression of PhANGs – new evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. J. Plant Physiol. 168, 714–721. doi: 10.1016/j.jplph.2010.10.016
Keywords: chloroplast, gene expression, photosynthesis, plastid, retrograde signaling, signal transduction, transcription factor, tetrapyrrole biosynthesis
Citation: Jan M, Liu Z, Rochaix J-D and Sun X (2022) Retrograde and anterograde signaling in the crosstalk between chloroplast and nucleus. Front. Plant Sci. 13:980237. doi: 10.3389/fpls.2022.980237
Received: 28 June 2022; Accepted: 18 August 2022;
Published: 02 September 2022.
Edited by:
Deqiang Duanmu, Huazhong Agricultural University, ChinaReviewed by:
Shinji Masuda, Tokyo Institute of Technology, JapanLiangsheng Wang, China Agricultural University, China
Copyright © 2022 Jan, Liu, Rochaix and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuwu Sun, c3VueHV3dXNzZEBzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Masood Jan
Masood Jan Zhixin Liu1†
Zhixin Liu1† Jean-David Rochaix
Jean-David Rochaix Xuwu Sun
Xuwu Sun