
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 10 August 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.979801
This article is part of the Research Topic Multi-omics and Computational Biology in Horticultural Plants: From Genotype to Phenotype View all 32 articles
 Hongman Wang1,2
Hongman Wang1,2 Zuqi Dong1,2,3
Zuqi Dong1,2,3 Jianbing Chen4
Jianbing Chen4 Meng Wang4
Meng Wang4 Yuting Ding4
Yuting Ding4 Qingyun Xue1,2
Qingyun Xue1,2 Wei Liu1,2
Wei Liu1,2 Zhitao Niu1,2*
Zhitao Niu1,2* Xiaoyu Ding1,2*
Xiaoyu Ding1,2*Dendrobium officinale, an important orchid plant with great horticultural and medicinal values, frequently suffers from abiotic or biotic stresses in the wild, which may influence its well-growth. Heat shock proteins (Hsps) play essential roles in the abiotic stress response of plants. However, they have not been systematically investigated in D. officinale. Here, we identified 37 Hsp20 genes (DenHsp20s), 43 Hsp70 genes (DenHsp70s) and 4 Hsp90 genes (DenHsp90s) in D. officinale genome. These genes were classified into 8, 4 and 2 subfamilies based on phylogenetic analysis and subcellular predication, respectively. Sequence analysis showed that the same subfamily members have relatively conserved gene structures and similar protein motifs. Moreover, we identified 33 pairs of paralogs containing 30 pairs of tandem duplicates and 3 pairs of segmental duplicates among these genes. There were 7 pairs in DenHsp70s under positive selection, which may have important functions in helping cells withstand extreme stress. Numerous gene promoter sequences contained stress and hormone response cis-elements, especially light and MeJA response elements. Under MeJA stress, DenHsp20s, DenHsp70s and DenHsp90s responded to varying degrees, among which DenHsp20-5,6,7,16 extremely up-regulated, which may have a strong stress resistance. Therefore, these findings could provide useful information for evolutional and functional investigations of Hsp20, Hsp70 and Hsp90 genes in D. officinale.
Plants are commonly exposed to biotic and abiotic stresses, e.g., drought, heat, and various pathogens, which could cause adverse effects on their growth and development (Zhang et al., 2022). To overcome these difficulties, plants have evolved their own “stress tolerance system,” which attracted intense attention from botanists, leading to numerous functional genomic studies related to plant stress tolerance published, particularly in temperature stresses (Dangi et al., 2018). For example, Heat shock proteins (Hsps) help newly synthesized proteins to fold, or to protect proteins that might misfold and thereby lose their potential functional conformation during a heat stress event. However, stresses in nature rarely come alone. Heat stress is commonly associated with high light and drought, but also promotes the spreading of pathogens and pests, leading to serious harm to plants (Jacob et al., 2017). Thus, Hsps, which were proved to be involved in multiple stress resistance, especially in abiotic stresses, have an important effect on thermomorphogenesis (Lin et al., 2022). Heat shock proteins, originally only described in relation to heat shock (Ritossa, 1962), were actually induced by a wide variety of stresses, including exposure to cold, osmotic, drought, salt, UV, high light, wound healing, tissue remodeling, or biotic stresses (Lindquist and Craig, 1988; Vierling, 1991; Boston et al., 1996). Therefore, Hsps play a great role in alleviating the injury caused by stresses.
According to previous studies, Hsps can be grouped into six families including Hsp100s/ClpB, Hsp90s, Hsp70s/DnaK, Hsp60s, Hsp40s/DnaJ and Hsp20s based on their molecular weight and sequence homology (Wang et al., 2004; Waters, 2013). Among them, three gene families of Hsp20, Hsp70 and Hsp90 are the most important in plants. Hsp20, the most variable and diversified family in plants, was considered the most produced protein under heat stress conditions in many higher plants (Vierling, 2003; Charng et al., 2006; Basha et al., 2012). Hsp70, the most abundant heat shock proteins in eukaryotic cells, was expressed and accumulated under many stresses, such as drought stress (Cho and Choi, 2009), high-salinity stress (Wang et al., 2008), heavy metal stress (Tukaj et al., 2011) and virus stress (Aparicio et al., 2005), suggesting that Hsp70 can increase the resistance of plants to various biotic or abiotic stresses. Unlike Hsp20 and Hsp70 genes, which mainly play a role in plant growth and development and response to environmental stress, Hsp90s play essential roles in plant immunity (Shirasu, 2009; Kadota and Shirasu, 2012). For example, Wang et al. (2011) have found that wheat plants over-expressing the TaHsp90.2 and TaHsp90.3 from Hsp90 genes showed significant resistance to stripe rust. In rice, a chaperone complex consisting of cytosolic Hsp90 and its co-chaperone Hop/Sti1 participates in chitin responses and anti-fungal immunity (Chen et al., 2010).
In recent years, as more plant genomes have been assembled and reported, the Hsp20, Hsp70 and Hsp90 gene family were identified across many plant species, such as Arabidopsis thaliana (Krishna and Gloor, 2001; Lin et al., 2001; Scharf et al., 2001), Oryza sativa (Ouyang et al., 2009; Sarkar et al., 2012; Xu et al., 2012) and Triticum aestivum (Muthusamy et al., 2017; Lu et al., 2020; Lai et al., 2021). Dendrobium officinale Kimura et Migo, an endangered orchid endemic to China, has great horticultural and medicinal values (Niu et al., 2018; Zhu et al., 2018; Li et al., 2020). It grows in adverse conditions, e.g., epiphytic on cliffs or tree trunks, and distributed at high altitudes above 1,200 m. Moreover, D. officinale is susceptible to pests and diseases such as anthracnose, blackspot and phytophthora mealybug, which may have a negative influence on its well-growth. However, even in such harsh habitats, D. officinale still can grow well and accumulate important medicinal substances. Therefore, it is important to make clear the mechanism of its stress resistance, especially the response of Hsp genes to adversity. However, to date, the evolution and functions of Hsp20, Hsp70 and Hsp90 gene family in D. officinale still remains unclear. With the availability of chromosome-level genome sequence of D. officinale (Niu et al., 2020), it is now possible to conduct full study the three Hsp gene families in D. officinale.
Here, we used bioinformatics methods to identify Hsp20, Hsp70 and Hsp90 genes from D. officinale genome and uncover their sequence features, chromosomal positions, phylogenetic relationships, gene duplication events and syntenic analysis. Moreover, cis-elements, expression profiles, the protein three-dimensional (3D) structures and their protein–protein interaction (PPI) networks were all predicted to explore the possible biological functions of Hsp genes. The results would provide valuable information for further investigations of the Hsp20, Hsp70 and Hsp90 gene family in D. officinale.
The Hidden Markov models (HMMs) profiles of Hsp20 (PF00011), Hsp40 (PF00226), Hsp70 (PF00012) and Hsp90 (PF00183), were downloaded from the protein family database (Pfam).1 This analysis was used for the search to recognizing candidate proteins with an E-value of 1e-5 by HMMER v3.2.1. Meantime, Hsp20, Hsp40, Hsp60, Hsp70, Hsp90 and Hsp100 protein sequences of A. thaliana and O. sativa from Scharf (Krishna and Gloor, 2001; Lin et al., 2001; Scharf et al., 2001), Ouyang (Ouyang et al., 2009; Sarkar et al., 2012; Xu et al., 2012) and (Ratheesh et al., 2012)2 were used as queries to search for D. officinale proteins using BLASTP. Ultimately, the protein sequences were integrated by both the above methods. Here, we chose Hsp20, Hsp70 and Hsp90 gene family in D. officinale for further analysis. So, the output putative DenHsp20s, DenHsp70s and DenHsp90s were submitted to Pfam (see Footnote 1), NCBI-CDD3 and SMART4 to confirm conserved domains. In addition, the protein molecular weight (kDa), aliphatic index (AI), theoretical isoelectric point (pI) and grand average of hydropathicity (GRAVY) were estimated with ExPASy software (Wilkins et al., 1999). The chromosome locations were analyzed and displayed by TBtools v1.6 (Chen et al., 2020).
Multiple sequence alignments of Hsp20, Hsp70 and Hsp90 full-length amino acid sequences derived from D. officinale, A. thaliana and O. sativa were performed with MAFFT v7.487 software (Katoh and Dtandley, 2013). And the maximum likelihood phylogenetic trees were constructed by RAxML v1.3 with a bootstrap value of 1,000. The conserved motifs were determined by MEME,5 with default parameters, except that the number of motifs of 20 was specified. Additionally, the exon-intron structure of each sequence was displayed by GSDS software online.6 The different characteristic domains of the three gene families were aligned with previous studies (Krishna and Gloor, 2001; Lin et al., 2001; Scharf et al., 2001) and visualized by DNAMAN_9 software.
Firstly, the genomic DNA sequences of the Hsp20, Hsp70 and Hsp90 genes of D. officinale were aligned using BLASTN with an E-value of 1e-20. Then the gene duplications were identified with MCScanX using BLASTN results. The duplication events of these genes were visualized with Circos (Krzywinski et al., 2009). Besides, the syntenic blocks between D. officinale and other plant genomes were detected and displayed with MCScanX (cscore≥0.7). KaKs_Calculator 2.0 (Wang et al., 2010) was used to estimate Non-synonymous (Ka), synonymous (Ks) and Ka/Ks ratios. Commonly, Ka/Ks > 1 indicates positive selection, Ka/Ks < 1 indicates negative selection, and Ka/Ks = 1 indicates neutral selection (Wang et al., 2010).
The upstream 1,500 bp genomic DNA sequences of Hsp20, Hsp70 and Hsp90 genes were extracted as putative promoters. Then they were submitted to online PlantCare database7 to analyze the putative cis-elements. FIMO (Noble, 2011) in MEME software toolkit was used to predict heat shock responsive elements (HSEs) using sequence module nTTCnnGAAnnTTCn or nGAAnnTTCnnGAAn (Sarkar et al., 2009; Lopes-Caitar et al., 2013). Total cis-elements in promoter sequences were visualized by TBtools v1.6 software (Chen et al., 2020).
To analyze the expression patterns of these identified genes in D. officinale, we searched the NCBI SRA database8 for RNA-sequence data from four different tissues (root, stem, leaf, and flower) with the accession IDs SRR2014227, SRR2014230, SRR2014236, SRR2014246, SRR2014297, SRR2014325, SRR2014396, and SRR2014476 (Chen et al., 2017). Firstly, the download RNA-sequence data were converted to fastq format via fastq-dump of SRA toolkit.3.0.0. Then the clean reads were aligned to the D. officinale genome, and mapping by Hisat2 v2.2.1. And the data were sam to bam by SAMtools v1.14. The FPKM value of DenHsp20s, DenHsp70s and DenHsp90s were calculated by StringTie v2.2.0 to estimate the transcript abundances. The heat map was constructed by the pheatmap package in RStudio v1.4.1717 to visualize the expression.
Total RNA was extracted by an EASY spin Plant RNA Kit (Aidlab, China). First–strand cDNAs were synthesized using HiScript® III–RT SuperMix for qPCR (Vazyme, China). Primers (Supplementary Table S6) were designed using Snapgene software. qRT-PCR was performed using ABI-7500 Connect Real-Time PCR Detection System. cDNAs were diluted to 200 ng with 1 μl template in a reaction volume of 20 μl, run in three technical replicates. PCR amplification programs were used as follows: 95°C for 30 s followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 60°C for 15 s. The expression data were calculated by the 2-∆∆CT method (Livak and Schmittgen, 2002).
The tertiary structures of Hsp20, Hsp70 and Hsp90 proteins in D. officinale were predicted with SWISS-MODEL.9 The Hsp20, Hsp70 and Hsp90 protein sequences were aligned to STRING database10 online to predict the relationships. Cytoscape v3.7.2 software (Shannon et al., 2003) was used to visualize the regulatory networks.
A total of 165 Hsp genes, with 37 Hsp20 genes, 70 Hsp40 genes, 7 Hsp60 genes, 43 Hsp70 genes, 4 Hsp90 genes and 4 Hsp100 genes, were identified from D. officinale genome sequence using HMMER and BLASTP method. Here, we chose Hsp20, Hsp70 and Hsp90 gene family in D. officinale for further analysis (Table 1; Supplementary Table S1). The characters among the three kinds of Hsp genes were variable. For example, Hsp70 proteins contains more variable amino acid numbers (from 45 of DenHsp70-21 to 657 of DenHsp70-8) than Hsp20 (from 73 of DenHsp20-14 to 294 of DenHsp20-10) and Hsp90 (from 428 of DenHsp90-4 to 510 of DenHsp90-3). The protein molecular weight (MW) of Hsp70 ranged from 5.1 kDa (DenHsp70-14) to 73.4 kDa (DenHsp70-8), higher than Hsp20 (from 8.4 kDa of DenHsp20-14 to 33.3 kDa of DenHsp20-10) and Hsp90 (from 49.4 kDa of DenHsp90-4 to 59.3 kDa of DenHsp90-3). In contrast, the isoelectric point (PI) of Hsp90 (from 78.98 of DenHsp90-2 to 84.95 of DenHsp90-4) was higher than that of Hsp70 (from 4.4 of DenHsp70-9 to 8.99 of DenHsp70-25) and Hsp20 (from 4.92 of DenHsp20-12 to 9.34 of DenHsp20-11; Table 1; Supplementary Table S1).
Among the 19 assembled chromosomes of D. officinale, DenHsp20s, DenHsp70s and DenHsp90s were distributed on 13 chromosomes, 13 chromosomes and 4 chromosomes, respectively. As shown in Figure 1, most Hsp20 genes were located on Chr2 and Chr11. Chr12 contained the most Hsp70 genes, although it was not the longest chromosome. Gene clusters could be observed on Chr2, Chr11 and Chr12. Remarkably, most Hsp genes belonging to the same subfamilies were mapped on the same chromosome. This shows that tandem duplication events played an important role during the expansion of the Hsp family in D. officinale.
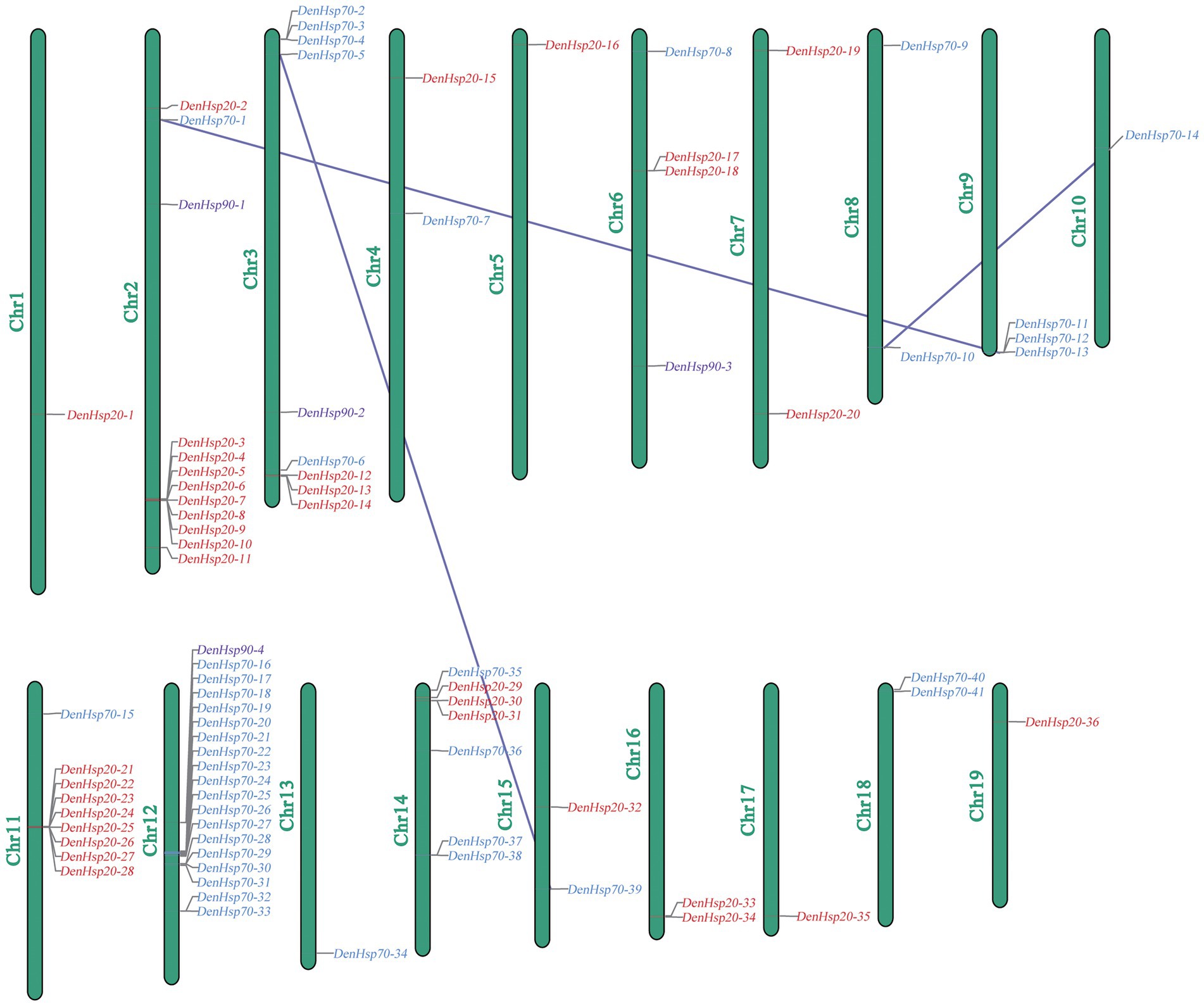
Figure 1. Chromosomal distributions of the identified Hsp20, Hsp70 and Hsp90 genes in Dendrobium officinale. Red, blue, purple colors represent Hsp20, Hsp70 and Hsp90 genes, respectively. Three lines represent three tandem duplicate pairs.
To explore the evolutionary relations of Hsp genes of D. officinale, phylogenetic trees of Hsp20, Hsp70 and Hsp90 genes were constructed, respectively, with Hsp genes from A. thaliana and O. sativa as outgroups (Figure 2).
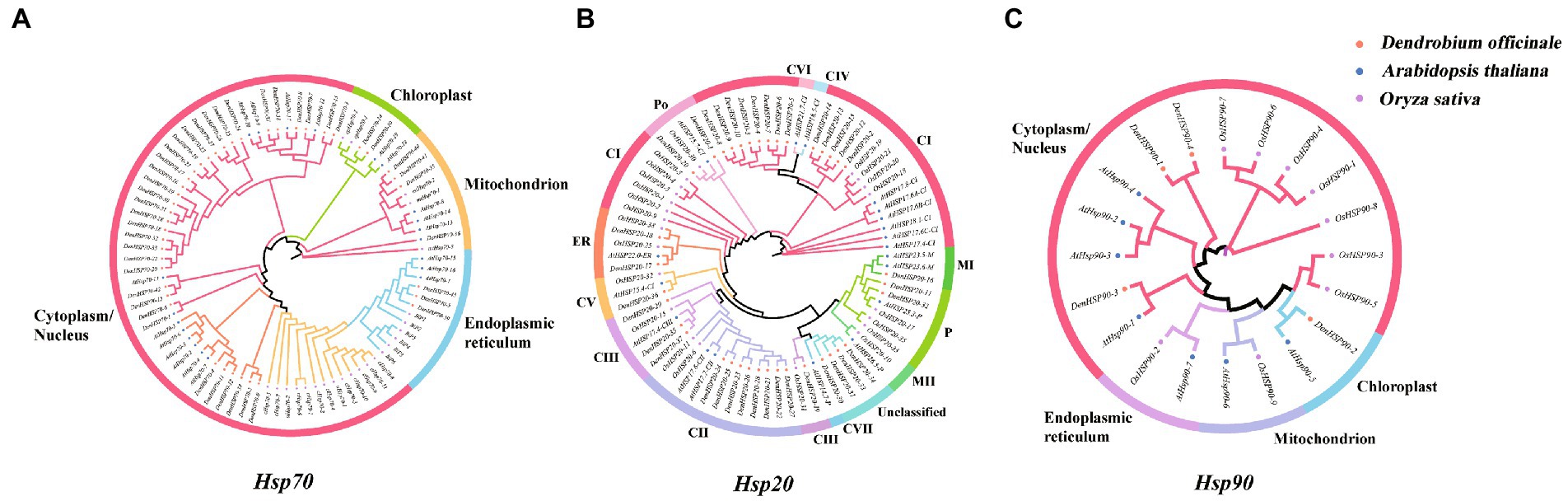
Figure 2. (A–C) represent the phylogenetic relationships of Hsp20, Hsp70 and Hsp90 genes in D. officinale, A. thaliana and O. sativa, respectively. Three maximum likelihood phylogenetic trees were constructed by RAxML with 1000 bootstraps. Orange, green, purple colors represent Hsp20, Hsp70 and Hsp90 protein sequences from D. officinale (Den), A. thaliana (At) and O. sativa (Os), respectively. Different subfamilies are shaded with different colors.
The 37 DenHsp20s were classified into 8 distinct subfamilies, including 13 CIs, 8 CIIs, 4 CIIIs, 1 CV, 2 ERs, 1 MI, 2 Ps, 2 Pos, and 4 unclassified, which were consistent with the results of previous studies where CI was the largest subfamily (Figure 2A). In addition to being consistent with the theory that the CIV subfamily exists only in dicotyledons as confirmed by previous studies, the CVI and CVII subfamilies of out phylogenetic tree also exist only in the dicotyledonous plant A. thaliana. The 43 DenHsp70s were classified into four subfamilies according to Wolf and CELLO subcellular localization prediction, 35 DenHsp70s in Cytoplasm/ Nucleus, 2 in Chloroplast, 3 in Mitochondrion, and 3 in Endoplasmic reticulum (Figure 2B). The DenHsp90s were divided into the same four subfamilies by the same method as DenHsp70s, and the four DenHsp90s were present in only two of these subfamilies, three in the Cytoplasm/ Nucleus and one in the Chloroplast (Figure 2C). By comparison, AtHsp90s was present in all four subfamilies, while OsHsp90s was present in three subfamilies except Chloroplast. Overall, the cytoplasm has the largest number of Hsp genes and is probably the main working region for heat shock proteins.
A phylogenetic tree was constructed from 84 amino acid sequences of DenHsp20s, DenHsp70s and DenHsp90s (Figure 3A). To resolve the motif composition of DenHsp20s, DenHsp70s and DenHsp90s, the 84 sequences were submitted to the MEME website. A total of 20 motifs were predicted with length ranging from 15 to 50 amino acids (Figure 3B). According to the detailed motif sequence, we found that the DenHsp20s, DenHsp70s and DenHsp90s had different motifs (As shown in Supplementary Figure 1). (i) Among the 20 motifs, Motif 2 and Motif 6 were widespread on almost all the DenHsp20s. Motif 14 and Motif 16 were specific to subfamilies CI and CII. (ii) The DenHsp70s had the most conserved motifs. Motif 3, Motif 5, Motif 8 and Motif 13 were widely widespread on the DenHsp70s.(iii) Motif 19 was widespread on all the DenHsp90s. (iiii) Especially, Motif 13 was on the DenHsp20-20 and DenHsp20-32 in addition to DenHsp70s.
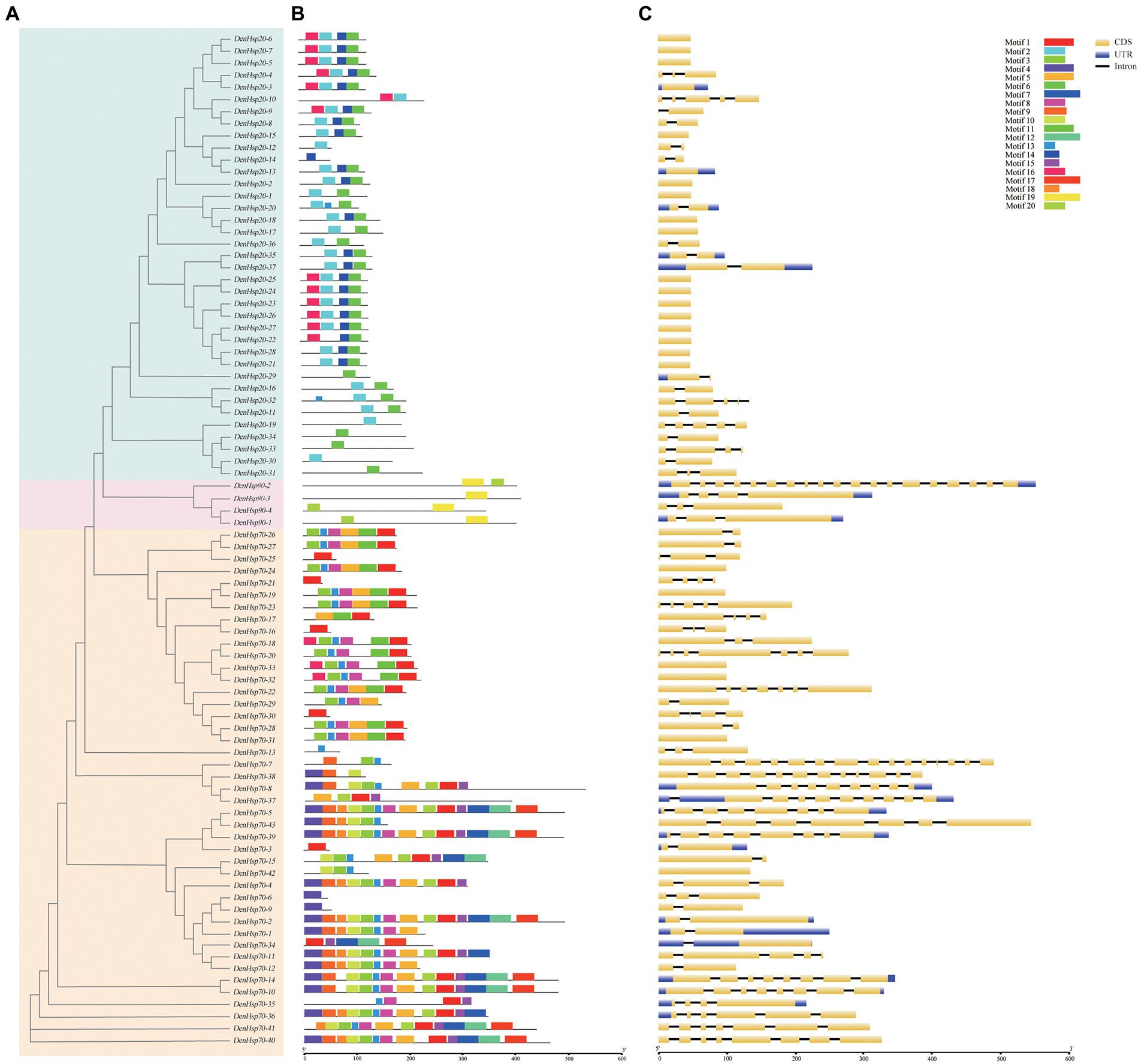
Figure 3. Phylogenetic relationships (A), conserved motifs (B) and exon-intron structures (C) of Hsp20, Hsp70 and Hsp90 genes in Dendrobium officinale. (A) Blue, red, orange colors represent Hsp20, Hsp90 and Hsp70 genes. (B) The conserved motifs of 84 proteins were identified using MEME and visualized by TBtools. Different colors represent 20 different motifs. (C) Yellow and blue boxes are, respectively, indicating CDS and UTR. And the black lines indicate introns.
Exon-intron structure provides an important clue for gene’s functional diversification. We investigated the exon-intron pattern of the 37 DenHsp20s, 43 DenHsp70s and 4 DenHsp90s according to D. officinale genome annotation information (Figure 3C). The results showed that the DenHsp20s were intron-less. It was obvious that genes in the same subfamily showed similar gene structures. Most members in subfamily CII of DenHsp20s had no introns. These results indicated that the structures of DenHsp20s were more conserved than those of DenHsp70s and DenHsp90s.
The basic structure of Hsps is conserved throughout the eukaryotic kingdom. Domain analysis helps to better understand different Hsp genes (Supplementary Figure 2). The Hsp20 sequences contain the central conserved domain, the α-crystallin domain (ACD). Hsp70 is composed of two domains: the ATPase domain, also referred as nucleotide binding domain, NBD (Flacherty et al., 1990), and the substrate binding domain (SBD; Zhu et al., 1996). Eukaryotic Hsp90 proteins contain 2 highly conserved domains: the adenosine triphosphate (ATP)-binding domain at the N-terminus and the highly charged (glutamic acid-rich) linker region (Xu et al., 2012).
Synteny analysis was conducted to the DenHsp20s, DenHsp70s and DenHsp90s using BLASTN and MCScanX to investigate gene duplication events. A total of 20 and 13 pairs of paralogous genes were detected among DenHsp20s and DenHsp70s, respectively (Figure 4A). Among them, there were 3 pairs of segmental duplications from DenHsp70s located on the D. officinale chromosomes (i.e., Chr2, Chr8, Chr10 and Chr15). While the remaining 30 pairs of tandem duplicates, with 20 pairs from DenHsp20s and 10 pairs from DenHsp70s, were focused on Chr2, Chr11 and Chr12 (Supplementary Table S3).
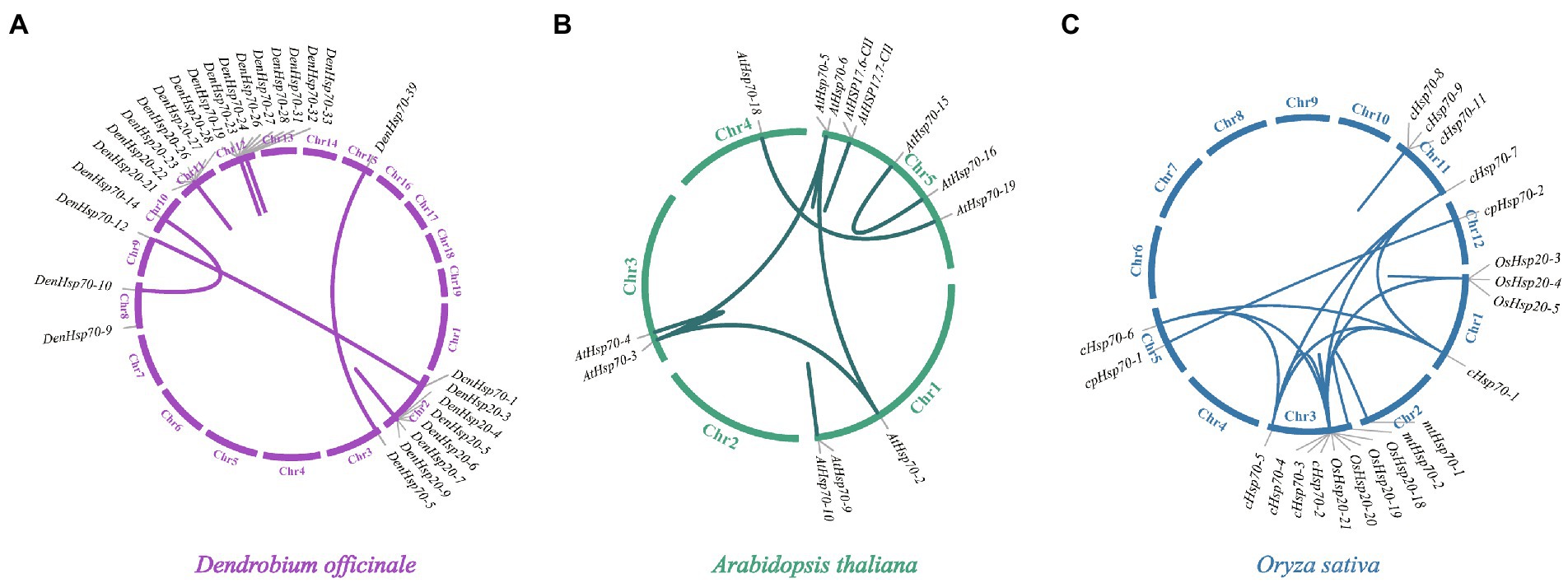
Figure 4. Schematic representations of the gene duplications of Hsp20, Hsp70 and Hsp90 genes from three different plants. (A–C) represent the syntenic gene pairs in D. officinale, A. thaliana and O. sativa, respectively.
Ka/Ks values of DenHsp20s’ and DenHsp70s’ duplicate gene pairs were calculated to evaluate the driving force underlying the HSP gene’s evolution. The results showed that Ka/Ks values of DenHsp20 duplicate genes ranged from 0.001 ~ 0.624, indicating that Hsp20 family was under negative selection during the evolution process (Supplementary Table S2). Among DenHsp70s paralogs, 7 pairs, DenHsp70-24 and DenHsp70-26, DenHsp70-26 and DenHsp70-27, DenHsp70-28 and DenHsp70-32, DenHsp70-28 and DenHsp70-33, DenHsp70-31 and DenHsp70-32, DenHsp70-31 and DenHsp70-33, DenHsp70-32 and DenHsp70-33, were positively selected, and the remaining pairs of genes experienced a negative selection (Table 2).
To further understand the replication event of the DenHsp20s and DenHsp70s, the replication events were compared between D. officinale and two other species (A. thaliana and O. sativa). The analysis also demonstrated that segmental duplication Hsp20 and Hsp70 gene pairs were found in genomes of A. thaliana (6 pairs) and O. sativa (12 pairs; Figures 4B,C).
Moreover, we analyzed the collinearity of Hsp20, Hsp70 and Hsp90 genes between D. officinale and four other plants (Figure 5). Collinearity analysis showed that the homologous genes between D. officinale and D. chrysotoxum were the most abundant, with 37 homologous gene pairs, followed by Vanilla planifolia (15 homologous gene pairs), O. sativa (15 homologous gene pairs) and A. thaliana (6 homologous gene pairs). DenHsp20-35 existed syntenic genes across the four species, and it is speculated that the gene may have originated from a common ancestor before the divergence of monocotyledons and dicotyledons with conserved and important functions. Excluding the dicotyledonous plant A. thaliana, DenHsp70-36, existed across the other three monocotyledons, may have been relatively conserved during monocotyledon evolution. In the collinearity analysis between D. officinale and D. chrysotoxum, the gene KAH0459475 of D. chrysotoxum had homologous pairs with three HSP70 genes (DenHsp70-17, DenHsp70-28, DenHsp70-31) in D. officinale, indicating that there was a duplication of the DenHsp70s.
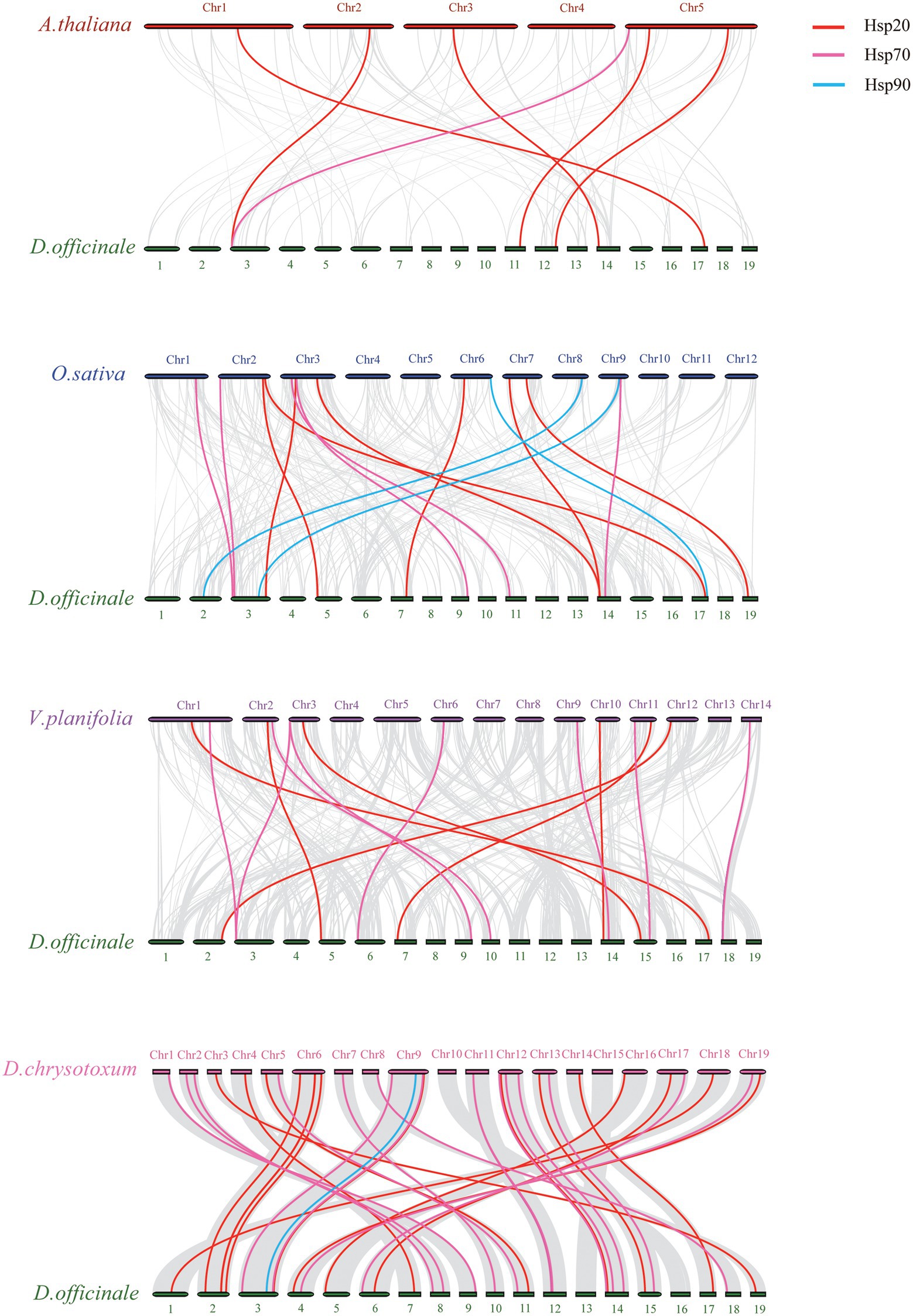
Figure 5. Collinearity analysis of Hsp20, Hsp70 and Hsp90 genes between Dendrobium officinale and four other plants, including A. thaliana, O. sativa, V. planifolia and D. chrysotoxum. Gray lines indicate the collinear blocks. Syntenic genes of Hsp20, Hsp70 and Hsp90 gene family are exhibited with red, pink and blue lines, respectively.
To better understand the potential function of DenHso20s, DenHsp70s and DenHsp90s, the cis-acting elements in the promoter regions were identified and analyzed. After removing non-functional terms, a total of 1,477 cis-acting elements in the promoter regions of DenHsp20s, DenHsp70s and DenHsp90s were classified into three categories of cis-elements, which are linked to plant growth and development (meristem expression, zein metabolism regulation, circadian control, etc.), stress responsiveness (light, anaerobic, low-temperature, drought, heat stress wound, and defense and stress), and phytohormone responsiveness (MeJA, abscisic acid, auxin, gibberellin and salicylic acid; Figure 6; Supplementary Figure 2; Supplementary Table S3).
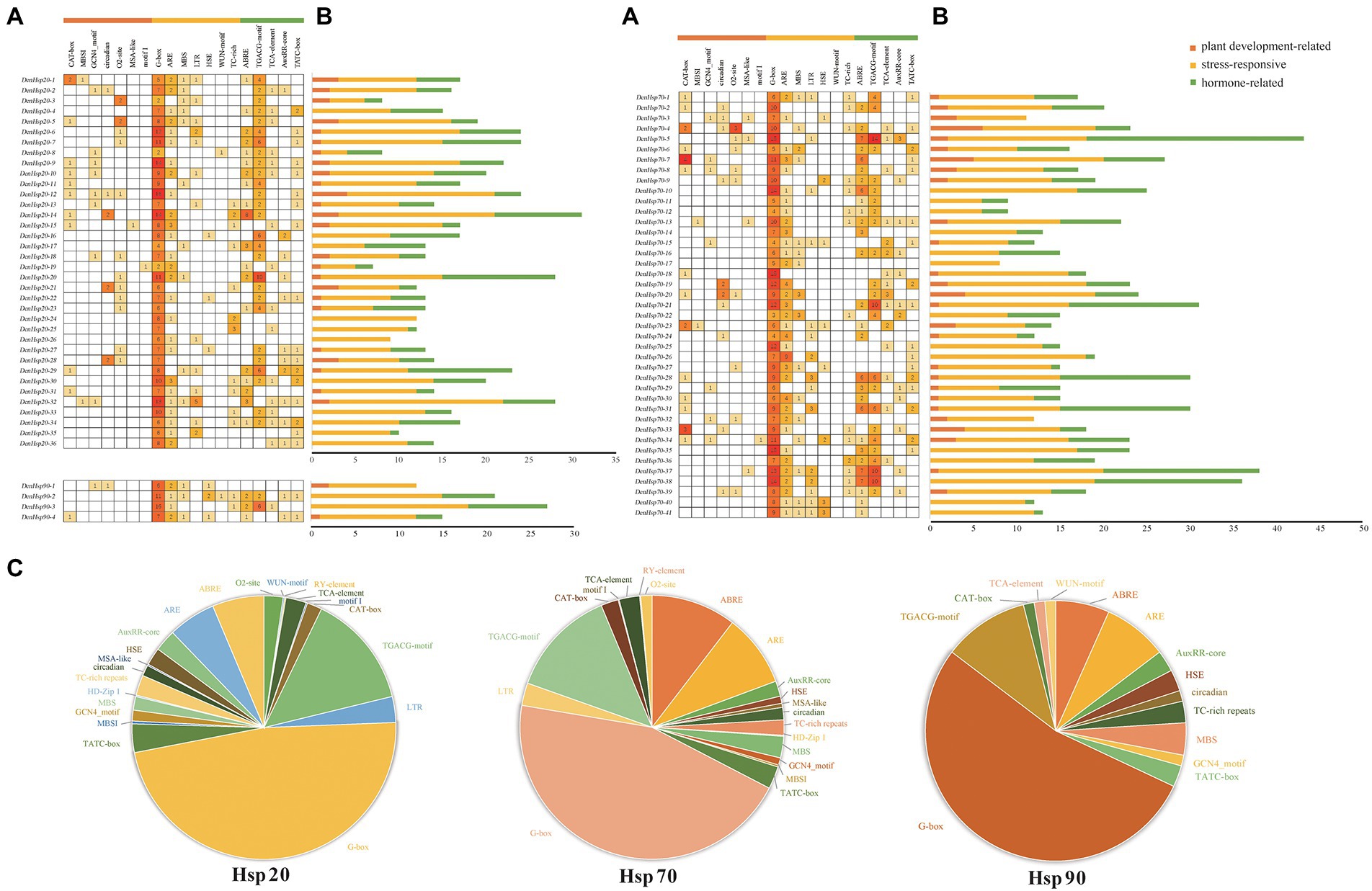
Figure 6. Information of cis-acting elements in Hsp20, Hsp70 and Hsp90 genes of Dendrobium officinale. (A) The gradient orange colors and numbers in the grid indicate the number of different cis-elements. (B) The different colors histogram indicates the number of cis-elements in each category. (C) The ratio of different cis-acting elements in Hsp20, Hsp70 and Hsp90 genes is shown as pie charts.
In DenHsp20s, most of the cis-elements were related to the stress responsiveness category (389/608), followed by the phytohormone responsiveness category (173/608) and plant growth and development category (46/608). The proportion of three categories in DenHsp70s and DenHsp90s were similar to those in DenHsp20s. Notably, 20 HSEs (accounting for 1.42%) were distributed across 3 DenHsp20s, 7 DenHsp70s and 3 DenHsp90s, most of which were located in DenHsp70s. As shown in Supplementary Figure 2, light responsive elements were the most abundant, accounting for 46.55% of all elements, which were related to the special photosynthetic pathway of D. officinale. MeJA responsive elements were the second abundant, accounting for 13.27% of all the cis-elements. In addition, there were a large number of stress responsive elements in drought, low-temperature and other stress environment. These results suggested that the ubiquitous cis-elements could be involved in DenHsp20s, DenHsp70s and DenHsp90s expression regulation in response to multiple abiotic stresses.
The tissue-specific expression of DenHsp20s, DenHsp70s and DenHsp90s were analyzed to further studying gene functions. (Figure 7A). Among the 84 Hsp genes of D. officinale, 45 genes were expressed, including 24 DenHsp20s, 18 DenHsp70s and 3 DenHsp90s. While these genes were expressed in different tissue. The results showed a diversified tissue-specific expression, e.g., most genes were highly expressed in flowers and leaves, lowly expressed in roots and stems of D. officinale. DenHsp70-36 was ubiquitously and highly expressed in every tissue, which was speculated that it may play a significant role in plant growth and development.
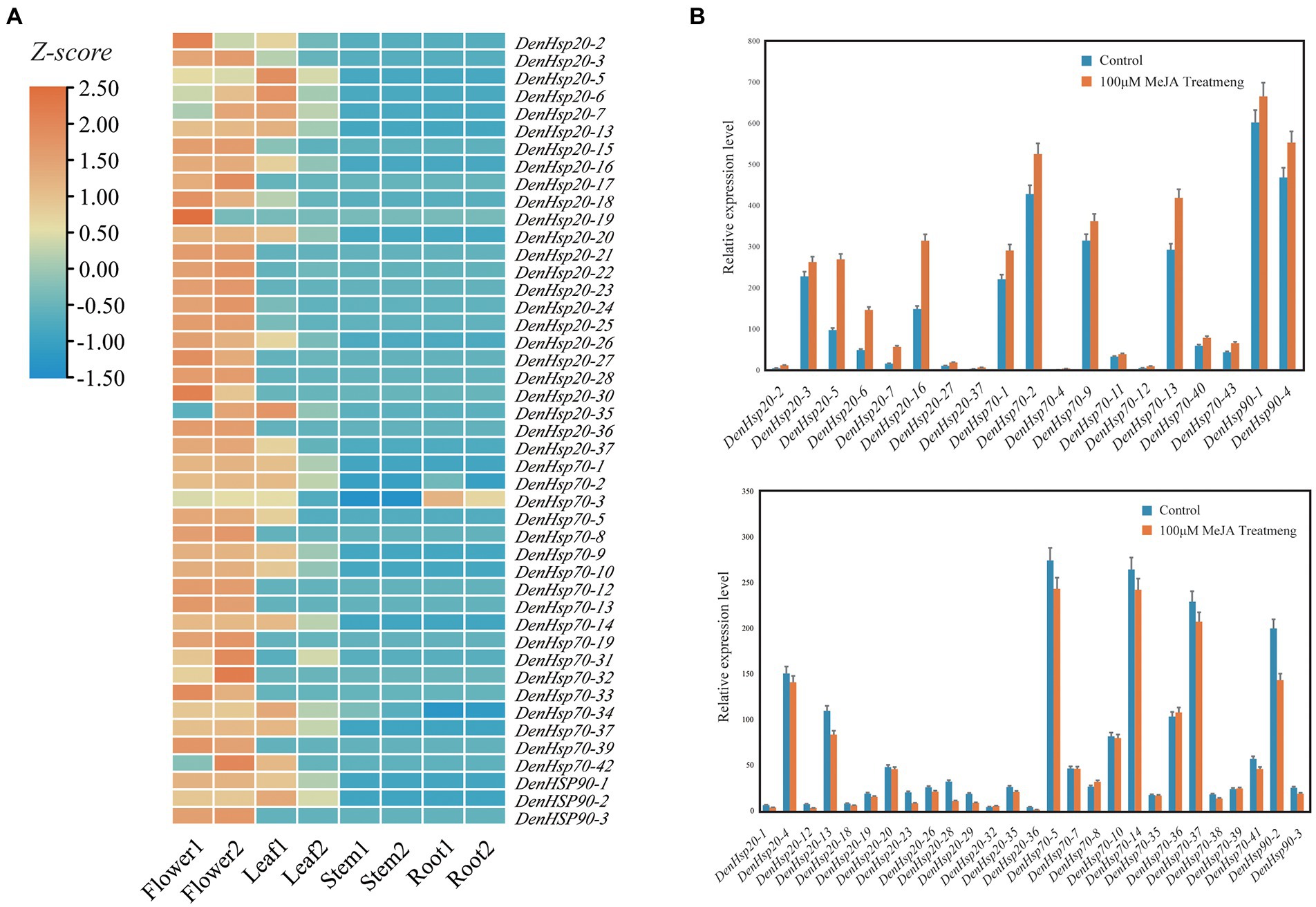
Figure 7. Expression analysis of DenHsp20s, DenHsp70s and DenHsp90s in different tissues and MeJA treat. (A) Expression profiles of Hsp20, Hsp70 and Hsp90 genes of Dendrobium officinale in different tissues including root, stem, leaf and flower. Z-score transformed FPKM values. (B) Relative expression levels of DenHsp20s, DenHsp70s and DenHsp90s under MeJA treatments.
Analysis of cis-elements indicated that most of Hsp genes in D. officinale contained MeJA response elements. To investigate the potential role of DenHsp20s, DenHsp70s and DenHsp90s under MeJA treatment, we determined the expression pattern by qRT-PCR (Figure 7B). Among them, 19 genes were significantly up-regulated, 24 were downregulated, and the remaining genes had no significant change. Moreover, in these 19 up-regulated genes, the relative expression levels of 4 genes (DenHsp20-5, DenHsp20-6, DenHsp20-7 and DenHsp70-4) were extremely up-regulated (more than 3-fold) under MeJA treatment, which may play a more important role in plant resistance to abiotic stresses.
Three-dimensional (3D) protein structures of DenHsp20 proteins, DenHsp70 proteins and DenHsp90 proteins were predicted with SWISS-MODEL. Subsequently, 62 successful models were defined by at least 30% identity of the target to a template, including 26 Hsp20 proteins, 16 Hsp70 proteins and 4 Hsp90 proteins (Supplementary Table S4). Compared with Hsp20 proteins (QMEAN DisCo Global from 0.60 to 0.78, GMQE from 0.42 to 0.78) and Hsp90 proteins (QMEAN DisCo Global from 0.74 to 0.78, GMQE from 0.75 to 0.82), the 3D structure models of Hsp70 proteins (QMEAN DisCo Global from 0.58 to 0.86, GMQE from 0.55 to 0.91) were of higher quality (Supplementary Table S4). A total of 46 different 3D structures, 26 for Hsp20 proteins, 4 for Hsp90 and 16 for Hsp70, were detected among Hsps, which indicated the diversified biological functions for Hsp genes in D. officinale.
To better understand the biological functions, the PPI networks were further analyzed to detect interactions among Hsp20 proteins, Hsp70 proteins and Hsp90 proteins and related proteins with the STRING website (Figure 8). Totally, 49 proteins and 661 connections were identified. Among the 661 connections, the Hsp90 proteins had the closest interaction with others, especially for the proteins of Hsp83. As shown in Supplementary Table S5, most of the proteins that interacted with Hsp proteins were the mediator of RNA polymerase II transcription subunits, such as MED14 and MED17. In addition, we also found that Hsp proteins may interact with stress-related transcription factors such as CYP and plant growth proteins like TCP. These results indicated that Hsp proteins of D. officinale have severe vital roles in multiple functions.
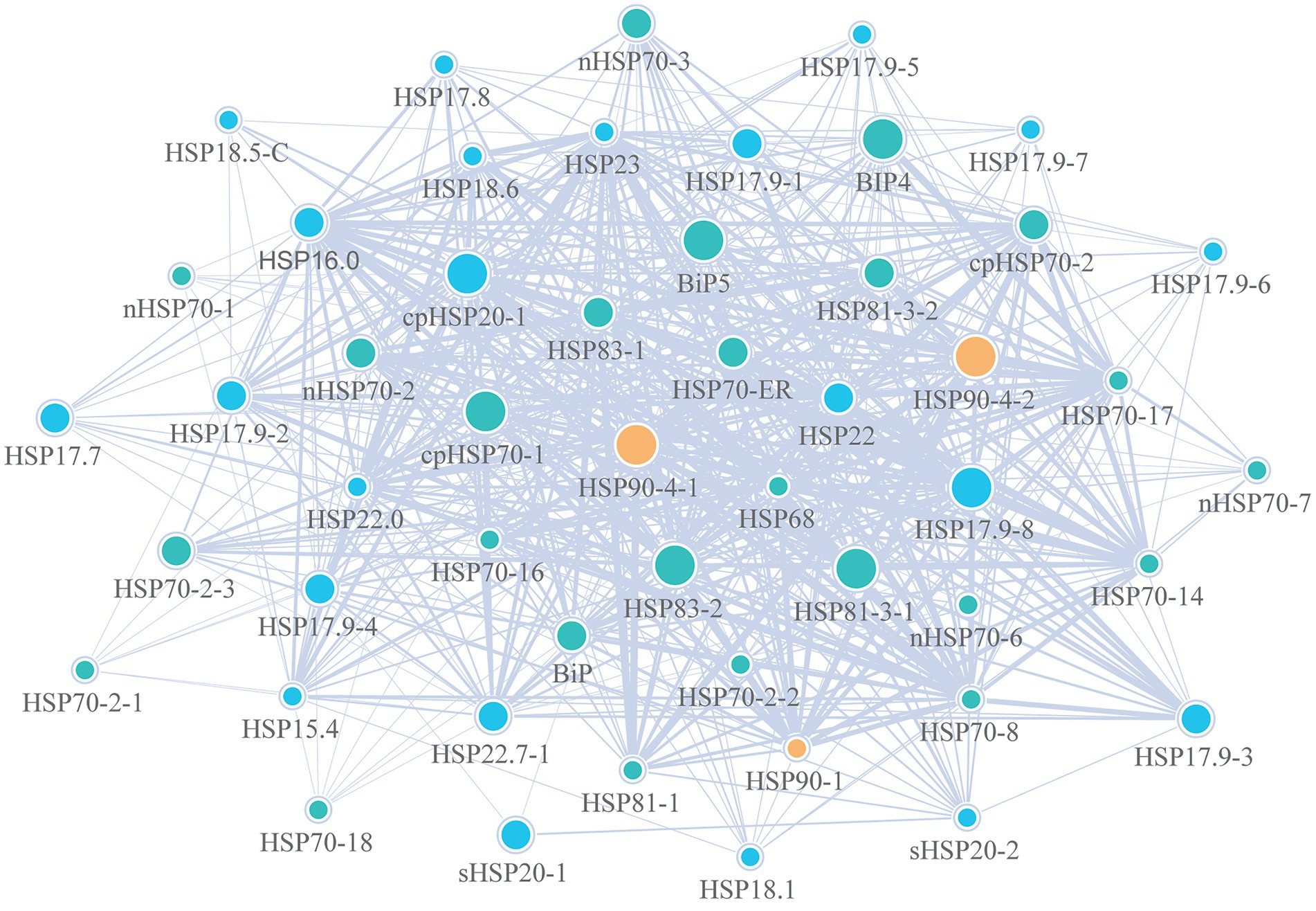
Figure 8. Protein–protein interaction (PPI) networks of Hsp20, Hsp70 and Hsp90 proteins in Dendrobium officinale. Blue, green and orange circles represent Hsp20, Hsp70 and Hsp90 proteins, respectively. The gradient circle size indicates the different degrees of importance.
The Hsp family, which plays an important role in the mechanism of plants responding to stresses and resisting the damage, is essential for plant species (Jiang et al., 2021). Though the sequences of Hsp genes were relatively conserved, the gene numbers of Hsp family varied greatly among plant lineages. For example, 17 genes of Hsp20 family were detected in A. thaliana (Scharf et al., 2001), while 39 and 117 genes were detected in O. sativa (Ouyang et al., 2009) and T. aestivum (Wang et al., 2017), respectively. In this study, a total of 84 Hsp genes, with 37 DenHsp20s, 43 DenHsp70s and 4 DenHsp90s, were identified from the chromosome-level genome sequences of D. officinale. We believed that three reasons may be responsible for the differences of different species and different Hsp families. Firstly, the varied gene number of Hsp family is correlated with the genome size variation. For example, A. thaliana, with a small genome size (125 Mb; Scharf et al., 2001) contained the lowest gene number (17 Hsp20 genes) of Hsp20 gene family, while T. Aestivum with large genome size (6.3 Gb; Wang et al., 2017), achieved the largest gene number (117 Hsp20 genes) of Hsp20 gene family. Secondly, gene duplication, which could multiply increase the quantity of genes, may be responsible to the gene number variation of Hsp gene family. In this study, a total of 30 gene duplications were investigated from the genome sequence, which increased the number of genes in the Hsp family by 25%. Moreover, genome expansion, especially for the expansion raised by WGD events may also cause the gene variation of the Hsp family. For example, the two rounds of WGD events that occurred in D. officinale have resulted in 3 genes increasing, which led to the number increase of the Hsp gene family. Compared with segmental duplications and tandem duplications accounted for a larger proportion in D. officinale, WGD was likely to enhance plant resistance by massively increasing the number of Hsp70 genes. Thus, we speculated that this may improve its adaption to environmental stresses. Thirdly, the different evolution patterns in different plant lineage may also resulted in the quantity variance. For example, in monocotyledonous plant lineage, Hsp20 genes undergone separate evolution after the divergence of monocots and dicots. As concluded from previous studies, the Hsp20 genes of A. thaliana can be divided into 12 subfamilies (CI, CII, CIII, CIV, CV, CVI, CVII, MI, MII, P, Po, ER; Scharf et al., 2001; Ma et al., 2006; Siddique et al., 2008). In this study, there were no CIV, CVI, CVII and MII subfamilies detected in D. officinale. The CIV subfamily was only present in dicotyledons in previous studies, which may lead to changes in the number of Hsp genes (Siddique et al., 2008; Zhao et al., 2018; Yao et al., 2020). Moreover, this could be a proof for the different evolutionary relationships in monocotyledons and dicots (Cui et al., 2021). We speculated that monocotyledons may require more Hsp20 genes to cope with environmental pressures. Therefore, genome size, gene duplication and different evolution patterns may be responsible for the quantity variance of Hsps.
Previous studies have shown that heat shock proteins play a critical role in the molecular mechanisms such as plant development and defense against abiotic (Jiang et al., 2021). However, different Hsp gene families have different functions. Even the same gene family has different functions. For instance, Hsp90 has been reported as a key regulator of normal growth and development in Nicotiana benthamiana and A. thaliana (Queitsch et al., 2002; Liu et al., 2004; Sangster and Queitsch, 2005; Sangster et al., 2007). While Zhang et al. (2013) analyzed the Hsp90 genes of Populus trichocarpa and found that Hsp90 proteins expression was observed in most organisms in response to stress. Inferred from our comparative analysis, we believed that protein structures, distribution and tissue-specific expression patterns and interactions of Hsps resulted in the diversity of biological functions.
Protein conformation is closely related to their biological functions. The diversity of protein structures may indicate the diversity of biological functions. In this study, a total of 46 different 3D structures among Hsp genes were detected, which indicated the diversified biological functions of Hsp genes. Moreover, the diversified distribution of Hsp genes, widely in the cytoplasm, nucleus and different organelles, may also be responsible for their diversified gene function. For example, previous studies have shown that (i) Hsps are mainly located in the cytoplasm and respond to abiotic and biotic stresses (Park and Seo, 2015). (ii) Hsp90C functions in a chaperone complex of the chloroplast matrix, facilitating membrane transport during protein entry into organelles (Inoue et al., 2013). (iii) Mitochondrial heat shock protein 70 (mtHsp70) functions in the unfolding, translocation, and folding of imported proteins (Voisine et al., 1999). In this study, in D. officinale, according to the result of subcellular localization, DenHsp20s were located in the cytosol, ER, mitochondria and chloroplast. While, DenHsp70s and DenHsp90s were located in Cytosol, ER, nucleus, mitochondria and chloroplast. These results implied that Hsp genes had various functions in D. officinale. For instance, we found that Hsp genes present tissue-specific expression patterns in D. officinale. Most Hsp genes were highly expressed in flowers and leaves, while they were rarely expressed in roots and stems.
Additionally, there are interactions between different proteins. For example, Hsp90 physically interacts with many cochaperones, including different Hsp families, to recruit and interact with diverse substrate proteins, leading to alteration of cellular processes. As expected, in this study, the integrated PPI network found that the majority of the Hsp20, Hsp70 and Hsp90 proteins were enriched. Among them, Hsp proteins were closely related to each other and might share biological functions. In addition to the Hsp proteins, some MED proteins and stress-related proteins were also enriched in the networks, which resulted in the diversity of biological functions.
Therefore, these results indicated that Hsps had various functions in D. officinale. Moreover, these also provided ideas for further research on the biological functions of Hsp genes.
Hsp genes were highly conserved in their gene sequences, especially for Hsp70 (Sharma and Masison, 2009). Thus, we evaluated that the evolution rate of protein coding genes of Hsp genes to see if they were undergone adaptive selection. Indeed, there were 6 gene pairs of DenHsp70s under positive selection (Ka/Ks > 1). We speculated that three reasons may possibly influence the evolution of DenHsp70s.
Firstly, previous studies showed that Hsp70 was the main and highly conserved protein activated by stress in living organisms, which could help the cells to withstand extreme stress (Usman et al., 2017). D. officinale grows in an adverse habitat, such as epiphytic on cliffs or tree trunks, and distributed at high altitude above 1,200 m (Yu et al., 2020). In extreme environments, plants often need Hsp70s to respond to stress quickly, so Hsp70s is often subjected to stronger selection pressure to maintain the stability of its protein structure.
Secondly, the special photosynthetic pathway of D. officinale may responsible to the adaptive evolution of DenHsp70s. D. officinale is a facultative CAM plant and the C3 pathway can be induced by controlling the growing environment (Zhang et al., 2014). Due to the importance of cis-elements in gene promoters for plant responses to environmental stresses (Yamaguchi-Shinozaki and Shinozaki, 2005), we further identified them in the putative promoter regions of DenHsp70s, and found that light-responsive elements were the most abundant. Since light is an important condition for photosynthesis, we believed that Hsp70s plays an important role in D. officinale photosynthesis, leading to the adaptive evolution of DenHsp70s.
Thirdly, it may partly enhance the heat dissipation capacity of plants. In previous studies, most of eleven chloroplast genome-encoded ndh genes (cp-ndh), which contribute to plant heat dissipation, were independently lost in D. officinale (Lin et al., 2017). However, Hsp70B has shown its abilities in the molecular protection of the photosystem reaction centers during photoinhibition and in the process of photosystem repair (Schroda et al., 1999). Moreover, a significant positive relationship between Hsp70 expression and the acquisition of thermotolerance has been identified (Lee et al., 2009), leading to increased heat and drought stress tolerance in plants (Alvim et al., 2001). Therefore, we speculated that Hsp70 genes play a more important role in adverse habitat, special photosynthetic pathway and heat dissipation.
In this study, a total of 37 Hsp20 genes (DenHsp20s), 43 Hsp70 genes (DenHsp70s) and 4 Hsp90 genes (DenHsp90s) were identified and confirmed in D. officinale genome. The DenHsp20s, DenHsp70s and DenHsp90s were randomly localized on different chromosomes, and they were classified into 8,4 and 2 subfamilies, respectively, based on the phylogenetic analysis and cellular locations. Moreover, gene structure, molecular evolution, interaction network and expression profiles were comprehensively reported. 13 duplicate gene pairs were identified in DenHsp70s, 7 of them were positively selected. These findings provided important information on the evolution of Hsp70 genes in D. officinale. The interaction network and expression profiles were analyzed to provide information on the function in stress response. This work would aid in elucidating the further functional characterizations of DenHsp20s, DenHsp70s and DenHsp90s in the future.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
XD, ZN, and JC designed the study. HW, QX, and WL performed the experiments. HW and ZD analyzed the data. MW and YD collected the materials. HW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Our work was funded by the National Natural Science Foundation of China (Grant nos. 32070353 and 31900268), Forestry Science and Technology Innovation and Promotion Project of Jiangsu Province [LYKJ(2021)12], Natural Science Foundation of Jiangsu Province (BK20190699), and Foundation of Key Laboratory of National Forestry and Grassland Administration for Orchid Conservation and Utilization (OC202102).
We would like to thank the College of Life Sciences, Nanjing Normal University for supporting this work. We are also grateful to the Jiangsu Provincial Engineering Research Center for Technical Industrialization for Dendrobiums for technical support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.979801/full#supplementary-material
SUPPLEMENTARY FIGURE 1 | Sequence logo of the Hsp20, Hsp70 and Hsp90 proteins motifs. The height of each amino acid represents the relative frequency of the amino acid at that position.
SUPPLEMENTARY FIGURE 2 | Multiple sequence alignment of members of the D. officinale Hsp20, Hsp70 and Hsp90 family. The multiple alignment was generated using DNAMAN_9. The positions of conserved functional domains are named above the align sequences.
SUPPLEMENTARY FIGURE 3 | Cis-acting elements in the promoter regions of Hsp20, Hsp70 and Hsp90 genes in D. officinale and statistical summary. (A) Cis-elements with similar functions are displayed in the same color. (B) The detailed percentages of each type of cis-elements. (C) The percentage of three kinds of cis-elements.
2. ^http://pdslab.biochem.iisc.ernet.in/hspir/
3. ^https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
4. ^http://smart.embl-heidelberg.de/
5. ^https://meme-suite.org/meme/
6. ^http://gsds.gao-lab.org/index.php
7. ^https://bioinformatics.psb.ugent.be/webtools/plantcare/html/
8. ^https://www.ncbi.nlm.nih.gov/sra
Alvim, F. C., Carolino, S. M., Cascardo, J. C., Nunes, C. C., Martinez, C. A., Otoni, W. C., et al. (2001). Enhanced accumulation of BiP in transgenic plants confers tolerance to water stress. Plant Physiol. 126, 1042–1054. doi: 10.1104/pp.126.3.1042
Aparicio, F., Thomas, C. L., Lederer, C., Niu, Y., Wang, D., and Maule, A. J. (2005). Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol. 138, 529–536. doi: 10.1104/pp.104.058958
Basha, E., O’Neill, H., and Vierling, E. (2012). Small heat shock proteins and α-crystallins: dynamic proteins with flexible functions. Trends Biochem. Sci. 37, 106–117. doi: 10.1016/j.tibs.2011.11.005
Boston, R. S., Viitanen, P. V., and Vierling, E. (1996). Molecular chaperones and protein folding in plants. Plant Mol. Biol. 32, 191–222. doi: 10.1007/BF00039383
Charng, Y. Y., Liu, H. C., Liu, N. Y., Hsu, F. C., and Ko, S. S. (2006). Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol. 140, 1297–1305. doi: 10.1104/pp.105.074898
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Hamada, S., Fujiwara, M., Zhu, T., Thao, N. P., Wong, H. L., et al. (2010). The hop/Sti1-Hsp90 chaperone complex facilitates the maturation and transport of a PAMP receptor in rice innate immunity. Cell Host Microbe 7, 185–196. doi: 10.1016/j.chom.2010.02.008
Chen, Z. H., Yuan, Y., Fu, D., and Yang, Y. J. (2017). Identification and expression profiling of the Auxin response factors in Dendrobium officinale under abiotic stresses. Int. J. Mol. Sci. 18:927. doi: 10.3390/ijms18050927
Cho, E. K., and Choi, Y. J. (2009). A nuclear-localized HSP70 confers thermoprotective activity and drought-stress tolerance on plants. Biotechnol. Lett. 31, 597–606. doi: 10.1007/s10529-008-9880-5
Cui, F. C., Taier, G., Wang, X. F., and Wang, K. H. (2021). Genome-wide analysis of the HSP20 gene family and expression patterns of HSP20 genes in response to abiotic stresses in Cynodon transvaalensis. Front. Genet. 12:732812. doi: 10.3389/fgene.2021.732812
Dangi, A. K., Sharma, B., Khangwal, I., and Shukla, P. (2018). Combinatorial interactions of biotic and abiotic stresses in plants and their molecular mechanisms: systems biology approach. Mol. Biotechnol. 60, 635–650. doi: 10.1007/s12033-018-0100-9
Flacherty, K. M., Luca-Flaherty, C., and McKay, D. B. (1990). Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346, 623–628. doi: 10.1038/346623a0
Inoue, H., Li, M., and Schnell, D. J. (2013). An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. 110, 3173–3178. doi: 10.1073/pnas.1219229110
Jacob, P., Hirt, H., and Bendahmane, A. (2017). The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 15, 405–414. doi: 10.1111/pbi.12659
Jiang, L. Y., Hu, W. J., Qian, Y. X., Ren, Q. Y., and Zhang, J. (2021). Genome-wide identification, classification and expression analysis of the Hsf and Hsp70 gene families in maize. Gene 770:145348. doi: 10.1016/j.gene.2020.145348
Kadota, Y., and Shirasu, K. (2012). The HSP90 complex of plants. Biochim. Biophys. Acta 1823, 689–697. doi: 10.1016/j.bbamcr.2011.09.016
Katoh, K., and Dtandley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Krishna, P., and Gloor, G. (2001). The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6, 238–246. doi: 10.1379/1466-1268(2001)006<0238:THFOPI>2.0.CO;2
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lai, D. L., Yan, J., Fan, Y., Li, Y., Ruan, J. J., Wang, J. Z., et al. (2021). Genome-wide identifcation and phylogenetic relationships of the Hsp70 gene family of Aegilops tauschii, wild emmer wheat (Triticum dicoccoides) and bread wheat (Triticum aestivum). 3 Biotech 11:301. doi: 10.1007/s13205-021-02639-5
Lee, S., Lee, D. W., Lee, Y., Mayer, U., Stierhof, Y. D., Lee, S., et al. (2009). Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, chip, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21, 3984–4001. doi: 10.1105/tpc.109.071548
Li, L. D., Jiang, Y., Liu, Y. Y., Niu, Z. T., Xue, Q. Y., Liu, W., et al. (2020). The large single-copy (LSC) region functions as a highly effective and efficient molecular marker for accurate authentication of medicinal Dendrobium species. Acta Pharm. Sin. B 10, 1989–2001. doi: 10.1016/j.apsb.2020.01.012
Lin, C. S., Chen, J. J. W., Chiu, C. C., Hsiao, H. C. W., Yang, C. J., Jin, X. H., et al. (2017). Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 90, 994–1006. doi: 10.1111/tpj.13525
Lin, J. Y., Shi, J. J., Zhang, Z. H., Zhong, B. J., and Zhu, Z. Q. (2022). Plant AFC2 kinase desensitizes thermomorphogenesis through modulation of alternative splicing. iScience 25:104051. doi: 10.1016/j.isci.2022.104051
Lin, B. L., Wang, J. S., Liu, H. C., Chen, R. W., Meyer, Y., Barakat, A., et al. (2001). Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6, 201–208. doi: 10.1379/1466-1268(2001)006<0201:GAOTHS>2.0.CO;2
Lindquist, S., and Craig, E. A. (1988). The heat-shock proteins. Annu. Rev. Genet. 22, 631–677. doi: 10.1146/annurev.ge.22.120188.003215
Liu, Y., Burch-Smith, T., Schiff, M., Feng, S., and Dinesh-Kumar, S. P. (2004). Molecular chaperone Hsp90 associates with resistance protein N and its signaling proteins SGT1 and Rar1 to modulate an innate immune response in plants. J. Biol. Chem. 279, 2101–2108. doi: 10.1074/jbc.M310029200
Livak, K. J., and Schmittgen, T. D. (2002). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopes-Caitar, V. S., Carvalho, M. C., Darben, L. M., Kuwahara, M. K., Nepomuceno, A. L., Dias, W. P., et al. (2013). Genome-wide analysis of the Hsp 20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics 14:577. doi: 10.1186/1471-2164-14-577
Lu, Y. Z., Zhao, P., Zhang, A. H., Ma, L. J., Xu, S. B., and Wang, X. M. (2020). Alternative splicing diversified the heat response and evolutionary strategy of conserved heat shock protein 90s in Hexaploid wheat (Triticum aestivum L.). Front. Genet. 11:577897. doi: 10.3389/fgene.2020.577897
Ma, C., Haslbeck, M., Babujee, L., Jahn, O., and Reumann, S. (2006). Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 141, 47–60. doi: 10.1104/pp.105.073841
Muthusamy, S. K., Dalal, M., Chinnusamy, V., and Bansal, K. C. (2017). Genome-wide identification and analysis of biotic and abiotic stress regulation of small heat shock protein (HSP20) family genes in bread wheat. J. Plant Physiol. 211, 100–113. doi: 10.1016/j.jplph.2017.01.004
Niu, Z. T., Pan, J. J., Xue, Q. Y., Zhu, S. Y., Liu, W., and Ding, X. Y. (2018). Plastome-wide comparison reveals new SNV resources for the authentication of Dendrobium huoshanense and its corresponding medicinal slice (Huoshan Fengdou). Acta Pharm. Sin. B 8, 466–477. doi: 10.1016/j.apsb.2017.12.004
Niu, Z. T., Zhu, F., Fan, Y. J., Li, C., Zhang, B. H., Zhu, S. Y., et al. (2020). The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharm. Sin. B 11, 2080–2092. doi: 10.1016/j.apsb.2021.01.019
Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Ouyang, Y. D., Chen, J. J., Xie, W. B., Wang, L., and Zhang, Q. F. (2009). Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol. Biol. 70, 341–357. doi: 10.1007/s11103-009-9477-y
Park, C. J., and Seo, Y. S. (2015). Heat shock proteins: A review of the molecular chaperones for plant immunity. Plant Pathol. J. 31, 323–333. doi: 10.5423/PPJ.RW.08.2015.0150
Queitsch, C., Sangster, T. A., and Lindquist, S. (2002). Hsp90 as a capacitor of phenotypic variation. Nature 417, 618–624. doi: 10.1038/nature749
Ratheesh, K. R., Nagarajan, N. S., Arunraj, S. P., Devanjan, S., Vinoth Babu, V. R., Vinoth Kumar, E., et al. (2012). HSPIR: a manually annotated heat shock protein information resource. Bioinformatics 28, 2853–2855. doi: 10.1093/bioinformatics/bts520
Ritossa, F. (1962). A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia 18, 571–573. doi: 10.1007/BF02172188
Sangster, T. A., Bahrami, A., Wilczek, A., Watanabe, E., Schellenberg, K., McLellan, C., et al. (2007). Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS One 2:e648. doi: 10.1371/journal.pone.0000648
Sangster, T. A., and Queitsch, C. (2005). The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 8, 86–92. doi: 10.1016/j.pbi.2004.11.012
Sarkar, N. K., Kim, Y. K., and Grover, A. (2009). Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics 10:393. doi: 10.1186/1471-2164-10-393
Sarkar, N. K., Kundnani, P., and Grover, A. (2012). Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 18, 427–437. doi: 10.1007/s12192-012-0395-6
Scharf, K. D., Siddique, M., and Vierling, E. (2001). The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing a-crystallin domains (Acd proteins). Cell Stress Chaperones 6, 225–237. doi: 10.1379/1466-1268(2001)006<0225:TEFOAT>2.0.CO;2
Schroda, M., Vallon, O., Wollman, F. A., and Beck, C. F. (1999). A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the Photoprotection and repair of photosystem II during and after Photoinhibition. Plant Cell 11, 1165–1178. doi: 10.1105/tpc.11.6.1165
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sharma, D., and Masison, D. C. (2009). Hsp70 structure, function, regulation and influence on yeast prions. Protein Pept. Lett. 16, 571–581. doi: 10.2174/092986609788490230
Shirasu, K. (2009). The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 60, 139–164. doi: 10.1146/annurev.arplant.59.032607.092906
Siddique, M., Gernhard, S., Koskull-Doring, P., Vierling, E., and Scharf, K. D. (2008). The plant sHSP superfamily: five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 13, 183–197. doi: 10.1007/s12192-008-0032-6
Tukaj, S., Bisewska, J., Roeske, K., and Tukaj, Z. (2011). Time-and dose-dependent induction of HSP70 in lemna minor exposed to different environmental stressors. Bull. Environ. Contam. Toxicol. 87, 226–230. doi: 10.1007/s00128-011-0339-3
Usman, M. G., Rafii, M. Y., Martini, M. Y., Yusuff, O. A., Ismail, M. R., and Miah, G. (2017). Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnol. Genet. Eng. Rev. 33, 26–39. doi: 10.1080/02648725.2017.1340546
Vierling, E. (1991). The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. & Plant Mol. Biol 42, 579–620. doi: 10.1146/annurev.pp.42.060191.003051
Vierling, E. (2003). The roles of heat shock proteins in plants. Annu. Rev. Plant Biol. 42, 579–620. doi: 10.1146/annurev.pp.42.060191.003051
Voisine, C., Craig, E. A., Zufall, N., Ahsen, O., Pfanner, N., and Voos, W. (1999). The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 97, 565–574. doi: 10.1016/S0092-8674(00)80768-0
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wang, X., Wang, R., Ma, C., Shi, X., Liu, Z., Wang, Z., et al. (2017). Massive expansion and differential evolution of small heat shock proteins with wheat (Triticum aestivum L.) polyploidization. Sci. Rep. 7:2581. doi: 10.1038/s41598-017-01857-3
Wang, G. F., Wei, X. N., Fan, R. C., Zhou, H. B., Wang, X. P., Yu, C. M., et al. (2011). Molecular analysis of common wheat genes encoding three types of cytosolic heat shock protein 90 (Hsp90): functional involvement of cytosolic Hsp90s in the control of wheat seedling growth and disease resistance. New Phytol. 191, 418–431. doi: 10.1111/j.1469-8137.2011.03715.x
Wang, X., Yang, P., Gao, Q., Liu, X., Kuang, T., Shen, S., et al. (2008). Proteomic analysis of the response to high-salinity stress in Physcomitrella patens. Planta 228, 167–177. doi: 10.1007/s00425-008-0727-z
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Geno. Proteom. Bioinform. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Waters, E. R. (2013). The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 64, 391–403. doi: 10.1093/jxb/ers355
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools on the ExPASy server. Methods Mol. Biol. 112, 531–552.
Xu, Z. S., Li, Z. Y., Chen, Y., Chen, M., Li, L. C., and Ma, Y. Z. (2012). Heat shock protein 90 in plants: molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 13, 15706–15723. doi: 10.3390/ijms131215706
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yao, F. W., Song, C. H., Wang, H. T., Song, S. W., Jiao, J., Wang, M. M., et al. (2020). Genome-wide characterization of the HSP20 gene family identifies potential members involved in temperature stress response in apple. Front. Genet. 11:609184. doi: 10.3389/fgene.2020.609184
Yu, Z. M., Zhao, C. H., Zhang, G. H., Silva, J. A. T., and Duan, J. (2020). Genome-wide identification and expression profile of TPS gene family in Dendrobium officinale and the role of DoTPS10 in linalool biosynthesis. Int. J. Mol. Sci. 21, 5419. doi: 10.3390/ijms21155419
Zhang, Z. J., He, D. X., Niu, G. H., and Gao, R. F. (2014). Concomitant CAM and C3 photosynthetic pathways in Dendrobium officinale plants. J. Am. Soc. Hortic. Sci. 139, 290–298. doi: 10.21273/JASHS.139.3.290
Zhang, J., Li, J. B., Liu, B. B., Zhang, L., Chen, J., and Lu, M. Z. (2013). Genome-wide analysis of the Populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genomics 14:532. doi: 10.1186/1471-2164-14-532
Zhang, H. M., Zhu, J. H., Gong, Z. Z., and Zhu, J. K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhao, P., Wang, D. D., Wang, R. Q., Kong, N. N., Zhang, C., Yang, C. H., et al. (2018). Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress. BMC Genomics 19:61. doi: 10.1186/s12864-018-4443-1
Zhu, S. Y., Niu, Z. T., Xue, Q. Y., Wang, H., Xie, X. Z., and Ding, X. Y. (2018). Accurate authentication of Dendrobium officinale and its closely related species by comparative analysis of complete plastomes. Acta Pharm. Sin. B 8, 969–980. doi: 10.1016/j.apsb.2018.05.009
Keywords: Dendrobium officinale, gene family, heat shock proteins, genome-wide analysis, expression profiles
Citation: Wang H, Dong Z, Chen J, Wang M, Ding Y, Xue Q, Liu W, Niu Z and Ding X (2022) Genome-wide identification and expression analysis of the Hsp20, Hsp70 and Hsp90 gene family in Dendrobium officinale. Front. Plant Sci. 13:979801. doi: 10.3389/fpls.2022.979801
Received: 28 June 2022; Accepted: 19 July 2022;
Published: 10 August 2022.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Ruisen Lu, Jiangsu Province and Chinese Academy of Sciences, ChinaCopyright © 2022 Wang, Dong, Chen, Wang, Ding, Xue, Liu, Niu and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhitao Niu, bml1emhpdGFvbmpAMTYzLmNvbQ==; Xiaoyu Ding, ZGluZ3h5bmpAMjYzLm5ldA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.