
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 August 2022
Sec. Plant Membrane Traffic and Transport
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.979066
This article is part of the Research Topic Regulation of Plant Organelle Biogenesis and Trafficking View all 8 articles
Eukaryotic cells possess endomembrane organelles equipped with specific sets of proteins, lipids, and polysaccharides that are fundamental for realizing each organelle’s specific function and shape. A tightly regulated membrane trafficking system mediates the transportation and localization of these substances. Generally, the secretory/exocytic pathway is responsible for transporting cargo to the plasma membrane and/or the extracellular space. However, in the case of oil body cells in the liverwort Marchantia polymorpha, the oil body, a liverwort-unique organelle, is thought to be formed by secretory vesicle fusion through redirection of the secretory pathway inside the cell. Although their formation mechanism remains largely unclear, oil bodies exhibit a complex and bumpy surface structure. In this study, we isolated a mutant with spherical oil bodies through visual screening of mutants with abnormally shaped oil bodies. This mutant harbored a mutation in a coat protein complex I (COPI) subunit MpSEC28, and a similar effect on oil body morphology was also detected in knockdown mutants of other COPI subunits. Fluorescently tagged MpSEC28 was localized to the periphery of the Golgi apparatus together with other subunits, suggesting that it is involved in retrograde transport from and/or in the Golgi apparatus as a component of the COPI coat. The Mpsec28 mutants also exhibited weakened stiffness of the thalli, suggesting impaired cell–cell adhesion and cell wall integrity. These findings suggest that the mechanism of cell wall biosynthesis is also involved in shaping the oil body in M. polymorpha, supporting the redirection of the secretory pathway inward the cell during oil body formation.
The trafficking system between endomembranous organelles, including the plasma membrane (PM) in eukaryotic cells, is responsible for the precise transportation and localization of proteins, lipids, and polysaccharides, and is referred to as membrane traffic. The membrane trafficking system mainly consists of the secretory, endocytic, and lysosomal/vacuolar transport pathways. The secretory, endocytic, and lysosomal/vacuolar transport pathways mediate transport from intracellular organelles to the extracellular space or PM, the extracellular space or PM to intracellular organelles, and to the lysosomes/vacuole, respectively (Kanazawa and Ueda, 2017; Aniento et al., 2022). A single round of trafficking between two compartments is initiated by sorting cargo and forming transport vesicles on a donor organelle membrane, generally mediated by SAR1/ARF GTPases and coat protein complexes, such as coat protein complexes I and II (COPI and COPII) and the clathrin coat (Dacks and Robinson, 2017; Bethune and Wieland, 2018; Dell'Angelica and Bonifacino, 2019). RAB GTPases mediate the targeting and tethering of transport vesicles to target organelle membranes. SNARE proteins execute membrane fusion between these membranes to unload cargo molecules (Kanazawa and Ueda, 2017; Aniento et al., 2022; Ito and Uemura, 2022). Among the coat protein complexes, COPI acts in retrograde transport from the Golgi apparatus to the endoplasmic reticulum (ER) and in intra-Golgi trafficking. The COPI coat is composed of hetero-heptameric subunits α, β, β′, γ, δ, ε, and ζ, which are highly conserved among eukaryotes (Gao et al., 2014; Ito et al., 2014; Dodonova et al., 2015; Dacks and Robinson, 2017).
Although the fundamental molecular framework and major membrane trafficking pathways are highly conserved among eukaryotic lineages, some lineages have also acquired lineage-specific organelles, which should be accompanied by the development of new trafficking pathways to or from these organelles. Comparative genomics analyses have inferred the molecular mechanisms underlying the emergence of new endomembranous organelles and membrane trafficking pathways and detected lineage-specific acquisition of key machinery components, such as coat protein complexes, RAB GTPases, and SNARE proteins. Given that practically all extant membrane trafficking pathways occur through the actions of these machinery components, acquiring a novel membrane trafficking pathway should be accompanied by obtaining a new set of these components. Attaining these components with differentiated functions is thought to be accomplished through gene duplication followed by neo- and/or sub-functionalization by accumulated mutations (Dacks and Field, 2007; Schlacht et al., 2014). Co-opting evolutionarily conserved machinery components with distinct membrane trafficking pathways would be another possibility to explore new trafficking pathways. These theoretical frameworks have been proposed for many years, based on trans-lineage phylogenetic analyses. However, empirical support for these frameworks is limited.
The liverwort oil body is a striking example of a lineage-unique organelle specifically acquired by distinctive eukaryotic lineages during evolution. Notably, the liverwort oil body is an organelle independent from the oil body in seed plants (also known as the oleosome or lipid body) in regard to its origin or function; the liverwort oil body is surrounded by the phospholipid bilayer membrane similar to endomembrane organelles and probably acts in chemical protection against microbes and herbivores, whereas the seed plant oil body is surrounded by a lipid monolayer derived from the ER and accumulates neutral lipids as energy sources (Huang, 1992; He et al., 2013; Huang et al., 2013; Kanazawa et al., 2020; Romani et al., 2020, 2022). The morphology and distribution patterns of the liverwort oil body, such as the shape, color, and number in a cell, vary depending on the liverwort species and have thus been used as an index for taxonomically classifying liverworts (Müller, 1939; He et al., 2013; Romani et al., 2022). In the liverwort Marchantia polymorpha, one oil body is formed in a specific idioblastic cell, referred to as the oil body cell. Recent analyses have revealed that at least two key transcription factors, MpC1HDZ and MpERF13, mediate oil body cell differentiation and oil body development in M. polymorpha (Kanazawa et al., 2020; Romani et al., 2020, 2022). The Qa-SNARE protein MpSYP12B, which is homologous to the PM-resident MpSYP13B protein, is targeted to the oil-body membrane (Kanazawa et al., 2016). Artificial secretory cargo, sec-mRFP, is transported to the oil body lumen when expressed under MpSYP12B promoter regulation in oil body cells, whereas sec-mRFP, driven by the MpSYP13B promoter, is secreted to the extracellular space (Kanazawa et al., 2020). Alongside other supporting evidence, it was proposed that cellular state-specific redirection of the secretory pathway forms the oil body.
Genome information and a series of versatile molecular genetic tools are available for M. polymorpha studies (Ishizaki et al., 2016; Bowman et al., 2017; Montgomery et al., 2020; Kohchi et al., 2021; Bowman, 2022; Bowman et al., in press), including the high-efficiency Agrobacterium-mediated transformation of sporelings (Ishizaki et al., 2008). Using this transformation method can generate several transgenic plants in which transfer DNA (T-DNA) is randomly inserted into the genome, enabling mutants defective in gene functions involved in various biological processes, such as air chamber formation (Ishizaki et al., 2013), rhizoid development (Honkanen et al., 2016, 2018; Proust et al., 2016), lateral organ development (Naramoto et al., 2019), and oil body formation (Kanazawa et al., 2020), to be screened. In this study, to obtain further insights into oil body biogenesis, we screened a T-DNA-mutagenized population of M. polymorpha for mutants with abnormally shaped oil bodies and isolated a mutant with oil bodies with increased circularity. By analyzing the gene responsible for this mutation and other related genes, we demonstrated that normal oil body formation in M. polymorpha requires COPI-mediated secretory activity.
Marchantia polymorpha accessions Takaragaike-1 (Tak-1, male) and Takaragaike-2 (Tak-2, female; Ishizaki et al., 2008) were used as the wild-type. Supplementary Table 1 lists the plants used in this study. The growth conditions and transformation methods have been previously described (Ishizaki et al., 2008; Kubota et al., 2013; Kanazawa et al., 2020). The thalli were grown asexually and maintained on 1/2 × Gamborg’s B5 medium containing 1.0% (w/v) agar at 22°C under continuous white light. The transition from vegetative to reproductive growth was induced by supplementation with far-red light to obtain spores by crossing male and female lines. Gemmae were cultured on 1/2 × Gamborg’s B5 medium plates containing 1.0% (w/v) sucrose at 22°C under continuous white light for 5 days, then transferred onto 1/2 × Gamborg’s B5 medium plates containing 20 μM β-estradiol (Fujifilm) or the equivalent volume [0.1% (v/v)] of DMSO (Fujifilm) as a mock control to evaluate the effect of MpSEC21 and MpRET1 knockdown on thallus development. The area of the thalli was measured using ImageJ software (version 1.53 k).1
We screened for mutants from T-DNA-inserted transgenic plants and identified the causal genes by TAIL-PCR as previously described by Kanazawa et al. (2020). Briefly, 48,825 T-DNA-inserted plants were obtained by the co-culture of M. polymorpha sporelings with agrobacteria harboring the binary vector pCAMBIA1300, followed by selection on hygromycin-supplemented medium plates. Hygromycin-resistant plants were stained with BODIPY 493/503 (4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene, Thermo Fisher), and mutants with abnormal oil-body morphology were examined under a fluorescent stereoscopic microscope (M165FC, Leica). The T-DNA flanking sequences were amplified by TAIL-PCR and sequenced, and the obtained sequences were subjected to a BLAST search using MarpolBase.2
Sample preparation and PCR for genotyping were performed as described by Kanazawa et al., 2020. Supplementary Table 2 lists the primers used.
Protein sequences were collected from Phytozome3 and the following genome portal sites: Amborella trichopoda v1.0, Arabidopsis thaliana TAIR10, Brachypodium distachyon v3.1, Chlamydomonas reinhardtii v5.5/v5.6, Klebsormidium nitens NIES-2285 V1.1,4 Medicago truncatula Mt4.0v1, Oryza sativa v7_JGI, Marchantia polymorpha v5.1,5 Physcomitrium patens v3.3, Populus trichocarpa v3.0/v4.1, Selaginella moellendorffii v1.0, Sphagnum fallax v0.5/v1.1, and Ostreococcus lucimarinus v2.0. A multiple sequence alignment was performed using the MUSCLE program version 3.8.31, with default parameters (Edgar, 2004a, 2004b). Alignment gaps were removed manually or with the Gblocks program version 0.91b (Castresana, 2000; Talavera and Castresana, 2007). The maximum likelihood phylogenetic analyses were performed using PhyML 3.0 under the LG model (Guindon et al., 2010). Bootstrap analyses were performed by resampling 1,000 sets.
Supplementary Tables 2, 3 list the oligonucleotides and plasmids used in this study, respectively. Open reading frames (ORFs) and genomic sequences were amplified by PCR from complementary DNA (cDNA) and genomic DNA prepared from M. polymorpha accession Tak-1, respectively, for entry vector construction. The amplified products were subcloned into pENTR/D-TOPO (Thermo Fisher) according to the manufacturer’s instructions. mCherry cDNA with a linker sequence was inserted into the AscI site of the pENTR vector containing the cDNA for MpSEC28 using the In-Fusion HD cloning kit (Clontech), according to the manufacturer’s instructions. The PCR-amplified DNA fragment for mCitrine was inserted into the SacI site of pMpGWB301 using an In-Fusion cloning kit to construct the pMpGWB301-mCitrine destination vector harboring the Gateway cassette. Target sequences for genome editing were selected from the CRISPRdirect website6 (Naito et al., 2015). Double-stranded DNA fragments of the target sequences were subcloned into pMpGE_En04 to construct dual-gRNA vectors (Koide et al., 2020). The dual-gRNA system was used to generate mutants with a large genomic sequence deletion (Sugano et al., 2018). However, gRNA destined for the 3′-end of the MpSEC28 gene did not work in this study, resulting in the mutations indicated in Figure 1F. For the amiRNA experiments, MpMIR160 was used as the backbone according to Flores-Sandoval et al., 2016. The amiRNA sequences were amplified by PCR and subcloned into the pENTR/D-TOPO vector.
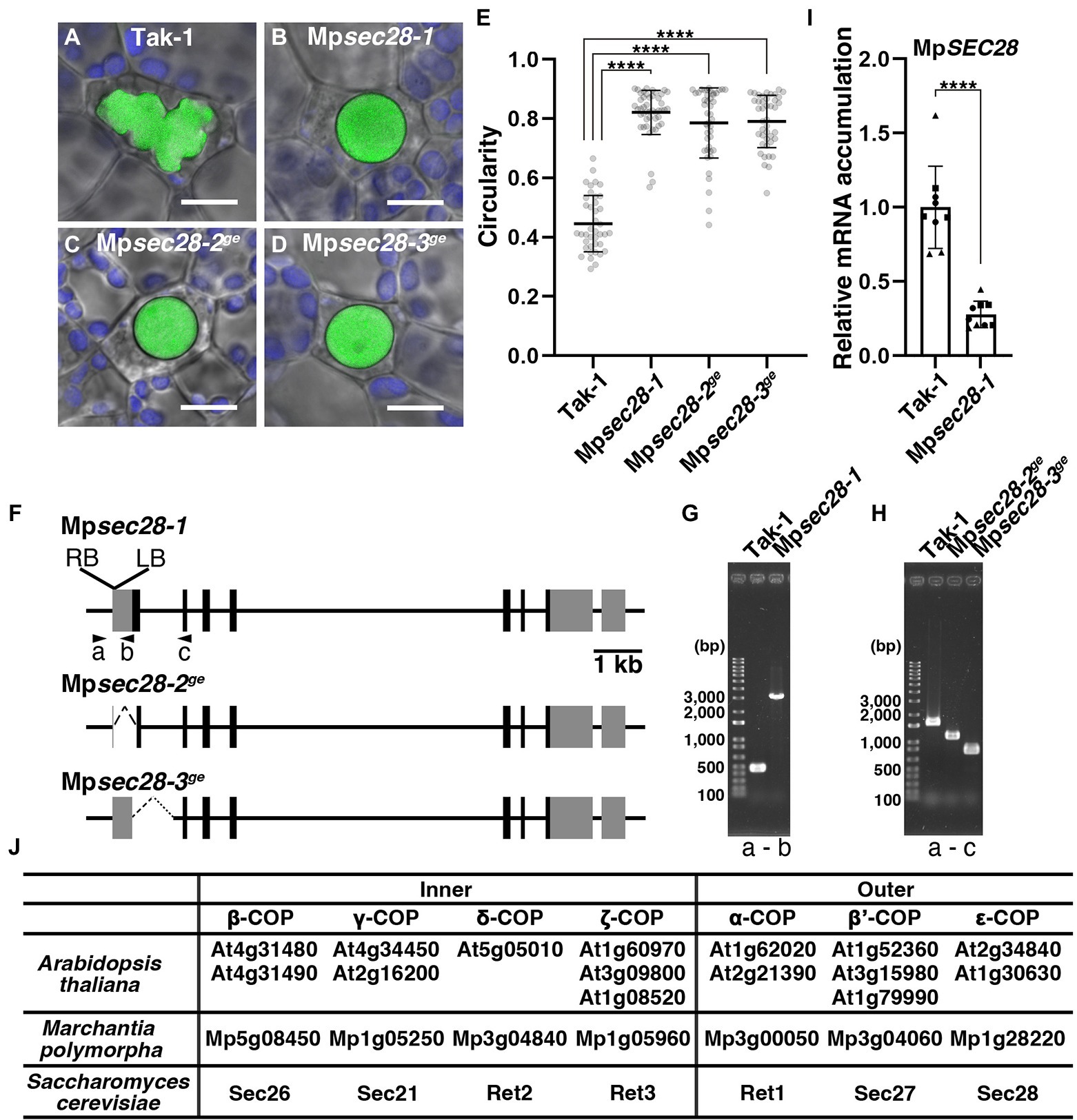
Figure 1. Abnormal oil body development in Mpsec28 mutants. (A–D) Confocal images merged with bright-field images of oil bodies in wild-type Tak-1 (A), Mpsec28-1 (B), Mpsec28-2ge (C), and Mpsec28-3ge (D). Green and blue pseudo-colors indicate the fluorescence from BODIPY and chlorophyll, respectively. Bars = 10 μm. (E) The circularity of the oil bodies in each genotype. Data are presented as the means ± s.d. Differences between wild-type and mutant plants were statistically significant based on a two-tailed Welch’s t-test. ****p < 0.001. The sample numbers were 40, 47, 44, and 42 oil bodies for Tak-1, Mpsec28-1, Mpsec28-2ge, and Mpsec28-3ge, respectively. p = 8.23 × 10−32 for Mpsec28-1, p = 2.73 × 10−24 for Mpsec28-2ge, and p = 4.40 × 10−28 for Mpsec28-3ge. (F) Schematic representation of the MpSEC28 gene structure and mutations generated in this study. Boxes indicate the UTRs (gray) and coding sequences (black). (G,H) PCR-based genotyping of Tak-1 and Mpsec28-1 (G), Tak-1, Mpsec28-2ge, and Mpsec28-3ge (H). (F) Shows the primer (A–C) annealing sites. (I) Relative MpSEC28 mRNA accumulation in Tak-1 and Mpsec28-1 measured by qRT-PCR. MpAPT was used as an internal reference. Data are presented as the means ± s.d. The difference between Tak-1 and Mpsec28-1 was statistically analyzed by a two-tailed Welch’s t-test. ****p < 0.001. Three biological replicates were prepared, and experiments were performed three times for each gene. Each biological replicate is indicated by a circle (replicate 1), triangle (replicate 2), or square (replicate 3). (J) Genes encoding COPI complex subunits in Arabidopsis thaliana, Marchantia polymorpha, and Saccharomyces cerevisiae.
The entry clones described above were transferred to destination vectors (pMpGWB101, pMpGWB301; Ishizaki et al., 2015), pMpGWB301-mCitrine, or pMpGWB301pro MpSYP2:GW-mCitrine for expression of chimeric proteins with fluorescent proteins, pMpGE011 for genome editing (Sugano et al., 2018), and pMpGWB368 (Furuya et al., 2022; Ishida et al., 2022) for β-estradiol-induced amiRNA expression using Gateway LR Clonase II Enzyme Mix (Thermo Fisher) according to the manufacturer’s instructions.
Five-day-old thalli were used for observation. For BODIPY staining, thalli were incubated in 200 nM BODIPY 493/503, dissolved in water for 30 min, washed twice in water, and used for microscopic observation. For β-estradiol treatment, gemmae were grown on a 1/2 × Gamborg’s B5 medium plate for 3 days, then soaked in a liquid medium containing 20 μM β-estradiol or 0.1% (v/v) DMSO for 48 h. The samples were mounted in a 1/2 × Gamborg’s B5 liquid medium and observed using an LSM780 confocal microscope (Carl Zeiss) equipped with an oil immersion lens (63 ×, numerical aperture = 1.4) and lambda and Airyscan detectors. For spectral imaging, the samples were excited at 488 nm (Argon 488) and 561 nm (DPSS 561–10), and emissions between 482 and 659 nm were collected. For high-resolution imaging using the Airyscan detector, samples were excited at 488 and 561 nm, and the emission was separated using a BP495-550 + BP570-630 filter. The images were acquired using line scanning. Spectral unmixing and Airy processing of the obtained images were performed using ZEN2.3 SP1 software (Carl Zeiss). We calculated the circularity of the oil bodies and analyzed colocalization using ImageJ and the Coloc_2 plugin in ImageJ Fiji (Schindelin et al., 2012), respectively.
The β-estradiol treatment was performed as described in the microscopy section. Total RNA was extracted as described by Kanazawa et al., 2013 and Kanazawa et al., 2020. cDNA was synthesized from total RNA using oligo dT(18) primers and SuperScript III Reverse Transcriptase (Thermo Fisher) according to the manufacturer’s instructions. qRT-PCR was performed with a LightCycler 96 (Roche) using the KAPA SYBR Fast qPCR Master Mix Kit (KAPA Biosystems) according to the manufacturer’s protocol. MpAPT was used as the housekeeping reference for normalization (Saint-Marcoux et al., 2015). Three biological replicates were prepared, and each experiment was repeated two or three times. Semi-quantitative RT-PCR was performed using KOD FX Neo (Toyobo), according to the manufacturer’s instructions.
We present representative confocal microscopy images of biological and technical replicates. The Source data list the results of quantitative image analyses and qRT-PCR, the number of generated transgenic lines, and the number of observed samples. Statistical analyses were performed using the Excel 365 software (ver. 2,202) or GraphPad Prism 9.3.1. (GraphPad Software), and graphs were drawn using GraphPad Prism 9.3.1.
Scheme of the assay is shown in Supplementary Figure 9. Twelve-day-old thalli were placed on 5 mm-gapped blocks. The blocks were placed on an electronic force balance, and the thalli were pressed using tweezers to record the force that severed the thalli.
The oil bodies of M. polymorpha can be visualized by staining with the lipophilic dye BODIPY 493/503 (Kanazawa et al., 2020). This allowed us to screen mutants with aberrantly shaped oil bodies. One of these mutants, Mpsec28-1 (identification of the responsible gene is described below), exhibited rounded oil body morphology. Oil bodies in wild-type plants exhibited a bumpy and complex morphology, with a mean circularity of 0.445 ± 0.0937 (mean ± s.d.; Figures 1A,E). The circularity of the oil body in the Mpsec28-1 mutant was 0.821 ± 0.0740, which was significantly higher than that of the wild-type (Figures 1B,E). This phenotype was heritable. The abnormal oil body phenotype was detected in 109 of 224 F1 plants obtained by crossing the Mpsec28-1 mutant with wild-type plants (p = 0.6885 in χ-squared test). This oil body phenotype was linked to the hygromycin resistance conferred by the HPT gene in the T-DNA, indicating that the phenotype was caused by a single nuclear mutation resulting from T-DNA insertion.
We performed a thermal asymmetric interlaced (TAIL)-polymerase chain reaction (PCR), sequenced the obtained DNA fragments, and conducted a BLAST search against the M. polymorpha genome sequence to identify the T-DNA insertion site. T-DNA was inserted into the 5′ untranslated region (UTR) of Mp1g28220 (Figures 1F,G). Mp1g28220 is annotated as the gene encoding the epsilon COPI subunit orthologous to Sec28 and ε-COP in Saccharomyces cerevisiae and mammals, respectively, which was supported by our maximum likelihood phylogenetic analysis (Figure 1J; Supplementary Figure 1; Hara-Kuge et al., 1994; Duden et al., 1998). Thus, we named this gene MpSEC28. The Mpsec28-1 allele was likely a knockdown allele, because mRNA accumulation was detected by qRT-PCR at a significantly reduced level compared to that in wild-type (Figure 1I). We generated other mutant alleles (Mpsec28-2ge and Mpsec28-3ge) using CRISPR-based genome editing, which should be knockout mutants because the start codon was deleted in these mutants (Figures 1F,H), to confirm that a mutation in MpSEC28 caused the abnormally shaped oil body phenotype. The mean circularities of the oil bodies in Mpsec28-2ge and Mpsec28-3ge were 0.785 ± 0.1173 and 0.790 ± 0.0875, respectively, similar to that of Mpsec28-1 (Figure 1E). These results indicated that normal oil body formation requires MpSEC28.
The COPI coat consists of α, β, β′, γ, δ, ε, and ζ subunits and mediates intra-Golgi transport and retrograde transport from the cis-Golgi to the ER (Figure 1J; Glick and Nakano, 2009; Ito et al., 2014; Aniento et al., 2022). We then examined whether the oil body phenotype was specifically caused by defective MpSEC28 function or whether defective functions of other COPI subunits exerted a similar effect. In S. cerevisiae, six genes encoding COPI subunits other than SEC28 are essential for viability (Hosobuchi et al., 1992; Duden et al., 1994, 1998; Letourneur et al., 1994; Cosson et al., 1996), which could also be the case for M. polymorpha COPI subunits. Therefore, we generated β-estradiol-inducible knockdown mutants of the α- and γ-subunits of the COPI-coat complex using artificial microRNA (amiRNA) for MpRET1 and MpSEC21, respectively (Supplementary Figures 2, 3). The constructs XVE > > amiR-MpRET1-1, XVE> > amiR-MpRET1-2, XVE> > amiR-MpSEC21-1, or XVE> > amiR-MpSEC21-2 were introduced into plants expressing mGFP-MpSYP12B, an oil-body membrane marker. We also introduced XVE > > amiR-Ctrl, in which the target sequence for MpRET1 or MpSEC21 was replaced with recognition sequences of SmaI and HindIII, into the mGFP-MpSYP12B plant as a negative control (Supplementary Figure 3C). After XVE > > amiR-MpRET1 and XVE > > amiR-MpSEC21 plants were treated with β-estradiol for 48 h, relative MpRET1 and MpSEC21 mRNA levels were significantly reduced compared to those in the mGFP-MpSYP12B plant (Figures 2A,B; Supplementary Figures 4A,B). In the knockdown plants, abnormally shaped oil bodies were frequently observed (Figures 2I,J; Supplementary Figure 4). A decrease in mRNA levels and abnormally shaped oil bodies upon β-estradiol treatment were not observed in the background and XVE > > amiR-Ctrl plants, nor in dimethyl sulfoxide (DMSO; mock)-treated XVE > > amiR-MpRET1 and XVE > > amiR-MpSEC21 plants (Figure 2; Supplementary Figure 4). These results strongly suggest that the abnormal oil body phenotype can be attributed to defective COPI function.
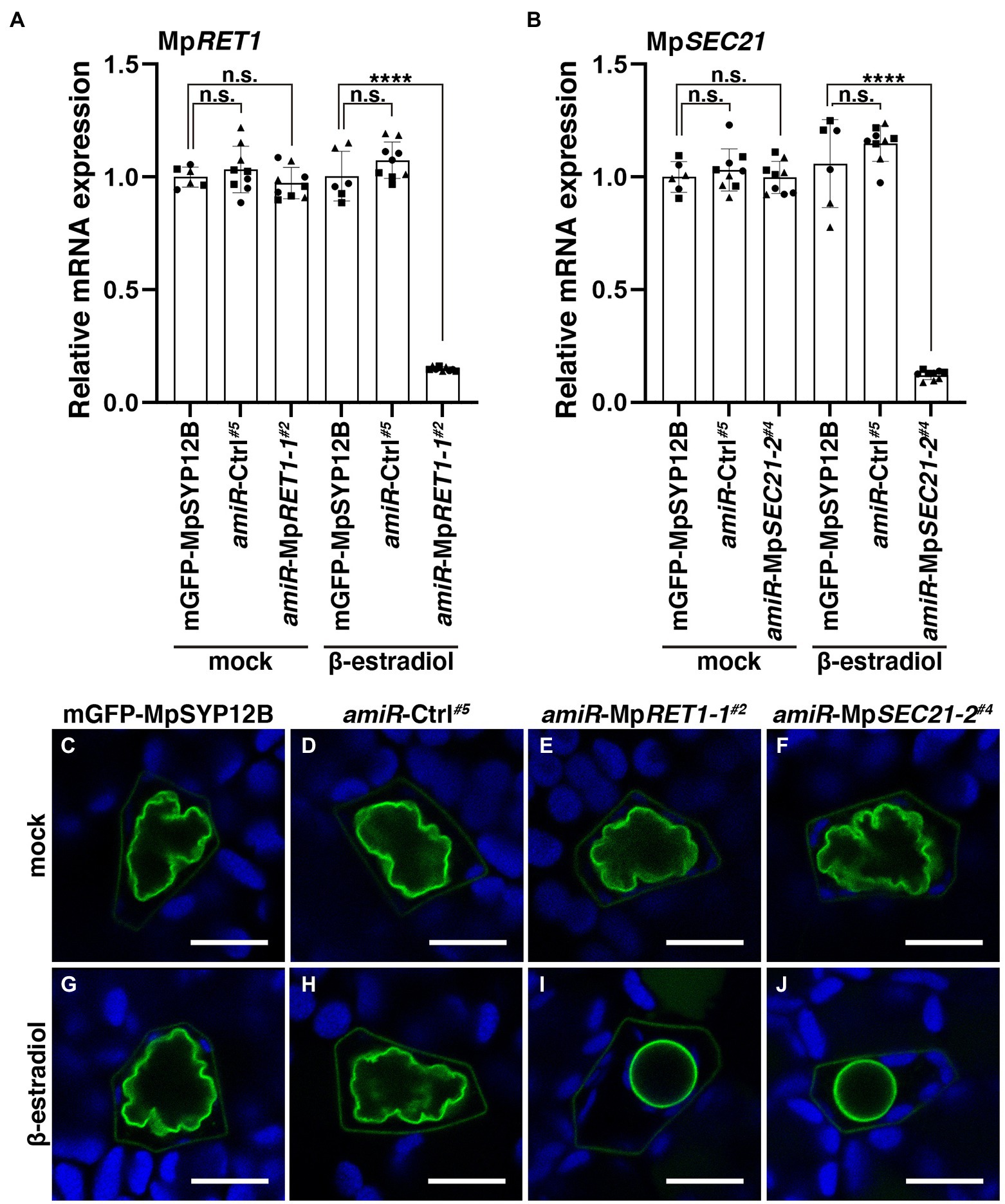
Figure 2. Defects in COPI function result in abnormally shaped oil bodies. (A,B) Relative MpRET1 (A) and MpSEC21 (B) mRNA expression levels in transgenic M. polymorpha plants with indicated transgenes measured by qRT-PCR. MpAPT was used as an internal reference. Data are presented as the means ± s.d. Significant differences between mGFP-MpSYP12B and each genotype were statistically analyzed by a two-tailed Welch’s t-test. n.s.: not significant; ****p < 0.001. Three biological replicates were prepared, and experiments were performed two or three times for each gene. Each biological replicate is indicated by a circle (replicate 1), triangle (replicate 2), or square (replicate 3). (C–J) Single confocal images of M. polymorpha thallus oil body cells in mGFP-MpSYP12B (C,G), amiR-Ctrl#5 (D,H), amiR-MpRET1-1#2 (E,I), and amiR-MpSEC21-2#4 (F,J) plants. Thalli were observed after 48 h of incubation with DMSO (0.1% v/v) (C–F) or 20 μM β-estradiol (G–J). Green and blue pseudo-colors indicate the fluorescence from mGFP and chlorophyll, respectively. Bars = 10 μm. Representative images are shown among replicates. The Source data lists the number of observed cells.
We then examined the effects of impairments in the function of COPI components on thallus growth. The thalli of Mpsec28 mutants were viable but were smaller than those of the wild-type (Figures 3A–E). MpRET1 and MpSEC21 knockdown conferred severe effects. Five-day-old thalli of XVE > > amiR-MpRET1 and XVE > > amiR-MpSEC21 plants grown on the normal medium exhibited no detectable abnormalities in thallus development. However, following 7 days of incubation on the medium containing β-estradiol, these plants showed severely impaired growth, whereas mock treatment did not affect thallus growth (Figures 3F–R and Supplementary Figure 5). The more severe effects of impairments in MpRET1 and MpSEC21 functions compared to the loss of function of MpSEC28 could reflect a difference in essentiality between these genes, as shown for orthologs in S. cerevisiae (Hosobuchi et al., 1992; Letourneur et al., 1994; Gerich et al., 1995; Duden et al., 1998; Ishii et al., 2016).
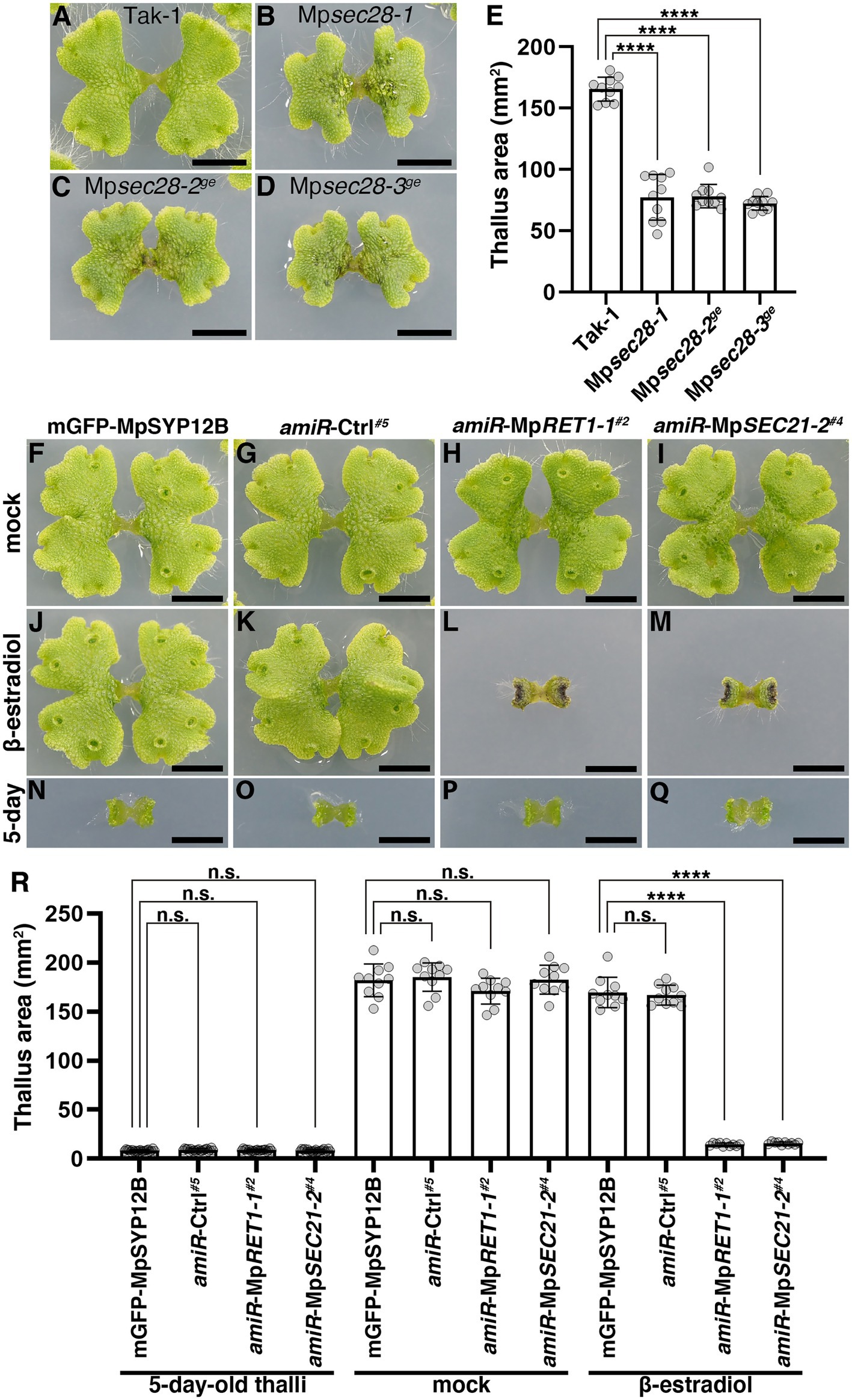
Figure 3. Effects of impaired COPI function on thallus growth. (A–D) Twelve-day-old M. polymorpha thalli of Tak-1 (A), Mpsec28-1 (B), Mpsec28-2ge (C), and Mpsec28-3ge (D). Bars = 5 mm. (E) Thallus areas of plants of the indicated genotypes. Data are presented as the means ± s.d. Significant differences between Tak-1 and each mutant were statistically analyzed by a two-tailed Welch’s t-test. ****p < 0.001. Ten thalli were analyzed for each genotype. p = 3.64 × 10−09 (Mpsec28-1), p = 8.78 × 10−14 (Mpsec28-2ge), and p = 1.85 × 10−13 (Mpsec28-3ge). (F–M) M. polymorpha thalli of mGFP-MpSYP12B (F,J,N), amiR-Ctrl#5 (G,K,O), amiR-MpRET1-1#2 (H,L,P), and amiR-MpSEC21-2#4 (I,M,Q) plants. Five-day-old thalli (N–Q) were transferred and grown on medium plates containing 0.1% (v/v) DMSO (F–I) or 20 μM β-estradiol (J–M) for 7 days. Bars = 5 mm. (R) The thallus area of 5-day-old plants with indicated genotypes (left) and plants treated with DMSO (mock, middle) or β-estradiol (right) for an additional 7 days. Data are presented as the means ± s.d. Significant differences between mGFP-MpSYP12B and each of the amiR plants were statistically analyzed using a two-tailed Welch’s t-test. n.s., not significant; ****p < 0.001. Twenty (5-day-old thalli) or ten (mock and β-estradiol) thalli were analyzed.
Immunoelectron microscopy using anti-γ-COP, anti-δ-COP, and anti-ε-COP antibodies revealed that these COPI subunits are localized to vesicles close to the Golgi apparatus in root cells of Arabidopsis and maize (Pimpl et al., 2000). We generated transgenic plants expressing mCherry-tagged MpSEC28 under the regulation of its own promoter to investigate the subcellular localization of MpSEC28 in M. polymorpha and observed its subcellular localization using a confocal laser scanning microscope equipped with lambda scan or Airyscan units for spectral imaging and high-resolution imaging, respectively. MpSEC28-mCherry was observed as a ring-shaped structure with faint dispersal localization in the cytosol of thallus non-oil body cells (Figure 4). MpSEC28-mCherry was also observed as ring-like structures without any detectable localization to the oil-body membrane in oil body cells (Supplementary Figure 6A). We compared its localization with that of other fluorescent markers [MpSEC21-mCitrine (COPI-coated vesicles), MpRET1-mCitrine (COPI-coated vesicles), mCitrine-MpGOS11 (Golgi apparatus), MpERD2-mCitrine (Golgi apparatus), mCitrine-MpSYP4 [trans-Golgi network (TGN)], mCitrine-MpRAB5 [multivesicular body (MVB)], and MpIDH1-mCitrine (mitochondria)] to help identify the MpSEC28-mCherry-positive structure (Kanazawa et al., 2016, 2020; Minamino et al., 2017; Norizuki et al., 2022). The data obtained using the lambda scan unit were analyzed for colocalization and indicated that MpSEC28-mCherry was highly colocalized with the COPI markers MpSEC21-mCitrine and MpRET1-mCitrine (Figures 4A,C; Supplementary Figure 7A). In contrast, Pearson colocalization coefficients with MpSEC28-mCherry and mCitrine-MpGOS11, MpERD2-mCitrine, mCitrine-MpSYP4, mCitrine-MpRAB5, or MpIDH1-mCitrine were significantly lower than that with MpSEC21-mCitrine (Figures 4B,C; Supplementary Figures 7B–E). Observations with the Airyscan unit also supported the colocalization of MpSEC28-mCherry with MpSEC21-mCitrine and MpRET1-mCitrine in ring-shaped structures (Figures 5A–D), which were associated with the periphery of the Golgi apparatus labeled by mCitrine-MpGOS11 (Figures 5E,F) or MpERD2-mCitrine (Figures 5G,H). MpSEC28 did not exhibit clear colocalization with mCitrine-MpSYP4 or mCitrine-MpRAB5 (Supplementary Figure 8). These results indicate that MpSEC28 is localized to COPI-coated vesicles that form at the periphery of the Golgi apparatus in M. polymorpha thallus cells.
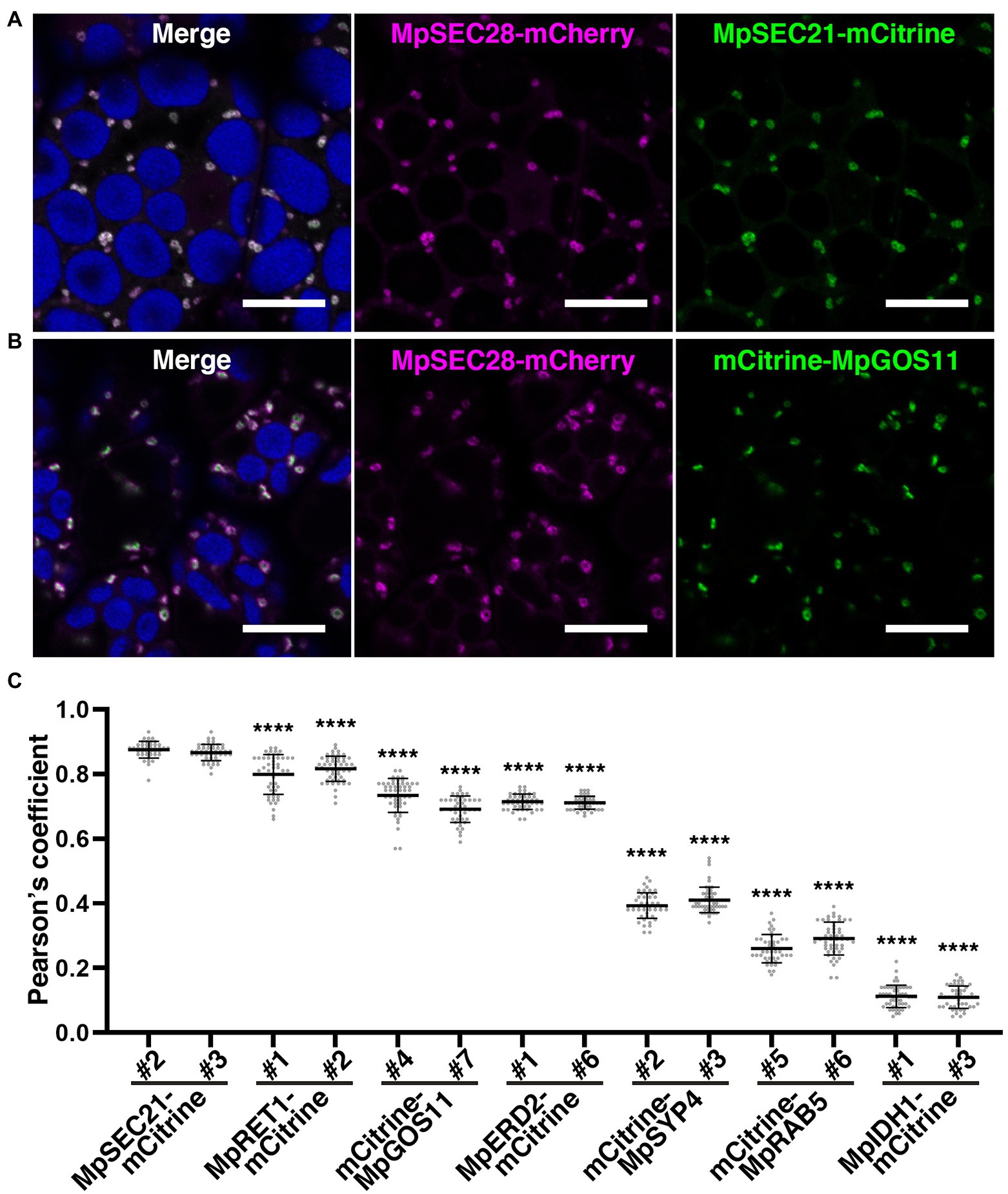
Figure 4. MpSEC28-mCherry colocalization with COPI components. (A–B) Single confocal images of M. polymorpha thallus cells expressing MpSEC28-mCherry and MpSEC21-mCitrine (A) or mCitrine-MpGOS11 (B). Magenta, green, and blue pseudo-colors indicate fluorescence from mCherry, mCitrine, and chlorophyll, respectively. Bars = 10 μm. (C) Pearson correlation coefficients between the fluorescence from MpSEC28-mCherry and organelle markers. Data are presented as the means ± s.d. Significant differences between MpSEC21-mCitrine#2 and each organelle marker were statistically analyzed using a two-tailed Welch’s t-test. ****p < 0.001. n = 50 and 51, and p = 2.33 × 10−11 and 2.25 × 10−13 for MpRET1-mCitrine#1 and #2, respectively. n = 52 and 44 and p = 6.71 × 10−28 and 6.80 × 10−38 for mCitrine-MpGOS11#4 and #7, respectively. n = 47 and 46 and p = 6.52 × 10−47 and 3.74 × 10−47 for MpERD2-mCitrine#1 and #6, respectively. n = 48 and 56 and p = 2.60 × 10−74 and 3.96 × 10−83 for mCitrine-MpSYP4#2 and #3, respectively. n = 45 and 50 and p = 7.07 × 10−73 and 5.64 × 10−71 for mCitrine-MpRAB5#5 and #6, respectively. n = 51 and 46 and p = 8.70 × 10−102 and 7.25 × 10−94 for MpIDH1-mCitrine#1 and #3, respectively.
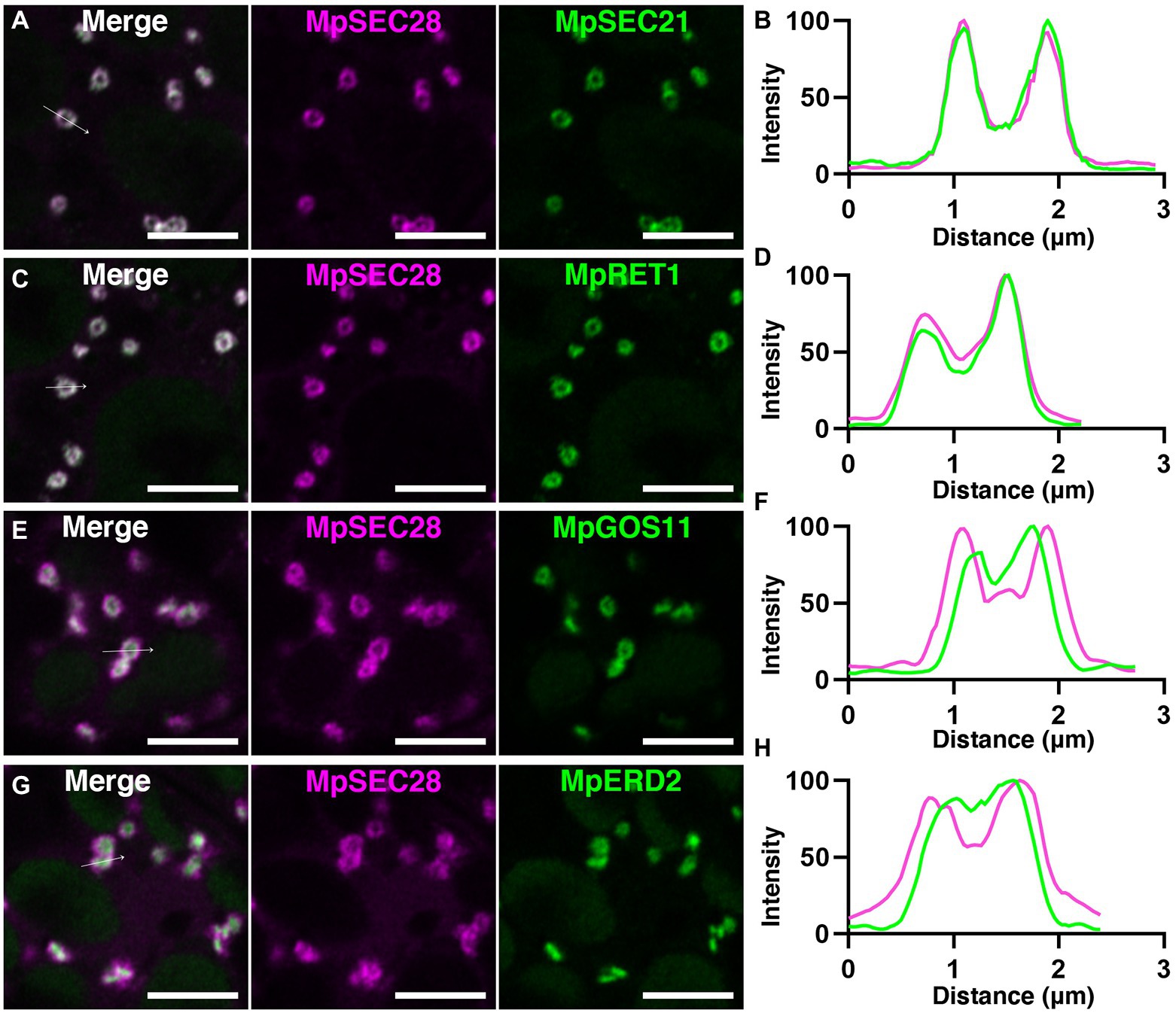
Figure 5. Golgi-periphery localization of MpSEC28-mCherry. (A,C,E,G) High spatial resolution images of M. polymorpha thallus cells expressing MpSEC28-mCherry and MpSEC21-mCitrine (A), MpRET1-mCitrine (C), mCitrine-MpGOS11 (E), or MpERD2-mCitrine (G). Magenta and green pseudo-colors indicate fluorescence from mCherry and mCitrine, respectively. Autofluorescence from chlorophyll in chloroplasts was also detected in the channel of mCitrine. Bars = 5 μm. (B,D,F,H) Line graphs showing relative fluorescence intensities from mCitrine and mCherry along the white arrows in A (B), C (D), E (F), and G (H).
MpSEC28-mCherry and MpSEC21-mCitrine were expressed in almost all cells in five-day-old thalli, including oil body cells (Supplementary Figure 6). The mRNA levels were not significantly changed in the Mperf13-1ge mutant, which possessed no oil body cells (Supplementary Figure 6C), indicating that MpSEC28 also acts in non-oil body cells. We noted that the thalli of Mpsec28 mutants were fragile during routine handling, suggesting that Mpsec28 mutations altered thalli stiffness. We performed a semi-quantitative chopping assay using 12-day-old thalli (Supplementary Figure 9). The mean force required to sever wild-type thalli was higher than that for Mpsec28 mutant thalli (Figure 6A), implying that Mpsec28 mutations affect cell wall integrity and cell–cell adhesion, probably due to the faulty secretion of substances required for cell wall integrity. Given that redirection of the secretory pathway is thought to form the oil body, transport to the oil body could also be compromised in Mpsec28 mutants. However, we did not detect markedly affected transport of a general secretion marker sec-mRFP driven by the MpSYP12B promoter or the oil-body-resident membrane protein mGFP-MpSYP12B to the oil body in Mpsec28 mutants (Figures 6B–G). These results suggest that partially defective COPI coat function confers selective effects on secretory activity, which might be required for cell wall integrity and oil body morphogenesis in M. polymorpha.
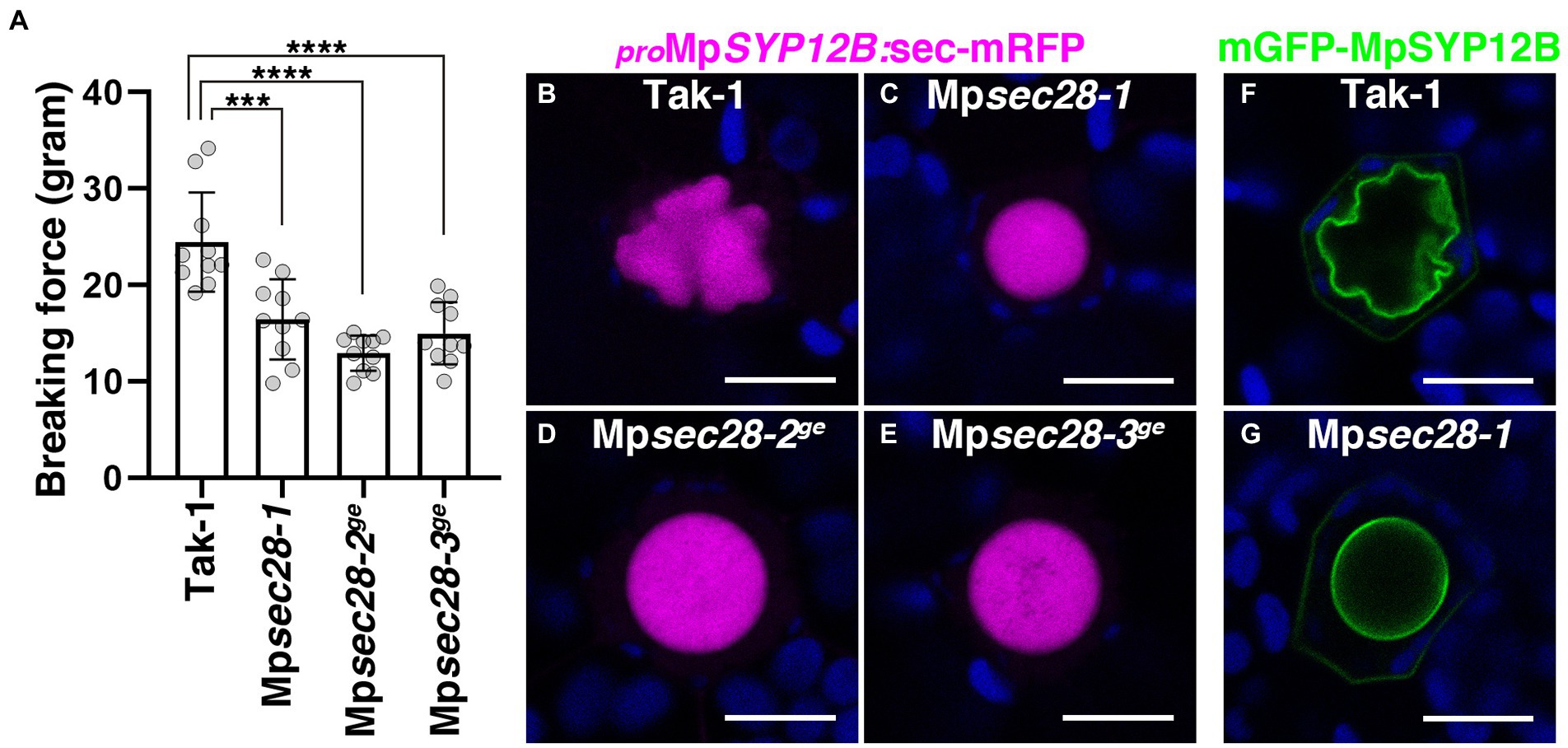
Figure 6. Effects of Mpsec28 mutations on thalli stiffness and transport to the oil body. (A) Semi-quantitative chopping assay. The force required to sever 12-day-old thalli was measured. Data are presented as the means ± s.d. Significant differences between Tak-1 and each genotype were statistically analyzed by a two-tailed Welch’s t-test. ***p < 0.005, ****p < 0.001. Ten thalli were analyzed for each genotype. p = 1.33 × 10−03 for Mpsec28-1, p = 3.21 × 10−05 for Mpsec28-2ge, and p = 1.79 × 10−04 for Mpsec28-3ge. (B–E) Single confocal images of M. polymorpha thallus cells expressing sec-mRFP in Tak-1 (B), Mpsec28-1 (C), Mpsec28-2ge (D), and Mpsec28-3ge (E) under the control of the MpSYP12B promoter. (F,G) Single confocal images of M. polymorpha thallus cells expressing mGFP-MpSYP12B under the control of its regulatory elements in Tak-1 (F) and Mpsec28-1 (G). Magenta, green, and blue pseudo-colors indicate fluorescence from mRFP, mGFP, and chlorophyll, respectively. Bars = 10 μm. Representative images are shown among replicates. The Source data lists the number of observed cells.
The secretory pathway begins with transport from the ER to the Golgi apparatus. Most de novo synthesized secretory and vacuolar proteins begin their journey from the ER, mediated by COPII-coated vesicles (Barlowe et al., 1994; Bethune and Wieland, 2018). Subpopulations of ER- and Golgi-resident proteins cycle between the ER and Golgi, and COPI-coated vesicles mediate retrograde transport from the Golgi to the ER (Matsuura-Tokita et al., 2006; Glick and Nakano, 2009; Ishii et al., 2016; Nakano, 2022). The COPI coat is also responsible for the intra-Golgi transport of Golgi-resident enzymes/proteins between Golgi cisternae. In this study, we isolated the Mpsec28-1 mutant with abnormally shaped oil bodies. MpSEC28 is orthologous to Sec28 and ε-COP, the epsilon subunits of the COPI complex in yeast and animals, respectively. The subcellular localization of MpSEC28-mCherry strongly suggested that MpSEC28 also acts as a component of the COPI coat. The mutant phenotype and responsible gene suggested that normal oil body formation requires COPI coat functionality. This was further supported by the knockdown of other putative COPI subunits, MpRET1 (α subunit) and MpSEC21 (γ subunit), which resulted in aberrant oil body morphology similar to that in Mpsec28 mutants. Intriguingly, transport of sec-mRFP and mGFP-MpSYP12B was not markedly affected by Mpsec28 mutations. This limited effect of mutations in MpSEC28 on transport of these cargos could reflect the limited requirement of MpSEC28 protein for the total secretory activity in M. polymorpha, which is consistent to what is known in yeast; Sec28 is an only subunit that is dispensable for viability among COPI subunits (Duden et al., 1998). Consistently, knockdown mutants of Mpret1 and Mpsec21 exhibited severer developmental inhibition than Mpsec28 mutants. Whereas the shared effect on the oil body morphology among mutations in COPI subunit genes suggests that the COPI activity is critical for normal oil body formation, we do not rule out the possibility that the severe inhibition of plant growth indirectly affected the oil body development when MpRET1 or MpSEC21 was knocked down.
It is interesting to ask why the mutants defective in COPI function exhibited abnormal oil body morphology. Mechanical support is required to shape and maintain the textured surface of the oil body in M. polymorpha. Given that the oil body lumen should be topologically equivalent to the extracellular space (Kanazawa et al., 2020), a cell wall-like structure might line the luminal side of the oil-body membrane, providing mechanical support to the oil body surface. Mutations in MpSEC28 resulted in thalli fragility suggesting that cell wall integrity requires COPI-mediated exocytic trafficking. The rounder shape of the oil body in mutants defective in COPI function could be due to the compromised integrity of the lining of the oil body lumen.
Arf GTPase activation triggers COPI coat assembly in yeast and animal cells and is catalyzed by Arf guanine nucleotide exchange factors (Arf GEFs; Stearns et al., 1990; Serafini et al., 1991; Waters et al., 1991; Donaldson et al., 1992; Hara-Kuge et al., 1994; Peyroche et al., 1996; Donaldson and Jackson, 2011; Nielsen, 2020). In Arabidopsis, two Arf GEFs with partially redundant functions, GNOM and GNL1, have been shown to act in the early secretory pathway by regulating COPI-coated vesicle formation (Richter et al., 2007; Teh and Moore, 2007; Naramoto et al., 2014; Singh et al., 2018). Calli of a mutant GNOM allele (emb30) exhibit defective cell–cell adhesion and increased fragility (Shevell et al., 2000), suggesting that cell–cell adhesion requires COPI-mediated trafficking in Arabidopsis. This is not surprising, given that the Golgi apparatus in plants is the site for pectic polysaccharide biosynthesis, which are crucial in cell–cell adhesion and the mechanical stiffness of plant tissues (Atmodjo et al., 2013; Hoffmann et al., 2021; Codjoe et al., 2022). In M. polymorpha, these Golgi apparatus functions should be conserved, which could account for the fragile nature of Mpsec28 mutants. Notably, major secretory activity, including the transport of PM proteins and general secretion, is redirected toward the oil body during oil body formation in oil body cells (Kanazawa et al., 2020). In such situations, the transport of cell wall-related enzymes and polysaccharides could also be redirected toward the oil body, accommodating the polysaccharide lining with the oil-body membrane to confer mechanical strength for the formation and/or maintenance of the complex and bumpy shape of the oil body. Verifying this possibility and unraveling the precise role of the COPI coat in oil body formation would be an interesting future project.
SEC28/ε-COP knockdown causes mislocalization of Golgi-resident Arabidopsis endomembrane protein 12 (AtEMP12), which harbors the KXD/E motif that mediates the interaction with the COPI coat at the C-terminal cytosolic tail (Woo et al., 2015). In this plant, the number of cisternae in the Golgi stack is significantly decreased compared to the Golgi in wild-type plants, suggesting that COPI is crucial in maintaining Golgi functions in plant cells. In our previous study, we proposed that the oil body is formed through redirection of the secretory pathway inward of the cell (Kanazawa et al., 2020), but the involvement of the Golgi apparatus and machinery components of membrane traffic in oil body formation remains elusive. In this study, we showed that the COPI coat is pivotal in normal oil body formation, strongly suggesting that the Golgi-dependent transport pathway substantially contributes to oil body formation. The results obtained in this study further support the proposal that the redirection of the secretory pathway forms the oil body in M. polymorpha. However, it is still possible that other subcellular activities dependent on the ER-Golgi trafficking are also involved in oil body formation. Further studies are needed to elucidate precise molecular and regulatory mechanisms of oil body formation in liverworts.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
TK performed the experiments and data analyses. RN designed and constructed vectors. TU supervised the study. TK and TU wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was financially supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [19H05670, 19H05675, 21H02515 (TU), and 22K06272 (TK)].
We thank Takayuki Kohchi, Keisuke Inoue, Yoshihiro Yoshitake, Keita Kinose, and Emi Hainiwa (Kyoto University), Kimitsune Ishizaki (Kobe University), and Shigeo S. Sugano (The National Institute of Advanced Industrial Science and Technology) for sharing vectors and plant materials. We thank Mayuko Yamamoto for supporting experiments. We also thank Kazuo Ebine, Naoki Minamino, and Takuya Norizuki (NIBB) for helpful discussions. Plant cultivation facilities were provided by the Model Plant Section, Model Organisms Facility, NIBB Trans-Scale Biology Center.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.979066/full#supplementary-material
3. ^https://phytozome-next.jgi.doe.gov/
4. ^http://www.plantmorphogenesis.bio.titech.ac.jp/~algae_genome_project/klebsormidium/
Aniento, F., de Medina, S., Hernandez, V., Dagdas, Y., Rojas-Pierce, M., and Russinova, E. (2022). Molecular mechanisms of endomembrane trafficking in plants. Plant Cell 34, 146–173. doi: 10.1093/plcell/koab235
Atmodjo, M. A., Hao, Z., and Mohnen, D. (2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64, 747–779. doi: 10.1146/annurev-arplant-042811-105534
Barlowe, C., Orci, L., Yeung, T., Hosobuchi, M., Hamamoto, S., Salama, N., et al. (1994). COPII: a membrane coat formed by sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907. doi: 10.1016/0092-8674(94)90138-4
Bethune, J., and Wieland, F. T. (2018). Assembly of COPI and COPII Vesicular coat proteins on membranes. Annu. Rev. Biophys. 47, 63–83. doi: 10.1146/annurev-biophys-070317-033259
Bowman, J. L. (2022). The liverwort Marchantia polymorpha, a model for all ages. Curr. Top. Dev. Biol. 147, 1–32. doi: 10.1016/bs.ctdb.2021.12.009
Bowman, J. L., Arteage-Vazquez, M., Berger, F., Briginshaw, L. N., Carella, P., Aguilar-Cruz, A., et al. (in press). The renaissance and enlightenment of Marchantia as a model system. Plant Cell.
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 171, 287–304.e15. doi: 10.1016/j.cell.2017.09.030
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Codjoe, J. M., Miller, K., and Haswell, E. S. (2022). Plant cell mechanobiology: greater than the sum of its parts. Plant Cell 34, 129–145. doi: 10.1093/plcell/koab230
Cosson, P., Demolliere, C., Hennecke, S., Duden, R., and Letourneur, F. (1996). Delta- and zeta-COP, two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO J. 15, 1792–1798. doi: 10.1002/j.1460-2075.1996.tb00528.x
Dacks, J. B., and Field, M. C. (2007). Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J. Cell Sci. 120, 2977–2985. doi: 10.1242/jcs.013250
Dacks, J. B., and Robinson, M. S. (2017). Outerwear through the ages: evolutionary cell biology of vesicle coats. Curr. Opin. Cell Biol. 47, 108–116. doi: 10.1016/j.ceb.2017.04.001
Dell'Angelica, E. C., and Bonifacino, J. S. (2019). Coatopathies: genetic disorders of protein coats. Annu. Rev. Cell Dev. Biol. 35, 131–168. doi: 10.1146/annurev-cellbio-100818-125234
Dodonova, S. O., Diestelkoetter-Bachert, P., von Appen, A., Hagen, W. J., Beck, R., Beck, M., et al. (2015). VESICULAR TRANSPORT. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science 349, 195–198. doi: 10.1126/science.aab1121
Donaldson, J. G., Finazzi, D., and Klausner, R. D. (1992). Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature 360, 350–352. doi: 10.1038/360350a0
Donaldson, J. G., and Jackson, C. L. (2011). ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12, 362–375. doi: 10.1038/nrm3117
Duden, R., Hosobuchi, M., Hamamoto, S., Winey, M., Byers, B., and Schekman, R. (1994). Yeast Beta-coat and Beta'-coat proteins (cop). 2 Coatomer subunits essential for endoplasmic reticulum-to-Golgi protein traffic. J. Biol. Chem. 269, 24486–24495. doi: 10.1016/S0021-9258(19)51110-3
Duden, R., Kajikawa, L., Wuestehube, L., and Schekman, R. (1998). Epsilon-COP is a structural component of coatomer that functions to stabilize alpha-COP. EMBO J. 17, 985–995. doi: 10.1093/emboj/17.4.985
Edgar, R. C. (2004a). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5, 113. doi: 10.1186/1471-2105-5-113
Edgar, R. C. (2004b). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Flores-Sandoval, E., Dierschke, T., Fisher, T. J., and Bowman, J. L. (2016). Efficient and inducible use of artificial MicroRNAs in Marchantia polymorpha. Plant Cell Physiol. 57, 281–290. doi: 10.1093/pcp/pcv068
Furuya, T., Nishihama, R., Ishizaki, K., Kohchi, T., Fukuda, H., and Kondo, Y. (2022). A glycogen synthase kinase 3-like kinase MpGSK regulates cell differentiation in Marchantia polymorpha. Plant Biotech. 39, 65–72. doi: 10.5511/plantbiotechnology.21.1219a
Gao, C., Cai, Y., Wang, Y., Kang, B. H., Aniento, F., Robinson, D. G., et al. (2014). Retention mechanisms for ER and Golgi membrane proteins. Trends Plant Sci. 19, 508–515. doi: 10.1016/j.tplants.2014.04.004
Gerich, B., Orci, L., Tschochner, H., Lottspeich, F., Ravazzola, M., Amherdt, M., et al. (1995). Non-Clathrin-coat protein-alpha is a conserved subunit of Coatomer and in saccharomyces-Cerevisiae is essential for growth. Proc. Natl. Acad. Sci. U. S. A. 92, 3229–3233. doi: 10.1073/pnas.92.8.3229
Glick, B. S., and Nakano, A. (2009). Membrane traffic within the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 25, 113–132. doi: 10.1146/annurev.cellbio.24.110707.175421
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hara-Kuge, S., Kuge, O., Orci, L., Amherdt, M., Ravazzola, M., Wieland, F. T., et al. (1994). En-bloc incorporation of Coatomer subunits during the assembly of cop-coated vesicles. J. Cell Biol. 124, 883–892. doi: 10.1083/jcb.124.6.883
He, X. L., Sun, Y., and Zhu, R. L. (2013). The oil bodies of liverworts: unique and important organelles in land plants. Crit. Rev. Plant Sci. 32, 293–302. doi: 10.1080/07352689.2013.765765
Hoffmann, N., King, S., Samuels, A. L., and McFarlane, H. E. (2021). Subcellular coordination of plant cell wall synthesis. Dev. Cell 56, 933–948. doi: 10.1016/j.devcel.2021.03.004
Honkanen, S., Jones, V. A. S., Morieri, G., Champion, C., Hetherington, A. J., Kelly, S., et al. (2016). The mechanism forming the cell surface of tip-growing rooting cells is conserved among land plants. Curr. Biol. 26, 3238–3244. doi: 10.1016/j.cub.2016.09.062
Honkanen, S., Thamm, A., Arteaga-Vazquez, M. A., and Dolan, L. (2018). Negative regulation of conserved RSL class I bHLH transcription factors evolved independently among land plants. elife 7:e38529. doi: 10.7554/eLife.38529
Hosobuchi, M., Kreis, T., and Schekman, R. (1992). Sec21 is a gene required for Er to Golgi protein-transport that encodes a subunit of a yeast Coatomer. Nature 360, 603–605. doi: 10.1038/360603a0
Huang, A. H. C. (1992). Oil bodies and oleosins in seeds. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 177–200. doi: 10.1146/annurev.pp.43.060192.001141
Huang, N. L., Huang, M. D., Chen, T. L., and Huang, A. H. (2013). Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol. 161, 1862–1874. doi: 10.1104/pp.112.212514
Ishida, S., Suzuki, H., Iwaki, A., Kawamura, S., Yamaoka, S., Kojima, M., et al. (2022). Diminished auxin signaling triggers cellular reprogramming by inducing a regeneration factor in the liverwort Marchantia polymorpha. Plant Cell Physiol. 63, 384–400. doi: 10.1093/pcp/pcac004
Ishii, M., Suda, Y., Kurokawa, K., and Nakano, A. (2016). COPI is essential for Golgi cisternal maturation and dynamics. J. Cell Sci. 129, 3251–3261. doi: 10.1242/jcs.193367
Ishizaki, K., Chiyoda, S., Yamato, K. T., and Kohchi, T. (2008). Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 49, 1084–1091. doi: 10.1093/pcp/pcn085
Ishizaki, K., Mizutani, M., Shimamura, M., Masuda, A., Nishihama, R., and Kohchi, T. (2013). Essential role of the E3 ubiquitin ligase NOPPERABO1 in schizogenous intercellular space formation in the liverwort Marchantia polymorpha. Plant Cell 25, 4075–4084. doi: 10.1105/tpc.113.117051
Ishizaki, K., Nishihama, R., Ueda, M., Inoue, K., Ishida, S., Nishimura, Y., et al. (2015). Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS One 10:e0138876. doi: 10.1371/journal.pone.0138876
Ishizaki, K., Nishihama, R., Yamato, K. T., and Kohchi, T. (2016). Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol. 57, 262–270. doi: 10.1093/pcp/pcv097
Ito, E., and Uemura, T. (2022). RAB GTPases and SNAREs at the trans-Golgi network in plants. J. Plant Res. 135, 389–403. doi: 10.1007/s10265-022-01392-x
Ito, Y., Uemura, T., and Nakano, A. (2014). Formation and maintenance of the Golgi apparatus in plant cells. Int. Rev. Cell Mol. Biol. 310, 221–287. doi: 10.1016/B978-0-12-800180-6.00006-2
Kanazawa, T., Era, A., Minamino, N., Shikano, Y., Fujimoto, M., Uemura, T., et al. (2016). SNARE molecules in Marchantia polymorpha: unique and conserved features of the membrane fusion machinery. Plant Cell Physiol. 57, 307–324. doi: 10.1093/pcp/pcv076
Kanazawa, T., Ishizaki, K., Kohchi, T., Hanaoka, M., and Tanaka, K. (2013). Characterization of four nuclear-encoded plastid RNA polymerase sigma factor genes in the liverwort Marchantia polymorpha: blue-light- and multiple stress-responsive SIG5 was acquired early in the emergence of terrestrial plants. Plant Cell Physiol. 54, 1736–1748. doi: 10.1093/pcp/pct119
Kanazawa, T., Morinaka, H., Ebine, K., Shimada, T. L., Ishida, S., Minamino, N., et al. (2020). The liverwort oil body is formed by redirection of the secretory pathway. Nat. Commun. 11, 6152. doi: 10.1038/S41467-020-19978-1
Kanazawa, T., and Ueda, T. (2017). Exocytic trafficking pathways in plants: why and how they are redirected. New Phytol. 215, 952–957. doi: 10.1111/nph.14613
Kohchi, T., Yamato, K. T., Ishizaki, K., Yamaoka, S., and Nishihama, R. (2021). Development and molecular genetics of Marchantia polymorpha. Annu. Rev. Plant Biol. 72, 677–702. doi: 10.1146/annurev-arplant-082520-094256
Koide, E., Suetsugu, N., Iwano, M., Gotoh, E., Nomura, Y., Stolze, S. C., et al. (2020). Regulation of photosynthetic carbohydrate metabolism by a Raf-like kinase in the liverwort Marchantia polymorpha. Plant Cell Physiol. 61, 631–643. doi: 10.1093/pcp/pcz232
Kubota, A., Ishizaki, K., Hosaka, M., and Kohchi, T. (2013). Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorphausing regenerating thalli. Biosci. Biotechnol. Biochem. 77, 167–172. doi: 10.1271/bbb.120700
Letourneur, F., Gaynor, E. C., Hennecke, S., Demolliere, C., Duden, R., Emr, S. D., et al. (1994). Coatomer is essential for retrieval of Dilysine-tagged proteins to the endoplasmic-reticulum. Cell 79, 1199–1207. doi: 10.1016/0092-8674(94)90011-6
Matsuura-Tokita, K., Takeuchi, M., Ichihara, A., Mikuriya, K., and Nakano, A. (2006). Live imaging of yeast Golgi cisternal maturation. Nature 441, 1007–1010. doi: 10.1038/nature04737
Minamino, N., Kanazawa, T., Nishihama, R., Yamato, K. T., Ishizaki, K., Kohchi, T., et al. (2017). Dynamic reorganization of the endomembrane system during spermatogenesis in Marchantia polymorpha. J. Plant Res. 130, 433–441. doi: 10.1007/s10265-017-0909-5
Montgomery, S. A., Tanizawa, Y., Galik, B., Wang, N., Ito, T., Mochizuki, T., et al. (2020). Chromatin Organization in Early Land Plants Reveals an ancestral association between H3K27me3, transposons, and constitutive heterochromatin. Curr. Biol. 30, 573–588.e7. doi: 10.1016/j.cub.2019.12.015
Müller, K. (1939). Untersuchungen über die Ölkörper der Lebermoose. Ber. Dtsch. Bot. Ges. 57, 326–370.
Naito, Y., Hino, K., Bono, H., and Ui-Tei, K. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. doi: 10.1093/bioinformatics/btu743
Nakano, A. (2022). The Golgi apparatus and its next-door neighbors. Front. Cell Dev. Biol. 10:884360. doi: 10.3389/fcell.2022.884360
Naramoto, S., Jones, V. A. S., Trozzi, N., Sato, M., Toyooka, K., Shimamura, M., et al. (2019). A conserved regulatory mechanism mediates the convergent evolution of plant shoot lateral organs. PLoS Biol. 17:e3000560. doi: 10.1371/journal.pbio.3000560
Naramoto, S., Otegui, M. S., Kutsuna, N., de Rycke, R., Dainobu, T., Karampelias, M., et al. (2014). Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26, 3062–3076. doi: 10.1105/tpc.114.125880
Nielsen, E. (2020). The small GTPase superfamily in plants: A conserved regulatory module with novel functions. Annu. Rev. Plant Biol. 71, 247–272. doi: 10.1146/annurev-arplant-112619-025827
Norizuki, T., Minamino, N., Sato, M., Tsukaya, H., and Ueda, T. (2022). Dynamic rearrangement and autophagic degradation of mitochondria during spermiogenesis in the liverwort Marchantia polymorpha. Cell Rep. 39:110975. doi: 10.1016/j.celrep.2022.110975
Peyroche, A., Paris, S., and Jackson, C. L. (1996). Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384, 479–481. doi: 10.1038/384479a0
Pimpl, P., Movafeghi, A., Coughlan, S., Denecke, J., Hillmer, S., and Robinson, D. G. (2000). In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12, 2219–2235. doi: 10.1105/tpc.12.11.2219
Proust, H., Honkanen, S., Jones, V. A., Morieri, G., Prescott, H., Kelly, S., et al. (2016). RSL class I genes controlled the development of epidermal structures in the common ancestor of land plants. Curr. Biol. 26, 93–99. doi: 10.1016/j.cub.2015.11.042
Richter, S., Geldner, N., Schrader, J., Wolters, H., Stierhof, Y. D., Rios, G., et al. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448, 488–492. doi: 10.1038/nature05967
Romani, F., Banic, E., Florent, S. N., Kanazawa, T., Goodger, J. Q. D., Mentink, R. A., et al. (2020). Oil body formation in Marchantia polymorpha is controlled by MpC1HDZ and serves as a defense against arthropod herbivores. Curr. Biol. 30, 2815–2828.e8. doi: 10.1016/j.cub.2020.05.081
Romani, F., Flores, J. R., Tolopka, J. I., Suarez, G., He, X., and Moreno, J. E. (2022). Liverwort oil bodies: diversity, biochemistry, and molecular cell biology of the earliest secretory structure of land plants. J. Exp. Bot. 73, 4427–4439. doi: 10.1093/jxb/erac134
Saint-Marcoux, D., Proust, H., Dolan, L., and Langdale, J. A. (2015). Identification of reference genes for real-time quantitative PCR experiments in the liverwort Marchantia polymorpha. PLoS One 10:e0118678. doi: 10.1371/journal.pone.0118678
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Schlacht, A., Herman, E. K., Klute, M. J., Field, M. C., and Dacks, J. B. (2014). Missing pieces of an ancient puzzle: evolution of the eukaryotic membrane-trafficking system. Cold Spring Harb. Perspect. Biol. 6:a016048. doi: 10.1101/cshperspect.a016048
Serafini, T., Orci, L., Amherdt, M., Brunner, M., Kahn, R. A., and Rothman, J. E. (1991). ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell 67, 239–253. doi: 10.1016/0092-8674(91)90176-y
Shevell, D. E., Kunkel, T., and Chua, N. H. (2000). Cell wall alterations in the arabidopsis emb30 mutant. Plant Cell 12, 2047–2059. doi: 10.1105/tpc.12.11.2047
Singh, M. K., Richter, S., Beckmann, H., Kientz, M., Stierhof, Y. D., Anders, N., et al. (2018). A single class of ARF GTPase activated by several pathway-specific ARF-GEFs regulates essential membrane traffic in Arabidopsis. PLoS Genet. 14, e1007795. doi: 10.1371/journal.pgen.1007795
Stearns, T., Willingham, M. C., Botstein, D., and Kahn, R. A. (1990). ADP-ribosylation factor is functionally and physically associated with the Golgi complex. Proc. Natl. Acad. Sci. U. S. A. 87, 1238–1242. doi: 10.1073/pnas.87.3.1238
Sugano, S. S., Nishihama, R., Shirakawa, M., Takagi, J., Matsuda, Y., Ishida, S., et al. (2018). Efficient CRISPR/Cas9-based genome editing and its application to conditional genetic analysis in Marchantia polymorpha. PLoS One 13:e0205117. doi: 10.1371/journal.pone.0205117
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Teh, O. K., and Moore, I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448, 493–496. doi: 10.1038/nature06023
Waters, M. G., Serafini, T., and Rothman, J. E. (1991). 'Coatomer': a cytosolic protein complex containing subunits of non-clathrin-coated Golgi transport vesicles. Nature 349, 248–251. doi: 10.1038/349248a0
Keywords: Marchantia polymorpha, oil body, membrane traffic, Golgi apparatus, secretory pathway, COPI coat
Citation: Kanazawa T, Nishihama R and Ueda T (2022) Normal oil body formation in Marchantia polymorpha requires functional coat protein complex I proteins. Front. Plant Sci. 13:979066. doi: 10.3389/fpls.2022.979066
Received: 27 June 2022; Accepted: 25 July 2022;
Published: 15 August 2022.
Edited by:
Qiong Zhao, East China Normal University, ChinaReviewed by:
Pengwei Wang, Huazhong Agricultural University, ChinaCopyright © 2022 Kanazawa, Nishihama and Ueda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takashi Ueda, dHVlZGFAbmliYi5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.