
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 October 2022
Sec. Plant Systematics and Evolution
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.978417
 Ran Chu1
Ran Chu1 Xuemin Xu1
Xuemin Xu1 Zhenwei Lu1
Zhenwei Lu1 Yonggui Ma2,3
Yonggui Ma2,3 Han Cheng1
Han Cheng1 Shixin Zhu1
Shixin Zhu1 Freek T. Bakker4
Freek T. Bakker4 M. Eric Schranz4
M. Eric Schranz4 Zhen Wei1*
Zhen Wei1*This study generated and analyzed complete plastome and internal transcribed spacer (ITS) data of 46 Lactuca species, 13 African endemic (AE) Lactuca species, and 15 species from eight related genera in Lactucinae. The new plastome and nuclear ITS sequences were then used to reconstruct the phylogenetic relationships of Lactuca species. The whole-plastome data were used to estimate divergence time and ancestral area reconstruction of the identified major Lactuca lineages. The results showed that Lactuca species are generally similar in plastome size, Guanine and Cytosine (GC) content, gene structure, and categories, although crop lettuce (Lactuca sativa L.) and its gene pool relatives were found to have one unique pseudogene (ψ ndhF), and accD, atpF, cemA, clpP, and rpl22 showed signs of positive selection. Our phylogenomic analysis demonstrated that Lactuca is monophyletic after excluding Lactuca alatipes Collett and Hemsl and AE Lactuca species. AE Lactuca species are morphologically distinct from core Lactuca lineage and need to be excluded from Lactua. The core Lactuca species most likely originated from Asia-Temperate W ~6.82 Mya and then dispersed globally and formed nine clades. Finally, the lettuce gene pool concept was amended according to the phylogenetic and historical biogeographic analyses. This study revised the circumscription of Lactuca, revealed robust phylogenetic relationships within the genus, and provided insights into Lactucinae phylogeny. The lettuce gene pool species could be used as potential genetic resources for lettuce breeding.
Lactuca L. contains cultivated lettuce (Lactuca sativa L.) and its wild relatives and is the second largest (~66 species) in Lactucinae (Asteraceae) (Kilian et al., 2009). Lactucinae taxa have been considered as complex and difficult to classify due to variable morphological characteristics and uncertain limitations among genera. In the last decade, the phylogenetic, molecular dating, and biogeographic analyses of Lactucinae at the subtribe level, based on non-coding nuclear marker [A44, internal transcribed spacer (ITS), external transcribed spacer (ETS)] and plastid DNA regions, revealed the general relationships and divergence among Lactuca and its allied genera (Wang et al., 2013; Kilian et al., 2017a; Kilian et al., 2017b; Wei et al., 2017; Güzel et al., 2021).
Lactuca wild species are distributed in Asia, Europe, Africa, and America (Lebeda et al., 2004a; Lebeda et al., 2004b; Lebeda et al., 2019). Wang et al. (2013) used extensive sampling of Chinese-centered Lactucinae taxa, based on ITS and five plastid DNA regions, and identified six core lineages in the subtribe, named Cicerbita Wall., Cicerbita II, Lactuca, Melanoseris Decne., Notoseris Shih, and Paraprenanthes Chang ex Shih. The “Cicerbita II lineage” was taxonomically referred to as Kovalevskiella Kamelin in the following study of Lactucinae, and Prenanthes purpurea L. and Lactuca triquetra Benth. and Hook.f. were both found to be monotypic genera (Kilian et al., 2017a; Kilian et al., 2017b). L. triquetra, endemic to Lebanon and Cyprus, was then transferred from Lactuca and revised as Astartoseris triquetra (Labill.) N. Kilian, Hand, Hadjik., Christodoulou and M. Bou Dagher-Kharrat (Kilian et al., 2017a). Wei et al. (2017) found that Lactuca species were polyphyletic, and African endemic (AE) Lactuca species + Asian Melanoseris bracteata (Hook.f. and Thomson ex C.B.Clarke) N.Kilian were sisters to a large polytomy including Lactuca and Melanoseris species. A diversification study of the Lactucinae using ITS and five non-coding plastid regions illustrated the Sub-Paratethyan origin of the subtribe and indicated close relationships between native AE Lactuca species and Melanoseris lineages (Kilian et al., 2017b). Jones et al. (2018) studied Northern Hemisphere disjunctions of Lactuca with a focus on the allopolyploid group endemic to America, using nuclear ribosomal DNA (ETS and ITS), a low-copy nuclear marker (A44) and five non-coding plastid markers. The phylogeny and systematics of the Lactucinae in SW Asia were analyzed and discussed based on ITS and five plastid regions, although the Lactucinae branch had many polytomies (Güzel et al., 2021). Phylogenetic relationships within Lactuca (~66 species) and the circumscription of Lactuca among its related genera were so far not well-resolved because of polytomies in the phylogenetic trees (Wang et al., 2013; Kilian et al., 2017b; Güzel et al., 2021).
High-throughput sequencing technology has helped resolve the origin and domestication history of cultivated lettuce and its most closely related wild species (Wei et al., 2021). However, the circumscription of Lactuca and the delimitation among Lactuca and other related lineages in Lactucinae were uncertain, and the phylogenetic relationships constructed using non-coding nrDNA or plastid regions were lacking robust support. A well-resolved phylogenetic tree could also help to identify lettuce gene pool species, meaning wild Lactuca could hybridize with cultivated lettuce (L. sativa) and generate fertile/sterile offspring or be partly cross-fertile with the crop (Zohary, 1991; van Treuren et al., 2012). In this study, we newly sampled plastomes of 46 wild Lactuca species plus 13 AE Lactuca species containing all of the biogeographic groups. Using these data, this paper aims to address four topics: 1) the general structure and characteristics of lettuce plastomes and the positive selection of plastid protein coding sequences (CDSs); 2) the taxonomic position and the circumscription of Lactuca and the main phylogenetic groups within the genus; 3) the origin of Lactuca and the historical biogeography of its ingroups; and 4) lettuce gene pool category revision.
Plant leaf materials were mainly collected from international herbaria, including B, BM, CSH, K, E, PE, WAG and ZZU (Figure 1; Supplementary Table S1; herbarium code referring to Index Herbariorum, http://sweetgum.nybg.org/science/ih/). All of the necessary permissions for using herbarium accessions were approved by the respective curators. Two fresh-collected leaf samples of the Centre for Genetic Resources, Netherlands (Supplementary Table S1), were provided by Dr. MJW (Marieke) Jeuken (Wageningen University).
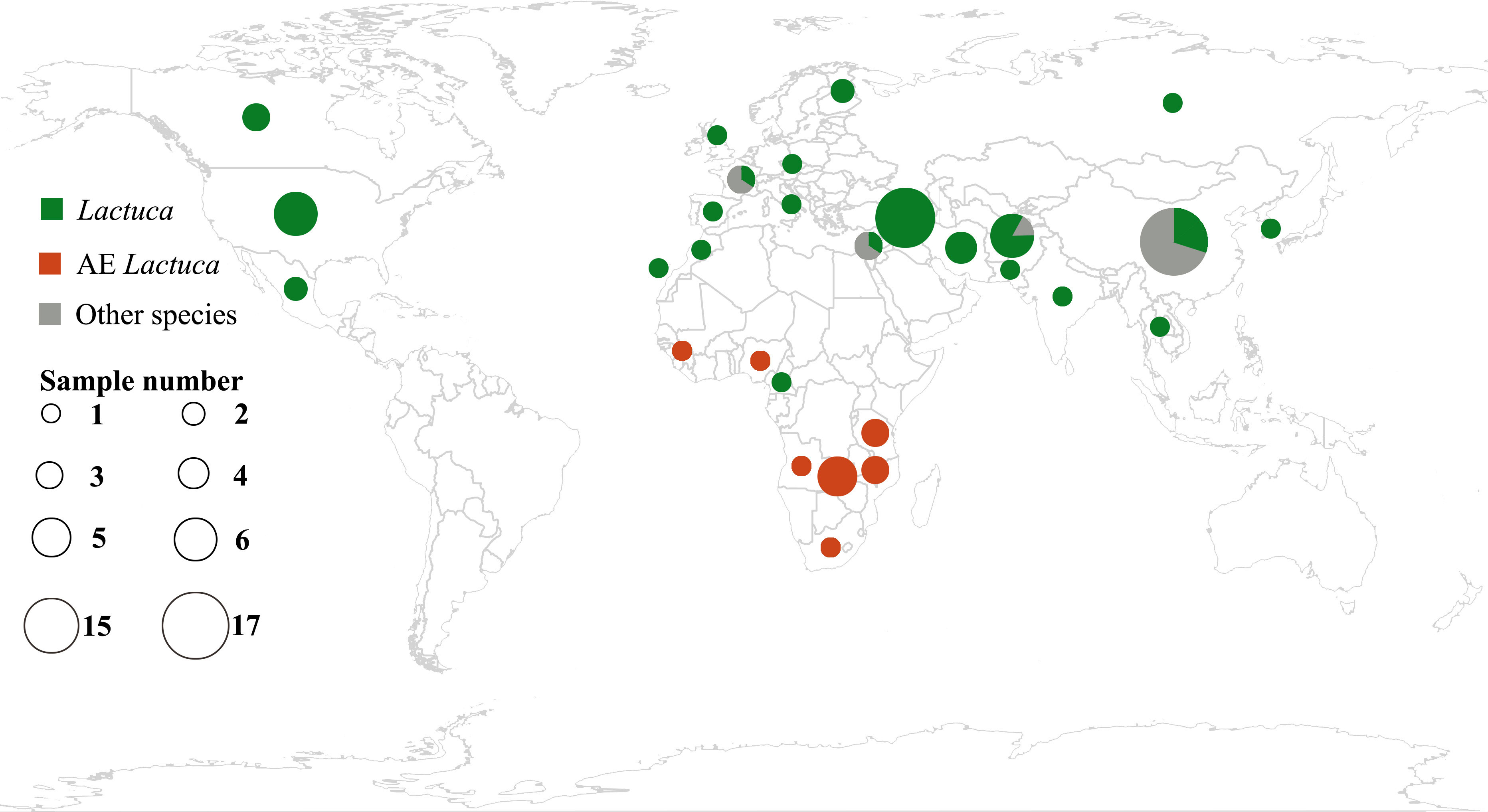
Figure 1 Geographic distribution of samples of Lactuca and related species using R package “rworldmap” (https://cran.r-project.org/web/packages/rworldmap/) and “ggplot2” (https://cran.r-project.org/web/packages/ggplot2/).
This study follows the concept of Lactuca and Lactucinae as indicated in the Cichorieae Portal (Kilian et al., 2009). After excluding species with ambiguous concepts or description, we identified 66 Lactuca species. Our Lactuca sampling (46 species) thus covers 70% of the genus and all geographical groups (Lebeda et al., 2004b; Kilian et al., 2017b). We also included species from eight related Lactucinae lineages: Astartoseris, Cicerbita, Faberia Hemsl. ex F. B. Forbes and Hemsl, Kovalevskiella [Cicerbita auriculiformis (C. Shih) N. Kilian and Cicerbita azurea (Ledeb.) Beauverd], Melanoseris [Melanoseris decipiens (C.B.Clarke) N.Kilian and Ze H.Wang and Lactuca crambifolia Boiss.], Notoseris, Paraprenanthes, and Prenanthes (Kilian et al., 2017b). In the taxon sampling, four complete plastomes were downloaded from NCBI, nine from China National GeneBank DataBase (https://db.cngb.org/), and two partial cp genomes were from a previous publication (Pouchon et al., 2018) (Supplementary Table S2). In total, this study comprised 111 accessions (96 of which are new), representing a total of 74 Lactucinae species and three outgroups (one species in Cichoriinae and two in Scorzonerinae) (Supplementary Tables S1, S2).
Total DNA was extracted from herbarium leaf material using a modified cetyltrimethyl-ammonium-bromide (CTAB) method (Doyle and Doyle, 1987) and purified with Wizard DNA Clean-Up System (Promega Corp.) (Wei et al., 2017). In total, DNA extracts were successfully obtained from 96 Lactuca and related taxa (Supplementary Table S1).
Eight-four DNA extracts were sequenced using Illumina HiSeq Xten/4000/Novaseq platforms (Novogene Inc., Tianjin, China). The other 12 samples were sequenced by the National High-Throughput DNA Sequencing Centre of the University of Copenhagen using Illumina HiSeq 2000 platform (http://seqcenter.ku.dk/facilities/). The clean data were assembled using NOVOPlasty v4.0 (Dierckxsens et al., 2016) with references AP007232 and DQ383816 and Get Organelle e-1.7.3.2 (Jin et al., 2020) with default settings and angiosperm plastome references. The assembled cp genomes were annotated and manually corrected in Geneious Prime 2020.1.2 (https://www.geneious.com). The annotated tRNA genes were checked using the tRNAscan-SE 2.0 Web Server (Chan et al., 2021). The physical maps of the cp genomes were visualized using OrganellarGenomeDRAW (Greiner et al., 2019). The same clean data of 96 samples were also used to assemble ITS sequences using Get Organelle e-1.7.3.2 (Jin et al., 2020) with reference ITS sequences of L. serriola (LT721932), L. georgica (LT722049), and L. altaica (LT722033). The ITS DNA regions were then extracted from the assembled sequences. Thirty-two previously published ITS sequences (Wang et al., 2013; Kilian et al., 2017b; Güzel et al., 2021) were downloaded and added to the phylogenetic analysis (Supplementary Table S3).
The CDSs of 108 plastomes of Lactucinae species were extracted and aligned using Geneious Prime 2022.1.1 (https://www.geneious.com). The variation rates, conserved sites, parsimony informative sites, singleton sites, and variation frequency were calculated using MEGA X (Kumar et al., 2018). The plastomes of Lactuca and AE Lactuca (59 species, one sequence per species) were uploaded to mVISTA, and the alignment and variation were visualized with Shuffle-LAGAN mode (Mayor et al., 2000). The annotation of L. sativa (AP007232) was used as a reference. The cp genes of Lactuca and AE Lactuca (59 species, one sequence per species) near the boundaries of the large single-copy region (LSC), small single-copy region (SSC), and two inverted repeat regions (IRs) were visualized and compared using IRscope (https://irscope.shinyapps.io/irapp/) (Amiryousefi et al., 2018).
The SSC inversions in plastomes were reversed first and then aligned with all plastome sequences in MAFFT version 7 (https://mafft.cbrc.jp/alignment/software/). Gelasia hirsuta (Gouan) Zaika, Sukhor. and N.Kilian, Pseudopodospermum hispanicum (L.) Zaika, Sukhor. and N.Kilian of Scorzonerinae and Cichorium intybus L. of Cichoriinae were used as outgroups. The phylogenetic analyses, using whole-plastome sequences, were constructed by maximum likelihood (ML) and Bayesian inference (BI) methods in CIPRES Science Gateway using the tool of RAxML-HPC2 on XSEDE 8.2.12 and MrBayes on XSEDE 3.2.7a (https://www.phylo.org/portal2/login!input.action). The RAxML tree was inferred with 1,000 bootstrap replicates using the GTR + GAMMA model for the nucleotide datasets; “print branch length (-k)” under “Configure bootstrapping” and other parameters used default settings. The parameters of BI analyses in MrBayes were as follows: lset nst = 6 rates = gamma; unlink statefreq = (all) revmat = (all) shape = (all) pinvar = (all); prset applyto = (all) ratepr = variable; mcmcp ngen = 10000000, relburnin = yes, burninfrac = 0.25, printfreq = 1000, samplefreq = 10000, nchains = 4 temp = 0.05. The burn-ins and effective sampling sizes (ESSs) of all parameters were checked for convergence between simultaneous runs in Tracer v.1.7.1 (Rambaut et al., 2018).
The rDNA ITS sequences from Lactuca and related species were also used for constructing phylogenetic trees following the same described methods as above for plastomes. All new ITS sequences were submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession number OP070064–OP070156. FigTree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) and iTOL v6 (Letunic and Bork, 2021) were used to visualize and edit the output trees.
The chromosome number data were downloaded and collected from the Chromosome Counts Database (http://ccdb.tau.ac.il/) and previous publications (Jones et al., 2018). The pseudogenes were manually checked and recorded in Geneious Prime 2022.1.1 (https://www.geneious.com). Chromosome numbers and the pseudogenes of Lactuca and related species were identified, analyzed, and plotted on the phylogenetic tree.
Selection pressures on the plastid CDS of 72 Lactuca taxa (representing 45 species) were analyzed in Datamonkey 2.0 (http://www.datamonkey.org/) using Single-Likelihood Ancestor Counting method with universal code (Sergei et al., 2005; Weaver et al., 2018). Lactuca alatipes Collett and Hemsl. was excluded in this analysis due to its phylogenetic position outside Lactuca in our phylogenetic analyses (see Results). The CDSs of 12 lettuce gene pool species (representing 23 taxa) (Zohary, 1991; van Treuren et al., 2012), L. sativa, Lactuca altaica Fisch. and C.A.Mey., Lactuca aculeata Boiss., Lactuca serriola L., Lactuca georgica Grossh., Lactuca scarioloides Boiss., Lactuca virosa L., Lactuca saligna L., Lactuca viminea (L.) J.Presl and C.Presl, Lactuca reviersii Litard. and Maire, Lactuca tetrantha B.L.Burtt and P.H.Davis, and Lactuca orientalis Boiss., were also checked for selection pressure analysis. After excluding pseudogenes, 61 plastid CDSs shared by all of the Lactuca taxa and 73 plastid CDSs shared by lettuce gene pool taxa were used in the analyses.
A total of 78 complete cp genomes (each plastome representing one species) of Lactuca and its related lineages were used in the divergence time estimation. The age of Lactuca and Lactucineae taxa was calculated with the Strict Clock model and GTR + Gamma site model in BEAST v2.6.0 (Bouckaert et al., 2019). The Calibrated Yule Model was selected as tree priors. Three nodes were calibrated: 1) The oldest reported Cichorieae fossil (early Miocene, 22.0–28.4 Mya) of the C. intybus type pollen, distributed widely in all Cichorieae subtribes except Scorzonerinae (Hochuli, 1978; Tremetsberger et al., 2012; Kilian et al., 2017b); 2) The secondary calibration prior was selected for the stem node of Lactuca lineage [8.0 Mya, 95% highest posterior density (95% HPD) 5.8–10.2 Mya] (Kilian et al., 2017b); 3) The third calibration prior was set for a group of four species (Lactuca intricata Boiss., Lactuca undulata Ledeb., Lactuca glauciifolia Boiss., and Lactuca perennis L.) within Lactuca (4.4 Mya) from Kilian et al. (2017b) calculated by plastid genes.
The Monte Carlo Markov chains (MCMCs) were run for 100,000,000 generations, and the tree was set to sample every 10,000 generations. The first 10% of trees were discarded as burn-ins. The burn-ins and ESSs of all parameters were checked for convergence between simultaneous runs in Tracer v.1.7.1 (Rambaut et al., 2018). The trees were resampled at lower frequency of 30,000 using LogCombiner in BEAST v2.6.0 (Bouckaert et al., 2019). TreeAnnotator in BEAST was used to summarize the most probable trees into a maximum clade credibility (MCC) tree with median node heights and visualized in FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
The geographic information of taxon distribution was downloaded from the Global Biodiversity Information Facility (GBIF, https://www.gbif.org/) database using R package “dismo” (https://cran.r-project.org/web/packages/dismo/) and compared with the native distribution of Lactuca on the Cichorieae Portal (Kilian et al., 2009). The ancestral area reconstruction was estimated using native distribution data in RASP V4.2 (Yu et al., 2020), and the Statistical Dispersal-Vicariance-Analysis (S-DIVA) model was used to reconstruct the ancestral range of each node using the tree files generated by BEAST v2.6.0 (Bouckaert et al., 2019). The regions of Lactuca and its related genera were counted and modified from Kilian et al. (2017b) and Jones et al. (2018) (A: N + Central + E Europe, B: SW +SE Europe, C: N Africa + Macaronesia, D: Tropical Africa, E: S Africa, F: Asia-Temperate W, G: Siberia + Russian Far East, H: Asia-Temperate E, I: Indian Subcontinent, J: Asia-Tropical E, K: N America, L: S America).
This study newly sequenced and assembled 96 plastomes of Lactuca and related taxa (representing 74 species, see Supplementary Table S1). The length of complete plastomes of Lactuca species (including native African ones) ranges from 152,076 to 152,947 bp (Figure 2A; Supplementary Figure S1), and the sizes of LSC, SSC, and IR of Lactuca species (including native African ones) are from 83,469 to 84,271 bp, 18,326 to 18,780 bp, and 24,849 to 25,106 bp (Supplementary Table S4). The GC contents of Lactuca species (including native African ones) are stable, ranging from 37.5% to 37.7% (Supplementary Table S4). The number of plastid genes of Lactuca species (including native African species) is from 126 to 134 due to the presence of pseudogenes. The information on pseudogenes was recorded and classified (Supplementary Table S5) for further comparison with phylogenetic analyses. The number of rRNA (8) and tRNA (37) genes were consistent across all taxa (Supplementary Table S4). Other Lactucinae species have similar cp genome lengths, GC contents, and numbers of genes (Supplementary Table S4).
Minor variations in plastome length of Lactuca and AE Lactuca (one sequence per species, in total 59 species) were observed, mostly in the SSC. Whereas it was relatively conserved across the IRs (Supplementary Figure S2). Photosynthesis-related genes were the most conserved genes, whereas ycf1, matK, accD, rpl22, ccsA, and rpl36 had the highest variation (Figure 2B). The genes with the largest number of parsimoniously informative sites were ycf1, rpoC2, matK, ycf2, ndhF, accD, and rpoB (Figure 2C).
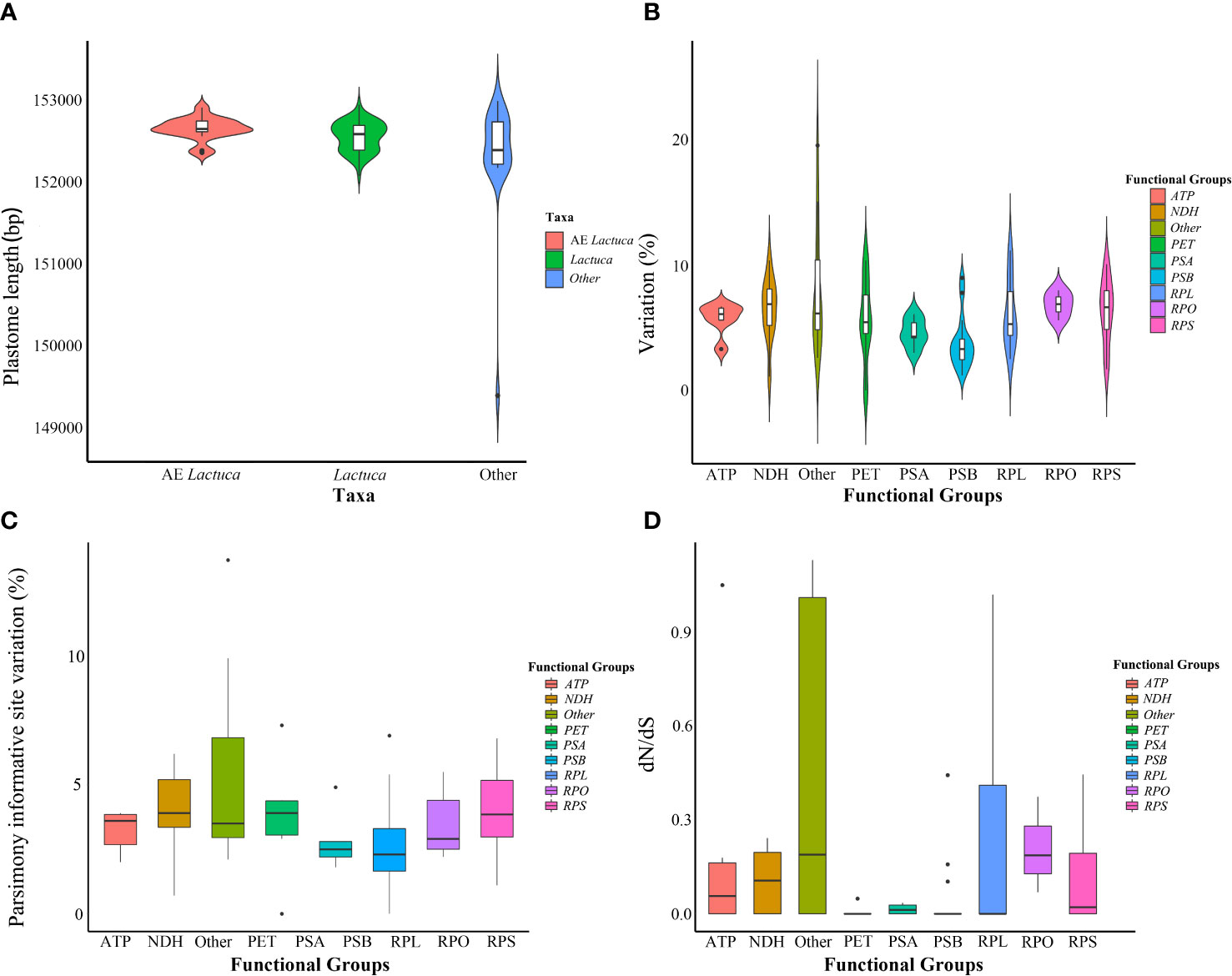
Figure 2 Information on plastomes of Lactuca and African endemic Lactuca species. (A) Plastome length. (B) Variation of different functional groups. (C) Parsimony informative site variation of different functional groups. (D) Non-synonymous/synonymous rate ratios (dN/dS) of protein-coding genes of different functional groups in cultivated lettuce gene pool species.
The same Lactuca and AE Lactuca taxa were used for the boundary comparison of LSC, SSC, and two IRs. The results showed slight contraction and expansion of the genes near the junction sites. Thirty species (34 taxa), 13 of which were AE Lactuca species, had one inverted SSC region (Supplementary Figure S3). The gene number and type of plastomes of those species with inverted SSC were similar to other plastomes with regular SSC.
The ML and BI trees generally shared the same topologies (Figures 3, 4). In the results, most of the nodes of the phylogenetic tree were fully supported with BS = 100 and PP = 1. Branches of the phylogenetic trees with bootstrap <50 were collapsed. Chromosome numbers and presence of pseudogenes of Lactuca and related species are indicated at the responding nodes (Figures 3, 4).
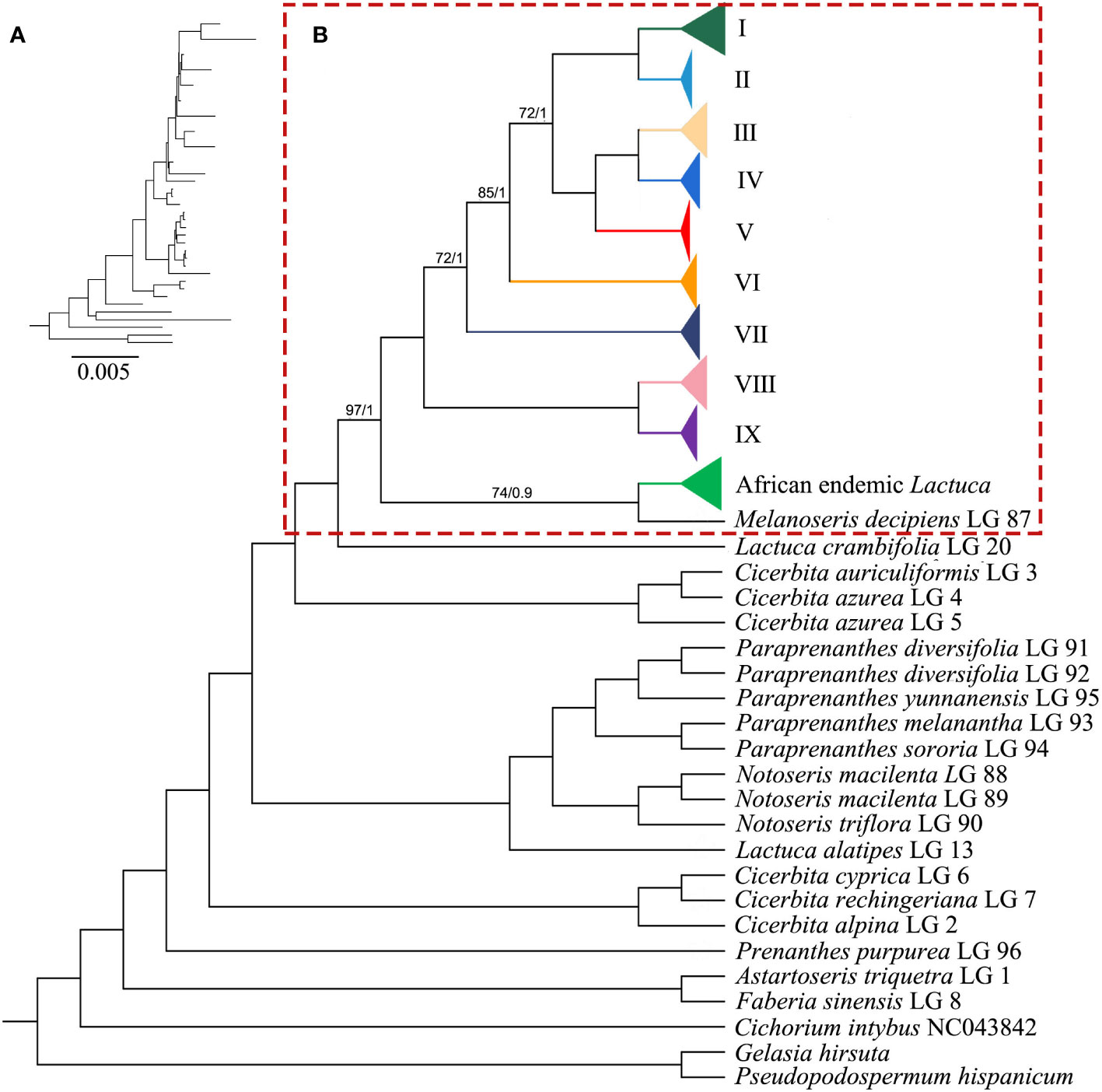
Figure 3 Phylogenetic cladogram of Lactuca and related genera based on complete plastomes. (A) Phylogenetic tree with branch length. (B) Phylogenetic cladogram with collapsed clades of Lactuca and AE Lactuca species. The width of the triangle represents the number of species. Support values are maximum likelihood bootstrap (BS) / Bayesian posterior probability (PP). No support values are shown for nodes with full support (BS = 100, PP = 1).
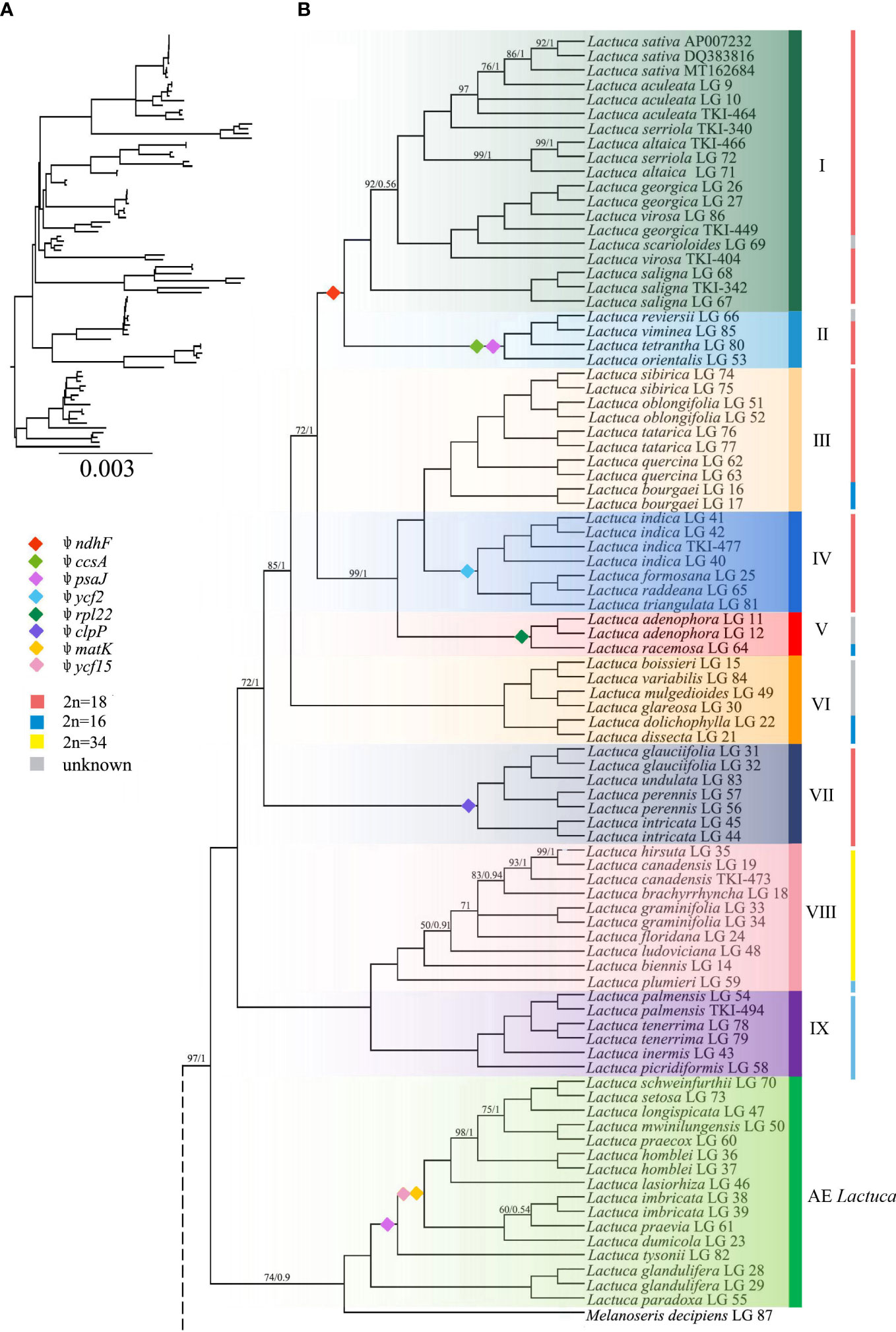
Figure 4 Phylogenetic tree of Lactuca and AE Lactuca species inferred from plastomes. (A) Phylogenetic tree with branch length. (B) ML/BI tree, the color is consistent with that in Figure 3. The nodes with full support (BS = 100, PP = 1) are not shown. The information on pseudogene and reverse SSC is indicated on the branches. The vertical line on the right represents the number of chromosomes. Support values are maximum likelihood bootstrap (BS) / Bayesian posterior probability (PP). No support values are shown for nodes with full support (BS = 100, PP = 1).
The tree was rooted with three outgroup species [G. hirsuta (Scorzonerinae), P. hispanicum (Scorzonerinae), and C. intybus (Cichoriinae)]. In Lactucinae, A. triquetra and Faberia sinensis Hemsl. are closely related and sisters to other Lactucinae species. The monotypic P. purpurea is a sister to three Cicerbita species and five other Lactucinae lineages. L. alatipes is found to be related to Notoseris and Paraprenanthes species other than Lactuca species. We downloaded the plastid DNA sequences of Paraprenanthes species from a previous publication (Wang et al., 2013), in which Lactuca parishii Craib ex Hosseus LAC 028 was treated as Paraprenanthes umbrosa (Dunn) Sennikov but later revised as P. parishii (Kilian et al., 2017b) and reconstructed a RAxML tree (Supplementary Figure S4; Supplementary Table S6). Three Kovalevskiella taxa (C. azurea LG 4, C. azurea LG 5, and C. auriculiformis LG 3) are the sisters to taxa of Melanoseris (L. crambifolia and M. decipiens), the AE Lactuca, and the core Lactuca (Clades I–IX).
The Melanoseris lineage (sensu in Kilian et al., 2017b) member L. crambifolia is a sister to M. decipiens + AE Lactuca Clade (Figure 4), containing 16 samples (13 species) endemic to Africa, and all of them are sisters to the core Lactuca clade. Thirteen taxa in this clade have reversed SSC regions, excluding Lactuca setosa Stebbins ex Jeffrey, Lactuca lasiorhiza (Hoffm.) Jeffrey, and Lactuca paradoxa Sch.Bip. ex A.Rich. Thirteen AE Lactuca species (excluding Lactuca glandulifera Hook.f. and L. paradoxa) share a common pseudogene (ψ psaJ), and 12 taxa, excluding L. glandulifera, L. paradoxa, and Lactuca tysonii (E.Phillips) C.Jeffrey, share two pseudogenes (ψ matK and ψ ycf15).
The core Lactuca was identified to comprise nine clades (Figures 3, 4). Clade IX and Clade VIII are sisters. Clade IX includes four species (six taxa) with a chromosome number of 16 (2n = 16). Clade VIII contains eight species, and seven of them are American native (especially N American endemic, NAE) Lactuca with a chromosome number of 34 (2n = 34). The only exception is Lactuca plumieri Gren. and Godr., distributed in Europe (2n = 16).
Most species in Clades I–VII have a chromosome number of 18 (2n = 18) despite some taxa with 16 or unknown chromosomes. The four Lactuca species (2n = 18) in Clade VII are sisters to all the other species in Clades I–VI. L. perennis LG 57, L. intricata LG 44, and L. intricata LG 45 have reverse SSC. Contraction and expansion of the junction sites of the cp genomes were observed in L. perennis LG 56, L. perennis LG 57, and L. intricata LG 44. Clades III, IV, V, and VI form a sister clade to a clade comprising Clade I (the crop lettuce clade) and Clade II. Clade VI contains six Lactuca species, and three of them have reverse SSC. Lactuca dolichophylla Kitam. LG 22 and Lactuca dissecta D.Don LG 21 are sister groups with the same chromosome number (2n = 16) and showed contraction and expansion near the junction sites of the cp genomes. Lactuca variabilis Bornm. LG 84, Lactuca mulgedioides Boiss. and Kotschy ex Boiss. LG 49, and L. dissecta LG 21 were observed to have reverse SSC.
The species in Clade V share one common pseudogene (ψ rpl22). Lactuca racemosa Willd. LG 64 (2n = 16) has a reversed SSC, and the genes near the junction sites of the cp genomes of Lactuca adenophora Boiss. and Kotschy LG 11 showed contraction to some extent. Clade IV includes four species (2n = 18), and they are sister groups to species in Clade III with mixed chromosome numbers (2n = 16, 18). In Clade IV, all of the species were Pterocypsela C.Shih and share a pseudogene (ψ ycf2), and Lactuca indica L. LG 41 has reverse SSC. In Clade III, Lactuca bourgaei Irish and Taylor LG 16 and LG 17 (2n = 16) are sisters to four other species (2n = 18). Some genes near the junction sites of the cp genomes of Lactuca oblongifolia Nutt. LG 51 and LG 52 and L. tatarica LG 76 and LG 77 showed expansion, and L. oblongifolia LG 51 has reverse SSC.
Clade I and Clade II are closely related with the same chromosome number (2n = 18). Clade II includes four species, and three have reverse SSC. All of the species share two pseudogenes, ψ ccsA and ψ psaJ. Clade I comprises 19 Lactuca taxa representing cultivated lettuce and its relatives. Three L. saligna accessions are sisters to all of the other Lactuca taxa in Clade I. L. georgica, L. virosa, and L. scarioloides are sisters to L. serriola, L. altaica, L. sativa, and L. aculeata. Reverse SSC was observed in L. aculeata LG 10, L. sativa DQ383816, L. georgica LG 26, and L. saligna LG 68.
Ninety-three ITS sequences from 73 species were successfully assembled. In the end, a total of 125 ITS sequences from 105 Lactuca and related species were used for phylogenetic analysis. The taxa sampling included 53 Lactuca species, accounting for 80.3% of the genus with a higher coverage than cp trees. However, many nodes of the ML tree support values were lower than 50%, and the BI tree had many polytomies. Therefore, those branches with low node support values were collapsed for confidence (Supplementary Figures S5, S6). As results, the phylogenetic positions of Lactucinae genera were not well resolved. The systematic position of P. purpurea was outside Lactucinae, and A. triquetra plus F. sinensis were parallel to the core groups of Lactucinae. Cicerbita, Kovalevskiella, Notoseris-Paraprenanthes, Melanoseris + L. bourgaei + AE Lactuca, and core Lactuca were in a large polytomy. L. alatipes is a sister to L. parishii in ITS trees, while in plastid trees, it is a sister to Notoseris-Paraprenanthes clade. L. bourgaei is close to Melanoseris clade, consisting of L. crambifolia and M. decipiens, but in chloroplast genome phylogenetic trees, it is in the core Lactuca.
Phylogenetic relationships of the core Lactuca lineages inferred from ITS sequences had similar clades as the plastome trees. The main incongruences between the two types of phylogenetic trees were in Clades III and VI. The two clades in the plastome tree were divided into two and three clades, respectively, in ITS trees. Clade III-1 is Lactuca quercina L., sister group to the crop lettuce and its relatives (Clade I). Clade III-2 contains L. oblongifolia, Lactuca sibirica Benth. ex Maxim., and L. tatarica and is a sister to Clade II. Clade III-3 is L. bourgaei within the Melanoseris Clade, a sister to AE Lactuca species. Clade VI was divided into two parts in ITS results, Clade VI-1 containing seven species and Clade VI-2 including L. dolichophylla and L. dissecta.
After excluding pseudogenes, 61 plastid CDSs shared by all of the Lactuca taxa and 73 plastid CDSs shared by lettuce gene pool taxa (Clade I and II) were used respectively to analyze the possible presence of positive selection. The results showed purifying selection on 61 common protein-coding genes of all Lactuca lineages (Supplementary Figure S7). The selection pressure analysis on 73 shared CDSs of the 12 lettuce gene pool species and cultivated lettuce demonstrated that five genes were under positive selection (dN/dS > 1), namely, accD, atpF, cemA, clpP, and rpl22 (Figure 2D; Supplementary Figure S8).
As the phylogenetic trees generated using ITS sequences contained polytomies, we only did further divergence time analysis using cp genomes. The topologies of phylogenetic trees constructed based on cp genomes using BEAST, RAxML, and MrBayes were similar despite the incongruent positions of M. decipiens and Clade VI. In ML and BI trees, M. decipiens is the sister group to the AE Lactuca species (Figure 3), while it is the sister to all of the Lactuca lineages (including AE Lactuca taxa) in the BEAST-dated tree (Figure 5). Most of the nodes in the dated phylogram of Lactuca and its related genera in Lactucinae were fully supported (PP = 1). The most recent common ancestor (MRCA) of Lactucinae and Cichoriinae started diverging around 18.35 Mya (Table 1). The MRCA of Lactucinae existed around 16.01 Mya in Asia-Temperate W (PP = 1; Figure 5). The core Lactucinae (excluding Astartoseris, Faberia, and Prenanthes) was estimated to diverge around 9.79 Mya with an ancestral area of Asia-Temperate W (PP = 1). Paraprenanthes and Notoseris lineages share an MRCA from Asia-Temperate E (PP = 1) around 2.04 Mya (Table 1).
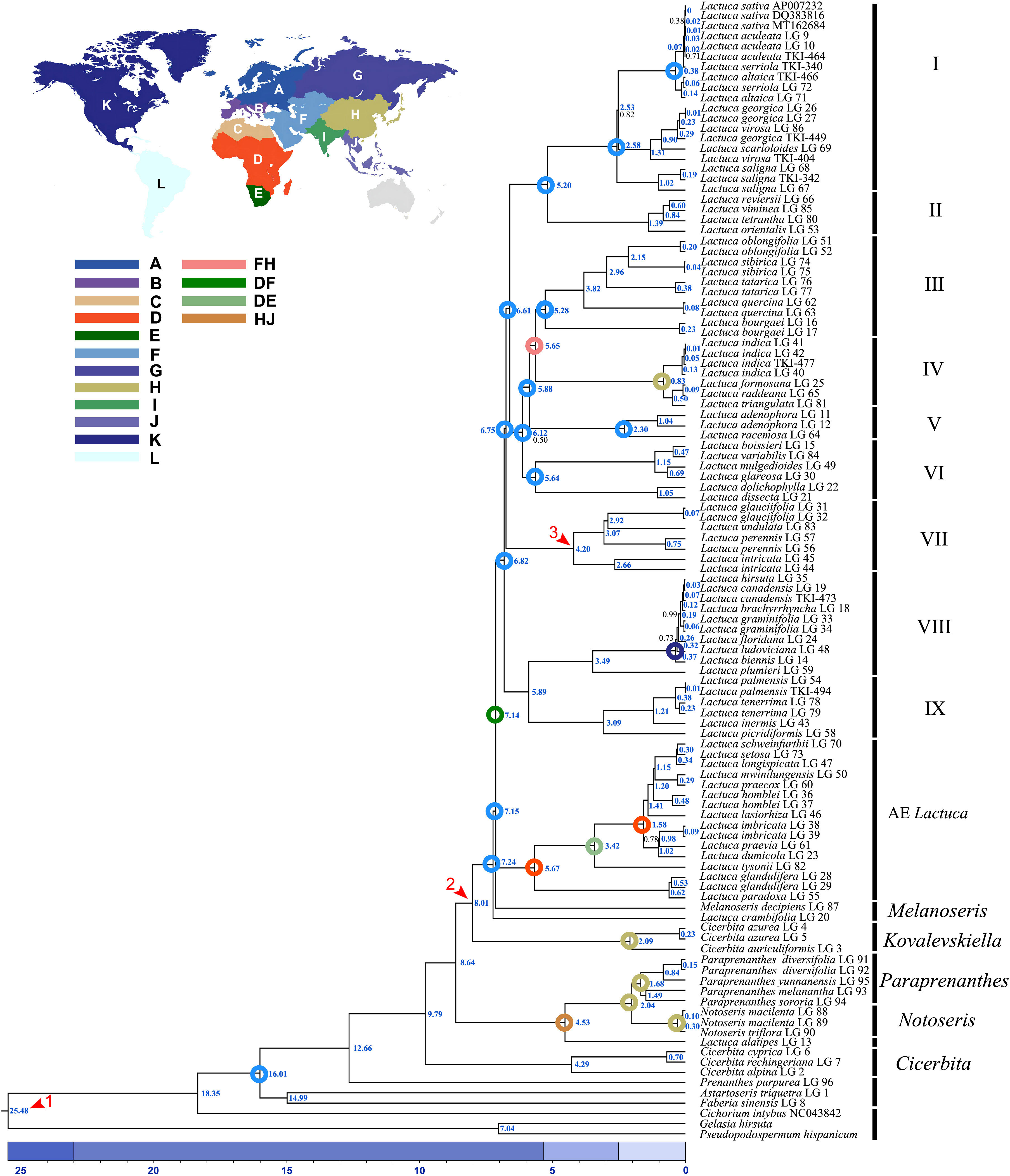
Figure 5 Dated phylogram of Lactuca and related genera based on complete plastomes. The red arrows are the calibration points of divergence time. The blue numbers indicate the median node age of divergence, and the black numbers indicate the support of Bayesian posterior probability (PP = 1 is not displayed; colored circles displayed on nodes are ancestral ranges with probability = 1, except the one of Lactuca lineages). Geographical areas are indicated with different colors: (A) N + Central + E Europe, (B) SW + SE Europe, (C) N Africa + Macaronesia, (D) Tropical Africa, (E) S Africa, (F) Asia-Temperate W, (G) Siberia + Russian Far East, (H) Asia-Temperate E, (I) Indian Subcontinent, (J) Asia-Tropical E, (K) N America, (L) S America.
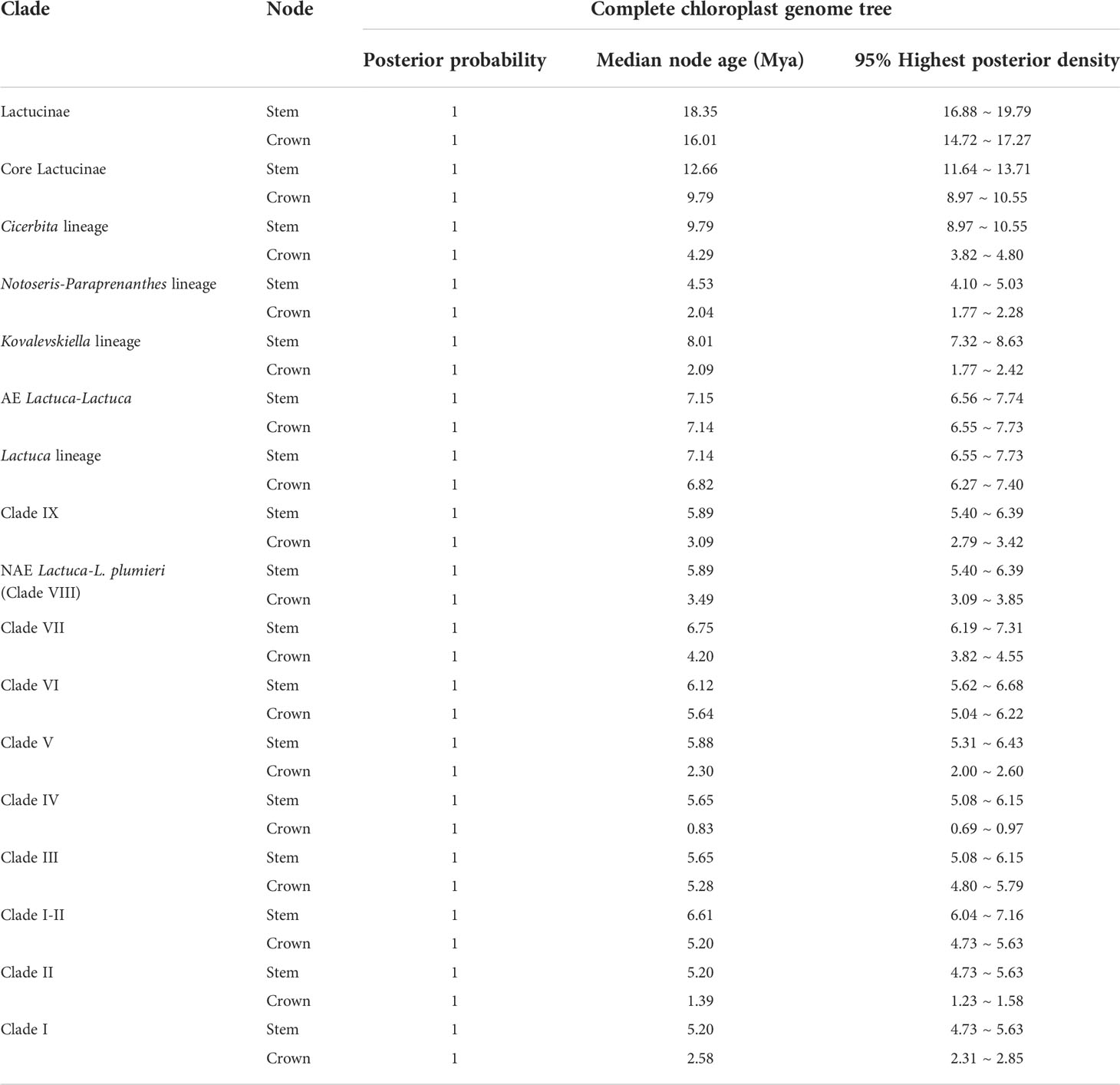
Table 1 Estimated divergence time of Lactuca and related genera based on the complete plastome sequences.
The divergence of the AE Lactuca and the core Lactuca lineage began from an MRCA distributed between Tropical Africa and Asia-Temperate W around 7.14 Mya. The MRCA of the AE Lactuca (Figure 5) very likely originated from Tropical Africa (PP = 1), and the recent speciation events occurred around 5.67 Mya (Table 1). After dispersal and vicariance events, the AE Lactuca lineage is now distributed in Tropical Africa (D) and S Africa (E).
Around 6.82 Mya, the non-African Lactuca lineage was inferred to have a possible MRCA from Asia-Temperate W (F). The Lactuca lineage then diffused to Europe, Asia, Indian Subcontinent, and North America after a series of dispersal, vicariance, and extinction events (Supplementary Figure S9; ancestral area estimation in Supplementary Material 1). Lactuca species in Clades I–VII share an MRCA originating from Asia-Temperate W with a probability of 1. The MRCA of the NAE Lactuca (excluding L. oblongifolia) and L. plumieri (Clade VIII) began to diverge around 3.49 Mya, and thereafter, the MRCA of NAE Lactuca diverged around 0.37 Mya and dispersed from N America to S America.
In this study, we used 46 Lactuca species plus 13 AE Lactuca species from all geographic (European, Asian, African, and American) groups to reconstruct deep-level phylogenetic relationships and estimate the historical biogeography of Lactuca. We will discuss four main issues: 1) the plastid genome evolution of cultivated lettuce and wild Lactuca species; 2) the delimitation among Lactuca and other lineages in Lactucinae and AE Lactuca species; 3) the origin, phylogeny, and historical biogeography of Lactuca and the main phylogenetic ingroups; and 4) revision to the lettuce gene pool category.
This study successfully sequenced, assembled, and annotated 96 complete plastomes of Lactuca lineages and related species. Generally speaking, the newly generated Lactuca plastomes showed similarities (Clade I to Clade IX) in plastome size, GC content, structure, and gene category, as well as published plastomes of other Asteraceae lineages (Pascual-Diaz et al., 2021).
The plastomes of Lactuca and AE Lactuca species (one sample per species) were used for variation analysis and boundary comparisons. The intergenic spacer regions, intron sequences, and genes (such as accD, matK, ndhF, rpl22, and ycf1) with high variation and parsimoniously informative sites could be used for further common DNA barcode selection and fast identification of Lactuca and Lactucinae species (Amar, 2020; Meena et al., 2020; Li et al., 2021; Song et al., 2022; Tang et al., 2022). The junction sites of LSC, SSC, and IRs of Lactuca and AE Lactuca were conserved. Reverse SSC regions in chloroplast genomes were detected in 30 Lactuca and related species (34 taxa), especially in AE Lactuca species (13 taxa). It has been demonstrated that plastome has two equimolar states that differ in the relative orientation of SSC regions (Palmer, 1983). Reverse SSC regions were identified in other Asteraceae plastome studies as well, such as cultivated lettuce, sunflower (Helianthus annuus L.), and Guizotia abyssinica Cass. (Timme et al., 2007; Dempewolf et al., 2010). It has been pointed out that heteroplasmy in SSC orientation of plastomes within individual plants should not be overlooked in phylogenetic analyses (Walker et al., 2015). In this study, isomer inversions were also observed in two different individuals of one species (e.g., L. saligna and L. georgica). Therefore, reverse SSC orientations among lineages could be considered as natural variations within and between individuals instead of phylogenetic signals.
There are two common pseudogenes (ψ rps19 and ψ ycf1) caused by the incomplete DNA fragments located on the IR boundaries of chloroplast genomes of Lactuca and AE Lactuca, which had been widely found in Asteraceae plastome studies (Wang et al., 2021). Another two common pseudogenes, ψ ndhF and ψ psaJ, were shared by certain related taxa. The pseudogenization of ψ ndhF was detected in 23 Lactuca taxa in Clades I and II, in which most of the species were considered as lettuce gene pool species (Koopman et al., 1998; Lebeda et al., 2007; van Treuren et al., 2012). Plastid NDH gene loss or pseudogenization has been reported in many angiosperms (Lin et al., 2017; Fu et al., 2021; Li et al., 2021). The pseudogenization of ψ psaJ occurred in four Lactuca and 13 AE Lactuca samples as results of early termination of genes. Photosystem I complex is composed of protein encoded by multiple chloroplast genes, including psaJ, psaA, psaB, psaC, and psaI, and nuclear genes (Varotto et al., 2002). Previous studies found that the growth rate and assimilation ability of psaJ-knockout mutants by chloroplast transformation in tobacco (Nicotiana tabacum) remained unchanged and were inhibited only under limited light intensity (Schottler et al., 2007). Pseudogenization of ψ ndhF and ψ psaJ in Lactuca and AE Lactuca does not seem to affect their growth significantly. A possible reason could be that functional copies of the genes have been transferred into the mitochondrial or nuclear genomes (Lin et al., 2015; Daniell et al., 2016).
The positive selection of Clade I and Clade II species (including L. sativa, L. altaica, L. aculeata, L. serriola, L. georgica, L. scarioloides, L. virosa, L. saligna, L. viminea, L. reviersii, L. tetrantha, and L. orientalis) was detected in five plastid-coding genes, accD, atpF, cemA, clpP, and rpl22.
In general, the plastid genes under positive selection of lettuce gene pool species were found to be involved in the processes of leaf and chloroplast development, photosynthesis, protein synthesis, fatty acid synthesis, other secondary metabolite synthesis, and protein differentiation and degradation, as well as energy metabolism (Hudson and Mason, 1988; Katoh et al., 1996; Majeran et al., 2000; Kode et al., 2005; Bock, 2007; Fleischmann et al., 2011; Olinares et al., 2011; Rodriguez-Concepcion et al., 2019; Caroca et al., 2021).
The concept of Lactuca has been revised multiple times during the last century. The global distribution of Lactuca species was summarized and classified into four geographic groups (European, Asian, African, and American) (Lebeda et al., 2004b; Lebeda et al., 2019). Using ITS and five plastid markers, the molecular phylogeny of Lactucinae focusing on the Chinese lineages identified six lineages (Cicerbita, Cicerbita II, Lactuca, Melanoseris, Notoseris, and Paraprenanthes) as a monophyletic core group within the subtribe, and Melanoseris and Lactuca were inferred as sister groups (Wang et al., 2013). A new genus of Lactucinae, Astartoseris, was proposed as monospecific, and L. triquetra was designated as the type species (Kilian et al., 2017a). The contradictory systematic positions of Astartoseris (L. triquetra), Faberia, and Prenanthes inferred from nuclear and plastid phylogenetics were observed and discussed in previous studies, and eventually they were retained in Lactucinae based on morphology, karyology, and potential origins discussed in Wang et al. (2013); Kilian et al. (2017a), and Kilian et al. (2017b). Therefore, the three lineages were not discussed further in this work.
The molecular phylogeny using nrDNA, plastid DNA regions, and plastomes of Lactuca, focusing on the AE Lactuca species, recognized Lactuca as a polyphyletic group (Wei, 2016; Wei et al., 2017). Although the supporting values were not very high, the results indicated that the AE Lactuca is very likely to be a sister to Melanoseris. Kilian et al. (2017b) studied the diversification of the Lactucinae (Compositae: Cichorieae) and referred to the “Cicerbita II lineage” as a new genus “Kovalevskiella.” All of the lineages of Lactucinae were recognized until then, although the phylogenetic relationships among Lactuca, AE Lactuca, Melanoseris, Notoseris, and Paraprenanthes were not well resolved due to weak support values.
Compared to non-coding plastid and nrDNA regions, complete plastomes have been shown to be valuable and robust for phylogeny reconstruction of angiosperm groups (Zhao et al., 2020; Su et al., 2021; Wen et al., 2021). The complete plastome sequences in this study illustrated that the core Lactuca lineage is monophyletic with robust support. Cicerbita species are sisters to all of the other lineages, including L. alatipes + Notoseris + Paraprenanthes, Kovalevskiella, and L. crambifolia + M. decipiens + AE Lactuca + core Lactuca lineages. This study is the first molecular sampling of L. alatipes, and the results indicated that this species is very likely to be Notoseris or Paraprenanthes. L. alatipes was described to have a glabrous stem, basal leaves hastate, margin sinuate, and more rarely lyrately pinnate with one pair of small ovate lateral lobes; paniculiform, with few to many capitula; capitula with nine florets; achene reddish, narrow, very compressed, prominently 4–5 ribs on each side (Collett and Hemsley, 1890). We downloaded plastid DNA markers from a previous publication (Wang et al., 2013) and reconstructed phylogenetic analysis of L. alatipes, Notoseris, and Paraprenanthes species (Supplementary Figure S4). L. parishii LAC 028 in Wang et al. (2013) was identified as P. umbrosa, but later corrected as P. parishii LAC 028 in Kilian et al. (2017b). The results of phylogenetic analysis showed that L. alatipes should be transferred from Lactuca to Paraprenanthes. L. alatipes (type from 1890) is conspecific to L. parishii (type from 1911), and both names should be synonymized under P. alatipes.
Our samples included two species of Kovalevskiella lineage endemic to the Pamiro-Alai and western Tian Shan mountain ranges. They were discussed in Kilian et al. (2017b); there will be no more discussion about them in this work.
The plastome phylogeny revealed that Lactuca currently circumscribed (when AE is included) is polyphyletic. AE Lactuca is paraphyletic with respect to Melanoseris species (M. decipiens and L. crambifolia). The taxonomic position of L. crambifolia was identified and discussed in the study of SW Asian-centered Lactucinae species and thus is not further discussed here (Güzel et al., 2021).
The MRCA of the AE Lactuca species and the core group of “true” Lactuca species diverged around 7.14 (6.55–7.73) Mya, and the MRCA of AE Lactuca Clade diverged about 5.67 Mya, and the 95% HPD interval of the node age was 5.16–6.24 Mya. All of the species in this clade are distributed in Africa, while the MRCA of the core Lactuca species diverged early in Asia around 6.82 (6.27–7.40) Mya.
Despite the geographic distribution and evolutionary history, AE Lactuca species also have distinctive morphological characteristics from Lactuca species: 1) the achene is usually broadly thickened at the margins, 1–3 (rarely 3–6–7) rounded longitudinal ribs on each side, sometimes with furrows (Stebbins, 1937; Jeffrey, 1966; Dethier, 1982; Kilian, 2001; Wei et al., 2022), but Lactuca achenes have no obvious thickened margins or prominent rounded ribs on each side; 2) AE Lactuca mostly have 3(4)–5 florets per capitulum, seldom 9–10 or 10–14 or ±20 florets, whereas Lactuca have a variable number of florets, usually 10–30 florets, sometimes 5 or 5–10 florets (Kilian et al., 2009).
Although the systematic positions of AE Lactuca species and Melanoseris species inferred from plastomes and ITS sequences were discovered close, no apparent morphological discontinuity between the two groups was observed on important taxonomic features, such as floret number per capitulum, floret color, or chromosome number. Lactuca attenuata Stebbins (2n = 32), L. glandulifera (2n = 16), and Lactuca homblei De Wild. (2n = 16) are the only three known AE Lactuca species with chromosome number, while Melanoseris species are generally diploid (2n = 16) and rarely tetraploid (Kilian et al., 2009; Shih and Kilian, 2011).
As discussed above, AE Lactuca species have different divergence times and distinctive features from the core Lactuca lineage and thus should be excluded from Lactuca. To well resolve phylogenetic relationships between AE Lactuca and Melanoseris, more taxon sampling, especially Asian Melanoseris, should be taken into account for high-throughput sequencing and further taxonomic revisions.
The MRCA of Lactuca most likely originated in Asia-Temperate W and diverged around 6.82 Mya. This time estimation is younger than previous research using plastid DNA regions (Kilian et al., 2017b), in which the Lactuca taxa were not monophyletic and mixed with some Melanoseris taxa into one clade. The Lactuca lineages can be divided into nine ingroups, Clades I–IX. The MRCA of Lactuca species in Clade VIII and IX diverged around 5.89 Mya, and the Lactuca species in these two clades are now in America, Europe, Africa + Macaronesia, Asia-Temperate W, and Indian Subcontinent. The MRCA of Lactuca species in Clades I–VII was from Asia-Temperate W, and its progenies are now in Asia, Europe, Africa, and widely widespread, such as L. serriola, L. altaica, L. virosa, L. saligna, and L. indica.
In this section, we will discuss the phylogenetic relationships within Lactuca lineages starting from the basal clades of the phylogenetic tree to the tips.
Clade IX. The MRCA of this clade diverged around 3.09 Mya. This clade (2n = 16) includes Lactuca palmensis Bolle, an endemic perennial herb from La Palma in the Canary Islands (Dias et al., 2018), Lactuca inermis Forssk., a perennial herb from Tropical Africa to SW Asia, L. tenerrima Pourr. from Western Mediterranean to NW Africa, and Lactuca picridiformis Boiss from Afghanistan, Iran, and Pakistan. The systematic position of L. picridiformis based on plastome and ITS results was inconsistent. The chromosome number and morphology of the species suggest that plastid phylogenetic results are more reliable, and the ITS results might be gene introgression caused by hybridization (Kilian et al., 2017b).
Clade VIII. The MRCA of the NAE Lactuca (allopolyploid, 2n = 34, excluding L. oblongifolia) and L. plumieri (2n = 16) differentiated around 3.49 Mya. Thereafter, the MRCA of NAE Lactuca diverged very recently during the late Pleistocene (0.37 Mya) with high support values, much later than that in a previous study (Kilian et al., 2017b). Lactuca watsoniana Trel. (2n = 34) is endemic to the Azores in Macaronesia (Watson, 1870) and systematically related to the NAE Lactuca in our ITS trees. The North American-Azorean clade was also resolved as a sister to SW European Alpine-Pyrenean L. plumieri in previous studies (Dias et al., 2018; Jones et al., 2018).
NAE Lactuca in Clade VIII was considered to be derived from ancestral hybridization events between species with basic chromosome numbers x = 8 and x = 9 (Babcock et al., 1937). The potential progenitors of the allopolyploid Lactuca with a basic chromosome number x = 8 could be L. plumieri (or its extinct ancestors), and x = 9 progenitors might be L. tatarica, L. quercina, L. sibirica, or L. indica (or their extinct ancestors of clade) (Jones et al., 2018). The morphological characteristics of florets and achenes among NAE Lactuca lineages, L. plumieri, L. sibirica, L. tatarica (Jones et al., 2018), and L. indica (Jones et al., 2018; Wei et al., 2022) also indicated similarities and possible hybridization events.
The MRCA of NAE Lactuca (excluding L. oblongifolia) and L. plumieri diverged around 5.89 Mya, very close to the divergence time (5.88 Mya) of MRCA of Clade III (including L. quercina, L. sibirica, L. tatarica, and L. oblongifolia), Clade IV (including L. indica), and Clade V. Just as Jones et al. (2018) proposed, the MRCAs of these two groups were likely to be once widespread concurrently from Europe, Africa, and Asia and migrated to America via North Atlantic land bridges (NALBs) (x = 8 progenitors) and Bering land bridges (BLBs) (x = 9 progenitors). The MRCAs of these two groups hybridized to produce x = 17 lineages in America, and then the diploid progenitors became extinct in America in late Pliocene, resulting in the contemporary geographic isolations of x = 8 and x = 9 Lactuca from NAE Lactuca.
Clade VII. The MRCA of L. glauciifolia-L. intricata clade (2n = 18) diverged from its sister Clades I–VI around 6.75 Mya, and all of the species shared a common ψ clpP. The species in this clade are now mainly distributed in the E Mediterranean and SW Asia and extend widely to Europe and Central Asia. Although the systematic positions of L. picridiformis, inferred plastome, and ITS data were contradicting, the plastome tree with high confidence values was supported by karyology and morphological characteristics (Kilian et al., 2009; Kilian et al., 2017b).
Clade VI. This clade includes perennial herbaceous species. The MRCA of Clade VI diverged from its sister Clades III–V around 6.12 Mya and originated from Asia-Temperate W and subsequently spread to Asia-Temperate E, Indian Subcontinent, and Asia-Tropical. Clade VI can be divided into two subclades, VI-1 and VI-2. Clade VI-1 includes Turkish endemic Lactuca species (Lactuca boissieri Rouy, L. variabilis, and Lactuca glareosa Schott and Kotschy), L. mulgedioides (distributed only in Syria, Lebanon, and Turkey) (Plastome data), and Lactuca fenzlii N.Kilian and Greuter endemic to Turkey, Lactuca aurea (Sch.Bip. ex Vis. and Pančić) Stebbins from Europe, and Lactuca deltoidea DC. ex C.A.Mey. near Caucasus (present in ITS trees). Clade VI-2 contains L. dissecta from SW to Central Asia and L. dolichophylla from SW Asia to Sino-Himalaya. The species composition of the clade was consistent with previous molecular and morphological results (Kilian et al., 2017b; Güzel et al., 2021).
Clade V. The MRCA of Clades III–V diverged around 5.88 Mya and originated from the Asia-Temperate W and then diffused globally. Clade V contains only L. adenophora and L. racemosa (2n = 16) in plastome trees, and these two species plus Lactuca macrophylla A. Gray present in ITS trees. L. macrophylla is morphologically similar to L. racemosa and L. adenophora and was reported to have some sterile individuals, indicating possible ancient reticulation, chloroplast capture, or introgressive hybridization among these species (Güzel et al., 2018; Güzel et al., 2021). Additionally, the achenes of L. macrophylla and L. racemosa are apically attenuate, very rare for Lactuca lineages, whereas the achene of L. adenophora is beaked (Güzel et al., 2018). The systematic positions of species in Clade V were both resolved within Lactuca lineages in our plastome and ITS trees with different sister clades. In Güzel et al. (2021), Clade V was part of Lactuca in ITS tree, but one of the Asian Melanoseris clades (not included in this study) was resolved as its sister group in the plastid tree. Therefore, these species should be retained in Lactuca.
Clade IV. This Clade includes species mainly in Asia-Temperate E, namely, L. indica, Lactuca raddeana Maxim., Lactuca formosana Maxim., and Lactuca triangulata Maxim. (2n = 18). The MRCA of Clades IV and III diverged in Asia Temperate regions about 5.65 Mya and spread to Central Asia and NE Asia. Jones et al. (2018) treated Lactuca jamaicensis Griseb, native to Jamaica, as a synonym of L. indica and considered its distribution as a consequence of human introduction. All of the species in Clade IV were once established as an independent genus Pterocypsela (Shih, 1988). The main characteristics of Pterocypsela are capitulum with 9–25 yellow florets, achenes black, ellipsoid and compressed, and broadly winged at the edges (Shih, 1988; Wei et al., 2022). Molecular evidence, including our plastome data, showed that this genus should be retained in Lactuca (Wang et al., 2013; Kilian et al., 2017b; Wei et al., 2017). Hybridization experiments showed that L. indica, L. raddeana, and L. tatarica were closely related and could produce sterile progeny (Thompson et al., 1941; Whitaker and Thompson, 1941).
Clade III. This clade includes L. bourgaei (2n = 16), L. quercina (2n = 18), and three closely related perennial herbaceous plants L. sibirica, L. tatarica, and L. oblongifolia (2n = 18). The phylogenetic relationships of these species are closely related in plastome data but dissected into three clades in nuclear data. Clade III-1 is L. quercina, closely related to the crop lettuce and its relatives (Clade I) in nuclear data but a sister of L. sibirica, L. tatarica, and L. oblongifolia in the plastome trees. L. quercina is different from L. sibirica, L. tatarica, and L. oblongifolia in the characteristics of lingual floret color and achenes: L. quercina inflorescences with 16–22 yellow florets, achenes base truncate, with completely annular carpopodium, corpus attenuate into a beak, beak 1.5–3.9 mm long, oblong-ellipsoid and compressed, black with 5–6 prominent ribs on each face (Güzel et al., 2018); the inflorescences of L. sibirica, L. tatarica, and L. oblongifolia generally have about 20 purple, blue, or nearly white florets, achenes narrowly ellipsoid, flattened and subcompressed, apically attenuate or with a ca. 1–2 mm beak, 4–5 or 5–7 narrow ribs on either side (Shih and Kilian, 2011). Clade III-2 contains L. oblongifolia, L. sibirica, and L. tatarica and is a sister of Clade II in ITS trees. L. sibirica and L. tatarica could produce fertile hybrid progeny (Koopman et al., 2001). The incongruence of relationships among L. sibirica, L. tatarica, and L. oblongifolia between plastomes and ITS sequences may reflect hybridization and introgression between L. sibirica and L. tatarica. Clade III-3 is L. bourgaei within the Melanoseris Clade in ITS trees. A previous study demonstrated the compatibility relationship between L. tatarica and L. bourgaei and produced sterile hybrids (Thompson, 1943). Therefore, this study retained L. bourgaei in Lactuca. Further studies with more molecular and genetic evidence are needed in the future.
Clade II. This clade is a sister of Clade I, and their MRCA originated from Asia-Temperate W and diverged around 5.20 Mya. Clade II contains four species (2n = 18): L. viminea, L. reviersii, L. tetrantha, and L. orientalis, as well as additional Lactuca acanthifolia Benth. and Hook.f. and Lactuca alpestris (Gand.) Rech.f. in the ITS trees. These species are natively distributed in Europe, N Africa + Macaronesia, Asia-Temperate, and Indian Subcontinent (Feráková and Májovský, 1977; Bano, 2009; Kilian et al., 2009; Bano and Qaiser, 2011). Some species are narrowly distributed, such as L. reviersii endemic to Morocco in NW Africa, L. tetrantha endemic to Cyprus (east Mediterranean island), L. acanthifolia endemic to Turkey and Greece, and L. alpestris endemic to Kriti island in Greece (https://portal.cybertaxonomy.org/flora-greece/intro).
Clade I. The MRCA of Clade I originated from Asia-Temperate W region and diverged around 2.58 Mya, and all of the species in the clade (excluding unknown L. scarioloides) have a chromosome number of 2n = 18. A recent whole-genome resequencing of 445 Lactuca accessions revealed that cultivated lettuce was domesticated near the Caucasus (Wei et al., 2021), included in our geographic region of Asia-Temperate W (F). Clade I contains the important economic vegetable cultivated lettuce (L. sativa) and lettuce gene pool species (Koopman et al., 1998). The primary lettuce gene pool includes L. aculeata, L. altaica, Lactuca azerbaijanica Rech. f., L. scarioloides, and L. serriola. The secondary gene pool includes L. saligna. The tertiary gene pool contains 13 wild Lactuca species, namely, L. acanthifolia., L. alpestrias, L. aurea, L. georgica, Lactuca longidentata Moris ex DC., L. orientalis, L. quercina, L. sibirica, L. tatarica, Lactuca taraxacifolia (Willd.) Schumach., L. viminea, L. virosa, and L. watsoniana (Zohary, 1991; Koopman et al., 1998; van Treuren et al., 2012; Wei et al., 2017; Wei et al., 2021).
There are three subclades in Clade I. The L. saligna clade is the sister of the other two clades, L. scarioloides-L. georgica-L. virosa clade and L. sativa-L. serriola-L. aculeata-L. altaica clade. The systematic positions of L. saligna were congruent between plastome and ITS trees. A previous study revealed similar mitochondrial genome sequences and structures between L. sativa and L. serriola, but that of L. saligna was shown to be significantly different (Kozik et al., 2019). L. saligna was placed between L. georgica-L. virosa clade and L. sativa-L. serriola-L. aculeata-L. altaica clade in the most recent coalescence-based phylogenetic tree inferred from 4,513 single-locus nuclear genes (Wei et al., 2021). L. saligna is a non-host for lettuce downy mildew (Bremia lactucae) and can hybridize with the crop lettuce L. sativa for improvement of genetic resistance to downy mildew (Lebeda and Reinink, 1994; Jeuken et al., 2001; Jeuken and Lindhout, 2002; Zhang et al., 2009). Therefore, L. saligna is genetically more closely related to cultivated lettuce and should be retained as the second gene pool species.
The phylogenetic relationship between L. georgica and L. virosa is very close, and they are morphologically similar: L. georgica is a perennial with solitary glabrous stems, capitula cylindrical with about 15 small florets, yellow corolla, achenes broadly obovoid, dark purple or nearly black, slightly flat or flattened, narrowly winged, 6–7 mm long, with a light or dark brown filamentous beak of about 2–2.5 mm, with usually seven distinct ribs on each side, distributed mainly in Asia-Temperate W; the latter has numerous heads with about 15 yellow ligulate florets, achenes broadly elliptical, narrowly winged, about 6–10 mm long, five ribs, distributed in S Europe and N Africa (Feráková and Májovský, 1977; Kilian et al., 2009; Beharav et al., 2018; Wei et al., 2022). The plastome of L. scarioloides is the first molecular sampling of the species, and the Irano-Turanian wild lettuce was reported to be morphologically similar to L. serriola, despite leaf shape and achene and capitula size (Zohary, 1991). The native distribution regions of L. georgica and L. scarioloides are overlapped, indicating a possible hybridization introgression or chloroplast capture between them, which may also explain the relationships among L. scarioloides, L. georgica, and L. virosa inferred from plastome data.
Another research based on mitochondrial genomes found that L. sativa ‘Salinas’ and L. virosa were more closely related than L. sativa ‘Wendel’ and L. serriola (Fertet et al., 2021), confirming the involvement of L. virosa in the breeding history of Salinas varieties (Mikel, 2007). The differences of the relationships among L. saligna, L. georgica, L. virosa, and L. sativa-L. serriola-L. aculeata-L. altaica clade, inferred from whole genomes and organelle (plastid and mitochondrion) genomes, are likely to be the consequences of the complex artificial domestication history of lettuce. In terms of hybridization introgression, the complete genome sequences of cultivated lettuce and its relatives are more reliable to reflect the true evolutionary history than organelle genomes. Thus, we think that L. georgica, L. scarioloides, L. virosa, and L. quercina (discussed in Clade III) should be placed in the tertiary gene pool.
The species in L. sativa-L. serriola-L. aculeata-L. altaica clade can cross with each other to produce progeny. These wild species belong to the primary gene pool of cultivated lettuce (Koopman et al., 1998; Koopman et al., 2001; Wei et al., 2021). Our plastome data showed that L. sativa and L. aculeata were closer than L. serriola and L. altaica, but the ITS tree indicated that L. serriola and L. altaica were closer to L. sativa than L. aculeata, consistent with genome sequences (Wei et al., 2021). Previous studies treated L. altaica and L. serriola as conspecific species (Koopman et al., 1998; Koopman et al., 2001; Wei et al., 2017), but the recent whole-gene sequencing study showed that they were two independent species with different morphological characteristics (Wei et al., 2021). L. altaica usually has 7 (9) to 15 (17) florets ligulate, achenes light brown oblanceolate compressed, 3.5 mm long and 1 mm wide, with 6–8 ribs on each side, apex acute filamentous beak 3 mm long. L. serriola has about (7) 10–30 (50) small florets, achenes oblong-ovate, brown or olive-gray to grayish, achene body 3.0–4.0 mm long and 1.2–1.5 mm wide, with 5–8 ribs on each side, beak white, filiform, 3.0–4.5 mm long (Feráková and Májovský, 1977; Shih, 1997; Shih and Kilian, 2011). L. azerbaijanica has not yet been studied using molecular evidence and is retained temporarily as a primary lettuce gene pool species.
The remaining species, closely related to lettuce but not reported to be compatible to L. sativa, were treated as an extension of the tertiary gene pool. L. indica has been cultivated for its edible leaves (Kadereit and Jeffrey, 2007) and also has some resistance to lettuce downy mildew (van Treuren et al., 2011). Hybridization experiments have shown that L. indica and L. sativa can produce fusiform brown achenes with strongly compressed body and filiform white beak. The fertility of F1 hybrid seedlings is ongoing being tested (unpublished data from Zhen Wei). Previous studies reported that somatic hybridization of L. sativa and L. tatarica produced fertile hybrids (Maisonneuve et al., 1995). Three wild species, L. aurea (2n = 16), L. longidentata (2n = 16), and L. watsoniana (2n = 34) have different chromosome numbers from L. sativa (2n = 18) (Feráková and Májovský, 1977; Dias et al., 2018) and therefore should be excluded from the lettuce gene pool. L. taraxacifolia has been renamed as Lactuca alaica Kovalevsk. and transferred to Kovalevskiella lineage (Kilian et al., 2017b). Considering phylogenetic positions, morphological features, and crossing experiments, we added L. indica, L. oblongifolia, L. reviersii, and L. tetrantha to the extension list of the tertiary gene pool. In total, 11 species in the extension list could be potential genetic resources for future lettuce breeding.
As we discussed above, the lettuce gene pool was modified as follows:
1) The primary gene pool: L. aculeata, L. altaica, L. serriola, and L. azerbaijanica
2) The secondary gene pool: L. saligna
3) The tertiary gene pool: L. georgica, L. scarioloides, and L. virosa
4) Extension of the tertiary gene pool: L. acanthifolia, L. alpestris, L. indica, L. oblongifolia, L. orientalis, L. quercina, L. reviersii, L. sibirica, L. tatarica, L. tetrantha, and L. viminea
Paraprenanthes alatipes (Collett and Hemsl.) Z. Wei & S.X. Zhu, comb. nov. ≡ Lactuca alatipes Collett & Hemsl. in J. Linn. Soc., Bot. 28: 79. 1890 – Type: Burma, Shan hills terai, 910 m, 1888, H. Collett 479.
= Lactuca parishii Craib in Kew Bull. 1911: 403. 1911, syn. nov. – Type: Burma, Moulmein, 1200-1500 m, 1862, Parish 423.
The datasets presented in this study can be found in the GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession number ON782474-ON782569 (plastome data) and OP070064-OP070156 (ITS data).
ZW, FTB and ES conceived the study. ZW conducted the experiments. RC, ZW, ZWL, HC, YGM and XMX analyzed the data. ZW and RC wrote the manuscript. FTB, ES, XMX, SXZ, ZWL, HC and YGM reviewed and edited the manuscript. SXZ did field investigation and material collection.
This work was supported by the National Natural Science Foundation of China (31700175; 31670188); Key Research and Development Project of Henan Province (221111110800); Fostering Project for Young Teachers of Zhengzhou University (JC21343014; JC21343016); Qinghai Provincial Department of Science and Technology (2020-ZJ-Y04); SYNTHESYS Joint Research Activities 4 (JRA4: Plants/fungi herbarium DNA).
We would like to thank all the curators, dr. ir. J.J. Wieringa (Naturalis Biodiversity Center, Leiden, The Netherlands), Dr. Alan Paton, Ms. Sally Dawson, and Ms. Marie Briggs (Royal Botanic Gardens, Kew, The United Kingdom), Ms. Suzanne Cubey (Royal Botanic Gardens, Edinburgh, The United Kingdom), Ms. Ranee Prakash (Natural History Museum, London, The United Kingdom), Dr. Xiaohua Jin (Institute of Botany, Chinese Academy of Sciences, China) for helping us obtain the herbarium samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.978417/full#supplementary-material
Amar, M. H. (2020). Ycf1-ndhF genes, the most promising plastid genomic barcode, sheds light on phylogeny at low taxonomic levels in Prunus persica. J. Genet. Eng Biotechnol. 18, 42. doi: 10.1186/s43141-020-00057-3
Amiryousefi, A., Hyvonen, J., Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Babcock, E. B., Stebhins, G. L., Jenkins, J. A. (1937). Chromosomes and phylogeny in some genera of the crepidinae. Cytologia Fujii Jubilaei, 188–210. doi: 10.1508/CYTOLOGIA.FUJIIJUBILAEI.188
Bano, R. (2009). Biosystematic studies of the genus lactuca l. (s. i.) lactuceae-asteraceae from Pakistan and Kashmir (Karachi: University of Karachi), 357.
Bano, R., Qaiser, M. (2011). A taxonomic revision of the genus lactuca l. (Cichorieae-asteraceae) from Pakistan and Kashmir. Pak. J. Bot. 43, 2259–2268.
Beharav, A., Hellier, B., Richardson, K. L., Lebeda, A., Kisha, T. (2018). Genetic relationships and structured diversity of Lactuca georgica germplasm from Armenia and the Russian federation among other members of Lactuca l., subsection Lactuca l., assessed by TRAP markers. Genet. Resour. Crop Ev 65, 1963–1978. doi: 10.1007/s10722-018-0669-7
Bock, R. (2007). “Structure, function, and inheritance of plastid genomes,” in Cell and molecular biology of plastids. Ed. Bock, R. (Berlin, Heidelberg: Springer Berlin Heidelberg), 29–63.
Bouckaert, R., Timothy, G. V., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., et al. (2019). BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PloS Comput. Biol. 15, e1006650. doi: 10.1371/journal.pcbi.1006650
Caroca, R., Howell, K. A., Malinova, I., Burgos, A., Tiller, N., Pellizzer, T., et al. (2021). Knockdown of the plastid-encoded acetyl-CoA carboxylase gene uncovers functions in metabolism and development. Plant Physiol. 185, 1091–1110. doi: 10.1093/plphys/kiaa106
Chan, P. P., Lin, B. Y., Mak, A. J., Lowe, T. M. (2021). tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 49, 9077–9096. doi: 10.1093/nar/gkab688
Collett, H., Hemsley, W. B. (1890). On a collection of plants from upper Burma and the shan states. J. Linn. Soc London Bot. 28, 1–150. doi: 10.1111/j.1095-8339.1890.tb01452.x
Daniell, H., Lin, C. S., Yu, M., Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
Dempewolf, H., Kane, N. C., Ostevik, K. L., Geleta, M., Barker, M. S., Lai, Z., et al. (2010). Establishing genomic tools and resources for Guizotia abyssinica (L.f.) cass.–the development of a library of expressed sequence tags, microsatellite loci, and the sequencing of its chloroplast genome. Mol. Ecol. Resour. 10, 1048–1058. doi: 10.1111/j.1755-0998.2010.02859.x
Dethier, D. (1982). Le genre Lactuca l. (Asteraceae) en afrique centrale. Bull. Jard Bot. Nat. Belg 52, 367–382. doi: 10.2307/3667889
Dias, E. F., Kilian, N., Silva, L., Schaefer, H., Carine, M., Rudall, P. J., et al. (2018). Phylogeography of the macaronesian lettuce species Lactuca watsoniana and l. palmensis (Asteraceae). Biochem. Genet. 56, 315–340. doi: 10.1007/s10528-018-9847-8
Dierckxsens, N., Mardulyn, P., Smits, G. (2016). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18–e18. doi: 10.1093/nar/gkw955
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phyt Bull. 19, 11–15. doi: 10.1016/0031-9422(80)85004-7
Feráková, V., Májovský, J. (1977). The genus lactuca l. @ in Europe (Bratislava: Univerzita Komenského).
Fertet, A., Graindorge, S., Koechler, S., de Boer, G. J., Guilloteau-Fonteny, E., Gualberto, J. M. (2021). Sequence of the mitochondrial genome of Lactuca virosa suggests an unexpected role in Lactuca sativa's evolution. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.697136
Fleischmann, T. T., Scharff, L. B., Alkatib, S., Hasdorf, S., Schottler, M. A., Bock, R. (2011). Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23, 3137–3155. doi: 10.1105/tpc.111.088906
Fu, P. C., Sun, S. S., Twyford, A. D., Li, B. B., Zhou, R. Q., Chen, S. L., et al. (2021). Lineage-specific plastid degradation in subtribe gentianinae (Gentianaceae). Ecol. Evol. 11, 3286–3299. doi: 10.1002/ece3.7281
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Güzel, M. E., Coşkunçelebi, K., Kilian, N., Makbul, S., Gültepe, M. (2021). Phylogeny and systematics of the lactucinae (Asteraceae) focusing on their SW Asian centre of diversity. Plant Syst. Evol. 307, 7. doi: 10.1007/s00606-020-01719-y
Güzel, M. E., Kilian, N., Gültepe, M., Kandemir, A., Coskuncelebi, K. (2018). Contributions to the taxonomy of Lactuca (Asteraceae) in Turkey. Turk J. Bot. 42, 197–207. doi: 10.3906/bot-1707-29
Hochuli, P. A. (1978). Palynologische untersuchungen im oligozän und untermiozän der zentralen und westlichen paratethys. Beitr Paläontol Österr 4, 1–132.
Hudson, G. S., Mason, J. G. (1988). The chloroplast genes encoding subunits of the h+-ATP synthase. Photosynth Res. 18, 205–222. doi: 10.1007/bf00042985
Jeuken, M., Lindhout, P. (2002). Lactuca saligna, a non-host for lettuce downy mildew (Bremia lactucae), harbors a new race-specific dm gene and three QTLs for resistance. Theor. Appl. Genet. 105, 384–391. doi: 10.1007/s00122-002-0943-z
Jeuken, M., van Wijk, R., Peleman, J., Lindhout, P. (2001). An integrated interspecific AFLP map of lettuce (Lactuca) based on two l. sativa × L. saligna F2 populations. Theor. Appl. Genet. 103, 638–647. doi: 10.1007/s001220100657
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., de Pamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi: 10.1186/s13059-020-02154-5
Jones, K. E., Schilling, E. E., Dias, E. F., Kilian, N. (2018). Northern hemisphere disjunctions in Lactuca (Cichorieae, asteraceae): independent Eurasia to north America migrations and allopolyploidization. Willdenowia 48, 259–284. doi: 10.3372/wi.48.48206
Kadereit, J. W., Jeffrey, C. (2007). “Asterales: introduction and conspectus,” in Flowering plants eudicots, vol. vol 8 . Ed. Kubitzki, K. (Berlin: Springer), 1–6.
Katoh, A., Lee, K. S., Fukuzawa, H., Ohyama, K., Ogawa, T. (1996). CemA homologue essential to CO2 transport in the cyanobacterium Synechocystis PCC6803. Proc. Natl. Acad. Sci. U S A 93, 4006–4010. doi: 10.1073/pnas.93.9.4006
Kilian, N. (2001). Lactuca stebbinsii (Lactuceae, compositae), a puzzling new species from Angola. Willdenowia 31, 71–78. doi: 10.3372/wi.31.31105
Kilian, N., Hand, R., Hadjikyriakou, G. N., Christodoulou, C. S., Dagher-Kharrat, M. B. (2017a). Astartoseris (Cichorieae, asteraceae), a new, systematically isolated monospecific genus accommodating Lactuca triquetra endemic to Lebanon and Cyprus. Willdenowia 47, 115–125. doi: 10.3372/wi.47.47203
Kilian, N., Hand, R., von Raab-Straube, E. (2009) Cichorieae systematics portal. Available at: http://cichorieae.e-taxonomy.net/portal/ (Accessed 30 April 2022).
Kilian, N., Sennikov, A., Wang, Z. H., Gemeinholzer, B., Zhang, J. W. (2017b). Sub-Paratethyan origin and middle to late Miocene principal diversification of the lactucinae (Compositae: Cichorieae) inferred from molecular phylogenetics, divergence-dating and biogeographic analysis. Taxon 66, 675–703. doi: 10.12705/663.9
Kode, V., Mudd, E. A., Iamtham, S., Day, A. (2005). The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44, 237–244. doi: 10.1111/j.1365-313X.2005.02533.x
Koopman, W. J. M., Guetta, E., Van De Wiel, C. C. M., Vosman, B., Van den Berg, R. G. (1998). Phylogenetic relationships among Lactuca (Asteraceae) species and related genera based on ITS-1 DNA sequences. Am. J. Bot. 85, 1517–1530. doi: 10.2307/2446479
Koopman, W. J. M., Zevenbergen, M. J., Van den Berg, R. G. (2001). Species relationships in lactuca s.l. (Lactuceae, asteraceae) inferred from AFLP fingerprints. Am. J. Bot. 10, 1881–1887. doi: 10.2307/3558364
Kozik, A., Rowan, B. A., Lavelle, D., Berke, L., Schranz, M. E., Michelmore, R. W., et al. (2019). The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PloS Genet. 15, e1008373. doi: 10.1371/journal.pgen.1008373
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lebeda, A., Doležalová, I., Astley, D. (2004a). Representation of wild lactuca spp. (Asteraceae, lactuceae) in world genebank collections. genet. resour. Crop Evol. 51, 167–174. doi: 10.1023/b:gres.0000020860.66075.f7
Lebeda, A., Doležalová, I., Feráková, V., Astley, D. (2004b). Geographical distribution of wild Lactuca species (Asteraceae, lactuceae). Bot. Rev. 70, 328–356. doi: 10.1663/0006-8101(2004)070[0328:GDOWLS]2.0.CO;2
Lebeda, A., Křístková, E., Doležalová, I., Kitner, M., Widrlechner, M. P. (2019). “Wild lactuca species in north America,” in North American crop wild relatives, vol. Vol 2 . Eds. Greene, S., Williams, K., Khoury, C., Kantar, M., Marek, L. (Cham: Springer), 131–194. doi: 10.1007/978-3-319-97121-6_5
Lebeda, A., Reinink, K. (1994). Histological characterization of resistance in Lactuca saligna to lettuce downy mildew (Bremia lactucae). Physiol. Mol. Plant Pathol. 44, 125–139. doi: 10.1016/S0885-5765(05)80106-7
Lebeda, A., Ryder, E., Grube, R., Dolezalova, I., Krıstkova, E. (2007). “Lettuce (Asteraceae; lactuca spp.),” in Genetic resources, chromosome engineering, and crop improvement series, vol. Vol 3377–472 . Ed. Singh, R. (Boca Raton: CRC Press).
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Lin, C. S., Chen, J. J. W., Chiu, C. C., Hsiao, H. C. W., Yang, C. J., Jin, X. H., et al. (2017). Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 90, 994–1006. doi: 10.1111/tpj.13525
Lin, C. S., Chen, J. J., Huang, Y. T., Chan, M. T., Daniell, H., Chang, W. J., et al. (2015). The location and translocation of ndh genes of chloroplast origin in the orchidaceae family. Sci. Rep. 5, 9040. doi: 10.1038/srep09040
Li, H., Xiao, W., Tong, T., Li, Y., Zhang, M., Lin, X., et al. (2021). The specific DNA barcodes based on chloroplast genes for species identification of orchidaceae plants. Sci. Rep. 11, 1424. doi: 10.1038/s41598-021-81087-w
Maisonneuve, B., Chupeau, M. C., Bellec, Y., Chupeau, Y. (1995). Sexual and somatic hybridization in the genus Lactuca. Euphytica 85, 281–285. doi: 10.1007/BF00023957
Majeran, W., Wollman, F. A., Vallon, O. (2000). Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b(6)f complex. Plant Cell 12, 137–150. doi: 10.1105/tpc.12.1.137
Mayor, C., Brudno, M., Schwartz, J. R., Poliakov, A., Rubin, E. M., Frazer, K. A., et al. (2000). VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047. doi: 10.1093/bioinformatics/16.11.1046
Meena, R. K., Negi, N., Uniyal, N., Shamoon, A., Bhandari, M. S., Pandey, S., et al. (2020). Chloroplast-based DNA barcode analysis indicates high discriminatory potential of matK locus in Himalayan temperate bamboos. 3 Biotech. 10, 534. doi: 10.1007/s13205-020-02508-7
Mikel, M. A. (2007). Genealogy of contemporary north American lettuce. Hortscience 42, 489–493. doi: 10.21273/hortsci.42.3.489
Olinares, P. D., Kim, J., van Wijk, K. J. (2011). The clp protease system; a central component of the chloroplast protease network. Biochim. Biophys. Acta 1807, 999–1011. doi: 10.1016/j.bbabio.2010.12.003
Palmer, J. (1983). Chloroplast DNA exists in two orientations. Nature 301, 92–93. doi: 10.1038/301092a0
Pascual-Diaz, J. P., Garcia, S., Vitales, D. (2021). Plastome diversity and phylogenomic relationships in asteraceae. Plants (Basel) 10, 2699. doi: 10.3390/plants10122699
Pouchon, C., Fernandez, A., Nassar, J. M., Boyer, F., Aubert, S., Lavergne, S., et al. (2018). Phylogenomic analysis of the explosive adaptive radiation of the Espeletia complex (Asteraceae) in the tropical Andes. Syst. Biol. 67, 1041–1060. doi: 10.1093/sysbio/syy022
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Rodriguez-Concepcion, M., D'Andrea, L., Pulido, P. (2019). Control of plastidial metabolism by the clp protease complex. J. Exp. Bot. 70, 2049–2058. doi: 10.1038/ncomms14953
Schottler, M. A., Flugel, C., Thiele, W., Stegemann, S., Bock, R. (2007). The plastome-encoded PsaJ subunit is required for efficient photosystem I excitation, but not for plastocyanin oxidation in tobacco. Biochem. J. 403, 251–260. doi: 10.1042/BJ20061573
Sergei, L., Kosakovsky, P., Simon, D. W., Frost (2005). Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22, 1208–1222. doi: 10.1093/molbev/msi105
Shih, C. (1988). Revision of Lactuca l. and two new genera of tribe lactuceae (Compositae) on the mainland of Asia. Acta Phytotaxon Sin. 26, 382–393.
Shih, C., Kilian, N. (2011). “Cichorieae [excl.Tragopogon, taraxacum, hieracium and pilosella],” in Flora of China (Beijing: Science Press & St Louis: Missouri Botanical Garden).
Song, W., Ji, C., Chen, Z., Cai, H., Wu, X., Shi, C., et al. (2022). Comparative analysis the complete chloroplast genomes of nine Musa species: Genomic features, comparative analysis, and phylogenetic implications. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.832884
Stebbins, G. L. (1937). The scandent species of Prenanthes and Lactuca in Africa. Bull. du Jardin botanique l'État Bruxelles 14, 333–352. doi: 10.2307/3666566
Su, C., Duan, L., Liu, P., Liu, J., Chang, Z., Wen, J. (2021). Chloroplast phylogenomics and character evolution of eastern Asian Astragalus (Leguminosae): Tackling the phylogenetic structure of the largest genus of flowering plants in Asia. Mol. Phylogenet. Evol. 156, 107025. doi: 10.1016/j.ympev.2020.107025
Tang, D., Lin, Y., Wei, F., Quan, C., Wei, K., Wei, Y., et al. (2022). Characteristics and comparative analysis of Mesona chinensis benth chloroplast genome reveals DNA barcode regions for species identification. Funct. Integr. Genomics. 22, 467–479. doi: 10.1007/s10142-022-00846-8
Thompson, R. C. (1943). Further studies on interspecific genetic relationships in Lactuca. J. Agric. Res. 66, 41–48.
Thompson, R. C., Whitaker, T. W., Kosar, W. F. (1941). Interspecific genetic relationships in Lactuca. J. Agric. Res. 63, 91–107.
Timme, R. E., Kuehl, J. V., Boore, J. L., Jansen, R. K. (2007). A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am. J. Bot. 94, 302–312. doi: 10.3732/ajb.94.3.302
Tremetsberger, K., Gemeinholzer, B., Zetzsche, H., Blackmore, S., Kilian, N., Talavera, S. (2012). Divergence time estimation in cichorieae (Asteraceae) using a fossil-calibrated relaxed molecular clock. Org. Divers. Evol. 13, 1–13. doi: 10.1007/s13127-012-0094-2
van Treuren, R., Coquin, P., Lohwasser, U. (2012). Genetic resources collections of leafy vegetables (lettuce, spinach, chicory, artichoke, asparagus, lamb’s lettuce, rhubarb and rocket salad): composition and gaps. Genet. Resour. Crop Ev 59, 981–997. doi: 10.1007/s10722-011-9738-x
van Treuren, R., van der Arend, A. J. M., Schut, J. W. (2011). Distribution of downy mildew (Bremia lactucae regel) resistances in a genebank collection of lettuce and its wild relatives. Plant Genet. Resour. 11, 15–25. doi: 10.1017/S1479262111000761
Varotto, C., Pesaresi, P., Jahns, P., Lessnick, A., Tizzano, M., Schiavon, F., et al. (2002). Single and double knockouts of the genes for photosystem I subunits G, K, and h of arabidopsis. effects on photosystem I composition, photosynthetic electron flow, and state transitions. Plant Physiol. 129, 616–624. doi: 10.1104/pp.002089
Walker, J. F., Jansen, R. K., Zanis, M. J., Emery, N. C. (2015). Sources of inversion variation in the small single copy (SSC) region of chloroplast genomes. Am. J. Bot. 102, 1751–1752. doi: 10.3732/ajb.1500299
Wang, C., Liu, J., Su, Y., Li, M., Xie, X., Su, J. (2021). Complete chloroplast genome sequence of Sonchus brachyotus helps to elucidate evolutionary relationships with related species of asteraceae. BioMed. Res. Int. 2021, 9410496. doi: 10.1155/2021/9410496
Wang, Z. H., Peng, H., Kilian, N. (2013). Molecular phylogeny of the Lactuca alliance (Cichorieae subtribe lactucinae, asteraceae) with focus on their Chinese centre of diversity detects potential events of reticulation and chloroplast capture. PloS One 8, e82692. doi: 10.1371/journal.pone.0082692
Watson, H. C. (1870). “Notes on the botany of the Azores” in The natural history of the Azores or Western Islands. Ed. Godman, F. C. (John Van Yoorbt Paternoster Row), 358.
Weaver, S., Shank, S. D., Spielman, S. J., Li, M., Muse, S. V., Kosakovsky, P. S. L. (2018). Datamonkey 2.0: A modern web application for characterizing selective and other evolutionary processes. Mol. Biol. Evol. 35, 773–777. doi: 10.1093/molbev/msx335
Wei, Z. (2016). Genetic diversity and evolution in lactuca l. (Asteraceae): From phylogeny to molecular breeding (Wageningen: Wageningen University).
Wei, Z., Chu, R., Luan, M., Lu, Z., Ma, Y., Luo, X., et al. (2022). Morphology and micro-morphology of achenes and their taxonomic implications to Lactuca species (Cichorieae; asteraceae). Taiwania 67, 171–180. doi: 10.6165/tai.2022.67.171
Wei, T., van Treuren, R., Liu, X., Zhang, Z., Chen, J., Liu, Y., et al. (2021). Whole-genome resequencing of 445 Lactuca accessions reveals the domestication history of cultivated lettuce. Nat. Genet. 53, 752–760. doi: 10.1038/s41588-021-00831-0
Wei, Z., Zhu, S. X., Van den Berg, R. G., Bakker, F. T., Schranz, M. E. (2017). Phylogenetic relationships within Lactuca l. (Asteraceae), including African species, based on chloroplast DNA sequence comparisons. Genet. Resour. Crop Ev 64, 55–71. doi: 10.1007/s10722-015-0332-5
Wen, J., Xie, D. F., Price, M., Ren, T., Deng, Y. Q., Gui, L. J., et al. (2021). Backbone phylogeny and evolution of apioideae (Apiaceae): New insights from phylogenomic analyses of plastome data. Mol. Phylogenet. Evol. 161, 107183. doi: 10.1016/j.ympev.2021.107183
Whitaker, T. W., Thompson, R. C. (1941). Cytological studies in Lactuca. Bulletin of the torrey botanical club 68, 388–394. doi: 10.2307/2481647
Yu, Y., Blair, C., He, X. (2020). RASP 4: Ancestral state reconstruction tool for multiple genes and characters. Mol. Biol. Evol. 37, 604–606. doi: 10.1093/molbev/msz257
Zhang, N. W., Lindhout, P., Niks, R. E., Jeuken, M. J. M. (2009). Genetic dissection of Lactuca saligna nonhost resistance to downy mildew at various lettuce developmental stages. Plant Pathol. 58, 923–932. doi: 10.1111/j.1365-3059.2009.02066.x
Zhao, D. N., Ren, Y., Zhang, J. Q. (2020). Conservation and innovation: Plastome evolution during rapid radiation of Rhodiola on the qinghai-Tibetan plateau. Mol. Phylogenet. Evol. 144, 106713. doi: 10.1016/j.ympev.2019.106713
Keywords: Lactuca phylogenetics, lettuce genetic resources, Lactucinae, Cichorieae, Compositae
Citation: Chu R, Xu XM, Lu ZW, Ma YG, Cheng H, Zhu SX, Bakker FT, Schranz ME and Wei Z (2022) Plastome-based phylogeny and biogeography of Lactuca L. (Asteraceae) support revised lettuce gene pool categories. Front. Plant Sci. 13:978417. doi: 10.3389/fpls.2022.978417
Received: 26 June 2022; Accepted: 12 September 2022;
Published: 12 October 2022.
Edited by:
Xiaohua Jin, Institute of Botany, (CAS), ChinaReviewed by:
Massoud Ranjbar, Bu-Ali Sina University, IranCopyright © 2022 Chu, Xu, Lu, Ma, Cheng, Zhu, Bakker, Schranz and Wei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen Wei, d2VpemhlbkB6enUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.