
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 06 October 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.976807
This article is part of the Research TopicConstruction of Regulatory Networks in Plant Abiotic Stress Responses, Volume IIView all 5 articles
 Tao Fan1,2†
Tao Fan1,2† Mehtab Muhammad Aslam3†
Mehtab Muhammad Aslam3† Jian-Li Zhou1
Jian-Li Zhou1 Mo-Xian Chen2
Mo-Xian Chen2 Jianhua Zhang3
Jianhua Zhang3 Shenxiu Du4
Shenxiu Du4 Kai-Lu Zhang2*
Kai-Lu Zhang2* Yun-Sheng Chen1*
Yun-Sheng Chen1*The circadian clock is an internal time-keeping mechanism that synchronizes the physiological adaptation of an organism to its surroundings based on day and night transition in a period of 24 h, suggesting the circadian clock provides fitness by adjusting environmental constrains. The circadian clock is driven by positive and negative elements that regulate transcriptionally and post-transcriptionally. Alternative splicing (AS) is a crucial transcriptional regulator capable of generating large numbers of mRNA transcripts from limited numbers of genes, leading to proteome diversity, which is involved in circadian to deal with abiotic stresses. Over the past decade, AS and circadian control have been suggested to coordinately regulate plant performance under fluctuating environmental conditions. However, only a few reports have reported the regulatory mechanism of this complex crosstalk. Based on the emerging evidence, this review elaborates on the existing links between circadian and AS in response to abiotic stresses, suggesting an uncovered regulatory network among circadian, AS, and abiotic stresses. Therefore, the rhythmically expressed splicing factors and core clock oscillators fill the role of temporal regulators participating in improving plant growth, development, and increasing plant tolerance against abiotic stresses.
The circadian clock permits an organism temporal coordination of the biological processes at specific times of the day or night, even in the absence of periodicity in the environment. The circadian clock is an endogenous rhythm mechanism in plants which integrates with several external factors to adjust physiological traits of the plants (Covington et al., 2008; Mizuno and Yamashino, 2008; Hsu and Harmer, 2012), providing an advantage to promote plant growth, survival, and adaption.
In the majority of the studies, the circadian clock is predicted to function as a biochemical oscillator with different functional modules, including input, oscillator, and output (Hsu and Harmer, 2014; Greenham and McClung, 2015). Input must be synchronized every day by the diurnal variations to environmental conditions, such as light, temperature, and hormones. (Bordage et al., 2016; Greenwood et al., 2019). Central oscillators are mutually regulated by feedback loops to generate the rhythms in their own expression through transcriptional and post-transcriptional regulation and also drive diverse physiological processes, including growth, flowering time, and stress responses (Figure 1A). Input-oscillator-output is clearly oversimplified as a classical figure view of the circadian system, and now it relies on a much more complex network and can be regulated by other associated life pathways (Harmer, 2009; McClung, 2019).
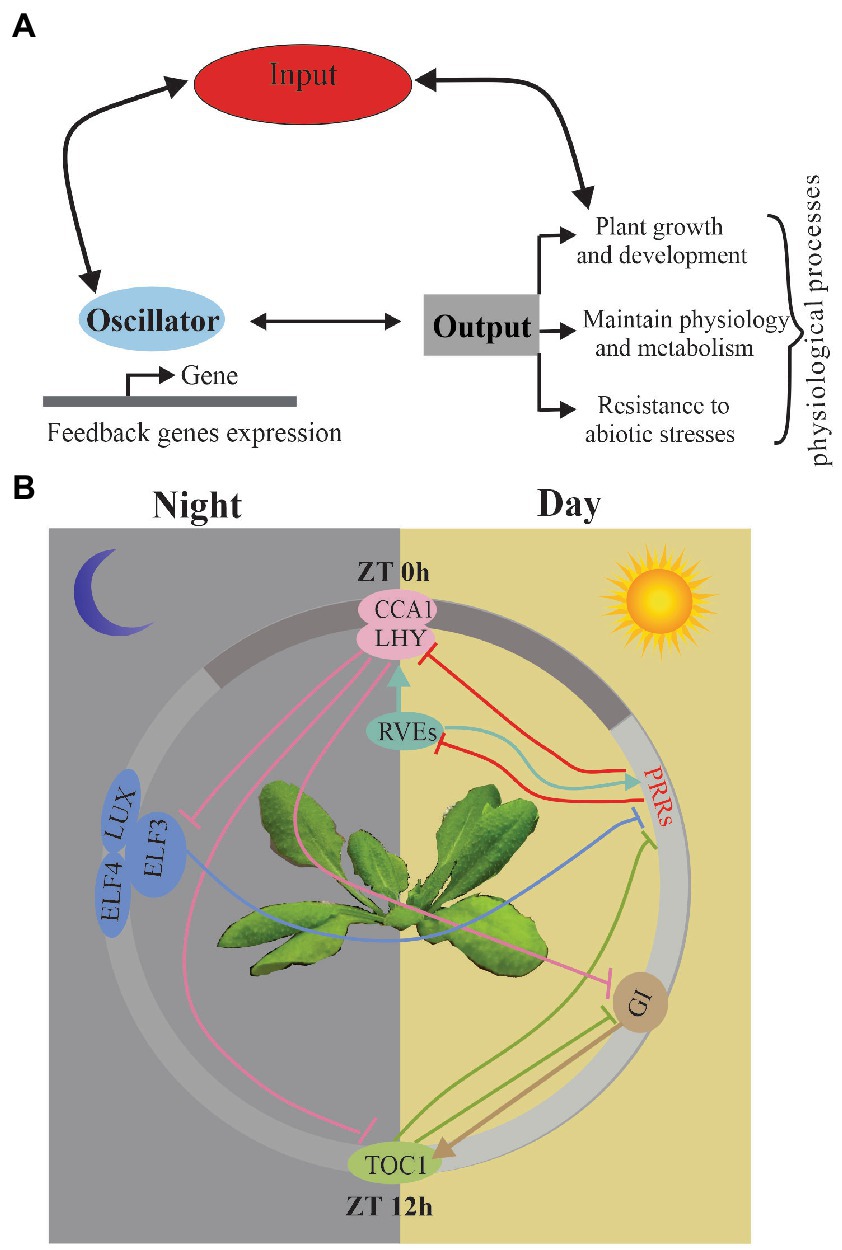
Figure 1. (A) Schematic of the circadian clock. A simplified version of the main functional modules of the circadian system; arrows indicate the relationship between modules. (B) The current model of A. thaliana circadian oscillator network with detailed model for transcriptional regulation among the clock components. It shows the relative time of action of each component from left to right in a circadian cycle. The white area indicates subjective day; gray area indicates subjective night. Arrows indicate induction, while perpendicular lines indicate transcriptional suppression.
Based on the importance of clock function, a tremendous amount of progress has been made on understanding the molecular mechanisms responsible for clock entrainment. In the past decades, extensive information on circadian synchronization induced by light has been found through studies on some plants, especially in Arabidopsis thaliana. In addition, a large amount of research indicated the involvement of temperature in circadian rhythms (Greenham and McClung, 2015; Philippou et al., 2019). Moreover, other signals such as hormones would entrain plant clock (Atamian and Harmer, 2016).
In A. thaliana, a model studying plant circadian clock, the oscillators are regulated transcriptionally and post-transcriptionally through AS, protein–protein interactions, and regulation of protein stability. Components of the circadian oscillator can be described based on the timing of their expression patterns with morning-phased (CCA1 and LHY), daytime-phased (PRR7, PRR9), afternoon-phased (RVE4, RVE6 and RVE8), and evening-phased genes (PRR5, TOC1, LUX, ELF3, ELF4, and GI) (Greenham and McClung, 2015; Philippou et al., 2019). The widespread feedback loops regulate the expression of each gene and their downstream targets all day (Harmer, 2009; Pokhilko et al., 2013; Fogelmark and Troein, 2014; Hsu and Harmer, 2014; McClung, 2014) (Figure 1B). In the morning phase, a motif exists in the TOC1 promoter named the evening element (EE), CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) repress the expression of an evening-phased component TIMING OF CAB EXPRESSION 1 (TOC1) (Endo et al., 2014). Recently, CCA1 and LHY have also been shown to repress several other evening genes, including GIGANTEA (GI), LUX ARRHYTHMO (LUX), EARLY FLOWERING 3 (ELF3), and ELF4 (McClung, 2011; Endo et al., 2014; Nolte and Staiger, 2015). CA1 and LHY repress evening genes depending on DEETIOLATED1 (DET1), a key repressor in photo morphogenesis (Romanowski and Yanovsky, 2015). In the day phase, PRRs (PRR9, PRR7 and PRR5) could repress the expression of the morning genes CCA1 and LHY (James et al., 2012). Recently, the PRRs have been shown to also repress expression of REVEILLE8 (RVE8) (McClung, 2014). In the afternoon phase, RVE8 is associated with PRR5 and TOC1 promoters (Salome et al., 2002; Harmer, 2009). RVE4 and RVE6, two close homologs of RVE8, also play the same roles as RVE8 in the circadian system (Harmer, 2009). In the evening phase, similar to TOC1 homologs PPRs, TOC1 represses CCA1 and LHY expression (Pokhilko et al., 2013; de Montaigu et al., 2015). Moreover, LUX, ELF3, and ELF4 represses the expression of the day-phased clock gene PRR9 (Hsu and Harmer, 2014). GI have been verified to regulate the TOC1 expression.
The central oscillator can generate rhythmic outputs, including growth and development, stress, and metabolism (Greenham and McClung, 2015; Malapeira et al., 2015; Atamian and Harmer, 2016). The circadian clock generates rhythms in plant growth and development processes such as hypocotyl growth, hormone signaling, germination, leaf and root growth, architecture, and flowering time (de Montaigu et al., 2010; Stitt and Zeeman, 2012; Kinmonth-Schultz et al., 2013; Johansson and Staiger, 2015). Moreover, plant metabolism is an important output, influencing starch metabolism, photosynthesis, fitness, biomass, metabolite profiling second messengers, micronutrients, and mineral homeostasis (Haydon et al., 2013; Malapeira et al., 2015). In addition, under circadian clock control, cold, drought, salinity, and reactive oxygen species (ROS) could also respond (Legnaioli et al., 2009; Grundy et al., 2015).
Abiotic stresses are the major environmental constraints that affect plant growth and productivity due to rapid fluctuations in ambient conditions. To adapt to abiotic stress challenges, plants can initiate appropriate molecular, cellular, and physiological adjustments to survive and reproduce (Figure 2). Plants transmit the stress signals within cells as well as between cells and tissues, and activate stress-related genes to respond to stress signals through transcriptional and post-transcriptional regulation, particularly splicing factor mediated pre-RNA splicing and DNA methylation (Bertolini et al., 2018; Zhang et al., 2018; Huertas et al., 2019; Chen et al., 2020). Moreover, hormones play a pivotal role in plant development and signaling networks regulating plant responses to a wide range of environmental conditions. Plant hormones fasten on the significant link between different hormones in response to stress, such as ABI5 in relation to other phytohormones involved in the abiotic stress responses, apocarotenoids which induce other phytohormones critical for plant growth, development, and stress response, and gibberellins which act as a promoter in primary root development (Skubacz et al., 2016; Verma et al., 2016; Felemban et al., 2019; Lopez-Ruiz et al., 2020). Indeed, the circadian clock regulates environmental stresses and plant biological processes, particularly the major circadian clock genes, such as PRR5 and PRR7 in A. thaliana, and LHY in A. thaliana and Glycine max (Kolmos et al., 2014; Nakamichi et al., 2016; Yoo et al., 2020; Wang K. et al., 2012). Moreover, OsPRR73, a circadian component, may confer salt tolerance by recruiting HDAC10 to repress OsHKT2;1, thus decreasing cellular Na(+) accumulation (Wei et al., 2021). In addition, the rice evening complex, composed of OsELF4a, OsELF3-1, and OsLUX, could regulate heading date and salt tolerance. Interestingly, in long day conditions, GI mutant osgi-101 showed salt tolerance and exhibits early heading phenotype. Compelling evidence shows that OsEC1 represses OsGI and thus links the circadian clock with salt tolerance (Wang X. et al., 2012). Moreover, DNA affinity purification sequencing (DAP-seq) coupled with transcriptome analysis indicates a direct transcriptional target of OsCCA1, substantially enriched in ABA signaling pathway. Rice CCA1 could transcriptionally regulate ABA signaling to confer tolerance to multiple abiotic stresses by a direct association of OsPP108 and OsbZIP46 promoters (Wei et al., 2022).
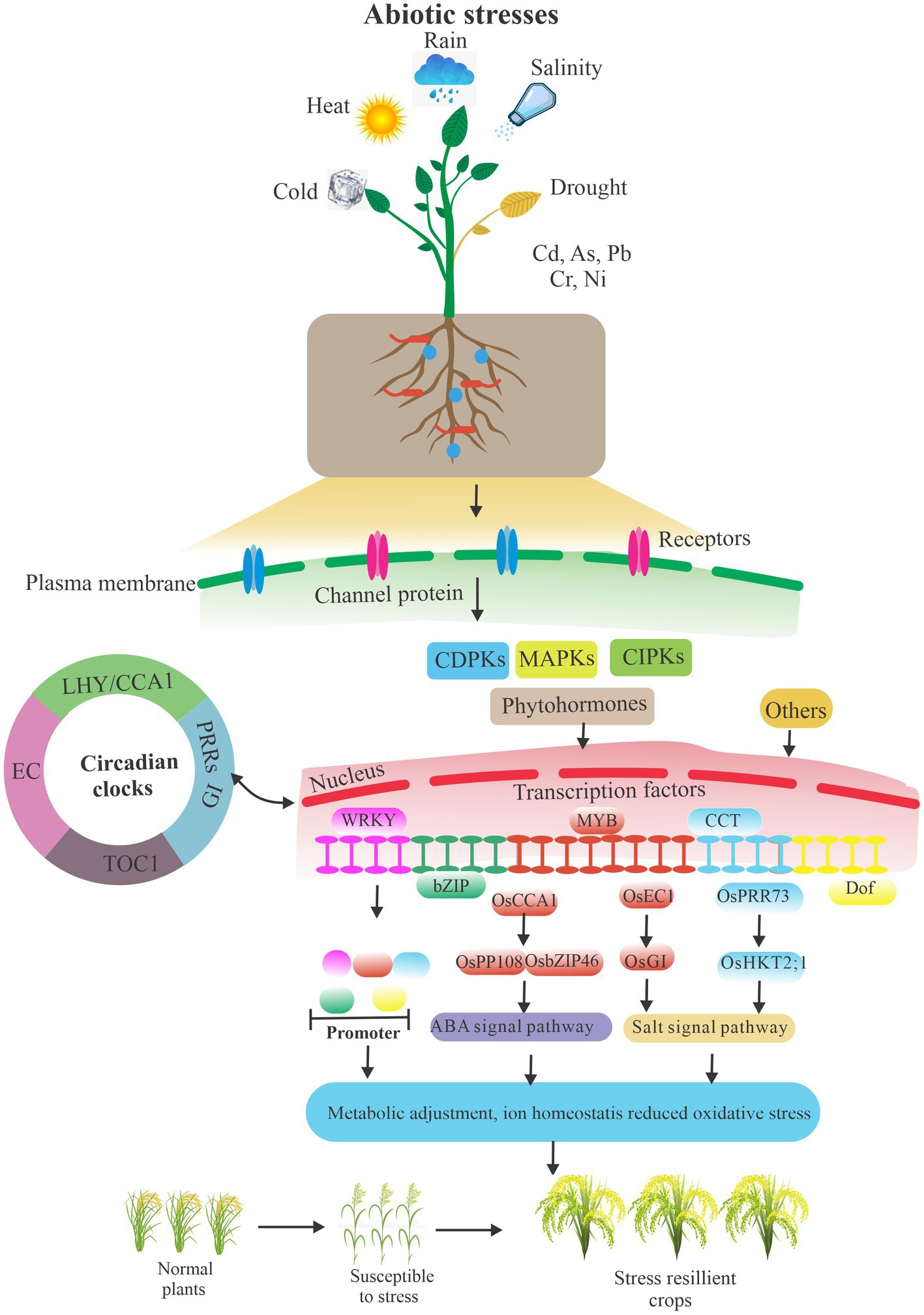
Figure 2. A simplified schematic of abiotic stresses and plant biology. Plants resist abiotic stresses by interacting with signaling pathways, circadian clocks, hormones, crucial genes transcriptional and post-transcriptional regulation, and other ways under environmental challenges. Others included endophytic fungi, a self-antioxidant defense system, proline accumulation, and so on. The arrows indicate the relationship between modules.
Plants adapt to various environmental stresses by endophytic fungi, a self-antioxidant defense system, proline accumulation, and so on (Khan et al., 2015; Wani et al., 2019; Hasanuzzaman et al., 2020). Interestingly, transcriptional regulation also plays an important role in plant biological processes from plant development to stress responses (Thatcher et al., 2016; Shang et al., 2017; Calixto et al., 2018; Szakonyi and Duque, 2018; Nimeth et al., 2020). In natural and agricultural backgrounds, plants also constantly suffer from environmental conditions. Stress mediates plant SR protein genes’ expression changing at the transcriptional and transcriptomic level (Zhang et al., 2020). Data certified that drought induced large developmental splicing changes in leaf and ear but relatively few in tassel.
To date, several studies documented the crosstalk between the circadian clock and AS, including splicing the clock to maintain and entrain circadian rhythms in Drosophila melanogaster (Shakhmantsir and Sehgal, 2019), chromatin remodeling, and AS of the A. thaliana circadian clock (Henriques and Mas, 2013). In this study, we paid more attention to the links between circadian and AS of plant life activities responsive to abiotic stresses, making it different from the published articles.
Research into what regulates circadian and AS offered more details about the fragile crosslink under environmental stresses, especially temperature changes (Figure 3).
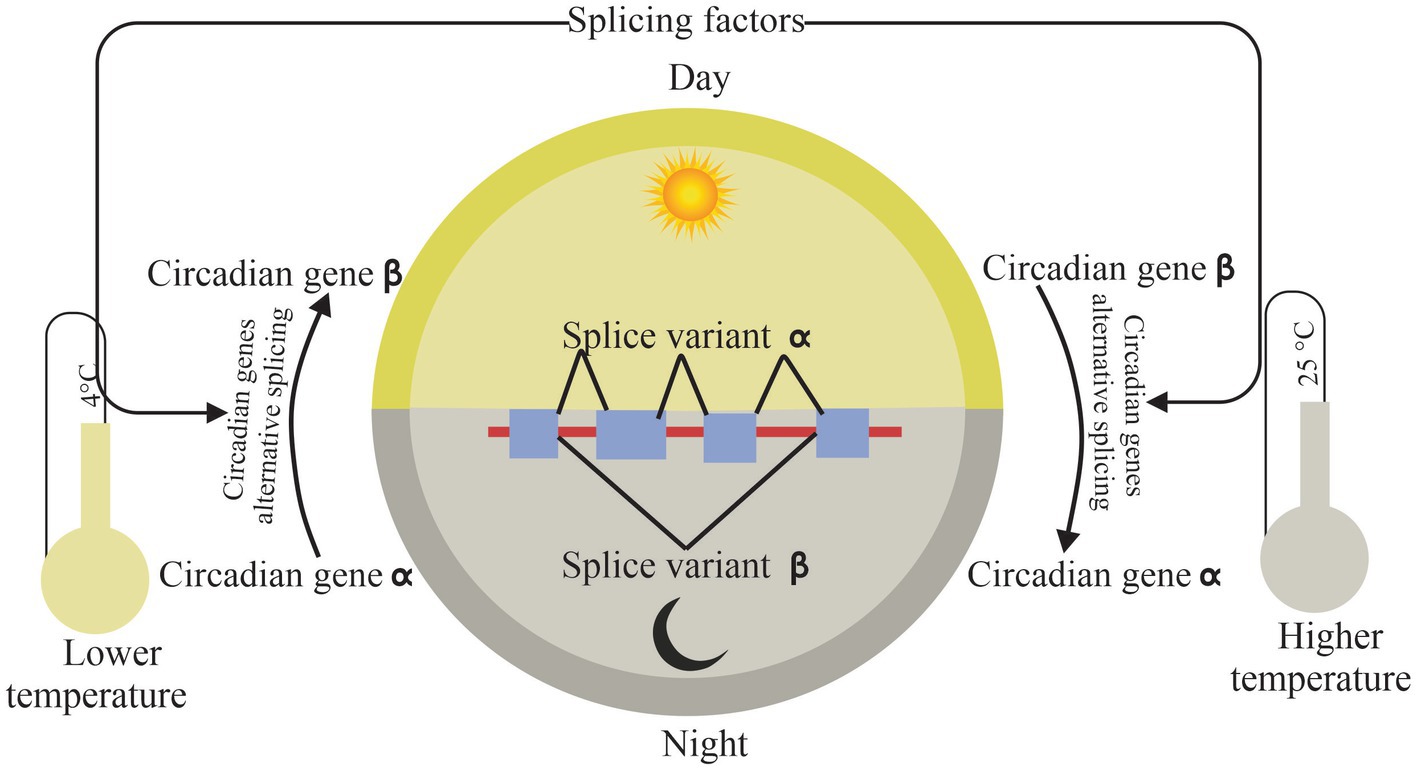
Figure 3. Connection of circadian and alternative splicing. Splicing factors affect circadian rhythm by self and core circadian genes splicing under temperature stress, while circadian-mediated alternative splicing according to rhythmic temperature changes in day and night. The arrows indicate the relationship between modules.
As a post-translational controller, AS plays a prominent role in the circadian clock, especially in spliceosome. SPLICEOSOMA L TIMEKEEPER LOCUS1 (STIPL1), a component of A. thaliana spliceosome, could affect the circadian period. There exists two STIPL splicing transcripts in A. thaliana. Interestingly, mutation of another STIPL2 does not cause splicing defects or changes in circadian rhythms in A. thaliana, diverging from STIPL1 (Bertoni, 2012), which may reveal the function of splicing factors in AS. SNW/Ski-interacting protein (SKIP, splicing factor), a component of the spliceosome, physically associates with the spliceosomal splicing factor Ser/Arg-rich protein45 and interacts with the pre-mRNA of clock genes, such as PRR9 and PRR7, involved in AS and mRNA maturation. Genome-wide investigations show that SKIP participate in regulating AS of several genes, possibly via modulating recognition or cleavage of 5′ and 3′ splice donor and acceptor sites. Therefore, SKIP could link AS and the circadian clock as post-transcriptional regulation in A. thaliana (Wang et al., 2012). Moreover, growing evidence suggests that some LSM genes encode core components of the spliceosome U6 complex which regulate circadian rhythms in both plants and mammals. The expression and AS of some core clock genes were different in A. thaliana lsm5 mutants. Later, expression analysis of either a weak lsm5 or a strong lsm4 mutant allele in A. thaliana displayed larger effects on AS than on constitutive splicing, including circadian clock genes. Notably, RNA-seq reveals large splicing defects were not observed in most of the introns evaluated in the strong lsm4 mutant allele, especially circadian clock genes. These findings support the idea that some LSM genes influence core clock genes AS, particularly intron retention events (Perez-Santangelo et al., 2014). Meanwhile, the spliceosome assembly factor GEMIN2 controls the AS of several clock genes and deadens caused by temperature on the circadian period in A. thaliana (Schlaen et al., 2015). Recently it has been discovered that PRMT5, a protein arginine methyl transferase, adds a new loop within the circadian clock of the A. thaliana by regulating AS of main clock genes. Meanwhile, investigations showed that PRMT5 has a role in the regulation of AS and the circadian network in D. melanogaster (Petrillo et al., 2011). Thus, these major spliceosomes can reveal several abiotic stress responses, and a link between AS and circadian rhythm changes.
Interestingly, the circadian clock could deal with the environmental changes by AS. Temperature variation can cause extensive dynamic changes in AS of clock genes and alternatively spliced transcripts. Temperature-associated AS is an additional loop to regulate the plant circadian clock (James et al., 2012). CIRCADIAN CLOCK-ASSOCIATED1 (CCA1), a core clock component, can be self-regulated by a splice variant CCA1β which is inhibited by low temperatures, while CCA1β suppresses the activities of functional CCA1α and LATE ELONGATED HYPOCOTYL (LHY) transcription factors by forming nonfunctional CCA1α-CCA1β and LHY-CCA1β heterodimers in A. thaliana (Park et al., 2012). While in control condition, CCA1α actively expresses without binging CCA1β. Moreover, frequency of AS, FRQ, a circadian clock gene, exhibits a robust circadian rhythm and regulates the response of the circadian clock to temperature changes. l-FRQ has a normal expression in condition temperatures. But the amount of l-FRQ increases significantly as temperatures rise, whereas s-FRQ levels increase while temperature is low. This leads to a variation in the l-FRQ to s-FRQ ratio as a function of temperature (Diernfellner et al., 2005, 2007). A later study investigated how the major clock genes, such as TIMING OF CAB EXPRESSION 1 (TOC1) and EARLY FLOWERING 3 (ELF3), undergo extensive AS under all sorts of environmental conditions, indicating AS creates a linkage between the circadian clock and environmental stress adaptation in plants (Kwon et al., 2014). Compared to normal conditions, TOC1 and ELF3 increase considerably in intron retention events under cold conditions. In recent years, new technology tools such as global circadian RNA-seq event centered, a splicing analysis tool, offer a new approach for study between the circadian clock and splicing events. Interestingly, expression of AS events of the circadian clock vary corresponding with the season and temperature in sugarcane (Dantas et al., 2019). That indicated plants deal with abiotic stress by major gene rhythmic expression which is regulated by AS.
Much evidence indicated some genes participating in abiotic stress responses, especially temperature stress, could undergo AS according to circadian rhythm changes. AtGRP7, a protein which is mediated by the circadian clock, increases stress tolerance under cold conditions and through this undergoes circadian oscillations. The protein can also autoregulate its expression by binding to its own pre-mRNA to influence its own AS (Staiger et al., 2003). Moreover, in D. melanogaster, splicing of an intron at the 3′ untranslated region of the period (per) mRNA is enhanced at cold temperatures, which reveals that daily fluctuations in the splicing of intron is regulated in a manner that depends on the photoperiod and temperature (Majercak et al., 2004). In addition, in plants, daily and circadian oscillated gene PROTEIN ARGININE METHYL TRANSFERASE 5 (PRMT5), involved in vernalization and carrying methyl groups to arginine residues present in histones and Sm spliceosome proteins, could link the circadian clock to controlling AS. Mutant Atprmt5 impairs several circadian rhythms and phenotypes resulting from AS of the core-clock gene PRR9. Further studies show that PRMT5 participates in the regulation of many pre-messenger-RNA splicing events, probably by modulating 5′-splice-site recognition, which indicates PRMT5 could link the circadian clock and AS to help organisms to synchronize physiological progresses to deal with daily changes in environmental conditions (Sanchez et al., 2010). There are few papers that focus on the changes of expression of key regulators of AS regulated by a circadian clock under both control and abiotic stress conditions, which provides us a novel viewpoint to explore in the future.
Numerous scholarly articles focus on AS, the circadian clock, and abiotic stress, but very little evidence pointed out how to deal with abiotic stresses via circadian-mediated AS in plants. The circadian clock regulates AS in a tissue-dependent manner and concurrent with circadian transcript abundance (McGlincy et al., 2012), found in different tissues and circadian transcript. One could hypothesize that the genes undergo regular splicing variant in tissues and development stages. Once the environment changes, the major splicing variant expressed more to respond to the particular stress. Recent research indicates the clock is a temporal regulator of AS (Genov et al., 2019), which may give us a possible explanation about temporal diversification of the proteome. More evidence indicates that AS events corresponding with circadian are widespread across mammalian tissues and might conduce to a temporal diversification of the proteome (El-Athman and Relogio, 2018). However, it remains to be determined which genes are the temporal regulator in plant life progress. The rhythmically expressed splicing factors and core clock oscillators fill the role of temporal regulators to participate in plant growth, development, and dealing with abiotic stresses. Those data present global circadian RNA-seq event, using a splicing analysis tool to shed light on the possible relationship between AS, circadian, and abiotic stresses.
Moreover, phytohormones play a critical role in plant adaptation to environmental stresses; the circadian clock could respond to these stresses through AS events (Figure 2). However, only a few studies have reported on how this process is mediated. A board range of studies investigated temperature variations, while only a few were conducted on drought and salinity stress. The abscisic acid (ABA), known as a plant stress hormone, plays a major role in abiotic stress responses, particularly under drought and salt stress (Zhang et al., 2006; Ma and Qin, 2014). Interestingly, the circadian clock regulates ABA signaling by circadian oscillator. There are bidirectional interactions between the circadian oscillator TIMING OF CAB2 EXPRESSION1 (TOC1) and ABA signaling (Legnaioli et al., 2009). ABA-inducible R2R3-type MYB transcription factor, MYB96, binds directly to the TOC1 promoter to activate its expression; TOC1 in turn regulates MYB96 expression possibly via CCA1. The whole complex CCA1-MYB96-TOC1 circuit connects circadian and ABA signaling to address abiotic stress (Lee et al., 2016). LATE ELONGATED HYPOCOTYL (LHY), a circadian oscillator, binds directly on the promoters of genes in ABA signal pathway, connecting circadian regulation with drought and salt stress tolerance through ABA signaling (Belbin and Dodd, 2018). More evidence indicates that AS regulates stress responses largely by targeting the ABA pathway. For instance, A. thaliana seedlings treated with ABA demonstrated varied conventional AS isoforms expression and increased non-conventional AS events number (Zhu et al., 2017). In agreement, plant mutant defective in splicing factors are severely impaired in their response to abiotic stress, such as STA1 (Lee et al., 2006), LSM4 (Zhang et al., 2011), and SKIP (Feng et al., 2015). These findings provide us a possible hypothesis: splicing factors may target circadian oscillator genes splicing or splicing factors via self-splicing to joint circadian oscillator genes to link ABA signaling pathway and respond to abiotic stress.
Auxin, another plant hormone, plays key roles in plant development and responses to environmental cues, and there is a new crosslink of the clock and auxin. The circadian clock core gene REVEILLE1 (RVE1) could regulate the expression of the auxin biosynthetic gene YUCCA8 (YUC8), suggesting a mechanism for coordinating plant growth with rhythmic changes in the variable environment (Rawat et al., 2009). CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) plays an essential role in gating auxin response (Xue et al., 2020). Interestingly, splice variant CCA1β inhibited by low temperature could suppress the function of another splice variant CCA1α, which exists as a new net between circadian and AS response to abiotic stresses through hormonal signaling pathways.
Y-SC and K-LZ: conceptualization. TF, MA, J-LZ, and M-XC: writing original draft preparation. TF, M-XC, J-LZ, SD, and Y-SC: writing review and editing. Y-SC: funding. All authors have read and agreed to the published version of the manuscript.
This work was supported by the Science Technology and Innovation Committee of Shenzhen (2021N062-JCYJ20210324115408023), the Natural Science Foundation of Jiangsu Province (SBK2020042924), the National Natural Science Foundation of China (32001452), and the Hong Kong Research Grant Council (AoE/M-403/16, GRF14160516, 12100318 and 12103220).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Atamian, H. S., and Harmer, S. L. (2016). Circadian regulation of hormone signaling and plant physiology. Plant Mol. Biol. 91, 691–702. doi: 10.1007/s11103-016-0477-4
Belbin, F. E., and Dodd, A. N. (2018). ABA signalling is regulated by the circadian clock component LHY. New Phytol. 220, 661–663. doi: 10.1111/nph.15473
Bertolini, E., Pe, M. E., and Mica, E. (2018). Transcriptional and posttranscriptional regulation of drought stress treatments in brachypodium leaves. Methods Mol. Biol. 1667, 21–29. doi: 10.1007/978-1-4939-7278-4_2
Bertoni, G. (2012). Circadian rhythms require proper RNA splicing. Plant Cell 24:3856. doi: 10.1105/tpc.112.241012
Bordage, S., Sullivan, S., Laird, J., Millar, A. J., and Nimmo, H. G. (2016). Organ specificity in the plant circadian system is explained by different light inputs to the shoot and root clocks. New Phytol. 212, 136–149. doi: 10.1111/nph.14024
Calixto, C., Guo, W., James, A. B., Tzioutziou, N. A., Entizne, J. C., Panter, P. E., et al. (2018). Rapid and dynamic alternative splicing impacts the Arabidopsis cold response transcriptome. Plant Cell 30, 1424–1444. doi: 10.1105/tpc.18.00177
Chen, H., Ruan, J., Chu, P., Fu, W., Liang, Z., Li, Y., et al. (2020). AtPER1 enhances primary seed dormancy and reduces seed germination by suppressing the ABA catabolism and GA biosynthesis in Arabidopsis seeds. Plant J. 101, 310–323. doi: 10.1111/tpj.14542
Covington, M. F., Maloof, J. N., Straume, M., Kay, S. A., and Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9:R130. doi: 10.1186/gb-2008-9-8-r130
Dantas, L., Calixto, C., Dourado, M. M., Carneiro, M. S., Brown, J., Hotta, C., et al. (2019). Alternative splicing of circadian clock genes correlates with temperature in field-grown sugarcane. Front. Plant Sci. 10:1614. doi: 10.3389/fpls.2019.01614
de Montaigu, A., Giakountis, A., Rubin, M., Toth, R., Cremer, F., Sokolova, V., et al. (2015). Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc. Natl. Acad. Sci. U.S.A. 112, 905–910. doi: 10.1073/pnas.1422242112
de Montaigu, A., Toth, R., and Coupland, G. (2010). Plant development goes like clockwork. Trends Genet. 26, 296–306. doi: 10.1016/j.tig.2010.04.003
Diernfellner, A., Colot, H. V., Dintsis, O., Loros, J. J., Dunlap, J. C., and Brunner, M. (2007). Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett. 581, 5759–5764. doi: 10.1016/j.febslet.2007.11.043
Diernfellner, A. C., Schafmeier, T., Merrow, M. W., and Brunner, M. (2005). Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev. 19, 1968–1973. doi: 10.1101/gad.345905
El-Athman, R., and Relogio, A. (2018). Escaping circadian regulation: an emerging hallmark of cancer? Cell Syst. 6, 266–267. doi: 10.1016/j.cels.2018.03.006
Endo, M., Shimizu, H., Nohales, M. A., Araki, T., and Kay, S. A. (2014). Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515, 419–422. doi: 10.1038/nature13919
Felemban, A., Braguy, J., Zurbriggen, M. D., and Al-Babili, S. (2019). Apocarotenoids involved in plant development and stress response. Front. Plant Sci. 10:1168. doi: 10.3389/fpls.2019.01168
Feng, J., Li, J., Gao, Z., Lu, Y., Yu, J., Zheng, Q., et al. (2015). SKIP confers osmotic tolerance during salt stress by controlling alternative gene splicing in Arabidopsis. Mol. Plant 8, 1038–1052. doi: 10.1016/j.molp.2015.01.011
Fogelmark, K., and Troein, C. (2014). Rethinking transcriptional activation in the Arabidopsis circadian clock. PLoS Comput. Biol. 10:e1003705. doi: 10.1371/journal.pcbi.1003705
Genov, N., Basti, A., Abreu, M., Astaburuaga, R., and Relogio, A. (2019). A bioinformatic analysis identifies circadian expression of splicing factors and time-dependent alternative splicing events in the HD-MY-Z cell line. Sci. Rep. 9:11062. doi: 10.1038/s41598-019-47343-w
Greenham, K., and McClung, C. R. (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16, 598–610. doi: 10.1038/nrg3976
Greenwood, M., Domijan, M., Gould, P. D., Hall, A., and Locke, J. (2019). Coordinated circadian timing through the integration of local inputs in Arabidopsis thaliana. PLoS Biol. 17:e3000407. doi: 10.1371/journal.pbio.3000407
Grundy, J., Stoker, C., and Carre, I. A. (2015). Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 6:648. doi: 10.3389/fpls.2015.00648
Harmer, S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60, 357–377. doi: 10.1146/annurev.arplant.043008.092054
Hasanuzzaman, M., Bhuyan, M., Zulfiqar, F., Raza, A., Mohsin, S. M., Mahmud, J. A., et al. (2020). Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants (Basel) 9(8), 618. doi: 10.3390/antiox9080681
Haydon, M. J., Hearn, T. J., Bell, L. J., Hannah, M. A., and Webb, A. A. (2013). Metabolic regulation of circadian clocks. Semin. Cell Dev. Biol. 24, 414–421. doi: 10.1016/j.semcdb.2013.03.007
Henriques, R., and Mas, P. (2013). Chromatin remodeling and alternative splicing: pre- and post-transcriptional regulation of the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 24, 399–406. doi: 10.1016/j.semcdb.2013.02.009
Hsu, P. Y., and Harmer, S. L. (2012). Circadian phase has profound effects on differential expression analysis. PLoS One 7:e49853. doi: 10.1371/journal.pone.0049853
Hsu, P. Y., and Harmer, S. L. (2014). Wheels within wheels: the plant circadian system. Trends Plant Sci. 19, 240–249. doi: 10.1016/j.tplants.2013.11.007
Huertas, R., Catala, R., Jimenez-Gomez, J. M., Mar, C. M., Crevillen, P., Pineiroet, M., et al. (2019). Arabidopsis SME1 regulates plant development and response to abiotic stress by determining spliceosome activity specificity. Plant Cell 31, 537–554. doi: 10.1105/tpc.18.00689
James, A. B., Syed, N. H., Bordage, S., Marshall, J., Nimmo, G. A., Jenkins, G. I., et al. (2012). Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24, 961–981. doi: 10.1105/tpc.111.093948
Johansson, M., and Staiger, D. (2015). Time to flower: interplay between photoperiod and the circadian clock. J. Exp. Bot. 66, 719–730. doi: 10.1093/jxb/eru441
Khan, A. L., Hussain, J., Al-Harrasi, A., Al-Rawahi, A., and Lee, I. J. (2015). Endophytic fungi: resource for gibberellins and crop abiotic stress resistance. Crit. Rev. Biotechnol. 35, 62–74. doi: 10.3109/07388551.2013.800018
Kinmonth-Schultz, H. A., Golembeski, G. S., and Imaizumi, T. (2013). Circadian clock-regulated physiological outputs: dynamic responses in nature. Semin. Cell Dev. Biol. 24, 407–413. doi: 10.1016/j.semcdb.2013.02.006
Kolmos, E., Chow, B. Y., Pruneda-Paz, J. L., and Kay, S. A. (2014). HsfB2b-mediated repression of PRR7 directs abiotic stress responses of the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 111, 16172–16177. doi: 10.1073/pnas.1418483111
Kwon, Y. J., Park, M. J., Kim, S. G., Baldwin, I. T., and Park, C. M. (2014). Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 14:136. doi: 10.1186/1471-2229-14-136
Lee, B. H., Kapoor, A., Zhu, J., and Zhu, J. K. (2006). STABILIZED1, a stress-upregulated nuclear protein, is required for pre-mRNA splicing, mRNA turnover, and stress tolerance in Arabidopsis. Plant Cell 18, 1736–1749. doi: 10.1105/tpc.106.042184
Lee, H. G., Mas, P., and Seo, P. J. (2016). MYB96 shapes the circadian gating of ABA signaling in Arabidopsis. Sci. Rep. 6:17754. doi: 10.1038/srep17754
Legnaioli, T., Cuevas, J., and Mas, P. (2009). TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 28, 3745–3757. doi: 10.1038/emboj.2009.297
Lopez-Ruiz, B. A., Zluhan-Martinez, E., Sanchez, M. P., Alvarez-Buylla, E. R., and Garay-Arroyo, A. (2020). Interplay between hormones and several abiotic stress conditions on Arabidopsis thaliana primary root development. Cells 9(12), 2576. doi: 10.3390/cells9122576
Majercak, J., Chen, W. F., and Edery, I. (2004). Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol. Cell. Biol. 24, 3359–3372. doi: 10.1128/MCB.24.8.3359-3372.2004
Malapeira, J., Benlloch, R., Henriques, R., and Mas, P. (2015). Plant circadian network: an integrative view. New York: Springer, 1–41.
Ma, Y., and Qin, F. (2014). “ABA regulation of plant responses to drought and salt stresses,” in, ed. D. Zhang (Springer Netherlands: Dordrecht), 315–336.
McClung, C. R. (2011). The genetics of plant clocks. Adv. Genet. 74, 105–139. doi: 10.1016/B978-0-12-387690-4.00004-0
McClung, C. R. (2014). Wheels within wheels: new transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep. 6:2. doi: 10.12703/P6-2
McClung, C. R. (2019). The plant circadian oscillator. Biology (Basel) 8. doi: 10.3390/biology8010014
McGlincy, N. J., Valomon, A., Chesham, J. E., Maywood, E. S., Hastings, M. H., and Ule, J. (2012). Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol. 13:R54. doi: 10.1186/gb-2012-13-6-r54
Mizuno, T., and Yamashino, T. (2008). Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 49, 481–487. doi: 10.1093/pcp/pcn008
Nakamichi, N., Takao, S., Kudo, T., Kiba, T., Wang, Y., Kinoshita, T., et al. (2016). Improvement of Arabidopsis biomass and cold, drought and salinity stress tolerance by modified circadian clock-associated pseudo-response regulators. Plant Cell Physiol. 57, 1085–1097. doi: 10.1093/pcp/pcw057
Nimeth, B. A., Riegler, S., and Kalyna, M. (2020). Alternative splicing and DNA damage response in plants. Front. Plant Sci. 11:91. doi: 10.3389/fpls.2020.00091
Nolte, C., and Staiger, D. (2015). RNA around the clock – regulation at the RNA level in biological timing. Front. Plant Sci. 6:311. doi: 10.3389/fpls.2015.00311
Park, M. J., Seo, P. J., and Park, C. M. (2012). CCA1 alternative splicing as a way of linking the circadian clock to temperature response in Arabidopsis. Plant Signal. Behav. 7, 1194–1196. doi: 10.4161/psb.21300
Perez-Santangelo, S., Mancini, E., Francey, L. J., Schlaen, R. G., Chernomoretz, A., Hogenesch, J. B., et al. (2014). Role for LSM genes in the regulation of circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 111, 15166–15171. doi: 10.1073/pnas.1409791111
Petrillo, E., Sanchez, S. E., Kornblihtt, A. R., and Yanovsky, M. J. (2011). Alternative splicing adds a new loop to the circadian clock. Commun. Integr. Biol. 4, 284–286. doi: 10.4161/cib.4.3.14777
Philippou, K., Ronald, J., Sanchez-Villarreal, A., Davis, A. M., and Davis, S. J. (2019). Physiological and genetic dissection of sucrose inputs to the Arabidopsis thaliana circadian system. Genes (Basel) 10(5), 334. doi: 10.3390/genes10050334
Pokhilko, A., Mas, P., and Millar, A. J. (2013). Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst. Biol. 7:23. doi: 10.1186/1752-0509-7-23
Rawat, R., Schwartz, J., Jones, M. A., Sairanen, I., Cheng, Y., Kinoshita, T., et al. (2009). REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. U.S.A. 106, 16883–16888. doi: 10.1073/pnas.0813035106
Romanowski, A., and Yanovsky, M. J. (2015). Circadian rhythms and post-transcriptional regulation in higher plants. Front. Plant Sci. 6:437. doi: 10.3389/fpls.2015.00437
Salome, P. A., Michael, T. P., Kearns, E. V., Fett-Neto, A. G., Sharrock, R. A., McClung, C. R., et al. (2002). The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol. 129, 1674–1685. doi: 10.1104/pp.003418
Sanchez, S. E., Petrillo, E., Beckwith, E. J., Zhang, X., Rugnone, M. L., Kinoshita, T., et al. (2010). A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116. doi: 10.1038/nature09470
Schlaen, R. G., Mancini, E., Sanchez, S. E., Perez-Santangelo, S., Rugnone, M. L., Simpsonet, C. J., et al. (2015). The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 112, 9382–9387. doi: 10.1073/pnas.1504541112
Shakhmantsir, I., and Sehgal, A. (2019). Splicing the clock to maintain and entrain circadian rhythms. J. Biol. Rhythm. 34, 584–595. doi: 10.1177/0748730419868136
Shang, X., Cao, Y., and Ma, L. (2017). Alternative splicing in plant genes: a means of regulating the environmental fitness of plants. Int. J. Mol. Sci. 18. doi: 10.3390/ijms18020432
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I. (2016). The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7, 1884. doi: 10.3389/fpls.2016.01884
Staiger, D., Zecca, L., Wieczorek, K.D.A., Apel, K., and Eckstein, L. (2003). The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. The Plant Journal: For Cell and Molecular Biology 33.
Stitt, M., and Zeeman, S. C. (2012). Starch turnover: pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15, 282–292. doi: 10.1016/j.pbi.2012.03.016
Szakonyi, D., and Duque, P. (2018). Alternative splicing as a regulator of early plant development. Front. Plant Sci. 9:1174. doi: 10.3389/fpls.2018.01174
Thatcher, S. R., Danilevskaya, O. N., Meng, X., Beatty, M., Zastrow-Hayes, G., Kinoshita, T., et al. (2016). Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 170, 586–599. doi: 10.1104/pp.15.01267
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wang, K., Bu, T., Cheng, Q., Dong, L., Su, T., Chen, Z., et al. (2021). Two homologous LHY pairs negatively control soybean drought tolerance by repressing the abscisic acid responses. New Phytol. 229, 2660–2675. doi: 10.1111/nph.17019
Wang, X., He, Y., Wei, H., and Wang, L. (2021). A clock regulatory module is required for salt tolerance and control of heading date in rice. Plant Cell Environ. 44, 3283–3301. doi: 10.1111/pce.14167
Wang, X., Wu, F., Xie, Q., Wang, H., Wang, Y., Yue, Y., et al. (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24, 3278–3295. doi: 10.1105/tpc.112.100081
Wani, A. S., Ahmad, A., Hayat, S., and Tahir, I. (2019). Epibrassinolide and proline alleviate the photosynthetic and yield inhibition under salt stress by acting on antioxidant system in mustard. Plant Physiol. Biochem. 135, 385–394. doi: 10.1016/j.plaphy.2019.01.002
Wei, H., Wang, X., He, Y., Xu, H., and Wang, L. (2021). Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2; 1-mediated sodium homeostasis. EMBO J. 40:e105086. doi: 10.15252/embj.2020105086
Wei, H., Xu, H., Su, C., Wang, X., and Wang, L. (2022). Rice CIRCADIAN CLOCK ASSOCIATED1 transcriptionally regulates ABA signaling to confer multiple abiotic stress tolerance. Plant Physiol. c196, kiac196. doi: 10.1093/plphys/kiac196
Xue, X., Sun, K., and Zhu, Z. (2020). CIRCADIAN CLOCK ASSOCIATED 1 gates morning phased auxin response in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 527, 935–940. doi: 10.1016/j.bbrc.2020.05.049
Yoo, Y. H., Jiang, X., and Jung, K. H. (2020). An abiotic stress responsive U-box e3 ubiquitin ligase is involved in OsGI-mediating diurnal rhythm regulating mechanism. Plants (Basel) 9(9), 1071. doi: 10.3390/plants9091071
Zhang, D., Chen, M. X., Zhu, F. Y., Zhang, J., and Liu, Y. G. (2020). Emerging Functions of Plant Serine/Arginine-Rich (SR) Proteins: Lessons from Animals. Critical Reviews in Plant Sciences, 39.
Zhang, H., Lang, Z., and Zhu, J. K. (2018). Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506. doi: 10.1038/s41580-018-0016-z
Zhang, J., Jia, W., Yang, J., and Ismail, A. M. (2006). Role of ABA in integrating plant responses to drought and salt stresses. Field Crop Res. 97, 111–119. doi: 10.1016/j.fcr.2005.08.018
Zhang, Z., Zhang, S., Zhang, Y., Wang, X., Li, D., Li, Q., et al. (2011). Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23, 396–411. doi: 10.1105/tpc.110.081356
Keywords: abiotic stress, circadian clock, plant, signal transduction, splicing regulation
Citation: Fan T, Aslam MM, Zhou J-L, Chen M-X, Zhang J, Du S, Zhang K-L and Chen Y-S (2022) A crosstalk of circadian clock and alternative splicing under abiotic stresses in the plants. Front. Plant Sci. 13:976807. doi: 10.3389/fpls.2022.976807
Received: 23 June 2022; Accepted: 05 August 2022;
Published: 06 October 2022.
Edited by:
Ji Huang, Nanjing Agricultural University, ChinaReviewed by:
Lei Wang, Institute of Botany (CAS), ChinaCopyright © 2022 Fan, Aslam, Zhou, Chen, Zhang, Du, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai-Lu Zhang, MTQyNTM1MjkyOUBxcS5jb20=; Yun-Sheng Chen, Y2hlbnl1bnNoZW5nbHdAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.