
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 02 September 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.975852
This article is part of the Research Topic Mechanisms of Abiotic Stress Responses and Tolerance in Plants: Physiological, Biochemical and Molecular Interventions, volume II View all 37 articles
 Raheela Jabeen1*
Raheela Jabeen1* Atia Iqbal2
Atia Iqbal2 Farah Deeba1
Farah Deeba1 Faisal Zulfiqar3*
Faisal Zulfiqar3* Ghulam Mustafa4
Ghulam Mustafa4 Haq Nawaz5
Haq Nawaz5 Ume Habiba2
Ume Habiba2 Muhammad Nafees3
Muhammad Nafees3 Abbu Zaid6
Abbu Zaid6 Kadambot H. M. Siddique7*
Kadambot H. M. Siddique7*Defensin genes form part of a plant’s defense system and are activated when exposed to biotic or abiotic stress. They play a vital role in controlling many signaling pathways involved in various plant defense mechanisms. This research aimed to isolate and characterize novel defensin genes from selected medicinally important plants to explore their signaling mechanisms and defense associated roles for breeding. The DNA of Albizia lebbeck and Moringa oleifera was subjected to PCR amplification using gene-specific primers of defensin genes. Two novel defensin genes were isolated in each species, with sequence lengths of 300 bp in A. lebbeck and 150 bp in M. oleifera. In-silico analysis undertaken to retrieve and align their orthologous sequences revealed 100% similarity of the A. lebbeck gene with the Musa acuminate peroxidase P7-like gene and 85% similarity of the M. oleifera gene with the Manihot esculenta GDP dissociation inhibitor gene. The reliability, stability and physiochemical properties of homology models of these sequences was confirmed through online computational studies. This preliminary study confirmed the presence of novel genes with peroxidase P7 and Rab GDP dissociation inhibitor gene-like activity in A. lebbeck and M. oleifera, respectively, and their potential defense role in plants. Thus, the defensin genes of both species could be used in the synthesis of transgenic self-defensive plants with increased disease resistance and as potential candidates for improved crop production and thraputic formulation in the future.
Plants are often exposed to biotic and abiotic stresses and use their natural defense system to circumvent them (Chojak-Koźniewska et al., 2018), producing several metabolites and secondary messengers to guard against these assaults (Reichling, 2018). Once the stress is detected, signaling pathways are triggered that induce gene expression to help plants acclimate to these conditions (Novaković et al., 2018). Among these, antimicrobial peptides (AMPs) form the first line of defense in a plant’s protective mechanisms (Mahlapuu et al., 2016) and are higly important part of innate immunity (Zharkova et al., 2019), serving an antimicrobial function (Rawat et al., 2017). Defensins belong to the predominant families of AMPs. They are small (45–55 amino acids), non-acidic, rich in cysteine residues, and highly similar to peptides in many other organisms (Al Akeel et al., 2014). Plant defensins involve mechanisms to inhibit fungi and bacteria, reportedly by the structural targets in the cell membrane of microbes by producing reactive oxygen species (Parisi et al., 2019). They can also protect the host by inducing and potentiating several other plant-defending mechanisms (Souza et al., 2017). The isolation and characterization of these defensins are vital for developing economically and medically important substances. Analysis of sequenced plant genomes revealed that defensins are present as multigene families and overrepresented in the genomes of some plant species (Kovaleva et al., 2017). Cowpea defensin Cp-thionin II was isolated and characterized, with its antifungal activity checked against F. culmorum (Schmidt et al., 2019). Similarly, two tomato defensin genes (SlyDF1 and SlyDF2) were cloned and characterized from Solanum lycopersicum (Cui et al., 2018). The AMPs are not only have their importance in plant defense system, but animals have also AMPs as their part of immune system, e.g., three novel β-defensin AMPs were also characterized in rainbow trout (Oncorhynchus mykiss; Casadei et al., 2009).
With the wealth of defensin nucleotide sequences available, gene isolation strategies coupled with recombinant production are used increasingly to characterize closely related plant defensin peptides. Many public databases and tools are available to study the structure and physicochemical properties of plant genes and transcriptomes (Lakshmi Priya and Sabu, 2015).
Albizzia lebbeck and Moringa oleifera have therapeutic potential against several diseases. Albizzia lebbeck bark extract is very much effective against speck disease (Arasu et al., 2022). The aquous extract of the same plant have reported anti-inflammatory activity (Kamala Lakshmi and Valarmathi, 2020). similarly the plant also possesses antidiabetic and anti lipedemic effect (Azam et al., 2022). Moringa oleifera also have several medicinal properties like anti diabetic, anti cancerous and anti inflammatory etc. (Kumar, 2017). Considering the importance of these medicinal plants, this study resulted in isolation and characterizion of defensin genes from leaf samples of Albizia lebbeck and Moringa oleifera. In silico analysis of these isolates revealed the properties and functional similarty of these novel genes with peroxidase P7 and Rab GDP dissociation inhibitor like gene activities, respectively. Physiochemical and homology modeling characterization studies revealed high similarity index of 100% and 85% for genes from A. lebbeck and M. oleifera with reliable structural stability properties. This preliminary study provides the data which may help us to understand the defense mechanism and transgenic plant breed optimization. Future experimental study of these genes may provide a pipeline for drug discovery project.
Isolation and characterization of peroxidase P7-like gene and Rab-GDI like gene from potential medicinal plants.
Leaf samples of A. lebbeck and M. oleifera were procured, identified, and confirmed from the Department of Botany, The Women University Multan, Pakistan.
Genomic DNA from leaf samples of two plant species was extracted using Thermo Scientific GeneJet Plant Genomic DNA Purification Mini kit and stored at −20°C for further investigation (Long et al., 2019). The extracted DNA was analyzed by agarose gel electrophoresis on 1% agarose gel in 1X TAE buffer.
DNA fragments of plants were amplified by PCR using forward primer 5′-CGCTGCCCCGACGCTTAC-3′ and reverse primer 5′-GACGACTTGGTAGTTGCTGT-3′ using an open reading frame of acknowledged plant defensin genes. The PCR profile for gene amplification was set as follows: 95°C for 3 min followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 48°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 7 min. The PCR products were resolved on 1% agarose gel using 1X TAE buffer at 80 V, followed by ethidium bromide staining. PCR products were purified using a Thermo Scientific GeneJET Gel Extraction Kit. Purified PCR products were sequenced at the DNA sequencing facility of the Center for Applied Molecular Biology, Lahore, Pakistan, under the Access to the Scientific Instrumentation Program of the Higher Education Commission of Pakistan.
All sequencing results were compiled using DNA Dragon software1 (Hepperle, 2011). The consensus sequences obtained from DNA Dragon were checked for their homology with reported sequences presenting nucleotide database using BLASTn and BLASTx programs of NCBI BLAST2 (Ingti et al., 2017). The sequences showing significant similarity with query sequences were retrieved and aligned using online Multiple Sequence Alignment tool CLUSTALW3 (Thompson et al., 1994). Phylogenetic diversity was analyzed using the neighbor-joining method (Kumar et al., 2016) in the Molecular Evolutionary Genetics Analysis (MEGA) Program, version X (Newman et al., 2016). The predicted amino acid sequence of plant defensin genes was subjected to the ProtParam tool4 on the ExPASy server to compute the physicochemical properties (Lim and Yoon, 2018).
The three-dimensional structures of defensin proteins were predicted using I-TASSER, SWISS-MODEL and RaptorX web tools (Fahim et al., 2018). The rough models obtained were subjected to GROMOS96 43B1 executed using Swiss-pdbViewer version 4.0.1 for energy minimization (Amir et al., 2019). PROCHECK and ProSA (Protein Structure Analysis) were used to assess the reliability of the homology models (Kumar et al., 2018). Possible docking sites and ligands were predicted using RaptorX (Wu et al., 2018). CD search and pfam analysis tools were used for the detection of structural and functional domains in plant defensins (Marchler-Bauer et al., 2017; Mistry et al., 2021).
The data were analyzed by simple regression rational analysis (Douwes et al., 2008).
A good quality DNA (Figure 1A) from leaf samples of both plants were used for defensen gene amplification by PCR. Amplicons appeared as unique bright bands on the agarose gel (Figure 1B). The gene sequence length was ~150 bp for Moringa oleifera and >300 bp for Albizia lebbeck, as expected.
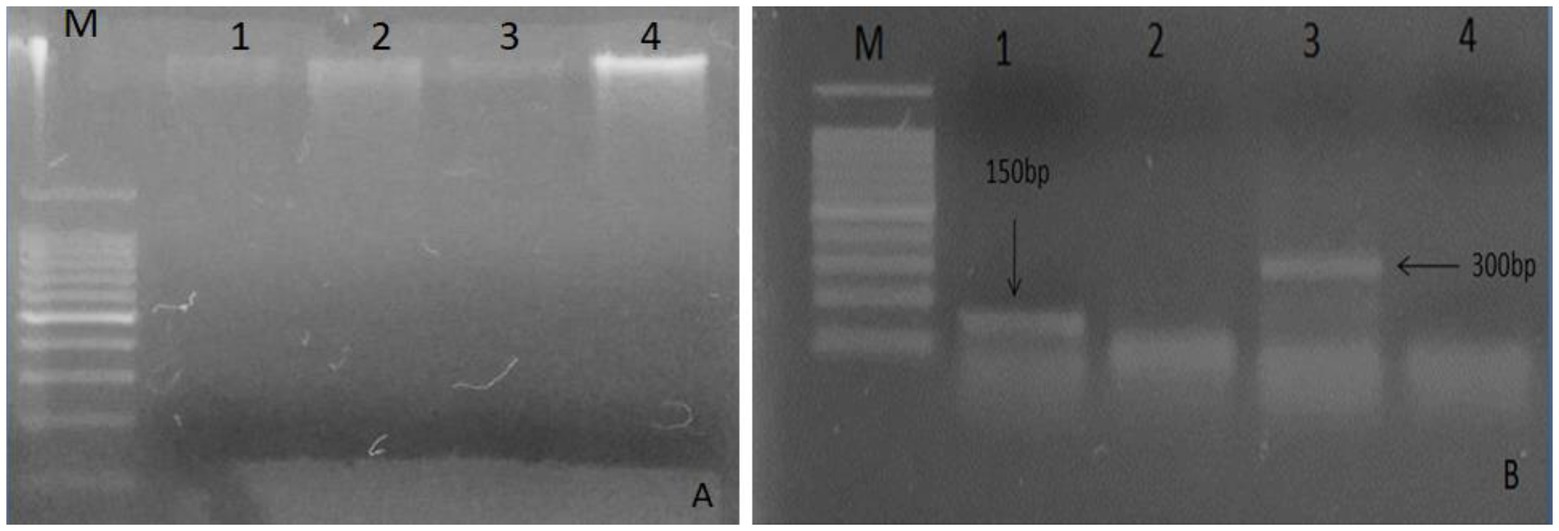
Figure 1. Analysis of total DNA extracted on 1% agarose from plant leaves. (A) Lane M: marker 100 bp, lanes 1 and 3 show DNA bands of Moringa oleifera, and lanes 2 and 4 show bands of Albizia lebbeck; (B) Lane M indicates 100 bp DNA ladder, lane 1 shows 150 bp gene of Moringa oleifera, and lane 3 shows 300 bp gene of Albizia lebbeck.
The sequenced fragments of A. lebbeck and M. oleifera were 300 bp and 317 bp in length containing open reading frames of 204 bp and 174 bp, respectively. Consensus sequences of both fragments were analyzed for their homology with sequences in the NCBI database using BLASTN and BLASTX on the BLAST server.5 The A. lebbeck defensin gene showed 100% similarity with predicted Musa acuminata subsp. malaccensis peroxidase P7-like gene (accession number XM_009390426.2) and protein sequence (accession number XP_009388701.1) and the M. oleifera defensin gene showed 85% and 91% similarity with predicted Manihot esculenta GDP dissociation inhibitor gene sequence (accession number XM_021748685.1) and protein sequence (accession number XP_021604377.1), respectively. The orthologous sequences of A. lebbeck and M. oleifera defensins were retrieved and subjected to multiple sequence alignment using the online CLUSTALW tool. Phylogenetic analyses retrieved the orthologous polypeptide sequences. Figure 2 shows the phylogenetic trees of A. lebbeck and M. oleifera defensins. For the defensin-deduced amino acid sequences, 14 published homologous sequences from different plants were used to construct a phylogenetic tree by neighbor-joining distance analysis. For A. lebbeck, the topology showed that the polypeptides from four different branches were highly homologous sequences. The A. lebbeck polypeptide is clustered with predicted Musa acuminata peroxidase P7-like protein, indicating that it is the most recent ancestor of A. lebbeck defensin (Figure 2A). The analysis for M. oleifera showed clustering of the M. oleifera defensin polypeptide with Manihot esculenta GDP dissociation inhibitor 1 (Figure 2B). As DNA sequences with more than 75% similarity and protein sequences with more than 25% similarity are considered homologous sequences, we retrieved the sequences of homologous proteins of A. lebbeck (M. acuminata peroxidase P7-like) and M. oleifera defensins (M. esculenta GDP dissociation inhibitor 1) to check the properties and structure prediction of novel defensins from understudied medicinal plants.
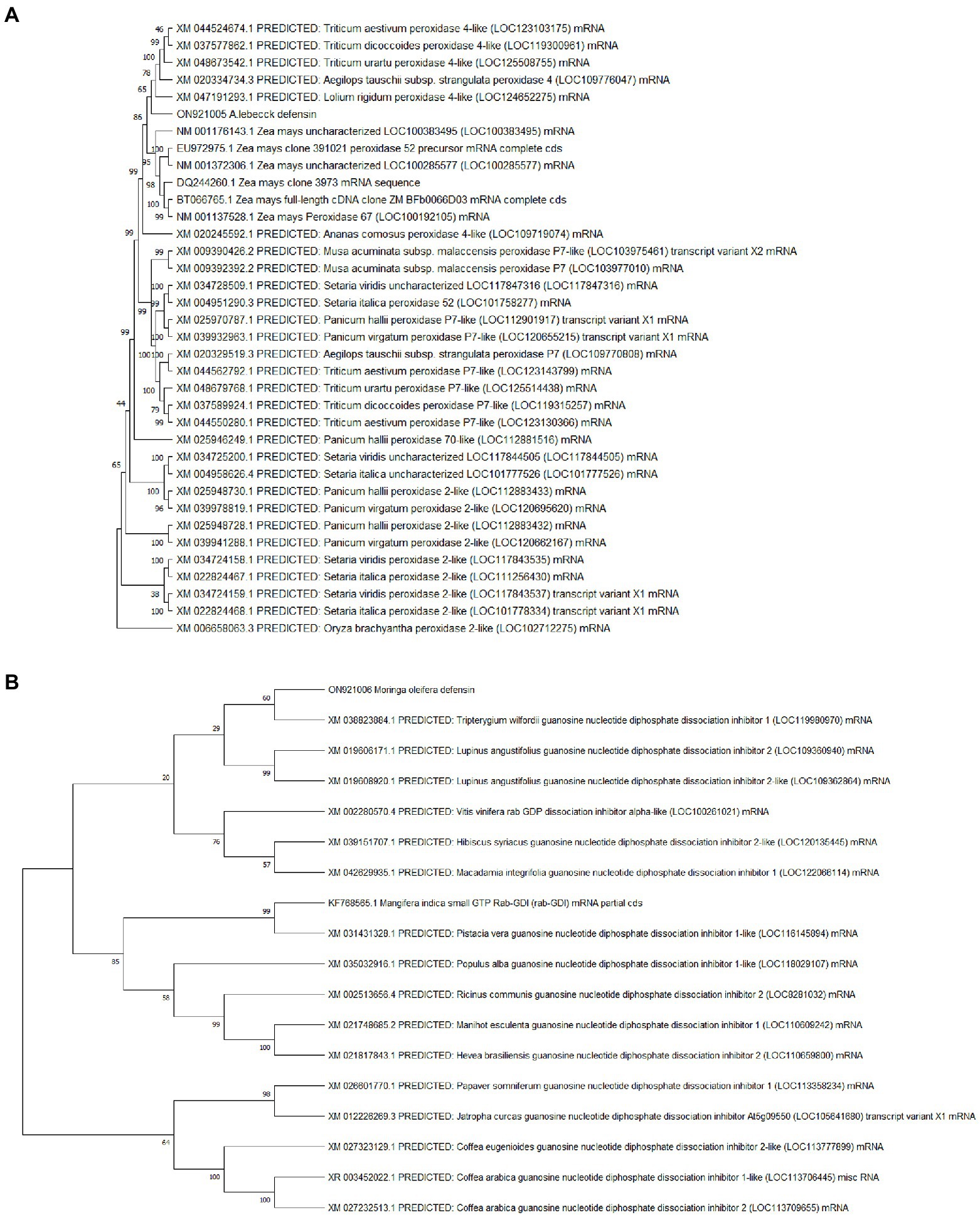
Figure 2. The phylogenetic analysis of (A) A. lebbeck defensin and (B) M. oleifera defensin with other plant species by MEGA X from CLUSTAL W alignment. The neighbor-joining method was used to construct the tree with p-distance.
Table 1 summarizes the ProtParam analysis of physicochemical properties of M. acuminata peroxidase P7-like and M. esculenta GDP dissociation inhibitor 1. Musa acuminata peroxidase P7-like peptide had a predicted molecular weight of ~48 kDa and is neutral with an isoelectric point of 7.02, whereas M. esculenta GDP dissociation inhibitor 1 had a molecular weight of ~50 kDa with an acidic isoelectric point of 5.53. Protein low instability and high aliphatic index values predicted that these proteins are heat stable. The grand average of hydropathicity (GRAVY) index indicated that M. esculenta GDP dissociation inhibitor 1 is more hydrophilic and has more surface accessibility to interact with water than M. acuminata peroxidase P7-like.
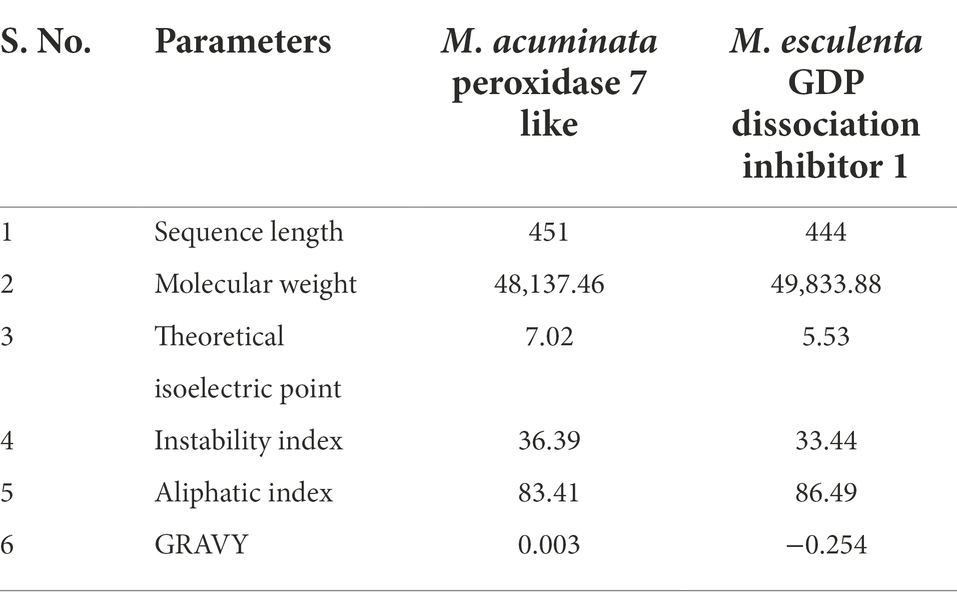
Table 1. Physicochemical properties of M. acuminata peroxidase 7 like and M. esculenta GDP dissociation inhibitor 1 by ProtParam.
The I-TASSER server with diverse threading templates was used for comparative homology modeling of M. acuminata peroxidase P7-like and M. esculenta GDP dissociation inhibitor 1. In I-TASSER, the C-score is used to evaluate the result from the consensus of top structural suits. The template modeling score (TM score) is used to measure the homology between structures of templates and models and determine the sequence identity in the structurally aligned area.
For M. acuminata peroxidase P7-like protein, the predicted TM score of the selected model was 0.49 ± 0.15, and RMSD (root mean square deviation) score was 11.6 ± 4.5 Å, observed in the correct configuration. Stereochemical properties were stabilized by energy minimization using Swiss-pdbViewer.6 Thepredicted homology model was authenticated using the ProSA and PROCHECK server. Figure 3A shows the Ramachandran plot in which% residues in the core allowed and disallowed regions tells about the acceptability of predicted structure. The maximum likelihood of finding protein residues (>90%) in the core/allowed regions indicatesthat the model built by I-TASSER was the good predicted structure. Figures 3B,C show the final modeled structure of M. acuminata peroxidase P7-like. Superimposition of predicted structure with template structure is represented in Figure 3D. PROCHECK and SWISS-MODEL revealed that all residues are in the core/allowed region, with none in the disallowed region, indicating the acceptability of Ramachandran plots (Table 2).
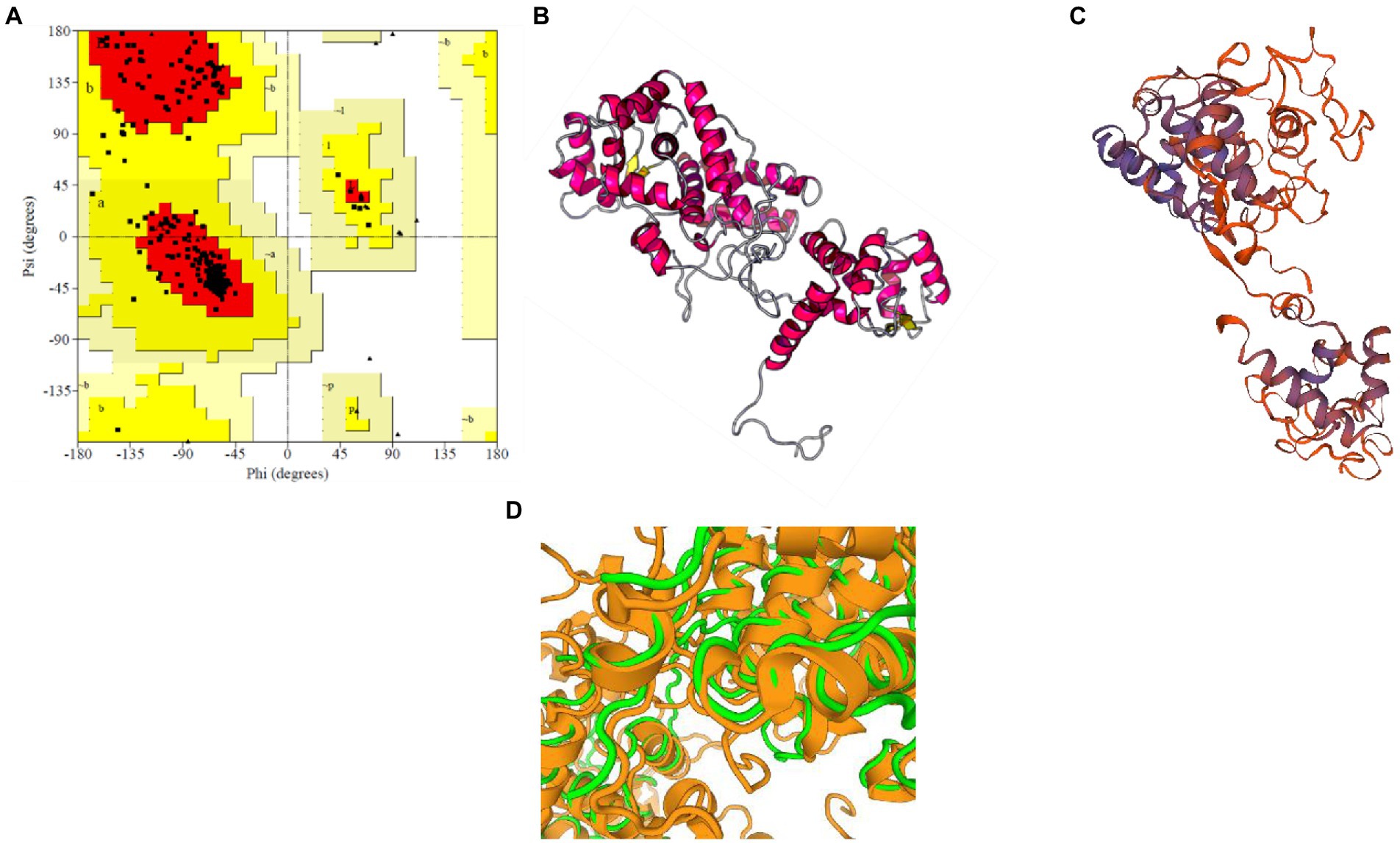
Figure 3. Prediction and validation of 3D structure of M. acuminata peroxidase P7-like protein. (A) Shows the Ramachandran plot and % residues in the core allowed and disallowed regions. (B,C) Show the final modeled structure of M. acuminata peroxidase P7-like predicted using I-Tasser and SWISS-MODEL, respectively. (D) The 3D structure superposition of template structure and predicted model is shown.
The structure of M. acuminata peroxidase P7-like protein was further evaluated using ProSA that mainly considers two predicted structure features: z-score and residue energies plot. Z-score reveals information on the model’s quality, which can be used to measure the amount of total energy of the structure deviating from the energy distribution derived from random conformations. Z-scores outside a range characteristic for native proteins indicate erroneous structures M. acuminata peroxidase P7-like had a z-score of −7.29 (Figure 4A), similar to the scores obtained for similar-sized proteins, indicating structure reliability. The residue energy plot depicted the local model quality by plotting energies as a function of amino acid sequence position. The negative values of the M. acuminata peroxidase P7-like structure indicates that the predicted model is error-free and acceptable (Figure 4B). To identify the presence of domain in M. acuminata protein, pfam and CD search was performed which showed the presence of secretory protein domain (Figure 4C).
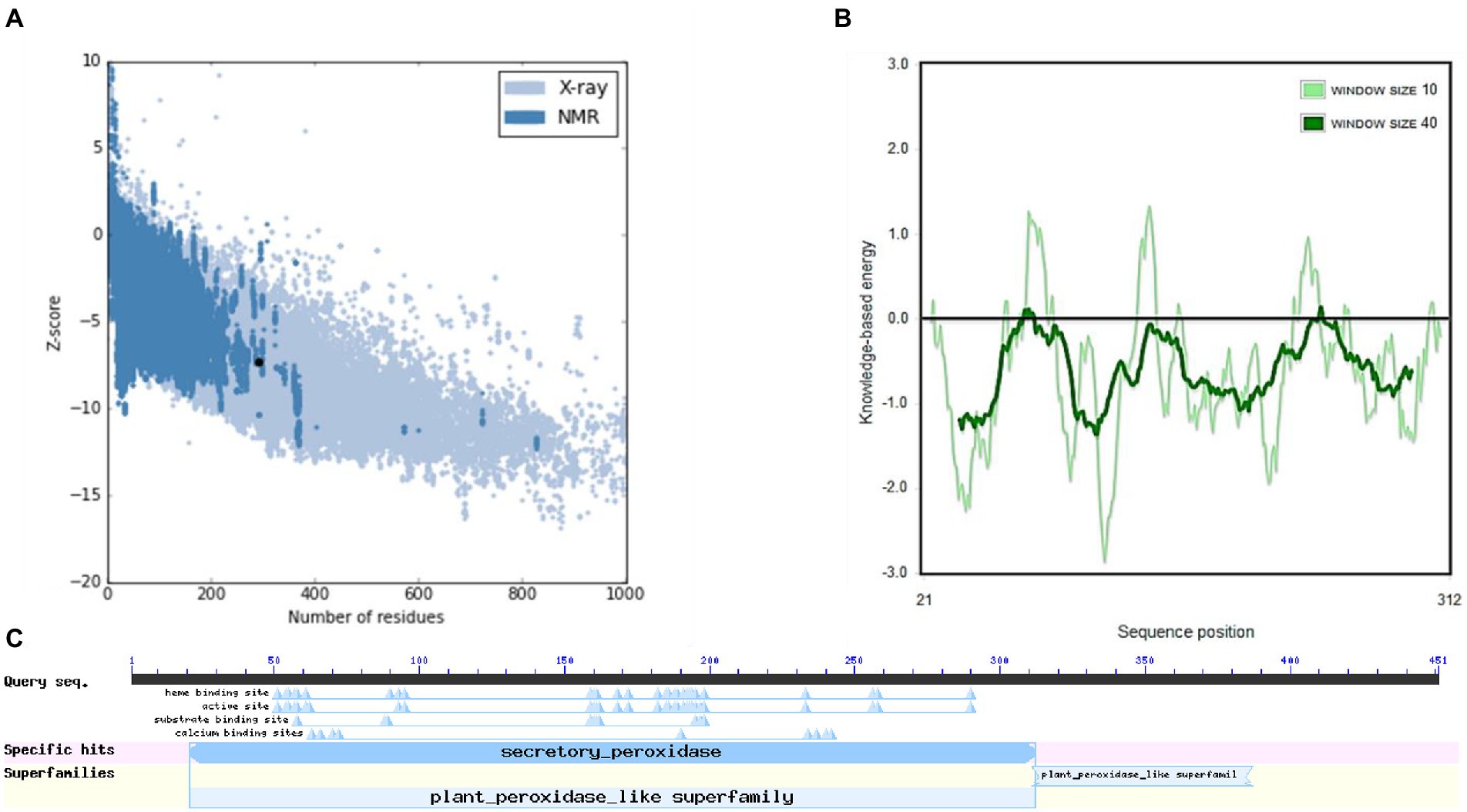
Figure 4. ProSA-web service analysis and domain search of M. acuminata peroxidase P7-like protein. (A) ProSA-web z-scores of the protein chains present in the Protein Data Bank refer to their length. Light and dark blue colors correspond to X-ray crystallography and NMR spectroscopy, respectively. Large dots highlight the z-scores of M. acuminata peroxidase P7-like protein. (B) Energy plot of M. acuminata peroxidase P7-like protein. (C) CD search and Pfam search shows the presence of secretory peroxidase domain in M. acuminata peroxidase P7-like protein.
The binding site of the targeted protein was predicted using the raptorX tool, with the 3D structure of M. acuminata peroxidase 7 shown in Figure 5. The input sequence was processed in two domains, with six binding pockets predicted. Possible ligands that bind with these pockets are HEM, pyridine, Ca, Na, and K. HEM and Ca bind to pockets in both domains, while pyridine, Na, and K bind to pockets in domain 1 only.
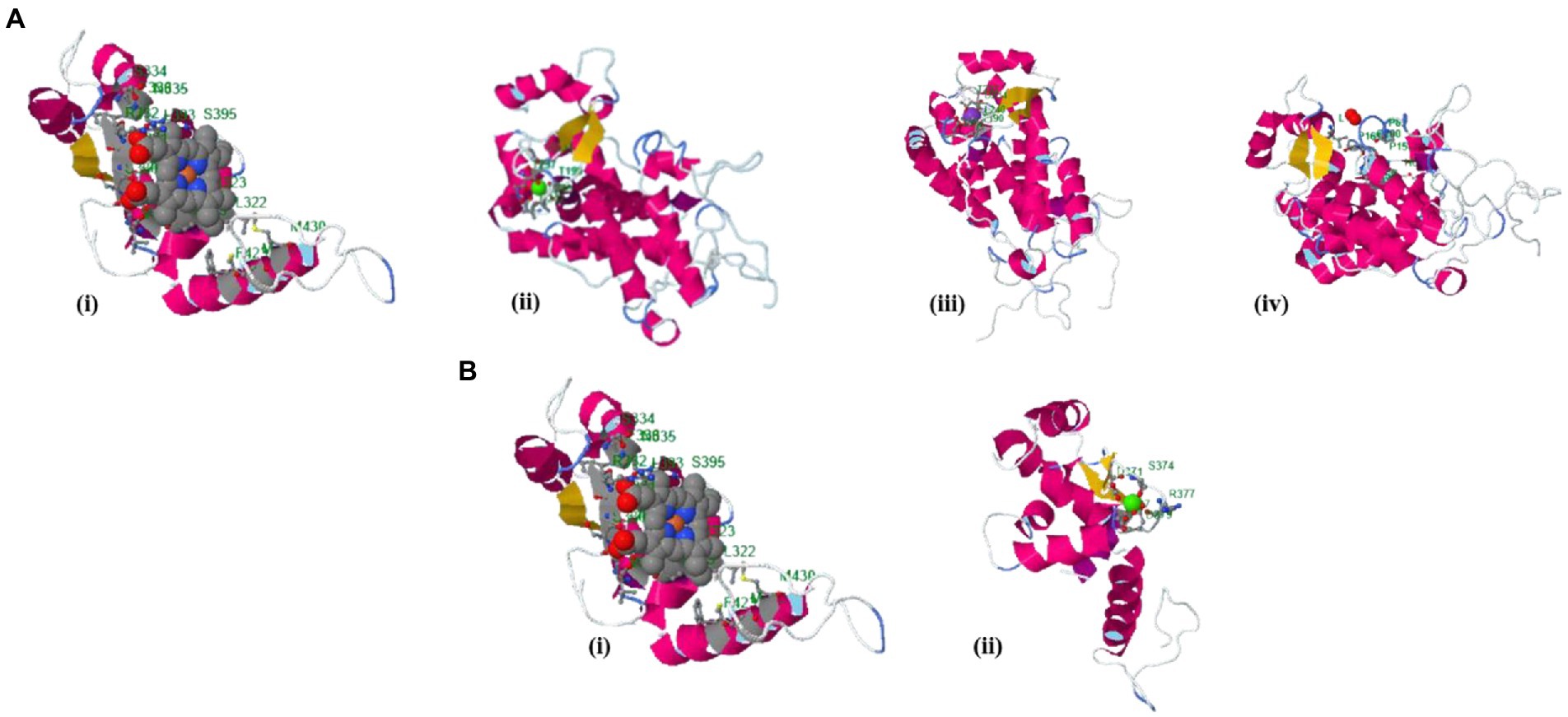
Figure 5. Identified binding sites of M. acuminata peroxidase P7-like 3D structure using the raptorX tool. (A) Ligand binding pockets identified in domain 1: (I) HEM; (ii) Calcium; (Ca) (iii) Potassium (K); (iv) Pyridine. (B) Binding pockets identified in domain 2: (i) HEM; (ii) Calcium (Ca).
The selected model of M. esculenta GDP dissociation inhibitor 1 had a predicted TM score of 0.89 ± 0.07 and RMSD score of 4.4 ± 2.9 Å, observed in the correct configuration. Stereochemical properties were stabilized by energy minimization using Swiss-pdbViewer. The predicted homology model was authenticated using ProSA and PROCHECK. Figure 6A shows the Ramachandran plot and % of the residues in the core allowed and disallowed regions. The maximum likelihood of finding protein residues (>90%) in the core/allowed regions indicates that the predicted model is of good stereochemical quality. PROCHECK revealed that all residues are in the core/allowed region, with none in the disallowed region, indicating the acceptability of Ramachandran plots (Table 3).
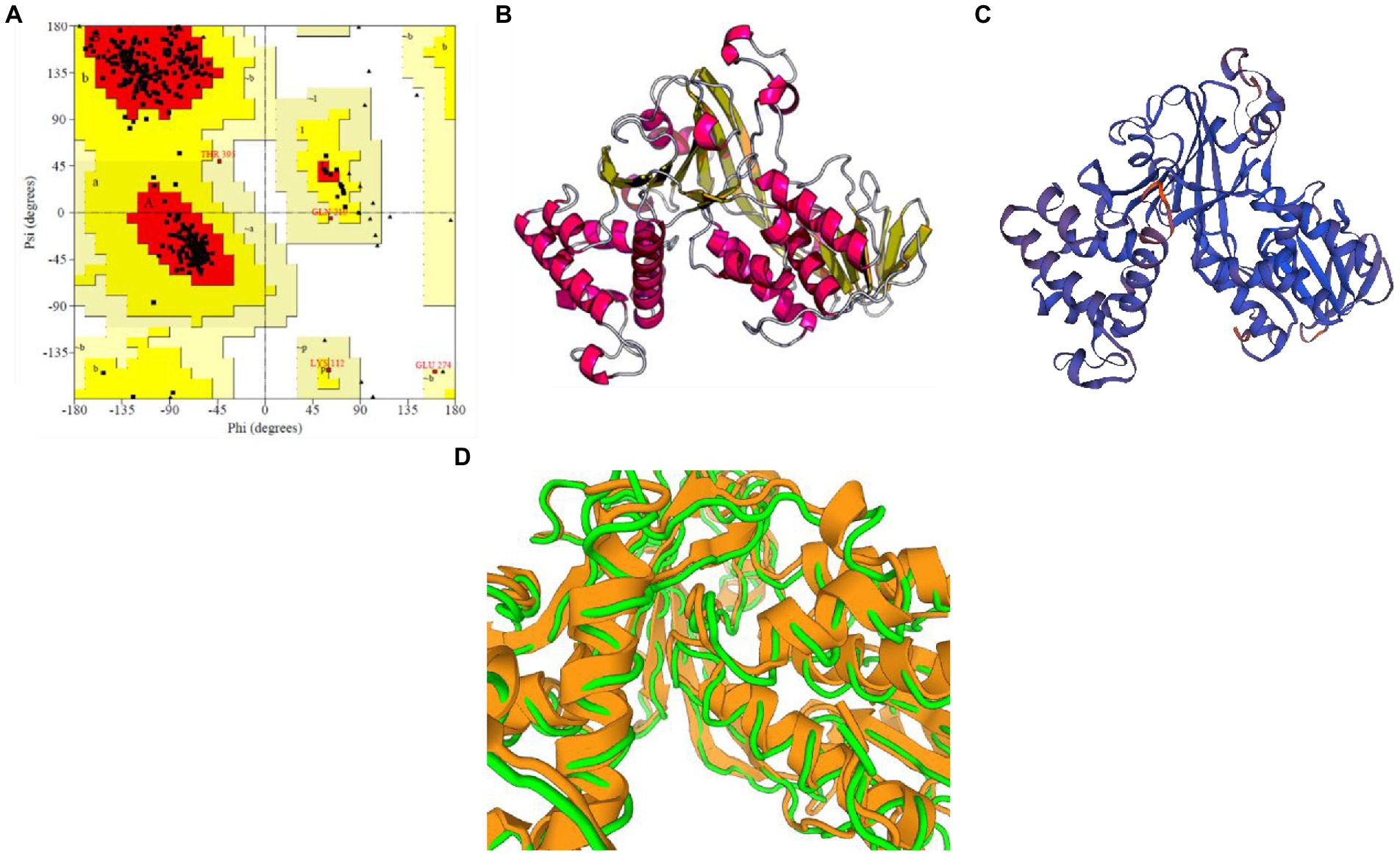
Figure 6. Prediction and validation of 3D structure of M. esculenta GDP dissociation inhibitor 1. (A) Ramachandran plot of M. esculenta GDP dissociation inhibitor 1 protein. The plot calculations for the 3D models were computed in PROCHECK. The most favored regions are in red, additional allowed, generously allowed, and disallowed regions are indicated in yellow, light yellow, and white, respectively. (B,C) The final 3D structure of M. esculenta GDP dissociation inhibitor 1 predicted using I-tasser and SWISS-MODEL, respectively. The α-helix is represented by red helix, β-sheet by yellow arrows, and loops by gray lines (D).
Figures 6B,C show the final modeled structure of M. esculenta GDP dissociation inhibitor 1, which was further evaluated using ProSA. Superimposition of predicted structure with template structure is represented in Figure 6D. Manihot esculenta GDP dissociation inhibitor 1 had a z-score of −10.11 (Figure 7A), similar to the scores of similar-sized proteins, indicating structure reliability. The residue energy plot depicted local model quality by plotting energies as a function of amino acid sequence position. The negative values of the residue energy plot of M. esculenta GDP dissociation inhibitor 1 structure indicate that predicted model is error-free and acceptable (Figure 7B). To identify the presence of domain in M. esculenta GDP dissociation inhibitor, pfam and CD search was performed which showed the presence of GDI domain (Figure 7C).
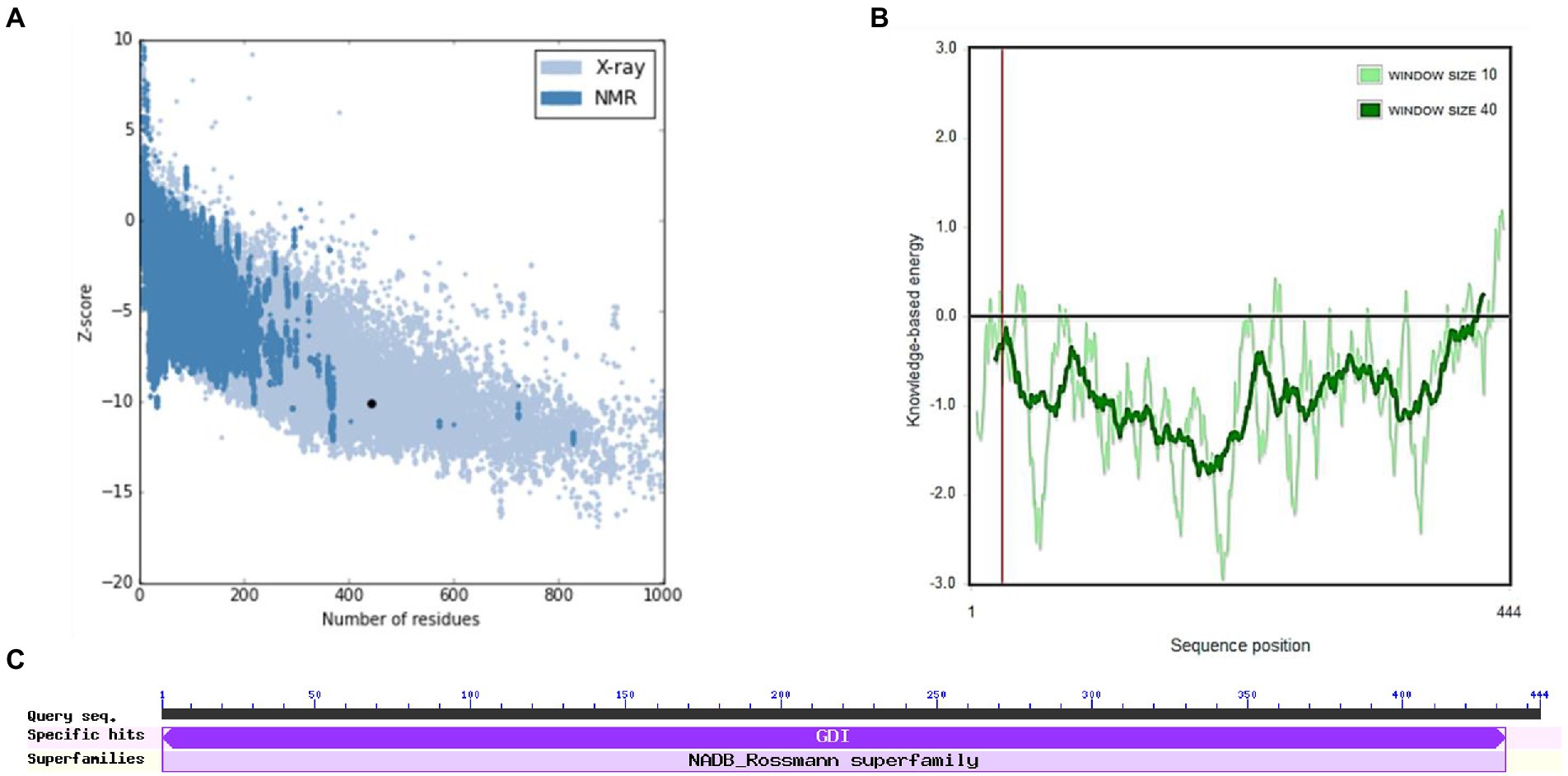
Figure 7. ProSA-web service analysis and domain search of M. esculenta GDP dissociation inhibitor 1. (A) ProSA-web z-scores of all protein chains present in the Protein Data Bank refer to their length. Light and dark blue colors correspond to X-ray crystallography and NMR spectroscopy, respectively. Large dots highlight the z-scores of M. esculenta GDP dissociation inhibitor 1. (B) Energy plot of M. esculenta GDP dissociation inhibitor 1. (C) CD search and Pfam search shows the presence of GDI domain in M. esculenta GDP dissociation inhibitor 1.
The binding site of the targeted protein was predicted using the raptorX tool, with the 3D structure of M. esculenta GDP dissociation inhibitor 1 shown in Figure 8. The input sequence was processed, with four binding pockets predicted. Possible ligands that bind with these pockets are dihydroflavine-adenine dinucleotide (FDA), sulfate ion (SO42−), chloride ion (Cl−), and galactose-uridine-5′-diphosphate (GDU).
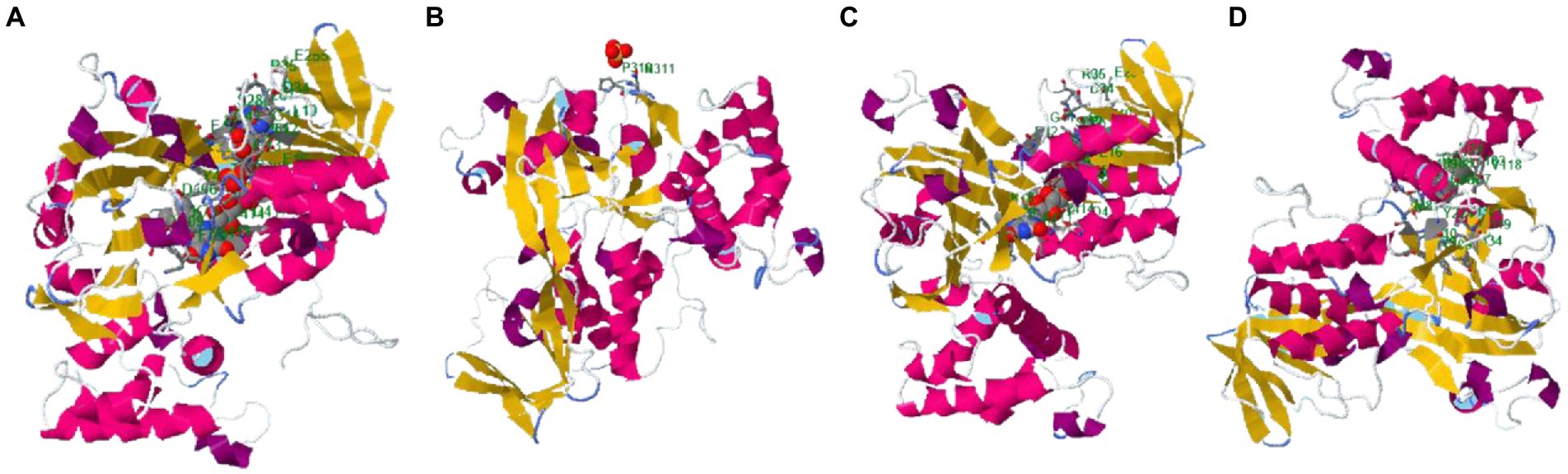
Figure 8. Identified binding sites of M. esculenta GDP dissociation inhibitor 1 using the raptorX tool. Four binding pockets were identified: (A) Dihydroflavine-adenine dinucleotide (FDA), (B) sulfate ion (SO42−), (C) chloride ion (Cl−), and (D) galactose-uridine-5′-diphosphate (GDU).
Plants have the ability to integrate divergent signaling pathways to allow appropriate defense responses against various stresses (Brenner et al., 2012; Overmyer et al., 2018) Therefore, overexpression of a single gene can have a negative (i.e., susceptibility) or positive effect (i.e., tolerance) on the plant response to other stresses. These genes also serve as a promising source for engineering disease resistance traits in plants, enhancing self-defense mechanisms and reducing the requirement for supplementary chemical microbicides (Boukhatem et al., 2014). In this study, in vitro and in silico methods were used to isolate and characterize novel plant defending family-specific antimicrobial genes from Albizia lebecck and Moringa oleifera. Same kind of study on defensin gene isolation from Saccharum officinarum was done by De-Paula and colleagues, who analyzed six sugar cane putative defensins (De-Paula et al., 2008). In our study, primers were designed on a consensus sequence of 3–5 aligned sequences retrieved from the NCBI database. Highly similar sequences that exhibited 75%–100% similarity with the primary sequence were selected for alignment through the BLAST tool. In a recent study on one class of defensin genes—biodefensin—Rawat et al. (2017) isolated the full length of this gene encoding about 80 amino acids from Brassica juncea (family Brassicaceae). The authors reported that the gene had high levels of similarity with gamma thionin and knottin families of plant AMPs. Our isolates also showed that the biodefensin had similarities with defense genes in other plants. Studies have shown that gene transcription is significantly enhanced under stress, confirming the inducible defense response of AMP in plants (Rawat et al., 2017). The plant defensins Rs-AFP1 and Rs-AFP2 from radish (Raphanus sativus) is an example of potent antifungal proteins. The exact process involved in the antifungal function of these genes needs further study, but they likely target the cell membranes of microbes. These genes may also initiate other mechanisms involved in plant defense (Souza et al., 2017).
We used many online bioinformatics tools to study the physicochemical and functional properties, and also structural analyses of our novel defensin genes. Same kind of comparative analysis on Kisspeptin Receptors and their physicochemical characterization was done by Mukesh Kumar and his co-workers (Kumar et al., 2018). We predicted the 3-D structures of defensin proteins using I-TASSER and RaptorX web tools. Our novel gene, isolated from Moringa oleifera that showing homology with the GDP dissociation inhibitor gene, may function in membrane transport, as found in the GDP dissociation inhibitor (Palme, 1997; Zarsky et al., 1997). The GDP dissociation inhibitor gene regulates nucleotide states and subcellular localization of Rab/Ypt proteins (Ueda et al., 1998). Rab3a-GDI (Rab guanosine nucleotide diphosphate dissociaton inhibitor protein), first isolated from bovine cytosol (Sasaki et al., 1991), inhibits the dissociation of GDP to Rab3a and the binding of GTP to Rab3a, and thus has a potential function in regulating Rab GTPases (Burstein and Macara, 1992) and secretory pathways (Andreeva et al., 1997). While we have not tested the function of our novel gene, but its homology with GDP dissociation inhibitor and function in membrane vesicular transport strongly suggest that it will also inhibit the entry of microbes across the membrane, thus preventing infection. Our other novel gene, isolated from Albizzia lebecck showed 100% similarity with predicted Musa acuminata subsp. malaccensis peroxidase P7-like gene. In a cold tolerance study of Musa spp. ‘Dajiao’, He et al. (2018) found that membrane-bound peroxidases potentially decrease lipid peroxidation and play a role in maintaining leaf cell water potential, contributing to plant defense against cold stress. Other researchers also found that membrane peroxidases are involved in the H2O2 scavenging system and ROS signaling networks (Passardi et al., 2004). The role of peroxidases in plant defense is elaborated by their activity on cell walls through the oxidation of phenolic compounds by reducing H2O2 under normal and stressed conditions (Almagro et al., 2009). Another study identified three defense enzymes—phenylalanine ammonia-lyase (PAL), peroxidase (PO), and polyphenol oxidase (PPO)—involved in phenol synthesis and oxidation in banana roots infected with Radopholussimilis, which strengthened cell walls. Wounded roots had significantly higher PO levels than control roots, they also reported enhanced PO activity in banana roots inoculated with fungus within 24 h of infection (Wuyts et al., 2006). These results and the homology of our isolated gene with Musa acuminata subsp. malaccensis peroxidase P7-like gene strongly suggests that Albizzin has the same functional potential in the defense mechanism of plants and can serve its function against fungal diseases.
No doubt huge efforts are being made on genetic researches all over the world to discover more potent medicinal plants with beneficial gene products. However, still a large number of medicinal plants are at the need to be investigated for their possible defensive values. This study suggests a strategy for isolation of effective candidate genes that are part of signal transduction and defense system of plants. Examining the efficiency of these genes in practice is key to understand its role in defense mechanism. Screening of defensin gene from M. oliefera and A. lebecck resulted in isolation of GDP dissociation inhibitor like gene and peroxidase P7-like gene, respectively. These genes are very important as their role in membrane transport, regulation of nucleotide states, subcellular localization, free radical sacavenging properties and signaling of ROS. Further investigation on physical characterization and interaction of gene with downstream stress-responsive and growth and developmental-related genes of plant breeds with these genes would be worth examining whether these plants have enhanced resistance or tolerance to multiple biotic and abiotic stresses. Hence these genes would be potential candidate for breeding of plants with high resistance and strong defense. Structural characterization and biological interactions of GDP dissociation inhibitor and peroxidase P7-like gene from M. oliefera and A. lebecck respectively, revealed in this study conclude that these genes can serve for the synthesis of transgenic plants that will be more stress resistant and can give better crop yield in the future.
The original contributions presented in the study are publicly available. This data can be found at: ON921005, ON921006.
Research work was supervised by RJ and AI. Material preparation, data collection, and analysis were performed by UH, RJ, and FD. The manuscript was finalized by HN and GM. Article review, editing, and proof reading was done by FZ, MN, AZ, and KS. All authors contributed to the article and approved the submitted version.
The research leading to these results received funding from Higher Education Commission, Government of Pakistan under grant agreement no. 21-1603/SRGP/R&D/HEC/2017.
We are highly thankful to the Higher Education Commission Pakistan. We heartily acknowledge The Women University Multan and Higher Education Commission Pakistan for the completion of this research and giving a platform to do this research work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. ^http://www.dna-dragon.com/
2. ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
3. ^http://www.genome.jp/tools/clustalw/
4. ^https://web.expasy.org/protparam/
Al Akeel, R., Al-Sheikh, Y., Mateen, A., Syed, R., Janardhan, K., and Gupta, V. (2014). Evaluation of antibacterial activity of crude protein extracts from seeds of six different medical plants against standard bacterial strains. Saudi J. Biol. Sci. 21, 147–151. doi: 10.1016/j.sjbs.2013.09.003
Almagro, L., Gómez Ros, L. V., Belchi-Navarro, S., Bru, R., Ros Barceló, A., and Pedreño, M. A. (2009). Class III peroxidases in plant defence reactions. J. Exp. Bot. 60, 377–390. doi: 10.1093/jxb/ern277
Amir, A., Kapoor, N., and Kumar, H. T. (2019). Target protein in human herpes virus 8 (HHV8). Biotech. Today Int. J. Biol. Sci. 9, 41–48. doi: 10.5958/2322-0996.2019.00005.X
Andreeva, A. V., Kutuzov, M. A., Evans, D. E., and Hawes, C. R. (1997). Rab-GDP dissociation inhibitor isoforms in Arabidopsis thaliana. J. Exp. Bot. 48, 2109–2110. doi: 10.1093/jxb/48.12.2109
Arasu, M. V., Al-Dhabi, N. A., Choi, K. C., Bensy, A. D., and Rajaselvam, J. (2022). Bioactive potential of Albizia lebbeck extract against phytopathogens and protective properties on tomato plant against speck disease in greenhouse. Physiol. Mol. Plant Pathol. 117:101750. doi: 10.1016/j.pmpp.2021.101750
Azam, S., Latif, A., Hussain, K., Shahbaz, H., Perveen, S., Ashfaq, I., et al. (2022). Anti-diabetic and anti-lipidepmic effect of Albizia lebbeck seeds against HepG2 cells. Pak. J. Pharm. Sci. 35, 793–800.
Boukhatem, M. N., Ferhat, M. A., Kameli, A., Saidi, F., and Kebir, H. T. (2014). Lemon grass (Cymbopogon citratus) essential oil as a potent anti-inflammatory and antifungal drugs. Libyan J. Med. 9:25431. doi: 10.3402/ljm.v9.25431
Brenner, W. G., Ramireddy, E., Heyl, A., and Schmulling, T. (2012). Gene regulation by cytokinin in arabidopsis. Front. Plant Sci. 3:8. doi: 10.3389/fpls.2012.00008
Burstein, E. S., and Macara, I. G. (1992). Characterization of a guanine nucleotide-releasing factor and a GTPase-activating protein that are specific for the ras-related protein p25rab3A. Proc. Natl. Acad. Sci. 89, 1154–1158. doi: 10.1073/pnas.89.4.1154
Casadei, E., Wang, T., Zou, J., Vecino, J. L., Wadsworth, S., and Secombes, C. J. (2009). Characterization of three novel β-defensin antimicrobial peptides in rainbow trout (Oncorhynchus mykiss). Mol. Immunol. 46, 3358–3366. doi: 10.1016/j.molimm.2009.07.018
Chojak-Koźniewska, J., Kuźniak, E., and Zimny, J. (2018). The effects of combined abiotic and pathogen stress in plants: insights from salinity and pseudomonas syringae pv lachrymans interaction in cucumber. Front. Plant Sci. 9:1691. doi: 10.3389/fpls.2018.01691
Cui, J., Jiang, N., Meng, J., Hou, X., Yang, G., and Luan, Y. (2018). Identification and characterization of defensin genes conferring Phytophthora infestans resistance in tomato. Physiol. Mol. Plant. Path 103, 28–35. doi: 10.1016/j.pmpp.2018.04.003
De-Paula, V., Razzera, G., Medeiros, L., Miyamoto, C., Almeida, M., Kurtenbach, E., et al. (2008). Evolutionary relationship between defensins in the Poaceae family strengthened by the characterization of new sugarcane defensins. Plant Mol. Biol. 68, 321–335. doi: 10.1007/s11103-008-9372-y
Douwes, E., Crouch, N. R., Edwards, T. J., and Mulholland, D. A. (2008). Regression analysis of southern African ethnomedicinal plants: informing the targeted selection of bioprospecting and pharmacological screening subjects. J. Ethnopharmacol. 119, 356–364. doi: 10.1016/j.jep.2008.07.040
Fahim, A., Rehman, Z., Bhatti, M. F., Ali, A., Virk, N., Rashid, A., et al. (2018). Structural insights and characterization of human Npas4 protein. PeerJ 6:e4978. doi: 10.7717/peerj.4978
He, W. D., Gao, J., Dou, T. X., Shao, X. H., Bi, F. C., Sheng, O., et al. (2018). Early cold-induced peroxidases and aquaporins are associated with high cold tolerance in Dajiao (Musa spp.‘Dajiao’). Front. Plant Sci. 9:282. doi: 10.3389/fpls.2018.00282
Hepperle, D. (2011). DNA dragon 1.4. 1-DNA sequence contig assembler software. Available at: www.Dna-dragon.Com (Accessed November 15, 2020).
Ingti, B., Laskar, M. A., Choudhury, S., Maurya, A. P., Paul, D., Talukdar, A. D., et al. (2017). Molecular and in silico analysis of a new plasmid-mediated AmpC β-lactamase (CMH-2) in clinical isolates of Klebsiella pneumoniae. Infect. Genet. Evol. 48, 34–39. doi: 10.1016/j.meegid.2016.12.009
Kamala Lakshmi, B., and Valarmathi, S. (2020). In vitro anti-inflammatory activity of aqueous extract of Albizia lebbeck leaf (l.). J. Phytopharmacol. 9, 356–360. doi: 10.31254/phyto.2020.9511
Kovaleva, V., Yusypovych, Y., and Gout, R. (2017). Scots pine defensins: structure, properties and biological functions. Біологічністудії 11, 66–67.
Kumar, S. (2017). Medicinal importance of Moringa oleifera: drumstick plant. Indian J. Sci. Res. 1, 129–133.
Kumar, M., Basumatary, G., Ram, R. K., Singh, D. K., and Udit, U. K. (2018). Comparative studies on Kisspeptin receptor and their physicochemical characterization. Int. J. Curr. Microbiol. App. Sci. 7, 43–47.
Kumar, A., Kumar, S., and SubirKundu, M. D. (2016). Homology modeling of outer membrane lipoprotein of Salmonella typhi. Res. Environ. Life Sci. 9, 900–902.
Lakshmi Priya, P. M., and Sabu, K. K. (2015). In Silico analysis of Transcriptomes of Catharanthus roseus and Rauvolfia serpentina, two potent medicinal plants using a pipeline developed from publicly available tools. Am. J. Bioinform. Res. 5, 21–25. doi: 10.5923/j.bioinformatics.20150502.01
Lim, S., and Yoon, T. (2018). “Genomic comparison of four Metapneumovirus strains using decision tree, Apriori algorithm, ClustalW, and phylogenetic reconstruction,” in Proceedings of the 2018 5th International Conference on Bioinformatics Research and Applications, 95–100.
Long, D. D., Bach, N. X., Thao, N. T., Nhung, P. T., Thom, V. T., and Huyen, P. T. (2019). Comparative analysis of different DNA extraction methods and preliminary analysis of genetic diversity of Hedera nepalensis K. Koch. in Vietnam based on GBSSI Marker. VNU J. Sci. Med. Pharmaceu Sci. 35, 88–95. doi: 10.25073/2588-1132/vnumps.4165
Mahlapuu, M., Håkansson, J., Ringstad, L., and Björn, C. (2016). Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6:194. doi: 10.3389/fcimb.2016.00194
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Newman, L., Duffus, A. L., and Lee, C. (2016). Using the free program MEGA to build phylogenetic trees from molecular data. Am. Biol. Teach. 78, 608–612. doi: 10.1525/abt.2016.78.7.608
Novaković, L., Guo, T., Bacic, A., Sampathkumar, A., and Johnson, K. L. (2018). Hitting the wall—sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plan. Theory 7:89. doi: 10.3390/plants7040089
Overmyer, K., Vuorinen, K., and Brosche, M. (2018). Interaction points in plant stress signaling pathways. Physiol. Plant. 162, 191–204. doi: 10.1111/ppl.12639
Palme, K. (1997). At-GDI1 from Arabidopsis thaliana encodes a rab-specific GDP dissociation inhibitor that complements the sec19 mutation of. FEBS Lett. 403, 303–308. doi: 10.1016/S0014-5793(97)00072-0
Parisi, K., Shafee, T. M., Quimbar, P., van der Weerden, N. L., Bleackley, M. R., and Anderson, M. A. (2019). The evolution, function and mechanisms of action for plant defensins. Semin. Cell. Dev. Biol. 88, 107–118. doi: 10.1016/j.semcdb.2018.02.004
Passardi, F., Penel, C., and Dunand, C. (2004). Performing the paradoxal: how plant peroxidases modify the cell wall. Trends Plant Sci. 9, 534–540. doi: 10.1016/j.tplants.2004.09.002
Rawat, S., Ali, S., Nayankantha, N. C., Chandrashekar, N., Mittra, B., and Grover, A. (2017). Isolation and expression analysis of defensin gene and its promoter from Brassica juncea. J. Plant Dis. Prot. 124, 591–600. doi: 10.1007/s41348-017-0103-y
Reichling, J. (2018). Plant–microbe interactions and secondary metabolites with antibacterial, antifungal and antiviral properties. Annual Plant Rev. 15, 214–347. doi: 10.1002/9781119312994.apr0420
Sasaki, T., Kaibuchi, K., Kabcenell, A. K., Novick, P. J., and Takai, Y. (1991). A mammalian inhibitory GDP/GTP exchange protein (GDP dissociation inhibitor) for smg p25A is active on the yeast SEC4 protein. Mol. Cell. Biol. 11, 2909–2912.
Schmidt, M., Arendt, E. K., and Thery, T. L. (2019). Isolation and characterisation of the antifungal activity of the cowpea defensin Cp-thionin II. Food Microbiol. 82, 504–514. doi: 10.1016/j.fm.2019.03.021
Souza, T. P., Dias, R. O., and Silva-Filho, M. C. (2017). Defense-related proteins involved in sugarcane responses to biotic stress. Genet. Mol. Biol. 40, 360–372. doi: 10.1590/1678-4685-gmb-2016-0057
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Ueda, T., Yoshizumi, T., Anai, T., Matsui, M., Uchimiya, H., and Nakano, A. (1998). AtGDI2, a novel Arabidopsis gene encoding a Rab GDP dissociation inhibitor. Gene 206, 137–143. doi: 10.1016/S0378-1119(97)00584-2
Wu, Q., Peng, Z., Zhang, Y., and Yang, J. (2018). COACH-D: improved protein–ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 46, W438–W442. doi: 10.1093/nar/gky439
Wuyts, N., Swennen, R., and De Waele, D. (2006). Activity of phenylalanine ammoni-lyase, peroxidise and polyphenol oxidase in roots of banana(Musa acuminate AAA, cvs Grande Naine and Yangambi km5) before and after infection with Radopholus similis. Nematol 8, 201–209. doi: 10.1163/156854106777998674
Zarsky, V., Cvrckova, F., BischofF, F., and Palme, K. (1997). At-GDI1 from Arabidopsis thaliana encodes a Rab-specific GDP dissociation inhibitor that complements the sec19 mutation of Saccharomyces cerevisiae. FEBS Lett. 403, 303–308. doi: 10.1016/S0014-5793(97)00072-0
Zharkova, M. S., Orlov, D. S., Golubeva, O. Y., Chakchir, O. B., Eliseev, I. E., Grinchuk, T. M., et al. (2019). Application of antimicrobial peptides of the innate immune system in combination with conventional antibiotics—a novel way to combat antibiotic resistance? Front. Cell. Infect. Microbiol. 9:128. doi: 10.3389/fcimb.2019.00128
Keywords: defensin gene, Albizia lebbeck, Moringa oleifera, in silico analysis, plant defense system
Citation: Jabeen R, Iqbal A, Deeba F, Zulfiqar F, Mustafa G, Nawaz H, Habiba U, Nafees M, Zaid A and Siddique KHM (2022) Isolation and characterization of peroxidase P7-like gene and Rab-GDI like gene from potential medicinal plants: A step toward understanding cell defense signaling. Front. Plant Sci. 13:975852. doi: 10.3389/fpls.2022.975852
Received: 23 June 2022; Accepted: 10 August 2022;
Published: 02 September 2022.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Anuj Kumar, Dalhousie University, CanadaCopyright © 2022 Jabeen, Iqbal, Deeba, Zulfiqar, Mustafa, Nawaz, Habiba, Nafees, Zaid and Siddique. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raheela Jabeen, ZHJyYWhlZWxhLjkwNTRAd3VtLmVkdS5waw==; Faisal Zulfiqar, Y2guZmFpc2FsLnp1bGZpcWFyQGdtYWlsLmNvbQ==; Kadambot H. M. Siddique, a2FkYW1ib3Quc2lkZGlxdWVAdXdhLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.