- 1School of Pharmacy, Nanjing University of Chinese Medicine, Nanjing, China
- 2State Key Laboratory of Dao-di Herbs, National Resource Center for Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing, China
- 3State Key Laboratory for Biology of Plant Diseases and Insert Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 4School of Traditional Chinese Medicine, Southern Medical University, Guangzhou, China
Salvia miltiorrhiza Bunge (Danshen in Chinese) is vulnerable to Fusarium wilt, which severely affects the quality of the crude drug. Mycorrhizal colonization enhances resistance to fungal pathogens in many plant species. In this study, pre-inoculation of S. miltiorrhiza with the arbuscular mycorrhizal fungi (AMF) Glomus versiforme significantly alleviated Fusarium wilt caused by Fusarium oxysporum. Mycorrhizal colonization protected S. miltiorrhiza from pathogen infection, thereby preventing a loss of biomass and photosynthesis. There were greater defense responses induced by pathogen infection in AMF pre-inoculated plants than those in non-treated plants. AMF pre-inoculation resulted in systemic responses upon pathogen inoculation, including significant increases in the protein content and activities of phenylalanine ammonia-lyase (PAL), chitinase, and β-1,3-glucanase in S. miltiorrhiza roots. In addition, mycorrhizal pre-inoculation caused upregulation of defense-related genes, and jasmonic acid (JA) and salicylic acid (SA) signaling pathway genes after pathogen infection. The above findings indicate that mycorrhizal colonization enhances S. miltiorrhiza resistance against F. oxysporum infection by enhancing photosynthesis, root structure, and inducing the expression of defense enzymes and defense-related genes on the other hand.
Introduction
Salvia miltiorrhiza Bunge, a diploid species belonging to the family Lamiaceae (Li et al., 2017), is an industrially important medicinal plant widely used for treatment of coronary and cerebrovascular diseases (Chen et al., 2018; Liu et al., 2018; Shi et al., 2018). With increasing demand for S. miltiorrhiza in domestic and international markets, the planting area has expanded in China (Shi et al., 2018). S. miltiorrhiza is mostly cultivated with large-scale continuous planting, but Fusarium wilt caused by Fusarium oxysporum is a major threat (Zhang et al., 2016). Fusarium wilt is a fast-spreading epidemic disease that causes severe damage to the quality and productivity of S. miltiorrhiza, similar to the damage experienced by crops such as cucumber, chickpeas, banana, and cotton (de Lamo and Takken, 2020; Ankati et al., 2021; Qi et al., 2022). It is estimated that up to 70% of continuously cropped S. miltiorrhiza is affected (Yang et al., 2013). Synthetic fungicides are commonly used to control Fusarium wilt, however their use causes environmental pollution and threatens human health (Neeraj and Singh, 2011). Therefore, there is an urgent need for the identification of new biological control methods to suppress Fusarium wilt in the agricultural production of S. miltiorrhiza (Hammad and El-Mohandes, 1999).
In response to fungus pathogens, plants have evolved a series of complex strategies to protect themselves from damage (Song et al., 2015). Symbisis between plant root systems and arbuscular mycorrhizal fungi (AMF) can be exploited for crop disease management (Ajit et al., 2017). Arbuscular mycorrhizal symbiosis can enhance plant resistance against various pathogens such as Alternaria spp., Rhizoctonia, Fusarium, Verticillium, and Thielaviopsis (Nair et al., 2015; Mustafa et al., 2017). The protective effects may result from a combination of diverse mechanisms (Dey and Ghosh, 2022). AMF induced plant defense response plays an important role in plant disease resistance (Jung et al., 2012). The defense responses of plants can be pre-axisting and induced (Xu et al., 2022). Plant physical structures and phytochemicals provide basic defense against fungal pathogens (Robert-Seilaniantz et al., 2011; Bellincampi et al., 2014; Ziv et al., 2018). After recognition of fungal pathogens, defense signaling is activated, leading to induction of immunity, local defense responses, and systemic defense signaling (Tian et al., 2016). Mycorrhiza-induced resistance is characterized by induction of root cell wall thickening, accumulation of phytoalexins, induced expression of plant defense genes, and stimulation of plant defense enzymes such as PAL, chitinase, and β-1,3-glucanase (Song et al., 2015; Eke et al., 2016; Bai et al., 2018).
Pathogen infection can reduce plant photosynthesis and damage the root system of plants (Dong et al., 2016). Reduced photosynthesis prevents plants from obtaining carbon nutrients, and root damage limits the absorption of nutrients and water (Serrano et al., 2016). In previous research, we observed that AMF increases photosynthesis and improves the root system of plants (Chen et al., 2017b). Therefore, we investigated if AMF can alleviate the photosynthesis and the root structure damage leading to reduced yield of S. miltiorrhiza caused by pathogen infection.
Previously, we found that arbuscular mycorrhizal symbiosis decreases the disease incidence of continuously cropped S. miltiorrhiza by nearly 75% (Yang et al., 2013). However, there is little known about the response of AMF-inoculated S. miltiorrhiza to F. oxysporum infection and mycorrhizal-induced defense mechanisms are poorly understood. In this study, we investigated the mechanisms of defense response in S. miltiorrhiza against F. oxysporum infection induced by pre-inoculation with AMF from two perspectives: the photosynthesis and root structure, and changes in expression of defense-related genes.
Materials and methods
Plant materials and fungal strains
S. miltiorrhiza seeds were collected from a planting base located in Laiwu, Shandong Province in North China (36°20’ N, 117°41’ E). The authors identified the seedlings as S. miltiorrhiza Bunge.
The AMF G. versiforme was originally provided by Professor Honggang Wang (Chinese Academy of Agricultural Sciences). and was propagated using Sorghum bicolor as the host. The spores, hyphae, colonized roots, and substrates were collected as AMF inocula. The AMF inocula was identified as G. versiforme following Wang et al. (2016) described (Figure S1).
The pathogen was isolated from roots of diseased S. miltiorrhiza that showed symptoms of Fusarium wilt and identified as F. oxysporum (Yang et al., 2013). The pathogen was cultured for five days in Armstrong Fusarium Medium Base (20.0 g glucose, 0.2 mg FeSO4, 1.6 g KCl, 0.4 g MgSO4·7H2O, 5.9 g Ca(NO3)2, 0.2 mg ZnSO4, 1.1 g KH2PO4, and 0.2 mg MnSO4 per liter, pH 7.0) at 28°C in darkness and on a shaker at 150 rpm. Three layers of sterile gauze were used to filtrate mycelia and the suspension concentration was 106 spores/ml in aseptic distilled water.
Cultivation substrate
Vermiculite was used as the germination substrate of S. miltiorrhiza seeds. After 30 days of germination, S. miltiorrhiza seedlings were transplanted to 1:1 (v/v) mixture of paddy soil and vermiculite. The paddy soil contained organic matter (0.49 g·kg-1), total N (3.85 g·kg-1), total P (8.43 g·kg-1), available P (2.27 mg·kg-1), total K (28.43 g·kg-1), available K (8.71 mg·kg-1), available Zn (0.07 mg·kg-1), available Mn (0.74 mg·kg-1), available Fe (1.6 mg·kg-1), and available Cu (0.13 mg·kg-1), with a pH value of 8.7. The substrate was sterilized at 121°C for 2 hours before use.
Experimental design
S. miltiorrhiza seeds were surface-sterilized in 75% ethanol for 1 min, soaked in 2% (V/V) NaClO for 10 min, and then rinsed with sterile water for 5 min. Germination substrate was autoclaved vermiculite. S. miltiorrhiza in AM treatment were pre-inoculated with G. versiforme, i.e., 100 g (equivalent to ~1250 spores) of AMF inoculum was mixed with 1 kg vermiculite. In NM treatment, an equal amount of autoclaved AMF inoculum was mixed with the vermiculite.
Thirty days after sowing, the mycorrhizal colonization of S. miltiorrhiza was assessed. S. miltiorrhiza seedlings were transplanted into square pots (7 cm × 7 cm), and inoculated with F. oxysporum. Four treatments were designed (NM-Fo, NM+Fo, AM-Fo, and AM+Fo): (1) NM-Fo: non-mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen; (2) NM+Fo: non-mycorrhizal S. miltiorrhiza inoculated with pathogen; (3) AM-Fo: mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen; (4) AM+Fo: mycorrhizal S. miltiorrhiza inoculated with pathogen. Each treatment included 60 pots. Seedlings were incubated in 5 mL spore suspension for 30min. Control S. miltiorrhiza were treated with 5 mL sterilized spore suspension for 30 min. Experiments were conducted in a greenhouse (30°C, 14L:10D photoperiod), with a photon flux density of 350 photon µmol·m−2·s−1 (photosynthetic active radiation).
Assessment of AMF colonization
AMF colonization was measured 30 days after germination. The roots of mycorrhizal S. miltiorrhiza were cut into 1 cm long sections and then stained with Trypan Blue following the protocol published previously (Phillips and Hayman, 1970). AMF colonization of S. miltiorrhiza was determined as described previously (Giovannetti and Mosse, 1980).
Disease incidence measured
Seven days after pathogen inoculation, disease incidence and disease index were measured. Disease incidence was calculated as the percentage of diseased S. miltiorrhiza. Disease severity was estimated using a Disease Index (DI) calculated as disease grades 0–5: 0, no symptoms; 1, growth delayed and no significant necrosis or atrophy of shoots and roots; 2, light chlorosis and necrosis on shoots and roots; 3, medium chlorosis and necrosis on shoots and roots; 4, high chlorosis and necrosis on shoots and roots; and 5, failed seedlings (Soudani et al., 2022). Disease incidence, disease index, and control efficacy were calculated using the following formulas:
Assessment of plant growth
Thirty days after pathogen inoculation, S. miltiorrhiza seedings were removed from the soil, the shoots and roots were separated, and the fresh weights of both the shoots and roots were recorded.
Root system measurement
Thirty days after pathogen inoculation, the roots of S. miltiorrhiza were scanned with an Epson Expression/STD 4800 scanner (Seiko Epson Corporation, Nagano, Japan), and the root length, root projArea, and root surfArea were derived with WinRHIZO image analysis software (Regent Instruments Inc., Quebec, QC, Canada).
Chlorophyll fluorescence measurement
The chlorophyll fluorescence parameters were determined 30 days after pathogen inoculation. A dual-PAM-100 device (Heinz Walz, Effeltrich, Germany) was used to measure the Chlorophyll fluorescence parameters of the two uppermost leaves of S. miltiorrhiza at 25°C according the previous published protocols (Ritchie and Bunthawin, 2010). Before measurement, the minimal fluorescence in the dark-adapted state (F0) was recorded after the plants were kept in the darkness for 30 min. The maximal fluorescence in the dark-adapted state (Fm), the maximal fluorescence (Fm’), the minimal fluorescence in the light-adapted state (F0’), and the steady-state fluorescence (Fs) of leaves were determined following the previously described methods (Gong et al., 2013). The chlorophyll fluorescence parameters ΦPSII, Fv/Fm, qP, and qN were as described (Zai et al., 2012; Sowik et al., 2016).
Chlorophyll measurement
Thirty days after pathogen inoculation, chlorophyll content was measured as described previously (Gregor and Maršálek, 2004). Approximately 0.05 g fresh leaves of S. miltiorrhiza were ground into fine powder and 8 mL 95% ethanol was added. Samples were stored in the dark for 48 h. The absorption of the continuation filtrate was measured at 665 nm, 649 nm, and 470 nm and the content of chlorophyll was calculated according to the following formulas:
Ca = 13.95A665 - 6.88A649, Cb = 24.96A649 - 7.32A665, CChl = Ca + Cb, CCar = (1000A470 - 2.05Ca - 114.8Cb)/245
Chlorophyll a content = Ca × V/W, Chlorophyll b content=Cb × V/W, Total Chlorophyll content = CChl × V/W, Carotenoid content = CCar × V/W
Content of soluble protein measurement
Thirty days after pathogen inoculation, soluble protein content was determined according to the previously published method (Yen and Pratap-Singh, 2021). A standard curve was constructed using different concentrations (0-2 mg·mL-1) of bovine serum albumin (BSA) to estimate of protein content.
Activities of defense-related enzymes
The activities of defense-related enzymes were detected five days following infection. Approximately 0.1 g root samples of S. miltiorrhiza were ground into fine powder in liquid nitrogen and were extracted with 2 mL 0.05 M sodium acetate buffer (pH 5.0). Extracts were centrifuged at 12,000 g for 15 min at 4°C and the supernatant fractions were used to assay enzyme activity. PAL activity was analyzed as Mozzetti et al. (1995) described. β-1,3-Glucanase activity was assayed by the laminarin-dinitro salicylic acid method (Pan, 1991). Chitinase activity was analyzed as Boller and Mauch (1988) described.
Expression of defense-related genes
The expression levels of defense-related genes, SmLOX (JX297420.1), SmAOS, SmAOC (HM156740.1), SmOPR (MN125491.1), SmJAR, SmPDF2.1 (OP066222), SmPAL (DQ408636.1), SmNPR1, SmPR1, and SmPR10 (KF877034.1), were measured by qRT-PCR three days after pathogen inoculation. To do this, 0.1 g root samples were ground into fine powder in liquid nitrogen and total RNA was extracted using the RNeasy Plus Mini kit (Qiagen, Germany). Reverse transcription was performed using PrimeScript™ Reverse Transcriptase (TaKaRa, Japan). Primer Premier 5 software used to design the primers as shown in Table S1 and qRT-PCR analysis was conducted using SYBR® Premix Ex Taq™ II (TaKaRa, Japan), with SmActin (DQ243702) as a reference gene using a LightCycler 480 real-time PCR system (Roche, Switzerland). CT values were calculated to analyze the relative expression levels using the 2-ΔΔCt method (Guo et al., 2016).
Statistical analysis
All data were analyzed using IBM SPSS Statistics 24. Results are presented as the mean values ± standard deviation (SD). Data were analyzed with two-way ANOVA followed by Tukey’s test and differences were reported as significant for values of P < 0.05.
Results
Induction of disease resistance by Mycorrhizal colonization
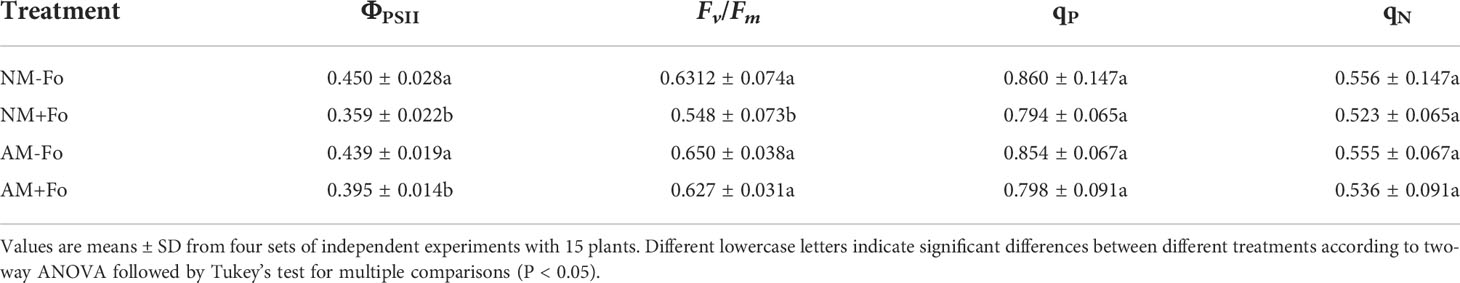
Mycorrhizal colonization was examined 30 days post-inoculation. Among the S. miltiorrhiza treated with G. versiforme (AM treatment), 83.33 ± 3% were successfully colonized by G. versiforme (Figures 1A, B, Table 1). There was no fungal structure in the roots of plants in the NM treatment. The results showed that S. miltiorrhiza was successfully colonized by the AMF and the pathogen could be inoculated later.
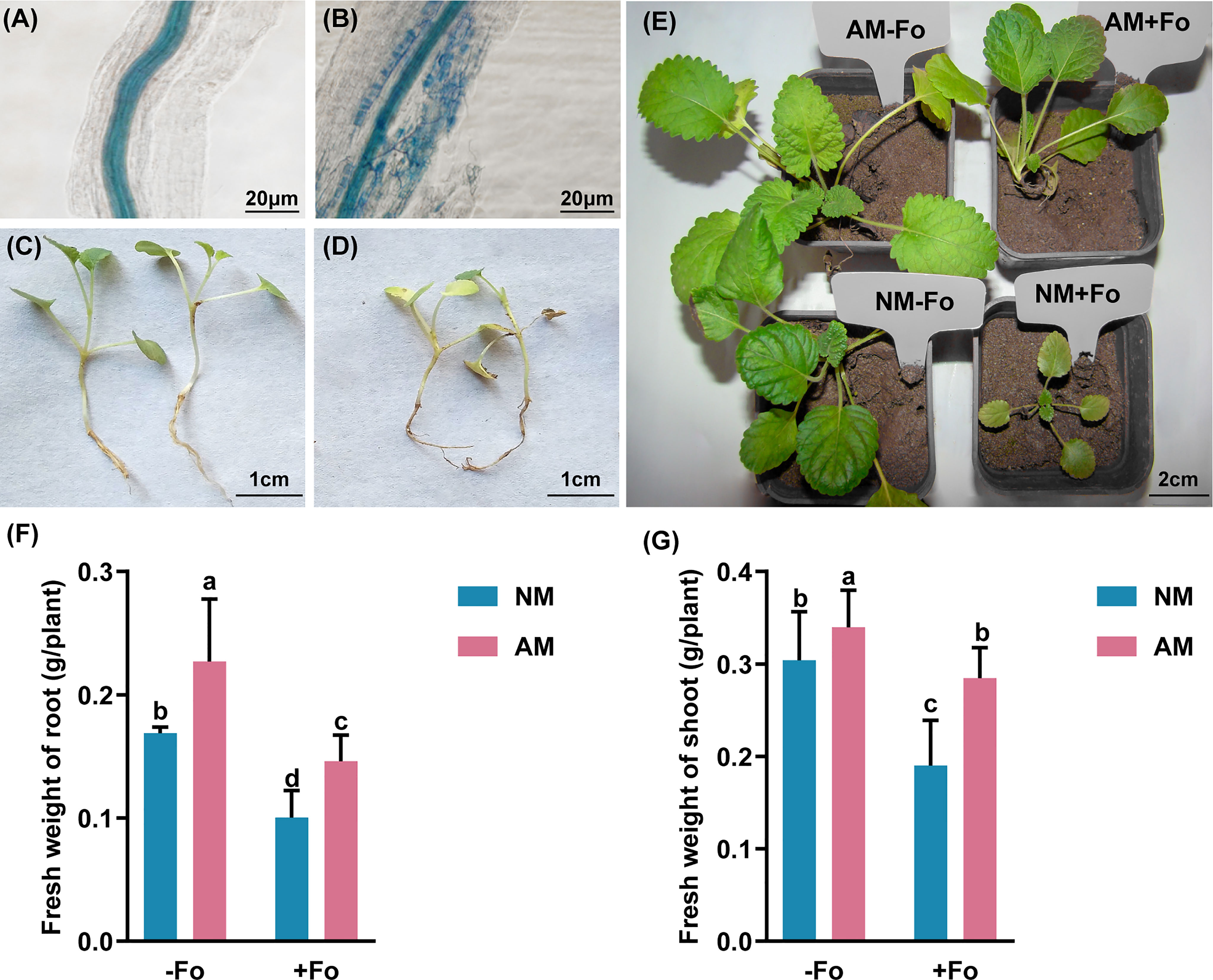
Figure 1 G versiforme alleviates disease of S. miltiorrhiza infected with F oxysporum. (A, B) Uncolonized roots (A) and colonized roots (B) by G versiforme. The photos were taken 30 days after mycorrhizal inoculation. (C, D) S. miltiorrhiza seedlings without pathogen inoculation (C) and diseased S. miltiorrhiza seedlings infected with the pathogen (D). The photos were taken 7 days after inoculating with F oxysporum. (E) S. miltiorrhiza plants of four treatments 30 days after inoculating with F oxysporum. (F, G) Fresh weight of shoot (F) and root (G) of S. miltiorrhiza 30 days after pathogen inoculation. Four treatments included: (1) NM-Fo: non-mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen inoculation; (2) NM+Fo: non-mycorrhizal S. miltiorrhiza inoculated with pathogen; (3) AM-Fo: mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen; (4) AM+Fo: mycorrhizal S. miltiorrhiza inoculated with pathogen. Values are means ± SD from four sets of independent experiments with 30 pots per treatment for each set of experiments. Different lowercase letters indicate significant differences between different treatments according to two-way ANOVA followed by Tukey’s test for multiple comparisons (P < 0.05).
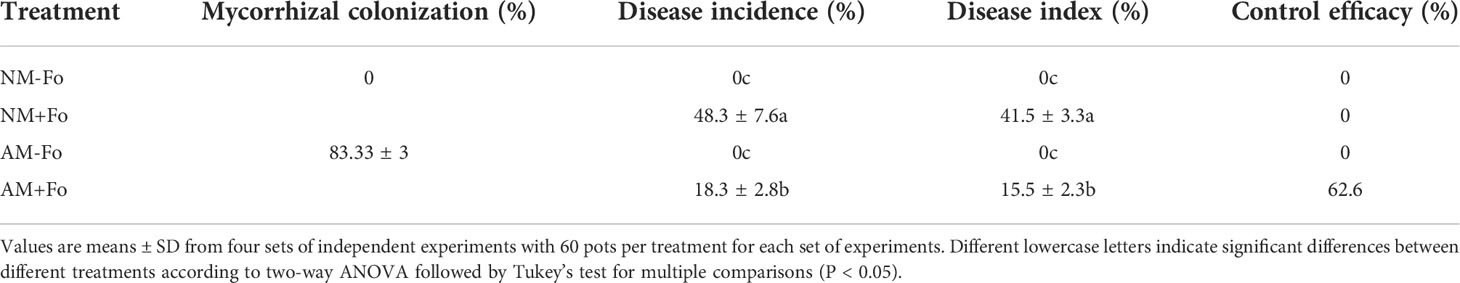
Table 1 Mycorrhizal colonization, disease incidences, and indices of S. miltiorrhiza inoculated with F. oxysporum.
No disease symptoms were found in the two groups without inoculation of the pathogen (Figures 1C, E). Disease symptoms of S. miltiorrhiza infected with F. oxysporum exhibited dwarfish stem, yellow and smallish leaves, and generally withered plants (Figures 1D, E). Pre-inoculation of S. miltiorrhiza with the G. versiforme significantly decreased the disease incidence and disease severity of Fusarium wilt compared to the plants in the NM+Fo treatment. The disease incidence and disease index of the NM+Fo treatment were 48.3% and 41.5%, while those of the AM+Fo treatment were only 18.3% and 15.5% after seven days of pathogen inoculation (Table 1). Disease incidence was reduced by 62.1% in mycorrhizal plants. Mycorrhizal plants had significantly decreased disease symptoms compared to non-mycorrhizal inoculated plants 45 days after pathogen infection (Figure 1E). The control efficacy of AMF pre-inoculation was 62.6% (Table 1).
G. versiforme alleviated the retarded growth of S. miltiorrhiza resulting from F. oxysporum infection
G. versiforme colonization significantly increased the fresh weight of shoots and roots by 11.74% and 34.56%, respectively (Figures 1F, G). In contrast, F. oxysporum decreased the shoot biomass and root biomass by 37.5% and 40.6%, respectively (Figures 1F, G). Mycorrhizal plants promoted the accumulation of plant biomass relative to non-mycorrhizal plants after inoculation with the pathogen (Figure 1E). Compared to NM+Fo treatment, pre-inoculation with AMF (AM+Fo treatment) increased the fresh weight of shoots and roots by 49.8% and 45.7%, respectively.
G. versiforme improved root morphology of S. miltiorrhiza infected with F. oxysporum
The results of root scanning showed that the F. oxysporum infection seriously damaged the root system of S miltiorrhiza, resulting in less fibrous roots and root vascular blocking, while mycorrhizal colonization greatly promoted the development of root system (Figure 2A). Mycorrhizal S. miltiorrhiza partially resisted root damage caused by pathogen infection (Figure 2A). G. versiforme colonization significantly increased the length of root by 32.80%, root projArea by 16.27%, and root surfArea by 18.18%, but pathogen infection decreased those of S. miltiorrhiza (Figures 2B–D). Pre-inoculating AMF decreased the loss of root biomass caused by pathogen infection. The length of root and root surfArea of S. miltiorrhiza in NM+Fo treatment were significantly lower than those of S. miltiorrhiza in AM+Fo treatment (Figures 2B, D).
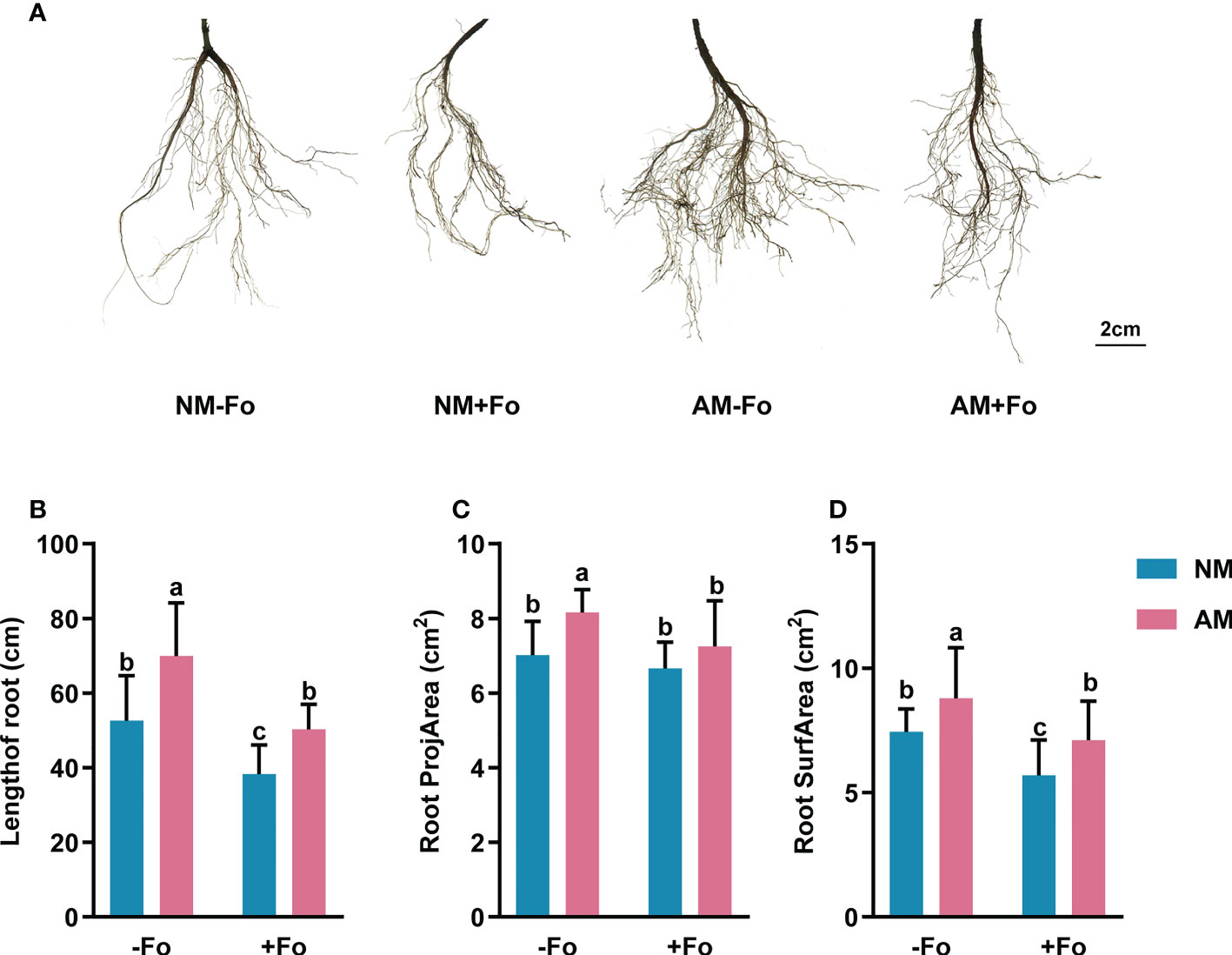
Figure 2 Root morphology of S. miltiorrhiza in the four treatments. (A) Root scans of S. miltiorrhiza in AM+Fo treatment and NM+Fo treatment. Length of root (B), root projArea (C), and root surfArea (D) of S. miltiorrhiza 30 days after pathogen inoculation. Four treatments included: (1) NM-Fo: non-mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen inoculation; (2) NM+Fo: non-mycorrhizal S. miltiorrhiza inoculated with pathogen; (3) AM-Fo: mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen; (4) AM+Fo: mycorrhizal S. miltiorrhiza inoculated with pathogen. Values are means ± SD from four sets of independent experiments with 30 pots per treatment for each set of experiments. Different lowercase letters indicate significant differences between different treatments according to two-way ANOVA followed by Tukey’s test for multiple comparisons (P < 0.05).
G. versiforme improved photosynthesis of S. miltiorrhiza infected with F. oxysporum
F. oxysporum infection decreased the photosynthesis-related parameters ΦPSII and Fv/Fm of non-mycorrhizal S. miltiorrhiza by 20.2% and 13%, respectively. While 10% decreased on ΦPSII and no significant difference in Fv/Fm (Table 2) of mycorrhizal S. miltiorrhiza. Pathogen inoculation also decreased the qP and qN, but there was no significant difference between the four treatments (Table 2).
The content of Chlorophyll a, Chlorophyll b, and total Chlorophyll of S. miltiorrhiza in the NM+Fo treatment were significantly decreased by 10%, 11%, and 15% compared with NM-Fo treatment. However, the above parameters were not decreased by pathogen inoculation in mycorrhizal S. miltiorrhiza (Table 3). In addition, AMF colonization significantly increased the content of carotenoid (Table 3).
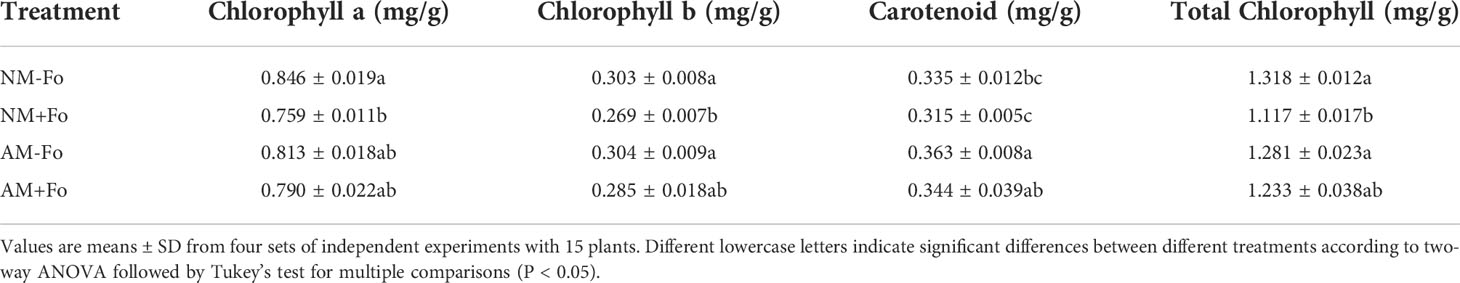
Table 3 Content of Chlorophyll a, Chlorophyll b, Carotenoid, and total Chlorophyll in leaves of S. miltiorrhiza seedlings.
G. versiforme improved the protein content of S. miltiorrhiza infected with F. oxysporum
Mycorrhizal colonization significantly reduced the content of soluble protein in the roots of S. miltiorrhiza by 22.6% compared with non-mycorrhizal plants (Figure 3). F. oxysporum infection significantly reduced the protein content in non-mycorrhizal S. miltiorrhiza by 72.4% but increased protein content by 48% in mycorrhizal S. miltiorrhiza (Figure 3).
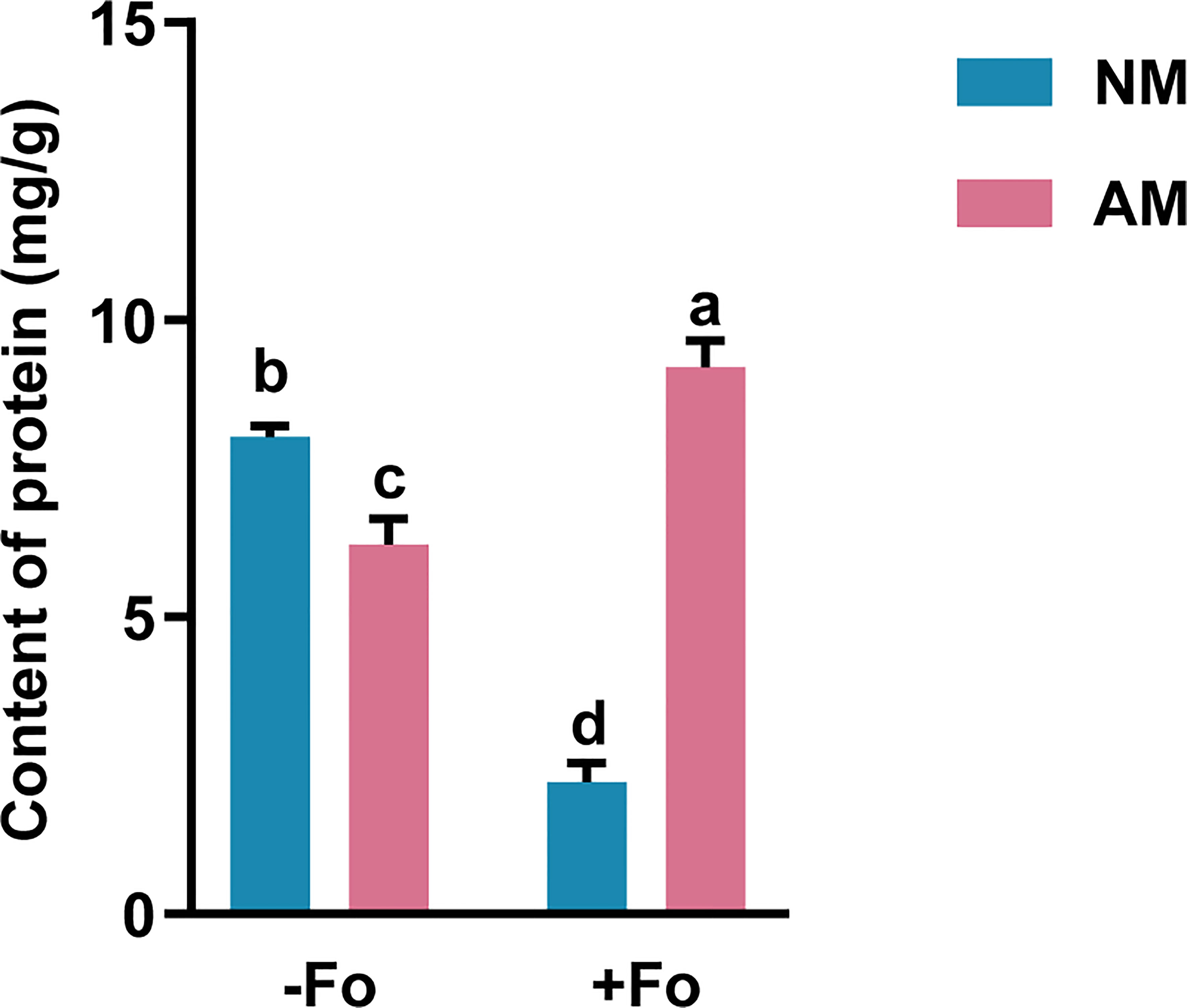
Figure 3 Protein content in the roots of S. miltiorrhiza from the four treatments. The protein content was measured 30 days after pathogen inoculation. Data are mean values ± SD; significant differences (P < 0.05 using Tukey’s test) among treatments in the same column are indicated by different letters. Four treatments included: (1) NM-Fo: non-mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen inoculation; (2) NM+Fo: non-mycorrhizal S. miltiorrhiza inoculated with pathogen; (3) AM-Fo: mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen; (4) AM+Fo: mycorrhizal S. miltiorrhiza inoculated with pathogen. Values are means ± SD from four sets of independent experiments with 15 plants per treatment. Different lowercase letters indicate significant differences between different treatments according to two-way ANOVA followed by Tukey’s test for multiple comparisons (P < 0.05).
Induction of defense-related enzymes in Mycorrhizal S. miltiorrhiza by pathogen infection
To determine the effects of AMF colonization on defense responses in S. miltiorrhiza, the levels of three defense-related enzymes, PAL, β-1,3-glucanase, and chitinase, were analyzed in the roots of S. miltiorrhiza after pathogen infection.
The PAL activity of mycorrhizal and pathogen-infected S. miltiorrhiza (AM+Fo treatment) was significantly increased by 39% compared with that of control NM treatment S. miltiorrhiza (Figure 4A). However, inoculation of S. miltiorrhiza with AMF or pathogen alone did not significantly enhance PAL activity in the roots of S. miltiorrhiza (Figure 4A).
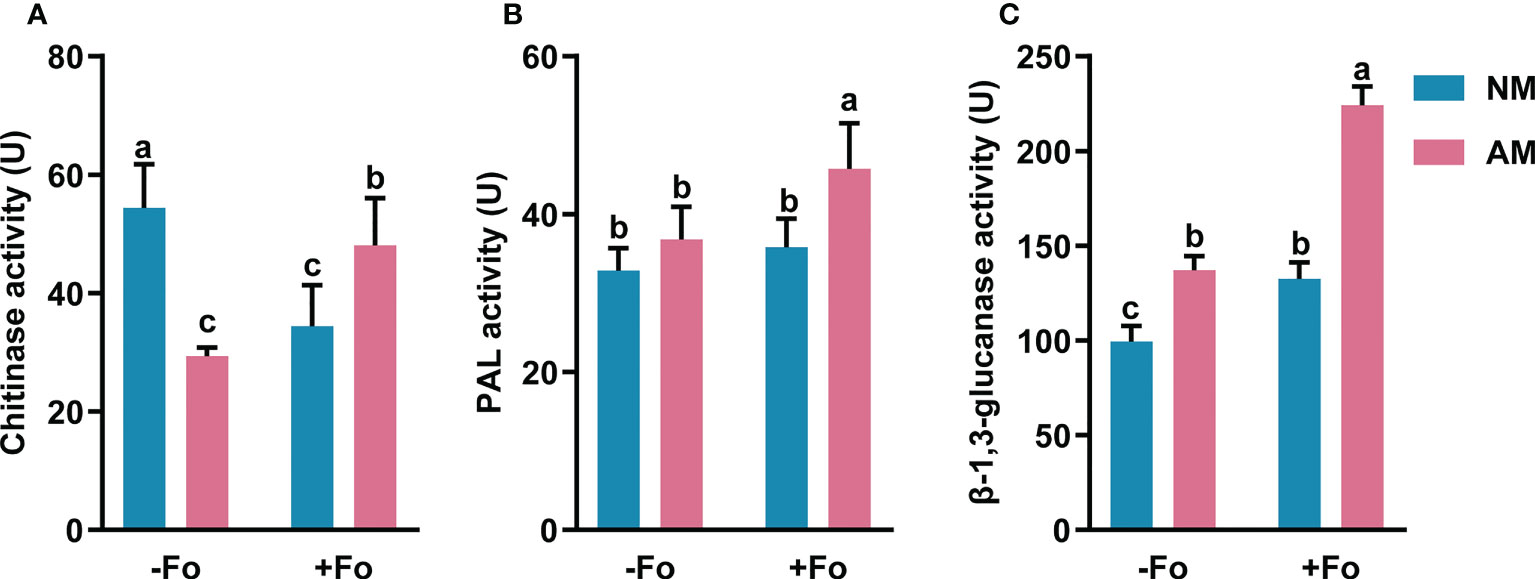
Figure 4 The activity of chitinase (A), PAL (B), and β-1,3-glucanase (C) in the roots of S. miltiorrhiza five days after pathogen inoculation. Four treatments included: (1) NM-Fo: non-mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen inoculation; (2) NM+Fo: non-mycorrhizal S. miltiorrhiza inoculated with pathogen; (3) AM-Fo: mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen; (4) AM+Fo: mycorrhizal S. miltiorrhiza inoculated with pathogen. Values are means ± SD from four sets of independent experiments with 15 plants per treatment. Different lowercase letters indicate significant differences between different treatments according to two-way ANOVA followed by Tukey’s test for multiple comparisons (P < 0.05).
Inoculation of AMF or pathogen alone significantly increased β-1,3-glucanase activity by 28% and 34%, respectively, while inoculation of S. miltiorrhiza with both AMF and pathogen increased β-1,3-glucanase activity by 125% (Figure 4B).
Unlike the increased activities of PAL and β-1,3-glucanase, chitinase activity significantly decreased by 39% and 45% after AMF colonization or pathogen infection, respectively. However, there was a smaller drop (11.55%) in the activity of chitinase after AMF and pathogen dual inoculation (Figure 4C).
Overall, mycorrhizal S. miltiorrhiza treatment showed higher increases in three enzymes activities after pathogen infection, especially PAL and β-1,3-glucanase, suggesting that mycorrhizal pre-inoculation enhanced the activities of these enzymes in the roots of S. miltiorrhiza upon pathogen infection.
Mycorrhizal colonization induced transcription of defense-related genes
To determine whether the transcript induction of defense-related genes was enhanced by mycorrhizal colonization, gene expression was analyzed from S. miltiorrhiza roots three days after pathogen inoculation using real-time RT-PCR. The amplification efficiency of the primer pairs ranged from 90 to 110% (Table S1, Figure S2). These primers were used for quantitive analysis of the transcriptional activity of defense-related genes. The JA synthesis pathway genes, SmLOX, SmAOS, SmAOC, and SmOPR, were significantly up-regulated by 443%, 653%, 178%, and 113%, respectively, in mycorrhizal S. miltiorrhiza roots after pathogen infection (Figures 5A–D). However, pathogen infection alone did not induce these gene transcription. Similarly, the JA signaling pathway gene, SmJAR, and the markers of the JA defense-response pathway, SmPDF2.1, were upregulated by 116% and 257%, respectively, in mycorrhizal S. miltiorrhiza roots after pathogen infection (Figures 5E, F). In addition, inoculation AMF alone up-regulated the transcripts of SmAOS, SmAOC, SmJAR, and SmPDF2.1. by 156%, 325%, 123%, and 163%, respectively (Figures 5B, C, E, F).
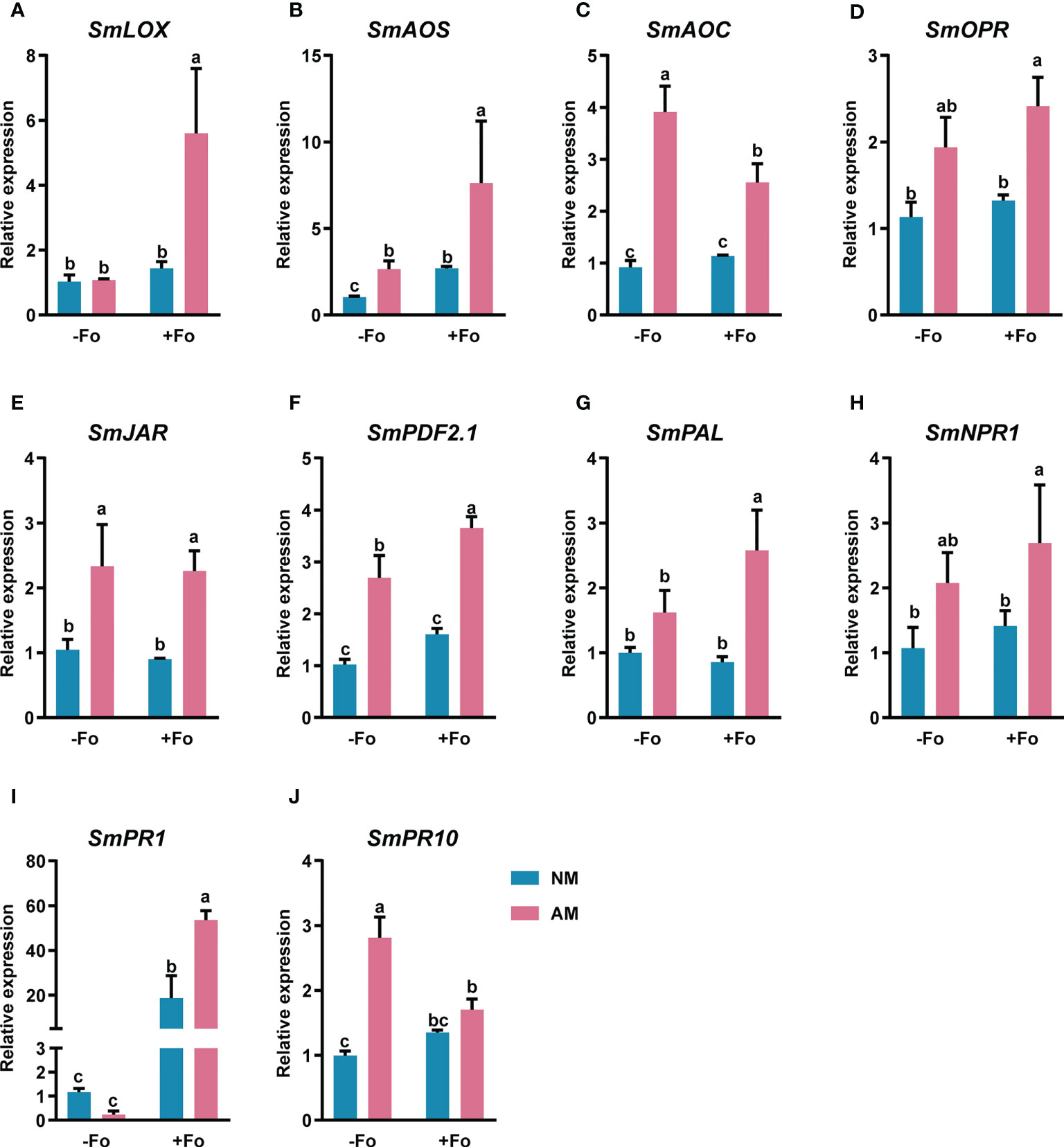
Figure 5 Relative expression levels of SmLOX (A), SmAOS (B), SmAOC (C), SmOPR (D), SmJAR (E), SmPDF2.1 (F), SmPAL (G), SmNPR1 (H), SmPR1 (I), and SmPR10 (J) in the roots of S. miltiorrhiza three days after pathogen inoculation. Four treatments included: (1) NM-Fo: non-mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen inoculation; (2) NM+Fo: non-mycorrhizal S. miltiorrhiza inoculated with pathogen; (3) AM-Fo: mycorrhizal S. miltiorrhiza inoculated with heat-killed pathogen; (4) AM+Fo: mycorrhizal S. miltiorrhiza inoculated with pathogen. Values are means ± SD from four sets of independent experiments with 15 plants per treatment. Different lowercase letters indicate significant differences between different treatments according to two-way ANOVA followed by Tukey’s test for multiple comparisons (P < 0.05).
SmPAL, the key gene involved in the biosynthesis of SA (Shine et al., 2016), and SmNPR1, a master regulator of SA (Tada et al., 2008), were significantly up-regulated by 156% and 151%, respectively, in mycorrhizal S. miltiorrhiza roots after pathogen infection (Figures 5G, H). However, there were no expression changes in response to either mycorrhizal colonization or pathogen infection alone. SmPR1 and SmPR10, encode pathogenesis-related proteins and were significantly up-regulated in mycorrhizal S. miltiorrhiza roots after pathogen infection (Figures 5I, J). After pathogen infection, SmPR1 was 45-fold up-regulated in mycorrhizal S. miltiorrhiza roots, while 15-fold up-regulated in non-mycorrhizal S. miltiorrhiza roots (Figure 5I).
Discussion
Fusarium wilt has become a major disease of S. miltiorrhiza and is a major limiting factor for cultivation. We showed that the Fusarium wilt caused by F. oxysporum can be alleviated through mycorrhizal pre-inoculation. Pre-inoculation of S. miltiorrhiza with G. versiforme significantly decreased disease incidence (from 48.3% to 18.3%) and disease index (from 41.5% to15.5%) of Fusarium wilt compared to S. miltiorrhiza without mycorrhizal colonization (Table 1). F. oxysporum infection reduced the shoot and root biomass of non-mycorrhizal S. miltiorrhiza by 37.5% and 40.6%, however, G. versiforme pre-inoculation reduced the loss of shoot biomass to 16.19% and root biomass to 35.68% (Figures 1F, G). The results were in accordance with previous reports that AMF colonization alleviates alfalfa leaf spots caused by Phoma medicaginis (Li et al., 2021), and that Funneliformis mosseae significantly alleviates early blight disease in tomato caused by Alternaria solani Sorauer (Song et al., 2015).
Roots allow plants to absorb nutrients and water. Infection of S. miltiorrhiza by F. oxysporum leads to Fusarium wilt, with symptoms including less fibrous roots and root vascular blocking, decreasing absorption of nutrients and water and resulting in plant wilting and death (Yang et al., 2013; Chen et al., 2017a). We found that G. versiforme increased the length of root by 32.80%, root projArea by 16.27%, and root surfArea by 18.18% of S. miltiorrhiza (Figures 2A–D). Root structure is key to determining a plant’s ability to effectively explore soils (Dorlodot et al., 2007). AMF improves plant nutrition, and this could contribute to increased plant tolerance and compensation for root damage caused by the pathogen (Cordier et al., 1998). The increased nutrition and fitness of mycorrhizal plants likely serve as systemic protection mechanisms against pathogen attack (Fritz et al., 2006). Many studies have shown that the major benefit of AMF colonization is its effect on the host root system (Gutjahr et al., 2009; Vos et al., 2013). Consistently, the improved root system structure we observed here appears to be a key factor in the disease resistance of S. miltiorrhiza induced by G. versiforme.
Photosynthesis not only provides nutrients for plant growth, but also produces defense-related substances to counter pathogens (Serrano et al., 2016). Suppressing photosynthesis is also a strategy for successful infection of pathogens. Fv/Fm and ΦPSII are important indicators of the photosynthetic apparatus and are widely used to assess plant-pathogen interactions (Wang et al., 2018). Consistent with our findings (Table 2), a previous study found that pathogen infection significantly reduced Fv/Fm and ΦPSII in plants not inoculated with AMF, but had no effect in mycorrhizal plants (Wang et al., 2018). Here, F. oxysporum infection decreased the photosynthesis-related parameters Fv/Fm and ΦPSII by 20.2% and 13% in non-mycorrhizal S. miltiorrhiza, respectively, while ΦPSII only decreased by 10% and Fv/Fm did not decrease in mycorrhizal S. miltiorrhiza (Table 2). In plants, carotenoids play vital roles in photosynthesis as light-harvesting pigments and photo-protective compounds (Gupta and Hirschberg, 2022). Previous study demonstrated a positive correlation between carotenoids and photosynthetic rate (Lobato et al., 2010). In this study, the pathogen infection significantly increased the carotenoid content in mycorrhizal S. miltiorrhiza. Together, these results suggest that the disease resistance of S. miltiorrhiza can be improved by improving photosynthesis.
The activity of defense-related enzymes (e.g., PAL, chitinase, and β-1,3-glucanase) can be enhanced when systemic resistance is activated (Hura et al., 2014; Jain and Choudhary, 2014; Eke et al., 2016; Gharbi et al., 2017). Here, we found that, after pathogen infection, the activities of PAL, chitinase, and β-1,3-glucanase showed greater increases in the roots of mycorrhizal S. miltiorrhiza than in non-mycorrhizal S. miltiorrhiza plants (Figures 4A–C). These enzymes are crucial components in plant resistance to biotic diseases (Funnell et al., 2004). PAL is the key enzyme in the biosynthesis of multiple antimicrobial compounds (phenolic acid, flavonoids), lignin (a rapidly deposited physical barrier), and salicylic acid, three compunds that are related to plant resistance (Wang et al., 2019). Chitinase and β-1,3-glucanase can degrade pathogenic fungal cellular components to inactivate fungi, and also produce monomers to further stimulate plant defense responses (Anguelova-Merhar et al., 2001; Doxey et al., 2007; Kumar et al., 2018). Our results showed that AMF can trigger the expression of defense enzymes in the host plant, which was similar to the response of F. oxysporum infection (Figures 4A–C). Inoculating AMF or pathogen alone significantly increased β-1,3-glucanase activity, inhibited chitinase activity, and did not affect PAL activity in S. miltiorrhiza (Figures 4A–C). These results indicate that inoculation of AMF or infection with pathogen alone can stimulate β-1,3-glucanase-related defense responses, but do not affect chitin- and PAL-related defense responses. This result is in agreement with the report that mycorrhizal fungi initially trigger plant defense mechanisms similarly to a biotrophic pathogen (Paszkowski, 2006). Song et al. (2015) found that AMF inoculation itself did not affect most enzyme activities, but after pathogen attack AMF pre-inoculation induces tomato plants to produce a defense response of four defense-related enzymes. Our results showed that upon pathogen attack (AM+Fo treatment), AMF pre-inoculation strongly induced the activities of PAL and β-1,3-glucanase by 39.23% and 125.18%, respectively (Figures 4A, B). PAL and β-1,3-glucanase activities in the AM+Fo treatment were the highest among all treatments. Pre-inoculation of AMF can alleviate the inhibitory effect on chitinase activity caused by the pathogen. Overall, pre-inoculation with G. versiforme inhibits pathogen infection by increasing the activities of PAL, β-1,3-glucanase, and chitinase.
Disease resistance in plants is tightly regulated through an interlinked network of JA and SA signaling pathways (Song et al., 2015). The JA signaling pathway plays an important role in plant defense response, and SmLOX, SmAOS, SmAOC, and SmOPR are important genes in JA biosynthes. Plant defensins (PDFs) are a family of small cysteine-rich basic proteins (García-Olmedo et al., 1998). SmPDF2.1, a gene encoding plant defensin, is a marker of the jasmonate (JA) defense-response pathway (Hanks et al., 2005). The stronger induction of these genes in mycorrhizal plants after pathogen infection suggested that mycorrhizal colonization activates the JA signaling pathway and enhances the resistance of S. miltiorrhiza to F. oxysporum. This is consistent with previous studies that mycorrhizal colonization enhances resistance to early blight in tomato by initiating a systemic defense response and that the JA signaling pathway is critical in the mycorrhizal-initiated disease resistance process (Song et al., 2015). SmPAL is a key gene involved in the biosynthesis of SA (Shine et al., 2016), and SmNPR1 is a master regulator of SA (Tada et al., 2008). The induction of SmPR1 and SmPR10 indicates that mycorrhizal colonization provokes SA signaling pathways upon pathogen attack. PR genes are usually used as marker genes of the acquisition of systemic resistance in plants (Mitsuhara et al., 2008) and the levels of PR proteins are used as an indicator of defense responses (Song et al., 2015). Pathogenesis-related 1 (PR1) protein is a commonly used reporter of SA-activated defense responses in plants (Pečenková et al., 2022). Consistent with our studies, many studies reported that mycorrhizal colonization induced the transcription of PR genes (Ismail and Hijri, 2012; Li et al., 2021).
Conclusion
Pre-inoculation of S. miltiorrhiza with the AMF, G. versiforme, enhanced resistance to Fusarium wilt by priming the systemic defense response. Mycorrhizal colonization improved the root structure and photosynthesis capacity of S. miltiorrhiza to reduce disease incidence. Infection with the pathogen alone could evade the PAL- and chitinase-related defense responses, however pre-inoculation of S. miltiorrhiza with AMF strongly induced PAL-, β-1,3-glucanase-, and chitinase-related defense responses upon pathogen attack. JA and SA signaling pathways are key components of the plant defense response, and were strongly activated by pre-inoculation of AMF upon pathogen attack.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
MC and GY designed the experiments, which were performed by CP, HZ, and SL. CP wrote the manuscript and analyzed the results. YG, ZC, WG, and YS revised the manuscript. MC and LH provided the funding. All authors contributed to the article and approved the submitted version.
Funding
The Scientific and Technological Innovation Project of China Academy of Chinese Medical Sciences (CI2021A03906) and the National Natural Science Foundation of China (81773849, 82173931, 81803658) supported this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.975558/full#supplementary-material
References
Ajit, V., Ram, P., Narendra, T. (2017). Mycorrhiza-Nutrient Uptake, Biocontrol, Ecorestoration. (Berlin: Springer Cham). C1–C1. doi: 10.1007/978-3-319-68867-1_27
Anguelova-Merhar, V. S., Westhuizen, A. J., Pretorius, Z. A. (2001). β-1,3-Glucanase and chitinase activities and the resistance response of wheat to leaf rust. J. Phytopathol. 149, 381–384. doi: 10.1111/j.1439-0434.2001.tb03866.x
Ankati, S., Srinivas, V., Pratyusha, S., Gopalakrishnan, S. (2021). Streptomyces consortia-mediated plant defense against fusarium wilt and plant growth-promotion in chickpea. Microb. Pathogenesis 157, 104961. doi: 10.1016/j.micpath.2021.104961
Bai, Y., Kissoudis, C., Yan, Z., Visser, R. G. F., van der Linden, G. (2018). Plant behaviour under combined stress: tomato responses to combined salinity and pathogen stress. Plant J. 93, 781–793. doi: 10.1111/tpj.13800
Bellincampi, D., Cervone, F., Lionetti, V. (2014). Plant cell wall dynamics and wall-related susceptibility in plant–pathogen interactions. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00228
Boller, T., Mauch, F. (1988). Biomass part b: Lignin, pectin, and chitin. Methods Enzymol. 161, 430–435. doi: 10.1016/0076-6879(88)61052-4
Chen, H., Wu, H., Yan, B., Zhao, H., Liu, F., Zhang, H., et al. (2018). Core microbiome of medicinal plant Salvia miltiorrhiza seed: A rich reservoir of beneficial microbes for secondary metabolism? Int. J. Mol. Sci. 19, 672. doi: 10.3390/ijms19030672
Chen, M., Yang, G., Liu, D., Li, M., Qiu, H., Guo, L., et al. (2017a). Inoculation with Glomus mosseae improves the growth and salvianolic acid b accumulation of continuously cropped Salvia miltiorrhiza. Appl. Sci. 7, 692. doi: 10.3390/app7070692
Chen, M., Yang, G., Sheng, Y., Li, P., Qiu, H., Zhou, X., et al. (2017b). Glomus mosseae inoculation improves the root system architecture, photosynthetic efficiency and flavonoids accumulation of liquorice under nutrient stress. Front. Plant Sci. 08. doi: 10.3389/fpls.2017.00931
Cordier, C., Pozo, M. J., Barea, J. M., Gianinazzi, S., Gianinazzi-Pearson, V. (1998). Cell defense responses associated with localized and systemic resistance to Phytophthora parasitica induced in tomato by an arbuscular mycorrhizal fungus. Mol. Plant-Microbe Interact. 11, 1017–1028. doi: 10.1094/mpmi.1998.11.10.1017
de Lamo, ,. F. J., Takken, F. L. W. (2020). Biocontrol by Fusarium oxysporum using endophyte-mediated resistance. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00037
Dey, M., Ghosh, S. (2022). Arbuscular mycorrhizae in plant immunity and crop pathogen control. Rhizosphere 22, 100524. doi: 10.1016/j.rhisph.2022.100524
Dong, X., Wang, M., Ling, N., Shen, Q., Guo, S. (2016). Potential role of photosynthesis-related factors in banana metabolism and defense against Fusarium oxysporum f. sp. cubense. Environ. Exp. Bot. 129, 4–12. doi: 10.1016/j.envexpbot.2016.01.005
Dorlodot, S., Forster, B., Pagès, L., Price, A., Tuberosa, R., Draye, X. (2007). Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci. 12, 474–481. doi: 10.1016/j.tplants.2007.08.012
Doxey, A. C., Yaish, M. W. F., Moffatt, B. A., Griffith, M., McConkey, B. J. (2007). Functional divergence in the arabidopsis β-1,3-Glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol. Biol. Evol. 24, 1045–1055. doi: 10.1093/molbev/msm024
Eke, P., Chatue, G. C., Wakam, L. N., Kouipou, R. M. T., Fokou, P. V. T., Boyom, F. F. (2016). Mycorrhiza consortia suppress the fusarium root rot (Fusarium solani f. sp. phaseoli) in common bean (Phaseolus vulgaris l.). Biol. Control 103, 240–250. doi: 10.1016/j.biocontrol.2016.10.001
Fritz, M., Jakobsen, I., Lyngkjær, M. F., Thordal-Christensen, H., Pons-Kühnemann, J. (2006). Arbuscular mycorrhiza reduces susceptibility of tomato to Alternaria solani. Mycorrhiza 16, 413–419. doi: 10.1007/s00572-006-0051-z
Funnell, D. L., Lawrence, C. B., Pedersen, J. F., Schardl, C. L. (2004). Expression of the tobacco β-1,3-glucanase gene, PR-2d, following induction of SAR with Peronospora tabacina. Physiol. Mol. Plant P 65, 285–296. doi: 10.1016/j.pmpp.2005.02.010
García-Olmedo, F., Molina, A., Alamillo, J. M., Rodríguez-Palenzuéla, P. (1998). Plant defense peptides. Pept. Sci. 47, 479–491. doi: 10.1002/(sici)1097-0282(1998)47:6<479::aid-bip6>3.0.co;2-k
Gharbi, Y., Barkallah, M., Bouazizi, E., Hibar, K., Gdoura, R., Triki, M. A. (2017). Lignification, phenols accumulation, induction of PR proteins and antioxidant-related enzymes are key factors in the resistance of Olea europaea to verticillium wilt of olive. Acta Physiol. Plant 39, 43. doi: 10.1007/s11738-016-2343-z
Giovannetti, M., Mosse, B. (1980). An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84, 489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x
Gong, M., Tang, M., Chen, H., Zhang, Q., Feng, X. (2013). Effects of two Glomus species on the growth and physiological performance of Sophora davidii seedlings under water stress. New For. 44, 399–408. doi: 10.1007/s11056-012-9349-1
Gregor, J., Maršálek, B. (2004). Freshwater phytoplankton quantification by chlorophyll a: a comparative study of in vitro, in vivo and in situ methods. Water Res. 38, 517–522. doi: 10.1016/j.watres.2003.10.033
Guo, J., Ma, X., Cai, Y., Ma, Y., Zhan, Z., Zhou, Y. J., et al. (2016). Cytochrome P450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytol. 210, 525–534. doi: 10.1111/nph.13790
Gupta, P., Hirschberg, J. (2022). The genetic components of a natural color palette: A comprehensive list of carotenoid pathway mutations in plants. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.806184
Gutjahr, C., Casieri, L., Paszkowski, U. (2009). Glomus intraradices induces changes in root system architecture of rice independently of common symbiosis signaling. New Phytol. 182, 829–837. doi: 10.1111/j.1469-8137.2009.02839.x
Hammad, A. M. M., El-Mohandes, M. A. O.. (1999). Controlling fusarium wilt disease of cucumber plants via antagonistic microorganisms in free and immobilized states. Microbiol Res. 154, 113–117. doi: 10.1016/s0944-5013(99)80002-0
Hanks, J. N., Snyder, A. K., Graham, M. A., Shah, R. K., Blaylock, L. A., Harrison, M. J., et al. (2005). Defensin gene family in Medicago truncatula: structure, expression and induction by signal molecules. Plant Mol. Biol. 58, 385–399. doi: 10.1007/s11103-005-5567-7
Hura, K., Hura, T., Dziurka, K., Dziurka, M. (2014). Biochemical defense mechanisms induced in winter oilseed rape seedlings with different susceptibility to infection with Leptosphaeria maculans. Physiol. Mol. Plant P 87, 42–50. doi: 10.1016/j.pmpp.2014.06.001
Ismail, Y., Hijri, M. (2012). Arbuscular mycorrhisation with Glomus irregulare induces expression of potato PR homologues genes in response to infection by Fusarium sambucinum. Funct. Plant Biol. 39, 236–245. doi: 10.1071/fp11218
Jain, S., Choudhary, D. K. (2014). Induced defense-related proteins in soybean (Glycine max l. Merrill) plants by Carnobacterium sp. SJ-5 upon challenge inoculation of Fusarium oxysporum. Planta 239, 1027–1040. doi: 10.1007/s00425-014-2032-3
Jung, S. C., Martinez-Medina, A., Lopez-Raez, J. A., Pozo, M. J. (2012). Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 38, 651–664. doi: 10.1007/s10886-012-0134-6
Kumar, M., Brar, A., Yadav, M., Chawade, A., Vivekanand, V., Pareek, N. (2018). Chitinases–potential candidates for enhanced plant resistance towards fungal pathogens. Agriculture-london 8, 88. doi: 10.3390/agriculture8070088
Li, B., Cui, G., Shen, G., Zhan, Z., Huang, L., Chen, J., et al. (2017). Targeted mutagenesis in the medicinal plant Salvia miltiorrhiza. Sci. Rep-uk 7, 43320. doi: 10.1038/srep43320
Li, Y., Duan, T., Nan, Z., Li, Y. (2021). Arbuscular mycorrhizal fungus alleviates alfalfa leaf spots caused by Phoma medicaginis revealed by RNA-seq analysis. J. Appl. Microbiol. 130, 547–560. doi: 10.1111/jam.14387
Liu, L., Yang, D., Xing, B., Zhang, H., Liang, Z. (2018). Salvia castanea hairy roots are more tolerant to phosphate deficiency than Salvia miltiorrhiza hairy roots based on the secondary metabolism and antioxidant defenses. Mol. J. Synthetic Chem. Nat. Prod. Chem. 23, 1132. doi: 10.3390/molecules23051132
Lobato, A., Gonçalves-Vidigal, M., Filho, P. V., Andrade, C., Kvitschal, M., Bonato, C. (2010). Relationships between leaf pigments and photosynthesis in common bean plants infected by anthracnose. New Zeal J. Crop Hort 38, 29–37. doi: 10.1080/01140671003619308
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genomics 279, 415–427. doi: 10.1007/s00438-008-0322-9
Mozzetti, C., Ferraris, L., Tamietti, G., Matta, A. (1995). Variation in enzyme activities in leaves and cell suspensions as markers of incompatibility in different phytophthora-pepper interactions. Physiol. Mol. Plant P 46, 95–107. doi: 10.1006/pmpp.1995.1008
Mustafa, G., Khong, N. G., Tisserant, B., Randoux, B., Fontaine, J., Magnin-Robert, M., et al. (2017). Defence mechanisms associated with mycorrhiza-induced resistance in wheat against powdery mildew. Funct. Plant Biol. 44, 443–454. doi: 10.1071/fp16206
Nair, A., Kolet, S. P., Thulasiram, H. V., Bhargava, S. (2015). Systemic jasmonic acid modulation in mycorrhizal tomato plants and its role in induced resistance against Alternaria alternata. Plant Biol. 17, 625–631. doi: 10.1111/plb.12277
Neeraj, Singh, K. (2011). Organic amendments to soil inoculated arbuscular mycorrhizal fungi and Pseudomonas fluorescens treatments reduce the development of root-rot disease and enhance the yield of Phaseolus vulgaris l. Eur. J. Soil Biol. 47, 288–295. doi: 10.1016/j.ejsobi.2011.07.002
Pan, S. Q. (1991). A technique for detection of chitinase, β -1,3-Glucanase, and protein patterns after a single separation using polyacrylamide gel electrophoresis or isoelectrofocusing. Phytopathology 81, 970. doi: 10.1094/phyto-81-970
Paszkowski, U. (2006). A journey through signaling in arbuscular mycorrhizal symbioses 2006. New Phytol. 172, 35–46. doi: 10.1111/j.1469-8137.2006.01840.x
Pečenková, T., Pejchar, P., Moravec, T., Drs, M., Haluška, S., Šantrůček, J., et al. (2022). Immunity functions of arabidopsis pathogenesis-related 1 are coupled but not confined to its c-terminus processing and trafficking. Mol. Plant Pathol. 23, 664–678. doi: 10.1111/mpp.13187
Phillips, J. M., Hayman, D. S. (1970). Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. T. Brit. Mycol. Soc. 55, 158–IN18. doi: 10.1016/s0007-1536(70)80110-3
Qi, D., Zou, L., Zhou, D., Zhang, M., Wei, Y., Li, K., et al. (2022). Biocontrol potential and antifungal mechanism of a novel Streptomyces sichuanensis against Fusarium oxysporum f. sp. cubense tropical race 4 in vitro and in vivo. Appl. Microbiol. Biot. 106, 1633–1649. doi: 10.1007/s00253-022-11788-3
Ritchie, R. J., Bunthawin, S. (2010). The use of pulse amplitude modulation (PAM) fluorometry to measure photosynthesis in a CAM orchid, Dendrobium spp. (D. cv. viravuth pink). Int. J. Plant Sci. 171, 575–585. doi: 10.1086/653131
Robert-Seilaniantz, A., Grant, M., Jones, J. D. G. (2011). Hormone crosstalk in plant disease and defense: More than just JASMONATE-SALICYLATE antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Serrano, I., Audran, C., Rivas, S. (2016). Chloroplasts at work during plant innate immunity. J. Exp. Bot. 67, 3845–3854. doi: 10.1093/jxb/erw088
Shi, M., Huang, F., Deng, C., Wang, Y., Kai, G. (2018). Bioactivities, biosynthesis and biotechnological production of phenolic acids in Salvia miltiorrhiza. Crit. Rev. Food Sci. 59, 1–40. doi: 10.1080/10408398.2018.1474170
Shine, M. B., Yang, J., El-Habbak, M., Nagyabhyru, P., Fu, D., Navarre, D., et al. (2016). Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 212, 627–636. doi: 10.1111/nph.14078
Song, Y., Chen, D., Lu, K., Sun, Z., Zeng, R. (2015). Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00786
Soudani, S., Poza-Carrión, C., Gómez, N. D., González-Coloma, A., Andrés, M. F., Berrocal-Lobo, M. (2022). Essential oils prime epigenetic and metabolomic changes in tomato defense against Fusarium oxysporum. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.804104
Sowik, I., Borkowska, B., Markiewicz, M. (2016). The activity of mycorrhizal symbiosis in suppressing verticillium wilt in susceptible and tolerant strawberry (Fragaria x ananassa duch.) genotypes. Appl. Soil Ecol. 101, 152–164. doi: 10.1016/j.apsoil.2016.01.021
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant immunity requires conformational charges of NPR1 via s-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Tian, S., Torres, R., Ballester, A. R., Li, B., Vilanova, L., González-Candelas, L. (2016). Molecular aspects in pathogen-fruit interactions: Virulence and resistance. Postharvest Biol. Tec 122, 11–21. doi: 10.1016/j.postharvbio.2016.04.018
Vos, C., Schouteden, N., Tuinen, D., Chatagnier, O., Elsen, A., Waele, D. D., et al. (2013). Mycorrhiza-induced resistance against the root–knot nematode meloidogyne incognita involves priming of defense gene responses in tomato. Soil Biol. Biochem. 60, 45–54. doi: 10.1016/j.soilbio.2013.01.013
Wang, R., Wang, G. L., Ning, Y. (2019). PALs: Emerging key players in broad-spectrum disease resistance. Trends Plant Sci. 24, 785–787. doi: 10.1016/j.tplants.2019.06.012
Wang, Y., Yin, Q., Qu, Y., Li, G., Hao, L. (2018). Arbuscular mycorrhiza-mediated resistance in tomato against Cladosporium fulvum-induced mould disease. J. Phytopathol. 166, 67–74. doi: 10.1111/jph.12662
Wang, Y., Zhang, S., Yin, X., Liu, J., Wu, F. (2016). Isolation and identification of arbuscular mycorrhizal fungi from mainland China. Microbiol. China 10, 2154–2165. doi: 10.13344/j.microbiol.china.150921
Xu, X., Chen, Y., Li, B., Zhang, Z., Qin, G., Chen, T., et al. (2022). Molecular mechanisms underlying multi-level defense responses of horticultural crops to fungal pathogens. Hortic. Res. 9, uhac066. doi: 10.1093/hr/uhac066
Yang, L., Miao, Z., Yang, G., Shao, A., Huang, L., Shen, Y., et al. (2013). Fusarium wilt of Salvia miltiorrhiza and its pathogenic bacteria. China J. Chin. Material Med. 38, 4040–4043. doi: 10.4268/cjcmm20132309
Yen, P. P., Pratap-Singh, A. (2021). Vacuum microwave dehydration decreases volatile concentration and soluble protein content of pea (Pisum sativum l.) protein. J. Sci. Food Agr. 101, 167–178. doi: 10.1002/jsfa.10627
Zai, X. M., Zhu, S. N., Qin, P., Wang, X. Y., Che, L., Luo, F. X. (2012). Effect of Glomus mosseae on chlorophyll content, chlorophyll fluorescence parameters, and chloroplast ultrastructure of beach plum (Prunus maritima) under NaCl stress. Photosynthetica 50, 323–328. doi: 10.1007/s11099-012-0035-5
Zhang, H., Jin, W., Zhu, X., Liu, L., He, Z., Yang, S., et al. (2016). Identification and characterization of Salvia miltiorrhiza in miRNAs in response to replanting disease. PLoS One 11, e0159905. doi: 10.1371/journal.pone.0159905
Keywords: arbuscular mycorrhizal fungi, disease resistance, Fusarium oxysporum, Fusarium wilt, Salvia miltiorrhiza
Citation: Pu C, Ge Y, Yang G, Zheng H, Guan W, Chao Z, Shen Y, Liu S, Chen M and Huang L (2022) Arbuscular mycorrhizal fungi enhance disease resistance of Salvia miltiorrhiza to Fusarium wilt. Front. Plant Sci. 13:975558. doi: 10.3389/fpls.2022.975558
Received: 22 June 2022; Accepted: 20 October 2022;
Published: 01 December 2022.
Edited by:
Linkun Wu, Fujian Agriculture and Forestry University, ChinaReviewed by:
Rupali Gupta, Volcani Center, IsraelWei Guo, Chinese Academy of Agricultural Sciences (CAAS), China
Minmin Li, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2022 Pu, Ge, Yang, Zheng, Guan, Chao, Shen, Liu, Chen and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meilan Chen, Y2hlbm1laWxhbkBucmMuYWMuY24=; Luqi Huang, aHVhbmdsdXFpMDFAMTI2LmNvbQ==
 Chunjuan Pu
Chunjuan Pu Yang Ge
Yang Ge Guang Yang
Guang Yang Han Zheng
Han Zheng Wei Guan
Wei Guan Zhi Chao
Zhi Chao Ye Shen2
Ye Shen2 Meilan Chen
Meilan Chen Luqi Huang
Luqi Huang