- Jiangsu Key Laboratory for the Research and Utilization of Plant Resources, Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (Nanjing Botanical Garden Mem. Sun Yat-Sen), Nanjing, China
As one of the biggest plant specific transcription factor (TF) families, basic helix–loop–helix (bHLH) protein, plays significant roles in plant growth, development, and abiotic stress responses. However, there has been minimal research about the effects of methyl jasmonate (MeJA) treatment on the bHLH gene family in Lycoris radiata (L’Her.) Herb. In this study, based on transcriptome sequencing data, 50 putative L. radiata bHLH (LrbHLH) genes with complete open reading frames (ORFs), which were divided into 20 bHLH subfamilies, were identified. The protein motif analyses showed that a total of 10 conserved motifs were found in LrbHLH proteins and motif 1 and motif 2 were the most highly conserved motifs. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of LrbHLH genes revealed their involvement in regulation of plant growth, jasmonic acid (JA) mediated signaling pathway, photoperiodism, and flowering. Furthermore, subcellular localization revealed that most LrbHLHs were located in the nucleus. Expression pattern analysis of LrbHLH genes in different tissues and at flower developmental stages suggested that their expression differed across lineages and might be important for plant growth and organ development in Lycoris. In addition, all LrbHLH genes exhibited specific spatial and temporal expression patterns under MeJA treatment. Moreover, protein-protein interaction (PPI) network analysis and yeast two-hybrid assay showed that numerous LrbHLHs could interact with jasmonate ZIM (zinc-finger inflorescence meristem) domain (JAZ) proteins. This research provides a theoretical basis for further investigation of LrbHLHs to find their functions and insights for their regulatory mechanisms involved in JA signaling pathway.
Introduction
Lycoris Herbert belongs to the family Amaryllidaceae, consisting of almost 20 species native to eastern Asia moist warm temperate woodlands (Shi et al., 2006). Plants in the genus Lycoris have been utilized as traditional medicine preparation, and more than 110 potent structurally distinct Amaryllidaceae alkaloids were isolated or identified for extensive pharmacological and phytochemical investigations (Cahlíková et al., 2020). For example, according to Compendium of Materia Medica, Lycoris plants are described as potent antidotes to poison, effective agents to alleviate pain and relieve inflammation, and diuretic drugs (Jin, 2011). Amaryllidaceae alkaloids from L. radiata bulbs were traditionally used for treating sore carbuncle, neurodegenerative diseases, poliomyelitis, suppurative wounds and ulcers (Feng et al., 2011; Chen et al., 2016; Shen et al., 2019).
Many genes potentially participated in Amaryllidaceae alkaloid and anthocyanin biosynthesis as well as sucrose degradation have been identified in Lycoris plants through transcriptome sequencing (Wang et al., 2017; Park et al., 2019; Yue et al., 2019; Li et al., 2020a; Wang et al., 2021). For example, genes involved in Amaryllidaceae alkaloid biosynthesis including phenylalanine ammonia lyase (PAL, Li et al., 2018), cinnamate 4-hydroxylase (C4H, Li et al., 2018), tyrosine decarboxylase (TYDC, Wang et al., 2019b); norbelladine synthase (NBS, Li et al., 2020a); norbelladine 4’-O-methyltransferase (N4OMT, Li et al., 2019a), and cytochrome P450 monooxygenase 96T1 (CYP96T1, Li et al., 2020a) have been characterized from Lycoris plants. Besides, key functional genes involved in anthocyanin metabolism during flower development in L. radiata have also been characterized (Wang et al., 2021). Moreover, a relatively active cell wall invertase (CWIN)-catalyzed apoplasmic sucrose cleavage pattern at the competence stage have been found to increase L. sprengeri bulblet regeneration (Ren et al., 2021), and the mode of sucrose degradation could be identified as a metabolic marker for early vegetative propagation in bulbs of Lycoris (Ren et al., 2022). Considering transcription factors (TF) are important gene expression switches that activate or repress the expression of specific target genes by interacting with cis-elements in the gene promoter region, regulating various biological processes including plant development, growth, biosynthesis of secondary metabolites, and response to stresses (Sun et al., 2018), it is necessary to investigate whether and how TFs participate the biological processes in Lycoris plants.
The bHLH TF family is the second largest class of TFs widely distributed in plants, animals, and microorganisms (Sun et al., 2018). The bHLH domain comprises approximately 60 amino acids with a basic amino acid region and a helix–loop–helix region (Toledo-Ortiz et al., 2003). The basic region is located on the N-terminal side of the bHLH domain and comprises around 15-20 amino acids (Feller et al., 2011; Sun et al., 2018). It forms a DNA-binding domain, which regulates G-box and E-box DNA binding activity on genes. In contrast, the HLH region contains approximately 40-50 amino acids in the C-terminal domain with two alpha helixes separated by a loop of variable length (Murre et al., 1989). It promotes the interaction with other bHLH proteins and the formation of homodimers and heterodimeric among bHLH TF. The basic region of plant bHLH domains comprises a highly conserved His5-Glu9-Arg13 sequence that binds to the E-box element (5′-CANNTG-3′) (Sun et al., 2018). bHLH proteins have been clarified to 15–26 subfamilies in numerous plants, including rice (Carretero-Paulet et al., 2010), Arabidopsis (Carretero-Paulet et al., 2010), Chinese cabbage (Song et al., 2014), tomato (Sun et al., 2015), Brachypodium distachyon (Niu et al., 2017), peanut (Gao et al., 2017), cotton (Lu et al., 2018), grape (Wang et al., 2018), maize (Zhang et al., 2018a), and jujube (Li et al., 2019b), Cucumis sativus L. (Li et al., 2020b), Capsicum (Liu et al., 2021).
In plants, bHLH TFs influence the development of shoot branches (Komatsu et al., 2001), microspores (Sorensen et al., 2003), fruits and flowers (Gremski et al., 2007), trichomes (Morohashi et al., 2007), stomata (Pillitteri et al., 2007), and roots (Menand et al., 2007). The bHLH TFs are also implicated in various signal transduction and anabolic pathways, including anthocyanin synthesis (Sakamoto et al., 2001), tryptophan production (Smolen et al., 2002), light signal transduction (Castillon et al., 2007), and gibberellin production (Arnaud et al., 2010). For example, the Arabidopsis bHLH protein AtPIF3 (PHYTOCHROME INTERACTING FACTOR3) and AtPIF4 (PHYTOCHROME INTERACTING FACTOR4) interact with phytochrome to control the expression of genes regulating light response (Toledo-Ortiz et al., 2003). In flavonoid biosynthesis, bHLH proteins serve as R2R3-MYB and WD40 cofactors in the synthesis of MYB-bHLH-WD40 (MBW) complex (Xie et al., 2012). TRANSPARENT TESTA8 (TT8, AtbHLH42) regulates anthocyanin and procyanidin synthesis in vegetative organs by forming MBW (TT2-TT8-TTG1) complexes (Baudry et al., 2004). The bHLH proteins also affect stress responses, such as drought response (Sun et al., 2018), heat response (Koini et al., 2009), salt response (Verma et al., 2020), and cold response (Feng et al., 2012). The expression level of AtbHLH92 is upregulated under salt, drought, and cold stresses (Jiang et al., 2009). In rice, OsbHLH148 is associated with jasmonic acid (JA) signaling and participates in both drought stress and trauma response (Seo et al., 2011). OsRERJ1 bHLH TF physically associates with OsMYC2 to participate in the transcriptional induction of JA-mediated stress responsive genes thus defensing against herbivory and bacterial infection (Valea et al., 2021). In apple, MdbHLH3 expression was induced at low temperature and responsible for anthocyanin accumulation and fruit coloring (Xie et al., 2012).
In this study, 50 LrbHLH genes were identified from L. radiata transcriptome data (Wang et al., 2021), and their motif pattern and phylogenetic relationship between Arabidopsis and L. radiata were analyzed. Subcellular localization analysis revealed that LrbHLH proteins were mainly localized in the nucleus. The expression levels of LrbHLH genes changed in different tissues and under methyl jasmonate (MeJA) treatment. The probable protein-protein interaction (PPI) of LrbHLHs were predicted and confirmed using yeast two-hybrid (Y2H) system. Our results provide important insights into the bHLH genes in L. radiata and lay a foundation for further investigation of bHLH gene function in the biological pathway.
Materials and methods
Plant materials and treatments
Lycoris radiata (L’Her.) Herb. was planted in Experimental Plantation of the Institute of Botany, Jiangsu Province and Chinese Academy of Sciences, Nanjing, China. The seedlings with the same or similar sizes (2.8– 3.2 cm) in diameter were transferred into plastic pots with a mixture of vermiculite and soil (1:1, v/v) and maintained in a plant growth chamber under the following conditions: 16 h light/8 h dark cycle at 25°C/22°C, and 120 μmol m-2 s-1 irradiation). After one week of maintenance, the seedlings were subjected to 100 μmol L−1 MeJA for 0 h, 6 h, 12 h, 24 h, and 36 h, and roots were sampled for gene expression analysis. Each treatment was replicated three times. Tissue-specific transcription profiles of 50 LrbHLH genes were explored in the petals, flower-stalks, gynoeciums, stamens, leaves, seeds, roots, and bulbs of these plants. All samples were immediately frozen in liquid nitrogen and stored at -80°C.
Transcriptome-wide identification and expression profiling of LrbHLH genes
The L. radiata transcriptome database during four flower development stage with 87,584 unigenes was used for potential LrbHLH searching (Wang et al., 2021). 162 AtbHLH proteins downloaded from TAIR (Arabidopsis Information Resource database, https://www.arabidopsis.org/) were utilized to determine sequence homology with L. radiata transcripts from the database by basic local alignment (BLASTn). The hidden Markov model (HMM) profile of the bHLH domain (protein family ID: PF00010) obtained from the Pfam protein family database (http://pfam.xfam.org; Mistry et al., 2021) was used to search candidate LrbHLH genes. Then, we verified the bHLH domain in the predicted LrbHLH transcription factors utilizing NCBI Batch CD-Search Tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) with default parameters. This characteristic was deemed to have a high-confidence association with the conserved domain. Sequences predicted as specific hits were retained for further analysis (Supplementary Table 1). Furthermore, PFAM and SMART (http://smart.embl-heidelberg.de/; Letunic et al., 2021) databases were used for verifying the bHLH domain in all candidate protein sequences. Finally, the ExPASy website (https://web.expasy.org/protparam; Artimo et al., 2012) was utilized to determine the full length of amino acid sequences, isoelectric points (PI), molecular sizes (MW), and protein instability index.
Phylogenetic tree and protein motif analyses of LrbHLH proteins
The phylogenetic tree of bHLHs from L. radiata and A. thaliana was constructed with MEGA7 using the neighbor-joining method (https://www.megasoftware.net/; Kumar et al., 2016a). LrbHLH proteins were then classified according to their phylogenetic relationships with AtbHLH proteins. The online tool MEME (Multiple EM for Motif Elicitation, version 5.1.1) was utilized to search for the conserved motifs of LrbHLH proteins (https://meme-suite.org/meme/tools/meme; Bailey et al., 2015), with motif number and width of 12-50 for each gene. Motifs were also searched in protein databases using the SMART program (http://smart.embl.de/).
The GO and KEGG annotation of LrbHLHs
Gene Ontology (GO) functional annotations were conducted using KOBAS (Bu et al., 2021). The top 20 functional terms for credibility in the three categories (biological processes, cellular components, and molecular function) were selected for visualization. KEGG annotations were completed utilizing KASS (Kanehisa et al., 2017).
Expression profiles analysis and quantitative real-time PCR (qRT-PCR) analysis of LrbHLH genes
RNA-seq data for the LrbHLH genes were obtained from previous studies on gene expression in different flower developmental stages (Wang et al., 2021) and MeJA treatment (Wang et al., 2017). LrbHLH gene expression profiles were evaluated utilizing the values of FPKM. TBtools (Chen et al., 2020) software was utilized to generate LrbHLH expression heatmaps. Total RNA was extracted with RNAprep Pure Plant Kit (TIANGEN, Beijing, China) according to the manufacturer’s protocol. cDNA was synthesized by using PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa Bio, Dalian, China) and utilized for qRT-PCR assays. Relative expression levels of genes were analyzed using qRT-PCR with SYBR® Premix Ex Taq™ II (Takara Bio, Dalian, China) on a Bio-Rad iQ5 Real-Time PCR System (Bio-Rad, CA, USA) in 15 μL reactions. Each reaction contained 7.5 μL 2×TransStart® Top Green qPCR SuperMix, 5.9 μL ddH2O, 1μL cDNA, and 0.6 μL of 10 μM forward and reverse primers. The RT-qPCR protocol included the following: the PCR reaction conditions were at 95°C for 5 min; denaturation 5 s at 95°C; 60°C for 30 s; 40 cycles. The LrTIP41 gene was utilized as an endogenous control to normalize relative expression levels based on the 2-ΔΔCt method (Schmittgen and Livak, 2008). The specific primer sequences used in this research are listed in Supplementary Table 1.
Gene cloning and construction of expression vectors
Cloning of LrbHLHs was based on putative ORFs of unigenes from the RNA-seq database. Primers (Supplementary Table 1) were synthesized for ORF sequence amplification using Tks GflexTM DNA Polymerase (Takara Bio, Dalian, China) from L. radiata petal cDNA. Reaction conditions were: 5 min of 95°C, 35 cycles for 30 s at 94°C, 30 s at 60°C, 1 min at 72°C, with extension at 72°C for 10 min. PCR products were cloned into pMD19-T simple vectors (Takara Bio, Dalian, China). Afterward, those T-vectors were transferred into DH5α competent cells (Takara Bio, Dalian, China) for colony PCR amplification and then validated by sequencing.
Subcellular localization analysis of LrbHLH proteins
Subcellular localization of LrbHLH proteins was predicted using WoLF PSORT (https://wolfpsort.hgc.jp; Horton et al., 2021), ProtComp 9.0 (http://linux1.softberry.com), and Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/). The coding region of each LrbHLH gene was PCR-amplified with specific primers (Supplementary Table 1) and inserted into the plant expression vector pBinGFP4 (digested with KpnI/BamHI) with ClonExpress® II One Step Cloning Kit (Vazyme, Nanjing, China). The recombinant vectors were then transformed into Agrobacterium tumefaciens EHA105 competent cells. The A. tumefaciens strains containing various constructs were cultivated, harvested, followed by resuspension in an invasive solution (10 mM MES, 0.2 mM Acetosyringone, and 10 mM MgCl2) with a final OD600 value of 0.6. Forty-day-old Nicotiana benthamiana plants were used for infiltration. After infiltration, plants were grown at 22°C in the dark and then transferred to normal growth conditions (25°C/16 h light and 22°C/8 h dark cycle) for three days. GFP fluorescent signals in the epidermal cells of N. benthamiana leaves were observed under a confocal microscope (Zeiss LSM900, Jena, Germany).
PPI network prediction of LrbHLH proteins
Potential PPI network was predicted using STRING server online database based on A. thaliana homologous proteins (https://cn.string-db.org; Szklarczyk et al., 2021). The protein sequences of 50 LrbHLHs and 7 LrJAZs were uploaded into the server selecting A. thaliana as the comparative organism. The LrbHLHs genes interaction network was constructed after blasting with the highest bitscore.
Analysis of LrbHLHs interactions utilizing yeast two-hybrid (Y2H) assay
The full-length coding regions (CDs) of LrbHLH28, LrbHLH31, LrbHLH48 and LrJAZs (Supplementary Table 4) were cloned into pGADT7 (AD) or pGBKT7 (BD) vector, using the ClonExpress® II One Step Cloning Kit (Vazyme, Nanjing, China) respectively. The auto-activation potentiality and interaction assay of LrbHLH and LrJAZ proteins were tested according to the Matchmaker Gold Y2H System (Takara Bio, Dalian, China). The positive clones were then sequentially selected on SD/-Ade/-His/-Leu/-Trp medium with 5-bromo-4-chloro-3-indoxyl α-D-galactoside (X-α-gal) and Aureobasidin A (AbA) to test the interactions between LrbHLHs and LrJAZs.
Statistical analysis
All experiments were independently duplicated at least three times. The results were represented as mean ± SD of biological triplicates. The student’s t-test was utilized for data significance analysis at p < 0.05 or p < 0.01 level.
Results
Identification and characterization of LrbHLH proteins in L. radiata
We used HMMER 3.0 and BLASTP to predict putative LrbHLH protein sequences in the L. radiata transcriptome database with an E-value threshold of < 1 e-5 (Wang et al., 2021). All candidate sequences were confirmed in NCBI and with SMART to further identify conserved complete bHLH domains. In total, 50 LrbHLH proteins (namely LrbHLH1-LrbHLH50) were identified. The CDs of LrbHLH genes varied from 258 bp to 1,662 bp. LrbHLH proteins ranged from 86 to 554 amino acids in size with molecular weight of 9.75 kDa to 74.73 kDa and isoelectric point of 4.74 to 9.30. Subcellular localization prediction showed that most LrbHLH proteins was localized in the nucleus, and few LrbHLH proteins located in the cytoplasm, chloroplast, or mitochondria (Supplementary Table 2a).
Phylogenetic analysis and classification of LrbHLH proteins
Phylogenetic tree was constructed with LrbHLHs and A. thaliana bHLH (AtbHLH) protein sequences to investigate their evolutionary relationships. Fifty LrbHLH proteins were clustered into 20 groups, which were designated as group Ia, II, IIIa+c, IIIb, IIId+e, IIIf, IVa, IV(b+c), Va, VI, VII, VIIIa, VIIIb, IX, X, XI, XII, XIII, XIV, and XV (Figure 1). Among them, group XII was the largest group containing 13 LrbHLH proteins. In contrast, only one LrbHLH was found in group II, IIIa+c, IVa, VI, VIIIa, and XI. None of LrbHLH protein was found in group orphans, VIII, Ib, Vb, and VIIIc.
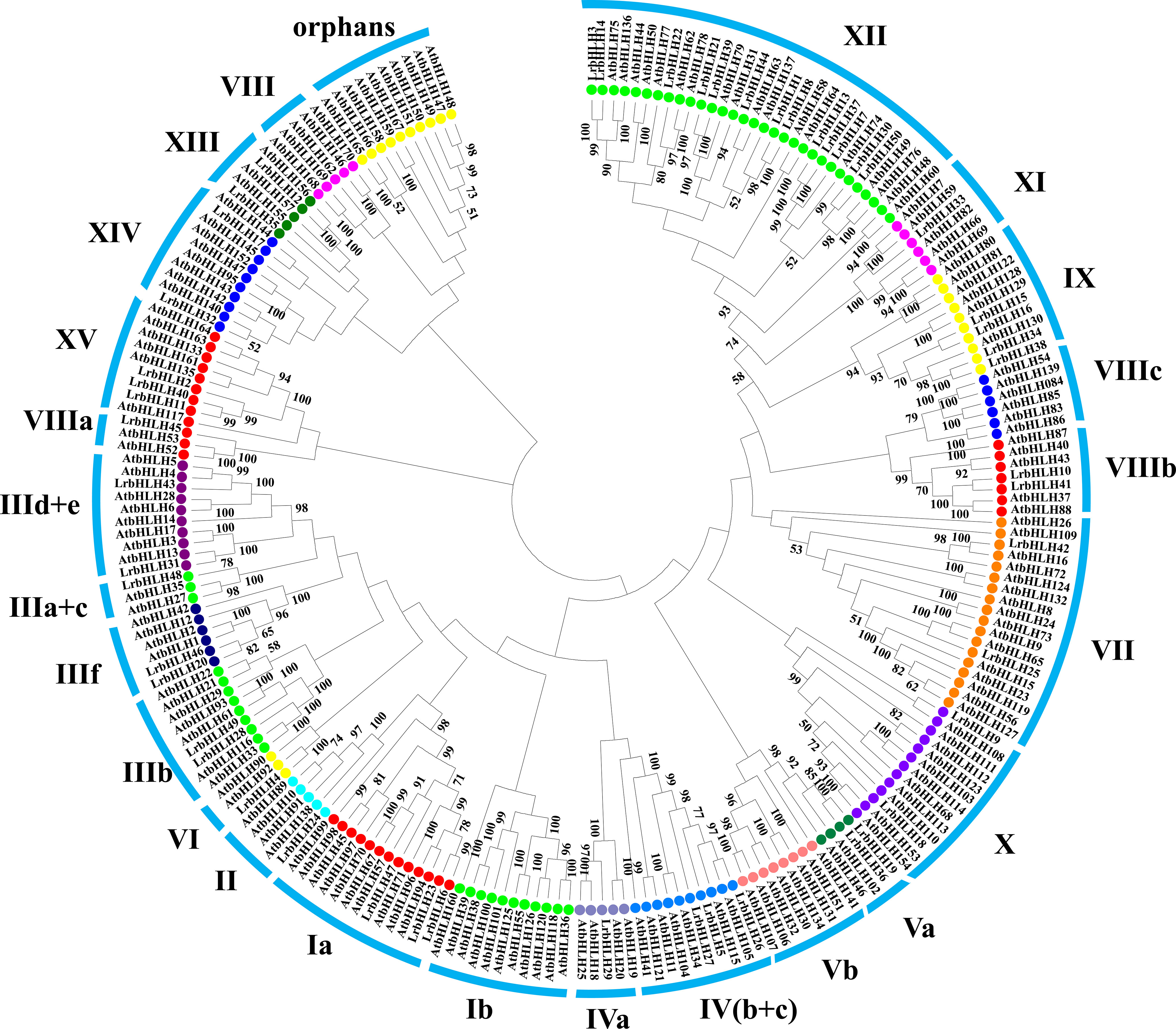
Figure 1 The phylogenetic analysis of L. radiata and Arabidopsis bHLH proteins. The neighbor-joining method was utilized to generate the phylogenetic tree relying on the bHLH domains alignment. These numbers are computed using 1000 bootstrap replicates to verify reliability. The replicate trees percentages over 50 are presented on branches. The tree shows the 25 subfamilies marked with blue font on a colored background.
Multiple sequence alignment, conserved motif, and bHLH domain analysis of LrbHLH proteins
The conserved motifs of LrbHLH proteins were further predicted, and a total of 10 conserved motifs were found (Figures 2, 3A). As shown in Figure 2, motif 1 was the most highly conserved motif present in almost all the LrbHLH proteins except LrbHLH17 and LrbHLH50. Motif 2 was the second most highly conserved motif. In addition, analysis of conserved amino acid residues in the bHLH domain showed that there are two helix regions, one basic region, and one loop region in LrbHLH proteins (Figure 3B and Supplementary Figure 1). Motif 1 was closer to the N-terminal and comprised with the basic region and the first helix region, whereas motif 2 mainly existed in the second helix region (Figure 3B). Moreover, motifs in LrbHLH proteins belonging to the same group were the same or structurally similar. For instance, motif 1, motif 2, and motif 3 were identified in all 11 LrbHLH proteins belonging to group XII. Motif 1, motif 2, motif 3, motif 5, and motif 10 were recognized in 4 LrbHLH proteins of group IX (Figure 2).
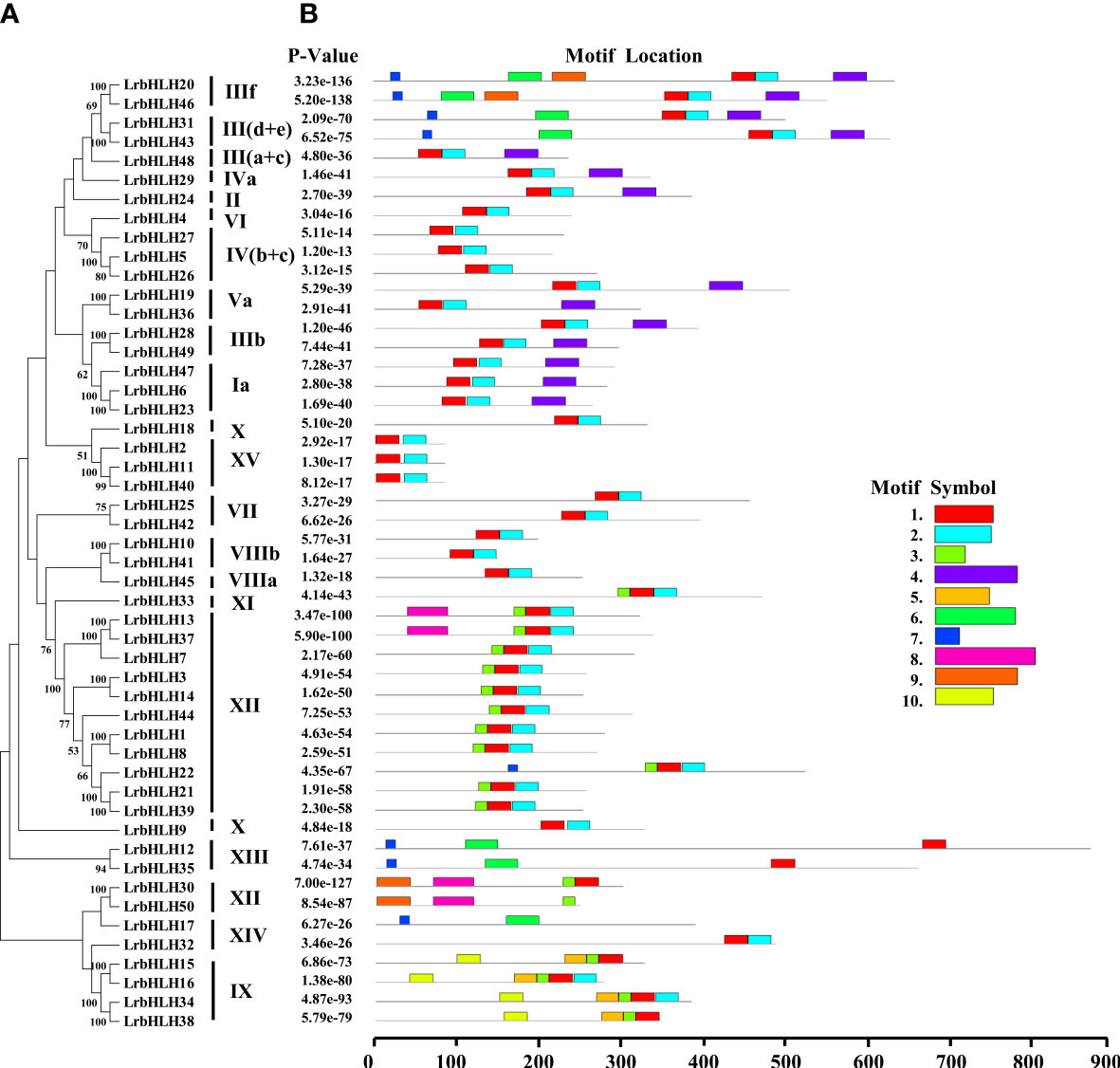
Figure 2 The phylogenetic relationships and conserved motifs analysis of LrbHLH proteins. (A) Neighbor-joining LrbHLHs phylogenetic tree (bootstrap values for 1000 replicates); (B) Conserved motifs distribution in LrbHLH proteins. Different motifs are represented with different colored boxes. The motif length is represented by the box length.
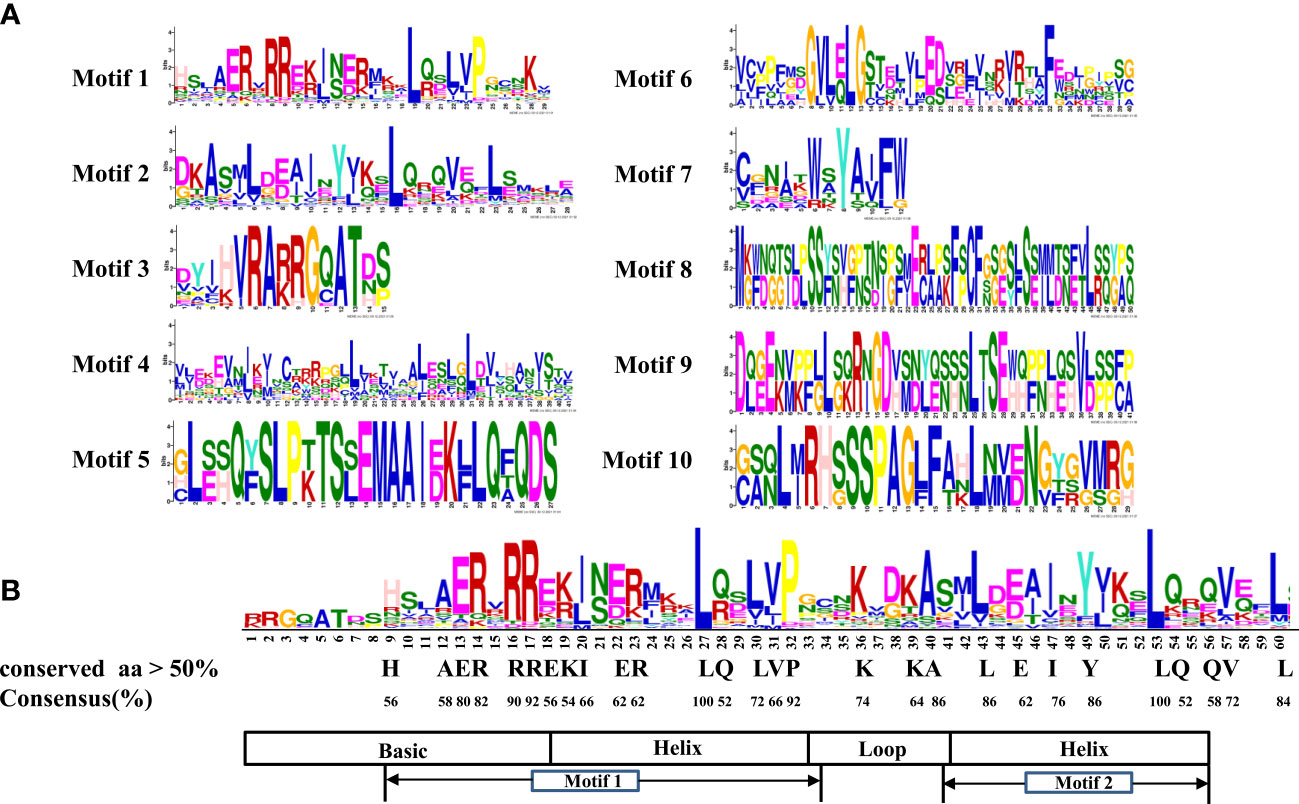
Figure 3 The conserved motifs analysis of LrbHLH proteins. (A) Conservation and diversity of the motifs in LrbHLH proteins. The schematic representation of ten motifs in LrbHLH proteins is elucidated by MEME; (B) LrbHLH proteins share a conserved bHLH domain. The amino acid height represents the sequence conservation. More than 50% amino acid conservation was presented with the capital letters, and the conserved amino acid percentages among bHLH domains are presented with numbers.
On the other hand, multiple sequence alignment of 50 LrbHLHs revealed that 28 amino acid residues were considerably conserved with a consensus ratio > 50%, and 11 of them were conserved with a consensus ratio > 75% (Figure 3B and Supplementary Figure 1). These 28 conserved residues were distributed among the basic regions (His-9, Ala-12, Glu-13, Arg-14, Arg-16, and Arg-17), the first helix region (Glu-18, Lys-19, Ile-20, Glu-22, Arg-23, Gln-28, Leu-27, Leu-30, Val-31, and Pro-32), the loop region (Lys-36 and Lys-39), and the second helix region (Ala-40, Leu-43, Glu-45, Ile-47, Tyr-49, Leu-53, Gln-54, and Gln-56). The DNA binding bHLH proteins were further categorized according to the residues Glu-13 and Arg-17 presence or absence in the basic regions with putative E-box and non-E-box binding proteins. Among LrbHLHs, 28 proteins containing His-9, Glu-13, Arg-14, Arg-16, and Arg-17 residues were confirmed to binding the G-box motif (CACGTG), whereas 12 LrbHLH proteins containing Glu-13 and Arg-17 residues were found to recognize the types of E-boxes (CANNTG) and were defined as non-G-box binders (Figure 3B and Supplementary Figure 1).
The GO and KEGG enrichment between LrbHLH genes
LrbHLH proteins were annotated to three main GO categories, including 36 biological process terms, 6 molecular function terms, and 4 cellular component terms. The top 20 GO terms of level two were visualized (Figure 4). In biological process terms, 36 LrbHLH proteins were implicated in the term ‘regulation of transcription, DNA-templated’. In cellular component terms, 49 LrbHLH proteins were components of the term ‘nucleus’, but few of them were components of the term ‘cytosol and cytoplasm’. In molecular function terms, 49 LrbHLH proteins and 36 LrbHLH proteins were categorized as exhibiting ‘protein dimerization activity’ and ‘DNA-binding transcription factor activity’, respectively (Figure 4).
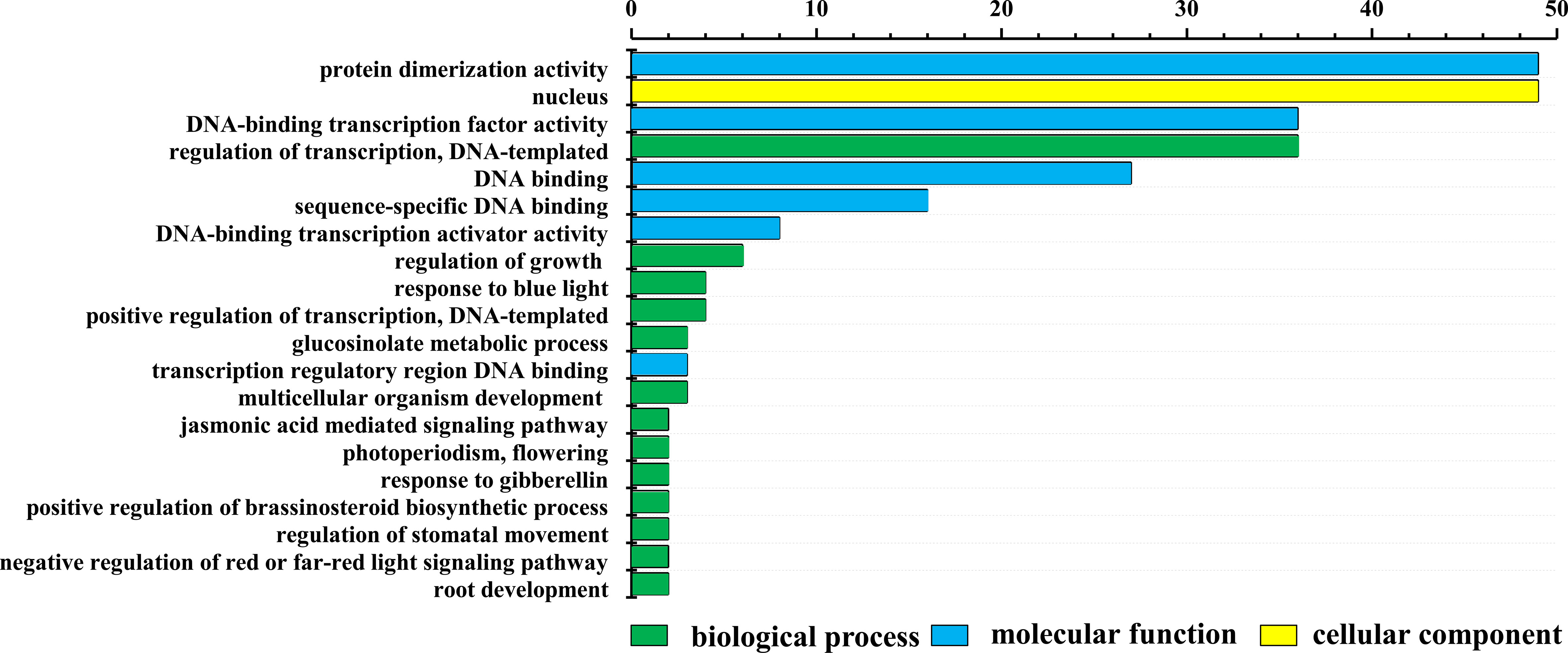
Figure 4 Gene ontology (GO) enrichment analysis of LrbHLH proteins. The top 20 GO terms of level 2 in biological process, cellular component, and molecular function were visualized. The X and Y axes represent the enriched protein numbers and the information on GO terms, respectively.
The KEGG pathway analysis showed that LrbHLH25, LrbHLH32, and LrbHLH43 were annotated to three KEGG homologous proteins and participated in three pathways. LrbHLH25 was homologous to PIF4 (KO: K16189) and implicated in plant hormone signal transduction pathway (ko04075). The LrbHLH32 was a homolog of PIF3 (KO: K12126) and participated in circadian rhythm-plant pathway (ko04712) and plant hormone signal transduction pathway (ko04075). LrbHLH43 was annotated to MYC2 (KO: K13422) that participated in plant hormone signal transduction pathway (ko04075) and the MAPK signaling pathway (ko04016). Therefore, PIF3, PIF4 and MYC2 are all involved in plant hormone signal transduction pathway. PIF3 and MYC2 mainly transmit gibberellin (GA) signal pathway and JA signal pathway, respectively.
Subcellular localization of LrbHLH proteins
Most LrbHLH proteins were predicted to be localized in the nucleus, whereas few of LrbHLHs were located in the cytoplasm and/or other organelles (Supplementary Table 2a). We then examined the subcellular localization of some LrbHLH proteins in N. benthamiana epidermal cells. As expected, LrbHLH4, LrbHLH7, LrbHLH14, LrbHLH18, LrbHLH19, LrbHLH28, LrbHLH36, LrbHLH37, LrbHLH43, LrbHLH47, and LrbHLH48 were localized in the nucleus, whereas LrbHLH22 were located in both the nucleus and membrane, and LrbHLH25 was present in the nucleus and cytoplasm (Figure 5).
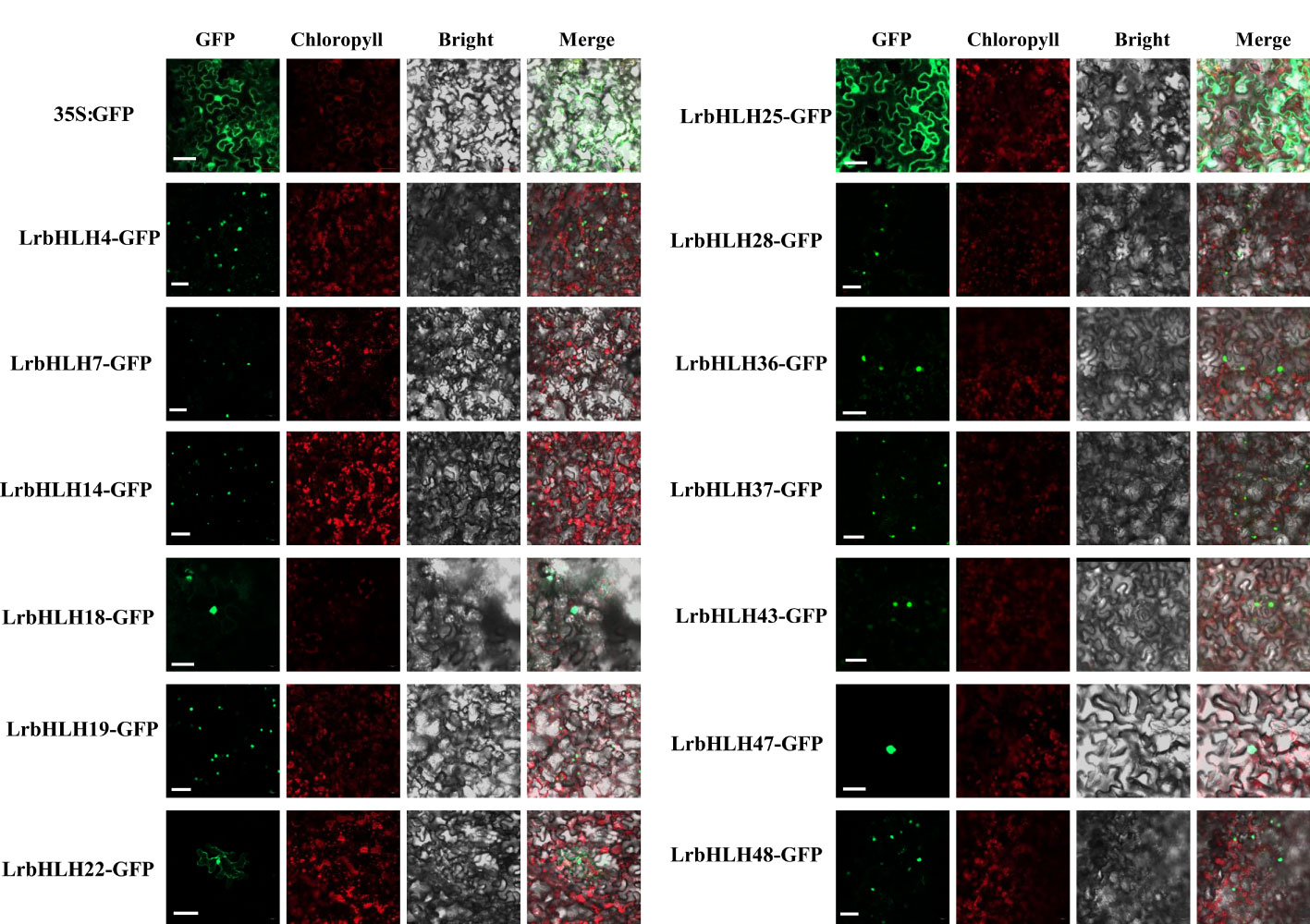
Figure 5 Subcellular localization of 15 LrbHLHs with GFP as a control, which was transiently expressed in N. benthamiana leaves. The photographs were taken using the green channel (GFP fluorescence), red channel (Chlorophyll represents chlorophyll auto fluorescence), bright channel, and their combination under a confocal microscope. Scale bar = 10 µm.
Expression patterns analysis of LrbHLH genes in different tissues and at different flower developmental stages
As the transcriptome analysis of different tissues (root, leaf, and bulb) has been revealed in L. longituba (Li et al., 2020a), we then searched the orthologous genes of LrbHLH by using local blast within L. longituba transcriptome data. 50 orthologous genes of LrbHLH were found in L. longituba (Supplementary Table 2B). As indicated in Supplementary Figure 2, some LrbHLH genes were exhibited differential expression in the three tissues, whereas other LrbHLH genes showed similar expression patterns in diverse tissues. For example, four LrbHLHs including LrbHLH14, LrbHLH25, LrbHLH32, and LrbHLH44 were relatively highly expressed in leaves, whereas LrbHLH5, LrbHLH12, and LrbHLH17 were preferentially expressed in roots. LrbHLH6, LrbHLH39 and LrbHLH49 had the highest relative expression levels in bulb. To further elucidate the biological function of LrbHLH proteins, qRT-PCR was utilized to determine the spatial specificity expression pattern of 50 LrbHLH genes in eight L. radiata organs. As indicated in Figure 6A, some LrbHLH genes were exhibited differential expression in the eight tissues, whereas other LrbHLH genes showed similar expression patterns in diverse tissues, which could be attributed to the functional differentiation of LrbHLH genes during plant development. For example, eight LrbHLHs (i.e., LrbHLH2, LrbHLH4, LrbHLH15, LrbHLH16, LrbHLH25, LrbHLH26, LrbHLH27, and LrbHLH47) were relatively highly expressed in leaves. LrbHLH1, LrbHLH20, LrbHLH41 and LrbHLH46 were preferentially expressed in petals. LrbHLH3, LrbHLH5 and LrbHLH8 showed highly expression levels in gynoecium, whereas LrbHLH24 and LrbHLH45 had relatively high expression levels in stamen. In addition, six LrbHLHs (i.e., LrbHLH23, LrbHLH33, LrbHLH34, LrbHLH35, LrbHLH38 and LrbHLH42) were highly expressed in flower-stalk tissue. In particular, LrbHLH6, LrbHLH7, LrbHLH14 and LrbHLH37 were predominantly expressed in root. LrbHLH9 and LrbHLH11 were relatively highly abundant in seed. LrbHLH12, LrbHLH13, LrbHLH17, LrbHLH18, LrbHLH36, LrbHLH40 and LrbHLH43 had the highest relative expression levels in bulb. Conversely, some genes were not expressed specifically. Furthermore, LrbHLH29, LrbHLH49 and LrbHLH50 showed low expression in all tissues. These results suggested that LrbHLHs might play the same role in the growth and development of L. radiata. Additionally, most LrbHLHs exhibited diverse tissue-specific expression patterns, implying their numerous functions in diverse organs.
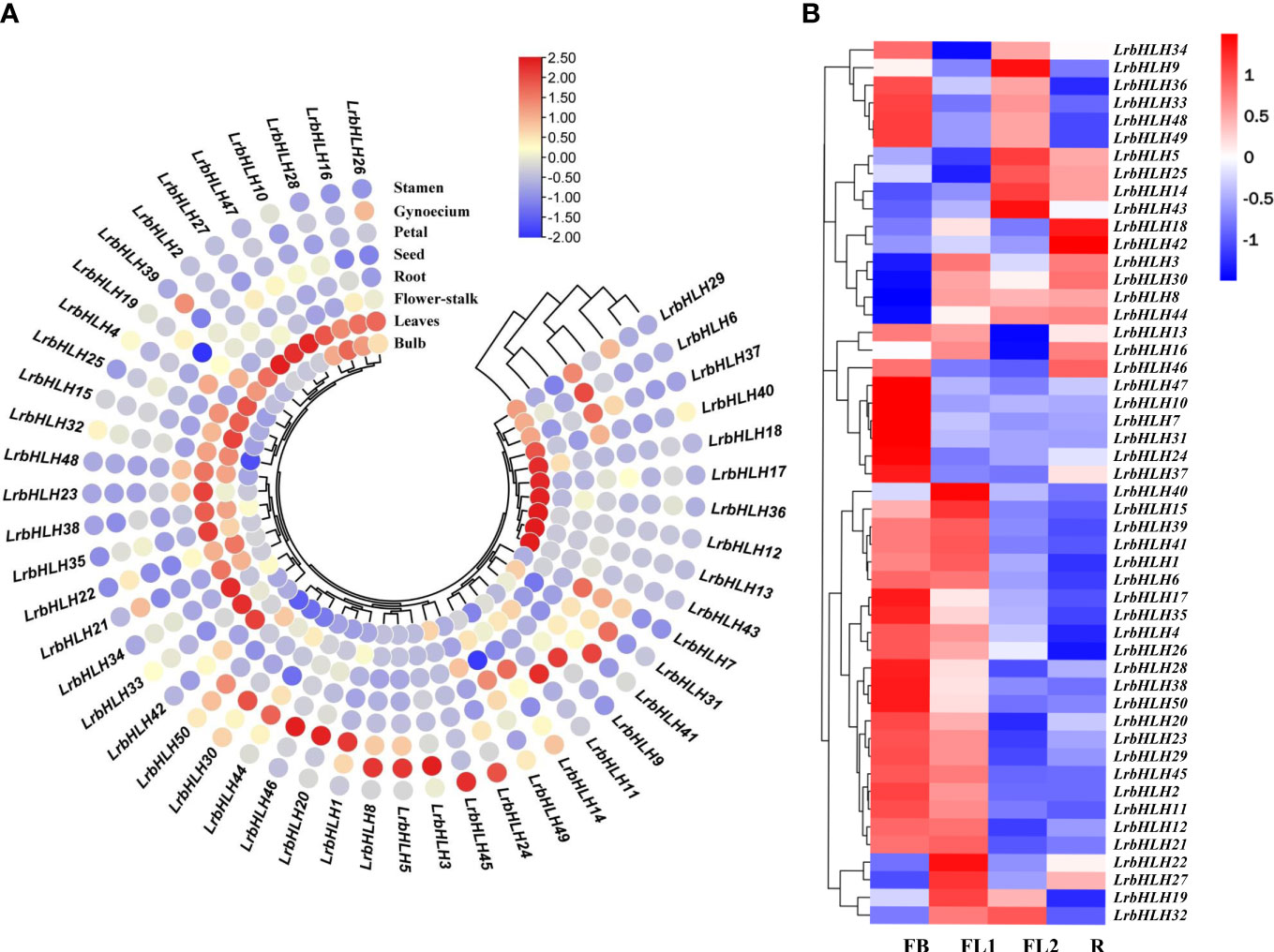
Figure 6 LrbHLH gene expression patterns in various L. radiata tissues. (A) Expression profile heatmap with hierarchal clustering of LrbHLHs in different tissues of L. radiata; (B) Expression profile heatmap with hierarchal clustering of LrbHLHs at different flower developmental stages of L. radiata. The relative expression for each genes is depicted by color intensity in each field. Higher values are represented by red whereas lower values are represented by blue. FB, flower-bud differentiation stage; FL1, partially opening flower stage; FL2, fully opened flower stage; R, senescent flower stage.
The expression of LrbHLHs was further observed at four flower development stages including flower-bud differentiation (FB) stage, partially opening flower (FL1) stage, fully opened flower (FL2) stage, and senescent flower (R) stage, based on our previously published RNA-seq data (Figure 6B, Wang et al., 2021). Over half of LrbHLH genes was highly expressed at FB stage and then markedly decreased at R stage. In addition, the expression of LrbHLH8 and LrbHLH44 exhibited a gradual increase during flower development. Remarkably, the expression of LrbHLH5, LrbHLH14, LrbHLH25 and LrbHLH43 genes exhibited opposite trends in the flowering developmental stages, indicating they may perform diverse functions. However, the expression of LrbHLH18 and LrbHLH42 were relatively higher at the late stages of flower development.
Expression patterns of LrbHLHs in response to MeJA treatment
Previous study of L. aurea transcriptome sequencing has revealed that MeJA treatment could induce the expression of LabHLH genes (Wang et al., 2017). Thus, the orthologous genes of LrbHLH in L. aurea transcriptome data were also searched. All the 50 LrbHLH genes were found to have orthologous transcripts in L. aurea transcriptome (Supplementary Table 2C). Among them, 22 homologous LrbHLHs were up-regulated, while 10 homologous LrbHLH genes were suppressed with MeJA treatment for 6 h (Supplementary Figure 3). In addition, homologous genes of LrbHLH6, LrbHLH23, LrbHLH34, LrbHLH35, and LrbHLH38 exhibited no expression changes after MeJA treatment. Furthermore, 9 LrbHLH genes including LrbHLH4, LrbHLH13, LrbHLH24, LrbHLH31, LrbHLH37, LrbHLH42, LrbHLH43, LrbHLH48 and LrbHLH49 were subjected to qRT-PCR for confirming their expression profiles under MeJA with different treatment time in L. radiata roots. Consequently, these LrbHLHs were significantly up-regulated, which were similar to their expression trends in RNA-Seq data (Figure 7). These findings revealed that LrbHLHs displayed different expression patterns in response to MeJA treatment and potentially participate in biological processes via JA signaling pathways.
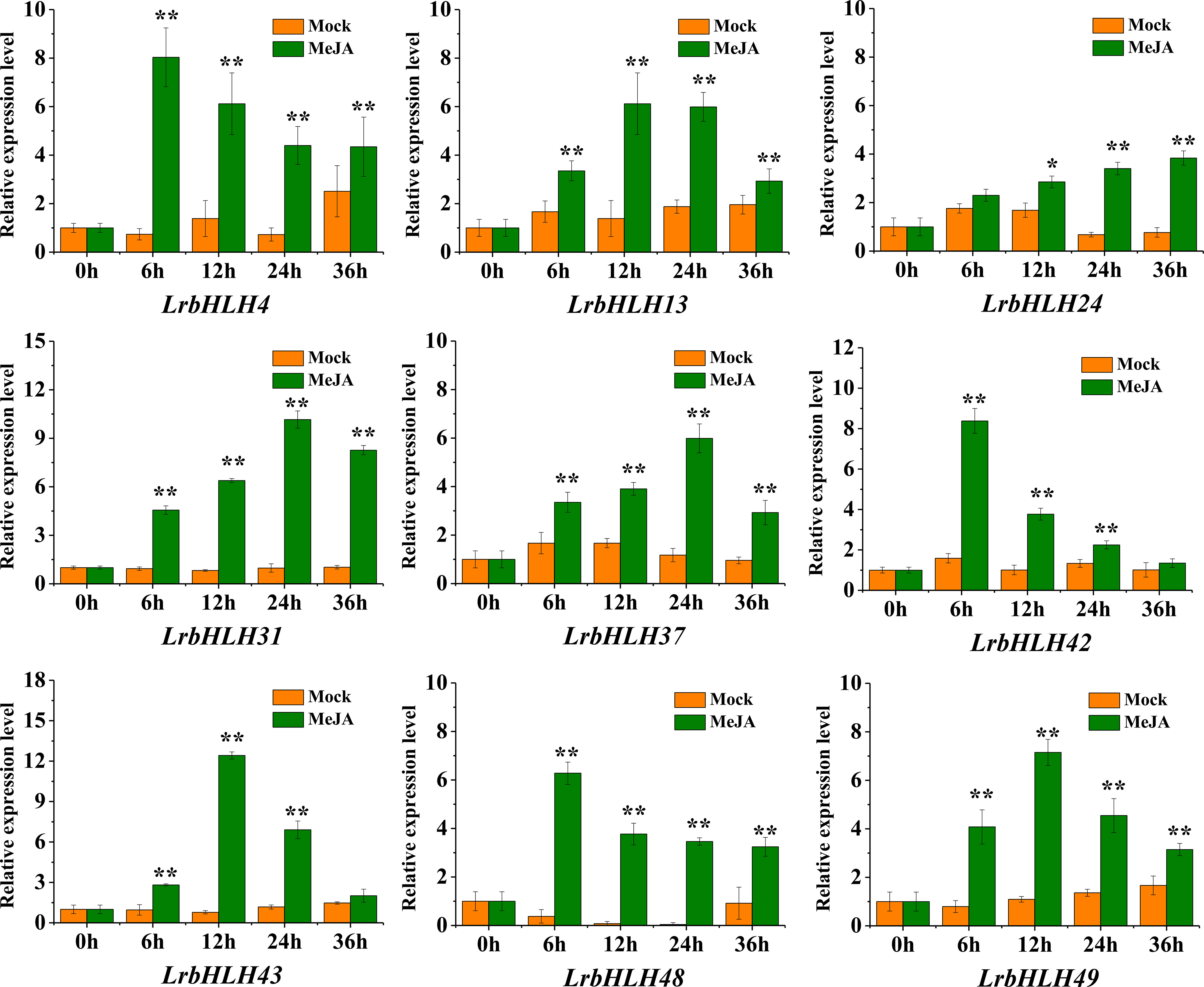
Figure 7 qRT-PCR analysis of LrbHLHs under MeJA treatment. The values represent means ± SD (n = 3). * and ** indicates significant difference between control and treatment according to student’s t-test at p < 0.05 and p < 0.01 level, respectively.
PPI networks of LrbHLH proteins
Based on the protein orthologs of Arabidopsis (Supplementary Table 3 and Table 4), LrbHLH proteins and LrJAZ proteins homologous to LaJAZs (Wang et al., 2020) were analyzed to determine functional and physical interactions using the STRING database (Supplementary Table 5). Generally, several key interactions were predicted, and some LrbHLH and LrJAZ proteins might interact with at least four proteins (Figure 8). For instance, LrbHLH48 (homologous to At5g57150) can interact with many LrbHLH proteins including LrbHLH4 (homologous to AtbHLH92), LrbHLH16 (homologous to At1g05805), LrbHLH24 (homologous to At1g06170), LrbHLH33 (homologous to AtLRL3), LrbHLH36 (homologous to AtBIM2), LrbHLH41 (homologous to AtHEC1), LrbHLH47 (homologous to At3g61950) and LrJAZ1 (homologous to AtJAZ1), acting as a center of the network node. Meanwhile, LrJAZ1 could also function as another center of the network node, showing the multiple interactions with LrbHLH and LrJAZ proteins. These findings confirm the functional diversity of LrbHLH genes.
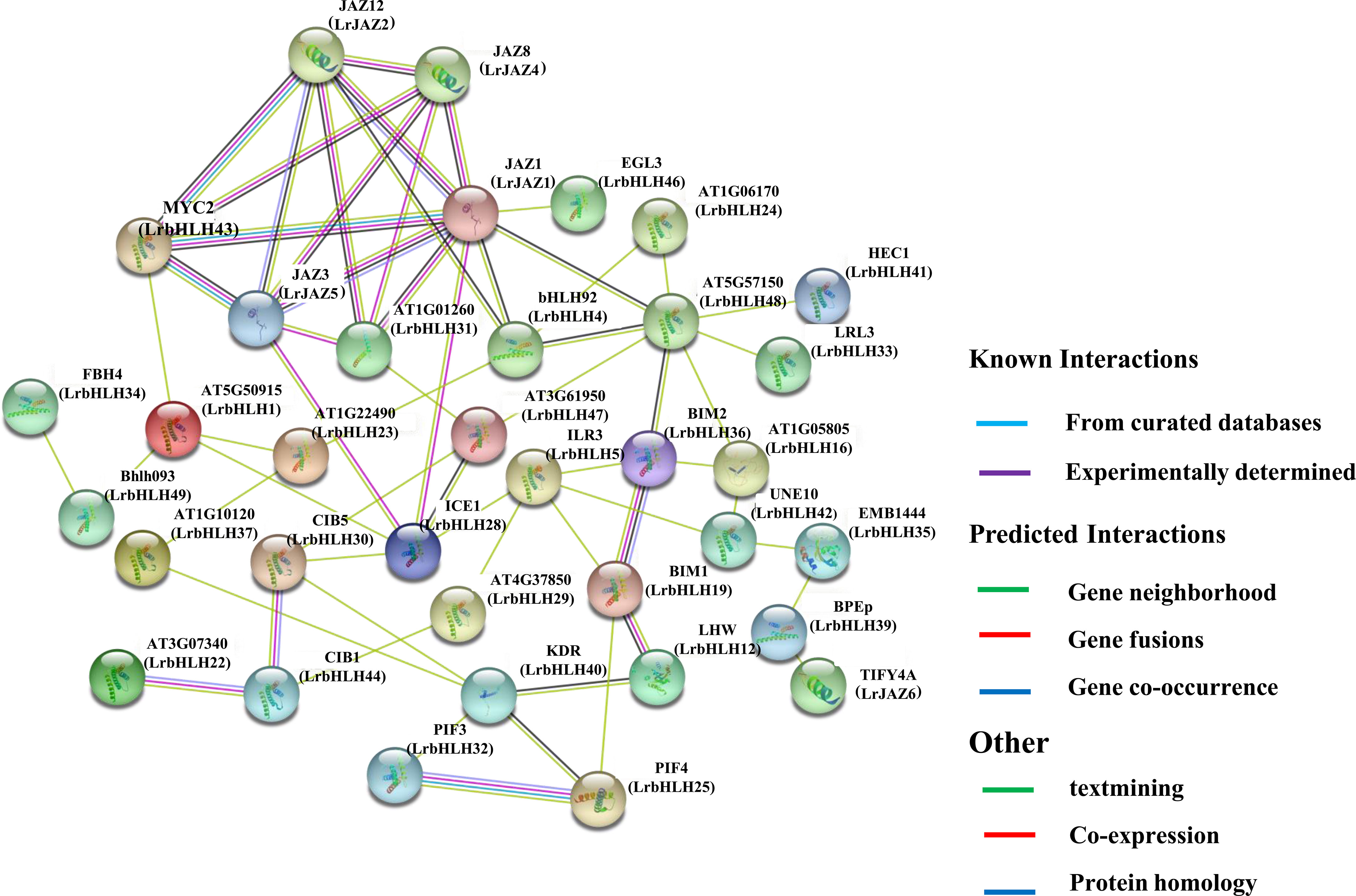
Figure 8 The predicted network of protein-protein interactions between LrbHLHs by STRING database. Different colors represent different interaction types. Arabidopsis bHLH names are marked, whereas their homologs in L. radiata are in parentheses.
To further verify protein-protein interactions in the predicted network, we randomly selected three LrbHLH (LrbHLH28, LrbHLH31 and LrbHLH48) genes and five LrJAZ (LrJAZ1, LrJAZ4, LrJAZ5, LrJAZ6 and LrJAZ7) genes to investigate their interactions using the Y2H technique in yeast (Figure 9). None of the three LrbHLH proteins could interact with the empty BD vector, while these LrbHLH proteins could interact with the empty AD, suggesting their self-activation. In addition, none of LrJAZ1, LrJAZ4, LrJAZ5, LrJAZ6, and LrJAZ7 proteins could interact with AD empty vector, indicating they have no self-activation (Figure 9). Among the combination of LrbHLH proteins and LrJAZ proteins, LrbHLH31 and JAZ1 showed a strong interaction. In addition, LrbHLH28 could interact with JAZ4, JAZ6, and JAZ7; whereas LrbHLH48 could interact with JAZ5, JAZ6, and JAZ7 respectively.
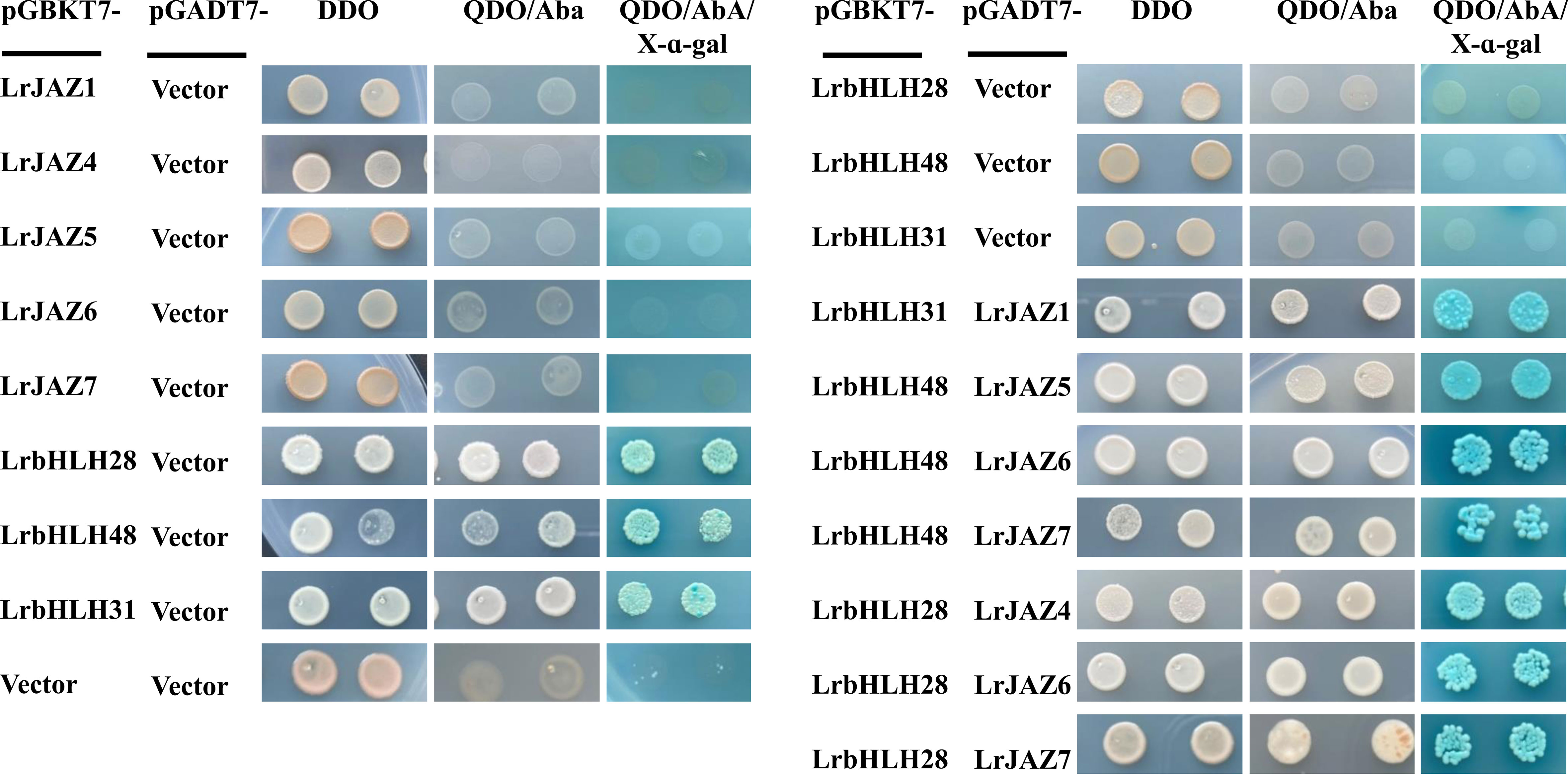
Figure 9 The protein-protein interactions between LrbHLHs and LrJAZs detected by yeast two-hybrid (Y2H) system. The coding sequences of LrbHLH28, LrbHLH31 and LrbHLH48 were ligated to the GAL4 activation domain (AD) and DNA-binding domain (BD), whereas the sequences of JAZ genes were fused to the GAL4 activation domain (AD). Positive control, negative control, and the fusion constructs were transformed into the Y2H strain and successively incubated in DDO (SD/-Leu/-Trp) medium, QDO (SD/-Ade/-Leu/-Trp/-His) medium supplemented with Aureobasidin A (AbA) and QDO medium supplemented with 5-bromo-4-chloro-3-indoxyl α-D-galactoside (X-α-Gal) and AbA.
Discussion
Characterization of the bHLH gene family in L. radiata
In plants, bHLH TF family is the second largest TFs family. For example, there are 162 bHLHs identified in A. thaliana (Carretero-Paulet et al., 2010), 167 bHLHs in rice (Carretero-Paulet et al., 2010), 230 bHLHs in Brassica rapa (Song et al., 2014), and 319 bHLHs in Glycine max (Hudson and Hudson, 2015). In this study, we identified 50 LrbHLH candidate genes through homolog search and domain analysis utilizing previously published transcriptome data (Supplementary Table 2; Wang et al., 2021). The LrbHLHs had similar properties in terms of the encoding amino acids number, isoelectric point, and motifs to bHLHs in A. thaliana and other plants (Carretero-Paulet et al., 2010). Plant bHLH members were grouped into several clades (or subfamilies), with the numbers of clades varying from 15 to 25 (Heim et al., 2003; Carretero-Paulet et al., 2010). We also found that 50 LrbHLH proteins in L. radiata could be classified into 20 clades (Figure 1). Interestingly, no LrbHLH proteins were in group orphans, VIII, Ib, Vb, VIIIc based on the Arabidopsis classification, showing L. radiata may lose homologs in these group throughout evolution. In addition, several LrbHLH proteins were grouped into functional clades of Arabidopsis, providing valuable insights for studying their biological roles. For example, LrbHLH20 and LrbHLH46 were grouped into subfamily IIIf and showed highly homologous to Arabidopsis MYC1, TT8, GLABROUS3 (GL3), and ENHANCER OF GLABRA3 (EGL3) proteins. It has been proved that AtTT8, AtGL3, and AtEGL3 could regulate anthocyanin and proanthocyanidins (PA) biosynthesis in Arabidopsis (Li, 2014). Besides, AtGL3 and AtEGL3 could modulate root hair patterning and trichome formation (Ramsay and Glover, 2005; Zhao et al., 2012). Thus, LrbHLH20 and LrbHLH46 might potentially modulate PA biosynthesis and trichome formation in L. radiata.
The conserved bHLH domain with estimated 60 amino acids comprised one basic, two helixs, and one loop regions (Li et al., 2006). This structure was also observed in L. radiata (Figure 3). Nineteen amino acid residues in the LrbHLH proteins are highly conserved, with a consensus ratio over 50%. These residues are highly conserved in bHLH TFs of Arabidopsis, Zea mays, and grape, suggesting the conservation of bHLH TFs families among different plants (Toledo-Ortiz et al., 2003; Zhang et al., 2018a; Wang et al., 2018). In addition, at least five basic residues in the basic region of the bHLH domain determine their DNA binding activity of bHLH TFs, and Glu-13 is important in specific recognition of the E-box DNA binding motif, whereas Arg-16 fixes and stabilizes the Glu-13 position. Moreover, His/Lys-9 and Arg-17 confer peculiarity to G-box discriminates, while Leu-27 is critical for dimer formation (Toledo-Ortiz et al., 2003). We also observed that Leu-27 was conserved in all LrbHLHs and homologous with AtbHLH proteins. Those results indicated that each LrbHLH protein have the basic functional property.
The GO annotation of LrbHLH genes indicating their potential functions
These enriched GO annotations of LrbHLH genes match to the known functional bHLH TFs. For instance, LrbHLH genes were predominantly involved in the ‘DNA binding transcription factor’, ‘transcription regulator’, ‘DNA binding’, and ‘protein dimerization activities’ in the molecular function section (Figure 4). This conformed that bHLH TFs regulate numerous genes implicated in different regulatory pathways (Castelain et al., 2012). For example, bHLH TFs NtMYC2a and NtMYC2b could form nuclear complexes with the NtJAZ1 repressor and modulate multiple jasmonate-inducible steps in nicotine biosynthesis (Zhang et al., 2012). bHLH TFs could also respond to Fe-deficient and salinity stresses (Huang and Dai, 2015; Babitha et al., 2015). TcMYC is highly expressed in leaves and xylem, unregulated by high-salinity and drought stresses in yew trees (Yang et al., 2018). The CsbHLH18 of sweet orange regulates the antioxidant gene, which helps in the modulation of cold tolerance (Geng and Liu, 2018).
The bHLH gene expression patterns facilitate their functional analysis
We systematically analyzed the expression patterns of LrbHLH genes in various tissues to investigate their physiological role in L. radiata biological processes. The homologous genes with similar expression profiles are conserved due to the dosage effect, whereas homologous genes with different expression profiles are retained by functionalization and subfunctionalization (Kim et al., 2014). We also observed that LrbHLH3/LrbHLH14, LrbHLH21/LrbHLH39, LrbHLH1/LrbHLH8, LrbHLH13/LrbHLH37, LrbHLH30/LrbHLH50, LrbHLH15/LrbHLH16, LrbHLH34/LrbHLH38, LrbHLH10/LrbHLH41, LrbHLH19/LrbHLH36, LrbHLH6/LrbHLH23, LrbHLH20/LrbHLH46 and LrbHLH11/LrbHLH40 were homologous genes (Figure 1). Among them, two gene pairs including LrbHLH20/LrbHLH46 and LrbHLH30/LrbHLH50 showed similar expression pattern, whereas other ten duplicated pairs had significantly different expression profiles in different tissues (Supplementary Figure 2; Figure 6). This suggests that there was a functional divergence among duplicated LrbHLH pairs at some point during evolution process. In addition, several LrbHLH genes highly expressed in specific tissues suggested their potential roles in organ development. For instance, LrbHLH1/LrbHLH8 were homolog to AtbHLH31, which participates in targeting cell expansion during petal growth (Varaud et al., 2011) exhibited higher expression level during flower development, suggesting LrbHLH1/LrbHLH8 may have the similar function in L. radiata. Meanwhile, we identified one putative MYC2 homolog gene (Dombrecht et al., 2007) LrbHLH43, which only exhibited higher expression level in bulb, suggesting its specific function in bulb. Otherwise, bHLH genes showed extensive expression levels in roots, including 78 bHLH genes in maize (Zhang et al., 2018a), five bHLH genes in wheat (Wang et al., 2019a), and two bHLH genes in Dendrobium officinale (Wang and Liu, 2020). We also observed that LrbHLH6, LrbHLH7, LrbHLH14, and LrbHLH37 genes were highly expressed in roots (Figure 6A), implying they could play important roles in L. radiata roots.
JA is a critical defense signaling molecule regulating different biological processes. Previous study revealed that MeJA could regulate the expression of bHLH family genes (Zander et al., 2020; Guo et al., 2021; Li et al., 2022). Recently, tomato bHLH gene SlJIG has been demonstrated to be induced by JA, and function in terpene biosynthesis and resistance to insects and fungus (Cao et al., 2022). We also analyzed the expression levels of bHLH genes in Lycoris under MeJA treatment. Most of the LrbHLH genes and orthologous genes of LrbHLH (LabHLH) were simultaneously affected by MeJA treatments (Supplementary Figure 3 and Figure 7). Those results indicate that the important biological functions of LrbHLHs may correlated with JA signaling pathway.
PPI network prediction and validation of LrbHLHs
The PPI networks were utilized for identifying the interactions between LrbHLHs. bHLH TF MYC2 and its homologs MYC3 and MYC4 acts as the master regulator of diverse JA responses (Breeze, 2019). Meanwhile, the plant-specific group of JAZ proteins plays a critical part in repressing the activity of MYC TFs (Howe et al., 2018). Subsequently, we analyzed the potential interactions among LrbHLH proteins and JAZ proteins in the anticipated network. Consequently, LrbHLHs and JAZ proteins were found to form a signal transmission network (Figure 8 and Supplementary Figure 4). This is consistent with former researches that the bHLH proteins binding activity is based on the homodimers or heterodimers formation among bHLH proteins (Pires and Dolan, 2010; Feller et al., 2011). FLOWERING BHLH 4 (FBH4, homolog of LrbHLH34 and LrbHLH38) and CIB1 (homolog of LrbHLH44) are implicated in the regulation of flowering time (Ito et al., 2012; Liu et al., 2013); HECATE (HEC, homolog of LrbHLH10 and LrbHLH41) interact with SPATULA (SPT) for the pistil development regulation through targeting cytokinins among other hormones (Schuster et al., 2015). SCREAM (SCRM, also known as ICE1; homolog of LrbHLH28) can interact with FAMA, SPEECHLESS (SPCH), and MUTE and regulate stomatal differentiation (Qi and Torii, 2018). Moreover, INDUCER OF CBP EXPRESSION 1 (ICE1) regulates lateral bud growth and plant stress response (Zhang et al., 2018b), whereas Lotus japonicas ROOTHAIRLESS LIKE (LRL3, homolog of LrbHLH33) regulates root hair development (Breuninger et al., 2016). MYC2 (homolog of LrbHLH43) is vital in the optical, GA, and JA signals as well as modulates many signal transmissions (Lorenzo et al., 2004; Dombrecht et al., 2007). PIF3 (homolog of LrbHLH32) interacts with phyB1 in light signal transduction (Kumar et al., 2016b). Additionally, LrbHLH25 may have the similar functions with PIF4 and PIF5 and play conserved roles in the response to phytochrome signaling in plants (Shi et al., 2018). Furthermore, we confirmed protein interactions between LrbHLH28, LrbHLH31, LrbHLH48, and LrJAZ proteins (Figure 9). LrbHLH28 has a highly expression level at flower developmental stage and belong to ICE1 branch, which could regulate the growth of lateral bud (Zhang et al., 2018b). Meanwhile, ICE1 could interact with the HOS1 protein, a crucial flowering time regulator (Liu et al., 2016). Flower bHLH transcriptional activator FBH4 may control expression of the photoperiodic flowering regulator CONSTANS to regulate flowering (Ito et al., 2012). LrbHLH34 and LrbHLH38 were homologs of FBH4, suggesting they might cause early flowering regardless of the photoperiod. Additionally, CRY2-interacting bHLH (CIB) proteins such as CIB1, CIB2, CIB3, CIB4, and CIB5 could activate Flowering Locus T (FT) transcription through the interaction with cryptochrome 2 (CRY2) protein for mediating flowering time regulation in Arabidopsis (Liu et al., 2013). Since CIB genes are conserved in plants (Liu et al., 2013), LrbHLH44 (CIB1 homologs) and LrbHLH30 (CIB5 homologs) might have the similar function in flower development. Additional research is necessary to demonstrate the comprehensive interaction network of the LrbHLH TFs during L. radiata growth and development.
Conclusion
In this study, 50 LrbHLH genes were identified from the transcriptome of L. radiata, and enriched by systematically analyzing basic biochemical information, phylogenetic relationships, conserved motifs, and gene expression profiling. Most LrbHLH genes were revealed to likely play various important roles in Lycoris growth and development, especially in flower development stages. The analysis of the expression pattern of LrbHLH genes in response to MeJA treatment suggested that the LrbHLH genes could be importantly involved in JA signaling. Meanwhile, by comprehensive analysis of the comparison of homologous functional proteins, the PPI prediction networks and the yeast two-hybrid system, LrbHLH28, LrbHLH31 and LrbHLH48 was demonstrated to interact with LrJAZ proteins and might functionally regulate biological processes in JA signaling pathway. These findings provide a serviceable opportunity to understand the biological characteristics and functions of LrbHLH genes in Lycoris.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
NW and ZW designed and conceived experiments, and wrote the manuscript. NW performed most of the experiments and data analysis. XS collected the experimental materials. FZ provided helpful comments on the manuscript. ZW provided guidance on the whole study and contributed with valuable discussions. All authors read and approved the final manuscript.
Funding
This research was financially supported by the Jiangsu Key Laboratory for the Research and Utilization of Plant Resources (Grant No. JSPKLB202020); Jiangsu Agricultural Science and Technology Innovation Fund (Grand No. CX(20)3171); Jiangsu Provincial Crop Germplasm Resource Bank for Conservation (Grand No. 2022-SJ-015); Projects of Independently Development of Jiangsu Provincial Department of Science and Technology (Grand No. BM2018021-5).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.975530/full#supplementary-material
Supplementary Figure 1 | Multiple sequence alignment of the bHLH conserved domain in L.radiata bHLH proteins. The yellow boxes indicate 30% identity of amino acids, the blue boxes indicate 50% identity of amino acids, and the red boxes indicate 75% identity of amino acids.
Supplementary Figure 2 | Expression profile heatmap with hierarchal clustering of LrbHLHs in different tissues of L. longituba. Red and blue represent high and low relative transcript abundance, respectively.
Supplementary Figure 3 | Heatmap of LrbHLH gene expression profiles with MeJA treatment. Red and blue represent high and low relative transcript abundance, respectively.
Supplementary Figure 4 | The predicted network of protein-protein interactions between LrbHLHs by STRING database.
References
Arnaud, N., Girin, T., Sorefan, K., Fuentes, S., Wood, T. A., Lawrenson, T., et al. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24, 2127–2132. doi: 10.1101/gad.593410
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Babitha, K. C., Vemanna, R. S., Nataraja, K. N., Udayakumar, M. (2015). Overexpression of EcbHLH57 transcription factor from Eleusine coracana l. @ in tobacco confers tolerance to salt, oxidative and drought stress. PLoS One 10, e0137098. doi: 10.1371/journal.pone.0137098
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Breeze, E. (2019). Master MYCs: MYC2, the jasmonate signaling “master switch”. Plant Cell 31, 9–10. doi: 10.1105/tpc.19.00004
Breuninger, H., Thamm, A., Streubel, S., Sakayama, H., Nishiyama, T., Dolan, L. (2016). Diversification of a transcription factor family led to the evolution of antagonistically acting genetic regulators of root hair growth. Curr. Biol. 26, 1622–1628. doi: 10.1016/j.cub.2016.04.060
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al. (2021). KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, W317–W325. doi: 10.1093/nar/gkab447
Cahlíková, L., Breiterová, K., Opletal, L. (2020). Chemistry and biological activity of alkaloids from the genus Lycoris (Amaryllidaceae). Molecules 25, 4797. doi: 10.3390/molecules25204797
Cao, Y., Liu, L., Ma, K., Wang, W., Lv, H., Gao, M., et al. (2022). The jasmonate-induced bHLH gene SlJIG functions in terpene biosynthesis and resistance to insects and fungus. J. Integr. Plant Biol. 64, 1102–1115. doi: 10.1111/jipb.13248
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Castelain, M., Hir., R., Bellini, C. (2012). The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiol. Plant 145, 450–460. doi: 10.1111/j.1399-3054.2012.01600.x
Castillon, A., Shen, H., Huq, E. (2007). Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12, 514–521. doi: 10.1016/j.tplants.2007.10.001
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G. L., Tian, Y. Q., Wu, J. L., Li, N., Guo, M. Q. (2016). Antiproliferative activities of amaryllidaceae alkaloids from Lycoris radiata targeting DNA topoisomerase I. Sci. Rep. 6, 38284. doi: 10.1038/srep38284
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., et al. (2007). MYC2 differentially modulates diversejasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245. doi: 10.1105/tpc.106.048017
Feller, A., Machemer, K., Braun, E. L., Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Feng, T., Wang, Y. Y., Su, J., Li, Y., Cai, X. H., Luo, X. D. (2011). Amaryllidaceae alkaloids from Lycoris radiata. Helv. Chim. Acta 94, 178–183. doi: 10.1002/hlca.201000176
Feng, X. M., Zhao, Q., Zhao, L. L., Qiao, Y., Xie, X. B., Li, H. F., et al. (2012). The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 12, 22. doi: 10.1186/1471-2229-12-22
Gao, C., Sun, J., Wang, C., Dong, Y., Xiao, S., Wang, X., et al. (2017). Genome-wide analysis of basic/helix-loop-helix gene family in peanut and assessment of its roles in pod development. PloS One 12, e0181843. doi: 10.1371/journal.pone.0181843
Geng, J. J., Liu, J. H. (2018). The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene. J. Exp. Bot. 69, 2677–2692. doi: 10.1093/jxb/ery065
Gremski, K., Ditta, G., Yanofsky, M. F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134, 3593–3601. doi: 10.1242/dev.011510
Guo, J., Sun, B., He, H., Zhang, Y., Tian, H., Wang, B. (2021). Current understanding of bHLH transcription factors in plant abiotic stress tolerance. Int. J. Mol. Sci. 22, 4921. doi: 10.3390/ijms22094921
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2021). WoLF PSORT: protein localization predictor. Int. J. Mol. Sci. 22, 11458. doi: 10.3390/ijms222111458
Howe, G. A., Major, I. T., Koo, A. J. (2018). Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 69, 20.1–20.29. doi: 10.1146/annurev-arplant-042817-040047
Huang, D. Q., Dai, W. H. (2015). Molecular characterization of the basic helix-loop-helix (bHLH) genes that are differentially expressed and induced by iron deficiency in Populus. Plant Cell Rep. 34, 1211–1124. doi: 10.1007/s00299-015-1779-8
Hudson, K. A., Hudson, M. E. (2015). A classification of basic helix-loop-helix transcription factors of soybean. Int. J. Genomics 2015, 603182. doi: 10.1155/2015/603182
Ito, S., Song, Y., Josephson-Day, A. R., Miller, R. J., Breton, G., Olmstead, R. G., et al. (2012). FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 3582–3587. doi: 10.1073/pnas.1118876109
Jiang, Y., Yang, B., Deyholos, M. K. (2009). Functional characterization of the arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Genet. Genomics 282, 503–516. doi: 10.1007/s00438-009-0481-3
Jin, Z. (2011). Amaryllidaceae and sceletium alkaloids. Nat. Prod. Rep. 28, 1126–1142. doi: 10.1039/c0np00073f
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361. doi: 10.1093/nar/gkw1092
Kim, J., Lee, J., Choi, J. P., Park, I., Yang, K., Kim, M. K., et al. (2014). Functional innovations of three chronological mesohexaploid Brassica rapa genomes. BMC Genomics 15, 606. doi: 10.1186/1471-2164-15-606
Koini, M. A., Alvey, L., Allen, T., Tilley, C. A., Harberd, N. P., Whitelam, G. C., et al. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19, 408–413. doi: 10.1016/j.cub.2009.01.046
Komatsu, M., Maekawa, M., Shimamoto, K., Kyozuka, J. (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231, 364–373. doi: 10.1006/dbio.2000.9988
Kumar, S., Stecher, G., Tamura, K. (2016a). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kumar, I., Swaminathan, K., Hudson, K., Hudson, M. E. (2016b). Evolutionary divergence of phytochrome protein function in Zea mays PIF3 signaling. J. Exp. Bot. 67, 4231–4240. doi: 10.1093/jxb/erw217
Letunic, I., Khedkar, S., Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, S. (2014). Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav. 9, e27522. doi: 10.4161/psb.27522
Li, C., Cai, X., Shen, Q., Chen, X., Xu, M., Ye, T., et al. (2022). Genome-wide analysis of basic helix-loop-helix genes in Dendrobium catenatum and functional characterization of DcMYC2 in jasmonate-mediated immunity to Sclerotium delphinii. Front. Plant Sci. 13. doi: 10.3389/fpls
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Li, H., Gao, W., Xue, C., Zhang, Y., Liu, Z., Zhang, Y., et al. (2019b). Genome-wide analysis of the bHLH gene family in Chinese jujube (Ziziphus jujuba mill.) and wild jujube. BMC Genomics 20, 568. doi: 10.1186/s12864-019-5936-2
Li, W., Qiao, C., Pang, J., Zhang, G., Luo, Y. (2019a). The versatile O-methyltransferase LrOMT catalyzes multiple O-methylation reactions in amaryllidaceae alkaloids biosynthesis. Int. J. Biol. Macromol. 141, 680–692. doi: 10.1016/j.ijbiomac.2019.09.011
Liu, Y., Li, X., Li, K., Liu, H., Lin, C. (2013). Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in arabidopsis. PloS Genet. 9, e1003861. doi: 10.1371/journal.pgen.1003861
Liu, X., Pan, T., Liang, W., Gao, L., Wang, X., Li, H., et al. (2016). Overexpression of an orchid (Dendrobium nobile) SOC1/TM3-like ortholog, DnAGL19, in Arabidopsis regulates HOS1-FT expression. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00099
Liu, R., Song, J., Liu, S., Chen, C., Zhang, S., Wang, J., et al. (2021). Genome-wide identification of the Capsicum bHLH transcription factor family: discovery of a candidate regulator involved in the regulation of species-specific bioactive metabolites. BMC Plant Biol. 21, 262. doi: 10.1186/s12870-021-03004-7
Li, J., Wang, T., Han, J., Ren, Z. (2020b). Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber. BMC Plant Biol. 20, 272. doi: 10.1186/s12870-020-02440-1
Li, Q., Xu, J., Yang, L., Zhou, X., Cai, Y., Zhang, Y. (2020a). Transcriptome analysis of different tissues reveals key genes associated with galanthamine biosynthesis in Lycoris longituba. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.519752
Li, W., Yang, Y., Qiao, C., Zhang, G., Luo, Y. (2018). Functional characterization of phenylalanine ammonia-lyase- and cinnamate 4-hydroxylase-encoding genes from lycoris radiata, a galanthamine-producing plant. Int. J. Biol. Macromol. 117, 1264–1279. doi: 10.1016/j.ijbiomac.2018.06.046
Lorenzo, O., Chico, J. M., Sánchez-Serrano, J. J., Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. doi: 10.1105/tpc.022319
Lu, R., Zhang, J., Liu, D., Wei, Y. L., Wang, Y., Li, X. B. (2018). Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum). BMC Plant Biol. 18, 304. doi: 10.1186/s12870-018-1523-y
Menand, B., Yi, K., Jouannic, S., Hoffmann, L., Ryan, E., Linstead, P., et al. (2007). An ancient mechanism controls the development of cells with a rooting function in land plants. Science 316, 1477–1480. doi: 10.1126/science.1142618
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Morohashi, K., Zhao, M., Yang, M., Read, B., Lloyd, A., Lamb, R., et al. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145, 736–746. doi: 10.1104/pp.107.104521
Murre, C., McCaw, P. S., Baltimore, D. (1989). A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777–783. doi: 10.1016/0092-8674(89)90682-X
Niu, X., Guan, Y., Chen, S., Li, H. (2017). Genome-wide analysis of basic helix-loop-helix (bHLH) transcription factors in Brachypodium distachyon. BMC Genomics 18, 619. doi: 10.1186/s12864-017-4044-4
Park, C. H., Yeo, H. J., Park, Y. E., Baek, S. A., Kim, J. K., Park, S. U. (2019). Transcriptome analysis and metabolic profiling of Lycoris radiata. Biology 8, 63. doi: 10.3390/biology8030063
Pillitteri, L. J., Sloan, D. B., Bogenschutz, N. L., Torii, K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505. doi: 10.1038/nature05467
Pires, N., Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Qi, X., Torii, K. U. (2018). Hormonal and environmental signals guiding stomatal development. BMC Biol. 16, 21. doi: 10.1186/s12915-018-0488-5
Ramsay, N. A., Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Ren, Z., Xu, Y., Lvy, X., Zhang, D., Gao, C., Lin, Y., et al. (2021). Early sucrose degradation and the dominant sucrose cleavage pattern influence Lycoris sprengeri bulblet regeneration in vitro. Int. J. Mol. Sci. 22, 11890. doi: 10.3390/ijms222111890
Ren, Z. M., Zhang, D., Jiao, C., Li, D. Q., Wu, Y., Wang, X. Y., et al. (2022). Comparative transcriptome and metabolome analyses identified the mode of sucrose degradation as a metabolic marker for early vegetative propagation in bulbs of Lycoris. Plant J. 112, 115–134. doi: 10.1111/tpj.15935
Sakamoto, W., Ohmori, T., Kageyama, K., Miyazaki, C., Saito, A., Murata, M., et al. (2001). The purple leaf (Pl) locus of rice: the pl (w) allele has a complex organization and includes two genes encoding basic helix-loop-helix proteins involved in anthocyanin biosynthesis. Plant Cell Physiol. 42, 982–991. doi: 10.1093/pcp/pce128
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schuster, C., Gaillochet, C., Lohmann, J. U. (2015). Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142, 3343–3350. doi: 10.1242/dev.120444
Seo, J. S., Joo, J., Kim, M. J., Kim, Y. K., Nahm, B. H., Song, S. I., et al. (2011). OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 65, 907–921. doi: 10.1111/j.1365-313X.2010.04477.x
Shen, C. Y., Xu, X. L., Yang, L. J., Jiang, J. G. (2019). Identification of narciclasine from Lycoris radiata (L’Her.) herb. and its inhibitory effect on LPS-induced inflammatory responses in macrophages. Food Chem. Toxicol. 125, 605–613. doi: 10.1016/j.fct.2019.02.003
Shi, S. D., Qiu, Y. X., Wu, L., Fu, C. X. (2006). Interspecific relationships of Lycoris (amaryllidaceae) inferred from inter-simple sequence repeat data. Sci. Hortic. 110, 285–291. doi: 10.1016/j.scienta.2006.07.011
Shi, Q., Zhang, H., Song, X., Ye, J., Liang, R., Li, G. (2018). Functional characterization of the maize phytochrome-interacting factors PIF4 and PIF5. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02273
Smolen, G. A., Pawlowski, L., Wilensky, S. E., Bender, J. (2002). Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161, 1235–1246. doi: 10.1093/genetics/161.3.1235
Song, X. M., Huang, Z. N., Duan, W. K., Ren, J., Liu, T. K., Li, Y., et al. (2014). Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genomics 289, 77–91. doi: 10.1007/s00438-013-0791-3
Sorensen, A. M., Kröber, S., Unte, U. S., Huijser, P., Dekker, K., Saedler, H. (2003). The Arabidopsis aborted microspores (AMS) gene encodes a MYC class transcription factor. Plant J. 33, 413–423. doi: 10.1046/j.1365-313x.2003.01644.x
Sun, H., Fan, H. J., Ling, H. Q. (2015). Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16, 9. doi: 10.1186/s12864-014-1209-2
Sun, X., Wang, Y., Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, 10800. doi: 10.1093/nar/gkab835
Toledo-Ortiz, G., Huq, E., Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 15, 1749–1770. doi: 10.1105/tpc.013839
Valea, I., Motegi, A., Kawamura, N., Kawamoto, K., Miyao, A., Ozawa, R., et al. (2021). The rice wound-inducible transcription factor RERJ1 sharing same signal transduction pathway with OsMYC2 is necessary for defense response to herbivory and bacterial blight. Plant Mol. Biol. 109, 651–666. doi: 10.1007/s11103-021-01186-0
Varaud, E., Brioudes, F., Szécsi, J., Leroux, J., Brown, S., Perrot-Rechenmann, C., et al. (2011). AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23, 973–983. doi: 10.1105/tpc.110.081653
Verma, D., Jalmi, S. K., Bhagat, P. K., Verma, N., Sinha, A. K. (2020). A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 287, 2560–2576. doi: 10.1111/febs.15157
Wang, R., Han, X., Xu, S., Xia, B., Jiang, Y., Xue, Y., et al. (2019b). Cloning and characterization of a tyrosine decarboxylase involved in the biosynthesis of galanthamine in Lycoris aurea. PeerJ 7, e6729. doi: 10.7717/peerj.6729
Wang, Y., Liu, A. (2020). Genomic characterization and expression analysis of basic Helix-Loop-Helix (bHLH) family genes in traditional chinese herb dendrobium officinale. Plants (Basel). 9, 1044. doi: 10.3390/plants9081044
Wang, N., Shu, X., Zhang, F., Zhuang, W., Wang, T., Wang, Z. (2021). Comparative transcriptome analysis identifies key regulatory genes involved in anthocyanin metabolism during flower development in Lycoris radiata. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.761862
Wang, P., Su, L., Gao, H., Jiang, X., Wu, X., Li, Y., et al. (2018). Genome-wide characterization of bHLH genes in grape and analysis of their potential relevance to abiotic stress tolerance and secondary metabolite biosynthesis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00064
Wang, L., Xiang, L., Hong, J., Xie, Z., Li, B. (2019a). Genome-wide analysis of bHLH transcription factor family reveals their involvement in biotic and abiotic stress responses in wheat (Triticum aestivum l.). 3 Biotech. 9, 236. doi: 10.1007/s13205-019-1742-4
Wang, R., Xu, S., Wang, N., Xia, B., Jiang, Y., Wang, R. (2017). Transcriptome analysis of secondary metabolism pathway, transcription factors, and transporters in response to methyl jasmonate in Lycoris aurea. Front. Plant Sci. 1971 7. doi: 10.3389/fpls.2016.01971
Wang, P., Yu, S., Han, X., Xu, J., He, Q., Xu, S., et al. (2020). Identification, molecular characterization and expression of JAZ genes in Lycoris aurea. PloS One 15, e0230177. doi: 10.1371/journal.pone.0230177
Xie, X. B., Li, S., Zhang, R. F., Zhao, J., Chen, Y. C., Zhao, Q., et al. (2012). The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 35, 1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x
Yang, Y. F., Zhang, K. K., Yang, L. Y., Lv, X., Wu, Y., Liu, H. W., et al. (2018). Identification and characterization of MYC transcription factors in taxus sp. Gene 675, 1–8. doi: 10.1016/j.gene.2018.06.065
Yue, Y., Liu, J., Shi, T., Chen, M., Li, Y., Du, J., et al. (2019). Integrating transcriptomic and GC-MS metabolomic analysis to characterize color and aroma formation during tepal development in Lycoris longituba. Plants 8, 53. doi: 10.3390/plants8030053
Zander, M., Lewsey, M. G., Clark, N. M., Yin, L., Bartlett, A., Guzmán, J. P. S., et al. (2020). Integrated multi-omics framework of the plant response to jasmonic acid. Nat. Plants 6, 290–302. doi: 10.1038/s41477-020-0605-7
Zhang, H. B., Bokowiec, M. T., Rushton, P. J., Han, S. C., Timko, M. P. (2012). Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol. Plant 5, 73–84. doi: 10.1093/mp/ssr056
Zhang, T. T., Lv, W., Zhang, H. S., Ma, L., Li, P. H., Ge, L., et al. (2018a). Genome-wide analysis of the basic helix-loop-helix (bHLH) transcription factor family in maize. BMC Plant Biol. 18, 235. doi: 10.1186/s12870-018-1441-z
Zhang, T., Mo, J., Zhou, K., Chang, Y., Liu, Z. (2018b). Overexpression of Brassica campestris BcICE1 gene increases abiotic stress tolerance in tobacco. Plant Physiol. Biochem. 132, 515–523. doi: 10.1016/j.plaphy.2018.09.039
Zhao, H., Wang, X., Zhu, D., Cui, S., Li, X., Cao, Y., et al. (2012). A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J. Biol. Chem. 287, 14109–14121. doi: 10.1074/jbc.M111.280735
Keywords: Lycoris radiata, bHLH transcription factors, expression patterns, MeJA treatment, regulatory networks
Citation: Wang N, Shu X, Zhang F and Wang Z (2023) Transcriptome-wide characterization of bHLH transcription factor genes in Lycoris radiata and functional analysis of their response to MeJA. Front. Plant Sci. 13:975530. doi: 10.3389/fpls.2022.975530
Received: 22 June 2022; Accepted: 12 December 2022;
Published: 10 January 2023.
Edited by:
Iftikhar Ali, State Key Laboratory of Molecular Developmental Biology (CAS), ChinaReviewed by:
Javed Iqbal, Quaid-i-Azam University, PakistanMuthusamy Ramakrishnan, Nanjing Forestry University, China
Yuanzhong Jiang, Sichuan University, China
Copyright © 2023 Wang, Shu, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong Wang, d2FuZ3pob25nMTlAMTYzLmNvbQ==
 Ning Wang
Ning Wang Xiaochun Shu
Xiaochun Shu Fengjiao Zhang
Fengjiao Zhang Zhong Wang
Zhong Wang