
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 11 August 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.975434
 Mauro Commisso†
Mauro Commisso† Stefano Negri†
Stefano Negri† Elisa Gecchele
Elisa Gecchele Emanuela Fazion
Emanuela Fazion Cecilia Pontoriero
Cecilia Pontoriero Linda Avesani*
Linda Avesani* Flavia Guzzo*
Flavia Guzzo*Tryptamine and serotonin are indolamines that fulfill diverse biological functions in all kingdoms of life. Plants convert L-tryptophan into tryptamine and then serotonin via consecutive decarboxylation and hydroxylation reactions catalyzed by the enzymes tryptophan decarboxylase (TDC) and tryptamine 5-hydroxylase (T5H). Tryptamine and serotonin accumulate to high levels in the edible fruits and seeds of many plant species, but their biological roles in reproductive organs remain unclear and the metabolic pathways have not been characterized in detail. We identified three TDC genes and a single T5H gene in tomato (Solanum lycopersicum L.) by homology-based screening and confirmed their activity by heterologous expression in Nicotiana benthamiana. The co-analysis of targeted metabolomics and gene expression data revealed complex spatiotemporal gene expression and metabolite accumulation patterns that suggest the involvement of the serotonin pathway in multiple biological processes. Our data support a model in which SlTDC1 allows tryptamine to accumulate in fruits, SlTDC2 causes serotonin to accumulate in aerial vegetative organs, and SlTDC3 works with SlT5H to convert tryptamine into serotonin in the roots and fruits.
Indolamines are ubiquitous tryptophan-derived indole alkaloids found in all kingdoms of life, albeit with extremely diverse distribution profiles and biological functions (Roshchina, 2010). The ability of many plants to produce the indolamines tryptamine and serotonin has driven research into the corresponding metabolic pathways, in many cases due to the role of these products as intermediates in the synthesis of specialized metabolites with pharmacological activity, such as antitumor indole alkaloids and melatonin (Arnao and Hernández-Ruiz, 2018). Recent literature provides evidence that plant indolamines are involved in various physiological processes, including development and biotic/abiotic stress responses, leading some authors to consider serotonin and melatonin as a new class of phytohormones (Bhatla, 2018; Arnao and Hernández-Ruiz, 2019; Erland et al., 2019; Akula and Mukherjee, 2020; Sun et al., 2021). In contrast, the potential biological functions of tryptamine have received far less attention because it is widely considered as an intermediate rather than an end-product, even though both tryptamine and serotonin accumulate to high levels in particular organs of some plant species (Negri et al., 2021). For example, melatonin is found in the fleshy fruits and seeds of some plants with concentrations in the ng g−1 FW range (Nawaz et al., 2016) whereas the concentration of tryptamine and serotonin can reach hundreds of μg g−1 FW (Huang and Mazza, 2011; Islam et al., 2016; Yılmaz et al., 2019). This 1,000-fold difference in abundance represents a significant investment of resources to ensure the accumulation of tryptamine and serotonin, which must therefore provide some advantage during the reproductive phase. Moreover, dynamic indolamine accumulation profiles have been reported throughout fruit development and ripening, with corresponding changes in gene expression profiles (Hano et al., 2017; Commisso et al., 2019).
In plants, tryptamine is synthesized from L-tryptophan by the enzyme tryptophan decarboxylase (TDC), a cytosolic pyridoxal-5′-phosphate-dependent type II aromatic L-amino acid decarboxylase (AADC). This diverts carbon and nitrogen from primary metabolism into the indole alkaloid pathway of specialized metabolism. The first plant TDC gene was found in the Madagascar periwinkle Catharanthus roseus (De Luca et al., 1989) and several orthologs have been identified by homology-based screening and functionally characterized by expression in Escherichia coli. Most plants have a single TDC gene, including C. roseus (Goddijn et al., 1994), whereas two TDC genes are found in Aegilops variabilis, Camptotheca acuminata, Capsicum annuum and Ophiorrhiza pumila (López-Meyer and Nessler, 1997; Yamazaki et al., 2003; Park et al., 2009; Li et al., 2016; You et al., 2020). Oryza sativa has three TDC genes (Kang et al., 2007a), and tryptamine is the first committed step in the rice indolamine pathway leading to serotonin and melatonin production at different subcellular levels (Back et al., 2016).
A cytochrome P450 monooxygenase located in the membrane of the endoplasmic reticulum (ER) is the tryptamine 5′-hydroxylase (T5H) that converts tryptamine into serotonin (5-hydroxytryptamine). Only the rice T5H gene has been characterized thus far, thanks to the discovery of sl mutants that accumulate tryptamine due to a deficit in T5H expression (Kang et al., 2007b; Fujiwara et al., 2010). Melatonin is then synthesized in a two-step process that involves the N-acetylation and methylation (in either order) of serotonin (Kang et al., 2011, 2013; Byeon et al., 2015). In parallel, other tryptamine and serotonin derivatives can be synthesized in plants including their N-cinnamoyl amides, in which the indolamine backbone is joined to the phenylpropanoid skeleton of cinnamic acid derivatives (mainly ferulic and coumaric acids) to generate powerful antioxidants with likely defensive functions against fungal pathogens and UV stress (Zeiss et al., 2020).
Despite the existence of alternative metabolic routes that produce melatonin via the hydroxytryptophan intermediate might not be excluded, the presence of TDC and T5H genes is a reasonable hypothesis in all plants that produce tryptamine and serotonin, or their downstream metabolites. For example, we recently identified TDC genes in green kiwifruit (Actinidia deliciosa) and yellow kiwifruit (Actinidia chinensis) based on the bulk accumulation of tryptamine and serotonin in both unripe and ripe berries (Commisso et al., 2019). However, given their arboreal nature, metabolic interventions to address the biological role of plant indolamines in the reproductive organs of these species might be challenging. As an alternative, tomato (Solanum lycopersicum) is a model fleshy fruit crop that is known to produce both tryptamine and serotonin. A putative TDC was purified from the cultivar Big Boy nearly 50 years ago (Gibson et al., 1972) but the molecular basis of tryptamine and serotonin biosynthesis in tomato was then largely ignored until more recent interest in melatonin. Two putative TDCs and one T5H were then postulated to exist in tomato (Hano et al., 2017), leading to the prediction of five putative TDC genes based on the protein sequences (Pang et al., 2018), and the very recent in vivo characterization of the isoform SlTDC1 (Tsunoda et al., 2021).
Here we report for the first time the identity of all biosynthetic genes involved in the tomato serotonin pathway, comprising three TDC genes and a single T5H gene. The genes were functionally characterized by transient expression in Nicotiana benthamiana, and the expression profiles were associated with untargeted metabolomics data as a means to link gene expression to enzymatic activity and metabolite accumulation. We also investigated the spatial and temporal profiles of TDC and T5H gene expression in relation to tryptamine and serotonin production in the whole tomato plant, providing insight into the physiological functions of these indolamines.
The protein sequences of functionally characterized plant TDCs and T5Hs were downloaded from NCBI (Supplementary Table 1) and used as blastp queries against tomato proteins deposited in SGNdb (ITAG release 3.20)1 with the parameters percentage identity >50%, alignment score > 500, and the presence of the protein candidates in the output list of all queries. All alignments of nucleotide and amino acid sequences were performed by using Clustal Omega with default parameters.2 To build the phylogenetic tree in which we reported the putative tomato TDCs and the plant AADCs listed in Supplementary Table 1 we used a MUSCLE alignment (MEGA X) applying the neighbor-joining method and bootstrap analysis (1,000 iterations).
Tomato plants (Solanum lycopersicum cv. Micro-Tom) were grown in a growth chamber at 25°C with a photoperiod 15 h: 9 h (light: dark), resulting in blooming after 1 month and a complete life cycle of about 4 months. Nicotiana benthamiana plants were grown in a growth chamber at 25°C with a photoperiod 13 h: 11 h (light: dark) until they reached about 30 cm in height before transient expression experiments. Both species were grown in a 4:1 mixture of peat and sand.
Total RNA was extracted from 50 to 100 mg frozen powdered plant material (tissue dependent) using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, St. Louis, United States). RNA quality was checked through NanoDrop 8,000 spectrophotometer (Thermo Fisher Scientific, Waltham, United States) and samples beyond the range Abs260/Abs280 = 1.8–2.2 and Abs260/Abs230 = 1.7–2.2 were mixed with one third of a volume of 7.5 M LiCl (Thermo Fisher Scientific) and resolubilized. RNA integrity was checked by 2% (w/v) agarose gel electrophoresis. Then, according to the manufacturers’ instructions, 2 μg total RNA was subjected to DNase treatment with Turbo DNase (Thermo Fisher Scientific) and used to prepare cDNA through the SuperScript™ III First-Strand Synthesis System (Thermo Fisher Scientific).
The tomato SlTDCs and SlT5H sequences were cloned by using specific primers (Supplementary Tables 2, 3) from the cDNA obtained from total RNA of leaves and flowers at anthesis sampled from 2-month-old Micro-Tom plants, and of mature-green fruits collected at 20 days post anthesis. The amplification products were directionally cloned into vector pENTR/D-TOPO for sequencing, and verified inserts were transferred to the binary overexpression vector pK7WG2 by LR recombination (Karimi et al., 2002).
Vectors containing the SlTDC and SlT5H sequences or the negative control marker GFP were introduced into Agrobacterium tumefaciens strain EHA105. A positive control containing the Actinidia chinensis TDC gene (AcTDC) was also available (Commisso et al., 2019) and the Oryza sativa T5H gene (GenBank AK071599) was prepared by GeneArt Gene Synthesis (Thermo Fisher Scientific). Transient expression was carried out as previously described (Gecchele et al., 2015). Briefly, three leaves of a 5-week-old N. benthamiana plant were syringe infiltrated with a bacterial suspension (OD600 = 0.9). Three plants were treated with each construct and one leaf (a biological replicate) was collected from each plant after 3 days, then another after 4 days, and another after 5 days. The leaves were immediately homogenized in liquid nitrogen, followed by extraction and untargeted metabolomics analysis by HPLC-ESI-MS as previously described (Commisso et al., 2019).
Chromatograms and mass spectra were processed using Data Analysis v3.2 (Bruker, Billerica, United States) and metabolites were identified based on m/z, retention time and MS/MS fragmentation pattern. The chromatograms were converted to netCDf files for peak alignment and area extraction using MZmine.3 The resulting feature matrix was analyzed using SIMCA v13.0 (Umetrics, Umea, Sweden). Pareto scaling was applied to all analytical methods. Unsupervised principal component analysis (PCA) was used to identify the major clusters defined by the samples before supervised orthogonal partial least squares discriminant analysis (OPLS-DA/O2PLS-DA). OPLS-DA/O2PLS-DA models were cross-validated by ANOVA with a threshold of p < 0.01.
Tomato plants at the flowering stage grown in the above-mentioned conditions were used for sampling roots, hypocotyl, cotyledons, first leaves following cotyledons, true leaves from six different nodes, the stem (separate internodes), flower buds and flowers at anthesis. Roots were further separated into the primary root, primary-proximal secondary root and primary-distal secondary root. Another group of plants was grown to complete the life cycle and, following fruit set, fruits were collected at various developmental and ripening phases based on phenological observation: immature-green (berry still growing), mature-green (fruit at maximum size), early breaker (green-white/yellow fruit), late breaker (red-orange turning), ripe and over-ripe. Immature-green and mature-green fruits were pooled for the analysis of exocarp (peel), mesocarp (flesh) and seeds, and the same strategy was applied to ripe and over-ripe fruits. All samples consisted of three biological replicates, each represented by a pool of material collected from five different plants. The samples were immediately frozen in liquid nitrogen and ground to a fine powder using an A11 basic analytical mill (IKA-Werke, Staufen, Germany). The powder was stored at −80°C.
We extracted 100 mg frozen powder representing each sample in 1 ml 2:1 (v/v) LCMS-grade chloroform-methanol (Honeywell, Charlotte, United States) then added 150 μl LCMS-grade ultrapure water (Honeywell) to allow phase separation. The samples were mixed vigorously for 30 s, sonicated at 40 kHz in a Sonica Ultrasonic Cleaner (Soltec, Milan, Italy) at 4°C for 15 min, and centrifuged (16,000 × g, 15 min, 4°C). The upper methanolic phase was recovered and the volume of each sample was recorded allowing appropriate dilution (according to the original tissue) in methanol and subsequent 1:2 dilution with an aqueous solution containing known amounts of the standards d4-tryptamine and d4-serotonin (Sigma-Aldrich). The final methanol-aqueous mixtures were passed through a Minisart RC4 filter (0.2 μm pores) and, for each extract, 1 μl was injected into an Acquity I Class UPLC system (Waters, Milford, United States) equipped with an Acquity UPLC BEH C18 column (Waters) kept at 30°C. The separation of the metabolites in the extracts was performed via reverse phase chromatography through establishment of a 20-min polarity gradient among two mobile phases (A: 0.1% formic acid in water, B: acetonitrile), which flowed at 0.350 ml min−1. The initial conditions were 99% A and 1% B, and the following elution profile was applied: 0–1 min, 1% B; 1 min – 10 min, 1–40% B; 10 min – 13 min 30 s, 40–70% B; 13 min 30 s – 14 min, 70–99% B; 14 min – 16 min, 99% B; 16 min – 16 min 6 s, 99–1% B (initial conditions). The system was equilibrated in 99% A and the elution was complete after 20 min.
The UPLC was connected online to a Xevo G2-XS QToF mass spectrometer (Waters) featuring an electrospray ionization (ESI) source operating in positive ionization mode within the range 50–2000 m/z. The ion source parameters were: capillary = 0.8 kV, sampling cone = 40 V, source offset = 80 V, source temperature = 120°C, desolvation temperature = 500°C, cone gas flow rate = 50 l h−1 and desolvation gas flow rate = 1,000 l h−1. Nitrogen gas was used for the nebulizer and desolvation, whereas argon was used to induce collision-induced dissociation. An MS method was created to acquire data in continuum mode using a fixed collision energy in two scan functions. In function 1, the low energy was disabled, whereas in function 2, the high energy was set to 35 V. In both functions, the Xevo G2-XS was set to perform the analysis with a scan time of 0.3 s. The lock mass solution used as “calibrator” to verify MS accuracy consisted of a 100 pg. μL−1 leucine-enkephalin solution (Waters) injected at a flow rate of 10 μl min−1, generating a signal of 556.2771 m/z in positive mode and 554.2615 m/z in negative mode. A quality control (QC) sample was prepared by mixing equal parts of each sample in order to check UPLC-QToF performance throughout the experiment. MassLynx v4.1 (Waters) was used to manually extrapolate the peak areas relative to the tryptamine and serotonin signals and the corresponding deuterated commercially authentic standards. Peak extrapolation was based on the following m/z values, previously chosen after analysis of the commercial standards and that corresponded to the highest in-source generated fragments detected in positive ionization mode: 160.0777 for serotonin and 164.1009 for d4-serotonin; 144.0863 for tryptamine and 148.1061 for d4-tryptamine. The concentrations of endogenous tryptamine and serotonin in the samples were determined from the area ratios of endogenous to corresponding stable isotope-labeled standards. Previously, to assess the linearity of the method, two calibration curves (R2 = 0.99) were built by analyzing different solutions with increasing amounts (1–1,000 pg) of d4-tryptamine and d4-serotonin.
Total RNA was extracted and cDNA synthesized as reported above. A 1:10 dilution of the cDNA solution was used for qRT-PCR analysis to quantify the levels of SlTDC1, SlTDC2, SlTDC3 and SlT5H mRNA (primers are reported in Supplementary Table 4). A 25-μL reaction was performed in triplicate for each sample in 96-well plates using the GoTaq qPCR Master Mix (Promega, Madison, United States) according to manufacturer’s instructions. A cDNA pool representing all samples was also tested to normalize expression data across plates. The qRT-PCR was performed in a StepOne Plus thermocycler (Applied Biosystems, Foster City, United States) setting the following cycling conditions: initial denaturation at 95°C for 2 min followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s and extension at 60°C for 30 s. The amplification process was followed by melting curve analysis (55–95°C) with temperature increments of 0.3°C every 15 s. Raw data were processed with LinRegPCR software to determine baseline and threshold cycles (Ct). Transcript levels were expressed as mean normalized expression (Muller et al., 2002) by Ct comparison with the SlCAC calibrator gene (Solyc08g006960.2.1; González-Aguilera et al., 2016). Gene expression and metabolomic data were integrated by evaluating the Pearson correlation coefficient in Microsoft Excel (Microsoft, Redmond, United States).
Several tomato TDC orthologs (SlTDCs) have been predicted and one of them, SlTDC1 (Solyc07g054860), was recently shown to produce tryptamine (Tsunoda et al., 2021). We used the amino acid sequences of well-characterized plant TDCs for homology-based screening of the Sol Genomics Network database (SGNdb) to further identify three tomato proteins characterized by high percentage identity scores (Table 1) that we annotated as SlTDC2 (Solyc07g054280), SlTDC3 (Solyc09g064430) and SlTDC4 (Solyc03g044120).
The alignment of SlTDCs candidates (Supplementary Figure 1) with other functionally characterized plant TDCs (Supplementary Table 1) revealed the presence of many conserved regions necessary to the decarboxylation reaction including the sites for substrate selectivity and cofactor (pyridoxal-5′-phosphate, PLP) binding (Torrens-Spence et al., 2020; Zhou et al., 2020). The histidine and tyrosine residues in the first and second putative catalytic sites are considered key requirements for L-tryptophan decarboxylation, and are highly conserved in SlTDC1, SlTDC2 and SlTDC3. Contrarily, the tyrosine was replaced by phenylalanine in SlTDC4, in which, moreover, the PLP-binding site was completely missing and the glycine in the putative substrate selection site was replaced with serine. Importantly, glycine at this site facilitates specific binding to indole structures whereas serine binds phenyl amino acids, therefore being necessary to distinguish between tryptophan and tyrosine substrates.
To investigate the phylogenetic relationships between the three putative SlTDCs, SlTDC1 and other plant AADCs, we constructed a phylogenetic tree (Supplementary Figure 2) including the known functionally annotated plant TDCs and tyrosine decarboxylases (TyDCs). SlTDC1 and SlTDC2 clustered in the exclusive TDC family clade, showing close relationships with TDCs from other solanaceous species, whereas SlTDC3 and SlTDC4 did not. SlTDC3 clustered with PepTDC2 and OsTDC2, but this group was closer to functionally characterized TyDCs than other TDCs and was the only group of functionally characterized TDCs excluded from the TDC clade. SlTDC4 clustered in another TyDC clade, close to TyDCs from Petroselinum crispum and Papaver somniferum.
The SlTDC1, SlTDC2 and SlTDC3 CDS that we cloned (Supplementary Information 1) from different Micro-Tom organs were found to be identical to those deposited for cv. Heinz 1706 in SGNdb (Table 2). In contrast, the amplification of SlTDC4 from Micro-Tom total cDNA did not yield a PCR product, suggesting this gene is not expressed in Micro-Tom plants under standard growing conditions. This hypothesis was supported by the negligible expression levels of SlTDC4 reported in Heinz 1706 microarray (Consortium, 2012) and Micro-Tom RNA-Seq (TomExpress) expression datasets (Supplementary Figure 3).4 The SlTDC4 sequence was therefore amplified from Micro-Tom genomic DNA (Supplementary Information 2).
To assess the function of the Micro-Tom SlTDC candidates, we developed an in vivo functional assay based on transient expression of the SlTDC genes in Nicotiana benthamiana, which does not produce detectable levels of tryptamine or serotonin in its aerial organs. The constructs harboring the CDS of SlTDC1, SlTDC2, SlTDC3 and SlTDC4 under the control of the strong constitutive cauliflower mosaic virus 35S (CaMV35S) promoter were introduced via Agrobacterium tumefaciens into N. benthamiana plants. Leaf methanol extracts were analyzed by HPLC-ESI-MS and two datasets corresponding to negative and positive ionization were created for multivariate statistical analysis. PCA and OPLS-DA of the negative data matrix revealed no clustering among the samples. In contrast, PCA (Figure 1A) and corresponding OPLS-DA (Figure 1B) of the positive data matrix revealed two distinct groups, with the SlTDC1 and SlTDC2 infiltrated plants in one cluster and the SlTDC3 and SlTDC4 infiltrated plants in the other, along with the negative control expressing green fluorescent protein (GFP). OPLS-DA highlighted specific metabolites that were highly correlated with each group (Figure 1C). The in-source fragment and molecular ion of tryptamine were the metabolites that correlated best with the SlTDC1 and SlTDC2 infiltrated plants (Figure 1D) followed by serotonin and an unidentified metabolite (Supplementary Figure 4). Conversely, the clustering of the SlTDC3 and SlTDC4 infiltrated plants with the GFP control indicated similar metabolic profiles in all three cases. Even so, the SlTDC3 infiltrated plants accumulated small amounts of tryptamine and serotonin, whereas neither of these metabolites was detected at significant levels in leaves transiently expressing SlTDC4 (Figure 1D; Supplementary Figure 4), suggesting that this gene does not function as a TDC (further evidence is presented in Supplementary Information 3).
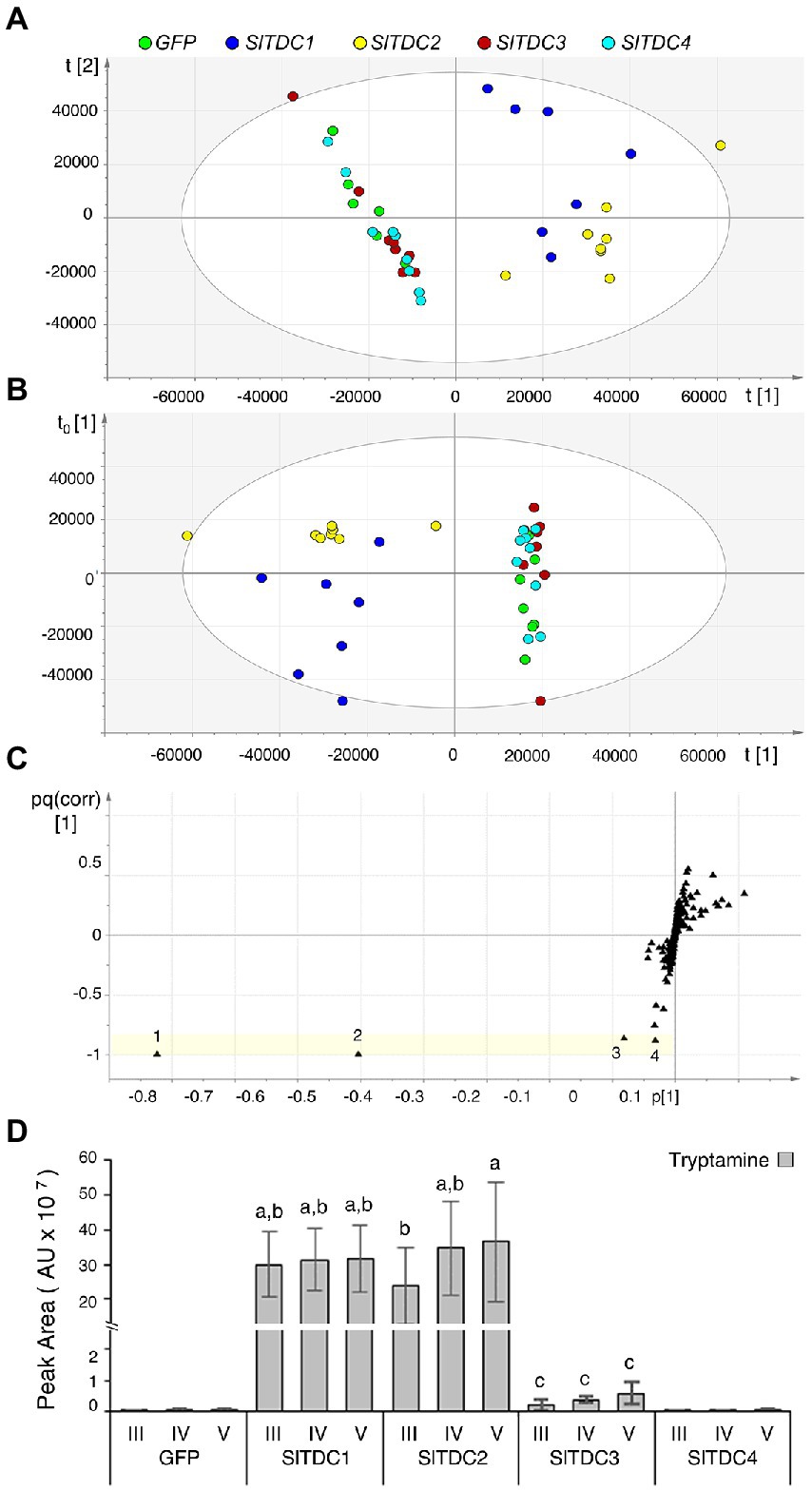
Figure 1. Multivariate statistical analysis of the untargeted metabolomics dataset (positive ionization) following the HPLC-ESI-MS analysis of Nicotiana benthamiana plants transiently expressing individual SlTDC genes or GFP as a negative control. (A) PCA score scatter plot. (B) OPLS-DA score scatter plot. (C) OPLS-DA S-loading plot, with each metabolite represented by a black triangle. The yellow area highlights metabolites characteristic of the SlTDC1/SlTDC2 cluster but not the GFP/SlTDC3/SlTDC4 cluster: 1 and 2 represent molecular and fragment ions of tryptamine (161 and 144 m/z, respectively), 3 is a molecular ion of serotonin (177 m/z), and 4 is an unidentified metabolite (177 m/z). (D) Levels of tryptamine in the infiltrated leaves. AU = arbitrary unit. Data are means ± standard deviations (n = 3). Different letters on bars indicate that means are significantly different according to post-hoc Tukey-ANOVA (p < 0.05).
The high levels of serotonin detected in tomato, as reported by recent literature, suggest the existence of a SlT5H that converts tryptamine to serotonin. We therefore sought candidate tomato T5H genes by using the previously characterized OsT5H (Fujiwara et al., 2010) as a blastp search query against SGNdb. This recovered Solyc09g014900.2 (SlT5H), which shares 55.3% identity with OsT5H and is annotated as a cytochrome P450 monooxygenase predicted by Predotar to be localized to the ER. Repeating the strategy deployed for TDC (see Supplementary Information 1), we used the SlT5H CDS as a blastn query against the KaFTom database and recovered a sequence with 99.93% identity (LEFL1021DE05). The sequences were translated in silico and the alignment revealed only one substitution. The SlT5H CDS deposited in SGNdb was cloned from cDNA prepared from mature-green Micro-Tom fruits (Table 2) and transferred to vector pK7WG2 for functional characterization in N. benthamiana plants.
Plants were infiltrated simultaneously with pK7WG2.SlTDC1 and pK7WG2.SlT5H whereas plants infiltrated with either of the individual genes were considered as negative controls. Plants infiltrated with SlTDC1 and OsT5H (the bona fide rice T5H gene) were used as positive controls. Methanolic extracts were analyzed by HPLC-ESI-MS and the untargeted metabolomics datasets were explored by PCA and O2PLS-DA as above. PCA of the positive data matrix resolved three distinct clusters: plants expressing SlTDC1; plants expressing SlT5H; and plants co-expressing SlTDC1 and OsT5H or SlTDC1 and SlT5H (Figure 2A). The O2PLS-DA score scatter plot (Figure 2B) confirmed the PCA clustering and identified molecules that correlated with each of the clusters (Figure 2C). Serotonin correlated solely with the SlTDC1 + OsT5H and SlTDC1 + SlT5H plants (Figure 2D), along with an unidentified metabolite (222 m/z) that was present at much lower levels (Supplementary Figure 5). Our data confirmed that SlT5H converts tryptamine to serotonin and can thus be considered as a bona fide T5H. As above, analysis of the negative data matrix revealed no clustering among the samples.
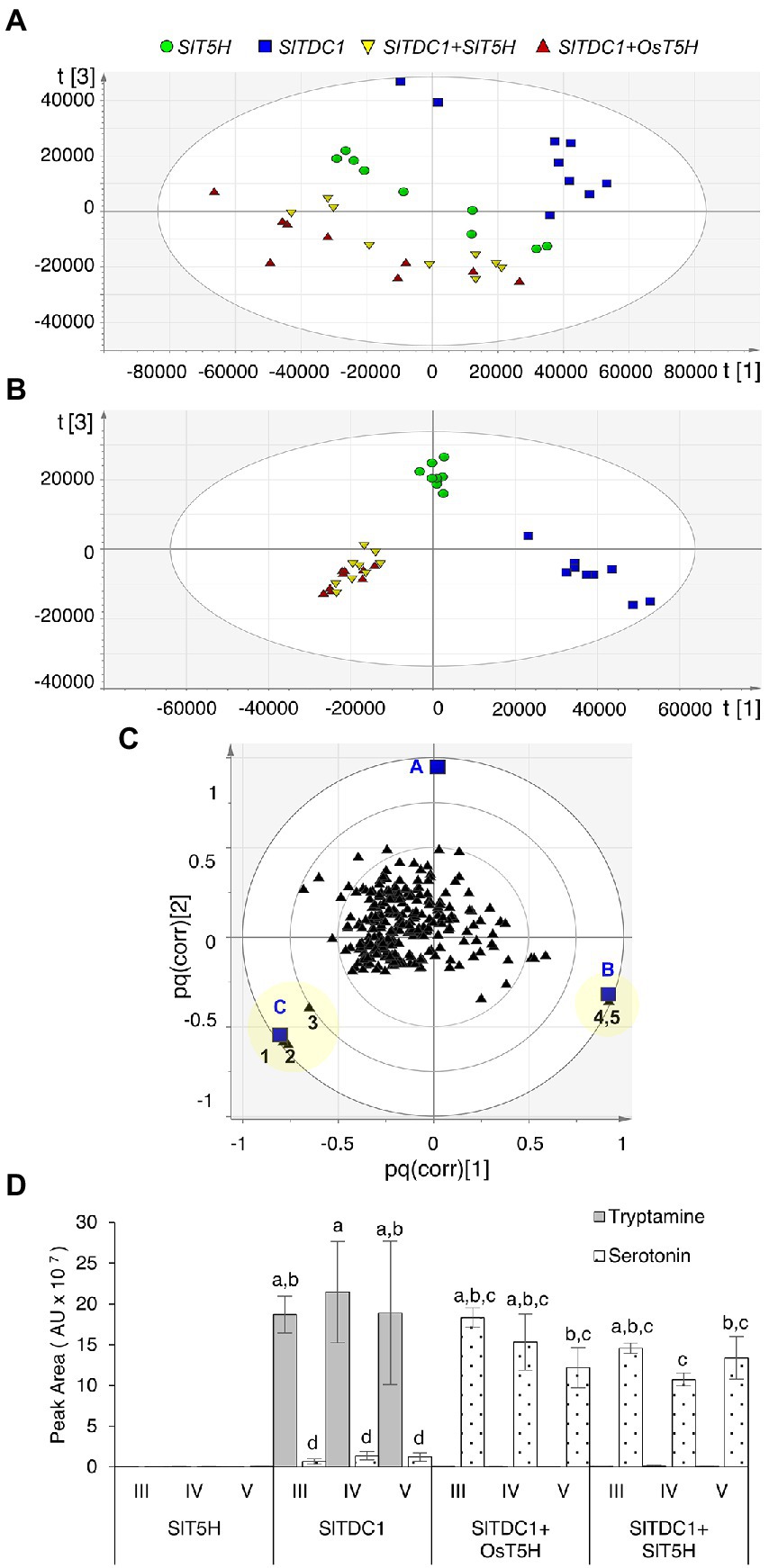
Figure 2. Multivariate statistical analysis of the untargeted metabolomics dataset (positive ionization) following the HPLC-ESI-MS analysis of Nicotiana benthamiana plants transiently expressing SlTDC1 and T5H genes. (A) PCA score scatter plot. (B) O2PLS-DA score scatter plot. (C) O2PLS-DA correlation loading plot, with each metabolite represented by a black triangle and classes by blue squares (A = SlT5H, B = SlTDC1, and C = SlTDC1 + OsT5H and SlTDC1 + SlT5H). The yellow area highlights metabolites characteristic of each class: 1 and 2 represent molecular and fragment ions of serotonin (177 and 160 m/z, respectively), 3 is an unidentified metabolite (222 m/z), and 4 and 5 are molecular and fragment ions of tryptamine (161 and 144 m/z, respectively). (D) Levels of tryptamine and serotonin. AU = arbitrary unit. Data are means ± standard deviations (n = 3). Different letters on bars indicate that means are significantly different according to post-hoc Tukey-ANOVA (p < 0.05).
Having discovered multiple TDC genes in tomato, we explored their functional diversification by gene expression profiling and the targeted analysis of tryptamine and serotonin in various Micro-Tom organs collected at different stages (Figure 3).
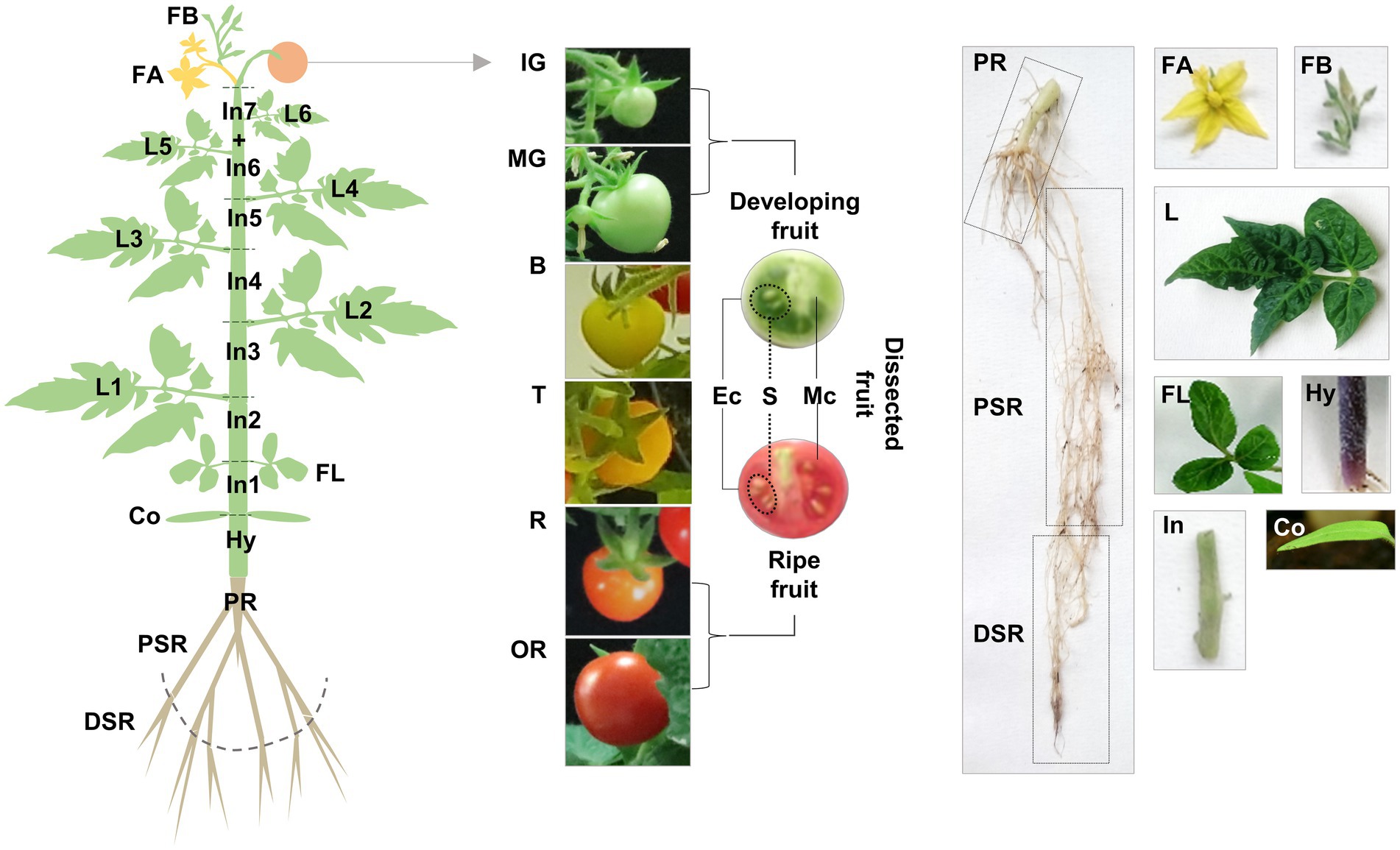
Figure 3. Schematic representation of our sampling strategy for gene expression profiling and targeted metabolomics. Vegetative organs: roots (PR, primary root; PSR, proximal secondary root; DSR, distal secondary root), stem (Hy, hypocotyl; In, internode), cotyledons (Co), leaves (FL, first leaves; L, true leaves). Reproductive organs: flowers (FB, flower buds; FA, flowers at anthesis), fruit (IG, immature-green; MG, mature-green; B, breaker; T, late breaker/turning; R, ripe; OR, over-ripe), dissected fruit (Ec, exocarp; Mc, mesocarp; S, seeds).
We observed major differences in the levels of both indolamines in the vegetative organs, revealing distinct accumulation patterns (Supplementary Table 5; Figure 4). Serotonin levels in some organs were up to 150-fold higher than tryptamine levels, with the total concentration ranging from 27.19 to 9322.02 ng g−1 FW for serotonin and 1.60 to 337.60 ng g−1 FW for tryptamine. The lowest levels of both compounds were detected in the roots, with opposing profiles (tryptamine increasing, serotonin decreasing) moving from the primary embryonal root to the distal part of the secondary roots. The highest levels of both compounds were detected in the leaves, with distinct trends in the older basal leaves and the younger ones close to the shoot apex. Tryptamine accumulation increased in a gradient along the longitudinal axis of the plant, from the first leaves to the leaves of the last upper node (Figure 4A), whereas serotonin accumulation increased until the leaves of the fourth node and then sharply dropped in the leaves of the fifth and sixth nodes (Figure 4B). Interestingly, a serotonin gradient opposite to that observed in the leaves was detected in the stem, with the levels declining from the first to the last internodes (Figure 4B). In flowers, serotonin was approximately six-fold more abundant than tryptamine, but we observed no significant change in abundance between the flower bud stage and fully-open flower stage.
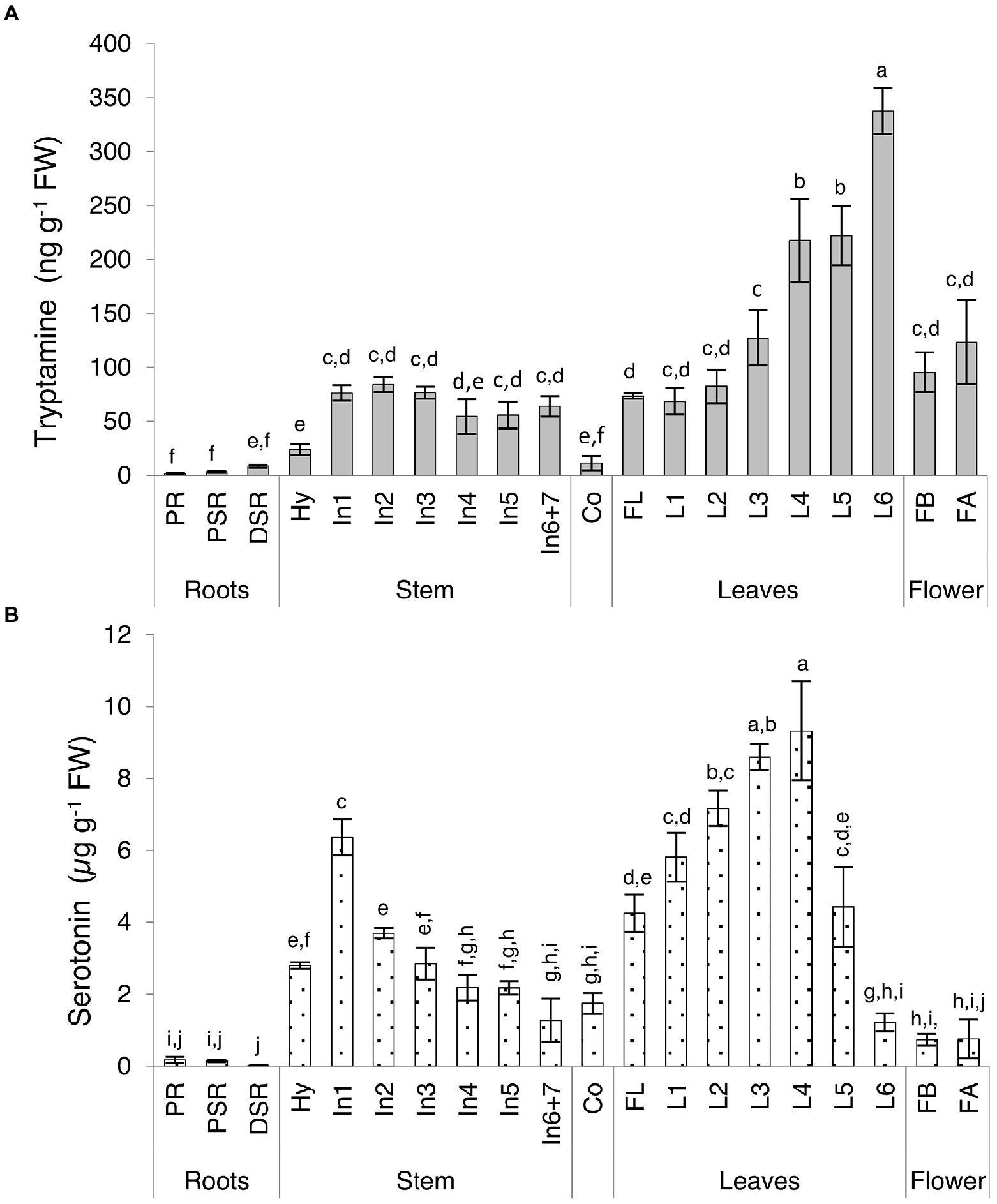
Figure 4. Quantities of (A) tryptamine and (B) serotonin detected in Micro-Tom vegetative organs and flowers by UPLC-ESI-MS analysis expressed as (A) ng g−1 FW or (B) μg g−1 FW. Data are means ± SD (n = 3). Different letters on bars indicate that means are significantly different according to post-hoc Tukey-ANOVA (p < 0.05). The abbreviations and the position of the samples are explained in Figure 3.
Tryptamine and serotonin reached similar levels in the Micro-Tom fruit, in contrast to the other organs (Supplementary Table 6; Figure 5A). Tryptamine was 10-fold more abundant than serotonin at the immature-green stage and levels slightly increased at the mature-green stage in concert with rapid fruit growth, before leveling off during ripening and declining slightly in the over-ripe fruit. Serotonin levels increased rapidly between the immature and mature-green fruit stages, becoming more or less equal to tryptamine in the mature-green fruit and then continuing to increase until the breaker stage (onset of ripening), then leveling off and remaining fairly constant until senescence. By normalizing the quantity of tryptamine and serotonin to the average fresh weight of the fruits at each stage, we were able to express the indolamine content per whole fruit (Supplementary Table 6; Figure 5B). The fully-ripe berry fruit contained ~5 μg of tryptamine and ~ 14 μg of serotonin.
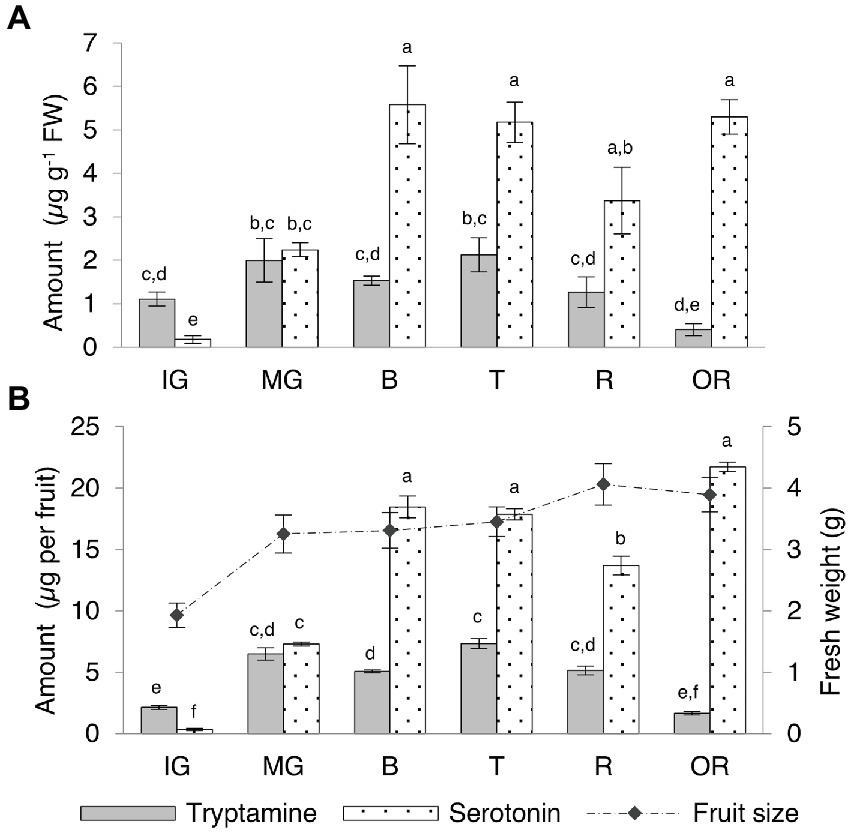
Figure 5. Quantities of tryptamine and serotonin detected by UPLC-ESI-MS in Micro-Tom fruits at different developmental and ripening stages expressed as (A) μg g−1 FW and as (B) μg per whole berry. Data are means ± SD (n = 3). Different letters on bars indicate that means are significantly different according to post-hoc Tukey-ANOVA (p < 0.05). The abbreviations and the position of the samples are explained in Figure 3.
We also divided the unripe (immature-green and mature-green) and fully-ripe (ripe and over-ripe) berries into different tissues (exocarp, mesocarp and seeds) for further investigation (Supplementary Table 7; Figure 6). Seeds contained the highest levels of tryptamine and serotonin and a slight increase in serotonin was observed as the fruit ripened. Intermediate and high levels of tryptamine and serotonin, respectively, in the mesocarp were observed to decrease during the ripening process, but serotonin was consistently twice as abundant as tryptamine. The lowest levels of both compounds were detected in the skin, but serotonin was four-fold more abundant than tryptamine at the unripe stage and twice as abundant in the ripe fruit.
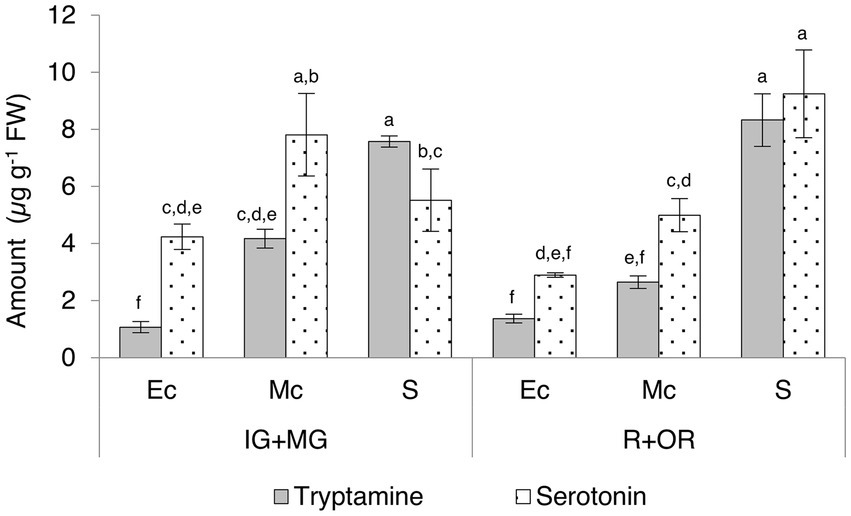
Figure 6. Quantities of tryptamine and serotonin detected by UPLC-ESI-MS in the skin (exocarp, Ec), flesh (mesocarp, Mc) and seeds (S) of unripe (IG + MG) and ripe (R + OR) Micro-Tom fruits, expressed as μg g−1 FW. Data are means ± SD (n = 3). Different letters on bars indicate that means are significantly different according to post-hoc Tukey-ANOVA (p < 0.05). The abbreviations and the position of the samples are explained in Figure 3.
RNA extracted from the same samples used for targeted metabolomics was used to determine the expression profiles of the SlTDC and SlT5H genes by quantitative real-time PCR (qRT-PCR). SlTDC1 was expressed solely in the reproductive organs (Figure 7A), with low levels of mRNA in the flowers and immature-green berry, but a burst of expression toward the end of the mature-green stage. This peak was followed by a drop from the breaker stage to the over-ripe fruit. The highest levels of SlTDC1 mRNA were present in the exocarp of the developing berry and, to a lesser extent, in the ripe fruit. The transcript levels in the seeds remained remarkably low.
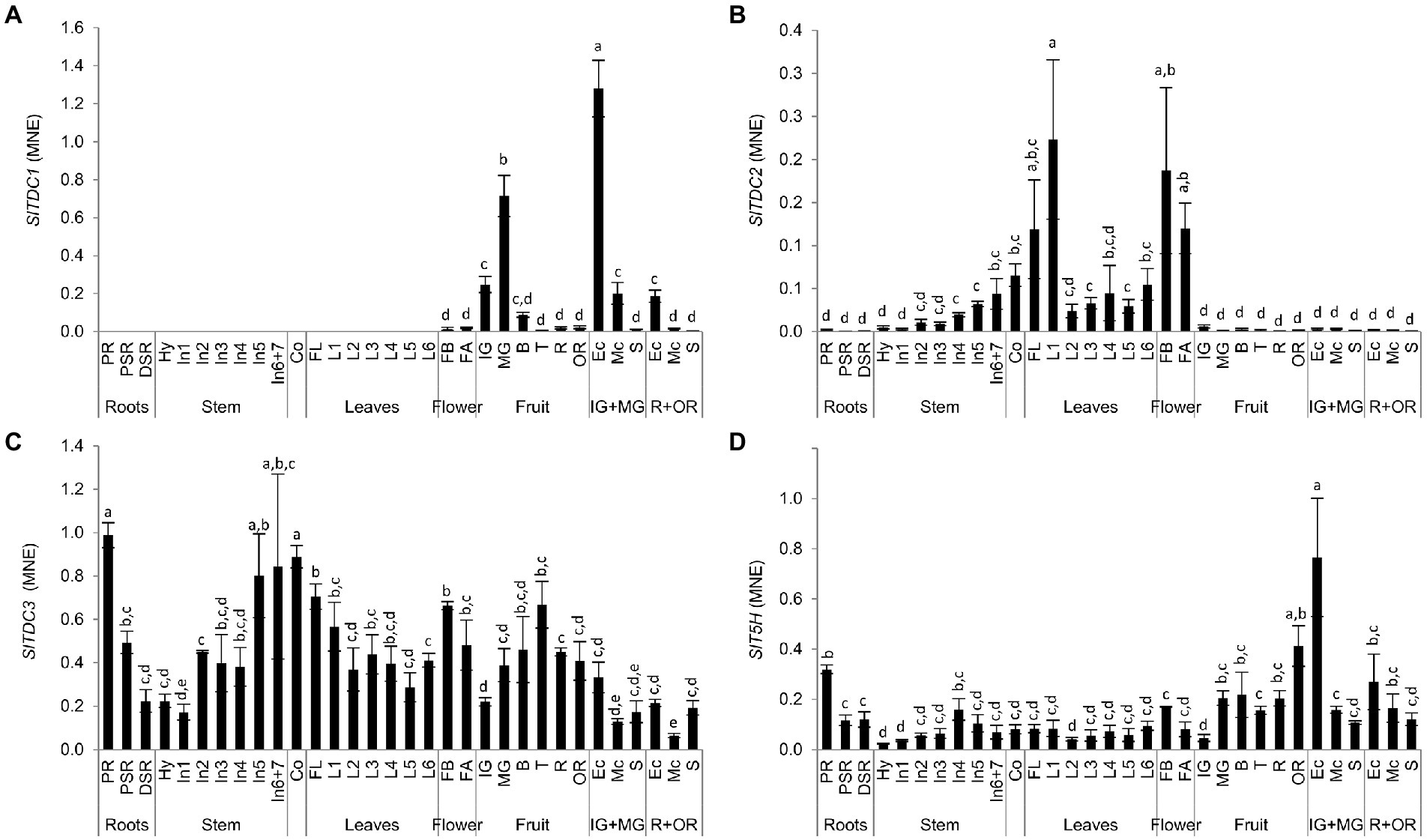
Figure 7. Analysis of (A) SlTDC1, (B) SlTDC2, (C) SlTDC3 and (D) SlT5H expression in Micro-Tom organs by qRT-PCR. Data are means ± SD (n = 3). MNE = Mean normalized expression. Different letters on bars indicate that means are significantly different according to post-hoc Tukey-ANOVA (p < 0.05). The abbreviations and the position of the samples are explained in Figure 3.
In contrast, SlTDC2 was expressed mainly in the aerial vegetative organs and flowers with minimal levels in the primary root and fruit (Figure 7B). Interestingly, we observed an SlTDC2 expression gradient in the stem, with the lowest levels in the hypocotyl and basal internodes and the highest levels in the last internodes. SlTDC2 was expressed strongly in the flowers, cotyledons and oldest leaves of the plant, followed by a sharp decrease immediately after the first node.
In contrast to the restricted profiles of SlTDC1 and SlTDC2, SlTDC3 was expressed in all plant organs (Figure 7C). However, we still observed opposing expression gradients in the stems and leaves, with increasing transcript levels from the hypocotyl to the upper internodes but higher levels in older than younger leaves. SlTDC3 was strongly expressed in the primary root and lower but still significant mRNA levels were detected in the secondary roots. SlTDC3 expression also increased in fruits following the end of berry development and peaked at the turning stage, complementing the low levels of SlTDC1 mRNA at this stage. SlTDC3 mRNA levels were higher in the skin and seeds of ripe berries than the flesh.
As SlTDC3, also SlT5H was ubiquitously expressed (Figure 7D). Among the vegetative organs, the highest SlT5H mRNA levels were observed in the primary root. A slight increase in expression was observed in the stem from the hypocotyl to the fourth internode, followed by a decrease in the younger stem portion, thus mirroring the distribution of serotonin in the leaves. SlT5H mRNA levels were similar in all leaves and the cotyledons, but were slightly higher in the flower buds than in the flower at anthesis. SlT5H was expressed throughout fruit development and ripening, peaking at the mature-green stage and then remaining constant until the over-ripe stage, when a further increase was observed. Notably, as observed for SlTDC1, the highest levels of SlT5H mRNA levels were detected in the exocarp of unripe berries.
To better represent these data, we generated heat map atlases of gene expression and indolamine levels in the vegetative organs (Figure 8A) and developing/ripening fruit (Figure 8B).
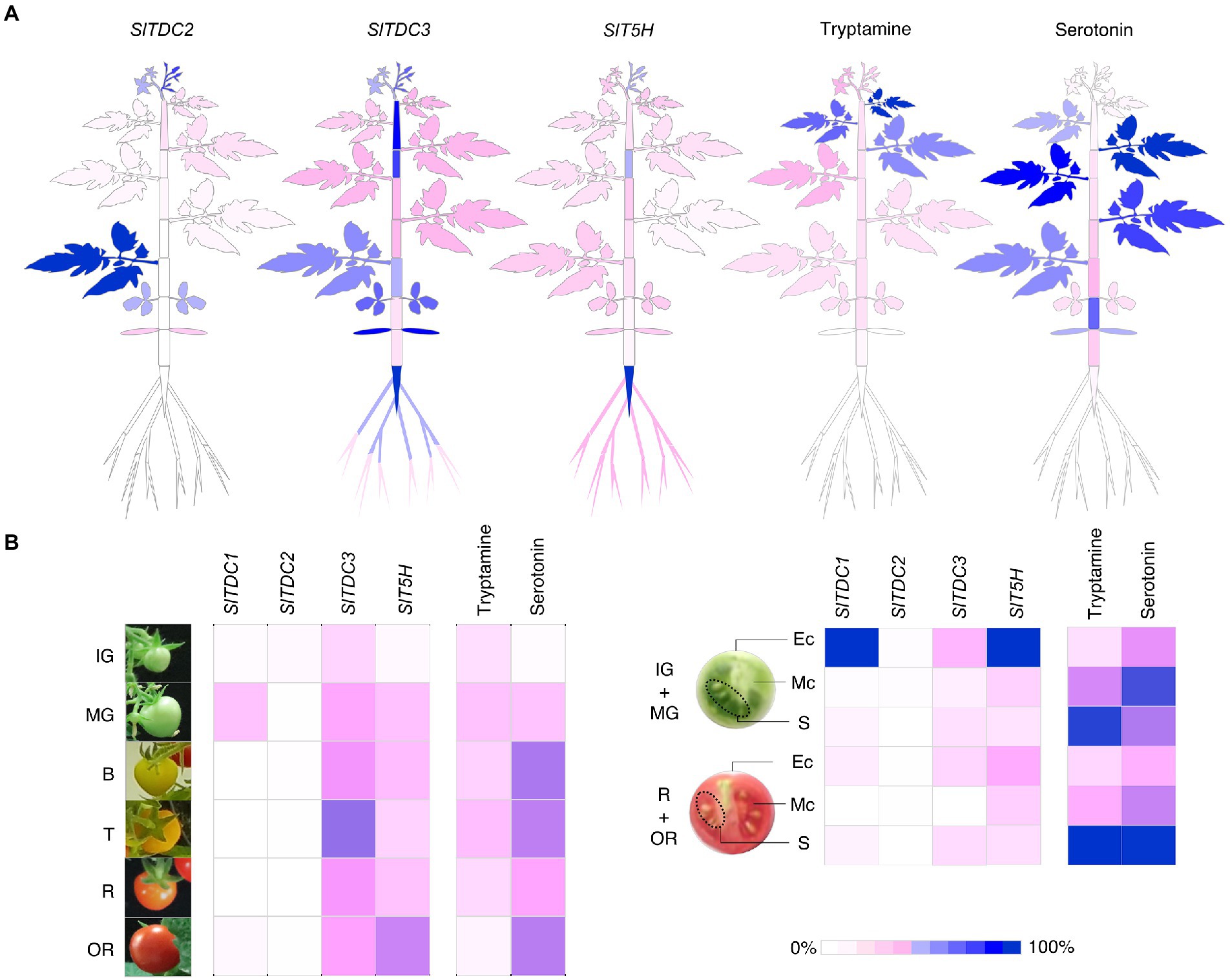
Figure 8. Heat maps representing the percentage relative expression of SlTDCs and SlT5H genes and the accumulation of tryptamine and serotonin in Micro-Tom plants (A) and fruits (B). Expression levels are shown as percentages relative to the organ with the highest expression (100%). Each organ and stage/tissue identified in this heat map refers to the sampling design in Figure 3.
To gain insight into the relationship between indolamine accumulation and TDC and T5H gene expression in tomato, we looked for linear correlations between the gene expression profiles and metabolites by calculating Pearson correlation coefficients (PCCs; Supplementary Figure 6). This revealed a positive correlation between SlTDC3 and SlT5H expression in the roots and the production of serotonin, and a negative correlation between these three factors and the accumulation of tryptamine, suggesting that SlTDC3 and SlT5H expression results in the depletion of tryptamine in the roots and its conversion to serotonin. In the basal leaves (from the cotyledons to the leaves of the third node), we observed a positive correlation between tryptamine and serotonin levels and between SlT5H and SlTDC2 expression. The levels of both indolamines increased along the shoot axis toward the apex although the tryptamine gradient was shallower than that of serotonin. These profiles showed a negative correlation with the expression of SlTDC3. On the other hand, in the upper leaves (between the fourth and sixth nodes), we observed a strong negative correlation between tryptamine and serotonin levels, reflecting the gradient of serotonin in leaves up to the fourth node followed by a rapid drop-off in leaves next to the shoot apex. In these organs, we also observed a positive correlation between SlTDC3 and SlT5H expression that appeared unrelated to indolamine accumulation. No clear relationships were observed in the fruit until PCCs were calculated by distinguishing the phases of fruit development and ripening. In unripe berries (analysis based on IG, MG and B samples), SlTDC1 expression showed strong positive correlation with the accumulation of tryptamine whereas SlTDC3 and SlT5H expression correlated with the accumulation of serotonin. During the ripening phase (analysis based on T, R and OR samples), tryptamine accumulation was strongly negatively correlated with SlTDC1 expression, which was near zero in all but the OR samples, and weakly positively correlated with the expression of SlTDC3. Tryptamine also showed a strong negative correlation with SlT5H expression. Interestingly, SlT5H expression was not positively correlated with serotonin.
In this work, we have provided new evidence for the functional characterization of tomato genes that were recently proposed to be involved in tryptamine and serotonin production.
Two in silico studies suggested the existence of two up to five TDCs in tomato (Hano et al., 2017; Pang et al., 2018), and the function of one isoform (SlTDC1) has already been assessed in this species (Tsunoda et al., 2021). By using homology-based screening we identified three SlTDCs candidates characterized by high percentage identities to SlTDC1 and other plant TDCs. With the exception of SlTDC4, all SlTDCs were found to share key residues required for PLP binding, tryptophan selection and catalytic activity (Torrens-Spence et al., 2020; Zhou et al., 2020). Phylogenetic analysis revealed that SlTDC1 and SlTDC2 clustered with other solanaceous TDCs whereas SlTDC4 was phylogenetically closer to functionally characterized TyDCs. However, these automatic homology-based classifications are not always reliable due to the similarity between TDCs and TyDCs (Abu-Zaitoon, 2014). For example, SlTDC3 clustered with the characterized TDCs from rice and bell pepper, which show closer phylogenetic proximity to TyDCs than to other plant TDCs. Homology-based screening is thus a useful but insufficient tool to assign gene functions. We therefore developed an in planta assay system for direct functional characterization of the SlTDC candidates to confirm their TDC activity.
The transient expression of SlTDCs in N. benthamiana, a solanaceous species that does not accumulate detectable levels of tryptamine or serotonin in its leaves, revealed that SlTDC1, SlTDC2 and SlTDC3 were bona fide TDCs because the infiltrated plants accumulated tryptamine. Interestingly, the lower tryptamine levels observed for SlTDC3 suggest lower enzyme activity or lower substrate affinity for this candidate. Although substrates other than tryptophan might be preferred by SlTDC3, we detected no metabolites exclusive to the SlTDC3 infiltrated plants, providing strong evidence for a specific TDC function.
Tomato plants accumulate serotonin in a spatiotemporally regulated manner suggesting the presence of an enzyme that synthesizes serotonin from tryptamine (Islam et al., 2016; Hano et al., 2017) orthologous to the OsT5H gene previously discovered in rice (Fujiwara et al., 2010). Both the OsT5H gene and its putative tomato ortholog represent the CYP71 group of cytochrome P450 monooxygenases. Through untargeted metabolomics analysis we revealed the accumulation of serotonin in N. benthamiana plants infiltrated with both SlTDC1 and SlT5H, thus confirming the function of SlT5H. Moreover, tryptamine was detected in only trace amounts as a residual substrate in these plants, indicating a high rate of conversion by SlT5H.
We investigated the spatiotemporal distribution of tryptamine and serotonin in Micro-Tom fruits as well as the vegetative and reproductive organs from early development to senescence, thus adding more details to the information already available for this species (Udenfriend et al., 1959; West, 1959; Feldman and Lee, 1985; Hano et al., 2017). Targeted UPLC-ESI-MS analysis revealed the organ-specific accumulation of these compounds in whole plants and their tissue-specific accumulation in fruits. Tryptamine accumulated to high levels only in the reproductive organs (up to ~8.33 μg g−1 FW in the seeds) whereas, in accordance with recent literature (Negri et al., 2021), levels were much lower in the rest of the plant (up to ~0.34 μg g−1 FW in younger leaves). In contrast, yet as commonly observed in many plant species (Erland et al., 2016), serotonin also accumulated to high levels in some vegetative organs (up to ~9.32 μg g−1 FW) hinting that these indolamines serve different tissue-specific purposes. The large quantities of serotonin detected in all plant tissues, and the accumulation of tryptamine in the flowers, fruit and seeds, suggest that neither indolamine is a mere intermediate but also an end-product with a specific biological function.
The measurement of tryptamine and serotonin levels in leaves at each internode along the longitudinal axis of Micro-Tom plants revealed a gradient of both molecules. Serotonin increased from low levels at the basal nodes, peaking at the fourth node and then sharply declining in the leaves close to the terminal bud, confirming the gradient is not just a consequence of dilution due to the expansion of older leaves. In contrast, tryptamine levels increased continuously and were highest in the top two leaves. Several reports suggest that tryptamine interferes with insect reproduction by inhibiting oviposition, feeding and/or larval development (Thomas et al., 1998, 1999; Gill et al., 2003). The higher tryptamine content in young tomato leaves may serve as a deterrent to protect these developing photosynthetic organs, which are more attractive to insects. Interestingly, although tryptamine levels were constant along the stem, we observed a gradient of serotonin with opposite polarity to the gradient in leaves. Concentration gradients in the stem have already been reported for phytohormones including auxin (Kojima et al., 2002), abscisic acid (Everat-Bourbouloux and Charnay, 1982) and cytokinins (Eklöf et al., 1996), yet at much lower levels than serotonin (10–0.1 ng g−1 FW).
Many berries accumulate tryptamine and serotonin at similar or higher levels than tomato (Erland et al., 2016; Islam et al., 2016) with some fruits, including banana, kiwifruit, grape and pineapple, showing no specific developmental trends in indolamine levels (Udenfriend et al., 1959; Foy and Parratt, 1960; Murch et al., 2010; Commisso et al., 2019). We found that serotonin and tryptamine accumulate in developing Micro-Tom fruits, with tryptamine reaching a plateau at the mature-green stage, just before the onset of ripening, and serotonin reaching a plateau at the breaker stage. Evidence that the indolamines do not accumulate merely as a consequence of development but are involved in its regulation comes from the overexpression of SlTDC1 in tomato, which produced serotonin-rich fruits with a shorter transition to the breaker stage compared to wild-type plants (Tsunoda et al., 2021). The authors proposed an interaction between serotonin and the ethylene pathway that regulates berry ripening in tomato, but further investigation is required.
We also measured the accumulation of indolamines in different parts of the unripe and ripe berry. The unripe and ripe seeds accumulated large amounts of tryptamine and serotonin whereas moderate levels were detected in the mesocarp and low levels in the exocarp. This partially agrees with previous studies (Feldman and Lee, 1985; Hano et al., 2017) with differences, in this case, attributed to the different cultivars analyzed or even to different growing practices, which have been recently demonstrated to affect indolamine content in tomato (Lee et al., 2021).
The gene expression profiling that we performed though our sophisticated experimental design revealed complementary expression trends among the SlTDC genes to coordinate tryptamine biosynthesis with an organ-specific distribution. SlTDC1 was expressed only during the reproductive phase. Conversely, SlTDC2 was strongly expressed in the aerial vegetative organs and flowers but not in the fruit. SlTDC3 was expressed at different levels in all organs, and was the only SlTDC gene active in the roots. It must be considered that SlTDC1 and SlTDC2 were much more efficient than SlTDC3 in the conversion of tryptophan to tryptamine in our heterologous system. Therefore, even the low levels of SlTDC1 and SlTDC2 expression in the flowers, shoots and fruit tissues may be more relevant to tryptamine production than the expression profiles suggest, whereas the ubiquitously expressed but less active SlTDC3 may play a different role in determining tryptamine levels. In this context, tryptamine levels in tomato appear to be finely regulated by different mechanisms, suggesting that it is not merely a metabolic intermediate for the production of serotonin, melatonin and their derivatives but may also perform multiple biological functions.
In plants with at least two TDC genes (e.g., bell pepper and Camptotheca acuminata), one of the genes tends to be responsive to stress (López-Meyer and Nessler, 1997; Park et al., 2009). In rice, very low level constitutive TDC gene expression is boosted by nutrient depletion, which induces senescence in the leaves (Kang et al., 2009). Adding to the previous work on TDC genes in tomato (Hano et al., 2017; Tsunoda et al., 2021), we show for the first time that the tomato TDC gene family is also regulated in a complex manner, with different paralogs showing spatiotemporally regulated expression profiles.
In contrast to the intricate regulation of TDC genes, we found that SlT5H was expressed in all Micro-Tom organs, as previously observed in tomato (Hano et al., 2017) and rice (Kang et al., 2007b). T5H activity is highest in the roots of both species, but only in rice does this correspond to the accumulation of large amounts of serotonin.
By combining the spatiotemporal gene expression and indolamine accumulation profiles, we were able to build a model (Figure 9) showing the dynamic regulation of indolamine biosynthesis and, in some organs, their channeling into downstream pathways, such as melatonin or feruloyl and coumaroyl derivatives (which should be measured in future studies).
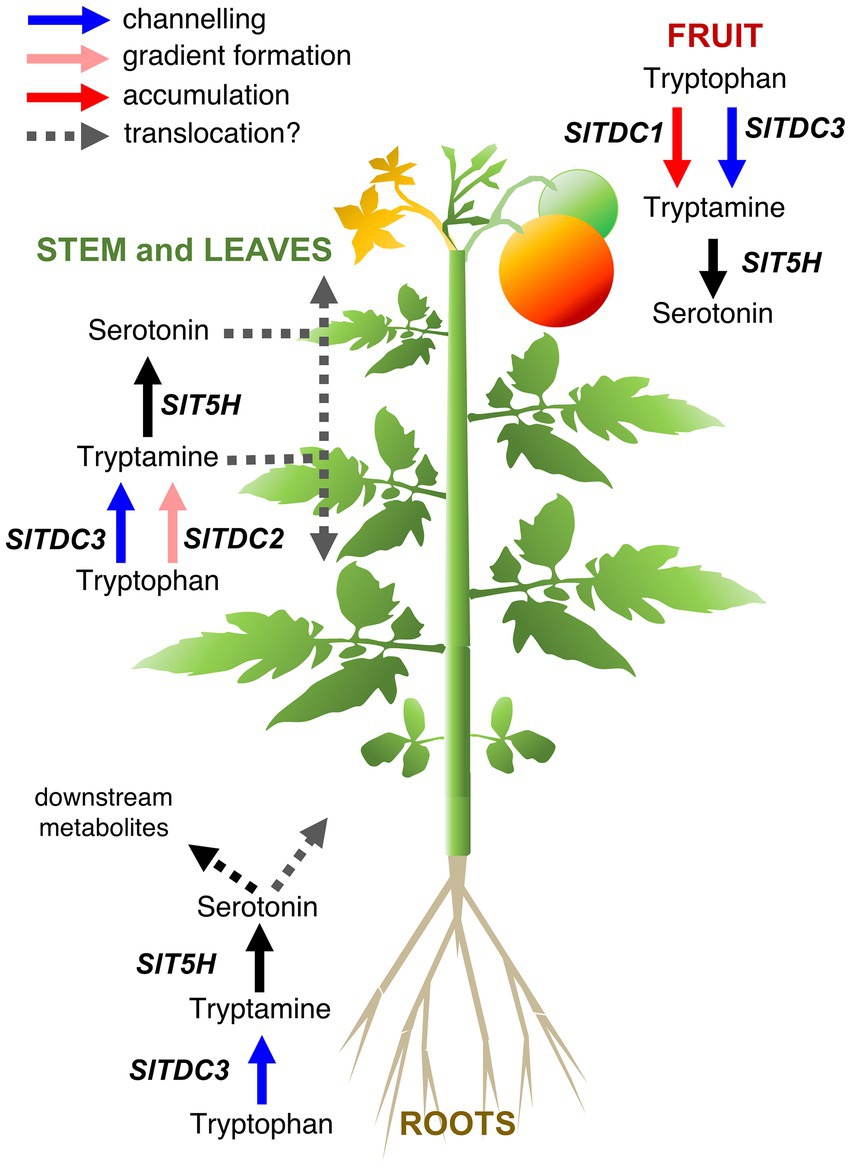
Figure 9. Proposed model for the functional interactions of tomato serotonin pathway genes in different organs of the Micro-Tom plant. Different functions of the TDC genes that we hypothesize are represented by arrows with different colors: SlTDC1 converts tryptophan to tryptamine for accumulation (red arrow) in the immature fruit, SlTDC3 works in cooperation with SlT5H to channel (blue arrows) tryptophan to serotonin in fruit, aerial organs and roots. SlTDC2 works together with SlT5H to generate the indolamine accumulation gradient along the aerial growth axis (pink arrows).
Pearson correlation analysis suggested that SlTDC3 and SlT5H may channel tryptamine into serotonin production in the roots. However, the absence of high serotonin levels in the roots suggests conversion to downstream products or translocation to other parts of the plant.
SlTDC1 and SlTDC3 were expressed in the fruit, suggesting that SlTDC1 might ensures the production of sufficient tryptamine for a specific function during berry development whereas SlTDC3 and SlT5H might work together to regulate the serotonin content. Interestingly, SlTDC1 and SlT5H were expressed strongly in the fruit but the tryptamine and serotonin content was lower than in some vegetative organs with minimal SlTDC1 and SlT5H expression. This may reflect the translocation of these indolamines to the berry flesh or even the seeds, which accumulated the highest levels of both indolamines despite the low gene expression levels.
The relationships between genes and metabolites in the rest of the plant are much more complex given the gradients of gene expression and indolamine accumulation in the leaves and internodes of the stem. SlTDC2 appears to play a fundamental role in the vegetative aerial organs of Micro-Tom plants, but the expression profile did not precisely match the distribution of tryptamine or serotonin in these organs. Indolamine levels in the root and along the longitudinal axis of the Micro-Tom plant, as well as the expression profiles of the SlTDC and SlT5H genes, are roughly consistent with the auxin gradients, as reported in Arabidopsis roots (Ljung et al., 2005; Petersson et al., 2009) and the tomato stem (Kojima et al., 2002). The discovery of tryptophan-dependent pathways that lead to auxin biosynthesis suggests the involvement of tryptamine as an intermediate, but this hypothesis is still under debate (Brumos et al., 2014). Mechanisms other than the direct involvement of tryptamine in auxin biosynthesis should also be considered, such as signaling via auxin receptors facilitated by the structural similarities between plant indolamines and indole acetic acid (Erland et al., 2015; Mukherjee, 2018; Pelagio-Flores et al., 2018).
In conclusion, the complex expression profiles of the three TDC genes therefore suggest unique biological roles despite the conserved catalytic function of the corresponding proteins. Similarly, the dynamic spatiotemporal concentration of the indolamines indicate they play unique and beneficial roles in tomato plants but are not indispensable, given the absence of these metabolites in some plant species.
The original contributions presented in the study are included in the article/Supplementary material; further inquiries can be directed to the corresponding authors.
FG and LA conceived and supervised the research. MC and SN performed the in silico analysis, targeted and untargeted metabolomics experiments, analyzed and interpreted data, and wrote the draft article. MC, SN, EF, and CP performed the transient expression experiments. EG and SN performed the gene expression experiments and analyzed and interpreted data. SN integrated data from the gene expression and metabolomics datasets. FG, LA, SN, and MC revised the draft. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.975434/full#supplementary-material
1. ^https://solgenomics.net/tools/blast/
2. ^https://www.ebi.ac.uk/Tools/msa/clustalo/
Abu-Zaitoon, Y. M. (2014). Phylogenetic analysis of putative genes involved in the tryptophan-dependent pathway of auxin biosynthesis in rice. Appl. Biochem. Biotechnol. 172, 2480–2495. doi: 10.1007/s12010-013-0710-4
Akula, R., and Mukherjee, S. (2020). New insights on neurotransmitters signaling mechanisms in plants. Plant Signal. Behav. 15:1737450. doi: 10.1080/15592324.2020.1737450
Arnao, M. B., and Hernández-Ruiz, J. (2018). Phytomelatonin, natural melatonin from plants as a novel dietary supplement: sources, activities and world market. J. Funct. Foods 48, 37–42. doi: 10.1016/j.jff.2018.06.023
Arnao, M. B., and Hernández-Ruiz, J. (2019). Melatonin: a new plant hormone and/or a plant master regulator? Trends Plant Sci. 24, 38–48. doi: 10.1016/j.tplants.2018.10.010
Back, K., Tan, D. X., and Reiter, R. (2016). Melatonin biosynthesis in plants: multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 61, 426–437. doi: 10.1111/jpi.12364
Bhatla, S. C. (2018). “Recently Discovered Plant Growth Regulators,” in Plant Physiology, Development and Metabolism. Springer, 681–728.
Brumos, J., Alonso, J. M., and Stepanova, A. N. (2014). Genetic aspects of auxin biosynthesis and its regulation. Physiol. Plant. 151, 3–12. doi: 10.1111/ppl.12098
Byeon, Y., Choi, G.-H., Lee, H. Y., and Back, K. (2015). Melatonin biosynthesis requires N-acetylserotonin methyltransferase activity of caffeic acid O-methyltransferase in rice. J. Exp. Bot. 66, 6917–6925. doi: 10.1093/jxb/erv396
Commisso, M., Negri, S., Bianconi, M., Gambini, S., Avesani, S., Ceoldo, S., et al. (2019). Untargeted and targeted metabolomics and tryptophan decarboxylase in vivo characterization provide novel insight on the development of kiwifruits (Actinidia deliciosa). Int. J. Mol. Sci. 20, 897. doi: 10.3390/ijms20040897
Consortium, T. G. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
De Luca, V., Marineau, C., and Brisson, N. (1989). Molecular cloning and analysis of cDNA encoding a plant tryptophan decarboxylase: comparison with animal dopa decarboxylases. Proc. Natl. Acad. Sci. 86, 2582–2586. doi: 10.1073/pnas.86.8.2582
Eklöf, S., Åstot, C., Moritz, T., Blackwell, J., Olsson, O., and Sandberg, G. (1996). Cytokinin metabolites and gradients in wild type and transgenic tobacco with moderate cytokinin over-production. Physiol. Plant. 98, 333–344. doi: 10.1034/j.1399-3054.1996.980215.x
Erland, L. A., Murch, S. J., Reiter, R. J., and Saxena, P. K. (2015). A new balancing act: The many roles of melatonin and serotonin in plant growth and development. Plant Signal. Behav. 10:e1096469. doi: 10.1080/15592324.2015.1096469
Erland, L. A., Turi, C. E., and Saxena, P. K. (2016). Serotonin: An ancient molecule and an important regulator of plant processes. Biotechnol. Adv. 34, 1347–1361. doi: 10.1016/j.biotechadv.2016.10.002
Erland, L. A., Turi, C. E., and Saxena, P. K. (2019). “Serotonin in plants: origin, functions, and implications,” in Serotonin. ed. P. M. Pilowsky (Elsevier), 23–46.
Everat-Bourbouloux, A., and Charnay, D. (1982). Endogenous abscisic acid levels in stems and axillary buds of intact or decapitated broad-bean plants (Vicia faba L.). Physiol. Plant. 54, 440–445. doi: 10.1111/j.1399-3054.1982.tb00705.x
Feldman, J. M., and Lee, E. M. (1985). Serotonin content of foods: effect on urinary excretion of 5-hydroxyindoleacetic acid. Am. J. Clin. Nutr. 42, 639–643. doi: 10.1093/ajcn/42.4.639
Foy, J., and Parratt, J. (1960). A note on the presence of noradrenaline and 5-hydroxytryptamine in plantain (Musa sapientum, var. Paradisiaca). J. Pharm. Pharmacol. 12, 360–364.
Fujiwara, T., Maisonneuve, S., Isshiki, M., Mizutani, M., Chen, L., Wong, H. L., et al. (2010). Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J. Biol. Chem. 285, 11308–11313. doi: 10.1074/jbc.M109.091371
Gecchele, E., Merlin, M., Brozzetti, A., Falorni, A., Pezzotti, M., and Avesani, L. (2015). A Comparative Analysis of Recombinant Protein Expression in different Biofactories: bacteria, insect cells and plant Systems. J. Vis. Exp. e52459. doi: 10.3791/52459
Gibson, R., Barrett, G., and Wightman, F. (1972). Biosynthesis and metabolism of indol-3yl-acetic acid: III. Partial purification and properties of a tryptamine-forming l-tryptophan dbcarboxylase from tomato shoots. J. Exp. Bot. 23, 775–786. doi: 10.1093/jxb/23.3.775
Gill, R. I., Ellis, B. E., and Isman, M. B. (2003). Tryptamine-induced resistance in tryptophan decarboxylase transgenic poplar and tobacco plants against their specific herbivores. J. Chem. Ecol. 29, 779–793. doi: 10.1023/A:1022983529555
Goddijn, O. J., Lohman, F. P., de Kam, R. J., and Hoge, J. H. C. (1994). Nucleotide sequence of the tryptophan decarboxylase gene of Catharanthus roseus and expression of tdc-gus A gene fusions in Nicotiana tabacum. Mol. Gen. Genet. 242, 217–225. doi: 10.1007/BF00391016
González-Aguilera, K. L., Saad, C. F., Chávez Montes, R. A., Alves-Ferreira, M., and de Folter, S. (2016). Selection of reference genes for quantitative real-time RT-PCR studies in tomato fruit of the genotype MT-Rg1. Front. Plant Sci. 7:1386. doi: 10.3389/fpls.2016.01386
Hano, S., Shibuya, T., Imoto, N., Ito, A., Imanishi, S., Aso, H., et al. (2017). Serotonin content in fresh and processed tomatoes and its accumulation during fruit development. Sci. Hortic. 214, 107–113. doi: 10.1016/j.scienta.2016.11.009
Huang, X., and Mazza, G. (2011). Application of LC and LC-MS to the analysis of melatonin and serotonin in edible plants. Crit. Rev. Food Sci. Nutr. 51, 269–284. doi: 10.1080/10408398.2010.529193
Islam, J., Shirakawa, H., Nguyen, T. K., Aso, H., and Komai, M. (2016). Simultaneous analysis of serotonin, tryptophan and tryptamine levels in common fresh fruits and vegetables in Japan using fluorescence HPLC. Food Biosci. 13, 56–59. doi: 10.1016/j.fbio.2015.12.006
Kang, S., Kang, K., Lee, K., and Back, K. (2007a). Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta 227, 263–272. doi: 10.1007/s00425-007-0614-z
Kang, S., Kang, K., Lee, K., and Back, K. (2007b). Characterization of tryptamine 5-hydroxylase and serotonin synthesis in rice plants. Plant Cell Rep. 26, 2009–2015. doi: 10.1007/s00299-007-0405-9
Kang, K., Kim, Y.-S., Park, S., and Back, K. (2009). Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol. 150, 1380–1393. doi: 10.1104/pp.109.138552
Kang, K., Kong, K., Park, S., Natsagdorj, U., Kim, Y. S., and Back, K. (2011). Molecular cloning of a plant N-acetylserotonin methyltransferase and its expression characteristics in rice. J. Pineal Res. 50, 304–309. doi: 10.1111/j.1600-079X.2010.00841.x
Kang, K., Lee, K., Park, S., Byeon, Y., and Back, K. (2013). Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 55, 7–13. doi: 10.1111/jpi.12011
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY™ vectors for agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Kojima, K., Ohtake, E., and Yu, Z. (2002). Distribution and transport of IAA in tomato plants. Plant Growth Regul. 37, 249–254. doi: 10.1023/A:1020857004069
Lee, J. H., Jayaprakasha, G. K., Enciso, J., Avila, C. A., Crosby, K. M., and Patil, B. S. (2021). Production system influences tomato phenolics and indoleamines in a cultivar-specific manner. Food Res. Int. 140:110016. doi: 10.1016/j.foodres.2020.110016
Li, L., Zheng, M., Long, H., Deng, G., Ishihara, A., Liu, F., et al. (2016). Molecular cloning and characterization of two genes encoding tryptophan decarboxylase from Aegilops variabilis with resistance to the cereal cyst nematode (Heterodera avenae) and root-knot nematode (Meloidogyne naasi). Plant Mol. Biol. Report. 34, 273–282. doi: 10.1007/s11105-015-0909-3
Ljung, K., Hull, A. K., Celenza, J., Yamada, M., Estelle, M., Normanly, J., et al. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17, 1090–1104. doi: 10.1105/tpc.104.029272
López-Meyer, M., and Nessler, C. L. (1997). Tryptophan decarboxylase is encoded by two autonomously regulated genes in Camptotheca acuminata which are differentially expressed during development and stress. Plant J. 11, 1167–1175. doi: 10.1046/j.1365-313X.1997.11061167.x
Mukherjee, S. (2018). Novel perspectives on the molecular crosstalk mechanisms of serotonin and melatonin in plants. Plant Physiol. Biochem. 132, 33–45. doi: 10.1016/j.plaphy.2018.08.031
Muller, P. Y., Janovjak, H., Miserez, A. R., and Dobbie, Z. (2002). Short technical report processing of gene expression data generated by quantitative real-time RT-PCR. BioTechniques 32, 1372–1379.
Murch, S. J., Hall, B. A., Le, C. H., and Saxena, P. K. (2010). Changes in the levels of indoleamine phytochemicals during véraison and ripening of wine grapes. J. Pineal Res. 49, 95–100. doi: 10.1111/j.1600-079X.2010.00774.x
Nawaz, M. A., Huang, Y., Bie, Z., Ahmed, W., Reiter, R. J., Niu, M., et al. (2016). Melatonin: current status and future perspectives in plant science. Front. Plant Sci. 6:1230. doi: 10.3389/fpls.2015.01230
Negri, S., Commisso, M., Avesani, L., and Guzzo, F. (2021). The case of tryptamine and serotonin in plants: a mysterious precursor for an illustrious metabolite. J. Exp. Bot. 72, 5336–5355. doi: 10.1093/jxb/erab220
Pang, X., Wei, Y., Cheng, Y., Pan, L., Ye, Q., Wang, R., et al. (2018). The tryptophan decarboxylase in Solanum lycopersicum. Molecules 23:998. doi: 10.3390/molecules23050998
Park, S., Kang, K., Lee, K., Choi, D., Kim, Y.-S., and Back, K. (2009). Induction of serotonin biosynthesis is uncoupled from the coordinated induction of tryptophan biosynthesis in pepper fruits (Capsicum annuum) upon pathogen infection. Planta 230, 1197–1206. doi: 10.1007/s00425-009-1015-2
Pelagio-Flores, R., López-Bucio, J. S., and López-Bucio, J. (2018). Serotonin and Melatonin in Root Morphogenesis. Amsterdam, New York: Elsevier. 103.
Petersson, S. V., Johansson, A. I., Kowalczyk, M., Makoveychuk, A., Wang, J. Y., Moritz, T., et al. (2009). An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21, 1659–1668. doi: 10.1105/tpc.109.066480
Roshchina, V. V. (2010). “Evolutionary considerations of neurotransmitters in microbial, plant, and animal cells,” in Microbial Endocrinology. ed. M. Lyte and P. P. E. Freestone (Springer), 17–52.
Sun, C., Liu, L., Wang, L., Li, B., Jin, C., and Lin, X. (2021). Melatonin: a master regulator of plant development and stress responses. J. Integr. Plant Biol. 63, 126–145. doi: 10.1111/jipb.12993
Thomas, J. C., Akroush, A. M., and Adamus, G. (1999). The indole alkaloid tryptamine produced in transgenic Petunia hybrida. Plant Physiol. Biochem. 37, 665–670. doi: 10.1016/S0981-9428(00)80096-0
Thomas, J. C., Saleh, E. F., Alammar, N., and Akroush, A. M. (1998). The indole alkaloid tryptamine impairs reproduction in Drosophila melanogaster. J. Econ. Entomol. 91, 841–846. doi: 10.1093/jee/91.4.841
Torrens-Spence, M. P., Chiang, Y. C., Smith, T., Vicent, M. A., Wang, Y., and Weng, J. K. (2020). Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins. Proc. Natl. Acad. Sci. 117, 10806–10817. doi: 10.1073/pnas.1920097117
Tsunoda, Y., Hano, S., Imoto, N., Shibuya, T., Ikeda, H., Amagaya, K., et al. (2021). Physiological roles of tryptophan decarboxylase revealed by overexpression of SlTDC1 in tomato. Sci. Hortic. 275:109672. doi: 10.1016/j.scienta.2020.109672
Udenfriend, S., Lovenberg, W., and Sjoerdsma, A. (1959). Physiologically active amines in common fruits and vegetables. Arch. Biochem. Biophys. 85, 487–490. doi: 10.1016/0003-9861(59)90516-8
Yamazaki, Y., Sudo, H., Yamazaki, M., Aimi, N., and Saito, K. (2003). Camptothecin biosynthetic genes in hairy roots of Ophiorrhiza pumila: cloning, characterization and differential expression in tissues and by stress compounds. Plant Cell Physiol. 44, 395–403. doi: 10.1093/pcp/pcg051
Yılmaz, C., Taş, N. G., Kocadağlı, T., and Gökmen, V. (2019). Determination of serotonin in nuts and nut containing products by liquid chromatography tandem mass spectrometry. Food Chem. 272, 347–353. doi: 10.1016/j.foodchem.2018.08.064
You, D., Feng, Y., Wang, C., Sun, C., Wang, Y., Zhao, D., et al. (2020). Cloning, characterization, and enzymatic identification of a new tryptophan decarboxylase from Ophiorrhiza pumila. Biotechnol. Appl. Biochem. 8, 381–389. doi: 10.1002/bab.1935
Zeiss, D. R., Piater, L. A., and Dubery, I. A. (2020). Hydroxycinnamate amides: intriguing conjugates of plant protective metabolites. Trends Plant Sci. 26, 184–195. doi: 10.1016/j.tplants.2020.09.011
Keywords: tryptamine, serotonin, fruit indolamines, tryptophan decarboxylase, tryptamine 5-hydroxylase, biosynthesis pathway, Solanum lycopersicum
Citation: Commisso M, Negri S, Gecchele E, Fazion E, Pontoriero C, Avesani L and Guzzo F (2022) Indolamine accumulation and TDC/T5H expression profiles reveal the complex and dynamic regulation of serotonin biosynthesis in tomato (Solanum lycopersicum L.). Front. Plant Sci. 13:975434. doi: 10.3389/fpls.2022.975434
Received: 22 June 2022; Accepted: 27 July 2022;
Published: 11 August 2022.
Edited by:
Danièle Werck, UPR2357 Institut de biologie moléculaire des plantes (IBMP), FranceReviewed by:
Heribert Warzecha, Darmstadt University of Technology, GermanyCopyright © 2022 Commisso, Negri, Gecchele, Fazion, Pontoriero, Avesani and Guzzo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda Avesani, bGluZGEuYXZlc2FuaUB1bml2ci5pdA==; Flavia Guzzo, ZmxhdmlhLmd1enpvQHVuaXZyLml0
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.