
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 23 September 2022
Sec. Plant Systems and Synthetic Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.975356
This article is part of the Research TopicSystems biology of maturation and senescence in horticultural plantsView all 10 articles
 Chunzhen Cheng1†
Chunzhen Cheng1† Ziwei Guo1†
Ziwei Guo1† Hua Li2†
Hua Li2† Xiaopeng Mu1
Xiaopeng Mu1 Pengfei Wang1
Pengfei Wang1 Shuai Zhang1
Shuai Zhang1 Tingzhen Yang3
Tingzhen Yang3 Huacheng Cai3
Huacheng Cai3 Qian Wang3
Qian Wang3 Peitao Lü2*
Peitao Lü2* Jiancheng Zhang1*
Jiancheng Zhang1*Fruit ripening is accompanied by a wide range of metabolites and global changes in gene expression that are regulated by various factors. In this study, we investigated the molecular differences in red apple ‘Hongmantang’ fruits at three ripening stages (PS1, PS5 and PS9) through a comprehensive analysis of metabolome, transcriptome and chromatin accessibility. Totally, we identified 341 and 195 differentially accumulated metabolites (DAMs) in comparison I (PS5_vs_PS1) and comparison II (PS9_vs_PS5), including 57 and 23 differentially accumulated flavonoids (DAFs), respectively. Intriguingly, among these DAFs, anthocyanins and flavonols showed opposite patterns of variation, suggesting a possible competition between their biosynthesis. To unveil the underlying mechanisms, RNA-Seq and ATAC-Seq analyses were performed. A total of 852 DEGs significantly enriched in anthocyanin metabolism and 128 differential accessible regions (DARs) significantly enriched by MYB-related motifs were identified as up-regulated in Comparison I but down-regulated in Comparison II. Meanwhile, the 843 DEGs significantly enriched in phenylalanine metabolism and the 364 DARs significantly enriched by bZIP-related motifs showed opposite trends. In addition, four bZIPs and 14 MYBs were identified as possible hub genes regulating the biosynthesis of flavonols and anthocyanins. Our study will contribute to the understanding of anthocyanins and flavonols biosynthesis competition in red apple fruits during ripening.
The old saying ‘An apple a day keeps the doctor away’ can be explained by the fact that apples can help people to stay healthy and away from the doctor by reducing the risk of numerous chronic diseases due to its high antioxidant content. As a major contributor to the total antioxidant capacity, flavonoids were hence considered as one of the most important phytonutrients that apples nourish people (Lee et al., 2003). Apples are identified as one of the main diet sources of flavanols and flavonols for people (Vrhovsek et al., 2004). Moreover, the anthocyanins accumulation in apples skin and flesh affects greatly the external quality and health care values of apples (Martin, 2013). Therefore, the study of flavonoids, especially anthocyanin biosynthesis, has been an attention hotspot for apple studies.
To date, there are more than 10,000 known flavonoids that have been identified. According to their structures, they can be divided into six groups: dihydroflavones, flavones, isoflavones, flavonols, flavanols and anthocyanidins. The main synthetic pathway of flavonoids are well established, i.e., dihydro-flavonoids are first synthesized, and then flavonoids, isoflavones, flavonols, flavanols and anthocyanidins are further synthesized through a series of branching pathways catalyzed by enzymes encoded by structural genes, such as phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone-3-hydroxylase (F3H), flavonoid-3’-hydroxylase (F3’H), flavonoid-3’-5’-hydroxylase (F3’5’H), flavonol synthase (FLS), dihydroflavonol reductase (DFR), anthocyanidin synthase (ANS), leucocyanidin reductase (LAR), anthocyanidin reductase (ANR), etc. (Gutierrez et al., 2017). In addition to structural genes, the flavonoids biosynthesis is also regulated by many transcription factors (TFs), which are directly or indirectly modulating the spatiotemporal expression of structural genes (Zhang et al., 2021). Among them, MYB has long been considered as one of the most important TFs regulating the flavonoids synthesis in apple (Takos et al., 2006). Some other TFs have also contributed greatly to the regulation of flavonoids biosynthesis, mainly through interacting with various MYB members. For example, EIL3 (EIN3-LIKE1) can bind to the promoter of MdMYB1 to activate MdMYB1 gene expression and to induce anthocyanin accumulation and fruit color transformation (An et al., 2018c); MdERF1B interacts with MdMYB9, MdMYB1 and MdMYB11 and can bind to the promoters of their encoded genes to affect anthocyanin biosynthesis (Zhang et al., 2018).
Recently, integrated metabolomic and transcriptomic analyses have been successfully applied in the exploration of the key factors regulating metabolites accumulations in many fruits (Xu H. et al., 2020; Zhang et al., 2020; Fu et al., 2022). However, the two methods can only reflect changes in gene expression and metabolites accumulation at the RNA and metabolite levels, respectively, and the factors influencing gene expression cannot be directly deciphered. Access to open chromatin can reveal the regulation mechanisms of gene expression, which is a hot topic in epigenetic research. ATAC-seq (Assay for Transpoase-accessible Chromatin with High Throughput sequencing), an innovative technology developed by Buenrostro et al. (2013), is an epigenetic research technique for analyzing Tn5 transposase accessibility chromatin region using high-throughput sequencing and is considered as a potential tool for identifying cis-regulatory regions such as TF binding sites (Grandi et al., 2022). Based on the ATAC-Seq data, all the open chromatin regions in a specific spatiotemporal condition could be obtained. Through investigating the accessible chromatin regions (ACRs) in root, leaf bud, flower, flower bud, developing seed, and pod of soybean using ATAC-Seq, Huang et al. (2022) reported that the ACRs occupied more than 3% of the soybean genome. By integrating the RNA-seq and TF ChIP-seq results, they found that ACRs were tightly associated with gene expressions and TF binding capacities. In Vitis vinifera leaves, more than 16,000 ACRs were identified, with nearly 5,000 distal enhancer candidates located in intergenic regions > 2 kb from the nearest transcription start site (TSS), which were found to be enriched in TCP family binding sites by motif search, suggesting that these regions may be regulated by TCP (Schwope et al., 2021). Through the combined ATAC-Seq and RNA-Seq analyses, nine TF genes, including RAV1, were identified as responsible for high cold hardiness of V. amurensis (Ren et al., 2021).
Red-flesh apples, containing anthocyanins in both fruit skin and flesh, are popular for their anthocyanin-rich characteristics. The red-flesh apple ‘Hongmantang’, selected from Malus pumila Mill. × M. baccata (L.) Borkh hybrids, is of purple-red flower and blood-red flesh. Compared with its parent M. pumila Mill., its total anthocyanins and total polyphenols contents in fruits are much higher, suggesting that it can be used as a suitable material for the flavonoids biosynthesis studies of Malus species (Yang et al., 2015). In our previous study, through analyzing the changes in total anthocyanins contents during fruit ripening, we found that there were two anthocyanin content peaks (at PS5 and PS9, respectively) throughout fruit ripening stage (Guo et al., 2021) (Figure S1). In this study, to reveal the mechanisms underlying the bioactive compounds changes during fruit ripening, we investigated the metabolome, transcriptome and chromatin accessibility changes in ‘Hongmantang’ fruits at three ripening stages, PS1 (at about 7 weeks post flowering with the lowest anthocyanin content), PS5 (at about 15 weeks post flowering with the highest anthocyanins content) and PS9 (at about 23 weeks post flowering, the second anthocyanins accumulation peak), by using the ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS)-based widely targeted metabolome, RNA-Seq and ATAC-Seq technologies, respectively. The results obtained in this study could provide a basis for the comprehensive understanding of the metabolites changes during fruit ripening in red apple.
The red apple ‘Hongmantang’ used in this study is an ornamental red apple with purple-red flower and blood-red flesh, and was selected from M. pumila Mill. × M. baccata (L.) Borkh interspecific hybrids by Shanxi Academy of Agricultural Sciences. In this study, the ‘Hongmantang’ fruits were harvested at three key developmental stages (PS1: at about 7 weeks post flowering; PS5: at about 15 weeks post flowering; PS9: at about 23 weeks post flowering) and stored in a -80°C freezer for further use.
Briefly, 50 mg of red apple fruit samples were added to 1 mL of 70% methanol solution, homogenized at 30 Hz for 3 min, vortexed for 1 min, placed on ice for 15 min, centrifuged at 12,000 rpm at 4°C for 10 min, and then the supernatant was collected for subsequent LC-MS/MS analysis at Wuhan MetWare Biotechnology Co., Ltd. (Wuhan, China). The analysis was performed using a liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS/MS) system according to the method described by Chen et al. (2013). The column of Agilent SB-C18 (1.8 µm, 2.1 mm * 100 mm) and the mobile phase consisted of solvent A (ultrapure water with 0.04% acetic acid) and solvent B (acetonitrile with 0.04% acetic acid) were used in the analysis. The gradient program was set as follows: 95% solvent A and 5% solvent B at 0 min; a liner gradient to 5% solvent A and 95% solvent B within 9 min and kept for 1 min; and adjustment to 95% solvent A and 5% solvent B within 1.1 min and kept for 2.9 min (Li et al., 2022). The flow rate was 0.35 mL/min with injection volume of 4 μL, and the column temperature was 40°C. The effluent was connected to an ESI-triple quadrupole-linear ion trap (QTRAP)-MS (AB4500 Q TRAP UPLC/MS/MS System) equipped with an ESI Turbo Ion-Spray interface. Analyst 1.6.3 software (AB Sciex) was used for controlling the positive and negative ion modes. The MetWare database (MWDB) together with several public databases were used for metabolites annotation (Li et al., 2022). And metabolites were quantified using the multiple reaction monitoring method. Three biological replicates were performed on fruits at each selected stage (PS1, PS5 and PS9). The OPLS-DA (Orthogonal Projection to Latent Structures Discriminant Analysis) model was established using multiple supervision methods and examined using the Variable Importance in Projection (VIP) parameter. For the identification of differentially accumulated metabolites (DAMs) in fruits of the three ripening stages, VIP ≥ 1 and fold change ≥ 2 or ≤ 0.5 were used as criteria (Yuan et al., 2018; Zhang et al., 2020).
Total RNA of red apple fruits was extracted using Trizol (Invitrogen, CA, USA). After digestion of DNA contamination with DNase I, the RNA quality and quantity were detected using NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and a Bioanalyzer 2100 system (Agilent Technologies, CA, USA). High-quality total RNA was used for RNA-Seq on a MGI-SEQ 2000 platform to generate 150 bp paired-end reads at Wuhan Frasergen Bioinformatics Co., Ltd. (Wuhan, China). The raw clean reads were mapped to the M. baccata (L.) Borkh. genome [downloaded from https://www.ncbi.nlm.nih.gov/genome/?term=Malus_baccata_v1.0] for gene transcriptional level studies using hisat2 software. For each stage, three biological replicates were performed, with at least ten fruits being mixed in each replicate. Differentially expressed genes (DEGs) were identified using DESeq2 with criteria of log2(fold change) > 1 or < -1 and corrected q-value < 0.05. The RNA-Seq raw data has been deposited at NCBI under the BioProject number PRJNA861071 (https://www.ncbi.nlm.nih.gov/sra/PRJNA861071).
ATAC-Seq libraries were constructed and sequenced according to the methods described by Buenrostro et al. (2013) and Corces et al. (2017). For fruits at each stage, two biological replicates were performed. Briefly, total DNA from Tn5 digestion was purified by phenol/chloroform extraction and precipitation, electrophoresed on a 2% agarose gel, amplified using PCR, purified and sequenced by Illumina method. clean reads were mapped to the M. baccata genome using Bowtie2 (Langmead and Salzberg, 2012), and redundancy was removed from the obtained comparison files using the Picard program, followed by peak detection by MACS2. The distribution of peaks in each sample relative to gene transcription start sites (TSSs) and transcription end sites (TESs) was statistically analyzed and the proportion of peaks at various locations in the genome, including 5’UTR, 3’UTR, 1st exon, other exon, 1st intron, other intron, downstream (≤ 300 bp) and distal intergenic, was calculated. For the identification of differential peaks, DESeq2 was used with a threshold of FDR (false discovery rate) < 0.05 and fold change > 1.5 or < -1.5. The GenomicFeatures (R packages) was used to extract the genes nearby the differential peaks. For the enrichment analysis of genes involved in differential accessible regions (DARs), GO and KEGG enrichment analyses were performed with P (classicFisher) ≤ 0.05 and corrected P-value < 0.05, respectively. The ATAC-Seq raw data has been deposited at NCBI under BioProject number PRJNA860789 (https://www.ncbi.nlm.nih.gov/sra/PRJNA860789).
Complementary DNA (cDNA) of different samples was synthesized separately using the PrimeScript RT Master Mix (Perfect Real Time) kit (Takara, Dalian, China). Gene specific primers for selected genes were designed using Primer 3 (https://bioinfo.ut.ee/primer3-0.4.0/) according to their CDS (coding sequence) sequences (Table S1). qRT-PCR experiment was performed on Efficiency T96 real-time quantitative fluorescent PCR instrument (Beijing Leadaeon Technology Co. Ltd, Beijing, China). The qRT-PCR conditions were set as follows: pre-denaturation at 95°C for 30 s; denaturation at 95°C for 10 s, annealing at 54.8~58.8°C for 20 s, extension at 72°C for 20 s, 50 cycles. qRT-PCR reaction system consisted of 10 µL SYBR Premix ExTaq™ (TaKaRa) fluorescent dye (containing Oligo (dT) and random primers), 7.4 µL ddH2O, 0.8 µL of each upstream and downstream primer, and 1 µL cDNA template. Their relative expression levels in different samples were calculated using the 2−ΔΔCt method with actin as internal reference gene (Jiang et al., 2019).
The UPLC-MS/MS-based untargeted metabolome approach enables the detection of many primary and secondary metabolic compounds from red apple fruit samples. Totally, we identified 753 metabolites from red apple fruits at three ripening stages, including 399 primary metabolites and 354 secondary metabolites (Table S2). These metabolites could be further classified into 11 groups, including phenolic acids, flavonoids, lipids, amino acids and derivatives, organic acids, nucleotides and derivatives, alkaloids, terpenoids, tannins, lignans and coumarins, and others (Figure S2). Metabolites belonging to amino acids and derivatives, flavonoids, alkaloids, phenolic acids, nucleotides and derivatives, lignans and coumarins, and tannins mostly showed high accumulation at PS1 but low accumulation at PS5. However, many terpenoids and some lipids and phenolic acids accumulated highly at PS9. Moreover, some metabolites, such as saccharides and alcohols, vitamin and stilbene, were classified into ‘others’ group and showed very low or even no accumulation at PS1, but their contents increased with fruit ripening.
PCA analysis of the metabolites among red apple fruits at three different ripening stages revealed that their metabolomes were clearly different (Figure 1A). A total of 341 differentially accumulated metabolites (DAMs) were identified, including 69 increased and 272 decreased DAMs in comparison PS5_vs_PS1 (Comparison I) (Table S3; Figure 1B). There were much less increased metabolites than decreased metabolites for DAMs belonging to phenolic acids (16 increased and 54 decreased), flavonoids (5 increased and 52 decreased), lipids (7 increased and 41 decreased), amino acids and derivatives (4 increased and 37 decreased), organic acids (9 increased and 16 decreased), alkaloids (3 increased and 16 decreased), terpenoids (1 increased and 13 decreased), nucleotides and derivatives (2 increased and 9 decreased), tannins (only 9 decreased) and lignans and coumarins (2 increased and 5 decreased). However, for the saccharides and alcohols related DAMs, a larger proportion of increased metabolites (17 increased and 14 decreased) was observed. Notably, among the 57 different accumulated flavonoids (DAFs) (Table S4; Figure 1B), two anthocyanins, cyanidin-3-O-galactoside and cyanidin-3-O-glucoside (Kuromanin), increased in PS5 for 1.57- and 2.31-fold, respectively.
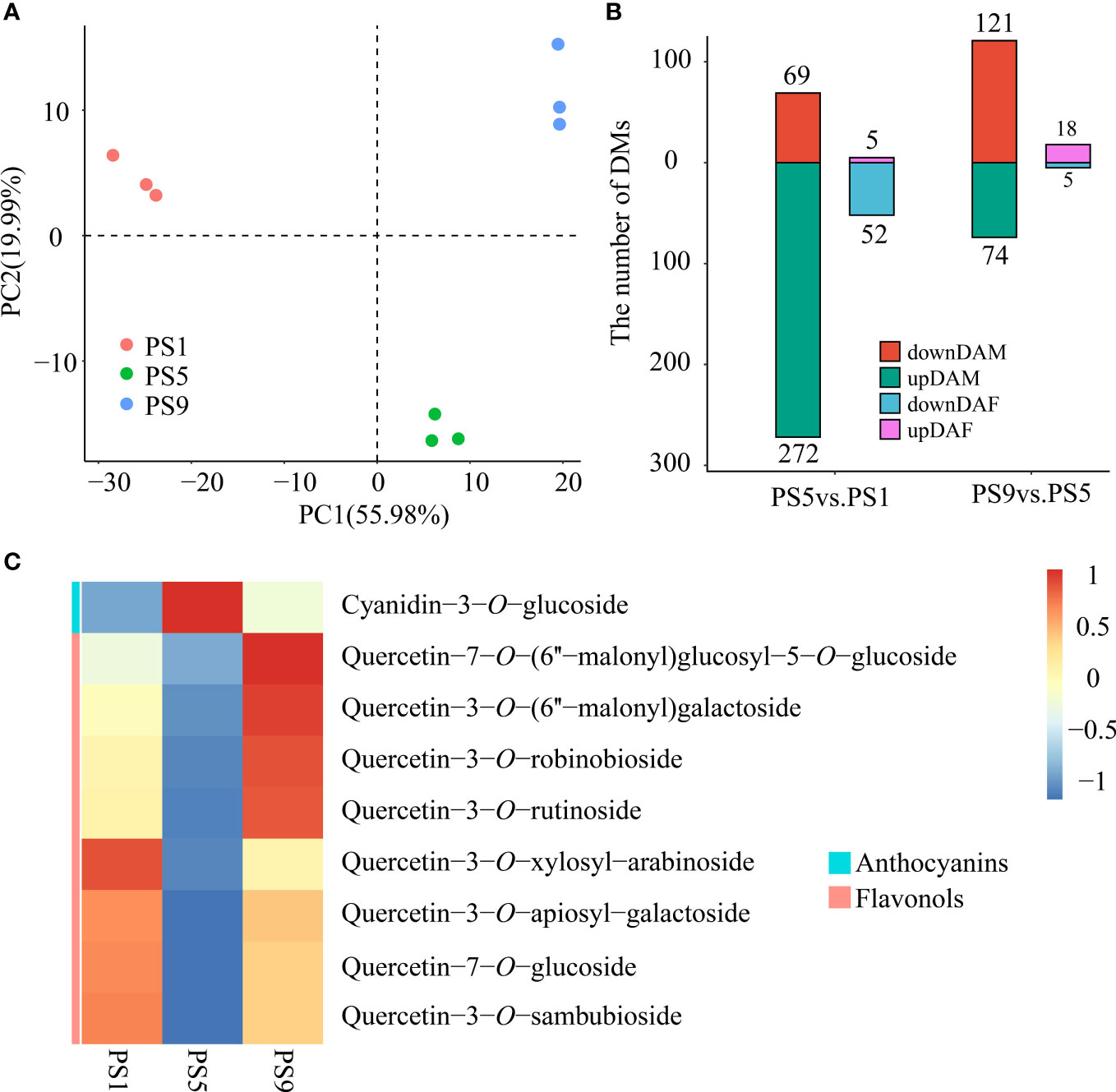
Figure 1 Metabolome differences in red apple fruits at three different ripening stages. (A) PCA analysis of red apple fruit metabolome data at PS1, PS5 and PS9; (B) The numbers of differentially accumulated metabolites and flavonoids in two comparisons. DAM, differentially accumulated metabolites; DAF, differentially accumulated flavonoids; (C) Differential accumulation was commonly identified in comparisons of PS5_vs_PS1 and PS9_vs_PS5 of anthocyanins and flavonols.
A total of 195 DAMs were identified in PS9_vs_PS5 (Comparison II), including 121 increased and 74 decreased (Table S5). There were more increased than decreased metabolites for DAMs belonging to phenolic acids (24 increased and 6 decreased), flavonoids (18 increased and 5 decreased), lipids (23 increased and 8 decreased), terpenoids (24 increased and 2 decreased) and lignans and coumarins (5 increased and 2 decreased). However, more metabolites were decreased than increased for DAMs belonging to amino acids and derivatives (6 increased and 19 decreased), organic acids (8 increased and 9 decreased), alkaloids (1 increased and 6 decreased), nucleotides and derivatives (4 increased and 7 decreased) and tannins (only 1 decreased).
For the DAMs belonging to amino acids and derivatives, organic acids, alkaloids, nucleotides and derivatives and tannins, the number of decreased DAMs was higher than that of the increased ones in both comparisons, suggesting that the accumulations of these kinds of metabolites mostly reduced as fruit ripened. In contrast, the number of the increased saccharides and alcohols related metabolites was both higher than that of the decreased ones, which is in consistent with the sugar accumulation during fruit ripening. Moreover, the numbers of increased DAMs belonging to phenolic acids, flavonoids, lipids, terpenoids, and lignans and coumarins were much lower than the that of the decreased ones in Comparison I, but much higher than the numbers of decreased ones in Comparison II, suggesting that the accumulation of these kinds of metabolites reduced in the early fruit development stages but increased in the late fruit ripening stages.
Interestingly, we found that in Comparison I, all differentially accumulated anthocyanins increased, while all differentially accumulated quercetins decreased (Table S4). In Comparison II, we found a decrease in all differentially accumulated anthocyanins (Table S6), and an increase in all quercetins except quercetin-3-O-rhamnosyl (1→2) arabinoside (Table S6). In both comparisons, Cyanidin-3-O-glucoside and the eight quercetins were found to be DAMs, and showed completely opposite expression trends (Figure 1C). Therefore, we conclude that there is dynamic competition between anthocyanins and flavonols biosynthesis during red apple fruit development and ripening.
To further explore the molecular mechanisms regulating the dynamic of anthocyanins and flavonols, especially quercetins, during red apple fruit ripening, we performed RNA-seq analysis on red apple fruits at PS1, PS5 and PS9 stages. PCA analysis of transcriptome data showed that red apple fruits at the three ripening stages were well distinguished from each other (Figure 2A). The mapping ratios of all transcriptome data from the nine cDNA libraries against the reference genome were all above 79% (Table S7). By using |log2 (Fold change)| ≥ 1 and q-value ≤ 0.05 as criteria, we identified 8,089 differentially expressed genes (DEGs, 2,579 up-regulated and 5,510 down-regulated) in PS5_vs_PS1, and 11,681 DEGs (4,345 up-regulated and 7,336 down-regulated) in PS9_vs_PS5 (Figure 2B). Among them, 852 DEGs were up-regulated in PS5_vs_PS1 but down-regulated in PS9_vs_PS5 (Figure 2C; Table S8), which showed the same dynamic patterns as anthocyanins accumulation during fruit ripening. KEGG enrichment analysis showed that these 852 common DEGs were involved in phenylpropanoid biosynthesis, phenylalanine metabolism, flavonoid biosynthesis, flavone and flavonol biosynthesis and anthocyanin biosynthesis pathways (Figure 2D). Moreover, we identified 843 DEGs up-regulated in PS9_vs_PS5 but down-regulated in PS5_vs_PS1 (showing the same dynamic patterns as most flavonols, especially quercetins) (Figure 2C; Table S9). KEGG enrichment analysis showed that these genes were significantly enriched in phenylalanine metabolism, phenylpropanoid biosynthesis, etc. (Figure 2D). Among the 852 DEGs that were found to be up-regulated in PS5_vs_PS1 but down-regulated in PS9_vs_PS5 and the 843 DEGs up-regulated in PS9_vs_PS5 but down-regulated in PS5_vs_PS1, there were many flavonoid biosynthesis structural genes, including seven PALs, fifteen 4CLs, two CHSs, four CHIs, three F3Hs, three F3’5’Hs, five F3’Hs, four FLSs, two DFRs, one ANS, three LARs, four ANRs, and two UFGTs (Figure S3). Interestingly, almost all the DEGs encoding enzymes catalyzing the reactions from dihydroflavonols to anthocyanidins, such as F3’5’H, F3’H, DFR and ANS, showed the lowest expression at PS9, whereas the two genes encoding FLS enzyme catalyzing the conversion of dihydroflavonols to flavonols showed the highest expression at PS9.
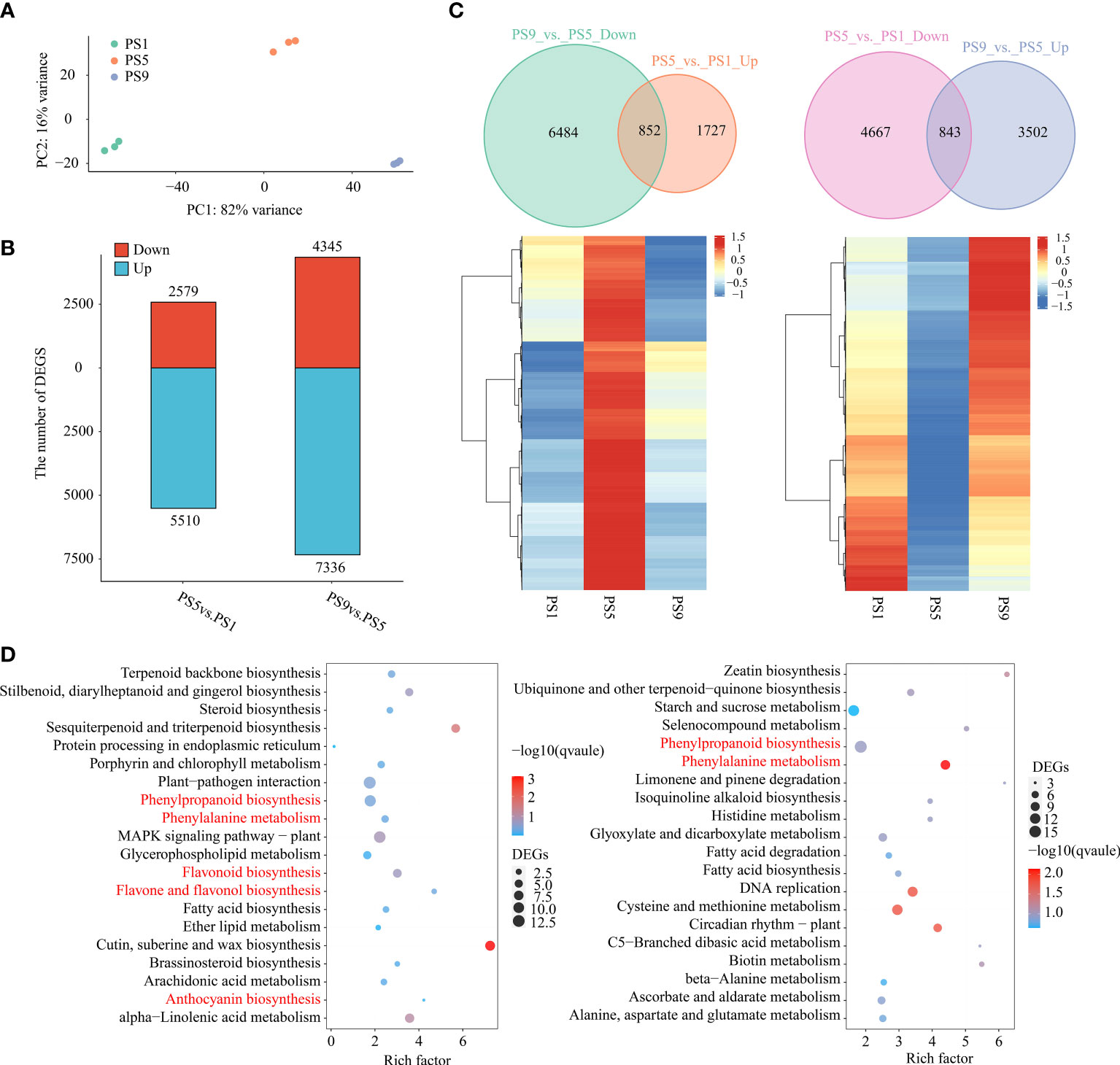
Figure 2 Comparative transcriptome analysis of red apple fruits at three different ripening stages. (A) PCA analysis result of red apple transcriptome data for PS1, PS5 and PS9; (B) Number of differentially expressed genes; (C) Venn diagram and heatmap of DEGs up-regulated in PS5_vs_PS1 but down-regulated in PS9_vs_PS5 (left) and DEGs down-regulated in PS5_vs_PS1 but up-regulated in PS9_vs_PS5 (right); (D) KEGG enrichment analysis of DEGs showing the same pattern of changes in anthocyanins (left) and flavonols (right) accumulations during fruit ripening.
Quantitative real time PCR (qRT-PCR) results showed that all the expression patterns of our selected flavonoids biosynthetic structural genes were identical to our transcriptome data, suggesting that our transcriptome data are reliable. The lowest expression of DFR was also found at PS9, indicating that its low expression might be closely related to the anthocyanins reduction at this stage. The FLS gene, however, showed the lowest expression at PS5 but the highest expression at PS9 according to our transcriptome and qRT-PCR results, which was the same pattern as the change in flavonols content (Figure S4).
ATAC-Seq was applied to investigate the chromatin accessibility changes during ripening in red apple (Table S10). PCA analysis of the ATAC-Seq data showed a clear distinction in chromatin accessibility among the three ripening stages in red apple fruits (Figure 3A). We identified 41,292 to 58,869 peaks in each ATAC-Seq library (Table S11; Figure S5). Relative location analysis of peaks showed that they were mainly distributed in distal intergenic region (75.78%), promoter (≤ 1 kb) region (12.83%) and promoter (1-2 kb) region (5.11%) (Figure 3B), with their distributions peaked around the TSSs (Figure 3C). By using FDR < 0.05 and FC > 1.5 or FC < -1.5 as criteria, we identified 908 up-regulated differential accessible regions (DARs) and 662 down-regulated DARs in PS5_vs_PS1, and 3,347 up-regulated and 816 down-regulated DARs in PS9_vs_PS5 (Figure 3D, Table S12).
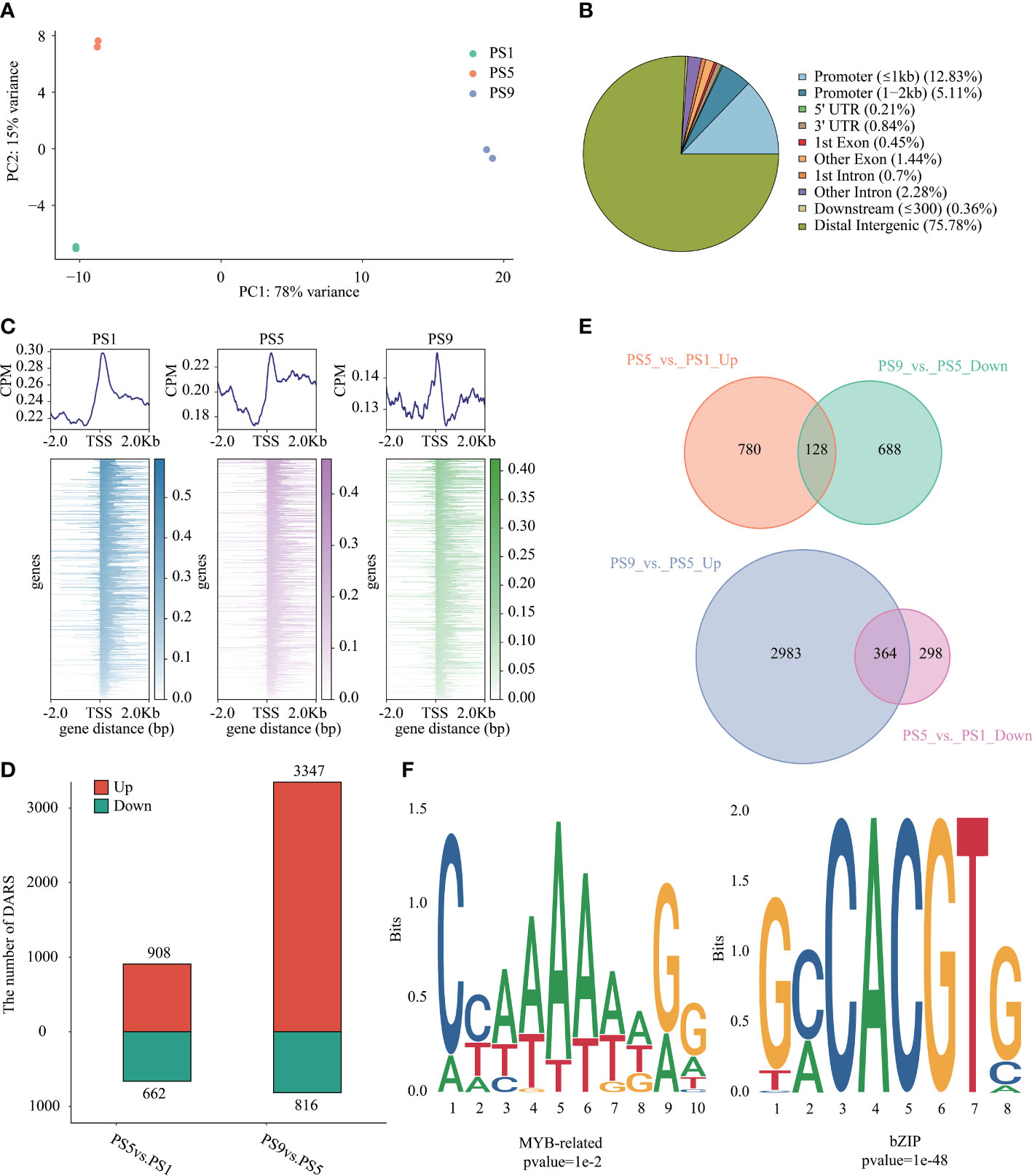
Figure 3 Chromatin accessibility features of red apple fruit during ripening. (A) PCA analysis result of red apple ATAC-Seq data at PS1, PS5 and PS9; (B) Relative location of the identified peaks in different gene regions; (C) Relative location of peaks to red apple genes in the upstream of 2 Kb and downstream of 2 Kb in the TSSs region; (D) Numbers of DARs in different comparisons; (E) Venn diagrams of DARs up-regulated in PS5_vs_PS1 but down-regulated in PS9_vs_PS5 (top) and DARs down-regulated in PS5_vs_PS1 but up-regulated in PS9_vs_PS5 (bottom); (F) Motif enrichment analysis results of MYB related motifs in DARs up-regulated in PS5_vs_PS1 but down-regulated in PS9_vs_PS5 and bZIP motifs in DARs up-regulated in PS9_vs_PS5 but down-regulated in PS5_vs_PS1.
To show the possible regulation of chromatin accessibility on the expression of genes related to anthocyanins and flavonols biosynthesis, we further focused on DARs showing opposite patterns of change between PS5_vs_PS1 and PS9_vs_PS5. In total, we identified 128 DARs up-regulated in the former comparison but down-regulated in the latter one (consistent with the pattern of change for differentially accumulated anthocyanins) (Figure 3E, Table S13), and 364 DARs up-regulated in the later comparison but down-regulated in the former one (consistent with the pattern of change for differentially accumulated quercetins) (Figure 3E, Table S14). The MYB-bHLH-WD40 (MBW) regulatory complex has been frequently discovered to function in regulating flavonoids biosynthesis (Zhao et al., 2019). Among the DARs up-regulated in PS5_vs_PS1 and down-regulated in PS9_vs_PS5, we identified two MYB genes (C1H46_025763 and C1H46_027060) and one bHLH gene (C1H46_000912) (Figure S6A). Moreover, ten MYB genes, eleven bHLH genes and one WD40 gene were found to be down-regulated in the DARs of PS5_vs_PS1 and up-regulated in PS9_vs_PS5 (Figure S6B). Interestingly, two bHLH genes (C1H46_035986 and C1H46_021331) are identified in the common DARs. A motif enrichment analysis using HOMER showed that MYB-related motifs were significantly enriched in the DARs with the same change pattern as the differentially accumulated anthocyanins (Figure 3F, Table S15), while bZIP-related motifs were significantly enriched in the DARs with the same change pattern as differentially accumulated flavonols (Figure 3F, Table S16).
To further reveal the potential regulatory roles of TFs on the expression of genes related to anthocyanins and flavonols biosynthesis, PlantTFDB was applied to predict the genes encoding TFs in the above common DEGs. The results showed that among the 852 common DEGs that were up-regulated in PS5_vs_PS1 but down-regulated in PS9_vs_PS5, genes encoding MYB, ERF, WRKY, bHLH and NAC accounted for more than 10% of all TF genes, with the percentages of 14.29%, 14.29%, 14.29%, 13.27% and 12.24%, respectively (Figure 4A; Table S17). In contrast, among the 843 common DEGs up-regulated in PS9_vs_PS5 but down-regulated in PS5_vs_PS1, genes encoding TF family proteins such as MYB, ERF, TCP, bZIP, Dof, bHLH and ZF-LD accounted for a larger proportion of all TF-encoding DEGs, accounting for 15.38%, 9.62%, 9.62%, 7.69%, 7.69%, 5.77% and 5.77%, respectively (Figure 4A; Table S18).
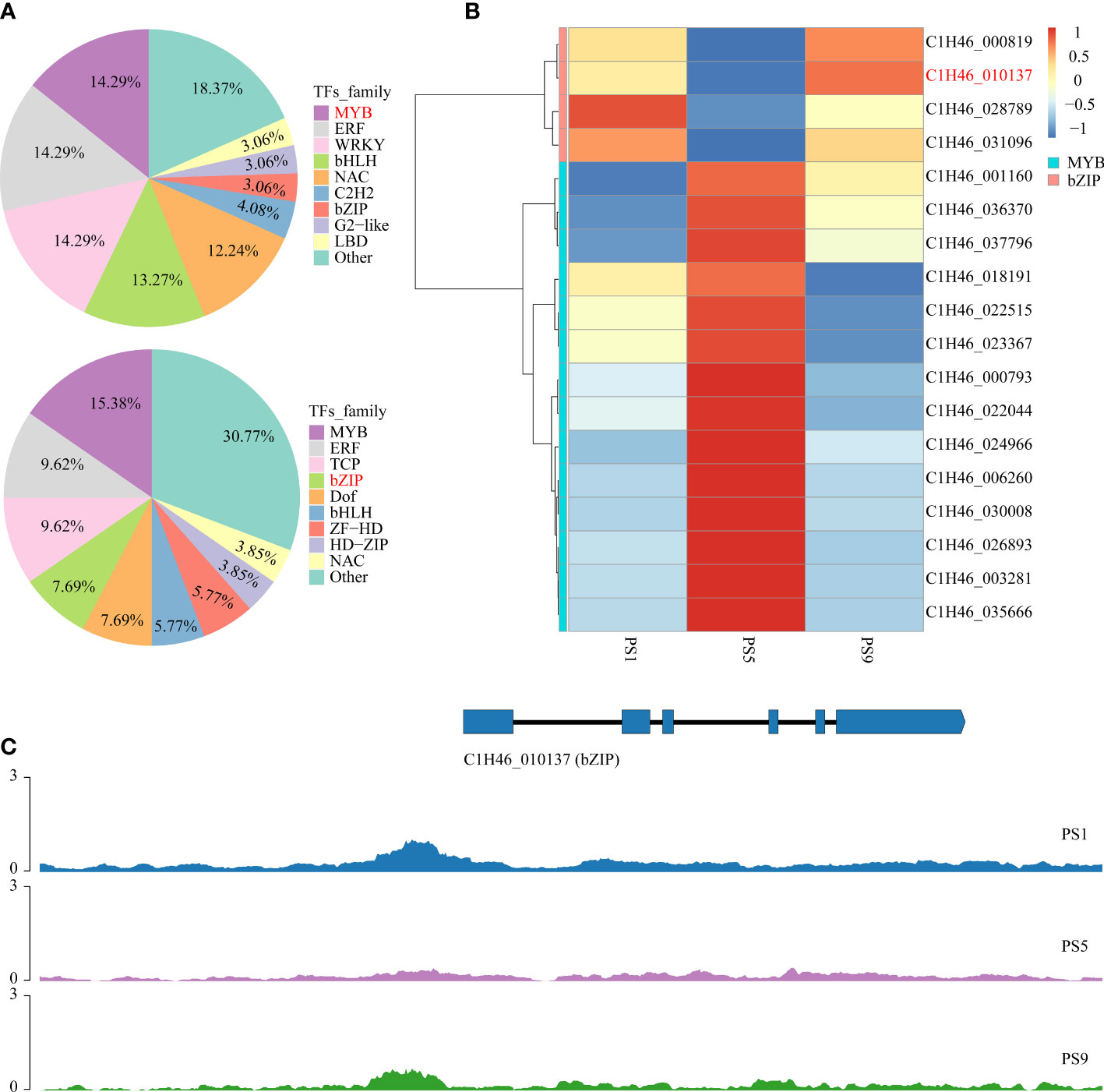
Figure 4 Candidate TF genes that play roles in the competition for anthocyanins and quercetins biosynthesis. (A) Frequencies of the transcription factor gene families in all the differentially expressed transcription factor genes; (B) Heatmap of differentially expressed MYB and bZIP genes that showed adverse change patterns in comparison PS5_vs_PS1 and comparison PS9_vs_PS5. (C) Chromatin accessibilities of a bZIP gene (C1H46_010137) at three different ripening stages.
By integrating the results of metabolome and motif enrichment analysis, it was predicted that TF MYBs have many functions and different MYB members play various roles in regulating both anthocyanins and flavonols biosynthesis, while bZIP transcription factors might play important roles in regulating flavonols biosynthesis during fruit ripening in red apple. Among these MYB and bZIP genes, four bZIPs and 14 MYBs showed significant positive correlations with anthocyanins and quercetins contents, respectively (Figure 4B; Table 1), and they could be considered as hub genes regulating the dynamics of anthocyanins and quercetins, respectively. Among these bZIP and MYB genes, some of their homologous genes, including Arabidopsis vip1, pear MYB10, apple MYB24-like and MYB11 have been reported to be associated with anthocyanin biosynthesis (Stracke et al., 2001; Sun et al., 2019; Wang et al., 2019a; Li et al., 2020b; Land et al., 2021). Notably, the chromatin accessibility in the promoter region of one bZIP gene (C1H46_010137) was found to be positively correlated with its gene expression level at the three stages (Figure 4C; Tables S19, S20), and its function in the competition for anthocyanins and flavonols biosynthesis needs to be further explored in future studies. Moreover, our qRT-PCR results showed that the expression patterns of the two selected MYB and three bZIP genes were identical to our transcriptome data (Figure S4), which well supported our transcriptome data and indicated that these TFs might contribute greatly to the competition between anthocyanins and flavonols biosynthesis.
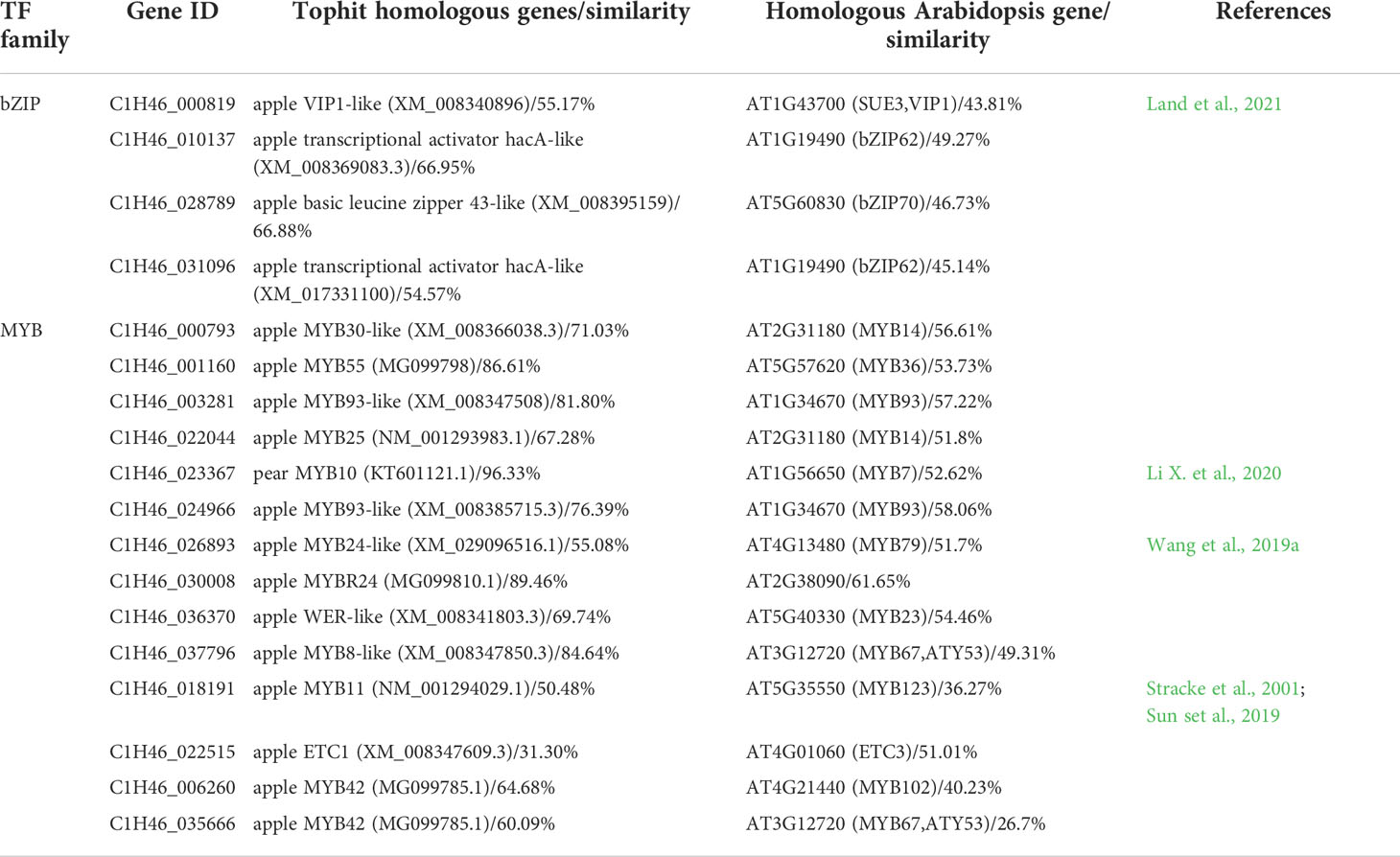
Table 1 The identified differentially expressed bZIP and MYB genes that showed opposite change patterns between PS5_vs_PS1 and PS9_vs_PS5 comparisons.
Fruit ripening is a complex process accompanied by extensive metabolites and global changes in gene expression controlled by a variety of factors, including developmental and environmental factors. These changes varied greatly in different plant species and different cultivars of the same plant species. Malus spp. is rich in flavonoids, especially in their flowers and fruits. In this study, we investigated the metabolome, transcriptome and chromatin accessibility changes of apple ‘Hongmantang’ with purple-red flower and blood-red flesh during fruit ripening, through integrative analyses of the datasets of UPLC-MS/MS-based widely targeted metabolome, RNA-Seq and ATAC-Seq from three ripening stages (PS1, PS5 and PS9).
In the study by Xu et al. (2020), more than half of the metabolites identified in apple (M. domestica) fruits showed high accumulation at early ripening stages. In our present study, we found that most metabolites belonging to amino acids and derivatives, organic acids, nucleotides and derivatives and tannins accumulated the highest in red apple fruits at PS1, and the numbers of decreased DAMs were both higher than that of the increased DAMs in the two comparisons, suggesting that the accumulations of these kinds of metabolites mostly reduced as fruit ripened. However, the numbers of the increased saccharides and alcohols related metabolites were both higher than the decreased ones, which is in consistent with the sugar accumulation during fruit ripening (Xu et al., 2020). Moreover, the numbers of increased DAMs belonging to phenolic acids, flavonoids, lipids, terpenoids, lignans and coumarins were much lower than the decreased ones in the comparison PS5_vs_PS1, but were much higher than the increased ones in the comparison PS9_vs_PS5, suggesting that the accumulations of these kinds of metabolites are associated with the red apple ripening process.
Given that they share the same biosynthesis substrates, competition occurred between anthocyanins and flavonols biosynthesis pathways in many plants (Luo et al., 2016; Zhong et al., 2020; Wong et al., 2022). For example, a comparison of anthocyanins and flavonols contents in the petals of ten Rhododendron species revealed that species with low anthocyanin contents had high flavonols (Liu et al., 2016); in Malus, competition between anthocyanins and kaempferol glycosides was found to be related to pollen tube growth and seed set (Chen et al., 2021); in bilberry, procyanidins and quercetins were identified as the major flavonoids in the early fruit ripening stage, but anthocyanins accounted for the largest fraction in the later ripening stages (Jaakola et al., 2002). In our present study, we found that many flavonoids were identified as DAMs during red apple fruit ripening. Moreover, in both PS5_vs_PS1 and PS9_vs_PS5 comparisons, cyanidin-3-O-glucoside and eight quercetins were found to be DAMs, but showed completely opposite accumulation trends. Therefore, it is hypothesized that there is a dynamic competition between anthocyanins and flavonols biosynthesis in red apple fruits during fruit development and ripening.
The accumulations of anthocyanins and flavonols largely contribute to the color changes during fruit ripening (Jaakola et al., 2002). Dihydroflavonols, the intermediates and branch point of the anthocyanins and flavonols biosynthetic pathways, are required for anthocyanins and flavonols productions and as substrates for their competition (Davies et al., 2003). This competitive process is controlled by many biosynthetic genes, especially DFRs and FLSs (Chen et al., 2021). In tomato, the anthocyanin without mutant, harboring a defect in DFR, displayed elevated flavonols and anthocyanins deficiency, and another mutant, anthocyanin reduced (are) has been reported to be caused by the mutation in a FLS, a gene encoding an enzyme catalyzing the first step of flavonol biosynthesis (Maloney et al., 2014). By using the white-flowered, flavonol accumulating Mitchell line of petunia as material, Davies et al. (2003) found that overexpression both of 35SCaMV-DFR sense and/or FLS antisense transgene led to the anthocyanins production and resulted in a pink-flowered phenotype. Constitutive overexpression of FLS genes from Rosa rugosa, Prunus persica, and Petunia hybrida in tobacco all resulted in white flowers, while the heterologous overexpression of DFR genes resulted in increased anthocyanins accumulations and redder flowers (Luo et al., 2016; Wong et al., 2022). Liu et al. (2019b) reported that heterologous expression of the Muscari aucheri MaFLS gene in tobacco increased NtFLS expression, but inhibited the expression of tobacco anthocyanins biosynthesis genes, such as NtDFR, NtANS and NtAN2, and reduced the pigmentation of tobacco petals. Similarly, Park et al. (2020) reported that the overexpression of the onion FLS gene (AcFLS-HRB) increased the flavonols contents in tobacco petals but decreased the anthocyanins accumulation and produced lighter-pink flowers. In our study, almost all the DEGs encoding enzymes that catalyze the reactions from dihydroflavonols to anthocyanidins were least expressed at PS9, which is in consistent with the relatively low accumulation of anthocyanins in red apple fruits at this stage. Moreover, two red apple FLS genes had the highest expression at PS9, which well explains the high accumulation of flavonols in red apple fruit at the late stage. It is therefore concluded that the differential expression of these structural genes contributes greatly to the accumulations of different kinds of flavonoids, and that altering their expression by genetic methods may have great potential for balancing anthocyanins and flavonols production (Davies et al., 2003).
In addition to structural genes, the biosynthesis of flavonoids is also controlled by many TFs (Martins et al., 2013; Zhang et al., 2021). The regulatory roles of MBW complex (consisted of MYB, bHLH and WD40 TFs) in flavonoid biosynthesis are well known (Zhao et al., 2019). Consistently, we identified many MBW genes, especially MYB and bHLH genes, as DEGs during ripening in red apple, indicating that these TFs play important roles in flavonoid biosynthesis in red apple. Moreover, we identified more than twenty MBW genes in the DARs that showed opposite change patterns between PS5_vs_PS1 and PS9_vs_PS5 comparisons. For example, the mutant of Arabidopsis vip1, a homologous gene of the bZIP (C1H46_000819), showed much less anthocyanin accumulation under phosphate-limiting conditions (Land et al., 2021); the pear MYB10, a homologous gene of the red apple MYB (C1H46_023367) has been proved to play roles in regulating anthocyanin biosynthesis and transport in red-skinned pears (Li et al., 2020); and the apple MYB24-like (homologous to red apple MYB (C1H46_030008)) and MYB11 (homologous to red apple MYB (C1H46_01819)) genes, were both proved to be involved in flavonoids biosynthesis (Sun et al., 2019; Wang et al., 2019a). As one of the most important TFs regulating the flavonoids synthesis, the regulatory roles of MYBs in both flavonols and anthocyanins biosynthesis have been continuously demonstrated in many fruit trees, such as apple (Takos et al., 2006; An et al., 2018a; An et al., 2018b; Wang et al., 2019a; Wang et al., 2019b; Jia et al., 2020; Xu H. et al., 2020), pear (Li et al., 2020a; Li et al., 2020), grape (Deluc et al., 2006), kiwifruit (Wang et al., 2019; Li et al., 2020a) and blueberry (Han et al., 2021) and so on. These reports showed that MYBs can be extensively involved in regulating flavonoids biosynthesis by interacting with some other TFs and with structural genes of flavonoids biosynthesis, thus affecting the color formation and development of plant organs such as leaves, petals and fruits.
Moreover, there are evidences that MYBs paly diverse roles in regulating flavonols and anthocyanins biosynthesis. For example, overexpression of apple MdMYB10 gene increased the anthocyanins accumulation but decreased the kaempferol 3-O-glycosides contents in flowers (Chen et al., 2021). Overexpression of a Gerbera hybrida R2R3-MYB transcription factor gene (GhMYB1a) resulted in decreased anthocyanins but increased flavonols accumulations in both gerbera and tobacco (Zhong et al., 2020). In our study, genes encoding MYB transcription factors were found to account for the largest fraction of differentially expressed TFs, which showed positive or negative expression patterns with the accumulations of anthocyanins and flavonols. Furthermore, ATAC-Seq analysis revealed that MYB-related motifs were significantly enriched in DARs, with the same change pattern as anthocyanins, which again indicates that MYBs contributed greatly to the regulation of both anthocyanins and flavonols biosynthesis in red apple.
Some other TFs also contribute greatly to the biosynthesis of flavonoids in plant, mainly through interacting with MYBs. Similarly, in our study, we also identified many other TF genes that showed positive or negative expression patterns with anthocyanins and flavonols accumulations, such as ERFs, bHLHs, WRKYs, NACs, bZIPs, TCPs and so on. And there were many evidences for the roles of these TFs in flavonoids biosynthesis (An et al., 2018c; An et al., 2020; Li C. et al., 2020; Li et al., 2020b; Ma et al., 2021; Zhang et al., 2021; Tu et al., 2022). Notably, bZIP-related motifs were identified to be significantly enriched in DARs that showed the same change pattern with differentially accumulated anthocyanins, suggesting that such TFs also play a major role in regulating flavonoids biosynthesis in red apple. In consistent with our study, numerous evidences demonstrate that the combinatorial action of bZIP and MYB in controlling the flavonoids biosynthesis in many plants (Stracke et al., 2010; Shin et al., 2013; Zhao et al., 2021). In Arabidopsis, bZIP TFs were shown to be functional in promoting anthocyanins accumulation by interacting with MYBL2 (Nguyen et al., 2015), PFG1/MYB12 (Stracke et al., 2010) and MYB75 (Shin et al., 2013) TFs and anthocyanin biosynthetic genes. In soybean, GmMYB and GmbZIP5 interact synergistically in controlling flavonoids biosynthesis. In tea plants, the CsbZIP1-CsMYB12 interaction mediates flavonols production via a coordinated activator-repressor network (Zhao et al., 2021).
Notably, most of the bZIP family genes reported to be involved in the regulation of anthocyanins biosynthesis were identified as positive regulators of anthocyanins biosynthesis (Han et al., 2021; Tu et al., 2022), which is also consistent with our results. Arabidopsis HY5 promotes anthocyanin accumulation by upregulating the expression of the DFR gene (Zhang et al., 2011). Tomato HY5 has been reported to be functional in mediating the CRY1a-induced anthocyanins biosynthesis (Liu et al., 2018; Qiu et al., 2019), and SlHY5 frameshift mutation showed decreased anthocyanin accumulation (Qiu et al., 2019). Soybean bZIP TF G/HBF-1 plays a key role in increasing phytoalexin accumulation by binding to the promoter of CHS gene (Dröge-Laser et al., 1997). The pear bZIP transcription factor PybZIPa has the ability to bind to the promoter of PyUFGT and functions in promoting the light-induced anthocyanin accumulation (Liu et al., 2019a). Grapevine VvHY5 promotes flavonol biosynthesis by activating the expression of FLS (Loyola et al., 2016); another grapevine bZIP, VvibZIPC22, can promote the accumulation of several kinds of flavonoids by regulating the expression of the CHS and FLS genes; and VvHY5 promotes flavonol biosynthesis by activating the expression of an FLS gene (Malacarne et al., 2016). In the study by Wang et al. (2022), two pomegranate bZIP genes, PgbZIP16 and PgbZIP34, which are associated with anthocyanin biosynthesis, were expressed at much higher levels in red petals than in white petals, and their heterologous transient overexpression in tobacco leaves both resulted in increased anthocyanin accumulation. In apple, An et al. (2018d) reported that the ABA inducible MdbZIP44 functioned in the ABA-induced anthocyanin accumulation by enhancing the binding activity of MdMYB1 to the promoters of anthocyanin biosynthetic genes. All these reports suggest that the bZIP TFs identified in our present study might play important roles in regulating the biosynthesis of flavonoids in red apple.
In summary, through combined analyses of metabolome, transcriptome and chromatin accessibility datasets, we investigated the molecular changes in red apple ‘Hongmantang’ fruits at three ripening stages (PS1, PS5 and PS9) (Figure 5). Metabolic analysis showed that most of the metabolites belonging to amino acids and derivatives, organic acids, nucleotides and their derivatives, and tannins decreased, while most metabolites belonging to saccharides and alcohols, vitamins and stilbene increased with fruit ripening. Notably, a dynamic competition between anthocyanins and flavonols biosynthesis was found. The differential expression of structural genes for flavonoids biosynthesis, such as FLSs and DFRs, played a major role in the accumulations of the two kinds of flavonoids. Moreover, chromatin accessibility and changes in gene expression of MYB and bZIP genes were predicted to play dominant regulatory roles in balancing the anthocyanins and flavonols biosynthesis.
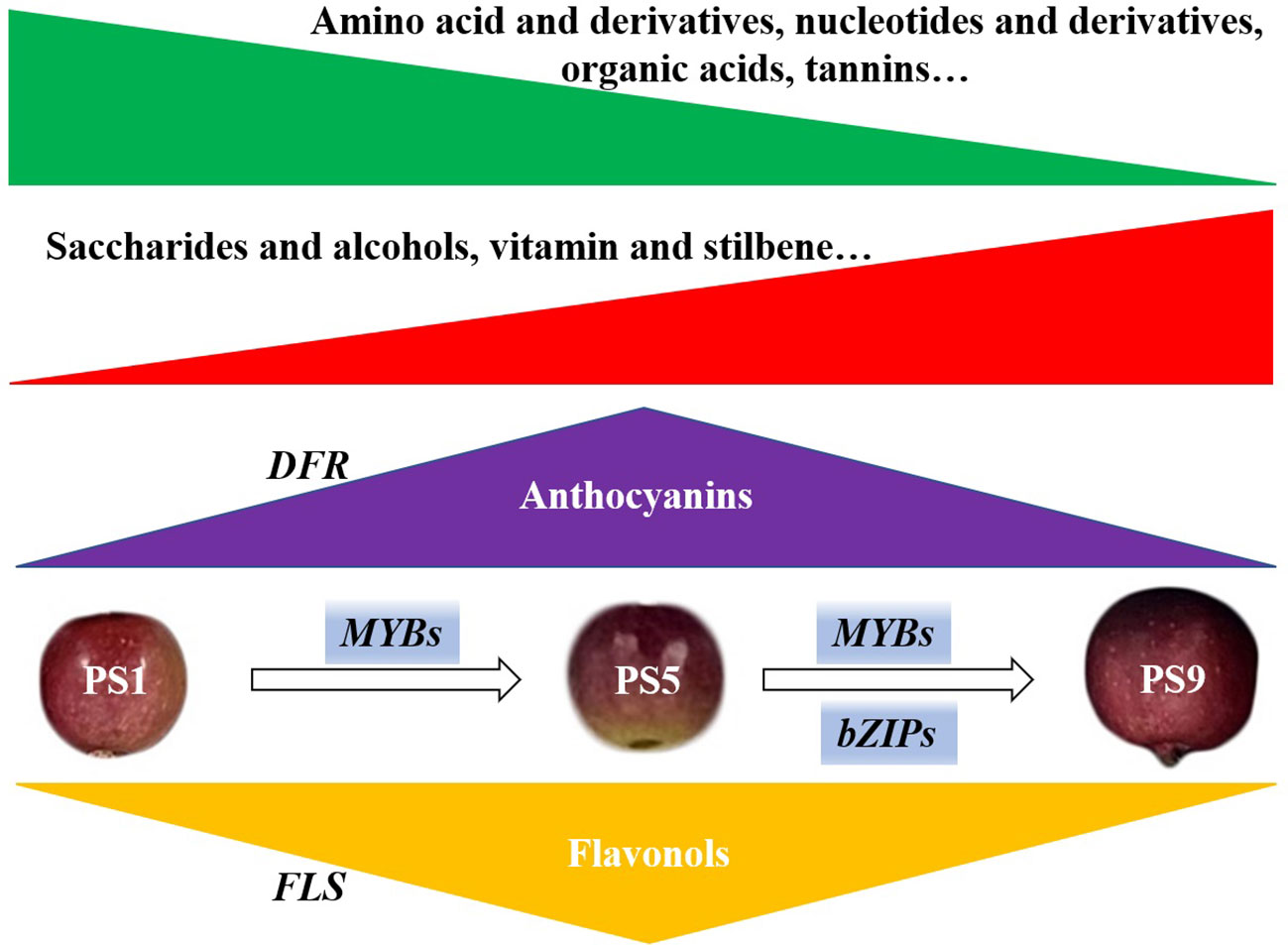
Figure 5 Schematic diagram of the molecular changes in red apple fruits during ripening. The green and red triangle represent the decreased and increased change pattern of metabolites. The accumulations of metabolites mostly belonging to amino acids and derivatives, organic acids, nucleotides and derivatives, and tannins reduced with fruit ripening, while the accumulations of metabolites mostly belonging to saccharides and alcohols, vitamins and stilbene increased with fruit ripening. The accumulation of anthocyanins peaked at PS5 and decreased at PS9. However, the flavonols accumulated least at PS5 but increased at PS9. The differential expression of DFRs and FLSs, functioned greatly to the accumulations of anthocyanins and flavonols, respectively. Changes in chromatin accessibility and expression of MYBs and bZIPs suggest that they play dominant regulatory roles in the competition for anthocyanins and flavonols biosynthesis.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA860789 & PRJNA861071.
JZ, PL and CC conceived and designed the experiments. CC, ZG, HL, JZ and PL wrote the original draft. CC, ZG, HL, XM, PW, SZ, TY, HC and QW performed the experiments. CC, ZG, HL and PL analyzed the results. CC, HL, XM, PL and JZ reviewed and edited the manuscript. All authors read and approved the final manuscript. The authors have no conflict of interest to declare.
This work was supported by the National Key Research and Development Program of China (2018YFD1000200), the earmarked fund for Modern Agro-industry Technology Research System of Shanxi Province, the Reward Fund for PhDs and Postdoctors of Shanxi Province (SXBYKY2022004), the Fund for High-level Talents of Shanxi Agricultural University (2021XG010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.975356/full#supplementary-material
Supplementary Figure 1 | Changes in phenotype (A) and anthocyanin accumulation (B) of red apple during fruit ripening. PS1 to P10 represent fruits collected at 7, 9, 11, 13, 15, 17, 19, 21, 23 and 25 weeks post flowering, respectively. There were two anthocyanins accumulation peaks in red apple fruits during ripening, i.e., at PS5 (15 weeks post flowering) and PS9 (23 weeks post flowering), respectively.
Supplementary Figure 2 | Heatmap of clustering of metabolites in red apple fruits at three ripening stages. PS1, PS5 and P9 represent fruits collected at 7, 15 and 23 weeks post flowering, respectively. For each stage, three replicates were performed. Red and green colors represent high and low accumulations, respectively.
Supplementary Figure 3 | Differentially expressed flavonoids biosynthesis-related genes in red apple fruits at different ripening stages. PAL, phenylalanine ammonia-lyase; 4CL, 4‐coumarate‐CoA ligase; CHS, chalcone synthase; CHI, Chalcone isomerase; F3H, flavonone-3-hydroxylase; F3’H, flavonoid‐3’‐hydroxylase; F3’5’H, flavonoid-3’-5’‐hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol reductase; ANS, anthocyanidin synthase; LAR, leucocyanidin reductase; ANR, anthocyanidin reductase; UFGT, anthocyanidin 3-O-glucosyltransferase; PA, proanthocyanidins. Red and blue colors represent high expression levels of genes.
Supplementary Figure 4 | Quantitative real time PCR (qRT-PCR) analysis of some differentially expressed flavonoids biosynthetic structural genes and transcription factor genes. C1H46_001201: PAL (phenylalanine ammonia-lyase); C1H46_013998: 4CL (4‐coumarate‐CoA ligase); C1H46_010805: CHS (chalcone synthase); C1H46_009496: CHI (Chalcone isomerase); C1H46_033494: F3H (flavonone-3-hydroxylase); C1H46_012818: F3’H (flavonoid-3’-5’‐hydroxylase); C1H46_020507: DFR (dihydroflavonol reductase); C1H46_005520: FLS (flavonol synthase); C1H46_043502: UFGT (anthocyanidin 3-O-glucosyltransferase); C1H46_023367 and C1H46_018191: MYB genes; C1H46_000819, C1H46_010137 and C1H46_028789: bZIP genes. The bar plots represent the relative expression levels of genes according to qRT-PCR analysis. The triangles with dashed lines represent the average FPKM values of the genes.
Supplementary Figure 5 | Insert size distribution, euclidean distance and venn diagrams of the ATAC-Seq data for fruits at three different ripening stages. For each stage of fruits, two biological replicates were performed.
Supplementary Figure 6 | Heatmap of the MBW genes identified in differential accessible regions (DARs) showing opposite change pattern in the comparisons of PS5_vs_PS1 and PS9_vs_PS5. A: The MBW genes identified in DARs up-regulated in PS5_vs_PS1 and down-regulated in PS9_vs_PS5. B: The MBW genes identified in DARs down-regulated in PS5_vs_PS1 and up-regulated in PS9_vs_PS5, the two bHLH genes in red were identified in the common DARs.
An, J. P., An, X. H., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2018a). BTB protein MdBT2 inhibits anthocyanin and proanthocyanidin biosynthesis by triggering MdMYB9 degradation in apple. Tree Physiol. 38 (10), 1578–1587. doi: 10.1093/treephys/tpy063
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., Hao, Y. J. (2018b). R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96 (3), 562–577. doi: 10.1111/tpj.14050
An, J. P., Wang, X. F., Li, Y. Y., Song, L. Q., Zhao, L. L., You, C. X., et al. (2018c). EIN3-LIKE1, MYB1, and ethylene response factor 3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 178 (2), 808–823. doi: 10.1104/PP.18.00068
An, J. P., Yao, J. F., Xu, R. R., You, C. X., Wang, X. F., Hao, Y. J. (2018d). Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 41 (11), 2678–2692. doi: 10.1111/pce.13393
An, J. P., Zhang, X. W., Bi, S. Q., You, C. X., Wang, X. F., Hao, Y. J. (2020). The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 101 (3), 573–589. doi: 10.1111/tpj.14555
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y., Greenleaf, W. J. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 10 (12), 1213–1218. doi: 10.1038/nmeth.2688
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant. 6 (6), 1769–1780. doi: 10.1093/mp/sst080
Chen, W., Xiao, Z., Wang, Y., Wang, J., Zhai, R., Lin-Wang, K., et al. (2021). Competition between anthocyanin and kaempferol glycosides biosynthesis affects pollen tube growth and seed set of Malus. Horticult. Res. 8 (1), 173. doi: 10.1038/s41438-021-00609-9
Corces, M. R., Trevino, A. E., Hamilton, E. G., Greenside, P. G., Sinnott-Armstrong, N. A., Vesuna, S., et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14 (10), 959–962. doi: 10.1038/nmeth.4396
Davies, K. M., Schwinn, K. E., Deroles, S. C., Manson, D. G., Lewis, D. H., Bloor, S. J., et al. (2003). Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica. 131 (3), 259–268. doi: 10.1023/A:1024018729349
Deluc, L., Barrieu, F., Marchive, C., Lauvergeat, V., Decendit, A., Richard, T., et al. (2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140 (2), 499–511. doi: 10.1104/pp.105.067231
Dröge-Laser, W., Kaiser, A., Lindsay, W. P., Halkier, B. A., Loake, G. J., Doerner, P., et al. (1997). Rapid stimulation of a soybean protein-serine kinase that phosphorylates a novel bZIP DNA-binding protein, G/HBF-1, during the induction of early transcription-dependent defenses. EMBO J. 16 (4), 726–738. doi: 10.1093/emboj/16.4.726
Fu, L., Ding, Z., Tie, W., Yang, J., Yan, Y., Hu, W. (2022). Integrated metabolomic and transcriptomic analyses reveal novel insights of anthocyanin biosynthesis on color formation in Cassava tuberous roots. Front. Nutr. 9. doi: 10.3389/fnut.2022.842693
Grandi, F. C., Modi, H., Kampman, L., Corces, M. R. (2022). Chromatin accessibility profiling by ATAC-seq. Nat. Protoc. 17 (6), 1518–1552. doi: 10.1038/s41596-022-00692-9
Guo, Z., Hou, W., Fu, H., Zhang, J., Wang, P., Mu, X., et al. (2021). Changes of phenolic substances and antioxidant capacity during fruit development of different apple varieties. Shandong Agric. Sci. 53 (11), 35–44. doi: 10.14083/j.issn.1001-4942.2021.11.006
Gutierrez, E., Velasco, A. G., Antonio Lucas, J., Gutierrez-Mañero, F. J., Ramos-Solano, B. (2017). The flavonol-anthocyanin pathway in blackberry and arabidopsis: State of the art. Flavonoids From Biosynthesis to Hum. Health. doi: 10.5772/67902
Han, T., Wu, W., Li, W. (2021). Transcriptome analysis revealed the mechanism by which exogenous ABA increases anthocyanins in blueberry fruit during veraison. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.758215
Han, H., Xu, F., Li, Y., Yu, L., Fu, M., Liao, Y., et al. (2021). Genome-wide characterization of bZIP gene family identifies potential members involved in flavonoids biosynthesis in Ginkgo biloba l. Sci. Rep. 11 (1), 23420. doi: 10.1038/s41598-021-02839-2
Huang, M., Zhang, L., Zhou, L., Yung, W. S., Wang, Z., Xiao, Z., et al. (2022). Identification of the accessible chromatin regions in six tissues in the soybean. Genomics. 114 (3), 110364. doi: 10.1016/j.ygeno.2022.110364
Jaakola, L., Määttä, K., Pirttilä, A. M., Törrönen, R., Kärenlampi, S., Hohtola, A. (2002). Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 130 (2), 729–739. doi: 10.1104/pp.006957
Jia, D., Li, Z., Dang, Q., Shang, L., Shen, J., Leng, X., et al. (2020). Anthocyanin biosynthesis and methylation of the MdMYB10 promoter are associated with the red blushed-skin mutant in the red striped-skin “changfu 2“ apple. J. Agric. Food Chem. 68 (15), 4292–4304. doi: 10.1021/acs.jafc.9b07098
Jiang, S., Chen, M., He, N., Chen, X., Wang, N., Sun, Q., et al. (2019). MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Horticult. Res. 6 (1), 40. doi: 10.1038/s41438-019-0118-6
Land, E. S., Cridland, C. A., Craige, B., Dye, A., Hildreth, S. B., Helm, R. F., et al. (2021). A role for inositol pyrophosphates in the metabolic adaptations to low phosphate in arabidopsis. Metabolites. 11, 601. doi: 10.3390/metabo11090601
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods. 9 (4), 357–359. doi: 10.1038/nmeth.1923
Lee, K. W., Kim, Y. J., Kim, D. O., Lee, H. J., Lee, C. Y. (2003). Major phenolics in apple and their contribution to the total antioxidant capacity. J. Agric. Food Chem. 51 (22), 6516–6520. doi: 10.1021/jf034475w
Li, Y., Cui, W., Qi, X., Lin, M., Qiao, C., Zhong, Y., et al. (2020a). MicroRNA858 negatively regulates anthocyanin biosynthesis by repressing AaMYBC1 expression in kiwifruit (Actinidia arguta). Plant Sci. 296, 110476. doi: 10.1016/j.plantsci.2020.110476
Li, H., Lv, Q., Liu, A., Wang, J., Sun, X., Deng, J., et al. (2022). Comparative metabolomics study of tartary (Fagopyrum tataricum (L.) gaertn) and common (Fagopyrum esculentum moench) buckwheat seeds. Food Chem. 371, 131125. doi: 10.1016/j.foodchem.2021.131125
Liu, C. C., Chi, C., Jin, L. J., Zhu, J., Yu, J. Q., Zhou, Y. H. (2018). The bZIP transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 41 (8), 1762–1775. doi: 10.1111/pce.13171
Liu, H., Su, B., Zhang, H., Gong, J., Zhang, B., Liu, Y., et al. (2019b). Identification and functional analysis of a flavonol synthase gene from grape hyacinth. Molecules. 24 (8), 1579. doi: 10.3390/molecules24081579
Liu, H., Su, J., Zhu, Y., Yao, G., Allan, A. C., Ampomah-Dwamena, C., et al. (2019a). The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Horticult. Res. 6 (1), 134. doi: 10.1038/s41438-019-0217-4
Liu, L., Zhang, L. Y., Wang, S. L., Niu, X. Y. (2016). Analysis of anthocyanins and flavonols in petals of 10 Rhododendron species from the sygera mountains in southeast Tibet. Plant Physiol. Biochem. 104, 250–256. doi: 10.1016/j.plaphy.2016.03.036
Li, C., Wu, J., Hu, K., Wei, S. W., Sun, H. Y., Hu, L. Y., et al. (2020). PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Horticult. Res. 7 (1), 37. doi: 10.1038/s41438-020-0254-z
Li, X., Wu, T., Liu, H., Zhai, R., Wen, Y., Shi, Q., et al. (2020). REVEILLE transcription factors contribute to the nighttime accumulation of anthocyanins in ‘red zaosu’ (Pyrus bretschneideri rehd.) pear fruit skin. Int. J. Mol. Sci. 21 (5), 1634. doi: 10.3390/ijms21051634
Li, Y., Xu, P., Chen, G., Wu, J., Liu, Z., Lian, H. (2020b). FvbHLH9 functions as a positive regulator of anthocyanin biosynthesis by forming a HY5-bHLH9 transcription complex in strawberry fruits. Plant Cell Physiol. 61 (4), 826–837. doi: 10.1093/pcp/pcaa010
Loyola, R., Herrera, D., Mas, A., Wong, D. C. J., Höll, J., Cavallini, E., et al. (2016). The photomorphogenic factors UV-b RECEPTOR 1, ELONGATED HYPOCOTYL 5, and HY5 HOMOLOGUE are part of the UV-b signalling pathway in grapevine and mediate flavonol accumulation in response to the environment. J. Exp. Bot. 67 (18), 5429–5445. doi: 10.1093/jxb/erw307
Luo, P., Ning, G., Wang, Z., Shen, Y., Jin, H., Li, P., et al. (2016). Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01257
Malacarne, G., Coller, E., Czemmel, S., Vrhovsek, U., Engelen, K., Goremykin, V., et al(2016). The grapevine VvibZIPC22 transcription factor is involved in the regulation of flavonoid biosynthesis. Journal of Experimental Botany, 67(11):3509–3522. doi: 10.1093/jxb/erw181
Maloney, G. S., DiNapoli, K. T., Muday, G. K. (2014). The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol. 166 (2), 614–631. doi: 10.1104/pp.114.240507
Martin, C. (2013). The interface between plant metabolic engineering and human health. In Curr. Opin. Biotechnol. 24 (2), 344–353. doi: 10.1016/j.copbio.2012.11.005
Martins, T. R., Berg, J. J., Blinka, S., Rausher, M. D., Baum, D. A. (2013). Precise spatio-temporal regulation of the anthocyanin biosynthetic pathway leads to petal spot formation in Clarkia gracilis (Onagraceae). New Phytol. 197 (3), 958–969. doi: 10.1111/nph.12062
Ma, H., Yang, T., Li, Y., Zhang, J., Wu, T., Song, T., et al. (2021). The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell. 33 (10), 3309–3330. doi: 10.1093/plcell/koab188
Nguyen, N. H., Jeong, C. Y., Kang, G. H., Yoo, S. D., Hong, S. W., Lee, H. (2015). MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in arabidopsis. Plant J. 84 (6), 1192–1205. doi: 10.1111/tpj.13077
Park, S., Kim, D. H., Yang, J. H., Lee, J. Y., Lim, S. H. (2020). Increased flavonol levels in tobacco expressing AcFLS affect flower color and root growth. Int. J. Mol. Sci. 21 (3), 1011. doi: 10.3390/ijms21031011
Qiu, Z., Wang, H., Li, D., Yu, B., Hui, Q., Yan, S., et al. (2019). Identification of candidate HY5-dependent and -independent regulators of anthocyanin biosynthesis in tomato. Plant Cell Physiol. 60 (3), 643–656. doi: 10.1093/pcp/pcy236
Ren, C., Li, H., Wang, Z., Dai, Z., Lecourieux, F., Xin, H., et al. (2021). Characterization of chromatin accessibility and gene expression upon cold stress reveals that the RAV1 transcription factor functions in cold response in Vitis amurensis. Plant Cell Physiol. 62 (10), 1615–1629. doi: 10.1093/pcp/pcab115
Schwope, R., Magris, G., Miculan, M., Paparelli, E., Celii, M., Tocci, A., et al. (2021). Open chromatin in grapevine marks candidate CREs and with other chromatin features correlates with gene expression. Plant J. 107 (6), 1631–1647. doi: 10.1111/tpj.15404
Shin, D. H., Choi, M., Kim, K., Bang, G., Cho, M., Choi, S. B., et al. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in arabidopsis. FEBS Lett. 587 (10), 1543–1547. doi: 10.1016/j.febslet.2013.03.037
Stracke, R., Favory, J. J., Gruber, H., Bartelniewoehner, L., Bartels, S., Binkert, M., et al. (2010). The arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-b radiation. Plant Cell Environ. 33 (1), 88–103. doi: 10.1111/j.1365-3040.2009.02061.x
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 (5), 447–456. doi: 10.1016/s1369-5266(00)00199-0
Sun, Q., Jiang, S., Zhang, T., Xu, H., Fang, H., Zhang, J., et al. (2019). Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 289, 110286. doi: 10.1016/j.plantsci.2019.110286
Takos, A. M., Jaffé, F. W., Jacob, S. R., Bogs, J., Robinson, S. P., Walker, A. R. (2006). Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 142 (3), 1216–1232. doi: 10.1104/pp.106.088104
Tu, M., Fang, J., Zhao, R., Liu, X., Yin, W., Wang, Y., et al. (2022). CRISPR/Cas9-mediated mutagenesis of VvbZIP36 promotes anthocyanin accumulation in grapevine (Vitis vinifera). Horticult. Res. 9, uhac022. doi: 10.1093/hr/uhac022
Vrhovsek, U., Rigo, A., Tonon, D., Mattivi, F. (2004). Quantitation of polyphenols in different apple varieties. J. Agric. Food Chem. 52 (21), 6532–6538. doi: 10.1021/jf049317z
Wang, Y., Liu, W., Jiang, H., Mao, Z., Wang, N., Jiang, S., et al. (2019a). The R2R3-MYB transcription factor MdMYB24-like is involved in methyl jasmonate-induced anthocyanin biosynthesis in apple. Plant Physiol. Biochem. 139, 273–282. doi: 10.1016/j.plaphy.2019.03.031
Wang, Y., Sun, J., Wang, N., Xu, H., Qu, C., Jiang, S., et al. (2019b). MdMYBL2 helps regulate cytokinin-induced anthocyanin biosynthesis in red-fleshed apple (Malus sieversii f. Niedzwetzkyana) callus. Funct. Plant Biol. 46 (2), 187–196. doi: 10.1071/FP17216
Wang, L., Tang, W., Hu, Y., Zhang, Y., Sun, J., Guo, X., et al. (2019). A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang Plant J. 99 (2), 359–378. doi: 10.1111/tpj.14330
Wang, S., Zhang, X., Li, B., Zhao, X., Shen, Y., Yuan, Z. (2022). Genome-wide identification and characterization of bZIP gene family and cloning of candidate genes for anthocyanin biosynthesis in pomegranate (Punica granatum). BMC Plant Biol. 22 (1), 170. doi: 10.1186/s12870-022-03560-6
Wong, D. C. J., Perkins, J., Peakall, R. (2022). Anthocyanin and flavonol glycoside metabolic pathways underpin floral color mimicry and contrast in a sexually deceptive orchid. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.860997
Xu, J., Yan, J., Li, W., Wang, Q., Wang, C., Guo, J., et al. (2020). Integrative analyses of widely targeted metabolic profiling and transcriptome data reveals molecular insight into metabolomic variations during apple (Malus domestica) fruit development and ripening. Int. J. Mol. Sci. 21 (13), 4797. doi: 10.3390/ijms21134797
Xu, H., Zou, Q., Yang, G., Jiang, S., Fang, H., Wang, Y., et al. (2020). MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Horticult. Res. 7 (1), 72. doi: 10.1038/s41438-020-0294-4
Yang, T., Gao, J., Wang, Q., Cai, H., Li, C., Du, X., et al. (2015). A new Malus ornamental variety ‘Hongmantang’. J. Fruit Sci. 32 (4), 727–729. doi: 10.13925/j.cnki.gsxb.20150130
Yuan, H., Zeng, X., Shi, J., Xu, Q., Wang, Y., Jabu, D., et al. (2018). Time-course comparative metabolite profiling under osmotic stress in tolerant and sensitive tibetian hulless barley. BioMed. Res. Int. 2018, 9415409. doi: 10.1155/2018/9415409
Zhang, J., Xu, H., Wang, N., Jiang, S., Fang, H., Zhang, Z., et al. (2018). The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol. Biol. 98 (3), 205–218. doi: 10.1007/s11103-018-0770-5
Zhang, Q., Wang, L., Liu, Z., Zhao, Z., Zhao, J., Wang, Z., et al. (2020). Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba mill. peel coloration. Food Chem. 312, 125903. doi: 10.1016/j.foodchem.2019.125903
Zhang, Y., Liu, F., Wang, B., Wu, H., Wu, J., Liu, J., et al. (2021). Identification, characterization and expression analysis of anthocyanin biosynthesis-related bHLH genes in blueberry (Vaccinium corymbosum l.). Int. J. Mol. Sci. 22 (24), 13274. doi: 10.3390/ijms222413274
Zhang, Y., Zheng, S., Liu, Z., Wang, L., Bi, Y.(2011). Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. Journal of Plant Physiology, 168(4):367–374. doi: 10.1016/j.jplph.2010.07.025
Zhao, M., Li, J., Zhu, L., Chang, P., Li, L., Zhang, L. (2019). Identification and characterization of MYB-bHLH-WD40 regulatory complex members controlling anthocyanidin biosynthesis in blueberry fruits development. Genes. 10 (7), 496. doi: 10.3390/genes10070496
Zhao, X., Zeng, X., Lin, N., Yu, S., Fernie, A. R., Zhao, J. (2021). CsbZIP1-CsMYB12 mediates the production of bitter-tasting flavonols in tea plants (Camellia sinensis) through a coordinated activator–repressor network. Horticult. Res. 8 (1), 110. doi: 10.1038/s41438-021-00545-8
Keywords: anthocyanin, flavonol, fruit ripening, expression regulation, omics
Citation: Cheng C, Guo Z, Li H, Mu X, Wang P, Zhang S, Yang T, Cai H, Wang Q, Lü P and Zhang J (2022) Integrated metabolic, transcriptomic and chromatin accessibility analyses provide novel insights into the competition for anthocyanins and flavonols biosynthesis during fruit ripening in red apple. Front. Plant Sci. 13:975356. doi: 10.3389/fpls.2022.975356
Received: 22 June 2022; Accepted: 09 September 2022;
Published: 23 September 2022.
Edited by:
Jose G. Vallarino, University of Malaga, SpainReviewed by:
Delphine Maya Pott, University of Tübingen, GermanyCopyright © 2022 Cheng, Guo, Li, Mu, Wang, Zhang, Yang, Cai, Wang, Lü and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiancheng Zhang, empjbmQwMDFAMTYzLmNvbQ==; Peitao Lü, cHRsdkBmYWZ1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.