
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 16 August 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.974598
This article is part of the Research Topic Regulation of Proteolysis and Proteome Composition in Plant Response to Environmental Stress View all 11 articles
Abiotic stresses are major environmental conditions that reduce plant growth, productivity and quality. Protein-protein interaction (PPI) approaches can be used to screen stress-responsive proteins and reveal the mechanisms of protein response to various abiotic stresses. Biotin-based proximity labeling (PL) is a recently developed technique to label proximal proteins of a target protein. TurboID, a biotin ligase produced by directed evolution, has the advantages of non-toxicity, time-saving and high catalytic efficiency compared to other classic protein-labeling enzymes. TurboID-based PL has been successfully applied in animal, microorganism and plant systems, particularly to screen transient or weak protein interactions, and detect spatially or temporally restricted local proteomes in living cells. This review concludes classic PPI approaches in plant response to abiotic stresses and their limitations for identifying complex network of regulatory proteins of plant abiotic stresses, and introduces the working mechanism of TurboID-based PL, as well as its feasibility and advantages in plant abiotic stress research. We hope the information summarized in this article can serve as technical references for further understanding the regulation of plant adaptation to abiotic stress at the protein level.
Plant life needs certain natural factors such as temperature, moisture and nutrition, while plants often suffer from environmental stresses during development including changes of temperature, salinity, water, light, nutrient availability, and toxic chemicals (Vanstraelen and Benkova, 2012; Skalak et al., 2021; Zhou et al., 2021). In response to these abiotic stresses, various adaptations have evolved in plants at the physiological, molecular, and cellular levels, which are regulated by complex signal transduction pathways (Zhu, 2016). In these important pathways, regulatory proteins play an essential role. Proteins rarely act on their own, while often function as complexes through protein-protein interactions (PPIs) (Struk et al., 2019). Therefore, the study of PPIs can not only infer the protein functions within the cell, but also uncover unidentified proteins from their interactions with known proteins (Zhang et al., 2010). There are two types of PPIs, namely constitutive and regulative, in the cell (Fujikawa et al., 2014). Constitutive PPIs are typically ubiquitous and strong interactions, whereas regulative PPIs occur only in certain cellular or developmental contexts or in response to specific incentives (Morsy et al., 2008). The dynamic changes of regulative PPIs confer cells with the ability to rapidly respond to intracellular and extracellular stimuli (Syafrizayanti et al., 2014). Regulative PPIs have the features of instantaneity, specificity and instability, which make them challenging to be studied (Lalonde et al., 2008).
Many classic PPI approaches, such as Yeast Two-Hybrid (Y2H), Co-Immunoprecipitation (Co-IP), Affinity Purification (AP), Pull-down, Bimolecular Fluorescence Complementation (BiFC), and Split Luciferase (Split-LUC), have been utilized for studying the protein interaction network in plant response to abiotic stresses. These techniques have identified many critical regulatory proteins involved in abiotic stress responses (Urano et al., 2010; Li et al., 2016; Zhang et al., 2016; Han et al., 2020; Qin L. et al., 2021). However, they also have many limitations, which hinder their applications, particularly in the analyses of regulative PPIs. Recently, a new PPI technique named TurboID-based proximity labeling (PL) has been applied in bio-research, which has a number of advantages especially for studying dynamic and transient PPIs (Bohnert et al., 2006; Branon et al., 2018; Zhang et al., 2019; Cho et al., 2020; Li et al., 2021). Although there are only a few cases of its application in plant research, we envision wide usage of this cutting-edge technique in dissecting the protein interaction network that regulates abiotic stress responses in plants.
Classic PPI approaches, including Y2H, Co-IP, AP, Pull-down and BiFC, have been widely utilized in plant studies and introduced in detail in a number of review articles (Drewes and Bouwmeester, 2003; Weinthal and Tzfira, 2009; Zhang et al., 2010; Braun et al., 2013; Ferro and Trabalzini, 2013; Rao et al., 2014). The Y2H system includes bait and prey proteins in frame with DNA-binding domain (BD) or a transactivation domain (AD), respectively. When AD and BD domains are in spatial proximity to each other, expression of the reporter gene is activated to demonstrate the interaction between bait and prey (Causier and Davies, 2002). Co-IP and AP are two in vivo PPI approaches under near-physiological conditions. Co-IP and AP have similar principles, which entail overexpression of the bait protein (with or without an affinity tag) in plant protoplast or tissue, and isolation of the bait with its interacting partners (prey) through purification based on antibody-antigen interactions. The isolated bait-prey complex can be analyzed by liquid chromatography tandem-MS (LC-MS/MS) to achieve high throughput analysis (Ransone, 1995; Masters, 2004; Xie et al., 2012; Xing et al., 2016). Pull-down is an in vitro method that can be used to detect or validate the direct interaction between bait and prey proteins. In Pull-down approach, bait or prey protein is usually expressed as a fusion protein with tags in bacteria, the immobilizing bait-tag fusion protein on tag specific column is used as affinity support to catch and purify the prey proteins that interact with bait protein, and these prey proteins can be detected by sodium-dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by western-blotting detection (Louche et al., 2017). BiFC and Split-LUC are based on the principle of fluorescent protein-fragment complementation assay (PCA). The individual N- or C- terminal part of a fluorescent protein normally has no fluorescence signal, but when N- and C- terminal parts are fused with two partner proteins, respectively, interaction of these two proteins will make the N- and C- parts close enough to regain the fluorescent protein structure and activity (Kerppola, 2006, 2008; Azad et al., 2014). The results of BiFC and Split-LUC can both be shown by the emission of the reconstructed fluorescent proteins, and Split-LUC signals can also be quantified by the luciferase activity assay (Kerppola, 2006, 2008; Azad et al., 2014).
Abiotic stresses such as temperature extremes, salinity, drought, reduced nutrient availability, and toxic chemicals are major limiting factors for plant development. Plants have evolved excellent defense mechanisms to protect themselves from abiotic stresses including stress sensing, signal transduction and transcriptional regulation, etc., (Zhang H. et al., 2022). The classic PPI approaches mentioned above are widely used to build up the protein interaction network and identify hub proteins, including transcription factors, signaling molecules and transporter proteins in the regulation of plant response to abiotic stresses (Supplementary Table 1). For example, in phosphorus (Pi) deficiency stress, the interaction of the Ubiquitin-Conjugating Enzyme PHO2 and the Pi transporter PHTs had been verified by Y2H and BiFC (Liu et al., 2012), and the feedback inhibition of SPX-domain proteins on PHOSPHATE STARVATION RESPONSE 1 (PHR1), the central transcriptional regulator of Pi Starvation Responses (PSR), had also been assessed by Y2H, Co-IP and BiFC (Lv et al., 2014). These results greatly contributed to the understanding of the genetic network that controls PSR in plants.
Although the classic PPI approaches have been successfully applied in many abiotic stress studies, their shortages cannot be overlooked. For example, Y2H has benefits of high sensitivity, maintaining the natural folding of fusion proteins and convenient operation that bypasses the complicated steps of protein extraction and purification. However, it also has obvious disadvantages, including high technical false positive rates due to the strong spontaneous activation of reporter gene transcription, and toxicity of plant proteins to yeast cells, which can cause false negative results. Also, Y2H often fails to detect protein interactions that rely on post-translational modifications (PTMs). Therefore, Y2H is more suitable for cDNA library screening rather than confirming protein interactions, and the Y2H results often need to be confirmed by other PPI approaches (Hamdi and Colas, 2012; Mehla et al., 2015). Unlike Y2H, Co-IP/AP-MS can be used to pull down protein complexes under native physiological conditions to reflect the in vivo binding. But these approaches also suffer from high false positive rates. Also, Co-IP/AP-MS need to overexpress the bait protein which may influence its physiological properties. In addition, the choice of lysis conditions may have strong influence on the result of Co-IP/AP-MS. Lysis conditions may break PPI, and the low solubility of some subcellular structures in normal lysis buffer, e.g., plasmalemma, cytoskeleton and nucleus, may lead to negative results as well. Another shortcoming of Co-IP/AP-MS is that the instantaneous interactions or weak interactions often fail to be detected, and it is unable to distinguish direct and indirect interactions between the examined proteins. Pull-down is an approach used to detect the direct interaction between two proteins in vitro, with the outstanding features of being quick, sensitive, and quantifiable. But there are some disadvantages of Pull-down, such as it cannot reflect the protein interactions in plant physiological conditions, and each experiment needs to be optimized to keep characterized interactions from artifacts (Struk et al., 2019). Comparing to Co-IP/AP-MS or Pull-down, BiFC and Split-LUC have the advantage in identifying weak and instantaneous interactions because of the stability of the reconstituted GFP/YFP or LUC complexes. BiFC can also reveal the cellular localization of the PPI complex, which is convenient for further cellular studies. However, BiFC and Split-LUC can only be used to investigate the interaction of two proteins, and the interaction might be influenced by protein conformation, which could be changed after the joining of the N- and C- terminus of fluorescent proteins. These factors all limit the application of BiFC and Split-LUC in high throughput PPI analyses (Kerppola, 2008).
Due to the above disadvantages of classic PPI approaches and to avoid spurious results from heterologous expression, instantaneous and weak interactions, and indistinguishable cellular localization of the target proteins, enzyme-catalyzed PL techniques have been developed as novel alternative approaches to study PPIs (Qin W. et al., 2021; Xu et al., 2021; Yang et al., 2021; Mair and Bergmann, 2022). TurboID, a biotin ligase, has been exploited as an important PL enzyme with the advantages of non-toxicity and high catalytic efficiency (May et al., 2020).
TurboID is a 35 kDa biotin ligase engineered by yeast display-based directed evolution, which has 15 mutations relative to the wild-type Escherichia coli biotin ligase (BirA) (Branon et al., 2018). By fusing TurboID with the target protein of interest and expressing it in cells, when biotin is supplied in the presence of ATP, TurboID catalyzes biotin and forms reactive biotinoyl-5′-AMP (bioAMP) from biotin and ATP. These free bioAMPs are released and diffused to the vicinity of the target protein, which can covalently bind to lysine residues of proteins that are in close proximity to the TurboID enzyme (Roux et al., 2012; Branon et al., 2018). The biotin-labeled proteins are enriched and affinity purified by streptavidin pulldown and subsequently identified by MS, so as to identify the proximal proteins of the target protein (Roux et al., 2012). In contrast to classic methods, TurboID-based PL adds covalently bound tag in living cells, such that spatial relationships and interaction networks are not disrupted. In addition, the TurboID-based PL system simply requires a supply of exogenous non-toxic biotin, which permits it to be applied in vivo without causing damage to living cells. Furthermore, TurboID has high catalytic efficiency and biotinylation of proximal proteins can be completed in living cells within 10 min at 25°C, which allows its quick application in plants grown under ambient conditions (Branon et al., 2018; Mair et al., 2019; May et al., 2020; Zhang et al., 2020). Most importantly, TurboID can identify weak and transient protein interactions in living cells, which frequently fail to be captured by classic Co-IP/AP approaches (Branon et al., 2018; Kim et al., 2019). Moreover, it can also identify rare protein complexes or local organelle proteomes in individual cell types of complex multicellular organisms (Branon et al., 2018; Mair et al., 2019).
TurboID-based PL techniques have been applied successfully in a number of biological studies. For example, TurboID-based PL has been used to map local proteomes and screen novel interactors in vivo in zebrafish (Xiong et al., 2021). TurboID biotin ligase can also efficiently tag the entire proteome of specific cell types in the mouse brain, and dynamically track and identify tissue-specific or stimulation-specific secretory proteins in living body (Chua et al., 2021; Kim et al., 2021; Sun et al., 2022). TurboID-based PL was also used to identify host proteins interacting with viruses including coronavirus and syndrome coronavirus-2 (SARS-CoV-2), which help to elucidate the mechanism of virus infection and provide resources for the development of antiviral drugs for coronavirus disease 2019 (COVID-19) treatment (V’Kovski et al., 2020; Zhang Y. et al., 2022).
In plant research, TurboID-based PL technology has also been utilized in a variety of systems to study PPIs. For example, TurboID-based PL was used to identify interactors of a plant immune receptor N, which is a Nucleotide-binding Leucine-rich Repeat (NLR) that confers plant resistance to Tobacco Mosaic Virus (TMV) in Nicotiana benthamiana. In this work, a new regulator Ubiquitin Protein Ligase E3 Component N-Recognin 7 (UBR7) was found, which directly interacts with N and mediates immunity against plant pathogens (Zhang et al., 2019). TurboID-based PL was also applied to identify partners of the stomatal-specific transcription factor FAMA and help to obtain the nuclear proteome of young guard cells in Arabidopsis thaliana seedlings, which demonstrate that TurboID-based PL can be used to detect interactions of low abundant proteins and local proteomes of rare plant cell types (Mair et al., 2019). In addition, TurboID-based PL was also used to characterize neighboring proteins of Brassinosteroid-Insensitive 2 (BIN2), the regulatory kinase of Brassinosteroid (BR) pathways. This study uncovered a suite of previously unidentified BIN2 proximal proteins, which further enriched BIN2-mediated BR signaling networks (Kim et al., 2019). Furthermore, TurboID-based PL was successfully used to identify multiple interacting proteomes in the cell suspension cultures of tomato (Solanum lycopersicum), N. benthamiana and Arabidopsis, which showed that this technology can effectively capture membrane-associated protein interactions in different plant model systems (Arora et al., 2020).
Abiotic stresses can be sensed by plants not only at the cell surface, such as by receptors at the cell wall and plasma membrane, but also in intracellular compartments, such as by signaling proteins in the cytoplasm and nucleus. Stress signaling triggers physical or chemical changes of biomolecules in the plant cell, which can lead to a cellular stress response (Zhang H. et al., 2022). Signal transduction in this process involves secondary messengers and regulatory proteins, and the interactions between the components of signaling pathways tend to be transient and dynamic. For example, many kinases in the Mitogen-Activated Protein Kinase (MAPK) signal transduction cascades can be rapidly activated by abiotic stresses. These kinases can affect their own activities by interacting with specific partner proteins and can also modulate the activities of substrates through transient kinase-substrate interactions, thus dynamically acting in various physiological processes and regulating plant tolerance to abiotic stresses (Mishra et al., 2006; Moustafa et al., 2014; Andrasi et al., 2019). In addition, the stress-related PTMs, including phosphorylation, glycosylation, ubiquitination, sumoylation, oxidation, carbonylation and nitrosylation, etc., are also modulated by transient enzyme-substrate interactions because of the rapid turnover of the corresponding enzymes (Wu et al., 2016). Understanding these dynamic and transient PPIs are one of the major challenges in the investigation of the stress signaling network in plants. In this sense, the high efficiency of TurboID-based PL in detecting dynamic and transient protein interactions will make it a particularly useful tool in studying abiotic stress responses in plants. Furthermore, as many regulatory proteins acting in stress signal transduction pathways, such as transcription factors, transmembrane receptors and kinases are in low abundance (Kosova et al., 2011; Abreu et al., 2013), the advantage of TurboID-based PL in capturing low-abundant proteins will also greatly contribute to the identification of stress related factors at the protein level.
Abiotic stress causes multilevel responses, including stress sensing, signal transduction, transcription, transcript processing, translation and PTMs (Zhang H. et al., 2022). These responses can be initiated in various cellular structures including plasma membrane, nucleus, mitochondria, chloroplast, endoplasmic reticulum (ER) and cell wall. The important functions of these cellular structures in stress responses and the involvement of protein interactions in the regulation of their activities suggest that organelle proteome analysis may provide key information of the cellular mechanisms of plant response to stresses (Couee et al., 2006; Nouri and Komatsu, 2010; Pang et al., 2010; Hüner et al., 2012; Komatsu et al., 2012; Yin and Komatsu, 2016). However, due to the dynamic state of organelles and their proteins, clarifying the subcellular distribution and expression of organelle proteins has always been a challenging task (Boisvert et al., 2010). Utilization of TurboID-based PL may open a new avenue for organ and subcellular proteome research. TurboID-based PL has been used to label proteins located in the cell membrane, mitochondrial matrix, cytoplasm, nucleus, and ER lumen/membrane in mammalian cells (Branon et al., 2018; May et al., 2020). It can also be used to efficiently and specifically detect the proteome of different subcellular components of plants, such as the nuclear proteome of stomatal guard cells in Arabidopsis (Mair et al., 2019). Compared with classic tools, TurboID-based PL technology can label the organelle proteome of interest in living cells without isolating tissues and organelles, therefore, it can be used to investigate non-membrane-enclosed organelles that cannot be purified by classic biochemical fractionation methods. Another important application of the TurboID-based PL technology is that it can be used in combination with fluorescent microscopy to assess the cellular compartmentalization information of protein interactions. Because one protein can display diverse functions depending on its subcellular localization (Kosova et al., 2018), this application is helpful for understanding the spatial-specific regulatory process in cells in response to abiotic stress.
Plants have evolved excellent defense mechanisms to protect themselves from abiotic stresses. Classic PPI approaches like Y2H, Co-IP/AP-MS, Pull-down, BiFC and Split-LUC have contributed to identification of stress-regulatory proteins in plants (Figure 1). However, due to their limitations in detecting weak instantaneous interactions, distinguishing cellular localizations and directly assessing protein interactions in subcellular organs, further application of classic PPIs in systematic studies of plant stress responses is largely hindered. As a recently developed PPI approach, TurboID-based PL has been applied in mapping PPIs in a variety of species and has proven especially useful in dissecting signaling pathways (Kim et al., 2019). TurboID-based PL has a number of advantages, such as high flexibility, easy implementation, and great efficiency in detecting protein interactors that are low abundant, transient and specifically expressed in organelles. All of these advantages may greatly help us in our study of the mechanisms of plant stress responses (Kim et al., 2019; Mair et al., 2019; Figure 1). However, TurboID-based PL also has its own limitations, which results in their non-applicability in some abiotic stress conditions. For instance, biotin ligase activity is markedly influenced by low temperature, so TurboID-based PL is not suitable for studying cold stress responses. Also, the proximity-dependent labeling method only provide information on which proteins are in proximity to each other, it does not show direct evidence for a physical interaction between these proteins. Therefore, TurboID-based PL may needs to be combined with classic PPI approaches to map protein interactions in plant responses, and new proximity labeling ligases should be developed to overcome these shortcomings of TurboID. Overall, with the continuous renovation of PPI approaches, we believe the future research on protein interactions will provide in-depth knowledge of systematic molecular mechanisms for plant abiotic stress responses.
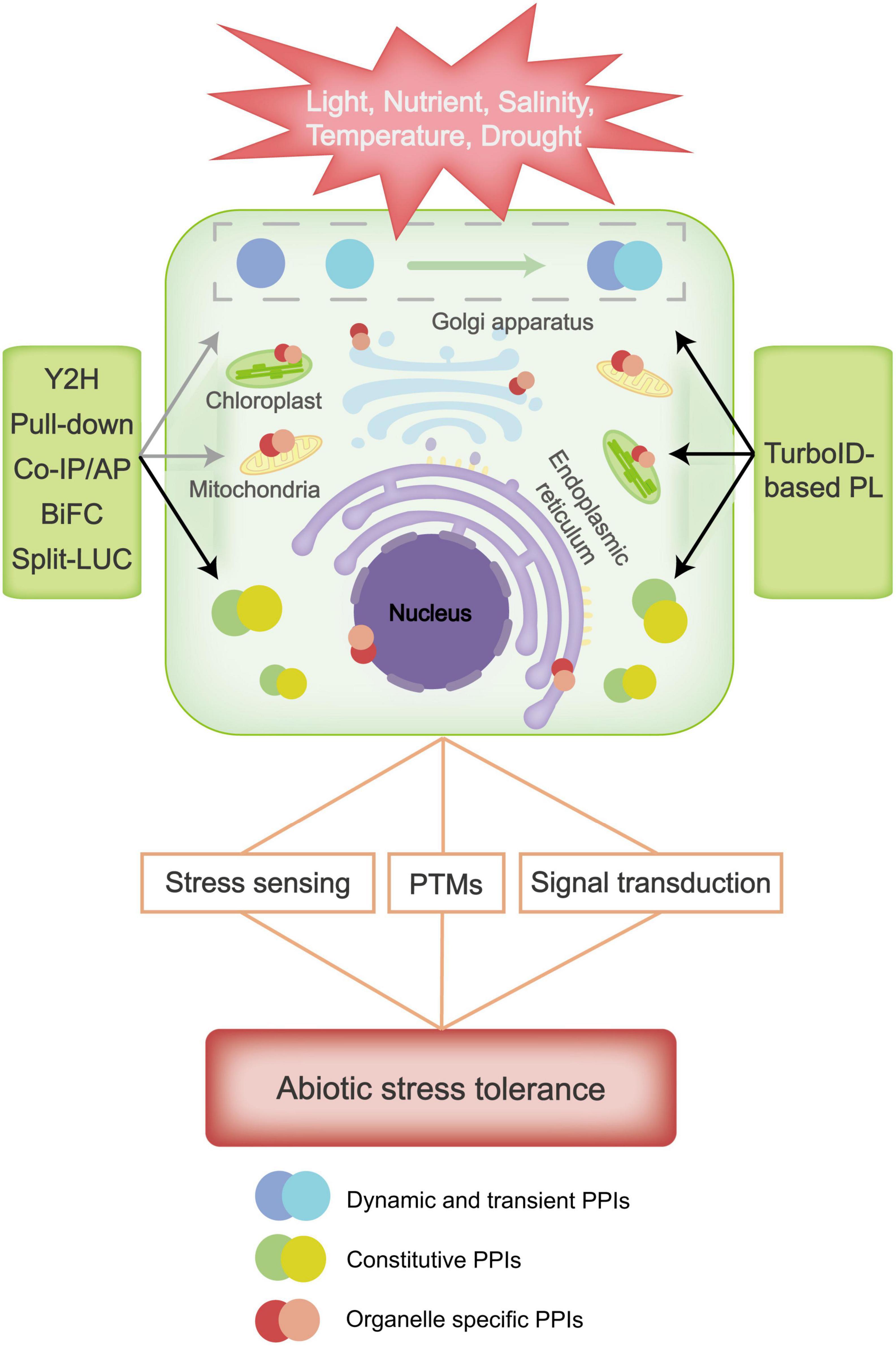
Figure 1. Plant resistance to abiotic stresses involves stress sensing, signal transduction and post-translational modifications (PTMs) of proteins, etc., in which many protein-protein interactions (PPIs) are involved. Classic methods such as Yeast Two Hybrid (Y2H), Pull-down, Co-Immunoprecipitation (Co-IP), Affinity Purification (AP), Bimolecular Fluorescence Complementation (BiFC), and Split Luciferase (Split-LUC) can easily detect stable protein interactions, such as constitutive PPIs that are typically macromolecular complex, but they are difficult to detect dynamic and organelle specific interacting proteins in response to abiotic stress. TurboID-based proximity labeling (PL) has great advantages in detecting dynamic, transient and organelle specific interacting proteins, and can be applied to study regulative PPIs under abiotic stress. Black and gray arrows indicate high and low applicability of the classic and TurboID-based technique(s) in detecting the corresponding PPIs.
KZ, YL, TH, and ZL were contributed to the writing of this review. All authors contributed to the article and approved the submitted version.
This studies on plant organogenesis in Huang lab have been supported by the Guangdong Basic and Applied Basic Research Foundation (2021A1515011035) and Guangdong Special Support Program for Young Talents in Innovation Research of Science and Technology (2019TQ05N940).
We thank the Instrumental Analysis Center of Shenzhen University (Lihu Campus) and Central Research Facilities of College of Life Sciences and Oceanography for assistance on our work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.974598/full#supplementary-material
Abreu, I. A., Farinha, A. P., Negrão, S., Gonçalves, N., Fonseca, C., Rodrigues, M., et al. (2013). Coping with abiotic stress: Proteome changes for crop improvement. J. Proteom. 93, 145–168. doi: 10.1016/j.jprot.2013.07.014
Andrasi, N., Rigo, G., Zsigmond, L., Perez-Salamo, I., Papdi, C., Klement, E., et al. (2019). The mitogen-activated protein kinase 4-phosphorylated heat shock factor A4A regulates responses to combined salt and heat stresses. J. Exp. Bot. 70, 4903–4918. doi: 10.1093/jxb/erz217
Arora, D., Abel, N. B., Liu, C., Van Damme, P., Yperman, K., Eeckhout, D., et al. (2020). Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell 32, 3388–3407. doi: 10.1105/tpc.20.00235
Azad, T., Tashakor, A., and Hosseinkhani, S. (2014). Split-luciferase complementary assay: Applications, recent developments, and future perspectives. Anal. Bioanal. Chem. 406, 5541–5560. doi: 10.1007/s00216-014-7980-8
Bohnert, H. J., Gong, Q., Li, P., and Ma, S. (2006). Unraveling abiotic stress tolerance mechanisms-getting genomics going. Curr. Opin. Plant Biol. 9, 180–188. doi: 10.1016/j.pbi.2006.01.003
Boisvert, F. M., Lam, Y. W., Lamont, D., and Lamond, A. I. (2010). A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell Proteom. 9, 457–470. doi: 10.1074/mcp.M900429-MCP200
Branon, T. C., Bosch, J. A., Sanchez, A. D., Udeshi, N. D., Svinkina, T., Carr, S. A., et al. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887. doi: 10.1038/nbt.4201
Braun, P., Aubourg, S., Van Leene, J., De Jaeger, G., and Lurin, C. (2013). Plant protein interactomes. Annu. Rev. Plant Biol. 64, 161–187. doi: 10.1146/annurev-arplant-050312-120140
Causier, B., and Davies, B. (2002). Analysing protein-protein interactions with the yeast two-hybrid system. Plant Mol. Biol. 50, 855–870. doi: 10.1023/a:1021214007897
Cho, K. F., Branon, T. C., Udeshi, N. D., Myers, S. A., Carr, S. A., and Ting, A. Y. (2020). Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat. Protoc. 15, 3971–3999. doi: 10.1038/s41596-020-0399-0
Chua, X. Y., Aballo, T., Elnemer, W., Tran, M., and Salomon, A. (2021). Quantitative interactomics of lck-TurboID in living human T cells unveils T cell receptor stimulation-induced proximal lck interactors. J. Proteome Res. 20, 715–726. doi: 10.1021/acs.jproteome.0c00616
Couee, I., Sulmon, C., Gouesbet, G., and El Amrani, A. (2006). Involvement of soluble sugars in reactive oxygen species balance and responses to oxidative stress in plants. J. Exp. Bot. 57, 449–459. doi: 10.1093/jxb/erj027
Drewes, G., and Bouwmeester, T. (2003). Global approaches to protein-protein interactions. Curr. Opin. Cell Biol. 15, 199–205. doi: 10.1016/s0955-0674(03)00005-x
Ferro, E., and Trabalzini, L. (2013). The yeast two-hybrid and related methods as powerful tools to study plant cell signalling. Plant Mol. Biol. 83, 287–301. doi: 10.1007/s11103-013-0094-4
Fujikawa, Y., Nakanishi, T., Kawakami, H., Yamasaki, K., Sato, M. H., Tsuji, H., et al. (2014). Split luciferase complementation assay to detect regulated protein-protein interactions in rice protoplasts in a large-scale format. Rice 7:11. doi: 10.1186/s12284-014-0011-8
Hamdi, A., and Colas, P. (2012). Yeast two-hybrid methods and their applications in drug discovery. Trends Pharmacol. Sci. 33, 109–118. doi: 10.1016/j.tips.2011.10.008
Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N., Qiu, N., et al. (2020). C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 11:115. doi: 10.3389/fpls.2020.00115
Hüner, N. P., Bode, R., Dahal, K., Hollis, L., Rosso, D., Krol, M., et al. (2012). Chloroplast redox imbalance governs phenotypic plasticity: The “grand design of photosynthesis” revisited. Front. Plant Sci. 3:255. doi: 10.3389/fpls.2012.00255
Kerppola, T. K. (2006). Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1, 1278–1286. doi: 10.1038/nprot.2006.201
Kerppola, T. K. (2008). Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487. doi: 10.1146/annurev.biophys.37.032807.125842
Kim, K. E., Park, I., Kim, J., Kang, M. G., Choi, W. G., Shin, H., et al. (2021). Dynamic tracking and identification of tissue-specific secretory proteins in the circulation of live mice. Nat. Commun. 12:5204. doi: 10.1038/s41467-021-25546-y
Kim, T. W., Chan, H. P., Hsu, C. C., Zhu, J. Y., and Wang, Z. Y. (2019). Application of TurboID-mediated proximity labeling for mapping a GSK3 kinase signaling network in Arabidopsis. bioRxiv [Preprint]. doi: 10.1101/636324
Komatsu, S., Kuji, R., Nanjo, Y., Hiraga, S., and Furukawa, K. (2012). Comprehensive analysis of endoplasmic reticulum-enriched fraction in root tips of soybean under flooding stress using proteomics techniques. J. Proteom. 77, 531–560. doi: 10.1016/j.jprot.2012.09.032
Kosova, K., Vitamvas, P., Prasil, I. T., and Renaut, J. (2011). Plant proteome changes under abiotic stress-contribution of proteomics studies to understanding plant stress response. J. Proteom. 74, 1301–1322. doi: 10.1016/j.jprot.2011.02.006
Kosova, K., Vitamvas, P., Urban, M. O., Prasil, I. T., and Renaut, J. (2018). Plant abiotic stress proteomics: The major factors determining alterations in cellular proteome. Front. Plant Sci. 9:122. doi: 10.3389/fpls.2018.00122
Lalonde, S., Ehrhardt, D. W., Loque, D., Chen, J., Rhee, S. Y., and Frommer, W. B. (2008). Molecular and cellular approaches for the detection of protein-protein interactions: Latest techniques and current limitations. Plant J. 53, 610–635. doi: 10.1111/j.1365-313X.2007.03332.x
Li, H., Frankenfield, A. M., Houston, R., Sekine, S., and Hao, L. (2021). Thiol-cleavable biotin for chemical and enzymatic biotinylation and its application to mitochondrial TurboID proteomics. J. Am. Soc. Mass Spectrom. 32, 2358–2365. doi: 10.1021/jasms.1c00079
Li, J., Ouyang, B., Wang, T., Luo, Z., Yang, C., Li, H., et al. (2016). HyPRP1 gene suppressed by multiple stresses plays a negative role in abiotic stress tolerance in tomato. Front. Plant Sci. 7:967. doi: 10.3389/fpls.2016.00967
Liu, T. Y., Huang, T. K., Tseng, C. Y., Lai, Y. S., Lin, S. I., Lin, W. Y., et al. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24, 2168–2183. doi: 10.1105/tpc.112.096636
Louche, A., Salcedo, S. P., and Bigot, S. (2017). Protein-protein interactions: Pull-down assays. Methods Mol. Biol. 1615, 247–255. doi: 10.1007/978-1-4939-7033-9_20
Lv, Q., Zhong, Y., Wang, Y., Wang, Z., Zhang, L., Shi, J., et al. (2014). SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26, 1586–1597. doi: 10.1105/tpc.114.123208
Mair, A., and Bergmann, D. C. (2022). Advances in enzyme-mediated proximity labeling and its potential for plant research. Plant Physiol. 188, 756–768. doi: 10.1093/plphys/kiab479
Mair, A., Xu, S. L., Branon, T. C., Ting, A. Y., and Bergmann, D. C. (2019). Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. eLife 8:e47864. doi: 10.7554/eLife.47864
Masters, S. C. (2004). Co-immunoprecipitation from transfected cells. Methods Mol. Biol. 261, 337–350
May, D. G., Scott, K. L., Campos, A. R., and Roux, K. J. (2020). Comparative application of BioID and TurboID for protein-proximity biotinylation. Cells 9:1070. doi: 10.3390/cells9051070
Mehla, J., Caufield, J. H., and Uetz, P. (2015). The yeast two-hybrid system: A tool for mapping protein-protein interactions. Cold Spring Harb. Protoc. 2015, 425–430. doi: 10.1101/pdb.top083345
Mishra, N. S., Tuteja, R., and Tuteja, N. (2006). Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452, 55–68. doi: 10.1016/j.abb.2006.05.001
Morsy, M., Gouthu, S., Orchard, S., Thorneycroft, D., Harper, J. F., Mittler, R., et al. (2008). Charting plant interactomes: Possibilities and challenges. Trends Plant Sci. 13, 183–191. doi: 10.1016/j.tplants.2008.01.006
Moustafa, K., AbuQamar, S., Jarrar, M., Al-Rajab, A. J., and Tremouillaux-Guiller, J. (2014). MAPK cascades and major abiotic stresses. Plant Cell Rep. 33, 1217–1225. doi: 10.1007/s00299-014-1629-0
Nouri, M. Z., and Komatsu, S. (2010). Comparative analysis of soybean plasma membrane proteins under osmotic stress using gel-based and LC MS/MS-based proteomics approaches. Proteomics 10, 1930–1945. doi: 10.1002/pmic.200900632
Pang, Q., Chen, S., Dai, S., Chen, Y., Wang, Y., and Yan, X. (2010). Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599. doi: 10.1021/pr100034f
Qin, L., Sun, L., Wei, L., Yuan, J., Kong, F., Zhang, Y., et al. (2021). Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress. Plant J. 105, 1010–1025. doi: 10.1111/tpj.15083
Qin, W., Cho, K. F., Cavanagh, P. E., and Ting, A. Y. (2021). Deciphering molecular interactions by proximity labeling. Nat. Methods 18, 133–143. doi: 10.1038/s41592-020-01010-5
Ransone, L. J. (1995). Detection of protein-protein interactions by coimmunoprecipitation and dimerization. Methods Enzymol. 254, 491–497. doi: 10.1016/0076-6879(95)54034-2
Rao, V. S., Srinivas, K., Sujini, G. N., and Kumar, G. N. (2014). Protein-protein interaction detection: Methods and analysis. Int. J. Proteomics 2014:147648. doi: 10.1155/2014/147648
Roux, K. J., Kim, D. I., Raida, M., and Burke, B. (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810. doi: 10.1083/jcb.201112098
Skalak, J., Nicolas, K. L., Vankova, R., and Hejatko, J. (2021). Signal integration in plant abiotic stress responses via multistep phosphorelay signaling. Front. Plant Sci. 12:644823. doi: 10.3389/fpls.2021.644823
Struk, S., Jacobs, A., Sanchez Martin-Fontecha, E., Gevaert, K., Cubas, P., and Goormachtig, S. (2019). Exploring the protein-protein interaction landscape in plants. Plant Cell Environ. 42, 387–409. doi: 10.1111/pce.13433
Sun, X., Sun, H., Han, X., Chen, P. C., Jiao, Y., Wu, Z., et al. (2022). Deep single-cell-type proteome profiling of mousebrain by nonsurgical AAV-mediated proximity labeling. Anal. Chem. 94, 5325–5334. doi: 10.1021/acs.analchem.1c05212
Syafrizayanti Betzen, C., Hoheisel, J. D., and Kastelic, D. (2014). Methods for analyzing and quantifying protein-protein interaction. Expert. Rev. Proteom. 11, 107–120. doi: 10.1586/14789450.2014.875857
Urano, K., Kurihara, Y., Seki, M., and Shinozaki, K. (2010). ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr. Opin. Plant Biol. 13, 132–138. doi: 10.1016/j.pbi.2009.12.006
Vanstraelen, M., and Benkova, E. (2012). Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28, 463–487. doi: 10.1146/annurev-cellbio-101011-155741
V’Kovski, P., Steiner, S., and Thiel, V. (2020). Proximity labeling for the identification of coronavirus-host protein interactions. Methods Mol. Biol. 2203, 187–204. doi: 10.1007/978-1-0716-0900-2_14
Weinthal, D., and Tzfira, T. (2009). Imaging protein-protein interactions in plant cells by bimolecular fluorescence complementation assay. Trends Plant Sci. 14, 59–63. doi: 10.1016/j.tplants.2008.11.002
Wu, X., Gong, F., Cao, D., Hu, X., and Wang, W. (2016). Advances in crop proteomics: PTMs of proteins under abiotic stress. Proteomics 16, 847–865. doi: 10.1002/pmic.201500301
Xie, G., Kato, H., and Imai, R. (2012). Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem. J. 443, 95–102. doi: 10.1042/BJ20111792
Xing, S., Wallmeroth, N., Berendzen, K. W., and Grefen, C. (2016). Techniques for the analysis of protein-protein interactions in vivo. Plant Physiol. 171, 727–758. doi: 10.1104/pp.16.00470
Xiong, Z., Lo, H. P., McMahon, K. A., Martel, N., Jones, A., Hill, M. M., et al. (2021). In vivo proteomic mapping through GFP-directed proximity-dependent biotin labelling in zebrafish. eLife 10:e64631. doi: 10.7554/eLife.64631
Xu, Y., Fan, X., and Hu, Y. (2021). In vivo interactome profiling by enzyme-catalyzed proximity labeling. Cell Biosci. 11:27. doi: 10.1186/s13578-021-00542-3
Yang, X., Wen, Z., Zhang, D., Li, Z., Li, D., Nagalakshmi, U., et al. (2021). Proximity labeling: An emerging tool for probing in planta molecular interactions. Plant Commun. 2:100137. doi: 10.1016/j.xplc.2020.100137
Yin, X., and Komatsu, S. (2016). Nuclear proteomics reveals the role of protein synthesis and chromatin structure in root tip of soybean during the initial stage of flooding stress. J. Proteome Res. 15, 2283–2298. doi: 10.1021/acs.jproteome.6b00330
Zhang, H., Li, W., Mao, X., Jing, R., and Jia, H. (2016). Differential activation of the wheat SnRK2 family by abiotic stresses. Front. Plant Sci. 7:420. doi: 10.3389/fpls.2016.00420
Zhang, H., Zhu, J., Gong, Z., and Zhu, J. K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhang, Y., Shang, L., Zhang, J., Liu, Y., Jin, C., Zhao, Y., et al. (2022). An antibody-based proximity labeling map reveals mechanisms of SARS-CoV-2 inhibition of antiviral immunity. Cell Chem. Biol. 29, 5–18. doi: 10.1016/j.chembiol.2021.10.008
Zhang, Y., Li, Y., Yang, X., Wen, Z., Nagalakshmi, U., and Dinesh-Kumar, S. P. (2020). TurboID-based proximity labeling for in planta identification of protein-protein interaction networks. J. Vis. Exp. 159:e60728. doi: 10.3791/60728
Zhang, Y., Song, G., Lal, N. K., Nagalakshmi, U., Li, Y., Zheng, W., et al. (2019). TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat. Commun. 10:3252. doi: 10.1038/s41467-019-11202-z
Zhang, Y. X., Gao, P., and Yuan, J. S. (2010). Plant protein-protein interaction network and interactome. Curr. Genom. 11, 40–46. doi: 10.2174/138920210790218016
Zhou, X., Joshi, S., Khare, T., Patil, S., Shang, J., and Kumar, V. (2021). Nitric oxide, crosstalk with stress regulators and plant abiotic stress tolerance. Plant Cell Rep. 40, 1395–1414. doi: 10.1007/s00299-021-02705-5
Keywords: plant, abiotic stress, protein interaction, TurboID, regulation network
Citation: Zhang K, Li Y, Huang T and Li Z (2022) Potential application of TurboID-based proximity labeling in studying the protein interaction network in plant response to abiotic stress. Front. Plant Sci. 13:974598. doi: 10.3389/fpls.2022.974598
Received: 21 June 2022; Accepted: 28 July 2022;
Published: 16 August 2022.
Edited by:
Zhiping Deng, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Andreas Bachmair, University of Vienna, AustriaCopyright © 2022 Zhang, Li, Huang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ziwei Li, bGl6aXdlaTE5ODlAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.