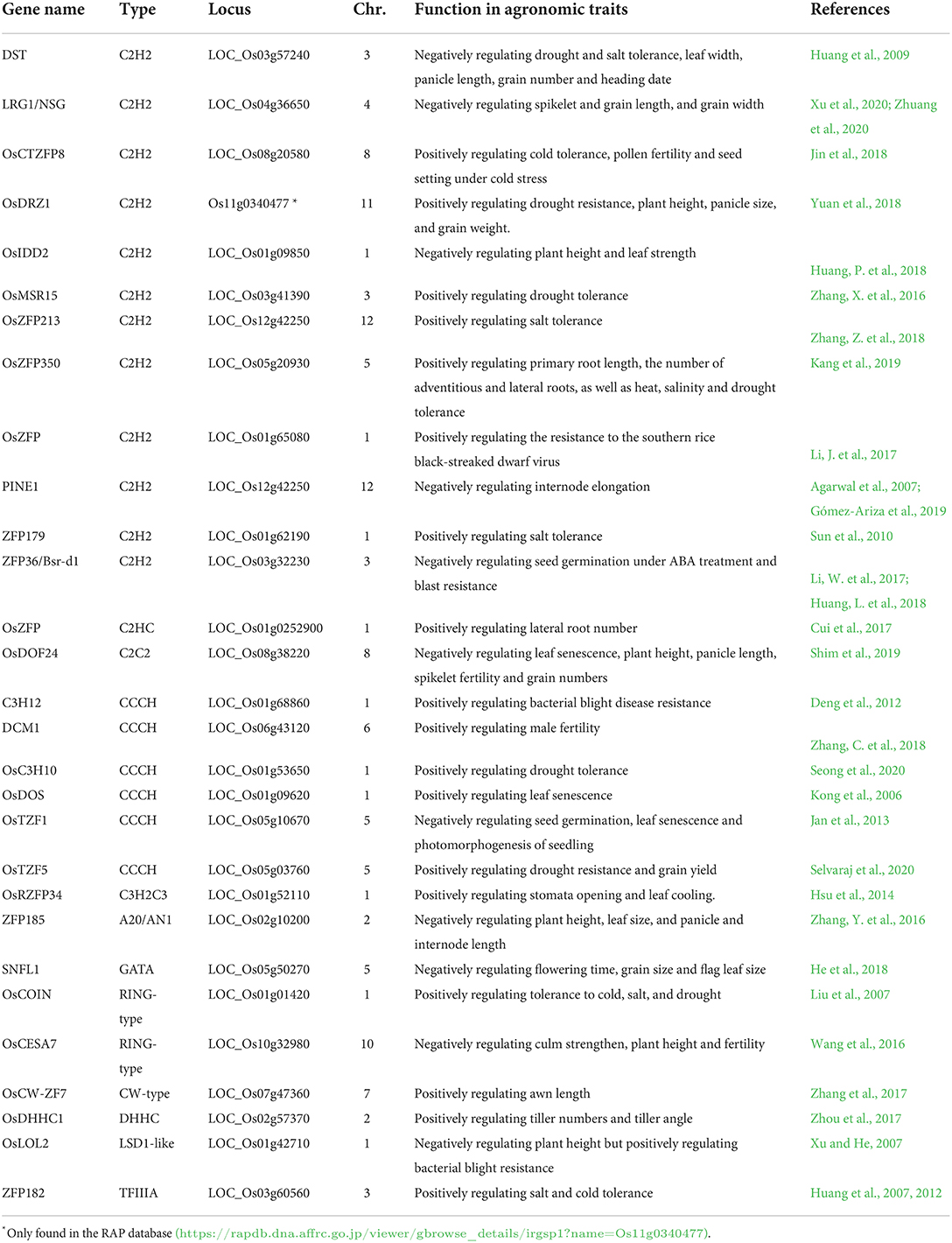
- 1Institute of Crop and Nuclear Technology Utilization, Zhejiang Academy of Agricultural Science, Hangzhou, China
- 2Guangdong Province Key Laboratory of Plant Molecular Breeding, Guangzhou, China
- 3Division of Integrative Bioscience and Biotechnology, Department of Life Sciences, Pohang University of Science and Technology (POSTECH), Pohang-si, South Korea
Rice is an important cereal crop, which provides staple food for more than half of the world's population. To meet the demand of the ever-growing population in the next few decades, an extra increase in rice yield is an urgent need. Given that various agronomic traits contribute to the yield of rice, deciphering the key regulators involved in multiple agronomic trait formation is particularly important. As a superfamily of transcription factors, zinc finger proteins participate in regulating multiple genes in almost every stage of rice growth and development. Therefore, understanding zinc finger proteins underlying regulatory network would provide insights into the regulation of agronomic traits in rice. To this end, we intend to summarize the current advances in zinc finger proteins, with emphasis on C2H2 and CCCH proteins, and then discuss their potential in improving rice yield.
Introduction
Rice crops are prone to various types of stresses, including biotic and abiotic stresses (Atkinson et al., 2013). Several gene families are induced and encode functional proteins (metabolic proteins) that directly protect rice crops against stresses or encode regulatory proteins that regulate the signal transduction in response to stresses (Jan et al., 2013). The regulatory proteins include transcription factors, protein kinases, protein phosphatases, and proteins involved in inorganic phosphate turnover (Han et al., 2020).
Zinc finger proteins that contain the zinc finger domain constitute one of the largest transcription factor families in eukaryotes (Seetharam and Stuart, 2013). The zinc finger was defined as the motifs in which cysteines and/or histidines coordinate a zinc atom(s) to form local peptide structures (Takatsuji, 1998). Zinc finger proteins participate in transcriptional regulation, RNA binding, regulation of apoptosis, and protein–protein interactions and play important roles in growth, development, and responses to environmental stresses (Takatsuji, 1998; Agarwal et al., 2007; Ciftci-Yilmaz and Mittler, 2008; Han et al., 2020).
Zinc finger proteins have been classified into different categories by different researchers. One of the classifications includes nine subfamilies, namely, Cys2/His2-type (C2H2), Cys3His (C3H), Cys3HisCys4 (C3HC4), Cys2HisCys5 (C2HC5), Cys4HisCys3 (C4HC3), Cys2HisCys (C2HC), Cys4 (C4), Cys6 (C6), and Cys8 (C8), based on their conserved cysteine (Cys) and histidine (His) residues that fold into a finger-like structure (Cassandri et al., 2017). Another classification includes six groups, namely, C2H2, C3H, C2C2, A20/AN1, C3H2C3, and C3HC4, based on the sequence characteristic of zinc finger conserved domain (Jin et al., 2018). Zinc finger is also classified into six groups, namely, C2H2, C2C2, C2HC, C2C2C2C2, C2HCC2C2, and CCCH, based on the number and order of the Cys and His residues that bind the zinc ion (Ciftci-Yilmaz and Mittler, 2008; Jan et al., 2013). The zinc domain types include C2H2, C8, C6, C3HC4, C2HC, C2HC5, C4, C3H, and C4HC3 (Berg and Shi, 1996).
Among different types of zinc finger proteins, the C2H2 zinc finger proteins constitute one of the largest families of transcriptional regulators in plants, with 176 members in Arabidopsis thaliana and 189 members in rice (Agarwal et al., 2007; Ciftci-Yilmaz and Mittler, 2008). The C2H2 proteins contain a conserved QALGGH sequence within their zinc finger domain (Takatsuji, 1998). The CCCH zinc finger proteins have been known to play important roles as RNA-binding proteins in animals (Wang, D. et al., 2008). A total of 67 and 68 CCCH zinc finger protein genes were found in rice and Arabidopsis, respectively (Wang, D. et al., 2008). There are 12 members of A20/AN1-type zinc finger proteins in rice, and 10 members in Arabidopsis (Huang et al., 2008). The classification of zinc finger protein genes can be different based on different family assignment rules (http://planttfdb.gao-lab.org/help_famschema.php). For example, japonica rice contains 135 C2H2 genes and 74 C3H genes according to the PlantTFDB v5.0 database (Tian et al., 2020).
The C2H2 zinc finger has a consensus sequence CX2-4CX3FX5LX2HX3-5H, where X represents any amino acid (Huang et al., 2012). The 3D structure of the zinc finger domain has been determined using Xenopus protein Xfin-31, which contains a YXCX2CX3FX5LX2HX3H sequence where the highly conserved residues are underlined (Lee et al., 1989). Most plant C2H2 zinc finger proteins have the highly conserved sequence QALGGH (Han et al., 2020). In this study, we use a rice C2H2 zinc finger transcription factor protein (GenBank: AAQ95583.1) to illustrate the zinc finger structure that contains two tandem finger motifs (Figures 1A,B), each motif having two b strands and one a-helix (Figure 1C) and (Figure 2A). The two b strands are arranged in a hairpin structure, accompanied by the a-helix and form a compact b b a domain in the presence of zinc (Wolfe et al., 2000). The zinc is tetrahedrally coordinated between two cysteines at the b-sheet (Figures 2B,C) and two histidines at the a-helix (Figures 2D,E).
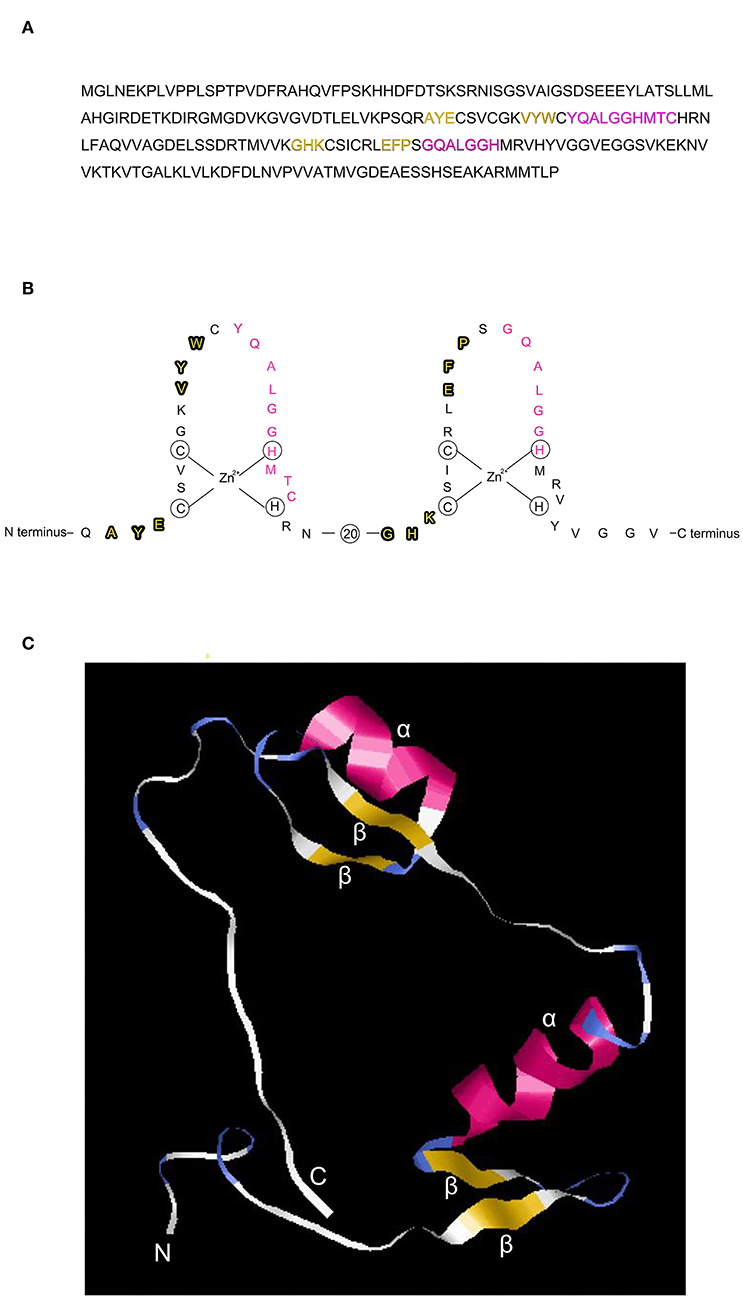
Figure 1. Structure of the C2H2-type zinc finger transcription factor ZFP39. (A) Amino acid sequence of the zinc finger protein ZFP39 (GenBank: AAQ95583.1) containing two tandem fingers, each finger has two β strands (in yellow) and one α helix (in pink). (B) Structure of the two tandem zinc finger motifs, 20 indicates 20 amino acid residues omitted between the two fingers for conciseness. The β strand and α helix of the zinc finger protein are highlighted in bold and pink, respectively. (C) 3D structure of the two tandem zinc fingers demonstrated using phyre2 (Protein Homology/analogY Recognition Engine V 2.0) (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) and shown by the RasMol software (http://www.rasmol.org). N and C indicate the N terminal and C terminal, respectively. β, β strand. α, α helix.
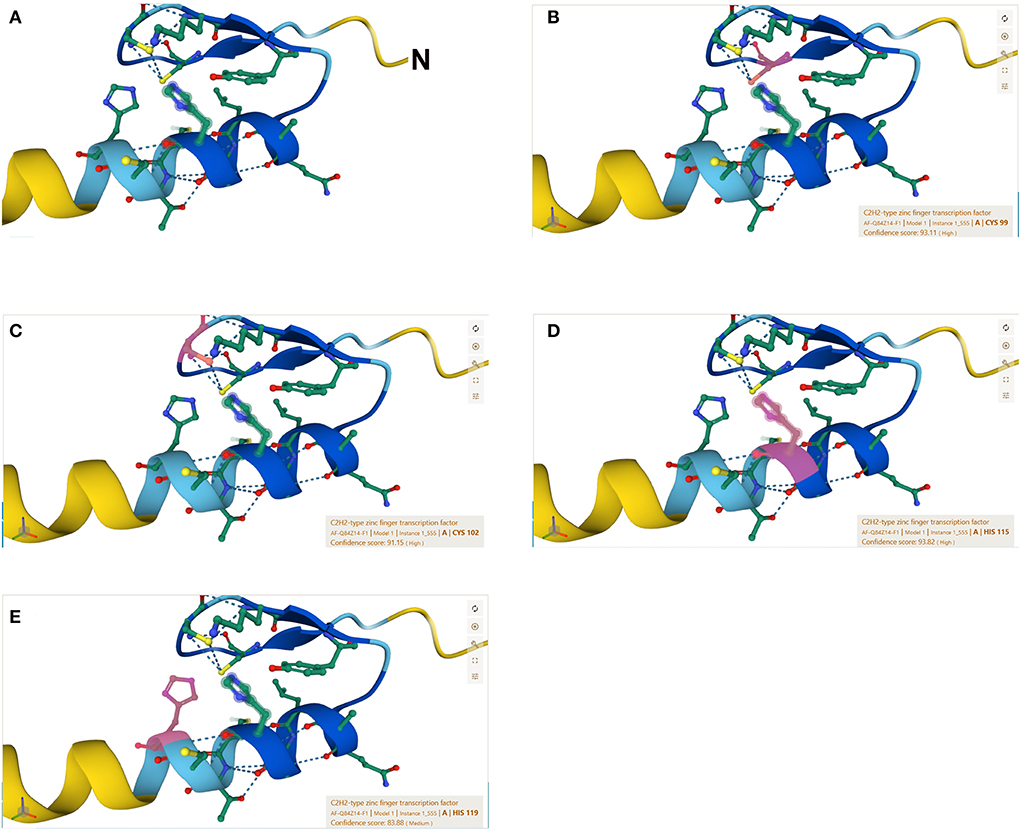
Figure 2. The ββα amino acid sequence of the ZFP39. (A) Particle ZFP39 sequence forming the ββα structure: N terminal-PSQRAYECSVCGKVYWCYQALGGHMTCHRNLFAQ-C terminal. See Figure 1A for the full amino acid sequence. (B) Pink color showing the 99th amino acid cysteine of the ZFP39. (C) Pink color showing the 102th amino acid cysteine of the ZFP39. (D) Pink color showing the 115th amino acid histidine of the ZFP39. (E) Pink color showing the 119th amino acid histidine of the ZFP39. The zinc is tetrahedrally coordinated between two cysteines at the β-sheet and two histidines at the α-helix.
From DNA recognition to biological function
The unfavorable environmental conditions will induce successive processes, including signal perception, signal transduction, transcription regulation, and stress-responsive gene expression (Baillo et al., 2019). In the process of transcription regulation, transcription factors bind with DNA elements and recruit additional proteins. Some transcription factors access and open the closed chromatin to permit other transcription factors bind with the specific DNA (Strader et al., 2022). The zinc finger protein Zif268, which contains three zinc fingers, binds the major groove of B-DNA and wraps the partway around the double helix (Pavletich and Pabo, 1991). DNA recognition usually requires two to four tandemly arranged zinc fingers and additional secondary structure elements (Wolfe et al., 2000). Studies have shown that an additional 20-residue proximal accessory region (PAR), at the N-terminal to the first zinc finger domain, is required for high-affinity DNA binding with C2H2 zinc finger protein ADR1, and the mutations in the PAR lead to a loss of high-affinity DNA binding (Bowers et al., 1999). When plants suffer from abiotic or biotic stress, the plant zinc finger proteins bind to the cis-elements of stress-related genes by DNA contact and recognition, subsequently recruiting additional proteins to enhance or suppress the transcription levels of stress-related genes, thus regulating the cell division, growth, and development.
Interplay with phytohormones to regulate agronomic traits
In this review, we mainly focus on the research advances in the involvement of zinc finger proteins in phytohormones-mediated agronomic traits in rice.
Gibberellin
Gibberellin (GA) is an important phytohormone governing several agronomic traits (Figure 3A). Knockout of the Swollen Anther Wall 1 (SAW1), encoding a CCCH-tandem zinc finger protein, resulted in male sterility. SAW1 directly targets the promoter of the GA biosynthetic gene OsGA20ox3, which triggers the synthesis of bioactive GA that is required for the induction of the anther-regulator gene OsGAMYB. The SAW1-OsGA20ox3-OsGAMYB module promotes the anther development (Wang et al., 2020). In contrast, another zinc finger protein ZFP207 (Cys2/His2 type) negatively regulates the GA synthesis by binding to the OsGA20ox2 (also termed as Green Revolution Gene SD1) promoter to repress its expression, which leads to dwarfism, short grain and panicle phenotypes in rice (Duan et al., 2020). Overexpression of ZFP207 caused a reduction in pollen viability (Duan et al., 2020). Another research showed that OsLOL1 (C2C2-type zinc finger protein) interacted with OsbZIP58 to regulate GA content by OsbZIP58-mediated activation of another GA biosynthetic gene OsKO2, which promotes the programmed cell death (PCD) and seed germination in rice (Wu et al., 2014). ZFP185 (A20/AN1-type) plays a distinct role in GA biosynthesis. Overexpression of ZFP185 in rice resulted in a decrease in endogenous GA3 content and then led to a semi-dwarfism phenotype (Zhang, Y. et al., 2016). A recent study revealed that a rice zinc finger protein PREMATURE INTERNODE ELONGATION 1 (PINE1) also negatively regulates GA before the reproductive switch, thereby restraining the elongation of the internode. Then, florigens repress PINE1 to promote stem responsiveness to GA and flowering (Gómez-Ariza et al., 2019). Furthermore, it is revealed that this PINE1 is identical to the DECELERATOR OF INTERNODE ELONGATION 1 (DEC1), which regulates the internode elongation antagonistically with an unknown function gene ACCELERATOR OF INTERNODE ELONGATION 1 (ACE1). ACE1 and DEC1 are highly conserved in the Gramineae family, rendering the plants environmental fitness (Nagai et al., 2020). Together, these findings elaborate the diverse roles of zinc finger proteins in the GA underlying relevant traits.
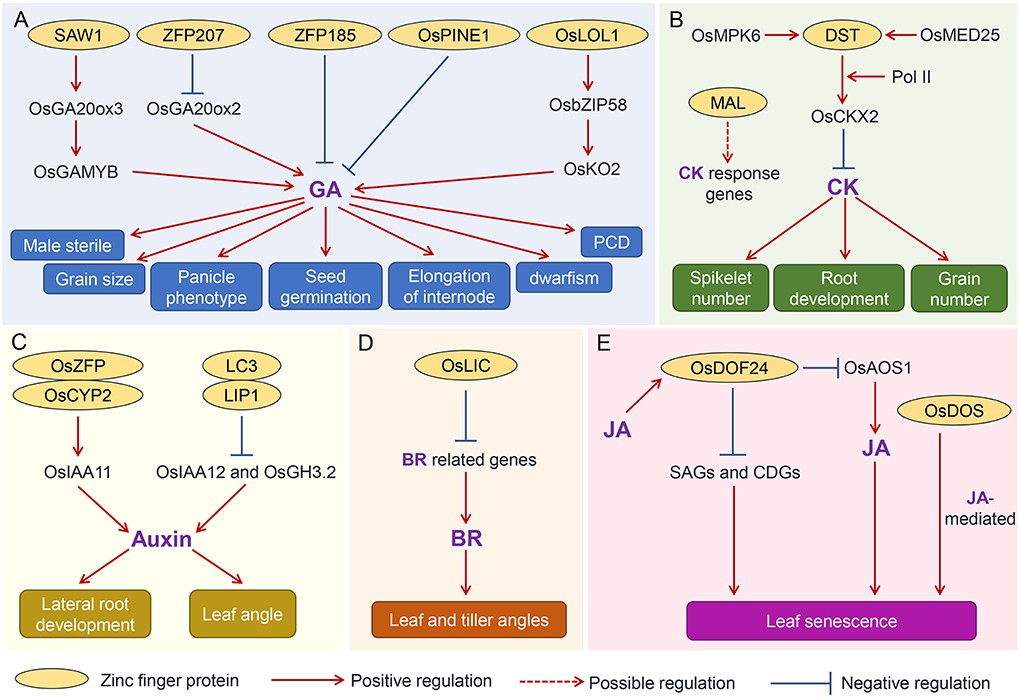
Figure 3. The proposed regulatory network of zinc finger proteins and phytohormones pathways in determining agronomic traits in rice. The regulatory network of zinc finger protein and GA (A), CK (B), auxin (C), BR (D), and JA (E). Positive and negative regulations of various agronomic traits mediated by different zinc finger proteins are indicated by solid lines, and the proposed regulation of zinc finger proteins on downstream targets is indicated by the dashed line. The proteins with cycle are the zinc finger proteins. GA, gibberellins; CK, cytokinin; BR, brassinosteroid; JA, jasmonic acid. SAW1, Swollen Anther Wall 1; OsGA20ox3, GA biosynthetic gene; OsGAMYB, anther-regulator gene; OsGA20ox2, GA biosynthetic gene; OSPINE1, premature internode elongation 1; OSKO2, GA biosynthetic gene; DST, drought and salt tolerance; OsMED25, mediator subunit; OsCKX2, cytokinin oxidase 2; MAL, meristem activityless; LC3, leaf inclination 3; LIP1, LC3-interacting protein 1; OsLIC, tiller angle increased controller; OsDOF24, DNA-binding one zinc finger 24; CDGs chlorophyll degradation genes; SAGs, senescence-associated genes; OsAOS1, JA biosynthetic gene.
Cytokinins
Cytokinin (CK) is a vital phytohormone that regulates the spikelet number and grain number of rice (Figure 3B). Gn1a/OsCKX2 (Grain number 1a/Cytokinin oxidase 2) has been proved to contribute to the grain yield. A C2H2-type zinc finger transcription factor DROUGHT AND SALT TOLERANCE (DST) was recently implicated to directly bind the promoter of OsCKX2 and then trigger its expression, consequently orchestrating CK level to increase grain number (Li et al., 2013). Subsequently, the OsMPK6 was found to interact with and phosphorylate DST to promote OsCKX2 expression and thus modulate spikelet number per panicle (Guo et al., 2020). In addition, the mediator subunit OsMED25 was also shown to act as a coactivator of DST, facilitating the recruitment of the RNA polymerase II (Pol II) to activate OsCKX2 transcription. This regulation regulates the level of CK, ultimately resulting in the change in spikelet number in rice (Lin et al., 2022). Considering the contribution of CK content to grain number, DST-OsCKX2 module provides a promising potential for improving rice yield by modifying different alleles combinations. In addition, a recent report also illustrated that the MERISTEM ACTIVITYLESS (MAL), encoding a RING-H2 finger domain (RFD)-containing protein, might also coordinate the CK signaling to modulate root development by regulating CK responsive genes (Jiang et al., 2020). However, the direct target of MAL is still unknown, which would be an quite interesting issue to be addressed in order to manipulate the root development for drought tolerance improvement.
Auxin
Auxin participates in the regulation of rice yield by regulating the grain yield too (Cao et al., 2020). However, the crosstalk between zinc finger proteins and auxin underlying this trait is unclear. But their relationships in regulating other traits have been explored (Figure 3C). For example, a C2HC-type zinc finger protein OsZFP interacted with the auxin-responsive gene OsCYP2 to modulate the lateral root development by orchestrating the expression of auxin signaling components, such as OsIAA11 (Cui et al., 2017). Leaf inclination regulation is one of the most significant features of auxin in regulating agronomic traits (Li et al., 2020). The leaf inclination3 (LC3)-interacting protein 1 (LIP1), a HIT zinc finger domain-containing protein, directly binds to the promoter regions of OsIAA12 and OsGH3.2 and then suppresses the auxin signaling, resulting in the change of leaf angle (Chen et al., 2018). The regulation of zinc finger proteins coordinating auxin to modulate yield and quality-related agronomic traits is still elusive and deserved to be uncovered in the future.
Brassinosteroids
Brassinosteroids (BRs) are the dominant phytohormone responsible for the regulation of leaf angle (Figure 3D). The Oraza sativa leaf and tiller angle increased controller (OsLIC) encodes a CCCH-type zinc finger protein, participating in modulating leaf and tiller angles. Microarray profiling revealed that expressions of BRs-related genes were significantly activated in the knockdown lines. Furthermore, BR treatment induced the expression of OsLIC, and sterol levels were higher in oslic mutants (Wang, L. et al., 2008), indicating that it might integrate the BRs pathway to determine the leaf angle. Considering OsLIC is a transcription activator, it coordinates with another transcription factor to directly suppress BR biosynthesis and/or signaling regulator expressions to orchestrate the BR response.
Jasmonic acid
The phytohormone jasmonic acid (JA) modulates the timing of leaf senescence, which is an important agronomic trait contributing to rice yield, in particular during grain filling (Figure 3E). Recently, a rice DNA-Binding One Zinc Finger 24 (OsDOF24) has been implicated to negatively regulate leaf senescence in rice by repressing the expression of senescence-associated genes (Osl85, Osl57, and OsNAP) and chlorophyll degradation genes (NYC1, NYC3, and SGR). Further analysis revealed that OsDOF24 is specifically induced by methyl jasmonate (JA), and then, in turn, it inhibits JA biosynthesis by directly binding the promoter of JA biosynthetic gene OsAOS1 and then probably represses its expression to modulate leaf senescence (Shim et al., 2019). In addition, anther zinc finger genes, OsDOS, has also been reported to be involved in the JA-mediated leaf senescence (Kong et al., 2006). For example, overexpression of OsDOS produced a delay of leaf senescence, while knockdown of OsDOS caused an accelerated leaf senescence. Subsequently, microarray analyses indicated that there were a number of differentially expressed genes involved in the JA pathway, suggesting OsDOS coordinates with JA to regulate leaf senescence (Kong et al., 2006). In summary, the zinc finger genes reviewed in this study are listed (Table 1), and the relevant regulatory mechanisms underlying agronomic traits are proposed (Figure 3).
Impact on agronomic traits
Except those mentioned above, the regulation of other zinc finger genes that are involved in phytohormones underlying agronomic traits is still under-explored. For example, O. sativa SHORT INTERNODES1 (OsSHI1), a plant-specific transcription factor containing the conserved zinc-finger DNA-binding domain, competes with IDEAL PLANT ARCHITECTURE1 (IPA1) to activate the expressions of O. sativa TEOSINTE BRANCHED1 (OsTB1) and O. sativa DENSE AND ERECT PANICLE1 (OsDEP1), resulting in the change of tiller number and panicle size (Duan et al., 2019). However, it is unclear whether OsSHI1 mediated such regulation is related to phytohormone. A DHHC-type zinc finger gene, OsDHHC1, regulates plant architecture by altering rice tillering; overexpression of OsDHHC1 increased 40% of tiller numbers and 10% of grain yield (Zhou et al., 2017). Taking into account the effect of OsCKX2 on tiller number (Yeh et al., 2015), it raises a possibility that OsDHHC1 may be involved in the CK pathway to modulate the tiller development.
Given that the cell morphogenesis eventually alters rice agronomic traits, identification of corresponding zinc finger proteins responsible for the cell division, differentiation, and proliferation would extend our knowledge about their roles in agronomic traits. DCM1, a CCCH zinc finger protein, has been shown to play an essential role in pollen development. Knockout of DCM1 resulted in abnormal male meiotic cytokinesis, eventually leading to the disordered spindle orientation during meiosis and the formation of pollen grains (nearly sterile). Further analyses proposed that DCM1 might be involved in the mRNA processing or elimination by interacting with OsPABN1 and OsPABN2 to regulate callose metabolism during meiosis (Zhang, C. et al., 2018). Three cysteine-tryptophan (CW) domains containing zinc finger proteins have been reported to be involved in histone recognition, namely, OsCW-ZF3, OsCW-ZF5, and OsCW-ZF7. Among them, only knockout of OsCW-ZF7 was examined to cause defective development of awns (Zhang et al., 2017). Investigation on the interaction between OsCW-ZF7 with histone modification and TFIID20 suggested that TFIID20-OsCW-ZFs-H3K4me module may regulate the awn development by targeting certain awn-related genes (such as An-6/7/8/9/10) in the awn primordium (Chen et al., 2018). Another zinc finger protein, NSG1, may also employ histone modification to regulate cell development. It was shown that NSG1 can recruit the corepressor TOPLESS-RELATED PROTEIN to directly bind to the promoter of LHS1 and then inhibit the LHS1 expression by downregulating histone acetylation levels of the chromatin, resulting in the morphological alternation of lateral organ identities in the spikelet development (Zhuang et al., 2020). A GATA zinc finger domain-containing protein, SNFL1, regulates flag leaf size by modulating the number of small vascular bundles rather than the number of epidermal cells (He et al., 2018). Similarly, NECK LEAF1 (NL1), the allelic gene of SNFL1, was also involved in the regulation of organ size, such as leaf and panicles. However, the downstream regulators are still unknown. In addition, a ZOS4-06-C2H2 zinc-finger gene, LRG1, regulates spikelet development and influences grain size (Xu et al., 2020). In conclusion, zinc finger proteins comprehensively participate in the regulation of various agronomic traits. The remained issues are the identification of their targets, since the engineering of a single zinc finger would likely cause unknown side effects on other traits except the deserved ones.
Response to abiotic stresses
Zinc finger proteins have been proved to regulate the responses to various abiotic stresses (Figure 4A). For example, overexpression of ZFP179 increased the tolerance to salt and oxidative stress, enhanced the reactive oxygen species (ROS)-scavenging ability, exhibited hypersensitive to ABA, and induced the expressions of stress-related genes, such as OsDREB2A, suggesting ZFP179-mediated salt tolerance was involved both in the ABA-dependent and ABA-independent signaling pathways (Sun et al., 2010). Recently, salt-responsive ERF1 (SERF1) has been evident to bind to the promoter of ZFP179 in vitro and in vivo, and thus, overexpression of SERF1 confers high tolerance to salt stress (Schmidt et al., 2013). OsRZFP34 is also induced by ABA treatment, as well as high temperature, functioning in the control of stomata opening. Further analyses demonstrated that OsRZFP34 mediated drought response by regulating the Ca2+ sensing, K+ regulator, and ABA-responsive genes (Sun et al., 2010). Subsequently, OsPUB67, a U-box E3 ubiquitin ligase, is proposed to target OsRZFP34 for degradation (Qin et al., 2020), thereby modulating the drought tolerance. Another ABA-responsive zinc finger gene ZFP36 was also characterized in rice. Upregulation of ZFP36 improves the tolerance to water stress and oxidative stress through induction by ABA, superoxide dismutase (SOD), and ascorbate peroxidase (APX) (Zhang et al., 2014). Later, the direct target of ZFP36 is OsAPX1 (Huang, L. et al., 2018). Furthermore, a late embryogenesis abundant protein OsLEA5 was proved to interact with ZFP36 to co-regulate ABA-inhibited seed germination (Huang, L. et al., 2018). In addition, a heat shock protein OsDjC46 was also identified to bind ZFP36, and it positively regulates salinity and drought tolerance through the ABA pathway in rice (Huang, L. et al., 2018), suggesting ZFP36-OsDjC46 complex may function in fine-tuning the dynamic of ABA-dependent antioxidant defense. On the contrary, an Oryza sativa cold-inducible zinc finger gene, OsCOIN, can strongly be induced by low temperature. Overexpression of OsCOIN enhanced rice seedlings tolerance to cold, salt, and drought, accompanied by an upregulation of pyrroline-5-carboxylate synthetase (OsP5CS) and an increase in cellular proline level (Liu et al., 2007). Since OsCOIN is expressed in all the rice tissues (Liu et al., 2007), it is reasonable that it renders rice the low-temperature resistance at most developmental stages, which is supposed to be validated by a recent study that OsCOIN is also activated in response to the low temperature during the reproductive stage of rice (Guo et al., 2022). It is worthy to mention that as transcription factors, zinc finger proteins may modulate the adaptation to cold by enhancing or maintaining the growth and development of the organs. For example, overexpressing OsCTZFP8 rendered cold tolerance with higher pollen fertilities, as well as increased seed setting and higher yield under cold treatments (Jin et al., 2018). ZFP182 is another cold-induced zinc finger protein. Overexpression of ZFP182 promoted rice salt tolerance in rice (Huang et al., 2007). Further analyses illustrated that ZFP182 triggers the accumulation of compatible osmolytes, such as free proline and soluble sugars, by activating the expression of OsP5CS and OsLEA3 (late embryogenesis abundant protein gene) upon stress (Huang et al., 2012). Although a ubiquitin fused to ribosomal protein L40 (ZIURP1) was found to interact with ZFP182 (Huang et al., 2012), the underlying biological significance of this interaction is poorly understood yet. However, the OsMPK1 and OsMPK5 were proved to be required for the induction of ZFP182 in ABA-mediated antioxidant defense, while ZFP182 did not regulate the OsMPK1 and OsMPK5 (Zhang et al., 2012), suggesting that the ZFP182 may act downstream of the two kinases under stress condition. Additionally, it has been shown that a zinc finger protein gene OsZFP213 positively regulated salt tolerance in rice: overexpression of OsZFP213 displayed enhanced salt tolerance, probably due to the upregulation of antioxidant system genes, higher catalytic activity of ROS scavenging enzymes, and lower level of ROS accumulation. OsZFP213 can also interact with a kinase, OsMAPK3, which improves the transactivation activity of OsZFP213 to regulate salt tolerance (Zhang, Z. et al., 2018).
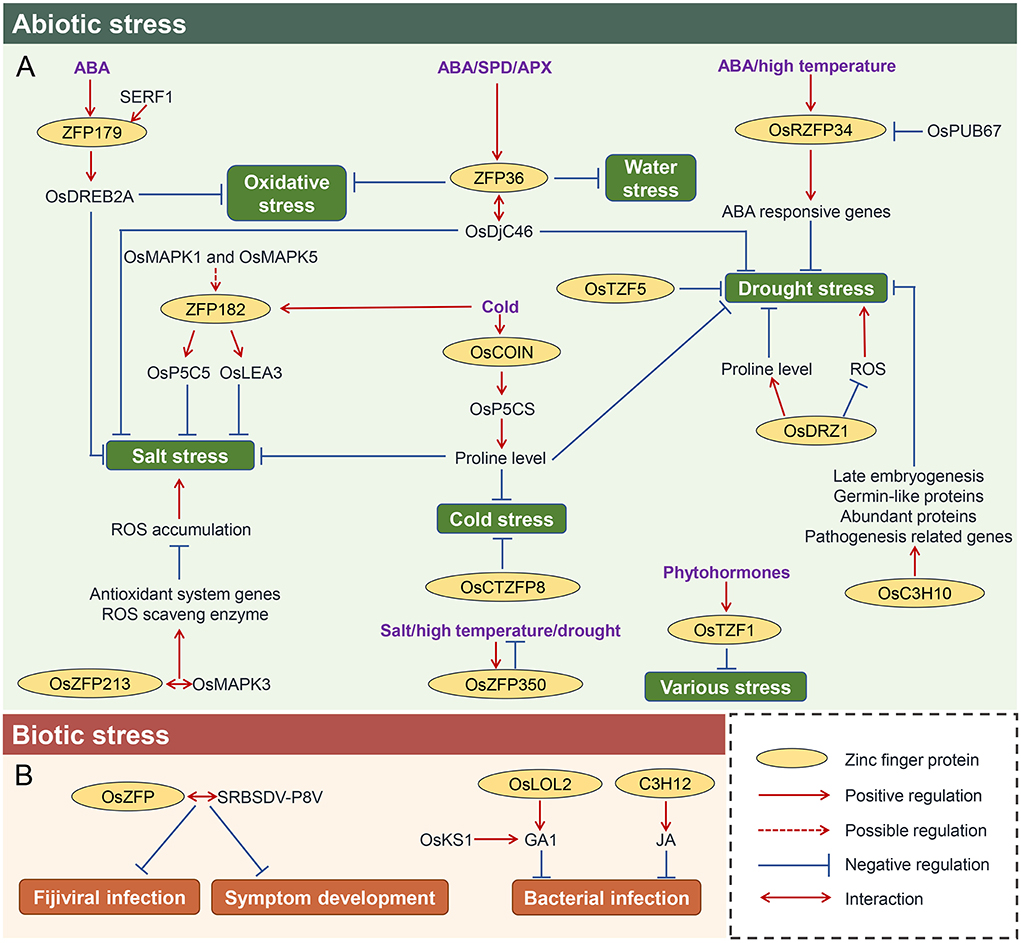
Figure 4. Regulations of various stresses response by zinc finger proteins in rice. The regulatory network of zinc finger protein in response to abiotic stress (A) and biotic stress (B). Positive and negative regulations of various stresses response mediated by different zinc finger proteins are indicated by solid lines, and the proposed regulation of zinc finger proteins on downstream targets is indicated by the dashed line. The proteins with cycle are the zinc finger proteins. Double arrow represents that the two proteins interact with each other. ABA, abscisic acid; SPD, spermidine; APX, ascorbate peroxidase; JA, jasmonic acid; GA, gibberellins; SERF1, salt-responsive ERF1; OsDREB2A, dehydration responsive element binding protein; OsDjC46, heat shock protein; OsP5CS, pyrroline-5-carboxylate synthetase; OsLEA3, Late embryogenesis abundant protein gene; OsPUB67, U-box E3 ubiquitin ligase; OsTZF5, CCCH-tandem zinc finger protein 5; OsDRZ1, drought-responsive zinc finger protein 1; OsCOIN, cold-inducible zinc finger gene; OsP5CS, pyrroline-5-carboxylate synthetase; SRBSDV-P8V, black-streaked dwarf virus protein P8; OsKS1, GA biosynthetic gene.
Accumulating studies unraveled the potential of zinc finger proteins in the drought response. For example, OsC3H10 is involved in the regulation of rice drought tolerance by modulating the expression of stress-related genes, which included LATE EMBRYOGENESIS ABUNDANT PROTEINs, PATHOGENESIS RELATED GENEs and GERMIN-LIKE PROTEINs (Xu et al., 2020). Notably, a drought-responsive zinc finger protein 1 (OsDRZ1) has been reported to play a critical role in modulating drought-responsive gene expression: overexpression of OsDRZ1 increased seedling drought tolerance and caused accumulation of more free proline but reduced ROS, while knockdown of OsDRZ1 led to the opposite trend (Yuan et al., 2018). Interestingly, this gene also plays a role in regulating plant architecture, implying it may orchestrate the trade-off between growth and stress.
In fact, zinc finger genes were broadly involved in multiple biological processes. For example, CCCH-type zinc finger gene, OsTZF1, can be induced by drought, high-salt stress, hydrogen peroxide, ABA, methylJA, and salicylic acid (SA), as well as red light (R) and far-red light (FR). Overexpression of OsTZF1 exhibited delayed seed germination, growth retardation at the seedling stage, and delayed leaf senescence and showed improved tolerance to high-salt and drought stresses (Jan et al., 2013). Using the RNA-binding assays, it was shown that the U-rich regions in the 3'-untranslated region of messenger RNAs could bind with OsTZF1 (Jan et al., 2013). Taken together, it could be proposed that OsTZF1 may be activated by various phytohormones and then directly bind to the downstream stress-responsive genes to modulate their expressions in response to various stresses. Similar case was also identified in another zinc finger gene, OsZFP350, which was upregulated by drought, salt, and high temperature. Overexpression of OsZFP350 increased primary root length and the number of adventitious and lateral roots, as well as increased the germination rate of seeds under abiotic stress, and attenuated the heat, salinity, and drought stress during root development (Kang et al., 2019). Similarly, the Oryza sativa CCCH-tandem zinc finger protein 5 (OsTZF5) also confers drought resistance and increases grain yield: introducing OsTZF5 in two commercial upland cultivars shows increased grain yield under confined field drought environments (Selvaraj et al., 2020), further suggesting that zinc finger genes play vital roles in maintaining the balance between stress and development in plants. Taken all above together, we proposed that zinc finger proteins are potential targets for simultaneously improving rice stress tolerance and yield.
Response to biotic stresses
Increasing evidence has been implicated that zinc finger proteins also participate in biotic response in rice (Figure 4B). For example, a rice C2H2 zinc finger protein OsZFP was found to interact with southern rice black-streaked dwarf virus (SRBSDV) protein P8, a minor core protein of SRBSDV, which may play an important role in fijiviral infection and symptom development (Li, J. et al., 2017), implying that P8 may execute the infection by interfering with the host OsZFP. LSD1-related proteins in Arabidopsis are well known as remarkable disease-defensed zinc finger proteins. Similarly, its ortholog in rice, termed OsLOL2, is also involved in disease response. Overexpression of OsLOL2 exhibited more resistance to rice bacterial blight than wild type, while decreased expression of OsLOL2 had similar or even sensitivity to bacterial blight. Interestingly, knockdown of OsLOL2 also resulted in dwarf phenotypes and decreased endogenous bioactive GA1 content, which can be restored by the application of GA or expression of GA biosynthetic gene OsKS1 (Xu and He, 2007). However, it is still unclear how the GA interplays with OsLOL2 in the culm development and whether their crosstalk is involved in the disease defense. A CCCH-type zinc finger protein, C3H12, regulates the rice-Xanthomonas oryzae pv. oryzae (Xoo) interaction: overexpression of C3H12 partially enhanced resistance to Xoo, while knockout or suppression of C3H12 resulted in partially increased susceptibility to Xoo. Since the JA levels and signaling genes were altered in c3h12 mutants (Deng et al., 2012), it was assumed that C3H12 may integrate into the JA pathway to induce the defense of rice.
Concluding remarks
Breeding stress-tolerant rice with better quality and higher yield by transgenic technologies is one of the most promising approaches to combat against multiple environmental stresses including drought, salt, and extreme temperatures. The zinc finger transcription factor genes are the candidates for genetic modification, as shown in many recent reports. For example, manipulation of ZFP182 or OsCOIN confers multiple stress resistances including salt, cold, and drought (Huang et al., 2007, 2012; Liu et al., 2007). Overexpression of OsZFP350 increased the germination rate of seeds under abiotic stress and increased the resistance to heat, salinity, and drought stress during root development (Kang et al., 2019). Two genes, OsTZF1 and DST, can be used to increase both salt and drought resistance (Huang et al., 2009; Jan et al., 2013). Overexpression of ZFP179 and OsZFP213 enhanced salt tolerance (Sun et al., 2010; Xu et al., 2020). OsDRZ1, OsMSR15, and OsTZF5 also exhibit potential in improving drought resistance in rice (Zhang, X. et al., 2016; Yuan et al., 2018; Selvaraj et al., 2020). Notably, some zinc finger genes play antagonistic roles between biotic and abiotic stress tolerances, and thus, overexpression of these specific zinc finger genes may alter the sensitivity to both of them. For example, overexpression of a rice A20/AN1-type zinc finger gene ZFP177 in tobacco conferred tolerance to both low- and high-temperature stresses, but increased sensitivity to salt and drought stresses (Huang et al., 2008). Therefore, ahead of extensive utilization in rice selected breeding production, these zinc finger protein genes must be tested strictly upon various biotic and abiotic stresses and then examined in the field conditions for certain years. On the contrary, because the responses of rice zinc finger proteins to stress conditions may be varied in different genetic backgrounds, it is also uncertain whether these zinc finger genes still possess the same effect capability on improving stress tolerance when they were employed in another genetic background or genotype.
Zinc finger transcription factors can be used to increase the tolerance to stresses and yield. Overexpressing OsCTZFP8 exhibited cold-tolerant phenotypes with higher pollen fertility, increased seed setting, and higher yield under cold treatments (Jin et al., 2018). Overexpression of OsDHHC1 increased by 40% of tiller numbers and 10% of grain yield (Zhou et al., 2017). These all indicated the potential utilization of zinc finger protein genes. It is quite interesting if and how many documented zinc finger genes are differentiated among various rice accessions. Using the database of re-sequencing of rice, we can identify their haplotypes and then evaluate their potential contributions to certain traits by integrative analyses of genome-wide association selection. As a consequence, the superior alleles would be the prime target or even directly selected for the genetic improvement of stress tolerance in rice breeding. Of the 2,408 transcription factor genes identified in japonica rice, there are a total of 189 C2H2 zinc finger protein genes and 74 C3H zinc finger protein genes, most of which have not been studied clearly. It is crucial to understand the regulatory mechanisms of these untested zinc finger genes before evaluating their potential in production.
Author contributions
YH, SY, and QY conceived the idea and led the writing of the manuscript. LD, MW, and QY helped in revising the manuscript and prepared the figures. YH, LD, SY, and QY reviewed, edited, and improved the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31971920) and the Guangdong Province Key Laboratory of Plant Molecular Breeding (GPKLPMB202204).
Acknowledgments
We are grateful to the support of the experimental platform and funding from the Institute of Crop and Nuclear Technology Utilization of Rice Breeding. We apologize in advance to colleagues whose valuable work was not cited due to article length considerations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agarwal, P., Arora, R., Ray, S., Singh, A. K., Singh, V. P., Takatsuji, H., et al. (2007). Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 65, 467–485. doi: 10.1007/s11103-007-9199-y
Atkinson, N. J., Lilley, C. J., and Urwin, P. E. (2013). Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 162, 2028–2041. doi: 10.1104/pp.113.222372
Baillo, E. H., Kimotho, R. N., Zhang, Z., and Xu, P. (2019). Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes. 10, 771. doi: 10.3390/genes10100771
Berg, J. M., and Shi, Y. (1996). The Galvanization of biology: a growing appreciation for the roles of zinc. Science. 271, 1081–1085. doi: 10.1126/science.271.5252.1081
Bowers, P. M., Schaufler, L. E., and Klevit, R. E. (1999). A folding transition and novel zinc finger accessory domain in the transcription factor ADR1. Nat. Struct. Biol. 6, 478–485. doi: 10.1038/8283
Cao, J., Li, G., Qu, D., Li, X., and Wang, Y. (2020). Into the seed: Auxin controls seed development and grain yield. Int. J. Mol. Sci. 21, 1662. doi: 10.3390/ijms21051662
Cassandri, M., Smirnov, A., Novelli, F., Pitolli, C., Agostini, M., Malewicz, M., et al. (2017). Zinc-finger proteins in health and disease. Cell Death Discoy. 3, 17071. doi: 10.1038/cddiscovery.2017.71
Chen, S. H., Zhou, L. J., Xu, P., and Xue, H. W. (2018). SPOC domain-containing protein Leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PLoS Genet. 14, e1007829. doi: 10.1371/journal.pgen.1007829
Ciftci-Yilmaz, S., and Mittler, R. (2008). The zinc finger network of plants. Cell. Mol. Life Sci. 65, 1150–1160. doi: 10.1007/s00018-007-7473-4
Cui, P., Liu, H., Ruan, S., Ali, B., Gill, R. A., Ma, H., et al. (2017). A zinc finger protein, interacted with cyclophilin, affects root development via IAA pathway in rice. J. Integr. Plant Biol. 59, 496–505. doi: 10.1111/jipb.12531
Deng, H., Liu, H., Li, X., Xiao, J., and Wang, S. (2012). A CCCH-type zinc finger nucleic acid-binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiol. 158, 876–889. doi: 10.1104/pp.111.191379
Duan, E., Wang, Y., Li, X., Lin, Q., Zhang, T., Wang, Y., et al. (2019). OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in Rice. Plant Cell 31, 1026–1042. doi: 10.1105/tpc.19.00023
Duan, M., Ke, X. J., Lan, H. X., Yuan, X., Huang, P., Xu, E. S., et al. (2020). A Cys2/His2 zinc finger protein acts as a repressor of the green revolution gene SD1/OsGA20ox2 in Rice (Oryza sativa L.). Plant Cell Physiol. 61, 2055–2066. doi: 10.1093/pcp/pcaa120
Gómez-Ariza, J., Brambilla, V., Vicentini, G., Landini, M., Cerise, M., Carrera, E., et al. (2019). A transcription factor coordinating internode elongation and photoperiodic signals in rice. Nat Plants 5, 358–362. doi: 10.1038/s41477-019-0401-4
Guo, T., Lu, Z. Q., Shan, J. X., Ye, W. W., Dong, N. Q., and Lin, H. X. (2020). ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell. 32, 2763–2779. doi: 10.1105/tpc.20.00351
Guo, Z., Cai, L., Liu, C., Chen, Z., Guan, S., Ma, W., et al. (2022). Low-temperature stress affects reactive oxygen species, osmotic adjustment substances, and antioxidants in rice (Oryza sativa L.) at the reproductive stage. Sci. Rep. 12, 6224. doi: 10.1038/s41598-022-10420-8
Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N., Qiu, N., et al. (2020). C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 11, 115. doi: 10.3389/fpls.2020.00115
He, P., Wang, X., Zhang, X., Jiang, Y., Tian, W., Zhang, X., et al. (2018). Short and narrow flag leaf1, a GATA zinc finger domain-containing protein, regulates flag leaf size in rice (Oryza sativa). BMC Plant Biol. 18, 273. doi: 10.1186/s12870-018-1452-9
Hsu, K. H., Liu, C. C., Wu, S. J., Kuo, Y. Y., Lu, C. A., Wu, C. R., et al. (2014). Expression of a gene encoding a rice RING zinc-finger protein, OsRZFP34, enhances stomata opening. Plant Mol. Biol. 86, 125–137. doi: 10.1007/s11103-014-0217-6
Huang, J., Sun, S., Xu, D., Lan, H., Sun, H., Wang, Z., et al. (2012). A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol. Biol. 80, 337–350. doi: 10.1007/s11103-012-9955-5
Huang, J., Wang, M. M., Jiang, Y., Bao, Y. M., Huang, X., Sun, H., et al. (2008). Expression analysis of rice A20/AN1-type zinc finger genes and characterization of ZFP177 that contributes to temperature stress tolerance. Gene. 420, 135–144. doi: 10.1016/j.gene.2008.05.019
Huang, J., Yang, X., Wang, M. M., Tang, H. J., Ding, L.-Y., Shen, Y., et al. (2007). A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim. Biophys. Acta. 1769, 220–227. doi: 10.1016/j.bbaexp.2007.02.006
Huang, L., Jia, J., Zhao, X., Zhang, M. Y., Huang, X. E., et al. (2018). The ascorbate peroxidase APX1 is a direct target of a zinc finger transcription factor ZFP36 and a late embryogenesis abundant protein OsLEA5 interacts with ZFP36 to co-regulate OsAPX1 in seed germination in Rice. Biochem. Biophys. Res. Commun. 495, 339–345. doi: 10.1016/j.bbrc.2017.10.128
Huang, P., Yoshida, H., Yano, K., Kinoshita, S., Kawai, K., Koketsu, E., et al. (2018). OsIDD2, a zinc finger and INDETERMINATE DOMAIN protein, regulates secondary cell wall formation. J. Integr. Plant Biol. 60, 130–143. doi: 10.1111/jipb.12557
Huang, X. Y., Chao, D. Y., Gao, J. P., Zhu, M. Z., Shi, M., and Lin, H. X. (2009). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23, 1805–1817. doi: 10.1101/gad.1812409
Jan, A., Maruyama, K., Todaka, D., Kidokoro, S., Abo, M., Yoshimura, E., et al. (2013). OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 161, 1202–1216. doi: 10.1104/pp.112.205385
Jiang, W., Zhou, S., Huang, H., Song, H., Zhang, Q., and Zhao, Y. (2020). MERISTEM ACTIVITYLESS (MAL) is involved in root development through maintenance of meristem size in rice. Plant Mol. Biol. 104, 499–511. doi: 10.1007/s11103-020-01053-4
Jin, Y. M., Piao, R., Yan, Y. F., Chen, M., Wang, L., He, H., et al. (2018). Overexpression of a new zinc finger protein transcription factor OsCTZFP8 improves cold tolerance in rice. Int. J. Genomics 2018, 5480617. doi: 10.1155/2018/5480617
Kang, Z., Qin, T., and Zhao, Z. (2019). Overexpression of the zinc finger protein gene OsZFP350 improves root development by increasing resistance to abiotic stress in rice. Acta Biochim. Pol. 66, 183–190.
Kong, Z., Li, M., Yang, W., Xu, W., and Xue, Y. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141, 1376–1388. doi: 10.1104/pp.106.082941
Lee, M. S., Gippert, G. P., Soman, K. V., Case, D. A., and Wright, P. E. (1989). Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 245, 635–637. doi: 10.1126/science.2503871
Li, J., Cai, N. J., Xue, J., Yang, J., Chen, J. P., and Zhang, H. M. (2017). Interaction between southern rice black-streaked dwarf virus minor core protein P8 and a rice zinc finger transcription factor. Arch. Virol. 162, 1261–1273. doi: 10.1007/s00705-017-3233-4
Li, S., Zhao, B., Yuan, D., Duan, M., Qian, Q., Tang, L., et al. (2013). Rice zinc finger protein DST enhances grain production through controlling GN1A/OsCKX2 expression. Proc Natl Acad Sci 110, 3167–3172. doi: 10.1073/pnas.1300359110
Li, W., Zhu, Z., Chern, M., Yin, J., Yang, C., Ran, L., et al. (2017). A natural allele of a transcription factor in rice confers Broad-Spectrum blast resistance. Cell. 170, 114–126. doi: 10.1016/j.cell.2017.06.008
Li, X., Wu, P., Lu, Y., Guo, S., Zhong, Z., Shen, R., et al. (2020). Synergistic interaction of phytohormones in determining leaf angle in crops. Int. J. Mol. Sci. 21, 5052. doi: 10.3390/ijms21145052
Lin, L., Du, M., Li, S., Sun, C., Wu, F., Deng, L., et al. (2022). Mediator complex subunit MED25 physically interacts with DST to regulate spikelet number in rice. J. Integr. Plant Biol. 64, 871–883. doi: 10.1111/jipb.13238
Liu, K., Wang, L., Xu, Y., Chen, N., Ma, Q., Li, F., et al. (2007). Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta. 226, 1007–1016. doi: 10.1007/s00425-007-0548-5
Nagai, K., Mori, Y., Ishikawa, S., Furuta, T., Gamuyao, R., Niimi, Y., et al. (2020). Antagonistic regulation of the gibberellic acid response during stem growth in rice. Nature. 584, 109–114. doi: 10.1038/s41586-020-2501-8
Pavletich, N. P., and Pabo, C. O. (1991). Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 252, 809–817. doi: 10.1126/science.2028256
Qin, Q., Wang, Y., Huang, L., Du, F., Zhao, X., Li, Z., et al. (2020). A U-box E3 ubiquitin ligase OSPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 102, 89–107. doi: 10.1007/s11103-019-00933-8
Schmidt, R., Mieulet, D., Hubberten, H. M., Obata, T., Hoefgen, R., Fernie, A. R., et al. (2013). Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131. doi: 10.1105/tpc.113.113068
Seetharam, A., and Stuart, G. W. (2013). A study on the distribution of 37 well conserved families of C2H2 zinc finger genes in eukaryotes. BMC Genom 14, 420. doi: 10.1186/1471-2164-14-420
Selvaraj, M. G., Jan, A., Ishizaki, T., Valencia, M., Dedicova, B., Maruyama, K., et al. (2020). Expression of the CCCH -tandem zinc finger protein gene OsTZF5 under a stress-inducible promoter mitigates the effect of drought stress on rice grain yield under field conditions. Plant Biotechnol. J. 18, 1711–1721. doi: 10.1111/pbi.13334
Seong, S. Y., Shim, J. S., Bang, S. W., and Kim, J. K. (2020). Overexpression of OSC3H10, a CCCH- Zinc finger, improves drought tolerance in rice by regulating Stress-Related genes. Plants 9, 1298. doi: 10.3390/plants9101298
Shim, Y., Kang, K., An, G., and Paek, N. C. (2019). Rice DNA-Binding one zinc finger 24 (OSDOF24) delays leaf senescence in a Jasmonate-Mediated pathway. Plant Cell Physiol. 60, 2065–2076. doi: 10.1093/pcp/pcz105
Strader, L., Weijers, D., and Wagner, D. (2022). Plant transcription factors - being in the right place with the right company. Curr. Opin. Plant Biol. 65, 102136. doi: 10.1016/j.pbi.2021.102136
Sun, S. J., Guo, S. Q., Yang, X., Bao, Y. M., Tang, H. J., Sun, H., et al. (2010). Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 61, 2807–2818. doi: 10.1093/jxb/erq120
Takatsuji, H.. (1998). Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 54, 582–596. doi: 10.1007/s000180050186
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., and Gao, G. (2020). PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Wang, B., Fang, R., Chen, F., Han, J., Liu, Y. G., Chen, L., et al. (2020). A novel CCCH-type zinc finger protein SAW1 activates OsGA20ox3 to regulate gibberellin homeostasis and anther development in rice. J. Integr. Plant Biol. 62, 1594–1606. doi: 10.1111/jipb.12924
Wang, D., Guo, Y., Wu, C., Yang, G., Li, Y., and Zheng, C. (2008). Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9, 44. doi: 10.1186/1471-2164-9-44
Wang, D., Qin, Y., Fang, J., Yuan, S., Peng, L., Zhao, J., et al. (2016). A missense mutation in the zinc finger domain of OSCESA7 deleteriously affects cellulose biosynthesis and plant growth in rice. PLoS ONE. 11, e153993. doi: 10.1371/journal.pone.0153993
Wang, L., Xu, Y., Zhang, C., Ma, Q., Joo, S. H., Kim, S. K., et al. (2008). OsLIC, a novel CCCH-type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS ONE. 3, e3521. doi: 10.1371/journal.pone.0003521
Wolfe, S. A., Nekludova, L., and Pabo, C. O. (2000). DNA recognition by Cys2his2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29, 183–212. doi: 10.1146/annurev.biophys.29.1.183
Wu, J., Zhu, C., Pang, J., Zhang, X., Yang, C., Xia, G., et al. (2014). OSLOL1, a C2C2-type zinc finger protein, interacts with Osbzip58 to promote seed germination through the modulation of gibberellin biosynthesis in Oryza sativa. The Plant J 80, 1118–1130. doi: 10.1111/tpj.12714
Xu, C., and He, C. (2007). The rice OsLOL2 gene encodes a zinc finger protein involved in rice growth and disease resistance. Mol. Genet. Genomics 278, 85–94. doi: 10.1007/s00438-007-0232-2
Xu, Q., Yu, H., Xia, S., Cui, Y., Yu, X., Liu, H., et al. (2020). The C2H2 zinc-finger protein lacking rudimentary glume 1 regulates spikelet development in rice. Sci Bull. 65, 753–764. doi: 10.1016/j.scib.2020.01.019
Yeh, S. Y., Chen, H. W., Ng, C. Y., Lin, C. Y., Tseng, T. H., Li, W. H., et al. (2015). Down-regulation of cytokinin oxidase 2 expression increases tiller number and improves rice yield. Rice. 8, 36. doi: 10.1186/s12284-015-0070-5
Yuan, X., Huang, P., Wang, R., Li, H., Lv, X., Duan, M., et al. (2018). A zinc finger transcriptional repressor confers pleiotropic effects on rice growth and drought tolerance by Down-Regulating Stress-Responsive genes. Plant Cell Physiol. 59, 2129–2142. doi: 10.1093/pcp/pcy133
Zhang, C., Shen, Y., Tang, D., Shi, W., Zhang, D., Du, G., et al. (2018). The zinc finger protein DCM1 is required for male meiotic cytokinesis by preserving callose in rice. PLoS Genet. 14, e1007769. doi: 10.1371/journal.pgen.1007769
Zhang, H., Liu, Y., Wen, F., Yao, D., Wang, L., Guo, J., et al. (2014). A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J. Exp. Bot. 65, 5795–5809. doi: 10.1093/jxb/eru313
Zhang, H., Ni, L., Liu, Y., Wang, Y., Zhang, A., Tan, M., et al. (2012). The C2H2-type zinc finger protein ZFP182 is involved in abscisic acid-induced antioxidant defense in rice. J. Integr. Plant Biol. 54, 500–510. doi: 10.1111/j.1744-7909.2012.01135.x
Zhang, X., Zhang, B., Li, M. J., Yin, X. M., Huang, L. F., Cui, Y. C., et al. (2016). OsMSR15 encoding a rice C2H2-type zinc finger protein confers enhanced drought tolerance in transgenic Arabidopsis. J. Plant Biol. 59, 271–281. doi: 10.1007/s12374-016-0539-9
Zhang, Y., Lan, H., Shao, Q., Wang, R., Chen, H., Tang, H., et al. (2016). An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa). J. Exp. Bot. 67, 315–326. doi: 10.1093/jxb/erv464
Zhang, Z., Liu, H., Sun, C., Ma, Q., Bu, H., Chong, K., et al. (2018). A C2H2 zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice. J. Plant Physiol. 229, 100–110. doi: 10.1016/j.jplph.2018.07.003
Zhang, Z., Zhang, F., Cheng, Z. J., Liu, L. L., Lin, Q. B., Wu, F. Q., et al. (2017). Functional characterization of rice CW-domain containing zinc finger proteins involved in histone recognition. Plant Sci. 263, 168–176. doi: 10.1016/j.plantsci.2017.06.013
Zhou, B., Lin, J. Z., Peng, D., Yang, Y. Z., Guo, M., Tang, D. Y., et al. (2017). Plant architecture and grain yield are regulated by the novel DHHC-type zinc finger protein genes in rice (Oryza sativa L.). Plant Sci. 254, 12–21. doi: 10.1016/j.plantsci.2016.08.015
Keywords: rice, transcription factor, zinc finger protein, yield, phytohormone, stress response
Citation: Huang Y, Du L, Wang M, Ren M, Yu S and Yang Q (2022) Multifaceted roles of zinc finger proteins in regulating various agronomic traits in rice. Front. Plant Sci. 13:974396. doi: 10.3389/fpls.2022.974396
Received: 21 June 2022; Accepted: 28 June 2022;
Published: 25 July 2022.
Edited by:
Quan Xu, Shenyang Agricultural University, ChinaReviewed by:
Chen Li, Guangdong Academy of Agricultural Sciences (GDAAS), ChinaJiang Hu, China National Rice Research Institute (CAAS), China
Xu Peng, Xishuangbanna Tropical Botanical Garden (CAS), China
Copyright © 2022 Huang, Du, Wang, Ren, Yu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qianying Yang, MTk5MDA4MjQzNkAxNjMuY29t; Shouwu Yu, eXVzd0B6YWFzLmFjLmNu
 Yifeng Huang1,2
Yifeng Huang1,2 Qianying Yang
Qianying Yang