- 1College of Environmental Science and Engineering, China West Normal University, Nanchong, China
- 2Integrative Science Center of Germplasm Creation in Western China (CHONGQING) Science City and Southwest University, Tibet Agriculture and Animal Husbandry College and Southwest University (TAAHC-SWU) Medicinal Plant Joint R&D Centre, School of Life Sciences, Southwest University, Chongqing, China
- 3Chongqing Academy of Science and Technology, Chongqing, China
- 4The Provincial and Ministerial Co-founded Collaborative Innovation Center for R&D in Tibet Characteristic Agricultural and Animal Husbandry Resources, Food Science College, Tibet Agriculture and Animal Husbandry University, Nyingchi, China
The bHLH transcription factors play important roles in the regulation of plant growth, development, and secondary metabolism. β-Caryophyllene, epi-cedrol, and β-farnesene, three kinds of sesquiterpenes mainly found in plants, are widely used as spice in the food industry and biological pesticides in agricultural production. Furthermore, they also have a significant value in the pharmaceutical industry. However, there is currently a lack of knowledge on the function of bHLH family TFs in β-caryophyllene, epi-cedrol, and β-farnesene biosynthesis. Here, we found that AabHLH112 transcription factor had a novel function to positively regulate β-carophyllene, epi-cedrol, and β-farnesene biosynthesis in Artemisia annua. Exogenous MeJA enhanced the expression of AabHLH112 and genes of β-caryophyllene synthase (CPS), epi-cedrol synthase (ECS), and β-farnesene synthase (BFS), as well as sesquiterpenes content. Dual-LUC assay showed the activation of AaCPS, AaECS, and AaBFS promoters were enhanced by AabHLH112. Yeast one-hybrid assay showed AabHLH112 could bind to the G-box (CANNTG) cis-element in promoters of both AaCPS and AaECS. In addition, overexpression of AabHLH112 in A. annua significantly elevated the expression levels of AaCPS, AaECS, and AaBFS as well as the contents of β-caryophyllene, epi-cedrol, and β-farnesene, while suppressing AabHLH112 expression by RNAi reduced the expression of the three genes and the contents of the three sesquiterpenes. These results suggested that AabHLH112 is a positive regulator of β-caryophyllene, epi-cedrol, and β-farnesene biosynthesis in A. annua.
Introduction
Artemisia annua, a traditional Chinese medicinal plant, not only produces the well-known artemisinin, but also produces a number of sesquiterpenes that play an important role in plants’ stress resistance, communication, and growth regulation (Chadwick et al., 2013). β-caryophyllene, epi-cedrol, and β-farnesene, three kinds of sesquiterpenes, are used as spice in the food industry and biological pesticides in agricultural production (Yu X. et al., 2012; Cheng et al., 2022). Besides, they also have a significant value in the pharmaceutical industry (Cheng et al., 2022). β-Caryophyllene was found to has good curative effect on colitis (Bento et al., 2011), cerebral ischemia (Chang et al., 2013), diabetes (Basha and Sankaranarayanan, 2014), anxiety and depression (Bahi et al., 2014), liver fibrosis (Mahmoud et al., 2014), and osteoarthritis (Rufino et al., 2015). In cancer studies, β-caryophyllene demonstrated synergy with the chemotherapy drug paclitaxel on human tumor cell lines, and alone it stimulates apoptosis and suppresses tumor growth (Legault and Pichette, 2007). epi-Cedrol is widely used in pharmaceutical industry for its sedative, anti-inflammatory, and cytotoxic activities (Zhang et al., 2016; Luo et al., 2019). β-farnesene, an important acyclic and volatile sesquiterpenes acting as the main component of the aphid alarm pheromones for many pest aphids, can be used to create new biological pesticides (Wang et al., 2014). Because of their pharmacological activity and commercial value, numerous studies have focused on improving the production of sesquiterpenes in A. annua.
β-Caryophyllene, epi-cedrol, and β-farnesene are biosynthesized via both the cytoplasmic mevalonate (MVA) pathway and plastidial methylerythritol phosphate (MEP) pathway (Navale et al., 2019). Plant cells use one molecule of dimethylallyl diphosphate (DMAPP) and two molecules of isopentenyl diphosphate (IPP) to produce farnesyl diphosphate (FPP) in a reaction catalyzed by FPP synthase (FPS) (Navale et al., 2019). Then the sesquiterpene synthase takes FPP as the substrate to synthesize a variety of sesquiterpenes (Figure 1).
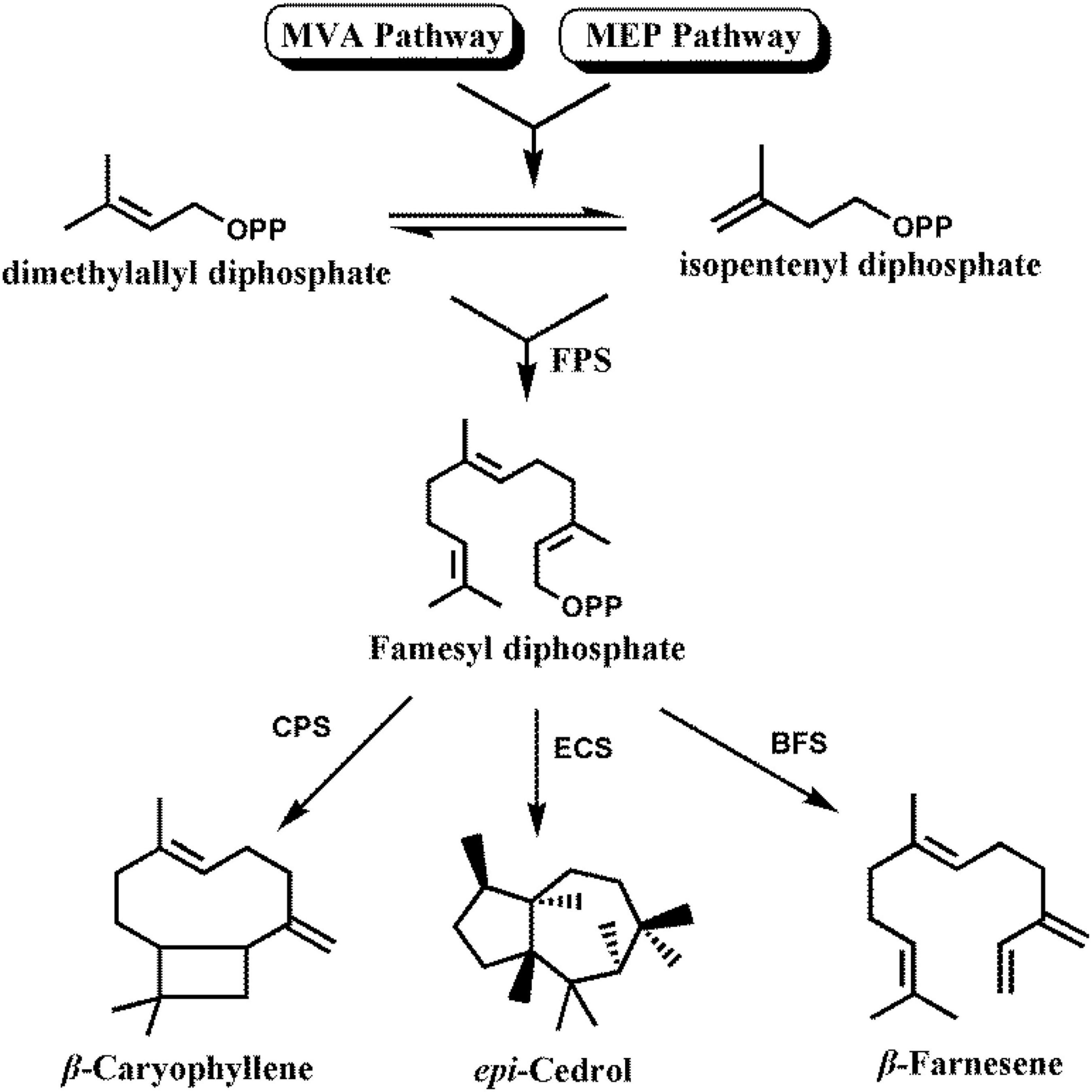
Figure 1. Biosynthetic pathway for sesquiterpenes in Artemisia annua. FPS, farnesyl diphosphate synthase; CPS, β-caryophyllene synthase; ECS, epi-cedrol synthase; BFS, β-farnesene synthase.
Transcription factors (TFs) are sequence-specific DNA-binding proteins which interact with the promoter regions of target genes and regulate their transcription (Yang et al., 2012). It is a useful tool to improve the production of secondary metabolites in pharmaceutically important plants, such as flavonoids or alkaloids (Gantet and Memelink, 2002). Recently, a number of AP2/ERF, WRKY, and bHLH transcription factors have been found to have global regulatory functions in pharmaceutical terpenoids biosynthesis (Lu et al., 2016; Li et al., 2017). In A. annua, overexpression of AaERF1 and AaERF2 significantly promoted the transcription of ADS and CYP71AV1, resulting in an increase of artemisinin content (Yu Z. et al., 2012). Overexpression of SmERF1L1 significantly increased tanshinones production in transgenic Salvia miltiorrhiza hairy roots (Huang et al., 2019). In addition, overexpression of SmWRKY1 significantly elevated the transcription of SmDXS and SmDXR, increasing tanshinone production in S. miltiorrhiza (Cao et al., 2018).
The bHLH family is the second largest transcription factor family in plants, which plays an important role in the regulation of pharmaceutical terpenoids biosynthesis (Ji et al., 2014). It was reported that SmbHLH3 acts as a transcription repressor for both phenolic acids and tanshinones biosynthesis in S. miltiorrhiza hairy roots (Zhang et al., 2020). In Arabidopsis, AtMYC2, a JA-responsive TF, can enhance sesquiterpenes biosynthesis by binding to the promoters of TPS11 and TPS21 (Hong et al., 2012). Chuang et al. (2018) reported that overexpression of PbbHLH4 markedly increased terpenes content in Phalaenopsis. JA-responsive SlMYC1 differentially regulates monoterpene and sesquiterpenes biosynthesis in tomato, which positively regulates monoterpene biosynthesis in leaf and stem but negatively regulates sesquiterpenes biosynthesis in stem (Xu et al., 2018). CrMYC2 directly activates the expression of ORCA genes through binding to the G-box in its promoter, and promotes vinblastine and vincristine biosynthesis in Catharanthus roseus (Zhang et al., 2011). BpMYC4 and BpbHLH9 from Betula platyphylla Suk are involved in regulating the biosynthesis of triterpenes (Yin et al., 2017). Furthermore, studies also showed that bHLH TFs could act as negative regulators in terpenes biosynthesis, including the biosynthesis of the diterpene paclitaxel and monoterpenoid indole alkaloid in Taxus cuspidata (Lenka et al., 2015) and Catharanthus roseus (Patra et al., 2018), respectively. One hundred and twenty two putative bHLH TFs were found in A. annua by genome-wide identification (Xiang et al., 2019) and only three bHLH TFs-AabHLH1, AabHLH112, and AaMYC2, respectively, have been reported to play regulatory roles in artemisinin biosynthesis (Ji et al., 2014; Shen et al., 2016; Xiang et al., 2019). Now, although our knowledge of the regulation of artemisinin biosynthesis is increasing, little is known about the regulation of non-artemisinin sesquiterpenes biosynthesis in A. annua. In this study, we found that the AabHLH112 has a novel function in improving the accumulation of β-caryophyllene, epi-cedrol, and β-farnesene in A. annua, and revealed the mechanism of AabHLH112-mediated upregulation of sesquiterpenes biosynthesis. Our results suggested that this transcription factor could be a useful tool to improve sesquiterpenes production in A. annua.
Materials and methods
Plant material and growth conditions
Seeds were harvested from wild-type A. annua grown in the experimental field of Southwest University (Chongqing, China) for this study. These seeds were surface-sterilized with 20% sodium hypochlorite solution for 20 min, and then washed three times with sterile water. Subsequently, seeds were germinated on 1/2 MS solid medium at 23 ± 2°C under a light period of 16-h-light/8-h-dark. All seedlings were grown in pots with organic substrates in an artificial climate room at 23 ± 2°C under a light period of 16-h-light/8-h-dark. Nicotiana benthamiana seeds were sown directly on solid and their growth conditions are consistent with A. annua. Nicotiana benthamiana plants grown for 30 days were used for dual-luciferase assays.
Exogenous methyl jasmonate treatment and sample collection
One-month old wild-type A. annua plants were treated according to the report (Xiang et al., 2015) with modifications. 300 μM exogenous MeJA solutions containing 0.4% ethanol was sprayed on the plant surfaces. Then the leaves of A. annua plants at 0, 3, 6, 12, and 24 h after exogenous MeJA treatment were collected respectively and then put in liquid nitrogen immediately for RNA extraction and determination of sesquiterpenes contents.
Artemisia annua transformation
The coding sequence of AabHLH112 was amplified and inserted into the pHB binary plasmid vector through BamHI and PstI sites to construct the plant overexpression vector, pHB-AabHLH112. Besides, the 352bp fragment from the AabHLH112 was amplified and inserted into the intermediate pHANNIBAL vector, and then the expression cassette was recombined into the pBin19 plasmid for constructing the AabHLH112-RNAi vector. pHB-AabHLH112 and pBin19-RNAi-AabHLH112 vectors were transferred into A. tumefaciens strain EHA105 to form engineering strains. These strains were grown on YEP solid medium containing related antibiotics for 48 h. Subsequently, positive monoclonal strain was inoculated into YEP liquid medium containing related antibiotics to culture OD600 = 0.5∼0.6. The supernatant was discarded after centrifugation, and then resuspended in the 1/2 MS liquid to OD600 = 0.3∼0.5. Then the culture was shaken at 200 rpm/28°C for 30 min. Cultured engineered strains were used to transform A. annua via Agrobacterium-mediated transformation as described previously (Xiang et al., 2019). After that, the transformed seedlings were transplanted into pots with organic substrates and cultured in an artificial climate room at 23 ± 2°C under a light period of 16 h-light/8 h-dark.
Quantitative real-time polymerase chain reaction
Quantitative real-time PCR was performed to analyze genes’ expression in this study. Total RNA of root, stem, leaf and flower was extracted using the total plant RNA Extract Kit (Tiangen, China), and then reversely transcribed into cDNA using Fast King RT Kit (Tiangen, China). Subsequently, the cDNA was used as the template for detecting the expression levels of AabHLH112 and sesquiterpene synthase genes AaCPS, AaECS, and AaBFS by qPCR experiment. The qPCR amplification conditions were 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 55°C for 20 s, and 72°C for 20 s. β-actin of A. annua was used as the reference gene in this study (Ma et al., 2018), and the relative expression levels were calculated using the 2–△△Ct method (Livak and Schmittgen, 2001). In addition, the expression levels of sesquiterpenes-related genes were detected in AabHLH112-overexpressing and RNAi-AabHLH112 transgenic A. annua using the above method. The primer sequences in qPCR experiment were listed in Supplementary Table 1.
Plant sesquiterpenes extraction and gas chromatography-mass spectrometry analysis
The sesquiterpenes of A. annua leaves were analyzed by gas chromatography-mass spectrometry (GC-MS) as described before with some modifications (Fu et al., 2017). The fresh leaves 1–8 from the main stem of 3 months old AabHLH112-overexpressing transgenic and wild-type A. annua plants were harvested, frozen immediately in liquid nitrogen, and then freeze-dried for 72 h at -80°C. 100 mg leaf powder was soaked in 4 mL n-hexane in 15 mL centrifuge tube with 35 μL trans-farnesol (80 μg/mL) as the internal standard, and ultrasonically extracted for 45 min at 28°C in an ultrasonic processor (KQ-500DE; Kunshan Ultrasonic Instrument Co. Ltd., Kunshan, China). Plant extract was centrifuged at 1,000 rpm for 10 min. The supernatant was filtered through 0.22 μm pore size filters and then analyzed by gas chromatography-mass spectrometry (GCMS-QP2010 Ultra; Shimadzu) with the temperature program: initial temperature of 70°C (1 min hold), increase to 160°C at 10°C/min, ramp to 240°C at 5°C/min, and finally increase to 280°C at 20°C/min (17 min hold). Helium was used as a carrier gas and 1 μL sample was injected in split mode; split rate, 2:1; ion source temperature, 230°C; ionization voltage, 70 eV with scanning from m/z 33 to 500. Qualitative analysis of all compounds was fulfilled by comparing with NIST (National Institute of Standards and Technology) database and Wiley libraries. The relative contents of sesquiterpenes were calculated by comparing peak areas with that of the internal standards. Trans-farnesol was purchased from Sigma-Aldrich in this study.
Cloning and analysis of the promoter regions of sesquiterpene synthase genes
To isolate the promoter fragments of sesquiterpene synthase genes (AaCPS, AaECS, and AaBFS), the total DNA of A. annua leaves was extracted as the template using cetyltrimethylammonium bromide (CTAB) method. All primer sequences were designed according to the genome of A. annua. To avoid non-specific amplification, forward and reverse primer sequences are located in their promoter and ORF regions, respectively. These promoter fragments were obtained by nested PCR using the Pro Taq DNA polymerase (Aikerui, Changsha, China) according to the manufacturer’s instructions. Agarose gel electrophoresis was used to detect PCR products. Subsequently, these fragments were inserted into the pJET2.1 vector (Thermo Fisher Scientific, Waltham, MA, United States) and sequenced respectively. The cis-elements of promoter region in AaCPS, AaECS, and AaBFS were predicted using plantCARE online website.1 The promoter sequences of sesquiterpene synthase genes were shown in Supplementary Data Sheet 1. Primers were listed in Supplementary Table 1.
Dual-luciferase assay
Dual-LUC assays were performed using the methods reported previously (Xiang et al., 2019). The promoter sequences of AaCPS (OP056317), AaECS (OP056318), and AaBFS (OP056319) genes were inserted into pGreenII 0800-LUC plasmid to generate pAaCPS:LUC, pAaECS:LUC, and pAaBFS:LUC constructs as the reporter vector, respectively. Subsequently, these reporter vectors were transferred into A. tumefaciens strain GV3101 together with the pSoup plasmid. The AabHLH112 (MG872820) was inserted into the pHB plasmid driven by CaMV 35S promoter as the effector vector and transferred into A. tumefaciens strain GV3101. Meanwhile, the pHB-YFP plasmid was transferred into GV3101 as a negative control. All engineering and control strains were inoculated into YEP liquid selective medium and cultured overnight at 28°C. The agrobacterium cells were collected by centrifuge at 4,500 rpm for 10 min and resuspended in the MS liquid to OD600 = 0.6 ± 0.05. The acetosyringone (As, 100 mM, 1:500, v:v) and 2-(N-morpholino) ethanesulfonic acid [MES, 0.5 M (pH = 5.7), 1:50, v:v] were added to the resuspension and then were injected into tobacco leaves after being placed for 4 h at room temperature. Tobacco plants injected with agrobacterium cells were exposed to weak light for 48–72 h. Infiltration and detection were performed as described previously (Ma et al., 2018). All experiments were repeated five times for each combination. Primers are listed in Supplementary Table 1.
Yeast one-hybrid assay
To investigate how AabHLH112 regulates the expression of AacCPS, AaECS, and AaBFS, yeast one-hybrid assays were fulfilled as described previously (Xiang et al., 2019). The coding sequence of AabHLH112 was amplified and inserted into pB42AD plasmid with the GAL4 activation domain (AD) through EcoRI and XhoI sites to generate pB42AD-AabHLH112 constructs as the prey. The 45∼55 bp fragments containing one G-box motifs from AaCPS, AaECS, and AaBFS promoters, named pAaCPS-G1 (−884∼−840), pAaCPS-G2 (−838∼−794), pAaCPS-G3 (−806∼−762), pAaCPS-G4 (−430∼−386), pAaECS-G1 (−1228∼−1178), pAaECS-G2 (−915∼−866), pAaECS-G3 (−385∼−336), pAaBFS-G1 (−1045∼−996), pAaBFS-G2 (−985∼−935), pAaBFS-G3 (−66∼−15) and the mutant of G-box were inserted into pLacZ plasmid through KpnI and XhoI sites as the bait, respectively. The pB42AD-AabHLH112 plasmid was co-transformed into yeast strain EGY48 with each individually bait constructs, respectively. The yeast cells were grown on SD-Ura-Trp selective medium for 48 h at 30°C. All independent yeast cells were shifted into SD-Ura-Trp liquid medium and cultured overnight at 30°C, and then these cells were collected by microcentrifugation and resuspended in 100 μL sterile water. Resuspended cells were grown on SD/-Ura-Trp medium with 5-Bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) for 24 48 h at 30°C. The empty pB42AD and pLacZ plasmids were used as the negative control. Five independent biological replicates were implemented for each experiment in this study. The primers are listed in Supplementary Table 1.
Results
Tissues expression patterns of AabHLH112 and three sesquiterpene synthase genes
To study the tissue expression patterns of AabHLH112 and sesquiterpenes biosynthesis genes, we measured their expression in various A. annua tissues including roots, stems, leaves, and flowers. AabHLH112 was highly expressed in flowers and leaves, and lowly expressed in roots and stems (Figure 2A). AaCPS and AaBFS genes’ expression has a tissue-specific mode, with a high expression in flowers and low expression in leaves, stems, and roots (Figures 2B,D). By comparison, AaECS was highly expressed in leaves and lowly expressed in roots and flowers (Figure 2C). Together, AabHLH112, AaCPS, AaECS, and AaBFS were expressed in all of the detected tissues.
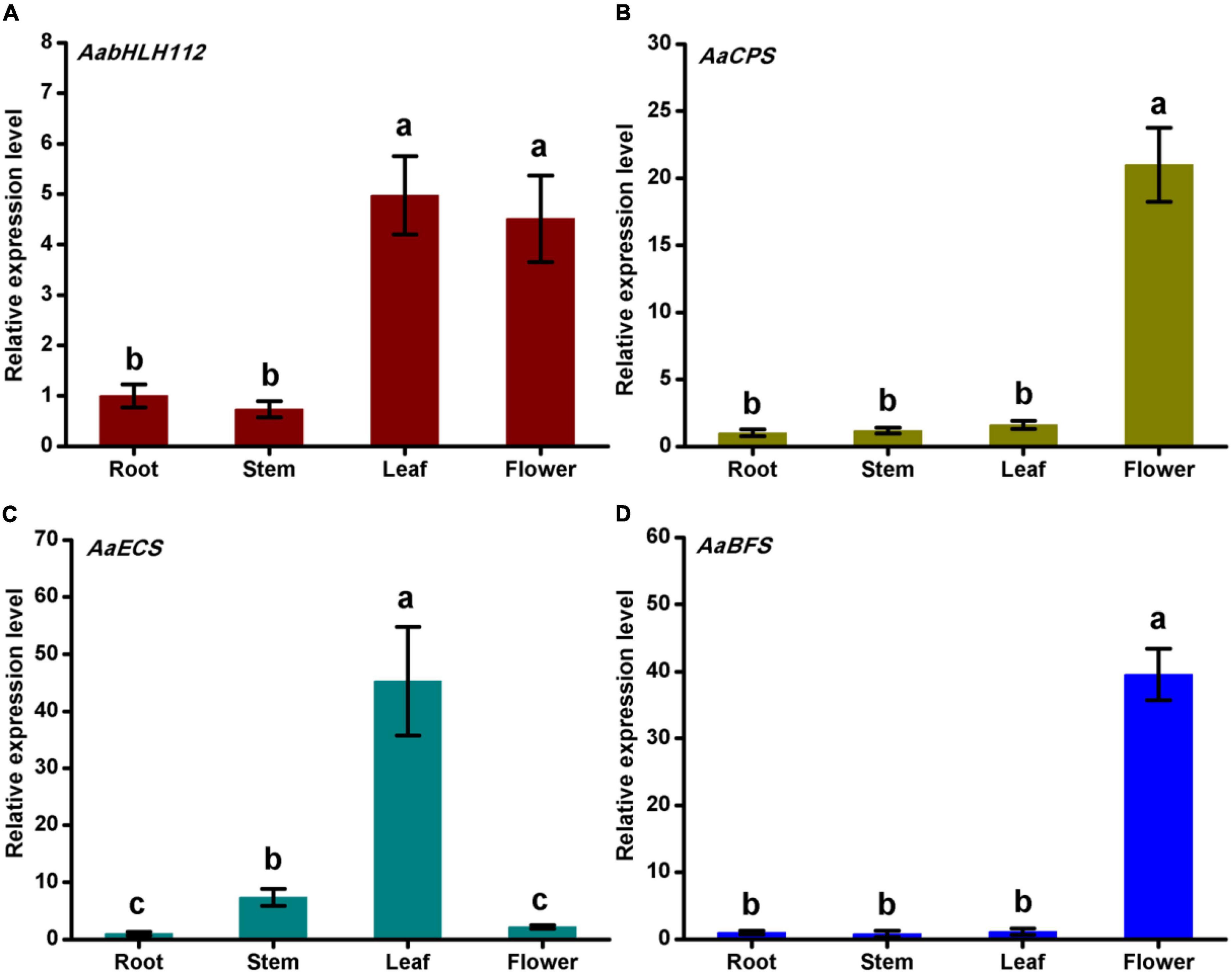
Figure 2. Genes expression levels of AabHLH112 (A), AaCPS (B), AaECS (C), and AaBFS (D) in root, stem, leaf, and flower of Artemisia annua. Bars are means ± SD from three independent biological replicates. One-way ANOVA was tested for significant differences among the means (indicated by different letters at p < 0.01).
Methyl jasmonate treatment induced the expression of AabHLH112 and three sesquiterpene synthase genes
The phytohormone JA plays a critical role in plant stress resistance and secondary metabolism (Wasternack and Song, 2017; Kianersi et al., 2021). To investigate the response of AabHLH112, AaCPS, AaECS, and AaBFS to MeJA treatment, wild type A. annua plants were treated with exogenous MeJA, and all the genes’ expression was quantified by qPCR. The expression of AabHLH112 was induced by MeJA, rapidly increased and peaked after 3 h of treatment, followed by a gradual decrease (Figure 3A). This result indicated that AabHLH112 is involved in MeJA response in A. annua, as in the case with the Arabidopsis homologous AtbHLH33 (Li et al., 2020). Several studies have shown that the expression of many genes involved in terpenes biosynthesis is significantly increased under JA treatment. Here, we found the expression levels of AaECS, AaCPS, and AaBFS were significantly upregulated under exogenous MeJA treatment (Figures 3B–D); besides, the accumulation of β-caryophyllene, epi-cedrol, and β-farnesene, direct products of the three enzymes, also showed an upward trend after MeJA treatment (Supplementary Figure 1). These results suggest that MeJA can induce the expression of these sesquiterpenes synthase genes, thus enhancing the biosynthesis of β-caryophyllene, epi-cedrol and β-farnesene in A. annua.
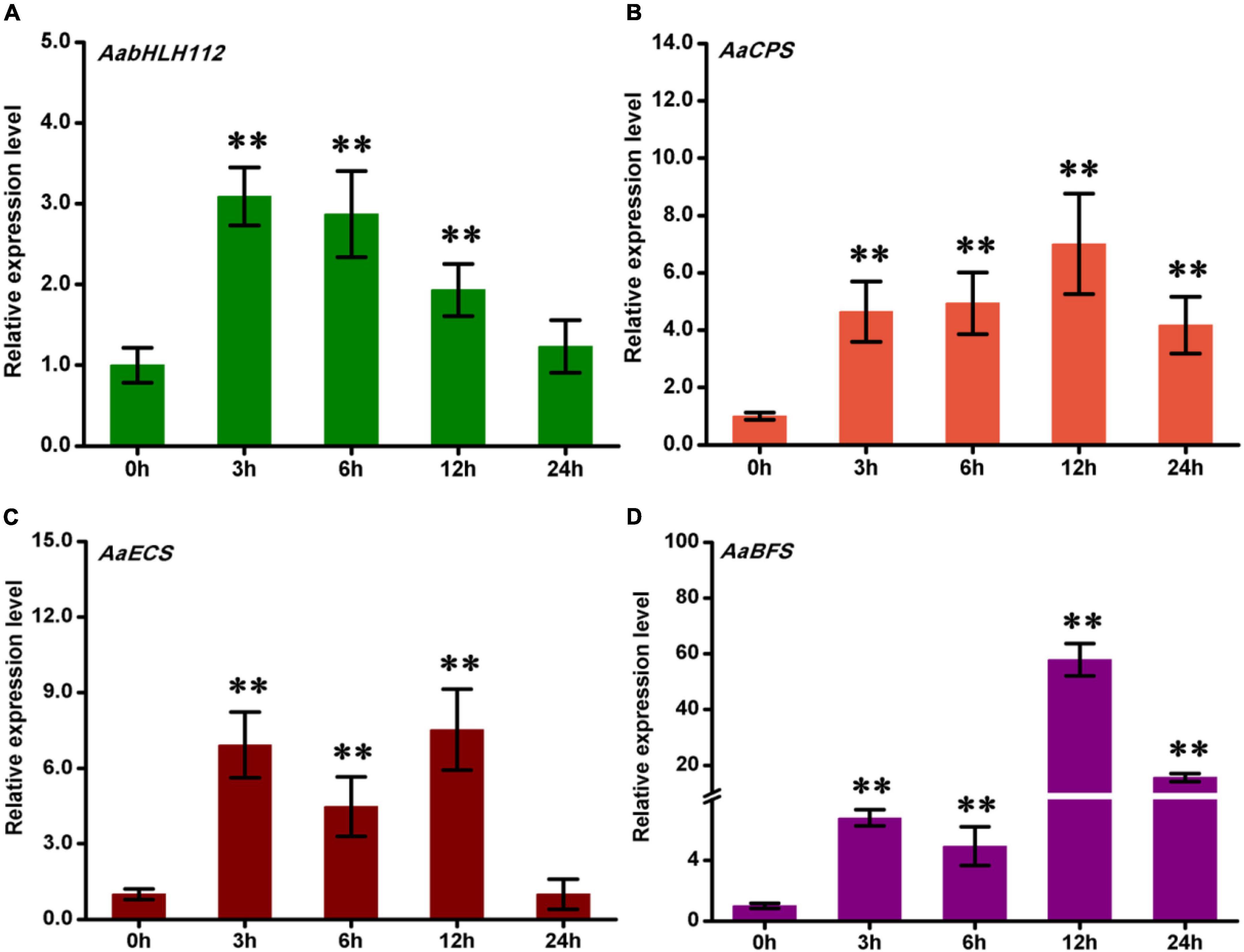
Figure 3. The expression of AabHLH112 (A), AaCPS (B), AaECS (C), and AaBFS (D) in response to MeJA treatment. Error bars represent the standard deviations of three technical replicates. Statistical significance was assessed with Student’s t-test (**p < 0.01).
AabHLH112 activates the expression of the AaECS, AaCPS, and AaBFS
Dual-LUC assays can be used to study the transcriptional regulation of transcription factors on target genes at the transcriptional level (Ma et al., 2018). The coding sequence of AabHLH112 was inserted into the plant overexpression vector pHB as an effector construct, and the promoter sequences of AaCPS (Supplementary Figure 2), AaECS (Supplementary Figure 3), and AaBFS (Supplementary Figure 4) genes were inserted into pGreenII 0800-LUC plasmid to generate pAaCPS:LUC, pAaECS:LUC, and pAaBFS:LUC constructs as the reporter vector, respectively. Meanwhile, the pHB-YFP (yellow fluorescent protein) plasmid was used as a negative control (Figure 4A). The effector and reporter constructs were transiently expressed in N. benthamiana leaves using Agrobacterium tumefaciens-mediated co-infiltration. The dual-LUC assay results showed that the promoter activity of the above sesquiterpene synthase genes AaCPS, AaECS, and AaBFS was significantly increased, with the LUC/REN value increased by 7.75, 5.58, and 4.05-folds than the YFP control (Figures 4B–D), respectively.
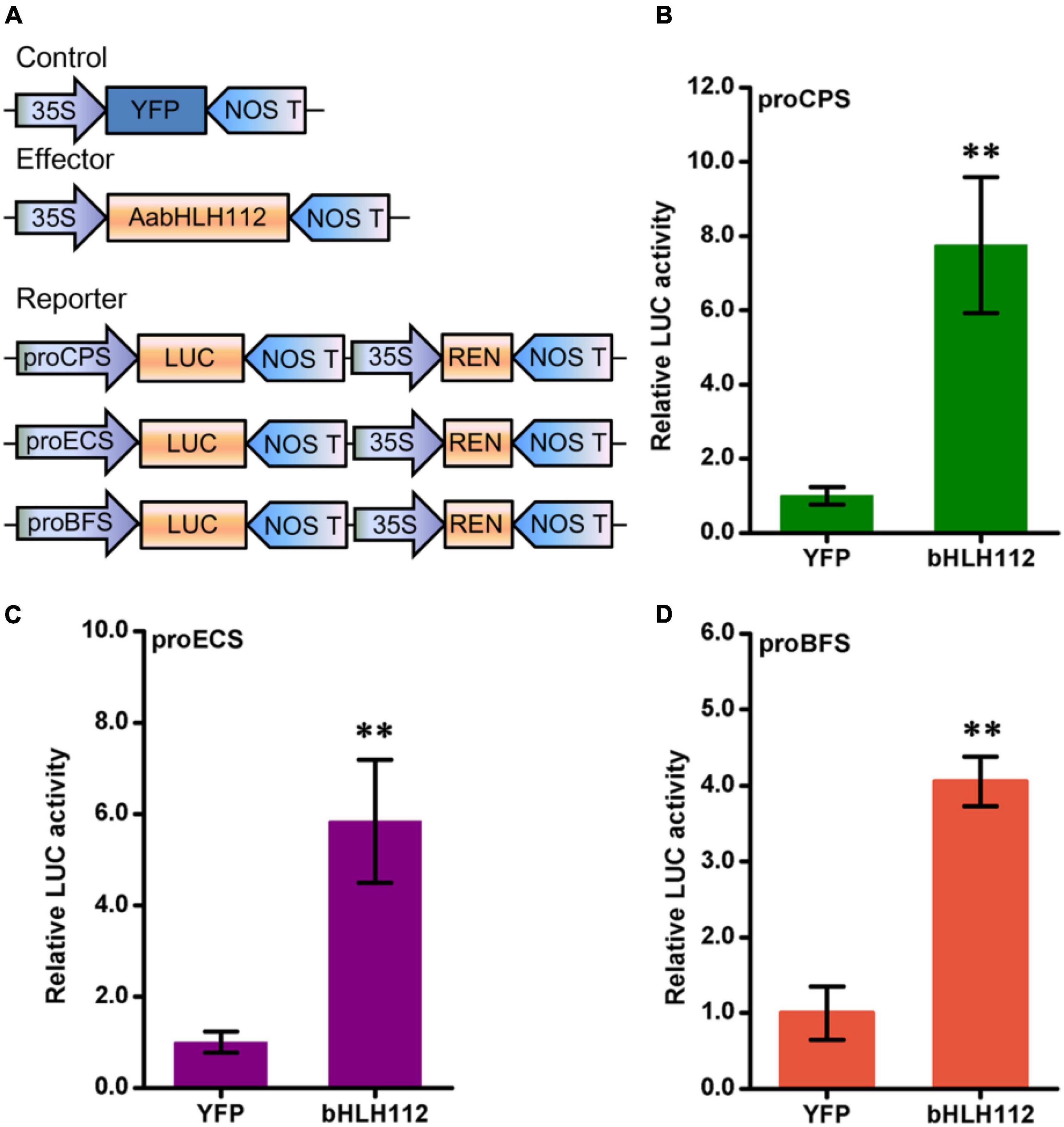
Figure 4. Biochemical assays between AabHLH112 and sesquiterpenes synthase genes. (A) Diagrams of reporter and effector constructs in transient dual-luciferase assays (REN, renilla luciferase; LUC, firefly luciferase). (B–D) Effects of AabHLH112 on activities of the AaCPS, AaECS, and AaBFS promoters in Nicotiana benthamiana leaves. The YFP effector was used as a negative control. The data represents the means ± SD (n = 3), **p < 0.01 in student’s t-test.
AabHLH112 bind to G-box cis-elements in the promoters of AaCPS and AaECS
The core DNA sequence motif recognized by bHLH proteins is a consensus hexanucleotide sequence known as the G-box (5-CANNTG-3). It was previously reported that AaMYC2 can specifically bind to the G-box cis-elements in the promoters of key enzyme genes of artemisinin biosynthesis, including AaCYP71AV1 and AaDBR2 (Shen et al., 2016). Recently, Xiang et al. (2019) reported that AabHLH112 can directly bind to G-box in the AaERF1 promoter and promote artemisinin biosynthesis. The above dual-LUC results showed that AabHLH112 could activate the transcription of AaCPS, AaECS, and AaBFS. In order to further investigate how AabHLH112 regulates the transcription of these sesquiterpene synthase genes, the promoter sequences of AaCPS, AaECS, and AaBFS were analysed by PlantCARE online website, and multiple G-box motifs were found in each promoter (Figures 5A,C,E). Following that, yeast one-hybrid assay was used to evaluate whether AabHLH112 can directly bind to the G-box motifs in these promoters. 45 50 bp fragments containing G-box motifs from each promoter were inserted into pLacZ plasmid as the bait, and AabHLH112 was inserted into pB42AD plasmid with the GAL4 activation domain to generate pB42AD-AabHLH112 constructs as the prey. The results indicated that AabHLH112 can directly bind to pAaCPS-G3 and pAaECS-G1 fragments containing the G-box in AaCPS and AaECS promoters (Figures 5B,D), but not the pAaCPS1-G1, pAaCPS-G2, pAaCPS-G4, pAaECS-G2, pAaECS-G3, and pAaBFS-G1, pAaBFS-G2, pAaBFS-G3 fragments. When the G-box cis-elements in the pAaCPS-G3 and pAaECS-G1 fragments were mutated to the 5’-AAAAA-3’ sequences resulting in pAaCPS-mG3 and pAaECS-mG1, respectively, the binding signals of AabHLH112 and pAaCPS-mG3 and pAaECS-mG1 disappeared (Figures 5B,D). These results demonstrate that AabHLH112 enhances AaCPS and AaECS transcription by directly binding to the specific G-box cis-elements in their promoters.
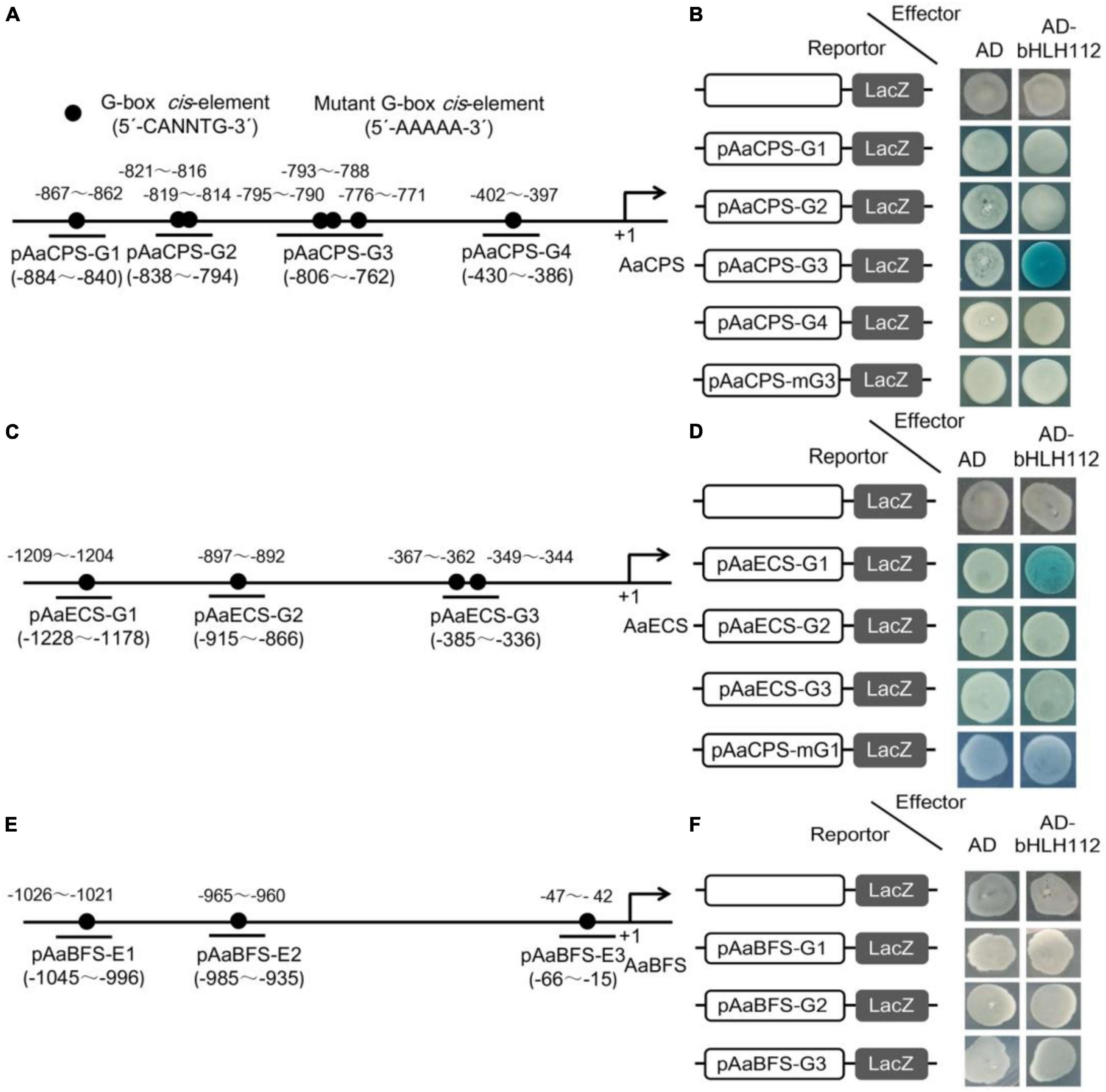
Figure 5. Yeast one-hybrid assays between AabHLH112 and G-box cis-elements of sesquiterpene synthase gene promoters, blue plaques indicate protein-DNA interactions. (A) Schematic diagram of the AaCPS promoter; (B) Y1H assay between AabHLH112 and AaCPS promoter; (C) Schematic diagram of the AaECS promoter; (D) Y1H assay between AabHLH112 and AaECS promoter; (E) Schematic diagram of the AaBFS promoter; (F) Y1H assay between AabHLH112 and AaBFS promoter; the black circles were the potential G-box elements in three promoters.
Generation of AabHLH112-overexpression and RNAi trangenic Artemisia annua
To better explore the function of AabHLH112 in sesquiterpenes biosynthesis in A. annua, AabHLH112-overexpressing and AabHLH112-RNAi transgenic A. annua plants were generated (Supplementary Figure 5). Primers designed according to the sequences of rubisco gene (rbc), AabHLH112 and marker gene (hygromycin-resistant) were used to determine the regeneration plants by genomic PCR. The 877bp fragment of the hygromycin-resistant gene and coding sequence of AabHLH112 with partial rbc terminator (1383bp) were specifically amplified from the transformed plants and the positive control (pHB-AabHLH112), while these specific fragments could not be amplified from wild-type plants (Supplementary Figures 5B,C). Besides, AabHLH112-RNAi transgenic plants were also generated and detected by genomic PCR. Specific detection primers were designed according to the sequences of AabHLH112, OCS terminator and NptII (kanamycin) genes. As shown in Supplementary Figures 5D,E, the partial AabHLH112 sequence and NptII gene were detected in the AabHLH112-RNAi transgenic plants.
AabHLH112 positively regulates the expression of AaCPS, AaECS, and AaBFS in Artemisia annua
The expression level of AabHLH112 in transgenic A. annua lines was determined by qPCR. Compared with the wild type (WT) A. annua, AabHLH112 presented higher expression level in AabHLH112-overexpression lines of OE-2, OE-7, and OE-8 with 3.44-fold, 5.56-fold, and 4.24-fold higher, respectively (Figure 6A). Meanwhile, the expression levels of AaCPS, AaECS, and AaBFS in the three lines (OE-2, OE-7, and OE-8) were approximately 3.35-7.34 fold, 2.65-9.15 fold, and 2.24-2.55 fold higher compared to the WT respectively (Figure 6A). On the contrary, The expression level of AabHLH112 were markedly decreased in AabHLH112-RNAi lines of Ri-4, Ri-8, and Ri-9 (Figure 6B), and the expression of AaCPS, AaECS, and AaBFS was also significantly reduced (Figure 6B) in the three AabHLH112-RNAi lines compared to WT plants. These results suggest that AabHLH112 positively regulates the expression of AaCPS, AaECS and AaBFS in A. annua.
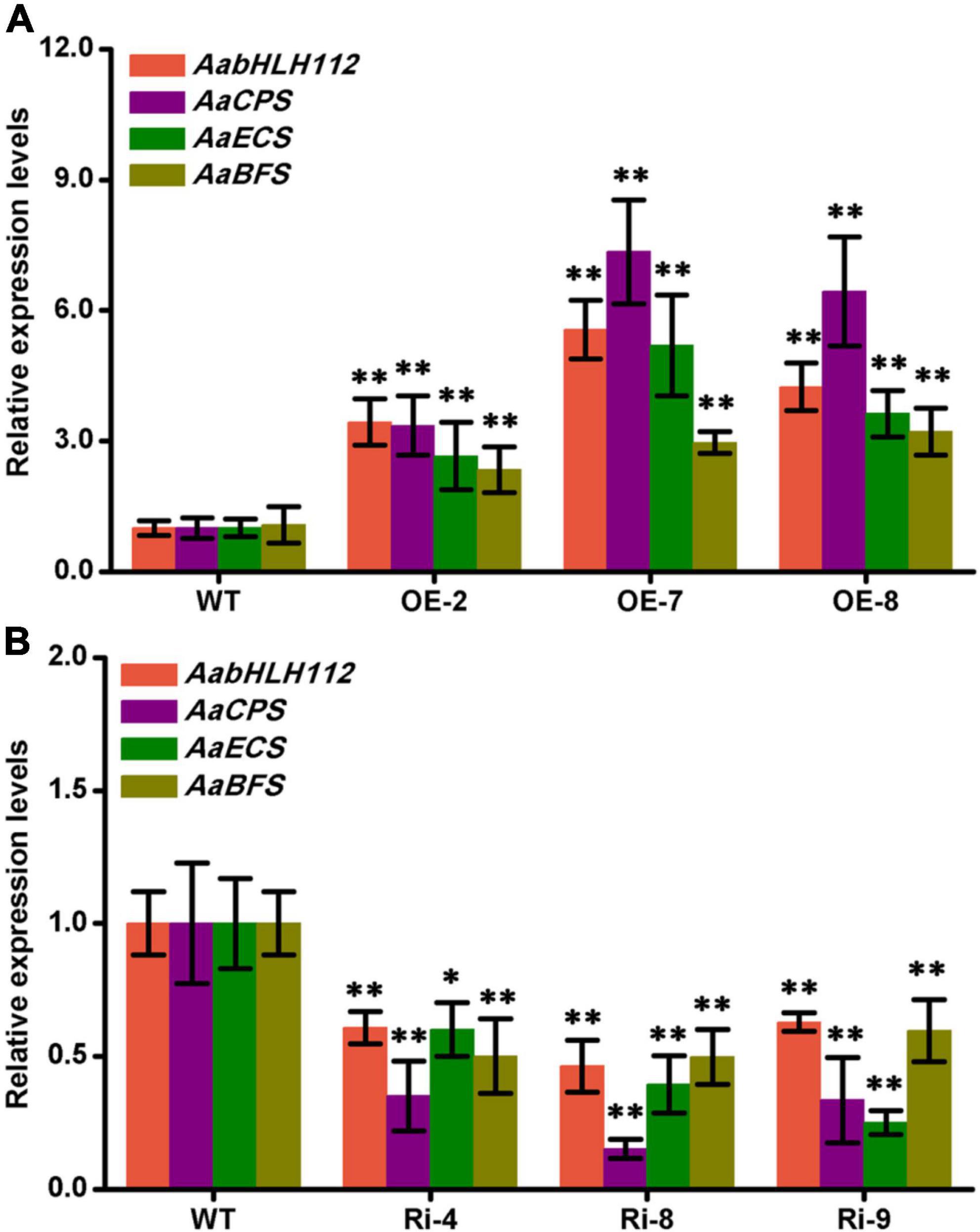
Figure 6. Genes relative expression levels in wild-type, AabHLH112-overexpression and AabHLH112-RNAi Artemisia annua plants. (A): Relative expression levels of AabHLH112, AaCPS, AaECS, and AaBFS in wild type and AabHLH112-overexpression transgenic plants; (B): Relative expression levels of AabHLH112, AaCPS, AaECS, and AaBFS in wild type and AabHLH112-RNAi transgenic plants. Error bars represent the standard deviations of three technical replicates. Statistical significance was assessed with Student’s t-test (*p < 0.05 and **p < 0.01).
AabHLH112 positively regulates β-caryophyllene, epi-cedrol, and β-farnesene biosynthesis in Artemisia annua
The contents of β-caryophyllene, epi-cedrol, and β-farnesene in the AabHLH112-overexpression and RNAi transgenic lines were detected by the GC-MS method. The wild type A. annua produced 1.87 mg/g FW (fresh weight) of β-caryophyllene, 1.07 mg/g FW of epi-cedrol and 0.53 mg/g FW of β-farnesene, while the three independent AabHLH112-overexpression lines (OE-2, OE-7, and OE-8) exhibited significant increase in sesquiterpenes contents. OE-2, OE-7, and OE-8 contained β-caryophyllene at levels of 2.34 mg/g FW, 2.45 mg/g FW, and 2.95 mg/g FW, respectively, representing 25.13, 31.01, and 57.75% increases in content compared with WT plants (Figure 7A). Compared to WT plants, the content of epi-cedrol was increased by 118, 176, and 200% in the three AabHLH112-overexpression lines respectively (Figure 7B); and the content of β-farnesene was increased by 341, 253, and 201% in the three AabHLH112-overexpression lines (Figure 7C). In the AabHLH112-RNAi lines of Ri-4, Ri-8 and Ri-9, the contents of β-caryophyllene, epi-cedrol and β-farnesene were significantly reduced, compared to that in WT plants. Ri-4, Ri-8, and Ri-9 lines contained β-caryophyllene at levels of 0.73 mg/g FW, 1.09 mg/g FW, and 0.94 mg/g FW respectively, representing 61.91, 42.28, and 50.54% reduction in content compared with WT plants (Figure 7D). Epi-cedrol content was reduced by 78.12, 88.45, and 76.82% in Ri-4, Ri-8 and Ri-9 lines respectively, relative to that in WT plants (Figure 7E). β-farnesene content in Ri-4, Ri-8, and Ri-9 lines was reduced by 84.27, 70.55, and 84.58% respectively, compared with that in WT plants (Figure 7F). These results showed that AabHLH112 is a positive regulator of β-caryophyllene, epi-cedrol, and β-farnesene biosynthesis in A. annua.
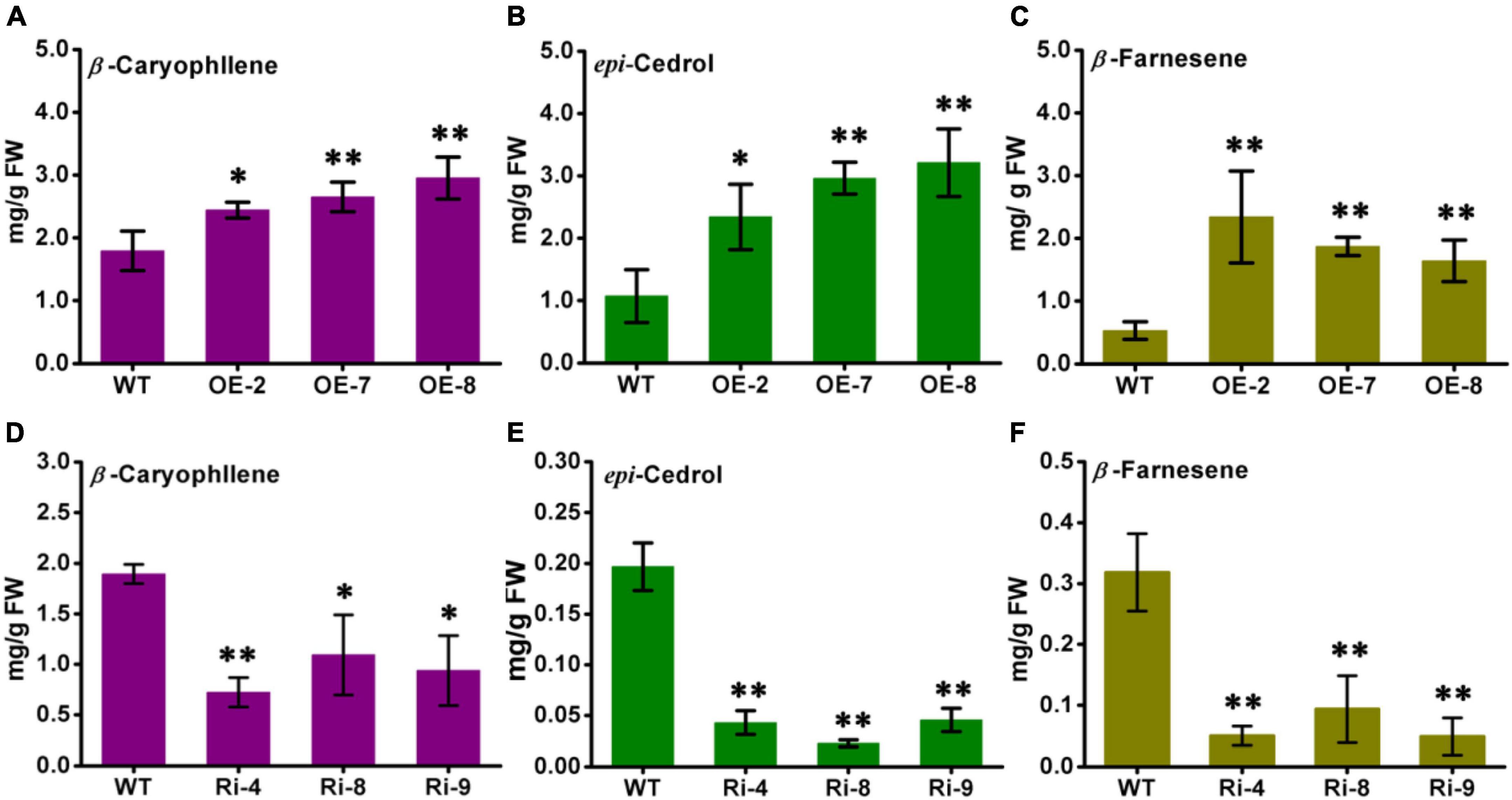
Figure 7. The contents of β-caryophyllene, epi-cedrol, and β-farnesene in wild-type, AabHLH112-overexpression, and AabHLH112-RNAi Artemisia annua plants. (A) β-caryophllene, (B) epi-cedrol, and (C) β-farnesene in AabHLH112-overexpressing lines, WT, wild type; OE-2,OE-7 and OE-8 are independent lines of AabHLH112-overexpressing lines; (D) β-caryophllene, (E) epi-cedrol and (F) β-farnesene in AabHLH112-RNAi lines, WT, wild type; Ri-4, Ri-8, and Ri-9 are independent lines of AabHLH112-RNAi lines, the data represents the means ± SD (n = 3), *p < 0.05, **p < 0.01 in student’s t-test.
Discussion
Artemisia annua is a traditional Chinese medicinal plant, not only producing the well-known drug artemisinin, but also producing other important sesquiterpenes of medicinal value, such as β-caryophyllene and epi-cedrol. The bHLH TFs are the second largest transcription factor family in the plants, which play an important role in plant growth, development, and secondary metabolism. Previous research revealed that bHLH TFs play an important part in the regulation of terpenoids biosynthesis. In tomato, overexpression of CubHLH1 resulted in a significant increase in carotenoid content (Endo et al., 2016). MYC, one subgroup of bHLH family, plays an important role in JA, ABA, and GA signaling and is involved in the regulation of terpenoids biosynthesis in plants (Hong et al., 2012; Du et al., 2017). In A. thaliana, AtMYC2 could directly bind to promoters of the sesquiterpene synthase genes TPS21 and TPS11 and activate their expression, thus increasing sesquiterpenes biosynthesis (Hong et al., 2012). AaMYC2 overexpression could activate the transcription of CYP71AV1 and DBR2, resulting in an increase of artemisinin content in A. annua (Shen et al., 2016). Besides, AabHLH1 positively regulated biosynthesis of artemisinin via activating the promoters of ADS and CYP71AV1 (Ji et al., 2014). In this study, we found that AbHLH112 positively regulates the biosynthesis of β-caryophyllene, epi-cedrol, and β-farnesene in A. annua besides upregulating artemisinin biosynthesis.
The gene expression was synchronous with its product. In this study, we measured the expression levels of AabHLHH112, AaCPS, AaECS, and AaBFS in roots, stems, leaves and flowers. And we found that AabHLH112 has the highest expression in leaves and flowers and the lowest expression in stems and roots, which has a similar expression pattern with AaADS involved in artemisinin biosynthesis (Pu et al., 2013). AaCPS and AaBFS, involved in β-caryophyllene and β-farnesene biosynthesis respectively, have a tissue-specific expression pattern, with the highest expression in flowers and the lowest expression in roots, stems and leaves. By contrast, AaECS is highly expressed in leaves and lowly expressed in roots and flowers, showing a tissue difference in the expression of these genes.
Methyl jasmonate (MeJA) is one of the most potent elicitors that can induce overaccumulation of many natural products in plants, such as artemisinin, tanshinones, vinblastine, and so on (Xiang et al., 2015; Shi et al., 2016; Zhang et al., 2018). Later studies showed that exogenous MeJA activated the transcription of some genes leading to the products’ overaccumulation (Wang et al., 2020). In this study, the 1-month-old A. annua seedlings were treated with MeJA for up to 24 h and the four genes expression were quantified by qPCR. The results showed that the genes detected had a higher expression after the MeJA treatment, indicating that exogenous MeJA treatment can induce the expression of sesquiterpene biosynthesis genes in A. annua. In addition, we found that the expression level of AabHLH112 was induced as early as 3 h after the treatment, and then began to decline, while the highest expression levels of AaECS and AaCPS were detected at 12 h after MeJA treatment. This result suggested that these sesquiterpenes synthase genes might be regulated by AabHLH112. To further explore the function of AabHLH112 in the regulation of sesquiterpene biosynthesis genes expression, dual-LUC assay was taken to determine the activation of AaCPS, AaECS, and AaBFS promoters by AabHLH112. The results indicated AabHLH112 can significantly enhance the transcriptional activity of AaCPS, AaECS, and AaBFS promoters in N. benthamiana leaves (Figures 4B–D).
It is known that G-box is a putative recognition site for bHLH TFs. We have reported in a previous study that AabHLH112 can bind to the G-box in the AaERF1 promoter and enhanced its transcription to upregulate artemisinin biosynthesis (Xiang et al., 2019). In this study, yeast one-hybrid assay (Y1H) showed that AabHLH112 can directly bind to pAaCPS-G3 and pAaECS-G1 fragments containing the G-box in the promoters of AaCPS and AaECS, but not bind to pAaCPS1-G1, pAaCPS-G2, pAaCPS-G4, pAaECS-G2, pAaECS-G3, and pAaBFS-G1, pAaBFS-G2, pAaBFS-G3 fragments (Figures 5B,D,F). Based on these results, we estimated that the flanking sequence of G-box cis-elements may affect the recognition of AabHLHL112 protein. In addition, there are three G-box cis-elements in the pAaCPS-G3 fragment and they are very close to each other. So, a profound study should be carried out to analyze which G-box is the real recognition site by AabHLH112. Interestingly, AabHLH112 positively regulated AaBFS expression to increase β-farnesene accumulation, but Y1H result showed there was no direct interaction between AabHLH112 and AaBFS promoter, suggesting that AabHLH112 upregulated β-farnesene biosynthesis through an indirect pathway, which deserves further exploration.
Taken together, our study indicated that AabHLH112, which is induced by MeJA, positively regulates the expression of AaCPS, AaECS, and AaBFS through directly binding to their promoters or an indirect pathway, thereby enhancing the biosynthesis of β-caryophyllene, epi-cedrol, and β-farnesene in Artemisia annua (Figure 8). Our study revealed the mechanism of AabHLH112-mediated upregulation of sesquiterpenes biosynthesis, and provided a potentially useful transcription factor that could be used to improve sesquiterpenes production in A. annua.
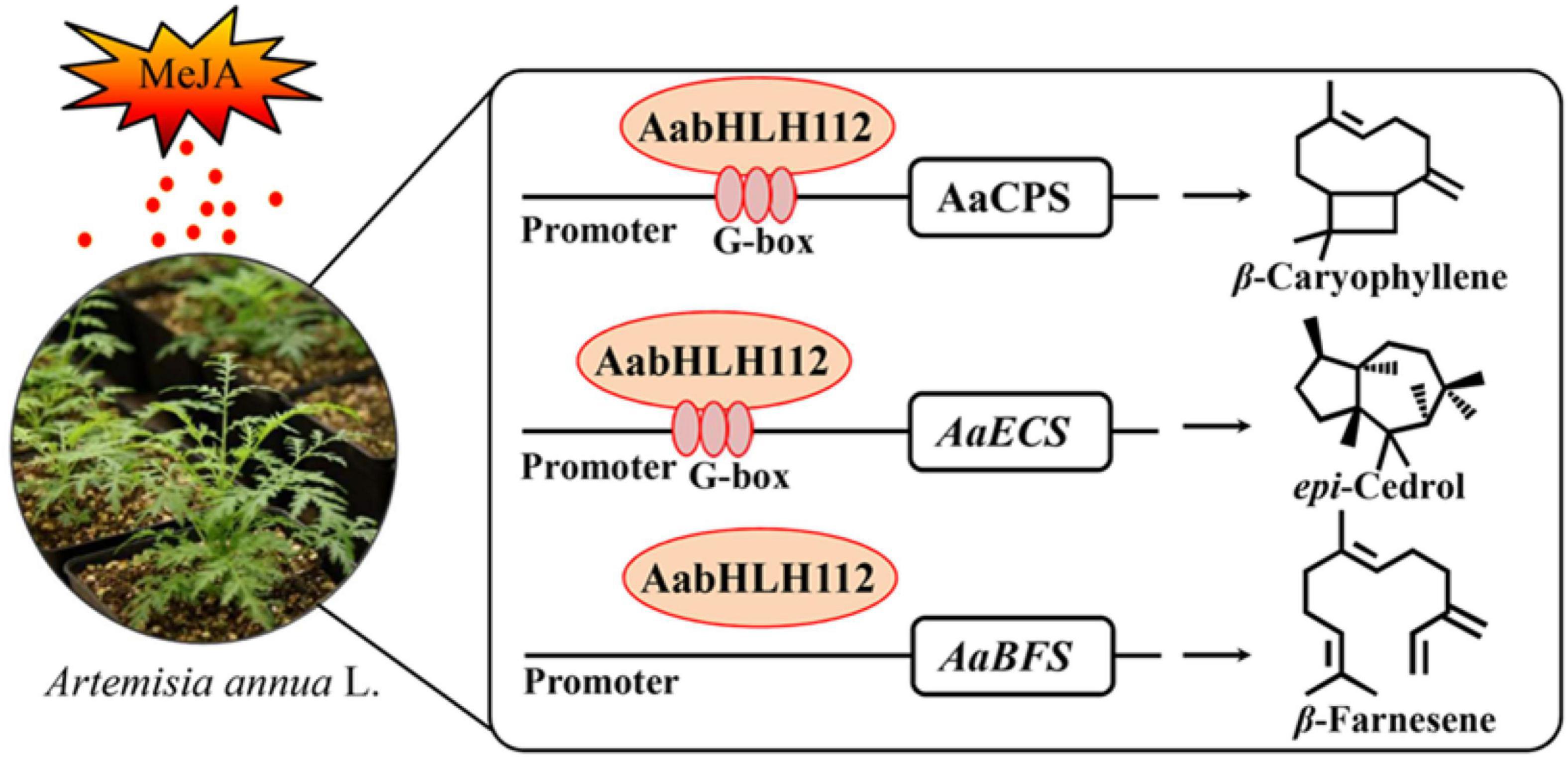
Figure 8. A simplified model for the regulation of sesquiterpenes biosynthesis by AabHLH112 in Artemisia annua.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
LX, ZL, and YT conceived and coordinated the study and wrote the manuscript. LX performed the gene cloning, vector construction, and plant transformation work. GS, MY, and MW did the gene expression, luciferase analysis, and Y1H experiments. GS, XL, and PH helped with the metabolites analysis by GC-MS. All authors reviewed the results and approved the final version of the manuscript.
Funding
This research was financially supported by the Science Funding of Sichuan Province (2020YJ0171, China), the NSFC projects (81973420 and 81803660), the National Key Research and Development Project (2019YFE0108700, China), and the Forth National Survey of Traditional Chinese Medicine Resources, Chinese or Tibet Medicinal Resources Investigation in Tibet Autonomous Region (20191217-540124, 20191223-540126, and 20200501-542329).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.973591/full#supplementary-material
Footnotes
References
Bahi, A., Mansouri, S., Memari, E., Ameri, M., Nurulain, S., and Ojha, S. (2014). β-Caryophyllene, a CB2 receptor agonist produces multiple behavioral changes relevant to anxiety and depression in mice. Physiol. Behav. 135, 119–124. doi: 10.1016/j.physbeh.2014.06.003
Basha, R., and Sankaranarayanan, C. (2014). β-Caryophyllene, a natural sesquiterpene, modulates carbohydrate metabolism in streptozotocin-induced diabetic rats. Acta Histochem. 116, 1469–1479. doi: 10.1016/j.acthis.2014.10.001
Bento, A., Marcon, R., Dutra, R., Claudino, R., Cola, M., Leite, D., et al. (2011). β-Caryophyllene inhibits dextran sulfate sodium-induced colitis in mice through CB2 receptor activation and PPARγ pathway. Am. J. Pathol. 178, 1153–1166. doi: 10.1016/j.ajpath.2010.11.052
Cao, W., Wang, Y., Shi, M., Hao, X., Zhao, W., Wang, Y., et al. (2018). Transcription factor SmWRKY1 positively promotes the piosynthesis of tanshinones in Salvia miltiorrhiza. Front. Plant Sci. 9:554. doi: 10.3389/fpls.2018.00554
Chadwick, M., Trewin, H., Gawthrop, F., and Wagstaff, G. (2013). Sesquiterpenoids lactones: benefits to plants and people. Int. J. Mol. Sci. 14, 12780–12805. doi: 10.3390/ijms140612780
Chang, H., Kim, J., Lee, J., Kim, W., and Chun, H. (2013). Protective effect of β-caryophyllene, a natural bicyclic sesquiterpene, against cerebral ischemic injury. J. Med. Food 16, 471–480. doi: 10.1089/jmf.2012.2283
Cheng, T., Zhang, K., Guo, J., Yang, Q., Li, T., Xiao, M., et al. (2022). Highly efficient biosynthesis of β-caryophyllene with a new sesquiterpene synthase from tobacco. Biotechnol. Biofuels Bioproducts 15:39. doi: 10.1186/s13068-022-02136-8
Chuang, Y., Hung, Y., Tsai, W., Chen, W., and Chen, H. (2018). PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 69, 4363–4377. doi: 10.1093/jxb/ery246
Du, M., Zhao, J., Tzeng, D., Liu, Y., Deng, W., Ynag, T., et al. (2017). MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 29, 1883–1906. doi: 10.1105/tpc.16.00953
Endo, T., Fujii, H., Sugiyama, A., Nakano, M., Nakajima, N., and Ikoma, Y. (2016). Overexpression of a citrus basic helix-loop-helix transcription factor (CubHLH1), which is homologous to Arabidopsis activation-tagged bri1 suppressor 1 interacting factor genes, modulates carotenoid metabolism in transgenic tomato. Plant Sci. 243, 35–48. doi: 10.1016/j.plantsci.2015.11.005
Fu, X., Shi, P., He, Q., Shen, Q., Tang, Y., Pan, Q., et al. (2017). AaPDR3, a PDR transporter 3, is involved in sesquiterpene β-caryophyllene transport in Artemisia annua. Front. Plant Sci. 8:723. doi: 10.3389/fpls.2017.00723
Gantet, P., and Memelink, J. (2002). Transcription factors: tools to engineer the production of pharmacologically active plant metabolite. Trends Pharmacol. Sci. 23, 563–569. doi: 10.1016/s0165-6147(02)02098-9
Hong, G., Xue, X., Mao, Y., Wang, L., and Chen, X. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant cell 24, 2635–2648. doi: 10.1105/tpc.112.098749
Huang, Q., Sun, M., Yuan, T., Wang, Y., Shi, M., and Lu, S. (2019). The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem. 274, 368–375. doi: 10.1016/j.foodchem.2018.08.119
Ji, Y., Xia, J., Shen, Y., Ma, D., Li, Z., and Pu, G. (2014). Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 55, 1592–1604. doi: 10.1093/pcp/pcu090
Kianersi, F., Pour-Aboughadareh, A., Majdi, M., and Poczai, P. (2021). Effect of methyl jasmonate on thymol, carvacrol, phytochemical accumulation, and expression of key genes involved in thymol/carvacrol biosynthetic pathway in some iranian thyme species. Int. J. Mol. Sci. 22:11124. doi: 10.3390/ijms222011124
Legault, J., and Pichette, A. (2007). Potentiating effect of β-caryophyllene on anticancer activity of α-humulene, isocaryophyllene and paclitaxel. J. Pharmacy Pharmacol. 59, 1643–1647. doi: 10.1211/jpp.59.12.0005
Lenka, S., Nims, N., Vongpaseuth, K., Boshar, R., Roberts, S., and Walker, E. (2015). Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1. TcJAMYC2, and TcJAMYC4. Front. Plant Sci. 6:115. doi: 10.3389/fpls.2015.00115
Li, X., Xu, Y., Shen, S., Yin, X., Klee, H., Zhang, B., et al. (2017). Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 68, 4929–4938. doi: 10.1093/jxb/erx316
Li, Y., Li, L., Ding, W., Li, H., Shi, T., Yang, X., et al. (2020). Genome-wide identification of osmanthus fragrans bHLH transcription factors and their expression analysis in response to abiotic stress. Environ. Exp. Bot. 172:103990.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, X., Tang, K., and Li, P. (2016). Plant metabolic engineering strategies for the production of pharmaceutical terpenoids. Front. Plant Sci. 7:1647. doi: 10.3389/fpls.2016.01647
Luo, F., Ling, Y., Li, Den, Tang, T., Liu, Y. C., Liu, Y., et al. (2019). Characterization of a sesquiterpene cyclase from the glandular trichomes of Leucosceptrum canum for sole production of cedrol in Escherichia coli and Nicotiana benthamiana. Phytochemistry 162, 121–128. doi: 10.1016/j.phytochem.2019.03.009
Ma, Y., Xu, D., Li, L., Zhang, F., Fu, X., Shen, Q., et al. (2018). Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua. Sci. Adv. 4:eaas9357. doi: 10.1126/sciadv.aas9357
Mahmoud, M., Swefy, S., Hasan, R., and Ibrahim, A. (2014). Role of cannabinoid receptors in hepatic fibrosis and apoptosis associated with bile duct ligation in rats. Eur. J. Pharmacol. 742, 118–124. doi: 10.1016/j.ejphar.2014.08.021
Navale, G., Sharma, P., Said, M., Ramkumar, S., Dharne, M., Thulasiram, H., et al. (2019). Enhancing epi-cedrol production in Escherichia coli by fusion expression of farnesyl pyrophosphate synthase and epi-cedrol synthase. Eng. Life Sci. 19, 606–616. doi: 10.1002/elsc.201900103
Patra, B., Pattanaik, S., Schluttenhofer, C., and Yuan, L. (2018). A network of jasmonate-responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 4, 1566–1581. doi: 10.1111/nph.14910
Pu, G., Ma, D., Wang, H., Ye, H., and Liu, B. (2013). Expression and localization of amorpha-4,11-diene synthase in Artemisia annua L. Plant Mol. Biol. Rep. 31, 32–37. doi: 10.1007/s11105-012-0472-0
Rufino, A., Ribeiro, M., Sousa, C., Judas, F., Salgueiro, L., Carlos Cavaleiro, C., et al. (2015). Evaluation of the anti-inflammatory, anti-catabolic and pro-anabolic effects of E-caryophyllene, myrcene and limonene in a cell model of osteoarthritis. Eur. J. Pharmacol. 750, 141–150. doi: 10.1016/j.ejphar.2015.01.018
Shen, Q., Lu, X., Yan, T., Fu, X., Lv, Z., Zhang, F., et al. (2016). The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 210, 1269–1281. doi: 10.1111/nph.13874
Shi, M., Zhou, W., Zhang, J., Huang, S., Wang, H., and Kai, G. (2016). Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins. Sci. Rep. 15:20919. doi: 10.1038/srep20919
Wang, G., Yu, D., Fan, J., Wang, C., and Xia, L. (2014). Expressing an (E)-β-farnesene synthase in the chloroplast of tobacco affects the preference of green peach aphid and its parasitoid. Plant Biol. J. Integr. 57, 770–782. doi: 10.1111/jipb.12319
Wang, J., Song, L., Gong, X., Xu, J., and Li, M. (2020). Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 21:1446. doi: 10.3390/ijms21041446
Wasternack, C., and Song, S. (2017). Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321. doi: 10.1093/jxb/erw443
Xiang, L., Jian, D., Zhang, F., Yang, C., Bai, G., Lan, X., et al. (2019). The cold-induced transcription factor bHLH112 promotes artemisinin biosynthesis indirectly via ERF1 in Artemisia annua. J. Exp. Bot. 70, 4835–4848. doi: 10.1093/jxb/erz220
Xiang, L., Zhu, S., Zhao, T., Zhang, M., Liu, W., Chen, M., et al. (2015). Enhancement of artemisinin content and relative expression of genes of artemisinin biosynthesis in Artemisia annua by exogenous MeJA treatment. Plant Growth Regul. 75, 435–441. doi: 10.1007/s10725-014-0004-z
Xu, J., Herwijnen, Z. O., Dräger, D., Sui, C., Haring, M., and Schuurink, R. (2018). SlMYC1 regulates type VI glandular trichome formation and terpene biosynthesis in tomato glandular cells. Plant Cell 30, 2988–3005. doi: 10.1105/tpc.18.00571
Yang, C., Fang, X., Wu, X., Mao, Y., Wang, L., and Chen, X. Y. (2012). Transcriptional regulation of plant secondary metabolism. J. Integr. Plant Biol. 54, 703–712. doi: 10.1111/j.1744-7909.2012.01161.x
Yin, J., Li, X., Zhan, Y., Li, Y., Qu, Z., Sun, L., et al. (2017). Cloning and expression of BpMYC4 and BpbHLH9 genes and the role of BpbHLH9 in triterpenoid synthesis in birch. BMC Plant Biol. 17:214. doi: 10.1186/s12870-017-1150-z
Yu, X., Pickett, J., Ma, Y., Bruce, T., Napier, J., Huw, D., et al. (2012). Metabolic engineering of plant-derived (E)-β-farnesene synthase genes for a novel type of aphid-resistant genetically modified crop plants. J. Integr. Plant Biol. 54, 282–299. doi: 10.1111/j.1744-7909.2012.01107.x
Yu, Z., Li, J., Yang, C., Hu, W., Wang, L., and Chen, X. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5, 353–365. doi: 10.1093/mp/ssr087
Zhang, C., Xing, B., Yang, D., Ren, M., Guo, H., Yang, S., et al. (2020). SmbHLH3 acts as a transcription repressor for both phenolic acids and tanshinone biosynthesis in Salvia miltiorrhiza hairy roots. Phytochemistry. 169:112183. doi: 10.1016/j.phytochem.2019.112183
Zhang, H., Hedhili, S., Montiel, G., Zhang, Y., Chatel, G., Pré, M., et al. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67, 61–71. doi: 10.1111/j.1365-313X.2011.04575.x
Zhang, X., Liu, J., Liu, Y., Wang, Y., Abozeid, A., Yu, Z., et al. (2018). Metabolomics Analysis reveals that ethylene and methyl jasmonate regulate different branch pathways to promote the accumulation of terpenoid indole alkaloids in Catharanthus roseus. J. Nat. Products 81, 335–342. doi: 10.1021/acs.jnatprod.7b00782
Keywords: Artemisia annua, AabHLH112, β-caryophyllene, epi-cedrol, β-farnesene, metabolic regulation
Citation: Xiang L, He P, Shu G, Yuan M, Wen M, Lan X, Liao Z and Tang Y (2022) AabHLH112, a bHLH transcription factor, positively regulates sesquiterpenes biosynthesis in Artemisia annua. Front. Plant Sci. 13:973591. doi: 10.3389/fpls.2022.973591
Received: 20 June 2022; Accepted: 05 August 2022;
Published: 02 September 2022.
Edited by:
Yongliang Liu, University of Kentucky, United StatesReviewed by:
Weimin Jiang, Hengyang Normal University, ChinaDongfeng Yang, Zhejiang Sci-Tech University, China
Copyright © 2022 Xiang, He, Shu, Yuan, Wen, Lan, Liao and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihua Liao, emhsaWFvQHN3dS5lZHUuY24=; Yueli Tang, dGFuZ3l1ZWxpXzk5OUAxNjMuY29t
 Lien Xiang
Lien Xiang Ping He3
Ping He3 Mingyuan Yuan
Mingyuan Yuan Xiaozhong Lan
Xiaozhong Lan Zhihua Liao
Zhihua Liao Yueli Tang
Yueli Tang