- College of Life Sciences, Linyi University, Linyi, China
Abiotic stress is an important factor affecting the normal growth and development of plants and crop yield. To reduce the impact of abiotic adversity on cotton growth and development, the material basis of cotton resistance and its physiological functions are analyzed at the molecular level. At the same time, the use of genetic engineering methods to recombine resistance genes has become a hot spot in cotton resistance research. This paper provides an overviews of the resistance mechanism of cotton against the threat of non-biological adversity, as well as the research progress of osmoregulation-related genes, protein-acting genes, and transcription regulatory factor genes in recent years, and outlines the explored gene resources in cotton resistance genetic engineering, with the aim to provide ideas and reference bases for future research on cotton resistance.
Introduction
Cotton is an important cash crop and occupies an important position in the industrial and agricultural economy of the country (Abdelraheem et al., 2019). Cotton fibers, seeds and stalks are used extensively in textile, food and feed processing, and papermaking. Currently, China is the world’s largest producer and consumer of raw cotton (Khan et al., 2020). However the shortage of water resources, ecological problems such as soil salinity, population expansion, environmental pollution and other human activities have made the phenomenon of land disputes over food and cotton are becoming more serious around the world (Solomon et al., 2007; Dos Reis et al., 2012; Ullah et al., 2017). The cultivation of cotton in china has paid more attention has been paid to improving yield and quality traits and developing disease and insect resistance (Cun et al., 2002; Davis et al., 2006). However, there is very little research on resistance and adaptability to abiotic adversity.
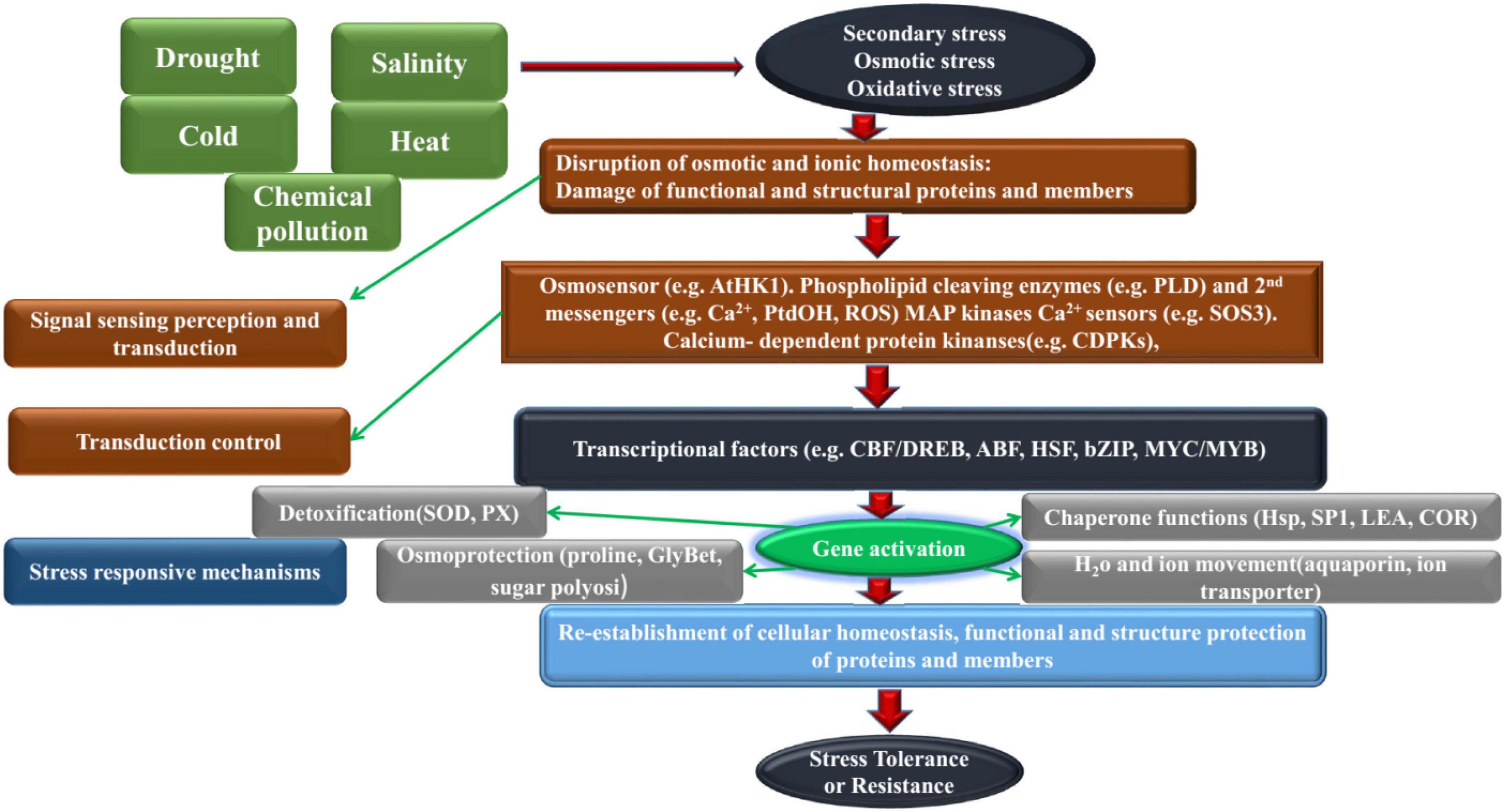
Abiotic stress mainly includes environmental factors that cause adverse effects such as extreme temperature, drought, salinity and alkali and low temperature usually cause a series of physiological and biochemical reactions in plants to adapt the adverse environment. These factors influence the development and growth of plants in addition to their morphological, physiological, biochemical and molecular processes (Liu et al., 2014; Zhang L. et al., 2014; Zhang et al., 2021). Plant responses to abiotic stress have been the subject of numerous studies worldwide. Initially, they were carried out on model plants and later on several important crops such as rice, corn, soybean, coffee and others (Ashraf, 2002; Gorham et al., 2010; Adams, 2011; Tiwari et al., 2013; Campbell et al., 2017). In order to adapt the environment and survive, plants have formed a series of resistance or tolerance to adverse environmental stresses during their long-term evolution resistance of plants. The resistance of plants is not just the result of a single mechanism. It is a very complex process (as shown in Figure 1). With the development of molecular biology, it has enabled people to understand the mechanism of plant resistance to stress at the molecular level of genetic composition, expression regulation, and signal transduction, especially in the molecular response of plants caused by abiotic stress. They will have severe impacts throughout the cotton growth and development stage, including a range of changes in plant morphology, physiology, quality and yield (Pei et al., 2020; Abro et al., 2022). Therefore, studying the adaptability of cotton to abiotic adversity and cultivating new varieties of resistant cotton are of great practical significance for cotton production.
The response pathway of cotton to abiotic stress can be roughly divided into four stages, ① the perception phase, the cotton senses the signal of abiotic adversity; ② the abiotic adversity signal is transmitted after the transmission stage; the reaction stage; the signal initiates a series of downstream molecular reactions ④ Expression stage; induces the expression of resistance-associated genes after going through the above four phases, cotton shows a specific response and resistance to the threat of non-biological adversity (Li et al., 2020). This paper summarizes, the research progress of cotton resistance response, development, utilization and molecular breeding of cotton resistant germplasm resources under abiotic stress, and proposed the development direction of cotton resistant breeding research in the future.
Effects of non-living stress on cotton growth and development
Non-biotic stress mainly includes drought, salinity-alkaline, polar temperature, soil nutrient deficiency and other unhealthy environmental conditions (Yao et al., 2011; Dongdong et al., 2016; He et al., 2016; Liang et al., 2016; Gao et al., 2018). The normal growth and development of cotton was inhibited by severe non-living stress, resulting in cotton plant yield reduction or even death.
Effects of abiotic stress on cotton growth and development cotton
Loss of crop production due to drought ranked first among all non-plant related pressures, and second among crop-related pressures. Drought can cause severe dehydration of plant cells; the normal cell membrane structure is destroyed, resulting in excessive closure of gas pores, which affects the absorption and uptake of CO2 and the decrease in photosynthesis in Figure 2. Chaves and Pereira (2003) and Onaga and Wydra (2016) investigated the influence of water stress on cotton seed germination, suggesting that cotton seeds under different drought stress were associated with decreases in water potential, germination rate, index, seedling height, root length, root-to stem ratio, dry weight and in associated with the fresh weight of young seedlings all decreased to varying degrees (Pettigrew, 2001; Fahad et al., 2017). Studying the growth and development of cotton in Jiangsu Province under drought conditions (Zhao et al., 2020), it was found that cotton growth was initially inhibited under drought conditions, and the plant height and leaf emergence rates were significantly slowed (Gao et al., 2020) found that when cotton was subjected to drought stress at different growth stages, its growth, development and yield were affected to some extent, and the influence from largest to smallest was the flower-boll, bud, maturity and seedling stage (Ullah et al., 2017; Ayele et al., 2020). The results show that drought resistant germplasm materials can effectively reduce water loss by increasing atmospheric pore resistance and reducing evapotranspiration intensity, thereby ensuring photochemical efficiency. Under drought conditions, root activity and proline accumulation of drought-resistant materials increased significantly (Liu et al., 2008).
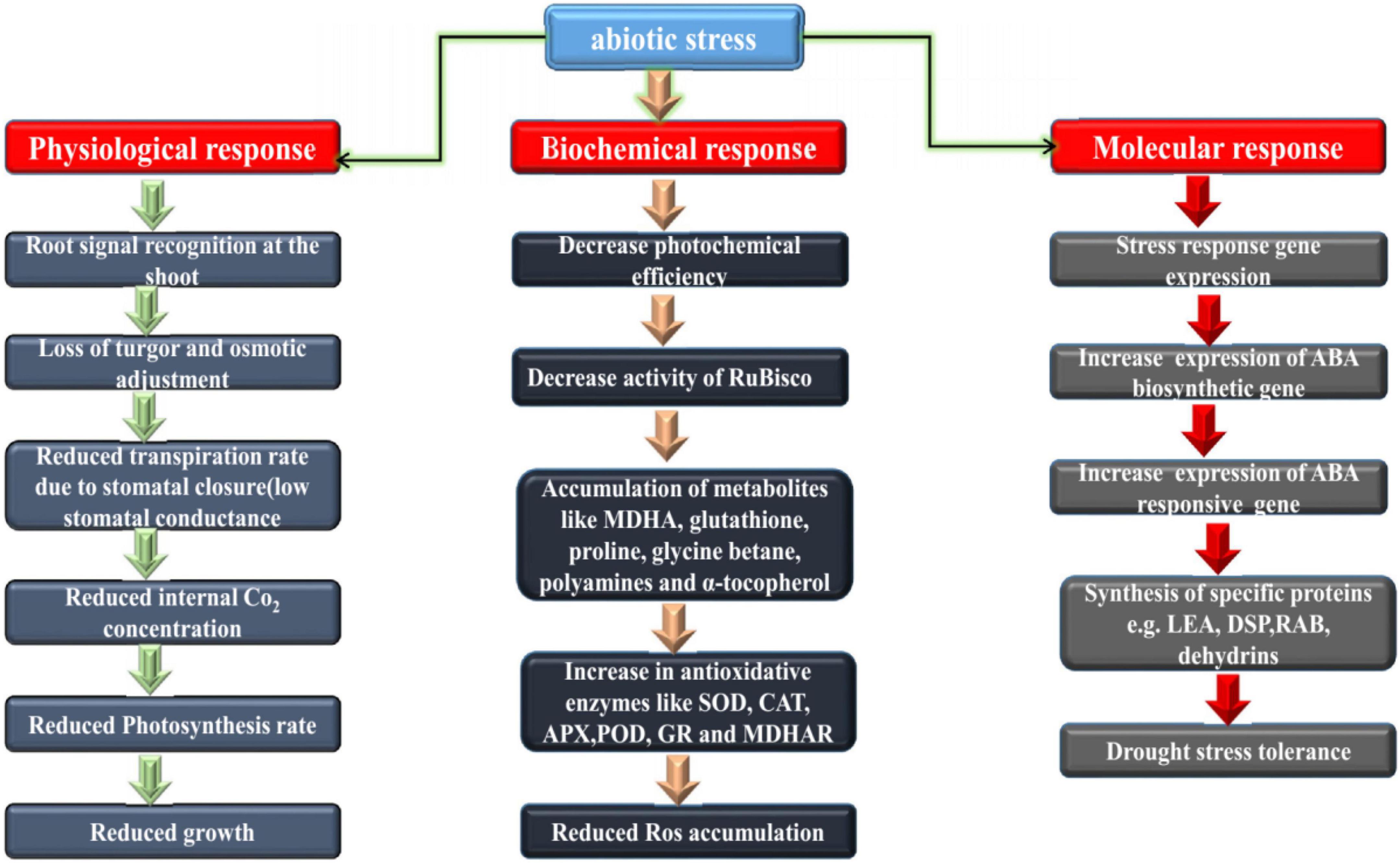
Figure 2. Physiological, biochemical and molecular basis of drought resistance in plants. Both major and minor changes that occur downstream of the transcriptional regulatory network.
Effects of salt and alkali on cotton growth and development
Salt damage is one of the most important impairments in agricultural production. The concentration of salt in an appropriate amount can be used as a nutrient component to promote the normal growth of cotton. However, high salt concentration leads to early leaf maturation or early decay, resulting in a reduction in cotton yield (Pettigrew, 2004; Ennahli and Earl, 2005; Davis et al., 2014; Zhang X. et al., 2014). Under normal conditions, salt stress occurs when soil salinity is between 0.20 and 0.25%. Ion toxicity caused by salt stress, osmotic stress and oxygenation stress cause cotton to become infected or even die. High soil salinity leads to water absorption, swelling and slow germination of seeds, low germination efficiency (Stroup et al., 2003). The lack of water in root system caused infiltration stress, which led to dehydration of the cells and loss of turgor pressure, which affected the physiological function of cotton. The higher salt concentration, the more severe osmotic stress (Zhang X. et al., 2014). At the same time, too many salt ions in the soil also create ion competition and inhibit the uptake of other ionites by plants. It was found that NaCL stress significantly reduced the levels of Ca2+ K+, Mg2+, P3, and Mn4+ in the leaves and roots of cotton seedlings, leading to nutritional imbalance of cotton (Du et al., 2017; Hu et al., 2017; Tränkner et al., 2018; Zhao et al., 2020). Plants have adapted to salt stress environments and evolved an adaptation mechanism. It is mainly divided into salt ion injury regulation and joint osmosis (Zhang L. et al., 2014; Iqbal et al., 2020a). The high content of K+/Na+ and Ca2+/Na+ in the fabric is an important indicator of the salt tolerance of cotton. The root system of salt-tolerant cultivar had some amount of Na+, and the K+/Na+ in the body was significantly higher than that of the less salt-tolerant cultivar (Marschner, 2012; Oosterhuis et al., 2014) reported that high salt concentration enriched, the leaves become more salt-sensitive varieties had more Cl– while the leaves of salt-tolerant varieties contained more K+, Ca2+, and K+/Na+. Armengaud et al. (2009) showed that selective potassium transport in the root could sustain the higher K+/Na+ content in the leaves, and this selective transport was an important feature of salt tolerance of cotton (Liu et al., 2008; Iqbal et al., 2020b; Zhao et al., 2020). Betaine is an important permeable modulating substance. When plants are subjected to environmental stress, betaine not only accumulates in cells to reduce osmotic potential, but also acts as a sort of preservative to maintain structure and maintaining integrity of macromolecules. At the same time, the membrane lipid peroxidation clearance system with high activity is very important for the stability of membrane structure and energy to maintain its normal physiological function (Fahad and Bano, 2012; Fahad et al., 2013), the activity of membrane lipid peroxidation removal system greatly increased and glutathione reducing enzyme activity by 55–101% compared to the photography increased 38–72%, and monosintin α content increased by 212∼320%, but the above indices did not change or decreased in salt-sensitive varieties.
Influence of temperature change on cotton growth and development
The change in pole temperature caused growth, breeding and injury of cotton. Polar temperature changes include low and high temperature stress, and low temperature stress is divided into cold damage and frost damage, which slow or stop plant activity (Nievola et al., 2017).
After the cotton suffered low temperature injury, and withered the whole plant or part of the dead. The damage degree of cotton was closely related to the impact of low temperature on cotton at the seedling stage (Zakaria, 2017), when the degree of frostbite was more than 15%, the growing period of cotton was delayed, and the yield and quality of cotton were affected (Fahad et al., 2014a,b). Under low temperature strips, the germination rate of cotton seeds slowed to the point of rotting. Even when the seeds were germinated, the growth rate of bud length and weight was significantly reduced, so the degree of firmness of cotton seedlings was significantly affected. Low temperature and low light stress leads to a decrease in photochemical activity inhibition of photoelectron transfer, and disruption of excitation energy distribution and transfer between PSI and PSII (Abdurakhmonov et al., 2010; Rantala and Tikkanen, 2018). The increase in nitrate reductase activity is an indication of the cold resistance of cotton. Under low temperature treatment, nitrate reductase activity of low temperature tolerant cultivars increased, and plant mass contained increased nitrogen content to withstand unfavorable low temperature environment (Iqbal et al., 2020b).
Cotton is native to low latitudes in the tropics, but due to the long period of human selection and domestication, its ability to withstand high temperature is not strong. High temperature stress can affect cotton seed germination and inhibit root line growth and plant growth. The optimal temperature ranges for germination and growth of cotton seeds were 28°–30°C, and the optimal temperature ranges for root growth were 30°–35°C and 22°–27°C (ii). High temperature stress (40°C during the day and 32° at night) reduced the rate of seed germination (Singh et al., 2007), impaired the uptake of water and nutrients in the root system, disrupted the formation and transport of pathogens and impaired the spread of the root system, as a result of which the root system led to a flatland formation. Tap and lateral roots are thin and poorly developed even under optimal water nutrient conditions (Ziska, 2000; Reddy et al., 2011). High temperature accelerate plant growth, shortens the boll growth period and carbon assimilation time, thereby reducing dry material accumulation, resulting in smaller bolls, lower yield, and worse cotton fiber quality (Thorp et al., 2020). Studies show that the relatively high temperature at night also affects the quantity and quality of buds and seed pods. In the reproductive growth phase, pollen germination is extremely sensitive to high temperature, and the optimum temperature is 28°C, while the pollen tube growth rate is linearly correlated with temperature below 37°C. At high temperatures, pollen viability, pollen germination rate and number of pollen grains at the column head decreased significantly, leading in pollen abortion (YunfeiJiang et al., 2017). The lowest, highest and optimal flower bud and tube growth temperature were determined by examining the flower bud and tube growth temperature of different cotton varieties (Kakani et al., 2005; Liu et al., 2006), at the same time, under high temperature stress, all organisms produce heat shock response (heat shock protein) to protect cells and organisms from serious damage (Craita, 2022). To regain normal cell viability and physiological viability. Therefore, the heat resistance of plants could be improved by increasing the surface level of heat stimulated protein (heat shock protein, HSPS).
Effects of soil nutrient stress on cotton growth and development
Nitrogen, phosphorus and potassium are three essential elements for plant growth, which play irreplaceable roles in plant life and have a major impact on cotton yield and quality. Under potassium deficiency has a major impact on yield and fiber quality (Yibati et al., 2022), potassium deficiency significantly inhibited the elongation and lateral rooting of the cotton root system. Potassium deficiency can inhibit the elongation and thickening of the main cotton stem, affecting the vegetative growth of cotton. It also inhibited the production of albumin content, chlorophyllin content and effective rate of photosynthesis in functional leaves of cotton main stem (Fontana et al., 2020). Cotton lack of potassium for a long time, they are prone to red petiole rot, so nutrition and reproductive growth will blocked, the growth potential of cotton plant will be weakened, early decline and other hair diseases (Zhang L. et al., 2014). Cotton cultivars with high potassium efficiency had a strong ability to transfer potassium to shoot, thereby preserving the physiological energy of chloroplasts in cotton leaves. With the decrease of potassium concentration, the number of root hairs in potassium efficient varieties increased, and site formation was closer to the root tip (Shahzad et al., 2019; Bamagoos et al., 2021). Xylem conduit differentiation has improved and tissue transport capability has been enhanced. With the increase in root-shoot ratio and length, the indexes of root absorption energy and root viability increased significantly, while the content of malondialdehyde (MDA) was lower than that of the low-effect varieties. Therefore, the stability of membrane system of the high-effect varieties was better and the resistance of the high-effect varieties was stronger under low potassium stress (Yang et al., 2021).
There are many adaptive changes in plants under low phosphorus stress, including root morphology, root system secretion, membrane and internal phosphorus transport system, etc. Phosphorus deficiency has the greatest impact on cotton growth and yield, resulting in delayed plant growth and development smaller leaves, thinner, weaker brittle stems, shorter than normal plants, lower root growth, delayed boll setting and maturation little boll, low yield, poor fiber quality (Hodge, 2004; Grossman and Rice, 2012). Under low phosphorus stress, main root growth was limited, but lateral root density and length were increased, and root hair the number and length were also increased (Linkohr et al., 2002). The plasma membrane is the decisive factor for phosphorus uptake efficiency in plants, and the plasma membrane of root cells first link to main transport phosphorus inverse concentration. The migration of mechanized phosphorus across the cell membrane into the cell is regulated by the same or different families of phosphorus transporting albumen, which can be subdivided into Pht1, Pht2, and Pht3, families according to sequential similarity. They are located on the membrane of the root system, the inner membrane of the plastids and the membrane of the mitochondria (Ma et al., 2001; He C. et al., 2005; He Z. et al., 2005). They are responsible for the uptake of soluble phosphorus from the soil solution, the transfer of inorganic phosphorus from cytoplasm to plastid, and the exchange of linear phosphorus (Hodge, 2004).
The amount of nitrogen fertilizer applied to our country’s agricultural fields is increasing year by year, but the loss of nitrogen fertilizer is becoming more and more serious. Nitrogen uptake efficiency and nitrogen use efficiency (NUE) isthe main cause of difference in nitrogen efficiency (Fahad et al., 2013). Under the low nitrogen conditions, the difference in nitrogen efficiency was mainly due to the difference in nitrogen utilization rate, while under the high nitrogen conditions, the nitrogen absorption efficiency was the main determinant. Under nitrogen stress, cotton root lines could absorb nitrogen in lower concentration by increasing the percentage of active absorption area, to increase growth and development of root lines and plants. The area of active uptake and harvesting area as well as the specific surface area were the main reasons for difference between varieties with different nitrogen efficiencies. When nitrogen is applied, HCO–3 or OH– was released into the rhizosphere and pH in the rhizosphere increased. In the absence of nitrogen, the morphological and physiological changes of cotton root could increase the utilization rate of nutrients by secreting protons, organic acids or increasing the activity of some enzymes (Fahad et al., 2013). Nitrogen efficient varieties adapted to lower nitrogen concentration and increased nutrient availability by increasing root shoot ratio, root density and root length. The results showed that the distribution ratio of nitrogen content in each organ of cotton from large to small leaf, stem and root, indicating that the cotton plant transported more nitrogen to the upper part of the soil, promoted photosynthesis (Niu et al., 2021).
Research progress on cotton stress resistance breeding
In recent years, the shortage of global water resources, soil salinity, frequent extreme weather conditions and so on, the demand for cotton side-effect resistance is getting higher and higher. Cotton breeding not only focuses on improving yield, quality and resistance to diseases and insects, but also pays increased attention to resistance to abiotic stress and efficient use of soil nutrients (Diouf et al., 2017; Abdelraheem et al., 2021). Explore the optimal resistance germplasm resources of cotton, separate and calibrate the target characteristic genes/QTL of resistance/resistance to different adversity, clone and functionally clarify the candidate genes related to resistance (Wang et al., 2003), carry out research on the mechanism of interaction between cotton and different adversity, molecular design and creation of new materials for resistant cotton, and lay the foundation for resistant breeding of cotton in Figure 1.
Exploration of cotton resistant and superior heterologous materials
Research on cotton resistance started late, however has progressed rapidly in recent years. At home and abroad, the collection, assessment and verification of cotton resistant germplasm resources on the non-biotic resistance of cotton (Nazeer et al., 2014; Yin et al., 2020) such as salt resistance, drought resistance, polar temperature tolerance, and soil trophic stress, and a number of important resistant germplasm resources were discovered. It is an important raw material for the resistance and breeding of cotton.
Exploration of drought-resistant and salt-resistant cotton resources
Cotton is recognized a salt and drought-resistant crop, and is the “pioneer crop” for desalination beach soil. It is an important source for increasing cotton acreage in the future to alleviate the conflict between grain and cotton and to utilize the reclaimed land. Selecting and breeding to salt-tolerant and drought-resistant cotton varieties to improve the adaptability of cotton to a salt-tolerant environment is one of the important ways to reduce the damage of salt-tolerant soils and to develop and utilize salt-tolerant land resources such as coastal beach (Sun et al., 2021; Zhang et al., 2022). The selection of drought-resistant and salt-tolerant raw materials is the basis for achieving this goal (Li et al., 2019). Seedling stage drought method to simulate drought environment to identify the drought resistance of 76 seedlings of cotton cultivars. 2 drought tolerant varieties (1193B and 18 short-season) and 5 sensitive varieties were selected (9811-36, 9810-3, 21025-2, China Cotton Institute 45 and 21021-8 (Hou et al., 2018). The above-mentioned varieties were further investigated for salt resistance, and it was found that low-NaCL concentration promotes seed germination while high concentration inhibits seed germination by evaluating salt resistance varieties and salt-sensitive varieties (Sun et al., 2018). The ship also screened three salt-resistant strains such as Salt 1032, 1129, and 2018. At the same time, it was found that the salt resistance of the new cotton variety Zhongmian Institute 76 significantly exceeded that of the comparison.
Exploration of cotton resources resistant to extreme temperature
Conductivity is an important indicator of cell membrane stability and fluidity and identifies the cold resistance of seedlings. The results showed that under low temperature stress, young cotton seedling electrical conductivity enzyme activity of peroxidants and soluble sugars increased, and root activity decreased, while young cotton seedling electrical conductivity and root activity decreased less in low temperature resistant varieties (Anastasia et al., 2010; Lin et al., 2021). The content of active and soluble sugars increased significantly. Thermal cell stability, proline, soluble sugar and photosynthetic rate of functional leaves of cotton main stem can be used as an important indicator for screening of cotton resources with high temperature tolerance (Hasanuzzaman et al., 2013). Currently, there is not much research on choosing high temperature resistant material for cotton pole. The results showed that anther development and pollen fertility of different cotton materials differed greatly in response to high temperature stress. The anther development and pollen fertility of high temperature resistant cotton materials were all normal, while the anther of sensitive cotton lines were dry, unruptured, and showed no male fertility (Bita and Gerats, 2013; Zahid et al., 2016). A group of raw materials with different tolerance to high temperature stress as sorted out. Using the above materials, the study on changes in physiological indicators of various high temperature tolerance cotton lines, such as SOD, POD, CAT, and MDA, was carried out. It laid the foundation for advancing separator research and breeding new cotton varieties in an environment of non-stop high temperature.
Exploitation of cotton resources with high effective nutrients
There were significant differences in nutrient uptake and availability between different cotton lines. Selecting the screening index of the combination and digging up nutrition and beneficial strains can improve the absorption and utilization efficiency of nutrients, improve the utilization rate of seed resources, reduce resources, protect the environment, and achieve sustainable agricultural development. Zhang et al. (2018) showed that the optimal seedling age for screening low potassium tolerance genotypes in cotton was 5 leaves. Echer et al. (2020) screened cotton varieties with high potassium efficiency genotypes under sand culture condition, and proposed that the potassium utilization index, potassium uptake and biomass were used as screening criteria for high potassium nutrition at the cotton seedling stage (Chattha et al., 2022). Five potassium efficient cultivars (602, Xinluzao 6, Jiaomian, 18-3 and Xinhai 13) and three potassium inefficient cultivars (Shi K7, Xinluzao 10, and Xinhai L4) were selected by the culture of the broth and two levels of potassium deficiency and appropriate potassium treatment, 86 cotton cultivars of different pedigree were screened for the potassium efficient basal type, and 4 varieties with high potassium efficiency and 2 varieties with low potassium efficiency were obtained. It is suggested that cotton lint yield and the potential to increase yield during potassium application are the ultimate indexes to judge whether cotton has high potassium efficiency. Through a hydroponics experiment (Zhang et al., 2021), two of high and low phosphorus treatments were established and 32 cotton varieties with high phosphorus genotypes were screened at seedling and bud stages from self-breeding or introduction in Xinjiang. Three phosphorus efficient and inefficient varieties were obtained. The results showed that the coefficient of variation of relative dry matter weight, relative phosphorus uptake and relative total plant phosphorus content were higher than those of other traits, that could be used as screening indices for phosphorus use efficiency at seedling and bud stages of cotton (Li et al., 2019), studied cotton varieties resistant to withered and verticillium wilt by vermiculite and irrigation with nutrient solution irrigation, and obtained two categories of varieties resistant to low phosphorus and sensitive to low phosphorus. The results showed that there were significant differences in cotton seedling dry matter, aerial fresh weight, total leaf area, chlorophyllin content, and dry matter weight the aboveground and phosphorus utilization rates between different phosphorus concentration and different varieties. It can be used as an index of assessment of low phosphorus tolerance of cotton at the seedling stage.
By determining the height, dry weight and total nitrogen content of cotton plants (Song et al., 2017), the seedling stages screening of nitrogen use efficiency of 48 cotton varieties self-bred or introduced in Xinjiang was completed to by 4 nitrogen-efficient varieties (602, Xinlu Early l2, Xinhai L2, and Xinlu Early 8) and 5 nitrogen-inefficient strains (Xinhai #. 14, Xinhai #. 2 and #. 1 Battery 1, Xinlu Early #. 2O and #. 18-3). Relative dry weight has a high degree of sensitivity to low nitrogen resistance of different cotton species, and correlates well with yield, which can be used as the primary index for screening high-efficiency cotton nitrogen. Since the relative nitrogen content, relative nitrogen uptake and relative dry weight showed a very significant negative correlation and a very significant positive correlation, it is recommended as an auxiliary index for screening of nitrogen-efficient varieties.
Research advances on molecular markers of important traits of cotton stress resistance
Molecular markers of important characteristic genes of cotton mainly concentrated on the screening of molecular markers such as yield and its constitutive factors, fiber quality traits, resistance to rot and wilt. There is less research on abiotic stress resistance, mainly focusing on QTL screening of drought resistance and salinity resistance traits.
Screening of molecular markers related to drought resistance of cotton
The high and low permeability of leaf pores is an indication of drought resistance crops. Mauricio et al. (2000) screened the high pore conductivity resource materials by disproportionation selection, and then configured the NM24016/TM-1 separation group to screen two QTL for blade pore conductivity (Wu et al., 2009; Li et al., 2021) (Zhongmian Institute #. 35 × Junmian #. 1) F2 to isolate the population. QTL screening for drought resistance was performed by observing the water loss capacity of leaves of cotton after 6 h of natural water loss. A QTL site was discovered on chromosome #16 that controls leaf water loss. The additive effect can explain the phenotypic variation of 12.2% [Lu Mian 97−8 × (12)] F2 isolated groups, and obtained data on the impact of constraints on cotton seed yield and plant height in each reproductive period by detecting the restraint index and coefficient, and then screened 7 plant heights in term of site drought resistance, with a contribution rate of 4.5%, 19–19.66%, including 4 plant heights QTL in bud phase, 1 plant height in flowering stage, and 2 plant heights in growth phase. Plant heights of different periods were distributed to different linkage groups, indicating that the same traits are controlled by different loci genes, and plant heights of different reproductive stages are controlled by different genes (Muhammad et al., 2011; Guo et al., 2021), used (FH one 901 × RH one 510) F to separate the group, and detected 7 drought-resistant related QTL. Three QTL were detected under drought conditions, one each for permeability potential, permeability adaptation and plant height, and2 QTL were detected after rehydration. The production of these QTL has laid the ground work for further use of drought-resistant molecular marker-assisted selection techniques and for creation of new drought-resistant strains.
Screening of molecular markers related to salt tolerance in cotton
Zhao et al. (2016) isolated the population with (Lu Yan 97−8 × Su 12) F2: and detected 4 QTL loci related to salt resistance, contributing 5.47–17.20%, 1 at middle seedling stage and 3 in bud stage (Xu et al., 2021) used two salt-tolerant materials (Zhongo 7 and Zhongmiansuo 35) and two salt-sensitive materials (Xinyan 96-48 and Zhongmiansuo 12) to screen the polymorphism of SSR molecules. These primers are expected to be candidate primers for molecular identification of salt tolerance of cotton (Yao et al., 2011; Zafar et al., 2022) analyzed the salt tolerance inheritance and gene effect of cotton salt tolerance mutant Shannong 011 and its hybrid offspring, and found that the salt tolerance of the mutant was enhanced by controlled nuclear genes, and their inheritance was dominated by additive action. The above research foundation for further research on salt-tolerant and alkali-tolerant cotton by submarker screening and auxiliary selection, and breeding of salt-tolerant varieties.
Study on the discovery and transformation of key factors of cotton resistance
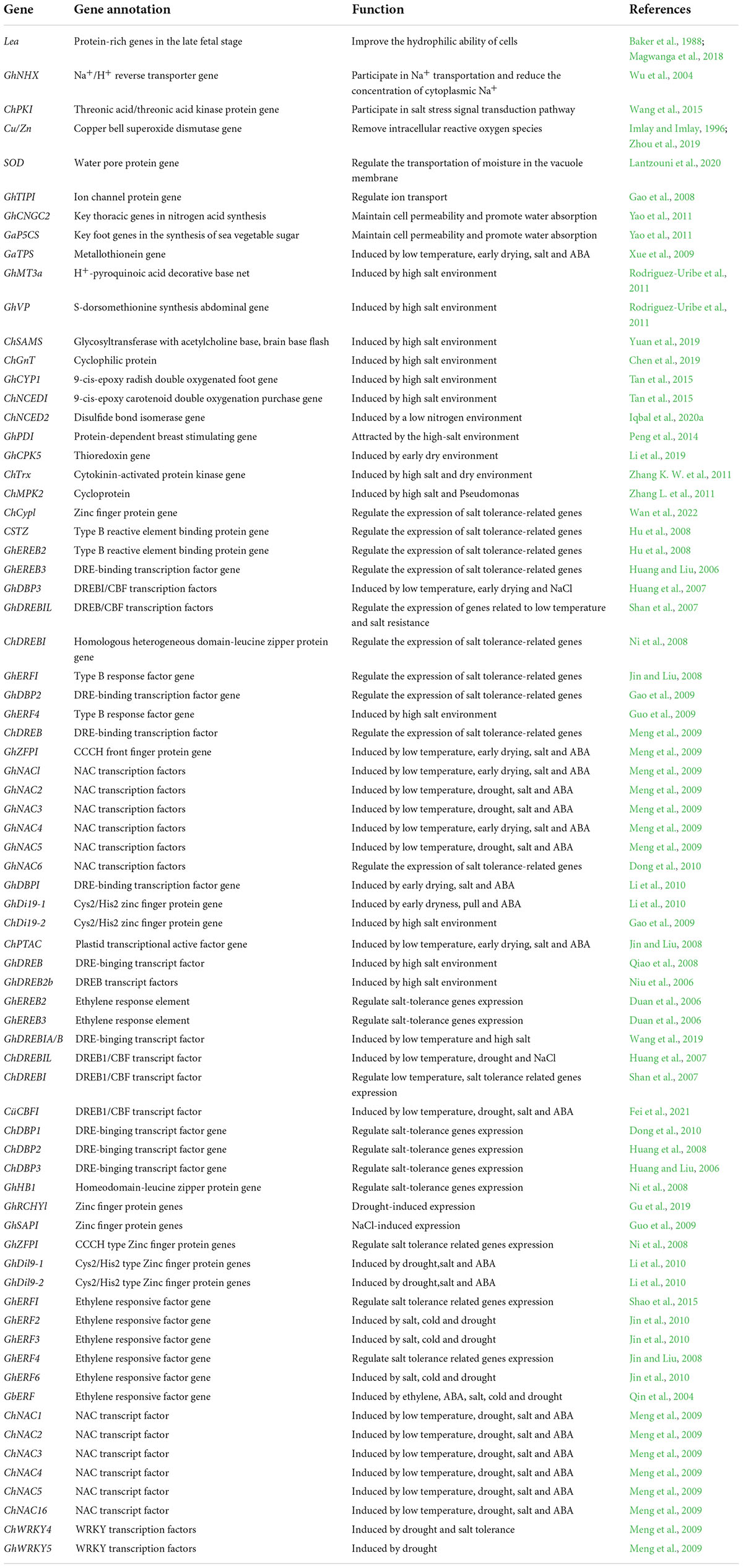
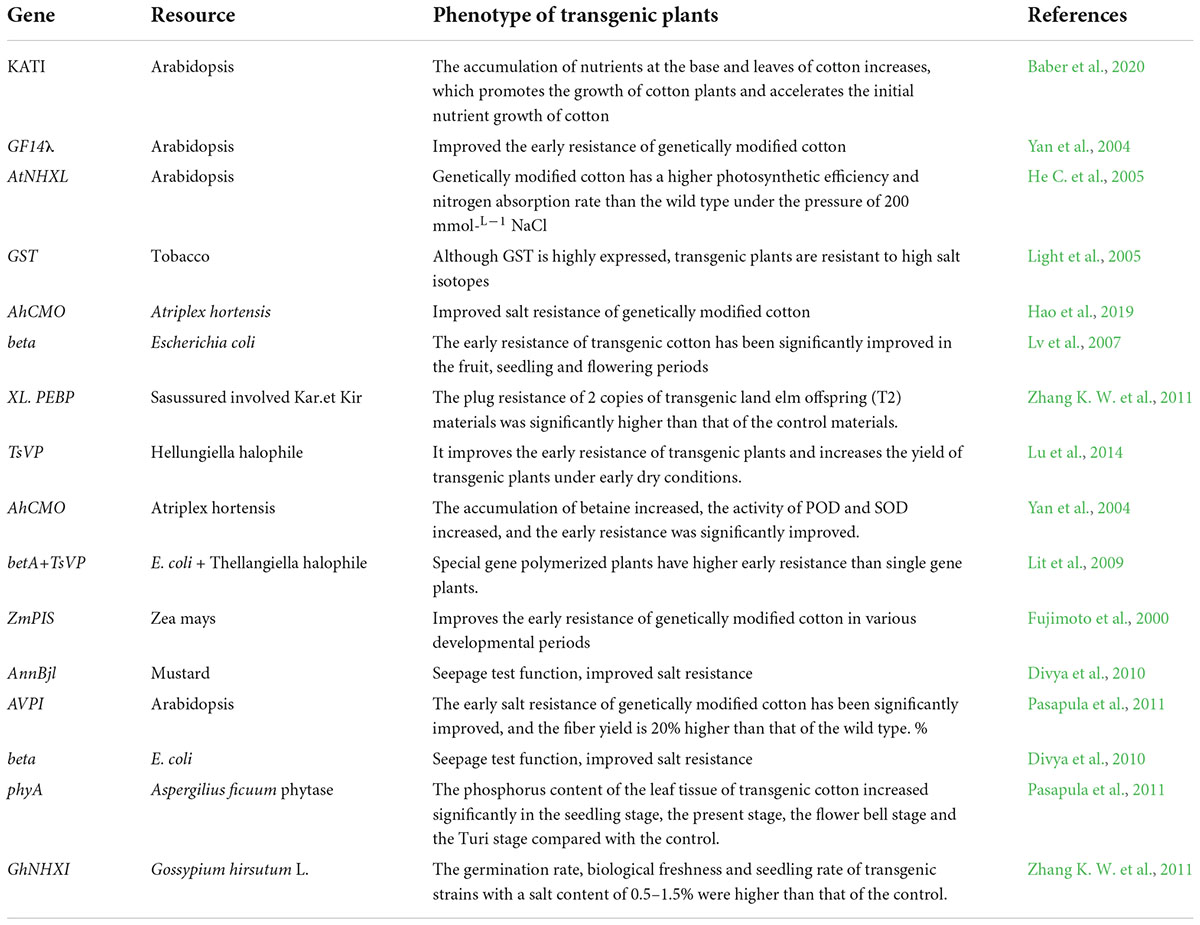
The gene’s stress response to stress is mainly manifested in the control of its own genetic material and epigenetic regulation under the influence of DNA methylation, RNA disruption or histone modification. There have been many studies on the inverse correlation of stress resistance non- living cotton and the analysis of its energy, which can be divided into two types: one is the functional factor which can enhance the ability of plants to adapt to adverse environments after induced expression by stress, such as GaP5CS, GaTPS, and Lea. The other is the transcription factor that regulates the expression of the underlying factor and the signal transduction under stress (Duan et al., 2006; Guo et al., 2010), such as GhERBL, GhERF1, GbCBF1. To date, more than 40 cotton resistance correlation factors have been isolated and identified (Table 1), and their functional characteristics have been preliminarily defined. Based on cloned endogenous or exogenous resistance-related genes, a batch of new cotton materials related to drought resistance and salt resistance have been successfully obtained using transgenic technology (Table 2) introduced Luomian #. 6 of B. buma DNA into two new salt-resistant cotton lines 91-11 and 91-15 with salt content of 0. The lint yield was 19.1% higher than recipient lumian 6 planted in 51% coastal saline area 7 and 237.8%. Combined the choline dehydrogenase gene betA with the mutant acetyl lactate synthesizing enzyme. Genes were introduced into three excellent varieties of cotton, and genetically modified plants and their offspring with significantly improved salt resistance were obtained, creating excellent materials for salt-resistant cotton is transmitted to cotton plants by the method of flowering powder tube guidance. The introduction of this gene increases the potassium accumulation of the stems and leaves of cotton, promotes the growth of cotton plants, and accelerates the nutrient growth rate of cotton in the early stage. However, the transgenic material is also manifested in the delayed reproductive period, and the position of stem node of the pregnant flower bud rises. The phenomenon needs to be further improved and studied drought tolerance measurement of the seedling stage, bud stage and flowering stage show that the introduction of exogenous ZmPIS genes improves the drought resistance of (GM) genetically modified cotton at different development periods, and after long-term drought stress during the flowering period, the yield of single seed cotton of GM strains is significantly higher than that of wild-type controls. Compared to the large-scale production and use of insect-resistant genetically modified cotton, the genetic engineering of cotton against abiotic stress has made some progress, however it is still in the experimental and research stage, and there is still some distance to commercial application. Compared with the production requirements, research progress needs to be further accelerated.
Cotton adversity stress transcription factor gene
Transcription factor (TF), also known as trans-acting factors, are a type of protein molecules that can specifically bind to cis-acting elements in the promoter region of eukaryotic genes to ensure that the target gene is expressed at a specific intensity at a specific time and place (Wu et al., 2021). Under the stress condition, the transcription factor associated with stress tolerance can regulate the expression of anti-retroviral motifs and transmit the stress signal, and the signal can be involved in the information transmission can be cascaded so that, the transcription factor can be used to improve the resistance of plants (Wang et al., 2020). In order achieve a more rational effect, cotton from land (Gossypium Hirsutum L.) is currently used. Among them, there are about 30 factors of reverse conversion of cotton resistance the energy properties of which have been preliminarily defined (Table 1).
Ethylene responsive element binding protein transfer factor
Ethylene responsive element binding protein (EREBP) belongs to the AP2/ERBP family of transcription factors and contains the AP2/ERBP domain (Riechmann and Meyerowitz, 1998). The cis-operating elements derived from EREBP transcription factors are different, which can be divided into two large subfamilies, DREB subfamily and the ERF subfamily member: The DREB subfamily can contain dehydration responsive elements/C repeat (DRE/CRT) identify (Saud et al., 2022) ERF sub-family member can identify GCC-box. Therefore, they play a very important role in the process of plant resistance to abiotic stress, and they are also a key regulator of abiotic stress resistance, the more is currently being introduced into adaptation to adversity (Gao et al., 2009). GhDREB -gene from cotton into wheat, and functional analysis showed the following: under drought, high salinity and freezing stress, transgenic plants accumulated a large amount of soluble sugar and the content of chlorophyllin increased significantly compared with the control, but the plant phenotype did not change much (Polge and Polge, 2007; Qiao et al., 2008) isolated GhRF1 gene from cotton and analyzed its function. GhRF1 gene transcription was found to rapidly accumulate under the stress of ethylene, ABA, high salinity, frost damage, and drought. This gene turned out play an important role in cotton stress. In this laboratory, the genes of anti-retroviral transcription factors PeDREB and HhERF from the psandophyte plant, A. chinensis and populus euphrica were cloned and the efficient plant expression vector was constructed (Jeong et al., 2010; Yadav et al., 2016). The pollen tube channel method was used to guide human cotton, and the transgenic lines were obtained. After drought and salt stress treatment, it was found that the drought tolerance of transgenic lines was increased, and the comprehensive analysis of various physiological indexes showed that the salt tolerance of transgenic lines was also increased.
NAC transfer factor
NAC transcription factors are a special type of transcription factors found in plants. Their common feature is the approximately 150 amino acid long NAC domain with a high degree of protection at the N-terminus, while the C-terminus is a highly variable transcriptional regulatory region. The results indicated that NAC transcription factor played an important role in plant resistance to drought and high salt stress through direct or inducible transcription factor and the expression of drought and high salt stress. It was found that rice tolerance to drought, high salinity and low temperature was increased when rice was overgrown in the vegetative growth stage. Xue et al. (2011) showed higher tolerance and yield in triticale under polyethylene glycol induction. Six NAC transcription factors have been isolated from upland cotton for the first time. They were expressed in roots, stems, leaves and fibers under drought, high salinity, cold and ABA stress, and the highest expression was found in leaves (Meng et al., 2009; Wang, 2010). However, due to the long transformation cycle of cotton, most cotton transcription factor genes are only in the stage of functional identification of gene clones in vitro and in model organisms.
Research and prospects
Problems in the research of cotton resistance breeding
Although the cultivars with abiotic stress resistance of cotton were selected, their development was slow compared to the research results of high yield, good quality and diseases and insect resistance. There is still a long way to go to meet the commercial need for stress resistant varieties mainly the following aspects of the problem.
(1) Lack of systematic identification and screening of cotton superior resistance to reverse germplasm resources. For a long time, there have been studies on yield, quality and disease and insect resistance in cotton breeding, but less research on non-biotic stress. Although the researchers have performed relevant work on the screening criteria of cotton resistance and identification of germplasm resources with resistance to resistance, the materials come from different sources of cotton, such as local varieties, extended varieties, introduced materials, and distant hybrid materials. Identification criteria and results from different antagonistic/sensory laboratory resources need to be integrated.
(2) Under tagging of deficiency and resistance elements is closely related, which limits the process of culturing new material aided by under tagging. Molecular markers were used to aid in the selection and polymerization of cotton. Research has mainly focused on improving of fiber quality and disease resistance of cotton. So far, i only preliminary studies of QTL mapping related to drought and salt tolerance have been performed in abiotic stresses (Darmanov et al., 2022), but there are still few sub-markers that are closely linked to the target and can be used directly as molecular markers to assist selection.
(3) The lack of an important value of anti-inverse factor. Although Kronn has a large number of inverse correlation factors of cotton resistance, they are mainly some stress response factors and transfer factors, and few of them have any real value. In meantime, the resistance mechanism also needs to be further elucidated. In addition, in the current cloning of cotton resistance to inverse factor, many in the drought resistance, salt resistance, and cold resistance, high temperature resistance, cotton nutrition efficient use of the target factor is not much.
(4) The theory of seed breeding and the method of sub-design must be thoroughly studied. In the past 10 years, the technology of seed breeding and seed breeding has been highly appreciated, and through the technology of gene transformation a batch of cotton materials with a wide cultivars has been created However, there is a need to do more research on the theory and method of organic combination of seed splitting breeding technology and conventional breeding technology, and the fruit of biomass technology should be used in cotton peanut production.
Prospects of the study on cotton seed resistance
Compared with the progress of research on resistance of rice, corn, wheat and other food crops, research on resistance of cotton is relatively stagnant, and there is still a certain gap in resistance breeding. In order to better use the research results of molecular biology in the practice of cotton resistance breeding as soon as possible, and speed up the process of cotton resistance breeding, future research, will be conducted based on the exploration and evaluation of excellent resources, the main effect of resistance QTL and the cloning of excellent key resistance genes, using MAS technology (marker assisted selection) and transgenic technology to polymerize resistance genes to achieve the analysis of resistance genes. Sub-design and breeding, to breed high-yielding, -quality, multi-resistant, and widely suitable new varieties of cotton (series). It is recommended to focus on the following aspects of work.
(1) The identification and evaluation of resistant gene-type varieties of cotton are carried out systematically. Through extensive collection of cotton resource materials from different sources, resistance biology identification of germplasm resources and analysis of genetic mechanism of target traits are carried out. Complete and clarify the resistance evaluation indicators under the threat of different adversities, combine the evaluation indicators with field trails, systematically and comprehensively identify and assess resistance gene varieties for cotton, and provide excellent resistance germplasm resources and gene sources for crop breeding practices as soon as possible.
(2) A batch of molecular markers of important resistance traits from cotton QTL were screened and used to aid in the selection of target traits. It is difficult to identify the resilience of different materials on large scale using morphological and physiological indicators. Molecular marker technology has been widely used in various studies of cotton to discover molecular markers closely linked to the characteristic target genes of resistance, and use MAS technology to conduct large-scale screening of the characteristic target genes to find marker resources to provide the design and future breeding of related resistance characteristic molecules.
(3) Explore anti-reverse heterogeneity resources and create new anti-reverse materials. Using omics and other techniques and biological information methods, cotton varieties that contain excellent resistance genes are being studied, and resistance-related genes are discovered and cloned. Functional verification of key genes for resistance is obtained, and their mechanism of action is clarified. Make full use of the research into other crop resistances, direct endogenous and exogenous resistance-genes to human cotton receptors, and create new transgenic materials containing excellent resistance genes to provide excellent genetic resources and target materials for resistance breeding.
(4) Strengthening the combination of basic research and seed breeding and strengthening multidisciplinary cooperation. Although a number of resistance related genes and resistance related transcription factors have been discovered, the endogenous or exogenous resistance related genes have been successfully transferred into cotton using genetic engineering, and the transformation materials with a significantly improved phase table have been obtained. However this work is limited to laboratory research and has not yet been applied to breeding practice. So far, the new varieties with improved non biotic resistance have not been used in production. Each step of cotton resistance breeding involves many scientific researches, such as developing new cotton varieties with high yield, quality, multi-resistance as well as high efficiency and utilization of nutrients through comprehensive assessment.
Author contributions
SS and LW: conceptualization, writing original draft, and writing review and editing. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Program of Shandong Provincial Natural Science Foundation of China (Grant No. ZR2021QC199), the State Key Laboratory of Cotton Biology Open Fund (Grant No. CB2018A20), the Linyi University Ph.D. Fund for Scientific Startup (Grant No. LYDX2018BS029), and College Students’ Innovative Training Program (Grant No. 2018104521).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelraheem, A., Esmaeili, N., O’Connell, M., and Zhang, J. (2019). Progress and perspective on drought and salt stress tolerance in cotton. Industr. Crops Product. 130, 118–129. doi: 10.1016/j.indcrop.2018.12.070
Abdelraheem, A., Thyssen, G. N., Fang, D. D., Jenkins, J. N., McCarty, J. C., Wedegaertner, T., et al. (2021). GWAS reveals consistent QTL for drought and salt tolerance in a MAGIC population of 550 lines derived from intermating of 11 upland cotton (Gossypium hirsutum) parents. Mol. Genet. Genom. 296, 119–129. doi: 10.1007/s00438-020-01733-2
Abdurakhmonov, L. Y., Abdukarimov, A., Rizaeva, S. M., Abdullaev, A., Barroso, P. A. V., Pádua, J. G., et al. (2010). Status of the global cotton germplasm resources. Crop. Sci. 50, 1161–1179. doi: 10.2135/cropsci2009.09.0551
Abro, S., Rizwan, M., Deho, Z. A., Abro, S. A., and Sial, M. A. (2022). Identification of heat tolerant cotton lines showing genetic variation in cell membrane thermostability, stomata, and trichome size and its effect on yield and fiber quality traits. Front. Plant Sci. 12:804315. doi: 10.3389/fpls.2021.804315
Adams, N. (2011). A Linkage Map and Drought Tolerance Analysis in a Backcross Inbred Line Population of Cotton. Ph. D, Thesis. New Mexico State University.
Anastasia, K., Kappas, I., Tsaftaris, A., Galanopoulou, D., and Farmaki, T. (2010). Cold acclimation and low temperature resistance in cotton: Gossypium hirsutum phospholipase Dα isoforms are differentially regulated by temperature and light. J. Exp. Bot. 61, 2991–3002. doi: 10.1093/jxb/erq124
Armengaud, P., Sulpice, R., Miller, A. J., Stitt, M., Amtmann, A., and Gibon, Y. (2009). Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol. 150, 772–785. doi: 10.1104/pp.108.133629
Ashraf, M. (2002). Salt tolerance of cotton: Some new advances. Crit. Rev. Plant Sci. 21, 1–30. doi: 10.1080/0735-260291044160
Ayele, A. G., Dever, J. K., Kelly, C. M., Sheehan, M., Morgan, V., and Payton, P. (2020). Responses of upland cotton (Gossypium hirsutum L.) lines to irrigated and rainfed conditions of texas high plains. Plants (Basel, Switzerland) 9:1598. doi: 10.3390/plants9111598
Baber, U., Nawaz, M. A., Arshad, U., Azhar, M. T., Atif, R. M., Golokhvast, K. S., et al. (2020). Transgenic crops for the agricultural improvement in Pakistan: A perspective of environmental stresses and the current status of genetically modified crops. GM Crops Food 11, 1–29. doi: 10.1080/21645698.2019.1680078
Baker, J., Van Dennsteele, C., and Dure, L. (1988). Sequence and characterization of 6 lea proteins and their genes from cotton. Plant Mol. Biol. 11, 277–291. doi: 10.1007/BF00027385
Bamagoos, A., Alharby, H., and Fahad, S. (2021). Biochar coupling with phosphorus fertilization modifies antioxidant activity, osmolyte accumulation and reactive oxygen species synthesis in the leaves and xylem sap of rice cultivars under high-temperature stress. Physiol. Mol. Biol. Plants. 27, 2083–2100. doi: 10.1007/s12298-021-01062-7
Bita, C. E., and Gerats, T. (2013). Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 4:273. doi: 10.3389/fpls.2013.00273
Campbell, B. T., Saha, S., Percy, R. G., Frelichowski, J., Jenkins, J. N., Park, W., et al. (2017). Screening tea varieties for nitrogen efficiency. J. Plant Nutr. 12, 1797–1804. doi: 10.1080/01904167.2016.1193605
Chattha, W. S., Patishtan, J., Shafqat, W., and Maathuis, F. J. M. (2022). Shoot potassium content provides a physiological marker to screen cotton genotypes for osmotic and salt tolerance. Int. J. Phytoremed. 24, 429–435. doi: 10.1080/15226514.2021.1951655
Chaves, M. M., and Pereira, J. M. (2003). Pereira Understanding plant responses to drought—from genes to the whole plant. Funct. Plant Biol. 30, 239–264. doi: 10.1071/FP02076
Chen, Q., Chen, Q. J., Sun, G. Q., Zheng, K., Yao, Z. P., Han, Y. H., et al. (2019). Genome-wide identification of cyclophilin gene family in cotton and expression analysis of the fibre development in Gossypium barbadense. Int. J. Mol. Sci. 20:349. doi: 10.3390/ijms20020349
Craita, E. B. (2022). Section plant sciences, institute for water and wetland research, radboud university nijmegen. Heyendaalseweg 135:6525AJ.
Cun, M., Guiliang, J., and Chuanlin, Z. (2002). The advances in cotton breeding resistance to fusarium and verticillium wilts in china during past fifty years. Zhongguo Nongye Kexue 35, 508–513.
Darmanov, M. M., Makamov, A. K., Ayubov, M. S., Khusenov, N. N., Buriev, Z. T., Shermatov, S. E., et al. (2022). Development of superior fibre quality upland cotton cultivar series ‘ravnaq’ using marker-assisted selection. Front. Plant Sci. 13:906472. doi: 10.3389/fpls.2022.906472
Davis, R. F., Earl, H. J., and Timper, P. (2014). Effect of simultaneous water deficit stress and meloidogyne incognita infection on cotton yield and fiber quality. J. Nematol. 46, 108–118.
Davis, R., Colyer, P., Rothrock, C., and Kochman, J. (2006). Fusarium wilt of cotton: Population diversity and implications for management. Plant Dis. 90, 692–703. doi: 10.1094/PD-90-0692
Diouf, L., Pan, Z., He, S. H., Gong, W. F., Jia, Y. H., Magwanga, R. O., et al. (2017). High-density linkage map construction and mapping of salt-tolerant QTLs at seedling stage in upland cotton using genotyping by sequencing (GBS). Int. J. Mol. Sci. 18:15038477. doi: 10.3390/ijms18122622
Divya, K., Jami, S. K., and Kira, P. B. (2010). Constitutive expression of mustard annexin,Artn8j1 enhances abiotic stress tolerance and fiber quality in cotton under stress. Plant Mol. Bio. 73, 293–308. doi: 10.1007/s11103-010-9615-6
Dong, C. J., Huang, B., and Liu, J. Y. (2010). The cotton dehydration-responsive element binding protein GhDBP1 contains an EAR-motif and is involved in the defense response of Arabidopsis to salinity stress. Funct. Plant Biol. 37, 64–73. doi: 10.1071/FP09100
Dongdong, L., Ming, Z., Lili, H., Xiaobo, C., Yang, G., Xingqi, G., et al. (2016). GhMAPKKK49, a novel cotton (Gossypium hirsutum L.) MAPKKK gene, is involved in diverse stress responses. Acta Physiol. Plant. 38:13. doi: 10.1007/s11738-015-2029-y
Dos Reis, S. P., Lima, A. M., and De Souza, C. R. B. (2012). Recent molecular advances on downstream plant responses to abiotic stress. Int. J. Mol. Sci. 13, 8628–8647. doi: 10.3390/ijms13078628
Du, Q., Zhao, X. H., Jiang, C. J., Wang, X. G., Han, Y., Wang, J., et al. (2017). Effect of potassium deficiency on root growth and nutrient uptake in maize (Zea mays L.). Agric. Sci. 8, 1263–1277. doi: 10.4236/as.2017.811091
Duan, H., Li, F., Wu, X., Ma, D., Wang, M., and Hou, Y. (2006). Cloning and characterization of two EREBP transcription factors from cotton (Gossypium hirsutum L.) [J]. Biochemistry 71, 285–293. doi: 10.1134/S0006297906030084
Echer, F. R., Peres, V. J. S., and Rosolem, C. A. (2020). Potassium application to the cover crop prior to cotton planting as a fertilization strategy in sandy soils. Sci. Rep. 10:20404. doi: 10.1038/s41598-020-77354-x
Ennahli, E., and Earl, H. J. (2005). Physiological limitations to photosynthetic carbon assimilation in cotton under water stress. Crop Sci. 45, 2374–2382. doi: 10.2135/cropsci2005.0147
Fahad, S., and Bano, A. (2012). Effect of salicylic acid on physiological and biochemical characterization of maize grown in saline area. Pak J. Bot. 44, 1433–1438. doi: 10.3389/fpls.2017.01147
Fahad, S., Bajwa, A. A., Nazir, U., Anjum, S. A., Farooq, A., Zohaib, A., et al. (2017). Crop production under drought and heat stress: Plant responses and management options. Front. Plant. Sci. 8:1147.
Fahad, S., Chen, Y., Saud, S., Wang, K., Xiong, D., Chen, C., et al. (2013). Ultraviolet radiation effect on photosynthetic pigments, biochemical attributes, antioxidant enzyme activity and hormonal contents of wheat. J. Food Agri. Environ. 11, 1635–1641.
Fahad, S., Hussain, S., Bano, A., Saud, S., Hassan, S., Shan, D., et al. (2014a). Potential role of phytohormones and plant growth-promoting rhizobacteria in abiotic stresses: Consequences for changing environment. Environ. Sci. Pollut. Res. 22, 4907–4921. doi: 10.1007/s11356-014-3754-2
Fahad, S., Hussain, S., Matloob, A., Khan, F. A., Khaliq, A., Saud, S., et al. (2014b). Phytohormones and plant responses to salinity stress: A review. Plant Growth Regul. 75, 391–404. doi: 10.1007/s10725-014-0013-y
Fei, J., Wang, Y. S., Cheng, H., Su, Y. B., Zhong, Y., and Zheng, L. (2021). Cloning and characterization of KoOsmotin from mangrove plant Kandelia obovata under cold stress. BMC Plant Biol. 21:10. doi: 10.1186/s12870-020-02746-0
Fontana, J. E., Wang, G., Sun, R., Xue, H., Li, Q., Liu, J., et al. (2020). Impact of potassium deficiency on cotton growth, development and potential microRNA-mediated mechanism. Plant Physiol. Biochem. 153, 72–80. doi: 10.1016/j.plaphy.2020.05.006
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Gao, M., Snider, J. L., Bai, H., Hu, W., Wang, R., Meng, Y., et al. (2020). Drought effects on cotton (Gossypium hirsutum L.) fibre quality and fibre sucrose metabolism during the flowering and boll-formation period. J. Agron. Crop Sci. 26, 309–321. doi: 10.1111/jac.12389
Gao, S. Q., Chen, M., and Xia, L. Q. (2009). A cotton (Gossypium hirsuturn) DRE-binding transcription factor gene, GhDREB, confers enhanced tolerance to drought, high salt,and freezing stresses in transgenic wheat. Plant Cell Rep. 28, 301–311. doi: 10.1007/s00299-008-0623-9
Gao, W., Xu, F.-C., Guo, D.-D., Zhao, J.-R., Liu, J., Guo, Y.-W., et al. (2018). Calcium-dependent protein kinases in cotton: Insights into early plant responses to salt stress. BMC Plant Biol. 18:1–15. doi: 10.1186/s12870-018-1230-8
Gao, Z., Li, B., Zheng, C., and Wang, G. (2008). Molecular detection of fungal communities in the hawaiian marine sponges suberites zeteki and mycale armata. Appl. Environ. Microbiol. 74, 6091–6101. doi: 10.1128/AEM.01315-08
Gorham, J., Läuchli, A., and Leidi, E. Q. (2010). “Plant responses to salinity,” in Physiology of Cotton, eds J. M. Stewart, D. M. Oosterhuis, J. J. Heitholt, and J. R. Mauney (Dordrecht, Netherlands: Springer), 129–141. doi: 10.1007/978-90-481-3195-2_13
Grossman, J. D., and Rice, K. J. (2012). Evolution of root plasticity responses to variation in soil nutrient distribution and concentration. Evol. Appl. 5, 850–857. doi: 10.1111/j.1752-4571.2012.00263.x
Gu, L., Ma, Q., and Zhang, C. (2019). The cotton GhWRKY91 transcription factor mediates leaf senescence and responses to drought stress in transgenic Arabidopsis thaliana. Front. Plant Sci. 10:1352. doi: 10.3389/fpls.2019.01352
Guo, A. H., Su, Y., and Huang, Y. (2021). QTL controlling fiber quality traits under salt stress in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 134, 661–685. doi: 10.1007/s00122-020-03721-x
Guo, H. M., Li, Z. C., and Zhang, H. (2010). Cloning of cotton CBF gene and its cold tolerance expression in transgenic tobacco. Acta Agron. Sin. 37, 286–293. doi: 10.3724/SP.J.1006.2011.00286
Guo, Y. H., Yu, Y. P., and Wang, D. (2009). GhZFP1, a novel CCCH¬type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZ1RD21A and GZIPR5. New Phytol. 183, 62–75. doi: 10.1111/j.1469-8137.2009.02838.x
Hao, Q., Zhang, L., Yang, Y., Shan, Z., and Zhou, X. A. (2019). Genome-wide analysis of the WOX gene family and function exploration of GmWOX18 in soybean. Plants (Basel) 8:215. doi: 10.3390/plants8070215
Hasanuzzaman, M., Nahar, K., Alam, M. M., Roychowdhury, R., and Fujita, M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684. doi: 10.3390/ijms14059643
He, C., Yan, J., and Shen, J. (2005). Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant Cell Physiol. 46, 1848–1854. doi: 10.1093/pcp/pci201
He, X., Zhu, L., Xu, L., Guo, W., and Zhang, X. (2016). GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 35, 2167–2179. doi: 10.1007/s00299-016-2027-6
He, Z., Ma, Z., Brown, K. M., and Lynch, J. P. (2005). Assessment of inequality of root hair density in Arabidopsis thaliana using the Gini coefficient: a close look at the effect of phosphorus and its interaction with ethylene. Ann. Bot. 95, 287–293. doi: 10.1093/aob/mci024
Hodge, A. (2004). The plastic plant: root responses to heterogeneous supplies of nutrients. New Phytol. 162, 9–24. doi: 10.1111/j.1469-8137.2004.01015.x
Hou, S., Zhu, G., Li, Y., Li, W., Fu, J., Niu, E., et al. (2018). Genome-wide association studies reveal genetic variation and candidates genes of drought stress related traits in cotton (Gossypium hirsutum L.). Front. Plant Sci. 9:1276. doi: 10.3389/fpls.2018.01276
Hu, W., Coomer, T. D., Loka, D. A., Oosterhuis, D. M., and Zhou, Z. (2017). Potassium deficiency affects the carbon-nitrogen balance in cotton leaves. Plant Physiol. Biochem. 115, 408–417. doi: 10.1016/j.plaphy.2017.04.005
Hu, Y. G., Lin, F. Y., Wang, S. Q., and He, B. R. (2008). [Cloning and expression analysis of drought-tolerant and wa-ter-saving related gene PmMYB in broomcorn millet]. Yi Chuan 30, 373–379. doi: 10.3724/SP.J.1005.2008.00373
Huang, B., and Liu, J. Y. (2006). Cloning and functional analysis of the novel gene GhDBP3 encoding a DRE-binding transcription factor from Gossypium hirsutum. Biochim. Biophy. Acta 1759, 263–269. doi: 10.1016/j.bbaexp.2006.04.006
Huang, B., Jin, L., and Liu, J. (2007). Molecular cloning and functional characterization of a DREB1/CBF-like gene (GhDREB1L) from cotton. Sci. China C Life Sci. 50, 7–14. doi: 10.1007/s11427-007-0010-8
Huang, B., Jin, L., and Liu, J. Y. (2008). Identification and characterization of the novel gene GhDBP2 encoding a DRE-binding protein from cotton (Gossypium hirsutum L.). J. Plant Physiol. 165, 214–223. doi: 10.1016/j.jplph.2006.11.003
Imlay, K. R., and Imlay, J. A. (1996). Cloning and analysis of sodC, encoding the copper-zinc superoxide dismutase of Escherichia coli. J. Bacteriol. 178, 2564–2571. doi: 10.1128/jb.178.9.2564-2571.1996
Iqbal, A., Dong, Q., Wang, X., Gui, H., Zhang, H., Zhang, X., et al. (2020a). High nitrogen enhance drought tolerance in cotton through antioxidant enzymatic activities. Nitr. Metab. Osmotic Adjust. Plants 9:178. doi: 10.3390/plants9020178
Iqbal, A., Dong, Q., Wang, X., Gui, H. P., Zhang, H., Pang, N., et al. (2020b). Nitrogen preference and genetic variation of cotton genotypes for nitrogen use efficiency. J. Sci. Food Agric. 100, 2761–2773. doi: 10.1002/jsfa.10308
Jeong, J. S., Kim, Y. S., and Back, K. H. (2010). Root-specific expression of osNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. doi: 10.1104/pp.110.154773
Jin, L. G., and Liu, J. Y. (2008). Molecular cloning, expression profile and promoter analysis of a novel ethylene responsive transcription factor gene GhERF4 from cotton (Gassypiurn hirstum). Plant Physiol. Biochern. 46:46. doi: 10.1016/j.plaphy.2007.10.004
Jin, L. G., Li, H., and Liu, J. Y. (2010). Molecular characterization of three ethylene responsive element binding factor genes from cotton. is Integr. Plant Biol. 52, 485–495. doi: 10.1111/j.1744-7909.2010.00914.x
Kakani, V. G., Reddy, K. R., Koti, S., Wallace, T. P., Prasad, P. V., Reddy, V. R., et al. (2005). Differences in in vitro pollen germination and pollen tube growth of cotton cultivars in response to high temperature. Ann. Bot. 96, 59–67. doi: 10.1093/aob/mci149
Khan, M. A., Wahid, A., Ahmad, M., Tahir, M. T., Ahmed, M., Ahmad, S., et al. (2020). “World cotton production and consumption: An overview,” in Cotton Production Uses, eds S. Ahmad and M. Hasanuzzaman (Singapore: Springer), 1–7. doi: 10.1007/978-981-15-1472-2_1
Lantzouni, O., Alkofer, A., Falter-Braun, P., and Schwechheimer, C. (2020). GROWTH-REGULATING FACTORS interact with DELLAs and regulate growth in cold stress. Plant Cell. 32, 1018–1034. doi: 10.1105/tpc.19.00784
Li, G., Tai, F. J., and Zheng, Y. (2010). Two cotton Cys2/His2-type zinc-finger proteins, Gh,Di19-1 and GhDi19-2, are involved in plant response to salt/drought stress and abscisic acid signaling. Plant Mod. Biol. 74, 437–452. doi: 10.1007/s11103-010-9684-6
Li, H. M., Liu, S. D., Ge, C. W., Zhang, X. M., Zhang, S. P., Chen, J., et al. (2019). Analysis of drought tolerance and associated traits in upland cotton at the seedling stage. Int. J. Mol. Sci. 20:3888. doi: 10.3390/ijms20163888
Li, H., Pan, Z., and He, S. (2021). QTL mapping of agronomic and economic traits for four F2 populations of upland cotton. J. Cotton. Res. 4:3. doi: 10.1186/s42397-020-00076-y
Li, S., Chen, H., Hou, Z., Li, Y., Yang, C., Wang, D., et al. (2020). Screening of abiotic stress-responsive cotton genes using a cotton full-length cDNA overexpressing Arabidopsis library. J. Int. Plant Biol. 62, 998–1016. doi: 10.1111/jipb.12861
Liang, C., Meng, Z., Meng, Z., Malik, W., Yan, R., Lwin, K. M., et al. (2016). GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci. Rep. 6, 1–14. doi: 10.1038/srep35040
Light, G. G., Mahan, J. R., and Roxas, V. P. (2005). Transgenic cotton(Gossypium hirsutum L. } seedlings expressing a tobacco glutathione S-transferase fail to provide improved stress tolerance. Planta 222, 346–354. doi: 10.1007/s00425-005-1531-7
Lin, M., Sun, S., Fang, J., Qi, X., Sun, L., Zhong, Y., et al. (2021). BSR-Seq analysis provides insights into the cold stress response of Actinidia arguta F1 populations. BMC Geno. 22:72. doi: 10.1186/s12864-021-07369-9
Linkohr, B. I., Williamson, L. C., Fitter, A. H., and Leyser, H. M. (2002). Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 29, 751–760. doi: 10.1046/j.1365-313X.2002.01251.x
Lit, S. L., Lian, L. J., and Tao, P. L. (2009). Overexpression of Thellungiella halophila H4 -PPase (Ts VP) in cotton enhances drought stress resistance of plants. Planta 229, 899–910. doi: 10.1007/s00425-008-0880-4
Liu, G., Li, X., Jin, S., Liu, X., and Zhu, L. (2014). Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS One 9:e86895. doi: 10.1371/journal.pone.0086895
Liu, L., Mishchenko, M. I., Cairns, B., Carlson, B. E., and Travis, L. D. (2006). Modeling single-scattering properties of small cirrus particles by use of a size-shape distribution of ice spheroids and cylinders. J. Quant. Spectrosc. Radiat. Trans. 101, 488–497. doi: 10.1016/j.jqsrt.2006.02.040
Liu, R.-X., Zhou, Z.-G., Guo, W.-Q., Chen, B.-L., and Oosterhuis, D. M. (2008). Effects of N fertilization on root development and activity of water-stressed cotton (Gossypium hirsutum L.) plants. Agric. Water Manage. 95, 1261–1270. doi: 10.1016/j.agwat.2008.05.002
Lu, J., Yuan, J.-G., Yang, J.-Z., and Yang, Z.-Y. (2014). Responses of morphology and drought tolerance of Sedum lineare to watering regime in green roof system: A root perspective. Urban Forestry Urban Green. 13, 682–688. doi: 10.1016/j.ufug.2014.08.003
Lv, S., Yang, A., and Zhang, K. (2007). Increase of glycinebetaine synthesis improves drought tolerance in cotton. Mol. Breed. 20, 233–248. doi: 10.1007/s11032-007-9086-x
Ma, Z., Bielenberg, D. G., Brown, K. M., and Lynch, J. P. (2001). Regulation of root hair density by phosphorus availability in Arabidopsis thaliana Plant. Cell Environ. 24, 459–467. doi: 10.1046/j.1365-3040.2001.00695.x
Magwanga, R. O., Lu, P., Kirungu, J. N., Lu, H., Wang, X., Cai, X., et al. (2018). Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 19:6. doi: 10.1186/s12863-017-0596-1
Marschner, H. (2012). Marschner’s Mineral Nutrition of Higher Plants. Cambridge, MA: Academic press.
Mauricio, U., Cantrell, R. G., Percy, R. G., Zeiger, E., and Lu, Z. (2000). QTL analysis of stomatal conductance and relationship to lint yield in an interspecific cotton. J. Cotton Sci. 4, 10–18.
Meng, C. M., Cai, C. P., and Zhang, T. Z. (2009). Characterization of six novel NAG genes and their responses to abiotic stresses in Gossypium hirsutum L.. Plant Sci. 176, 352–359. doi: 10.1016/j.plantsci.2008.12.003
Muhammad, S., Guo, W., Ullah, I., Tabbasam, N., Zafar, Y., Rahman, M., et al. (2011). QTL mapping for physiology, yield and plant architecture traits in cotton (Gossypium hirsutum L.) grown under well-watered versus water-stress conditions. Electr. J. Biotechnol. 14, 3–3.
Nazeer, W., Tipu, A. L., Ahmad, S., Mahmood, K., Mahmood, A., and Zhou, B. (2014). Evaluation of cotton leaf curl virus resistance in BC1, BC2, and BC3 progenies from an interspecific cross between Gossypium arboreum and Gossypium hirsutum. PLoS One 9:e111861. doi: 10.1371/journal.pone.0111861
Ni, Y., Wang, X., and Li, D. (2008). Novel cotton homeobox gene and its expression profiling in root development and in response to stresses and phytohorrnones. Acta Biochim. Biophys. Sin. 40, 78–84. doi: 10.1111/j.1745-7270.2008.00371.x
Nievola, C. C., Carvalho, C. P., Carvalho, V., and Rodrigues, E. (2017). Rapid responses of plants to temperature changes. Temperature (Austin) 4, 371–405. doi: 10.1080/23328940.2017.1377812
Niu, J. S., Zhang, L. N., Wang, Y. H., and Hong, D. F. (2006). Cloning, characterization and expression analysis of the receptor-like kinase gene (RLK) in common wheat. Zhi Wu Sheng Li Yu Fen Zi Sheng Wu Xue Xue Bao 32, 79–86.
Niu, J., Gui, H., Iqbal, A., Zhang, H., Dong, Q., and Pang, N. (2021). SN-use efficiency and yield of cotton (G. hirsutumn L.) are improved through the combination of n-fertilizer reduction and n-efficient cultivar. Agronomy 11: 55. agronomy11010055 doi: 10.3390/agronomy11010055
Onaga, G., and Wydra, K. (2016). “Advances in plant tolerance to abiotic stresses,” in Plant genomics, ed. I. Y. Abdurakhmonov (London: IntechOpen).
Oosterhuis, D., Loka, D., Kawakami, E., and Pettigrew, W. (2014). The physiology of potassium in crop production. Adv. Agron. 126, 203–234. doi: 10.1016/B978-0-12-800132-5.00003-1
Pasapula, V., Shen, G., and Kuppu, S. (2011). Expression of an Arabidopsis vacuolar 11+-pyrophosphatase gene(A VP,/) in cotton improves drought- and salt tolerance and increases fibre yield in the field conditions. Plant Biotech. 9, 88–99. doi: 10.1111/j.1467-7652.2010.00535.x
Pei, Y., Zhu, Y., Jia, Y., Ge, X., Li, X., Li, F., et al. (2020). Molecular evidence for the involvement of cotton GhGLP2, in enhanced resistance to Verticillium and Fusarium wilts and oxidative stress. Sci. Rep. 10, 1–15. doi: 10.1038/s41598-020-68943-x
Peng, Z., He, S., and Gong, W. (2014). Comprehensive analysis of differentially expressed genes and transcriptional regulation induced by salt stress in two contrasting cotton genotypes. BMC Genom. 15:760. doi: 10.1186/1471-2164-15-760
Pettigrew, W. T. (2001). Environmental effects on cotton fiber carbohydrate concentration and quality. Crop Sci. 41:1108. doi: 10.2135/cropsci2001.4141108x
Pettigrew, W. T. (2004). Pettigrew, physiological consequences of moisture deficit stress in cotton. Crop Sci. 44, 1265–1272. doi: 10.2135/cropsci2004.1265
Polge, T., and Polge, C. (2007). Thomas SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 12, 20–28. doi: 10.1016/j.tplants.2006.11.005
Qiao, Z. X., Huang, B., and Liu, J. Y. (2008). Molecular cloning and functional analysis of an ERF gene from cotton (Gossypium hirsatum) [J]. Machin Biophys. Acta 1779, 122–127. doi: 10.1016/j.bbagrm.2007.10.003
Qin, J., Zhao, J., and Zuo, K. (2004). Isolation and characterization of an ERF-like gene from Gossypium barbadense [J]. Plant Sci. 167, 1383–1389. doi: 10.1016/j.plantsci.2004.07.012
Rantala, S., and Tikkanen, M. (2018). Phosphorylation-induced lateral rearrangements of thylakoid protein complexes upon light acclimation. Plant Direct 2:e00039. doi: 10.1002/pld3.39
Reddy, A. S., Ali, G. S., Celesnik, H., and Day, I. S. (2011). Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23, 2010–2032. doi: 10.1105/tpc.111.084988
Riechmann, J. L., and Meyerowitz, E. M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646. doi: 10.1515/bchm.1998.379.6.633
Rodriguez-Uribe, L., Higbie, S. M., Stewart, J. M., Wilkins, T., Lindemann, W., Sengupta-Gopalan, C., et al. (2011). Identification of salt responsive genes using comparative microarray analysis in upland cotton (Gossypium hirsutum L.). Plant Sci. 180, 461–469. doi: 10.1016/j.plantsci.2010.10.009
Saud, S., Shi, Z., Xiong, L., Danish, S., Datta, R., Ahmad, I., et al. (2022). Recognizing the basics of phytochrome-interacting factors in plants for abiotic stress tolerance. Plant Stress 3:100050. doi: 10.1016/j.stress.2021.100050
Shahzad, A. N., Rizwan, M., and Asghar, M. G. (2019). Early maturing Bt cotton requires more potassium fertilizer under water deficiency to augment seed-cotton yield but not lint quality. Sci. Rep. 9:7378. doi: 10.1038/s41598-019-43563-2
Shan, D. P., Huang, J. G., and Yang, Y. T. (2007). Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid J1. New Phytol. 176, 70–81. doi: 10.1111/j.1469-8137.2007.02160.x
Shao, H., Wang, H., and Tang, X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6:902. doi: 10.3389/fpls.2015.00902
Singh, R. P., Prasad, P. V. V., Sunita, K., and Giri, S. N. (2007). influence of high temperature and breeding for heat tolerance in cotton. Adv. Agron. 93, 317–318. doi: 10.1016/S0065-2113(06)93006-5
Solomon, S., Manning, M., Marquis, M., and Qin, D. (2007). Climate Change 2007-the Physical Science Basis: Working Group I Contribution to the Fourth Assessment Report of the IPCC. Cambridge: Cambridge University Press.
Song, C., Du, L., Qi, L., Li, Y., Li, X., and Li, Y. (2017). Residual stress measurement in a metal microdevice by micro Raman spectroscopy. J. Micromech. Microeng. 27:105014.
Stroup, T. S., McEvoy, J. P., Swartz, M. S., Byerly, M. J., Glick, I. D., Canive, J. M., et al. (2003). The national institute of mental health clinical antipsychotic trials of intervention effectiveness (CATIE) project: Schizophrenia trial design and protocol development. Schizophr Bull. 29, 15–31. doi: 10.1093/oxfordjournals.schbul.a006986
Sun, F., Chen, Q., Chen, Q., Jiang, M., Gao, W., and Qu, Y. (2021). Screening of key drought tolerance indices for cotton at the flowering and boll setting stage using the dimension reduction method. Front. Plant Sci. 12:619926. doi: 10.3389/fpls.2021.619926
Sun, Z., Li, H., Zhang, Y., Li, Z., Ke, H., Wu, L., et al. (2018). Identification of SNPs and candidate genes associated with salt tolerance at the seedling stage in cotton (Gossypium hirsutum L.). Front. Plant Sci. 9:1011. doi: 10.3389/fpls.2018.01011
Tan, D. X., Tuong, H. M., Thuy, V. T. T., Son, L. V., and Mau, C. H. (2015). Cloning and overexpression of GmDREB2 gene from a vietnamese drought-resistant soybean variety. Brazilian Arch. Biol. Technol. 58, 651–657. doi: 10.1590/S1516-89132015050170
Thorp, K. R., Thompson, A. L., and Bronson, K. F. (2020). Irrigation rate and timing effects on arizona cotton yield, water productivity, and fiber quality. Agric. Water Manage. 234:106146. doi: 10.1016/j.agwat.2020.106146
Tiwari, R., Picchioni, G., Steiner, R., Hughus, S. E., Jones, D. C., and Zhang, J. F. (2013). Variation in salt tolerance during seed germination in a backcross inbreed line population and advanced breeding lines derived from upland cotton × pima cotton. Crop Sci. 53, 1974–1982. doi: 10.2135/cropsci2013.01.0028
Tränkner, M., Tavakol, E., and Jákli, B. (2018). Functioning of potassium and magnesium in photosynthesis, photosynthate translocation and photoprotection. Physiol. Plant. 163, 414–431. doi: 10.1111/ppl.12747
Ullah, A., Sun, H., Yang, X., and Zhang, X. (2017). Drought coping strategies in cotton: Increased crop per drop. Plant Biotechnol. J. 15, 271–284. doi: 10.1111/pbi.12688
Wan, F., Xu, Y., Wang, S., Gao, J., Lu, D., Zhou, C., et al. (2022). Identification and expression analysis of zinc finger A20/AN1 Stress-associated genes smsap responding to abiotic stress in eggplant. Horticulturae 8:108. horticulturae8020108 doi: 10.3390/horticulturae8020108
Wang, C., Ulloa, M., Shi, X., Yuan, X., Saski, C., Yu, J. Z., et al. (2015). Sequence composition of BAC clones and SSR markers mapped to Upland cotton chromosomes 11 and 21 targeting resistance to soil-borne pathogens. Front. Plant Sci. 6:791. doi: 10.3389/fpls.2015.00791
Wang, J. C. (2010). Cloning and Functional Study Of Five Adversity Associated Zinc Finger Protein Genes In Cotton [D], Nanjing; Nanjing Agricultural University Ph. D, Thesis.
Wang, W. X., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures towards genetic engineering for stress tolerance. Planta 128, 1–14. doi: 10.1007/s00425-003-1105-5
Wang, Y., Gao, Y., and Zang, P. (2020). Transcriptome analysis reveals underlying immune response mechanism of fungal (Penicillium oxalicum) disease in Gastrodia elata Bl. f. glauca S. chow (orchidaceae). BMC Plant Biol. 20:445. doi: 10.1186/s12870-020-02653-4
Wang, Y., Zhao, M., Xu, Z., Zhao, L., and Han, X. (2019). Cloning and expression analysis of TTG1 gene related to rosa rugosa trichomes formation. Am. J. Plant Sci. 10, 265–275. doi: 10.4236/ajps.2019.102020
Wu, H., Wang, X., Cao, Y., Zhang, H., Hua, R., Liu, H., et al. (2021). CpBBX19, a B-Box transcription factor gene of chimonanthus praecox, improves salt and drought tolerance in Arabidopsis. Genes (Basel) 12:1456. doi: 10.3390/genes12091456
Wu, J., Gutierrez, O. A., and Jenkins, J. N. (2009). Quantitative analysis and QTL mapping for agronomic and fiber traits in an RI population of upland cotton. Euphytica 165, 231–245. doi: 10.1007/s10681-008-9748-8
Wu, C. A., Yang, G.-D., Meng, Q. W., and Zheng, C. C. (2004). The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol. 45, 600–607. doi: 10.1093/pcp/pch071
Xu, P., Guo, Q., Meng, S., Zhang, X., Xu, Z., Guo, W., et al. (2021). Genome-wide association analysis reveals genetic variations and candidate genes associated with salt tolerance related traits in Gossypium hirsutum. BMC Genom. 22:26. doi: 10.1186/s12864-020-07321-3
Xue, G. P., Way, M., and Richardson, T. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up- regulated genes and dehydration tolerance in bread wheat. Mol. Plant 4, 697–712. doi: 10.1093/mp/ssr013
Xue, T., Li, X., Zhu, W., Wu, C., Yang, G., and Zheng, C. (2009). Cotton metallothionein GhMT3a, a reactive oxygen species scavenger, increased tolerance against abiotic stress in transgenic tobacco and yeast. J. Exp. Bot. 60, 339–349. doi: 10.1093/jxb/ern291
Yadav, V., Bharadi, V., and Yadav, S. K. (2016). Texture feature extraction using hybrid wavelet type I & II for finger knuckle prints for multi-algorithmic feature fusion. Proc. Comput. Sci. 79, 359–366. doi: 10.1016/j.procs.2016.03.047
Yan, J., He, C., and Wang, J. (2004). Overexpression of the Arabidopsis 14-3-3 protein GF14X in cotton leads to a” stay-green” phenotype and improves stress tolerance under moderate drought conditions. Plant Cell Physiol. 45, 1007–1014. doi: 10.1093/pcp/pch115
Yang, D., Li, F., Yi, F., Eneji, A. E., Tian, X., and Li, Z. (2021). Transcriptome analysis unravels key factors involved in response to potassium deficiency and feedback regulation of K+ uptake in cotton roots. Int. J. Mol. Sci. 22:3133. doi: 10.3390/ijms22063133
Yao, D., Zhang, X., Zhao, X., Liu, C., Wang, C., Zhang, Z., et al. (2011). Transcriptome analysis reveals salt-stress-regulated biological processes and key pathways in roots of cotton (Gossypium hirsutum L.). Genomics 98, 47–55. doi: 10.1016/j.ygeno.2011.04.007
Yibati, H., Zhang, Y., Li, Q., Xu, X., and He, P. (2022). Estimation of cotton nutrient uptake based on the quefts model in xinjiang. Agronomy 12:1427. doi: 10.3390/agronomy12061427
Yin, X., Zhan, R., He, Y., Song, S., Wang, L., Ge, Y., et al. (2020). Morphological description of a novel synthetic allotetraploid(A1A1G3G3) of Gossypium herbaceum L.and G.nelsonii Fryx. suitable for disease-resistant breeding applications. PLoS One 15:e0242620. doi: 10.1371/journal.pone.0242620
Yuan, Y., Xing, H., Zeng, W., Xu, J., Mao, L., Wang, L., et al. (2019). Genome-wide association and differential expression analysis of salt tolerance in Gossypium hirsutum L at the germination stage. BMC Plant Biol. 19:394. doi: 10.1186/s12870-019-1989-2
YunfeiJiang, R., Bueckert, A., Warkentin, T. D., and Davis, A. R. (2017). High temperature effects on in vitro pollen germination and seed set in field pea. Can. J. Plant Sci. 98, 71–80. doi: 10.1139/CJPS-2017-0073
Zafar, M. M., Jia, X., Shakeel, A., Sarfraz, Z., Manan, A., Imran, A., et al. (2022). Unraveling heat tolerance in upland cotton (Gossypium hirsutum L.) using univariate and multivariate analysis. Front. Plant Sci. 12:727835. doi: 10.3389/fpls.2021.727835
Zahid, K. R., Ali, F., Shah, F., Younas, M., Shah, T., Shahwar, D., et al. (2016). Response and tolerance mechanism of cotton Gossypium hirsutum L. to elevated temperature stress: A review. Front. Plant Sci. 7:937. doi: 10.3389/fpls.2016.00937
Zakaria, M. S. (2017). Cotton production and climatic factors: Studying the nature of its relationship by different statistical methods. Cogent Biol. 3:1292882. doi: 10.1080/23312025.2017.1292882
Zhang, H., Fu, X., and Wang, X. (2018). Identification and screening of nitrogen-efficient cotton genotypes under low and normal nitrogen environments at the seedling stage. J. Cotton. Res. 1:6. doi: 10.1186/s42397-018-0006-x
Zhang, H., Zhu, J., Gong, Z., and Zhu, J. K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhang, K. W., Guo, N., and Han, L. J. (2011). Improved salt tolerance and seed cotton yield in cotton(Gossypium hirsutum L.) by transformation with beta gene for glycinebetaine synthesis. Euphytica 181, 1–16. doi: 10.1007/s10681-011-0354-9
Zhang, L., Xi, D., Li, S., Gao, Z., Zhao, S., Shi, J., et al. (2011). A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Mol. Biol. 77, 17–31. doi: 10.1007/s11103-011-9788-7
Zhang, L., Ma, H., Chen, T., Pen, J., Yu, S., and Zhao, X. (2014). Morphological and physiological responses of cotton (Gossypium hirsutum L.) plants to salinity. PLoS One 9, e112807. doi: 10.1371/journal.pone.0112807
Zhang, X., Wang, L., Xu, X., Cai, C., and Guo, W. (2014). Genome-wide identification of mitogen-activated protein kinase gene family in Gossypium raimondii and the function of their corresponding orthologs in tetraploid cultivated cotton. BMC Plant Biol. 14:1–17. doi: 10.1186/s12870-014-0345-9
Zhang, Z., Zhu, L., and Li, D. (2021). In situ root phenotypes of cotton seedlings under phosphorus stress revealed through rhizopot. Front. Plant Sci. 12:716691. doi: 10.3389/fpls.2021.716691
Zhao, W., Dong, H., Zhou, Z., Wang, Y., and Hu, W. (2020). Potassium (K) application alleviates the negative effect of drought on cotton fiber strength by sustaining higher sucrose content and carbohydrates conversion rate. Plant Physiol. Biochem. 157, 105–113. doi: 10.1016/j.plaphy.2020.10.014
Zhao, Y. L., Wang, H. M., Shao, B. X., Chen, W., Guo, Z. J., Gong, H. Y., et al. (2016). SSR-based association mapping of salt tolerance in cotton (Gossypium hirsutum L.). Genet. Mol. Res. 25:15. doi: 10.4238/gmr.15027370
Zhou, C., Zhu, C., Fu, H., Li, X., Chen, L., Lin, Y., et al. (2019). Genome-wide investigation of superoxide dismutase (SOD) gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant (Camellia sinensis). PLoS One 14:e0223609. doi: 10.1371/journal.pone.0223609
Keywords: cotton, resistance, resource exploration, molecular marker, gene cloning
Citation: Saud S and Wang L (2022) Mechanism of cotton resistance to abiotic stress, and recent research advances in the osmoregulation related genes. Front. Plant Sci. 13:972635. doi: 10.3389/fpls.2022.972635
Received: 18 June 2022; Accepted: 25 July 2022;
Published: 17 August 2022.
Edited by:
Iftikhar Ali, State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology (CAS), ChinaReviewed by:
Xin He, Hunan Agricultural University, ChinaAyman E. L. Sabagh, Siirt University, Turkey
Copyright © 2022 Saud and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lichen Wang, bGljaGVud2FuZ0AxMjYuY29t
 Shah Saud
Shah Saud Lichen Wang
Lichen Wang