- 1Department of Molecular Biology, Sejong University, Seoul, South Korea
- 2Department of Bioindustry and Bioresource Engineering, Plant Engineering Research Institute, Sejong University, Seoul, South Korea
Triacylglycerol (TAG), which is a neutral lipid, has a structure in which three molecules of fatty acid (FA) are ester-bonded to one molecule of glycerol. TAG is important energy source for seed germination and seedling development in plants. Depending on the FA composition of the TAG, it is used as an edible oil or industrial material for cosmetics, soap, and lubricant. As the demand for plant oil is rising worldwide, either the type of FA must be changed or the total oil content of various plants must be increased. In this review, we discuss the regulation of FA metabolism by Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9, a recent genome-editing technology applicable to various plants. The development of plants with higher levels of oleic acid or lower levels of very long-chain fatty acids (VLCFAs) in seeds are discussed. In addition, the current status of research on acyltransferases, phospholipases, TAG lipases, and TAG synthesis in vegetative tissues is described. Finally, strategies for the application of CRISPR/Cas9 in lipid metabolism studies are mentioned.
Introduction
Fatty acids (FAs) are synthesized by the addition of two carbons by fatty acid synthase (FAS) in the plastid (Rawsthorne, 2002). FA biosynthesis is initiated by acetyl-CoA carboxylase, which converts acetyl-CoA to malonyl-CoA (Sasaki and Nagano, 2004). Malonyl-CoA is converted to malonyl-ACP by malonyl-CoA: acyl carrier protein (ACP) transacylase (Lessire and Stumpe, 1983). Malonyl-ACP is combined with acetyl-CoA by β-ketoacyl-acyl carrier protein synthase III (KAS III) to synthesize 4:0-ACP (Clough et al., 1992). KAS I is involved in the elongation from 4:0-ACP to 16:0-ACP and is synthesized as 18:0-ACP by KAS II (Shimakata and Stumpf, 1982). The 18:0-ACP is desaturated to 18:1-ACP by fatty acid biosynthesis 2 (FAB2; Lightner et al., 1994). Free FAs are removed from ACP by fatty acyl-ACP thioesterase A (FATA) and fatty acyl-ACP thioesterase B (FATB) and exit the plastid to form an acyl-CoA pool in the cytoplasm (Jones et al., 1995; Salas and Ohlrogge, 2002). Subsequently, acyl-CoAs are sequentially transferred to glycerol-3-phosphate (G3P) by acyltransferase enzymes in the endoplasmic reticulum (ER) to form triacylglycerol (TAG; Li-Beisson et al., 2013). To synthesize TAG, lysophosphatidic acid (LPA) is formed by attaching acyl-CoA at the sn-1 position of the G3P backbone by glycerol-3-phosphate acyltransferase (GPAT; Shockey et al., 2016). Lysophosphatidic acid acyltransferase (LPAT) then transfers acyl-CoA to the sn-2 position of LPA to form phosphatidic acid (PA). Phosphate at the sn-3 position of PA is cleaved by phosphatidate phosphatase (PAP) to form diacylglycerol (DAG; Carman and Han, 2006). Finally, TAG is produced by attaching acyl-CoA to the sn-3 position of DAG using diacylglycerol acyltransferase (DGAT; Cases et al., 1998; Zou et al., 1999; Figure 1).
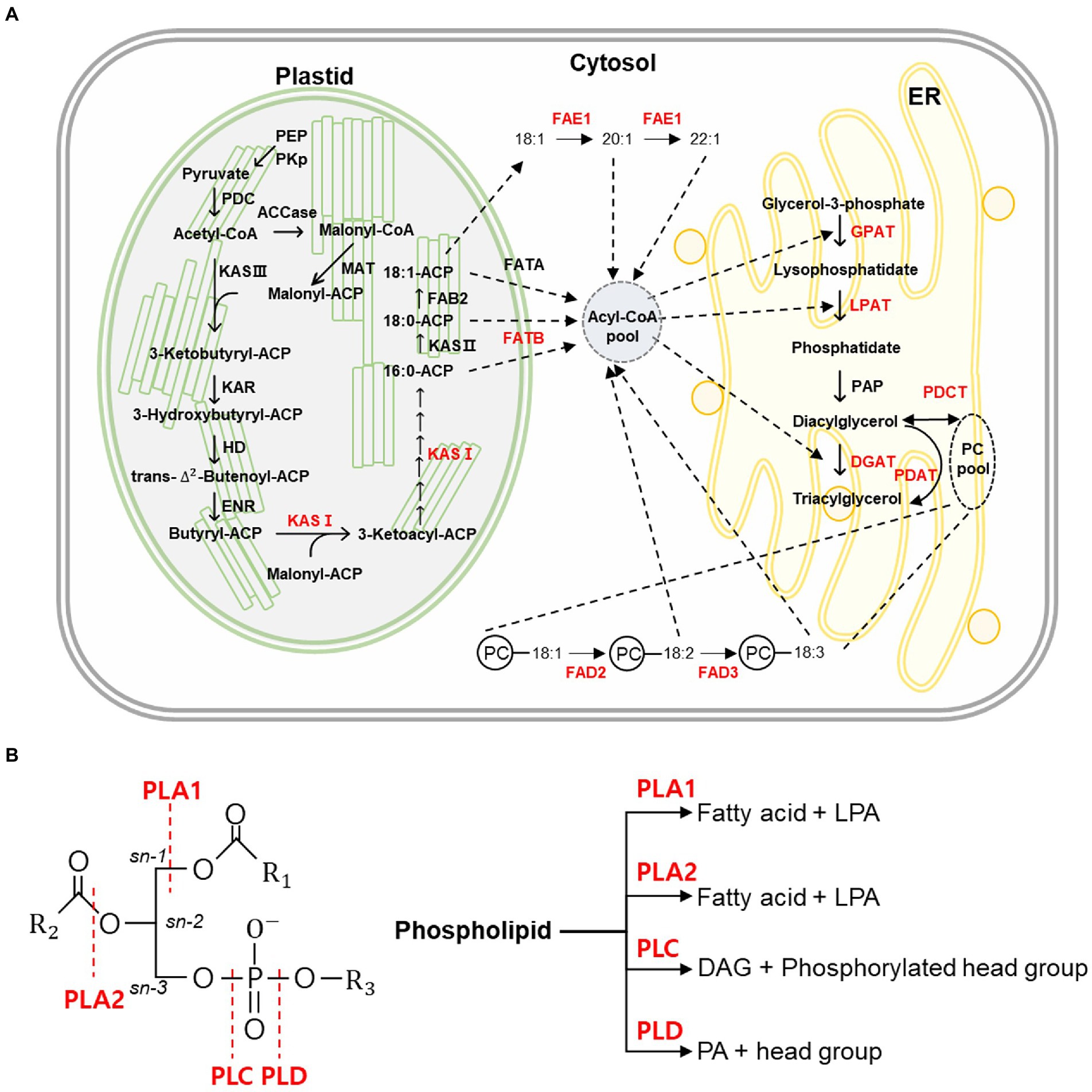
Figure 1. Fatty acid, triacylglycerol synthesis pathway, and function of phospholipase. (A) A schematic diagram of fatty acid and triacylglycerol synthesis pathway in plants. The figure illustrates the acyl-CoA synthesis pathway in the plastid and triacylglycerol (TAG) synthesis pathway by acyltransferase in endoplasmic reticulum (ER). Polyunsaturated fatty acids are synthesized in phosphatidylcholine (PC) by desaturase enzymes such as fatty acid desaturase 2 (FAD2) and FAD3. The FAE1 enzyme elongates the 18:1 fatty acid to 20:1 or 22:1, which are very long-chain fatty acids. Red-colored letters indicate the enzyme that was studied using Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (CRISPR/Cas9). The dotted lines represent the flow of the fatty acids in fatty acid and triacylglycerol synthesis. ACCase, acetyl-CoA carboxylase; ACP, acyl carrier protein; CoA, coenzyme A; DGAT, diacylglycerol acyltransferases; ENR, enoyl-ACP reductase; ER, endoplasmic reticulum; FAB2, fatty acid biosynthesis 2; FAD2, fatty acid desaturase 2; FAD3, fatty acid desaturase 3; FAE1, fatty acid elongase 1; FATA, fatty acyl-ACP thioesterase A; FATB, fatty acyl-ACP thioesterase B; GPAT, glycerol-3-phosphate acyltransferase; HD, 3-hydroxy acyl-ACP dehydratase; KAR, 3-ketoacyl-ACP reductase; KAS, β-ketoacyl-acyl carrier protein synthase; LPAT, lysophosphatidic acid acyltransferase; MAT, malonyl-CoA/ACP transacylase; PAP, phosphatidate phosphatase; PC, phosphatidylcholine; PDAT, phospholipid:diacylglycerol acyltransferase; PDC, pyruvate dehydrogenase complex; PDCT, phosphatidylcholine:diacylglycerol cholinephosphotransferase; PEP, phosphoenolpyruvate; and PKp, Plastidial pyruvate kinase. (B) The reaction of phospholipase in plants. Plants have four different forms of phospholipases (PLA1, PLA2, PLC, and PLD). Phospholipase is the enzyme that hydrolyzes phospholipids. The cleavage site of phospholipase is shown on the left figure and indicated by the red dotted lines. The right figure shows the product produced by phospholipase. DAG, diacylglycerol; LPA, lysophosphatidate; PA, phosphatidate; PLA1, phospholipase A1; PLA2, phospholipase A2; PLC, phospholipase C; and PLD, phospholipase D.
Polyunsaturated fatty acids (PUFAs) present in TAG are synthesized in phosphatidylcholine (PC), a membrane lipid (He et al., 2020). First, the oleic acid (18:1) of sn-2 in PC is converted to linoleic acid (18:2) by fatty acid desaturase 2 (FAD2), and then, linoleic acid (18:2) is converted to linolenic acid (18:3) by FAD3 (Lemieux et al., 1990; Browse et al., 1993; Dar et al., 2017). PUFAs are released from PC to form an acyl-CoA pool by reverse reaction of LPCAT (Lager et al., 2013). These acyl-CoAs are transferred into TAG through the acyl-CoA-dependent pathway by ER acyltransferases, as discussed above (Zou et al., 1999; Kim et al., 2005; Shockey et al., 2016). In addition, an acyl-CoA-independent pathway can directly synthesize TAG by transferring PUFAs in PC to DAG by phospholipid:diacylglycerol acyltransferase (PDAT; Dahlqvist et al., 2000; Figure 1A).
The discovery and functional studies of genes related to FA and TAG synthesis were carried out by forward genetics using mutants induced by ethyl methanesulfonate (EMS), and reverse genetics using T-DNA insertion mutants in Arabidopsis thaliana, a model plant (Lemieux et al., 1990; Browse et al., 1993; Lightner et al., 1994; Mcconn et al., 1994; Wu et al., 1994). Genetic studies on lipid metabolism in various crops have been conducted based on the insights from studies on Arabidopsis (Li-Beisson et al., 2013).
In most crops, FA composition consists of five common FAs: 16:0, 18:0, 18:1, 18:2, and 18:3 (Buchanan et al., 2015). However, some wild plants have unusual FAs (e.g., ω-hydroxy, 9,10-epoxy, caprylic acid, and ricinoleic acid) with specific functional groups on the FA carbon chain (Cahoon and Li-Beisson, 2020). Unusual FAs present in wild plants are industrially useful because they serve as raw materials for various polymers produced by chemical processes (Cahoon and Li-Beisson, 2020). Common FAs present in crops can also be useful in the food industry if the proportion of single types of FAs increases. For instance, vegetable oil with increased oleic acid content is suitable for frying and cooking oils (Przybylski and Aladedunye, 2012). In plant lipid metabolism engineering, strategies have mainly been used to control the FA pathway by overexpressing or mutating a specific gene to eliminate its function (Napier et al., 2014; Haslam et al., 2016). Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein 9 (CRISPR/Cas9), a recently emerged gene-editing tool, can easily and quickly edit the genome by precisely targeting a gene. Compared with traditional breeding process which requires removal of unfavored traits through repeated backcrossing and selection (Chen et al., 2019), CRISPR/Cas9 technology allows rapid development of a new cultivar with desirable traits. Besides, the conventional EMS mutagenesis is being replaced by CRISPR/Cas9 method because of its precision and completeness of mutation. In this review, we summarize the studies of CRISPR/Cas9-based knockout of mutants involved in lipid metabolism and discuss future directions in implementing this technology for the development of new oilseed crops.
CRISPR/Cas9 and lipid metabolic engineering
Transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), and CRISPR/Cas9 enable researchers to edit the genome in plants (Durai et al., 2005; Jinek et al., 2012; Joung and Sander, 2013). In TALENs, the Tal effector recognizes the DNA sequence and the FokI endonuclease cuts the DNA (Joung and Sander, 2013). In ZFNs, the Fok I endonuclease cuts DNA, however, unlike for TALEN, the zinc finger domain recognizes the DNA sequence (Durai et al., 2005). Both systems are widely used for genome editing; however, they are difficult to handle and require a long time for application to organisms compared to CRISPR/Cas9 (Chandrasegaran and Carroll, 2016). In contrast, CRISPR/Cas9 is less expensive and easy to use for any organism (Jinek et al., 2012; Chandrasegaran and Carroll, 2016).
CRISPR/Cas9 was first identified in the bacterial immune system (Brouns et al., 2008). The CRISPR/Cas9 system consists of two parts: a guide RNA (gRNA) and Cas9 protein. The gRNA has two parts: crispr RNA (crRNA) is a complementary sequence to the target gene, and trans-activating crispr RNA (tracrRNA) serves as a scaffold for linking with the Cas9 protein (Jinek et al., 2012). crRNA and tracrRNA are collectively called gRNA (Jinek et al., 2012; Ran et al., 2013). The Cas9 protein cuts double-stranded DNA, causing a double-strand break (DSB; Ran et al., 2013). The most used Cas9 protein is Streptococcus pyogenes Cas9 (SpCas9), which cuts the 3 bp position in front of the protospacer adjacent motif (PAM) corresponding to the NGG sequence in the DNA (Ran et al., 2013). Depending on the type of Cas9, Cas9 recognizes a PAM sequence that is different from NGG (Leenay and Beisel, 2017). Since the introduction of CRISPR/Cas9 technology, genome editing research has been conducted in various organisms, such as plants, humans, and microalgae (Jeon et al., 2017; Jaganathan et al., 2018; Subedi et al., 2020; Li et al., 2020b).
The mechanism of CRISPR/Cas9 involves the formation of DSB by Cas9, which leads to two DNA repair mechanisms: non-homologous end-joining (NHEJ) and homology-directed repair (HDR; Cong et al., 2013; Ran et al., 2013; Sander and Joung, 2014; Figure 2A). As the NHEJ process directly repairs through insertion or deletion, gene knockout follows where DSB has occurred (Jiang and Doudna, 2017). Compared to NHEJ, HDR leads to knock-in, in which a DNA fragment is inserted into the DSB region (Jiang and Doudna, 2017).
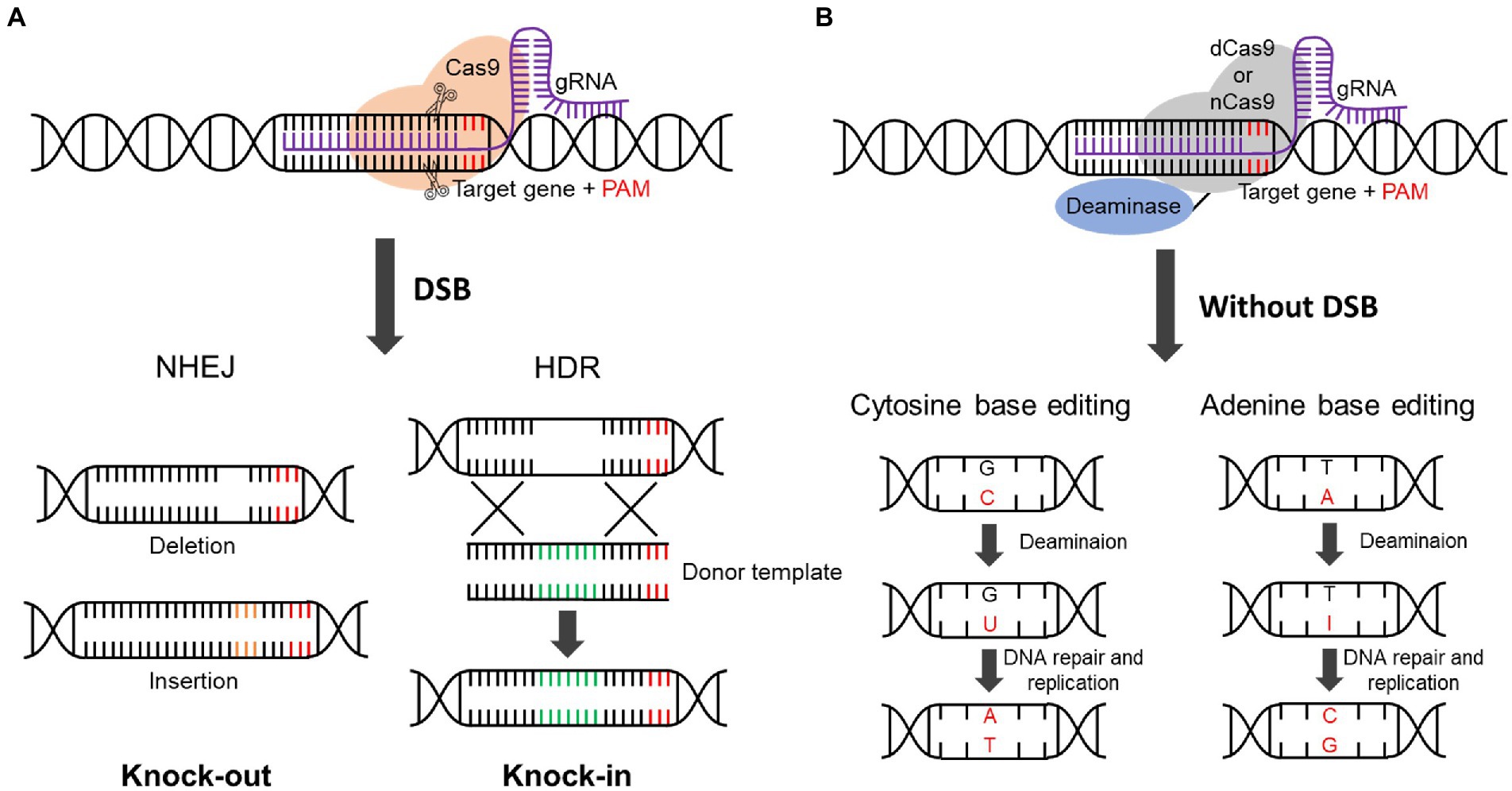
Figure 2. CRISPR/Cas9 and base editing mechanism. (A) Repair mechanism of CRISPR/Cas9. CRISPR/Cas9 is composed of Cas9 protein and gRNA. Cas9 recognizes the PAM sequence and cleaves 3 bp upstream of PAM to cause the DSB. When a DSB occurs, the cellular repair mechanism, including NHEJ and HDR processes, is initiated. NHEJ causes deletion or insertion, resulting in gene knockout due to a frameshift change. The donor template is inserted by HDR, and knockin occurs. DSB, Double-strand break; NHEJ, non-homologous end joining; HDR, homology-directed repair; PAM, protospacer adjacent motif. (B) Mechanism of base editing. The cytosine base editor or adenine base editor is made up of dead Cas9 (dCas9) or nickase Cas9 (nCas9) fused with cytosine deaminase or adenine deaminase. Cytosine deaminase removes the amine group from cytosine, resulting in a U-G mismatch. The U-G pair is converted to T-A by DNA repair and DNA replication. Adenine deaminase converts adenine to inosine by removing the amine group, resulting in an I-T mismatch. The I-T pair is converted to G-C by DNA repair and DNA replication.
CRISPR/Cas9-based technology for editing specific nucleotide sequence has also emerged recently. Dead Cas9 (dCas9) or nickase Cas9 (nCas9) fused with cytosine deaminase or adenine deaminase can convert specific nucleotides (C–T or A–G) without any DNA cleavage, it is referred to as the cytosine base editor or adenine base editor (Figure 2B; Komor et al., 2016; Nishida et al., 2016; Gaudelli et al., 2017). If one amino acid needs to be changed rather than knocked out, the base editor can be a powerful tool. For example, rice (Oryza sativa) has been modified to be herbicide resistant plant through base editing in the acetyl CoA carboxylase (ACCase) gene (Liu et al., 2020). A strategy for base editing technology in oil palm is also being introduced (Yarra et al., 2020). In addition to Cas9 and deaminase types, an online tool to design gRNA, and analysis methods to confirm mutation pattern are discussed (Yarra et al., 2020). As CRISPR/Cas9 technology develops rapidly, it has become easier and faster to knock out genes. One or two gRNA are generally used to generate single-gene mutants (Cong et al., 2013; Wang et al., 2015). More than two guide RNAs can be designed using cellular tRNA processing to target multiple genes (Xie et al., 2015). In plants, to avoid issues related to GMOs, the gene is mutated by directly injecting Cas9 protein and gRNA into the plant protoplast rather than introducing the Agrobacterium plasmid vector (Woo et al., 2015). The transient expression of Cas9 is also good strategy to develop transgene-free mutants because Cas9 DNA or RNA is degraded (Zhang et al., 2016). Currently, plant lipid metabolic engineering using CRISPR/Cas9 involves reduction of PUFAs that cause rancidity in oil, while increasing monounsaturated fatty acids (MUFAs) and inhibiting the synthesis of unhealthy saturated fatty acids (SFAs) and very long-chain fatty acids (VLCFAs).
Mutation in FAD2
Polyunsaturated fatty acids (18:2 and 18:3) were synthesized from PC in the ER (Ohlrogge and Browse, 1995). In PUFA synthesis, FAD2 desaturases 18:1 at the sn-2 position of PC into 18:2, and FAD3 desaturates from 18:2 to 18:3 (Lemieux et al., 1990; Browse et al., 1993; Dar et al., 2017; Figure 1A). As PUFAs are essential nutrients in humans, they can be beneficial to human health (Lee et al., 2016). However, it is easily oxidized at room temperature, and trans-fats are formed during deep-fat frying at high temperatures, they are unsuitable for salad dressings or cooking oils (Przybylski and Aladedunye, 2012; Saini and Keum, 2018). Therefore, making oil crops high in oleic acid is important in the food industry. Among oil crops, grape seed, sunflower, cotton, corn, soybean, camelina, perilla, and linseed have a high proportion of PUFAs in the oils (Dubois et al., 2007).
The function of FAD2 was first identified in the EMS and T-DNA mutants of Arabidopsis (Lemieux et al., 1990; Okuley et al., 1994). When FAD2 loses its activity, the PUFA content of the seed oil decreases, and the oleic acid content increases; however, it is sensitive to salt stress during seed germination and seedling growth (Zhang et al., 2012). Recently, various fad2 alleles have been reported to weaken the function of FAD2 through base editing, increase oleic acid content, and confer resistance to salt stress (Park et al., 2021).
Studies have reported the elimination of the FAD2 function using CRISPR/Cas9 in various crops (Table 1). In tobacco (Nicotiana tabacum L.), two homozygous ntfad2-2 mutants were found and their FA composition was checked. Consequently, the oleic acid content increased from 12 to 79% in mutants (Tian et al., 2020a). In rapeseed (Brassica napus), the FAD2 gene was knocked out using CRISPR/Cas9 in two cultivars (Okuzaki et al., 2018; Huang et al., 2020). First, compared with the wild type, which produces 74% oleic acid, BnFAD2_Aa of the cultivar Westar was knocked out to increase the oleic acid content to 80% (Okuzaki et al., 2018). Second, oleic acid of each knockout mutant of BnFAD2_A5 and BnaFAD2_C5 in cultivar J9707 was enhanced to 73–82%, whereas oleic acid was 66% in the wild type (Huang et al., 2020). FAD2 gene knockout studies of soybean (Glycine max) have been performed by several groups. In the cultivar Jinong38 (JN38), oleic acid content was 45–65% when GmFAD2-2 was knocked out (Al Amin et al., 2019). Oleic acid content increased to 34.47 and 40.45% in GmFAD2-1A and GmFAD2-2A mutants, respectively, and double mutants of GmFAD2-1A and GmFAD2-2A induced a high oleic acid content of up to 72% (Wu et al., 2020). In another group, they targeted both GmFAD2-1A and GmFAD2-1B in order to create double knockout mutants in the cultivar Maverick, and the oleic acid content was dramatically increased to 80% (Do et al., 2019).
In Camelina sativa, which is a hexaploid oil crop, when all three CsFAD2 were knocked out, oleic acid content increased up to 54–60% but showed a phenotype that did not grow properly in some studies (Jiang et al., 2017; Morineau et al., 2017; Lee et al., 2021). Four OsFAD2 copies have been identified in rice, and among them, OsFAD2-1 is most expressed in rice grains (Zaplin et al., 2013). Knockout of OsFAD2-1 from Japonica with CRISPR/Cas9 did not result in FA analysis (Bahariah et al., 2021), whereas oleic acid levels increased up to 80% in Nipponbare (Abe et al., 2018). FAD2 of peanut (Arachis hypogaea) was also mutated, but the seeds were not harvested; therefore, no FA analysis could be performed (Yuan et al., 2019). In cotton (Gossypium hirsutum), it was confirmed that among the eight FAD2 homologs, GhFAD2-1A and GhFAD2-1D are mostly expressed in the ovule. Therefore, GhFAD2-1A and GhFAD2-1D are simultaneously targeted by CRISPR/Cas9. Consequently, the oleic acid content was 75–77% (Chen et al., 2021). In addition, knockout of the FAD2 gene of pennycress (Thlaspi arvense) enhanced oleic acid from 12 to 35% in mutants but delayed the flowering and decreased the germination rate and seed weight (Jarvis et al., 2021).
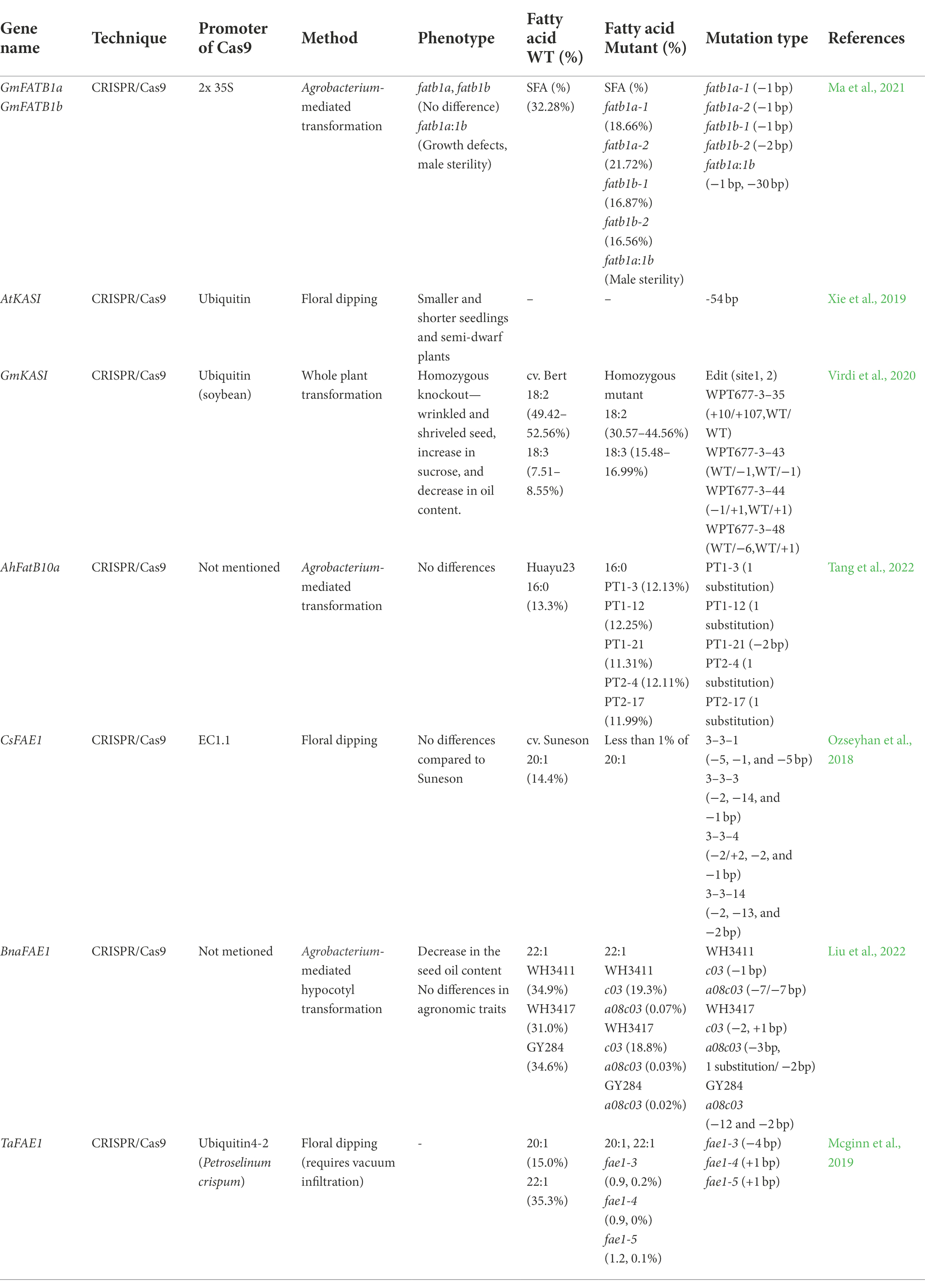
Mutation in FATB and KASI
It is important to reduce the content of SFAs in the food industry because high SFA intake can cause arteriosclerosis in humans (Siri-Tarino et al., 2010). The 16:0-ACP, 18:0-ACP, and 18:1-ACP synthesized from plastids are converted to their free-acyl forms by FATB and FATA which are then released into the cytoplasm and converted into acyl-CoA (Jones et al., 1995; Salas and Ohlrogge, 2002). Knockout of the FATB gene through CRISPR/Cas9 has been performed in soybean and peanut (Table 2; Ma et al., 2021; Tang et al., 2022). Soybeans have four GmFATB proteins, all of which have at least 78% homology with Arabidopsis FATB at the protein level. GmFATB2a and GmFATB2b are mainly expressed in flowers, and GmFATB1a and GmFATB1b are expressed in leaves and seeds. As a result of the simultaneous knockout of GmFATB1a and GmFATB1b expressed in seeds, the line in which both genes were disrupted showed male sterility. The SFA (palmitic acid and stearic acid) levels of the lines that lost only one of these two genes were 16–21%, but 32.2% in the wild type (Ma et al., 2021). In peanuts, gRNA was designed to target both AhFATB10a and AhFATB10b, but only a mutation in AhFATB10a occurred, which decreased palmitic acid content by approximately 1%, which was slightly lower than that of the wild type (13.3%; Tang et al., 2022).
Among the fatty acid synthases KASI, II, and III genes, CRISPR/Cas9 was mainly used for KASI knockout (Table 2). The in-frame deletion (−54 bp) of Arabidopsis KASI causes a semi-dwarf phenotype (Xie et al., 2019). In the KASI homozygous mutant of soybean, 18:2 level decreased by 8% and 18:3 level increased by 8.5% compared to the wild-type cultivar Bert. At the same time, the seeds of mutants were wrinkled and shriveled, and the sucrose content increased, while the oil content decreased (Virdi et al., 2020).
Mutation in FAE1
Fatty acids with 12–20 carbons are called long-chain fatty acids (LCFAs), and VLCFAs are longer than 22 carbons (Kihara, 2012). Eicosenoic acid (20:1) and erucic acid (22:1) are produced by the elongation of oleic acid by fatty acid elongase1 (FAE1; Millar and Kunst, 1997). The Arabidopsis FAE1 gene is mainly expressed in seed embryos (Rossak et al., 2001). Erucic acid, a VLCFA, is associated with myocardial infarction (Imamura et al., 2013). Therefore, researchers have studied the reduction of VLCFA using CRISPR/Cas9 in several plant oils (Table 2).
Simultaneously knocking out three FAE1 genes in Camelina (cultivar Suneson) decreased erucic acid content to less than 1% (Ozseyhan et al., 2018). In addition, seed weight, oil content, and seed shape were not significantly different from those of “Suneson” (Ozseyhan et al., 2018). In rapeseed, the erucic acid content of the a08c03 homozygous mutant was reduced to less than 0.1%, and the oil content decreased slightly, but there were no significant differences in other agronomic traits (Liu et al., 2022). In the case of a c03 single gene mutation, there was no decrease in oil content, and the content of erucic acid was 31–35% in the wild type but decreased by half in the mutant (Liu et al., 2022). In pennycress, the candidate gene of FAE1 with the highest homology to Arabidopsis FAE1 was mutated using CRISPR/Cas9. As a result, both 20:1 and 22:1 FAs decreased by less than 1% (Mcginn et al., 2019).
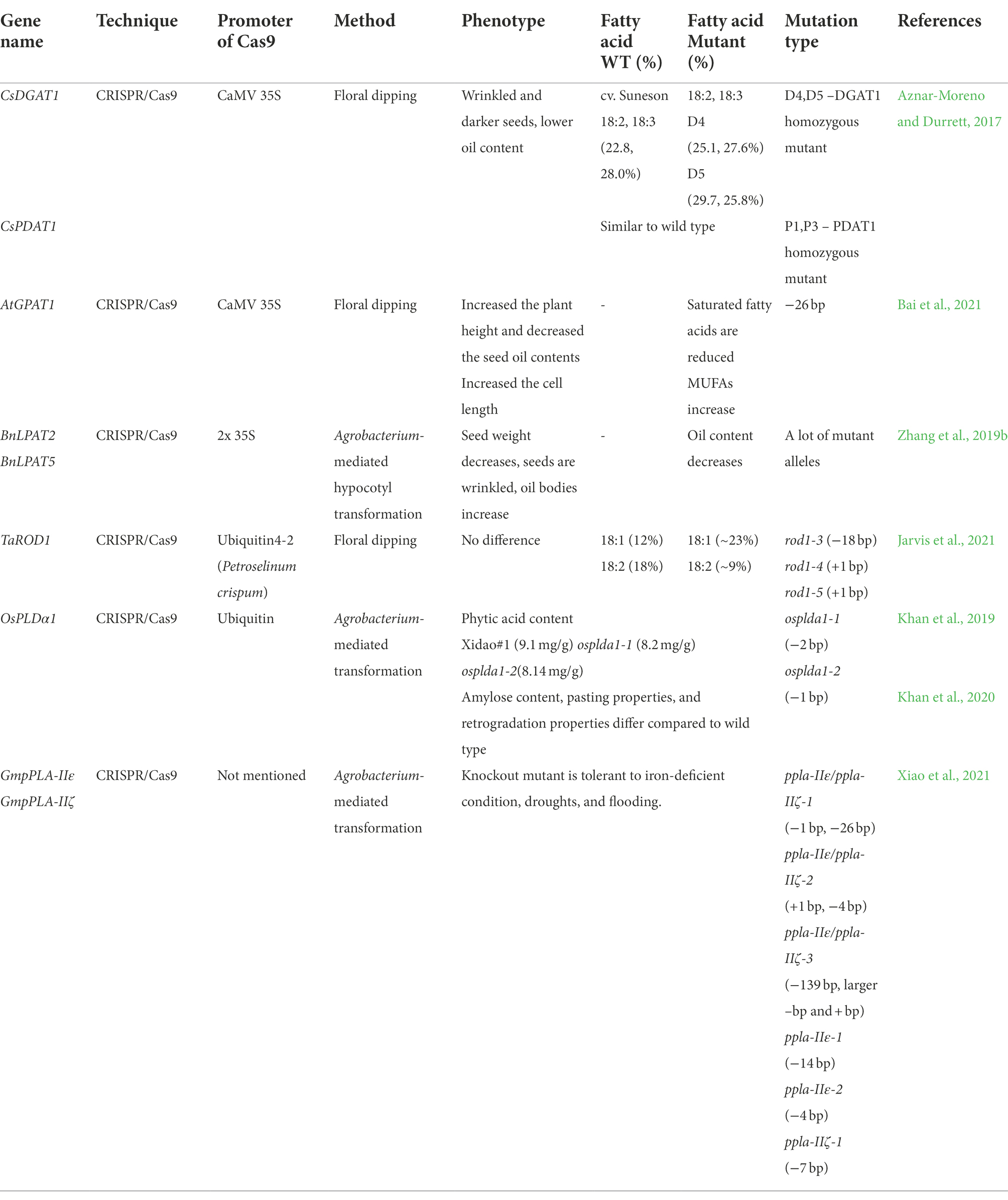
Mutation in acyltransferases
GPAT, LPAT, and DGAT are acyltransferase enzymes that synthesize TAG by transferring FA from the acyl-CoA pool to G3P (Chapman and Ohlrogge, 2012). In addition, PDAT transfers FA at the sn-2 position of PC to the sn-3 position of DAG to synthesize TAG (Dahlqvist et al., 2000). In Arabidopsis, there are 10 GPATs, five LPATs, and three DGATs (Zou et al., 1999; Kim and Huang, 2004; Yang et al., 2012; Zhou et al., 2013; Ayme et al., 2018). Among the acyltransferases, GPAT9, LPAT2, DGAT1, and PDAT1 are known to be involved in TAG synthesis (Zou et al., 1999; Banas et al., 2000; Kim et al., 2005; Shockey et al., 2016). Table 3 shows the results of acyltransferase gene editing by CRISPR/Cas9.
As a result of the deletion of Arabidopsis GPAT1, SFAs content decreased and MUFAs content increased (Bai et al., 2021). Plant height and cell length increased, but oil content decreased in gpat1 mutants (Bai et al., 2021). In rapeseed, Bnlpat2 and Bnlpat5 single mutants increased the content of 18:0 and 20:0, and decreased the content of 18:1, 18:2, and 18:3 (Zhang et al., 2019b). In the Bnlpat2 Bnlpat5 double mutant, 20:0 level was increased, while 18:2 and 18:3 levels were decreased. In all these mutants, seed weight decreased, while oil body size increased (Zhang et al., 2019b). In the camelina, DGAT1 and PDAT1 were knocked out using CRISPR/Cas9 (Aznar-Moreno and Durrett, 2017). In the csdgat1 homozygous mutant, 18:2 content was increased and 18:3 content was decreased, and the cspdat1 homozygous mutant showed an FA composition similar to that of the wild type (Aznar-Moreno and Durrett, 2017). Both mutants showed decreased oil content and the seeds were wrinkled and darkened (Aznar-Moreno and Durrett, 2017). There was no significant change in phenotype when the REDUCED OLEATE DESATURATION1 (ROD1) gene, which interconverts DAG and PC, was mutated in pennycress (Jarvis et al., 2021). FA analysis of mutant seeds showed that 18:1 content was increased and 18:2 content was decreased compared to the wild type (Jarvis et al., 2021).
Mutation in phospholipases
Phospholipids are plasma membrane lipids (Reszczynska and Hanaka, 2020). Phospholipase is one of the enzymes that hydrolyze phospholipids and is related to various cellular functions (Takac et al., 2019). Plant phospholipases can be categorized as phospholipases A, C, and D (Takac et al., 2019). There are two subtypes of phospholipase A: phospholipase A1 and phospholipase A2. Phospholipase A1 cleaves the acyl group at the sn-1 position, and phospholipase A2 cleaves the acyl group at the sn-2 position to release lysophospholipids (LPL; Ryu, 2004). Phospholipase C hydrolyzes phospholipids to release DAG and other phosphorylated head groups (Wang et al., 2012). Phospholipase D cleaves phosphate, releasing its head group and PA (Wang et al., 2012; Figure 1B).
Several studies have been conducted on phospholipase knockout or knockdown through genome editing or RNAi (Table 3; Li et al., 2006; Yamaguchi et al., 2009; Zhao et al., 2011; Guo et al., 2015; Zhang et al., 2019a). In the case of phospholipase research using CRISPR/Cas9, studies have only been conducted on soybeans and rice. In japonica rice cultivar Xidao#1, two independent knockout mutants were generated using CRISPR/Cas9. Analysis of the phytic acid and total phosphorous content of the grain showed a decrease of approximately 9 and 10%, respectively, in the ospldα1 mutants compared to the wild type (Khan et al., 2019). Additional experiments were performed using the same mutant (Khan et al., 2020). As the LPL content may affect the eating quality of rice, the LPL content in the mutants was checked (Liu et al., 2013). Except for LPC (14:0), the contents of LPC (16:0), LPC (18:1), LPE (14:0), LPE (16:0), and LPE (18:1) were increased by 11–32% in the ospldα1 mutant compared to the wild type (Khan et al., 2020). These mutants showed a decrease in amylose content, and consequently, low retrograded starch enthalpy, and high gelatinization enthalpy. The pasting property, peak viscosity, hot paste viscosity, breakdown, and cold paste viscosity all increased compared to the wild type, and only the setback viscosity decreased compared with the wild type (Khan et al., 2020). In soybean, three ppla-Iiε/ppla-Iiζ homozygous mutants, two ppla-Iiε mutants, and one ppla-Iiζ mutant were generated and studied (Xiao et al., 2021). Under P-deficient conditions, the main root length was longer in all mutant lines than in the wild type. In the Fe-deficient condition, all mutants had higher chlorophyll content, although there was a slight difference between mutants. The most shoot and root fresh weights of mutants were the same or higher than those of the wild type (Xiao et al., 2021).
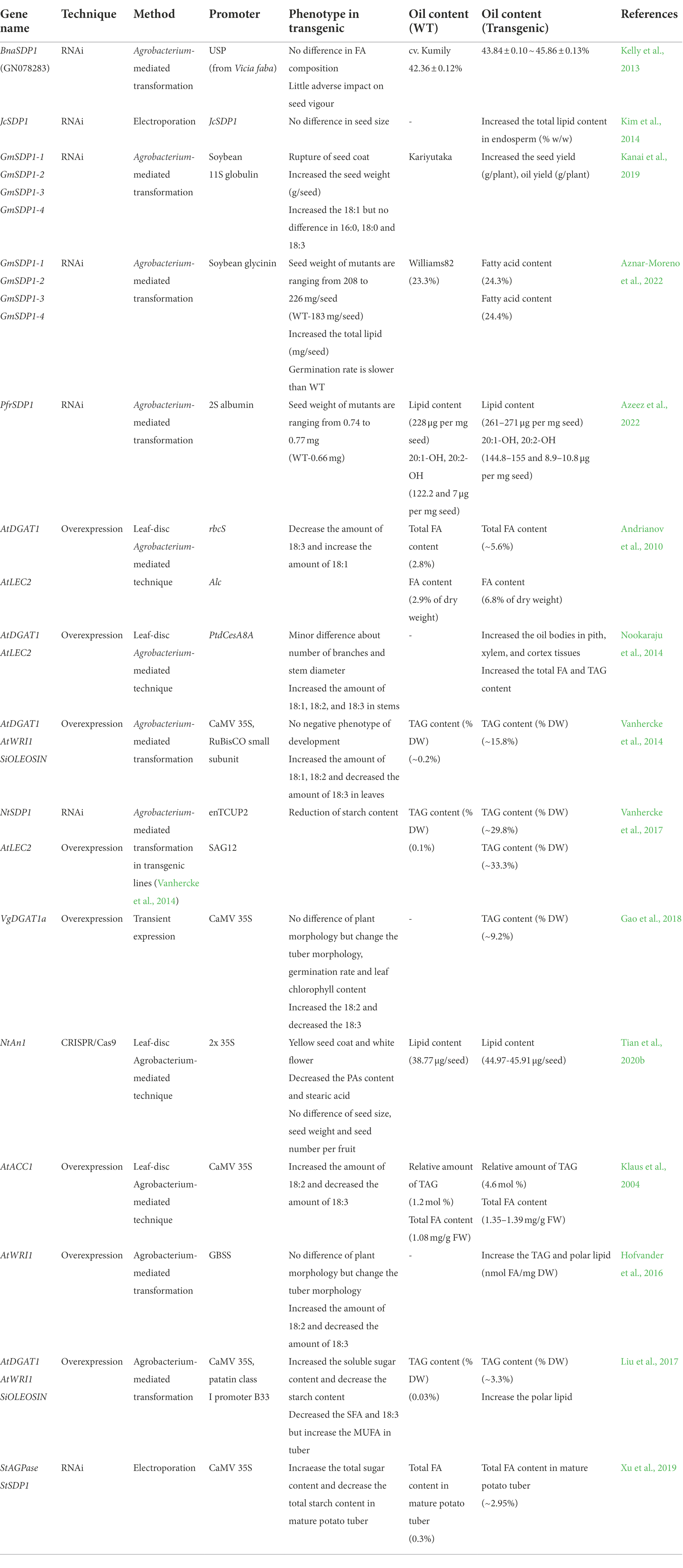
Mutation in TAG lipases
Triacylglycerol lipase catalyzes the hydrolysis of TAG to release G3P and FAs (Graham, 2008). TAGs are stored and mobilized in the form of lipid droplets (LDs), and oleosins play a role in maintaining the LD structure (Huang, 2018). In Arabidopsis, TAG is degraded by SUGAR DEPENDENT 1 (SDP1) to release FAs (Eastmond, 2006). In the sdp1 mutant, it was identified that SDP1-LIKE (SDP1L) has a function to hydrolyze the TAG (Kelly et al., 2011). In addition to SDP1, OIL BODY LIPASE 1 (OBL1) was discovered in castor (Ricinus communis) and it can hydrolyze the TAG (Eastmond, 2004). The OBL1 gene was also identified in Arabidopsis and tobacco, both of which are located in lipid droplets (LDs) and play an important role in pollen tube growth (Muller and Ischebeck, 2018). AtOBL1 has a lipase activity to TAG, DAG, and 1-MAG (Muller and Ischebeck, 2018). After the lipase degrades the TAG, FAs enter the peroxisome through PEROXISOMAL ABC-TRANSPORTER1 (PXA1), where the beta-oxidation process occurs, in which carbon is broken down by two to form acetyl-CoA (Poirier et al., 2006). Acetyl-CoA enters the TCA cycle to generate energy sources, such as ATP, NADH, and FADH2 (Martinez-Reyes and Chandel, 2020). Disruption of TAG lipase using CRISPR/Cas9 has not been attempted in various plants. However, it has been reported that SDP1 is disrupted using RNAi technology (Table 4; Kelly et al., 2013; Kim et al., 2014; Kanai et al., 2019; Azeez et al., 2022; Aznar-Moreno et al., 2022). When the SDP1 expression in rapeseed was decreased by RNAi, the oil yield (g/ m2m2 ) was further increased by 8% without affecting fatty acid composition (Kelly et al., 2013). The germination, shoot growth, and root growth are unaffected although the germination rate of seeds harvested 2 years ago decreased slightly (Kelly et al., 2013). When the SDP1 gene in Jatropha curcas was knockdown, the total lipid content of endosperm was increased compared to the control, but there was no significant difference in fatty acid composition (Kim et al., 2014). The knockdown of four GmSDP1 increased the seed weight, seed yield, and oil yield (g/plant), and oleic acid content was increased whereas linoleic acid content was decreased (Kanai et al., 2019). GmSDP1 was targeted by RNAi, which lead to enhance seed weight and overall lipid content but decreased the content of raffinose family oligosaccharides (Aznar-Moreno et al., 2022). Seed-specific silencing of the SDP1 gene in Physaria fendleri by RNAi increased seed weight and lipid content with the normal seedling establishment except for one line (Azeez et al., 2022). Based on previous results, it is possible to increase the oil content by knocking out TAG lipase using CRISPR/Cas9. Lipase has also been related to oil rancidity (Bhunia et al., 2021; Kumar et al., 2021). Rice bran oil (RBO) is abundant in nutrients but rapidly becomes rancid (Raghuram and Rukmini, 1995; Bhunia et al., 2021). Pearl millet seeds also have a high nutrition quality but the flour goes rancid rapidly (Kumar et al., 2021). Even though TAG is broken down by lipase and used as an energy source for germination, many fatty acids are released, which adversely affects rancidity (Kumar et al., 2021). The putative lipases were identified in rice and pearl millet (Bhunia et al., 2021; Kumar et al., 2021). In pearl millet, PgTAGLip1 and PgTAGLip2 polymorphisms were identified to cause loss-of-function mutation in an inbred line that low rancidity (Aher et al., 2022). Therefore, the disruption of lipase by CRISPR/Cas9 in rice or pear millet may be a key point in avoiding rancidity (Bhunia et al., 2021; Kumar et al., 2021).
Increase the TAG in vegetative tissue
In addition to plant seeds, TAG content can be increased in vegetative tissues such as leaf and tuber (Xu et al., 2018). In tobacco, oil enhancement is mainly achieved by overexpression of DGAT1 or positive transcription factors such as LEAFY COTYLEDON2 (LEC2) and WRINKLED1 (WRI1) (Table 4; Andrianov et al., 2010; Nookaraju et al., 2014; Vanhercke et al., 2014, 2017; Gao et al., 2018). TAG content was enhanced 20-fold in tobacco leaves when AtDGAT1 was expressed under the control of the ribulose-biphosphate carboxylase small subunit promoter (Andrianov et al., 2010). AtLEC2 was expressed under the control of the inducible Alc promoter (Andrianov et al., 2010). As a result, the FA content increased from 2.9% up to 6.8% (per dry weight) when treated with 1% acetaldehyde (Andrianov et al., 2010). Arabidopsis DGAT1 or LEC2 expression driven by the xylem-specific promoter in tobacco increases the FA and TAG content in the stem (Nookaraju et al., 2014). AtWRI1, AtDGAT1, and Sesamum indicum OLEOSIN (SiOLEOSIN) genes were transformed simultaneously into tobacco, and 15.8% of TAG was found in tobacco leaves (Vanhercke et al., 2014). AtLEC2 overexpression or silencing of SDP1 in transgenic tobacco (Vanhercke et al., 2014) accumulated the TAG up to 29.8 and 33.3% in the leaves, respectively (Vanhercke et al., 2017). Overexpression DGAT1a from Vernonia galamensis L. in tobacco, the TAG content of the leaves was enhanced up to 9.2% (per dry weight) without any deleterious phenotype (Gao et al., 2018). Two transgenic lines were generated using CRISPR/Cas9 by knocking out the NtAN1 gene, which regulates proanthocyanidins (PAs) and lipid accumulation in tobacco (Tian et al., 2020b). These mutants enhanced the lipid and protein content and also displayed yellow seed coat (Tian et al., 2020b).
Triacylglycerol enhancement research in the potato (Solanum tuberosum) tuber was also conducted (Table 4; Klaus et al., 2004; Hofvander et al., 2016; Liu et al., 2017; Xu et al., 2019). Arabidopsis ACCase was expressed in potato, the FA increased by 30% relative to the wild type, and showed a 5-fold increase in TAG accumulation compared to that of the wild type (Klaus et al., 2004). AtWRI1 was expressed under the control of the GBSS promoter. TAG increased 20-fold compared to that of wild type, and the polar lipid was also increased (Hofvander et al., 2016). Three genes were expressed simultaneously in the potato: AtDGAT1, AtWRI1, and SiOLEOSIN controlled by the 35S and B33 potato patatin promoters (Liu et al., 2017). As a result, TAG, which was 0.03% in the wild type, was enhanced up to 3.3% in the tuber (Liu et al., 2017). When potato ADP-glucose pyrophosphorylase (AGPase) and SDP1 were simultaneously silenced using RNAi, TAG content in the mature tuber of potato was increased by 16-fold compared to that of the wild type (Xu et al., 2019). TAG enhancement in vegetative tissue was mainly caused by overexpression or knockdown in tobacco and potato. In the future, if CRISPR is applied to enhance the TAG through lipid gene editing of tobacco or potato, it will be regarded as an important biofuel platform.
Conclusion and future perspective
Fatty acids constitute TAG, an energy source as well as a component of cell membranes and chloroplast membranes that are essential for plant cells (Kim, 2020). Since TAG in plant oil is a major source of food and industrial raw materials, attention has been focused on changing the FA composition and increasing the TAG content (Xu and Shanklin, 2016). So far, research has focused on creating high-oleic acid plant varieties by removing the FAD2 function (Table 1). Lately, research is underway to develop high-oleic acid varieties using multiple gRNAs to target both FAD2 and FATB genes in soybean (Kim et al., 2021). In rapeseed, researchers used CRISPR/Cas9 to target FAE1 and diminish the levels of 20:1 and 22:1 (Table 2). To lower the 20:1 and 22:1 content while further increasing the 18:1 content, FAD2 of pennycress was mutated in the fae1 knockout background (Jarvis et al., 2021). Moreover, CRISPR/Cas9 is commonly used in crops to investigate the roles of acyltransferase, phospholipase, and FAS genes (Tables 2, 3).
For future applications of CRISPR/Cas9 to lipid metabolism research, we suggest four possible strategies. First, CRISPR/Cas9 can be used to abolish the function of multiple lipid metabolism genes. Crops with high oleic acid content can be developed by simultaneously knocking out the FAE1 gene, which elongates the 18:1 to 20:1, 22:1, and the FAD2 gene, which desaturases the 18:1 to 18:2. Alternatively, it seems possible to develop crops with high oleic acid if FATB gene is mutated in fad2 or fae1 mutants (Figure 3A). Using CRISPR in mutants in which a specific gene has already been disrupted by EMS or RNAi may be a good strategy. For example, if fad2 is mutated using CRISPR/Cas9 in the fae1 EMS mutant line (Ozseyhan et al., 2018), a Camelina with higher oleic acid levels can also be developed. However, if there has no mutant background in which the lipid metabolism gene was disrupted by EMS or RNAi, multiple gRNAs can be used to target each distinct gene at once to abolish numerous gene functions simultaneously. Second, expression of the target gene can be controlled by removing the whole promoter region or cis-regulatory elements (CREs) using CRISPR/Cas9 (Figure 3B). For example, DGAT2 UPSTREAM GENE 1 (DUG1) which exists upstream of DGAT2 has a higher expression in leaves than DGAT2. Therefore, they deleted the 5′UTR region of DUG1 to the 5′UTR region of DGAT2 in Arabidopsis sdp1 mutant using CRISPR/Cas9 so that DGAT2 was controlled by the DUG1 promoter (Bhunia et al., 2022). As a result, total lipid content (% cell dry weight) in leaves increased 2-fold and TAG content (% CDW) increased 30-fold compared to sdp1 mutant (Bhunia et al., 2022). This method can be used only if the promoter direction of the upstream gene is appropriate (Bhunia et al., 2022). We think that the function of the upstream gene is irrelevant to plants since it will be eliminated by CRISPR/Cas9. It is also important to investigate the promoter expression level or tissue-specific expression of the upstream gene in advance. Deletion of CREs using CRISPR/Cas9 may be an effective strategy for regulating the transcription level of lipid genes (Figure 3B). Knockout causes complete loss of gene function, whereas deletion of CREs allows to fine-tune desirable traits more than knockout (Wolter et al., 2019). In fact, research was performed on the development of plants with agriculturally good traits by eliminating CREs using CRISPR/Cas9 (Li et al., 2020a, 2022; Wu et al., 2021). Third, it may be necessary to eliminate negative transcription factors that regulate the expression of lipid metabolism genes using CRISPR/Cas9 (Figure 3C). WRI1 (Baud et al., 2009), LEC1 (Mu et al., 2008), LEC2 (Kim et al., 2015), MYB96 (Lee et al., 2018), BASIC LEUCINE ZIPPER TF 67 (bZIP67; Mendes et al., 2013; Kim et al., 2022), ABSCISIC ACID INSENSITIVE 3 (ABI3; Giraudat et al., 1992), FUSCA3 (FUS3; Luerssen et al., 1998) have been reported as positive regulators of TAG biosynthesis. MYB89 (Li et al., 2017), WRKY6 (Song et al., 2020), MYB76 (Duan et al., 2017), TRANSPARENT TESTA GLABRA1 (TTG1; Chen et al., 2015), TRANSPARENT TESTA2 (TT2; Chen et al., 2012), and TT8 (Chen et al., 2014) have been reported as negative regulators. Therefore, knockout of the negative regulator using CRISPR/Cas9 in various oil crops may enhance the oil content. It is important to choose a negative TF that only increases oil content without any detrimental growth phenotype when the negative TF is eliminated because transcription factors can regulate not only lipid-related genes but also other genes. Finally, CRISPR/Cas9-based methods can be applied in plants by using dCas9 as a carrier, which is an inactive Cas9 (Figure 3D). Recently, DNMT3A, which methylates DNA, was fused with dCas9 to methylate DNA at a specific site to decrease gene expression, or TET was fused with dCas9 to demethylate DNA to increase gene expression (Xie et al., 2018). These techniques will be a new approach in the epigenetic study of genes involved in lipid metabolism. In addition, the base editor can be a good strategy to change one amino acid or mutate randomly in the plant genome (Figure 3D). For example, with random mutation of the FAD2 gene of Arabidopsis using base editing, the activity of FAD2 was weakened, and individuals with high oleic acid and resistance to salt stress were selected (Park et al., 2021).
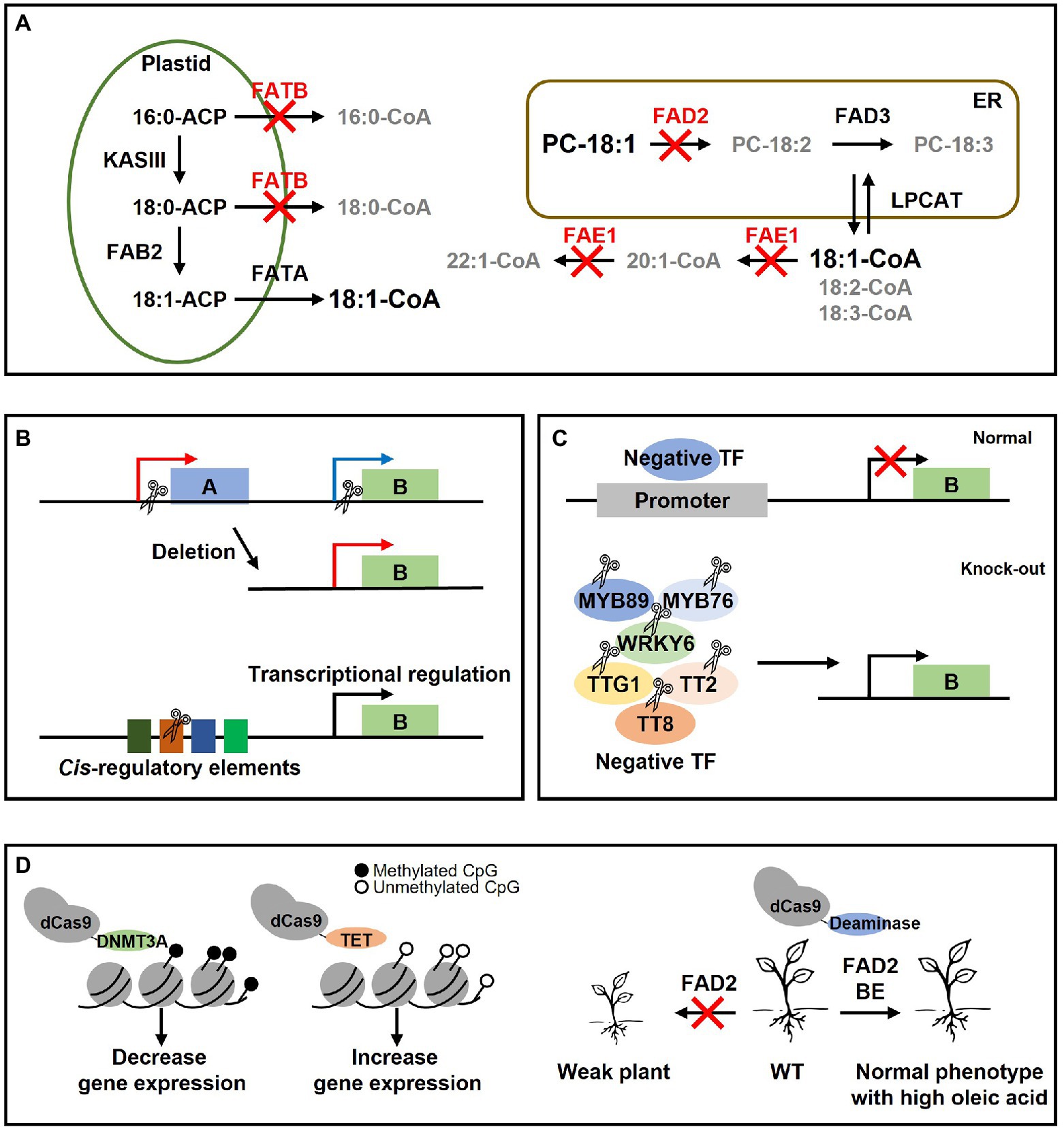
Figure 3. Future strategies of lipid metabolism research using CRISPR/Cas9 in this paper. (A) Schematic diagram of fatty acid synthesis for the development of plants with high oleic acid content in seeds. The flow of fatty acid synthesis is indicated by arrows. Red letters indicate three genes that may increase the oleic acid content if it is eliminated by CRISPR. Gray letters indicate fatty acids whose content is decreased when three genes (FATB, FAE1, and FAD2) are knocked out. It appears that plants with high oleic acid content may be created if three genes were deleted. (B) A study of promoter regulation using CRISPR/Cas9. B refers to the lipid gene and A is the upstream gene of the B gene. The scissor shape represents CRISPR/Cas9. By removing from the 5′UTR of the B gene to the 5′UTR of the A gene, the B gene can be controlled by the A promoter (Bhunia et al., 2022). In addition, the transcription level can be regulated by deleting the cis-regulatory elements of the B gene. (C) Knockout of the negative transcription factor in plant. The expression of lipid genes can be reduced in a normal plant by a variety of negative transcription factors. However, if the negative transcription factor is disrupted using CRISPR, the expression of the lipid gene can be increased. (D) CRISPR/Cas9-based technology. Epigenetic study of lipid gene seems possible if Cas9-based technique is used. For example, by methylation through dCas9-DNMT3A, lipid gene expression can be suppressed whereas demethylation via dCas9-TET can increase lipid gene expression. Alternatively, it is possible to develop a plant that slightly weakens the function of a specific protein by using base editing and has a normal phenotype than that of knockout mutants, but with a changed lipid composition.
There is a point to note when CRISPR/Cas9 is applied to mutate some genes. This is because mutations in lipid metabolism genes can alter lipid composition while also adversely affecting plant growth and development. For example, Camelina has three copies of the FAD2 gene because it is allohexaploid (Kang et al., 2011). If all three copy FAD2 genes are completely mutated, it adversely affects plant growth because this mutant is unable to synthesize polyunsaturated FAs, which are essential for maintaining the fluidity of cell membranes (Lee et al., 2021). To avoid such extreme phenotypes, it is necessary to specifically knockout only one or two FAD2 genes in camelina (Lee et al., 2021). In addition, in plants with a single copy of FAD2, it may be preferable to create a weak allele using the base editor (Park et al., 2021). A potential problem is that Cas9 can bind to unintended sites causing accidental mutations, or off-target mutations. However, these off-target mutations can be overcome in plants by backcrossing with wild type. In the future, oil crops that produce a large amount of useful FA for the industry should be developed by simultaneously controlling transcription factors and lipid metabolic genes using genome editing.
Author contributions
M-EP and HUK designed and structured the review, collected the information, organized the figures and tables, and wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Mid-Career Researcher Program of the National Research Foundation of Korea (NRF-2020R1A2C2008175), the New Breeding Technologies Development Program (project no. PJ016533), and the Next Generation BioGreen21 associated program (project no. PJ015714), and the Rural Development Administration, Republic of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, K., Araki, E., Suzuki, Y., Toki, S., and Saika, H. (2018). Production of high oleic/low linoleic rice by genome editing. Plant Physiol. Biochem. 131, 58–62. doi: 10.1016/j.plaphy.2018.04.033
Aher, R. R., Reddy, P. S., Bhunia, R. K., Flyckt, K. S., Shankhapal, A. R., Ojha, R., et al. (2022). Loss-of-function mutations in novel triacylglycerol lipase genes are associated with low rancidity in pearl millet flour. bioRxiv [Preprint]. doi: 10.1101/2022.04.02.486827
Al Amin, N., Ahmad, N., Wu, N., Pu, X., Ma, T., Du, Y., et al. (2019). CRISPR-Cas9 mediated targeted disruption of FAD2-2 microsomal omega-6 desaturase in soybean (Glycine max.L). BMC Biotechnol. 19:9. doi: 10.1186/s12896-019-0501-2
Andrianov, V., Borisjuk, N., Pogrebnyak, N., Brinker, A., Dixon, J., Spitsin, S., et al. (2010). Tobacco as a production platform for biofuel: overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotechnol. J. 8, 277–287. doi: 10.1111/j.1467-7652.2009.00458.x
Ayme, L., Arragain, S., Canonge, M., Baud, S., Touati, N., Bimai, O., et al. (2018). Arabidopsis thaliana DGAT3 is a [2Fe-2S] protein involved in TAG biosynthesis. Sci. Rep. 8:17254. doi: 10.1038/s41598-018-35545-7
Azeez, A., Parchuri, P., and Bates, P. D. (2022). Suppression of Physaria fendleri SDP1 increased seed oil and Hydroxy fatty acid content while maintaining oil biosynthesis through triacylglycerol remodeling. Front. Plant Sci. 13:931310. doi: 10.3389/fpls.2022.931310
Aznar-Moreno, J. A., and Durrett, T. P. (2017). Simultaneous targeting of multiple gene Homeologs to Alter seed oil production in Camelina sativa. Plant Cell Physiol. 58, 1260–1267. doi: 10.1093/pcp/pcx058
Aznar-Moreno, J. A., Mukherjee, T., Morley, S. A., Duressa, D., Kambhampati, S., Chu, K. L., et al. (2022). Suppression of SDP1 improves soybean seed composition by increasing oil and reducing Undigestible oligosaccharides. Front. Plant Sci. 13:863254. doi: 10.3389/fpls.2022.863254
Bahariah, B., Masani, M. Y. A., Rasid, O. A., and Parveez, G. K. A. (2021). Multiplex CRISPR/Cas9-mediated genome editing of the FAD2 gene in rice: a model genome editing system for oil palm. J. Genet. Eng. Biotechnol. 19:86. doi: 10.1186/s43141-021-00185-4
Bai, Y., Shen, Y., Zhang, Z., Jia, Q., Xu, M., Zhang, T., et al. (2021). A GPAT1 mutation in Arabidopsis enhances plant height but impairs seed oil biosynthesis. Int. J. Mol. Sci. 22, 1–18. doi: 10.3390/ijms22020785
Banas, A., Dahlqvist, A., Stahl, U., Lenman, M., and Stymne, S. (2000). The involvement of phospholipid:diacylglycerol acyltransferases in triacylglycerol production. Biochem. Soc. Trans. 28, 703–705. doi: 10.1042/bst0280703
Baud, S., Wuilleme, S., To, A., Rochat, C., and Lepiniec, L. (2009). Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J. 60, 933–947. doi: 10.1111/j.1365-313X.2009.04011.x
Bhunia, R. K., Menard, G. N., and Eastmond, P. J. (2022). A native promoter-gene fusion created by CRISPR/Cas9-mediated genomic deletion offers a transgene-free method to drive oil accumulation in leaves. FEBS Lett. 596, 1865–1870. doi: 10.1002/1873-3468.14365
Bhunia, R. K., Sinha, K., Kaur, R., Kaur, S., and Chawla, K. (2021). A holistic view of the genetic factors involved in triggering hydrolytic and oxidative rancidity of rice bran lipids. Food Rev. Int., 1–26. doi: 10.1080/87559129.2021.1915328
Brouns, S. J., Jore, M. M., Lundgren, M., Westra, E. R., Slijkhuis, R. J., Snijders, A. P., et al. (2008). Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964. doi: 10.1126/science.1159689
Browse, J., Mcconn, M., James, D., and Miquel, M. (1993). Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate. Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 268, 16345–16351. doi: 10.1016/S0021-9258(19)85427-3
Buchanan, B. B., Gruissem, W., and Jones, R. L. (2015). Biochemistry and Molecular Biology of Plants. (2nd Ed.). Wiley: Hoboken, NJ, USA.
Cahoon, E. B., and Li-Beisson, Y. (2020). Plant unusual fatty acids: learning from the less common. Curr. Opin. Plant Biol. 55, 66–73. doi: 10.1016/j.pbi.2020.03.007
Carman, G. M., and Han, G. S. (2006). Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 31, 694–699. doi: 10.1016/j.tibs.2006.10.003
Cases, S., Smith, S. J., Zheng, Y. W., Myers, H. M., Lear, S. R., Sande, E., et al. (1998). Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. U. S. A. 95, 13018–13023. doi: 10.1073/pnas.95.22.13018
Chandrasegaran, S., and Carroll, D. (2016). Origins of programmable nucleases for genome engineering. J. Mol. Biol. 428, 963–989. doi: 10.1016/j.jmb.2015.10.014
Chapman, K. D., and Ohlrogge, J. B. (2012). Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287, 2288–2294. doi: 10.1074/jbc.R111.290072
Chen, Y., Fu, M., Li, H., Wang, L., Liu, R., Liu, Z., et al. (2021). High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR/Cas9 system. Plant Biotechnol. J. 19, 424–426. doi: 10.1111/pbi.13507
Chen, K., Wang, Y., Zhang, R., Zhang, H., and Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Chen, M., Wang, Z., Zhu, Y., Li, Z., Hussain, N., Xuan, L., et al. (2012). The effect of TRANSPARENT TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol. 160, 1023–1036. doi: 10.1104/pp.112.202945
Chen, M., Xuan, L., Wang, Z., Zhou, L., Li, Z., Du, X., et al. (2014). TRANSPARENT TESTA8 inhibits seed fatty acid accumulation by targeting several seed development regulators in Arabidopsis. Plant Physiol. 165, 905–916. doi: 10.1104/pp.114.235507
Chen, M., Zhang, B., Li, C., Kulaveerasingam, H., Chew, F. T., and Yu, H. (2015). TRANSPARENT TESTA GLABRA1 regulates the accumulation of seed storage reserves in Arabidopsis. Plant Physiol. 169, 391–402. doi: 10.1104/pp.15.00943
Clough, R. C., Matthis, A. L., Barnum, S. R., and Jaworski, J. G. (1992). Purification and characterization of 3-ketoacyl-acyl carrier protein synthase III from spinach. A condensing enzyme utilizing acetyl-coenzyme a to initiate fatty acid synthesis. J. Biol. Chem. 267, 20992–20998. doi: 10.1016/S0021-9258(19)36787-0
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Dahlqvist, A., Stahl, U., Lenman, M., Banas, A., Lee, M., Sandager, L., et al. (2000). Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. U. S. A. 97, 6487–6492. doi: 10.1073/pnas.120067297
Dar, A. A., Choudhury, A. R., Kancharla, P. K., and Arumugam, N. (2017). The FAD2 gene in plants: occurrence, regulation, and role. Front. Plant Sci. 8:1789. doi: 10.3389/fpls.2017.01789
Do, P. T., Nguyen, C. X., Bui, H. T., Tran, L. T. N., Stacey, G., Gillman, J. D., et al. (2019). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and alpha-linolenic acid phenotype in soybean. BMC Plant Biol. 19:311. doi: 10.1186/s12870-019-1906-8
Duan, S., Jin, C., Li, D., Gao, C., Qi, S., Liu, K., et al. (2017). MYB76 inhibits seed fatty acid accumulation in Arabidopsis. Front. Plant Sci. 8:226. doi: 10.3389/fpls.2017.00226
Dubois, V., Breton, S., Linder, M., Fanni, J., and Parmentier, M. (2007). Fatty acid profiles of 80 vegetable oils with regard to their nutritional potential. Eur. J. Lipid Sci. Technol. 109, 710–732. doi: 10.1002/ejlt.200700040
Durai, S., Mani, M., Kandavelou, K., Wu, J., Porteus, M. H., and Chandrasegaran, S. (2005). Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 33, 5978–5990. doi: 10.1093/nar/gki912
Eastmond, P. J. (2004). Cloning and characterization of the acid lipase from castor beans. J. Biol. Chem. 279, 45540–45545. doi: 10.1074/jbc.M408686200
Eastmond, P. J. (2006). SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18, 665–675. doi: 10.1105/tpc.105.040543
Gao, C.-Y., Mao, X., Shang, H.-Q., Li, F., and Li, R.-Z. (2018). Enhanced oil accumulation in tobacco (Nicotiana tabacum L.) leaves by ectopic overexpression of VgDGAT1a for renewable production of biofuels. Curr. Sci. 114, 1234–1240. doi: 10.18520/cs/v114/i06/1234-1240
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of a*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi: 10.1038/nature24644
Giraudat, J., Hauge, B. M., Valon, C., Smalle, J., Parcy, F., and Goodman, H. M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. doi: 10.1105/tpc.4.10.1251
Graham, I. A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142. doi: 10.1146/annurev.arplant.59.032607.092938
Guo, Y., Abernathy, B., Zeng, Y., and Ozias-Akins, P. (2015). TILLING by sequencing to identify induced mutations in stress resistance genes of peanut (Arachis hypogaea). BMC Genomics 16:157. doi: 10.1186/s12864-015-1348-0
Haslam, R. P., Sayanova, O., Kim, H. J., Cahoon, E. B., and Napier, J. A. (2016). Synthetic redesign of plant lipid metabolism. Plant J. 87, 76–86. doi: 10.1111/tpj.13172
He, M., Qin, C. X., Wang, X., and Ding, N. Z. (2020). Plant unsaturated fatty acids: biosynthesis and regulation. Front. Plant Sci. 11:390. doi: 10.3389/fpls.2020.00390
Hofvander, P., Ischebeck, T., Turesson, H., Kushwaha, S. K., Feussner, I., Carlsson, A. S., et al. (2016). Potato tuber expression of Arabidopsis WRINKLED1 increase triacylglycerol and membrane lipids while affecting central carbohydrate metabolism. Plant Biotechnol. J. 14, 1883–1898. doi: 10.1111/pbi.12550
Huang, A. H. C. (2018). Plant lipid droplets and their associated proteins: potential for rapid advances. Plant Physiol. 176, 1894–1918. doi: 10.1104/pp.17.01677
Huang, H., Cui, T., Zhang, L., Yang, Q., Yang, Y., Xie, K., et al. (2020). Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. Theor. Appl. Genet. 133, 2401–2411. doi: 10.1007/s00122-020-03607-y
Imamura, F., Lemaitre, R. N., King, I. B., Song, X., Steffen, L. M., Folsom, A. R., et al. (2013). Long-chain monounsaturated fatty acids and incidence of congestive heart failure in 2 prospective cohorts. Circulation 127, 1512–1521. doi: 10.1161/CIRCULATIONAHA.112.001197
Jaganathan, D., Ramasamy, K., Sellamuthu, G., Jayabalan, S., and Venkataraman, G. (2018). CRISPR for crop improvement: an update review. Front. Plant Sci. 9:985. doi: 10.3389/fpls.2018.00985
Jarvis, B. A., Romsdahl, T. B., Mcginn, M. G., Nazarenus, T. J., Cahoon, E. B., Chapman, K. D., et al. (2021). CRISPR/Cas9-induced fad2 and rod1 mutations stacked with fae1 confer high oleic acid seed oil in pennycress (Thlaspi arvense L.). Front. Plant Sci. 12:652319. doi: 10.3389/fpls.2021.652319
Jeon, S., Lim, J. M., Lee, H. G., Shin, S. E., Kang, N. K., Park, Y. I., et al. (2017). Current status and perspectives of genome editing technology for microalgae. Biotechnol. Biofuels 10:267. doi: 10.1186/s13068-017-0957-z
Jiang, F., and Doudna, J. A. (2017). CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 46, 505–529. doi: 10.1146/annurev-biophys-062215-010822
Jiang, W. Z., Henry, I. M., Lynagh, P. G., Comai, L., Cahoon, E. B., and Weeks, D. P. (2017). Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 15, 648–657. doi: 10.1111/pbi.12663
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jones, A., Davies, H. M., and Voelker, T. A. (1995). Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. Plant Cell 7, 359–371. doi: 10.1105/tpc.7.3.359
Joung, J. K., and Sander, J. D. (2013). TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55. doi: 10.1038/nrm3486
Kanai, M., Yamada, T., Hayashi, M., Mano, S., and Nishimura, M. (2019). Soybean (Glycine max L.) triacylglycerol lipase GmSDP1 regulates the quality and quantity of seed oil. Sci. Rep. 9:8924. doi: 10.1038/s41598-019-45331-8
Kang, J., Snapp, A. R., and Lu, C. (2011). Identification of three genes encoding microsomal oleate desaturases (FAD2) from the oilseed crop Camelina sativa. Plant Physiol. Biochem. 49, 223–229. doi: 10.1016/j.plaphy.2010.12.004
Kelly, A. A., Quettier, A. L., Shaw, E., and Eastmond, P. J. (2011). Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol. 157, 866–875. doi: 10.1104/pp.111.181784
Kelly, A. A., Shaw, E., Powers, S. J., Kurup, S., and Eastmond, P. J. (2013). Suppression of the SUGAR-DEPENDENT1 triacylglycerol lipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.). Plant Biotechnol. J. 11, 355–361. doi: 10.1111/pbi.12021
Khan, M. S. S., Basnet, R., Ahmed, S., Bao, J., and Shu, Q. (2020). Mutations of OsPLDα1 increase Lysophospholipid content and enhance cooking and eating quality in rice. Plants (Basel) 9:390. doi: 10.3390/plants9030390
Khan, M. S. S., Basnet, R., Islam, S. A., and Shu, Q. (2019). Mutational analysis of OsPLDα1 reveals its involvement in phytic acid biosynthesis in rice grains. J. Agric. Food Chem. 67, 11436–11443. doi: 10.1021/acs.jafc.9b05052
Kihara, A. (2012). Very long-chain fatty acids: elongation, physiology and related disorders. J. Biochem. 152, 387–395. doi: 10.1093/jb/mvs105
Kim, H. U., and Huang, A. H. (2004). Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiol. 134, 1206–1216. doi: 10.1104/pp.103.035832
Kim, W. N., Kim, H. J., Chung, Y. S., and Kim, H. U. (2021). Construction of Multiple Guide RNAs in CRISPR/Cas9 Vector Using Stepwise or Simultaneous Golden Gate Cloning: Case Study for Targeting the FAD2 and FATB Multigene in Soybean. Plants (Basel) 10:2542. doi: 10.3390/plants10112542
Kim, H. U., Lee, K. R., Jung, S. J., Shin, H. A., Go, Y. S., Suh, M. C., et al. (2015). Senescence-inducible LEC2 enhances triacylglycerol accumulation in leaves without negatively affecting plant growth. Plant Biotechnol. J. 13, 1346–1359. doi: 10.1111/pbi.12354
Kim, I., Lee, K. R., Park, M. E., and Kim, H. U. (2022). The seed-specific transcription factor DPBF2 modulates the fatty acid composition in seeds. Plant Direct 6:e395. doi: 10.1002/pld3.395
Kim, H. U., Li, Y., and Huang, A. H. (2005). Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17, 1073–1089. doi: 10.1105/tpc.104.030403
Kim, M. J., Yang, S. W., Mao, H. Z., Veena, S. P., Yin, J. L., and Chua, N. H. (2014). Gene silencing of sugar-dependent 1 (JcSDP1), encoding a patatin-domain triacylglycerol lipase, enhances seed oil accumulation in Jatropha curcas. Biotechnol. Biofuels 7:36. doi: 10.1186/1754-6834-7-36
Klaus, D., Ohlrogge, J. B., Neuhaus, H. E., and Dormann, P. (2004). Increased fatty acid production in potato by engineering of acetyl-CoA carboxylase. Planta 219, 389–396. doi: 10.1007/s00425-004-1236-3
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi: 10.1038/nature17946
Kumar, R. R., Bhargava, D. V., Pandit, K., Goswami, S., Mukesh Shankar, S., Singh, S. P., et al. (2021). Lipase—the fascinating dynamics of enzyme in seed storage and germination—a real challenge to pearl millet. Food Chem. 361:130031. doi: 10.1016/j.foodchem.2021.130031
Lager, I., Yilmaz, J. L., Zhou, X. R., Jasieniecka, K., Kazachkov, M., Wang, P., et al. (2013). Plant acyl-CoA:lysophosphatidylcholine acyltransferases (LPCATs) have different specificities in their forward and reverse reactions. J. Biol. Chem. 288, 36902–36914. doi: 10.1074/jbc.M113.521815
Lee, K. R., Jeon, I., Yu, H., Kim, S. G., Kim, H. S., Ahn, S. J., et al. (2021). Increasing monounsaturated fatty acid contents in Hexaploid Camelina sativa seed oil by FAD2 gene knockout using CRISPR-Cas9. Front. Plant Sci. 12:702930. doi: 10.3389/fpls.2021.702930
Lee, H. G., Kim, H., Suh, M. C., Kim, H. U., and Seo, P. J. (2018). The MYB96 transcription factor regulates triacylglycerol accumulation by activating DGAT1 and PDAT1 expression in Arabidopsis seeds. Plant Cell Physiol. 59, 1432–1442. doi: 10.1093/pcp/pcy073
Lee, J. M., Lee, H., Kang, S., and Park, W. J. (2016). Fatty acid desaturases, polyunsaturated fatty acid regulation, and biotechnological advances. Nutrients 8:23. doi: 10.3390/nu8010023
Leenay, R. T., and Beisel, C. L. (2017). Deciphering, communicating, and engineering the CRISPR PAM. J. Mol. Biol. 429, 177–191. doi: 10.1016/j.jmb.2016.11.024
Lemieux, B., Miquel, M., Somerville, C., and Browse, J. (1990). Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor. Appl. Genet. 80, 234–240. doi: 10.1007/BF00224392
Lessire, R., and Stumpe, P. K. (1983). Nature of the fatty acid Synthetase Systems in Parenchymal and Epidermal Cells of Allium porrum L. leaves. Plant Physiol. 73, 614–618. doi: 10.1104/pp.73.3.614
Li, Q., Feng, Q., Snouffer, A., Zhang, B., Rodriguez, G. R., and Van Der Knaap, E. (2022). Increasing fruit weight by editing a Cis-regulatory element in tomato KLUH promoter using CRISPR/Cas9. Front. Plant Sci. 13:879642. doi: 10.3389/fpls.2022.879642
Li, D., Jin, C., Duan, S., Zhu, Y., Qi, S., Liu, K., et al. (2017). MYB89 transcription factor represses seed oil accumulation. Plant Physiol. 173, 1211–1225. doi: 10.1104/pp.16.01634
Li, C., Li, W., Zhou, Z., Chen, H., Xie, C., and Lin, Y. (2020a). A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 18, 313–315. doi: 10.1111/pbi.13217
Li, M., Qin, C., Welti, R., and Wang, X. (2006). Double knockouts of phospholipases Dζ1 and Dζ2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol. 140, 761–770. doi: 10.1104/pp.105.070995
Li, H., Yang, Y., Hong, W., Huang, M., Wu, M., and Zhao, X. (2020b). Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct. Target. Ther. 5:1. doi: 10.1038/s41392-019-0089-y
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism. Arabidopsis Book 11:e0161. doi: 10.1199/tab.0161
Lightner, J., Wu, J., and Browse, J. (1994). A mutant of Arabidopsis with increased levels of stearic acid. Plant Physiol. 106, 1443–1451. doi: 10.1104/pp.106.4.1443
Liu, Y., Du, Z., Lin, S., Li, H., Lu, S., Guo, L., et al. (2022). CRISPR/Cas9-targeted mutagenesis of BnaFAE1 genes confers low-erucic acid in Brassica napus. Front. Plant Sci. 13:848723. doi: 10.3389/fpls.2022.848723
Liu, Q., Guo, Q., Akbar, S., Zhi, Y., El Tahchy, A., Mitchell, M., et al. (2017). Genetic enhancement of oil content in potato tuber (Solanum tuberosum L.) through an integrated metabolic engineering strategy. Plant Biotechnol. J. 15, 56–67. doi: 10.1111/pbi.12590
Liu, X., Qin, R., Li, J., Liao, S., Shan, T., Xu, R., et al. (2020). A CRISPR-Cas9-mediated domain-specific base-editing screen enables functional assessment of ACCase variants in rice. Plant Biotechnol. J. 18, 1845–1847. doi: 10.1111/pbi.13348
Liu, L., Waters, D. L., Rose, T. J., Bao, J., and King, G. J. (2013). Phospholipids in rice: significance in grain quality and health benefits: a review. Food Chem. 139, 1133–1145. doi: 10.1016/j.foodchem.2012.12.046
Luerssen, H., Kirik, V., Herrmann, P., and Misera, S. (1998). FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J. 15, 755–764. doi: 10.1046/j.1365-313X.1998.00259.x
Ma, J., Sun, S., Whelan, J., and Shou, H. (2021). CRISPR/Cas9-mediated knockout of GmFATB1 significantly reduced the amount of saturated fatty acids in soybean seeds. Int. J. Mol. Sci. 22, 1–14. doi: 10.3390/ijms22083877
Martinez-Reyes, I., and Chandel, N. S. (2020). Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 11:102. doi: 10.1038/s41467-019-13668-3
Mcconn, M., Hugly, S., Browse, J., and Somerville, C. (1994). A mutation at the fad8 locus of Arabidopsis identifies a second chloroplast [omega]-3 desaturase. Plant Physiol. 106, 1609–1614. doi: 10.1104/pp.106.4.1609
Mcginn, M., Phippen, W. B., Chopra, R., Bansal, S., Jarvis, B. A., Phippen, M. E., et al. (2019). Molecular tools enabling pennycress (Thlaspi arvense) as a model plant and oilseed cash cover crop. Plant Biotechnol. J. 17, 776–788. doi: 10.1111/pbi.13014
Mendes, A., Kelly, A. A., Van Erp, H., Shaw, E., Powers, S. J., Kurup, S., et al. (2013). bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating FATTY ACID DESATURASE3. Plant Cell 25, 3104–3116. doi: 10.1105/tpc.113.116343
Millar, A. A., and Kunst, L. (1997). Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12, 121–131. doi: 10.1046/j.1365-313X.1997.12010121.x
Morineau, C., Bellec, Y., Tellier, F., Gissot, L., Kelemen, Z., Nogue, F., et al. (2017). Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 15, 729–739. doi: 10.1111/pbi.12671
Mu, J., Tan, H., Zheng, Q., Fu, F., Liang, Y., Zhang, J., et al. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148, 1042–1054. doi: 10.1104/pp.108.126342
Muller, A. O., and Ischebeck, T. (2018). Characterization of the enzymatic activity and physiological function of the lipid droplet-associated triacylglycerol lipase AtOBL1. New Phytol. 217, 1062–1076. doi: 10.1111/nph.14902
Napier, J. A., Haslam, R. P., Beaudoin, F., and Cahoon, E. B. (2014). Understanding and manipulating plant lipid composition: metabolic engineering leads the way. Curr. Opin. Plant Biol. 19, 68–75. doi: 10.1016/j.pbi.2014.04.001
Nishida, K., Arazoe, T., Yachie, N., Banno, S., Kakimoto, M., Tabata, M., et al. (2016). Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353:aaf8729. doi: 10.1126/science.aaf8729
Nookaraju, A., Pandey, S. K., Fujino, T., Kim, J. Y., Suh, M. C., and Joshi, C. P. (2014). Enhanced accumulation of fatty acids and triacylglycerols in transgenic tobacco stems for enhanced bioenergy production. Plant Cell Rep. 33, 1041–1052. doi: 10.1007/s00299-014-1582-y
Ohlrogge, J., and Browse, J. (1995). Lipid biosynthesis. Plant Cell 7, 957–970. doi: 10.1105/tpc.7.7.957
Okuley, J., Lightner, J., Feldmann, K., Yadav, N., Lark, E., and Browse, J. (1994). Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6, 147–158. doi: 10.1105/tpc.6.1.147
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Ozseyhan, M. E., Kang, J., Mu, X., and Lu, C. (2018). Mutagenesis of the FAE1 genes significantly changes fatty acid composition in seeds of Camelina sativa. Plant Physiol. Biochem. 123, 1–7. doi: 10.1016/j.plaphy.2017.11.021
Park, M. E., Yun, J. Y., and Kim, H. U. (2021). C-to-G Base editing enhances oleic acid production by generating novel alleles of FATTY ACID DESATURASE 2 in plants. Front. Plant Sci. 12:748529. doi: 10.3389/fpls.2021.748529
Poirier, Y., Antonenkov, V. D., Glumoff, T., and Hiltunen, J. K. (2006). Peroxisomal beta-oxidation—a metabolic pathway with multiple functions. Biochim. Biophys. Acta 1763, 1413–1426. doi: 10.1016/j.bbamcr.2006.08.034
Przybylski, O., and Aladedunye, F. A. (2012). Formation of trans fats during food preparation. Can. J. Diet. Pract. Res. 73, 98–101. doi: 10.3148/73.2.2012.98
Raghuram, T. C., and Rukmini, C. (1995). Nutritional significance of rice bran oil. Indian J. Med. Res. 102, 241–244.
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. doi: 10.1038/nprot.2013.143
Rawsthorne, S. (2002). Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 41, 182–196. doi: 10.1016/S0163-7827(01)00023-6
Reszczynska, E., and Hanaka, A. (2020). Lipids composition in plant membranes. Cell Biochem. Biophys. 78, 401–414. doi: 10.1007/s12013-020-00947-w
Rossak, M., Smith, M., and Kunst, L. (2001). Expression of the FAE1 gene and FAE1 promoter activity in developing seeds of Arabidopsis thaliana. Plant Mol. Biol. 46, 717–725. doi: 10.1023/A:1011603923889
Ryu, S. B. (2004). Phospholipid-derived signaling mediated by phospholipase A in plants. Trends Plant Sci. 9, 229–235. doi: 10.1016/j.tplants.2004.03.004
Saini, R. K., and Keum, Y. S. (2018). Omega-3 and omega-6 polyunsaturated fatty acids: dietary sources, metabolism, and significance: A review. Life Sci. 203, 255–267. doi: 10.1016/j.lfs.2018.04.049
Salas, J. J., and Ohlrogge, J. B. (2002). Characterization of substrate specificity of plant FatA and FatB acyl-ACP thioesterases. Arch. Biochem. Biophys. 403, 25–34. doi: 10.1016/S0003-9861(02)00017-6
Sander, J. D., and Joung, J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32, 347–355. doi: 10.1038/nbt.2842
Sasaki, Y., and Nagano, Y. (2004). Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 68, 1175–1184. doi: 10.1271/bbb.68.1175
Shimakata, T., and Stumpf, P. K. (1982). Isolation and function of spinach leaf beta-ketoacyl-[acyl-carrier-protein] synthases. Proc. Natl. Acad. Sci. U. S. A. 79, 5808–5812. doi: 10.1073/pnas.79.19.5808
Shockey, J., Regmi, A., Cotton, K., Adhikari, N., Browse, J., and Bates, P. D. (2016). Identification of Arabidopsis GPAT9 (At5g60620) as an essential gene involved in triacylglycerol biosynthesis. Plant Physiol. 170, 163–179. doi: 10.1104/pp.15.01563
Siri-Tarino, P. W., Sun, Q., Hu, F. B., and Krauss, R. M. (2010). Saturated fatty acids and risk of coronary heart disease: modulation by replacement nutrients. Curr. Atheroscler. Rep. 12, 384–390. doi: 10.1007/s11883-010-0131-6
Song, G., Li, X., Munir, R., Khan, A. R., Azhar, W., Yasin, M. U., et al. (2020). The WRKY6 transcription factor affects seed oil accumulation and alters fatty acid compositions in Arabidopsis thaliana. Physiol. Plant. 169, 612–624. doi: 10.1111/ppl.13082
Subedi, U., Ozga, J. A., Chen, G., Foroud, N. A., and Singer, S. D. (2020). CRISPR/Cas-mediated genome editing for the improvement of oilseed crop productivity. Crit. Rev. Plant Sci. 39, 195–221. doi: 10.1080/07352689.2020.1782568
Takac, T., Novak, D., and Samaj, J. (2019). Recent advances in the cellular and developmental biology of phospholipases in plants. Front. Plant Sci. 10:362. doi: 10.3389/fpls.2019.00362
Tang, Y., Huang, J., Ji, H., Pan, L., Hu, C., Qiu, X., et al. (2022). Identification of AhFatB genes through genome-wide analysis and knockout of AhFatB reduces the content of saturated fatty acids in peanut (Arichis hypogaea L.). Plant Sci. 319:111247. doi: 10.1016/j.plantsci.2022.111247
Tian, Y., Chen, K., Li, X., Zheng, Y., and Chen, F. (2020a). Design of high-oleic tobacco (Nicotiana tabacum L.) seed oil by CRISPR-Cas9-mediated knockout of NtFAD2-2. BMC Plant Biol. 20:233. doi: 10.1186/s12870-020-02441-0
Tian, Y., Liu, X., Fan, C., Li, T., Qin, H., Li, X., et al. (2020b). Enhancement of tobacco (Nicotiana tabacum L.) seed lipid content for biodiesel production by CRISPR-Cas9-mediated knockout of NtAn1. Front. Plant Sci. 11:599474. doi: 10.3389/fpls.2020.599474
Vanhercke, T., Divi, U. K., El Tahchy, A., Liu, Q., Mitchell, M., Taylor, M. C., et al. (2017). Step changes in leaf oil accumulation via iterative metabolic engineering. Metab. Eng. 39, 237–246. doi: 10.1016/j.ymben.2016.12.007
Vanhercke, T., El Tahchy, A., Liu, Q., Zhou, X. R., Shrestha, P., Divi, U. K., et al. (2014). Metabolic engineering of biomass for high energy density: oilseed-like triacylglycerol yields from plant leaves. Plant Biotechnol. J. 12, 231–239. doi: 10.1111/pbi.12131
Virdi, K. S., Spencer, M., Stec, A. O., Xiong, Y., Merry, R., Muehlbauer, G. J., et al. (2020). Similar seed composition phenotypes are observed from CRISPR-generated in-frame and knockout alleles of a soybean KASI Ortholog. Front. Plant Sci. 11:1005. doi: 10.3389/fpls.2020.01005
Wang, G., Ryu, S., and Wang, X. (2012). Plant phospholipases: an overview. Methods Mol. Biol. 861, 123–137. doi: 10.1007/978-1-61779-600-5_8
Wang, Z. P., Xing, H. L., Dong, L., Zhang, H. Y., Han, C. Y., Wang, X. C., et al. (2015). Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 16:144. doi: 10.1186/s13059-015-0715-0
Wolter, F., Schindele, P., and Puchta, H. (2019). Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 19:176. doi: 10.1186/s12870-019-1775-1
Woo, J. W., Kim, J., Kwon, S. I., Corvalan, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Wu, J., James, D. W., Dooner, H. K., and Browse, J. (1994). A mutant of Arabidopsis deficient in the elongation of palmitic acid. Plant Physiol. 106, 143–150. doi: 10.1104/pp.106.1.143
Wu, X., Liang, Y., Gao, H., Wang, J., Zhao, Y., Hua, L., et al. (2021). Enhancing rice grain production by manipulating the naturally evolved cis-regulatory element-containing inverted repeat sequence of OsREM20. Mol. Plant 14, 997–1011. doi: 10.1016/j.molp.2021.03.016
Wu, N., Lu, Q., Wang, P., Zhang, Q., Zhang, J., Qu, J., et al. (2020). Construction and analysis of GmFAD2-1A and GmFAD2-2A soybean fatty acid desaturase mutants based on CRISPR/Cas9 technology. Int. J. Mol. Sci. 21:1104. doi: 10.3390/ijms21031104
Xiao, Y., Karikari, B., Wang, L., Chang, F., and Zhao, T. (2021). Structure characterization and potential role of soybean phospholipases a multigene family in response to multiple abiotic stress uncovered by CRISPR/Cas9 technology. Environ. Exp. Bot. 188:104521. doi: 10.1016/j.envexpbot.2021.104521
Xie, X., Meesapyodsuk, D., and Qiu, X. (2019). Enhancing oil production in Arabidopsis through expression of a ketoacyl-ACP synthase domain of the PUFA synthase from Thraustochytrium. Biotechnol. Biofuels 12:172. doi: 10.1186/s13068-019-1514-8
Xie, K., Minkenberg, B., and Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U. S. A. 112, 3570–3575. doi: 10.1073/pnas.1420294112
Xie, N., Zhou, Y., Sun, Q., and Tang, B. (2018). Novel epigenetic techniques provided by the CRISPR/Cas9 system. Stem Cells Int. 2018, 1–12. doi: 10.1155/2018/7834175
Xu, C., and Shanklin, J. (2016). Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu. Rev. Plant Biol. 67, 179–206. doi: 10.1146/annurev-arplant-043015-111641
Xu, X., Vanhercke, T., Shrestha, P., Luo, J., Akbar, S., Konik-Rose, C., et al. (2019). Upregulated lipid biosynthesis at the expense of starch production in potato (Solanum tuberosum) vegetative tissues via simultaneous downregulation of ADP-glucose Pyrophosphorylase and sugar Dependent1 expressions. Front. Plant Sci. 10:1444. doi: 10.3389/fpls.2019.01444
Xu, X.-Y., Yang, H.-K., Singh, S. P., Sharp, P. J., and Liu, Q. (2018). Genetic manipulation of non-classic oilseed plants for enhancement of their potential as a biofactory for triacylglycerol production. Engineering 4, 523–533. doi: 10.1016/j.eng.2018.07.002
Yamaguchi, T., Kuroda, M., Yamakawa, H., Ashizawa, T., Hirayae, K., Kurimoto, L., et al. (2009). Suppression of a phospholipase D gene, OsPLDβ1, activates defense responses and increases disease resistance in rice. Plant Physiol. 150, 308–319. doi: 10.1104/pp.108.131979
Yang, W., Simpson, J. P., Li-Beisson, Y., Beisson, F., Pollard, M., and Ohlrogge, J. B. (2012). A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference, and evolution. Plant Physiol. 160, 638–652. doi: 10.1104/pp.112.201996
Yarra, R., Cao, H., Jin, L., Mengdi, Y., and Zhou, L. (2020). CRISPR/Cas mediated base editing: a practical approach for genome editing in oil palm. 3. Biotech 10:306. doi: 10.1007/s13205-020-02302-5
Yuan, M., Zhu, J., Gong, L., He, L., Lee, C., Han, S., et al. (2019). Mutagenesis of FAD2 genes in peanut with CRISPR/Cas9 based gene editing. BMC Biotechnol. 19:24. doi: 10.1186/s12896-019-0516-8
Zaplin, E. S., Liu, Q., Li, Z., Butardo, V. M., Blanchard, C. L., and Rahman, S. (2013). Production of high oleic rice grains by suppressing the expression of the OsFAD2-1 gene. Funct. Plant Biol. 40, 996–1004. doi: 10.1071/FP12301
Zhang, G., Bahn, S. C., Wang, G., Zhang, Y., Chen, B., Zhang, Y., et al. (2019a). PLDα1-knockdown soybean seeds display higher unsaturated glycerolipid contents and seed vigor in high temperature and humidity environments. Biotechnol. Biofuels 12:9. doi: 10.1186/s13068-018-1340-4
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7:12617. doi: 10.1038/ncomms12617
Zhang, J., Liu, H., Sun, J., Li, B., Zhu, Q., Chen, S., et al. (2012). Arabidopsis fatty acid desaturase FAD2 is required for salt tolerance during seed germination and early seedling growth. PLoS One 7:e30355. doi: 10.1371/journal.pone.0030355
Zhang, K., Nie, L., Cheng, Q., Yin, Y., Chen, K., Qi, F., et al. (2019b). Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus L.) using CRISPR-Cas9 system. Biotechnol. Biofuels 12:225. doi: 10.1186/s13068-019-1567-8
Zhao, J., Wang, C., Bedair, M., Welti, R., Sumner, L. W., Baxter, I., et al. (2011). Suppression of phospholipase Dγs confers increased aluminum resistance in Arabidopsis thaliana. PLoS One 6:e28086. doi: 10.1371/journal.pone.0028086
Zhou, X. R., Shrestha, P., Yin, F., Petrie, J. R., and Singh, S. P. (2013). AtDGAT2 is a functional acyl-CoA:diacylglycerol acyltransferase and displays different acyl-CoA substrate preferences than AtDGAT1. FEBS Lett. 587, 2371–2376. doi: 10.1016/j.febslet.2013.06.003
Keywords: acyltransferase, CRISPR/Cas9, FAD2, FAE1, FATB, KASI, lipase, TAG
Citation: Park M-E and Kim HU (2022) Applications and prospects of genome editing in plant fatty acid and triacylglycerol biosynthesis. Front. Plant Sci. 13:969844. doi: 10.3389/fpls.2022.969844
Edited by:
Anca Macovei, University of Pavia, ItalyReviewed by:
Rupam Kumar Bhunia, National Agri-Food Biotechnology Institute, IndiaNikolaos Tsakirpaloglou, Texas A&M University, United States
Copyright © 2022 Park and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyun Uk Kim, hukim64@sejong.ac.kr
 Mid-Eum Park
Mid-Eum Park