- 1Bioversity International, Parc Scientifique Agropolis II, Montpellier, France
- 2CIRAD, UMR AGAP Institut, Montpellier, France
- 3UMR AGAP Institut, Univ Montpellier, CIRAD, INRAE, Institut Agro, Montpellier, France
- 4Bioversity International, International Transit Centre, Leuven, Belgium
- 5Papua New Guinea (PNG) National Agricultural Research Institute, Southern Regional Centre, Laloki, Port Moresby, Papua New Guinea
Hybridization and introgressions are important evolutionary forces in plants. They contribute to the domestication of many species, including understudied clonal crops. Here, we examine their role in the domestication of a clonal crop of outmost importance, banana (Musa ssp.). We used genome-wide SNPs generated for 154 diploid banana cultivars and 68 samples of the wild M. acuminata to estimate and geo-localize the contribution of the different subspecies of M. acuminata to cultivated banana. We further investigated the wild to domesticate transition in New Guinea, an important domestication center. We found high levels of admixture in many cultivars and confirmed the existence of unknown wild ancestors with unequal contributions to cultivated diploid. In New Guinea, cultivated accessions exhibited higher diversity than their direct wild ancestor, the latter recovering from a bottleneck. Introgressions, balancing selection and positive selection were identified as important mechanisms for banana domestication. Our results shed new lights on the radiation of M. acuminata subspecies and on how they shaped banana domestication. They point candidate regions of origin for two unknown ancestors and suggest another contributor in New Guinea. This work feed research on the evolution of clonal crops and has direct implications for conservation, collection, and breeding.
Introduction
Domestication holds a special place in the long trajectory of plants evolution that spans over hundreds of millions of years (Morris et al., 2018). Domesticates have indeed emerged in the last 12.000 years as results of co-evolutionary interactions between plants and human populations (Meyer and Purugganan, 2013). This recent evolutionary history – with regards to plant life on earth - resulted in major phenotypic changes in crops and have fascinated biologists since the early beginning of evolutionary studies (Darwin, 1869). Domesticated plants have thus been widely used for developing and testing evolutionary theories (Ross-Ibarra et al., 2007; Meyer and Purugganan, 2013; Turcotte et al., 2014; Gaut, 2015). In addition, the increasing pressure on environment and on food systems pushed research towards a better understanding of the mechanisms underlying the transition from wild, often inedible, plants to high yielding nutritious crops. Logically, studies focusing on unravelling the origins and trajectories of crop species increased in the last decades.
Hybridization, which in plants is a starting point for hybrid speciation and enables the introduction of adaptive variation through introgressions (Soltis and Soltis, 2009; Abbott et al., 2016), appears to also be an important evolutionary force in domesticates. Advances in genetics and genomics have highlighted the prominent roles of hybridization, and of introgression in the creation of plant domesticates, and in their diversification (reviews in Arnold, 2004; Purugganan, 2019). Some of the most important crops on Earth are indeed hybrids between two or more species, such as wheat (Baidouri et al., 2017), sugarcane (Pompidor et al., 2021) or strawberry (Feng et al., 2021). Introgressions from related species, such as in sunflower (Baute et al., 2015), or from different genepools of same species, such as in maize (Gonzalez-Segovia et al., 2019) and Asian rice (Santos et al., 2019), also have contributed to shape domesticates diversity.
In the global picture of domestication studies, vegetatively propagated crops hold a special place, especially since their evolutionary history was long under-considered. However, many clonal crops, in addition to be of high economic importance, are also critical for food security in many developing countries. This is notably the case of cassava (Manihot esculenta Crantz), sweet potato (Ipomoea batatas L.), potato (Solanum tuberosum L.), yams (Dioscorea spp.) or bananas (Musa ssp.). The clonal nature of these crops has indeed led to the over-simplified assumption that most of them were resulting from the capture and clonal multiplication of interesting wild genotypes (McKey et al., 2010). However, a few in depth studies provided interesting insights, highlighting for some species the regular occurrence of sexual reproduction despite their vegetative mode of propagation, as in Ensete ventricosum (Shigeta, 1996), cassava (Pujol et al., 2005; Sardos et al., 2008) and yams (Dioscorea spp.) (Chaïr et al., 2010), or a hybrid nature for other species (Chaïr et al., 2016; Feng et al., 2021; Pompidor et al., 2021). Introgressions, who contributed to local adaptations were also identified, such as in apple (Cornille et al., 2012; Sun et al., 2020), and sometimes revealing complex schemes of crossing, such as in some citrus species (Wu et al., 2018; Ahmed et al., 2019). These studies also confirmed that not only the vegetatively propagated crops were under-studied, but their wild relatives were too (McKey et al., 2010), with the striking example of the greater yam (D. alata L.), a species of high importance in Africa and the Pacific, and for which no wild ancestral population was identified yet (Chaïr et al., 2016).
Banana (Musa spp.) is a vegetatively propagated crop native to a wide South-East Asia/Oceania region (Simmonds, 1962). It was domesticated from Musa acuminata (A genome), a wild species belonging to the monocots, more than 7,000 years ago, likely in New Guinea island (Denham et al., 2003). In this crop, the main traits selected during the wild-to-domesticate transition are parthenocarpy, i.e. the ability to set fruits without the need of prior pollination, and sterility (Simmonds, 1962; Denham et al., 2020). Together, parthenocarpy and sterility ensure the production of edible fleshy fruits that are free of seeds. They also make banana breeding quite challenging, as it is difficult to produce high-quality sterile and parthenocarpic improved varieties through recombination of fertile and non-parthenocarpic parents. Banana breeding is further hampered by the multiple levels of ploidy that can be found in cultivars (diploids, triploids, and tetraploids) and that reduce the number of potential diploid parents for crosses. In such context, the characterization of the diploid germplasm available and the better understanding of the wild-to-domesticate transition is key for the success of breeding schemes.
Cultivated bananas currently encompasses a wide diversity of cultivars of both dessert and cooking types. This diversity includes cultivars of pure M. acuminata ancestry and inter-specific hybrids between M. acuminata and a few other species (Heslop-Harrison and Schwarzacher, 2007). The simplistic view that interesting genotypes were captured and then conserved clonally does not apply to the domestication of bananas. First, back-crossing was confirmed in the making of interspecific hybrids (Baurens et al., 2019; Cenci et al., 2021). Second, intra-specific hybridization also occurred between different subspecies of M. acuminata (Carreel et al., 2002; Perrier et al., 2011), leading sometimes to complex genomic structures organized in sub-genomes mosaics (Martin et al., 2020b), and rising substantial questions on the adaptative traits and selective advantages provided by the introgressions of the different sub-species of M. acuminata involved in the setup of cultivars.
Musa acuminata is a complex of sub-species that are geographically segregated across a gradient spanning from East India and Sri Lanka to Papua New Guinea, and the northern tip of Queensland in Australia. Subspecies show unique features, morphologically (Simmonds, 1956), genetically (Hippolyte et al., 2012) and at the genome level (Martin et al., 2020a). The New Guinea subspecies M. acuminata ssp. banksii had a prominent role in the domestication of bananas and is believed to be the genepool at the origin of parthenocarpy (Simmonds, 1956). Four other subspecies are long known to have also contributed to the genetic set up of cultivated bananas: ssp. burmannica/siamea that is found from southern India and Sri Lanka to Cambodia, ssp. malaccensis located in the Malayan peninsula, ssp. zebrina in Java and ssp. errans in the Philippines (Carreel et al., 2002; Perrier et al., 2011). However, recent studies of small sets of banana cultivars with pure M. acuminata ancestries revealed the existence of several ancestral genepools contributing to the setup of banana cultivars and that are missing in the current representation of wild M. acuminata diversity (Martin et al., 2020b; Jeensae et al., 2021). Where these undefined genepools originated, how much they contributed to the diversity of banana cultivars, and what were their roles in the domestication and diversification of bananas is not known.
In the present paper, we used SNP markers generated by Restriction site associated DNA sequencing (RADseq; Davey and Blaxter, 2010) to explore the geographical patterns of diversity in a wide set of diploid bananas composed of 226 M. acuminata accessions and M. acuminata derived diploid cultivars obtained from genebanks and collecting missions. We aimed at (1) determine the diversity, distribution, and geographical radiation of M. acuminata in its natural range, (2) assess the contribution of the undefined genepools to the diversity of banana cultivars and identify candidate regions for their origins, and (3) clarify the processes of the transition between wild and domesticated plants by comparing the genetic diversity patterns of wild M. acuminata ssp. banksii and derived cultivated diploid bananas in New Guinea. This work has impact for Musa spp. genetic resources conservation and collection, banana breeding, and evolutionary biology research on vegetatively propagated crops.
Materials and methods
Plant materials
A set of 226 diploid banana accessions was selected (Table 1; Supplementary Table S1, Supplementary Material online). This set comprised 68 wild accessions belonging to subspecies of M. acuminata, 154 related edible diploid cultivars, three accessions of M. schizocarpa considered as outgroup and a hybrid between M. acuminata ssp. banksii and M. schizocarpa. These materials were provided by the ITC (170 samples Musa Germplasm Information System (MGIS - https://www.crop-diversity.org/mgis/) (Ruas et al., 2017), the banana collecting mission to the AROB (25 samples) (Sardos et al., 2018), CIRAD (24 samples) (Perrier et al., 2019), collecting missions to Indonesia (4 samples) (Sutanto et al., 2016) and EMBRAPA (3 samples).
Restriction-site-associated DNA sequencing
DNA from each accession was extracted following a 2X CTAB protocol (modified from Doyle and Doyle, 1990). Library for restriction-site-associated DNA sequencing (RADSeq; Davey & Blaxter, 2010) was built with the PstI restriction enzyme. The 300–500 short-insert libraries were sequenced with 91 bp paired-end reads using Illumina HiSeq2000 (Illumina, San Diego, CA, USA) by BGI Hong Kong. At BGI, the raw data were modified with the following two steps: (1) reads polluted by adapter sequences were deleted; and (2) reads that contained >50% low-quality bases (quality value ≤5) or >10% N bases were removed.
Read processing and SNP calling
Reads contained in raw FASTQ files (one per sample) were checked using FastQC and then cleaned to remove Illumina adapter sequences and low-quality ends (Phred score > 20) with Cutadapt (Martin, 2011). After trimming, reads inferior to 30 bp were discarded. Reads were then aligned against the Musa acuminata genome v2 downloaded on the Banana Genome Hub (Droc et al., 2013) using BWA-MEM (Li and Durbin, 2010). Re-alignment was done with the IndelRealigner module from GATK v4.1 (McKenna et al., 2010). We then followed the GATK pipeline recommended for a non-model organism by adding the recalibration step. It consisted of performing an initial round of SNP calling on the original uncalibrated data, selecting the SNPs with the highest confidence, and then executing a round of base recalibration on the original mapped reads files. The SNP calling was done with the GATK module HaplotypeCaller v4.1 to call SNPs and indels. Considering inter-sample variation, the SNP calling was done on all samples simultaneously. The pipeline used to perform those analyses is available at https://github.com/CathyBreton/Genomic_Evolution.
Genetic diversity analyses
For the initial set of 226 accessions, SNPs were filtered for missing data (5% as maximum allowed) and MAF (1%), yielding a total of 39,031 bi-allelic SNPs. At this stage, eight accessions were discarded as having more than 15% of missing data.
A dissimilarity matrix was calculated following the Simple-Matching index with a minimum of 70% of common sites between each pair of remaining individuals with DARwin 6 (Perrier and Jacquemoud-Collet, 2006). A weighted Neighbor-Joining (NJ) tree rooted using M. schizocarpa as outgroup was then constructed. This first tree allowed identifying cultivated accessions exhibiting identical or nearly identical genotypes and corresponding to duplicates or clonal varieties. To avoid potential bias in further analyses due to genotypes redundancy, we then allowed the presence of a single accession per Genotype Clusters, further reducing the set of accessions to 158 individuals. For each Genotype Clusters, selection of the unique representative kept for further analyses was based on the lowest rate of missing data per accession. For this subset of 158 representative accessions, we retrieved a new set of SNPs with a minor allele frequency of 0.01 (1%) or greater and allowing a maximum of 10% of missing data. With the 66,481 SNPs obtained, DARwin 6 was used to calculate a simple-matching distance matrix. A new weighted neighbour-joining tree rooted on M. schizocarpa was then constructed on the set of 158 accessions, eliminating potential distortion due to duplicated accessions.
Global population structure
Using VCFtools (Danecek et al., 2011), we generated a set of SNPs evenly distributed every 100 kb and not allowing missing data to reflect all chromosomal regions. This set involved 1278 SNPs used to investigate the structure of the 158 accessions of the pruned dataset. We used a Bayesian Markov Chain Monte Carlo (MCMC) approach implemented in the program STRUCTURE v2.3 (Pritchard et al., 2000). The admixture model with the assumption of correlated allele frequencies between groups (Falush et al., 2003) was chosen and 5 replicates of each value of k ranging from 1 to 15 were run with a burn-in-length of 50,000 followed by 150,000 iterations of each chain. The most likely true of the values of k was determined by examining DeltaK, an ad hoc quantity related to the second order rate of change of the log probability of data with respect to the number of clusters (Evanno et al., 2005) and plotted using STRUCTURE HARVESTER (Earl and vonHoldt, 2012). STRUCTURE was then run again following the same model for the best values of K identified with 5 replicates each and a burn-in length of 200,000 followed by 800,000 iterations of each chain.
Tests for introgression in cultivated bananas (AA)
The four taxon Patterson’s D test (Green et al., 2010; Durand et al., 2011) was developed to detect introgressions in closely related taxa. It considers an ancestral “A” allele and a derived “B” allele across the genome of four taxa with a tree topology (((P1,P2),P3),O). Under the hypothesis “without introgression” the two allelic patterns at the tip of the tree, “ABBA” or “BABA”, occur with equal frequency. An excess of “ABBA” or “BABA”, reflected by a D-statistic significantly different from zero, indicates potential gene flow between P2 and P3 or P1 and P3, respectively. Here, we used the derived statistic fd that is a more conservative estimator of introgression developed for small number of SNPs (Martin et al., 2015) implemented in https://github.com/simonhmartin/genomics_general. A R script allowed the calculation of the P-value based on jackknife for the null hypothesis that fd is 0. The procedure is available at https://github.com/CathyBreton/Genomic_Introgression_ABBA_BBAA_Test.
Two tests were performed. In the first one, 31 Papuan edible AAs closely related to the Papuan wild M. acuminata ssp. banksii were tested for introgression by the subspecies originating in SEA. In the second one, 23 edible AA from SEA were tested for introgression by the Papuan M. acuminata ssp. banksii. Accessions selected as representative for each wild taxon are presented in Supplementary Tables 2, 3.
Population differentiation between wild and cultivated Papuan bananas
Based on STRUCTURE outputs, we considered a sub-cluster of 31 AA from PNG with a M. acuminata ssp. banksii genomic background over 90%. This population and its wild counterpart are represented in our samples by 31 and 24 accessions respectively. A set of 238,357 SNPs was retrieved from GIGWA (Sempéré et al., 2019; Rouard et al., 2022) allowing a maximum of 50% of missing data for each of the two populations. Using VCFtools, we first assessed observed heterozygosity (Ho) and inbreeding coefficient (F) for each of these accessions. Then, considering 200 kb windows exhibiting more than one SNPs, we calculated the nucleotide diversity (π) and Tajima’s D for each population. Finally, we calculated weighted Fst between the cultivated accessions and their wild relative, genome-wide and for 200 kb windows along chromosomes. To better understand the nature of selection in the cultivated, we then considered the 1% lowest and highest Tajima’s D values and the 1% greater Fst values.
Results
Diversity analysis
The dissimilarity matrix and the NJ Tree obtained on the whole sample set comprising 226 individuals (Table 1 and Supplementary Figure 1) allowed the identification of 158 distinct genotypes including 26 genotype clusters (GC) and 132 unique genotypes (Supplementary Table 1). In the NJ tree constructed on this pruned dataset, the subspecies of M. acuminata: banksii, malaccensis, zebrina and burmannica/siamea formed segregated clusters. Three out of the four M. acuminata accessions (AMB007, AMB008 and Sup4), collected in Maluku islands (Indonesia, west of NG island), clustered at the margin of ssp. banksii while the fourth one AMB004 clustered within ssp. banksii from New Guinea. The philippino M. acuminata ssp. errans, represented by the accessions ‘errans’ and ‘UPLB’ (initially classified as ssp. banksii) also clustered at the margin of ssp. banksii, along with ‘Borneo’ classified as ssp. microcarpa and collected in Borneo. The subspecies sumatrana and truncata from Sumatra and the Malay Peninsula respectively, clustered with ssp. malaccensis while the seeded plants collected in East Africa clustered with ssp. zebrina (Figure 1A).
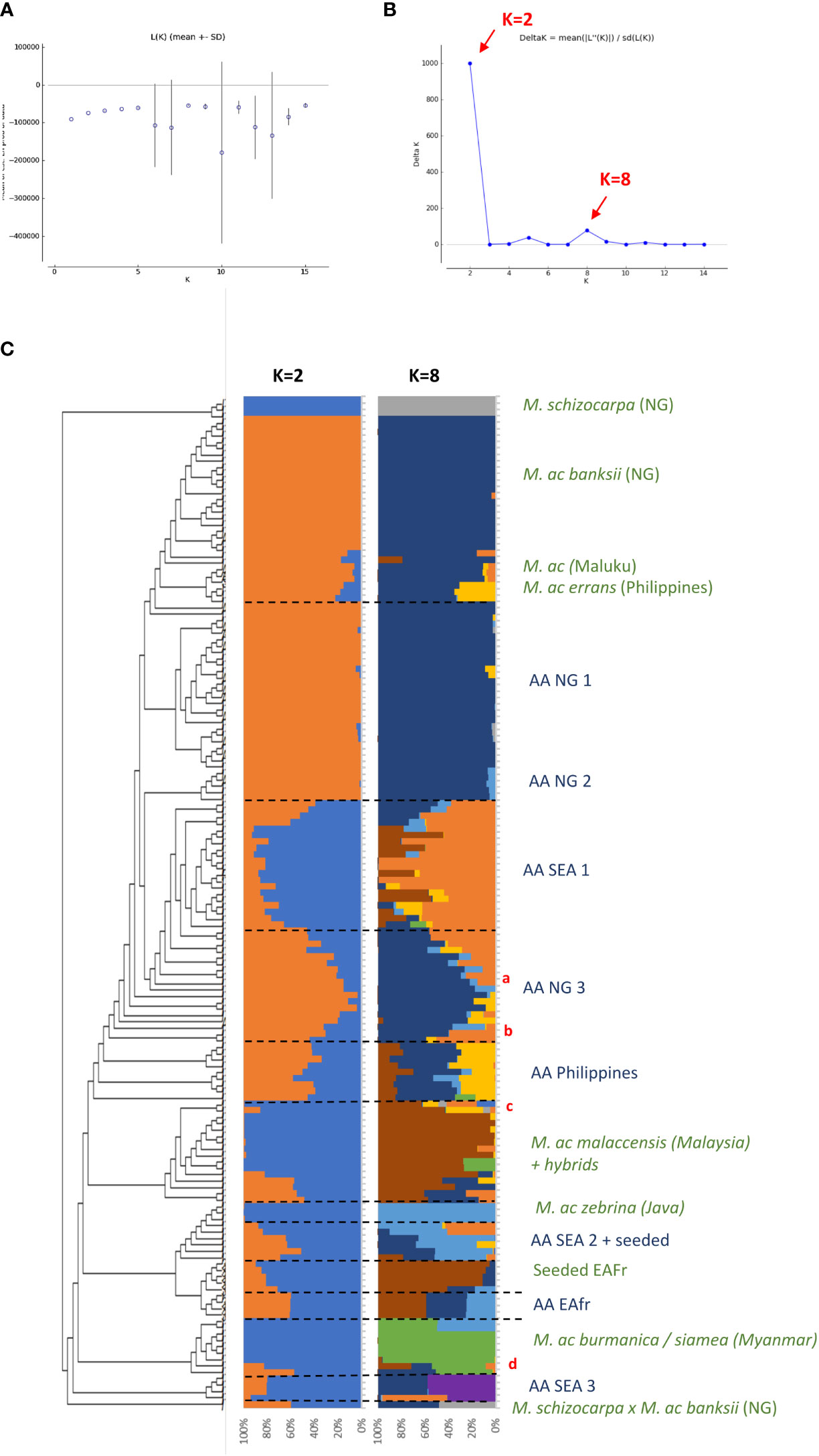
Figure 1 (A, B) present the results of the Bayesian clustering of the diploid bananas pruned sample (158 accessions) using STRUCTURE v2.3 (Pritchard et al., 2000) evaluated using STRUCTURE HARVESTER (Earl and vonHoldt, 2012) and based on the lnP(D)/K and DeltaK, respectively. (C) Presents the global genetic structure of the pruned sample of 158 genotypes. The cladogram was obtained from the NJ tree constructed with DARwin 6 (Perrier and Jacquemoud-Collet, 2006) on the Simple-Matching distance matrix calculated on 66,481 biallelic SNPs and using FigTree v1.4.3 (Rambaut, 2006-2016) and the R package ape (Paradis and Schliep, 2019). Bar plots represent STRUCTURE outputs for K=2 and K=8 as inferred from 1,278 SNPs distributed evenly across the genome, each bar corresponds to a genotype and colours correspond to the detected ancestral genepools. EAfr: East Africa; SEA: South-East Asia; NG: New-Guinea Island; a: ITC0299 ‘Guyod’ from the Philippines, b: ITC0447 ‘Pu-Te Wey’ from Malaysia, c: ITC1701 M. acuminata ssp. sumatrana from Sumatra and ITC0393 M. acuminata ssp. truncata from Malaysia, d: ITC1761 ‘Matti’ from India and ITC0610 ‘Tuu Gia’ from Vietnam.
Cultivated AA accessions are spread over the different clusters (Figure 1A). A first cluster composed only of edible AA accessions from New Guinea island (‘AA NG 1’) is tightly linked to the ssp. banksii cluster. A group of accessions from South-East Asia (‘AA SEA 2’), mixed with various seeded hybrids, clusters with ssp. zebrina, such as seeded and edible AA bananas from East Africa (‘Seeded EAfr’ and ‘AA EAfr’). Two accessions, ‘Matti’ and ‘Tuu Gia’ from India and Vietnam respectively, clustered with ssp. burmannica/siamea. The nine accessions from the Philippines form two clusters that are not linked to any wild representative, such as two groups of accessions from South-East Asia. The first one (‘SEA 1’) is located at the center of the tree while the second one (‘SEA 3’), composed of accessions classified as Pisang Jari Buaya and of an accession named ‘Pisang Madu’, clusters at the margin of the tree. Two additional groups of edible AA bananas from New Guinea (‘AA NG2’ and ‘AA NG3’) are spread between the ssp. banksii and the South-East Asian subspecies of M. acuminata.
Population structure
The two best values of k identified by STRUCTURE in the pruned dataset were k=2 and k=8 (Figures 1B, C). For k=2, the Bayesian analysis recognized two genepools corresponding roughly to New Guinea island and South-East Asia with a high number of admixed accessions. For k=8, this analysis confirmed discrete genepools for the M. acuminata taxa banksii, malaccensis, zebrina, burmannica/siamea and for M. schizocarpa. We noted two accessions from Thailand originally classified as ssp. malaccensis that seem to also hold some ssp. burmannica/siamea signature in their genome (‘Pa (Musore) n°2’ with Q burmannica/siamea = 27,1% and ‘THA018’ with Q burmannica/siamea = 27,3%). In addition, the genomic composition inferred for ‘truncata’ and ‘sumatrana’ accessions – belonging to eponymous subspecies - revealed patchworks of different genepools with a malaccensis dominance in both. However, their respective genomic profiles are slightly different (Figures 1A, 2).
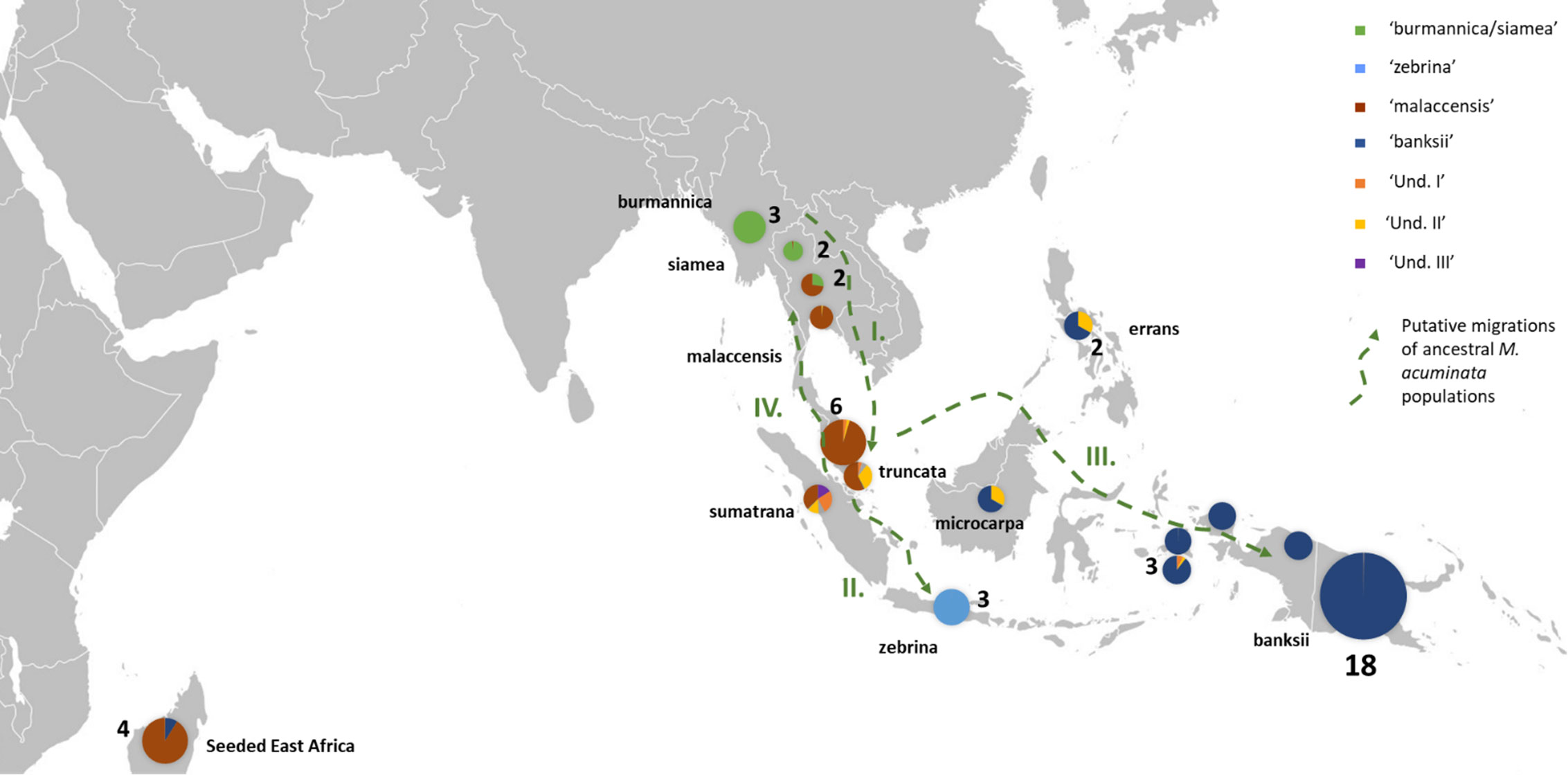
Figure 2 Distribution of the M. acuminata accessions of the sample. The pie charts illustrate genomic background (as inferred by STRUCTURE for k=8) and the numbers indicate the number of accessions sharing similar patterns. Putative dispersal roads of ancestral populations are represented by green dashed arrows. I. First dispersion from mainland South-East Asia towards the Malayan Peninsula and Sumatra, followed by II. Dispersal of populations to Java, III. dispersal to Borneo, the Philippines and New Guinea Island and IV. Secondary colonization of mainland South-East Asia. ‘Und.I’, ‘Und. II’ and ‘Und. III’ correspond to genepools for which no parental populations were identified in the samples.
Three undefined genepools, i.e. for which unadmixed individuals from the source populations are absent in the sample, were detected. The first one (denoted ‘Und-I’) (orange colored in Figures 1A, 2, 3) was very common in cultivated accessions. Three cultivars from Thailand and clustering within ‘AA SEA1’, are fully assigned (QUnd-I > 90%) to the Und-I genepool (‘Thong Dok Mak’, ‘Kluai Lep Mu Nang’ and ‘Sa’). The ‘Und-I’ alleles were detected in many cultivated accessions from SEA and in a few accessions of ‘AA NG3’. It was also inferred as introgressions in some wild specimens spread in different clusters, such as ‘sumatrana’ (QUnd-I = 26%) and three hybrids collected in Indonesian part of New Guinea in the 1960’s and clustering with ssp. malaccensis (‘Higa’ and ‘Hybrid’) and with ssp. banksii (‘Waigu’).
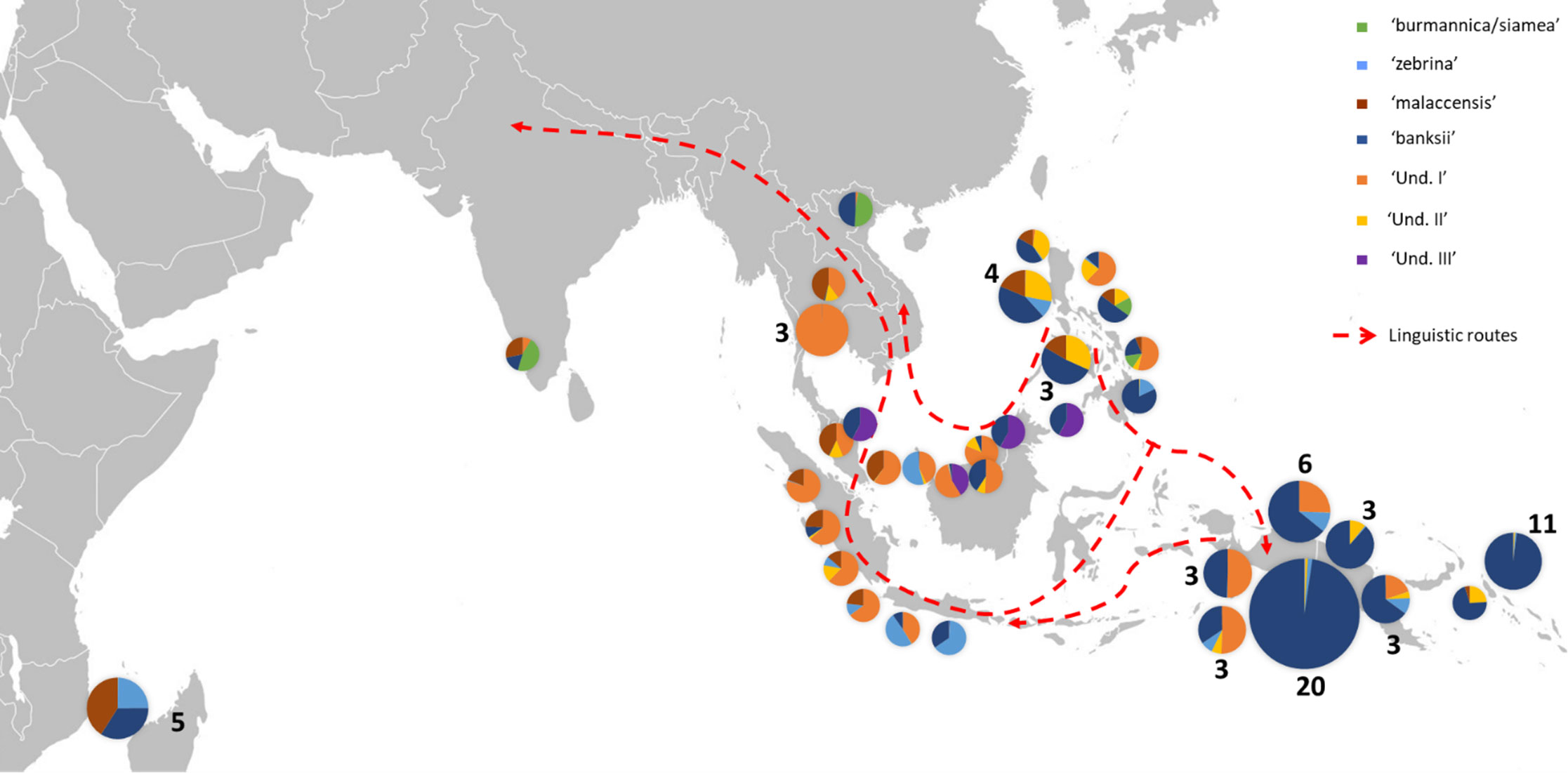
Figure 3 Distribution of cultivated AA accessions of the samples. The pie charts illustrate genomic background (as inferred by STRUCTURE for k=8) and the numbers indicate the number of accessions sharing similar patterns. Dashed red lines show linguistic paths for the words ‘banana’ in the region as inferred by Perrier et al. (2011). The historical movements of these terms reflect ancient spread of Musa cultivars in the region.
The two other undefined genepools were inferred as introgressions. The genepool ‘Und-II’ was detected in the two ssp. errans specimen and in the accession ‘Borneo’. These three accessions share similar profiles along with a partially common genetic background with ssp. banksii. ‘Und-II’ alleles were also detected in some of the cultivated accessions including the two ‘AA Philippines’ clusters (yellow color in Figure 1A). The third undefined genepool ‘Und-III’ was identified in the cluster ‘AA SEA3’ (purple color in Figure 1A) in which accessions were collected in the north coast of Borneo and on Sulu, an island located between Borneo and the main Philippines islands (Figure 3). The genepool ‘Und-III’ was also inferred as a small introgression in the subspecies sumatrana.
The inferred genomic composition of the cultivated AA bananas confirmed the hybrid status of most of them. Musa acuminata ssp. banksii from NG is a prominent contributor to these edible AAs, including those in South-East Asia, followed by the Javanese ssp. zebrina, ssp. malaccensis from the Malayan peninsula and then ssp. burmannica/siamea. The latter ranges from India to north Thailand and is the subspecies contributing the least to the edible samples. Interestingly the two undefined genepools ‘Und-I’ and ‘Und-II’ are important contributors to cultivated diploid bananas. Finally, we noted in this analysis that the 31 cultivated AA accessions in the clusters ‘AA NG1’ and ‘AA NG2’ were inferred with more than 90% of their genome belonging to ssp. banksii (Figures 1C, 3).
Detection of introgressions
Introgressions of South-East Asian subspecies into clusters ‘NG 1’ and ‘NG 2’
To enable testing the 31 cultivated AA accessions from New Guinea with Qbanksii > 90% for introgression by one or more of the six SE Asian subspecies of M. acuminata, we performed 186 tests. Only Patterson’s D tests for which the dominance of the BBAA pattern over ABBA and BABA were considered robust. Following this criteria, ten combinations which all exhibited BABA > BBAA were excluded. Within the remaining 176 tests, Patterson’s D was statistically significantly negative (Z score < -2) for 70 combinations, showing significant excess of BABA sites over ABBA sites and indicating a highest proximity between ssp. banksii and the SEA subspecies tested. At the contrary, for 11 combinations, involving 7 accessions, the D scores obtained were significantly positive (Z score > 2), revealing a significant bias towards ABBA pattern compared with BABA and reflecting possible introgression of given SEA subspecies. We noted that the sub-species zebrina was detected in the 7 accessions and was the only introgressing genepool for 4 of them. The subspecies ssp. burmanica/siamea was detected in 3 accessions. We noticed however that it was always coupled with suspected introgressions from other sub-species, zebrina (3 accessions), sumatrana (2 accession) and malaccensis (1 accession). For 95 combinations tested, D was not significantly departing from 0 (-2 < z-score < 2), therefore not showing significant differences between the counts of ABBA and BABA sites. Among those tests, nine accessions did not showcase any significant differences in the number of ABBA and BABA sites for any of the six SE Asian subspecies tested, suggesting that they may be truly unadmixed cultivated accessions (Supplementary Table 2).
Introgressions of subspecies banksii into South-East Asian cultivated AAs
We also performed Patterson’s D tests on 33 cultivated AA accessions originating in South-East Asia to check for their introgression by M. acuminata ssp. banksii. However, for most of the tests, the count of the different patterns showed topology discordance compared to assumption. That is to say that for eight accessions, both ABBA and BABA counts were dominant over BBAA, that for 18 accessions ABBA counts were dominant over BBAA and for one accession, namely ‘Malaysian Blood’, BABA was dominant over BBAA. For all the six tests for which no topology discordance was identified, statistically significant bias towards ABBA pattern were identified when compared to BABA, suggesting introgression of ssp. banksii in the accessions tested (Supplementary Table 3).
Pattern of differentiation between M. acuminata ssp. banksii and related cultivated AAs in New Guinea
The observed heterozygosity (Ho) of the accessions of the ‘banksii’ cluster collected in New Guinea island (Figure 4) ranged from 0.02 to 0.12. The four accessions collected in the region of Ambon and Seram, at the west of New Guinea island, had two profiles. The accession AMB004 was similar to the accessions from New Guinea (Ho=0.02) while the three others, AMB007, AMB008 and Sup04, had Ho ranging between 0.30 and 0.34. Inbreeding coefficient (F) confirmed significant excess of homozygous sites for the accessions from New Guinea and AMB004 but not for ‘AMB007’, ‘AMB008’ and ‘Sup04’. It is to be noted that none of the accessions collected in Ambon and Seram, including AMB004, was morphologically classified as belonging to the banksii subspecies at collection (Sutanto et al., 2016). In the 31 AA accessions from NG with Qbanksii > 0.90, heterozygosity ranged from 0.20 to 0.34 and inbreeding coefficient was reflecting excess of heterozygous sites for 22 accessions (Table 2).
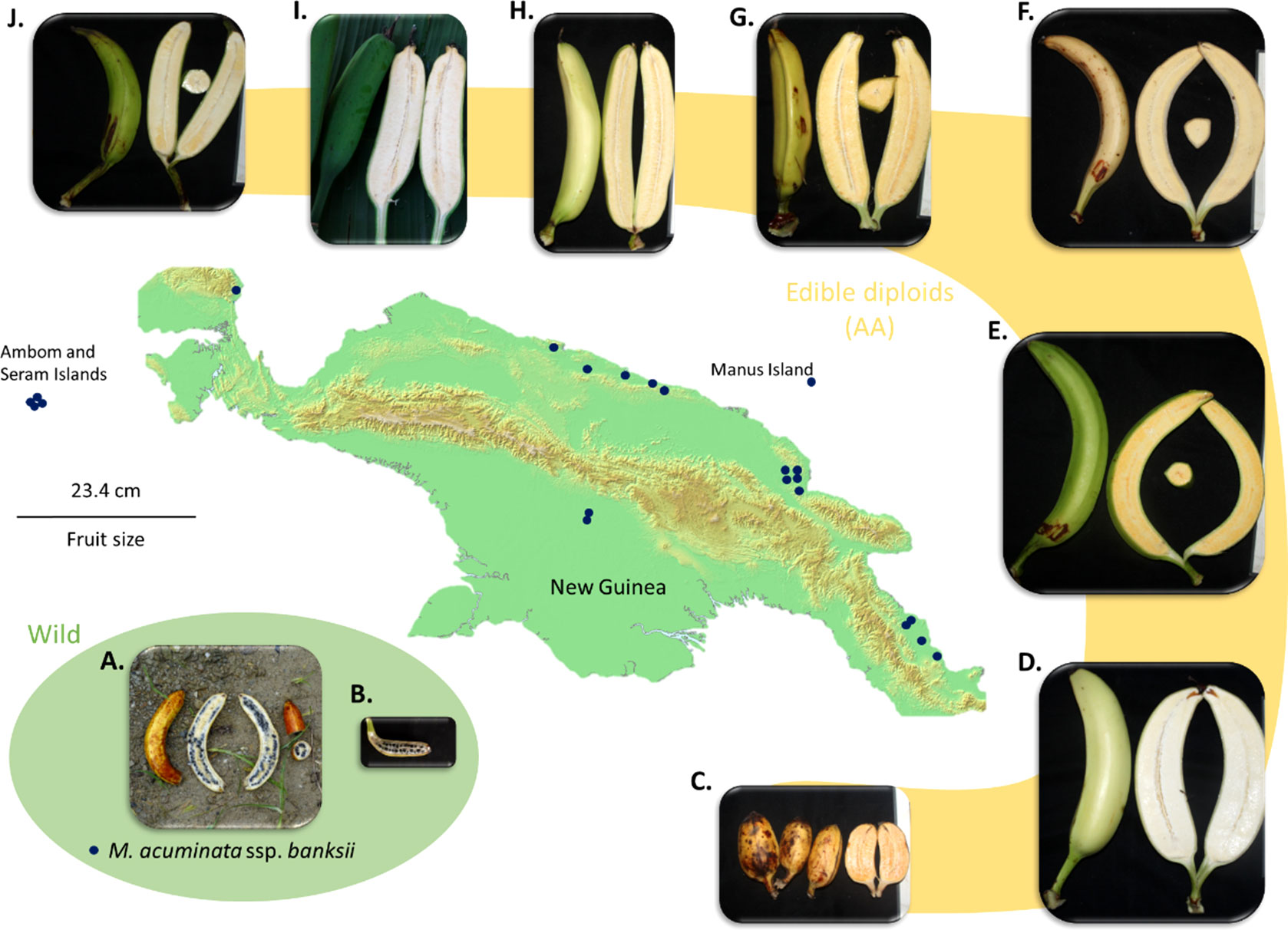
Figure 4 Map of New Guinea Island and examples of fruits of M. acuminata ssp. banksii (A, B) and of closely related cultivated AA accessions from Papua New Guinea (C). AROB003 ‘Mero Mero’, (D) AROB034 ‘Nesuri’, (E) AROB004 ‘Wiau’, (F) AROB035 ‘Talasea’, (G) AROB050 ‘Seseve’, (H) AROB047 ‘Tobaung’, (I) AROB019 ‘Tavilo’, and (J) AROB016 ‘Nape’e’). Dark blue dots on the map represent collection locations for accessions within the cluster ‘banksii’ (according to Sharrock, 1988; Sharrock et al., 1988; Sharrock et al., 1989, Sharrock 1989 and Sutanto et al., 2016). Photos: (A, B) taken by first author, (C-J) courtesy of NARI.
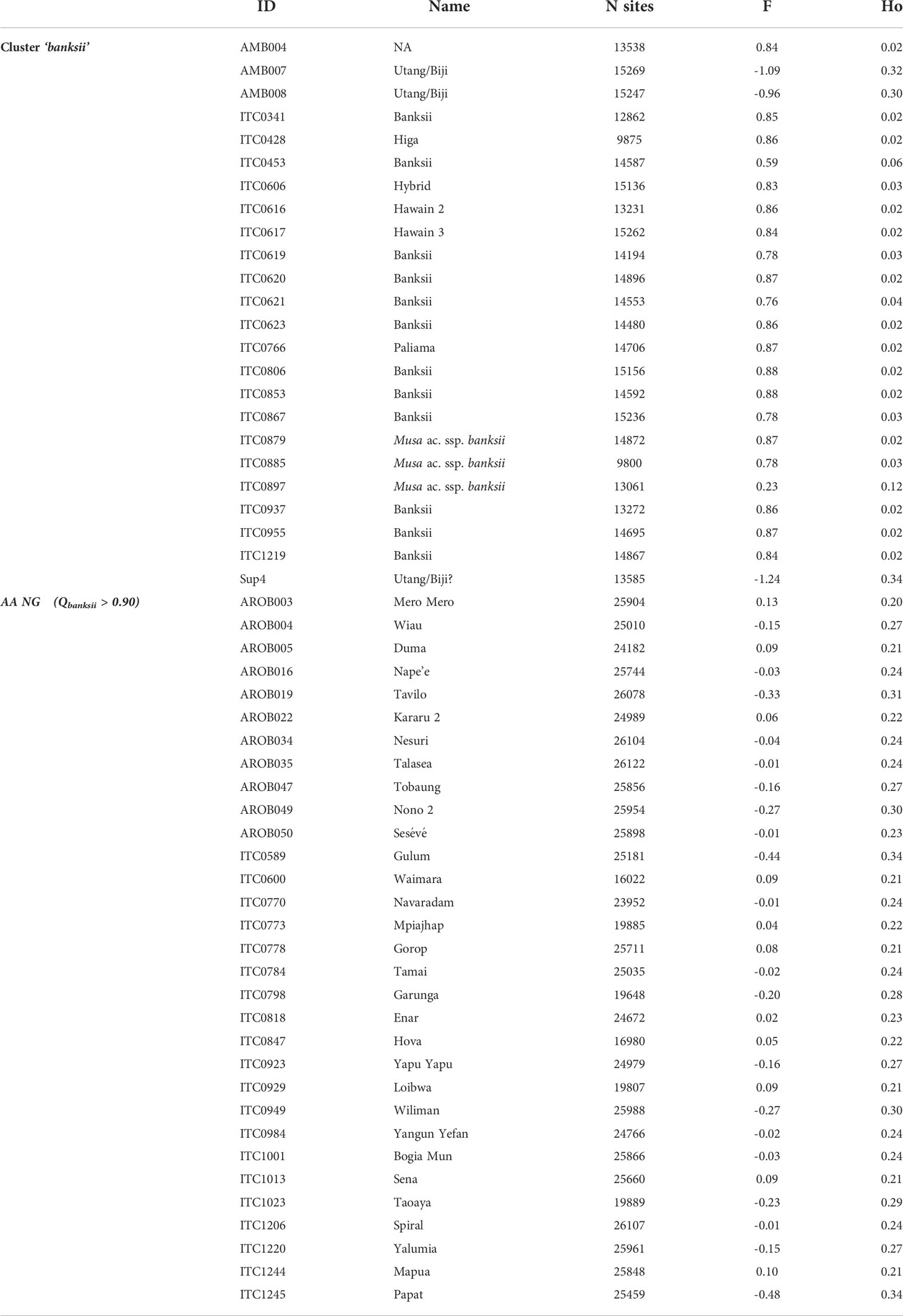
Table 2 Observed heterozygosity (Ho) and inbreeding coefficient (F) for the accessions of the clusters ‘banksii’ and ‘AA NG’.
For 24 accessions assigned to the subspecies banksii, we identified 1059 windows of 200kb size with more than 1 polymorphic SNP. For each window, Tajima’s D values were plotted against nucleotide diversity (π) (Figure 5A). Mean Tajima’s D in these windows was -0.28 (variance = 1.28) and the distribution was skewed towards negative values, reflecting a global excess of low frequency variants (Figures 5A–C), that can be interpreted as a signature for a recent expansion of the population after a bottleneck. The 31 cultivated AA diploids selected have a greater diversity as expressed by nucleotide diversity (π) and the highest number of 200kb windows with more than 1 variable SNP (1502). For these edible AA, mean Tajima’s D parameter was 0.50 and the distribution of the values obtained for the windows was somewhat bimodal (variance = 2.47) with the main peak being largely negative, reflecting excess of low frequency variants. The second peak is largely positive, reflecting excess of common variants (Figures 5B, C). Windows with Tajima’s D below the 1% lower limit (-2.39) and above the 1% upper limit (3.72) for the 31 AA from NG are presented in Table 3.
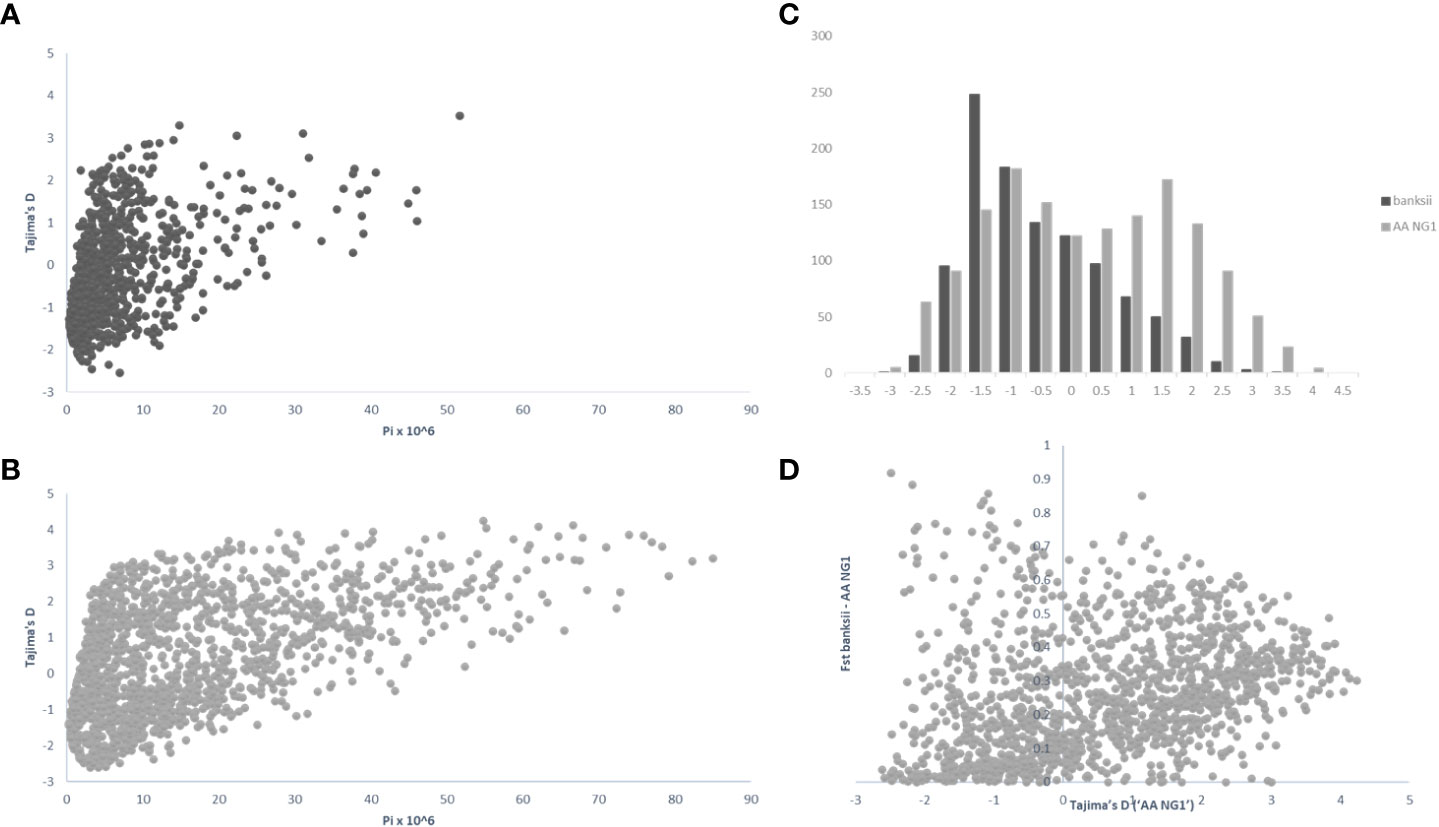
Figure 5 Distribution of pi (π), Tajima’s D and Fst calculated on 200 kb windows along the genomes for the clusters ‘banksii’ and ‘AA NG’ (Qbanksii>90%). Tajima’s D plotted against pi (π) for ‘banksii’ (A) and ‘AA NG’ (B); comparative distribution of Tajima’s D values (C) and Tajima’s D calculated for ‘AA NG’ plotted against Fst between ‘banksii’ and ‘AA NG’ (D).
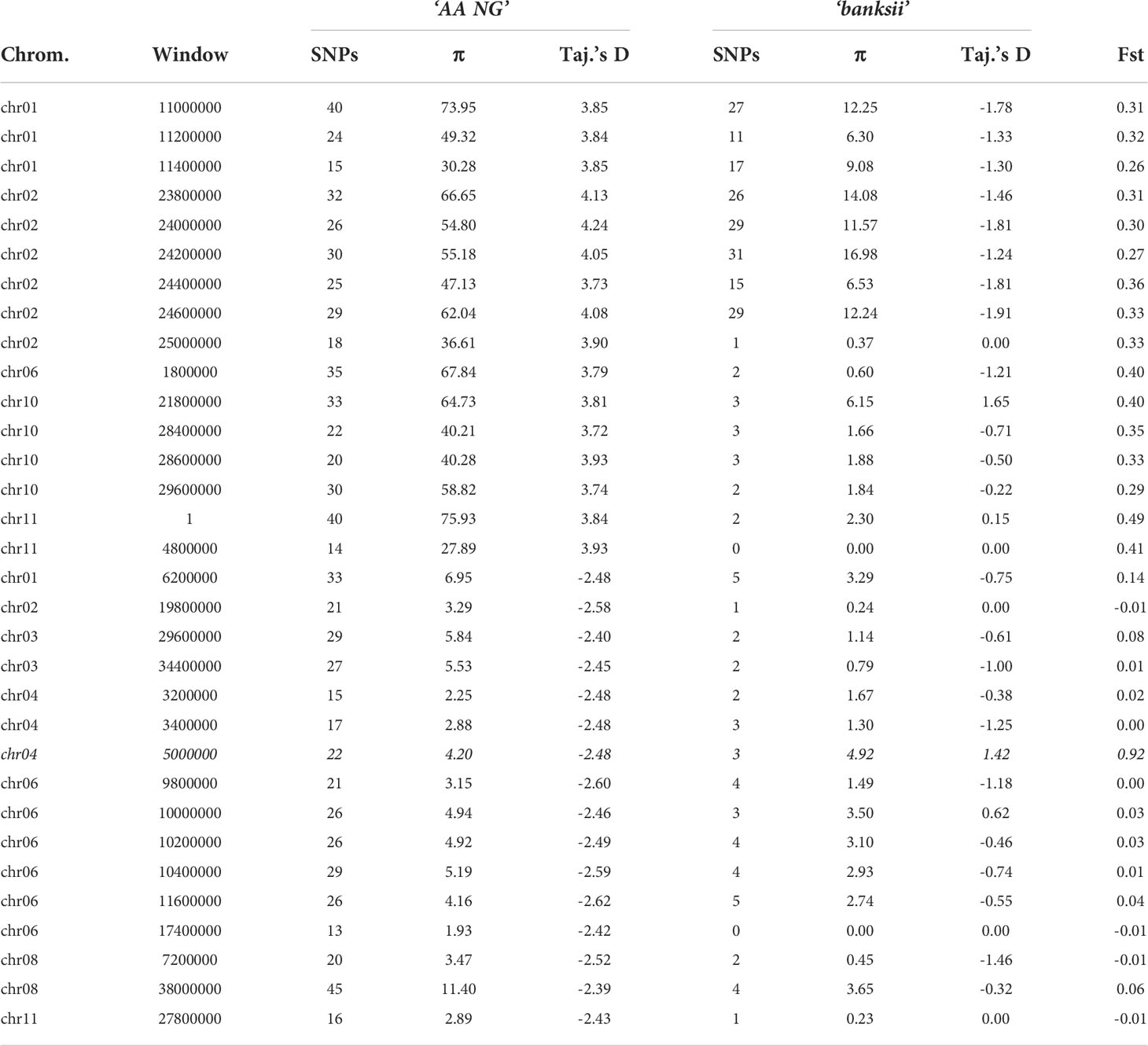
Table 3 Number of SNPs, Pi (π) and Fst between the clusters ‘banksii’ and ‘AA NG’ for the windows (200 kb) with the 1% lowest and 1% highest values of Tajima’s D calculated for ‘AA NG’.
Whole genome Fst calculated between ssp. banksii and the cultivated AA from NG was 0.30. Considering 200 kb windows exhibiting more than 1 polymorphic SNP, the highest Fst value was 0.92 calculated on 25 SNPs on chromosome 4 (bin start 5.000.000). Tajima’s D value for this genomic region was also among the 1% lowest Tajima’s D values calculated along the genome. Given that Tajima’s D values in the 31 AA from NG plotted against Fst shows that most windows with negative Tajima’s D exhibits also low Fst (Figure 5D and Supplementary Table 4), this genomic region on chromosome 4 is likely under strong selection.
Discussion
This study enables proposing hypotheses on the radiation of M. acuminata in South-East Asia and New Guinea and highlighting probable geographic regions of origins for the undefined genepools. Finally, by focusing on New Guinea Island we provide new insights on the wild-to-domesticates transition in the banana crop.
Secondary centers of radiation for M. acuminata are in Sumatra and the Malay peninsula
The geographic distribution of the diversity of the M. acuminata subspecies as detected in this study enables the proposition of a refined dispersal scenario. Musa acuminata arose about 10 million years ago, probably in the northern Indo-Burma region (Janssens et al., 2016). Phylogenetic and phylogeographic studies performed on different markers obtained discordant results on its diversification and its dispersal at the intraspecific level (Janssens et al., 2016; Rouard et al., 2018; Fu et al., 2022). Whole genome sequences of four M. acuminata subspecies, ssp. burmannica, zebrina, banksii and malaccensis, showed ssp. burmannica as the first of the sub-species to have diverged in the species tree (Rouard et al., 2018). This finding suggested that this early divergence occurred in the indo-burmese area, consistently with the inferred origin of M. acuminata in this region (Janssens et al., 2016). These genomic results also suggested secondary diversification and dispersal of the ancestors of the others M. acuminata subspecies through the Malayan peninsula to Java, and then to New Guinea island and back to the southern Indo-Malayan region (Rouard et al., 2018). Using chloroplast sequences, Fu et al. (2022) confirmed the early emergence of ssp. burmannica from the Indo-Burma region, as well as dispersal and emergence of the other subspecies towards Island South-East Asia and New Guinea but with a slightly different pattern. At the east, ssp. banksii emerged in New Guinea on the one hand while ssp. zebrina and spp. microcarpa appeared on Java and Borneo on the other hand. At the west, ssp. malaccensis and ssp. truncata diverged on the Malayan peninsula.
In this study, we confirmed the anticipated background described for the wild samples (Carreel et al., 1994; Perrier et al., 2011; Sardos et al., 2016; Christelova et al., 2017; Martin et al., 2020b) but we detected for the first time admixed profiles for ssp. truncata and for ssp. sumatrana. Although we cannot exclude that their under-representation, only one individual of each present in the set, influences the result, we hypothesize that these patterns result from shared ancestries with the different taxa inferred as introgressions, including the more distant M. schizocarpa. These shared ancestries are consistent with the proposed role of the Malayan peninsula and Sumatra as centers of secondary diversification and radiation for Musa section bananas (Janssens et al., 2016). Interestingly, ssp. microcarpa in Borneo and ssp. errans in the Philippines, which are both tightly linked to M. acuminata ssp. banksii in New Guinea, also share common ancestries with ssp. sumatrana and truncata. This pattern pleads for a dispersal road leading to ssp. banksii through Borneo and the Philippines rather than through Java. It also suggests independent subspeciation of ssp. zebrina in Java. Both ssp. sumatrana and ssp. truncata also share ancestry with ssp. malaccensis. It argues for the recolonization of the north of the peninsula from the secondary radiation center, as inferred from Rouard et al. (2018). Introgressions of ssp. burmannica/siamea into several M. acuminata ssp. malaccensis samples from Thailand confirm genetic contacts between both genepools in the region (Rouard et al., 2018; Martin et al., 2020b). Therefore, based on these results, we propose a refined dispersal scenario for M. acuminata with important secondary centers of radiation in Sumatra and the Malay peninsula from which three dispersion roads are inferred. The first one goes to Java, the second one passes through Borneo and the Philippines towards New Guinea and the third one goes back up to mainland South-East Asia (Figure 2).
Evidence for undefined ancestral genepools in cultivated diploids and their presumed origins
In congruence with Perrier et al. (2011), the genomic constitutions inferred for cultivated AA diploids showed high levels of admixture with patterns that follow the routes of linguistic diffusion in both directions. At the extremes of the species range, the eastern ssp. banksii signature was identified in the sole AA specimen from India and as far as in East Africa, while the Myanmar ssp. burmannica/siamea at the west, which contributes the least to cultivated diploids, was detected as introgressions in accessions from Vietnam and the Philippines (Figures 1A, 3). Our analysis also revealed three genepools for which no reference wild accessions were identified. It is consistent with recent findings of Martin et al. (2020b), who identified two cryptic ancestor populations co-existing within an accession called ‘Pisang Madu’, and of Jeensae et al. (2021), who reported a new genepool in some cultivated bananas. With the presence in our set of the clones ‘Pisang Madu’ and ‘Pisang Mas’, common to each study respectively, we assume ‘Und I’ and ‘Und III’ as the two cryptic ancestors inferred by Martin et al. (2020b) and ‘Und I’ as the unknown genepool discovered by Jeensae et al. (2021).
Our analysis applied to a wide set of accessions highlights the important contribution of the genepool ‘Und I’ to cultivated AA bananas. This genepool was indeed inferred in accessions of all origins, with the noticeable exception of the East Africa region. Based on linguistic and genetic evidence, the early origin of the East Africa bananas was pointed in the southeastern part of Indonesia, in a region between Java, Sulawesi and the western tip of New Guinea island (Perrier et al., 2011; Perrier et al., 2019), which could suggest that ‘Und I’ was not present in this region at this early time. Since the genepool ‘Und I’ is prominent in Thailand, Malaysia and Indonesia, we hypothesize its origin around the Gulf of Thailand and the west of the South China Sea, probably in mainland Southeast Asia.
On the contrary and according to our results, the genepool ‘Und III’ is rare in cultivated bananas and was only found in ‘Pisang Madu’ and in the accessions classified as belonging to the ‘Pisang Jari Buaya’ subgroup. The collection sites of these accessions plead for a potential origin of ‘Und-III’ in Island South-East Asia rather than on the continent, maybe in a region between north Borneo and the Philippines, but further investigation should be conducted (Figure 3).
Finally, the genepool ‘Und II’ inferred in our analysis appeared to be an important contributor to cultivated diploids. It was notably found as introgressions in nearly all cultivated accessions from the Philippines and in landraces from South-East Asia and from New Guinea (Figures 1A, 3). The genepool ‘Und. II’ was also inferred as introgressions in ssp. sumatrana, truncata, errans and in the ‘Borneo’ accession, classified as belonging to ssp. microcarpa (Figure 2). We therefore interpret it as a signature of shared ancestry between these wild accessions, as well as with the cultivated diploids affected. However, as wild populations of M. acuminata in Borneo were reported morphologically heterogenous (Häkkinen and Langhe, 2001), we cannot exclude the occurrence of an ancestral population corresponding to ‘Und. II’ on this island.
From seeded to edible bananas on New Guinea island
As confirmed in this study, M. acuminata ssp. banksii is a major contributor to cultivated bananas. By comparing a population of 31 closely related cultivated AA landraces from New Guinea to the sample of 24 M. acuminata ssp. banksii, we aimed at understanding the wild-to-cultivated transition. Unexpectedly, the subset of cultivated diploid AAs revealed higher levels of diversity than its wild relative, as expressed by pi and observed heterozygosity (Figure 5). It differs substantially from the standard scenario in which domestication is expected to induce a loss of genetic diversity (Meyer and Purugganan, 2013). Possible explanations for such pattern are multiple and non-exclusive: i) it could result from an insufficient sampling of the wild population, or from the loss of wild genepools after domestication; ii) it may reflect hybridization between genetically distant genepools at the origin of the cultivated population; iii) the accumulation of somatic mutations, a common phenomenon in clonally propagated crops, can also contribute to higher levels of diversity (Miller and Gross, 2011).
Regarding our sampling, M. acuminata ssp. banksii exhibited very low levels of diversity and a global excess of rare polymorphism as expressed by the distribution of Tajima’s D values. These results point towards a population under expansion after a bottleneck. This bottleneck, reflecting a drastic population reduction, could have been induced by different factors. First, M. acuminata ssp. banksii displays hermaphrodite flowers causing dominant selfing (Simmonds, 1956; Kallow et al., 2021), an adaptative trait that induce bottlenecks when it emerges in limited numbers of individuals (Foxe et al., 2009; Guo et al., 2009). Second, the last glacial period during which the climate was cooler and drier (Bowler et al., 1976; Hope et al., 2004) likely induced conditions less favourable for M. acuminata on the island. Third, early human activities have heavily impacted flora and fauna in this region (Fairbairn et al., 2006) and could have included the wild banana populations. Therefore, if first banana domesticates were extracted from the wild prior the bottleneck, their parental population(s) might have disappeared nowadays. Additionally, we cannot rule out a sampling effect to explain these results as most of the M. acuminata ssp. banksii accessions studied here were collected in the lowlands of Papua New Guinea (Figure 4), so we possibly captured only a portion of the diversity within the taxa. Specimens of M. acuminata ssp. banksii were also observed at a higher altitude (Eyland et al., 2021) and another genepool is suspected in Indonesian New Guinea (Simmonds, 1956; Argent, 1976), but these regions could not be explored.
Considering hybridization, this is not fully resolved. The Bayesian analysis run by STRUCTURE suggested M. acuminata ssp. banksii as unique ancestor for this set of cultivated accessions. The four taxa Patterson’s D test that was then run to refine these results revealed introgressions by South-East Asian M. acuminata subspecies in seven of the 31 cultivated banana of this set. These results suggest that Patterson’s D test is more sensitive than Bayesian clustering in detecting geneflow. However, it can only be run with taxa that are present in the setting. Therefore, we cannot totally rule out that genetically distant population - absent from the sampling - introgressed cultivated diploids from New Guinea.
Finally, the accumulation of somatic mutations through vegetative propagation creates diversity and increases heterozygosity in clonal crops (McKey et al., 2010). These mutations, as soon as they are not deleterious for the crop, occur and accumulate independently on both haplotypes. As a result, it creates rare diversity that, in a sterile crop, can only be transmitted to clonally derived landraces. Since duplicates and clonemates were removed from the set at the first step of the analysis, signatures of such accumulations can be found in higher heterozygosity levels and negative values of Tajima’s D (Figure 5) (further discussed below). However, they cannot explain all the diversity observed in the cultivated set.
The higher levels of diversity observed in the cultivated samples are due to a combination of these factors. Mutations have accumulated through vegetative propagation, increasing heterozygosity and rare alleles. It is also likely that a different wild population, extinct or not, contributed to the genetic make-up of these cultivated diploids.
Besides, in domesticated plants, selection is expected to induce either an excess of low frequency polymorphism due to post-domestication’s bottleneck expansion, as in selfing chickpea (Varshney et al., 2019), or a drop in rare alleles frequencies due to recent selection, such as in clonal African yams (Akakpo et al., 2017). In sugarcane, another clonal crop domesticated in New Guinea, coding regions of the genome were found with higher diversity and with moderate Fst values when comparing the cultivated to the wild. Authors suggested balancing selection and accumulation of mutations were jointly responsible for such pattern (Arro et al., 2016). The signal is not as clear in the edible AAs from New Guinea. The results, including higher diversity and the peak of positive values of Tajima’s D coupled to moderate Fst (Figure 5), suggest balancing selection at work. Balancing selection would also explain the higher heterozygosity identified in the cultivated samples. Farming practices could be responsible for such selection. In cassava, for example, farmers unconsciously favour heterozygous, more vigorous, plants that are then clonally propagated (Pujol et al., 2005). However, such as in grape vine, another clonal crop (Houel et al., 2010), balancing selection does not act alone in banana. We identified on chromosome 4 a genomic region that cumulates low Tajima’s D and high Fst values (Table 3; Figure 5). It strongly suggests positive selection and the fixation of an allele advantageous for the cultivated population. A gene of interest linked to domestication might be located in this region and should be further investigated.
To conclude, hybridization and introgressions have played a major role in the creation of banana domesticates. Undefined genepools contributed massively to the creation of the cultivated bananas of the sampling. In New Guinea, where cultivated diploids are tightly linked to the local wild relative M. acuminata ssp. banksii, a few introgressions were detected and an uncharacterized genepool is also suspected. The domestication and the diversification of banana therefore result from processes much more complex than expected for a clonal crop where, in theory, advantageous individuals are extracted from the wild and then maintained clonally (McKey et al., 2010).
This study revealed gaps in the knowledge of wild banana genepools that call for future actions. Further explorations of wild bananas species and populations are needed, in particular in the suggested areas of origins of the undefined ancestral genepools. The characterization of the entire wild banana genepool is a prerequisite to understand the diversification history of wild Musa species, including M. acuminata, and to develop efficient conservation plans for the taxa. It is also a key element for the resolution of the intra-specific hybridization patterns of cultivated bananas. Coupled to the investigation of the correlation between the different wild ancestors’ contributions to cultivars and selected agronomic traits, it will also enable the design of targeted and informed breeding strategies. Finally, with the latest availability of genomic resources and while clonal crops were understudied in the past, it may well be that cultivated bananas offer a unique ground for studying the evolutionary effects of hybridization.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/,PRJNA450532.
Author contributions
JS designed the study. JS and JP performed field collections in PNG and IH provided genebank samples. JS and MR coordinated genotyping experiments. JS and CB performed analyses with inputs from MR, XP and SC. NR supervised the study and acquired funding. JS, CB and MR wrote the manuscript with inputs and review from all authors. All authors approved the final manuscript.
Funding
This work was financially supported by CGIAR Fund, in particular the CGIAR Research Program, Roots, Tubers and Bananas and the Genebank Platform.
Acknowledgments
We thank BGI for their technical assistance and services for the RAD sequencing. This work was technically supported by the CIRAD - UMR AGAP HPC Data Centre of the South Green Bioinformatics platform (https://www.southgreen.fr/). We also acknowledge Robert Miller (University of Brasilia), Agus Sutanto (ICHORD), Jeff Daniells (Queensland DAF), Jaroslav Dolezel, Eva Hribova and Pavla Christelova (Institute of Experimental Botany) for providing some of the plant materials and DNA. Thank you also to Rachel Chase (Alliance of Bioversity and CIAT) for editing the manuscript. Finally, we acknowledge all contributors to the International Musa Germplasm Transit Centre (ITC). The ITC materials of this study, and more, are available for distribution under the strict terms of the International Treaty on Plant Genetic Resources for Food and Agriculture via MGIS (https://www.crop-diversity.org/mgis/).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.969220/full#supplementary-material
References
Abbott, R. J., Barton, N. H., Good, J. M. (2016). Genomics of hybridization and its evolutionary consequences. Mol. Ecol. 25, 2325–2332. doi: 10.1111/mec.13685
Ahmed, D., Comte, A., Curk, F., Costantino, G., Francois, L., Dereeper, A., et al. (2019). Genotyping by sequencing can reveal the complex mosaic genomes in gene pools resulting from reticulate evolution: A case study in diploid and polyploid citrus. Ann. Bot. 123, 1–21. doi: 10.1093/aob/mcz029
Akakpo, R., Scarcelli, N., Chaïr, H., Dansi, A., Djedatin, G., Thuillet, A.-C., et al. (2017). Molecular basis of African yam domestication: Analyses of selection point to root development, starch biosynthesis, and photosynthesis related genes. BMC Genomics 18 (1), 782. doi: 10.1186/s12864-017-4143-2
Argent, G. C. G. (1976)Wild bananas of Papua new Guinea. In: Notes from the royal botanic garden, Edinburgh. Available at: https://agris.fao.org/agris-search/search.do?recordID=US201302495245 (Accessed January 14, 2021).
Arnold, M. L. (2004). Natural hybridization and the evolution of domesticated, pest and disease organisms. Mol. Ecol. 13, 997–1007. doi: 10.1111/j.1365-294X.2004.02145.x
Arro, J., Park, J.-W., Wai, C. M., VanBuren, R., Pan, Y.-B., Nagai, C., et al. (2016). Balancing selection contributed to domestication of autopolyploid sugarcane (Saccharum officinarum l.). Euphytica 209, 477–493. doi: 10.1007/s10681-016-1672-8
Baidouri, M. E., Murat, F., Veyssiere, M., Molinier, M., Flores, R., Burlot, L., et al. (2017). Reconciling the evolutionary origin of bread wheat (Triticum aestivum). New Phytol. 213, 1477–1486. doi: 10.1111/nph.14113
Baurens, F.-C., Martin, G., Hervouet, C., Salmon, F., Yohomé, D., Ricci, S., et al. (2019). Recombination and Large structural variations shape interspecific edible bananas genomes. Mol. Biol. Evol. 36, 97–111. doi: 10.1093/molbev/msy199
Baute, G. J., Kane, N. C., Grassa, C. J., Lai, Z., Rieseberg, L. H. (2015). Genome scans reveal candidate domestication and improvement genes in cultivated sunflower, as well as post-domestication introgression with wild relatives. New Phytol. 206, 830–838. doi: 10.1111/nph.13255
Bowler, J. M., Hope, G. S., Jennings, J. N., Singh, G., Walker, D. (1976). Late quaternary climates of Australia and new Guinea. Quaternary Res. 6, 359–394. doi: 10.1016/0033-5894(67)90003-8
Carreel, F., Fauré, S., González De León, D., Lagoda, P., Perrier, X., Bakry, F., et al. (1994). Evaluation de la diversité génétique chez les bananiers diploïdes (Musa sp). Genet. Selection Evol. 26, 125s–136s.
Carreel, F., Gonzalez de Leon, D., Lagoda, P., Lanaud, C., Jenny, C., Horry, J. P., et al. (2002). Ascertaining maternal and paternal lineage within musa by chloroplast and mitochondrial DNA RFLP analyses. Genome 45, 679–692. doi: 10.1139/g02-033
Cenci, A., Sardos, J., Hueber, Y., Martin, G., Breton, C., Roux, N., et al. (2021). Unravelling the complex story of intergenomic recombination in ABB allotriploid bananas. Ann. Bot. 127, 7–20. doi: 10.1093/aob/mcaa032
Chaïr, H., Cornet, D., Deu, M., Baco, M. N., Agbangla, A., Duval, M. F., et al. (2010). Impact of farmer selection on yam genetic diversity. Conserv. Genet. 11, 2255–2265. doi: 10.1007/s10592-010-0110-z
Chaïr, H., Sardos, J., Supply, A., Mournet, P., Malapa, R., Lebot, V. (2016). Plastid phylogenetics of Oceania yams (Dioscorea spp., dioscoreaceae) reveals natural interspecific hybridization of the greater yam (D. alata). Botanical J. Linn. Soc. 180, 319–333. doi: 10.1111/boj.12374
Christelova, P., Langhe, E., Hribova, E., Cizkova, J., Sardos, J., Husakova, M., et al. (2017) Molecular and cytological characterization of the global musa germplasm collection provides insights into the treasure of banana diversity. Available at: http://biblio1.iita.org/handle/20.500.12478/1488 (Accessed January 14, 2021).
Cornille, A., Gladieux, P., Smulders, M. J. M., Roldán-Ruiz, I., Laurens, F., Cam, B. L., et al. (2012). New insight into the history of domesticated apple: Secondary contribution of the European wild apple to the genome of cultivated varieties. PloS Genet. 8, e1002703. doi: 10.1371/journal.pgen.1002703
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Darwin, C. (1869) The variation of animals and plants under domestication (London). Available at: http://darwin-online.org.uk/content/frameset?itemID=F880.1&viewtype=text&pageseq=1 (Accessed August 2, 2021). John Murray.
Davey, J. W., Blaxter, M. L. (2010). RADSeq: Next-generation population genetics. Brief Funct. Genomics 9, 416–423. doi: 10.1093/bfgp/elq031
Denham, T., Barton, H., Castillo, C., Crowther, A., Dotte-Sarout, E., Florin, S. A., et al. (2020). The domestication syndrome in vegetatively propagated field crops. Ann. Bot. 125, 581–597. doi: 10.1093/aob/mcz212
Denham, T. P., Haberle, S. G., Lentfer, C., Fullagar, R., Field, J., Therin, M., et al. (2003). Origins of agriculture at kuk swamp in the highlands of new Guinea. Science 301, 189–193. doi: 10.1126/science.1085255
Doyle, J. J., Doyle, J. L. (1990). A rapid total DNA preparation procedure for fresh plant tissue. Focus 12, 13–15 .
Droc, G., Larivière, D., Guignon, V., Yahiaoui, N., This, D., Garsmeur, O., et al. (2013). The banana genome hub. Database (Oxford) 2013, bat035. doi: 10.1093/database/bat035
Durand, E. Y., Patterson, N., Reich, D., Slatkin, M. (2011). Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252. doi: 10.1093/molbev/msr048
Earl, D. A., vonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the evanno method. Conserv. Genet. Resour. 4, 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Eyland, D., Breton, C., Sardos, J., Kallow, S., Panis, B., Swennen, R., et al. (2021). Filling the gaps in gene banks: Collecting, characterizing, and phenotyping wild banana relatives of Papua new Guinea. Crop Sci. 61, 137–149. doi: 10.1002/csc2.20320
Fairbairn, A. S., Hope, G. S., Summerhayes, G. R. (2006). Pleistocene occupation of new guinea’s highland and subalpine environments. World Archaeology 38, 371–386. doi: 10.1080/00438240600813293
Falush, D., Stephens, M., Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 164, 1567–1587.
Feng, C., Wang, J., Harris, A. J., Folta, K. M., Zhao, M., Kang, M. (2021). Tracing the diploid ancestry of the cultivated octoploid strawberry. Mol. Biol. Evol. 38, 478–485. doi: 10.1093/molbev/msaa238
Foxe, J. P., Slotte, T., Stahl, E. A., Neuffer, B., Hurka, H., Wright, S. I. (2009). Recent speciation associated with the evolution of selfing in Capsella. PNAS 106, 5241–5245.
Fu, N., Ji, M., Rouard, M., Yan, H.-F., Ge, X.-J. (2022). Comparative plastome analysis of musaceae and new insights into phylogenetic relationships. BMC Genomics 23, 223. doi: 10.1186/s12864-022-08454-3
Gaut, B. S. (2015). Evolution is an experiment: Assessing parallelism in crop domestication and experimental evolution: (Nei lecture, SMBE 2014, Puerto Rico). Mol. Biol. Evol. 32, 1661–1671. doi: 10.1093/molbev/msv105
Gonzalez-Segovia, E., Pérez-Limon, S., Cíntora-Martínez, G. C., Guerrero-Zavala, A., Janzen, G. M., Hufford, M. B., et al. (2019). Characterization of introgression from the teosinte zea mays ssp. mexicana to Mexican highland maize. PeerJ 7, e6815. doi: 10.7717/peerj.6815
Green, R. E., Krause, J., Briggs, A. W., Maricic, T., Stenzel, U., Kircher, M., et al. (2010). A draft sequence of the neandertal genome. Science 328, 710–722. doi: 10.1126/science.1188021
Guo, Y.-L., Bechsgaard, J. S., Slotte, T., Neuffer, B., Lascoux, M., Weigel, D., et al. (2009). Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. PNAS 106, 5246–5251.
Häkkinen, M., Langhe, E. (2001). Musa acuminata in Northern Borneo: preliminary report. INIBAP, Montpellier, France. 23p.
Heslop-Harrison, J. S., Schwarzacher, T. (2007). Domestication, genomics and the future for banana. Ann. Bot. 100, 1073–1084. doi: 10.1093/aob/mcm191
Hippolyte, I., Jenny, C., Gardes, L., Bakry, F., Rivallan, R., Pomies, V., et al. (2012). Foundation characteristics of edible musa triploids revealed from allelic distribution of SSR markers. Ann. Bot. 109, 937–951. doi: 10.1093/aob/mcs010
Hope, G., Kershaw, A. P., Kaars, S.v. d., Xiangjun, S., Liew, P.-M., Heusser, L. E., et al. (2004). History of vegetation and habitat change in the austral-Asian region. Quaternary Int. 118–119, 103–126. doi: 10.1016/S1040-6182(03)00133-2
Houel, C., Bounon, R., Chaïb, J., Guichard, C., Péros, J.-P., Bacilieri, R., et al. (2010). Patterns of sequence polymorphism in the fleshless berry locus in cultivated and wild vitis vinifera accessions. BMC Plant Biol. 10, 284. doi: 10.1186/1471-2229-10-284
Janssens, S. B., Vandelook, F., Langhe, E. D., Verstraete, B., Smets, E., Vandenhouwe, I., et al. (2016). Evolutionary dynamics and biogeography of musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of southeast Asia. New Phytol. 210, 1453–1465. doi: 10.1111/nph.13856
Jeensae, R., Kongsiri, N., Fluch, S., Burg, K., Boonruangrod, R. (2021). Cultivar specific gene pool may play an important role in musa acuminata colla evolution. Genet. Resour. Crop Evol 68, 1589–1601. doi: 10.1007/s10722-020-01088-y
Kallow, S., Panis, B., Vu, D. T., Vu, T. D., Paofa, J., Mertens, A., et al. (2021). Maximizing genetic representation in seed collections from populations of self and cross-pollinated banana wild relatives. BMC Plant Biol. 21 (1), 415. doi: 10.1186/s12870-021-03142-y
Li, H., Durbin, R. (2010). Fast and accurate long-read alignment with burrows–wheeler transform. Bioinformatics 26, 589–595. doi: 10.1093/bioinformatics/btp698
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200
Martin, G., Baurens, F., Hervouet, C., Salmon, F., Delos, J., Labadie, K., et al. (2020a). Chromosome reciprocal translocations have accompanied subspecies evolution in bananas. Plant J. 104, 1698–1711. doi: 10.1111/tpj.15031
Martin, G., Cardi, C., Sarah, G., Ricci, S., Jenny, C., Fondi, E., et al. (2020b). Genome ancestry mosaics reveal multiple and cryptic contributors to cultivated banana. Plant J. 102, 1008–1025. doi: 10.1111/tpj.14683
Martin, S. H., Davey, J. W., Jiggins, C. D. (2015). Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol. Biol. Evol. 32, 244–257. doi: 10.1093/molbev/msu269
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
McKey, D., Elias, M., Pujol, B., Duputié, A. (2010). The evolutionary ecology of clonally propagated domesticated plants. New Phytol. 186, 318–332. doi: 10.1111/j.1469-8137.2010.03210.x
Meyer, R. S., Purugganan, M. D. (2013). Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852. doi: 10.1038/nrg3605
Miller, A. J., Gross, B. L. (2011). From forest to field: Perennial fruit crop domestication. Am. J. Bot. 98, 1389–1414. doi: 10.3732/ajb.1000522
Morris, J. L., Puttick, M. N., Clark, J. W., Edwards, D., Kenrick, P., Pressel, S., et al. (2018). The timescale of early land plant evolution. PNAS 115, E2274–E2283.
Paradis, E., Schliep, K. (2019). Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in r. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Perrier, X., Jacquemoud-Collet, J.-P. (2006). DARwin software. CIRAD. Available at: https://darwin.cirad.fr/ as stated on the software webpage: https://darwin.cirad.fr/feedback.php
Perrier, X., Jenny, C., Bakry, F., Karamura, D., Kitavi, M., Dubois, C., et al. (2019). East African Diploid and triploid bananas: A genetic complex transported from south-East Asia. Ann. Bot. 123, 19–36. doi: 10.1093/aob/mcy156
Perrier, X., Langhe, E. D., Donohue, M., Lentfer, C., Vrydaghs, L., Bakry, F., et al. (2011). Multidisciplinary perspectives on banana (Musa spp.) domestication. PNAS 108, 11311–11318. doi: 10.1073/pnas.1102001108
Pompidor, N., Charron, C., Hervouet, C., Bocs, S., Droc, G., Rivallan, R., et al. (2021). Three founding ancestral genomes involved in the origin of sugarcane. Ann. Bot. 127, 827–840. doi: 10.1093/aob/mcab008
Pritchard, J. K., Stephens, M., Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Pujol, B., David, P., McKey, D. (2005). Microevolution in agricultural environments: how a traditional Amerindian farming practice favours heterozygosity in cassava (Manihot esculenta crantz, euphorbiaceae). Ecol. Lett. 8, 138–147. doi: 10.1111/j.1461-0248.2004.00708.x
Purugganan, M. D. (2019). Evolutionary insights into the nature of plant domestication. Curr. Biol. 29, R705–R714. doi: 10.1016/j.cub.2019.05.053
Rambaut (2006-2016) FigTree v1.4.3: Tree figure drawing tool. Available at: http://tree.bio.ed.ac.uk/software/figtree.
Ross-Ibarra, J., Morrell, P. L., Gaut, B. S. (2007). Plant domestication, a unique opportunity to identify the genetic basis of adaptation. PNAS 104, 8641–8648. doi: 10.1073/pnas.0700643104
Rouard, M., Droc, G., Martin, G., Sardos, J., Hueber, Y., Guignon, V., et al. (2018). Three new genome assemblies support a rapid radiation in musa acuminata (Wild banana). Genome Biol. Evol. 10, 3129–3140. doi: 10.1093/gbe/evy227
Rouard, M., Sardos, J., Sempéré, G., Breton, C., Guignon, V., V. den Houwe, I., et al. (2022). A digital catalog of high-density markers for banana germplasm collections. PLANTS PEOPLE PLANET, 4, 61–67. doi: 10.1002/ppp3.10187
Ruas, M., Guignon, V., Sempere, G., Sardos, J., Hueber, Y., Duvergey, H., et al. (2017). MGIS: Managing banana (Musa spp.) genetic resources information and high-throughput genotyping data. Database 2017. doi: 10.1093/database/bax046
Santos, J. D., Chebotarov, D., McNally, K. L., Bartholomé, J., Droc, G., Billot, C., et al. (2019). Fine scale genomic signals of admixture and alien introgression among Asian rice landraces. Genome Biol. Evol. 11, 1358–1373. doi: 10.1093/gbe/evz084
Sardos, J., Christelová, P., Čížková, J., Paofa, J., Sachter-Smith, G. L., Janssens, S. B., et al. (2018). Collection of new diversity of wild and cultivated bananas (Musa spp.) in the autonomous region of bougainville, Papua new Guinea. Genet. Resour. Crop Evol. 65, 2267–2286. doi: 10.1007/s10722-018-0690-x
Sardos, J., McKey, D., Duval, M. F., Malapa, R., Noyer, J. L., Lebot, V. (2008). Evolution of cassava (Manihot esculenta crantz) after recent introduction into a south pacific island system: The contribution of sex to the diversification of a clonally propagated crop. Genome 51, 912–921. doi: 10.1139/G08-080
Sardos, J., Perrier, X., Doležel, J., Hřibová, E., Christelová, P., Van den houwe, I., et al. (2016). DArT whole genome profiling provides insights on the evolution and taxonomy of edible banana (Musa spp.). Ann. Bot. 118, 1269–1278. doi: 10.1093/aob/mcw170
Sempéré, G., Pétel, A., Rouard, M., Frouin, J., Hueber, Y., De Bellis, F., et al. (2019). Gigwa v2–extended and improved genotype investigator. GigaScience 8 (5), giz051. doi: 10.1093/gigascience/giz051
Sharrock, S. (1988). Report on the first IBPGR-QDPI banana germplasm collecting mission to Papua New Guinea, 27 February to 22 March 1988 (Rome, Italy: IBPGR), 47.
Sharrock, S., Daniells, J. W., Kambuou, R. (1988). Report on the second IBPGR-QDPI banana germplasm collecting mission to Papua New Guinea, 22 October to 27 November 1988 (Rome, Italy: IBPGR), 29.
Sharrock, S., Jones, D. R., Banag, J. (1989). Report on the third IBPGR-QDPI banana germplasm collecting mission to Papua New Guinea, 15 February to 12 March 1989 (Rome, Italy: IBPGR), 17.
Shigeta, M. (1996). “Creating landrace diversity: The case of the ari people and ensete (Ensete ventricosum) in ethiopia,” in Redefining nature: ecology, culture and domestication (London: Routledge).
Simmonds, N. W. (1956). Botanical results of the banana collecting expedition, 1954-5. Kew Bull. 11, 463–489. doi: 10.2307/4109131
Simmonds, N. W. (1962)The evolution of the bananas. In: The evolution of the bananas. Available at: https://www.cabdirect.org/cabdirect/abstract/19630303919 (Accessed January 14, 2021).
Soltis, P. S., Soltis, D. E. (2009). The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60, 561–588. doi: 10.1146/annurev.arplant.043008.092039
Sun, X., Jiao, C., Schwaninger, H., Chao, C. T., Ma, Y., Duan, N., et al. (2020). Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 52, 1423–1432. doi: 10.1038/s41588-020-00723-9
Sutanto, A., Edison, H. S., Amril, R., Nasution, F., Hermanto, C., Cizkova, J., et al. (2016). Collecting banana diversity in eastern Indonesia. Acta Hortic. 1114, 19–26. doi: 10.17660/ActaHortic.2016.1114.3
Turcotte, M. M., Turley, N. E., Johnson, M. T. J. (2014). The impact of domestication on resistance to two generalist herbivores across 29 independent domestication events. New Phytol. 204, 671–681. doi: 10.1111/nph.12935
Varshney, R. K., Thudi, M., Roorkiwal, M., He, W., Upadhyaya, H. D., Yang, W., et al. (2019). Resequencing of 429 chickpea accessions from 45 countries provides insights into genome diversity, domestication and agronomic traits. Nat. Genet. 51, 857–864. doi: 10.1038/s41588-019-0401-3
Keywords: banana (Musa ssp.), domestication, hybridization, missing wild ancestor, Musa acuminata, selection, introgression
Citation: Sardos J, Breton C, Perrier X, Van den Houwe I, Carpentier S, Paofa J, Rouard M and Roux N (2022) Hybridization, missing wild ancestors and the domestication of cultivated diploid bananas. Front. Plant Sci. 13:969220. doi: 10.3389/fpls.2022.969220
Received: 14 June 2022; Accepted: 05 September 2022;
Published: 07 October 2022.
Edited by:
Wellington Ronildo Clarindo, Universidade Federal de Viçosa, BrazilReviewed by:
Zhongxiong Lai, Fujian Agriculture and Forestry University, ChinaMariana Cansian Sattler, Universidade Federal de Viçosa, Brazil
Copyright © 2022 Sardos, Breton, Perrier, Van den Houwe, Carpentier, Paofa, Rouard and Roux. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julie Sardos, j.sardos@cgiar.org
 Julie Sardos
Julie Sardos Catherine Breton
Catherine Breton Xavier Perrier
Xavier Perrier Ines Van den Houwe
Ines Van den Houwe Sebastien Carpentier4
Sebastien Carpentier4 Mathieu Rouard
Mathieu Rouard