
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 23 September 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.965878
 Muhammad Qudrat Ullah Farooqi1†
Muhammad Qudrat Ullah Farooqi1† Ghazala Nawaz2†
Ghazala Nawaz2† Shabir Hussain Wani3*†
Shabir Hussain Wani3*† Jeet Ram Choudhary4
Jeet Ram Choudhary4 Maneet Rana5
Maneet Rana5 Rameswar Prasad Sah6
Rameswar Prasad Sah6 Muhammad Afzal7
Muhammad Afzal7 Zahra Zahra8
Zahra Zahra8 Showkat Ahmad Ganie9
Showkat Ahmad Ganie9 Ali Razzaq10
Ali Razzaq10 Vincent Pamugas Reyes11
Vincent Pamugas Reyes11 Eman A. Mahmoud12
Eman A. Mahmoud12 Hosam O. Elansary13,14,15
Hosam O. Elansary13,14,15 Tarek K. Zin El-Abedin16
Tarek K. Zin El-Abedin16 Kadambot H. M. Siddique1*
Kadambot H. M. Siddique1*High-throughput sequencing technologies (HSTs) have revolutionized crop breeding. The advent of these technologies has enabled the identification of beneficial quantitative trait loci (QTL), genes, and alleles for crop improvement. Climate change have made a significant effect on the global maize yield. To date, the well-known omic approaches such as genomics, transcriptomics, proteomics, and metabolomics are being incorporated in maize breeding studies. These approaches have identified novel biological markers that are being utilized for maize improvement against various abiotic stresses. This review discusses the current information on the morpho-physiological and molecular mechanism of abiotic stress tolerance in maize. The utilization of omics approaches to improve abiotic stress tolerance in maize is highlighted. As compared to single approach, the integration of multi-omics offers a great potential in addressing the challenges of abiotic stresses of maize productivity.
Maize (Zea mays L.) is one of the most cultivated crops across the globe for food, animal feed, and as a source of biofuel (Ranum et al., 2014; Molla et al., 2019; Choudhary et al., 2020). Maize yield is highly dependent on a broad spectrum of climatic and soil conditions. However, abiotic stresses such as drought, salinity, high temperature, and cold, are restricting factors that affect its yield productivity (Wani et al., 2016; Figure 1).
Figure 1. Types of abiotic stresses that affect yield productivity in maize (https://biorender.com; accessed on 21 February 2022).
To date, drought is considered as a significant threat to crop growth depending on its severity and duration (Chen et al., 2012; Ganie and Ahammed, 2021). Edmeades et al. (2015) reported that maize is vulnerable to drought from flowering through to grain filling stage. It also directly affects the rate of photosynthesis activity within chloroplasts. In leaves, drought stress disturbs the concentration of abscisic acid (ABA), increasing various antioxidant enzymes such as GR (glutathione reductase), APX (ascorbate peroxidase), CAT (catalase), and SOD (superoxide dismutase) (Jiang, 2002; Mehla et al., 2017). On the other hand, salinity stress affects plants under irrigated and non-irrigated situations. A significant proportion of irrigated (50%) and all cultivated (20%) land is under salinity stress (Wang et al., 2017). This affects the growth and development of maize; however, the response of plants varies based on the degree of salinity and crop growth stage (Farooq et al., 2015). Short-term exposure of maize plants to salt stress influences its growth owing to osmotic stress. In salt-affected soils, excessive buildup of sodium and chloride ions in the rhizosphere leads to severe nutritional imbalances in maize due to strong interference of these ions with other essential mineral elements. Another type of stress, known as heat stress, reduces crop growth and productivity (Wahid et al., 2007; Fahad et al., 2017; El Sabagh et al., 2021). This affects cell metabolic activity and signals physiological networks, resulting in poor pollen dehiscence and fertility, stigma and silk emergence, seed set, and grain filling, reducing maize grain yield (Barnabás et al., 2007). Maize plants are also sensitive to low temperatures (<15°C) and can kill imbibed seeds and induce leaf senescence in maize (Miedema, 1982; Foyer et al., 2002). Additionally, chilling temperature (10°C) combined with excessive light stress reduces CO2 assimilation, leading to irreversible photosynthesis inhibition and cellular damage (Farooqi et al., 2016).
To withstand these abiotic stresses, plants can seize their growth activity under severe circumstances and develop various internal defense mechanisms (molecular, cellular, metabolic, and physiological) (Atif et al., 2019). For example, phenotypic stress adaptations have been observed in maize, such as reduced leaf angles, increased leaf wax, compacted tassels, and reduced evaporation rate in anthers which is crucial to preventing anther dehiscence (Shah et al., 2011). Similarly, genetic and metabolic networks are regulated in response to various abiotic stresses (Menezes-Benavente et al., 2004; Osmolovskaya et al., 2018).
The advances in genetics and molecular biology have led to the development of high-throughput sequencing technology. As a result, plant scientists were able to identify genes and genetic regions that are associated with traits of interest. Over the years, these genes and genetic information have been successfully used for crop improvement in terms of yield, biotic stresses, and abiotic stresses (Jena and Mackill, 2008; Angeles-Shim et al., 2020; Reyes et al., 2021a). Multi-omics approaches have aided in the understanding of maize crop growth, senescence, yield, and responses to biotic and abiotic stresses (Jeyasri et al., 2021; Kaur et al., 2021). As shown in Figure 2, omics approaches such as genomics and metabolomics are being utilized to identify loci and metabolite markers that are associated with abiotic stress tolerance. In addition to this, several approaches such as genome editing, and speed breeding can be applied to hasten the development of superior maize cultivars. To date several studies have utilized omics approaches for abiotic stress tolerance. For example, phenomics have aided in the development of non-destructive phenotyping approaches. Similarly, identification of regulatory networks that are associated with abiotic stress response are now possible due to transcriptomics. Collectively, omics approaches are beneficial in plant science research specially for understanding the genome, transcriptome, metabolome, and phenome of crops.
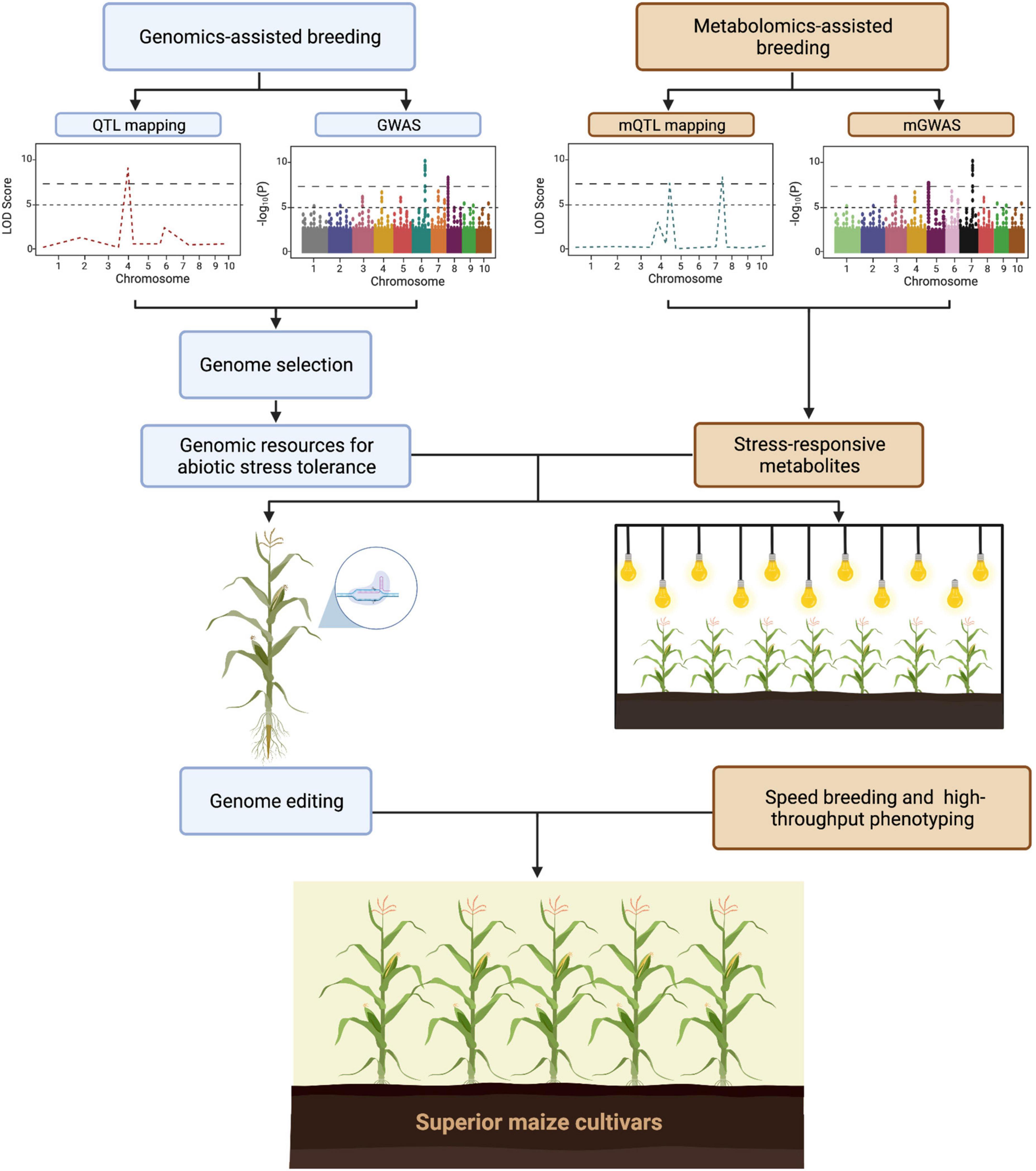
Figure 2. General workflow on the utilization of omics technologies in development of superior maize cultivars (https://biorender.com; accessed on 21 February 2022).
In this review, a holistic approach was used to discuss the recent developments on abiotic stress tolerance in maize. Firstly, we discussed the morphological, physiological, and molecular responses of maize to abiotic stresses. Then, we presented studies that used conventional and marker-assisted breeding approaches to understand the genetic mechanisms involved in stress adaptation were also. Lastly, we highlighted the use of “omic” tools and utilization of wild maize relatives for genetic resource development of stress tolerance in maize.
Maize has various physiological responses to abiotic stresses (Figure 3). In general, abiotic stresses such as high temperature, salt, and drought alter many physiological traits such as membrane permeability, net photosynthesis, osmolyte accumulation, respiration, osmotic potential, and mineral uptake in maize (Wahid et al., 2007; Turan et al., 2010; Waqas et al., 2017). For drought tolerance, several studies have demonstrated that photorespiration and raffinose (oligosaccharide) metabolism are essential but vary under various conditions (i.e., combined stresses or a single type of stress) (Obata et al., 2015; Sun et al., 2019; Rafique et al., 2020). Additionally, metabolic changes in metabolic profiles in maize results in changes via cell wall remodeling, maintaining metabolic homeostasis, and signaling mechanisms to tolerate multiple stress conditions.
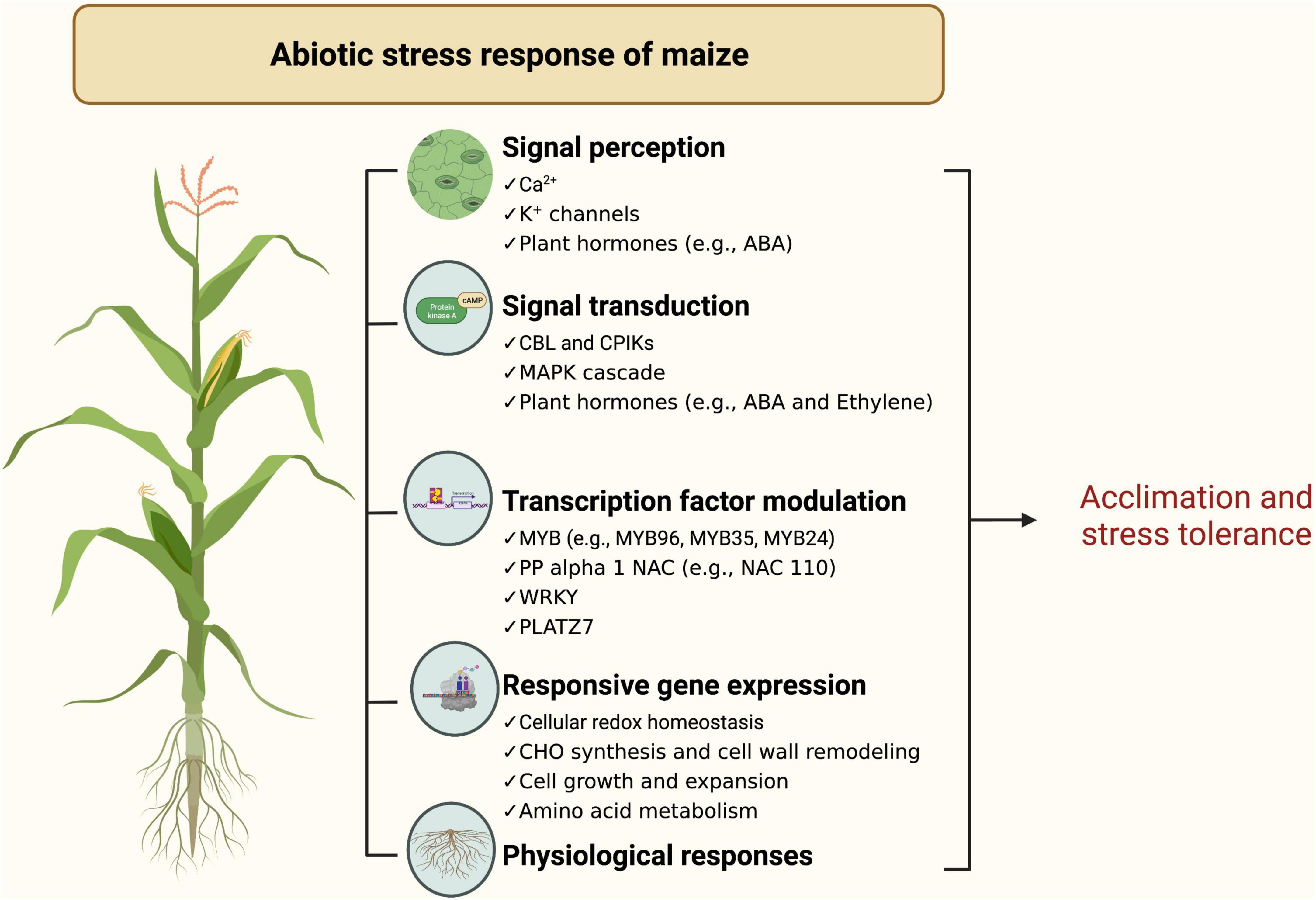
Figure 3. Schematic diagram of maize physiological and genetic response to abiotic stress (https://biorender.com; accessed on 21 February 2022).
Similarly, the level of reactive oxygen species (ROS) is accompanied by higher CAT, GR, glutathione-S-transferase activities, chlorophyll content, and membrane stability in maize seedlings under stress conditions (Zeid, 2009; Yadav et al., 2018). For instance, higher osmoprotectants are required for stress tolerance under salt stress conditions (Zeid, 2009). Furthermore, heat shock proteins, kinases, phosphatases, and a cascade of metabolic networks are activated under temperature stress, which induces heat tolerance in maize (Gill and Tuteja, 2010; Tiwari and Yadav, 2019). For a particular farming system, climate-smart agronomic techniques include those that improve farmer resilience to climate shocks and/or reduce productivity loss. To lessen the negative impacts of temperature fluctuations, these procedures are becoming more and more crucial. Plants may benefit from a change in planting time by avoiding the temperature extreme period during crucial growth stages by reducing the chance of heat and chilling damage during the silking and grain filling stages, respectively, the change in planting time greatly reduced the output losses (Waqas et al., 2017).
Since plants are easily exposed to various stresses, they have evolved avoidance mechanisms in response to stress. Several studies have observed induced specific morphological changes in stressful environments in maize. For example, the maize flowering stage is vulnerable to temperatures > 30°C; this results in pollen desiccation during anthesis and delays the silking interval, which negatively affects maize productivity (Parent and Tardieu, 2012; Leitner et al., 2014). Stresses initially inhibit cell proliferation, with prolonged exposure preventing cell expansion and mostly speeding up the vegetative growth and completing their life cycle before the onset of temperature stress (Kavar et al., 2008; Masuka et al., 2012; Godínez-Palma et al., 2013). Identifying the metabolic and physiological networks has provided an excellent foundation for adapting to abiotic stress. These physiological networks are controlled by genes and other molecular networks which serve as a great avenue for developing maize lines that are tolerant to wide arrays of abiotic stresses. Moreover, roots also play an essential role in maintaining plant water uptake for effective drought avoidance in maize. Adjustment of the hydraulic root architectural system, soil and water heterogeneity, and transpiration activities during the day can help maize plants endure heat and drought stresses (Leitner et al., 2014). Furthermore, stress-induced signal transduction proteins and enzymes such as APX, SOD, LEA, and heat shock (HSK) proteins are critical for enduring heat and drought stress, especially during grain filling in maize (Hasanuzzaman et al., 2013; Soliman et al., 2018).
The sequencing of the maize genome has identified various abiotic-responsive genes and expanded the genomic resources for exploring abiotic stress tolerance in its gene pool. To date, numerous studies have reported transcription factors, signal transduction genes, and several abiotic-stress-responsive cis-regulatory elements involved in abiotic stress adaptation.
Transcription factors (TFs) and transcription factor binding sites directly affect the transcriptional regulation of plant genes. Several studies have elucidated the roles of TFs in maize adaptation to abiotic stresses. For example, ZmNF-YB16 has been introduced into the inbred maize line (B104). The overexpression of this TF increased the expression of genes encoding antioxidant enzymes, antioxidant synthase, and molecular chaperones associated with the endoplasmic reticulum stress response. As a result, lines sustained greater photosynthesis, improved dehydration, and drought stress tolerance during vegetative and reproductive stages, increasing maize grain production under drought-stressed conditions (Wang et al., 2018a). Similarly, the WRKY genes in maize have been well associated with abiotic stress resistance. For example, the ZmWRKY40 and ZmNF-YB2 gene encoding a transcription factor has improved drought tolerance in maize through functional genomics studies (Nelson et al., 2007; Zhang et al., 2008; Wang et al., 2018b; Gangola and Ramadoss, 2020). The ABP9 gene in maize encodes the bZIP transcription factor in Arabidopsis, improving salt, drought, and cold tolerance by increasing oxidative enzyme levels (Zong et al., 2018). In addition, several MYB-related proteins in the maize genome have been identified, and 46 of them were already characterized in relation to different abiotic stresses. Among them, the expression of ZmMYB30 was observed to have increased remarkably under drought and high salinity conditions (Nelson et al., 2007; Wang et al., 2018a; Zong et al., 2018). The MYB TF, ZmMYB31, repressed isopalmitate biosynthesis, increased UV exposure sensitivity, decreased plant height, and activated several stress-responsive genes (Zm5H, C3H, ZmActin, and ZmCOMT) in transgenic maize plants (Fornalé et al., 2010). Interestingly, ZmSAPK8, an SnRK2 phosphokinase, was cloned from maize that confers salinity tolerance with transcriptional upregulation of stress-linked genes, including DREB2A, P5CS1, RAB18, RD29A, and RD29B under salt stress indicating ZmSAPK8 is involved in diverse signal transduction and has the potential to improve salt tolerance in crops (Ying et al., 2011). Other transcription factors like ZmNF-YB2 and ZmNAC3 were previously characterized through functional genomics and were described as stress-induced genes in maize that responds to physiological variations in photosynthesis and polysaccharide metabolism (Table 1).
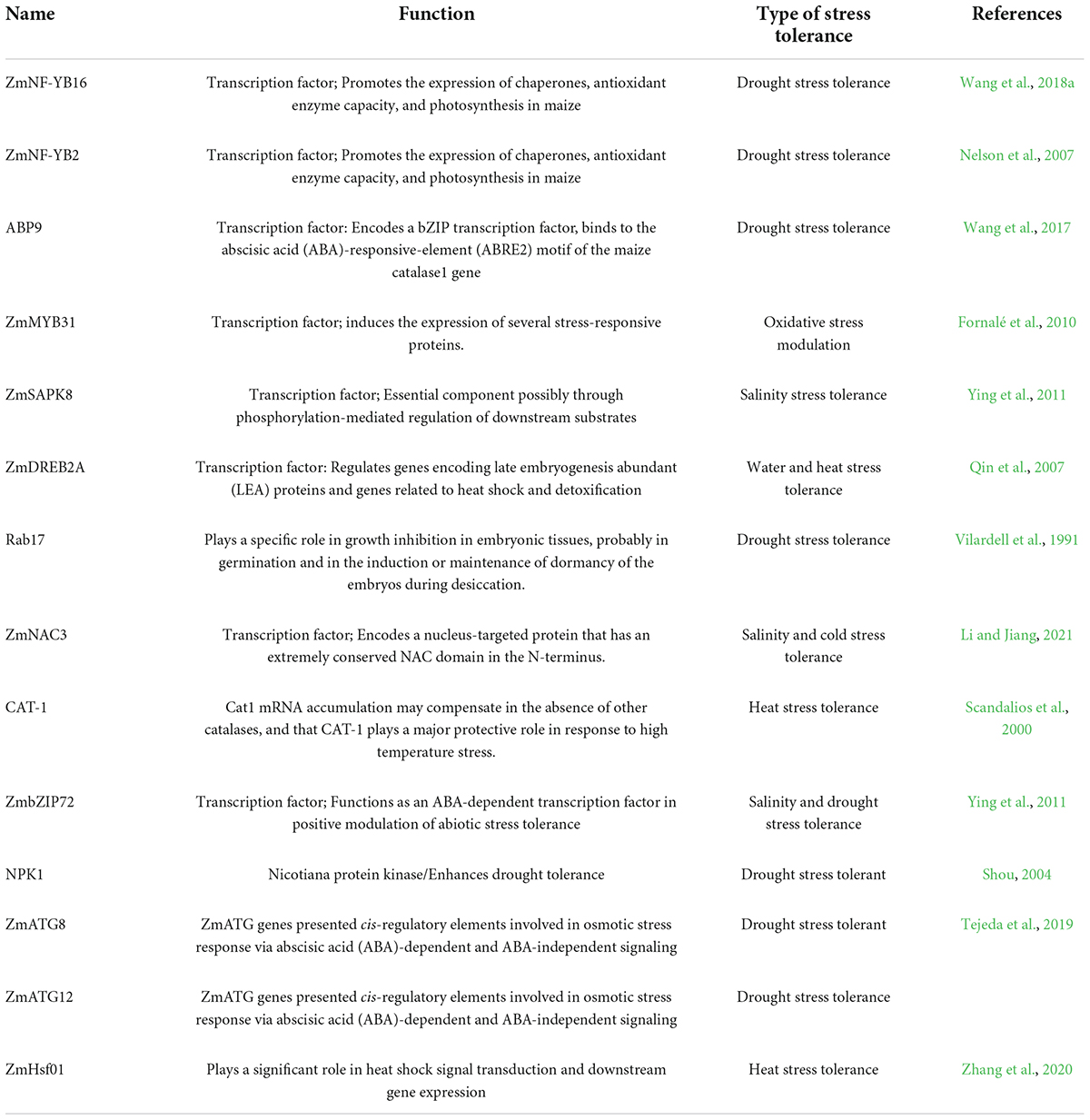
Table 1. Transcription factors and signal transduction genes associated with abiotic stress tolerance in maize.
Stress can also cause genome-wide transcriptome reprogramming in plants to respond to environmental stimuli. As a result, groups of genes linked to various physiological features and response pathways are controlled to counteract their negative impacts (Shao et al., 2021). Several transcriptomic studies have identified regulatory networks crucial to stress tolerance in maize. For example, a microarray-based technique was used to explore gene expression dynamics throughout seed development, which found 3445 genes with variable expression across samples from six different time points (Liu et al., 2008). Wang B. et al. (2019) demonstrated that drought-induced transcriptomic changes were strongly linked to developmental and physiological adaptation, affecting maize production. Similarly, the transcriptome sequencing of the maize roots grown in N, P, and K deficient environments identified a total of 2555 (N), 2340 (P), and 1173 (K) differentially expressed genes that are involved in nutrient utilization, plant hormones, and transcription factors (Ma et al., 2020).
Cis-acting elements function as stress signaling factors at terminal points in signal transduction pathways. In general, they impart several auxiliary functions to the plant systems, such as developmental regulation of growth-associated processes, morphological modifications, regulating senescence, and DNA damage repair mechanisms. Like TFs, Cis elements and signal transduction genes were also associated with abiotic stress response in maize. For example, in a recent study, the exogenous application of ABA-induced CAT1 (Catalase1) expression via the cis-element ABRE2 affects the enzymes that are related to ROS (H2O2) in maize (Guan et al., 2000). Similarly, several ectopic expressions of genes such as AtNHX1 and NPK1 were found to regulate transporters, ion channels, and oxidative signaling in response to salinity cold, heat, and salinity tolerance (Yin et al., 2004; Lin et al., 2014). Tejeda et al. (2019) characterized autophagy-related genes (ATG genes; ZmATG8 and ZmATG12) in maize landraces under osmotic stress and found them potential targets for functional characterization and development of osmotic-tolerant maize genotypes using molecular breeding strategies (Table 1).
MicroRNAs (miRNAs) were also documented to control abiotic stress tolerance in plants by binding to cis-regulatory elements or genes/transcripts. For example, miRNAs regulate target gene expression post-transcriptionally and play important roles in seed germination, ear development, and root architecture in maize (Spollen et al., 2008; Peng et al., 2018; Wani et al., 2020). Table 2 shows various miRNAs that are involved in maize abiotic stress tolerance.
Short tandem target mimics (STTM) is a technology that develops a resource for producing miRNA inactivation vectors and transgenic lines in model and crop plants (Peng et al., 2018). A recent study on a series of maize STTM166 transgenic plants identified 178 differentially expressed genes (60 downregulated and 118 upregulated genes). Most were involved in the cell membrane system, cell components, oxidation-reduction process, oxidoreductase activity, and carbohydrate metabolic processes. Several studies were also carried out to identify novel miRNA and mRNA interaction and their association with abiotic stress tolerance in maize. For example, a microarray expression analysis of a drought-associated study revealed 13 miRNAs families regulating 42 novel mRNAs target and 65 in maize (Li et al., 2020). Another study showed that 23 drought-responsive cis-regulatory elements and three TFs (GAMYB, HD-Zip III, and NAC) were associated with the target mRNAs (Aravind et al., 2017; Sepúlveda-García et al., 2020). The knock-down maize mutants of miR166 showed its association with adaptation through various phenotypic variations. It was observed that knock-down mutants had rolled leaves, inferior yield-related traits, epidermis structures, vascular patterns, enhanced abiotic stress resistance, elevated ABA levels, and reduced levels of indole acetic acid. The results shed light on the importance of ABA and auxin interaction in monocots and suggested that the specific mechanism differs from dicots (Aydinoglu, 2020; Li et al., 2020).
Du et al. (2016) performed overexpression studies to understand and improve the adaptive response of phosphorus (Pi) deficiency in maize and found a phosphorus-deficiency-induced long non-coding RNA1 (PILNCR1) that inhibits miR399 and thus miR399-mediated cleavage of ZmPHO2 [Zea mays PHOSPHATE2 (PHO2) pathway]. These findings indicate interactions among PILNCR1 and miR399 that could be potential targets for improving phosphorus efficiency in maize. Maize mutant lines for miR166 and its target gene Rld1 are involved in Rolled leaf 1 (Rld1) (a homologous gene of Arabidopsis HD-ZIP III transcription factor). The interaction of these two components is responsible for leaf polarity and exhibits various developmental defects such as delayed flowering, reduced stature, and curled leaf. Other mutants related to leaf polarity have been documented, including leaf-bladeless1 (lbl1), Arabidopsis SGS3 homolog, ragged seedling2 (rgd2), Arabidopsis ago7 homolog, milkweed pod1 (mwp1), and Arabidopsis KANADI homolog (Juarez et al., 2004; Douglas et al., 2010). Intriguingly, miR390-lbl1 and miR166-rld1 have been implicated synthesis, transport, and action in maintaining leaf developmental patterns (Nogueira et al., 2007, 2009; Itoh et al., 2008).
The current advances in sequencing have led to the development of genetic tools to understand complex traits in crops. Over the years, these tools were successfully used to develop new genetic resources that could be used to mine beneficial genes and marker-assisted breeding (Kitony et al., 2021; Reyes et al., 2021b). Investigating complex traits like abiotic stresses requires quantitative genetic approaches such as linkage mapping and association analysis using different mapping populations or association panels (Almeida et al., 2013; Samantara et al., 2021).
To date, QTL governing various abiotic stresses have been mapped using backcrossing, F2, RILs, introgression lines, and natural populations (Qiu et al., 2007; Mano and Omori, 2008; Osman et al., 2013; Zaidi et al., 2015; Frey et al., 2016). For cold tolerance, Hund et al. (2011) mapped important putative meta-QTL (MQTL Rt-6, MQTL Ax-2, MQTL Rt-7, and MQTL Ax-15) for root architecture traits during cold stress. Similarly, Hu et al. (2016) identified 12 QTL for germination rate and primary root length in maize under cold stress using B73 × Mo17 (IBM) derived population. Interestingly, the candidate gene GRMZM2G398807 governs cortical cell-delineating protein expression in the primary root, assisting in radicle protrusion. Goering et al. (2021) identified two QTL with high additive impact on chromosomes 1 and 4 using a panel of IBM Syn4 recombinant inbred lines. The genes with putative function related to auxin and gibberellin response were identified in the QTL region. Recently, Han et al. (2022) mapped five QTL clusters on chromosomes 1, 2, 3, 4, and 9 using an B73 x Mo17 (IBM) Syn10 double haploid population. From these clusters, 39 genes were extracted, and through RNA-seq, upregulated genes from B73 and Mo17 were identified.
For heat stress, Frey et al. (2016) identified two significant QTL for grain yield and one QTL for the leaf scorching trait. Similarly, McNellie et al. (2018) identified three major QTL, of which two major QTL were mapped on chromosome 3 (for plant death and leaf firing) and chromosome 1 (for leaf firing) in B73 × CML103 under heat stress at the late vegetative stage. The co-localized QTL for plant death and leaf firing reveals chromosome 3 as a potential region for heat stress tolerance. Inghelandt et al. (2019) identified QTL related to heat susceptibility index (HIS) of five traits on chromosome 2, 5, 9, and 10 using dent and flint maize inbred line populations. The study revealed an antagonistic pleiotropy between heat tolerance at seedling stage and adult stage. However, a low PVE was observed in these QTL making it not suitable for marker-assisted breeding applications.
Quantitative trait loci mapping under drought condition is associated with multiple traits such as anthesis–silking interval (ASI), root architecture, and grain yield traits. Giuliani et al. (2005) elucidated the role of Root-ABA1 on root architecture and leaf ABA concentration in response to different water deficit conditions. And in Landi et al. (2006) further validated the effect of this QTL under two environments (China and Italy). Almeida et al. (2013) mapped two significant constitutive QTL (chromosomes 2 and 6) and two adaptive QTL (chromosomes 5 and 7) for grain yield under drought stress. Semagn et al. (2013) identified four meta-QTL (mQTL2.2, mQTL6.1, mQTL7.5, and mQTL9.2) for grain yield under water-deficit and well-watered conditions. Zhao et al. (2019) identified three major QTL for drought stress tolerance (qKR-Ch.1-1 for kernel ratio, qEHPH-Ch.1-1 for ear height to plant height ratio, qGW-J1-1 for grain weight per ear, and qGW-Ch.4-1 for grain weight per ear) that could be targeted for introgression. Similarly, genetic regions that are associated with plant height, root length, and dry weight traits under salinity stress were mapped in the maize genome (Qiu et al., 2007; Osman et al., 2013).
Genome-wide associated studies (GWAS) is a powerful tool for dissecting genetic loci that significantly influence agronomic traits based on distinct phenotypes and extensive coverage of molecular markers. Many initiatives related to abiotic stress that include stress-linked genes in agricultural plants have used genome-wide association studies. For example, Liu et al. (2013) conducted GWAS using 368 maize varieties at the seedling stage and identified connections between the genetic variation in ZmDREB2.7 and the degree of drought tolerance in different maize varieties is highly dependent on DNA polymorphisms in the promoter region of ZmDREB2.7. In a study conducted by Thatcher et al. (2016), the genome-wide analysis showed significant changes in ears and leaves, magnifying that developmental splicing is linked to developmental stage, tissue type, and stress conditions. Song et al. (2016) identified cysteine-rich poly-comb-like (CPP) proteins in maize, and different expression levels of the ZmCPP gene under abiotic stresses (cold, salt, heat, and drought), indicated that ZmCPP functional characterization in maize could help understand maize growth and development. Using a genome-wide analysis of the AP2/ERF TF family, Zhou et al. (2012) showed that AP2/ERF gene family could participate in various stress responses such as drought and salinity. The same approach was carried out by Luo et al. (2019), wherein two promising genes, SAG4 and SAG6 (salt-tolerance-associated gene), were identified and have a great potential to develop salt-tolerant maize lines. Similarly, the GWAS approach was used to identify genetic regions associated with root-related traits, drought tolerance, and nitrogen use in maize (Wang N. et al., 2019; Guo et al., 2020). The success in GWAS utilization showed its great importance for studying multiple traits under drought, salt, and temperature stresses. GWAS studies in maize have identified novel gene candidates or genes responsible for abiotic stress, which can be used for maize breeding and designing climate-smart maize lines.
Maize (family Poaceae) comprises seven genera, including Chionachne, Sclerachne, Coix, Trilobachne, Polytoca, Tripsacum, and Zea. The genus Zea contains four species, but only Zea mays L. (2n = 20) is economically valuable. The other Zea species, referred to as teosintes, are essentially wild grasses native to Mexico and Central America (Doebley, 1990). The origin of maize (Zea mays L.) dates back more than 7,000 years and seems to have developed from annual teosinte (Zea mexicana) through gradual selection. The wild ancestor of modern maize has comparable plant architecture and growth forms to maize, while Tripsacum has higher chromosome numbers (2n = 36, 64, or 72), making it more challenging to hybridize with maize. These wild ancestors of maize have considerable genetic diversity and could be a new resource for germplasm enhancement (Maazou et al., 2016). For example, molecular evolution studies on maize have included genetic and genomic tools developed for Tripsacum (Blakey et al., 2007). Data from orthologous genes in maize and the Tripsacum genus revealed a distinct collection of genes with frequent non-synonymous substitutions in Tripsacum, which were prevalent when domestic maize was adapted to temperate regions through artificial selection. An intermediate metabolic route, phospholipid metabolism, was linked to cold and freezing tolerance. Anatomical descriptions such as aerenchyma tissue in roots and other properties in Tripsacum have contributed to survival under drought stress. Similar studies on root penetration and increased biomass in Tripsacum have revealed drought resistance. Physiological data revealed that the exceptional drought resistance of the Tripsacum genus is due to its high-water use efficiency and photosynthetic levels (Kemper et al., 1998). Later research revealed that the Tripsacum-introgressed cultivar grows better than modern maize under drought stress. Tripsacum introgression lines appear to have better-rooting systems that penetrate deeper into the soil and higher grain yields than modern maize cultivars (Gilker et al., 2002; Eubanks, 2006; Gitz et al., 2013). An evaluation of maize (Zea mays ssp. mays) × Tripsacum dactyloides L. hybrid (eastern gamagrass) calli revealed that hybrid plantlets had higher fresh weight than maize plants under salt stress and thus improved salt tolerance (Shavrukov and Sokolov, 2015). These hybrids retain sodium in their leaves, decreasing water potential and maintaining turgor pressure, vital for vegetative development (Pesqueira et al., 2003). Therefore, tapping the genetic potential of wild relatives is necessary to create new genetic resources that can serve as future accessions to develop maize lines with improved abiotic stress tolerance.
Traditional maize breeding can be used to develop high-yielding hybrids from selective parental mating based on specific combining abilities. The development of parental lines require 4–6 years and another 1–2 years for their hybrids, resulting in a long and somewhat complicated breeding cycle. To address this, speed breeding approaches were introduced (Hickey et al., 2017; Samantara et al., 2021). Several approaches can be used to shorten the breeding cycle in maize including (a) off-season nurseries, (b) double haploid (DH) technology, and (c) in vitro nurseries. The off-season nursery approach grows crops in a different location suited to their photoperiodism. DHs can be produced in two generations, significantly reducing the breeding cycle (Geiger and Gordillo, 2009). The most common method for maize DH uses the R1-nj color marker. In vitro nurseries are another option for breeding homozygous and homogenous lines, with a mix of DH technology (homozygosity per generation) and off-season nurseries (generations per year). These methods have several advantages and disadvantages. Off-season nurseries require quarantine clearance or other permissions to grow crops in a different location, extra time to transport genetic material and could introduce new pests and diseases. DH technology can have low success rates, hindering application for haploid induction and tissue culture adaptation. In addition, doubling is genotypic dependent and uses a carcinogenic agent (colchicine).
Genomic selection helps accumulate favorable genes with minor effects and improves multigenic traits, especially in stress-prone environments with a high environmental variation. The genomic selection focuses on estimating breeding values using many molecular markers that ideally cover the entire genome to predict the genetic value of the candidate for selection and using genetically estimated breeding values (GEBVs) to advance populations through rapid-cycle genomic selection without phenotyping in each cycle (Massman et al., 2013). Figure 4 is a schematic diagram integrating genomic selection and speed breeding for varietal and line development.
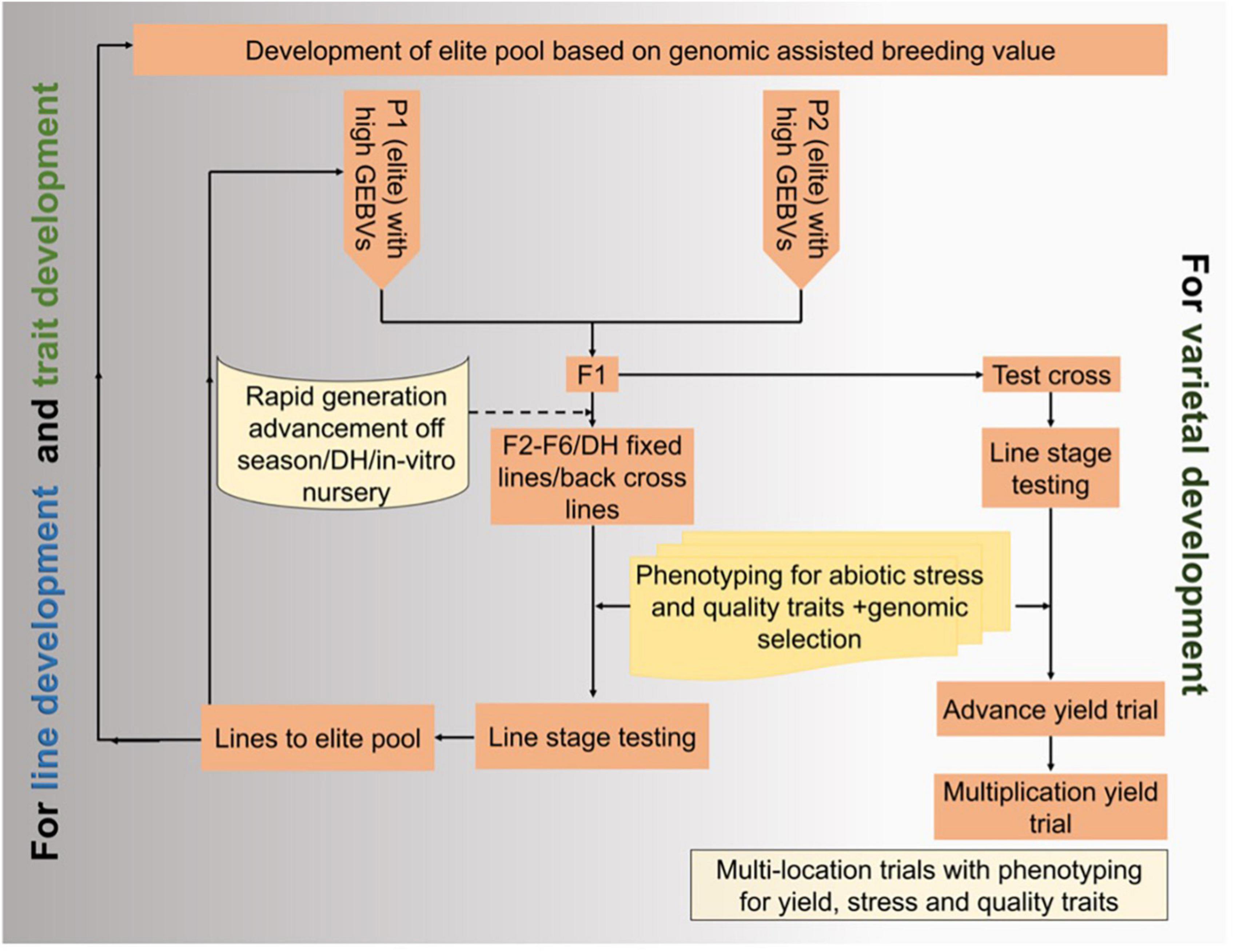
Figure 4. Integration of speed breeding and genomic selection in maize cultivar improvement and development.
Genomic selection improves polygenic traits, governed by small-effect genetic loci, such as plant yield. This tool uses marker effects across the genome to estimate GEBVs. Genomic selection produced higher stover and grain yield than MAS in a bi-parental maize population (Massman et al., 2013). Similarly, two fast cycles each year via genomic selection increased grain production by about 2% in a multi-parental maize population (Zhang et al., 2016). The accuracy of genomic predictions correlated with the test cross’s marker-predicted genotypic and phenotypic values in the validation population. The model’s genomic selection prediction accuracy was more accurate than genomic selection and marker-assisted selection. It could be used to capture alleles with fewer additive effects to increase genetic gain under drought. The predicted results differed slightly between the training and validation sets or linkage disequilibrium with causal variants in another study. The mean performance of the breeding populations differed from the underlying predicted traits (Windhausen et al., 2012). Genomics selection combined with MAS (marker-assisted selection, GS-MAS) had advantages over the QTL-MAS approach for drought-stressed growth characteristics (grain yield, anthesis, and plant height), evident from the differences between R2 (QTL-MAS) and prediction accuracies (GS-MAS) in the analysis (Tilman et al., 2011). Combining molecular marker data and phenotypic data as input variables delivered higher-quality estimates for grain production on average than phenotypic data alone (Trachsel et al., 2019).
Maize is susceptible to abiotic stresses, especially drought and waterlogging, allowing a higher degree of genotype × environmental interaction in a breeding program. To identify those with higher yield potential in the target environment, maize lines/hybrids were evaluated under targeted or managed stress conditions. However, most of the parents/hybrids/crosses were discarded due to poor performance in the field, which could be due to the low genetic value of the parents selected for crossing. Thus, parents of high GEBV should be used for crossing to accumulate the maximum number of favorable alleles for desirable traits in the progenies. Further, genetic gain is limited under abiotic stresses and high environmental error (Trachsel et al., 2019). Thus, combining genomic selection and speed breeding for abiotic stress tolerance would enhance the selection accuracy of parents to increase genetic gain and abiotic stress tolerance.
Genome editing tools are some of the major breakthroughs in molecular biology. These techniques offer a precise and efficient way to edit an organism’s genome. Some of the well-known genome editing tools are ZFNs (zinc-finger nucleases), TALENs (transcription activator-like effector nucleases), and clustered CRISPR (regularly interspaced short palindromic repeats)/Cas systems (Miller et al., 1985; Jinek et al., 2012; Joung and Sander, 2013). These tools use common sequence-specific nucleases to identify specific DNA sequences in the genome and generate desired double-stranded breaks (DSBs) (Mushtaq et al., 2021). ZFNs have been used to modify various crop plants, including maize. The ZFN-mediated targeted transgene integration was used to stack characteristic traits, especially for stress tolerance, in maize by combining several beneficial features (Ainley et al., 2013; Mushtaq et al., 2021). On the other hand, TALENs are a promising genetic tool for targeted gene mutagenesis in maize and other crops. This approach has been used to produce stable mutations at the maize glossy2 (gl2) locus that are heritable. The transgenic maize lines with mono-di-tri- allelic mutations conferred the glossy phenotype (Char et al., 2015). In most cases, integrated TALEN T-DNA separated independently from the new loss of function alleles, giving rise to null-segregant offspring in the T1 generation; thus, TALENs are an efficient tool for maize genome mutagenesis for identifying gene function and improving abiotic stress-related characteristics. Constructing TALEN repetitions is complex and TALEN gene targeting effectiveness varies.
CRISPR/Cas is a flexible genome editing technology with potential applications. Compared with ZFNs and TALENs, the CRISPR/Cas system has been adopted rapidly in plants due to its high efficiency, simplicity, cost-effectiveness, and ability to target multiple genes (Cong et al., 2013; Hsu et al., 2014; Razzaq et al., 2019). Several abiotic stress tolerance studies have been carried out using CRISPR/Cas. For example, drought-resistant characteristics have been incorporated into maize using CRISPR/Cas9 gene insertion and substitution techniques. To explore the targeted use of ARGOS8 native expression variation in drought tolerance breeding, CRISPR-Cas was used to generate novel variants of ARGOS8 (Shi et al., 2017). The low-expression gene ARGOS8 in maize, which negatively regulates ethylene responses, is crucial for plant growth. Field evaluation of lines showed that the ARGOS8 variations produced five extra bushels of grain production per acre under drought stress compared to the wild type. These findings show that CRISPR-Cas9 technology can create new allelic diversity in crops by substituting breeding drought-tolerant crops. In another study, CRISPR/Cas9 was used to unravel the abilities of the ZmWRKY40 gene encoding a transcription factor (Wang et al., 2018b), which improved drought tolerance, and resultant lines imparted drought tolerance in maize.
Therefore, genome editing techniques have a great potential to improve crops. After carefully selecting the genome-editing tool, target sequences can be designed and introduced into the most appropriate vectors. Suitable genetic cargo (DNA, RNA, or RNPs) is then selected for delivery by (i) modifying the targeted sequence, (ii) regenerating the edited calli, and (iii) producing the edited plants. Combining earlier genome editing tools with newly developed breakthroughs is speeding up genome editing in crop breeding to meet the world’s exponentially increasing need for food. In addition, climate change necessitates flexibility and ingenuity regarding crop resilience and production methods. Moreover, we must consider government restrictions and the public acceptability of these new breeding methods.
Numerous research digging into its systems biology, maize has benefited from the introduction of omics techniques. Because maize and its derivatives are used as both food and bioethanol, the metabolism of maize has received significant study. As reviewed by Medeiros et al. (2021) several aspects of maize metabolism have received significant attention, including (1) the role of the metabolome in basic molecular processes, responses to biotic and abiotic stresses, and beneficial biotic interactions; (2) the nutritional composition of maize kernels and the molecular mechanisms that underpin the production of specific metabolites; and (3) the mechanisms by which the metabolome and metabolic models link to leaf physiology. (4) The metabolic changes caused by genetic modifications; and (5) the degree of natural metabolic variance and its potential utility in breeding efforts.
In a study by Alvarez et al. (2008), changes in stomatal hormones like as ABA and cytokinins were discovered, underlining the importance of root-to-shoot signaling in maize under drought condition. Furthermore, an increase in the content of phenylpropanoid pathway intermediates was found. In a study by Casati et al. (2011), metabolomics was used to document the candidate signaling molecules associated with temporal effects of irradiation of canopy leaves in maize. The study identified myoinositol as a candidate molecule for UV-B responses. Witt et al. (2012) used the same approach to assess metabolome changes under drought condition. However, differential changes were not detected in their experiment which could possibly be attributed to experimental design such as the use of greenhouse instead of actual field setup. Metabolite profiling under salt stress conditions was carried out by Richter et al. (2015). Using two differently resistant maize hybrids, they were able to deduce that TCA cycle and sugar metabolism are highly affected biochemical pathways during salt stress. Sun et al. (2016) investigated the metabolic growth and response of maize to cyclic drought using a metabolomic approach. Their study demonstrated that distinct metabolic pathways in maize plants returned to normal at different speeds during recovery. However, metabolic study revealed quantitative differences between the two cycles, indicating the intricacy of metabolic processes launched by water cycle alterations.
Other than pathways, metabolomic studies have aided in showing which organs in maize are mostly affected by abiotic stress. For example, during drought condition, leaf blades were identified to have the greatest metabolic changes (Witt et al., 2012; Obata et al., 2015). And under high-salt conditions, metabolic changes were found to be greater in shoots than in roots (Gavaghan et al., 2011). In a study conducted by Ganie et al. (2015), metabolomic approach highlighted that under phosphorus starvation conditions, leaves are the main site for metabolic changes.
Conventional phenotyping uses destructive approaches to measure drought, heat, and salt tolerance characteristics. However, the development of high throughput phenotyping (HTP) platforms has greatly reduced the phenotyping issues that limit breeding programs. To date, several high-throughput phenotyping methods have been used to characterize maize phenomes, including penetrometers, electrical conductivity sensors to assess soil mapping variability, thermal imaging to monitor plant canopies, and spectral reflectance (Masuka et al., 2012). In addition, satellite imagery, mobile cameras, UAV imaging, and ground-based imaging platforms are widely used in maize plant stress assessment.
Typically, high-throughput plant phenotyping is accomplished by collecting photos that quantify attributes across a crop plant’s whole life cycle as traditional phenotyping is inefficient, expensive, and inaccurate (Arya et al., 2022). Nowadays, satellite imaging technology has become a powerful tool for collecting data in large agricultural practices, but few apply to small experimental breeding plots. For instance, satellite-driven NDVI (normalized differential vegetation index) strongly correlated with UAS multispectral imagery in maize plots (24 m2) (Sankaran et al., 2020). Unmanned aerial vehicle-red, green, and blue (UAV-RGB) images supplemented UAV thermal images for precise extraction of maize canopy temperature (Zhang et al., 2019a). In addition to general data measurement methods, advanced tools such as digital imagery, stable isotopes, spectral reflectance, and thermal imagery are used to improve data accuracy for soil and climate measurements. Maize studies have used SPAD meter, NDVI, and infrared gas analyzer (IRGA) to quantify environmental effects, digital imagery to measure early biomass in response to water stress, and thermal imagery to quantify leaf temperature during transpiration (Marti et al., 2007; Jones et al., 2009; Rorie et al., 2011). In several studies, infrared thermography (IRT) has been used to measure temperature differences between leaf, air, and canopies under heat and drought stress, spectroscopy to monitor the photosynthetic rate of leaves and canopies in response to the early onset of water stress, and fluorescence imaging to scrutinize plant growth during drought and low-temperature stress (Zia et al., 2013; Lee et al., 2015; Boote et al., 2016). Recently, fluorescence imaging was used to investigate plant chemical composition via spectral absorption and reflectance from cell to canopy level (Zhang et al., 2019a). The multispectral imaging techniques like X-ray-CT, MRI, and ultraviolet spectra (UV) can report changes in ionic balance, stomatal conductance, and transpiration, contributing to drought and high-temperature stress resistance in maize (Huang et al., 2016).
The fast-paced development of these HTPs have generated large number of datasets. And to date, one of the major challenges is the analytical approaches for these datasets. As previously reviewed, machine learning and deep learning techniques are some of the emerging techniques that can be used in the identification of hidden relationships between large datasets (Singh et al., 2018, 2021; Arya et al., 2022; Gill et al., 2022). In a review presented by Singh et al. (2018), both ML and DL are identified to be seamlessly integrated into data acquisition, data preprocessing, and data analytics for real-time HTP of plant traits in the field. Recently, Gill et al. (2022) highlighted that the current ML-based techniques focuses on a single stress or disease on a leaf or canopy, but in real-world situations, numerous diseases and stresses may appear on a single leaf or canopy. Therefore, ML platforms must be flexible and robust, with the ability to distinguish multiple disease symptoms on a single leaf or within the same plant canopy. Collectively, the quantity of data needed to train ML models is determined by the complexity of the problem and the complexity of the learning algorithm; thus, for wider applicability, the training data should really be continuously updated using techniques such as artificial learning to reflect the complexity of stress symptoms for the targeted crop.
This review concludes that the utilization of omics approaches is necessary to improve the abiotic stress tolerance in maize. The advancement in data utilization from high-throughput sequencing and phenotyping is necessary to exploit their potential in crop improvement fully. In addition studies on stress-tolerance-related mechanisms should consider the stress duration, which affects molecular and physiological reactions. However, short-term solutions are a more common practice among researchers. The molecular foundation of plant stress tolerance is influenced by stresses that occur under natural circumstances throughout the seasons. Thus, comparative studies on the expression and function of gene families under extreme conditions will help reduce the impact of abiotic stress in maize.
An opportunity for advancement in the omics approaches is also necessary. For example, integration of various omics approaches and in silico modeling can be employed to further dissect the genetics of abiotic stress tolerance in maize. These will widen our knowledge of molecular cascades and intracellular mechanisms governing stress adaptation. In addition, several modifications on the analytical approaches for the data generated can be implemented. For instance, HTS generates large amount of data. However, a bias in terms of analysis serves as a bottle neck to fully utilize the data. Using computer science and engineering, bioinformatics pipeline can be further fine-tuned to be more fitted for future analytical use. The same is true with HTPs, further refinement in the DL and ML algorithms is necessary to cater the needs of complex analyses.
SW, MF, and KHMS conceptualized the manuscript. MF, GN, SW, JC, MR, RS, MA, ZZ, SG, AR, VPR, EM, TE-A, HE, and KHMS contributed to the sections of the manuscript. VPR, MF, and SW contributed to the editing and revising of the manuscript. HE, SW, and KHMS supervised the writing and editing of the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ainley, W. M., Sastry-Dent, L., Welter, M. E., Murray, M. G., Zeitler, B., Amora, R., et al. (2013). Trait stacking via targeted genome editing. Plant Biotechnol. J. 11, 1126–1134. doi: 10.1111/pbi.12107
Almeida, G. D., Makumbi, D., Magorokosho, C., Nair, S., Borém, A., Ribaut, J.-M., et al. (2013). QTL mapping in three tropical maize populations reveals a set of constitutive and adaptive genomic regions for drought tolerance. Theor. Appl. Genet. 126, 583–600. doi: 10.1007/s00122-012-2003-2007
Alvarez, S., Marsh, E. L., Schroeder, S. G., and Schachtman, D. P. (2008). Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ. 31, 325–340. doi: 10.1111/j.1365-3040.2007.01770.x
Angeles-Shim, R. B., Reyes, V. P., del Valle, M. M., Lapis, R. S., Shim, J., Sunohara, H., et al. (2020). Marker-Assisted introgression of quantitative resistance gene pi21 confers broad spectrum resistance to rice blast. Rice Sci. 27, 113–123. doi: 10.1016/j.rsci.2020.01.002
Aravind, J., Rinku, S., Pooja, B., Shikha, M., Kaliyugam, S., Mallikarjuna, M. G., et al. (2017). Identification, characterization, and functional validation of drought-responsive MicroRNAs in subtropical maize inbreds. Front. Plant Sci. 8:941. doi: 10.3389/fpls.2017.00941
Arya, S., Sandhu, K. S., Singh, J., and kumar, S. (2022). Deep learning: as the new frontier in high-throughput plant phenotyping. Euphytica 218:47. doi: 10.1007/s10681-022-02992-2993
Atif, R. M., Shahid, L., Waqas, M., Ali, B., Rashid M, A. R., Azeem, F., et al. (2019). Insights on Calcium-Dependent Protein Kinases (CPKs) signaling for abiotic stress tolerance in plants. IJMS 20:5298. doi: 10.3390/ijms20215298
Aydinoglu, F. (2020). Elucidating the regulatory roles of microRNAs in maize (Zea mays L.) leaf growth response to chilling stress. Planta 251:38. doi: 10.1007/s00425-019-03331-y
Azahar, I., Ghosh, S., Adhikari, A., Adhikari, S., Roy, D., Shaw, A. K., et al. (2020). Comparative analysis of maize root sRNA transcriptome unveils the regulatory roles of miRNAs in submergence stress response mechanism. Environ. Exp. Bot. 171:103924. doi: 10.1016/j.envexpbot.2019.103924
Barnabás, B., Jäger, K., and Fehér, A. (2007). The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 31, 11–38. doi: 10.1111/j.1365-3040.2007.01727.x
Blakey, C. A., Costich, D., Sokolov, V., and Islam-Faridi, M. N. (2007). Tripsacum genetics: from observations along a river to molecular genomics. Maydica 52, 81–99.
Boote, B. W., Freppon, D. J., De La Fuente, G. N., Lübberstedt, T., Nikolau, B. J., and Smith, E. A. (2016). Haploid differentiation in maize kernels based on fluorescence imaging. Plant Breed. 135, 439–445. doi: 10.1111/pbr.12382
Casati, P., Campi, M., Morrow, D. J., Fernandes, J. F., and Walbot, V. (2011). Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genomics 12:321. doi: 10.1186/1471-2164-12-321
Char, S. N., Unger-Wallace, E., Frame, B., Briggs, S. A., Main, M., Spalding, M. H., et al. (2015). Heritable site-specific mutagenesis using TALENs in maize. Plant Biotechnol. J. 13, 1002–1010. doi: 10.1111/pbi.12344
Chen, J., Xu, W., Velten, J., Xin, Z., and Stout, J. (2012). Characterization of maize inbred lines for drought and heat tolerance. J. Soil Water Conservation 67, 354–364. doi: 10.2489/jswc.67.5.354
Choudhary, M., Singh, A., Gupta, M., and Rakshit, S. (2020). Enabling technologies for utilization of maize as a bioenergy feedstock. Biofuels, Bioprod. Bioref. 14, 402–416. doi: 10.1002/bbb.2060
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Ding, Y., Shi, Y., and Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Douglas, R. N., Wiley, D., Sarkar, A., Springer, N., Timmermans, M. C. P., and Scanlon, M. J. (2010). ragged seedling2 encodes an ARGONAUTE7-Like protein required for mediolateral expansion, but not dorsiventrality, of maize leaves. Plant Cell 22, 1441–1451. doi: 10.1105/tpc.109.071613
Du, H., Huang, M., and Liu, L. (2016). The genome wide analysis of GT transcription factors that respond to drought and waterlogging stresses in maize. Euphytica 208, 113–122. doi: 10.1007/s10681-015-1599-1595
Edmeades, G. O., Bolaños, J., Elings, A., Ribaut, J.-M., Bänziger, M., and Westgate, M. E. (2015). “The role and regulation of the anthesis-silking interval in maize,” in CSSA Special Publications, eds M. Westgate, Boote D. Knievel, and J. Kiniry (Madison, WI: Crop Science Society of America and American Society of Agronomy), 43–73. doi: 10.2135/cssaspecpub29.c4
El Sabagh, A., Hossain, A., Aamir Iqbal, M., Barutçular, C., Islam, M. S., Çið, F., et al. (2021). “Maize adaptability to heat stress under changing climate,” in Plant Stress Physiology, ed. A. Hossain (London: IntechOpen), doi: 10.5772/intechopen.92396
Eubanks, M. W. (2006). A genetic bridge to utilize Tripsacum germpiasm in maize improvement. Maydica 51, 315–327. doi: 10.1007/s00425-019-03136-z
Fahad, S., Bajwa, A. A., Nazir, U., Anjum, S. A., Farooq, A., Zohaib, A., et al. (2017). Crop production under drought and heat stress: plant responses and management options. Front. Plant Sci. 8:1147. doi: 10.3389/fpls.2017.01147
Farooq, M., Hussain, M., Wakeel, A., and Siddique, K. H. M. (2015). Salt stress in maize: effects, resistance mechanisms, and management. a review. Agron. Sustain. Dev. 35, 461–481. doi: 10.1007/s13593-015-0287-280
Farooqi, M. Q. U., Sa, K. J., Hong, T. K., and Lee, J. K. (2016). Bulk segregant analysis (BSA) for improving cold stress resistance in maize using SSR markers. Genet. Mol. Res 15, 1–12. doi: 10.4238/gmr15049326
Fornalé, S., Shi, X., Chai, C., Encina, A., Irar, S., Capellades, M., et al. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux: ZmMYB31 controls phenylpropanoids. Plant J. 64, 633–644. doi: 10.1111/j.1365-313X.2010.04363.x
Foyer, C. H., Vanacker, H., Gomez, L. D., and Harbinson, J. (2002). Regulation of photosynthesis and antioxidant metabolism in maize leaves at optimal and chilling temperatures: review. Plant Physiol. Biochem. 40, 659–668. doi: 10.1016/S0981-9428(02)01425-1420
Frey, F. P., Presterl, T., Lecoq, P., Orlik, A., and Stich, B. (2016). First steps to understand heat tolerance of temperate maize at adult stage: identification of QTL across multiple environments with connected segregating populations. Theor. Appl. Genet. 129, 945–961. doi: 10.1007/s00122-016-2674-2676
Gangola, M. P., and Ramadoss, B. R. (2020). “WRKY transcription factors for biotic and abiotic stress tolerance in plants,” in Transcription Factors for Abiotic Stress Tolerance in Plants, (Amsterdam: Elsevier), 15–28.
Ganie, A. H., Ahmad, A., Pandey, R., Aref, I. M., Yousuf, P. Y., Ahmad, S., et al. (2015). Metabolite profiling of Low-P tolerant and Low-P sensitive maize genotypes under phosphorus starvation and restoration conditions. PLoS One 10:e0129520. doi: 10.1371/journal.pone.0129520
Ganie, S. A., and Ahammed, G. J. (2021). Dynamics of cell wall structure and related genomic resources for drought tolerance in rice. Plant Cell Rep. 40, 437–459. doi: 10.1007/s00299-020-02649-2642
Gavaghan, C. L., Li, J. V., Hadfield, S. T., Hole, S., Nicholson, J. K., Wilson, I. D., et al. (2011). Application of NMR-based metabolomics to the investigation of salt stress in maize (Zea mays): application of NMR-based metabolomics to the investigation of salt stress in maize (Zea mays). Phytochem. Anal. 22, 214–224. doi: 10.1002/pca.1268
Geiger, H., and Gordillo, G. (2009). Double haploids in hybride maize breeding. Maydica 54, 485–499.
Gilker, R. E., Weil, R. R., Krizek, D. T., and Momen, B. (2002). Eastern gamagrass root penetration in adverse subsoil conditions. Soil Sci. Soc. Am. J. 66, 931–938. doi: 10.2136/sssaj2002.9310
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gill, T., Gill, S. K., Saini, D. K., Chopra, Y., de Koff, J. P., and Sandhu, K. S. (2022). A comprehensive review of high throughput phenotyping and machine learning for plant stress phenotyping. Phenomics 2, 156–183. doi: 10.1007/s43657-022-00048-z
Gitz, D. C., Baker, J. T., Stout, J. E., Brauer, D. K., Lascano, R. J., and Velten, J. P. (2013). Suitability of eastern gamagrass for in situ precipitation catchment forage production in playas. Agron. J. 105, 907–914. doi: 10.2134/agronj2012.0358
Giuliani, S., Sanguineti, M. C., Tuberosa, R., Bellotti, M., Salvi, S., and Landi, P. (2005). Root-ABA1, a major constitutive QTL, affects maize root architecture and leaf ABA concentration at different water regimes. J. Exp. Bot. 56, 3061–3070. doi: 10.1093/jxb/eri303
Godínez-Palma, S. K., García, E., Sánchez, M., de la, P., Rosas, F., and Vázquez-Ramos, J. M. (2013). Complexes of D-type cyclins with CDKs during maize germination. J. Exp. Bot. 64, 5661–5671. doi: 10.1093/jxb/ert340
Goering, R., Larsen, S., Tan, J., Whelan, J., and Makarevitch, I. (2021). QTL mapping of seedling tolerance to exposure to low temperature in the maize IBM RIL population. PLoS One 16:e0254437. doi: 10.1371/journal.pone.0254437
Guan, L. M., Zhao, J., and Scandalios, J. G. (2000). Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J. 22, 87–95. doi: 10.1046/j.1365-313x.2000.00723.x
Guo, J., Li, C., Zhang, X., Li, Y., Zhang, D., Shi, Y., et al. (2020). Transcriptome and GWAS analyses reveal candidate gene for seminal root length of maize seedlings under drought stress. Plant Sci. 292:110380. doi: 10.1016/j.plantsci.2019.110380
Han, Q., Zhu, Q., Shen, Y., Lee, M., Lübberstedt, T., and Zhao, G. (2022). QTL mapping low-temperature germination ability in the maize IBM Syn10 DH population. Plants 11:214. doi: 10.3390/plants11020214
Hasanuzzaman, M., Nahar, K., Alam Md Roychowdhury, R., and Fujita, M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. IJMS 14, 9643–9684. doi: 10.3390/ijms14059643
Hickey, L. T., Germán, S. E., Pereyra, S. A., Diaz, J. E., Ziems, L. A., Fowler, R. A., et al. (2017). Speed breeding for multiple disease resistance in barley. Euphytica 213:64. doi: 10.1007/s10681-016-1803-1802
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Hu, S., Lübberstedt, T., Zhao, G., and Lee, M. (2016). QTL mapping of low-temperature germination ability in the maize IBM Syn4 RIL population. PLoS One 11:e0152795. doi: 10.1371/journal.pone.0152795
Huang, M., He, C., Zhu, Q., and Qin, J. (2016). Maize seed variety classification using the integration of spectral and image features combined with feature transformation based on hyperspectral imaging. Appl. Sci. 6:183. doi: 10.3390/app6060183
Hund, A., Reimer, R., and Messmer, R. (2011). A consensus map of QTLs controlling the root length of maize. Plant Soil 344, 143–158. doi: 10.1007/s11104-011-0735-739
Inghelandt, D. V., Frey, F. P., Ries, D., and Stich, B. (2019). QTL mapping and genome-wide prediction of heat tolerance in multiple connected populations of temperate maize. Sci. Rep. 9:14418. doi: 10.1038/s41598-019-50853-50852
Itoh, J.-I., Hibara, K.-I., Sato, Y., and Nagato, Y. (2008). Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 147, 1960–1975. doi: 10.1104/pp.108.118679
Jena, K. K., and Mackill, D. J. (2008). Molecular markers and their use in marker-assisted selection in rice. Crop Sci. 48, 1266–1276. doi: 10.2135/cropsci2008.02.0082
Jeyasri, R., Muthuramalingam, P., Satish, L., Pandian, S. K., Chen, J.-T., Ahmar, S., et al. (2021). An overview of abiotic stress in cereal crops: negative impacts, regulation, biotechnology and integrated omics. Plants 10:1472. doi: 10.3390/plants10071472
Jiang, M. (2002). Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 53, 2401–2410. doi: 10.1093/jxb/erf090
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-Guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jones, H. G., Serraj, R., Loveys, B. R., Xiong, L., Wheaton, A., and Price, A. H. (2009). Thermal infrared imaging of crop canopies for the remote diagnosis and quantification of plant responses to water stress in the field. Funct. Plant Biol. 36:978. doi: 10.1071/FP09123
Joung, J. K., and Sander, J. D. (2013). TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55. doi: 10.1038/nrm3486
Juarez, M. T., Kui, J. S., Thomas, J., Heller, B. A., and Timmermans, M. C. P. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. doi: 10.1038/nature02363
Kaur, B., Sandhu, K. S., Kamal, R., Kaur, K., Singh, J., Röder, M. S., et al. (2021). Omics for the improvement of abiotic, biotic, and agronomic traits in major cereal crops: applications, challenges, and prospects. Plants 10:1989. doi: 10.3390/plants10101989
Kavar, T., Maras, M., Kidriè, M., Šuštar-Vozliè, J., and Megliè, V. (2008). Identification of genes involved in the response of leaves of Phaseolus vulgaris to drought stress. Mol. Breeding 21, 159–172. doi: 10.1007/s11032-007-9116-9118
Kemper, W., Alberts, E., Foy, C., Clark, R., Ritchie, J., and Zobel, R. (1998). Management of Carbon Sequestration in Soil, 1st Edn. Boca Raton, FL: CRC Press.
Kitony, J. K., Sunohara, H., Tasaki, M., Mori, J.-I., Shimazu, A., Reyes, V. P., et al. (2021). Development of an Aus-Derived Nested Association Mapping (Aus-NAM) population in rice. Plants 10:1255. doi: 10.3390/plants10061255
Kong, Y., Elling, A. A., Chen, B., and Deng, X. (2010). Differential expression of microRNAs in maize inbred and hybrid lines during salt and drought stress. AJPS 1, 69–76. doi: 10.4236/ajps.2010.12009
Landi, P., Sanguineti, M., Liu, C., Li, Y., Wang, T., Giuliani, S., et al. (2006). Root-ABA1 QTL affects root lodging, grain yield, and other agronomic traits in maize grown under well-watered and water-stressed conditions. J. Exp. Bot. 58, 319–326. doi: 10.1093/jxb/erl161
Lee, K.-M., Davis, J., Herrman, T. J., Murray, S. C., and Deng, Y. (2015). An empirical evaluation of three vibrational spectroscopic methods for detection of aflatoxins in maize. Food Chem. 173, 629–639. doi: 10.1016/j.foodchem.2014.10.099
Leitner, D., Meunier, F., Bodner, G., Javaux, M., and Schnepf, A. (2014). Impact of contrasted maize root traits at flowering on water stress tolerance - a simulation study. Field Crops Res. 165, 125–137. doi: 10.1016/j.fcr.2014.05.009
Li, J., Fu, F., An, M., Zhou, S., She, Y., and Li, W. (2013). Differential expression of MicroRNAs in response to drought stress in maize. J. Int. Agriculture 12, 1414–1422.
Li, N., Yang, T., Guo, Z., Wang, Q., Chai, M., Wu, M., et al. (2020). Maize microRNA166 inactivation confers plant development and abiotic stress resistance. IJMS 21:9506. doi: 10.3390/ijms21249506
Li, X., and Jiang, Y. (2021). Expression of ZmNAC3 responsive to various abiotic stresses in maize (Zea mays L.). Bangladesh J. Bot. 50, 141–146. doi: 10.3329/bjb.v50i1.52681
Lin, Y., Zhang, C., Lan, H., Gao, S., Liu, H., Liu, J., et al. (2014). Validation of potential reference genes for qPCR in maize across abiotic stresses, hormone treatments, and tissue types. PLoS One 9:e95445. doi: 10.1371/journal.pone.0095445
Liu, S., Wang, X., Wang, H., Xin, H., Yang, X., Yan, J., et al. (2013). Genome-Wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet 9:e1003790. doi: 10.1371/journal.pgen.1003790
Liu, X., Fu, J., Gu, D., Liu, W., Liu, T., Peng, Y., et al. (2008). Genome-wide analysis of gene expression profiles during the kernel development of maize (Zea mays L.). Genomics 91, 378–387. doi: 10.1016/j.ygeno.2007.12.002
Liu, X., Zhang, X., Sun, B., Hao, L., Liu, C., Zhang, D., et al. (2019). Genome-wide identification and comparative analysis of drought-related microRNAs in two maize inbred lines with contrasting drought tolerance by deep sequencing. PLoS One 14:e0219176. doi: 10.1371/journal.pone.0219176
Luan, M., Xu, M., Lu, Y., Zhang, Q., Zhang, L., Zhang, C., et al. (2014). Family-Wide survey of miR169s and NF-YAs and their expression profiles response to abiotic stress in maize roots. PLoS One 9:e91369. doi: 10.1371/journal.pone.0091369
Luo, M., Zhang, Y., Chen, K., Kong, M., Song, W., Lu, B., et al. (2019). Mapping of quantitative trait loci for seedling salt tolerance in maize. Mol. Breeding 39:64. doi: 10.1007/s11032-019-0974-977
Ma, N., Dong, L., Lü, W., Lü, J., Meng, Q., and Liu, P. (2020). Transcriptome analysis of maize seedling roots in response to nitrogen-, phosphorus-, and potassium deficiency. Plant Soil 447, 637–658. doi: 10.1007/s11104-019-04385-4383
Maazou, A., Tu, J., Qiu, J., and Liu, Z. (2016). Breeding for drought tolerance in maize (Zea mays L.). AJPS 07, 1858–1870. doi: 10.4236/ajps.2016.714172
Mano, Y., and Omori, F. (2008). Verification of QTL controlling root aerenchyma formation in a maize * teosinte “Zea nicaraguensis” advanced backcross population. Breed. Sci. 58, 217–223. doi: 10.1270/jsbbs.58.217
Marti, J., Bort, J., Slafer, G. A., and Araus, J. L. (2007). Can wheat yield be assessed by early measurements of normalized difference vegetation index? Ann. Appl. Biol. 150, 253–257. doi: 10.1111/j.1744-7348.2007.00126.x
Massman, J. M., Jung, H. G., and Bernardo, R. (2013). Genomewide selection versus marker-assisted recurrent selection to improve grain yield and stover-quality traits for cellulosic ethanol in maize. Crop Sci. 53, 58–66. doi: 10.2135/cropsci2012.02.0112
Masuka, B., Araus, J. L., Das, B., Sonder, K., and Cairns, J. E. (2012). Phenotyping for abiotic stress tolerance in maizef. J. Int. Plant Biol. 54, 238–249. doi: 10.1111/j.1744-7909.2012.01118.x
McNellie, J. P., Chen, J., Li, X., and Yu, J. (2018). Genetic mapping of foliar and tassel heat stress tolerance in maize. Crop Sci. 58, 2484–2493. doi: 10.2135/cropsci2018.05.0291
Medeiros, D. B., Brotman, Y., and Fernie, A. R. (2021). The utility of metabolomics as a tool to inform maize biology. Plant Commun. 2:100187. doi: 10.1016/j.xplc.2021.100187
Mehla, N., Sindhi, V., Josula, D., Bisht, P., and Wani, S. H. (2017). “An introduction to antioxidants and their roles in plant stress tolerance,” in Reactive Oxygen Species and Antioxidant Systems in Plants: Role and Regulation under Abiotic Stress, eds M. I. R. Khan and N. A. Khan (Singapore: An Introduction to Antioxidants and Their Roles in Plant Stress Tolerance), 1–23. doi: 10.1007/978-981-10-5254-5_1
Menezes-Benavente, L., Kernodle, S. P., Margis-Pinheiro, M., and Scandalios, J. G. (2004). Salt-induced antioxidant metabolism defenses in maize (Zea mays L.) seedlings. Redox Rep. 9, 29–36. doi: 10.1179/135100004225003888
Miller, J., McLachlan, A. D., and Klug, A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4, 1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x
Molla Md, S. H., Nakasathien, S., Ali Md, A., Khan, A., Alam Md, R., Hossain, A., et al. (2019). Influence of nitrogen application on dry biomass allocation and translocation in two maize varieties under short pre-anthesis and prolonged bracketing flowering periods of drought. Arch. Agronomy Soil Sci. 65, 928–944. doi: 10.1080/03650340.2018.1538557
Mushtaq, M., Dar, A. A., Basu, U., Bhat, B. A., Mir, R. A., Vats, S., et al. (2021). Integrating CRISPR-Cas and next generation sequencing in plant virology. Front. Genet. 12:735489. doi: 10.3389/fgene.2021.735489
Nelson, D. E., Repetti, P. P., Adams, T. R., Creelman, R. A., Wu, J., Warner, D. C., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. U S A. 104, 16450–16455. doi: 10.1073/pnas.0707193104
Nogueira, F. T. S., Chitwood, D. H., Madi, S., Ohtsu, K., Schnable, P. S., Scanlon, M. J., et al. (2009). Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet 5:e1000320. doi: 10.1371/journal.pgen.1000320
Nogueira, F. T. S., Madi, S., Chitwood, D. H., Juarez, M. T., and Timmermans, M. C. P. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21, 750–755. doi: 10.1101/gad.1528607
Obata, T., Witt, S., Lisec, J., Palacios-Rojas, N., Florez-Sarasa, I., Araus, J. L., et al. (2015). Metabolite profiles of maize leaves in drought, heat and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiol. 169, 2665–2683. doi: 10.1104/pp.15.01164
Osman, K. A., Tang, B., Wang, Y., Chen, J., Yu, F., Li, L., et al. (2013). Dynamic QTL analysis and candidate gene mapping for waterlogging tolerance at maize seedling stage. PLoS One 8:e79305. doi: 10.1371/journal.pone.0079305
Osmolovskaya, N., Shumilina, J., Kim, A., Didio, A., Grishina, T., Bilova, T., et al. (2018). Methodology of drought stress research: experimental setup and physiological characterization. IJMS 19:4089. doi: 10.3390/ijms19124089
Parent, B., and Tardieu, F. (2012). Temperature responses of developmental processes have not been affected by breeding in different ecological areas for 17 crop species. New Phytol. 194, 760–774. doi: 10.1111/j.1469-8137.2012.04086.x
Peng, T., Qiao, M., Liu, H., Teotia, S., Zhang, Z., Zhao, Y., et al. (2018). A resource for inactivation of MicroRNAs using short tandem target mimic technology in model and crop plants. Mol. Plant 11, 1400–1417. doi: 10.1016/j.molp.2018.09.003
Pesqueira, J., García, M. D., and Molina, M. C. (2003). NaCl tolerance in maize (Zea mays ssp. mays) x Tripsacum dactyloides L. hybrid calli in regenerated plants. Span. J. Agric. Res. 1:59. doi: 10.5424/sjar/2003012-2003021
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L.-S. P., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L: ZmDREB2A in drought and heat stress response. Plant J. 50, 54–69. doi: 10.1111/j.1365-313X.2007.03034.x
Qiu, F., Zheng, Y., Zhang, Z., and Xu, S. (2007). Mapping of QTL associated with waterlogging tolerance during the seedling stage in maize. Ann. Bot. 99, 1067–1081. doi: 10.1093/aob/mcm055
Rafique, S., Qadri, S., and Abdin, M. (2020). Comparative metabolic profile of maize genotypes reveals the tolerance mechanism associated in combined stresses. J. Pharmacogn. Phytochem. 9, 1217–1224.
Ranum, P., Peña-Rosas, J. P., and Garcia-Casal, M. N. (2014). Global maize production, utilization, and consumption. Ann. N.Y. Acad. Sci. 1312, 105–112. doi: 10.1111/nyas.12396
Razzaq, A., Saleem, F., Kanwal, M., Mustafa, G., Yousaf, S., Imran Arshad, H. M., et al. (2019). Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. IJMS 20:4045. doi: 10.3390/ijms20164045
Reyes, V. P., Angeles-Shim, R. B., Lapis, R. S., Shim, J., Sunohara, H., Jena, K. K., et al. (2021a). Improvement of Asian rice cultivars through marker-assisted introgression of yield QTLs grain number 1a (Gn1a) and Wealthy Farmer’s Panicle (WFP). Philipp. J. Biochem. Mol. Biol. 2:29. doi: 10.5555/pjbmb.ph.2021.02.02.23
Reyes, V. P., Angeles-Shim, R. B., Mendioro, M. S., Manuel, M. C. C., Lapis, R. S., Shim, J., et al. (2021b). Marker-Assisted introgression and stacking of major QTLs controlling grain number (Gn1a) and number of primary branching (WFP) to NERICA cultivars. Plants 10:844. doi: 10.3390/plants10050844
Richter, J. A., Erban, A., Kopka, J., and Zörb, C. (2015). Metabolic contribution to salt stress in two maize hybrids with contrasting resistance. Plant Sci. 233, 107–115. doi: 10.1016/j.plantsci.2015.01.006
Rorie, R. L., Purcell, L. C., Karcher, D. E., and King, C. A. (2011). The assessment of leaf nitrogen in corn from digital images. Crop Sci. 51, 2174–2180. doi: 10.2135/cropsci2010.12.0699
Samantara, K., Reyes, V. P., Agrawal, N., Mohapatra, S. R., and Jena, K. K. (2021). Advances and trends on the utilization of multi-parent advanced generation intercross (MAGIC) for crop improvement. Euphytica 217:189. doi: 10.1007/s10681-021-02925-2926
Sankaran, S., Zhang, C., Hurst, P., Marzougui, A., Veeranampalayam Sivakumar, A. N., Li, J., et al. (2020). “Investigating the potential of satellite imagery for high-throughput field phenotyping applications,” in Autonomous Air and Ground Sensing Systems for Agricultural Optimization and Phenotyping V, eds J. A. Thomasson and A. F. Torres-Rua (Bellingham, Was: SPIE).
Scandalios, J. G., Acevedo, A., and Ruzsa, S. (2000). Catalase gene expression in response to chronic high temperature stress in maize. Plant Sci. 156, 103–110. doi: 10.1016/s0168-9452(00)00235-1
Seeve, C. M., Sunkar, R., Zheng, Y., Liu, L., Liu, Z., McMullen, M., et al. (2019). Water-deficit responsive microRNAs in the primary root growth zone of maize. BMC Plant Biol. 19:447. doi: 10.1186/s12870-019-2037-y
Semagn, K., Beyene, Y., Warburton, M. L., Tarekegne, A., Mugo, S., Meisel, B., et al. (2013). Meta-analyses of QTL for grain yield and anthesis silking interval in 18 maize populations evaluated under water-stressed and well-watered environments. BMC Genomics 14:313. doi: 10.1186/1471-2164-14-313
Sepúlveda-García, E. B., Pulido-Barajas, J. F., Huerta-Heredia, A. A., Peña-Castro, J. M., Liu, R., and Barrera-Figueroa, B. E. (2020). Differential expression of maize and teosinte microRNAs under submergence, drought, and alternated stress. Plants 9:1367. doi: 10.3390/plants9101367
Shah, F., Huang, J., Cui, K., Nie, L., Shah, T., Chen, C., et al. (2011). Impact of high-temperature stress on rice plant and its traits related to tolerance. J. Agric. Sci. 149, 545–556. doi: 10.1017/S0021859611000360
Shao, A., Wang, W., Fan, S., Xu, X., Yin, Y., Erick, A., et al. (2021). Comprehensive transcriptional analysis reveals salt stress-regulated key pathways, hub genes and time-specific responsive gene categories in common bermudagrass (Cynodon dactylon (L.) Pers.) roots. BMC Plant Biol. 21:175. doi: 10.1186/s12870-021-02939-2931
Shavrukov, Y., and Sokolov, V. (2015). Maize-gamagrass interspecific hybrid, Zea mays x Tripsacum dactyloides, shows better salinity tolerance and higher Na+ exclusion than maize and sorghum. Int. J. Latest Res. Sci. Technol. 4, 128–133.
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Shou, H. (2004). Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. J. Exp. Bot. 55, 1013–1019. doi: 10.1093/jxb/erh129
Singh, A., Jones, S., Ganapathysubramanian, B., Sarkar, S., Mueller, D., Sandhu, K., et al. (2021). Challenges and opportunities in machine-augmented plant stress phenotyping. Trends Plant Sci. 26, 53–69. doi: 10.1016/j.tplants.2020.07.010
Singh, A. K., Ganapathysubramanian, B., Sarkar, S., and Singh, A. (2018). Deep learning for plant stress phenotyping: trends and future perspectives. Trends Plant Sci. 23, 883–898. doi: 10.1016/j.tplants.2018.07.004
Soliman, S., El-Keblawy, A., Mosa, K. A., Helmy, M., and Wani, S. H. (2018). “Understanding the phytohormones biosynthetic pathways for developing engineered environmental stress-tolerant crops,” in Biotechnologies of Crop Improvement, eds S. S. Gosal and S. H. Wani (Cham: Springer International Publishing), 417–450. doi: 10.1007/978-3-319-90650-8_15
Song, X. Y., Zhang, Y. Y., Wu, F. C., and Zhang, L. (2016). Genome-wide analysis of the maize (Zea may L.) CPP-like gene family and expression profiling under abiotic stress. Genet. Mol. Res. 15, 1–11. doi: 10.4238/gmr.15038023
Spollen, W. G., Tao, W., Valliyodan, B., Chen, K., Hejlek, L. G., Kim, J.-J., et al. (2008). Spatial distribution of transcript changes in the maize primary root elongation zone at low water potential. BMC Plant Biol. 8:32. doi: 10.1186/1471-2229-8-32
Sun, C., Gao, X., Chen, X., Fu, J., and Zhang, Y. (2016). Metabolic and growth responses of maize to successive drought and re-watering cycles. Agricultural Water Manag. 172, 62–73. doi: 10.1016/j.agwat.2016.04.016
Sun, Q., Wang, P., Li, W., Li, W., Lu, S., Yu, Y., et al. (2019). Genomic selection on shelling percentage and other traits for maize. Breed. Sci. 69, 266–271. doi: 10.1270/jsbbs.18141
Tejeda, L. H. C., Viana, V. E., Maltzahn, L. E., Busanello, C., Barros, L. M., da Maia, L. C., et al. (2019). Abiotic stress and self-destruction: ZmATG8 and ZmATG12 gene transcription and osmotic stress responses in maize. Biotechnol. Res. Innov. 3, 1–9. doi: 10.1016/j.biori.2019.12.001
Thatcher, S. R., Danilevskaya, O. N., Meng, X., Beatty, M., Zastrow-Hayes, G., Harris, C., et al. (2016). Genome-Wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 170, 586–599. doi: 10.1104/pp.15.01267
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U S A. 108, 20260–20264. doi: 10.1073/pnas.1116437108
Tiwari, Y. K., and Yadav, S. K. (2019). Temperature induced changes in anti-oxidative stress metabolism in maize. Israel J. Plant Sci. 66, 203–212. doi: 10.1163/22238980-20191072
Trachsel, S., Dhliwayo, T., Gonzalez Perez, L., Mendoza Lugo, J. A., and Trachsel, M. (2019). Estimation of physiological genomic estimated breeding values (PGEBV) combining full hyperspectral and marker data across environments for grain yield under combined heat and drought stress in tropical maize (Zea mays L.). PLoS One 14:e0212200. doi: 10.1371/journal.pone.0212200
Turan, M. A., Elkarim, A. H. A., Taban, N., and Taban, S. (2010). Effect of salt stress on growth and ion distribution and accumulation in shoot and root of maize plant. Afr. J. Agric. Res. 5, 584–588. doi: 10.1186/s12870-021-03237-6
Vilardell, J., Mundy, J., Stilling, B., Leroux, B., Pla, M., Freyssinet, G., et al. (1991). Regulation of the maizerab17 gene promoter in transgenic heterologous systems. Plant Mol. Biol. 17, 985–993. doi: 10.1007/BF00037138
Wahid, A., Gelani, S., Ashraf, M., and Foolad, M. (2007). Heat tolerance in plants: an overview. Environ. Exp. Bot. 61, 199–223. doi: 10.1016/j.envexpbot.2007.05.011
Wang, B., Li, Z., Ran, Q., Li, P., Peng, Z., and Zhang, J. (2018a). ZmNF-YB16 overexpression improves drought resistance and yield by enhancing photosynthesis and the antioxidant capacity of maize plants. Front. Plant Sci. 9:709. doi: 10.3389/fpls.2018.00709
Wang, C.-T., Ru, J.-N., Liu, Y.-W., Yang, J.-F., Li, M., Xu, Z.-S., et al. (2018b). The maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. IJMS 19:2580. doi: 10.3390/ijms19092580
Wang, B., Liu, C., Zhang, D., He, C., Zhang, J., and Li, Z. (2019). Effects of maize organ-specific drought stress response on yields from transcriptome analysis. BMC Plant Biol. 19:335. doi: 10.1186/s12870-019-1941-1945
Wang, N., Liu, B., Liang, X., Zhou, Y., Song, J., Yang, J., et al. (2019). Genome-wide association study and genomic prediction analyses of drought stress tolerance in China in a collection of off-PVP maize inbred lines. Mol. Breeding 39:113. doi: 10.1007/s11032-019-1013-1014
Wang, Y., Gu, W., Meng, Y., Xie, T., Li, L., Li, J., et al. (2017). γ-Aminobutyric acid imparts partial protection from salt stress injury to maize seedlings by improving photosynthesis and upregulating osmoprotectants and antioxidants. Sci. Rep. 7:43609. doi: 10.1038/srep43609
Wang, Y.-G., An, M., Zhou, S.-F., She, Y.-H., Li, W.-C., and Fu, F.-L. (2014). Expression profile of maize MicroRNAs corresponding to their target genes under drought stress. Biochem. Genet. 52, 474–493. doi: 10.1007/s10528-014-9661-x
Wani, S. H., Kumar, V., Khare, T., Tripathi, P., Shah, T., Ramakrishna, C., et al. (2020). miRNA applications for engineering abiotic stress tolerance in plants. Biologia 75, 1063–1081. doi: 10.2478/s11756-019-00397-397
Wani, S. H., Sah, S. K., Hossain, M. A., Kumar, V., and Balachandran, S. M. (2016). “Transgenic approaches for abiotic stress tolerance in crop plants,” in Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits, eds J. M. Al-Khayri, S. M. Jain, and D. V. Johnson (Cham: Springer International Publishing), 345–396. doi: 10.1007/978-3-319-22518-0_10
Waqas, M. A., Khan, I., Akhter, M. J., Noor, M. A., and Ashraf, U. (2017). Exogenous application of plant growth regulators (PGRs) induces chilling tolerance in short-duration hybrid maize. Environ. Sci. Pollut. Res. 24, 11459–11471. doi: 10.1007/s11356-017-8768-8760
Windhausen, V. S., Atlin, G. N., Hickey, J. M., Crossa, J., Jannink, J.-L., Sorrells, M. E., et al. (2012). Effectiveness of genomic prediction of maize hybrid performance in different breeding populations and environments. G3 Genes Genomes Genetics 2, 1427–1436. doi: 10.1534/g3.112.003699
Witt, S., Galicia, L., Lisec, J., Cairns, J., Tiessen, A., Araus, J. L., et al. (2012). Metabolic and phenotypic responses of greenhouse-grown maize hybrids to experimentally controlled drought stress. Mol. Plant 5, 401–417. doi: 10.1093/mp/ssr102
Xu, C., Yang, R.-F., Li, W.-C., and Fu, F.-L. (2010). Identification of 21 microRNAs in maize and their differential expression under drought stress. African J. Biotechnol. 9:13. doi: 10.3390/ijms16035714
Yadav, S. K., Tiwari, Y. K., Singh, V., Patil, A. A., Shanker, A. K., Jyothi Lakshmi, N., et al. (2018). Physiological and biochemical basis of extended and sudden heat stress tolerance in maize. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 88, 249–263. doi: 10.1007/s40011-016-0752-759
Yin, X.-Y., Yang, A.-F., Zhang, K.-W., and Zhang, J. (2004). Production and analysis of transgenic maize with improved salt tolerance by the introduction of AtNHX1 gene. Acta Botanica Sinica 46, 854–861.
Ying, S., Zhang, D.-F., Li, H.-Y., Liu, Y.-H., Shi, Y.-S., Song, Y.-C., et al. (2011). Cloning and characterization of a maize SnRK2 protein kinase gene confers enhanced salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 30, 1683–1699. doi: 10.1007/s00299-011-1077-z
Zaidi, P. H., Rashid, Z., Vinayan, M. T., Almeida, G. D., Phagna, R. K., and Babu, R. (2015). QTL mapping of agronomic waterlogging tolerance using recombinant inbred lines derived from tropical maize (Zea mays L) germplasm. PLoS One 10:e0124350. doi: 10.1371/journal.pone.0124350
Zeid, I. (2009). Trehalose as osmoprotectant for maize under salinity-induced stress. Res. J. Agriculture Biol. Sci. 5, 613–622.
Zhang, H., Li, G., Hu, D., Zhang, Y., Zhang, Y., Shao, H., et al. (2020). Functional characterization of maize heat shock transcription factor gene ZmHsf01 in thermotolerance. PeerJ 8:e8926. doi: 10.7717/peerj.8926
Zhang, L., Niu, Y., Zhang, H., Han, W., Li, G., Tang, J., et al. (2019a). Maize canopy temperature extracted from UAV thermal and RGB imagery and its application in water stress monitoring. Front. Plant Sci. 10:1270. doi: 10.3389/fpls.2019.01270
Zhang, M., An, P., Li, H., Wang, X., Zhou, J., Dong, P., et al. (2019b). The miRNA-Mediated post-transcriptional regulation of maize in response to high temperature. IJMS 20:1754. doi: 10.3390/ijms20071754
Zhang, X., Warburton, M. L., Setter, T., Liu, H., Xue, Y., Yang, N., et al. (2016). Genome-wide association studies of drought-related metabolic changes in maize using an enlarged SNP panel. Theor. Appl. Genet. 129, 1449–1463. doi: 10.1007/s00122-016-2716-2710
Zhang, Z., Wei, L., Zou, X., Tao, Y., Liu, Z., and Zheng, Y. (2008). Submergence-responsive MicroRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Ann. Bot. 102, 509–519. doi: 10.1093/aob/mcn129
Zhao, X., Zhang, J., Fang, P., and Peng, Y. (2019). Comparative QTL analysis for yield components and morphological traits in maize (Zea mays L.) under water-stressed and well-watered conditions. Breed. Sci. 69, 621–632. doi: 10.1270/jsbbs.18021
Zhou, M.-L., Tang, Y.-X., and Wu, Y.-M. (2012). Genome-Wide analysis of AP2/ERF transcription factor family in Zea mays. CBIO 7, 324–332. doi: 10.2174/157489312802460776
Zia, S., Romano, G., Spreer, W., Sanchez, C., Cairns, J., Araus, J. L., et al. (2013). Infrared thermal imaging as a rapid tool for identifying water-stress tolerant maize genotypes of different phenology. J. Agro Crop. Sci. 199, 75–84. doi: 10.1111/j.1439-037X.2012.00537.x
Keywords: genomics, miRNA, genome editing, phenomics, transcriptomics
Citation: Farooqi MQU, Nawaz G, Wani SH, Choudhary JR, Rana M, Sah RP, Afzal M, Zahra Z, Ganie SA, Razzaq A, Reyes VP, Mahmoud EA, Elansary HO, El-Abedin TKZ and Siddique KHM (2022) Recent developments in multi-omics and breeding strategies for abiotic stress tolerance in maize (Zea mays L.). Front. Plant Sci. 13:965878. doi: 10.3389/fpls.2022.965878
Received: 10 June 2022; Accepted: 23 August 2022;
Published: 23 September 2022.
Edited by:
Shah Fahad, The University of Haripur, PakistanReviewed by:
Karansher Singh Sandhu, Bayer Crop Science, United StatesCopyright © 2022 Farooqi, Nawaz, Wani, Choudhary, Rana, Sah, Afzal, Zahra, Ganie, Razzaq, Reyes, Mahmoud, Elansary, El-Abedin and Siddique. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shabir Hussain Wani, c2hhYmlyaHdhbmlAc2t1YXN0a2FzaG1pci5hYy5pbg==; Kadambot H. M. Siddique, a2FkYW1ib3Quc2lkZGlxdWVAdXdhLmVkdS5hdQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.