- 1Department of Biotechnology, Vignan’s Foundation for Science, Technology & Research (Deemed to be University), Guntur, India
- 2Department of Biological Sciences, Florida A&M University, Tallahassee, FL, United States
- 3Biochemistry Division, National Institute of Nutrition, Hyderabad, India
- 4International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, India
- 5Department of Biology, Florida State University, Tallahassee, FL, United States
Potassium (K+) is the most abundant cation that plays a crucial role in various cellular processes in plants. Plants have developed an efficient mechanism for the acquisition of K+ when grown in K+ deficient or saline soils. A total of 47 K+ transport gene homologs (27 HAKs, 4 HKTs, 2 KEAs, 9 AKTs, 2 KATs, 2 TPCs, and 1 VDPC) have been identified in Sorghum bicolor. Of 47 homologs, 33 were identified as K+ transporters and the remaining 14 as K+ channels. Chromosome 2 has been found as the hotspot of K+ transporters with 9 genes. Phylogenetic analysis revealed the conservation of sorghum K+ transport genes akin to Oryza sativa. Analysis of regulatory elements indicates the key roles that K+ transport genes play under different biotic and abiotic stress conditions. Digital expression data of different developmental stages disclosed that expressions were higher in milk, flowering, and tillering stages. Expression levels of the genes SbHAK27 and SbKEA2 were higher during milk, SbHAK17, SbHAK11, SbHAK18, and SbHAK7 during flowering, SbHAK18, SbHAK10, and 23 other gene expressions were elevated during tillering inferring the important role that K+ transport genes play during plant growth and development. Differential transcript expression was observed in different tissues like root, stem, and leaf under abiotic stresses such as salt, drought, heat, and cold stresses. Collectively, the in-depth genome-wide analysis and differential transcript profiling of K+ transport genes elucidate their role in ion homeostasis and stress tolerance mechanisms.
Introduction
Potassium (K+) is an essential macronutrient and most ubiquitous monovalent cation in plants. It contributes up to 10% of total plant dry weight and plays an overriding role in diverse cellular processes such as ion homeostasis, plant growth, development, transport, and signaling (Feng et al., 2019; Hussain et al., 2021). Despite its abundance, K+ is not readily available to plants since they absorb it in the ionic form only, and the concentrations at the root surface often fall below or up to μM range (Ashley et al., 2006). K+ uptake in plants is mediated by two mechanisms: a low affinity system that functions when extracellular K+ concentration is high (> 200 μM to mM) and a high affinity system that functions when extracellular concentration is low (20 μM Rb+) (Gierth et al., 2005). Na+ competes with K+ but does not fulfill the physiological functions, and higher K+/Na+ is critical in maintaining electro-neutrality of the cells (Hussain et al., 2021). K+ transport occurs through five major families, classified under 2 categories as K+ transporters and channels. The transporters include HAK (high-affinity K+)/KUP (K+ uptake)/KT (K+ transporter) family, Trk/HKT family, and KEA (K+ efflux anti-porter) family, while the K+ channels include the shakers/voltage-gated channels (AKT and KAT) and non-voltage-gated [tandem-pore K+ (TPK) and two-pore (TPC)] channels (Hedrich, 2012). HAK/KUP/KT transporters are critical in maintaining osmotic potential and salt tolerance (Feng et al., 2019) and are the largest family of K+ transporters (Ahn et al., 2004). While HKTs are involved in the uptake of K+ during short-term K+ starvation (Riedelsberger et al., 2021), KEAs are implicated in the regulation of thylakoid and stromal pH (Sánchez-McSweeney et al., 2021). Abiotic stresses like salt, drought, heat, and cold impair the final yields (Teklić et al., 2021). But K+ is a vital regulator of plant responses and imparts tolerance to the abiotic stresses (Hasanuzzaman et al., 2018; Sardans and Peñuelas, 2021). K+ reduces the adverse effects of drought stress tolerance alongside maintaining turgor pressure at low water potentials, alleviates salt stress by achieving homeostatic balance, enhances seed yield during heat stress by reducing the silique canopy, improves freezing tolerance by accumulation of osmolytes (Assaha et al., 2017; Aksu and Altay, 2020; Hu et al., 2021; Saadati et al., 2021).
Genome-wide analyses have been widely performed in HAK/KUP/KT family, but studies were limited to K+ transporters. Analysis of HAK/KUP/KT transporters on a genome scale has been carried out in Oryza sativa (Banuelos et al., 2002), Arabidopsis thaliana (Ahn et al., 2004), Populus trichocarpa (He et al., 2012), Zea mays (Zhang et al., 2012), Solanum lycopersicum (Hyun et al., 2014), Prunus persica (Song et al., 2015), Triticum aestivum (Cheng et al., 2018), Pyrus betulifolia (Li Y. et al., 2018), Manihot esculenta (Ou et al., 2018), Nicotiana tabacum (Song et al., 2019), Saccharum spontaneum (Feng et al., 2020b), Gossypium (Yang X. et al., 2020), and Ipomoea batatas (Jin et al., 2021). Genome-wide analysis of K+ transport gene family has been reported in Oryza sativa (Amrutha et al., 2007), Glycine max (Rehman et al., 2017), Cicer arietinum (Azeem et al., 2018), Cajanus cajan (Siddique et al., 2021), and Gossypium raimondii (Azeem et al., 2022) but not in Sorghum bicolor.
Sorghum, a moderately drought stress-tolerant crop is the fifth most important cereal. It is the staple food for human populations in arid regions and a good source of feed and fuel in the global agronomics and economics. It is a self-pollinated, C4 photosynthetic plant with a smaller genome size of 730 Mb (Paterson et al., 2009). In the present study, the discovery, and identification of K+ transport gene homologs in Sorghum bicolor were conducted including their expression profiles in different tissues under various abiotic stresses. Further, chromosomal locations, gene characterization, protein modeling, conserved motifs analysis, cellular localization, promoter analysis, evolutionary relationship, and protein-protein interactions were investigated, resulting in the characterization of candidate genes.
Materials and methods
In silico prediction, identification, and characterization of K+ transport gene homologs
All the 49 rice full-length cDNA sequences of K+ encoding genes were collected from rice (Amrutha et al., 2007). The homology search of the collected FASTA sequences was performed by BLASTN against Sorghum genome in the Gramene database with default settings.1 The coding sequences (CDS) and corresponding protein sequences were retrieved from the BLAST output using GENSCAN web server.2 To check the presence of the K+ domain, Conserved Domain Database search and SMART were employed.3, 4 For analysis of transmembrane, TMHMM (Moller et al., 2001), for prediction of gene structure, Gene Structure Display Server,5 for prediction of subcellular localization of the protein, WoLFPSORT,6 for isoelectric point (pI) molecular weight (MW), GRAVY (grand average of hydropathy) instability, and aliphatic indexes, ProtParam software,7 for identification of phosphorylation sites, NetPhos 3.18 for amino acid composition and net charge, pepcalc9 for conserved motifs, MEME10 with parameters like 10 number of motifs, 2–20 motif sites, 6–20 wide motif width were used. The genes on the chromosomes were mapped based on their physical location using MG2C11 tool.
Prediction of cis-elements, protein modeling and protein-protein interactions
Promoter elements were identified for all the transporter and channel genes by taking 2 kb sequence upstream to all the sorghum K+ transport homologs using PLACE tools.12 The 3D structures of all the K+ transport proteins were predicted using SWISS-MODEL server (Biasini et al., 2014). The predicted 3D structures of proteins were evaluated for stability using protein structure verification server (PSVS).13 The stability of the proteins was analyzed by Ramachandran plots. The predicted protein-protein interaction (PPI) map of sorghum K+ transport homologs was generated from the STRING database.14
Phylogenetic analysis and generation of synteny maps
A phylogenetic tree was constructed with amino acid sequences of Sorghum bicolor (Sb), Oryza sativa (Os), and Arabidopsis thaliana (At) using MEGA 10.0 software, by Neighbor-Joining method with 1000 bootstrap replicates (Kumar et al., 2018). Evolutionary analysis of orthologs and paralogs was performed by calculating synonymous (dS) and non-synonymous (dN) substitution rates using the PAL2NAL program.15 Synonymous (dS) and non-synonymous (dN) substitution rates were calculated by codeml in the PAML package. Synteny and collinearity were analyzed to identify K+ homologs using TBtools (Chen et al., 2020).
Digital and qRT-PCR analysis of K+ transport gene homologs under different abiotic stresses
For digital expression profiling of K+ transport genes, Genevestigator16 was used. The mRNA-seq data were used for analysis. The data are available for all the 46 genes (except SbHAK26) for 2 stress conditions (cold and drought) in 3 tissues (root, shoot, and leaf), and 4 developmental stages (milk stage, seedling stage, tillering stage, and flowering stage). Using hierarchical clustering, heat maps were generated separately for anatomical, developmental, and perturbations. Seeds of Sorghum bicolor L. BTx623 genotype were collected from the International Crops Research Institute for the Semi-Arid Tropics and used. Seventy-five-day-old seedlings maintained in green house at 28/20°C day/night temperatures were treated with salt (200 mM NaCl), drought (200 mM mannitol), heat (42°C), and cold (4°C) stresses for 4 h. Control (without any stress) plants were treated with tap water. Root, stem, and leaftissue samples were collected immediately, snap frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated from all the samples using Macherey-Nagel NucleoSpin RNA plant kit (740949.50) by following the instructions manual. Total RNA (2 μg) was taken as a template for first strand cDNA synthesis using RevertAid First Strand cDNA Synthesis Kit (#K1622, Thermo Scientific EU, Reinach, Switzerland). The relative expression levels of K+ gene homologs were studied using Mx3000p (Agilent) with 2X applied biosystems (ABI) Master Mix with gene specific primers (Supplementary Table 1) with the following thermal cycles: 1 cycle at 95°C for 10 min, followed by 40 cycles alternatively at 95°C for 15 s and 60°C for 1 min. The expression of each gene in various samples was normalized with ACTIN gene. The experiment was performed with two biological replicates and for each sample three technical replicates were used. The comparative 2–ΔΔCT method was used to calculate the relative quantities of each transcript in the samples (Schmittgen and Livak, 2008).
Results
Discovery, identification and characterization of K+ gene homologs
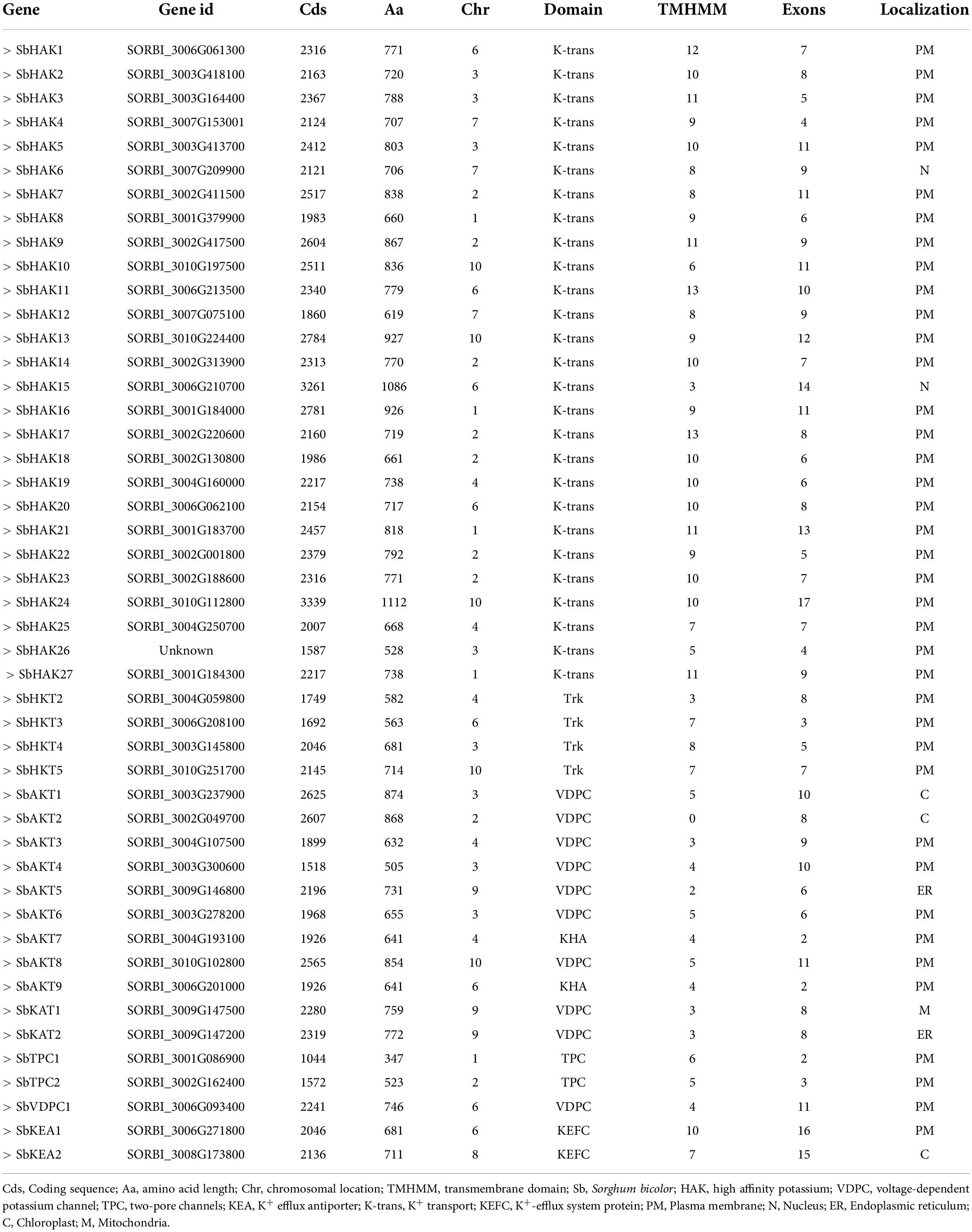
A total of 47 K+ transport homologs have been identified in sorghum (Table 1 and Supplementary Table 2). Of the 47 transport homologs 27 belong to HAKs (SbHAK1 to SbHAK27), 4 HKTs (SbHKT2, SbHAT3, SbHKT4, and SbHKT5), 9 AKTs (SbAKT1 to SbAKT9), 2 KEAs (SbKEA1 and SbKEA2), 2 KATs (SbKAT1 and SbKAT2), 2 TPKs (SbTPC1 and SbTPC2), and 1 VDPC (SbVDPC1) (Table 1). Homologs of OsHKTs (OsHKT1, OsHKT6, and OsHKT7) and OsKEA3 are not available in Sorghum bicolor. K+ transporter and channel domains like K-trans, TrK, voltage-dependent K+ channel, KHA, Two pore potassium channel, and K+-efflux system protein have been identified (Table 1). Predicted amino acid sequences were used to identify the number of transmembrane segments. While the number of transmembrane domains for K+ transporters varies from 0 (SbAKT2) to 13 (SbHAK11 and SbHAK17) (Table 1), the number of exons varies from 2 (SbAKT7, SbAKT9, and SbTPC1) to 17 (SbHAK24) in sorghum (Table 1 and Figure 1). Most of the K+ transport gene homologs are localized on the plasma membrane, followed by chloroplast, nucleus, endoplasmic reticulum, and mitochondria. SbHAK6 and HAK15 are localized on nucleus, SbAKT1, SbAKT2, and SbKEA on chloroplast, SbAKT5 and SbKAT2 on endoplasmic reticulum, and SbKAT1 on mitochondria (Table 1). The pI, MW, GRAVY, instability, aliphatic indexes (Supplementary Table 3), amino acid and net charge (Supplementary Table 4) have also been tabulated. Kinases play an important role in phosphorylation and K+ gene homologs showed 18 different serine, threonine, and tyrosine kinases (Supplementary Table 5). The consensus motif GVVYGDLGTSPLY was identified in all the HAK transporter proteins except HAK5, HAK12, and HAK22 (Figure 2). Another signature sequence like GGTFALYSLLCR has been observed in all the HAK transporters leaving out HAK7, HAK15, and HAK22. The motif SLVFWTLTLIPLLKYVFIVL has been detected in all the HAK transporters excluding HAK12 (Figure 2). The motifs, VEMEDFSSAQLLVLTLLM, FSVFTTVSTFSNCGFLPTNE, GEKLVNALFMAVNSRHAGE, and DLSTLASAILVLYVLMMYLP were noticed in all the sorghum HKT K+ transporter proteins (Figure 2). The KEA family displayed FFMTVGMSIDPKLLJREWP and KAFPNVKIFVRAKDLDH motifs (Figure 2). The sorghum K+ channel (AKT, KAT, TPK, and VDPC) proteins displayed the motif YWSITTLTTVGYGDLHAENP (Figure 2).
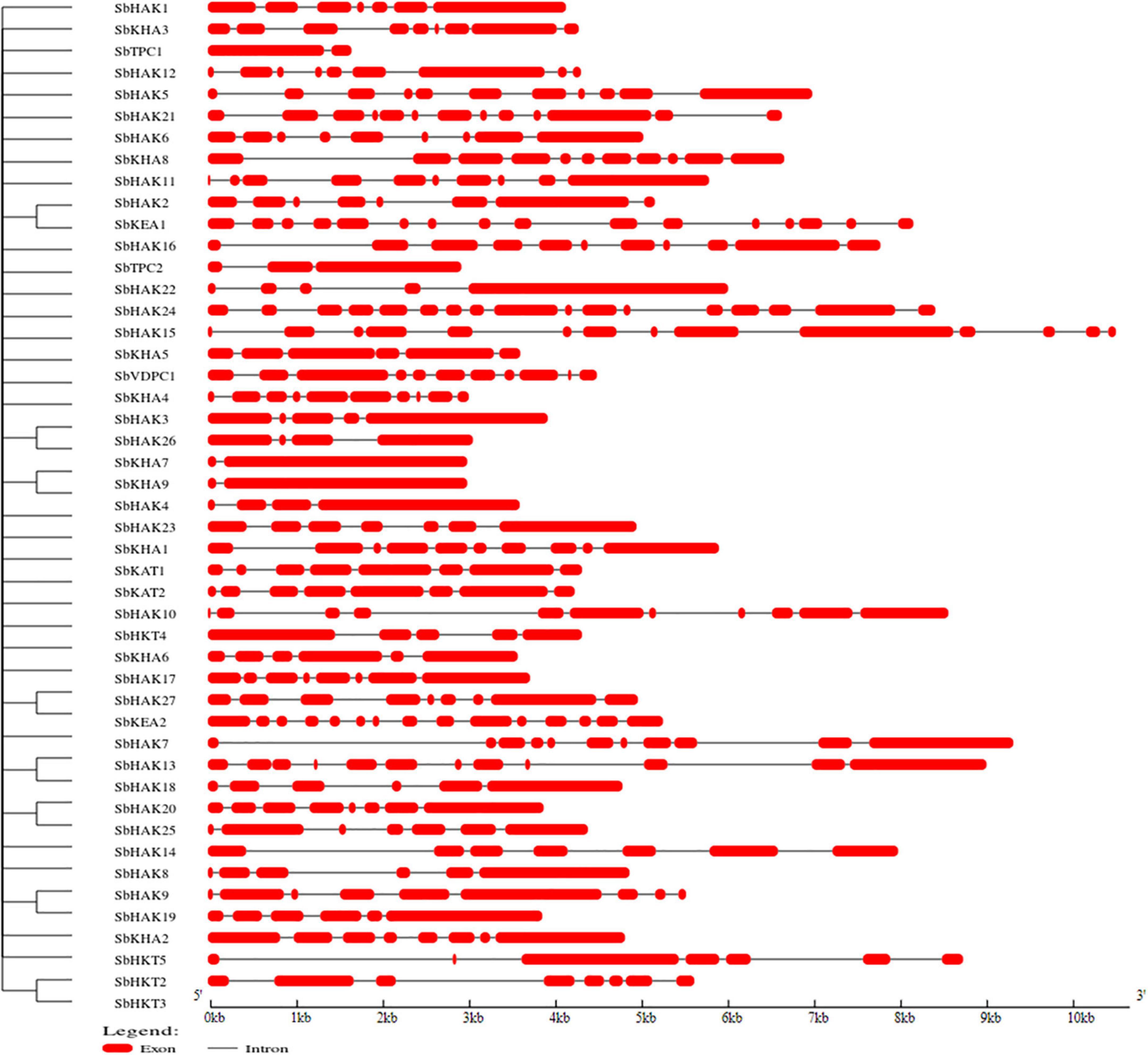
Figure 1. Characterization of K+ transporter gene homologs. Exons are represented as red boxes and introns as black lines. Sb, Sorghum bicolor; HAK, high affinity potassium; KEA, K+ efflux antiporter; VDPC, voltage-dependent potassium channel; TPC, two-pore channels.
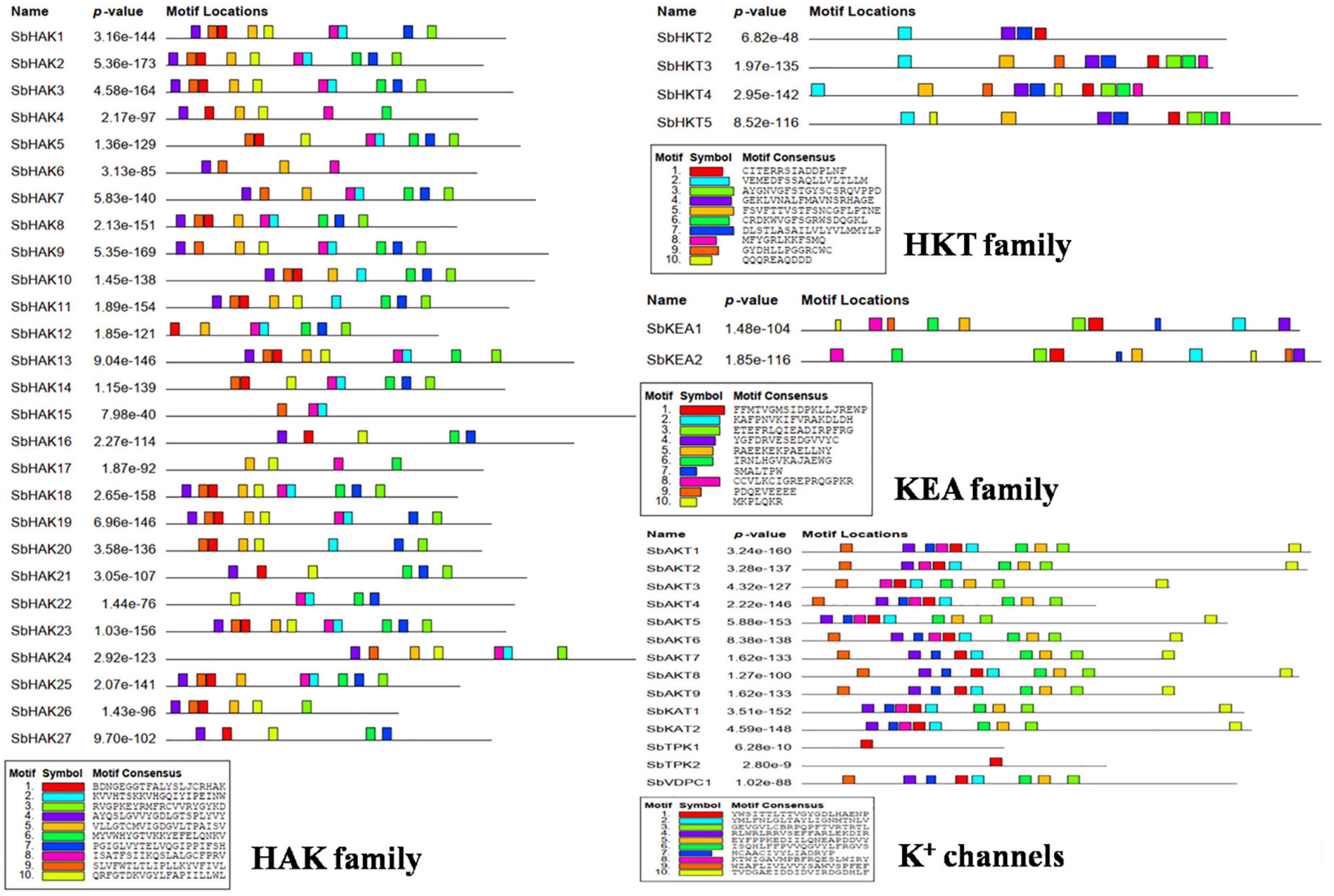
Figure 2. Conserved motif analysis of K+ transporter (HAK, HKT, and KEA) and channel (AKT, KAT, VDPC, and TPC) proteins. The consensus motif GVVYGDLGTSPLY was identified in all the HAK transporters except HAK5, HAK12, and HAK22. The motifs, VEMEDFSSAQLLVLTLLM, FSVFTTVSTFSNCGFLPTNE, GEKLVNALFMAVNSRHAGE, and LSTLASAILVLYVLMMYLP were observed in all the sorghum HKT transport proteins. The KEA family displayed FFMTVGMSIDPKLLJREWP and KAFPNVKIFVRAKDLDH motifs. All the channel proteins displayed the motif YWSITTLTTVGYGDLHAENP. Sb, Sorghum bicolor; HAK, high affinity potassium; KEA, K+ efflux antiporter; VDPC, voltage-dependent potassium channel; TPC, two-pore channels.
Promoter, 3D protein structures, and PPI analysis
Promoter analysis revealed the presence of biotic, abiotic, and phytohormone-responsive putative cis-elements (Supplementary Table 6). Different abiotic stress elements like DRE, CRT, CCAAT, MYB, MYC, LTRE, CBHFV, and IBOX have been identified (Figure 3). MYB represented the highest number of elements in all the transporters indicating their involvement in the stress tolerance. WBOX, the biotic stress-responsive element has also been recognized in all the K+ transporters. Most transporters have phytohormone-responsive elements like ABRE, WRKY, DPBF, ARR1, and GARE. ARR1, the cytokinin-responsive elements have been found as the highest number of elements among the phytohormone-responsive elements and identified in all the transporter and channel gene homologs (Figure 3). Such cis-element studies are essential since they might contribute to find out the functional regulation of KT/HAK/KUP gene family members in sorghum. 3D structures of K+ transport proteins were predicted with the best PDB templates (Figure 4). The template PDB id, template description, chain, model of the oligomer, and their structure validations are represented in the Supplementary Table 7. 3D structures of SbHAK26, SbAKT1, SbAKT2, SbAKT3, SbAKT4, SbAKT5, SbAKT6, SbAKT7, SbAKT8, SbAKT9, SbKAT1, SbKAT2, and SbVDPC1 proteins displayed significant sequence similarity percent ranging from 30.77% (SbHAK26) to 68.61% (SbAKT4). 3D structures of other K+ transport proteins did not show any significant (< 30%) sequence similarity (Supplementary Table 7). All the generated Ramachandran plots for structure validation are represented as Supplementary Figure 1. In the predicted PPI map, sorghum K+ proteins displayed interactions with several other K+ proteins. A total of 46 K+ proteins, except SbAKT9 were found in the PPI map (Figure 5). All the 46 proteins have been displayed as 46 nodes with 193 edges. Each protein showed more than one interactant (Supplementary Table 8). SbTPC1, SbTPC2, SbHAK23, SbHKT3, SbHKT4, SbAKT1, SbKEA1, SbHAK8, SbKAT1, and SbKAT2 have been found to be the major interacting proteins. SbHAK15 and SbVDPC1 did not show any interactions (Figure 5). All the STRING protein names used for PPIs are available in Supplementary Table 8.
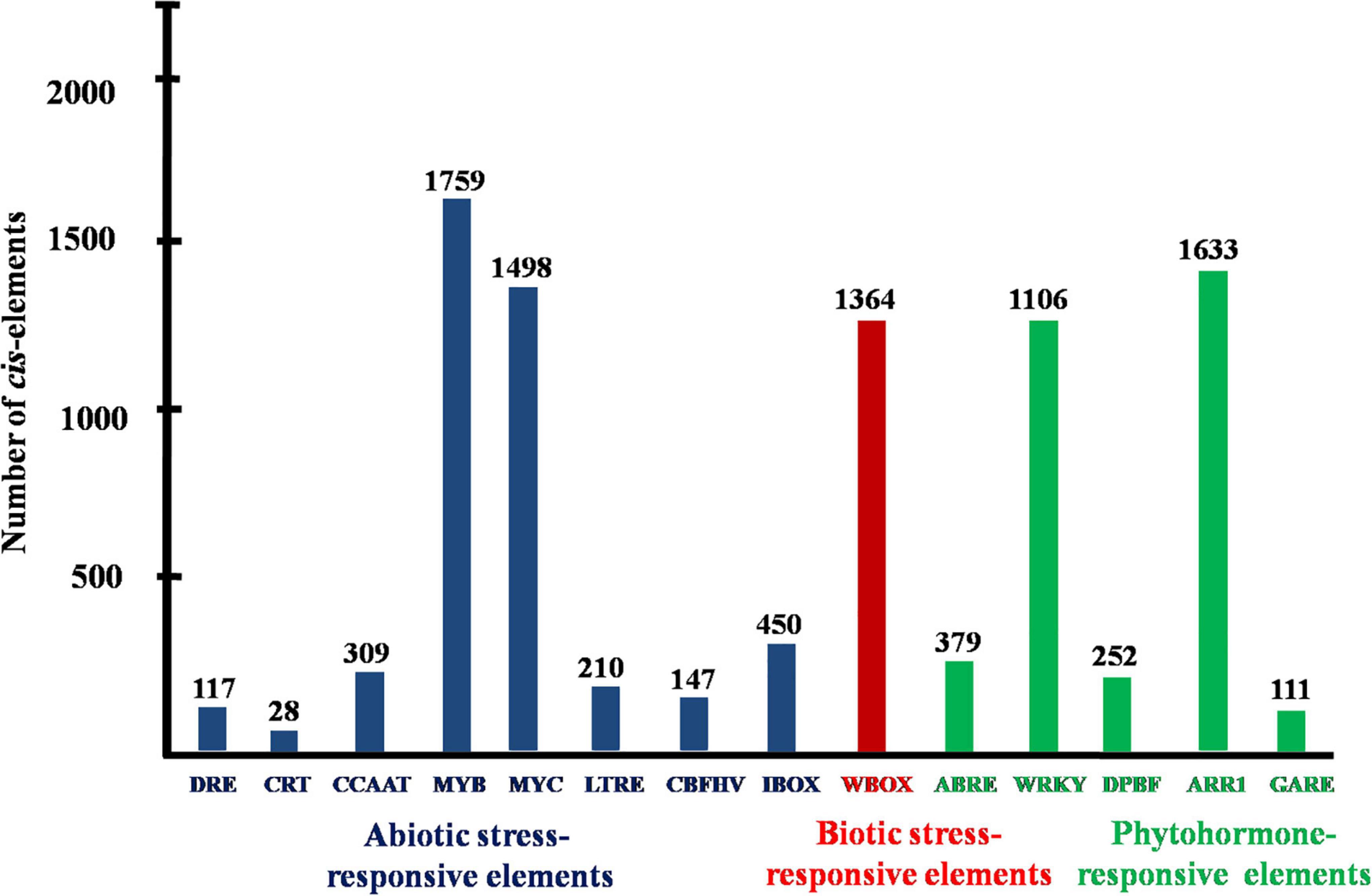
Figure 3. Conserved putative cis-acting elements of sorghum K+ transport gene homologs. DRE, dehydration-responsive elements; CRT, low-temperature responsive element; CCAAT, promoter of heat shock protein; MYB, responsive to drought and ABA; MYC, response to drought, cold and ABA; LTRE, low temperature and cold-responsive; CBFHV, dehydration-responsive element; IBOX, light regulation; WBOX, transcriptional repressor ERF3 gene; ABRE, early responsive to dehydration; WRKY, transcriptional repressor of the gibberellin; DPBF, ABA; AAR1, cytokinin-regulated transcription factor; GARE, gibberellic acid-responsive elements.

Figure 4. Structural analysis of 47 modeled sorghum K+ transport proteins. Sb, Sorghum bicolor; HAK, high affinity potassium; KEA, K+ efflux antiporter; VDPC, voltage-dependent potassium channel; TPC, two-pore channels.
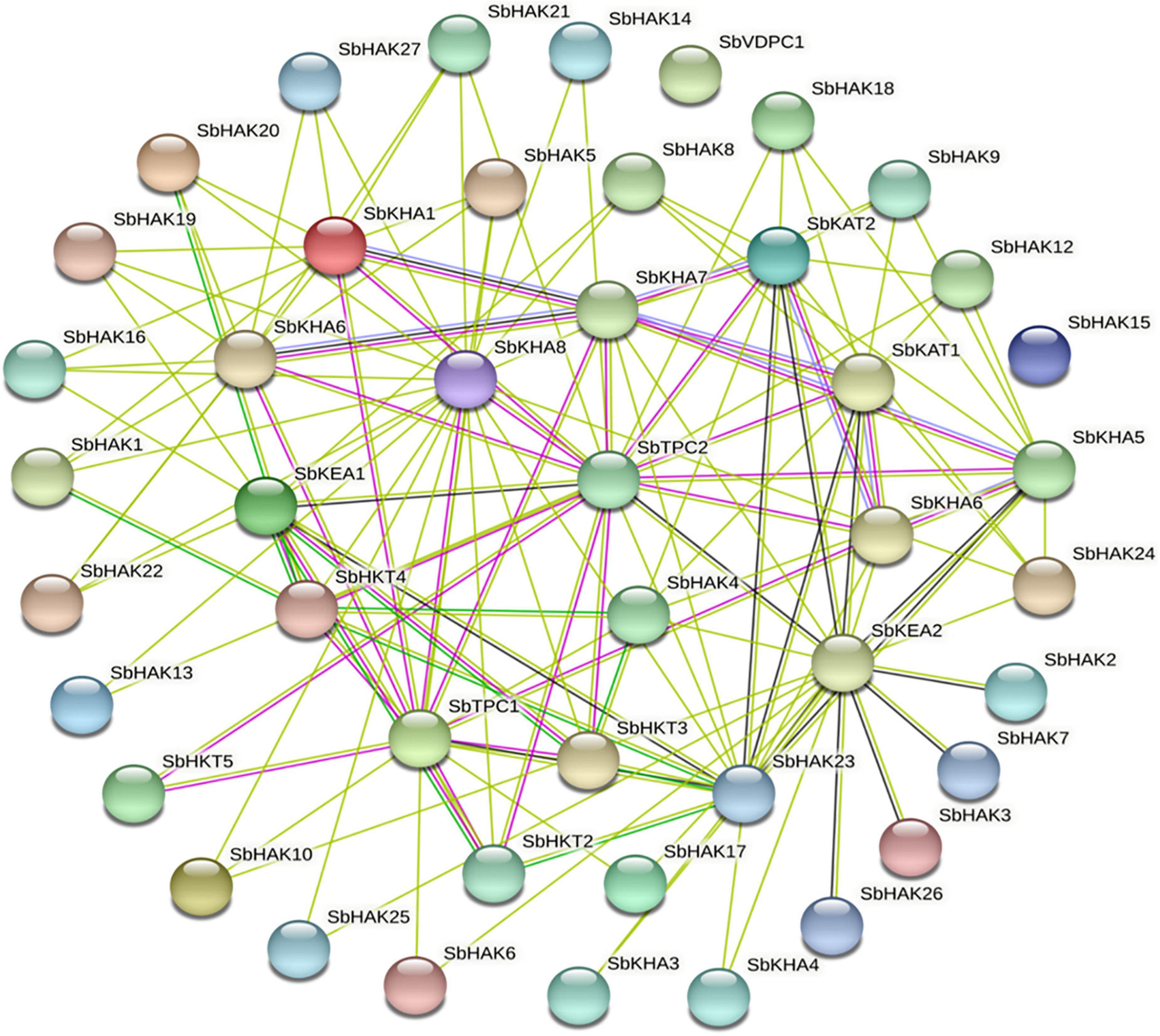
Figure 5. String analysis of sorghum K+ transport homologs. All the proteins displayed the interacting partners except SbHAK15 and SbVDPC1. Sb, Sorghum bicolor; HAK, high affinity potassium; KEA, K+ efflux antiporter; VDPC, voltage-dependent potassium channel; TPC, two-pore channels.
Evolutionary divergence, chromosomal location and synteny
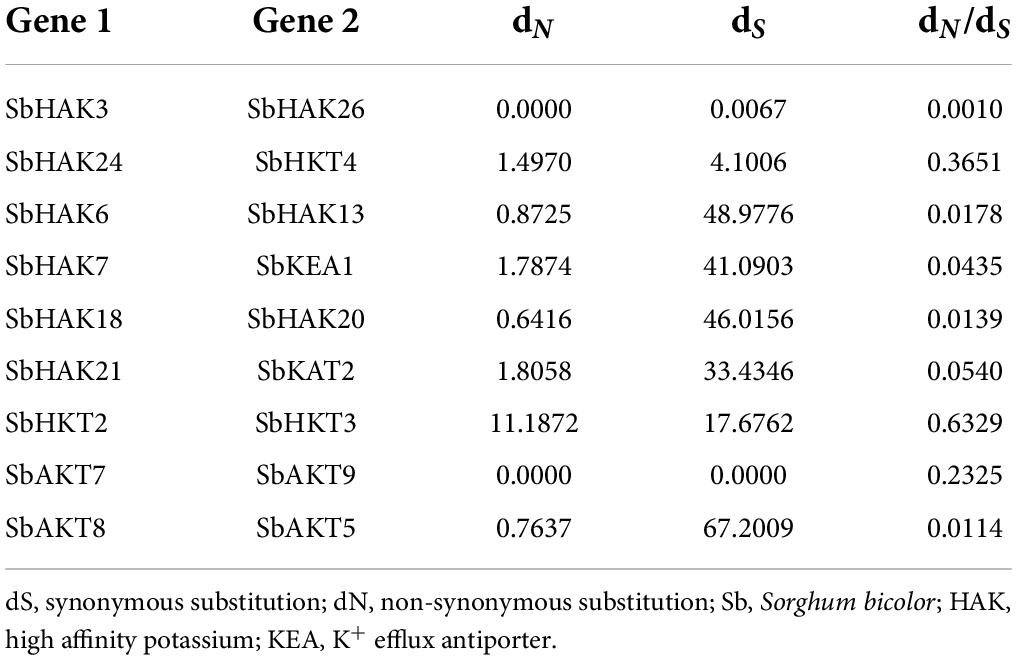
The phylogenetic tree revealed the evolutionary relationship of K+ transport homologs of Sorghum bicolor with Oryza sativa and Arabidopsis thaliana (Figure 6 and Supplementary Table 9). A total of 9 paralogs have been identified (Figure 7), 1 recognized as regional (SbHAK3 and SbHAK26) and 8 as segmental (SbHAK24 and SbHKT4, SbHAK6 and SbHAK13, SbHAK7 and SbKEA1, SbHAK18 and SbHAK20, SbHAK21 and SbKAT2, SbHKT2 and SbHKT3, SbAKT7 and SbAKT9, and SbAKT8 and SbAKT5) duplications. Sorghum showed 21 ortholog pairs (Figure 6), 18 with Oryza (SbHAK1 and OsHAK1, SbAKT2 and OsVDPC1, SbHAK2 and OsHAK16, SbHAK4 and OsHAK4, SbHAK5 and OsHAK26, SbHAK14 and OsHAK15, SbHAK10 and OsHAK13, SbHAK16 and OsAKT3, SbHAK12 and OsHAK20, SbHAK15 and OsHAK14, SbTPC1 and OsTPC1, SbHAK19 and OsHAK19, SbHAK27 and OsHAK27, SbKEA2 and OsKEA2, SbKAT1 and OsHKT2, SbAKT3 and OsAKT4, SbAKT4 and OsVDPC2, and SbAKT6 and OsHAK11) and 3 with Arabidopsis (SbHAK8 and AtHAK8, SbHAK23 and AtTPK3, and SbHAK22 and AtKEA1). All the 9 sorghum paralogs (SbHAK3 and SbHAK26, SbHAK24 and SbHKT4, SbHAK6 and SbHAK13, SbHAK7 and SbKEA1, SbHAK18 and SbHAK20, SbHAK21 and SbKAT2, SbHKT2 and SbHKT3, SbAKT7 and SbAKT9, and SbAKT8 and SbAKT5) display substitution rate < 1. The lowest dN/dS (0.0010) were observed in the regional paralog (SbHAK3 and SbHAK26) and the highest dN/dS (0.3651) in segmental paralog (SbHAK24 and SbHKT4) gene pairs (Table 2) respectively. Sorghum K+ transport gene homologs have been mapped onto Oryza (Figure 8). S. bicolor chromosome 2 displays 9 (highest) homologs followed by chromosome 3 and 6 with 8 homologs each, chromosome 1, 4, and 10 with 5 homologs each, chromosome 7 and 9 with 3 homologs each, and chromosome 8 with 1 homolog respectively. Similarly, O. sativa displays 10 homologs on chromosome 1, followed by chromosome 4 with 8 homologs, chromosomes 6 and 7 with 6 homologs each, chromosomes 2 and 3 with 5 homologs each, chromosomes 8 and 9 with 3 homologs each, chromosome 12 with 2 homologs, and chromosome 5 with 1 homolog. O. sativa and S. bicolor chromosomes 1 and 2 show the highest number of homologs with 10 and 9, respectively (Figure 8). Chromosome 5 of sorghum, 10 and 11 of rice do not contain any of the K+ gene homologs. A correspondence matrix was created and automated name-based and synteny maps were generated (Figure 8). The links on the chromosomes represent the gene homologs in sorghum and rice (Figure 8).
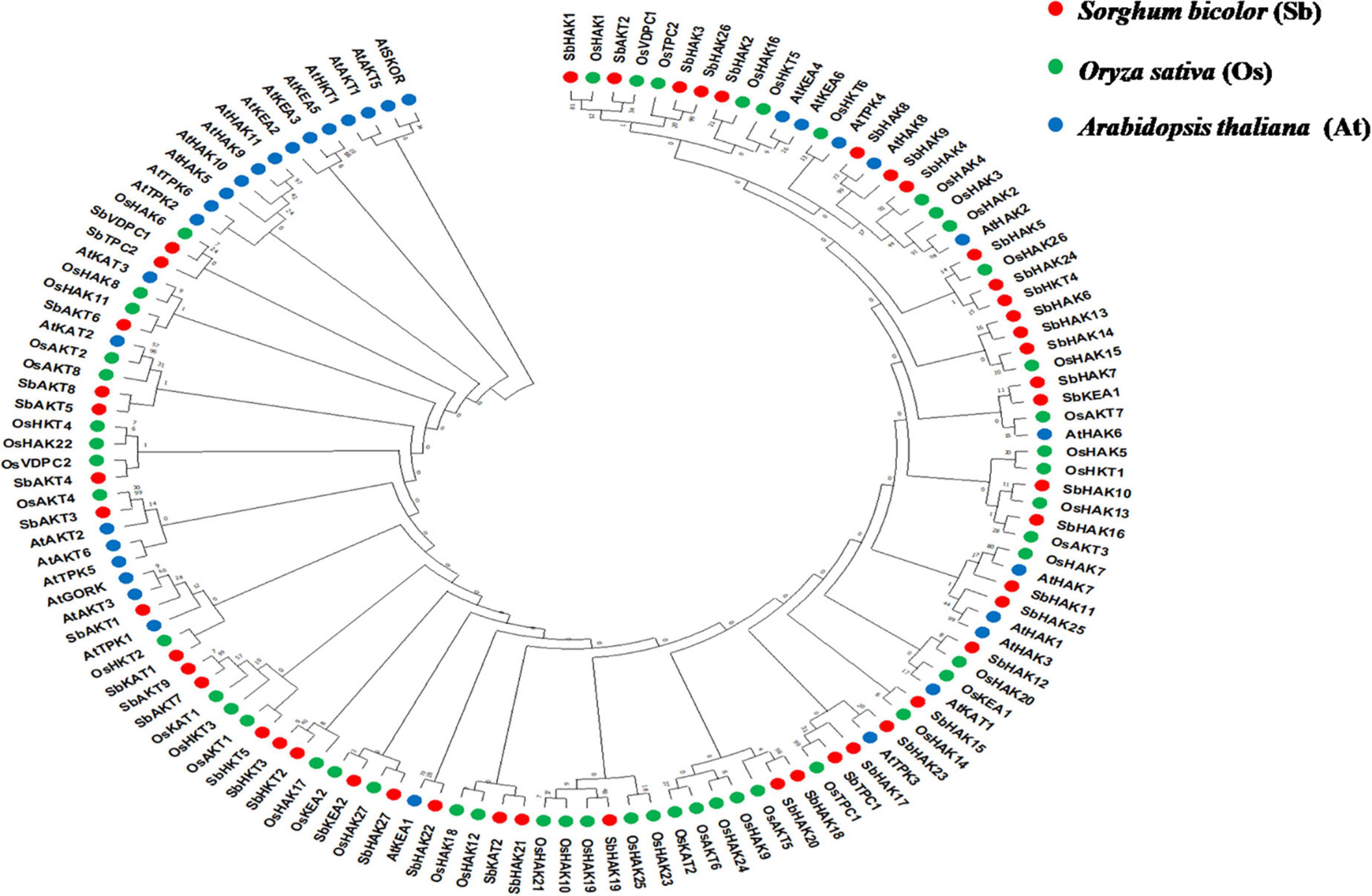
Figure 6. Phylogenetic tree of K+ transport proteins between Sorghum bicolor (Sb), Oryza sativa (Os), and Arabidopsis thaliana (At). Predicted amino acid sequences were used for construction of the tree. The tree was constructed by the Neighbor Joining method using MEGA-X. Values indicate the number of times (as a percentage) that each branch topology was found during bootstrap analysis. Sb, Sorghum bicolor; HAK, high affinity potassium; KEA, K+ efflux antiporter; VDPC, voltage-dependent potassium channel; TPC, two-pore channels.
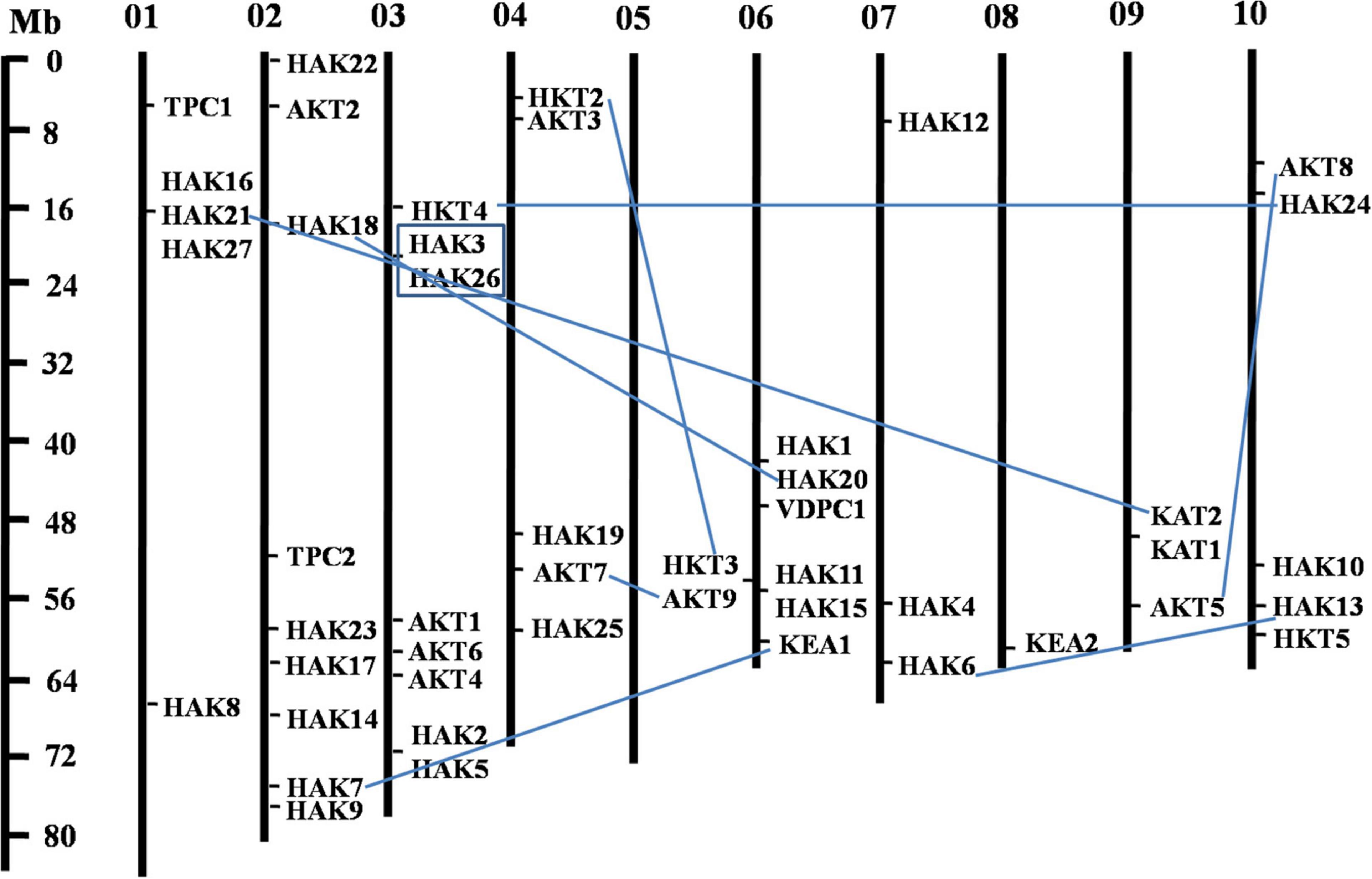
Figure 7. Physical mapping of sorghum K+ transport gene homologs. The 9 paralog gene pairs are represented in blue color. Of the 9 paralogs, 8 have been identified as segmental (SbHAK24 and SbHKT4, SbHAK6 and SbHAK13, SbHAK7 and SbKEA1, SbHAK18 and SbHAK20, SbHAK21 and SbKAT2, SbHKT2 and SbHKT3, SbAKT7 and SbAKT9, and SbAKT8 and SbAKT5) represented as lines and 1 as regional (SbHAK3 and SbHAK26) represented as box. Sb, Sorghum bicolor; HAK, high affinity potassium; KEA, K+ efflux antiporter; VDPC, voltage-dependent potassium channel; TPC, two-pore channels.
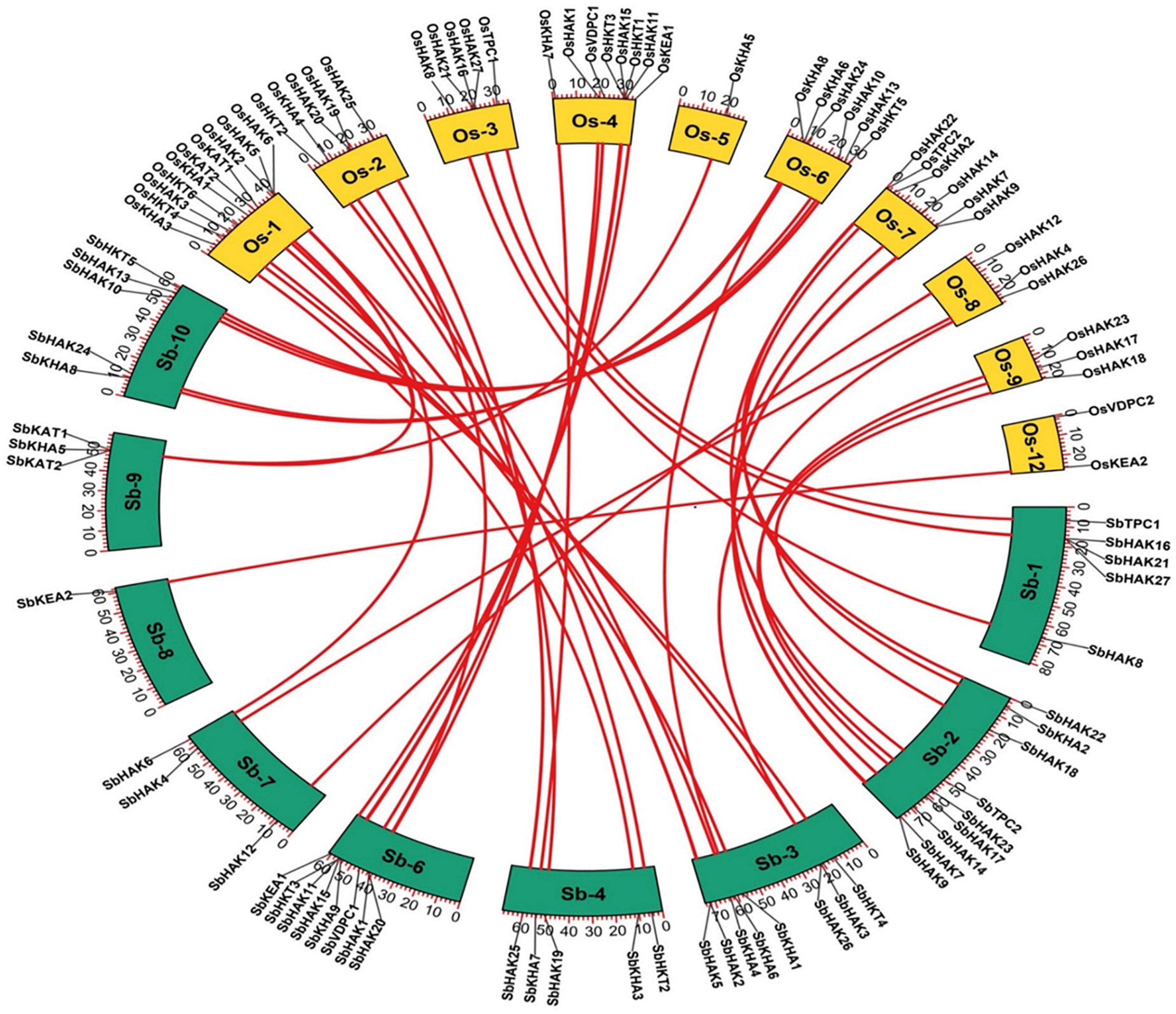
Figure 8. Synteny analysis of K+ transporter genes in Sorghum bicolor and Oryza sativa. The map was built with TB tools software. Sb, Sorghum bicolor; Os, Oryza sativa; HAK, high affinity K+; KEA, K+ efflux antiporter; VDPC, voltage-dependent K+ channel; TPC, two-pore channels.
Digital expressions and quantitative expression analysis of sorghum K+ transport gene homologs under abiotic stress conditions in different tissues
Digital expression of all the 46 K+ transport genes was analyzed in root, shoot, and leaf tissues exposed to cold and drought stress conditions. In anatomical tissues, high expression levels of K+ transport genes were noticed in root compared shoot, and leaf tissues (Figure 9A). In root tissues, SbHAK7, SbHAK18, SbHAK10, and SbHAK25, in shoot tissues, SbHAK11, SbKEA1, and SbHAK22, SbHAK11, SbKEA1, and SbKEA2 displayed high expressions respectively (Figure 9A). In developmental stages high expression levels of K+ transport genes were noticed in milk stage, flowering stage, tillering stage and seedling stage (Figure 9B). In milk stage SbHAK27 and SbKEA2, in the flowering stage SbHAK17, SbHAK11, SbHAK18, and SbHAK7, in tillering stage SbHAK18, SbHAK10, and 23 genes displayed elevated expression levels than other transport genes (Figure 9B). Differential expression profile of K+ transport genes was noticed in cold and drought conditions (Figure 9C). Higher expression levels were noticed in cold stress compared to drought stress. SbAKT1, SbHAK7, SbHKT5, SbHAK25, and SbAKT7 genes have higher expression levels in cold stress. SbHAK5 and SbHAK17 genes have higher expression in drought stress (Figure 9C).
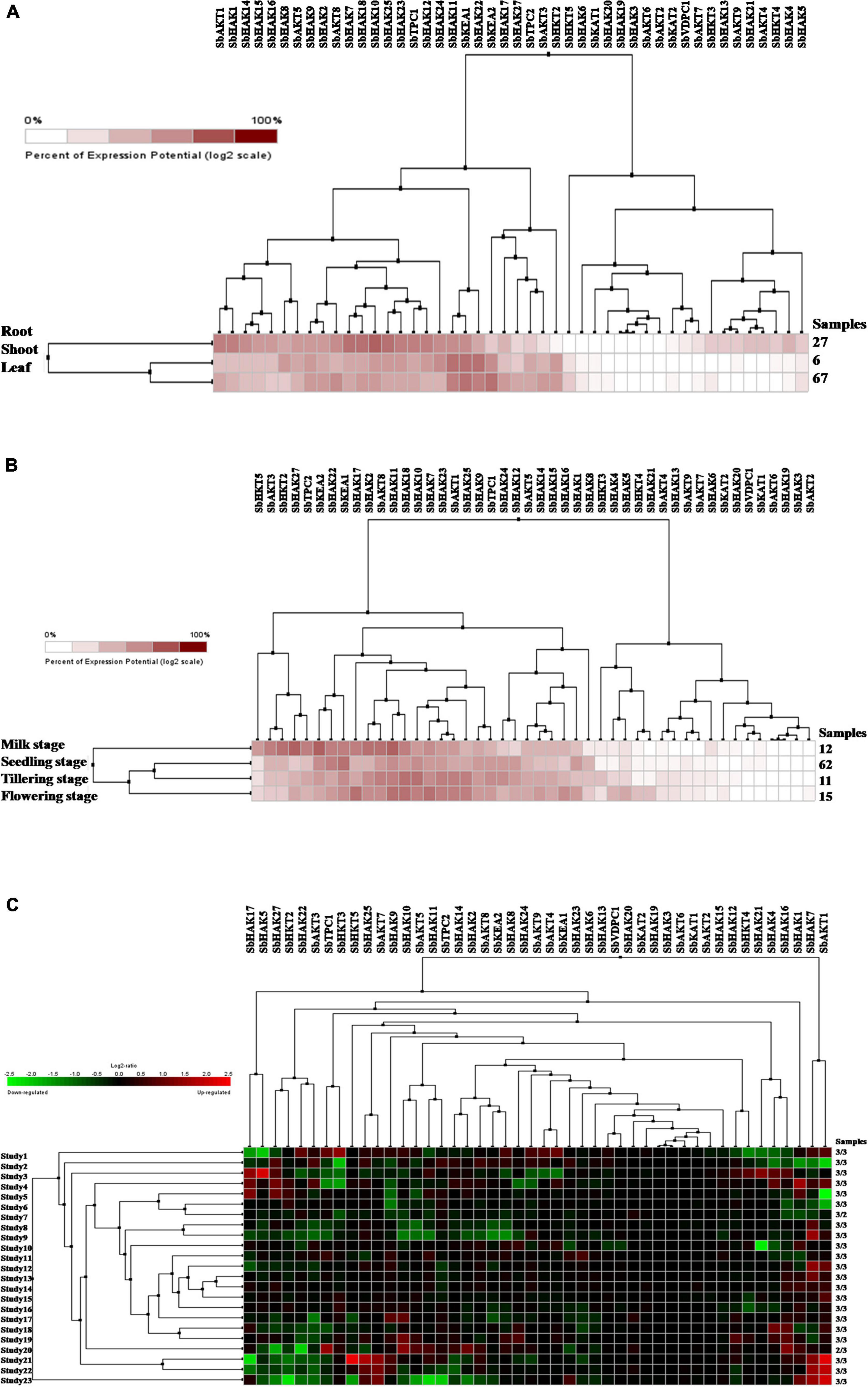
Figure 9. Digital expression profile of sorghum K+ transport genes in different tissues, developmental stages under cold and drought stresses (A) K+ transport in anatomical tissues, (B) K+ transport in developmental stages, (C) K+ transport gene expressions under cold and drought stresses. Sb, Sorghum bicolor; HAK, high affinity potassium; KEA, K+ efflux antiporter; VDPC, voltage-dependent potassium channel; TPC, two-pore channels.
Expression levels of only 32 K+ transport gene homologs (SbHAK1, SbHAK2, SbHAK3, SbHAK4, SbHAK5, SbHAK6, SbHAK7, SbHAK8, SbHAK9, SbHAK10, SbHAK11, SbHAK12, SbHAK13, SbHAK14, SbHAK15, SbHAK17, SbHAK18, SbHAK19, SbHAK20, SbHAK21, SbHAK22, SbHAK23, SbHAK24, SbHAK25, SbHAK26, SbHAK27, SbHKT2, SbHKT3, SbHKT4, SbHKT5, SbKEA1, and SbKEA2) were analyzed in sorghum root, stem, and leaf tissues subjected to salt, drought, heat, and cold stresses and shown in the heat map (Figure 10). qRT-PCR for other 15 K+ transport gene homologs could not be performed due to high sequence similarity. The homologs displayed differential gene expression in different tissues (Supplementary Table 10). SbHAK2, SbHAK20, SbHAK5, and SbHAK3 showed markedly increased expressions under salt, heat, drought and cold stresses respectively. Among the stress treatments, a 13.86-fold increase in transcript levels was observed in SbHAK3 in cold-stressed roots, followed by strong upregulation of SbHAK2 (13.39-folds increase) under salt stress in the leaves. SbHAK7 was enhanced by 11.95-folds in the leaves exposed to salt stress. Similarly, SbHAK12, SbHAK20, and SbHAK21 displayed 10.19, 10.48, and 10.14-folds enhanced activity in stem tissues subjected to salt (SbHAK12) and heat stresses (SbHAK20, SbHAK21) respectively. While SbHAK15 showed 9.36-fold increase in roots exposed to cold stress, SbHAK8, SbHAK14, and SbHAK10 exhibited higher activities in stems treated with salt and cold stresses respectively. Out of all gene homologs, SbHAK2, 3, 7, 8, 10, 12, 14, 15, 20, 21, and 14 recorded markedly high expressions in compared to other SbHAK, SbHKT, and SbKEA members. Transcripts SbHAK7, SbHAK8, SbHAK9, SbHAK10, SbHAK11, SbHAK12, SbHAK13, and SbHAK14 are highly upregulated under salt, heat, and cold, while SbHAK1, SbHAK4, SbHAK5, SbHAK18, SbHAK22, and SbHAK27 (4.55–7.94-folds) were well expressed under drought stress conditions. SbHAK17 has the lowest level of expression among all the genes across different stresses and tissues (Figure 10). Transcript expressions were increased in stem and leaf tissues subjected to high temperature stress especially in most of SbHAKs. Among the SbHKTs, the transcript level of SbHKT5 was superior (7.17-fold increase) in leaves exposed to salt stress. Similarly, SbKEA1 expression was significantly high (10.14-folds) under salt stress in the stems of Sorghum. Activity of SbKEA2 was 11.2-folds higher in salt-stressed leaves.
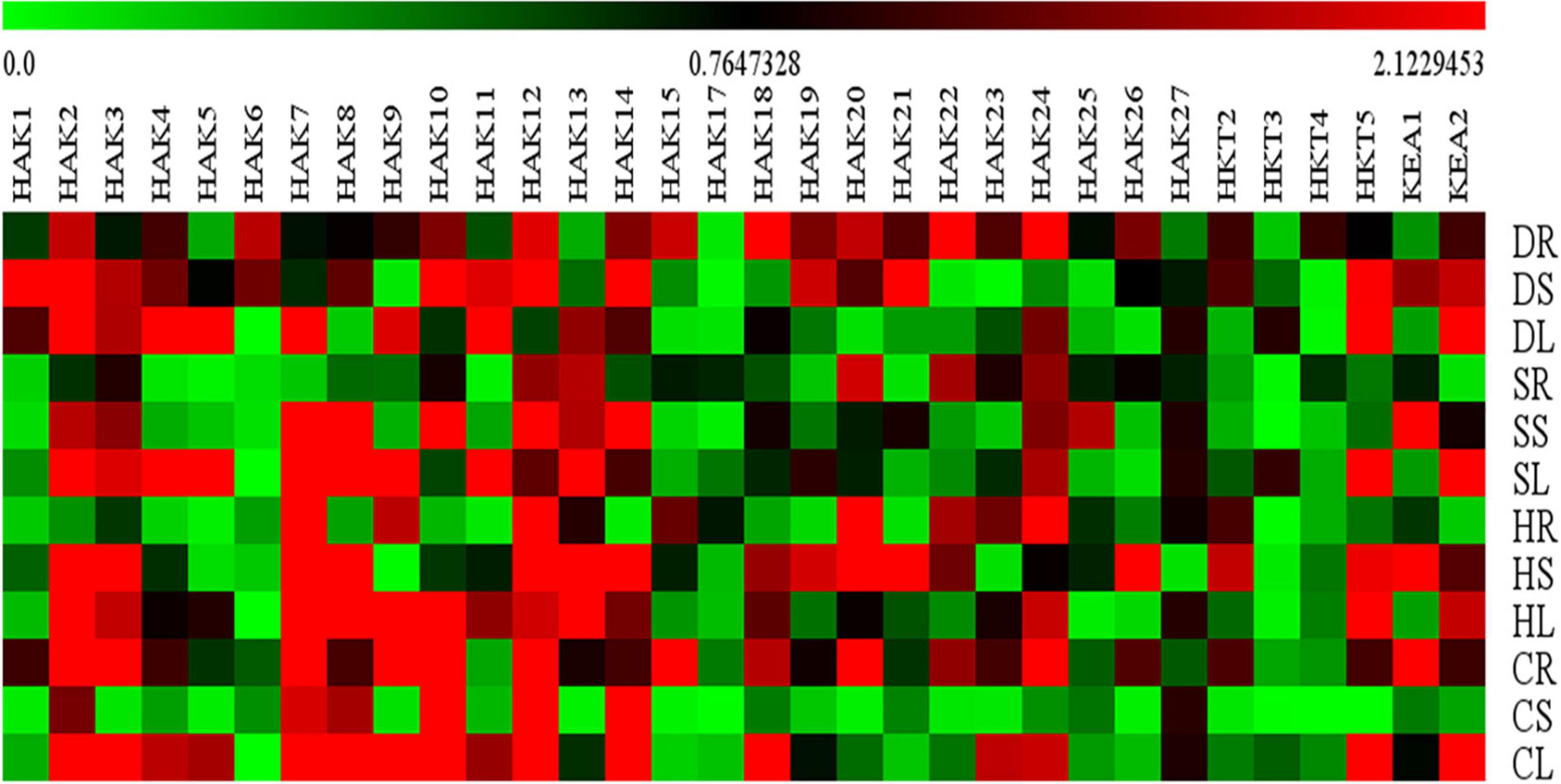
Figure 10. Relative expression analysis of sorghum K+ transporter gene homologs. Sorghum transporter expressions during salt, drought, heat, and cold stresses. Relative expression of transporters is shown during different stress conditions compared to its corresponding controls. Values represent the expression levels obtained after normalizing against control tissues. All samples were analyzed in triplicate, in two independent experiments. Names on the vertical axis indicate the tissues and the horizontal axis represents various genes. R, root; S, stem; L, leaf; S, salt; D. drought; H, heat; C, cold. Each color represents the relative expression levels of the transcripts. Sb, Sorghum bicolor; HAK, high affinity potassium; KEA, K+ efflux antiporter; VDPC, voltage-dependent potassium channel; TPC, two-pore channels.
Discussion
K+ plays a pivotal role as a constituent of the plant structure, in ion homeostasis, salt tolerance, plant growth, development, transport, aside from acting as a signaling molecule (Feng et al., 2019; Hussain et al., 2021). It has a regulatory function in many physiological and biochemical processes such as protein synthesis, and activation of enzymes (Hasanuzzaman et al., 2018). K+ is available to plants only in ionic form and higher K+/Na+ has been recognized unequivocally as a crucial molecule for maintaining electro-neutrality of the cells (Hussain et al., 2021). Under saline and water deficient conditions, K+ maintains ion homeostasis and modulates the osmotic balance. Further, K+ participates in stomatal regulation during drought stress and increases the antioxidative ability of the plants (Hasanuzzaman et al., 2018). Since many homologs have been detected in plants, we need to understand which of the homologs perform the crucial processes of plant growth, abiotic stress, and under K+ deprivation conditions (Hamamoto et al., 2008; Jiang et al., 2021). In the present investigation, a total of 47 K+ transporter gene homologs (Table 1) were discovered in all including 33 K+ transporters (27 HAKs, 4 HKTs, and 2 KEAs) and the remaining 14 (9 AKTs, 2 KATs, 2 TPCs, and 1 VDPC) as K+ channels in S. bicolor.
Characterization of K+ transport gene homologs in sorghum
Sorghum has 47 K+ transporter gene homologs (Table 1 and Figure 1) in comparison with 43 in Gossypium raimondii, (Azeem et al., 2022), 39 in Cajanus cajan (Siddique et al., 2021), and 36 in Cicer arietinum (Azeem et al., 2018). Nevertheless, these numbers are lower than that of Glycine max, where 70 homologs have been detected (Rehman et al., 2017), and 49 in Oryza sativa (Amrutha et al., 2007). The 27 genes encoding HAK transporters in Sorghum (Figure 1) are similar in the number of HAK encoding genes detected in Zea mays and Hordeum vulgare (Zhang et al., 2012; Cai et al., 2021). Triticum aestivum and Pyrus betulifolia have 56 HAK transporters each (Cheng et al., 2018; Li Y. et al., 2018) followed by 41 in Nicotiana tabacum (Song et al., 2019), 31 in Populus trichocarpa (He et al., 2012), 30 in Saccharum spontaneum (Feng et al., 2020b), 29 in Glycine max (Rehman et al., 2017), 26 in Oryza sativa (Amrutha et al., 2007), 22 in Salix purpurea and Ipomoea batatas (Liang et al., 2020; Jin et al., 2021), 21 in Populus trichocarpa, Prunus persica, Manihot esculenta, and Camellia sinensis (He et al., 2012; Song et al., 2015; Ou et al., 2018; Yang T. et al., 2020), 19 in Solanum lycopersicum (Hyun et al., 2014), 17 in Oryza sativa and Cajanus cajan (Banuelos et al., 2002; Siddique et al., 2021), 16 in Prunus persica and Gossypium raimondii (Song et al., 2015; Azeem et al., 2022), and 13 in Arabidopsis thaliana (Ahn et al., 2004). These studies indicate that K+ transport genes are highly conserved in plants during evolution. A total of 21, 24, 45, and 44 HAK/KUP/KT genes were identified in Gossypium hirsutum, Gossypium barbadense, Gossypium raimondii, and Gossypium arboretum genomes respectively (Yang X. et al., 2020). The higher number of K+ transporter homologs in Triticum is due to its ploidy nature (Kyriakidou et al., 2018). Sorghum has shared 4 HKT encoding genes with Glycine max (Rehman et al., 2017). Oryza sativa has 8 HKTs, followed by 4 in Sorghum bicolor and Glycine max, 2 HKTs in Gossypium raimondii, Cajanus cajan, and Cicer arietinum (Amrutha et al., 2007; Rehman et al., 2017; Azeem et al., 2018, 2022; Siddique et al., 2021). Glycine max has the highest number of KEA transporters (12) (Rehman et al., 2017), followed by 7 KEAs in Gossypium raimondii (Azeem et al., 2022), 6 KEAs in Cicer arietinum and Cajanus cajan (Azeem et al., 2018; Siddique et al., 2021), but 2 KEAs in sorghum, and 1 KEA in Oryza sativa (Amrutha et al., 2007). The number of K+ channels identified in sorghum corroborates the previously reported genomes. A total of 14 K+ channels were identified in sorghum (9 AKTs, 2 KATs, 2TPKs, and 1 VDPC), an equal number in Oryza sativa (14 AKTS) (Amrutha et al., 2007), belonging to the same family. But Cajanus cajan has 9 shakers and 5 TPKs (Siddique et al., 2021). However, 25 K+ channel gene homologs (16 VDPCs, 9 TPK/KCO) have been reported in Glycine max (Rehman et al., 2017) and 18 (11 shakers and 7 TPKs/KCO) in Gossypium raimondii (Azeem et al., 2022).
The conserved domains of K+ transporter system viz., K-trans, TrK, KEA, voltage-dependent K+ channel, KHA, and Two Pore Potassium (TPK) channel were identified in sorghum (Table 1) which corroborates the identified K+ genes in other plant genomes such as Oryza sativa (Amrutha et al., 2007), Glycine max (Rehman et al., 2017), Cicer arietinum (Azeem et al., 2018), Cajanus cajan (Siddique et al., 2021), and Gossypium raimondii (Azeem et al., 2022). The consensus motif GVVYGDLGTSPLY (Rodríguez-Navarro, 2000) was identified in all the sorghum HAK transporters except SbHAK5, SbHAK12, and SbHAK22 (Figure 2). Similar results were also reported in Oryza (Amrutha et al., 2007), Cicer arietinum (Azeem et al., 2018), Cajanus cajan (Siddique et al., 2021), and Gossypium raimondii (Azeem et al., 2022). The motif GGTFALYSLLCR was detected in Arabidopsis thaliana (Ahn et al., 2004), Cicer arietinum (Azeem et al., 2018), Cajanus cajan (Siddique et al., 2021), Gossypium (Azeem et al., 2022) was also noticed in sorghum inferring its evolutionary conservation. The conserved K+ channel motif GYGD (Kuang et al., 2015) has been observed in all the sorghum K+ channels (Figure 2), identical to that of Oryza sativa (Amrutha et al., 2007), Cicer arietinum (Azeem et al., 2018), Cajanus cajan (Siddique et al., 2021), and Gossypium (Azeem et al., 2022). Most of the sorghum HAK transporters are localized on plasma membrane (Table 1) akin to Triticum aestivum (Cheng et al., 2018), Saccharum spontaneum (Feng et al., 2020b), Salix purpurea (Liang et al., 2020), Camellia sinensis (Yang T. et al., 2020), Hordeum vulgare (Cai et al., 2021), and Ipomoea batatas (Jin et al., 2021). A. thaliana AtHAK5 has been associated with the uptake of Na+ (Wang Q. et al., 2015). This implies that these porters subscribe to the accumulation of Na+ under saline conditions. Since restricting Na+ uptake determines salt tolerance, care must be taken while breeding the crop plants for salt stress tolerance. Also, HKTs are involved in loading Na+ into the xylem. Zhu et al. (2016) also noticed a link between SOS1 and HKT pathways for salt stress in wheat. These studies indicate the HKTs critical role during salt stress tolerance. K+ efflux antiporters KEA1 and KEA2 have been found in inner envelop membrane in A. thaliana, and the loss of function mutants influence both ROS and reactive nitrogen species (RNS). Double knock-out mutants of kea1kea2 elicited an alteration of the ROS homeostasis. However, nitric oxide (NO) content has negatively affected photosynthesis increasing photorespiratory activity (Sánchez-McSweeney et al., 2021). The studies infer that KEAs maintain chloroplast osmotic balance. In sorghum, SbKEA1 is localized on chloroplast membranes and is perhaps involved in the regulation of thylakoid and stromal pH (Sánchez-McSweeney et al., 2021).
Analysis of putative cis-elements and 3D protein structures, and interactions
Hyun et al. (2014) and Assaha et al. (2017) reported the involvement of cis-regulatory elements in abiotic stress tolerance and in K+ homeostasis. Similarly, analysis of promoter sequences of K+ transport gene homologs in sorghum revealed the presence of cis-elements which may be involved in diverse abiotic stress tolerances. This prediction is in line with that of Cicer arietinum (Azeem et al., 2018), Cajanus cajan (Siddique et al., 2021), and Gossypium raimondii (Azeem et al., 2022). In the promoter regions, regulatory elements like ABRE, MYB, MYC, GARE, WBOX, LTRE, and CCAAT have been noticed in Cicer arietinum (Azeem et al., 2018), Pyrus betulifolia (Li Y. et al., 2018), Manihot esculenta (Ou et al., 2018), Nicotiana tabacum (Song et al., 2019), Salix purpurea (Liang et al., 2020), Camellia sinensis (Yang T. et al., 2020), Hordeum vulgare (Cai et al., 2021), Cajanus cajan (Siddique et al., 2021), Ipomoea batatas (Jin et al., 2021), and Gossypium species (Yang X. et al., 2020; Azeem et al., 2022) indicating the involvement of K+ transport gene homologs in abiotic stress tolerance. Aside abiotic stress-responsive elements, promoter analysis also revealed the presence of biotic stress-responsive and phytohormone stress-responsive elements in sorghum (Figure 3). The predicted elements indicate that K+ transporters are implicated in biotic stress response and their cross-talk with hormones during stress. Protein models (Figure 4) help to understand structure-function relationships (Rasheed et al., 2020). Protein-protein interactions (PPIs) of sorghum (Figure 5) displayed interaction with other K+ transporters and channels like what has been noticed in Cajanus cajan (Siddique et al., 2021). The SbHAK transporters also interacted with other SbHAK transporters like Ipomoea batatas HAK transporters (Jin et al., 2021).
Evolutionary divergence and comparative analysis
Phylogenetic analysis revealed the close relationship of Sorghum bicolor K+ transporters with Oryza sativa (Amrutha et al., 2007) than to Arabidopsis thaliana (Figure 6). A comparative phylogenetic analysis of K+ transporters has been carried out in Oryza sativa (Amrutha et al., 2007), Glycine max (Rehman et al., 2017), Cicer arietinum (Azeem et al., 2018), Cajanus cajan (Siddique et al., 2021), and Gossypium raimondii (Azeem et al., 2022). Similarly, comparative studies of HAK transporters have been reported in Oryza sativa (Banuelos et al., 2002), Arabidopsis thaliana (Ahn et al., 2004), Populus trichocarpa (He et al., 2012), Zea mays (Zhang et al., 2012), Solanum lycopersicum (Hyun et al., 2014), Prunus persica (Song et al., 2015), Glycine max (Rehman et al., 2017), Triticum aestivum (Cheng et al., 2018) and Pyrus betulifolia (Li Y. et al., 2018), Manihot esculenta (Ou et al., 2018), Nicotiana tabacum (Song et al., 2019), Saccharum spontaneum (Feng et al., 2020b), Salix purpurea (Liang et al., 2020), Hordeum vulgare (Cai et al., 2021), Ipomoea batatas (Jin et al., 2021), and Camellia sinensis (Yang T. et al., 2020). Most sorghum K+ homologs showed high gene conservation with Oryza sativa (Figure 6) as both share common ancestor (Wang X. et al., 2015). The tree indicated 9 paralog and 21 ortholog (18 with Oryza sativa and 3 with Arabidopsis thaliana) groups. Chromosomal distribution and synteny analyses revealed the presence of 1 regional (SbHAK3 and SbHAK26) and 8 segmental (SbHAK24 and SbHKT4, SbHAK6 and SbHAK13, SbHAK7 and SbKEA1, SbHAK18 and SbHAK20, SbHAK21 and SbKAT2, SbHKT2 and SbHKT3, SbAKT7 and SbAKT9, and SbAKT8 and SbAKT5) duplication gene pairs. Such gene duplication events have also been reported in Ipomoea batatas HAK transporters (Jin et al., 2021). All the 9 sorghum paralogs (SbHAK3 and SbHAK26, SbHAK24 and SbHKT4, SbHAK6 and SbHAK13, SbHAK7 and SbKEA1, SbHAK18 and SbHAK20, SbHAK21 and SbKAT2, SbHKT2 and SbHKT3, SbAKT7 and SbAKT9, and SbAKT8 and SbAKT5) have the dN/dS value < 1 (Table 2), indicating a purifying Darwinian selection during the evolution of HAK genes (Bowers et al., 2003). The HAK family of Saccharum spontaneum (Feng et al., 2020b) and Ipomoea batatas (Jin et al., 2021) have also showed the non-synonymous/synonymous value < 1. Uneven distributions of K+ transport gene homologs on different chromosomes have been observed in sorghum (Figure 7) similar to Oryza sativa (Amrutha et al., 2007), Glycine max (Rehman et al., 2017), Cicer arietinum (Azeem et al., 2018), Cajanus cajan (Siddique et al., 2021), and Gossypium raimondii (Azeem et al., 2022). Uneven distribution of HAK/KUP/KT homologs has been observed in angiosperms also (Nieves-Cordones et al., 2016). Gene order conservation of K+ gene homologs across sorghum and rice has been identified by circos (Figure 8). K+ transport homologs displayed very high conservation between sorghum and rice since they share a common ancestor (Wang X. et al., 2015). Chromosome 2 and 1 of sorghum and rice displayed the highest number of homologs with 9 and 10, respectively. Also, an equal number (3) of K+ transport gene homologs was observed on chromosome 9 of sorghum and rice (Figure 8).
Digital and qRT-PCR transcript patterns of sorghum K+ transport genes responding to abiotic stresses
Digital expression of K+ transport genes have been identified in different tissues like root, shoot, and leaf (Figure 9A) and in different developmental stages like milk stage, seedling stage, tillering stage, and flowering stage (Figure 9B). Expression of sorghum K+ transport genes has reported under cold and drought stresses (Figure 9C). Sorghum transcript analysis revealed that K+ transport genes are responsive to different abiotic stresses. qRT-PCR results indicated differential gene expression of sorghum K+ transport gene homologs in the root, stem, and leaf tissue treated with salt, drought, heat, and cold stresses (Figure 10). Such a differential gene expression of K+ transporters and channels in diverse tissues and under abiotic stresses have been reported in Triticum aestivum (Cheng et al., 2018), Cicer arietinum (Azeem et al., 2018), Saccharum spontaneum (Feng et al., 2020b), Cajanus cajan (Siddique et al., 2021), Ipomoea batatas (Jin et al., 2021), and Gossypium raimondii (Azeem et al., 2022). Reports exist that HAK/KUP/KT family members ameliorate the plants from salt stress. In cotton, GhPOT5, a homolog of OsHAK1 exhibited significantly higher expression under salt stress in comparison with other genes (Yang X. et al., 2020). Salinity reduces the uptake of K+ as evident in rice mutants Oshak1, when the levels were below 0.05 mM. Overexpression of HAK1 resulted in salt stress tolerance in rice (Chen et al., 2015). Hamamoto et al. (2015) noticed that AtHKT1 provides protection to the leaves under salt stress. In line with this, SbHKT5 displayed higher activity under salt stress in the present study. Similarly, K+ transporter genes were upregulated in the present study under water deficit conditions. Under drought conditions, root growth is restricted and diffusion of K+ toward the roots (Wang et al., 2013). Also, long-term exposure to water deficit conditions led to leaf damage due to ROS formation (Wang et al., 2013). In support of this, optimization of K+ supply mitigates the damage caused due to the oxidative stress in barley exposed to drought stress (Tavakol et al., 2021). Silencing HvAKT2 and HvHAK1 in barley enhanced the ROS (H2O2) production in PEG-treated leaves (Feng et al., 2020a). In rice, overexpression of OsHAK1 positively regulates drought stress, while the knockout lines accumulate less K+ and more H2O2 with stunted growth of the plants and less tolerance to drought stress (Chen et al., 2017). These results point out that K+ reduces the accumulation of H2O2 and thus helps the plants during drought stress. Earlier studies also revealed that HAK/KUP/KT family genes improve drought stress tolerance in plants (Wang et al., 2013; Li W. et al., 2018). Further, under water deficit conditions, K+ regulates opening of stomata and make the plants adaptive to drought (Tang et al., 2015). K+ increases the antioxidant defense in plants under abiotic stress conditions (Wang et al., 2013; Amanullah et al., 2016). Under extreme temperatures, osmolytes accumulate and K+ helps to maintain stomatal conductance and therefore avoids the damage (Azedo-Silva et al., 2004; Hasanuzzaman et al., 2018). These studies point out that K+ transporter genes play pivotal roles during environmental adversities and impart tolerance to multiple abiotic stresses.
In conclusion, genome-wide analysis of sorghum has led to the identification of 47 K+ transport gene homologs; 33 K+ transporters (27 HAKs, 4 HKTs, and 2 KEAs) and 14 K+ channels (9 AKTs, 2 KATs, 2 TPCs, and 1 VDPC). Gene characterization, conserved domains, motif identifications, localization, phylogenetic analysis revealed the close relation of Sorghum bicolor K+ transport gene homologs with its relative Oryza sativa. Identification of cis-acting elements would be helpful to explore further and to manipulate K+ porters as well as channels for designing better crops. Gene expression data indicate that such genes can be utilized effectively in breeding programs aimed at abiotic stress tolerance. The results bring forth precious information candidate gene identification for functional analyses and subsequent utilization in genetic engineering, and traditional breeding programs to improve sorghum for abiotic stress tolerance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
SAK and PBK designed the experiments. PHK, MN, TDD, AM, and PSR carried out the bioinformatics analysis. SAK and MN performed the qRT-PCR experiments. SAK, PHK, MN, TDD, PSR, and PBK analyzed the data. SAK, RK, and PBK prepared the manuscript and refined it. All authors have read and approved the manuscript.
Funding
RK and AM acknowledge the National Science Foundation REU (No. 15600349/2150087) grant.
Acknowledgments
SAK acknowledge the SERB-NPDF fellowship (PDF/2015/000929) and PBK was thankful to VFSTR for the Emeritus fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.965530/full#supplementary-material
Supplementary Figure 1 | Ramachandran plot validation for 3D structures of K+ transport proteins.
Supplementary Table 1 | List of qRT-PCR primers used for transcript profiling of Sb K+ transport genes.
Supplementary Table 2 | Nucleotide and protein sequences of Sb K+ transport homologs with ids.
Supplementary Table 3 | The pI, MW, GRAVY, instability, and aliphatic indexes of Sb K+ transport proteins.
Supplementary Table 4 | Amino acid composition and net charge of the Sb K+ transport genes.
Supplementary Table 5 | Types of protein kinases in the phosphorylation of Sb K+ transport genes.
Supplementary Table 6 | Conserved cis-acting regulatory elements in Sb K+ transport gene promoters.
Supplementary Table 7 | Modeled Sb K+ transport proteins using SWISS-MODEL server.
Supplementary Table 8 | STRING ids of Sb K+ transport proteins used for protein-protein interactions.
Supplementary Table 9 | List of K+ transport genes from Sorghum bicolor, Oryza sativa, and Arabidopsis thaliana used for construction of phylogenetic tree.
Supplementary Table 10 | Relative expression analysis of K+ transport gene homologs.
Footnotes
- ^ http://www.gramene.org/
- ^ http://genes.mit.edu/GENSCAN.html
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
- ^ http://smart.embl-heidelberg.de/
- ^ http://gsds.gao-lab.org/
- ^ https://wolfpsort.hgc.jp/
- ^ http://web.expasy.org/protparam/
- ^ https://services.healthtech.dtu.dk/service.php?NetPhos-3.1
- ^ https://pepcalc.com/
- ^ https://meme-suite.org/meme/tools/meme
- ^ http://mg2c.iask.in/mg2c_v2.1/
- ^ https://www.dna.affrc.go.jp/PLACE/?action=newplace
- ^ https://montelionelab.chem.rpi.edu/PSVS/PSVS/
- ^ https://string-db.org/
- ^ http://www.bork.embl.de/pal2nal/
- ^ https://genevestigator.com/gv
References
Ahn, S. J., Shin, R., and Schachtman, D. P. (2004). Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol. 134, 1135–1145. doi: 10.1104/pp.103.034660
Aksu, G., and Altay, H. (2020). The effects of potassium applications on drought stress in sugar beet. Sugar Tech 22, 1092–1102. doi: 10.26900/jsp.4.013
Amanullah Iqbal, A., and Hidayat, Z. (2016). Potassium management for improving growth and grain yield of maize (Zea mays L.) under moisture stress condition. Sci. Rep. 6, 1–12. doi: 10.1038/srep34627
Amrutha, R. N., Sekhar, P. N., Varshney, R. K., and Kavi Kishor, P. B. (2007). Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci. 172, 708–721. doi: 10.1016/j.plantsci.2006.11.019
Ashley, M. K., Grant, M., and Grabov, A. (2006). Plant responses to potassium deficiencies: A role for potassium transport proteins. J. Exp. Bot. 57, 425–436. doi: 10.1093/jxb/erj034
Assaha, D. V., Ueda, A., Saneoka, H., Al-Yahyai, R., and Yaish, M. W. (2017). The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 8:509. doi: 10.3389/fphys.2017.00509
Azedo-Silva, J., Osório, J., Fonseca, F., and Correia, M. J. (2004). Effects of soil drying and subsequent re-watering on the activity of nitrate reductase in roots and leaves of Helianthus annuus. Funct. Plant Biol. 31, 611–621. doi: 10.1071/fp04018
Azeem, F., Ahmad, B., Atif, R. M., Ali, M. A., Nadeem, H., Hussain, S., et al. (2018). Genome-wide analysis of potassium transport-related genes in chickpea (Cicer arietinum L.) and their role in abiotic stress responses. Plant Mol. Biol. Report. 36, 451–468. doi: 10.1007/s11105-018-1090-2
Azeem, F., Zameer, R., Rashid, M. A. R., Rasul, I., Ul-Allah, S., Siddique, M. H., et al. (2022). Genome-wide analysis of potassium transport genes in Gossypium raimondii suggest a role of GrHAK/KUP/KT8, GrAKT2. 1 and GrAKT1. 1 in response to abiotic stress. Plant Physiol. Biochem. 170, 110–122. doi: 10.1016/j.plaphy.2021.11.038
Banuelos, M. A., Garciadeblasm, B., Cuberom, B., and Rodrıguez-Navarro, A. (2002). Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 130, 784–795. doi: 10.1104/pp.007781
Biasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., et al. (2014). SWISS-MODEL:modeling protein tertiary and quaternary structure using evolutionary information. Nucl. Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340
Bowers, J. E., Chapman, B. A., Rong, J., and Paterson, A. H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. doi: 10.1038/nature01521
Cai, K., Zeng, F., Wang, J., and Zhang, G. (2021). Identification and characterization of HAK/KUP/KT potassium transporter gene family in barley and their expression under abiotic stress. BMC Genom. 22:317. doi: 10.1186/s12864-021-07633-y
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, G., Hu, Q., Luo, L. E., Yang, T., Zhang, S., Hu, Y., et al. (2015). Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ. 38, 2747–2765. doi: 10.1111/pce.12585
Chen, G., Liu, C., Gao, Z., Zhang, Y., Jiang, H., Zhu, L., et al. (2017). OsHAK1, a high-affinity potassium transporter, positively regulates responses to drought stress in rice. Front. Plant Sci. 8:1885. doi: 10.3389/fpls.2017.01885
Cheng, X., Liu, X., Mao, W., Zhang, X., Chen, S., Zhan, K., et al. (2018). Genome-wide identification and analysis of HAK/KUP/KT potassium transporters gene family in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 19:3969. doi: 10.3390/ijms19123969
Feng, H., Tang, Q., Cai, J., Xu, B., Xu, G., and Yu, L. (2019). Rice OsHAK16 functions in potassium uptake and translocation in shoot, maintaining potassium homeostasis and salt tolerance. Planta 250, 549–561. doi: 10.1007/s00425-019-03194-3193
Feng, X., Wang, Y., Zhang, N., Wu, Z., Zeng, Q., Wu, J., et al. (2020b). Genome-wide systematic characterization of the HAK/KUP/KT gene family and its expression profile during plant growth and in response to low-K+ stress in Saccharum. BMC Plant Biol. 20:20. doi: 10.1186/s12870-019-2227-7
Feng, X., Liu, W., Qiu, C. W., Zeng, F., Wang, Y., Zhang, G., et al. (2020a). HvAKT2 and HvHAK1 confer drought tolerance in barley through enhanced leaf mesophyll H+ homoeostasis. Plant Biotechnol. J. 18, 1683–1696. doi: 10.1111/pbi.13332
Gierth, M., Maäser, P., and Schroeder, J. I. (2005). The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol. 137, 1105–1114. doi: 10.1104/pp.104.057216
Hamamoto, S., Horie, T., Hauser, F., Deinlein, U., Schroeder, J. I., and Uozumi, N. (2015). HKT transporters mediate salt stress resistance in plants: From structure and function to the field. Curr. Opin. Biotechnol. 32, 113–120. doi: 10.1016/j.copbio.2014.11.025
Hamamoto, S., Marui, J., Matsuoka, K., Higashi, K., Igarashi, K., Nakagawa, T., et al. (2008). Characterization of a tobacco TPK-type K+ channel as a novel tonoplast K+ channel using yeast tonoplasts. J. Biol. Chem. 283, 1911–1920. doi: 10.1074/jbc.m708213200
Hasanuzzaman, M., Bhuyan, M. H. M., Nahar, K., Hossain, M. D., Mahmud, J. A., Hossen, M., et al. (2018). Potassium: A vital regulator of plant responses and tolerance to abiotic stresses. Agronomy 8:31. doi: 10.3390/agronomy8030031
He, C., Cui, K., Duan, A., Zeng, Y., and Zhang, J. (2012). Genome-wide and molecular evolution analysis of the poplar KT/HAK/KUP potassium transporter gene family. Ecol. Evol. 2, 1996–2004. doi: 10.1002/ece3.299
Hedrich, R. (2012). Ion channels in plants. Physiol. Rev. 92, 1777–1811. doi: 10.1152/physrev.00038.2011
Hu, W., Lu, Z., Meng, F., Li, X., Cong, R., Ren, T., et al. (2021). Potassium fertilization reduces silique canopy temperature variation in Brassica napus to enhance seed yield. Ind. Crops Prod. 168:113604. doi: 10.1016/j.indcrop.2021.113604
Hussain, S., Hussain, S., Ali, B., Ren, X., Chen, X., Li, Q., et al. (2021). Recent progress in understanding salinity tolerance in plants: Story of Na+/K+ balance and beyond. Plant Physiol. Biochem. 160, 239–256. doi: 10.1016/j.plaphy.2021.01.029
Hyun, T. K., Rim, Y., Kim, E., and Kim, J. S. (2014). Genome-wide and molecular evolution analyses of the KT/HAK/KUP family in tomato (Solanum lycopersicum L.). Genes Genom. 36, 365–374. doi: 10.1007/s13258-014-0174-0
Jiang, M., Chen, H., Liu, J., Du, Q., Lu, S., and Liu, C. (2021). Genome-wide identification and functional characterization of natural antisense transcripts in Salvia miltiorrhiza. Sci. Rep. 11, 1–14. doi: 10.1038/s41598-021-83520-6
Jin, R., Jiang, W., Yan, M., Zhang, A., Liu, M., Zhao, P., et al. (2021). Genome-wide characterization and expression analysis of HAK K+ transport family in Ipomoea. 3 Biotech 11, 1–18. doi: 10.1007/s13205-020-02552-3
Kuang, Q., Purhonen, P., and Hebert, H. (2015). Structure of potassium channels. Cell. Mol. Life Sci. 72, 3677–3693. doi: 10.1007/s00018-015-1948-5
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547. doi: 10.1093/molbev/msy096
Kyriakidou, M., Tai, H. H., Anglin, N. L., Ellis, D., and Strömvik, M. V. (2018). Current strategies of polyploid plant genome sequence assembly. Front. Plant Sci. 9:1660. doi: 10.3389/fpls.2018.01660
Li, Y., Peng, L., Xie, C., Shi, X., Dong, C., Shen, Q., et al. (2018). Genome-wide identification, characterization, and expression analyses of the HAK/KUP/KT potassium transporter gene family reveals their involvement in K+ deficient and abiotic stress responses in pear rootstock seedlings. Plant Growth Regul. 85, 187–198.
Li, W., Xu, G., Alli, A., and Yu, L. (2018). Plant HAK/KUP/KT K+ transporters: Function and regulation. Semin. Cell Dev. Biol. 74, 133–141. doi: 10.1016/j.semcdb.2017.07.009
Liang, M., Gao, Y., Mao, T., Zhang, X., Zhang, S., Zhang, H., et al. (2020). Characterization and expression of KT/HAK/KUP transporter family genes in willow under potassium deficiency, drought, and salt stresses. BioMed Res. Int. 2020:2690760. doi: 10.1155/2020/2690760
Moller, S., Croning, M. D., and Apweiler, R. (2001). Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17, 646–653. doi: 10.1093/bioinformatics/17.7.646
Nieves-Cordones, M., Ródenas, R., Chavanieu, A., Rivero, R. M., Martinez, V., Gaillard, I., et al. (2016). Uneven HAK/KUP/KT protein diversity among angiosperms: Species distribution and perspectives. Front. Plant Sci. 7:127. doi: 10.3389/fpls.2016.00127
Ou, W., Mao, X., Huang, C., Tie, W., Yan, Y., Ding, Z., et al. (2018). Genome-wide identification and expression analysis of the KUP family under abiotic stress in cassava (Manihot esculenta Crantz). Front. Physiol. 9:17. doi: 10.3389/fphys.2018.00017
Paterson, A. H., Bowers, J. E., Bruggmann, R., Dubchak, I., Grimwood, J., Gundlach, H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556. doi: 10.1038/nature07723
Rasheed, F., Markgren, J., Hedenqvist, M., and Johansson, E. (2020). Modeling to understand plant protein structure-function relationships-implications for seed storage proteins. Molecules 25:873. doi: 10.3390/molecules25040873
Rehman, H. M., Nawaz, M. A., Shah, Z. H., Daur, I., Khatoon, S., Yang, S. H., et al. (2017). In-depth genomic and transcriptomic analysis of five K+ transporter gene families in soybean confirm their differential expression for nodulation. Front. Plant Sci. 8:804. doi: 10.3389/fpls.2017.00804
Riedelsberger, J., Miller, J. K., Valdebenito-Maturana, B., Piñeros, M. A., González, W., and Dreyer, I. (2021). Plant HKT channels: An updated view on structure, function and gene regulation. Int. J. Mol. Sci. 22:1892. doi: 10.3390/ijms22041892
Rodríguez-Navarro, A. (2000). Potassium transport in fungi and plants. Biochim. Biophys. Acta Rev. Biomembr. 1469, 1–30. doi: 10.1016/s0304-4157(99)00013-1
Saadati, S., Baninasab, B., Mobli, M., and Gholami, M. (2021). Foliar application of potassium to improve the freezing tolerance of olive leaves by increasing some osmolyte compounds and antioxidant activity. Sci. Hortic. 276:109765. doi: 10.1016/j.scienta.2020.109765
Sánchez-McSweeney, A., González-Gordo, S., Aranda-Sicilia, M. N., Rodríguez-Rosales, M. P., Venema, K., Palma, J. M., et al. (2021). Loss of function of the chloroplast membrane K+/H+ antiporters AtKEA1 and AtKEA2 alters the ROS and NO metabolism but promotes drought stress resilience. Plant Physiol. Biochem. 160, 106–119. doi: 10.1016/J.PLAPHY.2021.01.010
Sardans, J., and Peñuelas, J. (2021). Potassium control of plant functions: Ecological and agricultural implications. Plants 10:419.
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Siddique, M. H., Babar, N. I., Zameer, R., Muzammil, S., Nahid, N., Ijaz, U., et al. (2021). Genome-wide identification, genomic organization, and characterization of potassium transport-related genes in Cajanus cajan and their role in abiotic stress. Plants 10:2238. doi: 10.3390/plants10112238
Song, Z. Z., Ma, R. J., and Yu, M. L. (2015). Genome-wide analysis and identification of KT/HAK/KUP potassium transporter gene family in peach (Prunus persica). Genet. Mol. Res. 14, 774–787. doi: 10.4238/2015.january.30.21
Song, Z., Wu, X., Gao, Y., Cui, X., Jiao, F., Chen, X., et al. (2019). Genome-wide analysis of the HAK potassium transporter gene family reveals asymmetrical evolution in tobacco (Nicotiana tabacum). Genome 62, 267–278. doi: 10.1139/gen-2018-0187
Tang, Z. H., Zhang, A. J., Wei, M., Chen, X. G., Liu, Z. H., Li, H. M., et al. (2015). Physiological response to potassium deficiency in three sweet potato (Ipomoea batatas [L.] Lam.) genotypes differing in potassium utilization efficiency. Acta Physiol. Plant. 37, 1–10. doi: 10.1007/s11738-015-1901-0
Tavakol, E., Jákli, B., Cakmak, I., Dittert, K., and Senbayram, M. (2021). Optimization of potassium supply under osmotic stress mitigates oxidative damage in barley. Plants 11:55. doi: 10.3390/plants11010055
Teklić, T., Paraðiković, N., Špoljarević, M., Zeljkovič, S., Lončarić, Z., and Lisjak, M. (2021). Linking abiotic stress, plant metabolites, biostimulants and functional food. Ann. Appl. Biol. 178, 169–191. doi: 10.1111/aab.12651
Wang, M., Zheng, Q., Shen, Q., and Guo, S. (2013). The critical role of potassium in plant stress response. Int. J. Mol. Sci. 14, 7370–7390. doi: 10.3390/ijms14047370
Wang, Q., Guan, C., Wang, P., Lv, M. L., Ma, Q., Wu, G. Q., et al. (2015). AtHKT1; 1 and AtHAK5 mediate low-affinity Na+ uptake in Arabidopsis thaliana under mild salt stress. Plant Growth Regul. 75, 615–623. doi: 10.1007/s10725-014-9964-2
Wang, X., Wang, J., Jin, D., Guo, H., Lee, T. H., Liu, T., et al. (2015). Genome alignment spanning major poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Mol. Plant 8, 885–898. doi: 10.1016/j.molp.2015.04.004
Yang, X., Zhang, J., Wu, A., Wei, H., Fu, X., Tian, M., et al. (2020). Genome-wide identification and expression pattern analysis of the HAK/KUP/KT gene family of cotton in fiber development and under stresses. Front. Genet. 11:566469. doi: 10.3389/fgene.2020.566469
Yang, T., Lu, X., Wang, Y., Xie, Y., Ma, J., Cheng, X., et al. (2020). HAK/KUP/KT family potassium transporter genes are involved in potassium deficiency and stress responses in tea plants (Camellia sinensis L.): Expression and functional analysis. BMC Genom. 21:556. doi: 10.1186/s12864-020-06948-6
Zhang, Z., Zhang, J., Chen, Y., Li, R., Wang, H., and Wei, J. (2012). Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol. Biol. Rep. 39, 8465–8473.
Keywords: HAK/KT/KUP, KEA, K+ channels, K+ transporters, Sorghum, Trk/HKT
Citation: Anil Kumar S, Hima Kumari P, Nagaraju M, Sudhakar Reddy P, Durga Dheeraj T, Mack A, Katam R and Kavi Kishor PB (2022) Genome-wide identification and multiple abiotic stress transcript profiling of potassium transport gene homologs in Sorghum bicolor. Front. Plant Sci. 13:965530. doi: 10.3389/fpls.2022.965530
Received: 09 June 2022; Accepted: 04 August 2022;
Published: 02 September 2022.
Edited by:
Sindhu Sareen, Indian Institute of Wheat and Barley Research (ICAR), IndiaReviewed by:
Ram Singh Purty, Guru Gobind Singh Indraprastha University, IndiaAdnan Aydin, Iğdır Üniversitesi, Turkey
Rakesh Singh, National Bureau of Plant Genetic Resources (ICAR), India
Copyright © 2022 Anil Kumar, Hima Kumari, Nagaraju, Sudhakar Reddy, Durga Dheeraj, Mack, Katam and Kavi Kishor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: P. B. Kavi Kishor, cGJrYXZpQHlhaG9vLmNvbQ==; Ramesh Katam, cmFtZXNoLmthdGFtQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share senior authorship
 S. Anil Kumar
S. Anil Kumar P. Hima Kumari
P. Hima Kumari Marka Nagaraju
Marka Nagaraju Palakolanu Sudhakar Reddy
Palakolanu Sudhakar Reddy T. Durga Dheeraj
T. Durga Dheeraj Alexis Mack
Alexis Mack Ramesh Katam
Ramesh Katam P. B. Kavi Kishor
P. B. Kavi Kishor