
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 07 September 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.965098
This article is part of the Research TopicWomen in Plant Biotechnology 2022View all 5 articles
A correction has been applied to this article in:
Corrigendum: The StBBX24 protein affects the floral induction and mediates salt tolerance in Solanum tuberosum
 Agnieszka Kiełbowicz-Matuk*
Agnieszka Kiełbowicz-Matuk* Klaudia Grądzka
Klaudia Grądzka Magdalena Biegańska
Magdalena Biegańska Urszula Talar
Urszula Talar Jagoda Czarnecka
Jagoda Czarnecka Tadeusz Rorat
Tadeusz RoratThe transition from vegetative growth to reproductive development is a critical developmental switch in flowering plants to ensure a successful life cycle. However, while the genes controlling flowering are well-known in model plants, they are less well-understood in crops. In this work, we generated potato lines both silenced and overexpressed for the expression of StBBX24, a clock-controlled gene encoding a B-box protein located in the cytosol and nuclear chromatin fraction. We revealed that Solanum tuberosum lines silenced for StBBX24 expression displayed much earlier flowering than wild-type plants. Conversely, plants overexpressing StBBX24 mostly did not produce flower buds other than wild-type plants. In addition, RT-qPCR analyses of transgenic silenced lines revealed substantial modifications in the expression of genes functioning in flowering. Furthermore, S. tuberosum lines silenced for StBBX24 expression displayed susceptibility to high salinity with a lower capacity of the antioxidant system and strongly decreased expression of genes encoding Na+ transporters that mediate salt tolerance, contrary to the plants with StBBX24 overexpression. Altogether, these data reveal that StBBX24 participates in potato flowering repression and is involved in salt stress response.
Proper timing of the transition from vegetative growth to reproductive development is essential for a successful plant life cycle and also has commercial significance for crop plants. Flowering time is determined by the interaction between endogenous genetic components as well as various environmental factors, especially day length and temperature (Ohto et al., 2001; Parcy, 2005; Amasino, 2010; Fornara et al., 2010; Seo et al., 2011; Srikanth and Schmid, 2011; Song et al., 2013). The transition from vegetative to floral development in higher plants is programmed by the simultaneous occurrence of several of these signals and underlies adaptation to different environments (Abelenda et al., 2014). While the mechanisms controlling flowering are well-known in the long-day (LD) plant Arabidopsis thaliana and short-day (SD) plant Oryza sativa, they are still little known in other plants.
The photoperiod-dependent mechanism promoting flowering in both LD- and SD-type plants is mainly based on the CO (CONSTANS) – FT (FLOWERING LOCUS T) regulon, where the CO protein is an inducer of FT expression (Johansson and Staiger, 2015). The transcriptional and post-translational factors regulating FT expression vary considerably in response to upstream environmental and endogenous signals among taxa (Ballerini and Kramer, 2011; Abelenda et al., 2014; Johansson and Staiger, 2015). In Arabidopsis, a facultative long-day species, the induction of FT expression for flowering during long days depends on the stability of the CO protein, which is regulated by the balanced action of GI (GIGANTEA), FKF1 (FLAVIN BINDING KELCH REPEAT F-BOX PROTEIN 1), and ZTL (ZEITLUPE; Pajoro et al., 2014; Song et al., 2014). The FT florigen protein complexes with FD, a bZIP transcription factor that activates the expression of several SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE transcription factors, which in turn induce FLORAL MERISTEM IDENTITY genes (Torti and Fornara, 2012).
Meanwhile, the regulation of flowering is more complex in species belonging to Solanaceae, such as potato (Solanum tuberosum) and tomato (Solanum lycopersicum). Floral promotion in these species is regulated by FT-like genes that function as inducers or repressors (Molinero-Rosales et al., 2004; Ballerini and Kramer, 2011; Abelenda et al., 2014, 2016). FT homologs in tomato and potato constitute a redundant family of transcription regulators that play important roles in flowering time, inflorescence architecture andin other developmental processes. They are divided into three major clades: FT-like, CEN/TFL-like, and MFT-like (Hedman et al., 2009). FT-like genes promote floral development in contrast to members of the CEN/TFL clade, which delay flowering and maintain indeterminacy in inflorescence meristem. MFT-like genes are implicated in seed development and germination (Hedman et al., 2009; Nakamura et al., 2011). Furthermore, the role of StBBX1 (StCOL1, previously named StCO) protein in potato flowering and tuberization has previously been reported (González-Schain et al., 2012; Abelenda et al., 2016). Potato StBBX1 protein controls StSP6A expression through direct activation of an additional FT family member, StSP5G, which in turn acts as a repressor of StSP6A transcription in leaves (Abelenda et al., 2016).
The CONSTANS (CO, also termed BBX1) protein belongs to the family of B-box type of zinc finger proteins named BBX. These proteins are transcription regulators forming complex families in plants (Grądzka and Kiełbowicz-Matuk, 2020; Talar and Kiełbowicz-Matuk, 2021), which widely differ in structure and fulfill distinct functions in the regulation of plant growth and development, including seedling photomorphogenesis (Datta et al., 2006, 2007, 2008; Indorf et al., 2007; Chang et al., 2008; Khanna et al., 2009; Holtan et al., 2011; Fan et al., 2012; Gangappa et al., 2013), photoperiodic regulation of flowering (Wenkel et al., 2006; Valverde, 2011), shade avoidance (Crocco et al., 2010) and responses to biotic and abiotic stresses (Nagaoka and Takano, 2003; Wang et al., 2013; Kiełbowicz-Matuk et al., 2014). As shown in Arabidopsis, several BBX proteins, in addition to CO participate in the control of flowering time through CO-dependent or CO-independent pathways. Thus, BBX6 (COL5) stimulates flowering by increasing CO expression under SD, whereas BBX7 (COL9) and BBX4 (COL3) suppress flowering by negatively regulating CO expression under LD and both LD and SD, respectively (Cheng and Wang, 2005; Datta et al., 2006; Ito et al., 2012; Gangappa and Botto, 2014). In addition, recent studies have revealed that other B-box members lacking the CCT domain regulate flowering via distinct overlapping or divergent functions. For instance, in Arabidopsis, overexpression of BBX24 promotes flowering in both LD and SD, while its deficiency delays flowering only in SD (Li et al., 2014).
We previously identified the BBX24 gene in two Solanum species and showed that its expression exhibited circadian cycling with significant increases in transcript and protein abundances after 8 h of light and a noticeable decrease with minima after 4 h in the darkness. Moreover, we revealed that the BBX24 gene was induced by high salinity being gated by the time of the day (Kiełbowicz-Matuk et al., 2014). In the present study, we explore the physiological function of StBBX24 in potato (currently called StBBX20, based on the present classification by Talar et al. (2017)) using silenced and overexpressed lines. We show that silenced lines flower earlier than wild-type plants, while lines with overexpression either do not produce flower buds or reveal late flowering. Interestingly, the lack of StBBX24 protein affects the expression pattern of critical genes associated with the floral transition process, unveiling a function of StBBX24 as a repressor of flowering in Solanum tuberosum. In other respects, we reveal that silenced lines display decreased tolerance to high salinity with a lower capacity of the antioxidant system and strongly decreased expression of genes encoding Na+ transporters, while the plants with overexpression of StBBX24 display the opposite response.
Solanum tuberosum L., cv. Desirée plants were propagated in vitro on solid MS medium from tuber single-eye plugs at 20/15°C (day/night) under a 150 μmol photon m−2 s−1 PFD and a 14-h photoperiod for 3 weeks. Four single-eye plugs from tubers were transferred on soil in plastic pots (diameter 21 cm) in the phytotron and grown under standard conditions (20 ± 1°C, 40% relative humidity and PFD of 350 μmol photons m−2 s−1) under 16-h-light/8-h-dark or 14-h-light/10-h-dark conditions. In these conditions, the term Zeitgeber time (ZT) refers to the experimental time (Johnson et al., 2003; Beneragama and Goto, 2010), and the ZT0 point corresponds to light on (initiation of experimental dawn). Two independent experiments and two independent plant samples from each experiment were used. The times of the first visible flower buds and the first flower opening were recorded. Tubers from twenty plants per genotype (four plants per plastic pot) were analyzed. The potato tubers were placed on a flat surface. The three mutually perpendicular axes of the potato tubers were designated and measured using a vernier caliper. The longest point of intersection depicted the length of the tuber, the longest intersection point perpendicular to the length represented the width of the tuber, and the longest intersection point perpendicular to the width and length represented the thickness of the tuber. The shape index of the measured tubers was calculated according to Ismail (1988).
For salt exposure, 3-week-old phytotron-grown plants were irrigated with an equal amount (per pot) of 0.15 M NaCl solution (−0.99 MPa osmotic potential) for 8 days. Plant material was harvested 8 h, 3 and 8 days after salt application.
The biomass of aerial parts was determined for plants grown under normal conditions or subjected to high salt. The samples were collected at ZT6, and excised shoots were dried at 80°C overnight before weighing. Sixteen plants per genotype and growth condition were analyzed.
One-cm diameter disks were excised from well-expanded leaves of plants grown under control conditions or subjected to high salt, then crushed in 2 ml 80% acetone. After storing 1 h in the dark at 4°C and centrifugation (8,000 g, 10 min), the content in chlorophylls a and b was measured spectrophotometrically and calculated according to (Lichtenthaler, 1987).
The total activity of Superoxide Dismutase and Peroxidase was assayed using the Superoxide Dismutase Activity Kit (MAK379, Merck) and Peroxidase Activity Assay Kit (MAK092, Merck), respectively, according to the protocols. Homogenates were prepared from pooled leaves (100 mg) in 0.1 M Tris–HCl, pH 7.4 containing 0.5% Triton X-100, 5 mM β-mercaptoethanol and 1 mM PMSF, incubated at 4°C for 30 min, and centrifuged at 14,000 g for 10 min. One Unit of SOD activity was defined as the amount of enzyme that inhibits the xanthine oxidase activity at 37°C. One unit of peroxidase activity was defined as the amount of enzyme that reduces 1.0 μmole of H2O2 per minute at 37°C. The soluble protein content was determined for all samples to normalize each enzyme activity between different trials. The activity measurements were performed spectrophotometrically at 410 nm and 570 nm wavelengths for SOD and peroxidase, respectively.
Total RNA was prepared from potato leaves and apical shoot parts using “TRI Reagent RT” (Molecular Research Center, Inc.) according to the manufacturer’s protocol. Equal RNA amounts were treated with RNase-free DNaseI (Ambion). The RNA concentration was measured using a Nano Drop photometer (Thermo Scientific). 2 μg of purified RNA was used for cDNA synthesis. The first DNA strand was prepared using the Thermo Scientific Maxima Reverse Transcriptase. Real-time qPCR was performed using a CFX96 Touch™Real-Time PCR detection system (Bio-Rad, Hercules, United States) and Maxima SYBR Green/ROX qPCR Master Mix (Thermo). Each assay using gene-specific primers (Supplementary Table S1) amplified a single product of the appropriate size with high PCR efficiency (90%–110%). The fluorescence data analysis was conducted using the CFX™ Software (Version 3.0, Bio-Rad, Hercules, United States). All RT-qPCR analyses were normalized using the threshold cycle (C1) values corresponding to the EF-1-α and/or the 18S reference genes (Nicot et al., 2005; Kiełbowicz-Matuk et al., 2014). The normalized expression of the target gene (ΔΔCq) was calculated as the relative quantity of target normalized to the quantities of the two reference genes, according to the manufacturer’s software. The values presented are the means of two technical replicates from two independent biological samples.
Nuclei-enriched fractions were isolated from 3-week-old S. tuberosum leaves at ZT6 as described by Kiełbowicz-Matuk et al. (2017). Further purification of nuclear proteins was performed by suspending the nuclear pellet in two volumes of 25 mM Tris–HCl, pH 8.0, 30 mM NaCl, 2 mM EDTA, and 1% Nonidet P-40 supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific) and incubation at 4°C for 30 min. Samples were then centrifuged at 13,000 g for 10 min to pellet the insoluble chromatin and collect the supernatant (nucleoplasm fraction). The chromatin pellet was washed and resuspended in 50 mM Tris–HCl, pH 8.0, 30 mM NaCl, 1 mM PMSF, 1% sodium deoxycholate, 1% SDS, 0.5% Triton X-100, and incubated at 4°C for 1 h before centrifugation at 14,000 g for 10 min. Proteins from the nucleoplasm and chromatin fractions were precipitated with four volumes of acetone.
Cytosolic proteins were prepared from young leaves of 3-week-old plants at ZT6 in a buffer containing 0.3 M sucrose, 50 mM Tris–HCl, pH 8.0, 10 mM MgCl2, 1 mM EDTA, and 1 mM PMSF, as described by Kiełbowicz-Matuk et al. (2014). The homogenate was filtered through a two-layer Miracloth (Calbiochem) and centrifuged at 3,000 g for 10 min at 4°C. Then, the cleared supernatant was centrifuged at 20,000 g for 20 min at 4°C and collected. Cytosolic proteins were precipitated with four volumes of acetone. Soluble proteins were extracted from young leaves of 3-week-old plants at ZT6 in a buffer containing 50 mM Tris–HCl, pH 8.0, 2% β-mercaptoethanol, and 1 mM PMSF.
Protein concentration was measured using a modified Lowry copper-based method (Pierce). Proteins were separated in SDS-PAGE gels containing 13% acrylamide, and then electroblotted onto a 0.22 μm nitrocellulose membrane (Schleicher and Schuell, Germany) using a semi-dry blotting apparatus (Bio-Rad). Membranes were stained with Ponceau red to ensure that equal protein amounts were transferred in each lane. The Western blot analysis was performed using the StBBX24 antiserum diluted 1:500. Sera raised against nuclear cap-binding protein subunit 2 (CBP20), histone H3, and UDP-glucose pyrophosphorylase (UGPase) were purchased from Agrisera (Vännäs, Sweden) and used diluted 1:500, 1:5,000, and 1:1,000, respectively. Sera raised against cytosolic cyclophilin (SsCyP) was diluted 1:500 (Kiełbowicz-Matuk et al., 2007). As a loading control to directly detect the total protein concentration for cytosolic fraction, antibodies raised against cytosolic marker UGPase protein were used. Anti-histone H3 antibodies were used as a loading control for nuclear proteins (Kiełbowicz-Matuk et al., 2017). Bound antibodies were detected using an anti-rabbit immunoglobulin G (horseradish peroxidase conjugate) diluted 1:10,000 (Roche) and a chemiluminescent substrate (Western Bright Quantum, Advansta). The Western blotting was performed using a G:BOX Imaging System (Syngene). Band intensity was analyzed using GeneSys software V1 6.5.0 (Syngene). Analyses were performed using protein extracts from two independent sets of plants per condition, and at least three replicates were conducted for each condition. Mean values ± SD from two biological and three technical replicates are presented.
To investigate the physiological function of StBBX24 in Solanum tuberosum plants, we designed artificial microRNAs (amiRNAs) targeting the StBBX24 mRNA to generate transgenic gene-silenced lines. Because cultivated potato species are autotetraploid and highly heterozygous, we sequenced multiple StBBX24 cDNA clones from the Desirée cultivar. StBBX24 mRNA fragments showing no allelic polymorphism were selected and analyzed to find the most effective target sites for RNA-induced silencing complex (RISC) slicing using Web MicroRNA Designer.1 Four StBBX24 allele variants were identified (Supplementary Figures S1A,B). The mRNA target sites within StBBX24 cDNA are marked in Supplementary Figure S1. Using the same platform, three amiRs were selected (Supplementary Table S1). Post-transcriptional silencing using artificial miRNA was performed as in Schwab et al. (2006) and Ossowski et al. (2008). Briefly, a set of overlapping PCRs was performed using the appropriate primers for the three constructs (Supplementary Table S1) to exchange miR319b/miR319b* for amiRBBX24.1/amiRBBX24.1*, amiRBBX24.2/amiRBBX24.2*, and amiRBBX24.3/amiRBBX24.3*. The DNA fragments carrying aMIRBBX24.1, aMIRBBX24.2, and aMIRBBX24.3 genes were cloned into the pHannibal vector (AJ311872; Wesley et al., 2001). The DNA constructs, including the CaMV 35S promoter and an OCS terminator, were transferred into a pART27 vector (Gleave, 1992). As a result, 92 transgenic lines harboring the T-DNAs containing the aMIRBBX24.1, aMIRBBX24.2 or aMIRBBX24.3 genes were generated. All lines were assayed for silencing efficiency using RT-qPCR (Supplementary Figure S2A). Forty lines exhibiting strong silencing of StBBX24 expression were chosen for the Western blot analysis. Among them, 10 lines harboring the aMIRBBX24.1 gene, i.e., amiRBBX24.1.1, amiRBBX24.1.5, amiRBBX24.1.9, amiRBBX24.1.10, amiRBBX24.1.12, amiRBBX24.1.13, amiRBBX24.1.15, amiRBBX24.1.17, amiRBBX24.1.25, and amiRBBX24.1.26, showed complete silencing at the protein level (Supplementary Figure S2B).
The StBBX24 overexpression lines (StBBX24-OE) were generated by recombining the StBBX24 full-length coding sequence from the pENTR clone with the pGWB402 binary vector via the Gateway LR Clonase system (Invitrogen). To generate the pENTR™/SD/D-TOPO® vector cDNA for StBBX24 was amplified using Phusion™ High-Fidelity DNA polymerase (Thermo Scientific) and gene-specific primer pairs (Supplementary Table S1), cloned into pENTR and then sequenced. pGWB402 was a gift from Tsuyoshi Nakagawa (Addgene plasmid #74796, http://n2t.net/addgene:74796;RRID:Addgene_74796; Nakagawa et al., 2007).
The plasmids were separately introduced into Agrobacterium tumefaciens strain LBA4404 and transformed into S. tuberosum by the Agrobacterium-mediated technique (Chen et al., 1994). Plants were regenerated as described by Mac et al. (2004).
Total genomic DNA and cellular RNA were isolated from potato leaves using a GeneJET Plant Genomic DNA Purification Kit (Thermo Scientific) and TRI Reagent RT (Molecular Research Center, Inc.), respectively. Transformed lines were analyzed using PCR and primers specific to the transgene. In addition, StBBX24 expression was assayed using quantitative real-time PCR and Western blot analysis, as described previously.
We previously reported that StBBX24 gene expression, was differentially regulated as a function of organ type during vegetative growth and reproductive development (Kiełbowicz-Matuk et al., 2014). In the present study, we further analyzed StBBX24 expression in young leaves (<2 cm in length) and apical shoot parts of 3-, 6-, and 9-week-old S. tuberosum plants grown under a 16 h photoperiod. At these stages, the stem length considerably varied (10, 20 and 35 cm at 3, 6, and 9 weeks, respectively) as well as the number of well-expanded composed leaves (0, 10, and 15, respectively). Most of the 9-week-old S. tuberosum plants exhibited emerged, but non open, flower buds. As shown in Figures 1A,B, similar StBBX24 transcript and protein amounts were observed in extracts from young leaves and apical shoot parts of 3- and 6-week-old plants. In comparison, the transcript and protein abundances in 9-week-old plants were much lower, two- to three-fold less, particularly in apical parts, revealing that the StBBX24 expression changes during the plant growth with a decreases in developing of floral organs.
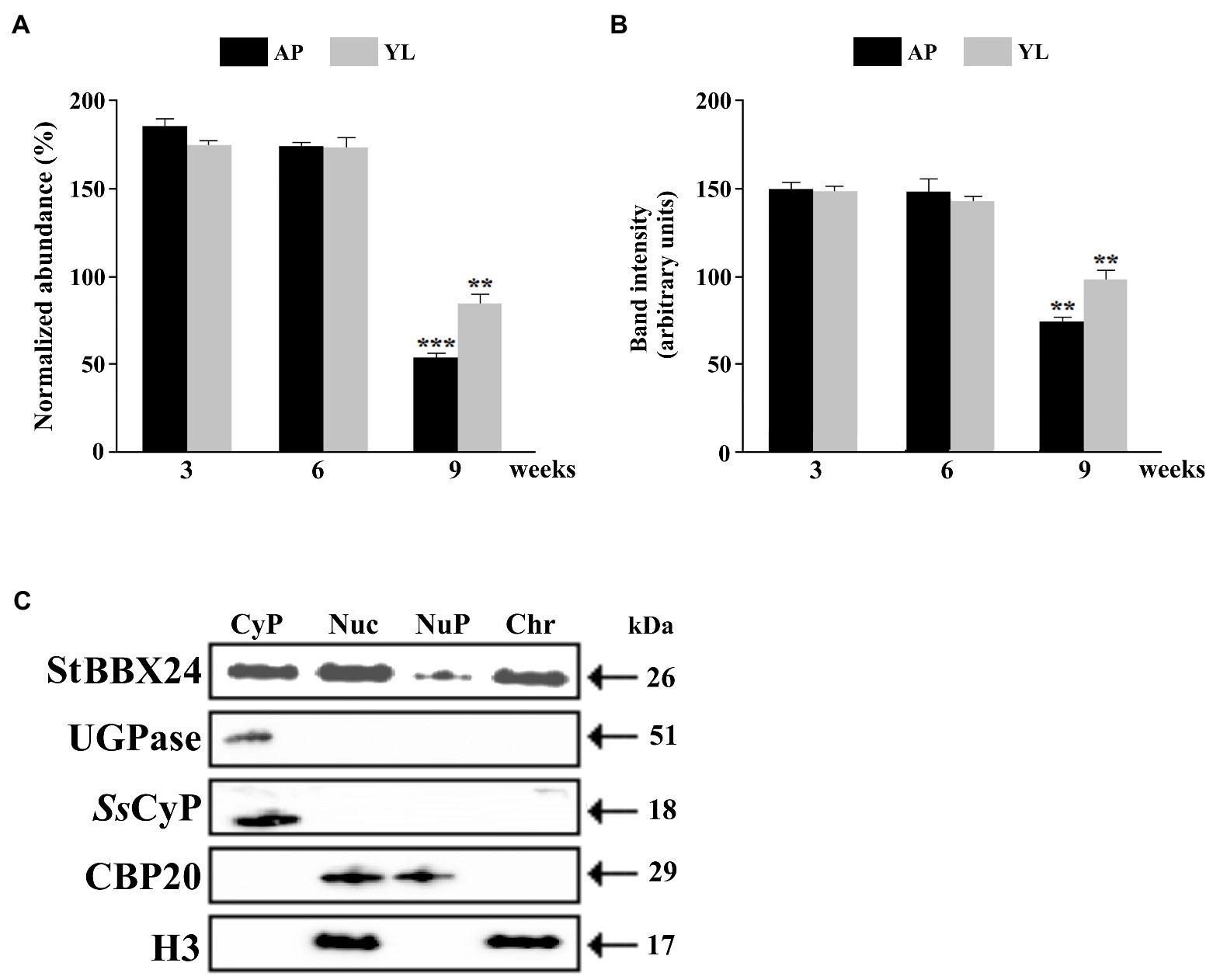
Figure 1. Expression of StBBX24 in young organs of S. tuberosum plants and nuclear localization of StBBX24. StBBX24 transcript (A) and protein (B) abundance in apical shoot parts and young leaves of phytotron-grown S. tuberosum plants. (A) Quantitative RT-PCR was performed as described in Materials and methods. (B) Western analysis of StBBX24 abundance was performed using sera raised against StBBX24 as described in Materials and methods. As a loading control of the total protein for cytosolic fraction, antibodies raised against cytosolic marker UGPase protein were used. AP – apical shoot part; YL – young leaves (including very young developing leaves). (C) Western blot analysis of proteins from cytosolic (Cyt), total nuclei (Nuc), nucleoplasm (NuP) and chromatin (Chr) fractions prepared from leaves of S. tuberosum plants. 10–40 μg proteins from nuclei-enriched and cytosolic fractions were set per lane. The analysis was carried out using sera raised against StBBX24, UDP-glucosepyrophosphorylase (UGPase), cytosolic cyclophilin (CyP), histone H3 and cap-binding protein subunit 2 (CBP20). Means ± SD of four values from two independent experiments (two replicates per experiment) are presented. Statistical analysis was performed using the t-test (Statistica program, version 10). ** and ***, values significantly different from the 3-week old plants value with p < 0.01 and p < 0.001, respectively.
We have previously shown that the StBBX24 protein is presented both in the nucleus and cytosol, and its distribution depends on the time of day (Kiełbowicz-Matuk et al., 2014). Here, we investigated its subnuclear localization in nucleoplasm and chromatin fractions from leaves of 3-week-old S. tuberosum plants at ZT6 when the amount of StBBX24 is the highest in nucleus (Kiełbowicz-Matuk et al., 2014). To validate the fraction purity, a Western blot analysis of proteins restricted to nucleoplasm or chromatin, i.e., cap-binding protein CBP20 and histone H3, respectively, was performed (Agrisera). As shown in Figure 1C, histone H3 was present in the chromatin fraction, whereas CBP20 was localized in the nucleoplasm fraction. Interestingly, the serum raised against recombinant StBBX24 specifically detected the protein in both chromatin and nucleoplasm fractions. However, the band intensity was higher in chromatin than in nucleoplasm, clearly showing that StBBX24 was mainly localized in the chromatin within nucleus.
To gain insight into the role of StBBX24 in cultivated potato development, we studied five silenced lines (amiRBBX24.1.1, amiRBBX24.1.5, amiRBBX24.1.9, amiRBBX24.1.13, and amiRBBX24.1.17) and wild-type plants grown under a 16-h photoperiod. We did not observe any difference concerning the plant architecture, but we did notice slight changes in the number and size of tubers in transgenic lines as compared with wild-type plants (Supplementary Figures S3A,B; Supplementary Tables S2, S3). However, most significantly, a pronounced phenotype was observed regarding flowering time in transgenic lines. Indeed, we noticed that the lack of StBBX24 protein strongly promoted flowering, with the opening of flower buds occurring ~16, 12, and 10 days earlier in amiRBBX24.1.5, amiRBBX24.1.9, amiRBBX24.1.17, amiRBBX24.1.1, and amiRBBX24.1.13 plants, respectively, as compared with in WT (Figures 2A,B). Meanwhile, when potato plants were grown under a shorter photoperiod (12 h), conditions preventing the flower bud differentiation process, no difference was noticed between WT and transgenic lines that both exhibited only vegetative development (Supplementary Figure S3C). These results reveal that StBBX24 may function as a regulator affecting the control of reproductive development in potatoes, likely through a role at the flower bud initiation stage.
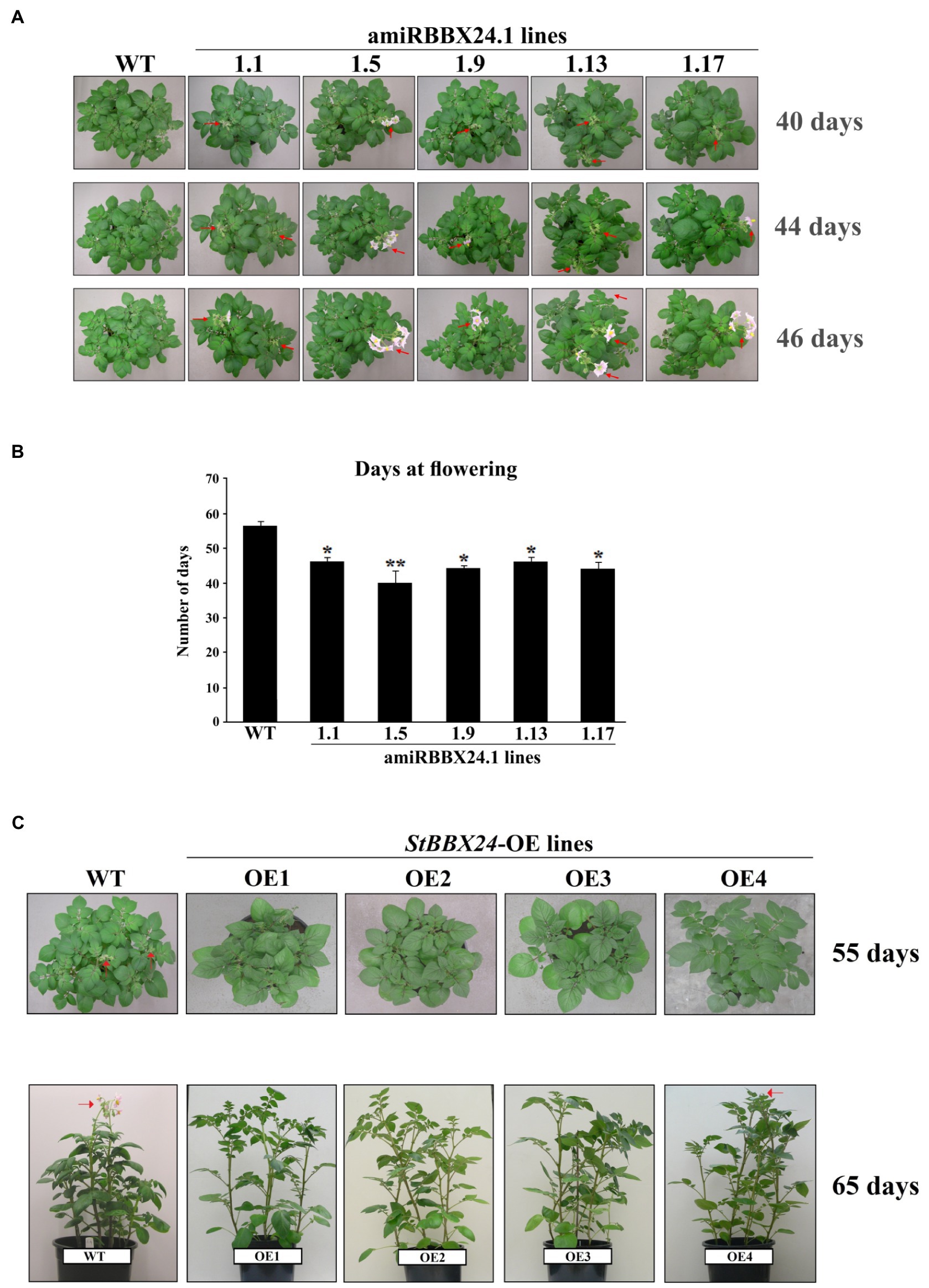
Figure 2. Flowering phenotype of S. tuberosum plants silenced for StBBX24 expression and overexpressing StBBX24. (A) Phenotype of WT, amiRBBX24.1.1, amiRBBX24.1.5, amiRBBX24.1.9, amiRBBX24.1.13 and amiRBBX24.1.17 transgenic plants at the pre-flowering and flowering stages. Sixteen to twenty plugs from potato tubers per genotype were grown in plastic pots (diameter 21 cm) in the growth room in standard conditions under a 16-h photoperiod. The red arrows indicate the presence of flower buds and open flowers. (B) Number of days at flowering in WT and lines silenced for StBBX24 expression. * and **, values significantly different from the WT value (t-test) with p < 0.05 and p < 0.01, respectively. WT, wild-type. (C) Phenotype of WT, StBBX24-OE1, StBBX24-OE2, StBBX24-OE3 and StBBX24-OE4 transgenic plants at the pre-flowering and flowering stages. Twenty plugs from potato tubers per genotype were grown in plastic pots (diameter 21 cm) in the growth room in standard conditions under a 16-h photoperiod. The red arrows indicate the presence of flower buds and open flowers.
Then we investigated the reproductive development of four StBBX24 overexpressing potato lines (StBBX24-OE1, StBBX24-OE2, StBBX24-OE3, and StBBX24-OE4) and the wild-type control grown under a 16-h photoperiod (Figure 2C). All transgenic lines were assayed for StBBX24 expression level using Western blot (Supplementary Figure S4). We did not notice any substantial alterations in plant architecture and size of tubers, and the number of tubers, between transgenic plants and their wild type (Supplementary Figures S5A,B; Supplementary Tables S2, S3.). Meanwhile, significant differences in phenotype were observed concerning flowering time in the overexpressing lines compared to the wild-type. We found that the StBBX24-OE1, StBBX24-OE2, and StBBX24-OE3 lines did not produce flower buds. Meanwhile, in the StBBX24-OE4 line, only a few flower buds occurred ~10 days later than in WT (Figure 2C). Our data provide evidence that StBBX24 protein may affect the success of potato reproduction.
Based on the accelerated flowering of silenced lines and lack or deleted flowering of overexpressed lines, we paid more attention to genes involved in regulating flowering time. We studied the expression patterns as a function of the photoperiod of the key flowering-related genes in amiRBBX24.1.13 and amiRBBX24.1.17 and StBBX24-OE3 and StBBX24-OE4 lines using RT-qPCR. In the lines with silenced StBBX24 gene expression, we observed significantly increased expression levels of floral activators, including StFKF1 (Flavin-binding, Kelch repeat, and F box 1), StGI (GIGANTEA), StGID (Gibberellin Insensitive Dwarf 1), and StSOC1 (Suppressor of Overexpression of CO1). As shown in Figure 3A, the transcript levels of StFKF1, StGI, StSOC1, and StGID were considerably higher than in WT. Of note, the expression patterns of these genes were still periodic, but the periods were shifted to the early light phase for StSOC1 and StGID1, as compared with those in WT (Figure 3A). Additionally, we analyzed the expression of critical floral repressors, such as StEBS (Early Bolting in Short days), StSVP, StCDF1, and StCDF2 (CYCLING DOF FACTORS 1 and 2), and beyond StEBS, we observed a significant decrease in their transcript abundance in silenced lines as compared with WT (Figure 3B). Regarding StEBS, the transcript abundance was considerably higher in both silenced lines, but the oscillation pattern was significantly affected (Figure 3B). On the contrary, in StBBX24 overexpressing lines, we noticed a decrease in the transcript level of some genes encoding flowering activators, including StFKF1 and StGI; Meanwhile, transcript amount for some repressors, such as StEBS was increased concerning the wild-type (Figures 4A,B). Notably, the StSOC and StSVP, displayed impaired expression patterns in both the light and dark phases of the 24-h diurnal cycle, as compared with those in WT (Figures 4A,B).
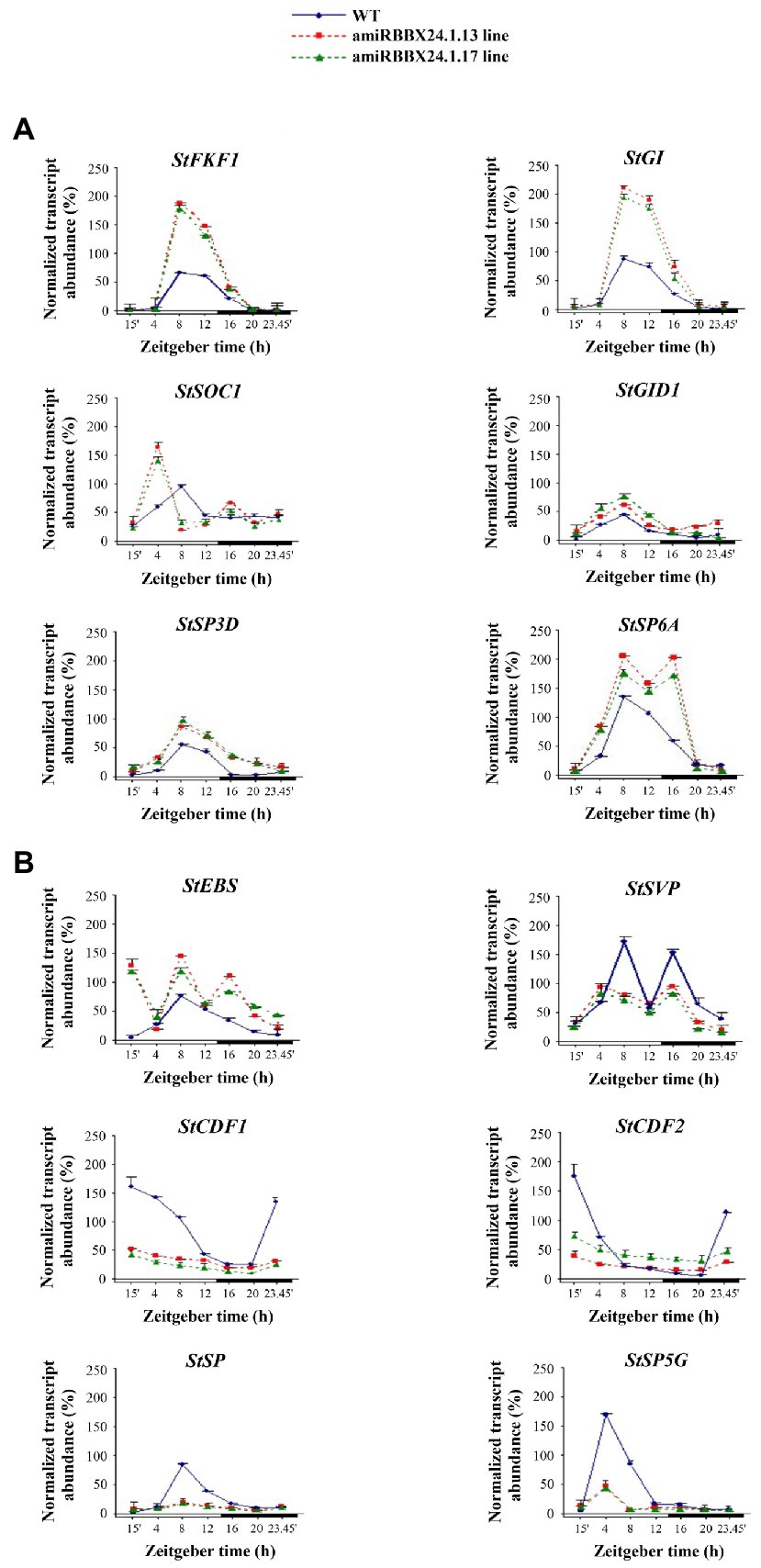
Figure 3. Expression profiles of flowering-related genes in S. tuberosum plants silenced for StBBX24 expression. Quantitative RT-PCR analysis of StFKF1 (ID: PGSC0003DMG400019971), StGI (ID: PGSC0003DMG400018791), StSOC1 (ID: PGSC0003DMG401024252), StGID1 (ID: PGSC0003DMG400000139), StSP3D (ID: PGSC0003DMG400016179), StSP6A (ID: PGSC0003DMG400023365) (A) and StEBS, StSVP (ID: PGSC0003DMG400016203), StCDF1 (ID: PGSC0003DMT400047370), StCDF2 (ID: PGSC0003DMT400003359), StSP (ID: PGSC0003DMG400007111), StSP5G (ID: PGSC0003DMG400023365) (B) transcript levels in leaves of WT, amiRBBX24.1.13 and amiRBBX24.1.17 3-week old plants under a 14-h photoperiod at different time points (15′, 4, 8, 12, 16, 20, 23h45′) for 24 h during the light and dark phases. Zeitgeber time (ZT) refers to experimental time where the ZT0 point corresponds to light-on (initiation of experimental dawn). The bars represent the subjective light and night conditions. RT-qPCR analyses were normalized using the threshold cycle (C1) values corresponding to the 18S and EF-1-α reference genes. The normalized expression of the target gene (ΔΔCq) was calculated as the relative quantity of target normalized to the quantities of the two reference genes according to the manufacturer’s software. Means ± SD of four values from two independent experiments (two replicates per experiment) are presented.
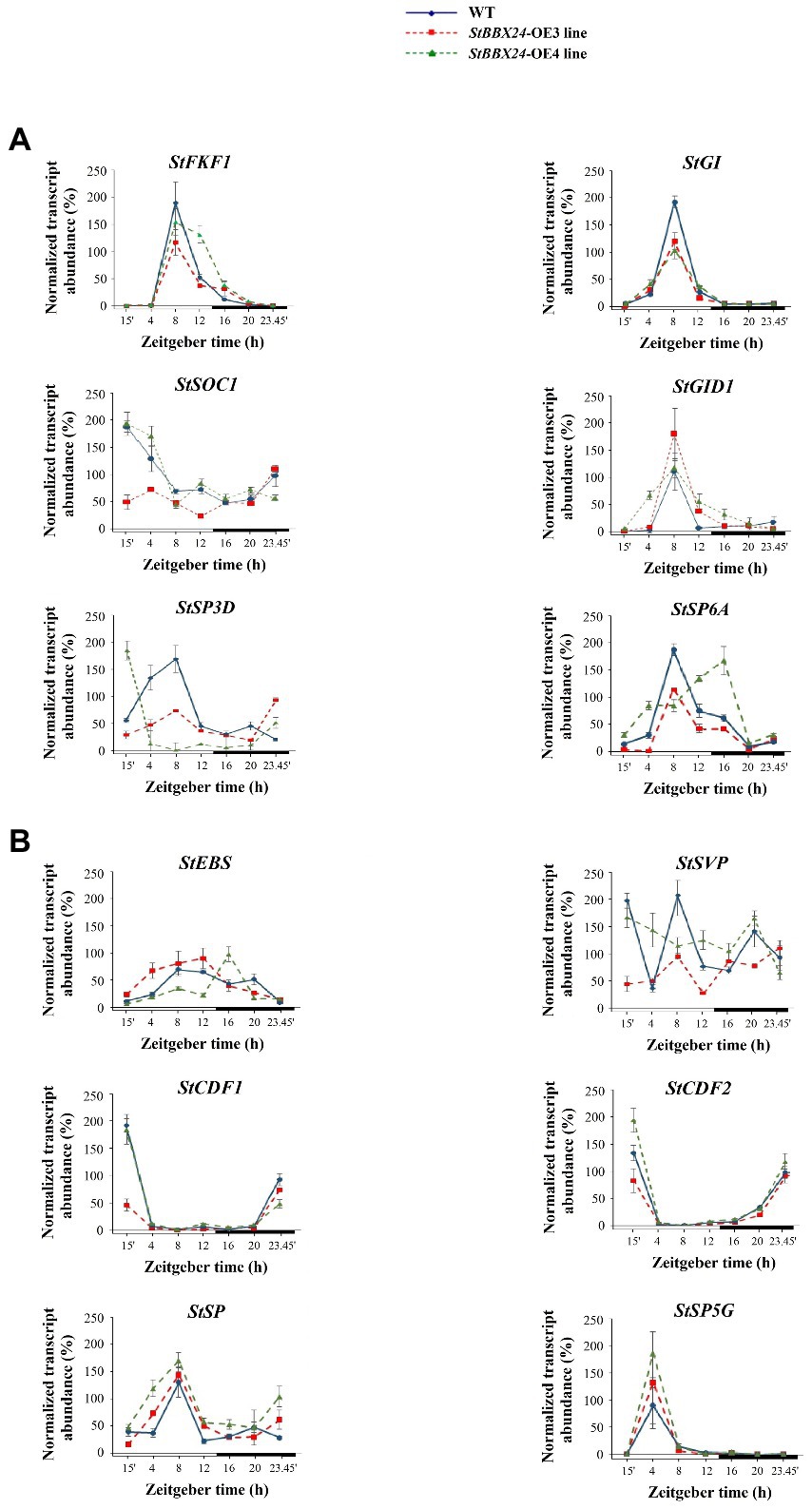
Figure 4. Expression profiles of flowering-related genes in S. tuberosum plants overexpressing StBBX24. Quantitative RT-PCR analysis of StFKF1 (ID: PGSC0003DMG400019971), StGI (ID: PGSC0003DMG400018791), StSOC1 (ID: PGSC0003DMG401024252), StGID1 (ID: PGSC0003DMG400000139), StSP3D (ID: PGSC0003DMG400016179), StSP6A (ID: PGSC0003DMG400023365) (A) and StEBS, StSVP (ID: PGSC0003DMG400016203), StCDF1 (ID: PGSC0003DMT400047370), StCDF2 (ID: PGSC0003DMT400003359), StSP (ID: PGSC0003DMG400007111), StSP5G (ID: PGSC0003DMG400023365) (B) transcript levels in leaves of WT, StBBX24-OE3 and StBBX24-OE4 3-week old plants under a 14-h photoperiod at different time points (15′, 4, 8, 12, 16, 20, 23h45′) for 24 h during the light and dark phases. The bars represent the subjective light and night conditions. RT-qPCR analyses were normalized using the threshold cycle (C1) values corresponding to the EF-1-α reference genes. The normalized expression of the target gene (ΔΔCq) was calculated as the relative quantity of target normalized to the quantities of the two reference genes according to the manufacturer’s software. Means ± SD of four values from two independent experiments (two replicates per experiment) are presented. Zeitgeber time (ZT) refers to experimental time where the ZT0 point corresponds to light-on (initiation of experimental dawn).
In Solanaceae, the participation of FLOWERING TIME-like genes, including StSP6A (SELF PRUNING 6A) and StSP3D (SELF PRUNING 3D) as inducers and StSP5G (SELF PRUNING 5G) and StSP (SELF PRUNING) as repressors of floral development has been reported. Thus, we investigated their expression in leaves of StBBX24-silenced plants and observed that StSP6A and StSP3D transcript levels were substantially up-regulated as compared with WT (Figure 3A), while those of StSP5G and StSP were significantly decreased (Figure 3B). Significant changes in transcript accumulation were also observed in plants with StBBX24 overexpression compared with non-transformed plants (Figures 4A,B). We noted that transcript levels of StSP6A and StSP3D were substantially down-regulated; in the meantime, StSP and StSP5G were elevated as compared with WT. Furthermore, it was noted that while the regular expression of StSP and StSP5G genes was intact, it was disrupted for StSP3D and StSP6A genes (Figures 4A,B). These findings reveal that the StBBX24 protein is an important regulator of flowering time in cultivated potato.
As StBBX24 encodes a protein participating in salt stress responses in S. tuberosum and sodium chloride (0.15 and 0.2 M) affect the circadian-regulated amount of BBX24 protein (Kiełbowicz-Matuk et al., 2014), we assessed the influence of StBBX24 on salt stress tolerance in potato by characterising the phenotype of three silenced lines and three overexpressed lines in high salinity conditions. Initially, we performed a phenotypic characterization of StBBX24-silenced and -overexpressed representative lines during the vegetative development under normal growth conditions, i.e., amiRBBX24.1.1, amiRBBX24.1.13, amiRBBX24.1.17, and StBBX24-OE2, StBBX24-OE3, and StBBX24-OE4 to evaluate their growth and morphological divergences. No significant difference was noticed in the rate of vegetative growth, the number of nodes and the number, shape and size of leaves (Supplementary Figures S3A, S5A).
When plants were watered with a nutrient solution containing 0.15 M NaCl, there was no difference between wild-type and StBBX24 overexpressing lines for 8 days (Figure 5C). Similarly, there were no noticeable changes between the wild type and StBBX24-silenced plants during the first 3 days of exposure to salt stress (Figure 5A). However, after 8 days of exposure to high salinity, pale green areas and severe chlorosis appeared in both well-expanded and old leaves of silenced plants (Figure 5A). When measuring the biomass of aerial parts and the leaf chlorophyll content following 8 days in high salt conditions, we noticed that amiRBBX24.1.17, amiRBBX24.1.13, and amiRBBX24.1.1 lines displayed reduced chlorophyll contents by ca. 38%, 21%, and 19%, respectively, compared to wild type and overexpressing lines (Figures 5B,D). As shown in Figure 5B, the degradation of photosynthetic structures under high salinity was associated with a reduction in the biomass of aerial parts that was significant for amiRBBX24.1.17 plants (ca. 25%) compared to wild type. This is illustrated by the data in Figure 5B showing that 3-week-old WT and silenced plants display similar weights of aerial parts and close chlorophyll contents, except for one line that exhibits a significantly lower content. Then, we investigated the impact of StBBX24 silencing and overexpression on the activities of antioxidant enzymes during 8 days of exposure to salt stress. As shown in Figure 6, superoxide dismutase and peroxidase activity were stimulated under salinity in both types of transgenic lines compared to the control condition. However, the increment of their activity in plants overexpressing StBBX24 was greater compared to amiRBBX24.1.1, amiRBBX24.1.13, and amiRBBX24.1.17 lines following 8 days in high salt conditions.
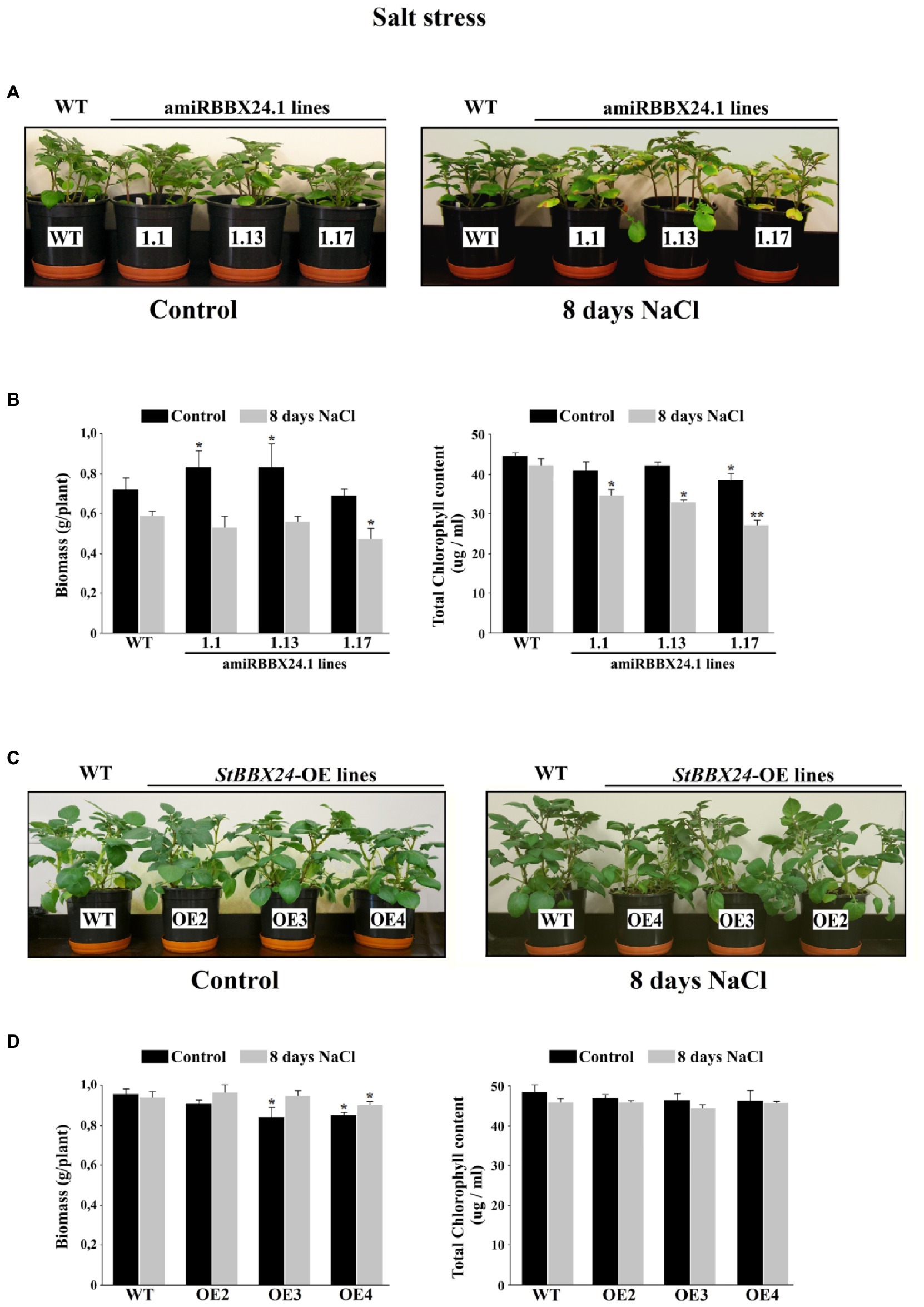
Figure 5. Phenotypic characterization of the StBBX24-silenced and -overexpressing S. tuberosum lines in response to salinity. (A,C) Phenotype of phytotron-grown S. tuberosum WT, amiRBBX24.1.1, amiRBBX24.1.13 and amiRBBX24.1.17 plants, and StBBX24-OE2, StBBX24-OE3 and StBBX24-OE4 lines in high salt conditions (0.15 M NaCl). Plugs from potato tubers were grown for 3-weeks in plastic pots in control conditions in a phytotron and were then watered with a solution containing 0.15 M NaCl in the same conditions for 8 days. (B,D) Biomass of aerial parts and leaf chlorophyll content of plants grown in control conditions or subjected to high salt for 8 days. Leaf biomass values are mean ± SD from 16 independent measurements per genotype. The chlorophyll content was measured using three 1-cm leaf disks from young well-expanded leaves. Statistical analysis was performed using the t-test (Statistica program, version 10). * and **, values significantly different from the WT value with p < 0.05 and p < 0.01, respectively. WT, wild-type.
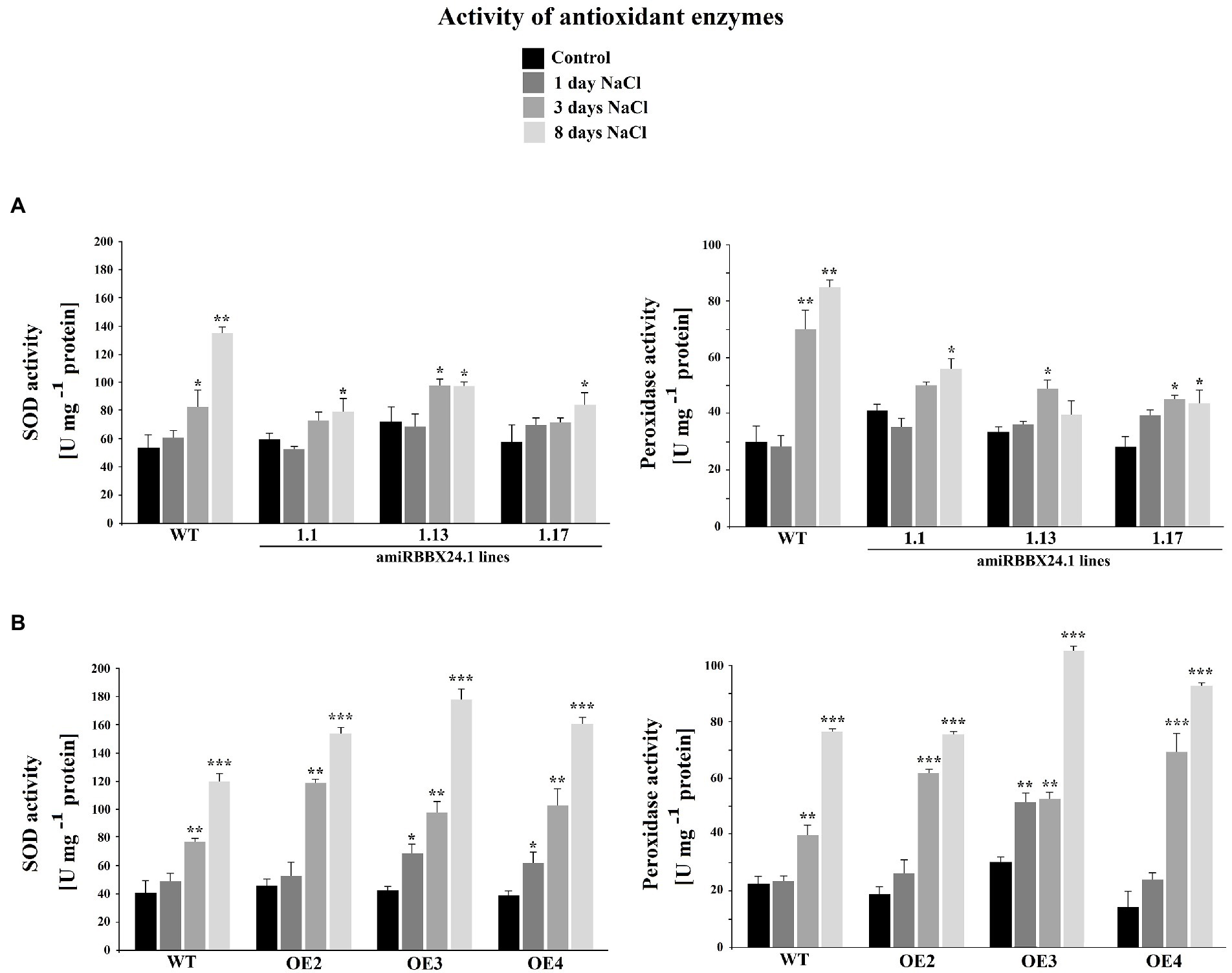
Figure 6. The activity of superoxide dismutase (SOD, EC 1.15.1.1) and peroxidase (POX, EC 1.11.17) enzymes in StBBX24-silenced (A) and -overexpressing (B) S. tuberosum lines subjected to salt stress. Antioxidant enzymes activity of phytotron-grown S. tuberosum WT, amiRBBX24.1.1, amiRBBX24.1.13 and amiRBBX24.1.17 plants, and StBBX24-OE2, StBBX24-OE3 and StBBX24-OE4 lines in high salt conditions (0.15 M NaCl). Measurement of superoxide dismutase and peroxidase activity was performed as described in Materials and methods. Error bars represent the bar errors. Statistical analysis was performed using the t-test (Statistica program, version 10). *, ** and ***, values significantly different from the control (C) value with p < 0.05, p < 0.01 and p < 0.001, respectively. WT, wild-type.
In order to gain better insight regarding the observed phenotype, we investigated the impact of StBBX24 silencing and overexpression on the alterations in expression profile for the essential genes involved in responses to salinity stress. In our research, we focused on genes encoding Na+ transporters that mediate tolerance to salt stress, i.e., HKT1 that selectively and directly unloads sodium from xylem vessels to xylem parenchyma cells, and two major factors maintaining low cytoplasmic Na+ concentrations in plant cells, the tonoplast-localized Na+/H+ NHX3 exchanger and the plasma membrane Na+/H+ SALT OVERLY SENSITIVE antiporters, SOS1, SOS2, and SOS3 (Blumwald, 2000; Deinlein et al., 2014). As shown in Figures 7A,B, substantial increases in transcript amounts of StHKT1, StNHX3, StSOS1, StSOS2, and StSOS3 were observed in wild-type compared to control conditions, and maximal levels were reached following 8 days of salt treatment. In two silenced lines analyzed (amiRBBX24.1.13 and amiRBBX24.1.17), the transcript levels of StHKT1, StNHX3, StSOS1, StSOS2, and StSOS3 genes were higher than in WT in control conditions. But, a gradual reduction in the abundance of StSOS1, StSOS2, and StNHX3 transcripts was noticed from the beginning of the salt treatment, with minimal expression levels observed on the eighth day. For instance, at this stage, the StSOS1, StSOS2, and StNHX3 transcript amounts were 3- to 4-fold lower in silenced lines than in wild-type (Figure 7A). The increased expression of StSOS3 and StHKT1 genes was noticed in transgenic lines compared to WT after 1 day of treatment, but then the abundance of these transcripts markedly decreased, reaching levels much lower than those in WT after 8 days of exposure (Figure 7). Significant changes in transcript accumulation were also observed in plants with StBBX24-OE3 and StBBX24-OE4 lines compared to control conditions, and maximal transcript abundance was reached on the eighth day after salt treatment (Figure 7B). These results indicate that the decreased sensitivity of silenced lines to salinity is accompanied by a much lower expression of genes encoding major Na+/H+ antiporters involved in salt tolerance.
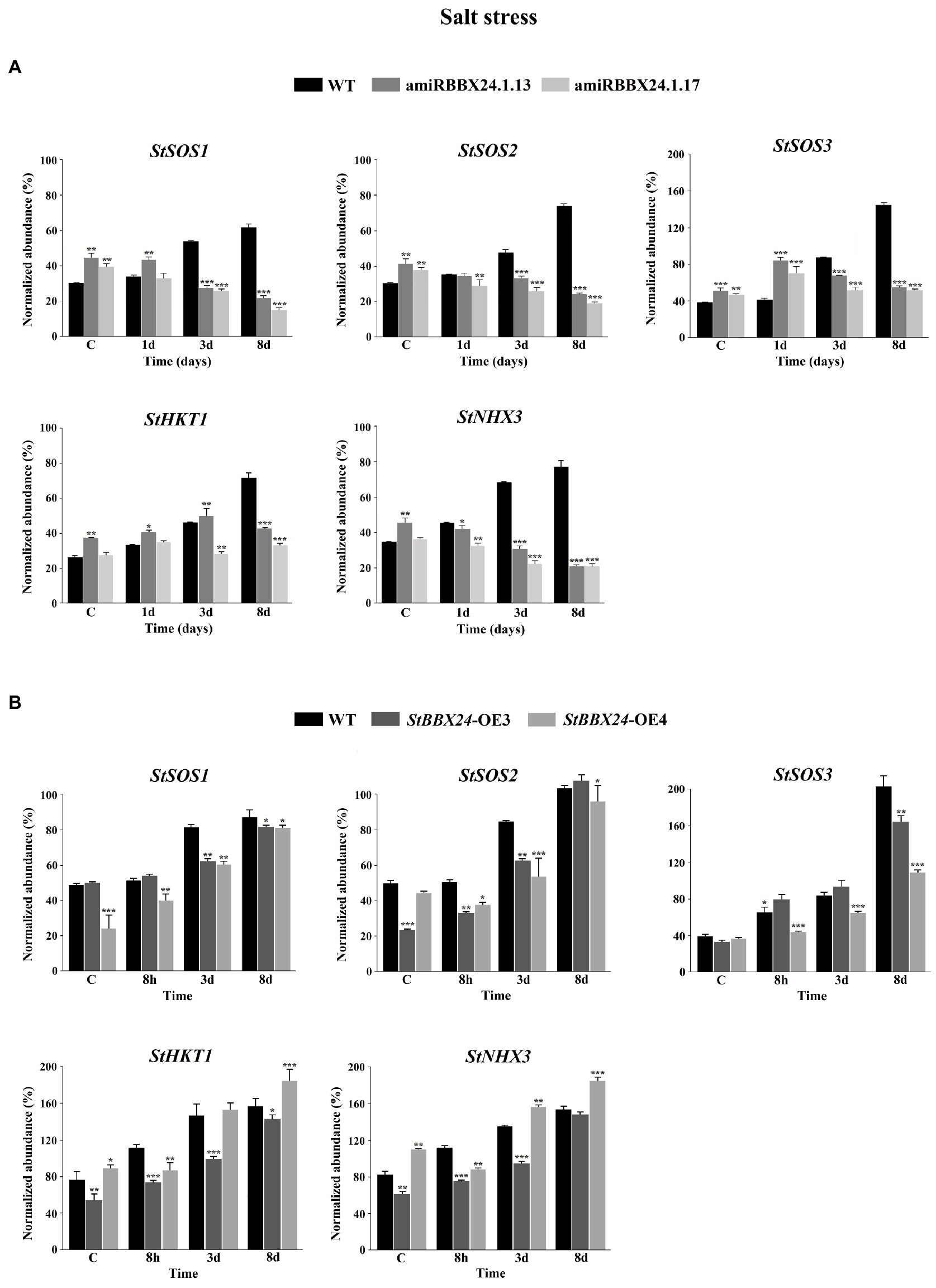
Figure 7. Changes in the expression of StSOS1, StSOS2, StSOS3, StHKT1 and StNHX3 genes in control conditions and plants subjected to high salt for 8 h, 3 and 8 days. (A) Quantitative RT-PCR analysis of StSOS1 (ID: PGSC0003DMG400010630), StSOS2 (ID: PGSC0003DMG400006384), StSOS3 (ID: PGSC0003DMG400019601), StHKT1 (ID: PGSC0003DMT400077052) and StNHX3 (ID: PGSC0003DMG400034953) transcript abundance in leaves of 3-week-old phytotron-grown WT, amiRBBX24.1.13 and amiRBBX24.1.17 plants. (B) Quantitative RT-PCR analysis of StSOS1 (ID: PGSC0003DMG400010630), StSOS2 (ID: PGSC0003DMG400006384), StSOS3 (ID: PGSC0003DMG400019601), StHKT1 (ID: PGSC0003DMT400077052) and StNHX3 (ID: PGSC0003DMG400034953) transcript abundance in leaves of 3-week-old phytotron-grown WT, StBBX24-OE3 and StBBX24-OE4 lines. Quantitative RT-PCR was performed as described in Materials and methods. Means ± SD of four values from two independent experiments (two replicates per experiment) are presented. *, ** and ***, values significantly different from the WT value (t-test) with p < 0.05, p < 0.01 and p < 0.001, respectively. WT, wild-type.
In this work, we show that the clock-controlled StBBX24 protein belonging to B-box proteins in potato is associated with regulating the timing of floral induction. Solanum tuberosum lines silenced for StBBX24 expression displayed much earlier flowering, while overexpression of StBBX24 resulted in a lack of flowering or delayed production of flower buds as compared with the wild type. Furthermore, RT-qPCR analyses of StBBX24-silenced lines revealed that StBBX24 participates in the reproductive development of potato very likely through the regulation of the expression level of some genes essential in this process. We also reveal that silenced lines display decreased tolerance to high salinity and strongly decreased expression of genes encoding Na+ transporters and the activity of antioxidant enzymes. Altogether, these data unveil the role of StBBX24 in the regulation of floral induction in potato and salinity tolerance.
The phenotypic analysis of S. tuberosum BBX24-silenced lines showed that they exhibited early flowering as compared with WT (Figures 2A,B), while plants overexpressing StBBX24 mostly did not produce flower buds (Figure 2C). Moreover, the transgenic plants did not display visible changes with respect to growth rate and leaves, with the exception of slight changes in tuber numbers (Figures 2A,C, Supplementary Figures S3A–C, S5A,B; Suplementary Tables S2, S3). Very interestingly, on the one hand, in Chrysanthemum, transgenic lines suppressed for BBX24 expression also flowered earlier than WT in long-day conditions (Yang et al., 2014), while in Arabidopsis, the loss of BBX24/STO delayed flowering in short-day, but not, long-day conditions (Li et al., 2014). On the other hand, the same authors noted that as compared with WT plants, BBX24-OX Chrysanthemum lines did not exhibit delayed flowering, whereas a delay was seen when Cm-BBX24 was overexpressed in A. thaliana (Li et al., 2014; Yang et al., 2014). These data reveal that BBX24 proteins play a crucial role in controlling flowering initiation in plants through a mechanism related to day length depending on the species. RT-qPCR analyses revealed that the expression of key genes in the flowering regulatory pathway was affected in BBX24-silenced plants (Figures 3A,B). Indeed, in these lines, the expression of genes promoting flowering, i.e., StFKF1, StGI, StSOC1, StGID1, StSP3D, and StSP6A was upregulated as compared with wild-type, while several genes encoding flowering repressors, i.e., StSVP, StCDF1, StCDF2, and StSP exhibited reduced transcript levels in full agreement with the flowering phenotype observed (Figures 3A,B). In Arabidopsis, GI and FKF1 function as circadian clock components involved in the LD-dependent degradation of CDFs that repress CO expression (Johansson and Staiger, 2015). The lack of StBBX24 in silenced lines, which flower earlier, correlates with the decreased expression of StCDF genes, indicating that a CDF-dependent mechanism regulating the expression of CO homolog(s) is likely to occur in potato similar to Arabidopsis. On the other hand, in Arabidopsis the expression of FT and SOC1 genes is repressed by two types of factors, FLC and SVP. In potato BBX24-silenced plants, we observed a significant reduction of StSVP expression. This is associated with upregulation of floral transition genes like StSP3D and StSOC1 (Figures 3A,B) that could induce FLORAL MERISTEM IDENTITY (FMI) genes. Conversely, the fact that BBX24 overexpressing lines do not substantially modify transcript levels of some main activators and repressors of flowering, i.e., StGI/StGID1 and StCDF1/StCDF2 respectively, cannot support this model (Figures 4A,B). Studies aiming at isolating StBBX24-interacting partners will allow better understanding the function of BBX24 in the complex network underlying reproductive development, and provide important insights into the mode of action of B-box proteins.
In Solanaceae, the SELF PRUNING (SP) protein is a main repressor of flowering functioning at the level of the SINGLE-FLOWER TRUSS (SFT) gene, a homolog of the FT gene. In addition, note also that SP belongs to a separate clade of FLOWERING TIME regulators in Solanaceae, termed CEN/TFL-like (Ballerini and Kramer, 2011) and acts antagonistically to the SFT floral inducer. In the S. tuberosum cultivar Desirée, StSP expression was substantially down-regulated in BBX24-silenced lines. In contrast, in lines with StBBX24 overexpression, the cyclicity of StSP expression was significantly affected, suggesting that, in these lines, the expression of the potato SFT homolog, encoding a floral inducer, could be less inhibited by SP, leading to earlier flowering. Ballerini and Kramer (2011) and Abelenda et al. (2014) suggested that StSP3D could be another inducer of SFT. Altogether, these data indicate that the early flowering phenotype of StBBX24-silenced lines can be associated with substantially modified expression of several genes controlling flowering. Although in this study we have focused on flowering, it is worth while mentioning that the slight changes in the tuber numbers in transformed plants as compared with the wild type, correspond to changes in transcript levels of two proteins engaged in tuber formation: StSP6A as an activator and StSP5G as a repressor of this process. Therefore, in the future, it is worth considering the potential role of StBBX24 in tuber forming as one of the factors in this complex process.
In other respects, we observed that the StBBX24 abundance was strongly decreased in leaves and shoot apex of plants exhibiting flower buds (Figures 1A,B). These data conclude that StBBX24 acts during vegetative growth as a negative regulator of genes promoting floral induction and as a positive regulator of those repressing this process, thus contributing to maintain indeterminacy of the shoot apical meristem. This role extends our knowledge about the functions of B-box proteins in plants; only a few having been described at the present time to fulfill critical roles in the control flowering. Indeed, in Arabidopsis, the key coordinator of light input is BBX1/CONSTANS/CO which promotes flowering under long-day conditions via triggering FT expression (Valverde, 2011). In potato, the BBX1 protein has been reported to fulfil a key role in tuber formation, but not in the flowering process (Abelenda et al., 2016). Our data reveal that another BBX protein in this species plays a critical function in the development of reproductive organs. Thus, we can assume that StBBX24 very likely interacts with one or several nuclear factors and modulates the expression of many genes notably involved in floral induction.
Our previous data showed that StBBX24 was localized in the cytosol and nucleus during the light phase (Kiełbowicz-Matuk et al., 2014), which was consistent with the nuclear localization reported for the Arabidopsis BBX24/STO-eGFP and Chrysanthemum Cm-BBX24-GFP fusion proteins (Indorf et al., 2007; Yang et al., 2014). Furthermore, the nuclear localization of the Arabidopsis BBX24 protein has been associated with the presence of a four-residue motif (KKPR) that acts as a functional NLS (Nuclear Localization Signal; Yan et al., 2011). Such a sequence is also present in the StBBX24 protein. Moreover, transient expression studies using Arabidopsis protoplasts have revealed nuclear localization of seven tomato BBX proteins, including SlBBX24 (Chu et al., 2016). Similarly, transgenic Arabidopsis lines expressing BBX24 proteins tagged with GFP under the control of a 35S promoter have also shown a precise nuclear localization of fusion protein (Job et al., 2018). In our work, we subsequently reveal that StBBX24 is mainly present in the chromatin fraction (Figure 1C) in accordance with its essential function in controlling gene expression at the transcript level. Because there is no DNA binding domain in StBBX24, we presume that it interacts with other proteins within the chromatin complex via the B-box domains and VPDLG (VP) motif, as suggested for Arabidopsis BBX24 (Gangappa and Botto, 2014). This interaction could modulate the transcriptional activity of BBX24 partners and confer BBX24 a regulatory function in transcription. In agreement with this hypothesis, several Arabidopsis BBX proteins interact with the HY5 transcription factor (ELONGATED HYPOCOTYL 5) individually or together (Datta et al., 2007, 2008; Gangappa et al., 2013). Moreover, some of them including AtBBX22, AtBBX24, and AtBBX25 interact also with HYH (HOMOLOG OF HY5) transcription factors (Datta et al., 2007, 2008; Gangappa et al., 2013).
We previously reported that BBX24 expression is induced in Solanum plants in response to high salinity and that this induction is gated by the time of day, uncovering the interaction of photoperiod and osmotic stress in the control of BBX24 expression (Kiełbowicz-Matuk et al., 2014). Here, we observed that S. tuberosum lines silenced for StBBX24 expression exhibited severe symptoms of growth impairment under high salinity conditions (Figures 5A,B). Interestingly, the BBX24/STO (SALT TOLERANCE) protein was originally described as a protein that confers salt tolerance when it was ectopically expressed in yeast (Lippuner et al., 1996). Meanwhile, in Arabidopsis, high salinity does not induce STO/BBX24 expression (Lippuner et al., 1996; Nagaoka and Takano, 2003). Nevertheless, overexpression of this gene enhances salt tolerance since it was found to cause increased root growth under high salinity conditions (Nagaoka and Takano, 2003). These results are consistent with our findings in silenced lines, but no evidence of salt susceptibility was observed in Chrysanthemum morifolium transgenic lines down-regulated for CmBBX24 expression (Yang et al., 2014).
In this case, the very harsh treatment (400 mM NaCl for 4 weeks) compared with that here (0.15 M for 8 days) may have masked the modified response to lower salt levels. In summary, these data indicate that BBX24 proteins play an important role in response to high salinity in different plant species. However, the mechanism by which they act remains to be elucidated. Several classes of Na+ transporters playing major roles in the alleviation of ionic stress by sequestration of Na+ in vacuole (NHX), Na+ extrusion from cells (SOS1, 2 and 3) and facilitated Na+ circulation (Class 1 HKT1) have been characterized (Blumwald, 2000; Sunarpi et al., 2005; Yamaguchi et al., 2013; Deinlein et al., 2014). Our results revealed that S. tuberosum lines silenced for StBBX24 displayed strongly decreased expression of all these genes following 8 days of salt application concomitantly to the stress symptoms appearing on leaves (Figure 5A). On the contrary, a substantial increase in the transcript abundance of these transporters was noticed in WT plants in response to salt consistent with their preserved growth (Figure 7). These findings are in agreement with reports showing improved salt tolerance in transgenic Arabidopsis, tomato, apple, poplar, Chrysanthemum and barley lines overexpressing SOS2, SOS1, or NHX (Guo et al., 2004; Huertas et al., 2012; Wang et al., 2012; Adem et al., 2014; An et al., 2014; Yang et al., 2015). Thus, our data revealing that the expression of these transporters is compromised in potato in the absence of StBBX24 indicate that the mechanisms leading to ion homeostasis under salinity are impaired, consequently causing inhibition of many cellular and photosynthetic processes due to Na+ over-accumulation within all cell sub-compartments. They prompt us to propose that StBBX24 is a down-stream component in the signaling pathway triggered by high salinity and regulating the expression of the major genes involved in cell sodium homeostasis.
Our results characterized the StBBX24 protein as an essential component in the network controlling reproductive development and in salt responses. One major aim for future research will be to identify the signals and actors involved in StBBX24 expression. We recently reported that the circadian rhythmicity of its expression was maintained by the StZPR1 nuclear factor that plays a fine tuning role downstream in the clock signaling network (Kiełbowicz-Matuk et al., 2017). The presence of various potential cis-elements associated with light, circadian, hormones or stress responses in the promoter region of StBBX24 suggests that multiple factors participate in the regulation of its expression. In other respects, studies aiming at isolating StBBX24-interacting partners will allow better understanding the function of BBX24 in the complex network underlying reproductive development and stress responses, and provide important insights into the mode of action of B-box proteins.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
AK-M and TR: conceptualization and funding acquisition. KG, MB, UT, JC, and AK-M: performed the experiments. AK-M, UT, and TR: writing–original draft preparation. AK-M, KG, and UT: writing–review and editing. AK-M, KG, UT, and MB: visualization. All authors contributed to the article and approved the submitted version.
This work was supported by the Polish National Science Centre under grants no. 2014/15/B/NZ9/04809 and 2018/29/B/NZ9/01457.
The authors wish to thank prof. Zofia Szweykowska-Kulińska and dr. Marcin Pieczyński of the Adam Mickiewicz University, Faculty of Biology, Institute of Molecular Biology and Biotechnology, for providing us with the pENTR/D-TOPO vector carrying the MIR319b gene and the pART27 binary vector.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.965098/full#supplementary-material
Abelenda, J. A., Navarro, C., and Prat, S. (2014). Flowering and tuberization: a tale of two night shades. Trends Plant Sci. 19, 115–122. doi: 10.1016/j.tplants.2013.09.010
Abelenda, J. A., Cruz-Oró, E., Franco-Zorrilla, J. M., and Prat, S. (2016). Potato St CONSTANS-like1 suppresses storage organ formation by directly activating the FT-like StSP5G repressor. Curr. Biol. 26, 872–881. doi: 10.1016/j.cub.2016.01.066
Adem, G. D., Roy, S. J., Zhou, M., Bowman, J. P., and Shabala, S. (2014). Evaluating contribution of ionic, osmotic and oxidative stress components towards salinity tolerance in barley. BMC Plant Biol. 14, 113. doi: 10.1186/1471-2229-14-113
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
An, J., Song, A., Guan, Z., Jiang, J., Chen, F., Lou, W., et al. (2014). The over-expression of Chrysanthemum crassum CcSOS1 improves the salinity tolerance of chrysanthemum. Mol. Biol. Rep. 41, 4155–4162. doi: 10.1007/s11033-014-3287-2
Ballerini, E. S., and Kramer, E. M. (2011). In the light of evolution: a reevaluation of conservation in the CO-FT regulon and its role in photoperiodic regulation of flowering time. Front. Plant Sci. 2:81. doi: 10.3389/fpls.2011.00081
Beneragama, C. K., and Goto, K. (2010). When does subjective day come under 24-h light/dark cycles? The case of circadian rhythms of UV-C resistance and timing of cell division in Euglena gracilis. Int. J. Botany 6, 28–34. doi: 10.3923/ijb.2010.28.34
Blumwald, E. (2000). Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 12, 431–434. doi: 10.1016/s0955-0674(00)00112-5
Chang, C. S., Li, Y. H., Chen, L. T., Chen, W. C., Hsieh, W. P., Shin, J., et al. (2008). LZF1, a HY5-regulated transcriptional factor, functions in Arabidopsis de-etiolation. Plant J. 54, 205–219. doi: 10.1111/j.1365-313X.2008.03401.x
Chen, H., Nelson, R. S., and Sherwood, J. L. (1994). Enhanced recovery of transformants of agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Bio Techniques 16, 664–668.
Cheng, X. F., and Wang, Z. Y. (2005). Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 43, 758–768. doi: 10.1111/j.1365-313X.2005.02491.x
Chu, Z., Wang, X., Li, Y., Yu, H., Li, J., Lu, Y., et al. (2016). Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front. Plant Sci. 7:1552. doi: 10.3389/fpls.2016.01552
Crocco, C. D., Holm, M., Yanovsky, M. J., and Botto, J. F. (2010). AtBBX21 and COP1 genetically interact in the regulation of shade avoidance. Plant J. 64, 551–562. doi: 10.1111/j.1365-313X.2010.04360.x
Datta, S., Hettiarachchi, G. H., Deng, X. W., and Holm, M. (2006). Arabidopsis CONSTANS LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18, 70–84. doi: 10.1105/tpc.105.038182
Datta, S., Hettiarachchi, C., Johansson, H., and Holm, M. (2007). SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19, 3242–3255. doi: 10.1105/tpc.107.054791
Datta, S., Johansson, H., Hettiarachchi, C., Irigoyen, M. L., Desai, M., Rubio, V., et al. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20, 2324–2338. doi: 10.1105/tpc.108.061747
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Fan, X. Y., Sun, Y., Cao, D. M., Bai, M. Y., Luo, X. M., Yang, H. J., et al. (2012). BZS1, a B-box protein, promotes photomorphogenesis downstream of both brassinosteroid and light signaling pathways. Mol. Plant 5, 591–600. doi: 10.1093/mp/sss041
Fornara, F., de Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550, 550.e1–550, 550.e2. doi: 10.1016/j.cell.2010.04.024
Gangappa, S. N., and Botto, J. F. (2014). The BBX family of plant transcription factors. Trends Plant Sci. 19, 460–470. doi: 10.1016/j.tplants.2014.01.010
Gangappa, S. N., Crocco, C. D., Johansson, H., Datta, S., Hettiarachchi, C., Holm, M., et al. (2013). The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25, 1243–1257. doi: 10.1105/tpc.113.109751
Gleave, A. P. (1992). A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. doi: 10.1007/BF00028910
González-Schain, N. D., Díaz-Mendoza, M., Zurczak, M., and Suárez-López, P. (2012). Potato CONSTANS is involved in photoperiodic tuberization in a graft-transmissible manner. Plant J. 70, 678–690. doi: 10.1111/j.1365-313X.2012.04909.x
Grądzka, K., and Kiełbowicz-Matuk, A. (2020). B-box proteins in plants – structural and functional heterogeneity. Postępy Biol. Komorki 47, 225–246.
Guo, Y., Qiu, Q. S., Quintero, F. J., Pardo, J. M., Ohta, M., Zhang, C., et al. (2004). Transgenic evaluation of activated mutant alleles of SOS2 reveals a critical requirement of its kinase activity and C-terminal regulatory domain for salt tolerance in Arabidopsis. Plant Cell 16, 435–449. doi: 10.1105/tpc.019174
Hedman, H., Kallman, T., and Lagercrantz, U. (2009). Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 70, 359–369. doi: 10.1007/s11103-009-9478-x
Holtan, H. E., Bandong, S., Marion, C. M., Adam, L., Tiwari, S., Shen, Y., et al. (2011). BBX32, an Arabidopsis B-box protein, functions in light signaling by suppressing HY5-regulated gene expression and interacting with STH2/BBX21. Plant Physiol. 156, 2109–2123. doi: 10.1104/pp.111.177139
Huertas, R., Olías, R., Eljakaoui, Z., Gálvez, F. J., Li, J., De Morales, P. A., et al. (2012). Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato. Plant Cell Environ. 35, 1467–1482. doi: 10.1111/j.1365-3040.2012.02504.x
Indorf, M., Cordero, J., Neuhaus, G., and Rodríguez-Franco, M. (2007). Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J. 51, 563–574. doi: 10.1111/j.1365-313X.2007.03162.x
Ismail, Z. E. (1988). Some of the physio-mechanical properties for potato planters. J. Agric. Sci. 13, 2259–2270.
Ito, S., Song, Y. H., Josephson-Day, A. R., Miller, R. J., Breton, G., Olmstead, R. G., et al. (2012). FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 109, 3582–3587. doi: 10.1073/pnas.1118876109
Job, N., Yadukrishnan, P., Bursch, K., Datta, S., and Johansson, H. (2018). Two B-box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol. 176, 2963–2976. doi: 10.1104/pp.17.00856
Johansson, M., and Staiger, D. (2015). Time to flower: interplay between photoperiod and the circadian clock. J. Exp. Bot. 66, 719–730. doi: 10.1093/jxb/eru441
Johnson, C. H., Elliott, J. A., and Foster, R. (2003). Entrainment of circadian programs. Chronobiol. Int. 20, 741–774. doi: 10.1081/cbi-120024211
Khanna, R., Kronmiller, B., Maszle, D. R., Coupland, G., Holm, M., Mizuno, T., et al. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21, 3416–3420. doi: 10.1105/tpc.109.069088
Kiełbowicz-Matuk, A., Rey, P., and Rorat, T. (2007). The abundance of a single domain cyclophilin in Solanaceae is regulated as a function of organ type and high temperature and not by other environmental constraints. Physiol. Plant. 131, 387–398. doi: 10.1111/j.1399-3054.2007.00968.x
Kiełbowicz-Matuk, A., Rey, P., and Rorat, T. (2014). Interplay between circadian rhythm, time of the day and osmotic stress constraints in the regulation of the expression of a Solanum double B-box gene. Ann. Bot. 113, 831–842. doi: 10.1093/aob/mct303
Kiełbowicz-Matuk, A., Czarnecka, J., Banachowicz, E., Rey, P., and Rorat, T. (2017). Solanum tuberosum ZPR1 encodes a light-regulated nuclear DNA-binding protein adjusting the circadian expression of StBBX24 to light cycle. Plant Cell Environ. 40, 424–440. doi: 10.1111/pce.12875
Li, F., Sun, J., Wang, D., Bai, S., Clarke, A. K., and Holm, M. (2014). The B-box family gene STO (BBX24) in Arabidopsis thaliana regulates flowering time in different pathways. PLoS One 9:e87544. doi: 10.1371/journal.pone.0087544
Lichtenthaler, H. (1987). Chlorophylls and carotenoids: pigments of photosynthetic membranes. Meth. Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Lippuner, V., Cyert, M. S., and Gasser, C. S. (1996). Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 271, 12859–12866. doi: 10.1074/jbc.271.22.12859
Mac, A., Krzymowska, M., Barabasz, A., and Hennig, J. (2004). Transcriptional regulation of the glu B promoter during plant response to infection. Cell. Mol. Biol. Lett. 9, 843–853.
Molinero-Rosales, N., Latorre, A., Jamilena, M., and Lozano, R. (2004). SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 218, 427–434. doi: 10.1007/s00425-003-1109-1
Nagaoka, S., and Takano, T. (2003). Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 54, 2231–2237. doi: 10.1093/jxb/erg241
Nakagawa, T., Suzuki, T., Murata, S., Nakamura, S., Hino, T., Maeo, K., et al. (2007). Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71, 2095–2100. doi: 10.1271/bbb.70216
Nakamura, S., Abe, F., and Kawahigashi, H. (2011). A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23, 3215–3229. doi: 10.1105/tpc.111.088492
Nicot, N., Hausman, J. F., Hoffmann, L., and Evers, D. (2005). Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914. doi: 10.1093/jxb/eri285
Ohto, M., Onai, K., Furukawa, Y., Aoki, E., Araki, T., and Nakamura, K. (2001). Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 127, 252–261. doi: 10.1104/pp.127.1.252
Ossowski, S., Schwab, R., and Weigel, D. (2008). Gene silencing in plants Rusing artificial microRNAs and other small RNAs. Plant J. 53, 674–690. doi: 10.1111/j.1365-313X.2007.03328.x
Pajoro, A., Biewers, S., Dougali, E., Leal Valentim, F., Mendes, M. A., Porri, A., et al. (2014). The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: a two-decade history. J. Exp. Bot. 65, 4731–4745. doi: 10.1093/jxb/eru233
Parcy, F. (2005). Flowering: a time for integration. Int. J. Dev. Biol. 49, 585–593. doi: 10.1387/ijdb.041930fp
Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133. doi: 10.1105/tpc.105.039834
Seo, P. J., Ryu, J., Kang, S. K., and Park, C. M. (2011). Modulation of sugar metabolism by an Arabidopsis. Plant J. 65, 418–429. doi: 10.1111/j.1365-313X.2010.04432.x
Song, Y. H., Ito, S., and Imaizumi, T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/j.tplants.2013.05.003
Song, Y. H., Estrada, D. A., Johnson, R. S., Kim, S. K., Lee, S. Y., Mac Coss, M. J., et al. (2014). Distinct roles of FKF1, GIGANTEA, and ZEITLUPE proteins in the regulation of CONSTANS stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. U. S. A. 111, 17672–17677. doi: 10.1073/pnas.1415375111
Srikanth, A., and Schmid, M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68, 2013–2037. doi: 10.1007/s00018-011-0673-y
Sunarpi, H. T., Motoda, J., and Kubo, M. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter- induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44, 928–938. doi: 10.1111/j.1365-313X.2005.02595.x
Talar, U., and Kiełbowicz-Matuk, A. (2021). Beyond Arabidopsis: BBX regulators in crop plants. Int. J. Mol. Sci. 22, 2906. doi: 10.3390/ijms22062906
Talar, U., Kiełbowicz-Matuk, A., Czarnecka, J., and Rorat, T. (2017). Genome-wide survey of B-box proteins in potato (Solanum tuberosum) – identification, characterization and expression patterns during diurnal cycle, etiolation and de-etiolation. PLoS One 12:e0177471. doi: 10.1371/journal.pone.0177471
Torti, S., and Fornara, F. (2012). AGL24 acts in concert with SOC1 and FUL during Arabidopsis floral transition. Plant Signal. Behav. 7, 1251–1254. doi: 10.4161/psb.21552
Valverde, F. (2011). CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 62, 2453–2463. doi: 10.1093/jxb/erq449
Wang, Q., Tu, X., Zhang, J., Chen, X., and Rao, L. (2013). Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol. Biol. Rep. 40, 2679–2688. doi: 10.1007/s11033-012-2354-9
Wang, R. K., Li, L. L., Cao, Z. H., Zhao, Q., Li, M., Zhang, L. Y., et al. (2012). Molecular cloning and functional characterization of a novel apple MdCIPK6L gene reveals its involvement in multiple abiotic stress tolerance in transgenic plants. Plant Mol. Biol. 79, 123–135. doi: 10.1007/s11103-012-9899-9
Wenkel, S., Turck, F., Singer, K., Gissot, L., Le Gourrierec, J., Samach, A., et al. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. doi: 10.1105/tpc.106.043299
Wesley, S. V., Helliwell, C. A., Smith, N. A., Wang, M. B., Rouse, D. T., Liu, Q., et al. (2001). Construct design for efficient, effective and high-through put gene silencing in plants. Plant J. 27, 581–590. doi: 10.1046/j.1365-313x.2001.01105.x
Yamaguchi, T., Hamamoto, S., and Uozumi, N. (2013). Sodium transport system in plant cells. Front. Plant Sci. 4:410. doi: 10.3389/fpls.2013.00410
Yan, H., Marquardt, K., Indorf, M., Jutt, D., Kircher, S., Neuhaus, G., et al. (2011). Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol. 156, 1772–1782. doi: 10.1104/pp.111.180208
Yang, Y., Ma, C., Xu, Y., Wei, Q., Imtiaz, M., Lan, H., et al. (2014). A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis. Plant Cell 26, 2038–2054. doi: 10.1105/tpc.114.124867
Keywords: antioxidant enzymes, double B-box protein, flowering time, flowering-related genes, salinity, sodium transporters, Solanum tuberosum
Citation: Kiełbowicz-Matuk A, Grądzka K, Biegańska M, Talar U, Czarnecka J and Rorat T (2022) The StBBX24 protein affects the floral induction and mediates salt tolerance in Solanum tuberosum. Front. Plant Sci. 13:965098. doi: 10.3389/fpls.2022.965098
Received: 09 June 2022; Accepted: 10 August 2022;
Published: 07 September 2022.
Edited by:
Bin Xu, Nanjing Agricultural University, ChinaReviewed by:
Lu Liu, Shanghai Jiao Tong University, ChinaCopyright © 2022 Kiełbowicz-Matuk, Grądzka, Biegańska, Talar, Czarnecka and Rorat. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnieszka Kiełbowicz-Matuk, YWtpZUBpZ3IucG96bmFuLnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.