- 1Key Laboratory of Digital Dry Land Crops of Zhejiang Province, Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2National Barley Improvement Centre, Hangzhou, China
- 3Institute of Crop Science, Zhejiang University, Hangzhou, China
Calmodulin (CaM) and calmodulin-like (CML) proteins are Ca2+ relays and play diverse and multiple roles in plant growth, development and stress responses. However, CaM/CML gene family has not been identified in barley (Hordeum vulgare). In the present study, 5 HvCaMs and 80 HvCMLs were identified through a genome-wide analysis. All HvCaM proteins possessed 4 EF-hand motifs, whereas HvCMLs contained 1 to 4 EF-hand motifs. HvCaM2, HvCaM3 and HvCaM5 coded the same polypeptide although they differed in nucleotide sequence, which was identical to the polypeptides coded by OsCaM1-1, OsCaM1-2 and OsCaM1-3. HvCaMs/CMLs were unevenly distributed over barley 7 chromosomes, and could be phylogenetically classified into 8 groups. HvCaMs/CMLs differed in gene structure, cis-acting elements and tissue expression patterns. Segmental and tandem duplication were observed among HvCaMs/CMLs during evolution. HvCML16, HvCML18, HvCML50 and HvCML78 were dispensable genes and the others were core genes in barley pan-genome. In addition, 14 HvCaM/CML genes were selected to examine their responses to salt, osmotic and low potassium stresses by qRT-PCR, and their expression were stress-and time-dependent. These results facilitate our understanding and further functional identification of HvCaMs/CMLs.
Introduction
As sessile organisms, plants are constantly exposed to fluctuating environments and are prone to suffer from abiotic stresses. During evolution, plants have developed sophisticated mechanisms to perceive and respond to environmental cues, in order to maintain normal growth and development. Calcium is an essential element for plants not only as a structural component for cell wall and membrane stability, but also as an important second messenger in signaling networks (Thor, 2019). Upon exposure to environmental stimuli, Ca2+ enters into cytosol through Ca2+-permeable cation channels from apoplast or intracellular compartments (such as the endoplasmic reticulum and vacuoles), which leads to an increase in cytosolic calcium concentration ([Ca2+]cyt). Then the cytosolic Ca2+ are transported out of cells or compartmented into calcium pools by Ca2+-ATPases and H+/Ca2+ antiporters, and [Ca2+]cyt is recovered to resting levels ranging from 50 to 200 nM (Lee and Seo, 2021). Ca2+ signatures, defined as spatial and temporal characteristics of transient Ca2+ influxes varying in amplitude, frequency, and intracellular distribution, encode unique information of external stimuli (Zhu et al., 2015; Leitão et al., 2019). Ca2+-binding proteins (Ca2+ sensors) decode, relay and amplify the information of Ca2+ signatures, and then regulate downstream signal transduction and initiate specific cellular responses (Zhu et al., 2015; Thor, 2019). Ca2+ sensors include calmodulins (CaMs) and CaM-like proteins (CMLs), Ca2+-dependent protein kinases (CDPKs or CPKs) and Ca2+- and Ca2+/CaM-dependent protein kinase (CCaMK), and calcineurin B-like proteins (CBLs; Galon et al., 2010). These proteins contain one or more EF-hand motifs and can bind Ca2+ reversibly to provoke conformational changes for binding and regulating downstream targets, and then propagate Ca2+ signals (McCormack et al., 2005; Zhu et al., 2015). CDPKs and CCaMK are sensor responders (with Ca2+-binding and kinase activities), while CaMs, CMLs and CBLs are sensor relay proteins (with only Ca2+-binding ability, except AtCaM7; Reddy et al., 2011; Vadassery et al., 2012). CBLs specifically interact with CBL-interacting protein kinases (CIPKs), whereas CaMs and CMLs interact with diverse proteins (Reddy et al., 2011). CaM is highly conserved during evolution and exists in all eukaryotes, whereas CMLs are restricted to plants and lower protists (Cheval et al., 2013). Generally, CaM proteins consist of 149 amino acid residues and contain 4 typical Ca2+-binding EF-hand motifs, while CML proteins considerably vary in length and possess 1 to 6 EF-hand motifs (Aleynova et al., 2020). Both CaMs and CMLs lack any other functional domains or intrinsic activities (Vadassery et al., 2012; Aleynova et al., 2020).
CaM and CML genes play diverse and multiple roles in plant growth, development and stress responses. Totally, 7 CaMs and 50 CMLs have been identified in Arabidopsis thaliana (McCormack and Braam, 2003; McCormack et al., 2005), and their functions have been analyzed in detail. CaM1 positively regulates ROS production, leaf senescence and ABA response (Dai et al., 2018). Together with CaM4, CaM1 confers salt resistance by promoting NO accumulation (Zhou et al., 2016). CaM4 negatively regulates freezing tolerance by interacting with CaM-binding protein PATL1 (Chu et al., 2018). NO functions as a signal and acts upstream of CaM3 in thermotolerance, and CaM3 inhibits NO accumulation and improves thermotolerance (Xuan et al., 2010; Zhang et al., 2020). CaM7 is a component of photomorphogenesis and multiple signaling pathways are involved in plant immunity, biotic and abiotic stress responses, and hormonal responses (Basu et al., 2021). On the other hand, CML24 plays important roles in pollen germination and pollen tube extension (Yang et al., 2014), circadian oscillation (Martí Ruiz et al., 2018), root mechanoresponses (Wang et al., 2011), Al resistance (Zhu et al., 2022), as well as seed germination, floral transition, ion stress and the sensing of photoperiod (Delk et al., 2005). CML42 participated in trichome morphology (Dobney et al., 2009), insect herbivory defense and drought stress responses (Vadassery et al., 2012; Scholz et al., 2015). CML37, CML38 and CML39 are involved in responses to H2O2 or MeJA treatments as well as drought, salinity, wounding and bacterial infection (Vanderbeld and Snedden, 2007; Scholz et al., 2015). Interestingly, CML37 and CML42 are proved to act antagonistically in the regulation of induced defense responses, and the double knock-out line, cml37 × cml42, shows wild-type phenotypes upon stresses (Heyer et al., 2021). CML8 and CML9 are implicated in both abiotic and biotic stress responses (Magnan et al., 2008; Zeng et al., 2015; Zhu et al., 2017, 2021). CML20 is a negative regulator in guard cell ABA signaling and drought tolerance (Wu et al., 2017). In rice, 5 CaMs and 32 CMLs have been identified (Boonburapong and Buaboocha, 2007). OsCaM1-1 mediates the expression of downstream target genes and contributed to rice thermotolerance (Wu et al., 2012). OsCML4 and OsDSR-1 improves drought tolerance of rice through ROS scavenging (Yin et al., 2015, 2017). OsCML16 plays important roles in OsERF48 regulation of root growth and drought tolerance (Jung et al., 2017). OsMSR2, a novel rice calmodulin-like gene, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis (Xu et al., 2011). In wheat, overexpression of TaCML20 enhances water soluble carbohydrate accumulation and yield (Kalaipandian et al., 2019). TaCML36 positively participates in an immune response to Rhizoctonia cerealis by modulating the expression of defense-associated genes (Lu et al., 2019). In tomato, SlCML37 interacts with proteasome maturation factor SlUMP1 and plays a role in fruit chilling stress tolerance (Tang et al., 2021). SlCML39 acts negatively in high temperature tolerance possibly through ABA signaling pathway (Ding et al., 2021). Overexpression of ShCML44 enhances tolerance to cold, drought and salinity stress by higher antioxidant enzymes activity, less membrane damage, better gas exchange and plant water relations in Solanum habrochaites (Munir et al., 2016). MtCML40 negatively regulates salt tolerance through targeting MtHKT-dependent Na+ accumulation in Medicago truncatula (Zhang et al., 2019). MtCML42 positively regulates cold tolerance through regulating MtCBF1 and MtCBF4 expression, and regulates flowering time through sequentially downregulation of MtABI5 and upregulation of MtFTa1 (Sun et al., 2021). Alfalfa (Medicago sativa) MsCML10 regulates cold tolerance through activating MsGSTU8 and MsFBA6, leading to improved maintenance of ROS homeostasis and increased accumulation of sugars for osmoregulation, respectively (Yu et al., 2022). MsCML46 enhances tolerance to abiotic stresses through alleviating osmotic stress and oxidative damage in transgenic tobacco (Du et al., 2021).
Although less well-studied than in Arabidopsis thaliana and rice, the CaM and CML gene family has been characterized in some other plant species, such as Brassica napus (He et al., 2020), wheat (Liu et al., 2022), papaya (Ding et al., 2018), wild tomato (Shi and Du, 2020) and grapevine (Vandelle et al., 2018). However, similar research has not been conducted in barley. In the present study, 5 HvCaM and 80 HvCML genes were identified in barley genome, and their phylogenetic relationships, gene structures, cis-acting elements, syntenic relationships, tissue expression patterns and responses to abiotic stresses were analyzed. These results facilitate our understanding and further functional identification of HvCaMs/CMLs.
Materials and methods
Identification of CaM/CML genes in barley
The protein sequences of CaM/CML genes in Arabidopsis thaliana (McCormack and Braam, 2003; McCormack et al., 2005) and rice (Boonburapong and Buaboocha, 2007) were retrieved from TAIR1 and RAP-DB,2 respectively. Then these protein sequences were used as queries (Blastp, E-value <1e-5) to search against barley genome database Morex v2 (Monat et al., 2019a). Meanwhile, ‘IPR002048’ was used as criterion to search against barley genome database in EnsemblPlants.3 The putative HvCaM/CML genes were further verified using IntertPro database4 (Blum et al., 2021), and those containing domains other than ‘EF-hand domain’ were discarded.
Physicochemical properties and subcellular localizations
The theoretical molecular weights (MW) and isoelectric points (pI) of HvCaMs/CMLs were calculated using ExPASy.5 The subcellular localizations of HvCaMs/CMLs were predicted using BUSCA6 (Savojardo et al., 2018).
Phylogenetic and syntenic relationships
The CaM/CML protein sequences of Arabidopsis (7 AtCaMs, 50 AtCMLs; McCormack and Braam, 2003; McCormack et al., 2005) and rice (5 OsCaMs, 32 OsCMLs; Boonburapong and Buaboocha, 2007) were downloaded from TAIR and RAP-DB, respectively. CaM1 from Amborella trichopoda was set as outgroup. CaM/CML protein sequences of Arabidopsis, rice and barley were aligned with MAFFT7 (Madeira et al., 2019), and the phylogenetic tree was constructed using MEGA X with maximum-likelihood (ML) method and 1,000 bootstrap replicates (Kumar et al., 2018). The genome sequences and annotations of Brassica napus was downloaded from BnPIR,8 the genomic data of Arabidopsis, rice and wheat were downloaded from EnsemblPlants,9 and their syntenic relations with barley were analyzed and visualized using TBtools (Chen et al., 2020).
Sequence, chromosomal location and duplication analyses
Conserved motifs of HvCaMs/CMLs were analyzed using MEME10 (Bailey et al., 2009) with following parameters: classic motif discovery mode, any number of repetitions (anr), motif number was set to 10. Number of EF-hand and calcium binding bites were analyzed using InterPro database (see Footnote 4; Blum et al., 2021). Motifs, gene structures and gene duplication events were visualized using TBtools (Chen et al., 2020).
Cis-acting elements
The 2000 bp upstream of coding sequences of HvCaMs/CMLs were extracted for cis-acting element analysis using PlantCARE database11 (Lescot et al., 2002).
Tissue expression patterns
Transcriptomic data (FPKM) were downloaded from BARLEX12 and normalized with log10(FPKM+1) transform. The expression heatmap of different tissues was drawn using TBtools (Chen et al., 2020).
HvCaMs/CMLs in pan-genome context
Assemblies of 20 barley genotypes were downloaded (Jayakodi et al., 2020), and nucleotide sequences of 85 HvCaM/CML genes were used as queries to search against each assembly. The Blastn results were verified for presence/absence variation and chromosomal location.
Plant materials, growth conditions and abiotic stress treatments
Barley (Hordeum vulgare cv. Golden Promise) seeds were sterilized with 10% commercial NaClO, rinsed with tap water and then germinated in BSM solution (0.5 mM KCl + 0.1 mM CaCl2) for 2 days. Afterwards BSM was changed to 1/5 Hoagland solution for another 5 days with a photoperiod of 14/10 h, light intensity of 200 ± 25 μmol·m−2·s−1, temperature of 23/18°C (day/night) and relative humidity of 60% (Cai et al., 2019). After growth for 7 days, seedlings were subjected to salt stress (200 mM NaCl), osmotic stress (20% PEG8000) and potassium deficiency (0.01 mM K+; Cai et al., 2021). Plants grown in 1/5 Hoagland solution were set as control. Solutions were renewed every 2 days. RNA extraction and qRT-PCR were performed on barley roots under both control and abiotic stress conditions after treatment for 1 h, 3 h, 6 h, 1 day, 3 days, and 6 days. All samples were collected in three replicates.
qRT-PCR
Total RNA was extracted using Easy Plant RNA Extraction Kit (DR0406050, Easy-Do, China). The cDNA was synthesized from total RNA using PrimeScript RT Master Mix (RR036A, TaKaRa, Japan) and was then used for qRT-PCR amplification. qRT-PCR was performed with LightCycler 480 II (Roche, Basel, Switzerland) using ChamQ Universal SYBR qPCR Master Mix (Q711, Vazyme, China). The relative gene expression was calculated with 2−△△CT method (Livak and Schmittgen, 2001) using actin as the internal standard. The primer sequences were listed in Supplementary File 1.
Results
Identification of CaM/CML genes in barley
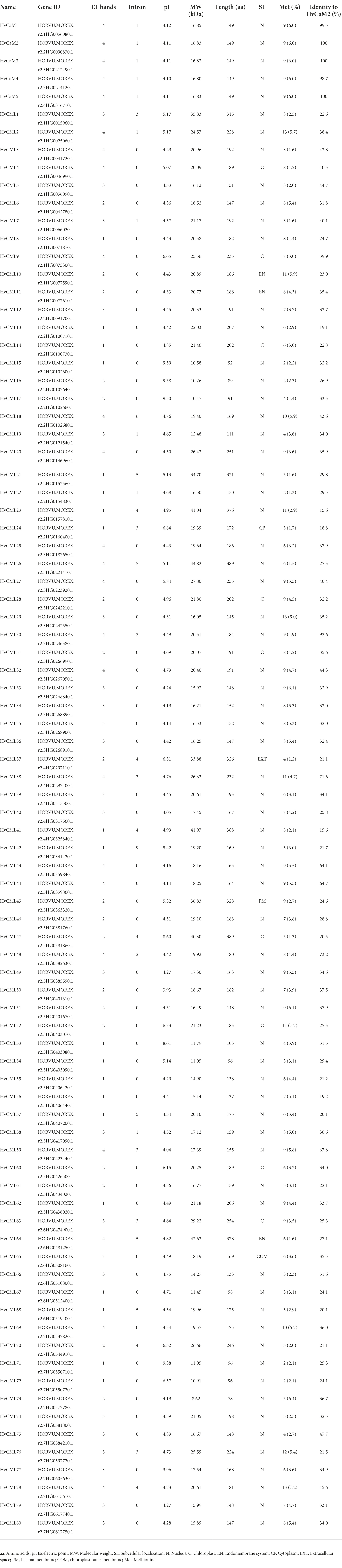
A total of 5 HvCaM and 80 HvCML genes were identified in barley and were named in the order of their chromosomal locations (viz. HvCaM1 to HvCaM5 and HvCML1 to HvCML80), respectively (Table 1). All HvCaMs shared the same protein length (149 aa), methionine percentage (6.0%), number of introns (1) and EF-hands with calcium-binding ability (4), as well as subcellular localization (nucleus). Their isoelectric points (pI) and theoretical molecular weights (MW) ranged from 4.10 to 4.12 and 16.80 to 16.85 kDa, respectively (Table 1). Notably, HvCaM2/3/5 differed in nucleotide sequence but coded the same peptide (Table 1; Supplementary File 2). The protein sequence identity of HvCaM1 and HvCaM4 to HvCaM2 were also as high as 99.3 and 98.7%, respectively (Table 1). Compared with HvCaMs, characteristics of HvCMLs were more diverged. The protein length, pI, MW and methionine percentage of HvCMLs ranged from 78 to 389 aa, 3.93 to 9.59, 8.62 to 44.82 kDa and 1.23 to 8.97%, respectively (Table 1). Twenty-three HvCMLs contained 3 EF-hand domains, followed by 20, 19 and 18 HvCMLs containing 1, 4 and 2 domains, respectively. All HvCMLs retained at least one EF-hand domain with calcium-binding ability except HvCML57 and HvCML68, whose sole EF-hand has lost the ability to bind calcium (Table 1; Supplementary File 3). Most of HvCMLs (53, 66.3%) did not contain introns, and the others had 1 to 9 introns. Likewise, most of the HvCMLs (64, 80.0%) were localized in nucleus, followed by 9 and 3 HvCMLs were in chloroplast and endomembrane system, respectively. HvCML24, HvCML37, HvCML45 and HvCML65 were localized in cytoplasm, extracellular space, plasma membrane and chloroplast outer membrane, respectively (Table 1). Sequence identity of all HvCMLs to HvCaM2 were lower than 50% except 6 HvCMLs (HvCML30, HvCML48, HvCML38, HvCML59, HvCML44 and HvCML43).
Phylogenetic and gene structure analysis of HvCaMs/CMLs
The protein sequence of 7 AtCaMs and 50 AtCMLs in Arabidopsis, 5 OsCaMs and 32 OsCMLs in rice, 5 HvCaMs and 80 HvCMLs in barley, together with CaM1 from Amborella trichopoda (outgroup), were used to construct phylogenetic tree (Figure 1). CaMs/CMLs were grouped into 9 clusters according to their phylogenetic relationships. Cluster I comprised 17 CaM members. Cluster VIII contained 28 members and 10 of them were HvCMLs. Cluster II and V each contained 27 CML genes (with 10 and 24 HvCMLs, respectively). Cluster IX and VII contained 26 and 24 CMLs, respectively (with 14 and 6 HvCMLs, respectively). Cluster III, VI and IV consisted of 13, 10 and 7 CML genes, respectively (HvCMLs in each cluster being 4, 7, and 5).
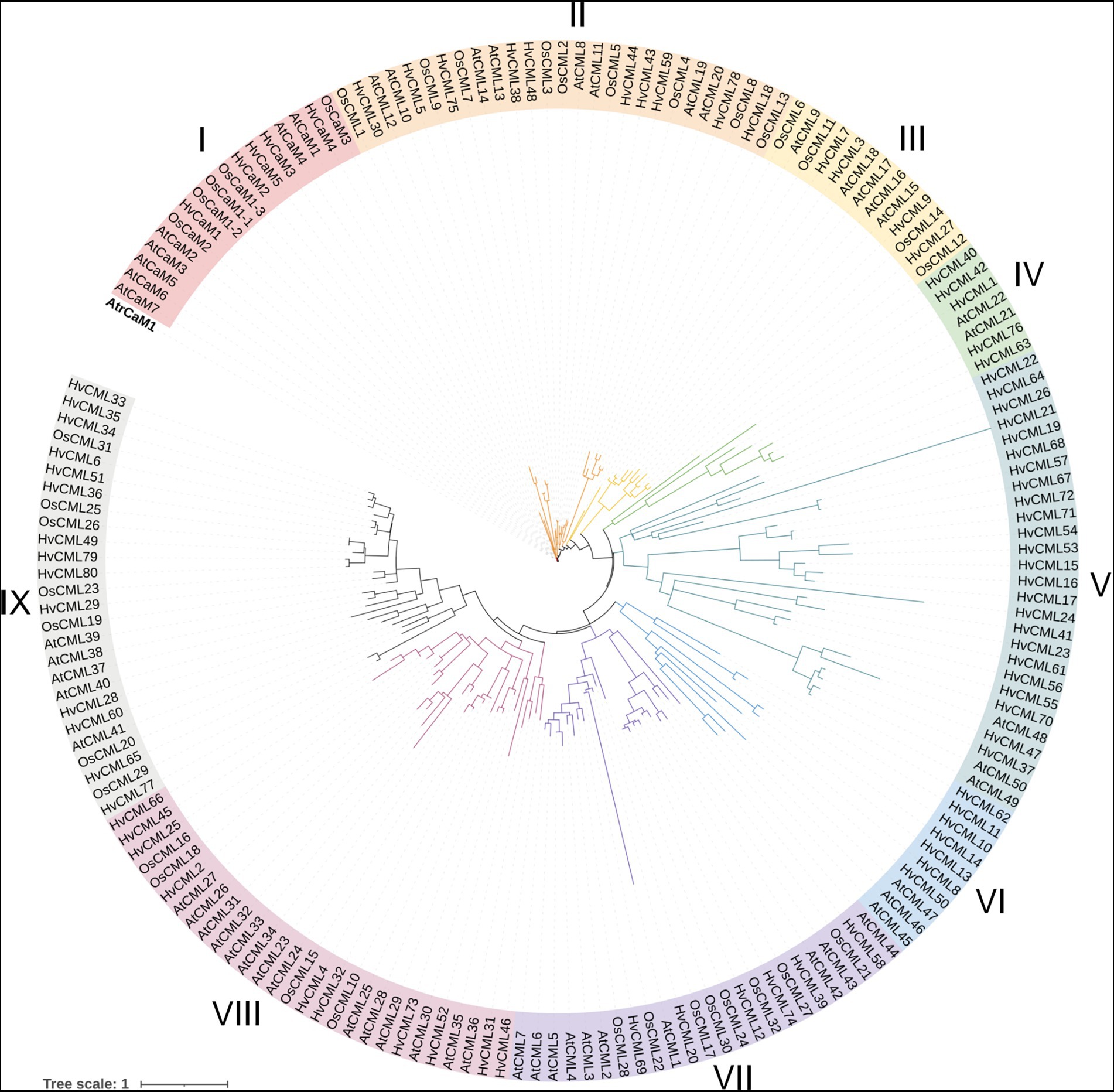
Figure 1. Phylogenetic tree of CaM and CML family proteins from barley, Arabidopsis and rice. CaM1 from Amborella trichopoda was used as outgroup. Atr, Amborella trichopoda; Hv, Hordeum vulgare; Os, Oryza sativa; At, Arabidopsis thaliana.
Phylogeny, conserved motifs and gene structures of HvCaMs/CMLs were further comprehensively analyzed (Figure 2). HvCaMs/CMLs were classified into 9 groups (Figure 2A). Group I consisted of 5 HvCaMs, all of which had one phase 0 intron and the same conserved motif arrangements (Figure 2). Group IX incorporated the most HvCML genes (14), followed by group VII and VIII with 12 HvCML members in each. Group II and VI each contained 10 HvCMLs, and group III, IV and V contained 7, 7 and 8 genes, respectively. HvCMLs with close phylogenetic relatives showed similar motif arrangements, though considerable variation was observed between genes from group V and VIII (Figure 2B). More than half genes from group VIII, II and III were intron-rich (91.7, 60.0 and 57.1%, respectively), followed by group IX, VI and V, in which the percentage was 21.4, 20.0 and 12.5%, respectively (Figure 2C). However, all HvCMLs in group IV and VII were intron-free (Figures 2A,C). Overall, HvCaMs were more conserved than HvCMLs in phylogeny and gene structure (Figures 1, 2; Table 1).
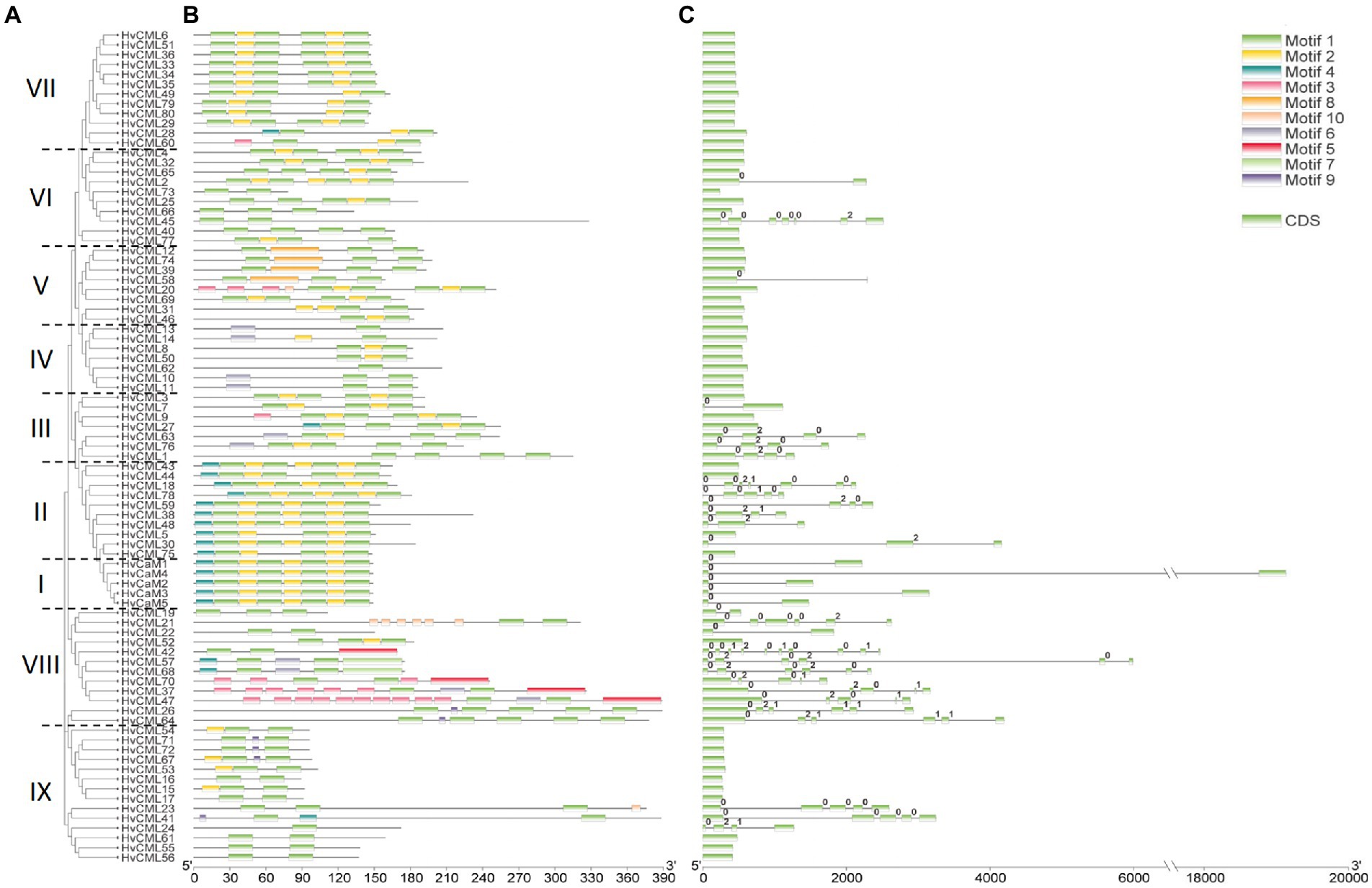
Figure 2. Phylogenetic relationship and sequence characteristics of HvCaMs/CMLs. (A) Phylogenetic tree of HvCaM/CML proteins; (B) Conserved motifs of HvCaM/CML proteins; (C) Gene structure of HvCaMs/CMLs.
Chromosomal distribution and duplication analysis of HvCaMs/CMLs
HvCaMs/CMLs were unevenly distributed over 7 chromosomes, with chromosome 5 containing 20 HvCML genes (HvCML43 to HvCML62), chromosome 2 (HvCaM2, HvCML12 to HvCML24) and chromosome 3 (HvCaM3 to HvCaM4, HvCML25 to HvCML36) each containing 14 genes, chromosome 1 (HvCaM1, HvCML1 to HvCML11) and chromosome 7 (HvCML69 to HvCML80) each containing 12 genes, and chromosome 4 (HvCaM5, HvCML37 to HvCML42) and chromosome 6 (HvCML63 to HvCML68) containing 7 and 6 genes, respectively (Figure 3A).
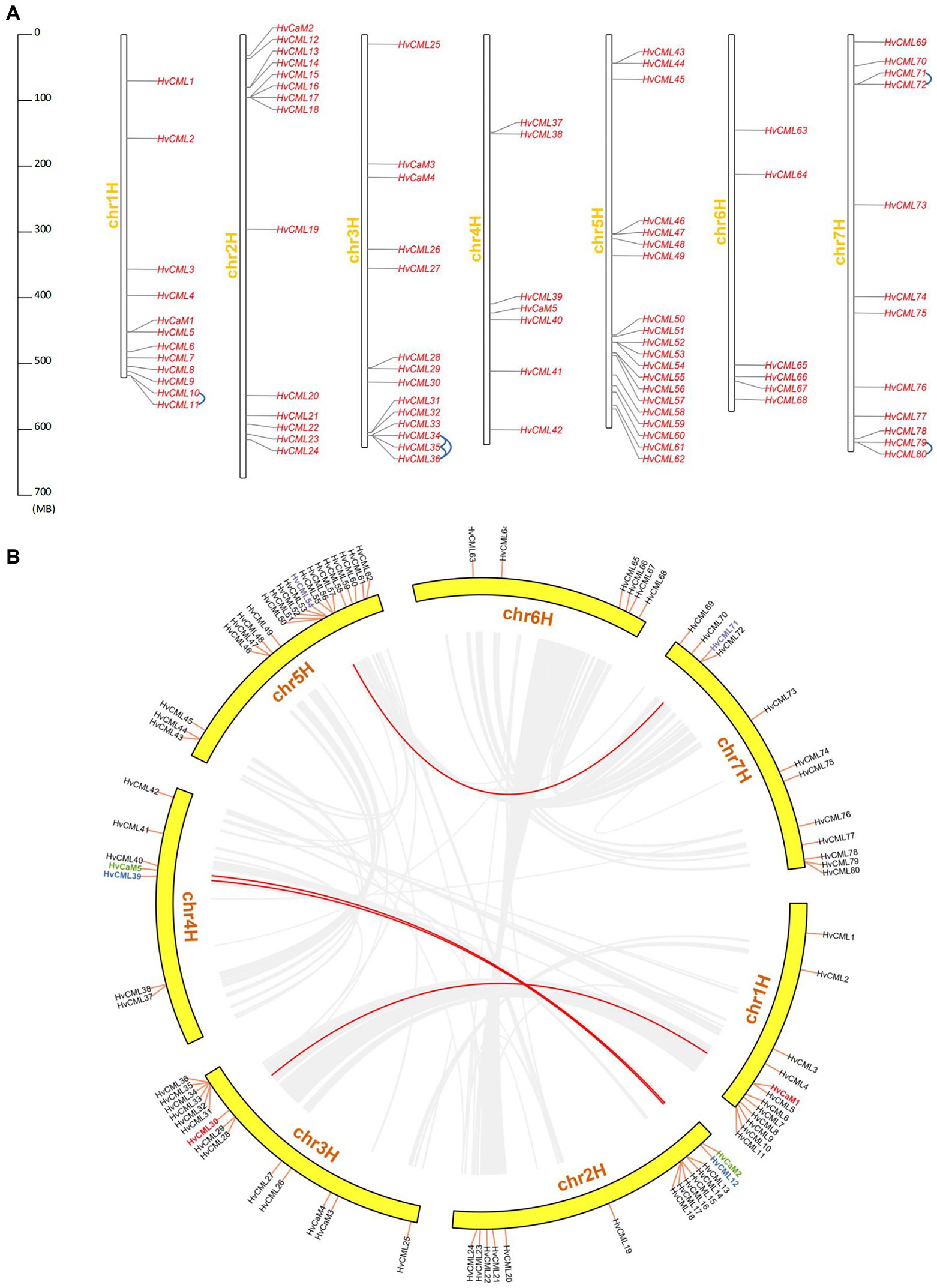
Figure 3. Chromosomal distribution and duplication of HvCaMs/CMLs. (A) Chromosomal distribution and tandem duplication of HvCaMs/CMLs; (B) Segmental duplication of HvCaMs/CMLs.
Tandem and segmental duplications are considered as the main driving forces expanding gene families during evolution (Kuo et al., 2019). In this study, tandem duplicated HvCML genes were defined as follows: (1) on a single chromosome with no more than one intervening gene; (2) similarity of aligned regions more than 70% (Zhu et al., 2014; Vatansever et al., 2016). In total, 9 genes (6 pairs) were involved in tandem duplication events (HvCML10 and HvCML11, HvCML34 and HvCML35, HvCML35 and HvCML36, HvCML34 and HvCML36, HvCML71 and HvCML72, HvCML79 and HvCML80; Figure 3A). On the other hand, 8 genes (4 pairs) were implicated in segmental duplication events (HvCaM1 and HvCML30, HvCaM2 and HvCaM5, HvCML12 and HvCML39, HvCML54 and HvCML71; Figure 3B).
Cis-acting elements analysis
The 2 kb upstream sequences of HvCaMs/CMLs coding regions were retrieved for cis-acting elements analysis. Totally, 69 cis-acting elements were identified and could be fractionalized into 6 types based on functional annotations (Figure 4; Supplementary File 4). Nearly half of the cis-acting elements (31, 44.9%) were in ‘light responsiveness’ category, followed by ones in ‘hormone response’ (11, 15.9%), ‘promoter/enhancer element’ (11, 15.9%), ‘development/tissue specificity’ (8, 11.6%) and ‘stress’ category (7, 10.1%). Only one kind of elements was identified in ‘circadian control’ category (Figure 4). CAAT-box and TATA-box in ‘promoter/enhancer element’ category, which are binding sites of RNA polymerase and responsible for transcription efficiency, were ubiquitously detected in all HvCaMs/CMLs, indicating their potential critical roles in controlling the transcription initiation and expression levels of HvCaMs/CMLs. ABRE, CGTCA-motif and TGACG-motif (in ‘hormone response’ category), which are involved in ABA and methyl jasmonate (MeJA) responsiveness, respectively, were widely present in HvCaMs/CMLs (Figure 4). Further, G-box in ‘light responsiveness’ category was also extensively distributed in promoter regions of HvCaMs/CMLs (Figure 4). On the other hand, the presence of other cis-acting elements was comparatively gene-specific. These results suggest that HvCaMs/CMLs might play pivotal roles in hormone metabolism and environmental responses, while the response patterns and expression profiling of HvCaMs/CMLs might be different.
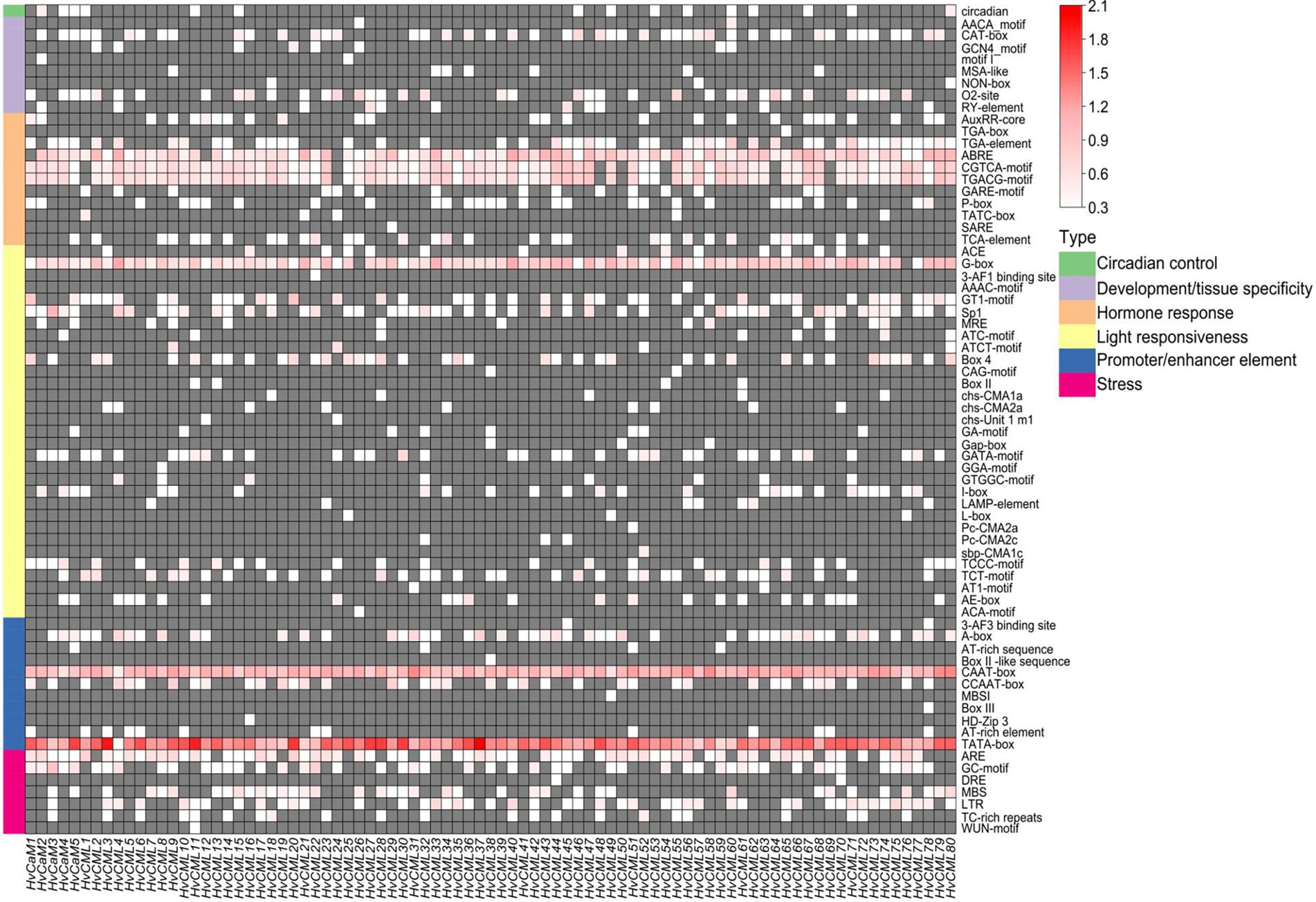
Figure 4. Cis-acting elements in promoter regions of HvCaMs/CMLs. Cis-acting elements were predicted based on 2 kb sequences upstream of coding sequences of HvCaMs/CMLs. The quantity of cis-acting elements was normalized by log10(number + 1) and then used for heatmap construction.
Synteny analysis of HvCaMs/CMLs
The syntenic relationships of CaMs/CMLs between barley and four plant species were investigated (Figure 5; Supplementary File 5). Only one pair of orthologous genes was identified between Arabidopsis and barley (AtCML41 and HvCML60). Similarly, only two pair of orthologous genes was identified between Brassica napus and barley (Supplementary File 5). This might be caused by evolutionally far genetic relationships between dicots and monocots. However, 60 pairs of orthologous genes were observed between barley and rice, involving 46 HvCaMs/CMLs (5 HvCaMs and 41 HvCMLs) and 49 rice genes (including 5 OsCaMs, 21 OsCMLs and 23 ‘other genes’). Nine HvCaMs/CMLs (3 HvCaMs and 6 HvCMLs) each had 2 orthologous genes in rice, while 5 OsCaMs/CMLs (2 OsCaMs and 3 OsCMLs) each had 2 orthologous genes in barley. Besides, OsCaM1-1 was orthologous to 3 HvCaM genes (HvCaM1, HvCaM2 and HvCaM5). Wheat was evolutionally closer to barley than rice, as expected, much more orthologous gene pairs (189 pairs) were detected between barley and wheat (Supplementary File 5).
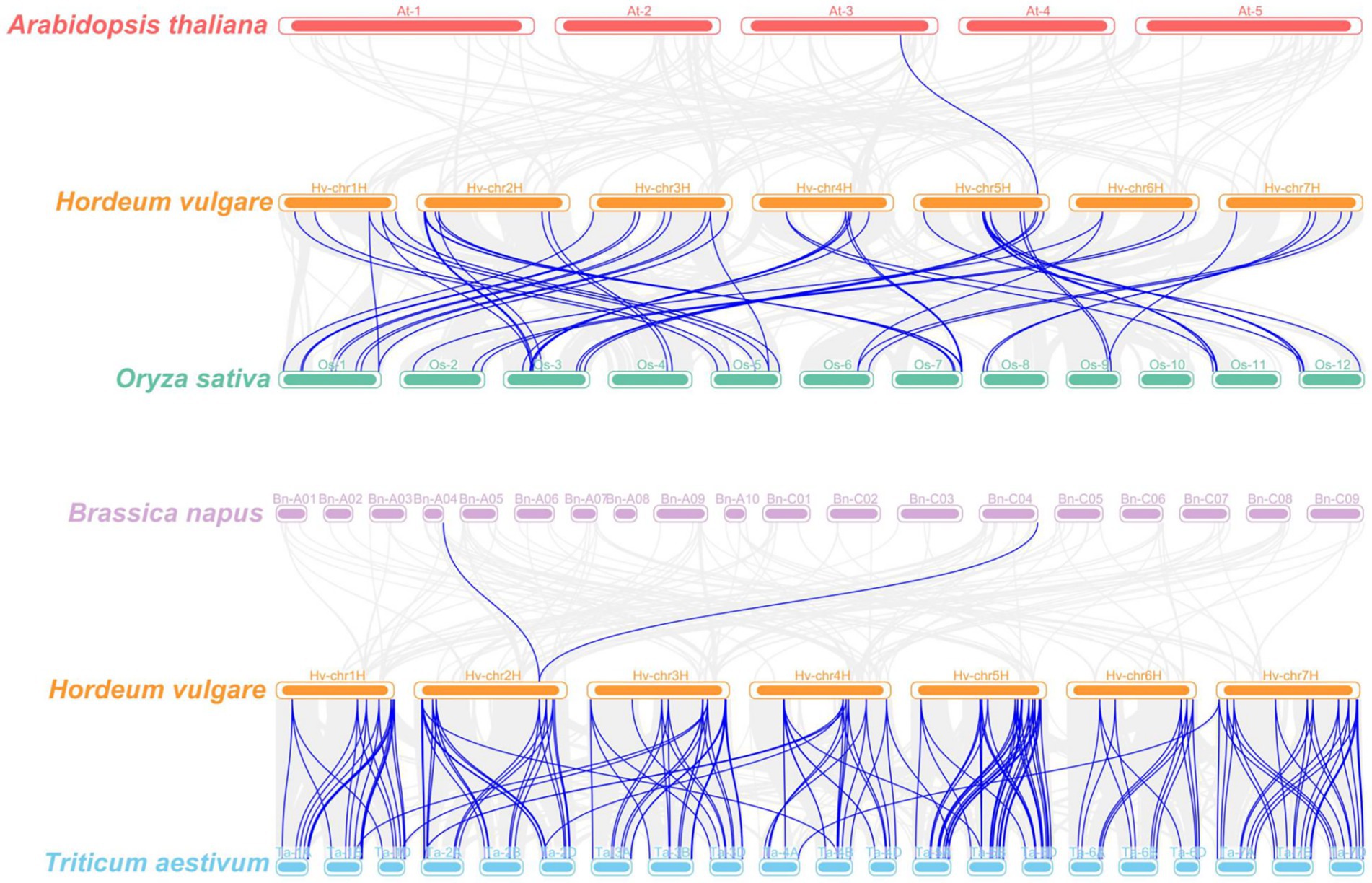
Figure 5. Synteny analyses of CaMs/CMLs between barley and four plant species (Arabidopsis thaliana, Oryza sativa, Brassica napus, and Triticum aestivum). Gray lines indicated collinear blocks and blue lines highlighted syntenic CaMs/CMLs gene pairs.
HvCaMs/CMLs in different genotypes
Pan-genome refers to a species-wide catalog of genic presence/absence variation or structural variation that affects (potentially non-coding) sequences of 50 or more base pairs in size (Jayakodi et al., 2020). The genic presence/absence variation of HvCaMs/CMLs was investigated in the context of the first-generation barley pan-genome comprising 20 varieties (Figure 6; Supplementary File 6; Jayakodi et al., 2020). The pan-genome can be divided into core genome and dispensable genome, the former comprises genes present in all genotypes while the latter comprises genes absent from some genotypes, which refers to genes showing presence/absence variation (Tettelin et al., 2005; Monat et al., 2019b). Accordingly, 81 of all 85 HvCaMs/CMLs (95.3%) were in the category of core genome and only 4 genes (HvCML16, HvCML18, HvCML50 and HvCML78) were in dispensable-genome category. B1K-04-12 was a wild barley genotype and displayed the least members (19 HvCaMs/CMLs) identical in nucleotide sequences to Morex (Figure 6). In the other 18 genotypes, 29 to 46 HvCaMs/CMLs were identical to those in Morex. Two HvCaMs (HvCaM3 and HvCaM4) and 3 HvCMLs (HvCML39, HvCML49 and HvCML73) were identical in nucleotide sequence among 20 genotypes. However, sequences of HvCaM1 and 6 HvCMLs (HvCML22, HvCML55, HvCML56, HvCML70, HvCML79 and HvCML80) in reference barley cultivar Morex differed from those in all the other 19 genotypes. Furthermore, the sequence identity of HvCaMs and HvCMLs between reference cultivar Morex and the other 19 genotypes ranged from 97.8 to 100% and from 90.0 to 100%, respectively. The biggest difference was in HvCML7 and HvCML6, whose sequence identity between Morex and the other genotypes averaged 93.7 and 96.0%, respectively.
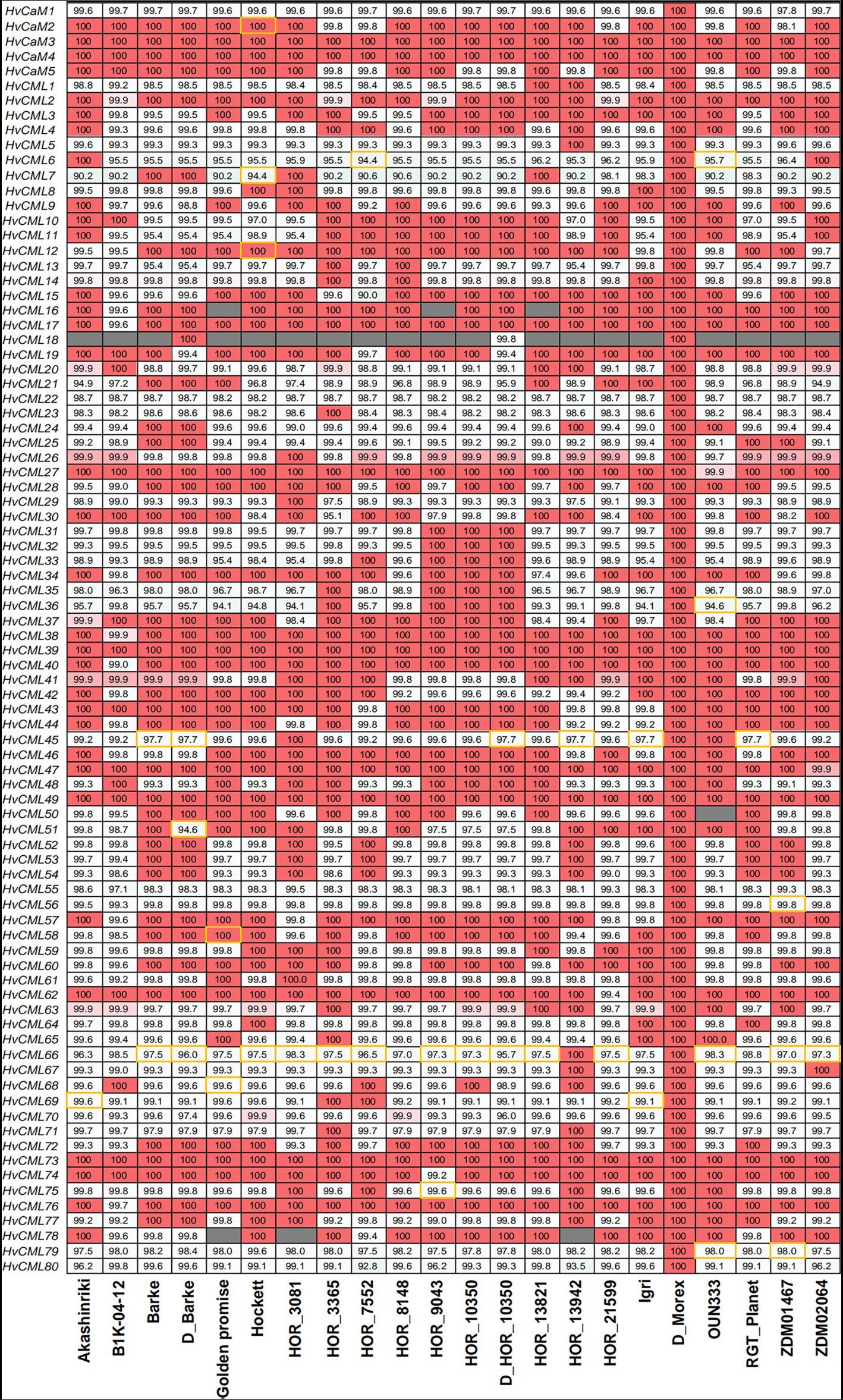
Figure 6. Nucleotide sequence identity of HvCaMs/CMLs between barley genotypes. Sequences of HvCaMs/CMLs in Morex were used as Blastn queries and the percent identity was designated in grids. Grids filled with gray indicated gene absence variation and those framed with yellow indicated discrepancies of chromosomal location between query and best hit.
Tissue expression patterns of HvCaMs/CMLs
Transcriptomic data of HvCaMs/CMLs were downloaded from BARLEX database to analyze their expression patterns in 14 tissues (Figure 7; Supplementary File 7). Five HvCaMs in group I ubiquitously expressed in all 14 tissues with relatively high levels. Most genes in group VII (15/19, 79.0%) also expressed in all tissues, although with comparatively low levels. Besides, there were 4 genes in group II, 5 genes in group III, 1 gene in group IV, 3 genes in group V and 4 genes in group VIII showing ubiquitous expression. Notably, the expression of 5 genes (HvCML7 and HvCML66 in group III, HvCML6 and HvCML51 in group 6 and HvCML19 in group VII) failed to be detected in any determined tissues under normal conditions, indicating that these genes might be luxury genes. Expression of the rest 43 (50.6%) genes could be detected in at least one tissues, showing high ratio of tissue-specific genes in HvCaM/CML gene family. Furthermore, 3 HvCMLs (HvCML73, HvCML16 and HvCML53) expressed in seedling roots but not in roots at 28 days after pollination. On the contrary, 4 genes (HvCML12, HvCML74, HvCML29 and HvCML36) were silent in roots at seedling stage but expressed at 28 days after pollination. Similar situation was also observed for 8 HvCMLs in developing grains at 5 days or 15 days after pollination. Thus, the expression of HvCMLs is not only tissue-specific but also development phase-dependent.
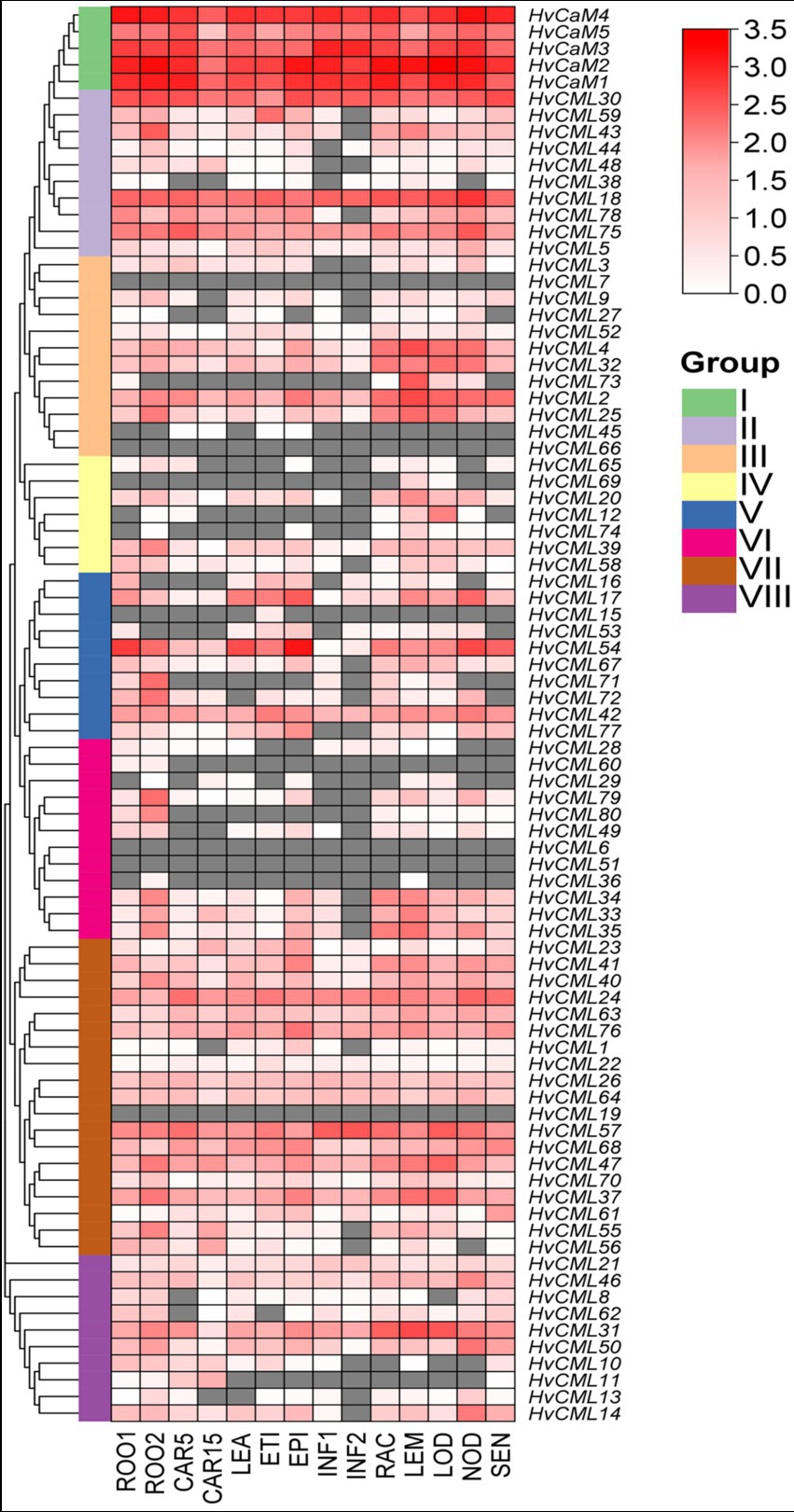
Figure 7. Expression profiling of HvCaMs/CMLs in 14 tissues based on transcriptomic data. FPKM values were normalized by log10(FPKM+1) transformation. ROO1, roots from seedlings (10 cm shoot stage); ROO2, roots (28 DAP); CAR5, developing grain (5 DAP); CAR15, developing grain (15 DAP); LEA, shoots from seedlings (10 cm shoot stage); ETI, etiolated seedling, dark condition (10 DAP); EPI, epidermal strips (28 DAP); INF1, young developing inflorescences (5 mm); INF2, developing inflorescences (1–1.5 cm); RAC, inflorescences, rachis (35 DAP); LEM, inflorescences, lemma (42 DAP); LOD, inflorescences, lodicule (42 DAP); NOD, developing tillers, 3rd internode (42 DAP); SEN, senescing leaves (56 DAP).
Expression of HvCaMs/CMLs in response to abiotic stresses
To investigate the response of HvCaMs/CMLs to abiotic stresses, the expression changes of 14 genes (5 HvCaMs and 9 HvCMLs) were examined through qRT-PCR after salt (200 mM NaCl), potassium deficiency (0.01 mM K+) and osmotic (20% PEG8000) treatments (Figure 8). HvCaM1, HvCaM4 and HvCaM5 displayed similar response patterns to salt stress (Figure 8A). They were down-regulated after salt treatment for 1 h to 1 day and were up-regulated after treatment for 3 days (Figure 8A), indicating the possibility of synergistic response to salt stress. The expression levels of HvCML37, HvCML42 and HvCML54 were significantly higher after salt treatment for 3 days, while HvCML17 and HvCML30 were up-regulated significantly after salt treatment for 3 h and 6 h, respectively (Figure 8A). These results suggested that response patterns to salt stress differed between HvCMLs and were time-dependent. Under potassium deficiency conditions, the expression levels of HvCaMs were all increased after treatment for 1–6 days and were comparatively higher than those after treatment for 1–6 h. HvCMLs displayed similar response patterns to potassium deficiency (Figure 8B). On the other hand, although the expression levels of HvCaMs/CMLs fluctuated, no obvious patterns were observed in response to osmotic stress, and no significant difference was observed between different treatments for 11 of 14 examined HvCaMs/CMLs (Figure 8C). These results indicated that HvCaMs/CMLs might respond to abiotic stress in a synergistic way, and respo.
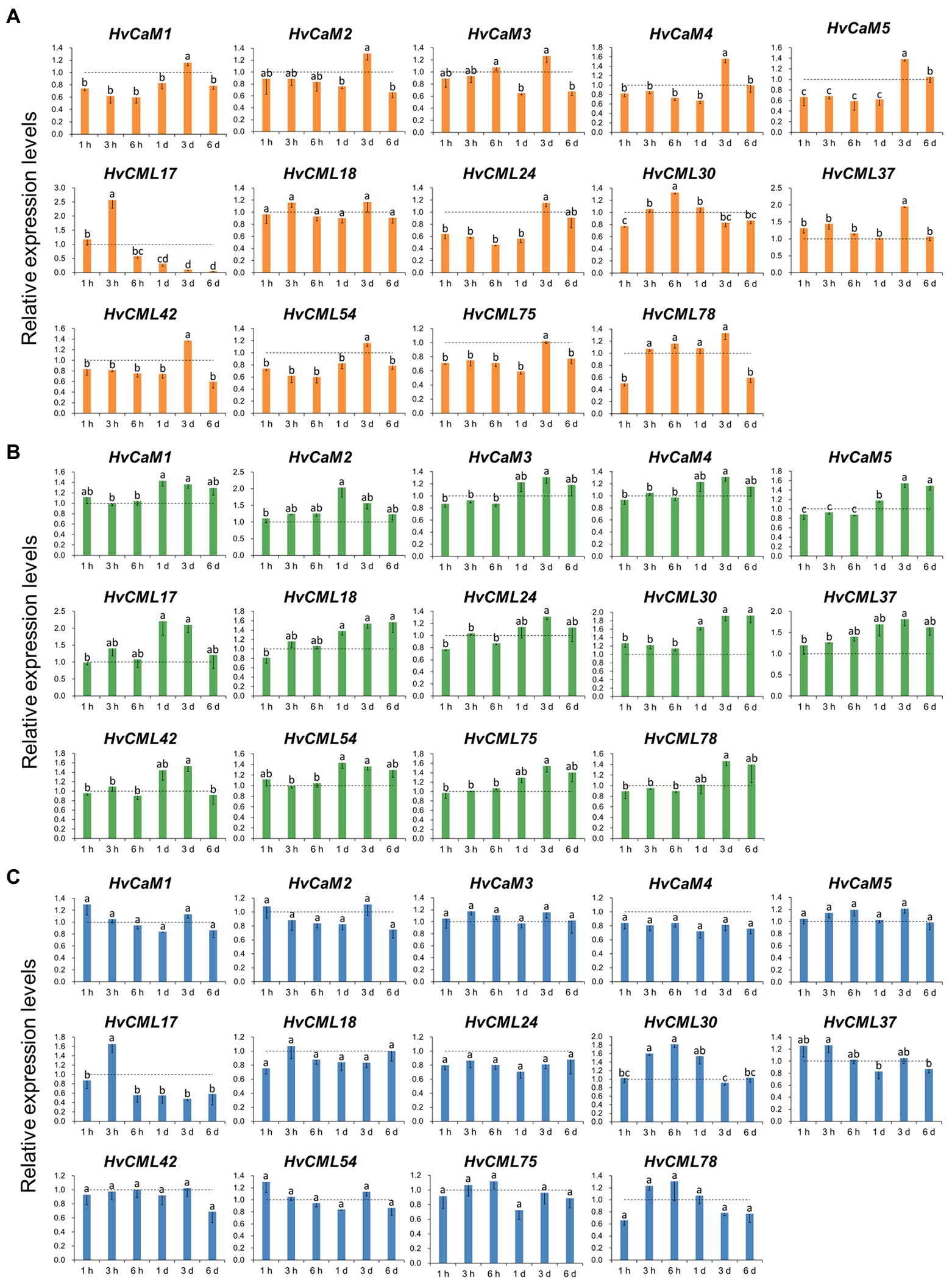
Figure 8. Expression levels of 14 HvCaMs/CMLs in response to salt stress (A), potassium deficiency (B) and osmotic stress (C) at seedling stage. Dotted lines indicated the expression levels of HvCaMs/CMLs in control seedlings. Lowercase letters indicated the significant difference at p < 0.05.
Discussion
CaM and CML gene family has been identified and analyzed in many plant species, such as Arabidopsis (McCormack and Braam, 2003), rice (Boonburapong and Buaboocha, 2007), wheat (Liu et al., 2022), Brassica napus (He et al., 2020) and papaya (Ding et al., 2018). Compared with relatively well-studied in Arabidopsis and rice, CaMs/CMLs gene family in barley remains largely unknown. So far, only HvCaM1 has been functionally characterized, which negatively regulated salt tolerance probably via interaction with HvCAMTA4 to modulate the expression of HvHKT1;5 and HvHKT1;1 in barley (Shen et al., 2020). In this work, we identified 5 HvCaMs and 80 HvCMLs in barley, and then investigated their phylogenetic relationships, sequence characteristics, syntenic relationships, presence/absence variation in pan-genome, expression patterns in different tissues and in response to abiotic stresses.
Identification of HvCaMs/CMLs in barley
A total of 179 genes were identified as CaM/CML gene family candidates in barley through preliminary search. As calcium sensor relays, CaMs and CMLs do not have any functional domains and catalytic activities other than EF-hand motifs (Reddy et al., 2011). Based on this criterion, 96 candidate genes were further screened in barley genome. It is noteworthy that CBLs are also sensor relays, and the abovementioned criterion fails to distinguish CBLs and CaMs/CMLs. Thus, the putative HvCaMs/CMLs, together with sensor relay genes in Arabidopsis (7 CaMs, 50 CMLs and 10 CBLs) and rice (5 OsCaMs, 32 OsCMLs and 10 OsCBLs), were phylogenetically analyzed (Kolukisaoglu et al., 2004; McCormack et al., 2005; Boonburapong and Buaboocha, 2007). The amino acid sequences of these genes were aligned with MAFFT and the phylogenetic tree was constructed with maximum likelihood method. According to phylogenetic evidence, the 96 candidate genes in barley were classified into 5 HvCaMs, 80 HvCMLs and 11 HvCBLs (Supplementary File 8). The 85 HvCaMs/CMLs were retrieved for further characterization. Phylogeny-assistant identification was also adopted for CaMs/CMLs identification in papaya (Ding et al., 2018).
Totally, 57 AtCaMs/CMLs (7 AtCaMs and 50 AtCMLs), 37 OsCaMs/CMLs (5 OsCaMs and 32 OsCMLs) and 248 TaCaMs/CMLs (18 TaCaMs and 230 TaCMLs) were identified in Arabidopsis, rice and wheat, respectively (McCormack and Braam, 2003; McCormack et al., 2005; Boonburapong and Buaboocha, 2007; Liu et al., 2022). In this research, 85 HvCaMs/CMLs (5 HvCaMs and 80 HvCMLs) were identified in barley. CML genes were much more than CaMs in Arabidopsis, rice, wheat and barley, similar results were also observed in other species from lower plants to higher plants (Zhu et al., 2015). Wheat is hexaploid with a genome size of about 17 Gb, which is nearly threefold the genome size of barley (~5.3 Gb). And the number of CaM/CML family members in wheat is also nearly three times of that in barley.The genome size of rice (~500 Mb) is 3.7-fold that of Arabidopsis (~135 Mb), while fewer CaMs and CMLs were identified. Notably, OsCMLs were absent in cluster IV, V and VI (Figure 1). Besides, orthologous gene pairs between barley and rice involved 49 rice genes, in which 23 genes were not identified as OsCMLs (Figure 5; Supplementary File 5). In addition, 53 OsCMLs were identified in another research on CaMs/CMLs evolution (Zhu et al., 2015). Thus, some members might be left out in previous identification of OsCMLs.
Similarities and differences of CaMs/CMLs among barley, Arabidopsis and rice
CaMs are highly conserved and ubiquitous in all eukaryotes (Halling et al., 2016). HvCaMs were phylogenetically closer to OsCaMs than AtCaMs (Figures 1, 5). All these CaM genes were interrupted by one phase 0 intron and coded polypeptides with 149 amino acids, which possessed 4 EF-hand motifs and contained 9 methionine residues (6.0%, Table 1; McCormack and Braam, 2003; Boonburapong and Buaboocha, 2007). Seven AtCaMs in Arabidopsis and 5 OsCaMs in rice coded 4 isoforms (AtCaM1/4, AtCaM2/3/5, AtCaM6 and AtCaM7) and 3 isoforms (OsCaM1-1/1–2/1–3, OsCaM2 and OsCaM3), respectively. Similarly, 5 HvCaMs in barley coded 3 isoforms (HvCaM2/3/5, HvCaM1 and HvCaM4; Supplementary File 9). Besides, the amino acid sequence coded by HvCaM2 was identical to that by OsCaM1-1 (Supplementary File 9). On the other hand, CMLs were more diversified in length, gene structure and methionine percentage. The length of amino acid sequences of AtCMLs and HvCMLs are similar, ranging from 83 to 354 and 78 to 389, respectively (McCormack and Braam, 2003; McCormack et al., 2005; Boonburapong and Buaboocha, 2007), while the length of OsCMLs varies from 146 to 250. Most of AtCMLs (31, 62.0%) and OsCMLs (20, 62.5%) contained 4 EF-hand motifs. By contrast, EF-hand motifs in HvCMLs varied from 1 to 4 (being 20, 18, 23 and 19, respectively; Table 1). Intron-free CML genes were predominant in all these three plants (74.0, 75.0 and 66.3% in Arabidopsis, rice and barley, respectively). Six HvCMLs, together with 6 AtCMLs and 6 OsCMLs, had higher methionine percentage than CaMs (6.0%).
HvCaMs/CMLs in different barley genotypes
The genomic information revealed by reference genome assembly was partly limited to the genotype sequenced and failed to capture the full complement of a species. Pan-genome enabled the characterization of the genetic diversity present in a species. In plants, core genes are often associated with essential metabolic processes, while dispensable genes are related to adaptive functions such as disease resistance and stress responses (Danilevicz et al., 2020; Yocca and Edger, 2022). HvCaM/CML was a large gene family, and according to the released first-generation barley pan-genome data (Jayakodi et al., 2020), 81 (95.3%) HvCaMs/CMLs were core genes (Figure 6). These results indicated that although the functions of HvCaMs/CMLs were largely unknown, they might play diverse and essential roles and were indispensable. The wild barley genotype B1K-04-12 had more HvCaMs/CMLs members differed in sequences from Morex than other cultivars and landraces (Figure 6), however, only HvCML18 was absent in this genotype. Comparatively, there were 3 HvCMLs absent in cultivar Golden promise. Thus, there was no necessary connection between evolutionary relationship and presence/absence variation. Recently, it has been found that allelic changes of cis-regulatory elements of RAP2.12 are responsible for differentially regulating tolerance to drought and flooding in Arabidopsis (Lou et al., 2022). Therefore, in addition to presence/absence variation, variation in cis-acting elements should be taken into consideration when characterizing certain CaM/CML genes. Notably, HvCML18 was absent in assembly projection of cultivar Barke, but was present in de novo annotation with the evidence from RNA-Seq and PacBio Iso-Seq data. Similar phenomena were also observed for HvCML18 in landrace HOR_10350. On the other hand, although HvCaM2 and HvCML12 in Hockett and HvCML58 in Golden Promise were identical to those in Morex in nucleotide sequence, they failed to be anchored on certain chromosomes (Supplementary File 6). Discrepancy in chromosomal location of genes was also observed between Morex and other 17 genotypes (Supplementary File 6), indicating more efforts were needed for genome annotation improvement.
Expression of HvCaMs/CMLs in different tissues and in response to abiotic stress
HvCaMs were ubiquitously expressed in all examined tissues with high levels, while HvCMLs displayed different expression patterns and levels among tissues (Figure 7; Supplementary File 7), which was consistent with tissue expression patterns of CaMs/CMLs in Brassica napus (He et al., 2020). HvCaM2 and HvCML30 were involved in segmental duplication and displayed similar tissue expression patterns and levels (Figures 3B, 6). HvCML39 and HvCML12, HvCML54 and HvCML71 were another two segmental duplication gene pairs, however, HvCML39 and HvCML54 expressed in all examined tissues, whereas HvCML12 and HvCML71 only expressed in certain tissues (Figures 3B, 6). HvCML34, HvCML35 and HvCML36 were tandem duplicated genes, HvCML34 and HvCML35 displayed the same tissue expression patterns and similar expression levels, while HvCML36 and above two genes differed in tissue expression patterns and levels (Figures 3A, 6). The same was true for the other three pairs of tandem duplicated CMLs (HvCML10 and HvCML11, HvCML71 and HvCML72, HvCML79 and HvCML80). These results suggested that gene expression patterns might not interrelate with duplication events. On the other hand, expression of HvCMLs were affected by development stages. For example, the expression of HvCML53 and HvCML73 were detected in seedling roots but were undetectable in roots at 28 days after pollination (Figure 7). By contrast, HvCML29 and HvCML36 did not express in roots from seedlings but expressed in roots at 28 days after pollination (Figure 7). Similar results were observed for expression of HvCMLs in developing grains and inflorescences (Figure 7). These results revealed that the expression of HvCMLs was more variable than HvCaMs, and was tissue-and development stage-dependent.
A series of cis-acting elements were identified in the upstream of HvCaMs/CMLs coding sequences, including 11 hormone-responsive elements and 7 stress-responsive ones (Figure 4; Supplementary File 4). Thus, the expression of HvCaMs/CMLs under abiotic stress treatments was investigated. Salt, osmotic and potassium deficiency stresses all affected the transcription levels of HvCaMs/CMLs in a time-dependent way (Figure 8). The expression response of HvCaMs to salt stress (up-regulation after 3 days) and HvCaMs/CMLs to potassium deficiency (up-regulation after 1–6 days, and relatively higher than that after 1–6 h) displayed similar patterns, indicating these genes might respond in a synergistic manner. Besides, no significant difference was observed in transcription levels of 11 HvCaMs/CMLs during osmotic stress (Figure 8), which was also observed in expression of OsCML1, OsCML3 and OsCML13 under osmotic stress (Chinpongpanich et al., 2012). These results indicated that expression patterns of HvCaMs/CMLs were stress-dependent.
Conclusion and prospects
In the current study, 5 HvCaMs genes and 80 HvCMLs were identified in barley. Eighty-one HvCaMs/CMLs were core genes and only 4 HvCMLs were dispensable genes based on the first generation of barley pan-genome. The expression of HvCaMs/CMLs varied with plant tissues, abiotic stresses and exposure time of stresses. The obtained results will be helpful for further understanding of CaM/CML family in barley.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
KC and JW designed the experiments and wrote the paper. KC analyzed the data. LK, WY, SX, and XX performed the experiments. GZ analyzed the data and revised the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (2021C02064-3-2), China Agriculture Research System of MOF and MARA (CARS-05-01A-06), the National Natural Science Foundation of China (32101642), and Natural Science Foundation of Zhejiang Province (LQ21C130006).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer GC declared a shared affiliation with the authors KC, WY, XX, and JW at the time of the review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.964888/full#supplementary-material
Footnotes
1. ^https://www.arabidopsis.org/
2. ^https://rapdb.dna.affrc.go.jp/
3. ^http://plants.ensembl.org/
4. ^https://www.ebi.ac.uk/interpro/
5. ^https://web.expasy.org/compute_pi/
6. ^http://busca.biocomp.unibo.it/
7. ^https://www.ebi.ac.uk/Tools/msa/mafft/
8. ^http://cbi.hzau.edu.cn/bnapus/index.php
9. ^http://plants.ensembl.org/info/data/ftp/index.html
10. ^https://meme-suite.org/meme/tools/meme
11. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
12. ^https://apex.ipk-gatersleben.de/apex/f?p=284:49:::::P49_GENE_CHOICE:4
References
Aleynova, O. A., Kiselev, K. V., Ogneva, Z. V., and Dubrovina, A. S. (2020). The grapevine calmodulin-like protein gene cml21 is regulated by alternative splicing and involved in abiotic stress response. Int. J. Mol. Sci. 21, 1–19. doi: 10.3390/ijms21217939
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Basu, R., Dutta, S., Pal, A., Sengupta, M., and Chattopadhyay, S. (2021). Calmodulin7: recent insights into emerging roles in plant development and stress. Plant Mol. Biol. 107, 1–20. doi: 10.1007/s11103-021-01177-1
Blum, M., Chang, H. Y., Chuguransky, S., Grego, T., Kandasaamy, S., Mitchell, A., et al. (2021). The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354. doi: 10.1093/nar/gkaa977
Boonburapong, B., and Buaboocha, T. (2007). Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 7:4. doi: 10.1186/1471-2229-7-4
Cai, K., Gao, H., Wu, X., Zhang, S., Han, Z., Chen, X., et al. (2019). The ability to regulate transmembrane potassium transport in root is critical for drought tolerance in barley. Int. J. Mol. Sci. 20:4111. doi: 10.3390/ijms20174111
Cai, K., Zeng, F., Wang, J., and Zhang, G. (2021). Identification and characterization of HAK/KUP/KT potassium transporter gene family in barley and their expression under abiotic stress. BMC Genomics 22, 317–314. doi: 10.1186/s12864-021-07633-y
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheval, C., Aldon, D., Galaud, J. P., and Ranty, B. (2013). Calcium/calmodulin-mediated regulation of plant immunity. Biochim. Biophys. Acta Mol. Cell Res. 1833, 1766–1771. doi: 10.1016/j.bbamcr.2013.01.031
Chinpongpanich, A., Limruengroj, K., Phean-O-Pas, S., Limpaseni, T., and Buaboocha, T. (2012). Expression analysis of calmodulin and calmodulin-like genes from rice, Oryza sativa L. BMC Res. Notes 5:625. doi: 10.1186/1756-0500-5-625
Chu, M., Li, J., Zhang, J., Shen, S., Li, C., Gao, Y., et al. (2018). AtCaM4 interacts with a Sec14-like protein, PATL1, to regulate freezing tolerance in Arabidopsis in a CBF-independent manner. J. Exp. Bot. 69, 5241–5253. doi: 10.1093/jxb/ery278
Dai, C., Lee, Y., Lee, I. C., Nam, H. G., and Kwak, J. M. (2018). Calmodulin 1 regulates senescence and ABA response in Arabidopsis. Front. Plant Sci. 9:803. doi: 10.3389/fpls.2018.00803
Danilevicz, M. F., Tay Fernandez, C. G., Marsh, J. I., Bayer, P. E., and Edwards, D. (2020). Plant pangenomics: approaches, applications and advancements. Curr. Opin. Plant Biol. 54, 18–25. doi: 10.1016/j.pbi.2019.12.005
Delk, N. A., Johnson, K. A., Chowdhury, N. I., and Braam, J. (2005). CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 139, 240–253. doi: 10.1104/pp.105.062612
Ding, H., Qian, Y., Fang, Y., Ji, Y., Sheng, J., and Ge, C. (2021). Characteristics of slcml39, a tomato calmodulin-like gene, and its negative role in high temperature tolerance of Arabidopsis thaliana during germination and seedling growth. Int. J. Mol. Sci. 22:11479. doi: 10.3390/ijms222111479
Ding, X., Zhang, L., Hao, Y., Xiao, S., Wu, Z., Chen, W., et al. (2018). Genome-wide identification and expression analyses of the calmodulin and calmodulin-like proteins reveal their involvement in stress response and fruit ripening in papaya. Postharvest Biol. Technol. 143, 13–27. doi: 10.1016/j.postharvbio.2018.04.010
Dobney, S., Chiasson, D., Lam, P., Smith, S. P., and Snedden, W. A. (2009). The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J. Biol. Chem. 284, 31647–31657. doi: 10.1074/jbc.M109.056770
Du, B., Chen, N., Song, L., Wang, D., Cai, H., Yao, L., et al. (2021). Alfalfa (Medicago sativa L.) MsCML46 gene encoding calmodulin-like protein confers tolerance to abiotic stress in tobacco. Plant Cell Rep. 40, 1907–1922. doi: 10.1007/s00299-021-02757-7
Galon, Y., Finkler, A., and Fromm, H. (2010). Calcium-regulated transcription in plants. Mol. Plant 3, 653–669. doi: 10.1093/mp/ssq019
Halling, D. B., Liebeskind, B. J., Hall, A. W., and Aldrich, R. W. (2016). Conserved properties of individual Ca2+-binding sites in calmodulin. Proc. Natl. Acad. Sci. U.S.A. 113, E1216–E1225. doi: 10.1073/pnas.1600385113
He, X., Liu, W., Li, W., Liu, Y., Wang, W., Xie, P., et al. (2020). Genome-wide identification and expression analysis of CaM/CML genes in Brassica napus under abiotic stress. J. Plant Physiol. 255:153251. doi: 10.1016/j.jplph.2020.153251
Heyer, M., Scholz, S. S., Reichelt, M., Kunert, G., Oelmüller, R., and Mithöfer, A. (2021). The Ca2+ sensor proteins CML37 and CML42 antagonistically regulate plant stress responses by altering phytohormone signals. Plant Mol. Biol. 109, 611–625. doi: 10.1007/s11103-021-01184-2
Jayakodi, M., Padmarasu, S., Haberer, G., Bonthala, V. S., Gundlach, H., Monat, C., et al. (2020). The barley pan-genome reveals the hidden legacy of mutation breeding. Nature 588, 284–289. doi: 10.1038/s41586-020-2947-8
Jung, H., Chung, P. J., Park, S. H., Redillas, M. C. F. R., Kim, Y. S., Suh, J. W., et al. (2017). Overexpression of OsERF48 causes regulation of OsCML16, a calmodulin-like protein gene that enhances root growth and drought tolerance. Plant Biotechnol. J. 15, 1295–1308. doi: 10.1111/pbi.12716
Kalaipandian, S., Xue, G. P., Rae, A. L., Glassop, D., Bonnett, G. D., and McIntyre, L. C. (2019). Overexpression of TaCML20, a calmodulin-like gene, enhances water soluble carbohydrate accumulation and yield in wheat. Physiol. Plant. 165, 790–799. doi: 10.1111/ppl.12786
Kolukisaoglu, Ü., Weinl, S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi: 10.1104/pp.103.033068
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuo, Y. T., Chao, Y. T., Chen, W. C., Shih, M. C., and Chang, S. B. (2019). Segmental and tandem chromosome duplications led to divergent evolution of the chalcone synthase gene family in phalaenopsis orchids. Ann. Bot. 123, 69–77. doi: 10.1093/aob/mcy136
Lee, H. J., and Seo, P. J. (2021). Ca2+ talyzing initial responses to environmental stresses. Trends Plant Sci. 26, 849–870. doi: 10.1016/j.tplants.2021.02.007
Leitão, N., Dangeville, P., Carter, R., and Charpentier, M. (2019). Nuclear calcium signatures are associated with root development. Nat. Commun. 10:4865. doi: 10.1038/s41467-019-12845-8
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Liu, Y., Chen, W., Liu, L., Su, Y., Li, Y., Jia, W., et al. (2022). Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in wheat (Triticum aestivum L.). Plant Signal. Behav. 17:e2013646. doi: 10.1080/15592324.2021.2013646
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lou, S., Guo, X., Liu, L., Song, Y., Zhang, L., Jiang, Y., et al. (2022). Allelic shift in cis-elements of the transcription factor RAP2.12 underlies adaptation associated with humidity in Arabidopsis thaliana. Sci Adv. 8:eabn8281. doi: 10.1126/sciadv.abn8281
Lu, L., Rong, W., Zhou, R., Huo, N., and Zhang, Z. (2019). TaCML36, a wheat calmodulin-like protein, positively participates in an immune response to Rhizoctonia cerealis. Crop J. 7, 608–618. doi: 10.1016/j.cj.2019.02.001
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Magnan, F., Ranty, B., Charpenteau, M., Sotta, B., Galaud, J.-P., and Aldon, D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56, 575–589. doi: 10.1111/j.1365-313X.2008.03622.x
Martí Ruiz, M. C., Hubbard, K. E., Gardner, M. J., Jung, H. J., Aubry, S., Hotta, C. T., et al. (2018). Circadian oscillations of cytosolic free calcium regulate the Arabidopsis circadian clock. Nat. Plants 4, 690–698. doi: 10.1038/s41477-018-0224-8
McCormack, E., and Braam, J. (2003). Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 159, 585–598. doi: 10.1046/j.1469-8137.2003.00845.x
McCormack, E., Tsai, Y. C., and Braam, J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Monat, C., Padmarasu, S., Lux, T., Wicker, T., Gundlach, H., Himmelbach, A., et al. (2019a). TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol. 20, 284–218. doi: 10.1186/s13059-019-1899-5
Monat, C., Schreiber, M., Stein, N., and Mascher, M. (2019b). Prospects of pan-genomics in barley. Theor. Appl. Genet. 132, 785–796. doi: 10.1007/s00122-018-3234-z
Munir, S., Liu, H., Xing, Y., Hussain, S., Ouyang, B., Zhang, Y., et al. (2016). Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci. Rep. 6, 1–20. doi: 10.1038/srep31772
Reddy, A. S. N., Ali, G. S., Celesnik, H., and Day, I. S. (2011). Coping with stresses: roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell 23, 2010–2032. doi: 10.1105/tpc.111.084988
Savojardo, C., Martelli, P. L., Fariselli, P., Profiti, G., and Casadio, R. (2018). BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 46, W459–W466. doi: 10.1093/nar/gky320
Scholz, S. S., Reichelt, M., Vadassery, J., and Mithöfer, A. (2015). Calmodulin-like protein CML37 is a positive regulator of ABA during drought stress in Arabidopsis. Plant Signal. Behav. 10:e1011951. doi: 10.1080/15592324.2015.1011951
Shen, Q., Fu, L., Su, T., Ye, L., Huang, L., Kuang, L., et al. (2020). Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiol. 183, 1650–1662. doi: 10.1104/pp.20.00196
Shi, J., and Du, X. (2020). Identification, characterization and expression analysis of calmodulin and calmodulin-like proteins in Solanum pennellii. Sci. Rep. 10, 7474–7417. doi: 10.1038/s41598-020-64178-y
Sun, Q., Huang, R., Zhu, H., Sun, Y., and Guo, Z. (2021). A novel Medicago truncatula calmodulin-like protein (MtCML42) regulates cold tolerance and flowering time. Plant J. 108, 1069–1082. doi: 10.1111/tpj.15494
Tang, M., Xu, C., Cao, H., Shi, Y., Chen, J., Chai, Y., et al. (2021). Tomato calmodulin-like protein SlCML37 is a calcium (Ca2+) sensor that interacts with proteasome maturation factor SlUMP1 and plays a role in tomato fruit chilling stress tolerance. J. Plant Physiol. 258-259:153373. doi: 10.1016/j.jplph.2021.153373
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. U. S. A. 102, 13950–13955. doi: 10.1073/pnas.0506758102
Thor, K. (2019). Calcium—nutrient and messenger. Front. Plant Sci. 10:440. doi: 10.3389/fpls.2019.00440
Vadassery, J., Reichelt, M., Hause, B., Gershenzon, J., Boland, W., and Mithöfer, A. (2012). CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159, 1159–1175. doi: 10.1104/pp.112.198150
Vandelle, E., Vannozzi, A., Wong, D., Danzi, D., Digby, A. M., Dal Santo, S., et al. (2018). Identification, characterization, and expression analysis of calmodulin and calmodulin-like genes in grapevine (Vitis vinifera) reveal likely roles in stress responses. Plant Physiol. Biochem. 129, 221–237. doi: 10.1016/j.plaphy.2018.06.003
Vanderbeld, B., and Snedden, W. A. (2007). Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol. Biol. 64, 683–697. doi: 10.1007/s11103-007-9189-0
Vatansever, R., Koc, I., Ozyigit, I. I., Sen, U., Uras, M. E., Anjum, N. A., et al. (2016). Genome-wide identification and expression analysis of sulfate transporter (SULTR) genes in potato (Solanum tuberosum L.). Planta 244, 1167–1183. doi: 10.1007/s00425-016-2575-6
Wang, Y., Wang, B., Gilroy, S., Wassim Chehab, E., and Braam, J. (2011). CML24 is involved in root mechanoresponses and cortical microtubule orientation in Arabidopsis. J. Plant Growth Regul. 30, 467–479. doi: 10.1007/s00344-011-9209-9
Wu, H. C., Luo, D. L., Vignols, F., and Jinn, T. L. (2012). Heat shock-induced biphasic Ca2+ signature and OsCaM1-1 nuclear localization mediate downstream signalling in acquisition of thermotolerance in rice (Oryza sativa L.). Plant Cell Environ. 35, 1543–1557. doi: 10.1111/j.1365-3040.2012.02508.x
Wu, X., Qiao, Z., Liu, H., Acharya, B. R., Li, C., and Zhang, W. (2017). CML20, an Arabidopsis calmodulin-like protein, negatively regulates guard cell ABA signaling and drought stress tolerance. Front. Plant Sci. 8:824. doi: 10.3389/fpls.2017.00824
Xu, G. Y., Rocha, P. S. C. F., Wang, M. L., Xu, M. L., Cui, Y. C., Li, L. Y., et al. (2011). A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta 234, 47–59. doi: 10.1007/s00425-011-1386-z
Xuan, Y., Zhou, S., Wang, L., Cheng, Y., and Zhao, L. (2010). Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings. Plant Physiol. 153, 1895–1906. doi: 10.1104/pp.110.160424
Yang, X., Wang, S. S., Wang, M., Qiao, Z., Bao, C. C., and Zhang, W. (2014). Arabidopsis thaliana calmodulin-like protein CML24 regulates pollen tube growth by modulating the actin cytoskeleton and controlling the cytosolic Ca2+ concentration. Plant Mol. Biol. 86, 225–236. doi: 10.1007/s11103-014-0220-y
Yin, X., Huang, L., Wang, M., Cui, Y., and Xia, X. (2017). OsDSR-1, a calmodulin-like gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Mol. Breed. 37, 1–13. doi: 10.1007/s11032-017-0668-y
Yin, X. M., Huang, L. F., Zhang, X., Wang, M. L., Xu, G. Y., and Xia, X. J. (2015). OsCML4 improves drought tolerance through scavenging of reactive oxygen species in rice. J. Plant Biol. 58, 68–73. doi: 10.1007/s12374-014-0349-x
Yocca, A. E., and Edger, P. P. (2022). Machine learning approaches to identify core and dispensable genes in pangenomes. Plant Genome 15, e20135–e20111. doi: 10.1002/tpg2.20135
Yu, S., Wu, J., Sun, Y., Zhu, H., Sun, Q., Zhao, P., et al. (2022). A calmodulin-like protein (CML10) interacts with cytosolic enzymes GSTU8 and FBA6 to regulate cold tolerance. Plant Physiol. 1–13. doi: 10.1093/plphys/kiac311
Zeng, H., Xu, L., Singh, A., Wang, H., Du, L., and Poovaiah, B. W. (2015). Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6:600. doi: 10.3389/fpls.2015.00600
Zhang, X., Wang, W., Kang, X., and Zhao, L. (2020). Arabidopsis CaM3 inhibits nitric oxide accumulation and improves thermotolerance by promoting S-nitrosoglutathione reductase via direct binding. Plant Growth Regul. 90, 41–50. doi: 10.1007/s10725-019-00552-9
Zhang, X., Wang, T., Liu, M., Sun, W., and Zhang, W. H. (2019). Calmodulin-like gene MtCML40 is involved in salt tolerance by regulating MtHKTs transporters in Medicago truncatula. Environ. Exp. Bot. 157, 79–90. doi: 10.1016/j.envexpbot.2018.09.022
Zhou, S., Jia, L., Chu, H., Wu, D., Peng, X., Liu, X., et al. (2016). Arabidopsis CaM1 and CaM4 promote nitric oxide production and salt resistance by inhibiting S-nitrosoglutathione reductase via direct binding. PLoS Genet. 12, e1006255–e1006228. doi: 10.1371/journal.pgen.1006255
Zhu, X., Dunand, C., Snedden, W., and Galaud, J. P. (2015). CaM and CML emergence in the green lineage. Trends Plant Sci. 20, 483–489. doi: 10.1016/j.tplants.2015.05.010
Zhu, X., Mazard, J., Robe, E., Pignoly, S., Aguilar, M., San Clemente, H., et al. (2021). The same against many: AtCML8, a Ca2+ sensor acting as a positive regulator of defense responses against several plant pathogens. Int. J. Mol. Sci. 22:10469. doi: 10.3390/ijms221910469
Zhu, X., Perez, M., Aldon, D., and Galaud, J.-P. (2017). Respective contribution of CML8 and CML9, two arabidopsis calmodulin-like proteins, to plant stress responses. Plant Signal. Behav. 12:e1322246. doi: 10.1080/15592324.2017.1322246
Zhu, X., Wang, P., Bai, Z., Herde, M., Ma, Y., Li, N., et al. (2022). Calmodulin-like protein CML24 interacts with CAMTA2 and WRKY46 to regulate ALMT1-dependent Al resistance in Arabidopsis thaliana. New Phytol. 233, 2471–2487. doi: 10.1111/nph.17812
Keywords: calmodulin, calmodulin-like, barley, expression, pan-genome, abiotic stress
Citation: Cai K, Kuang L, Yue W, Xie S, Xia X, Zhang G and Wang J (2022) Calmodulin and calmodulin-like gene family in barley: Identification, characterization and expression analyses. Front. Plant Sci. 13:964888. doi: 10.3389/fpls.2022.964888
Edited by:
Zhong-Hua Chen, Western Sydney University, AustraliaReviewed by:
Yanhao Xu, Hubei Academy of Agricultural Sciences, ChinaGuang Chen, Zhejiang Academy of Agricultural Sciences, China
Copyright © 2022 Cai, Kuang, Yue, Xie, Xia, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junmei Wang, MjF3YW5nam1Ac2luYS5jb20=
 Kangfeng Cai
Kangfeng Cai Liuhui Kuang
Liuhui Kuang Wenhao Yue
Wenhao Yue Shanggeng Xie
Shanggeng Xie Xue Xia
Xue Xia Guoping Zhang
Guoping Zhang Junmei Wang
Junmei Wang