
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 29 July 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.964621
This article is part of the Research Topic Heterologous Production of High Value Metabolites in Plants and Microbes View all 7 articles
VviNAC17 is a grapevine transcription factor activated by ABA. Because ABA has been proposed as the main signal modulating the secondary metabolism in grape berry skins, here we postulated VviNAC17 as a positive regulator of secondary metabolism in grape cells. To validate the hypothesis, VviNAC17 was constitutively and stably overexpressed in grape berry suspension-cultured cells of Gamay Fréaux cv. by Agrobacterium-mediated transformation. Targeted transcriptional analyses by qPCR showed that several genes involved the phenylpropanoid (VviPAL1), stilbenoid (VviSTS1) and flavonoid pathways (VviDFR, VviLAR1, VviANR, VviLDOX, and VviUFGT1), as well as anthocyanin vacuolar transport and accumulation (VviGST4 and VvMATE1) were significantly upregulated in VviNAC17-overexpressing transgenic cells, which translated in the stimulation of a number of enzymatic activities in those pathways. This was the case of phenylalanine ammonia lyase (PAL) and UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) that were about 2-fold and 3.5-fold higher in VviNAC17-overexpressing cells than in control cells. VviNAC17-overexpressing cells accumulated significantly higher amounts of anthocyanins, proanthocyanidins, total flavonoids and total phenolics. These findings confirmed that VviNAC17 is an important positive regulator of secondary metabolism in grapevine contributing to the accumulation of important berry quality-related secondary metabolites.
Grapevine (Vitis vinifera L.) is a fruit crop with enormous economic importance worldwide (Bouquet et al., 2006). Wine industry is fundamental from both an economic and social standpoint, so, increasing knowledge about the biochemical and physiological features underlying the quality of grape berries, as well as about strategies for its improvement, is always a constant demand. This is especially important in the context of the current climate changes, responsible for alterations in the viticultural map but also for increasing environmental constraints. These have increasingly been tentatively mitigated through practical short-term strategies such as exogenous application of protective compounds as the kaolin for instance (Conde et al., 2016, 2018; Garrido et al., 2019), but also through long-term strategies as plant breeding (Ollat et al., 2018).
Plant primary metabolism is an essential biochemical process in the regulation of plant nutrition, while secondary metabolism is strongly involved in plant defense processes, such as biotic and abiotic stress responses. Secondary metabolites are very important in the modulation of fruit quality characteristics and, consequently, in the quality of wine. In this regard, phenolics, including their subgroups as flavonoids (e.g., anthocyanins and proanthocyanidins), are crucial to obtaining a variety of wine attributes such as color/pigmentation, aroma, flavor, astringency, and texture (Teixeira et al., 2013). The initial step of phenolic compounds synthesis occurs via the general phenylpropanoid pathway (GPP) and is followed by a series of metabolic reactions. The products originating from GPP are used as substrates by three distinctive pathways and converted into stilbenes, flavonoids, and phenolic acids (Deng and Lu, 2017; Rienth et al., 2021). The primary substrate of GPP, phenylalanine, is converted to p-coumaroyl-CoA via PAL, cinnamic acid-4-hydroxylase (C4H) and p-coumaric:CoA ligase (4CL). The product of GPP, p-coumaroyl-CoA, is further used as a substrate by stilbene synthase (STS) and chalcone synthase (CHS) in the first steps of the stilbenoid and flavonoid pathway, respectively. In the grapevine genome 45 stilbene synthases genes (STS) are currently described (Vannozzi et al., 2012). The biosynthesis of stilbenes occurs under the control of VviMYB14 and VviMYB15, two R2R3 MYB TFs (Höll et al., 2013; Rienth et al., 2021). In a competitive manner with stilbene synthases, chalcone synthases (CHS) are responsible for the first step of the flavonoid synthesis pathway to produce flavanones, such as naringenin (Ferrer et al., 2008). Flavanones are converted by flavanone-3β-hydroxylase (F3H) into dihydroflavonols. This class of compounds is used to produce flavonol aglycones via flavonol synthase (FLS) and it is regulated by the light-induced transcription factor VviMYBF1 (Downey et al., 2003; Czemmel et al., 2009; Revilla et al., 2018). By the competitive action of dihydroflavonol-4-reductase (DFR), dihydroflavonols are reshuffled away from flavonol biosynthesis branch and transformed into leucoanthocyanidins, which might be either converted by leucoanthocyanidin dioxygenase (LDOX) or leucoanthocyanidin reductase (LAR) into anthocyanidins or flavan-3-ols, respectively (Bogs et al., 2005; Czemmel et al., 2012). In grapevine, the regulation of VviLAR1 and VviANR occurs consistently with the expression of two transcription factors, VviMYBPA1 and VviMYBPA2 (Bogs et al., 2005; Terrier et al., 2009; Rienth et al., 2021). Since the anthocyanidins are highly unstable molecules, these are immediately transformed either into anthocyanins or epicatechin by the competitive action of UDP-glucose: flavonoid-3-O-glucosyltransferase (UFGT) and anthocyanidin reductase (ANR; Bogs et al., 2005; Walker et al., 2007; Czemmel et al., 2012). The two most important transcriptional regulators of berry anthocyanin biosynthesis are the TFs VviMYBA1 and VviMYBA2 (Kobayashi et al., 2004; Walker et al., 2007; Rienth et al., 2021). The produced anthocyanins are transported across the tonoplast by primary or secondary transporters and accumulated in the vacuole. ATP-binding cassette (ABC), as is the case of VviABCC1, is responsible for the translocation of glycosylated anthocyanins (Francisco et al., 2013), and multidrug and toxic extrusion (MATE) transporters, like VviMATE1, are associated with the translocation of anthocyanin-acylglucosides (Gomez et al., 2009). Furthermore, glutathione S-transferases (VviGSTs), such as VviGST1 and VviGST4, have been implicated in the stabilization and transport of anthocyanins to the vacuole (Pérez-Díaz et al., 2016).
The secondary metabolism is strongly linked to abiotic stress, especially to abscisic acid (ABA)-mediated responses (Ferrandino and Lovisolo, 2014). The mechanisms involving the abscisic acid perception and signaling in grapevine have been continuously studied over the last decade, leading to the identification of several associated molecular mechanisms such as ABA receptors, PP2Cs and SnRK2 kinases, NAC transcription factors (TFs) and AREB/ABF (ABF1, AREB1/ABF2, ABF3, AREB2/ABF4; Kang et al., 2002; Gambetta et al., 2010; Boneh et al., 2012a,b; Yoshida et al., 2015; Pilati et al., 2017; Ju et al., 2020).
NAC proteins are plant-specific transcriptional regulators implicated in the development of shoot apical meristems, floral organs and lateral shoots, as well as, in a very important manner, in the plant hormonal regulation, including ABA signaling and transduction and ABA-related defensive mechanisms (Ernst et al., 2004). These proteins include the NAC domain, an N-terminal module of ca. 160 amino acids, that is accompanied by diverse C-terminal transcriptional activation domains (Aida et al., 1997). The interaction between NAC regulatory proteins and hormone signaling is very complex, with numerous studies describing interactions with abscisic acid, jasmonic acid, salicylic acid, and ethylene-stress responses (Sun et al., 2013; Sakuraba et al., 2020; Figueroa et al., 2021). Therefore, NAC TFs are key intermediates responsible for the regulation of many downstream ABA-regulated targets. In grapevine, VviNAC17 promoter is induced by ABA via VviABF2 and, as VviABF2, and this TF, using transgenic Arabidopsis as a model, was shown to play as an activator of several molecular mechanisms regulated by ABA, as those involved in abiotic-stress tolerance responses (Pilati et al., 2017; Ju et al., 2020).
As ABA signaling is directly linked to the stimulation of several secondary metabolic pathways, in this study we hypothesized that VviNAC17 TF is a positive regulator of secondary metabolism in grapevine, stimulating the phenylpropanoid, flavonoid and stilbenoid pathways. To test this hypothesis, VviNAC17 was constitutively and stably overexpressed in grape berry suspension-cultured cells of Gamay Fréaux cv., as a homologous grape model, through Agrobacterium-mediated transformation, and its function in the overproduction of target metabolites in those pathways and in the regulatory mechanisms was assessed.
Grape berry cells of V. vinifera L. cv. Gamay Fréaux cv. Teinturier, previously obtained and established from dedifferentiation of grape berry mesocarp and kindly provided by Prof. Serge Delrot (ISVV, Université de Bordeaux), were maintained in both solid and liquid cultures containing 0.1% (w/v) Gamborg B5 medium, 3% (w/v) sucrose, 250 mg L−1 casein hydrolysate, 0.2 mg L−1 kinetin and 0.1 mg L−1 naphthaleneacetic acid (NAA), in 250 ml flasks with shaking at 100 rpm at 23°C, under a 8 h dark/ 16 h light (200 μmol photons m−2 s−1) photoperiod. Cell suspensions were maintained by sub-culturing every 7 days by the transfer of 10 ml of cultures into 40 ml of fresh culture medium. As solid culture, calli were routinely sub-cultured every month.
The open reading frame (ORF) of VviNAC17 was cloned from grape berries of cv. Touriga Nacional. Its sequence was constituted by 1,002 bp and encoded 334 amino acids according to annotated transcripts of GSVIVG01014403001 (https://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/). VviNAC17 was cloned using Gateway® technology (Invitrogen). Primers with the attB sequences (Supplementary Table 1) for site-specific recombination with the entry plasmid pDONR221 were utilized for PCR amplification. The stop codon was removed in the cloning process for allowing the subsequent VviNAC17 C-terminus fusion with egfp in the destination vector, for later confirmation of successful transformation and constitutive expression, as well as confirmation of subcellular localization studies. Subsequently, recombination of VviNAC17 containing attB sites with the entry plasmid was performed using BP clonase. VviNAC17 in the entry plasmid was then recombined into the pH7FWG2 using LR clonase. The correct assembly of the C-terminally fused to GFP construct pH7FWG2:VviNAC17 was confirmed by sequencing.
Agrobacterium strain GV3101 was made chemically competent and transformed with the binary construct pH7FWG2-VviNAC17 (Höfgen and Willmitzer, 1988). Bacterial cell culture of Agrobacterium tumefaciens containing the construct pH7FWG2-VviNAC17 was cultivated on LB solid medium at 30°C for 48 h. A single colony was inoculated in 50 ml of liquid LB medium that contained 25 mg L−1 rifampicin and 100 mg L−1 spectinomycin and was incubated overnight at 28°C with shaking (180 rpm). Fifty ml of LB medium, supplemented with the same antibiotics, was inoculated with 1 ml of this 1-day-old culture and grown at 30°C with shaking (with shaking at 180 rpm) until OD600 reached around ≈ 0.1–0.4 (Genesys™ 20, Thermo Scientific™), as described in the literature (Wise et al., 2006). The bacterial medium was removed by centrifugation at 5,000 rpm for 5 min and the resulting bacterial pellet was washed and suspended in 1 ml of liquid Gamborg B5 medium. The transformation of Gamay cells was performed according to Martínez-Márquez et al. (2015) with some modifications. A 7-day-old Gamay cell suspension-cultured in Gamborg B5 medium was used to promote an efficient infection ratio between A. tumefaciens and Gamay cells healthy cells.
The liquid plant cell preparation was supplemented with 100 mg L−1acetosyringone. Subsequently, the bacterial suspension was added, and the infected culture was incubated on a shaker (100 rpm) for 30 min in the dark at 24°C. The suspension was then filtered, and the retained biomass washed with cold Gamborg B5 medium. Gentle vacuum was applied to remove excess of medium. The biomass was transferred to solid Gamborg B5 medium containing 100 mg L−1acetosyringone. After 2 days of co-culture incubation in the dark at 25°C, cells were transferred to solid Gamborg B5 medium containing 250 mg L−1cefotaxime and 5 mg L−1of hygromycin and kept at 25°C in the dark. Routine subculturing of the growing callus colonies was performed using decreasing cefotaxime concentrations. After 6 months, cefotaxime was eliminated from the medium and the transformed Gamay cells were grown in 8 mg L−1 of hygromycin.
To analyze the subcellular localization of the VviNAC17 protein, transformed Gamay cells with the construct pH7FWG2-VviNAC17-EGFP were observed using fluorescence microscopy. Green fluorescence signals were observed and registered using the fluorescence microscope Leica BM5000B with an excitation wavelength of 488 nm and emission wavelength of 507 nm.
The extraction of anthocyanins was performed using 100 mg of grape berry cells from each experimental condition as described in Vale et al. (2021). The extraction process was performed using 90% (v/v) methanol and 10% (v/v) deionized water. The suspensions were vigorously vortexed for 10 min and left overnight at 4°C for improved extraction. The samples were centrifuged at 18000 × g for 20 min. The supernatants were recovered and 200 μl of each extract were mixed with 1.8 ml of 25 mM KCl (pH = 1.0). The absorbance of samples for each experimental condition was measured at 520 and 700 nm with a Shimadzu UV-160A spectrophotometer (Kyoto, Japan). Total anthocyanin concentration was assessed as cyanidin-3-glucoside (C-3-G) equivalents, as described in the following equation:
where MW is the molecular weight of C-3-G (449.2 g mol−1), DF corresponds to the dilution factor and ε is the molar extinction coefficient of C-3-G (26 900 M−1 cm−1). The concentration was subsequently presented per mg of dry weight (DW) of used cells.
Quantification of flavonoids was performed using the aluminum chloride colorimetric method described by Matejić et al. (2013) with some modifications. An assay mixture containing 20 μl of standard solutions (quercetin) or methanolic extracts from the experimental conditions, 30% (v/v) of ethanol, 0.2% (w/v) of aluminum chloride and 20 mM of potassium acetate was used. Following incubation at room temperature during 40 min in the dark, the absorbance of the reaction mixtures was measured at 415 nm with a Shimadzu UV-160A spectrophotometer (Kyoto, Japan). Total flavonoid concentration in methanolic extracts was calculated using a quercetin calibration curve (5–100 μg mL−1) and expressed as quercetin equivalents per gram of DW.
Proanthocyanidins (PAs) concentration was quantified resorting to an adapted colorimetric DMAC assay based on Wallace and Giusti (2010). For extraction, 1 ml of absolute methanol was added to 5 mg of lyophilized grape berry cells and vigorously shaken for 30 min followed by centrifugation at 18000 × g for 30 min. DMAC reagent was prepared immediately before use by dissolving 2% DMAC (w/v) in a 1:1 ratio of 6 N H2SO4 and methanol absolute solution. The reaction mixture containing 1.175 ml of methanol and 50 μl of 2% (w/v) DMAC was initiated by adding 10 μl of extract of each experimental condition. Each reaction was incubated at room temperature during 15 min in the dark and the absorbance was determined at 640 nm using a Shimadzu UV-160A spectrophotometer (Kyoto, Japan). Total proanthocyanidins concentration was determined using a calibration curve with (+)-catechin standard solutions (50–500 μg mL−1) and expressed as catechin equivalents per gram of DW.
The concentration of phenolics was quantified by Folin–Ciocalteu colorimetric method as previously performed in Conde et al. (2016). Total phenolics were extracted in 1.5 ml of 90% (v/v) methanol from 20 mg of lyophilized control and transformed grape berry cells. The homogenates were vigorously shaken for 30 min and left overnight at 4°C. Subsequently, each experimental condition was centrifuged at 18000 × g for 20 min. Twenty μl of each supernatant were mixed with 1.58 ml of deionized water and 100 μl of Folin reagent, shaken and incubated in the dark for 5 min before the addition of 300 μl of 2 M sodium carbonate. After 2 h of incubation in the dark, the absorbance was quantified at 765 nm (Shimadzu UV-160A spectrophotometer, Kyoto, Japan). Total phenolic concentrations were determined as gallic acid equivalents (GAE) per gram of dry weight.
Approximately 200 mg of fresh control and VviNAC17-overexpressing Gamay cells were used for total RNA extraction, via the method described by Reid et al. (2006) in combination with the GRS Plant Total RNA kit (GRISP). The extraction buffer was modified to 2% (w/v) cethyl-trimethylammonium bromide (CTAB), 2% (w/v) soluble polyvinylpyrrolidone (PVP) K-30, 300 mM Tris–HCl (pH 8.0), 25 mM EDTA, 2 M NaCl and 40 mM dithiothreitol (DTT). Following an in-column DNase I (Thermo Scientific™) treatment, the RNA integrity was confirmed in a 1% (w/v) agarose gel. Complementary DNA (cDNA) was synthesized from 1 μg of total RNA using the Xpert cDNA Synthesis Master-mix Kit (GRISP) according to the manufacturer’s instructions. RNA purity and concentration were assessed using NanoDrop.
Quantitative Real-time PCR analyses (qPCR) were performed using Xpert Fast SYBR Blue (GRISP) and a CFX96 Real-Time Detection System (Bio-Rad) using 1 μl of cDNA in a reaction volume of 10 μl per well. Experiments were carried out in biological triplicates using VviACT1(actin) and VviGAPDH (glyceraldehyde-3-phosphate dehydrogenase) as reference genes, verified as stable and ideal for qPCR normalization in grapevine (Reid et al., 2006). Melting curves were performed after each run to confirm the absence of unspecific and primer-dimer amplification. Gene expression values were normalized by the average of expression of the reference genes, as described by Pfaffl (2001), and analyzed using the CFX Manager Software 2.0 (Bio-Rad). Specific primer pairs used in this work are listed in Supplementary Table 2.
Total protein extraction from grape berry cells was performed as in Conde et al. (2016). Sample powder was vigorously mixed with extraction buffer at a1:2 (v/v) powder: buffer ratio. Protein extraction buffer contained 50 mm Tris–HCl pH 8.9, 5 mm MgCl2, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride (PMSF), 5 mm dithiothreitol (DTT), 0.1% (v/v) Triton X-100 and 1% of poly(vinylpolypyrrolidone; PVPP). Homogenates were vigorously mixed for 20 min and centrifuged at 18000 × g for 20 min at 4°C. The supernatants were kept on ice and used for all enzymatic assays. Protein concentration of the crude extracts was quantified by the Bradford method (Bradford, 1976), with bovine serum albumin as standard.
Maximum velocity (Vmax) of UDP-glucose:flavonoid 3-O-glucosyltransferase (UFGT) activity was determined as in Lister et al. (1996) and adapted by Conde et al. (2016). The assay mixture contained 300 mm Tris–HCl buffer pH 8.0 (optimal activity), 1 mM UDP glucose and 200 μl of enzyme extract, and 5 mm of DTT in a reaction volume of 1 ml. The reaction was started with 1 mm quercetin as substrate, thus at a saturating concentration. The reaction mixture was kept in the dark under gentle shaking for 30 min. Following incubation, dilutions were prepared with 100 μl of each assay mixture and 900 μl of Tris–HCl buffer and the production of quercetin-3-glucoside was assessed at 350 nm (ε = 21,877 M−1 cm−1) with a Shimadzu UV-160A spectrophotometer (Kyoto, Japan).
Maximum velocity (Vmax) of phenylalanine ammonia lyase (PAL) activity was measured as in Conde et al. (2016). The assay mixture contained 0.2 ml of enzyme crude extract and 3.6 mm NaCl in 50 mm Tris–HCl pH 8.9 of reaction buffer in a reaction volume of 1 ml. Reactions were started by adding 25 mm of L-phenylalanine. The reaction mixture was kept at 41°C in the dark under gentle shaking for 30 min and the production of trans-cinnamic acid was assessed at 290 nm (ε = 17,400 M−1 cm−1).
Except for the results pictured in Figure 1D, for which a one-way ANOVA test and Tukey’s post-hoc test were used, results were analyzed by Student’s t-test using GraphPad Prism 9 software (GraphPad Software Inc., United States). The results are presented as mean ± standard deviation of the mean (SD), and statistical comparisons were made with a 95% confidence interval. Parametric testing was used as all data sets have Gaussian distributions. Statistical significance is indicated in the figures according to value of p, with p < 0.05 (*), p < 0.01 (**), p < 0.001(***) or p < 0.0001(****). For every experimental approach, at least three biological replicates were used.
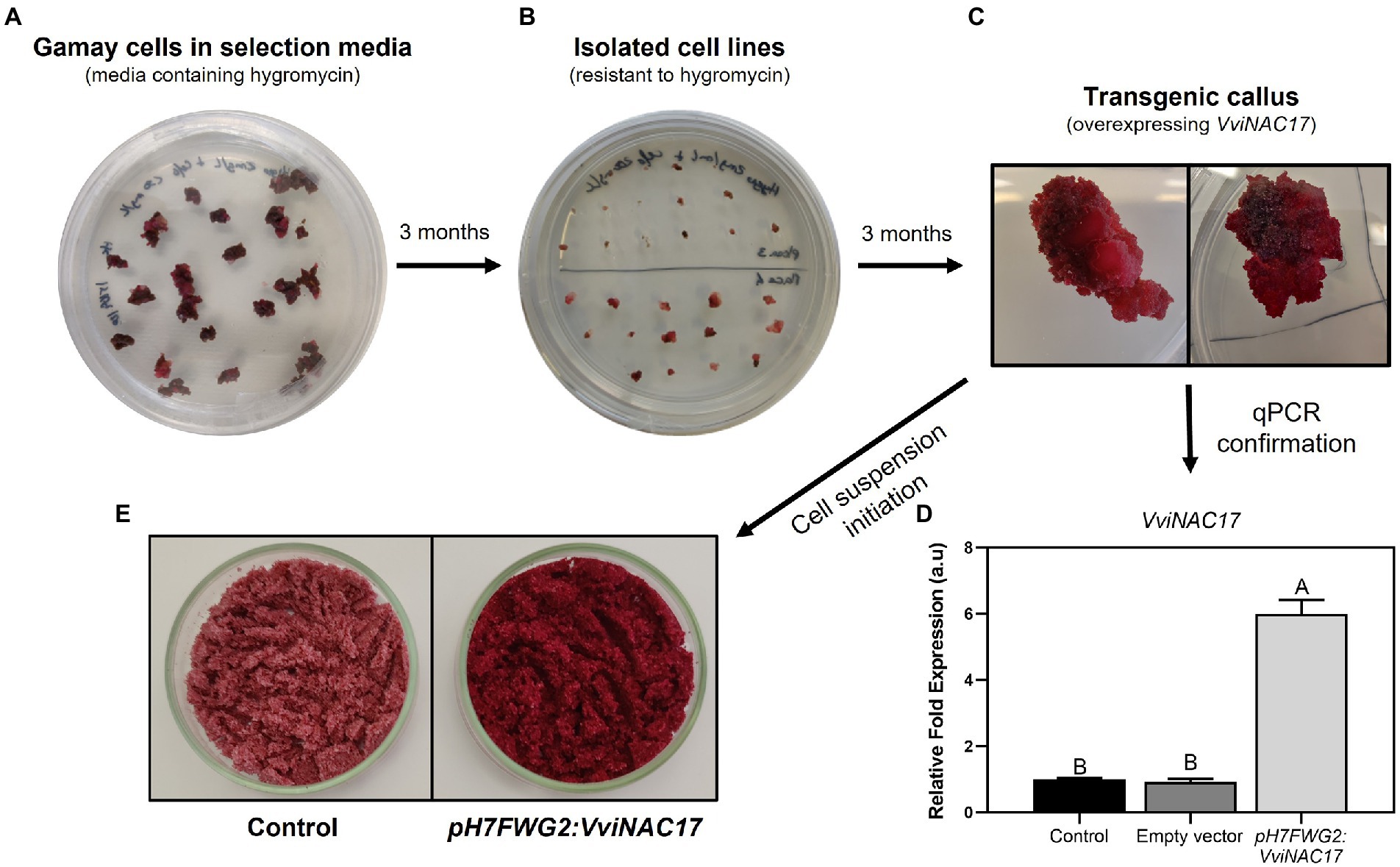
Figure 1. Transformation of non-embryogenic cell culture of Vitis vinifera Gamay overexpressing the transcription factor VviNAC17. (A) Transformed callus obtained after infection with Agrobacterium tumefaciens in selection media containing hygromycin and cefotaxime. (B) Multiple isolated cell lines obtained after 3 months of growth in selection media containing hygromycin and cefotaxime. (C) Transgenic callus after 3 months of growth. (D) Real-time qPCR demonstrating the upregulation of VviNAC17 in pH7FWG2-VviNAC17 Gamay cells. Statistical differences are denoted by different letters and were determined by one-way ANOVA and Tukey’s post hoc test. (E) Cell suspensions of control and pH7FWG2-VviNAC17-containing Gamay cells filtered after 7 days of growth in Gamborg B5 medium.
The transgenic grape cell line constitutively expressing VviNAC17, under the control of 35S cauliflower mosaic virus (CaMV) promoter, was obtained and established after transformation with the A. tumefaciens GV3101 strain, previously transformed with the construct pH7FWG2:VviNAC17 (Figures 1A–C). Six months after transformation, Hygromycin-selected Gamay transgenic and non-transformed calli were checked for plant genome T-DNA integration of VviNAC17 by PCR amplification using a specific primers pair for amplification of the promoter 35S (Supplementary Table 1), and its detection was confirmed to occur only in transgenic clones (Supplementary Figure 1). A transcriptional analysis by real-time qPCR confirmed the overexpression of VviNAC17 in the transgenic callus transformed with pH7FWG2:VviNAC17 (Figure 1D). The transgenic callus with the highest VviNAC17 transcripts abundance was selected for further studies. Six months after the initial transformation and selection in media containing hygromycin and cefotaxime, sufficient transgenic callus was obtained to initiate a suspension culture (Figure 1E).
Besides evident phenotypic changes marked by a deep and more intense purple color than control Gamay cells, the transgenic VviNAC17-overexpressing suspension culture interestingly exhibited a substantial increase in cell growth rate when compared to non-transformed cell lines when cultivated in Gamborg B5 medium (Supplementary Figure 2).
To further confirm VviNAC17-GFP recombinant protein synthesis in the transgenic cultures, a small fraction of tissue from transgenic calli was observed in a fluorescence microscope. As shown in Figure 2, green fluorescence was clearly observed in the nucleus of transgenic VviNAC17-overexpressing Gamay cells confirming the constitutive expression of VviNAC17 in transgenic cells, and also indicating its subcellular localization in the nucleus as expected. The fluorescence signal was absent in non-transformed cells.
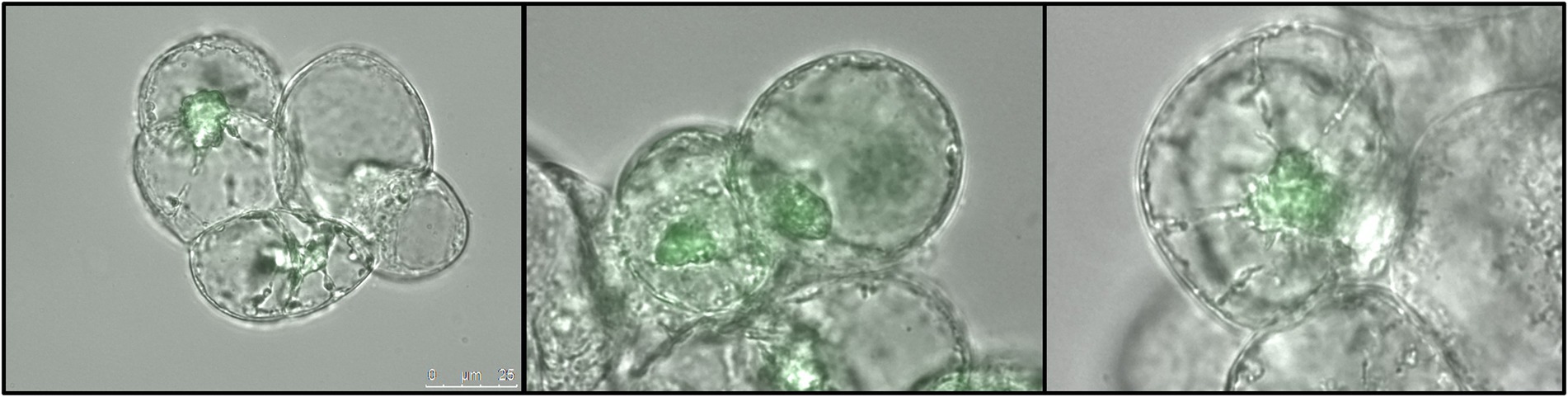
Figure 2. Subcellular localization of transcription factor VviNAC17 in V. vinifera Gamay transgenic cells. Distribution of the transcription factor VviNAC17 through the nucleus of transgenic VviNAC17-overexpressing Gamay cells 6 months after transformation with A. tumefaciens with the binary vector pH7FWG2-VviNAC17. Green fluorescence observed by fluorescence microscopy indicated the presence and expression of VviNAC17-GFP fusion protein in the nucleus and further confirmed the successful transformation of the cells. Bar of photographs—25 μm.
These analyses revealed a significant increase in the concentration of these compounds in the transgenic VviNAC17-overexpressing cells. Indeed, the concentration of anthocyanins in pH7FWG2-VviNAC17 berry cells was 19.96 mg of cyanidin-3-glucoside equivalents per gram of cells dry weight compared to 7.88 mg of cyanidin-3-gluoside equivalents per gram of dry weight (DW) in control cells, which corresponded approximately to a 2.5-fold increase (Figure 3A). The concentration of proanthocyanidins in control cells was 7.22 mg of (+)-catechin equivalents per gram of DW compared to 23.88 mg of (+)-catechin equivalents per gram of DW in VviNAC17-overexpressing cells (Figure 3B; 3.5-fold higher). VviNAC17-overexpressing cells also had a 2.5-fold higher concentration of flavonoids, reaching 4.88 mg of quercetin equivalents per gram of DW (Figure 3C). The concentration of total phenolics in transformed cells was also increased by approximately 2.5-fold, reaching 219.84 mg of gallic acid equivalents per gram of DW (Figure 3D).
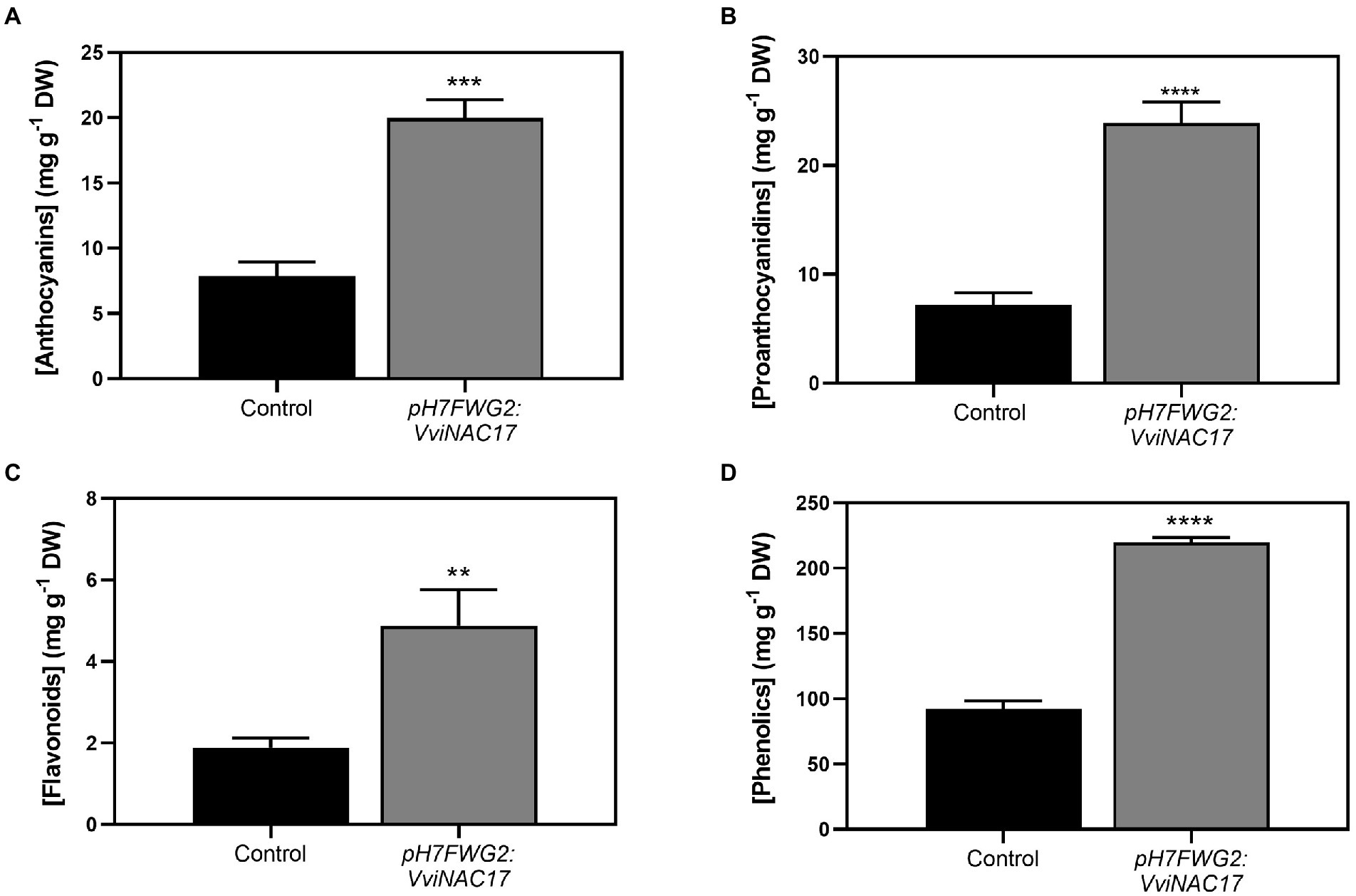
Figure 3. Effect of the overexpression of VviNAC17 on total anthocyanin (A), proanthocyanidins (B) flavonoid (C) and phenolics (D) content in transgenic cell suspensions of Gamay. Total anthocyanin concentration is represented by cyanidin-3-glucoside equivalents per g of dry weight (DW). Total proanthocyanidin concentration is represented by (+)-catechin per g of dry weight (DW). Total flavonoids concentration is represented by quercetin equivalents per g of dry weight (DW). Total phenolics concentration is represented by gallic acid equivalents (GAE) per g of dry weight (DW). Values are the mean ± SD of three biological replicates. Asterisks indicate statistical significance between non-transformed cells (control) and transformed Gamay cells (pH7FWG2-VviNAC17). (Student’s t-test; **p ≤ 0.01, ***p ≤ 0,001 and ****p ≤ 0.0001).
VviNAC17-overexpressing Gamay cells exhibited a 2-fold higher PAL specific activity when compared to control cells (Figure 4A), reaching a Vmax of 0.009 μmol min−1mg protein−1. In accordance with the biochemical activity of PAL, the expression of phenylalanine ammonia lyase 1 (VviPAL1) gene was approximately 3.5-fold higher in transgenic Gamay cells than in control cells (Figure 4B). The transcription of stilbene synthase 1 (VviSTS1), the most expressed STS gene in grapevine (Ciaffi et al., 2019), increased up to 4-fold in VviNAC17-overexpressing Gamay cells, in synchrony with the increase in the expression of the transcription factors VviMYB14 and VviMYB15, both positive regulators of VviSTS1 and other stilbene synthases in grapevine (Höll et al., 2013; Figure 5).
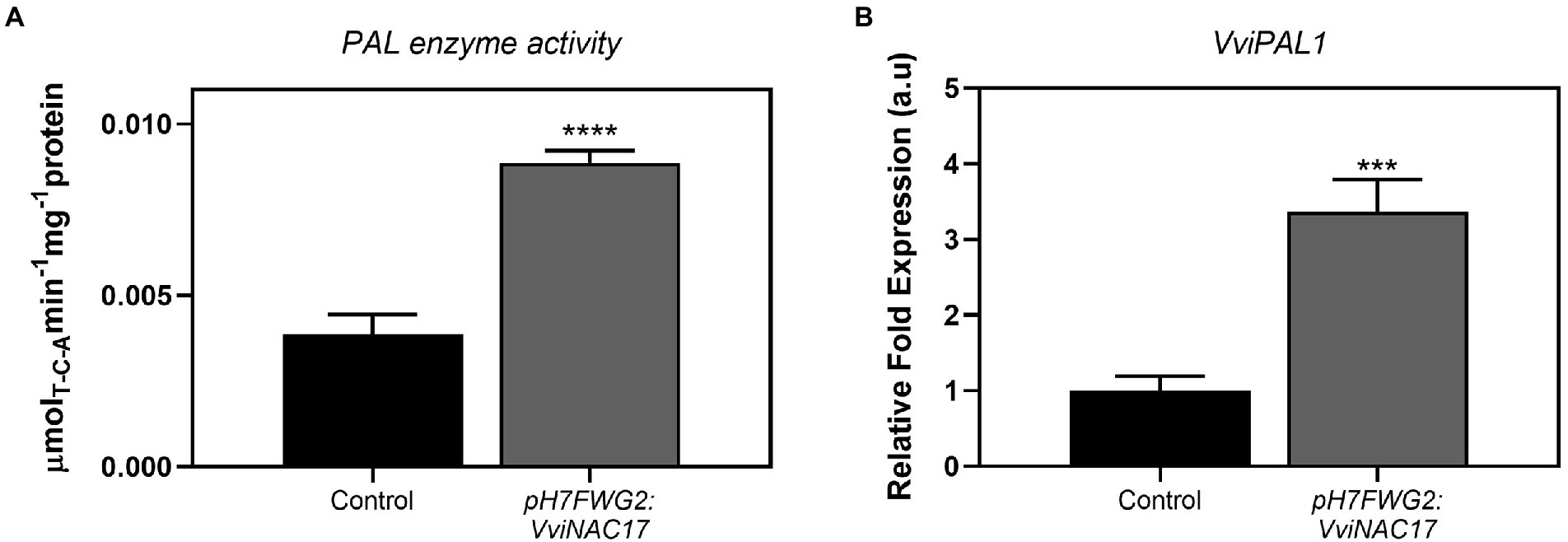
Figure 4. Stimulatory effect of the overexpression of VviNAC17 in Gamay cells on the phenylpropanoid pathway. (A) Phenylalanine ammonia lyase (PAL) enzyme activity determined as Vmax in non-transformed (control) and VviNAC17-overexpressing Gamay cells (pH7FWG2-VviNAC17). Values are the mean ± SD of five biological replicates. Asterisks indicate statistical significance (Student’s t-test; ****p ≤ 0.0001). (B) Transcript abundance of phenylalanine ammonia lyase 1 (VviPAL1) in non-transformed (control) and VviNAC17-overexpressing Gamay cells (pH7FWG2:VviNAC17). Gene expression analysis was performed by real-time qPCR in non-transformed (control) and VviNAC17-overexpressing Gamay cells (pH7FWG2:VviNAC17). VviPAL1 relative expression levels were obtained after normalization with the expression of the reference genes VviACT1 and VviGAPDH. Values are the mean ± SD of three biological replicates. Asterisks indicate statistical significance (Student’s t-test; ***p ≤ 0.001, ****p ≤ 0.0001).
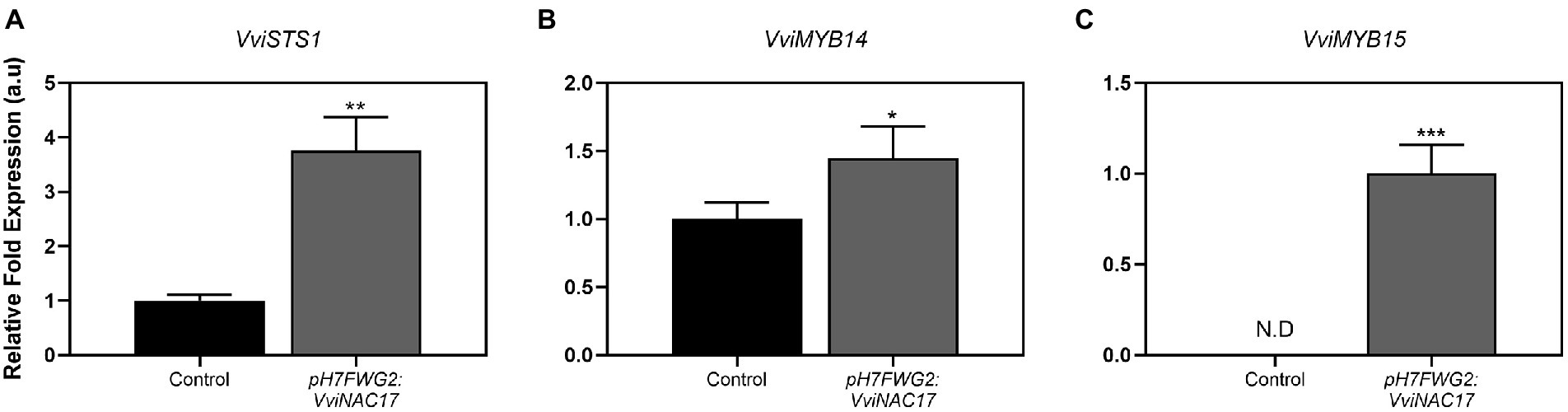
Figure 5. Effect of VviNAC17 overexpression in the transcript levels of grapevine stilbene synthase 1 (VviSTS1) (A) and transcription factors VviMYB14 (B) and VviMYB15 (C). Gene expression analysis was performed by real-time qPCR in non-transformed (control) and VviNAC17-overexpressing Gamay cells (pH7FWG2:VviNAC17). VviSTS1, VviMYB14 and VviMYB15 relative expression levels were obtained after normalization with the expression of the reference genes VviACT1 and VviGAPDH. Values are the mean ± SD of three biological replicates. Asterisks indicate statistical significance (Student’s t-test; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
The activity of UFGT enzyme was 3-fold higher in cells overexpressing VviNAC17 when comparing with control cells, reaching a Vmax of 0.003 μmol min−1mg protein−1 (Figure 6A), in agreement with the significant increase in the total concentration of anthocyanins previously depicted in Figure 3A. In the same way, the expression of VviUFGT1 was strongly stimulated in VvNAC17-overexpressing Gamay cells, reflected by a 4-fold increase in the transcripts abundance (Figure 6B). Two transcription factors associated with the positive regulation of VviUFGT1, VviMYBA1 and VviMYBA2, also displayed a significantly higher expression in transformed cells, by 2.5-and 3-fold, respectively (Gollop et al., 2002; Figures 6C,D).
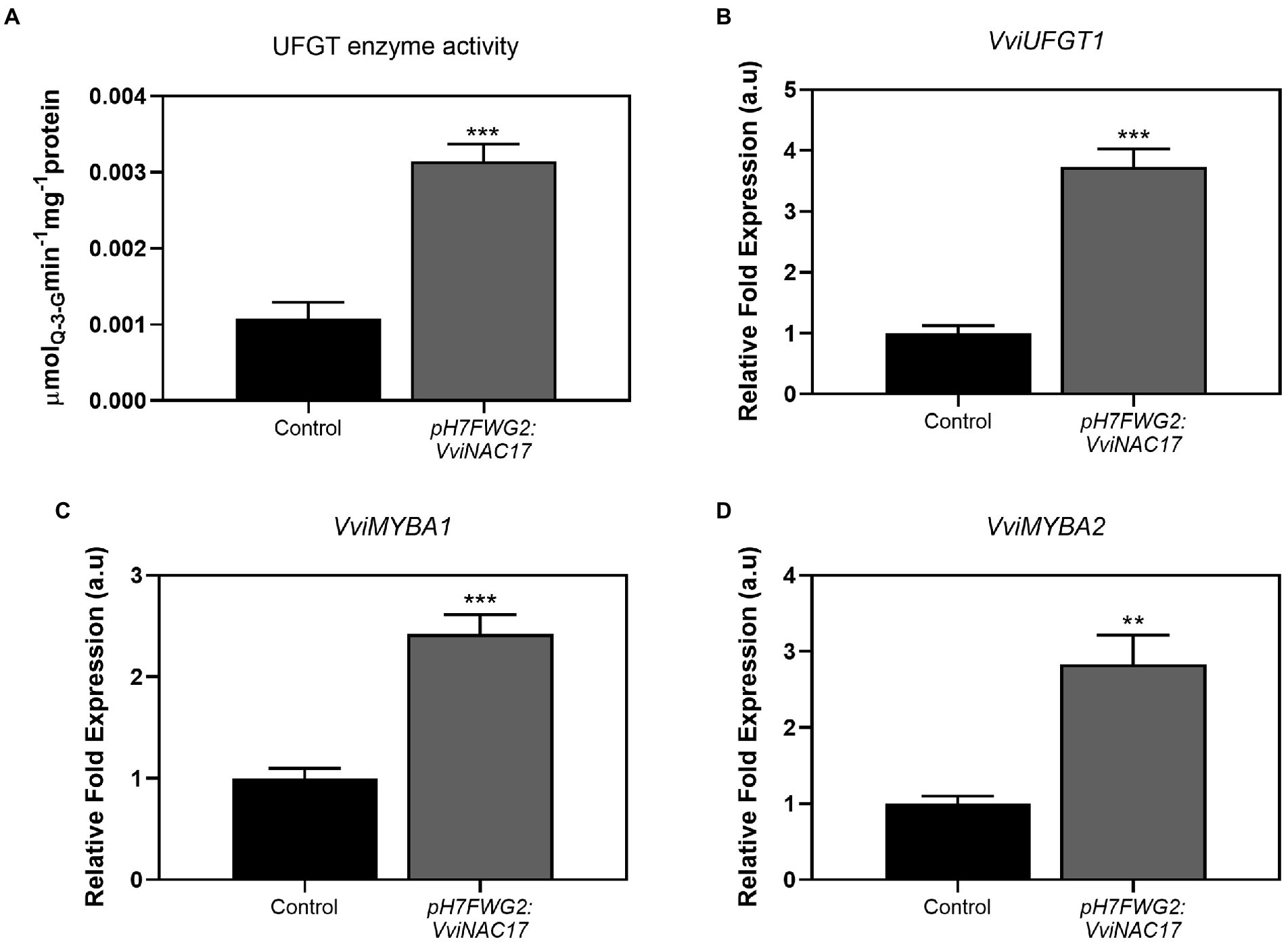
Figure 6. Stimulatory effect of the overexpression of VviNAC17 on the synthesis of anthocyanins. (A) UDP-glucose:flavonol 3-O-glucosyltransferase (UFGT) total enzyme activity determined as Vmax in non-transformed (control) and VviNAC17-overexpressing Gamay cells (pH7FWG2:VviNAC17). Values are mean ± SD of the five biological replicates. Asterisks indicate statistical significance (Student’s t-test; ***p ≤ 0.001). (B) Transcript levels of UDP-glucose:flavonol 3-O-glucosyltransferase (VviUFGT1), VviMYBA1 (C) and VviMYBA2 (D) from R2R3 MYB family in non-transformed (control) and VviNAC17-overexpressing Gamay cells. VviUFGT1, VviMYBA1 and VviMYBA2 relative expression levels were obtained after normalization with the expression of the reference genes VviACT1 and VviGAPDH. Values are presented as mean ± SD of three biological replicates. Asterisks indicate statistical significance (Student’s t-test; **p ≤ 0.01, ***p ≤ 0.001).
A significant stimulation of VviDFR transcripts, corresponding a 2-fold increase, was observed in transformed Gamay cells when compared to the control (Figure 7A). The transcripts abundance of VviLDOX were also significantly higher in transformed cells compared to control cells, corresponding a 2.5-fold increment in cells overexpressing VviNAC17 (Figure 7B). Specific targets associated with the synthesis of flavan-3-ols were also upregulated in transgenic cells, namely VviLAR1 and VviANR as shown in Figures 7C,D. The expression of the gene encoding for the transcription factor VvMYBPA1, involved in the activation of LAR and ANR was also strongly upregulated in VviNAC17-overexpressing cells (Figure 7E).
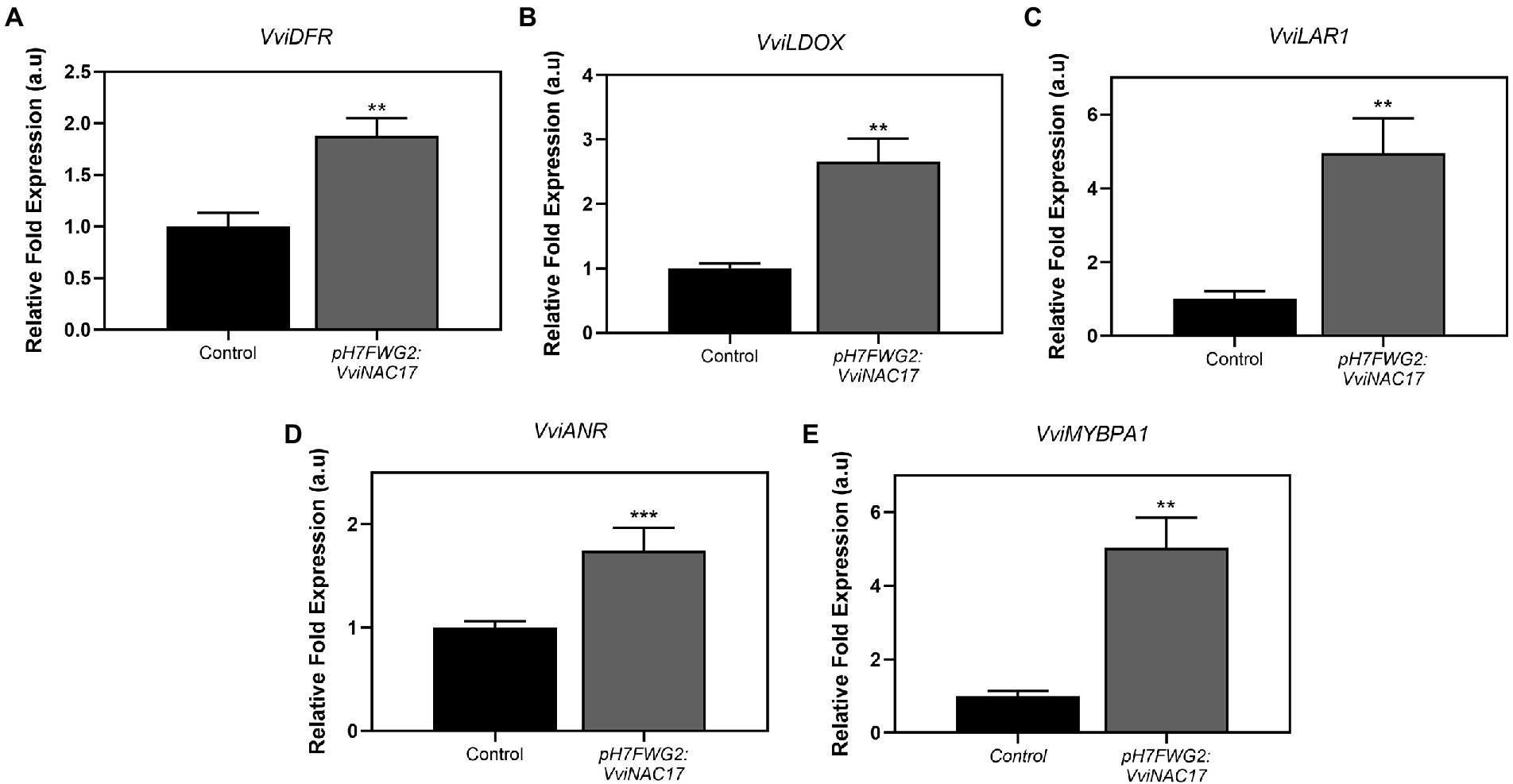
Figure 7. Stimulatory effect of the overexpression of VviNAC17 in the transcript levels of grapevine dihydroflavonol-4-reductase (VviDFR) (A), leucoanthocyanidin dioxygenase (VviLDOX) (B), leucoanthocyanidin reductase 1 (VviLAR1) (C), anthocyanidin reductase (VviANR) (D) and VviMYBPA1 (E). VviDFR, VviLDOX, VviLAR1, VviANR and VviMYBPA1 relative expression levels in non-transformed (control) and VviNAC17-overexpressing Gamay cells were obtained after normalization with the expression of the reference genes VviACT1 and VviGAPDH. Values are presented as mean ± SD of three biological replicates. Asterisks indicate statistical significance (Student’s t-test; **p ≤ 0.01).
The expression of VviGST4 was significantly higher in transformed cells, with a 2.5-fold increase in transgenic cells compared to control cells (Figure 8A). This enzyme represents a crucial step in anthocyanin stabilization through the conjugation with a reduced form of glutathione (GSH), essential for most of the anthocyanin vacuolar transport (Conn et al., 2008). Gene expression of the tonoplast anthocyanin transporter VviMATE1 was also highly upregulated in cells overexpressing VviNAC17, by 2-fold, compared to control cells (Figure 8B).
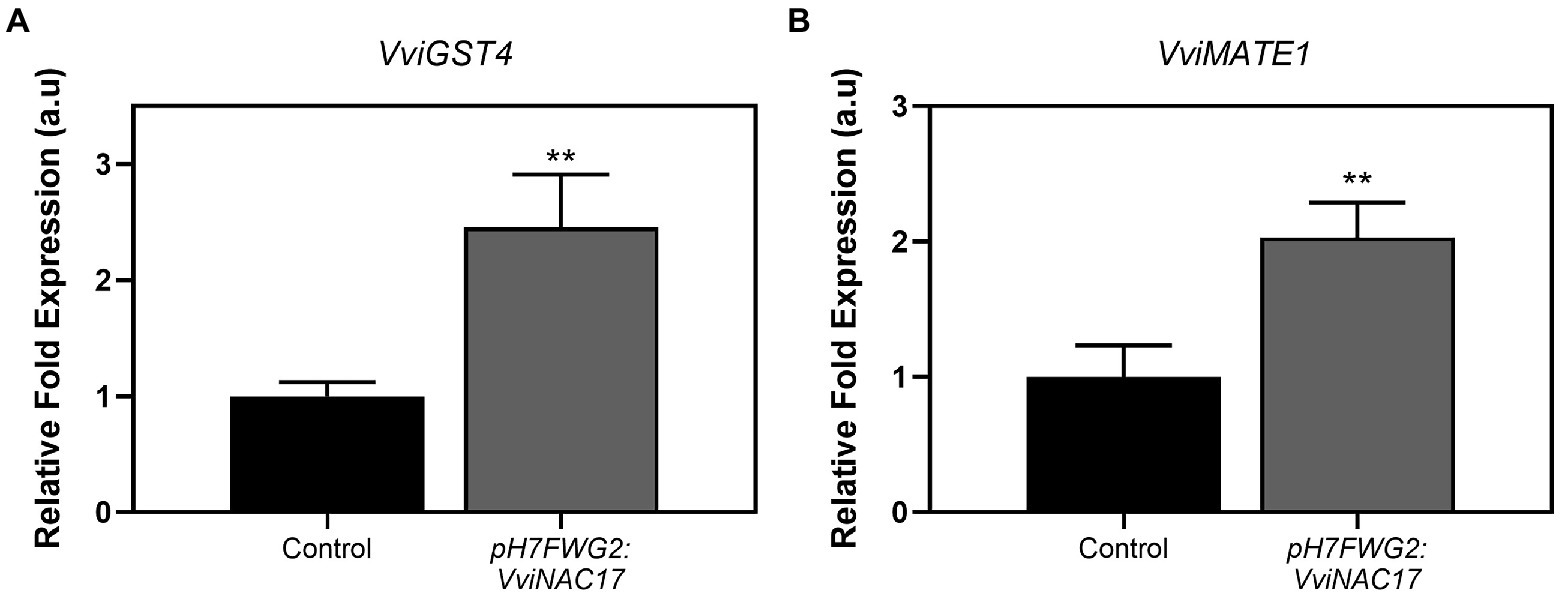
Figure 8. Stimulatory effect of the overexpression of VviNAC17 in the transcript levels of genes involved in anthocyanin S-conjugation (VviGST4) (A) and vacuolar transport capacity (VviMATE1) (B). VviGST4 and VviMATE1 relative expression levels in non-transformed (control) and VviNAC17-overexpressing Gamay cells were obtained after normalization with the expression of the reference genes VviACT1 and VviGAPDH. Values are presented as mean ± SD of three biological replicates. Asterisks indicate statistical significance (Student’s t-test; **p ≤ 0.01).
As mentioned in the Introduction section, using Arabidopsis as a model, VviNAC17 was recently described as a stress-responsive gene regulated by ABA and implicated in resistance to abiotic stress, by activating several stress-responsive genes (Pilati et al., 2017; Ju et al., 2020). However, the role of VviNAC17 in the regulation of secondary metabolism, hence, several ABA-regulated grape berry quality-related molecular mechanisms was so far unknown. Here, using grape berry cells of Gamay Fréuax cv. as homologous model, by successfully constitutively and stably expressing VviNAC17, we demonstrated that this TF is a positive regulator of several secondary metabolism biochemical steps through the activation of key genes involved in phenylpropanoid and flavonoid pathways, using transcriptional and biochemical analysis. Indeed, the synthesis and concentration of some of the main classes of secondary metabolites synthesized in the above-mentioned biochemical pathways, namely, total phenolics, flavonoids and, among them, total anthocyanins and total proanthocyanidins, were significantly increased in Gamay grape berry cells constitutively overexpressing VviNAC17. In fact, VviNAC17-overexpressing Gamay cells exhibited an impressive concentration of phenolic compounds of over 200 mg g DW−1 in a clear demonstration of a very active secondary metabolism induced by VviNAC17. This concentration is significantly higher than, for instance, the 40 mg g DW−1 phenolics, previously observed in Gamay suspension-cultured cells (Noronha et al., 2020). Also remarkable was the fact that despite the notorious increase in the concentration of these secondary metabolites, VviNAC17-overexpressing cells displayed a higher growth rate than control cells. This demonstrates that the stimulation of secondary metabolism did not result from a tradeoff with a slower cell growth. While this is apparently surprising, it may be an indicator that the uptake of carbon/energy sources and/or primary metabolism was also enhanced by VviNAC17 overexpression, a hypothesis that needs to be further explored.
Among this extensive secondary metabolism upregulation, the stimulation of the phenylpropanoid pathway was notorious in pH7FWG2:VviNAC17-overexpressing cells as confirmed by the higher expression of VviPAL1 compared to non-transformed cells, together with the higher PAL enzyme activity in transgenic cells, that is the result from the joint activity of all isozymes. Also, the upregulation of genes associated with the biosynthesis of stilbenes was evident in cells overexpressing VviNAC17, particularly the VviSTS1, and two TFs associated with the positive regulation of this particular pathway, VviMYB14 and VviMYB15 (Höll et al., 2013). While the upregulated expression of VviSTS1 and of its activating TFs VviMYB14 and VviMYB15 is suggestive of a complete stimulation of the stilbenoid pathway in VviNAC17-overexpressing cells that translates into increased amounts of stilbenes, this cannot be unequivocally concluded, as the total concentration of stilbenes was not quantified in this study and transcriptional modifications are not always translated into metabolic changes.
As referred to above, the total amount of flavonoids was higher in VviNAC17-overexpressing cells. This group of compounds includes anthocyanins, a group of water-soluble flavonoid compounds produced by almost vascular plants, and known by a diverse range of biological functions (Rinaldo et al., 2015). In grapevine, anthocyanins are responsible for berry color, one of the most important quality traits of fruit and red wines production. The increased content of anthocyanins in VviNAC17-overexpressing cells can be explained by the higher UFGT enzyme activity and VviUFGT1 expression levels in transgenic cells, a gene involved in anthocyanin biosynthesis by conversion of glycolates anthocyanidins into anthocyanins. Also, the initiation of UFGT expression is specifically regulated by two transcription factors, VviMYBA1 and VviMYBA2 (Walker et al., 2007). These two TFs were also upregulated in transgenic cells, which also corroborates with the higher expression levels of VviUFGT1 and with the higher anthocyanin content in cells overexpressing VviNAC17. The regulatory mechanism between VviNAC17 and MYB TFs is not well described, however several are the studies indicating a complex signaling network between members of NAC and MYB family (Ye et al., 2018; Chen et al., 2021).
The enzyme DFR is specific for the anthocyanin/proanthocyanidins pathway through the production of leucoanthocyanidins. This product is used as substrate by the enzyme LDOX, by converting leucoanthocyanidins to anthocyanidins. By transcriptional analysis, we demonstrated an upregulation of these two genes on transgenic cells, which supports the idea that the VviNAC17 is intrinsically associated to the regulation of secondary metabolism, namely on those targets implicated in the synthesis of anthocyanins.
The third main class of flavonoids family, the proanthocyanidins (flavan-3-ols), have been demonstrated to be very active as defense molecules to protect plants against stress but also have beneficial health effects for humans (Dixon et al., 2005). Also, grapevine berry mesocarp and seed derived proanthocyanidins greatly affect the organoleptic properties of the red wine – associated with increased content, degree of polymerization and galloylation of proanthocyanidins (Vidal et al., 2003; Yu et al., 2019). In this study, transgenic cells overexpressing VviNAC17 transcription factor exhibited upregulated levels of key genes implicated in proanthocyanidins biosynthesis, namely VviLAR1, VviANR and VviMYBPA1, but, even more importantly, a significantly increased concentration of PAs.
Anthocyanins are accumulated in the vacuole via primary transporters from the ABC family (Francisco et al., 2013) and tonoplast secondary transporters like VviMATE1 of multidrug and toxic extrusion family (Gomez et al., 2009). Also, members of multigenic GST family have been described to be crucial for anthocyanin accumulation. In grapevine, only VviGST1 and VviGST4 are involved in anthocyanin accumulation in the vacuole (Pérez-Díaz et al., 2016). Namely, VviGST4 is crucial for anthocyanin stabilization and transport, by promoting anthocyanin S-conjugation with reduced glutathione (Conn et al., 2008). In this study, we observed an increased expression of genes related with stabilization and transportation of anthocyanins to the vacuole, specifically, VviMATE1 and VviGST4, respectively. Cutanda-Perez et al. (2009) demonstrated that the ectopic expression of Vitis labrusca VlMYBA1-2 activates the gene expression linked to anthocyanin MATE transporter and GST. For this reason, the upregulation of VviMYBA1 and VviMYBA2 might also explain the increased levels of transcripts related with anthocyanin transport. On the other hand, the expression of VviABCC1 was not affected by the overexpression of VviNAC17 in transgenic cells.
In sum, we demonstrated that VviNAC17 is a transcription factor associated with a broad positive regulation of secondary metabolism in grapevine, particularly on the pathways regulated by ABA. We observed that Gamay cells overexpressing VviNAC17, together with significantly higher concentrations of total phenolics, flavonoids, anthocyanins and proanthocyanidins, have an upregulation of the expression of genes associated with phenylpropanoid, stilbene and flavonoid pathways. Also, genes encoding to the machinery involving the accumulation of anthocyanins in the vacuole were also upregulated in transgenic cells (Figure 9). The proper regulation of these metabolic pathways is crucial to ensure better berry quality, that consequently is translated into better wine quality and production of better products by the winemaking industry.
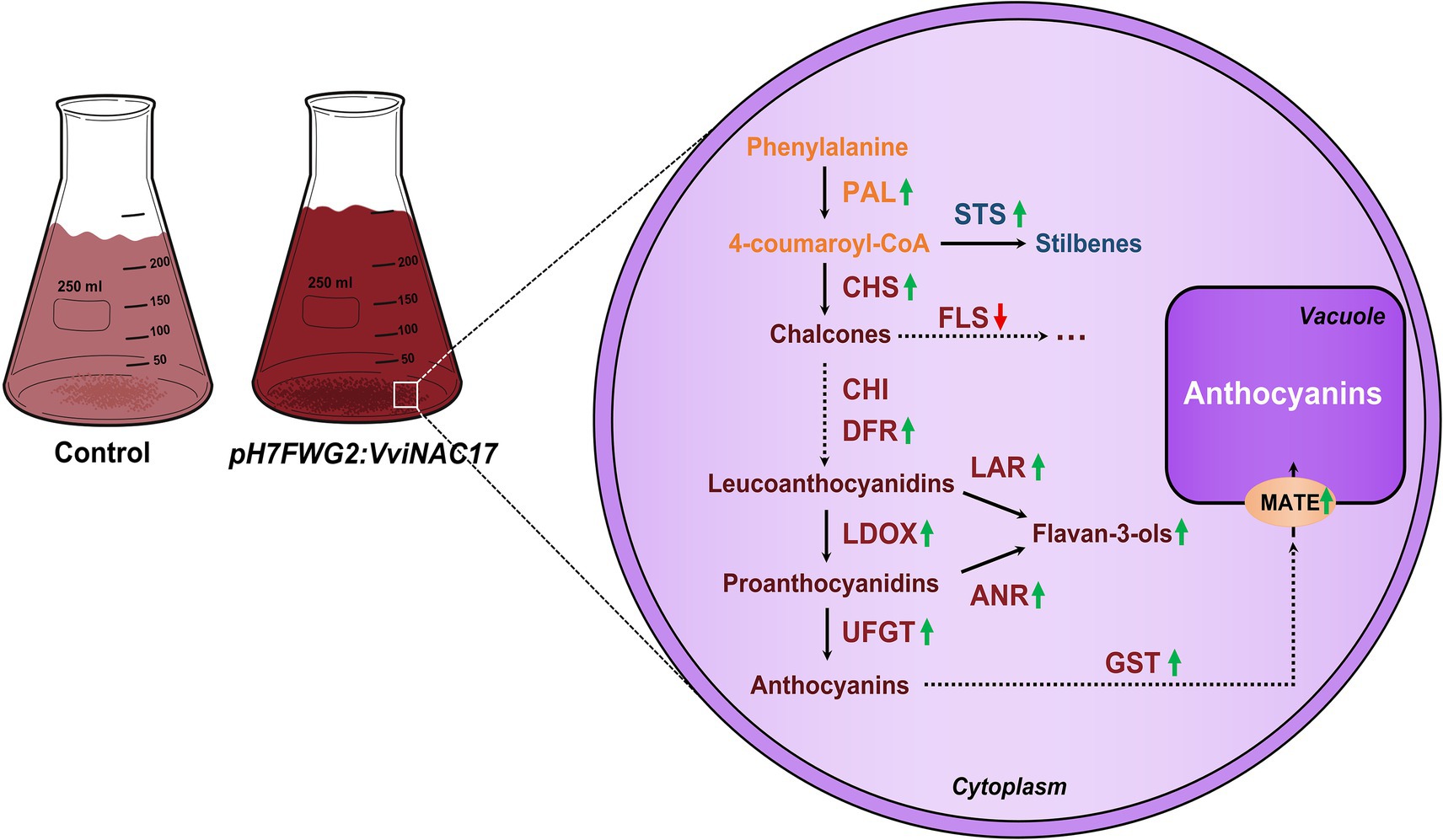
Figure 9. VviNAC17 plays a role in stimulating the synthesis of anthocyanins and flavan-3-ols (PAs) in grape berry cells. The modulation of secondary metabolism, namely phenylpropanoid (PAL), stilbene (STS) and flavonoid (CHS, CHI, DFR, LAR, LDOX, ANR and UFGT) pathways, is positively regulated by the transcription factor VviNAC17. Also, genes involved in the vacuolar transport and accumulation of anthocyanins (GST and MATE) are positively regulated by VviNAC17.
Despite the importance of these findings, some challenges remain. For instance, regenerating grapevine plants from these non-embryogenic VviNAC17-overexpressing cells might prove to be a difficult task. This work would also have benefit from an assessment of a direct physical interaction of VviNAC17 with its target genes, for instance via binding assays such as EMSA (Irfan et al., 2014).
Indeed, the accumulation of these secondary metabolites and the underlying upregulation of their biosynthetic pathways via the action of VviNAC17 is very relevant from an agronomic and/or nutritional standpoint. Anthocyanin and proanthocyanidin concentrations, for instance, are very important quality traits of wines. Also, the future perspective of having transgenic grapevines overexpressing VviNAC17, in addition to a higher grape berry quality scenario, might represent an opportunity to cultivate plants more able to resist to biotic stress due to their significantly increased synthesis of stilbenes and other phenolics, metabolites known to have an important role against pathogen attacks (Latouche et al., 2013; Viret et al., 2018). Parallel to this important agronomical added value, the function of VviNAC17, via its overexpression, might be harnessed to develop and optimize an in vitro strategy to overproduce these secondary metabolites with important bioactive and nutraceutical properties using Gamay Fréaux grape berry cells, that can in the future lead to their sustainable production, something that would greatly benefit agrifood, nutraceuticals and even the cosmetics industry.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
HB performed the experiments and wrote the manuscript. MV and MC performed the experiments. AG and HG advised, wrote, and reviewed the manuscript. AC conceptualized the work, performed the experiments, and wrote and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Fundação para a Ciência e Tecnologia (FCT), under the strategic program UIDB/BIA/04050/2020. This work was also supported by FCT and European Funds (FEDER/POCI/COMPETE2020) through the research project “MitiVineDrought—Combining ‘omics’ with molecular, biochemical, and physiological analyses as an integrated effort to validate novel and easy-to-implement drought mitigation strategies in grapevine while reducing water use” with ref. PTDC/BIA-FBT/30341/2017 and ref. POCI-01-0145-FEDER-030341; AC was supported with a post-doctoral researcher contract/position within the project “MitiVineDrought” (ref. PTDC/BIA-FBT/30341/2017 and ref. POCI-01-0145-FEDER-030341). HB was supported by a Ph.D. fellowship funded by FCT (SFRH/BD/144638/2019). MV was supported by a Ph.D. fellowship funded by FCT (SFRH/BD/144637/2019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.964621/full#supplementary-material
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Bogs, J., Downey, M. O., Harvey, J. S., Ashton, A. R., Tanner, G. J., and Robinson, S. P. (2005). Proanthocyanidin synthesis and expression of genes encoding Leucoanthocyanidin Reductase and Anthocyanidin Reductase in developing grape berries and grapevine leaves. Plant Physiol. 139, 652–663. doi: 10.1104/pp.105.064238
Boneh, U., Biton, I., Schwartz, A., and Ben-Ari, G. (2012a). Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Sci. 187, 89–96. doi: 10.1016/j.plantsci.2012.01.015
Boneh, U., Biton, I., Zheng, C., Schwartz, A., and Ben-Ari, G. (2012b). Characterization of potential ABA receptors in Vitis vinifera. Plant Cell Rep. 31, 311–321. doi: 10.1007/s00299-011-1166-z
Bouquet, A., Torregrosa, L., Iocco, P., and Thomas, M. R. (2006). Grapevine (Vitis vinifera L.) Agrobacterium Protocols. Vol. 2. Berlin: Springer, 273–285.
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chen, L., Wu, F., and Zhang, J. (2021). NAC and MYB families and lignin biosynthesis-related members identification and expression analysis in Melilotus albus. Plan. Theory 10:0303. doi: 10.3390/plants10020303
Ciaffi, M., Paolacci, A. R., Paolocci, M., Alicandri, E., Bigini, V., Badiani, M., et al. (2019). Transcriptional regulation of stilbene synthases in grapevine germplasm differentially susceptible to downy mildew. BMC Plant Biol. 19:404. doi: 10.1186/s12870-019-2014-5
Conde, A., Neves, A., Breia, R., Pimentel, D., Dinis, L.-T., Bernardo, S., et al. (2018). Kaolin particle film application stimulates photoassimilate synthesis and modifies the primary metabolome of grape leaves. J. Plant Physiol. 223, 47–56. doi: 10.1016/j.jplph.2018.02.004
Conde, A., Pimentel, D., Neves, A., Dinis, L.-T., Bernardo, S., Correia, C. M., et al. (2016). Kaolin foliar application has a stimulatory effect on Phenylpropanoid and flavonoid pathways in grape berries. Front. Plant Sci. 7:1150. doi: 10.3389/fpls.2016.01150
Conn, S., Curtin, C., Bézier, A., Franco, C., and Zhang, W. (2008). Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 59, 3621–3634. doi: 10.1093/jxb/ern217
Cutanda-Perez, M.-C., Ageorges, A., Gomez, C., Vialet, S., Terrier, N., Romieu, C., et al. (2009). Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 69, 633–648. doi: 10.1007/s11103-008-9446-x
Czemmel, S., Heppel, S. C., and Bogs, J. (2012). R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 249, 109–118. doi: 10.1007/s00709-012-0380-z
Czemmel, S., Stracke, R., Weisshaar, B., Cordon, N., Harris, N. N., Walker, A. R., et al. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates Flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530. doi: 10.1104/pp.109.142059
Deng, Y., and Lu, S. (2017). Biosynthesis and regulation of Phenylpropanoids in plants. Crit. Rev. Plant Sci. 36, 257–290. doi: 10.1080/07352689.2017.1402852
Dixon, R. A., Xie, D. Y., and Sharma, S. B. (2005). Proanthocyanidins--a final frontier in flavonoid research? New Phytol. 165, 9–28. doi: 10.1111/j.1469-8137.2004.01217.x
Downey, M. O., Harvey, J. S., and Robinson, S. P. (2003). Synthesis of flavonols and expression of flavonol synthase genes in the developing grape berries of shiraz and chardonnay (Vitis vinifera L.). Aust. J. Grape Wine Res. 9, 110–121. doi: 10.1111/j.1755-0238.2003.tb00261.x
Ernst, H. A., Nina Olsen, A., Skriver, K., Larsen, S., and Lo Leggio, L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303. doi: 10.1038/sj.embor.7400093
Ferrandino, A., and Lovisolo, C. (2014). Abiotic stress effects on grapevine (Vitis vinifera L.): focus on abscisic acid-mediated consequences on secondary metabolism and berry quality. Environ. Exp. Bot. 103, 138–147. doi: 10.1016/j.envexpbot.2013.10.012
Ferrer, J. L., Austin, M. B., Stewart, C. Jr., and Noel, J. P. (2008). Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 46, 356–370. doi: 10.1016/j.plaphy.2007.12.009
Figueroa, N., Lodeyro, A. F., Carrillo, N., and Gómez, R. (2021). Meta-analysis reveals key features of the improved drought tolerance of plants overexpressing NAC transcription factors. Environ. Exp. Bot. 186:104449. doi: 10.1016/j.envexpbot.2021.104449
Francisco, R. M., Regalado, A., Ageorges, A., Burla, B. J., Bassin, B., Eisenach, C., et al. (2013). ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-Glucosides. Plant Cell 25, 1840–1854. doi: 10.1105/tpc.112.102152
Gambetta, G. A., Matthews, M. A., Shaghasi, T. H., McElrone, A. J., and Castellarin, S. D. (2010). Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta 232, 219–234. doi: 10.1007/s00425-010-1165-2
Garrido, A., Serôdio, J., De Vos, R., Conde, A., and Cunha, A. (2019). Influence of foliar kaolin application and irrigation on photosynthetic activity of grape berries. Agronomy 9:0685. doi: 10.3390/agronomy9110685
Gollop, R., Even, S., Colova-Tsolova, V., and Perl, A. (2002). Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region1. J. Exp. Bot. 53, 1397–1409. doi: 10.1093/jexbot/53.373.1397
Gomez, C., Terrier, N., Torregrosa, L., Vialet, S., Fournier-Level, A., Verriès, C., et al. (2009). Grapevine MATE-type proteins act as vacuolar H+-dependent Acylated anthocyanin transporters. Plant Physiol. 150, 402–415. doi: 10.1104/pp.109.135624
Höfgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16:9877. doi: 10.1093/nar/16.20.9877
Höll, J., Vannozzi, A., Czemmel, S., D'Onofrio, C., Walker, A. R., Rausch, T., et al. (2013). The R2R3-MYB transcription factors MYB14 and MYB15 regulate Stilbene biosynthesis in Vitis vinifera. Plant Cell 25, 4135–4149. doi: 10.1105/tpc.113.117127
Irfan, M., Ghosh, S., Kumar, V., Chakraborty, N., Chakraborty, S., and Datta, A. (2014). Insights into transcriptional regulation of β-D-N-acetylhexosaminidase, an N-glycan-processing enzyme involved in ripening-associated fruit softening. J. Exp. Bot. 65, 5835–5848. doi: 10.1093/jxb/eru324
Ju, Y. L., Yue, X. F., Min, Z., Wang, X. H., Fang, Y. L., and Zhang, J. X. (2020). VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 146, 98–111. doi: 10.1016/j.plaphy.2019.11.002
Kang, J.-Y., Choi, H.-I., Im, M.-I., and Kim, S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. doi: 10.1105/tpc.010362
Kobayashi, S., Goto-Yamamoto, N., and Hirochika, H. (2004). Retrotransposon-induced mutations in grape skin color. Science 304:982. doi: 10.1126/science.1095011
Latouche, G., Bellow, S., Poutaraud, A., Fau-Meyer, S., and Cerovic, Z. G. (2013). Influence of constitutive phenolic compounds on the response of grapevine (Vitis vinifera L.) leaves to infection by Plasmopara viticola. Planta 237, 351–361. doi: 10.1007/s00425-012-1776-x
Lister, C. E., Lancaster, J. E., and Walker, J. R. L. (1996). Developmental changes in enzymes of flavonoid biosynthesis in the skins of red and green apple cultivars. J. Sci. Food Agric. 71, 313–320. doi: 10.1002/(SICI)1097-0010(199607)71:3<313::AID-JSFA586>3.0.CO;2-N
Martínez-Márquez, A., Morante-Carriel, J., Ramírez-Estrada, K., Cusido, R., Sellés-Marchart, S., Palazon, J., et al. (2015). A reliable protocol for the stable transformation of non-embryogenic cells cultures of grapevine (Vitis vinifera L.) and Taxus x media. Journal of biological. Methods 2:e21. doi: 10.14440/jbm.2015.51
Matejić, J. S., Džamić, A. M., Mihajilov-Krstev, T. M., Ranđelović, V. N., Krivošej, Z. D., and Marin, P. D. (2013). Total phenolic and flavonoid content, antioxidant and antimicrobial activity of extracts from Tordylium maximum. J. App. Pharmaceutical Sci. 3, 055–059. doi: 10.7324/JAPS.2013.30110
Noronha, H., Silva, A., Mitani-Ueno, N., Conde, C., Sabir, F., Prista, C., et al. (2020). The grapevine NIP2;1 aquaporin is a silicon channel. J. Exp. Bot. 71, 6789–6798. doi: 10.1093/jxb/eraa294
Ollat, N., Cookson, S. J., Destrac-Irvine, A., Lauvergeat, V., Ouaked-Lecourieux, F., Marguerit, E., et al. (2018). “Grapevine Adaptation to abiotic Stress: An Overview,” in XII International Conference on Grapevine Breeding and Genetics (July 15, 2018), 1248, 497–512. doi: 10.17660/ActaHortic.2019.1248.68
Pérez-Díaz, R., Madrid-Espinoza, J., Salinas-Cornejo, J., González-Villanueva, E., and Ruiz-Lara, S. (2016). Differential roles for VviGST1, VviGST3, and VviGST4 in Proanthocyanidin and anthocyanin transport in Vitis vinífera. Front. Plant Sci. 7:1166. doi: 10.3389/fpls.2016.01166
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pilati, S., Bagagli, G., Sonego, P., Moretto, M., Brazzale, D., Castorina, G., et al. (2017). Abscisic acid is a major regulator of grape berry ripening onset: new insights into ABA signaling network. Front. Plant Sci. 8:1093. doi: 10.3389/fpls.2017.01093
Reid, K. E., Olsson, N., Schlosser, J., Peng, F., and Lund, S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6:27. doi: 10.1186/1471-2229-6-27
Revilla, E., Arroyo-Garcia, R., Bellido, A., Carrasco, D., Puig, A., and Ruiz-Garcia, L. (2018). Fingerprints of Anthocyanins and Flavonols in wild grapes (Vitis vinifera L. ssp. sylvestris (Gmelin) Hegi). Grapes and Wines: Adv. Prod. Proces. Analy. Valorization 23. doi: 10.5772/intechopen.70861
Rienth, M., Vigneron, N., Darriet, P., Sweetman, C., Burbidge, C., Bonghi, C., et al. (2021). Grape berry secondary metabolites and their modulation by abiotic factors in a climate change scenario–A review. Front. Plant Sci. 12:643258. doi: 10.3389/fpls.2021.643258
Rinaldo, A. R., Cavallini, E., Jia, Y., Moss, S. M. A., McDavid, D. A. J., Hooper, L. C., et al. (2015). A grapevine anthocyanin Acyltransferase, transcriptionally regulated by VvMYBA, can produce Most Acylated Anthocyanins present in grape skins. Plant Physiol. 169, 1897–1916. doi: 10.1104/pp.15.01255
Sakuraba, Y., Kim, D., Han, S.-H., Kim, S.-H., Piao, W., Yanagisawa, S., et al. (2020). Multilayered regulation of membrane-bound ONAC054 is essential for Abscisic acid-induced leaf senescence in Rice. Plant Cell 32, 630–649. doi: 10.1105/tpc.19.00569
Sun, L., Zhang, H., Li, D., Huang, L., Hong, Y., Ding, X. S., et al. (2013). Functions of rice NAC transcriptional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 81, 41–56. doi: 10.1007/s11103-012-9981-3
Teixeira, A., Eiras-Dias, J., Castellarin, S. D., and Gerós, H. (2013). Berry phenolics of grapevine under challenging environments. Int. J. Mol. Sci. 14, 18711–18739. doi: 10.3390/ijms140918711
Terrier, N., Torregrosa, L., Ageorges, A. S., Vialet, S., Verriès, C., Cheynier, V. R., et al. (2009). Ectopic expression of VvMybPA2 promotes Proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Vale, M., Rodrigues, J., Badim, H., Gerós, H., and Conde, A. (2021). Exogenous application of non-mature miRNA-encoded miPEP164c inhibits Proanthocyanidin synthesis and stimulates anthocyanin accumulation in grape berry cells. Front. Plant Sci. 12:706679. doi: 10.3389/fpls.2021.706679
Vannozzi, A., Dry, I. B., Fasoli, M., Zenoni, S., and Lucchin, M. (2012). Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol. 12:130. doi: 10.1186/1471-2229-12-130
Vidal, S., Francis, L., Guyot, S., Marnet, N., Kwiatkowski, M., Gawel, R., et al. (2003). The mouth-feel properties of grape and apple proanthocyanidins in a wine-like medium. J. Sci. Food Agric. 83, 564–573. doi: 10.1002/jsfa.1394
Viret, O., Spring, J.-L., and Gindro, K. (2018). Stilbenes: biomarkers of grapevine resistance to fungal diseases. OENO one 52, 235–241. doi: 10.20870/oeno-one.2018.52.3.2033
Walker, A. R., Lee, E., Bogs, J., McDavid, D. A., Thomas, M. R., and Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. doi: 10.1111/j.1365-313X.2006.02997.x
Wallace, T. C., and Giusti, M. M. (2010). Evaluation of parameters that affect the 4-dimethylaminocinnamaldehyde assay for flavanols and proanthocyanidins. J. Food Sci. 75, C619–C625. doi: 10.1111/j.1750-3841.2010.01734.x
Wise, A. A., Liu, Z., and Binns, A. N. (2006). Three Methods for the Introduction of Foreign DNA into Agrobacterium Protocols, Berlin: Springer, 43–54.
Ye, Y., Wu, K., Chen, J., Liu, Q., Wu, Y., Liu, B., et al. (2018). OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice 11:36. doi: 10.1186/s12284-018-0228-z
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Keywords: VviNAC17, homologous expression, anthocyanins, flavonoids, phenolics, secondary metabolites, grape berry cells
Citation: Badim H, Vale M, Coelho M, Granell A, Gerós H and Conde A (2022) Constitutive expression of VviNAC17 transcription factor significantly induces the synthesis of flavonoids and other phenolics in transgenic grape berry cells. Front. Plant Sci. 13:964621. doi: 10.3389/fpls.2022.964621
Received: 08 June 2022; Accepted: 08 July 2022;
Published: 29 July 2022.
Edited by:
Suprasanna Penna, Bhabha Atomic Research Centre (BARC), IndiaReviewed by:
Sirsha Mitra, Savitribai Phule Pune University, IndiaCopyright © 2022 Badim, Vale, Coelho, Granell, Gerós and Conde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Artur Conde, YXJ0dXJjb25kZUBiaW8udW1pbmhvLnB0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.