
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 July 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.963873
This article is part of the Research TopicCreation and utilization of crop germplasm resourcesView all 18 articles
Cytokinins (CTKs) are a major phytohormone group that are significant in the promotion of cellular division, growth, and divergence. Isopentenyl transferase (IPT) regulates a rate-limiting step in plant CTK synthesis, promotes the synthesis of isopentenyl adenonucleotides from 5-AMP and isopentenyl pyrophosphate, and then converts both these chemicals into various CTKs. Here, the full-length cDNA of ZmIPT2, which encodes 322 amino acids, was isolated and was introduced into a maize inbred line by Agrobacterium-mediated transformation. In both controlled environments and field experiments, the overexpression of ZmIPT2 gene in the transformed plants delayed leaf senescence. Compared to the receptor line, the transgenic maize lines retained higher chlorophyll levels, photosynthetic rates, and cytokinin content for an extended period of time, and produced significantly higher grain yield by a margin of 17.71–20.29% under normal field planting conditions. Subsequently, ten possible genes that interacted with ZmIPT2 were analyzed by qRT-PCR, showing that the expression pattern of GRMZM2G022904 was consistent with ZmIPT2 expression. Through comprehensive analysis, we screened for transgenic lines with stable inheritance of ZmIPT2 gene, clear functional efficiency, and significant yield improvement, in order to provide theoretical basis and material support for the breeding of new high-yield transgenic maize varieties.
Senescence is a degradative process of plant tissue structures (Rivero et al., 2007). To improve overall plant health, leaves should be engineered for delayed senescence to permit the capture of sunlight energy for a longer duration, allowing for more efficient transformation of light into photosynthates (Sykorova et al., 2008). Additionally, a delay in senescence would also allow tissues to degenerate more slowly. In doing so, the stored resources can be systematically released to the appropriate sink tissues. Advantages of deferred senescence involve a better maintenance of photosynthetic rate, raised plant biomass, improved drought tolerance, and higher seed yield (McCabe et al., 2001; Chang et al., 2003; Rivero et al., 2010).
Cytokinins (CTKs) were thought to have evolved to coordinate the endogenous developmental processes and adaptive responses in plants (Li et al., 2016). Several links have been established between CTK level, pool strength, and source-pool conversion by regulating photoassimilate distribution (Brenner and Cheikh, 1995; Roitsch and Rainer, 2000; Kuiper, 1993). It is generally believed that the CTK-mediated delay of leaf senescence was related to sink/source regulation, and that part of this relationship is mediated by the cell wall invertase (CWINV) protein (Roitsch and Rainer, 2000). Scientists found that leaf senescence was delayed and expression level of CWINV was increased in PSAG12:IPT transgenic tobacco, indicating that CWINV was closely related to senescence. The CWINV inhibitor protein was expressed under the CTK inducible promoter, and there was no delay in leave senescence after CTK treatment (Balibrea Lara et al., 2004), indicating that the increased presence of CWINV was a necessary condition for CTK to delay senescence. This, at least, partially explained the molecular mechanisms of CTK in delaying leaf senescence (Peng et al., 2021).
The function of CTKs in deferring leaf senescence has been reported for several plant species (Gan and Amasino, 1995; Robson et al., 2004; Rivero et al., 2007; Sykorova et al., 2008; Peleg et al., 2011; Kant et al., 2015). Gan and Amasino were the first to apply the senescence-specific promoter, SAG12, fused with the IPT gene to explore the function of CTKs in deferring leaf senescence. The transgenic tobacco plants that were created in these studies reported higher biomass and yield (Gan and Amasino, 1995). Further studies on transgenic tobacco found that the expression of the phosphoenolpyruvate gene was regulated by PSARK, a gene specifically induced under plant aging and stress conditions (Delatorre et al., 2012). Regulation of the phosphoenolpyruvate gene could maintain a high concentration of cytokinin in plants under water stress, and improve the tolerance of plants to water stress and prolong the photosynthetic life of plant leaves (Rivero et al., 2007). Decreased local CTK production in roots or leaves, which are typically plentiful sources of CTKs, were assumed to initiate leaf senescence (Singh, 1992; Hirose et al., 2008; Kudo et al., 2010). Isopentenyladenine (iP), and one of its glucosides, iP9G, were both capable of delaying chlorophyll degradation in detached cotyledons senescence assays (Hallmark and Rashotte, 2020).
From an agronomic point of view, several studies link increased CTKs levels to seed yield in rice, soybean, and maize. In some varieties of rice, the cytokinin oxidase gene (OsCTkx2) was associated with the quantitative trait locus (QTL) for grain production (Ashikari et al., 2005). In soybean, CTKs have been shown to play a significant function on pod growth and flowering (Huff and Dybing, 1980; Ghiasi et al., 1987; Wiebold and Panciera, 1990; Westgate and Peterson, 1993; Nagel, 2001). In maize, CTKs’ exogenous application has been similarly revealed to increase the yield stability of heat stressed plants (Cheikh and Jones, 1994). Therefore, CTKs are involved in a large amount of quantitative and qualitative components of yield. Their participation in a huge quantity of regulatory roles in plants give much incentive to unravel biosynthesis of CTKs and signal pathways (Kieber and Schaller, 2014).
The production of isopentenyl transferase (IPT), a key enzyme of CTK biosynthesis, catalyzes the first step of de novo synthesis of cytokinin, promotes the synthesis of isopentenyl adenonucleotides from 5-AMP and isopentenyl pyrophosphate, and then converts both these chemicals into various CTKs. The IPT gene was first identified in Agrobacterium tumefaciens (Gan and Amasino, 1995). Since the discovery of the IPT gene, there have been a large number of reports using transgenic technology to study the use of this gene to improve yield, anti-aging, and disease resistance (Noh and Amasino, 1999). The mechanism of inducing the IPT gene expression to produce endogenous CTK to delay leaf senescence may be related to promoting the accumulation of stress response related proteins and antioxidant enzymes (Peng et al., 2021). Studies have shown that the heterologous expression of IPT under the transcription control of senescence -associated receptor-like kinase (SARK) promoter in wheat can improve plant drought tolerance by delaying cell senescence. Overexpressing IPT showed a water tolerance phenotype in different environments: transgenic TR1 and TR4 wheat showed delayed senescence and increased yield under sufficient water conditions (Beznec et al., 2021), similar to the reported effect of IPT expression in rice, tobacco, and maize (Rivero et al., 2007; Peleg et al., 2011; Qin et al., 2011; Decima Oneto et al., 2016). Interestingly, in transgenic cauliflower containing the IPT gene, a tetrapeptide-like repeat was found, which could induce protein folding, SOD and stress response. The protein folding and carbon fixation related protein levels of this transgenic cauliflower were higher than those of non-transgenic cauliflower, and the iron SOD and APX activities were also significantly higher than those of non-transgenic cauliflower (Liu et al., 2011). The transgenic plants had an increased number of stress-related proteins and molecular chaperones, induced the expression of stress-related genes, and protected cells during aging in contemporary plant breeding programs (Peng et al., 2021).
These techniques are considered new strategies for engineering CTKs for better crop production in agricultural systems (Jameson and Song, 2016; Kieber and Schaller, 2018). In addition, Arabidopsis researchers have shown that the manipulation of CTK levels via IPTs can enhance salt stress tolerance and drought (Li et al., 2016), laying the groundwork for similar studies in agriculturally significant plants. In addition, a growing number of studies have shown that during stress, the expression of the IPT gene can induce plants to produce various resistant or active substances, which can produce CTK responses, coordinate, and interact with other hormones. The production of various resistance and active substances in plants can enhance photosynthesis. Various metabolites can be synthesized to promote the response of plant cells to various adverse environments or diseases and insect pests. Moreover, agronomic traits have been significantly improved by the induction of the ITP gene (Li and Zhao, 2011), which provided a novel base for future molecular breeding of crop plants (Jameson and Song, 2016).
In crops such as lettuce (McCabe et al., 2001), rice (Jin et al., 2002), wheat (Sykorova et al., 2008), and ramie (An et al., 2017), CTK manipulation not only delays the aging of leaves, but also improves agronomic traits and stress resistance of crops. Gan and Amasino transferred the chimeric genes SAG12 and IPT into tobacco and found that the transgenic plants showed higher biomass and yield (Gan and Amasino, 1995). In rice, IPT-induced CTKs synthesis reduced environmental stress consequences on photosynthesis and yield and maintained nitrogen (N) acquisition (Reguera et al., 2013). Overexpression of OsIPT9 in rice can increase the CTK level of caryopsis development, resulting in enhanced grain filling of large and multi-panicle rice varieties, thus increasing yield (Panda et al., 2018). Transgenic wheat containing the IPT gene driven by AtMYB2xs promoter had improved yield under both sufficient water and water stress conditions (Joshi et al., 2019). Regulating IPT by using the AtMYB32 promoter can improve rape yield under both drought and conventional irrigation conditions (Kant et al., 2015). Multiple species showed improved yield by increasing IPT expression levels under drought conditions, indicating that if the expression of ITP can be fully controlled, it could be a key driver of yield in crop systems.
In maize, eight IPT genes have been found in the genome, all of which have complete open reading frames (ORFs) except ZmIPT3. Among the completed ORFs, the ZmIPT2 gene encodes a 322 amino acid protein and is stably expressed at the transcriptional level, in addition to being the highest expressed protein (Takei et al., 2001). Some studies have proved that in maize tissue, the content of CTKs was directly proportional to the expression level of ZmIPT2. When the expression level of ZmIPT2 increased, the CTKs content also increased. Therefore, the expression of this gene was considered to be related to the content of cytokinin. The ZmIPT2 gene played a very important role in overall seed development, from endosperm cell division to embryo development, and played a role in germplasm strength. It can also delay leaf senescence and improve yield (Brugiere et al., 2008).
The endeavor to comprehend the molecular basis of this increase in yield owning to deferred senescence and the possible involvement of CTKs in this process will be useful in aiding breeders and scientists alike. Our laboratory cloned ZmIPT2 gene from early maize, and transformed the maize using Agrobacterium, showing that the transgenic offspring held their leaves during the regular leaf senescence stage, and maintained a brighter green color in their leaves. The transgenic offspring also had an increased yield, although the genetic instability phenomenon was observed. Therefore, we reconstructed the monocotyledonous plant expression vector of this gene, and transformed maize inbred lines by Agrobacterium tumefaciens infection of young embryos to obtain T2 transgenic lines. We clarified the basic characteristics of this gene through bioinformatic analysis, examined the expression pattern under different growth stages, and analyzed the physiological indexes and yield traits of transgenic lines. The interacting genes of ZmIPT2 were then screened by qRT-PCR. The transgenic maize line that we have generated may be of great value as a genetic resource for further high-yield maize breeding.
The maize inbred line Zheng 58 was used to clone the target gene and the maize inbred line C01 was used as the receptor material for genetic transformation. The material for Zheng 58 was obtained from the Maize Research Institute of Northeast Agricultural University. The inbred line C01 used as a transgenic receptor was provided by the Life Science and Technology Center of China Seed Group Co., Ltd. According to the relevant laws of China, after the experiment the seeds were preserved on-site and the plants were properly disposed of.
Agrobacterium tumefaciens strain LBA4404 came from our laboratory. Escherichia coli strain DH5a came from Beijing Quanshijin company. The basic plant expression vector used in the transformation experiments was pEAST-T1, with the ubiquitin promotor, T-NOS terminator, and the CaMV35S promoter driven bar gene selection marker, which was provided by the Institute of Crop Research, Chinese Academy of Agricultural Sciences. Taq DNA polymerase was purchased from Beijing Quanshijin biological Co., Ltd.
The ZmIPT2 gene (Gene ID: GRMZM2G084462) was localized in bin 2.04 region based on B73 RefGen_v2 genome-wide data. Analysis of the ZmIPT2 ORF was performed with ORF finder1. The second-order structure of the ZmIPT2 protein was predicted using SOMPA2, with SWISS – MODELL3 to predict the protein tertiary structure. The Conserved Domain Database website4 was used for protein structure analysis, and Phytozome V 12.1 was used for the other species of IPT family protein sequences. The system evolution tree was constructed by using ClustalX 1.83 and MEGA 5 software, with the bootstrap value adjusted to 1000. The physicochemical properties and hydrophobicity of the ZmIPT2 protein were analyzed with protparam5 and ProtScale6. The subcellular localization of ZmIPT2 protein was predicted using Softberry7. The cis-acting elements of the ZmIPT2 gene flanking sequences were identified using the PlantCARE program8.
Using the accession number EU263126.1 on GenBank, the mRNA sequence of ZmIPT2 gene was downloaded and PCR primers were designed. Total RNA was extracted from leaves of maize inbred line Zheng 58 with the plant RNA Kit (Beijing Tiangen, China) and reverse transcribed to obtain the first strand cDNA. Under the following conditions, the cDNA was amplified by PCR with specific primers of ZmIPT2 using KOD plus NEO (Toyobo) polymerase mixture. The recovered PCR products were sent to Shanghai Sangong for sequencing.
Total RNA was extracted from leaves of the inbred maize line Zheng 58 with the Plant RNA Kit (Beijing Tiangen, China) following the manufacturer’s protocols. Extracted RNA was treated with DNase I (Omega, GA, United States) to remove residual DNA. The first-strand cDNA was synthesized from 1 μg of total RNA using 1 μL (200 U) PrimeScriptTM RT Enzyme Mix I (TaKaRa) according to the manufacturer’s protocol, to produce the full-length cDNA of ZmIPT2. PCR was performed on the cDNA using specific primers for ZmIPT2 using the KOD-Plus-Neo (TOYOBO) polymerase mix under the following conditions. The amplification product was purified from 1.0% agarose gel and cloned into the pEASY-T1 Simple vector (Transgen) to be sent for sequencing. The primers are listed in Supplementary Table 1. The full sequence of the ZmIPT2 cDNA clone used in this study can be found in GenBank (GenBank Accession No. EU263126.1).
For generating the transgenic lines overexpressing ZmIPT2, the coding sequence of ZmIPT2 was subcloned from the pEAST-T1 vector into the pEC-Ubi-Tnos-Bar vector, which consisted of a maize constitutive ubiquitin promoter, Bar gene with the CaMV 35S promoter, and a Tnos terminator (Sambrook et al., 1982), using to the standard protocol for the In-Fusion Cloning Kit [Takara Biomedical Technology (Beijing) Co., Ltd]. The recombinant plasmid was finally transformed into Agrobacterium tumefaciens LBA4404 using the freeze-thaw method (Supplementary Figure 1).
In this study, maize inbred line C01 was used as the starting genetic material. Maize transformation was performed according to the method published by Li et al. (2008). C01 seeds were surface sterilized and cultured on sterile MS agar at 28°C until etiolated seedlings grew to 3.0–4.0 cm. The maize shoot tip with exposed meristem was immersed in the transformed Agrobacterium suspension, which was grown a logarithmic stage, and then diluted to an OD600 of 0.8. The buds were soaked at 0.05 MPa for 10 min and then co-cultured on modified MS agar in the dark at 24°C for 3 days. The seedlings were then transplanted into pots and transferred to the greenhouse to continue to grow. Transgenic seedlings sprayed with 0.1% herbicide Basta were detected by PCR, and seeds were harvested from positive plants (T0). T1 plants were grown from harvested seeds and self-pollinated to produce the T2 generation. Because of the vector contains a screening marker gene Bar gene, the positive transgenic plants of each generation had the integration and stability of the ZmIPT2 gene by verified by PCR and Bar protein test strip. The expression level of ZmIPT2 and copy number in transgenic plants was confirmed by qRT–PCR.
The leaf of DNA was extracted by CTAB method (Murray and Thompson, 1980), and specific primers were designed according to the target gene sequence and terminator sequence. One end of the primer was located inside the target gene, and the other end was located inside the terminator. The primers of ZmIPT2 gene were F (5′-TTTAGCCCTGCCTTCA-3′), R (5′-AACCCATCTCATAAATAACG-3′), and bar gene specific primers BF1 (5′-CCATCTCAACCACTACATCG-3′) and BR1 (5′-AGCTGCCAGAAACCCCACGT-3′). PCR reactions were set up in volumes of 20 μL, as recommended by the manufacturer. The thermal cycling conditions were 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 50°C for 30 s, 72°C for 70 s, and 72°C for 10 min. Total RNA was extracted using Trizol reagent (Tiangen, Beijing, China) from 100 mg of maize seedling leaves. The RNA was DNase-treated, and 500 ng of this treated RNA was used for inverse transcription with the RT Reagent Kit (Quanshijin, Beijing, China), following the manufacturer’s protocols. The generated cDNA samples were diluted fivefold to serve as templates for the subsequent PCR. The primers Actin-F (5′-GTTGGGCGTCCTCGTCA-3′) and Actin-R (5′-TGGGTCATCTTCTCCCTGTT-3′) were designed for qRT-PCR, and three technical replicates were used for each of the RT-PCR experiments in this research.
The V6 (sixth leaf), R1 (silking), R2 (blister stage), and R6 (physical maturity) leaves of maize were collected at different growth stages. The Trizol reagent (Quanshijin, Beijing, China) was used to extract total RNA. For each sample, the Transscript® One step gDNA Removal Kit mix (Transgen Biotech, Beijing, China) was combined with 500 ng RNA to synthesize cDNA. qRT-PCR was performed using the TransStart Tip Green qPCR SuperMix fluorescent quantitative Kit (Genetically Modified Biotechnology Company, Beijing, China). The Actin1 and ZmIPT2 gene-specific primer sequences used for qRT-PCR are located Supplementary Table 1. The gene expression levels were calculated using the 2–ΔΔCt formula method (Livak and Schmittgen, 2001). For each sample, three biological replicates were included, with three technical replicates for each. The subsequent analysis was performed by using the obtained qRT-PCR data.
The transgene copy number was detected by qRT-PCR. Actin1 (Gene ID: 100282267), which is a single copy gene in maize, was used as the endogenous reference gene. According to the principle that the CT value obtained by PCR reaction is linearly inversely proportional to the logarithm of the number of initial templates, a quantitative standard curve with a correlation coefficient of more than 0.99 between the internal reference gene actin and the target gene ZmIPT2 was prepared. After diluting the DNA concentration of each sample to be tested by five times, the qRT- PCR reaction was conducted to obtain CT values and calculate the number of initial templates of each sample according to the standard curve. Since the actin gene is a single copy in maize, the ratio of the logarithm of the initial template number of ZmIPT2 gene and actin gene is the copy number of the target gene in maize (Han et al., 2016).
The relative chlorophyll content, photosynthetic efficiency, and dynamic changes of cytokinin content in different growth stages of the transgenic maize lines and receptor control lines with high target gene expression were quantified.
To measure photosynthetic efficiency, photosynthetic rate was measured using a Li-6400 portable photosynthetic system (Li COR, Lincoln, NE, United States). In order to reduce the laboratory environment and the gradient in the tube of the gas exchange system, the concentration of CO2 in the sample was set to 500 μmol–1, the relative humidity in the tube was set to 65%, and the temperature of the blade in the tube was set to 28°C. Conventional methods were used to estimate outdoor in situ gas exchange between 8:30AM and 11:30AM. The environmental conditions in the colorimeter were as described above and were set prior to recording the gas exchange data. No less than 20 min was required to achieve a stable gas exchange measurement, i.e., photosynthetic rate change < 0.5% in 1 min.
To measure chlorophyll content, leaves [100 mg fresh weight (FW)] were incubated in 10 ml tubes containing 5 ml 80% acetone at room temperature in darkness until the tissues turned white. Total chlorophyll content was set by measuring the absorbance at 645 and 663 nm applying the formula 20.2 A645 + 8.02 A663 (Chory et al., 1994). The extraction and purification of isopentenyl adenine standard (IP) and zeatin in leaves to measure the dynamic changes of cytokinin content were performed according to of the protocol described by Dong et al. (2008). The cytokinin content was decided by enzyme-linked immunosorbent assay (ELISA) protocol (Wu et al., 1988). Three samples were taken from each line in each period for mixed treatment, and three technical replicates were performed for each sample.
The seeds of transgenic plants and recipient inbred line C01 were planted in the transgenic experimental practice base of Northeast Agricultural University. The field experiment was performed by using the comparison method. The row spacing for each plot was 65 cm, the length was 2.0 m, and the plant spacing was 2 cm. The agronomic characteristics of each plot during maize growth were considered similar to a conventionally managed field. Three consecutive plants were selected in the middle row of each plot to avoid the effects of the border plants, and the identification method of agronomic traits was adopted from the Description and Data Standard of Maize Germplasm Resources. The agronomic traits in field investigation included ear length, ear diameter, axis diameter, grain length, grain width, grain thickness, bare tip length, 100-grain weight, and plot yield. Excel statistical data was used to test agronomic traits and green-holding traits, SPSS 22.0 was used for the analysis of data processing, and the T-test was used to test the significance of data difference.
The amino acid sequence of the ZmIPT2 gene was submitted to STRING9 to predict the proteins that interacted with the putative ZmIPT2 protein. These gene sequences were downloaded from NCBI database. The primers for these proteins coding sequences were designed using Primer5 and are listed in Table 3. The relative expression of these interacting proteins in maize leaves at different growth stages were verified by RT-qPCR. Then, the interacting genes were further determined by bioinformatic analysis.
The ZmIPT2 gene encoded a putative protein of 322 amino acids with a predicted molecular mass of 34.49 kDa, and a predicted isoelectric point (PI) of 5.11. The amino acid sequence comparison of the ZmIPT2 protein via ExPASy Protparam is shown in Table 1. Subcellular localization predictions showed that the protein was most likely to be localized on the cytoplasm with a confidence level of 0.478 and may also be distributed in the nucleus and mitochondria but with lower confidence (Supplementary Table 1). The results of cross-membrane domain analysis found that the ZmIPT2 protein did not exist in the transmembrane domain and was located in the extramembrane region with the greatest probability (probability = 1.1), indicating that the ZmIPT2 protein did not carry out transmembrane transport, but directly exercised its function, and that it belonged to the non-transmembrane protein class (Figure 1A). The results of signal peptide analysis showed that there was no signal peptide in the protein sequence. The ZmIPT2 protein was not a secretory protein and therefore could not migrate in cells (Figure 1B). Hydrophobic structure analysis showed that the ZmIPT2 protein contained a typical hydrophobic region at about 16 amino acids (Figure 1C). Meanwhile, the ZmIPT2 promoter indicated that it had core elements such as a TATA box and a CAAT box, as well as a large number of abiotic stress related elements referred to such as ABRE, an auxin response element, and a cis-acting regulatory CAT box that was involved in meristem expression (Figure 1D). It is speculated that the ZmIPT2 promoter may be a stress-inducible promoter. Its predicted Secondary Structure includes alpha-helices (55.85%), random coils (31.06%), extended strands (9.90%), And β-helices (7.76%) (Figure 1E). Its three-dimensional homology model is a 3A8T. 1 protein sequence with a similarity of protein 40.58% (Figure 1F).
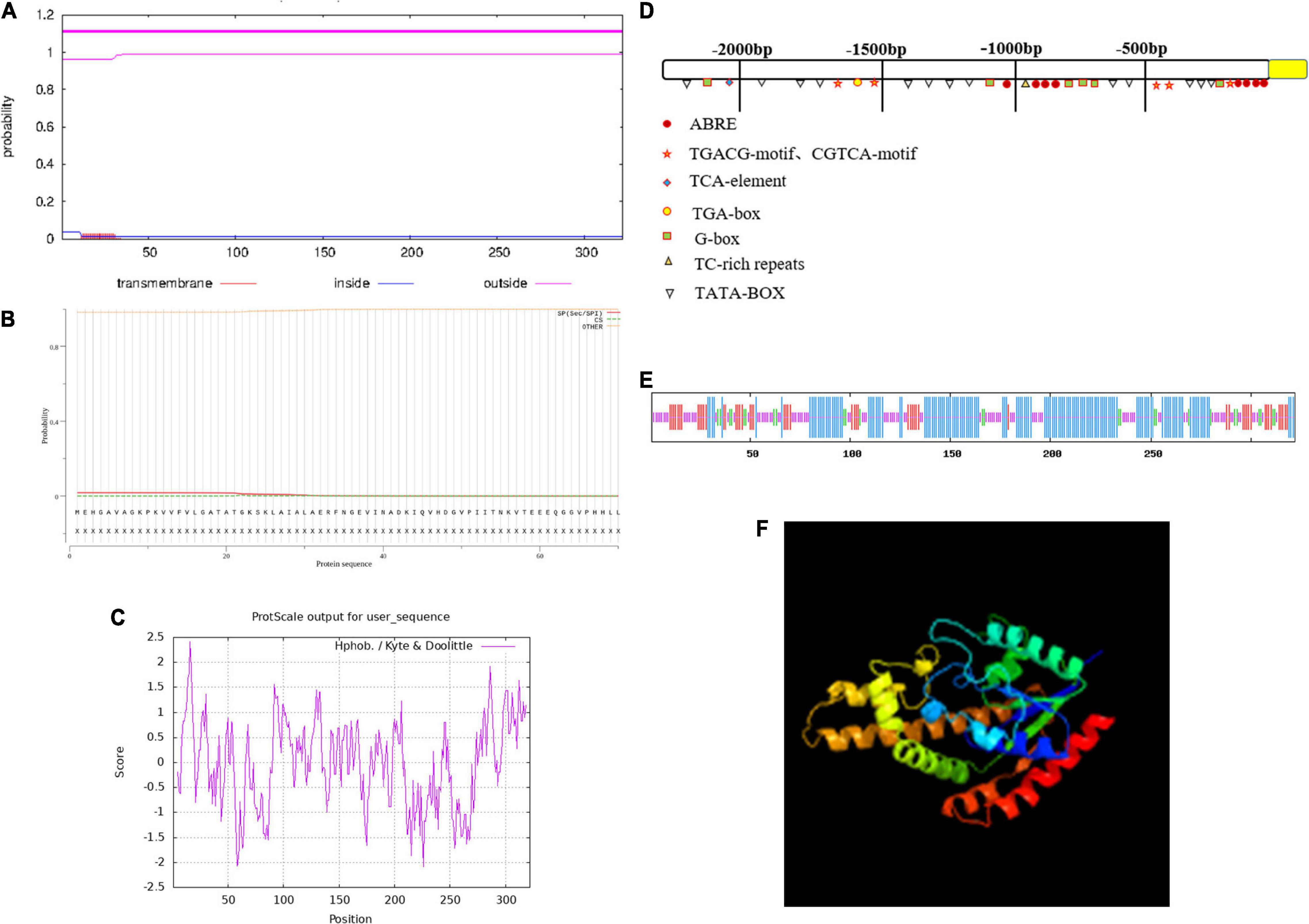
Figure 1. The characterization of the ZmIPT2 protein. (A) Transmembrane domain prediction of ZmIPT2. (B) Prediction of signal peptide and splice siteS. (C) Hydrophobicity analysis of ZmIPT2 coding protein. (D) Analysis of flanking sequence of ZmIPT2 gene. (E) Secondary structure analysis of the ZmIPT2. The helix (h), extended strand (e), coil(c), and turn (t) are indicated in different color as blue, red, yellow, and green, respectively. (F) 3-D structure model of ZmIPT2.
The CD search results indicated that the ZmIPT2 protein contained a conserved P-loop NTPase domain (Supplementary Figure 2). A BLAST search of the NCBI protein database showed that the predicted amino acid sequence of ZmIPT2 had different degrees of similarity with other known proteins. IPT protein sequences of 35 different species were downloaded, including those of Sorghum bicolor, Panicum miliaceum, Panicum hallii, and Dichanthelium oligosanthes (Supplementary Figure 3A). The results of the constructed evolutionary tree showed that ZmIPT2, SbIPT3, and DoIPT3 were in the same branch, among which ZmIPT2 and SbIPT3 had the closest genetic relationship with an amino acid consistency of 86.18% (Supplementary Figure 3B). The evolutionary grouping of proteins may reveal similarities and differences in function.
RT-PCR was performed using RNA from the maize line C01 as template for the synthesis of cDNA. The amplified cDNA fragment showed a 969-bp band in a 1% agarose gel and was ligated into the pEasy-T1 Simple vector. Sequencing showed that the plasmid contained the same sequence as the candidate gene in MaizeGDB10. The reconstructed full length of the cDNA sequence, which was 1339-bp long, was identified as the sequence of the full-length ZmIPT2 gene, including the 969 bp CDS, 155 bp 5′-UTR, and 216 bp 3′-UTR (Supplementary Figure 4).
Over 30 T0 independent transgenic plant lines overexpressing the ZmIPT2 gene from Agrobacterium tumefaciens were established and propagated in the greenhouse. The primer pair ZmIPT2-F/ZmIPT2-R was shown to specifically recognize transgenic maize ZmIPT2 by PCR amplification (Supplementary Figures 5A,B). Three transgenic maize lines (DNIPT2-C14, DNIPT2-C33, and DNIPT2-C34) with a single ZmIPT2 integration event were verified by copy number analysis of the T3 generation (Table 2).
Quantitative real-time PCR (qRT-PCR) was used to monitor the expression of ZmIPT2 in the leaves of transgenic lines at different growth stages (Figures 2A,B). The overexpressed ZmIPT2 gene first increased and then decreased with the development of the maize plants. The relative expression level of the gene reached its peak at the R1 stage, and then decreased in the following stages. The stability of ZmIPT2 in T2 and T3 plants was monitored by RT-PCR. No expression of the ZmIPT2 gene was detected in receptor line C01 (Supplementary Figures 5C,D).
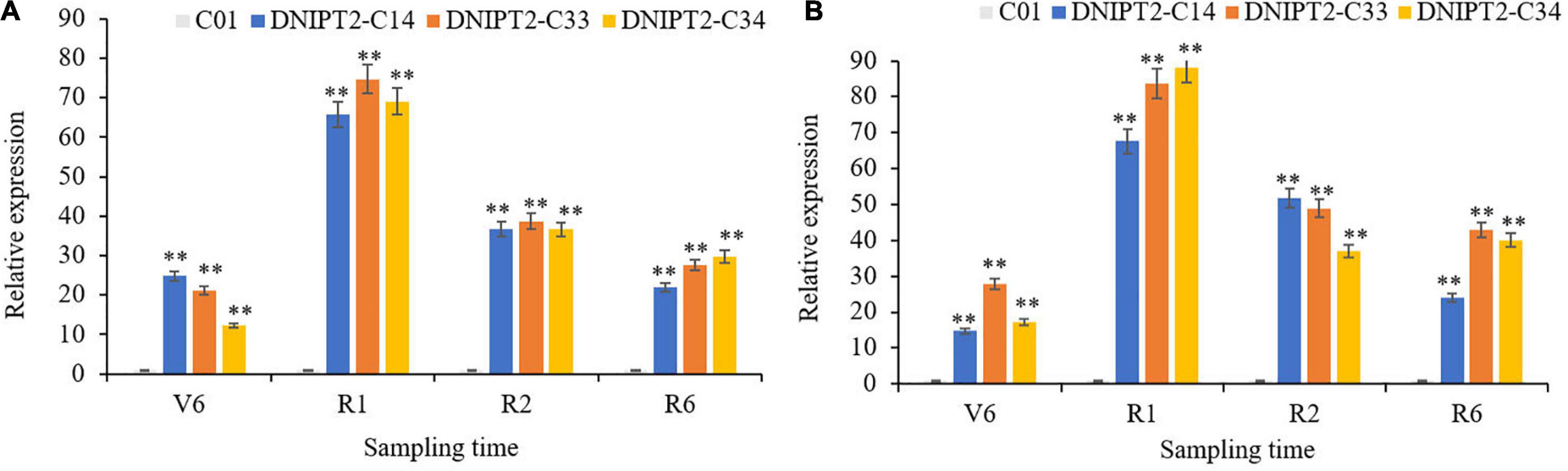
Figure 2. Relative expression levels of transgenic ZmIPT2 gene in maize leaves at different stages in T2 and T3 generation. (A) Shows Relative expression levels of transgenic ZmIPT2 gene in maize leaves at different stages in T2 generation, V6 (sixth leaf), R1 (silking), R2 (blister stage) and R6 (physical maturity); (B) shows Relative expression levels of transgenic ZmIPT2 gene in maize leaves at different stages in T3 generation. The error bars denote standard deviations of the qRT-PCR signals (n = 3). There was significant difference or extremely significant difference in the expression level of ZmIPT2 (**P < 0.01, Student t-test).
The most direct manifestations of maize leaf senescence are a decrease in photosynthetic rate and a decrease in chlorophyll content. Chlorophyll content was recorded at multiple growth stages, and the data showed that the chlorophyll levels in the transgenic lines were significantly higher in the four growth stage of the transgenic line compared to the receptor line. The transgenic plants maintained better canopy coverage and higher chlorophyll levels than recipient plants. In addition, the relative chlorophyll content of transgenic lines was the lowest at V6 (jointing stage), reached the peak at R1 (flowering stage), then decreased slowly for the remainder of the plants’ life (Figure 3A).
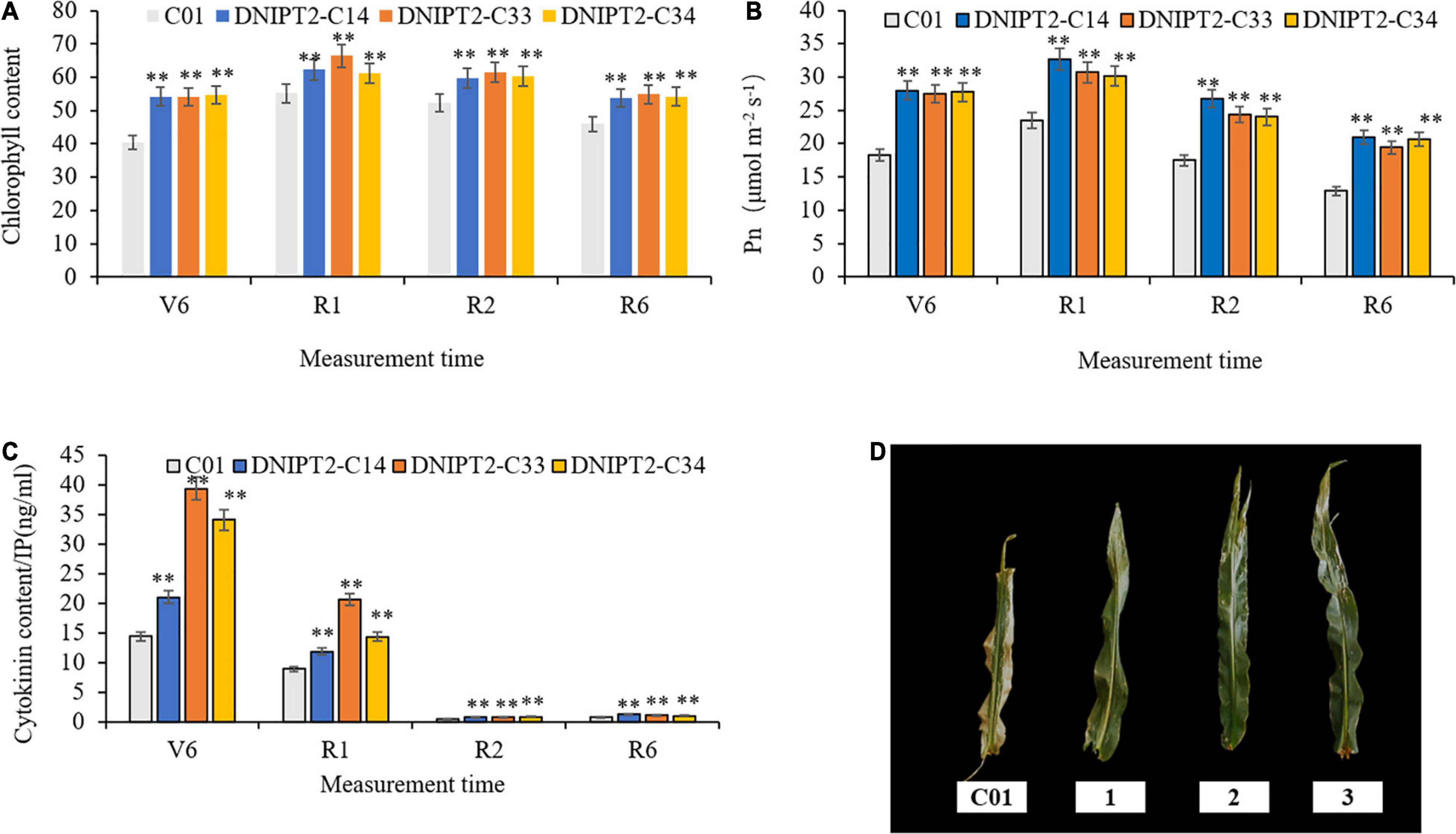
Figure 3. Detection of physiological and biochemical indexes of transgenic maize lines with ZmIPT2 gene. (A) Shows the relative chlorophyll content of transgenic lines at different growth stages of maize, V6 (sixth leaf), R1 (silking), R2 (blister stage) and R6 (physical maturity); (B) shows the photosynthetic rate content of transgenic lines at different growth stages of maize, V6 (sixth leaf), R1 (silking), R2 (blister stage) and R6 (physical maturity); (C) shows the content of cytotaxon (CTK) of transgenic lines at different growth stages of maize, V6 (sixth leaf), R1 (silking), R2 (blister stage) and R6 (physical maturity). (D) The comparison of leaf greenness in T3 transgenic maize lines with ZmIPT2 gene at generation at mature stage, C01, negative control; 1, DNIPT2-C14; 2, DNIPT2-C33; 3, DNIPT2-C34. There was significant difference or extremely significant difference in the expression level of ZmIPT2 (**P < 0.01, Student t-test).
It was found that among the maize plants, the transgenic lines showed a large green leaf area and obvious “stagnant green” phenomenon, while the receptor line showed a greater proportion of leaf edges withered and turned yellow (Figure 3D), thus proving that the overexpression of ZmIPT2 gene postponed the process of leaf senescence. In addition, when compared with the receptor line, the photosynthetic rate of the three transgenic lines had significantly increased by 22.16–39.56% in the four growth stages. With the process of leaf senescence, the photosynthetic efficiency of all lines showed a downward trend after flowering. Compared with the receptor control, the photosynthetic efficiency of the transgenic lines decreased more slowly, maintained high photosynthetic efficiency for a longer time, and the plants accumulated more photosynthetic products (Figure 3B).
The CTKs content in all plants reached its highest at growth phase R1, and then began to decrease (Figure 3C). In R6, the CTKs content of the receptor line was 0.81. The CTKs content of DNIPT2-C14, DNIPT2-C33, and DNIPT2-C34 were significantly higher than that of C01 (P < 0.01) at 1.29, 1.18, and 1.07, respectively. Although the CTKs content of the transgenic line was lower than that of the first three stages, they were still significantly higher than that of the receptor line C01, indicating that the overexpression of ZmIPT2 gene in the transgenic strain resulted in the increase of endogenous CTKs content in leaves and delayed the decline rate of CTKs.
The important agronomic traits and yield-related traits of T2 and T3 transgenic lines were quantified (Table 3). Compared with the recipient control line, the ear length, ear thickness, grain length, grain width, and grain thickness of the transgenic lines increased significantly (Figure 4). The average plot yield of T2 and T3 transgenic plants under normal field planting conditions ranged from 12.49 to 13.05%, with the yield of the DNIPT2-C14 line as the highest. Although the important agronomic traits of the three transgenic strains were improved compared with the recipient control line C01, the difference was not statistically significant, indicating that the introduction of ZmIPT2 gene had no adverse effect on the growth and development of maize plants.
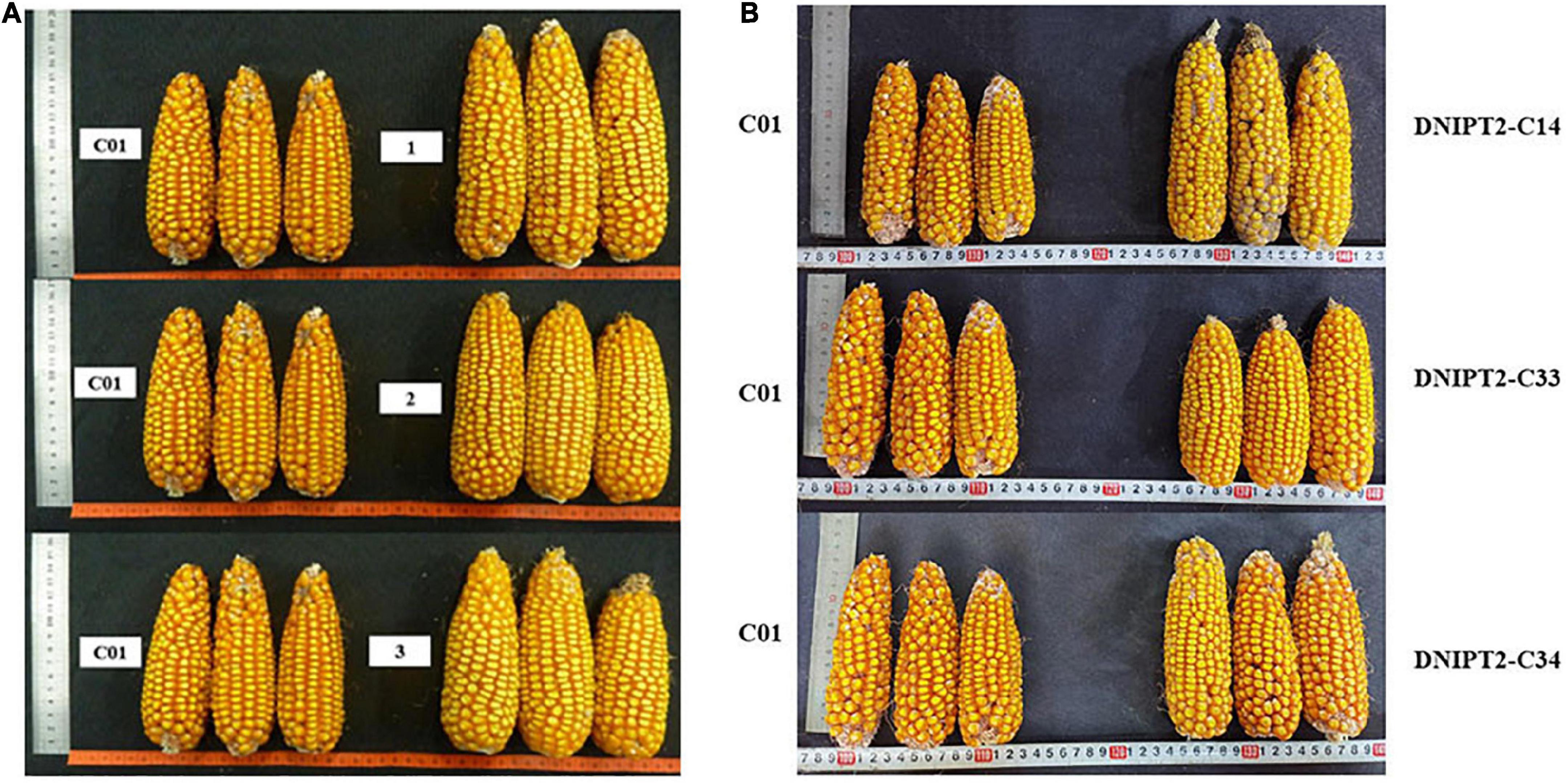
Figure 4. The ear and grain of transgenic maize lines with ZmIPT2 gene and the receptor line C01. (A) The ear of T2 transgenic maize lines with ZmIPT2 gene and the receptor line C01. (B) The ear of T3 transgenic maize lines with ZmIPT2 gene and the receptor line C01. C01, negative control; 1, DNIPT2-C24; 2, DNIPT2-C33; 3, DNIPT2-C34.
Ten proteins were predicted to interact with ZmIPT2 (Figure 5), including GRMZM2G022904, GRMZM2G076936, GRMZM2G168681, GRMZM2G06198, GRMZM2G133082, GRMZM2G14772, GRMZM2G10828, GRMZM2G145029, GRMZM2G098569, and GRMZM2G027059. GRMZM2G022904 and GRMZM2G076936 were cytochrome P450 superfamily proteins, which participate in the decomposition and anabolism of plant hormones. The other interacting proteins were involved in related reactions of IPT. qRT-PCR showed that the temporal and spatial expression patterns of GRMZM2G168681, GRMZM2G147721, and GRMZM2G022904 in transgenic lines were consistent with those of ZmIPT2 gene and reached their maximum at the flowering stage (Figures 5A–C). GRMZM2G022904, GRMZM2G168681, and GRMZM2G147721 were localized in bin 7.02, 8.03, and 8.06, respectively, in regions based on B73 RefGen_v2 genome-wide data analysis. The CD-Search results indicated that their protein contains conserved P450 superfamily and isoprenoid biosynthesis superfamily sequences (Supplementary Figure 6). In addition, GRMZM2G022904 encoded a cytokinin hydroxylase, which can participate in the regulation of isoprenoid CK and affect the development process of plants. However, GRMZM2G168681 and GRMZM2G147721 have not been fully proven to interact with the ZmIPT2 gene. The results of qRT-PCR, bioinformatics, and research reports preliminarily determined that the GRMZM2G022904 gene may interact with ZmIPT2 gene to participate in the molecular regulation mechanisms of anti-aging and increasing yield of maize.
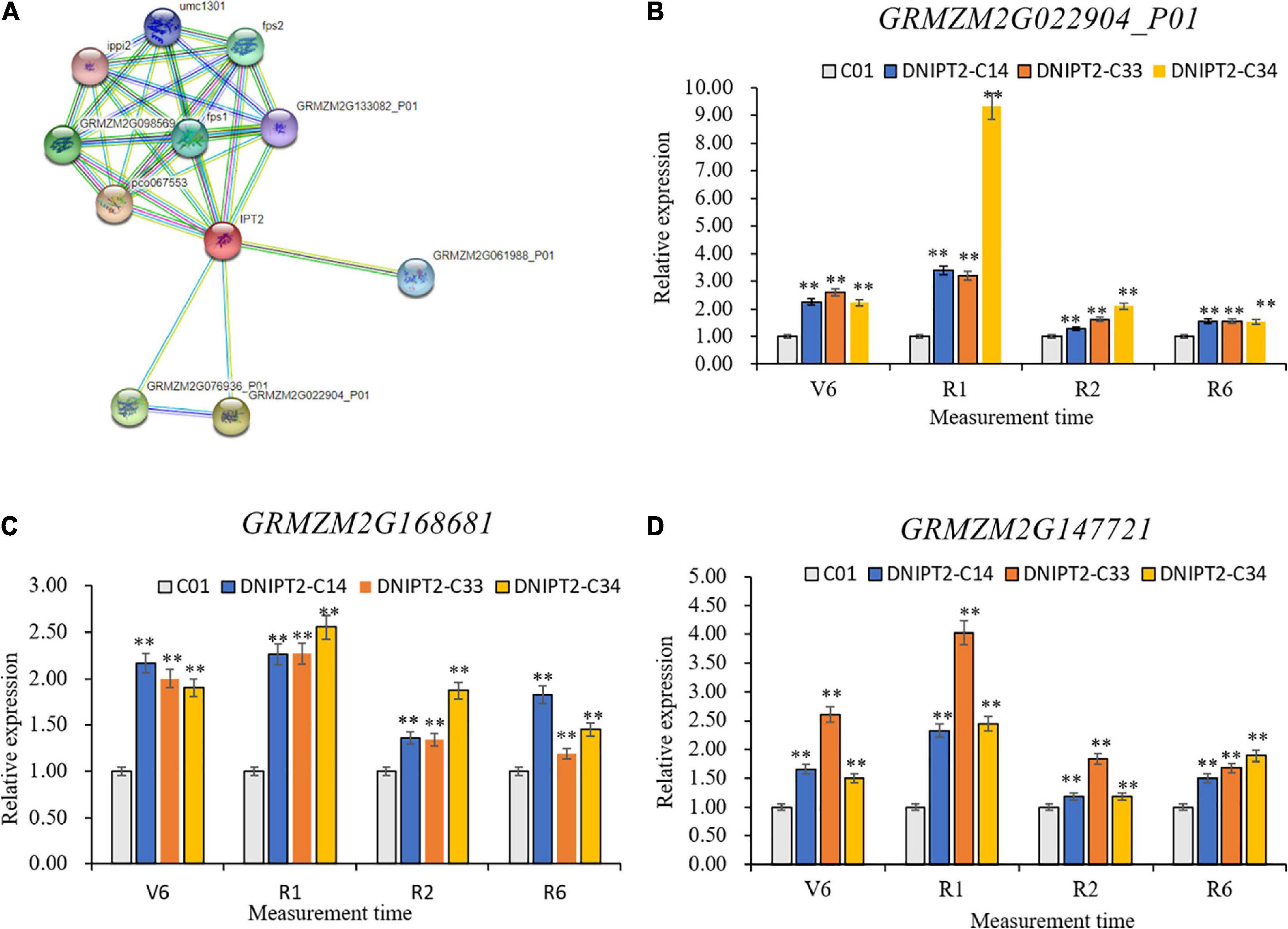
Figure 5. Gene analysis of ZmIPT2 interaction. (A) The predictive and analysis of ZmIPT2 interaction protein. (B) Relative expression levels of transgenic GRMZM2G022904_P01 gene in maize leaves at different stages. (C) Relative expression levels of transgenic GRMZM2G168681 gene in maize leaves at different stages. (D) Relative expression levels of transgenic GRMZM2G147721 gene in maize leaves at different stages. There was significant difference or extremely significant difference in the expression level (**P < 0.01, Student t-test).
Cytokinins are known to play an important function in controlling leaf senescence and increasing grain yield of plants. There are two main approaches to regulating endogenous CTKs, which are utilized by up regulating or down regulating two kinds of genes. One of these genes encodes for IPT, which is the rate-limiting enzyme in CTK synthesis. The other is CTK oxidase/dehydrogenase, which are related to its degradation. These genes can oxidize the side chain groups of CTKs and irreversibly inactivate them.
In rice, the number of grains was increased by the accumulation of CTKs in the inflorescence meristem per panicle. Rice gene Gn1a encodes an oxidase/dehydrogenase (OsCKX2), which plays a role in the degradation of CTKs. The 11 bp deletion in the coding region of OsCKX2 resulted in its early termination, which reduced the expression of OsCKX2 and increased the number of grains (Ashikari et al., 2005). In wheat, the direct homolog of rice OsCKX2, TaCKX6-D1, was correlated with 1000-grain weight significantly through association analysis and linkage mapping (Zhang et al., 2012). Therefore, the regulation pathway of CTKs may play a crucial role in grain yield.
In general, leaf senescence is accompanied by the degradation of chlorophyll, proteins, and nucleic acids, as well as a decrease in photosynthetic rate. CTKs can lessen sugar accumulation, increase chlorophyll synthesis, and prolong the leaf photosynthetic period (Wu et al., 2021). Many studies have shown that the IPT gene plays an important role in the regulation of endogenous CTK activity required for cell division (Qin et al., 2011) and can delay leaf senescence. Guo and Gan (2011) found that the expression of the IPT gene at axillary bud sites was increased in Arabidopsis mutant MYB2, resulting in the increase of the CTKs content, and a significant delay in the aging of the whole plant (Brugiere et al., 2008).
Currently, PSAG12:IPT, a carrier inducing IPT expression by the specific promoter of leaf senescence, has been transferred to a variety of grains and cash crops such as rapeseed, wheat, tomato, cauliflower, cotton, tobacco, and corn, and has shown to significantly delay the aging of leaves and other organs, affecting plant growth and development (Guo and Gan, 2011). PSAG12 drives IPT gene expression, increases cytokinin content, and delays leaf senescence. The increase of cytokinin in leaves can cause feedback into the inhibition of promoter activity and turn off the IPT gene, thus playing a role of self-regulation. The cytokinins synthesized by mature leaves of transgenic plants cannot be transported to other parts of the plant, thus they do not affect the normal development the plant, solving the problem of abnormal morphology of IPT constitutive overexpression transgenic plants. The construction of the chimeric gene PSAG12:IPT provides the possibility for the practical application of cytokinin biosynthesis related enzyme genes to regulate the senescence process of plant leaves (Liang et al., 2006).
Although the physiological function of CTK in delaying leaf senescence has been elucidated, the downstream molecular mechanisms of CTK in regulating leaf senescence is still not well understood. At present, it is generally believed that the CTK two-component system (TCS) pathway is involved in the regulation of CTK on leaf senescence. TCS is a multistep phosphate delivery system that includes histidine protein kinase (HKS) and downstream type A and type B response regulators (RR). The CTK response regulator (CRR) is located downstream of the signal and participates in the regulation of the physiological functions of CTK (Sakakibara, 2006). The first resolved downstream molecular mechanism of CTK regulating leaf senescence revealed that AHK3 played an important role in the CTK delay of leaf senescence. The functional deletion mutation of AHK3 delayed the CTK dependent leaf senescence and reduced the plants’ sensitivity to CTK, and removed the CTK dependent phosphorylation of the downstream response factor ARR2 of type B CTK, thus regulating leaf senescence (Kim et al., 2006). AHK2/AHK3 can be used as a receptor kinase combination to participate in the accumulation and regulation of plastid coding gene transcripts. The double mutant showed phenotypes such as fewer leaf cells and low chlorophyll content, both of which significantly affected development process (Riefler et al., 2006). In addition, ARR16 could significantly induce leaf senescence through overexpression of type A CTK response factor, and the senescence process could not be inhibited by 6-BA, indicating that ARR16 was involved in the CTK regulation of leaf senescence (Ren et al., 2009).
In maize kernels, CTKs levels peaked at approximately 10 days after pollination (DAP) (Brugiere et al., 2003). Our results revealed that the chlorophyll content, CTKs, and photosynthetic rate rose substantially throughout the development of transgenic plants. Additionally, the CTK content reached its maximum in the R1 period, which was consistent with previous reports, indicating that R1 is an important period for ZmIPT2 to regulate CTKs, which in turn affected the later grain formation stage. Although CTKs content showed an overall decrease in the R6 period, transgenic strains still had a higher CTKs content than the receptor control. In the later stages of development, the leaves of the transgenic strains remained consistently green, and the yields of DNIPT2-C14, DNIPT2-C33, and DNIPT2-C34 were significantly higher than that of the receptor control. These results show that ZmIPT2 gene could up-regulate the expression of CTKs content in the later stages of growth and development, improve the photosynthetic rate and chlorophyll content, and play a significant role in delaying leaf senescence, which were consistent with the results of previous studies.
Genetic improvement of grain yield is of great importance to insure food security. A large number of significant agronomic characteristics including yield showed continuous phenotypic variation. As a complex quantitative character, grain yield is determined many factors, such as grain number and grain weight (Melchinger et al., 1998; Yano, 2001). Various studies have proved that IPT gene has the function of improving crop yield and quality (Yang et al., 2017).
There were two types of IPT, one of which is an adenine modified tRNA, called tRNA IPT (EC. 2.5.1.8), which catalyzes the transfer of isopentenyl of dimethene diphosphate to the adenine residue of the precursor tRNA molecule to form a mature tRNA molecule. The modified nucleotide was located adjacent to the anti-codon, affecting the fidelity and efficiency of transcription. The other type of isopentyl transferase catalyzes the formation of IPMP, and is called adenylate isopentenyl transferase (IPT: ec2.5.1.27) which has been identified in Agrobacterium tumefaciens with a structure similar to tRNA IPT.
Studies based on ATP/ADP mutants and tRNA IPT showed that ATP/ADP IPTs are involved in the synthesis of the bulk of isopentenyladenine and trans-zeatin (t-Z) type CTK, while tRNA IPTs are required for cis-zeatin (cis-Z) type CTK production (Miyawaki et al., 2006). An ATP/ADP-PpIPTs study found that the overexpression of PpIPT1, PpIPT3, PpIPT5, and PpIPT7 genes in Arabidopsis can increase cytokinin content in transgenic plants and improve salt resistance (Li et al., 2018). CRISPR/Cas9 targeted gene knockout Phtheirospermum japonicum (Pj)IPT1a inhibits parasite induced CTK response in host, revealing the important role of PjIPT1a in Arabidopsis phloem-induced CTK response (Greifenhagen et al., 2021). In the transgenic Camellia sinensis containing IPT5, the 3’ UTR variant 2 (3AS2) was found to be the main transcript which participated in the regulation of tea axillary bud germination and shoot branching, providing gene resources for improving the plant type of woody plants (Zhang et al., 2021). In addition, the triple mutant of TaIPT8-5a/5b/5d showed a decreased t-Z type CTK level and reduced drought tolerance under both normal and drought conditions. In contrast, drought induced TaIPT8 transgenic wheat plants showed stronger drought tolerance, indicating that CTK plays a beneficial role in drought tolerance by regulating the redox state of cells (Wang et al., 2022).
Previous studies have elucidated the contribution of CTKs to crop yield (Jameson and Song, 2016; Chen et al., 2020). The specific role of IPTs in crop yield and its role in controlling grain production should be emphasized. Many efforts have been made to modify the spatial-temporal expression of IPTs, using promoters with different drivers to improve crop yield. For example, the IPT gene is responsible for delayed foliar senescence under the control of the SAG12 promoter (Gan and Amasino, 1995), which increased plant productivity (Kant et al., 2015). Plants that contained IPT driven by the SARK promoter had higher yield than the non-transgenic control (Leta et al., 2016).
Strategies for increasing yield should be focused on transgenic approaches based on the specific expression of IPT genes in early grain development (Sykorova et al., 2008). Cytokinin accumulation was increased in rice inflorescence meristems by decreasing the expression of the cytokinin degrading enzyme, cytokinin oxidase/dehydrogenase (OSCKX2), which in turn increased grain yield due to the increased number of reproductive organs (Ashikari et al., 2005). With the discovery of biosynthetic enzymes IPT1 to IPT10 (Irie et al., 2004; Brugiere et al., 2008; Vyroubalova et al., 2009) and degrading enzymes CKX1 to CKX12, the role of CTKs in maize kernel development has been made clearer (Morris et al., 1999; Brugiere et al., 2003; Massonneau et al., 2004; Smehilova et al., 2009; Vyroubalova et al., 2009). In IPTs, IPT1, and IPT10 are abundantly constitutively expressed in all organs, but other IPT transcripts show different spatiotemporal expression patterns (Vyroubalova et al., 2009). In addition, another study showed that the modified cytokinin-degrading enzyme Oslogl5 plants had significantly increased seed setting rate, total grains, fuller grains per panicle, and 1000-grain weight under drought conditions (Wang et al., 2020). Cheikh and Jones (1994) found that in the vegetative organs of maize, the content of CTKs varied with the expression level of the related gene. When the expression level of ZmIPT2 reached the maximum, the cytokinin also reached the maximum, which showed that the expression level of ZmIPT2 gene in the cell core was closely related to the content of cytokinin. In maize kernels, the endosperm, especially the basal metastatic cell layer (BETL), was the main expression site of ZmIPT2 at 8–10 days after maize pollination, and its expression in BETL and endosperm continued until late development, suggesting that this gene plays an important role in CTK biosynthesis (Brugiere et al., 2008).
In vitro studies showed that ZmIPT2-thymine (T) with ADP, ATP, or AMP as substrate, had higher IPT activity than ZmIPT2-cytosine (C). It was found that a favorable ZmIPT2-T allele was related to grain weight (Weng et al., 2013). Yang recently reported that overexpression of IPT2 gene in maize could regulate cytokinin content. Compared to the control, the photosynthetic rate and chlorophyll content of transgenic maize were significantly increased, delaying leaf senescence and increasing yield (Yang et al., 2017). These results indicated that regulating the metabolism of CTKs in reproductive organs may be an effective way to increase crop yield by increasing the flow of assimilates from source to sink or increasing sink capacity (Weng et al., 2013).
In addition, the high yield of crops has been shown to be closely related to the amount of source material on the plant. Leaves are the main source tissue for plants to carry out photosynthesis (Andrade et al., 2000). Therefore, crop yield can be roughly determined by the number of leaf sources, as well as the photosynthetic capacity of these leaf sources. With increasing age, leaf senescence also increases. By prolonging the functional period of leaves by delaying senescence, grain yield and plant biomass can be significantly improved (Dong et al., 2000; Thomas and Smart, 2010). Waggoner and Berger (1987) successfully used the functional period of green leaf area to express the correlation between maize leaf greenness and yield. However, some scholars believe that there is no obvious correlation between maize greenness and yield (Jiang et al., 2004; Massonneau et al., 2004; Wada and Wada, 2008).
This study discovered that the green maintenance capability of the leaves of the overexpression ZmIPT2 strain after silking was positively correlated with the plot yield. The plot yield increased by an average of 12.49, 13.05, and 12.90%, respectively, compared to the yield of the receptor control strain. In this experiment, the ZmIPT2 gene was transformed into maize to increase the yield, which was a rapid method when compared with other high-yield breeding methods. Three genetically stable T3 generation transgenic lines were obtained from this study. Our results showed that the overexpression of ZmIPT2 gene could delay leaf senescence and significantly or extremely significantly improve yield traits such as ear length, ear diameter, 100 grain weight, and plot yield by significantly increasing plant CTKs content, net photosynthetic rate, and relative chlorophyll content. Among the transgenic lines, the DNIPT2-C14 line showed significantly increased yield traits when compared to the control, with a plot yield increase of 20.29% compared with the control. The field experiment was conducted for only 2 years: yield increase degree takes many years/field experiments to further confirm, so the results of this study, though promising, may be inaccurate. The three single-copy high-yield transgenic lines obtained in this will be screened out by backcross transfer method, and the target genes will be transferred into the backbone maize inbred lines for the preparation of hybrid combinations to breed new varieties of high-yield transgenic maize.
The study of IPT, the main regulator of plant yield, provides important insights for crop breeding such as yield improvement by way of abiotic stress tolerance. Recent studies have shown that CYP735A1 and CYP735A2 are cytochrome P450 monooxygenases (P450), which catalyze the biosynthesis of t-Z (Takei et al., 2004). In Arabidopsis, it was found that that the co-expression of adenosine monopentenyltransferase AtIPT4 and CYP735A enabled yeast to excrete t-Z and nucleosides into the culture medium. Similarly, the GRMZM2G022904 gene from this study is a cytochrome P450 (CYP450) superfamily protein, namely cytokinin hydroxylase, which can participate in the decomposition and anabolism of plant hormones and synthesis and metabolism of terpenoids, phenylpropanoids, alkaloids, sterols, and fatty acids in plants. Its mechanism of action in catalytic reaction is diverse and complex, and it is referred to as a universal catalyst (Schuler and Werck-Reichhart, 2003). The expression level of the GRMZM2G022904 gene was positively correlated with the expression of ZmIPT2 gene and positively regulated the ZmIPT2 gene to participate in the IPT reaction and delay leaf senescence. However, our study only preliminarily verified that GRMZM2G022904 was a key gene involved in the isopentenyl transferase pathway and the interaction between ZmIPT2 and GRMZM2G022904 by qRT-PCR. The relationship and mode of action between these two genes remains to be further studied.
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
ZW and HD contributed to the conception and design of the work. YS, CL, and YZ performed the experiment. PG, QW, and LZ analyzed the data. YS and CL wrote the manuscript. All authors read and approved the final manuscript.
This research was supported by the Genetically Modified Organisms Breeding Major Projects of China (2016ZX08003003).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the strains and carriers presented by Lixinhai, researcher of Crop Research Institute, Chinese Academy of Agricultural Sciences.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.963873/full#supplementary-material
An, X., Zhang, J., Liao, Y., Liu, L., Peng, D., and Wang, B. (2017). Senescence is delayed when ramie (Boehmeria nivea L.) is transformed with the isopentyl transferase (ipt) gene under control of the SAG12 promoter. FEBS Open Bio 7, 636–644. doi: 10.1002/2211-5463.12191
Andrade, F. H., Otegui, M. E., and Vega, C. (2000). Intercepted radiation at flowering and kernel number in Maize. Agron. J. 92, 92–97. doi: 10.2134/agronj2000.92192x
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Balibrea Lara, M. E., Gonzalez Garcia, M. C., Fatima, T., Ehness, R., Lee, T. K., Proels, R., et al. (2004). Extracellular invertase is an essential component of cytokinin-mediated delay of senescence. Plant Cell 16, 1276–1287. doi: 10.1105/tpc.018929
Beznec, A., Faccio, P., Miralles, D. J., Abeledo, L. G., Oneto, C. D., Garibotto, M. B., et al. (2021). Stress-induced expression of IPT gene in transgenic wheat reduces grain yield penalty under drought. J. Genet. Eng. Biotechnol. 19, 67–84. doi: 10.1186/s43141-021-00171-w
Brenner, M. L., and Cheikh, N. (1995). “The role of hormones in photosynthate partitioning and seed filling,” in Plant Hormones, ed. P. J. Davies (Dordrecht: Springer), 649–670. doi: 10.1007/978-94-009-3585-3_25
Brugiere, N., Humbert, S., Rizzo, N., Bohn, J., and Habben, J. E. (2008). A member of the maize isopentenyl transferase gene family, Zea mays isopentenyl transferase 2 (ZmIPT2), encodes a cytokinin biosynthetic enzyme expressed during kernel development. Cytokinin biosynthesis in maize. Plant Mol. Biol. 67, 215–229. doi: 10.1007/s11103-008-9312-x
Brugiere, N., Jiao, S., Hantke, S., Zinselmeier, C., Roessler, J. A., Niu, X., et al. (2003). Cytokinin oxidase gene expression in maize is localized to the vasculature, and is induced by cytokinins, abscisic acid, and abiotic stress. Plant Physiol. 132, 1228–1240. doi: 10.1104/pp.102.017707
Chang, H., Jones, M. L., Banowetz, G. M., and Clark, D. G. (2003). Overproduction of cytokinins in petunia flowers transformed with P(SAG12)-IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiol. 132, 2174–2183. doi: 10.1104/pp.103.023945
Cheikh, N., and Jones, R. J. (1994). Disruption of Maize Kernel Growth and Development by Heat Stress (Role of Cytokinin/Abscisic Acid Balance). Plant Physiol. 106, 45–51. doi: 10.1104/pp.106.1.45
Chen, L., Zhao, J., Song, J., and Jameson, P. E. (2020). Cytokinin dehydrogenase: a genetic target for yield improvement in wheat. Plant Biotechnol. J. 18, 614–630. doi: 10.1111/pbi.13305
Chory, J., Reinecke, D., Sim, S. S., Washburn, T. T., and Brenner, M. (1994). A role for cytokinins in De-Etiolation in Arabidopsis (det Mutants Have an Altered Response to Cytokinins). Plant Physiol. 104, 339–347. doi: 10.2307/4275627
Decima Oneto, C., Otegui, M. E., Baroli, I., Beznec, A., Faccio, P., Bossio, E., et al. (2016). Water deficit stress tolerance in maize conferred by expression of an isopentenyltransferase (IPT) gene driven by a stress- and maturation-induced promoter. J. Biotechnol. 220, 66–77. doi: 10.1016/j.jbiotec.2016.01.014
Delatorre, C. A., Cohen, Y., Liu, L., Peleg, Z., and Blumwald, E. (2012). The regulation of the SARK promoter activity by hormones and environmental signals. Plant Sci. 193-194, 39–47. doi: 10.1016/j.plantsci.2012.05.005
Dong, H., Niu, Y., Li, W., and Zhang, D. (2008). Effects of cotton rootstock on endogenous cytokinins and abscisic acid in xylem sap and leaves in relation to leaf senescence. J. Exp. Bot. 59, 1295–1304. doi: 10.1093/jxb/ern035
Dong, S. T., Wang, K. J., and Hu, C. H. (2000). Development of canopy apparent photosynthesis among maize varieties from different Eras. Acta Agron. Sin. 26, 200–204. doi: 10.3321/j.issn:0496-3490.2000.02.012
Gan, S., and Amasino, R. M. (1995). Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270, 1986–1988. doi: 10.1126/science.270.5244.1986
Ghiasi, H., Paech, C., and Dybing, C. D. (1987). Free amino acid content and metabolic activities of setting and aborting soybean ovaries. Plant Physiol. 85, 91–95. doi: 10.1104/pp.85.1.91
Greifenhagen, A., Braunstein, I., Pfannstiel, J., Yoshida, S., Shirasu, K., Schaller, A., et al. (2021). The Phtheirospermum japonicum isopentenyltransferase PjIPT1a regulates host cytokinin responses in Arabidopsis. New Phytol. 232, 1582–1590. doi: 10.1111/nph.17615
Guo, Y., and Gan, S. (2011). AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 156, 1612–1619. doi: 10.1104/pp.111.177022
Hallmark, H. T., and Rashotte, A. M. (2020). Cytokinin isopentenyladenine and its glucoside isopentenyladenine-9G delay leaf senescence through activation of cytokinin-associated genes. Plant Direct 4:e00292. doi: 10.1002/pld3.292
Han, Q., Liu, R. F., Lu, L. H., and Shou, H. X. (2016). Detection of transgene copy number in transgenic soybean by real-time fluorescence quantitative PCR. J. Nucl. Agric. Sci. 30, 646–653. doi: 10.11869/j.issn.100-8551.2016.04.0646
Hirose, N., Takei, K., Kuroha, T., Kamada-Nobusada, T., Hayashi, H., and Sakakibara, H. (2008). Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59, 75–83. doi: 10.1093/jxb/erm157
Huff, A., and Dybing, C. D. (1980). Factors affecting shedding of flowers in Soybean (Glycine max (L.) Merrill). J. Exp. Bot. 31, 751–762. doi: 10.1093/jxb/31.3.751
Irie, Y., Itokazu, N., Anjiki, N., Ishige, A., Watanabe, K., and Keung, W. M. (2004). Eugenol exhibits antidepressant-like activity in mice and induces expression of metallothionein-III in the hippocampus. Brain Res. 1011, 243–246. doi: 10.1016/j.brainres.2004.03.040
Jameson, P. E., and Song, J. (2016). Cytokinin: a key driver of seed yield. J. Exp. Bot. 67, 593–606. doi: 10.1093/jxb/erv461
Jiang, G. H., He, Y. Q., Xu, C. G., Li, X. H., and Zhang, Q. (2004). The genetic basis of stay-green in rice analyzed in a population of doubled haploid lines derived from an indica by japonica cross. Theor. Appl. Genet. 108, 688–698. doi: 10.1007/s00122-003-1465-z
Jin, J., Ran, Q., Zhang, X. P., Chen, M., and Chen, C. X. (2002). Investigation of the A2Σ- state of CuO by laser-induced fluorescence. Chin. Phys. 11, 481–485. doi: 10.1088/1009-1963/11/5/313
Joshi, S., Choukimath, A., Isenegger, D., Panozzo, J., Spangenberg, G., and Kant, S. (2019). Improved wheat growth and yield by delayed leaf senescence using developmentally regulated expression of a cytokinin biosynthesis gene. Front. Plant Sci. 10:1285. doi: 10.3389/fpls.2019.01285
Kant, S., Burch, D., Badenhorst, P., Palanisamy, R., Mason, J., and Spangenberg, G. (2015). Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.). PLoS One 10:e0116349. doi: 10.1371/journal.pone.0116349
Kieber, J. J., and Schaller, G. E. (2014). Cytokinins. Arabidopsis Book 12:e0168. doi: 10.1199/tab.0168
Kieber, J. J., and Schaller, G. E. (2018). Cytokinin signaling in plant development. Development 145:dev149344. doi: 10.1242/dev.149344
Kim, H. J., Ryu, H. J., Hong, S. H., Woo, H. R., Lim, P. O., Lee, I. C., et al. (2006). Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabido. Proc. Natl. Acad. Sci. U.S.A. 103, 814–819. doi: 10.1073/pnas.0505150103
Kudo, T., Kiba, T., and Sakakibara, H. (2010). Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52, 53–60. doi: 10.1111/j.1744-7909.2010.00898.x
Kuiper, D. (1993). Sink strength: established and regulated by plant growth regulators. Plant Cell Environ. 16, 1025–1026. doi: 10.1111/j.1365-3040.1996.tb02052.x
Leta, T. B., Miccah, S. S., Steven, M. R., Wondyifraw, T., Charless, M., Clet, W. M., et al. (2016). Drought tolerant tropical maize (Zea mays L.) developed through genetic transformation with isopentenyltransferase gene. Afr. J. Biotechnol. 15, 2447–2464. doi: 10.5897/ajb2016.15228
Li, B., Wei, A., Song, C. X., Li, N., and Zhang, J. R. (2008). Heterologous expression of the TsVP gene improves the drought resistance of maize. Plant Biotechnol. J. 6, 146–159. doi: 10.1111/j.1467-7652.2007.00301.x
Li, H. L., and Zhao, D. G. (2011). IPT gene expression increased the aphid-resistant of transgenic plant in oilseed. Mol. Plant Breed. 9, 343–349. doi: 10.3969/mpb.009.000343
Li, M. J., Wei, Q. P., Peng, F. T., Yu, W., Luo, J. J., and Zhao, Y. F. (2018). Identification and Characterization of ATP/ADP Isopentenyltransferases (ATP/ADP PpIPTs) Genes in Peach. J. Plant Growth Regul. 38, 416–430. doi: 10.1007/s00344-018-9851-6
Li, W., Herrera-Estrella, L., and Tran, L. P. (2016). The Yin-Yang of cytokinin homeostasis and drought acclimation/adaptation. Trends Plant Sci. 21, 548–550. doi: 10.1016/j.tplants.2016.05.006
Liang, Q. X., Cao, G. Q., Su, M. J., and Qin, G. Y. (2006). Research progress on plant leaf senescence. Chin. Agric. Sci. Bull. 22, 282–285. doi: 10.3969/j.issn.1000-6850.2006.08.072
Liu, M. S., Li, H. C., Chang, Y. M., Wu, M. T., and Chen, L. F. (2011). Proteomic analysis of stress-related proteins in transgenic broccoli harboring a gene for cytokinin production during postharvest senescence. Plant Sci. 181, 288–299. doi: 10.1016/j.plantsci.2011.06.005
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Massonneau, A., Houba-Herin, N., Pethe, C., Madzak, C., Falque, M., Mercy, M., et al. (2004). Maize cytokinin oxidase genes: differential expression and cloning of two new cDNAs. J. Exp. Bot. 55, 2549–2557. doi: 10.1093/jxb/erh274
McCabe, M. S., Garratt, L. C., Schepers, F., Jordi, W. J. R. M., Stoopen, G. M., Davelaar, E., et al. (2001). Effects of PSAG12-IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol. 127, 505–516. doi: 10.1104/pp.010244
Melchinger, A. E., Friedrich Utz, H., and Schön, C. C. (1998). Quantitative Trait Locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL Effects. Genetics 149, 383–403. doi: 10.1016/1369-5266(88)80015-3
Miyawaki, K., Tarkowski, P., Matsumoto-Kitano, M., Kato, T., Sato, S., Tarkowska, D., et al. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 16598–16603. doi: 10.1073/pnas.0603522103
Morris, R. O., Bilyeu, K. D., Laskey, J. G., and Cheikh, N. N. (1999). Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem. Biophys. Res. Commun. 255, 328–333. doi: 10.1006/bbrc.1999.0199
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant ONA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Nagel, L. (2001). Cytokinin regulation of flower and pod Set in Soybeans (Glycine max(L.) Merr.). Ann. Bot. 88, 27–31. doi: 10.1006/anbo.2001.1423
Noh, Y. S., and Amasino, R. M. (1999). Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 41, 181–194. doi: 10.1023/A:1006342412688
Panda, B. B., Sekhar, S., Dash, S. K., Behera, L., and Shaw, B. P. (2018). Biochemical and molecular characterisation of exogenous cytokinin application on grain filling in rice. BMC Plant Biol. 18:89. doi: 10.1186/s12870-018-1279-4
Peleg, Z., Reguera, M., Tumimbang, E., Walia, H., and Blumwald, E. (2011). Cytokinin-mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water-stress. Plant Biotechnol. J. 9, 747–758. doi: 10.1111/j.1467-7652.2010.00584.x
Peng, K. X., Zhang, W., Zhu, X. X., and Zhang, K. W. (2021). Research progress on the mechanisms of cytokinin-inhibited leaf senescence. Plant Physiol. J. 57, 12–18. doi: 10.13592/j.cnki.ppj.2020.0149
Qin, H., Gu, Q., Zhang, J., Sun, L., Kuppu, S., Zhang, Y., et al. (2011). Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiol. 52, 1904–1914. doi: 10.1093/pcp/pcr125
Reguera, M., Peleg, Z., Abdel-Tawab, Y. M., Tumimbang, E. B., Delatorre, C. A., and Blumwald, E. (2013). Stress-induced cytokinin synthesis increases drought tolerance through the coordinated regulation of carbon and nitrogen assimilation in rice. Plant Physiol. 163, 1609–1622. doi: 10.1104/pp.113.227702
Ren, B., Liang, Y., Deng, Y., Chen, Q. G., Zhang, J., Yang, X. H., et al. (2009). Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 19, 1178–1190. doi: 10.1038/cr.2009.88
Riefler, M., Novak, O., Strnad, M., and Schmulling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54. doi: 10.1105/tpc.105.037796
Rivero, R. M., Gimeno, J., Van Deynze, A., Walia, H., and Blumwald, E. (2010). Enhanced cytokinin synthesis in tobacco plants expressing PSARK:IPT prevents the degradation of photosynthetic protein complexes during drought. Plant Cell Physiol. 51, 1929–1941. doi: 10.1093/pcp/pcq143
Rivero, R. M., Kojima, M., Gepstein, A., Sakakibara, H., Mittle, R., Gepstein, S., et al. (2007). Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. U.S.A. 104, 19631–19636. doi: 10.1073/pnas.0709453104
Robson, P. H., Donnison, I. S., Wang, K., Frame, B., Pegg, S. E., Thomas, H., et al. (2004). Leaf senescence is delayed in maize expressing the Agrobacterium IPT gene under the control of a novel maize senescence-enhanced promoter. Plant Biotechnol. J. 2, 101–102. doi: 10.1111/j.1467-7652.2004.00054.x
Roitsch, T., and Rainer, E. (2000). Regulation of source/sink relations by cytokinins. Plant Growth Regul. 32, 359–367. doi: 10.1023/A:1010781500705
Sakakibara, H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Sambrook, J., Fritsch, F. E., and Maniatis, T. (1982). Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. BioScience 33, 721–722. doi: 10.2307/1309366
Schuler, M. A., and Werck-Reichhart, D. (2003). Functional genomics of P450s. Annu. Rev. Plant Biol. 54, 629–667. doi: 10.1146/annurev.arplant.54.031902.134840
Singh, R. P. (1992). Association between gene Lr34 for leaf rust resistance and leaf tip necrosis in wheat. Crop Sci. 32, 874–878. doi: 10.2135/cropsci1992.0011183X003200040008x
Smehilova, M., Galuszka, P., Bilyeu, K. D., Jaworek, P., Kowalska, M., Sebela, M., et al. (2009). Subcellular localization and biochemical comparison of cytosolic and secreted cytokinin dehydrogenase enzymes from maize. J. Exp. Bot. 60, 2701–2712. doi: 10.1093/jxb/erp126
Sykorova, B., Kuresova, G., Daskalova, S., Trckova, M., Hoyerova, K., Raimanova, I., et al. (2008). Senescence-induced ectopic expression of the A. tumefaciens ipt gene in wheat delays leaf senescence, increases cytokinin content, nitrate influx, and nitrate reductase activity, but does not affect grain yield. J. Exp. Bot. 59, 3773–3787. doi: 10.1093/jxb/erm319
Takei, K., Sakakibara, H., and Sugiyama, T. (2001). Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 276, 26405–26410. doi: 10.1074/jbc.M102130200
Takei, K., Yamaya, T., and Sakakibara, H. (2004). Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J. Biol. Chem. 279, 41866–41872. doi: 10.1074/jbc.M406337200
Thomas, H., and Smart, C. M. (2010). Crops that stay greed1. Ann. Appl. Biol. 123, 193–219. doi: 10.1111/j.1744-7348.1993.tb04086.x
Vyroubalova, S., Vaclavikova, K., Tureckova, V., Novak, O., Smehilova, M., Hluska, T., et al. (2009). Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiol. 151, 433–447. doi: 10.1104/pp.109.142489
Wada, Y., and Wada, G. (2008). Varietal difference in leaf senescence during ripening period of advanced Indica Rice. Jpn. J. Crop Sci. 60, 529–536. doi: 10.1626/jcs.60.529
Waggoner, P.E., and Berger, R.D. (1987). Defoliation, Disease, and Growth. Phytopathology. 77, 393–398.
Wang, C., Wang, G., Gao, Y., Lu, G., Habben, J. E., Mao, G., et al. (2020). A cytokinin-activation enzyme-like gene improves grain yield under various field conditions in rice. Plant Mol. Biol. 102, 373–388. doi: 10.1007/s11103-019-00952-5
Wang, N., Chen, J., Gao, Y., Zhou, Y. B., Chen, M., Xu, Z. S., et al. (2022). Genomic analysis of isopentenyltransferase genes and functional characterization of TaIPT8 indicates positive effects of cytokinins on drought tolerance in wheat. Crop J. doi: 10.1016/j.cj.2022.04.010
Weng, J. F., Li, B., Liu, C. L., Yang, X. Y., Wang, H. W., Hao, Z. F., et al. (2013). A non-synonymous SNP within the isopentenyl transferase 2 locus is associated with kernel weight in Chinese maize inbreds (Zea mays L.). BMC Plant Biol. 13:98. doi: 10.1186/1471-2229-13-98
Westgate, M. E., and Peterson, C. M. (1993). Cytokinin regulation of flower and pod set in Soybeans (Glycine max(L.) Merr.). J. Exp. Bot. 44, 109–117. doi: 10.1006/anbo.2001.1423
Wiebold, W. J., and Panciera, M. T. (1990). Vasculature of Soybean racemes with altered intraraceme competition. Crop Sci. 30, 1089–1093. doi: 10.2135/cropsci1990.0011183X003000050026x
Wu, S. R., Chen, W. F., and Zhou, X. (1988). Enzyme linked immunosorbent assay for endogenous plant hormones. Plant Physiol. Commun. 5, 53–57.
Wu, W., Du, K., Kang, X., and Wei, H. (2021). The diverse roles of cytokinins in regulating leaf development. Hortic. Res. 8:118. doi: 10.1038/s41438-021-00558-3
Yang, F., Liu, C. H., Yang, X. Y., Weng, J. F., Zhou, Y., Wang, Z. H., et al. (2017). Transformation of ZmIPT2 gene related to leaf senescence into maize and function identification. J. Maize Sci. 25, 45–51. doi: 10.13597/j.cnki.maize.science.20170108
Yano, M. (2001). Genetic and molecular dissection of naturally occurring variation. Curr. Opin. Plant Biol. 4, 130–135. doi: 10.1016/S1369-5266(00)00148-5
Zhang, L., Zhao, Y. L., Gao, L. F., Zhao, G. Y., Zhou, R. H., Zhang, B. S., et al. (2012). TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol. 195, 574–584. doi: 10.1111/j.1469-8137.2012.04194.x
Keywords: maize, leaf senescence, ZmIPT2 gene, grain yield, overexpression
Citation: Song Y, Li C, Zhu Y, Guo P, Wang Q, Zhang L, Wang Z and Di H (2022) Overexpression of ZmIPT2 gene delays leaf senescence and improves grain yield in maize. Front. Plant Sci. 13:963873. doi: 10.3389/fpls.2022.963873
Received: 08 June 2022; Accepted: 28 June 2022;
Published: 19 July 2022.
Edited by:
Xia Xin, Institute of Crop Sciences (CAAS), ChinaReviewed by:
Jianfeng Weng, Institute of Crop Sciences (CAAS), ChinaCopyright © 2022 Song, Li, Zhu, Guo, Wang, Zhang, Wang and Di. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenhua Wang, emhlbmh1YXdhbmdfMjAwNkAxNjMuY29t; Hong Di, ZGlob25nZGhAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.