- 1The Key Laboratory of Biology and Genetic Improvement of Oil Crops, The Ministry of Agriculture and Rural Affairs of PRC, Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, Wuhan, China
- 2Hainan University, Haikou, China
As one of the largest classes of lectins, legume lectins have a variety of desirable features such as antibacterial and insecticidal activities as well as anti-abiotic stress ability. The Sclerotinia disease (SD) caused by the soil-borne fungus Sclerotinia sclerotiorum is a devastating disease affecting most oil crops such as Brassica napus. Here, we identified 130 legume lectin (LegLu) genes in B. napus, which could be phylogenetically classified into seven clusters. The BnLegLu gene family has been significantly expanded since the whole-genome duplication (WGD) or segmental duplication. Gene structure and conserved motif analysis suggested that the BnLegLu genes were well conserved in each cluster. Moreover, relative to those genes only containing the legume lectin domain in cluster VI–VII, the genes in cluster I–V harbored a transmembrane domain and a kinase domain linked to the legume lectin domain in the C terminus. The expression of most BnLegLu genes was relatively low in various tissues. Thirty-five BnLegLu genes were responsive to abiotic stress, and 40 BnLegLu genes were strongly induced by S. sclerotiorum, with a most significant up-regulation of 715-fold, indicating their functional roles in SD resistance. Four BnLegLu genes were located in the candidate regions of genome-wide association analysis (GWAS) results which resulted from a worldwide rapeseed population consisting of 324 accessions associated with SD. Among them, the positive role of BnLegLus-16 in SD resistance was validated by transient expression in tobacco leaves. This study provides important information on BnLegLu genes, particularly about their roles in SD resistance, which may help targeted functional research and genetic improvement in the breeding of B. napus.
Introduction
Biotic and abiotic stresses severely disrupt the homeostasis of the plant immune system and cause great negative effects on plant growth, development, and yield (Kosová et al., 2011). Plant lectins are widely distributed in nature, and play important roles in the plant immune system (Katoch and Tripathi, 2021). Numerous induced lectins and lectin receptor-like kinases have been reported to be involved in various stress responses (Van Damme, 2022). All lectins possess at least one non-catalytic domain, which binds reversibly to a specific mono- or oligosaccharide (Nivetha et al., 2021). Legume lectins, one of the largest lectin families, are particularly abundant in legume seeds and vegetative tissues according to the early technology (Tsaneva and Van Damme, 2020). However, with the characterization of increasing legume lectins in various species, a great number of lowly expressed or induced legume lectin homologs have been discovered in non-leguminous species such as there was 54 legume lectin genes in A. thaliana (Lagarda-Diaz et al., 2017), suggesting their ubiquitous presence in plants. Interestingly, classical Fabaceae lectins only contain the representative legume lectin domain (250 aa), while the ubiquitous legume lectins additionally include a long (60–150 aa) extension or an extended receptor-like kinase (RLK) domain in the C terminus, such as LegLus in A. thaliana (Damme et al., 2008; Tsaneva and Van Damme, 2020; De Coninck and Van Damme, 2021). In addition, numerous classical legume lectins have been specifically characterized at the structure level (Manoj and Suguna, 2001). Among these legume lectins, ConA (concanavalin A) in Canavalia gladiata (Jacq.) DC. is the first lectin whose primary and 3D structure have been resolved (Edelman et al., 1972; Hardman and Ainsworth, 1972; Cavada et al., 2019). Despite of the conserved primary, secondary and tertiary structure of the legume lectin domain, its quaternary structure shows considerable variations. Most classical legume lectins can form homodimers or homo-tetramers (“dimers of dimers”; Lagarda-Diaz et al., 2017), and the monomer structure displays a jelly-roll tertiary fold like a β-sandwich (a flattened six-stranded β-sheet and a curved seven-stranded β-sheet interconnected by several loops) of 25–30 kDa, which contains a carbohydrate recognition domain (CRD) and metal binding sites for divalent cations (Ca2+ and Mn2+; Gautam et al., 2018b; Katoch and Tripathi, 2021). One loop important for carbohydrate binding is highly variable, leading to changes in carbohydrate specificity (Katoch and Tripathi, 2021). However, it remains quite difficult to study the structure of LecRLK (receptor kinases with a legume lectin domain) due to glycosylation. For instance, crystallization and preliminary x-ray studies of AtLecRK-I.9 (At5g60300) failed to clearly elucidate its structure, owing to the rapid crystal decay during X-ray diffraction (Li et al., 2016).
Generally, legume lectins have vital functions in plant growth and development, including the storage of carbohydrates and hormones, and cell–cell interaction via binding to cell surface receptors (Lagarda-Diaz et al., 2017; Gautam et al., 2018b). In recent years, there has been increasing evidence demonstrating that a certain number of legume lectins are involved in the response to various stimuli (such as hormones and abiotic stress), and plant-insect, bacterial and fungal interactions (Bellande et al., 2017). For example, ConA inhibits the development of tomato moth (Lacanobia oleracea) and peach-potato aphid (Myzus persicae) when overexpressed in transgenic plants (Gatehouse et al., 1999). Besides, ConA is toxic to grain aphids through induction of gut cell apoptosis (Sprawka et al., 2015). The lectins from Egyptian Pisum sativum seeds could inhibit the growth of Aspergillus flavus, Trichoderma viride and Fusarium oxysporum (Li et al., 2012). In P. sativum, PslecRLK (pea lectin receptor-kinase) promotes the tolerance to high salinity stress by alleviating osmotic and ionic stresses and upregulating stress-responsive genes (Joshi et al., 2010), and overexpression of PslecRLK in rice could improve the salinity tolerance by inhibiting sodium accumulation (Passricha et al., 2019). In Glycine max, GmLecRLK contributes to salt stress tolerance by regulating salt-responsive genes in soybean (Zhang et al., 2022). In A. thaliana, the A4 (LecRKA4.1/At5g01540, LecRKA4.2/At5g01550 and LecRKA4.3/At5g01560) subfamily of lectin-receptor kinases negatively regulates abscisic acid response in seed germination (Xin et al., 2009). Mechanically, the antifungal activity of legume lectins can be mainly ascribed to the indirect effect of their binding with cell wall components, which can affect fungal survival or other activities like spore germination (Leal et al., 2012). Some legume seed proteins can bind with carbohydrate components of the bacterial cell wall or extracellular glycans and prevent the entry of microorganisms into the cytoplasm (Fonseca et al., 2022). Previous studies have demonstrated that lectins from chickpea have antibacterial activity against some bacterial pathogens such as Escherichia coli, Bacillus subtilis, Salmonella marcescens, and Pseudomonas syringae (Gautam et al., 2018a). In addition, the lectins from Egyptian P. sativum seeds can inhibit the growth of Aspergillus flavus, Trichoderma viride and Fusarium oxysporum (Sitohy et al., 2007). Furthermore, some recent studies have revealed that several lecRKs play important roles in stress signal transduction in plant immunity (Desclos-Theveniau et al., 2012; Singh and Zimmerli, 2013). AtLecRK-V.5 (AT3G59700) represses stomatal immunity induced by P. syringae (P. syringae; Arnaud et al., 2012; Desclos-Theveniau et al., 2012). AtLecRK-IX.2 (AT5G65600) not only contributes to plant resistance to P. syringae (Luo et al., 2017), but also plays an important role in plant immunity by phosphorylating the bacterial effector AvrPtoB at S335 and thereby potentially decreasing its virulence (Xu et al., 2020). Moreover, AtLecRK-VI.2 (AT5G01540) is a potential primary eNAD (P)+-binding receptor, and plays a central role in the biological induction of SAR (systemic acquired resistance; Wang et al., 2019). AtLecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana (Nicotiana. L) pattern-triggered immunity. AtLecRK-I.9 is involved in protein–protein interaction with proteins containing RGD (arginine-glycine-aspartic acid) as a ligand, and plays a structural and signaling role at the plant cell surface against pathogens (Bouwmeester et al., 2011; Huang et al., 2014; Wang et al., 2016). In addition, AtLecRK-I.9 acts as an extracellular ATP receptor in A. thaliana (Wang et al., 2018b), which plays an important role in systemic wound response activation (Pham et al., 2020; Myers et al., 2022). AtLecRK-I.9 also affects jasmonate signaling to contribute to resistance to P. syringae (Balagué et al., 2017).
Sclerotinia stem rot caused by Sclerotinia sclerotiorum is one of the devastating diseases affecting the yield and quality of oil crops, including B. napus. Several genes or factors contributing to plant SD resistance have been reported to date, such as OsPGIP2 (Wang et al., 2018a), GDSL1 (Ding et al., 2020), BnF5H (Cao et al., 2022), Cinnamoyl-CoA Reductase 2 (Liu et al., 2021a), and WRKY15 and WRKY33 (Liu et al., 2018), which have provided some insights into plant defense against S. sclerotiorum. However, the SD resistance of B. napus belongs to quantitative resistance, and thus its genetic mechanism remains largely unknown. Considering the potent antibacterial and antifungal activities of legume lectins, they may be promising targets to be utilized for the improvement of SD resistance. Here, we performed a genome-wide characterization of the legume lectin genes in B. napus, and then phylogenetically and structurally categorized the BnLegLu members. The potential contribution of BnLegLu genes to SD resistance was investigated based on the spatiotemporal expression profiles under abiotic stimulation and S. sclerotiorum induction, and the candidate genes were further screened by genome-wide association study (GWAS) with a rapeseed population comprising 324 accessions collected all over the world. A transgenic study with transient expression of BnLegLu-16 in tobacco (Nicotiana L) leaves validated its positive role in regulating SD resistance under S. sclerotiorum inoculation. The findings of the present study may lay a solid foundation for further exploring the role of LegLus in rapeseed defense against SD.
Results
Identification and characterization of LegLus in Brassica napus
A total of 130 legume lectin genes with an intact legume lectin domain in B. napus were identified using PF00139 as a query. The detailed information of each BnLegLu gene is presented in Supplementary Table S1. A total of 72 genes belonged to the A subgenome, whereas 58 genes belonged to the C subgenome (Supplementary Table S1). The protein length varied from 223 aa (amino acids; BnaA06g01170D) to 941 aa (BnaC09g35640D), with an average length of 528 aa, and the molecular weight (MW) varied from 24.96 to 103.59 kDa. The pI (isoelectric points) ranged from 4.68 to 9.99. Overall, the number of exons ranged from one to six, with an average of two exons, and 57 of the BnLegLu members only had one exon. Moreover, the instability index ranged from 13.76 to 51.48, and 72% (93 out of 130) BnLegLu members had an instability index lower than 40, indicating the stability of these proteins (Supplementary Table S1). Based on prediction with DeepTMHMM and CELLO server, 123 BnLegLu proteins contained SP (signal peptide) in their N-terminus; 107 members harbored the transmembrane domain (TM); and 103 members were localized in the plasma membrane (Supplementary Table S1), implying their possible functions in the plant-pathogen interaction on the cell surface. In addition, there are several genes containing multiple TM domains, such as BnaC09g35640D, BnaC04g27370D and BnaA04g04820D, which had three TM domains, and BnaA03g26180D, which had two TM domains (Supplementary Table S1). Moreover, the BnLegLu genes exhibited an uneven distribution across the chromosomes. More BnLegLu genes were distributed on chromosomes A03 (14 genes), C03 (12 genes), A10 (12 genes), C09 (10 genes), and A06 (eight genes), while fewer genes were found on chromosome A01 (two genes), C01 (two genes) and C07 (one gene; Supplementary Table S1; Figure 1). Duplication events of the BnLegLu genes were detected based on BLAST and MCScanX analyses of the genome of B. napus (Supplementary Table S1; Figure 1). There was a cluster of eight tandem repeats (BnaA10g13070D to BnaA10g13180D) on chromosome A10. A local duplication event was also identified on chromosome C09 (BnaC09g35610D to BnaC09g35670D; Figure 1). In total, 77.69% (101/130) of the BnLegLu genes originated from whole-genome duplication (WGD) or segmental duplication and 18 BnLegLu members were derived from dispersed duplication. Additionally, nine genes were derived from tandem duplication and two genes came from proximal gene duplication (Supplementary Table S1). Furthermore, 106 duplicated BnLegLu pairs between the two subgenomes were found in B. napus (Figure 1; Supplementary Table S2). Collectively, 130 BnLegLu genes in B. napus were identified and their detailed information, including gene characteristics and chromosomal distribution, was further elucidated.
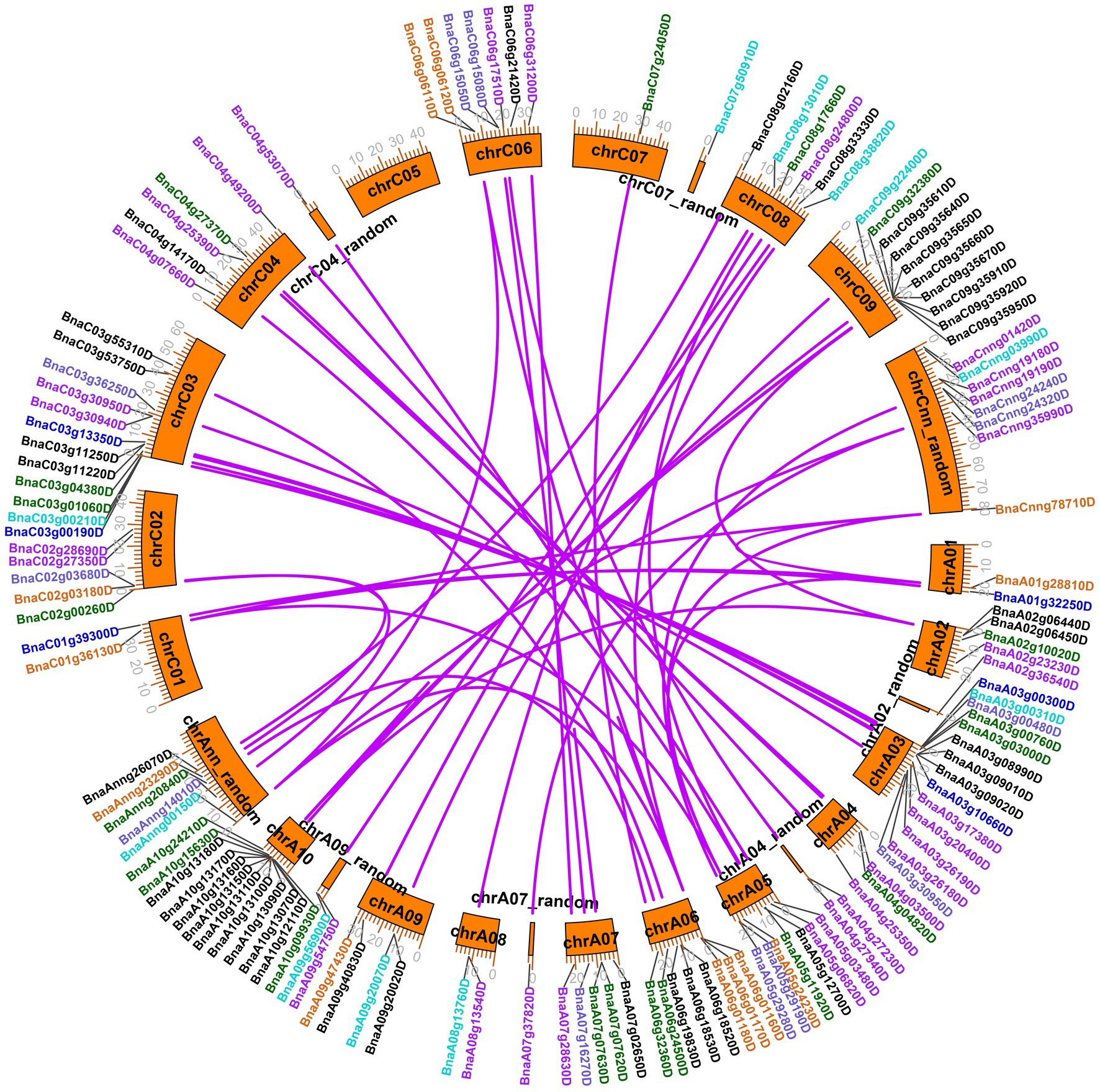
Figure 1. Chromosomal locations and duplicated gene analysis of BnLegLu genes in Brassica napus. The chromosomal locations of all BnLegLu genes are represented on different chromosomes, and different colors represent different BnLegLu subfamily genes. Subfamilies I to VII are indicated by black, purple, medium blue, dark turquoise, dark green, chocolate and slate blue color, respectively. Rose red lines are used to highlight the duplicated BnLegLu gene pairs.
Phylogenetic analysis of BnLegLus in Brassica napus
To determine the phylogenetic classification of the BnLegLu genes, we constructed a phylogenetic tree of legume lectins by combing the LegLus proteins in A. thaliana (54; Eggermont et al., 2017) with the NJ (neighbor-joining) method (Figure 2). Seven (I–VII) clusters were categorized with well-supported bootstraps indication. The gene number varied significantly among the seven clusters, with the largest number of genes (54) in cluster I and the smallest number (9) in cluster III (Figure 2). Several well-studied AtLegLu genes were categorized into different clusters with BnLegLu genes, indicating that they possibly have distinct functions (Figure 2). Both AtLecRK-I.9 (AT5G60300) and AtLecRK-I.8 have been reported as immune elicitors in A. thaliana by binding NAD+, and inducing the expression of disease resistance genes (Wang et al., 2017). Nine genes (BnaA1013160D, BnaC09g35650D, BnaA10g13170D, BnaC09g35660D, BnaC03g11250D, BnaA03g09010D, BnaA10g13150D, BnaC09g35950D and BnaC09g35640D) and two genes (BnaC09g35920D and BnaA10g13110D) were located in the same clade with AtLecRK-I.9 and AtLecRK-I.8, respectively (Figure 2). AtLecRK-I.3 (AT3G45410) has been reported to be responsive to salt stress mediated by the ethylene signaling pathway (He et al., 2004). Phylogenetically, BnaA06g18530D and BnaA07g02650D were close to AtLecRK-I.3 (Figure 2). AtLecRK-V.5 (AT3G59700) and AtLECRK-V.2 (AT1G07130) were found be involved in stomatal immunity (Arnaud et al., 2012; Desclos-Theveniau et al., 2012). In cluster II, BnaC06g17510 and BnaA04g27230 were grouped with AtLecRK-V.5, while BnaC06g31200D and BnaA07g28630D were grouped with AtLECRK-V.2 (Figure 2). BnaC09g22400D and BnaA09g20070D in cluster III were closely related to AtLecRK-VII.1 (AT4G04960), which controls stomatal immunity and jasmonate-mediated stomatal closure (Yekondi et al., 2018). The BnLegLu genes in cluster III were grouped with LecRKA4.2 (AT5G01550), LecRKA4.1 (AT5G01540), and LecRKA4.3 (AT5G01560; Xin et al., 2009), indicating their possible functions in seed germination. Further, seven BnLegLu genes in cluster V (BnaA07g07630D, BnaA07g07620D, BnaA05g11920D, BnaC08g17660D, BnaA06g24500D, BnaAnng20840D and BnaA02g10020D) were clustered together with AtLecRK-IX.2 (AT5G65600), which functions in plant PAMP-triggered immunity (PTI; Wang et al., 2015; Luo et al., 2017). In cluster VI, BnaCnng78710D, BnaA05g24230D, BnaA01g28810D and BnaC01g36130D were phylogenetically close to AT3G16530 and AtLEC (AT3G15356), and the expression of AT3G15356 is up-regulated by multiple stimuli such as developmental signal, wounding, jasmonate, ethylene, and chitin elicitor (Lyou et al., 2009). BnaC02g03180D and BnaAnng23290D were homologs of AtSAI-LLP1 (AT5G03350), which is induced by salicylic acid (SA) and acts as a component of the SA-mediated defense processes associated with the effector-triggered immunity response (Armijo et al., 2013; Figure 2).
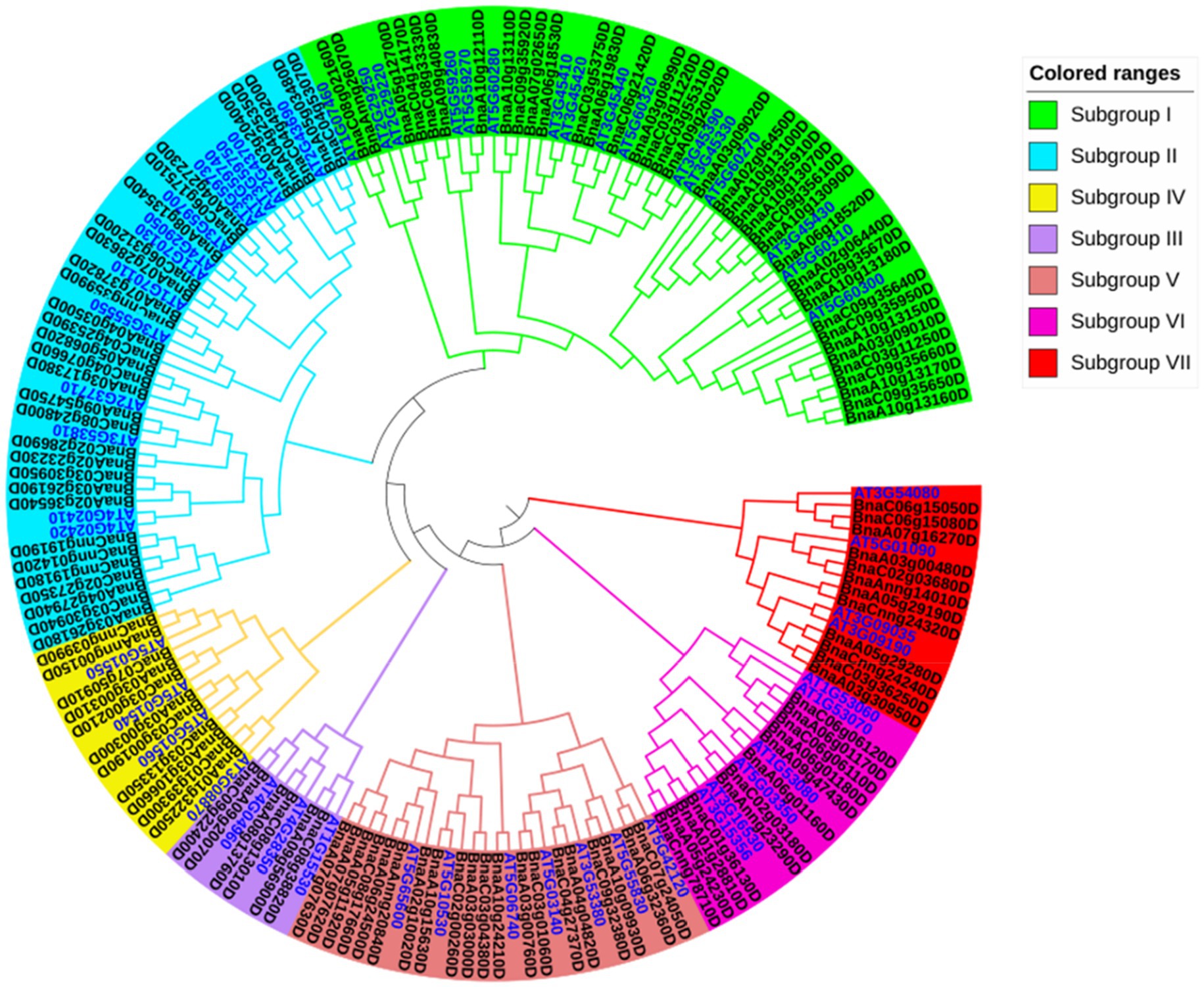
Figure 2. Phylogenetic analysis of BnLegLu proteins in A. thaliana and B. napus. All legume lectin proteins were clustered into seven subfamilies (I–VII) with differently colored branches (I, green; II, blue; III, purple; IV, yellow; V, dark red; VI, rose red; VII, red). The gene IDs for B. napus are black, the gene IDs for A. thaliana are bright blue.
Conserved structure of BnLegLus
To explore the evolutionary characteristics of BnLegLus, we further compared the gene structure and conserved motifs among the seven clades. The structures of most genes in cluster VI–VII were well conserved, and none of them contained any intron, except for BnaA09g47430D with one intron (Supplementary Table S1; Supplementary Figure S1). However, the genes in cluster I–V showed remarkable variations in intron number from zero to five as well as significant variations in intron length, with BnaC09g35950D and BnaC03g11250D having particularly long introns (Supplementary Table S1; Supplementary Figure S1).
Subsequently, we scanned the conserved motifs in all BnLegLu proteins and arranged them according to the gene phylogenic category (Supplementary Figure S2). The motifs 9, 6, 7 and 4, which correspond to the legume lectin domain, were highly conserved among all BnLegLu genes, while motifs1, 2, 3, 10 were annotated as kinase-related motifs and motifs 5 and 8 were unknown motifs (Supplementary Figure S2; Supplementary Table S3). Motif 7 was present in all the BnLegLu proteins except for BnaC06g3120D and BnaC04g25930D. Additionally, most genes contained motif 9 (124 out of 130) and motif 4 (120 out of 130; Supplementary Figure S2). The motifs in clusters VI and VII were well conserved without any kinase-related motif. All BnLegLu proteins in cluster VI contained four full motifs of legume lectin, while all BnLegLu proteins in cluster VII had no motif 6, and BnaAnng14010D, BnaC02g03680D and BnaA03g00480D only contained motif 9 and motif 7 (Supplementary Figure S2). All BnLegLu proteins in cluster III had full motifs of the protein kinase domain. These specific features implied the functional diversity of BnLegLu genes.
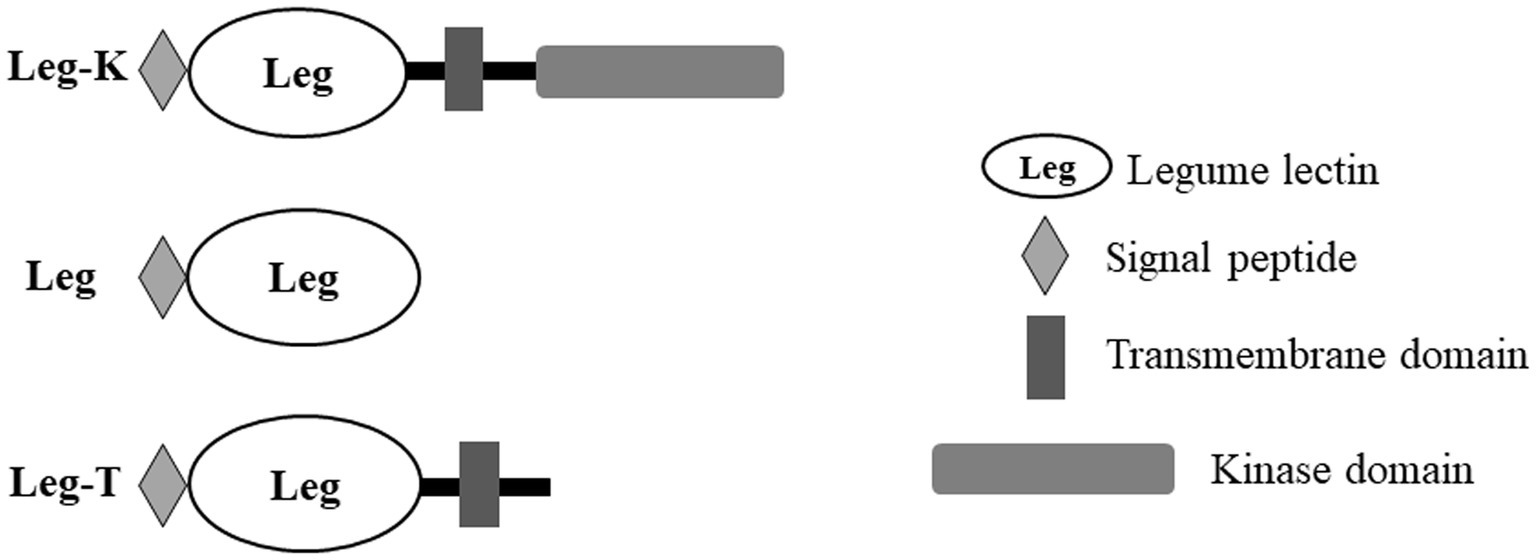
Moreover, an analysis of BnLegLu proteins with the domain search program of the Conserved Domain Database (NCBI)1 revealed that the BnLegLu proteins have relatively simple domain architecture, which could be divided into three categories: Leg-K representing BnlegLu proteins that contained SP, TM, legume domain and kinase domain, such as majority genes in cluster I–V; Leg representing BnlegLu proteins that only contained SP and legume domain, such as all genes in cluster VI except for BnaA06g01170D, which had no SP; Leg-T representing BnlegLu proteins that contained SP, TM and legume domain, like genes in cluster VII except for BnaCnng24320D and BnaA05g29190D, which had no TM (Figure 3; Supplementary Tables S1, S3). In addition, 92 out 130 of BnLegLus contained a kinase domain. All proteins that contained a protein kinase domain were synthesized with a signal peptide and a transmembrane domain, except for four genes (BnaA10g13090D, BnaC09g35610D, BnaA03g26180D and BnaA05g03480D) did not contain a signal peptide (Figure 3; Supplementary Tables S1, S3).
Cis-element and protein interaction analysis of BnLegLu genes
To explore the possible regulatory mechanism of BnLegLu genes, we further analyzed the cis-elements in their promoter regions and the possible interactive proteins with the available public databases.2 The cis-elements in the 2-kb upstream region of BnLegLus were analyzed through the PlantCARE database. The results indicated the enrichment of stress- and hormone-related elements in these promoters, such as DRE core/DRE1 (involved in dehydration response), MBS (MYB binding site for drought stress), LTR (responsive to low temperature), TC-rich (defense responsive element), WUN-motif (responsive to wounds), ABRE (ABRE2/3a/4; responsive to abscisic acid), CGTCA/TGACG-motif (responsive to methyl jasmonic acid, JA), ERE (response to ethylene), p-box/TATC-box/GARE-motif (responsive to gibberellin), TGA-element/AuxRR-core (responsive to auxin), and TCA-element (responsive to salicylic acid; Supplementary Figure S3; Supplementary Table S4). Among the cis-elements, ABRE and JA-related elements were predominant in BnLegLus, as 97 and 101 out of the 130 BnLegLu genes contained ABRE and JA-related elements, respectively (Supplementary Figure S3; Supplementary Table S4). The category of cis-elements showed distinct patterns among different clusters. The elements of ABRE, JA-related, ERE and WUN-motif were abundant in the BnLegLu genes of cluster III and cluster VI, but the auxin-related, DRE-core, MBS and TC-rich elements were enriched in cluster III members. The BnLegLu genes in cluster I contained relatively more LTR elements, with a maximum of eight (BnaC09g35640D); while the BnLegLu genes in cluster V had more JA-related elements, with a maximum of 12 (BnaA10g24210D). The BnLegLu genes in cluster VII contained more GA-related and WUN-motif elements (Supplementary Figure S3; Supplementary Table S4). These results indicated the important functions of BnLegLu genes in response to environmental stress, which is possibly mediated by the hormone pathways.
Based on the protein interaction networks in A. thaliana, we attempted to identify the potential interactive proteins of BnLegLus. By using the orthologs of BnLegLus in A. thaliana as the query, 4,606 putative interactive proteins were identified, which corresponded to 14,848 proteins in B. napus. Most BnLegLu proteins had interactions with each other (Figure 4A). To further elucidate the functional category of BnLegLu interactive proteins, we performed gene ontology (GO) and a Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. Interestingly, significant enrichment was found for the GO in terms of response to biotic and mechanical stimuli, insects, molecules of bacterial origin and the defense response by callose deposition, suggesting their important roles in plant defense (Figure 4B; Supplementary Table S5). In addition, salicylic acid-mediated signaling pathways, regulation of DNA replication and multi-organism process were also remarkably enriched, implying their important roles in plant development and signal transduction (Figure 4B; Supplementary Table S5). The KEGG enrichment analysis revealed that these interactive proteins participate in photosynthesis, glycolysis/gluconeogenesis and fundamental metabolisms such as glutathione metabolism, inositol phosphate metabolism and glyoxylate and dicarboxylate metabolism (Supplementary Figure S5).
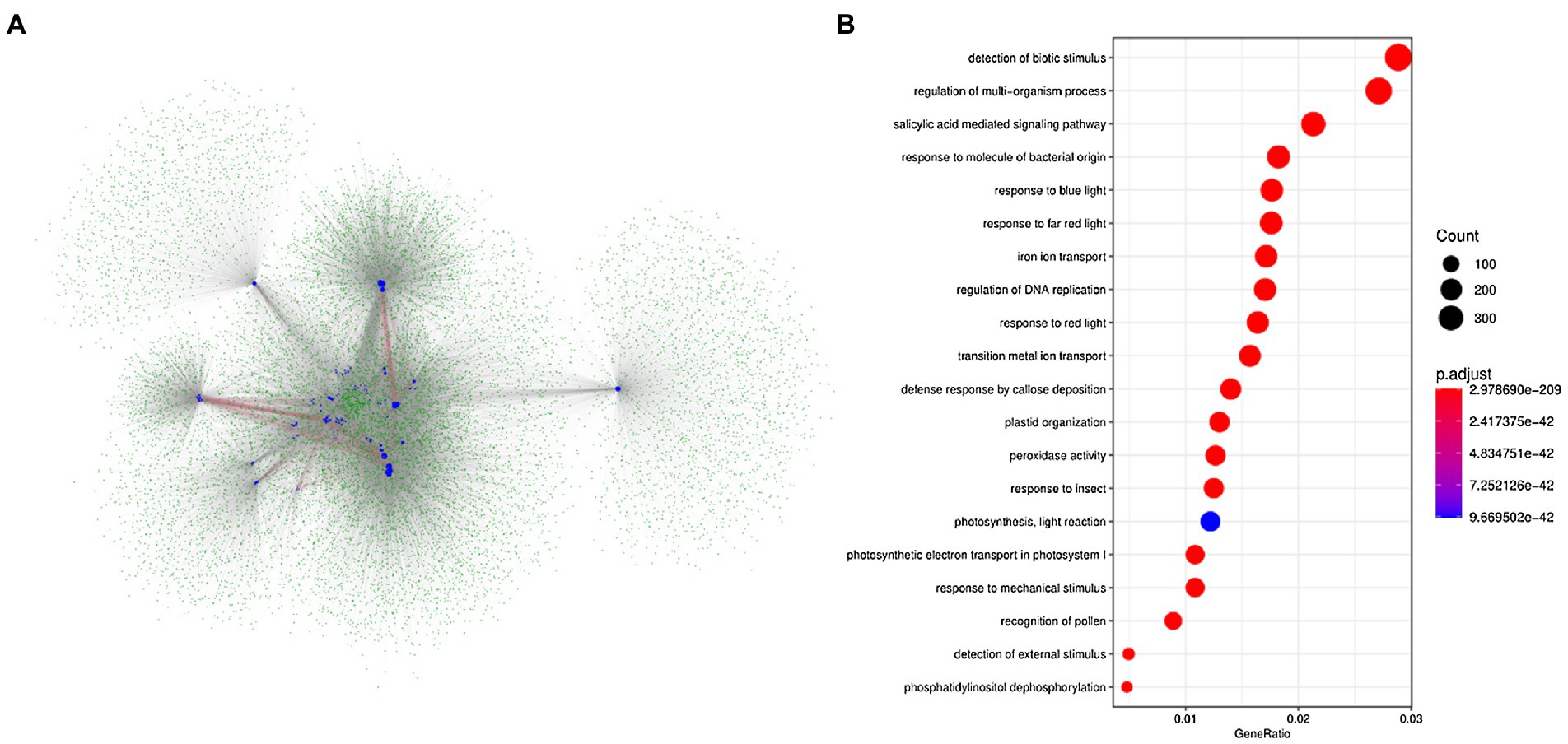
Figure 4. Analysis of the network for the interaction of BnLegLu proteins in B. napus. (A) Protein–protein interaction network of BnLegLu. The BnLegLu proteins are indicated by medium blue circles, and the green circles represent the proteins that interact with BnLegLu proteins. The red lines indicate the interaction between BnLegLu proteins, and the gray lines represent the interaction between BnLegLus and other proteins. (B) GO analysis of proteins interacting with BnLegLu proteins.
Expression analysis of BnLegLus in various tissues during different developmental stages
To investigate the expression patterns of BnLegLus, comprehensive expression analysis was performed by using the published transcriptome information resource (Liu et al., 2021b).3 This database collected 92 tissue samples at three distinct developmental stages (leaf, silique wall and seed), including the cotyledon, root, stem peel, leaf, bud, flower, silique, silique wall and seed. The FPKM (fragments per kilobase million) values for each BnLegLu gene are presented in Supplementary Table S6 and the expression profiles are clustered in Supplementary Figure S5. In general, 25 out of the 130 genes were barely expressed across tissues (FPKM < 0.5). Moreover, most BnLegLu genes showed moderate expression (FPKM < 15), and only several genes were highly expressed in some specific tissues (Supplementary Figure S5; Supplementary Table S6). A comparison of the expression levels among different tissues revealed that BnLegLus had high expression in roots, old leaves and mature silique walls, but were barely expressed in the pollen. For example, four genes of BnaA05g24230D, BnaC04g07660D, BnaCnng24320D and BnaC06g06120D showed the highest expression in the root, old leaf, silique wall and seed (Figure 5; Supplementary Table S6). In addition, the expression of BnLegLu genes in cluster I–V in old leaves and silique walls continuously increased along with development, except for some unexpressed or lowly expressed genes (Supplementary Figure S5; Supplementary Table S6), while those genes in cluster VI–VII exhibited high expression in the early stage of seed development. In summary, the expression of BnLegLus varied greatly among different tissues, implying their functional diversity in the development of B. napus.
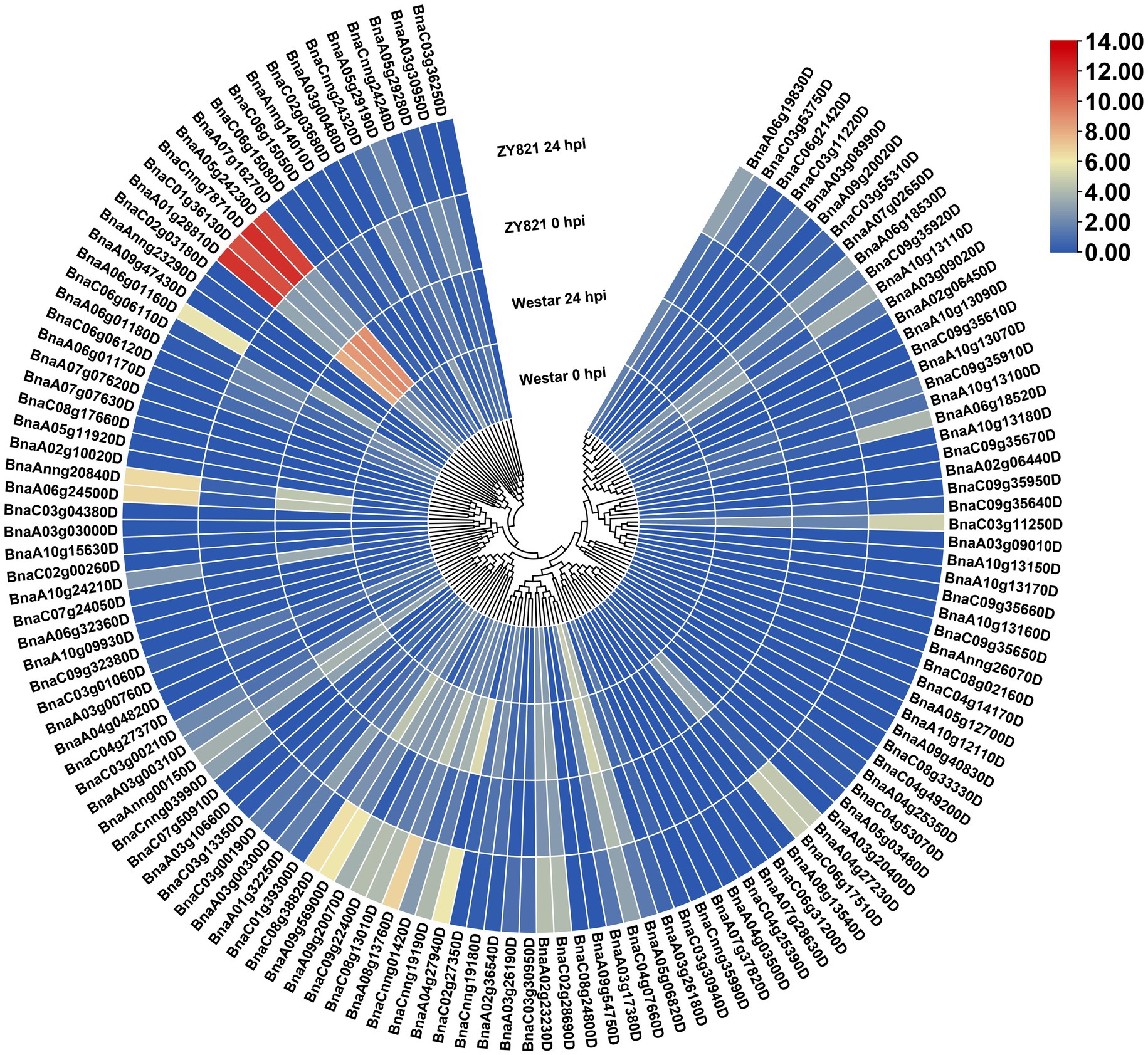
Figure 5. Expression patterns of the BnLegLu genes in Westar and ZY821 cultivars at 0 and 24 h after Sclerotinia sclerotiorum inoculation. The heatmap was generated by taking log2 fold of FPKM values. The color bar shows the relative expression from low (blue) to high (red).
Expression analysis of BnLegLus under abiotic stress and Sclerotinia sclerotiorum induction
To gain insights into the response of BnLegLus to environmental stimuli, we examined the expression fluctuations of BnLegLus under various abiotic stresses (dehydration, cold, ABA and salinity treatments; Zhang et al., 2019). The majority of BnLegLu genes had very low levels of expression in the control, which was inconsistent with tissue expression data. Abiotic stress significantly changed the expression levels of 35 BnLegLu genes (≥2-fold and FPKM ≥ 2), among which 30 were up-regulated and five were down-regulated (Supplementary Figure S6; Supplementary Table S7). The BnLegLu genes in cluster I and cluster IV, which showed low expression in different tissues, exhibited minor fluctuations in the transcript after induction by abiotic stress (Supplementary Figure S6; Supplementary Table S7). However, six BnLegLu genes in cluster II (BnaC04g07660D), VI (BnaA01g28810D, BnaC01g36130D, BnaCnng78710D and BnaA05g24230D) and VII (BnaCnng24320D) showed a significant response to all stress conditions (Supplementary Figure S6; Supplementary Table S7). Compared with that in the control, the expression of BnaC04g07660D and BnaCnng24320D was increased to 6.4-fold and 53-fold in 24 h of cold treatment; that of BnaA01g28810D and BnaC01g36130D was elevated to 70-fold and 24-fold under salt stress in 24 h; that of BnaCnng78710D and BnaA05g24230D was increased to 3.1-fold and 3.7-fold in 4 h of cold treatment, but their expression was significantly down-regulated after 24 h of ABA treatment and NaCl treatment (Supplementary Figure S6; Supplementary Table S7). Furthermore, we also analyzed the expression changes of BnLegLus under the induction of S. sclerotiorum by utilizing the public transcriptome data (Girard et al., 2017). More than half of the BnLegLu genes showed very low levels of expression (FPKM < 0.5) in the two cultivars of Zhouyou821 (resistant to SD) and Westar (susceptible to SD), except for BnaC04g07660D, which had a FPKM value higher than 15 (Figure 5; Supplementary Table S8). A total of 40 BnLegLu genes showed significant changes in expression under the induction of S. sclerotiorum (≥2-fold and FPKM ≥ 2), including 30 up-regulated genes and 10 down-regulated genes at 24 h post-inoculation (hpi) at least in one cultivar (Figure 5; Supplementary Table S8). Compared with the down-regulation, the up-regulation seemed to be more significant, moreover, the expression levels of most BnLegLu genes changed more significantly in ZY821 than those of Westar (Figure 5; Supplementary Table S8). Relative to those in cluster I, II, IV, V and VII, the genes in cluster III and VI showed more remarkable up-regulation of expression in response to S. sclerotiorum. Six genes (BnaA08g13760D, BnaC08g13010D, BnaC09g22400D, BnaA09g20070D, BnaA09g56900D and BnaC08g38820D) in cluster III were induced by S. sclerotiorum and more significantly induced that of four genes (BnaA01g28810D, BnaC01g36130D, BnaCnng78710D and BnaA05g24230D) in cluster VI, with the most significant up-regulation reaching 715-fold (Figure 5; Supplementary Table S8).
Next, we selected 16 candidate BnLegLu genes with particular increases in expression after S. sclerotiorum induction for real-time PCR (qRT-PCR) analysis, including BnaA04g27940D, BnaA04g27230D, BnaC06g17510D, BnaA09g56900D, BnaC08g38820D, BnaA08g13760D, BnaC08g13010D, BnaC09g22400D, BnaA09g20070D, BnaA06g24500D, BnaAnng20840D, BnaA06g01160D, BnaA01g28810D, BnaC01g36130D, BnaCnng78710D and BnaA05g24230D, which were renamed as BnLegLu-1 to BnLegLu-16. We collected the leaves of Zhouyou821 after inoculation with S. sclerotiorum at different time points (0, 12, 24, 36 and 48 h; Figure 6). As a result, four genes (BnLegLu-4, BnLegLu-5, BnLegLu-6 and BnLegLu-9) showed complex changes in expression. After inoculation, the expression of BnLegLu-4 was generally up-regulated all the time with a slight decrease at 24 and 48 hpi, while that of the other three genes was down-regulated at 12 hpi first, and then continuously increased, but slightly decreased at 36 hpi (Figure 6). The remaining 12 genes were continuously up-regulated at all times after infection, which was consistent with the transcriptome data. The expression of BnLegLu-10 to BnLegLu-16 was very remarkably up-regulated, even by up to 50,000-fold (BnLegLu-13; Figure 6). These results indicated that these genes may play potential roles in the SD resistance of B. napus.
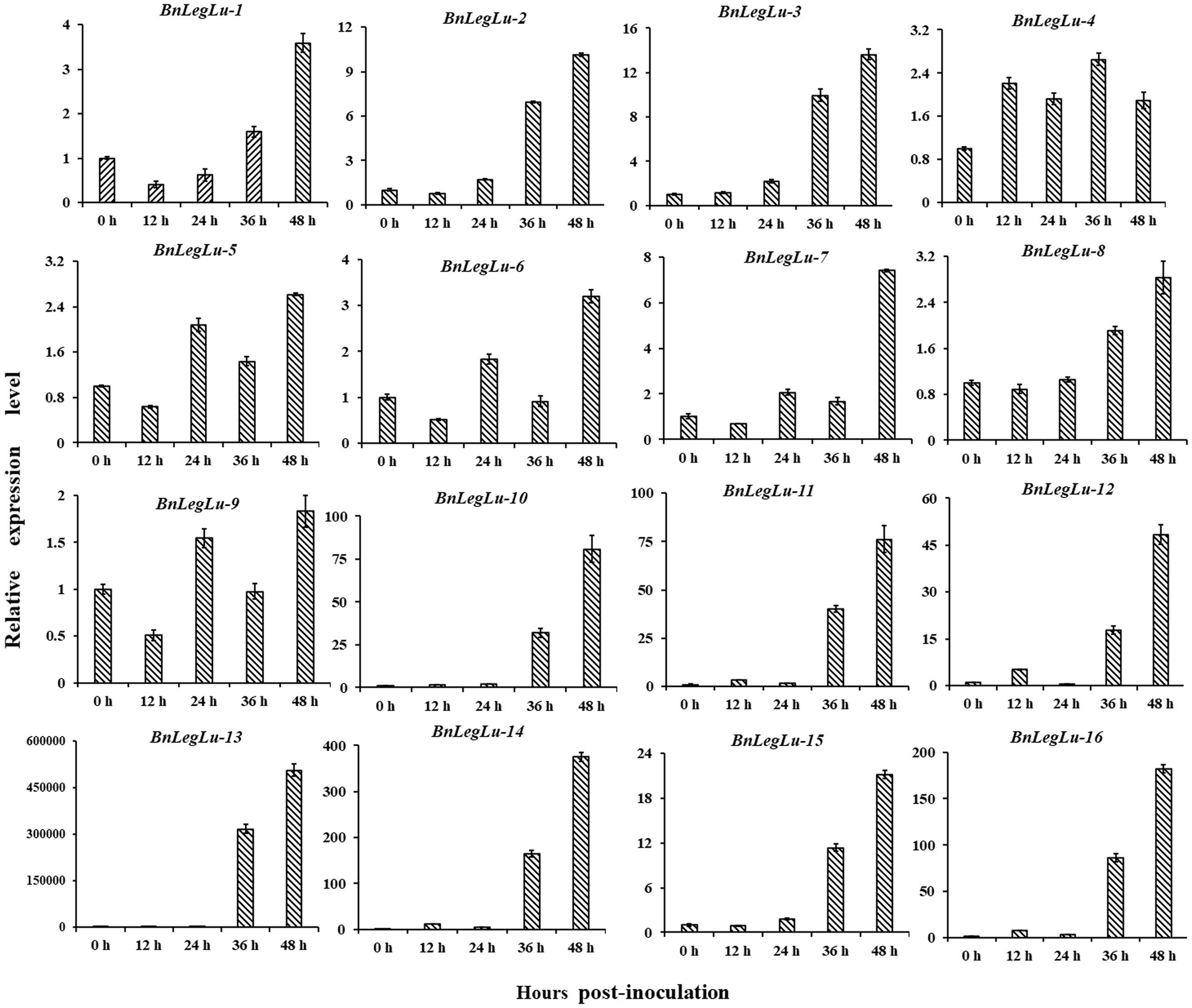
Figure 6. Expression validation of 16 candidate BnLegLu genes in response to S. sclerotiorum determined by qRT-PCR. The time points 0, 12, 24, 36 and 48 h represent hours after inoculation with S. sclerotiorum. The error bars show the standard error of three replicates.
Preliminary validation of candidate BnLegLus for SD resistance in Brassica napus
Based on the available GWAS data in our laboratory for SD disease in the panel of 324 accessions collected worldwide (Tang, 2019; Ding et al., 2020), we detected four BnLegLu genes located in the region of significant SNPs associated with SD, including BnLegLu-1 (81 kb near the associated SNP of 391,255 bp on chromosome A04, BnLegLu-2 (88 kb near the associated SNP of 92,114 bp on chromosome A04), BnLegLu-5 (14 kb near the associated SNP of 34,896,206 bp on chromosome C08), BnLegLu-16 (77 kb near the associated SNP of 18,259,474 bp on chromosome A05; Figure 7A; Supplementary Figure S7). We also analyzed the haplotype of BnLegLu-16 and three haplotypes divided by the SNPs in BnLegLu-16 is displayed in Figure 7B and Supplementary Table S10. The result showed that the disease lesion size in haplotype I and III exhibited significant difference (p = 0.02). Furthermore, we transiently overexpressed BnLegLu-16 in tobacco leaves, and inoculated them with the S. sclerotiorum hyphal plugs on leaves. Compared to the control, the disease lesion size was significantly small on leaves with overexpression of BnLegLu-16, and the difference was increasing obviously along with the time post-inoculation (Figures 7D,E). We further examined the subcellular localization by detecting the fluorescence under confocal microscopy. Results showed that the GFP-tagged BnLegLu-16 (BnLegLu-16–GFP) displayed an obvious signal in the plasma membrane, while the GFP-only control produced a fluorescence signal throughout the cell (Figure 7C). Furthermore, BnLegLu-16-GFP co-localized with the mCherry-tagged H+-ATPase (Sun et al., 2021), which was used as a marker for plasma membrane location (Figure 7C). The results showed that the fluorescence was specifically present on the cell membrane (Figure 7C), indicating the possible function compartment of BnLegLu-16.
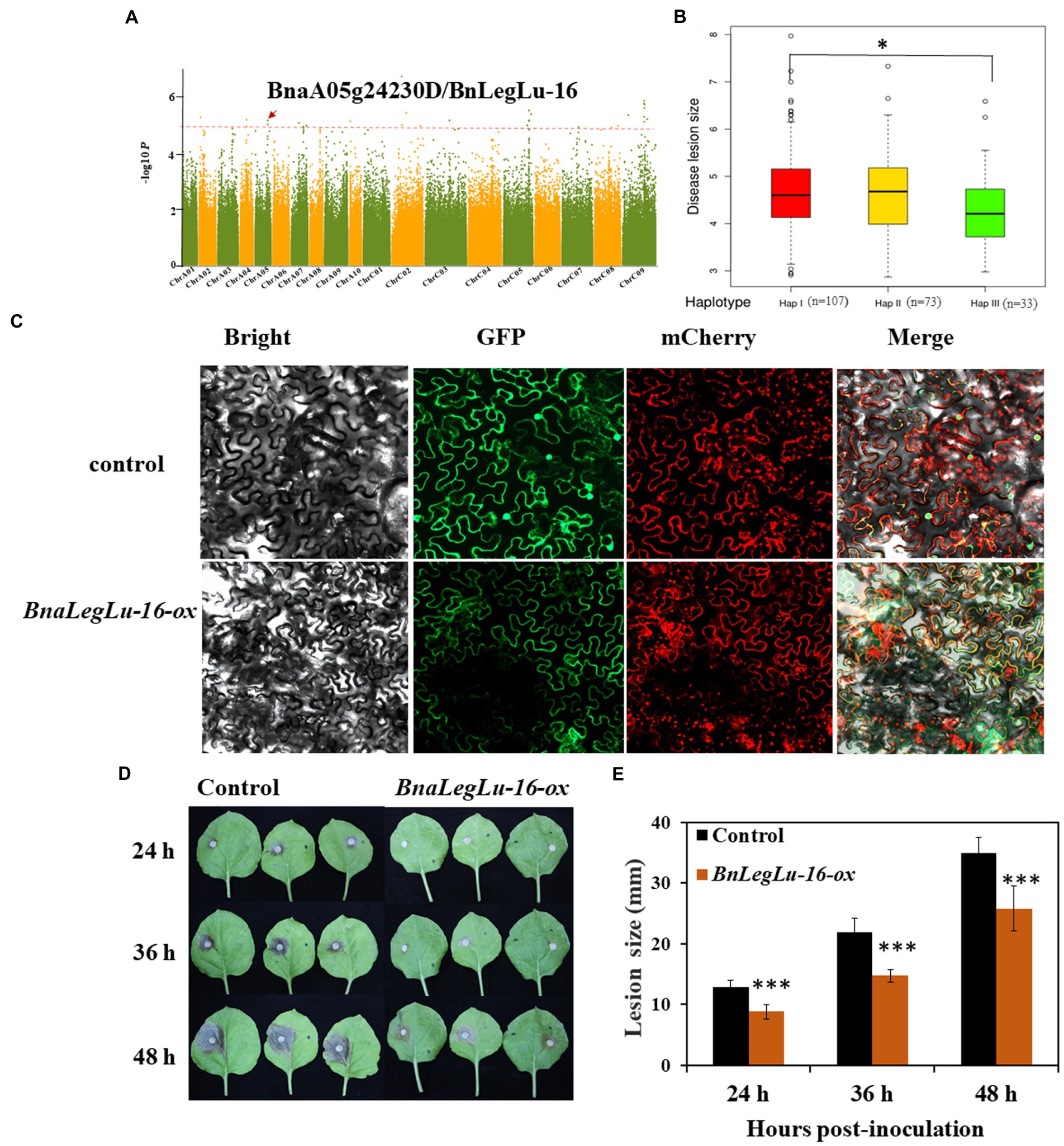
Figure 7. Functional validation of BnLegLu-16 for SD resistance. (A) Genome-wide association analysis (GWAS) for SD resistance in a B. napus population comprising 324 accessions and Manhattan plots of SD resistance from GWAS; Manhattan plots of the disease after 36 h; the red dashed line shows GWAS threshold (1/SNP number). (B) Haplotype analysis of BnLegLu-16. (C) Subcellular localization of BnLegLu-16-ox in tobacco, BnLegLu-16-GFP fusion protein was transiently co-expressed with the mCherry–H+-ATPase, which was used as the plasma membrane marker in tobacco leaves. GFP and mCherry were transiently co-expressed as the control. After 48 h post-incubation, the tobacco leaves were observed with confocal microscope (ZEISS LSM710, Germany). (D) Disease lesion sizes on leaves at 24, 36 and 48 hpi with S. sclerotiorum, which was transiently injected through agrobacterium containing BnLegLu-16-ox and the control into tobacco leaves. (E) Disease lesion sizes statistically analyzed by comparing the overexpression BnLegLu-16-ox to the control. The data represent the means ± 2 SD from three independent experiments, with each containing 20 leaves. Significant differences in lesion size between BnLegLu-16-ox and the control are indicated (Student’s t-test) as follows: ***p < 0.001). The error bars show the standard error of three replicates.
Discussion
Legume lectins were extensively distributed in the plant, and display a variety of functions including antimicrobial and insecticidal activities (Lagarda-Diaz et al., 2017; Katoch and Tripathi, 2021). Increasing evidence has suggested that insect and pathogen infestation of crops is one of the crucial factors threatening global agriculture production, whose impacts will further increase in the future due to increasing demand for food and changing climatic conditions (Fedoroff et al., 2010). Besides, crops will be confronted with stress more often and different stresses will occur in combination more frequently (Saijo and Loo, 2020). Hence, as potential candidates for resistance to susceptible crops, legume lectins are worth in-depth and extensive investigation. In the last two decades, much research attention has been paid to legume lectins that are weakly expressed in non-storage tissues (Katoch and Tripathi, 2021; Van Damme, 2022). It has been found that AtLecRK-V.5, AtLecRK-IX.2 and AtLecRK-I.9 contribute to plant resistance to P. syringae (Desclos-Theveniau et al., 2012; Balagué et al., 2017), and AtLecRK-VI.2 and AtLecRK-I.9 play important roles in plant immune response (Huang et al., 2014). The interaction between S. sclerotiorum and its host B. napus is very complex due to the facultative parasitic nature of S. sclerotiorum (Ding et al., 2021). In addition, no immune germplasm has been identified in B. napus or its close relatives so far, bringing about great challenges to the breeding of B. napus for SD resistance (Wu et al., 2016). Numerous studies have shown that there are many potential genes with the ability to resist SD in B. napus. Moreover, the heritability of the resistance of B. napus to S. sclerotiorum is moderate (60–75%; Ding et al., 2021). These resistance genes can be mainly divided into three categories: (i) Some genes can produce phytoalexins (secondary metabolites) to kill the pathogen or prevent pathogen infection and spread during induction of S. sclerotiorum, such as flavonoids, terpenoids, indole alkaloid camalexin, glucosinolates, lignin (Cao et al., 2022), and brassinosteroids (BR; Liang and Rollins, 2018). (ii) Some genes can generate activating enzymes or antimicrobial peptides to degrade the cell wall of S. sclerotiorum or block or interfere with its pathogenicity factors, such as BnPGIPs, which could inhibit hyphal extension and plant cell wall degradation through PG secreted by S. sclerotiorum (Wang et al., 2018a). (iii) Some genes can activate the defense signaling pathways involving the MAPK (mitogen-activated protein kinase) cascade reaction, ROS (reactive oxygen species), SA (salicylic acid), and JA (jasmonic acid) to activate the resistance to S. sclerotiorum (Nováková et al., 2014), such as BnWRKY33, BnWRKY70, BnMPK3, BnMPK4 and BnMPK6 (Liu et al., 2018). In general, great progress has been made in improving SD resistance in rapeseed, but the key genes for the resistance remain to be explored. The LegLu genes are an important disease resistant gene family with antimicrobial and insecticidal activities, but their role in SD resistance remains largely unknown. In this study, we performed a comprehensive analysis of BnLegLus at the genome level and analyzed their evolutionary characteristics and functional impact on SD resistance. The findings provide new insights into this gene family and help to predict its potential function in plant stress response, particularly in SD resistance.
In this study, we identified 130 BnLegLu genes in B. napus by using the Darmor-bzh v4.1 genome sequence information (Supplementary Table S1). According to the phylogenetic analysis, the BnLegLu genes could be classified into seven clusters together with AtLegLus (Figure 2). The majority of BnLegLu genes in cluster I–V contained extended RLKs domains, which could explain the higher molecular weight of BnLegLu genes in these clusters than those in cluster VI–VII. Although BnLegLu genes in both cluster VI and VII contained the legume lectin domain, the members in cluster VII generally had higher average molecular weights than those in cluster VI due to the additional transmembrane domain. In terms of size polymorphism, the BnLegLu genes also showed variations in exon number, pI, instability index and subcellular localization in three categories (Figure 3; Supplementary Table S1). All BnLegLu genes in Leg and Leg-T only contained one exon except for BnaA09g47430D, while the number of exons ranged from one to six in Leg-K (Supplementary Table S1; Supplementary Figure S1). The average pI of BnLegLu proteins in Leg-K, Leg and Leg-T was 6.88, 8.08 and 6.89, and the instability index was 42.4, 21.4 and 37.98, respectively. Prediction of subcellular localization revealed that the BnLegLu genes in Leg-K, Leg and Leg-T were mainly located in the plasma membrane, extracellular, and chloroplast/plasma membrane, respectively (Figure 3; Supplementary Table S1). The observed variations in the characteristics of BnLegLus are probably the result of a series of evolutionary processes. Tandem and segmental duplications have been identified as the major mechanisms driving the expansion of lectins in soybean and rice (Jiang et al., 2010; Van Holle and Van Damme, 2015), which was also observed in B. napus. Most of BnLegLu genes (101 of 130) originated from WGD or segmental duplication, implying that this gene family is associated with the adaptation to various environmental stresses through duplication and divergence (Figure 1; Supplementary Table S1). In general, the fusion of protein and extended kinase domains represents the development of plant adaptation to stress (Bailey et al., 2018). All identified types of LecRLK can be considered as plant defense-related proteins as they might act as a receptor at the level of the cell wall/plasma membrane of the plant cell during a pathogen attack (Bellande et al., 2017). However, the functionality of these lectins needs to be investigated in more detail in the future.
Given that some lowly expressed lectins are not constitutively expressed but induced by environmental stresses, we investigated the spatiotemporal expression patterns of BnLegLu genes. Although the expression level of most BnLegLu genes was low, it showed great differentiation during the growth and development of B. napus (Supplementary Figure S5). A considerable number of genes play important roles in leaf development, silique wall development and seed development, such as some BnLegLu genes in cluster II, III and IV, whose expression level continuously increased at the leaf and silique wall developmental stage; some BnLegLu genes in cluster VI and VII had high expression in the early stage of seed development (Supplementary Figure S5). An in silico analysis of BnLegLus under abiotic and SD induction revealed that nearly 30% of BnLegLu genes are responsive to at least one stress (Figure 5; Supplementary Figure S6). Among these genes, those in cluster III and VI exhibited relatively high expression after induction by at least one stress (Figure 5; Supplementary Figure S6). Moreover, 16 BnLegLu genes were confirmed to be responsive to S. sclerotiorum as indicated by real-time PCR (qRT-PCR) analysis (Figure 6). In addition, cis-element promoter analysis also demonstrated that the BnLegLu genes in cluster III and VI contained more hormone-related and plant defense-related elements such as auxin-related, DRE-core, MBS and TC-rich elements (Supplementary Figure S3). Furthermore, the GO and KEGG pathway analysis predicted that the BnLegLu proteins are involved in response to biotic, mechanical stimuli and insects (Figure 4; Supplementary Figure S4). Overall, the expression of BnLegLus varied greatly among various stages of plant development, implying their essential roles in the development of B. napus, particularly in SD resistance, which may be an important adaptation strategy to the environment.
The mechanism for the response of BnLegLus to stress may be different in Leg-K, Leg and Leg-T. According to the phylogenetic analysis with the already known functions of AtLegLu, the BnLegLu genes in Leg-K may play important roles in plant immune response (Yekondi et al., 2018), while the BnLegLu genes in Leg may function in SA-mediated defense processes associated with the effector-triggered immunity response (Armijo et al., 2013; Figure 2). The role of BnLegLus in Leg-T remains unclear, as the function of their homologs in A. thaliana is unknown (Figure 2). The BnLegLu genes containing the kinase domain in cluster III may respond to S. sclerotiorum through stress signal transduction (Bellande et al., 2017); while the four BnLegLus genes in cluster VI whose expression was induced by SD may recognize and respond to S. sclerotiorum hyphae as a function of classical lectins (De Coninck and Van Damme, 2021; Fonseca et al., 2022; Figure 2). Previous studies have shown that lectins can bind to hyphae, resulting in the swelling of hyphae, vacuolation of the cell content, destruction of the nutrient absorption of the fungi, interference with spore germination, and even cell wall lysis (Lagarda-Diaz et al., 2017; Katoch and Tripathi, 2021). Here, subcellular localization analysis revealed that BnLegLus-16 was localized on the cell membrane, and its overexpression enhanced SD resistance in tobacco (Figures 7C–E), which provides a causal gene contributing to SD resistance in the plant. To evaluate the function of BnLegLus-16 on SD resistance in B. napus, the stable transformants with overexpression of BnLegLus-16 in B. napus and its close relative of A. thaliana is needed. In addition, the elaborate work of BnLegLus-16 on cell membrane during S. sclerotiorum-B. napus interaction also needs to be further emphasized.
Materials and methods
Identification of BnLegLu gene family in Brassica napus
In order to identify the BnLegLu, we used PF00139 as a query from Pfam database4 (Mistry et al., 2021) to search for LegLu genes in all proteins of B. napus (the E-value was set to 1e-5) by HMMER3.0.5 Then, the SMART database6 and Conserved Domain Database (NCBI; Lu et al., 2020)7 were used for verification of the candidate genes. The redundant BnLegLu genes were manually excluded. The current genome sequence and annotation information of rapeseed cultivar “Darmor-bzh” was obtained from the Brassicaceae Database (Chalhoub et al., 2014).8 The legume lectin genes in A. thaliana were downloaded from the Tair database.9
Peptide length, molecular weight, isoelectric point and instability index of each BnLegLu protein were calculated with the online ExPasy program.10 Signal peptide and transmembrane domain were predicted by online DeepTMHMM program (Singh et al., 2011).11 Subcellular localization was predicted by online CELLO v.2.5 program (Yu et al., 2004).12 The physical locations of BnLegLus on the chromosomes were obtained from the annotation of the B. napus genome. To identify the gene duplication events, BLASTP was used with the E-value of 1 × 10−10 to align the sequence and MCScanX was used to detect the duplication patterns, including segmental and tandem duplication. The chromosomal locations and duplication events were visualized using the TBtools software (Chen et al., 2020).
Phylogenetic analysis of BnLegLu gene family
The legume lectin protein sequences of A. thaliana and B. napus were subjected to multiple sequence alignment using the ClustalW2 (Larkin et al., 2007) to acquire insights into the evolutionary relationships between BnLegLu members. Phylogenetic trees were generated with the MEGA7 (Kumar et al., 2016) program using the Neighbor-Joining (NJ) method with 1,000 bootstrap replications. The tree was visualized using iTOL v6.5.2.13
Analysis of gene structures and protein conserved domains of BnLegLu gene family
The information of gene structures for BnLegLus was obtained from GFF files. Multiple Expectation Maximization for MEME (Motif Elicitation 5.4.1; Bailey et al., 2009) was used to analyze the conserved motifs in BnLegLu proteins. To this end, the following parameters were calibrated: maximum 10 motifs, with an optimal width of 6–50 amino acids. The remaining parameters were set to their default values. The identified motifs were annotated by using the Pfam database.14 The Conserved Domain Database (NCBI)15 was used to analyze the conserved domain of BnLegLu proteins. The TBtools was used to visualize the gene and motif structures (Chen et al., 2020).
Cis-element and protein interaction analysis of BnLegLus
The promoters of BnLegLus (2-kb upstream sequences from the initial codon) were extracted to identify the cis-acting regulatory elements using PlantCARE.16 The protein–protein interaction (PPI) of BnLegLu proteins in A. thaliana was downloaded from STRING (Szklarczyk et al., 2019).17 The functional association networks of the BnLegLu proteins in B. napus were predicted based on the homologs in A. thaliana, and Cytoscape (Shannon et al., 2003) was used to display the interactions. The genes interacting with the BnLegLu proteins were taken for gene ontology and KEGG enrichment analysis using the cluster Profiler in R (Yu et al., 2012).
Expression analysis of BnLegLus under different conditions
The transcriptome data from 92 tissue samples (eight different tissues covering ZS11 and three distinct developmental stages, including cotyledon, root, stem peel, leaf, bud, flower, silique, silique wall and seed; Liu et al., 2021b), four treatments (dehydration, cold, ABA and salinity; Zhang et al., 2019) and two cultivars (susceptible B. napus cv. Westar and tolerant B. napus cv. Zhongyou 821) induced by S. Sclerotiorum (Girard et al., 2017) were used in this study. The expression levels of BnLegLu genes were calculated with stringtie. Finally, the FPKM values were converted to log2 fold, and heat maps of all data were displayed by TBtools (Chen et al., 2020).
The seeds of ZY821 (from our laboratory) were germinated and grown in a growth room at 22°C with a 16-h light and 8-h dark photoperiod. S. Sclerotiorum strains (separated from the stubbles of oilseed rape in the fields of Yangluo, Wuhan, designated as WH-13) were used in this study. First, the fungal strains preserved at 4°C were sub-cultured on potato dextrose agar medium. Then, the new marginal hyphae were excised using a 7-mm puncher and closely upended onto the adaxial surface of healthy leaves (five-leaf stage). The inoculated plants were placed in a humidification chamber to keep the humidity above 85%. Each plant was inoculated with three leaves, and samples were taken every 12 h and immediately stored in liquid nitrogen. For each biological replicate, lesions were pooled from a minimum of three different plants and ground to a powder in liquid nitrogen. Total RNA was extracted by using Invitrogen TRIZOL Reagent.18 First-strand complementary DNA (cDNA) was synthesized by TaKaRa reverse transcription kit.19 Quantitative RT-PCR (qPCR) was performed using Bio Supermix20 following the manufacturer’s instructions. Reaction steps were performed with the following program: 95°C for 3 min; 40 cycles of 95°C for 15 s; 56°C for 15 s followed by 65°C for 5 s and 95°C for 5 s. The B. napus β-actin gene (AF111812) was used as a reference standard. All experiments were performed in three biological replicates. The relative expression was calculated using the 2 − ∆∆Ct method (Livak and Schmittgen, 2001).
Functional analysis of BnLegLus by genome-wide association analysis and transgenic strategy
To screen the potential BnLegLu genes responsible for SD resistance, the candidate genes based on the GWAS results obtained previously in our laboratory were checked (Tang, 2019; Ding et al., 2020). The GWAS population included 324 rapeseed accessions with different resistance levels collected worldwide. Resequencing of the GWAS accessions was performed by the commercial Illumina HiSeqXTen service (BGI-Shenzhen, China). For SD resistance determination, the leaves of plants grown in a field in Wuhan in 2015 at the stage of three-to-four leaves were excised and incubated in a growth room after inoculation with S. Sclerotiorum. The disease lesion size was examined at 12, 24, 36 and 48 hpi.
Open reading frame of BnLegLus-16 was cloned from ZY821 into pCambia2300-GFP at Bam-HI and KpnI sites using the ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., LTD, Nanjing, China; primers are shown in Supplementary Table S9). Agrobacterium (GV3101 competent cell) containing corresponding plasmids was infiltrated into four-week-old tobacco (Nicotiana. L) leaves for transient expression of fusion protein. Every tobacco was injected with three leaves and each plasmid was injected with 15 plants. These plants were incubated at 22°C in a growth room with a 16-h light and 8-h dark photoperiod for 60 h (Sparkes et al., 2006). For an accurate subcellular localization, the mCherry–H+-ATPase fusion protein, a marker of plasma membrane protein (Sun et al., 2021), was co-expressed with the BnLegLu-16–GFP, the fluorescence of tobacco leaves was monitored by confocal microscopy (ZEISS LSM710, Germany). The fluorescence emissions were at 510–540 nm for eGFP or at 600–650 nm for mCherry and excitations were at 450–490 nm for eGFP or at 523–588 nm for mCherry. Then, the leaves (expressed BnLegLus-16-ox and empty GFP vector, respectively) were excised and inoculated with S. Sclerotiorum hyphal plugs on detached leaves in the chamber with the humidity >85% and cultured in darkness. Each leaf was inoculated with one mycelium block. The size of disease spots was measured and photographed every 12 h. Then, the data were statistically analyzed, and three replicates were set for each experiment.
Conclusion
In this study, we conducted detailed characterization and investigation of BnLegLu genes. In total, 130 BnLegLu genes were identified and phylogenetically categorized into seven clusters. The genes in cluster I–V were variable in terms of gene number, structure and conserved motif, while those in cluster VI–VII were highly conserved. The majority of BnLegLu genes in cluster I–V contained an extended protein kinase at the C-terminal. Furthermore, most BnLegLu genes were lowly expressed and some BnLegLu genes were induced by stress, and some BnLegLus were particularly induced by S. sclerotiorum. Four BnLegLu genes were selected based on transcriptional variations in response to SD stimulation and significant sites in GWAS for SD resistance. Furthermore, we experimentally validated that the up-regulated expression of BnLegLus-16 could inhibit the spread of SD. In addition, we also discussed the possible mechanism for the response of BnLegLus to S. sclerotiorum, and proposed some directions for further study of BnLegLus-16. In summary, this study explores the function of BnLegLus in SD resistance, and also provides some clues for further exploration of the role of BnLegLus in SD resistance. The results are of great significance for further research on the function of BnLegLu gene family for the genetic improvement of agronomic traits or stress tolerance in B. napus.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, and further inquiries can be directed to the corresponding author.
Author contributions
RZ and ZB: conceptualization and writing—review and editing. RZ and MX: data curation. ZB and SL: funding acquisition. FG: investigation. RZ, MX, JL, and MT: methodology. XC: project administration. RZ: software and writing—original draft. FG and YL: validation. YL: germplasm resources. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Central Public-interest Scientific Institution Basal Research Fund, grant numbers 2021-2060302-061-027 and 2021-2060302-061-029; China Agriculture Research System of MOF and MARA (CARS-12); the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-OCRI); and the National Key Research and Development Program of China grant number U20A2034.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.963263/full#supplementary-material
Footnotes
1. ^https://www.ncbi.nlm.nih.gov/cdd
2. ^https://www.string-db.org/
3. ^http://yanglab.hzau.edu.cn/BnTIR
7. ^https://www.ncbi.nlm.nih.gov/cdd2
8. ^https://www.genoscope.cns.fr/brassicanapus/
9. ^https://www.arabidopsis.org/
11. ^https://dtu.biolib.com/DeepTMHMM
12. ^http://cello.life.nctu.edu.tw/
14. ^http://pfam.xfam.org/search
15. ^https://www.ncbi.nlm.nih.gov/cdd
16. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
17. ^https://www.string-db.org/
18. ^https://www.thermofisher.com
References
Armijo, G., Salinas, P., Monteoliva, M. I., Seguel, A., García, C., Villarroel-Candia, E., et al. (2013). A salicylic acid-induced lectin-like protein plays a positive role in the effector-triggered immunity response of Arabidopsis thaliana to Pseudomonas syringae Avr-Rpm1. Mol. Plant Microbe Interact. 26, 1395–1406. doi: 10.1094/mpmi-02-13-0044-r
Arnaud, D., Desclos-Theveniau, M., and Zimmerli, L. (2012). Disease resistance to Pectobacterium carotovorum is negatively modulated by the Arabidopsis lectin receptor kinase LecRK-V.5. Plant Signal. Behav. 7, 1070–1072. doi: 10.4161/psb.21013
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bailey, P. C., Schudoma, C., Jackson, W., Baggs, E., Dagdas, G., Haerty, W., et al. (2018). Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 19, 23. doi: 10.1186/s13059-018-1392-6
Balagué, C., Gouget, A., Bouchez, O., Souriac, C., Haget, N., Boutet-Mercey, S., et al. (2017). The Arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 18, 937–948. doi: 10.1111/mpp.12457
Bellande, K., Bono, J. J., Savelli, B., Jamet, E., and Canut, H. (2017). Plant lectins and lectin receptor-like kinases: how do they sense the outside? Int. J. Mol. Sci. 18:1164. doi: 10.3390/ijms18061164
Bouwmeester, K., de Sain, M., Weide, R., Gouget, A., Klamer, S., Canut, H., et al. (2011). The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 7:e1001327. doi: 10.1371/journal.ppat.1001327
Cao, Y., Yan, X., Ran, S., Ralph, J., Smith, R. A., Chen, X., et al. (2022). Knockout of the lignin pathway gene BnF5H decreases the S/G lignin compositional ratio and improves Sclerotinia sclerotiorum resistance in Brassica napus. Plant Cell Environ. 45, 248–261. doi: 10.1111/pce.14208
Cavada, B. S., Osterne, V. J. S., Lossio, C. F., Pinto-Junior, V. R., Oliveira, M. V., Silva, M. T. L., et al. (2019). One century of ConA and 40 years of ConBr research: A structural review. Int. J. Biol. Macromol. 134, 901–911. doi: 10.1016/j.ijbiomac.2019.05.100
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Damme, E., Lannoo, N., and Peumans, W. J. (2008). Plant Lectins. Adv. Bot. Res. 48, 107–209. doi: 10.1016/S0065-2296(08)00403-5
De Coninck, T., and Van Damme, E. J. M. (2021). Review: The multiple roles of plant lectins. Plant Sci. 313:111096. doi: 10.1016/j.plantsci.2021.111096
Desclos-Theveniau, M., Arnaud, D., Huang, T. Y., Lin, G. J., Chen, W. Y., Lin, Y. C., et al. (2012). The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. Tomato DC3000. PLoS Pathog. 8:e1002513. doi: 10.1371/journal.ppat.1002513
Ding, L. N., Li, T., Guo, X. J., Li, M., Liu, X. Y., Cao, J., et al. (2021). Sclerotinia stem rot resistance in rapeseed: recent progress and future prospects. J. Agric. Food Chem. 69, 2965–2978. doi: 10.1021/acs.jafc.0c07351
Ding, L. N., Li, M., Guo, X. J., Tang, M. Q., Cao, J., Wang, Z., et al. (2020). Arabidopsis GDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus. Plant Biotechnol. J. 18, 1255–1270. doi: 10.1111/pbi.13289
Edelman, G. M., Cunningham, B. A., Reeke, G. N. Jr., Becker, J. W., Waxdal, M. J., and Wang, J. L. (1972). The covalent and three-dimensional structure of concanavalin A. Proc. Natl. Acad. Sci. U. S. A. 69, 2580–2584. doi: 10.1073/pnas.69.9.2580
Eggermont, L., Verstraeten, B., and Van Damme, E. J. M. (2017). Genome-wide screening for lectin motifs in Arabidopsis thaliana. Plant. Genome 10:2. doi: 10.3835/plantgenome2017.02.0010
Fedoroff, N. V., Battisti, D. S., Beachy, R. N., Cooper, P. J., Fischhoff, D. A., Hodges, C. N., et al. (2010). Radically rethinking agriculture for the 21st century. Science 327, 833–834. doi: 10.1126/science.1186834
Fonseca, V. J. A., Braga, A. L., Filho, J. R., Teixeira, C. S., da Hora, G. C. A., and Morais-Braga, M. F. B. (2022). A review on the antimicrobial properties of lectins. Int. J. Biol. Macromol. 195, 163–178. doi: 10.1016/j.ijbiomac.2021.11.209
Gatehouse, A. M. R., Davison, G. M., Stewart, J. N., Gatehouse, L. N., Kumar, A., Geoghegan, I. E., et al. (1999). Concanavalin A inhibits development of tomato moth (Lacanobia oleracea) and peach-potato aphid (Myzus persicae) when expressed in transgenic potato plants. Mol. Breed. 5, 153–165. doi: 10.1023/A:1009681705481
Gautam, A. K., Gupta, N., Narvekar, D. T., Bhadkariya, R., and Bhagyawant, S. S. (2018a). Characterization of chickpea (Cicer arietinum L.) lectin for biological activity. Physiol. Mol. Biol. Plants 24, 389–397. doi: 10.1007/s12298-018-0508-5
Gautam, A. K., Shrivastava, N., Sharma, B., and Bhagyawant, S. S. (2018b). Current scenario of legume lectins and their practical applications. J. Crop. Sci. Biotechnol. 21, 217–227. doi: 10.1007/s12892-018-0002-0
Girard, I. J., Tong, C., Becker, M. G., Mao, X., Huang, J., de Kievit, T., et al. (2017). RNA sequencing of Brassica napus reveals cellular redox control of Sclerotinia infection. J. Exp. Bot. 68, 5079–5091. doi: 10.1093/jxb/erx338
Hardman, K. D., and Ainsworth, C. F. (1972). Structure of concanavalin A at 2.4-A resolution. Biochemistry 11, 4910–4919. doi: 10.1021/bi00776a006
He, X. J., Zhang, Z. G., Yan, D. Q., Zhang, J. S., and Chen, S. Y. (2004). A salt-responsive receptor-like kinase gene regulated by the ethylene signaling pathway encodes a plasma membrane serine/threonine kinase. Theor. Appl. Genet. 109, 377–383. doi: 10.1007/s00122-004-1641-9
Huang, P. Y., Yeh, Y. H., Liu, A. C., Cheng, C. P., and Zimmerli, L. (2014). The Arabidopsis LecRK-VI.2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity. Plant J. 79, 243–255. doi: 10.1111/tpj.12557
Jiang, S. Y., Ma, Z., and Ramachandran, S. (2010). Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol. Biol. 10, 79. doi: 10.1186/1471-2148-10-79
Joshi, A., Dang, H. Q., Vaid, N., and Tuteja, N. (2010). Pea lectin receptor-like kinase promotes high salinity stress tolerance in bacteria and expresses in response to stress in planta. Glycoconj. J. 27, 133–150. doi: 10.1007/s10719-009-9265-6
Katoch, R., and Tripathi, A. (2021). Research advances and prospects of legume lectins. J. Biosci. 46:104. doi: 10.1007/s12038-021-00225-8
Kosová, K., Vítámvás, P., Prášil, I. T., and Renaut, J. (2011). Plant proteome changes under abiotic stress--contribution of proteomics studies to understanding plant stress response. J. Proteomics 74, 1301–1322. doi: 10.1016/j.jprot.2011.02.006
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lagarda-Diaz, I., Guzman-Partida, A. M., and Vazquez-Moreno, L. (2017). Legume lectins: proteins with diverse applications. Int. J. Mol. Sci. 18:1242. doi: 10.3390/ijms18061242
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Leal, A. F., Lopes, N. E., Clark, A. T., de Pontes Filho, N. T., Beltrão, E. I., and Neves, R. P. (2012). Carbohydrate profiling of fungal cell wall surface glycoconjugates of Aspergillus species in brain and lung tissues using lectin histochemistry. Med. Mycol. 50, 756–759. doi: 10.3109/13693786.2011.631946
Li, Z., Chakraborty, S., and Xu, G. (2016). X-ray crystallographic studies of the extracellular domain of the first plant ATP receptor, DORN1, and the orthologous protein from Camelina sativa. Acta Crystallogr. F Struct. Biol. Commun. 72, 782–787. doi: 10.1107/s2053230x16014278
Li, T., Yin, X., Liu, D., Ma, X., Lv, H., and Sun, S. (2012). Isolation and characterization of a novel lectin with antifungal and antiproliferative activities from Sophora alopecuroides seeds. Acta Biochim. Biophys. Sin. Shanghai 44, 606–613. doi: 10.1093/abbs/gms037
Liang, X., and Rollins, J. A. (2018). Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 108, 1128–1140. doi: 10.1094/phyto-06-18-0197-rvw
Liu, F., Li, X., Wang, M., Wen, J., Yi, B., Shen, J., et al. (2018). Interactions of WRKY15 and WRKY33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnol. J. 16, 911–925. doi: 10.1111/pbi.12838
Liu, D., Wu, J., Lin, L., Li, P., Li, S., Wang, Y., et al. (2021a). Overexpression of Cinnamoyl-CoA reductase 2 in Brassica napus increases resistance to Sclerotinia sclerotiorum by affecting lignin biosynthesis. Front. Plant Sci. 12:732733. doi: 10.3389/fpls.2021.732733
Liu, D., Yu, L., Wei, L., Yu, P., Wang, J., Zhao, H., et al. (2021b). BnTIR: an online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol. J. 19, 1895–1897. doi: 10.1111/pbi.13665
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–d268. doi: 10.1093/nar/gkz991
Luo, X., Xu, N., Huang, J., Gao, F., Zou, H., Boudsocq, M., et al. (2017). A lectin receptor-like kinase mediates pattern-triggered salicylic acid signaling. Plant Physiol. 174, 2501–2514. doi: 10.1104/pp.17.00404
Lyou, S. H., Park, H. J., Jung, C., Sohn, H. B., Lee, G., Kim, C. H., et al. (2009). The Arabidopsis AtLEC gene encoding a lectin-like protein is up-regulated by multiple stimuli including developmental signal, wounding, jasmonate, ethylene, and chitin elicitor. Mol. Cells 27, 75–81. doi: 10.1007/s10059-009-0007-1
Manoj, N., and Suguna, K. (2001). Signature of quaternary structure in the sequences of legume lectins. Protein Eng. 14, 735–745. doi: 10.1093/protein/14.10.735
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–d419. doi: 10.1093/nar/gkaa913
Myers, R. J., Fichman, Y., Stacey, G., and Mittler, R. (2022). Extracellular ATP plays an important role in systemic wound response activation. Plant Physiol. 189, 1314–1325. doi: 10.1093/plphys/kiac148
Nivetha, R., Meenakumari, M., Bhuvaragavan, S., Hilda, K., and Janarthanan, S. (2021). In silico analysis of carbohydrate-binding pockets in the lectin genes from various species of Canavalia. Comput. Biol. Chem. 92:107477. doi: 10.1016/j.compbiolchem.2021.107477
Nováková, M., Sašek, V., Dobrev, P. I., Valentová, O., and Burketová, L. (2014). Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum—reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol. Biochem. 80, 308–317. doi: 10.1016/j.plaphy.2014.04.019
Passricha, N., Saifi, S. K., Kharb, P., and Tuteja, N. (2019). Marker-free transgenic rice plant overexpressing pea LecRLK imparts salinity tolerance by inhibiting sodium accumulation. Plant Mol. Biol. 99, 265–281. doi: 10.1007/s11103-018-0816-8
Pham, A. Q., Cho, S. H., Nguyen, C. T., and Stacey, G. (2020). Arabidopsis Lectin receptor kinase P2K2 is a second plant receptor for extracellular ATP and contributes to innate immunity. Plant Physiol. 183, 1364–1375. doi: 10.1104/pp.19.01265
Saijo, Y., and Loo, E. P. (2020). Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 225, 87–104. doi: 10.1111/nph.15989
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Singh, N. K., Goodman, A., Walter, P., Helms, V., and Hayat, S. (2011). TMBHMM: a frequency profile based HMM for predicting the topology of transmembrane beta barrel proteins and the exposure status of transmembrane residues. Biochim. Biophys. Acta 1814, 664–670. doi: 10.1016/j.bbapap.2011.03.004
Singh, P., and Zimmerli, L. (2013). Lectin receptor kinases in plant innate immunity. Front. Plant Sci. 4:124. doi: 10.3389/fpls.2013.00124
Sitohy, M., Doheim, M., and Badr, H. (2007). Isolation and characterization of a lectin with antifungal activity from Egyptian Pisum sativum seeds. Food Chem. 104, 971–979. doi: 10.1016/j.foodchem
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Sprawka, I., Goławska, S., Parzych, T., Sytykiewicz, H., and Czerniewicz, P. (2015). Apoptosis induction by concanavalin A in gut cells of grain aphid. Arthropod. Plant Interact. 9, 133–140. doi: 10.1007/s11829-015-9356-1
Sun, Z., Zang, Y., Zhou, L., Song, Y., Chen, D., Zhang, Q., et al. (2021). A tomato receptor-like cytoplasmic kinase, SlZRK1, acts as a negative regulator in wound-induced jasmonic acid accumulation and insect resistance. J. Exp. Bot. 72, 7285–7300. doi: 10.1093/jxb/erab350
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–d613. doi: 10.1093/nar/gky1131
Tang, M.Q. (2019). Population genome variations and subgenome asymmetry in Brassica napus L. Huazhong Agricultural University Wuhan, China, D50–D59.
Tsaneva, M., and Van Damme, E. J. M. (2020). 130 years of plant Lectin research. Glycoconj. J. 37, 533–551. doi: 10.1007/s10719-020-09942-y
Van Damme, E. J. M. (2022). 35 years in plant lectin research: a journey from basic science to applications in agriculture and medicine. Glycoconj. J. 39, 83–97. doi: 10.1007/s10719-021-10015-x
Van Holle, S., and Van Damme, E. J. (2015). Distribution and evolution of the lectin family in soybean (Glycine max). Molecules 20, 2868–2891. doi: 10.3390/molecules20022868
Wang, Y., Cordewener, J. H., America, A. H., Shan, W., Bouwmeester, K., and Govers, F. (2015). Arabidopsis lectin receptor kinases LecRK-IX.1 and LecRK-IX.2 are functional analogs in regulating phytophthora resistance and plant cell death. Mol. Plant Microbe Interact. 28, 1032–1048. doi: 10.1094/mpmi-02-15-0025-r
Wang, C., Huang, X., Li, Q., Zhang, Y., Li, J. L., and Mou, Z. (2019). Extracellular pyridine nucleotides trigger plant systemic immunity through a lectin receptor kinase/BAK1 complex. Nat. Commun. 10, 4810. doi: 10.1038/s41467-019-12781-7
Wang, Y., Nsibo, D. L., Juhar, H. M., Govers, F., and Bouwmeester, K. (2016). Ectopic expression of Arabidopsis L-type lectin receptor kinase genes LecRK-I.9 and LecRK-IX.1 in Nicotiana benthamiana confers Phytophthora resistance. Plant Cell Rep. 35, 845–855. doi: 10.1007/s00299-015-1926-2
Wang, Z., Wan, L., Xin, Q., Chen, Y., Zhang, X., Dong, F., et al. (2018a). Overexpression of OsPGIP2 confers Sclerotinia sclerotiorum resistance in Brassica napus through increased activation of defense mechanisms. J. Exp. Bot. 69, 3141–3155. doi: 10.1093/jxb/ery138
Wang, L., Wilkins, K. A., and Davies, J. M. (2018b). Arabidopsis DORN1 extracellular ATP receptor; activation of plasma membrane K(+) -and Ca(2+) -permeable conductances. New Phytol. 218, 1301–1304. doi: 10.1111/nph.15111
Wang, C., Zhou, M., Zhang, X., Yao, J., Zhang, Y., and Mou, Z. (2017). A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. Elife 6:e25474. doi: 10.7554/eLife.25474
Wu, J., Zhao, Q., Liu, S., Shahid, M., Lan, L., Cai, G., et al. (2016). Genome-wide association study identifies new loci for resistance to Sclerotinia stem rot in Brassica napus. Front. Plant Sci. 7:1418. doi: 10.3389/fpls.2016.01418
Xin, Z., Wang, A., Yang, G., Gao, P., and Zheng, Z. L. (2009). The Arabidopsis A4 subfamily of lectin receptor kinases negatively regulates abscisic acid response in seed germination. Plant Physiol. 149, 434–444. doi: 10.1104/pp.108.130583
Xu, N., Luo, X., Wu, W., Xing, Y., Liang, Y., Liu, Y., et al. (2020). A plant Lectin receptor-like kinase phosphorylates the bacterial effector AvrPtoB to dampen its virulence in Arabidopsis. Mol. Plant 13, 1499–1512. doi: 10.1016/j.molp.2020.09.016
Yekondi, S., Liang, F. C., Okuma, E., Radziejwoski, A., Mai, H. W., Swain, S., et al. (2018). Nonredundant functions of Arabidopsis LecRK-V.2 and LecRK-VII.1 in controlling stomatal immunity and jasmonate-mediated stomatal closure. New Phytol. 218, 253–268. doi: 10.1111/nph.14953
Yu, C. S., Lin, C. J., and Hwang, J. K. (2004). Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. doi: 10.1110/ps.03479604
Yu, G., Wang, L. G., Han, Y., and He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287. doi: 10.1089/omi.2011.0118
Zhang, Y., Ali, U., Zhang, G., Yu, L., Fang, S., Iqbal, S., et al. (2019). Transcriptome analysis reveals genes commonly responding to multiple abiotic stresses in rapeseed. Mol. Breed. 39, 158. doi: 10.1007/s11032-019-1052-x
Keywords: legume lectin, Brassica napus, Sclerotinia sclerotiorum, phylogenetic analysis, genome-wide association study
Citation: Zuo R, Xie M, Gao F, Liu J, Tang M, Cheng X, Liu Y, Bai Z and Liu S (2022) Genome-wide identification and functional exploration of the legume lectin genes in Brassica napus and their roles in Sclerotinia disease resistance. Front. Plant Sci. 13:963263. doi: 10.3389/fpls.2022.963263
Edited by:
Lin Huang, Sichuan Agricultural University, ChinaReviewed by:
Guangda Ding, Huazhong Agricultural University, ChinaLiezhao Liu, Southwest University, China
Copyright © 2022 Zuo, Xie, Gao, Liu, Tang, Cheng, Liu, Bai and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zetao Bai, YmFpemV0YW9AY2Fhcy5jbg==
 Rong Zuo
Rong Zuo Meili Xie
Meili Xie Feng Gao1
Feng Gao1 Minqiang Tang
Minqiang Tang Shengyi Liu
Shengyi Liu