- 1State Key Laboratory of Rice Biology, Key Laboratory of Rice Biology and Breeding, Ministry of Agriculture, China National Rice Improvement Centre, China National Rice Research Institute, Hangzhou, China
- 2Rice Research Institute, Shengyang Agricultural University, Shenyang, China
The stigma exsertion rate (SER) is a complex agronomy phenotype controlled by multiple genes and climate and a key trait affecting the efficiency of hybrid rice seed production. Using a japonica two-line male sterile line (DaS) with a high SER as the donor and a tropical japonica rice (D50) with a low SER as the acceptor to construct a near-isogenic line [NIL (qSE4DaS)]. Populations were segregated into 2,143 individuals of BC3F2 and BC4F2, and the stigma exsertion quantitative trait locus (QTL) qSE4 was determined to be located within 410.4 Kb between markers RM17157 and RM17227 on chromosome 4. Bioinformatic analysis revealed 13 candidate genes in this region. Sequencing and haplotype analysis indicated that the promoter region of LOC_Os04g43910 (ARF10) had a one-base substitution between the two parents. Further Reverse Transcription-Polymerase Chain Reaction (RT-PCR) analysis showed that the expression level of ARF10 in DaS was significantly higher than in D50. After knocking out ARF10 in the DaS background, it was found that the SER of arf10 (the total SER of the arf10-1 and the arf10-2 were 62.54 and 66.68%, respectively) was significantly lower than that of the wild type (the total SER was 80.97%). Transcriptome and hormone assay analysis showed that arf10 had significantly higher auxin synthesis genes and contents than the wild type and the expression of auxin signaling-related genes was significantly different, Similar results were observed for abscisic acid and jasmonic acid. These results indicate that LOC_Os04g43910 is mostly likely the target gene of qSE4, and the study of its gene function is of great significance for understanding the molecular mechanisms of SER and improving the efficiency of hybrid seed production.
Introduction
Rice (Oryza sativa L.) is a staple food for billions of people worldwide (Khush, 2005). High yield has always been the primary goal of rice breeding in China for the past few decades. The successful development of hybrid rice was another milestone in enhancing rice yield following dwarf breeding. Rice is a typical self-pollinating crop (Virmani and Athwal, 1973), but the male sterile line with pollen abortion cannot self-fertilize, so the male sterile line is the key line for hybrid rice breeding. Stigma exsertion is an important agronomic trait for male sterile lines to successfully receive restorers’ pollen. Studies have shown that flowering does not mean fertilization (Kato and Namai, 1987), and the exposed stigma can maintain vigor for 4 to 6 days, which greatly improves the outcrossing rate of rice; thereby improving the seed production efficiency of hybrid rice (Yan and Li, 1987; Bi and Tan, 1988; Tian et al., 2004). Therefore, the stigma exsertion rate (SER) of male sterile lines is a key factor of hybrid rice. Increasing the SER is beneficial to increase the yield of hybrids and promote their commercialization (Virmani, 1994).
In the past two decades, dozens of QTLs related to the SER, distributed on 12 chromosomes of rice, have been detected by various germplasm resources and methods (Miyata et al., 2007; Yan et al., 2009; Liu et al., 2019; Tan et al., 2020). For this research, male sterile lines, maintainer lines, and wild rice with high stigma exsertion are commonly used. From the two-line male sterile line DaS, qPES3, qPDES4, and qPES12 were mapped on chromosomes 3, 4, and 12, respectively (Li et al., 2017). Using the single-segment substitution line created by the maintainer line IR66897B, qSER-2a, qSER2b, and qSER3a, qSER3b were mapped on chromosomes 2 and 3, respectively (Tan et al., 2021). From the maintainer XieqingzaoB, qSE7 (Zhang et al., 2018) and qSE11 (Rahman et al., 2017) were mapped on chromosomes 7 and 11, respectively. Many QTLs for SER, such as qPEST-5, qPEST-8, qRES-5 (Uga et al., 2003), qRES-10 (Uga et al., 2003), qSER-2 (Bakti and Tanaka, 2019), qSER-3 (Bakti and Tanaka, 2019), qPES-9, and qSER-1a (Tan et al., 2020) have been found in wild rice, which is an important germplasm resource (Marathi et al., 2015; Marathi and Jena, 2015). qES3 is a QTL of SER that has been mapped multiple times (Miyata et al., 2007) and later confirmed as the grain size gene GS3 (Takano-Kai et al., 2011), which affects the SER by controlling the length of the stigma. Another gene, qSTL3, which also controls stigma length, was mapped from the indica rice Kasalath (Liu et al., 2015). However, neither of these two genes were cloned with SER as the mapping feature trait; only LOC_Os07g15370 has been cloned using SER as the mapping feature trait (Liu et al., 2019). Unfortunately, this gene has not been confirmed to affect SER in sterile lines. Despite substantial research, most genes only stay in the mapping stage because the stigma exsertion is not only affected by genes, but also by environmental factors (e.g., temperature, moisture, wind; Yan et al., 2009). In addition, most populations used in these studies were either F2 populations, recombinant inbred lines, backcross inbred lines, or double haploid lines. It is difficult to exclude epistatic effects of different chromosome segments in these populations, further challenging QTL cloning of SER (Liu et al., 2015). Therefore, we presently have a poor understanding of the molecular mechanisms of SER in rice, and it is urgent to clone SER-related genes.
In our previous study, the F2 population was constructed using the japonica two-line male sterile line, DaS, and tropical japonica, D50, to map QTL of SER, qPES3 on Chr3, qPDES4 on Chr4, and qPES12 on Chr12, and the contribution rates were 25.6, 17.86, and 16.98%, respectively (Li et al., 2017). In this study, we further constructed a near-isogenic line NIL (qSE4DaS), whose SER was significantly higher than that of D50. The qSE4 gene was narrowed within 410.4 kb on chromosome 4 using the backcross inbred populations of BC3F2 and BC4F2. Within this region, a possible stigma exsertion-related gene was identified, and the function of the gene was preliminarily studied. These results provide a significant breakthrough for molecular level research on the SER of rice and have important value for the hybrid rice with high and stable yield in seed production.
Materials and methods
Population and field experiments
The process of constructing the mapping population is shown in Figure 1. In our previous study, QTL qSE4 was detected for a dual SER using an F2 population derived from a cross between D50 and DaS (Li et al., 2017). D50 is a tropical japonica with a low SER. DaS is a japonica two-line male sterile line with a high SER selected from the offspring of indica-japonica hybrids.
We fine-mapped qSE4 by constructing a NIL with respect to qSE4. To this end, an F2:3 line with the DaS genotype in the qSE4 region was selected to successively backcross with D50 for four generations. The SSR markers RM17157 and RM17303 were used for marker-assisted selection (MAS) of each generation among the segregating progenies. Finally, we used the BC3F2 and BC4F2 populations to map qSE4 and selected two BC4F2 individuals as NILs. To test recovery rate of the NILs, we used 134 pairs of polymorphic SSR markers between DaS and D50, which evenly distributed on 12 rice chromosomes. It was found that the genotypes of all the markers of the two NILs were the same as those of D50 except for RM17207 and RM17227.
The qSE4 knockout vector was constructed using the BGK03 vector of the Biogle kit and sent to Wuhan Biorun Company for the transformation of the parental DaS.
All materials were grown at the experimental field of China National Rice Research Institute or the test field in Lingshui, Hainan Province, China. Standard crop management practices were followed.
Phenotypic evaluation
After full flowering of the rice plants, 6 main panicles were randomly selected from each line, and the single (SSE), dual (DSE), and total (TSE) SER were investigated. SSE (%) = single stigma exposed spikelets / total spikelets × 100%; DSE (%) = dual stigma exposed spikelets/total spikelets × 100%; TSE (%) = SSE + DSE.
DNA extraction and molecular marker screening
The genomic DNA was extracted from fresh leaves using the Sodium Dodecyl Sulfate (SDS) method (Orjuela et al., 2010). Polymerase Chain Reaction (PCR) was performed in 12 μl reaction volumes containing 2 template DNA, 5 μl of 2 × T5 Super PCR Mix (PAGE; TSINGKE, Beijing, China), 1 μl of 10 μmol/μL primer pairs, and 4 μl of double distilled H2O. Target DNA segments were amplified with the following program, 95°C for 5 min, followed by 35 cycles of 95°C for 10 s, 55°C for 10 s, and 72°C for 10 s, and a final extension of 72°C for 8 min. The PCR products were separated by 8% non-denaturing polyacrylamide gel electrophoresis, and bands were detected using the silver staining methods described by Wang et al. (2015). Molecular markers are provided by the preservation of our laboratory.
RNA extraction and qRT-PCR analysis
Total RNA was isolated from young panicles at the pre-heading stage (Liu et al., 2019) using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was reverse transcribed to generate cDNA using the Rever Tra Ace qPCR RT Kit (Toyobo, Osaka, Japan). Gene expression was measured by qRT-PCR using the OsActin (LOC_Os03g50885) as an internal control. PCR was performed via the Applied Biosystems Quant Studio 3 Real-Time PCR System (Thermo Fisher Scientific, USA). The PCR program was 95°C for 5 min, followed by 40 cycles at 95°C for 5 s, 60°C for 34 s, followed at 95°C for 15 s, 60°C for 1 min, 95°C for 15 s.
RNA-seq analysis
Young panicles of DaS and arf10 at the pre-heading stage were harvested and immediately frozen in liquid nitrogen. RNA-seq analysis was performed by Novogene (Beijing, China). After the RNA was reversed to double-stranded cDNA, qualified libraries were constructed, then sequenced by the Illumina NovaSeq 6,000. Differential expression analysis between the two comparisons was performed using the DESeq2 R package (1.20.0). The resulting p-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Thresholds of padj < 0.05 and |log2foldchange| > 1 were set for significant differential expression. GO enrichment analysis and KEGG enrichment analysis of differentially expressed genes were both analyzed by the cluster Profiler R package (3.8.1).
Measurement of free IAA, ABA, and JA content
Young panicles of DaS and arf10 at the pre-heading stage were harvested and immediately frozen in liquid nitrogen. The samples (100 mg) were resuspended with liquid nitrogen and homogenized with 400 μl of acetonitrile (50%) which contained mixed internal standards and extracted at 4°C. Then centrifuged at 12,000 rpm for 10 min. The supernatant passed through the HLB sorbent (first flow-through fraction) and then was eluted subsequently with 500 μl of acetonitrile (30%; second flow-through fraction). These two fractions were combined into the same centrifuge tube and mixed well. Finally, these solutions were injected into the LC-MS/MS system for analysis.
Statistical analysis
One-way ANOVA was used to test the statistically significant differences among tested varieties, which was performed using SPSS 22.0 software (IBM Inc.).
Results
Development of the near isogenic line for qSE4
Based on previous research, qSE4 was mapped between markers RM17157 and RM17303 (Li et al., 2017; Figure 2A). One line from the F2:3 population was selected for four rounds of backcrossing with D50, and near-isogenic lines (NILs) were used to isolate qSE4 (Figure 1). The markers RM17157 and RM17303 were used for marker-assisted selection in the segregating progenies carrying the DaS qSE4 allele during each backcross generation. After continuous backcrossing for four generations and selfing, the genetic background became similar to that of the recurrent parent D50, except for the substituted target segments. Two individuals with the highest recovery rate to the recurrent parent were selected among the segregating progeny, i.e., NIL (qSE4DaS)-1 and NIL (qSE4DaS)-2, which carry the homozygous allele of DaS in the region of the qSE4.
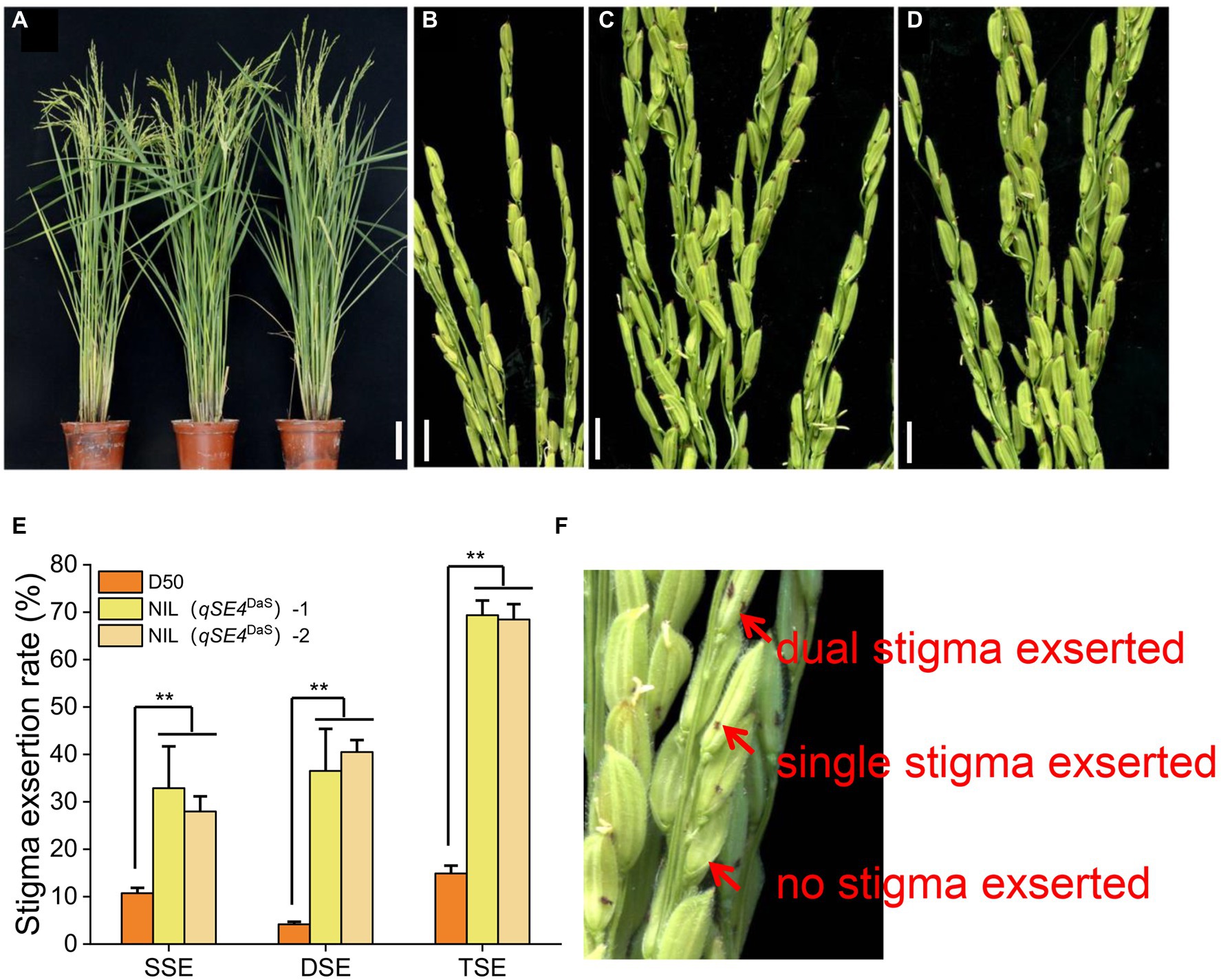
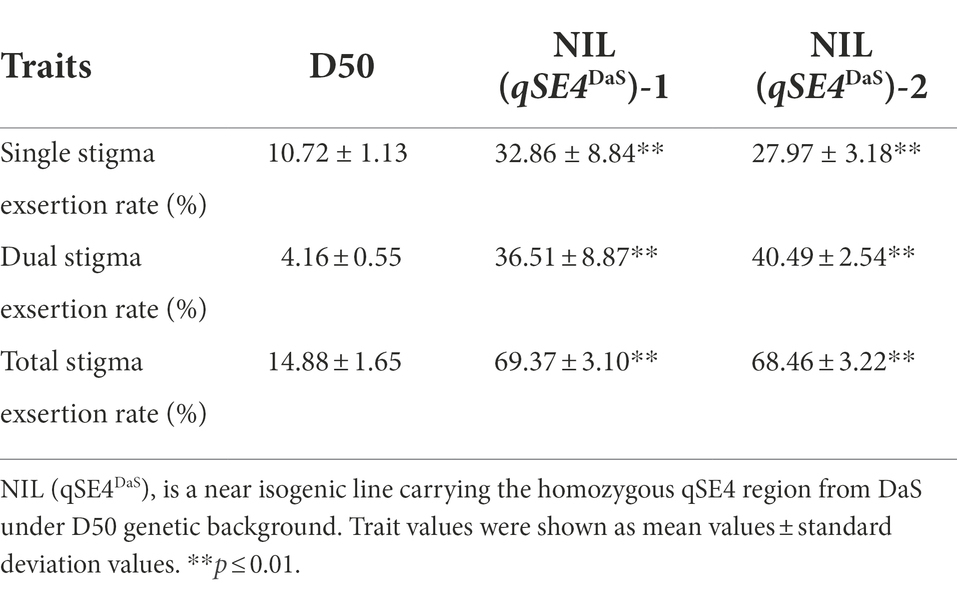
Figure 2. Phenotype of parental D50 and NIL. (A) D50, NIL (qSE4DaS)-1 and NIL (qSE4DaS)-2 from left to right during heading, bar = 10 cm. (B) Stigma exsertion phenotype of D50, bar = 1 cm. (C) Stigma exsertion phenotype of NIL (qSE4DaS)-1, bar = 1 cm. (D) Stigma exsertion phenotype of NIL (qSE4DaS)-2, bar = 1 cm. (E) Stigma exertion rate for D50, NIL (qSE4DaS)-1 and NIL (qSE4DaS)-2. SSE: single stigma exsertion rate, DSE: dual stigma exsertion rate, TSE: total stigma exsertion rate. (F) Stigma exsertion phenotype in rice, contains dual, single, and no stigma exsertion. The data represent the mean values ± SD (n = 6), **p ≤ 0.01.
The NIL (qSE4DaS)-1 had increased exsertion rates of 22.14, 32.35, and 54.49% for single stigma exsertion rate (SSE), dual stigma exsertion rate (DSE), and total stigma exsertion rate (TSE), respectively, when compared to the recurrent parent D50. The NIL (qSE4DaS)-2 had increased exsertion rates of 17.25, 36.33, and 53.58% for SSE, DSE, and TSE, respectively (Table 1; Figure 3). This indicated that qSE4 is responsible for the high stigma exsertion rate in NIL (qSE4DaS).
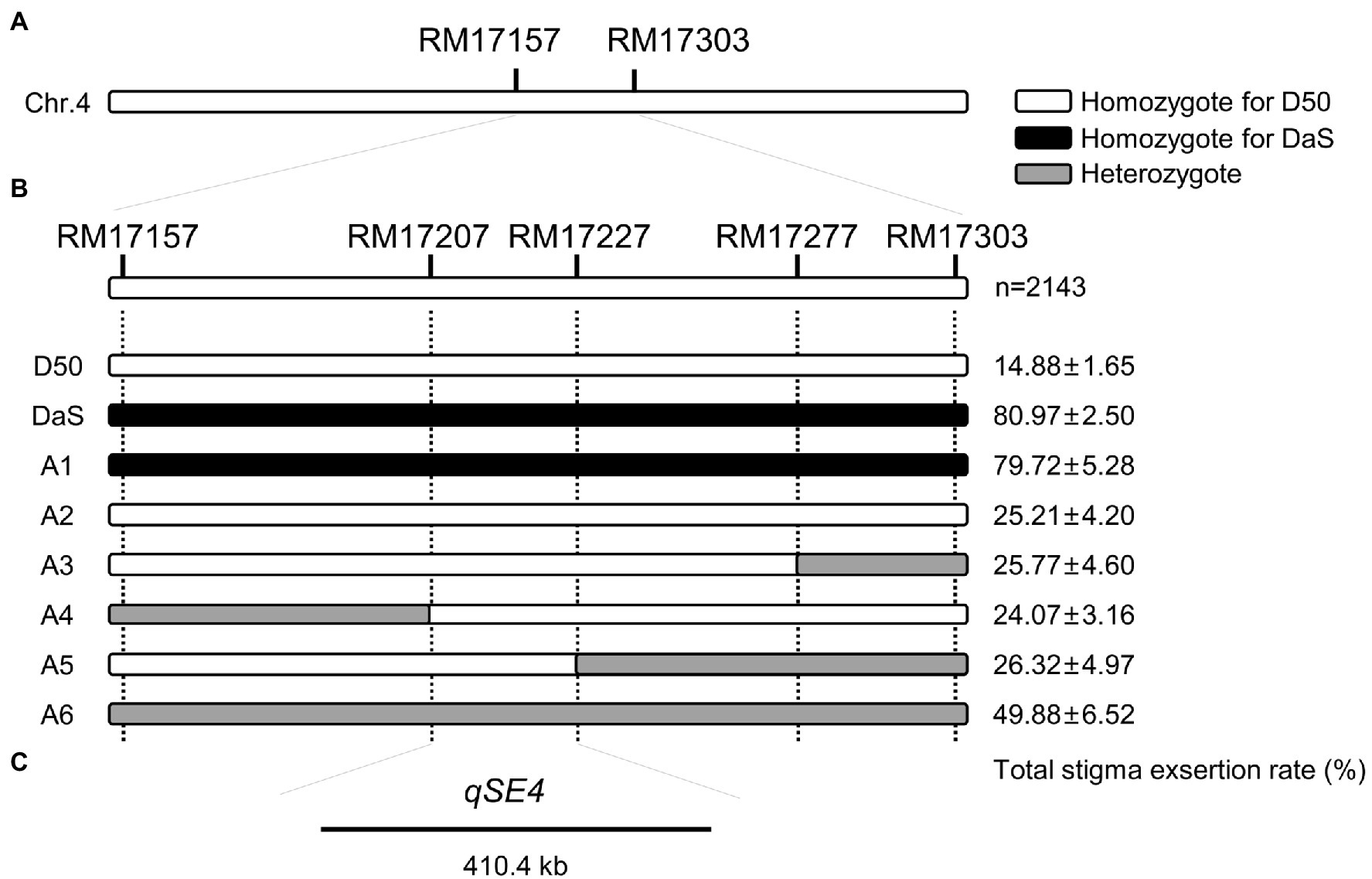
Figure 3. Fine mapping of qSE4. (A) qSE4 is located between chromosome 4 markers RM17157 and RM17303. (B) Genotype and phenotype of D50, DaS and six recombinant individuals (A1–A6). The data represent the mean values ± SD (n = 6). (C) qSE4 was fine mapped within 410.4 Kb between markers RM17207 and RM17227.
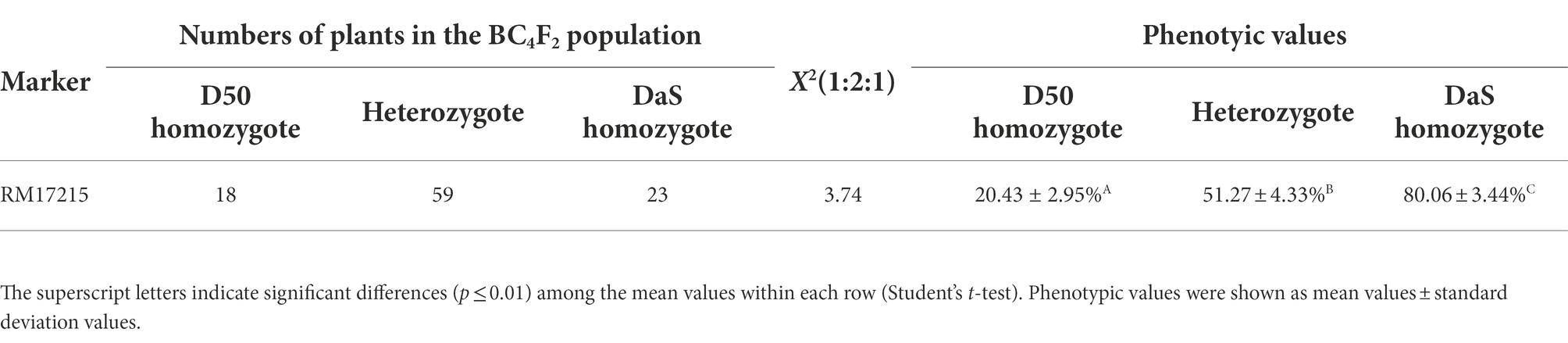
Among the random selection of 100 individuals in the BC4F2 population, the marker RM17157 was used to validate the effect of qSE4. The Chi-squared test revealed the phenotypic separation ratio was fitted to 1:2:1 (X2 = 3.74 < X20.05,2 = 5.99), suggesting that the effect of qSE4 is likely controlled by one genetic locus (Table 2).
Fine mapping of qSE4
Selfing of some BC3F1 individual plants to produce BC3F2, and one heterozygote recombinant from the BC3F1 population that carried the target QTL region from DaS was backcrossed with D50 to produce a larger BC4F2 population. A total of 2,143 individuals were used for fine mapping the qSE4 using three additional polymorphic SSR markers between DaS and D50. Five homozygous recombinant lines and one heterozygote line in the QTL region were analyzed for fine mapping (Figure 2B). The phenotypic performance of the SERs varied from 24.07 to 79.72% in the recombinant lines. The total SERs of A2, A3, A4, and A5 were similar to that of D50 and significantly lower than that of A1 which was similar to DaS. The total SER of heterozygous A6 was between that of the two parents (Figure 2B). Based on genetic and phenotypic analysis, the location of qSE4 was narrowed to a 410.4-kb region between the RM17207 and RM17227 (Figure 2C).
Candidate gene analysis of qSE4
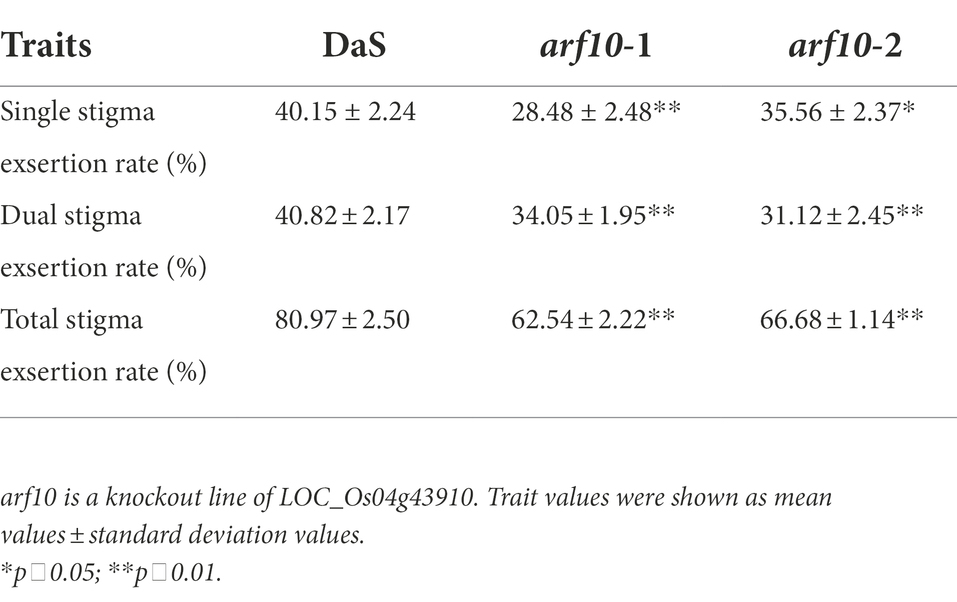
After removing unknown retrotransposons, transposons, and putative genes (Li et al., 2021) within the 410.4-kb region, 13 genes remained (Figure 4A) according to the Rice Genome Annotation Database (rice.plantbiology.msu.edu/, MSU-version_7.0). Sequencing analysis found that the two parents had one base difference in the promoter region of LOC_Os04g43910 (A for DaS and G for D50), but no difference in the coding region (Figure 4B). Gene expression analysis showed that the expression level of LOC_Os04g43910 in parental DaS was significantly higher than in D50. The expression level of lines carrying the DaS allele in progenies was also significantly higher than that carrying the D50 allele (Figure 4C). The differences of LOC_Os04g43910 among different cultivars were further analyzed, and it was found that the cultivars of haplotype G were mostly wide-grained cultivars with lower SERs. The haplotype A cultivars were mostly slender grains with higher SERs (Figure 4D). Therefore, LOC_Os04g43910 is predicted as the candidate gene. Further, LOC_Os04g43910 (ARF10) was knocked out in the DaS background using CRISPR/Cas9 technology, two mutants which named arf10-1 and arf10-2 were obtained. The arf10-1 and arf10-2 mutant plants contained 1-bp deletion and 1-bp insertion, respectively, in the exon of ARF10 (Figure 5A). These two mutations caused the premature appearance of stop codons in the ARF10 gene, resulting in a significant decrease in the expression of ARF10 (Figure 5B). The SERs were significantly reduced in both knockout lines compared to the wild type (Figures 5C–H; Table 3). These results further confirmed that ARF10 was most likely to be the causal gene for the qSE4 influencing the SER in rice.
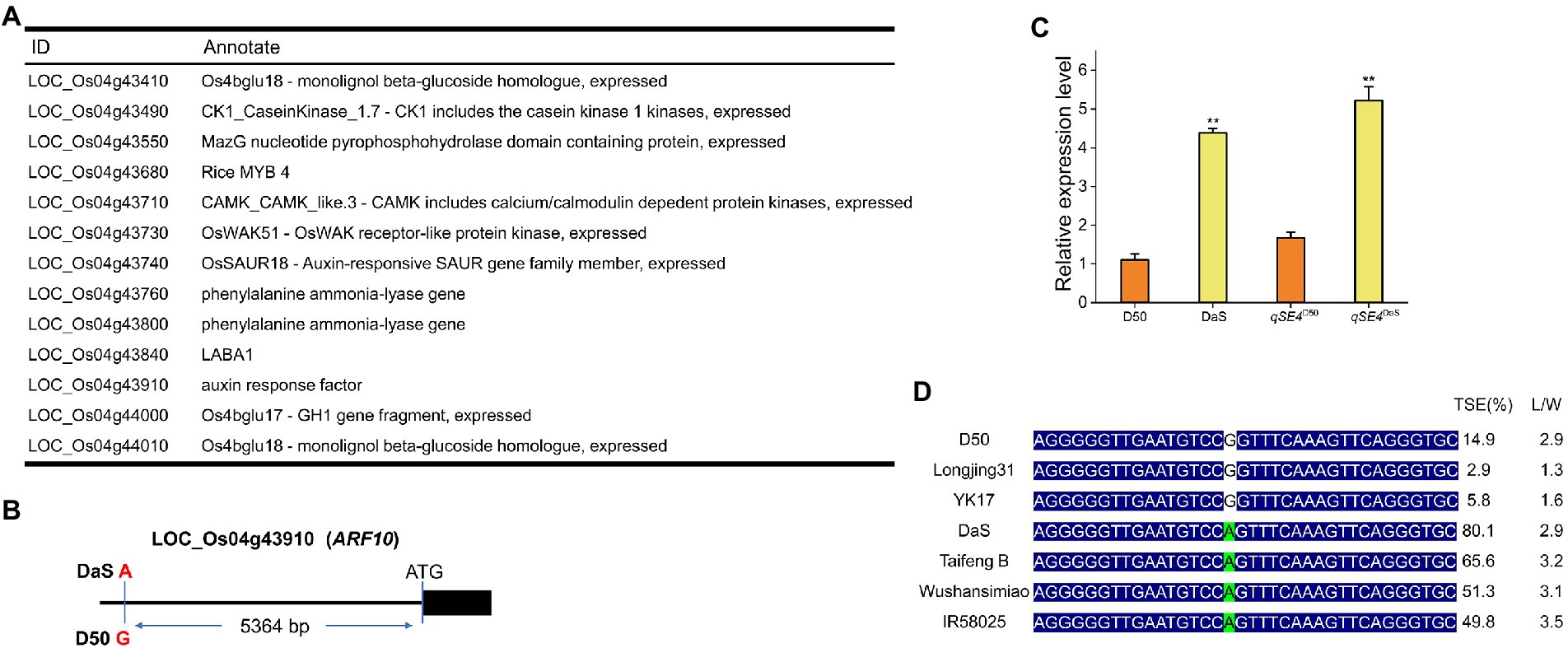
Figure 4. Identification of candidate genes for qSE4. (A) Thirteen candidate genes for qSE4 (http://rice.plantbiology.msu.edu/, MSU- version_7.0). (B) At 5364 bp upstream of the ATG initiation codon of the gene ARF10, there was one base different between the two parental DaS and D50, DaS was A, and D50 was G. (C) Relative expression of ARF10 in DaS, D50, RIL (qSE4D50) and RIL (qSE4DaS). (D) The relationship between the haplotype in the 5,364 base upstream of ATG and stigma exsertion phenotype in different rice varieties. TSE: total stigma exsertion rate; L/W: length/width. The data represent the mean values ± SD (n = 6), **p ≤ 0.01.
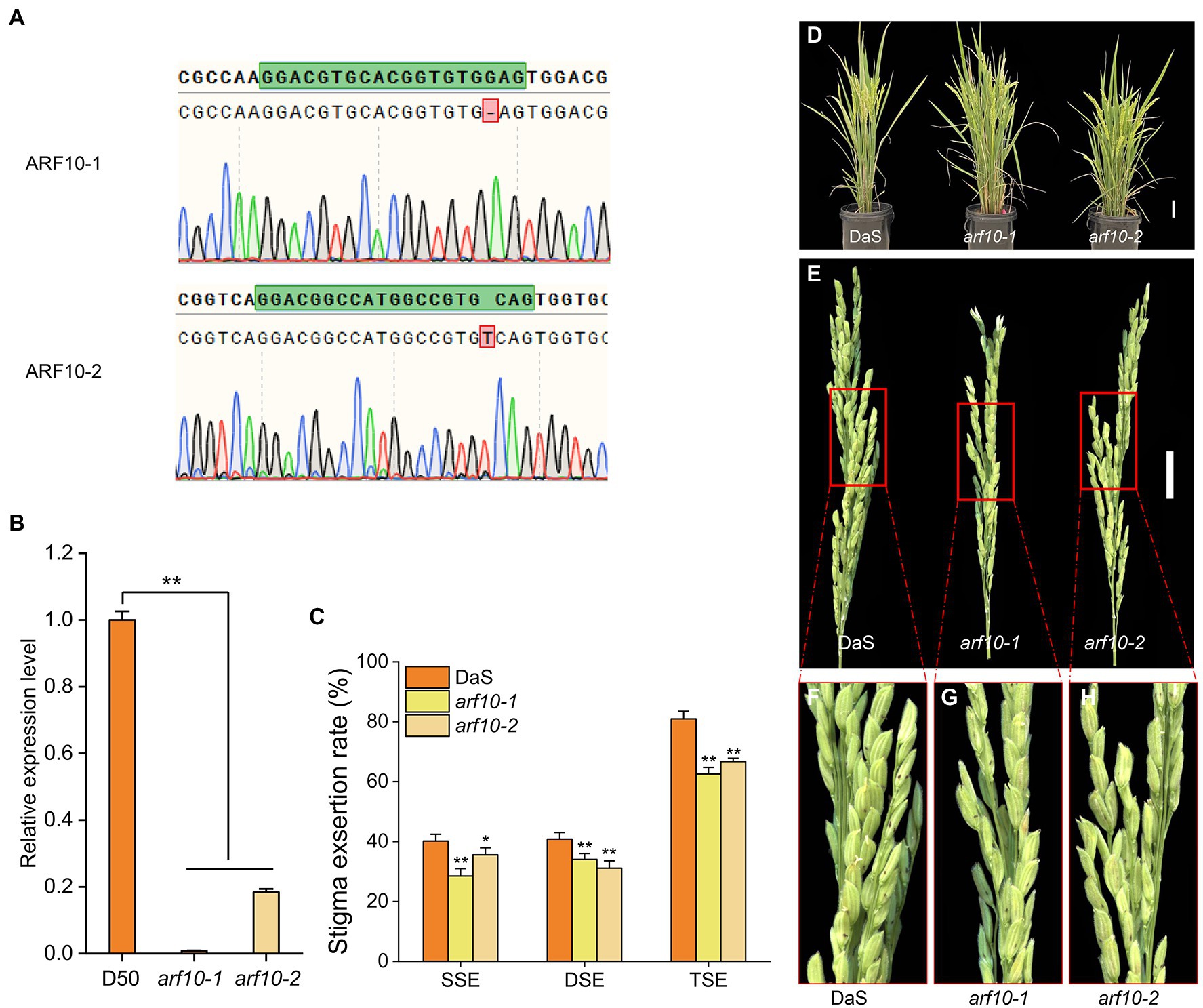
Figure 5. Stigma exsertion phenotype of knockout line arf10 and the wild type DaS. (A) ARF10 mutant sites of two knockout lines arf10. (B) Relative expression levels of ARF10 in two knockout lines and the wild type DaS (n = 3). (C) Plant stigma exsertion phenotypes of the two knockout lines and the wild type DaS at the heading stage. (D) Stigma exsertion phenotypes at heading in the two knockout lines and the wild DaS, bar = 10 cm. (E) Stigma exsertion rate of two knockout lines and the wild type DaS at heading stage, bar = 5 cm. The data represent the mean values ± SD (n = 6), *p ≤ 0.05, **p ≤ 0.01.
Transcriptome analysis of ARF10 knockout plants
RNA-sequencing (RNA-seq) was conducted to show the transcriptomic variation in the young panicle between the wild-type DaS and the mutant arf10 (Figure 6). A total of 10,414 differentially expressed genes (DEGs) were detected (padj <0.05), of which 53.7% (5,597 genes) were up-regulated and 46.3% (4,817 genes) were down-regulated in the arf10 plant compared with the expression levels in DaS plant (Figure 6A; Supplementary Figure 1). We randomly selected 8 genes to analyze their expression levels via qRT-PCR. The results were consistent with the results of the transcriptome (Supplementary Figure 2). GO analysis reveals DEGs enrichment in biological processes, cellular components, and molecular functions. Metabolic process, biochemical reaction, and biosynthetic process represent 41.32, 16.51, and 11.85% of total DEGs, respectively, and were the largest subcategories in the biological process. Ribosome, cell periphery, and external encapsulating structure represent 30.17, 21.55, and 14.08% of total DEGs, respectively, and were the largest subcategories in the cellular component. Enzyme activity, binding, and transmembrane transporter activity represent 43.67, 28.85, and 3.90% of total DEGs, respectively, and were the largest subcategories in the molecular function (Supplementary Figure 3). KEGG pathway analysis showed that ribosome, phenylpropanoid biosynthesis, and plant hormone signal transduction were the most significantly enriched pathways (padj <0.05) of DEGs (Figure 6D).
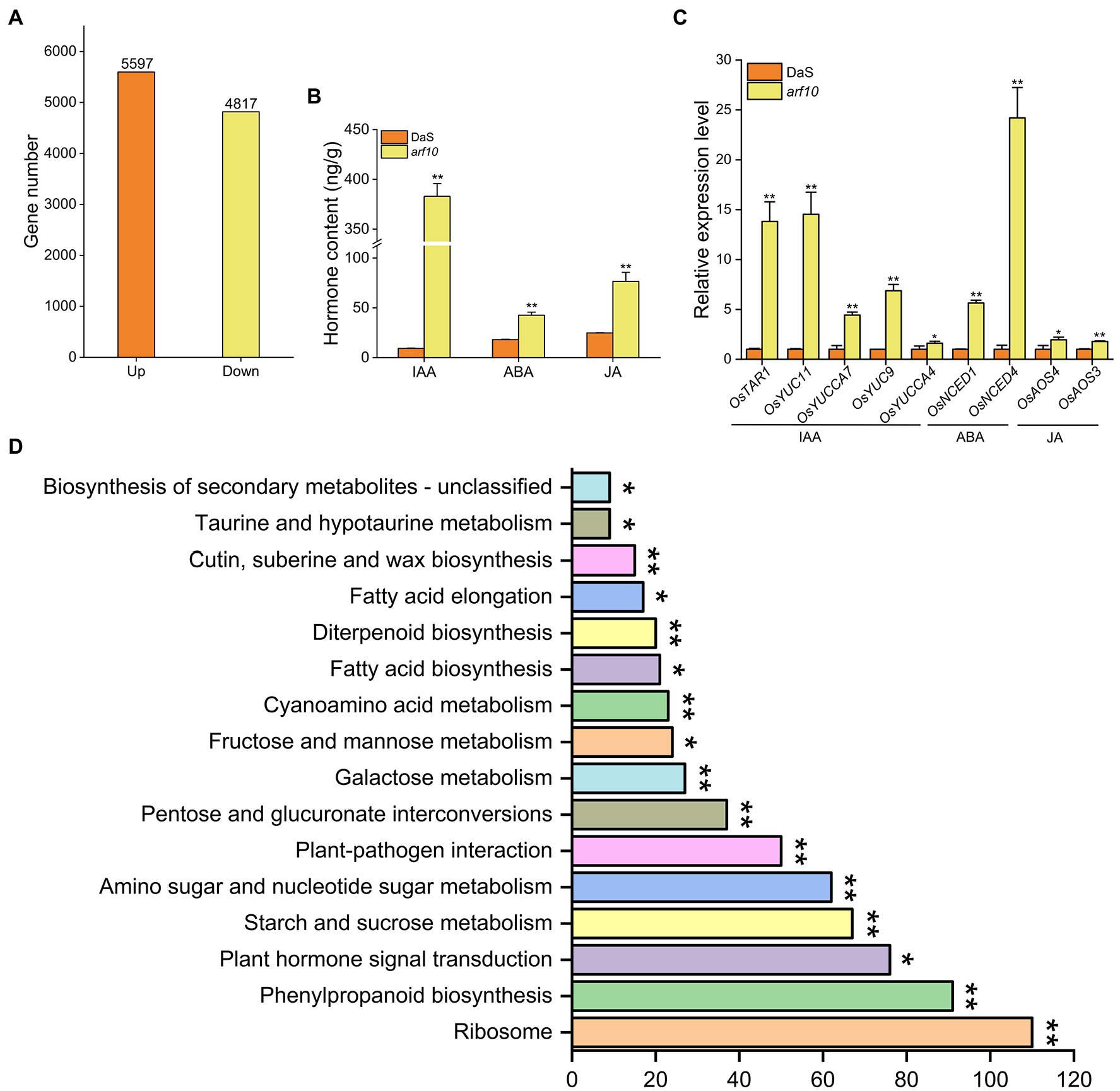
Figure 6. Transcriptome analysis of knockout lines arf10 and DaS. (A) Number of up- and down-regulated genes (padj <0.05) in young panicle at the pre-heading stage of arf10 compared with DaS. (B) KEGG pathway enrichment analysis of DEGs. (C) Contents of auxin, abscisic acid, and jasmonic acid in arf10 and DaS at the pre-heading stage. (D) Relative expression of auxin, abscisic acid, and jasmonic acid synthesis genes of arf10 and DaS at the pre-heading stage. The data represent the mean values ± SD (n = 3), *p ≤ 0.05 and **p ≤ 0.01.
Since ARF10 is an auxin-responsive factor, we detected the hormone content of DaS and arf10 and analyzed the expression of hormone synthesis and signaling-related genes. Compared with wild-type DaS, the content of IAA, ABA, and JA in the knockout line arf10 was significantly increased (Figure 6B). It was further found that the genes OsTAR1, OsYUC11, OsYUCCA7, OsYUC9, and OsYUCCA4 for the IAA synthesis pathway; OsNCED1 and OsNCED4 for the ABA synthesis pathway; OsAOS3 and OsAOS4 for the JA synthesis pathway were significantly higher in arf10 than in DaS (Figure 6C). In addition, the genes of IAA, ABA, and JA signaling pathways were also expressed significantly differently in arf10 and DaS (Supplementary Figure 4). These results suggested that hormones are key factors affecting the SER.
Discussion
The application of hybrid rice has greatly improved rice yields, but the low efficiency of hybrid seed production limits its promotion in Southeast Asia (Xie, 2008). The increase of stigma exsertion can increase hybrid seed production and promote the commercialization of hybrid rice (Zhou et al., 2017). In the past two decades, dozens of QTLs of the SER have been mapped and found to be distributed on 12 chromosomes in rice. However, only two were located on chromosome 4. One is qSER4, a QTL that reduces the SER identified from wild rice ‘W0120’, which is close to marker C5-indel3632 (physical location Chr4:5050622; Bakti and Tanaka, 2019). The other is PES4, which was identified from the upland rice maintainer Huhan1B, which was located between the markers RM6909 and RM1113 on chromosome 4. In this study, we used DaS, a japonica two-line sterile line with a high SER, and D50, a tropical japonica rice with a low SER, to construct a backcross inbred mapping population and mapped the major QTL: qSE4, which controls the SER, between the markers RM17207 and RM17227 on Chromosome 4 (Figure 2C). qSE4 is far from qSER4, but within the same interval of RM6909 and RM1113 as PES4. Thus, qSE4 is a QTL that controls SERs and can be detected reproducibly across different germplasm resources.
Because it is beneficial to the application of heterosis, stigma exsertion is a research hotspot for many crops. So far, only a few genes related to stigma exsertion have been cloned from limited crops. In tomato, the transcription factor Style2.1 can regulate cell elongation, and the difference in the promoter region of Style2.1 among different varieties leads to different levels of gene expression, determining whether the tomato stigma is exposed or inserted (Chen et al., 2007). This is different from the GS3 gene in rice, where mutations in GS3 promote an increase in the number of cells, leading to elongation of the stigma (Takano-Kai et al., 2011). The zinc-finger transcription factor, SE3.1 in tomatoes, shows another newly discovered regulatory mechanism. SE3.1 controls stigma exsertion, which is not related to its expression level but is related to its gene function. The loss of SE3.1 function promotes tomato stigma retraction, while Style2.1 and SE3.1 are key genes that regulate the two-step process of stigma from exsertion to insert during tomato domestication (Shang et al., 2021). Recently, there was a major breakthrough in the study of stigma exsertion in legumes. It was found that SGE1 controls the stigma exsertion of Medicago truncatula by regulating the secretion of keratin and wax of flowers (Zhu et al., 2020). In this study, qSE4 was fine-mapped within 410.4 kb, and 13 candidate genes were identified via bioinformatics analysis (Figure 4A). qRT-PCR and sequencing analysis showed that a nucleotide change at 5364 bp of the promoter of LOC_Os04g43910 (ARF10) could affect the expression of this gene, thereby affecting the stigma exsertion rate (Figures 4B,C). Haplotype analysis further indicated that 5364G was associated with low stigma exsertion and 5364A was associated with high stigma exsertion (Figure 4D). The way ARF10 regulates stigma exsertion is similar to that of Style2.1, and the stigma exsertion is determined by the change in expression level, so ARF10 is likely to be a candidate gene of qSE4. We further validated its function by knocking out ARF10 in the DaS background. The expressions of ARF10 in the two knockout lines was significantly lower than of the wild type (Figure 5B), and the SER was also significantly lower than that of the wild type (Figure 5C). These results point to ARF10 as a target gene of qSE4.
Auxin response factors (ARFs) are a class of transcription factors that regulate the response to plant auxins (Wang et al., 2007). Twenty-five ARFs family member genes were found in rice, which plays important functions in rice disease resistance (Qin et al., 2020; Zhang et al., 2020; Zhao et al., 2020), root growth (Qi et al., 2012; Shen et al., 2013; Shao et al., 2019), and plant morphology (Attia et al., 2009; Sakamoto et al., 2013; Zhang et al., 2015; Huang et al., 2016, 2021; Chen et al., 2018; Liu et al., 2018). ARF25 may regulate the expression of the auxin synthesis gene OsYUCCAs to affect the auxin content of rice roots (Qi et al., 2012), while ARF16 may regulate auxin redistribution (Shen et al., 2015). Compared with wild DaS, the auxin content and five auxin synthesis genes in arf10 increased significantly (Figures 6B,C), suggesting that ARF10 may also regulate the expression of auxin synthesis genes to affect the auxin content in rice young panicles. Auxin has a dual effect on plant growth, low concentrations promote growth and high concentrations inhibits growth, therefore, the high concentration of auxin in arf10 may be the reason for its reduced SER. Consistent with this notion, low concentrations of IAA were found in tobacco to promote stigma growth while high concentrations inhibited stigma growth (Chen and Zhao, 2008).
Various biochemical processes in plants are regulated by hormone crosstalk. Auxin also performs some functions in conjunction with other hormones. Auxin and abscisic acid jointly control seed dormancy in Arabidopsis thaliana (Liu et al., 2013), and together with brassinolide regulate plant height and leaf angle (Liu et al., 2018). The contents of ABA and JA in arf10 were significantly higher than those of the wild type (Figure 6B), and their synthetic genes also increased significantly (Figure 6C), suggesting that ARF10 may be involved in the synthesis of ABA and JA. KEGG analysis showed that the differential genes were enriched in the hormone signaling pathway (Figure 6D; Supplementary Figure 4). These results all indicated that IAA, ABA, and JA may jointly regulate the degree of stigma exsertion in rice.
The development of NILs is an effective strategy for studying QTLs as the interference of the noise from the background is significantly reduced (Ding et al., 2011). In our study, NIL of the qSE4 carrying DaS allele in the D50 background significantly increased the stigma exsertion rate compared with the recurrent parent, D50 (Figure 3E), suggesting that qSE4 is responsible for the increased SER in NIL. This not only provides the basis for the subsequent cloning of the qSE4 target gene but also provides molecular markers linked to the qSE4 target gene. Molecular markers can be applied to gene pyramid breeding in rice with a high SER. Cultivating rice male sterile lines with a high SER can increase the yield of hybrid rice seed production, reduce the production cost of hybrid rice, and promote hybrid rice development with health and sustainability.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA850518.
Author contributions
HP and SZ designed experiments. GN, WY, CW, TS, AR, WX, HS, TS, SG, JG, XL and WL performed experiments. GN analyzed data and compiled figures. GN wrote the manuscript. SZ edited the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the China Natural Science Foundation (no. 31871597), Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (2021C02063-2), and the Key Research and Development Program of China National Rice Research Institute (CNRRI-2020-02).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank AiMi Academic Services (www.aimieditor.com) for the English language editing and review services.
Supplementary materials
The Supplementary materials for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.959859/full#supplementary-material
References
Attia, K., Abdelkhalik, A., Ammar, M., Wei, C., Yang, J., Lightfoot, D., et al. (2009). Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Curr. Issues Mol. Biol. 11, i29–i34. doi: 10.21775/9781912530069.04
Bakti, C., and Tanaka, J. (2019). Detection of dominant QTLs for stigma exsertion ratio in rice derived from Oryza rufipogon accession 'W0120'. Breed. Sci. 69, 143–150. doi: 10.1270/jsbbs.18139
Bi, X. Y., and Tan, S. Z. (1988). Receptivity of exserted stigmas [China]. Int. Rice Res. Newsletter 13, 7–8.
Chen, K., Cong, B., Wing, R., Vrebalov, J., and Tanksley, S. (2007). Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science 318, 643–645. doi: 10.1126/science.1148428
Chen, D., and Zhao, J. (2008). Free IAA in stigmas and styles during pollen germination and pollen tube growth of Nicotiana tabacum. Physiol. Plant. 134, 202–215. doi: 10.1111/j.1399-3054.2008.01125.x
Chen, S. H., Zhou, L. J., Xu, P., and Xue, H. W. (2018). SPOC domain-containing protein leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PLoS Genet. 14:e1007829. doi: 10.1371/journal.pgen.1007829
Ding, X., Li, X., and Xiong, L. (2011). Evaluation of near-isogenic lines for drought resistance QTL and fine mapping of a locus affecting flag leaf width, spikelet number, and root volume in rice. Theor. Appl. Genet. 123, 815–826. doi: 10.1007/s00122-011-1629-1
Huang, G., Hu, H., van de Meene, A., Zhang, J., Dong, L., Zheng, S., et al. (2021). AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell 33, 3120–3133. doi: 10.1093/plcell/koab175
Huang, J., Li, Z., and Zhao, D. (2016). Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of Auxin signaling in Rice. Sci. Rep. 6:29938. doi: 10.1038/srep29938
Kato, H., and Namai, H. (1987). Floral characteristcs and environmental factors for increasing natural outcrossing rate for F1 hybrid seed production of rice Oryza sativa L. Breed. Sci. 37, 318–330. doi: 10.1270/jsbbs1951.37.318
Khush, G. S. (2005). What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 59, 1–6. doi: 10.1007/s11103-005-2159-5
Li, W., Sheng, Z., Zhu, Z., Wei, X., Shi, L., Wu, Y., et al. (2017). QTL mapping of japonica Rice stigma Exsertion rate. Chin. J. Rice Sci. 31, 23–30. doi: 10.16819/j.1001-7216.2017.6043
Li, W., Yang, B., Xu, J., Peng, L., Sun, S., Huang, Z., et al. (2021). A genome-wide association study reveals that the 2-oxoglutarate/malate translocator mediates seed vigor in rice. Plant J. 108, 478–491. doi: 10.1111/tpj.15455
Liu, Q., Qin, J., Li, T., Liu, E., Fan, D., Edzesi, W. M., et al. (2015). Fine mapping and candidate gene analysis of qSTL3, a stigma length-conditioning locus in Rice (Oryza sativa L.). PLoS One 10:e0127938. doi: 10.1371/journal.pone.0127938
Liu, X., Yang, C. Y., Miao, R., Zhou, C. L., Cao, P. H., Lan, J., et al. (2018). DS1/OsEMF1 interacts with OsARF11 to control rice architecture by regulation of brassinosteroid signaling. Rice 11:46. doi: 10.1186/s12284-018-0239-9
Liu, Y., Zhang, A., Wang, F., Kong, D., Li, M., Bi, J., et al. (2019). Fine mapping a quantitative trait locus, qSER-7, that controls stigma exsertion rate in rice (Oryza sativa L.). Rice 12:46. doi: 10.1186/s12284-019-0304-z
Liu, X., Zhang, H., Zhao, Y., Feng, Z., Li, Q., Yang, H. Q., et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 110, 15485–15490. doi: 10.1073/pnas.1304651110
Marathi, B., and Jena, K. K. (2015). Floral traits to enhance outcrossing for higher hybrid seed production in rice: present status and future prospects. Euphytica 201, 1–14. doi: 10.1007/s10681-014-1251-9
Marathi, B., Ramos, J., Hechanova, S. L., Oane, R. H., and Jena, K. K. (2015). SNP genotyping and characterization of pistil traits revealing a distinct phylogenetic relationship among the species of Oryza. Euphytica 201, 131–148. doi: 10.1007/s10681-014-1213-2
Miyata, M., Yamamoto, T., Komori, T., and Nitta, N. (2007). Marker-assisted selection and evaluation of the QTL for stigma exsertion under japonica rice genetic background. Theor. Appl. Genet. 114, 539–548. doi: 10.1007/s00122-006-0454-4
Orjuela, J., Garavito, A., Bouniol, M., Arbelaez, J., Moreno, L., Kimball, J., et al. (2010). A universal core genetic map for rice. Theor. Appl. Genet. 120, 563–572. doi: 10.1007/s00122-009-1176-1
Qi, Y., Wang, S., Shen, C., Zhang, S., Chen, Y., Xu, Y., et al. (2012). OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 193, 109–120. doi: 10.1111/j.1469-8137.2011.03910.x
Qin, Q., Li, G., Jin, L., Huang, Y., Wang, Y., Wei, C., et al. (2020). Auxin response factors (ARFs) differentially regulate rice antiviral immune response against rice dwarf virus. PLoS Pathog. 16:e1009118. doi: 10.1371/journal.ppat.1009118
Rahman, M. H., Zhang, Y., Zhang, K., Rahman, M. S., Barman, H. N., Riaz, A., et al. (2017). Genetic dissection of the major quantitative trait locus (qSE11), and its validation As the major influence on the rate of stigma Exsertion in Rice (Oryza sativa L.). front. Plant Sci. 8:1818. doi: 10.3389/fpls.2017.01818
Sakamoto, T., Morinaka, Y., Inukai, Y., Kitano, H., and Fujioka, S. (2013). Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 73, 676–688. doi: 10.1111/tpj.12071
Shang, L., Song, J., Yu, H., Wang, X., Yu, C., Wang, Y., et al. (2021). A mutation in a C2H2-type zinc finger transcription factor contributed to the transition toward self-pollination in cultivated tomato. Plant Cell 33, 3293–3308. doi: 10.1093/plcell/koab201
Shao, Y., Zhou, H. Z., Wu, Y., Zhang, H., Lin, J., Jiang, X., et al. (2019). OsSPL3, an SBP-domain protein, regulates crown root development in Rice. Plant Cell 31, 1257–1275. doi: 10.1105/tpc.19.00038
Shen, C., Wang, S., Zhang, S., Xu, Y., Qian, Q., Qi, Y., et al. (2013). OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 36, 607–620. doi: 10.1111/pce.12001
Shen, C., Yue, R., Sun, T., Zhang, L., Yang, Y., and Wang, H. (2015). OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.). Plant Sci. 231, 148–158. doi: 10.1016/j.plantsci.2014.12.003
Takano-Kai, N., Doi, K., and Yoshimura, A. (2011). GS3 participates in stigma exsertion as well as seed length in rice. Breed. Sci. 61, 244–250. doi: 10.1270/jsbbs.61.244
Tan, Q., Wang, C., Luan, X., Zheng, L., Ni, Y., Yang, W., et al. (2021). Dissection of closely linked QTLs controlling stigma exsertion rate in rice by substitution mapping. Theor. Appl. Genet. 134, 1253–1262. doi: 10.1007/s00122-021-03771-9
Tan, Q., Zou, T., Zheng, M., Ni, Y., Luan, X., Li, X., et al. (2020). Substitution mapping of the major quantitative trait loci controlling stigma Exsertion rate from Oryza glumaepatula. Rice 13:37. doi: 10.1186/s12284-020-00397-1
Tian, D., Huang, S., Duan, Y., and Wang, Y. (2004). The relationship between flowering and pollination time and outcrossing rate of male sterile lines in hybrid rice seed production. Hybrid Rice 19, 50–54. doi: 10.16267/j.cnki
Uga, Y., Fukuta, Y., Cai, H. W., Iwata, H., Ohsawa, R., Morishima, H., et al. (2003). Mapping QTLs influencing rice floral morphology using recombinant inbred lines derived from a cross between Oryza sativa L. and Oryza rufipogon Griff. Theor. Appl. Genet. 107, 218–226. doi: 10.1007/s00122-003-1227-y
Virmani, S. S. (1994). Heterosis and Hybrid Rice Breeding, Germany: Springer Science & Business Media.
Virmani, S. S., and Athwal, D. S. (1973). Genetic variability in floral characteristics influencing outcrossing in Oryza sativa L. Crop Sci. 13, 66–67. doi: 10.2135/cropsci1973.0011183X001300010019x
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, F., Yang, T., Burlyaeva, M., Li, L., Jiang, J., Fang, L., et al. (2015). Genetic diversity of grasspea and its relative species revealed by SSR markers. PLoS One 10:e0118542. doi: 10.1371/journal.pone.0118542
Xie, F. (2008). “Priorities of IRRI hybrid rice breeding,” in International Symposium on Hybrid Rice; September 11, 2008.
Yan, W. G., and Li, S. F. (1987). Study on out-crossing characteristics among male sterile lines containing same nucleus in rice. Hybrid Rice 4, 8–11.
Yan, W. G., Li, Y., Agrama, H. A., Luo, D., Gao, F., Lu, X., et al. (2009). Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol. Breed. 24, 277–292. doi: 10.1007/s11032-009-9290-y
Zhang, H., Li, L., He, Y., Qin, Q., Chen, C., Wei, Z., et al. (2020). Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc. Natl. Acad. Sci. U. S. A. 117, 9112–9121. doi: 10.1073/pnas.1918254117
Zhang, S., Wang, S., Xu, Y., Yu, C., Shen, C., Qian, Q., et al. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38, 638–654. doi: 10.1111/pce.12397
Zhang, K., Zhang, Y., Wu, W., Zhan, X., Anis, G. B., Rahman, M. H., et al. (2018). qSE7 is a major quantitative trait locus (QTL) influencing stigma exsertion rate in rice (Oryza sativa L.). Sci. Rep. 8:14523. doi: 10.1038/s41598-018-32629-2
Zhao, Z. X., Feng, Q., Cao, X. L., Zhu, Y., Wang, H., Chandran, V., et al. (2020). Osa-miR167d facilitates infection of Magnaporthe oryzae in rice. J. Integr. Plant Biol. 62, 702–715. doi: 10.1111/jipb.12816
Zhou, H., Li, P., Xie, W., Hussain, S., Li, Y., Xia, D., et al. (2017). Genome-wide association analyses reveal the genetic basis of stigma Exsertion in Rice. Mol. Plant 10, 634–644. doi: 10.1016/j.molp.2017.01.001
Keywords: stigma exsertion rate, QTL, near-isogenic line, hormone, rice
Citation: Guo N, Wang Y, Chen W, Tang S, An R, Wei X, Hu S, Tang S, Shao G, Jiao G, Xie L, Wang L, Sheng Z and Hu P (2022) Fine mapping and target gene identification of qSE4, a QTL for stigma exsertion rate in rice (Oryza sativa L.). Front. Plant Sci. 13:959859. doi: 10.3389/fpls.2022.959859
Edited by:
Hanwei Mei, Shanghai Agrobiological Gene Center, ChinaReviewed by:
Zhi Gang Zhao, Nanjing Agricultural University, ChinaYunlong Pang, Shandong Agricultural University, China
Copyright © 2022 Guo, Wang, Chen, Tang, An, Wei, Hu, Tang, Shao, Jiao, Xie, Wang, Sheng and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peisong Hu, cGVpc29uZ2h1QDEyNi5jb20=; Zhonghua Sheng, c2hlbmd6aG9uZ2h1YTY2NkAxNjMuY29t
†These authors have contributed equally to this work
 Naihui Guo
Naihui Guo Yakun Wang1†
Yakun Wang1† Xiangjin Wei
Xiangjin Wei Gaoneng Shao
Gaoneng Shao Guiai Jiao
Guiai Jiao Peisong Hu
Peisong Hu