- Fungal Genomics Laboratory, Department of Entomology and Plant Pathology, Center for Integrated Fungal Research, North Carolina State University, Raleigh, NC, United States
Rice blast disease caused by the hemi-biotrophic fungus Magnaporthe oryzae is the most destructive disease of rice world-wide. Traditional disease resistance strategies for the control of rice blast disease have not proved durable. HIGS (host induced gene silencing) is being developed as an alternative strategy. Six genes (CRZ1, PMC1, MAGB, LHS1, CYP51A, CYP51B) that play important roles in pathogenicity and development of M. oryzae were chosen for HIGS. HIGS vectors were transformed into rice calli through Agrobacterium-mediated transformation and T0, T1 and T2 generations of transgenic rice plants were generated. Except for PMC1 and LHS1, HIGS transgenic rice plants challenged with M. oryzae showed significantly reduced disease compared with non-silenced control plants. Following infection with M. oryzae of HIGS transgenic plants, expression levels of target genes were reduced as demonstrated by Quantitative RT-PCR. In addition, treating M. oryzae with small RNA derived from the target genes inhibited fungal growth. These findings suggest RNA silencing signals can be transferred from host to an invasive fungus and that HIGS has potential to generate resistant rice against M. oryzae.
Introduction
With finite agricultural resources and an increasing human population (Ramankutty et al., 2018), new approaches are in high demand to support sustainable food production. Rice (Oryza sativa L.), which is the staple food for over half of the world’s population and provides more than 1/5 of the calories consumed by humans, is crucial for global food security (Seck et al., 2012). Worldwide production of milled rice was forecasted at a record of 510.7 million tons in 2021/22 (Childs and LeBeau, 2021). However, diseases severely threaten rice production. Rice blast disease, caused by the hemi-biotrophic fungus Magnaporthe oryzae (teleomorph of Pyricularia oryzae), leads to annual yield losses worth billions of dollars, enough to feed 60 million people (Dean et al., 2005; Talbot and Wilson, 2009). In addition to rice, the fungus also infects approximately fifty grass and sedge species (Ou, 1985). Eighty-five countries throughout the world have reported the presence of this disease, and it is likely to present practically everywhere that rice is grown commercially (Ou, 1985). More effective control of blast disease would constitute a major contribution to ensuring global food security.
Rice blast disease is initiated by a three-celled tear shaped conidium which germinates once attached to the plant surface. A melanized appressorium at the tip of the germ tube forms a penetration peg, which pushes through the plant cuticle and cell wall. Primary hyphae differentiate as bulbous hyphae in the epidermal cells before emerging as invasive hyphae, spreading and causing lesions on aerial tissues. Within 5–7 days lesions discharge numerous conidia enabling new infections (Hamer et al., 1988; Dean et al., 2005; Talbot and Wilson, 2009). The pathogen can infect and cause lesions on all above-ground plant tissues such as leaves, nodes, culms, panicles, peduncles and grains. Infection can occur at the seedling stage, the entire vegetative phase as well as the reproductive phase. To manage this disease, crop rotation, proper fertilization in addition to irrigation are highly recommended. Chemical fungicides are also used, but none are considered to be highly successful. Genetic resistance is the most effective way for rice blast disease control, however, resistance strategies employed to date are generally not durable (Ou, 1980; Zeigler et al., 1994; Dean et al., 2012).
Plants, unlike animals, do not possess circulating immune cells, but are equipped with two interconnected tiers of receptors for the recognition of pathogens and subsequent defense responses. The first tier governed by pattern recognition receptors (PRRs) is triggered by pathogen-associated molecular patterns (PAMPs) and is known as PAMP-triggered immunity (PTI). Successful pathogens further secrete effectors to subvert PTI and facilitate infection. In response, host plants have evolved resistance (R) genes that recognize pathogen effectors and induce the second tier of defense: effector-triggered immunity (ETI) (Jones and Dangl, 2006; Dangl et al., 2013). Breeding rice varieties carrying R genes are used for blast disease control. Over 100 major blast R genes have been identified and more than 30 of them have been cloned (Wang et al., 2017). Major blast R genes such as Pita, Ptr, Pi-z, Pi-b and Pikh, Pikm, Piks have been deployed in US rice varieties since 1960 (Jia et al., 2019). Although incorporation of R genes into rice has proved to be an effective and environmentally friendly strategy for blast resistance breeding (Singh et al., 2012; Wang and Valent, 2017), many years are required to develop new cultivars. Moreover, the pathogen is capable of rapid genetic changes through mechanisms such as active transposable elements, random mutation and recombination to “break-down” R gene resistance within 2 or 3 years after planting (Wang and Valent, 2017). Furthermore, many R genes may also be associated with yield loss or nutrition defects and consequently may not be commercially popular (Brown, 2002; Burdon and Thrall, 2003). Therefore, innovative strategies are needed for durable and effective resistance. The concept of parasite-derived resistance (PDR) which confers host resistance by expressing parasite genes in the host plant was proposed decades ago as a radical approach but only since the discovery of small RNA silencing mechanism is this now a possibility (Sanford and Johnston, 1985).
With several notable exceptions such as budding yeast, RNA interference (RNAi) and its role in chromatin regulation are largely conserved throughout all eukaryotes (Shabalina and Koonin, 2008; Drinnenberg et al., 2009). Small non-coding RNA can induce silencing of target genes at the transcriptional and post-transcriptional levels (Ghildiyal and Zamore, 2009). It is usually initiated by processing double stranded RNA (dsRNA) into short RNA duplexes through the action of “Dicer” (RNase III), followed by the cleavage, and degradation or translational repression of targeted mRNA through the RNA-induced Silencing Complex (RISC) (Ghildiyal and Zamore, 2009). RNAi mechanisms have been widely exploited and used as a powerful genetic tool to silence genes of interest in target organisms (Tang and Galili, 2004; Tan and Yin, 2005; Saksmerprome et al., 2009; Das and Sherif, 2020).
Conceptually, host-induced gene silencing (HIGS) is performed by expressing an RNAi construct in a host plant resulting in production of an RNA silencing signal that will be absorbed by the invading pathogen and silence the target pathogen gene. This concept was successful for control of root-knot nematodes and of lepidopteran and coleopteran insects feeding on transgenic plants carrying RNAi constructs targeting corresponding pest genes (Huang et al., 2006; Baum et al., 2007; Mao et al., 2007). HIGS has been demonstrated in several different fungal patho-systems. HIGS was first reported by Nowara and coworkers in 2010, where HIGS in barley (Hordeum vulgare) was evaluated against the obligate pathogen powdery mildew fungus Blumeria graminis. Host-induced gene silencing of the effector gene Avra10 resulted in reduced fungal development in the host plant (Nowara et al., 2010). In addition to being effective against obligate biotrophic and hemi-biotrophic fungi and Oomycetes, where intimate contact is established with living plant cells, HIGS is also reported to be effective against necrotrophic fungal pathogens such as Fusarium oxysporum and Rhizoctonia solani (Govindarajulu et al., 2015; Hu et al., 2015; Sanju et al., 2015; Zhou et al., 2016). HIGS has also been shown to reduce the synthesis of toxic secondary metabolites, demonstrating its potential for precise engineering for food security (Thakare et al., 2017). Resistance conferred by HIGS may be more durable due to the vital function of genes that can be targeted such as genes encoding chitin or cellulose synthase, cytochrome P450 lanosterol C14a-demethylase in fungi (Koch et al., 2013; Cheng et al., 2015; Jahan et al., 2015; Andrade et al., 2016).
Though HIGS has been shown to be successful in a number of patho-systems (Nowara et al., 2010; Panwar et al., 2013; Hu et al., 2015; Zhou et al., 2016; Qi et al., 2018; Song and Thomma, 2018; Xu et al., 2018; Dou et al., 2020; Raruang et al., 2020), its applicability for control of rice blast disease has not been extensively explored (Guo et al., 2019; Li et al., 2020). This study was conducted to assess the effectiveness of targeting 6 key genes regulating growth and pathogenicity using HIGS for rice blast disease control. To accelerate these studies, we employed a vector that enabled visual detection of transgenic material. As small silencing RNAs have been illustrated to transfer bi-directionally between plants and fungi (Weiberg et al., 2013; Wang et al., 2016; Cai et al., 2018), the mechanism(s) of how they achieve cross-kingdom transportation is still unclear (Wang and Dean, 2020). In this study, we also investigated the hypothesis that RNA silencing signals may be absorbed by M. oryzae.
Materials and methods
Vector construction
Six genes of M. oryzae were selected as targets for HIGS (Table 1). Gene annotation and sequence information were obtained from the database ‘‘EnsemblFungi’’1. Invitrogen online RNAi designer tools were used to choose the optimal silencing fragment for target gene (Invitrogen2). Selected fragments were checked using the BLAST tool of the rice database ‘‘Rice genome annotation project’’ to confirm the absence of identical sequence (maximum length of identity less than 19nt) in the rice genome to ensure target fragments were specific to M. oryzae3. Threshold of expected value (E-value) is 10.
Primers were designed to generate PCR amplicons around 200- to 500- base pair (bp) length corresponding to exons of selected genes. The primer sequences used for the preparation of HIGS constructs are listed in Supplementary table 1. Flanking sequence and sequence of attB recombination sites were added to 5’end of each primer.
Magnaporthe oryzae strain 70-15 was cultured in liquid complete media (1L contains 10 g sucrose, 6 g casamino acid, 6 g yeast extract and 1 ml Aspergillus nidulans trace elements liquid) for 3 days and then mycelia were collected for RNA extraction. RNA was isolated using the RNeasy plant mini kit (Qiagen, Hilden, Germany) following the manual instruction. Total RNA was then pretreated with RNase-free DNase I (NEB, Ipswich, MA, United States) and subjected to reverse transcription using the Reverse Transcription System (Promega, Madison, WI, United States) based on manual instruction.
PCR reaction was performed in a total volume of 25 μL using standard Taq polymerase from NEB company to obtain target gene fragments. PCR parameters (primer and template annealing time, annealing temperature, final extension time) were optimized for each candidate gene. PCR mix with total volume of 25 μL contained 50 ng of M. oryzae cDNA, 0.4 μM of forward primer and 0.4 μM of reverse primer, 200 μM of dNTPs, 2.5 μL of 10X buffer, Nuclease free water and 0.625 units of Taq DNA polymerase. The PCR amplification conditions were as follows: 95°C for 5 min; 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds and 72°C for 45 seconds; 72°C for 10 min and hold at 4°C (annealing temperature ranged from 53–58°C). For genes combination CYP51A and CYP51B, two amplicons were linked into one by double joint PCR. PCR products were subsequently confirmed by size-fractionation on 1.5% agarose gels and were purified by kit, eluted with 30 μl of water (QIAquick Gel Extraction Kit, QIAGEN).
Vector pDONOR221 was used as ENTRY vector for the “Gateway cloning system”. After purification of the PCR products, target fragments of each M. oryzae gene were inserted to the entry vector pDONR221 (Invitrogen) through BP recombination cloning individually. Then the pDONR221 carrying two recombination sites for “LR Clonase reaction” was transformed to Escherichia coli DB3.1 competent cell. Plasmids purified by a minipreparation kit (QIAprep Spin Miniprep Kit, QIAGEN) were further confirmed by PCR amplifying corresponding target regions.
For HIGS expression, pBDL03 (gift from Dr. Shaohong Qu) was used as the destination vector (Zhao et al., 2021). This vector is derived from the pANDA vector (Okano et al., 2008) and designed so that each target fragment is inserted into two regions flanked by recombination sites in opposite directions, resulting in inverted repeat sequences linked by Gus gene (Fragments were separated by Gus linker, Gus linker sequence can be found on website4), controlled by maize (Zea mays) ubiquitin promoter and T3A terminator (Miki and Shimamoto, 2004; Okano et al., 2008). The vector contains the hygromycin resistance gene under control of “Cauliflower Mosaic Virus (CaMV) 35S promoter” and 35S terminator for selection in rice (Zhao et al., 2021). In addition, the vector contains mCherry gene (Subach et al., 2009) to facilitate visual confirmation of transgenic plants (Zhao et al., 2021). This gene was under control of a CaMV 35S promoter and ‘‘Agrobacterium Nopaline Synthase terminator’’ (tNOS). The target sequences were inserted in the destination vector through ‘‘LR recombination’’ using ‘‘Gateway LR clonase II enzyme Mix’’ (Invitrogen/life technologies). Manufacture’s guide was followed while the incubation time for recombination was extended to overnight. Plasmids obtained from the LR reaction were transformed to E. coli DB3.1 competent cells. Positive plasmids purified by minipreparation (QIAprep Spin Miniprep Kit, QIAGEN) were confirmed by amplifying corresponding target fragments as well as size fraction check on agarose gel after restriction enzyme (KpnI and SacI from NEB company) digestion. DNA sequencing was applied for final confirmation and MEGA45 software was used for sequence processing and alignment.
Agrobacterium tumefaciens EHA101 was cultured in liquid and solid YEP media (1L contains 10 g yeast extract, 10 g peptone, 5 g NaCl, 15 g agar for solid media). Positive plasmids were transformed into agrobacteria by electroporation.
Rice transformation
Rice (Oryzae sativa ssp japonica cv Nipponbare) was used in this study. Rice callus was induced from the embryos of mature seeds and transformation was conducted using the Agrobacterium-mediated transformation method (Patel et al., 2013).
Callus induction
Husk from rice seeds were removed, and seeds were then sterilized by using 75% ethanol for 1min, then 50% bleach (3% sodium hypochlorite) for 30 min, followed by washing 3 times with sterile distilled water. Seeds without husks were cultured on callus induction media (4g/L N6 salts, 1 mg/L Thiamine-HCL, 250 mg/L Myo-inositol, 1 g/L Casein hydrolyzate, 690 mg/L Proline, 30 g/L Maltose, 5 mg/L 2,4-D, 1 mg/L BA, pH 5.9). Friable calli were transferred to fresh callus induction medium every 2–3 weeks.
Transgenic rice generation
The following media were used. Infection media: 2.2 g/L MS salts, 1 mg/L Thiamine-HCL, 250 mg/L Myo-inositol, 1 g/L Casein hydrolyzate, 690 mg/L Proline, 30 g/L Glucose, 1 mg/L 2,4-Dichlorophenoxyacetic acid (2,4-D), 200 uM acetosyringone, pH 5.4; Co-culture media: same as callus induction media with 60 mg/L maltose and 200 μM of acetosyringone; Selection media: same as callus induction medium with 200 mg/L Timentin and appropriate selective agent (Hygromycin 50 mg/L) and reduced 2,4-D (2 mg/L); Regeneration media: 2.2 g/L MS salts, 1 mg/L benzyl adenine (BA), 30 g/L Maltose, 3 g/L Phytagel with 200 mg/L Timentin and appropriate selective agent (reduced concentration, Hygromycin 20 mg/L); Rooting media: 2.2 g/L MS salts, 30 g/L Maltose, 3 g/L Phytagel with 200 mg/L Timentin and appropriate selective agent (Hygromycin 50 mg/L).
Agrobacteria carrying the destination vector were cultured in 5 mL YEP medium at 28°C overnight followed by transfer to 45 mL MS infection media. Agrobacteria were grown at 28°C for another 2–3 h until the optimal OD value was achieved (OD600: 0.5–0.6). Then 2 to 4 months old calli were mixed with the Agrobacteria culture for 3 min at 42°C followed by 10 min incubation at room temperature. After incubation with Agrobacteria, callus was blotted on three layers of sterile filter papers to get rid of excessive Agrobacteria suspension and then cultured onto co-culture medium, incubated at 25°C in a growth chamber without light for three days. Callus were further transferred to selection medium and incubated in a growth chamber at 25°C in dark for 2 weeks. Callus on hygromycin-based selection medium were sub-cultured every 3 weeks. Calli were visually inspected as well as by epi-fluorescence microscopy (SMZ1000, Nikon, Melville, NY, United States) for mCherry signal. After 2–3 times of sub-culture, calli were transferred to shoot and root regeneration media. Rooted plantlets were transplanted to soil and grown in the greenhouse.
Plant materials and growth conditions
Transgenic rice plants as well as non-transgenic control plants (Nipponbare wild type) were grown in a Greenhouse at North Carolina State University with a temperature range from 20°C (night) to 38°C (day). Plants were also maintained in a growth chamber in the NCSU Phytotron at 26°C with a 12-h light/dark photoperiod. The transgenic plants (T0) were self-pollinated and T1 seeds were collected. T1 plants were grown to obtain T2 progeny.
For rice growth, seeds of the T0, T1, and T2 transgenic plants as well as wild type plants were always de-husked first and sterilized by immersion in 75% ethanol for 1 min followed by 3% sodium hypochlorite for 30 min. After three times washing with sterile water, seeds were germinated on half-strength Murashige and Skoog media and incubated for around 10 days before transferring to pots.
Genomic DNA isolation and genotyping confirmation of transgenic rice
Transgenic plants were initially evaluated under fluorescence microscope (SMZ1000, Nikon, Melville, NY, United States) to screen for positive transgenic rice. Transformation was confirmed in all transgenic lines using gDNA PCR to amplify the target fragments presenting with opposite direction on the left and right sides of Gus linker gene on the destination vector. Primer pairs “Gus3 & corresponding target gene primer” were used to amplify fragment inserted on the left border of Gus gene while primer pairs “Gus4 & corresponding target gene primer” were used to amplify fragment inserted on the right border of Gus gene.
Genomic DNA was extracted from young leaves of transgenic rice using the CTAB method. PCR analysis was performed in a final volume of 25 μL containing 1 μL of forward primer (Gus3 primer or Gus4 primer) (10 μM), 1 μL of reverse primer (target gene reverse primer) (10 μM), 1 μL of dNTPs (10 μM), 2.5 μL of 10 × buffer and 0.125 μL of Taq DNA polymerase from NEB company. Fifty ng of genomic DNA was used as template. The PCR amplification conditions were as follows: 95°C for 5 min; 30 cycles of 95°C for 30 seconds, 55°C for 30 seconds and 72°C for 45 seconds; 72°C for 10 min and hold at 4°C.
Pathogenicity assay and disease assessment
Rice seeds were de-husked and surface sterilized before cultured on half MS media for germination. Seedlings were screened using fluorescence microscope (SMZ1000, Nikon, Melville, NY, United States) for positive transgenic plants based on the mCherry marker gene carried by the destination vector. Seedlings with roots and shoots showing red fluorescence under fluorescence microscope were selected as positive transgenic plants. Genomic DNA PCR was further conducted to confirm positive transgenic plants and non-transgenic segregants. Positive transgenic plants were used for subsequent “pathogenicity assay” while non-transgenic segregants were used as negative control.
Rice seedlings were then transferred to pots and incubated at 26°C with a 12-h light/dark photoperiod. All inoculations and disease evaluation were conducted in the Phytotron facilities at North Carolina State University. Pathogenicity assay was conducted for both the T1 generation and T2 generation rice plants. At least 15 plants per transgenic line were used each time and at least 5 leaves were inoculated for each rice plant.
Strain M. oryzae KJ-201 was used for inoculation. M. oryzae was grown on oatmeal agar media (Difco, Detroit, MI, United States) for 2–3 weeks at 24°C in the dark. Conidial production was induced by scratching the plate surface with a sterilized loop and placing the plate after scratching under continuous fluorescence light for 1 week for sporulation.
Two-month-old rice plants were inoculated with M. oryzae spore suspension at the concentration of 5 × 105spores/mL containing 0.05% Tween-20 (Park et al., 2012). Fully expanded rice leaves were selected for inoculation. A 2- to 3-inch-long piece of transparent Scotch tape (3M company, MN, United States) was placed perpendicular on the rice leaf. Leaves were wounded using a mouse ear punch (Kent Scientific Corporation, CT, United States) in the middle of the leaf at the site of tape attachment. Then, a 10 μL of spore suspension was added to the press-injured site. Both sides of the inoculated site were wrapped with tape to hold the spore suspension and maintain humidity. Inoculated plants were placed in black plastic bags in a growth chamber to block light and retain humidity at 25°C. Bags were removed after 24 h. Plants were then incubated in the growth chamber under 12 h light, 12 h dark at 25°C.
Disease lesions were recorded around 2 weeks after inoculation and photographed. Necrotic tissue size was measured by analyzing the photographs with ‘‘ImageJ’’ software6 (Park et al., 2012). Data was analyzed statistically using ‘‘Graphpad-Prism’’ software7.
RNA isolation, cDNA synthesis and quantitative real-time PCR (qRT-PCR)
To examine fungal target gene’s transcription following infection of transgenic rice plants, leaves from transgenic rice inoculated with M. oryzae strain KJ-201 were collected for quantitative RT-PCR analysis. Two-month-old T1 and T2 generation plants were inoculated with M. oryzae, and total RNA was extracted from infected leaves 2 weeks after inoculation. Three to four leaves from the same infected plant were ground to a fine powder with liquid nitrogen using a precooled sterile mortar and pestle. Total RNA was extracted using the RNeasy plant mini kit (QIAGEN) or using TRIzol reagent (Invitrogen) according to the manufacturer’s instruction. RNA was pretreated with RNase-free DNase I (Invitrogen) to remove genomic DNA contamination, then first-strand of cDNA was synthesized using the reverse transcription system (Promega).
The cDNA was then diluted 20 times with distilled water and used as template for qRT-PCR, which was performed in a 96-well plate using the iQ5 real-time PCR detection system (Bio-Rad). Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) reagent was used. The reaction included 5 μL of SYBRGreen qPCR SuperMix, 1 μL of cDNA template from each sample, 0.5 μL of 10 μM primer, and 3 μL of distilled water. The thermal cycle was split into three stages: initiated by one cycle of 95°C for 20 seconds (hold stage), followed by 40 cycles of 95°C for 15 seconds, 60°C for 30 seconds (PCR stage), with a final melt curve stage: 95°C for 15 seconds, 60°C for 1 min and 95°C for 15 seconds.
Magnaporthe oryzae β-Tubulin gene and M. oryzae ILV5 gene were tested as internal controls (Fudal et al., 2007; Wilson et al., 2010). Primers for target genes were also redesigned (sequence information in Supplementary Table 1). Three independent amplifications were performed for each primer set as technical replication.
Relative fungal target gene transcription was calculated using the 2–ΔΔCT method. ΔΔCT represents (CT, target - CT, reference) transgenic rice - (CT, target -CT, reference) nipponbare. Relative gene transcription was then calculated as a ratio represented by the equation 2–ΔΔCT (Livak and Schmittgen, 2001; Park et al., 2012). Statistical analysis was conducted using “GraphPad-Prism” software.
Synthesis of artificial dsRNA
Entry vectors were used as templates to obtain PCR products for target genes. PCR products of control gene EGFP were obtained by using cDNA of M. oryzae strain KJ201-EGFP as template. The T7 promoter sequence was added to genes’ forward and reverse primers (Supplementary Table 1). The PCR products were further used as templates for corresponding dsRNA synthesis. Synthesis of dsRNA in vitro used the high-scribe T7 in vitro transcription system (Fermentas, Waltham, MA, United States). Template DNA was removed after in vitro transcription by using DNase I. RNA transcripts were purified and annealed in TE buffer at 65°C followed by snap cooling. The dsRNA concentration was determined by Nanodrop spectrophotometry. Synthesized dsRNA was stored at −80°C.
Preparation of diced-siRNA pool
The dsRNA was digested with Short Cut RNase III (New England Biolabs, United States) to prepare diced-siRNA pools according to the manufacturer’s instruction. Briefly, 10 μg dsRNA were digested with Short Cut RNase III in a 100 μL reaction system (dH2O, 10X ShortCut Reaction Buffer: 10 μL, dsRNA: 10 μg, ShortCut RNase III: 10 μL, 10X MnCl2: 10 μL) at 37°C for 20 min. Then 10 μL 10X EDTA was added to stop the reaction. The diced small RNAs were purified by precipitation using nuclease-free glycogen, one-tenth volume of 3M sodium acetate (pH 5.2) and 3 volumes of cold ethanol. Diced siRNA were stored at -80°C (Kalleda et al., 2013).
Treatment of siRNA from the target gene inhibited growth of Magnaporthe oryzae
Magnaporthe oryzae strain KJ-201 was cultured on oatmeal media and sporulation was induced using the method described above. Spores were washed from plates and filtered using two layers of cheesecloth. Concentration of spore suspension was adjusted to 200 spores/μL using distilled water and incubated for 12 h at room temperature. Synthetic siRNA was then added to diluted spore suspension to a final concentration of 30 nM of siRNA with 4 spores/μl. Suspension was incubated for another 24 h at room temperature and 100 μL of suspension was then inoculated onto solid complete media. Plates were incubated at 25°C for 5 days without light and then photographed. The diameter of fungal colonies was measured. For each synthetic siRNA treatment, a minimum of three replicates were included and the experiments were repeated three times.
Results
Selection of target fungal genes and design of experimental workflow
To investigate the potential of HIGS against M. oryzae in rice, six genes with different biological functions in M. oryzae were selected as candidate targets (Table 1). Calcium as an intracellular signaling molecule plays important roles in cellular development and pathogenicity in fungi. Silencing calcineurin encoding genes PsCNA1/PsCNB1 in Puccinia striiformis f. sp. tritici through HIGS has been demonstrated to lead to slower extension of fungal hyphae and reduced production of urediospores (Zhang et al., 2012). In M. oryzae, the transcription factor CRZ1 acts as a downstream regulator in calcium-dependent signaling pathway, and knockout mutants result in hypersensitivity to Ca2+, reduced numbers of conidia, abnormal appressoria and attenuated pathogenicity (Choi et al., 2009; Zhang et al., 2009). Systematic RNA silencing has also been conducted in M. oryzae, and 37 calcium-signaling related genes were targeted (Kadotani et al., 2003). Among those target genes, silencing of PMC1 which is involved in a vacuolar membrane located Ca2+ pump can lead to reduced vegetative growth as well as sporulation (Kadotani et al., 2003). Thus, CRZ1 and PMC1 were selected for HIGS assay in this study.
Regulatory guanine nucleotide-binding proteins (G proteins) are involved in the signal transduction cascades which are important for proper regulation of cell function, division and differentiation (Liu and Dean, 1997). The G protein α subunit gene MAGB of M. oryzae is involved in signal transduction pathways controlling vegetative growth, conidiation, conidium attachment, appressorium formation, mating as well as pathogenicity (Fang and Dean, 2000). This gene was also selected as a promising candidate for HIGS.
Host colonization requires secretion of effector proteins into plant cells to suppress host immunity and promote pathogen growth. However, individual effector genes are not good candidates for silencing because pathogens usually secrete multiple effectors with overlapping function. Silencing conserved fungal genes that have essential biological functions, such as genes essential for effector secretion are more likely to confer host durable resistance. In M. oryzae, there are two distinct effector secretion pathways. First is the conventional Endoplasmic Reticulum (ER)-Golgi secretion pathway secreting apoplastic effectors (effectors that do not enter host cells, and generally remain within the extra invasive hyphae membrane (EIHM) compartment) (Zhang and Xu, 2014). The ER chaperone LHS1 is important for protein translocation across ER membrane as well as extracellular activities. Mutant lhs1 M. oryzae strain exhibit severely impaired conidiation, penetration and biotrophic invasion in susceptible rice (Yi et al., 2009). Consequently, LHS1 was chosen as one of our HIGS candidates.
Fungal Cytochrome P450 lanosterol C-14a- demethylase (CYP51) genes are essential for ergosterol biosynthesis. Silencing of these genes can restrict fungal growth. CYP51 is also a target for systemic fungicides acting as demethylation inhibitors (DMIs). Fungicides like tebuconazole and triadimefon can bind CYP51 and interfere with fungal membrane integrity (Yoshida, 1993; Deng et al., 2007; Koch et al., 2013). It has been shown that silencing CYP51 in Arabidopsis and barley confers strong resistance to Fusarium species (Koch et al., 2013). As M. oryzae contains two homologous CYP51 genes (designated as CYP51A and CYP51B), they were both targeted simultaneously for our HIGS assay.
Figure 1 describes the experimental workflow that was applied to this research. Certain fragments of target fungal genes were cloned and inserted into HIGS destination vectors individually. Five HIGS vectors (one with both CYP51A and CYP51B fragments) were then transformed to rice calli respectively to generate transgenic rice. Molecular and phenotype characterization of HIGS plants were conducted to confirm positive transgenic rice and evaluate resistance to M. oryzae. M. oryzae was further co-cultured with small RNA to investigate the potential mechanism that underlies HIGS (Figure 1).
Figure 1. Schematic overview of host-induced gene silencing (HIGS) experimental workflow. Specific fragments were chosen and inserted into HIGS vectors to target important genes of Magnaporthe oryzae. Transgenic rice was generated by transforming HIGS vectors into rice calli. Transgenic rice was then subjected to molecular and phenotype characterization. Positive transgenic rice was first confirmed through genotyping. Then transgenic rice was inoculated with M. oryzae for fungal resistance evaluation. Relative transcription of fungal target genes from infected transgenic rice leaves was analyzed through qRT-PCR. Furthermore, M. oryzae was co-cultured with sRNA derived from target genes for fungal growth assay.
Five host-induced gene silencing vectors were constructed targeting six fungal genes respectively
Target fragments from six candidate fungal genes were selected based on the criteria of presence of certain sequence motifs, optimal GC content, differential thermal stability, absence of poly-asparagine (polyN) sequence and low complexity. Specificity of those fragments was confirmed through a sequence similarity search on the rice genome annotation database8, and no homologous sequences were found. Information on candidate genes and target fragments is shown in Table 1.
PCR products were obtained for six target fragments by using cDNA of M. oryzae strain 70-15 as template. Fragments of CYP51A and CYP51B were linked together through joining PCR (Supplementary Figure 1). In total, five fragments were prepared for HIGS vector construction (Table 1). Using the “Gateway cloning system,” all were first cloned into the entry vector pDONR221 and then moved into the HIGS destination vector individually (Zhao et al., 2021). After recombination cloning, each target fragment was inserted into two regions of the destination vector flanked by recombination sites. Those insertions were designed to be in opposite directions, enabling the formation of inverted repeats (Miki and Shimamoto, 2004). In total, five different HIGS vectors targeting 6 fungal genes were constructed for rice transformation.
Generation of transgenic rice plants expressing host-induced gene silencing constructs
Rice calli were induced from Nipponbare seeds (Figures 2A–C). Following Agrobacterium mediated transformation (Figures 2D,E), transgenic rice seedlings were generated (Figure 2F,G, H,I). mCherry fluorescence signal was visibly detected in positive transgenic calli as well as in regenerated rice shoots and roots (Figure 3). T0 generation transgenic rice were grown in the green house, and the presence of the inserted target fragments was confirmed by genomic DNA PCR. Inverted repeat sequences on both sides of Gus linker were amplified from transgenic T0 plants using different primer sets (Figure 4). All transgenic rice plants showed normal growth pattern compared to wild type Nipponbare.
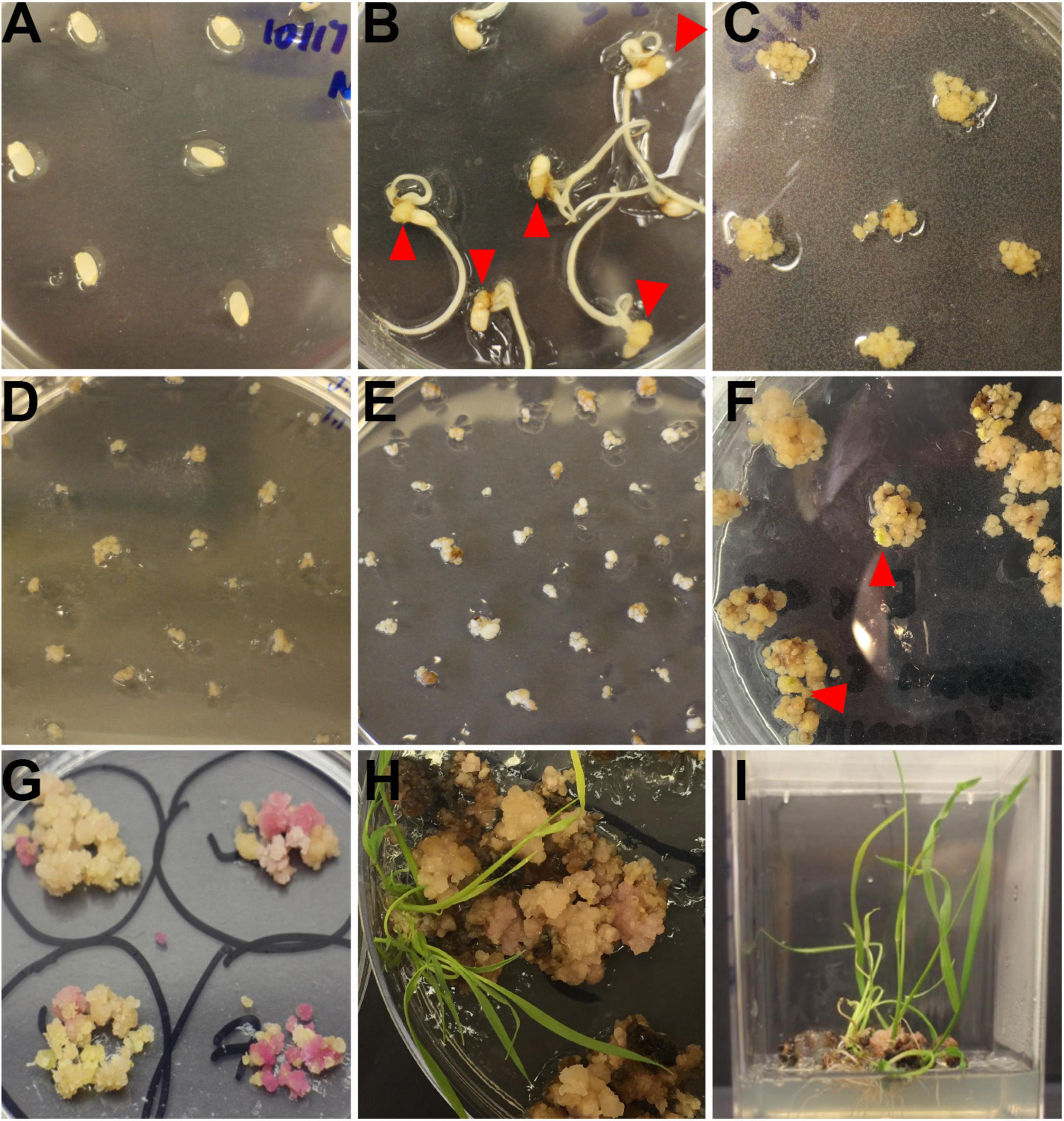
Figure 2. Workflow of rice transformation. (A) Seeds of Nipponbare on callus induction media. (B) Induced rice calli (red triangle) on callus induction media. (C) Rice calli used for transformation on fresh callus induction media. (D) Transformed calli co-cultured with Agrobacteria, calli were covered by Agrobacteria. (E) Rice calli on hygromycin selection media. Non-transformed calli turned dark brown and died while transformed calli grew fresh white colored calli. (F) Rice calli on shoot regeneration media. Some calli were ready to regenerate rice shoots (red triangle). (G) Rice calli on shoot regeneration media. Positively transformed calli turned red and were ready to generate shoots. (H) Regenerated rice shoots from transformed calli on shoot reaeration media. (I) Rice calli with regenerated shoots and roots on root regeneration media.
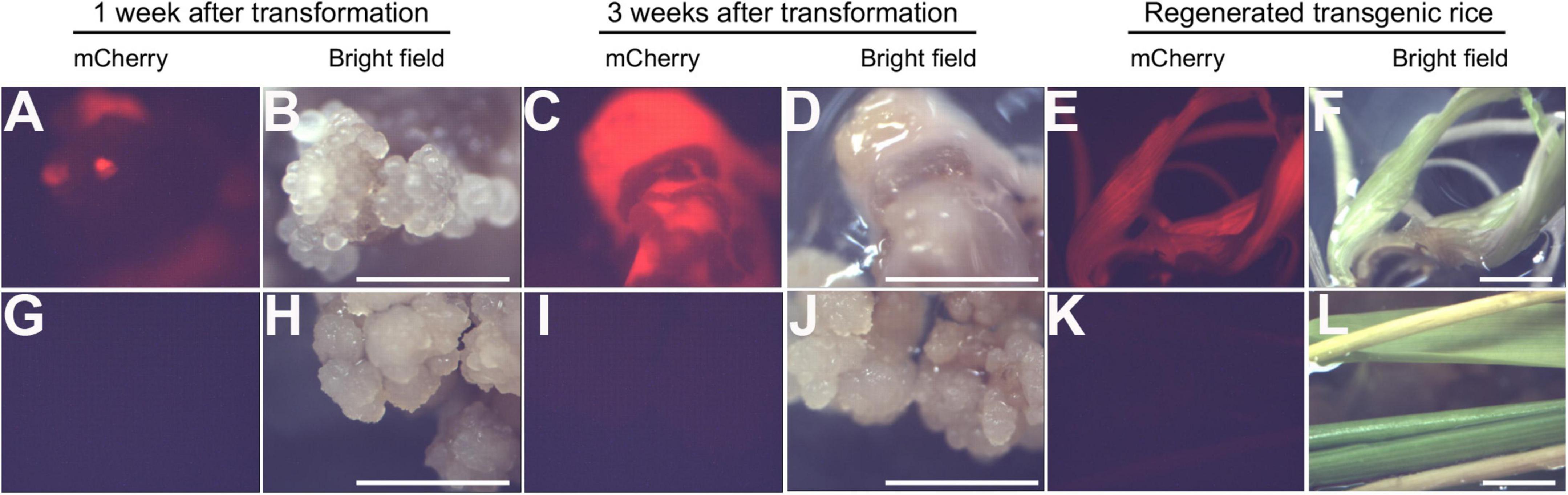
Figure 3. Positive transgenic rice confirmed through epifluorescence microscopy. (A–F) Transgenic rice calli (A–D) and seedling (E,F) after 1 week of transformation (A,B), 3 weeks of transformation (C,D), and shoot regeneration (E,F). (G–L). Non-transformed control rice calli (G–J) and seedling (K,L) after 1 week of transformation (G,H), 3 weeks of transformation (I,J), and shoot regeneration (K,L). Scale bar = 1 cm.
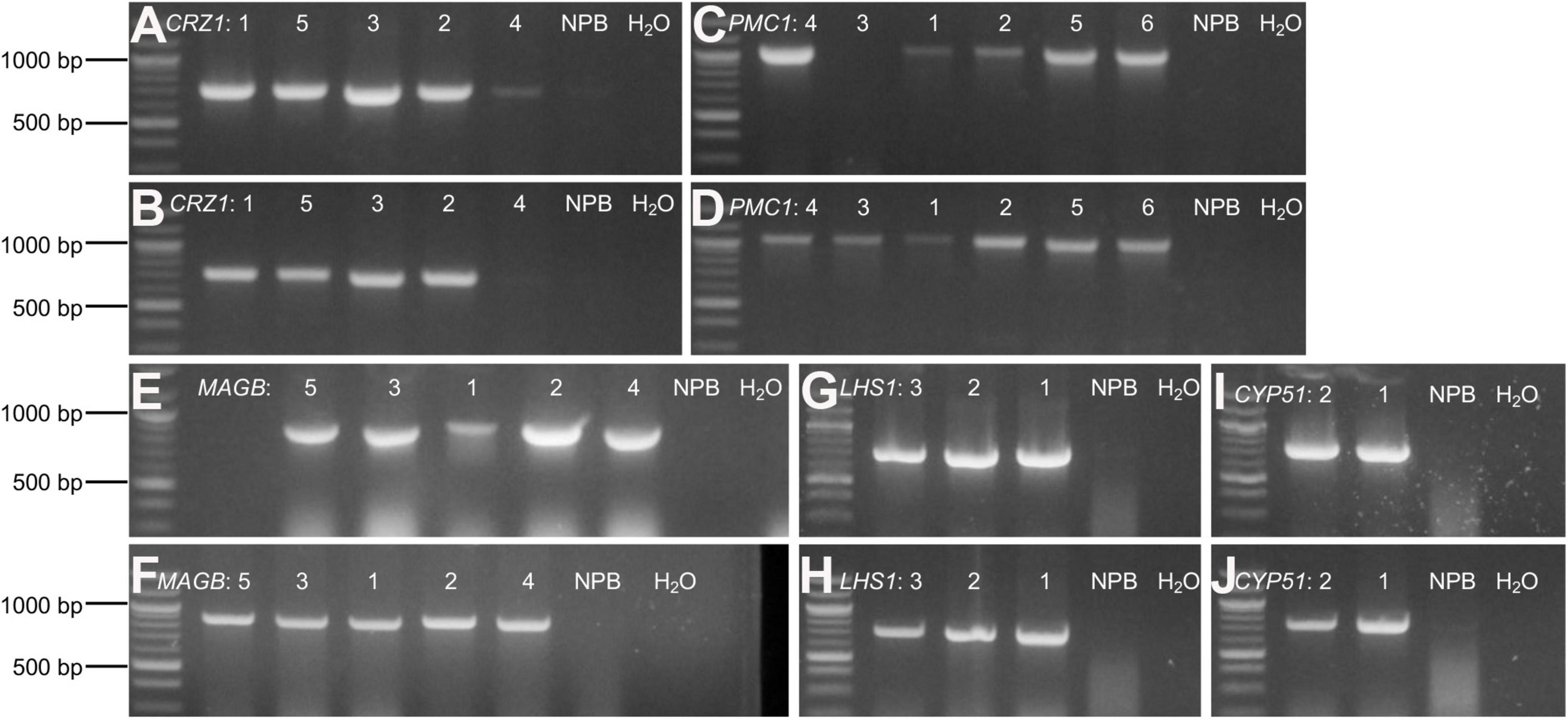
Figure 4. Confirm positive T0 transgenic rice through gDNA PCR. (A–J) Agarose gel electrophoresis for amplified target fragments of candidate HIGS gene CRZ1 (A,B), PMC1 (C,D), MAGB (E,F), LHS1 (G,H), and CYP51 (I,J). Genomic DNA isolated from T0 transgenic rice leaves was used as templates. Primer pairs of “gus3 + corresponding target gene reverse primer” (A,C,E,G,I) and “gus4 + corresponding target gene reverse primer” (B, D, F, H, J) were used to amplify target fragments on both sides of Gus linker. Quick-Load Purple 1 kb Plus DNA Ladder from NEB was used as the DNA ladder. Arabic numbers were assigned to different rice plant lines representing individual transgenic event. Wild type Nipponbare rice plants as well as distilled water were used as negative control. The same assays were conducted to screen positive transgenic T1 and T2 generation rice plants.
Expression of specific host-induced gene silencing vectors in transgenic rice confers resistance to Magnaporthe oryzae
Following self-pollination, T1 and T2 generation rice plants were obtained for pathogenicity assays. All transgenic rice derived from different HIGS constructs appeared healthy and demonstrated normal growth phenotype. Positive transgenic rice plants were confirmed through mCherry fluorescence detection as well as genomic DNA PCR. Negative plants segregating from each line without carrying the corresponding HIGS fragments were used as negative controls, as were wild type Nipponbare plants. Fully expanded rice leaves were infected with M. oryzae strain KJ-201 under favorable conditions, and necrotic lesion size analyzed a week later.
For T1 generation rice derived from HIGS vectors aiming to silence the fungal genes CRZ1, MAGB and CYP51, the lesion size on positive transgenic plants were significantly smaller compared to non-transgenic (negative) segregants plants and the wild type Nipponbare control (Figure 5). Five different lines were tested for HIGS plants derived from CRZ1 gene (lines 1, 2, 3, 4, 5), five lines for MAGB gene (lines 1, 2, 3, 4, and 5) and two lines for CYP51 gene (lines 1 and 2). For CRZ1, the lesion area was reduced 56.0%, 69.6%, 53.7% and 53.9%, respectively, for intra-line comparisons of lines “1”, “2”, “3” and crossline comparison between “4 and 5”. The lesions were 78.5% smaller for positive MAGB lines “1”, “2”, “3” compared with wild type and 65.0% and 63.5% smaller for intra-line (positive vs negative) comparison for line “4” and “5”, respectively. In addition, the CYP51 lines “1” and “2” also showed a 67.1% and 69.7% reduction of lesion area compared with the non-transgenic segregants. However, for transgenic rice derived from HIGS vectors aiming to silence PMC1 and LHS1, no significant differences for lesion size among positive transgenic rice, non-transgenic segregants and wild type Nipponbare were observed (Figure 5).
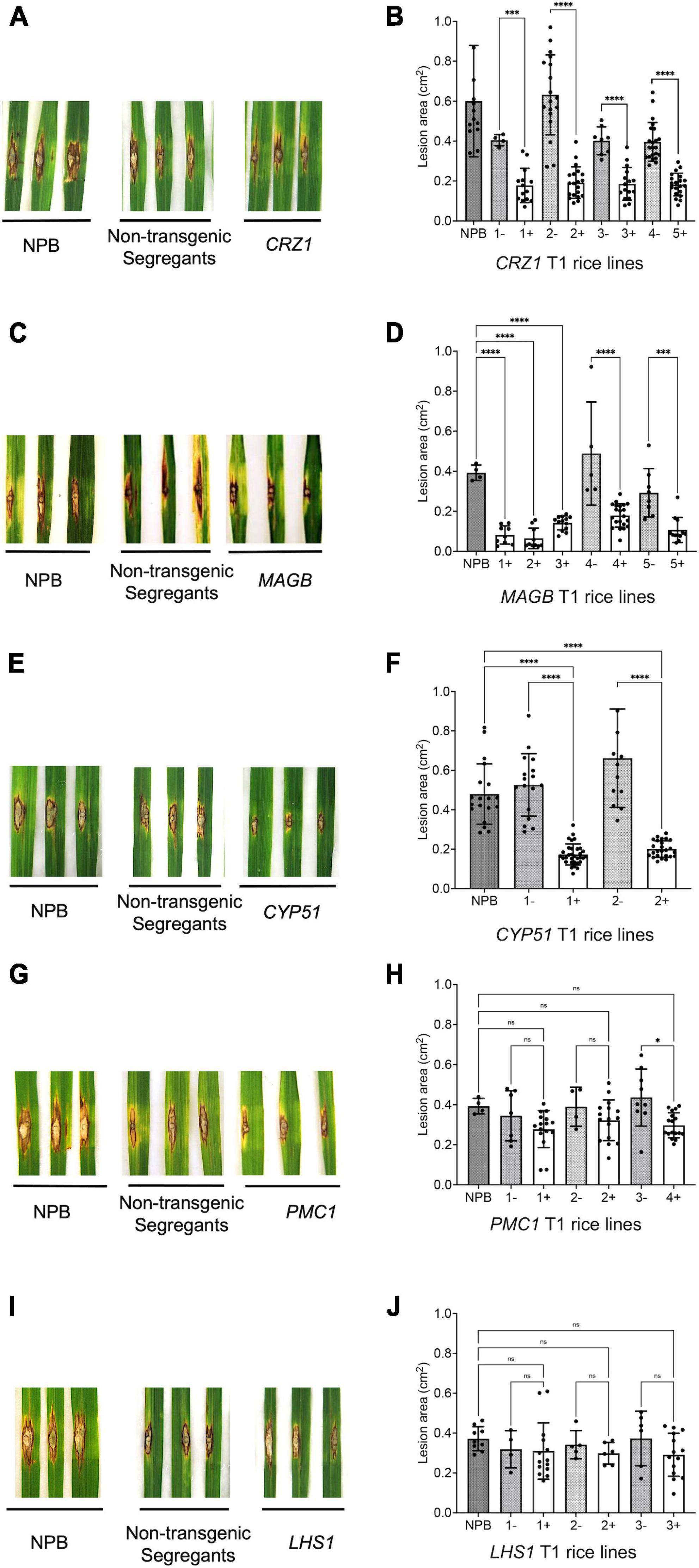
Figure 5. Certain host-induced gene silencing (HIGS) transgenic rice (T1 generation) showed resistance to Magnaporthe oryzae. (A–J) Punch inoculation of HIGS transgenic rice plants targeting M. oryzae gene CRZ1 (A,B), MAGB (C,D), CYP51 (E,F), PMC1 (G,H) and LHS1 (I,J). Wild type Nipponbare (NPB) and non-transgenic segregants derived from corresponding T0 plants were used as negative control. Lesion area was measured and analyzed statistically (B,D,F,H,J) using multiple rice lines. The symbol “+” represents positive transgenic plants while “-” represents non-transgenic segregants. Ordinary one-way ANOVA and Turkey’s multiple comparison were used for statistics analysis. *P < 0.0332, ***P < 0.0002, ****P < 0.0001, ns: not significant.
To confirm resistance phenotype obtained from the T1 generation, T2 rice plants targeting CRZ1, MAGB and CYP51 were grown and inoculated with M. oryzae again. Similar results were obtained (Figure 6). For CRZ1 transgenic plants, the lesion area of line “1” were 52.7% smaller than the negative line “4” while the intra-line reductions were 32.2% for line “2” and 65.4% for line “3”. For MAGB transgenic plants, the lesion area was reduced 80.5% when comparing line “1” with wild type. The reduction of lesion areas was 51.5%, 66.4%, 68.5% and 75.1% when compared positive transgenic rice line “2”, “3”, “4”, and “5” with their non-transgenic segregants respectively. For CYP51 transgenic rice, the lesions were 50.5% and 60.0% smaller for line “1” and “2” when compared with corresponding non-transgenic segregants (Figure 6).
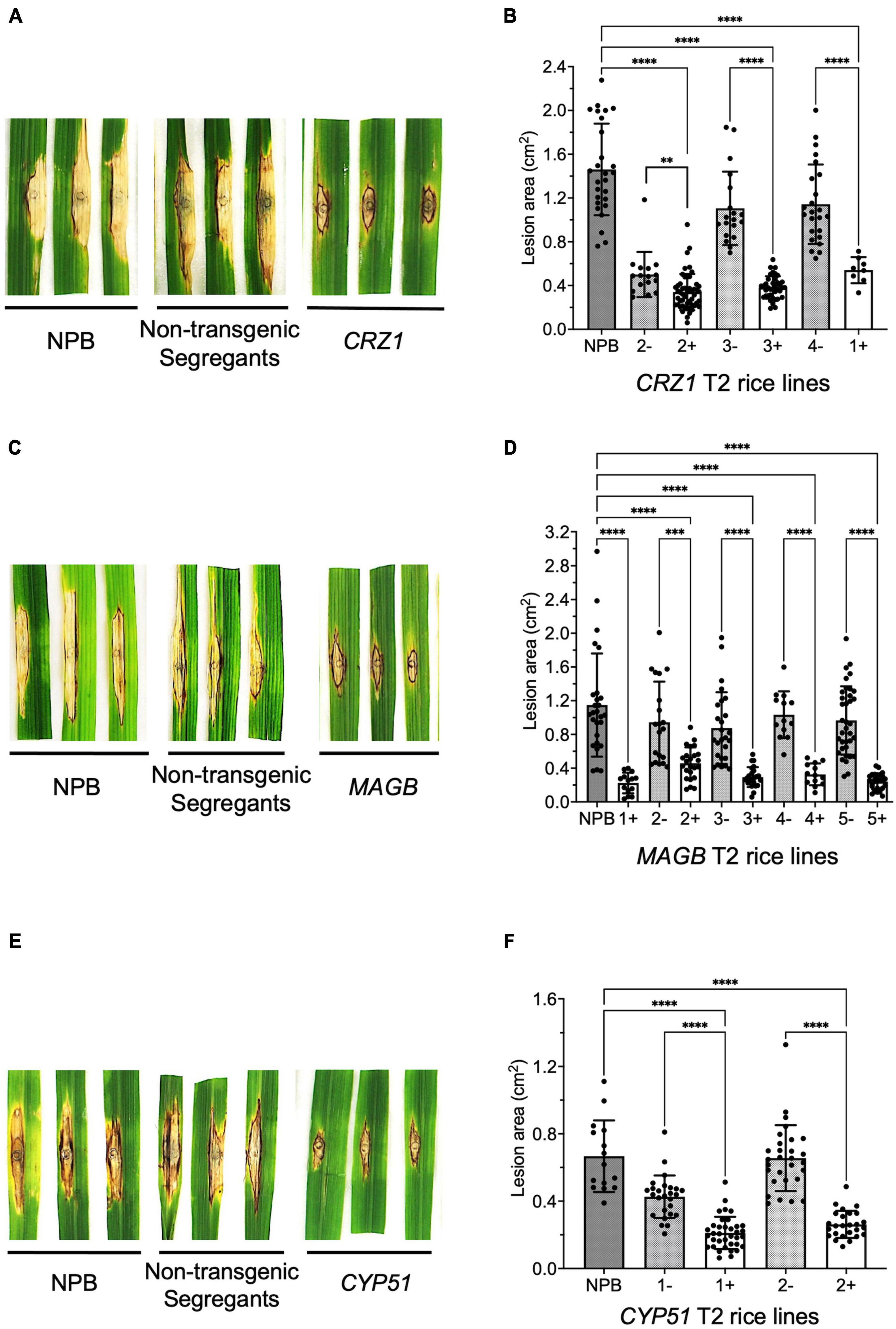
Figure 6. Confirm resistant host-induced gene silencing (HIGS) transgenic rice in T2 generation. (A–F) Punch inoculation of HIGS transgenic rice plants targeting Magnaporthe oryzae gene CRZ1 (A,B), MAGB (C,D), CYP51 (E,F). Wild type Nipponbare (NPB) and non-transgenic segregants derived from corresponding T1 plants were used as negative control. Lesion area was measured and analyzed statistically (B,D,F) using multiple rice lines. The symbol “+” represents positive transgenic plants while “-” represents non-transgenic segregants. Ordinary one-way ANOVA and Turkey’s multiple comparison were used for statistics analysis. ***P < 0.0002, ****P < 0.0001.
Target fungal genes are repressed in Magnaporthe oryzae infected host-induced gene silencing-expressing transgenic rice plants
Quantitative real time-PCR analysis was carried out to verify silencing of target fungal genes. M. oryzae was inoculated on rice leaves and fungal transcripts were analyzed 14 days after inoculation (Figure 7). Results showed reduced levels of CRZ1 and MAGB endogenous transcripts in positive transgenic rice compared to non-transgenic segregants as well as wild type Nipponbare. For gene CRZ1, the transcription level was reduced by 63.1, 54.6, and 66.5% when positive transgenic line “1”, “2”, and 3” were compared with non-transgenic segregants (Figure 7A). For gene MAGB, the transcription was reduced 89.1% in line “2” and 67.0% in line “3” using ILV5 as the internal reference, while the reductions were 79.9 and 61.7%, respectively, when β-Tubulin was used as fungal reference. However, reduced MAGB transcription was not detected for line 5 (Supplementary Figure 2A and Figure 7B). In addition, no significant difference was observed for CYP51 transcription between transgenic rice plants and wildtype control (Supplementary figure 2B). Overall, for HIGS plants targeting CRZ1 and MAGB, the qRT-PCR results are consistent with the hypothesis that impairment of the pathogen’s ability to cause disease on rice leaves is related to reduced fungal gene expression induced by HIGS.
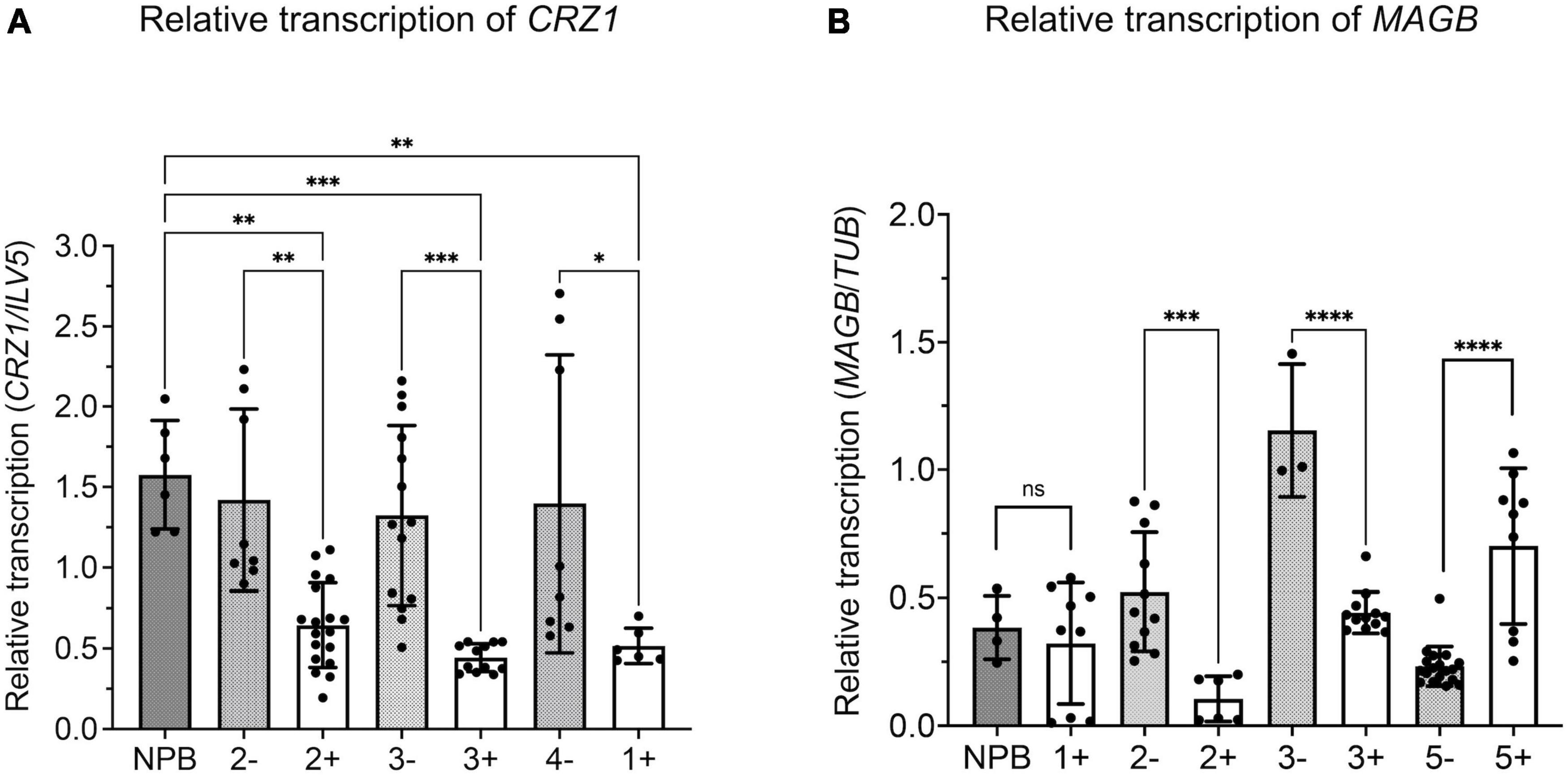
Figure 7. Relative transcription of target gene reduced in Magnaporthe oryzae invading host-induced gene silencing (HIGS) transgenic rice targeting CRZ1 and MAGB. A,B Analysis of relative transcription of fungal gene CRZ1 (A) and MAGB (B) through quantitative RT-PCR. Fungal gene ILV5 (A) or β-Tubulin (B) were used as the reference gene. Multiple rice lines inoculated with M. oryzae were tested. “+” represents positive transgenic rice while “-” represents non-transgenic segregants. cDNA was generated from total RNA isolated from M. oryzae-inoculated rice leaves after 14 days. Bars represent mean values ± SDs of at least three independent sample collections. (*P < 0.0332, **P < 0.0021, ***P < 0.0002, ****P < 0.0001; Tukey test).
Incubation with small RNA derived from fungal genes inhibits growth of Magnaporthe oryzae
We show HIGS can confer rice resistance to M. oryzae, however, how are small RNA transferred from the plant cell to fungal cell and trigger silencing of target genes is not fully understood. To investigate whether fungal cells can take up dsRNA and siRNA targeting the fungal genes, dsRNA derived from fragments of PMK1, MPG1 as well as CYP51, CRZ1, and MAGB were synthesized in vitro using the T7 RNA polymerase transcription system (Supplementary Figure 3). Furthermore, siRNA targeting each gene were processed using short cut RNase III (Supplementary Figure 3). Fungal spore suspensions were incubated with synthesized dsRNA or siRNA and then subjected to morphological evaluation. PMK1 and MPG1 are genes important for appressorium formation in M. oryzae. PMK1 is a mitogen-activated protein (MAP) kinase gene regulating formation of appressorium and infectious hyphal growth of M. oryzae (Bruno et al., 2004). MPG1 is a hydrophobin-encoding gene. Loss of function mutant shows reduced virulence, conidiation and appressorium formation (Soanes et al., 2002). siRNA derived from M. oryzae PMK1 and MPG1 dramatically interfered with appressorium formation and reduced formation by around 70% (Figures 8A–C). Total number of fungal colonies on dsRNA and siRNA treated plates were not significantly different from control plates (Figure 8D and Supplementary Figure 4). Fungi treated with buffer solution as well as dsRNA/siRNA derived from eGFP were used as controls. dsRNA as well as siRNA derived from CRZ1, MAGB and CYP51 greatly reduced fungal colony radius (Figure 8E and Supplementary Figure 4), indicating that both naked dsRNA and siRNA may move into fungal cells and induce RNAi, resulting in growth inhibition.
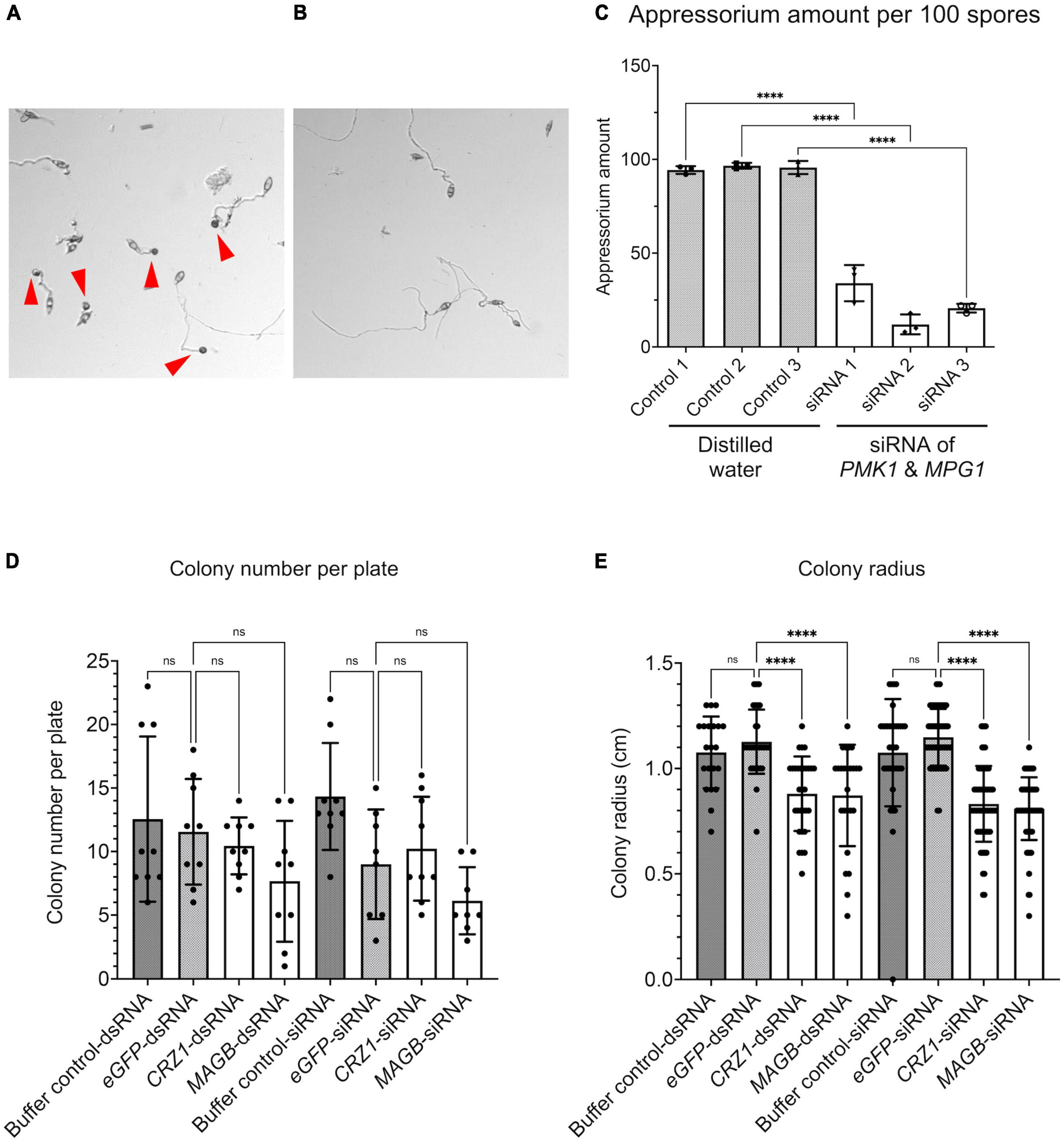
Figure 8. RNA silencing signals can be absorbed by Magnaporthe oryzae resulting in functional collapse. (A) Spore suspension treated with distilled water. Appressoria (red arrows point to appressoria) were formed at tips of germ tube. (B) Spore suspension treated with siRNA derived from PMK1&MPG1, less appressoria were formed. (C) Appressorium amount after water treatment and PMK1&MPG1 siRNA treatment. Ordinary one-way ANOVA and Turkey’s multiple comparison were used for statistics analysis. (D) No statistical difference was observed considering fungal colony number when spore suspensions were treated with buffer control, dsRNA/siRNA derived from eGFP, CRZ1 or MAGB. (E) Treatment of dsRNA/siRNA derived from CRZ1/MAGB reduced fungal colony radius when compared to spore suspensions treated with buffer control or eGFP-dsRNA/siRNA. Buffer solution was negative control solution that went through the whole process of dsRNA/siRNA synthesis but without templates. **** P < 0.0001, ns: not significant.
Discussion
As a novel RNAi based technology for efficient control of fungal pathogens and other pests, our results show that HIGS can be effective at significantly reducing rice blast disease caused by M. oryzae. In general, we were able to achieve greater than 50% reductions in symptoms and in some cases 80% reduction in lesion size when certain genes were targeted. M. oryzae as a hemibiotroph pathogen initially absorbs nutrients from living cells and then kills host cells in order to live necrotrophically on the dead tissues. We do not know which infection stage is most effective for HIGS, but during both stages, RNA silencing signals likely have the opportunity to move from plant cells to invading fungal cells and induce silencing of target genes. Choosing the right fungal gene to silence and gene segment is vital. Genes sharing duplicate function with others, performing minor effects for fungal growth and infection or genes capable of rapid evolution of substitution such as effectors are not likely to be appropriate HIGS candidates. Previous HIGS studies (Nowara et al., 2010; Yin et al., 2011; Zhang et al., 2012; Panwar et al., 2013; Hu et al., 2015; Zhou et al., 2016) have targeted many key fungal genes playing important roles in growth and development such as genes encoding calcineurin, MAP kinases, F-box proteins, chitin synthases, RNA polymerases, ubiquitin E3 ligases and others all of which represented good HIGS candidates. HIGS technology has been evaluated for rice blast disease. Zhu et al. used Brome mosaic virus (BMV) mediated HIGS to show that fungal pathogenicity genes: MoABC1, MoMAC1 and MoPMK1 are good targets for HIGS conferred disease resistance. However, the assay was analyzed through a transient expression system (Zhu et al., 2017). Targeting bZIP transcription factor encoding gene MoAP1 as well as the actin cross-linking protein Fimbrin encoding gene MoFim1 also reduced rice blast disease (Guo et al., 2019; Li et al., 2020). Here we tested 6 other target genes and found that CRZ1, MAGB and CYP51 genes important for signal transduction and membrane integrity, are also viable and effective targets for HIGS. In the future, multiple fungal genes could be targeted together through co-transformation or crossing rice lines to provide rice with more durable and robust resistance to M. oryzae.
It is also important to identify a suitable fragment of the candidate gene as the target sequence because different gene fragments may perform with different efficiency to generate siRNA leading to RNAi (Baldwin et al., 2018). In this study, transgenic rice derived from HIGS vectors targeting LHS1 and PMC1 did not exhibit resistance to M. oryzae. One reason may be that proper gene function is only affected when the gene expression is totally eliminated. However, it may be also due to inappropriate selection of target fragments for those two genes such that no or very little silencing was accomplished to generate an observable phenotype. Another aspect of note was for HIGS plants targeting fungal gene CYP51. While disease resistance phenotype was observed in both T1 and T2 generation (Figures 5, 6), the relative transcription of CYP51 in infected HIGS plants was not altered (Supplementary Figure 2B). This may have resulted from several issues such as DNA contamination during RNA extraction, interference in the qPCR process or small RNA off targeting in the HIGS system. However, the most likely reason may be poor design of CYP51 primers used in RT-qPCR analysis. CYP51 is one of the most ancient and conserved P450s across kingdoms (Yoshida et al., 1997; Debeljak et al., 2003; Kim et al., 2005; Becher and Wirsel, 2012). Rice also contains multiple CYP51 genes (Nelson et al., 2004; Lepesheva and Waterman, 2007). Although we carefully chose the fungal specific CYP51 fragment to construct the HIGS vector, the primers used in qPCR analysis may not have been specific enough to amplify the fungal CYP51 fragment exclusively. Rice CYP51 may also have been amplified causing the unexpected qPCR results. Every consideration needs to be taken when choosing genes conserved among plants and fungi as HIGS targets. On the other hand, targeting a conserved fragment across different pathogenic fungi may confer HIGS plants with broad spectrum resistance to pathogens.
Although several studies have demonstrated the possibility of using HIGS as a novel strategy for disease control, few have provided insight into the underlying signal mechanism(s). What is the silencing signal (dsRNA or siRNA)? How do small RNA signals get sorted and transported between plant to pathogen without degradation? As demonstrated by this study, siRNA and dsRNA can be absorbed by fungal mycelium and may induce silencing of target genes. Similar results have also been obtained for Aspergillus nidulans (Kalleda et al., 2013) as well as Rhizoctonia solani (Zhou et al., 2016). For M. oryzae, silencing of MoAP1 can be achieved through in vitro feeding of artificial small RNA targeting MoAP1. This resulted in impaired fungal development (Guo et al., 2019). Those interesting results further point out the potential for using small RNAs as fungicide for disease control.
Recently, Hailing Jin’s group has demonstrated that extracellular vesicles carrying siRNA can move between fungal and plant cells (Jin, 2008; Weiberg et al., 2013; Wang et al., 2016; Cai et al., 2018). Nevertheless, the evidence is still limited, and it is not clear how such vesicles are directed to cross into invading pathogen cells and maintain optimal concentrations to induce RNAi. Important proteins involved in silencing signal sorting, transportation, recognition, absorption and protection from degradation are worthy of further research. Using fluorescence or radioactively labeled siRNA may help to decipher the entire process of HIGS. Inhibitors such as secramine that block plant vesicle secretion may further confirm the role of vesicles for small RNA communication (Pelish et al., 2006; von Kleist and Haucke, 2012). For practical application, artificial vesicles containing siRNA as well as synthesized naked siRNA (Guo et al., 2019) may be suitable as bio-fungicides to control plant diseases. For example, spraying dsRNA targeting CYP51 in Fusarium graminearum provided some disease control on barley (Koch et al., 2016). Spray induced gene silencing (SIGS) has also been demonstrated for rice-M. oryzae interaction system. Spray of dsRNA targeting fungal MoDES1 induced silencing of MoDES1 and conferred significant resistance against blast disease (Sarkar and Roy-Barman, 2021). Above all, our study demonstrates the effectiveness of using HIGS in rice blast disease control and highlights that targets and silencing fragments need to be carefully evaluated. Moreover, to facilitate effective application, further studies are needed to address the underlying mechanism(s) for cross-kingdom siRNA communication.
Data availability statement
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MW and RD designed the experiments. MW carried out experiments and analyzed data. MW and RD wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by Agriculture and Food Research Initiative Competitive Grant no. 2013-68004-20378 from the USDA National Instituteof Food and Agriculture.
Acknowledgments
Special thanks to Shaohong Qu (Zhejiang Academy of Agricultural Sciences, China) for sharing the pBDL03 vector. Thanks to Rongda Qu from NCSU Crop Science department for helping with the rice transformation. Special thanks to NCSU greenhouse and phytotron staff. Many thanks to Tim Sit from NCSU Entomology and Plant Pathology for helping with the qRT-PCR assay. Thanks to Yeonyee Oh for all the support and help during this research project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.959641/full#supplementary-material
Footnotes
- ^ fungi.ensembl.org/index.html
- ^ rnaidesigner.thermofisher.com/rnaiexpress/design.do
- ^ rice.uga.edu/analyses_search_blast.shtml
- ^ bsw3.naist.jp/simamoto/pANDA/real/gus_linker.htm
- ^ https://www.megasoftware.net/mega4/mega.html
- ^ rsbweb.nih.gov/ij/
- ^ www.graphpad.com/
- ^ rice.uga.edu/analyses_search_blast.shtml
References
Andrade, C., Tinoco, M., Rieth, A., Maia, F., and Aragão, F. (2016). Host-induced gene silencing in the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Pathol. 65, 626–632.
Baldwin, T., Islamovic, E., Klos, K., Schwartz, P., Gillespie, J., Hunter, S., et al. (2018). Silencing efficiency of dsRNA fragments targeting Fusarium graminearum TRI6 and patterns of small interfering RNA associated with reduced virulence and mycotoxin production. PLoS One 13:e0202798. doi: 10.1371/journal.pone.0202798
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326.
Becher, R., and Wirsel, S. G. (2012). Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl. Microbiol. Biotechnol. 95, 825–840. doi: 10.1007/s00253-012-4195-9
Brown, J. K. (2002). Yield penalties of disease resistance in crops. Curr. Opin. Plant Biol. 5, 339–344.
Bruno, K. S., Tenjo, F., Li, L., Hamer, J. E., and Xu, J. R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot Cell 3, 1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004
Burdon, J. J., and Thrall, P. H. (2003). The fitness costs to plants of resistance to pathogens. Genome Biol. 4:227.
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F.-M., Palmquist, J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. doi: 10.1126/science.aar4142
Cheng, W., Song, X. S., Li, H. P., Cao, L. H., Sun, K., Qiu, X. L., et al. (2015). Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to F usarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13, 1335–1345. doi: 10.1111/pbi.12352
Childs, N., and LeBeau, B. (2021). Rice outlook: October 2021, RCS-21I. Washington, DC: Department of Agriculture, Economic Research Service.
Choi, J., Kim, Y., Kim, S., Park, J., and Lee, Y.-H. (2009). MoCRZ1, a gene encoding a calcineurin-responsive transcription factor, regulates fungal growth and pathogenicity of Magnaporthe oryzae. Fungal Genet. Biol. 46, 243–254. doi: 10.1016/j.fgb.2008.11.010
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
Das, P. R., and Sherif, S. M. (2020). Application of exogenous dsRNAs-induced RNAi in agriculture: challenges and triumphs. Front. Plant Sci. 11:946. doi: 10.3389/fpls.2020.00946
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430.
Dean, R. A., Talbot, N. J., Ebbole, D. J., Farman, M. L., Mitchell, T. K., Orbach, M. J., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434, 980–986.
Debeljak, N., Fink, M., and Rozman, D. (2003). Many facets of mammalian lanosterol 14α-demethylase from the evolutionarily conserved cytochrome P450 family CYP51. Arch. Biochem. Biophys. 409, 159–171. doi: 10.1016/s0003-9861(02)00418-6
Deng, J., Carbone, I., and Dean, R. A. (2007). The evolutionary history of cytochrome P450 genes in four filamentous Ascomycetes. BMC Evol. Biol. 7:30. doi: 10.1186/1471-2148-7-30
Dou, T., Shao, X., Hu, C., Liu, S., Sheng, O., Bi, F., et al. (2020). Host-induced gene silencing of Foc TR4 ERG6/11 genes exhibits superior resistance to Fusarium wilt of banana. Plant Biotechnol. J. 18, 11–13. doi: 10.1111/pbi.13204
Drinnenberg, I. A., Weinberg, D. E., Xie, K. T., Mower, J. P., Wolfe, K. H., Fink, G. R., et al. (2009). RNAi in budding yeast. Science 326, 544–550.
Fang, E. G., and Dean, R. A. (2000). Site-directed mutagenesis of the magB gene affects growth and development in Magnaporthe grisea. Mol. Plant Microbe Interact. 13, 1214–1227. doi: 10.1094/MPMI.2000.13.11.1214
Fudal, I., Collemare, J., Böhnert, H. U., Melayah, D., and Lebrun, M. H. (2007). Expression of Magnaporthe grisea avirulence gene ACE1 is connected to the initiation of appressorium-mediated penetration. Eukaryotic cell 6, 546–554. doi: 10.1128/EC.00330-05
Ghildiyal, M., and Zamore, P. D. (2009). Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 10, 94–108.
Govindarajulu, M., Epstein, L., Wroblewski, T., and Michelmore, R. W. (2015). Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 13, 875–883. doi: 10.1111/pbi.12307
Guo, X.-Y., Li, Y., Fan, J., Xiong, H., Xu, F.-X., Shi, J., et al. (2019). Host-induced gene silencing of MoAP1 confers broad-spectrum resistance to Magnaporthe oryzae. Front. Plant Sci. 10:433. doi: 10.3389/fpls.2019.00433
Hamer, J. E., Howard, R. J., Chumley, F. G., and Valent, B. (1988). A mechanism for surface attachment in spores of a plant pathogenic fungus. Science 239, 288–290.
Hu, Z., Parekh, U., Maruta, N., Trusov, Y., and Botella, J. R. (2015). Down-regulation of Fusarium oxysporum endogenous genes by host-delivered RNA interference enhances disease resistance. Front. Chem. 3:1. doi: 10.3389/fchem.2015.00001
Huang, G., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2006). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. 103, 14302–14306. doi: 10.1073/pnas.0604698103
Jahan, S. N., Åsman, A. K., Corcoran, P., Fogelqvist, J., Vetukuri, R. R., and Dixelius, C. (2015). Plant-mediated gene silencing restricts growth of the potato late blight pathogen Phytophthora infestans. J. Exp. Bot. 66, 2785–2794. doi: 10.1093/jxb/erv094
Jia, Y., Jia, M. H., Wang, X., and Zhao, H. (2019). “A toolbox for managing blast and sheath blight diseases of rice in the United States of America,” in Protecting rice grains in the post-genomic Era, ed. Y. Jia (London: IntechOpen), 35–52.
Jin, H. (2008). Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett. 582, 2679–2684.
Kadotani, N., Nakayashiki, H., Tosa, Y., and Mayama, S. (2003). RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol. Plant Microbe Interact. 16, 769–776.
Kalleda, N., Naorem, A., and Manchikatla, R. V. (2013). Targeting fungal genes by diced siRNAs: A rapid tool to decipher gene function in Aspergillus nidulans. PLoS One 8:e75443. doi: 10.1371/journal.pone.0075443
Kim, H. B., Schaller, H., Goh, C.-H., Kwon, M., Choe, S., An, C. S., et al. (2005). Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity. Plant Physiol. 138, 2033–2047. doi: 10.1104/pp.105.061598
Koch, A., Biedenkopf, D., Furch, A., Weber, L., Rossbach, O., Abdellatef, E., et al. (2016). An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 12:e1005901. doi: 10.1371/journal.ppat.1005901
Koch, A., Kumar, N., Weber, L., Keller, H., Imani, J., and Kogel, K.-H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase–encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. U.S.A. 110, 19324–19329. doi: 10.1073/pnas.1306373110
Lepesheva, G. I., and Waterman, M. R. (2007). Sterol 14α-demethylase cytochrome P450 (CYP51), a P450 in all biological kingdoms. Biochim. Biophys. Acta 1770, 467–477. doi: 10.1016/j.bbagen.2006.07.018
Li, Y.-B., Xu, R., Liu, C., Shen, N., Han, L.-B., and Tang, D. (2020). Magnaporthe oryzae fimbrin organizes actin networks in the hyphal tip during polar growth and pathogenesis. PLoS Pathog. 16:e1008437. doi: 10.1371/journal.ppat.1008437
Liu, S., and Dean, R. A. (1997). G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant Microbe Interact. 10, 1075–1086. doi: 10.1094/MPMI.1997.10.9.1075
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408.
Mao, Y.-B., Cai, W.-J., Wang, J.-W., Hong, G.-J., Tao, X.-Y., Wang, L.-J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495. doi: 10.1093/pcp/pch048
Nelson, D. R., Schuler, M. A., Paquette, S. M., Werck-Reichhart, D., and Bak, S. (2004). Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135, 756–772. doi: 10.1104/pp.104.039826
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Okano, Y., Miki, D., and Shimamoto, K. (2008). Small interfering RNA (siRNA) targeting of endogenous promoters induces DNA methylation, but not necessarily gene silencing, in rice. Plant J. 53, 65–77.
Ou, S. (1980). Pathogen variability and host resistance in rice blast disease. Ann. Rev. Phytopathol. 18, 167–187.
Panwar, V., McCallum, B., and Bakkeren, G. (2013). Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the Barley stripe mosaic virus. Plant Mol. Biol. 81, 595–608. doi: 10.1007/s11103-013-0022-7
Park, C.-H., Chen, S., Shirsekar, G., Zhou, B., Khang, C. H., Songkumarn, P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern–triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Patel, M., Dewey, R. E., and Qu, R. (2013). Enhancing Agrobacterium tumefaciens-mediated transformation efficiency of perennial ryegrass and rice using heat and high maltose treatments during bacterial infection. Plant cell Tissue Organ Cult. 114, 19–29.
Pelish, H. E., Peterson, J. R., Salvarezza, S. B., Rodriguez-Boulan, E., Chen, J.-L., Stamnes, M., et al. (2006). Secramine inhibits Cdc42-dependent functions in cells and Cdc42 activation in vitro. Nat. Chem. Biol. 2, 39–46. doi: 10.1038/nchembio751
Qi, T., Zhu, X., Tan, C., Liu, P., Guo, J., Kang, Z., et al. (2018). Host-induced gene silencing of an important pathogenicity factor P s CPK 1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol. J. 16, 797–807. doi: 10.1111/pbi.12829
Ramankutty, N., Mehrabi, Z., Waha, K., Jarvis, L., Kremen, C., Herrero, M., et al. (2018). Trends in global agricultural land use: Implications for environmental health and food security. Annu. Rev. Plant Biol. 69, 789–815.
Raruang, Y., Omolehin, O., Hu, D., Wei, Q., Han, Z.-Q., Rajasekaran, K., et al. (2020). Host induced gene silencing targeting Aspergillus flavus aflM reduced aflatoxin contamination in transgenic maize under field conditions. Front. Microbiol. 11:754. doi: 10.3389/fmicb.2020.007
Saksmerprome, V., Charoonnart, P., Gangnonngiw, W., and Withyachumnarnkul, B. (2009). A novel and inexpensive application of RNAi technology to protect shrimp from viral disease. J. Virol. Methods 162, 213–217. doi: 10.1016/j.jviromet.2009.08.010
Sanford, J., and Johnston, S. (1985). The concept of parasite-derived resistance—deriving resistance genes from the parasite’s own genome. J. Theor. Biol. 113, 395–405.
Sanju, S., Siddappa, S., Thakur, A., Shukla, P. K., Srivastava, N., Pattanayak, D., et al. (2015). Host-mediated gene silencing of a single effector gene from the potato pathogen Phytophthora infestans imparts partial resistance to late blight disease. Funct. Integr. Genomics 15, 697–706. doi: 10.1007/s10142-015-0446-z
Sarkar, A., and Roy-Barman, S. (2021). Spray-induced silencing of pathogenicity gene MoDES1 via exogenous double-stranded RNA can confer partial resistance against fungal blast in Rice. Front. Plant Sci. 12:733129. doi: 10.3389/fpls.2021.733129
Seck, P. A., Diagne, A., Mohanty, S., and Wopereis, M. (2012). Crops that feed the world 7: Rice. Food Secur. 4, 7–24.
Shabalina, S. A., and Koonin, E. V. (2008). Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 23, 578–587.
Singh, V. K., Singh, A., Singh, S., Ellur, R. K., Choudhary, V., Sarkel, S., et al. (2012). Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker assisted backcross breeding. Field Crops Res. 128, 8–16.
Soanes, D. M., Kershawus Magnaporthe, M. J., Cooley, R. N., and Talbot, N. J. (2002). Regulation of the MPG1 hydrophobin gene in the rice blast fung grisea. Mol. Plant Microbe Interac 15, 1253–1267. doi: 10.1094/MPMI.2002.15.12.1253
Song, Y., and Thomma, B. P. (2018). Host-induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis. Mol. Plant Pathol. 19, 77–89. doi: 10.1111/mpp.12500
Subach, F. V., Patterson, G. H., Manley, S., Gillette, J. M., Lippincott-Schwartz, J., and Verkhusha, V. V. (2009). Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153–159. doi: 10.1038/nmeth.1298
Talbot, N. J., and Wilson, R. A. (2009). Under pressure: investigating the biology of plant infection by Magnaporthe oryza. Nat Rev Microbiol 7, 185–195. doi: 10.1038/nrmicro2032
Tan, F. L., and Yin, J. Q. (2005). Application of RNAi to cancer research and therapy. Front. Biosci. 10:1946–1960. doi: 10.2741/1670
Tang, G., and Galili, G. (2004). Using RNAi to improve plant nutritional value: From mechanism to application. Trends Biotechnol. 22, 463–469. doi: 10.1016/j.tibtech.2004.07.009
Thakare, D., Zhang, J., Wing, R. A., Cotty, P. J., and Schmidt, M. A. (2017). Aflatoxin-free transgenic maize using host-induced gene silencing. Sci. Adv. 3:e1602382. doi: 10.1126/sciadv.1602382
von Kleist, L., and Haucke, V. (2012). At the crossroads of chemistry and cell biology: Inhibiting membrane traffic by small molecules. Traffic 13, 495–504. doi: 10.1111/j.1600-0854.2011.01292.x
Wang, B.-h, Ebbole, D. J., and Wang, Z.-h (2017). The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. J. Integr. Agric. 16, 2746–2760.
Wang, M., and Dean, R. A. (2020). Movement of small RNAs in and between plants and fungi. Mol. Plant Pathol. 21, 589–601.
Wang, M., Weiberg, A., Lin, F.-M., Thomma, B. P., Huang, H.-D., and Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. plants 2, 1–10. doi: 10.1038/nplants.2016.151
Weiberg, A., Wang, M., Lin, F.-M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Wilson, R. A., Gibson, R. P., Quispe, C. F., Littlechild, J. A., and Talbot, N. J. (2010). An NADPH-dependent genetic switch regulates plant infection by the rice blast fungus. Proc. Natl. Acad. Sci. 107, 21902–21907.
Xu, J., Wang, X., Li, Y., Zeng, J., Wang, G., Deng, C., et al. (2018). Host-induced gene silencing of a regulator of G protein signalling gene (Vd RGS 1) confers resistance to Verticillium wilt in cotton. Plant Biotechnol. J. 16, 1629–1643. doi: 10.1111/pbi.12900
Yi, M., Chi, M.-H., Khang, C. H., Park, S.-Y., Kang, S., Valent, B., et al. (2009). The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell 21, 681–695. doi: 10.1105/tpc.107.055988
Yin, C., Jurgenson, J. E., and Hulbert, S. H. (2011). Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 24, 554–561. doi: 10.1094/MPMI-10-10-0229
Yoshida, Y. (1993). “Lanosterol 14α-demethylase (cytochrome P45014DM),” in in Cytochrome P450, (Berlin: Springer), 627–639.
Yoshida, Y., Noshiro, M., Aoyama, Y., Kawamoto, T., Horiuchi, T., and Gotoh, O. (1997). Structural and evolutionary studies on sterol 14-demethylase P450 (CYP51), the most conserved P450 monooxygenase: II. Evolutionary analysis of protein and gene structures. J. Biochem. 122, 1122–1128. doi: 10.1093/oxfordjournals.jbchem.a021870
Zeigler, R. S., Leong, S., and Teng, P. S. (1994). Rice blast disease. Los Banos: International Rice Research Institute.
Zhang, H., Guo, J., Voegele, R. T., Zhang, J., Duan, Y., Luo, H., et al. (2012). Functional characterization of calcineurin homologs PsCNA1/PsCNB1 in Puccinia striiformis f. sp. tritici using a host-induced RNAi system. PLoS One 7:e49262. doi: 10.1371/journal.pone.0049262
Zhang, H., Zhao, Q., Liu, K., Zhang, Z., Wang, Y., and Zheng, X. (2009). MgCRZ1, a transcription factor of Magnaporthe grisea, controls growth, development and is involved in full virulence. FEMS Microbiol. Lett. 293, 160–169. doi: 10.1111/j.1574-6968.2009.01524.x
Zhang, S., and Xu, J.-R. (2014). Effectors and effector delivery in Magnaporthe oryzae. PLoS Pathog. 10:e1003826.
Zhao, M., Wang, C., Wan, J., Li, Z., Liu, D., Yamamoto, N., et al. (2021). Functional validation of pathogenicity genes in rice sheath blight pathogen Rhizoctonia solani by a novel host-induced gene silencing system. Mol. Plant Pathol. 22, 1587–1598. doi: 10.1111/mpp.13130
Zhou, B., Bailey, A., Niblett, C., and Qu, R. (2016). Control of brown patch (Rhizoctonia solani) in tall fescue (Festuca arundinacea Schreb.) by host induced gene silencing. Plant Cell Rep. 35, 791–802. doi: 10.1007/s00299-015-1921-7
Keywords: rice blast, host-induced gene silencing, transgenic plant, RNA silencing, cross-kingdom communication
Citation: Wang M and Dean RA (2022) Host induced gene silencing of Magnaporthe oryzae by targeting pathogenicity and development genes to control rice blast disease. Front. Plant Sci. 13:959641. doi: 10.3389/fpls.2022.959641
Received: 01 June 2022; Accepted: 25 July 2022;
Published: 11 August 2022.
Edited by:
Mahmut Tör, University of Worcester, United KingdomReviewed by:
Laurence Veronique Bindschedler, Royal Holloway, University of London, United KingdomAnton Hartmann, Ludwig Maximilian University of Munich, Germany
Copyright © 2022 Wang and Dean. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralph A. Dean, cmFscGhfZGVhbkBuY3N1LmVkdQ==
 Mengying Wang
Mengying Wang Ralph A. Dean
Ralph A. Dean