- School of Life Sciences, Jiangsu University, Zhenjiang, Jiangsu, China
The plant-specific SHORT INTERNODES/STYLISH (SHI/STY) proteins belong to a family of transcription factors that are involved in the formation and development of early lateral roots. However, the molecular evolution of this family is rarely reported. Here, a total of 195 SHI/STY genes were identified in 21 terrestrial plants, and the Brassica species is the focus of our research. Their physicochemical properties, chromosome location and duplication, motif distribution, exon-intron structures, genetic evolution, and expression patterns were systematically analyzed. These genes are divided into four clades (Clade 1/2/3/4) based on phylogenetic analysis. Motif distribution and gene structure are similar in each clade. SHI/STY proteins are localized in the nucleus by the prediction of subcellular localization. Collinearity analysis indicates that the SHI/STYs are relatively conserved in evolution. Whole-genome duplication is the main factor for their expansion. SHI/STYs have undergone intense purifying selection, but several positive selection sites are also identified. Most promoters of SHI/STY genes contain different types of cis-elements, such as light, stress, and hormone-responsive elements, suggesting that they may be involved in many biological processes. Protein–protein interaction predicted some important SHI/STY interacting proteins, such as LPAT4, MBOATs, PPR, and UBQ3. In addition, the RNA-seq and qRT-PCR analysis were studied in detail in rape. As a result, SHI/STYs are highly expressed in root and bud, and can be affected by Sclerotinia sclerotiorum, drought, cold, and heat stresses. Moreover, quantitative real-time PCR (qRT-PCR) analyses indicates that expression levels of BnSHI/STYs are significantly altered in different treatments (cold, salt, drought, IAA, auxin; ABA, abscisic acid; 6-BA, cytokinin). It provides a new understanding of the evolution and expansion of the SHI/STY family in land plants and lays a foundation for further research on their functions.
Introduction
Gene families play important roles in the evolution of plants, but their functions and evolutionary history are not well understood. Ancient SHORT INTERNODES/STYLISH (SHI/STY) protein family of transcription factors have been found in all sequenced land plant species, from Physcomitrella patens to flowering plants, like Arabidopsis thaliana, Brassica napus, Zea mays, and Glycine max. The extensive presence of these proteins in terrestrial plants suggests that they may participate in important functions for plant growth and development (Kuusk et al., 2006). In Arabidopsis, ten SHI/STY members were reported, containing SHI, STY1, STY2, LATERAL ROOT PRIMORDIUM1 (LRP1), SHI RELATED SEQUENCE3 (SRS3), SRS4, SRS5, SRS6, SRS7, SRS8 (Singh et al., 2020). They have a highly conserved IGGH domain that covers a RING-like zinc finger domain (CX2CX7CX4CX2C2X6C) for binding to RNA, protein, and lipid substrates (Fridborg et al., 2001; Kuusk et al., 2006). The zinc finger domain is composed of one or several small protein motifs, which can contact the target molecules by forming stable finger-like projections (Klug, 1999; Laity et al., 2001).
The cysteine arrangement in SHI/STYs (about 111–139 amino acids), H-XI2-C-XX2C-Xlo-14-C-X2-C-X4-H-X2-C-X ~ -C or C-X2-CXI2-C-X2-C-X7-C (the X could be any amino acid), is conserved in the Histidine and Cysteine domain of the Zinc-binding site of protein kinase C in their activation domain (Hubbard et al., 1991). Furthermore, the IGGH domain of the SHI/STY proteins contains acidic amino acids and can act as a transcriptional activator (Fridborg et al., 2001; Singh et al., 2020).
SHI/STY genes not only regulate the growth and development of plants, but also respond to hormones and some abiotic stresses (Zhao et al., 2020). Several studies have shown that SHI/STY genes are associated with auxin signaling pathways in terrestrial plants and play important functions in the regulation of plants tissues and other hormonal pathways (Eklund et al., 2010; Zawaski et al., 2011; Islam et al., 2013; Youssef et al., 2017). In addition, some members are involved in auxin biosynthesis by regulating genes related to photomorphogenesis (Eklund et al., 2010; Staldal et al., 2012; Baylis et al., 2013; Yuan et al., 2018). The phenotype of atshi mutant is similar to that of the gibberellin (GA) biosynthesis deficient mutant, suggesting that AtSHI may take part in the GA signaling pathway (Fridborg et al., 1999). Overexpression of the SHI gene in Kalanchoe and Poinsettia results in a compact phenotype (Lutken et al., 2010; Islam et al., 2013). SHI, STY1, and STY2 genes can synergistically promote the development of pistil, stamen, and leaf, and their overexpression can also inhibit stem elongation and tapetal dehiscence (Kuusk et al., 2006; Kim et al., 2010). Rice LRP is related to plant height through the GA signaling pathway (Duan et al., 2019). SHI/STYs are vital for the formation and development of the early lateral root in Arabidopsis (Smith and Fedoroff, 1995). Furthermore, LRP1 can form complexes with SRS6, SHI, SRS3, SRS7, and STY1 and participate in auxin signal transduction and chromatin modification during lateral root development (De Smet et al., 2008; De Rybel et al., 2012). SWIRM domain PAO protein (SWP1) can inhibit LRP1 through histone deacetylation of chromatin. Insertion mutagenesis of SWP1 or overexpression of LRP1 can reduce this inhibition and increase root elongation (Krichevsky et al., 2009). In addition, LRP1 and STY1 regulate the expression of YUCCA 4 (YUC4) during auxin synthesis (Eklund et al., 2010; Singh et al., 2020). It is reported that STY1 (SRS1) not only regulates auxin biosynthesis and affects the apical pattern of stamen, but also functions in cell proliferation and flowering (Eklund et al., 2010; Staldal et al., 2012).
About 12–20 million years ago, the segregation between Brassica species and Arabidopsis occurred (Yang et al., 1999; Town et al., 2006). Brassica species have achieved triploidization of the whole genome about 5–15 million years ago (Beilstein et al., 2010; Wang et al., 2011). After that, Brassica oleracea and Brassica rapa were separated about 4.6 million years ago (Liu et al., 2014). B. napus (AACC 2n = 38) is an allotetraploid, produced by the hybridization of B. rapa (AA 2n = 20) and B. oleracea (CC 2n = 18) naturally, about 7,500 years ago (Chalhoub et al., 2014). It has been reported that every Brassica species has undergone gene duplication events during evolution. The whole-genome sequencing and assembly had been completed in B. napus. 101,040 gene models were generated from 35.5 Gb of sequencing data. The assembled C subgenome (525.8 Mb) was larger than the A subgenome (314.2 Mb; Xie et al., 2020, Yang et al., 2020).
In this study, 195 members of the SHI/STY gene family were identified from the 21 land plants, including P. patens and Selaginella moellendorffii, and a variety of monocotyledons and dicotyledons in the Phytozome database1 (Goodstein et al., 2012). Based on the analysis of the molecular evolution characteristics of the SHI/STY gene family in terrestrial plants, an evolutionary map was also proposed.
Materials and methods
Retrieval of SHI/STYs in land plants
Here, all Arabidopsis SHI/STY proteins sequences were used to perform a BLAST search in the Phytozome database2 (Goodstein et al., 2012) and the Genoscope database for B. napus (Chalhoub et al., 2014).3 Secondly, the Batch CD-Search4 (Marchler-Bauer and Bryant, 2004) was used to screen again the conserved domain of SHI/STYs, and only the sequences with a RING-like zinc-finger domain (DUF702) were selected for further analysis (Fridborg et al., 2001). The physicochemical properties of these SHI/STY proteins were predicted with the ProtParam (Gasteiger et al., 2003). Their subcellular locations were predicted with the CELLO (Chen et al., 2011, 2017).
Phylogenetic analysis and characterization of SHI/STYs
To investigate the evolutionary patterns of these plant SHI/STYs, the phylogenetic trees were constructed with the neighbor joining (NJ) method in MEGA7.0.21 (Tamura et al., 2013). The reliability of these trees was evaluated by 1,000 bootstrap replications. Multiple Em for Motif Elicitation (MEME, http://meme-suite.org/tools/meme; Bailey et al., 2006) was used to analyze the conserved motifs of these SHI/STY proteins. The Amazing Optional Gene Viewer in TBtools (Chen et al., 2020) was used to present the distribution of the conserved motifs and the exon-intron structures of SHI/STYs.
Cis-acting element analysis
To analyze the cis-acting elements of the SHI/STY promoters, TBtools (Chen et al., 2020) was used to obtain the 2,000 bp promoter sequences from the upstream coding sequence (CDS). All the promoters of SHI/STY genes in the Brassica species were acquired from NCBI.5 Three tools, the Gtf/Gff3 Sequence Extractor, the Fasta Extractor, and Amazing Fasta Extractor in TBtools were used to extract the information of promoters. The distribution of cis-acting elements in SHI/STYs was analyzed with Plantcare6 (Lescot et al., 2002).
Chromosomal location, identification of paralogous SHI/STY Genes
The information for the length and location of SHI/STYs in the Brassica species were obtained from the Phytozome7 (Goodstein et al., 2012) and the Genoscope,8 respectively. Multiple collinear scanning toolkits (MCScanX) in TBtools (Chen et al., 2020) were used to analyze synteny relationships and gene duplication events among B. napus, A. thaliana, B. oleracea, and B. rapa. The Ks and Ka values of gene pairs were calculated in Tbtools. Duplication time of these gene pairs can be calculated by the frequency (λ) of 1.4 × 10−8 homogeneous substitutions per site each year according to T = Ks/2λ (Wang et al., 2011).
SHI/STYs expression analysis in Brassica napus
To investigate the expression pattern of SHI/STYs in B. napus, their RNA-seq data were obtained from the Genoscope database9 (Chalhoub et al., 2014). Mev in SOURCEFORGE10 was used to analyze their tissue expression levels (Sullivan et al., 2011). In addition, RNA-seq data under the Sclerotinia sclerotiorum stress, drought, and heat treatment were obtained from NCBI (GSE169299 and GSE156029), respectively (Clough and Barrett, 2016). These data were standardized based on the log2 scale and clustered and visualized with TBtools.
Selective pressure analysis
Likelihood-ratio test was used to compare the implemented models. Here, M8 (ωs ≥ 1), M8a (ωs = 1), M7 (beta), and M5 (gamma) evolution models were used to evaluate selective pressure (Chalhoub et al., 2014). CDS sequences of SHI/STY genes were used to calculate selective pressure with Selecton (Chalhoub et al., 2014).11 In addition, I-TASSER12 (Yang et al., 2015) was used to predict the tertiary structures of BnaA04g04120D, BnaA07g12710D, BnaA03g59180D, and Potri.003G85901.1.p proteins, which were selected as query sequences in different clades in the selective pressure analysis.
Protein–protein interaction network analysis of SHI/STYs
The protein–protein interaction network of SHI/STYs in B. napus was predicted with STRING (version 11.0; https://string-db.org/cgi/input.pl; Szklarczyk et al., 2019). There is no relevant data on B. napus in STRING, but B. rapa is highly homologous to B. napus. Here, all BnSHI/STYs protein sequences were used as the basis of the query to search for the interaction network of homologous proteins in B. rapas. The full network type was used to complete the analysis, and the edges represent the physical and functional protein associations. The minimum interaction score required was set to intermediate confidence (0.400).
Plant material RNA extraction and qRT-PCR
Rape seeds were germinated on Petri dishes in the greenhouse under the conditions of 50 μmol/m2/s light intensity and 70% relative humidity and then grown at 20 ± 5°C, 16 h light /8 h dark. Two-week-old seedlings were treated with Hoagland liquid medium containing plant hormone (50 μM ABA, 10 μM IAA, 75 μM 6-BA) for 24 h. Meanwhile, two-week-old seedlings were treated with cold (4°C), salt (200 mM NaCl) and drought (15%PEG) for 24 h. The whole treated seedlings were then collected and immediately stored at −80°C for RNA extraction. The extraction of total RNA and subsequent synthesis of cDNA were described by Sarwar et al. (Sarwar et al., 2021). Actin gene (GenBank ID: XM_013858992) was used as an internal reference. Relative expression level of the BnSHI/STYs was measured by the 2−∆∆Ct method. Rape seedlings without any treatment were used as the control. The significant difference between different treatments was measured by T-test, and the results were visualized by GraphPad Prism7.4 software (Drafor et al., 2021). All gene-specific primers (Supplementary Table S1) used in this study were designed with BrassicaEDB.13
Results
Identification and classification of SHI/STY proteins
To identify SHI/STY members in land plants, the amino acid sequences of 10 SHI/STYs in Arabidopsis were used as queries to perform the BLAST search in the Phytozome database14 (Goodstein et al., 2012) and the Genoscope database15 (Chalhoub et al., 2014), respectively. As a result, 195 SHI/STY genes were identified in 21 land plants. In addition, the DUF702 domain was predicted in all SHI/STY protein sequences by Batch CD-Search16 (Marchler-Bauer and Bryant, 2004).
The physical and chemical properties of these identified members were analyzed (Supplementary Table S2). These genes encoded 95–603 amino acids with 9.35 to 61.83 KD and their isoelectric points ranged from 5.56 to 10.12. Subcellular location revealed that 91.8% of SHI/STY proteins were predicted to only locate in the nucleus. In addition, ten members were predicted as nuclear and extracellular localization proteins. This is consistent with the previous report that SHI/STYs are mainly localized in nuclear (Zhang et al., 2015).
In order to study their evolutionary relationship in land plants, these SHI/STY protein sequences were used to construct phylogenetic trees with the NJ method (Supplementary Figure S1). Combined with the results of conserved motif analysis and phylogenetic tree analysis, SHI/STY proteins in all land plants evolved into four clades (Figure 1; Supplementary Figure S1). Clade1 includes 63 SHI/STYs in these 21 land plants. In Clade 1, two and four members existed in P. patens and S. moellendorffii, respectively, which constitute the earliest SHI/STY members of terrestrial plants. Clade 2 is the largest one, which contains 77 members and all of them are dicotyledons. In addition, Clade 3 consists of 46 SHI/STYs, all of which come from both monocotyledons and dicotyledons. Clade 4 is a new evolutionary clade. Compared to the other three clades, it only contains 9 SHI/STY genes of dicotyledons.
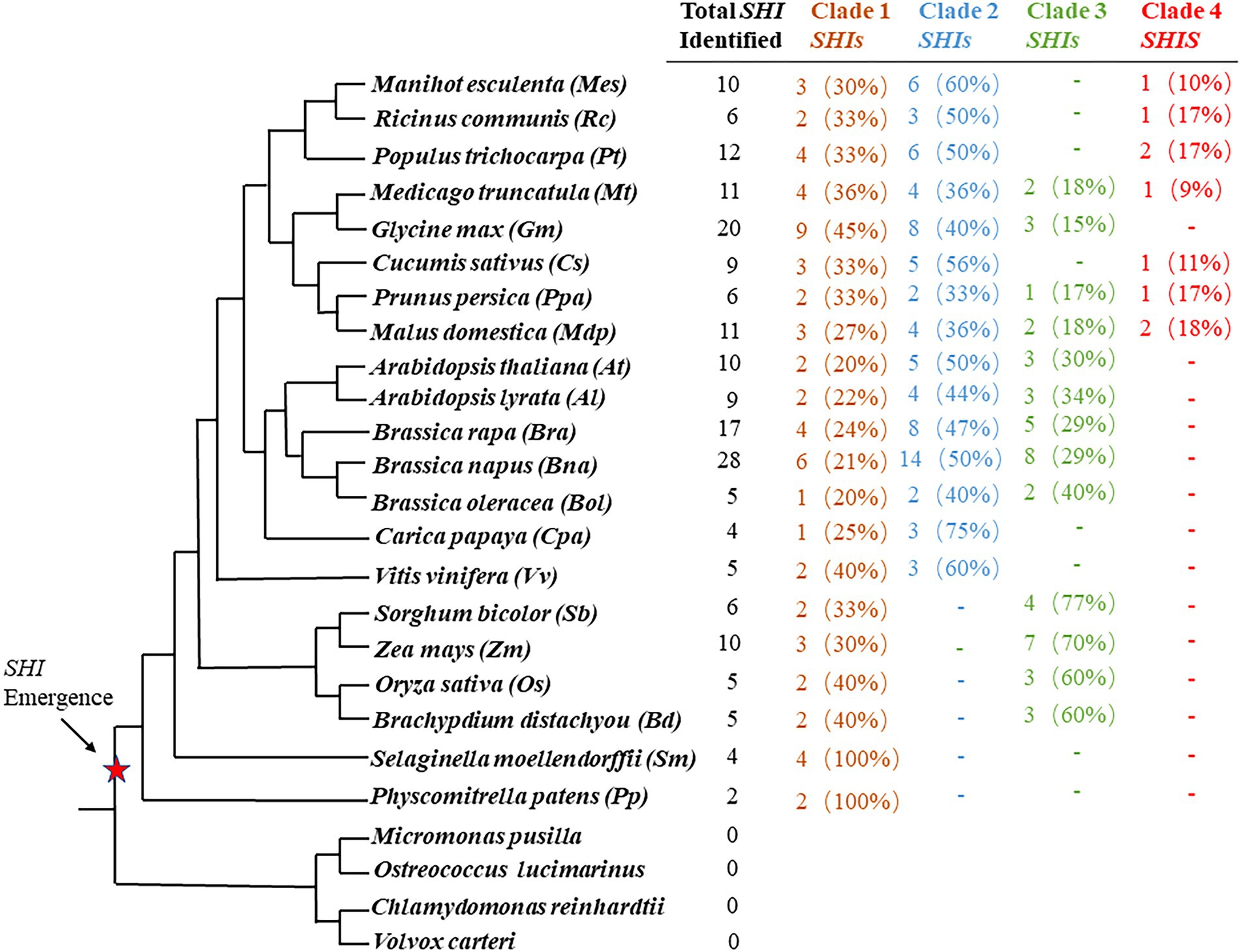
Figure 1. SHI/STY genes are identified in land plants. The number of SHI/STY genes identified in each sequenced plant genome was listed in sequence. Percentages for each type are listed in parentheses. The red star marks indicate when SHI/STY appeared during the evolution of land plants. “-” indicates that there is no SHI/STY in this species.
Motif analyses of SHI/STY proteins in land plants
To study the homology domain and conservation degree of the SHI/STY proteins in land plants, MEME was used to perform motif analyses. As a result, 16 conserved motifs were identified (Figure 2; Supplementary Figure S2). The motif distribution of SHI/STY proteins was similar in the same clade. The RING-like zinc finger domain (Motif 1) and the IGGH domain (Motif 2) are conserved in most of the SHI/STYs. Besides, Motif 14 and Motif 4 are conserved in SHI/STYs. In addition, different motif distribution is found (Figure 3). For example, Motif 10 and Motif 16 are only identified in Clade 1, and Motif 8 and Motif 15 are conserved in Clade 2. The number of motifs is the highest in Clade 2 and the lowest in Clade 4. It can be predicted that the SHI/STY proteins might lost or obtain some motifs in the long process of genetic evolution.
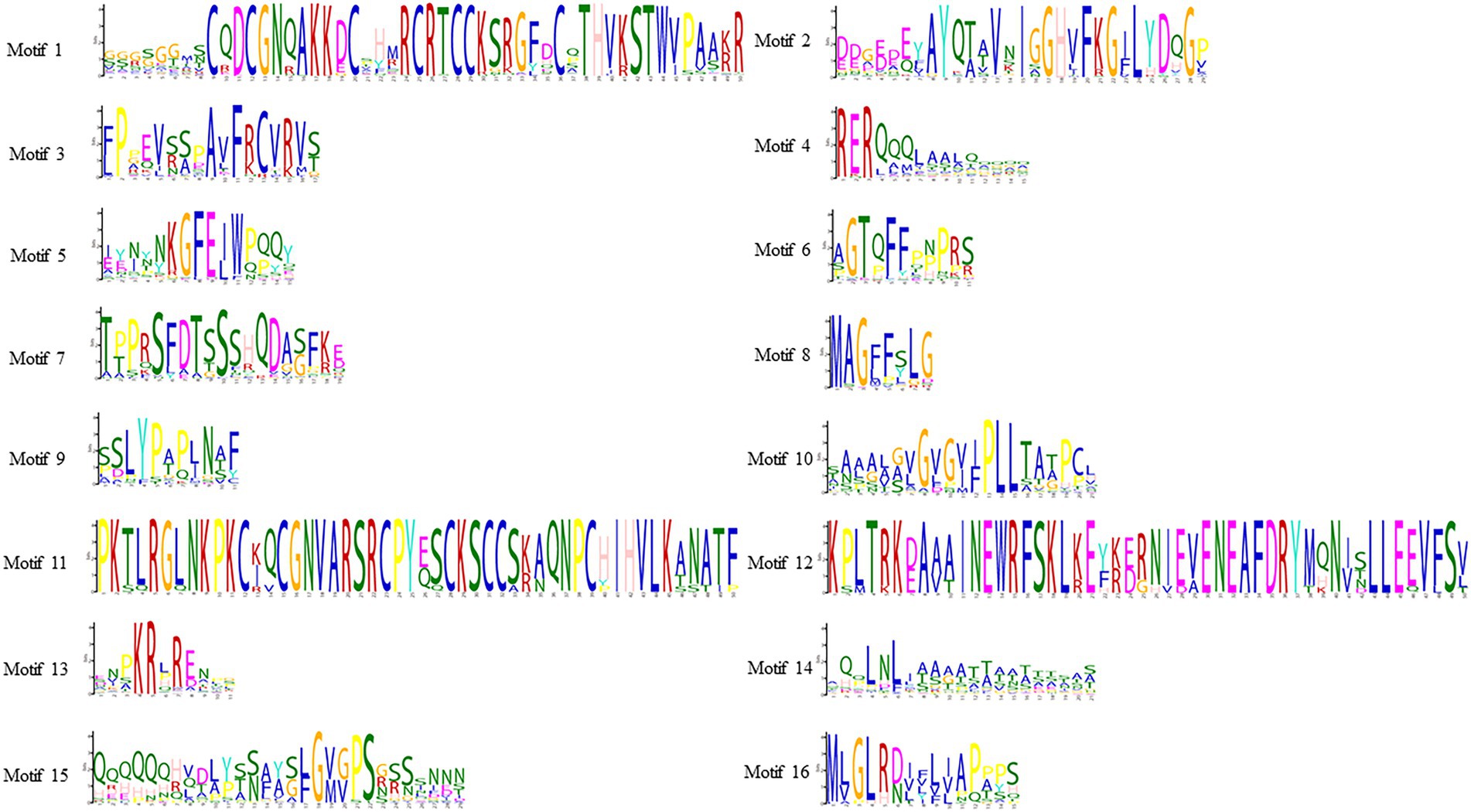
Figure 2. Sequence logos of conserved motifs in SHI/STY proteins. This logo represents the sequence of 16 motifs. The letters represent any amino acid, and the size represents how well it has been preserved at that location. The predicted motif sequence was obtained from the MEME website based on the sequences of SHI/STYs (http://meme-suite.org/tools/meme).
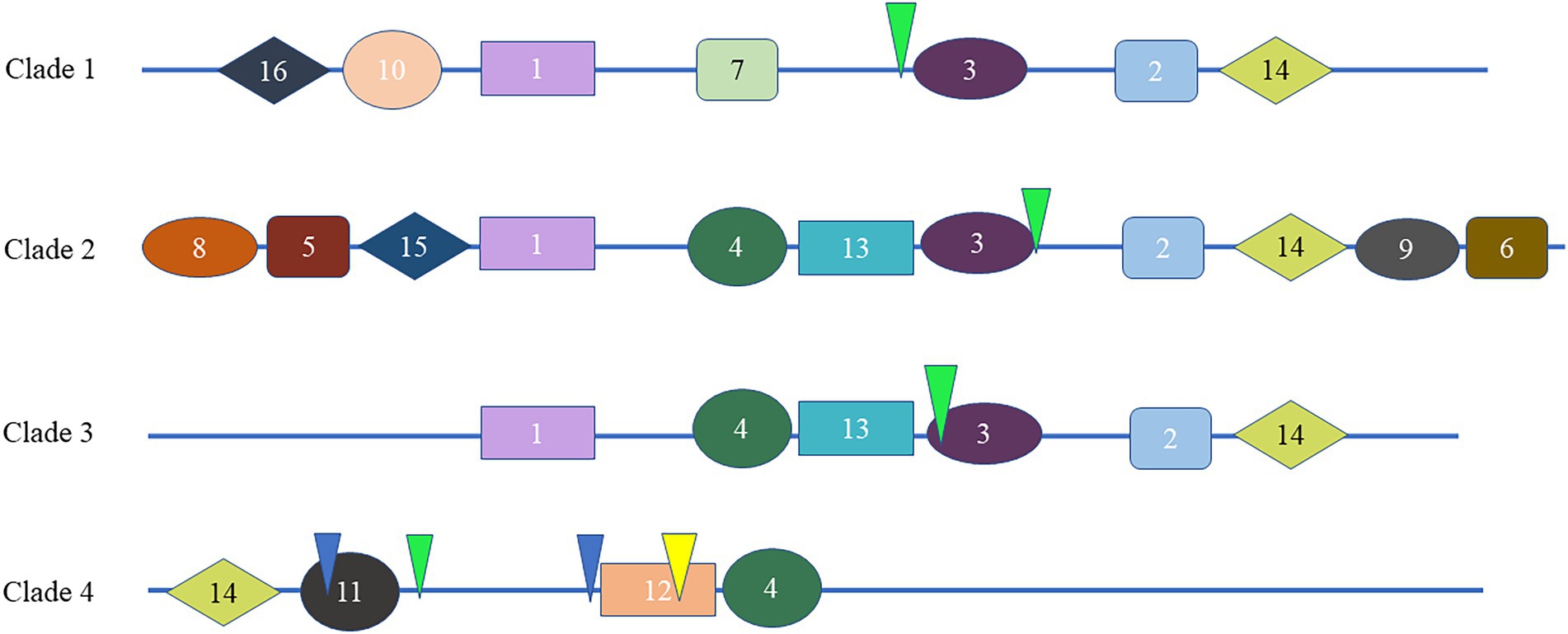
Figure 3. Different motif distribution of the SHI/STY proteins and exon–intron organization in different clades. The different colorful patterning represents the relative positions of motifs on the protein sequence in the different clades. The inverted triangles with yellow, green and blue marks indicate the insertion positions of phase 0, 1 and 2 introns, respectively.
To understand the diversity of SHI/STYs, the gene organization and intron phase distribution were analyzed (Figure 3). Genes with the same phylogenetic group usually have similar exon-intron structures. Statistical analysis showed that only one intron was conserved in Clades 1, 2, and 3, while four conserved introns were found in Clade 4. All eukaryotes have phase 0, phase 1 and phase 2 introns, among which phase 0 introns account for the highest proportion and have the highest conservation (Long et al., 1995). It is clear that the phase 1 introns were conserved in most SHI/STYs, while introns in Clade 4 are different from the others. The conserved introns in Clades 1–3 suggest that the SHI/STY gene family is relatively conserved during the evolution of land plants. However, the new intron pattern in Clade 4 reflects the structure change of SHI/STY genes during the evolution from lower plants to higher ones.
Chromosomal distribution and duplication of SHI/STY genes
It was reported that duplication occurred in both A and C subgenomes of B. napus (Chalhoub et al., 2014; Panchy et al., 2016). To analyze duplication events of SHI/STYs, MCScanX of TBtools was used to investigate their duplication events in the Brassica species (Figures 4, 5). BnSHI/STYs were first located onto the chromosomes of B. napus. The result shows that 13 BnSHI/STYs were located on C chromosomes, while 12 BnSHI/STYs were located on A chromosomes. Chromosome C07 contained the largest number of BnSHI/STY genes (four genes), but no BnSHI/STY was identified on Chromosomes C03, C05, A03, A05 and A08.
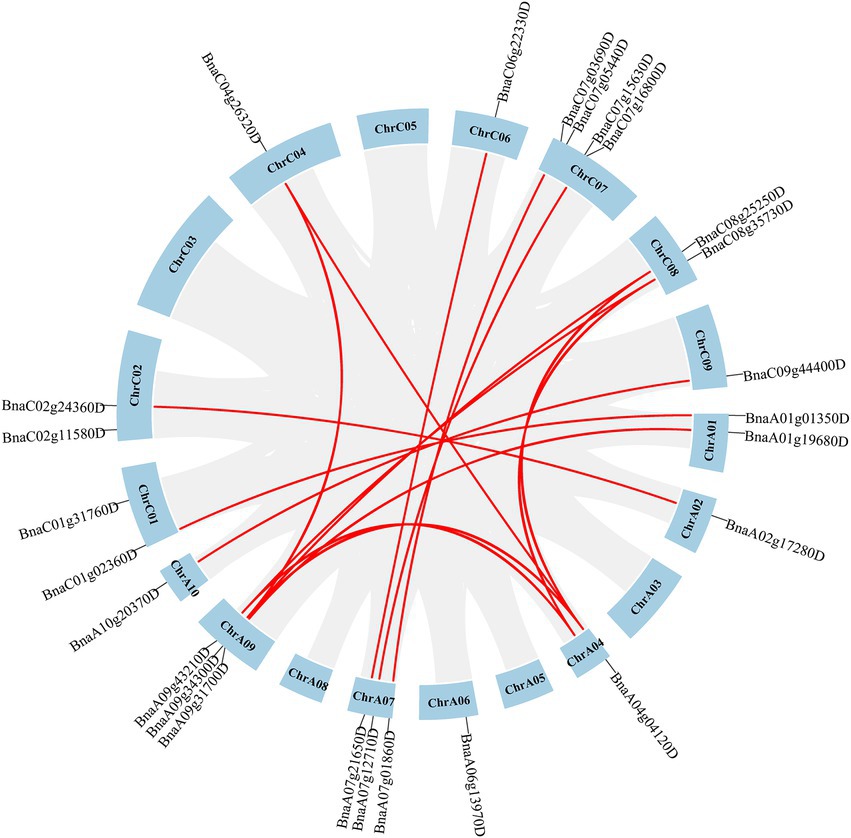
Figure 4. Chromosome distribution of BnSHI/STYs. 25 BnSHI/STYs are distributed on 19 chromosomes of Brassica napus. In addition, 3 BnSHI/STYs are not mapped to chromosomes. The duplicated BnSHI/STY gene pairs were indicated in red lines. The number of chromosomes is displayed on each chromosome.
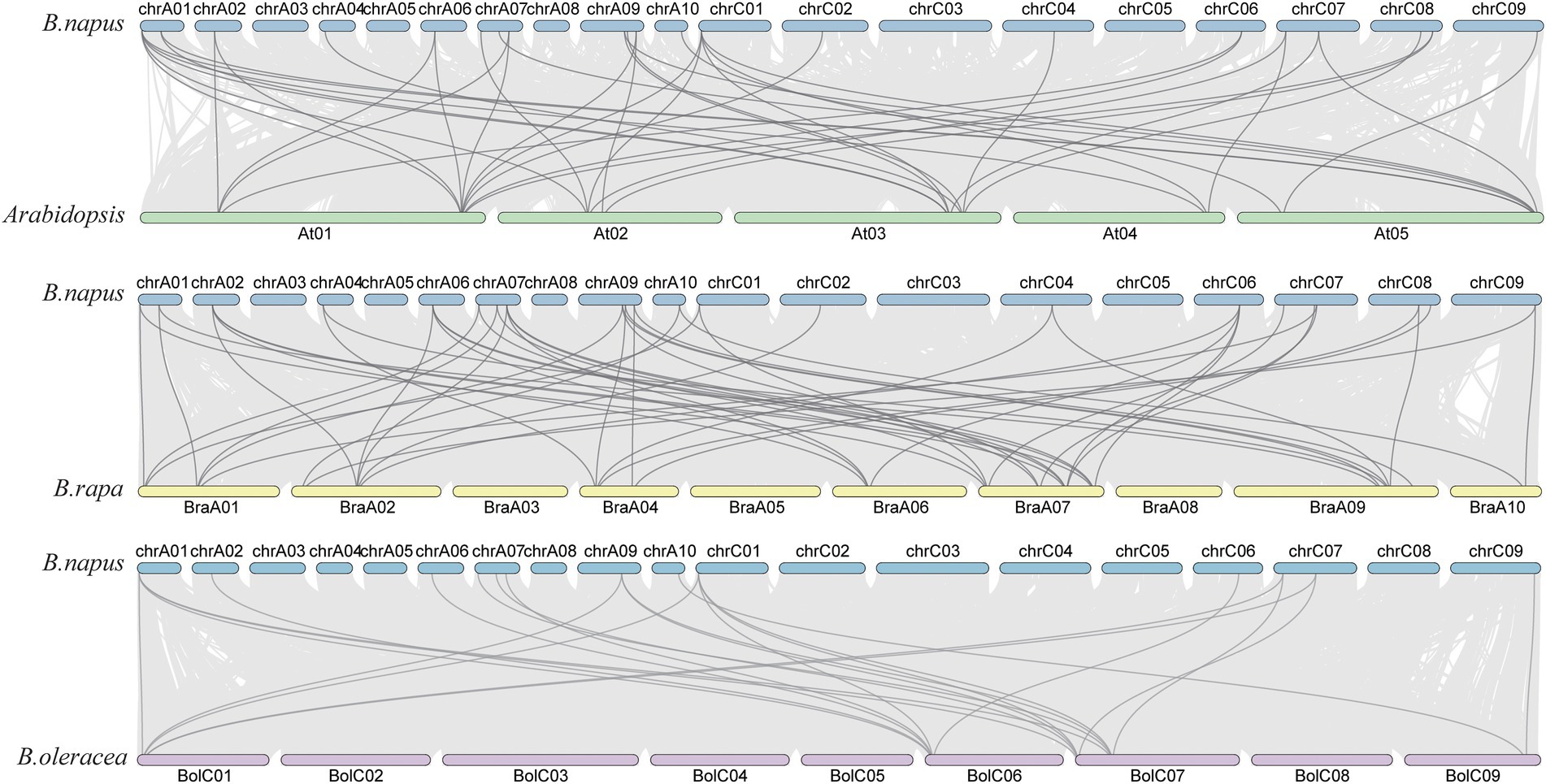
Figure 5. The collinear relationship of SHI/STYs on the chromosome. A chromosome is represented by a bar. The green, yellow, and purple bars represent the chromosomes of Arabidopsis, B. rapa, and B. oleracea, respectively. The collinear blocks (between the genomes of B. napus and Arabidopsis, B. napus and B. rapa, B. napus and B. oleracea) were indicated in the gray lines.
Next, syntenic relationships of the SHI/STYs between B. napus and other three species (B. rapa, B. oleracea, and Arabidopsis) were analyzed to investigate their evolutionary relationship. Their location maps are shown in Figure 5. Collinearity analysis revealed that B. napus had more orthologous SHI/STYs than Arabidopsis, B. rapa, and B. oleracea. B. rapa has the most syntenic SHI/STY genes with B. napus, followed by Arabidopsis. B. oleracea has the least syntenic SHI/STY genes with B. napus. Next, we calculated the divergence time of duplicated gene pairs in these four species (Supplementary Table S3). Among these SHI/STYs, 20 pairs of duplicate genes were identified, including 3 pairs in Arabidopsis, 11 pairs in B. napus, 5 pairs in B. olerace and 1 pair in B. rapa. SHI/STYs duplicated earliest at 113 million years ago in Arabidopsis, and followed by B. rapa and B. napus about 54 and 27 million years ago. In B. napus, 81.8% of duplicated gene pairs were caused by WGD. These results indicated that gene duplication promoted the amplification of SHI/STY genes in B. napus genome to a large extent, and WGD or segmental duplication was the main driver.
Analysis of the cis-acting element of SHI/STYs
To identify the functional differences of SHI/STYs in four Brassica species, we analyzed the cis-elements in the promoters of SHI/STY genes with PlantCARE software (Figure 6). three kinds of cis-acting elements were found in the promoter region of the SHI/STY gene family. Most cis-elements of BnSHI/STYs promoters are involved in plant growth and development, hormonal responsive and abiotic stress responsive. Some cis-elements, like meristem expression, endosperm expression, seed-specific regulation and light responsive are the main cis-elements involved in plant growth and development regulation. Light responsive related elements are most common in the SHI/STY promoters, indicating that the SHI/STY gene family could be induced by light to regulate plant growth and development. Low-temperature responsiveness and wound responsive are the main members of the regulation of abiotic stress. At the same time, we also found that elements related to hormone response, including salicylic acid responsiveness, MeJA-responsiveness, and gibberellin-responsiveness, accounted for a large proportion. These results suggest that SHI/STYs not only regulate plant growth and development, but also play an important role in hormone response and abiotic stress. However, an element related to endosperm expression was found only in the promoters of B. napus and B. rapas. MeJA-related responsiveness and salicylic acid (SA) responsiveness were identified mainly in the second category. We found that the number of MeJA responsiveness elements was the largest, followed by the SA responsiveness elements. In addition, some cis-acting elements related to abiotic stresse responses were identified, such as wound-responsive and low-temperature responsiveness.
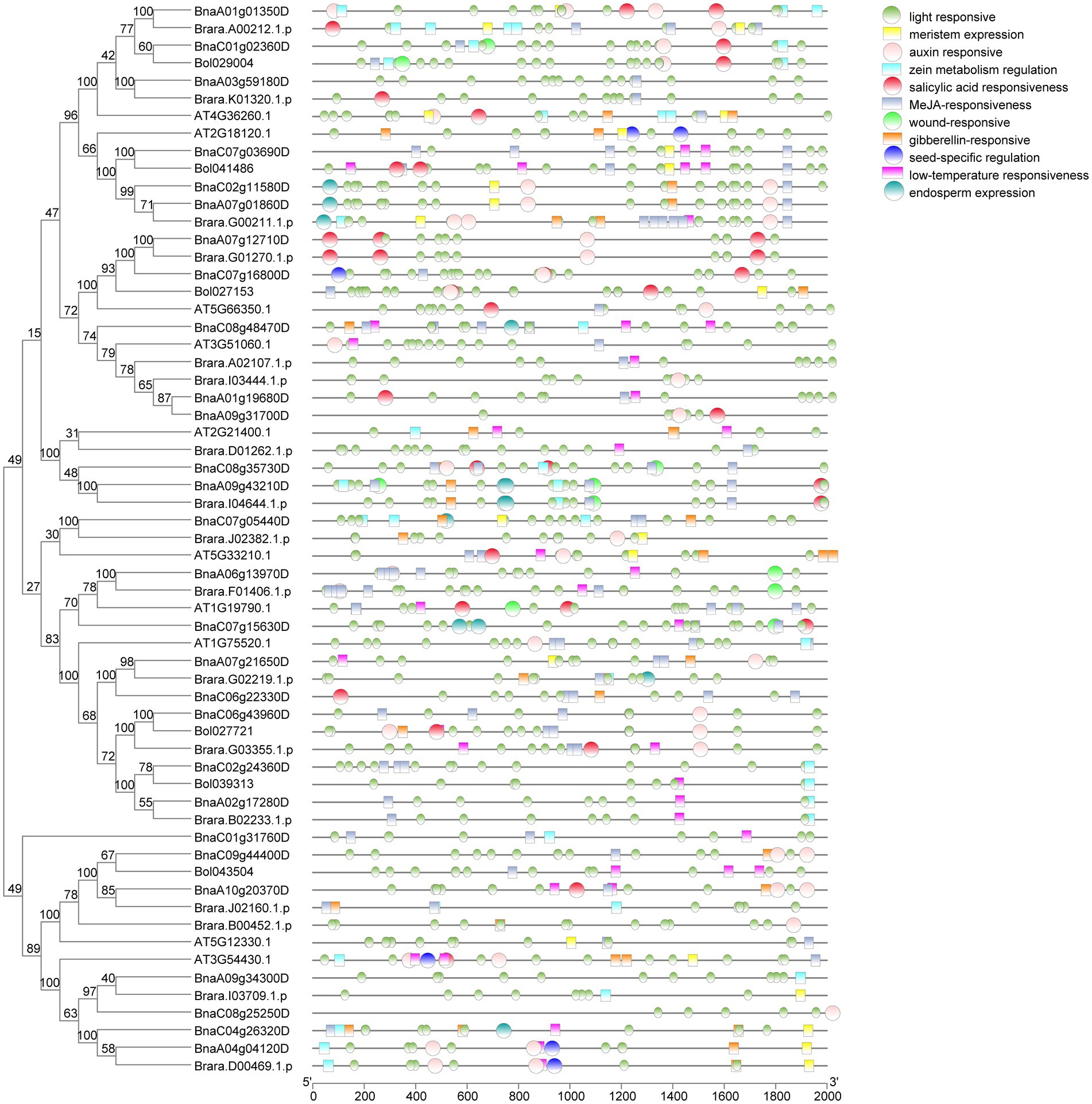
Figure 6. Cis-element analysis in the promoter regions of the SHI/STY genes. The PlantCARE software was used to determine the presence of different cis-acting elements. The different cis-elements were represented in different colored boxes.
Differences of the cis-elements in the promoter region between homologous genes may appear during the process of gene duplication, and the difference in cis-element might lead to different expressions of duplicated genes and increase the divergence of gene function. For example, there are more low-temperature responsiveness related elements in duplicated gene BnaA10g20370D than in BnaC09g44400D (Figure 6), and it could be found that the former were up-regulated when B. napus suffered cold. The cis-elements in the duplicated gene (BnaC07g12710D/BnaA06g13970D) were different. These two genes are expressed in different tissues, and BnaC07g12710D is highly expressed in the stem, but the other is expressed in the bud. Therefore, the difference in SHI/STY gene expression is caused by the difference in cis-elements.
Positive selection in SHI/STY gene family
To describe the selective pressure on different clades during the evolutionary process, the Ka/Ks values were calculated. As a result, the Ka/Ks values of the four clades were all less than 0.5, suggesting that the SHI/STY gene family could have experienced intense purifying selection in the long evolutionary process (Table 1). The Ka/Ks value of Clade 3 is the largest, ranging from 0.45 to 0.49, while the Ka/Ks value of Clade 1 is the smallest, ranging from 0.25 to 0.27. It indicates that the purification effect of Clade 1 is stronger than others. Here, four models (M7, M8, M8a, and M5) were used to calculate their Ka/Ks values. And only M8 and M5 models identified some positive selection sites in Clade 1, Clade 3 and Clade 4. Next, the tertiary structure of SHI/STY proteins was predicted to locate the positive selection sites (Table 1; Figure 7). The results showed that most of the positive selection sites were located in Motif 1 and Motif 2. These two motifs code the highly conserved IGGH domain and RING-like zinc finger domain, respectively. Its ability to bind to RNA, protein, and lipid substrates demonstrates that it functions as a transcriptional activator (Fridborg et al., 2001; Kuusk et al., 2006; Zhao et al., 2020). Amino acids in these two motifs are involved in maintaining the structure and functions of the motifs. The changes in these amino acids may have a significant effect on the functions of SHI/STY proteins.
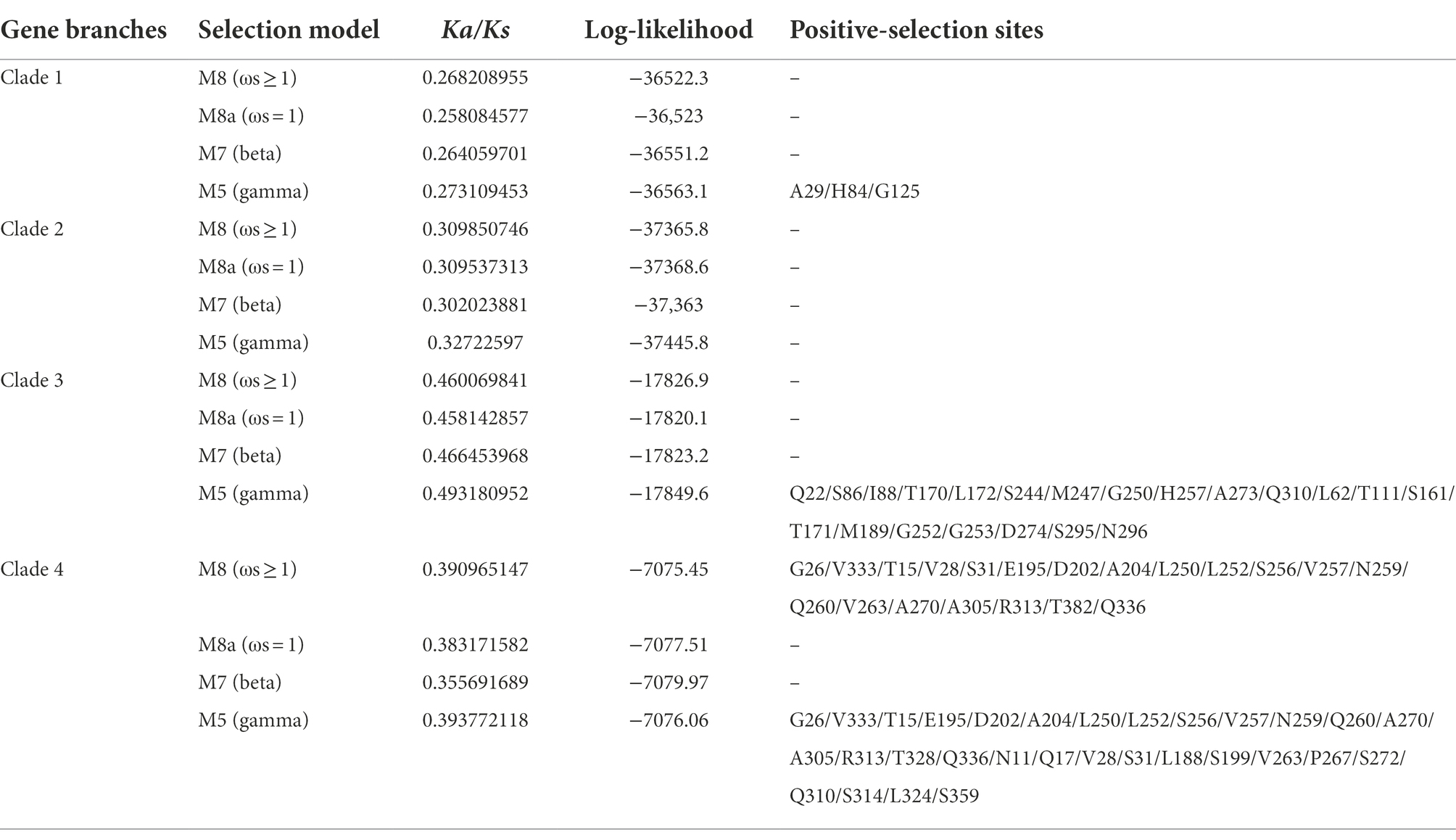
Table 1. Likelihood values and parameter estimates of positive selection in codons of SHI/STY gene family in 21 land plants.
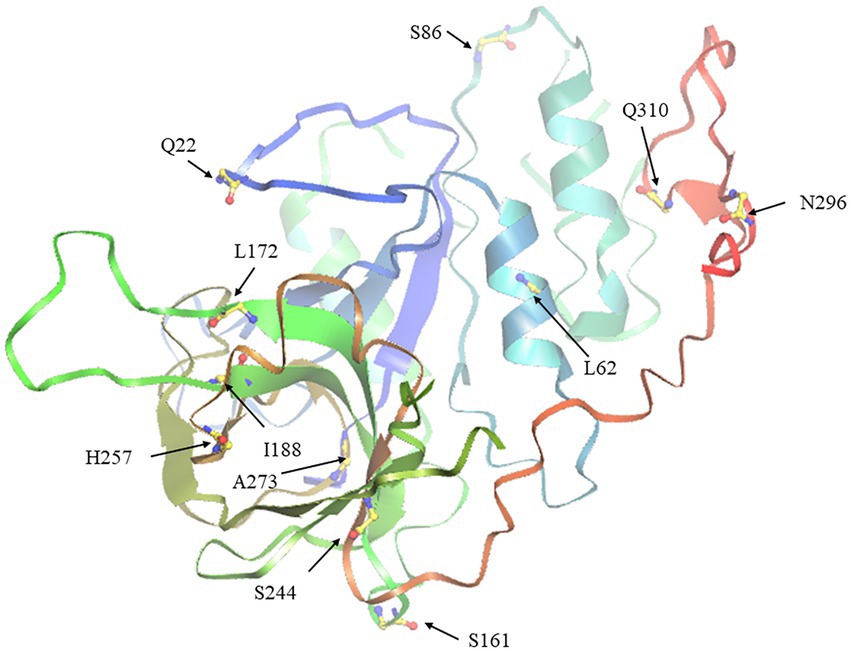
Figure 7. The tertiary spatial structure of BnSHI/STY protein in Clade 3. The eleven predicted positive selection sites were marked with arrows.
The predicted protein–protein interaction network of SHI/STYs
Protein–protein interaction networks are a useful and convenient tool for the systematic study of complex biological activities in cells (Hao et al., 2016). To fully understand the function of SHI/STYs in B. napus, the interaction network of SHI/STYs was predicted using the STRING (Figure 8). The result indicated that the SHI/STYs interacted with many different proteins like lysophosphatidic acid acyltransferase (LPAT4), membrane-bound O-acyltransferases (MBOATs), Pentatricopeptide repeat (PPR), ubiquitins (UBQ3), low-density lipoprotein (LDL), calcineurin B subunit-related (CnB) and peptidyl-prolyl cis-trans isomerase.
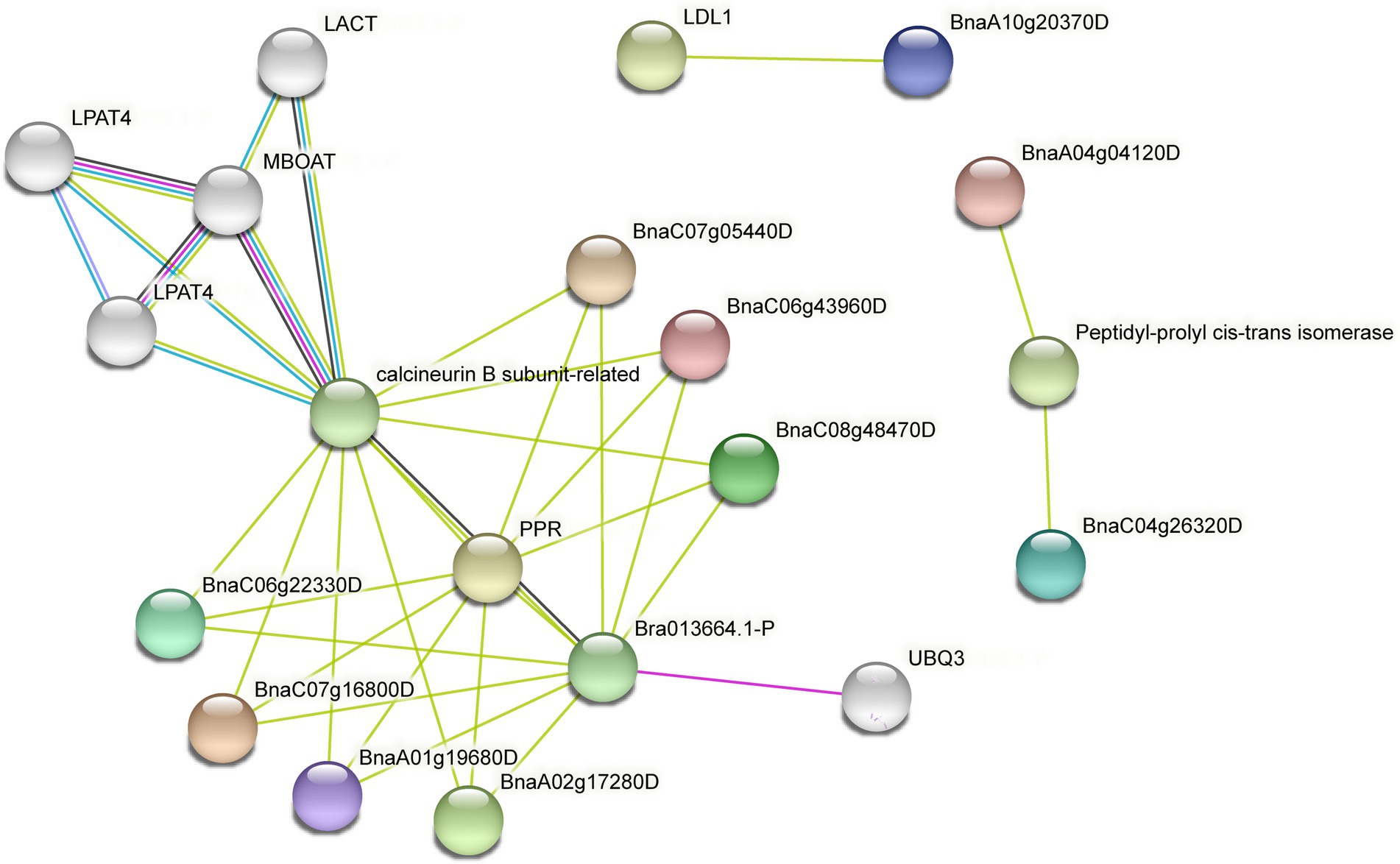
Figure 8. The protein–protein interaction network of SHI/STYs. Eleven SHI/STYs were involved in the construction of the protein–protein interaction network. Fifty-three protein–protein interactions were found between SHI/STYs and some other proteins. The colorful balls represent different proteins and the line between proteins indicates the interaction between the two proteins.
LPAT is crucial to the biosynthesis of phospholipids and TAG due to its strong flux control of the de novo phospholipid biosynthesis pathway (Angkawijaya et al., 2019). In Arabidopsis, LPAT4 plays a role in the N starvation response by playing a function in endoplasmic-reticulum-localized denovo of glycerolipid biosynthesis (Angkawijaya et al., 2019). MBOAT is a large family of integral transmembrane enzymes that function in lipid biosynthesis or phospholipid remodeling and cell surface protection (Hofmann, 2000; Chang et al., 2006; Chang and Magee, 2009; Liu et al., 2012). The result indicated that SHI/STYs in B. napus interact indirectly with these two proteins. Thus, SHI/STYs might be involved in phospholipid and TAG biosynthesis, lipid synthesis, phospholipid remodeling, and cell surface protection. The CnB can regulate the proteasome pathway (Li et al., 2011). The plant PPR proteins may be an evolutionary adaption to complicated RNA metabolism in mitochondria and chloroplasts (Germain et al., 2013). Some interaction networks between proteins (CnB, PPR) and BnSHI/STYs (BnaC06g22330D, BnaC07g05440D, etc.) suggested that BnSHI/STYs might be involved in the proteasome pathway and RNA metabolism. Furthermore, the expression pattern of UBQ3 significantly changed when subjected to cold stress (Ding et al., 2020), suggesting that UBQ3 is associated with cold stress. It can be speculated that these BnSHI/STYs interacting with UBQ3 may also be involved in the cold stress response. Peptidyl-prolyl cis-trans isomerase can regulate the phosphorylation of protein and plays role in cell proliferation and transformation (Chen et al., 2018). In our protein interaction network, BnaA04g04120D and BnaC04g26320D interact with this protein, which means that they may also play homologous functions in cell proliferation and transformation. Above all, by influencing regulatory pathways and biological processes, the SHI/STY proteins are crucial for plant growth and development.
Analysis of the expression patterns of Brassica SHI/STY genes
To fully study the expression patterns of BnSHI/STYs in different tissues and stress conditions, TBtools was used to analyze their expression levels with the obtained RNA-seq data from the NCBI database (Figure 9). The expression pattern of BnSHI/STYs can be roughly divided into three types according to their expression levels. The first type is the majority, such as BnaC08g35730D, BnaA06g13970D, and BnaC01g02360D, which are mainly expressed in roots and buds, and some of them are expressed in the stem. The second type is mainly expressed in the stem, such as BnaC04g26320D, BnaC07g16800D, and BnaA07g12710D, etc. The BnSHI/STYs in the third type are mainly expressed in buds, filaments, and roots, such as BnaA07g21650D, BnaC06g43960D, and BnaC02g24360D, etc. Some of them are also expressed in stems and petals such as BnaC06g22330D. Combined with the expression levels, BnSHI/STYs within the first type mainly belong to Clades 1 and 3, indicating that they are mainly involved in root growth and development. The second type was mainly distributed in Clade 2 and was related to the growth and development of the stem.
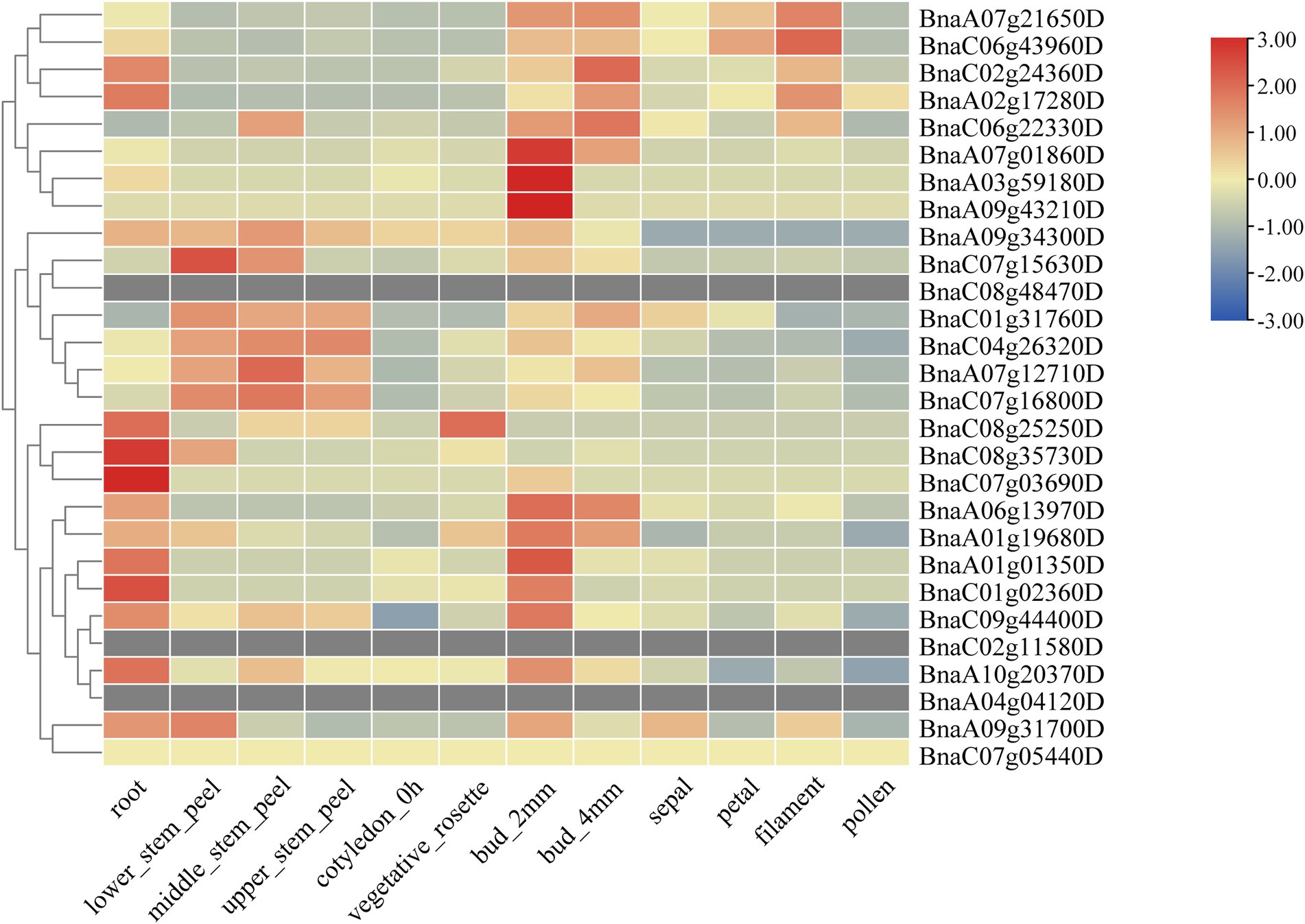
Figure 9. The expression patterns of BnSHI/STYs in different tissues. BnSHI/STYs are clustered by hierarchical clustering. The different color box represents expression quantity in different tissues, red to blue means expression lever from high to lower.
Next, the expression patterns of BnSHI/STYs infected by S. sclerotiorum, and under drought, cold and heat stress were also analyzed. 5, 10 and 3 BnSHI/STYs were up-regulated in the epidermis, mesophyll, and vascular, respectively, compared with control (Figure 10A). Moreover, 7 BnSHI/STYs were up-regulated under high temperature, and another 7 BnSHI/STYs were up-regulated when B. napus suffered drought treatment (Figures 10B,C). Some SHI/STY genes such as BnaC08g48470D, BnaA01g19680D and BnaC07g16800D, etc., were up-regulated under both high-temperature and drought. This result indicates that these BnSHI/STYs may play important functions when plants suffered biological and abiotic stress. We also found that about 53.6% of SHI/STYs were down-regulated under cold stress, such as BnaA01g19680D, BnaA06g13970D and BnaC08g48470D, etc. And most of the down-regulated members belong to the Clade 3, and up-regulated SHI/STYs belong to the Clades 1 and 2, suggesting that SHI/STYs in Clades 1 and 2 might help plants to respond to cold stress.
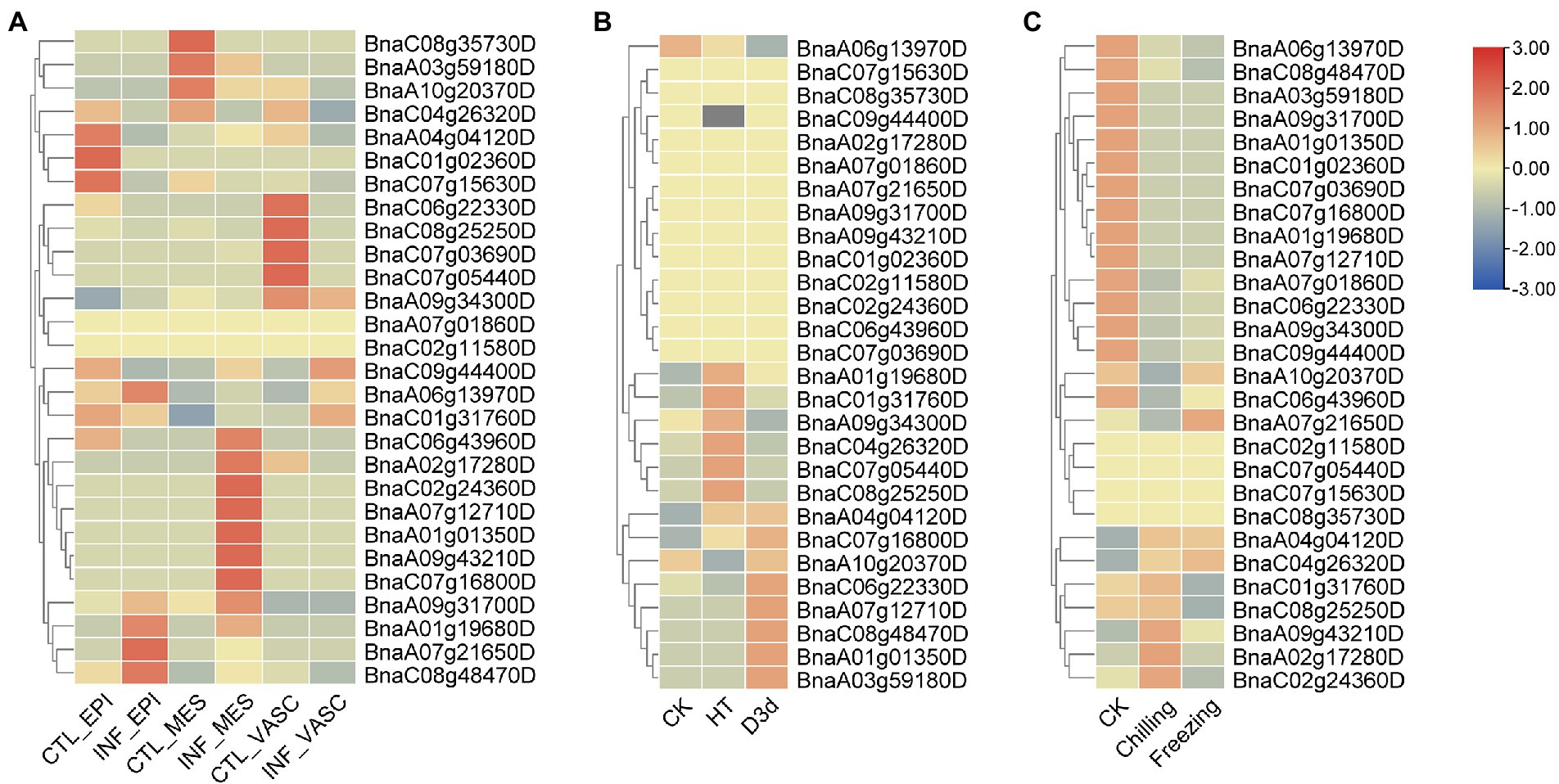
Figure 10. The expression patterns of BnSHI/STYs. (A) The expression pattern of BnSHI/STYs after infection by Sclerotinia sclerotiorum. (B) The expression pattern of BnSHI/STY in B. napus under abiotic stress. (C) The expression pattern of BnSHI/STYs in cold stress. RNA sequence data that were treated with cold shock at temperatures of chill (4°C) and freezing (4°C) were used to complete this figure, and “CK” was treated at 25°C. Genes with high expression levels are shown in red, and genes with low expression levels are shown in blue. The gray box indicates that the expression data of this gene were not found in the database.
The expression patterns of duplicated genes were also investigated under different stress treatments. For example, BnaA07g21650D was up-regulated under cold and infected by S. sclerotiorum, but was down-regulated under drought. While its duplicated gene (BnaC06g22330D) presented the opposite expression pattern. This indicates that the functions of duplicated genes have diverged during evolution.
Expression patterns of BnSHI/STY genes under different stress and hormone treatments
To further explore and understand the possible functions of BnSHI/STYs under biotic and abiotic stresses, eight BnSHI/STYs were selected randomly and their expression levels in rape seedlings treated with different abiotic stresses and hormones were measured by qRT-PCR (Figure 11). Overall, the expression pattern of BnSHI/STYs was different under different stress treatments. For example, BnaC09g44400D was significantly upregulated under all stresses except cold and 6-BA. The expression level of BnaA09g34300D was up-regulated under various stresses, but significantly down-regulated under IAA treatment. BnaC04g26320D was significantly upregulated in all stresses except 6-BA. Under abiotic stress, 5 BnSHI/STY genes were upregulated under cold stress, seven genes were upregulated under drought stress, while five genes were upregulated under salt stress. Compared with CK, salt stress had the least effect on the expression level of BnSHI/STYs selected, and the expression level of BnaA09g34300D was up-regulated by about 1.5 times. BnaA09g43210D was most affected by abiotic stress, and the expression level of BnaA09g43210D increased about 16 times under cold stress, and about 31 times under drought stress. In the experiment of IAA, ABA, 6-BA hormone treatment, the expression level of BnSHI/STYs in six selected samples was up-regulated by IAA, and the expression level of BnaA09g34300D was down-regulated. Six selected BnSHI/STYs were up-regulated by ABA and BnaA01g19680D down-regulated. The expression pattern of BnaA09g43210D was most up-regulated by IAA and ABA, which was about 11 and 15 times that of CK. The expression levels of 4 selected BnSHI/STYs were up-regulated by 6-BA, but no BnSHI/STY was down-regulated by 6-BA. As a result, our data further suggest that SHI/STYs are involved in plant stress and hormone responsive.
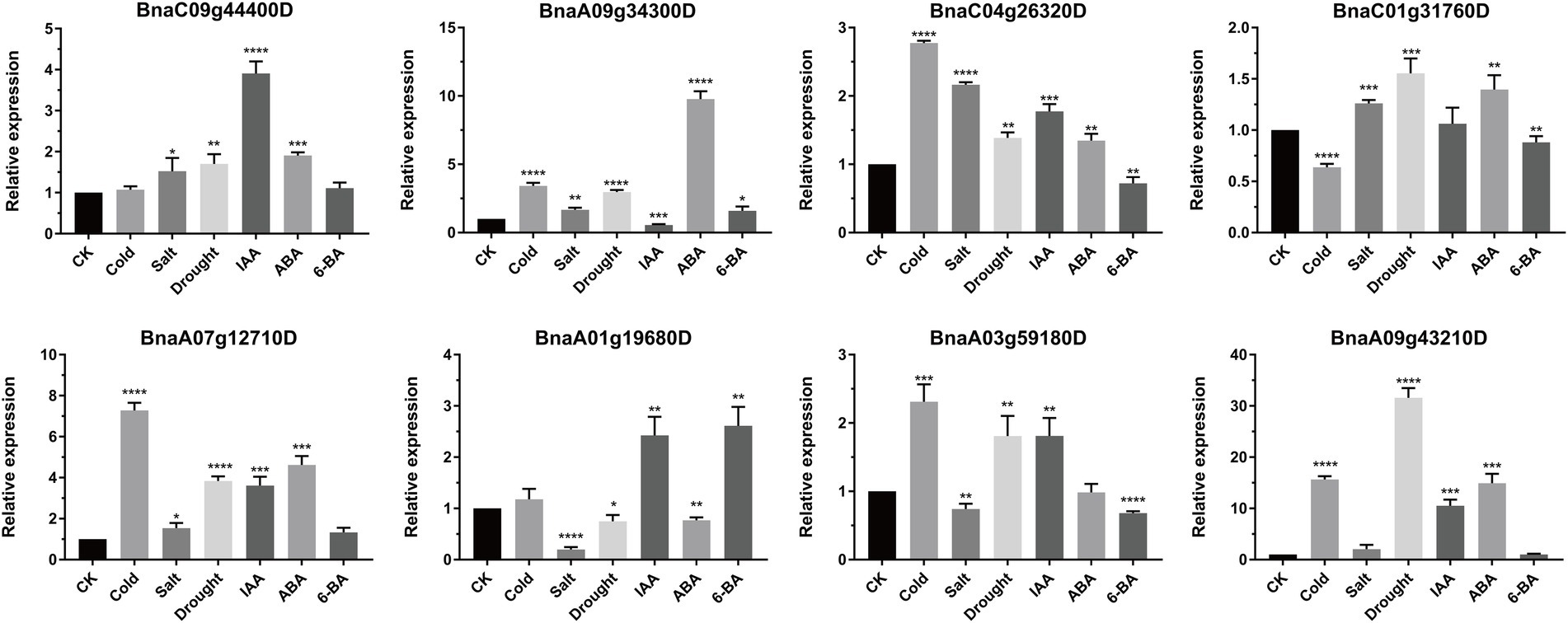
Figure 11. Expression levels of eight BnSHI/STYs under different abiotic stresses and hormone treatments. The selected BnSHI/STYs were normalized concerning the reference gene (Actin). The X-axis corresponds to different abiotic stress treatments and hormone treatments. The values on the Y-axis represent the mean ± SD of three biological replicates. Asterias on vertical bar shows significant difference at *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
In Arabidopsis, the SHI/STY gene family has been extensively studied in recent years (Singh et al., 2020). The SHI/STY protein contains a zinc finger-like RING domain that is conserved at the N-terminal (Fridborg et al., 2001). Members of SHI/STY gene family were important for the plant by playing functions in the formation and development of lateral roots (Smith and Fedoroff, 1995). The function of SHI/STY has been reported in some plants as well, for example, by binding to cis-element in YUCCA4 gene promoter directly, AtLRP1 has transcriptional activity in auxin biosynthesis (Singh et al., 2020). Consequently, the SHI/STY genes play different functions in the coordination of multiple hormonal pathways that regulate growth and development (Yang et al., 2020). Therefore, the study of the evolution of the SHI/STY gene family is of great significance. In the aggregate, 195 SHI/STY genes were identified in 21 land plants, and divided into 4 clades (Figure 1). SHI/STYs in Clade 1 appear earliest and have been identified in P. patens and S. moellendorffii. SHI/STY genes in monocotyledons are only present in Clade 1 and Clade 3. The members of Clade 2 were identified from dicotyledons, and the members of Clade 4 were identified only from higher dicotyledons. Based on the distribution of SHI/STY members and their conserved motifs in different clades, a model was speculated to describe the evolution scenario of the SHI/STY gene family in land plants (Figure 12). The first SHI/STYs containing conserved Motif 1 appeared in P. patrella and S. moellendorffii. Among monocotyledons, some SHI/STYs obtained Motif 10 and Motif 16 and were conserved in dicotyledons, forming Clade 1. Another group of SHI/STYs acquired Motif 4, and these SHI/STYs evolved in three different directions. Members of dicotyledons and monocotyledons with similar motif structures are divided into Clade 3, some SHI/STY acquired Motif 8 in the evolution process, and this kind of SHI/STY is divided into Clade 2. Clade 4 evolved from Clade 3, and in the evolution process, conserved Motif 1 was lost in SHI/STYs in Clade 4, but a new conserved motif was obtained. There are more SHI/STYs in B. napus than in other species. The gene structure and distribution pattern of conserved motifs are consistent with the position of genes in the phylogenetic tree. Hence, SHI/STY proteins might not be conserved between the species, but are more likely to have experienced gene duplication events to expand their numbers in land plants.
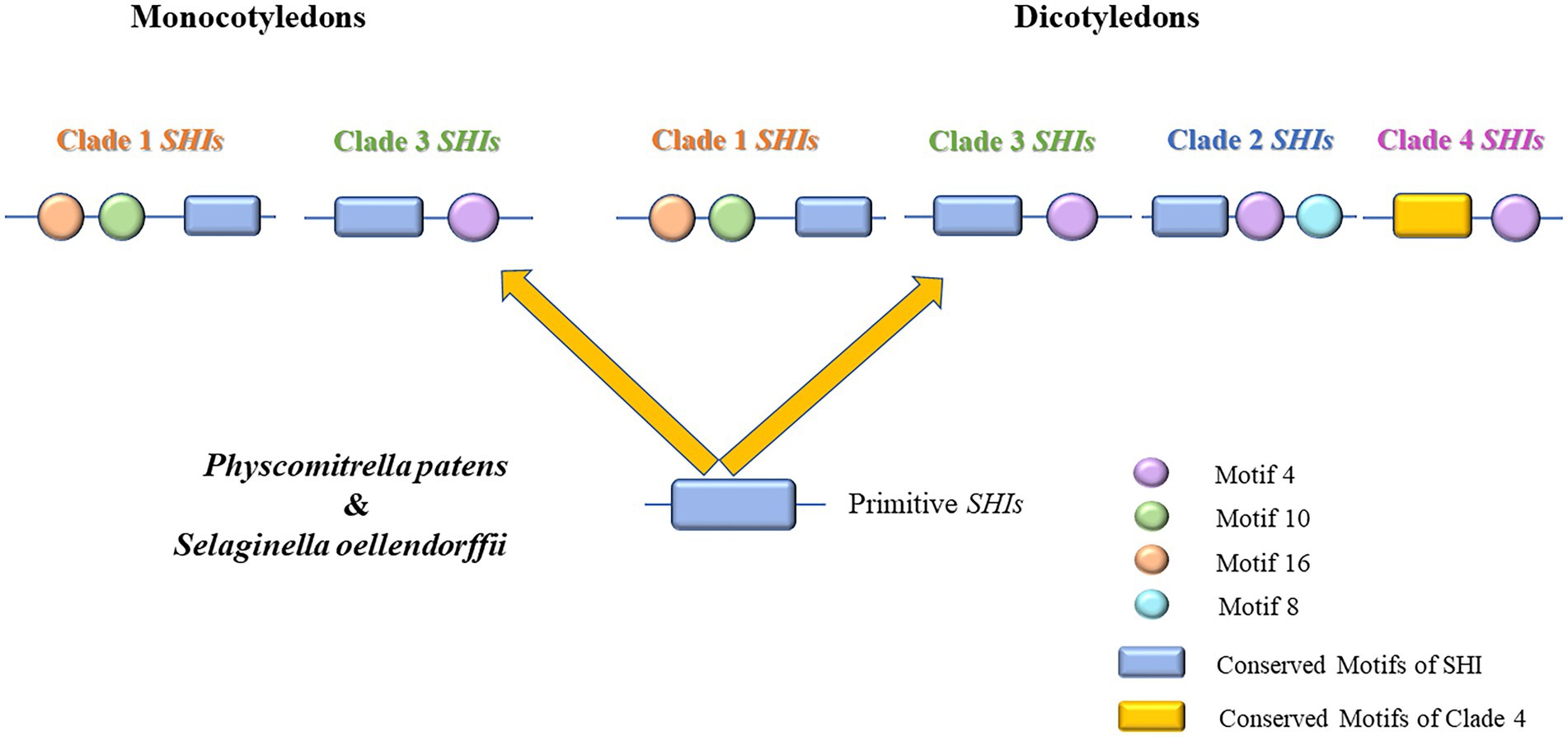
Figure 12. Evolution scenario of the SHI/STY gene family in land plants. In Physcomitrella patens and Selaginella oellendorffii, the SHI/STY gene family emerged. The SHI/STY gene family is formed by adding different motifs at both ends of conserved domains to form Clade 1 and Clade 3 in monocotyledons. In dicotyledons, the Clade 3 gene was further duplicated and gradually evolved into three branches through loss or acquisition of new motifs (some retained the characteristics of Clade 3, some acquired motif 8 and evolved into Clade 2, while the conserved motif in Clade 4 was lost).
The zinc finger domain similar to RING (CX2CX7CX4CX2C2X6C, Motif 1) is a type of RING domain (C3H2C3 or C3HC4), which was identified as a DNA binding motif (Kaulen et al., 1991). This domain can play an important role in many physiological and biochemical processes by binding to RNA, protein, and lipid substrates (Berg and Shi, 1996; Elenbaas et al., 1996). The IGGH domain (motif 2) is positioned in the C-terminal and contains four highly conserved residues in the region, which plays the function of a transcriptional activator (Fridborg et al., 2001). In addition, these results suggest that SHI/STYs may play an important role in regulating gene transcription. Within each clade, the variety and number of conserved motifs are similar. As a result, it can be found that the difference in motifs between clades might cause the divergence of gene function in the SHI/STY gene family.
Previous research suggests that intron-rich genes might be ancestral and that intron-poor genes might be caused by intron losing (Roy, 2006). Genes with more introns provide variant proteins that may play different roles in biological processes by providing more opportunities for alternative splicing (Min et al., 2015). In Arabidopsis and rice, genes with fewer introns are more likely to involve in resistance to salt stress and drought response than gene families with more introns (Liu et al., 2021). Our results show that Clade 4 has more introns than the other three branches, and Clade 4 is relatively new. Naturally, it could be predicted that SHI/STYs in Clade 4 might obtain introns to adapt to the environmental change in the long evolutionary process.
Gene duplication is quite common and crucial for all species and plays an important role in species evolution (Lynch and Conery, 2000). It promotes the diversity of gene families greatly, and increases the complexity and the size of the genome (Kong et al., 2007; Panchy et al., 2016). Tandem duplication WGD are two main mechanisms of gene duplication (Zhu et al., 2014). Since most plants have undergone polyploidization events during their evolution, and WGD and segmental duplication are prevalent in plant genomes (Cannon et al., 2004; Parkin et al., 2005). Previous studies have shown that Brassica plants have experienced extensive gene duplication events during evolution (Cheng et al., 2013; Chalhoub et al., 2014). B. napus is an allopolyploid production which produced naturally by the hybridization of B. rapa and B. oleracea (Chalhoub et al., 2014). About 13 million years ago the Brassica genome was tripled (Yang et al., 1999; Lysak et al., 2005). Studies have revealed that Brassica species like B. oleracea and B. rapa diverged from A. thaliana during a triplication of the whole genome between 20 and 40 million years ago (Cheng et al., 2013; Chalhoub et al., 2014; Dun et al., 2014). In the diploid ancestors of B. napus, the divergence time of the A and C genome segments spanned about 0.12–1.37 million years ago (Cheung et al., 2009). The SHI/STY genes might undergo duplication events during the evolutionary process because of their similar motif structure and exon-intron pattern in each subgroup. The common mutation rate (1.4 × 10−8 synonym substitutions per site per year) was used to estimate the time of BnSHI/STYs divergence (Wang et al., 2011). It was found that most of all BnSHI/STYs were caused by gene duplication events (Supplementary Table S4). 20 BnSHI/STYs of them were derived from WGD or segmental duplications, and the rest of the 8 BnSHI/STY genes were caused by dispersed duplications. All BnSHI/STYs had been duplicated before the divergence of the A and C genomes, except three gene pairs (BnaA07g01860D/BnaC02g11580D, BnaA07g21650D/BnaC06g22330D, and BnaC02g24360D/BnaA02g17280D; Supplementary Table S4). The ratio of the non-synonymous substitution rate to the synonymous substitution rate, namely Ka/Ks, is an important parameter to analyze sequence evolution (Sueoka, 1992; Hurst, 2002). Ka/Ks > 1 is potential evidence of positive selection during gene differentiation, while Ka/Ks < 1 usually denotes negative selection/purified selection (Guo et al., 2017). Our study found that Ka/Ks ratios of all clades were less than 1, suggesting that purification selection plays a role after SHI/STYs differentiation. Most of the SHI/STYs duplicated after triploids of the whole genome but before the divergence of A and C genomes. The result of collinearity analysis indicated that there are many homologous SHI/STYs between B. napus and Arabidopsis, B. napus and B. rapa, B. napus and B. oleracea, respectively. 71.4 percent of BnSHI/STYs are generated by the WGD. Therefore, WGD is the main driver of SHI/STYs expansion in the genome of B. napus.
Cis-acting elements in promoters play crucial roles in regulating gene expression by binding to transcription factors (Maire et al., 1989; Otsuki and Yamamoto, 2020). The cis-acting elements in the promoter regions of genes are closely related to the expression pattern of genes (Chow et al., 2018). In addition, the same regulatory elements in gene promoters suggest that they may have similar regulation functions (Xie et al., 2020). Light responsive related elements are most common in all SHI/STYs promoters, indicating that light signals may be involved in SHI/STYs regulating the growth and development of the Brassica species. There are no same cis-acting elements in duplicated genes. Most of the duplicated genes in these four Brassica species have different cis-acting elements pattern. For example, Bol027721 contains a zein metabolism regulation element, which does not exist in its duplicated gene Bol039313. Low-temperature responsiveness-related element exists in most SHI/STYs except BnaA03g59180D, BnaC07g05440D. It suggests that not all cis-acting elements were conserved in each promoter of duplicated SHI/STYs. The variety of cis-acting elements in the promoters, indicates that these SHI/STYs might be associated with abundant regulatory pathways and biological processes. The diversity of cis-acting elements in duplicated genes indicates that the responses of genes to biotic and abiotic stresses have changed, which is the molecular basis of gene functional diversity. The results suggest that SHI/STY could play an important role in regulating hormone responses and responding to abiotic stresses to impact growth and development in Brassica species.
Abiotic stress changes plant physiological processes by affecting gene expression, RNA or protein stability, and ion transport (Kosova et al., 2015). Temperature and drought are two serious factors that jeopardize plant growth and development. Over time, plants have evolved many survival mechanisms to respond to stress, such as developing specific morphological characteristics and regulating metabolic pathways (Vijayaraghavareddy et al., 2020). The first plant organ to sense salt stress is the root, which immediately transmits this signal to the leaves through the stem, leading to stomatal closure and reducing water loss (Cosgrove and Hedrich, 1991; Christmann et al., 2013). Previous research has shown that SHI/STYs play a crucial role in the growth, development, and responses to biotic and abiotic stress of plants (Yang et al., 2020). Plant hormones are necessary for plant growth and development and play an important role in stress response (Shu et al., 2016). It can find that most of all BnSHI/STYs were highly expressed in root and bud, and most of them were up-regulated under drought stress, such as BnaC03g59180D, BnaA01g01350D, BnaC08g48470D, et al. Some SHI/STYs such as BnaC01g31760D, BnaA07g12710D, BnaC07g16800D were expressed in the stem. Duplicated gene pairs (BnaA06g13970D/BnaC01g31760D) have different expression patterns (Figure 10).
When the plant suffered an infection of S. sclerotiorum., some SHI/STYs were down-regulated. Some members of them, such as BnaA03g59180D, BnaA10g20370D, BnaA04g04120D and BnaC06g22330D, were up-regulated under drought, while BnaC04g26320D, BnaC08g25250D, BnaC07g05440D and BnaA09g34300D were up-regulated under high-temperature. Furthermore, the expression level of the SHI/STY genes in Clade 1 was higher than in other clades, indicating that the SHI/STY genes in Clade 1 could be more crucial for the growth and development of Brassica. Differences in expression patterns of these genes may be caused by changes in the upstream cis-acting elements of allopolyploid genes formed by natural polyploids and their adaptation to the environment. As a result, it can be summarized that BnSHI/STYs can not only influence the growth and development of B. napus but can help the B. napus to adapt to biological and abiotic stress such as cold, heat, and drought. The qRT-PCR data further showed that BnSHI/STYs expression levels were affected by different abiotic stresses and different hormone treatments (Figure 11). Under cold and drought stress, the expression lever of some SHI/STYs was raised, such as BnaA07g12710D, BnaA03g59180D, and BnaA09G43210D, which is consistent with the results of Figure 10B. Under the treatment of plant hormones, the expression of most SHI/STYs was up-regulated. The results of homeopathic element analysis showed that these SHI/STYs have low-temperature responsiveness and Auxin responsiveness. These results suggest that members of the SHI/STY gene family are involved in plant response to external stress.
Conclusion
In this paper, 195 SHI/STY genes were identified in 21 land plants. According to phylogenetic analysis, gene structure, and motif distribution, these genes were further divided into four clades. The difference in motifs between clades might cause the divergence of gene function in the SHI/STY gene family. Collinearity analysis of four Brassica shows that the SHI/STY gene family is relatively conserved in the evolution of B. rapa, Arabidopsis, B. oleracea, and B. napus. WGD is the main factor of SHI/STYs expansion. The SHI/STY genes might have occurred intense purifying selection in the evolutionary process. The different types of cis-element in most members of SHI/STY gene family indicate that they might be associated with abundant regulatory pathways and biological processes. This is consistent with the result of the protein–protein interaction network analysis. The expression analysis indicated that SHI/STYs were expressed in different tissues and regulated by various abiotic and biological stresses. The qRT-PCR data further suggest that SHI/STYs are involved in plant stress and hormone response and play an important role in plant growth and development. Finally, an evolutionary model was proposed based on the evolution of SHI/STY gene family.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
DF is the executor of the study, mainly completing the basic analysis of data and writing the first draft of the paper. WZ, XC, FH, and ZY participated in the analysis of the results. JC is the project leader, directing the research design and final revision of data analysis results, as well as writing and revising the paper. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the National Science Foundation of China (No. 31871655 and 32072019).
Acknowledgments
This paper was polished by Rehman Sarwar.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.958964/full#supplementary-material
Abbreviations
SHI/STY, SHORT INTERNODES/STYLISH; LRP1, LATERAL ROOT PRIMORDIUM1; SRS, SHI RELATED SEQUENCE3; GA, Gibberellin; NJ, Neighbor joining; LPAT4, Lysophosphatidic acid acyltransferase; MBOATs, Membrane-bound O-acyltransferases; PPR, Pentatricopeptide repeat; UBQ3, Ubiquitins; LDL, Low-density lipoprotein; CnB, Calcineurin B subunit-related; ABA, Abscisic acid; IAA, 3-Indoleacetic acid; 6-BA, 6-Benzylaminopurine.
Footnotes
1. ^https://phytozome.jgi.doe.gov/pz/portal
2. ^https://phytozome.jgi.doe.gov/pz/portal
3. ^http://www.genoscope.cns.fr/brassicanapus
4. ^https://www.ncbi.nlm.nih.gov/cdd
5. ^https://www.ncbi.nlm.nih.gov/blast
6. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html
7. ^https://phytozome.jgi.doe.gov/pz/portal
8. ^https://www.genoscope.cns.fr/brassicanapus/
9. ^http://www.genoscope.cns.fr/brassicanapus
10. ^http://sourceforge.net/projects/mev-tm4/files/mev-tm4/MeV%204.8.1/
11. ^http://selecton.tau.ac.il/
12. ^https://zhanglab.ccmb.med.umich.edu/I-TASSER/
13. ^https://brassica.biodb.org/
14. ^https://phytozome.jgi.doe.gov/pz/portal
References
Angkawijaya, A. E., Nguyen, V. C., and Nakamura, Y. (2019). LYSOPHOSPHATIDIC ACID ACYLTRANSFERASES 4 and 5 are involved in glycerolipid metabolism and nitrogen starvation response in Arabidopsis. New Phytol. 224, 336–351. doi: 10.1111/nph.16000
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Baylis, T., Cierlik, I., Sundberg, E., and Mattsson, J. (2013). SHORT INTERNODES/STYLISH genes, regulators of auxin biosynthesis, are involved in leaf vein development in Arabidopsis thaliana. New Phytol. 197, 737–750. doi: 10.1111/nph.12084
Beilstein, M. A., Nagalingum, N. S., Clements, M. D., Manchester, S. R., and Mathews, S. (2010). Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 107, 18724–18728. doi: 10.1073/pnas.0909766107
Berg, J. M., and Shi, Y. (1996). The galvanization of biology: a growing appreciation for the roles of zinc. Science 271, 1081–1085. doi: 10.1126/science.271.5252.1081
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chang, T. Y., Chang, C. C., Ohgami, N., and Yamauchi, Y. (2006). Cholesterol sensing, trafficking, and esterification. Annu. Rev. Cell Dev. Biol. 22, 129–157. doi: 10.1146/annurev.cellbio.22.010305.104656
Chang, S. C., and Magee, A. I. (2009). Acyltransferases for secreted signalling proteins (review). Mol. Membr. Biol. 26, 104–113. doi: 10.1080/09687680802706432
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Huang, H., and Wu, C. H. (2011). Protein bioinformatics databases and resources. Methods Mol. Biol. 694, 3–24. doi: 10.1007/978-1-60761-977-2_1
Chen, C., Huang, H., and Wu, C. H. (2017). Protein bioinformatics databases and resources. Methods Mol. Biol. 1558, 3–39. doi: 10.1007/978-1-4939-6783-4_1
Chen, Y., Wu, Y. R., Yang, H. Y., Li, X. Z., Jie, M. M., Hu, C. J., et al. (2018). Prolyl isomerase Pin1: a promoter of cancer and a target for therapy. Cell Death Dis. 9, 883. doi: 10.1038/s41419-018-0844-y
Cheng, F., Mandakova, T., Wu, J., Xie, Q., Lysak, M. A., and Wang, X. (2013). Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25, 1541–1554. doi: 10.1105/tpc.113.110486
Cheung, F., Trick, M., Drou, N., Lim, Y. P., Park, J. Y., Kwon, S. J., et al. (2009). Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell 21, 1912–1928. doi: 10.1105/tpc.108.060376
Chow, C. N., Chiang-Hsieh, Y. F., Chien, C. H., Zheng, H. Q., Lee, T. Y., Wu, N. Y., et al. (2018). Delineation of condition specific Cis- and trans-acting elements in plant promoters under various Endo- and exogenous stimuli. BMC Genomics 19, 85. doi: 10.1186/s12864-018-4469-4
Christmann, A., Grill, E., and Huang, J. (2013). Hydraulic signals in long-distance signaling. Curr. Opin. Plant Biol. 16, 293–300. doi: 10.1016/j.pbi.2013.02.011
Clough, E., and Barrett, T. (2016). The gene expression omnibus database. Methods Mol. Biol. 1418, 93–110. doi: 10.1007/978-1-4939-3578-9_5
Cosgrove, D. J., and Hedrich, R. (1991). Stretch-activated chloride, potassium, and calcium channels coexisting in plasma membranes of guard cells of Vicia faba L. Planta 186, 143–153. doi: 10.1007/BF00201510
De Rybel, B., Audenaert, D., Xuan, W., Overvoorde, P., Strader, L. C., Kepinski, S., et al. (2012). A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat. Chem. Biol. 8, 798–805. doi: 10.1038/nchembio.1044
De Smet, I., Vassileva, V., De Rybel, B., Levesque, M. P., Grunewald, W., Van Damme, D., et al. (2008). Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322, 594–597. doi: 10.1126/science.1160158
Ding, A., Bao, F., Zhang, T., Yang, W., Wang, J., Cheng, T., et al. (2020). Screening of optimal reference genes for qRT-PCR and preliminary exploration of cold resistance mechanisms in Prunus mume and Prunus sibirica varieties. Mol. Biol. Rep. 47, 6635–6647. doi: 10.1007/s11033-020-05714-x
Drafor, G., Duah, E., Ankamah, N. A., Kpene, G. E., and Mante, P. K. (2021). Investigating the anticonvulsant properties of aqueous Ethanolic extracts of the leaves, roots, and fruits of Jatropha gossypifolia L(Euphorbiaceae). Adv. Pharmacol. Pharm. Sci. 2021:5547353. doi: 10.1155/2021/5547353
Duan, E., Wang, Y., Li, X., Lin, Q., Zhang, T., Wang, Y., et al. (2019). OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell 31, 1026–1042. doi: 10.1105/tpc.19.00023
Dun, X., Shen, W., Hu, K., Zhou, Z., Xia, S., Wen, J., et al. (2014). Neofunctionalization of duplicated Tic40 genes caused a gain-of-function variation related to male fertility in Brassica oleracea lineages. Plant Physiol. 166, 1403–1419. doi: 10.1104/pp.114.246470
Eklund, D. M., Staldal, V., Valsecchi, I., Cierlik, I., Eriksson, C., Hiratsu, K., et al. (2010). The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 22, 349–363. doi: 10.1105/tpc.108.064816
Elenbaas, B., Dobbelstein, M., Roth, J., Shenk, T., and Levine, A. J. (1996). The MDM2 oncoprotein binds specifically to RNA through its RING finger domain. Mol. Med. 2, 439–451. doi: 10.1007/BF03401903
Fridborg, I., Kuusk, S., Moritz, T., and Sundberg, E. (1999). The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 11, 1019–1031. doi: 10.1105/tpc.11.6.1019
Fridborg, I., Kuusk, S., Robertson, M., and Sundberg, E. (2001). The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiol. 127, 937–948. doi: 10.1104/pp.010388
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Germain, A., Hotto, A. M., Barkan, A., and Stern, D. B. (2013). RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA 4, 295–316. doi: 10.1002/wrna.1161
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Guo, Y., Liu, J., Zhang, J., Liu, S., and Du, J. (2017). Selective modes determine evolutionary rates, gene compactness and expression patterns in Brassica. Plant J. 91, 34–44. doi: 10.1111/tpj.13541
Hao, T., Peng, W., Wang, Q., Wang, B., and Sun, J. (2016). Reconstruction and application of protein-protein interaction network. Int. J. Mol. Sci. 17, 907–925. doi: 10.3390/ijms17060907
Hofmann, K. (2000). A superfamily of membrane-bound O-acyltransferases with implications for wnt signaling. Trends Biochem. Sci. 25, 111–112. doi: 10.1016/S0968-0004(99)01539-X
Hubbard, S. R., Bishop, W. R., Kirschmeier, P., George, S. J., Cramer, S. P., and Hendrickson, W. A. (1991). Identification and characterization of zinc binding sites in protein kinase C. Science 254, 1776–1779. doi: 10.1126/science.1763327
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18, 486–487. doi: 10.1016/S0168-9525(02)02722-1
Islam, M. A., Lutken, H., Haugslien, S., Blystad, D. R., Torre, S., Rolcik, J., et al. (2013). Overexpression of the AtSHI gene in poinsettia, Euphorbia pulcherrima, results in compact plants. PLoS One 8:e53377. doi: 10.1371/journal.pone.0053377
Kaulen, H., Pognonec, P., Gregor, P. D., and Roeder, R. G. (1991). The Xenopus B1 factor is closely related to the mammalian activator USF and is implicated in the developmental regulation of TFIIIA gene expression. Mol. Cell. Biol. 11, 412–424.
Kim, S. G., Lee, S., Kim, Y. S., Yun, D. J., Woo, J. C., and Park, C. M. (2010). Activation tagging of an Arabidopsis SHI-RELATED SEQUENCE gene produces abnormal anther dehiscence and floral development. Plant Mol. Biol. 74, 337–351. doi: 10.1007/s11103-010-9677-5
Klug, A. (1999). Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 293, 215–218. doi: 10.1006/jmbi.1999.3007
Kong, H., Landherr, L. L., Frohlich, M. W., Leebens-Mack, J., Ma, H., and dePamphilis, C. W. (2007). Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 50, 873–885. doi: 10.1111/j.1365-313X.2007.03097.x
Kosova, K., Vitamvas, P., Urban, M. O., Klima, M., Roy, A., and Prasil, I. T. (2015). Biological networks underlying abiotic stress tolerance in temperate crops--a proteomic perspective. Int. J. Mol. Sci. 16, 20913–20942. doi: 10.3390/ijms160920913
Krichevsky, A., Zaltsman, A., Kozlovsky, S. V., Tian, G. W., and Citovsky, V. (2009). Regulation of root elongation by histone acetylation in Arabidopsis. J. Mol. Biol. 385, 45–50. doi: 10.1016/j.jmb.2008.09.040
Kuusk, S., Sohlberg, J. J., Magnus Eklund, D., and Sundberg, E. (2006). Functionally redundant SHI family genes regulate Arabidopsis gynoecium development in a dose-dependent manner. Plant J. 47, 99–111. doi: 10.1111/j.1365-313X.2006.02774.x
Laity, J. H., Lee, B. M., and Wright, P. E. (2001). Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46. doi: 10.1016/S0959-440X(00)00167-6
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, N., Zhang, Z., Zhang, W., and Wei, Q. (2011). Calcineurin B subunit interacts with proteasome subunit alpha type 7 and represses hypoxia-inducible factor-1alpha activity via the proteasome pathway. Biochem. Biophys. Res. Commun. 405, 468–472. doi: 10.1016/j.bbrc.2011.01.055
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930. doi: 10.1038/ncomms4930
Liu, H., Lyu, H. M., Zhu, K., Van de Peer, Y., and Max Cheng, Z. M. (2021). The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 105, 1072–1082. doi: 10.1111/tpj.15088
Liu, Q., Siloto, R. M., Lehner, R., Stone, S. J., and Weselake, R. J. (2012). Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog. Lipid Res. 51, 350–377. doi: 10.1016/j.plipres.2012.06.001
Long, M., Rosenberg, C., and Gilbert, W. (1995). Intron phase correlations and the evolution of the intron/exon structure of genes. Proc. Natl. Acad. Sci. U. S. A. 92, 12495–12499. doi: 10.1073/pnas.92.26.12495
Lutken, H., Jensen, L. S., Topp, S. H., Mibus, H., Muller, R., and Rasmussen, S. K. (2010). Production of compact plants by overexpression of AtSHI in the ornamental Kalanchoe. Plant Biotechnol. J. 8, 211–222. doi: 10.1111/j.1467-7652.2009.00478.x
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Lysak, M. A., Koch, M. A., Pecinka, A., and Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15, 516–525. doi: 10.1101/gr.3531105
Maire, P., Wuarin, J., and Schibler, U. (1989). The role of cis-acting promoter elements in tissue-specific albumin gene expression. Science 244, 343–346. doi: 10.1126/science.2711183
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Min, X. J., Powell, B., Braessler, J., Meinken, J., Yu, F., and Sablok, G. (2015). Genome-wide cataloging and analysis of alternatively spliced genes in cereal crops. BMC Genom. 16, 721. doi: 10.1186/s12864-015-1914-5
Otsuki, A., and Yamamoto, M. (2020). Cis-element architecture of Nrf2-sMaf heterodimer binding sites and its relation to diseases. Arch. Pharm. Res. 43, 275–285. doi: 10.1007/s12272-019-01193-2
Panchy, N., Lehti-Shiu, M., and Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Parkin, I. A., Gulden, S. M., Sharpe, A. G., Lukens, L., Trick, M., Osborn, T. C., et al. (2005). Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171, 765–781. doi: 10.1534/genetics.105.042093
Sarwar, R., Jiang, T., Ding, P., Gao, Y., Tan, X., and Zhu, K. (2021). Genome-wide analysis and functional characterization of the DELLA gene family associated with stress tolerance in B. napus. BMC Plant Biol. 21, 286. doi: 10.1186/s12870-021-03054-x
Shu, K., Chen, Q., Wu, Y., Liu, R., Zhang, H., Wang, P., et al. (2016). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. 85, 348–361. doi: 10.1111/tpj.13109
Singh, S., Yadav, S., Singh, A., Mahima, M., Singh, A., Gautam, V., et al. (2020). Auxin signaling modulates LATERAL ROOT PRIMORDIUM1 (LRP1) expression during lateral root development in Arabidopsis. Plant J. 101, 87–100. doi: 10.1111/tpj.14520
Smith, D. L., and Fedoroff, N. V. (1995). LRP1, a gene expressed in lateral and adventitious root primordia of arabidopsis. Plant Cell 7, 735–745.
Staldal, V., Cierlik, I., Chen, S., Landberg, K., Baylis, T., Myrenas, M., et al. (2012). The Arabidopsis thaliana transcriptional activator STYLISH1 regulates genes affecting stamen development, cell expansion and timing of flowering. Plant Mol. Biol. 78, 545–559. doi: 10.1007/s11103-012-9888-z
Sueoka, N. (1992). Directional mutation pressure, selective constraints, and genetic equilibria. J. Mol. Evol. 34, 95–114. doi: 10.1007/BF00182387
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Town, C. D., Cheung, F., Maiti, R., Crabtree, J., Haas, B. J., Wortman, J. R., et al. (2006). Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18, 1348–1359. doi: 10.1105/tpc.106.041665
Vijayaraghavareddy, P., Vemanna, R. S., Yin, X., Struik, P. C., Makarla, U., and Sreeman, S. (2020). Acquired traits contribute more to drought tolerance in wheat Than in Rice. Plant Phenomics 2020:5905371. doi: 10.34133/2020/5905371
Wang, X., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Xie, T., Zeng, L., Chen, X., Rong, H., Wu, J., Batley, J., et al. (2020). Genome-wide analysis of the lateral organ boundaries domain gene family in Brassica Napus. Genes (Basel) 11, 280–300. doi: 10.3390/genes11030280
Yang, Y. W., Lai, K. N., Tai, P. Y., Ma, D. P., and Li, W. H. (1999). Molecular phylogenetic studies of Brassica, rorippa, arabidopsis and allied genera based on the internal transcribed spacer region of 18S-25S rDNA. Mol. Phylogenet. Evol. 13, 455–462. doi: 10.1006/mpev.1999.0648
Yang, J., Xu, P., and Yu, D. (2020). Genome-wide identification and characterization of the SHI-related sequence gene family in Rice. Evol. Bioinform. Online 16:1176934320941495. doi: 10.1177/1176934320941495
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-TASSER suite: protein structure and function prediction. Nat. Methods 12, 7–8. doi: 10.1038/nmeth.3213
Youssef, H. M., Eggert, K., Koppolu, R., Alqudah, A. M., Poursarebani, N., Fazeli, A., et al. (2017). VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 49, 157–161. doi: 10.1038/ng.3717
Yuan, T. T., Xu, H. H., Zhang, Q., Zhang, L. Y., and Lu, Y. T. (2018). The COP1 target SHI-RELATED SEQUENCE5 directly activates Photomorphogenesis-promoting genes. Plant Cell 30, 2368–2382. doi: 10.1105/tpc.18.00455
Zawaski, C., Kadmiel, M., Ma, C., Gai, Y., Jiang, X., Strauss, S. H., et al. (2011). SHORT INTERNODES-like genes regulate shoot growth and xylem proliferation in Populus. New Phytol. 191, 678–691. doi: 10.1111/j.1469-8137.2011.03742.x
Zhang, Y., von Behrens, I., Zimmermann, R., Ludwig, Y., Hey, S., and Hochholdinger, F. (2015). LATERAL ROOT PRIMORDIA 1 of maize acts as a transcriptional activator in auxin signalling downstream of the aux/IAA gene rootless with undetectable meristem 1. J. Exp. Bot. 66, 3855–3863. doi: 10.1093/jxb/erv187
Zhao, S. P., Song, X. Y., Guo, L. L., Zhang, X. Z., and Zheng, W. J. (2020). Genome-wide analysis of the Shi-related sequence family and functional identification of GmSRS18 involving in drought and salt stresses in soybean. Int. J. Mol. Sci. 21, 1810–1827. doi: 10.3390/ijms21051810
Keywords: SHI/STY gene family, molecular evolution, gene duplication, Cis-acting elements, land plants
Citation: Fang D, Zhang W, Cheng X, Hu F, Ye Z and Cao J (2022) Molecular evolutionary analysis of the SHI/STY gene family in land plants: A focus on the Brassica species. Front. Plant Sci. 13:958964. doi: 10.3389/fpls.2022.958964
Edited by:
Laura Emma Maria Morello, National Research Council (CNR), ItalyReviewed by:
Wei Li, Cotton Research Institute (CAAS), China Xiaqing Yu, Nanjing Agricultural University, ChinaCopyright © 2022 Fang, Zhang, Cheng, Hu, Ye and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Cao, Y2ppbmZvckAxNjMuY29t
 Da Fang
Da Fang Weimeng Zhang
Weimeng Zhang Jun Cao
Jun Cao