- 1College of Physical Education, Changsha University, Changsha, China
- 2Department of Pratacultural Sciences, College of Agronomy, Hunan Agricultural University, Changsha, China
- 3CAS Key Laboratory of Plant Germplasm Enhancement and Specialty Agriculture, Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 4Grassland Research Center of Hunan Province, Changsha, China
Salt stress is one of the major abiotic factors limiting the productivity of bermudagrass (Cynodon dactylon). However, the role of hormonal reprogramming and crosstalk in regulating root growth and salt tolerance in bermudagrass was not reported. Here, we examined the physiological and hormonal responses of two contrasting bermudagrass genotypes; ‘C43,’ salt-tolerant ‘C198’ salt-sensitive. Under salt stress, ‘C43’ had better membrane stability and higher photosynthetic activity than the ‘C198.’ Salt stress promoted root growth and improved root/shoot ratio and root activity in ‘C43,’ but the root growth of ‘C198’ was inhibited by salt stress, leading to diminished root activity. The two bermudagrass genotypes also showed critical differences in hormonal responses, especially in the roots. The root contents of indole-3-acetic acid (IAA), cytokinin derivatives, such as trans-zeatin riboside (tZR) and dihydrozeatin riboside (DHZR) were increased in ‘C43,’ but decreased in ‘C198’ when exposed to salt stress. The root growth rate was positively correlated with the root IAA, tZR and DHZR, indicating their crucial role in root growth under salt stress. The expressions of TAA/YUCCA and CYP735A involved in IAA and tZR biosynthesis were induced by salt stress in ‘C43,’ but inhibited in ‘C198,’ leading to reduced hormone accumulations. Salt stress decreased the iP, tZ, and DHZ content in the roots of both genotypes, and no significant difference was observed between the two genotypes. Salt stress reduced the content of GA3 in both genotypes by inhibiting GA20ox and GA2ox genes, which could be attributed to the reduced shoot growth in both genotypes. The increased ABA level by salt stress was significantly higher in ‘C198’ than ‘C43.’ Furthermore, there were positive and negative correlations between different hormones and root growth, suggesting that root growth could be regulated by complex hormonal reprogramming and crosstalk. This study provides a foundation for understanding the underlying mechanisms of hormonal-mediated root growth and salt tolerance in bermudagrass.
Introduction
Salt stress is a severe global problem that adversely affects plant growth, development, and productivity (Munns and Tester, 2008; Gupta and Huang, 2014). The primary effects of salt stress are caused by osmotic stress, which results from the accumulation of ions in the rhizosphere (Ismail et al., 2014). The accumulation of ions (mainly Na+) in the plant cells further causes ion toxicity and inhibits various developmental processes (Munns and Tester, 2008; Deinlein et al., 2014). Salt stress induces the overproduction of reactive oxygen species (ROSs), including H2O2 (hydrogen oxide), ⋅O2– (superoxide anion radical), ⋅OH (hydroxyl radical) in different organelles and results in oxidative stress (Munns and Tester, 2008), which further disturbs cellular metabolism, and inhibit enzyme activity and photosynthesis, and cause an eventual death of plants (Yu D. et al., 2020).
Roots are the essential organs that absorb water and nutrients from the soil and supply them to the shoots (Zou et al., 2021). Depending on the severity of the stress and plant species, salt stress could impair root development by inhibiting root length and root branching (Gupta and Huang, 2014; Zou et al., 2021). However, root growth can be promoted under salt stress in halophytes (Rahman et al., 2021) and a few salt-tolerant plant species, such as bermudagrass (Hu et al., 2012, 2015). Such responses could be one of the strategies for salt stress acclimation in plants. When exposed to salt stress, plants undergo a dynamic change in the root system architecture to maintain growth and development, thereby enhancing salt tolerance (Zou et al., 2021). Root system architecture modification, such as decreasing lateral root initiation, limiting root growth rates, and redirection of root growth, are crucial mechanisms to cope with osmotic stress and maintain normal root activities under salt stress (Zou et al., 2021). Salt stress-induced increase in root/shoot ratio as a result of improved root growth and inhibition of shoot growth (Albacete et al., 2008) could be an adaptive response to recover the functional equilibrium between below- and above-ground organs, which allows the roots to obtain more water and nutrients from the root zone in saline soils (Pérez-Alfocea et al., 2010).
As a sessile organism, plants develop various complex tolerance mechanisms to cope with the adverse effects of salt stress at the morphological, physiological, biochemical, and molecular levels (Gupta and Huang, 2014). Different metabolites and hormones are produced in different organs to provide stress tolerance mechanisms. Roots are sites for the biosynthesis of various hormones and metabolites involved in salt tolerance (Lu et al., 2019).
Phytohormones regulate plant growth and development and abiotic stress responses (Ryu and Cho, 2015; Yu Z. et al., 2020). The five major plant hormones are ethylene, ABA, gibberellin (GA), auxins such as indole-3-acetic acid (IAA), and cytokinin (CK). These hormones could play vital roles in the salt tolerance of plants individually or collectively through hormonal crosstalk (Ryu and Cho, 2015; Yu Z. et al., 2020). Auxin is a crucial hormone involved in almost all stages of plant development in all organs (Wolters and Jürgens, 2009). IAA is the most active form of auxin that play a crucial role in plant growth and development, including stress response (Cao et al., 2019). ABA is recognized as a stress hormone involved in stress adaptation by inducing stomatal closure and reducing water loss (Mehrotra et al., 2014; Sah et al., 2016). Additionally, ABA could enhance root growth in a dose-dependent manner; a higher concentration inhibits root elongation while promoting root elongation at a moderate concentration (Ghassemian et al., 2000). ABA inhibited the root growth of Arabidopsis by increasing ethylene biosynthesis (Luo et al., 2014).
Cytokinin is another essential plant hormone involved in plant growth and development. CK derivatives, such as tZR and iPA facilitate the responses to delay both stomatal closure and leaf senescence under abiotic stresses, and early CK-induced metabolic disorder may be one of the reasons for the inhibition of shoot growth and leaf senescence under abiotic stress (Ryu and Cho, 2015). CKs can also play positive and negative roles in the stress tolerance of plants (Hai et al., 2020). GA is a critical growth hormone involved in seed germination, stem and root elongation, leaf expansion, flower, and seed development (Ryu and Cho, 2015). GA stimulates root growth and development by monitoring cell proliferation and elongation. The deficiency in endogenous GAs could cause the development of shorter roots and smaller root meristems (Achard et al., 2009).
Hormonal crosstalk is crucial to regulating plant growth and development (Pacifici et al., 2015). For example, ABA plays a role in root growth with auxin crosstalk (Wang et al., 2017; Sun et al., 2018). Auxin could interact with cytokinin and negatively regulate cytokinin accumulation by rapidly inhibiting cytokinin biosynthesis. In contrast, cytokinin overproduction had a slower effect on auxin to regulate plant development (Nordstrom et al., 2004). Despite the role of these hormones have been studied in different plant species (Ryu and Cho, 2015), how these hormones are reprogrammed and the role of their complex interactions in regulating root growth and salt tolerance in bermudagrass was not reported.
Bermudagrass (Cynodon dactylon (L). Pers.) is a warm-season turfgrass species widely used to establish sports fields, lawns, and golf courses (Pessarakli, 2015). The growth and development of bermudagrass are affected by different abiotic stresses, including salt stress (Hu et al., 2015). Our previous study showed that improving the root/shoot ratio in plants exposed to salt stress is one of the salt tolerance mechanisms in bermudagrass (Hu et al., 2012). However, the detailed mechanisms associated with the differences in root growth and salt tolerance in bermudagrass, especially at hormonal level, were not reported. Because roots are the sites of hormone synthesis, we hypothesized that the difference in root growth between salt-tolerant and salt-sensitive bermudagrass genotypes could be related to the accumulation of different phytohormones and their specific crosstalk.
In this study, we used liquid chromatography coupled with MS (LC-MS) to characterize the accumulations of different phytohormones in two contrasting bermudagrass genotypes (‘C43,’ salt-tolerant; ‘C198’ salt-sensitive) in response to salt stress. Furthermore, to understand how phytohormones are regulated at the molecular level, we investigated the mRNA expression levels of critical genes involved in different phytohormones’ metabolism. Overall, this study shed light on how hormonal reprogramming and crosstalk are involved in root growth and salt tolerance in bermudagrass.
Materials and methods
Plant materials and growth conditions
Plant materials were prepared according to our previous studies (Hu et al., 2012, 2015). Briefly, uniform stolons with two nodes (5 cm long) of bermudagrass, ‘C43’ (salt-tolerant) and ‘C198’ (salt-sensitive), were planted in solid growth substances (2 peat soil: 1 sand, v/v). Two weeks after planting, an equal amount of plants was transplanted to plastic pots (7 cm diameter and 9 cm tall) filled with coarse silica sand as the plant anchor medium. Pots were suspended over tubs containing 46 L of aerated half-strength Hoagland solution (Hoagland and Arnon, 1950). The tubs were refilled every other day and renewed weekly. The bottom of the pots had a coarse nylon screen to allow the root growth into the solutions. Plants were grown in an environmentally controlled walk-in growth chamber with a temperature regime of 30/25°C (day/night) and photosynthetically active radiation levels of 800 μmol m–2 s–1 at canopy height for 14 h for 4 weeks. During the establishment period of shoot and root, the shoots were cut weekly at 4-cm height, and roots were clipped back to the bottoms of the pots at the beginning of the salt treatment to allow the plants to reach full maturity and develop uniform and equal size roots and shoots.
Treatment and experiment design
After 4 weeks of cultivation, plants were subjected to 0 mM (control), 180 mM and 300 mM NaCl (salt stress) for 10 days and then canopy height and root length were determined. Roots elongated from nylon screen into nutrient solution were sampled after 4 h light. Plants were removed from the nutrient solution and roots were gently washed for 30 s with distilled water. The root tips, encompassing the meristem and the elongation zone, were excised with a scalpel from the remaining root system and immediately frozen in liquid nitrogen and stored at −80°C for further analysis. Four independent biological replicates, each containing a pool of twenty different plants, were sampled for hormone and qPCR analysis. The salt treatments and grass genotypes were arranged in a randomized complete block design with four replicates.
Measurements
Shoot and root growth rate analysis
Shoot and root growth rates were determined as the difference in growth before and after salt treatment (Hu et al., 2012). The root growth rate was determined only for roots extending from the nylon screen at the pot bottom.
Root viability
The root activity was detected by the TTC method as described previously (Lutts et al., 2004). Briefly, 0.5 g of fresh roots were cut into pieces and quickly rinsed in deionized water containing 0.06% Tween 20 and incubated at 30°C in darkness in tubes containing 6 ml of 0.5% TTC 50 mM K2HPO4, pH 7.0 for 20 h. The colored root samples were blotted dry and rinsed in deionized water, then incubated in 10 ml of 95% ethanol at 60°C for 4 h under gentle agitation during the extraction. The absorbance of the extract solution was recorded at 530 nm and root activity was calculated according to the standard curve with the following equation:
Where D represents the deoxidizing amount of the TTC (mg); W is the fresh weight of roots (g); t stands for the time of coloration (h).
Cell membrane stability
Cell membrane stability was determined as electrolyte leakage (EL) according to our previous study (Hu et al., 2009). Briefly, about 0.1 g plant tissues were washed with deionized water three times and placed into plastic tubes filled with 15 ml deionized water after being cut to 1 cm long segments and then shaken for 24 h. The initial conductivity (Ci) was determined using a conductivity meter (JENCO-3173, Jenco Instruments, Inc., San Diego, CA, United States). Subsequently, the tubes were autoclaved at 121°C for 20 min, and the second conductivity (Cmax) was determined after the solution cooled to room temperature. Relative EL was calculated using the formula: EL (%) = (Ci/Cmax) × 100.
Photosynthetic parameters
Photosynthetic gas exchange measurements were performed in the third fully expanded leaves using a portable infrared gas analyzer (Li-6400, LICOR, Inc., Lincoln, NE, United States) with the controlled atmosphere (400 μmol mol–1 CO2, 500 μmol s–1 flow rate) and a Licor 6400 LED external light source providing a photosynthetic photon flux density of 600 μmol m–2s–1. Net photosynthetic rate (Pn) and stomatal conductance (Gs) were measured for three subsamples in each pot, with each subsample consisting of five leaves (Hu et al., 2009).
Hormone extraction and fractionation for high performance liquid chromatography-mass spectrometry analysis
Hormones were analyzed as previously described (Albacete et al., 2008). Cytokinins (tZ, ZR, DHZ, DHZR, iP, iPA), auxins (IAA, IBA), gibberellic acid (GA3), and ABA were extracted and purified according to the method of Dobrev and Kamínek (2002). Briefly, about 0.5 g frozen tissues were ground into a fine powder using a pre-chilled mortar and pestle and then soaked in 5 ml of cold extraction mixture of methanol/water/formic acid (15/4/1, v/v, pH 2.5). After 24 h of extraction at −20°C, solids were separated by centrifugation at 12,000 ×g for 15 min, and re-extracted for 1 h in an additional 2 ml of the same extraction solution at 4°C. After centrifugation, extracts were combined with the first supernatant. Pooled supernatants were passed through a Sep-Pak tC18 cartridge (SepPak Plus, Waters, Milford, MA, United States) on an automated solid-phase extraction system (SPE215; Gilson, Middleton, WI, United States) to remove interfering lipids and plant pigments. The column was washed with 0.3 ml of extraction solvent and then combined with elutes. The combined elute evaporated in a vacuum concentrator at 40°C (MiniVac Beta, LABOGENE, Copenhagen, Denmark) and then reconstituted with 2 ml of 1 M formic acid. The hormone-containing fraction was passed through an Oasis MCX 96-Well Plate 150 mg (Waters) preconditioned with 2 ml of methanol followed by 2 ml of 1 M formic acid. After washing with 1 M formic acid, ABA and auxins were eluted with 2 ml of methanol, and cytokinins were eluted with 2 ml of 0.35 M ammonia in 60% (v/v) methanol as indicated in Dobrev and Kamínek (2002). Each fraction was evaporated to dryness in a vacuum concentrator, and the residues were dissolved in a 500 μl of water/methanol (70/30, v/v) mixture. Before injection, the dissolved fraction was filtered through 13-mm-diameter Millex filters with a 0.22-μm-pore nylon membrane (Millipore, Bedford, MA, United States) and the samples were transferred to 2-ml HPLC vials.
Analyses were carried out on a high-performance liquid chromatography (HPLC)-mass spectrometry (MS) system (Accela; Thermo Fisher Scientific, San Jose, CA, United States) coupled to a triple quadrupole mass spectrometer (TSQ Quantum Access MAX, Thermo Fisher Scientific, San Jose, CA, United States) and equipped with a heated electrospray ionization source (HESI-I). For each sample, 10 μl was injected into a Zorbax SB-C18 HPLC column (3.5 μm, 150 × 2.1 mm, Agilent Technologies) maintained at 35°C and eluted at a flow rate of 200 μl min–1. Mobile phase A consisted of water with 0.1%formic acid, and mobile phase B (methanol), was used for the chromatographic separation. The elution program maintained 70% A, 30% B for 3 min, then a linear gradient from 30 to 70% B in 7 min, followed by another linear gradient from 70 to 95% B in 10 min, and finally 95% B maintained for another 5 min. The column was equilibrated with the starting composition of the mobile phase for 30 min before each analytical run. The eluting ions were subjected to multiple reaction monitoring (MRM). The mass spectrometer was operated in the positive mode for cytokinins and IAA, and negative mode for ABA with a capillary spray voltage of 3500 V and a scan speed of 22,000 (m/z)/s from 50 to 600 m/z. Instrument control, data acquisition and processing were performed with Xcalibur 2.1 software (Thermo Fisher Scientific, San Jose, CA, United States). The level of plant hormones (tZ, t-ZR, DHZ, DHZR, iP, iPA, ABA, GA3, IBA, and IAA) in the samples were quantified using the calibration curves constructed for each analyzed component (0.5, 1.0, 2.0, 5.0, and 10.0 μg.ml–1) and corrected with the labeled forms of each compounded as internal standards at 0.1 μg.ml–1. The recovery percentages ranged between 90 and 92%.
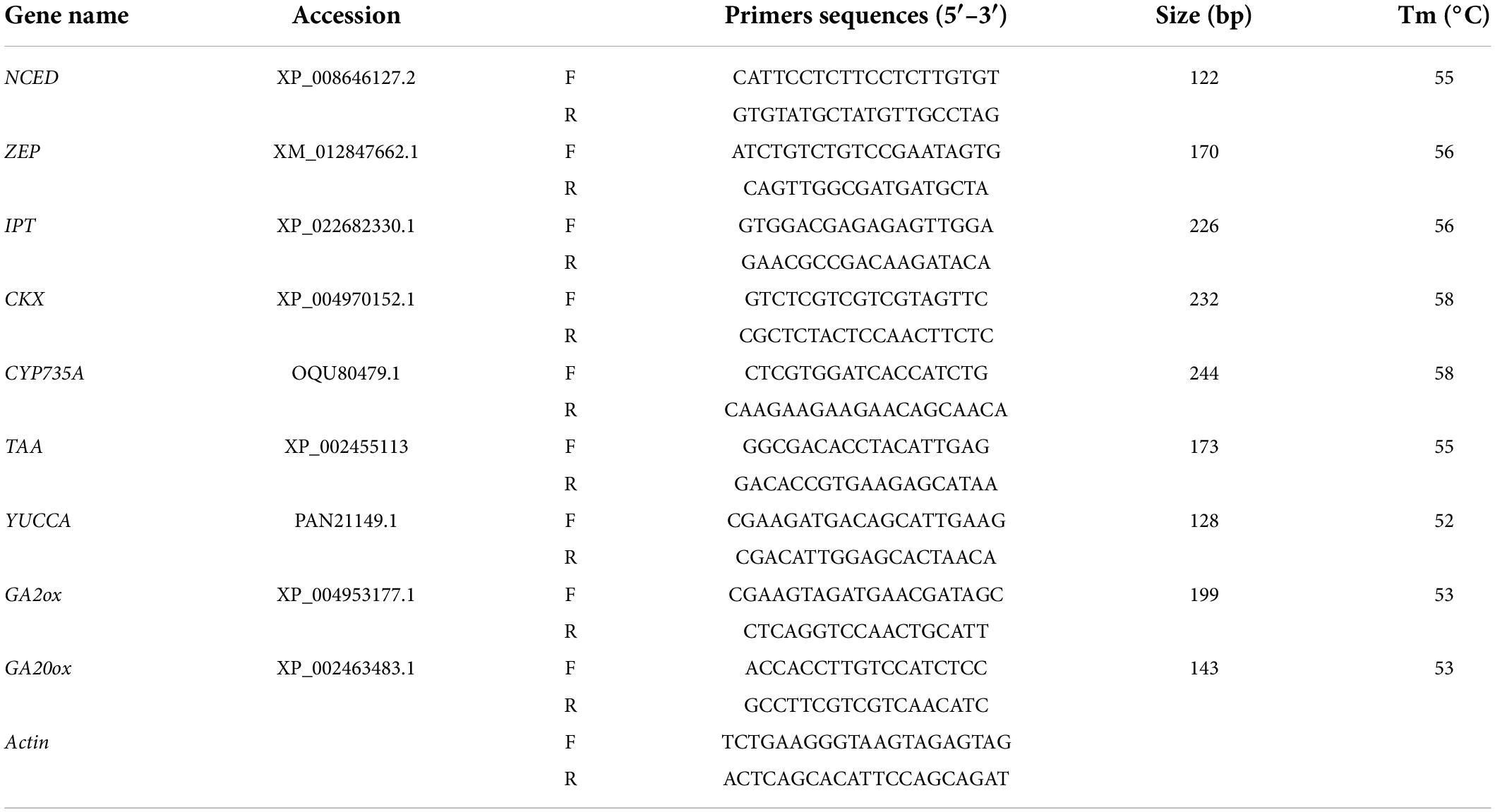
qRT-PCR analysis of genes involved in hormone biosynthesis
Total RNA was extracted from fresh leaves and roots using the Trizol reagent (Invitrogen, Carlsbad, CA, United States) according to the user’s manual. After extraction, the RNA pellet was dissolved in 100 μl of RNase-free water. RNase-free DNase I was added to the total RNA to remove DNA contamination. The total RNA concentration was then determined by absorbance at 260 nm and RNA quality was evaluated on a 0.8% agarose gel. The first-strand cDNA fragments were synthesized from 2 μg of total RNA using oligo(dT)12-18 primer using a cDNA synthesis kit (Fermentas, Burlington, ON, Canada). The gene-specific primers (Table 1) were designed based on the target gene sequences using Primer 5 software (Hu et al., 2015). The actin gene was used as an internal standard. The qRT-PCRs were performed with ABI7500 in a final volume of 20 μl, with each containing 2 μl of cDNA, 10 μl of 2 × SYBR Green qPCR Mix (Takara, Otsu, Shiga, Japan) and 2 μM of the forward and reverse primers. Three independent biological replicates of each sample and two technical replicates of each biological replicate were used for real-time PCR analysis. The thermal cycling conditions were as follows: 40 cycles of 95°C denaturation for 5 s, and 52∼55°C annealing and extension for 20 s. After the PCR, a melting curve was generated by gradually increasing the temperature to 95°C to test the amplicon specificity. The relative expression of genes was calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Statistical analysis
All data were subjected to two-way ANOVA(analysis of variance) according to the general linear model procedure of SAS (SAS Institute, Cary, NC, United States) to determine the effects of genetypes, salinity and their interactions. Treatment means were separated using the Duncan’s multiple range test at the P = 0.05 level of probability.
Results
Phenotypic responses of bermudagrass to salinity stress
Salt stress caused severe damage to shoot growth in both bermudagrass genotypes, but the effect was more pronounced in the ‘C43’ genotype, as evidenced by severe leaf damage and reduced shoot growth (Figure 1). Both genotypes showed a higher shoot growth rate than root growth under normal conditions, and the salt-sensitive genotype had significantly higher shoot and root growth rates than the salt-tolerant (Figure 1). Interestingly, root growth was promoted in the salt-tolerant genotype ‘C43,’ but inhibited in the salt-sensitive ‘C198’ genotype (Figure 1C). Although both genotypes exhibited a high root-to-shoot ratio when exposed to salt stress, the tolerant genotype had a significantly higher root/shoot ratio and root activity than the salt-sensitive genotype up on salt stress (Figures 1D, 2).
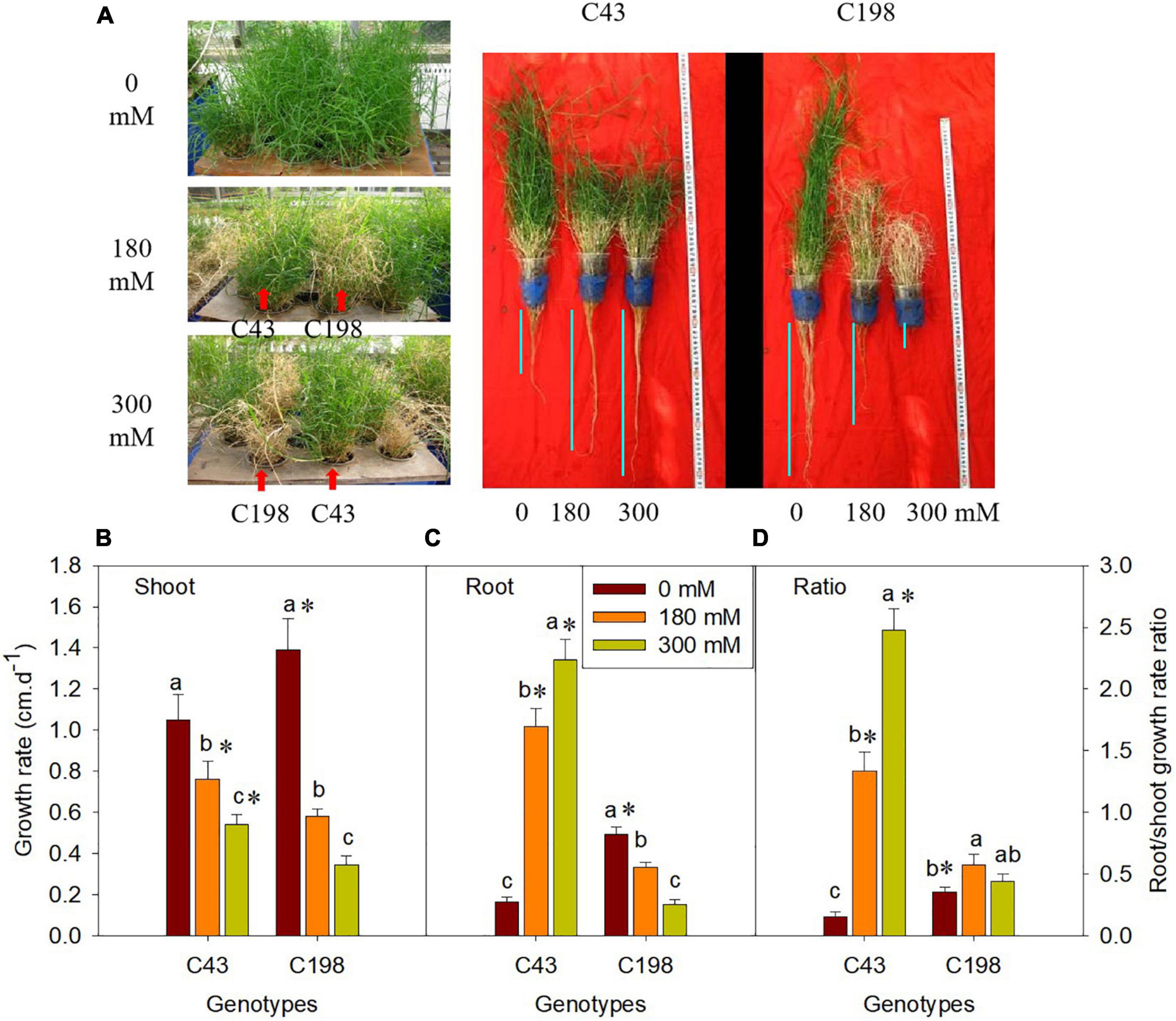
Figure 1. Effect of salt stress on phenotype of bermudagrass (A; C43, tolerant genotype; C198, sensitive genotype), and on the shoot growth rate (B), root growth rate (C) and root/shoot ratio (D) in two genotypes of bermudagrass. Bars marked by the same letters are not significant at P < 0.05 (Tukey’s test) for the comparison of different salt concentrations within a given genotype. Bars marked with star (*) indicate significance at P < 0.05 (Tukey’s test) between the two genotypes at a given salt concentration. Data represent the mean ± SD of four independent biological replicates.
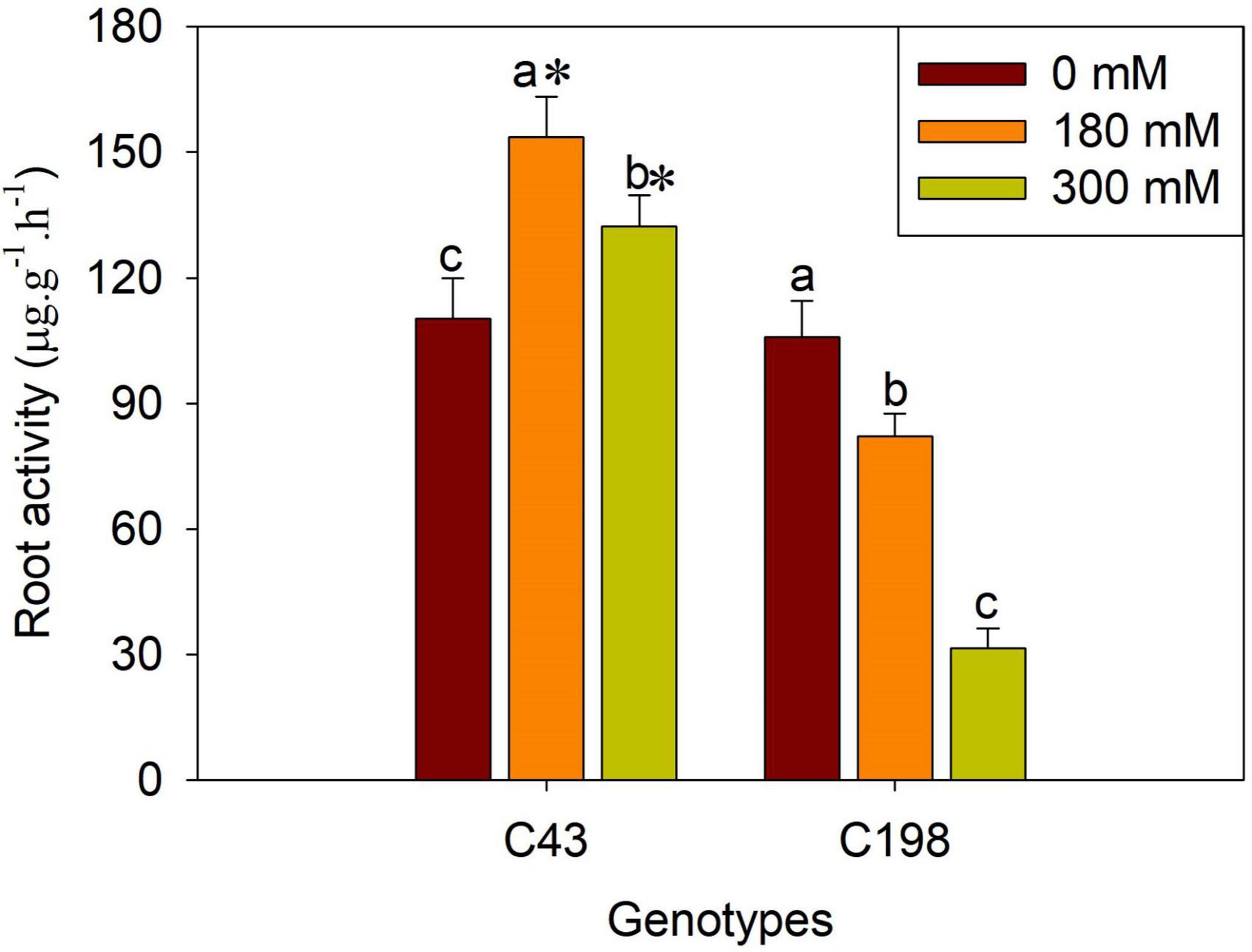
Figure 2. Effects of salt stress on root activity in the two bermudagrass genotypes (C43, tolerant; C198, sensitive). Bars marked by the same letters are not significant at P < 0.05 (Tukey’s test) for the comparison of different salt concentrations within a given genotype. Bars marked with star (*) indicate significance at P < 0.05 (Tukey’s test) between the two genotypes at a given salt concentration. Data represent the mean ± SD of four independent biological replicates.
Physiological responses of bermudagrass to salinity stress
We measured electrolyte leakage in the leaves and roots of the two bermudagrass genotypes to investigate the degree of salt stress-induced membrane damage (Figure 3). The two genotypes did not show a significant difference under control conditions. However, salt stress exposure significantly increased ion leakage in the leaves and roots of both genotypes, more pronounced in the salt-sensitive genotype (Figure 3). Notably, salt stress-induced electrolyte leakage was lower in the roots than in the leaves in both genotypes (Figure 3).
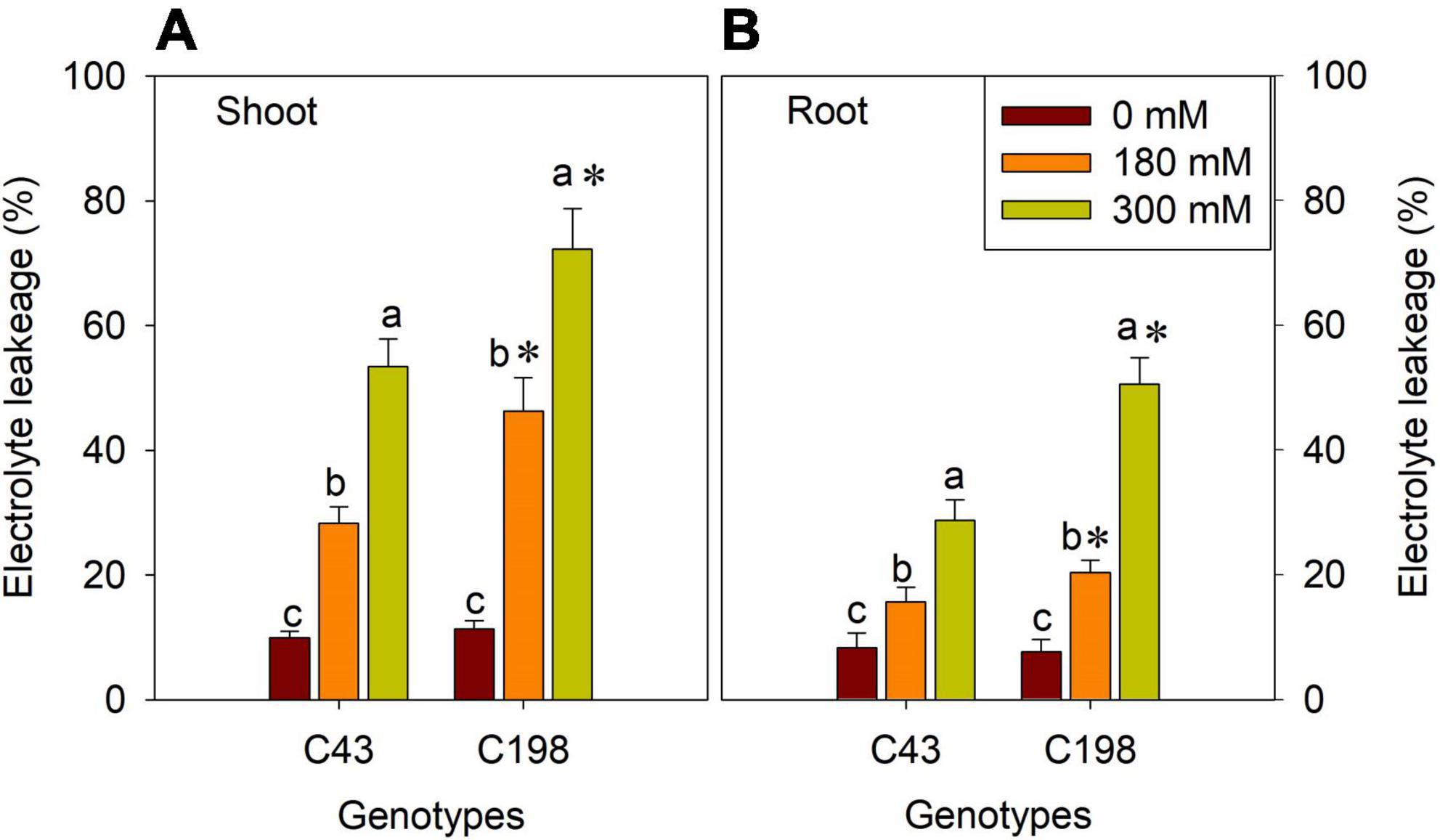
Figure 3. Effects of salt stress on shoot (A) and root (B) membrane stability in the two bermudagrass genotypes (C43, tolerant; C198, sensitive). Bars marked by the same letters are not significant at P < 0.05 (Tukey’s test) for the comparison of different salt concentrations within a given genotype. Bars marked with star (*) indicate significance at P < 0.05 (Tukey’s test) between two genotypes at a given salt concentration. Data represent the mean ± SD of four independent biological replicates.
Furthermore, to get insights into the effect of salt stress on the photosynthetic process of bermudagrass, we measured net Pn and Gs in the leaves of both genotypes after salt stress. There were no significant differences in these parameters between the two bermudagrass under normal conditions. But, salt stress significantly reduced Pn (Figure 4A) and Gs (Figure 4B) in both genotypes compared to the control. The decrease in these photosynthetic parameters was aggravated with increasing salt concentration. Interestingly, the salt-tolerant genotype ‘C43’ showed significantly higher Pn and Gs than the salt-sensitive bermudagrass, indicating its tolerance to salt stress (Figures 4A,B).
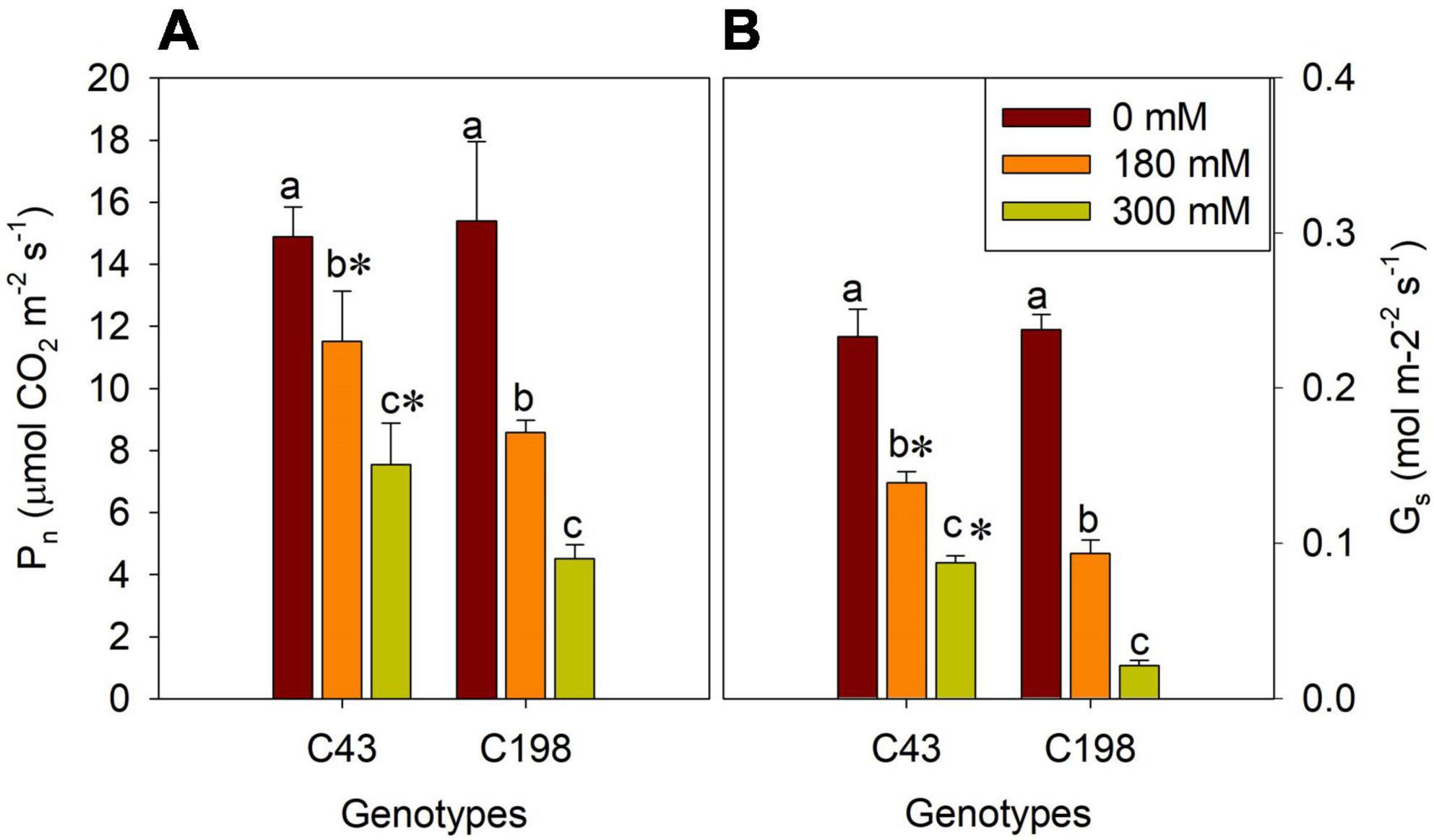
Figure 4. Effects of salt stress on leaf net photosynthetic rate (Pn, A) and stomatal conductance (Gs, B) in the two genotypes of bermudagrass (C43, tolerant; C198, sensitive). Bars marked by the same letters are not significant at P < 0.05 (Tukey’s test) for the comparison of different salt concentrations within a given genotype. Bars marked with star (*) indicate significance at P < 0.05 (Tukey’s test) between the two genotypes at a given salt concentration. Data represent the mean ± SD of four independent biological replicates.
Hormonal responses to salinity stress
Given the role of phytohormones in salt tolerance, an HPLC-MS/MS approach was used to quantify the accumulations of various phytohormones in both genotypes after salt exposure. The representative chromatogram of hormone standards and tissue extracts showed that ABA, auxin (IAA, IBA), cytokinins (tZ, tZR, DHZ, DHZR, iP, iPA) and GA3 had clear separations (Supplementary Figure 1). Under the control condition, a significant difference was observed for DHZR between the roots of two genotypes (Figure 5A). Salt stress decreased the iP, tZ and DHZ content in the roots of both genotypes compared to the control, and no significant difference was observed between ‘C43’ and ‘C198’ under both salt stress levels. Despite salt stress decreased the content of root iPA in both genotypes, ‘C43’ had significantly higher root iPA content than ‘C198’ at 180 and 300 mM salt treatments. Interestingly, the root tZR content was significantly increased in the salt-treated ‘C43,’ but decreased in ‘C198’ compared to the control (Figure 5A).
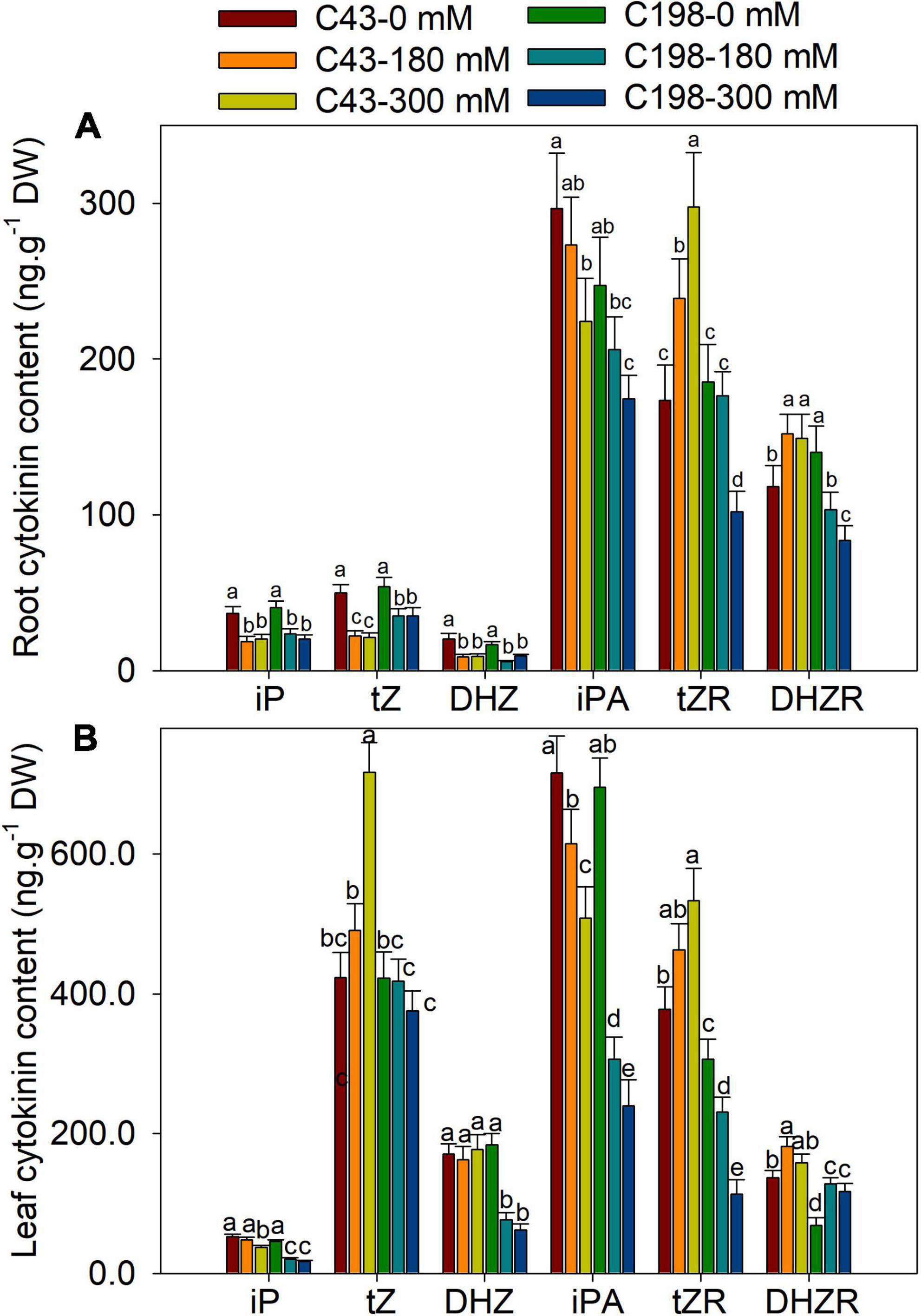
Figure 5. Effects of salt stress on root (A) and leaf (B) cytokinin content in the two genotypes of bermudagrass (C43, tolerant; C198, sensitive). Bars marked by the same letters are not significant at P < 0.05 (Tukey’s test) for the comparison of different salt concentrations in two genotypes. Data represent the mean ± SD of four independent biological replicates.
Both genotype’s leaf iP and iPA content showed decreasing trends with increasing NaCl concentrations (Figure 5B). Relative to the control, the leaf iP content of ‘C43’ decreased to 91% and 71% and that of ‘C198’ to 45% and 37% under 180 and 300 mM NaCl, respectively. In ‘C43’ bermudagrass, 180 and 300 mM NaCl decreased the leaf iPA content to 85% and 71%, but to 44% and 34% l in ‘C198,’ respectively, relative to the control (Figure 5B). Under control conditions, the leaf tZR and DHZR content were higher in ‘C43’ than ‘C198.’ Salt stress caused a significant decline in leaf DHZ content in ‘C198’ but had no effect in ‘C43.’ Compared to the control, salt stress significantly increased leaf tZR content in ‘C43’ by 1.3- and 1.4-fold under 180 and 300 mM NaCl levels, respectively. In contrast, salt stress significantly decreased leaf tZR content in ‘C198’ to 75% and 37% relative to the control (Figure 5B). Salt stress significantly increased leaf DHZR content in both genotypes, with a higher level in ‘C43’ than ‘C198’ bermudagrass.
Salt stress increased the accumulation of ABA in the roots of both genotypes when compared to the control, which was increased by 80% and 267% in ‘C43’ and by 166% and 412% in ‘C198’ after exposure to 180 mM and 300 mM NaCl levels, respectively (Figure 6A). Under the control condition, the root GA3 content was higher in ‘C198’ than in ‘C43.’ The root GA3 content significantly decreased with salt treatments in both genotypes (Figure 6A). Salt stress caused a significant reduction in leaf IAA content in ‘C198,’ whereas the leaf IAA content significantly increased in ‘C43’ after salt stress. Relative to the control, the root IAA content was increased by 1.4- and 2.1-fold in ‘C43,’ but decreased to 67% and 54% in the salt-sensitive bermudagrass under 180 and 300 mM NaCl levels, respectively (Figure 6A). The root IBA content was higher in ‘C43’ than ‘C198’ under control conditions and showed an opposite trend in both genotypes under salt stress compared to the IAA content. Increasing the salt stress level decreased root IBA content in ‘C43’ and significantly increased in ‘C198’ compared to the control plants (Figure 6A).
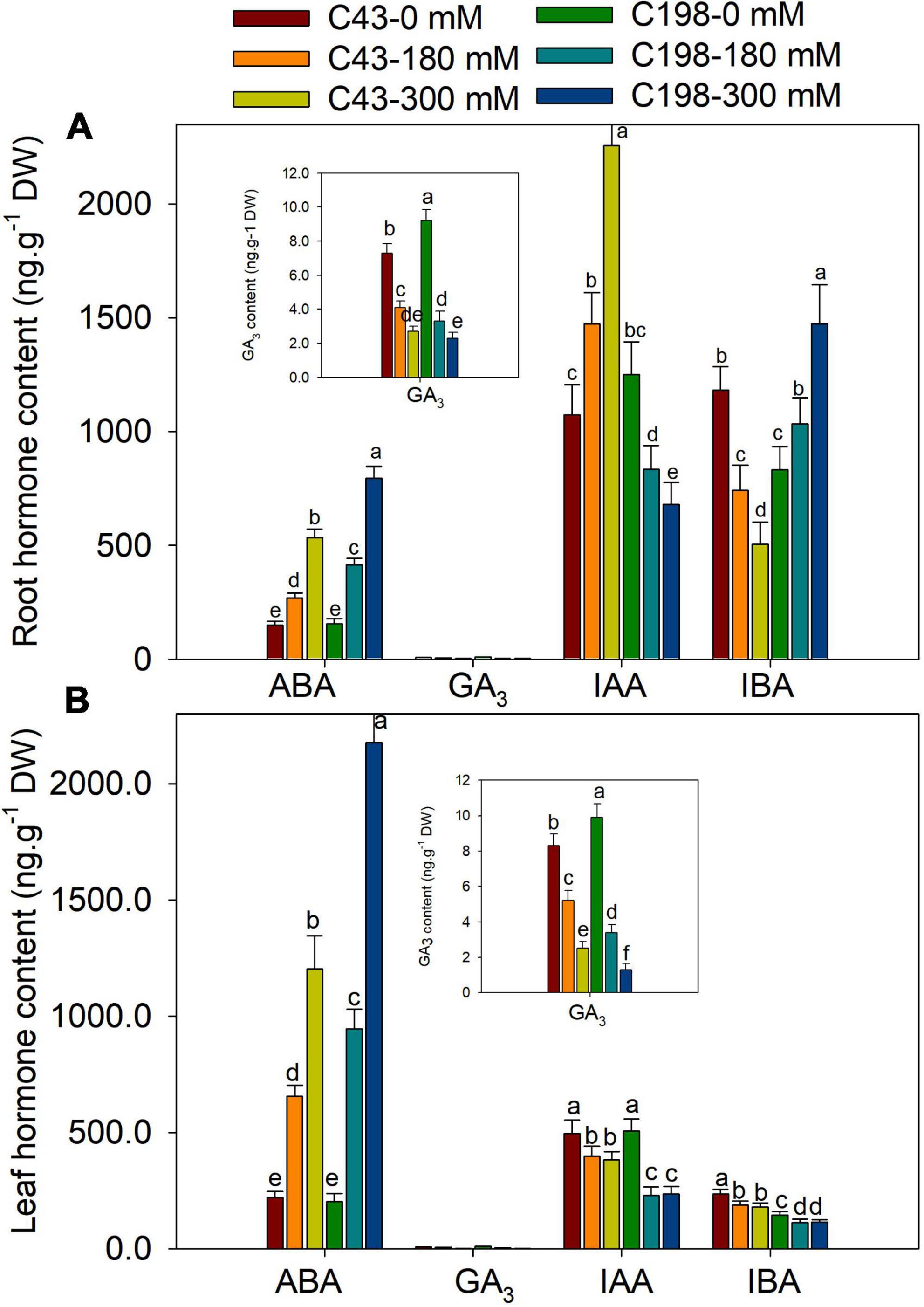
Figure 6. Effects of salt stress on root (A) and leaf (B) cytokinin content in the two genotypes of bermudagrass (C43, tolerant; C198, sensitive). Bars marked by the same letters are not significant at P < 0.05 (Tukey’s test) for the comparison of different salt concentrations in two genotypes. Data represent the mean ± SD of four independent biological replicates.
Salt stress also induced a remarkable accumulation of ABA in the leaves of both genotypes compared to the control. Exposure of plants to 180 mM and 300 mM NaCl increased ABA content by 2.9- and 5.4-fold in ‘C43’ and 4.6- and 10.7-fold in ‘C198’ relative to the control, respectively (Figure 6B). The salt-sensitive bermudagrass exhibited higher leaf GA3 content than the salt-tolerant under control conditions. The leaf GA3 content significantly decreased with an increasing salt concentration in both genotypes (Figure 6B). Salt stress caused a significant reduction in leaf IAA content in ‘C198’ but significantly increased in ‘C43’ after salt stress exposure. We observed 1.4- and 2.1-fold increase in leaf IAA content in ‘C43’ under 180 and 300 mM NaCl, respectively. In comparison, leaf IAA content was significantly decreased to 67% and 54% in ‘C198’ after 180 and 300 mM NaCl, respectively (Figure 6B).
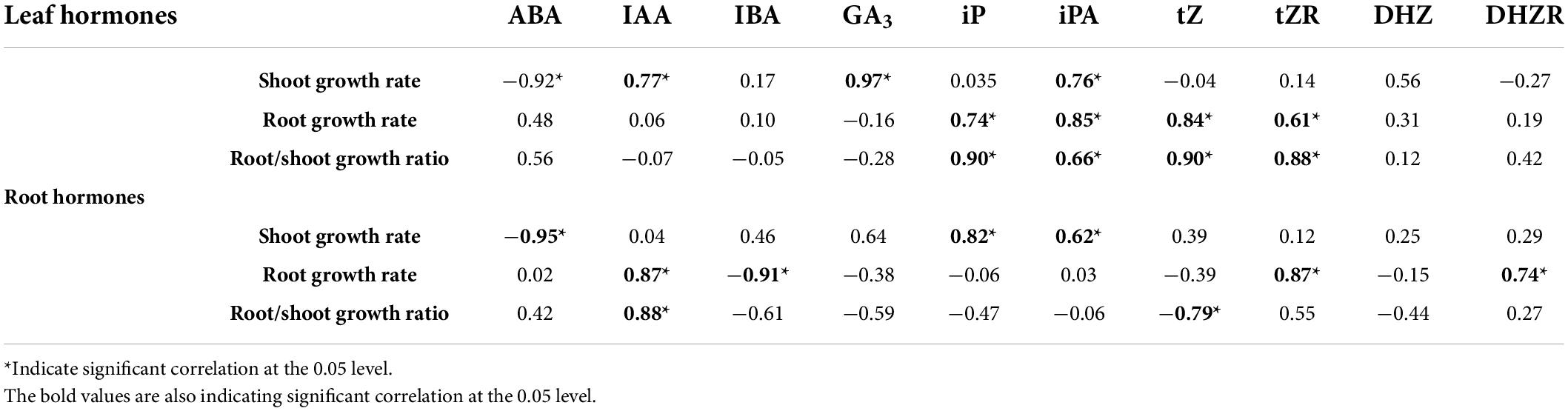
Furthermore, correlation analysis was performed to understand the relationship between different traits. The result showed that leaf and root ABA contents were negatively correlated with the shoot growth rate under salt stress. The shoot growth rate was positively correlated with the leaf IAA, GA3 and iPA, and root iP and iPA content. The root growth rate was positively correlated with the leaf iP, iPA, tZ and tZR, and root IAA, tZR and DHZR content under salt stress (Table 2).
Hormones related gene expressions in bermudagrass
To further examine whether salt regulation of root and shoot growth was associated with hormonal responses at the transcription level, the expression of genes involved in the biosynthesis of ABA, GA, cytokinins, and IAA were measured using quantitative RT-PCR. We investigated the transcription levels of two ABA biosynthesis genes, Zeaxanthin epoxidase (ZEP) and 9-cis-epoxycarotenoid dioxygenase (NCED). Salt stress did not affect the expression of ZEP in both genotypes. However, the expression of NCED was markedly increased in the roots of the two bermudagrass genotypes, more profoundly in ‘C198’ than in ‘C43’ (Figure 7A). We also analyzed the expression levels of the isopentenyl transferase (IPT) and CYP735A genes, encoding the key enzymes involved in the biosynthesis of tZR. Salt stress did not affect the expression level of IPT in the roots of both genotypes. Interestingly, CYP735A exhibited contrasting expression between the two bermudagrass in response to salt stress, increased in ‘C43’ and decreased in ‘C198’ (Figure 7A). We examined the changes in the expression of genes involved in the degradation of cytokinins (cytokinin oxidase/dehydrogenases, CKX). Salt stress caused a remarkable increase in the expression level of CKX in ‘C198,’ particularly under severe salt stress (300 mM). In comparison, only a slight increase in the expression level of CKX was observed in ‘C43’ under severe salt stress (300 mM) (Figure 7A).
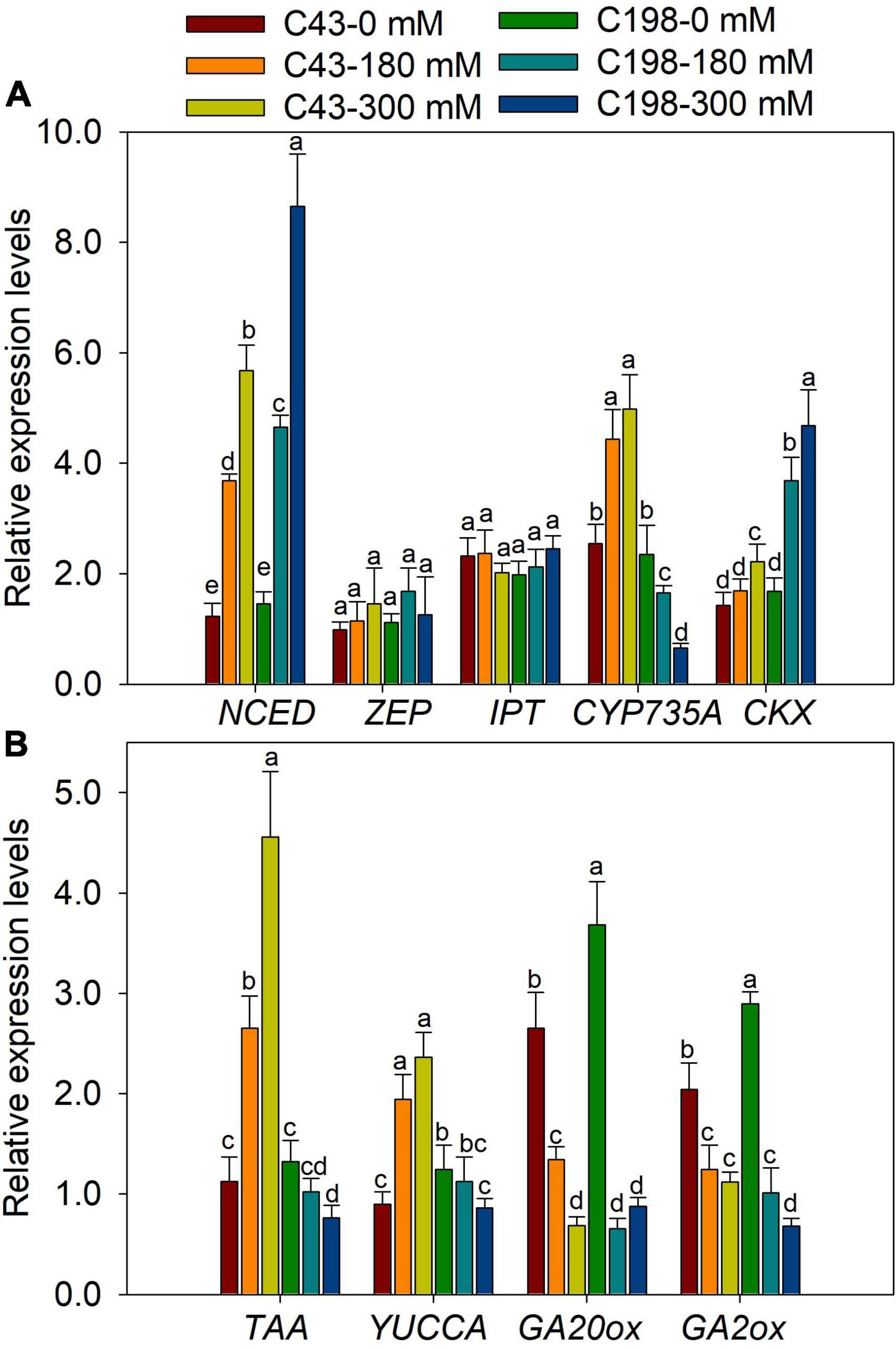
Figure 7. Gene expression levels in roots of the two bermudagrass genotypes differing in salt tolerance (C43: tolerant; C198: sensitive). (A) genes related to ABA and cytokinin biosynthesis; (B) genes relate to auxin and GA biosynthesis. Bars marked by the same letters are not significant at P < 0.05 (Tukey’s test) for the comparison of different treatments. Data represent the mean ± SD of four independent biological replicates.
The genes encoding the key enzymes for IAA biosynthesis, TAA and YUCCA, were up-regulated in ‘C43’ and down-regulated in ‘C198’ following salt stress (Figure 7B). The expression level of GA-synthesis genes (GA20ox) and GA-degradation genes (GA2ox) were significantly down-regulated under salt stress in the roots of the two genotypes (Figure 7B). Salt stress induced the expression of NCED in the leaves of the two bermudagrass genotypes, while the ZEP was markedly increased in ‘C43’ but decreased in ‘C198’ under salt stress (Figure 8A). Contrastingly, the expression levels of IPT and CYP735A were increased in the leaves of ‘C43’ but decreased in ‘C198’ after salt stress (Figure 8A). Salt stress increased the expression level of CKX only in the leaves of ‘C198’ (Figure 8A). TAA and YUCCA expression levels were significantly decreased in leaves of both genotypes under salt stress conditions, with a pronounced decrease in ‘C198’ than in ‘C43’ compared to the control (Figure 8B). Similarly, the expression levels of GA20ox and GA2ox involved in GAs metabolism were significantly decreased with an increasing salt concentration in the leaves of both genotypes (Figure 8B).
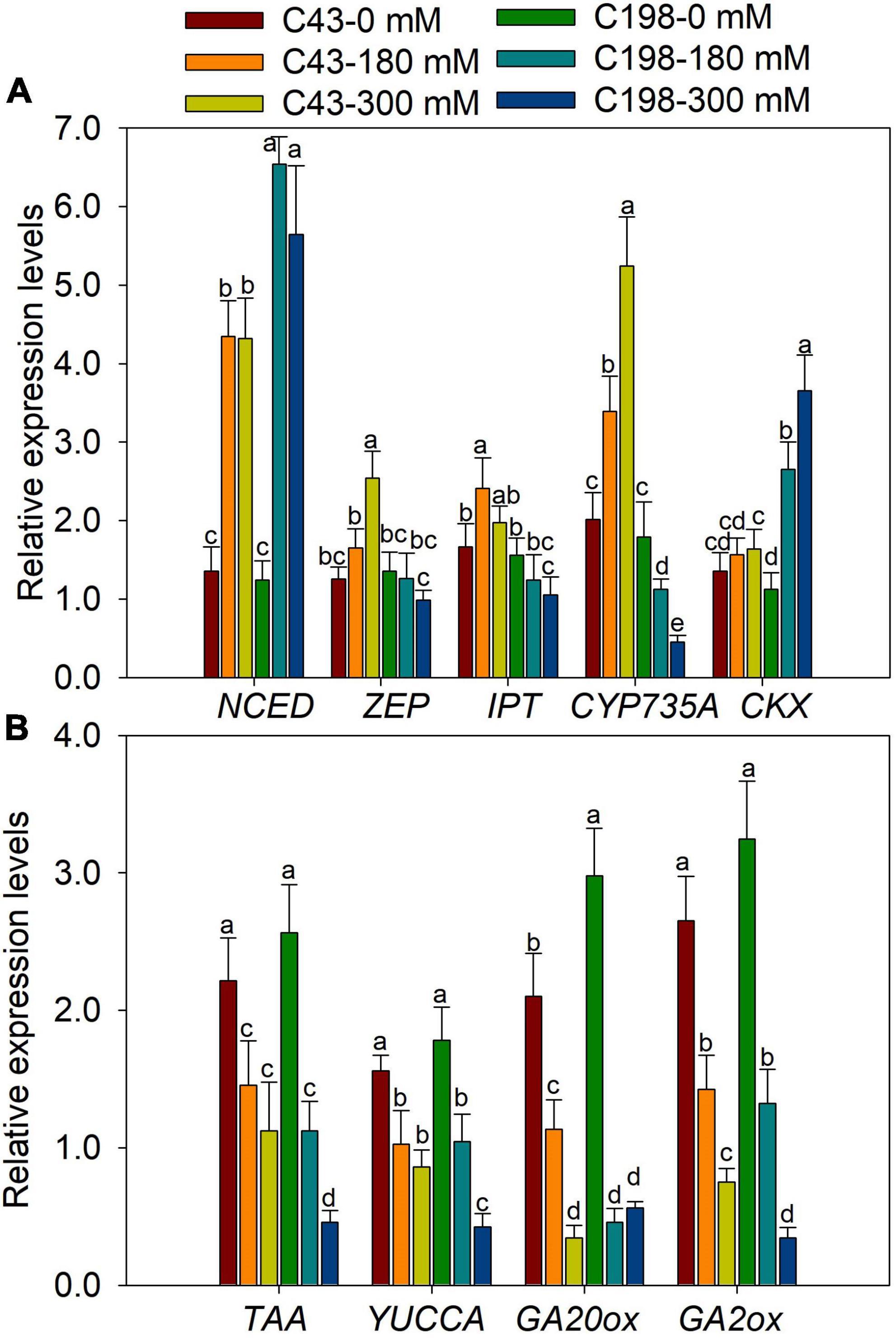
Figure 8. Gene expression levels in leaves of the two bermudagrass genotypes differing in salt tolerance (C43: tolerant; C198: sensitive). (A) genes related to ABA and cytokinin biosynthesis; (B) genes relate to auxin and GA biosynthesis. Bars marked by the same letters are not significant at P < 0.05 (Tukey’s test) for the comparison of different treatments. Data represent the mean ± SD of four independent biological replicates.
Discussion
The adverse impact of salt stress on plant growth and development and related tolerance mechanisms have been widely reported in various plant species (Zelm et al., 2020; Zhao et al., 2021). However, how plants reprogram different phytohormones in response to salt stress and mechanisms associated with phytohormones-mediated root growth and salt tolerance are poorly understood, especially in bermudagrass. This study investigated the morphological, physiological and hormonal responses of two contrasting bermudagrass genotypes; salt-tolerant ‘C43’ and susceptible ‘C43’ (Hu et al., 2012, 2015).
The deleterious effect of salt stress is intensely associated with osmotic stress and ion toxicity due to the accumulation of toxic ions in plant cells (Munns and Tester, 2008). Increasing evidences revealed that salt stress reduces membrane integrity, water content, enzyme activity, and photosynthesis (Shahid et al., 2020; Fang et al., 2021). When exposed to moderate and high salt stress levels, the two bermudagrass genotypes showed a clear phenotypic difference. Under salt stress, the salt-tolerant bermudagrass ‘C43’ showed better green leaves, membrane stability, and photosynthetic activity than the salt-sensitive ‘C198.’
Plant roots are a primary organ for nutrient and water uptake from the soil. Thus, the root system could play a key role in plant salt tolerance (Zou et al., 2021). The literature revealed that salt stress harms the growth and development of plant roots. Meanwhile, studies also indicated that salt stress could promote root growth in salt-tolerant grasses/halophytes (Meng et al., 2018; Rahman et al., 2021). In the present study, the two genotypes showed a contrasting root growth under salt stress. The root growth of the salt-tolerant bermudagrass ‘C43’ was promoted by salt stress, resulting in the higher root/shoot ratio and root activity. Conversely, the root growth of the salt-sensitive bermudagrass was significantly inhibited by salt stress, leading to a lower root/shoot ratio and diminished root activity.
It is well established that an improved root growth under salt stress could retain more toxic ions and prevent translocation to the shoot, providing a crucial mechanism of salt stress tolerance (Cassaniti et al., 2009). Additionally, better root growth could help plants uptake more water and nutrients from the soil under salt-induced osmotic stress conditions (Zou et al., 2021). Here, the salt-tolerant genotype appeared to allocate more energy to root growth, as evidenced by promoted root growth in response to salt stress, which could be an adaptive response to recover the functional equilibrium under new environmental conditions (Li and Li, 2017). Overall, the better green leaves, membrane stability, and higher photosynthetic activity in ‘C43’ could result from improved root growth that retains more toxic ions and/or maintains the water and nutrient supply to the shoots.
Cytokinin is involved in bermudagrass root growth and salt tolerance
Cytokinin is the most crucial hormone that plays a crucial role in the plant cell cycle and various developmental processes, including cell division, cell elongation, transportation of nutrients, root growth, and abiotic stress response (Wu et al., 2021). In this study, the three derivatives of CK, tZ, tZR and DHZR were notably increased in the salt-tolerant genotype but decreased in the salt-sensitive genotype, suggesting that CK may play roles in bermudagrass root growth and salt stress tolerance. Similar results have been reported in Suaeda salsa under salinity stress (Guo et al., 2020). Adenosine phosphate-isopentenyl transferase (IPT) is a key enzyme that catalyzes the de novo synthesis of CKs and the biosynthesis of the major derivatives of CKs, iP- and tZ is catalyzed by IPT. Overexpression of IPT has enhanced the biosynthesis of CKs (Galichet et al., 2008). In our study, the expression of IPT was notably upregulated in the leaves of ‘C43’ genotype, which could be attributed to the higher accumulation of tZ in ‘C43’ leaves. But, IPT was decreased in ‘C198,’ resulting in a decrease in tZ content. CK oxidase/dehydrogenases (CKX), an enzyme that catalyzes the degradation of CK, was upregulated by salt stress in both genotypes, but ‘C198’ had significantly higher expression in both leaves and roots, resulting in lower CK contents in both tissues. The upregulation of IPT in the salt-tolerant bermudagrass was responsible for the accumulation of CK, especially tZ, and may be involved in the salt tolerance of bermudagrass (Figure 9).
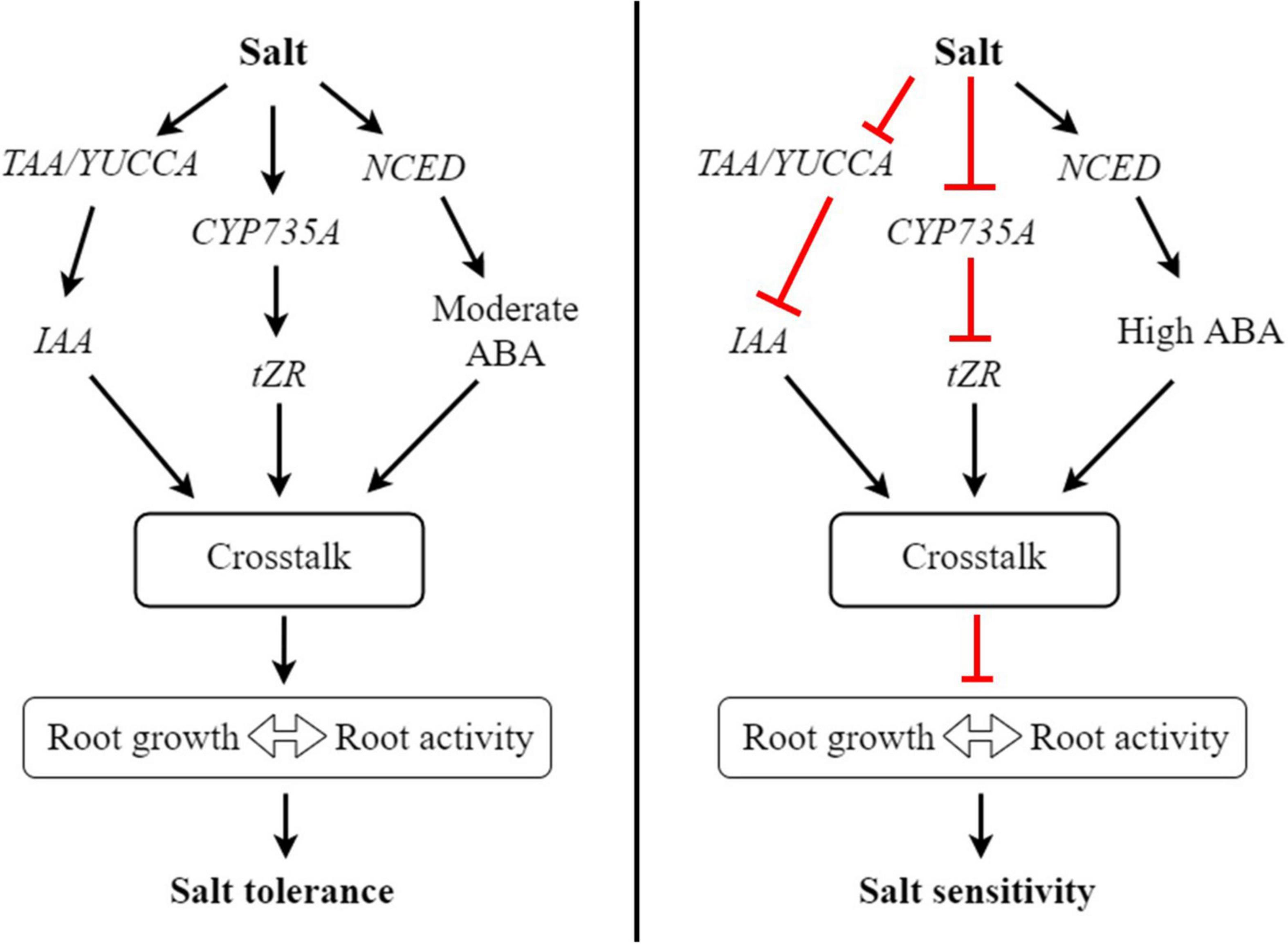
Figure 9. A proposed working model of how hormonal reprogramming and crosstalk are involved in root growth and salt tolerance in bermudagrass. In roots, salt stress induces the expression of TAA/YUCC, CYP735A, and NCED genes involved in auxin, cytokinin, and ABA biosynthesis, respectively. The activation of these genes increases the accumulation of respective hormones in roots. Subsequently, these hormones could work together through hormonal crosstalk and increase root growth and root activity, ultimately improving salt tolerance in bermudagrass. In contrast, salt stress inhibits the expression of TAA/YUCC and CYP735A genes, leading to reduced auxin and cytokinin levels in roots, respectively. On the other hand, salt stress strongly induces the expression of NCED, leading to the accumulation of a high level of ABA in roots. The reduced levels of auxin and cytokinin and high ABA level undergo hormonal crosstalk in roots and inhibit root growth, thus diminishing root activity and increasing salt sensitivity in bermudagrass.
CYP735A is one of the critical enzymes involved in the CK biosynthesis pathway and is required for shoot growth in Arabidopsis (Kiba et al., 2013). Mutation of CYP735A significantly reduced the contents of tZ and tZR, leading to a pronounced growth defect in Jatropha plants (Cai et al., 2018). In our study, the concentration of tZR was significantly higher in the roots of ‘C43’ genotype, suggesting that the accumulation of this CK derivative could play a significant role in bermudagrass salt tolerance by promoting root growth. Interestingly, the expression of CYP735A was remarkably increased in the roots of ‘C43,’ but decreased in ‘C43’ after salt stress. Collectively, these results demonstrate that the upregulation of crucial CK biosynthesis genes could play an invaluable role in salt tolerance by increasing the accumulation of CK derivatives, thereby promoting root growth and salt tolerance in bermudagrass (Figure 9).
The role of auxin in root growth and salt tolerance in bermudagrass
Auxin is a critical plant hormone involved in growth, development, and stress response (Zhao, 2018; Blakeslee et al., 2019; Cao et al., 2019). Auxin can be synthesized through the tryptophan aminotransferase (TAA) and YUCCA (YUC) enzymes-mediated (TAA)/YUCCA (YUC) pathway. In Arabidopsis, the inactivation of several YUC genes dramatically decreased endogenous IAA (Chen et al., 2014). Additionally, the endogenous level of IAA in yuc1 yuc2 yuc6 triple mutants was decreased, leading to increased drought sensitivity in Arabidopsis (Shi et al., 2014). In this study, the concentration of IAA in the roots of the salt-tolerant bermudagrass ‘C43’ was significantly higher than that of ‘C43’ after salt exposure, suggesting that IAA could play a role in root growth. Similarly, salt stress-induced accumulation of IAA has been reported in the roots of a halophyte species, Prosopis strombulifera (Llanes et al., 2014).
On the other hand, salt stress reduced the expression of TAA and YUCCA genes in the leaves of both genotypes. Interestingly, the expression levels of these genes were dramatically increased in the roots of ‘C43,’ but conversely reduced in ‘C198,’ which was consistent with the higher accumulation of IAA in ‘C43.’ It is well established that the higher expression of auxin biosynthesis genes results in higher IAA production in root tips (Brumos et al., 2018). Accumulation of IAA promoted root growth in tea plants exposed to aluminum stress (Gao et al., 2022). Thus, we can speculate that root growth and salt tolerance in bermudagrass could be related to auxin accumulation in the root due to the increased expression of TAA and YUCC genes by salt stress, as shown in the model (Figure 9).
Abscisic acid function in root growth and salt tolerance in bermudagrass
It is well documented that salt-induced osmotic stress could increase the concentration of ABA in both roots and shoots of salt-treated plants, providing a mechanism of salt stress tolerance (Ryu and Cho, 2015; Yu Z. et al., 2020; Huang et al., 2021). Here, salt stress increased the ABA content in the leaves and roots of both genotypes, more prominently in the salt-sensitive genotype. Zeaxanthin epoxidase (ZEP) and 9-cis-epoxycarotenoid dioxygenase (NCED) are critical enzymes in the biosynthesis of ABA. In this study, salt stress did not affect the expression of ZEP in the roots of both genotypes.
Additionally, the expression of NCED was induced by salt stress in bermudagrass. The expression was significantly higher in the roots and leaves of the C198 genotype, consistent with the elevated levels of ABA in both tissues of the salt-sensitive genotype. The accumulation of ABA is primarily associated with the upregulation of NCED genes (Leng et al., 2013). Thus, the accumulation of ABA in bermudagrass is related to the pronounced expression of NCED by salt stress (Figure 9). Because ABA is often recognized as a negative regulator of plant growth (Brookbank et al., 2021), the reduction of shoot growth in the two bermudagrass could have resulted from the increased accumulation of endogenous ABA in the salt-treated leaves.
Interestingly, the increased ABA content was positively correlated with the reduced shoot growth (Table 2). More importantly, ABA regulates root growth and lateral root branching in plants (Sah et al., 2016; Wang et al., 2017). ABA was reported to promote root growth at low concentrations, but it could inhibit root formation at high concentrations (Rowe et al., 2016; Li et al., 2017). In the present study, a higher concentration of ABA was observed in the salt-sensitive genotype with retarded root growth and poor root activity. The increased levels of ABA in the roots and leaves of ‘C198’ could be one reason for the dramatic inhibition of shoot and root growth, increased salt sensitivity.
The role of gibberellin in bermudagrass root growth and salt tolerance
Gibberellin is crucial for plant growth and development, and its deficiency is associated with plant dwarfism (El-Sharkawy et al., 2012; Lee et al., 2014). Stress conditions could inhibit GA biosynthesis and/or increase its degradation, disturbing normal plant metabolism and growth (Llanes et al., 2016). Here, the two bermudagrass genotypes showed a reduced level of GA3 in response to salt stress, leading to shoot growth reduction. GA20-oxidase (GA20-ox) and GA2-oxidase (GA2ox) are key enzymes involved in the biosynthesis and degradation of GAs, respectively. Overexpression of OsGA2ox5 improved rice tolerance to salt stress (Shan et al., 2014).
GA20ox2 was involved in root elongation and root branching by regulating IAA synthesis and transport in Arabidopsis (Lv et al., 2018). Thus, the decrease in GA3 in bermudagrass could be related to the downregulation of GA2ox and GA20ox by salt stress. Under normal conditions, the GA3 content was higher in the salt-sensitive genotype, but relatively ‘C43’ had significantly higher GA3 content under salt stress. At optimal concentration, GA3 is beneficial for the physiology and metabolism of plants under abiotic stresses (Iqbal et al., 2011). These results suggest that the salt-tolerant genotype could produce optimum GA3 to maintain normal functioning, thus improving root growth and salt tolerance.
Hormonal crosstalk is essential for root growth, shoot source activity and salt tolerance in bermudagrass
When exposed to a stressful condition, plants reprogram the biosynthesis and transport of hormones in a way to cope with the stress. Hormonal crosstalk is a crucial process for the understanding of the role of phytohormones, because hormones may not act independently to regulate plant growth and development (Pacifici et al., 2015). To provide a stress defense mechanism and regulate root development, hormonal crosstalk could occur at multiple levels (Murphy, 2015; Aerts et al., 2020). During lateral root formation, cytokinin has been reported to play antagonistic role with auxin in regulating shoot branching (Shimizu-Sato et al., 2009), and lateral root growth (Durbak et al., 2012).
Studies have shown that the crosstalk and regulation of hormones by each other occurs at the transcription level through regulating biosynthesis and degradation genes. For instance, auxin could inhibit IPT expression and conversely increase the expression of CKX to inhibit CK biosynthesis (Nordstrom et al., 2004). Despite they work antagonistically at low and moderate concentrations however, they auxin and CK can work synergistically at higher concentration (Kurepa et al., 2019). In our study, the contents of auxin and cytokinin were higher in the salt tolerant genotype, suggesting that these hormones may work together to positively regulate root growth and shoot source activity in bermudagrass under salt stress (Figures 9, 10). Additionally, the expression of IPT was higher and CKX was lower in the ‘C43,’ suggesting that auxin may not antagonistic to CK in bermudagrass.
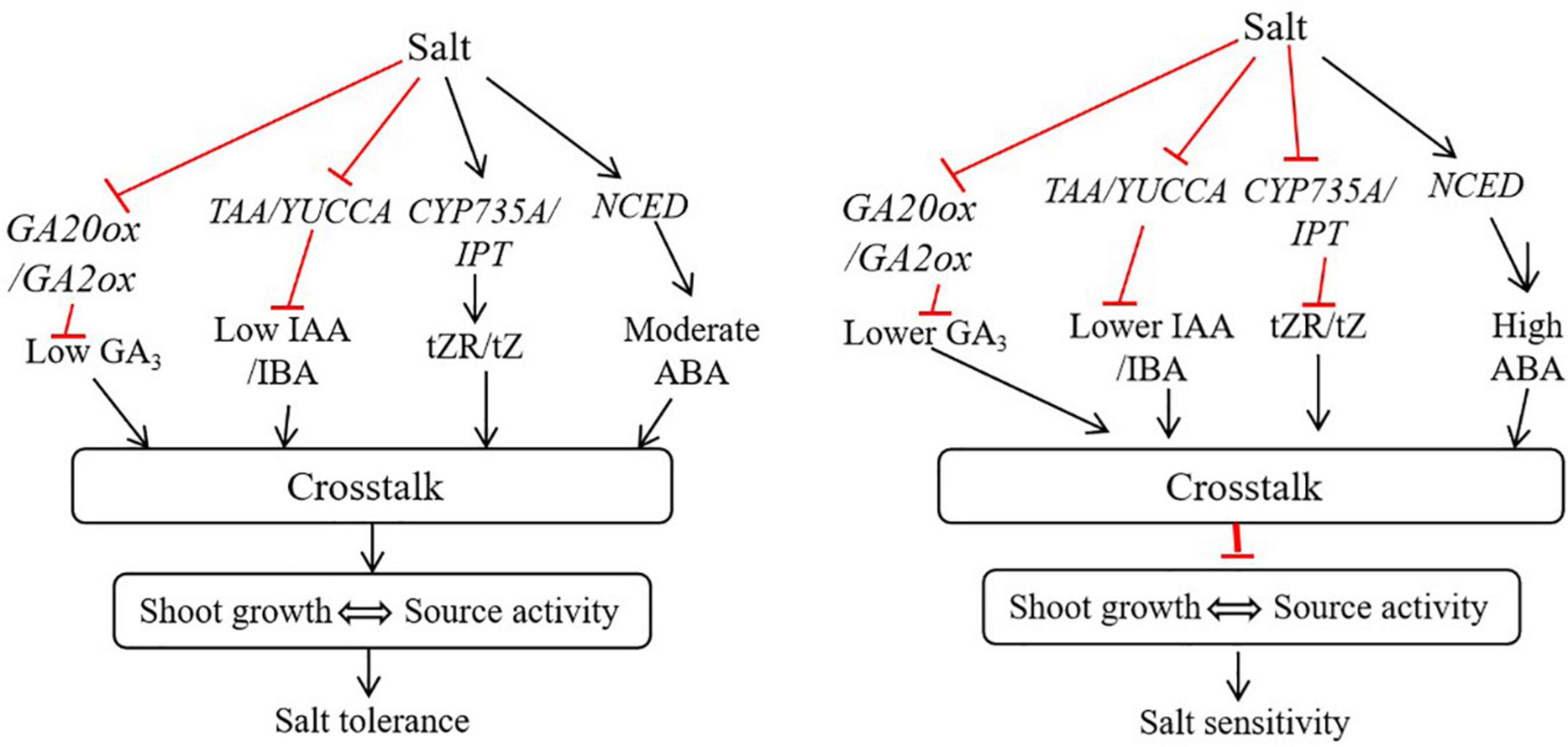
Figure 10. A proposed working model of how hormonal reprogramming and crosstalk are involved in shoot growth and salt tolerance in bermudagrass. In shoots, salt stress induces the expression of CYP735A/IPT and NCED genes involved in tZR, tZ and ABA biosynthesis. The activation of these genes increases the accumulation of respective hormones in shoots. Subsequently, these hormones could work together through hormonal crosstalk and maintain shoot growth and source activity, ultimately improving salt tolerance in bermudagrass. In contrast, salt stress inhibits the expression of TAA/YUCC, CYP735A/IPT, and GA20ox/GA2ox genes, leading to reduced auxin, cytokinin and gibberellin levels in shoot, respectively. On the other hand, salt stress strongly induces the expression of NCED, leading to the accumulation of a high level of ABA in shoot. The reduced levels of auxin, gibberellin and cytokinin and high ABA level undergo hormonal crosstalk in shoot and inhibit shoot growth, thus diminishing source activity and increasing salt sensitivity in bermudagrass.
The negative effect of ABA in seed germination was improved by GAs through antagonistic crosstalk (Verma et al., 2016). In this study, the adverse effect of accumulated ABA in salt tolerant genotype bermudagrass could be masked by accumulation of GAs through hormonal crosstalk, leading to improved growth and salt tolerance (Figures 9, 10). The increase in root growth rate during stress can be regulated by ABA, osmotic stress-induced ABA accumulation could induce auxin transport, thereby improve root elongation under osmotic stress (Xu et al., 2013). Here, the moderately increased level of ABA as a result of salt-induced osmotic stress could be involved in auxin transport to regulate root growth and shoot source activity in bermudagrass. CKs can interact with other hormones, including auxin (IAA) and ABA, to regulate the growth and development of roots (Nguyen et al., 2018). Treatment of plants with exogenous IAA and GA3 could improve tolerance to salt stress by regulating various physiological and biochemical traits (Khalid and Aftab, 2020).
The contents of IAA, GA3 and IBA were decreased in the leaves of the two bermudagrass in response to salt stress. Interestingly, IAA, GA3 and IBA contents were significantly higher in ‘C43.’ Moreover, we found a significant positive correlation between the contents of these hormones and shoot growth under control and salt stress conditions (Table 2). Thus, the decrease in shoot growth in both genotypes could be attributed to the decrease in these hormones. The negative effect of increased ABA level in the salt-tolerant genotype could be masked by other hormones through hormonal crosstalk, leading to better shoot growth and salt tolerance. Therefore, the crosstalk between different phytohormones could be crucial in bermudagrass root growth and salt tolerance.
Due to the complex nature of plant hormone interactions, integrating physiological and molecular levels to the whole plant response is a key to uncovering the detailed mechanisms of hormonal crosstalk (Ross et al., 2011). It has been suggested that combining experimental and modeling studies is a crucial way to dissect the complexity of hormonal crosstalk in regulating root development (Liu et al., 2014). In maize, a higher level of ABA resulting from salt stress has been shown to disturb auxin transport and distribution and inhibit lateral root growth (Lu et al., 2019). A higher salt level could lead to a higher ABA accumulation, which negatively regulates auxin distribution and root development, while moderate salt stress has the opposite effect (Yu Z. et al., 2020). Therefore, in bermudagrass, the decrease in root growth could be associated with a significantly higher level of ABA caused by salt stress, which further inhibits auxin biosynthesis and distribution.
Conclusion
This study demonstrates the role of hormonal reprogramming and crosstalk in regulating root growth and salt tolerance in bermudagrass. In addition, to the morphological and physiological differences, the two contrasting genotypes showed an apparent hormonal regulation, including biosynthesis and degradation in response to salt stress. With improved root growth and root activity by salt stress, the salt-tolerant genotype ‘C43’ had a higher accumulation of IAA, CK, and GA3 in the roots, which are crucial for plant growth and development. The expressions of hormones related genes involved in the biosynthesis of these hormones were induced by salt stress in the salt-tolerant genotype. The antagonistic interaction of two hormones might be affected by the third hormone to synergistically improve root growth and salt tolerance in bermudagrass. Moreover, the crosstalk of hormones and the outcomes depend on the stress condition, the plant species, and the hormones concentration. Thus, further study is necessary to elucidate how hormonal crosstalk regulates root growth and salt tolerance in bermudagrass at the molecular level.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
YY, QX, and L-XH conceived and designed the experiments. YY and MW performed the experiments and wrote the manuscript. YY, MW, and N-FL analyzed the data. HD and Y-BZ helped with manuscript reviewing. QX and L-XH revised the manuscript. All authors read, revised and approved the final manuscript.
Funding
This work was financially supported by the Key Projects of Hunan Provincial Education Department (Grant No. 20A046), the Natural Science Foundation of Hunan Province (Grant No. 2020JJ5245), and Changsha Municipal Science and Technology Project (Grant No. KC1809004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.956410/full#supplementary-material
Supplementary Figure 1 | Representative chromatogram of hormone standards and tissue extracts of bermudagrass under salt stress (A) cytokinin standards; (B) cytoinin in tissues; (C) ABA, auxin andGA3 standards; (D) ABA, auxin and GA3 in tissues.
Abbreviations
ABA, abscisic acid; DHZ, dihydrozeatin; GA, gibberellic acid; iP, isopentenyl; tZ, trans-zeatin; iPA, isopentenyl adenosine; DHZR, dihydrozeatin riboside; IAA, indole acetic acid; tZR, trans-zeatin riboside; Pn, net photosynthetic rate.
References
Achard, P., Gusti, A., Cheminant, S., Alioua, M., Dhondt, S., Coppens, F., et al. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188–1193. doi: 10.1016/j.cub.2009.05.059
Aerts, N., Mendes, M. P., and Van Wees, S. C. M. (2020). Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 105, 489–504. doi: 10.1111/tpj.15124
Albacete, A., Ghanem, M. E., Martínez-Andújar, C., Acosta, M., Sánchez-Bravo, J., Martínez, V., et al. (2008). Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinised tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 59, 4119–4131. doi: 10.1093/jxb/ern251
Blakeslee, J. J., Spatola Rossi, T., and Kriechbaumer, V. (2019). Auxin biosynthesis: Spatial regulation and adaptation to stress. J. Exp. Bot. 70, 5041–5049. doi: 10.1093/jxb/erz283
Brookbank, B. P., Patel, J., Gazzarrini, S., and Nambara, E. (2021). Role of basal ABA in plant growth and development. Genes 12:1936. doi: 10.3390/genes12121936
Brumos, J., Robles, L. M., Yun, J., Vu, T. C., Jackson, S., Alonso, J. M., et al. (2018). Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 47, 306–318.e5. doi: 10.1016/j.devcel.2018.09.022
Cai, L., Zhang, L., Fu, Q., and Xu, Z. F. (2018). Identification and expression analysis of cytokinin metabolic genes IPTs, CYP735A and CKXs in the biofuel plant Jatropha curcas. PeerJ 6:e4812. doi: 10.3390/ijms20246343
Cao, X., Yang, H., Shang, C., Ma, S., Liu, L., and Cheng, J. (2019). The roles of auxin biosynthesis YUCCA gene family in plants. Int. J. Mol. Sci. 20:6343. doi: 10.3390/ijms20246343
Cassaniti, C., Leonardi, C., and Flowers, T. J. (2009). The effects of sodium chloride on ornamental shrubs. Sci. Hort. 122, 586–593. doi: 10.1016/j.scienta.2009.06.032
Chen, Q., Dai, X., DePaoli, H., Cheng, Y., Takebayashi, Y., Kasahara, H., et al. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55, 1072–1079. doi: 10.1093/pcp/pcu039
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Dobrev, P. I., and Kamínek, M. (2002). Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J. Chrom. A. 950, 21–29. doi: 10.1016/S0021-9673(02)00024-9
Durbak, A., Yao, H., and McSteen, P. (2012). Hormone signaling in plant development. Curr. Opin. Plant Biol. 15, 92–99. doi: 10.1016/j.pbi.2011.12.004
El-Sharkawy, I., Kayal, W., Prasath, D., Fernandez, H., Bouzayen, M., Svircev, A. M., et al. (2012). Identification and genetic characterization of a gibberellin 2-oxidase gene that controls tree stature and reproductive growth in plum. J. Exp. Bot. 63, 1225–1239. doi: 10.1093/jxb/err345
Fang, S., Hou, X., and Liang, X. (2021). Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 12:667458. doi: 10.3389/fpls.2021.667458
Galichet, A., Hoyerova, K., Kaminek, M., and Gruissem, W. (2008). Farnesylation directs atipt3 subcellular localization and modulates cytokinin biosynthesis in Arabidopsis. Plant Physiol. 146, 1155–1164. doi: 10.1104/pp.107.107425
Gao, Y., Wang, M., Shi, Y., Yang, L., Hu, J., and Fan, K. (2022). IAA accumulation promotes the root growth of tea plants under aluminum. Agronomy 12:1110. doi: 10.3390/agronomy12051110
Ghassemian, M., Nambara, E., Cutler, S., Kawaide, H., Kamiya, Y., and McCourt, P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell. 12, 1117–26.
Guo, J., Lu, C., Zhao, F., Gao, S., and Wang, B. (2020). Improved reproductive growth of euhalophyte Suaeda salsa under salinity is correlated with altered phytohormone biosynthesis and signal transduction. Funct. Plant Biol. 47, 170–183. doi: 10.1071/FP19215
Gupta, B., and Huang, B. (2014). Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014:701596. doi: 10.1155/2014/701596
Hai, N. N., Chuong, N. N., Tu, N. H. C., Kisiala, A., Hoang, X. L. T., and Thao, N. P. (2020). Role and regulation of cytokinins in plant response to drought stress. Plants 9:422. doi: 10.3390/plants9040422
Hoagland, D. R., and Arnon, D. I. (1950). The water-culture method for growing plants without soil. Calif. Agr. Expt. Stn. Circ. 347:32. doi: 10.1016/S0140-6736(00)73482-9
Hu, L., Li, H., Chen, L., Lou, Y. H., Amombo, E., and Fu, J. M. (2015). RNA-seq for gene identification and transcript profiling in relation to root growth of bermudagrass (Cynodon dactylon) under salinity stress. BMC Genom. 16:575. doi: 10.1186/s12864-015-1799-3
Hu, L. X., Huang, Z. H., Liu, S. Q., and Fu, J. M. (2012). Growth response and gene expression in antioxidant-related enzymes in two bermudagrass genotypes differing in salt tolerance. J. Am. Soc. Hortic. Sci. 137, 134–143. doi: 10.21273/JASHS.137.3.134
Hu, L. X., Wang, Z. L., and Huang, B. R. (2009). Photosynthetic responses of bermudagrass to drought stress associated with stomatal and metabolic limitations. Crop Sci. 49, 1902–1909. doi: 10.2135/cropsci2008.12.0697
Huang, Y., Zhou, J., Li, Y., Quan, R., Wang, J., Huang, R., et al. (2021). Salt stress promotes abscisic acid accumulation to affect cell proliferation and expansion of primary roots in rice. Int. J. Mol. Sci. 22:10892. doi: 10.3390/ijms221910892
Iqbal, N., Nazar, R., Iqbal, M. R. K., Masood, A., and Nafees, A. K. (2011). Role of gibberellins in regulation of source sink relations under optimal and limiting environmental conditions. Curr. Sci 100, 998–1007.
Ismail, A., Takeda, S., and Nick, P. (2014). Life and death under salt stress: Same players, different timing? J. Exp. Bot. 65, 2963–2979. doi: 10.1093/jxb/eru159
Khalid, A., and Aftab, F. (2020). Effect of exogenous application of IAA and GA3 on growth, protein content, and antioxidant enzymes of Solanum tuberosum L. grown in vitro under salt stress. In Vitro Cell. Dev. Biol. Plant 56, 377–389. doi: 10.1007/s11627-019-10047-x
Kiba, T., Takei, K., Kojima, M., and Sakakibara, H. (2013). Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell 27, 452–461. doi: 10.1016/j.devce1.2013.10.004
Kurepa, J., Shull, T. E., and Smalle, J. A. (2019). Antagonistic activity of auxin and cytokinin in shoot and root organs. Plant Direct 3:e00121. doi: 10.1002/pld3.121
Lee, D. H., Lee, I. C., Kim, K. J., Kim, D. S., Na, H. J., Lee, I. J., et al. (2014). Expression of gibberellin 2-oxidase 4 from Arabidopsis under the control of a senescence-associated promoter results in a dominant semi-dwarf plant with normal flowering. J. Plant Biol. 57, 106–116. doi: 10.1007/s12374-013-0528-1
Leng, P., Yuan, B., and Guo, Y. (2013). The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 65, 4577–4588. doi: 10.1093/jxb/eru204
Li, W., and Li, Q. (2017). Effect of environmental salt stress on plants and the molecular mechanism of salt stress tolerance. Int. J. Environ. Sci. Nat. Res. 7:555714. doi: 10.19080/IJESNR.2017.07.555714
Li, X., Chen, L., Forde, B. G., and Davies, W. J. (2017). The biphasic root growth response to abscisic acid in Arabidopsis involves interaction with ethylene and auxin signalling pathways. Front. Plant Sci. 8:1493. doi: 10.3389/fpls.2017.01493
Liu, J., Rowe, J., and Lindsey, K. (2014). Hormonal crosstalk for root development: A combined experimental and modeling perspective. Front. Plant Sci. 5:116. doi: 10.3389/fpls.2014.00116
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Llanes, A., Andrade, A., Alemano, S., and Luna, V. (2016). Alterations of endogenous hormonal levels in plants under drought and salinity. Am. J. Plant Sci. 7, 1357–1371. doi: 10.4236/ajps.2016.79129
Llanes, A., Masciarelli, O., Ordoñez, R., Isla, M. I., and Luna, V. (2014). Differential growth responses to sodium salts involve different ABA catabolism and transport in the halophyte Prosopis strombulifera. Biol. Plant 58, 80–88. doi: 10.1007/s10535-013-0365-6
Lu, C., Chen, M. X., Liu, R., Zhang, L., Hou, X., Liu, S., et al. (2019). Abscisic acid regulates auxin distribution to mediate maize lateral root development under salt stress. Front. Plant Sci. 10:716. doi: 10.3389/fpls.2019.00716
Luo, X., Chen, Z., Gao, J., and Gong, Z. (2014). Abscisic acid inhibits root growth in Arabidopsis through ethylene biosynthesis. Plant J. 79, 44–55. doi: 10.1111/tpj.12534
Lutts, S., Almansouri, M., and Kinet, J. M. (2004). Salinity and water stress have contrasting effects on the relationship between growth and cell viability during and after stress exposure in durum wheat callus. Plant Sci. 167, 9–18. doi: 10.1016/j.plantsci.2004.02.014
Lv, S. F., Yu, D. Y., Sun, Q. Q., and Jiang, J. (2018). Activation of gibberellin 20-oxidase 2 undermines auxin-dependent root and root hair growth in NaCl-stressed Arabidopsis seedlings. Plant Growth Regul. 84, 225–236. doi: 10.1007/s10725-017-0333-9
Mehrotra, R., Bhalothia, P., Bansal, P., Basantani, M. K., Bharti, V., and Mehrotra, S. (2014). Abscisic acid and abiotic stress tolerance - different tiers of regulation. J. Plant Physiol. 171, 486–496. doi: 10.1016/j.jplph.2013.12.007
Meng, X., Zhou, J., and Sui, N. (2018). Mechanisms of salt tolerance in halophytes: Current understanding and recent advances. Open Life Sci. 13, 149–154. doi: 10.1515/biol-2018-0020
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nguyen, T. N., Tuan, P. A., Mukherjee, S., Son, S. H., and Ayele, B. T. (2018). Hormonal regulation in adventitious roots and during their emergence under waterlogged conditions in wheat. J. Exp. Bot. 69, 4065–4082. doi: 10.1093/jxb/ery190
Nordstrom, A., Tarkowski, P., Tarkowska, D., Norbaek, R., Astot, C., Dolezal, K., et al. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: A factor pf potential importance for auxin cytokinin regulated development. Proc. Nat. Acad. Sci. U.S.A. 101, 8039–8044. doi: 10.1073/pnas.0402504101
Pacifici, E., Polverari, L., and Sabatini, S. (2015). Plant hormone crosstalk: The pivot of root growth. J. Exp. Bot. 66, 1113–1121. doi: 10.1093/jxb/eru534
Pérez-Alfocea, F., Albacete, A., Ghanem, M. E., and Dodd, I. C. (2010). Hormonal regulation of source-sink relations to maintain crop productivity under salinity: A case study of root-to-shoot signalling in tomato. Funct. Plant Biol. 37, 592–603. doi: 10.1071/FP10012
Pessarakli, M. (2015). Using bermudagrass (Cynodon dactylon L.) in urban desert landscaping and as a forage crop for sustainable agriculture in arid regions and combating desertification. Int. J. Water Resour. Arid Environ. 4, 8–14.
Rahman, M. M., Mostofa, M. G., Keya, S. S., Siddiqui, M. N., Ansary, M. M. U., Das, A. K., et al. (2021). Adaptive mechanisms of halophytes and their potential in improving salinity tolerance in plants. Int. J. Mol. Sci. 22:10733. doi: 10.3390/ijms221910733
Ross, J. J., Weston, D. E., Davidson, S. E., and Reid, J. B. (2011). Plant hormone interactions: How complex are they? Physiol. Plant 141, 299–309. doi: 10.1111/j.1399-3054.2011.01444.x
Rowe, J. H., Topping, J. F., Liu, J., and Lindsey, K. (2016). Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 211, 225–239. doi: 10.1111/nph.13882
Ryu, H., and Cho, Y. G. (2015). Plant hormones in salt stress tolerance. J. Plant Biol. 58, 147–155. doi: 10.1007/s12374-015-0103-z
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. doi: 10.3389/fpls.2016.00571
Shahid, M. A., Sarkhosh, A., Khan, N., Balal, R. M., Ali, S., Rossi, L., et al. (2020). Insights into the physiological and biochemical impacts of salt stress on plant growth and development. Agronomy 10:938. doi: 10.3390/agronomy10070938
Shan, C., Mei, Z., Duan, J., Chen, H., Feng, H., and Cai, W. (2014). OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One 9:e87110. doi: 10.1371/journal.pone.0087110
Shi, H., Chen, L., Ye, T., Liu, X., Ding, K., and Chan, Z. (2014). Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 82, 209–217. doi: 10.1016/j.plaphy.2014.06.008
Shimizu-Sato, S., Tanaka, M., and Mori, H. (2009). Auxin–cytokinin interactions in the control of shoot branching. Plant Mol. Biol. 69, 429–435. doi: 10.1007/s11103-008-9416-3
Sun, L. R., Wang, Y. B., He, S. B., and Hao, F. S. (2018). Mechanisms for abscisic acid inhibition of primary root growth. Plant Signal. Behav. 13:e1500069. doi: 10.1080/15592324.2018.1500069
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wang, T., Li, C., Wu, Z., Jia, Y., Wang, H., Sun, S., et al. (2017). Abscisic acid regulates auxin homeostasis in rice root tips to promote root hair elongation. Front. Plant Sci. 8:1121. doi: 10.3389/fpls.2017.01121
Wolters, H., and Jürgens, G. (2009). Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10, 305–317. doi: 10.1038/nrg2558
Wu, Y., Liu, H., Wang, Q., and Zhang, G. (2021). Roles of cytokinins in root growth and abiotic stress response of Arabidopsis thaliana. Plant Growth Regul. 94, 151–160. doi: 10.1007/s10725-021-00711-x
Xu, W., Jia, L., Shi, W., Liang, J., Zhou, F., Li, Q., et al. (2013). Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol. 197, 139–150. doi: 10.1111/nph.12004
Yu, D., Boughton, B. A., Hill, C. B., Feussner, I., Roessner, U., and Rupasinghe, T. W. T. (2020). Insights into oxidized lipid modification in barley roots as an adaptation mechanism to salinity stress. Front. Plant Sci. 11:1. doi: 10.3389/FPLS.2020.00001
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., and Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Zelm, E. V., Zhang, Y., and Testerink, C. (2020). Salt tolerance mechanisms of plants. Ann. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Zhao, S., Zhang, Q., Liu, M., Zhou, H., Ma, C., and Wang, P. (2021). Regulation of plant responses to salt stress. Int. J. Mol. Sci. 22:4609. doi: 10.3390/ijms22094609
Zhao, Y. (2018). Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 69, 417–435. doi: 10.1146/annurev-arplant-042817-040226
Keywords: bermudagrass, gene expression, hormone, root growth, salt stress
Citation: Yang Y, Wassie M, Liu N-f, Deng H, Zeng Y-b, Xu Q and Hu L-x (2022) Genotypic-specific hormonal reprogramming and crosstalk are crucial for root growth and salt tolerance in bermudagrass (Cynodon dactylon). Front. Plant Sci. 13:956410. doi: 10.3389/fpls.2022.956410
Received: 30 May 2022; Accepted: 18 July 2022;
Published: 04 August 2022.
Edited by:
Jing Zhang, Nanjing Agricultural University, ChinaReviewed by:
Zhou Li, Sichuan Agricultural University, ChinaJibiao Fan, Yangzhou University, China
Kun Zhang, Qingdao Agricultural University, China
Copyright © 2022 Yang, Wassie, Liu, Deng, Zeng, Xu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Xu, eHEyMDA1MjAwNUAxMjYuY29t; Long-xing Hu, Z3Jhc3NAaHVuYXUuZWR1LmNu
†These authors have contributed equally to this work
 Yong Yang
Yong Yang Misganaw Wassie
Misganaw Wassie Ning-fang Liu2
Ning-fang Liu2 Qian Xu
Qian Xu Long-xing Hu
Long-xing Hu