- State Key Laboratory of Subtropical Silviculture, National Forestry and Grassland Innovation Alliance on Dendrobium catenatum, SFGA Engineering Research Center for Dendrobium catenatum, Zhejiang A&F University, Hangzhou, China
Dendrobium catenatum, belonging to the Orchidaceae, is a precious Chinese herbal medicine. Sclerotium delphinii (P1) is a broad-spectrum fungal disease, which causes widespread loss in the near-wild cultivation of D. catenatum. Thus, resistance breeding of D. catenatum has become the key to solve this problem. The basic helix–loop–helix (bHLH) gene family is closely related to plant resistance to external stresses, but the related research in D. catenatum is not deep enough yet. Phylogenetic analysis showed that 108 DcbHLH genes could be divided into 23 subgroups. Promoter cis-acting elements revealed that DcbHLHs contain a large number of stress-related cis-acting elements. Transcriptome analysis of MeJA and P1 treatment manifested that exogenous MeJA can change the expression pattern of most bHLH genes, especially the IIIe subgroup, including inhibiting the expression of DcbHLH026 (MYC2a) and promoting the expression of DcbHLH027 (MYC2b). Subcellular localization indicated that they were located in the nucleus. Furthermore, exogenous MeJA treatment significantly delayed disease time and reduced lesion size after infection with P1. DcMYC2b-overexpression Arabidopsis lines showed significantly smaller lesions after being infected with P1 than the wild type, indicating that DcMYC2b functions as an important positive regulator in D. catenatum defense against P1. Our findings shed more insights into the critical role of the DcbHLH family in plants and the resistance breeding of D. catenatum.
Introduction
Dendrobium catenatum (D. officinale), a perennial herb of the genus Dendrobium in the Orchidaceae family, is a valuable traditional Chinese medicinal herb used for relieving upset stomach, promoting body fluid production, enhancing immunity, nourishing “yin,” and exerting antioxidant and anti-tumor effects (Chen et al., 2020b). In the past, D. catenatum has undergone a cross from natural wild to facility cultivation followed by near-wild cultivation (Chen et al., 2020b). Although the renewal of cultivation patterns has brought about species conservation and the rise of industry, resulting in great ecological and economic benefits (Si et al., 2017), it is susceptible to diseases in its large-scale cultivation, particularly southern blight disease, which causes substantial economic loss (Chen et al., 2019a). Southern blight disease is a broad-spectrum disease caused by the necrotrophic pathogen Sclerotium delphinii (P1) and Sclerotium rolfsii (Iquebal et al., 2017; Uji et al., 2017), and the former is more pathogenic (Uji et al., 2017). In warm and high humidity environments, S. delphinii infects the host and secretes numerous cell wall-degrading enzymes (CWDEs), leading to the degradation of the host cell wall (Li et al., 2017), followed by hyphal penetration of plant tissues, causing local necrosis and the formation of massive white mycelium and rapeseed-like sclerotia (Uji et al., 2017).
The family comprising basic helix–loop–helix (bHLH) transcription factors (TFs) is the second-largest family in eukaryotes (Ledent and Vervoort, 2001; Toledo-Ortiz et al., 2003), with a highly conserved bHLH domain, which can be divided into two functional regions: the basic region and the HLH region (Li et al., 2006). The former is located at the N-terminal end of the bHLH domain and associated with DNA binding, which enables the recognition of an E-Box (5′-CANNTG-3′) or G-box (5′-CACGTG-3′) upstream of a specific DNA sequence for the expression of genes involved in physiological and developmental processes and stress responses (Wang et al., 2015). The C-terminal HLH region is characterized by two amphipathic α-helices with a linking loop and is responsible for dimerization and interactions with other bHLH proteins (Heim et al., 2003). With the release of a vast number of genome sequences, more and more bHLH genes have been identified (Heim et al., 2003; Li et al., 2006, 2019; Yang et al., 2020; Zhou et al., 2020). In plants, the classification of bHLH proteins is usually based on the sequence homology of bHLH domains (Buck and Atchley, 2003; Pires and Dolan, 2010), with the number of subgroups ranging from 15 to 26. Some atypical bHLH proteins are even classified into 32 subgroups after phylogenetic analysis (Carretero-Paulet et al., 2010). There is a special subgroup within the family of bHLH TFs called myelocytomatosis oncogenes (MYC). Members of this subgroup contain a bHLH_MYC_N domain at their N-terminal end in addition to a bHLH domain at their C-terminal end (Chen et al., 2019b). Among them, the bHLH_MYC_N domain contains a JAZ interaction domain (JID) and a putative transcriptional activation domain (TAD). Previous studies have demonstrated that the family of bHLH TFs plays important role in plant growth and development (Hao et al., 2021). For example, 77 bHLH proteins in Gossypium hirsutum may be involved in brassinosteroid (BR) hormone signaling pathways to promote cotton fiber cell elongation (Lu et al., 2018). In Arabidopsis, MADS-Box and bHLH TFs coordinate transmitting-tract development, as well as bHLH IIIe subfamily member AtMYC2 functions synergistically with AtMYC3 and AtMYC4 in regulating leaf senescence (Di Marzo et al., 2020). Meanwhile, there are many bHLH genes involved in plant responses to biotic and abiotic stresses (Hao et al., 2021). MdbHLH130, an apple bHLH TFs, can improve plant tolerance to water deficit stress (Zhao et al., 2020). Overexpression of Vitis amurensis VabHLH1 and Vitis vinifera VvbHLH1 in Arabidopsis enhanced tolerance to low-temperature stress (Xu et al., 2014). In Arabidopsis, four IVc bHLH TFs (bHLH34, bHLH104, bHLH105/ILR3-IAA-LEUCINE RESISTANT3, and bHLH115) formed homodimers and heterodimers to regulate the Fe deficiency response and the Fe homeostasis (Zhang et al., 2015; Li et al., 2016; Liang et al., 2017; Tissot et al., 2019). Arabidopsis bhlh104 mutant was sensitive to cadmium (Cd) stress, whereas plants overexpressing bHLH104 exhibited enhanced Cd tolerance (Yao et al., 2018). The jasmonate (JA) signaling pathway primarily responds to plant immunity to necrotrophic fungal pathogens and herbivorous insects in the plant (Yan and Xie, 2015). AtMYC2, one of the core TFs of the JA signaling pathway, plays a crucial step in plant responses to biotic stresses (Dombrecht et al., 2007). For instance, AtMYC2, AtMYC3, and AtMYC4 act to decrease the feeding of Spodoptera littoralis by regulating glucosinolate biosynthesis (Schweizer et al., 2013). Moreover, AtMYC2 can directly bind to the promoters of terpenoid synthesis genes (TPS10, TPS11, and TPS21) and activate their expression to promote terpenoid synthesis, thereby enhancing terpenoid-mediated direct and indirect plant resistance to pests (Fernandez-Calvo et al., 2011; Adams et al., 2021). In Oryza sativa, the OsMYC2-RNAi lines showed enhanced resistance against bacterial pathogen Xanthomonas oryzae pv. oryzae (Giri et al., 2017). In tomatoes, SlMYC2 positively regulates both wounding-responsive genes by activating JA2L and pathogen-responsive genes by activating ERF.C3 (Du et al., 2017). Botrytis cinerea infection assay of tomato presented significantly larger necrotic lesions in MYC2-RNAi plants than in the wild type (Du et al., 2017). Interestingly, AtMYC2 negatively regulated resistance to B. cinerea and Fusarium oxysporum (Song et al., 2014). However, no systematic characterization of bHLH TFs in D. catenatum and their functions under biotic and abiotic stresses have been reported yet.
In the present study, 108 DcbHLH genes were identified. Phylogenetic analysis of bHLH proteins with Arabidopsis, O. sativa, Phalaenopsis equestris, and Apostasia shenzhenica showed that DcbHLHs were divided into 23 subgroups. Meanwhile, the gene structure, physical and chemical properties, conserved motifs of proteins, and the promoter cis-acting element were analyzed. Besides, the role of JA in enhancing D. catenatum resistance to P1 was investigated. At the transcriptome level, the expression of DcbHLH026(MYC2a)/027(MYC2b) was significantly changed after MeJA and P1 treatment, indicating that it might be a negative or positive regulator of the JA-mediated plant response to P1. Overexpression of DcMYC2b enhanced the tolerance to P1 in transgenic Arabidopsis plants, suggesting that DcMYC2b-mediated JA signaling pathway plays a positive regulatory role in D. catenatum response to P1. In brief, our study revealed a potential regulatory mechanism of DcMYC2b involved in the D. catenatum defense against P1, which provides an important theoretical basis for future resistance breeding of D. catenatum.
Materials and methods
Plant materials and growth conditions
Dendrobium catenatum clonal cultivar was placed in a modified 1/2 Murashige and Skoog (MS) medium, cultivated in a growth room with a 12-h light/12-h dark photoperiod, 60% relative humidity, and 25°C temperature.
The Columbia accession (Col-0) of Arabidopsis was used as wild type (WT). Arabidopsis mutants coi1-1 (Xu et al., 2002), myc3 (GK445B11; Fernandez-Calvo et al., 2011), and myc4 (GK491E10; Fernandez-Calvo et al., 2011) were provided by Professor Xie Daoxin of Tsinghua University and Professor Song Susheng of Capital Normal University. myc2 mutant (SALK_017005c) was obtained from Arashare.1 Arabidopsis plants were grown on soil under long-day photoperiod conditions (16-h light/8-h dark).
Identification and analysis of bHLH genes in Dendrobium catenatum, Phalaenopsis equestris, and Apostasia shenzhenica
Dendrobium catenatum, P. equestris, and A. shenzhenica genome sequences were retrieved from NCBI Genome.2 The hidden Markov model (HMM) file of the bHLH domain (PF00010) was downloaded from the Pfam database.3 The HMMER program was used to search for bHLH proteins with a cut-off E-value of 0.001 using PF00010 as a query. We obtained 141 suspected bHLH genes in D. catenatum, of which the 60 with the lowest e-value were selected to construct species-specific HMM, which was used to check again. The obtained potential bHLH genes were further confirmed using Pfam4 and SMART databases.5 The acquired sequences were submitted to ExPASy6 to calculate the molecular weight (MW) and theoretical isoelectric point (pI).
Bioinformatic analysis of bHLH genes
Multiple sequence alignment of bHLH proteins was performed using ClustalW with default parameters. MEGA 7.0 software was used to construct neighbor-joining (NJ) distance trees with the bHLH proteins of D. catenatum, P. equestris, A. shenzhenica, Arabidopsis (Toledo-Ortiz et al., 2003), and O. sativa (Li et al., 2006). Evolutionary trees were constructed with the following parameters: 1,000 bootstrap replications, p-distance model or method, and pairwise deletion for gaps or missing data. The phylogenetic tree was subsequently visualized with EvolView.7 Subfamily grouping of the DcbHLH proteins was performed according to the classification scheme of the AtbHLH proteins (Heim et al., 2003).
We used GSDS8 to predict the number of untranslated regions and coding domain sequences (CDSs) of the DcbHLH genes. The conserved motifs were investigated by the MEME version 5.3 online tool.9
To investigate putative genes in the promoter regions of DcbHLH genes, 1,500-bp genomic DNA sequences upstream of the transcription start site were retrieved and screened against the Plant CARE database.10 The gene structure, motif, and composition of cis-acting elements were visualized using the TBtools (Chen et al., 2020a).
The expression patterns of DcbHLH genes
In order to analyze the expression patterns of DcbHLH genes, we downloaded the RNA-Seq from the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra), which contains the data for different organs/tissues (Zhang et al., 2017), drought (Zou et al., 2018), salt (Zhang et al., 2021), and cold stresses (Wu et al., 2016), and S. delphinii (MN061040.1, named P1) inoculation assays (Li et al., 2021).
The expression data of different organs/tissues were provided by Zhang et al. (2017), including the expression levels in 10 organs/tissues [leaf (SRR4431601), stem (SRR4431600), root (SRR5722140), green root tip (SRR4431599), white part of the root (SRR4431598), flower bud (SRR4431603), sepal (SRR4431597), labellum (SRR4431602), pollinia (SRR5722145), and gynostemium (SRR4431596)].
For the drought and rewatering experiments, the protocol followed was as follows: watering on the 1st day, simulating drought on the 2nd–7th day, rewatering on the 8th day, and then watering every 2 days at 15:30 h (Zou et al., 2018). The expression data were obtained from the leaves that were collected at both 06:30 and 18:30 h on the 2nd [DR5 (SRR7223299) and DR8 (SRR7223300)], 7th [DR6 (SRR7223298) and DR10 (SRR7223296)], and 9th [DR7 (SRR7223301) and DR15 (SRR7223297)] days, respectively, and at 18:30 h on the 8th day [DR11 (SRR7223295)].
The salt expression data were provided by Zhang et al. (2021). RNA-Seq data included D. catenatum plants treated with 250 mM NaCl for 0 h (SRR13986990, SRR13986991, and SRR13986992), 4 h (SRR13987000, SRR13987001, and SRR1387002), and 12 h (SRR13986997, SRR13986998, and SRR1398699) for leaves, and 0 h (SRR13986996, SRR13987005, and SRR13987006), 4 h (SRR13986993, SRR13986994, and SRR13986995), and 12 h (SRR13986989, SRR13987003, and SRR13987004) for roots.
The expression data of leaves under cold stress treatments in 4-month-old D. catenatum plants, containing 20°C control (SRR3210630, SRR3210635, and SRR3210636) and 0°C cold acclimations (SRR3210613, SRR3210621, and SRR3210626) for 20 h, were obtained from NCBI provided by Wu et al. (2016).
The P1 inoculation and methyl jasmonate (MeJA) pre-treatment transcriptome data were from our previous study (Li et al., 2021), which consists of four treatments, including CK (no treatment: SRR14635793, SRR14635796, and SRR14635797), JA (pre-treated by MeJA for 4 h: SRR14635790, SRR14635791, and SRR14635792), P1 (inoculated by P1 for 24 h: SRR14635787, SRR14635788, and SRR14635789), and P1 + JA (pre-treated by MeJA for 4 h and then infected by P1 for 24 h: SRR14635786, SRR14635794, and SRR14635795). The expression abundance of DcbHLH genes was calculated using the fragments per kilobase of transcript per million fragments mapped (FPKM) values. Using TBtools, we generated a heat map of DcbHLH genes (Chen et al., 2020a).
Pathogen inoculation assays
Sclerotium delphinii was grown on the PDA medium at 25°C for 6 days. Five 5-mm agar disks containing mycelia were collected and cultured in 200 ml PDB medium for 6 days at 25°C with constant shaking at 180 rpm. After diluting to 1/2 of the original concentration, it was used for the subsequent inoculation assay of D. catenatum and Arabidopsis.
The 4-month-old D. catenatum plantlets with the same growth state and size were selected and divided into six groups [CK, P1, Phenidone (Phe), JA, Phe + P1, and JA + P1]. Phenidone decreased endogenous JA synthesis by inhibiting the activity of 13-lipoxygenase (LOX) in plants (Bruinsma et al., 2010). The plantlets were pre-treated with sterile water (mock) or a solution containing Phe (16 mmol/L) or MeJA (100 μmol/L) for 4 h. After that, they were washed with sterile water and put into a sterile bottle with sterile water. Among them, P1, Phe + P1, and JA + P1 were evenly sprayed with 2 ml of mycelia suspensions (the others were spotted with 0.25% ethanol solution). The infection ratings and disease index were calculated as described previously at each time point (0, 24, 30, 38, 48, and 72 h; Li et al., 2021). The infection experiment was repeated three times with 10 plantlets each (30 plantlets per infection). The leaves of each treatment were collected at 24 h post-inoculation (hpi) for subsequent analysis.
The leaves with relatively consistent growth status and size in Arabidopsis Col-0, mutants coi1-1, myc2, myc3, and myc4, and transgenic lines grown for 20 days were placed on tetrad Petri dishes containing filter paper moistened with sterile water, and each leaf was inoculated with 5 μl of sterile water (mock) or mycelia suspensions in the middle position on the front of each leaf, with three replicates for at least 10 leaves in each experiment. The size of the leaf lesion was calculated at 48 hpi using ImageJ.11
Determination of JA content using LC–MS/MS
The content of JA, (3R,7S)-jasmonoyl-L-isoleucine (JA-Ile), and 12-oxophytodienoic acid (OPDA) were measured by LC–MS/MS. In short, the leaves of D. catenatum preserved at ultralow temperature after inoculating by P1 for 24 h were ground with mixer mil (30 Hz, 1 min). Then, 50 mg of leaf samples were extracted with methanol/water/formic acid (15:4:1, V/V/V) at 4°C (vortex, 10 min), and then centrifuged at 12,000 rpm for 5 min. The supernatant was evaporated and concentrated with nitrogen at room temperature, reconstituted in 100 μl of 80% (v/v) methanol, and filtered (SCAA-104, 0.22 μm pore size; ANPEL, Shanghai, China). It was placed in the injection bottle for LC–MS/MS (Ultra-performance liquid chromatography (UPLC), ExionLC™ AD, https://sciex.com.cn/; and Tandem mass spectrometry (MS/MS), QTRAP® 6,500+, https://sciex.com.cn/). Three replicates of each assay were performed. Based on the MWDB (Metware database) constructed by the standard, the mass spectrometry data were analyzed qualitatively. Multiple reaction monitoring (MRM) of triple quadrupole mass spectrometry and the internal standard method were used for quantitative analysis. Analyst 1.6.3 software was used to process MS data.
RNA extraction and quantitative real-time PCR
The total RNA was extracted using the MiniBEST Plant RNA Extraction Kit (TaKaRa, Japan). The cDNA was reverse-transcribed with the PrimerScript RT Enzyme Mix I kit (TaKaRa, Japan). qPCR analysis was performed with SYBR® Premix Ex Taq II (TaKaRa, Japan) on CFX96 Touch™ Real-Time PCR System (BIO-RAD, United States). Three independent biological replicates were performed on each analysis with three technical replicates. The DcACTIN was used as the internal control gene according to the previous study (Li et al., 2021). The relative expression levels were evaluated using the 2-ΔΔCT method [3]. The gene-specific primers were designed by Primer Premier 5 (Supplementary Table S1).
Protein secondary structure and tertiary structure prediction
Online software SOPMA 2.012 was used to predict the secondary structure of the four DcbHLH IIIe proteins, that is, DcbHLH025/026/027/028 proteins. SWISS-MODEL online homology modeling13 was applied to predict the tertiary structure of each of the above four proteins, and the predicted protein tertiary structure models were assessed for Model Credibility using SAVES online software.14 DcbHLH025/026/027/028 was renamed as DcMYC2d/2a/2b/2c, respectively, which is used throughout the rest of the article.
Subcellular localization of DcMYC2a and DcMYC2b
The CDS of DcMYC2a and DcMYC2b were cloned into the plant expression vector pMDC43 to construct 35S: DcMYC2a:GFP and 35S: DcMYC2b:GFP fusion construct, respectively. Subsequently, the control plasmid (empty vector) and fusion plasmids were transiently expressed in tobacco leaves. After 3 days of post-infiltration, the GFP fluorescence was observed using a confocal microscope (Zeiss, LSM 880) with 488 and 594 nm argon lasers.
Generation of transgenic plants and transgene analysis
Arabidopsis (Col-0) plants were used for Agrobacterium tumefaciens-mediated transformation to generate DcMYC2a/2b-overexpressing (DcMYC2a/2b-OE) plants. The recombinant plasmids pMDC43-DcMYC2a-GFP and pMDC43-DcMYC2b-GFP were transformed into A. tumefaciens GV3101, which was used to transform Arabidopsis plants by the floral-dip method. Regenerated seedlings were selected on MS medium with 20 mg.L−1 hygromycin. The expression of DcMYC2a/2b in transgenic Arabidopsis plants was analyzed by quantitative real-time PCR (qRT-PCR). Homozygous T3 generation transgenic lines were used for further experiments.
Results
Identification and characterization of DcbHLH, PebHLH, and AsbHLH genes
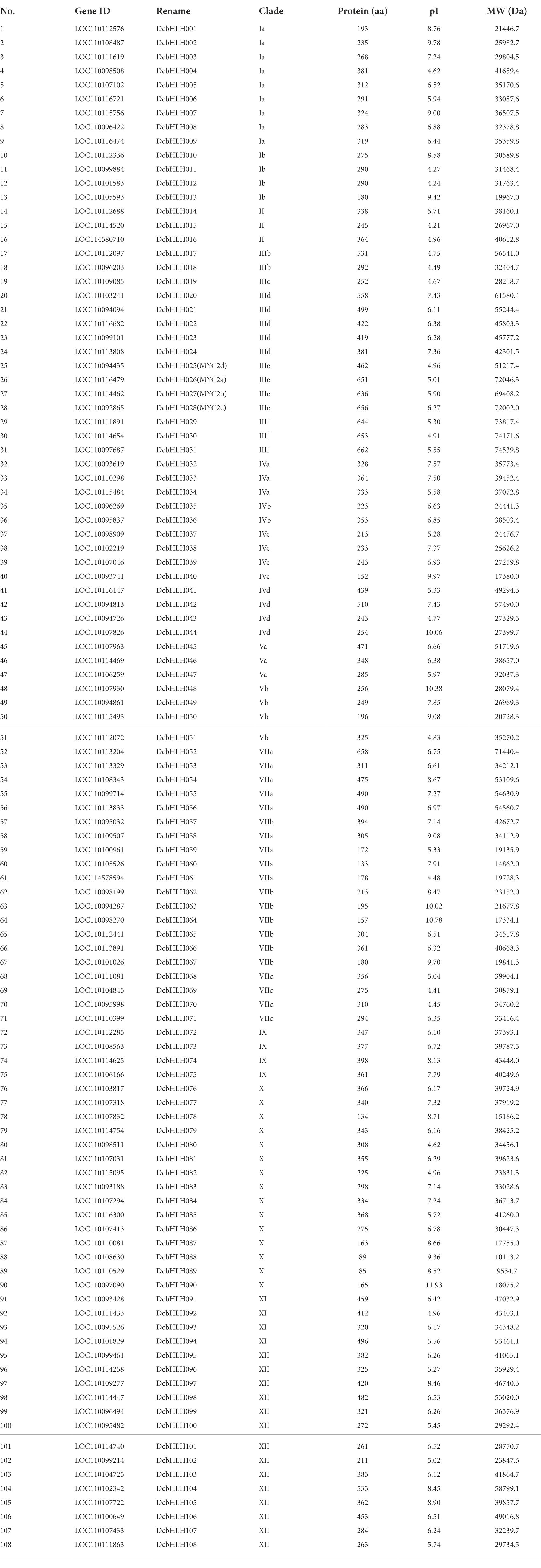
We conducted a genome-wide search for D. catenatum, P. equestris, and A. shenzhenica, which together belong to the Orchidaceae family. A total of 108 DcbHLH, 83 AsbHLH, and 109 PebHLH proteins were obtained after removing redundant proteins. To further characterize these bHLH proteins, we analyzed the physicochemical properties of these proteins (Table 1; Supplementary Tables S2, S3). The amino acids of DcbHLH ranged from 85 (DcbHLH089) to 662 (DcbHLH031) in length and from 9534.7 (DcbHLH089) to 74539.8 (DcbHLH031) in molecular weight, the amino acid length of PebHLH ranged from 85 (PebHLH086) to 916 (PebHLH091) and the molecular weight size from 9578.7 (PebHLH086) to 100520.6 (PebHLH091), and the amino acid length of AsbHLH ranged from 67 (AsbHLH042) to 687 (AsbHLH026) and the molecular weight size from 7702.9 (AsbHLH042) to 74504.6 (AsbHLH026). The pI ranged from 4.21 (DcbHLH015) to 11.93 (DcbHLH090) for the DcbHLH proteins, from 4.47 (AsbHLH009) to 12.28 (AsbHLH042) for the AsbHLH proteins, and from 4.15 (PebHLH011) to 12.13 (PebHLH087) for the PebHLHs. The large differences in molecular weight, amino acid length, and pI among the DcbHLH, PebHLH, and AsbHLH members indicated that the DcbHLH, PebHLH, and AsbHLH gene family members may have undergone a long historical evolution and participated in different biological processes.
Phylogenetic analysis of bHLH genes
To better study the classification and evolutionary relationships of the DcbHLH proteins and to further predict the function of the DcbHLH proteins, we constructed a phylogenetic tree based on bHLH protein sequences from five species (Arabidopsis, O. sativa, D. catenatum, P. equestris, and A. shenzhenica) using the neighbor-joining (NJ) method (Figure 1). According to the clade support values and the classification of Arabidopsis, the 108 DcbHLH proteins were clustered into 23 subgroups, and the other two species belonging to the Orchidaceae cluster similarly to D. catenatum, where 109 AsbHLH proteins cluster into 24 subgroups and 83 PebHLH proteins cluster into 24 subgroups. The results showed that the largest number of members of the X subgroup contained 12 DcbHLH proteins, 12 AsbHLH proteins, and 21 PebHLH proteins. Interestingly, none of the DcbHLH, AsbHLH, or PebHLH proteins clustered in subgroup VI, where DcbHLH was also not identified in subgroup IIIa. It is possible that D. catenatum has lost the genes of IIIa and VI subgroups during evolution. Arabidopsis bHLH genes, belonging to four subgroups of the III subfamily, are involved in JA signal transduction pathways, including subgroup IIIe which positively regulates JA response, subgroup IIId which negatively regulates JA response, subgroup IIIf which is involved in JA-mediated anthocyanin synthesis and epidermal hair initiation, and subgroup IIIb which is associated with JA-induced freezing tolerance (Heim et al., 2003). We predicted that the DcbHLH, PebHLH, and AsbHLH genes, which are in these four subgroups, might also be involved in the JA signal transduction pathway.
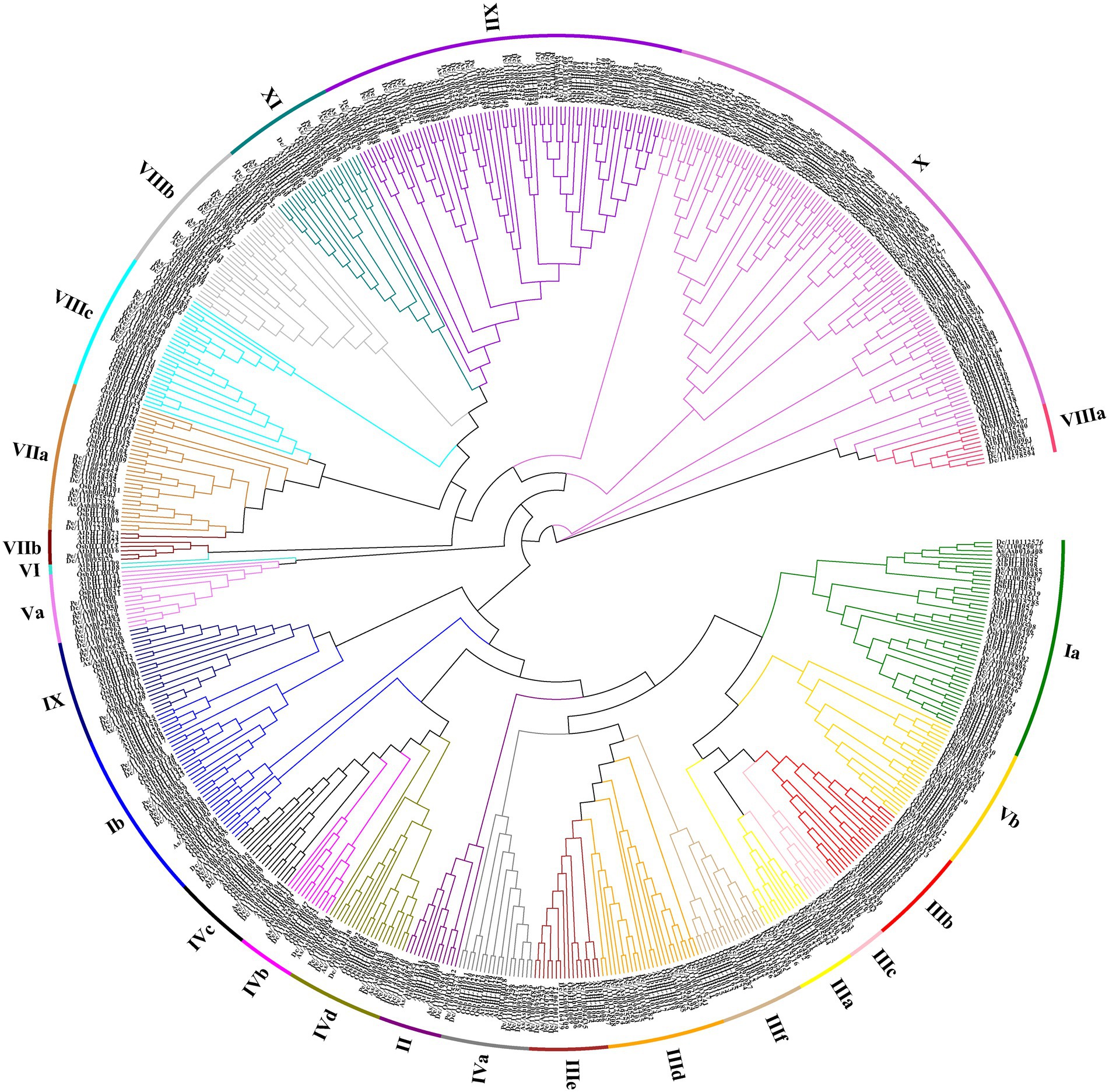
Figure 1. Phylogenetic analysis of the basic helix–loop–helices (bHLHs) from Dendrobium catenatum, Phalaenopsis equestris, Apostasia shenzhenica, Arabidopsis, and Oryza sativa genome. The neighbor-joining (NJ) tree was drawn using MEGA 7.0 with 1,000 bootstrap replicates. Dc, D. catenatum; Pe, P. equestris; As, A. shenzhenica; At, Arabidopsis; and Os, O. sativa.
Conserved motif, gene structure, and Cis-acting element analysis of DcbHLH genes
In order to verify the accuracy of phylogenetic analysis and further study the structure of DcbHLH proteins, we identified 20 conserved motifs using the MEME program (Figures 2A,B). DcbHLH proteins in the same subgroup have similar conserved motifs, which indicate that these DcbHLH proteins may have similar functions. The vast majority of DcbHLH proteins contain motif 1 and motif 2, suggesting that the DcbHLH genes are functionally similar in some ways. In addition, some subfamilies contain specific motifs. For instance, motif 6, motif 9, motif 11, and motif 12 are unique to subfamily III. It is possible that these motifs are involved in JA signal transduction pathways in the plant.

Figure 2. Phylogenetic relationships, architecture of conserved protein motifs, and gene structure in bHLHs of Dendrobium catenatum. (A) The phylogenetic tree of bHLH proteins. It was constructed according to the NJ method by MEGA 7.0 with 1,000 bootstrap replicates. Different color boxes represent different clades. (B) The conserved motif composition of bHLH proteins. Solid boxes of different colors represent different motifs, and the legend is on the right side of the figure. (C) UTR-coding domain sequence (CDS) structure of bHLH genes. The green box represents UTRs, and the yellow box represents CDSs.
Exon-intron structural diversity is considered to play an important role in the evolution of bHLH genes. We mapped the gene structure based on the sequence information of the DcbHLH gene and its CDS (Figures 2A,C). The number of introns contained in the DcbHLH gene ranges from 0 to 10, of which 20 DcbHLH genes do not contain introns, nine DcbHLH genes contain one intron, and the other genes contain two or more introns. Members belonging to the same subgroups had similar patterns of intron-exon distribution. For instance, members belonging to subgroups IIId and IIIe contained no intron, but all contained one exon, and members belonging to subgroup IIIf contained both seven introns and eight exons.
Many bHLH genes play important roles in plant growth and development as well as in the response to various stresses (Mao et al., 2017; Hao et al., 2021). In order to further predict the function of the DcbHLH genes, we made a prediction of cis-acting elements for the sequence 1,500 bp upstream of the start codon of the DcbHLH genes (Supplementary Figure S1), and cis-acting elements can be classified into three categories according to functions. The first category comprises phytohormone-responsive elements, including methyl jasmonate (MeJA-responsive, 13.79%), abscisic acid (ABA-responsive, 8.69%), salicylic acid (SA-responsive, 1.75%), gibberellins (GA-responsive, 2.93%), and auxins-responsive (2.57%) elements, and this category consists of TGACG-motif, CGTCA-motif, ABRE, TCA-element, TATC-box, P-box, GARE-motif, TGA-element, TGA-box, AuxRE, and AuxRR-core. The second category belongs to the growth and development class of elements (8.08%), including cell differentiation, circadian control, cell cycle regulation, meristem expression, and flavonoid synthesis, and this class consists of ARE, AT-rich sequence, HD-Zip-1, RY-element, GCN4_motif, AACA_motif, circadian, MSA-like, CAT-box, and MBSI. The third category comprises stressor elements that are involved in adaptation to adversity and include light-responsive (45.83%), anaerobic induction (6.22%), low-temperature responsive (2.47%), drought-inducible (2.11%), defense and stress-responsive (2.06%), anoxic specific induction (0.41%), and wound-responsive (3.09%) elements; this category consists of ACE, G-box, GT1-motif, 3-AF1 binding site, Sp1, AAAC-motif, Box 4, ATC-motif, ATCT-motif, CAG-motif, GATA-motif, Box II, TCT-motif, chs-CMA1a, Pc-CMA2c, LAMP-element, I-box, GA-motif, Gap-box, TCCC-motif, chs-Unit 1 m1, L-box, LS7, AE-box, ARE, LTR, MBS, TC-rich repeats, GC-motif, and WUN-motif. Different types and numbers of regulatory elements were identified in the promoter regions of different DcbHLH genes, indicating that DcbHLH genes might have different functions in stress resistance, growth, and development.
The expression patterns of DcbHLH genes
We analyzed the temporal and spatial expression patterns of 108 DcbHLH genes in different tissues or organs (leaf, stem, root, green root tip, white part of the root, flower bud, sepal, labellum, pollinia, and gynostemium; Figure 3; Supplementary Table S4). The expression of some DcbHLH genes in various tissues or organs is very low or basically not expressed, such as DcbHLH001/064/070, which indicates that these genes may not be necessary for these tissues or organs to perform their functions at this stage. Some genes also show high expression in various tissues or organs, such as DcbHLH039/040. Most genes are highly expressed in one or more tissues or organs. In addition, the expression of DcbHLHs can be roughly divided into four groups based on the similarity of gene expression in different tissues and organs: I (pollinia), II (leaf, stem, and labellum), III (root, green root tip, and white part of the root), and IV (flower bud, sepal, and gynostemium). The above results showed that the DcbHLHs family is widely involved in plant growth and development.
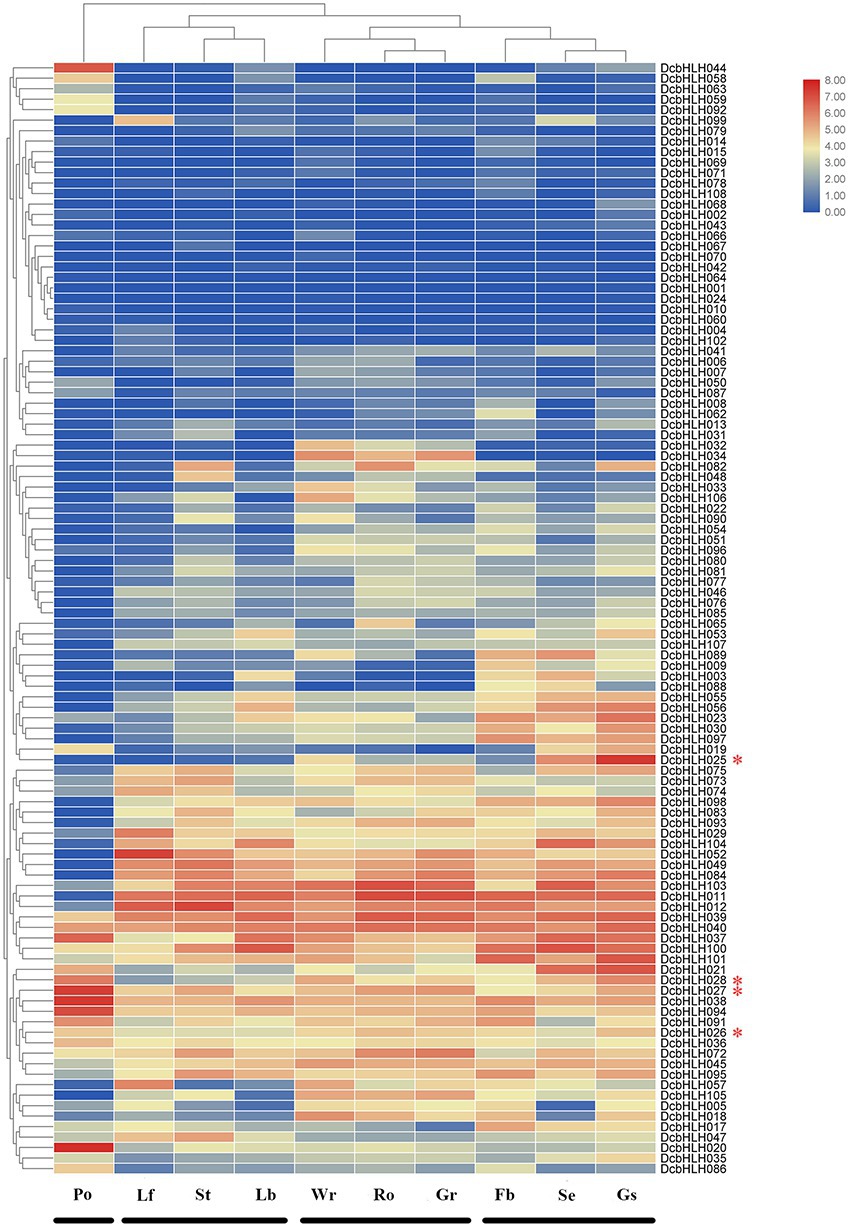
Figure 3. Relative expression analysis of the DcbHLH genes in different tissues. Expression patterns of DcbHLH genes in 10 Dendrobium catenatum tissues and organs. Lf, leaf; Ro, root; Gr, green root tip; Wr, the white part of the root; St, stem; Fb, flower bud; Se, sepal; Lb, labellum (lip); Po, pollinia; and Gs, gynostemium (column).
In the low-temperature treatment (Figure 4A; Supplementary Table S5), compared with the control group, the expression of DcbHLH009/017/018/025(MYC2d)/029/052/057/099/106 was upregulated significantly, while the expression of DcbHLH023/027(MYC2b)/065/082 was downregulated significantly. Among them, DcbHLH IIId subgroup member DcbHLH017 was highly homologous with Arabidopsis Inducer of CBF Expression 1 (AtICE1), which is a positive regulator of plant resistance to cold stress (Chinnusamy et al., 2003), indicating that DcbHLH017 is likely to positively regulate plant cold resistance.
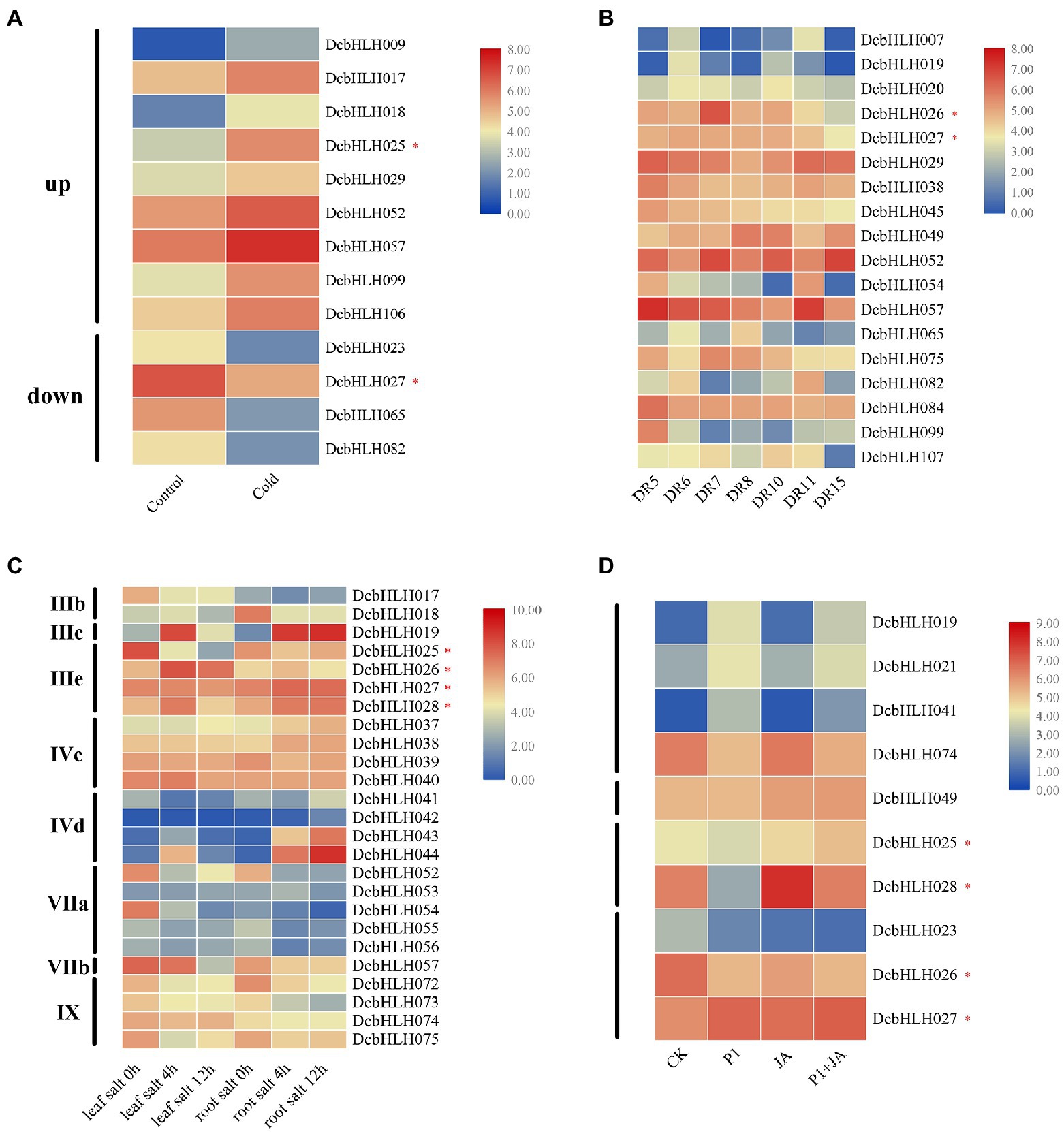
Figure 4. Relative expression analysis of the DcbHLH genes under different stress conditions. (A) Low-temperature treatment (CK: 20°C, 20 h; Cold: 0°C, 20 h). (B) Drought treatment [Drought stage: DR5/DR6 (the next day 06:30 and 18:30 h), Dr7/DR8 (06:30 and 18:30 h on the seventh day)]; Rehydration stage: DR10 (18:30 h on the eighth day), and DR11/DR15 (06:30 and 18:30 h on the ninth day). (C) Salt stress treatment. Leaf salt 0/4/12 h: Leaf treated with 250 mM NaCl for 0/4/12 h. Root salt 0/4/12 h: Root treated with 250 mM NaCl for 0/4/12 h. (D) Expression profiles of DcbHLH genes to S. delphinii after pre-treatment with MeJA (CK: control. P1: 24 h after post-inoculation with S. delphinii. JA: pre-treated with MeJA for 4 h and then inoculated with sterile distilled water for 24 h. P1 + JA: pre-treated with MeJA for 4 h and then inoculated with S. delphinii for 24 h).
In the drought–rewatering assay, most genes showed non-significant trends, but a small number of genes showed significant changes (Figure 4B; Supplementary Table S6). At the DR5-DR8 stage (continuous drought stage), the expression levels of DcbHLH019/026 (MYC2a) were upregulated first and then decreased, those of DcbHLH029/038/045/054/057/082/084/099 were downregulated, that of DcbHLH049 was upregulated gradually, that of DcbHLH052 downregulated first, rapidly upregulated, and then sharply downregulated, whereas the expression of DcbHLH065 showed the opposite trend. At the DR10-DR15 stage (the rewatering stage), the expression of DcbHLH007/020/026(MYC2a)/029/082/107 was upregulated first and then downregulated, while DcbHLH049 showed the opposite trend. The expressions of DcbHLH027(MYC2b)/065/075 gradually decreased, and the expression of DcbHLH052 was upregulated first, then decreased, and then increased, while DcbHLH054/057 showed the opposite trend.
In the high salt environment (Figure 4C; Supplementary Table S7), we found that there were several subgroups whose expression in leaves or roots changed significantly after plants were subjected to salt stress, including subgroups IIIb, IIIc, IIIe, IVc, IVd, VIIa, VIIb, and IX. In particular, the expression of the genes of subgroup IIIe, that is, DcbHLH025(MYC2d)/026(MYC2a)/027(MYC2b)/028(MYC2c), which are related to JA signaling pathway, was found to be significantly altered. These results suggested that some subgroups of DcbHLH TFs may play important roles in plant resistance to salt stress and that the JA signal transduction pathway may have been induced after plants were subjected to salt stress. The expression of the DcbHLH gene in the stem and leaves of D. catenatum under salt stress conditions is significantly different (Figures 3, 4C), which may be due to the tissue specificity of gene expression.
Given that some bHLH genes play pivotal roles in plant response to biotic stress (Dombrecht et al., 2007; Hao et al., 2021), we designed a set of experimental conditions to determine their functions after infection with P1 and treatment with the resistance hormone JA (Li et al., 2021). Compared with the CK group, the expression of some DcbHLH genes in the three treatment samples (P1, JA, and P1 + JA) changed significantly (Figure 4D; Supplementary Table S8). In addition, DcbHLH019/021/041/074 showed significant changes in the expression following P1 induction but were unaffected by exogenous JA, indicating that these genes were specifically induced by P1. Some genes, such as DcbHLH049, were induced only by JA, suggesting that these genes did not respond to P1, which might be involved in specific resistance or development pathways triggered by high concentrations of JA. Some genes, such as DcbHLH025(MYC2d)/028(MYC2c), were upregulated by JA but repressed by P1, suggesting that these two genes might be involved in JA-regulated growth and development pathways. The transcript levels of DcbHLH023/026(MYC2a) were significantly downregulated in P1, JA, and P1 + JA treatment samples, which indicated that P1 might be a negative regulator of the JA-mediated plant response. In contrast to the DcMYC2a, the DcbHLH027 (MYC2b) was significantly upregulated in all treatment samples, indicating that it may play a positive regulatory role.
JA enhanced the resistance of Dendrobium catenatum to P1
In order to explore the effect of JA on the resistance of D. catenatum to P1 infection, we carried out a P1 inoculation experiment. Different responses to the disease infection were observed in MeJA- and phe-pre-treated, and control plantlets (Figures 5A,B; Supplementary Table S9). At 24 hpi, the symptoms, including water-stained and brown necrotic lesions, were much more severe in the plantlets pre-treated with phe than in the control plantlets, whereas the plants pre-treated with exogenous MeJA remained disease-free (Figures 5A,B; Supplementary Table S9). The disease index increased with prolonged hpi, and the MeJA-pre-treated plantlets showed the slowest disease development in early stage (24–30 hpi), showing a certain resistance to P1, while the plantlets pre-treated with phe showed a rapid development. The plantlets treated with the above three treatments (P1, Phe + P1, and JA + P1) were in the same disease situation at about 38 hpi. To better understand the mechanism of D. catenatum resistance to P1, we measured the endogenous JA content in D. catenatum, including 12-oxophytodienoic acid (OPDA), the receptor-active ligand (3R,7S)-jasmonoyl-L-isoleucine (JA-Ile), and JA at 24 hpi (Figure 5D; Supplementary Table S10). The results showed that a large amount of endogenous JAs was synthesized after P1 infection, which suggested that P1 activated the plant JA signaling pathway. The content of all three JAs was significantly decreased in the treatment P1 + Phe, but not completely suppressed, indicating that phenidone only partially suppresses the activity of LOX and plants still show partial JA-related resistance. There was no significant difference in OPDA content between P1 and P1 + JA samples, indicating that they had approximately equal rates of endogenous JA synthesis, and the difference in JA content between these two samples might be caused by exogenous MeJA demethylation. Interestingly, the levels of JA-lle, the active form of JA in planta, were not significantly different between P1 and P1 + JA samples. It is possibly due to the fact that the catalytic activity of Jasmonoyl-isoleucine synthetase (JAR1), which catalyzes JA synthesis of JA-lle, reached its maximum at an earlier time point, leading to the stronger resistance of JA + P1 sample at the pre-stage and preventing pathogen infiltration. To further explore the reasons for the differences in various treatments, we conducted qRT-PCR of the marker genes of the JA signaling pathway. The results indicated that the expression of DcPR3 was significantly induced by P1 infection or exogenous JA treatment (Figure 5C), while that of DcPR3 was inhibited in the samples pre-treated by Phe. The expression of DcLOX2 was significantly different in P1, JA, and P1 + JA treatments compared to CK, and it was significantly inhibited in Phe and Phe + P1 samples but significantly induced in P1, JA, and P1 + JA samples. More importantly, the expression level of DcLOX2 was significantly higher in the P1 + JA sample than in the P1 sample. These results suggest that the JA pathway plays a key role in the resistance of D. catenatum to P1.
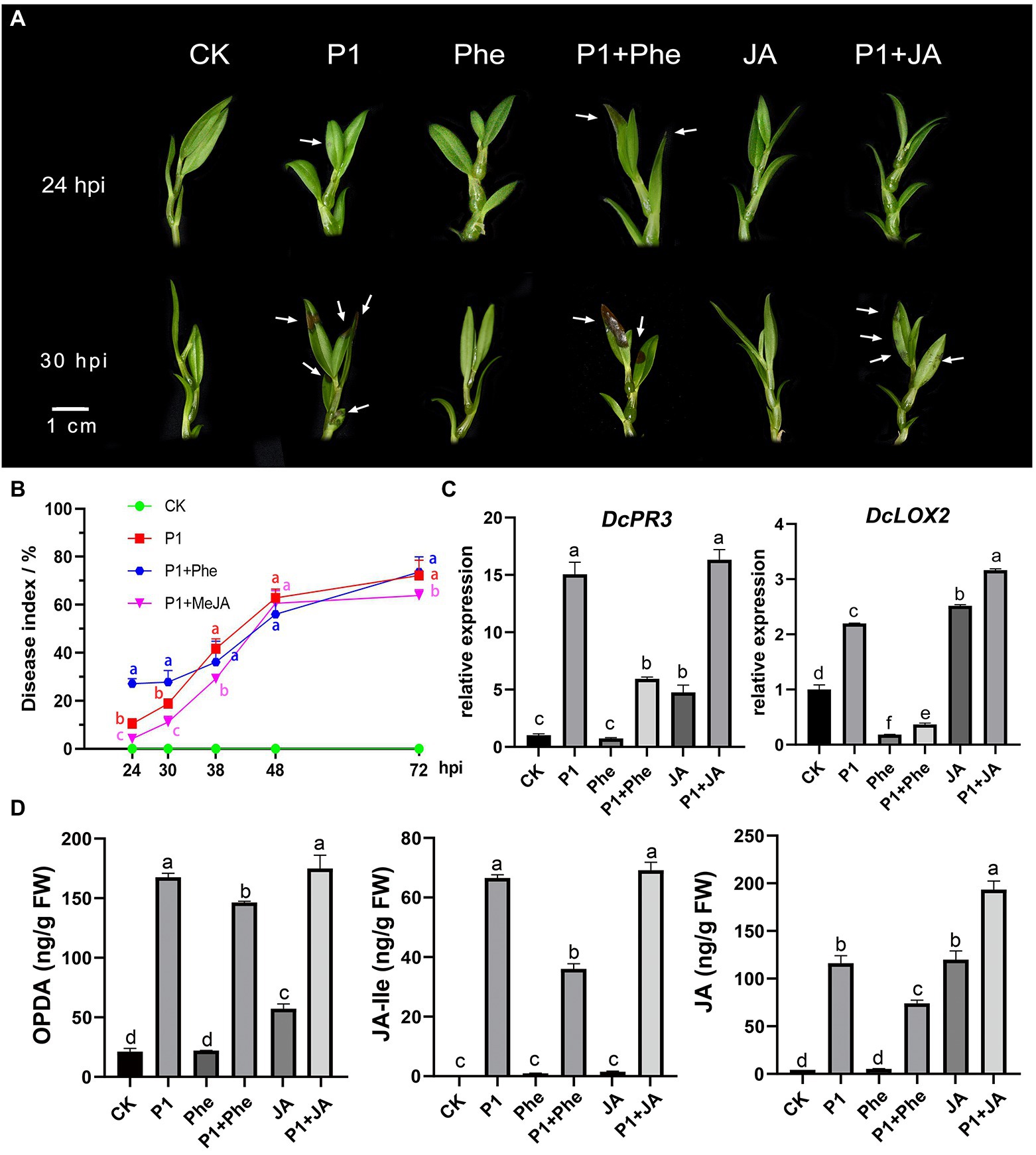
Figure 5. Enhanced resistance of D. catenatum to P1 infection by JA. (A) Disease symptoms of D. catenatum plantlets pre-treated with Phe (16 mmol/L) or MeJA (100 μmol/L) following inoculation with S. delphinii at 24 and 30 hpi. Arrows point to the lesion. Bar = 1 cm. (B) Disease indexes line charts of the MeJA- or Phe-pre-treated D. catenatum were determined from 24 hpi (1 dpi) to 72 hpi (3 dpi). The values are the mean ± SD; n = 10. “abc” indicates a significant difference at p = 0.05. (C) The expression of important downstream gene DcPR3, DcLOX2 of JA signaling pathway under different treatments through RT-qPCR assay. CK: control. P1: 24 h post-inoculation with S. delphinii. JA: pre-treated with MeJA for 4 h and then inoculated with sterile distilled water for 24 h. P1 + JA: pre-treated with MeJA for 4 h and then inoculated with S. delphinii for 24 h. Phe: pre-treated with MeJA for 4 h and then inoculated with sterile distilled water for 24 h. Phe + P1: pre-treated with Phe for 4 h and then inoculated with S. delphinii for 24 h. The actin gene of D. catenatum was used as an internal control. The error bars indicate SD from three independent experiments. The “abcdef” showed a significant difference at p = 0.05 compared with the CK. (D) Content of oxophytodienoic acid (OPDA), JA-lle, and JA in the leaves of D. catenatum after inoculation with S. delphinii for 24 h. The abscissa represents the different treatments, and the ordinate represents the content of the multitest substance (ng/g FW). “abcdef” indicates a significant difference at p = 0.05.
Protein structure prediction and subcellular localization of DcbHLH IIIe genes
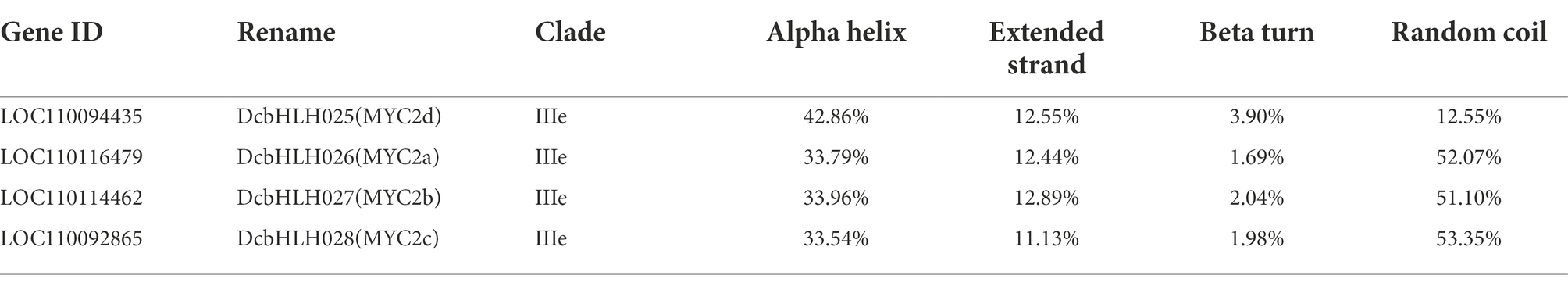
The infection of P1 and exogenous JA significantly changed the expression of D. catenatum bHLH IIIe subgroup members. In order to better predict their functions, we predicted the protein secondary and tertiary structures of the DcbHLH IIIe subgroup members DcbHLH025(MYC2d)/026(MYC2a)/027(MYC2b)/028(MYC2c) (Table 2; Figure 6A). The protein secondary structure and tertiary structure analyses showed that DcMYC2a/2b/2c/2d had similar secondary structures, contained a large number of irregular curls and alpha helices, and each member contained a highly similar three-dimensional structure, which indicated that the members of this subgroup are functionally similar or redundant. Subcellular localization of DcMYC2a/2b was examined with the tobacco leaf transient expression system. Both proteins were able to detect GFP fluorescence signals in the nuclei of N. tabacum epidermal cells (Figure 6B), demonstrating DcMYC2a/2b encoded nuclear proteins.
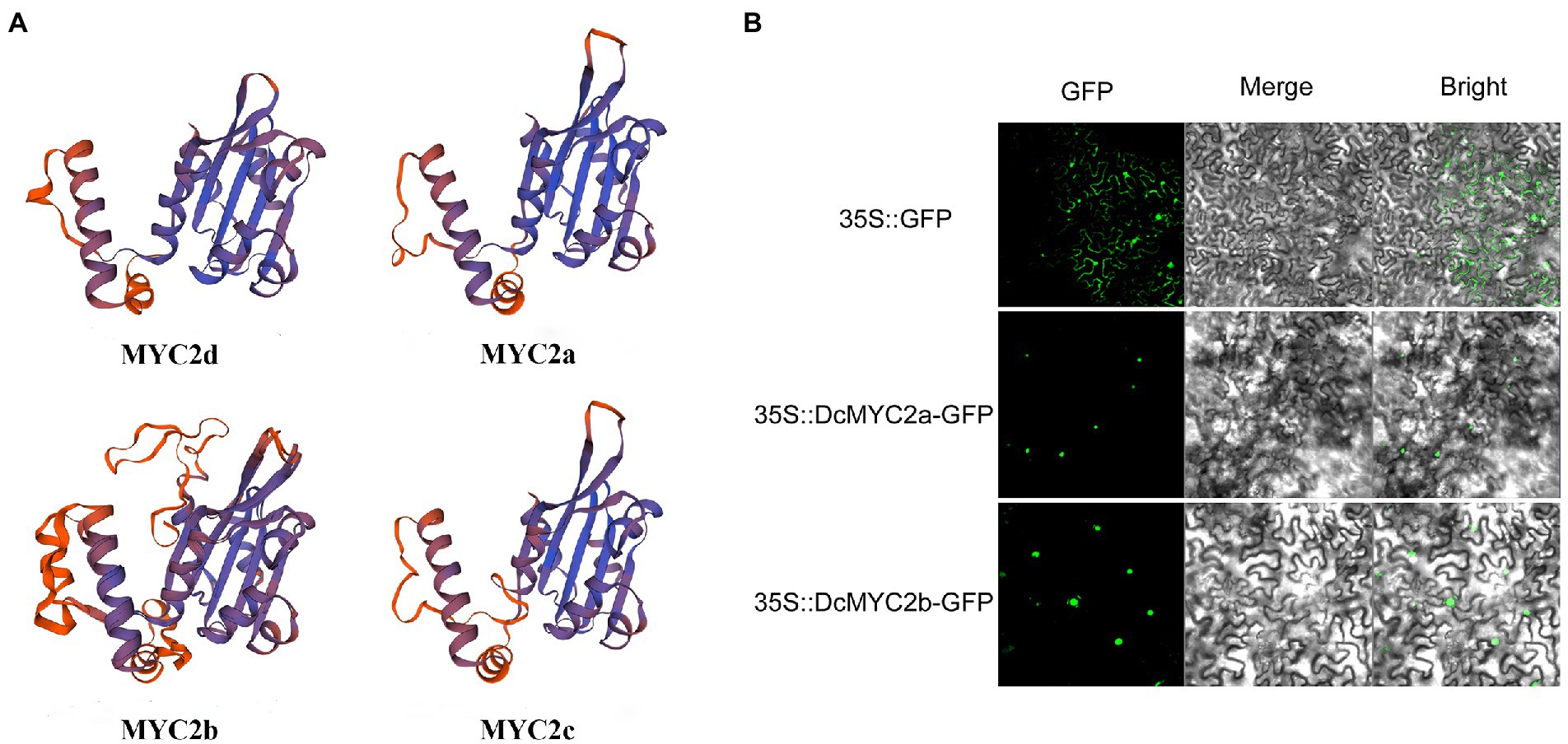
Figure 6. Prediction model of protein tertiary structure of subgroup IIIe members and subcellular localization. (A) Protein tertiary structure of DcMYC2a/2b/2c/2d. (B) Subcellular localization of DcMYC2a/2b in tobacco cells.
Overexpression of DcMYC2b improved P1 tolerance in yransgenic Arabidopsis
As Arabidopsis is also the host of P1, in order to further explore the function of MYC TFs, we obtained Arabidopsis mutants myc2/3/4 and DcMYC2a/b-OE plants of Arabidopsis and infected their isolated leaves with P1. The results showed that Arabidopsis mutants myc2/myc3/myc4 leaves had significantly smaller lesion size (24.01/24.21/20.42 mm2) than Col-0 (29.15 mm2) after P1 infection, while the leaves of coi1-1 displayed bigger lesion size (33.33 mm2, Figures 7A,B; Supplementary Table S11), suggesting that AtMYC2/3/4 function in Arabidopsis as negative regulators of downstream genes of JA signaling pathway related to P1 defense, which is consistent with the result of inoculation assays of necrotrophic pathogen B. cinerea in Arabidopsis myc2 mutant (Song et al., 2014). Three independent homozygous lines of DcMYC2a/b were selected (OE3/4/6 for DcMYC2a and OE3/4/6 DcMYC2b) with high gene expression levels in the T3 generation. DcMYC2a and DcMYC2b have higher expression than Col-0 in their respective DcMYC2a-OE and DcMYC2b-OE lines (Figure 7C), indicating that DcMYC2a-OE and DcMYC2b-OE lines were successfully constructed. After infection with P1, the lesion size of DcMYC2a-OE lines (18.31 mm2) was not significantly different from that of Col-0 (20.67 mm2), while the lesion size of DcMYC2b-OE lines (15.30 mm2) was significantly smaller than that of Col-0 (Figures 7D,E; Supplementary Table S12), suggesting that DcMYC2b may be a positive regulator in the JA-mediated resistance of D. catenatum to P1.
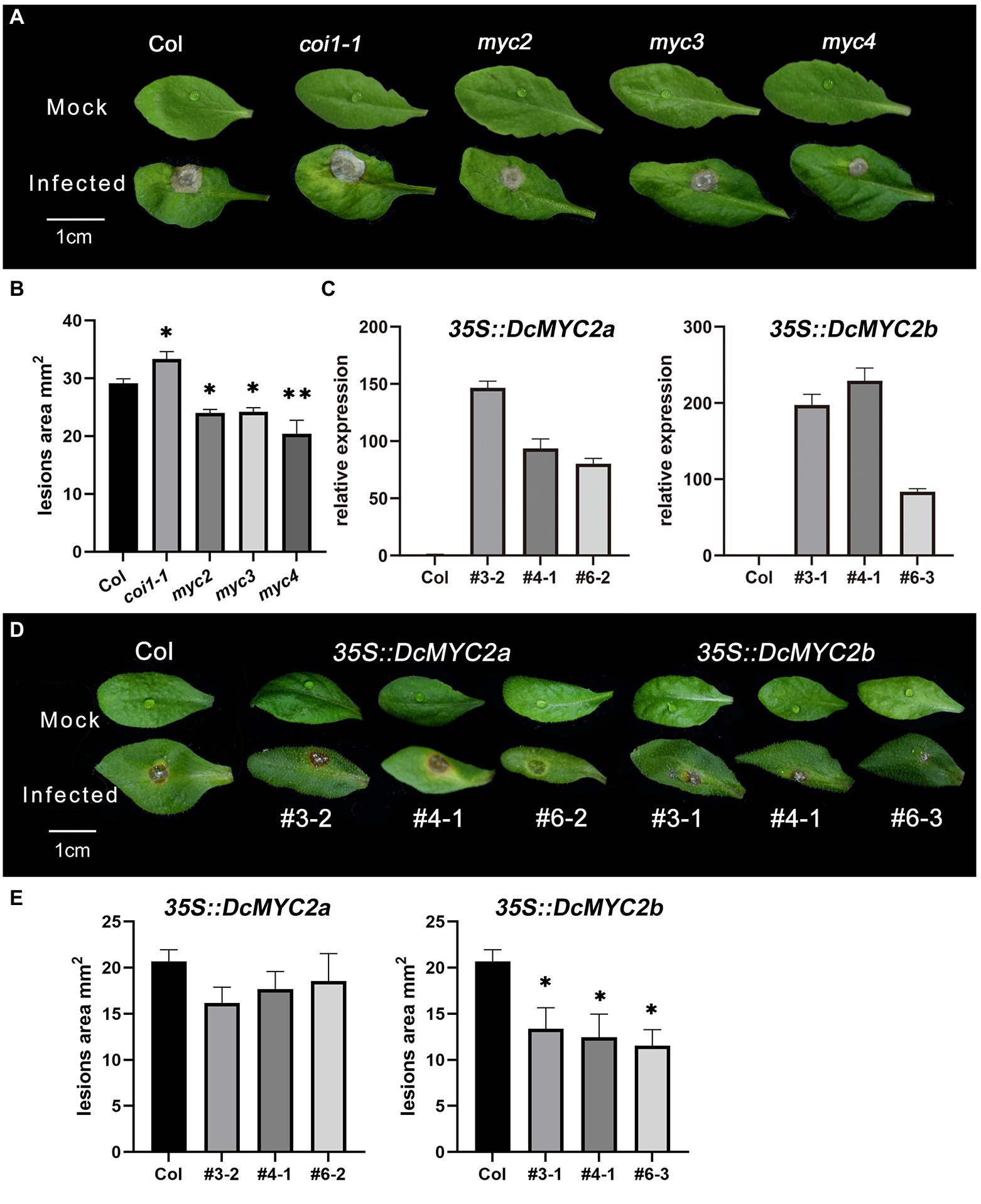
Figure 7. Functional validation of DcMYCs and AtMYCs genes. (A) Disease symptoms of leaves of Arabidopsis myc2, myc3, and myc4 mutants following inoculation with Sclerotium delphinii at 48 hpi. Bar = 1 cm. (B) Lesion size of leaves of Arabidopsis myc2, myc3, and myc4 mutants at 48 hpi. Significant difference (compared with the Col, *p < 0.05, **p < 0.01). (C) Relative expression of DcMYCs in Arabidopsis DcMYCs-OE strain. The abscissa represents different lines of DcMYCs-OE. (D) Disease symptoms of leaves of Arabidopsis DcMYC2a/2b-OE at 48 hpi. Bar = 1 cm. The # represents different lines. (E) Lesion size of leaves of Arabidopsis DcMYC2a/2b-OE at 48 hpi. The * shows a significant difference at p < 0.05 compared with the Col.
Discussion
Dendrobium catenatum is an important economical medicinal plant under the forest. The quality of near-wild cultivation is significantly better than that of facility cultivation (Chen et al., 2020b). However, D. catenatum cultivated in the near wild is vulnerable to harsh environmental conditions, most prominently southern blight disease (Chen et al., 2019a). Searching for effective strategies to improve the resistance of D. catenatum under near-wild conditions becomes a difficult problem. The members of the family bHLH TFs play vital roles in plant growth, development, light signal transduction, and stress responses (Abe et al., 2003; Castelain et al., 2012; Liu et al., 2013; Yao et al., 2018; Zhang et al., 2020b). They are also involved in the crosstalk of hormone signaling, including abscisic acid (ABA), JA, brassinosteroid (BR), salicylic acid (SA), and ethylene (ET) (Murre et al., 1989; Heim et al., 2003; Simionato et al., 2007; Feller et al., 2011), and they are pivotal for the plant growth and survival in the environment. In our study, we constructed an evolutionary tree by combining DcbHLH with AtbHLH and OsbHLH, which are taxonomically well-defined, as well as bHLH (PebHLH and AsbHLH) of two D. catenatum relatives. A total of 108 DcbHLH genes were identified and divided into 12 subfamilies and 23 subgroups according to the phylogenetic analysis. However, DcbHLH was not identified in subgroup IIIa. We speculated that bHLH genes of subgroup IIIa may not be necessary to maintain the normal growth and development of D. catenatum, and it was lost in the evolution of D. catenatum.
An important regulation pathway is that transcription factors interact with cis-acting elements to express the genes involved in stress response (Mao et al., 2017) and developmental processes specifically (Li et al., 2012; Chen et al., 2018). Although functional studies of bHLH genes are still lacking in D. catenatum, the gene function can be predicted by the upstream cis-acting elements and the expression pattern of these genes, and the published bHLH gene functions of other species. The cis-acting element analysis pointed out that the DcbHLH genes contain a large number of cis-acting elements related to plant growth and development, and response to hormones and external stress, which indicated the importance of bHLH genes in plant growth and development as well as response to stress. For example, overwhelmingly members of subfamily VII belong to the PIF family (Paik et al., 2017). In Arabidopsis, PIFs interact with phytochrome (PFR) and crosstalk with other signaling pathways to regulate plant growth and response to environmental signals, including light and stress (Castillon et al., 2007). In our study, members of the bHLH VII subfamily of D. catenatum contain a large number of light-responsive elements, suggesting the potential functions of the members of subfamily VII in responding to light signals and participating in growth and defense mechanisms. Arabidopsis bHLH IIIe TF MYC5 has redundant functions with MYC2, MYC3, and MYC4, which interact with MYB21 and MYB24 to form the bHLH–MYB complex to regulate stamen development (Qi et al., 2015). The homologous genes of AtMYC2/3/4/5 and DcMYC2a/2b/2c show high expression levels in pollens except for DcMYC2d, which illustrated that DcbHLH IIIe TFs may affect the pollen development of D. catenatum. Besides, AtMYC5 is a positive regulator of signal transduction under salt stress (Ramsay and Glover, 2005). We found that DcMYC2a and DcMYC2c were strongly induced by salt stress, which may have redundant functions in regulating plants to adapt to high salt environments. In Arabidopsis, ICE1 encodes an MYC-type bHLH TF that directly binds the promoter of the C-Repeat Binding Factor/Dehydration-Responsive-Element-Binding protein (CBF/DREB1) protein-encoding gene and activates its transcription to improve cold tolerance through an ABA-independent pathway (Stockinger et al., 1997). DcbHLH017 is highly homologous with AtICE1 and is highly expressed in plants under cold stress. We speculated that it may play an important role in D. catenatum response to cold stress.
The phytohormone JA plays a vital role in plant development and the response to various stresses. Exogenous JA could enhance the resistance of D. catenatum to P1 and delay the disease time. Phe, as an inhibitor of LOX activity and a key gene in the JA synthesis pathway, significantly reduced the resistance to P1 and inhibited the expression of DcPR3 and DcLOX2, which are marker genes for JA-regulated pathogen responses. In addition, the expression of DcLOX2 in the P1 + JA sample was significantly higher than in the P1 sample. These results demonstrated the importance of the JA signaling pathway in the resistance of D. catenatum to P1 infection. The MYC2 TFs are core regulators of the JA signaling pathway (Kazan and Manners, 2013). In previous studies, the resistance of MYC2 to a necrotrophic pathogen exhibited species specificity (Song et al., 2014; Du et al., 2017). In Arabidopsis, the MYC2 plays a negative regulatory role in plant resistance to the necrotrophic pathogen. Consistently, myc2 mutants are more resistant to the necrotrophic pathogen, such as Botrytis cinerea, than the wild type, due to the fact that the MYC2 interacts with EIN3 and inhibits the function of EIN3, thereby inhibiting ethylene-mediated plant resistance to a pathogen (Song et al., 2014). Our experiments also proved that the Arabidopsis myc2 mutant exhibited a bigger lesion size than Col-0 when infected with P1. Arabidopsis MYC3 and MYC4 are homologous genes to MYC2, and they have redundant functions and mediate a variety of physiological processes regulated by JA (Kazan and Manners, 2013; Schweizer et al., 2013; Zhang et al., 2020a). In our study, the phenotype of Arabidopsis myc3 and myc4 mutants infected by P1 was consistent with myc2 mutants, indicating that they had certain similarities in function. When one MYC gene loses its function, other myc genes cannot completely replace its function, indicating that they had unique characteristics in JA signaling pathway. However, in Solanum lycopersicum, SlMYC2 positively regulates both wound and pathogen response genes, but it cannot change the increased disease resistance of the Arabidopsis myc2-2 mutant, and thus, SlMYC2 and its Arabidopsis homologs have different action modes in regulating pathogen response genes (Du et al., 2017). The functional differences between AtMYC2 and SlMYC2 TFs may not be due to their different binding capacities, but rather to their different transcriptional regulatory activities, as SlMYC2 and AtMYC2 contain a highly conserved basic region that mediates binding to G-boxes of their target gene promoters (Toledo-Ortiz et al., 2003; Kazan and Manners, 2013). SlMYC2 and AtMYC2 may recruit different partner proteins while regulating pathogen-regulated gene transcription. DcMYC2a/2b/2c/2d had close homology with AtMYC2/3/4/5, and the expression of DcMYC2a/2b/2c/2d was significantly changed after the plants were treated with exogenous JA and infected with P1. However, the expression patterns of these four duplicated DcMYC2 genes showed significant differences. DcMYC2a exhibited negative regulation under both JA and P1, but there was no significant difference in the lesion size between the DcMYC2a-OE and wild-type (Col-0) plants, possibly due to the fact that DcMYC2a was involved in other JA-mediated pathways. Instead, the DcMYC2b expression pattern suggested that it may be a positive regulator of the JA-related defense pathway against pathogens. It was confirmed by P1 inoculation assays of Arabidopsis DcMYC2b-OE plants, whose lesion size was significantly smaller than Col-0. DcMYC2b and AtMYC2 have opposite regulatory effects in the process of plant response to P1, which may be caused by their recruitment of different chaperones through some subtle differences between the secondary and tertiary structures of proteins. The expression patterns of DcMYC2c and DcMYC2d were upregulated by JA, but repressed by P1, suggesting that these two genes might be involved in JA-regulated growth and development pathways. We performed secondary and tertiary structure predictions of the above four proteins and found similar structures, but there were some differences. It is possible that these differences enable them to recruit different partner proteins in order to perform different functions in the JA signaling pathway. As the key step in JA signaling pathway, the mechanism by which MYC2 genes and their downstream genes form a transcriptional regulatory network that affects disease resistance in plants remains to be further studied.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
CL: conceptualization, experimentation, writing of the original draft, reviewing, and editing. XiC: conceptualization, experimentation, reviewing, and editing. QS: experimentation and data curation. XuC: data curation. MX and TY: experimentation. DS: data curation and editing. LW and DC: writing, reviewing, and editing. ZH and JS: conceptualization, writing, reviewing, and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31901235), the Major Science and Technology Projects of Yunnan Province (202102AE090042), and the Scientific R&D Foundation for Talent Start-up Project of Zhejiang A&F University (2019FR010).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.956210/full#supplementary-material
Supplementary Figure S1 | Cis-acting elements in the 1,500 bp promoter regions of the DcbHLH genes. Different color boxes represent different clades. Solid boxes of different colors represent different cis-acting elements, and the legend is on the right side of the figure.
Footnotes
1. ^https://www.arashare.cn/index/Product/index.html
2. ^https://www.ncbi.nlm.nih.gov/genome/
5. ^http://smart.emblheidelberg.de/
6. ^https://web.expasy.org/protparam/
7. ^https://evolgenius.info/evolview-v2/#login
8. ^http://gsds.cbi.pku.edu.cn/
9. ^http://meme-suite.org/tools/meme
10. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
11. ^https://imagej.nih.gov/ij/download.html?utm_source=ld246.com
12. ^https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Adams, K. L., Aljohani, A., Chavez, J., and de Roode, J. C. (2021). Effects of cardenolides of milkweed plants on immunity of the monarch butterfly. Arthropod Plant Interact. 15, 249–252. doi: 10.1007/s11829-021-09812-w
Bruinsma, M., van Broekhoven, S., Poelman, E. H., Posthumus, M. A., Muller, M. J., van Loon, J. J., et al. (2010). Inhibition of lipoxygenase affects induction of both direct and indirect plant defences against herbivorous insects. Oecologia 162, 393–404. doi: 10.1007/s00442-009-1459-x
Buck, M. J., and Atchley, W. R. (2003). Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 56, 742–750. doi: 10.1007/s00239-002-2449-3
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martinez-Garcia, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Castelain, M., Le Hir, R., and Bellini, C. (2012). The non-DNA-binding bHLH transcription factor PRE3/bHLH135/ATBS1/TMO7 is involved in the regulation of light signaling pathway in Arabidopsis. Physiol. Plant. 145, 450–460. doi: 10.1111/j.1399-3054.2012.01600.x
Castillon, A., Shen, H., and Huq, E. (2007). Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12, 514–521. doi: 10.1016/j.tplants.2007.10.001
Chen, Q. Y., Chen, D. H., Shi, Y., Si, W. S., Wu, L. S., and Si, J. P. (2019a). Occurrence regularity of Dendrobium catenatum southern blight disease. Chin. J. Chin. Mat. Med. 44, 1789–1792. doi: 10.19540/j.cnki.cjcmm.20190129.005
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H. C., Cheng, W. H., Hong, C. Y., Chang, Y. S., and Chang, M. C. (2018). The transcription factor OsbHLH035 mediates seed germination and enables seedling recovery from salt stress through ABA-dependent and ABA-independent pathways, respectively. Rice 11:50. doi: 10.1186/s12284-018-0244-z
Chen, D. H., Qiu, H. L., Huang, Y., Zhang, L., and Si, J. P. (2020b). Genome-wide identification and expression profiling of SET DOMAIN GROUP family in Dendrobium catenatum. BMC Plant Biol. 20:40. doi: 10.1186/s12870-020-2244-6
Chen, S., Zhao, H., Luo, T., Liu, Y., Nie, X., and Li, H. (2019b). Characteristics and expression pattern of MYC genes in Triticum aestivum, Oryza sativa, and Brachypodium distachyon. Plan. Theory 8:274. doi: 10.3390/plants8080274
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Di Marzo, M., Roig-Villanova, I., Zanchetti, E., Caselli, F., Gregis, V., Bardetti, P., et al. (2020). MADS-box and bHLH transcription factors coordinate transmitting tract development in Arabidopsis thaliana. Front. Plant Sci. 11:526. doi: 10.3389/fpls.2020.00526
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245. doi: 10.1105/tpc.106.048017
Du, M., Zhao, J., Tzeng, D. T. W., Liu, Y., Deng, L., Yang, T., et al. (2017). MYC2 orchestrates a hierarchical transcriptional Cascade That regulates Jasmonate-mediated plant immunity in tomato. Plant Cell 29, 1883–1906. doi: 10.1105/tpc.16.00953
Feller, A., Machemer, K., Braun, E. L., and Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66, 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Fernandez-Calvo, P., Chini, A., Fernandez-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., Geerinck, J., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Giri, M. K., Gautam, J. K., Rajendra Prasad, V. B., Chattopadhyay, S., and Nandi, A. K. (2017). Rice MYC2 (OsMYC2) modulates light-dependent seedling phenotype, disease defence but not ABA signalling. J. Biosci. 42, 501–508. doi: 10.1007/s12038-017-9703-8
Hao, Y., Zong, X., Ren, P., Qian, Y., and Fu, A. (2021). Basic Helix-loop-Helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. Int. J. Mol. Sci. 22:7152. doi: 10.3390/ijms22137152
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Iquebal, M. A., Tomar, R. S., Parakhia, M. V., Singla, D., Jaiswal, S., Rathod, V. M., et al. (2017). Draft whole genome sequence of groundnut stem rot fungus Athelia rolfsii revealing genetic architect of its pathogenicity and virulence. Sci. Rep. 7:5299. doi: 10.1038/s41598-017-05478-8
Kazan, K., and Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Ledent, V., and Vervoort, M. (2001). The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 11, 754–770. doi: 10.1101/gr.177001
Li, X., Chen, L., Hong, M., Zhang, Y., Zu, F., Wen, J., et al. (2012). A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa. PLoS One 7:e44145. doi: 10.1371/journal.pone.0044145
Li, X., Duan, X., Jiang, H., Sun, Y., Tang, Y., Yuan, Z., et al. (2006). Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 141, 1167–1184. doi: 10.1104/pp.106.080580
Li, H., Gao, W., Xue, C., Zhang, Y., Liu, Z., Zhang, Y., et al. (2019). Genome-wide analysis of the bHLH gene family in Chinese jujube (Ziziphus jujuba mill.) and wild jujube. BMC Genomics 20:568. doi: 10.1186/s12864-019-5936-2
Li, Y., He, F., Lai, H., and Xue, Q. (2017). Mechanism of in vitro antagonism of phytopathogenic Scelrotium rolfsii by actinomycetes. Eur. J. Plant Pathol. 149, 299–311. doi: 10.1007/s10658-017-1177-x
Li, C., Shen, Q., Cai, X., Lai, D., Wu, L., Han, Z., et al. (2021). JA signal-mediated immunity of Dendrobium catenatum to necrotrophic southern blight pathogen. BMC Plant Biol. 21:360. doi: 10.1186/s12870-021-03134-y
Li, X., Zhang, H., Ai, Q., Liang, G., and Yu, D. (2016). Two bHLH transcription factors, bHLH34 and bHLH104, regulate Iron homeostasis in Arabidopsis thaliana. Plant Physiol. 170, 2478–2493. doi: 10.1104/pp.15.01827
Liang, G., Zhang, H., Li, X., Ai, Q., and Yu, D. (2017). bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 68, 1743–1755. doi: 10.1093/jxb/erx043
Liu, Y., Li, X., Li, K., Liu, H., and Lin, C. (2013). Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9:e1003861. doi: 10.1371/journal.pgen.1003861
Lu, R., Zhang, J., Liu, D., Wei, Y. L., Wang, Y., and Li, X. B. (2018). Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum). BMC Plant Biol. 18:304. doi: 10.1186/s12870-018-1523-y
Mao, K., Dong, Q., Li, C., Liu, C., and Ma, F. (2017). Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 8:480. doi: 10.3389/fpls.2017.00480
Murre, C., McCaw, P. S., and Baltimore, D. (1989). A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777–783. doi: 10.1016/0092-8674(89)90682-x
Paik, I., Kathare, P. K., Kim, J. I., and Huq, E. (2017). Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10, 1035–1046. doi: 10.1016/j.molp.2017.07.002
Pires, N., and Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Qi, T., Huang, H., Song, S., and Xie, D. (2015). Regulation of Jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell 27, 1620–1633. doi: 10.1105/tpc.15.00116
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Schweizer, F., Fernandez-Calvo, P., Zander, M., Diez-Diaz, M., Fonseca, S., Glauser, G., et al. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25, 3117–3132. doi: 10.1105/tpc.113.115139
Si, J. P., Wang, Q., Liu, Z. J., Liu, J. J., and Luo, Y. B. (2017). Breakthrough in key science and technologies in Dendrobium catenatum industry. Chin. J. Chin. Mat. Med. 42, 2223–2227. doi: 10.19540/j.cnki.cjcmm.2017.0102
Simionato, E., Ledent, V., Richards, G., Thomas-Chollier, M., Kerner, P., Coornaert, D., et al. (2007). Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol. Biol. 7:33. doi: 10.1186/1471-2148-7-33
Song, S., Huang, H., Gao, H., Wang, J., Wu, D., Liu, X., et al. (2014). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26, 263–279. doi: 10.1105/tpc.113.120394
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U. S. A. 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Tissot, N., Robe, K., Gao, F., Grant-Grant, S., Boucherez, J., Bellegarde, F., et al. (2019). Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 223, 1433–1446. doi: 10.1111/nph.15753
Toledo-Ortiz, G., Huq, E., and Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15, 1749–1770. doi: 10.1105/tpc.013839
Uji, Y., Akimitsu, K., and Gomi, K. (2017). Identification of OsMYC2-regulated senescence-associated genes in rice. Planta 245, 1241–1246. doi: 10.1007/s00425-017-2697-5
Wang, J., Hu, Z., Zhao, T., Yang, Y., Chen, T., Yang, M., et al. (2015). Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genomics 16:39. doi: 10.1186/s12864-015-1249-2
Wu, Z. G., Jiang, W., Chen, S. L., Mantri, N., Tao, Z. M., and Jiang, C. X. (2016). Insights from the cold Transcriptome and Metabolome of Dendrobium officinale: global reprogramming of metabolic and gene regulation networks during cold acclimation. Front. Plant Sci. 7:1653. doi: 10.3389/fpls.2016.01653
Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W. L., Ma, H., et al. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. doi: 10.1105/tpc.003368
Xu, W., Zhang, N., Jiao, Y., Li, R., Xiao, D., and Wang, Z. (2014). The grapevine basic helix-loop-helix (bHLH) transcription factor positively modulates CBF-pathway and confers tolerance to cold-stress in Arabidopsis. Mol. Biol. Rep. 41, 5329–5342. doi: 10.1007/s11033-014-3404-2
Yan, C., and Xie, D. (2015). Jasmonate in plant defence: sentinel or double agent? Plant Biotechnol. J. 13, 1233–1240. doi: 10.1111/pbi.12417
Yang, M., Zhou, C., Yang, H., Kuang, R., Huang, B., and Wei, Y. (2020). Genome-wide analysis of basic helix-loop-helix transcription factors in papaya (Carica papaya L.). PeerJ. 8:e9319. doi: 10.7717/peerj.9319
Yao, X., Cai, Y., Yu, D., and Liang, G. (2018). bHLH104 confers tolerance to cadmium stress in Arabidopsis thaliana. J. Integr. Plant Biol. 60, 691–702. doi: 10.1111/jipb.12658
Zhang, C., Lei, Y., Lu, C., Wang, L., and Wu, J. (2020a). MYC2, MYC3, and MYC4 function additively in wounding-induced jasmonic acid biosynthesis and catabolism. J. Integr. Plant Biol. 62, 1159–1175. doi: 10.1111/jipb.12902
Zhang, J., Liu, B., Li, M., Feng, D., Jin, H., Wang, P., et al. (2015). The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27, 787–805. doi: 10.1105/tpc.114.132704
Zhang, G.-Q., Liu, K.-W., Li, Z., Lohaus, R., Hsiao, Y.-Y., Niu, S.-C., et al. (2017). The Apostasia genome and the evolution of orchids. Nature 549, 379–383. doi: 10.1038/nature23897
Zhang, M., Yu, Z., Zeng, D., Si, C., Zhao, C., Wang, H., et al. (2021). Transcriptome and Metabolome reveal salt-stress responses of leaf tissues from Dendrobium officinale. Biomol. Ther. 11:736. doi: 10.3390/biom11050736
Zhang, T., Yuan, Y., Zhan, Y., Cao, X., Liu, C., Zhang, Y., et al. (2020b). Metabolomics analysis reveals Embden Meyerhof Parnas pathway activation and flavonoids accumulation during dormancy transition in tree peony. BMC Plant Biol. 20:484. doi: 10.1186/s12870-020-02692-x
Zhao, Q., Fan, Z., Qiu, L., Che, Q., Wang, T., Li, Y., et al. (2020). MdbHLH130, an apple bHLH transcription factor, confers water stress resistance by regulating stomatal closure and ROS homeostasis in transgenic tobacco. Front. Plant Sci. 11:543696. doi: 10.3389/fpls.2020.543696
Zhou, X., Liao, Y., Kim, S. U., Chen, Z., Nie, G., Cheng, S., et al. (2020). Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep. 10:13723. doi: 10.1038/s41598-020-69305-3
Keywords: Dendrobium catenatum, jasmonate, Sclerotium delphinii, bHLH transcription factor, DcMYC2b
Citation: Li C, Cai X, Shen Q, Chen X, Xu M, Ye T, Si D, Wu L, Chen D, Han Z and Si J (2022) Genome-wide analysis of basic helix–loop–helix genes in Dendrobium catenatum and functional characterization of DcMYC2 in jasmonate-mediated immunity to Sclerotium delphinii. Front. Plant Sci. 13:956210. doi: 10.3389/fpls.2022.956210
Edited by:
Youxiong Que, Fujian Agriculture and Forestry University, ChinaReviewed by:
Xin He, Hunan Agricultural University, ChinaDong-Liang Huang, Guangxi Academy of Agricultural Sciences, China
Copyright © 2022 Li, Cai, Shen, Chen, Xu, Ye, Si, Wu, Chen, Han and Si. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigang Han, aGFuemdAemFmdS5lZHUuY24=; Jinping Si, bHNzanBAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Cong Li
Cong Li Xiang Cai†
Xiang Cai† Xueliang Chen
Xueliang Chen Mengxi Xu
Mengxi Xu Lingshang Wu
Lingshang Wu Donghong Chen
Donghong Chen Jinping Si
Jinping Si