- Institute for Biology, Department of Genetics, Martin Luther University Halle-Wittenberg, Halle (Saale), Saxony-Anhalt, Germany
The Gram-negative plant-pathogenic bacterium Xanthomonas euvesicatoria is the causal agent of bacterial spot disease in pepper and tomato plants. Pathogenicity of X. euvesicatoria depends on a type III secretion (T3S) system which translocates effector proteins into plant cells and is associated with an extracellular pilus and a translocon in the plant plasma membrane. Effector protein translocation is activated by the cytoplasmic T3S chaperone HpaB which presumably targets effectors to the T3S system. We previously reported that HpaB is controlled by the translocated regulator HpaA which binds to and inactivates HpaB during the assembly of the T3S system. In the present study, we show that translocation of HpaA depends on the T3S substrate specificity switch protein HpaC and likely occurs after pilus and translocon assembly. Translocation of HpaA requires the presence of a translocation motif (TrM) in the N-terminal region. The TrM consists of an arginine-and proline-rich amino acid sequence and is also essential for the in vivo function of HpaA. Mutation of the TrM allowed the translocation of HpaA in hpaB mutant strains but not in the wild-type strain, suggesting that the recognition of the TrM depends on HpaB. Strikingly, the contribution of HpaB to the TrM-dependent translocation of HpaA was independent of the presence of the C-terminal HpaB-binding site in HpaA. We propose that HpaB generates a recognition site for the TrM at the T3S system and thus restricts the access to the secretion channel to effector proteins. Possible docking sites for HpaA at the T3S system were identified by in vivo and in vitro interaction studies and include the ATPase HrcN and components of the predicted cytoplasmic sorting platform of the T3S system. Notably, the TrM interfered with the efficient interaction of HpaA with several T3S system components, suggesting that it prevents premature binding of HpaA. Taken together, our data highlight a yet unknown contribution of the TrM and HpaB to substrate recognition and suggest that the TrM increases the binding specificity between HpaA and T3S system components.
Introduction
The Gram-negative plant-pathogenic bacterium Xanthomonas euvesicatoria (also known as Xanthomonas campestris pv. vesicatoria) is the causal agent of bacterial spot disease on pepper and tomato plants and causes significant yield losses due to necrotic lesions and defoliation (Potnis et al., 2015). Pathogenicity of X. euvesicatoria depends on a type III secretion (T3S) system which is conserved in many Gram-negative plant-and animal-pathogenic bacteria and translocates bacterial effector proteins (T3Es, type III effectors) into eukaryotic cells (Büttner and Bonas, 2002; An et al., 2020; Timilsina et al., 2020). T3S systems span both bacterial membranes and are associated with an extracellular pilus-like structure which serves as a transport channel for secreted proteins to eukaryotic cells (Büttner, 2012; Deng et al., 2017; Hajra et al., 2021). Translocation of T3Es is mediated by a bacterial T3S translocon in the eukaryotic plasma membrane (Büttner, 2012; Ji and Dong, 2015; Dey et al., 2019). T3Es interfere with various cellular pathways to the benefit of the pathogen, thus allowing bacterial multiplication and the formation of disease symptoms in susceptible plants. In resistant plants, however, individual T3Es are directly or indirectly recognized by the plant immune system (Büttner, 2016; Khan et al., 2018; Schreiber et al., 2021). Corresponding plant resistance proteins subsequently activate defence responses which often lead to the induction of a hypersensitive response (HR), a rapid local cell death at the infection site that restricts bacterial multiplication (Khan et al., 2016; Ngou et al., 2022).
The T3S system from X. euvesicatoria is encoded by a chromosomal hrp (HR and pathogenicity) gene cluster which contains 25 genes organized in eight operons (Büttner and Bonas, 2002). Eleven gene products are conserved in animal-and/or plant-pathogenic bacteria and presumably constitute the core elements of the secretion apparatus. They are, therefore, referred to as Hrc for Hrp conserved (Hueck, 1998). Homologous proteins from animal-pathogenic bacteria are designated Sct (secretion and cellular translocation) followed by a letter corresponding to the nomenclature of T3S system components from Yersinia spp. (Hueck, 1998; Büttner, 2012; Wagner and Diepold, 2020). In several animal-pathogenic bacteria, the contribution of Sct proteins to the formation of membrane-associated and cytoplasmic core components of the T3S system has been intensively studied (Deng et al., 2017; Wagner et al., 2018; Lara-Tejero and Galan, 2019; Milne-Davies et al., 2021). Structural and electron microscopy studies revealed that the T3S system consists of multimeric rings in the inner membrane (IM) and outer membrane (OM) which are connected by a periplasmic inner rod (Worrall et al., 2016; Hu et al., 2017, 2018, 2019; Torres-Vargas et al., 2019; Lunelli et al., 2020). The IM rings surround the export apparatus which is presumably situated above the IM in the periplasm and is assembled by SctR, SctS, SctT, SctU and SctV proteins (Dietsche et al., 2016; Zilkenat et al., 2016; Kuhlen et al., 2018; Johnson et al., 2019). The IM proteins SctU and SctV contain large cytoplasmic domains which are likely involved in substrate docking (Büttner, 2012; Sal-Man et al., 2012; Portaliou et al., 2017; Wagner et al., 2018; Xing et al., 2018; Milne-Davies et al., 2021). The IM rings also associate with the cytoplasmic sorting platform which is a dynamic structure and consists of the ATPase complex, six spoke-like structures formed by SctL dimers and six pods assembled by members of the SctQ protein family (Hu et al., 2015, 2017; Lara-Tejero, 2019; Berger et al., 2021).
The sorting platform presumably engages T3S substrates, however, the mechanisms underlying substrate recognition and substrate docking are not yet understood (Akeda and Galan, 2005; Spaeth et al., 2009; Lara-Tejero et al., 2011). Studies in animal-pathogenic bacteria suggest that substrate recruitment by the sorting platform involves T3S chaperones which bind to and stabilize secreted proteins and facilitate the entry of their interaction partners into the secretion channel (Büttner, 2012; Deng et al., 2017; Wagner et al., 2018). Substrates of T3S systems include extracellular components of the T3S system such as pilus and translocon proteins as well as T3Es. Given the architecture of the T3S system, secretion of pilus and translocon proteins presumably precedes translocation of T3Es, suggesting a hierarchy in T3S (Deng et al., 2005; Winnen et al., 2008; Lara-Tejero et al., 2011; Portaliou et al., 2017). Recognition of T3S substrates by the T3S system depends on the presence of a secretion signal which is often located in the N-terminal region and is not conserved on the amino acid level (Sory et al., 1995; Rüssmann et al., 2002; Löwer and Schneider, 2009; Samudrala et al., 2009; Wang et al., 2013). In addition to the T3S signal, T3Es contain a translocation signal for delivery across the eukaryotic cell membrane (Schesser et al., 1996; Cheng et al., 1997; Boyd et al., 2000; Stebbins and Galan, 2001; Schechter et al., 2004).
In X. euvesicatoria, T3S substrate specificity is regulated by the control protein HpaC which promotes secretion of translocon and effector proteins and inhibits the secretion of the predicted inner rod protein HrpB2 (Lorenz et al., 2008b; Lorenz and Büttner, 2011; Schulz and Büttner, 2011; Hartmann et al., 2012). The HpaC-mediated substrate specificity switch is likely controlled by a conformational change in the cytoplasmic domain of the export apparatus component HrcU, HrcUC. A point mutation in HrcUC, which presumably mimics the predicted conformational change, restores T3S in the absence of HpaC, suggesting that the contribution of HpaC to T3S is mediated by HrcUC (Lorenz and Büttner, 2011). Given that HrcUC interacts with the N-terminal region of HrpB2, which contains the T3S signal, a conformational change in HrcUC might alter substrate recognition by the T3S system (Lorenz and Büttner, 2011; Scheibner et al., 2016).
It is still unknown how T3Es are recognized by the T3S system after the HpaC-mediated T3S substrate specificity switch. T3S system-mediated delivery of T3Es depends on translocation signals which have been identified in the N-terminal 100–200 amino acids (Büttner et al., 2006; Schulze et al., 2012; Scheibner et al., 2017, 2018). Furthermore, many T3Es contain an N-terminal characteristic amino acid pattern, termed translocation motif (TrM), which harbours arginine-and proline-rich sequences (Escolar et al., 2001; Prochaska et al., 2018). The TrM was previously identified as a membrane targeting signal in the T3E XopB and was shown to contribute to the translocation of XopB, AvrBs1 and AvrBsT (Escolar et al., 2001; Prochaska et al., 2018). Interestingly, mutation of the TrM in XopB abolished membrane localization and facilitated translocation of XopB in the absence of the T3S chaperone HpaB (Prochaska et al., 2018). It was, therefore, proposed that HpaB releases XopB from the membrane in order to facilitate its export by the T3S system. HpaB was previously shown to interact with and to promote the translocation of different T3Es (Büttner et al., 2004; Schulze et al., 2012; Lonjon et al., 2017; Scheibner et al., 2018; Gan et al., 2021). Notably, however, HpaB also interacts with several cytoplasmic components of the T3S system, suggesting an association with the sorting platform (Büttner et al., 2004; Lorenz and Büttner, 2009). HpaB-effector complexes are presumably dissociated by the ATPase HrcN, which might thus facilitate the entry of effectors into the inner transport channel (Lorenz and Büttner, 2009; Gan et al., 2021).
We previously suggested that the contribution of HpaB to T3E delivery is controlled by HpaA which binds to HpaB and acts as a secreted regulator (Lorenz et al., 2008a). HpaA contributes to T3S and pathogenicity and is itself translocated into the plant cell where it localizes to the nucleus (Huguet et al., 1998; Lorenz et al., 2008a). Nuclear localization of HpaA, however, is likely dispensable for its virulence function because the insertion of nuclear exclusion signals, which abolished nuclear localization of HpaA, did not affect the in vivo activity of HpaA (Lorenz et al., 2008a). We, therefore, speculate that the virulence function of HpaA is attributed to its role as regulator of HpaB. Previous interaction studies showed that the C-terminal region of HpaA contains a binding site for HpaB and is essential for protein function (Lorenz et al., 2008a). The interaction between HpaA and HpaB likely prevents an inhibitory effect of HpaB on T3S during the assembly of the T3S system. This hypothesis is supported by the previous finding that deletion of hpaB in a hpaA mutant restored T3S of extracellular components of the T3S system (Lorenz et al., 2008a). Furthermore, N-terminal deletion derivatives of HpaA, which are secretion-deficient but interact with HpaB, still promote secretion of translocon proteins but not of T3Es (Lorenz et al., 2008a). Thus, secretion and translocation of HpaA after assembly of the T3S system is presumably required to liberate HpaB and to activate HpaB-dependent T3E delivery. The timing of HpaA secretion and translocation is, therefore, a key event for the control of T3E delivery in X. euvesicatoria.
In the present study, we analysed the translocation signal and TrM of HpaA and its contribution to the docking of HpaA to T3S system components. We show that the TrM is essential for HpaA translocation and in vivo function. Interaction studies revealed that the TrM interferes with the efficient binding of HpaA to T3S system components, suggesting that it controls the binding specificity of HpaA. Interestingly, mutation of the TrM allowed HpaA translocation in the absence but not the presence of HpaB. We, therefore, propose that HpaB is involved in the recognition of the TrM by the T3S system.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Escherichia coli strains were grown at 37°C in lysogeny broth (LB) medium, and X. euvesicatoria was grown at 30°C in nutrient-yeast extract-glycerol (NYG) medium or minimal medium A (MA; 12 mM K2HPO4, 6.6 mM KH2PO4, 1.52 mM (NH4)2SO4, 0.34 mM Na-citrate*2H2O, 1 mM MgSO4; pH 7.0) supplemented with sucrose (10 mM) and casamino acids (0.3%). Antibiotics were added to the media at the following final concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; rifampicin, 100 μg/ml; spectinomycin, 100 μg/ml, gentamycin, 15 μg/ml; streptomycin, 25 μg/ml and nalidixic acid, 15 μg/ml. Plasmids were introduced into E. coli by transformation and into X. euvesicatoria by electroporation or triparental conjugation, using pRK2013 as helper plasmid.
Plant material and plant infections
For infection studies, X. euvesicatoria strains were resuspended in 1 mM MgCl2 at an optical density (OD600nm) of 0.1 [corresponding to 1 × 108 colony-forming units (CFU) ml−1] if not stated otherwise and infiltrated into leaves of the near-isogenic pepper cultivars Early Cal Wonder (ECW), ECW-10R and ECW-30R using a needleless syringe. Infected plants were incubated in growth chambers for 16 h of light at 28°C and 8 h of darkness at 22°C. Disease symptoms and the hypersensitive response (HR) were documented over a period of one to 9 days post inoculation (dpi). For a better visualization of the HR, infected leaves were bleached in 70% ethanol. Results of the infection experiments were reproduced at least two times.
Generation of expression constructs
For the generation of hpaA expression constructs, hpaA and derivatives were amplified by PCR from X. euvesicatoria strain 85–10 and cloned into the Golden Gate-compatible expression vector pBRM using the type IIs restriction enzyme BsaI in a single restriction/ligation reaction (Engler et al., 2008). Vector pBRM allows the expression of genes under control of a lac promoter in frame with a 3 x c-Myc epitope-encoding sequence. Similarly, hpaA and derivatives thereof were cloned into the BsaI sites of plasmid pBR356, which contains the avrBs3∆2 reporter gene (Scheibner et al., 2016). To introduce the mutations in the TrM, the 5′ and 3′ regions of hpaA were amplified as separate fragments using primers that annealed back-to-back at the TrM-encoding sequence and contained the mutations. For bacterial adenylate cyclase-based two-hybrid (BACTH) assays, hpaA and derivatives thereof were cloned into the Golden Gate-compatible vectors pUT18GG, pUT18CGG, pKT25GG and pKNT25GG. Additionally, we amplified hpaB, hpaC and hrcUC by PCR from X. euvesicatoria strain 85–10, subcloned the corresponding amplicons into pICH41021 as blunt-end fragments after SmaI digest and ligation, and cloned the resulting inserts into the BsaI sites of vectors pUT18GG, pUT18CGG, pKT25GG and pKNT25GG using Golden Gate cloning. Primers used in this study are listed in Supplementary Table S2.
Analysis of in vitro T3S
In vitro T3S assays with X. euvesicatoria strains were performed as described previously (Rossier et al., 1999). For this, bacteria were grown overnight in minimal medium A (MA) (pH 7.0) supplemented with sucrose (10 mM) and casamino acids (0.3%) and shifted to MA (pH 5.3; T3S permissive) containing 50 μg ml−1 bovine serum albumin (BSA) and 10 μM thiamine at an optical density at 600 nm (OD600) of 0.2 for 4 h. Cultures were incubated on a rotary shaker at 30°C, and bacterial cells and secreted proteins from 6 ml-cultures were separated by filtration. Proteins in 4 ml of the culture supernatants were precipitated with trichloroacetic acid and resuspended in 20 μl of Laemmli buffer. Total cell extracts and culture supernatants were analysed by SDS-PAGE and immunoblotting, using antibodies directed against the c-Myc epitope as well as against HrpF and HrcJ, respectively (Rossier et al., 1999; Büttner et al., 2002). Horseradish peroxidase-labelled anti-mouse and anti-rabbit antibodies were used as secondary antibodies. Binding of antibodies was visualized using a chemiluminescence imager (Vilber Fusion FX Edge). Experiments were performed three times with similar results.
Glutathione S-transferase (GST) pull-down assays
For GST pull-down assays, E. coli BL21 (DE3) cells expressing the potential interaction partners under control of the lac promoter were grown in LB medium until an OD600 of 0.6–0.8 and gene expression was induced in the presence of IPTG (isopropyl-β-D-thiogalactopyranoside; 2 mM final concentration) for 2 h at 37°C. Bacterial cells were harvested by centrifugation, broken with a French press and cell debris were removed by an additional centrifugation step. GST and GST fusion proteins present in the cell lysates were immobilized on a glutathione sepharose matrix according to the manufacturer’s instructions (GE Healthcare). After washing of the matrix, immobilized GST and GST fusion proteins were incubated with bacterial lysates containing the predicted interaction partner for 2 h at 4°C on an overhead shaker. Unbound proteins were removed by washing the sepharose matrix and bound proteins were eluted with Laemmli buffer. Cell lysates and eluted proteins were analysed by SDS-PAGE and immunoblotting using antibodies directed against the c-Myc epitope and GST, respectively. Horseradish peroxidase-labelled anti-mouse and anti-goat antibodies were used as secondary antibodies. Binding of antibodies was visualized using a chemiluminescence imager (Vilber Fusion FX Edge). Experiments were performed three times with similar results.
Fluorescence microscopy analyses
X. euvesicatoria strains were grown overnight in MA medium (pH 7.0) supplemented with sucrose (10 mM) and casamino acids (0.3%). Cells were resuspended in MA medium (pH 5.3) supplemented with BSA and thiamine at an OD600 of 0.15 and the cultures were incubated on a tube rotator at 30°C for at least 1 h. Bacteria were transferred to a microscopy slide on top of a pad of 1.5% agarose dissolved in MA medium (pH 5.3) as described (Hausner et al., 2019). Fluorescence was inspected using a confocal laser scanning microscope (Leica STELLARIS 8) with a 60 x magnification objective and 5 x digital magnification. The fluorophores were excited with WLL (white light laser) according to their wave lengths (sfGFP excitation at 485 nm; mCherry excitation at 587 nm). Fluorescence signals were detected with HyD S detectors with adjusted detection windows corresponding to emission of the fluorophores (sfGFP emission at 510 nm; mCherry emission at 610 nm). Pictures were taken from approximately 100 cells of three transconjugants for each strain and were processed with the Leica LIGTHNING detection concept to reveal or sharpen fine structures and details. Protein synthesis was analysed by SDS-PAGE and immunoblotting using GFP- (Roche), mCherry- (Abcam), HrpB1-and HrcC-specific antibodies (Wengelnik et al., 1996; Rossier et al., 2000).
BACTH assays
For BACTH assays, expression constructs encoding T25 and T18 fusion proteins were transferred to the BTH101 E. coli reporter strain and transformants were selected on LB medium containing kanamycin and gentamycin. At least three colonies per strain were used to inoculate LB overnight cultures with appropriate antibiotics, which were incubated at 30°C on a rotary shaker. Two microlitres of the overnight cultures were spotted on LB plates containing appropriate antibiotics, X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranosid; 40 μg/ml) and 2 mM IPTG. Plates were photographed after five to 6 days of incubation at room temperature. The results of all BACTH assays were reproduced at least two times with new transformants; one representative colony is shown. To detect T18 and T25 fusions, corresponding expression constructs were transferred to JM109 E. coli cells and bacterial cultures were induced with IPTG at an OD600 of 0.6–0.8 and incubated on a rotary shaker for 2 h at 37°C. Bacterial cells were collected by centrifugation, resuspended in Laemmli buffer and analysed by immunoblotting, using a FLAG epitope-specific antibody.
Coimmunoprecipitation experiments in X. euvesicatoria
For coimmunoprecipitation studies, bacteria were grown over night in 50 ml MA medium (pH 5.3) supplemented with sucrose (10 mM) and casamino acids (0.3%). Similar amounts of bacterial cells adjusted to the same optical density were harvested by centrifugation, resuspended in 1 ml PBS supplemented with proteinase inhibitors (PIC, proteinase inhibitor cocktail, Roche) and broken with a french press. To dissolve proteins, 0.1% Nonidet-P40 was added to the cell lysate. After 15 min of incubation on ice, cell debris were removed by centrifugation and the supernatant was incubated with 20 μl of protein G agarose beads (Roche) for 1 h at 4°C. The mixture was centrifuged for 90 s at 700 × g and the supernatants were incubated with 60 μl of protein G agarose in the presence or absence of 4 ng anti-c-Myc antibody over night at 4°C on an overhead shaker. The protein G agarose was washed three times with PBS buffer and resuspended in 40 μl of Laemmli buffer. Cell lysates and immunoprecipitated proteins were analysed by immunoblotting, using c-Myc epitope-and HrcN-specific antibodies.
Generation of modular hrp gene cluster constructs for the analysis of HpaA-sfGFP
The modular hrp gene cluster constructs used for fluorescence microscopy were generated as described previously (Hausner et al., 2019) using the modular cloning system (Weber et al., 2011) based on the Golden Gate cloning method (Engler et al., 2009). For the generation of the hpaA-2xLinker-sfgfp module, hpaA was amplified with primers hpaA-NTM-MoClo-F and hpaA-NTM-MoClo-R and the amplicon was cloned into vector pAGM9121 using BpiI and ligase, thus resulting in the level-2 construct pAGB1302. This module was subsequently cloned using BsaI and ligase into vector pAGM1311 to create the level-1 construct pAGB1304, which finally allows the generation of translational fusions of hpaA with genes encoding C-terminal fusion partners. In the next step, hpaA (construct pAGB1304), a linker sequence (2 × AKLEGPAGL, construct pAGB1000) and sfgfp (construct pAGB997) were cloned using BpiI and ligase into vector pICH41308, thus generating the level 0 construct pAGB1328, which contains hpaA-2xLinker-sfgfp. The insert of pAGB1328 (hpaA-2xLinker-sfgfp) was transferred together with the inserts of constructs pAGB249 (PhrpD) and pAGB232 (Xcv term2) into the destination vector pICH47742 using BsaI and ligase and resulting in the level 1 construct pAGB1330 (PhrpD hpaA-2xLinker-sfgfp). The level 0 construct pAGB232 was generated by amplifying the terminator (Xcv term2) with primers termX2-euk-MoClo-F and termX2-euk-MoClo-R and cloning the PCR product into the vector pICH41276 using BpiI and ligase. The insert of pAGB1330 (PhrpD hpaA-2xLinker-sfgfp) was then ligated with the inserts of pAGB157 (xopA + hpaH), pAGB160 (hrpX), pAGB163 (hrpG*), pICH54066 (dummy pos. 6) and pICH50881 (end linker) into the BpiI sites of the level M vector pAGM8079, leading to the level M construct pAGB1332.
To introduce a hpaA deletion and hpaA/hpaB double deletion into the modular gene cluster, a hpaA deletion module was generated by PCR using primers hpaA-Del-MoClo-F and hrcD-Del-MoClo-R3. The resulting PCR fragment (contains hrcD and ∆hpaA) was cloned into vector pAGM9121 using BpiI and ligase, resulting in the level-2 construct pAGB1323 (hrcD, ∆hpaA). For the generation of the hpaB deletion, the original module pAGB208 (hrpE, hpaB) was separated by amplifying hrpE with primers hpaB-Del-MoClo-I-F and hpaB-Del-MoClo-I-R and the fragment containing the deletion in hpaB with primers hpaB-Del-MoClo-II-F and hpaB-Del-MoClo-II-R. The PCR fragments were individually cloned using BpiI and ligase into pAGM9121 to generate the level-2 constructs pAGB649 (hrpE) and pAGB650 (∆hpaB). For the introduction of a deletion in hpaA, the insert of pAGB1323 (hrcD, ∆hpaA) was ligated with the inserts of pAGB205 (hrcS), pAGB207 (hrpD6), pAGB208 (hrpE, hpaB) and pAGB209 (hpaE) using BsaI and ligase into vector pAGM1311 to create the level-1 construct pAGB1324. For the hpaA/hpaB double deletion the same modules were inserted into vector pAGM1311, except for the pAGB208 construct, which was replaced by constructs pAGB649 (hrpE) and pAGB650 (∆hpaB) resulting in the level-1 construct pAGB1359. The inserts of the resulting constructs pAGB1324 (hrcS to hpaE region with a deletion in hpaA) and pAGB1359 (hrcS to hpaE region with a deletion in hpaA and hpaB) were individually assembled with the insert of construct pAGB210 (PhrpC, hrcU to hrcR) into the vector pICH41331 using BpiI and ligase to generate the level 0 constructs pAGB1325 (hrpC to hpaB operons with a deletion in hpaA) and pAGB1360 (hrpC to hpaB operons with a deletion in hpaA and hpaB). In a following step, each of these inserts was transferred using BsaI and ligase into the level 1 destination vector pICH47751, resulting in the level 1 constructs pAGB1326 (hrpC to hpaB operons with a deletion in hpaA) and pAGB1361 (hrpC to hpaB operons with a deletion in hpaA and hpaB). The inserts of pAGB1326 or pAGB1361 were subsequently cloned together with the inserts of pAGB154 (hrpA to hrpB operons), pAGB156 (hrpF operon), pICH54011 (dummy pos.1), pICH50900 (end linker) into the BpiI sites of vector pAGM8031 to generate the level M constructs pAGB1327 (hrp gene cluster with a deletion in hpaA) and pAGB1362 (hrp gene cluster with a deletion in hpaA and hpaB).
Finally, the level M construct pAGB1332 (contains xopA, hpaH, hrpX, hrpG* and hpaA-2xLinker-sfgfp) was assembled with the end linker pICH79264 and the level M construct pAGB1327 (hrp gene cluster with a deletion in hpaA) or pAGB1362 (hrp gene cluster with a deletion in hpaA and hpaB) in the level P vector pICH75322 using BsaI and ligase, thus resulting in the level P constructs pAGB1334 (contains hpaA-2xLinker-sfgfp and a deletion in hpaA) and pAGB1374 (contains hpaA-2xLinker-sfgfp and a deletion in hpaA and hpaB).
Generation of modular hrp gene cluster constructs for the analysis of peripheral protein localization in bacterial cells
As control for peripheral protein localization in bacterial cells (periplasm or membrane-associated), a hrcC-2xLinker-mCherry module was generated. For this, hrcC was amplified with primers hrcC-NTM-MoClo-F and hrcC-NTM-MoClo-R and the resulting amplicon was cloned using BpiI and ligase into vector pAGM9121 to generate the level-2 construct pAGB611. The insert of pAGB611 was subsequently transferred into vector pAGM1311 using BsaI and ligase to create the level-1 construct pAGB474. This module allows the generation of translational fusions of hrcC to genes encoding potential C-terminal fusion partners. mCherry as potential fusion partner was amplified using primers mCherry-CTM-woL-F and mCherry-CTM-woL-R, and the resulting PCR fragment was cloned into vector pICH41021 using SmaI and ligase to generate the level-1 construct pAGB1048. The inserts of constructs pAGB474 (hrcC), pAGB1048 (mCherry) and pAGB1000 (encodes linker sequence 2 × AKLEGPAGL) were cloned using BpiI and ligase into vector pICH41308, thus resulting in the level 0 construct pAGB1059. In the next step, the inserts of constructs pAGB1059 (hrcC-2xLinker-mcherry), pAGB232 (Xcv term2) and pAGB511 (PhrpA) were assembled in the destination vector pICH47781 using BsaI and ligase, thus generating the level 1 construct pAGB1063. The native hrpA promoter (PhrpA) was amplified with primers PhrpA-MoClo-F and PhrpA-MoClo-R and the resulting PCR product was cloned as blunt-end fragment using SmaI and ligase into vector pICH41021 to generate the level-1 construct pAGB614. The insert of pAGB614 was subsequently inserted into the BpiI sites of vector pICH41295 to generate the level 0 construct pAGB511 (PhrpA). The insert of pAGB1063 (PhrpA hrcC-2xLinker-mcherry) was assembled with the inserts of pAGB157 (xopA + hpaH), pAGB160 (hrpX), pAGB163 (hrpG*), pICH54022 (dummy pos. 2′) and pICH50881 (end linker) in vector pAGM8079 using BpiI and ligase, resulting in the level M construct pAGB1067.
For the analysis of HrcC-mCherry localization, hrcC was deleted in the modular hrp gene cluster. For this, we generated a hrcC deletion module by PCR using the phosphorylated primers P-hrcC-Del-MoClo-F and P-hrcC-Del-MoClo-R and pAGB196 (hrcC) as template. Ligation of the PCR product resulted in the level-2 construct pAGB515. The insert of pAGB515 (∆hrcC) was then assembled with the inserts of constructs pAGB192 (hrcL), pAGB193 (hrcN), pAGB194 (hrpB7) and pAGB195 (hrcT) into vector pAGM1311 using BsaI and ligase to create the level-1 construct pAGB676. The insert of construct pAGB676 (contains hrcL to hrcT and hrcC with a deletion) was subsequently assembled with the insert of pAGB197 (contains hrpB1 to hrpB4) in the BpiI sites of vector pICH41331, resulting in the level 0 construct pAGB680. The insert of pAGB680 (hrpA and hrpB operon with a hrcC deletion) was transferred into the destination vector pICH47811 using BsaI and ligase, thus resulting in the level 1 construct pAGB683. To generate the level M construct containing the hrp gene cluster with a hrcC deletion, the inserts of constructs pAGB683 (hrpA to hrpB operons with a deletion in hrcC), pAGB155 (hrpC, hrpD, hrpE and hpaB operons), pAGB156 (hrpF operon), pICH54011 (dummy pos.1) and pICH50900 (end linker) were assembled in the BpiI sites of vector pAGM8031, resulting in the level M construct pAGB739. In a final cloning step, the inserts of constructs pAGB739 (hrp gene cluster with a hrcC deletion), pAGB1067 (hrcC-2xLinker-mcherry) and pICH79264 (end linker) were cloned into the BsaI sites of vector pICH75322, thus resulting in the level P construct pAGB1075.
Results
The N-terminal 70 amino acids of HpaA including the TrM are essential for translocation
We previously showed that the N-terminal 70 amino acids of HpaA are sufficient to target a reporter protein for translocation (Lorenz et al., 2008a). This region contains a characteristic amino acid pattern (46-PRLRPAPPRRRRRGI-61), designated TrM, with a Pro/Arg-rich sequence, which is also present in the N-terminal regions of T3Es from X. euvesicatoria (Figure 1A). In HpaA, the TrM overlaps with an N-terminal predicted nuclear localization (NLS1) motif (amino acids 54–60) which is one out of two putative NLS motifs and presumably involved in nuclear targeting of HpaA after translocation into the plant cells (Figure 1A; Huguet et al., 1998; Lorenz et al., 2008a).
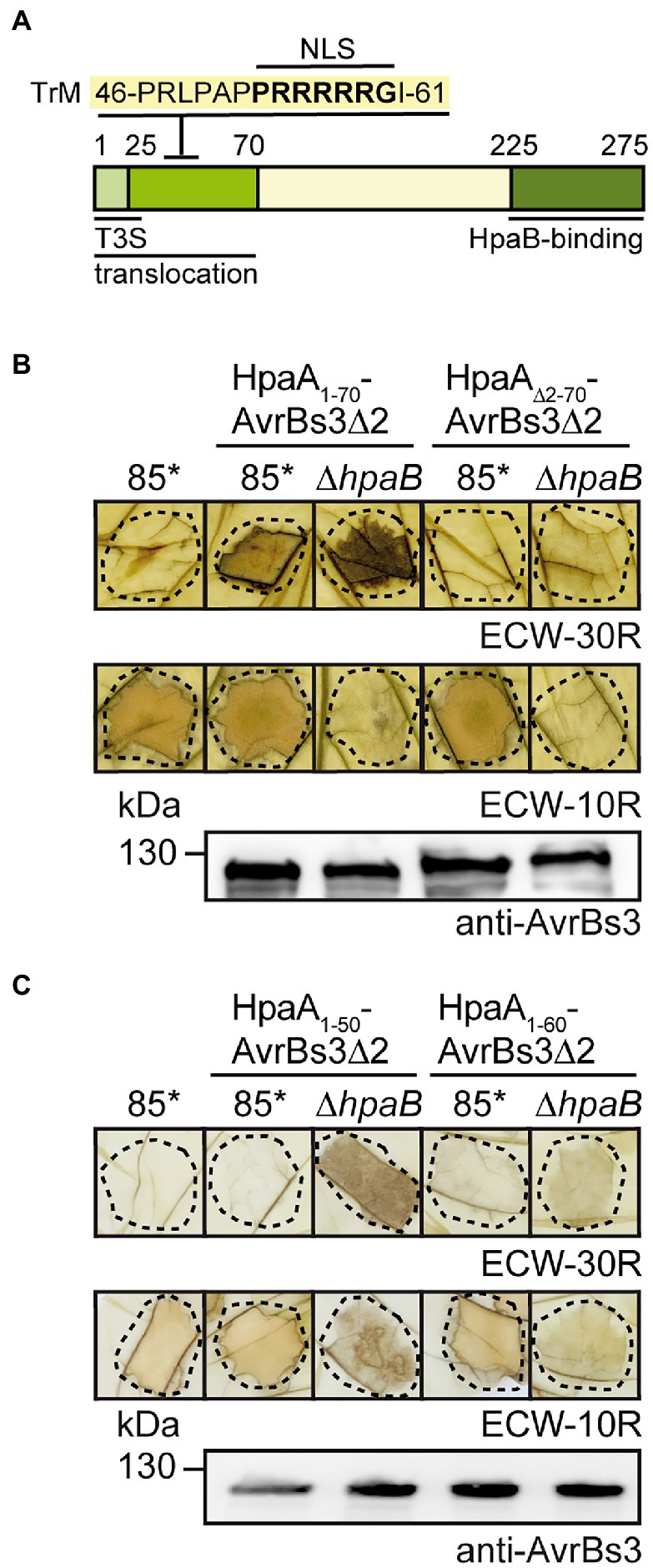
Figure 1. The N-terminal region is essential for HpaA translocation. (A) Schematic representation of functional regions in HpaA. Letters refer to the amino acid sequence of the N-terminal TrM, numbers indicate amino acid positions. The localization of the N-terminal T3S and translocation signal and the C-terminal HpaB-binding site is indicated (Lorenz et al., 2008a). TrM, translocation motif. (B) The N-terminal 70 amino acids are essential for HpaA translocation. X. euvesicatoria strains 85* and 85*∆hpaB with or without expression constructs encoding HpaA1-70-AvrBs3∆2 or HpaA∆2-70-AvrBs3∆2 as indicated were infiltrated into leaves of AvrBs3-responsive ECW-30R and AvrBs1-responsive ECW-10R pepper plants. Leaves were bleached in ethanol 2 dpi. Dashed lines indicate the infiltrated areas. For protein analysis, bacterial strains were grown in nutrient-yeast extract-glycerol (NYG) medium and equal amounts of protein extracts were analysed by immunoblotting using an AvrBs3-specific antiserum. (C) The N-terminal 50 amino acids of HpaA contain a minimal translocation signal. X. euvesicatoria strains 85* and 85*∆hpaB with or without expression constructs encoding HpaA1-50-AvrBs3∆2 or HpaA1-60-AvrBs3∆2 as indicated were infiltrated into leaves of AvrBs3-responsive ECW-30R and AvrBs1-responsive ECW-10R pepper plants. HR reactions and protein synthesis were documented as described in (B).
To further study the translocation signal of HpaA, we generated fusion proteins between HpaA derivatives and the reporter protein AvrBs3∆2, which is an N-terminal deletion derivative of the TAL effector AvrBs3 and lacks the T3S export signal. AvrBs3∆2 is only translocated when fused to a functional translocation signal and, hence, induces the HR in AvrBs3-responsive pepper plants, which contain the R gene Bs3 (Szurek et al., 2002; Noël et al., 2003). We first investigated whether the N-terminal 70 amino acids are essential for translocation of HpaA. For this, an AvrBs3∆2 reporter fusion containing HpaA deleted in amino acids 2–70 was analysed in X. euvesicatoria strain 85-10hrpG* (85*). Strain 85* contains a constitutively active version of the key regulator HrpG, which allows the analysis of in vitro T3S and the elicitation of accelerated plant reactions (Wengelnik et al., 1999). When bacteria were infiltrated into leaves of AvrBs3-responsive pepper plants, HpaA1-70-AvrBs3∆2 but not HpaA∆2-70-AvrBs3∆2 induced a visible HR, suggesting that the N-terminal 70 amino acids are sufficient and essential for translocation of HpaA (Figure 1B). Immunoblot analysis of bacterial cell extracts showed that HpaA∆2-70-AvrBs3∆2 was stably synthesized (Figure 1B).
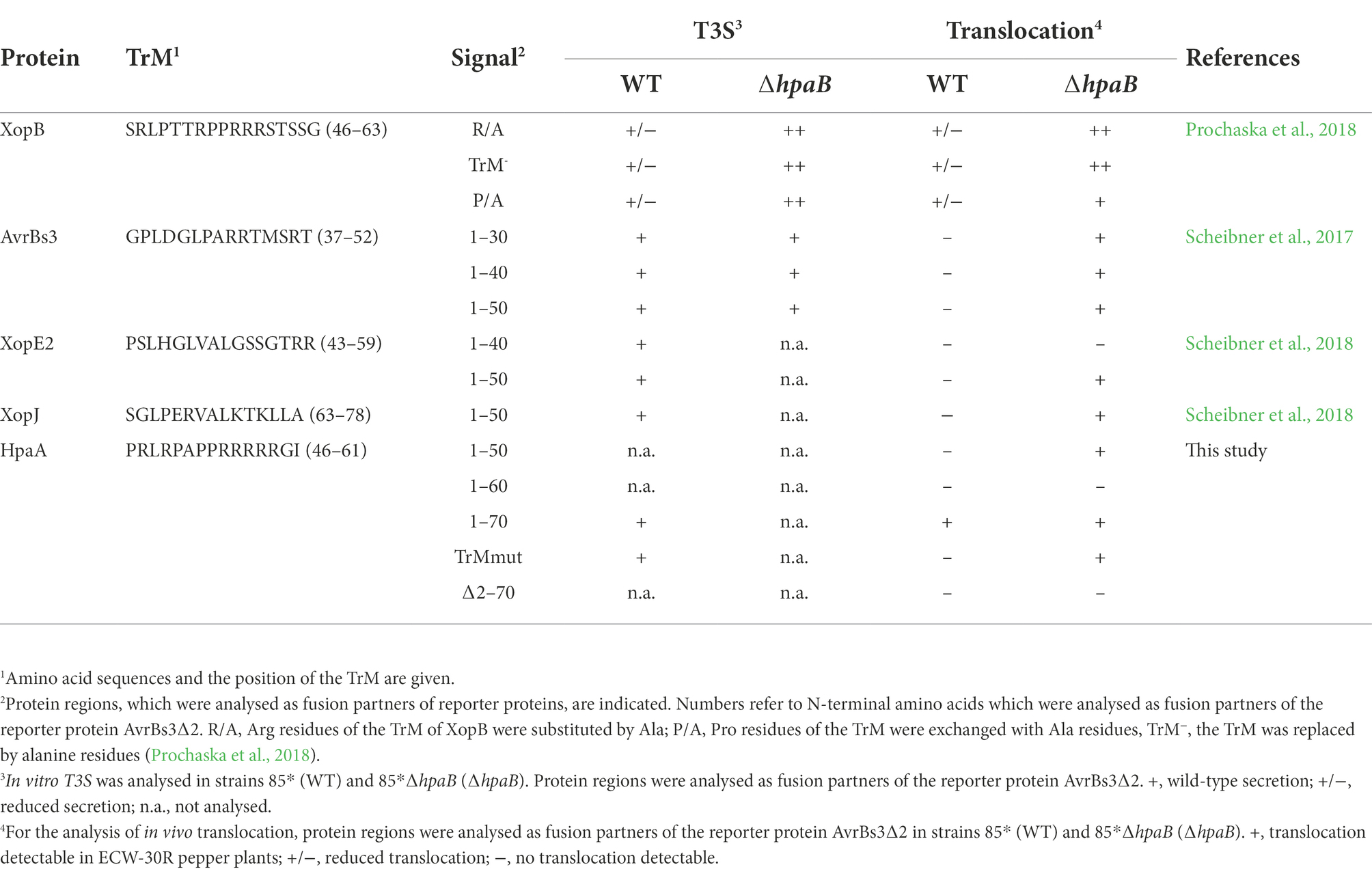
We also generated expression constructs containing the N-terminal 50 (lacking the TrM) or 60 (including the Pro/Arg-rich sequence) amino acids of HpaA as fusion partners of AvrBs3∆2. When analysed in strain 85*, HpaA1-50-AvrBs3∆2 and HpaA1-60-AvrBs3∆2 were stably synthesized but did not elicit a visible HR in AvrBs3-responsive pepper plants (Figure 1C). This is in contrast to HpaA1-70-AvrBs3∆2, which is translocated by the wild-type strain (Lorenz et al., 2008a), suggesting that the N-terminal 50 and 60 amino acids of HpaA and thus the Pro/Arg-rich sequence were not sufficient to target the AvrBs3∆2 reporter for translocation. It can, however, not be excluded that additional amino acids present in HpaA1-70-AvrBs3∆2 are required for a correct exposure of the TrM on the protein surface and thus its recognition by the T3S system. Notably, when analysed in the hpaB deletion mutant strain 85*∆hpaB, HpaA1-50-AvrBs3∆2 induced the AvrBs3-specific HR, indicating that the N-terminal 50 amino acids of HpaA contain a minimal translocation signal which is recognized in the absence of HpaB but not in the wild-type strain (Figure 1C). Similar findings were previously observed for N-terminal regions of the effector proteins AvrBs3, XopE2 and XopJ (Scheibner et al., 2017, 2018). Interestingly, N-terminal minimal translocation signals do not include the TrM (Table 1).
The contribution of the TrM to translocation of HpaA depends on the T3S chaperone Hpab
To further investigate the contribution of the TrM to HpaA translocation, we introduced point mutations leading to the exchange of amino acids 54-PRRRRRG-60 to 54-PAAAAAG-60 (hereafter referred to as TrMmut) in HpaA-AvrBs3∆2 and HpaA1-70-AvrBs3∆2. These mutations abolished the detectable translocation of both fusion proteins by strain 85* but did not interfere with protein stability as was shown by immunoblot analyses of bacterial cell extracts (Figures 2A,B). Interestingly, however, the TrM mutation promoted translocation of HpaA-AvrBs3∆2 fusions in strain 85*∆hpaB (Figures 2A,B). This is in agreement with the above observation that HpaA1-50-AvrBs3∆2 lacking the TrM is translocated by strain 85*∆hpaB but not by the wild-type strain (Figure 1C). Notably, the contribution of the TrM to HpaA translocation is independent of HpaB binding because the HpaB-binding site is located in the C-terminal 50 amino acids of HpaA (Lorenz et al., 2008a). Our findings are reminiscent of the previous observation that mutation of the TrM in the effector protein XopB interfered with XopB translocation in the wild-type strain but led to enhanced translocation of XopB reporter fusions in the absence of HpaB (Prochaska et al., 2018; Table 1). The authors postulated that the TrM mutation abolished the membrane localization of XopB and thus bypassed the need of XopB for the predicted HpaB-mediated release from the membrane (Prochaska et al., 2018). It is, however, unknown whether the TrM also serves as a membrane localization signal in other T3Es.
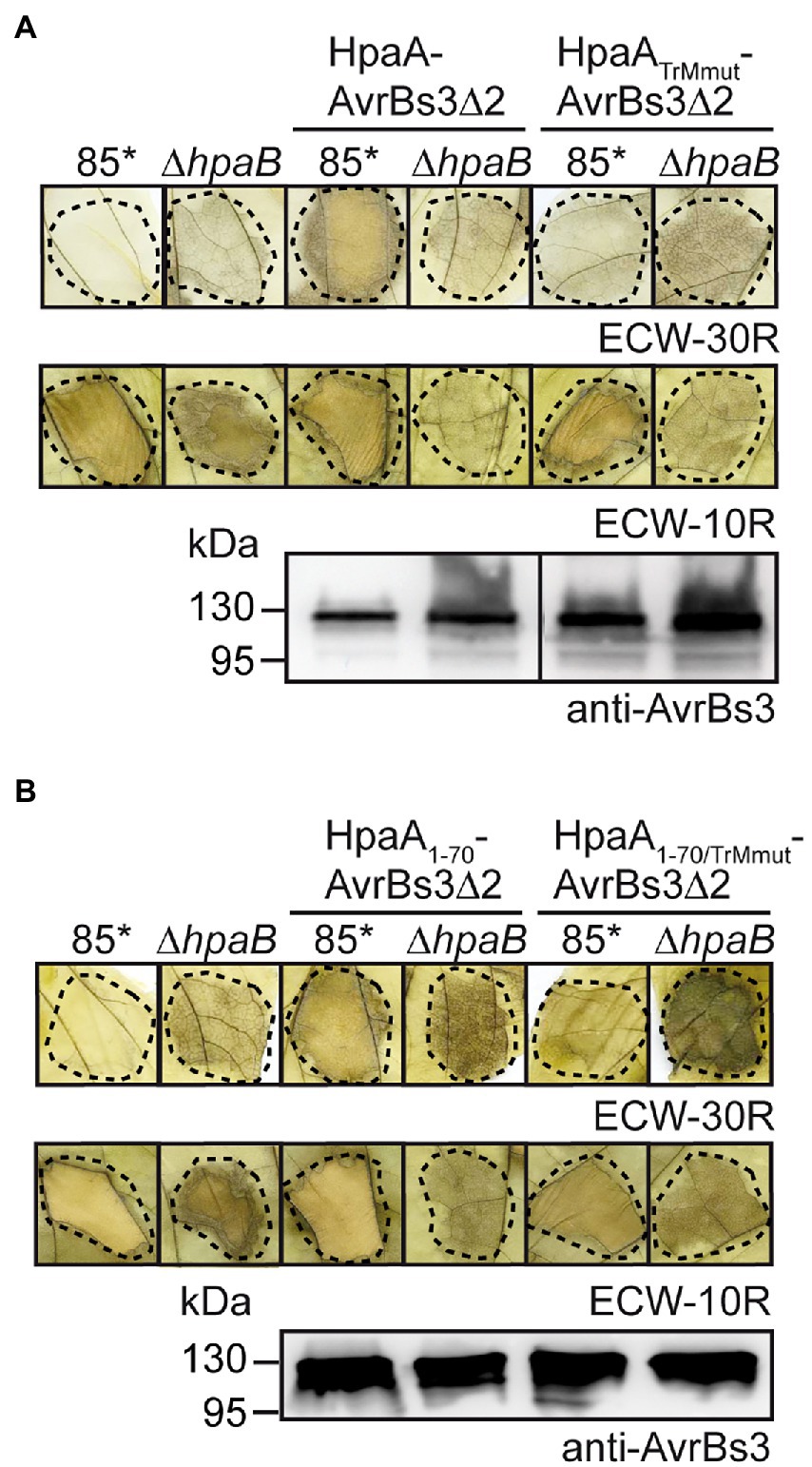
Figure 2. The N-terminal TrM determines HpaB-dependent translocation of HpaA. (A) The TrM is essential for HpaA translocation. X. euvesicatoria strains 85* and 85*∆hpaB with or without expression constructs encoding HpaA-AvrBs3∆2 or HpaATrMmut-AvrBs3∆2 as indicated were infiltrated at a density of 4 × 108 cfu ml−1 into leaves of AvrBs3-responsive ECW-30R and AvrBs1-responsive ECW-10R pepper plants. Leaves were bleached in ethanol 2 dpi. Dashed lines indicate the infiltrated areas. For protein analysis, bacteria were grown in NYG medium, and equal amounts of protein extracts were analysed by immunoblotting using an AvrBs3-specific antiserum. (B) Mutation of the TrM promotes translocation of HpaA in the absence of HpaB. X. euvesicatoria strains 85* and 85*∆hpaB with or without expression constructs encoding HpaA1-70-AvrBs3∆2 or HpaA1-70/TrMmut-AvrBs3∆2 as indicated were infiltrated into leaves of AvrBs3-responsive ECW-30R and AvrBs1-responsive ECW-10R pepper plants. HR reactions and protein synthesis were documented as described in (A).
HpaA does not localize to the bacterial membranes
To investigate whether HpaA localizes to the bacterial membranes as was suggested for XopB, we analysed a HpaA–sfGFP fusion protein in X. euvesicatoria by fluorescence microscopy. For this, we used a modular expression construct containing the T3S gene cluster, regulatory (hrpG*, hrpX) and accessory genes (the putative translocon gene xopA and the lytic transglycosylase gene hpaH), which allows the assembly and activation of the T3S system in X. euvesicatoria (Noël et al., 2002; Hausner et al., 2017, 2019). The modular T3S gene cluster was assembled from single modules containing promoters and genes using the Golden Gate-based modular cloning system as described previously (Hausner et al., 2019). For this, promoter and gene modules were assembled in different cloning steps by alternating the use of the type IIs restriction enzymes BsaI and BpiI. This led to level 0 constructs containing single or multiple operons, which were flanked by identical fusion sites. For further assembly, level 0 modules were transferred to different level 1 vectors which allowed subsequent positional cloning into a level M vector. Two level M modules corresponding to the hrp gene cluster and the regulatory and accessory genes, respectively, were finally assembled in a level P vector (Hausner et al., 2019).
The modular design of the construct facilitates the rapid deletion and insertion of single gene and promoter modules and thus the analysis of reporter fusions. For the experiments with HpaA-sfGFP, the corresponding gene fusion was inserted into the flanking region of the modular T3S gene cluster which was deleted in the native hpaA gene (Figure 3A). The resulting construct was transferred to X. euvesicatoria strain 85*∆hrp_ fsHAGX which lacks the chromosomal T3S gene cluster and contains mutations in the native hrpG, hrpX, xopA and hpaH genes (Hausner et al., 2019). To analyse whether HpaA-sfGFP was functional, bacteria were infiltrated into leaves of resistant and susceptible pepper plants. As expected, the wild-type strain induced the HR and water-soaked lesions, respectively, whereas plant reactions were significantly reduced after infection with a strain containing the modular T3S gene cluster deleted in hpaA (Figure 3B). The wild-type phenotype was restored upon insertion of hpaA-sfGFP into the modular construct, suggesting that the sfGFP fusion partner did not significantly interfere with HpaA function (Figure 3B). Immunoblot analysis of bacterial cell extracts showed that HpaA-sfGFP was stably synthesized (Supplementary Figure S1).
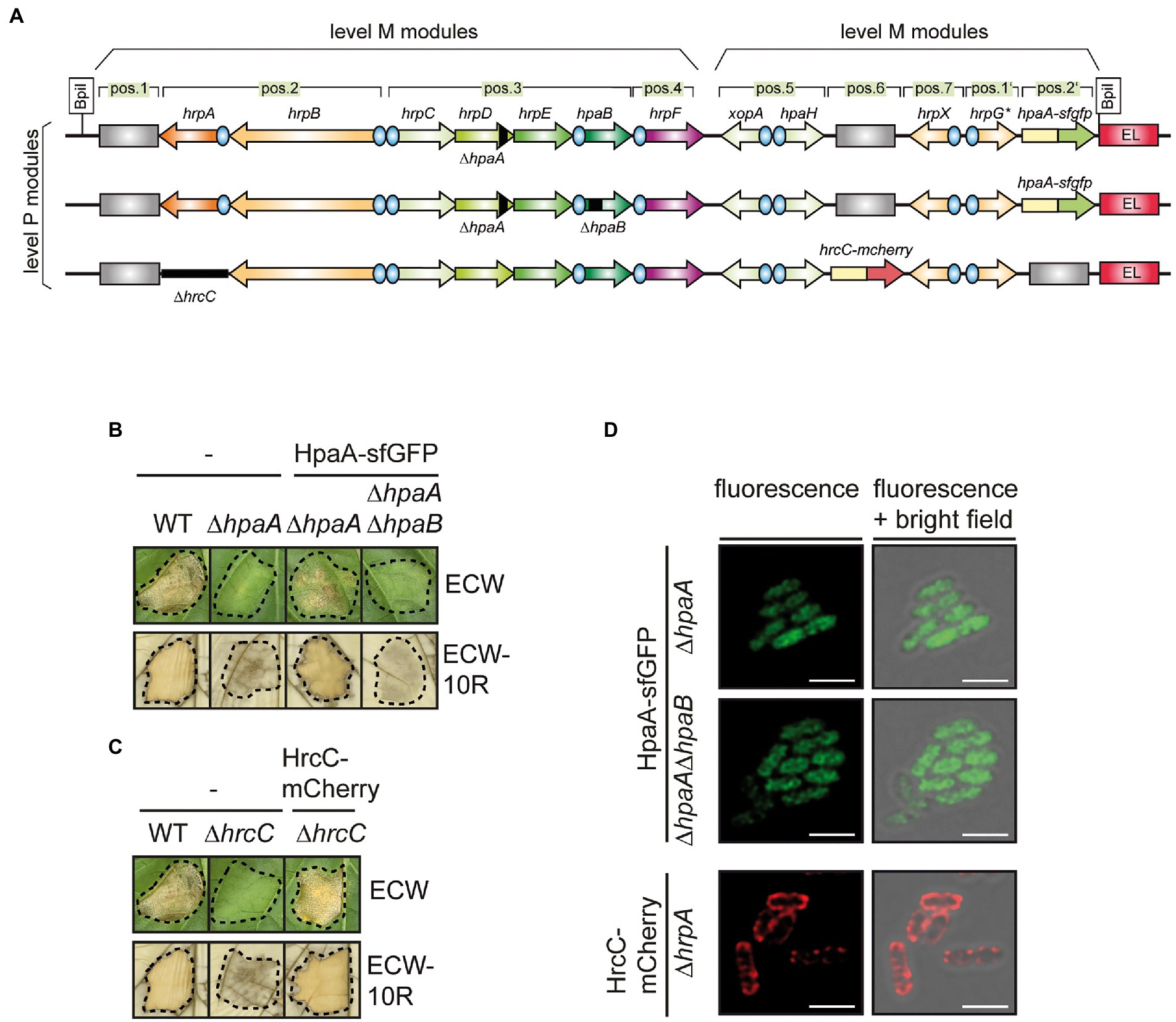
Figure 3. Localization studies with a HpaA-sfGFP fusion protein. (A) Schematic representation of modular T3S gene cluster constructs. Genes and operons are represented by arrows, promoters by blue circles. Grey rectangles represent dummy modules that were replaced by reporter fusions. Deletions are indicated with black rectangles. The names of single operons and genes is given above the arrows. The modular level P constructs resulted from the assembly of two level M modules. (B) Complementation studies with HpaA-sfGFP. Strain 85*∆hrp containing the wild-type modular T3S gene cluster (WT) or a derivative thereof with a deletion in hpaA (∆hpaA) and strain 85*∆hrp_fsHAGX containing modular T3S gene cluster constructs with deletions in hpaA or both hpaA and hpaB (∆hpaAB) and encoding HpaA-sfGFP in the flanking region of the T3S gene cluster as depicted in (A) were infiltrated into leaves of susceptible ECW and resistant ECW-10R pepper plants. Dashed lines indicate the infiltrated areas. Disease symptoms were photographed 8 dpi. For the better visualization of the HR, leaves were bleached in ethanol 2 dpi. HpaA-sfGFP was stably synthesized as is shown in Supplementary Figure S1. (C) HrcC-mCherry complements the hrcC mutant phenotype. Strain 85*∆hrp containing the wild-type modular T3S gene cluster (WT) or a derivative thereof with a deletion in hrcC (∆hrcC) and strain 85*∆hrp_fsHAGX containing the modular T3S gene cluster deleted in hrcC and encoding HrcC-mCherry in the flanking region of the T3S gene cluster as depicted in (A) were infiltrated into leaves of susceptible ECW and resistant ECW-10R pepper plants. Plant reactions were monitored as described in (B). HrcC-mCherry was stably synthesized as is shown in Supplementary Figure S1. (D) HpaA-sfGFP is evenly distributed in bacterial cells. X. euvesicatoria strain 85*∆hrp_fsHAGX with plasmids containing the modular T3S gene cluster, accessory and regulatory genes with deletions in hpaA, hpaA and hpaB or hrcC and hpaA-sfgfp or hrcC-mcherry inserted into the flanking region of the modular T3S gene cluster as indicated was incubated under T3S-permissive conditions and analysed by fluorescence microscopy. One representative image for every strain is shown. The size bar corresponds to 2 μm. The pictures in the right panels result from an overlay of the signals from the fluorescent channel for GFP with the bright field images. For better visualisation of fine structures, the confocal pictures were processed with the LIGHTNING technology of Leica. Experiments were performed with different transconjugants for each strain three times with similar results. The results from one representative experiment are shown.
For localization studies, bacteria were incubated under T3S-permissive conditions and analysed by fluorescence microscopy. The GFP fluorescence was evenly distributed in all cells (Figure 3D). A similar localization pattern was observed upon additional deletion of hpaB, suggesting that HpaB does not significantly affect the localization of HpaA (Figure 3D). As control for the localization studies, we generated an additional construct expressing the modular T3S gene cluster deleted in the native secretin gene hrcC and containing a hrcC-mCherry fusion in the flanking region (Figure 3A). mCherry is a red fluorescent protein suitable for the analysis of periplasmic proteins (Shaner et al., 2004; Meiresonne et al., 2017, 2019). The secretin HrcC contains a Sec signal for the transport across the IM and forms an oligomeric protein channel in the OM (Wengelnik et al., 1996; Silva et al., 2020). Infection and immunoblot experiments showed that HrcC-mCherry was stably synthesized and complemented the hrcC mutant phenotype in planta (Figure 3C and Supplementary Figure S1). When analysed by fluorescence microscopy, HrcC-mCherry localized to the periphery of the bacterial cells as expected and also formed fluorescent spots likely corresponding to assembled secretin channels in the OM (Figure 3D). The clear difference in the fluorescence signals of HrcC-mCherry and HpaA-sfGFP indicates that both proteins do not colocalize at the cell periphery. We, therefore, conclude that HpaA does not primarily localize to the bacterial membranes as was reported for XopB. The absence of detectable fluorescent foci for HpaA-sfGFP, which would indicate an association with assembled T3S systems, suggests that HpaA is not exclusively attached to the secretion apparatus but also present in the bacterial cytoplasm.
The TrM is dispensable for secretion but essential for the in vivo function of HpaA
The finding that the TrM is essential for HpaA translocation prompted us to analyse its contribution to HpaA secretion. For this, we introduced expression constructs encoding HpaA-c-Myc and HpaATrMmut-c-Myc into strain 85*∆hpaA. For the analysis of in vitro T3S, bacteria were incubated under T3S-permissive conditions, and total cell extracts and culture supernatants were analysed by immunoblotting. Figure 4A shows that HpaA-c-Myc and HpaATrMmut-c-Myc were both detected in the culture supernatant, suggesting that the TrM mutation did not interfere with HpaA secretion. As expected, HpaATrMmut-c-Myc restored secretion of the translocon protein HrpF in strain 85*∆hpaA (Figure 4A). This is in agreement with our previous observation that HpaA derivatives, which contain the C-terminal HpaB-binding site, promote secretion of translocon proteins (Lorenz et al., 2008a).
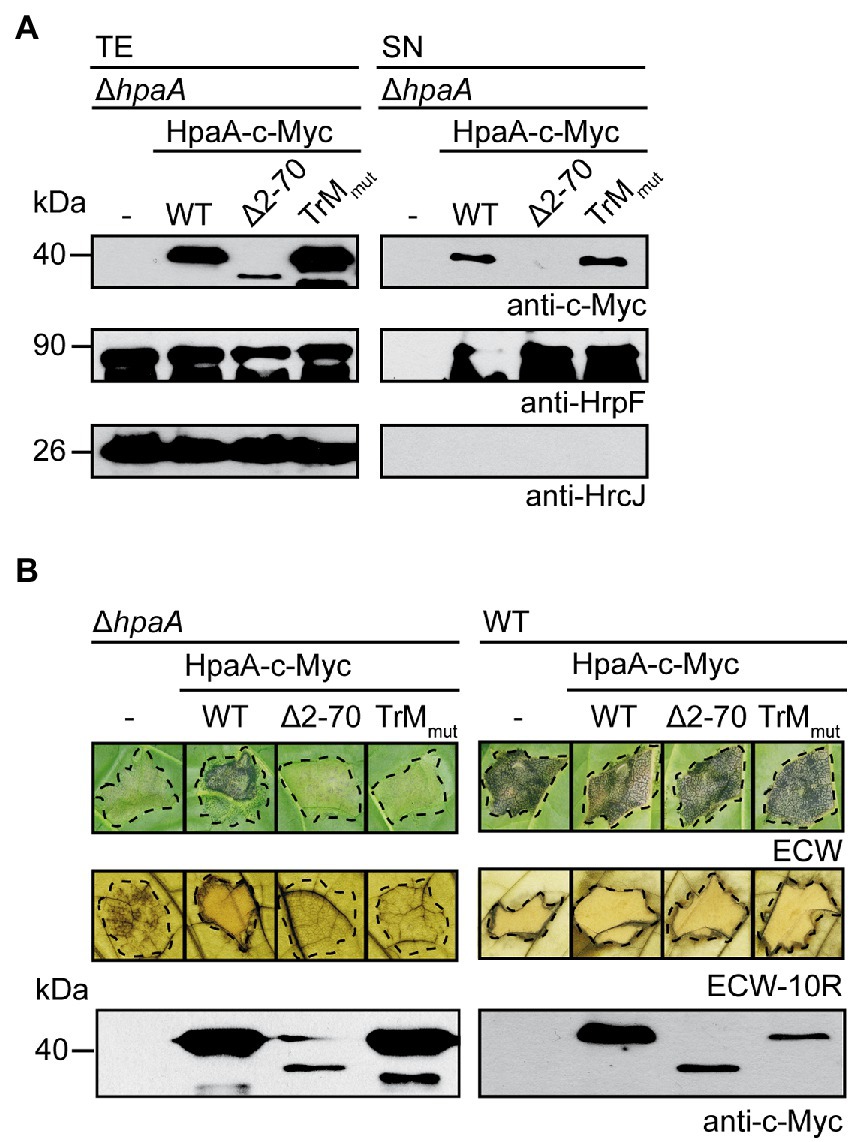
Figure 4. The TrM is essential for the in vivo function of HpaA. (A) Mutation of the TrM does not interfere with T3S of HpaA. X. euvesicatoria strain 85*∆hpaA (∆hpaA) without expression construct (−) or containing HpaA-c-Myc (WT), HpaA∆2-70-c-Myc (∆2–70) or HpaATrMmut-c-Myc (TrMmut) as indicated was incubated under T3S-permissive conditions. Total cell extracts (TE) and culture supernatants (SN) were analysed by immunoblotting using antibodies directed against the c-Myc epitope, the translocon protein HrpF and the IM protein HrcJ, respectively. Experiments were performed three times with similar results.
Interestingly, when bacteria were infiltrated into leaves of susceptible and resistant pepper plants, HpaA-c-Myc but not HpaATrMmut-c-Myc restored pathogenicity of strain 85*∆hpaA, suggesting that the TrM mutation abolished the in vivo function of HpaA (Figure 4B). The lack of function was presumably not caused by a dominant-negative effect of HpaATrMmut-c-Myc because ectopic expression of HpaA derivatives in strain 85* did not significantly interfere with pathogenicity and HR induction in susceptible and resistant plants, respectively (Figure 4B). When analysed in strain 85–10, however, both HpaA-c-Myc and HpaATrMmut-c-Myc led to reduced plant reactions after infection of susceptible and resistant pepper plants (Supplementary Figure S2). We assume that enhanced levels of HpaA bind to and thus inhibit the function of HpaB. In agreement with this hypothesis, ectopic expression of HpaA deleted in the C-terminal HpaB-binding site did not significantly interfere with pathogenicity of strain 85–10 (Supplementary Figure S2). The observed dominant-negative effect of hpaA-c-myc and hpaATrMmut-c-myc expression constructs likely did not interfere with protein function because ectopic expression of hpaA-c-myc in strain 85*∆hpaA restored the wild-type phenotype (Figure 4B). In summary, the results of our in vitro and in vivo experiments suggest that the TrM is dispensable for the in vitro secretion of HpaA but essential for the translocation and in planta activity of HpaA.
Recognition of the translocation signal of HpaA depends on the HpaC-mediated T3S substrate specificity switch
Given the predicted function of HpaA as regulator of HpaB, we assumed that the translocation of HpaA occurs after the assembly of the T3S system. In X. euvesicatoria, secretion of translocon and T3E proteins is activated by the T3S4 protein HpaC which binds to and likely induces a conformational change in the cytoplasmic domain of the IM protein HrcU (Lorenz et al., 2008b; Lorenz and Büttner, 2011). HpaC also interacts with T3S substrates and the T3S chaperone HpaB, suggesting that it is involved in substrate recognition (Büttner et al., 2006). To investigate a possible interaction of HpaC with HpaA, we performed in vitro GST pull-down assays. For this, GST and a GST-HpaC fusion protein were immobilized on glutathione sepharose and incubated with an E. coli lysate containing HpaA-c-Myc. When eluted proteins were analysed by immunoblotting, HpaA-c-Myc was detected in the eluate of GST-HpaC but not of GST alone (Figure 5A). Similar findings were obtained for HpaA derivatives lacking the C-terminal 50 amino acids or amino acids 2–70, suggesting that the N-and C-terminal regions of HpaA are dispensable for HpaC binding (Figures 5A,B). As expected, HpaA∆2-70-c-Myc interacted with GST-HpaB (Figure 5B). When HpaA derivatives were analysed as GST fusions, HpaC-c-Myc coeluted with GST-HpaA and GST-HpaA∆C50 but not with GST (Figure 5C). We conclude from these experiments that the HpaB-binding site, which is located in the C-terminal 50 amino acids of HpaA (Lorenz et al., 2008a), is dispensable for HpaC binding, suggesting that HpaA, HpaB and HpaC can form a common protein complex.
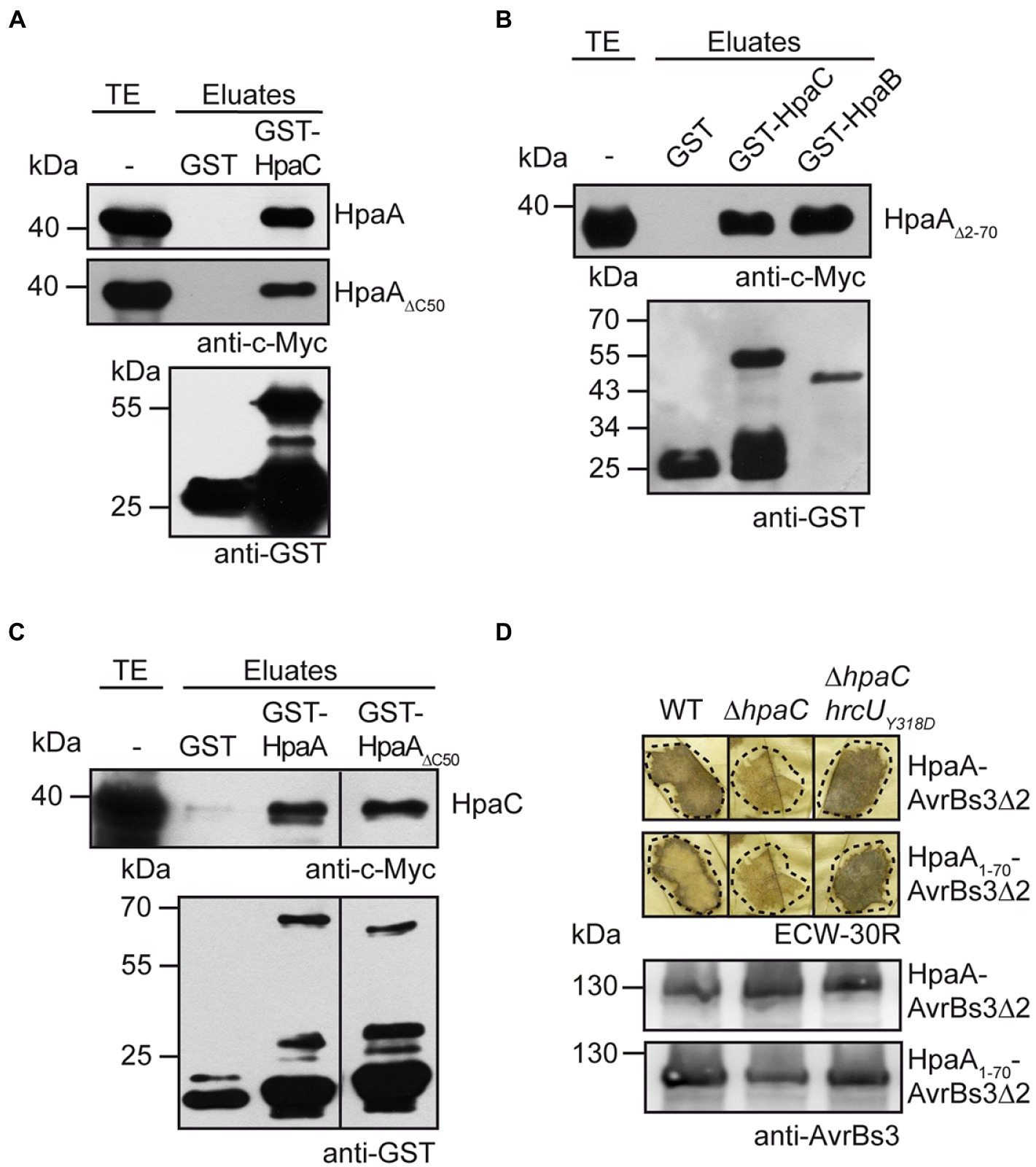
Figure 5. HpaA interacts with the T3S4 protein HpaC and depends on the predicted HpaC-mediated conformational change in HrcUC for efficient translocation. (A) HpaA interacts with HpaC independently of the C-terminal HpaB-binding site. GST and GST-HpaC were immobilized on glutathione sepharose and incubated with bacterial lysates containing HpaA-c-Myc or HpaA∆C50-c-Myc as indicated. Total cell extracts (TE) and eluted proteins (eluates) were analysed by immunoblotting using c-Myc epitope- or GST-specific antibodies. (B) The N-terminal 70 amino acids of HpaA are dispensable for the interaction with HpaC. GST, GST-HpaC and GST-HpaB were immobilized on glutathione sepharose and incubated with a bacterial lysate containing HpaA∆2-70-c-Myc. TE and eluates were analysed as described in (A). (C) The C-terminal HpaB-binding site in HpaA is dispensable for the interaction with HpaC. GST, GST-HpaA and GST-HpaA∆C50 were immobilized on glutathione sepharose and incubated with a bacterial lysate containing HpaC-c-Myc. TE and eluates were analysed as described in (A). (D) Efficient translocation of HpaA depends on HpaC. X. euvesicatoria strains 85* (WT), 85*∆hpaC (∆hpaC) and 85*∆hpaChrcUY318D (∆hpaChrcUY318D) containing HpaA-AvrBs3∆2 or HpaA1-70-AvrBs3∆2 as indicated were infiltrated into leaves of AvrBs3-responsive ECW-30R pepper plants. For the better visualization of the HR, leaves were bleached in ethanol 2 dpi. Dashed lines indicate the infiltrated areas. For protein analysis, bacterial cell extracts were analysed by immunoblotting, using an AvrBs3-specific antibody.
To investigate the contribution of HpaC to HpaA translocation, we performed in vivo translocation assays. Translocation of HpaA-AvrBs3∆2 and HpaA1-70-AvrBs3∆2 was significantly reduced in strain 85*∆hpaC when compared with strain 85*, suggesting that the N-terminal translocation signal targets HpaA for HpaC-dependent translocation (Figure 5D). We also analysed translocation of HpaA-AvrBs3∆2 fusions in strain 85*∆hpaChrcUY318D which contains a genomic point mutation (Y318D amino acid exchange) in the C-terminal region of HrcU (HrcUC). The Y318D mutation in HrcUC promotes the secretion of middle and late substrates in the absence of HpaC, presumably by mimicking a predicted HpaC-induced conformational change (Lorenz and Büttner, 2011). Infection experiments showed that translocation of HpaA-AvrBs3∆2 and HpaA1-70-AvrBs3∆2 was restored in strain 85*∆hpaChrcUY318D, suggesting that the HpaC-dependent translocation of HpaA depends on a certain conformation of the cytoplasmic domain of HrcU rather than on the direct interaction of HpaA with HpaC (Figure 5D). Our data hence indicate that the predicted HpaC-induced conformational change in HrcUC restores recognition of the N-terminal translocation signal of HpaA in a hpaC deletion mutant.
HpaA interacts with components of the predicted sorting platform and the export apparatus
The importance of the TrM for HpaA translocation suggests a possible contribution of this motif to the docking of HpaA to the T3S system. Potential T3S substrate docking sites include the ATPase HrcN, the cytoplasmic domains of the export apparatus components HrcU and HrcV as well as components of the postulated sorting platform such as HrcQ and HrcL (Lorenz and Büttner, 2009, 2011; Lorenz et al., 2012; Hartmann and Büttner, 2013). To investigate a possible docking of HpaA to components of the sorting platform and the export apparatus, we performed in vitro GST pull-down assays. For this, GST fusions of the ATPase HrcN, the predicted sorting platform components HrcQ and HrcL as well as the export apparatus components HrcU and the cytoplasmic domain of HrcV were immobilized on glutathione sepharose and incubated with bacterial lysates containing HpaA-c-Myc. Figure 6A shows that HpaA-c-Myc coeluted with all GST fusion proteins but not with GST alone. Similar findings were observed for a HpaA-c-Myc derivative deleted in amino acids 2–70, suggesting that the N-terminal region is dispensable for the interaction of HpaA with the analysed T3S system components (Figure 6B). In reciprocal experiments, c-Myc epitope-tagged derivatives of HrcQ, HrcL and the cytoplasmic domain of HrcV (HrcVC) coeluted with GST-HpaA but not with GST (Figure 6C). Due to protein instabilities, HrcU, HrcV and HrcN were not included in these experiments. The interaction of HpaA with HrcN was further analysed by in vivo co-immunoprecipitation experiments in X. euvesicatoria strain 85*. For this, strain 85* with or without an expression construct encoding HpaA-c-Myc was incubated under T3S-permissive conditions and HpaA-c-Myc was precipitated using a c-Myc epitope-specific antibody. Immunoblot analysis of precipitated proteins showed that HrcN specifically co-immunoprecipitated with HpaA-c-Myc, suggesting that both proteins are part of a common complex (Figure 6D).
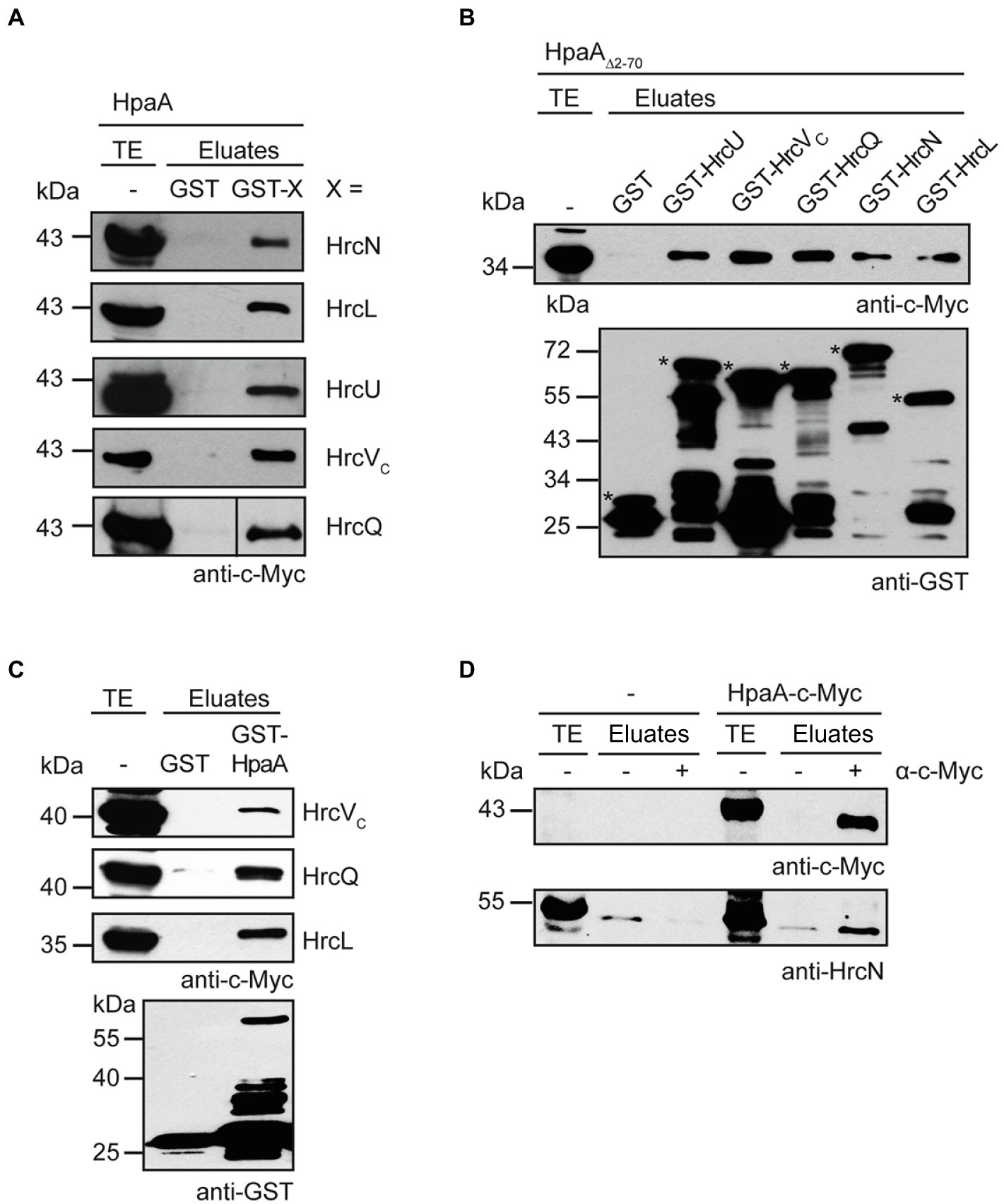
Figure 6. HpaA interacts with cytoplasmic components of the T3S system. (A) In vitro interaction studies with HpaA and cytoplasmic components of the T3S system. GST and GST fusions of HrcN, HrcL, HrcU, HrcVC and HrcQ were immobilized on glutathione sepharose and incubated with a bacterial lysate containing HpaA-c-Myc. Total cell extracts (TE) and eluted proteins (eluates) were analysed by immunoblotting using a c-Myc epitope-specific antibody. GST and GST fusions were stably synthesized as is shown in Supplementary Figure S3. (B) The N-terminal region of HpaA is dispensable for the in vitro interaction with cytoplasmic components of the T3S system. GST and GST fusions as described in (A) were immobilized on glutathione sepharose and incubated with a bacterial lysate containing HpaA∆2-70-c-Myc. TE and eluates were analysed by immunoblotting using c-Myc epitope-and GST-specific antibodies. Asterisks indicate the signals corresponding to GST and GST fusion proteins. Additional signals represent degradation products. (C) GST-HpaA interacts with HrcVC, HrcQ and HrcL. GST and GST-HpaA were immobilized on glutathione sepharose and incubated with bacterial lysates containing HrcVC-c-Myc, HrcQ-c-Myc and HrcL-c-Myc as indicated. TE and eluates were analysed as described in (B). (D) In vivo interaction studies with HpaA and HrcN. Cell lysates of X. euvesicatoria strain 85* without expression construct (−) or encoding HpaA-c-Myc as indicated were incubated in the absence (−) or presence (+) of a c-Myc epitope-specific antibody. TE and precipitated proteins (eluates) were analysed by immunoblotting using c-Myc epitope-and HrcN-specific antibodies. The experiment was performed three times with similar results.
The TrM is dispensable for the interaction of HpaA with components of the T3S system
In addition to GST pull-down assays and co-immuno- precipitation experiments, we performed bacterial two hybrid-based interaction studies to confirm the interaction of HpaA with T3S system components and to further investigate the contribution of the TrM to the identified interactions. For this, we used the BACTH system which depends on the association of two subdomains (T18 and T25) of the catalytic domain of the adenylate cyclase (Cya) for the production of cAMP. The interaction of T18 and T25 fusion proteins leads to the reconstitution of Cya, and the subsequent cAMP production activates the expression of the lac operon in E. coli strains lacking the native cya gene (Karimova et al., 1998; Battesti and Bouveret, 2012). For BACTH assays, we generated expression constructs encoding HpaA and components of the T3S system as fusion partners of the T18 or the T25 domain. HrcN, HrcL and HrcUC were analysed as C-terminal fusion partners of the T18 domain in the E. coli reporter strain BTH101 for interaction with HpaA-T25. When bacteria were grown on indicator plates, we observed an interaction of HpaA-T25 with T18-HrcN and a weak interaction of HpaA-T25 with T18-HrcL. In contrast, no interaction between HpaA-T25 and T18-HrcUC was detected (Figure 7A). Vice versa, T25-HrcUC did not detectably interact with HpaA-T18 or T18-HpaA (Figures 7B,C). Similar findings were observed for HrcUC derivatives with C-terminal T18 or T25 fusion partners, suggesting that the C-terminal domain of HrcU does not interact with HpaA in BACTH assays (Figure 7D). Notably, we previously reported that HrcUC interacts in vitro with the early T3S substrate HrpB2 but not with the putative translocon protein XopA or the effector protein XopC, suggesting that HrcUC is not a general T3S substrate docking site (Lorenz and Büttner, 2011). To analyse the interaction between HpaA and HrcQ, we used the alternative translation product HrcQC which corresponds to the C-terminal domain of HrcQ (Otten et al., 2021). We observed an interaction between HrcQC-T25 and HpaA-T18, suggesting that HpaA interacts with the C-terminal domain of HrcQ (Figure 7B).
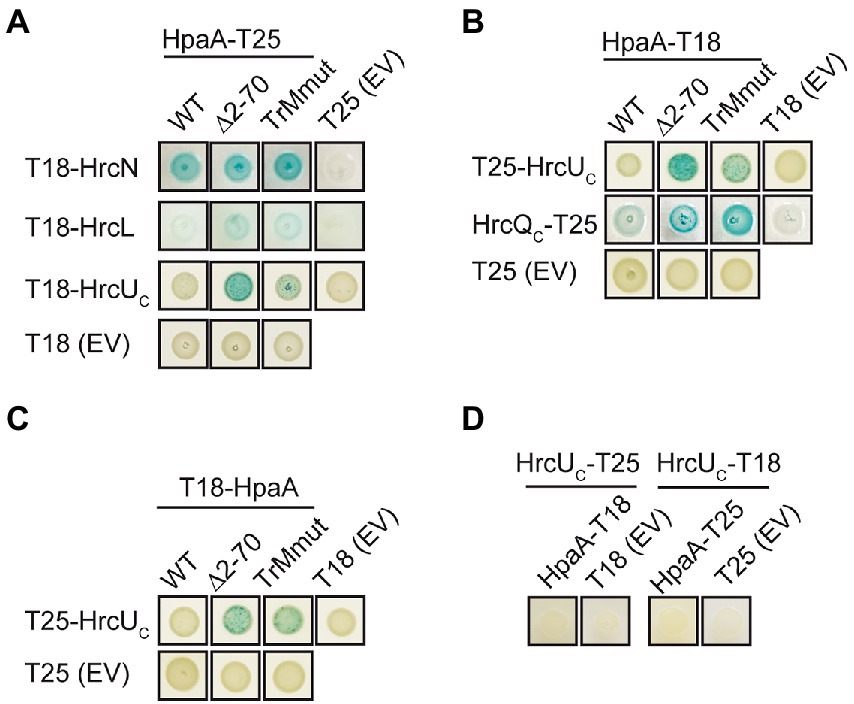
Figure 7. Interaction studies with HpaA using the BACTH system. (A) Interaction studies with HpaA, HrcN, HrcL and the cytoplasmic domain of HrcU. Expression constructs encoding T18 fusions of HrcN, HrcL and the cytoplasmic domain of HrcU (HrcUC) and T25 fusions of HpaA, HpaA∆2–70 or HpaATrMmut were transferred to E. coli BTH101 cells as indicated. As controls, bacteria were cotransformed with T18 or T25 fusion constructs and the corresponding empty vectors (EV) encoding T18 or T25 as indicated. Bacterial cultures were grown on indicator plates and photographs were taken after 5 days. One representative colony per interaction is shown. Cotransformations were performed three times. At least three different transformants for every combination were analysed in every experiment with similar results. All proteins were stably synthesized as is shown in Supplementary Figure S4. (B) Mutations in the N-terminal region of HpaA promote the interaction with HrcUC and HrcQC. Interactions between T18 and T25 fusions containing HpaA, HrcUC and HrcQC as indicated were analysed in BTH101 cells. Bacterial cultures were grown on indicator plates and the results of the interaction assays were documented as described in (A). (C) An N-terminal HpaA deletion derivative interacts with HrcUC. BTH101 cells were cotransformed with expression constructs encoding T25, T25-HrcUC, T18 and T18 fusions of HpaA, HpaA∆2–70 or HpaATrMmut as indicated. Bacterial cultures were grown on indicator plates and the results of the interaction assays were documented as described in (A). (D) Interaction studies with HpaA and HrcUC. BTH101 cells were cotransformed with expression constructs encoding T25, T18 or corresponding fusions containing HrcUC and HpaA as indicated. Bacterial cultures were grown on indicator plates and the results of the interaction assays were documented as described in (A).
We also performed BACTH assays with HpaA derivatives deleted in amino acids 2–70 or mutated in the TrM. Both derivatives interacted with HrcL and HrcN, suggesting that the N-terminal region of HpaA is dispensable for the binding of HpaA to the ATPase and HrcL (Figure 7A). Unexpectedly, deletion of the N-terminal region or mutation of the TrM enhanced the interaction of HpaA with HrcQC and HrcUC whereas no interactions with T18 and T25 domains were detected (Figure 7A–C). This suggests that the N-terminal region including the TrM interferes with the efficient binding of HpaA to HrcQC and HrcUC. To our knowledge, a similar negative influence of a translocation signal on the interaction between a T3S substrate and components of the T3S system has not yet been described. Taken together, our data suggest that the TrM promotes the binding specificity of HpaA and might thus prevent a premature interaction of HpaA with components of the sorting platform.
Discussion
In the present study, we analysed T3S and translocation of HpaA, which is a regulator of the T3S chaperone HpaB and contributes to the activity of the T3S system (Lorenz et al., 2008a). Binding of HpaA presumably prevents an inhibitory effect of HpaB on T3S during the assembly of the T3S system and thus promotes the secretion of pilus and translocon proteins (Büttner et al., 2004; Lorenz et al., 2008a). According to this model, secretion and translocation of HpaA liberates HpaB and activates the HpaB-mediated translocation of T3Es (Lorenz et al., 2008a; Figure 8). Given the essential role of HpaA as regulator of HpaB, HpaA is likely the first protein to be translocated into the plant cell after assembly of the translocon in the plant plasma membrane. The in planta activity of HpaA remains to be investigated. As reported previously, HpaA contains an N-and C-terminal NLS and localizes to the plant cell nucleus when transiently expressed in Nicotiana benthamiana (Lorenz et al., 2008a). The contribution of HpaA to T3S, however, is likely attributed to its function as a regulator of HpaB in the bacterial cytosol because nuclear exclusion of HpaA did not interfere with its virulence function (Lorenz et al., 2008a).
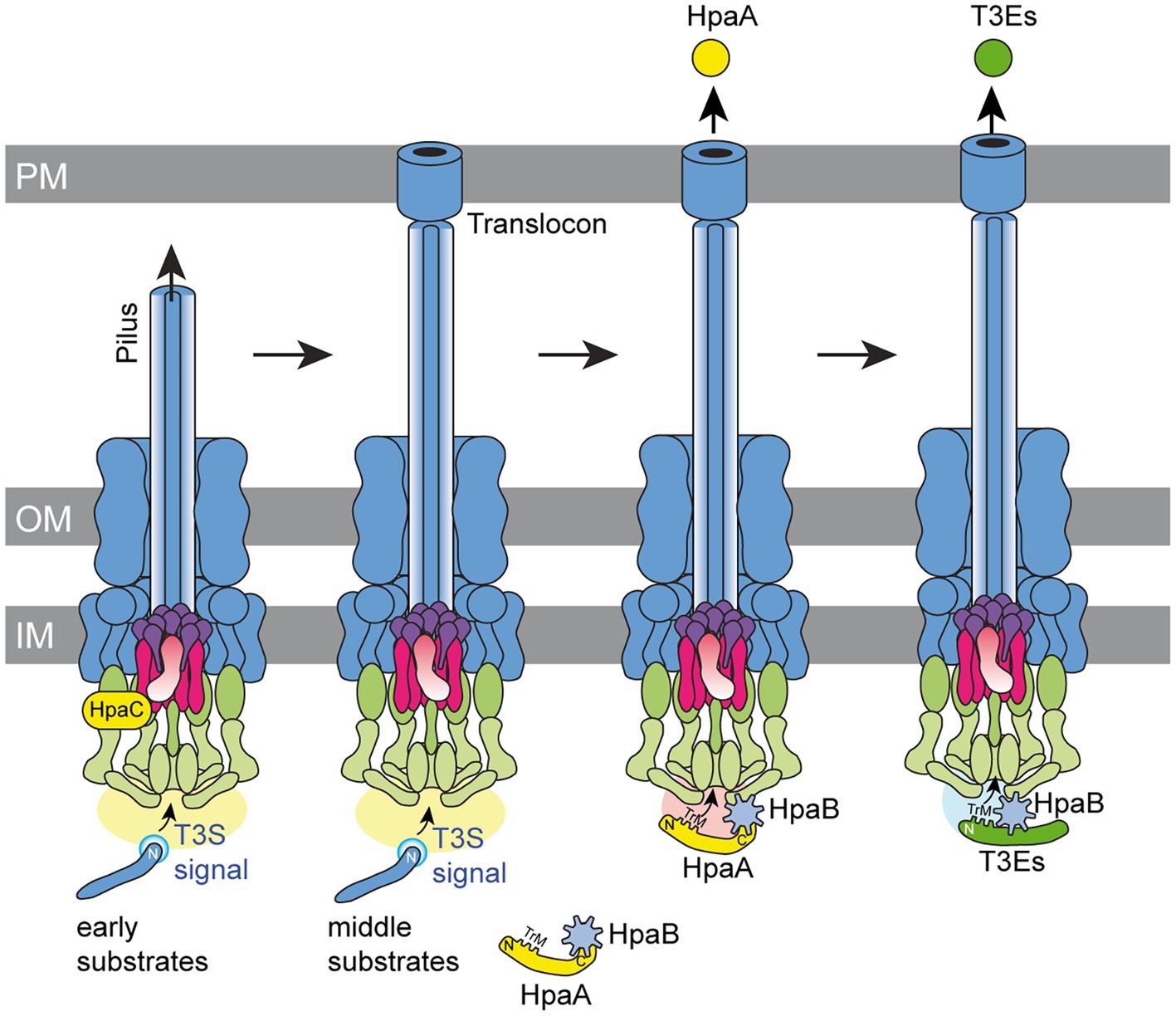
Figure 8. Model of the interplay between HpaA and HpaB during T3S in X. euvesicatoria. The T3S system assembles in both bacterial membranes and contains ring structures in the outer membrane (OM) and inner membrane (IM). The IM rings surround the export apparatus (shown in purple) and are connected to the cytoplasmic sorting platform (shown in green). The assembly and activation of the T3S system leads to the secretion of early substrates (i.e., the predicted inner rod protein HrpB2) and allows formation of the extracellular pilus. A switch in T3S substrate specificity induced by the T3S4 protein HpaC presumably involves a conformational change in the cytoplasmic domain of the export apparatus component HrcU (shown in pink) and activates the secretion of middle substrates such as components of the translocon in the plant plasma membrane (PM). The T3S chaperone HpaB is inactivated during assembly of the pilus and the translocon by the interaction with HpaA. A second predicted switch in T3S substrate specificity activates translocation of HpaA, which requires the N-terminal TrM for efficient export. Translocation of HpaA depends on HpaB, which binds to components of the T3S system and allows the specific recognition of the TrM, independently of its interaction with the C-terminal region of HpaA. Translocation of HpaA liberates HpaB and thus allows the translocation of T3Es, which also depend on the presence of a TrM for efficient recognition by the T3S system.
The regulatory function of HpaA is reminiscent of the activity of the gatekeeper protein YopN (Yersinia outer protein N) from Yersinia spp., which inhibits T3E delivery when present in the cytoplasm (Iriarte et al., 1998; Amer et al., 2016). Similarly to HpaA, YopN is translocated, yet, the function of YopN in host cells remains to be determined. In the bacterial cytoplasm, YopN acts in complex with the regulator TyeA and the SycN/YscB chaperones (Cheng et al., 2001). While binding of SycN/YscB chaperones to the N-terminal region of YopN is required for its entry into the T3S system, binding of TyeA to the C-terminal region of YopN inhibits its transport across the bacterial membranes (Cheng et al., 2001). Host cell contact leads to the dissociation of the YopN-TyeA complex and activates secretion of YopN (Cheng and Schneewind, 2000; Cheng et al., 2001; Amer et al., 2016). Interestingly, an additional secreted control protein, YscX, was identified in Yersinia pseudotuberculosis and likely inhibits secretion of inner rod, needle and effector proteins when present in the cytoplasm (Gurung et al., 2022). The striking similarities between T3S control mechanisms involving YopN and YscX in Yersinia spp. and HpaA in X. euvesicatoria suggest that animal-and plant-pathogenic bacteria use common strategies to activate T3E delivery after assembly of the T3S system.
In case of HpaA, we have not yet identified a negative regulator which prevents secretion of HpaA during the formation of the T3S system. We assume that delivery of HpaA is activated by a yet unknown signal after assembly of the pilus and the translocon (Figure 8). In agreement with this hypothesis, we show that translocation of HpaA depends on the HpaC-mediated T3S substrate specificity switch which promotes secretion of translocon and effector proteins after pilus formation (Lorenz et al., 2008b). The results of our protein–protein interaction studies suggest that HpaC interacts with HpaA, yet, the importance of this interaction remains to be investigated. It is possible that the interaction of HpaA with HpaC controls the HpaC-mediated T3S substrate specificity switch. Notably, however, the contribution of HpaC to HpaA transport was independent of a direct protein–protein interaction and likely caused by the conformational change in HrcUC which is induced by HpaC (Lorenz et al., 2008b; Lorenz and Büttner, 2011). We assume that the HpaC-mediated conformational change in HrcUC first promotes secretion of translocon proteins, which are essential for translocon formation and thus T3E delivery. HpaA translocation might be activated by an additional yet unknown signal after translocon formation. It remains to be investigated whether the membrane insertion of the translocon during the natural infection or a yet unknown trigger under in vitro conditions generates a signal that is transmitted to the base of the T3S system and activates HpaA export. Interestingly, in animal-pathogenic bacteria, host cell contact likely leads to the transmission of a signal through the extracellular needle to components of the secretion apparatus in the bacterial membranes and thus activates T3E delivery (Martinez-Argudo and Blocker, 2010; El Hajjami et al., 2018; Guo et al., 2019).
We previously reported that delivery of HpaA depends on a translocation signal which is located in the N-terminal 70 amino acids (Lorenz et al., 2008a). Here, translocation studies with HpaA-reporter fusions showed that the N-terminal TrM is essential for HpaA translocation. The TrM is not highly conserved in different T3Es but contains a certain amino acid composition enriched in arginine and proline residues (Escolar et al., 2001; Prochaska et al., 2018). In the T3E XopB from X. euvesicatoria, the TrM was described as membrane targeting motif required for the localization of XopB at the bacterial cell membranes (Prochaska et al., 2018). The contribution of the TrM to HpaB-dependent secretion and translocation of XopB was, therefore, proposed to result from a HpaB-dependent release of XopB from the membranes. According to this model, mutation of the TrM interferes with the membrane localization of XopB and thus facilitates its secretion and translocation in the absence of HpaB (Prochaska et al., 2018). In HpaA, however, the TrM does not confer membrane localization as was revealed by the analysis of a HpaA-sfGFP fusion by fluorescence microscopy. Furthermore, as reported previously, secretion and translocation of HpaA is independent of the presence of the HpaB-binding site, suggesting that the interaction with HpaB is dispensable for efficient HpaA delivery (Lorenz et al., 2008a). This finding indicates that HpaB is not required to target HpaA to the T3S system.
Interestingly, our data revealed that the TrM is essential for translocation but not for in vitro T3S of HpaA, suggesting that HpaA can be recognized by the T3S system in the absence of the TrM. However, we assume that TrM-independent secretion of HpaA into the extracellular milieu does not occur during the natural infection. Given that pilus and translocon likely form a continuous transport channel between bacterium and plant cell, the completed assembly of the T3S system would leave HpaA with no other option than to be translocated. In agreement with this hypothesis, the mutation of the TrM abolished the delivery of HpaA under natural conditions. The lack of translocation results in a loss of the in vivo function of HpaA because cytoplasmic HpaA presumably remains attached to HpaB and thus inhibits HpaB-mediated T3E delivery. Notably, mutation of the TrM enhanced translocation of HpaA in hpaB mutants, suggesting that the essential contribution of the TrM to HpaA translocation depends on the presence of HpaB. This observation is reminiscent of the previous finding that the N-terminal regions of T3Es, which lack the TrM, target the AvrBs3∆2 reporter for translocation in hpaB mutants but not in the wild-type strain (see Table 1). The signals, which allow translocation in the absence of HpaB, were previously referred to as minimal translocation signals because they are not sufficient to promote translocation in the wild-type strain (Büttner et al., 2004; Hartmann and Büttner, 2013; Scheibner et al., 2017, 2018). We speculate that HpaB generates a docking site for the TrM after assembly of the T3S system and thus restricts translocation to proteins containing a TrM.
HpaB was previously shown to interact with cytoplasmic components of the T3S system such as components of the predicted sorting platform and the export apparatus, and might thus promote the interaction of T3Es with the T3S system (Lorenz and Büttner, 2009; Lorenz et al., 2012; Drehkopf et al., 2020). It is, however, still unknown how T3S substrates are recognized. One potential substrate docking site is HrcUC which interacts with the N-terminal region of the putative inner rod protein HrpB2 (Lorenz and Büttner, 2011). Notably, however, HrcUC does not interact with T3Es or the putative translocon protein XopA when analysed in vitro, suggesting that it is not a general T3S substrate docking site (Lorenz and Büttner, 2011). In the present study, we identified HrcVC, HrcQC, HrcN and HrcL as potential docking sites for HpaA using in vitro and in vivo approaches. Furthermore, we show that HpaA and the ATPase HrcN are part of a common protein complex, suggesting that HpaA interacts with various components of the T3S system. Unexpectedly, the interaction of HpaA with HrcQC and HrcUC appeared to be enhanced upon mutation of the TrM. It is, therefore, possible that the TrM—rather than being essential for the docking of HpaA to components of the T3S system—prevents an unspecific premature interaction of HpaA with components of the T3S system. A similar contribution of the TrM to the interaction of other T3Es with T3S system components remains to be investigated. Our finding sheds new light on the contribution of export signals to T3S. In conclusion, we propose that the TrM targets T3Es for translocation by promoting their specific interaction with T3S system components in the presence of HpaB and possibly preventing a premature unspecific docking to the sorting platform. Notably, a negative influence of the N-terminal region of HpaA was not observed for the interaction of HpaA with HrcN, the ATPase of the T3S system. HrcN presumably forms a hexameric channel through which T3S substrates including HpaA are transported (Lorenz and Büttner, 2009; Büttner, 2012). A negative influence of the N-terminal region of HpaA on the interaction with HrcN would, therefore, generally interfere with HpaA export.
Taken together, our data suggest that the translocation of HpaA is activated after assembly of the T3S system and depends on the TrM and HpaB. The precise timing of HpaA export is presumably essential for efficient T3E translocation and thus for pathogenicity of X. euvesicatoria. We propose that the interaction of HpaB with components of the T3S system generates a docking site which restricts the transit through the secretion channel to T3S substrates with a TrM after assembly of the T3S system (Figure 8). The mechanisms which activate HpaA export and control HpaB-dependent recognition of HpaA by the T3S system remain a challenging subject for future studies. In depth analysis of protein–protein interactions between HpaA, T3S system components and HpaB will help to unravel the molecular mechanisms underlying HpaA translocation and thus HpaB-dependent effector protein delivery.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
SD, CO and DB designed and performed the experiments and analysed the data. DB wrote the manuscript with contributions from SD and CO. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the Deutsche Forschungsgemeinschaft (BU2145/9-2 and BU2145/10-1) to DB.
Acknowledgments
We thank M. Jordan for technical assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary materials
The Supplementary materials for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls. 2022.955776/full#supplementary-material
References
Akeda, Y., and Galan, J. E. (2005). Chaperone release and unfolding of substrates in type III secretion. Nature 437, 911–915. doi: 10.1038/nature03992
Amer, A. A., Gurung, J. M., Costa, T. R., Ruuth, K., Zavialov, A. V., Forsberg, A., et al. (2016). YopN and TyeA hydrophobic contacts required for regulating Ysc-Yop type III secretion activity by Yersinia pseudotuberculosis. Front. Cell. Infect. Microbiol. 6, 66. doi: 10.3389/fcimb.2016.00066
An, S. Q., Potnis, N., Dow, M., Vorhölter, F. J., He, Y. Q., Becker, A., et al. (2020). Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 44, 1–32. doi: 10.1093/femsre/fuz024
Battesti, A., and Bouveret, E. (2012). The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58, 325–334. doi: 10.1016/j.ymeth.2012.07.018
Berger, C., Ravelli, R. B. G., Lopez-Iglesias, C., Kudryashev, M., Diepold, A., and Peters, P. J. (2021). Structure of the Yersinia injectisome in intracellular host cell phagosomes revealed by cryo FIB electron tomography. J. Struct. Biol. 213:107701. doi: 10.1016/j.jsb.2021.107701
Boyd, A. P., Lambermont, I., and Cornelis, G. R. (2000). Competition between the Yops of Yersinia enterocolitica for delivery into eukaryotic cells: role of the SycE chaperone binding domain of YopE. J. Bacteriol. 182, 4811–4821. doi: 10.1128/JB.182.17.4811-4821.2000
Büttner, D. (2012). Protein export according to schedule – architecture, assembly and regulation of type III secretion systems from plant and animal pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. doi: 10.1128/MMBR.05017-11
Büttner, D. (2016). Behind the lines - actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev. 40, 894–937. doi: 10.1093/femsre/fuw026
Büttner, D., and Bonas, U. (2002). Getting across-bacterial type III effector proteins on their way to the plant cell. EMBO J. 21, 5313–5322. doi: 10.1093/emboj/cdf536
Büttner, D., Gürlebeck, D., Noel, L. D., and Bonas, U. (2004). HpaB from Xanthomonas campestris pv. vesicatoria acts as an exit control protein in type III-dependent protein secretion. Mol. Microbiol. 54, 755–768. doi: 10.1111/j.1365-2958.2004.04302.x
Büttner, D., Lorenz, C., Weber, E., and Bonas, U. (2006). Targeting of two effector protein classes to the type III secretion system by a HpaC-and HpaB-dependent protein complex from Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 59, 513–527. doi: 10.1111/j.1365-2958.2005.04924.x
Büttner, D., Nennstiel, D., Klüsener, B., and Bonas, U. (2002). Functional analysis of HrpF, a putative type III translocon protein from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 184, 2389–2398. doi: 10.1128/JB.184.9.2389-2398.2002
Cheng, L. W., Anderson, D. M., and Schneewind, O. (1997). Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24, 757–765. doi: 10.1046/j.1365-2958.1997.3831750.x
Cheng, L. W., Kay, O., and Schneewind, O. (2001). Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J. Bacteriol. 183, 5293–5301. doi: 10.1128/JB.183.18.5293-5301.2001
Cheng, L. W., and Schneewind, O. (2000). Yersinia enterocolitica TyeA, an intracellular regulator of the type III machinery, is required for specific targeting of YopE, YopH, YopM, and YopN into the cytosol of eukaryotic cells. J. Bacteriol. 182, 3183–3190. doi: 10.1128/JB.182.11.3183-3190.2000
Deng, W., Li, Y., Hardwidge, P. R., Frey, E. A., Pfuetzner, R. A., Lee, S., et al. (2005). Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect. Immun. 73, 2135–2146. doi: 10.1128/IAI.73.4.2135-2146.2005
Deng, W., Marshall, N. C., Rowland, J. L., McCoy, J. M., Worrall, L. J., Santos, A. S., et al. (2017). Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337. doi: 10.1038/nrmicro.2017.20
Dey, S., Chakravarty, A., Guha Biswas, P., and De Guzman, R. N. (2019). The type III secretion system needle, tip, and translocon. Protein Sci. 28, 1582–1593. doi: 10.1002/pro.3682
Dietsche, T., Tesfazgi Mebrhatu, M., Brunner, M. J., Abrusci, P., Yan, J., Franz-Wachtel, M., et al. (2016). Structural and functional characterization of the bacterial type III secretion export apparatus. PLoS Pathog. 12:e1006071. doi: 10.1371/journal.ppat.1006071
Drehkopf, S., Otten, C., Hausner, J., Seifert, T., and Büttner, D. (2020). HrpB7 from Xanthomonas campestris pv. vesicatoria is an essential component of the type III secretion system and shares features of HrpO/FliJ/YscO family members. Cell. Microbiol. 22:e13160. doi: 10.1111/cmi.13160
El Hajjami, N., Moussa, S., Houssa, J., Monteyne, D., Perez-Morga, D., and Botteaux, A. (2018). The inner-rod component of Shigella flexneri type 3 secretion system, MxiI, is involved in the transmission of the secretion activation signal by its interaction with MxiC. Microbiology 7, e00520. doi: 10.1002/mbo3.520
Engler, C., Gruetzner, R., Kandzia, R., and Marillonnet, S. (2009). Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4:e5553. doi: 10.1371/journal.pone.0005553
Engler, C., Kandzia, R., and Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One 3:e3647. doi: 10.1371/journal.pone.0003647
Escolar, L., Van den Ackerveken, G., Pieplow, S., Rossier, O., and Bonas, U. (2001). Type III secretion and in planta recognition of the Xanthomonas avirulence proteins AvrBs1 and AvrBsT. Mol. Plant Pathol. 2, 287–296. doi: 10.1046/j.1464-6722.2001.00077.x
Gan, Y. L., Yang, L. Y., Yang, L. C., Li, W. L., Liang, X. L., Jiang, W., et al. (2021). The C-terminal domain of the type III secretion chaperone HpaB contributes to dissociation of chaperone-effector complex in Xanthomonas campestris pv. campestris. PLoS One 16:e0246033. doi: 10.1371/journal.pone.0246033
Guo, E. Z., Desrosiers, D. C., Zalesak, J., Tolchard, J., Berbon, M., Habenstein, B., et al. (2019). A polymorphic helix of a Salmonella needle protein relays signals defining distinct steps in type III secretion. PLoS Biol. 17:e3000351. doi: 10.1371/journal.pbio.3000351
Gurung, J. M., Amer, A. A. A., Chen, S., Diepold, A., and Francis, M. S. (2022). Type III secretion by Yersinia pseudotuberculosis is reliant upon an authentic N-terminal YscX secretor domain. Mol. Microbiol. 117, 886–906. doi: 10.1111/mmi.14880
Hajra, D., Nair, A. V., and Chakravortty, D. (2021). An elegant nano-injection machinery for sabotaging the host: role of type III secretion system in virulence of different human and animal pathogenic bacteria. Phys Life Rev 38, 25–54. doi: 10.1016/j.plrev.2021.05.007
Hartmann, N., and Büttner, D. (2013). The inner membrane protein HrcV from Xanthomonas is involved in substrate docking during type III secretion. Mol. Plant-Microbe Interact. 26, 1176–1189. doi: 10.1094/MPMI-01-13-0019-R
Hartmann, N., Schulz, S., Lorenz, C., Fraas, S., Hause, G., and Büttner, D. (2012). Characterization of HrpB2 from Xanthomonas campestris pv. vesicatoria identifies protein regions that are essential for type III secretion pilus formation. Microbiology 158, 1334–1349. doi: 10.1099/mic.0.057604-0
Hausner, J., Hartmann, N., Jordan, M., and Büttner, D. (2017). The predicted lytic transglycosylase HpaH from Xanthomonas campestris pv. vesicatoria associates with the type III secretion system and promotes effector protein translocation. Infect Immunol. 85, e00788–e00716. doi: 10.1128/IAI.00788-16
Hausner, J., Jordan, M., Otten, C., Marillonnet, S., and Büttner, D. (2019). Modular cloning of the type III secretion gene cluster from the plant-pathogenic bacterium Xanthomonas euvesicatoria. ACS Synth. Biol. 8, 532–547. doi: 10.1021/acssynbio.8b00434
Hu, B., Lara-Tejero, M., Kong, Q., Galan, J. E., and Liu, J. (2017). In situ molecular architecture of the Salmonella type III secretion machine. Cell 168:e10, 1065–1074.e10. doi: 10.1016/j.cell.2017.02.022
Hu, B., Morado, D. R., Margolin, W., Rohde, J. R., Arizmendi, O., Picking, W. L., et al. (2015). Visualization of the type III secretion sorting platform of Shigella flexneri. Proc. Natl. Acad. Sci. U. S. A. 112, 1047–1052. doi: 10.1073/pnas.1411610112
Hu, J., Worrall, L. J., Hong, C., Vuckovic, M., Atkinson, C. E., Caveney, N., et al. (2018). Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat. Commun. 9, 3840. doi: 10.1038/s41467-018-06298-8
Hu, J., Worrall, L. J., Vuckovic, M., Hong, C., Deng, W., Atkinson, C. E., et al. (2019). T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly. Nat. Microbiol. 4, 2010–2019. doi: 10.1038/s41564-019-0545-z
Hueck, C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. doi: 10.1128/MMBR.62.2.379-433.1998
Huguet, E., Hahn, K., Wengelnik, K., and Bonas, U. (1998). hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol. 29, 1379–1390. doi: 10.1046/j.1365-2958.1998.01019.x
Iriarte, M., Sory, M. P., Boland, A., Boyd, A. P., Mills, S. D., Lambermont, I., et al. (1998). TyeA, a protein involved in control of Yop release and in translocationof Yersinia Yop effectors. EMBO J. 17, 1907–1918. doi: 10.1093/emboj/17.7.1907
Ji, H., and Dong, H. (2015). Key steps in type III secretion system (T3SS) towards translocon assembly with potential sensor at plant plasma membrane. Mol. Plant Pathol. 16, 762–773. doi: 10.1111/mpp.12223
Johnson, S., Kuhlen, L., Deme, J. C., Abrusci, P., and Lea, S. M. (2019). The structure of an Injectisome export gate demonstrates conservation of architecture in the core export gate between Flagellar and virulence type III secretion systems. mBio 10, 10. doi: 10.1128/mBio.00818-19
Karimova, G., Pidoux, J., Ullmann, A., and Ladant, D. (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95, 5752–5756. doi: 10.1073/pnas.95.10.5752
Khan, M., Seto, D., Subramaniam, R., and Desveaux, D. (2018). Oh, the places they'll go! A survey of phytopathogen effectors and their host targets. Plant J. 93, 651–663. doi: 10.1111/tpj.13780
Khan, M., Subramaniam, R., and Desveaux, D. (2016). Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr. Opin. Microbiol. 29, 49–55. doi: 10.1016/j.mib.2015.10.006
Kuhlen, L., Abrusci, P., Johnson, S., Gault, J., Deme, J., Caesar, J., et al. (2018). Structure of the core of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 25, 583–590. doi: 10.1038/s41594-018-0086-9
Lara-Tejero, M. (2019). The type III secretion system sorting platform. Curr. Top. Microbiol. Immunol. 427, 133–142. doi: 10.1007/82_2019_167
Lara-Tejero, M., and Galan, J. E. (2019). The injectisome, a complex nanomachine for protein injection into mammalian cells. EcoSal Plus 8, 245–259. doi: 10.1128/ecosalplus.ESP-0039-2018
Lara-Tejero, M., Kato, J., Wagner, S., Liu, X., and Galan, J. E. (2011). A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331, 1188–1191. doi: 10.1126/science.1201476
Lonjon, F., Lohou, D., Cazale, A. C., Büttner, D., Ribeiro, B. G., Peanne, C., et al. (2017). HpaB-dependent secretion of type III effectors in the plant pathogens Ralstonia solanacearum and Xanthomonas campestris pv. vesicatoria. Sci. Rep. 7, 4879. doi: 10.1038/s41598-017-04853-9
Lorenz, C., and Büttner, D. (2009). Functional characterization of the type III secretion ATPase HrcN from the plant pathogen Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 191, 1414–1428. doi: 10.1128/JB.01446-08
Lorenz, C., and Büttner, D. (2011). Secretion of early and late substrates of the type III secretion system from Xanthomonas is controlled by HpaC and the C-terminal domain of HrcU. Mol. Microbiol. 79, 447–467. doi: 10.1111/j.1365-2958.2010.07461.x
Lorenz, C., Hausner, J., and Büttner, D. (2012). HrcQ provides a docking site for early and late type III secretion substrates from Xanthomonas. PLoS One 7:e51063. doi: 10.1371/journal.pone.0051063
Lorenz, C., Kirchner, O., Egler, M., Stuttmann, J., Bonas, U., and Büttner, D. (2008a). HpaA from Xanthomonas is a regulator of type III secretion. Mol. Microbiol. 69, 344–360. doi: 10.1111/j.1365-2958.2008.06280.x
Lorenz, C., Schulz, S., Wolsch, T., Rossier, O., Bonas, U., and Büttner, D. (2008b). HpaC controls substrate specificity of the Xanthomonas type III secretion system. PLoS Pathog. 4:e1000094. doi: 10.1371/journal.ppat.1000094
Löwer, M., and Schneider, G. (2009). Prediction of type III secretion signals in genomes of gram-negative bacteria. PLoS One 4:e5917. doi: 10.1371/journal.pone.0005917
Lunelli, M., Kamprad, A., Burger, J., Mielke, T., Spahn, C. M. T., and Kolbe, M. (2020). Cryo-EM structure of the Shigella type III needle complex. PLoS Pathog. 16:e1008263. doi: 10.1371/journal.ppat.1008263
Martinez-Argudo, I., and Blocker, A. J. (2010). The Shigella T3SS needle transmits a signal for MxiC release, which controls secretion of effectors. Mol. Microbiol. 78, 1365–1378. doi: 10.1111/j.1365-2958.2010.07413.x
Meiresonne, N. Y., Consoli, E., Mertens, L. M. Y., Chertkova, A. O., Goedhart, J., and den Blaauwen, T. (2019). Superfolder mTurquoise2(ox) optimized for the bacterial periplasm allows high efficiency in vivo FRET of cell division antibiotic targets. Mol. Microbiol. 111, 1025–1038. doi: 10.1111/mmi.14206
Meiresonne, N. Y., van der Ploeg, R., Hink, M. A., and den Blaauwen, T. (2017). Activity-related conformational changes ind,d-Carboxypeptidases revealed by in vivo periplasmic Förster resonance energy transfer assay in Escherichia coli. mBio 8, 8. doi: 10.1128/mBio.01089-17
Milne-Davies, B., Wimmi, S., and Diepold, A. (2021). Adaptivity and dynamics in type III secretion systems. Mol. Microbiol. 115, 395–411. doi: 10.1111/mmi.14658
Ngou, B. P. M., Jones, J. D. G., and Ding, P. (2022). Plant immune networks. Trends Plant Sci. 27, 255–273. doi: 10.1016/j.tplants.2021.08.012
Noël, L., Thieme, F., Gäbler, J., Büttner, D., and Bonas, U. (2003). XopC and XopJ, two novel type III effector proteins from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 185, 7092–7102. doi: 10.1128/JB.185.24.7092-7102.2003
Noël, L., Thieme, F., Nennstiel, D., and Bonas, U. (2002). Two novel type III system-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184, 1340–1348. doi: 10.1128/JB.184.5.1340-1348.2002
Otten, C., Seifert, T., Hausner, J., and Büttner, D. (2021). The contribution of the predicted sorting platform component HrcQ to type III secretion in Xanthomonas campestris pv. vesicatoria depends on an internal translation start site. Front. Microbiol. 12:752733. doi: 10.3389/fmicb.2021.752733
Portaliou, A. G., Tsolis, K. C., Loos, M. S., Balabanidou, V., Rayo, J., Tsirigotaki, A., et al. (2017). Hierarchical protein targeting and secretion is controlled by an affinity switch in the type III secretion system of enteropathogenic Escherichia coli. EMBO J. 36, 3517–3531. doi: 10.15252/embj.201797515
Potnis, N., Timilsina, S., Strayer, A., Shantharaj, D., Barak, J. D., Paret, M. L., et al. (2015). Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant Pathol. 16, 907–920. doi: 10.1111/mpp.12244
Prochaska, H., Thieme, S., Daum, S., Grau, J., Schmidtke, C., Hallensleben, M., et al. (2018). A conserved motif promotes HpaB-regulated export of type III effectors from Xanthomonas. Mol. Plant Pathol. 19, 2473–2487. doi: 10.1111/mpp.12725
Rossier, O., Van den Ackerveken, G., and Bonas, U. (2000). HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38, 828–838. doi: 10.1046/j.1365-2958.2000.02173.x
Rossier, O., Wengelnik, K., Hahn, K., and Bonas, U. (1999). The Xanthomonas Hrp type III system secretes proteins from plant and mammalian pathogens. Proc. Natl. Acad. Sci. U. S. A. 96, 9368–9373. doi: 10.1073/pnas.96.16.9368
Rüssmann, H., Kubori, T., Sauer, J., and Galan, J. E. (2002). Molecular and functional analysis of the type III secretion signal of the Salmonella enterica InvJ protein. Mol. Microbiol. 46, 769–779. doi: 10.1046/j.1365-2958.2002.03196.x
Sal-Man, N., Deng, W., and Finlay, B. B. (2012). EscI: a crucial component of the type III secretion system forms the inner rod structure in enteropathogenic Escherichia coli. Biochem. J. 442, 119–125. doi: 10.1042/BJ20111620
Samudrala, R., Heffron, F., and McDermott, J. E. (2009). Accurate prediction of secreted substrates and identification of a conserved putative secretion signal for type III secretion systems. PLoS Pathog. 5:e1000375. doi: 10.1371/journal.ppat.1000375
Schechter, L. M., Roberts, K. A., Jamir, Y., Alfano, J. R., and Collmer, A. (2004). Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186, 543–555. doi: 10.1128/JB.186.2.543-555.2004
Scheibner, F., Hartmann, N., Hausner, J., Lorenz, C., Hoffmeister, A. K., and Büttner, D. (2018). The type III secretion chaperone HpaB controls the translocation of effector and non-effector proteins from Xanthomonas campestris pv. vesicatoria. Mol. Plant-Microbe Interact. 31, 61–74. doi: 10.1094/MPMI-06-17-0138-R
Scheibner, F., Marillonnet, S., and Büttner, D. (2017). The TAL effector AvrBs3 from Xanthomonas campestris pv. vesicatoria contains multiple export signals and can enter plant cells in the absence of the type III secretion translocon. Front. Microbiol. 8, 2180. doi: 10.3389/fmicb.2017.02180
Scheibner, F., Schulz, S., Hausner, J., Marillonnet, S., and Büttner, D. (2016). Type III-dependent translocation of HrpB2 by a non-pathogenic hpaABC mutant of the plant-pathogenic bacterium Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 82, 3331–3347. doi: 10.1128/AEM.00537-16
Schesser, K., Frithz-Lindsten, E., and Wolf-Watz, H. (1996). Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178, 7227–7233. doi: 10.1128/jb.178.24.7227-7233.1996
Schreiber, K. J., Chau-Ly, I. J., and Lewis, J. D. (2021). What the wild things do: mechanisms of plant host manipulation by bacterial type III-secreted effector proteins. Microorganisms 9:1029. doi: 10.3390/microorganisms9051029
Schulz, S., and Büttner, D. (2011). Functional characterization of the type III secretion substrate specificity switch protein HpaC from Xanthomonas. Infect. Immun. 79, 2998–3011. doi: 10.1128/IAI.00180-11
Schulze, S., Kay, S., Büttner, D., Egler, M., Eschen-Lippold, L., Hause, G., et al. (2012). Analyses of new type III effectors from Xanthomonas uncover XopB and XopS as suppressors of plant immunity. New Phytol. 195, 894–911. doi: 10.1111/j.1469-8137.2012.04210.x
Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N., Palmer, A. E., and Tsien, R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572. doi: 10.1038/nbt1037
Silva, Y. R. O., Contreras-Martel, C., Macheboeuf, P., and Dessen, A. (2020). Bacterial secretins: mechanisms of assembly and membrane targeting. Protein Sci. 29, 893–904. doi: 10.1002/pro.3835
Sory, M. P., Boland, A., Lambermont, I., and Cornelis, G. R. (1995). Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. U. S. A. 92, 11998–12002. doi: 10.1073/pnas.92.26.11998
Spaeth, K. E., Chen, Y. S., and Valdivia, R. H. (2009). The Chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog. 5:e1000579. doi: 10.1371/journal.ppat.1000579
Stebbins, C. E., and Galan, J. E. (2001). Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature 414, 77–81. doi: 10.1038/35102073
Szurek, B., Rossier, O., Hause, G., and Bonas, U. (2002). Type III-dependent translocation of the Xanthomonas AvrBs3 protein into the plant cell. Mol. Microbiol. 46, 13–23. doi: 10.1046/j.1365-2958.2002.03139.x
Timilsina, S., Potnis, N., Newberry, E. A., Liyanapathiranage, P., Iruegas-Bocardo, F., White, F. F., et al. (2020). Xanthomonas diversity, virulence and plant-pathogen interactions. Nat. Rev. Microbiol. 18, 415–427. doi: 10.1038/s41579-020-0361-8
Torres-Vargas, C. E., Kronenberger, T., Roos, N., Dietsche, T., Poso, A., and Wagner, S. (2019). The inner rod of virulence-associated type III secretion systems constitutes a needle adapter of one helical turn that is deeply integrated into the system's export apparatus. Mol. Microbiol. 112, 918–931. doi: 10.1111/mmi.14327
Wagner, S., and Diepold, A. (2020). A unified nomenclature for injectisome-type type III secretion systems. Curr. Top. Microbiol. Immunol. 427, 1–10. doi: 10.1007/82_2020_210
Wagner, S., Grin, I., Malmsheimer, S., Singh, N., Torres-Vargas, C. E., and Westerhausen, S. (2018). Bacterial type III secretion systems: a complex device for the delivery of bacterial effector proteins into eukaryotic host cells. FEMS Microbiol. Lett. 365, fny201. doi: 10.1093/femsle/fny201
Wang, Y., Sun, M., Bao, H., and White, A. P. (2013). T3_MM: a Markov model effectively classifies bacterial type III secretion signals. PLoS One 8:e58173. doi: 10.1371/journal.pone.0058173
Weber, E., Engler, C., Gruetzner, R., Werner, S., and Marillonnet, S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS One 6:e16765. doi: 10.1371/journal.pone.0016765
Wengelnik, K., Marie, C., Russel, M., and Bonas, U. (1996). Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J. Bacteriol. 178, 1061–1069. doi: 10.1128/jb.178.4.1061-1069.1996
Wengelnik, K., Rossier, O., and Bonas, U. (1999). Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 181, 6828–6831. doi: 10.1128/JB.181.21.6828-6831.1999
Winnen, B., Schlumberger, M. C., Sturm, A., Schupbach, K., Siebenmann, S., Jenny, P., et al. (2008). Hierarchical effector protein transport by the Salmonella Typhimurium SPI-1 type III secretion system. PLoS One 3:e2178. doi: 10.1371/journal.pone.0002178
Worrall, L. J., Hong, C., Vuckovic, M., Deng, W., Bergeron, J. R. C., Majewski, D. D., et al. (2016). Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature 540, 597–601. doi: 10.1038/nature20576
Xing, Q., Shi, K., Portaliou, A., Rossi, P., Economou, A., and Kalodimos, C. G. (2018). Structures of chaperone-substrate complexes docked onto the export gate in a type III secretion system. Nat. Commun. 9, 1773. doi: 10.1038/s41467-018-04137-4
Zilkenat, S., Franz-Wachtel, M., Stierhof, Y. D., Galan, J. E., Macek, B., and Wagner, S. (2016). Determination of the stoichiometry of the complete bacterial type III secretion needle complex using a combined quantitative proteomic approach. Mol. Cell. Proteomics 15, 1598–1609. doi: 10.1074/mcp.M115.056598
Keywords: type III secretion, plant pathogen, chaperone, translocation signal, sorting platform, gatekeeper
Citation: Drehkopf S, Otten C and Büttner D (2022) Recognition of a translocation motif in the regulator HpaA from Xanthomonas euvesicatoria is controlled by the type III secretion chaperone HpaB. Front. Plant Sci. 13:955776. doi: 10.3389/fpls.2022.955776
Edited by:
Vladimir Orbovic, University of Florida, United StatesReviewed by:
Doron Teper, Agricultural Research Organization (ARO), IsraelSang-Wook Han, Chung-Ang University, South Korea
Copyright © 2022 Drehkopf, Otten and Büttner. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniela Büttner, ZGFuaWVsYS5idWV0dG5lckBnZW5ldGlrLnVuaS1oYWxsZS5kZQ==
 Sabine Drehkopf
Sabine Drehkopf Christian Otten
Christian Otten Daniela Büttner
Daniela Büttner