
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 September 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.955199
This article is part of the Research Topic Adaptation Mechanisms of Grass and Forage Plants to Stressful Environments View all 42 articles
The homeodomain-leucine zipper (HD-Zip) family participates in plant growth, development, and stress responses. Here, 40 HD-Zip transcription factors of Lotus japonicus were identified and gave an overview of the phylogeny and gene structures. The expression pattern of these candidate genes was determined in different organs and their response to abiotic stresses, including cold, heat, polyethylene glycol and salinity. The expression of the LjHDZ7 was strongly induced by abiotic stress, especially salt stress. Subsequently, LjHDZ7 gene was overexpressed in Arabidopsis. The transgenic plants grew obviously better than Col-0 plants under salt stress. Furthermore, LjHDZ7 transgenic lines accumulated higher proline contents and showed lower electrolyte leakage and MDA contents than Col-0 plants under salt stress. Antioxidant activities of the LjHDZ7 overexpression lines leaf were significantly higher than those of the Col-0 plants under salt stress. The concentration of Na+ ion in LjHDZ7 overexpression lines was significantly lower than that of Col-0 in leaf and root parts. The concentration of K+ ion in LjHDZ7 overexpression lines was significantly higher than that of Col-0 in the leaf parts. Therefore, these results showed that overexpression of LjHDZ7 increased resistance to salt stress in transgenic Arabidopsis plants, and certain genes of this family can be used as valuable tools for improving abiotic stresses.
Homeodomain-leucine zipper (HD-Zip) contains a conserved homology domain (HD) motif and a leucine zipper (LZ) motif (Ariel et al., 2007). HD-Zip is a unique transcription factor (TF) that is present in a variety of plants (Henriksson et al., 2005; Sakuma et al., 2013). HD-Zip is involved in various regulatory processes of plant growth and development and responses to various external environmental signals (Wang et al., 2013; Ribone et al., 2015). HD-Zip is expressed in various tissues and organs of plants and plays an important role in different growth stages (Brant et al., 2014). Excluding the conserved HD-Zip domain, these family members have different amino acid sequence lengths and protein structures (Arce et al., 2011).
The HD-Zip gene family can be divided into four subfamilies, I to IV, according to the amino acid sequence (Capella et al., 2015). Each subfamily member has a unique function and forms a complex interaction network. HD-Zip I positively responds to various environmental stresses, such as drought, salty, pathogens, chilling injury, light signal response and hormone response (Romani et al., 2016). The expression of HD-Zip II genes is affected by light. The TFs of this family play important roles in the development of plant organs, shade avoidance response and hormone response (Turchi et al., 2015; Carabelli et al., 2018; He et al., 2021). HD-Zip III has obvious effects on the development of plant embryos, the formation of microtubule tissue, the differentiation of apical provinces and the polar transport of auxin (Yang S. et al., 2018). HD-Zip IV promotes the differentiation of epidermal cells in plant organs and the formation of anthocyanins (Nakamura et al., 2006; Zalewski et al., 2013).
Plants often face various biological and abiotic stresses during growth and development. Under various stresses, plants form a complex signal transduction network to resist adversity. The HD-Zip gene family has been reported to participate in adversity stress in many species (Zhang J. et al., 2020). HD-Zip genes play a crucial role in plant protection against pathogens and abiotic stresses (Gao et al., 2015). Overexpression of the wheat TaHDZipI-5 gene improved transgenic wheat freezing and drought resistance (Yang et al., 2017). Overexpression of PsnHDZ63 could increase reactive oxygen species scavenging ability and enhanced salt stress tolerance in transgenic Populus simonii × P. nigra (Guo et al., 2021). Overexpression of the rice HD-Zip I gene OsHOX24 imparted higher sensitivity to stress hormones, ABA, and abiotic stresses in transgenic Arabidopsis plants than in wild-type plants (Annapurna et al., 2016; Tang et al., 2019). These studies showed that the HD-Zip gene family could play an important role in plant resistance to abiotic stress.
Homeodomain-leucine zipper genes from model plants to higher plants have been identified and analyzed. For example, the model plants Arabidopsis thaliana (Nakamura et al., 2006), rice (Oryza sativa L.) (Zhang et al., 2012), maize (Zea mays L., Gramineae family), moso bamboo (Phyllostachys edulis) (Gao et al., 2021), potato (Solanum tuberosum L., Solanaceae family) (Li et al., 2019), and pear (Pyrus pyrifolia, rose family) (Yang Q. et al., 2018). Many families and genera of the HD-Zip gene family in these species have been identified and analyzed (Yang S. et al., 2022). The functions of a number of HD-Zip genes have been studied (Zhao et al., 2014), and the regulatory mechanisms of certain HD-Zip genes have been clarified (Zhang et al., 2012). However, the evolution of these genes in specific species and groups remains elusive; moreover, limited information is available on the HD-Zip family in Lotus japonicus.
Lotus japonicus is an important leguminous forage that is used as a protein feed source, biological nitrogen fixation resource and ecological conservation species. In this study, we identified 40 HD-Zip TFs of Lotus japonicus. To provide insights on HD-Zip gene evolution and function, we performed analyses of the phylogeny, proteins, gene structure, promoter cis-elements, and tissue and stress expression patterns. We found that the expression of LjHDZ7 was strongly induced by salt stress. Further studies showed that overexpression of LjHDZ7 increased the salt resistance of transgenic Arabidopsis plants. Our results will be helpful in understanding the evolutionary relationships, proteins, gene structures, and biological functions of HD-Zip TFs in Lotus japonicus and will provide a foundation for further elucidating the salt resistance mechanism of the LjHDZ7 gene.
The protein sequences of the Lotus japonicus, Medicago truncatula, Trifolium pretense, and Arabidopsis thaliana HD-Zip family were downloaded from the plant TFDB V4.0 database1. The candidate protein sequences were screened and identified using the CDD2 and SMART3, and manual elimination was performed to remove redundant sequences and sequences without typical domains. The HD-Zip family was finally determined by the protein sequence of the conserved domain.
To investigate the phylogenetic relationships of the HD-Zip members among different plants, multiple HD-Zip amino acid sequences were aligned via Clustal 2.1 software using the default settings to examine the evolutionary relationships among the sequences. A phylogenetic tree was constructed with MEGA7.0 software with 1000 bootstrap replicates according to the maximum-likelihood (ML) method (Li et al., 2022).
The Lotus japonicus HD-Zip protein structural characteristics, isoelectric point (pI), molecular weight (MW) and Grand average of hydropathicity (GRAVY) values were determined using the ExPASy ProtParam tool4. The subcellular localization of HD-Zip genes was predicted using the online tools UniProt5 and WoLF PSORT6.
Conserved protein motifs of the HD-Zip family of Lotus japonicus were predicted using MEME Suite7 with the default settings. The details of the top 10 predicted motifs were obtained from the MEME suite. The conserved domains of the HD-Zip family of Lotus japonicus were predicted using NCBI-CDD (see text footnote 2). The distribution of the conserved domain and motif were drawn via the visualization tool in TBtools software. The three-dimensional structure (3D) of LjHD-Zip proteins was predicted in the Phyre28 using the default advanced settings.
The DNA sequences of the HD-Zip gene family of Lotus japonicus were obtained from an online transcriptome and genome database9. These gene structures were displayed by server GSDS2.010, and the intron/exon distribution pattern of each LjHD-Zip is illustrated. These gene chromosome localizations were illustrated using MG2C_V2.111.
The 2000 bp upstream sequence of the initiation codon of each LjHD-Zip gene was extracted from the corresponding scaffolds (see text footnote 10). Then, the cis-elements in the promoters of each LjHD-Zip gene were predicted using the PlantCARE server12. The distribution of the cis-elements in the promoter was drawn via the visualization tool for cis-elements in TBtools software.
Lotus japonicus ecotype “MG20” was used in this study. We selected seeds with full grains, planted them in pots with perlite, grew them in a controlled environment with a photoperiod of 16 h/8 h (light/dark) at 24°C and 70% relative humidity in a growth chamber for 60 days, and then started the treatment. For the drought treatment, we removed the original culture solution and then watered the solution with 15% polyethylene glycol (PEG) to simulate drought stress. The leaf sample material was measured at 0, 3, 6, 12, and 24 h. For the salt treatment, the original culture solution was removed and the solution was watered with 250 mM NaCl. The leaf sample material was measured at 0, 3, 6, 12, and 24 h. For the low-temperature treatment, the seedlings were transported to an incubator at 4°C and the leaf sample material was measured at 0, 3, 6, 12, and 24 h. For the high-temperature treatment, the seedlings were transported to an incubator at 42°C and the leaf sample material was measured at 0, 3, 6, 12, and 24 h. Each treatment included three replicates. Each replicate included 10 plants.
Seed germination in the salt stress treatment was evaluated by cultivating seeds of the transgenic Arabidopsis line and Col-0 were grown on MS medium containing 150 mM NaCl for 7 days. Leaf growth and root length were assessed by cultivating the seeds of Col-0 and overexpression LjHDZ7 transgenic Arabidopsis in MS medium for 7 days, transferring the seedlings to vertical plate culture with MS medium containing 150 mM NaCl, and then cultivating the seedlings for 10 days before investigation. For the salt stress treatments, seeds of Col-0 and overexpression LjHDZ7 transgenic Arabidopsis were grown in MS medium for 7 days, transplanted into nutrient soil, grown for 35 days, and watered with 300 mM NaCl. Col-0 and transgenic Arabidopsis shoot were harvested and oven dried at 80°C for 24 h to obtain dry weights (DW) (Wu et al., 2018).
Total RNA was extracted from leaves using plant isolation kits (Sangon Biotech, Shanghai, China, Cat. #B518631). cDNA (complementary DNA) was prepared from total RNA with random primers using cDNA synthesis kits (Vazyme, Nanjing, China, Cat. #R312-02) (Nie et al., 2022). The resulting cDNA was used as a template for qPCR (quantitative real-time PCR) analysis using SYBR Green real-time PCR master mix (Vazyme). The qPCR assays were performed in triplicate using a real-time PCR system (Bio-Rad, Hercules, CA, United States) based on the manufacturer’s instructions (Wang et al., 2022b). The primers used for the qRT–PCR sets are listed in Supplementary Table 7. Lotus japonicus LjUbi (Lotus ubiquitin gene) was used as a reference gene, and then expression analyses with different treatment times were performed to determine the gene expression under different stress treatments, with 0 h used as a reference. The relative gene expression was quantified using the 2–△ △ CT method (Livak and Schmittgen, 2001). The PCR program was as follows: 1 cycle of 95°C for 30 s, followed by 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72° for 30 s (Wang et al., 2022a). The resulting clusters were visualized with MeV software.
The protein interaction network was analyzed and predicted using STRING software13, using Arabidopsis as the background and the default advanced settings. Functional enrichment analysis was performed using STRING.
Electrolyte leakage was determined by using the conductivity meter method (Su et al., 2015). The leaves were cut into pieces and placed in deionized water in the dark for 12 h, and then the electrical conductivity (EC1) was measured. The sample leaves were boiled for 30 min in water, and after cooling to room temperature, the electrical conductivity (EC2) was measured. Leakage was calculated by the formula:
Relative electrolyte leakage (%) = EC1/EC2 × 100%, where EC1 and EC2 refer to electric conductivity of live leaves and boiled leaves, respectively.
The malondialdehyde (MDA) contents were determined using the thiobarbituric acid reaction method (He et al., 2018). Fresh leaf samples were powdered in liquid nitrogen and homogenized with trichloroacetic acid (TCA). The supernatant was centrifuged and extracted and then mixed with thiobarbituric acid (TBA). The mixture was boiled and centrifuged, and then the absorbance was measured at wavelengths of 450, 532, and 600 nm. The contents were calculated by the formula:
Malondialdehyde concentration (C) (μmol/L) = 6.45 × (OD532 nm – OD600 nm) – 0.56 OD450 nm. OD450, OD532, and OD600 represented the absorbance value at 450, 532, and 600 nm, respectively. MDA content (μmol/g) = CV/1000 W. C: MDA concentration; V, volume extract; W, fresh weight.
The proline content was determined using the acidic ninhydrin reaction method (Bai et al., 2017). Fresh leaf samples were powdered in liquid nitrogen, homogenized in sulfosalicylic acid and heated in boiling water. The samples were centrifuged, and the supernatant was mixed with acidic ninhydrin and glacial acetic acid. After cooling, the samples were mixed with toluene, the supernatant was collected, and then the absorbance was determined at 520 nm. The content was calculated by the formula:
Proline contents (μg/g) = (C × VT/VS)/W. C: The standard curve for absorbance value at 520 nm (A520), A520 = 0.0512C + 0.001, R2 = 0.9955; VT, volume extract; VS, supernatant volume; W, fresh weight.
Superoxide dismutase (SOD), peroxidase (POD) and Catalase (CAT) activity was determined following the protocols proposed by the corresponding assay kit (Geruisi Biotechnology, Suzhou, China), respectively. The homogenate mixed with 0.1 g fresh sample and 1 mL extraction solution was centrifuged at 8,000 g for 10 min at 4°C, after which the supernatant was used to assay the activity of antioxidant enzymes. One unit of SOD was defined as the amount of enzyme that caused 50% of NBT photochemical reduction at 450 nm. One unit of POD was the change of 0.01 in a 1 mL reaction mixture at 470 nm per minute and per 1 g tissue. One unit of CAT was the 1 nmol H2O2 degradation at 510 nm per minute and per 1 mg tissue (Dong et al., 2021; Lü et al., 2022; Zhang et al., 2022).
The leaves and roots from Col-0 and LjHDZ7 overexpression plants were separately harvested, dried for 48 h at 80°C and then ground to powder. The same mass leaves and roots powder was extracted in concentrated (69%, v/v) HNO3 for 24 h at room temperature (Li et al., 2016). Na+ and K+ concentrations were determined by using a flame photometer (Sherwood flame photometer-410, Cambridge, United Kingdom).
The data in this paper were analyzed using SPSS version 17.0 and the least significant difference (LSD) test. The three biological replicates were performed. Mean and standard error values were calculated for the variable comparisons. Values of p < 0.05 were considered statistically significant.
A phylogenetic tree was constructed using full-length protein sequences obtained for Lotus japonicus, Medicago truncatula, Trifolium pretense, and Arabidopsis thaliana. HD-Zip genes were divided into four subfamilies (Figure 1). The number of HD-Zip genes in subgroup I was the largest (Supplementary Figure 1A), and the subfamily genes accounted for 35.42, 32.69, 36.96, and 32.50% of the whole family in Arabidopsis thaliana, Medicago truncatula, Trifolium pretense, and Lotus japonicus, respectively (Supplementary Figure 2). Longer amino acid sequences were obtained for HD-Zip III and HD-Zip IV, while shorter sequences were obtained for HD-Zip I and HD-Zip II (Supplementary Figures 1B–E).
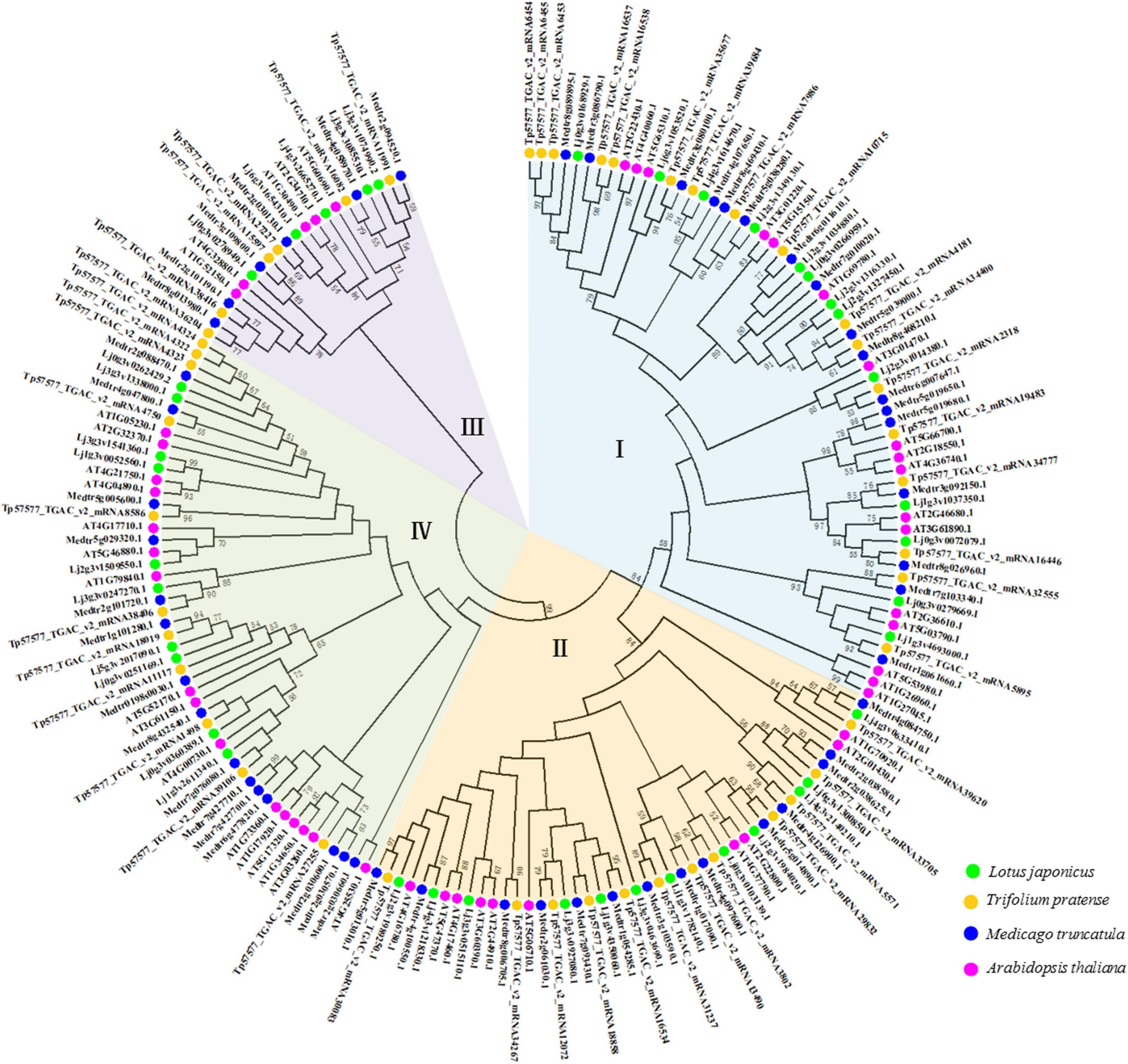
Figure 1. Phylogenetic tree of the HD-Zip proteins of three leguminous forage plants and Arabidopsis thaliana. Relationship among HD-Zip proteins constructed using the maximum-likelihood method. Different colored branches (blue, yellow, purple, and green) represent different groups (I, II, III, and IV). Labels above the nodes represent HD-Zip proteins from the source species.
The physicochemical properties of the amino acids encoded in the three leguminous forage species were analyzed (Supplementary Tables 1–4). The largest and smallest proteins were Tp57577_TGAC_v2_mRNA16082 of the HD-Zip III subfamily and Lj4g3v0633410.1 of the HD-Zip II subfamily, respectively, which had peptide chain lengths of 896 aa and 117 aa, respectively. The pI of 70.07% of the HD-Zip family proteins was less than 7, and they were weakly acidic. The pI ranged from 4.44 to 9.80, and the span was very large, indicating that the subfamily members came from different physiological environments. The physicochemical characteristics of Lotus japonicus were further predicted. The total average hydropathicity GRAVY values were negative and ranged from –1.186 to –0.116, thus implying their hydrophilic nature (Supplementary Table 1). The subcellular localization of Lotus japonicus HD-Zip proteins was in the nucleus, as indicated by UniProt software and WoLF PSORT (Supplementary Tables 1, 5).
The structure of HD-Zip proteins was analyzed in Lotus japonicus (Supplementary Figure 3). The multiple sequence alignment showed that HD-Zip I and HD-Zip II have higher sequence similarity and HD-Zip III and HD-Zip IV have a closer homologous evolution relationship. We examined the LjHD-Zip proteins for discovery motifs using the MEME program, and the top 10 enriched motifs were identified (Supplementary Table 6). The structural characteristics of the motif are shown in Supplementary Figure 4. From the conservative domain and motif analysis of LjHD-Zip proteins, HD-Zip I and HD-Zip II had the same conservative domain while HD-Zip III and HD-Zip IV had more similar structures. The advanced structure of plant proteins is related to their biological function and activity. The three-dimensional (3D) structure of LjHD-Zip proteins was studied by modeling predictions (Supplementary Figure 5). HD-Zip I and HD-Zip II were highly similar in their α-helix and β-sheet structures, while HD-Zip III and HD-Zip IV had complex protein structures.
We analyzed and drew the exon and intron structure diagram of HD-Zip genes in Lotus japonicus (Supplementary Figure 6A). All LjHD-Zip gene members have introns, and the number of exons in subgroup I and II ranged from 2 to 4, those in subgroup III ranged from 12 to 18 except for one, and those in subgroup IV ranged from 7 to 11 (Supplementary Figure 6B). The number of introns in each subgroup was relatively stable, and the gene structure of subgroup III and IV was relatively complex.
All 40 LjHD-Zip gene members were investigated for chromosome localization. They were unevenly distributed on 6 chromosomes (Supplementary Figure 7A), and the genes of each subgroup were unevenly distributed on each chromosome (Supplementary Figure 7B). The unbalanced distribution of HD-Zip genes on the chromosomes was related to the functional characteristics of the genes over the process of evolution.
The 2000 bp upstream sequence of the LjHD-Zip gene was intercepted, and the regulatory elements of each gene promoter were obtained by PlantCARE (Supplementary Figure 8). The promoter region of the HD-Zip genes had a number of elements involved in different metabolic regulatory elements, and a total of 95 cis-elements were detected in Lotus japonicus. We classified these elements into 6 categories and analyzed the regulatory elements of each subgroup. All identified cis-elements had stress and hormone response elements, which accounted for 10% and 16% of Lotus japonicus HD-Zip I, respectively (Figure 2A). The 13-gene promoter of HD-Zip I contained 14 types of stress-related cis-elements, including ABA, GA, MeJA, ETH, SA, drought, temperature, and salt stress cis-elements (Figure 2B). The Lj1g3v1037350.1, Lj0g3v0266959.1, Lj2g3v1034880.1, Lj2g3v1316330.1, Lj2g3v1327450.1, and Lj0g3v0072079.1 of HD-Zip I were selected for intensive study according to the ABA and salt stress-related cis-elements. We further analyzed the regulatory elements of each gene promoter of HD-ZIP II, III, and IV all genes (Supplementary Figure 9). Three genes of each HD-Zip family were selected for analysis expression level under different stress types and stress time points.
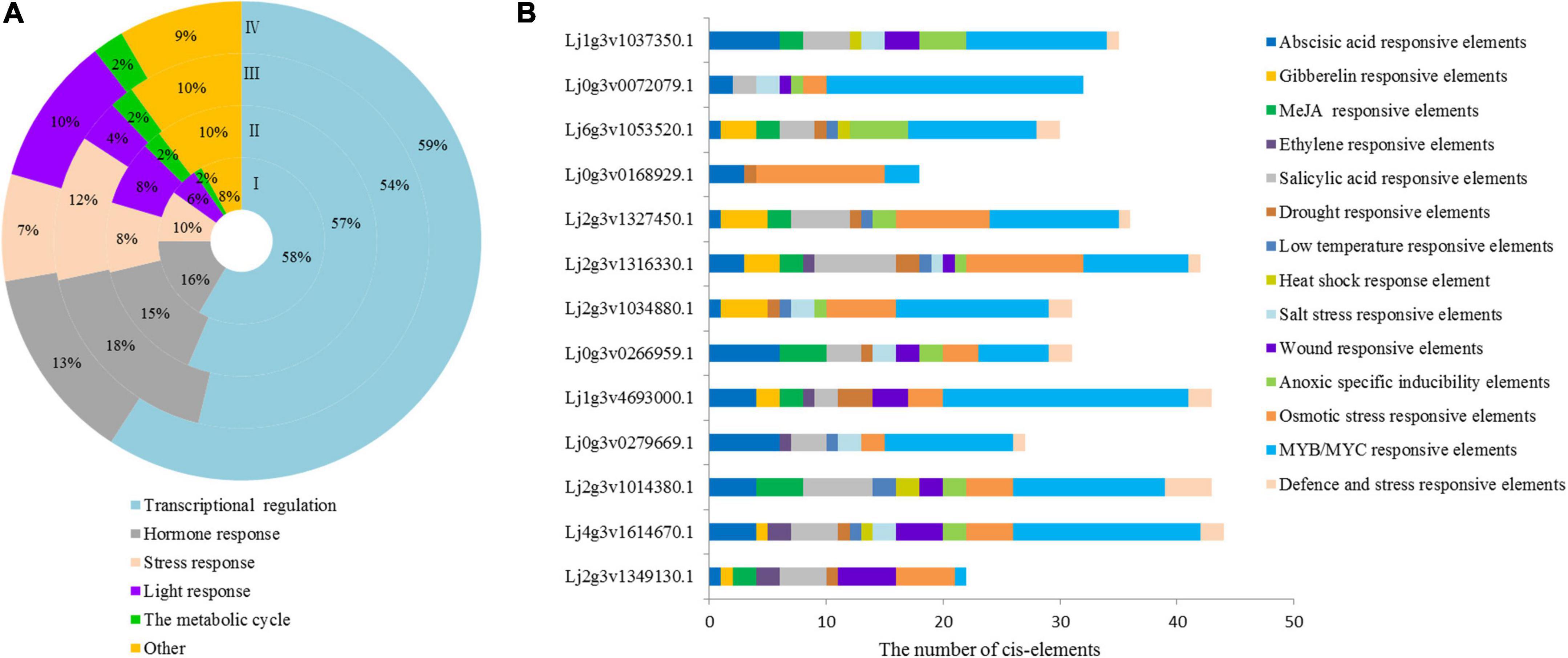
Figure 2. Cis-element statistical analysis of HD-Zip promoters of Lotus japonicus. (A) Different cis-element statistical analysis of four subfamilies. (B) Cis-element statistical analysis of HD-Zip I family gene promoters related to stress and hormone responses. Different cis-elements with similar functions are shown in the same color.
The expression of 6 HD-Zip I genes in different tissues (root, stem, leaf) was analyzed by qRT–PCR (Figure 3A). The relative expression of Lj2g3v1327450.1, Lj2g3v1316330.1, and Lj0g3v0072079.1 was higher than that of the other three genes in the stems. The expression of Lj1g3v1037350.1, Lj0g3v0266959.1, and Lj0g3v0072079.1 was higher than that of the other genes in the roots. After stress treatment for 24 h, these seedlings showed different degrees of wilting and damage (Figure 3E). The electrolyte leakage of the 4 and 42°C treatment leaves was significantly higher than that of the control leaves (Figure 3C). Our results showed that these six genes had differential expression abundance under different stress types and stress time points (Figure 3D). Similarly, nine genes of HD-ZIP II, III, and IV had also differential expression abundance under different stress types and stress time points (Supplementary Figure 10). The expression of Lj0g3v0072079.1 was strongly induced by the stress treatments, especially salt stress. The expression of Lj0g3v0072079.1 rapidly increased and then decreased after the 42°C and PEG treatments for 3 h, while it decreased after the 4°C and salt treatments for 6 h (Figure 3B). Therefore, Lj0g3v0072079.1 may play an important role in stress responses.
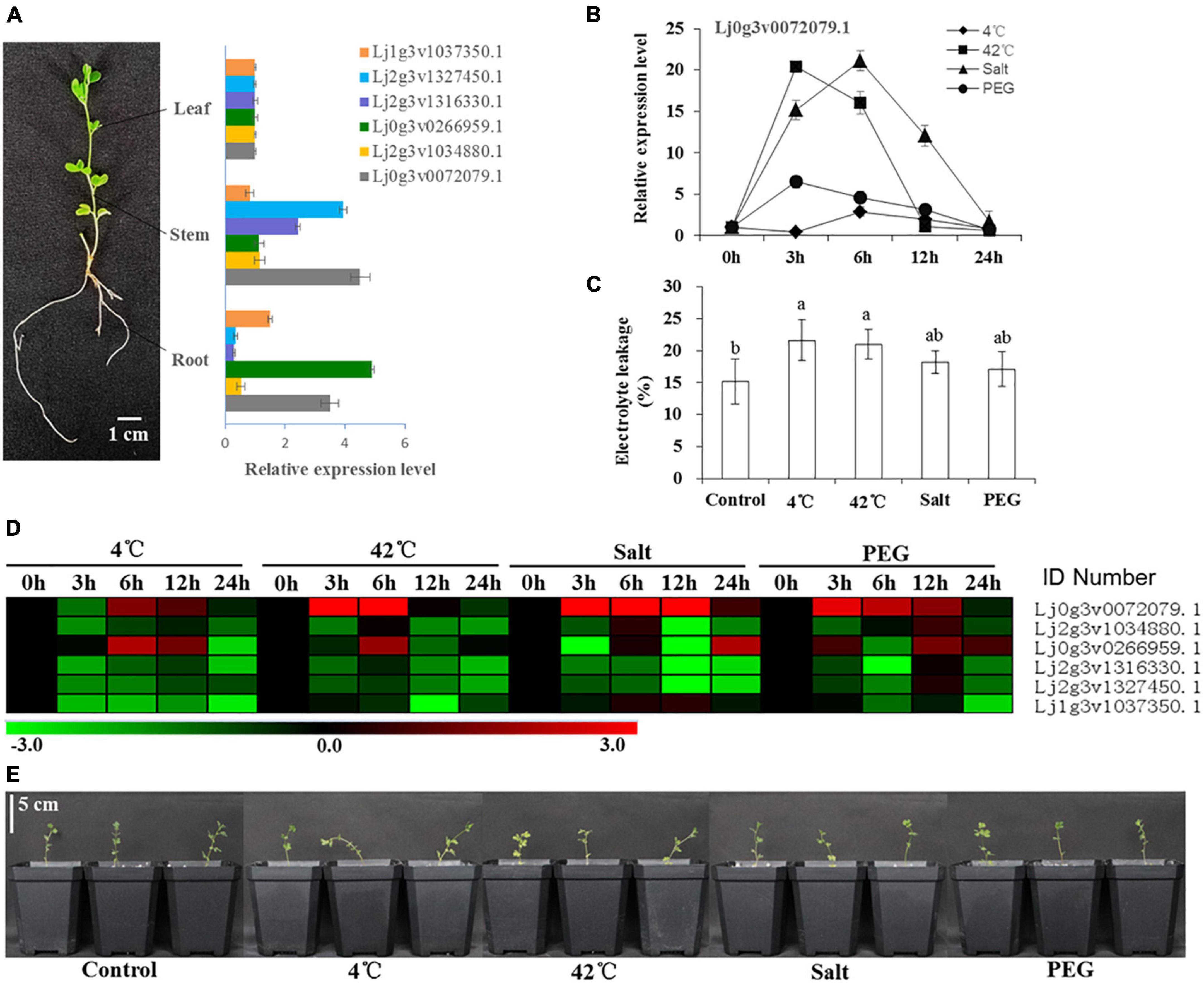
Figure 3. Expression levels of six HD-Zip I genes under various stresses in different times. (A) Tissue specificity analysis of six HD-Zip I genes and mRNA expression levels in leaves, stems and roots of Lotus japonicus. (B) Expression level of Lj0g3v0072079.1 during different stresses and times. (C) Electrolyte leakage of Lotus japonicus under different stresses at 24 h. (D) Heatmap of six HD-Zip I genes under 4°C, 42°C, salt and PEG stresses at different times. (E) Phenotypes of Lotus japonicus seedlings under different stresses at 24 h. Data values are the means ± SD of three independent biological samples. Different letters indicate significance using the LSD test (p < 0.05).
The interacting proteins of Lj0g3v0072079.1 were predicted (Supplementary Figure 11A). The red node query protein was interacted with the stress response proteins RD26 and WRKY and ABA signaling-related proteins ABI (ABA-insensitive), HAB (hypersensitive to ABA), PYL (regulatory component of ABA receptors) and PP2C (protein phosphatase 2C). We further predicted the interaction protein networks of Lj0g3v0072079.1, with Medicago truncatula as the background, and obtained stress-related proteins (Supplementary Figure 12). We performed a Gene Ontology analysis and found that the functional enrichment network of Lj0g3v0072079.1 proteins participated in the ABA-activated signaling pathway (Supplementary Table 8). The Lotus japonicus PP2C gene families Lj4g3v3044980.2 and Lj6g3v1211810.1 were found through homology comparisons (Supplementary Figure 13). The degrees of up- or down-regulation of Lj4g3v3044980.2 and Lj6g3v1211810.1 were different under different stresses (Supplementary Figure 11B). These results show that Lj0g3v0072079.1 may be involved in stress responses by regulating the ABA signaling pathway.
Through sequence comparisons, we identified the homology between Lj0g3v0072079.1 and AtHB7 (Supplementary Figure 14); therefore, we named the Lj0g3v0072079.1 gene LjHDZ7 and subsequently overexpressed LjHDZ7 in Arabidopsis. Our results showed that the seed germination of the LjHDZ7 overexpression line was better than that of Col-0 under salt stress (Figure 4A). The root growth of the transgenic lines was greater than that of the Col-0 line under salt stress (Figure 4B). The leaves of Col-0 gradually turned yellow and died, whereas the leaves of the transgenic lines showed obviously better growth under salt stress (Figure 4C). The shoot drought weight (DW) of the transgenic lines was greater than that of the Col-0 line under salt stress (Figure 5A). The electrolyte leakage and MDA contents of Col-0 plants leaf were higher than those of the LjHDZ7 overexpression lines (Figures 4D,E). The proline contents of the LjHDZ7 overexpression lines leaf were higher than those of the Col-0 plants under salt stress (Figure 4F). Furthermore, the SOD, peroxidase (POD) and catalase (CAT) activities of the LjHDZ7 overexpression lines leaf were significantly higher than those of the Col-0 plants under salt stress (Figures 4G–I). Similarly, MDA contents of Col-0 plants root was higher than that of the LjHDZ7 overexpression lines, while proline contents and SOD activities of the LjHDZ7 overexpression lines root were significantly higher than those of the Col-0 plants under salt stress (Figures 5B–F). These results showed that overexpressing LjHDZ7 could increase the salt resistance of plants.
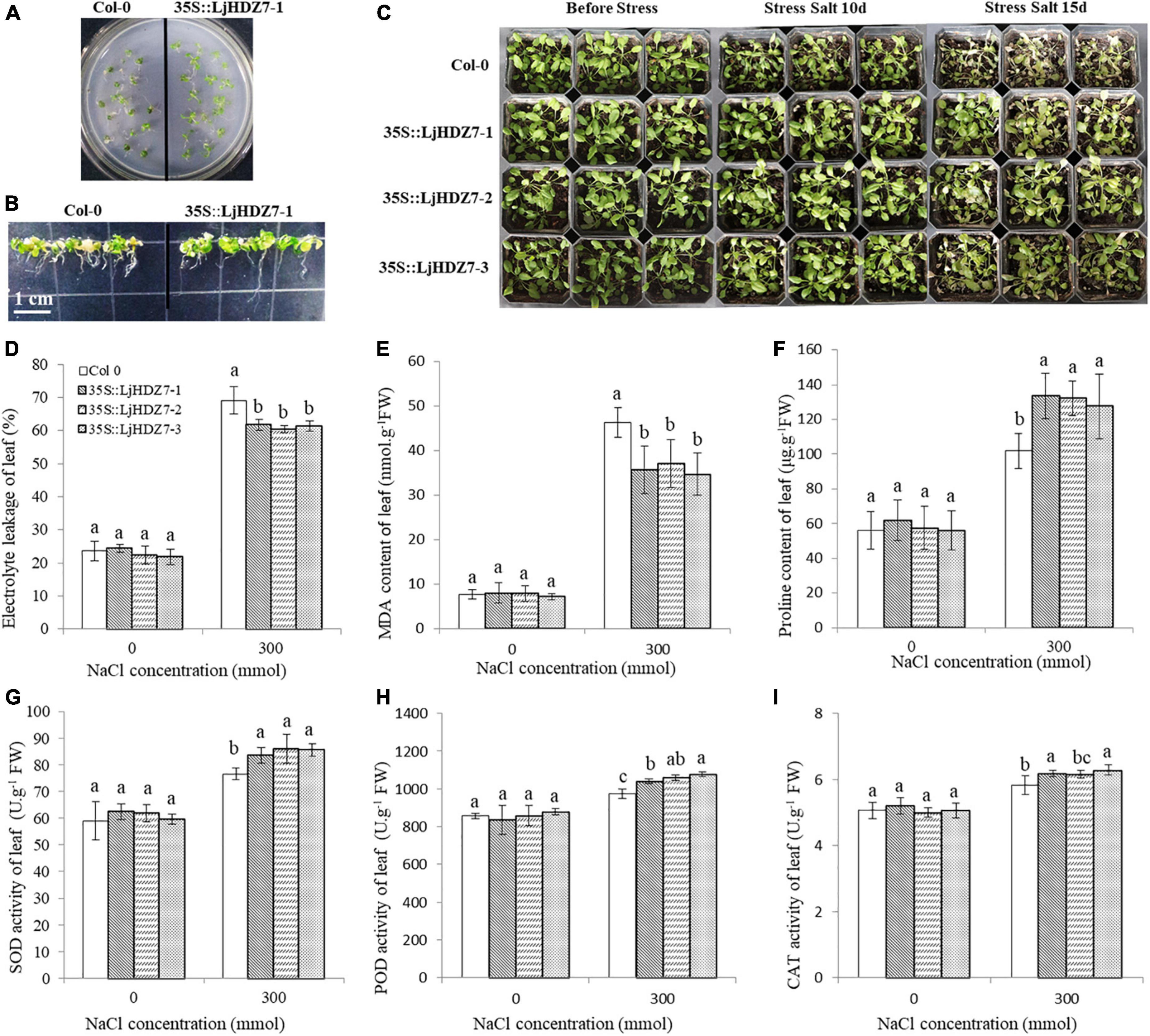
Figure 4. Overexpression of LjHDZ7 in transgenic Arabidopsis improved salt tolerance. (A) Seed germination of wild-type (Col-0) and 35S:LjHDZ7-1 transgenic Arabidopsis plants in MS medium containing 150 mM NaCl. (B) Seeds of Col-0 and 35S:LjHDZ7-1 transgenic Arabidopsis were planted in MS medium for 7 days, and seedlings were transferred to MS medium containing 150 mM NaCl for 10 days before investigation. (C) Phenotypic analysis of Col-0 and 35S:LjHDZ7 transgenic Arabidopsis lines under salt tolerance. (D) Electrolyte leakage of leaf. (E) MDA content of leaf. (F) Proline content of leaf. (G) Superoxide dismutase (SOD) activity of leaf. (H) Peroxidase (POD) activity of leaf. (I) Catalase (CAT) activity of leaf. Data values are the means ± SD of three independent biological samples. Different letters indicate significance differences based on the LSD test (p < 0.05).
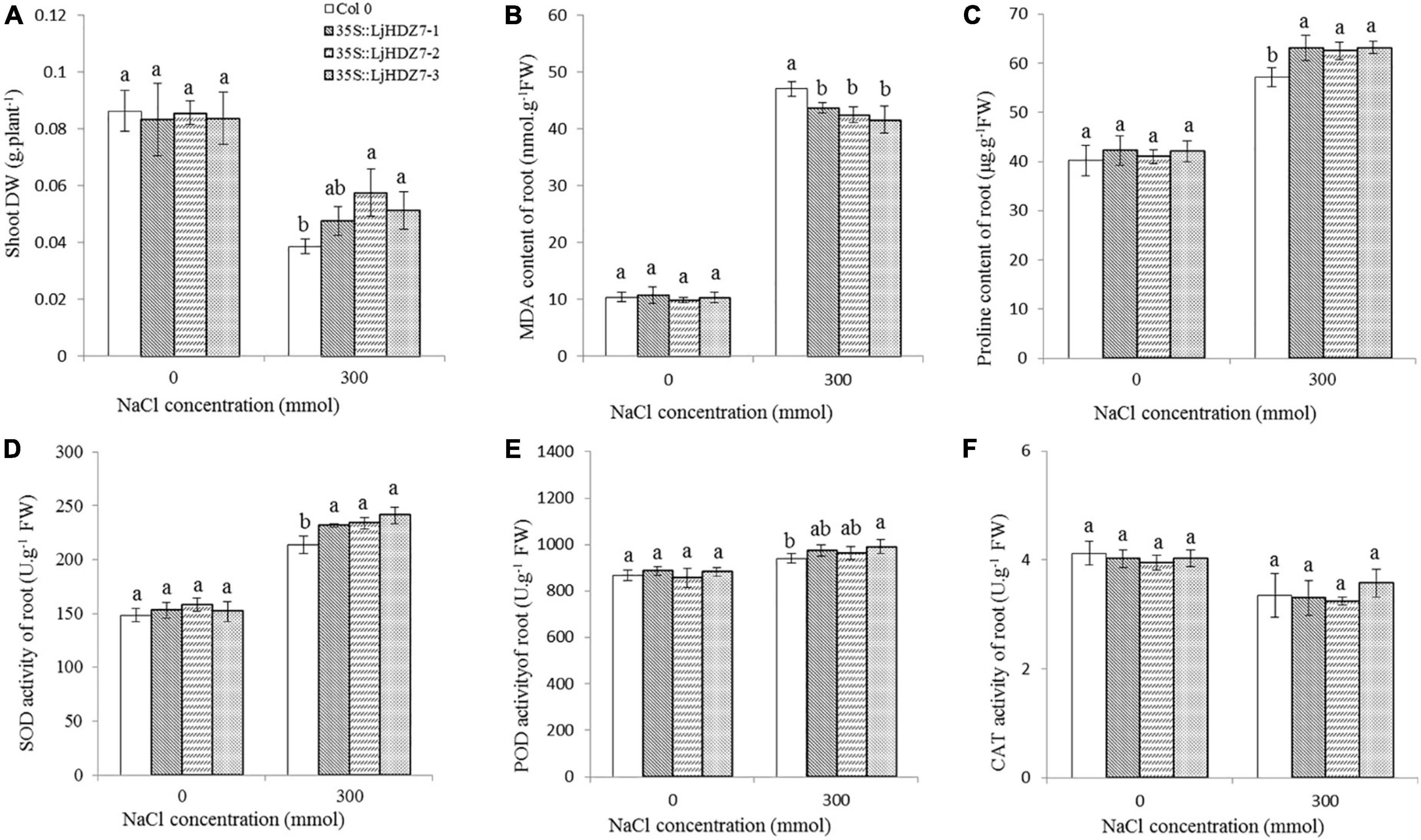
Figure 5. The effect of physiological index in Col-0 and LjHDZ7 overexpression lines. (A) The shoot DW of Col-0 and LjHDZ7 overexpression lines. (B) MDA content of root. (C) Proline content of root. (D) SOD activity of root. (E) POD activity of root. (F) CAT activity of root. Data values are the means ± SD of three independent biological samples. Different letters indicate significance differences based on the LSD test (p < 0.05).
In this study, the concentrations of Na+ and K+ ions in leaves and roots were measured (Figure 6). Under 300 mM NaCl treatment, the concentration of Na+ ions in all plants significantly increased in the leaf and root parts, while the concentration of Na+ ions in LjHDZ7 overexpression lines was significantly lower than that of Col-0 in both leaf and root parts (Figures 6A,C). Under 300 mM NaCl treatment, the concentration of K+ ions in all plants was significantly decreased in the leaf and root parts, but the concentration of K+ ions in LjHDZ7 overexpression lines was significantly higher than that of Col-0 in the leaf parts (Figures 6B,D). Under 300 mM NaCl treatment, the expression of the ethylene biosynthesis genes AtACS5 and AtACS7 was obviously increased in Col-0 and transgenic plants, while the expression of AtACS5 was not significantly different between Col-0 and the LjHDZ7 overexpression lines. The expression of AtACS7 in the two transgenic lines was significantly lower than that in Col-0 (Figures 7A,B). Under 300 mM NaCl treatment, the expression of the auxin biosynthesis genes AtYUC1 and AtYUC2 was slightly decreased in Col-0 and transgenic plants, but the expression of AtYUC1 in LjHDZ7 overexpression plants was slightly higher than that in Col-0, and one transgenic line reached the level of significance. The expression of AtYUC2 in the two transgenic lines was significantly higher than that in Col-0 (Figures 7C,D).
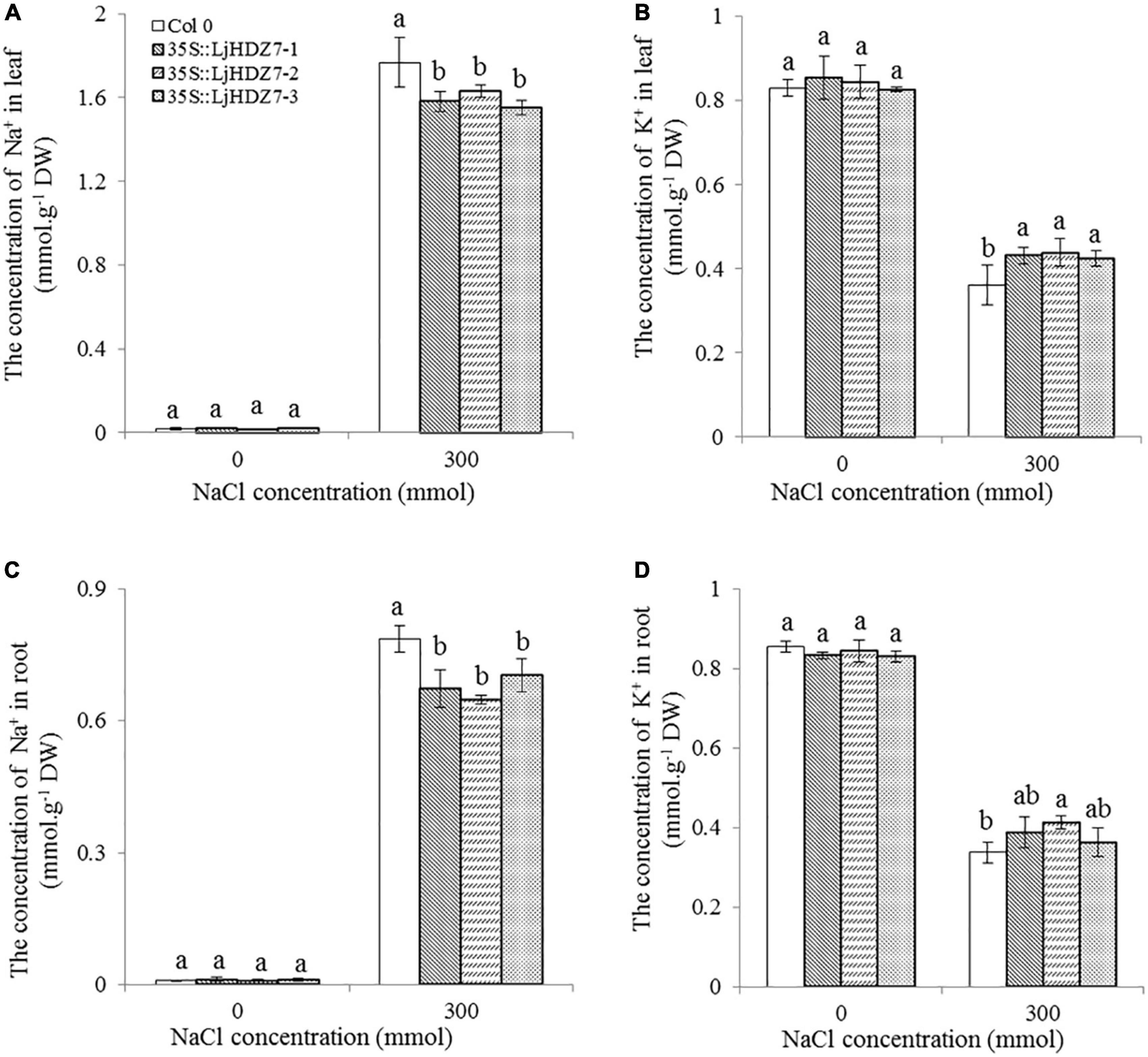
Figure 6. The concentrations of Na+ and K+ in the leaves and roots of Col-0 and LjHDZ7-overexpressing lines. The concentration of Na+ in leaves (A), the concentration of K+ in leaves (B), the concentration of Na+ in roots (C) and the concentration of K+ in roots (D) of Col-0 and LjHDZ7 overexpression lines under control conditions and 300 mM NaCl. Data values are the means ± SD of three independent biological samples. Different letters indicate significant differences based on the LSD test (p < 0.05).
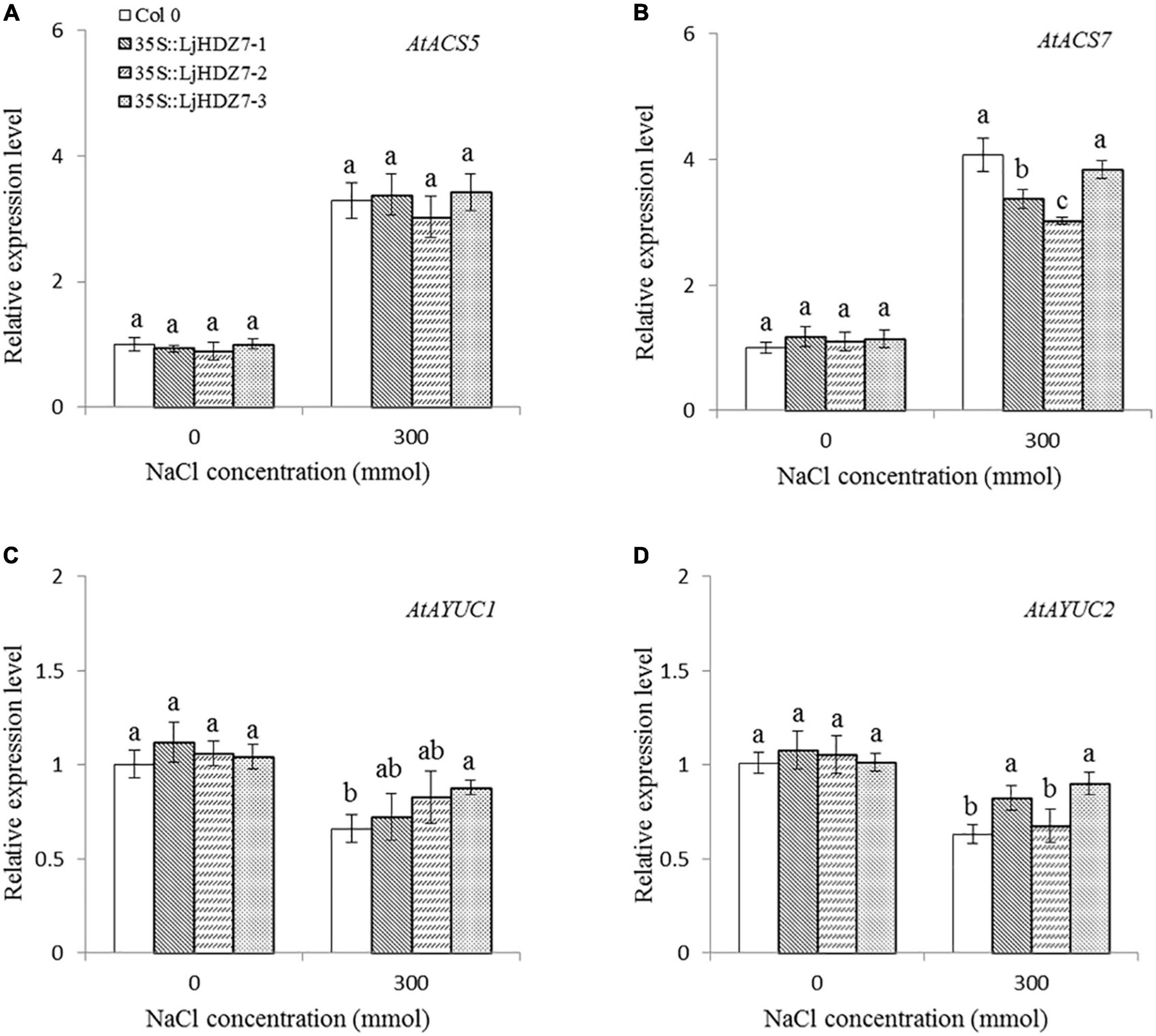
Figure 7. The expression levels of auxin and ethylene biosynthesis genes. The expression level of AtACS5 (A), the expression level of AtACS7 (B), the expression level of AtYUC1 (C) and the expression level of AtYUC2 (D) in Col-0 and LjHDZ7 overexpression lines under control conditions and 300 mM NaCl. Data values are the means ± SD of three independent biological samples. Different letters indicate significant differences based on the LSD test (p < 0.05).
Phylogenetic trees represent an established method for determining evolutionary changes and functional relationships (Li et al., 2015). The HD-Zip protein family has been identified in many species, from mosses to higher plants, such as Ceratopteris richardii (Aso et al., 1999), Physcomitrium patens (Yip et al., 2016), angiosperms and gymnosperms (Prigge and Clark, 2010). The phylogenetic analysis indicated that the HD-Zip gene family originated before the differentiation of vascular plants and moss lineages (Sakakibara et al., 2001). In our phylogenetic tree, a total of 186 HD-Zip proteins from three leguminous forage species and Arabidopsis were identified and divided into four subfamilies (Figure 1). This result was consistent with research reports on HD-Zip proteins in other species (Turchi et al., 2015). In addition, we also found that the three legumes had different numbers of HD-Zip genes and HD-Zip amino acid sequences (Supplementary Figure 1). Following species divergence and in conjunction with genome evolution, some members of the HD-Zip family diverged from their common ancestral genes (Jiang et al., 2015). Earlier findings showed that the exceptional increase in the sizes of TF families in higher plants is often related to WGD (whole-genome duplication) events (Lehti-Shiu et al., 2017). Legumes have undergone two WGDs within their MRCA (the Most Recent Common Ancestor). In particular, a WGD event resulted in major TF family expansions within the legumes (approximately 58 million years ago). Furthermore, TF family expansions occurred at the Glycine genus after a WGD (approximately 13 million years ago), which explains of the increase in most TF families in this genus (Moharana and Venancio, 2020). A study of legume genome evolution through the Medicago truncatula and Lotus japonicus genomes demonstrated WGD events early in legume evolution (Cannon et al., 2006). Therefore, the molecular evolution of the HD-Zip gene family in legumes shows that legume genomes have undergone extensive rearrangement, including translocation and inversion (Li et al., 2014).
Previous experiments have proven that the HD-Zip gene family has a variety of motifs and structures in plants that participate in various functions (Capella et al., 2014). Great differences were observed in the number and length of exons in each subgroup, suggesting that different gene structures indicated the different gene functions (Prigge et al., 2005). In our results, Lotus japonicus HD-Zip family proteins were all hydrophilic and localized to the nucleus (Supplementary Table 1), and they had different motifs, conserved domains, protein 3D structures (Supplementary Figures 3–5) and intron and exon numbers (Supplementary Figure 6). HD-Zip I and II were highly similar in protein structure, while HD-Zip III and IV had complex protein structures. Furthermore, HD-Zip proteins and the gene structure of each subfamily were very similar. The function of each subfamily was related to their structure. Each subfamily HD-Zip gene may have evolved from the same ancestor. HD-Zip I members have been proven to play critical roles in the regulation of plant developmental processes, signaling networks and responses to environmental stresses (Gong et al., 2019). HD-Zip I gene LpHOX21 may act as a positive transcriptional regulator for heat tolerance in perennial ryegrass (Wang et al., 2019). Overexpression of the wheat TaHDZipI-5 gene improved transgenic wheat freezing and drought resistance (Yang et al., 2017).
The response sequence of the gene promoter determines the specific expression pattern of genes and can reflect the function of genes (Butler and Kadonaga, 2002). In this study, a total of 95 cis-acting elements were detected in the promoter of Lotus japonicus HD-Zip genes (Supplementary Figure 8). We further found that 95 cis-acting elements contained stress-related elements (MBS, DRE, ERE, etc.) and hormone response elements (TCA-element, ABRE, TGA-element, et al.). Stress- and hormone-related elements are related to the stress function of genes (Zhou et al., 2021). All identified cis-elements were classified, and the stress and hormone response elements accounted for 10 and 16% in Lotus japonicus HD-Zip I, respectively (Figure 2A). The HD-Zip I gene promoters were classified into 14 types of stress-related cis-elements, including ABA- and salt stress-related cis-elements (Figure 2B). Therefore, the Lotus japonicus HD-Zip I gene promoter could respond to stress factors and HD-Zip I members could play an important role in plant abiotic stress.
Transcription factor can regulate plant responses to hormones and environmental factors by regulating gene expression (Nath et al., 2019). Studies have shown that the expression of HD-Zip subfamily I TFs is induced by drought, high salt, ABA and chilling injury (Manavella et al., 2008). Here, our results showed that the expression of six selected HD-Zip I members was induced by different stresses (Figure 3D). These six genes had differential expression abundance in different stress types and stress time points; therefore, they may be involved in different adversity pathways. Expression of the HD-Zip family HaHB11 was induced by ABA, NaCl and water deficit in sunflower (Cabello et al., 2016). Arabidopsis AtHB13 was upregulated by low temperatures and played a key role in cold, drought, and salinity stresses (Cabello and Chan, 2012; Cabello et al., 2015). We further found that the expression of the LjHDZ7 gene was strongly induced by abiotic stress, especially salt stress (Figure 3B). Therefore, LjHDZ7 may play an important role in salt stress.
Homeodomain-leucine zipper genes participate in the hormone signaling pathway and regulate plant cell expansion, division and differentiation by interacting with hormone pathway genes and downstream genes to improve plant stress tolerance (Agalou et al., 2008). In our study, many proteins related to the stress response were predicted to interact with LjHDZ7, such as RD26 (Fujita et al., 2004) and WRKY (Singh et al., 2017), ABA receptor protein, ABA signaling ABI and HAB (Supplementary Figure 11A). Furthermore, the homologous family of PP2C was identified and showed different expression levels under different stressors in Lotus japonicus (Supplementary Figure 11B). PYL and PP2C are key factors in the ABA pathway (Cheng et al., 2017). As previously reported for HD-Zip proteins in Arabidopsis thaliana, AtHB12 is involved in ABA signaling by regulating PP2C and ABA receptor gene activities (Valdés et al., 2012). Therefore, our results indicated that LjHDZ7 may regulate the stress response by the ABA signaling pathway, which needs to be further investigated.
The combination of modern molecular biology and traditional breeding methods can improve breeding efficiency. A drought-induced HD-Zip I gene, Zmhdz10, was isolated from maize and positively regulated drought and salt tolerance in both genetically transformed rice and Arabidopsis (Zhao et al., 2014). Overexpression of the wheat TaHDZipI-5 gene increased transgenic wheat freezing and drought resistance (Yang et al., 2017). We cloned the LjHDZ7 gene and obtained LjHDZ7-overexpressing transgenic Arabidopsis plants. Under salt stress, the growth of the transgenic lines was obviously better than that of the Col-0 line (Figures 4A–C). The electrolyte leakage and MDA contents of Col-0 plants were higher than those of the LjHDZ7 overexpression lines (Figures 4D,E, 5B,C). Furthermore, the proline contents, SOD, POD, and CAT activities of LjHDZ7 overexpression lines were higher than those of Col-0 plants (Figures 4F–I, 5D–F). These results showed that the overexpression of LjHDZ7 could increase the salt resistance of plants. Overexpression of PsnHDZ63 resulted in improved reactive oxygen species scavenging ability and enhanced salt stress tolerance in transgenic Populus simonii × P. nigra (Guo et al., 2021). HD-Zip I gene BpHOX2 could bind to different cis-acting elements to regulate gene expression, thus improving osmotic tolerance in birch (Tan et al., 2020). Therefore, the HD-Zip gene family may become valuable to improve stress tolerance in molecular breeding methods.
Excessive Na+ in plants could result in ion toxicity, which causes an imbalance in ion uptake and nutritional disorders and finally leads to growth inhibition and even plant death (Wu et al., 2019; Zhang H. et al., 2020). Maintaining K+ and Na+ ion homeostasis is important for a series of plant physiological and biochemical processes and for resistance to salinity (Ma et al., 2017). In the present study, the concentration of Na+ ions in LjHDZ7 overexpression lines was significantly lower than that in Col-0 in both leaf and root parts (Figures 6A,C). Furthermore, the concentration of K+ ions in LjHDZ7 overexpression lines was significantly higher than that in Col-0 in the leaf parts (Figures 6B,D). These results indicated that LjHDZ7 overexpression lines may have a stronger selective absorption and transportation capacity for K+ and Na+ and could maintain a higher concentration of K+ under saline conditions.
The phytohormone ethylene plays essential roles in plant abiotic stress adaptation. ACS (ACC synthase) is the key enzyme in ethylene biosynthesis. A previous study showed that the expression of AtACS7 was regulated by salt stress (Dong et al., 2011). In this study, under 300 mM NaCl treatment, the expression of the ethylene biosynthesis genes AtACS5 and AtACS7 was obviously increased in both Col-0 and LjHDZ7 overexpression lines, while there was no significant difference between Col-0 and LjHDZ7 overexpression lines (Figures 7A,B). There may be differences in the expression of other AtACS genes between Col-0 and LjHDZ7 overexpression lines. Auxin regulates many of the key processes in plant growth and stress responses. The YUCCA (YUC) family is the key limiting enzyme in the auxin biosynthetic pathway (Liu et al., 2015). A previous study showed that plant IAA levels were significantly decreased under salt stress (Hofmann, 2011). Our study showed that the expression of the AtYUC1 and AtYUC2 genes was downregulated in Col-0 and LjHDZ7 overexpression plants under salt treatment. The expression of AtYUC2 in two transgenic lines was significantly higher than that in Col-0 plants (Figures 7C,D). These results showed that LjHDZ7 could change endogenous ethylene and auxin levels by regulating the expression of ethylene and auxin biosynthesis genes under salt treatment.
We performed an evolutionary analysis of the HD-Zip family in three leguminous forage species to obtain comprehensive information on this family, including the protein features and phylogenetic relationships. Furthermore, the motifs and domains of the proteins, gene structures, promoter cis-elements and stress expression patterns of HD-Zip genes were analyzed in Lotus japonicus. We further found that the expression of the LjHDZ7 gene was strongly induced by abiotic stress, especially salt stress. Furthermore, our results showed that overexpression of LjHDZ7 could increase salt resistance in transgenic Arabidopsis plants. Our results are important because they provide a theoretical basis for further functional studies of the HD-Zip gene family in Lotus japonicus and resources for prospective applications for stress-resistant breeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
DW and SN wrote the main manuscript text and contributed to the conception of the study. DW, YG, YL, and SN performed the experiments and the data analyses. All authors have read and agreed to the published version of the manuscript.
This research was funded the Fundamental Research Funds of China West Normal University (19E052). The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.955199/full#supplementary-material
Supplementary Figure S1| Statistical analysis of the HD-Zip subfamily in each plant species. (A) Number of HD-Zip genes found in each subfamily of Lotus japonicus, Medicago truncatula, Trifolium pratense, and Arabidopsis thaliana. (B–E) Amino acid sequences of HD-Zip genes in Arabidopsis thaliana, Medicago truncatula, Trifolium pretense, and Lotus japonicus, respectively. Genes corresponding to (B–E) X-axis NO were in Supplementary Tables S1–S4, respectively.
Supplementary Figure S2| Distribution of HD-Zip subfamilies within four plant species.
Supplementary Figure S3| Conserved motif and domain analysis of HD-Zip proteins from Lotus japonicus. The phylogenetic tree was generated based on the protein sequences of HD-Zip proteins. Different colored boxes represent different types of motifs and domains.
Supplementary Figure S4| Sequence information for Motif 1- Motif 10.
Supplementary Figure S5| Three-dimensional modeling structure of HD-Zip proteins in Lotus japonicus.
Supplementary Figure S6| (A) Chromosome localization of HD-Zip in Lotus japonicus. Location of HD-Zip genes in Lotus japonicus. (B) Number of four distinct HD-Zip subfamilies per chromosome.
Supplementary Figure S7| Gene structure of the HD-Zip family of Lotus japonicus. (A) Exon/intron structures of HD-Zip genes of Lotus japonicus. Exons and introns are represented by yellow bones and gray lines, respectively. (B) Exon and gene numbers of four HD-Zip subfamilies of Lotus japonicus.
Supplementary Figure S8| Cis-element identification in the promoters of HD-Zip from Lotus japonicus.
Supplementary Figure S9| Promoters cis-element analysis of HD-Zip of Lotus japonicus. Cis-element statistical analysis of (A) HD-Zip II, (B) HD-Zip III, and (C) HD-Zip IV family gene promoters related to stress and hormone responses. Different cis-elements with similar functions are shown in the same color.
Supplementary Figure S10| Expression levels of nine HD-Zip II, III and HD-Zip IV genes under various stresses in different times. (A) Expression level of Lj4g3v2140210.1 during different stresses and times. (B) Lj2g3v1989250.1. (C) Lj3g3v0463690.1. (D) Lj4g3v2665270.1. (E) Lj3g3v1074990.2. (F) Lj0g3v0278949.1. (G) Lj1g3v2611340.1. (H) Lj0g3v0251169.1. (I) Lj0g3v0262429.2.
Supplementary Figure S11| Predicted protein interaction networks of Lj0g3v0072079.1. (A) Protein interaction network of Lj0g3v0072079.1 was generated using the STRING online database, with Arabidopsis thaliana as the background. Edges represent protein–protein associations. The nodes represent proteins, and red nodes represent query proteins. The black, green, blue, light sky blue and purple lines represent co-expression, text mining, gene co-occurrence, protein homology, and experimental determination, respectively. (B) Relative expression levels of Lj4g3v3044980.2 and Lj6g3v1211810.1 under different stresses at 24 h.
Supplementary Figure S12| Predicted protein interaction networks of Lj0g3v0072079.1, with Medicago truncatula as the background.
Supplementary Figure S13| Amino acid sequence alignment of the protein phosphatase 2C (PP2C) family. Including Lj4g3v3044980.2 and Lj6g3v1211810.1 from Lotus japonicus and ATABI1, ATABI2, ATHAB1, and ATHAB2 from Arabidopsis thaliana.
Supplementary Figure S14| Amino acid sequence alignment of LjHDZ7, ATHB-7, and AET01987.
Supplementary Table S2| Basic physicochemical properties of the HD-Zip family in Arabidopsis thaliana.
Supplementary Table S3| Basic physicochemical properties of the HD-Zip family in Medicago truncatula.
Supplementary Table S4| Basic physicochemical properties of the HD-Zip family in Trifolium pratense.
HD-Zip, the homeodomain leucine zipper; TF, transcription factor; Mw, molecular weight; pI, isoelectric point; SA, salicylic acid; JA, jasmonic acid; MeJA, methyl jasmonate; GA, gibberellin; q-PCR, quantitative polymerase chain reaction; ABA, abscisic acid; MDA, malonaldehyde; PEG6000, polyethylene glycol 6000; NCBI, National Center for Biotechnology Information.
Agalou, A., Purwantomo, S., OVernaS, E., Johannesson, H., Zhu, X., Estiati, A., et al. (2008). A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 66, 87–103. doi: 10.1007/s11103-007-9255-7
Annapurna, B., Khurana, J. P., and Mukesh, J. (2016). Characterization of Rice Homeobox Genes, OsHOX22 and OsHOX24, and Over-expression of OsHOX24 in Transgenic Arabidopsis Suggest Their Role in Abiotic Stress Response. Front. Plant Sci. 7:627. doi: 10.3389/fpls.2016.00627
Arce, A., Raineri, J., Capella, M., Cabello, J., and Chan, R. (2011). Uncharacterized conserved motifs outside the HD-Zip domain in HD-Zip subfamily I transcription factors; a potential source of functional diversity. BMC Plant Biol. 11:42. doi: 10.1186/1471-2229-11-42
Ariel, F. D., Manavella, P. A., Dezar, C. A., and Chan, R. (2007). The true story of the HD-Zip family. Trends Plant Sci. 12, 419–426. doi: 10.1016/j.tplants.2007.08.003
Aso, K., Kato, M., Banks, J. A., and Hasebe, M. (1999). Characterization of homeodomain-leucine zipper genes in the fern Ceratopteris richardii and the evolution of the homeodomain-leucine zipper gene family in vascular plants. Mol. Biol. Evol. 16, 544–552. doi: 10.1093/oxfordjournals.molbev.a026135
Bai, J., Mao, J., Yang, H., Khan, A., Fan, A., Liu, S., et al. (2017). Sucrose non-ferment 1 related protein kinase 2 (SnRK2) genes could mediate the stress responses in potato (Solanum tuberosum L.). BMC Genet. 18:41. doi: 10.1186/s12863-017-0506-6
Brant, R., Cabedo, M., Xie, Y., and Wenkel, S. (2014). Homeodomain leucine-zipper proteins and their role in synchronizing growth and development with the environment. J. Integr. Plant Biol. 56, 518–526. doi: 10.1111/jipb.12185
Butler, J. E., and Kadonaga, J. T. (2002). The RNA polymerase II core promoter: A key component in the regulation of gene expression. Genes. Dev. 16, 2583–2592. doi: 10.1101/gad.1026202
Cabello, J. V., Arce, A. L., and Chan, R. (2015). The homologous HD-Zip I transcription factors HaHB1 and AtHB13 confer cold tolerance via the induction of pathogenesis-related and glucanase proteins. Plant J. 69, 141–153. doi: 10.1111/j.1365-313X.2011.04778.x
Cabello, J. V., and Chan, R. L. (2012). The homologous homeodomain-leucine zipper transcription factors HaHB1 and AtHB13 confer tolerance to drought and salinity stresses via the induction of proteins that stabilize membranes. Plant Biotechnol. J. 10, 815–825. doi: 10.1111/j.1467-7652.2012.00701.x
Cabello, J. V., Giacomelli, J. I., Piattoni, G. V., Iglesias, A. A., and Chan, R. L. (2016). The sunflower transcription factor HaHB11 improves yield, biomass and tolerance to flooding in transgenic Arabidopsis plants. J. Biotec. 222, 73–83. doi: 10.1016/j.jbiotec.2016.02.015
Cannon, S. B., Sterck, L., Rombauts, S., Sato, S., Cheung, F., Gouzyf, J., et al. (2006). Legume genome evolution viewed through the Medicago truncatula and Lotus japonicus genomes. Proc. Natl. Acad. Sci. U.S.A. 103, 14959–14964. doi: 10.1073/pnas.0603228103
Capella, M., Ré, D., Arce, A. L., and Chan, R. L. (2014). Plant homeodomain-leucine zipper I transcription factors exhibit different functional AHA motifs that selectively interact with TBP or/and TFIIB. Plant Cell Rep. 33, 955–967. doi: 10.1007/s00299-014-1576-9
Capella, M., Ribone, P. A., Arce, A. L., and Chan, R. L. (2015). Arabidopsis thaliana HomeoBox 1 (AtHB1), a Homedomain-Leucine Zipper I (HD-Zip I) transcription factor, is regulated by PHYTOCHROME-INTERACTING FACTOR 1 to promote hypocotyl elongation. New Phytol. 207, 669–682. doi: 10.1111/nph.13401
Carabelli, M., Possenti, M., Sessa, G., Ruzza, V., Morelli, G., and Ruberti, I. (2018). Arabidopsis HD-Zip II proteins regulate the exit from proliferation during leaf development in canopy shade. J. Exp. Bot. 69, 5419–5431. doi: 10.1093/jxb/ery331
Cheng, C., Wang, Z., Ren, Z., Zhi, L., Yao, B., Su, C., et al. (2017). SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PLoS Genet. 13:e1006947. doi: 10.1371/journal.pgen.1006947
Dong, H., Zhen, Z., Peng, J., Chang, L., Gong, Q., and Wang, N. (2011). Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J. Exp. Bot. 62, 4875–4887. doi: 10.1093/jxb/err143
Dong, T., Zhang, R., Liu, J., Fowler, J., Miller, T., and Xu, X. (2021). Warming alters sex-specific responses in leaf defense against insect herbivory in Populus cathayana. Environ. Exp. Bot. 189:104557. doi: 10.1016/j.envexpbot.2021.104557
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Gao, Y., Gao, S., Xiong, C., Yu, G., Chang, J., Ye, Z., et al. (2015). Comprehensive analysis and expression profile of the homeodomain leucine zipper IV transcription factor family in tomato. Plant Physiol. Biochem. 96, 141–153. doi: 10.1016/j.plaphy.2015.07.025
Gao, Y., Liu, H., Zhang, K., Li, F., Wu, M., and Xiang, Y. (2021). A moso bamboo transcription factor, Phehdz1, positively regulates the drought stress response of transgenic rice. Plant Cell Rep. 40, 187–204. doi: 10.1007/s00299-020-02625-w
Gong, S., Ding, Y., Hu, S., Ding, L., Chen, Z., and Zhu, C. (2019). The role of HD-Zip class I transcription factors in plant response to abiotic stresses. Physiol. Plant 167, 516–525. doi: 10.1111/ppl.12965
Guo, Q., Jiang, J., Yao, W., Li, L., Zhao, K., Cheng, Z., et al. (2021). Genome-wide analysis of poplar HD-Zip family and over-expression of PsnHDZ63 confers salt tolerance in transgenic Populus simonii×P.nigra. Plant Sci. 311:111021. doi: 10.1016/j.plantsci.2021.111021
He, A., Niu, S., Zhao, Q., Li, Y., Gou, J., Gao, H., et al. (2018). Induced Salt Tolerance of Perennial Ryegrass by a Novel Bacterium Strain from the Rhizosphere of a Desert Shrub Haloxylon ammodendron. Int. J. Mol. Sci. 19:469. doi: 10.3390/ijms19020469
He, G., Liu, P., Zhao, H., and Sun, J. (2021). The HD-ZIP II Transcription Factors Regulate Plant Architecture through the Auxin Pathway. Int. J. Mol. Sci. 21:3205. doi: 10.3390/ijms21093250
Henriksson, E., Olsson, A., Johannesson, H., Johansson, H., Hanson, J., Engström, P., et al. (2005). Homeodomain Leucine Zipper Class I Genes in Arabidopsis. Expression Patterns and Phylogenetic Relationships. Plant Physiol. 139, 509–518. doi: 10.1104/pp.105.063461
Hofmann, N. (2011). YUC and TAA1/TAR proteins function in the same pathway for auxin biosynthesis. Plant Cell 23:3869. doi: 10.1105/tpc.111.231112
Jiang, H., Jing, J., Liu, H., Dong, Q., Yan, H., and Gan, D. (2015). Genome-wide analysis of HD-Zip genes in grape (Vitis vinifera). Tree Genet. Genom. 11:827. doi: 10.1007/s11295-014-0827-9
Lehti-Shiu, M. D., Panchy, N., Wang, P., Uygun, S., and Shiu, S. H. (2017). Diversity, expansion, and evolutionary novelty of plant DNA-bindingtranscription factor families. Biochim. Biophys. Acta Gene. Regul. Mech. 1860, 3–20. doi: 10.1016/j.bbagrm.2016.08.005
Li, H., Gan, Y., Yue, L., Han, Q., Chen, J., Liu, Q., et al. (2022). Newly Isolated Paenibacillus monticola sp. nov., a Novel Plant Growth-Promoting Rhizobacteria Strain From High-Altitude Spruce Forests in the Qilian Mountains. China. Front. Microbiol. 13:833313. doi: 10.3389/fmicb.2022.833313
Li, P., Huang, J., Yu, S., Li, Y., Sun, P., Wu, C., et al. (2016). ArabidopsisYL1/BPG2 Is Involved in Seedling Shoot Response to Salt Stress through ABI4. Sci. Rep. 6:30163. doi: 10.1038/srep30163
Li, Q., Cao, C., Zhang, C., Zheng, S., Wang, Z., and Wang, L. (2015). The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J. Exp. Bot. 66, 2515–2526. doi: 10.1093/jxb/erv046
Li, W., Dong, J., Cao, M., Gao, X., Wang, D., Liu, B., et al. (2019). Genome-wide identification and characterization of HD-ZIP genes in potato. Gene 697, 103–117. doi: 10.1016/j.gene.2019.02.024
Li, Z., Jiang, H., Zhou, L., Deng, L., Lin, Y., Peng, X., et al. (2014). Molecular evolution of the HD-ZIP I gene family in legume genomes. Gene 533, 218–228. doi: 10.1016/j.gene.2013.09.084
Liu, W., Li, R., Han, T., Cai, W., Fu, Z., and Lu, Y. (2015). Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 168, 343–356. doi: 10.1104/pp.15.00030
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-△ △ CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lü, X., Shao, K., Xu, J., Li, J., Ren, W., Chen, J., et al. (2022). A heat shock transcription factor gene (HaHSFA1) from a desert shrub, Haloxylon ammodendron, elevates salt tolerance in Arabidopsis thaliana. Environ. Exp. Bot. 201:104954. doi: 10.1016/j.envexpbot.2022.104954
Ma, Q., Hu, J., Zhou, X. R., Yuan, H. J., Kumar, T., Luan, S., et al. (2017). ZxAKT1 is essential for K+ uptake and K+/Na+ homeostasis in the succulent xerophyte Zygophyllum xanthoxylum. Plant J. 90, 48–60. doi: 10.1111/tpj.13465
Manavella, P. A., Dezar, C. A., Ariel, F. D., Drincovich, M. F., and Chan, R. L. (2008). The sunflower HD-Zip transcription factor HAHB4 is up-regulated in darkness, reducing the transcription of photosynthesis-related genes. J. Exp. Bot. 59:3143. doi: 10.1093/jxb/ern170
Moharana, K. C., and Venancio, T. M. (2020). Polyploidization events shaped the transcription factor repertoires in legumes (Fabaceae). Plant J. 103, 726–741. doi: 10.1111/tpj.14765
Nakamura, M., Katsumata, H., Abe, M., Yabe, N., Komeda, Y., Yamamoto, K. T., et al. (2006). Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol. 141, 1363–1375. doi: 10.1104/pp.106.077388
Nath, V. S., Mishra, A. K., Kumar, A., Matoušek, J., and Jakše, J. (2019). Revisiting the Role of Transcription Factors in Coordinating the Defense Response Against Citrus Bark Cracking Viroid Infection in Commercial Hop (Humulus Lupulus L.). Viruses 11:419. doi: 10.3390/v11050419
Nie, S., Yang, Z., Xiao, C., Yang, R., and Wang, D. (2022). Physiological and Transcriptomic Analyses of the Effects of SlBRI1 Expression Levels on the Drought Tolerance of Tomato Seedlings. J. Plant Growth. Regul. doi: 10.1007/s00344-022-10587-4
Prigge, M. J., and Clark, S. E. (2010). Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 8, 350–361. doi: 10.1111/j.1525-142X.2006.00107.x
Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Drews, G. N., and Clark, S. E. (2005). Class III Homeodomain-Leucine Zipper Gene Family Members Have Overlapping, Antagonistic, and Distinct Roles in Arabidopsis Development. Plant Cell 17, 61–76. doi: 10.1105/tpc.104.026161
Ribone, P. A., Capella, M., and Chan, R. L. (2015). Functional characterization of the homeodomain leucine zipper I transcription factor AtHB13 reveals a crucial role in Arabidopsis development. J. Exp. Bot. 66, 5929–5943. doi: 10.1093/jxb/erv302
Romani, F., Ribone, P. A., Capella, M., Miguel, V. N., and Chan, R. L. (2016). A matter of quantity: Common features in the drought response of transgenic plants overexpressing HD-Zip I transcription factors. Plant Sci. 251, 139–154. doi: 10.1016/j.plantsci.2016.03.004
Sakakibara, K., Nishiyama, T., Kato, M., and Hasebe, M. (2001). Isolation of Homeodomain–Leucine Zipper Genes from the Moss Physcomitrella patens and the Evolution of Homeodomain–Leucine Zipper Genes in Land Plants. Mol. Biol. Evol. 18, 491–502. doi: 10.1093/oxfordjournals.molbev.a003828
Sakuma, S., Pourkheirandish, M., Hensel, G., Kumlehn, J., Stein, N., Tagiri, K., et al. (2013). Divergence of expression pattern contributed to neofunctionalization of duplicated HD-Zip I transcription factor in barley. New Phytol. 197, 939–948. doi: 10.1111/nph.12068
Singh, K. A., Kumar, R. S., Dwivedi, V., Rai, A., Pal, S., Shasany, A., et al. (2017). A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 215, 1115–1131. doi: 10.1111/nph.14663
Su, L., Dai, Z., Li, S., and Xin, H. (2015). A novel system for evaluating drought–cold tolerance of grapevines using chlorophyll fluorescence. BMC Plant Biol. 15:82. doi: 10.1186/s12870-015-0459-8
Tan, Z., Wen, X., and Wang, Y. (2020). Betula platyphylla BpHOX2 transcription factor binds to different cis-acting elements and confers osmotic tolerance. J. Integr. Plant Biol. 62, 1762–1779. doi: 10.1111/jipb.12994
Tang, Y., Bao, X., Wang, S., Liu, Y., Tan, J., Yang, M., et al. (2019). A Physic Nut Stress-Responsive HD-Zip Transcription Factor, JcHDZ07, Confers Enhanced Sensitivity to Salinity Stress in Transgenic Arabidopsis. Front. Plant Sci. 10:942. doi: 10.3389/fpls.2019.00942
Turchi, L., Baima, S., Morelli, G., and Ruberti, I. (2015). Interplay of HD-Zip II and III transcription factors in auxin-regulated plant development. J. Exp. Bot. 66, 5043–5053. doi: 10.1093/jxb/erv174
Valdés, A., Overnäs, E., Johansson, H., Rada-Iglesias, A., and Engström, P. (2012). The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 80, 405–418. doi: 10.1007/s11103-012-9956-4
Wang, D., Cui, B., Guo, H., Liu, Y., and Nie, S. (2022a). Genome-wide identification and expression analysis of the CBF transcription factor family in Lolium perenne under abiotic stress. Plant Signal. Behav. 17:2086733. doi: 10.1080/15592324.2022.2086733
Wang, D., Yang, Z., Wu, M., Wang, W., Wang, Y., and Nie, S. (2022b). Enhanced brassinosteroid signaling via the overexpression of SlBRI1 positively regulates the chilling stress tolerance of tomato. Plant Sci. 320:111281. doi: 10.1016/j.plantsci.2022.111281
Wang, H., Li, G., Zhang, D., Lin, J., Sheng, B., Han, J., et al. (2013). Biological functions of HD-Zip transcription factors. Hereditas 35:1179. doi: 10.3724/SP.J.1005.2013.01179
Wang, J., Zhuang, L., Zhang, J., Yu, J., Yang, Z., and Huang, B. (2019). Identification and characterization of novel homeodomain leucine zipper (HD-Zip) transcription factors associated with heat tolerance in perennial ryegrass. Environ. Exp. Bot. 160, 1–11. doi: 10.1016/j.envexpbot.2018.12.023
Wu, H., Shabala, L., Zhou, M., Su, N., Wu, Q., Ul-Haq, T., et al. (2019). Root vacuolar Na+ sequestration but not exclusion from uptake correlates with barley salt tolerance. Plant J. 100, 55–67. doi: 10.1111/tpj.14424
Wu, Q., Tang, Y., Dong, T., Liao, Y., Li, D., He, X., et al. (2018). Additional AM fungi inoculation increase Populus cathayana intersexual competition. Front. Plant Sci. 9:607. doi: 10.3389/fpls.2018.00607
Yang, Q., Niu, Q., Li, J., Zheng, X., Ma, Y., Bai, S., et al. (2018). PpHB22, a member of HD-Zip proteins, activates PpDAM1 to regulate bud dormancy transition in ‘Suli’ pear (Pyrus pyrifolia White Pear Group). Plant Physiol. Biochem. 127, 355–365. doi: 10.1016/j.plaphy.2018.04.002
Yang, S., Olena, P., and Tobias, S. (2018). ALTERED MERISTEM PROGRAM1 restricts shoot meristem proliferation and regeneration by limiting HD-ZIP III-mediated expression of RAP2.6L. Plant Physiol. 177, 1580–1594. doi: 10.1104/pp.18.00252
Yang, S., Wang, Y., Zhu, H., Zhang, M., Wang, D., Xie, K., et al. (2022). A novel HD-Zip I/C2H2-ZFP/WD-repeat complex regulates the size of spine base in cucumber. New Phytol. 233, 2643–2658. doi: 10.1111/nph.17967
Yang, Y., Luang, S., Harris, J., Riboni, M., Li, Y., Bazanova, N., et al. (2017). Overexpression of the class I homeodomain transcription factor TaHDZipI-5 increases drought and frost tolerance in transgenic wheat. Plant Biotechnol. J. 16, 1227–1240. doi: 10.1111/pbi.12865
Yip, H. K., Floyd, S. K., Sakakibara, K., and Bowman, J. L. (2016). Class III HD-Zip activity coordinates leaf development in Physcomitrella patens. Dev. Biol. 419, 184–197. doi: 10.1016/j.ydbio.2016.01.012
Zalewski, C. S., Floyd, S. K., Furumizu, C., Sakakibara, K., and Bowman, J. L. (2013). Evolution of the Class IV HD-Zip Gene Family in Streptophytes. Mol. Biol. Evol. 30, 2347–2365. doi: 10.1093/molbev/mst132
Zhang, H., Feng, H., Zhang, J., Ge, R., Zhang, L., Wang, Y., et al. (2020). Emerging crosstalk between two signaling pathways coordinates K+ and Na+ homeostasis in the halophyte Hordeum brevisubulatum. J. Exp. Bot. 71, 4345–4358. doi: 10.1093/jxb/eraa191
Zhang, J., Wu, J., Guo, M., Aslam, M., and Cao, S. (2020). Genome-wide characterization and expression profiling of Eucalyptus grandis HD-Zip gene family in response to salt and temperature stress. BMC Plant Biol. 20:451. doi: 10.1186/s12870-020-02677-w
Zhang, M., Bai, R., Nan, M., Ren, W., Wang, C., Shabala, S., et al. (2022). Evaluation of salt tolerance of oat cultivars and the mechanism of adaptation to salinity. J. Plant Physiol. 273:153708. doi: 10.1016/j.jplph.2022.153708
Zhang, S., Haider, I., Kohlen, W., Li, J., and Ouwerkerk, P. (2012). Function of the HD-Zip I gene OsHOX22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 80, 571–585. doi: 10.1007/s11103-012-9967-1
Zhao, Y., Ma, Q., Jin, X., Peng, X., Liu, J., Deng, L., et al. (2014). A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol. 55, 1142–1156. doi: 10.1093/pcp/pcu054
Keywords: HD-ZIP, Lotus japonicus, gene expression, LjHDZ7, salt stress
Citation: Wang D, Gong Y, Li Y and Nie S (2022) Genome-wide analysis of the homeodomain-leucine zipper family in Lotus japonicus and the overexpression of LjHDZ7 in Arabidopsis for salt tolerance. Front. Plant Sci. 13:955199. doi: 10.3389/fpls.2022.955199
Received: 28 May 2022; Accepted: 12 August 2022;
Published: 14 September 2022.
Edited by:
Jin-Lin Zhang, Lanzhou University, ChinaReviewed by:
Rahat Sharif, Yangzhou University, ChinaCopyright © 2022 Wang, Gong, Li and Nie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuming Nie, bmllc2h1bWluZ0AxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.