- 1Department of Entomology and Plant Pathology, North Carolina State University, Raleigh, NC, United States
- 2Department of Plant Sciences, University of the Free State, Bloemfontein, South Africa
- 3Syngenta (Pty.) Ltd., Sensako Research Station, Napier, South Africa
- 4Global Wheat Program, International Maize and Wheat Improvement Center (CIMMYT), Ankara, Turkey
- 5Food and Agriculture Organization, Riyadh, Saudi Arabia
- 6Agrotechnological Department, Omsk State Agrarian University, Omsk, Russia
- 7Biotrigo Genética, Passo Fundo, Brazil
- 8Department of Crop and Soil Sciences, North Carolina State University, Raleigh, NC, United States
- 9United States Department of Agriculture, Agricultural Research Service, Raleigh, NC, United States
The globally distributed causal agent of powdery mildew on wheat, Blumeria graminis f. sp. tritici (Bgt), is one of the most rapidly adapting plant pathogens and requires monitoring for shifts in virulence to wheat resistance (Pm) genes. Virulence frequencies were assessed in a total of 346 Bgt isolates from several countries that had either lately recorded increasing powdery mildew epidemics (Brazil, South Africa, and Australia) or not recently been surveyed (Turkey and Russia). The results were compared to previously published surveys of United States and Egyptian Bgt (390 isolates). Many of the Pm genes that have potentially been employed longer (Pm1a–Pm17) were shown to have lost effectiveness, and the complexity of virulence to those genes was higher among Brazilian isolates than those from any other country. Some cases of high virulence frequency could be linked to specific Pm gene deployments, such as the widespread planting of cultivar Wyalkatchem (Pm1a) in Australia. Virulence was also assessed to a set of Pm genes recently introgressed from diploid and tetraploid wheat relatives into a hexaploid winter wheat background and not yet commercially deployed. The isolate collections from Fertile Crescent countries (Egypt and Turkey) stood out for their generally moderate frequencies of virulence to both the older and newer Pm genes, consistent with that region’s status as the center of origin for both host and pathogen. It appeared that the recently introgressed Pm genes could be the useful sources of resistance in wheat breeding for other surveyed regions.
Introduction
Wheat (Triticum aestivum) is one of the most important global staple crops in area (218 million hectares), as primary calorie source (18%), and as protein source (19%) (FAOSTAT, 2017), and the diseases of wheat have become a global concern, causing ∼13% annual losses in yield (Oerke, 2005; Savary et al., 2019). While the area of wheat cultivation has not increased significantly over the last 50 years, global wheat production tripled during that time (FAOSTAT, 2017). This increase was due to the widespread application of “Green Revolution” practices, such as synthetic fertilizers, pesticides, and irrigation (Tilman et al., 2001; Zhu and Chen, 2002; Chen et al., 2007; Bradley and Thomas, 2019), as well as genetic gains made with modern wheat cultivars (Lantican et al., 2005; Reynolds and Borlaug, 2006). Such changes have resulted in large-scale, dense plantings of genetically uniform, semi-dwarf wheat cultivars, and increasing risk of epidemics caused by several pathogens (Bennett, 1984; Imani et al., 2002; Van de Wouw et al., 2009; McDonald and Stukenbrock, 2016).
Powdery mildew, caused by the ascomycete Blumeria graminis f. sp. tritici (Bgt), is among the top seven pathogens that affect global wheat (Triticum spp.) production (Savary et al., 2019). Depending on the climate zone, Bgt epidemics commence with the release of conidia from mycelium on infected host plants or ascospores from sexual fruiting bodies (chasmothecia) that allow the pathogen to perennate during hot dry seasons or cold winters (Glawe, 2008; Zou et al., 2018b). With the start of milder weather in autumn or spring, primary Bgt inoculum is dispersed to newly sown wheat crops. An increase in humidity provides optimal conditions for the development of asexually produced conidia and secondary infections that enable polycyclic disease development and larger epidemics (Te Beest et al., 2008).
Whereas powdery mildew has long been common in temperate regions and areas with humid, mild climates, it has recently become increasingly important in drier and hotter areas due to intensified agricultural production and in areas with shifting climates that favor earlier disease development (Tilman et al., 2001; Lantican et al., 2005; Oerke, 2005; Morgounov et al., 2011; Abdelrhim et al., 2018; Tadesse et al., 2019). The recent reports of more frequent powdery mildew epidemics in South Africa, Brazil, and parts of Turkey, Russia, Kazakhstan, and Australia exemplify this trend (D. Lesch, Z. Pretorius, W. Boshoff, P. Kuhnem, A. Morgounov, personal observation; Costamilan, 2005; Morgounov et al., 2011; Golzar et al., 2016).
The obligate-biotrophic lifestyle of Bgt is characterized by the infection of epidermal leaf cells via a specialized infection structure, the appressorium, and retrieval of nutrients employing a specialized feeding structure, a haustorium (Zhang et al., 2005). This highly specialized form of parasitism requires biotrophic pathogens such as Blumeria to employ a set of secreted effector proteins to interfere with the host’s immune system and facilitate colonization (Wicker et al., 2013; Mueller et al., 2019). Although the exact function of these effectors is not known, genetic mapping and characterization of a subset of these genes, such as AvrPm1a, AvrPm2, and AvrPm3, has shown that they are the triggers of plant immune reactions following their recognition by cognate host resistance genes (Bourras et al., 2015, 2019; Praz et al., 2017; Hewitt et al., 2021).
Compared to the highly effective, durable powdery mildew resistance conferred by the mlo gene in barley (Acevedo-Garcia et al., 2017; Kusch and Panstruga, 2017), powdery mildew resistance in wheat is limited to partial, race-non-specific resistance and race-specific, nucleotide-binding leucine-rich repeat-type resistance genes, referred to as Pm (powdery mildew) genes (Johnson et al., 2003; Krattinger et al., 2009; Alam et al., 2011; Cowger et al., 2012; Moore et al., 2015; Kang et al., 2020). Although over 100 Pm alleles have been mapped to 63 different loci in wheat (Lu et al., 2020), a much smaller number of Pm genes have been deployed commercially, likely due to fungicide availability, a breeding emphasis on yield, and in some cases linkage drag.
Due to the high capacity of B. graminis to overcome host resistance (McDonald and Linde, 2002; Spanu, 2012; Mueller et al., 2019), many Pm genes have lost effectiveness against the pathogen a few years after their deployment; e.g., Pm3a (Niewoehner and Leath, 1998) and Pm4a (Parks et al., 2008) in the mid-Atlantic region of the United States. In that same region, another example was a shift in the Bgt population to Pm17 virulence that occurred within 5 years of widespread planting of cultivars bearing that gene (Cowger et al., 2009, 2018).
Pm genes remain a key component of sustainable disease control in wheat. Thus, management strategies to counter the appearance of virulent Bgt strains have relied on the introgression of novel Pm genes from wild relatives into the germplasm of breeding programs, and the replacement of cultivars containing ineffective Pm genes with others bearing previously unused resistance genes (Xu et al., 2015; Wiersma et al., 2017; Zou et al., 2018a; Li et al., 2019b; Tan et al., 2019; Alemu et al., 2021).
The goal of this study was to assess the virulence of Bgt in several major wheat-growing countries, including parts of Brazil, South Africa, and Australia, where the disease has become an emerging problem. Also included were regions of Turkey, Russia, and Kazakhstan where the disease has long been endemic, but the pathogen population had not been thoroughly surveyed in the past decade. Bgt populations were tested for virulence against a differential set of 19 wheat lines, each carrying a single Pm gene, including Pm genes that had potentially been in commercial use for a relatively longer time and Pm genes that were recently introgressed into hexaploid wheat from diploid and tetraploid wild relatives. The goals were to compare the efficacy of Pm genes among geographic regions and to determine whether and where the new genes could be useful sources of resistance for breeding programs.
Materials and Methods
Sampling of Isolates
Samples of infected wheat bearing chasmothecia of Bgt were collected from 33 locations in Brazil, South Africa, Turkey, Russia, Kazakhstan, and Australia from 2016 to 2019 (Table 1; Supplementary Table 1; Figure 1). At each location, field samples were collected at several places within a field, with each field sample consisting of between 8 and 15 wheat leaves. Exceptions to this occurred at Omsk, Russia, where each sample was collected from a plot of a breeding line within a larger breeding nursery, and in Australia, where only one or two single-pustuled isolates were derived from conidial samples per region (Table 1 and Supplementary Table 1).
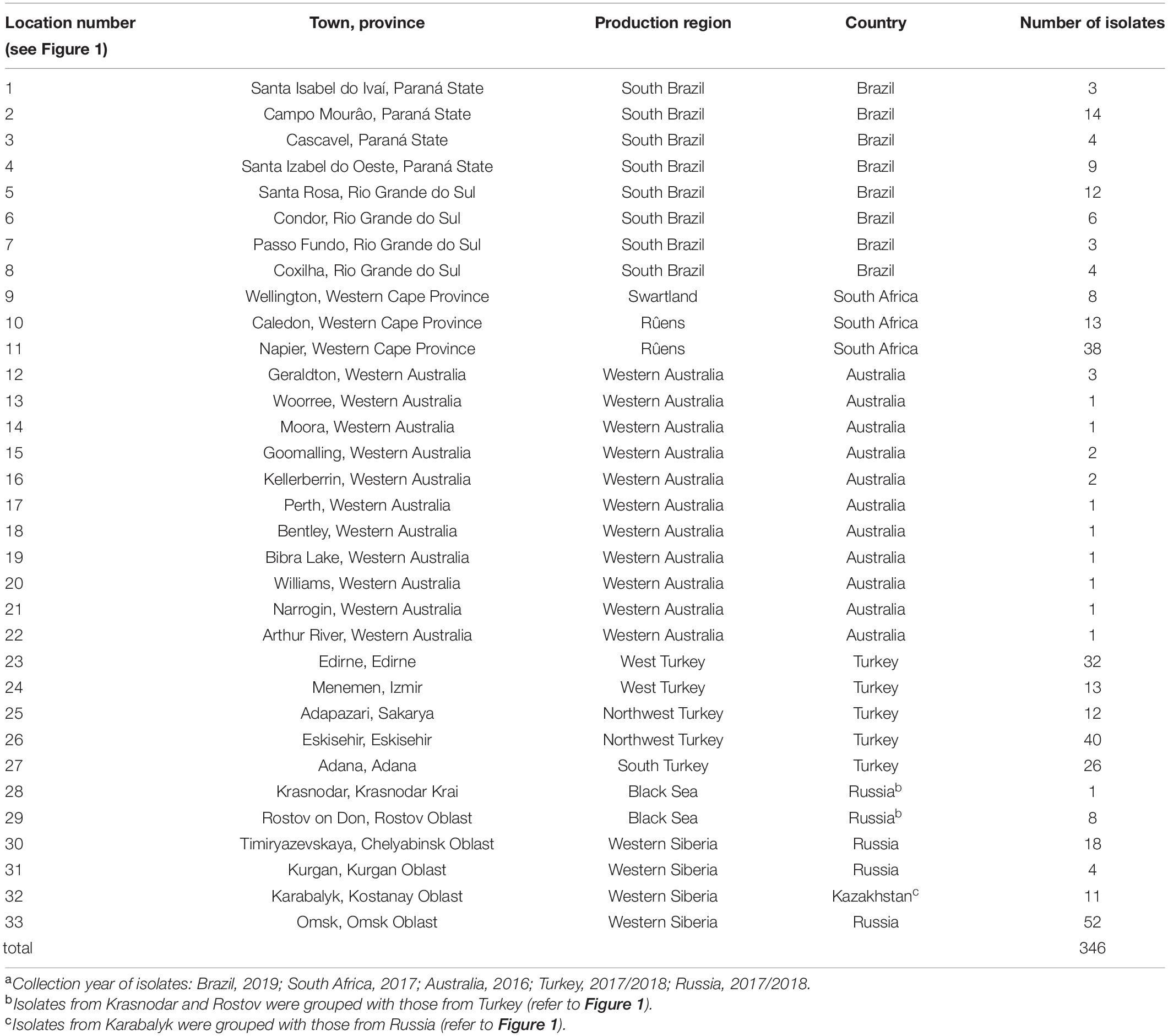
Table 1. Origins of 346 Blumeria graminis f. sp. tritici isolates evaluated for virulence, including closest town and province, region, country of the collected field samples, and number of isolates recovered.a
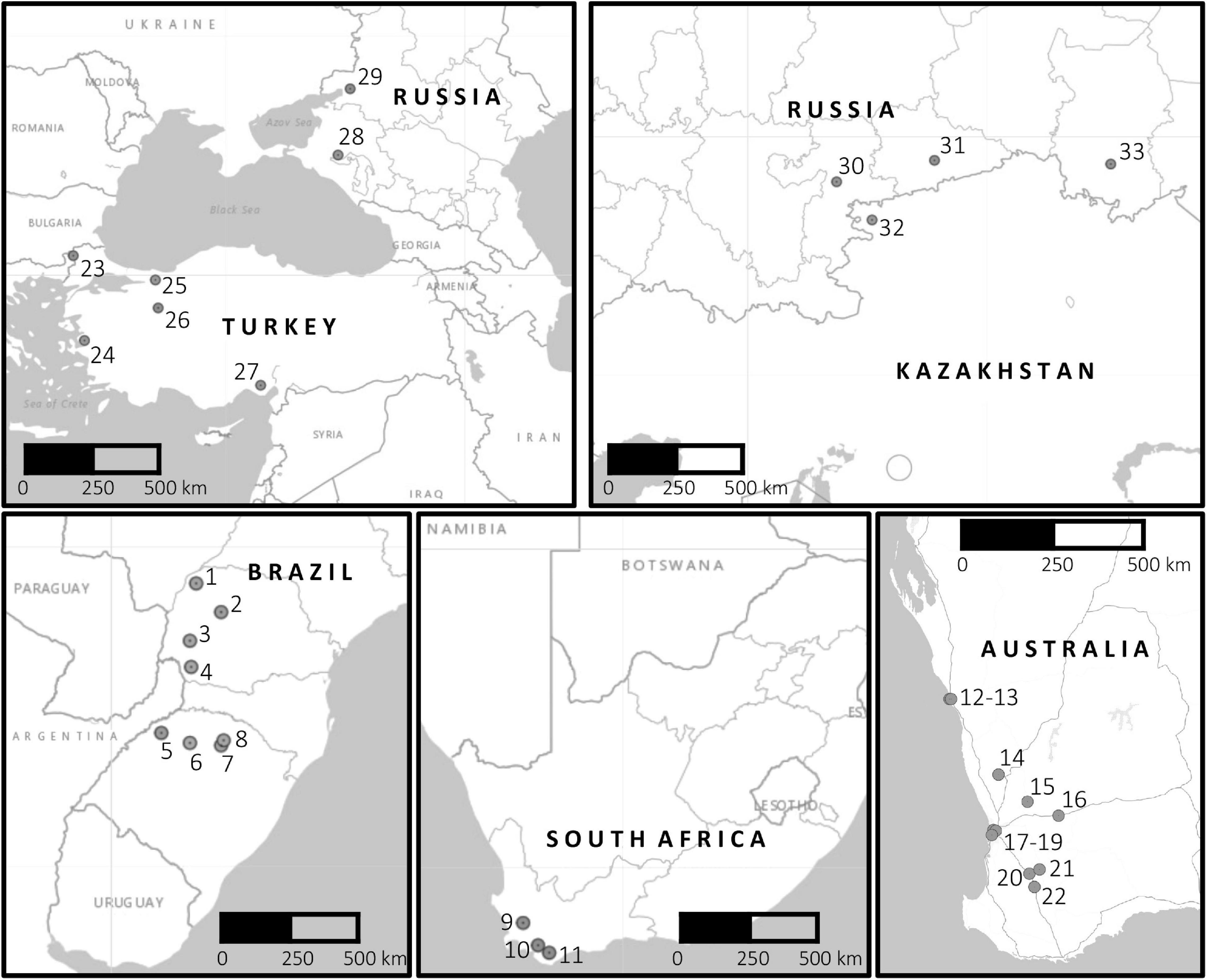
Figure 1. Locations of origin of 346 Blumeria graminis f. sp. tritici isolates from six countries tested for virulence. Location numbers on the map correspond to the numbers in Table 1.
Recovery of Isolates
Field samples were shipped in paper envelopes and after receipt were stored at 18°C for 30 days to facilitate maturation of chasmothecia (Legler et al., 2012). For the derivation of pure cultures, the protocol of Abdelrhim et al. (2018) was followed: infected leaf samples were cut into 5-cm-long segments, which were then placed in a moist chamber for differentiation of chasmothecia, followed by the ejection of ascospores onto detached segments of 10-day-old primary leaves of susceptible cultivar Jagalene floated on 0.5 w/w% water-agar containing 0.0053 mg L–1 benzimidazole in a 60 mm x 15 mm petri dish. Plates with infected Jagalene leaf segments were incubated in a growth chamber for 10 days at 17°C with a 12-h photoperiod, using fluorescent lamps.
After pustules appeared on infected Jagalene, pure cultures were obtained in two consecutive rounds of single-spore isolation, using an autoclaved paintbrush and subculturing on fresh Jagalene leaves under the conditions described above. Up to three single-spored isolates were obtained from each of the 5-cm leaf segments if the colonies that arose from the ascospore infections were at least 2 cm apart. This was done because the viability of chasmothecia in some samples was low, and only a fraction of the 5-cm leaf samples yielded successful primary ascospore infections. The pure isolates were cultured and increased every 10 days for several rounds on Jagalene by dispersing conidiospores from an infected leaf onto a fresh set of leaves with the aid of a spore-settling plastic cup.
Virulence Assay
The virulence profile of each isolate was determined by inoculating leaf segments of 19 differential plant lines, each bearing a single Pm gene. The plant lines represented two sets of Pm genes. The first was a set of genes that were introgressed and/or identified in common wheat earlier (1960s–2008) and may have been in commercial use relatively longer; these will hereafter be referred to as “older Pm genes” (Pm1a–Pm17, Table 2). The second was a set of genes exclusively sampled from wild, diploid, and tetraploid wheat relatives, originating in the Fertile Crescent, and more recently introgressed into North Carolina soft red winter wheat germplasm in the program of Dr. Paul Murphy, North Carolina State University. These genes will be referred to here as “newer Pm genes” (Pm25–Pm53, Table 2).
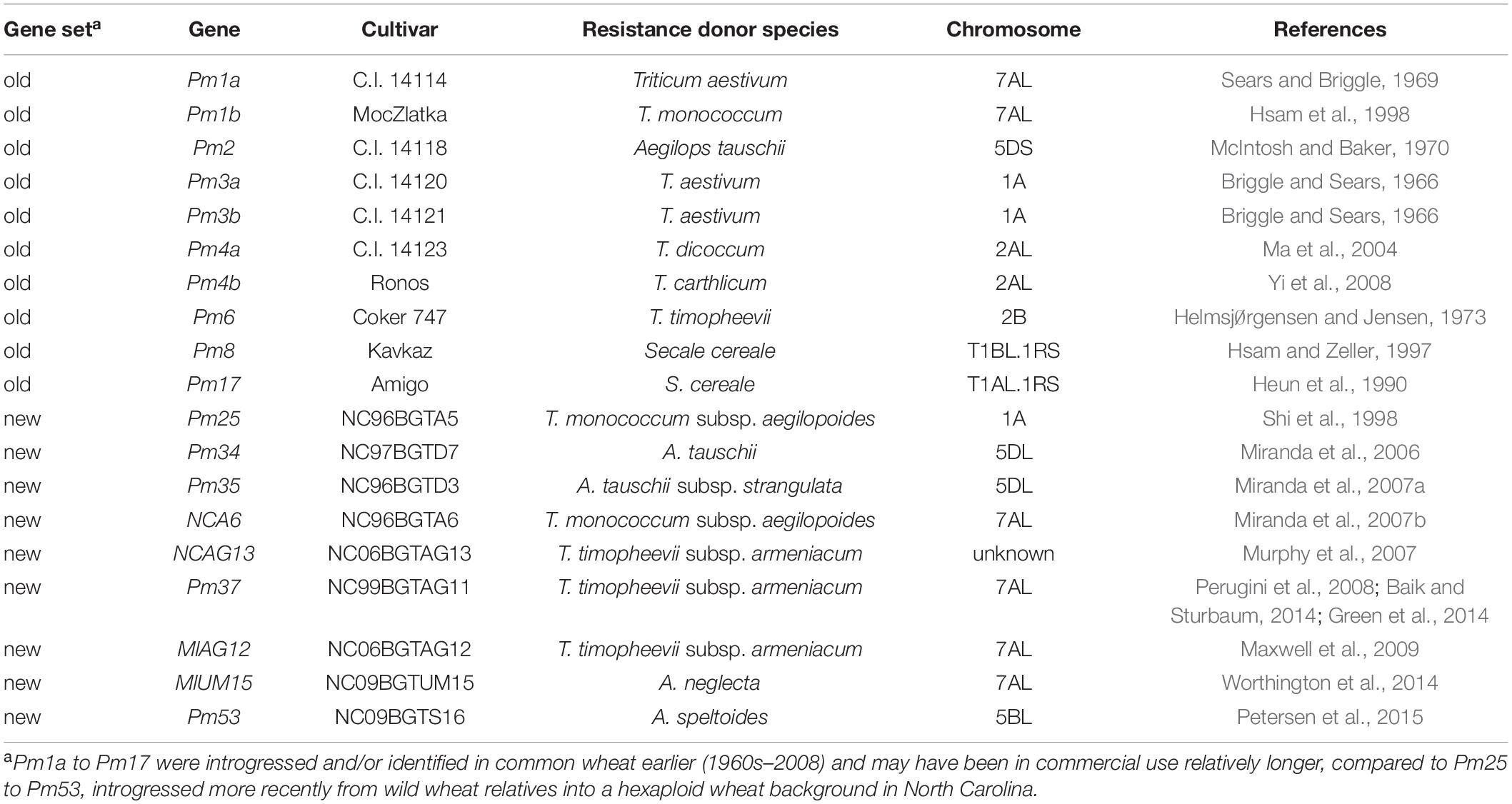
Table 2. Pm genes and differential cultivars used in virulence tests of 346 Blumeria graminis f. sp. tritici isolates from six countries.
Two 2.5-cm primary leaf segments of each 10-day-old differential line and four segments of Jagalene were placed alongside each other on large water-agar petri dishes. Jagalene leaf segments near the perimeter of the dish served as a susceptible control. Two dishes were prepared for each isolate, such that there were four replicate leaf segments for each combination of isolate and differential line. After 10 days of incubation under the conditions described above, the differential lines were scored for their disease reaction on a 0–9 scale adapted from Moseman et al. (1984): 0 = no symptoms, 1 = faint discoloration, 2 = necrotic lesions, 3 = necrotic lesions and traces of mycelium, 4 = chlorotic lesions with mycelium, 5 = one to two pustules of conidia, 6 = less than 20% of a leaf area covered with pustules, 7 = 20–50% coverage with pustules, 8 = medium-sized pustules and 50–90% coverage, 9 = large pustules and 100% coverage. Based on an average over the four leaf segments, each isolate was rated as avirulent for average scores of 0–4 (differential line was resistant), intermediately virulent for scores of 5–6, and fully virulent for scores of 7–9 (differential line was fully susceptible).
Assessment of Clonality
For the purpose of this study, over-representation of virulence haplotypes due to clonality from polycyclic increase in individual genotypes was undesirable. Isolate clonality was possible because sampling resolution was in some cases as high as two isolates being derived from the same 5-cm leaf segment of an infected wheat leaf sample. Potential clonality was assessed by comparing isolates from a given location for an exact match in their virulence profiles to the 19 Pm genes (Supplementary Table 1). If one or more isolates originated from the same plant sample and matched in their virulence profile, all but one of those isolates were excluded. A total of 47 isolates were matched in virulence profiles with at least one other isolate derived from the same location. Of them, 11 isolates were derived from the same plant as other isolates, and thus, two isolates from Brazil, six from South Africa, two from Turkey, and one from Australia were excluded.
Data Analysis
To make geographically broader inferences about the effectiveness of Pm genes, additional data matching the present Pm gene set were taken from recently published surveys in Egypt and the United States, which were conducted in the same laboratory as the current one, using the same methods. Data from Egypt were included to represent a wheat-growing region near the Fertile Crescent (Abdelrhim et al., 2018). Data from the United States consisted of virulence profiles of isolates of the Southeast, Mid-Atlantic, and Great Lakes regions, the United States wheat-growing regions most conducive to Bgt epidemics; these were thus a subset of the isolates in the previous report (Cowger et al., 2018). Because the sampled location Karabalyk, Kazakhstan was close to the two Russian locations Timiryazevskaya and Kurgan (Figure 1), all of which are in a spring wheat region, data from these three locations were grouped together. Isolates from Russian locations Krasnodar and Rostov on Don were grouped with those from Turkey, because these two locations were representative of the winter wheat region close to Turkey and remote from the sampled locations in the Russian spring wheat region.
For a given country (as defined above and shown in Table 1), frequencies of virulence to each Pm gene were calculated as the percentage of isolates virulent to each Pm-gene line. Global frequencies of virulence to Pm genes were estimated as unweighted means using countries’ mean virulence frequencies, recognizing the limitations imposed by unsampled geographic regions and varying sample sizes. Virulence complexity was calculated as the percentage of Pm genes to which an isolate was virulent. To test whether mean virulence complexity was significantly higher for the older set of Pm genes than for the newer set, mean virulence complexity of each country was calculated separately for older and newer genes and then compared in one-tailed Student’s t-test (α = 0.01), using the function “t.test” in R. Virulence complexity values allowed a within-country comparison of older and newer sets of Pm genes for their effectiveness, as well as an overall comparison between countries. For the comparison of countries’ mean virulence complexity, residual analysis was applied to test for a linear relationship between the country as the independent variable and virulence complexity as the dependent variable. Arcsine-transformed data fitted a linear regression model best and were chosen for testing whether means differed significantly. The data were fitted to a linear model using the modeling function “lm” in R. For statistical separation of countries’ virulence complexity means, a Dunnett–Tukey–Kramer pairwise multiple comparison test, implemented in the DTK package in R, was used with a familywise error rate of α = 0.05.
Genotyping
To test for the presence of certain Pm genes in cultivars from which isolates were sampled and/or that have been popular in Brazil and Australia, a genetic evaluation was conducted on 10 Brazilian spring wheat cultivars, provided by Biotrigo Genetica, and two Australian winter wheat cultivars provided by InterGrain through the Australian Grains GeneBank (Supplementary Table 2). DNA was extracted from three primary leaves per cultivar as biological replicates, using the EZNA HP Plant DNA Mini Kit D2485-02 (Omega Bio-Tek, Norcross, GA, United States), following the manufacturer’s protocol for dried specimens. DNA samples were tested by endpoint genotyping using the PCR Allelic Competitive Extension (PACE) assay (3cr bioscience) and kompetitive allele-specific PCR (KASP) markers to detect polymorphisms linked to Pm1a, Pm6, and Pm8 (Supplementary Table 3A; Sarinelli, 2017; Sarinelli et al., 2019). Gene-specific markers were unavailable for the other Pm genes used in this study.
To provide positive controls, DNA was extracted from the following genotypes: C.I. 14114 for Pm1a (Sears and Briggle, 1969), Coker 747 for Pm6 (Helmsj∅rgensen and Jensen, 1973), and Kavkaz for Pm8 (Hsam and Zeller, 1997). The cultivar Shirley was included as a control representing a modern United States cultivar and for testing the KASP markers for Pm1a and Pm8, which that cultivar is believed to possess (Hsam and Zeller, 1997; Baik and Sturbaum, 2014; Green et al., 2014). DNA from universally susceptible cultivar Jagalene was used as a negative control for all genes.
For endpoint genotyping, each DNA sample was analyzed in two independent reactions amplified on a QuantStudio 3 real-time PCR cycler (Thermo Fischer Scientific, Waltham, MA, United States), using thermal cycling conditions recommended by the manufacturer in a reaction volume of 10 μl (Supplementary Table 3B). Each reaction was composed of 10 ng of sample DNA, 0.138 μl PACE assay mix, 1.5 μM ROX as a reference dye, 30 μM of a common reverse primer, and each 12 μM of two allele-specific primers. The allele-specific primers contained unique 5′ tail sequences that corresponded to either the HEX (resistance allele) or FAM (susceptible allele) fluorescent cassettes present in the master mix. The excitation maxima (FAM = 493 nm and HEX = 538 nm) and emission maxima (FAM = 517 nm and HEX = 554 nm) of the dyes deviated by similar amounts from the wavelength spectrum of the QuantStudio 3 machine for excitation [channel x1 = 470 nm (FAM), channel x2 = 520 (HEX)] and emission [channel m1 = 520 nm (FAM), channel m2 = 558 (HEX)]. Real-time PCR reporter fluorescence, measured at 33–35 cycles and normalized to ROX, was used for allele discrimination using the “analyze” function in Design and Analysis Software 2.6.0 (Thermo Fischer Scientific, Waltham, MA, United States). Assays for Pm6 and Pm8 were rerun on additional DNA samples and new reagents to exclude any source of contamination. The two independent assays showed similar allele discrimination patterns for the tested samples.
Results
In total, 346 isolates were derived from samples originating in six countries: 55 from Brazil, 59 from South Africa, 123 from Turkey, 83 from Russia, 11 from Kazakhstan, and 15 from Australia (Table 1 and Figure 1). As described in the methods, previously published data on virulence to the same set of 19 Pm genes from 70 Egyptian isolates and 320 United States isolates (Abdelrhim et al., 2018; Cowger et al., 2018) were compared with the data obtained in this survey. Virulence to Pm genes varied from complete fixation (all isolates were virulent) to total absence of virulence across countries and between Pm genes (Table 3).
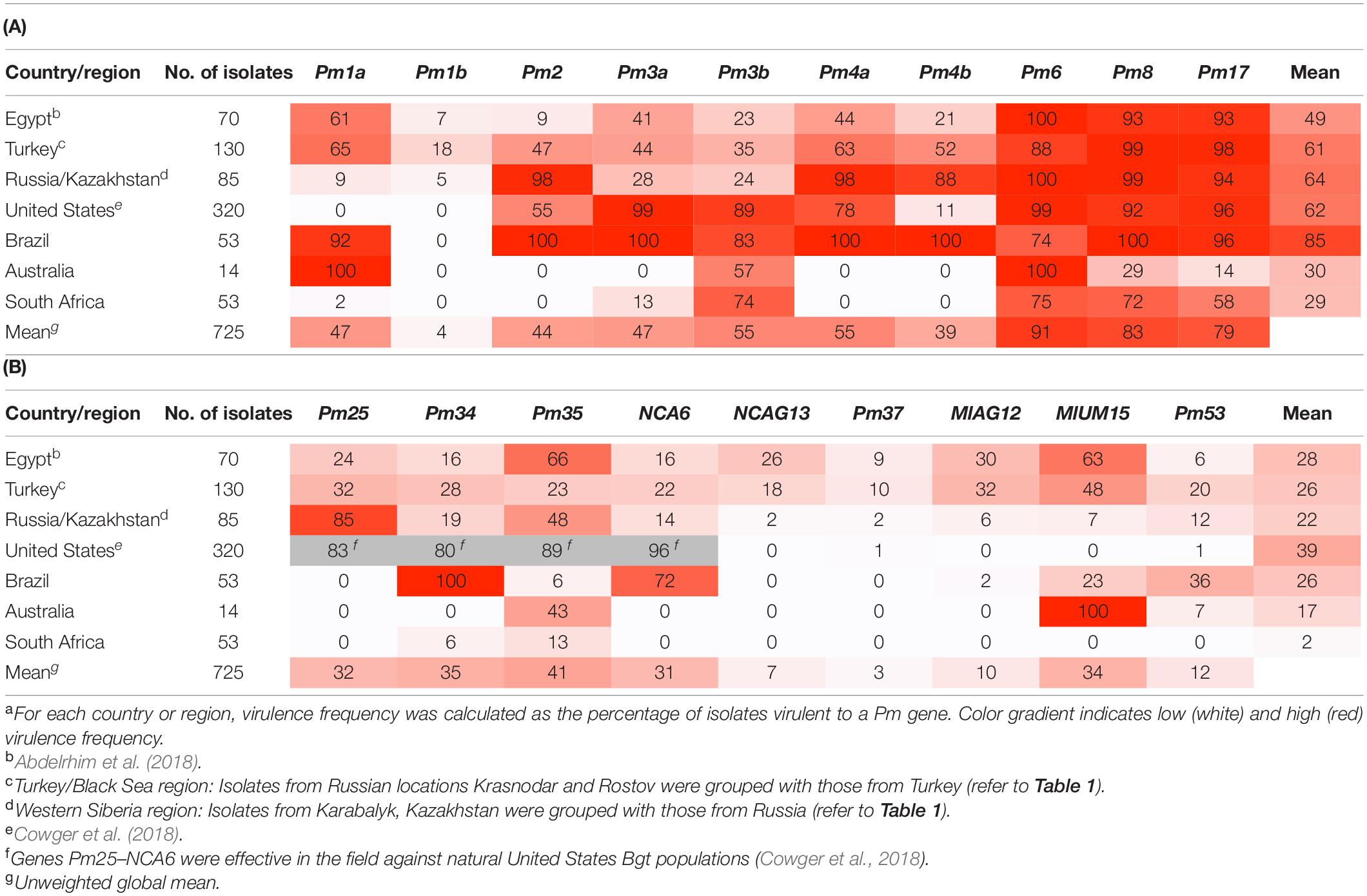
Table 3. Virulence frequencies of Blumeria graminis f. sp. tritici isolates collected from six countries, compared to previously published frequencies in the United States and Egypt, for (A) ten older Pm genes, identified earlier and potentially in use internationally, and (B) nine newer Pm genes, more recently introgressed into hexaploid wheat in North Carolina.a
Older Pm Genes (Pm1a–Pm17)
Among the older Pm genes, generally high frequencies of virulence were observed, although this varied considerably by country (Table 3A). Across all countries, the highest virulence frequencies were recorded for Pm6, Pm8, and Pm17, with most isolates virulent to at least one if not all of those genes. Beginning around the center of origin of Bgt, isolates from Turkey and the southern Russian Black Sea region (locations 28 and 29, Figure 1) exhibited comparatively moderate frequencies of virulence to the older Pm genes (35–65%), except for Pm1b, Pm6, Pm8, and Pm17 (Table 3A). Virulence to Pm1b was low in all surveyed populations, but the frequency was highest in Turkish isolates at 18%, in line with the generally intermediate levels of virulence among these isolates. Also in the Fertile Crescent, a pattern of mostly moderate virulence frequencies had been observed among Egyptian isolates previously tested on these older Pm genes (Abdelrhim et al., 2018).
Moving farther afield, isolates from the spring wheat region of Russia and Kazakhstan (Western Siberia) exhibited a different pattern than that found for Turkey and the Black Sea winter wheat region. Isolates from these Central Asian regions possessed near-complete virulence to Pm2, Pm4a, and Pm4b, but their virulence to Pm1a, Pm1b, Pm3a, and Pm3b was low (<30%).
In the diaspora of countries most remote from the Fertile Crescent, frequencies of virulence to the older Pm genes tended to be either high or low. For example, Pm1a was either virtually ineffective (Brazil, Australia) or fully effective (United States, South Africa). Among Brazilian isolates, virulence frequencies to the older genes were all ≥75% except for Pm1b, and for six of those ten genes (Pm2, Pm3a, Pm4a, Pm4b, Pm8, and Pm17), virulence was ≥95%, indicating a likely complete loss of effectiveness when confronted with natural Bgt populations in the field.
In South Africa, virulence to the majority of older Pm genes was absent or low (0–13%); however, as mentioned above, Pm3b, Pm6, Pm8, and Pm17 showed moderate to high virulence frequencies (58–75%). A distinctive virulence pattern was seen in the Australian sample, with Pm1a and Pm6 being completely ineffective, whereas virulence to Pm8 and Pm17 was relatively low (14 and 29%, respectively). Overall, virulence to seven of the ten older Pm genes was absent or <30% in Australia. However, these results should be treated with caution as the Australian sample was small.
For the older Pm genes, average virulence complexity (the percentage of Pm genes to which isolates of a country were virulent) was significantly higher in Brazil (85%) than in any other country (Figure 2A). South Africa and Australia had the lowest mean complexity of virulence to the older genes, whereas complexity in Egyptian, Turkish, Russian, and United States collections was intermediate.
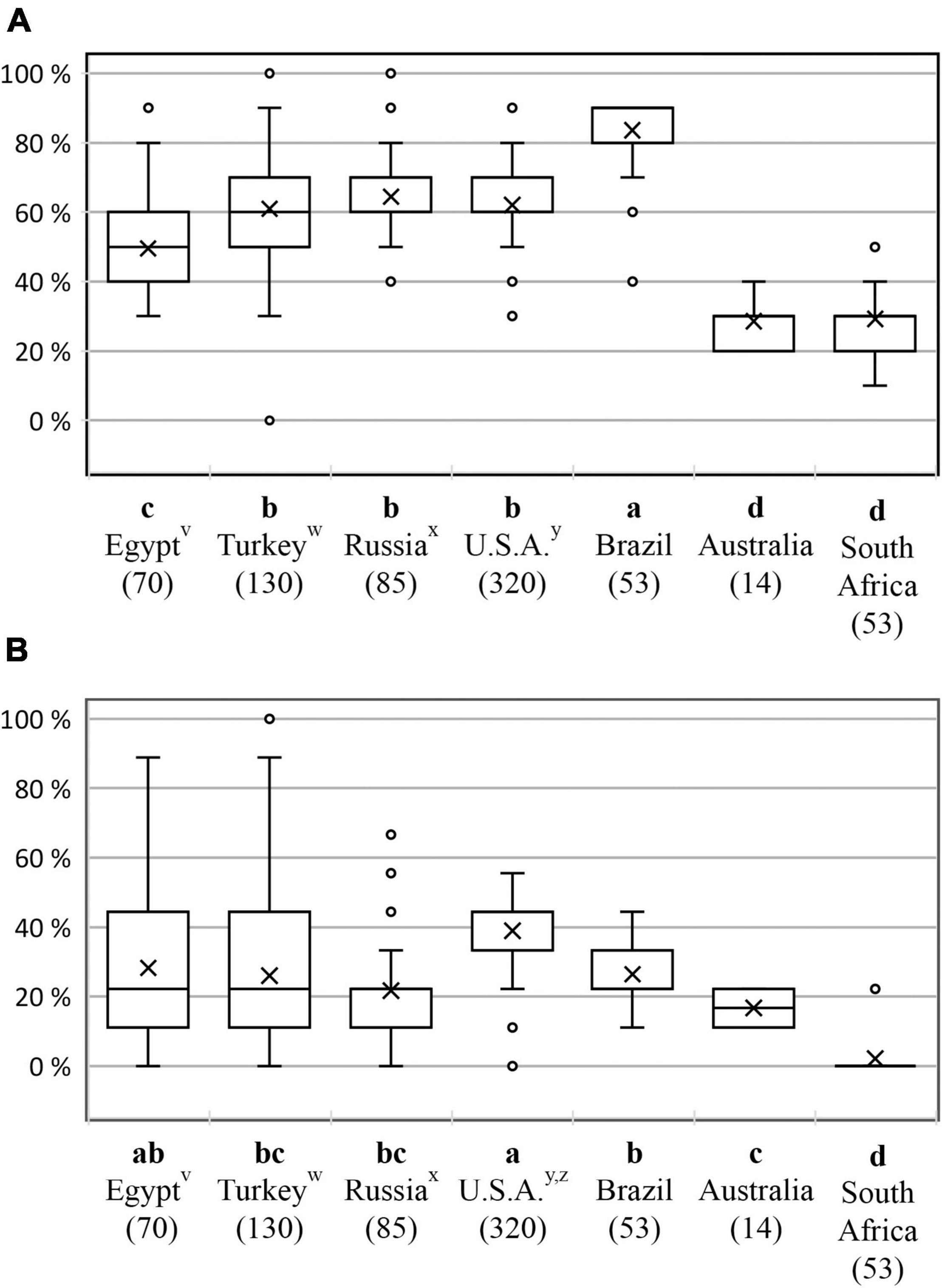
Figure 2. Complexity of virulence in recently phenotyped Blumeria graminis f. sp. tritici populations from several countries compared to previously published virulence data for the United States and Egypt, for (A) 10 lower-numbered, older Pm genes (Pm1a–Pm17) and (B) nine newer Pm genes recently introgressed into hexaploid wheat from wild relatives (Pm25–Pm53). Virulence complexity was calculated as mean percentage of Pm genes to which isolates were virulent. Within a panel, countries topped by the same letter did not have significantly different virulence complexities (p < 0.05). Within each box, medians of virulence complexity are indicated by lines and means by crosses. vAbdelrhim et al. (2018). wTurkey/Black Sea region: isolates from Russian locations Krasnodar and Rostov were grouped with those from Turkey (refer to Table 1). xWestern Siberia region: isolates from Karabalyk, Kazakhstan were grouped with those from Russia (refer to Table 1). yCowger et al. (2018). zGenes Pm25–NCA6 were effective in the field against a natural United States Bgt population (Cowger et al., 2018); see text.
Because isolates from Brazil had comparatively high frequencies of virulence to the older Pm genes, available KASP markers were employed to test whether cultivars from which isolates were derived possessed Pm1a, Pm6, or Pm8. Likewise, the fixation of virulence to Pm1a and Pm6 observed in the Australian Bgt collection was investigated by assessing whether two popular Australian cultivars (Wyalkatchem and Mace) contained Pm1a or Pm6. Endpoint genotyping for Pm1a using three diagnostic KASP markers confirmed the presence of a Pm1a allele conferring resistance to powdery mildew in C.I. 14114 (Pm1a control), Wyalkatchem and Shirley, whereas all other cultivars possessed the alternate allele, indicating the absence of Pm1a (Supplementary Table 2 and Supplementary Figure 1). Tests for Pm6 and Pm8 showed that only Shirley and the positive controls (cultivars Coker 747 and Kavkaz, respectively) had the resistance allele, whereas all other cultivars possessed the alternate allele.
Newer Pm Genes (Pm25–Pm53)
Overall, virulence to the recently introgressed set of Pm genes was low relative to the older genes (Table 3B). Across all countries, unweighted mean virulence frequencies were 31–41% for five Pm genes (Pm25, Pm35, Pm34, NCA6, and MlUM15) and ≤12% for an additional four genes (NCAG13, Pm37, MlAG12, and Pm53).
Isolates from the Fertile Crescent countries Egypt and Turkey (along with the adjacent Russian winter wheat locations 28 and 29) tended to exhibit low to intermediate frequencies of virulence to the newer Pm genes. For example, among Turkish isolates, some virulence was evident to all newer Pm genes, with virulence frequencies of 10–48% (Table 3B). The consistently moderate virulence in Turkish isolates was comparable to the virulence profiles of Egyptian isolates, although in the latter virulence to Pm35 and MlUM15 exceeded 60%.
Moving northeast to the Russian spring wheat region and Kazakhstan, virulence frequencies were still in the low to moderate range for Pm34, Pm35, NCA6, and Pm53, but high for Pm25 and very low for the four remaining genes (NCAG13, Pm37, MlAG12, and MlUM15). Virulence to the newer genes was generally low in isolates from countries where wheat powdery mildew epidemics have only recently increased. In Australia, virulence to the newer Pm genes was virtually absent, with the exceptions of virulence to Pm35 (43%) and MIUM15 (100%). Similarly, in South Africa, isolates were mostly avirulent to the newer Pm genes, except for Pm35 with 13%. Whereas Brazilian isolates displayed virulence frequencies ≤6% to five new Pm genes (Pm25, Pm35, NCAG13, Pm37, and MlAG12), these isolates were fixed for virulence to Pm34, and 72% were also virulent to NCA6.
Across countries, mean virulence complexity was significantly lower for the newer set of Pm genes than for the older set, as would be expected (p < 0.05; Figure 2B). The mean virulence complexity to the newer set was similar for Egypt, Turkey, Russia, Brazil, and Australia and ranged from 17 to 28%, whereas South Africa could be statistically separated from all countries with a virulence complexity of 2%. In the eastern US, the mean virulence complexity of 39% to the newer Pm genes was nearly exclusively due to the virulence frequencies ≥83% for Pm25, Pm34, Pm35, and NCA6 (Table 3B and Supplementary Figure 2A), although as previously reported (Cowger et al., 2018), those genes were fully effective against field populations in North Carolina and remain effective (C. Cowger, personal observations). The reason for the difference between the results of the detached leaf assay and field performance of these genes remains unknown. In contrast, there was a virtual absence of virulence (≤11%) among the United States isolates to the remaining five newer genes (Table 3B and Supplementary Figure 2B).
Of the isolates that were found to be virulent to newer Pm genes in each country, more than half showed intermediate levels of virulence, except for Brazil and Australia (Supplementary Figure 3B). Particularly, a high proportion of intermediate virulence was recorded for virulent South African isolates to older and newer Pm genes (65 and 100%, respectively), suggesting there was some level of resistance conferred by these Pm genes that had not been completely overcome.
It was noteworthy that in isolates of most countries (Egypt, Australia, United States, and South Africa), there was a near-perfect co-segregation of virulence/avirulence to Pm1a and MlUM15 (Supplementary Table 4). In contrast, however, more frequent recombination between these two virulence traits was observed in populations from Brazil and Turkey, with 70 and 19% recombinants, respectively; i.e., some isolates were virulent to Pm1a but not to MlUM15, and vice versa. MlUM15 mapped close to the multi-allelic Pm1 locus on wheat chromosome 7AL (Worthington et al., 2014).
Discussion
Frequencies of virulence to powdery mildew resistance genes were investigated and compared for several global wheat production regions across the world where the disease was either relatively long endemic or recently emergent. The results indicated a widespread loss of effectiveness of many older Pm genes, likely in at least some cases because of greater exposure of pathogen populations to these Pm genes, or their genetic linkage to other resistance genes that have been deliberately used in commercial cultivars to control other diseases, such as stem rust and leaf rust. Fortunately, judging from the present results, a number of newer Pm genes more recently introgressed from wild relatives of wheat would likely be effective across most surveyed countries. However, many of these newly introgressed genes were already associated with moderate frequencies of Bgt virulence in countries closer to the Fertile Crescent landscapes where accessions contributing the genes were collected. This is likely due to standing variation for virulence in the Bgt populations that have co-evolved with those wild wheat relatives.
While it is nearly impossible to determine the relative area and duration of use of a Pm gene, widespread use of particular Pm genes appears to have driven the selection of virulent strains in local Bgt populations, such as virulence to Pm3a, Pm4a, Pm6, and Pm17 in the United States (Bennett, 1984; Niewoehner and Leath, 1998; Cowger et al., 2009, 2018). Moreover, the gene pool giving rise to commercial wheat cultivars has become smaller and is nowadays extensively shared among countries (Balfourier et al., 2019).
It is conceivable that some older Pm genes were introduced to the surveyed countries in common wheat lines distributed through efforts of the International Maize and Wheat Improvement Center (CIMMYT) and collaborating national agricultural research institutions (Evenson and Gollin, 2003; Lantican et al., 2005; Joukhadar et al., 2017; Kirsten and Nhemachena, 2017). Cultivars sampled in Turkish locations Menemen, Adapazari, and Adana represented mostly CIMMYT material (A. Morgounov, personal observation). Most of those wheat lines were developed for rust resistance, which may have led to an indirect selection of Pm genes in linkage (Lillemo et al., 2010). For example, selection for the rust resistance gene complex Sr31/Lr26/Yr9, translocated from rye into the B genome of hexaploid wheat, has been common in CIMMYT material and likely led to a co-selection of Pm8 (Hsam and Zeller, 1997; Mago et al., 2005).
However, the influence of CIMMYT germplasm is not the only factor contributing to the high levels of virulence to the older Pm genes. Winter wheat in Russian locations Krasnodar and Rostov often utilizes the Bezostaya-1 background, which contains the Lr34/Yr18/Sr57/Pm38 complex of genes (Morgounov et al., 2011; Rinaldo et al., 2017), as well as Pm8 in cultivars Kavkaz and Aurora derived from Bezostaya-1. This common background has probably helped to select for virulence at least to Pm8 (Bennett, 1984; Singrün et al., 2004). CIMMYT germplasm has not been important in cultivar development in the Russian-Kazakh spring wheat region (A. Morgounov, personal observation), yet virulence frequencies to genes such as Pm6, Pm8, and Pm17 were as high as those in Turkey. This indicated that these Pm genes may have also been present in germplasm used for breeding in this spring wheat region.
Historically, South African wheat germplasm has contained a low proportion of CIMMYT material (Byerlee and Moya, 1993; Trethowan et al., 2005). However, it is possible that high virulence frequencies to certain older genes, such as Pm8 and Pm17, were driven by the few cultivars derived from CIMMYT germplasm and released in that country. For example, cultivar Kavkaz (Pm8) and Amigo (Pm17) were used in breeding South African cultivars released in the 1980s and 1990s, such as Gamtoos (Kavkaz/Buho’S’//Kalyansona/Bluebird), Letaba (Warrior5*/Agent//Kavkaz), and Riemland (Flamink/Amigo) (Heisey et al., 2002; Smit et al., 2010; Kirsten and Nhemachena, 2017).
The development of Australian cultivars has drawn heavily from CIMMYT material since 1960 (Joukhadar et al., 2017), again with a focus on introgression of rust resistance genes, some of which are linked to Pm genes (Cuddy et al., 2016). For example, the Lr20-Sr15-Pm1 locus has tightly linked alleles for resistance to leaf rust and powdery mildew (Neu et al., 2002). A succession of popular wheat cultivars that has dominated the Western Australian wheat market since 2008 was thought to have this resistance gene complex, although responses to powdery mildew or the presence of resistance alleles were heterogeneous (Cuddy et al., 2016; Park et al., 2020; GRIS, 2021). First released in 2003, Wyalkatchem dominated over 30% of the area sown to wheat until 2010, when it was replaced by its descendant Mace out of the cross Wyalkatchem/Stylet//Wyalkatchem. Mace occupied over 50% of wheat plantings from 2013 to 2016 (Zaicou-Kunesch et al., 2017). Third in this succession was Scepter, a cultivar related to Mace (A. Milgate, personal communication), which was released in 2015 and overtook Mace by market share in 2018 (Shackley et al., 2019).
In Western Australia, wheat powdery mildew has been recognized as a disease that requires annual control and monitoring for virulence to Pm genes (R. Lopez and A. Milgate, personal communication). Surveys from 2011 to 2014 detected high virulence to Pm1a, Pm6, Pm8, and Pm17, whereas virulence was low for Pm2, Pm3a, and Pm4a (Golzar et al., 2016). Although Golzar et al. (2016) rated mean disease scores on single-gene differential seedlings using a composite inoculum from 40 plant samples originating in various locations across Western Australia, their study and the present one found similar levels of virulence to Pm1a, Pm2, Pm3a, and Pm4a, whereas virulence was higher to Pm6 and lower to Pm8 and Pm17 in the current collection. Discrepancies between the two studies could be due to the lower sample size in the present one or to a virulence shift in the population. Although Golzar et al. (2016) proposed that most Australian cultivars lack Pm genes, Pm1a was identified in Wyalkatchem in this study using KASP markers. At the time of those authors’ survey in 2011–2014, when they observed high Bgt susceptibility in both Wyalkatchem and the Pm1a differential line Thew, the Australian Bgt population had probably already been selected for virulence to Pm1a due to widespread planting of Wyalkatchem. The cultivar Mace, the dominant wheat cultivar in Western Australia in 2013–2016, was found in this study to not possess Pm1a. Interestingly, the apparent fixation of Pm1a virulence in the present sample of Australian isolates from 2016 was apparently maintained despite reduced exposure to Pm1a after Wyalkatchem declined in market share starting in 2010.
While certain Pm genes are apparently ineffective in Australia and South Africa, the generally low virulence frequencies among Bgt populations in those countries to other Pm genes are likely due to a combination of three factors. First, as the relatively hot and dry climate of those regions is less conducive to cereal powdery mildew epidemics, Bgt populations have historically been small, leading to a lower probability of the appearance and establishment of virulent isolates (Tadesse et al., 2019). Second, most commercial wheat cultivars in South Africa and Australia seem to lack known Pm genes, other than Pm1a, so that selective pressure for virulence has likely been generally low (Sensako, 2022; A. Milgate, personal communication). And third, a founder effect may have occurred when a subsample of the ancestral Bgt population (Troch et al., 2014) was disseminated to those countries, probably with wheat when it was introduced in Australia by colonists (Joukhadar et al., 2017). This is supported by findings on B. graminis f. sp. hordei, the forma specialis that pathogenizes barley (Hordeum vulgare), for which genetic variability was low in Australian subpopulations (Kominkova et al., 2016).
On the other hand, it is possible that high levels of virulence to particular Pm genes in countries where wheat powdery mildew is emergent, such as Australia and South Africa, can be attributed to standing variation; i.e., the presence of particular virulence alleles in the founding Bgt populations. The Bgt gene pool has maintained a high degree of genetic diversity, even through the shift from wild to domesticated wheat genotypes (Wicker et al., 2013), and this may include virulence alleles. For example, virulence alleles of AvrPm17 were present in Bgt populations prior to commercial deployment of Pm17, which led to a rapid selection of virulent isolates and loss of effectiveness of the resistance gene in wheat (Mueller et al., 2021).
In contrast to other older Pm genes, globally low frequencies of virulence to Pm1b indicate that this gene has likely not been widely used or has only been used in combination with quantitative resistance or other Pm genes, which would render it more durable. Pm1b originated from a Triticum monococcum accession collected in the Suweida province of southern Syria (Valkoun and Mamluk, 1993). Exposure to T. monococcum (einkorn wheat) or its wild progenitor T. boeoticum (Harlan and Zohary, 1966; Heun et al., 1997) likely explains the finding of some Pm1b virulence (7–18%) in Bgt populations from Turkey and Egypt, Syria’s neighbors in the Fertile Crescent.
Interestingly, isolate collections from Turkey and Egypt stood out for their generally moderate levels of virulence to both the older and the newer Pm genes, even though the newer Pm genes have likely never been commercially deployed there. The newer genes were introgressed mainly in the past two decades from accessions of wild wheat relatives, e.g., NCA6 and Pm25 from wild einkorn wheat T. monococcum subsp. aegilopoides collected in the Karacadag Mountain range of Iraq (Shi et al., 1998; Miranda et al., 2007b), MlUM15 from Aegilops neglecta collected in Karamusa, Central Turkey (Worthington et al., 2014), and Pm34 from Aegilops tauschii collected in Iran (Murphy et al., 1999). NCAG13 and MlAG12 were detected in T. timopheevii subsp. armeniacum accessions, which are native to the Transcaucasian region, including Azerbaijan and Iraq (Murphy et al., 2007; Maxwell et al., 2009). These Pm genes are likely not yet widely present outside their regions of origin and the southeastern US, where they were introgressed into regionally adapted winter wheat germplasm. Thus, finding low to moderate frequencies of virulence in Turkey and Egypt suggests previous exposure of B. graminis strains to these Pm genes in these geographic regions.
Moderate frequencies are the signatures of a stabilizing or balancing selection for virulence, often found in natural ecosystems where wild host plants and pathogen populations have coevolved in a reciprocal genotype frequency-dependent manner (Leonard et al., 2004; Brown and Tellier, 2011). Our findings suggest that the isolates from Turkey and Egypt make up a genetically diverse population and resemble part of the ancestral, natural population of Bgt in the Fertile Crescent (Troch et al., 2014), the center of genetic diversity of wheat and its wild progenitors, which Bgt can infect (Dubcovsky and Dvorak, 2007; Ben-David et al., 2016). Even though wild wheat relatives comprise a small proportion of the wheat population in the Fertile Crescent nowadays (Özkan et al., 2010), intermixing occurs occasionally between B. graminis from wild grasses and Bgt from commercial wheat fields, enhancing allelic diversity in the Bgt population (Parks et al., 2009; Ben-David et al., 2016). This contrasts to subpopulations on other continents that may have experienced genetic drift and a loss of virulence diversity after migration to locations remote from their center of genetic diversity.
Estimates of virulence to Pm genes based on laboratory detached leaf assays should be considered carefully, since in a previous study, Pm25, Pm34, Pm35, and NCA6 were rated susceptible to Bgt in the laboratory assay although they were and remain effective in the field against North Carolina Bgt populations (Cowger et al., 2018; C. Cowger, personal observation). In addition, large proportions of isolates with intermediate virulence to these four newer Pm genes were recorded for many countries in this study, such as Turkey (Supplementary Figure 3B). In cases where this study indicated high virulence frequencies to some of the newer genes, these genes may still be effective in the field and should be tested there (e.g., Pm34 in Brazil). Also, intermediate reactions may indicate that virulence has been developing; i.e., to Pm genes that have been deployed widely in the sampling area but relatively recently, or genes that have only been deployed in some parts of the sampling area (Supplementary Table 5). In a similar vein, resistance to the Australian Bgt population conferred by Pm6 and Pm8 was confined to the adult-plant stage and was not observed at the seedling stage (Golzar et al., 2016). This also illustrates the importance of phenotyping in the field to complement laboratory studies with seedling leaf tissue.
Surprisingly, Brazilian isolates registered significantly higher complexity of virulence to the older Pm genes than those from any other country sampled, despite the relatively recent emergence of serious powdery mildew epidemics in Brazil. Sampling locations for this study targeted the main wheat growing states in the south of Brazil, Parana and Rio Grande do Sul, where powdery mildew has been reported as a common disease (Costamilan, 2005). Surveys across those two states from 1995 to 2003 determined virulence levels to some older Pm genes, including a low frequency of virulence to Pm1a and Pm2, moderate levels to Pm4a and Pm6, and high frequencies to Pm3a, Pm4b, and Pm8 (Costamilan and Linhares, 2002; Costamilan, 2005). In general, wheat powdery mildew was not a major problem in Brazil until 2015 (P. Kuhnem, personal observation). However, in the past 6 years, it became the most prevalent wheat disease in southern Brazil (states of Parana, Santa Catarina, and Rio Grande do Sul). This was likely due to mild, humid winters conducive to Bgt epidemics, and the widespread use of susceptible cultivars (P. Kuhnem, personal observation).
From the present survey, virulence in Parana and Rio Grande appears to have increased dramatically in the past 15 years, such that now all the older Pm genes (except Pm1b) evidently have lost effectiveness against Bgt. Over 70% of the registered cultivars in Brazil had CIMMYT material as background in 2002 (Lantican et al., 2005), but the dominance of that background in current Brazilian cultivars has declined (P. Kuhnem, personal observation). Genes used in Brazilian breeding nurseries included Pm4a from cultivar Khapli (Reis et al., 1979; Bennett, 1984) and Pm17 in the 1A.1R wheat-rye translocation (Byerlee and Moya, 1993). Another factor that could have contributed to the high complexity of Bgt virulence in Brazil is the use of Pm genes in neighboring countries, such as Paraguay, where widespread planting of cultivar Timgalen in the 1970s caused an upsurge in virulence to Pm6 (Scott et al., 1980). Genotyping several wheat cultivars recently popular in South Brazil with KASP markers for Pm1a, Pm6, and Pm8 indicated that none of the cultivars had any of those genes.
A generally low level of virulence to the newer Pm genes in isolates from countries with recently escalating wheat powdery mildew epidemics suggested that these newer Pm genes would likely be effective there. An exception was MlUM15, which is tightly linked or allelic to Pm1 or another Pm gene on chromosome 7AL (Hsam et al., 1998; Perugini et al., 2008; Maxwell et al., 2009; Worthington et al., 2014). In this study, virulence to Pm1a and to MlUM15 was linked in most isolates except those from Brazil and Turkey. If Pm1a and MlUM15 were tightly linked, they would be expected to place simultaneous selection pressure on both AvrPm1a (Hewitt et al., 2021) and the Avr gene corresponding to MlUM15. In that case, Bgt isolates would require mutations in both Avr genes to become virulent to both Pm genes. If instead Pm1a and MlUM15 are allelic to one another, as Worthington et al. (2014) proposed, how could co-segregation of virulence/avirulence to the two alleles be explained? The wheat alleles Pm3a and Pm3f were shown to have different but overlapping recognition specificities for fungal effector alleles of AvrPm3a (Stirnweis et al., 2014; Bourras et al., 2015). It is possible that similarly, Pm1a and MlUM15 have different but overlapping recognition spectra for alleles of AvrPm1a and AvrMlUM15, such that in most Bgt populations, both Pm genes recognize both AvrPm1a and AvrMlUM15. However, perhaps, Brazilian and Turkish Bgt populations have private, unique forms of AvrPm1a and AvrMlUM15 that are recognized by MlUM15 but not Pm1a. This would explain the larger fractions of isolates virulent to Pm1a but avirulent to MlUM15 in Bgt populations of those two countries. Nonetheless, virulence to this gene appears to be linked to Pm1a virulence in most populations, and thus, the new gene would likely not be useful in those countries. In Brazil, where MlUM15 might have value, further testing of this gene in the field is advisable. Mapping and characterization of the Avr gene that is recognized by MlUM15 would also be useful.
As new Pm genes are made available, it is important to exercise responsible stewardship when they are deployed to help to prevent a rapid appearance of virulent isolates and limit the development of a larger, virulent population of Bgt (Li et al., 2019a; Tan et al., 2019; Simeone et al., 2020). Besides cultivar and crop rotations, it is recommended to introgress effective Pm genes in combinations of two or more into breeding lines with a quantitative resistance background (Brown, 2015; Rimbaud et al., 2018; Aravindh et al., 2020). Ideally, good stewardship should also include the stacking of resistance genes with complementary function, which can be elucidated by their molecular characterization (Hafeez et al., 2021). A coordinated, international effort to track deployed resistance genes in current and future cultivars with molecular markers and the periodic monitoring of pathogen populations for virulence frequencies would allow the timely identification of breakdowns in resistance gene stacks in the field and illuminate which genes need to be replaced. Seed of germplasm lines used in this study is freely available and can be obtained from the authors.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
TK helped design the study, cultured, phenotyped, and genetically analyzed isolates and cultivars, conducted statistical analyses, and drafted the manuscript. WB and ZP collected samples in South Africa. BA, AM, and VS collected samples in Turkey, Russia, and Kazakhstan. PK collected the samples in Brazil. PM created germplasm lines containing introgressed genes Pm25–Pm53. CC conceptualized and supervised the study and helped to prepare the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded in part by the United States Department of Agriculture-Agricultural Research Service (USDA-ARS) including ARS project number 6070-22000-018-00D.
Conflict of Interest
DL was employed by Syngenta (Pty.) Ltd. PK was employed by Biotrigo Genética.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Michael Elliott, Matthew Hargrove, Emily Meyers, and Rebecca Whetten for help with culture maintenance and virulence assays and Logan Clark for DNA extractions. We would also like to thank Francisco Lopez Ruiz for providing isolates from Australia and Dr. Andrew Milgate and Sally Norton for discussion and help in obtaining seed.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.954958/full#supplementary-material
Supplementary Figure 1 | Allele discrimination plots of kompetitive allele-specific PCR (KASP) markers linked to Pm gene resistance loci. KASP markers are diagnostic of alleles linked to the presence (red) or the absence (blue) of (A) Pm1a, (B) Pm6z, and (C) Pm8. Fluorescence values of reporter cassettes measured at 44 cycles were normalized to readings of the ROX reference dye. Gray lines indicate the real-time fluorescence signal of cycles 1–50. zDespite lower confidence in allele discrimination for Pm6 and Pm8 with medium FAM fluorescence signal (>60,000 relative fluorescence units) detected in all samples (including the no-template control, yellow), positive controls for both alleles separated into distinct clusters, so that all tested cultivar samples could be distinguished from samples with a resistance allele.
Supplementary Figure 2 | Complexity of virulence to wheat resistance genes among Blumeria graminis f. sp. tritici populations from several countries compared to previously published virulence data for the United States and Egypt, for recently introgressed genes subsetted into (A) four Pm genes that conferred susceptibility to Bgt in detached leaf assays, but were effective against United States Bgt populations in the fieldd (Pm25–NCA6), and (B) five Pm genes which were effective both in lab assays and in the field (NCAG13–Pm53). Virulence complexity was calculated as mean percentage of Pm genes to which isolates were virulent. Within a panel, countries topped by the same letter did not have significantly different virulence complexities (p < 0.05). Within each box, medians are indicated by lines and means of virulence complexity by crosses. vAbdelrhim et al. (2018). wTurkey/Black Sea region: Isolates from Russian locations Krasnodar and Rostov were grouped with those from Turkey (refer to Table 1). xWestern Siberia region: Isolates from Karabalyk, Kazakhstan were grouped with those from Russia (refer to Table 1). yCowger et al. (2018). zGenes Pm25–NCA6 were effective in the field against a natural United States Bgt population (Cowger et al., 2018).
Supplementary Figure 3 | Complexity of virulence to wheat resistance genes among Blumeria graminis f. sp. tritici populations from five global regions compared to previously published data from the United States and Egypt, for (A) ten older Pm genes, in commercial use for a longer time (Pm1a–Pm17) and (B) nine newer Pm genes, more recently introgressed into hexaploid wheat from wild relatives (Pm25–Pm53). Virulence complexity was calculated as mean percentage of Pm genes to which isolates were intermediately virulent (yellow) and fully virulent (red). Within a panel, countries topped by the same letter did not have significantly different virulence complexities (P < 0.05); numbers below countries indicate the number of isolates analyzed. Error bars indicate standard error of the means. vAbdelrhim et al. (2018). wTurkey/Black Sea region: Isolates from Russian locations Krasnodar and Rostov were grouped with those from Turkey (refer to Table 1). xWestern Siberia region: Isolates from Karabalyk, Kazakhstan were grouped with those from Russia (refer to Table 1). yCowger et al. (2018). zNewer Pm genes Pm25–NCA6 were effective in the field against natural United States Bgt population (Cowger et al., 2018).
References
Abdelrhim, A., Abd-Alla, H. M., Abdou, E. S., Ismail, M. E., and Cowger, C. (2018). Virulence of Egyptian Blumeria graminis f. sp. tritici population and response of Egyptian wheat cultivars. Plant Dis. 102, 391–397. doi: 10.1094/PDIS-07-17-0975-RE
Acevedo-Garcia, J., Spencer, D., Thieron, H., Reinstadler, A., Hammond-Kosack, K., Phillips, A. L., et al. (2017). mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 15, 367–378. doi: 10.1111/pbi.12631
Alam, M. A., Xue, F., Wang, C., and Ji, W. (2011). Powdery mildew resistance genes in wheat: identification and genetic analysis. J. Mol. Biol. Res. 1, 20–39. doi: 10.5539/jmbr.v1n1p20
Alemu, A., Brazauskas, G., Gaikpa, D. S., Henriksson, T., Islamov, B., Jorgensen, L. N., et al. (2021). Genome-wide association analysis and genomic prediction for adult-plant resistance to septoria tritici blotch and powdery mildew in winter wheat. Front. Genet. 12:661742. doi: 10.3389/fgene.2021.661742
Aravindh, R., Sivasamy, M., Ganesamurthy, K., Jayaprakash, P., Gopalakrishnan, C., Geetha, M., et al. (2020). Marker assisted stacking/pyramiding of stem rust, leaf rust and powdery mildew disease resistance genes (Sr2/Lr27/Yr30, Sr24/Lr24 and Sr36/Pm6) for durable resistance in wheat (Triticum aestivum L.). Electron. J. Plant Breed. 11, 907–915. doi: 10.37992/2020.1103.148
Baik, B., and Sturbaum, A. (2014). “2014 soft wheat quality laboratory annual report,” in 60th Annual Research Review.
Balfourier, F., Bouchet, S., Robert, S., De Oliveira, R., Rimbert, H., Kitt, J., et al. (2019). Worldwide phylogeography and history of wheat genetic diversity. Sci. Adv. 5:eaav0536. doi: 10.1126/sciadv.aav0536
Ben-David, R., Parks, R., Dinoor, A., Kosman, E., Wicker, T., Keller, B., et al. (2016). Differentiation among Blumeria graminis f. sp. tritici isolates originating from wild versus domesticated triticum species in Israel. Phytopathology 106, 861–870. doi: 10.1094/PHYTO-07-15-0177-R
Bennett, F. G. A. (1984). Resistance to powdery mildew in wheat: a review of its use in agriculture and breeding programmes. Plant Pathol. 33, 279–300. doi: 10.1111/j.1365-3059.1984.tb01324.x
Bourras, S., Kunz, L., Xue, M., Praz, C. R., Mueller, M. C., Kalin, C., et al. (2019). The AvrPm3-Pm3 effector-NLR interactions control both race-specific resistance and host-specificity of cereal mildews on wheat. Nat. Commun. 10:2292. doi: 10.1038/s41467-019-10274-1
Bourras, S., McNally, K. E., Ben-David, R., Parlange, F., Roffler, S., Praz, C. R., et al. (2015). Multiple avirulence loci and allele-specific effector recognition control the Pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 27, 2991–3012. doi: 10.1105/tpc.15.00171
Bradley, J., and Thomas, G. (2019). Wheat Powdery Mildew Epidemiology and Crop Management Options. South Perth, WA: Grains Research & Development Corporation. Available online at: https://grdc.com.au/resources-and-publications/grdc-update-papers/tab-content/grdc-update-papers/2019/02/wheat-powdery-mildew-epidemiology-and-crop-management-options
Briggle, L. W., and Sears, E. R. (1966). Linkage of resistance to Erysiphe graminis f. sp. tritici (Pm3) and hairy glume (Hg) on chromosome 1A of wheat. Crop Sci. 6, 559–561.
Brown, J. K. (2015). Durable resistance of crops to disease: a Darwinian perspective. Annu. Rev. Phytopathol. 53, 513–539.
Brown, J. K., and Tellier, A. (2011). Plant-parasite coevolution: bridging the gap between genetics and ecology. Annu. Rev. Phytopathol. 49, 345–367. doi: 10.1146/annurev-phyto-072910-095301
Byerlee, D., and Moya, P. (1993). Impacts of international wheat breeding research in the developing world, 1966–90. Mexico: CIMMYT.
Chen, Y., Zhang, F., Tang, L., Zheng, Y., Li, Y., Christie, P., et al. (2007). Wheat powdery mildew and foliar N concentrations as influenced by N fertilization and belowground interactions with intercropped faba bean. Plant Soil 291, 1–13. doi: 10.1007/s11104-006-9161-9
Costamilan, L. M. (2005). Variability of the wheat powdery mildew pathogen Blumeria graminis f. sp. tritici in the 2003 crop season. Fitopatol. Brasil. 30, 420–422.
Costamilan, L. M., and Linhares, W. I. (2002). Efetividade de genes de resistência de trigo a oídio. Fitopatol. Brasil. 27, 621–625.
Cowger, C., Miranda, L., Griffey, C., Hall, M., Murphy, J. P., and Maxwell, J. (2012). “Wheat Powdery Mildew,” in Disease Resistance in Wheat, ed. I. Sharma (Wallingford: CAB International), 84–119.
Cowger, C., Mehra, L., Arellano, C., Meyers, E., and Murphy, J. P. (2018). Virulence differences in Blumeria graminis f. sp. tritici from the central and eastern United States. Phytopathology 108, 402–411. doi: 10.1094/PHYTO-06-17-0211-R
Cowger, C., Parks, R., and Marshall, D. (2009). Appearance of powdery mildew of wheat caused by Blumeria graminis f. sp. tritici on Pm17-bearing cultivars in North Carolina. Plant Dis. 93:1219. doi: 10.1094/PDIS-93-11-1219B
Cuddy, W., Park, R. F., Bansal, U., Bariana, S., Singh, D., Roake, J., et al. (2016). Expected responses of Australian wheat, triticale and barley varieties to the cereal rust diseases and genotypic data for oat varieties. Cereal Rust Rep. 14, 1–8.
Dubcovsky, J., and Dvorak, J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316, 1862–1866. doi: 10.1126/science.1143986
Evenson, R. E., and Gollin, D. (2003). Crop Variety Improvement and its Effect on Productivity : The Impact of International Agricultural Research. Wallingford: CABI Publishing.
FAOSTAT (2017). FAOSTAT Statistical Database. Rome: Food and Agriculture Organization of the United Nations.
Glawe, D. A. (2008). The powdery mildews: a review of the world’s most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 46, 27–51. doi: 10.1146/annurev.phyto.46.081407.104740
Golzar, H., Shankar, M., and D’Antuono, M. (2016). Responses of commercial wheat varieties and differential lines to western Australian powdery mildew (Blumeria graminis f. sp. tritici) populations. Austral. Plant Pathol. 45, 347–355. doi: 10.1007/s13313-016-0420-9
Green, A. J., Berger, G., Griffey, C. A., Pitman, R., Thomason, W., and Balota, M. (2014). Genetic resistance to and effect of leaf rust and powdery mildew on yield and its components in 50 soft red winter wheat cultivars. Crop Protect. 64, 177–186. doi: 10.1016/j.cropro.2014.06.023
GRIS (2021). Genetic Resources Information System for Wheat and Triticale - Passport Data for 171 000 Varieties and Lines. Available online at: http://wheatpedigree.net
Hafeez, A. N., Arora, S., Ghosh, S., Gilbert, D., Bowden, R. L., and Wulff, B. B. H. (2021). Creation and judicious application of a wheat resistance gene atlas. Mol. Plant 14, 1053–1070. doi: 10.1016/j.molp.2021.05.014
Harlan, J. R., and Zohary, D. (1966). Distribution of wild wheats and barley. Science 153, 1074–1080. doi: 10.1126/science.153.3740.1074
Heisey, P. W., Lantican, M. A., and Dubin, H. J. (2002). Impacts of International Wheat Breeding Research in Developing Countries, 1966-97. Impact Studies 7653. Mexico: CIMMYT.
Helmsj∅rgensen, J., and Jensen, C. J. (1973). Gene Pm6 for resistance to powdery mildew in wheat. Euphytica 22:423. doi: 10.1007/bf00022656
Heun, M., Friebe, B., and Bushuk, W. (1990). Chromosomal location of the powdery mildew resistance gene of Amigo wheat. Phytopathology 80, 1129–1133.
Heun, M., Schäfer-Pregl, R., Klawan, D., Castagna, R., Accerbi, M., Borghi, B., et al. (1997). Site of Einkorn wheat domestication identified by DNA fingerprinting. Science 278, 1312–1314. doi: 10.1126/science.278.5341.1312
Hewitt, T., Mueller, M. C., Molnar, I., Mascher, M., Holusova, K., Simkova, H., et al. (2021). A highly differentiated region of wheat chromosome 7AL encodes a Pm1a immune receptor that recognizes its corresponding AvrPm1a effector from Blumeria graminis. New Phytol. 229, 2812–2826. doi: 10.1111/nph.17075
Hsam, S. L. K., and Zeller, F. J. (1997). Evidence of allelism between genes Pm8 and Pm17 and chromosomal location of powdery mildew and leaf rust resistance genes in the common wheat cultivar’Amigo’. Plant Breed. 116, 119–122. doi: 10.1111/j.1439-0523.1997.tb02164.x
Hsam, S. L. K., Huang, X. Q., Ernst, F., Hartl, L., and Zeller, F. J. (1998). Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 5. Alleles at the Pm1 locus. Theor. Appl. Gene. 96, 1129–1134. doi: 10.1007/s001220050848
Imani, Y., Ouassou, A., and Griffey, C. A. (2002). Virulence of Blumeria graminis f. sp. tritici populations in Morocco. Plant Dis. 86, 383–388. doi: 10.1094/PDIS.2002.86.4.383
Johnson, J. W., Ge, Y. F., Roberts, J. J., Raymer, P., and Seo, Y. (2003). Adult-Plant Resistance to Powdery Mildew in Knox 62 Wheat. Cereal Res. Commun. 31, 281–288.
Joukhadar, R., Daetwyler, H. D., Bansal, U. K., Gendall, A. R., and Hayden, M. J. (2017). Genetic diversity, population structure and ancestral origin of Australian Wheat. Front. Plant Sci. 8:2115. doi: 10.3389/fpls.2017.02115
Kang, Y., Zhou, M., Merry, A., and Barry, K. (2020). Mechanisms of powdery mildew resistance of wheat – a review of molecular breeding. Plant Pathol. 69, 601–617. doi: 10.1111/ppa.13166
Kirsten, J., and Nhemachena, C. R. (2017). A historical assessment of sources and uses of wheat varietal innovations in South Africa. South Afr. J. Sci. 113, 1–8. doi: 10.17159/sajs.2017/20160008
Kominkova, E., Dreiseitl, A., Maleckova, E., Dolezel, J., and Valarik, M. (2016). Genetic diversity of Blumeria graminis f. sp. hordei in Central Europe and its comparison with Australian population. PLoS One 11:e0167099. doi: 10.1371/journal.pone.0167099
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Kusch, S., and Panstruga, R. (2017). mlo-based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant Microbe. Interact. 30, 179–189. doi: 10.1094/MPMI-12-16-0255-CR
Lantican, M. A., Dubin, J., and Morris, M. L. (2005). Impacts of International Wheat Breeding Research in the Developing World, 1998-2002. Mexico: CIMMYT.
Legler, S. E., Caffi, T., and Rossi, V. (2012). A nonlinear model for temperature-dependent development of Erysiphe necator chasmothecia on grapevine leaves. Plant Pathol. 61, 96–105. doi: 10.1111/j.1365-3059.2011.02498.x
Leonard, K. J., Anikster, Y., and Manisterski, J. (2004). Patterns of virulence in natural populations of Puccinia coronata on wild oat in Israel and in agricultural populations on cultivated oat in the United States. Phytopathology 94, 505–514. doi: 10.1094/PHYTO.2004.94.5.505
Li, G., Cowger, C., Wang, X., Carver, B. F., and Xu, X. (2019b). Characterization of Pm65, a new powdery mildew resistance gene on chromosome 2AL of a facultative wheat cultivar. Theor. Appl. Genet. 132, 2625–2632. doi: 10.1007/s00122-019-03377-2
Li, F., Upadhyaya, N. M., Sperschneider, J., Matny, O., Nguyen-Phuc, H., Mago, R., et al. (2019a). Emergence of the Ug99 lineage of the wheat stem rust pathogen through somatic hybridisation. Nat. Commun. 10:5068. doi: 10.1038/s41467-019-12927-7
Lillemo, M., Singh, R. P., and van Ginkel, M. (2010). Identification of Stable Resistance to Powdery Mildew in Wheat Based on Parametric and Nonparametric Methods. Crop Sci. 50, 478–485. doi: 10.2135/cropsci2009.03.0116
Lu, P., Guo, L., Wang, Z., Li, B., Li, J., Li, Y., et al. (2020). A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat. Commun. 11:680. doi: 10.1038/s41467-020-14294-0
Ma, Z. Q., Wei, J. B., and Cheng, S. H. (2004). PCR-based markers for the powdery mildew resistance gene Pm4a in wheat. Theor. Appl. Genet. 109, 140–145. doi: 10.1007/s00122-004-1605-0
Mago, R., Miah, H., Lawrence, G. J., Wellings, C. R., Spielmeyer, W., Bariana, H. S., et al. (2005). High-resolution mapping and mutation analysis separate the rust resistance genes Sr31, Lr26 and Yr9 on the short arm of rye chromosome 1. Theor. Appl. Genet. 112, 41–50. doi: 10.1007/s00122-005-0098-9
Maxwell, J. J., Lyerly, J. H., Cowger, C., Marshall, D., Brown-Guedira, G., and Murphy, J. P. (2009). MlAG12: a Triticum timopheevii-derived powdery mildew resistance gene in common wheat on chromosome 7AL. Theor. Appl. Genet. 119, 1489–1495. doi: 10.1007/s00122-009-1150-y
McDonald, B. A., and Linde, C. (2002). Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. doi: 10.1146/annurev.phyto.40.120501.101443
McDonald, B. A., and Stukenbrock, E. H. (2016). Rapid emergence of pathogens in agro-ecosystems: global threats to agricultural sustainability and food security. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 371, 20160026. doi: 10.1098/rstb.2016.0026
McIntosh, R. A., and Baker, E. P. (1970). Cytogenetical studies in wheat. IV. Chromosome location and linkage studies involving the Pm2 locus for powdery mildew resistance. Euphytica 19, 71–77.
Miranda, L. M., Murphy, J. P., Marshall, D., and Leath, S. (2006). Pm34: a new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L.). Theor. Appl. Genet. 113, 1497–1504. doi: 10.1007/s00122-006-0397-9
Miranda, L. M., Perugini, L., Srnic, G., Brown-Guedira, G., Marshall, D., Leath, S., et al. (2007b). Genetic mapping of a Triticum monococcum-derived powdery mildew resistance gene in common wheat. Crop Sci. 47, 2323–2329.
Miranda, L. M., Murphy, J. P., Marshall, D., Cowger, C., and Leath, S. (2007a). Chromosomal location of Pm35, a novel Aegilops tauschii derived powdery mildew resistance gene introgressed into common wheat (Triticum aestivum L.). Theor. Appl. Genet. 114, 1451–1456. doi: 10.1007/s00122-007-0530-4
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Morgounov, A., Tufan, H. A., Sharma, R., Akin, B., Bagci, A., Braun, H.-J., et al. (2011). Global incidence of wheat rusts and powdery mildew during 1969–2010 and durability of resistance of winter wheat variety Bezostaya 1. Eur. J. Plant Pathol. 132, 323–340. doi: 10.1007/s10658-011-9879-y
Moseman, J. G., Nevo, E., Morshidy, M. A. E., and Zohary, D. (1984). Resistance of Triticum dicoccoides to infection with Erysiphe graminis tritici. Euphytica 33, 41–47. doi: 10.1007/bf00022748
Mueller, M. C., Kunz, L., Schudel, S., Kammerecker, S., Isaksson, J., Wyler, M., et al. (2021). Standing genetic variation of the AvrPm17 avirulence gene in powdery mildew limits the effectiveness of an introgressed rye resistance gene in wheat. BioRxiv [Preprint]. 2021.2003.2009.433749. doi: 10.1101/2021.03.09.433749
Mueller, M. C., Praz, C. R., Sotiropoulos, A. G., Menardo, F., Kunz, L., Schudel, S., et al. (2019). A chromosome-scale genome assembly reveals a highly dynamic effector repertoire of wheat powdery mildew. New Phytol. 221, 2176–2189. doi: 10.1111/nph.15529
Murphy, J. P., Leath, S., Huynh, D., Navarro, R. A., and Shi, A. (1999). Registration of NC96BGTA4, NC96BGTA5 and NC96BGTA6 wheat germplasm. Crop Sci. 39, 883–884.
Murphy, J. P., Navarro, R. A., Marshall, D., Cowger, C., Cox, T. S., Kolmer, J. A., et al. (2007). Registration of NC06BGTAG12 and NC06BGTAG13 powdery mildew–resistant wheat germplasm. J. Plant Reg. 1, 75–77.
Neu, C., Stein, N., and Keller, B. (2002). Genetic mapping of the Lr20-Pm1 resistance locus reveals suppressed recombination on chromosome arm 7AL in hexaploid wheat. Genome 45, 737–744. doi: 10.1139/g02-040
Niewoehner, A. S., and Leath, S. (1998). Virulence of Blumeria graminis f. sp. tritici on winter wheat in the eastern United States. Plant Dis. 82, 64–68. doi: 10.1094/PDIS.1998.82.1.64
Özkan, H., Willcox, G., Graner, A., Salamini, F., and Kilian, B. (2010). Geographic distribution and domestication of wild emmer wheat (Triticum dicoccoides). Gene. Res. Crop Evol. 58, 11–53. doi: 10.1007/s10722-010-9581-5
Park, R. F., Bansal, U., Bariana, S., and Singh, D. (2020). Rust resistance genotypes and expected rust responses of Australian common wheat, durum wheat and triticale varieties. Cereal Rust Rep. 17:10.
Parks, R., Carbone, I., Murphy, J. P., and Cowger, C. (2009). Population genetic analysis of an Eastern U.S. wheat powdery mildew population reveals geographic subdivision and recent common ancestry with U.K. and Israeli populations. Phytopathology 99, 840–849. doi: 10.1094/PHYTO-99-7-0840
Parks, R., Carbone, I., Murphy, J. P., Marshall, D., and Cowger, C. (2008). Virulence structure of the eastern U.S. wheat powdery mildew population. Plant Dis. 92, 1074–1082. doi: 10.1094/PDIS-92-7-1074
Perugini, L. D., Murphy, J. P., Marshall, D., and Brown-Guedira, G. (2008). Pm37, a new broadly effective powdery mildew resistance gene from Triticum timopheevii. Theor. Appl. Genet. 116, 417–425. doi: 10.1007/s00122-007-0679-x
Petersen, S., Lyerly, J. H., Worthington, M. L., Parks, W. R., Cowger, C., Marshall, D. S., et al. (2015). Mapping of powdery mildew resistance gene Pm53 introgressed from Aegilops speltoides into soft red winter wheat. Theor. Appl. Genet. 128, 303–312. doi: 10.1007/s00122-014-2430-8
Praz, C. R., Bourras, S., Zeng, F., Sanchez-Martin, J., Menardo, F., Xue, M., et al. (2017). AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. New Phytol. 213, 1301–1314. doi: 10.1111/nph.14372
Reis, E. M., Minella, E., Baier, A. C., and Santos, H. P. D. (1979). Reaction of wheat cultivars and lines to Erysiphe graminis (DC) f. sp tritici Marchall. Summa phytopathol. 5, 54–64.
Reynolds, M. P., and Borlaug, N. E. (2006). Impacts of breeding on international collaborative wheat improvement. J. Agr. Sci. 144, 3–17. doi: 10.1017/s0021859606005867
Rimbaud, L., Papaix, J., Rey, J. F., Barrett, L. G., and Thrall, P. H. (2018). Assessing the durability and efficiency of landscape-based strategies to deploy plant resistance to pathogens. PLoS Comput. Biol. 14:e1006067. doi: 10.1371/journal.pcbi.1006067
Rinaldo, A., Gilbert, B., Boni, R., Krattinger, S. G., Singh, D., Park, R. F., et al. (2017). The Lr34 adult plant rust resistance gene provides seedling resistance in durum wheat without senescence. Plant Biotechnol. J. 15, 894–905. doi: 10.1111/pbi.12684
Sarinelli, J. M., Murphy, J. P., Tyagi, P., Holland, J. B., Johnson, J. W., Mergoum, M., et al. (2019). Training population selection and use of fixed effects to optimize genomic predictions in a historical USA winter wheat panel. Theor. Appl. Genet. 132, 1247–1261. doi: 10.1007/s00122-019-03276-6
Sarinelli, M. J. (2017). Genomic Selection and Association Mapping with a Historical Data Set of Southeastern USA Soft Red Winter Wheat. Raleigh: N.C.S. University.
Savary, S., Willocquet, L., Pethybridge, S. J., Esker, P., McRoberts, N., and Nelson, A. (2019). The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 3, 430–439. doi: 10.1038/s41559-018-0793-y
Scott, P. R., Johnson, R., Wolfe, M. S., Lowe, H. J. B., and Bennett, F. G. A. (1980). Host-specificity in cereal parasites in relation to their control. Appl. Biol. 5, 349–393.
Sears, E. R., and Briggle, L. W. (1969). Mapping the gene Pm1 for resistance to Erysiphe graminis f. sp. tritici on Chromosome 7A of wheat. Crop Sci. 9, 96–97. doi: 10.2135/cropsci1969.0011183X000900010033x
Sensako (2022). Western Cape Winter Rainfall Region Wheat Disease Resistance. Bethelem: Sensako Performance Seed. Available online at: https://www.sensako.co.za/Products/ProductDetail/76
Shackley, B., Paynter, B., Troup, G., Bucat, J., Seymour, M., and Blake, A. D. (2019). 2020 Western Australian Crop Sowing Guide. Available online at: https://www.agric.wa.gov.au/barley/2020-western-australian-crop-sowing-guide
Shi, A. N., Leath, S., and Murphy, J. P. (1998). A major gene for powdery mildew resistance transferred to common wheat from wild einkorn wheat. Phytopathology 88, 144–147. doi: 10.1094/PHYTO.1998.88.2.144
Simeone, R., Piarulli, L., Nigro, D., Signorile, M. A., Blanco, E., Mangini, G., et al. (2020). Mapping powdery mildew (Blumeria graminis f. sp. tritici) resistance in wild and cultivated tetraploid wheats. Int. J. Mol. Sci. 21:7910. doi: 10.3390/ijms21217910
Singrün, C., Rauch, P., Morgounov, A., Hsam, S., and Zeller, F. (2004). Identification of powdery mildew and leaf rust resistance genes in common wheat (Triticum aestivum L.). Wheat varieties from the Caucasus, Central and Inner Asia. Gene. Res. Crop Evol. 51, 355–370. doi: 10.1023/B:GRES.0000023455.48325.9e
Smit, H. A., Tolmay, V. L., Barnard, A., Jordaan, J. P., Koekemoer, F. P., Otto, W. M., et al. (2010). An overview of the context and scope of wheat (Triticum aestivum) research in South Africa from 1983 to 2008. South Afr. J. Plant Soil 27, 81–96. doi: 10.1080/02571862.2010.10639973
Spanu, P. D. (2012). The genomics of obligate (and nonobligate) biotrophs. Annu. Rev. Phytopathol. 50, 91–109. doi: 10.1146/annurev-phyto-081211-173024
Stirnweis, D., Milani, S. D., Jordan, T., Keller, B., and Brunner, S. (2014). Substitutions of two amino acids in the nucleotide-binding site domain of a resistance protein enhance the hypersensitive response and enlarge the PM3F resistance spectrum in wheat. Mol. Plant Microbe. Interact. 27, 265–276. doi: 10.1094/MPMI-10-13-0297-FI
Tadesse, W., Bishaw, Z., and Assefa, S. (2019). Wheat production and breeding in Sub-Saharan Africa. Int. J. Clim. Change strategies Manage. 11, 696–715. doi: 10.1108/ijccsm-02-2018-0015
Tan, C., Li, G., Cowger, C., Carver, B. F., and Xu, X. (2019). Characterization of Pm63, a powdery mildew resistance gene in Iranian landrace PI 628024. Theor. Appl. Genet. 132, 1137–1144. doi: 10.1007/s00122-018-3265-5
Te Beest, D. E., Paveley, N. D., Shaw, M. W., and van den Bosch, F. (2008). Disease-weather relationships for powdery mildew and yellow rust on winter wheat. Phytopathology 98, 609–617. doi: 10.1094/PHYTO-98-5-0609
Tilman, D., Fargione, J., Wolff, B., D’Antonio, C., Dobson, A., Howarth, R., et al. (2001). Forecasting agriculturally driven global environmental change. Science 292, 281–284. doi: 10.1126/science.1057544
Trethowan, R. M., Hodson, D., Braun, H.-J., and Pfeiffer, W. H. (2005). “Wheat breeding environments,” in Impacts of International Wheat Breeding Research in the Developing World, 1988–2002, eds M. A. Lantican, J. Dubin, and M. L. Morris (Mexico: CIMMYT), 4–11.
Troch, V., Audenaert, K., Wyand, R. A., Haesaert, G., Hofte, M., and Brown, J. K. (2014). Formae speciales of cereal powdery mildew: close or distant relatives? Mol. Plant Pathol. 15, 304–314. doi: 10.1111/mpp.12093
Valkoun, J., and Mamluk, O. F. (1993). Disease Resistance and Agronomic Performance of Durum and Bread Wheat Lines Derived from Crosses with Triticum Monococcum. Chichester: John Wiley & Sons.
Van de Wouw, M., Kik, C., van Hintum, T., van Treuren, R., and Visser, B. (2009). Genetic erosion in crops: concept, research results and challenges. Plant Gene. Res. 8, 1–15. doi: 10.1017/s1479262109990062
Wicker, T., Oberhaensli, S., Parlange, F., Buchmann, J. P., Shatalina, M., Roffler, S., et al. (2013). The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 45, 1092–1096. doi: 10.1038/ng.2704
Wiersma, A. T., Pulman, J. A., Brown, L. K., Cowger, C., and Olson, E. L. (2017). Identification of Pm58 from Aegilops tauschii. Theor. Appl. Genet. 130, 1123–1133. doi: 10.1007/s00122-017-2874-8
Worthington, M., Lyerly, J., Petersen, S., Brown-Guedira, G., Marshall, D., Cowger, C., et al. (2014). MlUM15: an Aegilops neglecta-derived Powdery Mildew Resistance Gene in Common Wheat. Crop Sci. 54, 1397–1406. doi: 10.2135/cropsci2013.09.0634
Xu, H., Yi, Y., Ma, P., Qie, Y., Fu, X., Xu, Y., et al. (2015). Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor. Appl. Genet. 128, 2077–2084. doi: 10.1007/s00122-015-2568-z
Yi, Y. J., Liu, H. Y., Huang, X. Q., An, L. Z., Wang, F., and Wang, X. L. (2008). Development of molecular markers linked to the wheat powdery mildew resistance gene Pm4b and marker validation for molecular breeding. Plant Breed. 127, 116–120. doi: 10.1111/j.1439-0523.2007.01443.x
Zaicou-Kunesch, C., Trainor, G., Shackley, B., Curry, J., Nicol, D., Shankar, M., et al. (2017). Wheat Variety Fact sheets for Western Australia. Western Australia: Department of Agriculture and Food, Bulletin 4881. Available online at: https://grdc.com.au/resources-and-publications/all-publications/publications/2017/07/wheat-variety-fact-sheets-for-western-australia-2017
Zhang, Z., Henderson, C., Perfect, E., Carver, T. L., Thomas, B. J., Skamnioti, P., et al. (2005). Of genes and genomes, needles and haystacks: Blumeria graminis and functionality. Mol. Plant Pathol. 6, 561–575.
Zhu, Z. L., and Chen, D. L. (2002). Nitrogen fertilizer use in China – contributions to food production, impacts on the environment and best management strategies. Nutrient Cycl. Agroecosyst. 63, 117–127. doi: 10.1023/a:1021107026067
Zou, Y.-F., Qiao, H.-B., Cao, X.-R., Liu, W., Fan, J.-R., Song, Y.-L., et al. (2018b). Regionalization of wheat powdery mildew oversummering in China based on digital elevation. J. Integrat. Agr. 17, 901–910. doi: 10.1016/s2095-3119(17)61851-3
Keywords: common wheat, powdery mildew, resistance genes, virulence frequency, virulence complexity, genetic diversity, Blumeria graminis f. sp. tritici, durability
Citation: Kloppe T, Boshoff W, Pretorius Z, Lesch D, Akin B, Morgounov A, Shamanin V, Kuhnem P, Murphy P and Cowger C (2022) Virulence of Blumeria graminis f. sp. tritici in Brazil, South Africa, Turkey, Russia, and Australia. Front. Plant Sci. 13:954958. doi: 10.3389/fpls.2022.954958
Received: 27 May 2022; Accepted: 20 June 2022;
Published: 01 August 2022.
Edited by:
Peng Zhang, The University of Sydney, AustraliaReviewed by:
Zhiyong Liu, Institute of Genetics and Developmental Biology (CAS), ChinaVolker Mohler, Bayerische Landesanstalt für Landwirtschaft (LfL), Germany
Copyright © 2022 Kloppe, Boshoff, Pretorius, Lesch, Akin, Morgounov, Shamanin, Kuhnem, Murphy and Cowger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christina Cowger, Y2hyaXN0aW5hLmNvd2dlckB1c2RhLmdvdg==
 Tim Kloppe
Tim Kloppe Willem Boshoff
Willem Boshoff Zacharias Pretorius
Zacharias Pretorius Driecus Lesch3
Driecus Lesch3 Alexey Morgounov
Alexey Morgounov Vladimir Shamanin
Vladimir Shamanin Christina Cowger
Christina Cowger