
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 July 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.952856
This article is part of the Research Topic Generating Useful Genetic Variation in Crops by Induced Mutation, Volume II View all 6 articles
 Backki Kim1
Backki Kim1 Yoonjung Lee2
Yoonjung Lee2 Ji-Young Nam1
Ji-Young Nam1 Gileung Lee3
Gileung Lee3 Jeonghwan Seo4
Jeonghwan Seo4 Dongryung Lee5
Dongryung Lee5 Yoo-Hyun Cho6
Yoo-Hyun Cho6 Soon-Wook Kwon7*
Soon-Wook Kwon7* Hee-Jong Koh1*
Hee-Jong Koh1*Morphological and biochemical changes accompanying embryogenesis and seed development are crucial for plant survival and crop productivity. Here, we identified a novel yellowish-pericarp embryo lethal (yel) mutant of the japonica rice cultivar Sindongjin (Oryza sativa L.), namely, yel-sdj. Seeds of the yel-sdj mutant showed a yellowish pericarp and black embryo, and were embryonic lethal. Compared with wild-type seeds, the yel-sdj mutant seeds exhibited significantly reduced grain size, grain weight, and embryo weight, and a remarkably lower rate of embryo retention in kernels subjected to milling. However, the volume of air space between embryo and endosperm, density of embryo, and total phenolic content (TPC) and antioxidant activity of mature grains were significantly higher in the yel-sdj mutant than in the wild type. Genetic analysis and mapping revealed that the yel-sdj mutant was non-allelic to the oscop1 null mutants yel-hc, yel-cc, and yel-sk, and its phenotype was controlled by a single recessive gene, LOC_Os01g01484, an ortholog of Arabidopsis thaliana DE-ETIOLATED 1 (DET1). The yel-sdj mutant carried a 7 bp deletion in the second exon of OsDET1. Seeds of the osdet1 knockout mutant, generated via CRISPR/Cas9-based gene editing, displayed the yel mutant phenotype. Consistent with the fact that OsDET1 interacts with CONSTITUTIVE PHOTOMORPHOGENIC 10 (OsCOP10) and UV-DAMAGED DNA BINDING PROTEIN 1 (OsDDB1) to form the COP10-DET1-DDB1 (CDD), seeds of oscop10 and osddb1 knockout mutants also showed the yel phenotype. These findings will enhance our understanding of the functional roles of OsDET1 and the CDD complex in embryogenesis and flavonoid biosynthesis in rice seeds.
DE-ETIOLATED 1 (DET1) encodes a nuclear-localized protein that presumably acts downstream of multiple photoreceptors to modulate the light-mediated signaling pathways (Pepper et al., 1994). DET1 plays an important role in regulating the expression of development-related genes and is highly conserved across higher eukaryotes (Schroeder et al., 2002). DET1 was first identified in dark-grown Arabidopsis thaliana seedlings, which showed a de-etiolated phenotype characterized by the expansion of leaves, inhibition of hypocotyl elongation, and accumulation of anthocyanins (Chory et al., 1989). Previously, genetic screening studies identified a class of mutants displaying de-etiolated or constitutive photomorphogenic phenotypes in the dark and uncovered that DET1 acts as a crucial regulator of light signaling during seedling development in Arabidopsis (Pepper et al., 1994; Mayer et al., 1996). Besides its function in seedling photomorphogenesis, DET1 plays an important role in the developmental and environmental responses of plants, as exemplified by its role in chloroplast development (Chory and Peto, 1990), circadian period regulation (Millar et al., 1995; Lau et al., 2011), flowering time regulation (Kang et al., 2015), UV tolerance (Castells et al., 2010), fruit pigmentation (Mustilli et al., 1999), and seed germination (Shi et al., 2015). Collectively, DET1 is a central regulator that integrates the light signal with various developmental and biosynthetic pathways in plants.
DET1 interacts with CONSTITUTIVE PHOTOMORPHOGENIC 10 (COP10) and UV-DAMAGED DNA BINDING PROTEIN 1 (DDB1) to form the COP10-DET1-DDB1 (CDD) complex, which acts as a ubiquitination-promoting factor to regulate photomorphogenesis in Arabidopsis (Schroeder et al., 2002; Yanagawa et al., 2004). Although the underlying molecular mechanism of the CDD complex has not been fully elucidated, some possible models for role of the CDD complex and its relationship have been established at the molecular level (see review for details, Lau and Deng, 2012). COP10, a ubiquitin-conjugating enzyme (E2) variant (UEV) protein, was identified as a negative regulator of photomorphogenic development in the dark (Wei et al., 1994; Suzuki et al., 2002). COP10 has the ability to enhance the activity of multiple E2 enzymes and directly interacts with both COP1 and the COP9 signalosome to mediate the repression of photomorphogenesis by degrading ELONGATED HYPOCOTYL 5 (HY5) (Osterlund et al., 2000; Suzuki et al., 2002; Yanagawa et al., 2004; Lau and Deng, 2009). DDB1, on the other hand, is highly conserved among eukaryotes and was originally identified in human as a recognition protein that counteracts UV-induced DNA damage, and plays a role in nucleotide excision repair (NER) (Chu and Chang, 1988). The Arabidopsis genome encodes two DDB1 homologs, DDB1a and DDB1b, which are 91% identical at the amino acid level (Schroeder et al., 2002). DDB1 functions as an adapter linking the substrate receptors to CULLIN4 (CUL4)-based E3 ligases for ubiquitination (He et al., 2006; Lee and Zhou, 2007). In Arabidopsis, COP10- or DDB1-containing complexes cooperate with CUL4 to form an E3 ligase machinery, which is involved in photomorphogenesis and ubiquitin-mediated protein degradation (Yanagawa et al., 2004; Chen et al., 2006; Ganpudi and Schroeder, 2013). However, unlike Arabidopsis, the molecular functions of COP10 and DDB1, not only as independent proteins but also as CDD complex components, remain unclear in rice.
In plants, successful embryogenesis is a prerequisite for proper seed germination and early vegetative growth. Furthermore, interaction between embryo and endosperm affects the agronomically important traits of plants, such as starch composition and endosperm size, which determine grain yield and quality (Lafon-Placette and Kohler, 2014; An et al., 2020). Thus, given the biological and agricultural importance of embryogenesis, the molecular mechanisms underlying this process have been of considerable interest. Over the last several decades, numerous embryonic lethal or embryo-defective mutants have been identified in the model dicot plant, Arabidopsis (Meinke and Sussex, 1979a,b), and in model monocots, maize (Clark and Sheridan, 1991; Sheridan and Clark, 1993) and rice (Nagato et al., 1989; Kitano et al., 1993; Hong et al., 1995). These mutants exhibit a wide range of phenotypes, such as no embryo, incomplete embryo organs, colorless embryo, albino, and pigmented cotyledons (Meinke and Sussex, 1979a; Meinke, 1985; Hong et al., 1995; Satoh et al., 1999). Among them, Arabidopsis “fusca” (fus) mutants display purple coloration in cotyledons, because of high-level anthocyanin accumulation, and exhibit seedling lethality and defective photomorphogenesis. Interestingly, some of the fus mutants we re revealed to be allelic to cop/det mutants exhibiting the “fusca” phenotype (Castle and Meinke, 1994; Misera et al., 1994). Previously, we reported the “fusca”-like cop1 null mutants in rice, which we named as yellowish-pericarp embryo lethal (yel), and showed that the corresponding gene, OsCOP1, regulates flavonoid biosynthesis and embryo development. However, the effect of COP1 mutation on embryo development and pigmentation in rice was different from that in Arabidopsis; unlike the Arabidopsis fus mutants, which exhibited seedling lethality and anthocyanin accumulation in the cotyledons, the rice yel mutants showed embryonic lethality and flavonoid accumulation in the embryo and pericarp (Kim et al., 2018, 2021).
In the present study, we characterized a novel rice yel mutant displaying yellowish-pericarp and embryonic lethality, and identified OsDET1, an ortholog of Arabidopsis DET1, as the causal gene. Furthermore, using the CRISPR/Cas9 gene editing tool, we confirmed that oscop10 and osddb1 knockout mutants also showed the yel phenotype. Although the role of DET1 in photomorphogenic development has been extensively studied in Arabidopsis, limited information is available in rice. Therefore, characterization of the osdet1, oscop10, and osddb1 null mutants conducted in this study provides new insights into the regulation of flavonoid biosynthesis and embryo development in rice.
The novel yel mutant was derived from the japonica rice (Oryza sativa L. ssp. japonica) cultivar Sindongjin (SDJ) by gamma ray (γ-ray) irradiation, and named yel-sdj. Given its embryonic lethality, the yel-sdj mutant maintained as a heterozygote. An F2 mapping population was developed from a cross between the heterozygous yel-sdj mutant and a Korean indica rice accession, Milyang 23 (M.23). Additional F2 populations were derived by crossing the heterozygous yel-sdj mutant with SDJ (wild-type [WT]) and other yel mutants (yel-hc, yel-cc, and yel-sk) to calculate segregation ratios and perform the allelism test. The F2 populations as well as WT and yel-sdj mutant plants were cultivated in a paddy field at the Experimental Farm of Seoul National University, Suwon, South Korea. Knockout transgenic plants generated using the CRISPR/Cas9 technology were grown in the Living Modified Organism (LMO) experimental field (RDA-GA-AB-2011-014) at the Experimental Farm of Seoul National University, Suwon, South Korea.
Rice grains were air-dried after harvesting, and moisture content was reduced to approximately 13%. Grains were stored in an environmentally controlled room at 10°C for 2 months, and then dehusked and hand-selected to eliminate cracked or abnormally developed seeds. The length, width, and thickness of a total of 90 mature dehulled rice grains (30 seeds × 3 replicates) of each genotype were measured using digimatic calipers (Mitutoyo, Japan). Hundred-grain, -endosperm, and -embryo weights (100 seeds × 3 replicates; 10% water content) were measured using an analytical balance (CAS Corporation, NJ, United States). To measure embryo and endosperm weight, embryos were excised from the grains and weighed separately. Phenotypic data collected from yel-sdj mutant and WT genotypes were statistically analyzed using SAS version 9.4 (SAS Institute Inc., Cary, NC, United States).
The rate of embryo retention in kernels was measured using a small-scale grain polisher (Kett, Tokyo, Japan). A total of 100 brown rice kernels, with uniform appearance, were selected from yel-sdj mutant and WT seeds, respectively, mixed, and polished together under the same milling conditions for 5 s. Subsequently, kernels completely devoid of the embryo and those with a retained embryo were counted for each genotype. Five replications were conducted and the average trait value was used for data analysis.
Five dehulled kernels each of WT and yel-sdj mutant genotypes were randomly selected, and then scanned using SkyScan 1272 (Bruker, Belgium, Kontich), with pixel size set to 5 μm. The image acquisition process was carried out using the X-ray tube, with the following settings: voltage, 60 kV; current, 166 μA; exposure time, 0.45 s; four-frame averaging; rotation step, 0.40°; rotation angle, 180°. The scan duration was approximately 25 min. Following scanning, the raw images were converted to three-dimensional (3D) structures using the NRecon reconstruction software (SkyScan, Belgium), with the following settings: smoothing, 2; ring artifact correction, 24; beam hardening reduction, 50%. The resulting images were saved in bitmap (.bmp) format. The reconstructed images of the grains were then analyzed using 3D Slicer (v.4.13.0) (Fedorov et al., 2012).
To analyze the seed phenolic content and antioxidant activity, the extracts of WT and yel-sdj mutant seeds were prepared as described by Chung et al. (2017). Briefly, the seeds of each genotype were ground to a fine powder. Then, 1 g of each powdered sample was extracted with 10 mL of acetonitrile (ACN) and 2 mL of 0.1 N HCl, and sonicated using JAC-5020 Ultrasonic cleaner ABS (U1tech, Gyeonggi-Do, South Korea) at 40 kHz and room temperature for 20 min. After centrifugation at 1,962 × g and 4°C for 5 min, the supernatant was collected in a round-bottomed flask. The above process was repeated 3 and 13 times for WT and yel-sdj seed extracts, respectively. The final extracts were concentrated in a rotary vacuum evaporator (EYELA SB-1200; Tokyo Rikakikai Co., Ltd., Tokyo, Japan) at 35°C. The residue was reconstituted with 5 mL of 80% methanol and filtered through a 0.22 μm polytetrafluoroethylene (PTFE) syringe filter (CHOICE 13 mm; Thermo Scientific, Waltham, MA, United States).
Total phenolic content (TPC) was determined using a spectrophotometric assay based on the Lowry method, with slight modifications (Winters and Minchin, 2005). Briefly, 10 μL of each sample was mixed either with 990 μL of distilled water (blank) or with 990 μL of buffer (790 μL of distilled water, 50 μL of 1 N Folin-Ciocalteu reagent, and 150 μL of Na2CO3 in saturated NaOH solution). Following 1 h incubation at room temperature, absorbance was measured at 765 nm using a UV-Vis spectrophotometer. TPC was calculated from the calibration curve of gallic acid and expressed as micrograms of gallic acid equivalents per gram of dry weight (μg GAE/g DW). All samples were analyzed in triplicate.
Antioxidant enzyme activity in seeds was determined by performing the 2,2-diphenyl-1-picrylhydrazyl (DPPH) free-radical scavenging assay, as described previously (Kim et al., 2020). Briefly, 50 μL of the seed extract was added to 950 μL of 0.1 mM DPPH in methanol. The mixture was transferred to a 4 mL cuvette (10 mm × 10 mm × 45 mm; Ratiolab GmbH, Dreieich, Germany) and allowed to stand at room temperature in the dark for 30 min. Then, absorbance was measured at 517 nm using the OPTIZEN POP UV spectrophotometer (Mecasys Co., Daejeon, South Korea). The DPPH free-radical scavenging activity was calculated as inhibition percentage using the following equation:
where ODsample and ODcontrol represent the absorbance of the seed sample and DPPH standard solution, respectively.
Genomic DNA was extracted from the yel mutant-type seeds and leaves of 549 F2 individuals derived from the yel-sdj × M.23 cross. To identify the gene responsible for the yel-sdj mutant phenotype, bulked segregant analysis (BSA) was performed using a set of single nucleotide polymorphism (SNP) markers developed previously by designing primers based on nucleotide sequence differences between indica and japonica rice accessions (Seo et al., 2020). To fine-map the yel-sdj locus, sequence-tagged site (STS) primers were designed with Primer3 (version 0.4.0),1 based on the available rice genome sequence data.2 Primers designed and used in this study are listed in Supplementary Table 1.
Full-length sequence of the OsDET1 gene was amplified from WT and yel-sdj mutant seeds by performing overlapping extension PCR. The amplified products were purified using a PCR purification kit (iNtRON Biotechnology, South Korea), cloned into the pGEM-T Easy Vector (Promega, United States), and transformed into Escherichia coli strain DH5α. The inserts were sequenced, and sequences were compared using the CodonCode Aligner software (version 1.6.3; CodonCode Corporation, MA, United States).
To knock out the OsDET1, OsCOP10, and OsDDB1 genes, CRISPR/Cas9 vectors were constructed as described previously (Lowder et al., 2015). Briefly, guide RNAs (gRNAs) targeting each gene were designed using web-based tools, CRISPR RGEN Tools3 (Park et al., 2015) and CRISPRdirect4 (Naito et al., 2015). OsDET1 was targeted using two gRNAs, whereas OsCOP10 and OsDDB1 were each targeted using a single gRNA. The OsDET1-targeting gRNAs were cloned separately into two different gRNA expression vectors, pYPQ131C (Addgene plasmid #69284) and pYPQ132C (Addgene plasmid #69285), while OsCOP10- and OsDDB1-targeting gRNAs were cloned separately into pYPQ141C (Addgene plasmid #69292) under the expression of the OsU6 promoter. The gRNA expression cassettes were then assembled into the Golden Gate recipient vector pYPQ142 (Addgene plasmid #69294). pYPQ165 (Addgene #109327) was used as a Cas9 entry vector, which contained an egg cell-specific promoter. Finally, a Gateway assembly LR reaction was performed using the Cas9 entry vector (pYPQ165), gRNA cassettes (pYPQ141C or pYPQ142), and pMDC99 binary vector to generate the T-DNA binary vectors.
The final constructs were transformed into the seeds of the japonica cultivar Dongjin via Agrobacterium-mediated transformation using the LBA4404 strain, as described previously (Nishimura et al., 2006), with slight modifications. Primers used for vector construction and genotyping are listed in Supplementary Table 1.
All fresh plant samples were flash-frozen in liquid nitrogen. Total RNA was extracted from the leaf, leaf sheath, root, and young panicle (3 cm) of WT plants, and from 7-day-old seeds of both WT and yel-sdj mutant seeds, in three biological replicates, using TaKaRa MiniBEST Plant RNA Extraction Kit (Takara, Japan) according to the manufacturer’s instructions. Total RNA samples were subjected to first-strand cDNA synthesis using M-MLV reverse transcriptase (Promega, Madison, WI, United States), and qRT-PCR was performed using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (Takara Bio, Japan) on a CFX96™ Real-time PCR Detection System (Bio-Rad, Hercules, CA, United States) according to the manufacturer’s instructions. Primers used for qRT-PCR analysis are listed in Supplementary Table 1. Expression levels of genes were normalized relative to that of ACTIN, a housekeeping gene. Data were analyzed using the comparative Ct method. Expression levels were compared using two-tailed Student’s t-test.
The novel yel mutant, yel-sdj, was derived by γ-ray irradiation of the japonica rice cultivar SDJ. The main distinctive feature of the yel-sdj mutant seed was its embryo and pericarp color; the embryo was black, and the pericarp was yellowish or mixed yellow-purple (Figure 1A). Homozygous yel-sdj mutant seeds failed to germinate in the standard germination test (data not shown). Additionally, the yel-sdj mutant grains exhibited significantly reduced length, width, and thickness compared with WT grains, although no significant difference was detected between the length-to-width ratio of dehulled yel-sdj mutant and WT grains (Table 1). Hundred-grain weight was significantly lower in yel-sdj than in the WT. Consistently, the hundred-endosperm and -embryo weights were also significantly lower in the yel-sdj mutant than in the WT (Table 1). These results indicate that the overall development of seed is affected in the yel-sdj mutant, resulting in altered pigmentation of embryo and pericarp, and reduced weight of grain, endosperm, and embryo.
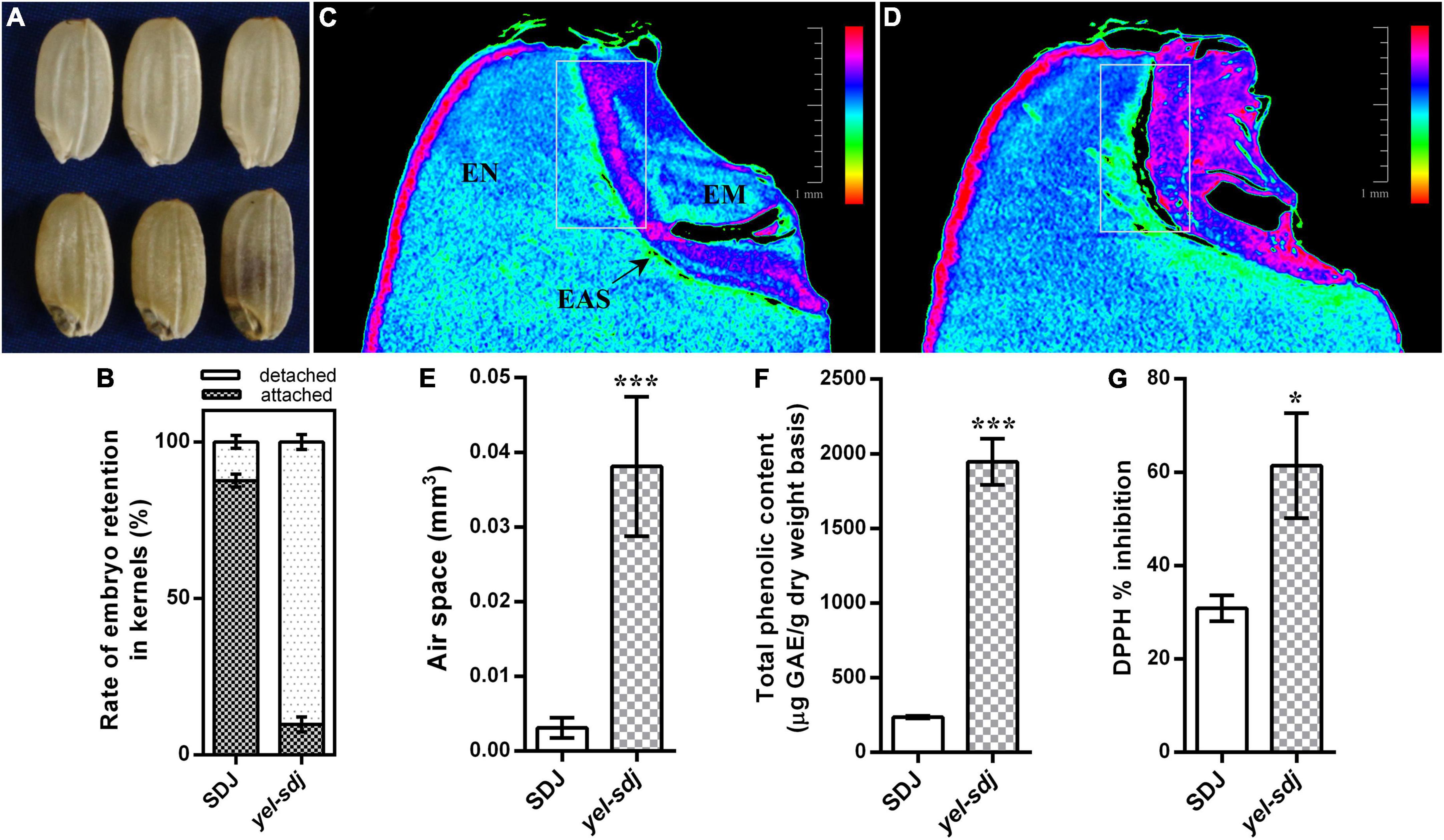
Figure 1. Comparison of the grain characteristics of wild-type (WT; Sindongjin [SDJ]) and yel-sdj mutant rice. (A) Evaluation of the morphology of WT (upper panel) and yel-sdj mutant (lower panel) grains. (B) Rate of embryo retention in kernels after milling for 5 s. Data represent the mean ± standard deviation (SD) of five biological replicates. (C,D) Longitudinal cross section of WT (C) and yel-sdj mutant (D) grains using computed tomography (CT). White rectangles indicate the area coinciding with the air space found between the embryo and endosperm (scale bar = 1 mm). The color scale represents embryo density. EN, endosperm; EM, embryo; EAS, endosperm adjacent to the scutellum. (E–G) Volume of air space between the embryo and endosperm (E), content of total phenolics (F), and antioxidant activity (G) in WT and yel-sdj mutant grains. Data represent the mean ± SD of three biological replicates. Asterisks indicate statistical significance, as determined by Student’s t-test (*p < 0.05, ***p < 0.001).
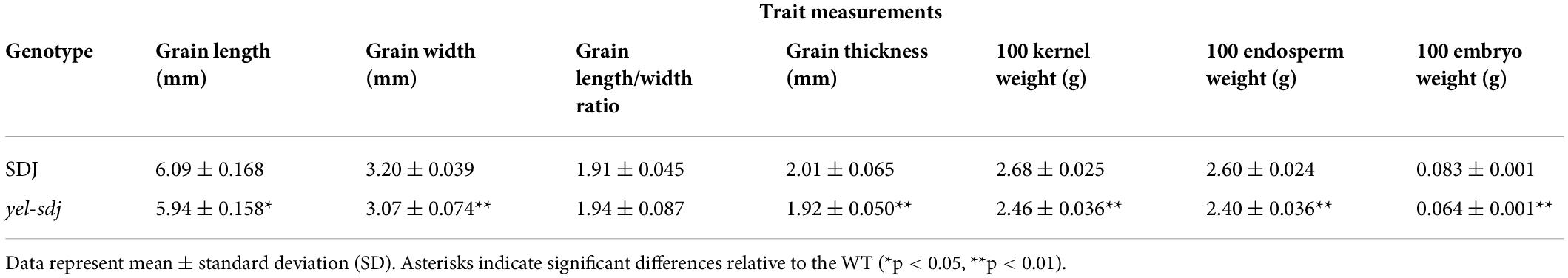
Table 1. Measurement of grain related traits in the wild-type (WT) rice cultivar Sindongjin (SDJ) and yel-sdj mutant.
While detaching embryos from grains to measure the embryo weight, we empirically found that the removal of yel-sdj embryos from grains was easier than that of WT embryos. Therefore, we investigated the rate of embryo retention in the kernels using a small-scale grain polisher. The results revealed that the rate of embryo retention was remarkably lower in the yel-sdj mutant than in the WT; while approximately 88% of WT kernels contained embryos after milling for 5 s, only 10% of yel-sdj kernels retained the embryos (Figure 1B). To understand why the strength of embryo attachment differed between yel-sdj and WT seeds, we examined the histological properties and internal morphology of WT and yel-sdj grains by CT. Interestingly, significantly greater volume of air space was observed between the embryo and endosperm in yel-sdj mutant seeds than in WT seeds (Figures 1C,D and Supplementary File 1). The volume of air space between the embryo and endosperm in the yel-sdj mutant (0.0381 ± 0.00834 mm3) was approximately 12 times higher than that in the WT (0.0031 ± 0.00121 mm3) (Figure 1E). In addition, the density of embryo was relatively higher, and the area of endosperm adjacent to the scutellum (EAS) was wider in the yel-sdj mutant than in the WT (Figures 1C,D). These results suggest that the formation of air space along the border between the embryo and endosperm is responsible for the easy detachment of embryo from the kernel in the yel-sdj mutant.
Seed TPC and antioxidant activity were assessed in the yel-sdj mutant and WT using grain extracts. The TPC of yel-sdj grains (1,946 μg GAE/g DW) was approximately eightfold higher than that of WT grains (236 μg GAE/g DW), a significant difference (p < 0.001) (Figure 1F). Additionally, the DPPH radical scavenging activity in yel-sdj grains (61.4% inhibition) was approximately twofold higher than that in WT grains (30.8% inhibition) (p < 0.05; Figure 1G). These results suggest that the high TPC of yel-sdj grains leads to increased antioxidant activity.
Since the homozygous yel-sdj mutant was embryo lethal, we crossed a heterozygous yel-sdj mutant plant with the WT cultivar SDJ and used the resultant F1 and F2 populations for genetic analysis. The F2 seeds showed a WT:yel-sdj segregation ratio of 3:1 (Table 2). Additionally, to determine whether the yel-sdj mutant allele is novel, we conducted an allelism test by crossing the yel-sdj mutant with three oscop1 null mutants, yel-hc, yel-cc, and yel-sk. Since homozygous oscop1 mutants are embryo lethal (like the yel-sdj mutant), heterozygous plants of each oscop1 mutant were used in these crosses. All F1 seeds obtained from the three crosses showed WT phenotype, and the F2 seeds of only some F1 plants showed a WT:yel segregation ratio of 9:7 (Table 2). Therefore, we conclude that the yel-sdj mutant phenotype was controlled by a single recessive gene, and yel-sdj and oscop1 null mutants (yel-hc, yel-sk, and yel-cc) were non-allelic, indicating that a novel locus is responsible for the yel-sdj phenotype.
An F2 population derived from a cross between a yel-sdj heterozygous mutant plant and M.23 was used to map the locus responsible for the yel-sdj phenotype. To conduct preliminary genetic mapping, BSA was performed using the 96 SNP array, and the yel-sdj locus was mapped to a region between the start of chromosome 1 and id1004256 (Figure 2A). To refine the flanking region, an F2 population of 549 individuals was genotyped using newly designed markers (Supplementary Table 1). Finally, the yel-sdj locus was mapped to an approximately 114 kb region between the S01002 and RM3252 markers, and 13 candidate genes were identified within this region (Figure 2A). Among these 13 genes, LOC_Os01g01484 (Os01g0104600), an ortholog of AtDET1, was selected as a strong candidate gene associated with the yel-sdj phenotype, given its previously reported role in embryonic lethality and anthocyanin accumulation in Arabidopsis. Sequence analysis in SDJ and yel-sdj mutant revealed a deletion of 7 bp (TATGAGA, where the A of ATG is +1 bp) at position +365 to +371 bp in the second exon of locus LOC_Os01g01484 in the yel-sdj mutant (Figure 2B). The 7 bp deletion was predicted to cause a frameshift and consequently a premature stop codon at the 45th amino acid, resulting in aberrant protein production (Figure 2C).
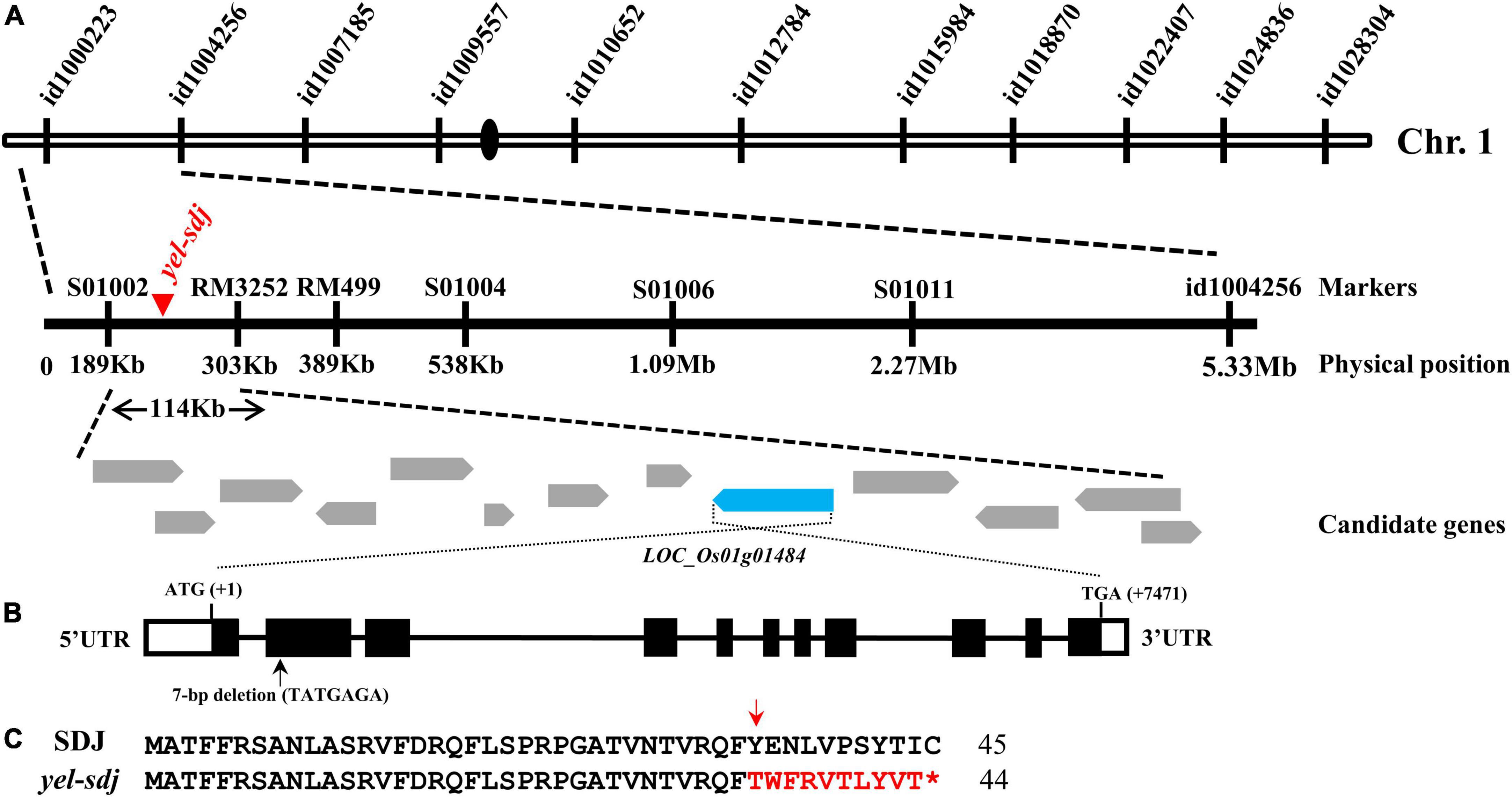
Figure 2. Map-based cloning of the gene responsible for the yel-sdj mutant phenotype. (A) Schematic showing the physical position of the causal locus on rice chromosome 1, as identified by map-based cloning. (B) Gene structure of OsDET1. Black lines, white solid boxes, and black solid boxes indicate introns, untranslated regions, and exons, respectively. The 7 bp deletion is indicated with a black arrow. ATG and TGA indicate the initiation and termination codons, respectively. (C) Comparison of the predicted amino acid sequence of the mutated region of gene between the WT (SDJ) and yel-sdj mutant. The 7 bp deletion resulted in a frameshift (red arrow) and premature stop (red asterisk) in yel-sdj. Amino acids in the frameshift region are indicated in red.
To confirm the association of OsDET1 with the yel phenotype, the protein-coding sequence of this gene was edited using the CRISPR/Cas9 system. Two gRNAs complimentary to the sequence located near the 7 bp deletion were designed to target the coding sequence of OsDET1, and an egg cell-specific Cas9 promoter was used for vector construction to overcome lethality during tissue culture in the homozygous T0 plant regeneration. A total of 24 positive T0 transgenic plants were obtained, and T1 seeds exhibiting the yel mutant phenotype were identified, although these seeds showed variable pericarp color (Figure 3A). Mutations in target regions in T1 seeds displaying the yel mutant phenotype were confirmed by PCR and Sanger sequencing. Sequence analysis revealed a variety of insertions and deletions at the two target sites in all yel phenotype seeds. Target site 1 (gRNA1) showed relatively higher frequency of mutations than target site 2 (gRNA2) (Figure 3B). All insertions and deletions introduced into the OsDET1 gene in yel phenotype seeds were predicted to lead to frameshift mutations and premature stop codons. These results demonstrated that the yel mutant phenotype was caused by the loss-of-function of OsDET1. Additionally, embryo development was severely compromised in the osdet1 null mutant, indicating that OsDET1 is essential for maintaining normal embryogenesis in rice.
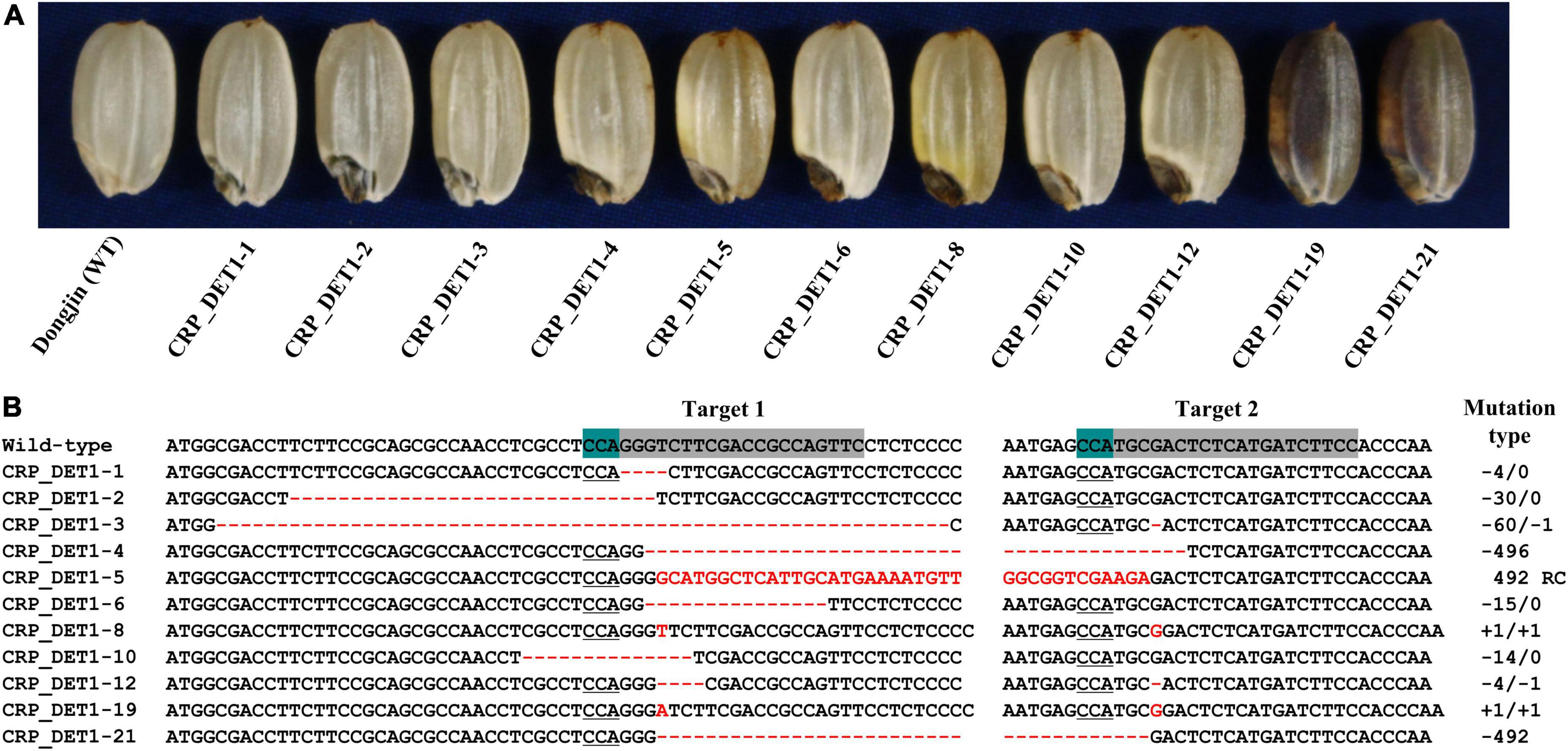
Figure 3. Grain phenotypic analysis and sequence analysis of CRISPR/Cas9-induced knockout yel mutants generated by targeting the OsDET1 gene. (A) Grain phenotype of Dongjin (WT) and transgenic seeds. Grains showing the yel phenotype were randomly selected from each positive T0 transgenic plant. (B) Comparison of the OsDET1 nucleotide sequence targeted by CRISPR/Cas9 in WT and mutant plants. Targets 1 and 2 represent the first and second exons, respectively, of OsDET1 (LOC_Os01g01484/Os01g0104600). Red dashes and letters indicate deletions and insertions, respectively, in transgenic lines. The protospacer adjacent motif (PAM) is highlighted in green in the WT and is underlined in mutant lines. Sequences of targets 1 and 2 are highlighted in gray in the WT. Mutation types are shown to the right of each mutated sequence (-, deletion; +, insertion; RC, reverse complementary).
To determine if the genes encoding OsCOP10 and OsDDB1, which form the CDD complex together with OsDET1, are also involved in embryo development and flavonoid biosynthesis in rice, we mutated OsCOP10 and OsDDB1 using the CRISPR/Cas9 technology. CRISPR/Cas9 vectors designed to target the first exon of OsCOP10 (LOC_Os07g38940/Os07g0577400) or second exon of OsDDB1 (LOC_Os05g51480/Os05g0592400) were used for rice transformation, and seeds appearing phenotypically similar to those with the yel phenotype were collected from T0 positive transgenic plants (Figures 4A,B). Analysis of sequences targeted by OsCOP10- and OsDDB1-specific gRNAs revealed that all seeds exhibiting the yel phenotype carried mutations at the target sites (Figure 4C). This result indicates that loss-of-function mutations of OsCOP10 and OsDDB1 result in embryo lethality and altered pigmentation of the embryo and pericarp. Furthermore, this result implies that OsDET1 associates with OsCOP10 and OsDDB1 to form the CDD complex, and genes encoding all three proteins participate together in pathways regulating embryogenesis and flavonoid biosynthesis in rice.
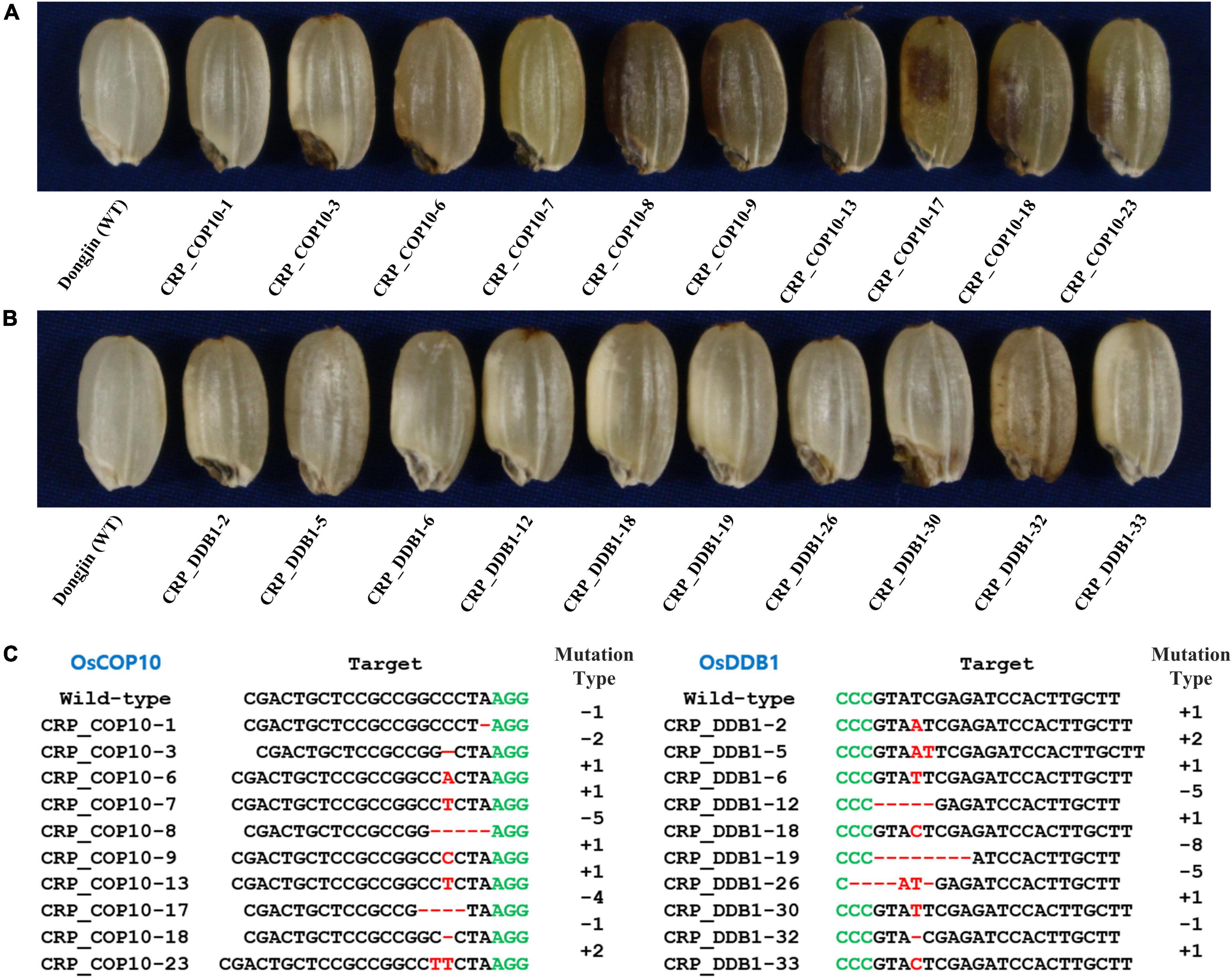
Figure 4. Grain phenotypic analysis and sequence analysis of CRISPR/Cas9-induced knockout yel mutants generated by targeting OsCOP10 and OsDDB1 genes. (A,B) Grain phenotype of Dongjin (WT) seeds and CRISPR mutant seeds generated by targeting OsCOP10 (A) and OsDDB1 (B). Grains showing the yel phenotype were randomly selected from each positive T0 transgenic plant. (C) Sequence comparison of OsCOP10 (LOC_Os07g38940/Os07g0577400) and OsDDB1 (LOC_Os05g51480/Os05g0592400) target regions in the WT and mutants. The first exon of OsCOP10 and second exon of OsDDB1 were targeted to induce mutations. Red dashes and letters indicate deletions and insertions, respectively, in transgenic lines. Black and green letters indicate the target sequence and PAM, respectively. Mutation types are shown to the right of each mutated sequence (-, deletion; +, insertion).
The expression pattern of OsDET1 in different plant organs was investigated by qRT-PCR. We found that OsDET1 was predominantly expressed in the leaf and relatively higher in the leaf sheath and developing seed than in the root and young panicle in WT plants (Figure 5A). Since the yel phenotype was observed in seeds carrying mutations in photomorphogenesis-related genes, we examined the expression levels of genes encoding the CDD complex components and OsCOP1 in yel-sdj mutant seeds at 7 days after pollination (DAP). The relative expression levels of OsDET1 and OsDDB1 were lower, whereas that of OsCOP1 was significantly higher in developing yel-sdj mutant seeds than in WT seeds. No significant difference was detected in expression level of OsCOP10 between the WT and yel-sdj mutant (Figure 5B).
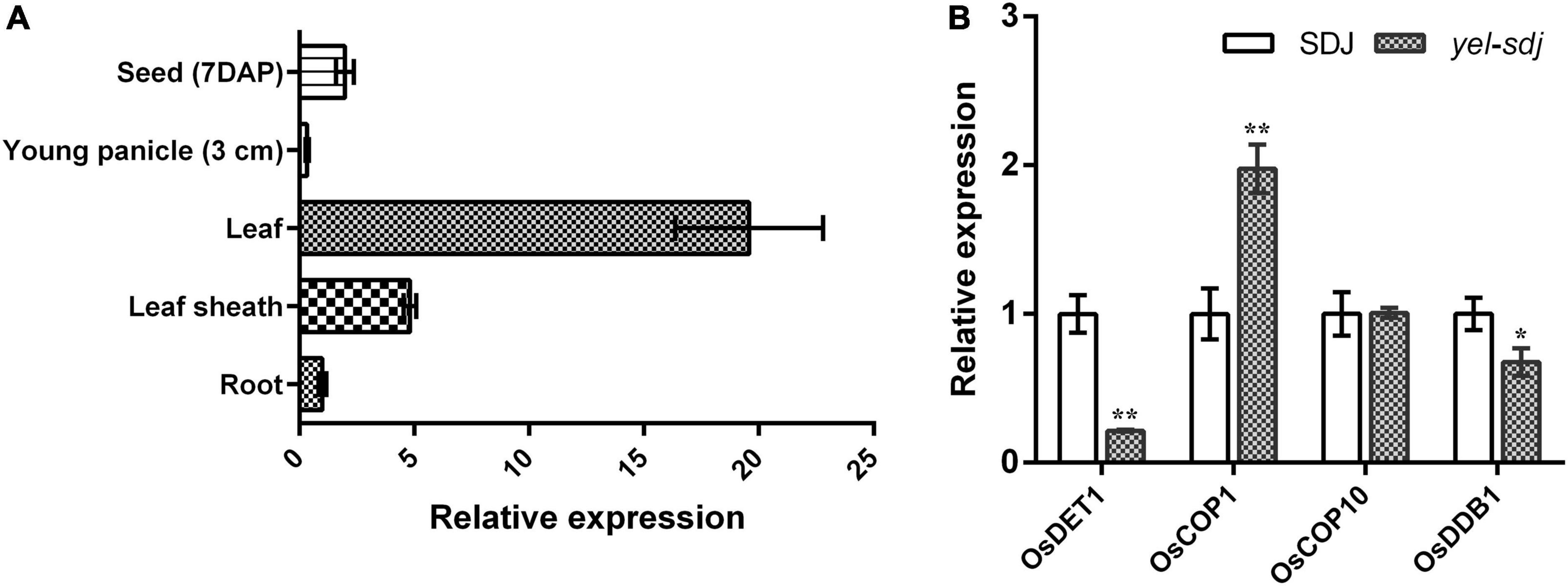
Figure 5. Expression analysis of OsDET1 and yel phenotype-associated genes. (A) Quantitative real-time PCR (qRT-PCR) analysis of OsDET1 in various organs of SDJ (WT). (B) Relative expression levels of yel phenotype-associated genes in the developing seeds of SDJ (WT) and yel-sdj mutant at 7 days after pollination (DAP). Expression level of genes was normalized relative to that of ACTIN. Data represent mean ± SD of three biological replicates. Asterisks indicate statistical significance, as determined by Student’s t-test (*p < 0.05, **p < 0.01).
DET1, a key negative regulator of light signaling, has been extensively studied as a repressor of photomorphogenesis, together with COP1, in Arabidopsis. However, unlike in Arabidopsis, only a few det1 mutants have been reported in rice. Genetic complementation analysis demonstrated that a single nucleotide mutation in OsDET1 can increase the leaf chlorophyll content in rice (Huang et al., 2013). Furthermore, OsDET1 influences rice seed germination and seedling growth, and triggers dark-induced leaf senescence, by modulating the signaling pathway and biosynthesis of abscisic acid (ABA) (Zang et al., 2016). In the present study, we identified a novel det1 null mutant of rice that displayed yellow and black pigmentation in the pericarp and embryo, respectively, and exhibited embryonic lethality (Figure 1A). The yel-sdj mutant, unlike previously reported rice mutants harboring a weak allele or transgenic rice plants generated by RNA interference (RNAi), is likely to be complete a loss-of-function mutant, based on the severity of its phenotype. This presumption is supported by the yel mutant phenotype of T1 transgenic seeds of the osdet1 knockout mutant generated using the CRISPR/Cas9 system (Figure 3). Furthermore, severe developmental defects, leading to seedling lethality and failure to germinate, have been observed in OsDET1 RNAi transgenic plants, which expression level was reduced markedly at the vegetative growth stage and T1 transgenic seeds, respectively (Zang et al., 2016). These results indicate that yel-sdj is a null mutant, and its yel phenotype is caused by the loss-of-function of OsDET1. In addition, we previously identified three Arabidopsis “fusca”-like cop1 null mutants (yel-hc, yel-sk, and yel-cc) in rice. These mutants carry loss-of-function alleles of OsCOP1 and share several phenotypic characteristics with the yel-sdj mutant, including embryonic lethality and high-level flavonoid accumulation in the pericarp and embryo (Kim et al., 2021). Similarly, the Arabidopsis det1 null mutant displays the phenotypic characteristics of strong cop1 mutant alleles, such as short hypocotyls, opened cotyledons, and anthocyanin accumulation (McNellis et al., 1994; Misera et al., 1994; Pepper et al., 1994). Thus, the phenotype of the osdet1 null mutant is similar to that of oscop1 null mutants, which implies a molecular link between OsDET1 and OsCOP1, both of which are involved in the regulation of embryo development and flavonoid biosynthesis in rice.
The role of the CDD complex in plants has been discovered through mutant screens. In Arabidopsis, both DET1 and COP10 have been identified as one of the pleiotropic COP/DET/FUS loci that act to repress photomorphogenic development of seedlings in the dark. Additionally, the cop10-1 and det1-6 T-DNA insertion mutants exhibit seedling lethality and fus phenotypic characteristics (such as anthocyanin accumulation) (Pepper et al., 1994; Wei et al., 1994). Consistent with these observations, complementation tests determined that det1 and cop10 are allelic to fus2 and fus9, respectively (Castle and Meinke, 1994; Misera et al., 1994). However, mutations in DDB1, which encodes another component of the CDD complex, result in different phenotypic changes compared with mutations in DET1 and COP10. For example, the ddb1a null mutant shows no obvious phenotype, whereas the det1-1 ddb1a double mutant exhibits enhanced det1 null mutant phenotype. By contrast, the loss-of-function ddb1b mutants exhibit both embryo lethal and viable phenotypes (Schroeder et al., 2002; Bernhardt et al., 2010). In tomato (Solanum lycopersicum), the High Pigment (HP) genes, HP1 (Liu et al., 2004) and HP2 (Mustilli et al., 1999), encode DDB1 and DET1 homologs of Arabidopsis, respectively; however, the phenotypic characteristics of tomato hp1 and hp2 mutants differ from those of Arabidopsis ddb1 and det1 mutants, respectively. Tomato hp1 and hp2 mutants show no obvious phenotype in the dark; however, when grown under light, both display high-level anthocyanin accumulation in seedlings, short, and dark plants, dark-green immature fruits (due to the overproduction of chlorophyll), and increased flavonoid and carotenoid production in ripe fruits (Yen et al., 1997; Mustilli et al., 1999; Liu et al., 2004). The phenotypes of LeCOP1LIKE RNAi seedlings and fruits are similar to those of light-grown hp1 and hp2 mutants (Mustilli et al., 1999; Liu et al., 2004). Besides phenotypic similarities among the mutants of CDD components, the evidence that the genes forming CDD complex are associated with each other has been revealed at the molecular level. OsDET1 interacts physically with OsDDB1 and OsCOP10 in rice. (Zang et al., 2016), and DET1 and DDB1 interact with each other to suppress photomorphogenesis in Arabidopsis (Schroeder et al., 2002). In addition, COP10 interacts with COP1 to promote the degradation of photomorphogenesis-regulating proteins in Arabidopsis (Suzuki et al., 2002; Yanagawa et al., 2004). In this study, we demonstrated that all seeds displaying the yel phenotype carried mutations in OsDET1, OsCOP10, and OsDDB1 genes (Figures 1A, 3A, 4A,B). Given the phenotype of our knockout mutants, this result implies that genes encoding the CDD components in rice, OsDET1, OsCOP10, and OsDDB1, are essential for the functional roles or share molecular pathways or genetic signals during embryo development and flavonoid biosynthesis. Furthermore, the osdet1 null mutant was phenotypically highly similar to the oscop1 null mutants (yel-hc, yel-cc, and yel-sk), although COP1 and DET1 seem to regulate ubiquitination independently through distinct multimeric units. This result suggests the possibility that the CDD complex functions together with COP1 to regulate flavonoid biosynthesis and embryogenesis in rice. These findings suggest that OsDET1, OsCOP10, OsDDB1, and OsCOP1 perform a common function, and therefore the loss-of-function mutation of any of these genes results in the yel phenotype in rice seeds. Taken together, these findings explain why different yel mutants, harboring mutations in different genes, exhibit phenotypic similarities.
In Arabidopsis, COP1 is a RING E3 ubiquitin ligase that works in complex with SUPPRESSOR OF PHYA-105 (SPA1) protein, and the COP1/SPA1 complex targets photomorphogenic-promoting transcription factors, such as HY5, HY5 HOMOLOG (HYH), LONG HYPOCOTYL IN FAR-RED 1 (HFR1) for ubiquitination, and protein degradation in the dark (Hoecker and Quail, 2001; Holm et al., 2002; Jang et al., 2005). Furthermore, the CDD complex, consisting of DET1, COP10, and DDB1, acts together with the COP1 E3 ligase complex for the COP1-mediated protein degradation (Osterlund et al., 2000; Yanagawa et al., 2004; Canibano et al., 2021). In addition, the CDD complex forms CUL4-CDD E3 ubiquitin ligase through DDB1 and enhances E3 activity, which is required for the degradation of key regulators and other substrates (Chen et al., 2006; Lau and Deng, 2012). Among the photomorphogenesis-promoting transcription factors, it is well known that HY5, a bZIP transcription factor, is a central regulator of photomorphogenesis and positively regulates anthocyanin biosynthesis by binding transcription factors, such as PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1), MYB12, MYB111, which further activate the regulatory genes and structural genes (Holm et al., 2002; Stracke et al., 2010; Gangappa and Botto, 2016). However, unlike Arabidopsis, little is known about the role of OsDET1, OsCOP10, and OsDDB1, which affect flavonoid biosynthesis in rice. Thus, the possible mechanism could be that the OsDET1, OsCOP10, and OsDDB1 mutations inhibit HY5 ubiquitination and degradation, and the resulting yel phenotype may support the transcriptional regulatory role where the COP1 and CDD complex negatively regulates HY5, a positive regulator of flavonoid biosynthesis. Namely, upregulated HY5 activates transcription factors which regulates flavonoid biosynthesis genes, resulting in flavonoid accumulation in embryo and pericarp of rice grain. The molecular function and mechanism of OsDET1, OsCOP10, and OsDDB1, CDD complex components, in flavonoid biosynthesis still remain unclear in rice. Therefore, further efforts are required to better understand the role of the CDD complex in flavonoid biosynthesis including embryo development.
Interaction between the embryo and endosperm affects not only the growth of embryo and endosperm itself but also seed development. It has been reported that maize embryo is separated from the endosperm by fibrous layer and EAS (Doll and Ingram, 2022). In particular, the EAS, which originates from the starchy endosperm cell layer adjacent to the scutellum, is the region where the cell death and the accumulation of crushed cell walls happen, allowing the embryo expansion as the embryo grows toward the endosperm (Doll et al., 2020; Doll and Ingram, 2022). In addition, transcriptome analysis revealed that the genes involved in sugar and amino acid transport such as the SWEET family and UMAMIT family genes are strongly activated in the EAS to provide nutrition to the embryo (Doll et al., 2020). In the present study, we found that the embryo of yel-sdj mutant is easily detached from the kernels (Figure 1B), and micro-CT analysis revealed that a dramatically wide area of EAS was observed in matured yel-sdj mutant seed (Figures 1C,D). Although the molecular mechanism associated with the development of EAS by the mutation of OsDET1 is still unclear, it may be assumed that the degradation of starch or cell death in the broad EAS region reduced EAS density, resulting in loosening of the starch granules packing and embryo-endosperm interface tissue. Subsequently, as the seeds mature, its water content decreases thus, embryo split from the adjacent endosperm and forms air space between embryo and endosperm. This abnormal embryo-endosperm adhesion enables the embryo of yel-sdj to detach easily from the kernels. Taken together, the elucidation of OsDET1 function, which affects EAS formation, will provide novel insights into seed development, especially the embryo-endosperm interaction of monocots, and expand our understanding of the molecular mechanisms during embryogenesis in higher plants.
Overall, we identified a novel mutant (yel-sdj) exhibiting yellowish pericarp and embryonic lethality, and showed that OsDET1 plays a crucial role in flavonoid biosynthesis and embryogenesis in rice seed. In addition, we demonstrated that mutations in OsCOP10 and OsDDB1, which encode members of the CDD complex, cause phenotypes similar to those exhibited by the typical yel mutants, such as yel-hc, yel-cc, yel-sk, and yel-sdj. Additionally, our results demonstrated that modification of the light signal transduction machinery could have a significant effect on flavonoid biosynthesis and embryo development in rice seed. Further examination of mutations in other light signal transduction machinery genes, whose proteins associate with OsDET1 and OsCOP1, will facilitate a better understanding of the common molecular mechanisms and metabolic pathways involved in embryo development and flavonoid biosynthesis in rice seed.
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
BK designed and performed the research, analyzed the data, and wrote the manuscript. Y-HC provided the material. BK, YL, J-YN, GL, and JS carried out experiments. DL validated the data. S-WK designed and supervised the experiment. H-JK designed and supervised the experiment and revised the manuscript. All authors have read and agreed to the final version of the manuscript.
This work was supported by the New Breeding Technologies Development Program (Project No. PJ 01654601), Rural Development Administration, South Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.952856/full#supplementary-material
An, L., Tao, Y., Chen, H., He, M., Xiao, F., Li, G., et al. (2020). Embryo-endosperm interaction and its agronomic relevance to rice quality. Front. Plant Sci. 11:587641. doi: 10.3389/fpls.2020.587641
Bernhardt, A., Mooney, S., and Hellmann, H. (2010). Arabidopsis DDB1a and DDB1b are critical for embryo development. Planta 232, 555–566. doi: 10.1007/s00425-010-1195-9
Canibano, E., Bourbousse, C., Garcia-Leon, M., Garnelo Gomez, B., Wolff, L., Garcia-Baudino, C., et al. (2021). DET1-mediated COP1 regulation avoids HY5 activity over second-site gene targets to tune plant photomorphogenesis. Mol. Plant 14, 963–982. doi: 10.1016/j.molp.2021.03.009
Castells, E., Molinier, J., Drevensek, S., Genschik, P., Barneche, F., and Bowler, C. (2010). det1-1-induced UV-C hyposensitivity through UVR3 and PHR1 photolyase gene over-expression. Plant J. 63, 392–404. doi: 10.1111/j.1365-313X.2010.04249.x
Castle, L. A., and Meinke, D. W. (1994). A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell 6, 25–41. doi: 10.1105/tpc.6.1.25
Chen, H., Shen, Y., Tang, X., Yu, L., Wang, J., Guo, L., et al. (2006). Arabidopsis CULLIN4 Forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18, 1991–2004. doi: 10.1105/tpc.106.043224
Chory, J., Peto, C., Feinbaum, R., Pratt, L., and Ausubel, F. (1989). Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell 58, 991–999. doi: 10.1016/0092-8674(89)90950-1
Chory, J., and Peto, C. A. (1990). Mutations in the DET1 gene affect cell-type-specific expression of light-regulated genes and chloroplast development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 87, 8776–8780. doi: 10.1073/pnas.87.22.8776
Chu, G., and Chang, E. (1988). Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 242, 564–567. doi: 10.1126/science.3175673
Chung, I. M., Oh, J. Y., and Kim, S. H. (2017). Comparative study of phenolic compounds, vitamin E, and fatty acids compositional profiles in black seed-coated soybeans (Glycine Max (L.) Merrill) depending on pickling period in brewed vinegar. Chem. Cent. J. 11, 64. doi: 10.1186/s13065-017-0298-9
Clark, J. K., and Sheridan, W. F. (1991). Isolation and characterization of 51 embryo-specific Mutations of Maize. Plant Cell 3, 935–951. doi: 10.1105/tpc.3.9.935
Doll, N. M., and Ingram, G. C. (2022). Embryo-Endosperm Interactions. Annu. Rev. Plant Biol. 73, 293–321. doi: 10.1146/annurev-arplant-102820-091838
Doll, N. M., Just, J., Brunaud, V., Caius, J., Grimault, A., Depege-Fargeix, N., et al. (2020). Transcriptomics at Maize Embryo/Endosperm Interfaces Identifies a Transcriptionally Distinct Endosperm Subdomain Adjacent to the Embryo Scutellum([OPEN]). Plant Cell 32, 833–852. doi: 10.1105/tpc.19.00756
Fedorov, A., Beichel, R., Kalpathy-Cramer, J., Finet, J., Fillion-Robin, J. C., Pujol, S., et al. (2012). 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 30, 1323–1341. doi: 10.1016/j.mri.2012.05.001
Gangappa, S. N., and Botto, J. F. (2016). The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353–1365. doi: 10.1016/j.molp.2016.07.002
Ganpudi, A. L., and Schroeder, D. F. (2013). Genetic interactions of Arabidopsis thaliana damaged DNA binding protein 1B (DDB1B) with DDB1A, DET1, and COP1. G3 3, 493–503. doi: 10.1534/g3.112.005249
He, Y. Z. J., Mccall, C. M., Hu, J., Zeng, Y. X., and Xiong, Y. (2006). DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 20, 2949–2954. doi: 10.1101/gad.1483206
Hoecker, U., and Quail, P. H. (2001). The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis. J. Biol. Chem. 276, 38173–38178. doi: 10.1074/jbc.M103140200
Holm, M., Ma, L. G., Qu, L. J., and Deng, X. W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. doi: 10.1101/gad.969702
Hong, S. K., Aoki, T., Kitano, H., Satoh, H., and Nagato, Y. (1995). Phenotypic diversity of 188 Rice Embryo Mutants. Dev. Genet. 16, 298–310. doi: 10.1002/dvg.1020160403
Huang, J., Qin, F., Zang, G., Kang, Z., Zou, H., Hu, F., et al. (2013). Mutation of OsDET1 increases chlorophyll content in rice. Plant Sci. 210, 241–249. doi: 10.1016/j.plantsci.2013.06.003
Jang, I. C., Yang, J. Y., Seo, H. S., and Chua, N. H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19, 593–602. doi: 10.1101/gad.1247205
Kang, M. Y., Yoo, S. C., Kwon, H. Y., Lee, B. D., Cho, J. N., Noh, Y. S., et al. (2015). Negative regulatory roles of DE-ETIOLATED1 in flowering time in Arabidopsis. Sci. Rep. 5:9728. doi: 10.1038/srep09728
Kim, B., Piao, R., Lee, G., Koh, E., Lee, Y., Woo, S., et al. (2021). OsCOP1 regulates embryo development and flavonoid biosynthesis in rice (Oryza sativa L.). Theor. Appl. Genet. 134, 2587–2601. doi: 10.1007/s00122-021-03844-9
Kim, B., Woo, S., Kim, M. J., Kwon, S. W., Lee, J., Sung, S. H., et al. (2018). Identification and quantification of flavonoids in yellow grain mutant of rice (Oryza sativa L.). Food Chem. 241, 154–162. doi: 10.1016/j.foodchem.2017.08.089
Kim, S. H., Yang, Y. J., and Chung, I. M. (2020). The effect of degree of milling on the nutraceutical content in ecofriendly and conventional rice (Oryza sativa L.). Foods 9:1297. doi: 10.3390/foods9091297
Kitano, H., Tamura, Y., Satoh, H., and Nagato, Y. (1993). Hierarchical regulation of organ differentiation during embryogenesis in rice. Plant J. 3, 607–610. doi: 10.1046/j.1365-313X.1993.03040607.x
Lafon-Placette, C., and Kohler, C. (2014). Embryo and endosperm, partners in seed development. Curr. Opin. Plant Biol. 17, 64–69. doi: 10.1016/j.pbi.2013.11.008
Lau, O. S., and Deng, X. W. (2009). Effect of Arabidopsis COP10 ubiquitin E2 enhancement activity across E2 families and functional conservation among its canonical homologues. Biochem. J. 418, 683–690. doi: 10.1042/BJ20081943
Lau, O. S., and Deng, X. W. (2012). The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 17, 584–593. doi: 10.1016/j.tplants.2012.05.004
Lau, O. S., Huang, X., Charron, J. B., Lee, J. H., Li, G., and Deng, X. W. (2011). Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell 43, 703–712. doi: 10.1016/j.molcel.2011.07.013
Lee, J., and Zhou, P. (2007). DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 26, 775–780. doi: 10.1016/j.molcel.2007.06.001
Liu, Y. S., Roof, S., Ye, Z. B., Barry, C., Van Tuinen, A., Vrebalov, J., et al. (2004). Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl. Acad. Sci. U.S.A. 101, 9897–9902. doi: 10.1073/pnas.0400935101
Lowder, L. G., Zhang, D., Baltes, N. J., Paul, J. W. III, Tang, X., Zheng, X., et al. (2015). A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. doi: 10.1104/pp.15.00636
Mayer, R., Raventos, D., and Chua, N. H. (1996). det1, cop1, and cop9 mutations cause inappropriate expression of several gene sets. Plant Cell 8, 1951–1959. doi: 10.1105/tpc.8.11.1951
McNellis, T. W., Von Arnim, A. G., Araki, T., Komeda, Y., Misera, S., and Deng, X. W. (1994). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6, 487–500. doi: 10.1105/tpc.6.4.487
Meinke, D. W. (1985). Embryo-lethal mutants of Arabidopsis thaliana: analysis of mutants with a wide range of lethal phases. Theor. Appl. Genet. 69, 543–552. doi: 10.1007/BF00251102
Meinke, D. W., and Sussex, I. M. (1979b). Isolation and characterization of six embryo-lethal mutants of Arabidopsis thaliana. Dev. Biol. 72, 62–72. doi: 10.1016/0012-1606(79)90098-8
Meinke, D. W., and Sussex, I. M. (1979a). Embryo-lethal mutants of Arabidopsis thaliana: a model system for genetic analysis of plant embryo development. Dev. Biol. 72, 50–61. doi: 10.1016/0012-1606(79)90097-6
Millar, A. J., Straume, M., Chory, J., Chua, N. H., and Kay, S. A. (1995). The regulation of circadian period by phototransduction pathways in Arabidopsis. Science 267, 1163–1166. doi: 10.1126/science.7855596
Misera, S., Muller, A. J., Weiland-Heidecker, U., and Jurgens, G. (1994). The FUSCA genes of Arabidopsis: negative regulators of light responses. Mol. Gen. Genet. 244, 242–252. doi: 10.1007/BF00285451
Mustilli, A. C., Fenzi, F., Ciliento, R., Alfano, F., and Bowler, C. (1999). Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11, 145–157. doi: 10.1105/tpc.11.2.145
Nagato, Y., Kitano, H., Kamijima, O., Kikuchi, S., and Satoh, H. (1989). Developmental mutants showing abnormal organ differentiation in rice embryos. Theor. Appl. Genet. 78, 11–15. doi: 10.1007/BF00299746
Naito, Y., Hino, K., Bono, H., and Ui-Tei, K. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. doi: 10.1093/bioinformatics/btu743
Nishimura, A., Aichi, I., and Matsuoka, M. (2006). A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 1, 2796–2802. doi: 10.1038/nprot.2006.469
Osterlund, M. T., Hardtke, C. S., Wei, N., and Deng, X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. doi: 10.1038/35013076
Park, J., Bae, S., and Kim, J. S. (2015). Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31, 4014–4016. doi: 10.1093/bioinformatics/btv537
Pepper, A., Delaney, T., Washburn, T., Poole, D., and Chory, J. (1994). DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell 78, 109–116. doi: 10.1016/0092-8674(94)90577-0
Satoh, N., Hong, S. K., Nishimura, A., Matsuoka, M., Kitano, H., and Nagato, Y. (1999). Initiation of shoot apical meristem in rice: characterization of four SHOOTLESS genes. Development 126, 3629–3636. doi: 10.1242/dev.126.16.3629
Schroeder, D. F., Gahrtz, M., Maxwell, B. B., Cook, R. K., Kan, J. M., Alonso, J. M., et al. (2002). De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12, 1462–1472. doi: 10.1016/s0960-9822(02)01106-5
Seo, J., Lee, G., Jin, Z., Kim, B., Chin, J. H., and Koh, H. J. (2020). Development and application of indica-japonica SNP assays using the Fluidigm platform for rice genetic analysis and molecular breeding. Mol. Breed. 40:39. doi: 10.1007/s11032-020-01123-x
Sheridan, W. F., and Clark, J. K. (1993). Mutational analysis of morphogenesis of the maize embryo. Plant J. 3, 347–358. doi: 10.1111/j.1365-313X.1993.tb00186.x
Shi, H., Wang, X., Mo, X., Tang, C., Zhong, S., and Deng, X. W. (2015). Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc. Natl. Acad. Sci. U.S.A. 112, 3817–3822. doi: 10.1073/pnas.1502405112
Stracke, R., Favory, J. J., Gruber, H., Bartelniewoehner, L., Bartels, S., Binkert, M., et al. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33, 88–103. doi: 10.1111/j.1365-3040.2009.02061.x
Suzuki, G., Yanagawa, Y., Kwok, S. F., Matsui, M., and Deng, X. W. (2002). Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16, 554–559. doi: 10.1101/gad.964602
Wei, N., Kwok, S. F., Von Arnim, A. G., Lee, A., Mcnellis, T. W., Piekos, B., et al. (1994). Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell 6, 629–643. doi: 10.1105/tpc.6.5.629
Winters, A. L., and Minchin, F. R. (2005). Modification of the Lowry assay to measure proteins and phenols in covalently bound complexes. Anal. Biochem. 346, 43–48. doi: 10.1016/j.ab.2005.07.041
Yanagawa, Y., Sullivan, J. A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., et al. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18, 2172–2181. doi: 10.1101/gad.1229504
Yen, H. C., Shelton, B. A., Howard, L. R., Lee, S., Vrebalov, J., and Giovannoni, J. J. (1997). The tomato high-pigment (hp) locus maps to chromosome 2 and influences plastome copy number and fruit quality. Theor. Appl. Genet. 95, 1069–1079. doi: 10.1007/s001220050664
Keywords: yellowish-pericarp embryo lethal (yel) mutant, OsDET1, CDD complex, embryo development, CRISPR/Cas9, rice (Oryza sativa)
Citation: Kim B, Lee Y, Nam J-Y, Lee G, Seo J, Lee D, Cho Y-H, Kwon S-W and Koh H-J (2022) Mutations in OsDET1, OsCOP10, and OsDDB1 confer embryonic lethality and alter flavonoid accumulation in Rice (Oryza sativa L.) seed. Front. Plant Sci. 13:952856. doi: 10.3389/fpls.2022.952856
Received: 25 May 2022; Accepted: 06 July 2022;
Published: 25 July 2022.
Edited by:
Apichart Vanavichit, Kasetsart University, ThailandReviewed by:
Yanfeng Ding, Nanjing Agricultural University, ChinaCopyright © 2022 Kim, Lee, Nam, Lee, Seo, Lee, Cho, Kwon and Koh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soon-Wook Kwon, c3drd29uQHB1c2FuLmFjLmty; Hee-Jong Koh, aGVlamtvaEBzbnUuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.