
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 August 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.951824
This article is part of the Research TopicPlant-derived natural compounds in drug discovery: The prism perspective between plant phylogeny, chemical composition, and medicinal efficacyView all 13 articles
The genetic relationships among the species in Scutellaria genus remain unclear because of the variation in the number of species and complex trait. The usage of S. baicalensis and its four substitute medicinal species (S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula) in traditional medicines make their specialized metabolism important in China, but interspecific genetic and chemical differences have rarely been reported for these species. In this study, the chloroplast genomes of four substitute species for S. baicalensis were assembled, and comparative and phylogenetic analyses were performed with these species and other Scutellaria relatives. In addition, metabolomics analyses were performed and the contents of the main active compounds were determined to reveal the interspecific chemical diversity of S. baicalensis and its four substitute species. The full lengths of their chloroplast genomes ranged from 151,574 to 151,816 bp with an average GC content of 38.34%, and a total of 113 genes were annotated. In the chloroplast genomes of S. baicalensis and its four substitutes, one hypervariable region (petA-psbL) is proposed as a potential DNA barcode. Phylogenetic analysis showed that the subdivision of the genus Scutellaria should be reconsidered. The metabolomics and content determination analyses showed that the four species exhibit a metabolism similar to that of S. baicalensis in different parts. Except for the roots of S. likiangensis, all parts of the substitute species showed high contents of baicalin. Genetic and chemical analyses of four substitute medicinal species for S. baicalensis were performed here for the first time, and their pharmacophylogenetic relationships were further explored, providing a scientific basis for the subsequent development of the medicinal value and resource utilization of Scutellaria.
Scutellaria is a significant genus in the family Lamiaceae, which includes approximately 360–469 accepted species worldwide (Ranjbar and Mahmoudi, 2017; Safikhani et al., 2018; Zhao et al., 2020; Shen et al., 2021). According to records, 72 species in this genus have been used with a long history as traditional herbal medicines to treat various diseases, such as cancer and inflammatory, hepatic, gastric, neurological, and cardiovascular diseases (Marsh et al., 2014; Grzegorczyk-Karolak et al., 2016; Wang et al., 2018, 2020). Scutellaria species are rich in flavonoids and terpenoids, and the main compounds baicalein, baicalin, and wogonin have been evaluated in clinical trials (Shen et al., 2021).
In China, S. baicalensis is the most widely studied species in Scutellaria. Scutellariae Radix, consisting of the roots of S. baicalensis, has been used as a traditional Chinese medicine for thousands of years and is officially recorded in successive editions of the Chinese Pharmacopoeia (Wang et al., 2018). According to research, the roots of some closely related species, such as S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula, are similar in shape to the roots of S. baicalensis, and they have abundant natural medicinal resources and have been widely used as substitutes for Scutellariae Radix in China (Shen et al., 2021). These alternatives have been used as herbal folk medicines for a long time and are listed in some local flora and local standards for traditional Chinese medicine. In the current Sichuan Standard for Traditional Chinese Medicine (2010), Scutellariae Amoenae Radix (Chuan Huang Qin) originating from the roots of S. amoena, S. hypericifolia, and S. tenax var. patentipilosa is used to prevent miscarriage and bleeding. In addition, in the current standard Quality standard of Chinese and ethnic medicinal materials in Guizhou Province (2003), the root of S. amoena is listed as Scutellariae Amoenae Radix (Xi’nan Huangqin) and described as useful for treating fever, cough, hemoptoe, jaundice, diarrhea, and carbuncles and for preventing miscarriage. Furthermore, the root of S. viscidula was listed in the Drug Standard of Jilin Province (1977) as Radix Scutellariae Viscidula and described as useful for treating cough, jaundice, diarrhea, and swelling and pain of the eye and for preventing miscarriage. Despite their important medicinal value, the genetic and chemical diversity of these substitutes for S. baicalensis remains unclear.
According to the Flora of China (Wu and Li, 1977), S. baicalensis and its substitute species (S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula) all belong to the Scutellaria subgen. Scutellaria sect. Stachymacris subsect. Angustifoliae clade, which has high similarity in terms of morphology and root shape. To date, only one study has used the plastid molecular marker trnL-F to discriminate between S. baicalensis and its adulterants (S. amoena and S. viscidula) (Wang et al., 2012). In recent years, chloroplast (cp) genomes have played an essential role in identifying closely related plant species and performing phylogenetic analysis since cp DNA is more discriminating and versatile than nuclear DNA sequences (Cui et al., 2019a; Zhao et al., 2020). The cp is an essential photosynthetic organelle in plants (Daniell et al., 2016). The cp is a uniparentally inherited plastid that contains circular double-stranded DNA (Zhao et al., 2020). A typical angiosperm cp genome has a four-part structure that includes two identical regions in opposite orientations called inverted repeats (IRs), separated by large single-copy (LSC) and small single-copy (SSC) regions (Ruhlman and Jansen, 2014). Scutellaria is a very isolated genus with unsatisfactory traditional divisions, and complete cp genomes have been published for only 13 species in the genus (Jiang et al., 2017; Lee and Kim, 2019; Zhao et al., 2020). In addition, little is known regarding the plastome structural variation among S. baicalensis and its substitutes. Thus, sequencing the cp genomes of these four Scutellariae Radix substitute species is beneficial not only for accurately identifying closely related species but also for greatly contributing to medicinal cp genetic development in Scutellaria. In addition, until now, among the substitute species for S. baicalensis, only one complete cp genome (S. amoena) of the subsect. Angustifoliae clade has been published. The availability of complete cp genome sequences of S. hypericifolia, S. likiangensis, and S. viscidula will help reveal the evolutionary pattern of the complete cp genome in the subsect. Angustifoliae clade.
Plant secondary metabolites usually provide the ultimate features for distinguishing among similar species (Nguyen et al., 2016). The phytochemical diversity of plant metabolites could provide a basis for our understanding of the evolution and classification of different plant species (Wink, 2015). Currently, metabolomics is considered the most common method for profiling and comparing phytochemical compositions among species (Kushalappa and Gunnaiah, 2013). To date, a large number of chemical components have been identified from the genus Scutellaria, and flavonoids and diterpenes have been confirmed to be the two main groups of active constituents in this genus. Based on the pharmacophylogenetic theory, plants with similar therapeutic effects are phylogenetically related and often similar in chemical composition. However, due to the lack of adequate chemical composition and content determination studies, additional evidence is needed to confirm the relationship between chemical diversity and phylogeny in Scutellaria (Shen et al., 2021). Previous studies have indicated that the major flavonoids baicalin, baicalein, wogonoside, and wogonin are 4′-deoxyflavones unique to the roots of S. baicalensis with a wide range of pharmacological activities, and high concentrations of these compounds have also been detected in many Scutellaria plants. However, only two substitutes (S. amoena and S. viscidula) for S. baicalensis have been reported to contain compounds similar to those in S. baicalensis, such as baicalein, baicalin, and wogonin (Wang et al., 2012; Ling et al., 2016). Moreover, the chemical profiles of these four substitutes (S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula) for S. baicalensis remain to be completely explored. Currently, metabolomics is considered the most common method for profiling and comparing phytochemical compositions among species (Kushalappa and Gunnaiah, 2013). Due to the significant differences in chemical composition between the aerial parts and roots of S. baicalensis previously reported (Shen et al., 2019), in this study of the four species mentioned above, we divided S. baicalensis and its four substitute species into aerial parts and roots for separate metabolomics determination.
In this study, we first sequenced the complete cp genomes of S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula by using Illumina sequencing technology. Comparative analyses were conducted to explore the different characteristics of the cp genomes of these four substitutes for S. baicalensis. In addition, a phylogenetic tree was constructed to reveal the phylogenetic positions of these medicinal plants in Scutellaria. Furthermore, the chemical profiles of metabolites in different plant parts were determined by ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS), and chemometric analyses (PCA, PLS-DA, and HCA) were conducted. In addition, high-performance liquid chromatography (HPLC) analysis was used to determine the 15 main flavonoids in different parts of S. baicalensis and its four substitute species. Analyses of phylogenetic relationships and phytochemical diversity among the four substitute species for S. baicalensis, in this study, can expand our understanding of the genetics and chemistry of the genus Scutellaria.
Fresh whole plants of S. amoena, S. baicalensis, S. hypericifolia, S. likiangensis, and S. viscidula were collected from the Gucheng District (Lijiang City, Yunnan Province), Baihua Mountain (Beijing), Luoshage (Dali Bai Autonomous Prefecture, Yunnan Province), Wenhai Road (Yulong Naxi Autonomous County, Lijiang City, Yunnan Province), and Wulan Halaga (West Ujimqin, Inner Mongolia Autonomous Region) in China, respectively. Three samples were collected at each site in August 2019 and were divided into two parts: aerial parts and roots. The samples were authenticated by Professor Qiang Wang (State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences), and voucher specimens (HQS-01, S. amoena; HQS-02, S. baicalensis; HQS-03, S. hypericifolia; HQS-04, S. likiangensis; and HQS-05, S. viscidula) were deposited in the pharmacophylogeny research center (Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing, China).
Fresh leaves were collected and dried using silica gel for DNA extraction. Genomic DNA was extracted using the Plant Genomic DNA Kit (Huayueyang, Beijing, China) according to the instructions. Then, 1% (w/v) agarose gel electrophoresis was used to test DNA integrity, and concentrations were determined using a NanoPhotometer® spectrophotometer (IMPLEN, Munchen, Germany) and a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States). Finally, high-quality DNA was used for library construction and sequencing on any Illumina platform, and 150-bp paired-end reads were generated.
K-values of 21, 55, 85, and 115 were obtained using GetOrganelle software (Jin et al., 2018) to obtain the desired results, and the entire genome of S. baicalensis Georgi (NC_027262.1) from the National Center for Biotechnology Information (NCBI) was used as the reference for cp genome assembly.
Chloroplast annotation of the assembled sequences was performed using GeSeq1 (Michael et al., 2017). These sequences were compared with the known database, and sequences of the protein-coding gene BLAT and the rRNA/tRNA gene BLAT were identified based on similarity values exceeding 85%. In addition to the default known database in GeSeq, the cp genome annotation process also included the addition of the entire genome of S. baicalensis from the NCBI database. Based on the GeSeq annotation results, the software used to create a circular physical map of the cp genome was OGDRAW2 (Stephan et al., 2019).
The GC content of the cp genome was analyzed using MEGA-X (Kumar et al., 2018). The MISA Perl script3 (Beier et al., 2017) was used to identify simple sequence repeats (SSRs). The definition of microsatellites (unit size/minimum repeats) was (1/10) (2/5) (3/4) (4/3) (5/3) (6/3), and the maximum sequence length between two SSRs was set at 100 (Zhao et al., 2020). REPuter (Kurtz et al., 2001) was used to identify the sizes and locations of repeat sequences, including forwarding, palindromic, reverse, and complement repeats, in the cp genomes. The minimal size for all repeat types was 30 bp, with a Hamming distance of 3 (Zhao et al., 2020). The spliced cp genome sequence file was uploaded to the Tandem Repeats Finder v4.09 website4 to identify the tandem repeats in cp DNA with default parameters (Benson, 1999).
Thirteen reported complete cp genomes of Scutellaria were downloaded from the NCBI database: S. baicalensis (MF521633), S. calcarata (MN128385), S. indica var. coccinea (MN047312), S. insignis (KT750009), S. lateriflora (KY085900), S. mollifolia (MN128384), S. orthocalyx (MN128383), S. quadrilobulata (MN128381), S. kingiana (MN128389), S. altaica (MN128387), S. przewalskii (MN128382), S. scordifolia (MT712016), and S. tsinyunensis (MT544405). The mVISTA web interface5 was used to align and compare the five cp genome [S. amoena, S. baicalensis (MF521633), S. hypericifolia, S. likiangensis, S. viscidula] sequences (Mayor et al., 2000). Shuffle-LAGAN was selected as the alignment program for detecting the rearrangements and inversions in sequences. Multiple sequence alignment of cp genomes was conducted using Condon Code Aligner V.9.0.1 and MAFFT v.7.4716 (Frazer et al., 2004). The divergence among different cp genomes and the identification of mutational hotspots were performed by quantifying nucleotide variability in DnaSP v6.12.03 (Rozas et al., 2017). The window length was set to 600 bp, with a 200-bp step size. IRscope7 was used to visualize the gene differences at the boundaries of the junction sites of the 14 cp genomes (Amiryousefi et al., 2018). In this study, in addition to S. baicalensis and its four substitute species (S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula), an additional 12 cp genomes were downloaded from NCBI to construct a cp phylogenetic tree. Maximum likelihood (ML) phylogenetic inference was performed using IQ-TREE software (Nguyen et al., 2015) with 1000 bootstrap replicates based on the TVM+F+R6 nucleotide substitution model to assess branch support. MrModeltest 2.3 (Nylander, 2004) was applied to calculate the best-fitting model based on Akaike’s information criterion. Bayesian inference (BI) phylogenetic trees were analyzed in MrBayes version v3.2.7 (Ronquist and Huelsenbeck, 2003) based on the complete cp genome tree.
Dividing each whole plant into the aerial part and root part, all dried samples of the two parts were crushed and screened through a 40-mesh sieve. Samples (25 mg) were macerated with MeOH (25 ml) and placed in an ultrasonic water bath for 30 min. After cooling to room temperature and complementing the reduced volume, each extract was filtered through a 0.25-μm membrane filter, and 1 μL extracts were injected for each analysis. Three biological replicates of the samples were completed using different individuals of the same species.
The UPLC-QTOF-MS analysis was performed on a Waters Acquity Ultra High-Performance LC system (Waters Co., Milford, MA, United States) coupled with a Waters Xevo G2-XS TOF mass spectrometer (Waters Co., Milford, MA, United States) equipped with an electrospray ionization interface. Chromatographic separations were conducted on a Waters Acquity BEH C18 column (2.1 mm × 100 mm, 1.7 μm). The mobile phase was comprised of acidified water with 0.1% formic acid (A) and MeOH (B). The column temperature was maintained at 35°C. The elution program was as follows: 0–3 min: 10–35% B; 3–9 min: 35–70% B; 9–12 min: 70–100% B; and 12–14 min: 100% B. The flow rate was 0.3 mL/min, and the photodiode array detector scanning range was from 200 to 400 nm. The negative mode was used for ionizing the metabolites, and the mass spectra were scanned from 1,000 to 1,200 m/z. The parameters were set as follows: the cone voltage and capillary voltage were set at 40 V and 2.4 kV, respectively. The source temperature and the desolvation temperature were 100°C and 350°C, respectively. The desolvation gas and cone gas rates were 900 and 50 L/h, respectively. Leucine-enkephalin was used as the lock mass in the analyses (negative ion mode: [M-H] – = 554.2615).
The aerial parts and root samples were all processed according to a previously reported method (Shen et al., 2019). These samples were also analyzed following our previously established protocol (Shen et al., 2019). Three biological replicates of the samples were completed using different individuals of the same species.
Progenesis QI 2.3 software (Waters, Milford, MA, United States) was used to process the centroid MSE raw data from UPLC-QTOF-MS, including import data, alignment review, experimental design setup, peak selection, deconvolution, and compound identification. The MetaboAnalyst 5.08 webserver was used for multivariate statistical analyses, including principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) in the Pareto scaling mode, hierarchical clustering analysis (HCA) with Euclidean distances, and heatmap analysis.
Using Illumina HiSeq/MiSeq sequencing platforms, we obtained 3137–3953 M (Illumina raw reads) for four substitute medicinal species of S. baicalensis (S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula), and 3127–3838 M reads were finally assembled to generate complete cp genomes. The complete cp genomes of the four species were 151,574–151,816 bp in total size (Supplementary Table S1). The lengths were similar in S. amoena and S. likiangensis. S. viscidula was the longest, and S. hypericifolia was the shortest. The cp genomes all exhibited the typical quadripartite structure with a large single-copy region (LSC) (83,810–83,995 bp), a small single-copy region (SSC) (17,321–17,326 bp), and a pair of inverted repeats (IRs) (25,253–25,265 bp) (Supplementary Table S1 and Figure 1). The sizes of the SSC and IR regions were identical among S. amoena, S. hypericifolia, and S. likiangensis but differed in S. viscidula. The average GC content of the genomes was 38.34%, which is similar to values in other Scutellaria species (Lukas and Novak, 2013; Zhao et al., 2020). The GC content was the highest (43.61–43.64%) in the IR regions, the lowest (32.68–32.72%) in the SSC region, and was in the range of 36.34–36.36% in the LSC region (Supplementary Table S1).
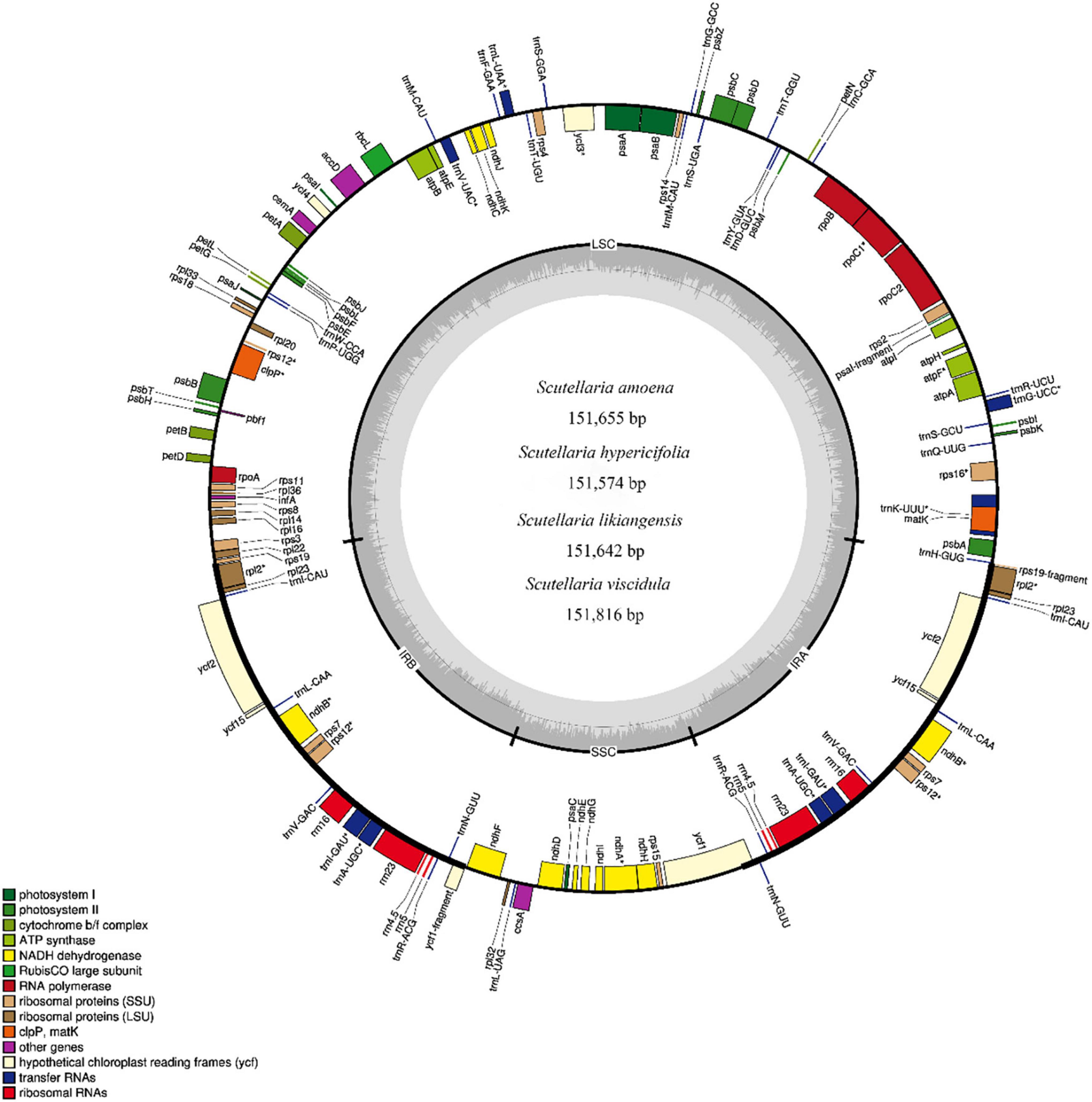
Figure 1. Gene map of the complete cp genomes of S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula. Only one map is shown here due to the negligible differences among the four species. Genes inside the circle are transcribed clockwise, while genes outside the circle are transcribed counterclockwise. The different colors indicate different functional groups of genes. Dark gray areas inside the circle indicate the GC content, while light gray areas indicate the AT content of the genome.
In four Scutellaria species, a total of 113 genes were identified, including 80 protein-coding genes, 29 tRNA genes, and 4 rRNA genes. Of these, 18 genes were duplicated in the IR regions, including seven protein-coding genes (rps12, rps7, ndhB, ycf15, ycf2, rpl23, and rpl2), seven tRNAs (trnN-GUU, trnR-ACG, trnA-UGC, trnI-GAU, trnV-GAC, trnL-CAA, and trnI-CAU), and four rRNAs (rrn5, rrn4.5, rrn23, and rrn16) (Supplementary Table S2). There were five SSR types (mono-, di-, tri-, tetra-, and pentanucleotides) in these four Scutellaria species (Supplementary Figure S1) and 40, 43, 40, and 43 repeat sequences in the S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula cp genomes, respectively. The number of mononucleotide repeats was the highest, accounting for 58.1% of the total number of SSRs, and the number of pentanucleotide repeats was the lowest, accounting for 3.73%. On average, there were approximately 22 tandem repeats, 13 forward repeats, and 17 palindrome repeats. The highest total number of repetitive sequences was found in S. viscidula, with 61 duplicate sequences, and the lowest total was found in S. hypericifolia, with 50 repetitive sequences (Supplementary Figure S1).
Interspecific comparisons of sequence identity among the cp genomes of S. baicalensis and its four substitute medicinal species were conducted with mVISTA, and the annotated S. baicalensis sequence was used as a reference. The mVISTA results indicated that the cp genomes of the species of Scutellaria all showed a high degree of conservation (Supplementary Figure S2). The variation was more pronounced in the single copy region than in the IR region, and the protein-coding region was more conserved in terms of sequence than the non-coding region. In these five cp genomes of Scutellaria species, the non-coding region was more variable than the coding region, and the LSC and SSC regions were more variable than the IR region. Based on the mVISTA results, 6 highly variable regions were found in four species of Scutellaria: trnK (UUU)-rps16, rps16-trnQ (UUG), atpH-atpl, petN-psbM, trnT (GGU)-psbD, and petA-psbJ (Supplementary Figure S2). The average nucleotide variability (PI) of the cp genomes of the four Scutellaria species was 0.00136 (Supplementary Figure S3), and mutation hotspots with high PI values (>0.01) were distributed in the LSC region with a maximum PI value of 0.0155 (petA-psbL). The mutational hotspots within these Scutellaria species were commonly located in the LSC and SSC regions, as well as the IR region with PI values lower than 0.002. The development and utilization of these highly variable sites are beneficial to phylogenetic research on the subspecies and related genera of Scutellaria (Supplementary Figure S3).
The expansion and contraction of IR regions in the cp genomes of these five Scutellaria species and 12 other species of Scutellaria were compared (Supplementary Figure S4). In all species, the IRa/LSC junction was located within the rps19 gene. The rps19 gene had 41–46 bp projections into the IRa region, resulting in the presence of a portion of the rps19 gene (rps19 fragment) in the IRb region. The ycf1 gene straddles the IRb/SSC boundary, but only a small fraction of the gene (771–783 bp) is located in the IRb region, and the pseudogene ycf1 was detected in the IRa region. The length of trnH from the IRa/LSC border was 0 bp.
In this study, based on complete cp genome sequences, 17 species (including the four Scutellaria cp genomes newly sequenced in this study) from Scutellaria were used for the construction of an ML tree and BI trees (with Pogostemon cablin as an outgroup, Figure 2 and Supplementary Figure S5). All species of Scutellaria were recovered in two subclades (ML/BS 100, BI/PP 1) (Figure 2). Subclade I (100, 1) comprised eight species. Subclade I is divided into Subclade I1 and Subclade I2, with 100% support, and these clustered species all have well-developed roots in their morphology. Subclade I1 contains two species, S. altaica and S. przewalskii, both of which are perennial subshrubs with developed woody rhizomes and belong to Subg. Scutellaria Sect. Lupulinaria in the Flora of China (Wu and Li, 1977). Subclade I2 consists of six species S. amoena, S. likiangensis, S. hypericifolia, S. baicalensis, S. viscidula, and S. kingiana. In the maximum likelihood tree constructed based on the cp genome, S. kingiana was found to have high support (100, 1) for clustering these five species into one, indicating a close affinity. Moreover, S. kingiana is a dwarf perennial herb with creeping woody developed rhizomes and belongs to Subgen. Anaspis in the Flora of China (Wu and Li, 1977). The species of S. amoena, S. baicalensis, S. hypericifolia, S. likiangensis, and S. viscidula are clustered together and are all perennial herbs with developed fleshy rhizomes and belong to Scutellaria subgen. Scutellaria sect. Stachymacris subsect. Angustifoliae clade in the flora of China (Wu and Li, 1977; Lou and Qin, 1995; He et al., 2012). In addition, S. viscidula and S. baicalensis clustered in one clade with high support (BS value 100), which indicated that they are more closely related. Scutellaria amoena, S. likiangensis, and S. hypericifolia were clustered in one clade with high support (100,1). Moreover, as substitutes for S. baicalensis, the roots of S. amoena, S. likiangensis, S. hypericifolia, and S. viscidula are yellow and well developed (Lou and Qin, 1995; Li et al., 2003; He et al., 2012). Subclade II (100, 1) consists of nine species: S. calcarata, S. lateriflora, S. indica var. coccinea, S. insignis, S. mollifolia, S. orthocalyx, S. quadrilobulata, S. tsinyunensis, and S. scordifolia (Figure 2). In Subclade II, all cp sequences from the same clade had high support, and as in Subclade I, the positions of these nine species in phylogenetic trees are also different from the traditional Chinese plant classification system in Flora of China. In addition, according to the morphological description of the Flora of China (Wu and Li, 1977), most of them are perennial herbs and have no well-developed roots (Figure 2). Therefore, not only the taxonomic positions of the subgenera but also the taxonomic positions of sections and series need further confirmation in the genus Scutellaria.
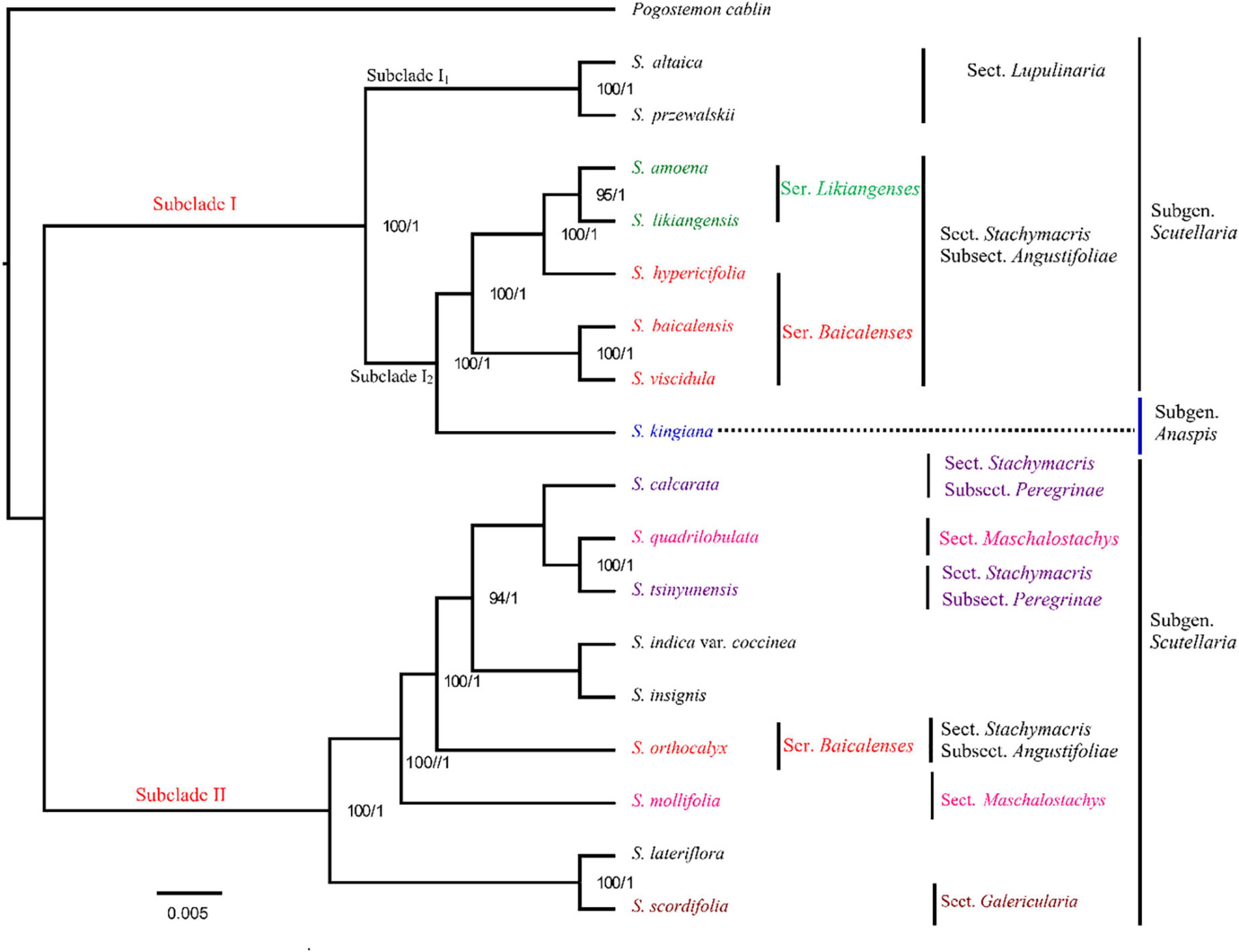
Figure 2. Phylogenetic relationships of the 17 species by maximum likelihood (ML) and Bayesian inference (BI) analyses (Pogostemon cablin as an outgroup) based on alignments of complete chloroplast genomes. The numbers above nodes are supported values with ML bootstrap values on the left and Bayesian posterior probability (PP) values on the right.
The UPLC-QTOF-MS analyses of the aerial parts and roots of S. baicalensis and its four substitute medicinal species (S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula) were performed in negative ion mode using electrospray ionization (ESI–). Progenesis QI is the most commonly used and reliable software for the online identification of metabolomics data (Gu et al., 2019; Wang et al., 2019). In this study, the chemical composition of these five species was tentatively identified by an online search of an in-house library of the genus Scutellaria and a Progenesis MetaScope internal repository through Progenesis QI software and by comparison with literature and standards, as shown in Table 1. Progenesis QI was used with m/z and typical fragment ions through comparison with candidate compounds from various databases. For example, in the online identification by Progenesis QI, baicalin was identified based on molecular weight, adduct ion (m/z 467.0583, M+Na-2H), and typical fragment ion (m/z 269.0431, C15H9O5–). The compound apigenin was identified by two adduct ions, M-H (C15H10O5–, m/z 269.0699) and 2M-H (C30H18O10–, m/z 539.0989), and the fragment ion was identified as C7H3O4– (m/z 151.0039). We further demonstrated the reliability of Progenesis QI identification with reference substances. In the negative ion mode, most of the compounds exhibited deprotonation. The most common compounds were -H2O and -CO2. Compounds containing carboxylic acid groups commonly lost CO2 by negative ionization. Flavonoid glycosides were mainly identified by the loss of character classes of molecules, including the loss of CO and the loss of molecules, by cleavage through the RDA reaction. The loss of sugars such as rhamnose, arabinose, and glucuronide mainly occurred in flavonoid glycosidic compounds. However, similar to other plant species, the species of Scutellaria also have specific metabolites, which may contain many isomeric compounds. Given their similar or identical molecular weights, mass spectrometry fragmentation behavior, and minor differences in retention times, it remains challenging to resolve these structural isomers using only mass spectrometry-based metabolomics approaches. Therefore, when identifying compounds by searching for isomers online with Progenesis QI, this study identified compounds based on their polarity, ion fragmentation patterns, and UV absorption wavelengths and referred to literature reports for further confirmation. For example, compounds with retention times of 8.52 min, M-H– (C18H15O7–) ions, and 343.0809 m/z were tentatively identified as nevadensin (Liang et al., 2018). The major fragment ions of nevadensin were 241.0496 (M-2CH2O- CH2-CO), 313.0352 (M-CH3), and 328.0572 (M-H-CH3). All compounds were identified by the same method. Finally, a total of 75 compounds were putatively identified in negative ion mode, 16 of which were identified by Progenesis QI in agreement with the results of the standards comparison. These compounds included 1 aromatic aldehyde, 2 phenylethanol glycosides, 4 pyrone glycosides, 9 diterpenoids, 55 flavonoids, and 4 other compounds.
Principal component analysis was first applied to analyze the chemical diversity of different parts among the tested Scutellaria species (Figure 3A). In this model, differences in the chemical composition of the five Scutellaria species were observed. The three substitute species other than S. likiangensis (S. amoena, S. hypericifolia, and S. viscidula) were similar in distance from S. baicalensis for both the aerial parts and roots, but S. likiangensis was more distant. In addition, the distance between S. hypericifolia and S. amoena was shorter for aerial parts.
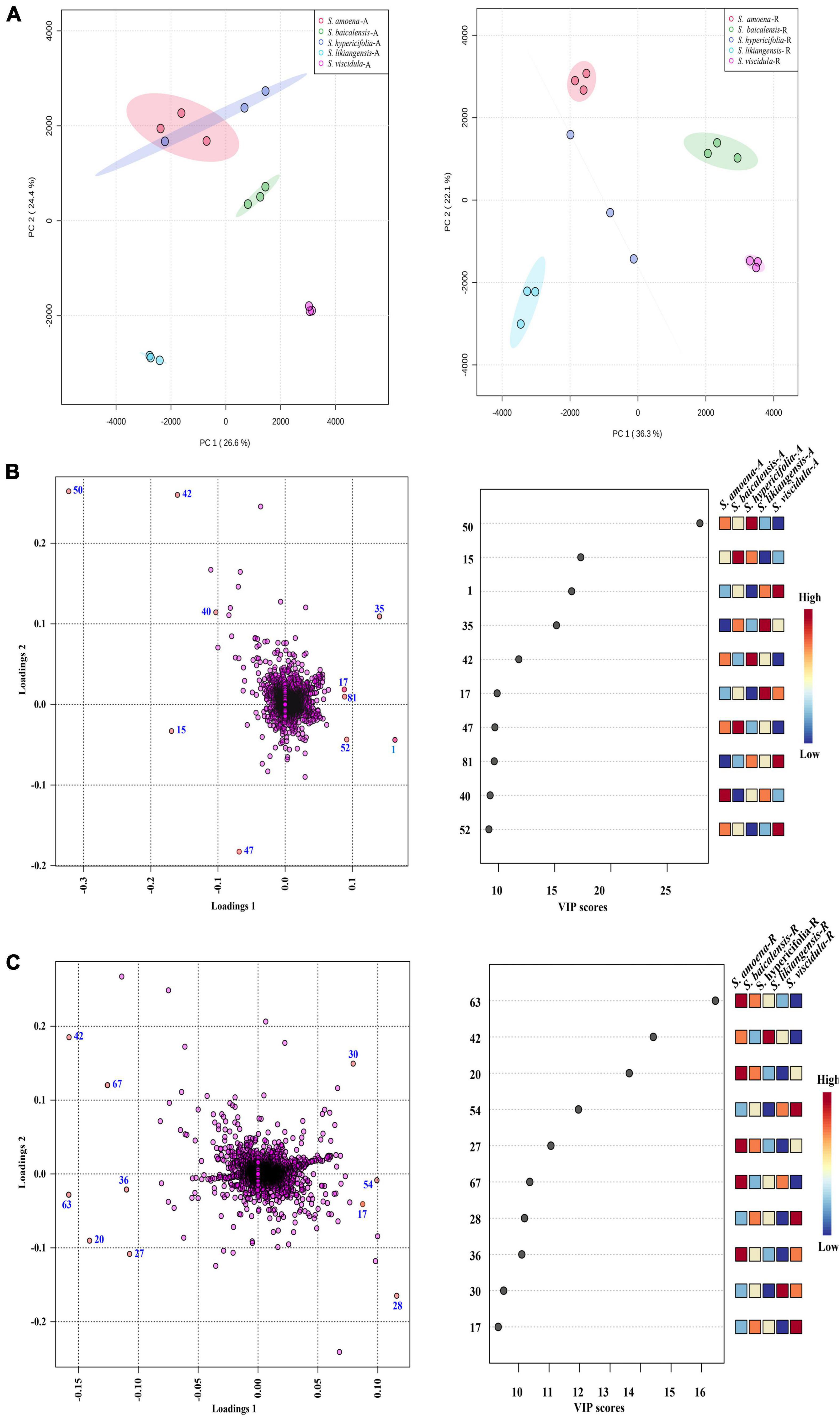
Figure 3. The multivariate analysis of the metabolite data was derived from five Scutellaria species. (A) PCA score map of the different parts of Scutellaria species (-A: aerial parts, -R: roots). (B) The loading plot and variable importance in the projection of the PLS-DA model for the aerial parts of Scutellaria species. (C) The loading plot and variable importance in the projection of the PLS-DA model for the roots of Scutellaria species. (The colored boxes on the right indicate the relative concentrations of the corresponding metabolite in each group under study; the number of the metabolites corresponds to Table 1).
Furthermore, partial least squares discriminant analysis (PLS-DA) was used to better describe the different chemical metabolic profiles of S. baicalensis and its four substitute species in different parts (Figure 3). The variable importance in the project (VIP) value reflects the influence of metabolites on classification. Therefore, metabolites with VIP values ranking in the top 10 were selected as potential markers to distinguish species (Figures 3B,C). In the aerial parts, the PLS-DA loading plot revealed that the separation of the species was mainly caused by pyrone glycoside, flavonoids, and diterpenes. Moreover, the compound scutevulin-2′-O-β-D-glucuronopyranoside (40) showed the highest relative content in S. amoena, the compounds apigenin-6-C-glucoside-8-C-arabinoside (15) and isoscutellarein-8-glucuronide (47) were more abundant in S. baicalensis, and the compounds luteolin-4′-O-glucoside (42) and chrysin-7-O-D-glucuronopyranoside (50) displayed the highest content in S. hypericifolia. The compounds isocarthamidin-7-O-D-glucuronide (17) and 5,7,2′-trihydroxy-6-methoxyflavanone-7-O-β-D-glucuronide (35) were more abundant in S. likiangensis, and the compounds scusalvioside A (1), scutebarbatine H (52) and unknown (81) demonstrated relatively high contents in S. viscidula. In the PLS-DA plot for the roots, S. amoena showed relatively high concentrations of baicalein (63), unknown (20), chrysin 6-C-alpha-L-arabinopyranoside-8-C-glucoside (27), 5,7,2′-trihydroxy-6′-methoxyflavone (67), and unknown (36); S. hypericifolia showed the highest concentration of luteolin-4′-O-glucoside (42); S. likiangensis showed the highest concentration of chrysin-8-C-D-glucopyranoside (30); and S. viscidula displayed the highest concentrations of isocarthamidin-7-O-D-glucuronide (17) and unknown (28, 54). The unknown metabolites 20, 28, 36, 54, and 81 were not annotated because their m/z and possible molecular formulas did not match any of the previously reported compounds.
The HPLC analytical method described in our previous reports (Shen et al., 2019) was used to simultaneously quantify 15 compounds in every sample. The analytical results are shown in Figure 4 and Supplementary Table S3. In the aerial parts of five Scutellaria species, the contents of isocarthamidin-7-O-D-glucuronide (17), carthamidin-7-O-D-glucuronide (19), scutellarin (22), apigenin-7-O-β-D-glucopyranoside (33), isoscutellarein-8-glucuronide (47), and apigenin (59) were higher than those in roots. Of these, compounds 17, 19, and 22 were higher in the aboveground parts of the five species of Scutellaria. In addition, the contents of some compounds were consistent with the relative abundances of compounds identified in the PLS-DA pattern. In the aerial parts, the concentration of isocarthamidin-7-O-D-glucuronide (17) was significantly lower in S. hypericifolia (53.6195 ± 18.9497 mg/g) and that of isoscutellarein-8-glucuronide (47) was significantly higher in S. baicalensis (5.1336 ± 1.6451 mg/g). In addition, carthamidin-7-O-D-glucuronide (19, 120.5610 ± 5.0985 mg/g) and scutellarin (22, 26.5083 ± 3.4493 mg/g) displayed the highest contents in S. likiangensis. Apigenin-7-O-β-D-glucopyranoside (33) showed the lowest content (1.6602 ± 0.2706 mg/g) in S. viscidula. Apigenin (59) displayed a relatively high content in S. hypericifolia (0.0802 ± 0.0175 mg/g).
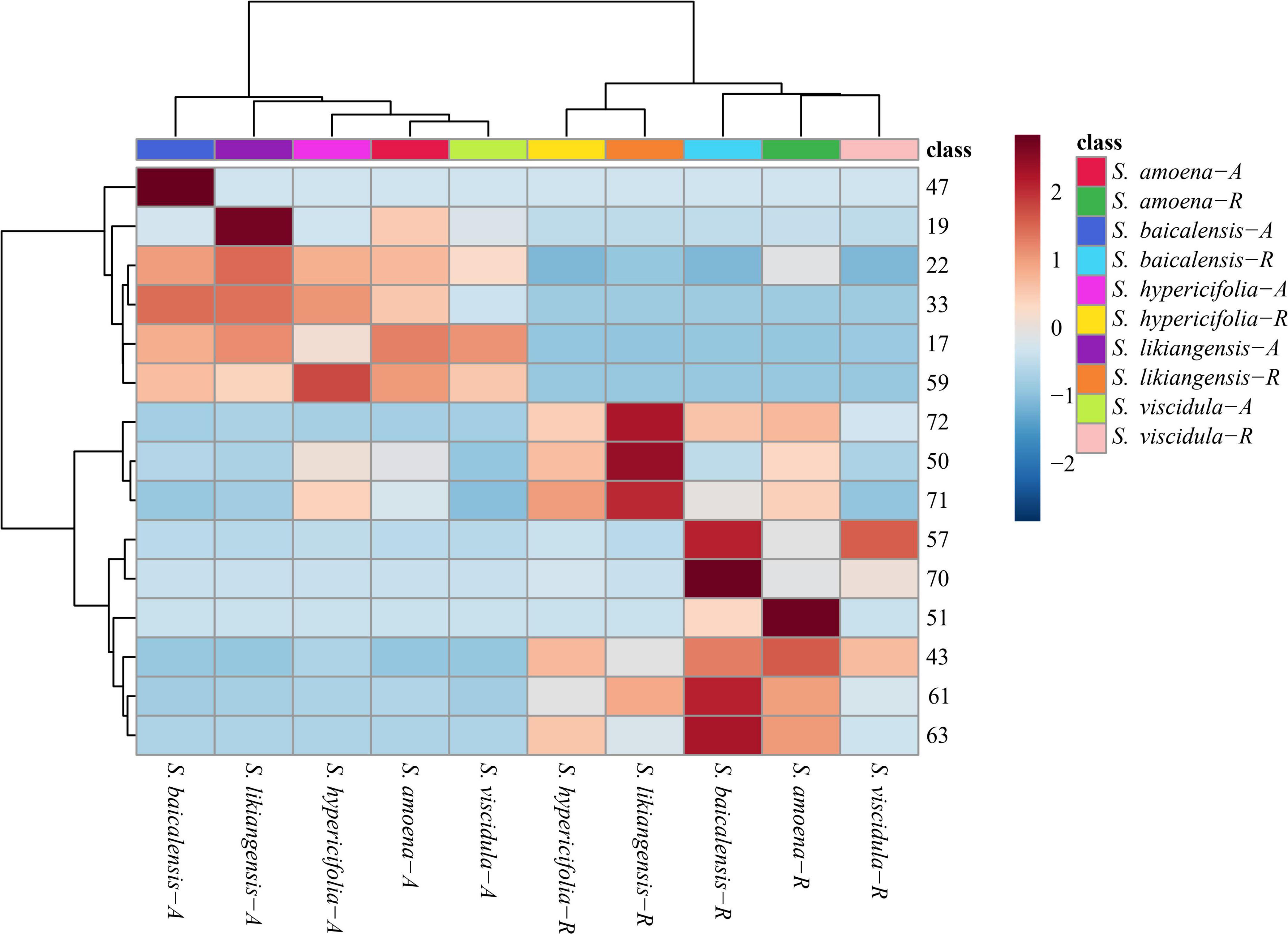
Figure 4. Clustering heatmap of 15 compounds in different parts of five Scutellaria species (the number of analytes corresponds to Table 1; -A: aerial parts, -R: roots).
Nine compounds, baicalin (43), chrysin-7-O-D-glucuronopyranoside (50), luteolin (51), wogonoside (57), alpinetin (61), baicalein (63), wogonin (70), chrysin (71), and oroxylin A (72), were more abundant in the roots of these five Scutellaria species. In addition, the contents of chrysin-7-O-D-glucuronopyranoside (50), chrysin (71), and oroxylin A (72) in S. likiangensis were higher at 69.7029 ± 19.0448, 1.6112 ± 0.6629, and 2.7468 ± 0.6434 mg/g, respectively. Wogonoside (57), alpinetin (61), baicalein (63), and wogonin (70) were more abundant in S. baicalensis, with contents of 31.6445 ± 0.9172, 1.8570 ± 0.1488, 14.6276 ± 0.9966, and 0.0077 ± 0.0021 mg/g, respectively. The compounds baicalin (43) and luteolin (51) were more abundant in S. amoena. Furthermore, the dried root of S. baicalensis (Scutellariae Radix) is a typical Chinese materia medica. According to the Chinese pharmacopoeia (2020 edition), the content of baicalin in Scutellariae Radix should not be less than 9.0%. In this study, in contrast to the content in S. likiangensis (50.1253 ± 14.5487 mg/g), the contents of baicalin (43) were higher than 9.0% in the roots of S. amoena (148.8557 ± 47.1488 mg/g), S. baicalensis (130.6066 ± 9.3707 mg/g), S. hypericifolia (96.1738 ± 28.3923 mg/g), and S. viscidula (93.5047 ± 8.3265 mg/g) (Supplementary Table S3).
In this study, the complete cp genomes of four species of Scutellaria (S. amoena, S. hypericifolia, S. likiangensis, S. viscidula) were assembled for the first time, and comparative and phylogenetic analyses with other related Scutellaria species were also performed (Lee and Kim, 2019; Liu et al., 2020; Zhao et al., 2020). The four Scutellaria cp genomes have high similarities in genome structure and size, gene number, and the distribution of repeat sequences. Furthermore, one high-frequency sequence variation intergenic region (petA-psbJ) was found among the above four Scutellaria species and S. baicalensis. The fragment petA-psbJ has been used in previous phylogenetic studies (Latvis et al., 2017) but not in Lamiaceae. petA-psbJ may represent a potential barcode specific to these Scutellaria species that are currently available. In addition, the phenomena of expansion and contraction of the IR region of the among the species of Scutellaria have been observed. Changes in the boundaries of LSCs and IRs are relatively common in plants (Li et al., 2018; Yan et al., 2019; Zhao et al., 2020). Similar to the findings of Zhao et al. (2020), the changes in the LSC region were larger, which indicates that divergence in LSC length leads to variation in the size of the cp genomes based on IR contraction or expansion. During the evolution of cp genomes, differences in the four IR boundaries among species have been frequently observed, leading to further variation in cp genome size. Thus, although IR regions are the most conserved regions in the cp genome sequence, they have been used to explain differences in size among cp genomes due to contraction and expansion at their boundaries (Liu et al., 2019; Xue et al., 2019). In addition, the location of the IR boundary can be used to study phylogeny; in closely related species, small expansions or contractions tend to have similar endpoints (Cui et al., 2019b).
Morphological polymorphisms make the classification of Scutellaria extremely difficult, and it is also challenging to define species, groups, and phylogenetic relationships within the genus. Scutellaria is a very isolated genus with unsatisfactory traditional divisions. Currently, the commonly used classification method in China is based on the Flora of China (1977) (Wu and Li, 1977). In this study, phylogenetic analyses showed that the cp-based phylogenetic tree and traditional plant classification of Scutellaria in China were incongruent. According to the Flora of China (1977), the species of S. hypericifolia, S. baicalensis, and S. viscidula are grouped, while in the cp-based phylogenetic tree, S. hypericifolia is clustered with S. likiangensis and S. viscidula. In addition, it was observed that S. baicalensis and its substitute species may be a special group in the genus Scutellaria.
Phylogenetically closely related species often share similar chemical characteristics and even similar clinical efficacy, which is the core idea of pharmacophylogeny (Gong et al., 2020; Hao and Xiao, 2020). The 17 species of genus Scutellaria involved in the cp genome phylogenetic tree are classified into two main branches, where the presence or absence of a well-developed root is a significant morphological difference between Subclade I and Subclade II. Subclade I can be divided into two clades, Subclade I1 and Subclade I2. Subclade I1 contains the species S. altaica and S. przewalskii, both of which are perennial subshrubs with developed woody rhizomes and no phytochemical studies. In Subclade I2, S. kingiana is a dwarf perennial herb and has not been chemically studied. The species of S. amoena, S. baicalensis, S. hypericifolia, S. likiangensis, and S. viscidula are clustered together and are all erect or ascending erect perennial herbs with fleshy rhizomes. As in previous reports of S. baicalensis, there are significant differences in the chemical composition between their aerial parts and roots (Shen et al., 2019), while both are reported to have various benefits for human health, such as anti-aging, anti-inflammation, and antitumor activities, which are mainly attributed to flavonoids (Gao et al., 2016; Zhang et al., 2017; Shen et al., 2019, 2020; Song et al., 2020). In this study, we further explored the chemical characteristics of different parts (aerial parts and roots) of these species. The four substitute medicinal species and S. baicalensis share similarity in chemical composition both in aerial parts and roots. It is reported that the 15 accessed chemical components were the active or main compounds in the species of S. baicalensis. Among these compounds, 19, 22, 33, 43, 47, 50, 51, 57, and 59 were all detected in the aerial parts of the four substitute medicinal species for S. baicalensis. The compounds isocarthamidin-7-O-D-glucuronide (17), scutellarin (22), and apigenin (59) showed higher contents and various activities (Li et al., 2017; Wang and Ma, 2018; Oaarowski et al., 2019) in the aerial parts. Moreover, the content of scutellarin (22, 26.5083 ± 3.4493) was highest in the aerial parts of S. likiangensis, which could be a good choice as a natural source of scutellarin for making cerebrovascular and cardiovascular drugs (Breviscapin) (Wen et al., 2021). In particular, it has been reported that the pharmacological activities of Scutellariae Radix are closely related to flavones lacking 4’-OH groups on the B-rings (Zhao et al., 2018; Song et al., 2020; Sun et al., 2020), such as baicalin (43), wogonoside (57), baicalein (63), wogonin (70), chrysin (71), and oroxylin A (72). The roots of the four substitute species all contain these special compounds. The compounds in the roots of S. amoena were similar to those in the roots of S. baicalensis. In addition, according to the standard of the Chinese pharmacopoeia (baicalin > 9%), S. likiangensis could not be used as a substitute species for S. baicalensis. Subclade II consists of nine species: S. calcarata, S. lateriflora, S. indica var. coccinea, S. insignis, S. mollifolia, S. orthocalyx, S. quadrilobulata, S. tsinyunensis, and S. scordifolia, and all of them have short roots and are small plants. Among them, only S. lateriflora and S. scordifolia have had limited chemical studies. S. lateriflora contains both flavonoids and diterpenoids, and only flavonoids have been reported for S. scordifolia. Currently, flavonoids and diterpenes are thought to be the two main active constituent groups in the genus Scutellaria. The relationship of patterns of major chemical diversity to the known phylogeny of these Scutellaria species are still unclear due to the lack of adequate studies on phylogeny, chemical composition and constituent contents. In further studies, additional evidence is needed to confirm the relationship between the chemical diversity and the known phylogeny in Scutellaria.
The species S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula have been traditionally used for a long time as substitutes for S. baicalensis in their places of origin. In recent years, this substitution has gradually decreased due to the expansion of areas of artificial cultivation of S. baicalensis and declines in the availability of wild resources. However, as these substitutes for S. baicalensis have been included in the local standards, long-term use habits are not applied differently. Considering the differences in geographical distribution, growth environment, and species genetics, our attention to the possible nuances in efficacy is still needed. Although the present study revealed that their primary chemical composition is similar, there are differences in the types of compounds and relative contents of the main active ingredients. Therefore, the similarities and differences in their pharmacological activities still need to be further investigated to ensure the validity and stability of the quality of the medicinal herbs.
In this study, the complete cp genomes of four species of Scutellaria (S. amoena, S. hypericifolia, S. likiangensis, and S. viscidula) were sequenced by high-throughput sequencing. In addition, the complete cp genomes and chemical analysis of S. hypericifolia, S. likiangensis, and S. viscidula are reported for the first time. Through comparison, we found that the cp genomes of these four species have similar structural characteristics and the typical four-part structure of other land plants. The nucleotide variation analysis showed that the mutational hot spot of petA-psbL could be used as an effective molecular marker for distinguishing S. baicalensis from its substitutes (S. amoena and S. hypericifolia, S. likiangensis, and S. viscidula). The phylogenetic analysis showed that the traditional morphological classification method in China is different from the cp-based phylogenetic tree, which suggests that the subdivision of the genus Scutellaria should be reconsidered. Additionally, the chemical distributions of four Scutellaria species determined using UPLC-QTOF-MS and HPLC analyses revealed that each Scutellaria plant exhibits a chemical metabolism similar to that of S. baicalensis. Further investigations are needed to reveal the links between the genetic and chemical relationships of Scutellaria species; however, information on these four complete cp genomes of Scutellaria will provide a valuable medicinal genetic resource for Scutellaria species authentication and phylogenetic analysis, and this comparative study will be a basis for further studies on the genus Scutellaria.
The data presented in this study are deposited in GenBank, accession numbers MW286269–MW286272.
JS wrote the manuscript. CH and PX systemically revised the manuscript for important content. JS, YW, and PL helped to complete the data analysis. JS, YL, and KY completed the figures and tables. QW helped to collect and identify the samples. HY and QW reviewed and edited the original manuscript. CH proposed the conception. JS designed the structure of the manuscript. All authors read and approved the final manuscript.
This research was funded by the Innovation Team and Talents Cultivation Program of the National Administration of Traditional Chinese Medicine (Grant No: ZYYCXTD-D-202005) and the CAMS Innovation Fund for Medical Sciences (CIFMS) ID: 2021-I2M-1–071. The Major Scientific and Technological Special Project for The Drug Innovation Major project (No. 2018ZX09711001-008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.951824/full#supplementary-material
Amiryousefi, A., Hyvonen, J., and Poczai, P. (2018). IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Beier, S., Thiel, T., Munch, T., Scholz, U., and Mascher, M. (2017). MISA-web: A web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Benson, G. (1999). Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Cui, Y., Chen, X., Nie, L., Sun, W., Hu, H., Lin, Y., et al. (2019a). Comparison and phylogenetic analysis of chloroplast genomes of three medicinal and edible Amomum species. Int. J. Mol. Sci. 20:4040. doi: 10.3390/ijms20164040
Cui, Y., Nie, L., Sun, W., Xu, Z., Wang, Y., Yu, J., et al. (2019b). Comparative and phylogenetic analyses of ginger (Zingiber officinale) in the Family Zingiberaceae based on the complete chloroplast genome. Plants 8:283. doi: 10.3390/plants8080283
Daniell, H., Lin, C. S., Yu, M., and Chang, W. J. (2016). Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 17:134. doi: 10.1186/s13059-016-1004-2
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M., and Dubchak, I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, W273–W279.
Gao, L., Duan, D. D., Zhang, J. Q., Zhou, Y. Z., Qin, X. M., and Du, G. H. (2016). A bioinformatic approach for the discovery of antiaging effects of baicalein from Scutellaria baicalensis Georgi. Rejuvenation Res. 19, 414–422. doi: 10.1089/rej.2015.1760
Gong, X., Yang, M., He, C. N., Bi, Y. Q., Zhang, C. H., Li, M. H., et al. (2020). Plant pharmacophylogeny: Review and future directions. Chin. J. Integr. Med. 28, 567–574. doi: 10.1007/s11655-020-3270-9
Grzegorczyk-Karolak, I., Golab, K., Gburek, J., Wysokinska, H., and Matkowski, A. (2016). Inhibition of advanced glycation end-product formation and antioxidant activity by extracts and polyphenols from Scutellaria alpina L. and S. altissima L. Molecules 21, 739–839. doi: 10.3390/molecules21060739
Gu, R., Rybalov, L., Negrin, A., Morcol, T., Long, W., Myers, A. K., et al. (2019). Metabolic profiling of different parts of Acer truncatum from the Mongolian Plateau using UPLC-QTOF-MS with comparative bioactivity assays. J Agric. Food Chem. 67, 1585–1597. doi: 10.1021/acs.jafc.8b04035
Hao, D. C., and Xiao, P. G. (2020). Pharmaceutical resource discovery from traditional medicinal plants: Pharmacophylogeny and pharmacophylogenomics. Chin. Herb. Med. 12, 104–117.
He, C. N., Peng, Y., Xiao, W., and Xiao, P. G. (2012). The ethnopharmacological investigation of Chinese Scutellaria plants. Mod. Chin. Med. 14, 16–20.
Jiang, D., Zhao, Z., Zhang, T., Zhong, W., Liu, C., Yuan, Q., et al. (2017). The chloroplast genome sequence of Scutellaria baicalensis provides insight into intraspecific and interspecific chloroplast genome diversity in Scutellaria. Genes 8:277. doi: 10.3390/genes8090227
Jin, J., Yu, W., Yang, J., Song, Y., dePamphilis, C. W., Yi, T., et al. (2018). GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genome. bioRxiv [Preprint]. doi: 10.1101/256479
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kurtz, S., Choudhuri, J. V., Ohlebusch, E., Schleiermacher, C., Stoye, J., and Giegerich, R. (2001). REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Kushalappa, A. C., and Gunnaiah, R. (2013). Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci. 18, 522–531. doi: 10.1016/j.tplants.2013.05.002
Latvis, M., Mortimer, S. M. E., Morales-Briones, D. F., Torpey, S., Uribe-Convers, S., Jacobs, S. J., et al. (2017). Primers for Castilleja and their utility across Orobanchaceae: I. Chloroplast primers. Appl. Plant Sci. 5:1700038. doi: 10.3732/apps.1700020
Lee, Y., and Kim, S. (2019). The complete chloroplast genome of Scutellaria indica var. coccinea (Lamiaceae), an endemic taxon in Korea. Mitochondrial DNA B Resour. 4, 2539–2540. doi: 10.1080/23802359.2019.1640649
Li, K., Fan, H., Yin, P., Yang, L., Xue, Q., Li, X., et al. (2017). Structure-activity relationship of eight high content flavonoids analyzed with a preliminary assign-score method and their contribution to antioxidant ability of flavonoids-rich extract from Scutellaria baicalensis shoots. Arab. J. Chem. 11, 159–170.
Li, X., Huang, L. Q., Shao, A. J., Lin, S. F., and Zheng, X. (2003). A survey of studies on germplasm resources of Scutellaria baicalensis Georgi. World Sci. Technol. 5, 54–58.
Li, X., Li, Y., Zang, M., Li, M., and Fang, Y. (2018). Complete chloroplast genome sequence and phylogenetic analysis of Quercus acutissima. Int. J. Mol. Sci. 19:2443. doi: 10.3390/ijms19082443
Liang, C., Zhang, X., Diao, X., Liao, M., Sun, Y., and Zhang, L. (2018). Metabolism profiling of nevadensin in vitro and in vivo by UHPLC-Q-TOF-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1084, 69–79. doi: 10.1016/j.jchromb.2018.03.032
Ling, G., Chengkang, L., Fanli, Y., Chongyang, D., and Chengke, B. (2016). Similarity and diversity evaluation of bioactive ingredients in S. baicalensis and S. viscidula by HPLC. Northwest Pharm. 31, 115–118.
Liu, X., Chang, E.-M., Liu, J.-F., Huang, Y.-N., Wang, Y., and Yao, N. (2019). Complete chloroplast genome sequence and phylogenetic analysis of Quercus bawanglingensis Huang, Li et Xing, a vulnerable oak tree in China. Forests 10:587.
Liu, X., Zuo, Y., Lin, L., Li, W., Wang, Q., and Deng, H. (2020). The complete chloroplast genome of Scutellaria tsinyunensis (Lamiaceae), an endemic species from China. Mitochondrial DNA B. Resour. 5, 2568–2570. doi: 10.1080/23802359.2020.1781562
Lou, Z., and Qin, B. (1995). Species systematization and quality evaluation of commonly used Chinese traditional drugs, Vol. 2. Beijing: Peking Union Medical College and Beijing Medical University Press.
Lukas, B., and Novak, J. (2013). The complete chloroplast genome of Origanum vulgare L. (Lamiaceae). Gene 528, 163–169. doi: 10.1016/j.gene.2013.07.026
Marsh, Z., Yang, T., Nopo-Olazabal, L., Wu, S., Ingle, T., Joshee, N., et al. (2014). Effect of light, methyl jasmonate and cyclodextrin on production of phenolic compounds in hairy root cultures of Scutellaria lateriflora. Phytochemistry 107, 50–60. doi: 10.1016/j.phytochem.2014.08.020
Mayor, C., Brudno, M., Schwartz, J. R., Poliakov, A., Rubin, E. M., Frazer, K. A., et al. (2000). VISTA : Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046–1047. doi: 10.1093/bioinformatics/16.11.1046
Michael, T., Pascal, L., Tommaso, P., Ulbricht-Jones, E. S., Axel, F., Ralph, B., et al. (2017). GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W1–W11. doi: 10.1093/nar/gkx391
Nguyen, H. T., Lee, D. K., Lee, W. J., Lee, G., Yoon, S. J., Shin, B. K., et al. (2016). UPLC-QTOFMS based metabolomics followed by stepwise partial least square-discriminant analysis (PLS-DA) explore the possible relation between the variations in secondary metabolites and the phylogenetic divergences of the genus Panax. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1012–1013, 61–68. doi: 10.1016/j.jchromb.2016.01.002
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nylander, J. A. A. (2004). MrModeltest v.2. Program distributed by the author. Uppsala: Uppsala University.
Oaarowski, M., Kujawski, R., Mikoajczak, P. A., Wielgus, K., Klejewski, A., Wolski, H., et al. (2019). In vitro and in vivo activities of flavonoids – apigenin, baicalin, chrysin, scutellarin – in regulation of hypertension – a review for their possible effects in pregnancy-induced hypertension. Herb. Polonica 65, 55–70.
Ranjbar, M., and Mahmoudi, C. (2017). A taxonomic revision of Scutellaria sect. Lupulinaria subsect. Lupulinaria (Lamiaceae) in Iran. Feddes Repertorium 128, 63–101.
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574.
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Ruhlman, T. A., and Jansen, R. K. (2014). The plastid genomes of flowering plants. Methods Mol. Biol. 1132, 3–38.
Safikhani, K., Jamzad, Z., and Saeidi, H. (2018). Phylogenetic relationships in Iranian Scutellaria (Lamiaceae) based on nuclear ribosomal ITS and chloroplast trnL-F DNA data. Plant Syst. Evol. 304, 1077–1089.
Shen, J., Li, P., He, C.-N., Liu, H.-T., Liu, Y.-Z., Sun, X.-B., et al. (2019). Simultaneous determination of 15 flavonoids from different parts of Scutellaria baicalensis and its chemometrics analysis. Chin. Herb. Med. 11, 20–27.
Shen, J., Li, P., Liu, S., Liu, Q., Li, Y., Sun, Y., et al. (2021). Traditional uses, ten-years research progress on phytochemistry and pharmacology, and clinical studies of the genus Scutellaria. J. Ethnopharmacol. 265:113198. doi: 10.1016/j.jep.2020.113198
Shen, J., Li, P., Liu, S., Liu, Q., Li, Y., Zhang, Z., et al. (2020). The chemopreventive effects of Huangqin-tea against AOM-induced preneoplastic colonic aberrant crypt foci in rats and omics analysis. Food Funct. 11, 9634–9650.
Song, J. W., Long, J. Y., Xie, L., Zhang, L. L., Xie, Q. X., Chen, H. J., et al. (2020). Applications, phytochemistry, pharmacological effects, pharmacokinetics, toxicity of Scutellaria baicalensis Georgi. and its probably potential therapeutic effects on COVID-19: A review. Chin. Med. 15:102. doi: 10.1186/s13020-020-00384-0
Stephan, G., Pascal, L., and Ralph, B. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Sun, C., Zhang, M., Dong, H., Liu, W., Guo, L., and Wang, X. (2020). A spatially-resolved approach to visualize the distribution and biosynthesis of flavones in Scutellaria baicalensis Georgi. J. Pharm. Biomed. Anal. 179:113014. doi: 10.1016/j.jpba.2019.113014
Wang, L., Chen, W., Li, M., Zhang, F., Chen, K., and Chen, W. (2020). A review of the ethnopharmacology, phytochemistry, pharmacology, and quality control of Scutellaria barbata D. Don. J. Ethnopharmacol. 254:112260. doi: 10.1016/j.jep.2019.112260
Wang, L., and Ma, Q. (2018). Clinical benefits and pharmacology of scutellarin: A comprehensive review. Pharmacol. Ther. 190, 105–127. doi: 10.1016/j.pharmthera.2018.05.006
Wang, X. G. H., M, R., and Liu, T. (2012). Analysis of trnL-F sequences of the medicinal plants Scutellaria baicalensis and its adulterants. Chin. Agric. Sci. Bull. 28, 173–177.
Wang, Z., Zhu, C., Liu, S., He, C., Chen, F., and Xiao, P. (2019). Comprehensive metabolic profile analysis of the root bark of different species of tree peonies (Paeonia Sect. Moutan). Phytochemistry 163, 118–125. doi: 10.1016/j.phytochem.2019.04.005
Wang, Z. L., Wang, S., Kuang, Y., Hu, Z. M., Qiao, X., and Ye, M. (2018). A comprehensive review on phytochemistry, pharmacology, and flavonoid biosynthesis of Scutellaria baicalensis. Pharm. Biol. 56, 465–484. doi: 10.1080/13880209.2018.1492620
Wen, L., He, T., Yu, A., Sun, S., Li, X., Wei, J., et al. (2021). Breviscapine: A review on its phytochemistry, pharmacokinetics and therapeutic effects. Am. J. Chin. Med. 49, 1369–1397. doi: 10.1142/S0192415X21500646
Wink, M. (2015). “Introduction: Biochemistry, physiology and ecological functions of secondary metabolites,” in Biochemistry of plant secondary metabolism, Vol. 40, ed. M. Wink (Chichester: Wiley-Bleckwell). doi: 10.1080/10256016.2015.1014355
Wu, C. Y., and Li, X. W. (1977). “Scutellaria L,” in Flora reipublicae popularis sinicae, Vol. 65, eds C. Y. Wu and X. W. Li (Beijing: Science Press), 124–248.
Xue, S., Shi, T., Luo, W., Ni, X., Iqbal, S., Ni, Z., et al. (2019). Comparative analysis of the complete chloroplast genome among Prunus mume, P. armeniaca, and P. salicina. Hortic. Res. 6:89.
Yan, M., Zhao, X., Zhou, J., Huo, Y., Ding, Y., and Yuan, Z. (2019). The complete chloroplast genomes of Punica granatum and a comparison with other species in Lythraceae. Int. J Mol. Sci. 20:2886. doi: 10.3390/ijms20122886
Zhang, F., Zhang, B., Shen, R., Xu, X., Guo, L., Wang, Y., et al. (2017). The Scutellaria baicalensis stem-leaf total flavonoid regulates the balance of Th17/Treg in EAE rats. Int. J. Clin. Exp. Med. 10, 2408–2418.
Zhao, F., Li, B., Drew, B. T., Chen, Y. P., Wang, Q., Yu, W. B., et al. (2020). Leveraging plastomes for comparative analysis and phylogenomic inference within Scutellarioideae (Lamiaceae). PLoS One 15:e0232602. doi: 10.1371/journal.pone.0232602
Keywords: Scutellaria baicalensis, pharmacophylogeny, substitute medicinal species, chloroplast genomics, metabolomics
Citation: Shen J, Li P, Wang Y, Yang K, Li Y, Yao H, Wang Q, Xiao P and He C (2022) Pharmacophylogenetic study of Scutellaria baicalensis and its substitute medicinal species based on the chloroplast genomics, metabolomics, and active ingredient. Front. Plant Sci. 13:951824. doi: 10.3389/fpls.2022.951824
Received: 24 May 2022; Accepted: 18 July 2022;
Published: 17 August 2022.
Edited by:
Wansheng Chen, Second Military Medical University, ChinaReviewed by:
Ruibing Chen, Naval Medical University, ChinaCopyright © 2022 Shen, Li, Wang, Yang, Li, Yao, Wang, Xiao and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunnian He, Y25oZUBpbXBsYWQuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.