
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 September 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.951737
This article is part of the Research TopicAbiotic Stress: Molecular Genetics and Genomics, Volume IIView all 32 articles
The well-developed root system enables plant survival under various environmental stresses. WUSCHEL-RELATED HOMEOBOX GENE 11 (WOX11) plays a critical role in adventitious root formation and development in rice, Arabidopsis, and easy-to-root tree poplar. However, in difficult-to-root trees, the knowledge of WOX11 during adventitious root formation and development remains scarce. In this study, the JrWOX11 gene was isolated from a difficult-to-root tree walnut and heterologously expressed in the “84K” poplar. The results showed that JrWOX11 contained a similar structure and sequence to the homologous genes in rice, Arabidopsis, and poplar, but had different numbers and types of motifs and cis-elements. JrWOX11 lacked the motif GGAIQY compared to that in easy-to-root trees. In addition, JrWOX11 expression was induced by ABA, PEG, and NaCl treatments. Overexpression of JrWOX11 in poplar promoted root initiation and significantly increased adventitious root (ARs) number, lateral roots (LRs) number, and root hair (RH) length. Furthermore, the aboveground biomass was notably increased under NaCl and PEG treatments in transgenic plants. When NaCl and PEG were removed, the survival rate, aerial shoot development, and de novo root organogenesis were also markedly enhanced in transgenic shoot cuttings. The study provides valuable information on the differences between JrWOX11 and the homologous genes in rice, Arabidopsis, and poplar, and supports the critical role of JrWOX11 in the formation of AR and tolerance to salt and osmotic stresses.
– WOX11 in walnut lacks the motif GGAIQY compared to that in easy-to-root trees.
– JrWOX11 overexpression enhanced stress tolerance by promoting the rooting capacity and regulating root system architecture.
Walnut (Juglans regia L.) is the most widespread nut tree species globally and is thus economically important (Zhang et al., 2020). It is also a difficult-to-root tree (Pei and Gu, 2002). In difficult-to-root trees, such as some Eucalyptus, Pinus, Juglans, Carya, Quercus, and Castanea (Basheer-Salimia, 2007; Amissah et al., 2008; Liu H. et al., 2018), the capacity of cuttings to form AR is relatively low, especially when the mature plant organs, such as shoots, were used as explants (Pijut et al., 2011). This is a significant limitation in the clonal propagation of commercial germplasm in difficult-to-root trees.
To overcome the problems associated with the loss of, or reduced, the competence of difficult-to-root trees to form AR, extensive studies have been conducted (Klerk, 2002; Gonin et al., 2019). For easy-to-root plants, a high auxin concentration above the cut site (due to auxin biosynthesis and auxin flow induced by wounding) is generally enough for adventitious root formation (Da Costa et al., 2013; Zhang et al., 2019). The detached leaves of Arabidopsis thaliana placed on a B5 medium initiate adventitious root formation at the cut site without any exogenous plant hormones (Chen et al., 2014). By contrast, in difficult-to-root trees, rejuvenation treatment and exogenous auxin are two prerequisites for rooting (Pei et al., 2004). Some previous studies have suggested that the rejuvenation treatment could reduce the thickness and density of sclerenchyma between the cortex and phloem, which is beneficial to rooting (Maynard and Bassuk, 1990; Liu H. et al., 2018). Rejuvenation treatment also induced the expression of the WOX11 gene in walnut (Chang et al., 2020). Given the role WOX11/12 plays in the process of adventitious root formation in difficult-to-root trees, it is reasonable to hypothesize that there might be some key differences in WOX11/12 between easy-to-root plants and difficult-to-root trees.
The WOX11/12 gene is one member of the intermediate clade in the WUSCHEL-related homeobox (WOX) family (Liu and Xu, 2018). In Arabidopsis, the expression of AtWOX11 and AtWOX12 can be induced by auxin, and AtWOX11 acts redundantly with AtWOX12 to stimulate cell fate transition to root founder cells (Liu J. et al., 2014). In rice, OsWOX11, OsWOX12A, and OsWOX12B could respond rapidly to auxin, cytokinin, and abiotic stress stimuli of drought, salt, and cold (Cheng et al., 2014; Jiang et al., 2017). Overexpression of OsWOX11 in rice improves crown root emergence and growth, with the well-developed root system enabling rice survival under drought stress (Zhao et al., 2009; Cheng et al., 2016). In easy-to-root tree species poplar, such as P. tomentosa, “Nan-lin895” (P. deltoides × P. euramericana) and “84K” poplar (P. alba × P. glandulosa), WOX11/12s not only promoted adventitious root formation and increased the number of AR on the cuttings but also induced ectopic roots in the aerial parts of transgenic poplars (Liu B. et al., 2014; Xu et al., 2015; Wang et al., 2020). A new study has also reported that PagWOX11/12a enhanced the plant response to salt stress through the control of redox metabolic pathways (Wang et al., 2021). Despite the functional importance of WOX11/12 protein in adventitious root formation and development and stress resistance in easy-to-root plants, the current knowledge regarding the WOX11/12 gene in difficult-to-root trees is scarce.
In this work, the JrWOX11 gene was cloned from hybrid walnut “Zhongningsheng” (J. hindsii × J. regia, “ZNS”). The JrWOX11 gene contained a similar structure and conserved domain sequence to the homologous genes in rice, Arabidopsis, and poplar, but had different numbers and types of cis-elements. Expression analysis showed that the JrWOX11 transcript was expressed at significantly higher levels in roots than in other tissues and was induced by abscisic acid (ABA), salt (NaCl), and polyethylene glycol (PEG). Additionally, the overexpression of JrWOX11 promoted adventitious root formation in poplar. The AR number, LR number, LR length, and RH length were also increased. Moreover, the tolerance to salt and osmotic stress (NaCl and PEG) in transgenic poplar (P. alba × P. glandulosa clone “84K”) plants was enhanced. Our results demonstrated that JrWOX11 plays a crucial role in adventitious root formation and confers salt and osmotic stress tolerance in walnut.
Hybrid poplar (P. alba × P. glandulosa) clone “84K” was used for transformation. The sterile seedlings of “84K” were cultured on half MS medium supplemented with 0.05 mg⋅L–1 of indole-3-butyric acid (IBA) and 0.02 mg⋅L–1 of naphthylacetic acid (NAA) in the tissue culture lab of the China Academy of Forestry, Beijing, China. The growth conditions were kept at 23∼25°C with 16 h light/8 h dark photoperiod. The shoots were subcultured on the same amount of fresh medium every 4 weeks.
Tobacco plants (Nicotiana benthamiana) were grown in pots under an 8 h day/16 h night photoperiod at 23°C. After ∼5 weeks, when 7∼10 tobacco leaves became expanded, the top leaves (third∼fifth) were used for transient expression.
The 1-year-old seedlings of hybrid walnut “ZNS,” a walnut stock cultivar characterized by high yields, good adaptability, and high graft compatibility, were cultured in the greenhouse of the China Academy of Forestry, Beijing, China. The growth condition was kept at ∼25°C with a humidity of 50∼60%.
An RNeasy Plant Mini Kit and an RNase-free DNase I kit (Qiagen, Hilden, Germany) were used to extract total RNA from the rejuvenated stem of “ZNS.” Complementary DNA (cDNA) synthesized by the SuperScript II reverse transcriptase (Thermo Fisher Scientific, Vilnius, Lithuania) was used as the template for cloning the coding sequence (CDS) of the JrWOX11 gene. The putative sequence of WOX11 online (GenBank accession no. XM_018977839) was used for primers design. The PCR product was cloned into pDONR222 and then recombined into the pMDC32 vector and pEarleyGate104 vector (ABRC stock DB3-686) to produce 35S:JrWOX11 and 35S:YFP-JrWOX11 constructs, respectively. Both of the above constructs were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation.
Genomic DNA was extracted from the leaves of 1-year-old seedlings of “ZNS” walnut via a Plant DNA Isolation Reagent kit (Takara, Kyoto, Japan) and was used as the template for cloning JrWOX11 promoter (-2,000 bp to + 206 bp). The PCR product was cloned into the pCAMBI1031 vector. All the above gene-specific primers are shown in Supplementary Table 1.
The protein sequences of WOX11/12 homologous genes in monocotyledon Oryza sativa, dicotyledonous herb Arabidopsis thaliana, easy-to-root trees Populus trichocarpa, P. tomentosa, “Nan-lin895” (P. deltoides × P. euramericana), “84K” poplar (P. alba × P. glandulosa), Salix purpurea, Malus domestica, Morus notabilis, Durio zibethinus, and difficult-to-root trees J. regia, Carya illinoinensis, Quercus suber, Quercus lobata, and Castanea mollissima were downloaded from NCBI and phytozome website. The motif enrichment and sequence distances compared to JrWOX11 protein were analyzed by MegAlign software (DNASTAR. Madison, WI, United States) and MEME v4.9.0 online software.1 The gff3 files of JrWOX11 (refer to XM_018977839) in walnut and OsWOX11, OsWOX12a, OsWOX12b, AtWOX11, AtWOX12, PrtWOX11/12a, and PrtWOX11/12b in model plants were used to analyze gene structure by Gene Structure Display Server.2
The upstream regions (2.0 kb) of the translation initiation sites (ATG) of WOX11/12 homologous genes in O. sativa, A. thaliana, and P. trichocarpa were used as promoter fragments for the cis-elements analysis compared with that of JrWOX11 gene using the program PlantCARE online.3
To evaluate JrWOX11 localization within cells, we utilized a 35S:YFP-JrWOX11 construct, such that the JrWOX11 coding region was fused to the YFP C-terminus. The A. tumefaciens GV3101 colony carrying the 35S:YFP construct, RFP-H2B (a nucleus localization signal marker tagged with RFP) and OsSP1-RFP (a plasma membrane localization marker) were individually cultured to OD600 = 0.4–0.6, re-suspended with the 10 mM EMS, 10 mM MgCl2 (pH 5.8) and 150 μM acetosyringone to OD600 = 0.2 (1:1:1 v/v/v), and co-infiltrated into N. benthamiana leaves as a control. The A. tumefaciens GV3101 cells containing the 35S:YFP-JrWOX11 construct and RFP-H2B were individually cultured, re-suspended, mixed, and co-infiltrated into N. benthamiana leaves as described above to clarify the subcellular localization of JrWOX11. After about 48∼56 h incubation at 23°C under an 8 h day/16 h night photoperiod, N. benthamiana leaves were collected to observe the excitation of YFP and RFP using an LSM 510 confocal laser scanning microscope (Carl Zeiss AG, Oberkochen, Germany) with fluorescence imaging at 488 and 561 nm, respectively, and emission being captured between 500 and 550 nm.
To reveal expression profiles of JrWOX11 gene in different organs, including roots (R), stem (S), leaf (L), female flower (FF), immature fruit (IF), and zygotic embryo (ZE), they were collected from 23-year-old “ZNS” trees from Luoning county, Henan Province. Six samples of each tissue from the same tree were mixed as one biological replicate, and three replicate trees were sampled. All samples were snap-frozen in liquid nitrogen and stored at −80°C for the expression analysis of JrWOX11 as described in the previous study (Chang et al., 2020).
To investigate the expression profiles of JrWOX11 under salt and osmotic stress, the 1-year-old “ZNS” seedlings were irrigated with 200 mM NaCl and 10% (w/v) PEG 6000 solution, and the roots were collected after seven treatment durations (0, 1, 3, 6, 9, 12, and 24 h). For ABA treatment, the top fourth–sixth walnut leaves were sprayed with 100 μM ABA and collected after 0, 1, 3, 6, 9, 12, and 24 h. Ten leaf sections were used for each treatment, and experiments were repeated three times. All samples were snap-frozen in liquid nitrogen and stored at −80°C for the expression analysis of JrWOX11.
Total RNA was isolated using the method described above (Section “RNA isolation, DNA isolation, gene cloning, and vector construction”). The synthesized cDNA was diluted 10-fold with ddH2O to serve as a template for qRT-PCR on a Roche LightCycler 480 (Roche Applied Science, Penzberg, Upper Bavaria, Germany). Expression levels were normalized relative to the control (GAPDH) using the 2−△△Ct method. The qRT-PCR primers are shown in Supplementary Table 1.
An A. tumefaciens strain GV3101 containing the 35S:JrWOX11 construct was used for the transformation of poplar “84K” as described previously (Liu B. et al., 2014; Li et al., 2017). The discs from the top fourth–fifth leaves from 4-week-old “84K” were notched and incubated with A. tumefaciens GV3101 cells containing the 35S:JrWOX11 construct for about 3 days. The hygromycin-resistant shoots were obtained from the leaf explants after 2–4 weeks on the MS medium supplemented with 0.5 mg⋅L–1 of 6-BA, 0.05 mg⋅L–1 of NAA, 3 mg⋅L–1 of hygromycin, and 200 mg⋅L–1 of timentin. Roots were induced on root induction medium (RIM: 1/2 MS + 0.05 mg⋅L–1 of IBA + 0.02 mg⋅L–1 of NAA) + 3 mg⋅L–1 of hygromycin. The resistant plants were detected by qRT-PCR of the JrWOX11 gene expression. The plants with high expression levels of the JrWOX11 gene were subcultured for later experiments.
To determine the role of the JrWOX11 gene in root formation, a stereo microscope (Leica, Wetzlar, Germany) was used to observe the stem samples from the control and transgenic (OE6#) “84K” after 0, 1, 2, 3, 4, 5, and 6 days on RIM. Cross-sections (30 μm thick) of the stem base were prepared with a VT-1000S vibrating blade microtome (VT-1000S, Leica, Wetzlar, Germany) and stained with 2% w/v toluidine blue O solution for visualizing root primordia under a BX51 digital microscope (Olympus, Tokyo, Japan). Five seedlings of each line per replicate were used, and two independent experiments were performed.
To analyze the rooting index, including the AR number, LR number, AR length, LR length, and root hair (RH) length, the shoots (about 2.5 cm high) were cut from 4-week-old non-transgenic “84K” (CK) and transgenic seedlings (OE#3 and OE#6) and moved to RIM under 16 h light/8 h dark for about 2 and 4 weeks, respectively. Ten individuals per replicate were used for each line, and three independent experiments were performed.
To evaluate salt and osmotic stress tolerance, 2-week-old non-transgenic “84K” (CK) and transgenic (OE3# and OE6#) surface-sterilized plants were grown on RIM supplemented with 0 (control) or 200 mM NaCl (salt treatment) or 5% w/v PEG 6000. Three weeks later, the plant height and fresh weight (FW) were measured, and the dry weight (DW) was determined after oven drying at 80°C for 72 h. Six biological replicates of each line per treatment were used, and two independent experiments were performed.
To examine the rooting capacity under salt and osmotic stress, shoot segments (about 2.5 cm high) of 4-week-old non-transgenic “84K” (CK) and transgenic (OE3# and OE6#) surface-sterilized plants were cultivated on RIM supplemented with 0 (control) or 200 mM NaCl (salt treatment) or 5% w/v PEG 6000 for 4 weeks and then transferred to RIM for 2 weeks. The phenotypes of the root systems were photographed every week, and the survival rate, the number of axillary buds regenerated, and the number of ARs were quantified. Six biological replicates of each line per treatment were used, and two independent experiments were performed.
Significant differences between means of CK and transgenic lines were determined using the SPSS statistical package (version 16.0; SPSS Inc., Chicago, IL, United States) at a significance level of ∗P < 0.05 and ∗∗P < 0.01 (Student’s t-test).
In “ZNS” walnut, only one WOX11/12 gene was isolated and submitted to NCBI’s GenBank (JrWOX11, NCBI Accession No: ON979687) (Figure 1A). The full-length sequence of JrWOX11 cDNA is 747 nucleotides and encodes a deduced protein of 248 amino acids (aa) residues, with a predicted molecular mass of 27.05 kDa and a theoretical pI of 5.42 (Supplementary Table 2). However, there were 3, 2, and 2 WOX11/12 genes in rice, Arabidopsis, and P. trichocarpa, respectively (Figure 1A) (Zhang et al., 2010; Cheng et al., 2014). Twenty-six WOX11/12 protein sequences from 15 species, namely, two monocotyledons (maize and rice), a dicotyledonous herb (A. thaliana), eight easy-to-root trees (P. trichocarpa, P. tomentosa, Nan-lin895 poplar, “84K” poplar, S. purpurea, Malus domestica, Morus notabilis, and D. zibethinus), and four difficult-to-root trees (Carya illinoinensis, Q. suber, Q. lobata, and Castanea mollissima), were downloaded from NCBI and characterized. The protein length ranged from 236 to 356 aa, the pIs from 5.42 to 8.67, and the molecular mass from 25.92 to 36.47 kDa. Multialignment of WOX11/12 protein sequences (by MegAlign software) showed that CiWOX11 (96.37%) in Carya illinoinensis, QlWOX11 (82.57%) in Q. lobata, QsWOX11 (81.82%) in Q. suber, and CmWOX11 (82.23%) in Castanea mollissima shared high sequence identity with JrWOX11 in “ZNS” walnut. The WOX11/12 in Populus, S. purpurea, Malus domestica, Morus notabilis, and D. zibethinus had relatively moderate sequence identities with JrWOX11, ranging from 61.48 to 74.44%. The sequence of WOX11/12 in Z. mays, O. sativa and A. thaliana had relatively low sequence identities with JrWOX11, ranging from 45.22 to 57.01% (Supplementary Table 2). The simplified neighbor-joining (NJ) phylogenetic tree of the related species also showed that JrWOX11 was similar to PtrWOX11/12a and PtrWOX11/12b (Figure 1B).
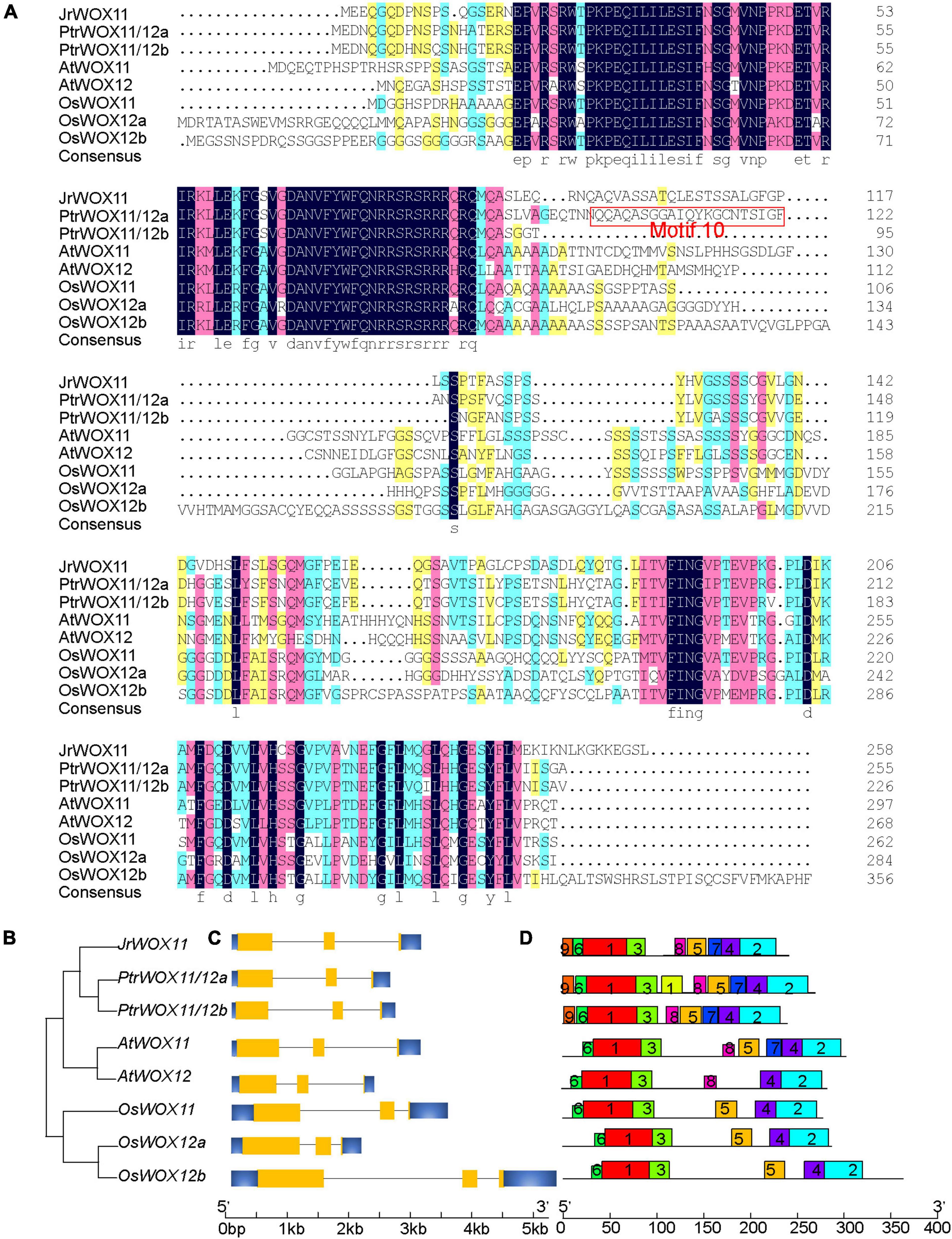
Figure 1. Identification and characterization of cloning of the JrWOX11 gene. (A) Sequence alignment of homologous WOX11/12 proteins. Protein sequences were obtained from NCBI and are shown in Supplementary Table 2. (B) Phylogenetic tree based on full-length coding sequences of WOX11/12 genes. (C) Gene structure analysis of WOX11/12 genes. (D) Motifs analysis of WOX11 proteins. MEME software was used to search motifs, and then redraw the map with the software TBtools (Chen et al., 2020). Motif10 was marked with a red box in panel (A).
Gene structure analysis revealed that the homologous WOX11 genes had the same gene structure, such as three exons and two introns (Figure 1C). Motif enrichment analysis revealed five prominent motifs (Figure 1D). Among them, motif1, motif3, and motif6 were related to the homeodomain, whereas motif2 and motif4 were associated with the C-terminus domain of intermediate clade WOXs (Zhang et al., 2010). Motif7 and motif8 were observed only in dicotyledons, and motif9 was conserved only in the easy-to-root trees. Even though AtWOX11 and AtWOX12 have a similar function in adventitious root formation, motif5 was missing in AtWOX12. Furthermore, motif10 was only found in easy-to-root trees, such as P. trichocarpa, Ptr; S. purpurea, Sp; Malus domestica, Md; Morus notabilis, Mn; and D. zibethinus, Dz; but was not present in difficult-to-root trees, such as J. regia, Jr; Carya illinoinensis, Ci; Quercus suber, Qs; Quercus lobata, Ql, and Castanea mollissima, Cm. The sequence of motif10 in PtrWOX11/12a was “QQAQASGGAIQYKGCNTSIGF,” among which “GGAIQY” was extremely conserved (Figures 1A,D and Supplementary Figure 2). To sum up, there might be differences in the number of WOX11/12 proteins and the motifs between easy-to-root trees and difficult-to-root trees.
The 2 kb sequence upstream of WOX11/12s CDS was used to identify cis-acting elements. The results showed that the identified cis-elements could be divided into four categories: (1) growth- and development-responsive elements, (2) plant hormone-responsive, (3) abiotic stress-responsive elements, and (4) WOX-consensus motif (Figure 2 and Supplementary Table 3). There were 3, 1, 2, 5, 5, 5, 3, and 2 root-specific elements in the promoter region of OsWOX11, OsWOX12a, OsWOX12b, AtWOX11, AtWOX12, PtrWOX11/12a, PtrWOX11/12b, and JrWOX11, respectively. Plant hormone-related elements were also identified, such as 2, 5, 4, 5, 3, 2, 3, and 10 auxin-responsive elements (ARFAT and TGA elements), 4, 1, 5, 8, 9, 8, 8, and 5 cytokinin-responsive element (ARR1AT), 1, 1, 0, 1, 3, 1, 0, and 2 gibberellin-responsive element (GARE1OSREP1), and 2, 3, 0, 0, 3, 1, 2, and 7 abscisic acid-responsive element (ABRE), in the promoter region of OsWOX11, OsWOX12a, OsWOX12b, AtWOX11, AtWOX12, PtrWOX11/12a, PtrWOX11/12b, and JrWOX11, respectively. A few elements responsive to various abiotic stresses were also identified, mainly including DRE, MBS, G-box, and W-box, and the total numbers were 0, 3, 3, 4, 1, 3, 4, and 1 in the above sequence of WOX genes. Compared to rice, Arabidopsis, and poplar, the promoter region of WOX11 in walnut contained fewer root motifs and stress-responsive elements but contained more abscisic acid-responsive elements and AuxRE.
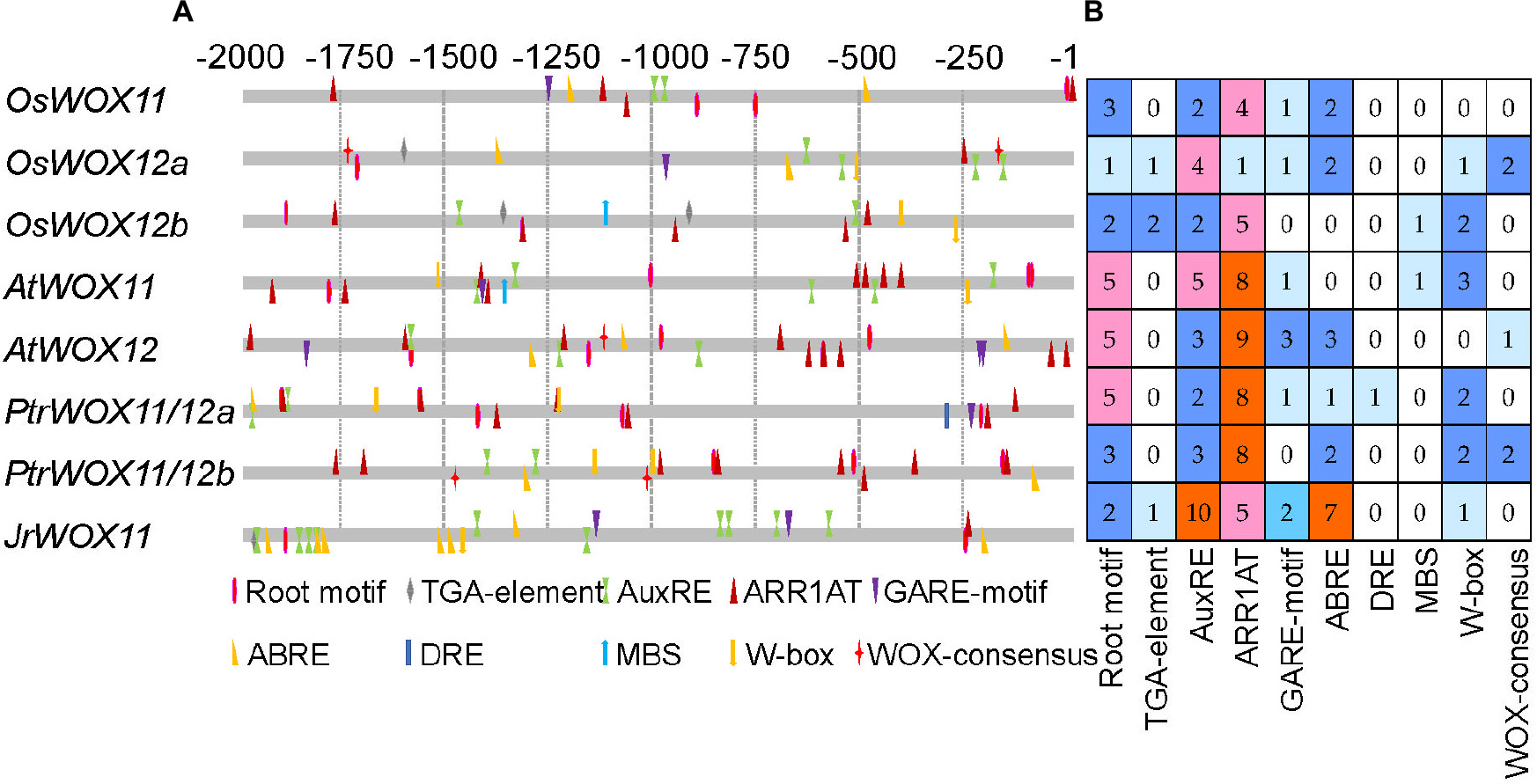
Figure 2. The cis-element distribution and number analysis of the promoter region of WOX11/12 genes in Oryza sativa, Arabidopsis thaliana, Populus trichocarpa, and Juglans hindsii × Juglans regia cv. “ZNS.” (A) The position of the cis-element in the upstream 2,000 bp region of WOX11/12 genes. (B) The number of each cis-element.
WOX11 is shown to specifically bind to the WOX-consensus “TTAATGG/C” motif (Lohmann et al., 2001; Zhao et al., 2009, 2015; Liu B. et al., 2018). Within the OsWOX12, AtWOX12, and PtrWOX11/12b genes, we identified one or two WOX-consensus motifs common to their promoters. However, walnut did not harbor any WOX11/WOX12b genes, suggesting that WOX11-binding genes might differ in different plants (Figure 2 and Supplementary Table 3).
In our previous research, JrWOX11 was predicted to be localized in the nucleus (Chang et al., 2020). To validate this prediction, the 35S:YFP-JrWOX11 construct was transiently co-expressed with the nuclear marker in tobacco leaves. The 35S:YFP construct mixed with the nucleus and the membrane markers was transformed as a control. The nucleus and plasma membrane were marked by RFP staining. As shown in Figures 3e–h, the 35S:YFP-JrWOX11 fluorescent signals and nucleus marker were observed in the same location, indicating that JrWOX11 is localized in the nucleus. This was consistent with the prediction. By contrast, in cells transformed with the positive control 35S:YFP, the fusion protein was distributed throughout the cell, including the nucleus and cytoplasm, which were marked by the nuclear and plasma membrane RFP marker staining (Figures 3a–d).
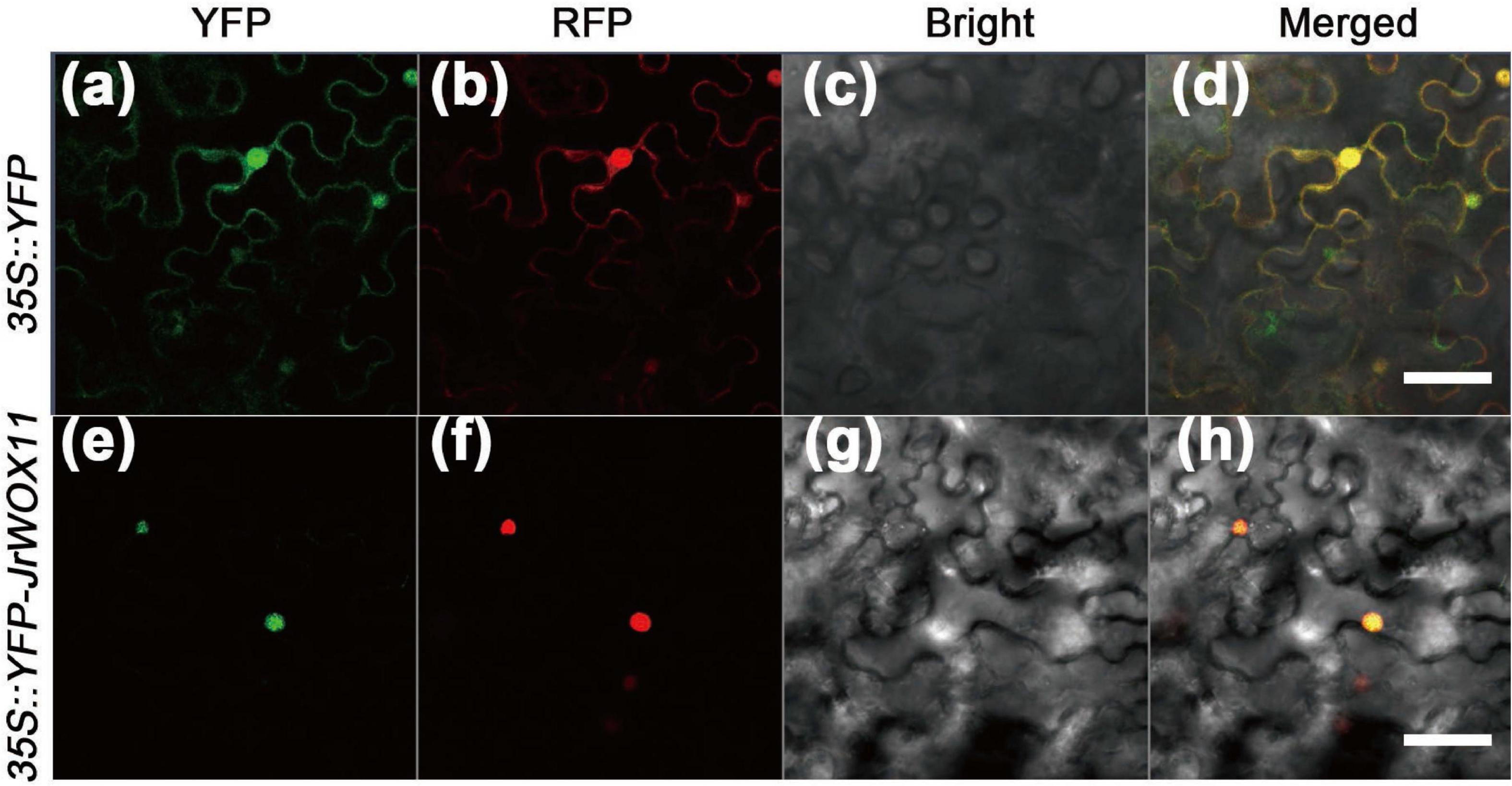
Figure 3. Subcellular localization of JrWO11 protein. 35S::YFP and 35S::YFP-JrWO11 fusion proteins were expressed transiently into Nicotiana benthamiana leaf epidermal cells 48∼56 h after vacuum injection. The YFP channel (a,e), the RFP channel (b,f), the bright field (c,g), and the merged images (d,h). Scale bar: 50 μm.
The spatial and temporal expression patterns of the JrWOX11 gene in different tissues were analyzed using qRT-PCR. As shown in Figure 4A and our previous study (Chang et al., 2020), the JrWOX11 showed relatively high expression in roots (R), but relatively low expression in stems (S). Previous studies reported that the WOX11 gene was involved in drought and salt resistance (Cheng et al., 2016; Wang et al., 2018, 2020). Here, the transcription levels of JrWOX11 in response to ABA, NaCl, and PEG were analyzed. As shown in Figures 4B–D, JrWOX11 transcription was rapidly induced after treatment with ABA, NaCl, and PEG. The transcription levels increased with prolonged treatments, peaking at 9, 6, and 6 h, respectively, then slightly decreased after that. It could be concluded that the expression of JrWOX11 is responsive to ABA, NaCl, and PEG.
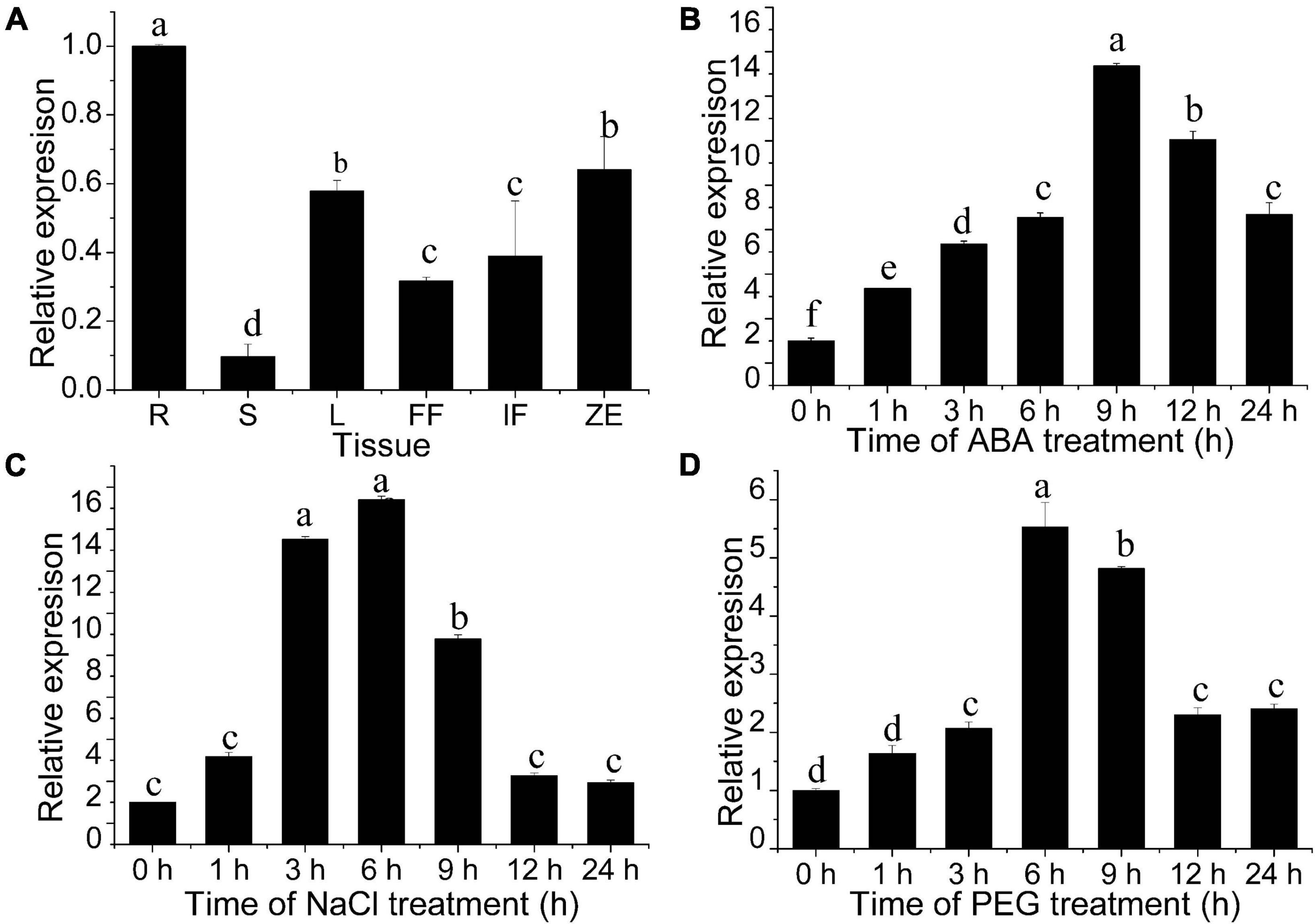
Figure 4. Expression patterns of JrWOX11 as assessed by qRT-PCR. (A) Spatial and temporal expression of JrWOX11 in different tissues. (B–D) Relative expression of JrWOX11 under 100 μM ABA, salt (200 mM NaCl) and 10% (w/v) PEG 6000 solution. Error bars = SEM. Statistical differences were analyzed using Student’s t-test, n = 3. Different letters labeled indicate significant differences.
The JrWOX11 had the highest expression in roots, which may play a crucial role in root growth and development. To verify this, JrWOX11 was transformed into “84K” poplar, resulting in 14 transgenic lines (Supplementary Figure 1). Lines OE3# and OE6# with high expression of JrWOX11 were used in the work reported here. The phenotypes of the stem base of control (CK) and the OE6# line were analyzed by stereo microscopy and sectioning (Figures 5A,B). The results showed that on day 5, small protrusions from the stem base were observed for the first time in control stems, which might have represented a bump of AR primordium. On day 6, the skin of the original protrusion at the base of the control stem ruptured and ARs appeared. By contrast, ARs in OE lines occurred on day 3, 2 days earlier than in the control. Therefore, the JrWOX11 gene is considered to promote the emergence of adventitious roots.
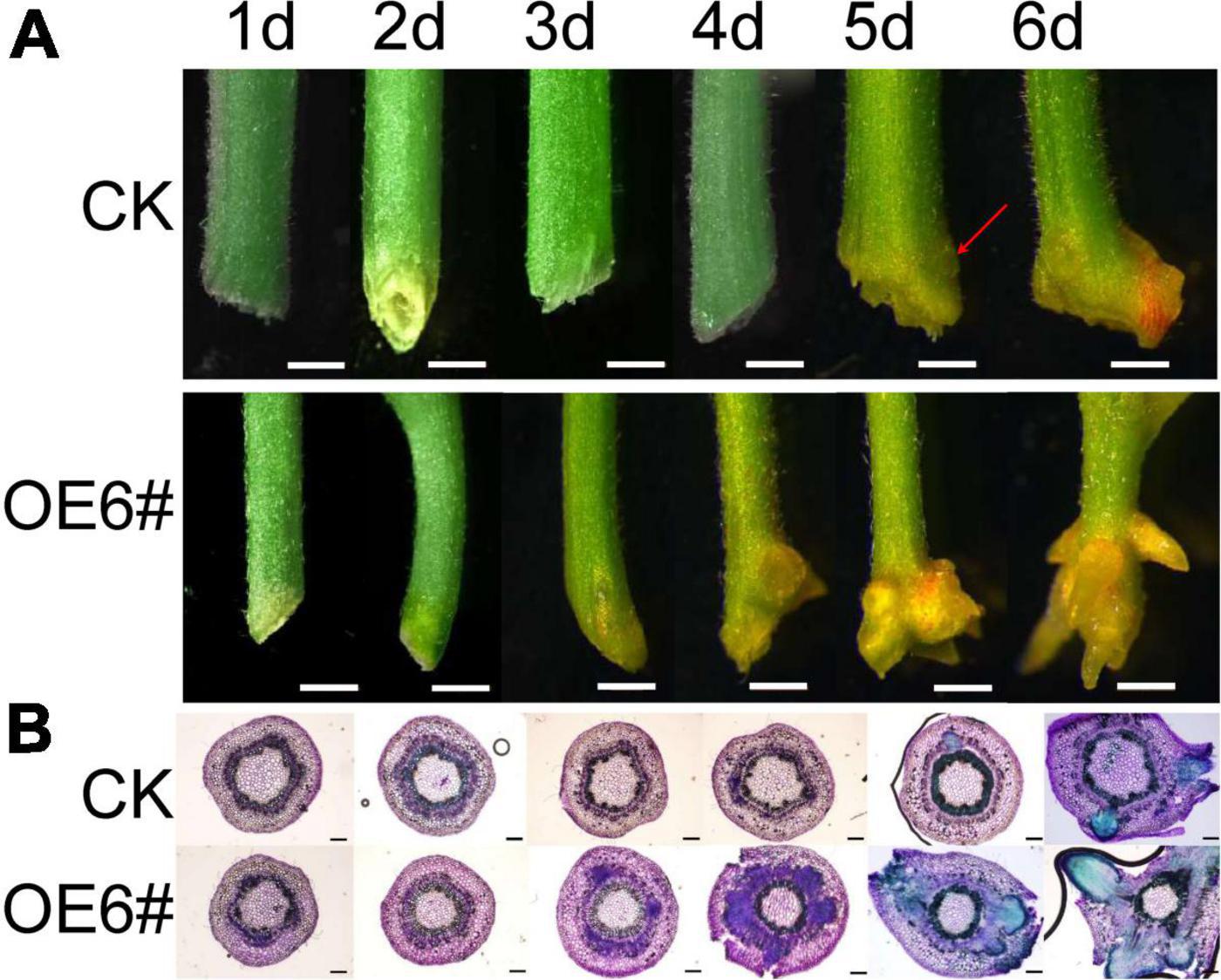
Figure 5. Overexpression of JrWOX11 promotes root formation. (A) The phenotype investigation of the poplar stem base during rooting 1–6 days. Bar = 1 mm. (B) The histological investigation of the poplar stem base during rooting 1–6 days. Bar = 200 μm.
To further verify whether JrWOX11 was involved in the development of the AR system, the root phenotypes of two OE lines (3# and 6#) were analyzed (Figures 6A–C). In 2-week-old seedlings, the AR number, length, and LR length were significantly higher in JrWOX11-transgenic lines (OE3# and OE6#) than controls (CK1 and CK2) (Figures 6H,I). Similarly, these root indices in 4-week-old seedlings of OE3# and OE6# lines were also significantly increased compared with controls (Figures 6E,F). Interestingly, transgenic plants overexpressing JrWOX11 had significantly increased root hair length (Figures 6D,G). These results suggested that JrWOX11 might regulate root development and root hair growth.
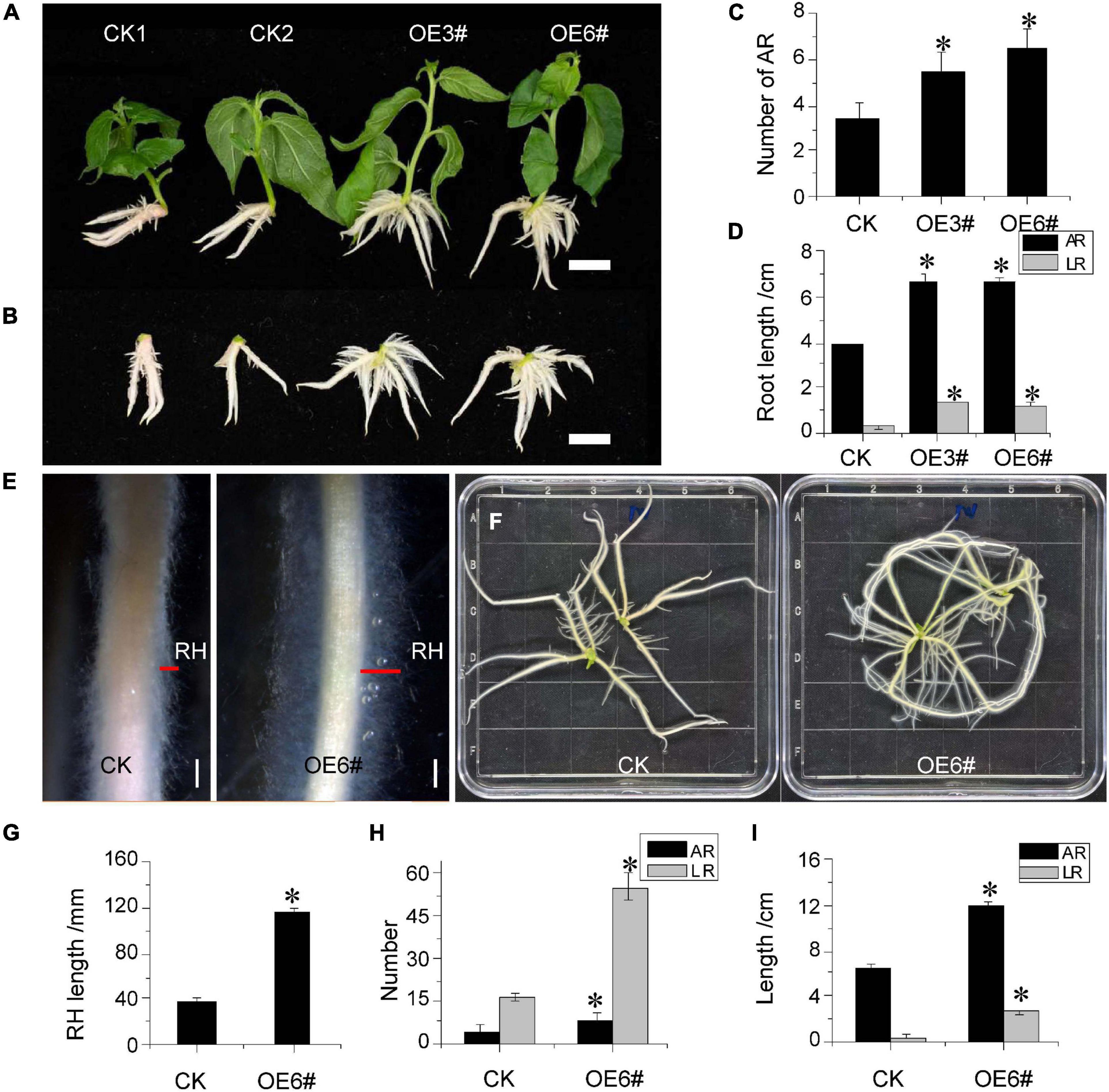
Figure 6. Overexpression of JrWOX11 regulated root system architecture. (A) Comparison of AR number between CK and OE 3# and OE 3# 6# (2 weeks old). (B) The vertical view of ARs of CK and overexpressing lines 3# and 6# (2 weeks old). Bar = 1 cm. (C,D) Average AR length, AR number, and LR length in Control and the overexpressing lines (2 weeks old). (E) Root hairs of 4-week-old seedlings. Bars = 2 mm. (F) Root system phenotypes of the control group (CK) and WOX11 transgenic plants (OE#6) (4 weeks old). Bar = 1 cm. (G–I) Length of RH, average root length, and root number in Control and the overexpressing line (OE#6) (4 weeks old). Asterisk indicates a significant difference (*P < 0.05) between transgenic lines and WT plants.
To validate the effectiveness of root system architecture in NaCl and osmotic stress tolerance, 2-week-old OE (3# and 6#) and control (CK) seedlings were subjected to no-stress treatment (Control), 200 mM NaCl, and 5% w/v PEG6000 treatment in sterile seed tubes (Figures 7A,B). Three weeks later, the plant height, the aboveground fresh weight, and dry weight of OE3# and OE6# were superior to CK in the control treatment, suggesting that the overexpression of the JrWOX11 gene improved the growth of transgenic plants. Under salt and osmotic stress, the aboveground fresh weight and dry weight were significantly increased in JrWOX11-overexpressing plants compared with CK plants, indicating that JrWOX11 improved tolerance to NaCl and osmotic stress by promoting root growth.
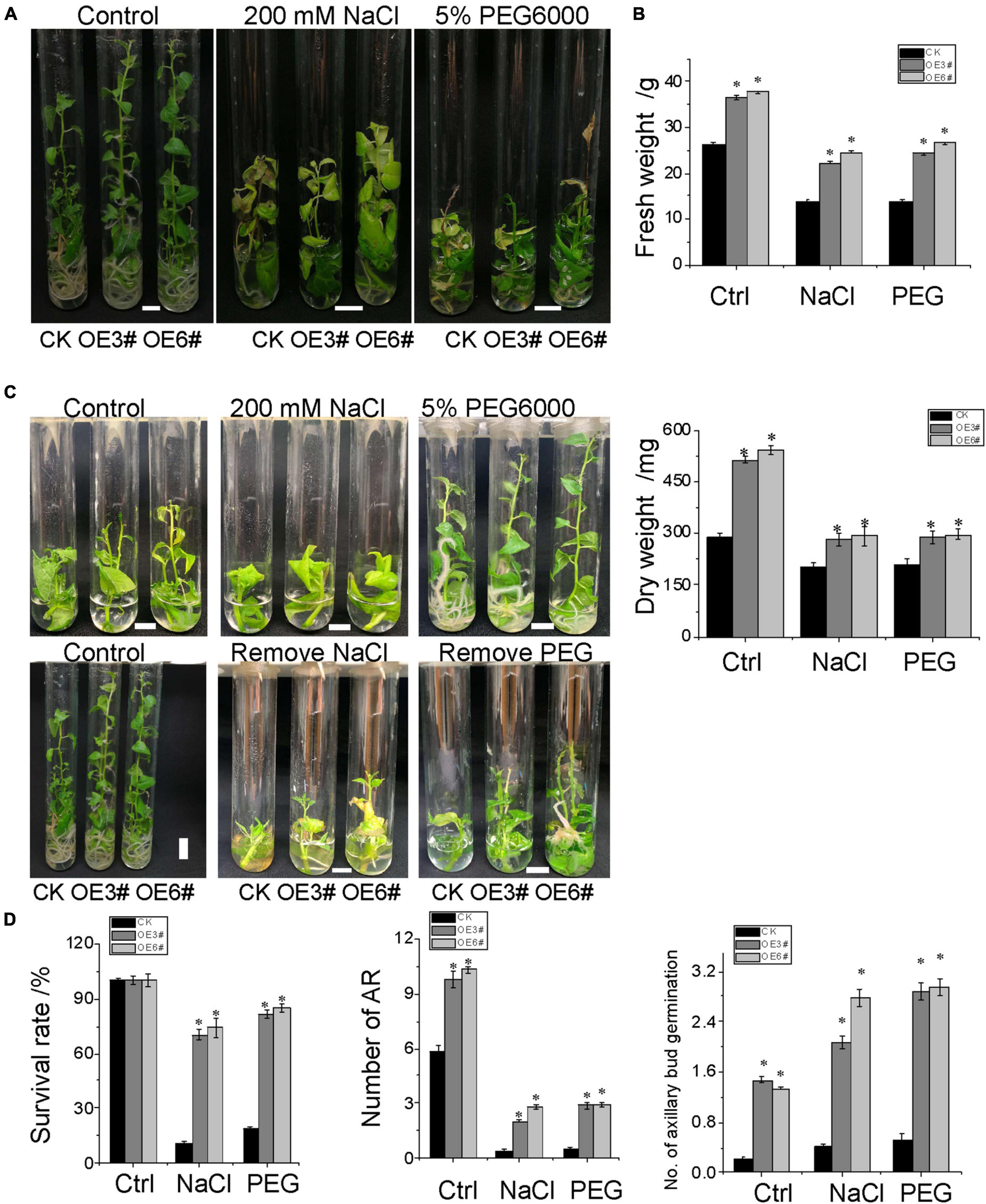
Figure 7. Stress response of JrWOX11-transgenic “84K” plants. (A) The growth and phenotype of JrWOX11-transgenic and WT poplar plants. (B) Effects of salt and osmotic stress on fresh weight and dry weight. Four-week-old seedlings were cultured vertically on the liquid RIM medium (1/2MS + 0.01 mg⋅L– 1 of IBA + 30 g⋅L– 1 of sucrose) supplemented with 200 mM NaCl or 5% PEG6000 for 2 weeks. (C) The photographs were taken, and the fresh weight and dry weight of each plant were measured. (D) The survival rate and regeneration response were observed when NaCl and PEG were removed. About 2.5 cm shoot cuttings were cultured vertically on liquid RIM medium supplemented with 200 mM NaCl or 5% PEG6000 for 4 weeks, then were transferred to liquid RIM medium without NaCl and PEG6000 for 2 weeks. The photographs were taken, the survival rate and the number of axillary bud regeneration, and the number of ARs of each plant were measured. All the above data were represented as the mean ± SD of at least three independent replications. Asterisk indicates a significant difference (*P < 0.05) between transgenic lines and WT plants.
Adventitious root formation is a de novo organogenesis process that enables plants to cope with environmental stresses. To evaluate the growth and root regeneration response of JrWOX11-overexpressing plants and control type (CK) to NaCl and osmotic stress, shoot cuttings were subjected to 200 mM NaCl and 5% w/v PEG6000 stress for 4 weeks. The root and aboveground biomass were significantly improved in JrWOX11-overexpressing plants compared with CK shoot cuttings under no-stress conditions (Figure 7C and Supplementray Figure 3). Additionally, the transgenic shoot cuttings showed significantly improved salt and osmotic tolerance compared to CK plants (Figure 7C). The survival rate, the number of axillary buds regenerated, and the number of ARs were increased substantially 2 weeks after plants were transferred to media without 200 mM NaCl and 5% w/v PEG6000 (Figure 7D). These findings suggested that JrWOX11 enhanced tolerance to salt and osmotic stress by improving the de novo root organogenesis and shoot development (Liu J. et al., 2014, Liu B. et al., 2018; Hu and Xu, 2016; Zhou et al., 2017).
Recalcitrance to adventitious root formation from stem cuttings is a major limitation for clonal propagation in difficult-to-root trees (Macdonald, 1990). Given the energy and time required for maintaining difficult-to-root trees in a juvenile state in various environments, the development of rootstock varieties that are easy-to-root and tolerant to environmental stresses through molecular breeding is crucial. However, there is little knowledge about the genes conferring rooting capacity and abiotic stress tolerance. Previous studies have shown that the WOX11/12 gene, encoding a transcription factor, can be induced by salt and drought stress and that it participates in root formation and development in rice, Arabidopsis, and poplar (Zhao et al., 2009, 2015; Liu B. et al., 2014; Liu J. et al., 2014; Hu and Xu, 2016). Here, we isolated and characterized a WOX11 gene in “ZNS” walnut, a difficult-to-root tree. In addition, in transgenic “84K” poplar overexpressing JrWOX11, the capacity to form adventitious roots and to develop the root system has been improved. Transgenic “84K” poplar overexpressing JrWOX11 also showed increased NaCl and osmotic tolerance.
Previous studies have shown that WOX11 is a crucial factor that controls normal cells shifting to root primordium cells (Liu J. et al., 2014). This process mainly depends on auxin biosynthesis and auxin transport (Sarkar et al., 2007; Chen et al., 2016; Wei et al., 2019). In Arabidopsis, AtWOX11 binds directly to the promoter of a LATERAL ORGAN BOUNDARIES DOMAIN transcription factor (AtLBD16) to result in the formation of LR root primordium cells (Wu et al., 2018). In the present study, overexpression of JrWOX11 in “84K” poplar promoted the formation of root primordium cells 2s day earlier than in the control plants (Figure 5). It could be speculated that the JrWOX11 gene regulated adventitious root formation via the auxin-dependent pathway.
Overexpression of WOX11/12 genes, such as OsWOX11, PtoWOX11, PeWOX11a, PeWOX11b, and PagWOX11/12a resulted in increased numbers of adventitious and ectopic roots, which contributed to plant resistance to drought (Cheng et al., 2016; Wang et al., 2020). Furthermore, Wang et al. (2021) reported that PagWOX11/12a enhanced plant response to salt stress by controlling the redox metabolic pathways. Consistent with the previous studies in rice and poplar, JrWOX11 not only increased the numbers of ARs, LRs, ectopic roots, and the length of RH but also improved the NaCl and osmotic tolerance in overexpressing “84K” poplar (Figure 7 and Supplementary Figure 3). Notwithstanding its limitations, the study suggests that JrWOX11 increases “84K” poplar salt and osmotic tolerance by controlling adventitious root formation and root system development.
To investigate further the potential differences in WOX11/12 between easy-to-root plants and difficult-to-root trees, we compared JrWOX11 to 26 WOX11/12 proteins from 15 species. In rice, Arabidopsis, and some easy-to-root trees, more than one WOX11/12 protein was found (Figure 1). However, “ZNS” walnut contained only one WOX11 protein, JrWOX11. In terms of protein distance matrix to JrWOX11, the descending order of similarity was: difficult-to-root trees (96.37 to 81.21%) > easy-to-root trees (74.44 to 61.48%) > Arabidopsis (57.01 and 54.67%) > rice (55.04 to 52.36%) > maize (49.78 and 45.22%). These results were consistent with the evolutionary relationships of the 27 WOX11/12 proteins analyzed. Furthermore, the MEME result of WOX11/12 pointed out another difference between easy-to-root trees and difficult-to-root trees, with motif10 structure (GGAIQY motif) lacking in difficult-to-root trees (Figure 1 and Supplementary Figure 2). Given that JrWOX11 improves the capacity for adventitious root formation in transgenic “84K” poplar, a lack of the GGAIQY motif was not a determining factor for rooting difficulty in difficult-to-root trees. However, when the expression of JrWOX11 was suppressed, the rooting capacity was significantly diminished (Chang et al., 2022). Age and associated sclerenchyma in difficult-to-root trees should also be considered (Sun and Zhu, 2020). It was reported that WOX11/WOX12 could be suppressed by age-regulated ETHYLENE INSENSITIVE 3 (EIN3) in Arabidopsis (Li et al., 2020). In walnut, rejuvenation diminished the extent of sclerenchyma, which is beneficial to auxin transport and rooting (Liu H. et al., 2018).
We also analyzed the differences in the cis-element of WOX11/12 genes between easy-to-root trees and difficult-to-root trees. Compared with OsWOX11, OsWOX12a, OsWOX12b, AtWOX11, AtWOX12, PtrWOX11/12a, and PtrWOX11/12b, the promoter of JrWOX11 in “ZNS” walnut and J. regia (Chang et al., 2020) harbored more AuxRE and ABREs but fewer MBS, G-box, W-box, and DRE motifs (Figure 2 and Supplementary Figure 4), which are generally known as elements that respond to abiotic stresses (Shi et al., 2011; Lee et al., 2013; Baruah et al., 2021). The result might imply that JrWOX11 enhanced the stress tolerance of transgenic “84K” poplar via the ABA-dependent pathways (Putterill et al., 2021). Further work is needed to test this suggestion.
In the present study, we successfully cloned full-length cDNA and genomic sequences of JrWOX11 from “ZNS” walnut; its expression was induced by ABA, NaCl, and PEG treatment. Compared to easy-to-root plants, walnut harbored fewer WOX11/12 genes, the JrWOX11 protein sequence lacked the “GGAIKY” motif, and the promoter of JrWOX11 harbored more AuxRE and ABREs but fewer MBS, G-box, W-box, and DRE motifs. Additionally, the overexpression of JrWOX11 significantly improved the adventitious root formation capacity, increased the number of AR and LR, and elongated the LR and RH lengths in “84K” poplar overexpressing JrWOX11. Moreover, the tolerance to NaCl and osmotic stress in transgenic “84K” poplar plants and shoot cuttings was also enhanced. This likely reflects the fact that JrWOX11 promotes de novo root organogenesis and root system development, thereby regulating NaCl and osmotic tolerance. The differences in WOX11/12 between easy-to-root plants and difficult-to-root trees provide the foundation for the molecular breeding of difficult-to-root trees.
The data presented in this study are deposited in the GenBank repository, accession number: ON979687.
DP and QZ contributed substantially to the experimental design, conceived the study, and approved the final manuscript. XS contributed to funding, interpreted the data, authored or reviewed the draft, and revised the manuscript. ML gave substantial suggestions to the manuscript. YC, PZ, and XL carried out mainly experiments, finished the draft of the manuscript, and comprehensively analyzed the data from all experimental results. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (Grant No. 32101479), the State Key Laboratory of Tree Genetics and Breeding program (Grant No. CAFYBB2019ZY001), and the Postdoctoral Initiative Foundation of Henan Normal University (Grant No. 5101049470215).
We are grateful to the following investigators for helping with the tissue culture: Ms. Jiaqi Zhang and Bi Liang at Zhejiang A& F University. We would like to express our gratitude to EditSprings (https://www.editsprings.cn) for the expert linguistic services provided.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.951737/full#supplementary-material
Supplementary Figure 1 | The detection of transgenic 84K poplar. (a) DNA detection of HPT gene in WT and transgenic line OE1#∼OE14 #. (b) The expression of the JrWOX11 gene in stems of WT and transgenic line OE1#∼OE14 #5 microshoots.
Supplementary Figure 2 | Sequence alignment of homologous WOX11/12 proteins. (a) Phylogenetic tree based on full-length coding sequences of WOX11/12 genes using the NJ method. (b) Motifs analysis of WOX11/12 proteins. MEME software was used to search motifs, and then redraw the map with the software TBtools (c). Zm, Zea mays; Os, Oryza sativa; At, Arabidopsis thaliana; Ptr, Populus trichocarpa; Pto, Populus tomentosa; Pe, P. deltoides × P. euramericana cv. “Nan-lin895,” Pag, P. alba × P. glandulosa cv. “84K,” Sp, Salix purpurea; Md, Malus domestica; Mn, Morus notabilis; Dz, Durio zibethinus; Jr, Juglans hindsii × J. regia cv. “ZNS,” Ci, Carya illinoinensis; Qs, Quercus suber; Ql, Quercus lobuta; Cm, Castanea mollissima.
Supplementary Figure 3 | Plant phenotype determination during NaCl and PEG treatment. (a) Shoot cuttings of CK, OE3#, and OE6 # were cultured vertically on the liquid RIM medium (1/2MS + 0.01 mg⋅L–1 of IBA + 30 g⋅L–1 of sucrose) as Control. (b,c) Shoot cuttings of CK, OE3#, and OE6 were cultured vertically on liquid RIM medium supplemented with 100 and 200 mM NaCl. (d,e) Shoot cuttings of CK, OE3#, and OE6 were cultured vertically on liquid RIM medium supplemented with 5 and 10% PEG6000. Photographic pictures were made once a week. 1∼4 w: 1∼4 weeks after the shoot cuttings were cultured on the corresponding medium. R1W∼R2W: 1∼4 weeks after the corresponding stress is removed.
Supplementary Figure 4 | Promoter sequence alignment of WOX11 gene in “ZNS” walnut and Juglans regia.
Amissah, J. N., Paolillo, D. J., and Bassuk, N. (2008). Adventitious root formation in stem cuttings of Quercus bicolor and Quercus macrocarpa and its relationship to stem anatomy. J. Am. Soc. Hortic. Sci. 133, 479–486. doi: 10.1117/12.665960
Baruah, P. M., Kashyap, P., Krishnatreya, D. B., Bordoloi, K. S., and Agarwala, N. (2021). Identification and functional analysis of drought responsive lncRNAs in tea plant. Plant Gene 27:100311. doi: 10.1016/j.plgene.2021.100311
Basheer-Salimia, R. (2007). Juvenility, Maturity, and rejuvenation in woody plants. Hebron Univ. Res. J. 3, 17–43.
Chang, Y., Song, X., Zhang, Q., Liu, H., Bai, Y., Lei, X., et al. (2020). Genome-wide identification of WOX gene family and expression analysis during rejuvenational rhizogenesis in walnut (Juglans regia L.). Forests 11:16. doi: 10.3390/f11010016
Chang, Y., Song, X., Zhang, Q., Zhang, P., Lei, X., and Pei, D. (2022). Robust CRISPR/Cas9 mediated gene editing of JrWOX11 manipulated adventitious rooting and vegetative growth in a nut tree species of walnut. Sci. Hortic. 303:111199. doi: 10.1016/j.scienta.2022.111199
Chen, C., Chen, H., Zhang, Y., Thomas, H., Frank, M., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06
Chen, L., Tong, J., Xiao, L., Ruan, Y., Liu, J., Zeng, M., et al. (2016). YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 67, 4273–4284. doi: 10.1093/jxb/erw213
Chen, X., Qu, Y., Sheng, L., Liu, J., Huang, H., and Xu, L. (2014). A simple method suitable to study de novo root organogenesis. Front. Plant Sci. 5:208. doi: 10.3389/fpls.2014.00208
Cheng, S., Huang, Y., Zhu, N., and Zhao, Y. (2014). The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 549, 266–274. doi: 10.1016/j.gene.2014.08.003
Cheng, S., Zhou, D.-X., and Zhao, Y. (2016). WUSCHEL -related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 11:e1130198. doi: 10.1080/15592324.2015.1130198
Da Costa, C., de Almeida, M., Ruedell, C. M., Schwambach, J., Maraschin, F. S., and Fett-Neto, A. G. (2013). When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 4:133. doi: 10.3389/fpls.2013.00133
Gonin, M., Bergougnoux, V., Nguyen, T. D., Gantet, P., and Champion, A. (2019). What makes adventitious roots? Plants 8:240. doi: 10.3390/plants8070240
Hu, X., and Xu, L. (2016). Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 172, 2363–2373. doi: 10.1104/pp.16.01067
Jiang, W., Zhou, S., Zhang, Q., Song, H., Zhou, D.-X., and Zhao, Y. (2017). Transcriptional regulatory network of WOX11 is involved in the control of crown root development, cytokinin signals, and redox in rice. J. Exp. Bot. 68, 2787–2798. doi: 10.1093/jxb/erx153
Klerk, G. (2002). Rooting of microcuttings:theory and practice. Vitr. Cell. Dev. Biol. Plant 38, 415–422. doi: 10.1079/IVP2002335
Lee, S. C., Kim, S. H., and Kim, S. R. (2013). Drought inducible OsDhn1 promoter is activated by OsDREB1A and OsDREB1D. J. Plant Biol. 56, 115–121. doi: 10.1007/s12374-012-0377-3
Li, H., Yao, L., Sun, L., and Zhu, Z. (2020). ETHYLENE INSENSITIVE 3 suppresses plant de novo root regeneration from leaf explants and mediates age-regulated regeneration decline. Development 147:dev179457. doi: 10.1242/dev.179457
Li, J., Zhang, J., Jia, H., Liu, B., Sun, P., Hu, J., et al. (2017). The WUSCHEL-related homeobox 5a (PtoWOX5a) is involved in adventitious root development in poplar. Tree Physiol. 38, 139–153. doi: 10.1093/treephys/tpx118
Liu, B., Wang, L., Zhang, J., Li, J., Zheng, H., Chen, J., et al. (2014). WUSCHEL-related homeobox genes in Populus tomentosa: diversified expression patterns and a functional similarity in adventitious root formation. BMC Genomics 15:296. doi: 10.1186/1471-2164-15-296
Liu, J., Sheng, L., Xu, Y., Li, J., Yang, Z., Huang, H., et al. (2014). WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26, 1081–1093. doi: 10.1105/tpc.114.122887
Liu, B., Zhang, J., Yang, Z., Matsui, A., Seki, M., Li, S., et al. (2018). PtWOX11 acts as master regulator conducting the expression of key transcription factors to induce de novo shoot organogenesis in poplar. Plant Mol. Biol. 98, 389–406. doi: 10.1007/s11103-018-0786-x
Liu, H., Gao, Y., Song, X. B., Ma, Q. G., Zhang, J. P., and Pei, D. (2018). A novel rejuvenation approach to induce endohormones and improve rhizogenesis in mature Juglans tree. Plant Methods 14:13. doi: 10.1186/s13007-018-0280-0
Liu, J., Hu, X., Qin, P., Prasad, K., Hu, Y., and Xu, L. (2018). The WOX11 - LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol. 59, 734–743. doi: 10.1093/pcp/pcy010
Liu, W., and Xu, L. (2018). Recruitment of IC-WOX genes in root evolution. Trends Plant Sci. 23, 490–496. doi: 10.1016/j.tplants.2018.03.011
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., et al. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. doi: 10.1016/S0092-8674(01)00384-1
Macdonald, B. (1990). Practical Woody Plant Propagation for Nursery Growers. Portland, OR: Timber Press.
Maynard, B. K., and Bassuk, N. L. (1990). Stockplant etiolation, shading, and stem banding effects on the auxin dose-response of rooting in softwood stem cuttings of Carpinus betulus ‘fastigiata’. J. Am. Soc. Hortic. Sci. 25:1126a. doi: 10.21273/HORTSCI.25.9.1126a
Pei, D., and Gu, R. S. (2002). Shoot rooting in vitro for walnut cultivars. Sci. Silvae Sin. 38, 32–37. doi: 10.3321/j.issn:1001-7488.2002.02.007
Pei, D., Zhang, J. P., and Xu, H. Z. (2004). Cutting propagation method for Juglans: China, CN ZL200410009201.4.
Pijut, P. M., Woeste, K. E., and Michler, C. H. (2011). Promotion of adventitious root formation of difficult-to-root aardwood tree species. Hortic. Rev. 38, 213–251. doi: 10.1002/9780470872376.ch6
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (2021). The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Shi, J., Zhang, L., An, H., Wu, C., and Guo, X. (2011). GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol. Biol. 12:22. doi: 10.1186/1471-2199-12-22
Sun, L., and Zhu, Z. (2020). The Molecular basis of age-modulated plant de novo root regeneration decline in Arabidopsis thaliana. Plant Cell Physiol. 62, 3–7. doi: 10.1093/pcp/pcaa134
Wang, L., Zhang, C., Wang, Y., Wang, Y., Yang, C., Lu, M., et al. (2018). Tamarix hispida aquaporin ThPIP2;5 confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis. Environ. Exp. Bot. 152, 158–166. doi: 10.1016/j.envexpbot.2017.05.018
Wang, L. Q., Li, Z., Wen, S. S., Wang, J. N., Zhao, S. T., and Lu, M. Z. (2020). WUSCHEL-related homeobox gene PagWOX11/12a responds to drought stress by enhancing root elongation and biomass growth in poplar. J. Exp. Bot. 71, 1503–1513. doi: 10.1093/jxb/erz490
Wang, L. Q., Wen, S. S., Wang, R., Wang, C., Gao, B., and Lu, M. Z. (2021). PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant Biotechnol. J. 19, 2249–2260. doi: 10.1111/pbi.13653
Wei, K., Ruan, L., Wang, L., and Cheng, H. (2019). Auxin-induced adventitious root formation in nodal cuttings of Camellia sinensis. Int. J. Mol. Sci. 20:4817. doi: 10.3390/ijms20194817
Wu, L., Jie, Y., Yachao, G., Peng, Q., and Lin, X. (2018). Pivotal role of LBD16 in root and root-like organ initiation. Cell. Mol. Life Sci. 75, 3329–3338. doi: 10.1007/s00018-018-2861-5
Xu, M., Xie, W., and Huang, M. (2015). Two WUSCHEL-related homeobox genes, PeWOX11a and PeWOX11b, are involved in adventitious root formation of poplar. Physiol. Plant. 155, 446–456. doi: 10.1111/ppl.12349
Zhang, G., Zhao, F., Chen, L., Pan, Y., Sun, L., Bao, N., et al. (2019). Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants 5, 491–497. doi: 10.1038/s41477-019-0408-x
Zhang, J., Zhang, W., Ji, F., Qiu, J., Song, X., Bu, D., et al. (2020). A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication. Plant Biotechnol. J. 18, 1848–1850. doi: 10.1111/pbi.13350
Zhang, X., Zong, J., Liu, J., Yin, J., and Zhang, D. (2010). Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and Poplar. J. Integr. Plant Biol. 52, 1016–1026. doi: 10.1111/j.1744-7909.2010.00982.x
Zhao, Y., Cheng, S., Song, Y., Huang, Y., Zhou, S., Liu, X., et al. (2015). The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 27, 2469–2483. doi: 10.1105/tpc.15.00227
Zhao, Y., Hu, Y., Dai, M., Huang, L., and Zhou, D.-X. (2009). The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell 21, 736–748. doi: 10.1105/tpc.108.061655
Keywords: JrWOX11, overexpression, adventitious root formation, abiotic stress tolerance, motif-”GGAIQY”
Citation: Chang Y, Song X, Li M, Zhang Q, Zhang P, Lei X and Pei D (2022) Characterization of walnut JrWOX11 and its overexpression provide insights into adventitious root formation and development and abiotic stress tolerance. Front. Plant Sci. 13:951737. doi: 10.3389/fpls.2022.951737
Received: 24 May 2022; Accepted: 29 July 2022;
Published: 06 September 2022.
Edited by:
Prasanta Kumar Subudhi, Louisiana State University, United StatesReviewed by:
Boon Chin Tan, University of Malaya, MalaysiaCopyright © 2022 Chang, Song, Li, Zhang, Zhang, Lei and Pei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dong Pei, cGVpLmRvbmdAY2FmLmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.