- Anhui Provincial Key Laboratory of Conservation and Exploitation of Important Biological Resources, College of Life Sciences, Anhui Normal University, Wuhu, China
Polyamines (PAs) play a critical role in growth and developmental processes and stress responses in plants. Polyamine oxidase (PAO) is a flavin adenine dinucleotide (FAD)-dependent enzyme that plays a major role in PA catabolism. Here, for the first time, PAO genes in maize were screened for the whole genome-wide and nine ZmPAO genes were identified in this study, named as ZmPAO1-9. Based on structural characteristics and a comparison of phylogenetic relationships of PAO gene families from seven representative species, all nine PAO proteins in maize were categorized into three distinct subfamilies. Further, chromosome location and schematic structure revealed an unevenly distribution on chromosomes and evolutionarily conserved structure features of ZmPAO genes in maize, respectively. Furthermore, transcriptome analysis demonstrated that ZmPAO genes showed differential expression patterns at diverse developmental stages of maize, suggesting that these genes may play functional developmental roles in multiple tissues. Further, through qRT-PCR validation, these genes were confirmed to be responsive to heat, drought and salinity stress treatments in three various tissues, indicating their potential roles in abiotic stress responses. Eventually, to verify the biological function of ZmPAO genes, the transgenic Arabidopsis plants overexpressing ZmPAO6 gene were constructed as a typical representative to explore functional roles in plants. The results demonstrated that overexpression of ZmPAO6 can confer enhanced heat tolerance through mediating polyamine catabolism in transgenic Arabidopsis, which might result in reduced H2O2 and MDA accumulation and alleviated chlorophyll degradation under heat stress treatment, indicating that ZmPAO6 may play a crucial role in enhancing heat tolerance of transgenic Arabidopsis through the involvement in various physiological processes. Further, the expression analysis of related genes of antioxidant enzymes including glutathione peroxidase (GPX) and ascorbate peroxidase (APX) demonstrated that ZmPAO6 can enhance heat resistance in transgenic Arabidopsis through modulating heat-induced H2O2 accumulation in polyamine catabolism. Taken together, our results are the first to report the ZmPAO6 gene response to heat stress in plants and will serve to present an important theoretical basis for further unraveling the function and regulatory mechanism of ZmPAO genes in growth, development and adaptation to abiotic stresses in maize.
Introduction
Polyamines (PAs), largely including spermidine (Spd), spermine (Spm), and putrescine (Put), are small molecular aliphatic amine (Knott et al., 2007; Hussain et al., 2011). PAs have been detected and studied in many actively growing tissues and play crucial roles in nitrogen metabolism (Moschou et al., 2012), dark-induced senescence (Sobieszczuk-Nowicka et al., 2015), flowering (Ahmed et al., 2017), fruit ripening (Handa and Mattoo, 2010; Fortes and Agudelo-Romero, 2018), chloroplast chemical penetration (Ioannidis et al., 2016), callus growth (Liu and Moriguchi, 2007), regulation of protein synthesis (Igarashi and Kashiwagi, 2000), signal molecules (Pál et al., 2019) and especially in the oxidative balance of cells in plants (Murray Stewart et al., 2018). Moreover, increasing studies have elucidated that PAs are involved extensively in various abiotic and biotic stress responses (Alcázar et al., 2006; Kusano et al., 2008; Moschou et al., 2008b; Toumi et al., 2010; Tiburcio et al., 2014; Liu et al., 2015; Corpas et al., 2019). For instance, under salt stress, the application of spermine reduced the harmful effects on the growth and gas exchange of Tropaeolum majus, increased the activities of CAT and POD under severe salt stress, and the activities of POD and APX under moderate salt stress (da Silva et al., 2022). Foliar application of putrescine on cucumber alleviated the decrease of stomatal aperture and photosynthesis caused by salt stress treatment, and promoted the growth of cucumber (Ma et al., 2022). Moreover, exogenous putrescine significantly improved ROS scavenging ability and promoted the metabolism of endogenous PAs so as to provide better drought resistance for Cabernet Sauvignon (Zhao et al., 2021). Treatment of seeds of mung bean with PAs can enhance the drought resistance of mung bean plants by accumulating osmoprotectants, improving water status and chlorophyll value, and reducing oxidative damage (Sadeghipour, 2019). Therefore, PAs play a crucial role in maintaining normal growth and development and resisting adverse environmental stresses in plants.
The levels of cellular endogenous PAs largely depend on the dynamic balance of biosynthesis and catabolism of PAs. Polyamine biosynthesis has been thoroughly studied in animals and plants. Generally, the biosynthesis of PAs begins with the synthesis of putrescine. L-ornithine is used for precursor to synthesize Put in animals, whereas L-ornithine and L-arginine serve as precursors to generate Put in plants (Wu et al., 2005; Pegg and Casero, 2011). In plants, arginine decarboxylase (ADC) and ornithine decarboxylase (ODC) are required for catalyzing the synthesis of Put, respectively. Then, Put can be converted into Spd via Spd synthase (SPDS). Finally, Spd can be further converted into Spm under the catalysis of Spm synthase (SPMS; Moschou et al., 2008a). In contrast to the anabolism of PAs, the catabolism of PAs in plants is mainly dependent on the catalyzation from two major categories of amine oxidases: diamine oxidases (DAOs) containing divalent Cu2+, also known as copper amine oxidases (CuAOs), and flavin-containing polyamine oxidases (PAOs; Pegg and Casero, 2011; Planas-Portell et al., 2013). As far as subcellular localization is concerned, some CuAO and PAO proteins are localized in peroxisomes (Wang et al., 2019). These proteins contain a C-terminal peroxisomal targeting type I (PTS1), a tripeptide consisting of the consensus sequence (S/A/C)(K/R/H)(L/M; Hao et al., 2018). For example, two CuAO (AtCuAO2 and AtCuA03) and three PAO (AtPAO2-AtPAO4) from Arabidopsis (Fincato et al., 2011; Planas-Portell et al., 2013), two PAO (CsPAO2 and CsPAO3) from sweet orange (Wang and Liu, 2015), four PAO (SLPAO2-SLPAO 5) from tomato (Hao et al., 2018), and three PAO (OsPAO3-OsPAO5) from rice (Ono et al., 2012) all carry a putative type-I peroxisomal targeting signal in their C-termini. The function of CuAOs is to catalyze the oxidation of Put and cadaverine (Cad) to 4-aminobutyraldehyde, H2O2 and ammonia (Moschou et al., 2012). In contrast, according to functional differentiation in PAOs involved in polyamine catabolism, PAOs are mainly categorized into two groups in plants. The PAOs of the first group in plants are required for participating in PA terminal catabolism and perform the oxidation and decomposition of Spd and Spm to produce H2O2, 1, 3-diaminopropane (DAP), and 4-aminobutanal (Spd catabolism) or N-(3-aminopropyl)-4-aminobutanal (Spm catabolism; Angelini et al., 2010; Moschou et al., 2012; Mo et al., 2015), whereas the PAOs in the second group resembles the mammalian Spm oxidases (SMOs), which are mainly involved in back-conversion reactions of PAs by catalyzing Spm to Spd and Spd to Put, in a reverse reaction of PA synthesis and with concomitant production of 3-aminopropanal and H2O2 (Angelini et al., 2010; Moschou et al., 2012).
In plants, genes encoding PAOs have been identified and characterized in several species including Arabidopsis (Fincato et al., 2011), rice (Sagor et al., 2017), sweet orange (Wang and Liu, 2015), tomato (Hao et al., 2018), and tea plant (Li et al., 2020). Most of the identified PAO genes in plants are largely involved in PA back-conversion pathway. For example, five PAO genes (AtPAO1-AtPAO5) in Arabidopsis have been revealed to encode PAO proteins (Fincato et al., 2011), all of which are involved in catalyzing PA back-conversion reactions (Tavladoraki et al., 2006; Kamada-Nobusada et al., 2008). Also, a total of seven PAO genes (OsPAO1-OsPAO7) have been identified in rice. Among them, there are four OsPAO genes (OsPAO1, OsPAO3-OsPAO5) encoding PAO proteins that are involved in the back-conversion reactions of PAs (Ono et al., 2012). Moreover, there are six PAO genes (CsPAO1-CsPAO6) identified to encode their proteins in sweet orange. Among them, the CsPAO3 gene has been implicated in participating in the back-conversion reactions of PAs (Wang and Liu, 2015, 2016).
The catabolism of PA generally results in the accumulation of H2O2 (Liu et al., 2015). H2O2 can be regarded as a crucial signal molecule at low levels, whereas at high levels it is a toxic compound that causes damage to plant cells (Quan et al., 2008). Therefore, the ratio of PA catabolism to biosynthesis has been considered to be a vital factor in inducing tolerance responses or plant cell death under biotic and abiotic stresses (Moschou et al., 2008a). This suggests that H2O2 produced by PA catabolism may play a crucial role in maintaining ROS homeostasis, which is closely related to the stress response of plants. For example, the overexpressing of MdSPDS1 in sweet orange enhanced the activity of PAOs to catalyze polyamine catabolism and thereby result in the accumulation of H2O2, which likely triggered the hypersensitivity or activation of defense-related genes and eventually significantly reduced the sensitivity of transgenic plants to canker disease (Fu et al., 2011). Under salt stress, the contents of the apoplastic Spm and Spd in maize increased significantly and then the oxidation of free apoplast polyamine catalyzed by polyamine oxidase promoted maize leaf elongation (Rodríguez et al., 2009). In addition, Spd could outflow into the apoplast, and then Spd was decomposed by PAOs to produce H2O2, which led to stress tolerance response or induction of programmed cell death (PCD) under salt stress in tobacco (Moschou et al., 2008a). Similarly, the ABA signaling pathway integrated PAs and PAOs to regulate the H2O2 accumulation, which can also cause further stress response or PCD in grape (Toumi et al., 2010). Moreover, H2O2 produced by Spd oxidation that was catalyzed by PAOs as a signal molecule could trigger the opening of hyperpolarized Ca2+ channels to result in Ca2+ influx, and thereby regulate the growth of pollen tubes in Arabidopsis (Wu et al., 2010). Therefore, H2O2 produced by PAOs catalyzing the catabolism of PAs may play a vital role in growth, development and response to abiotic stresses in plants via modulating the dynamic balance of H2O2.
Maize (Zea mays L.), as one of the most important crop species in the world, has become one of the important model monocot species for functional genomics analysis. It has been of great significance to the study of molecular biology in plants. Meanwhile, maize is a vital part of the food system (Palacios-Rojas et al., 2020). However, biotic and abiotic stresses have significant adverse effects on maize yield and quality. Since PAOs play a vital role in responding to various stress treatments, the study of the roles of PAOs may provide a new way to enhance the stress resistance in plants. However, little is known about identification and functional analysis of polyamine oxidase gene family in maize up to now. In this study, the relevant information on ZmPAO genes for the whole genome-wide and the role of ZmPAO6 under heat stress treatment were further clarified. Taken together, this study may contribute to an in-depth comprehension of the evolution of PAO gene family in maize and their crucial roles in abiotic stress resistance.
Materials and Methods
Identification of PAO Family Genes From Maize
To obtain all putative PAO family genes in maize, taking the reported PAO gene sequences of Arabidopsis, rice, and maize as queries, the programs were used to search the maize genome databases, including the Maize Genome Browser (http://www.maizesequence.org), Phytozome (https://phytozome-next.jgi.doe.gov/), Maize Genetics and Genomics Database (MaizeGDB, http://www.maizegdb.org/), and NCBI (http://www.ncbi.nlm.nih.gov; Zhang et al., 2022). The information about the coding sequence length, amino acid length, and chromosomal localization of each gene was also obtained from NCBI. Furthermore, the molecular weight and theoretical isoelectric point (PI) value of the ZmPAO proteins were reckoned within the online Expasy Bioinformatics Resource Portal (http://web.expasy.org/compute_pi/).
Sequence Alignment and Phylogenetic Analysis
To determine the phylogenetic relationships of the PAO proteins, full-length amino acid sequences of PAO proteins identified in maize, rice, Arabidopsis, barley, tomato, sweet orange, and cotton were aligned by the Clustal W program using their default settings. The information of PAO genes in these plants were listed in Supplementary Table S1. Based on the aligned protein sequences, the phylogenetic tree was constructed through the neighbor-joining (NJ) method and examined by bootstrap analyses (1,000 replicates) with MEGA 7.0 (Kumar et al., 2016). Furthermore, the amino acid sequences of PAO proteins in maize were aligned using DNAMAN 7.0 software for multiple protein sequences alignment.
Analysis of Gene Structure and Conserved Motif of Maize PAO Proteins
The conserved motifs in the full-length amino acid sequences of ZmPAO proteins were identified by the motif analysis tool MEME Suite version 4.0.0 (MEME; http://alternate.meme-suite.org/tools/meme; Bailey et al., 2009). The parameters were set as the following: (1) the maximum number of patterns was set to identify ten motifs; (2) the optimal width of the motif was set between 10 and 50; (3) the rest were executed according to the default parameters (Hu et al., 2021). Furthermore, to analyze the exon and intron structures of PAO genes, the structures of PAO genes were analyzed by the Gene Structure Display Server database (GSDS, http://gsds.cbi.pku.edu.cn/; Hu et al., 2015).
Chromosomal Localization and Prediction of cis-Acting Elements in Maize PAO Genes
The chromosomal location image of ZmPAO genes was accomplished by MapInspect (https://mapinspect.software.informer.com/) based on the information from the Maize Genetics and Genomics Database (http://www.maizegdb.org/). To analyze the cis-regulatory elements of the ZmPAO genes in response to abiotic stresses, the promoter sequence in the upstream 2,000 bp of each ZmPAO gene start codon was submitted to the PlantCare database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) to predict stress response and hormone-related cis-acting elements (Chang et al., 2008).
Expression Profile Analysis of Maize PAO Genes in Different Tissues
To explore the spatiotemporal expression patterns of ZmPAO genes, the tissue-specific expression patterns of ZmPAO genes were analyzed using the genome-wide gene expression atlas of maize inbred line B73 that was reported previously (Sekhon et al., 2011). Microarray analysis was completed to determine the expression patterns of ten representative tissues (seed, root, seedling, stem, shoot tip, silks, leaf, tassel, husk, and endosperm). The signal values for maize PAO genes in the ten tissues were list in Supplementary Table S2. The Normalized expression values of the ten tissues were averaged. The gene expression levels were presented as log2 values which were used to generate the heat map of the expressions of ZmPAO genes using HEML1.0.3.3 (HeatMap Illustrator, http://ccd.biocuckoo.org/).
Maize Growth Conditions and Abiotic Stress Treatment
The maize B73 inbred seedlings were grown at 28°C with long-day conditions of 15 h of light and 9 h of dark and an environmental humidity of 60%. After 3 weeks, the seedlings were subjected to heat (42°C), drought, and salt treatments (200 mM NaCl) for 1, 2, 4, and 8 h, respectively. Notably, to impose the drought stress, the roots of the seedlings were pulled out from the soil at 28°C. Three biological replicates were performed for each sample. All collected samples were rapidly frozen in liquid nitrogen and stored at –80°C until RNA extraction.
Generation of ZmPAO6 Transgenic Arabidopsis Plants
To create ZmPAO6 overexpression constructs, the coding sequence of ZmPAO6 was PCR-amplified, with maize B73 cDNA used the templates to clone with high-fidelity polymerase (Trans Start® Fast Pfu DNA Polymerase). The PCR product was purified and cloned into the PHB vector with the EasyGeno Assembly Cloning kit (TIANGEN, China). The vectors were transformed into Agrobacterium strain for transformation into Arabidopsis by the floral dip method. Seeds of the T0 and T1 generations were selected on 1/2 MS plates supplemented with hygromycin (30 μg/ml) and confirmed by PCR with primers (Hyg-F: GGTCGCGGAGGCTATGGATGC; Hyg-R: GCTTCTGC GGGCGATTTGTGT). All genes or clones were confirmed by sequencing.
Heat Tolerance Assay of Transgenic Arabidopsis
To evaluate the tolerance for heat stress, the seeds of WT and transgenic Arabidopsis homozygous T3 lines were grown at 22°C under long-day conditions of 16 h of light and 8 h of dark and an environmental humidity of 60% for 7 days. The three-week-old seedlings were exposed to heat stress (42°C) in the incubator for 36 h. Phenotypic records and physiological parameters analysis were completed at the designed processing time.
RNA Extraction and Quantitative Real-Time PCR Analysis
According to the manufacturer’s instruction, total RNA was extracted from the collected samples using Trizol RNA isolation (United States of America). For quantitative real-time PCR, cDNA from three distinct biological samples was used for analysis. The PCR condition was performed as follows: pre-denaturation for 15 min at 95°C, 40 cycles at 95°C for 10 s, 55°C for 30 s, and 72°C for 30 s. The gene expression levels were calculated using the 2−ΔΔCt method (Rao et al., 2013). Each experiment was conducted in the form of at least three technologies and biological replication. All the primers in this study were listed in Supplementary Table S3.
Physiological Parameter Determination
3, 3′-Diaminobenzidine (DAB) was used to dye the leaves to observe the accumulation of H2O2. The contents of H2O2 and MDA were measured using the H2O2 assay kit (A064-1-1) and MDA assay kit (A003-1-1; Nanjing Jiancheng, Nanjing, China). Moreover, the contents of chlorophyll and polyamines were detected by the spectrophotometer and high-performance liquid chromatography, respectively. The experiment was repeated three times at least.
Results
Identification and Analysis of PAO Genes in Maize
In this study, a total of nine ZmPAO genes (namely ZmPAO1-ZmPAO9) were identified. The related information of ZmPAO genes was shown in Table 1, including the chromosome location, the length of coding sequences and corresponding information about the nine proteins. The length of CDS sequences of these putative genes ranged from 504 bp (ZmPAO7) to 1,590 bp (ZmPAO4), and the amino acid sequences of these proteins ranged from 167 (ZmPAO7) residues to 529 (ZmPAO4) residues, respectively. The MWs of ZmPAO proteins varied from 19.0 kDa (ZmPAO7) to 57.9 kDa (ZmPAO1) and pIs from 5.21 (ZmPAO3) to 9.89 (ZmPAO7).
Phylogenetic Analysis and Sequence Alignment of ZmPAO Proteins
To identify the candidate PAO proteins in maize, the sequences of PAO proteins from several various species including maize, Arabidopsis, rice, barley, tomato, sweet orange and cotton were collected to construct a phylogenetic tree by MEGA 7.0 (Figure 1). The phylogenetic tree revealed that these proteins can be categorized into four groups (I–IV). However, the ZmPAO proteins in maize were only clustered into three groups (I, II, and IV). The Group I was consisted of 22 PAO proteins, including six PAO proteins (ZmPAO3, ZmPAO4, ZmPAO6, ZmPAO7, ZmPAO8, and ZmPAO9). ZmPAO2 protein was clustered into the Group II, and the two remaining proteins including ZmPAO1 and ZmPAO5 were clustered into the Group IV. Multiple sequence alignment was performed using the amino acid sequences of ZmPAO proteins by DNAMAN software (Figure 2). The results demonstrated that ZmPAO4, ZmPAO6, and ZmPAO9 proteins contain SRL sequences. Moreover, ZmPAO3 protein contains CRT sequences, which share high homology with rice OsPAO4 protein. Thus, these results indicated that ZmPAO proteins in maize shared highly conserved homology with other plants in evolution.
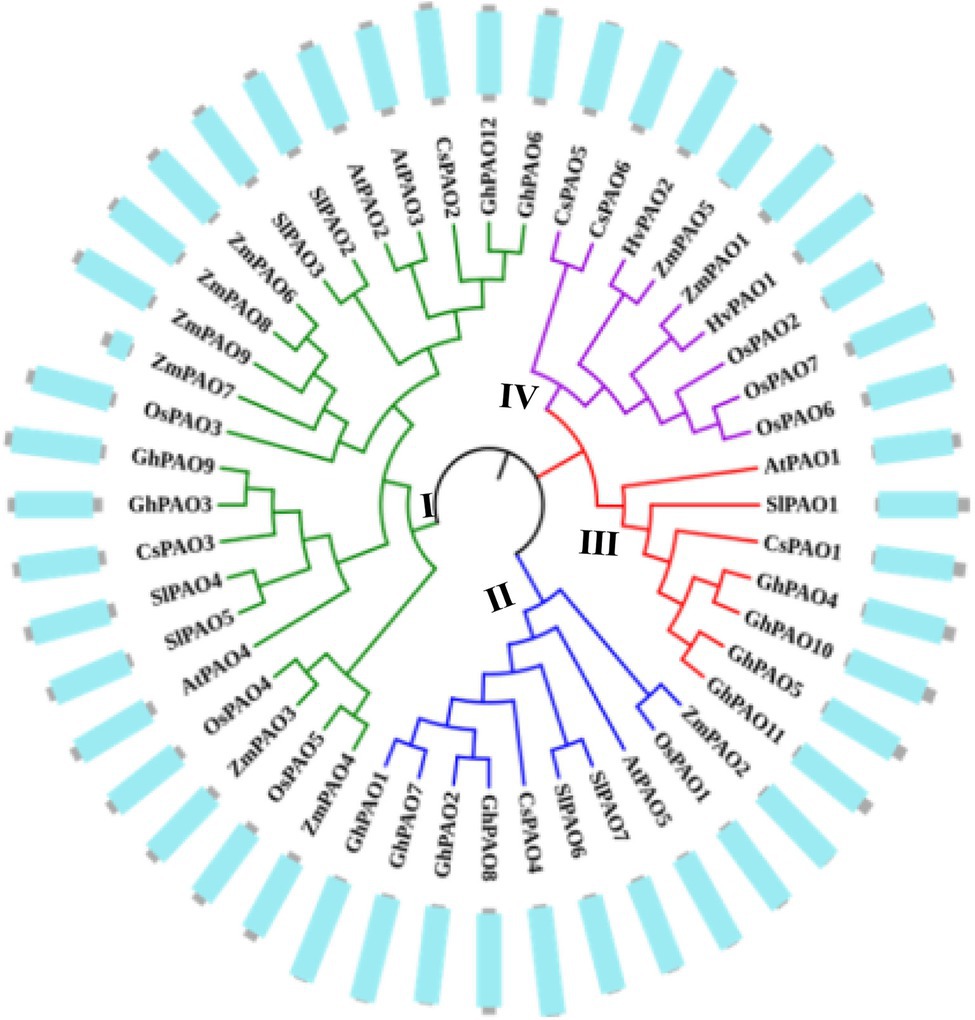
Figure 1. Phylogenetic analysis among the polyamine oxidase (PAO) proteins from seven representative species. The accession numbers of the genes were listed in Supplementary Table 1. The colored branch of the tree indicated that the total PAO proteins were classed into four groups, and nine ZmPAO proteins were classed into three groups according to the amino acid sequence alignment by MEGA 7.0 software with the Neighbor-joining method. The conserved amine-oxidase domains of these proteins were exhibited in the outer ring. Gray represents the length of the amino acid sequence and blue represents the sequence of the conserved amine-oxidase domain.

Figure 2. Alignment of the amino acid sequences of maize PAO proteins. Sequence alignment was performed by DNAMAN software. Identical and similar residues are shaded in black or pink and blue background, respectively. The peroxisomal targeting signal of ZmPAO3, ZmPAO4, ZmPAO6, and ZmPAO9 are indicated in the red box.
Conserved Motifs of ZmPAO Proteins and Gene Structure of ZmPAO Genes
To further gain more insights into the evolutionarily conserved structure features of PAO gene family in maize, a total of ten conserved motifs in ZmPAO, OsPAO, and AtPAO proteins were identified by the MEME website (Figure 3A). In the Group I, all ZmPAO proteins contain the Motifs 1–9 except ZmPAO7 protein, which lacks the Motifs 2–8. All members of the Groups III and IV include the Motifs 3, 5, 8, 9, and 10. Meanwhile, to further perceive the structure of the PAO gene family in maize, the intron–exon structures of ZmPAO genes were analyzed by GSDS software (Figure 3B). ZmPAO3, ZmPAO6, and ZmPAO9 genes all contain nine introns. Secondly, both ZmPAO4 and ZmPAO8 genes contain eight introns. Notably, there are some gene structures without introns in ZmPAO2, OsPAO1, and AtPAO5 genes. In addition, ZmPAO1, ZmPAO5, and ZmPAO7 genes contain 7, 5, and 3 introns, respectively. Overall, these results further demonstrated that the PAO gene family in maize exhibited highly conserved structure features during the evolution of plants.
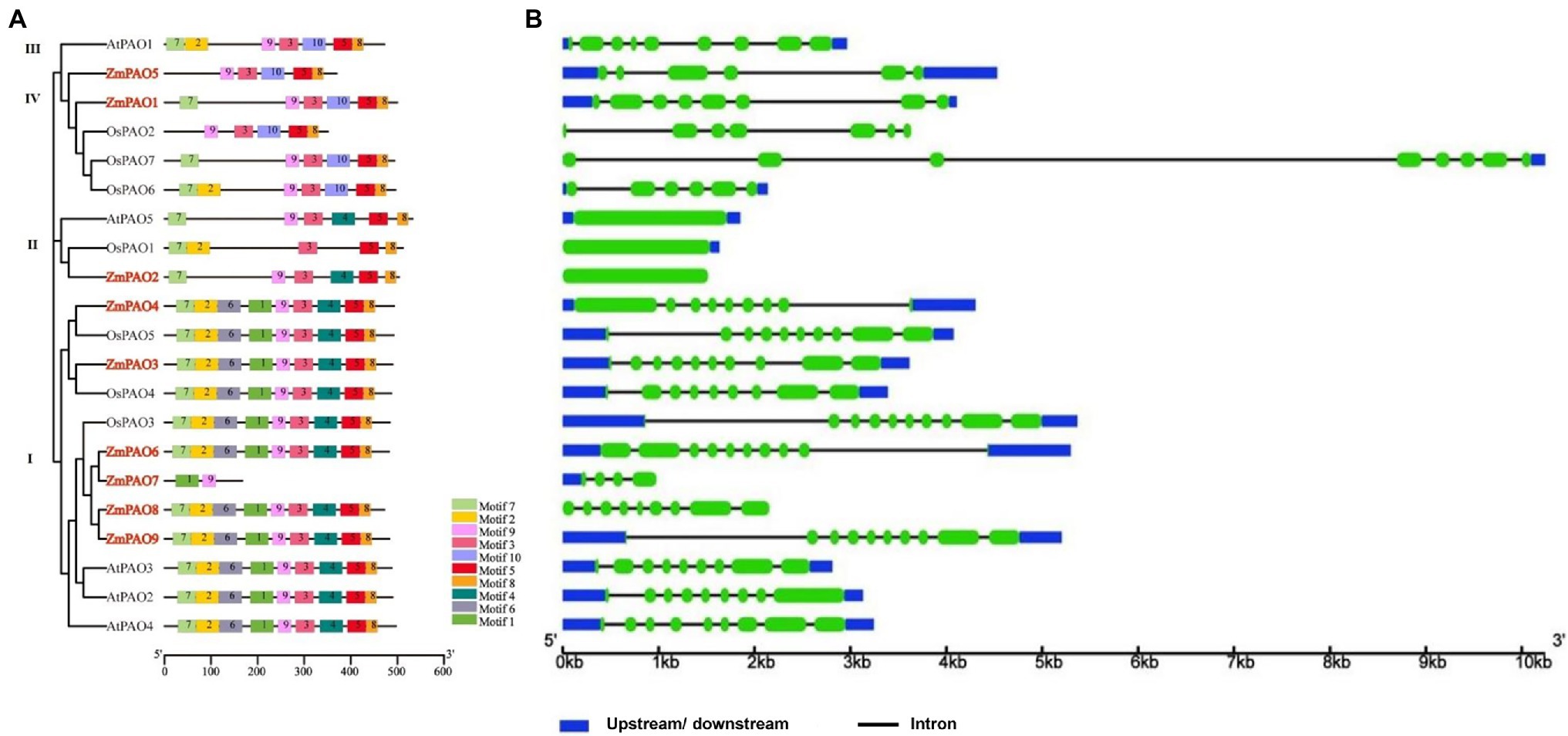
Figure 3. Conserved motifs of PAO proteins and gene structure of PAO genes in maize, rice and Arabidopsis. (A) Distribution of all motifs in maize, rice and Arabidopsis were identified by MEME. Different motifs are indicated in various colored boxes. (B) Exon/intron structures of PAO genes in maize, rice and Arabidopsis. Exons are indicated with green boxes, and introns are shown as black lines. The upstream/downstream region of PAO genes are indicated in blue boxes. The length of each PAO gene inferred by the scale at the bottom.
Chromosomal Location and Gene Duplication of PAO Gene Family in Maize
To generate the chromosome location graphics of ZmPAO genes, the physical positions of ZmPAO genes were investigated by analyzing the genomic distribution of ZmPAO genes on chromosomes in maize. The results indicated that the ZmPAO genes were distributed unevenly across five of all the ten chromosomes in the maize genome (Figure 4). Among them, the Chromosome 10 contained the largest number of ZmPAO genes with three members (namely ZmPAO1, ZmPAO4, and ZmPAO6), whereas ZmPAO5 and ZmPAO1 were located on the Chromosomes 1 and 3, respectively. Moreover, ZmPAO3 and ZmPAO9 were distributed on the Chromosome 2. Similarly, ZmPAO7 and ZmPAO8 were distributed on the Chromosome 6. Moreover, gene duplication events were investigated to explore the evolutionary patterns of the PAO gene family in maize. Two PAO gene pairs (ZmPAO3/ZmPAO4 and ZmPAO6/ZmPAO9) were revealed to be involved in maize segmental duplication for they were exhibited to have very high homology in the sequences by analyses of sequence alignment (Figure 2). This result indicated that gene duplication events might have occurred during their process of evolution.
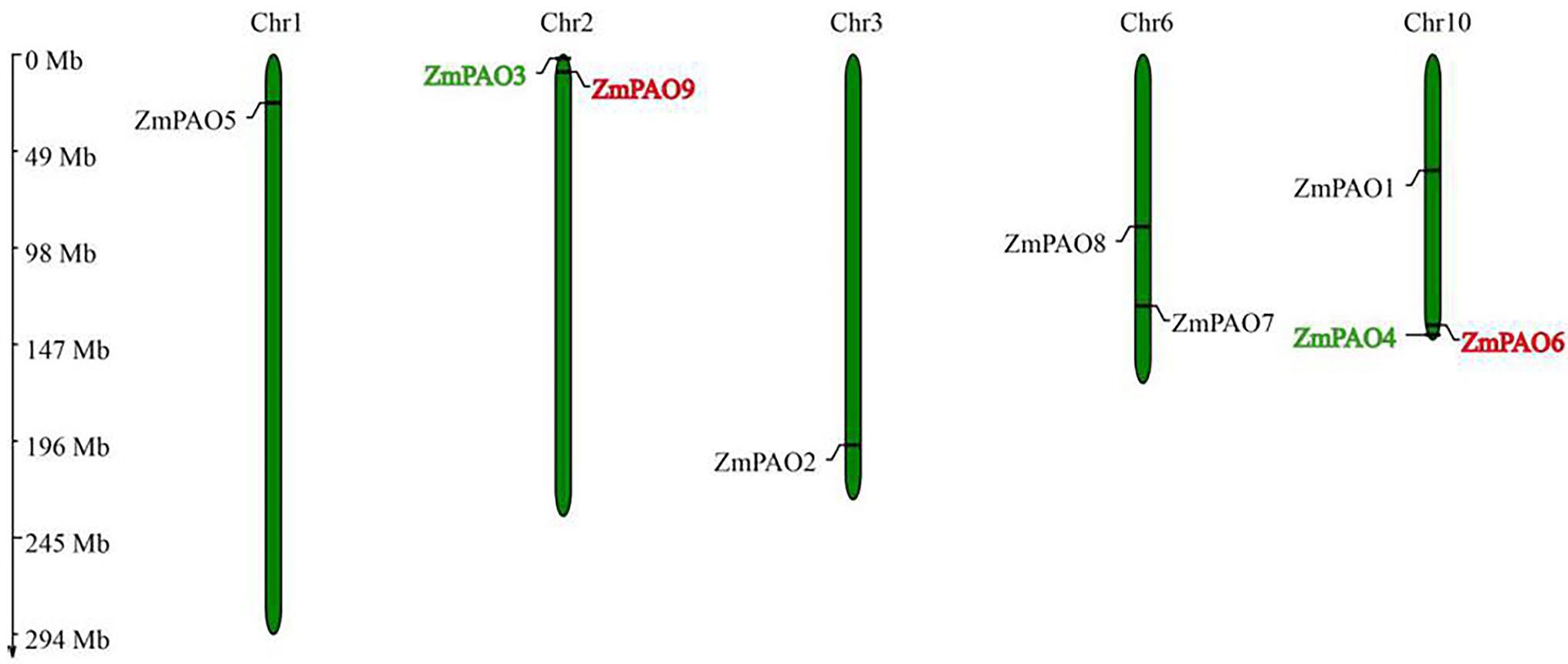
Figure 4. Chromosomal location and gene duplication of the PAO gene family in maize. The gene names on each chromosome correspond to the approximate locations of each ZmPAO genes. Green and red gene names represent tandem duplicated gene pairs. The size of a chromosome is indicated by its relative length. The chromosome numbers are indicated at the top of each bar. The scale on the left is in megabases.
The cis-Acting Regulatory Elements in the Promoter of Maize PAO Genes
To further explore the potential regulatory mechanisms of ZmPAO genes in response to abiotic stresses in maize, the promoter sequences were analyzed using the PLACE database to identify cis-regulatory elements in the promoter region. The results indicated that 11 types of stress- and hormone-related cis-acting regulatory elements were detected in the promoters of maize PAO genes: five hormone-related elements (ABRE, CGTCA-motif, TGACG-motif, GARE-motif, and P-box) and six stress-related elements (TC-rich repeats, MBS, LTR, ERE, WUN-motif, and W-box; Figure 5). It is worth noting that the ABRE (a cis-element involved in ABA responsiveness) were detected in the promoter regions of all the ZmPAO genes. Except for ZmPAO3, MeJA-responsive elements were discovered in the promoters of the other eight ZmPAO genes. In addition, ethylene-responsive element (ERE) was uncovered in the promoter regions of the ZmPAO3 and ZmPAO5 genes. Low temperature-responsive element (LTR) was predicted in the promoter regions of the ZmPAO3, ZmPAO4, ZmPAO5, and ZmPAO6 genes, while WUN-motif (wound-responsive element) was discovered in the promoter region of the ZmPAO7 gene. Meanwhile, the promoter regions of most ZmPAO genes contain the W-box element. TC-rich repeats as defense and stress-responsive elements were predicted to exist in the promoters of the ZmPAO2 and ZmPAO8 genes. This result indicated that ZmPAO genes might be mainly involved in hormone regulation and adversity stress.
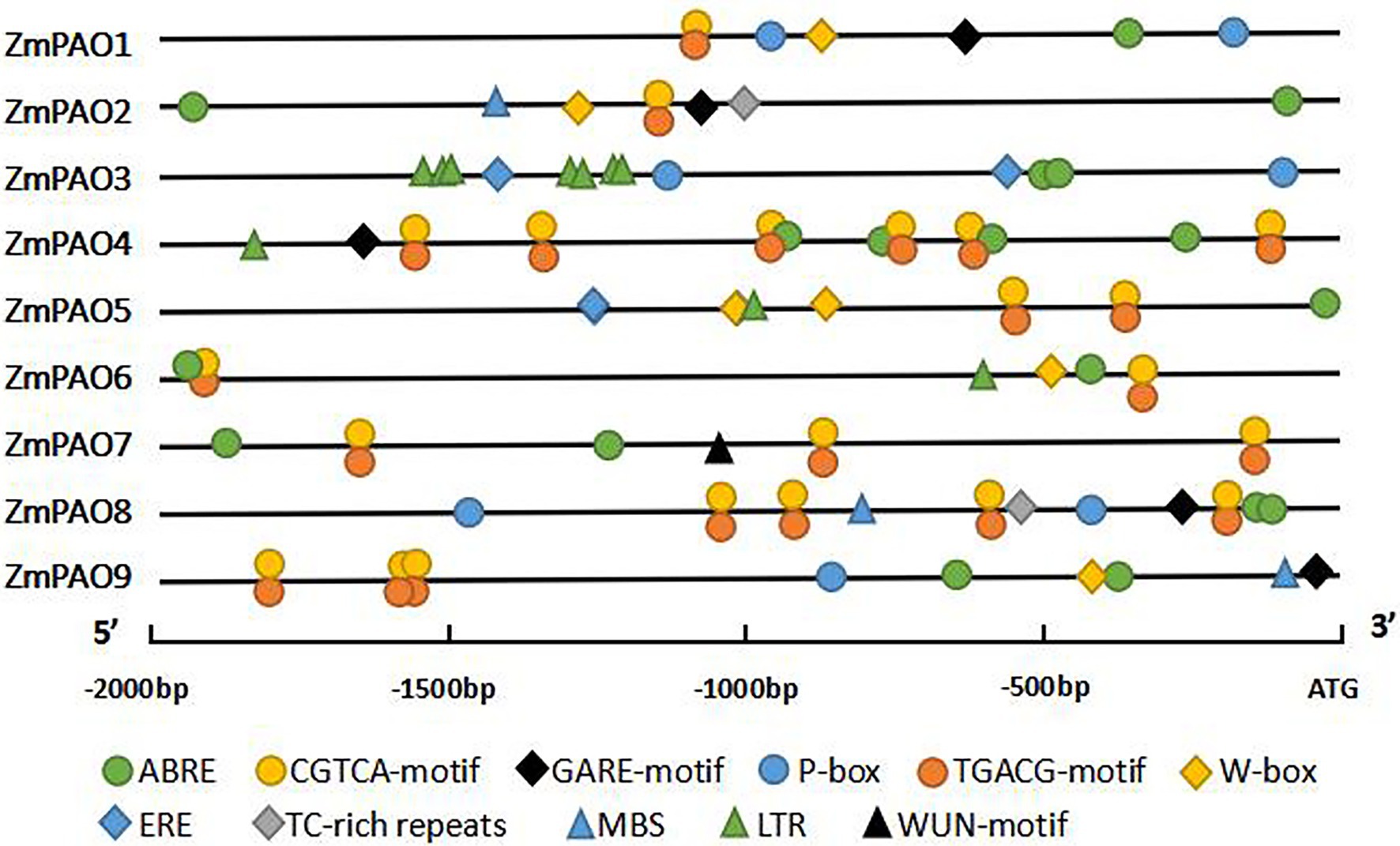
Figure 5. Distribution of major stress-related cis-elements in the promoter sequences of the 9 ZmPAO genes. The putative core sequences of these cis-acting elements are represented by different symbols, as shown in the figure key at the bottom. The cis elements distributed on the sense chain and the reverse chain are above and below the black lines, respectively. The 2,000 bp sequences upstream of the initiation codon (ATG) of the PAO genes can be estimated using the scale per 500 bp at the bottom. ABRE, ABA responsive element; ERE, ethylene-responsive element; GARE-motif and P-box, gibberellin responsive element; CGTCA-motif and TGACG-motif, MeJA-responsive element; LTR, low temperature-responsive element; TC-rich repeats, defense and stress-responsive element; WUN-motif, wound-responsive element; MBS, MYB binding site. W-box, wound and pathogen responsive element.
Analysis of Microarray Expression Profile of Maize PAO Genes
To further investigate the roles of maize PAO genes in plant growth and development, the expression profiles analysis was accomplished using the available transcriptomic data of maize B73 (Sekhon et al., 2011). The signal values for all these maize PAO genes are given in Supplementary Table S2. The expression heat map of all the ZmPAO genes from ten developmental stages was constructed using Helm software (Figure 6). The results revealed that all nine ZmPAO genes were expressed at varying levels in all tissues. Compared with the other ZmPAO genes examined, the transcripts of the ZmPAO9 gene showed relatively high expression levels at all stages, while the transcripts of the ZmPAO5 gene maintained low expression levels in all tissues. Interestingly, some tissue-specific genes were discovered, such as high expression levels of the ZmPAO1 gene in seedlings and high expression levels of the ZmPAO6 gene in roots and stems, suggesting that those genes may play a vital role in plant growth and development processes of specific tissues. In general, the expression patterns in the same paralogous gene pairs (ZmPAO3/ZmPAO9, ZmPAO4/ZmPAO6) were similar, indicating they might be formed by segmental duplication and retain their functions. It is noteworthy that the expression patterns of four ZmPAO genes (ZmPAO2, ZmPAO5, ZmPAO8, and ZmPAO9) in all tissues were relatively stable, implying that these genes might be involved in the basic metabolism. Furthermore, the hierarchically clustered heat map could be divided into two clusters. The first cluster included six members (ZmPAO1, ZmPAO2, ZmPAO4, ZmPAO6, ZmPAO8, and ZmPAO9), while the second cluster included three members (ZmPAO3, ZmPAO5, and ZmPAO7). Notably, the changing trend of gene expression levels of the members of the same cluster was similar in the ten tissues. Overall, the results demonstrated that these identified ZmPAO genes showed differential expression patterns at diverse developmental stages of maize, suggesting that these genes may function in multiple tissues.
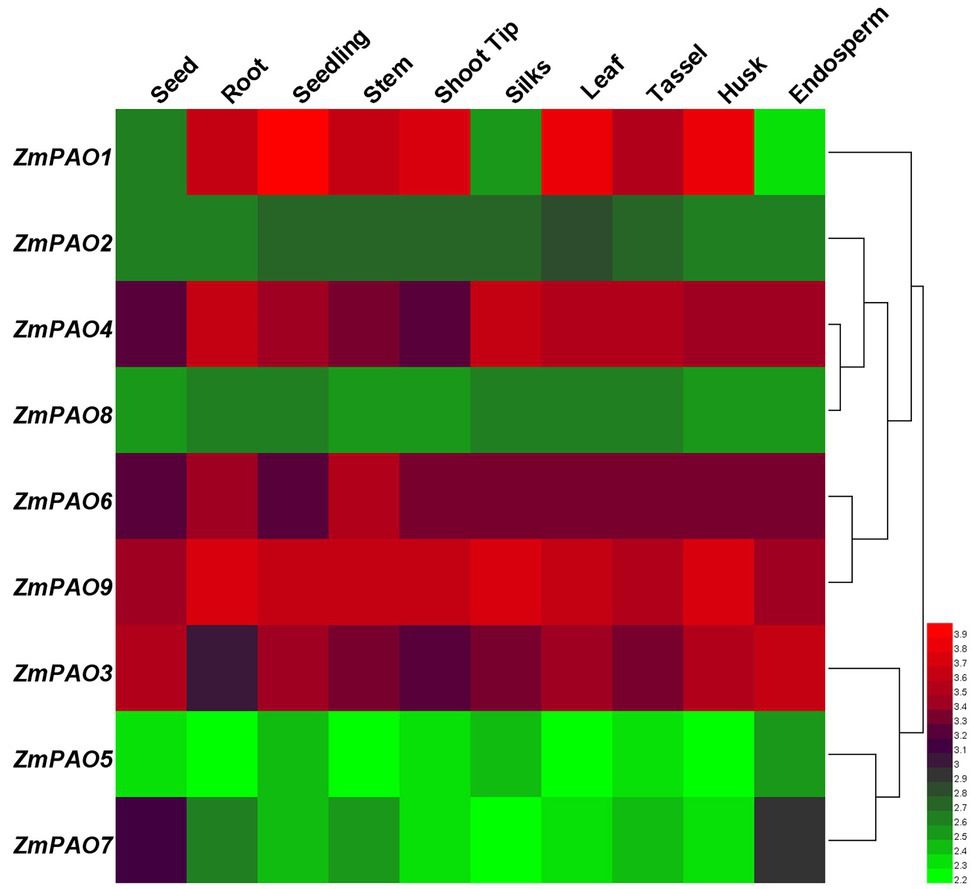
Figure 6. Hierarchical clustering of expression profiles of ZmPAO gene family in ten tissues. Ten tissues from different developmental stages including seed, root, seedling, stem, shoot tip, silks, leaf, tassel, husk, and endosperm were investigated. The different tissues are marked at the top of each channel. Cluster dendrograms are displayed on the right. The color scale representing the value of the log 2 signal is shown on the right. Green represents low expression levels and red indicates high expression levels of the transcript abundance.
Expression Profile Analyses of Maize PAO Genes Under Abiotic Stress Treatment
To survey if these predicted genes were expressed in maize and to further confirm their stress-responsiveness to abiotic stresses, the expression profiles of the nine ZmPAO genes in roots, stems, and leaves under heat, salt, and drought treatments were determined by qRT-PCR (Figure 7). The results indicated that the expression levels of the nine ZmPAO genes were different in three distinct tissues under varied abiotic stress conditions. The expression levels of most ZmPAO genes changed significantly before and after abiotic stress treatments, which indicated that these genes were responsive to these stress treatments and could be involved in distinct stress response pathways. Firstly, the detection of gene expression levels at different time points after heat stress revealed that most ZmPAO genes were sensitive to heat stress (Figure 7A). The gene expression levels of ZmPAO3, ZmPAO4, and ZmPAO6 changed significantly in all tissues after heat stress, which indicated that they could be involved in heat stress. Notably, the expression of ZmPAO1 was down-regulated at all time points in other tissues except that it was up-regulated at 8 h after heat stress in the roots. Moreover, most members exhibited low expression levels in the stems (Figure 7B). Secondly, under the treatment of 200 mM NaCl concentration, the expression level of ZmPAO gene changed significantly at different time points (Figure 8). In the leaves (Figure 8A), the expression levels of ZmPAO2, ZmPAO3, ZmPAO4, ZmPAO7, and ZmPAO9 genes were significantly down-regulated at all time points. In the stems (Figure 8B), the expression levels of ZmPAO1 and ZmPAO5 genes under salt treatment were both down-regulated compared with the control group. In the roots (Figure 8C), the expression levels of 6 genes (ZmPAO2-ZmPAO6 and ZmPAO9) were significantly up-regulated in the early stage of salt stress. Moreover, the expression level of ZmPAO1 gene was inhibited during the whole salt stress period, and only the expression level of ZmPAO6 gene was higher than the control group during the salt stress period. Finally, under drought treatment, the expression changes of ZmPAO genes at different times were analyzed (Figure 9). In the leaves (Figure 9A), the expression levels of ZmPAO2, ZmPAO3, ZmPAO4, ZmPAO6, ZmPAO7, and ZmPAO9 genes decreased as a whole, among which the expression levels of ZmPAO3 and ZmPAO4 genes decreased the most. In the stems (Figure 9B), the expression levels of most ZmPAO genes were decreased under drought treatment compared with the control group. In the roots (Figure 9C), the expression levels of ZmPAO3 and ZmPAO8 genes were similar, which were slowly up-regulated at the beginning of drought treatment, reached the maximum at 2 h, and decreased at 4 h. Taken together, the results demonstrated that most of these predicted genes of the PAO gene family in maize were responsive to various abiotic stress treatments, suggesting their potential roles in abiotic stress responses.
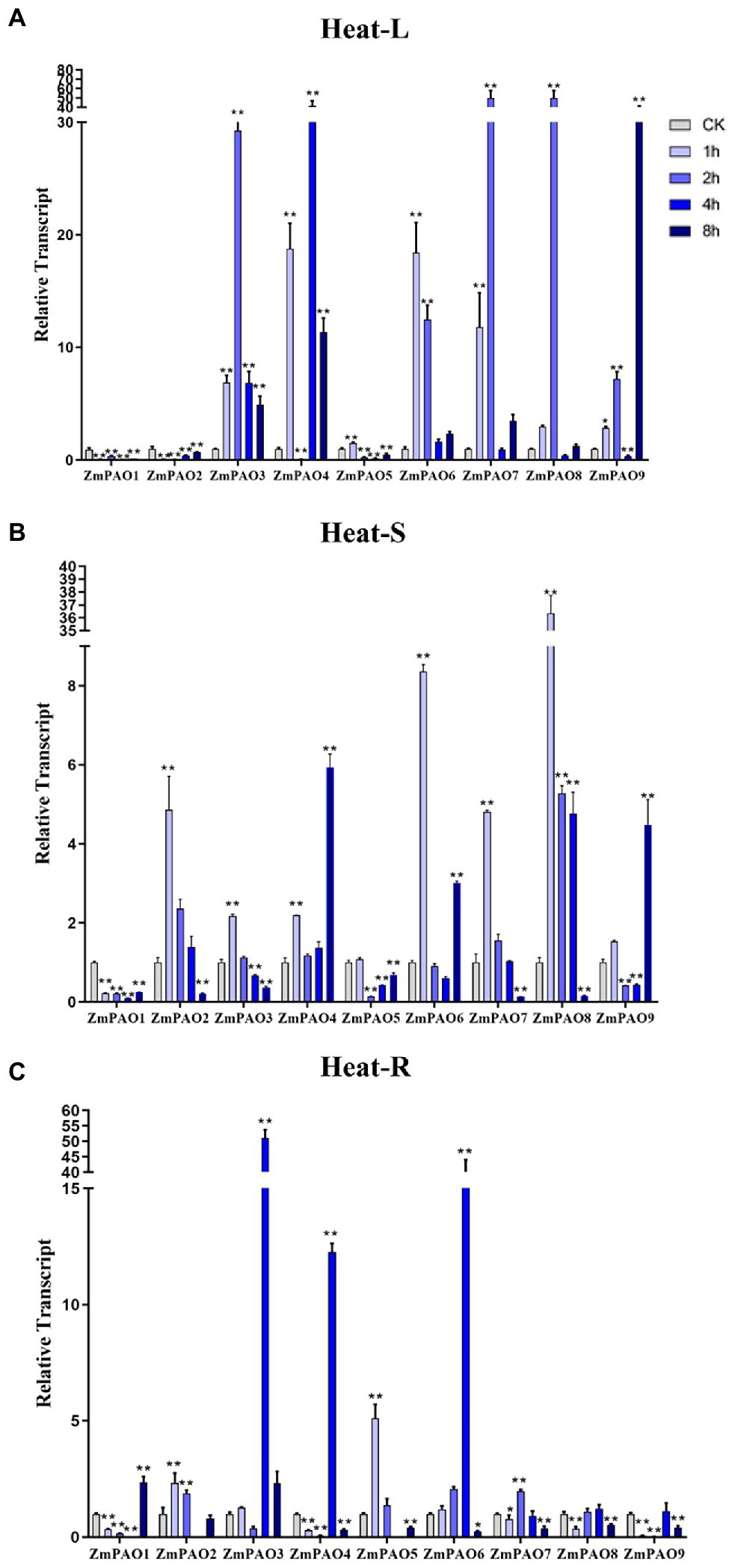
Figure 7. Expression profiles of ZmPAO genes under heat stress treatment in different tissues. qRT-PCR data was normalized using ZmActin gene. (A–C) Expression profiles of ZmPAO genes under heat treatment (42°C) in leaves, stems, and roots, respectively. Error bars are caused by three biological replicates. Asterisks above the error bars represent statistical significance using one-way ANOVA and a Fisher’s least significant difference (LSD). * Significantly different at p < 0.05; ** significantly different at p < 0.01.
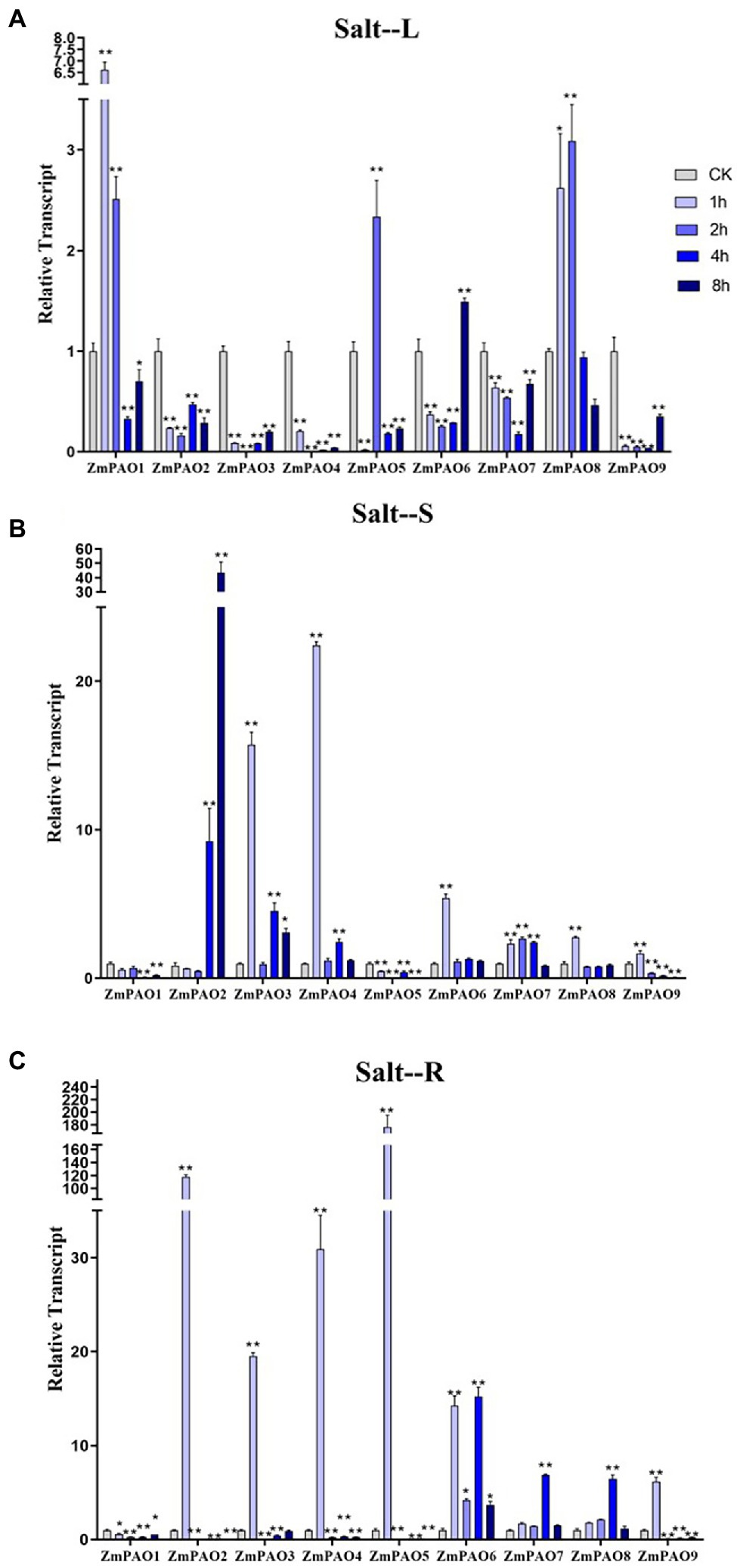
Figure 8. Expression profiles of ZmPAO genes under salt stress treatment in different tissues. qRT-PCR data was normalized using ZmActin gene. (A–C) Expression profiles of ZmPAO genes under salt treatment with 200 mM NaCl in leaves, stems, and roots, respectively. Error bars are caused by three biological replicates. Asterisks above the error bars represent statistical significance using one-way ANOVA and a Fisher’s LSD. * Significantly different at p < 0.05; ** significantly different at p < 0.01.
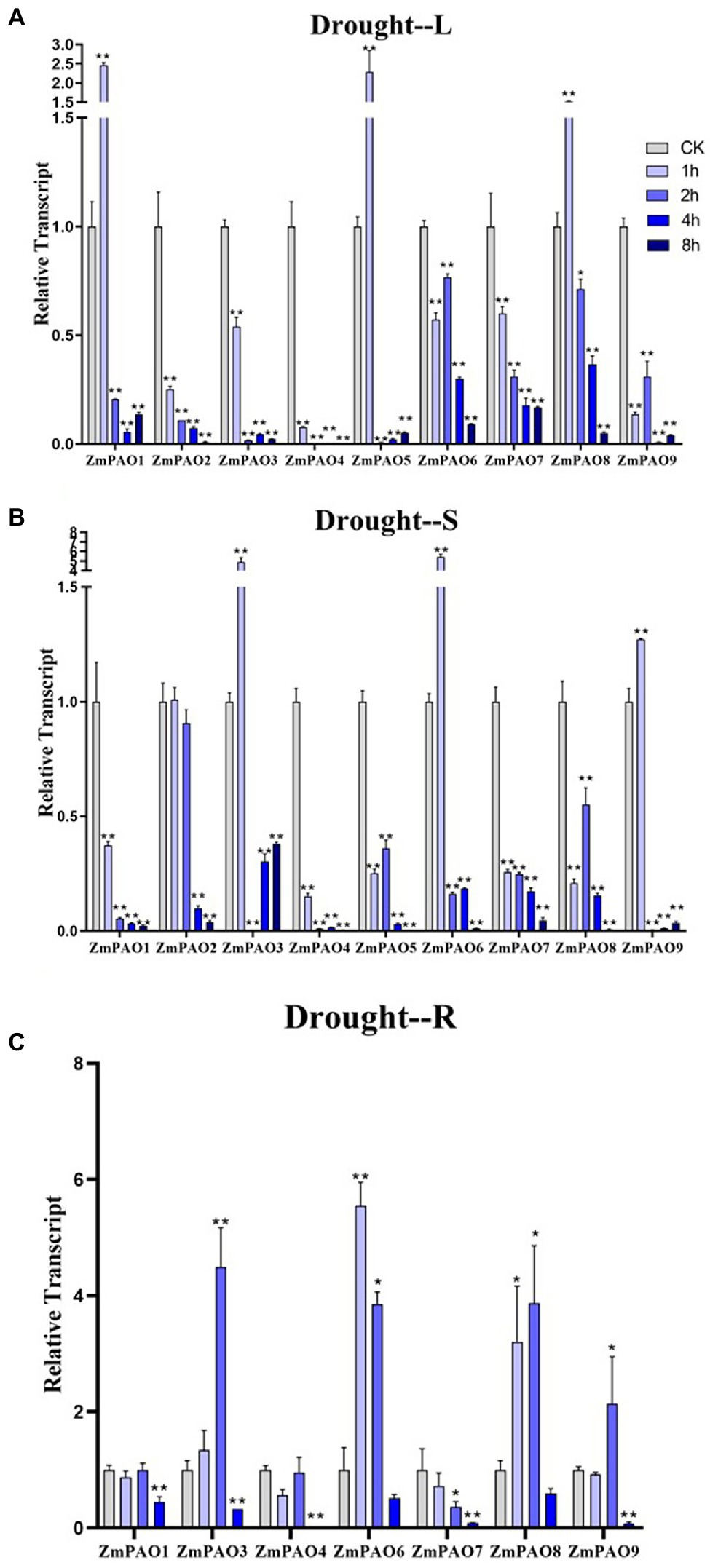
Figure 9. Expression profiles of ZmPAO genes under drought stress treatment in different tissues. qRT-PCR data was normalized using ZmActin gene. (A–C) Expression profiles of ZmPAO genes under drought stress in leaves, stems, and roots, respectively. Error bars are caused by three biological replicates. Asterisks above the error bars represent statistical significance using one-way ANOVA and a Fisher’s LSD. * Significantly different at p < 0.05; ** significantly different at p < 0.01.
Genetic Transformation and Overexpression Analysis of ZmPAO6 Gene in Arabidopsis
The ZmPAO6 clone map and the double enzyme (BamH I and Xba I) digestion map of the recombinant plasmid were shown in Supplementary Figures 1A,B, respectively. To investigate the role of ZmPAO6 gene under heat stress, the recombinant vector with the ZmPAO6 gene was transformed into the Arabidopsis line using Agrobacterium-mediated method by dipping the Arabidopsis floral to obtain the overexpressed Arabidopsis plants. Consequently, a total of 14 transgenic lines were generated in this study (Supplementary Figure 2). In addition, according to the analysis of the phylogenetic tree, the ZmPAO6 gene is located in the Group I, and its orthologous genes in Arabidopsis (AtPAO2 and AtPAO3) and rice (OsPAO3) are both confirmed to be located in peroxisomes and participate in the reverse conversion pathway of polyamine catabolism. Thus, the amino acid sequence of ZmPAO6 gene and these three orthologous PAO genes were compared, and the results elaborated that the sequence homology of both ZmPAO6 and OsPAO3, AtPAO2, or AtPAO3 are 88.28, 69.29, and 68.28%, respectively (Supplementary Figure 3). Peroxisome targeting sequence (PSTI) SRL was also presented at the end of ZmPAO6 protein. The function of ZmPAO6 gene may resemble the orthologous OsPAO3 gene, which encompasses high homology, locates in subcellular peroxisome and participates in the reverse conversion pathway of decomposing PAs. Thus, this result indicated that the ZmPAO6 gene may function in growth, development and adversity stress in maize.
Detection of Polyamine and Hydrogen Peroxide Contents in Transgenic Plants
To further confirm the function of ZmPAO6 gene in transgenic Arabidopsis, two representative transgenic lines (#3 and #14) were selected and identified for further functional analysis (Figure 10A). According to the metabolic pathways and products of PAs, PAs can be decomposed by polyamine oxidase to produce H2O2 (Moschou et al., 2012). The leaves of the WT Arabidopsis and transgenic Arabidopsis lines were firstly stained by DAB, and the H2O2 content was detected as shown in Figure 10B. The leaves of transgenic lines # 3 and # 14 were stained in a darker color and larger area compared with the WT Arabidopsis, indicating that the content of H2O2was higher in transgenic plants, which may be due to the overexpression of ZmPAO6 in Arabidopsis may promote the catabolism of PAs. Therefore, these results suggested that ZmPAO6 gene may be involved in the catabolism of plant polyamines. To confirm the effect of overexpressed ZmPAO6 gene on its polyamine catabolism in transgenic Arabidopsis plants, the content changes of PAs in the WT and transgenic lines with the overexpression of ZmPAO6 gene were determined by high-performance liquid chromatography, respectively (Figure 10C). The contents of Put and Spd in the transgenic lines were higher than those of the WT Arabidopsis, especially the content of Spd. Meanwhile, compared with the WT Arabidopsis, the content of Spm in the transgenic lines were reduced and the line #14 had the lowest content of Spm. The results further revealed that the overexpressed ZmPAO6 gene could be involved in decomposing Spm into Spd and further converting Spd into Put through catalyzing the back-conversion reactions in the catabolism of polyamines in the transgenic Arabidopsis plants. Therefore, these results suggested that the ZmPAO6 gene may be implicated in promoting specific PA degradation mainly via participating in back-conversion reaction of PAs.
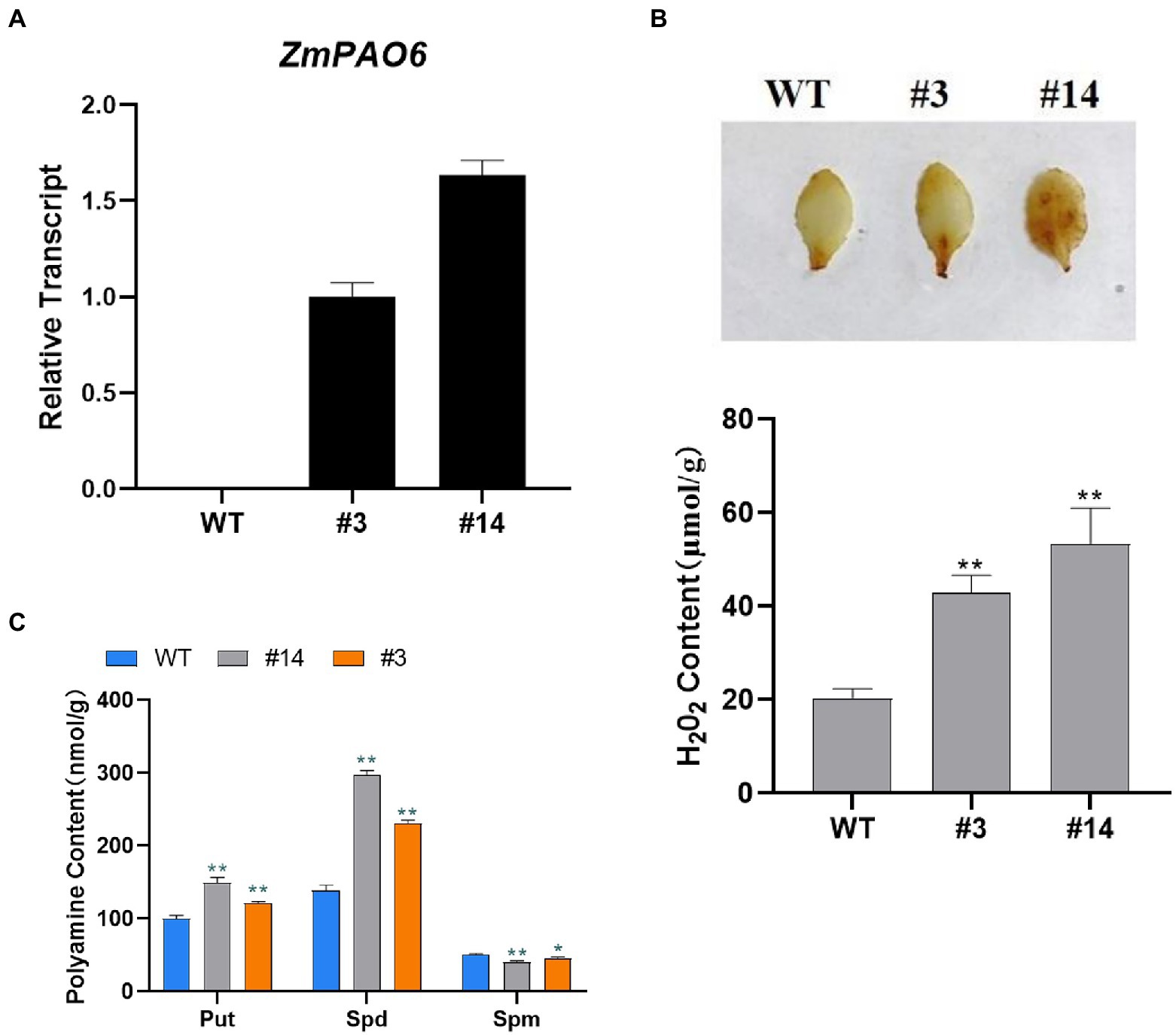
Figure 10. Overexpression of ZmPAO6 gene leads to changes in characteristics of transgenic Arabidopsis. (A) Molecular characterization of transgenic lines by qRT-PCR analysis. (B) H2O2 contents in WT and transgenic lines. (C) PAs contents in WT Arabidopsis and transgenic lines detected by HPLC. Error bars are caused by three biological replicates. Asterisks above the error bars represent statistical significance using one-way ANOVA and a Fisher’s LSD. * Significantly different at p < 0.05; ** significantly different at p < 0.01.
Overexpression of ZmPAO6 Gene Enhances Heat Tolerance in Transgenic Arabidopsis
To detect the tolerance of transgenic Arabidopsis in response to heat stress, 3-week-old seedlings were chosen and treated for 36 h at 42°C. From the phenotypic observation, the leaves of WT Arabidopsis showed obvious withering and curling compared with those of the transgenic Arabidopsis lines (Figures 11A,B). However, the leaves of the line #14 were slightly curled, and the leaves of the line #3 were not significantly damaged. The results indicated that the damage of transgenic plants under heat stress was lower than that of the WT Arabidopsis. The expression level changes of ZmPAO6 gene in the lines # 3 and # 14 before and after heat stress treatment were determined by qRT-PCR (Figure 11C). The results revealed that the expression levels of ZmPAO6 gene in the lines # 3 and # 14 were up-regulated significantly after heat stress, indicating that the overexpression of ZmPAO6 gene could enhance thermotolerance of transgenic Arabidopsis plants.
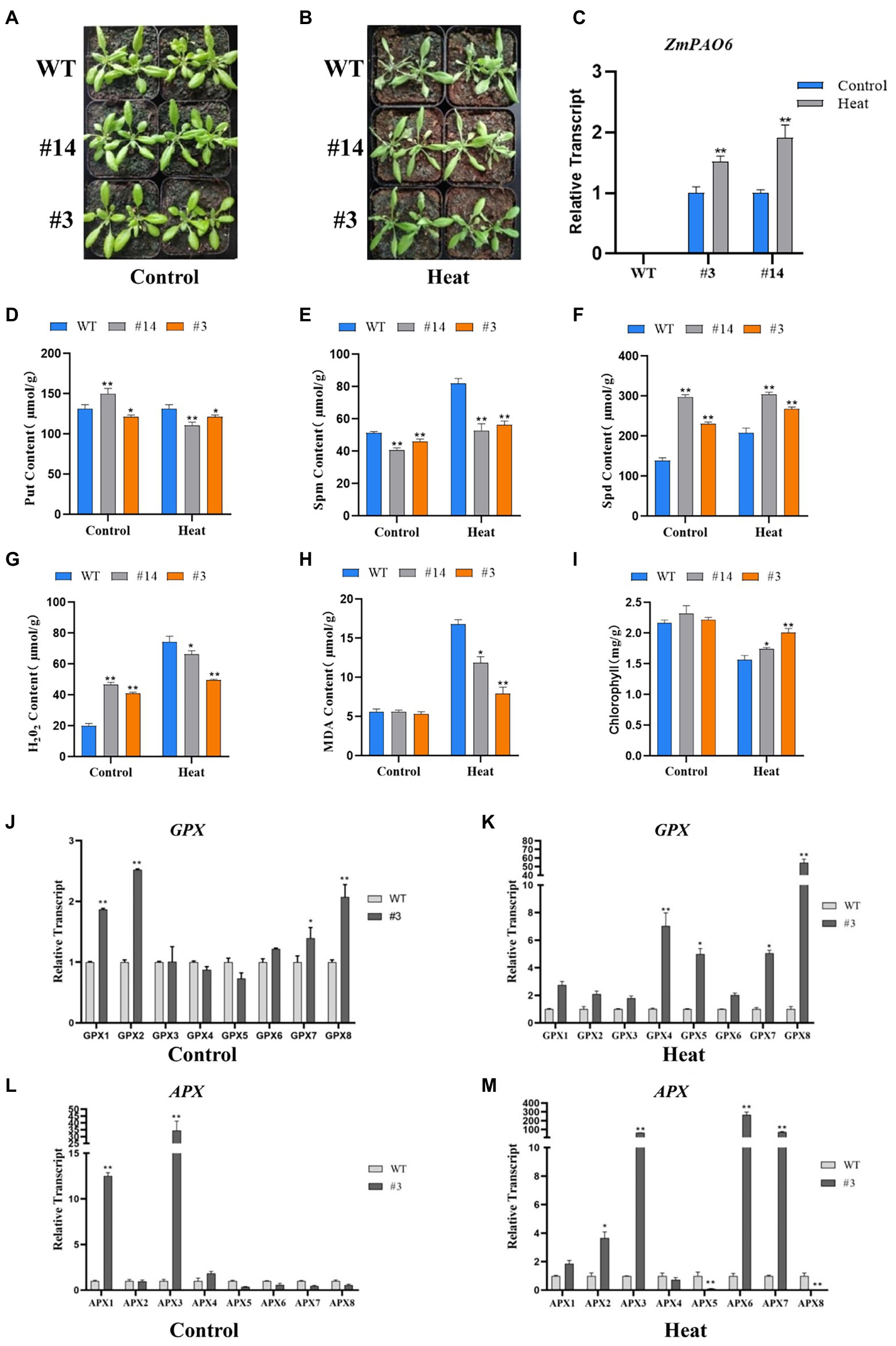
Figure 11. Overexpression of ZmPAO6 gene enhances heat tolerance in Arabidopsis. (A,B) The growth status of WT Arabidopsis and transgenic lines during heat stress treatment. (C) Expression profiles of ZmPAO6 gene in WT Arabidopsis and the transgenic lines under control and heat stress treatment. (D–F) Analysis of Put, Spm and Spd contents in WT Arabidopsis and the transgenic lines under control and heat stress treatment. (G–I) The change of H2O2, MDA, and Chlorophyll contents in seedlings leaves. (J–M) The expression levels of AtGPX1-8 and AtAPX1-8 under control and heat stress treatment. Error bars are caused by three biological replicates. Asterisks above the error bars represent statistical significance using one-way ANOVA and a Fisher’s LSD. * Significantly different at p < 0.05; ** significantly different at p < 0.01.
After heat stress treatment, the PA contents of the WT and transgenic plants were further determined (Figures 11D–F). Compared with the control, the Put content of transgenic plants decreased slightly or remained unchanged, while the Put content of WT plants increased, which were slightly higher than those of transgenic plants (Figure 11D). The Spm content of the WT and transgenic Arabidopsis increased, but the increasing of Spm content in WT Arabidopsis was higher than that in transgenic Arabidopsis plants (the lines # 3 and # 14; Figure 11E). In addition, the Spd content of all plants increased after heat treatment. Notably, the Spd content of the WT Arabidopsis were much lower than those of the transgenic plants (Figure 11F). This result further indicated that ZmPAO6 gene is likely to mainly participate in back-conversion reaction of PAs. Furthermore, to explore molecular mechanism of heat tolerance of transgenic Arabidopsis in response to heat stress, the analyses of H2O2, MDA and chlorophyll contents were further determined according to the physiological methods (Figures 11G–I). The results demonstrated that the contents of H2O2 in the WT and transgenic plants increased under heat stress treatment compared with those under the control condition. However, the content of H2O2 in the WT Arabidopsis was obviously higher than those in the transgenic Arabidopsis lines. Therefore, the ZmPAO6 gene could be involved in modulating heat-induced H2O2 accumulation through the catabolism pathways of polyamines in transgenic Arabidopsis plants to maintain the dynamic balance of H2O2 in the cell of plants and thereby enhancing thermotolerance of plants. In addition, the MDA contents both in WT Arabidopsis and in transgenic plants also increased after heat stress treatment, but the MDA content of WT Arabidopsis was higher than those of the transgenic Arabidopsis lines, indicating that the cells in the WT Arabidopsis were damaged more deeply than those in the transgenic Arabidopsis plants. Notably, the chlorophyll contents of the WT and transgenic Arabidopsis lines decreased after heat stress treatment. However, the chlorophyll contents of the transgenic Arabidopsis lines were higher than those of the WT Arabidopsis, suggesting that an enhanced heat resistance could occur in the transgenic Arabidopsis lines compared with in the WT Arabidopsis.
To further confirm the role of the ZmPAO6 gene in regulating the dynamic balance of H2O2 in the transgenic Arabidopsis plants, the expression levels of some antioxidase genes (AtGPX1-8 and AtAPX1-8) published in the previous studies after heat stress treatment were further determined in this study (Figures 11J–M). Under the control condition, the expression levels of most of AtGPX genes in the transgenic Arabidopsis plant (line #3) were higher than that of the WT Arabidopsis. However, under heat stress treatment, the expression levels of all AtGPX genes in the transgenic Arabidopsis plant (line #3) were significantly enhanced. Notably, the expression level of AtGPX8 gene in the transgenic Arabidopsis plant was higher at more than 60 times than that in the WT Arabidopsis. Likewise, in the control group, the expression levels of only three genes (AtAPX1, AtAPX3, and AtAPX4) in the transgenic Arabidopsis plant were significantly higher than those in the WT Arabidopsis, while the expression levels of the other AtAPX genes in the transgenic Arabidopsis plant were the same as or lower than those in the WT Arabidopsis. However, after heat stress treatment, the expression levels of some genes such as AtAPX3, AtAPX6, and AtAPX7 were significantly up-regulated in the transgenic Arabidopsis plant compared with those in the WT Arabidopsis, indicating that the expression levels of AtAPX3, AtAPX6, and AtAPX7 genes can be induced intensively in the transgenic Arabidopsis plants in response to heat stress to further enhance the scavenging ability of hydrogen peroxide in cells of plants. Taken together, these results demonstrated that the ZmPAO6 gene can enhance heat resistance in transgenic Arabidopsis through modulating heat-induced H2O2 accumulation in polyamine catabolism.
Discussion
In plants, PAs play a vital role in regulating the growth and development and protecting the plants from adverse environment including biotic and abiotic stresses. However, the catabolism of PAs in maize is not well understood. The purpose of this study is to screen key ZmPAO genes that are more sensitive to abiotic stresses and to lay a foundation for further revealing the regulatory mechanism of PAO genes in maize. Meanwhile, the possible mechanism of ZmPAO6 gene conferring the thermotolerance in the transgenic Arabidopsis plants in response to heat stress was also determined in this study.
Characterization of PAO Genes in Maize and Evolution of ZmPAO Proteins
An increasing number of studies have been devoted to elucidating the function of plant PAO genes in growth and stress responses. Until now, 5, 6, and 7 PAO genes have been identified in Arabidopsis, sweet orange, and rice, respectively (Fincato et al., 2011; Ono et al., 2012; Wang and Liu, 2015). In this study, a total of nine ZmPAO genes were firstly identified from the latest maize B73 genome database. The number of PAO genes in maize was similar to that in rice. Phylogenetic analysis demonstrated that the plant PAO genes were generally classified into four clades, whereas the members of maize were mainly divided into the Groups I, II, and IV but not into the Group III. Notably, the PAO genes in maize were closer to that in rice in phylogenetic relationship compared with those in the other species, indicating that the evolution of the PAO genes may occur after divergence of monocots and dicots. Furthermore, it was revealed that the result of multiple sequence alignment was consistent with that of phylogenetic relationship analysis and that among the members of the same group of PAO proteins in these plants higher identities were shared for each other, indicating that the plant PAO proteins might be highly conversed in evolution. Interestingly, AtPAO2-4 and OsPAO3-5 were localized in peroxisomes based on possessing (S/A/C)(K/R/H)(L/M) in their C-termini (Fincato et al., 2011; Ono et al., 2012). In this study, presence of SRL sequence in the C-termini of maize ZmPAO4, ZmPAO6, and ZmPAO9 and presence of CRL sequence in the C-termini of maize ZmPAO3 suggest that in the Group I, all the PAO proteins from rice, Arabidopsis and maize carry peroxisome targeting signals at their C-termini. These data suggested that these ZmPAO proteins in the Group I may be localized in peroxisomes. Notably, the ZmPAO2 gene had no introns in gene structure, suggesting that ZmPAO2 likely has different functions than the other ZmPAO genes. In addition, the analysis of the conserved domain of ZmPAO proteins was performed to confirm that all nine ZmPAO proteins contained amine-oxidase domains, indicating that these proteins were highly conserved. The analysis of gene structures and conserved motifs of ZmPAO proteins demonstrated that the members in the same subfamily have similar structures and even the same or similar motifs. The analysis of gene duplication events of ZmPAO genes further elaborated that two pairs of genes (ZmPAO3/ZmPAO4 and ZmPAO6/ZmPAO9) could be resulted from segmental duplication during their process of evolution, indicating that segmental duplication events might play a vital role in the evolution of PAO gene family in maize.
Expression Profiles Analysis of ZmPAO Genes in Response to Abiotic Stresses
The dynamic balance of intracellular polyamines was attributed to the regulation of PA biosynthesis and catabolism. Previous studies have shown that the dynamic balance of PAs could be challenged under adverse environmental conditions. For instance, the excessive accumulation of PAs caused by various abiotic stresses requires more PAO proteins to rebalance the dynamic balance of polyamines (Wang et al., 2011; Minocha et al., 2014). It was reported that a spaceflight induced wheat mutant, named salinity tolerance 1 (st1), is a salinity-tolerant line. In the salt-treated wheat mutant st1, genes encoding polyamine oxidase showed higher expression compared with the salt-treated WT, indicating that these genes may play a crucial role in salinity tolerance in st1 mutant (Xiong et al., 2017). Moreover, previous studies have reported that polyamine oxidase encoding extracellular enzyme in rice may play a key role in the protection of plant cells (Sagor et al., 2021). Therefore, it is necessary to analyze the expression patterns of ZmPAO genes under various abiotic stresses. This study demonstrated that the expression levels of most ZmPAO genes were up-regulated under heat and salt treatments. In addition, except for the up-regulation of expression levels of ZmPAO1, ZmPAO5, and ZmPAO8 genes in the early stage of stress, almost all ZmPAO genes in stems and leaves were down-regulated under drought treatment. These results suggested that maize PAO genes could be involved in various abiotic stress responses.
Ectopic Overexpression of ZmPAO6 Enhances Heat Tolerance in Transgenic Arabidopsis
To adequately comprehend the role of PAO genes in plant development and abiotic stresses, it is crucial to conduct in-depth research by overexpressed transgenic plants, which will be helpful in further exploring the biochemical and physiological roles of these PAO genes in maize. PAOs have been proved to be the pivotal enzymes in regulating the levels of intracellular PAs, and the dynamic balance of PAs is essential for plant development (Handa and Mattoo, 2010; Tiburcio et al., 2014). The stress resistance of plants mainly depends on stress-responsive genes, and the overexpression of these genes could improve the adaptability of plants to various environmental stresses (Alcázar et al., 2006). According to the phylogenetic analysis and expression patterns of ZmPAO genes under various abiotic stresses in this study, the ZmPAO6 gene was selected for performing the preliminary functional verification experiments under heat stress treatment to further reveal if this newly-identified gene could function in improving heat tolerance in transgenic Arabidopsis plants. Thus, the ZmPAO6 gene was overexpressed in Arabidopsis by genetic transformation. Under control condition, overexpression of ZmPAO6 resulted in an increased production of H2O2. Similarly, a previous study showed that overexpression of AtPAO3 resulted to an increased production of H2O2 in Arabidopsis (Andronis et al., 2014). Moreover, ectopic overexpression of AtPAO5 enhanced the conversion of lateral root primordia into shoots and influenced the PA homeostasis during this period, and resulted in elevated ROS level in the lateral root primordia of Arabidopsis (Kaszler et al., 2021). Therefore, overexpression of ZmPAO6 in transgenic Arabidopsis may also share similar functional effects in this study. Moreover, high temperature is a vital environmental factor that affects plant growth and crop yield (Iba, 2002). In this study, compared with the WT Arabidopsis, the leaves of transgenic Arabidopsis plants maintained a better green and stretched state. After heat stress treatment, the phenotype and physiological indicators of the WT and transgenic Arabidopsis plants exhibited apparent different changes. Further studies demonstrated that the transgenic Arabidopsis plants had better growth vitality under heat stress treatment, indicating that the overexpression of ZmPAO6 gene in the transgenic Arabidopsis conferred its tolerance to heat stress. The enhancement of adaptability of these transgenic plants in response to heat stress was required for the synchronous adaptation of external morphological and biochemical levels.
In previous studies, the plant protection depended on increased levels of H2O2 produced by Spm oxidation, implying that the catabolism of PAs mainly including Spm catalyzed by PAOs has been considered to be a crucial component of the plant defense response (Gonzalez et al., 2011). In this study, under normal and heat stress conditions, Arabidopsis plants overexpressing the ZmPAO6 gene showed lower Spm contents compared with WT Arabidopsis, indicating that ZmPAO6 may be involved in the back-conversion reactions of PAs, which is mainly involved in the oxidation of Spm to Spd. Accordingly, the Spd content of transgenic Arabidopsis was higher than those of WT Arabidopsis. Therefore, the results in this study suggested that plant-specific PA degradation (mainly Spm) may play a vital role in plant defense. Furthermore, MDA is used as a widely marker of lipid oxidation damage at the physiological level of plants, which reflects the level of membrane lipid peroxidation in plants to measure the degree of oxidative stress (Davey et al., 2005). In this study, less accumulation of MDA was detected in the ZmPAO6-overexpressed transgenic Arabidopsis lines compared with that in the WT Arabidopsis, which indicated that the overexpression of ZmPAO6 gene could maintain the stability of lipid membrane structure under heat stress treatment.
In addition, the plants are likely to regulate stress resistance by complex signal regulatory pathways under various abiotic stresses. Among them, one of the consequences of abiotic stresses is to increase the concentration of intracellular ROS (Apel and Hirt, 2004). H2O2 was initially considered to be a toxic reactive oxygen species because of its damage to a variety of cell structures. In addition, H2O2 complements, cooperates or antagonizes many cellular regulatory circuits through positive interactions with other signals and plant hormones in growth, development and stress responses (Quan et al., 2008; Petrov and Van Breusegem, 2012). H2O2 is a vital molecule involved in signal transduction and plant defense responses, which is constantly generated from a variety of sources, including PA catabolism (Moschou et al., 2008a; Cheng et al., 2015; Jasso-Robles et al., 2020). Therefore, before the experiment, we hypothesized that the H2O2 content of transgenic Arabidopsis was higher than that of WT Arabidopsis under heat stress, and this could in turn affect the heat resistance of transgenic Arabidopsis. However, in this study, contrary to our expectations, H2O2 contents in transgenic Arabidopsis lines were significantly lower than that in WT Arabidopsis under heat stress treatment. A previous study showed that ABA enhanced the accumulation of PA in grape and induced the PA oxidation pathway (Toumi et al., 2010). The accumulated PAs was mainly oxidized in the apoplast to produce H2O2 which integrated the signal pathway. The PAO generated apoplastic H2O2 resulting from osmotic stress acted as a secondary messenger in the signaling pathway and/or induces PCD in grape (Toumi et al., 2010). Under normal and stress conditions, maintaining the appropriate balance of PA catabolic pathways with the H2O2 dual action is helpful to clarify the adaptation mechanisms of plants (Yoda et al., 2009; Andronis et al., 2014). Therefore, we have reason to speculate that the overexpression of ZmPAO6 gene may maintain the balance between PA catabolism and H2O2 in transgenic Arabidopsis under heat stress treatment. Taking together, although the H2O2 levels of WT and transgenic Arabidopsis plants increased significantly under heat stress treatment, the H2O2 contents in the transgenic plants were lower than those of the WT Arabidopsis, indicating that overexpression of ZmPAO6 gene can result in the reduced accumulation of heat-induced H2O2 content, which suggested that maize PAO proteins might be crucial for modulating the dynamic balance of H2O2 in transgenic Arabidopsis.
To further explore the underlying molecular mechanism of H2O2 pathways involved in heat stress, the expression levels of glutathione peroxidase (GPX) and ascorbate peroxidase (APX) genes were investigated in this study, which belong to the main antioxidant enzymes for H2O2 scavenging and can catalyze the reduction of H2O2 to prevent the potential cell damage caused by H2O2 (Ozyigit et al., 2016). In this study, the expression levels of antioxidant enzyme genes (AtGPX1-8 and AtAPX1-8) in the WT and transgenic Arabidopsis plants were determined under the control and heat stress treatment conditions, respectively. In this study, after heat stress treatment, the expression levels of most AtGPX and AtAPX genes in transgenic Arabidopsis plants were apparently greater than that of the WT Arabidopsis, especially the AtGPX genes. In addition, it has been revealed that the exogenous H2O2 increased the expression levels of APX genes in tomato seedlings under salinity-alkalinity stress (Xu et al., 2021). Therefore, the increased expression of most AtAPX and AtGPX genes in transgenic Arabidopsis lines may be resulted from the enhanced H2O2 contents exhibited in these lines. We speculate that the overexpressed ZmPAO6 gene in transgenic Arabidopsis produced the corresponding polyamine oxidase to catalyze the decomposition of PA accumulated under heat stress treatment to produce H2O2. Moreover, compared with the WT Arabidopsis, the expression levels of antioxidant enzymes genes in transgenic plants were up-regulated. Studies have shown that all eight APX genes of Arabidopsis participated in the processes of growth and development and abiotic stress response (Li et al., 2019). Recent evidence suggested that GPXs can protect cells from stress-induced oxidative damages and function as a vital component of growth and development in plants (Bela et al., 2015). Moreover, the accumulation of ROS is regulated by a complex antioxidant scavenging system, including GRX, SOD, APX, etc., that function as antioxidants and allow the transmission of oxidative signals (Singh et al., 2022). Therefore, these data suggested that increased the activity of some key antioxidant enzymes and reduced heat-induced H2O2 accumulation, thus enhancing stress resistance of the transgenic plants. In this study, the H2O2 contents of transgenic Arabidopsis plants were lower than those of WT Arabidopsis, perhaps due to the increased activity of AtAPX and AtGPX enzymes, which is likely to scavenge excess H2O2. However, further studies are required for clarifying whether the increased expression levels of these antioxidant enzyme genes control the H2O2 content or whether the H2O2 content controls the induction of these genes or enzymes. Although it may be involved in some complicated metabolic processes, it is worth studying carefully to further understand the regulation of H2O2 homeostasis. In conclusion, alterations in PA catabolism affect the levels of H2O2 and the expression levels of AtAPX and AtGPX genes that enhance the antioxidant defense of the plants, suggesting a link between the ZmPAO6 gene and other enzymes involved in H2O2 homeostasis in transgenic Arabidopsis, and indicating that this gene can enhance heat resistance in transgenic Arabidopsis through modulating heat-induced H2O2 accumulation in polyamine catabolism.
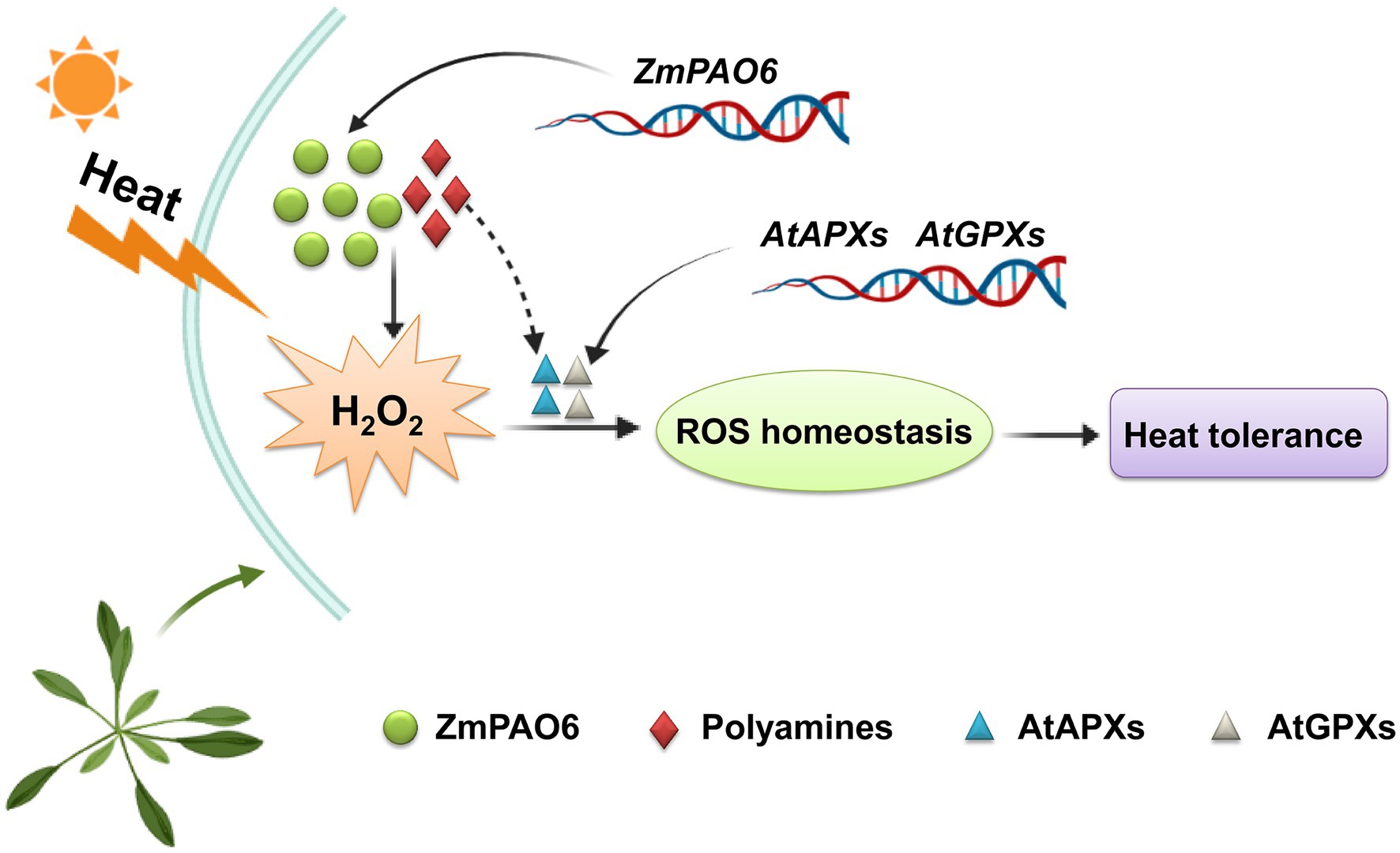
Furthermore, we proposed a model of the ZmPAO6 gene involved in PA catabolism and H2O2 homeostasis in transgenic Arabidopsis (Figure 12). Under heat stress treatment, the expression level of ZmPAO6 gene was up-regulated, and the corresponding activity of polyamine oxidase was also increased, which may promote the decomposition of PAs (mainly Spm) to produce H2O2. In addition, H2O2 may be used as a signal molecule to increase the expression levels of AtAPX and AtGPX genes to accelerate the degradation of H2O2. On the other hand, heat stress causes the accumulation of H2O2 in transgenic Arabidopsis, which may also act on related antioxidant enzyme genes. In addition, Arabidopsis plants overexpressing the ZmPAO6 gene showed increased the expression levels of AtAPX and AtGPX genes, which represented an attempt to scavenge excess H2O2 generated from the oxidation of PAs catalyzed by potential ZmPAO6 protein. However, the exact mechanism is unknown for maintaining ROS homeostasis in regulating the heat stress response in transgenic Arabidopsis. Therefore, the positive regulatory mechanism of ZmPAO6 gene in response to heat stress remains to be further elucidated in future.
Conclusion
In our study, a total of nine PAO genes were identified in maize, which were distributed on five chromosomes at different densities. These PAO proteins were mainly categorized into three distinct subfamilies based on the similarities of the amino acid sequences. The motif composition and exon/intron distribution were considerably conserved among the members of the same group or subgroup. The close phylogenetic relationship among PAO proteins of different species in the same subgroup provides insights into their putative functions. A comprehensive expression profile of all members of PAO gene family in maize suggests that these genes play functional developmental roles in different tissues. Furthermore, the expression profiles of ZmPAO genes were up-regulated or down-regulated under three various abiotic stress treatments, indicating that these PAO genes may play a crucial role in resisting adverse environment of maize. Eventually, the functional analysis of the maize representative ZmPAO6 gene in response to heat stress demonstrated that overexpression of ZmPAO6 in transgenic Arabidopsis can confer enhanced heat tolerance through modulating heat-induced H2O2 accumulation in polyamine catabolism. Taken together, our results are the first to report the ZmPAO6 gene response to heat stress in plants and will serve to present an important theoretical basis for further unraveling the function and regulatory mechanism of ZmPAO genes in growth, development and adaptation to abiotic stresses in maize.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
These methods were carried out in accordance with relevant guidelines and regulations including the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. We confirm that all experimental protocols were approved by Anhui Agriculture University and Anhui Normal University.
Author Contributions
YQ, YX, WH, YZ, and XL designed the studies, carried out all the experimental analyses, and prepared all figures and tables. YX and WH drafted the manuscript. YQ assisted in explaining the results and revised the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (NSFC; grant no. 31571673) and the open fundings of National Engineering Laboratory of Crop Stress Resistance Breeding (grant no. KNZJ1023) and Anhui Provincial Key Laboratory of the Conservation and Exploitation of Biological Resources (grant no. Swzy202003) and Anhui Provincial Academic Funding Project for Top Talents in Disciplines (Majors; grant no. gxbjZD2021044). The funders had no role in the study design, collection, analysis, and interpretation of data, or in the writing of the report or decision to submit the article for publication.
Acknowledgments
The authors are grateful to the editor and reviewers for critically evaluating the manuscript and providing constructive comments for its improvement.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.950064/full#supplementary-material
References
Ahmed, S., Ariyaratne, M., Patel, J., Howard, A. E., Kalinoski, A., Phuntumart, V., et al. (2017). Altered expression of polyamine transporters reveals a role for spermidine in the timing of flowering and other developmental response pathways. Plant Sci. 258, 146–155. doi: 10.1016/j.plantsci.2016.12.002
Alcázar, R., Marco, F., Cuevas, J. C., Patron, M., Ferrando, A., Carrasco, P., et al. (2006). Involvement of polyamines in plant response to abiotic stress. Biotechnol. Lett. 28, 1867–1876. doi: 10.1007/s10529-006-9179-3
Andronis, E. A., Moschou, P. N., Toumi, I., and Roubelakis-Angelakis, K. A. (2014). Peroxisomal polyamine oxidase and NADPH-oxidase cross-talk for ROS homeostasis which affects respiration rate in Arabidopsis thaliana. Front. Plant Sci. 5:132. doi: 10.3389/fpls.2014.00132
Angelini, R., Cona, A., Federico, R., Fincato, P., Tavladoraki, P., and Tisi, A. (2010). Plant amine oxidases "on the move": an update. Plant Physiol. Biochem. 48, 560–564. doi: 10.1016/j.plaphy.2010.02.001
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bela, K., Horváth, E., Gallé, Á., Szabados, L., Tari, I., and Csiszár, J. (2015). Plant glutathione peroxidases: emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 176, 192–201. doi: 10.1016/j.jplph.2014.12.014
Chang, W. C., Lee, T. Y., Huang, H. D., Huang, H. Y., and Pan, R. L. (2008). PlantPAN: Plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genom. 9:561. doi: 10.1186/1471-2164-9-561
Cheng, W. H., Wang, F. L., Cheng, X. Q., Zhu, Q. H., Sun, Y. Q., Zhu, H. G., et al. (2015). Polyamine and its metabolite H2O2 play a key role in the conversion of embryogenic callus into somatic embryos in upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 6:1063. doi: 10.3389/fpls.2015.01063
Corpas, F. J., Del Río, L. A., and Palma, J. M. (2019). Plant peroxisomes at the crossroad of NO and H(2) O(2) metabolism. J. Integr. Plant Biol. 61, 803–816. doi: 10.1111/jipb.12772
da Silva, T. I., Dias, M. G., De Araújo, N. O., De Sousa Santos, M. N., Cruz, R. R. P., Dias, T. J., et al. (2022). Spermine reduces the harmful effects of salt stress in Tropaeolum majus. Physiol. Mol. Biol. Plants 28, 687–696. doi: 10.1007/s12298-022-01165-9
Davey, M. W., Stals, E., Panis, B., Keulemans, J., and Swennen, R. L. (2005). High-throughput determination of malondialdehyde in plant tissues. Anal. Biochem. 347, 201–207. doi: 10.1016/j.ab.2005.09.041
Fincato, P., Moschou, P. N., Spedaletti, V., Tavazza, R., Angelini, R., Federico, R., et al. (2011). Functional diversity inside the Arabidopsis polyamine oxidase gene family. J. Exp. Bot. 62, 1155–1168. doi: 10.1093/jxb/erq341
Fortes, A. M., and Agudelo-Romero, P. (2018). Polyamine metabolism in climacteric and non-climacteric fruit ripening. Methods Mol. Biol. 1694, 433–447. doi: 10.1007/978-1-4939-7398-9_36
Fu, X. Z., Chen, C. W., Wang, Y., Liu, J. H., and Moriguchi, T. (2011). Ectopic expression of MdSPDS1 in sweet orange (Citrus sinensis Osbeck) reduces canker susceptibility: involvement of H₂O₂ production and transcriptional alteration. BMC Plant Biol. 11:55. doi: 10.1186/1471-2229-11-55
Gonzalez, M. E., Marco, F., Minguet, E. G., Carrasco-Sorli, P., Blázquez, M. A., Carbonell, J., et al. (2011). Perturbation of spermine synthase gene expression and transcript profiling provide new insights on the role of the tetraamine spermine in Arabidopsis defense against Pseudomonas viridiflava. Plant Physiol. 156, 2266–2277. doi: 10.1104/pp.110.171413
Handa, A. K., and Mattoo, A. K. (2010). Differential and functional interactions emphasize the multiple roles of polyamines in plants. Plant Physiol. Biochem. 48, 540–546. doi: 10.1016/j.plaphy.2010.02.009
Hao, Y., Huang, B., Jia, D., Mann, T., Jiang, X., Qiu, Y., et al. (2018). Identification of seven polyamine oxidase genes in tomato (Solanum lycopersicum L.) and their expression profiles under physiological and various stress conditions. J. Plant Physiol. 228, 1–11. doi: 10.1016/j.jplph.2018.05.004
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, W., Ren, Q., Chen, Y., Xu, G., and Qian, Y. (2021). Genome-wide identification and analysis of WRKY gene family in maize provide insights into regulatory network in response to abiotic stresses. BMC Plant Biol. 21:427. doi: 10.1186/s12870-021-03206-z
Hussain, S. S., Ali, M., Ahmad, M., and Siddique, K. H. (2011). Polyamines: natural and engineered abiotic and biotic stress tolerance in plants. Biotechnol. Adv. 29, 300–311. doi: 10.1016/j.biotechadv.2011.01.003
Iba, K. (2002). Acclimative response to temperature stress in higher plants: approaches of gene engineering for temperature tolerance. Annu. Rev. Plant Biol. 53, 225–245. doi: 10.1146/annurev.arplant.53.100201.160729
Igarashi, K., and Kashiwagi, K. (2000). Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 271, 559–564. doi: 10.1006/bbrc.2000.2601
Ioannidis, N. E., Malliarakis, D., Torné, J. M., Santos, M., and Kotzabasis, K. (2016). The Over-expression of the plastidial transglutaminase from maize in Arabidopsis increases the activation threshold of photoprotection. Front. Plant Sci. 7:635. doi: 10.3389/fpls.2016.00635
Jasso-Robles, F. I., Gonzalez, M. E., Pieckenstain, F. L., Ramírez-García, J. M., Guerrero-González, M. L., Jiménez-Bremont, J. F., et al. (2020). Decrease of Arabidopsis PAO activity entails increased RBOH activity, ROS content and altered responses to Pseudomonas. Plant Sci. 292:110372. doi: 10.1016/j.plantsci.2019.110372
Kamada-Nobusada, T., Hayashi, M., Fukazawa, M., Sakakibara, H., and Nishimura, M. (2008). A putative peroxisomal polyamine oxidase, AtPAO4, is involved in polyamine catabolism in Arabidopsis thaliana. Plant Cell Physiol. 49, 1272–1282. doi: 10.1093/pcp/pcn114
Kaszler, N., Benkő, P., Bernula, D., Szepesi, Á., Fehér, A., and Gémes, K. (2021). Polyamine metabolism is involved in the direct regeneration of shoots from Arabidopsis lateral root primordia. Plants (Basel) 10:305. doi: 10.3390/plants10020305
Knott, J. M., Römer, P., and Sumper, M. (2007). Putative spermine synthases from Thalassiosira pseudonana and Arabidopsis thaliana synthesize thermospermine rather than spermine. FEBS Lett. 581, 3081–3086. doi: 10.1016/j.febslet.2007.05.074
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kusano, T., Berberich, T., Tateda, C., and Takahashi, Y. (2008). Polyamines: essential factors for growth and survival. Planta 228, 367–381. doi: 10.1007/s00425-008-0772-7
Li, Z. Q., Li, J. T., Bing, J., and Zhang, G. F. (2019). The role analysis of APX gene family in the growth and developmental processes and in response to abiotic stresses in Arabidopsis thaliana. Yi Chuan 41, 534–547. doi: 10.16288/j.yczz.19-026
Li, M., Lu, J., Tao, M., Li, M., Yang, H., Xia, E. H., et al. (2020). Genome-wide identification of seven polyamine oxidase genes in Camellia sinensis (L.) and their expression patterns under various abiotic stresses. Front. Plant Sci. 11:544933. doi: 10.3389/fpls.2020.544933
Liu, J. H., and Moriguchi, T. (2007). Changes in free polyamine titers and expression of polyamine biosynthetic genes during growth of peach in vitro callus. Plant Cell Rep. 26, 125–131. doi: 10.1007/s00299-006-0223-5
Liu, J. H., Wang, W., Wu, H., Gong, X., and Moriguchi, T. (2015). Polyamines function in stress tolerance: from synthesis to regulation. Front. Plant Sci. 6:827. doi: 10.3389/fpls.2015.00827
Ma, S., Zhou, X., Jahan, M. S., Guo, S., Tian, M., Zhou, R., et al. (2022). Putrescine regulates stomatal opening of cucumber leaves under salt stress via the H(2)O(2)-mediated signaling pathway. Plant Physiol. Biochem. 170, 87–97. doi: 10.1016/j.plaphy.2021.11.028
Minocha, R., Majumdar, R., and Minocha, S. C. (2014). Polyamines and abiotic stress in plants: a complex relationship. Front. Plant Sci. 5:175. doi: 10.3389/fpls.2014.00175
Mo, H., Wang, X., Zhang, Y., Zhang, G., Zhang, J., and Ma, Z. (2015). Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae. Plant J. 83, 962–975. doi: 10.1111/tpj.12941
Moschou, P. N., Paschalidis, K. A., Delis, I. D., Andriopoulou, A. H., Lagiotis, G. D., Yakoumakis, D. I., et al. (2008a). Spermidine exodus and oxidation in the apoplast induced by abiotic stress is responsible for H2O2 signatures that direct tolerance responses in tobacco. Plant Cell 20, 1708–1724. doi: 10.1105/tpc.108.059733
Moschou, P. N., Paschalidis, K. A., and Roubelakis-Angelakis, K. A. (2008b). Plant polyamine catabolism: The state of the art. Plant Signal. Behav. 3, 1061–1066. doi: 10.4161/psb.3.12.7172
Moschou, P. N., Wu, J., Cona, A., Tavladoraki, P., Angelini, R., and Roubelakis-Angelakis, K. A. (2012). The polyamines and their catabolic products are significant players in the turnover of nitrogenous molecules in plants. J. Exp. Bot. 63, 5003–5015. doi: 10.1093/jxb/ers202
Murray Stewart, T., Dunston, T. T., Woster, P. M., and Casero, R. A. Jr. (2018). Polyamine catabolism and oxidative damage. J. Biol. Chem. 293, 18736–18745. doi: 10.1074/jbc.TM118.003337
Ono, Y., Kim, D. W., Watanabe, K., Sasaki, A., Niitsu, M., Berberich, T., et al. (2012). Constitutively and highly expressed Oryza sativa polyamine oxidases localize in peroxisomes and catalyze polyamine back conversion. Amino Acids 42, 867–876. doi: 10.1007/s00726-011-1002-3
Ozyigit, I., Filiz, E., Vatansever, R., Kurtoglu, K. Y., Koc, I., Öztürk, M. X., et al. (2016). Identification and comparative analysis of H2O2-scavenging enzymes (ascorbate peroxidase and glutathione peroxidase) in selected plants employing bioinformatics approaches. Front. Plant Sci. 7:301. doi: 10.3389/fpls.2016.00301
Pál, M., Ivanovska, B., Oláh, T., Tajti, J., Hamow, K., Szalai, G., et al. (2019). Role of polyamines in plant growth regulation of Rht wheat mutants. Plant Physiol. Biochem. 137, 189–202. doi: 10.1016/j.plaphy.2019.02.013
Palacios-Rojas, N., Mcculley, L., Kaeppler, M., Titcomb, T. J., Gunaratna, N. S., Lopez-Ridaura, S., et al. (2020). Mining maize diversity and improving its nutritional aspects within agro-food systems. Compr. Rev. Food Sci. Food Saf. 19, 1809–1834. doi: 10.1111/1541-4337.12552
Pegg, A. E., and Casero, R. A. Jr. (2011). Current status of the polyamine research field. Methods Mol. Biol. 720, 3–35. doi: 10.1007/978-1-61779-034-8_1
Petrov, V. D., and Van Breusegem, F. (2012). Hydrogen peroxide-a central hub for information flow in plant cells. AoB Plants 2012:pls014. doi: 10.1093/aobpla/pls014
Planas-Portell, J., Gallart, M., Tiburcio, A. F., and Altabella, T. (2013). Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana. BMC Plant Biol. 13:109. doi: 10.1186/1471-2229-13-109
Quan, L. J., Zhang, B., Shi, W. W., and Li, H. Y. (2008). Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 50, 2–18. doi: 10.1111/j.1744-7909.2007.00599.x
Rao, X., Huang, X., Zhou, Z., and Lin, X. (2013). An improvement of the 2ˆ(−delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 3, 71–85.
Rodríguez, A. A., Maiale, S. J., Menéndez, A. B., and Ruiz, O. A. (2009). Polyamine oxidase activity contributes to sustain maize leaf elongation under saline stress. J. Exp. Bot. 60, 4249–4262. doi: 10.1093/jxb/erp256
Sadeghipour, O. (2019). Polyamines protect mung bean [Vigna radiata (L.) Wilczek] plants against drought stress. Biol. Futur. 70, 71–78. doi: 10.1556/019.70.2019.09
Sagor, G. H. M., Inoue, M., Kusano, T., and Berberich, T. (2021). Expression profile of seven polyamine oxidase genes in rice (Oryza sativa) in response to abiotic stresses, phytohormones and polyamines. Physiol. Mol. Biol. Plants 27, 1353–1359. doi: 10.1007/s12298-021-01006-1
Sagor, G. H. M., Kusano, T., and Berberich, T. (2017). Identification of the actual coding region for polyamine oxidase 6 from rice (OsPAO6) and its partial characterization. Plant Signal. Behav. 12:e1359456. doi: 10.1080/15592324.2017.1359456
Sekhon, R. S., Lin, H., Childs, K. L., Hansey, C. N., Buell, C. R., De Leon, N., et al. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66, 553–563. doi: 10.1111/j.1365-313X.2011.04527.x
Singh, A., Mehta, S., Yadav, S., Nagar, G., Ghosh, R., Roy, A., et al. (2022). How to cope with the challenges of environmental stresses in the era of global climate change: an update on ROS Stave off in plants. Int. J. Mol. Sci. 23:1995. doi: 10.3390/ijms23041995
Sobieszczuk-Nowicka, E., Kubala, S., Zmienko, A., Małecka, A., and Legocka, J. (2015). From accumulation to degradation: reprogramming polyamine metabolism facilitates dark-induced senescence in barley leaf cells. Front. Plant Sci. 6:1198. doi: 10.3389/fpls.2015.01198
Tavladoraki, P., Rossi, M. N., Saccuti, G., Perez-Amador, M. A., Polticelli, F., Angelini, R., et al. (2006). Heterologous expression and biochemical characterization of a polyamine oxidase from Arabidopsis involved in polyamine back conversion. Plant Physiol. 141, 1519–1532. doi: 10.1104/pp.106.080911
Tiburcio, A. F., Altabella, T., Bitrián, M., and Alcázar, R. (2014). The roles of polyamines during the lifespan of plants: from development to stress. Planta 240, 1–18. doi: 10.1007/s00425-014-2055-9
Toumi, I., Moschou, P. N., Paschalidis, K. A., Bouamama, B., Ben Salem-Fnayou, A., Ghorbel, A. W., et al. (2010). Abscisic acid signals reorientation of polyamine metabolism to orchestrate stress responses via the polyamine exodus pathway in grapevine. J. Plant Physiol. 167, 519–525. doi: 10.1016/j.jplph.2009.10.022
Wang, W., and Liu, J. H. (2015). Genome-wide identification and expression analysis of the polyamine oxidase gene family in sweet orange (Citrus sinensis). Gene 555, 421–429. doi: 10.1016/j.gene.2014.11.042
Wang, W., and Liu, J. H. (2016). CsPAO4 of Citrus sinensis functions in polyamine terminal catabolism and inhibits plant growth under salt stress. Sci. Rep. 6:31384. doi: 10.1038/srep31384
Wang, W., Paschalidis, K., Feng, J. C., Song, J., and Liu, J. H. (2019). Polyamine catabolism in plants: a universal process with diverse functions. Front. Plant Sci. 10:561. doi: 10.3389/fpls.2019.00561
Wang, J., Sun, P. P., Chen, C. L., Wang, Y., Fu, X. Z., and Liu, J. H. (2011). An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. J. Exp. Bot. 62, 2899–2914. doi: 10.1093/jxb/erq463
Wu, G., Bazer, F. W., Hu, J., Johnson, G. A., and Spencer, T. E. (2005). Polyamine synthesis from proline in the developing porcine placenta. Biol. Reprod. 72, 842–850. doi: 10.1095/biolreprod.104.036293
Wu, J., Shang, Z., Wu, J., Jiang, X., Moschou, P. N., Sun, W., et al. (2010). Spermidine oxidase-derived H₂O₂ regulates pollen plasma membrane hyperpolarization-activated Ca(2+) -permeable channels and pollen tube growth. Plant J. 63, 1042–1053. doi: 10.1111/j.1365-313X.2010.04301.x
Xiong, H., Guo, H., Xie, Y., Zhao, L., Gu, J., Zhao, S., et al. (2017). RNAseq analysis reveals pathways and candidate genes associated with salinity tolerance in a spaceflight-induced wheat mutant. Sci. Rep. 7:2731. doi: 10.1038/s41598-017-03024-0
Xu, J., Kang, Z., Zhu, K., Zhao, D., Yuan, Y., Yang, S., et al. (2021). RBOH1-dependent H(2)O(2) mediates spermine-induced antioxidant enzyme system to enhance tomato seedling tolerance to salinity-alkalinity stress. Plant Physiol. Biochem. 164, 237–246. doi: 10.1016/j.plaphy.2021.04.017
Yoda, H., Fujimura, K., Takahashi, H., Munemura, I., Uchimiya, H., and Sano, H. (2009). Polyamines as a common source of hydrogen peroxide in host- and nonhost hypersensitive response during pathogen infection. Plant Mol. Biol. 70, 103–112. doi: 10.1007/s11103-009-9459-0
Zhang, J., Liao, J., Ling, Q., Xi, Y., and Qian, Y. (2022). Genome-wide identification and expression profiling analysis of maize AP2/ERF superfamily genes reveal essential roles in abiotic stress tolerance. BMC Genom. 23:125. doi: 10.1186/s12864-022-08345-7
Keywords: polyamine oxidases, polyamines, Zea mays L., stress response, functional analysis
Citation: Xi Y, Hu W, Zhou Y, Liu X and Qian Y (2022) Genome-Wide Identification and Functional Analysis of Polyamine Oxidase Genes in Maize Reveal Essential Roles in Abiotic Stress Tolerance. Front. Plant Sci. 13:950064. doi: 10.3389/fpls.2022.950064
Edited by:
Ji-Hong Liu, Huazhong Agricultural University, ChinaReviewed by:
Chenliang Yu, Zhejiang Agriculture and Forestry University, ChinaPhotini V. Mylona, Hellenic Agricultural Organization (HAO), Greece
Copyright © 2022 Xi, Hu, Zhou, Liu and Qian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yexiong Qian, cXl4MjAxMUBhaG51LmVkdS5jbg==
†These authors have contributed equally to this work
 Yan Xi†
Yan Xi† Yexiong Qian
Yexiong Qian