- 1College of Horticulture, China Agricultural University, Beijing, China
- 2Department of Fruit Tree Sciences, Agricultural Research Organization, The Volcani Center, Bet Dagan, Israel
- 3State Key Laboratory of Agrobiotechnology, China Agricultural University, Beijing, China
WD40 proteins serve as crucial regulators in a broad spectrum of plant developmental and physiological processes, including anthocyanin biosynthesis. However, in fig (Ficus carica L.), neither the WD40 family nor any member involved in anthocyanin biosynthesis has been elucidated. In the present study, 204 WD40 genes were identified from the fig genome and phylogenetically classified into 5 clusters and 12 subfamilies. Bioinformatics analysis prediction localized 109, 69, and 26 FcWD40 proteins to the cytoplasm, nucleus and other cellular compartments, respectively. RNA-seq data mining revealed 127 FcWD40s expressed at FPKM > 10 in fig fruit. Most of these genes demonstrated higher expression in the early stages of fruit development. FcWD40-97 was recruited according to three criteria: high expression in fig fruit, predicted nuclear localization, and closest clustering with TTG1s identified in other plants. FcWD40-97, encoding 339 amino acids including 5 WD-repeat motifs, showed 88.01 and 87.94% amino acid sequence similarity to apple and peach TTG1, respectively. The gene is located on fig chromosome 4, and is composed of 1 intron and 2 exons. Promoter analysis revealed multiple light-responsive elements, one salicylic acid-responsive element, three methyl jasmonate-responsive elements, and one MYB-binding site involved in flavonoid biosynthesis gene regulation. FcWD40-97 was in the FPKM > 100 expression level group in fig fruit, and higher expression was consistently found in the peel compared to the flesh at the same development stages. Expression level did not change significantly under light deprivation, whereas in leaves and roots, its expression was relatively low. Transient expression verified FcWD40-97’s localization to the nucleus. Yeast two-hybrid (Y2H) and biomolecular fluorescence complementation (BiFC) assays revealed that FcWD40-97 interacts with FcMYB114, FcMYB123, and FcbHLH42 proteins in vitro and in vivo, showing that FcWD40-97 functions as a member of the MYB–bHLH–WD40 (MBW) complex in anthocyanin-biosynthesis regulation in fig. We therefore renamed FcWD40-97 as FcTTG1. Our results provide the first systematic analysis of the FcWD40 family and identification of FcTTG1 in fig pigmentation.
Introduction
WD40 proteins, also called WD-repeat (WDR) proteins, constitute one of the largest protein families in eukaryotic organisms (Stirnimann et al., 2010). WD40 proteins were first defined in bovine β-transducin, with an approximately 43-residue fragment containing a glycine–histidine (GH) pair at the N terminus and a tryptophan–aspartate (WD) pair at the C terminus (Fong et al., 1986). WD40 proteins are usually regarded as adaptors, recruiting other factors to form protein complexes or protein–DNA complexes (Jain and Pandey, 2018; Dayebgadoh et al., 2019). There is accumulating evidence of WD40s’ versatile roles in a wide spectrum of biochemical, developmental and physiological processes, including signal transduction, chromatin assembly, cytoskeleton assembly, cell division, flowering and abiotic stress responses (Smith, 2008; Hu et al., 2020; Tan et al., 2021).
WD40s have been grouped in different ways by different researchers, according to evolutionary relationships or comparisons with Arabidopsis. In Arabidopsis thaliana, 237 AtWD40 genes with at least 4 WDR motifs were classified into 33 groups, and in Cucumis sativus, 191 CsWD40 genes were classified into 21 groups (Li et al., 2014). In Solanum tuberosum, 178 StWD40 genes and in Mangifera indica, 315 MiWD40 genes were classified into 14 and 11 groups, respectively (Liu et al., 2020; Tan et al., 2021). In Triticum aestivum, Prunus persica, Rosa chinensis, and Ginkgo biloba, 743 TaWD40 genes, 220 PpWD40 genes, 187 RcWD40 genes, and 167 GbWD40 genes were each clustered into 5 groups (Hu et al., 2018; Feng et al., 2019; Sun et al., 2020; Zheng et al., 2021a,b).
The most well-known function of WD40 is as part of the MYB–bHLH–WD40 (MBW) complex in the regulation of anthocyanin/proanthocyanidin biosynthesis. TRANSPARENT TESTA GLABRA1 (TTG1) is a WD40 protein that was first identified in Arabidopsis; AtTTG1 encodes 341 amino acids; Atttg1 mutants completely lack anthocyanins in the epidermis and in subepidermal layers of leaves and stems (Walker et al., 1999). Moreover, AtTTG1, AtMYB123, and AtbHLH42 can form a ternary complex that synergistically activates the expression of BANYULS (anthocyanidin reductase) and promotes proanthocyanidin biosynthesis (Baudry et al., 2004). TTG1 has been identified in other plants as well; Fragaria × ananassa (strawberry) FaTTG1, Camellia sinensis CsWD40 and Vitis vinifera (grape) VvWDR1 encode 344, 342, and 336 amino acids, respectively (Schaart et al., 2013; Liu et al., 2018; Jiu et al., 2021). Overexpression of TTG1-like CsWD40 in tobacco resulted in a significant increase in anthocyanin content in the petals of the transgenic plants; when it was coexpressed with CsMYB5e in tobacco, both anthocyanin and proanthocyanidin content increased (Liu et al., 2018). VvWDR1 increased anthocyanin accumulation in transgenic tobacco by interacting with the VvMYBA2r–VvMYCA1 complex (Jiu et al., 2021).
Fig (Ficus carica L.) is one of the most ancient cultivated fruit trees in the world. Fig fruit development presents a typical double-sigmoid growth curve, i.e., a fast–slow–fast growth pattern (Kislev et al., 2006; Stover et al., 2007; Flaishman et al., 2008). Fig fruit have a high content of anthocyanins, proanthocyanidins and other flavonoids (Solomon et al., 2006; Wang et al., 2019). The pigmentation of fig peel and flesh tissue follows an obvious tissue-specific pattern. The fig peel can be green, yellow or red/black-purple at ripening, whereas the flesh can be amber or from light to deep red at harvest.
A series of anthocyanin biosynthesis-related genes have been functionally characterized in fig, including FcANS (Cao et al., 2016), FcMADS9 (Li et al., 2021) and FcHY5 (Wang et al., 2019); in addition, three MYB genes—FcMYB21, FcMYB114 and FcMYB123 (Wang et al., 2019; Li et al., 2020)—and bHLH regulator FcbHLH42 (Song et al., 2021) were also functionally proven to participate in anthocyanin and flavonoid biosynthesis. Although a FcWD40 gene was isolated (Lama et al., 2020), there has been no systematic analysis of the FcWD40 gene family, nor has FcTTG1 been identified. Based on the published fig genome (Usai et al., 2020), we screened 204 putative FcWD40 genes and studied their structure, cellular localization and expression patterns in fig fruit. FcWD40-97 was identified by closest sequence similarity to TTG1s from other plants and its interaction with fig anthocyanin-biosynthesis regulators FcMYBs and FcbHLH. The result provides us with the first global view of FcWD40s and reveals TTG1 in fig.
Materials and Methods
Search for FcWD40s
The whole-genome sequence of fig was downloaded from the NCBI online database (1 Usai et al., 2020); data of Ficus microcarpa and Ficus hispida were downloaded from the Genome Warehouse (GWH) and Sequence Archive (GSA) database (Zhang et al., 2020). Protein data of A. thaliana Araport11 were downloaded from the TAIR database2. The Hidden Markov Model (HMM) profile of the WD40 domain (PF00400) was retrieved from Pfam3. Candidate genes with typical WD40 domains in the fig genome were preliminarily screened using the HMM and SMART online tools (Eddy, 1998; Letunic and Bork, 2018). Systematic WD40 domain (PF00400) analysis was performed using the Pfam database (Finn et al., 2014). The redundant sequences were deleted utilizing HMMER software4 with a default E-value < 10–5, then sequences with less than 150 amino acids were discarded; this was followed by a high-standard (E-value < 10–10) screening of the conserved WD40 domain using NCBI CDD5. Finally, all of the candidate sequences were confirmed by Pfam and the conserved domain database (Marchler-Bauer and Bryant, 2004).
Bioinformatics Analysis
ClustalW software with default parameters was used to carry out multiple-sequence alignments of the predicted WD40 protein sequences of fig and Arabidopsis (Thompson et al., 2002). An unrooted phylogenetic tree was constructed using the neighbor-joining method by MEGA 7 with the following parameters: Poisson model, pairwise deletion, 1,000 bootstrap iterations.
The coding sequences and genomic sequences of the identified FcWD40 genes were obtained. The location of each FcWD40 on the 13 fig chromosomes was determined by searching WD40 sequences to fig chromosome using Blast programs. An intron/exon structure map of the FcWD40s was drawn using TBtools (Chen et al., 2020). The ExPASy Server tool6 was applied to predict the molecular mass and theoretical isoelectric point (pI) of the WD40 proteins. The PSORT II database7 was used to examine the subcellular localization of FcWD40 proteins. In addition, the WD40 domain and motif were identified in the genes.
MCScanX was used to analyze synteny of the WD40 genes between fig and other plants (Wang et al., 2012), and collinearity relationship was drawn by Circos and TBtool software (Krzywinski et al., 2009). Ka/Ks was calculated with KaKs Calculator 2.08 (Wang et al., 2010). The gene duplications were dated using the formula T = Ks/2r; r, which is the rate of divergence for nuclear genes, was taken to be 7 × 10–9 synonymous substitutions per site per year according to a previous report on A. thaliana (Ossowski et al., 2010). The potential cis-regulatory elements on promoter regions of WD40 genes were identified by Plant CARE (Lescot et al., 2002).
RNA-Seq Data Mining
Two fig fruit RNA-seq libraries which were established and uploaded to NCBI by our laboratory were re-mined. PRJNA7237339 is a library of fruit flesh and peel at six stages covering the whole fruit development process of cv. “Purple-Peel,” which has a purple peel and red flesh (Song et al., 2021). PRJNA49494510 is a transcriptome library of light-deprived “Purple-Peel” fig fruit and a control (Wang et al., 2019). The number of mapped clean reads for each transcript was normalized to FPKM (fragments per kilobase of exon model per million mapped reads) by RSEM11. Genes that were differentially expressed under light deprivation and their corresponding controls were analyzed by edgeR12 (Robinson et al., 2010).
Cloning of FcTTG1 and qRT-PCR Analysis
Fig root, shoot, young leaf, old leaf and fruit were collected from greenhouse-grown “Purple-Peel” fig trees at the Shangzhuang Experimental Station of China Agricultural University at Beijing. The fig trees were planted with 3.5 m × 4 m spacing in 2017. Fig fruit were separated into peel and flesh using a scalpel. All of the plant material was frozen using liquid nitrogen and stored for later use. DNA was extracted from fig leaves using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980), and RNA was isolated following protocols developed in our laboratory and used in previous publications (Chai et al., 2014).
The FcTTG1 gene sequence was cloned from ‘Purple-Peel’ fig flesh. The expression of FcWD40-97 was validated by qRT-PCR. PrimeScript™ RT reagent kit (RR037Q, Takara, Dalian, China) was used to reverse-transcribe the total RNA. 18S was used in parallel as an internal reference gene to normalize the expression levels. The qRT-PCR was performed with ChamQ Universal SYBR qPCR Master Mix (Q711-02, Vazyme, Nanjing, China). A 10-μL reaction mixture was added to each well. The PCR program was as follows: 95°C for 30 s, and 40 cycles of: 95°C for 10 s, 60°C for 60 s. Each sample was analyzed in three technical replicates, and the 2–ΔΔCt method was applied for relative quantification analysis (Livak and Schmittgen, 2001). The expression of each gene was determined using three biological replicates. Statistical analysis of one-way ANOVA followed by Duncan’s new multiple range test and Student’s t-test were performed with IBM SPSS Statistics Version 23.0. The significance level was set to P < 0.05. All primers used for qRT-PCR are listed in Supplementary Table 1.
Subcellular Localization and Prediction of Protein–Protein Interactions for FcTTG1
The full-length cDNA of FcTTG1 was cloned for subcellular localization using primers containing restriction sites XbaI and BamHI, and inserted into binary vector pBI121 to generate fusion construct 35S:FcTTG1-GFP. Then, 35S:FcTTG1-GFP and the control vector were transferred into Agrobacterium tumefaciens strain GV3101 by heat shock. Four-week-old Nicotiana tabacum leaves were agroinfiltrated (Kumar and Kirti, 2010) with the bacterial suspension (OD600 = 0.8) and then kept in an incubator at 28°C for 72 h, followed by live-cell imaging using an inverted confocal laser-scanning microscope (Olympus FV3000, Tokyo, Japan).
The STRING 11.5 server13 was employed to illustrate the protein–protein interaction network. The exhibited specifications were used with confidence values of 0.4 for network, 10 for edge evidence and maximum number of interactions.
Yeast Two-Hybrid and Bimolecular Fluorescence Complementation Assays
The interaction of FcTTG1 with FcMYBs and FcbHLHs was explored using the Matchmaker Gold Yeast Two-Hybrid System (Clontech, USA). FcTTG1 was ligated to a pGBKT7 vector used as bait, and FcMYB114, FcbHLH1, FcbHLH42, and FcMYC2 were ligated to a pGADT7 vector and used as prey proteins. Plasmids pGBKT7-53 plus pGADT7-T and pGADT7-lam plus pGADT7-T plasmids were used as positive and negative controls, respectively. Yeast cells were cultured on the SD/-Leu-Trp (DDO) medium, then transformed colonies were plated on SD/-Leu-Trp-His-Ade (QDO) with 20 mM X-α-gal and 125 ng/mL AbA medium for further verification of the actual interactions. The yeast strains were co-transformed with FcTTG1 plus FcMYB114, FcMYB123, FcbHLH1, FcbHLH42, and FcMYC2, respectively. The primers used for Y2H vector construction are presented in Supplementary Table 1.
The full-length cDNA of FcTTG1 was inserted into a pSPYCE-35S (YCE) vector, and those of FcMYB114, FcMYB123 and FcbHLH42 were respectively subcloned into pSPYNE-35S (YNE) vectors. All of the constructs and control vectors (pSPYNE-35S and pSPYCE-35S) were transformed into A. tumefaciens strain GV3101, and 4-week-old N. benthamiana plants leaves were transiently co-transformed as described by Song et al. (2022). Fluorescence signals were observed 72 h after transfection using a confocal laser-scanning microscope (Olympus FV3000, Tokyo, Japan). The primers used for the BiFC vector construction are presented in Supplementary Table 1.
Results
Phylogenetic Analysis
A total of 204 WD40 proteins with at least 1 WDR motif were recruited from F. carica after removing repetitive sequences. For convenience, the genes were numbered from FcWD40-1 to FcWD40-204. Among them, 190 genes had at least 4 WDR motifs. FcWD40 proteins varied from 189 (FcWD40-204) to 3628 (FcWD40-28) amino acids, and the predicted molecular mass ranged from 20.67 kDa (FcWD40-204) to 403.52 kDa (FcWD40-28). Theoretical pI ranged from 4.29 to 10.46, with 120 acidic and 84 alkaline pI. Predicted subcellular localization showed 109 FcWD40 proteins in the cytoplasm, 69 in the nucleus, 10 in the endoplasmic reticulum, 10 in the mitochondria, 3 in the plasma membrane, 2 in the Golgi, and 1 in the vacuole (Supplementary Table 2).
A phylogenetic tree was generated using the 204 FcWD40s and 233 AtWD40s. The FcWD40s were categorized into five clusters (I–V), and cluster V had three subclusters (Va, Vb, and Vc). Cluster IV was the largest subgroup with 137 members, followed by cluster I with 107 members; cluster II was the smallest, with 37 members (Figure 1A).
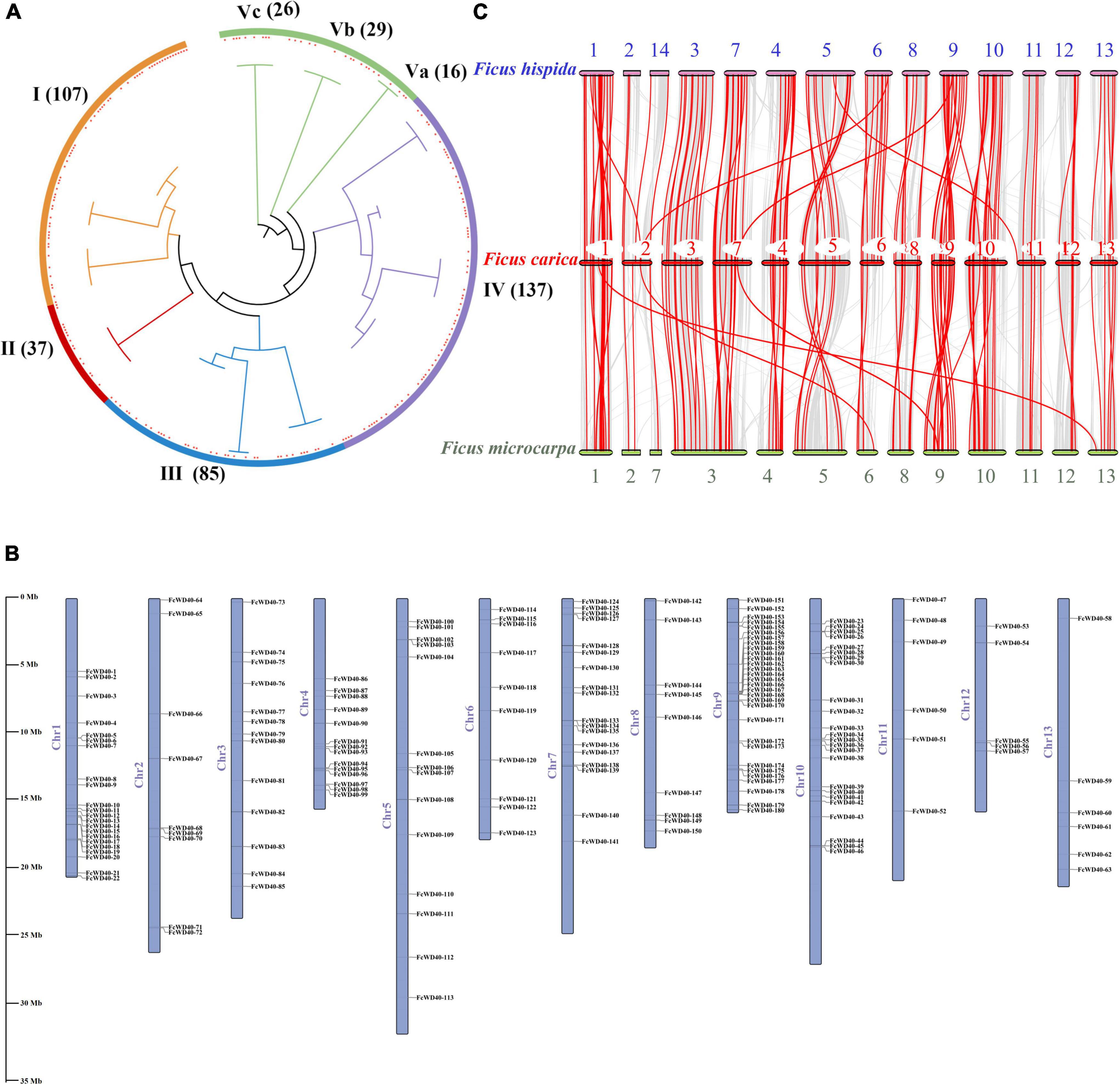
Figure 1. Chromosomal locations, phylogenetic relationships, and collinearity analysis of the FcWD40 gene family. (A) Phylogenetic tree of 437 WD40 genes identified in Ficus carica and Arabidopsis thaliana genomes. Orange, red, blue, purple, and green highlighted areas show clusters I–V. Red squares represent fig WD40. The numbers on the branch points represent total number of WD40 gene; scale length represents genetic distance. (B) FcWD40 genes are marked on chromosomes; scale bar on the left indicates length of fig chromosome (Mb). (C) Collinearity relationship of WD40 genes among Ficus carica, Ficus hispida, and Ficus microcarpa. Identified collinear genes are linked by red lines.
Chromosomal Distribution and Evolution
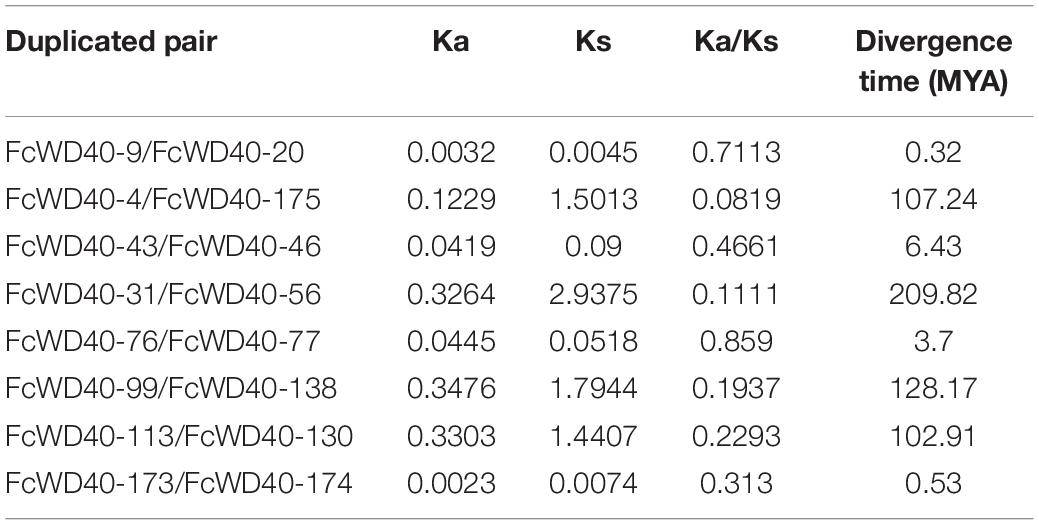
We mapped 180 FcWD40s to 13 chromosomes; positions of the other 24 genes were not found. FcWD40s were widespread and unevenly distributed on the fig chromosomes (Figure 1B). Chromosome 3 hosted 30 FcWD40s, followed by chromosome 9 with 24 genes; only 5 genes were mapped to chromosome 12. Three pairs of genes—FcWD40-15/FcWD40-16, FcWD40-44/FcWD40-45, FcWD40-154/FcWD40-155—were identified as tandem duplications, and eight pairs of genes might have resulted from segmental duplication events (Supplementary Figure 1).
The substitution rate of non-synonymous (Ka) and synonymous (Ks) is the basis for evaluating the positive selection pressure of duplication events. We identified the eight duplicated gene pairs in the FcWD40 family (Table 1). The Ka/Ks ratios of the eight duplicated gene pairs ranged from 0.08 to 0.71, suggesting negative selective pressure on them. Duplication of these eight WD40 gene pairs was calculated to have occurred between 0.32 and 209.82 million years ago.
Comparative syntenic maps were constructed using FcWD40s associated with those of F. hispida and F. microcarpa to reveal phylogenetic relationships between species (Figure 1C). The collinearity analysis identified 184 and 206 WD40 orthologs in F. hispida and F. microcarpa, respectively; 110 FcWD40 genes showed a collinear relationship with those of the two other Ficus species, whereas 34 FcWD40 genes did not, suggesting unique WD40s in the evolution of F. carica. Further analysis demonstrated that F. hispida WD40 genes had the most homologous gene pairs with the WD40 genes of F. carica (134), followed by F. microcarpa (125), indicating similar evolutionary distances between the edible fig and the two evergreen Ficus species. Moreover, in comparison with F. microcarpa, F. hispida genes showed a closer phylogenetic relationship with FcWD40 genes. Detailed results of the analysis are shown in Supplementary Table 3.
Gene Structure and Promoter Analysis
FcWD40s were categorized into 12 subfamilies based on their domain compositions. Sixty-three FcWD40 genes containing only WD40 domains were classified as subfamily A; among them, 23 had predicted nuclear localization. Eighty-seven FcWD40 genes containing another unknown domain were classified as subfamily L, among them 34 genes were with predicted nuclear localization. The remaining 54 FcWD40s, comprising other domains, were grouped into subfamilies B, comprising other 2 genes containing the zinc finger domain; (C) 4 genes with the CAF1 domain, including 2 genes with predicted nuclear localization; (D) 16 genes with the LisH domain, including 4 genes with predicted nuclear localization; (E) 4 genes with the UTP12/UTP15/UTP21 domains; FcWD40s with (F) the coatomer WD/COP1 domain, (G) katanim_80, (H) beach, (I) protein kinase, (J) NLE, and (K) F-box-like domain. Transcription regulation occurs in the nucleus, and at least 4 WDR motifs are required to form a β-propeller (Chothia et al., 1997). Therefore, the 66 FcWD40s with predicted nuclear localization and with at least 4 WDR motifs were selected for analysis; their domains are shown in Figure 2A, and a phylogenetic tree was generated (Supplementary Figure 2).
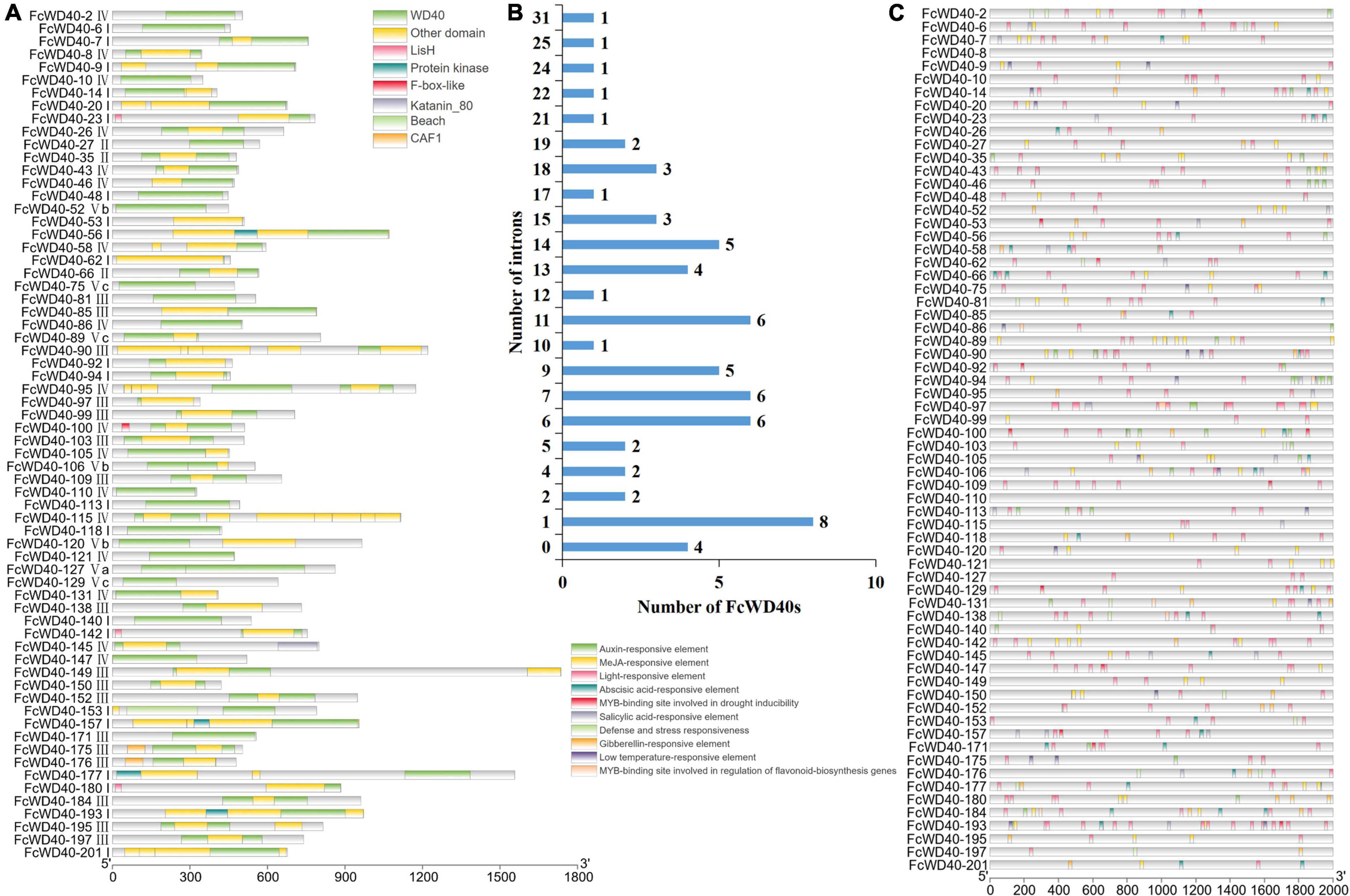
Figure 2. Analysis of domain structure and promoter of FcWD40s. (A) Distribution of primary domains of 66 FcWD40 proteins with nuclear localization and genes expressed in fig fruit. (B) Introns number of 66 FcWD40s. (C) Promoter analysis of FcWD40s by Plant CARE, based on 2000-bp sequence upstream of the genes.
Gene structure analysis of the FcWD40s revealed wide variation in the number of exons and introns. The maximum number of predicted exons, 40, was found in FcWD40-128, whereas only 1 exon and no intron were found in 9 FcWD40 genes (Supplementary Table 2). The gene structures of the aforementioned 66 FcWD40s were also examined. Among them, FcWD40-8, FcWD40-48, FcWD40-113, and FcWD40-118 had no intron, whereas the other 62 FcWD40s had 1–31 introns (Figure 2B).
Ten conserved motifs were identified in the FcWD40 proteins, designated motif 1 to 10. Motifs 1–5 were identified as the WD40 domain. All 204 FcWD40 proteins had motif 1, which contained 15 amino acids, and 202 FcWD40 proteins had motif 2; 19 FcWD40 proteins had motif 3; 198 and 197 FcWD40 proteins had motif 4 and motif 5, respectively. Eight FcWD40 proteins had motif 6 or motif 7. Five FcWD40 proteins had motif 8, and motifs 9 and 10 were the least common (Supplementary Table 4).
A variety of cis-elements were revealed in the 2-kb promoter region of the 66 FcWD40s with predicted nuclear localization. Cis-elements related to hormones, such as auxin, methyl jasmonate (MeJA), abscisic acid, salicylic acid and gibberellin were rather common, suggesting that FcWD40 gene expression could be responsive to a variety of plant hormones. Furthermore, light, defense and stress, low temperatures, and a MYB-binding site involved in drought inducibility were identified (Figure 2C). Notably, only FcWD40-97 had a MYB-binding site involved in the regulation of flavonoid gene biosynthesis among the 66 putatively nuclear-localized FcWD40 genes, and it had 3 MeJA-responsive elements as well (Figure 2C).
Expression Pattern in Fruit Development
One-hundred and twenty-seven FcWD40s were expressed at FPKM > 10 in at least one of the six samples covering the whole fig fruit development process. Three expression groups (S1–S3) were assembled based on the genes’ highest transcription levels: S1, at least one sample of FPKM ≥ 100; S2, FPKM between 99 and 50; and S3, FPKM between 49 and 10. There were 14, 21, and 92 FcWD40s in groups S1, S2, and S3, respectively (Figure 3). In group S1, FcWD40-11/68/78/80/97/101 were highly expressed in stages 1–4, while FcWD40-16/18/22/54 were significantly upregulated in stages 5 or 6, in both flesh and peel.
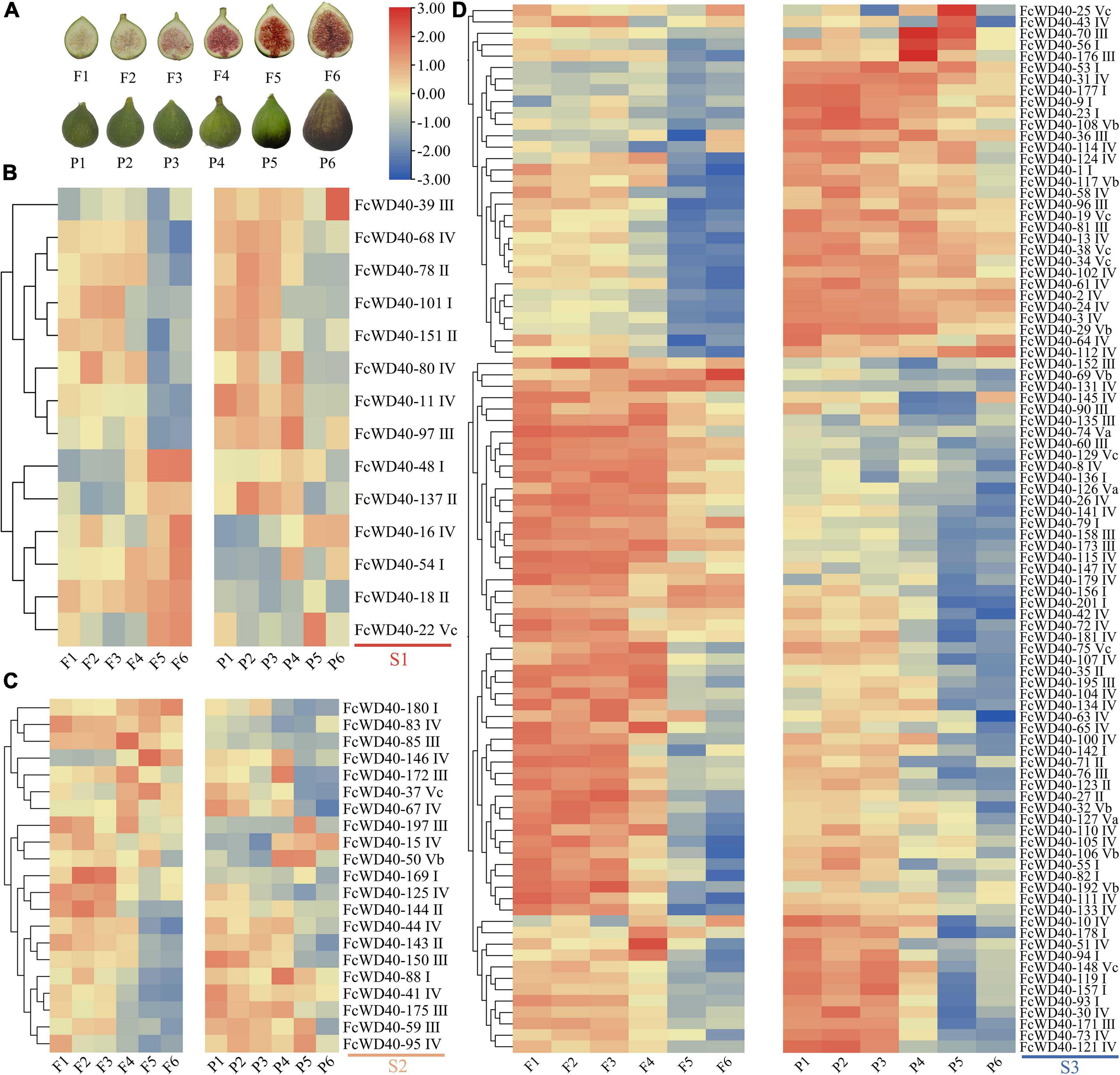
Figure 3. Expression profiles of FcWD40 genes in the flesh and peel tissue of developing and ripening fruit. (A) Photographs of fig flesh and peel color at different stages. (B) Members of FcWD40 in group S1 (FPKM ≥ 100). (C) Members of FcWD40 in group S2 (100 > FPKM ≥ 50). (D) Members of FcWD40 in group S3 (50 > FPKM > 10). Color bars represent level of downregulation (blue) and upregulation (red).
Different expression patterns of FcWD40s were found in the peel and flesh. FcWD40-48 and FcWD40-137 were highly expressed in stage 5 and 6 in the flesh, but had lower expression levels in the corresponding peel samples. FcWD40-39 was specifically expressed in the peel. Most of the group S2 and S3 FcWD40s had higher expression in stages 1–4 (Figure 3). Of the 66 FcWD40s predicted to be located in the nucleus, 43 showed expression of FPKM > 10 in fig fruit. These included FcWD40-48 and FcWD40-97 in group S1, 6 genes: FcWD40-85/95/150/175/180/197 in group S2, and 35 genes in group S3.
Six FcWD40s in fig fruit showed a significant change in expression under light deprivation (FPKM > 10, | log2(fold change)| ≥ 2, p-adjust < 0.05): FcWD40-22 and FcWD40-28 showed differential expression in the peel; FcWD40-16, FcWD40-48, and FcWD40-79 showed differential expression in the flesh, respectively, moreover, FcWD40-113 showed differential expression in both tissue types (Figure 4). Among these 6 FcWD40s, FcWD40-48 and FcWD40-113 were predicted to be located in the nucleus; 3 of the 6 FcWD40s were from group S1, i.e., 11 members of group S1 FcWD40s did not respond to fruit light deprivation.
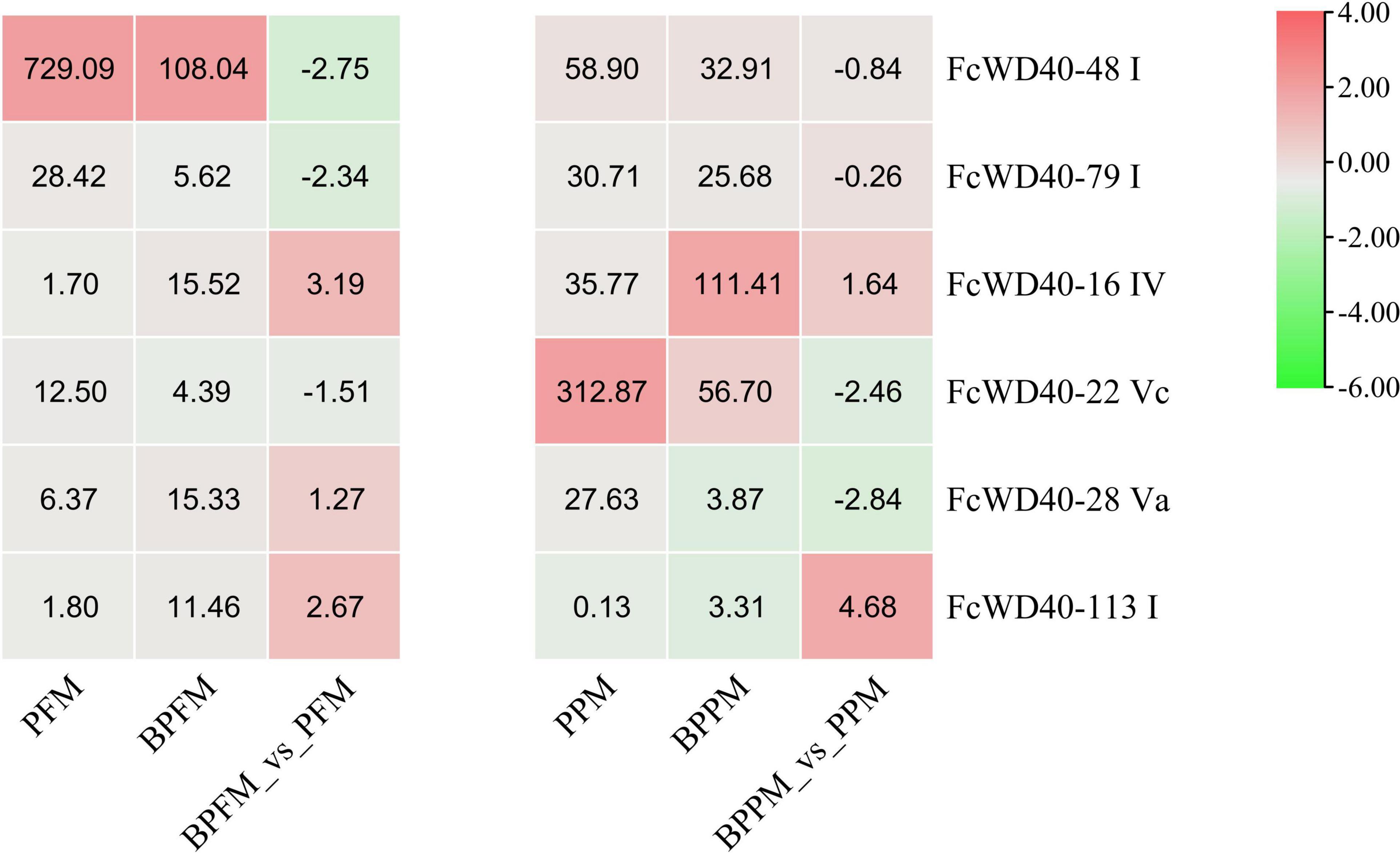
Figure 4. Expression profiles of FcWD40 genes under light deprivation. PFM and PPM represent the mature-stage flesh and peel of “Purple-Peel,” respectively; BPFM and BPPM represent the bagging mature-stage flesh and peel of ‘Purple-Peel’ fruit, respectively.
Screen for FcTTG1 and Expression Analysis
To find the WD40s associated with fig fruit color development, a phylogenetic tree was constructed using the 43 fruit-expressed FcWD40 genes with predicted nuclear localization and with at least 4 WDR motifs, together with 8 TTG1 proteins identified from other plants (Figure 5A). Another phylogenetic tree was constructed to further analyze the evolutionary relationships of all FcWD40s and 8 TTG1 proteins (Supplementary Figure 3). FcWD40-97 showed the highest sequence similarity to the TTG1s. The deduced amino acid sequence of FcWD40-97 was 79.88% similar to TTG1 from Arabidopsis, 88.01 and 87.94% similar to apple and peach TTG1, respectively, and 85.67 and 84.88% similar to CsWD40 and FaTTG1, respectively. The four hypothetical amino acid residue pairs (WD, FD, LD, and WE) were also highly conserved among the TTG1 proteins involved in anthocyanin or proanthocyanidin biosynthesis in different plants (Figure 5B). Therefore, we suggested that FcWD40-97 is a FcTTG1.
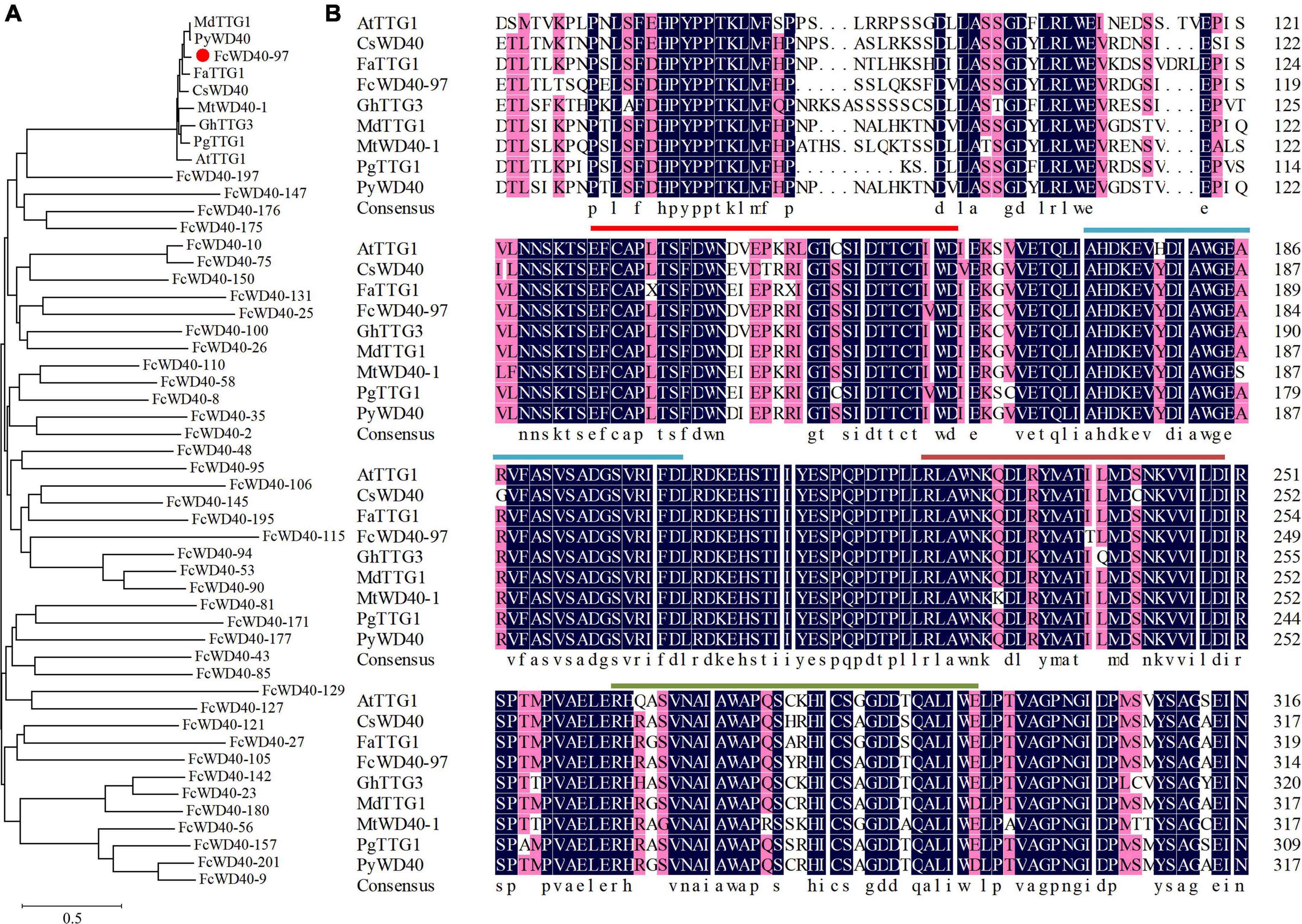
Figure 5. Phylogenetic analysis of 43 FcWD40 proteins and TTG1s of other species. (A) Phylogenetic relationships of TTG1 proteins. GenBank accession numbers: Arabidopsis thaliana AtTTG1 (NM_122360), Malus domestica MdTTG1 (GU173813), Pyrus pyrifolia PyWD40 (HQ641374.1), Fragaria × ananassa FaTTG1 (JQ989287), Camellia sinensis CsWD40 (MH618664), Punica granatum PgTTG1 (HQ199314), Gossypium hirsutum GhTTG3 (AAM95645), and Medicago truncatula, MtWD40-1 (EU040206.1). FcWD40-97 is indicated with red dot. (B) Alignment of deduced amino acid sequences of FcWD40 proteins and other known TTG1 homologs. The four conserved amino acid residue pairs (WD, FD, LD, and WE) are marked by different-colored straight lines.
Expression of FcWD40-97 in different tissues of the fig tree was determined by qRT-PCR. It was expressed in all six tissues examined, with significantly higher transcript levels in the flesh and peel, followed by shoots; its expression level in young and mature leaves and roots was markedly lower (Figure 6A). In the fruit, FcWD40-97 was more highly expressed at developmental stage 4 in both flesh and peel (Figures 6B,C), which was largely consistent with our fig fruit RNA-seq results.
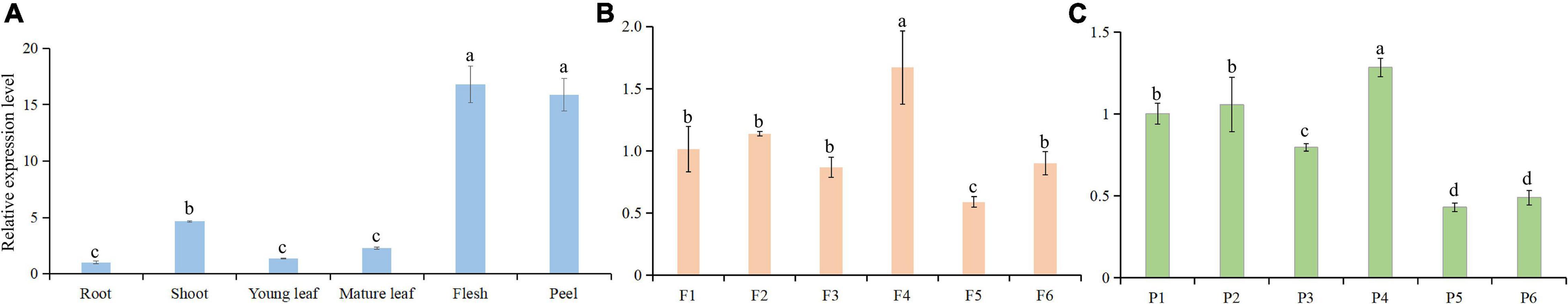
Figure 6. Relative expression levels of FcTTG1 in different tissues and at different developmental stages in Ficus carica. (A) Fig plant tissues; flesh and peel are from ripe fruit. (B) Fig fruit flesh at developmental stages 1–6 corresponding to Figure 3A. (C) Fig fruit peel at developmental stages 1–6 corresponding to Figure 3A. Relative expression levels are shown as mean ± SD from three replications. Different letters between bars indicate significant difference at p < 0.05.
Subcellular Localization and Interaction Network of FcWD40-97
The predicted nuclear localization of FcWD40-97 was validated by fusing its full-length ORF to the N terminus of green fluorescent protein (GFP) driven by the CaMV 35S promoter. By fluorescence microscopy, the 35S:FcTTG1–GFP fusion proteins were detected exclusively in the cell nucleus, whereas the control was uniformly dispersed throughout the cell (Figure 7A).
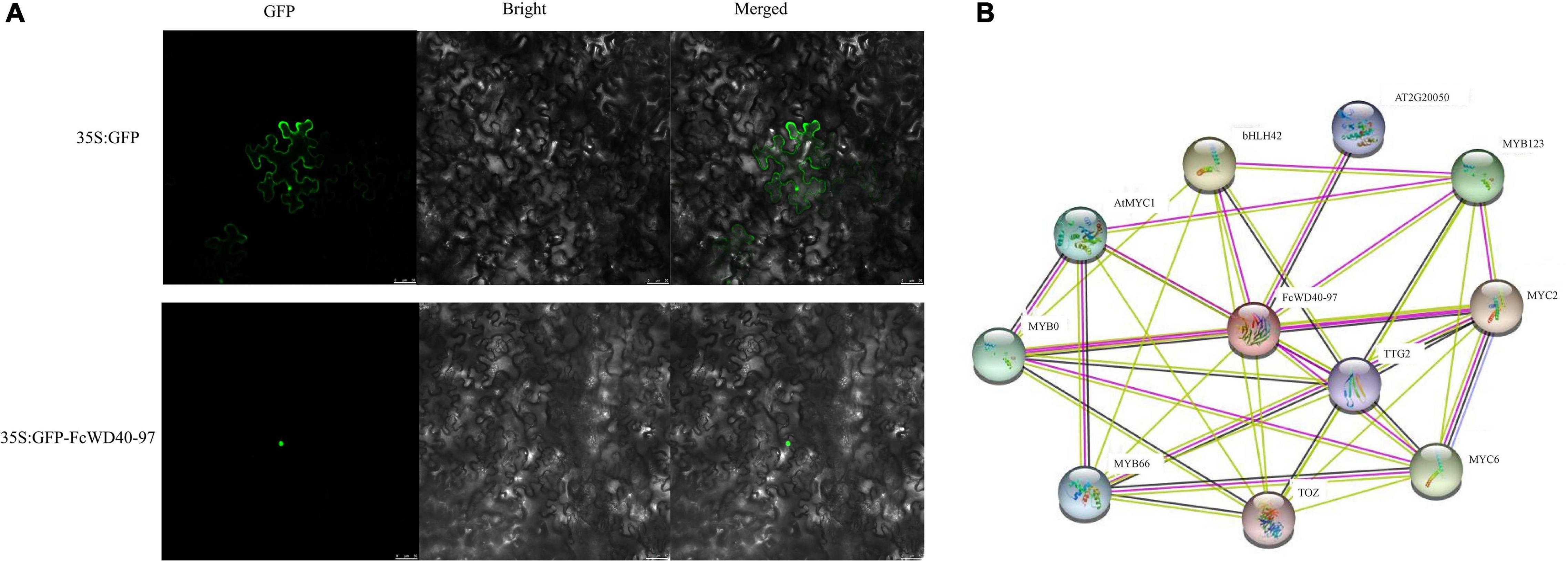
Figure 7. Subcellular localization and interaction network prediction of FcTTG1. (A) Subcellular localization of FcTTG1 in Nicotiana benthamiana. Images under fluorescence (left), bright field (middle) and merged (right). Bars = 50 μm. (B) Predicted interaction network for FcTTG1. Pink lines – experimentally determined physical interactions; yellow lines – interactions by text mining; black lines – co-expression; purple lines – protein homology.
Computational analysis of protein–protein interactions was performed to predict FcWD40-97’s function. The interaction network was observed among several proteins, including FcWD40-97’s interaction with 4 bHLH proteinsork was observed among several proteins, including predict FcWD40-97’s function. detected exclusively in, AT2G20050 (PP2C) and TOZ (WD40), depicting the physical interactions, co-expression, and homology of the proteins (Figure 7B).
Y2H-based protein-interaction tests were conducted between FcWD40-97 and FcMYBs/FcbHLHs. Yeast cells of co-transformants with four combinations: BD–FcWD40-97 plus AD–FcMYB114, BD–FcWD40-97 plus AD–FcMYB123, BD–FcWD40-97 plus AD–FcbHLH1, BD–FcWD40-97 plus AD–FcbHLH42, BD–FcWD40-97 plus AD–FcMYC2 grew on the DDO medium. Moreover, they also grew on QDO/X-α-gal/AbA and the colonies turned blue (Figure 8A).
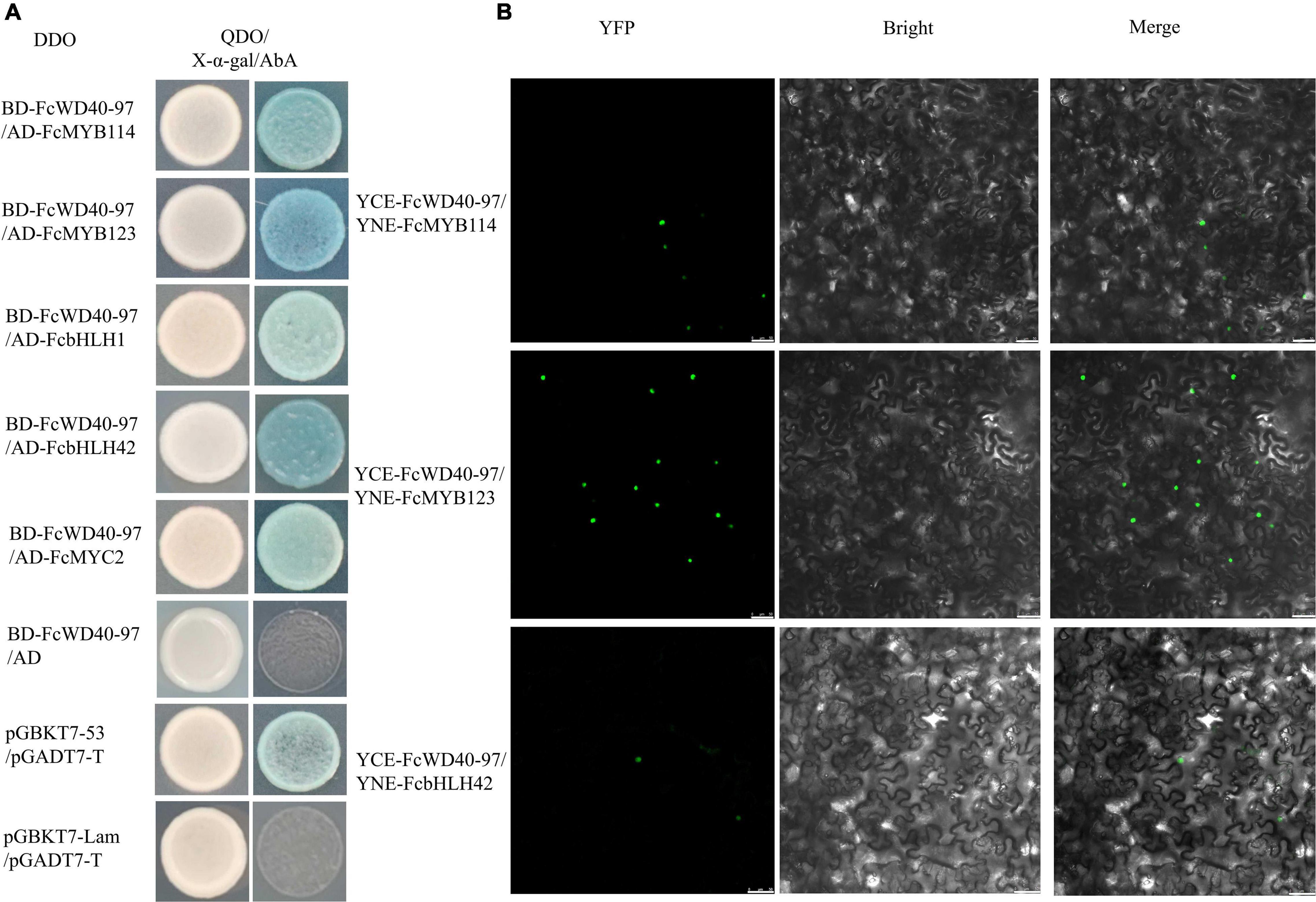
Figure 8. Interaction of FcTTG1 with FcMYBs and FcbHLHs in vitro and in vivo. (A) Yeast two-hybrid test to determine the in vitro interaction with different constructs. (B) In vivo verification of interactions between FcTTG1 and FcMYB114, FcMYB123, and FcbHLH42. Bars = 50 μm.
Three anthocyanin biosynthesis-related proteins were selected for further validation by BiFC. Yellow fluorescent protein (YFP) fluorescence was observed in the nuclei of tobacco leaf cells co-transformed with YCE–FcWD40-97 plus YNE–FcMYB114, YCE–FcWD40-97 plus YNE–FcMYB123, and YCE–FcWD40-97 plus YNE–FcbHLH42 (Figure 8B). No YFP fluorescence was observed in the control combinations, i.e., empty YNE vector plus YCE– FcWD40-97, and empty YCE plus YNE–FcMYB114, YNE–FcMYB123, or YNE–FcbHLH42. The results demonstrated that FcWD40-97 interacts with FcMYB114, FcMYB123, and FcbHLH42 in vivo, and is potentially able to form the MBW transcription complex in fig. This confirmed the identity of FcWD40-97 as FcTTG1.
Discussion
FcWD40 Family
There are around 200 putative WD-containing proteins in diploid plants which serve important roles in a wide spectrum of physiological functions, such as growth, the cell cycle, signal transduction, chromatin remodeling, transcriptional regulation, and others. WD40 domain-containing proteins have also been identified in prokaryotes, as well as lower eukaryotes. It has been postulated that the WD40 repeats arose from intragenic duplication and recombination events (Andrade et al., 2001); however, recently, the common ancestry of WD40 genes has been questioned (Jain and Pandey, 2018).
Tandem and segmental duplications are primary driving forces in generating members of a gene family during evolution (Cannon et al., 2004). In our study, 3 and 8 pairs of FcWD40s among the 204 WD40-containing proteins in fig were predicted as resulting from tandem duplications and segmental duplication events, respectively. This ratio is significantly lower than that of two other gene families that we previously studied: among 118 identified FcbHLHs, 11 and 4 pairs were predicted to originate from tandem replication and fragment replication, respectively (Song et al., 2021), and among 31 FcPLCP genes, 6 duplicated gene pairs were predicted (Zhai et al., 2021).
A similar frequency of tandem and segmental duplications has been predicted for WD40 genes in other plants. Among the 230 WD40 genes of Arabidopsis, 13 were identified as involved in tandem duplication events (Li et al., 2014). In the 178 WD40 genes of potato, 3 pairs of genes were identified as tandemly duplicated, and 14 pairs of genes were related to segmental duplication (Liu et al., 2020). Among of the 187 WD40 genes in rose, 11 members were identified as stemming from tandem duplication (Sun et al., 2020). In the hexaploid wheat, 743 WD40 genes were identified, and 39 and 46 pairs of WD40s were distinguished as tandem and segmental duplication genes, respectively (Hu et al., 2018). In 184 and 206 WD40 genes of F. hispida and F. microcarpa, 9 and 14 pairs of tandem duplications were identified, and 15 and 13 genes were related to segmental duplication, respectively (Zhang et al., 2020). In our study, all FcWD40 tandem duplications originated from subfamily IV, and segmental duplication gene pairs were mainly from subfamily III. Our findings suggest tandem and segmental duplication as important driving forces for expansion in specific FcWD40 subfamilies, whereas they are less crucial in the evolution of the FcWD40 family as a whole.
FcTTG1 Homologs and Expression Characteristics
Genes with similar amino acid sequences usually have similar functions, and vice versa (Liu et al., 2022). Eight TTG1 proteins from different plants were found to be 78.2%–99.12% homologous, indicating that the functions of TTG1s are conserved in different species. In the present study, FcWD40-97 showed the highest similarity to TTG1s of other plants, with a deduced amino acid sequence that had 79.88% similarity to AtTTG1 (Li et al., 2014), 88.01 and 87.94% to apple TTG1 and peach WD40, respectively (An et al., 2012; Cui et al., 2021), 85.67, 85.17, and 85.59% to CsWD40, FaTTG1, and Punica granatum (Pg) TTG1, respectively (Ben-Simhon et al., 2011; Schaart et al., 2013; Liu et al., 2018), and 82.90 and 80.52% to Gossypium hirsutum (Gh) TTG3 and Medicago truncatula (Mt) WD40-1 (Humphries et al., 2005; Pang et al., 2009), respectively. Moreover, FcWD40-97 shared the four highly conserved WDR motifs with other TTG1s, reported to be related to anthocyanin biosynthesis. Therefore we suggest that FcWD40-97 is FcTTG1.
FcTTG1 encoded 339 amino acids, which is more than grape TTG1 (336), but less than TTG1 of Arabidopsis (341), strawberry (344) and Camellia sinensis (342). In addition, FcTTG1 has 5 WDR motifs, similar to strawberry (Schaart et al., 2013), tea tree (Liu et al., 2018), grape (Jiu et al., 2021) and pear (Cui et al., 2021), whereas Arabidopsis (Baudry et al., 2004), Chinese bayberry (Liu et al., 2013) and pomegranate (Ben-Simhon et al., 2011) each have 4 WDR motifs.
Many studies have reported that TTG1 genes are mainly expressed in anthocyanin-accumulating tissues. Apple TTG1 showed the highest transcript level in the peel (An et al., 2012). The expression level of PgWD40 was higher in red-colored leaves and stems than in their green-colored counterparts (Ben-Simhon et al., 2011). Chinese bayberry WD40-1 showed the highest transcription level in fruit (Liu et al., 2013). Freesia × hybrida TTG1 was mainly expressed in petals (Shan et al., 2019). In our study, FcTTG1 was preferentially expressed in fruit flesh and peel, and highest expression in these two tissues was found at developmental stage 4. The expression pattern in fig flesh was in line with the high expression of FcMYB114 and FcbHLH42, which are the major transcription regulators in fig pigmentation (Song et al., 2021); in fig peel, FcTTG1 was highly expressed before FcMYB114 and FcbHLH42.
Conservation and Pleiotropy of FcTTG1 in the Isoflavone-Biosynthesis Pathway
The TTG1-dependent MBW transcriptional complex is necessary for the regulation of anthocyanin/proanthocyanidin biosynthesis among plant species (Lloyd et al., 2017). In Arabidopsis, the ttg1 mutant lacks seed-coat pigmentation, resulting in a transparent testa (tt) phenotype; absence of anthocyanin biosynthesis was also observed in the epidermis and subepidermal layers of ttg1 mutant leaves and stems (Walker et al., 1999). The rice homozygous ttg1 mutant exhibits significantly decreased anthocyanin accumulation in various organs (Yang et al., 2021), validating the crucial role of TTG1 in the MBW complex.
A small group of MYBs and bHLHs have been identified as components of the MBW complex. In Arabidopsis, PAP1, PAP2, MYB113, MYB114, and TT2 are MYBs that regulate anthocyanin and proanthocyanidin biosynthesis (Nesi et al., 2001; Gonzalez et al., 2008). In apple, MYB1, MYBA, MYB3, MYB110a, and MYB10 regulate anthocyanin biosynthesis in the peel, flesh and leaves (Ban et al., 2007; Espley et al., 2007; Chagné et al., 2013; Vimolmangkang et al., 2013), while MYB23 regulates proanthocyanidin biosynthesis (An et al., 2018). In grape, MYBA1, MYBA2, MYBA5, MYBA6, and MYBA7 were identified as regulators of anthocyanin biosynthesis (Matus et al., 2017), while MYBPAR regulates proanthocyanidin biosynthesis (Koyama et al., 2014). Similar redundance has been found with bHLHs in anthocyanin/proanthocyanidin biosynthesis. Arabidopsis bHLHs GL3, EGL3, and TT8 were reported as components of MBW in flavonoid-biosynthesis regulation (Nesi et al., 2001; Gonzalez et al., 2008; Xu et al., 2015).
In contrast to the small group of MYBs and bHLHs involved in isoflavone biosynthesis, less complexity has been found with TTG1. TTG1 demonstrates high conservation and pleiotropy in different MBW complexes. In our study, FcTTG1 could interact with FcMYB114, FcMYB123, and FcbHLH42, indicating the generality of FcTTG1 in fig anthocyanin and proanthocyanidin biosynthesis. Similarly, strawberry TTG1 was reported to control proanthocyanidin biosynthesis together with FaMYB9/FaMYB11 and FabHLH3 (Schaart et al., 2013). In mango, TTG1 was found to interact with MYB0, TT8 and bHLH1 (Tan et al., 2021).
The constitutive nature of FcTTG1 in the MBW complex is further supported by its expression pattern under different light treatments. FcTTG1 expression was not significantly changed when fig peel anthocyanin accumulation was almost completely inhibited by light deprivation, in line with the rice wild-type TTG1 which is not significantly affected by light vs. dark (Yang et al., 2021). Nevertheless, light deprivation led to 1.59-fold downregulation of FcMYB114 and 0.82-fold downregulation of FcbHLH42 in the peel (Wang et al., 2019; Song et al., 2021). Similarly, Arabidopsis TTG1 is rather stably expressed in both seedlings and rosette leaves when treated with white light, whereas MYBs (PAP1 and PAP2) and bHLHs (TT8, EGL3 and GL3) are upregulated (Cominelli et al., 2008).
Similar to other TTG1s, FcTTG1 serves as an essential and constitutive component of MBW complexes, that is, variable R2R3–MYBs and bHLHs share the same TTG1 constituents. The tissue-specific pattern of anthocyanin and proanthocyanidin biosynthesis and the response to environmental stimuli rely more on R2R3–MYB factors, whereas TTG1 is an indispensable and constant regulator in the MBW complexes.
Conclusion
We identified 204 FcWD40 genes in the fig genome, and described their chromosome distribution, evolutionary characteristics, gene structure, and cellular localization. The expression patterns of FcWD40s during fig fruit development and in response to light deprivation were grouped. Agglomerative hierarchical clustering recruited FcWD40-97 as the ideal candidate FcTTG1, as further confirmed by a set of molecular biology validations. FcWD40 was revealed as a constitutive component of the MBW complex regulating anthocyanin biosynthesis of fig fruit. This result uncovers the specific FcWD40 in fig anthocyanin biosynthesis and extends our knowledge on fig fruit color development. Further functional studies should be performed to confirm the role of FcTTG1 in fig fruit development.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJNA723733 and PRJNA494945.
Author Contributions
ZF, YZ, and YW performed the experiments. LZ analyzed the data. ZF prepared the first draft of the manuscript. MF and HM revised the manuscript. HM supervised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31372007) and 111 Project (B17043).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.948084/full#supplementary-material
Supplementary Figure 1 | Collinearity analysis of FcWD40 genes; circle plot created with MCScanX tool. Identified collinear genes are linked by colored lines.
Supplementary Figure 2 | Phylogenetic analysis of 66 FcWD40s with predicted nuclear localization and at least 4 WD-repeat (WDR) motifs.
Supplementary Figure 3 | Phylogenetic analysis of FcWD40s and TTG1s of Arabidopsis thaliana, Malus domestica, Pyrus pyrifolia, Fragaria × ananassa, Camellia sinensis, Punica granatum, Gossypium hirsutum, and Medicago truncatula. FcWD40-97 is indicated with red dot. Blue color background indicates TTG1 subgroups, and red star indicates potential TTG1 homolog in fig.
Supplementary Figure 4 | Interaction of control combinations in vivo.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/Traces/wgs/VYVB01?display=history%EF%BC%89
- ^ http://arabidopsis.org
- ^ http://pfam.xfam.org/family/PF00400
- ^ http://hmmer.org/
- ^ https://www.ncbi.nlm.nih.gov/cdd/
- ^ http://web.expasy.org/compute_pi/
- ^ https://psort.hgc.jp/form2.html
- ^ https://sourceforge.net/projects/kakscalculator2/
- ^ https://www.ncbi.nlm.nih.gov/bioproject/PRJNA723733
- ^ https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA494945
- ^ http://deweylab.biostat.wisc.edu/rsem/
- ^ http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html
- ^ https://cn.string-db.org/cgi/network?taskId=bvuGEO8PZi4D&sessionId=b9QKtOaUD9B4
References
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., and Hao, Y. J. (2018). R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96, 562–577. doi: 10.1111/tpj.14050
An, X. T., Tian, Y., Chen, K. Q., Wang, X. F., and Hao, Y. J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169, 710–717. doi: 10.1016/j.jplph.2012.01.015
Andrade, M. A., Perez-Iratxeta, C., and Ponting, C. P. (2001). Protein repeats: structures, functions, and evolution. J. Struct. Biol. 134, 117–131. doi: 10.1006/jsbi.2001.4392
Ban, Y., Honda, C., Hatsuyama, Y., Igarashi, M., Bessho, H., and Moriguchi, T. (2007). Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 48, 958–970. doi: 10.1093/pcp/pcm066
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Ben-Simhon, Z., Judeinstein, S., Nadler-Hassar, T., Trainin, T., Bar-Ya Akov, I., Borochov-Neori, H., et al. (2011). A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 234, 865–881. doi: 10.1007/s00425-011-1438-4
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Cao, L., Xu, X., Chen, S., and Ma, H. (2016). Cloning and expression analysis of Ficus carica anthocyanidin synthase 1 gene. Sci. Hortic. 211, 369–375. doi: 10.1016/j.scienta.2016.09.015
Chagné, D., Lin-Wang, K., Espley, R. V., Volz, R. K., How, N. M., Rouse, S., et al. (2013). An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 161, 225–239. doi: 10.1104/pp.112.206771
Chai, L., Li, Y., Chen, S., Perl, A., Zhao, F., and Ma, H. (2014). RNA sequencing reveals high resolution expression change of major plant hormone pathway genes after young seedless grape berries treated with gibberellin. Plant Sci. 229, 215–224. doi: 10.1016/j.plantsci.2014.09.010
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chothia, C., Hubbard, T., Brenner, S., Barns, H., and Murzin, A. (1997). Protein folds in the all-β and all-α classes. Annu. Rev. Bioph. Biom. Struct. 26, 597–627. doi: 10.1146/annurev.biophys.26.1.597
Cominelli, E., Gusmaroli, G., Allegra, D., Galbiati, M., Wade, H. K., Jenkins, G. I., et al. (2008). Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana[J]. J. Plant Physiol. 165, 886–894. doi: 10.1016/j.jplph.2007.06.010
Cui, D., Zhao, S., Xu, H., Allan, A. C., Zhang, X., Fan, L., et al. (2021). The interaction of MYB, bHLH and WD40 transcription factors in red pear (Pyrus pyrifolia) peel. Plant Mol. Biol. 106, 407–417. doi: 10.1007/s11103-021-01160-w
Dayebgadoh, G., Sardiu, M. E., Florens, L., and Washburn, M. P. (2019). Biochemical reduction of the topology of the diverse WDR76 protein interactome. J. Proteome Res. 18, 3479–3491. doi: 10.1021/acs.jproteome.9b00373
Eddy, S. R. (1998). Profile hidden Markov models. Bioinformatics 14, 755–763. doi: 10.1093/bioinformatics/14.9.755
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Feng, R., Zhang, C., Ma, R., Cai, Z., Lin, Y., and Yu, M. (2019). Identification and characterization of WD40 superfamily genes in peach. Gene 710, 291–306. doi: 10.1016/j.gene.2019.06.010
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: The protein families database. Nucleic Acids Res. 42:D222–D230. doi: 10.1093/nar/gkt1223
Flaishman, M. A., Rodov, V., and Stover, E. (2008). The fig: botany, horticulture, and breeding. Hort. Rev. 34, 113–196. doi: 10.1002/9780470380147.ch2
Fong, H. K. W., Hurley, J. B., Hopkins, R. S., Miake-Lye, R., Johnson, M. S., and Doolittle, R. F. (1986). Repetitive segmental structure of the transducin beta subunit: Homology with the CDC4 gene and identification of related mRNAs. Proc. Natl. Acad. Sci. U.S.A. 83:2162. doi: 10.1073/pnas.83.7.2162
Gonzalez, A., Zhao, M., Leavitt, J. M., and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Hu, R., Xiao, J., Gu, T., Yu, X., Zhang, Y., Chang, J., et al. (2018). Genome-wide identification and analysis of WD40 proteins in wheat (Triticum aestivum L.). BMC Genomics 19:803. doi: 10.1186/s12864-018-5157-0
Hu, R., Xiao, J., Zhang, Q., Gu, T., Chang, J., Yang, G., et al. (2020). A light-regulated gene, TaLWD1L-A, affects flowering time in transgenic wheat (Triticum aestivum L.). Plant Sci. 299:110623. doi: 10.1016/j.plantsci.2020.110623
Humphries, J. A., Walker, A. R., Timmis, J. N., and Orford, S. J. (2005). Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol. Biol. 57, 67–81. doi: 10.1007/s11103-004-6768-1
Jain, B. P., and Pandey, S. (2018). WD40 repeat proteins: signalling scafold with diverse functions. Protein J. 37, 391–406. doi: 10.1007/s10930-018-9785-7
Jiu, S., Guan, L., Leng, X., Zhang, K., Haider, M. S., Yu, X., et al. (2021). The role of VvMYBA2r and VvMYBA2w alleles of the MYBA2 locus in the regulation of anthocyanin biosynthesis for molecular breeding of grape (Vitis spp.) skin coloration. Plant Biotechnol. J. 19, 1216–1239. doi: 10.1111/pbi.13543
Kislev, M. E., Hartmann, A., and Bar-Yosef, O. (2006). Early domesticated fig in the Jordan Valley. Science 312, 1372–1374.
Koyama, K., Numata, M., Nakajima, I., Goto-Yamamoto, N., Matsumura, H., and Tanaka, N. (2014). Functional characterization of a new grapevine MYB transcription factor and regulation of proanthocyanidin biosynthesis in grapes. J. Exp. Bot. 65, 4433–4449. doi: 10.1093/jxb/eru213
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, K. R. R., and Kirti, P. B. (2010). A mitogen-activated protein kinase, AhMPK6 from peanut localizes to the nucleus and also induces defense responses upon transient expression in tobacco. Plant Physiol. Bioch. 48, 481–486. doi: 10.1016/j.plaphy.2010.03.010
Lama, K., Harleva, G., Shafrana, H., Peera, R., and Flaishman, M. A. (2020). Anthocyanin accumulation is initiated by abscisic acid to enhance fruit color during fig (Ficus carica L.) ripening. J. Plant Physiol. 251:153192. doi: 10.1016/j.jplph.2020.153192
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Li, J., An, Y., and Wang, L. (2020). Transcriptomic analysis of Ficus carica peels with a focus on the key genes for anthocyanin biosynthesis. Int. J. Mol. Sci. 21:1245. doi: 10.3390/ijms21041245
Li, J., Ma, N., An, Y., and Wang, L. (2021). FcMADS9 of fig regulates anthocyanin biosynthesis. Sci. Hortic. 278:109820. doi: 10.1016/j.scienta.2020.109820
Li, Q., Zhao, P., Li, J., Zhang, C., Wang, L., and Ren, Z. (2014). Genome-wide analysis of the WD-repeat protein family in cucumber and Arabidopsis. Mol. Genet. Genomics 289, 103–124. doi: 10.1007/s00438-013-0789-x
Liu, J. L., Zhang, C. X., Han, J. Y., Fang, X. Y., Xu, H. P., and Liang, C. L. (2022). Genome-wide analysis of KNOX transcription factors and expression pattern of dwarf-related KNOX genes in pear. Front. Plant Sci. 13:806765. doi: 10.3389/fpls.2022.806765
Liu, X. F., Feng, C., Zhang, M. M., Yin, X. R., Xu, C. J., and Chen, K. S. (2013). The MrWD40-1 gene of Chinese Bayberry (Myrica rubra) interacts with MYB and bHLH to enhance anthocyanin accumulation. Plant Mol. Biol. Rep. 31, 1474–1484. doi: 10.1007/s11105-013-0621-0
Liu, Y., Hou, H., Jiang, X., Wang, P., Dai, X., Chen, W., et al. (2018). A WD40 repeat protein from Camellia sinensis regulates anthocyanin and proanthocyanidin accumulation through the formation of MYB-bHLH-WD40 ternary complexes. Int. J. Mol. Sci. 19:1686. doi: 10.3390/ijms19061686
Liu, Z., Liu, Y., Coulter, J. A., Shen, B., Li, Y., Li, C., et al. (2020). The WD40 gene family in potato (Solanum tuberosum L.): Genome-wide analysis and identification of anthocyanin and drought-related WD40s. Agronomy 10:401. doi: 10.3390/agronomy10030401
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32:W327–W331. doi: 10.1093/nar/gkh454
Matus, J. T., Cavallini, E., Loyola, R., Höll, J., Finezzo, L., Santo, S. D., et al. (2017). A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J. 91, 220–236. doi: 10.1111/tpj.13558
Murray, M., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326.
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. doi: 10.1105/tpc.010098
Ossowski, S., Schneeberger, K., Lucas-Lledó, J. I., Warthmann, N., Clark, R. M., Shaw, R. G., et al. (2010). The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327, 92–94. doi: 10.1126/science.1180677
Pang, Y. Z., Wenger, J. P., Saathoff, K., Peel, G. J., Wen, J. Q., Huhman, D., et al. (2009). A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 151, 1114–1129. doi: 10.1104/pp.109.144022
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). EdgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26, 139–140. doi: 10.1093/bioinformatics/btp616
Schaart, J. G., Dubos, C., Fuente, I. R. D. L. F., van Houwelingen, A. M. M. L., de Vos, R. C. H., Jonker, H. H., et al. (2013). Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria×ananassa) fruits. New Phytol. 197, 454–467. doi: 10.1111/nph.12017
Shan, X. S., Li, Y. Q., Yang, S., Gao, R. F., Zhou, L. D., Bao, T. T., et al. (2019). A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiol. Bioch. 141, 60–72. doi: 10.1016/j.plaphy.2019.05.015
Smith, T. F. (2008). Diversity of WD-repeat proteins. Subcell. Biochem. 48, 20–30. doi: 10.1007/978-0-387-09595-0_3
Solomon, A., Golubowicz, S., Yablowicz, Z., Grossman, S., Bergman, M., Gottlieb, H. E., et al. (2006). Antioxidant activities and anthocyanin content of fresh fruits of common fig (Ficus carica L.). J. Agric. Food Chem. 54, 7717–7723. doi: 10.1021/jf060497h
Song, M. Y., Wang, H. M., Ma, H. Q., and Zheng, C. L. (2022). Genome-wide analysis of JAZ family genes expression patterns during fig (Ficus carica L.) fruit development and in response to hormone treatment. BMC Genomics 23:170. doi: 10.1186/s12864-022-08420-z
Song, M. Y., Wang, H. M., Wang, Z., Huang, H. T., Chen, S. W., and Ma, H. Q. (2021). Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in fig (Ficus carica L.). Front. Plant Sci. 12:730692. doi: 10.3389/fpls.2021.730692
Stirnimann, C. U., Petsalaki, E., Russell, R. B., and Müller, C. W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574. doi: 10.1016/j.tibs.2010.04.003
Stover, E., Aradhya, M., Ferguson, L., and Crisosto, C. H. (2007). The fig: Overview of an ancient fruit. Hortscience 42, 1083–1087.
Sun, Y. B., Zhang, X. J., Zhong, M. C., Dong, X., Yu, D. M., Jiang, X. D., et al. (2020). Genome-wide identification of WD40 genes reveals a functional diversification of COP1-like genes in Rosaceae. Plant Mol. Biol. 104, 81–95. doi: 10.1007/s11103-020-01026-7
Tan, L., Salih, H., Htet, N., Azeem, F., and Zhan, R. (2021). Genomic analysis of WD40 protein family in the mango reveals a TTG1 protein enhances root growth and abiotic tolerance in Arabidopsis. Sci. Rep. 11:2266. doi: 10.1038/s41598-021-81969-z
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2002). Multiple sequence alignment using clustalw and clustalx. Curr. Protoc. Bioinform 2:2.3. doi: 10.1002/0471250953.bi0203s00
Usai, G., Mascagni, F., Giordani, T., Vangelisti, A., Bosi, E., Zuccolo, A., et al. (2020). Epigenetic patterns within the haplotype phased fig (Ficus carica L.) genome. Plant J. 102, 600–614. doi: 10.1111/tpj.14635
Vimolmangkang, S., Han, Y. P., Wei, G. C., and Korban, S. S. (2013). An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 13:176. doi: 10.1186/1471-2229-13-176
Walker, A. R., Davison, P. A., Bolognesi-Winfiel, A. C., James, C. M., Srinivasan, N., Blundel, T. L., et al. (1999). The TRANSPARENT TESTA GLABRA1 Locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 11, 1337–1349. doi: 10.1105/tpc.11.7.1337
Wang, D. P., Zhang, Y. B., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteo. Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, Z., Song, M., Li, Y., Chen, S., and Ma, H. (2019). Differential color development and response to light deprivation of fig (Ficus carica L.) syconia peel and female flower tissues: Transcriptome elucidation. BMC Plant Biol. 19:217. doi: 10.1186/s12870-019-1816-9
Xu, W., Dubos, C., and Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Yang, X. H., Wang, J. H., Xia, X. Z., Zhang, Z. Q., He, J., Nong, B. X., et al. (2021). OsTTG1, a WD40 repeat gene, regulates anthocyanin biosynthesis in rice. Plant J. 107, 198–214. doi: 10.1111/tpj.15285
Zhai, Y. L., Cui, Y. Y., Song, M. Y., Vainstein, A., Chen, S. W., and Ma, H. Q. (2021). Papain-Like cysteine protease gene family in fig (Ficus carica L.): Genome-wide analysis and expression patterns. Front. Plant Sci. 12:681801. doi: 10.3389/fpls.2021.681801
Zhang, X., Wang, G., Zhang, S. C., Chen, S., Wang, Y. B., Wen, B., et al. (2020). Genomes of the banyan tree and pollinator wasp provide insights into fig-wasp coevolution. Cell 183, 875–889. doi: 10.1016/j.cell.2020.09.043
Zheng, J. R., Liao, Y. L., Xu, F., Zhou, X., Ye, J. B., Fu, M. Y., et al. (2021a). Genome-wide identification of WD40 superfamily genes and prediction of WD40 genes involved in flavonoid biosynthesis in Ginkgo biloba. Not. Bot. Horti. Agrobo. 49:803. doi: 10.15835/nbha49212086
Keywords: fig (Ficus carica L.), WD40, expression profile, MBW complex, TTG1, anthocyanin biosynthesis
Citation: Fan Z, Zhai Y, Wang Y, Zhang L, Song M, Flaishman MA and Ma H (2022) Genome-Wide Analysis of Anthocyanin Biosynthesis Regulatory WD40 Gene FcTTG1 and Related Family in Ficus carica L. Front. Plant Sci. 13:948084. doi: 10.3389/fpls.2022.948084
Received: 19 May 2022; Accepted: 23 June 2022;
Published: 14 July 2022.
Edited by:
Jeanne M. E. Jacobs, AgResearch Ltd., New ZealandReviewed by:
Yuhui Liu, Gansu Agricultural University, ChinaHelen H. Tai, Agriculture and Agri-Food Canada, Canada
Copyright © 2022 Fan, Zhai, Wang, Zhang, Song, Flaishman and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiqin Ma, aHFtYUBjYXUuZWR1LmNu
 Zhiyi Fan
Zhiyi Fan Yanlei Zhai
Yanlei Zhai Yuan Wang1
Yuan Wang1 Miaoyu Song
Miaoyu Song Moshe A. Flaishman
Moshe A. Flaishman Huiqin Ma
Huiqin Ma