- 1College of Food Science, Tea Research Institute, Southwest University, Chongqing, China
- 2Chongqing Key Laboratory of Speciality Food Co-built by Sichuan and Chongqing, Southwest University, Chongqing, China
- 3College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 4Key Laboratory of Tea Science in Universities of Fujian Province, College of Horticulture, Fujian Agriculture and Forestry University, Fuzhou, China
Golden2, ARR-B, Psr1 (GARP) proteins are plant-specific transcription factors that play vital and diverse roles in plants. However, systematic research on the GARP gene family in plants, including tea plant (Camellia sinensis), is scarce. In this study, a total of 69 GARP genes were identified and characterized from the tea plant genome based on the B-motif sequence signature. The CsGARP genes were clustered into five subfamilies: PHR1/PHL1, KAN, NIGT1/HRS1/HHO, GLK and ARR-B subfamilies. The phylogenetic relationships, gene structures, chromosomal locations, conserved motifs and regulatory cis-acting elements of the CsGARP family members were comprehensively analyzed. The expansion of CsGARP genes occurred via whole-genome duplication/segmental duplication, proximal duplication, and dispersed duplication under purifying selective pressure. The expression patterns of the CsGARP genes were systematically explored from various perspectives: in different tissues during different seasons; in different leaf color stages of tea plant; under aluminum treatment and nitrogen treatment; and in response to abiotic stresses such as cold, drought and salt and to biotic stress caused by Acaphylla theae. The results demonstrate that CsGARP family genes are ubiquitously expressed and play crucial roles in the regulation of growth and development of tea plant and the responses to environmental stimuli. Collectively, these results not only provide valuable information for further functional investigations of CsGARPs in tea plant but also contribute to broadening our knowledge of the functional diversity of GARP family genes in plants.
Introduction
During their lifetimes, plants often encounter unfavorable or stressful conditions, including abiotic stresses such as drought, cold, salt, and nutrient deficiency and biotic stresses such as pathogen infection and herbivore attack. To respond to and resist these adverse conditions, plants have evolved several specific regulatory networks (Zhu, 2016; Zhang et al., 2021). It is well recognized that transcription factors (TFs) play fundamental roles in the regulation of plant growth and development and stress responses. In Arabidopsis, 2296 TF-encoding genes classified into 58 TF families have been identified and broadly investigated. Among these TF families, the Golden2, ARR-B, Psr1 (GARP) family members, which are mostly related to MYB or MYB-like TFs, compose a plant-specific TF family (Safi et al., 2017). GARP TFs contain a conserved domain named the B-motif, which is the signature motif of GARP members. However, GARPs are often mischaracterized as MYB or MYB-like TFs since their B-motifs and the MYB-like domains are highly similar. In contrast, sequence alignment showed that certain sequences of GARP and MYB-related proteins are not conserved; for example, GARP proteins contain a Ser-His-Leu-Gln-Lys/Met-Tyr/Phe (SHLQ(K/M)(Y/F)) consensus sequence, MYB-related proteins have a Ser-His-Ala-Gln-Lys-Tyr/Phe-Phe (SHAQK(Y/F)F) motif in their DNA-binding region, and all three tryptophan (W) residues are not identical between the B-motif and MYB domain (Riechmann et al., 2000; Hosoda et al., 2002; Safi et al., 2017). GARP TFs can therefore be separated from MYB or MYB-related superfamily TFs according to these specific sites. In Arabidopsis, 56 GARP family genes composing the phosphorus starvation response 1 (PHR1)/PHR1-like 1 (PHR1/PHL1), KANADI (KAN), NITRATE-INDUCIBLE, GARP-TYPE TRANSCRIPTIONAL REPRESSOR1/HYPERSENSITIVE TO LOW Pi-ELICITED PRIMARY ROOT SHORTENING1/HRS1 homolog protein (NIGT1/HRS1/HHO), GOLDEN2-LIKE (GLK) and type-B authentic response regulator (ARR-B) subfamilies have been identified (Riechmann et al., 2000; Safi et al., 2017); however, GARP family members have rarely been identified in other plant species, and their detailed information is largely unknown.
GARP family TFs are involved in very diverse functions in across physiological processes and stress responses in plants, such as growth and development, chloroplast development, nutrient sensing, floral transition, hormone signaling, and stress responses (Safi et al., 2017). Specifically, GLK genes in different species are well known to control chloroplast structure formation, and mutations in these genes cause albino leaf phenotypes and affect photosynthesis by reducing chlorophyll accumulation (Fitter et al., 2002; Powell et al., 2012; Nadakuduti et al., 2014; Chen et al., 2016; Nagatoshi et al., 2016; Lupi et al., 2019; Taketa et al., 2021). NIGT1/HRS1/HHO subgroup TFs constitute a type of NO3–-inducible TF-encoding genes in plants and function as negative regulators in the Arabidopsis response to nitrogen starvation by repressing the expression of other key NO3–-inducible genes, including NRT2.1 (Sawaki et al., 2013; Medici et al., 2015; Kiba et al., 2018; Maeda et al., 2018; Ueda et al., 2020b; Wang et al., 2020d). Additionally, NIGT1/HRS1/HHO TFs can bind to the promoters of inorganic phosphorus (Pi)-responsive genes, including PHT1;1, PHT1;4, and SYG1/Pho81/XPR1 (SPX) family members, to directly modulate their transcription in response to Pi deficiency (Maeda et al., 2018; Ueda et al., 2020a; Wang et al., 2020d), indicating that NIGT1/HRS1/HHO genes play important roles in the absorption and utilization of nitrogen and Pi. Similarly, PHR1/PHL1 subgroup members are broadly recognized to be involved in the plant response to Pi starvation and act as central regulators by regulating the transcription of Pi-responsive genes (Puga et al., 2014; Wang et al., 2014, 2018; Sun et al., 2016; Osorio et al., 2019). By functioning together with the phytohormone auxin, KAN has been shown to regulate organ polarity in leaves and gynoecia (Kerstetter et al., 2001; Pires et al., 2014), leaf structure (Candela et al., 2008; Zhang et al., 2009) and the shoot apical meristem (Huang et al., 2014; Xie et al., 2015; Bhatia et al., 2019; Ram et al., 2020). ARR-B TFs have usually been reported to function as positive regulators of cytokinin signaling and play crucial roles in plant responses to abiotic stress and developmental regulation (Argyros et al., 2008; Kim et al., 2013; Cortleven et al., 2016; Worthen et al., 2019; Wang et al., 2020a).
Tea plant (Camellia sinensis), a perennial evergreen woody crop species that is one of the world’s most consequential plant species, is globally cultivated in more than 50 countries (Xia et al., 2020). Recently, tea plant has gained more attention from a wide array of aspects, such as its growth and development, stress response and metabolite regulation. Many functional genes involved in the biosynthesis pathways of polyphenols, caffeine, and theanine, and in the stress response and formation of aromatic compounds have been thoroughly studied (Xia et al., 2020). Particularly, since the completion of tea plant genome sequence (Wei et al., 2018; Wang et al., 2020c), studies of tea plant genes have greatly advanced. A number of gene families whose members play critical roles in tea plant growth and development and response to stress, such as phosphate transporters (Cao et al., 2021), amino acid permeases (AAPs) (Duan et al., 2020), natural resistance-associated macrophage proteins (NRAMPs) (Li J. et al., 2021), and xyloglucan endotransglycosylases/hydrolases (XTHs) (Wu et al., 2021), and several TF family genes, such as R2R3-MYBs (Chen et al., 2021), Dofs (Yu et al., 2020), and Hsfs (Zhang et al., 2020), have been identified throughout the genome and provide a foundation for further exploring their functions in tea plant. Recently, two CsGLK genes were isolated from tea plant, and silencing their expression disrupted chloroplast and photosynthesis-related development in that species (Hao et al., 2020). Besides, no other CsGARP genes have been reported in tea plant, and little is known about the GARP TF family. Given that GARP TFs are essential in all aspects of plant developmental processes and stress responses, a thorough genome-wide investigation of tea plant is warranted.
In this study, 69 CsGARP TF genes were identified and characterized from the tea plant genome. Moreover, their phylogenetic relationships, gene structures, chromosomal locations, conserved motifs, gene duplication information and regulatory cis-acting elements were comprehensively analyzed. The subcellular localization detection showed that CsNIGT1b, CsKAN1b, and CsBOA1 were localized in cell nucleus. The expression patterns of CsGARP genes in different tissues during different seasons, in different leaf color stages, under aluminum (Al) treatment and nitrogen treatment, and in response to abiotic and biotic stresses were also investigated. The results provide valuable information for further functional studies of CsGARP genes in tea plant.
Materials and Methods
Collection of Sequence Data
The Camellia sinensis genome sequence and 18 additional representative genome sequences were used for comparative analyses. The species included 2 basal angiosperms, 4 monocots and 13 dicots (5 superasterids and 8 superrosids). The cds files and generic feature format (gff) files and the genome data for tea plant (cultivar Suchazao) were downloaded from the Tea Plant Information Archive (TPIA1) (Xia et al., 2019), and the data for Actinidia chinensis was downloaded from the Kiwifruit Genome Database (KGB2) (Yue et al., 2020). The protein sequences of another 17 species were downloaded from Phytozome v12.13 (Goodstein et al., 2012). The species information is detailed in Supplementary Table 1.
GARP Gene Family Identification and Phylogenetic and Gene Structure Analysis
According to the report by Zhu (2016), the amino acid sequences of 56 GARP gene family members in Arabidopsis thaliana were retrieved from The Arabidopsis Information Resource (TAIR4) and used to perform local BLASTP searches (E-value-5) to identify homologous genes in tea plant and other plant species. GARPs form a distinct group of TFs distantly related to MYB superfamily TFs, and MYB and MYB-like TFs were also included in the original BLASTP results. To separate GARP family member sequences from MYB-related sequences, phylogenetic analyses were conducted inclusive of all candidate sequences screened above and those from Arabidopsis thaliana. The MAFFT program (with the default parameters) was used to perform multiple sequence alignments of the GARP protein sequences, and the preliminary tree construction of the aligned GARP protein sequences was based on the maximum likelihood method and performed via FastTree software. Moreover, to further ensure the reliability of the identified GARPs, the sequences were aligned using ClustalX to determine the conserved sites of the GARP-specific B-motif in tea plant, after which all candidate sequences and those from Arabidopsis were confirmed by comparison with GARP member sequences through searches of the NCBI Conserved Domain database and the SMART database. Apart from those of Arabidopsis thaliana, the sequences of GARPs from other plant species were identified as described above.
A phylogenetic tree was also constructed via the same strategy including GARP from tea plant, Vitis vinifera and Arabidopsis thaliana, and the tree was constructed by the use of the Evolview website5. Based on the well-identified GARPs from Arabidopsis thaliana, the phylogenetic tree was divided into five branches. To visualize the gene structure of the exon–intron distribution of the CsGARP genes, the coding DNA sequence (CDS) of each CsGARP gene and its corresponding genomic DNA sequence were uploaded and analyzed using the online Gene Structure Display Server6.
Chromosomal Locations of GARP Genes in Tea Plant
To analyze the chromosomal locations of the CsGARP gene family members in tea plant, the gff files and IDs of the tea plant CsGARP were used to map the locations of CsGARP genes by TBtools software (Chen et al., 2020).
Conserved Motif Analysis
The conserved motifs of the CsGARP protein sequences in tea plant were predicted through the MEME online program7 with the following parameters: number of repetitions = any; maximum number of motifs = 15; and optimum motif length = 6–200 residues.
Gene Synteny and Evolutionary Rate Analyses
CsGARP gene duplication information was investigated using BLASTP with filters including a percent identity > 75% and a query coverage > 75% of the query length. Gene pairs with less than 100 kb on the same chromosome and > 100 kb distance on the same or even different chromosomes were considered tandemly duplicated (TD) and segmentally duplicated (SD) gene pairs, respectively. Synonymous (Ks) and non-synonymous (Ka) substitution rates were estimated using KaKs_Calculator 2.0 (Wang et al., 2010), and the diversion year was calculated according to T = Ks/2λ, where the clock-like rate (λ) was assumed to be 6.5 synonymous substitutions per 10–9 years (Wang et al., 2021). Genome and protein sequence information for Vitis vinifera, Arabidopsis thaliana and Actinidia chinensis were downloaded from Phytozome v12.1 (see text footnote 3), TAIR (see text footnote 4), and the KGD (see text footnote 2), respectively. The synteny of CsGARP genes was predicted and represented using MCScanX (Wang et al., 2012), with the default parameters. The orthologous/homologous gene pairs were displayed as chord diagrams using the Perl circos package (Naquin et al., 2014).
Analysis of Regulatory Cis-Acting Elements
To further analyze the regulatory mechanisms of the CsGARP genes in tea plant in response to stress and growth and development, the sequence of the region 1500 bp upstream of the translation start site of the CsGARP genes was downloaded from the TPIA database8, and the putative cis-acting elements were identified through the PlantCARE program9. The cis-acting elements involved in abiotic stress responses, biotic stress responses, tissue-specific expression, light responses, the circadian rhythm and the cell cycle, as well as core promoter elements, were summarized and analyzed.
Expression Pattern Analysis
A total of five publicly available RNA sequencing (RNA-seq) datasets were downloaded from the Short Read Archive of the NCBI database for expression analysis in different tissues of tea plant (project accession number: PRJEB39502), at different leaf color stages (project accession number: PRJNA277458), in response to cold stress (project accession number: PRJNA411886), in response to salinity and drought stress (project accession number: PRJEB11522), and in response to Al treatment (project accession number: PRJNA517582). The sequence database of RNA in response to Acaphylla theae attack was maintained in our laboratory. The raw read counts for each transcript were calculated using the HISAT2 and featureCounts pipeline and then normalized to transcripts per million (TPM). A heatmap was generated and visualized using TBtools software. The color scale shown with the heatmap represents the TPM counts, and the ratios were log2 transformed.
Nitrogen Treatment and Gene Expression Analysis via qRT–PCR
Since tea plants preferentially take up ammonium (NH4+) rather than NO3–, NH4+ treatment was employed to investigate the expression patterns of CsGARP genes in response to nitrogen stimuli. The expression levels of genes in tea plant were measured in response to different doses of nitrogen treatment. One-year-old tea plants (Camellia sinensis cv. Longjing 43) were grown hydroponically as previously described by Tang et al. (2020). Nutrient solutions consisting of 0, 1.5, 3, and 6 mmol (NH4)2SO4 were used to cultivate tea plants under controlled growth conditions of 26/22°C (light/dark: 14/10 h) air temperature, 70% relative humidity and a light intensity of 200 mmol m–2 s–1 for three weeks. Afterward, young leaves were sampled, and their total RNA was extracted using an RNAprep Pure Plant Kit (Tiangen, Beijing, China). First-strand cDNA was synthesized using a Prime-Script™ RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). qRT–PCR primers of 20 genes were designed with Primer-BLAST10 and are listed in Supplementary Table 2. qRT–PCR amplifications were performed on the LightCycler 480 II Real-Time PCR System (Roche, Switzerland) using 10 μL reaction mixtures consisting of 5 μL of SYBR qPCR Master Mix (High ROX Premixed, Vazyme, Nanjing, China), 1 μL of cDNA, 1 μL of each primer (10 mM), and 3 μL of RNase-free water, as described by Yue et al. (2019). Two technical replicates and three biological replicates were carried out for all qRT–PCR quantifications. The transcript results were calculated according to the 2–ΔΔCt method, with the CsPTB gene serving as a housekeeping gene.
Subcellular Localization Assay
The CsNIGT1b, CsKAN1b, and CsBOA1 were selected to detect subcellular localizations of their encoded proteins on tobacco leaves. The open reading frames (ORFs) of these genes were amplified and inserted into the pCAMBIA2300-35Spro-GFP vector. The recombinant plasmids were transformed into Agrobacterium GV3101 and then transfected into the 4- to 6-week-old tobacco leaves. The infiltrated plants were grown under dark condition for 2 to 3 day and the subcellular localization were observed with the fluorescence confocal microscopy (Carl Zeiss, Germany). The primers used for ORFs cloning were listed in Supplementary Table 2.
Results
Identification of CsGARP Transcription Factors in Tea Plant
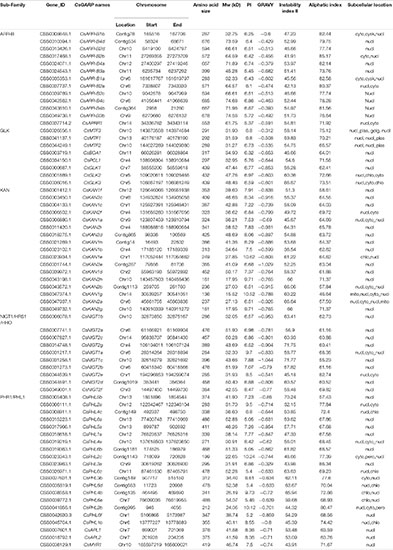
Since GARP proteins are highly similar to MYB or MYB-like TFs in terms of both sequence and structure, candidate genes with typical identical GARP amino acids were compared and screened according to the methods of Safi et al. (2017). In total, 69 GARP genes were identified in tea plant after the putative MYB or MYB-like sequences were removed, and the GARP genes were named based on their subfamily location. The lengths of the sequences of the GARP proteins ranged from 136 (CsKAN1g) to 685 (CsARR-B4c) amino acids, and accordingly, the molecular weight varied from 15.52 kDa to 74.68 kDa. The theoretical isoelectric point (pI) ranged from 5.05 (CsPHL6b) to 10.62 (CsKAN1e). All the proteins had grand average of hydropathy (GRAVY) values below zero, suggesting that they are all hydrophilic proteins. Detailed information concerning these genes, including their name, chromosome position, instability index II value, aliphatic index value and predicted subcellular localization, is listed in Table 1. The GARP proteins were predicted to be most likely located in the nucleus, while several were mainly predicted to be located in the cytoplasm (CsARR-B1b, CsARR-B2b, CsARR-B1a, CsPHL2c and CsPHL3b), mitochondrion (CsKAN1g), nucleus and cytoplasm (CsMTF3), and chloroplast (CsKAN1e, CsPHL4b, and CsPHL1b). Additionally, CsNIGT1b, CsKAN1b, and CsBOA1 were selected to perform subcellular localization assay; as showed in Figure 1, all proteins were localized in cell nucleus as predicted.
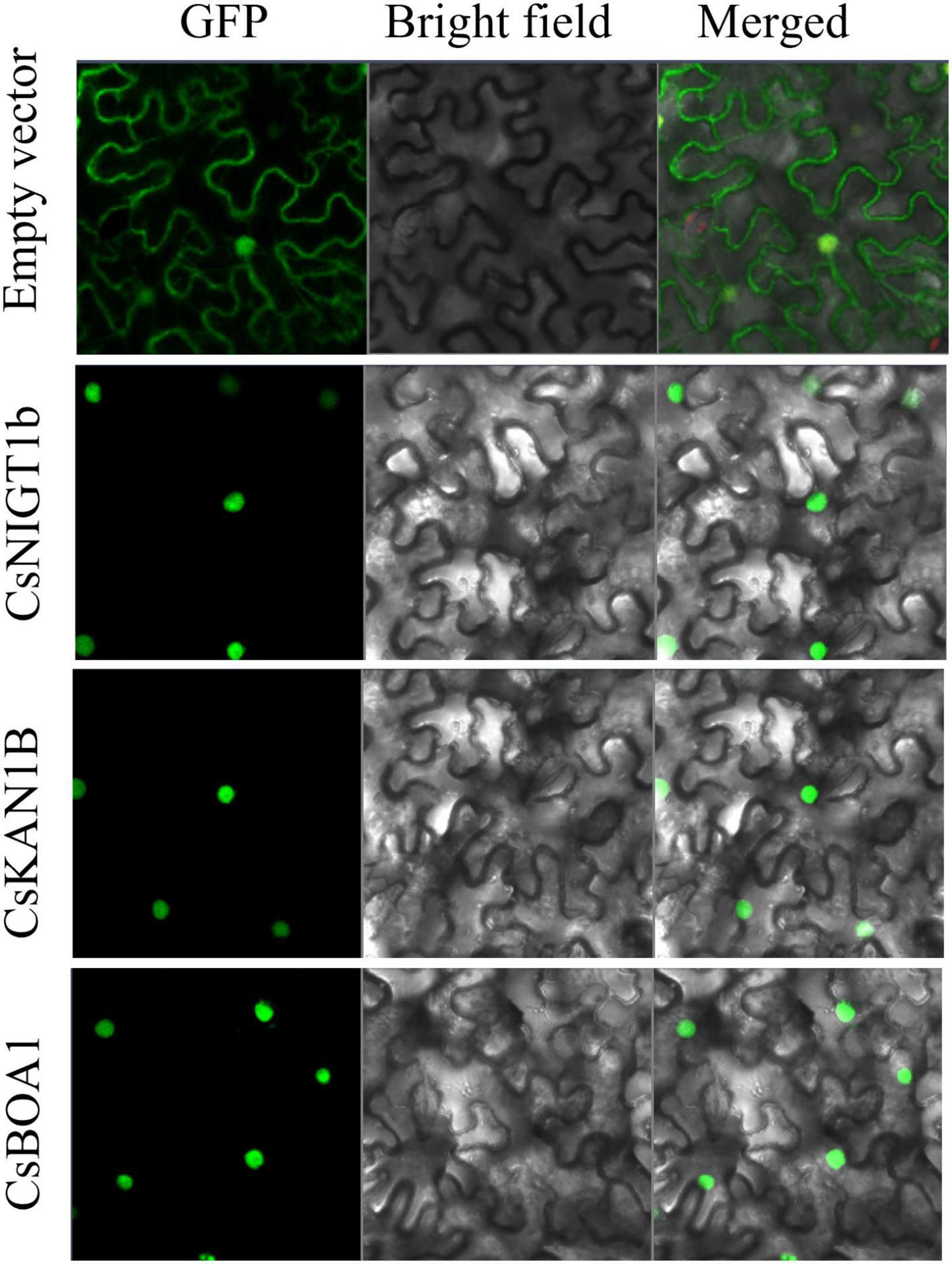
Figure 1. Subcellular localization of CsNIGT1b, CsKAN1b, and CsBOA1 proteins. The free GFP and recombinant GFP proteins of CsNIGT1b, CsKAN1b, and CsBOA1 were transiently expressed in tobacco leaves and observed using a fluorescence confocal microscopy.
A total of 992 GARP genes were also identified and compared among 19 plant species, such as Amborella trichopoda, Nymphaea colorata, Oryza sativa, Sorghum bicolor, Eucalyptus grandis, Arabidopsis thaliana, and Vitis vinifera (Supplementary Table 1). The numbers of GARP genes differed among these plant species, of which Glycine max contained the most abundant GARP genes, while the numbers in Amborella trichopoda, Colin coffee and Ananas comosus were less than 35. In addition, there were more genes in the KAN, PHR1/PHL1 and ARR-B subfamilies than in the NIGT1/HRS1/HHO and GLK subfamilies.
Phylogenetic and Gene Structure Analysis
To investigate the evolutionary relationships between GARPs of tea plant and GARP proteins from other species, a maximum likelihood phylogenetic tree was constructed based on the multiple alignments of the GARP amino acid sequences of 19 plant species. The GARPs could be clearly clustered into five subfamilies, i.e., PHR1/PHL1, KAN, NIGT1/HRS1/HHO, GLK and ARR-B subfamilies, as shown for Arabidopsis (Supplementary Figure 1). A GARP phylogenetic tree of tea plant, Arabidopsis and Vitis vinifera was also constructed and analyzed (Figure 2). In tea plant, 21 members belong to PHR1/PHL1s, 17 to KANs, 12 to ARR-Bs, 10 to NIGT1/HRS1/HHOs and 9 to GLKs. The GLK group was the smallest, while the PHR1/PHL1 group was the largest.
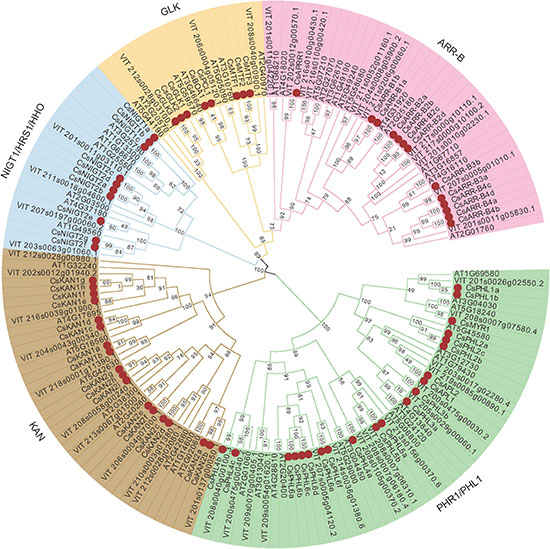
Figure 2. Phylogenetic tree analysis of GARP proteins from tea plant, Arabidopsis and Vitis vinifera constructed by the maximum likelihood (ML) method. The CsGARPs are marked by red dots.
We also analyzed the gene structures of the GARP family members based on the tea plant genome data. As shown in Figure 3, except for CsMTF1 and CsMTF2, which had no introns, all the GARP genes had one to 11 introns. Interestingly, CsMTF3, CsBOA1 and CsPCL1 contained one intron, and six introns were identified in CsGLK2, showing that GLK subfamily genes had high variability in terms of their gene structure. Similarly, two and 11 introns were identified in CsARR-B3a and CsAPRR1, respectively, and the rest of the ARR-B subfamily members had high similarity in terms of gene structure, having 4 to 6 exons, although the length of the genes was highly variable. The NIGT1/HRS1/HHO subfamily genes had 3 to 5 introns, and their gene structure was conserved. Although the lengths of genes in the PHR1/PHL1 and KAN subfamilies were highly variable, the numbers of introns were mainly three to four and five to seven, respectively.
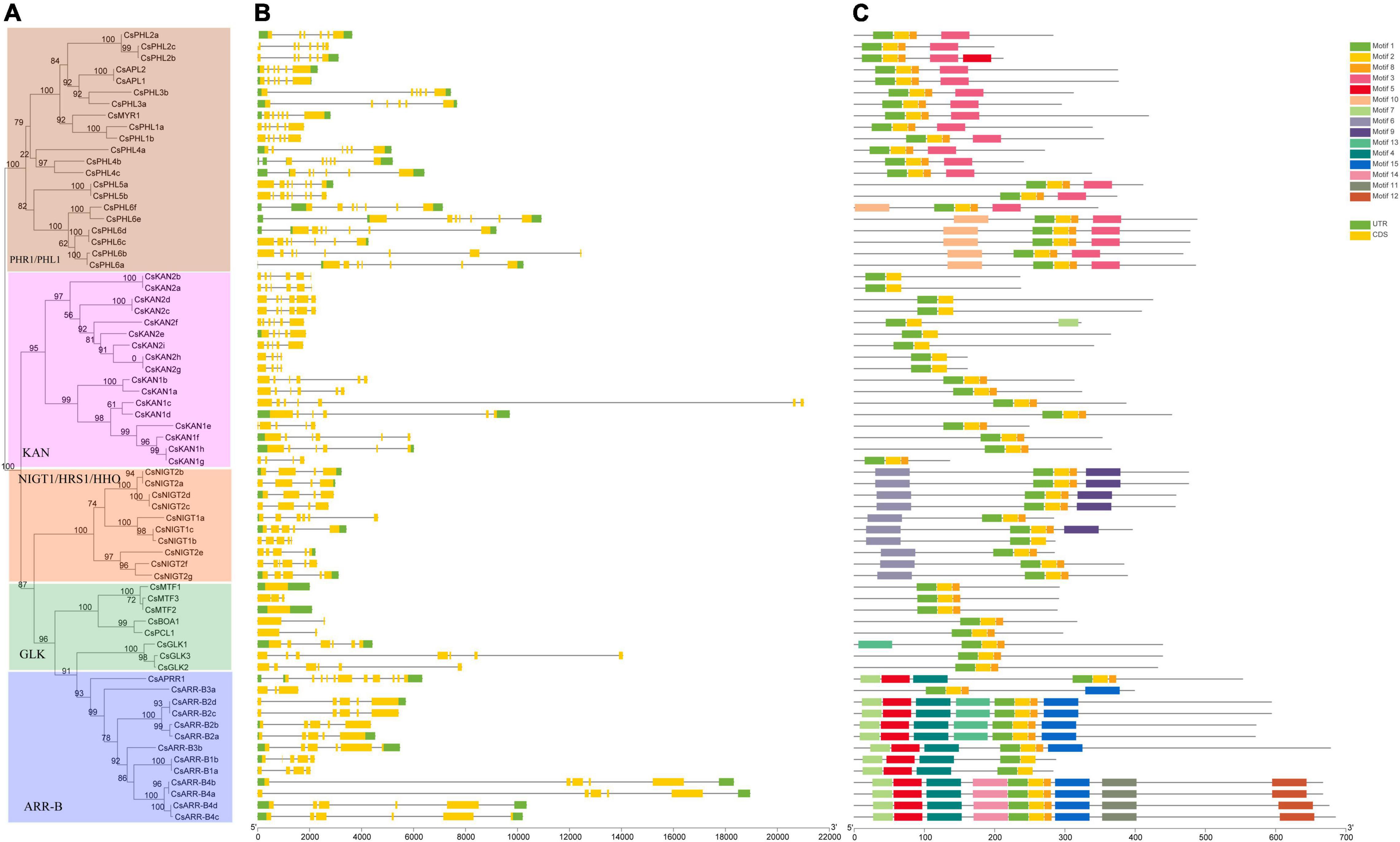
Figure 3. Phylogenetic relationships, gene structure and conserved protein motifs of CsGARP TF genes. (A) The phylogenetic tree was constructed using the ML method; (B) gene structure of CsGARP TF genes; (C) motif composition of CsGARP proteins. The motifs are colored with different boxes.
Conserved Motif Analysis
Sequence alignment of the 69 CsGARPs was conducted using ClustalX, and the results were visualized with Jalview. As shown in Supplementary Figure S2, these sequences had a low degree of conservation within the N- and C-terminal sequences, whereas they were highly conserved in the B-motif sequence, which is the signature motif of GARP proteins and is highly similar to the MYB-like domain. In particular, we observed that the sites of Trp (W), Leu-His (LH), Phe (F), Pro (P), Asn (M), and Ser-His-Leu-Gln (SHLQ) were totally conserved, and other residues, such as Arg (R), Leu-Gly-Gly (LGG), Ala (A), Lys (K), Leu (L), Thr (T) and Tyr (Y), had a high degree of similarity within the B-motif (Figure 4).
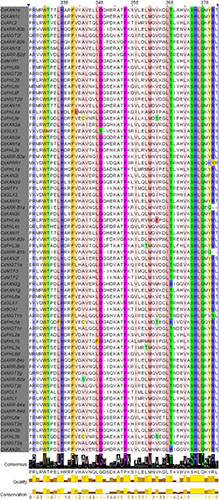
Figure 4. Multiple sequence alignment of the B-motif of 69 CsGARP proteins. The conserved B-motif sequences of 69 CsGARPs were determined using ClustalX, and visual analysis was performed using Jalview.
Additionally, motif analysis was conducted using MEME, and 15 types of conserved motifs were identified (Figure 3C). All the GARP proteins had motif 1 and motif 2, which referred to the B-motif of the GARP signature motif, and except for the KAN2 subgroup and CsARR-B1a/b proteins, all the rest of the proteins contained a short motif 8 after motif 2. In particular, all PHR1/PHL1 members had motif 3, motif 10 was specifically predicted to lie within the N-terminus of PHL6 subgroup members, and motif 6 was identified in all members of the NIGT1/HRS1/HHO subfamily, of which motif 9 was present within the C-terminal sequence of several proteins. Except for CsGLK1, all the GLK proteins contained only motifs 1, 2 and 8. Unexpectedly, more motif members were identified within the ARR-B subfamily members, in which motifs 4, 5, 7, 10, 11, 12, 13, 14 and 15 were present, and apart from CsARR-B3a, almost all the members had motifs 4, 5, and 7 within their N-terminal sequence, while most of them had motif 15 within their C-terminal sequence (Figure 3C).
Chromosomal Distribution and Synteny Analysis
In total, 54 out of the 69 GARP genes were unevenly distributed on 14 chromosomes of the tea plant genome, while the other 15 genes were not mapped to the chromosomes. Chromosome 10 contained 10 genes, which was the most, and chromosome 6 had 8 genes. Five genes were located on chromosome 4. Chromosomes 1, 7, 11, 12, and 13 each contained 4 genes. Only one gene was mapped onto chromosomes 2, 3, and 8, and chromosome 15 had no GARP genes (Table 1 and Supplementary Figure S3).
To analyze the evolution of the CsGARP TF family in tea plant, gene duplication events were analyzed. As shown in Figure 5 and Supplementary Table 3, 40 (57.97%) CsGARP genes were members of 23 gene pairs derived from whole-genome duplication (WGD)/segmental duplication events across chromosomes, eight genes were derived from proximal duplication, and 21 genes (30.43%) were derived from dispersed duplication. Additionally, none of the CsGARP genes were found to be derived from tandem duplication events. These results indicated that tea plant CsGARP genes were mainly generated by gene duplication during evolution. The/Ka/Ks ratio was also calculated for these duplicated gene pairs (Supplementary Table 4), all of which had a Ka/Ks ratio less than 1 (ranging from 0.23 to 0.60), suggesting purifying selective pressure occurred during GARP gene family evolution and a conserved function shared by these genes. Moreover, the divergence of these genes was estimated by the Ks values; the results showed that the duplication period of most of these genes was approximately 25−61 million years ago (MYA), and several genes duplication events occurred 100 MYA (Supplementary Table 4).
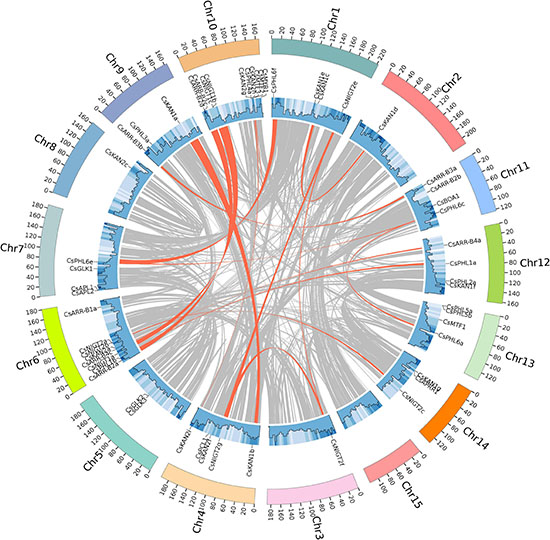
Figure 5. Gene location, duplication, and collinearity analysis of CsGARPs. The gray curves show the details of collinear regions in the tea plant (Suchazao) genome, and the red curves show the gene pairs that have undergone segmental duplication.
To better understand the evolution of GARP genes, a comparative synteny map of Camellia sinensis with Arabidopsis, Vitis vinifera and Actinidia eriantha was constructed. As shown in Figure 6, tea plant GARP genes shared 53 syntenic gene pairs with Arabidopsis and 58 with Vitis vinifera. Additionally, 92 syntenic gene pairs were identified between tea plant and Actinidia eriantha, which constituted the largest number of background collinear blocks. Interestingly, 21 genes were found between tea plant and other plant comparative synteny maps, and these collinear gene pairs were highly conserved within several syntenic blocks, such as CsARR-B2c, CsKAN2g, CsMYR1 and CsNIGT1b on Chr10 and CsARR-B3a, CsBOA1 and CsPHL6c on Chr11, suggesting that these orthologous pairs may be important for plant evolution and that these GARP family genes may play fundamental roles.
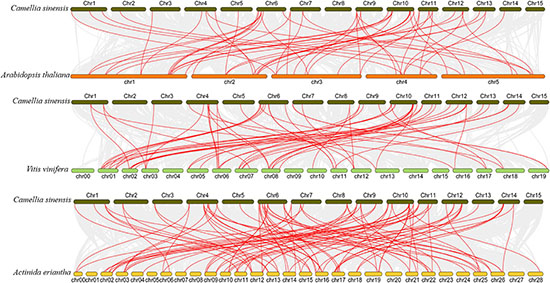
Figure 6. Collinearity analysis of GARP genes between the tea plant genome and Arabidopsis, Vitis vinifera and Actinidia eriantha genomes. The gray lines in the background indicate the collinearity blocks between species. The collinear gene pairs are linked with red lines.
Identification of Cis-Acting Elements
To investigate the regulation of tea plant GARP gene expression, cis-acting elements in the 1500 bp upstream DNA sequence of the genes were identified and analyzed. As shown in Figure 7, the cis-acting elements in the GARP gene promoters could be clustered into seven classes: abiotic stress-responsive elements, biotic stress-responsive elements, tissue-specific expression elements, light-responsive elements, core promoter elements, circadian elements and cell cycle elements. All the gene promoters contained a large number of core elements, especially CAAT-boxes and TATA-boxes. Twenty-one types of light-responsive elements, such as G-boxes, B-boxes, and LAMP elements, were predicted to be present within GARP promoter regions. In addition, numerous stress-related elements, such as AREs, which are associated with anaerobic condition responsiveness, MBSs, which are involved in drought responsiveness, CGTCA motifs, which are related to methyl jasmonate (MeJA) responsiveness, and AREBs, which are involved in abscisic acid responsiveness, were widely distributed among the GARP gene members in tea plant. In particular, the stress-related cis-acting elements were highly clustered within the GARP gene promoter regions, especially in the ARR-B and GLK gene members, suggesting that GARP genes play critical roles in regulating abiotic and biotic stress responses. Additionally, certain elements involved in plant tissue-specific expression, such as O2_sites, GCN4 motifs, and RY-elements, were identified in the promoter of GARP genes, and more than half of the promoters of GARPs (36) contained at least one CAT-box, which is related to meristem expression, indicating that GARP genes are important for tea plant growth and development. Moreover, several genes had one predicted circadian-related or MSA-like element in their promoter region.
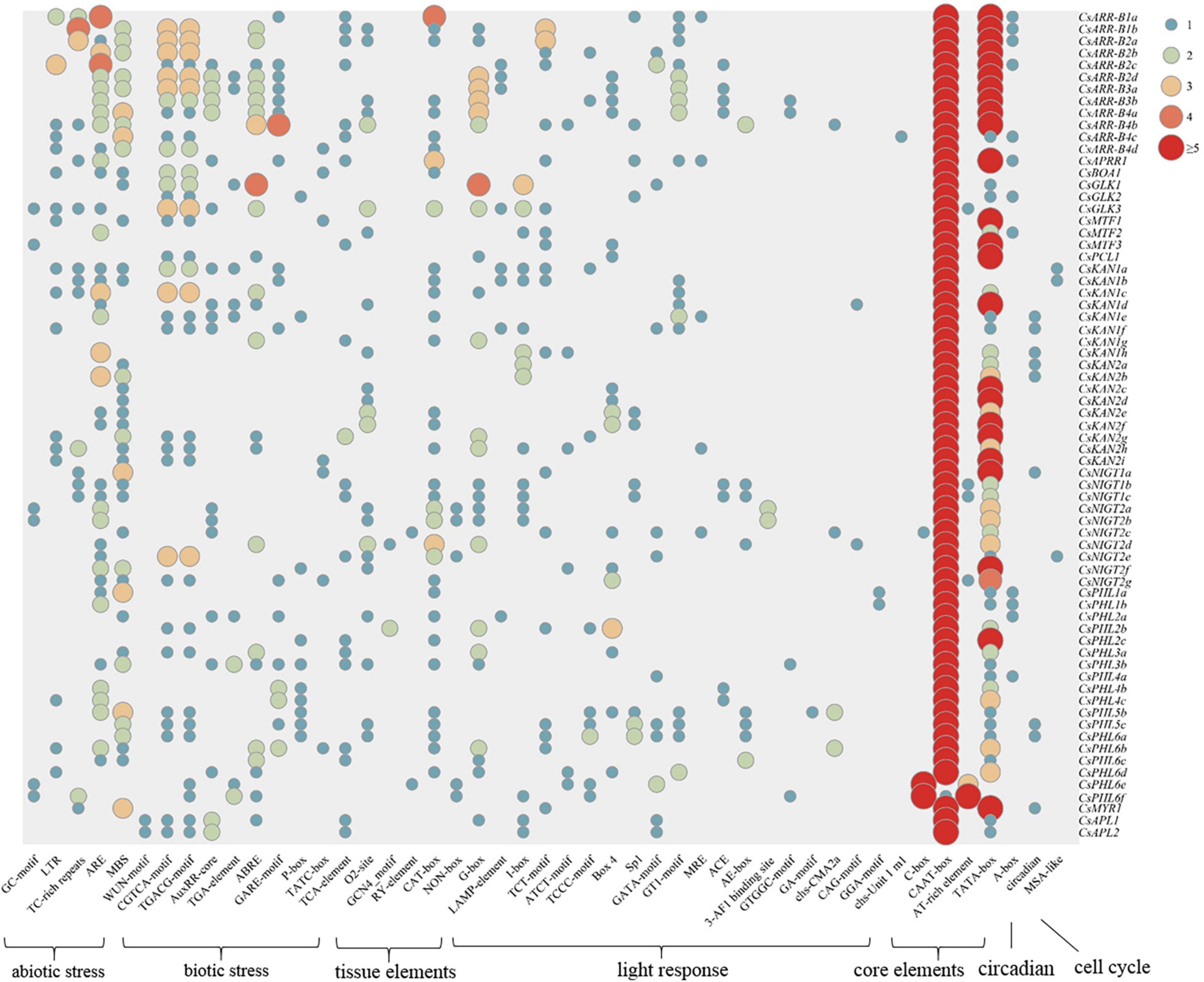
Figure 7. Analysis of cis-acting elements of 69 CsGARP genes. The cis-acting elements were classified into 7 types: abiotic stress-responsive elements, biotic stress-responsive elements, tissue-specific expression elements, light-responsive elements, core promoter elements, circadian elements and cell cycle elements.
Expression Patterns in Different Tissues During Different Seasons
The expression patterns of GARP genes in the roots, leaves, stems, buds and flower tissues at different seasons across years were determined and analyzed based on tea plant RNA-seq data (Wang et al., 2020c). A heatmap was generated and showed that these genes could be clustered into six groups according to their expression profiles (Figure 8). The genes of group 1 (11 genes), which were mainly KAN and NIGT1/HRS1/HHO subfamily genes, had low expression levels among the tested tissues. Similarly, the genes clustered into group 2 had low or undetectable transcript levels in the leaf, stem, bud and flower tissues, but they exhibited high levels in the roots, suggesting that these genes, especially those of several KAN subfamily members, had strong root-specific expression. On the other hand, several genes of group 6 had lower expression levels in the roots than in other tissues. Interestingly, 38 genes belonging to groups 3, 4 and 5, especially the group 3 genes, were highly expressed in all of these tissues. For instance, five PHR1/PHL1 subfamily genes in group 3, i.e., CsPHL4b, CsPHL3b, CsPHL4c, CsPHL6e and CsPHL6f, presented high transcript abundance in the roots, leaves, stems, buds and flowers, implying that they might play critical roles in tea plant growth and development. Interestingly, we found that the expression of CsGLK1 and CsGLK2 was lower in the roots than in the other tissues and that a small group of KAN genes (CsKAN1d/e/f/g/h) had relatively high expression levels in the stems and buds.
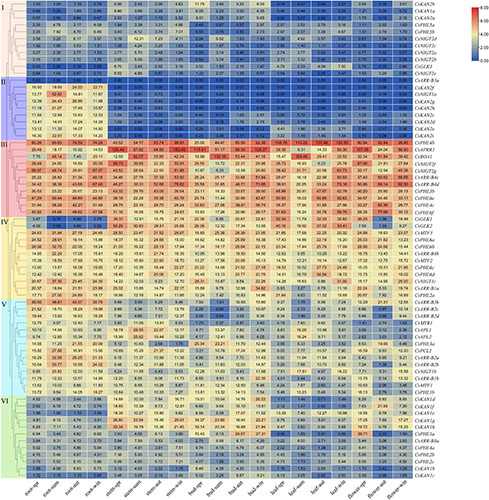
Figure 8. Expression analysis of CsGARPs in different tissues in different seasons. The CsGARPs were clustered into 6 groups with different colors according to their expression levels calculated with TPM values. The red and blue colors represent high and low expression levels, respectively.
Moreover, the tissue expression patterns of target genes in the spring, summer, autumn and winter seasons were investigated, and their patterns fluctuated with the seasons. For instance, in all the tested tissues, CsAPRR1, CsGLK1, CsGLK2, CsGLK3 and CsNIGT2f had high transcript levels in the spring; CsAPL1/WDY1, CsAPL2/WDY2 and CsBOA1 had high transcript levels in the summer; CsPHL6e, CsPHL6f and CsMTF2 had high transcript levels in autumn; and CsARR-B1a, CsARR-B2c, CsARR-B4c, CsARR-B4d and CsNIGT2g had high transcript levels in the winter. CsPHL4b was expressed at high levels in both autumn and winter.
Expression Patterns in Different Leaf Color Stages of Tea Plant
We investigated the expression patterns in the white-, yellow–green and green-leaf stages during tea plant young leaf development based on RNA-seq data obtained from a unique albino cultivar named Anjibaicha. More than half of the genes had low or undetectable transcript levels among these three test samples, and most of the gene expression levels were higher in the green stage than in the white and yellow-green stages (Figure 9). In particular, we found that several genes, such as CsGLK1, CsGLK2, CsPHL6f, CsPHL6d, and CsNIGT1b, were significantly upregulated in the green stage but were expressed at low levels in the white and yellow-green stages, suggesting that these genes are important for tea plant leaf color development. On the other hand, ten genes, including CsBOA1, CsPCL1 and CsAPRR1, presented higher expression levels in the white and/or yellow-green stages than in the green stage; this was especially the case for CsBOA1, whose expression was highest in the yellow-green stage followed by the white stage, indicating that this gene might be involved in leaf color changes from the white stage to the green stage. The changes in GARP gene expression levels in different leaf color stages implied that these genes may function in regulating the transformation of tea leaves from being white to becoming green in color.
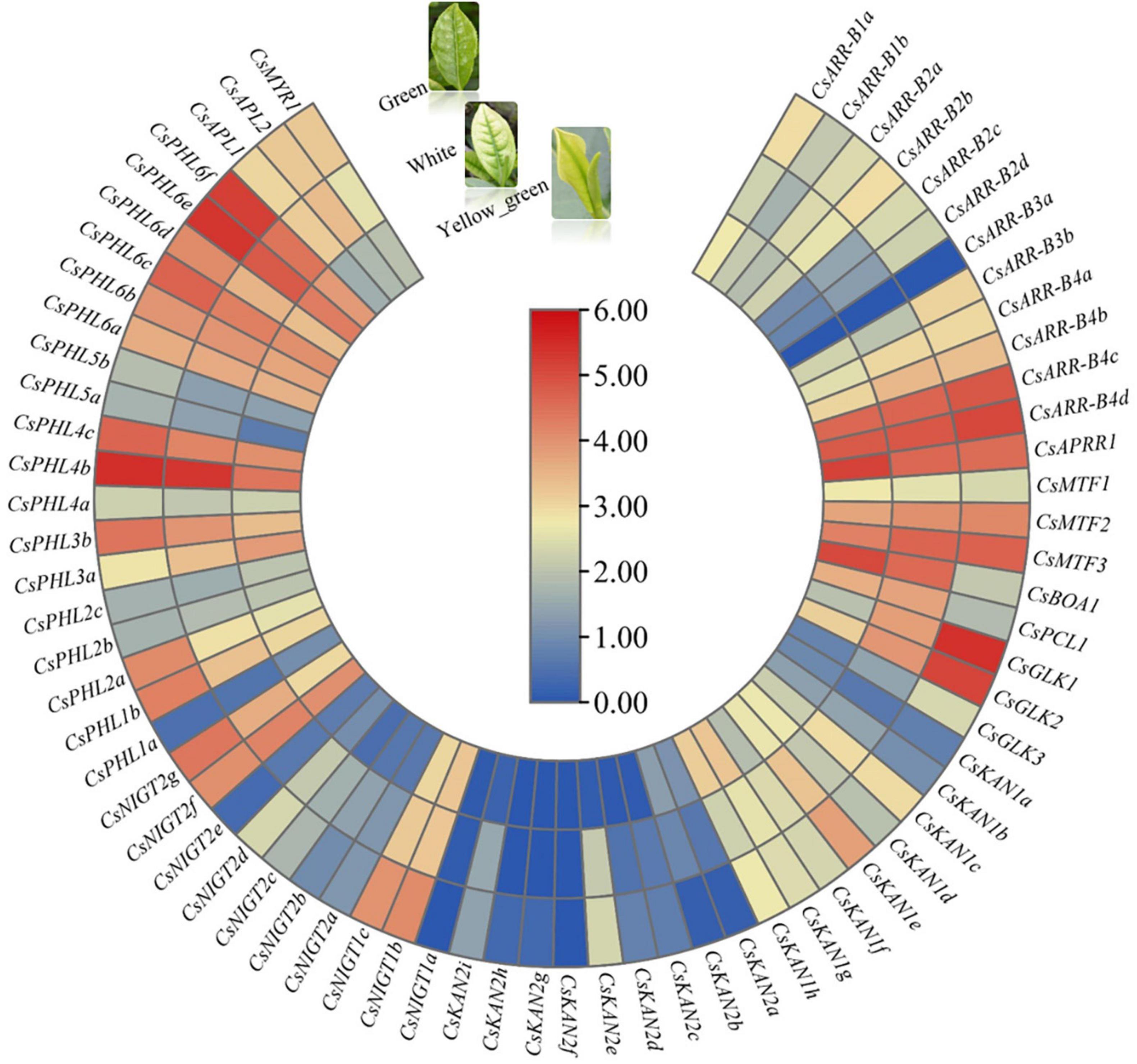
Figure 9. Expression analysis of CsGARPs in different tea plant leaf color stages. The expression levels of CsGARPs were calculated with TPM values. The red and blue colors represent high and low expression levels, respectively.
Expression Patterns During Cold Stress
We investigated and compared the expression patterns of GARP genes in response to cold stress between the fish leaf (FL) and the two-leaf-and-one-bud (TAB) stages (Hao et al., 2018). As shown in Figure 10, several genes, including CsAPRR, CsARR-B1b, CsBOA1, CsGLK1, CsGLK2, CsKAN1d, CsKAN1e, CsNIGT2d, CsNIGT1b, CsNIGT1c, CsPCL1, CsPHL2b, and CsPHL4a, exhibited different patterns between the FL and TAB stages upon cold treatment, suggesting that these genes may be associated with the difference in responsiveness of tissues under cold stress; the rest of the genes exhibited a similar tendency in response to cold stress. Interestingly, we found that certain ARR-B (5), GLK (2), KAN (5), NIGT1/HRS1/HHO (2), and PHR1/PHL1 (2) subfamily genes were considerably upregulated, whereas most differentially expressed PHR1/PHL1 subfamily genes were downregulated.

Figure 10. Expression analysis of CsGARPs in response to cold stress in FL and TAB tissues. The expression levels of CsGARPs were calculated with TPM values. The red and blue colors represent high and low expression levels, respectively.
Expression Patterns in Response to Salinity and Drought Stress
To evaluate the putative roles of GARP genes in the tea plant response to drought and salt stress, transcriptome data collected during continuous salinity and drought stress were isolated and analyzed (Figure 11). Most CsKAN subfamily member genes (11/17) had low or undetectable transcript levels among the tested samples. However, both CsKAN2h and CsKAN2g were dramatically upregulated under drought and salt treatment, whereas the expression of CsKAN1b, CsKAN1c, CsKAN1d and CsKAN1e was repressed. Interestingly, we found that the expression of CsGLK1, CsGLK2, CsGLK3, CsPCL1, and CsMTF1 was significantly downregulated in response to drought and/or salt stress, whereas CsMTF2, CsMTF3 and CsAPRR1 displayed upregulation expression patterns at several points during the stress treatment. In addition, most ARR-B subfamily member genes, such as CsARR-B2b, CsARR-B2c and CsARR-B3b, were downregulated under stress, while CsARR-B4c and CsARR-B4d were induced by stress. Notably, several of NIGT subfamily member genes, including CsNIGT1b, CsNIGT1c, CsNIGT2g and CsNIGT2f, were upregulated in response to drought and salt treatments, while the expression of CsNIGT2a-e was suppressed. In addition, the expression levels of most genes in the PHR1/PHL1 subfamily, such as CsPHL1b, CsPHL3b and CsPHL6b, were downregulated under drought and salt stress, while certain members—especially CsPHL6f—were considerably upregulated.
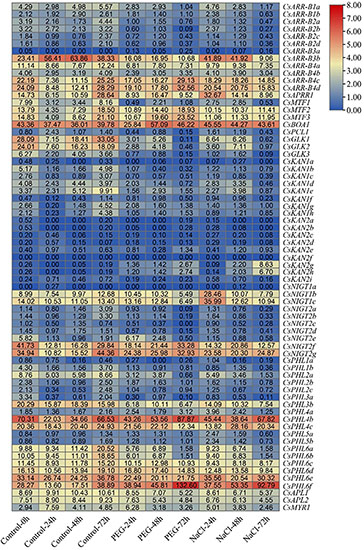
Figure 11. Expression analysis of CsGARPs in response to drought and salt stress treatments. The expression levels of CsGARPs were calculated with TPM values. The red and blue colors represent high and low expression levels, respectively. N: control conditions, PEG-N: drought stress imposed via 25% polyethylene glycol (PEG)6000, NaCl-N: salt stress imposed via 200 mM NaCl.
Expression Patterns Under AL Treatment
Tea plant is well recognized as having high Al tolerance. In this study, the expression patterns of GARP genes in the roots of tea plant treated with different Al doses (0, 0.4, 1 and 4 mmol L–1) were analyzed (Figure 12A). With an increase in Al concentration, 19 and 24 genes were downregulated and upregulated, respectively, and the expression of the remaining genes was not affected by Al treatment. For instance, the expression levels of CsAPRR1, CsGLK1, CsKAN2a, and CsNIGT2b were gradually downregulated with an increasing Al dose from 0 to 4 mmol L–1, and the transcript levels of CsARR-B3b, CsBOA1, CsKAN2d and CsKAN2e increased, suggesting that the expression of these genes somewhat depended on the Al dose. Additionally, we found that the expression of CsNIGT2g and CsPHL4b was high in the tested root tissues, but the expression of these genes did not significantly change under Al treatment, indicating that these genes highly expressed in the roots may play important roles in tea plant root growth and development.
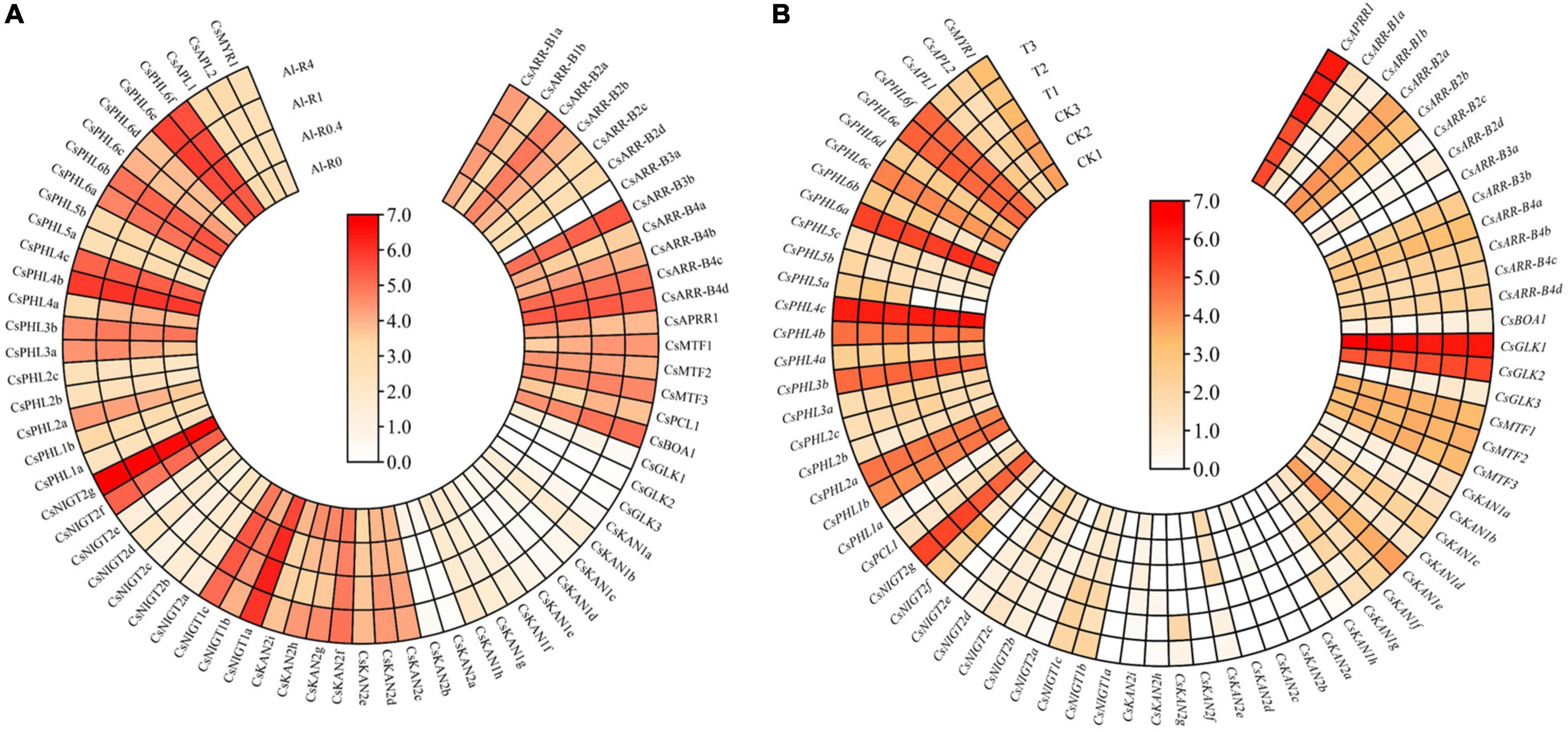
Figure 12. Expression analysis of CsGARPs under Al treatment (A) and Acaphylla theae attack (B). The expression levels of CsGARPs were calculated with TPM values. The red and blue colors represent high and low expression levels, respectively.
Expression Patterns in Response to Acaphylla theae Attack
We investigated the expression patterns of GARP genes in response to Acaphylla theae attack. As shown in Figure 12B, the expression of most genes in leaves in response to Acaphylla theae infestation was not affected compared with that in healthy leaves. The expression levels of CsKAN1h, CsNIGT1c, CsNIGT2f, and CsPHL6b were significantly upregulated after Acaphylla theae attack, whereas the transcript levels of CsKAN2e and CsNIGT2c strongly decreased, suggesting that these differentially expressed genes might be involved in the tea plant response to biotic stress, such as attack from Acaphylla theae.
Gene Expression via qRT–PCR in Response to Nitrogen Stimuli
Tea plants preferentially take up NH4+ rather than NO3–. Using qRT–PCR, we further explored the expression patterns of 20 CsGARP genes in response to NH4+ treatment. As shown in Figure 13, the expression of six out of eight NIGT1/HRS1/HHO members (CsNIGT1b, CsNIGT1c, CsNIGT2c, CsNIGT2e, CsNIGT2f, and CsNIGT2g) significantly increased under NH4+ treatment, while the transcription of CsNIGT1a was repressed, and NH4+ treatment did not affect the expression of CsNIGT2a. After NH4+ treatment, the expression level of CsGLK1 also significantly increased, whereas the low NH4+ condition repressed CsBOA1 transcription. The expression levels of CsARR-B4c and CsKAN1d were not affected by NH4+ treatment. Additionally, the expression of CsPHR1/PHL1 genes was upregulated under certain treatments, but the relative expression level fold-changes were less than 2.0. These findings could provide putative gene candidates for the functional characterization and improvement of nitrogen absorption and utilization in future tea plant breeding programs.

Figure 13. Expression analysis of CsGARPs under nitrogen treatment. The relative expression levels of CsGP genes were determined using qRT–PCR, and the results were calculated using the 2–ΔΔCt method and normalized to those of the CsPTB housekeeping control gene.
Discussion
The members of the GARP TF family play crucial roles in various physiological processes and stress responses in plants. According to the phylogenetic tree and conserved domain analysis, GARP TFs can be classified into PHR1/PHL1, KAN, ARR-B, NIGT1/HRS1/HHO and GLK subfamilies (Safi et al., 2017). GARP TFs contain a signature conserved sequence called the B-motif, which is highly similar to the MYB-like domain (Hosoda et al., 2002; Safi et al., 2017); hence, GARP TFs have been somewhat inaccurately categorized as MYB or MYB-like superfamily TFs in previous reports. The functions of GARP TF genes have been well elucidated in model plant species and other plant species, and 56 GARP members have been identified in Arabidopsis; however, a systematic genome-wide identification and analysis of GARP TFs in plants is currently lacking. In recent years, several subfamily members, such as ARR-Bs in peach (Zeng et al., 2017) and tomato (Wang et al., 2020a) and GLKs in rice (Bhutia et al., 2020), have been identified and investigated across the genome; nonetheless, there have been no comprehensive investigations of the GARP gene family, and the functions of the GARPs are largely unknown in plants. In this study, we performed a genome-wide identification of the CsGARP gene family based on the feature sites of the B-motif that differed from those of the MYB-like domain. Sixty-nine CsGARP genes were identified in tea plant (Table 1), and their gene structure, chromosome location, phylogenetic relationships, conserved domains, cis-acting elements and tissue-specific expression patterns under various treatments were investigated.
We constructed a phylogenetic tree to perform a systematic analysis of GARP family genes in plants, which included tea plant and 18 other plant species. A total of 992 GARP proteins composed the tree and clustered into five subtrees, which is consistent with results for Arabidopsis (Safi et al., 2017), suggesting that the GARP TF family is ubiquitous in plants. Among these selected species, tea plant contains more GARP TFs than most other plant species do. Although tea plant has the largest genome size (3.1 Gb) among the species composing the tree, the number of GARP genes in tea plant was lower than the number in Glycine max, Actinidia chinensis and Populus trichocarpa, whose genome size was 0.994 Gb, 0.604 Gb and 0.434 Gb (Supplementary Table 1), respectively, indicating that the GARP numbers had no apparent relationship with genome size in plants. Generally, the numbers of GARP genes are mainly increased in certain subfamilies controlling developmental processes and the response to Pi, indicating that these pathways are important for plant evolution. We also found that the orthologous GARP genes between tea plant and Arabidopsis thaliana, between tea plant and Actinidia chinensis, and between tea plant and Vitis vinifera were primarily clustered into several syntenic blocks, such as on chromosome 4, chromosome 6, chromosome 10, and chromosome 11 of tea plant, and tea plant and Actinidia eriantha shared 92 syntenic gene pair relationships within the GARP family, indicating that tea plant and Actinidia eriantha are closely evolutionarily related. Wei et al. (2018) indicated that tea plant experienced two WGD events after the core-eudicot whole-genome triplication-γ (WGT-γ) event, which was shared one time with Actinidia chinensis. Recently, Wang et al. (2021) showed that tea plant experienced a Polemonioids-Primuloids-core Ericales (PPC)-WGD (∼63 MYA) event, which resulted in the expansion of genes involved in flavor compound biosynthesis and the stress response. Among the CsGARP TF family member genes, 40 out of 69 mainly resulted from segmental duplication events, 21 were derived from dispersed duplication events, and eight genes were derived from proximal duplication events (Figure 5 and Supplementary Table 3). We also estimated the divergence years of CsGARP genes and found that most of the genes originated approximately 25−61 MYA, and some originated approximately 100 MYA (Supplementary Table 4). Although the WGD events and the exact timepoint at which they occur in tea plant are inconclusive, most CsGARP genes had duplicated around the time of the most recent WGD event. Furthermore, Ka/Ks analysis showed that all of the CsGARP genes were under purifying selection, indicating that gene duplication events were exposed to strong purifying constraints during evolution and that duplication events, except for tandem duplications, are the main driving force in the expansion of CsGARP genes in tea plant.
Although GARP TFs are usually clustered into the MYB superfamily and share a MYB-like domain, the functional domain of the B-motif is distinctly different from that of the MYB domain. Safi et al. (2017) indicated that, unlike MYB proteins, GARP TFs contain only the first of three regularly spaced tryptophan residues (W) and have a consensus amino acid sequence SHLQ (K/M) (Y/F), while MYB TFs instead have the conserved sequence SHAQK(Y/F)F. These differences are present within the functional sites, implying that GARP TFs do not belong to the MYB superfamily and that GARP TFs can be separated from the MYB superfamily TFs according to the phylogenetic tree and functional domain analysis. All CsGARP proteins were predicted to contain motif 1 and motif 2, which are components of the B-motif and are important for DNA binding. In addition, different subfamily members had specific motifs. CsPHR1/PHL1 TFs have a motif 3 consistent with a coiled-coil domain, which is involved in protein–protein interactions, after the MYB-related domain; hence, these subfamily members were usually called MYB-CC family members in several previous studies (Sun et al., 2016; Wang et al., 2018). CsNIGT1 subfamily TFs contain motif 6 at the N-terminus, which is rich in hydrophobic amino acids and named the hydrophobic and globular domain (HGD), the function of which is unknown, and several members contain motif 9, which is predicted to be an EAR-like motif critical for inhibiting gene expression (Wang et al., 2020b; Li Q. et al., 2021). Among these five subfamilies, ARR-B proteins have more, unique motifs (Figure 3), implying that they may have diverse functions. Except for the MYB-like DNA-binding domain, ARR-B proteins contain a key functional receiver (REC) domain at their N-terminus, which functions as a phosphorylation-mediated switch involved in regulation and plays important roles in cytokinin-mediated plant growth and development (Hosoda et al., 2002; Cho et al., 2016). On the other hand, although the lengths of CsGARP genes are different, the structures of genes in the same subfamily are conserved, and each subfamily can also be classified into different subgroups. Therefore, the consensus motifs and structures of genes in each subfamily are strongly associated with the results of the subfamily classification, and the different and unique conserved motifs and sites between GARP proteins and MYB-like proteins show that GARP and MYB TFs should not be clustered into MYB superfamily TFs.
In tea plant, CsKAN2 genes were mainly transcribed in the roots, while CsKAN1s had higher levels in the stems, buds and flowers (Figure 8), indicating that CsKAN1 and CsKAN2 may play different roles among these tissues. KAN1 is a transcriptional repressor and plays key roles in the regulation of abaxial identity, leaf development, and meristem formation by mediating the auxin response in plants (Kerstetter et al., 2001; Candela et al., 2008; Huang et al., 2014; Xie et al., 2015; Bhatia et al., 2019; Ram et al., 2020). Recently, we observed that several CsKAN1 genes were significantly upregulated in zigzag-shaped stems compared with normal stems (Cao et al., 2020). High transcription of CsKAN1s in the stems and buds during the four seasons showed that KAN1 genes are involved in stem formation and bud development. Additionally, by interacting with ULTRAPETALA trxG genes, KAN1 also regulates gynoecium patterning in Arabidopsis (Pires et al., 2014). CsKAN1s exhibited relatively high transcription in the flowers, showing that gynoecium development in tea plant is also controlled by CsKAN1s genes. Given that KAN1 and KAN2 have partially similar functions, the regulatory functions of KAN2 are not clear. Most CsKAN2s genes were predominantly expressed in tea plant roots, and previous reports showed that KAN genes play crucial roles in lateral root development (Hawker and Bowman, 2004). Considering that KAN genes play critical functions in the plant auxin response, we speculate that CsKAN2s genes may be essential for root development through integration with the auxin pathway. On the other hand, we found that the expression levels of certain CsKAN2s genes, such as CsKAN2a/b/c/d, were upregulated in tea plant leaves in response to cold stress (Figure 10), that CsKAN2g/h was induced upon drought and salt stresses (Figure 11), that CsKAN2a/b/g/h was repressed and CsKAN2c/d was upregulated in response to Al treatment, and that CsKAN2e was downregulated in response to biotic stress, indicating that these genes might also play important roles in the tea plant response to stress stimuli.
It has been well recognized that NIGT1/HRS1/HHO and PHR1/PHL1 are key regulatory TFs involved in the plant response to nitrogen and Pi. In this study, we found that most CsNIGT and CsPHR1/PHL1 genes were specifically or primarily expressed in the roots (Figure 8). In Arabidopsis, NIGT1 genes (HRS1 and HHO1) are expressed in the transition domain of the root apical meristem and in the elongation zone (Medici et al., 2015), and PHR1/PHL1-related genes are primarily expressed in the roots and shoots (Rubio et al., 2001; Sun et al., 2016). These tissues play important roles in the plant response to nutrients, including nitrogen and Pi. NIGT1/HRS1/HHO TFs have been widely reported to be induced by NO3–. Tea plant is a high nitrogen- and Pi-consuming plant species and is considered to preferentially take up NH4+ (Tang et al., 2020). In this study, we determined the expression patterns of 20 CsGARP genes, i.e., 8 CsNIGT1/HRS1/HHO genes, 7 CsPHR1/PHL1 genes, 2 CsGLK genes, 2 CsARR-B genes and CsKAN1d gene, in response to NH4+ treatment (Figure 13). Interestingly, although NIGT1/HRS1/HHO genes have been characterized as NO3–-induced genes (Sawaki et al., 2013; Maeda et al., 2018), 6 out of 8 CsNIGT1/HRS1/HHO genes were significantly upregulated in response to NH4+ treatment, indicating that CsNIGT1s are also involved in the tea plant response to NH4+ treatment and might play important roles in nitrogen absorption and utilization in tea plant. Recently, numerous differentially expressed genes were identified via transcriptome sequencing in tea plant in response to nitrogen treatment (Li et al., 2017; Ruan et al., 2021), and our results could provide valuable candidate genes for further studies on the transcriptional regulatory mechanism of the tea plant response to nutrients. Additionally, in this study, we found that the expression levels of several CsNIGT1/HRS1/HHO and CsPHR1/PHL1 genes were also regulated by stress treatments. Environmental stresses, including drought, cold and salt, impact plant growth and physiological processes by modulating nutrient metabolism, such as carbon nitrogen and Pi metabolism (Ding et al., 2018). Recently, the occurrence of cross-talk among nutrient utilization, normal growth and development and stress response has been well established (Wu et al., 2020; Han et al., 2021; Sevanthi et al., 2021), yet the putative functions of NIGT1/HRS1/HHO and PHR1/PHL1 genes and their mediation of the absorption and utilization of nitrogen and Pi under stress are worthy of further investigation.
Generally, CsARR-B genes were also expressed at relatively high levels in the roots (Figure 7), suggesting that this subfamily of TFs may be involved in root growth regulation. Salvi et al. (2020) indicated that, by interacting with auxin and PLETHORA, the ARR-B gene ARR1 controls expansion of the division zone in Arabidopsis roots. Given that ARR-Bs serve as key regulators involved in cytokinin signal transduction, increasing evidence has shown that ARR-Bs also play important roles in the signaling of other phytohormones, such as auxin and zeatin (Safi et al., 2017; Xiang et al., 2021). Additionally, genes such as CsAPRR1 were expressed at high levels in the stems, buds and flowers (Figure 8). In these tissues, the phytohormones cytokinin, auxin and zeatin accumulate at high levels and facilitate cell division and expansion. Moreover, the transcript levels of several of these genes, including CsARR-B4c and CsARR-B4d, were maintained at high levels among different tissues, implying that these genes may play fundamental roles in tea plant development and growth. In addition, we found that the transcript levels of several genes, such as CsARR-B1b, CsAPRR1, and CsARR-B4c, were significantly regulated in response to stress treatments, including cold, drought, salt and Al (Figures 10, 11, 12A). Correspondingly, it has been widely shown that ARR-B genes are upregulated in response to cold and heat stress (Peng et al., 2015; Xiang et al., 2021) and are considered key cold-responsive genes (Gong et al., 2018). Therefore, by modulating cytokinin, auxin and zeatin signaling, these differentially expressed CsARR-B genes might participate in the tea plant response to stress at some level.
GLK genes have been shown to function extensively in chloroplast development, photosynthesis regulation, the stress response, and fruit development (Fitter et al., 2002; Powell et al., 2012; Nadakuduti et al., 2014; Nagatoshi et al., 2016; Lupi et al., 2019). In particular, GLK genes were the first genes identified to control chloroplast structure, as defects in this gene resulted in pale green leaf blade color (Hall et al., 1998). In the present study, CsGLK genes were expressed at high levels in the leaves (Figure 8), which is associated with their functions in chloroplast and photosynthesis of leaves. Interestingly, we also found that the expression levels of CsGLK1/2 genes were lower in white and yellow-green leaves than in green leaves (Figure 9). Numerous studies have demonstrated aberrant morphology of chloroplast ultrastructure and low chlorophyll contents in white, yellow, and yellow-green tea leaves. Recently, Hao et al. (2020) identified pale green branches with chloroplast defects from tea plant and found that CsGLKs were significantly downregulated in these mutant tissues. Unexpectedly, in the present study the expression levels of CsBOA1 and CsPCL1 were upregulated in both the white and the yellow-white leaves and downregulated in the green leaves (Figure 9). However, the functions of BOA1 and PCL1 are largely unknown in plants. Therefore, to some extent, we propose that the albinism and etiolation of tea plants mainly resulted from the downregulation of CsGLKs. In addition, we found that this subfamily of genes was regulated by stress treatments; this was especially the case for CsBOA1, which was significantly induced in response to Al treatment and drought (Figures 11, 12A), whereas it was downregulated in mature leaves in response to cold but upregulated in young leaves (Figure 10), implying that CsGLK genes are involved in tea plant stress responsiveness. Notably, Ahmad et al. (2019) showed that the GLK1/2-WRKY40 transcription module plays a negative regulatory role in the abscisic acid (ABA) response, indicating that GLK genes are also involved in ABA signaling. Overall, the current study provides insights into the diverse functions of CsGARP genes and lays a foundation for future research to determine the functions of CsGARP genes in tea plant.
Data Availability Statement
The original contributions presented in this study are publicly available. This data can be found here: PRJNA843954.
Author Contributions
CY conceived and designed the experiments and wrote the original draft. CY, QC, and JH performed the experiments. CL, JH, and QC analyzed the results. LL and LZ reviewed and edited the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (SWU-KR22016), the National Natural Science Foundation of China (31600555), the Germplasm Creation Research Program of Southwest University, Tea resources development and Utilization Function Laboratory 2021 (8).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.947072/full#supplementary-material
Supplementary Figure 1 | Phylogenetic tree of GARP proteins from 19 species constructed by the ML method.
Supplementary Figure 2 | Overview of multiple sequence alignment of 69 CsGARP proteins. The sequence alignment of 69 CsGARPs was conducted using ClustalX, and visual analysis was performed through Jalview.
Supplementary Figure 3 | Chromosomal locations of 69 CsGARP genes.
Footnotes
- ^ http://tpia.teaplant.org
- ^ http://kiwifruitgenome.org/organism/5
- ^ https://phytozome-next.jgi.doe.gov/pz/portal.html
- ^ https://www.arabidopsis.org/
- ^ http://evolgenius.info/#/
- ^ http://gsds.gao-lab.org/index.php
- ^ http://meme-suite.org/tools/meme
- ^ http://tpdb.shengxin.ren/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ https://www.ncbi.nlm.nih.gov/tools/primer-blast/
References
Ahmad, R., Liu, Y., Wang, T. J., Meng, Q., Yin, H., Wang, X., et al. (2019). GOLDEN2-LIKE transcription factors regulate WRKY40 expression in response to abscisic acid. Plant Physiol. 179, 1844–1860. doi: 10.1104/pp.18.01466
Argyros, R. D., Mathews, D. E., Chiang, Y. H., Palmer, C. M., Thibault, D. M., Etheridge, N., et al. (2008). Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 20, 2102–2116. doi: 10.1105/tpc.108.059584
Bhatia, N., Åhl, H., Jönsson, H., and Heisler, M. G. (2019). Quantitative analysis of auxin sensing in leaf primordia argues against proposed role in regulating leaf dorsoventrality. Elife 8:e39298. doi: 10.7554/eLife.39298
Bhutia, K. L., Nongbri, E. L., Gympad, E., Rai, M., and Tyagi, W. (2020). In silico characterization, and expression analysis of rice golden 2-like (OsGLK) members in response to low phosphorous. Mol. Biol. Rep. 47, 2529–2549. doi: 10.1007/s11033-020-05337-2
Candela, H., Johnston, R., Gerhold, A., Foster, T., and Hake, S. (2008). The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell 20, 2073–2087. doi: 10.1105/tpc.108.059709
Cao, D., Liu, Y., Linlong, M., Liu, Z., Li, J., Beibei, W., et al. (2021). Genome-wide identification and characterization of phosphate transporter gene family members in tea plants (Camellia sinensis L. O. kuntze) under different selenite levels. Plant physiology and biochemistry. 166, 668–676. doi: 10.1016/j.plaphy.2021.06.038
Cao, H., Wang, F., Lin, H., Ye, Y., Zheng, Y., Li, J., et al. (2020). Transcriptome and metabolite analyses provide insights into zigzag-shaped stem formation in tea plants (Camellia sinensis). BMC Plant Biol. 20:98. doi: 10.1186/s12870-020-2311-z
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, M., Ji, M., Wen, B., Liu, L., Li, S., Chen, X., et al. (2016). GOLDEN 2-LIKE transcription factors of plants. Front. Plant Sci. 7:1509. doi: 10.3389/fpls.2016.01509
Chen, X., Wang, P., Gu, M., Lin, X., Hou, B., Zheng, Y., et al. (2021). R2R3-MYB transcription factor family in tea plant (Camellia sinensis): genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 113, 1565–1578. doi: 10.1016/j.ygeno.2021.03.033
Cho, L. H., Yoon, J., Pasriga, R., and An, G. (2016). Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 170, 2159–2171. doi: 10.1104/pp.15.01723
Cortleven, A., Marg, I., Yamburenko, M. V., Schlicke, H., Hill, K., Grimm, B., et al. (2016). Cytokinin regulates the etioplast-chloroplast transition through the two-component signaling system and activation of chloroplast-related genes. Plant Physiol. 172, 464–478. doi: 10.1104/pp.16.00640
Ding, L., Lu, Z., Gao, L., Guo, S., and Shen, Q. (2018). Is nitrogen a key determinant of water transport and photosynthesis in higher plants upon drought stress? Front. Plant Sci. 9:1143. doi: 10.3389/fpls.2018.01143
Duan, Y., Zhu, X., Shen, J., Xing, H., Zou, Z., Ma, Y., et al. (2020). Genome-wide identification, characterization and expression analysis of the amino acid permease gene family in tea plants (Camellia sinensis). Genomics 112, 2866–2874. doi: 10.1016/j.ygeno.2020.03.026
Fitter, D. W., Martin, D. J., Copley, M. J., Scotland, R. W., and Langdale, J. A. (2002). GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 31, 713–727. doi: 10.1046/j.1365-313x.2002.01390.x
Gong, X. X., Yan, B. Y., Hu, J., Yang, C. P., Li, Y. J., Liu, J. P., et al. (2018). Transcriptome profiling of rubber tree (Hevea brasiliensis) discovers candidate regulators of the cold stress response. Genes Genomics 40, 1181–1197.
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Hall, L. N., Rossini, L., Cribb, L., and Langdale, J. A. (1998). GOLDEN 2: a novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10, 925–936. doi: 10.1105/tpc.10.6.925
Han, M. L., Lv, Q. Y., Zhang, J., Wang, T., Zhang, C. X., Tan, R. J., et al. (2021). Decreasing nitrogen assimilation under drought stress by suppressing DST-mediated activation of Nitrate Reductase 1.2 in rice. Mol. Plant 15, 167–178. doi: 10.1016/j.molp.2021.09.005
Hao, X., Tang, H., Wang, B., Yue, C., Wang, L., Zeng, J., et al. (2018). Integrative transcriptional and metabolic analyses provide insights into cold spell response mechanisms in young shoots of the tea plant. Tree Physiol. 38, 1655–1671. doi: 10.1093/treephys/tpy038
Hao, X., Zhang, W., Liu, Y., Zhang, H., Ren, H., Chen, Y., et al. (2020). Pale green mutant analyses reveal the importance of CsGLKs in chloroplast developmental regulation and their effects on flavonoid biosynthesis in tea plant. Plant Physiol. Biochem. 146, 392–402. doi: 10.1016/j.plaphy.2019.11.036
Hawker, N. P., and Bowman, J. L. (2004). Roles for Class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135, 2261–2270. doi: 10.1104/pp.104.040196
Hosoda, K., Imamura, A., Katoh, E., Hatta, T., Tachiki, M., Yamada, H., et al. (2002). Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 14, 2015–2029. doi: 10.1105/tpc.002733
Huang, T., Harrar, Y., Lin, C., Reinhart, B., Newell, N. R., Talavera-Rauh, F., et al. (2014). Arabidopsis KANADI1 acts as a transcriptional repressor by interacting with a specific cis-elementent and regulates auxin biosynthesis, transport, and signaling in opposition to HD-ZIPIII factors. Plant Cell 26, 246–262. doi: 10.1105/tpc.113.111526
Kerstetter, R. A., Bollman, K., Taylor, R. A., Bomblies, K., and Poethig, R. S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. doi: 10.1038/35079629
Kiba, T., Inaba, J., Kudo, T., Ueda, N., Konishi, M., Mitsuda, N., et al. (2018). Repression of nitrogen starvation responses by members of the Arabidopsis GARP-Type transcription factor NIGT1/HRS1 Subfamily. Plant Cell 30, 925–945. doi: 10.1105/tpc.17.00810
Kim, H. J., Chiang, Y. H., Kieber, J. J., and Schaller, G. E. (2013). SCF(KMD) controls cytokinin signaling by regulating the degradation of type-B response regulators. Proc. Natl. Acad. Sci. U.S.A. 110, 10028–10033. doi: 10.1073/pnas.1300403110
Li, J., Duan, Y., Han, Z., Shang, X., Zhang, K., Zou, Z., et al. (2021). Genome-wide identification and expression analysis of the NRAMP family genes in tea plant (Camellia sinensis). Plants 10:1055. doi: 10.3390/plants10061055
Li, Q., Zhou, L., Li, Y., Zhang, D., and Gao, Y. (2021). Plant NIGT1/HRS1/HHO transcription factors: key regulators with multiple roles in plant growth, development, and stress responses. Int. J. Mol. Sci. 22:8685. doi: 10.3390/ijms22168685
Li, W., Xiang, F., Zhong, M., Zhou, L., Liu, H., Li, S., et al. (2017). Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis). Sci. Rep. 7:1693. doi: 10.1038/s41598-017-01949-0
Lupi, A. C. D., Lira, B. S., Gramegna, G., Trench, B., Alves, F. R. R., Demarco, D., et al. (2019). Solanum lycopersicum GOLDEN 2-LIKE 2 transcription factor affects fruit quality in a light- and auxin-dependent manner. PLoS One 14:e0212224. doi: 10.1371/journal.pone.0212224
Maeda, Y., Konishi, M., Kiba, T., Sakuraba, Y., Sawaki, N., Kurai, T., et al. (2018). A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 9:1376. doi: 10.1038/s41467-018-03832-6
Medici, A., Marshall-Colon, A., Ronzier, E., Szponarski, W., Wang, R., Gojon, A., et al. (2015). AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6:6274. doi: 10.1038/ncomms7274
Nadakuduti, S. S., Holdsworth, W. L., Klein, C. L., and Barry, C. S. (2014). KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 78, 1022–1033. doi: 10.1111/tpj.12529
Nagatoshi, Y., Mitsuda, N., Hayashi, M., Inoue, S., Okuma, E., Kubo, A., et al. (2016). GOLDEN 2-LIKE transcription factors for chloroplast development affect ozone tolerance through the regulation of stomatal movement. Proc. Natl. Acad. Sci. U.S.A. 113, 4218–4223. doi: 10.1073/pnas.1513093113
Naquin, D., d’Aubenton-Carafa, Y., Thermes, C., and Silvain, M. (2014). CIRCUS: a package for Circos display of structural genome variations from paired-end and mate-pair sequencing data. BMC Bioinformatics 15:198. doi: 10.1186/1471-2105-15-198
Osorio, M. B., Ng, S., Berkowitz, O., De Clercq, I., Mao, C., Shou, H., et al. (2019). SPX4 acts on PHR1-dependent and -independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol. 181, 332–352. doi: 10.1104/pp.18.00594
Peng, X., Wu, Q., Teng, L., Tang, F., Pi, Z., and Shen, S. (2015). Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 15:108. doi: 10.1186/s12870-015-0489-2
Pires, H. R., Monfared, M. M., Shemyakina, E. A., and Fletcher, J. C. (2014). ULTRAPETALA trxG genes interact with KANADI transcription factor genes to regulate Arabidopsis gynoecium patterning. Plant Cell 26, 4345–4361. doi: 10.1105/tpc.114.131250
Powell, A. L., Nguyen, C. V., Hill, T., Cheng, K. L., Figueroa-Balderas, R., Aktas, H., et al. (2012). Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336, 1711–1715. doi: 10.1126/science.1222218
Puga, M. I., Mateos, I., Charukesi, R., Wang, Z., Franco-Zorrilla, J. M., de Lorenzo, L., et al. (2014). SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 14947–14952. doi: 10.1073/pnas.1404654111
Ram, H., Sahadevan, S., Gale, N., Caggiano, M. P., Yu, X., Ohno, C., et al. (2020). An integrated analysis of cell-type specific gene expression reveals genes regulated by REVOLUTA and KANADI1 in the Arabidopsis shoot apical meristem. PLoS Genetic. 16:e1008661. doi: 10.1371/journal.pgen.1008661
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Ruan, L., Wei, K., Li, J., He, M., Wu, L., Aktar, S., et al. (2021). Responses of tea plants (Camellia sinensis) with different low-nitrogen tolerances during recovery from nitrogen deficiency. J. Sci. Food Agric. 102, 1405–1414. doi: 10.1002/jsfa.11473
Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Safi, A., Medici, A., Szponarski, W., Ruffel, S., Lacombe, B., and Krouk, G. (2017). The world according to GARP transcription factors. Curr. Opin. Plant Biol. 39, 159–167. doi: 10.1016/j.pbi.2017.07.006
Salvi, E., Rutten, J. P., Di Mambro, R., Polverari, L., Licursi, V., Negri, R., et al. (2020). A self-organized PLT/Auxin/ARR-B network controls the dynamics of root zonation development in Arabidopsis thaliana. Dev. Cell 53, 431.e–443.e. doi: 10.1016/j.devcel.2020.04.004
Sawaki, N., Tsujimoto, R., Shigyo, M., Konishi, M., Toki, S., Fujiwara, T., et al. (2013). A nitrate-inducible GARP family gene encodes an auto-repressible transcriptional repressor in rice. Plant Cell Physiol. 54, 506–517. doi: 10.1093/pcp/pct007
Sevanthi, A. M., Sinha, S. K., Rani, M., Saini, M. R., Kumari, S., Kaushik, M., et al. (2021). Integration of dual stress transcriptomes and major QTLs from a pair of genotypes contrasting for drought and chronic nitrogen starvation identifies key stress responsive genes in rice. Rice 14:49. doi: 10.1186/s12284-021-00487-8
Sun, L., Song, L., Zhang, Y., Zheng, Z., and Liu, D. (2016). Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol. 170, 499–514. doi: 10.1104/pp.15.01336
Taketa, S., Hattori, M., Takami, T., Himi, E., and Sakamoto, W. (2021). Mutations in a Golden2-Like gene cause reduced seed weight in barley albino lemma 1 mutants. Plant Cell Physiol. 62, 447–457. doi: 10.1093/pcp/pcab001
Tang, D., Liu, M. Y., Zhang, Q., Ma, L., Shi, Y., and Ruan, J. (2020). Preferential assimilation of NH4(+) over NO3(-) in tea plant associated with genes involved in nitrogen transportation, utilization and catechins biosynthesis. Plant Sci. 291:110369. doi: 10.1016/j.plantsci.2019.110369
Ueda, Y., Nosaki, S., Sakuraba, Y., Miyakawa, T., Kiba, T., Tanokura, M., et al. (2020b). NIGT1 family proteins exhibit dual mode DNA recognition to regulate nutrient response-associated genes in Arabidopsis. PLoS Genetic. 16:e1009197. doi: 10.1371/journal.pgen.1009197
Ueda, Y., Kiba, T., and Yanagisawa, S. (2020a). Nitrate-inducible NIGT1 proteins modulate phosphate uptake and starvation signalling via transcriptional regulation of SPX genes. Plant J. 102, 448–466. doi: 10.1111/tpj.14637
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., and Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Wang, X., Wang, H. F., Chen, Y., Sun, M. M., Wang, Y., and Chen, Y. F. (2020d). The transcription factor NIGT1.2 modulates both phosphate uptake and nitrate influx during phosphate starvation in Arabidopsis and maize. Plant Cell 32, 3519–3534. doi: 10.1105/tpc.20.00361
Wang, J., Xia, J., Song, Q., Liao, X., Gao, Y., Zheng, F., et al. (2020a). Genome-wide identification, genomic organization and expression profiles of SlARR-B gene family in tomato. J. Appl. Genet. 61, 391–404. doi: 10.1007/s13353-020-00565-5
Wang, X., Feng, H., Chang, Y., Ma, C., Wang, L., Hao, X., et al. (2020c). Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 11:4447. doi: 10.1038/s41467-020-18228-8
Wang, W., Wang, X., Wang, Y., Zhou, G., Wang, C., Hussain, S., et al. (2020b). SlEAD1, an EAR motif-containing ABA down-regulated novel transcription repressor regulates ABA response in tomato. GM Crops Food 11, 275–289. doi: 10.1080/21645698.2020.1790287
Wang, Y., Chen, F., Ma, Y., Zhang, T., Sun, P., Lan, M., et al. (2021). An ancient whole-genome duplication event and its contribution to flavor compounds in the tea plant (Camellia sinensis). Hortic. Res. 8:176.
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, Z., Ruan, W., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111, 14953–14958. doi: 10.1073/pnas.1404680111
Wang, Z., Zheng, Z., Song, L., and Liu, D. (2018). Functional characterization of Arabidopsis PHL4 in plant response to phosphate starvation. Front. Plant Sci. 9:1432. doi: 10.3389/fpls.2018.01432
Wei, C., Yang, H., Wang, S., Zhao, J., Liu, C., Gao, L., et al. (2018). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. U.S.A. 115, 4151–4158. doi: 10.1073/pnas.1719622115
Worthen, J. M., Yamburenko, M. V., Lim, J., Nimchuk, Z. L., Kieber, J. J., and Schaller, G. E. (2019). Type-B response regulators of rice play key roles in growth, development and cytokinin signaling. Development 146:dev174870. doi: 10.1242/dev.174870
Wu, K., Wang, S., Song, W., Zhang, J., Wang, Y., Liu, Q., et al. (2020). Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 367:eaaz2046. doi: 10.1126/science.aaz2046
Wu, Z., Cui, C., Xing, A., Xu, X., Sun, Y., Tian, Z., et al. (2021). Identification and response analysis of xyloglucan endotransglycosylase/hydrolases (XTH) family to fluoride and aluminum treatment in Camellia sinensis. BMC Genomics. 22:761. doi: 10.1186/s12864-021-08056-5
Xia, E. H., Li, F. D., Tong, W., Li, P. H., Wu, Q., Zhao, H. J., et al. (2019). Tea plant information archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol. J. 17, 1938–1953. doi: 10.1111/pbi.13111
Xia, E. H., Tong, W., Wu, Q., Wei, S., Zhao, J., Zhang, Z. Z., et al. (2020). Tea plant genomics: achievements, challenges and perspectives. Hortic. Res. 7:7. doi: 10.1038/s41438-019-0225-4
Xiang, N., Hu, J. G., Yan, S., and Guo, X. (2021). Plant hormones and volatiles response to temperature stress in sweet corn (Zea mays L.) seedlings. J. Agric. Food Chem. 69, 6779–6790. doi: 10.1021/acs.jafc.1c02275
Xie, Y., Straub, D., Eguen, T., Brandt, R., Stahl, M., Martínez-García, J. F., et al. (2015). Meta-analysis of Arabidopsis KANADI1 direct target genes identifies a basic growth-promoting module acting upstream of hormonal signaling pathways. Plant Physiol. 169, 1240–1253. doi: 10.1104/pp.15.00764
Yu, Q., Li, C., Zhang, J., Tian, Y., Wang, H., Zhang, Y., et al. (2020). Genome-wide identification and expression analysis of the Dof gene family under drought stress in tea (Camellia sinensis). PeerJ 8:e9269. doi: 10.7717/peerj.9269
Yue, C., Cao, H., Lin, H., Hu, J., Ye, Y., Li, J., et al. (2019). Expression patterns of alpha-amylase and beta-amylase genes provide insights into the molecular mechanisms underlying the responses of tea plants (Camellia sinensis) to stress and postharvest processing treatments. Planta 250, 281–298. doi: 10.1007/s00425-019-03171-w
Yue, J., Liu, J., Tang, W., Wu, Y. Q., Tang, X., Li, W., et al. (2020). Kiwifruit Genome Database (KGD): a comprehensive resource for kiwifruit genomics. Hortic. Res. 7:117. doi: 10.1038/s41438-020-0338-9
Zeng, J., Zhu, X., Haider, M. S., Wang, X., Zhang, C., and Wang, C. (2017). Genome-wide identification and analysis of the type-B authentic response regulator gene family in peach (Prunus persica). Cytogenet Genome Res. 151, 41–49. doi: 10.1159/000458170
Zhang, G. H., Xu, Q., Zhu, X. D., Qian, Q., and Xue, H. W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21, 719–735. doi: 10.1105/tpc.108.061457
Zhang, H., Zhu, J., Gong, Z., and Zhu, J.-K. (2021). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119.
Zhang, X., Xu, W., Ni, D., Wang, M., and Guo, G. (2020). Genome-wide characterization of tea plant (Camellia sinensis) Hsf transcription factor family and role of CsHsfA2 in heat tolerance. BMC Plant Biol. 20:244. doi: 10.1186/s12870-020-02462-9
Keywords: tea plant, GARP transcription factor, expression analysis, stress response, nitrogen response
Citation: Yue C, Chen Q, Hu J, Li C, Luo L and Zeng L (2022) Genome-Wide Identification and Characterization of GARP Transcription Factor Gene Family Members Reveal Their Diverse Functions in Tea Plant (Camellia sinensis). Front. Plant Sci. 13:947072. doi: 10.3389/fpls.2022.947072
Received: 18 May 2022; Accepted: 02 June 2022;
Published: 30 June 2022.
Edited by:
Yuhua Wang, Nanjing Agricultural University, ChinaReviewed by:
Raj Kumar Joshi, Rama Devi Women’s University, IndiaChunzhen Cheng, Shanxi Agricultural University, China
Mei-Ya Liu, Tea Research Institute (CAAS), China
Copyright © 2022 Yue, Chen, Hu, Li, Luo and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuan Yue, eWNjeXl4QDE2My5jb20=; Liang Zeng, emVuZ2xpYW5nYmFieUAxMjYuY29t
 Chuan Yue
Chuan Yue Qianqian Chen
Qianqian Chen Juan Hu
Juan Hu Congcong Li
Congcong Li Liyong Luo
Liyong Luo Liang Zeng
Liang Zeng