
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 28 July 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.946827
 Yan Zhao1,2*
Yan Zhao1,2* Guanghui Zhang1
Guanghui Zhang1 Qingyan Tang1
Qingyan Tang1 Wanling Song1
Wanling Song1 Qingqing Gao1,2
Qingqing Gao1,2 Guisheng Xiang1
Guisheng Xiang1 Xia Li1,2
Xia Li1,2 Guanze Liu1
Guanze Liu1 Wei Fan1
Wei Fan1 Xiaoning Li1
Xiaoning Li1 Shengchao Yang1*
Shengchao Yang1* Chenxi Zhai3*
Chenxi Zhai3*Erigeron breviscapus, a traditional Chinese medicinal plant, is enriched in flavonoids that are beneficial to human health. While we know that R2R3-MYB transcription factors (TFs) are crucial to flavonoid pathway, the transcriptional regulation of flavonoid biosynthesis in E. breviscapus has not been fully elucidated. Here, EbMYBP1, a R2R3-MYB transcription factor, was uncovered as a regulator involved in the regulation of flavonoid accumulation. Transcriptome and metabolome analysis revealed that a large group of genes related to flavonoid biosynthesis were significantly changed, accompanied by significantly increased concentrations of the flavonoid in EbMYBP1-OE transgenic tobacco compared with the wild-type (WT). In vitro and in vivo investigations showed that EbMYBP1 participated in flavonoid biosynthesis, acting as a nucleus-localized transcriptional activator and activating the transcription of flavonoid-associated genes like FLS, F3H, CHS, and CHI by directly binding to their promoters. Collectively, these new findings are advancing our understanding of the transcriptional regulation that modulates the flavonoid biosynthesis.
Flavonoids are one of the most abundant secondary metabolites and are found in plants across a wide range of morphological classes (Buer et al., 2010; Tohge et al., 2013). There are several main classes of derivatives produced by the phenylpropanoid pathway, such as flavonols, anthocyanins, and proanthocyanidins (Routaboul et al., 2012; Saito et al., 2013). Flavonols have excellent antioxidant properties and are thought to perform key functions as UV filters, exhibiting important roles during plant evolution (Pollastri and Tattini, 2011). Additionally, flavonoids are known to exhibit anti-inflammatory, anti-proliferative, and antioxidative properties, preventive properties toward cardiovascular disease as well as diabetes (Williams et al., 2004; Perez-Vizcaino and Duarte, 2010; Zhang et al., 2015; Zakaryan et al., 2017).
Flavonoids are synthesized through the phenylpropanoid pathway in plants, initially catalyzed by phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H) and 4-hydroxy-cinnamoyl CoA ligase (4CL) (Ferreyra et al., 2021). Great progress has been made to elucidate the structural genes of the flavonoid biosynthesis pathway. It was found that CHS catalyzes the first step in flavonoid biosynthesis, converting p-coumaryl-CoA and malonyl-CoA to naringenin chalcone (Pandith et al., 2016). CHS further converts the initial product into many flavonoids, including flavonol, flavone, flavanone and anthocyanidin. Another enzyme, flavonol synthase (FLS), a key enzyme in the flavonol pathway, contributes to the synthesis of flavonols. It competes with dihydroflavonol 4-reductase (DFR) for the same substrate, dihydroflavonol (Wellmann et al., 2002; Davies and Schwinn, 2003; Luo et al., 2008; Sheng et al., 2020). In sum, numerous regulatory proteins including transcription factors (TFs) are associated with flavonoid biosynthesis.
Flavonoid biosynthesis has been studied in many plant species. A wide variety of transcription factors (TFs) are implicated in flavonoid biosynthesis, including MYB, basic helix-loop-helix (bHLH), basic leucine zipper (bZIP), WD40, and zinc finger proteins (Chen et al., 2019; Naik et al., 2022). In plants, MYB TFs control the transcription of flavonoid biosynthesis genes, a process which has been extensively studied. MYB genes are a major transcription factor family in plants and are involved in various biological processes. MYB proteins, including MYB12, MYB11, and MYB111, have been identified in Arabidopsis as flavonol-specific factors based on sequence similarity (Mehrtens et al., 2005; Stracke et al., 2007). Furthermore, the SG7 MYBs are found in grapes (Czemmel et al., 2009; Matus et al., 2009), apples (Wang et al., 2017), pears (Zhai et al., 2019; Premathilake et al., 2020), peaches (Cao et al., 2019) and other plants (Ballester et al., 2010; Liu et al., 2016). Additionally, AgMYB12, located in the S7 subgroup of the R2R3-MYB family, displayed as a positive regulator of apigenin biosynthesis, activating the expression of AgFNS gene in celery (Wang et al., 2022). These results showed that MYB TFs play key regulatory roles in flavonoid biosynthesis.
Erigeron breviscapus (E. breviscapus) is a medicinal plant in the Compositae family with great medicinal potential, comprised of 25 kinds of flavonoids, 46 kinds of caffeoyl compounds, 78 kinds of volatile oils, and nearly 40 kinds of other compounds (Wu et al., 2021). Previously, a total of 108 R2R3-MYB transcription factors have been identified in the E. breviscapus genome (He et al., 2021; Song et al., 2021). Phylogenetic analysis suggests possible involvement of several MYBs in phenylpropanoid metabolism including flavonoids, flavonols, and anthocyanins (Song et al., 2021). However, no transcription factor involved in flavonoid biosynthesis has been identified in E. breviscapus. Thus, it is imperative to identify MYB TFs in E. breviscapus that can specifically and positively regulate flavonoid biosynthesis, the findings for which will provide us with a greater understanding of the flavonoid biosynthesis pathway. Here, EbMYBP1, a flavonoid biosynthesis regulator, was isolated from E. breviscapus. Transcriptome and metabolome analysis showed that overexpression of EbMYBP1 resulted in an increased flavonoid content in transgenic tobacco lines through upregulation of flavonoid-related biosynthesis genes FLS, F3H, CHS, and CHI. Our finding will provide valuable insights to elucidate the roles of EbMYBP1 in positively regulating flavonoid biosynthesis in E. breviscapus.
In this study, Erigeron breviscapus cv. Long Jin NO. 1 (LJ1) and Nicotiana tabacum cv. Yunyan87 (Y87) were used as plant materials, which were obtained from Longjin Biotech Co., Ltd. (Xuanwei, Yunnan, China) and Yunnan Agricultural University (Kunming, Yunnan, China), respectively. Seeds of LJ1 were sowed in nutrient soils and grown in a greenhouse in Yunnan Agricultural University. Seeds of Y87 were surface sterilized with 75% alcohol for 30 s, 10% H2O2 for 10 min, washing five times with sterile H2O, and placing the seeds on MS medium for 2–3 weeks. Sterilized plants were prepared for use in transgenic plants.
The roots, stems, leaves, flowers samples of E. breviscapus were classified, and three biological replicates were set for each tissue. After dried at a constant temperature at 45°C, the samples were ground into a fine powder using a high-speed grinder (MM400, Zhongxingwy, Beijing, China). Then, 30 mL of 80% methanol was added into 1 g of root, stem, leaf, flower powder. To obtain the filtrate, the sample was kept at 4°C overnight, then centrifuged for 12 min at 6,000 × g. Extracted samples were analyzed using UPLC (UPLC, Agilent 1260 Infinity). The analysis was performed using a binary mobile phase composed of A: water + 0.1% phosphoric acid and B: 100% acetonitrile. The following conditions were used in the analysis: UPLC column, SunFire C18 (5 μm, 4.6 × 250 mm); mobile phase solvent A, water with 0.1% phosphoric acid, solvent B acetonitrile with 0.1% phosphoric acid. The following gradient conditions were used to achieve separation: starting at 80% A at 0 min, 80% A at 3 min, 74.5% A at 17 min, 56% A at 28 min, 38% A at 30 min, 38% A at 32 min and holding for 5 min. The flow rate was 1 mL/min, the column temperature was 40°C, and the injection volume was 8 μL.
Total genomic DNA and RNA were isolated from the leaf tissues of E. breviscapus using a DNA/RNA isolation kit (TIANGEN, China). Based on the manufacturer’s instructions, cDNA was synthesized using a PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan). Amplification of full length EbMYBP1 cDNA with gene specific primers (see Supplementary Table 1) was performed by PCR, after which the sequence was generated. The generated sequence was checked against GenBank by BLASTX and BLASTp. Amino acid sequence alignments were performed by ClustalX, and a phylogenetic tree of EbMYBP1 and of closely related proteins in other plant species was constructed by MEGA 7.0.
The full-length cDNA sequence of EbMYBP1 was inserted into pCAMBIA1300-35S-N-GFP and pCAMBI A1300-35S-GFP-C vectors, resulting in pCAMBIA1300-35S-EbMYBP1-GFP (EbMYBP1-GFP) and pCAMBIA1300-35S-GFP-EbMYBP1 (GFP-EbMYBP1) fusion constructs, which were subsequently transformed into A. tumefaciens GV3101 by the electroporation method. The pCAMBIA1300-35S-N-GFP and pCAMBIA1300-35S-GFP-C vectors were used as controls. Agrobacterium was cultured on YEB agar supplemented with selection antibiotics, and then incubated at 28°C for 2∼3 day. The confluent Agrobacterium containing the target vector was resuspended in an infiltration buffer (0.5 × MS, 10 mM MES (pH 5.6), 150 μg/mL acetosyringone) to an OD600 of 0.5 and incubated at room temperature without shaking for 2 h before infiltration. Approximately 500 μL of the Agrobacterium mixture was then infiltrated into 3–4 young leaves of each plant, with at least two points for each leaf. The subcellular localization assay was performed 48 h after inoculation. Confocal images were taken by using a Zeiss LSM 880 confocal laser scanning microscope (Zeiss, Germany).
The ORF sequence of EbMYBP1 was amplified and ligated into pGBKT7 containing a GAL4 DNA-binding domain, generating the pGBKT7-EbMYBP1. The pGBKT7-EbMYBP1, negative control (pGBKT7 or pGADT7), and positive control (pGADT7 + pGBKT7-53) vectors were transformed into Y2HGold yeast cells. The yeast cells were cultivated on SD/-Trp medium or SD–Trp/His/Ade medium. Transcriptional activity was assayed by the growth status. The primers used are listed in Supplementary Table 1.
The CDS of EbMYBP1 was cloned from leaves of 8-week-old E. breviscapus. The target fragments were subcloned into the PC1300-35S vector between the SmaI and XbaI sites to generate a PC1300-35S-EbMYBP1 construct using the primers listed in Supplementary Table 1. Then, the plasmids were introduced into Agrobacterium tumefaciens GV3101, which was then used in tobacco cultivar Yunyan87 (Nicotiana tabacum) transformation by the leaf disc methods. Regenerated shoots and healthy resistant shoots were grown on selective shooting medium and rooting medium, respectively, which both contained 50 mg⋅L–1 kanamycin and 250 mg⋅L–1 carbenicillin. Well-developed rooted plants were transferred to soil and then grown in a growth room at 25 ± 2°C, with 65–70% relative humidity.
Leaves were harvested from 7-week-old plants. The samples from Yunyan87 (wild-type, WT) and homozygous T2 lines of transgenic tobacco plants (EbMYBP1-OE), were collected for transcriptome and metabolome sequencing. All samples were frozen immediately in liquid nitrogen and stored at −80°C. The leaves from six individual plants were sampled as one biological replicate, and three biological replicates were used in this study.
The leaves of tobacco from six different plants that grew for about 7 weeks were collected to quantify the total flavonoids content. Flavonoids were extracted and quantified using the method (Wang et al., 2016). The absorbance of the samples was determined at 535 nm with a spectrophotometer (UV-1800, Shimadzu), and methanol with 1% HCl was used as a blank control. The total flavonoids content was measured with (mg/g FW) = (1/958 × A535 × 10,000 × V)/fresh weight (g), where the V indicates the total volume of the extract (mL). At least three biological replicates were used for each sample.
Metabolite profiling was performed using an LC–ESI–MS/MS system (UPLC; Shim-pack UFLC SHIMADZU CBM30A, Shimadzu, Kyoto, Japan; MS/MS, Applied Biosystems 6500 QTRAP, Applied Biosystems, Foster City, CA, United States) (Chen et al., 2013). The data from six samples (EbMYBP1-OE and WT × three biological replicates) were processed by OPLS-DA and PCA to detect differences in metabolic composition between the EbMYBP1-OE and WT tobacco.
The seven-week-old tobacco leaves of the WT and EbMYBP1-OE were used for transcriptome analysis, and three biological replicates were made for each sample. Firstly, RNA was prepared, and reverse transcribed into the cDNA library. In order to ensure the quality of the data, fastp was used to control the raw data. 1% agarose gel, Nanophotometer spectrophotometer and Agilent 2100 Bioanalyzer were used to analyze the purity of the RNA and detect the integrity of the RNA. These data were subsequently used to analyze base composition and mass distribution to confirm the accuracy of this set of data. We used HISAT2 software for alignment analysis with the reference genome, assembled the transcripts of the new genes with Stringtie, analyzed the gene expression level with FPKM, screened the significantly different accumulation genes according to FDR < 0.05 and fold change ≥ 2 with DESeq2, and enriched GO and KEGG with Tbtools software (Chen et al., 2020) and the KEGG database. The transcriptome data of WT and EbMYBP1-OE have been deposited to national center for biotechnology information database as a sequence read archive under BioProject ID (PRJNA836017).
The instantaneous color determination experiment was performed on the leaves of 4-week-old tobacco seedlings. The promoter regions of tobacco cultivar Yunyan87 (Nicotiana tabacum) genes NtFLS2, NtF3H, NtFLS1, NtDFR, NtCHI, and NtCHS were amplified and cloned into pGreenII 0800-LUC vector making pGreen 0800:FLS2, pGreen 0800:F3H, pGreen 0800:FLS1, pGreen 0800:DFR, pGreen 0800:CHI, and pGreen 0800:CHS reporter constructs. The EbMYBP1 ORF was amplified and cloned into the pGreenII 62-SK vector to obtain the 35S:EbMYBP1 effector construct. LUC and REN luciferase activities were determined using a dual luciferase assay kit (Promega, United States) according to the manufacturer’s instructions. These plasmids were transiently infiltrated in tobacco leaves by Agrobacterium. Three biological repeats were included in each combination.
The ORF sequence of EbMYBP1 was cloned and fused with the GST tag in pGEX-4T-1 vectors (GE Healthcare Life Science) and expressed in Escherichia coli strain Rosetta (DE3) by induction with 0.5 mM isopropyl-β-D-thiogalactopyranoside for 6 h at 20°C. The recombinant proteins were purified with a GST-tagged protein purification kit (Clontech). Probes containing MYB-specific cis-elements derived from the promoters of FLS, CHS, CHI, and F3H genes were labeled with 5′6-FAM (FITC) fluorescent dye. Unlabeled probes were used in competition assays. MYB-specific cis-elements within probes were used in mutation assays. Two biological experiments were performed with similar results. All primer sequences used are listed in Supplementary Table 1.
Total RNA of 7-week-old WT and EbMYBP1-OE tobacco leaves was extracted using a total RNA small extraction kit (Magen, China). RNA was reverse transcribed into cDNA with a reverse transcription Kit (Takara, China). A 20 μL reaction system was constructed according to qPCR SYBR Green Master Mix (Vazyme, China). PCR was performed on a QuantStudio 5 (ABI) instrument (Thermo Fisher, Singapore). All primers used in this study are listed in Supplementary Table 1. The NtActin gene was used as an internal control, thus obtaining true differences in genes of interest-specific expression. Thermo Fisher Expression of genes was used to calculate the relative expression by the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Statistical analysis of data was conducted using one-way analysis of variance, followed by Tukey’s comparison tests, or using Student’s t-test in GraphPad Prism 9 software. Differences were considered statistically significant at P < 0.05.
The full-length cDNA sequence for EbMYBP1 was cloned, containing an open reading frame (ORF) of 1,014 bp, encoding 337 amino acid residues, and with a predicted molecular weight of 38.29 kDa. Sequence alignment showed that the R2R3 domains in EbMYBP1 were highly conserved (Figure 1A). EbMYBP1 contained both SG7 motif-1 ([K/R][R/x][R/K]xGRT[S/x][R/G]xx[M/x]K) and SG7 motif-2 ([W/x][L/x]LS) (Figure 1A), suggesting their putative role in regulating flavonoid biosynthesis.
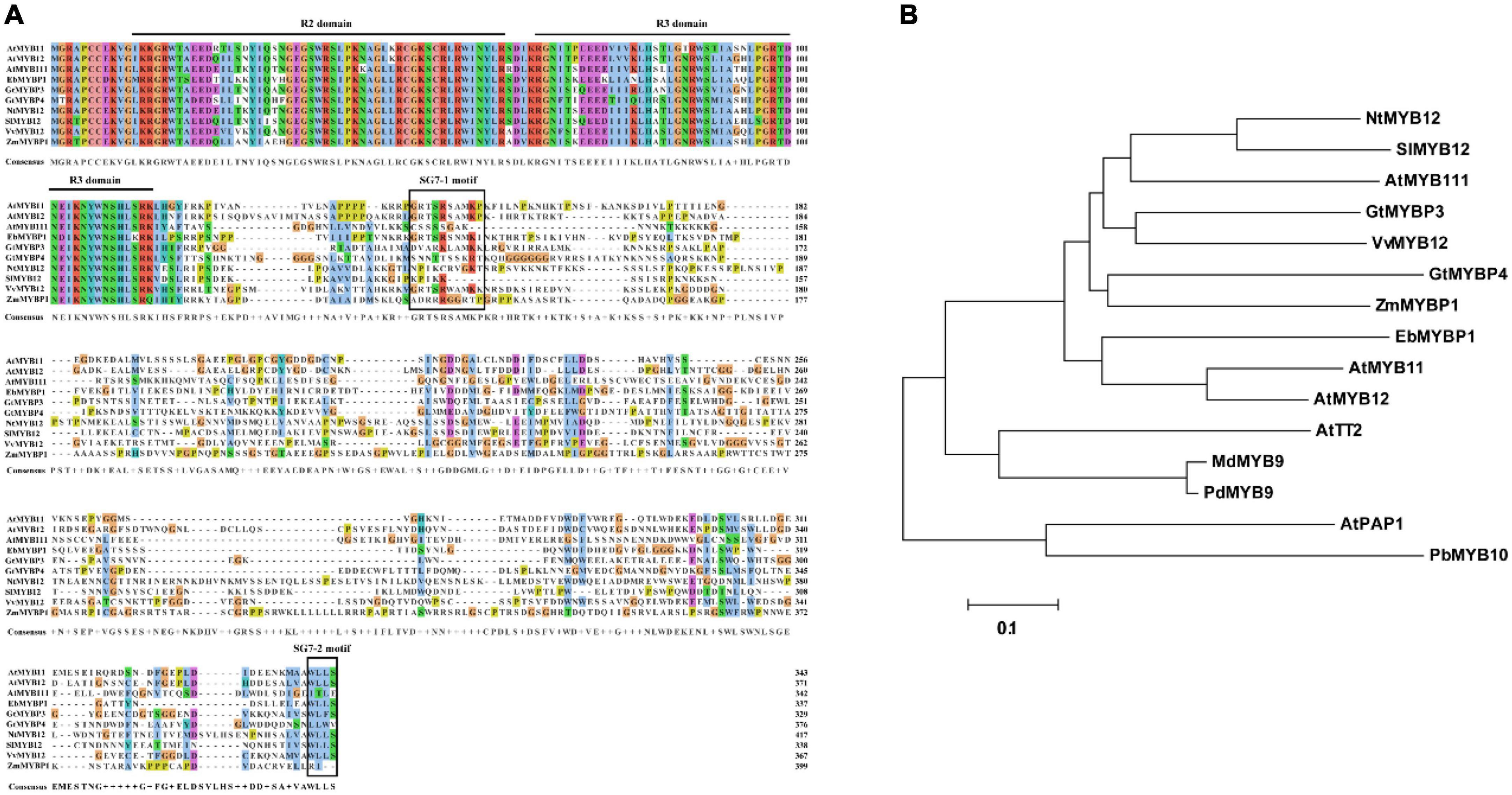
Figure 1. Multiple sequence alignment and phylogenetic relationship of EbMYBP1. (A) Multiple sequence alignment of EbMYBP1. The R2/R3 repeats and SG7-1/SG7-2 are indicated in black frame. (B) Phylogenetic analysis of EbMYBP1 and other 14 homologs R2R3-MYB proteins. The tree was constructed using MEGA7 software based on the neighbor-jointing method.
Next, MYB TFs from different species were selected for phylogenetic analysis. Phylogenetic analysis showed that EbMYBP1 is clustered with some SG7 family proteins, particularly flavonoid regulators such as Arabidopsis (A. thaliana) AtMYB11, AtMYB12, and AtMYB111; tobacco (Nicotiana tabacum) NtMYB12; tomato (Solanum lycopersicum L.) SlMYB12; Japanese gentian (Gentiana triflora) GtMYBP3, GtMYBP4; grape (Vitis vinifera) VvMYB12; and maize (Zea mays) ZmMYBP (Figure 1B). EbMYBP1 was most closely related to AtMYB12 and AtMYB11 in A. thaliana as shown in the phylogenetic tree.
To determine if EbMYBP1 expression is associated with flavonoid accumulation in E. breviscapus, the expression profiles of EbMYBP1 in different tissues of E. breviscapus were examined. In the transcriptome data of four tissues (flowers, stems, leaves and roots) in our laboratory (Song et al., 2021), EbMYBP1 was expressed ubiquitously in all four tissues, the highest expression was found in the leaf tissues and the lowest expression was found in the roots by RT-qPCR (Figure 2B). Moreover, the genes in the flavonoid biosynthesis pathway included EbPAL, EbCHS, and EbCHI, all of which indicated high expression level in the leaf. Next, we analyzed the flavonoid contents of these four tissues. In all four tissues, four main flavonoids (scutellarin, apigenin-7-O-glucuronide, scutellarein, apigenin) were detected, and their levels varied among tissues (Supplementary Figure 1). Scutellarin accumulation was highest in leaves followed by flowers, stems and roots (Figure 2C). The results showed a positive correlation between EbMYBP1 expression level and scutellarin accumulation.
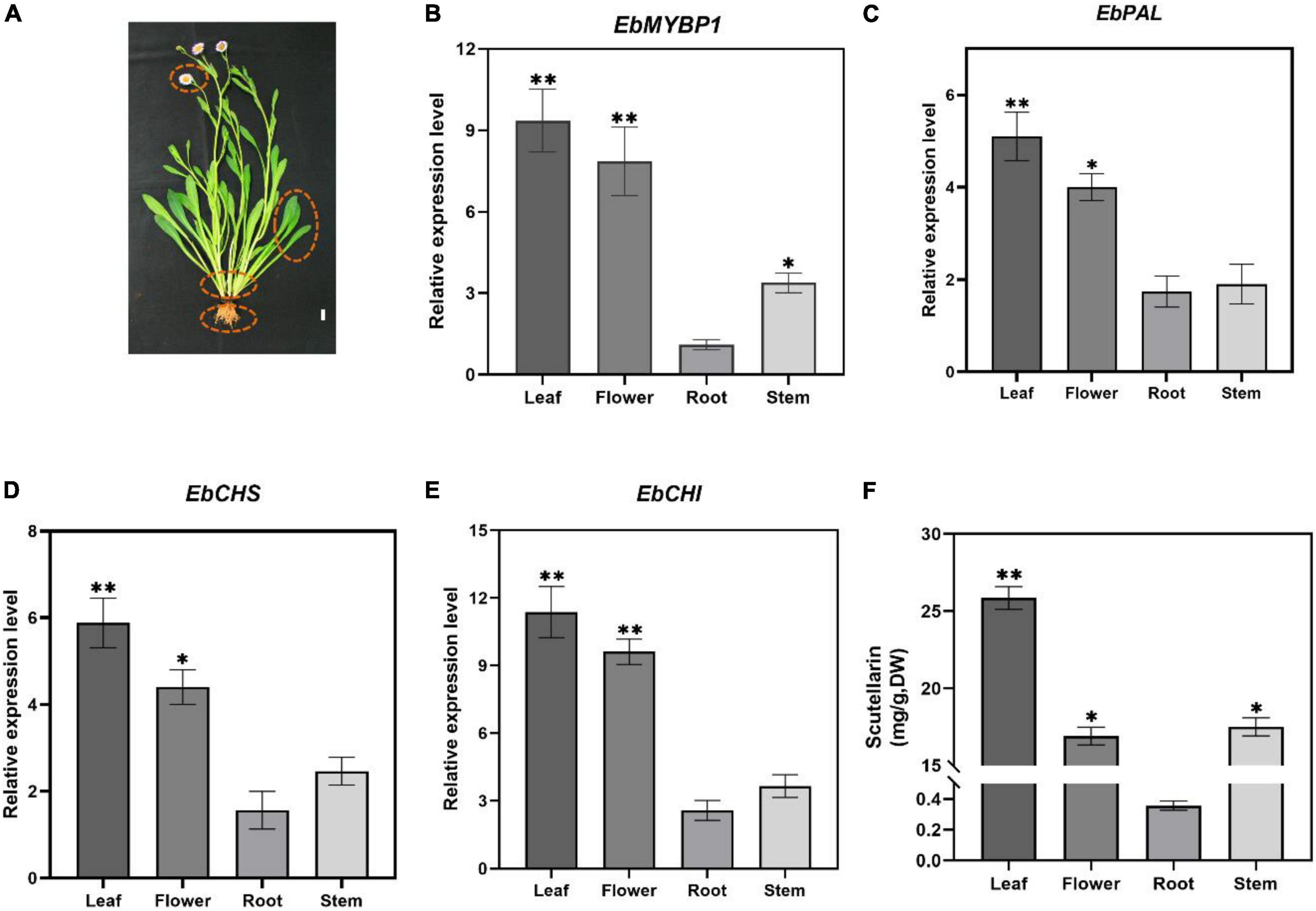
Figure 2. Relative expression of EbMYBP1 in E. breviscapus. (A) Flowering E. breviscapus plants were used in the experiment. Red circles represent flowers, leaves, stems and roots, respectively, which are used in RT-qPCR of EbMYBP1 and contents of scutellarin analysis, Bar = 1 cm. (B–E) Relative expression of EbMYBP1, EbPAL, EbCHS, and EbCHI in different tissues of E. breviscapus. (F) Contents of scutellarin in different tissues of E. breviscapus. Data represent mean ± SD of three biological replicates. Asterisks indicate significant differences (*P < 0.05; **P < 0.01).
To analyze the basic characteristic of EbMYBP1 as a transcription factor, two fusion proteins of EbMYBP1-GFP and GFP-EbMYBP1 were constructed for transient transformation via Agrobacterium infiltration methodology. The transient expression results indicated that the fluorescence of EbMYBP1-GFP and GFP-EbMYBP1 was exclusively localized in the nucleus of epidermal cells from young N. benthamiana leaves, whereas that of the control GFP protein was distributed throughout the cell (Figure 3A). Thus, the results revealed that EbMYBP1 is a nuclear-localized protein, colocalized in the nucleus.
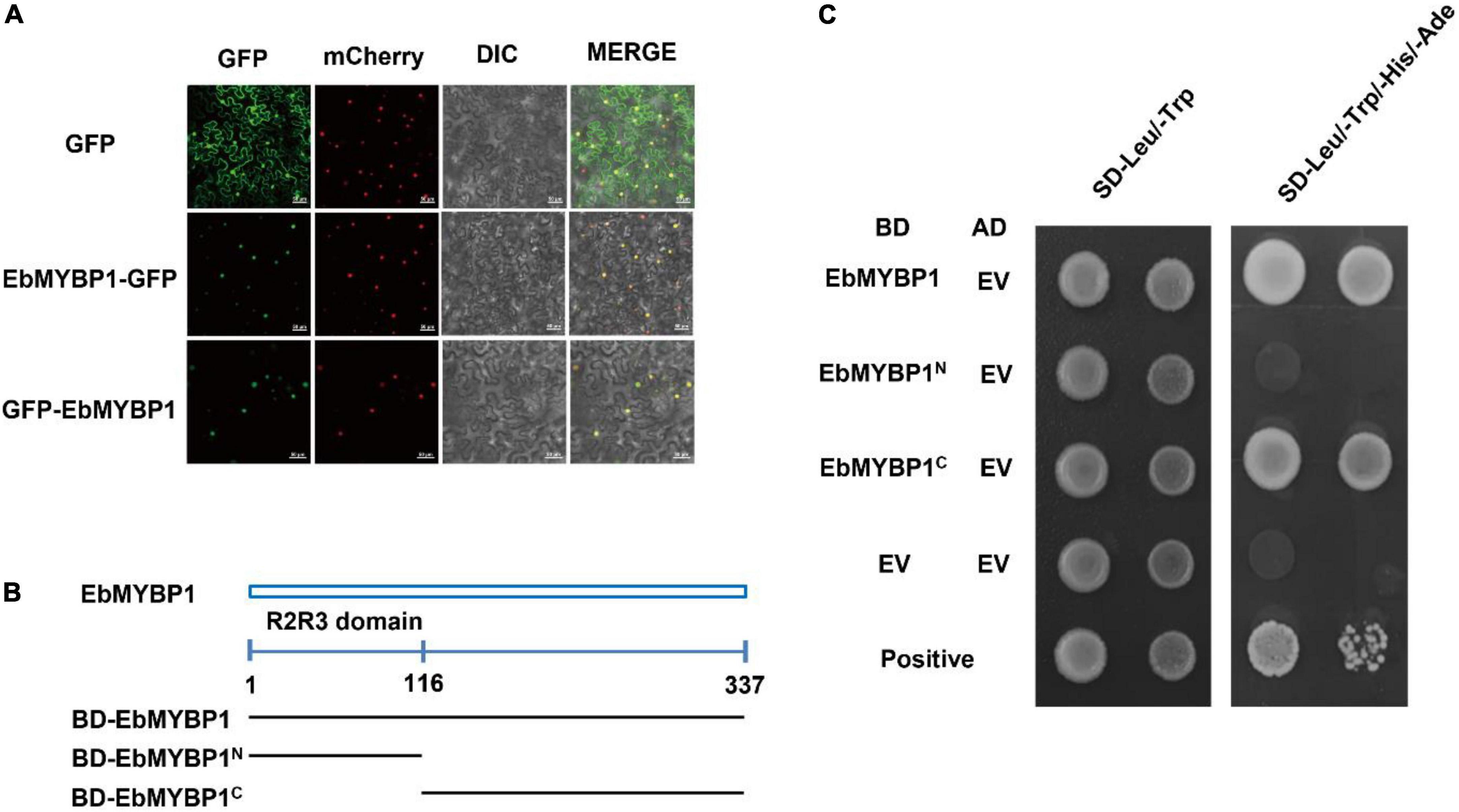
Figure 3. Analysis of the subcellular localization and transactivation activity of EbMYBP1. (A) Subcellular localization of EbMYBP1 in N. benthamiana leaves. EbMYBP1 fused to the N-terminal of GFP (middle) or C-terminal of GFP (lower) under the control of the 35S promoter. The HY5-mCherry was used as nucleus marker. Differential interference contrast (DIC) and GFP, mCherry fluorescence images as well as merged images are shown. At least three individual experiments were performed for each combination with the similar results. Bars = 50 μm. (B) Schematic diagrams illustrate the different portions of EbMYBP1 ORF that were fused to the yeast vector pGBKT7. (C) Transactivation activity of the EbMYBP1 protein in yeast. The transformed cells were selected on SD/-Trp medium (left panel) and SD/-Trp/-His medium (right panels). Three biological experiments produce similar results.
The transactivation activity of EbMYBP1 was verified in yeast. The complete and various truncated EbMYBP1 ORFs were cloned into pGBKT7 plasmid, generating GAL4BD- EbMYBP1 recombinants. The recombinants were transformed into the Y2HGold strain, determining the transactivation ability of EbMYBP1. On the SD/-Trp medium, all the transformants and the negative control pGBKT7 grew well. On the SD-Trp/His/Ade medium, EbMYBP1 containing N-terminal regions did not grow. However, transformants containing the EbMYBP1 C-terminal regions grew well on SD–Trp/His/Ade medium (Figure 3C). These results indicated that EbMYBP1, as reported in the case of several other transcription factors, exhibited transactivation activity and its activation domain is located in the C-terminal region.
To investigate the possible role of EbMYBP1 in flavonoid biosynthesis, the complete ORF of EbMYBP1 was cloned in PC1300-35S to generate the 35S:EbMYBP1 and transformed into tobacco. Three independent transgenic lines EbMYBP1-OE-8, EbMYBP1-OE-10, and EbMYBP1-OE-15 (OE8, OE10, and OE15) were used for phenotype analysis (Supplementary Figure 2). Phenotypic analysis showed no obvious phenotypic changes compared with those of WT plants (Figures 4A,C). To further investigate whether EbMYBP1 expression is associated with total flavonoid accumulation in tobacco, the EbMYBP1 expression level and total flavonoid content was analyzed in the leaves of transgenic and WT plants. The total flavonoid content increased significantly by 1.89-, 1.31-, and 1.54- fold in the transgenic lines (OE8, OE10, and OE15 respectively) as compared to that of the WT (P < 0.01; Figure 4B). Therefore, OE8 was chosen and used for further transcriptome and metabolome analysis. These results suggested that the EbMYBP1 expression level was positively related to total flavonoids accumulation.
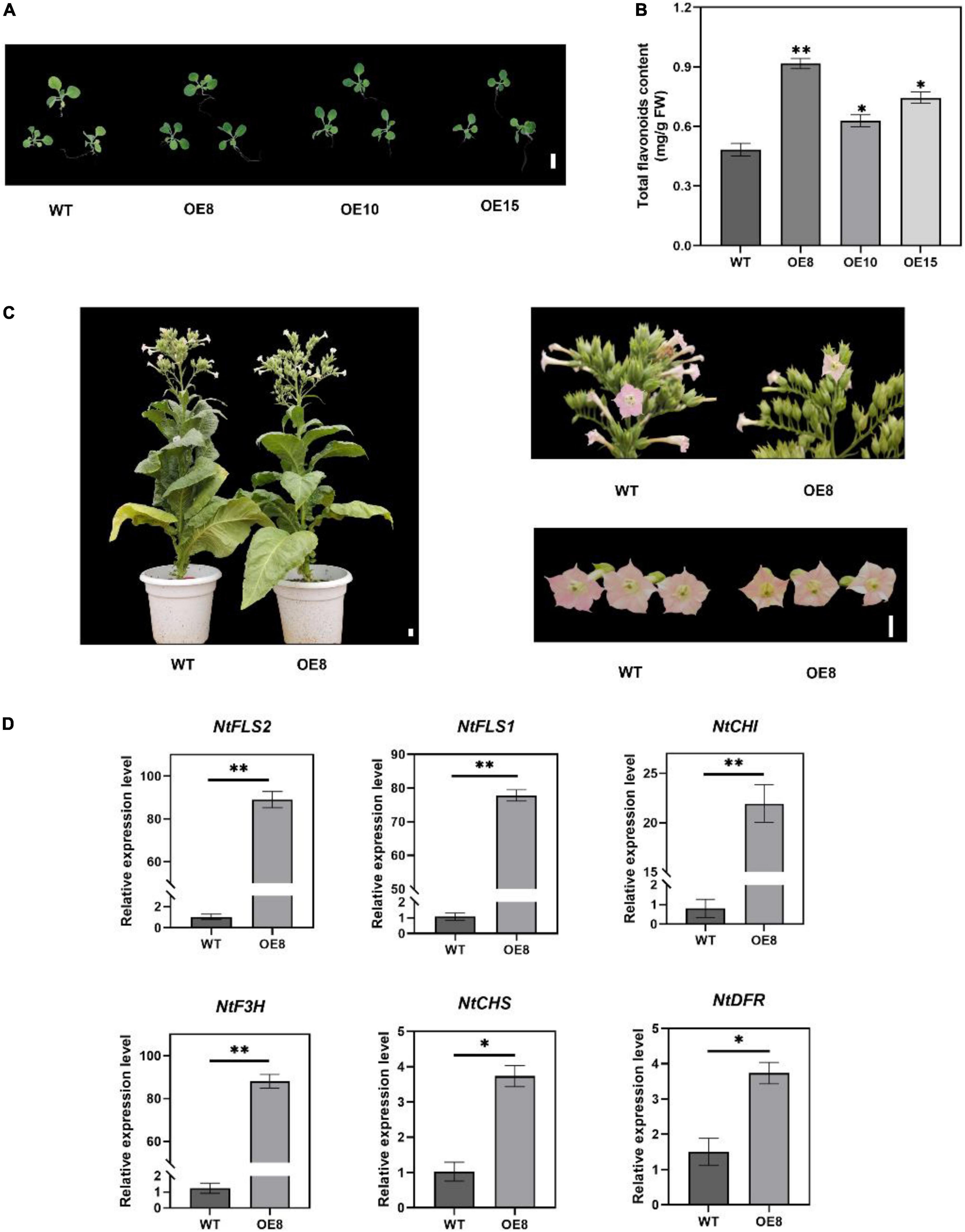
Figure 4. Characterization of transgenic tobacco lines overexpressing EbMYBP1. (A) Phenotype of transgenic and WT tobacco plants. (B) Contents of total flavonoids in the petals of transgenic and WT plants. (C) OE8 and WT tobacco plants. (D) RT-qPCR assay of the flavonoid pathway genes between WT and OE8. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone-3-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol 4-reductase. Data represent mean ± SD of three biological replicates. Asterisks indicate significant differences (*P < 0.05; **P < 0.01). Bar = 1 cm.
To further investigate the flavonoid-associated metabolic regulatory network in tobacco overexpressing EbMYBP1, 7-week-old leaves of OE8 were selected for metabolite analysis using UPLC-ESI-MS/MS. PCA analysis was conducted for all samples in WT and OE8. The results showed that these metabolites could be clearly divided into two groups (Supplementary Figure 3). The Partial least squares discriminant analysis (PLS-DA) and Orthogonal projection to latent structures-discriminant analysis (OPLS-DA) model, indicated that the data was reliable and meaningful (Supplementary Figure 4).
A total of 422 metabolites were obtained using a widely targeted metabolomics method (Supplementary Table 2). Furthermore, 98 flavonoid-related metabolites were identified, including 41 flavonols, 29 flavonoids, 7 flavonoid carbonosides, 5 dihydro flavonoids, 4 isoflavones, 4 anthocyanins, 3 dihydroflavonols, 3 flavanols, and 2 chalcones. A clear separation could be observed between WT and OE8, indicating distinct flavonoid profiles in WT and OE8 samples.
Differentially accumulated metabolites (DAMs) were selected according to a fold change ≥2 or ≤0.5 (P-value < 0.05). 243 DAMs were identified, and their accumulation was significantly different between WT and OE8, and 27 flavonoids metabolites displayed significantly higher content in the OE8 than in the WT. Moreover, the most prominently differential metabolites were 3,7-Di-O-methylquercetin and kaempferol 3-O-rutinoside in flavonol based on fold change ≥ 2 or ≤ 0.5 and VIP ≥ 1 in the OPLS-DA model (Supplementary Figure 5B and Supplementary Table 3). These results suggest that EbMYBP1 play an important role in regulation of flavonol biosynthesis.
To investigate the genetic basis of flavonoid-associated metabolism and its relationship to EbMYBP1 overexpression, RNA-Seq profiling was performed using the leaves of wild-type and EbMYBP1-OE (OE8) line. The libraries had 40,655,986–58,533,428 clean reads, and 95.00–96.50% reads were successfully mapped to the tobacco genome. Comparative analysis of the leaf transcriptome identified a total of 8,147 differentially expressed genes (DEGs) between the wild-type and OE8, of which 4,203 genes were upregulated, and 3,944 genes were downregulated (Supplementary Table 4). The PCA and cluster dendrogram of the transcriptome also supported the classification of gene expression patterns into two groups (Supplementary Figure 6A).
The DEGs were annotated to 44 GO terms (Supplementary Figure 6C), among which the three categories with the largest number of DEGs included “metabolic process” (1,148 genes), “catalytic activity” (1,074 genes), and “cell” (568 genes). KEGG pathway and enrichment analysis showed that DEGs were significantly assigned to 130 enriched pathways. The top five pathways in order of smallest to largest q value were “biosynthesis of secondary metabolites,” “metabolic pathways,” “MAPK signaling pathway,” “phenylalanine metabolism” and “flavonoid biosynthesis.” 664, 1,015, 114, 45, and 46 related genes were enriched in these pathways, respectively (Supplementary Figure 6D).
In the plant flavonoid-related pathway there were 46 DEGs. These include key enzyme-coding structural genes involved in flavonoid biosynthesis such as six phenylalanine ammonia lyases (PALs), one cinnamate 4-hydroxylases (C4Hs), six 4-coumarate-CoA ligases (4CLs), four chalcone synthases (CHSs), four chalcone isomerases (CHIs), two flavanone-3-hydroxylases (F3Hs), two flavonol synthase (FLSs), three flavonoid 3′-hydroxylases (F3’Hs), two dihydroflavonol reductases (DFRs) (Supplementary Figure 7). In addition, 343 transcription factors were found in the DEGs, of which 202 were upregulated and 141 were downregulated in the OE8. A total of 33 DEGs were found in the bHLH family, followed by 25 DEGs in the MYB family.
To test the reliability of transcriptome data, RT-qPCR assays were performed on six flavonoid structural genes that were differentially expressed in OE8. Gene expression as elucidated by RT-qPCR exhibited similar trends to that of the transcriptome data. RT-qPCR analysis showed that the expression of the flavonoid biosynthesis-related genes FLS2, FLS1, DFR, F3H, CHI, and CHS was upregulated in the OE8 lines compared to the control (Figure 4D). These results indicated that EbMYBP1 positively regulates flavonoid biosynthesis, and EbMYBP1 regulates genes expression directly or indirectly in the flavonoid synthesis pathway.
To determine whether EbMYBP1 directly regulate flavonoid biosynthesis related genes, we analyzed the cis-elements of the above six structural gene promoters. These promoter sequences were cloned, containing at least one conserved MYB-recognition element, indicating that EbMYBP1 can bind to their promoters. We then investigate whether EbMYBP1 directly affects the transcriptional activity of the promoters of the six representative genes using a transient transactivation assay in tobacco leaves. The cis-elements, bound to MYB proteins within the promoter region of these six genes, were identified using PlantCARE. MYB-binding elements were found in the promoters of all six genes involved in flavonoid biosynthesis pathway (Supplementary Figure 8). Therefore, dual-LUC assays were performed to identify whether EbMYBP1 activates the promoters of these genes in vivo. The LUC activity levels from the pGreen 0800:FLS2, pGreen 0800:F3H, pGreen 0800:FLS1, pGreen 0800:DFR, pGreen 0800:CHI, and pGreen 0800:CHS reporters were 10.31-, 10.73-, 6.54-, 4.14-, 2.94-, and 2.24-fold higher in EbMYBP1 than in the presence of the control, respectively (Figure 5).
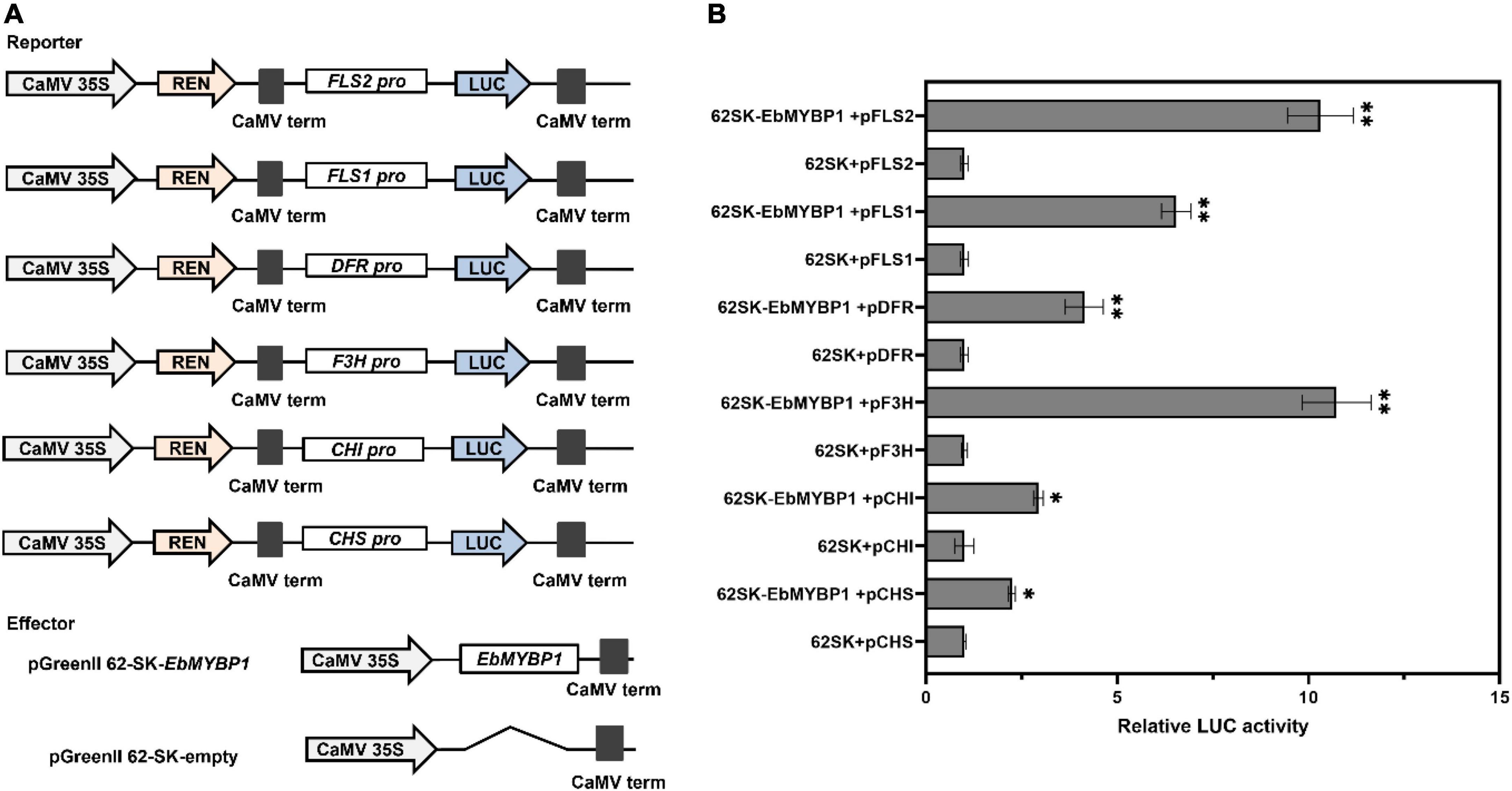
Figure 5. EbMYBP1 activated the expression of flavonoid–biosynthesis genes. (A) Diagrams of the reporter and effector constructs used in the dual-luciferase reporter assay. (B) Transient dual-luciferase reporter assays show that the activation of FLS2, FLS1, DFR, F3H, CHI, and CHS expression is activated by EbMYBP1. Transient expression assays used the promoter fragments of the FLS2, FLS1, DFR, F3H, CHI, and CHS genes. The luciferase luminescence intensities were quantitated following transfection with different vectors: 62SK represents empty pGreenII 62-SKvector; EbMYBP1 represents the pGreenII 62-SK-EbMYBP1 vector; FLS2, FLS1, DFR, F3H, CHI, and CHS represent pGreenII 0800-LUC-FLS2, pGreenII 0800-LUC-FLS1, pGreenII 0800-LUC-DFR, pGreenII 0800-LUC-F3H, pGreenII 0800-LUC-CHI, and pGreenII 0800-LUC-CHS vectors, respectively. Renilla luciferase (REN) was used for normalization. Data represent mean ± SD of three biological replicates. Asterisks indicate significant differences (*P < 0.05; **P < 0.01).
To further investigate if EbMYBP1 bind to the MYB-binding cis-elements in the FLS2, F3H, CHI, and CHS promoters, EMSA was performed, the results of which confirmed that EbMYBP1 binds to their promoter regions (Figure 6). These findings indicate that EbMYBP1 activates FLS2, F3H, CHI, and CHS expression both in vivo and in vitro, possibly by binding directly to the cis-element of their promoters.
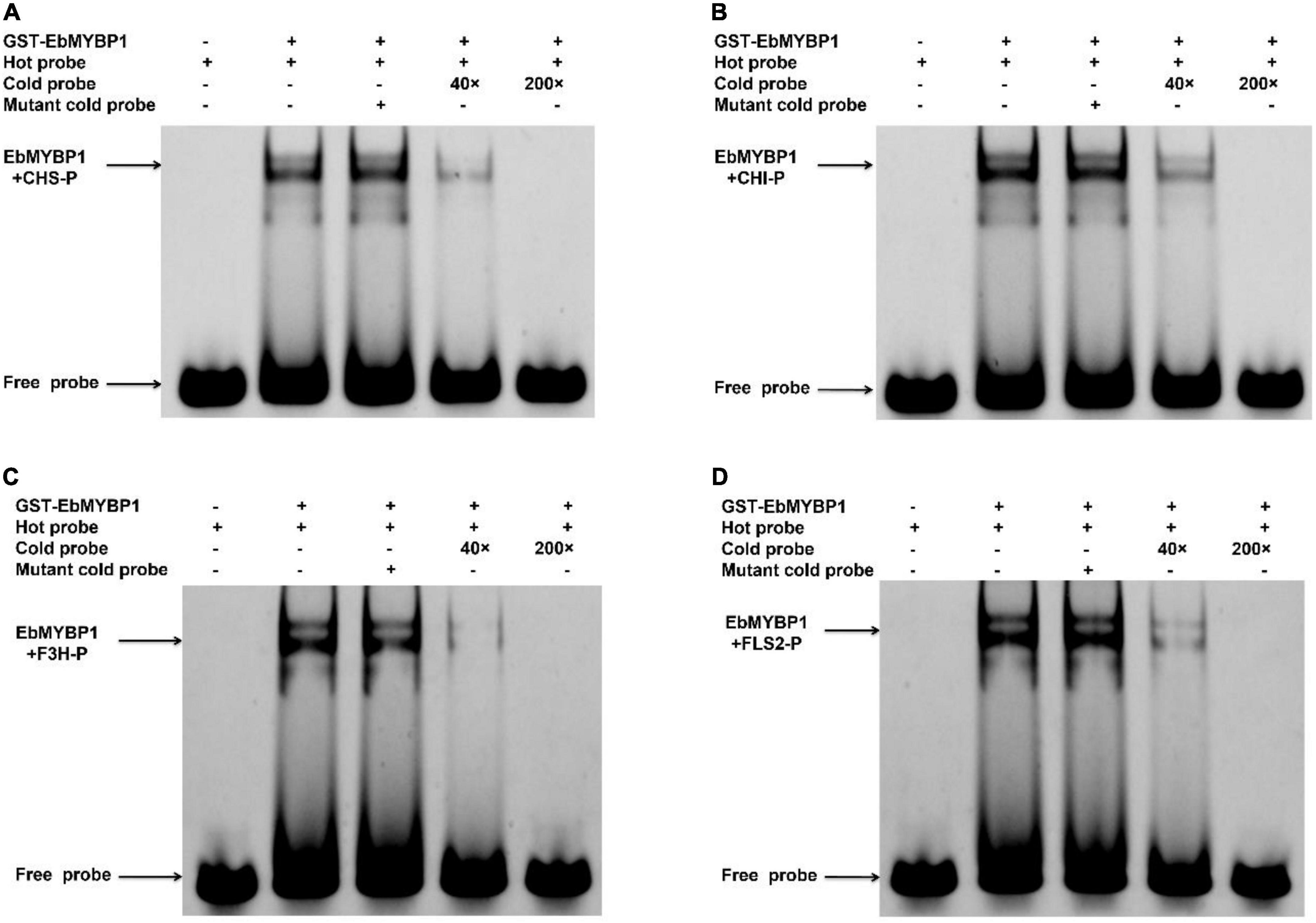
Figure 6. Electrophoretic mobility shift assay (EMSA) of EbMYBP1 binding to the CHS (A), CHI (B), F3H (C), and FLS2 (D) promoter region harboring EbMYBP1-binding sites in vitro.
Flavonoids and flavonoid glycosides are abundant active components in E. breviscapus (Wu et al., 2021). However, the regulatory mechanism underlying flavonoid and flavonoid glycoside metabolism remains unknown. Here, a novel R2R3-MYB transcription factor was identified in E. breviscapus, revealing the highly conserved R2R3 domain typical of MYB transcription factors at its N-terminus (Figure 1A). Similar to the AtMYB11, AtMYB12, and AtMYB111 from Arabidopsis and MYB protein P from maize, EbMYBP1 does not contain the conserved motif for interacting with bHLH protein (Grotewold et al., 1994; Mehrtens et al., 2005; Stracke et al., 2007). Additionally, EbMYBP1 has the SG7-1 and SG7-2 motifs, which are employed as identifiers for transcription factors related to flavonoid production (Stracke et al., 2001; Czemmel et al., 2009; Wang et al., 2022), clustering with proteins as flavonoid-specific activators. Many studies have demonstrated that TFs in the same subgroup shares similar functions (Pandey et al., 2014).
Thus, we hypothesized that EbMYBP1 also participates in the flavonoid metabolic pathway in E. breviscapus. Our findings suggested that the EbMYBP1 expression level correlated with the accumulation of flavonoids in different tissues in E. breviscapus. Our results showed that scutellarin are mainly abundant in the leaves of E. breviscapus (Figure 2C). The expression pattern of EbMYBP1 analysis revealed a correlation between EbMYBP1 expression and scutellarin accumulation in different tissues, indicating that EbMYBP1 may play a key role in flavonoid biosynthesis pathway. To further elucidate the function of EbMYBP1, EbMYBP1 was transformed and overexpressed in tobacco via the agrobacterium method.
The metabolome analysis showed that EbMYBP1 overexpression in tobacco resulted in significant flavonoid accumulation. This indicates that EbMYBP1 is involved in the regulation flavonoid biosynthesis in leaves compared to the WT. Meanwhile, the total flavonoids content was significantly higher in transgenic tobacco plants than that of WT (Figure 4B). The reason might be that the overexpression of EbMYBP1 significantly enhanced those genes expression involved in flavonoid biosynthesis. Therefore, the flavonoid metabolism has been expanded, resulting in increasing flavonoid accumulation. Additionally, flavonols are synthesized in all organs, however, other active components like anthocyanins, are accumulated in specific cell types (Tan et al., 2019). Consequently, there is a potential competition between flavonols and anthocyanins. Our results showed that the accumulation of kaempferol and its derivatives was increased.
In this study, the metabolome analysis identified 98 flavonoid-related metabolites. A dramatically higher level of 3,7-Di-O-methylquercetin and kaempferol 3-O-rutinoside, which were the flavonol metabolites that differed significantly, were found in leaves of OE8 compared to those in WT plants (Figure 2). Meanwhile, increased amounts of anthocyanin 3-O-beta-D-glucoside and cyanidin 3-O-galactoside in anthocyanin were detected within the leaves of OE8 relative to the WT (Supplementary Table 2). Previous studies showed that in Gerbera hybrida, GhMYB1a overexpression activates FLS expression, causing an increased in kaempferol and decrease in anthocyanins (Zhong et al., 2020). Overexpression of PpMYB15 or PpMYBF1 from Prunus persica L. Batsch led to significant accumulation of quercetin and kaempferol in their transgenic tobacco flowers (Cao et al., 2019). Similar results showed that overexpression of AtMYB12 (Luo et al., 2008; Misra et al., 2010), GtMYBP4 (Nakatsuka et al., 2012), AtMYB11 (Pandey et al., 2015), EsMYBF1 (Huang et al., 2016), and CcMYB12 (Blanco et al., 2018) led to accumulation of quercetin and kaempferol levels in transgenic tobacco flowers. Moreover, overexpression of grape MYBF1 or apple MYB22 complemented the deficient phenotype in flavonols in an Arabidopsis mutant (Czemmel et al., 2009; Wang et al., 2017).
Subsequently, transcriptomic analysis indicated that many genes related to flavonoid biosynthesis, from PAL to ANS genes, could be either directly or indirectly up-regulated by EbMYBP1. Moreover, the transcript abundances of the structural genes involved in flavonoid biosynthesis coincided with corresponding higher accumulated metabolite levels detected (Figure 3). RT-qPCR analysis of six flavonoid biosynthesis-related genes (FLS2, FLS1, CHI, F3H, CHS, and DFR) was further performed in transgenic tobacco. The RT-qPCR results showed that the expression levels of FLS2, FLS1, CHI, CHS, and F3H were significantly upregulated. Furthermore, DFR was also upregulated to a certain extent compared with the levels in WT. Previous studies showed that overexpression of the MYB gene not only upregulated expression of PAL, CHS, and CHI, but also anthocyanin biosynthesis genes such as DFR in the tobacco plants over-expressing EsMYB90 (Qi et al., 2020). In previous studies, it has been demonstrated that many R2R3-MYB transcription factors regulate flavonoid biosynthesis by interacting with the promoters of the targeted structural genes (Wang et al., 2017). The sequence of gene promoters also varies greatly in different plant species. Thus, the MYB TFs might exhibit different regulatory functions in flavonoid biosynthesis in different species (Liu et al., 2016).
Previous studies showed that AtMYB12 is involved in flavonoid biosynthesis and regulates the expression of AtCHS, AtCHI, AtF3H, and AtFLS (Mehrtens et al., 2005; Czemmel et al., 2009; Stracke et al., 2010). It has been demonstrated that upregulation of these genes gives rise to increased flavonoid accumulation in Arabidopsis (Guo et al., 2015; Wang et al., 2016, 2018). Flavonol synthase (FLS) and dihydroflavonol4-reductase (DFR) generally target the same substrate dihydroflavonol, leading to competition between flavonol and anthocyanin (Davies and Schwinn, 2003). It was found that strong upregulation of NtFLS and NtF3H may be crucial in promoting the branch of flavonol biosynthesis, whereas weak upregulation of NtDFR would decrease the branch of the anthocyanin biosynthesis. Moreover, CHS, CHI, F3H, and FLS genes are related to flavonoid biosynthesis, primarily being activated by the SG7 MYB factors like MYB11, MYB12, and MYB111, although SG7 MYB-independent flavonol biosynthesis was reported in pollen grains and siliques (Stracke et al., 2010). In this study, EbMYBP1 overexpression also activated the expression of NtDFR. Of these, DFR is related to anthocyanin and proanthocyanidin biosynthesis, and anthocyanidins are synthesized by ANS from leucoanthocyanidin produced by DFR. Meanwhile, EbMYBP1 activated the promoter of NtDRF, the gene that is thought to underlie anthocyanin synthesis.
In the flavonoid biosynthesis pathway, structural genes such as CHS, CHI, F3H, FLS2, FLS1, and DFR participated in flavonoid biosynthesis. Multiple cis-acting element were identified in these promoter regions, including MYB-binding elements, light-responsive elements, hormone-responsive elements, and elements for low-temperature responses (Supplementary Figure 8). These analyses indicated that various environmental and genetic factors may affect the biosynthesis of flavonoids. Thus, the promoters of these flavonoid biosynthesis genes were selected as potential targets of EbMYBP1 transcription activation. Dual-luciferase assay indicated that EbMYBP1 could bind and significantly activate the expression of FLS2, F3H, and FLS1, rather than DFR, CHI, and CHS (Figure 5). A similar promoter assay result was observed for AtMYB12/11/111, which was capable of activating CHS, CHI, F3H, and FLS promoters as well (Mehrtens et al., 2005; Stracke et al., 2007). Nevertheless, both VvMYBF1 and MdMYB22 have been shown to activate the promoters of VvCHI, VvFLS, and MdFLS in grapes and apples, respectively (Czemmel et al., 2009; Wang et al., 2017). Transient tobacco expression studies have reported that CsMYBF1 can only activate the CsCHS and CsFLS promoters (Liu et al., 2016). The EMSA assay showed that EbMYBP1 could specifically activate the expression of FLS2, F3H, CHI, and CHS genes, which function in flavonoid biosynthesis, leading to high levels of flavonoid production.
Based on our present findings, EbMYBP1, could regulate flavonoid biosynthesis by directly activating the expression of FLS2, CHI, CHS, and F3H genes. In conclusion, we identified a transcriptional activator EbMYBP1 that is positively involved in flavonoid biosynthesis by directly activating flavonoid-related genes. The findings reported here expanded our understanding of the intricate transcriptional regulatory network of flavonoid biosynthesis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
YZ, CZ, and SY conceived, designed the experiments, and wrote the article. YZ, QT, and QG performed the experiments. GZ, WS, GX, XL, GL, WF, and XL analyzed the data. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 81760694) and the Major Science and Technique Programs in Yunnan Province (No. 2019ZF011-1).
We are grateful to Chi Shan and Hannah Dahl for reading and commenting on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.946827/full#supplementary-material
Ballester, A.-R., Molthoff, J., de Vos, R., Hekkert, B. T. L., Orzaez, D., Fernaݩndez-Moreno, J.-P., et al. (2010). Biochemical and molecular analysis of pink tomatoes: deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 152, 71–84. doi: 10.1104/pp.109.147322
Blanco, E., Sabetta, W., Danzi, D., Negro, D., Passeri, V., Lisi, A. D., et al. (2018). Isolation and characterization of the flavonol regulator CcMYB12 from the globe artichoke [Cynara cardunculus var. scolymus (L.) Fiori]. Front. Plant Sci. 9:941. doi: 10.3389/fpls.2018.00941
Buer, C. S., Imin, N., and Djordjevic, M. A. (2010). Flavonoids: new roles for old molecules. J. Integ. Plant Biol. 52, 98–111. doi: 10.1111/j.1744-7909.2010.00905.x
Cao, Y., Xie, L., Ma, Y., Ren, C., Xing, M., Fu, Z., et al. (2019). PpMYB15 and PpMYBF1 transcription factors are involved in regulating flavonol biosynthesis in peach fruit. J. Agric. Food Chem. 67, 644–652. doi: 10.1021/acs.jafc.8b04810
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Zhang, K., Khurshid, M., Li, J., He, M., Georgiev, M. I., et al. (2019). MYB transcription repressors regulate plant secondary metabolism. Crit. Rev. Plant Sci. 38, 159–170. doi: 10.1080/07352689.2019.1632542
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. doi: 10.1093/mp/sst080
Czemmel, S., Stracke, R., Weisshaar, B., Cordon, N., Harris, N. N., Walker, A. R., et al. (2009). The Grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530. doi: 10.1104/pp.109.142059
Davies, K. M., and Schwinn, K. E. (2003). Transcriptional regulation of secondary metabolism. Funct. Plant Biol. 30, 913–925. doi: 10.1071/fp03062
Ferreyra, M. L. F., Serra, P., and Casati, P. (2021). Recent advances on the roles of flavonoids as plant protective molecules after UV and high light exposure. Physiol. Plantar. 173, 736–749. doi: 10.1111/ppl.13543
Grotewold, E., Drummond, B. J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553. doi: 10.1016/0092-8674(94)90117-1
Guo, J., Zhou, W., Lu, Z., Li, H., Li, H., and Gao, F. (2015). Isolation and functional analysis of chalcone isomerase gene from purple-fleshed sweet potato. Plant Mol. Biol. Rep. 33, 1451–1463. doi: 10.1007/s11105-014-0842-x
He, S., Dong, X., Zhang, G., Fan, W., Duan, S., Shi, H., et al. (2021). High quality genome of Erigeron breviscapus provides a reference for herbal plants in Asteraceae. Mol. Ecol. Resour. 21, 153–169. doi: 10.1111/1755-0998.13257
Huang, W., Khaldun, A. B. M., Chen, J., Zhang, C., Lv, H., Yuan, L., et al. (2016). A R2R3-MYB Transcription Factor regulates the flavonol biosynthetic pathway in a traditional chinese medicinal plant, Epimedium sagittatum. Front. Plant Sci. 7:1089. doi: 10.3389/fpls.2016.01089
Liu, C., Long, J., Zhu, K., Liu, L., Yang, W., Zhang, H., et al. (2016). Characterization of a Citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Sci. Rep. 6:25352. doi: 10.1038/srep25352
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, J., Butelli, E., Hill, L., Parr, A., Niggeweg, R., Bailey, P., et al. (2008). AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol. Plant J. 56, 316–326. doi: 10.1111/j.1365-313X.2008.03597.x
Matus, J. T., Loyola, R., Vega, A., Peña-Neira, A., Bordeu, E., Arce-Johnson, P., et al. (2009). Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J. Exp. Bot. 60, 853–867. doi: 10.1093/jxb/ern336
Mehrtens, F., Kranz, H., Bednarek, P., and Weisshaar, B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. doi: 10.1104/pp.104.058032
Misra, P., Pandey, A., Tiwari, M., Chandrashekar, K., Sidhu, O. P., Asif, M. H., et al. (2010). Modulation of transcriptome and metabolome of tobacco by arabidopsis transcription factor, AtMYB12, leads to insect resistance. Plant Physiol. 152, 2258–2268. doi: 10.1104/pp.109.150979
Naik, J., Misra, P., Trivedi, P. K., and Pandey, A. (2022). Molecular components associated with the regulation of flavonoid biosynthesis. Plant Sci. 317:111196. doi: 10.1016/j.plantsci.2022.111196
Nakatsuka, T., Saito, M., Yamada, E., Fujita, K., Kakizaki, Y., and Nishihara, M. (2012). Isolation and characterization of GtMYBP3 and GtMYBP4, orthologues of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J. Exp. Bot. 63, 6505–6517. doi: 10.1093/jxb/ers306
Pandey, A., Misra, P., Bhambhani, S., Bhatia, C., and Trivedi, P. K. (2014). Expression of Arabidopsis MYB transcription factor, AtMYB111, in tobacco requires light to modulate flavonol content. Sci. Rep. 4:5018. doi: 10.1038/srep05018
Pandey, A., Misra, P., and Trivedi, P. K. (2015). Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 34, 1515–1528. doi: 10.1007/s00299-015-1803-z
Pandith, S. A., Dhar, N., Rana, S., Bhat, W. W., Kushwaha, M., Gupta, A. P., et al. (2016). Functional promiscuity of two divergent paralogs of Type III Plant Polyketide Synthases. Plant Physiol. 171, 2599–2619. doi: 10.1104/pp.16.00003
Perez-Vizcaino, F., and Duarte, J. (2010). Flavonols and cardiovascular disease. Mol. Aspects Med. 31, 478–494. doi: 10.1016/j.mam.2010.09.002
Pollastri, S., and Tattini, M. (2011). Flavonols: old compounds for old roles. Ann. Bot. 108, 1225–1233. doi: 10.1093/aob/mcr234
Premathilake, A. T., Ni, J., Bai, S., Tao, R., Ahmad, M., and Teng, Y. (2020). R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit. Planta 252:59. doi: 10.1007/s00425-020-03473-4
Qi, Y., Gu, C., Wang, X., Gao, S., Li, C., Zhao, C., et al. (2020). Identification of the Eutrema salsugineum EsMYB90 gene important for anthocyanin biosynthesis. BMC Plant Biol. 20:186. doi: 10.1186/s12870-020-02391-7
Routaboul, J.-M., Dubos, C., Beck, G., Marquis, C., Bidzinski, P., Loudet, O., et al. (2012). Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J. Exp. Bot. 63, 3749–3764. doi: 10.1093/jxb/ers067
Saito, K., Yonekura-Sakakibara, K., Nakabayashi, R., Higashi, Y., Yamazaki, M., Tohge, T., et al. (2013). The flavonoid biosynthetic pathway in arabidopsis: structural and genetic diversity. Plant Physiol. Biochem. 72, 21–34. doi: 10.1016/j.plaphy.2013.02.001
Sheng, H., Sun, X., Yan, Y., Yuan, Q., Wang, J., and Shen, X. (2020). Metabolic engineering of microorganisms for the production of flavonoids. Front. Bioeng. Biotechnol. 8:589069. doi: 10.3389/fbioe.2020.589069
Song, W., Xiang, G., Lu, Y., Zhang, G., Yang, S., and Zhao, Y. (2021). Identification and expression profiling of R2R3-MYB transcription factors in Erigeron breviscapus. China J. Chin. Mater. Medica 46, 6149–6162. doi: 10.19540/j.cnki.cjcmm.20210924.101
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Jahns, O., Keck, M., Tohge, T., Niehaus, K., Fernie, A. R., et al. (2010). Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 188, 985–1000. doi: 10.1111/j.1469-8137.2010.03421.x
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Tan, H., Man, C., Xie, Y., Yan, J., Chu, J., and Huang, J. (2019). A crucial role of GA-regulated flavonol biosynthesis in root growth of Arabidopsis. Mol. Plant 12, 521–537. doi: 10.1016/j.molp.2018.12.021
Tohge, T., Watanabe, M., Hoefgen, R., and Fernie, A. (2013). Shikimate and Phenylalanine Biosynthesis in the Green Lineage. Front. Plant Sci. 4:62. doi: 10.3389/fpls.2013.00062
Wang, F., Kong, W., Wong, G., Fu, L., Peng, R., Li, Z., et al. (2016). AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genomics 291, 1545–1559. doi: 10.1007/s00438-016-1203-2
Wang, F., Ren, G., Li, F., Qi, S., Xu, Y., Wang, B., et al. (2018). A chalcone synthase gene AeCHS from Abelmoschus esculentus regulates flavonoid accumulation and abiotic stress tolerance in transgenic Arabidopsis. Acta Physiol. Plant 40:97. doi: 10.1007/s11738-018-2680-1
Wang, H., Liu, J.-X., Feng, K., Li, T., Duan, A.-Q., Liu, Y.-H., et al. (2022). AgMYB12, a novel R2R3-MYB transcription factor, regulates apigenin biosynthesis by interacting with the AgFNS gene in celery. Plant Cell Rep. 41, 139–151. doi: 10.1007/s00299-021-02792-4
Wang, N., Xu, H., Jiang, S., Zhang, Z., Lu, N., Qiu, H., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292. doi: 10.1111/tpj.13487
Wellmann, F., Lukačin, R., Moriguchi, T., Britsch, L., Schiltz, E., and Matern, U. (2002). Functional expression and mutational analysis of flavonol synthase from Citrus unshiu. Eur. J. Biochem. 269, 4134–4142. doi: 10.1046/j.1432-1033.2002.03108.x
Williams, R. J., Spencer, J. P. E., and Rice-Evans, C. (2004). Flavonoids: antioxidants or signalling molecules? Free Rad. Biol. Med. 36, 838–849. doi: 10.1016/j.freeradbiomed.2004.01.001
Wu, R., Liang, Y., Xu, M., Fu, K., Zhang, Y., Wu, L., et al. (2021). Advances in chemical constituents, clinical applications, pharmacology, pharmacokinetics and toxicology of Erigeron breviscapus. Front. Pharmacol. 12:656335. doi: 10.3389/fphar.2021.656335
Zakaryan, H., Arabyan, E., Oo, A., and Zandi, K. (2017). Flavonoids: promising natural compounds against viral infections. Arch. Virol. 162, 2539–2551. doi: 10.1007/s00705-017-3417-y
Zhai, R., Zhao, Y., Wu, M., Yang, J., Li, X., Liu, H., et al. (2019). The MYB transcription factor PbMYB12b positively regulates flavonol biosynthesis in pear fruit. BMC Plant Biol. 19:85. doi: 10.1186/s12870-019-1687-0
Zhang, X., Huang, H., Zhao, X., Lv, Q., Sun, C., Li, X., et al. (2015). Effects of flavonoids-rich Chinese bayberry (Myrica rubra Sieb. et Zucc.) pulp extracts on glucose consumption in human HepG2 cells. J. Funct. Foods 14, 144–153. doi: 10.1016/j.jff.2015.01.030
Keywords: Erigeron breviscapus, MYB transcription factor, flavonoid biosynthesis, secondary metabolism, scutellarin
Citation: Zhao Y, Zhang G, Tang Q, Song W, Gao Q, Xiang G, Li X, Liu G, Fan W, Li X, Yang S and Zhai C (2022) EbMYBP1, a R2R3-MYB transcription factor, promotes flavonoid biosynthesis in Erigeron breviscapus. Front. Plant Sci. 13:946827. doi: 10.3389/fpls.2022.946827
Received: 18 May 2022; Accepted: 07 July 2022;
Published: 28 July 2022.
Edited by:
Wei Ma, Nanyang Technological University, SingaporeReviewed by:
Sitakanta Pattanaik, University of Kentucky, United StatesCopyright © 2022 Zhao, Zhang, Tang, Song, Gao, Xiang, Li, Liu, Fan, Li, Yang and Zhai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Zhao, emhhb3lhbmttQDEyNi5jb20=; Shengchao Yang, c2hlbmdjaGFveWFuZ0AxNjMuY29t; Chenxi Zhai, Y2hlbnhpemhhaUBvdXRsb29rLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.