
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 July 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.945272
This article is part of the Research Topic Dissection of Physiological and Genetics Essences of Stress Resistance and High Yield in Triticeae Crops View all 8 articles
The development and production of bread wheat (Triticum aestivum L.) are widely affected by drought stress worldwide. Many NAC transcription factors (TFs) of stress-associated group (SNAC) are functionally proven to regulate drought tolerance. In this study, we identified 41 TaSNACs that were classified into 14 groups, and the expression of TaSNAC4-3D was induced in the leaf tissue via osmotic or abscisic acid (ABA) treatment. TaSNAC4-3D was localized to the nucleus through the transient expression assay, and the C-terminal region exhibited transcriptional activity via transactivation assays. TaSNAC4-3D was overexpressed in common wheat. The wheat plants with TaSNAC4-3D overexpression was more sensitive to drought stress compared with wild-type (WT) plants. The water loss rate showed no difference between transgenic lines and WT plants. However, drought stress increased H2O2 and O2– accumulation and promoted programmed cell death (PCD) in the leaf tissue of TaSNAC4-3D overexpression lines compared with WT plants. RNA-seq analysis was performed under well-watered and drought conditions, and four strong potential target genes, encoding senescence regulators, were identified by analyzing their promoters containing the NAC recognition sequence (NACRS). Based on these results, our findings revealed that TaSNAC4-3D negatively regulates drought tolerance by inducing oxidative damage in bread wheat.
Drought stress is a common abiotic stress in crop development and yield formation, and drought conditions have become more severe in recent years. Owing to their sessile lifestyle, crops have developed various strategies to cope with the adverse effects by drought stress. Transcriptional regulation is a crucial mechanism that regulates drought response, and many transcription factors (TFs) have been functionally verified to regulate drought tolerance in crops (Fujita et al., 2004; Ma et al., 2017; Li Y. et al., 2019; Wu et al., 2019).
The NAC (i.e., NAM, ATAF, and CUC2) TFs are among the largest TF families, with 105 NAC TFs in Arabidopsis, 151 in rice, and 283 in cotton (Ooka et al., 2003; Sun et al., 2018), several of which were functionally verified to regulate drought tolerance. Three AtNAC TFs, namely, ANAC019, ANAC055, and ANAC072, specifically bind to the CATGTG motif and were found to positively regulate drought stress (Tran et al., 2004). The expression of ATAF1, an NAC gene in Arabidopsis, was induced by drought and ABA treatments, and the Arabidopsis plants with ataf1 mutant showed high tolerance to drought stress (Lu et al., 2007). The AtNTL4, an NAC TF, was found to regulate the expression of several genes for reactive oxygen species (ROS) biosynthesis by binding to their promoters to promote ROS accumulation in Arabidopsis, which results in hypersensitivity to drought stress (Lee et al., 2012). ANAC016 inhibits the expression of AREB1 by binding to its promoter, thereby negatively regulating drought stress in Arabidopsis (Sakuraba et al., 2015). OsNAC2 was found to bind to the promoters of OsLEA3 and OsSAPK1, and the knockdown of OsNAC2 improved the rice yield under drought condition (Shen et al., 2017). OsNAC066 expression was induced by different abiotic stresses, and the rice plants with OsNAC066 knockout showed hypersensitivity to drought stress (Wang et al., 2020). OsNAC14 binds to the promoter of OsRAD51A1 and was proposed to positively regulate drought tolerance via DNA damage repair in rice (Sung et al., 2018). In cotton, GhirNAC2 binds to the promoters of GhNCED3s, which are responsible for ABA biosynthesis, and cotton plants with GhirNAC2s co-suppression are drought-sensitive (Shang et al., 2020). The expression of GhJUB1L1, a NAC gene in cotton, was induced by drought stress, and silencing it reduced drought tolerance (Chen Q. et al., 2021). Furthermore, many drought-responsive NAC TFs were functionally identified in tomatoes, maize, soybeans, and other plants (Peng et al., 2009; Mao et al., 2015; Tak et al., 2017; Nguyen et al., 2018; Thirumalaikumar et al., 2018). The aforementioned studies proved the critical roles of NAC TFs in regulating drought tolerance.
As a main food crop, bread wheat (Triticum aestivum L.) is the primary source of food for 30–40% of the population (Ray et al., 2013). In recent years, drought frequency and severity have worsened greatly affecting wheat production. Daryanto et al. (2016) examined wheat yield in response to drought stress using the field experiment data and found that drought stress induced a reduction of approximately 21% in wheat yield from 1980 to 2015. Therefore, screening of drought-responsive genes is crucial for further molecular breeding. Many TaNACs have been found to regulate drought response (Ma et al., 2022). TaSNAC4-3A expression is induced by drought stress, and its overexpression in Arabidopsis significantly increased drought tolerance by regulating stomatal aperture (Mei et al., 2021). TaNAC69 was overexpressed in wheat, and the transgenic wheat plants showed a higher biomass compared with WT plants under drought stress (Xue et al., 2011). TaSNAC8-6A overexpression in wheat stimulated the development of lateral roots to enhance drought tolerance, and chip-seq revealed several targeted genes of TaSNAC8-6A involved in auxin signaling pathways (Mao et al., 2020). TaNAC47 could bind to the ABA-responsive element (ABRE) cis-element, and the Arabidopsis plants with TaNAC47 overexpression showed hypersensitivity to ABA and higher tolerance to drought stress (Zhang L. et al., 2016). TaNAC071-A was overexpressed in wheat and was found to positively regulate drought tolerance modulated by TaMYBL1 (Mao et al., 2021). In our previous study, we collected a total of 12 drought-responsive TaNACs; interestingly, 10 of them were TaSNACs (Ma et al., 2022). Therefore, analyzing and selecting drought-responsive TaSNACs is of great significance. In this study, we identified 41 TaSNACs via genome-wide analysis by combining the previous studies and characterized a new drought-responsive NAC transcription factor of stress-associated group from wheat (TaSNAC) gene, named TaSNAC4-3D, which was found to negatively regulate drought tolerance by increasing oxidative damage in transgenic wheat plants with TaSNAC4-3D overexpression.
The wheat cv. Chinese Spring was used for expression analysis, and wheat seedlings were cultured as described by Ma et al. (2022). At the two-leaf stage, the wheat seedlings were transferred into Hogland solution and were then divided into control and Hogland solution containing 15% PEG-6,000 and 100 μM abscisic acid (ABA) treatment groups, respectively. The leaf and root tissues from the control and treated wheat seedlings were collected with three biological replications for each time point. For drought stress in soil, wheat seedlings were cultured as described by Ma et al. (2022). The leaf tissue was sampled at the point of relative water content (RWC) of soil of 50 and 40% with three biological replications, and the well-watered samples were regarded as the control group. All samples were stored at –80°C until RNA extraction.
The specific primers for TaSNAC4-3D cloning were designed using the genome data (Supplementary Table 1), and the PCR product was inserted into the pMD18-T vector (TaKaRa, Japan). The plasmids of the positive clones were sequenced to confirm the sequence of TaSNAC4-3D (Sangon, Shanghai, China). The full-length protein sequences of selected TaSNACs were used for multiple alignment using ClustalW, and a phylogenetic tree was constructed using the MEGA 5.11 software by the neighbor-joining method (Tamura et al., 2021), and bootstrap analysis was performed for 1,000 replicates. The gene structure of TaSNACs was determined using the Gene Structure Display Server 2.0 (GSDS, Hu et al., 2015).
TRIzol (TaKaRa, Japan) reagent was used for total RNA extraction, which was further used for first-strand cDNA synthesis using a PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Japan). The specific primers were designed for qPCR (Supplementary Table 1) on an ABI 7500 real-time PCR system (Applied Biosystems, United States) using the SYBR Premix Ex Taq Kit (TaKaRa, Japan). As the internal control, the wheat Tubulin gene was used to calculate the expression level by the 2–ΔΔCt method.
The TaSNAC-N (1–160 aa) and TaSNAC-C (161–316 aa) sequences were confirmed according to the location of the NAM domain, and primers were designed (Supplementary Table 1). Transcriptional activity assay was performed as described previously (Ma et al., 2022). For the subcellular localization assay, the specific primers without stop codons were used to obtain the complete coding sequence (CDS) of TaSNAC4-3D, and the product was inserted into the pJIT163-GFP vector to construct recombinant plasmids between the HindIII and BamHI sites. The recombinant plasmid and control pJIT163-GFP vectors were transformed into Arabidopsis protoplasts, which were prepared using the Arabidopsis Protoplast Preparation and Transformation Kit (Coolaber, Beijing, China). Green fluorescent protein (GFP) fluorescence was monitored using a laser scanning confocal microscopy (TCS SP8, Leica, Germany).
To obtain TaSNAC4-3D transgenic wheat plants, the complete CDS of TaSNAC4-3D was inserted into the pMWB110 vector. An Agrobacterium-mediated transformation system was used to introduce genetic transformation of TaSNAC4-3D in bread wheat (cv. Fielder). PCR amplification and qRT-PCR were performed to screen transgenic plants (Supplementary Table 1). Homozygous T3 transgenic seeds were obtained for further analysis.
The aboveground part of each plant under drought condition was weighed to evaluate the fresh weight and dried in an oven at 80°C for 24 h for dry weight. Each experiment included six replications. About 0.3 g of leaf tissue for each sample was used to determine the malondialdehyde (MDA) content (Zheng et al., 2008), which was determined by the absorption values at 600, 532, and 450 nm. For the determination of the proline content, about 0.3 g of leaf tissue for each sample was homogenized with 3% (w/v) sulfosalicylic acid, which was centrifuged at 10,000 g for 5 min. The supernatant (2 ml) was mixed with 2 ml of glacial acetic acid and 2 ml of acid ninhydrin, which was incubated in boiling water for 60 min. After cooling in ice, 4 ml of toluene was added to the mixture to analyze the absorbance of the chromophore. A calibration curve was established to determine the profile content using an UV spectrometer (UV-2600, Shimadzu, Kyoto, Japan) at 520 nm (Bates et al., 1973). Notably, 10 leaves from each line under well-watered condition were used for the determination of the water loss rate as described by Mega et al. (2019) and as described in our previous study (Ma et al., 2022). The O2– content and H2O2 content were determined using the detection kits (Solarbio, Beijing, China), which have been used in a previous study (Chen J. et al., 2021). Soil RWC was measured as our previous description (Ma et al., 2022). All data were obtained from three biological replications.
The wild-type (WT) and transgenic leaves under drought condition were dehydrated in paraffin, and the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) was performed using the TUNEL assay kit, as described by Li Y. B. et al. (2019). The fluorescence microscope at 590 nm was used to obtain photograph.
The leaves with TaSNAC4-3D overexpression and that of WT plants were used for RNA-seq analysis, and total RNA was extracted. Each library was constructed using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, Ipswich, United States) and sequenced on an Illumina HiSeq-PE150. After removing low-quality reads, the clean reads were used for alignment with the genome data of IWGSC RefSeq v1.1 (International Wheat Genome Sequencing Consortium (IWGSC), 2018) by Hisat 2 (version 2.0.5). Differential expression analysis was performed using DEseq2. The raw data were submitted to NCBI with the BioProject ID of PRJNA838079.
The high-confidence protein database of wheat genome (IWGSC RefSeq v2.1) was used for a Hidden Markov Model (HMM) searching for the NAC TFs (TaNACs, PF02365) based on the HMM profile (Mistry et al., 2021). Totally, 452 TaNACs were identified by further confirmation in SMART platform (Letunic et al., 2021). Detailed information on the 452 TaNACs, including annotation and ID conversion, is provided in Supplementary Table 2 (International Wheat Genome Sequencing Consortium (IWGSC), 2014; Clavijo et al., 2017; International Wheat Genome Sequencing Consortium (IWGSC), 2018). Similar to the previous study, NAC TFs were divided into several groups, including SNAC, ANAC34, NEO, SND, NAC22, NAC1, NAM/CUC3, OMNAC, and TIP (Nuruzzaman et al., 2010). To obtain the TaSNACs, the protein sequences of 452 TaNACs and 12 NACs from Arabidopsis for subgroup classification were used to construct a phylogenetic tree. From this phylogenetic tree, 40 TaSNACs were identified (Supplementary Figure 1 and Supplementary Table 3). After a sequence analysis with TaSNACs reported by Mao et al. (2020), TaSNAC8-6B was deleted as no NAM domain was found after HMM searching, and TaSNAC4-3A, which has been functionally verified (Mei et al., 2021), was collected to our research resulting in a total of 41 TaSNACs.
A phylogenetic tree was constructed using the 41 TaSNACs, which were divided into 14 homologous groups (Figure 1A). To avoid confusion with the further studies on TaSNACs, they were denoted as described by Mao et al. (2020), and three new members joined, namely, TaSNAC14-3A, TaSNAC14-3B, and TaSNAC14-3D (Supplementary Table 3). The exon–intron structure of these 41 TaSNACs was examined using the online GSDS server to understand the evolution of these genes (Hu et al., 2015), and a similar exon–intron structure was observed within the same homologous group of TaSNACs (Figure 1B). The structure of the relevant genes differed between the groups, indicating that these genes may have different functions during their evolution.
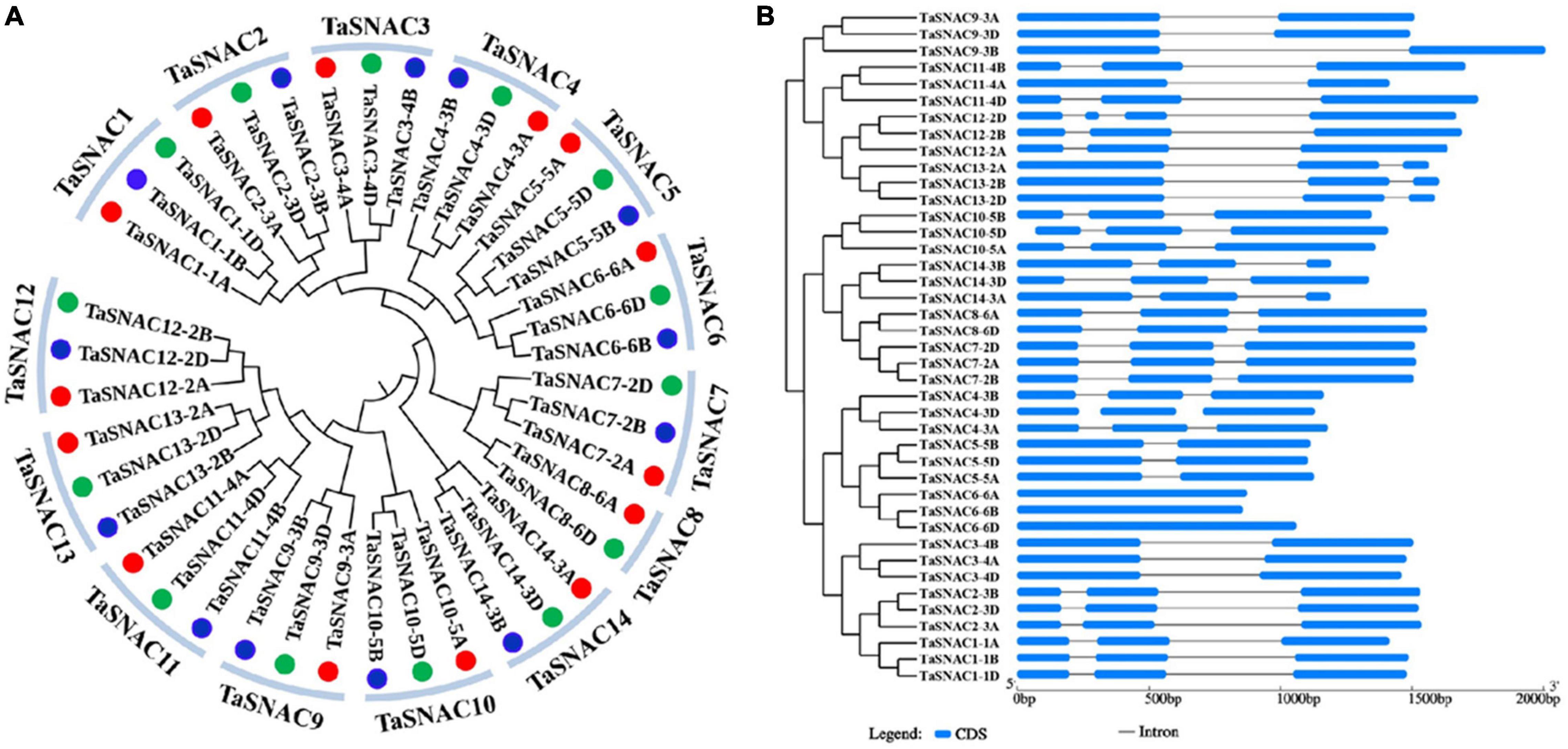
Figure 1. Phylogenetic tree and gene structure analysis of TaSNACs. (A) A total of forty-one TaSNACs were classified into 14 homologous groups, TaSNAC1 to TaSNAC14, through phylogenetic tree analysis. (B) The coding and genome sequences of 41 TaSNACs were obtained from the genome database and were used for gene structure analysis in the GSDS platform.
We examined the expression patterns of 14 groups of TaSNACs under 15% PEG-6000, drought, and ABA treatments. Eight groups of TaSNACs were upregulated under 15% PEG-6000 for 6 and 24 h (Figure 2A), seven groups of TaSNACs were upregulated under drought treatment with 50 and 40% soil RWC (Figure 2B), and eight groups of TaSNACs were upregulated under 100 μM ABA treatment for 6 and 24 h (Figure 2C). More importantly, we found that the group of TaSNAC4 was upregulated with more than 2-fold changes under the three treatments, which got our attention.
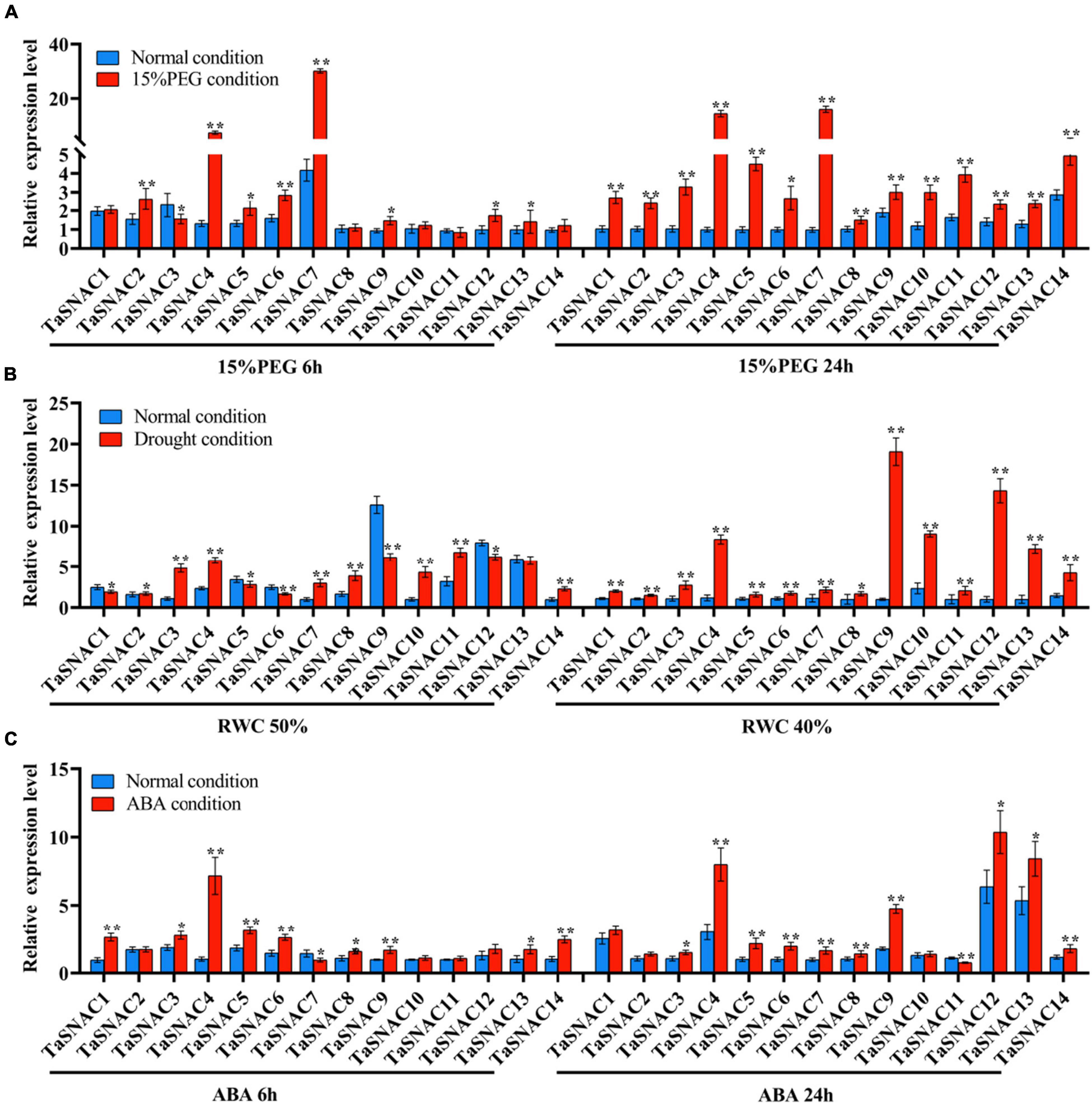
Figure 2. Expression pattern analysis of TaSNACs in 14 homologous groups under osmotic, drought, and abscisic acid (ABA) treatments. Wheat seedlings at the two-leaf stage were subjected to osmotic stress with 15% PEG-6000 for 6 and 24 h, and the expression pattern of 14 groups of TaSNACs was examined in the leaf tissue (A). Wheat seedlings at the two-leaf stage were subjected to drought stress by discontinuing watering, and the expression patterns of 14 groups of TaSNACs were examined in the leaf tissue when the soil RWC was 50 and 40% (B). Wheat seedlings at the two-leaf stage were subjected to 100 μM ABA treatment for 6 and 24 h, and the expression patterns of 14 groups of TaSNACs were examined in the leaf tissue (C). The value is shown as mean ± SD. The t-test was performed to analyze the difference between treatment and corresponding control, and the asterisk indicates a significant difference (*P < 0.05; **P < 0.01).
The WheatOmics platform collected the RNA-seq data about wheat (Ma S. et al., 2021), and the RNA-seq data of cold (Li et al., 2015), drought (Liu et al., 2015), osmotic (PRJNA306536), and salt (Zhang Y. et al., 2016) stresses was used to analyze the expression pattern of 41 TaSNACs. Differentially expressed TaSNACs were identified by a 1.5-fold change between the treatment and corresponding control. Among them, 15 differentially expressed TaSNACs were downregulated under cold stress. Under drought stress, 31 differentially expressed TaSNACs were identified, among which 9 TaSNACs were upregulated. After PEG-6000 treatment using cv. Giza168 and Gemmiza10, 22 differentially expressed TaSNACs in Giza168 and 30 differentially expressed TaSNACs in Gemmiza10 were identified. Under salt stress using cv. Chinese spring and QM6, 28 differentially expressed TaSNACs, including 22 downregulated TaSNACs, were identified in Chinese spring; 28 differentially expressed TaSNACs, including 23 downregulated TaSNACs, were identified in QM6. We found that TraesCS3D03G0748100 (TaSNAC4-3D) was differentially expressed under all abiotic stresses (Figure 3). Therefore, TaSNAC4-3D was selected as the candidate gene for further functional analysis.
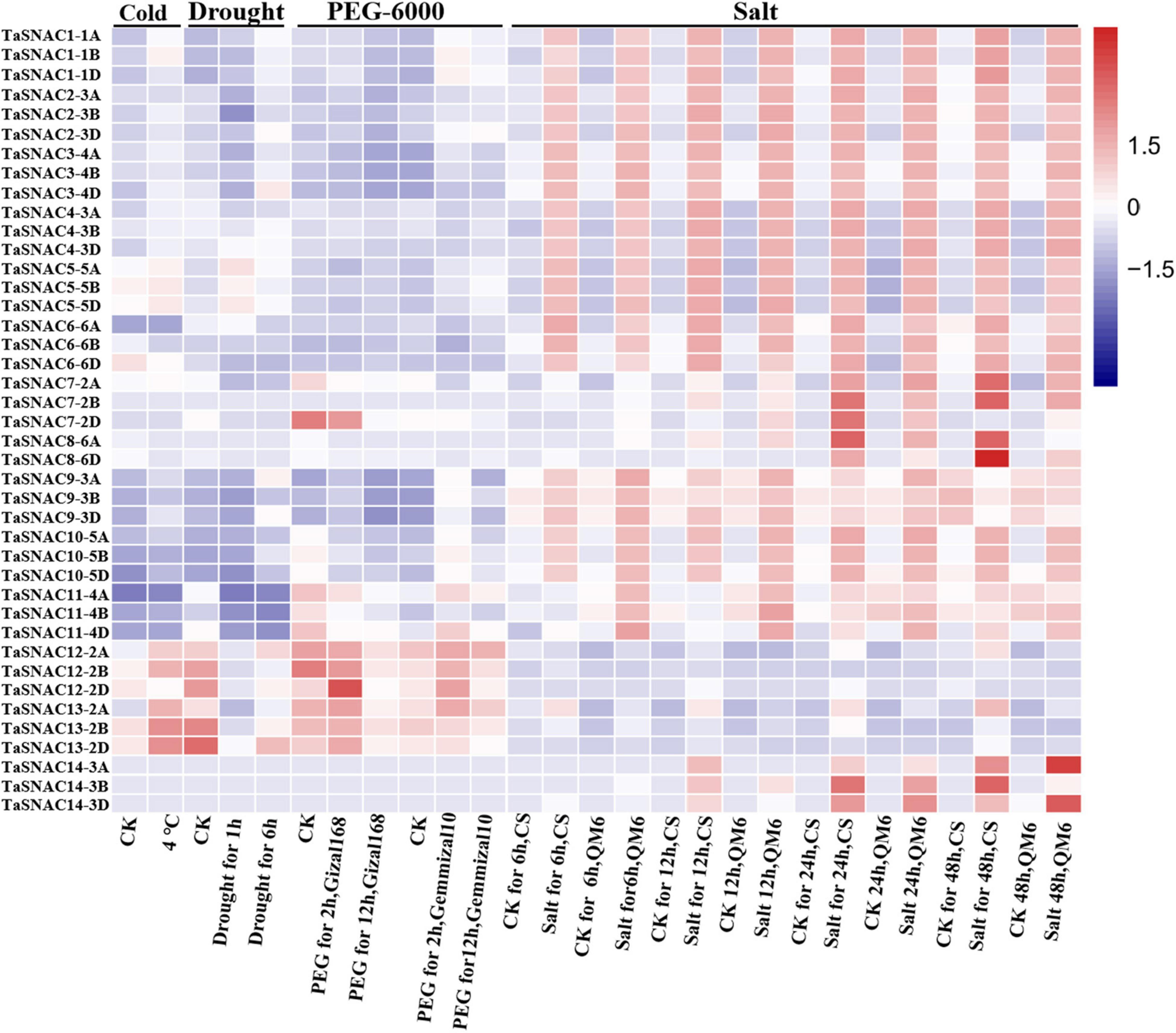
Figure 3. Gene expression heatmap of 41 TaSNACs. The transcriptomic data of abiotic stress, including cold, drought, osmotic, and salt stresses, was used to analyze the expression pattern of 41 TaSNACs, and the fragments per kilobase million value was used to represent the gene expression level. The red color and blue color represent high and low transcript abundance, respectively.
The expression pattern of TaSNAC4-3D under osmotic and ABA treatments was characterized via qRT-PCR. We used 15% PEG-6,000 to simulate osmotic stress. TaSNAC4-3D was significantly upregulated in the leaf tissue from 6 to 72 h of osmotic stress (Figure 4A) and in the root tissue from 12 to 48 h of drought stress (Figure 4B). The transcript levels of TaSNAC4-3D peaked at 12 h. Under ABA treatment, TaSNAC4-3D was upregulated in the leaf tissue for the first 6 h (Figure 4C) and downregulated in the root tissue for the first 3 h (Figure 4D). Therefore, we speculated that TaSNAC4-3D might participate in drought response in wheat.
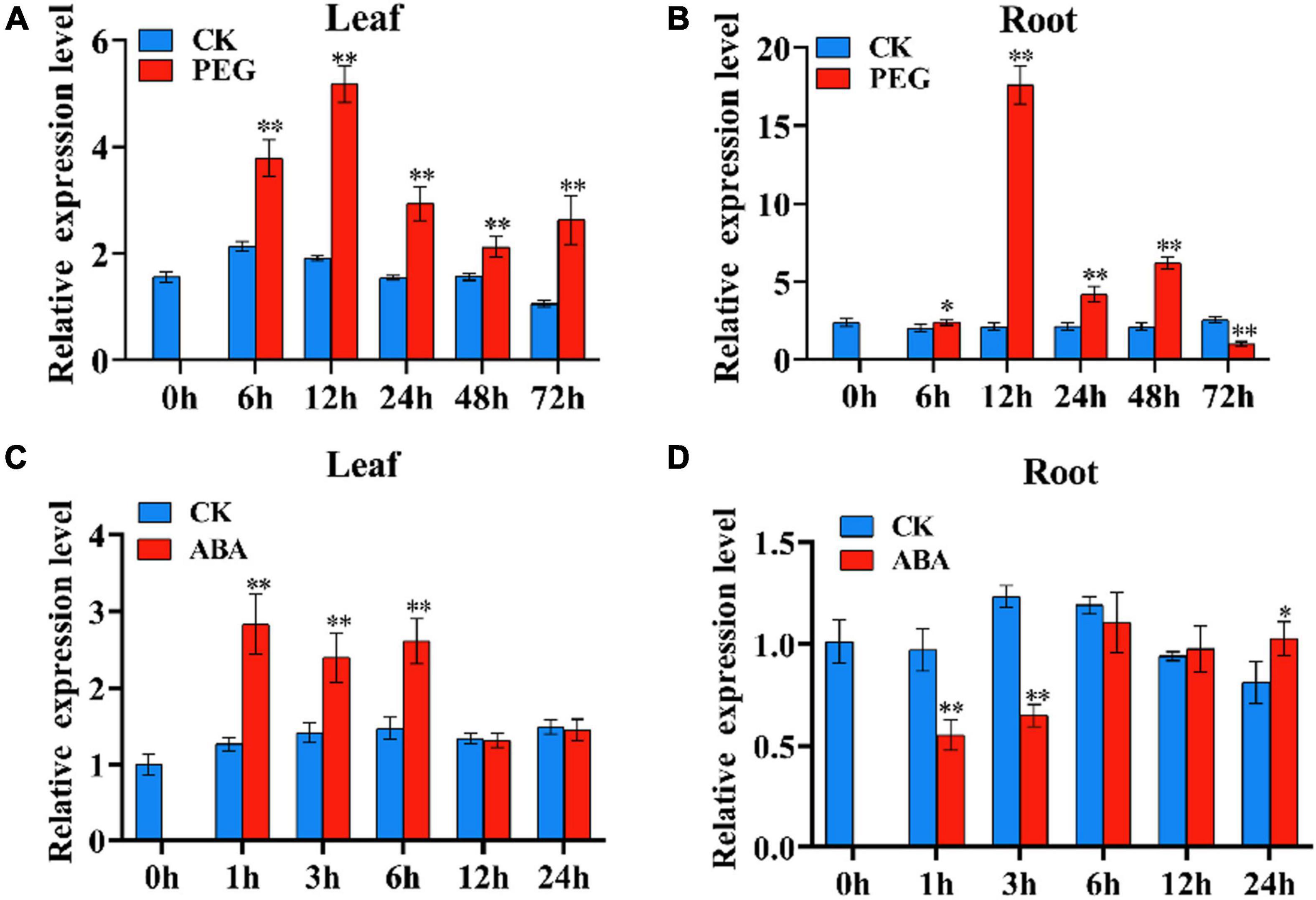
Figure 4. The expression pattern of TaNAC4-3D under osmotic stress and ABA treatment in wheat seedlings. The wheat seedlings were subjected to osmotic stress using 15% PEG-6000 for 6, 12, 24, 48, and 72 h at the two-leaf stage, and the corresponding controls were cultured in Hoagland solution. Under osmotic stress, the expression pattern of TaNAC4-3D was analyzed in the leaf (A) and root (B) tissues. The wheat seedlings at the two-leaf stage were cultured in Hoagland solution with 100 μM ABA and Hoagland solution as the control, respectively, for 1, 3, 6, 12, and 24 h. For ABA response, the expression pattern of TaNAC4-3D was analyzed in the root (C) and leaf (D) tissues. The value is shown as mean ± SD. The t-test was performed to analyze the difference between treatment and corresponding control, and the asterisk indicates a significant difference (*P < 0.05; **P < 0.01).
TaSNAC4-3D-GFP was transiently expressed in Arabidopsis protoplasts to investigate the subcellular localization of TaSNAC4-3D. GFP fluorescence was distributed throughout the cytoplasm and nucleus in the Arabidopsis protoplasts harboring control pJIT163-GFP vectors, and the red fluorescence was distributed on the vacuole membrane (Figure 5A). However, GFP fluorescence was specifically observed on the nucleus in Arabidopsis protoplasts when the fusion protein of TaSNAC4-3D-3D was expressed (Figure 5A), which confirmed that TaSNAC4-3D is localized to the nucleus.
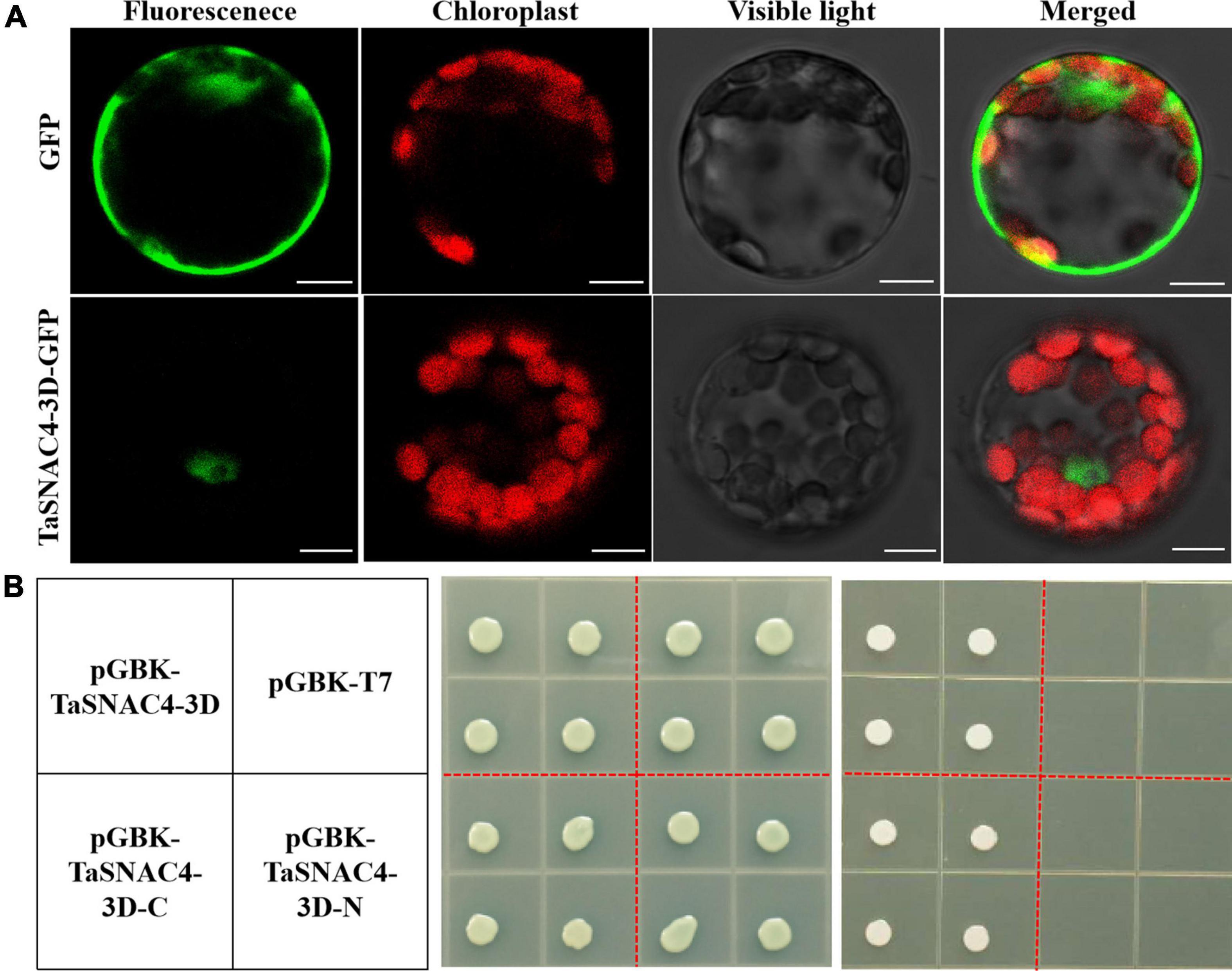
Figure 5. Subcellular localization and transcriptional activity analysis of TaSNAC4-3D. The recombinant plasmid and the control pJIT163-GFP vector were transformed into Arabidopsis protoplasts, and TaSNAC4-3D is localized to the nucleus (A). Scale bar = 10 μm. Three recombinant plasmids and empty pGBKT7 vector were transformed into the yeast strain. The growth status of transformed yeasts on SD/-Trp and SD/-Trp/-His/-Ade medium was observed, respectively (B).
For transcriptional activity analysis of TaSNAC4-3D, the plasmids of pGBKT7-TaSNAC4-3D, pGBKT7-TaSNAC4-3D-N (1–160 aa), pGBKT7-TaSNAC4-3D-C (161–316 aa), and empty pGBKT7 vector (negative control) were transformed into yeast (AH109), respectively. The transformants grew well on the SD medium without tryptophan (SD/-Trp), indicating the successful transformation of four vectors (Figure 5B), while only the transformants with the pGBKT7-TaSNAC4-3D and pGBKT7TaSNAC4-3D-C grew well on the SD medium without tryptophan, histidine, and adenine (SD/-Trp-His-Ade). These results suggested that the transactivation domain is located at the C-terminus of TaSNAC4-3D.
To investigate the functions of TaSNAC4-3D under drought stress, we obtained transgenic wheat plants with TaSNAC4-3D overexpression, which were detected by PCR amplification (Supplementary Figure 2). To assess the function of TaSNAC4-3D in drought regulation, we selected three independent lines (i.e., OE1, OE2, and OE3) with high transcript levels and WT plants for further analysis (Figures 6A,B). Under well-watered conditions, the growth status of the TaSNAC4-3D overexpression lines and WT plants showed no difference. The transgenic and WT plants were subjected to drought stress by discontinuing watering at the two-leaf stage. After 30 days of drought treatment, the TaSNAC4-3D overexpression lines exhibited a moderate stress phenotype, and the WT plants grew well. After 35 days of drought treatment, most leaves of the TaSNAC4-3D overexpression lines were wilted, whereas WT plants exhibited a moderate stress phenotype. All plants were rewatered on day 36. After 5 days, normal growth could not be restored in the transgenic plants, whereas the WT plants resumed their growth (Figure 6A).
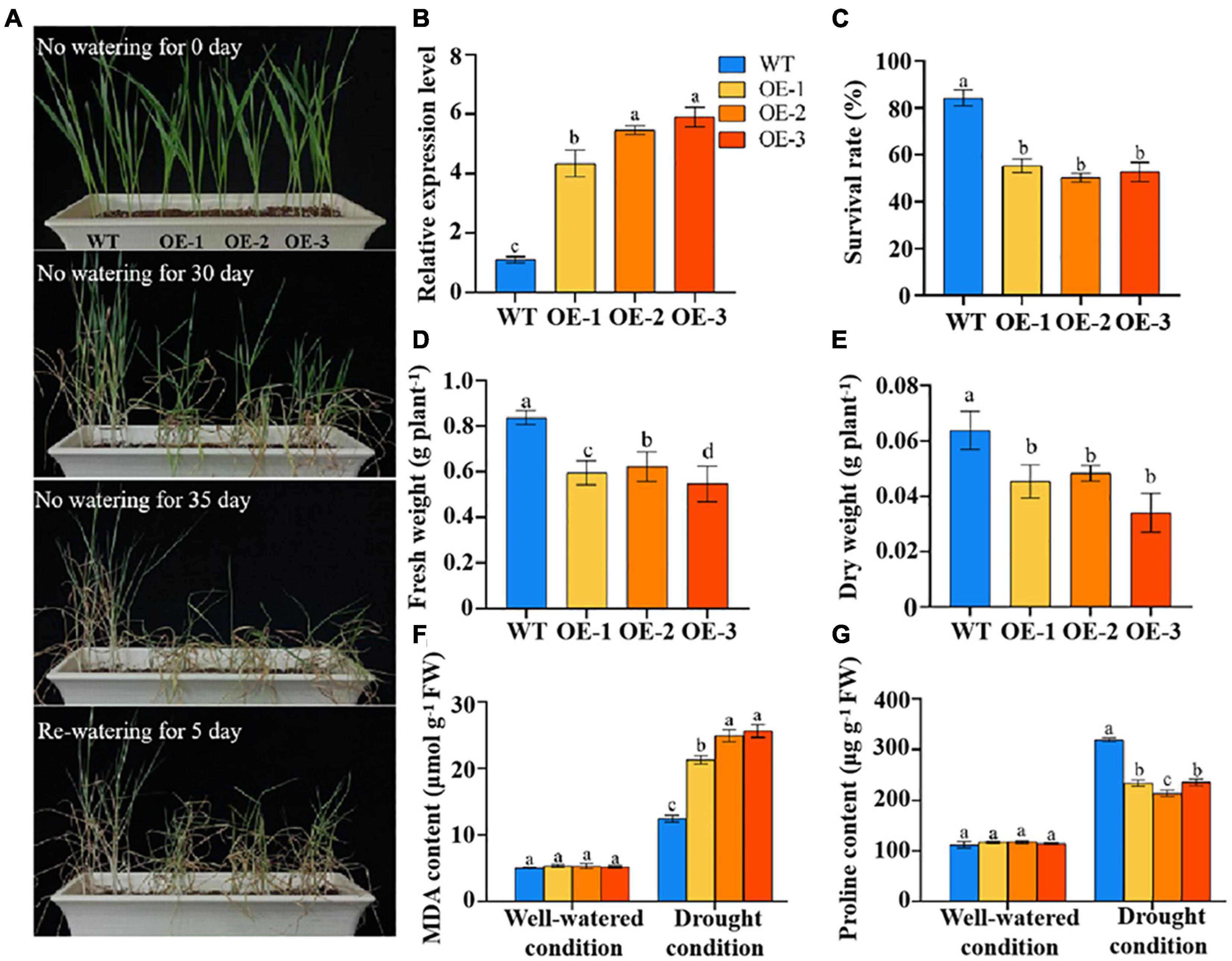
Figure 6. Overexpression of TaSNAC4-3D negatively regulates drought tolerance. The phenotype of three transgenic lines and wild-type (WT) plants under drought stress (A). The expression level of TaSNAC4-3D in three transgenic lines and WT plants (B). The survival rate (C), fresh weight (D), and dry weight (E) were measured using the three transgenic lines and WT plants under drought stress. The contents of MDA (F) and proline (G) were measured using the three transgenic lines and WT plants under well-watered and drought conditions, respectively. The value is shown as mean ± SD. Analysis of variance was used to analyze the difference, and the different letters mean the difference at 0.05 level.
The survival rate, fresh weight, and dry weight were investigated under drought stress, which were higher in WT plants than three transgenic lines (Figures 6C–E). The MDA and profile content showed no difference under well-watered condition, and significant difference under drought condition between the transgenic and WT plants (Figures 6F,G). From these results, we found that TaSNAC4-3D overexpression reduced drought tolerance in wheat seedlings.
Regulation of water loss is an important physiological process for alleviating drought stress. However, the water loss rate showed no difference between the transgenic lines and WT plants (Figure 7A). Oxidative damage is caused by drought stress. In this study, the contents of H2O2 and O2– were measured. The two indicators showed no difference between the transgenic lines and WT plants under normal conditions and were significantly higher in the transgenic lines than in WT plants under drought stress (Figures 7B,C). These results suggested that TaSNAC4-3D negatively regulates drought tolerance by increasing oxidative damage in wheat seedlings.
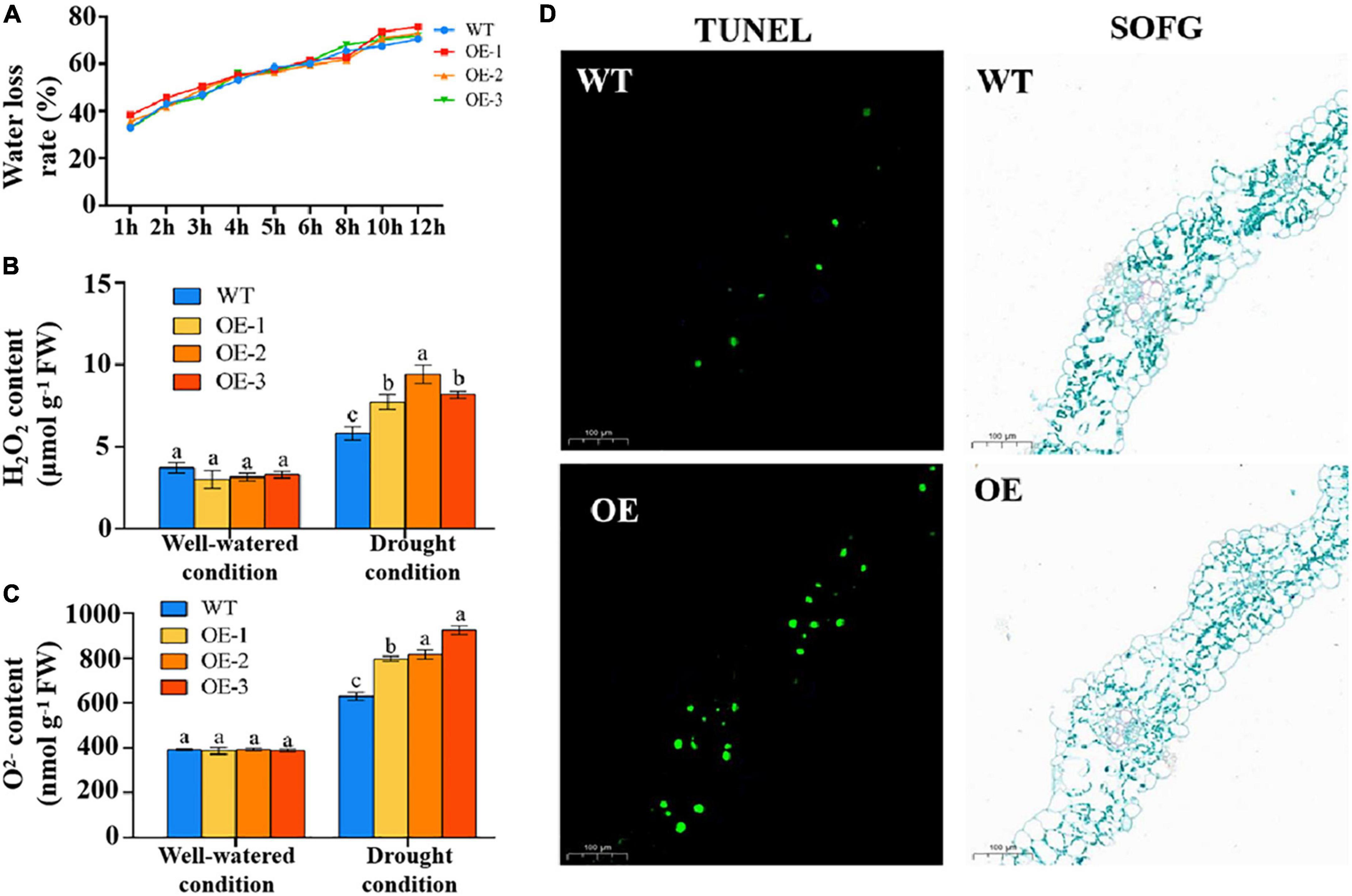
Figure 7. Overexpression of TaSNAC4-3D accelerated oxidative damage. The water loss rate was measured using the leaves of three transgenic lines and WT plants (A). The contents of H2O2 (B) and O2– (C) were measured using the leaves of three transgenic lines and WT plants under well-watered and drought conditions, respectively. TUNEL staining was performed using the leaves of transgenic and WT plants indicated that overexpression of TaSNAC4-3D accelerated PCD under drought stress (D). Scale bar = 100 μm, and FW represents fresh weight. Analysis of variance was used to analyze the difference, and the different letters mean the difference at 0.05 level.
Reactive oxygen species are important signaling molecules for PCD. Therefore, we speculated that PCD might be a factor causing plant wilting. To determine whether wheat leaves undergo PCD under drought stress, TUNEL staining was performed. The results showed that a greater number of green nuclei were observed in transgenic plants compared with WT plants (Figure 7D). These results indicated that TaSNAC4-3D overexpression induced excessive production of ROS under drought stress, thereby accelerating PCD.
The above results showed that overexpression of TaSNAC4-3D in wheat mediates drought tolerance. To better investigate the potential mechanism underlying the effects of TaSNAC4-3D, we analyzed and compared the RNA-Seq data using the transgenic and WT plants under well-watered and drought conditions. Three independent biological replications were set for each treated sample, and 12 libraries were obtained. The clean reads, average base sequencing error rate, Q20, Q30, and other information regarding these libraries are provided in Supplementary Table 4, and all these data indicate the high quality of 12 libraries.
The differentially expressed genes (DEGs) were selected using the parameters of more than twofold changes and a q-value < 0.05. Totally, 1,429 DEGs were obtained from plants with TaSNAC4-3D overexpression under well-watered condition (Figures 8A,B), including 712 upregulated and 717 downregulated genes, compared with WT plants, and 1,163 DEGs were obtained from plants with TaSNAC4-3D overexpression under drought condition (Figures 8A,B), including 921 upregulated and 242 downregulated genes. Detailed information on gene expression is provided in Supplementary Table 5. Among these DEGs, we found 53 persistently upregulated and 42 persistently downregulated genes under the two conditions (Figure 8B). Functional annotation revealed that the expression of genes, encoding the glycosyl hydrolases family, no apical meristem (NAM) protein, peptidase family, phosphatidylethanolamine-binding protein, protein kinase domain, raffinose synthase or Sip1, rhodanese-like domain, senescence regulator, zinc finger, and others, was upregulated, whereas the expression of genes, encoding fn3-like domain from purple acid phosphatase, lipoxygenase, major facilitator superfamily, NB-ARC domain, O-methyltransferase domain, subtilase family, UDP-glucoronosyl and UDP-glucosyl transferase, and others, was downregulated in the transgenic plants. We further analyzed the promoter of these selected 95 DEGs to find the NACRS with CACG core motif, and 93 persistently DEGs were selected (Supplementary Table 6). Interestingly, the expression of four senescence regulators genes, namely, TraesCS2A02G222500, TraesCS6B02G258900, TraesCS6A02G222200, and TraesCS6D02G212800, was upregulated in transgenic plants, and each contained more than ten NACRSs, which may be the strong potential target genes of TaSNAC4-3D.
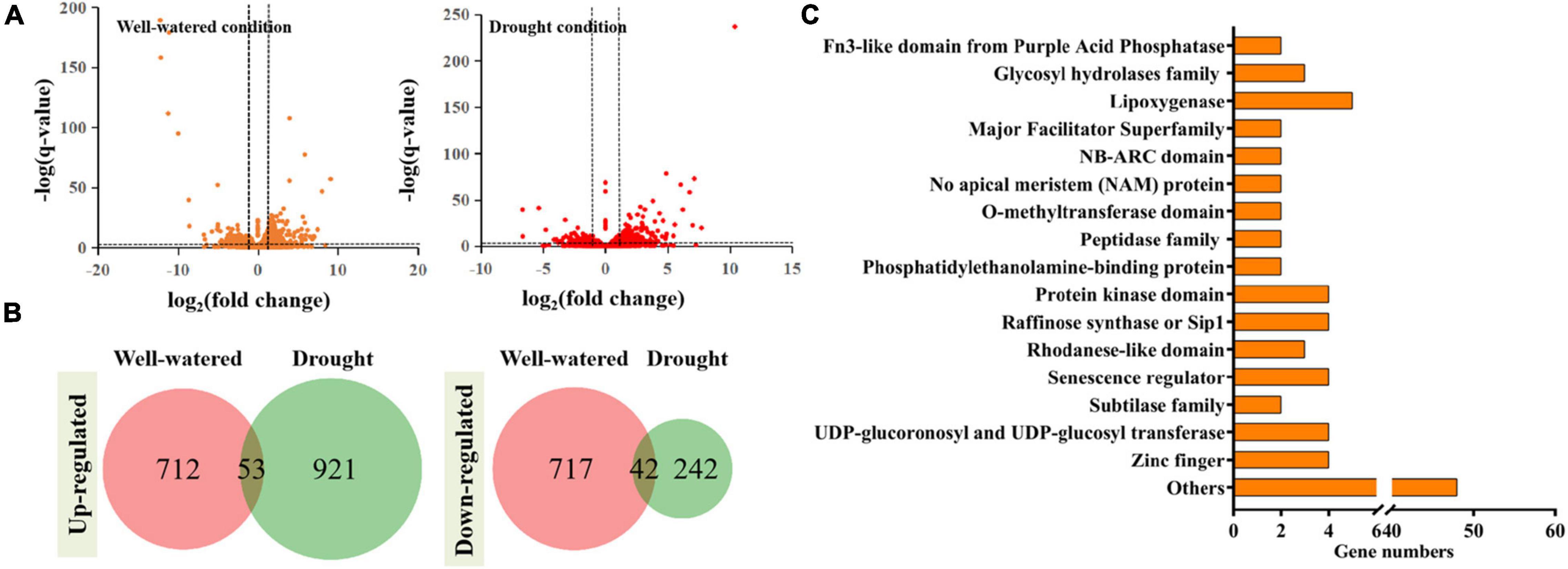
Figure 8. RNA-Seq analysis was performed using the leaves of transgenic and WT plants under well-watered and drought conditions. The DEGs were identified using the parameters: q-value < 0.05 and absolute fold change > 2 (A). 53 DEGs were upregulated and 42 DEGs were downregulated under two conditions (B). 93 genes contained the NACRS in their promoters (C).
NAC TFs are plant-specific proteins, and many members were functionally proven to regulate abiotic and biotic stress (Jensen and Skriver, 2014; Kim et al., 2016; Yuan et al., 2019). Many studies performed research on TaNAC identification along with the completion of bread wheat genome sequencing. Totally, 453 TaNACs were identified by Borrill et al. (2017) using the wheat genome by TGAC wheat assembly, and Ma J. et al. (2021) identified 462 TaNACs using IWGS RefSeq v1.0 database. In this study, we identified 452 TaNACs using the latest version of genome database (IWGSC RefSeq v2.1, Supplementary Table 2). This provides important information for further functional analysis. As one of the most important crops, about 50% of bread wheat is grown in arid and semiarid regions, and drought is a major abiotic stress for wheat production (Sternberg, 2011). Therefore, studying the regulatory mechanisms of wheat under drought stress is necessary to further improve its drought resistance. TaNACs were divided into several groups, including SNAC group (Nuruzzaman et al., 2010). In wheat, 12 TaNACs were functionally verified to be involved in drought response; 10 of them were found to be the members of TaSNACs (Ma et al., 2022), including nine positive regulators and one negative regulator. Therefore, TaSNACs are important drought regulators in wheat. A previous study identified 39 TaSNACs using the version of IWGS RefSeq v1.1 database (Mao et al., 2020). In this study, we collected the result of Mao et al. and finally identified 41 TaSNACs, which were divided into 14 groups (Figure 1 and Supplementary Table 3). Through expression pattern analysis, we found that most of them were differentially expressed under abiotic stress, and TaSNAC4 upregulated more than 2-fold changes under three abiotic stresses (Figure 2). By analyzing the RNA-seq data about abiotic stresses in wheat, TaSNAC4-3D showed differentially expressed under all abiotic stresses (Figure 3). Therefore, TaSNAC4-3D was selected for functional analysis.
Plants have developed several complex regulatory processes to adapt to drought stress. Under drought stress, water loss is reduced through stomatal closure, and several TaSNACs, including TaNAC2D, TaNAC2, and TaNAC48, were proved to regulate water loss under drought stress (Mao et al., 2012; Huang and Wang, 2016; Chen J. et al., 2021). However, the water loss rate was comparable between TaSNAC4-3D overexpression lines and WT plants. We further examined the content of H2O2 and O2– and found that they were significantly higher in TaSNAC4-3D overexpression lines than in WT plants (Figure 7). OsNAC4, a homolog of TaSNAC4-3D in rice, was functionally verified to promote PCD (Kaneda et al., 2009), and ANAC081, a homolog of TaSNAC4-3D in Arabidopsis, was proven to promote leaf senescence (Nagahage et al., 2020). These studies indicated that TaSNAC4-3D may be a negatively regulator in response to abiotic stress. However, TaSNAC4-3A, which belongs to the TaSNAC4 group, was found to positively regulate drought tolerance through its overexpression in Arabidopsis (Mei et al., 2021). In this study, TaSNAC4-3D was overexpressed in wheat, and wheat plants with TaSNAC4-3D overexpression were hypersensitive to drought stress (Figure 6). We further found that the transgenic plants accumulated more ROS content than WT plants under drought stress (Figure 7), which functioned similar to that of homologous genes in rice and Arabidopsis. Therefore, TaSNAC4-3D was confirmed to negatively regulate drought stress in wheat.
Programmed cell death is an important mechanism to deal with environmental stress (Wu et al., 2016), and excess ROS accumulation usually promotes PCD (Gechev et al., 2012). We observed the significant accumulation of H2O2 and O2– content in transgenic plants under drought stress compared with WT plants (Figure 7). In addition, overexpression of TaSNAC4-3D accelerated PCD under drought stress, which should be closely related to oxidative damage. Researchers have attempted to analyze the potential mechanism of TaNACs using RNA-seq (Mao et al., 2020; Mei et al., 2021), and the NACRS in the promoter was considered to be the binding core of the NAC TFs. In this study, we obtained 95 reliable DEGs using RNA-seq under well-watered and drought growth conditions, of which 93 DEGs contained NACRS in their promoters as the potential target genes (Figure 8 and Supplementary Table 5). As the homolog of TaSNAC4-3D in Arabidopsis was found to positively regulate leaf senescence (Nagahage et al., 2020), four upregulated senescence regulators in transgenic plants got our attention. Furthermore, the promoter of each senescence regulator gene contains more than 10 NACRSs (Supplementary Table 6). They should be the strong potential target genes of TaSNAC4-3D and play important roles in negatively regulating drought stress. Further studies are required to elucidate a more detailed mechanism.
In summary, we identified 452 TaNACs and 41 TaSNACs from the latest version of genome database (IWGSC RefSeq v2.1, Supplementary Table 2). From the expression pattern analysis of TaSNAC genes, a drought-responsive TaSNAC, TaSNAC4-3D, was selected. TaSNAC4-3D was overexpressed in wheat, and the transgenic wheat plants showed hypersensitivity to drought stress compared with WT plants that was possibly induced by oxidative damage. Moreover, four genes encoding senescence regulators were found to be the strong potential target genes of TaSNAC4-3D through RNA-seq and functional analysis.
The original contributions presented in this study are publicly available. This data can be found here: NCBI, PRJNA838079.
JM and LJ conceived and designed the research. JM, MZ, WL, XT, DZ, LW, and CL performed the experiments. JM, MZ, and XT analyzed the data. JM, MZ, XT, and DZ prepared the figures and provided the materials. JM and XT wrote the manuscript. All authors have read and approved the final manuscript.
This research was funded by the National Key Research and Development Program of China (2017YFD0301101 and 2016YFD0300203-3) and the Key Project of Science and Technology of Henan Province of China (182102110148).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.945272/full#supplementary-material
Supplementary Figure 1 | The phylogenetic tree using 452 TaNACs and 12 NACs from Arabidopsis for TaSNAC identification.
Supplementary Figure 2 | Identification of TaSNAC4-3D overexpression plants using PCR amplification.
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Borrill, P., Harrington, S. A., and Uauy, C. (2017). Genome-wide sequence and expression analysis of the NAC transcription factor family in polyploid wheat. Genes Genomes Genet. 7, 3019–3029. doi: 10.1534/g3.117.043679
Chen, J., Gong, Y., Gao, Y., Zhou, Y., Chen, M., Xu, Z., et al. (2021). TaNAC48 positively regulates drought tolerance and ABA responses in wheat (Triticum aestivum L.). Crop J. 9, 785–793. doi: 10.1016/j.cj.2020.09.010
Chen, Q., Bao, C., Xu, F., Ma, C., Huang, L., Guo, Q., et al. (2021). Silencing GhJUB1L1 (JUB1-like 1) reduces cotton (Gossypium hirsutum) drought tolerance. PLoS One 16:e0259382. doi: 10.1371/journal.pone.0259382
Clavijo, B. J., Venturini, L., Schudoma, C., Accinelli, G. G., Kaithakottil, G., Wright, J., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 2017, 885–896. doi: 10.1101/gr.217117.116
Daryanto, S., Wang, L., and Jacinthe, P. A. (2016). Global synthesis of drought effects on maize and wheat production. PLoS One 11:e0156362. doi: 10.1371/journal.pone.0156362
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Gechev, T. S., Dinakar, C., Benina, M., Toneva, V., and Bartels, D. (2012). Molecular mechanisms of desiccation tolerance in resurrection plants. Cell. Mol. Life Sci. 69, 3175–3186. doi: 10.1007/s00018-012-1088-0
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, Q., and Wang, Y. (2016). Overexpression of TaNAC2D displays opposite responses to abiotic stresses between seedling and mature stage of transgenic Arabidopsis. Front. Plant Sci. 7:1754. doi: 10.3389/fpls.2016.01754
International Wheat Genome Sequencing Consortium (IWGSC) (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788. doi: 10.1126/science.1251788
International Wheat Genome Sequencing Consortium (IWGSC) (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Jensen, M. K., and Skriver, K. (2014). NAC transcription factor gene regulatory and protein–protein interaction networks in plant stress responses and senescence. IUBMB Life 66, 156–166. doi: 10.1002/iub.1256
Kaneda, T., Taga, Y., Takai, R., Iwano, M., Matsui, H., Takayama, S., et al. (2009). The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 28, 926–936. doi: 10.1038/emboj.2009.39
Kim, H. J., Nam, H. G., and Lim, P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56. doi: 10.1016/j.pbi.2016.06.002
Lee, S., Seo, P. J., Lee, H. J., and Park, C. M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70, 831–844. doi: 10.1111/j.1365-313X.2012.04932.x
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, Q., Zheng, Q., Shen, W., Cram, D., Fowler, D. B., Wei, Y., et al. (2015). Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 27, 86–103. doi: 10.1105/tpc.114.134338
Li, Y. B., Cui, D. Z., Sui, X. X., Huang, C., Huang, C. Y., Fan, Q. Q., et al. (2019). Autophagic survival precedes programmed cell death in wheat seedlings exposed to drought stress. Int. J. Mol. Sci. 20:5777. doi: 10.3390/ijms20225777
Li, Y., Zhang, S., Zhang, N., Zhang, W., Li, M., Liu, B., et al. (2019). MYB-CC transcription factor, TaMYBsm3, cloned from wheat is involved in drought tolerance. BMC Plant Biol. 19:143. doi: 10.1186/s12870-019-1751-9
Liu, Z., Xin, M., Qin, J., Peng, H., Ni, Z., Yao, Y., et al. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15:152. doi: 10.1186/s12870-015-0511-8
Lu, P. L., Chen, N. Z., An, R., Su, Z., Qi, B. S., Ren, F., et al. (2007). A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 63, 289–305. doi: 10.1007/s11103-006-9089-8
Ma, J., Gao, X., Liu, Q., Shao, Y., Zhang, D., Jiang, L., et al. (2017). Overexpression of TaWRKY146 increases drought tolerance through inducing stomatal closure in Arabidopsis thaliana. Front. Plant Sci. 8:2036. doi: 10.3389/fpls.2017.02036
Ma, J., Tang, X., Sun, B., Wei, J., Ma, L., Yuan, M., et al. (2022). A NAC transcription factor, TaNAC5D-2, acts as a positive regulator of drought tolerance through regulating water loss in wheat (Triticum aestivum L.). Environ. Exp. Bot. 195:104805. doi: 10.1016/j.envexpbot.2022.104805
Ma, J., Yuan, M., Sun, B., Zhang, D., and Jiang, L. (2021). Evolutionary divergence and biased expression of NAC transcription factors in hexaploid bread wheat (Triticum aestivum L.). Plants 10:382. doi: 10.3390/plants10020382
Ma, S., Wang, M., Wu, J., Guo, W., Chen, Y., Li, G., et al. (2021). WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14, 1965–1968. doi: 10.1016/j.molp.2021.10.006
Mao, H., Li, S., Chen, B., Jian, C., Mei, F., Zhang, Y., et al. (2021). Variation in cis-regulation of a NAC transcription factor contributes to drought tolerance in wheat. Mol. Plant. 15, 276–292. doi: 10.1016/j.molp.2021.11.007
Mao, H., Li, S., Wang, Z., Cheng, X., Li, F., Mei, F., et al. (2020). Regulatory changes in TaSNAC8-6A are associated with drought tolerance in wheat seedlings. Plant Biotechnol. J. 18, 1078–1092. doi: 10.1111/pbi.13277
Mao, H., Wang, H., Liu, S., Li, Z., Yang, X., Yan, J., et al. (2015). A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 6:8326. doi: 10.1038/ncomms9326
Mao, X., Zhang, H., Qian, X., Li, A., Zhao, G., and Jing, R. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. doi: 10.1093/jxb/err462
Mega, R., Abe, F., Kim, J. S., Tsuboi, Y., Tanaka, K., Kobayashi, H., et al. (2019). Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 5, 153–159. doi: 10.1038/s41477-019-0361-8
Mei, C., Chen, B., Li, F., Zhang, Y., Kang, Z., Wang, X., et al. (2021). Overexpression of the wheat NAC transcription factor TaSNAC4-3A gene confers drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 160, 37–50. doi: 10.1016/j.plaphy.2021.01.004
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Nagahage, I., Sakamoto, S., Nagano, M., Ishikawa, T., and Yamaguchi, M. (2020). An Arabidopsis NAC domain transcription factor, ataf2, promotes age-dependent and dark-induced leaf senescence. Physiol. Plant. 170, 299–308. doi: 10.1111/ppl.13156
Nguyen, K. H., Mostofa, M. G., Li, W., Ha, C. V., Watanabe, Y., Le, D. T., et al. (2018). The soybean transcription factor GmNAC085 enhances drought tolerance in Arabidopsis. Environ. Exp. Bot. 151, 12–20. doi: 10.1016/j.envexpbot.2018.03.017
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. doi: 10.1016/j.gene.2010.06.008
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Peng, H., Cheng, H. Y., Chen, C., Yu, X. W., Yang, J. N., Gao, W. R., et al. (2009). A NAC transcription factor gene of Chickpea (Cicer arietinum), CarNAC3, is involved in drought stress response and various developmental processes. J. Plant Physiol. 166, 1934–1945. doi: 10.1016/j.jplph.2009.05.013
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insuffificient to double global crop production by 2050. PLoS One 8:e66428. doi: 10.1371/journal.pone.0066428
Sakuraba, Y., Kim, Y. S., Han, S. H., Lee, B. D., and Paek, N. C. (2015). The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27, 1771–1787. doi: 10.1105/tpc.15.00222
Shang, X., Yu, Y., Zhu, L., Liu, H., and Guo, W. (2020). A cotton nac transcription factor ghirnac2 plays positive roles in drought tolerance via regulating aba biosynthesis. Plant Sci. 296:110498. doi: 10.1016/j.plantsci.2020.110498
Shen, J., Lv, B., Luo, L., He, J., Mao, C., Xi, D., et al. (2017). The nac-type transcription factor osnac2 regulates aba-dependent genes and abiotic stress tolerance in rice. Sci. Rep. 7:40641. doi: 10.1038/srep40641
Sternberg, T. (2011). Regional drought has a global impact. Nature 472, 169–169. doi: 10.1038/472169d
Sun, H., Hu, M., Li, J., Chen, L., Li, M., Zhang, S., et al. (2018). Comprehensive analysis of NAC transcription factors uncovers their roles during fiber development and stress response in cotton. BMC Plant Biol. 18:1–15. doi: 10.1186/s12870-018-1367-5
Sung, S. J., Nuri, O., Joong, C. P., Shic, K. Y., Do, C. Y., and Ju-Kon, K. (2018). Overexpression of OsNAC14 improves drought tolerance in rice. Front. Plant Sci. 9:310. doi: 10.3389/fpls.2018.00310
Tak, H., Negi, S., and Ganapathi, T. R. (2017). Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 254, 803–816. doi: 10.1007/s00709-016-0991-x
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evolu. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thirumalaikumar, V. P., Devkar, V., Mehterov, N., Ali, S., Ozgur, R., Turkan, I., et al. (2018). NAC transcription factor JUNGBRUNNEN 1 enhances drought tolerance in tomato. Plant Biotechnol. J. 16, 354–366. doi: 10.1111/pbi.12776
Tran, L. S. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.2307/3872386
Wang, B., Zhong, Z., Wang, X., Han, X., and Zhang, Y. (2020). Knockout of the OsNAC006 transcription factor causes drought and heat sensitivity in rice. Int. J. Mol. Sci. 21:2288. doi: 10.3390/ijms21072288
Wu, H., Zheng, Y., Liu, J., Zhang, H., and Chen, H. (2016). Heme oxygenase-1 delays gibberellin-induced programmed cell death of rice aleurone layers subjected to drought stress by interacting with nitric oxide. Front. Plant Sci. 6:1267. doi: 10.3389/fpls.2015.01267
Wu, J., Jiang, Y., Liang, Y., Chen, L., Chen, W., and Cheng, B. (2019). Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol. Biochem. 137, 179–188. doi: 10.1016/j.plaphy.2019.02.010
Xue, G. P., Way, H. M., Richardson, T., Drenth, J., Joyce, P. A., and McIntyre, C. L. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 4, 697–712. doi: 10.1093/mp/ssr013
Yuan, X., Wang, H., Cai, J., Li, D., and Song, F. (2019). NAC transcription factors in plant immunity. Phytopathol. Res. 1, 1–13. doi: 10.1186/s42483-018-0008-0
Zhang, L., Zhang, L., Xia, C., Zhao, G., Jia, J., and Kong, X. (2016). The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front. Plant Sci. 6:1174. doi: 10.1111/j.1399-3054.2011.01539.x
Zhang, Y., Liu, Z., Khan, A. A., Lin, Q., Han, Y., Mu, P., et al. (2016). Expression partitioning of homeologs and tandem duplications contribute to salt tolerance in wheat (Triticum aestivum L.). Sci. Rep. 6:21476. doi: 10.1038/srep21476
Keywords: Triticum aestivum L., TaSNAC4-3D, drought stress, hypersensitive, transgenic wheat
Citation: Ma J, Zhang M, Lv W, Tang X, Zhao D, Wang L, Li C and Jiang L (2022) Overexpression of TaSNAC4-3D in Common Wheat (Triticum aestivum L.) Negatively Regulates Drought Tolerance. Front. Plant Sci. 13:945272. doi: 10.3389/fpls.2022.945272
Received: 16 May 2022; Accepted: 09 June 2022;
Published: 04 July 2022.
Edited by:
Shuaifeng Geng, Institute of Crop Sciences (CAAS), ChinaCopyright © 2022 Ma, Zhang, Lv, Tang, Zhao, Wang, Li and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhui Ma, Y3JpY2Fhc0AxNjMuY29t; Lina Jiang, amxuMDQxMDQwQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.