
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 16 August 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.944763
This article is part of the Research TopicThe Brassicaceae — Agri-Horticultural and Environmental Perspectives, Volume IIView all 19 articles
Heightening the resistance of plants to microbial infection is a widely concerned issue, especially for economical crops. Receptor-like proteins (RLPs), typically with tandem leucine-rich repeats (LRRs) domain, play a crucial role in mediating immune activation, being an indispensable constituent in the first layer of defense. Based on an analysis of orthologs among Brassica rapa, Brassica oleracea, and Brassica napus using Arabidopsis thaliana RLPs as a reference framework, we found that compared to A. thaliana, there were some obvious evolutionary diversities of RLPs among the three Brassicaceae species. BnRLP encoding genes were unevenly distributed on chromosomes, mainly on chrA01, chrA04, chrC03, chrC04, and chrC06. The orthologs of five AtRLPs (AtRLP3, AtRLP10, AtRLP17, AtRLP44, and AtRLP51) were highly conserved, but retrenchment and functional centralization occurred in Brassicaceae RLPs during evolution. The RLP proteins were clustered into 13 subgroups. Ten BnRLPs presented expression specificity between R and S when elicited by Sclerotinia sclerotiorum, which might be fabulous candidates for S. sclerotiorum resistance research.
Integrating exterior information with intrinsic cues is indispensable for all organisms, which is particularly crucial for plants due to their sessile lifestyle. Plants employ a large number and a vast variety of receptors for transducing signals from the extracellular matrix to the cell interior. Receptor-like proteins (RLPs), known as a kind of pattern recognition receptors (PRRs), are the main components of the first layer of plant immunity and a vital group of transmembrane receptors, which are generally composed of a short cytoplasmic domain, an extracellular leucine-rich repeat (LRR) domain, and a transmembrane domain (Wang et al., 2008). To date, 57 AtRLPs (Wang et al., 2008), 90 RLPs in rice (Fritz-Laylin et al., 2005), 82 RLPs in poplar (Petre et al., 2014), 144 RLPs in cotton (Chen et al., 2015), 176 RLPs in tomato (Kang and Yeom, 2018), and 228 RLPs in Brassica juncea (Yang et al., 2020) have been identified, and pan-genome prediction of resistance gene analogs (RGAs) in Brassica oleracea and also across the Brassicaceae were conducted (Bayer et al., 2019; Tirnaz et al., 2020). Currently, the enormous variation in RLPs between or within species has been discussed. RLPs are much more diverse and function in various pathways, involving both plant developmental regulation and immune response, even possessing dual roles. Indeed, most RLPs with assigned functions are involved in disease resistance. The identification of tomato Cf and Ve proteins that provide resistance against Cladosporium fulvum and Verticillium spp., respectively, started an era of RLP research (Jones et al., 1994; Kawchuk et al., 2001).
Subsequently, RLPs, for example, apple HcrVfs that confer resistance to fungal pathogens Cladosporium fulvum as well as Venturia inaequalis (Belfanti et al., 2004; Malnoy et al., 2008); tomato LeEIX proteins that function as a receptor to mediate recognition of the ethylene-inducing xylanase of Trichoderma viride (Ron and Avni, 2004); Nicotiana benthamiana elicitor-inducible leucine-rich repeat receptor-like protein (EILP) that is an ortholog of tomato Cf protein involved in bamboo mosaic virus movement (Chen et al., 2017); and Gbvdr6 that was homologous to Ve conferring resistance to Verticillium dahliae Kleb through the regulation of the JA/ET and SA signaling pathways in upland cotton were identified successively (Yang et al., 2017).
In Arabidopsis, of the 57 putative RLPs, approximately a quarter of them have been functionally characterized. CLAVATA2 (CLV2)/AtRLP10 and TOO MANY MOUTHS (TMM)/AtRLP17 are the first two AtRLPs that are experimentally validated implicating in the development functions and are involved in regulating the meristem and organ development (Jeong et al., 1999), as well as stomatal patterning and distribution (Nadeau and Sack, 2002), respectively. FASCIATED EAR2, an ortholog of CLV2 in maize, also plays an important role in the regulation of stem cell homeostasis (Taguchi-Shiobara et al., 2001). It is noteworthy that functional redundancy exists among AtRLPs. AtRLP2 and AtRLP12, identified as homologs of CLV2, rescue the clv2 meristem defects when driven by the endogenous promoter of CLV2 (Wang et al., 2010). Besides, the overexpression of AtRLP3 and AtRLP11 also rescued the phenotype of the clv2-1 mutant (Wu et al., 2016a). Nevertheless, Steidele and Stam clustered AtRLPs into two superclades (basal RLPs and pathogen-responsive RLPs) and referred to nine AtRLPs (AtRLP4, 10/CLV2, 17/TMM, 29, 44, 46, 51, 55, and 57) as putative developmental orthologs (PDOs) based on differences in transcript and protein abundance or clustering at the genomic loci (Steidele and Stam, 2021). Interestingly, CLV2/AtRLP10 plays a role both in developmental and defense-related processes.
Over the years, more AtRLPs have been shown to preferentially fulfill a role in pathogen defense. Among them, six AtRLPs (RLP1, 3, 23, 30, 32, and 42) were validated as pathogen-responsive RLPs (Steidele and Stam, 2021). AtRLP1/ReMAX is required for the perception of eMAX, which is a kind of MAMPs from Xanthomonas (Jehle et al., 2013a,b). AtRLP3/RFO implicates in resistance to the vascular wilt fungus Fusarium oxysporum (Shen and Diener, 2013). AtRLP23 perceives a conserved fragment found in most necrosis and ethylene-inducing peptide1-like proteins (NLPs) and thereby activates the immune responses, which are involved in pre-invasive resistance to the pathogen Botrytis cinerea (Albert et al., 2015; Ono et al., 2020). AtRLP30 and AtRLP18 impact the susceptibility of Pseudomonas syrinegae pv. phaseolicola to non-host resistance (Wang et al., 2008). Additionally, AtRLP30 involves resistance toward necrotrophic fungal pathogen Sclerotinia sclerotiorum as well (Zhang et al., 2013). AtRLP32 is confirmed as the receptor of proteobacterial translation initiation factor 1 (IF1) (Fan et al., 2022). AtRLP42/RBPG1 recognizes endopolygalacturonases from the plant pathogen Botrytis cinerea and saprotroph Aspergillus niger (Zhang et al., 2014). AtRLP44 activates brassinosteroid signaling through interaction with BAK1 (Wolf et al., 2014). AtRLP51/SNC2 and AtRLP55 implicate in basal defense against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (Zhang et al., 2010). AtRLP52 is suggested to influence resistance toward the powdery mildew pathogen Erysiphe cichoracearum (Ramonell et al., 2005). Previous studies indicated that disease resistance genes are more likely to be duplicated and underwent functional divergence compared to growth-related genes, and the most homologous AtRLP genes anchored at the same or adjacent locus and underwent massive duplications (Fritz-Laylin et al., 2005; Wang et al., 2008; Wu et al., 2016a). Generally, RLPs need to constitutively interact with additional components, such as RLKs, to activate cellular responses. For example, SUPPRESSOR OF BAK1-INTERACTING RECEPTOR-LIKE KINASE 1 (SOBIR1) is required for RLP-mediated resistance to bacterial, fungal, and oomycete pathogens. SOBIR1 physically interacts with Cf-4, Ve1, AtRLP1, AtRLP23, AtRLP30, AtRLP32, and AtRLP42, indicating the existence of crosstalk among different RLP signaling pathways (Wang et al., 2008; Jehle et al., 2013a; Liebrand et al., 2013; Zhang et al., 2013, 2014; Albert et al., 2015; Ma and Borhan, 2015; Catanzariti et al., 2017). Apart from the involvement in biotic stress responses, RLP was also engaged in abiotic stress tolerance. Currently, AtRLP28 is the only one found to play a role in salt stress tolerance (Wu et al., 2016a).
Brassica napus is one of the most economically important oil crops in the world. Its yield and quality, however, are affected remarkably by multiple external stimuli, especially the invasion of major Brassica pathogenic fungi, such as S. sclerotiorum and Leptosphaeria maculans. Brassica napus (AACC) are an allotetraploid generated by recombination of two diploid genomes, Brassica rapa (AA) and B. oleracea (CC). Brassica napus and its progenitor species share significant homology with Arabidopsis thaliana (Nagaharu, 1935). Currently, several pathogen-responsive RLPs were identified across Brassica species. Among them, LepR3 and Rlm2, which co-localized within the same genetic interval of blackleg resistance locus, were cloned from B. napus and rendered race-specific resistance to fungal pathogen L. maculans upon recognition of different cognate Avr proteins, AvrLm1 and AvrLm2, respectively, and both of them require SOBIR1 for their functions (Larkan et al., 2013, 2015; Ma and Borhan, 2015). Another two RLP genes identified from B. rapa are Bra032747 and Bra032746, which confer resistance to Brassica downy mildew (Zhang et al., 2018). Pathogen-associated cell-surface receptors are important for perceiving immunogenic signals in the challenged host (Kourelis and van der Hoorn, 2018). However, only a limited batch of RLPs was functionally characterized in Brassica species. Here, we performed the identification and phylogenetic analysis of RLPs in B. napus and its progenitors, B. rapa and B. oleracea, which are highly homologous to AtRLPs, and sorted out their distributions on chromosomes. To further assume the possible functions of identified RLPs, we investigated gene expression profiles of B. napus elicited by S. sclerotiorum utilizing the published pathogen-induced RNA-seq datasets. Taken together, these results will sketch out the basic information across identified Cruciferae RLPs, contributing to deciphering genome evolution and duplication, as well as elucidation of RLP gene function in detail.
Arabidopsis thaliana genomic and annotation data were downloaded from the TAIR (http://www.arabidopsis.org). Genomic and annotation data of B. napus and B. rapa were downloaded from BRAD (http://brassicadb.cn/; Chen et al., 2022). Brassica oleracea genomic and annotation data were downloaded from Ensemblplants (https://plants.ensembl.org/index.html).
In A. thaliana, a typical RLP protein consists of tandem LRR motifs, along with SP (signal peptide) and TM (transmembrane) in two terminals (Wang et al., 2008). But these LRR motifs vary in number and distribution, thus they are not competent as a good query for protein screening. Given this, two methods were used for the identification of RLP-encoding genes in this work. First, as queries, 55 identified Arabidopsis RLP protein sequences (two pseudogenes, AtRLP18 and AtRLP49, excluded in this work) were obtained and used to search analogs in three Brassica species using the BLASTP program based on a comparative genomics approach, thereby obtaining the first part of the candidate RLP proteins. Furthermore, RLP-encoding genes were also screened through Hidden Markov Model (HMM) profiles built by the model corresponding to 55 AtRLP proteins using the HMMSearch program (https://www.ebi.ac.uk/Tools/hmmer/). Subsequently, the two sets of candidates were merged and subjected to NCBI-BLASTP using UniProtKB/SwissoPrpt, and their annotations were downloaded for further screening.
The exon–intron organization of the RLP-encoding genes was depicted using GSDS2.0 (Hu et al., 2015), and the conserved motifs in RLPs were searched using the MEME suite (Bailey and Elkan, 1994). The SP and TM were predicted using SignalP-5.0 Server (http://www.cbs.dtu.dk/services/SignalP/), SMART (http://smart.embl-heidelberg.de/), TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), and Pfam (http://pfam.xfam.org/). Approximately 2,000-bp upstream flanking fragments of the RLP genes were derived from the genome and used for promoter cis-element prediction using PlantCARE (Lescot et al., 2002). The visualization was completed using TBtools (Chen et al., 2020). The phylogenetic analyses of the selected B. napus and Arabidopsis RLP proteins were generated using MEGA7.0 with the maximum likelihood (ML) algorithm. Bootstrap analysis with 1,000 replications was performed to assess group support.
The chromosome location of Brassicaceae RLPs was obtained from the genome annotation files. The distribution and tandem duplication of Brassicaceae RLP genes on chromosomes was generated and depicted using TBtools (Chen et al., 2020).
The RNA-seq data were obtained from NCBI. Data calibrations were performed, and some data with big deviations were removed. A heat map was drawn by HemI 1.0.3.3 (Deng et al., 2014). A mid-resistant B.napus variety Zhongyou821 was used for S. sclerotiorum inoculation and assessment of BnRLP gene expression. Plants were grown in a greenhouse with artificial irrigation. At the flowering stage, stems and leaves were inoculated with mycelia on living plants, harvested after 48 h inoculation, flash-frozen in liquid nitrogen, and stored at −80°C. Three plants with the closest phenotype and growth status were harvested, and harvesting was repeated three independent times. qRT-PCR was performed as described above with slight modifications (Li et al., 2016). Total RNA samples were isolated from rapeseed tissues using the Plant RNAprep Pure Kit (Tiangen, Beijing). RNA was quantified on a NanoDrop 1000 (NanoDrop Technologies, Inc.), and RNA integrity was evaluated on a 1% agarose gel. RNA was transcribed into cDNA using a GoScript RT Reagent Kit (Promega, USA). Primers used for qRT-PCR were designed using the Primer Premier 5.0 program to target the ORF of each gene with an amplicon sized between 80 and 200 bp (Supplementary Table 3). UBC9 served as a reference gene (Chen et al., 2010). qRT-PCR was performed using 10-fold diluted cDNA and a Universal SYBR Green Supermix Kit (Bio-RAD, USA) on a CFX96 real-time PCR machine (Bio-Rad, USA) according to the MIQE Guidelines (Minimum Information for Publication of Quantitative Real-Time PCR Experiments; Bustin et al., 2009).
Since A. thaliana is a close relative of B. napus (rapeseed), 55 AtRLP protein sequences (two pseudogenes, AtRLP18 and AtRLP49, excluded in this work) were downloaded from the Arabidopsis database (TAIR) and used as queries to identify the candidate RLP proteins in rapeseed by BLAST search against BRAD or Ensemblplants database. In addition, Hidden Markov Model (HMM) was used for further confirmation of candidate RLPs. To better understand the evolution of RLPs, B. rapa and B. oleracea genomes were also selected for the screening of orthologs. In total, 118, 173, and 276 RLPs were identified from B. rapa (AA), B. oleracea (CC), and B. napus (AACC), respectively (Supplementary Table 1). Although a pangenome investigation of resistance gene analogs (RGAs) across some B. oleracea varieties had identified 213 candidates (Bayer et al., 2019), we conducted a further fine selection to get a more reliable but attenuated set of BoRLPs. Then, all RLP CDSs were downloaded and aligned to corresponding genomic DNA to obtain high-confidence sequences. Sequence alignment revealed a great diversity of BnRLPs (Supplementary Figure 1). The number of introns ranged from 0 to 22 (BnaC06g23240D), and ~16% BnRLPs had no introns which were sharply attenuated compared to that of AtRLPs (70%). The orthologs of five AtRLPs (AtRLP3, AtRLP10, AtRLP17, AtRLP44, and AtRLP51) did not evolve any intron, showing high conservation. Apart from quantitative variation of BnRLP introns, some BnRLP genes possessed extra-long introns (more than 2,000 bp), like BnaA01g31040D (intron I, 5,336 bp), BnaA09g35430D (intron II, 4,756 bp; intron III, 5,018,bp), BnaC02g22740D (intron III, 6,474bp), and BnaC02g22760D (intron X, 5,030 bp). AtRLP9 (AT1G58190.1) also contained long introns (3,437 bp), but it had no corresponding orthologs in Brassicaceae species. Rapeseed amplified the RLP-encoding gene copies by a wide margin, and the tendency of increasing intron number and length variation was available for alternative splicing and further benefit for functional diversity.
Further analysis revealed that the composition and the number of conserved motifs or domains varied to a large extent. The length of amino acids ranged from 73 (BnaCnng19280D) to 2,736 aa (BnaC06g23240D; Supplementary Figure 2). Ten motifs were predicted in BnRLP proteins (Figure 1, Supplementary Figure 3). Motif 1, Motif 2, and Motif 9 were distributed on more than 80% BnRLP proteins, and the average number on each protein was 2.3, 1.7, and 2.2, respectively. More than 15% of BnRLP proteins possessed all 10 motifs, and 42 of the 276 had less than five types. Only a quarter of BnRLP proteins met the characteristics of canonical RLP protein, namely composed of the SP and TM domains at the ends and the tandem LRR domains in the middle (Supplementary Figure 1).
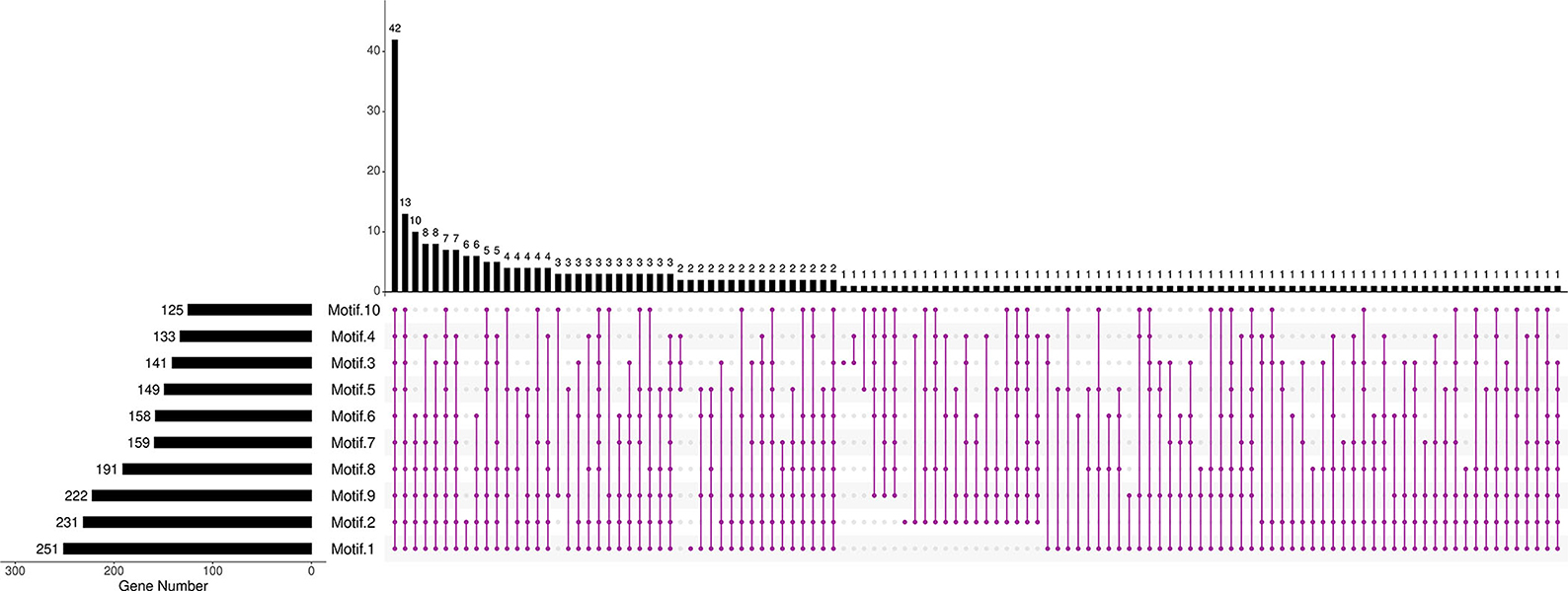
Figure 1. Distributions of motifs in BnRLPs. The Upset plot shows the distribution of motifs in the BnRLPs. The number chart above represents the number of genes contained in each type of RLP. The bar chart at the bottom left represents the number of events included in each type of motif. The dotted line shows the types of motifs contained in the group.
BnRLP encoding genes were unevenly distributed on eighteen chromosomes (Figure 2). The 116 and 148 BnRLPs were mapped onto chrA (Figure 2A) and chrC (Figure 2B) subgenome, respectively, and many of these genes reside in a cluster manner. The percentage of RLP genes on chromosomes in clusters in B. oleracea (51.22%) and B. rapa (49.14%) was lower than that of B. napus (56.06%). The numbers of genes in clusters ranged from two to six in B. napus, and the maximum gene number in B. oleracea and B. rapa were 7 and 5, respectively. In B. napus, 148 RLP genes were located in 55 clusters and the remaining 221 genes were singletons. Among these clusters, 12 with 29 genes were located on chrC04 (Figure 2B), which was similar to that in B. oleracea (seven clusters containing 23 genes on chromosome C04) (Figure 2D). The B. rapa genome carries 57 RLP genes in 23 clusters (Figure 2C). Compared to that in B.napus, the same number of clusters but fewer RLP genes (57 and 67 in B. rapa genome and B.napus A-subgenome) were observed in B. rapa. The cluster distribution between the two species was obviously different on chromosome A01 and chrA01, showing that B.napus evolved more tandem RLP genes on chrA01. Chromosome chrC04 contained the largest number of BnRLP genes (41 genes), followed by chrC03 (21 genes), chrC06 (19 genes), chrA04 (18 genes), and chrA01 (14 genes). Only one BnRLP gene was located on chrA10. RLP genes were assigned tandemly on most chromosomes, especially on chrA01, chrA04, chrA05, chrA07, chrC04, and chrC06, which presented higher distribution densities. Generally, the distribution of BnRLP genes on the A or C subgenome was similar to that of BrRLP or BoRLP genes in their respective genomes.

Figure 2. Locations of RLP genes on chromosomes in Brassica napus, Brassica rapa and Brassica oleracea. (A, B) Locations of RLP genes on chromosomes in B. napus; (C) Locations of RLP genes on chromosomes in B. rapa. (D) Locations of RLP genes on chromosomes in B. oleracea. A01-A09, C01-C08, and chrA01-chrC09 represent the chromosome number in B. rapa, B. oleracea, and B. napus, respectively. The blue line represents tandem duplication.
To better reveal the expansion of RLP genes in the rapeseed genome, the duplication patterns of 276 BnRLP genes were predicted and analyzed by MCScanX (Wang et al., 2012) and visualized by TBtools (Chen et al., 2020), and the result showed that rapeseed had 36 times as many RLP gene pairs as Arabidopsis (Figure 3). Moreover, the possible syntenic relationship of RLP-encoding genes between A. thaliana and Brassica genomes was also investigated. Subsequently, we obtained 59 orthologous gene pairs between A. thaliana and B. oleracea, 50 between A. thaliana and B. rapa, 250 between B. oleracea and B. napus, and 223 between B. rapa and B. napus, which are shown in Supplementary Figure 4. Ka/Ks analysis was performed by TBtools (Chen et al., 2020) between A. thaliana and the other three Brassica species(B. napus, B. rapa and B. oleracea), respectively. The Ka/Ks value of all pairs was <1 (Supplementary Table 2), suggesting that the main force for the evolution of those RLP gene pairs was negative selection. Most orthologous AtRLP genes are located on chromosomes 1 and 2, and correspondingly, chromosomes A04/A07 and C04/C06 possessed the higher orthologous regions in B. rapa and B. oleracea, respectively. Approximately, the aliquot of orthologous BnRLP genes was resourced from B. rapa and B. oleracea and numerically distributed evenly on chrA and chrC subgenomes.
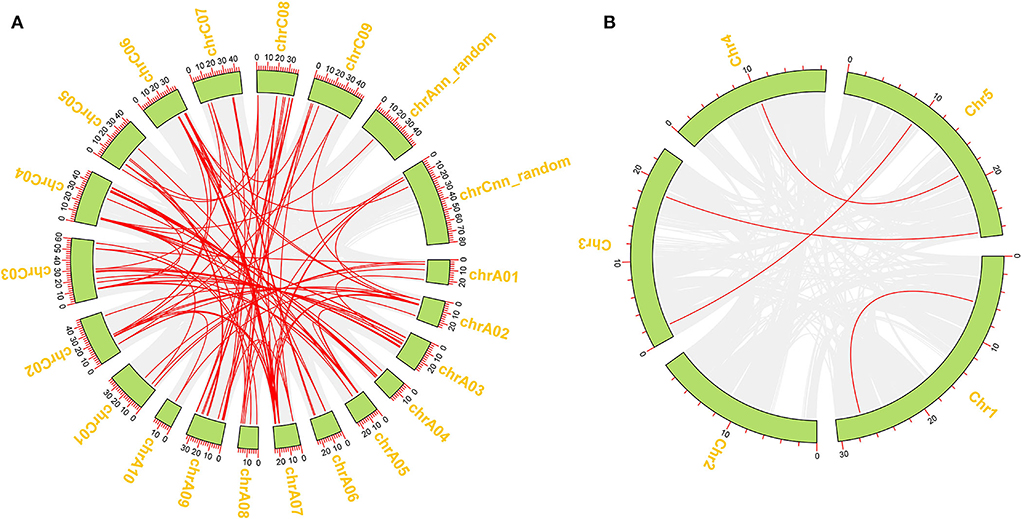
Figure 3. Duplication of BnRLP and AtRLP genes. (A) Duplication of RLP genes in Brassica napus. (B) Duplication of RLP genes in Arabidopsis thaliana. The red line represents the gene pairs and the green box represents the genome.
To better understand the evolving relationship between BnRLPs and AtRLPs, a phylogenetic tree was constructed by the ML method (Figure 4). The RLP proteins were clustered into 13 subgroups. Group I to Group IV consisted of no more than three proteins, respectively. The protein constructs of most BnRLPs in GroupV, X, and XI were canonical. More than half of BnRLPs in GroupVI had an SP domain or a TM domain. Most BnRLPs possessed only one type of domain, and two TM domains were predicted in four BnRLPs in GroupXII. In addition, besides a sub-clade of BnRLPs that only possessed the TM domain (one or more), the quantity of BnRLPs that the missing TM domain had increased to 69.23% in GroupXII, but most had an SP domain. Some RLP genes in GroupVII were more conserved since most members had no introns. Some bunches of tandem AtRLPs were especially prone to cluster together, which evolved independently with BnRLPs in Group XI and Group XIII. Other tandem ones evolved together with BnRLPs, like At1g17240 (AtRLP2) and At1g17250 (AtRLP3) in GroupVII, At2g32660 (AtRLP22) and At2g32680 (AtRLP23) in GroupXIII.
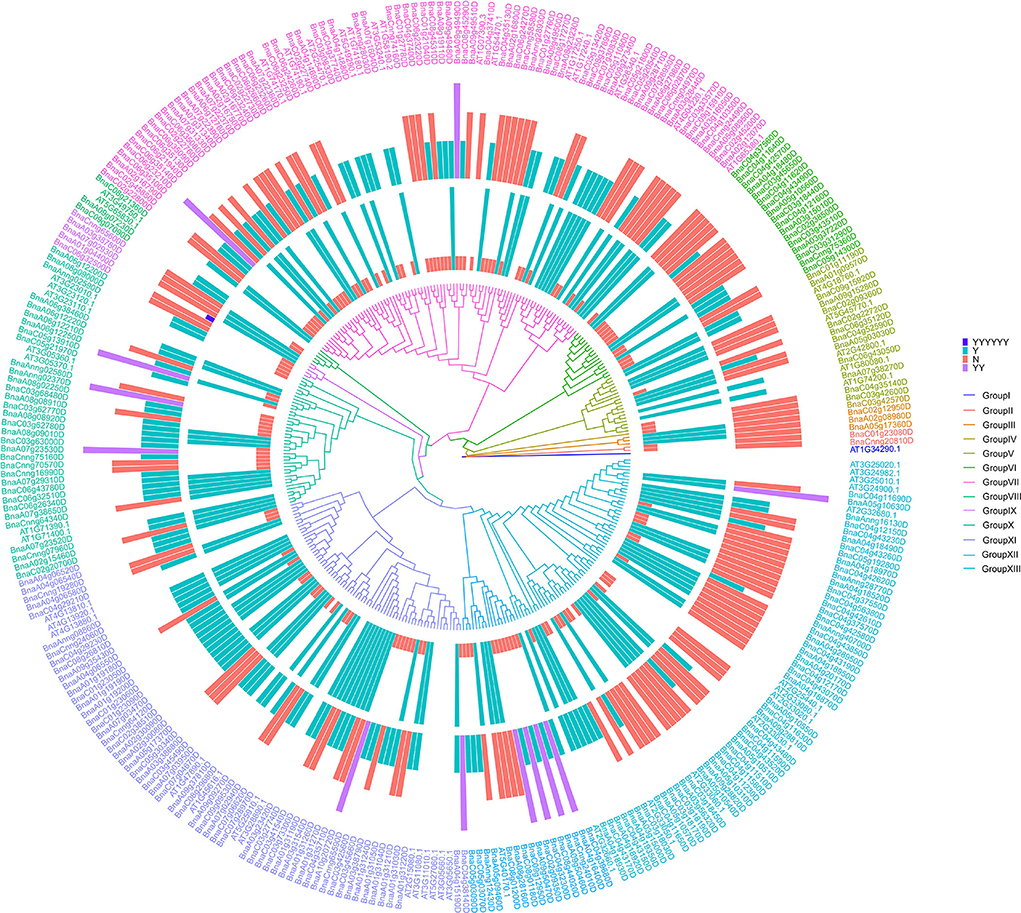
Figure 4. Phylogenetic trees of RLP proteins in Arabidopsis thaliana and Brassica napus. The phylogenetic tree was generated by the ML method with bootstrap analysis (1,000 bootstrap replicates) using an amino acid sequence alignment of RLP proteins from A. thaliana and B. napus by the MEGA 7.0 program. RLP proteins were clustered into 13 subgroups. The rings surrounded by the phylogenetic tree stand for two types of domains. The inner one represents the transmembrane domain and the outer represents the signal peptide. “Y” means that RLP contains one transmembrane domain/signal peptide, “YY” for two transmembrane domains, and “YYYYYY” for six transmembrane domains. “N” means that RLP does not contain transmembrane domain/signal peptide.
Cis-acting elements that are distributed in the promoter region can help predict the function of candidate genes. Most AtRLPs and BnRLPs were identified as related to stress responses, so the cis-acting elements related to stresses were further analyzed (Supplementary Figure 5, Figure 5). Concretely, cis-acting elements were categorized into MeJA-responsive (MeJA, name after), gibberellin-responsive (GA), abscisic acid-responsive (ABA), drought-inducible (drought), auxin-responsive (auxin), low temperature-responsive (low temperature), salicylic acid-responsive (SA), wound-responsive (wound), and defense- and stress-responsive (defense and stress) related. Approximately, 79.3 and 77.5% of the promoters contained MeJA and ABA-related cis-acting elements, followed by GA- and wound-related, which were found in half of the promoters of BnRLPs. Only 15 BnRLPs were related to wound responsiveness. Additionally, a quarter contained no more than three types of cis-acting elements. Among them, seven BnRLPs might exhibit functional specificity since they possessed single type of cis-acting element, such as BnaA09g37810D (drought), BnaA02g12070D (SA), BnaC02g20700D and BnaC09g12950D (MeJA), BnaC02g45850D and BnaC04g12150D (low temperature), BnaC09g15910D (defense and stress), and BnaC04g52590D (ABA).

Figure 5. Distributions of cis-acting elements in BnRLP promoter. The Upset plot shows the distribution of cis-acting elements in BnRLP promoter. The bar chart above represents the number of genes contained in each type of RLP. The bar chart at the bottom left represents the number of genes included in each type of cis-acting element. The dotted line shows the types of cis-acting elements contained in the group.
Sclerotinia sclerotiorum is one of the most common pathogens worldwide, which is the causal agent of stem rot disease in Brassica crops causing devastating decline in the economic value. To further unveil the protagonists in pathogen-defense responses, expression profiling data of RLP genes were derived from the GEO database. One series (accession No. SRP053361) (Wu et al., 2016b) was downloaded and analyzed. In total, 225 BnRLP genes identified showed expressional fluctuations during 24, 48, and 96 hpi (hour post-inoculated) by S. sclerotiorum in resistance (R) or susceptible (S) rapeseeds (Figure 6). To confirm the reliability of the dataset and further identify the genes that can be induced by S. sclerotiorum, seven upregulated BnRLP gene expressions were detected using real-time fluorescent quantitative PCR (Supplementary Figure 6). As we observed, besides inflorescence, the plant stem and leaves were also susceptible to S. sclerotiorum, and the infected stem becomes more prone to lodging at the flowering stage, so the fact that the RLPs are induced at a higher expression level at the flowering stage might have the potential to possess greater agricultural applicability. To this end, the stem and leaves were selected as target tissues to illuminate the tissue specificity under S. sclerotiorum infection. The results showed that those RLP genes can be induced significantly by S. sclerotiorum, but varied in expression level and pattern. Among them, BnaC07g28970D, BnaC02g22760D, and BnaA02g16770D accumulated more transcripts in the stem than that in the leaf, and the differences ranged from 2- to 4-fold. On the contrary, BnaA08g02250D, BnaA08g28160D, BnaC04g56380D, and BnaA04g14880D were induced more in the leaf. Moreover, BnaA03g56320D showed a similar expression level in stem and leaf, but at a relatively lower level compared to other genes. Overall, the results showed that all selected upregulated genes derived from the database could be induced by S. sclerotiorum.
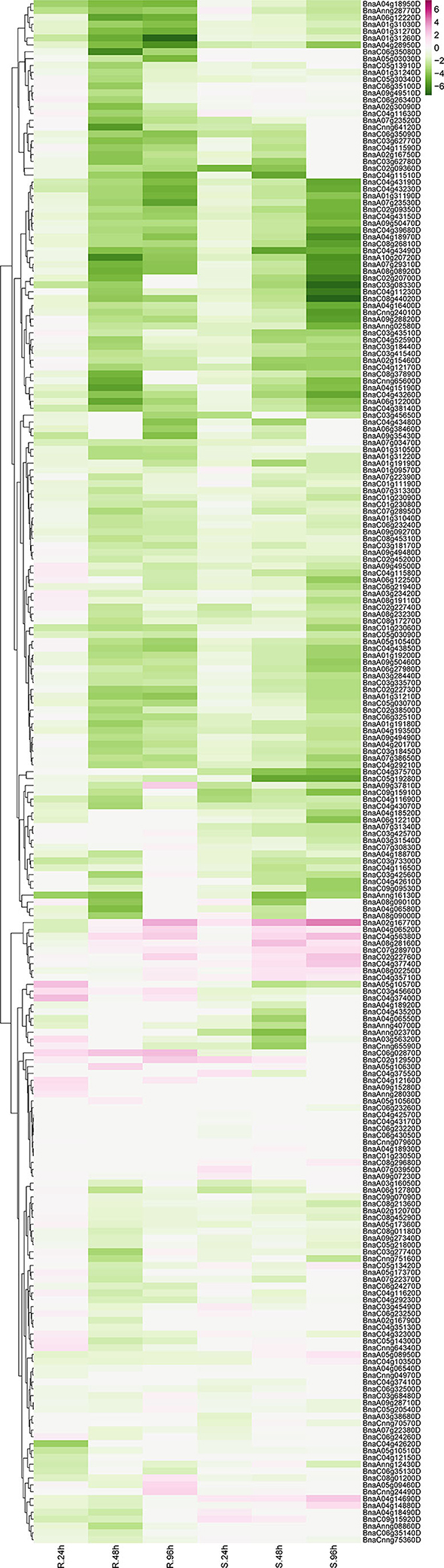
Figure 6. The expression heat map of BnRLP genes elicited by S. sclerotiorum. The expression heat map of BnRLP genes in the resistant (R) and susceptible (S) Brassica napus alleles at 24, 48, and 96 h post-inoculation, TPM value derived from the GEO database was used for heat map construction.
Unexpectedly, only 29 BnRLPs were upregulated (more than two times compared to mock) at a certain time point (Figure 7). Among them, some BnRLPs presented expression specificity between R and S, such as four BnRLPs (BnaA04g14880D, BnaA05g08950D, BnaA04g14690D, BnaC09g15920D) that were induced merely in S, and six BnRLPs (BnaA09g37810D, BnaA03g56320D, BnaA05g10570D, BnaC06g02870D, BnaC03g45660D, and BnaC04g37400D) in R. The expressional differences of some BnRLPs between R and S appeared due to the inconsistency of the induction time. For instance, transcript accumulations of BnaC07g28970D, BnaC02g22760D, and BnaA02g16770D in R were slower than S, and their expression levels at 96 hpi (R) were approximately equal to that at 48 hpi (S). Additionally, as results show, BnaC02g22760D and BnaA02g16770D were induced from 24hpi in S, and their expression levels continued increasing even at 96 hpi reaching a level of 13.85 and 31.03 times, respectively, but they were induced from 96 hpi in R. BnaA08g28160D and BnaC04g56380D possessed similar expression patterns between R and S. The transcript accumulations of BnaA08g28160D reached a peak at 48 hpi in both R and S. Furthermore, BnaC04g56380D was induced at 48 hpi and kept accumulating until 96 hpi in two rapeseed ecotypes. It is worth mentioning that LepR3/Rlm2 encodes a receptor-like protein triggered by the L. maculans effector AVRLM1 (Larkan et al., 2013, 2015), but the expression of BnaA10g20720D, which possessed 100% identity and was co-located in the same genetic interval with LepR3/Rlm2, was inhibited by S. sclerotiorum both in R and S (Figure 6).
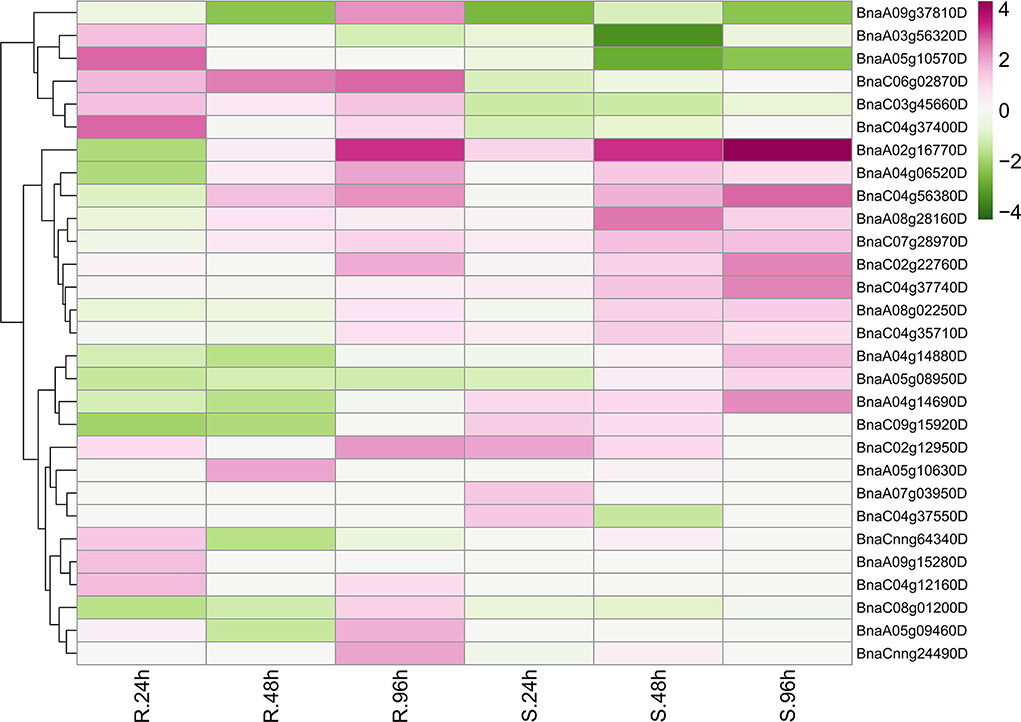
Figure 7. Upregulated BnRLPs elicited by Sclerotinia sclerotiorum. The expression heat map of upregulated BnRLP genes in the resistant (R) and susceptible (S) B. napus alleles at 24, 48, and 96 h post-inoculation by S. sclerotiorum, TPM value derived from the GEO database was used for heat map construction.
The fluctuation of gene expression profiles induced by various pathogens can be used as an indicator for functional assumption and further characterization. To investigate the possible functions of BnRLPs, we compared the transcriptional profiles of RLP genes with that of the functional assigned AtRLP genes. Intriguingly, AtRLP10 (CLV2, AT1G65380.1) was identified as a regulator of plant meristem and organ development (Jeong et al., 1999; Wang et al., 2011). As its homologs, the transcript abundance of BnaC02g45200D and BnaA02g12070D were decreased when there was a stimulus exerted by S. sclerotiorum. The expressional inclination of RLP10 genes indicated that their functions might be conserved among Brassica species and A. thaliana due to the trade-off between growth and defense response, which is widely known in plants. Another hint in favor of the possibility of functional conservation among RLP10 genes is that they shared high similarity/identity with AtRLP10 no matter in sequence (CDS and protein) or structural layer (Supplementary Figure 7). Unlike RLP10, a functional divergence occurred among the RLP30 genes as AtRLP30 (AT3G05360.1) was suggested to be involved in resistance toward S. sclerotiorum, while BnRLP30 genes (BnaAnng02580D and BnaAnng02370D) were not induced by it (Figure 6). The low similarity/identity between BnRLP30s and AtRLP30 also supported the probability of functional divergence in B. napus (Supplementary Figure 7). Instead, BnaA02g16770D and BnaC02g22760D, homologs of AtRLP15 (AT1G74190.1), might be fabulous candidates for S. sclerotiorum resistance research since they were proven to be few but induced to a greater extent.
The case that one single RLP gene is transcriptionally regulated by multiple external stimuli and involved in multiple biological processes has been reported in A. thaliana (Wu et al., 2016a). It was also a common phenomenon in B. napus, as exemplified by BnaA09g09270D, which can be induced by both S. sclerotiorum and L. maculans (Becker et al., 2017; Duke et al., 2017). The comparative analysis of BnRLPs phylogeny and their transcriptional profiles indicated that there was no significant correlation between BnRLPs phylogeny (predicted by sequence similarity) and the expression patterns, and this is in line with previous publications (Steidele and Stam, 2021). On the contrary, the diversification of gene expression patterns was observed among the duplicated BnRLPs paralogs. Gene duplication widely exists during the process of plant evolution in multiple forms, like tandem duplication, segmental duplication, and whole-genome duplication (Kondrashov et al., 2002; Conant and Wolfe, 2008). Retained duplicated genes can provide raw genetic resources for functional innovation, and the novel function can derive from several different ways, including gain-of-function mutations, the subdivision of ancestral functions, and gene dosage changes (Conant and Wolfe, 2008). In this study, tandem duplication was commonly found in BnRLPs, and some BnRLPs were amplified by a wide margin when compared with their counterparts in A. thaliana, such as homologs of AtRLP27 (AT2G33060.1) and AtRLP54 (AT5G40170.1).
As B. napus was hybridized by B. rapa and B. oleracea, in general, the evolution of BnRLP proteins was relatively conserved among the three Brassica species. However, because of gene duplications and translocations after hybridization, inaccurate assembly universally occurred during genetic recombination in plants, resulting in the loss-of-function of some genes (Schnable et al., 2012; Boutte et al., 2020). In this study, 5, 10, and 24 proteins were detected as containing no LRR domains in B. rapa, B. oleracea, and B. napus, respectively, but remained as RLPs. They possessed a high sequence identity and had a similar protein construct (consisted of SP and/or TM domain) with other typical RLPs. So those proteins with no LRR domains were also included to better reveal the evolution of BnRLPs.
When we did the phylogenetic analysis, the phylogeny of AtRLPs resembles that performed by previous publications (Wang et al., 2008; Steidele and Stam, 2021). In line with the classification by Steidele and Stam (2021), most upregulated BnRLPs were homologous with AtRLPs that were clustered into pathogen-responsive RLP clade. The exception is that BnaA02g16770D and BnaC02g22760D, homologs of AtRLP15 (AT1G74190.1) which were grouped into a basal sub-family but not the aforementioned pathogen-responsive RLP clade, upregulated after pathogen treatment to a great extent.
Based on the dataset, six BnRLP genes, namely BnaA09g37810D, BnaA03g56320D, BnaA05g10570D, BnaC06g02870D, BnaC03g45660D, and BnaC04g37400D, upregulated specifically in resistant rapeseed by S. sclerotiorum elicitation. Correspondingly, four BnRLPs (BnaA04g14880D, BnaA05g08950D, BnaA04g14690D, and BnaC09g15920D) showed preference for susceptible rapeseed. Those genes identified from incompatible interactions might be good candidates for further functional assignment of RLP genes against S. sclerotiorum infection. BnaA02g16770D and BnaC02g22760D, which could be induced universally by S. sclerotiorum, might also play a role in preventing fungal infection. Zhongyou821 was identified as a mid-resistant B.napus variety (Wang et al., 2004) and commonly used as a control cultivar for investigating disease indexes. In this study, we also used Zhongyou821 for infection and performed qRT-PCR to verify the expression level of some upregulated BnRLP genes, and the result showed that the transcript abundance in this study was dozens of times than that derived from the RNAseq datasets, but further confirmed they could be induced by S. sclerotiorum. The large difference might be due to the selection of different cultivars or ascribed to spatiotemporal expression differences. Overall, all selected upregulated BnRLPs genes derived from the database could be induced by S. sclerotiorum in Zhongyou821.
We conducted identification, gene and protein characterization, localization, evolution, and expression analysis of the highly conserved RLP members in B. napus and found that a majority of RLP gene families in this study were relatively conserved during the evolution of Brassicaceae. Exploring effective resistance genes against a vast variety of microbial pathogens is critical for crop breeding. Genome-wide identification and expression analysis of the RLP family members in B. napus provided an alternative strategy to reinforce the resistance against major pathogens in Brassicaceae. Our results provide important clues for further investigations of the function of Brassicaceae RLPs involved in the development and immune response and pave the way to molecular breeding of disease-resistant rapeseed.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
WL: formal analysis, writing the original draft, data curation, and validation. JL: investigation, resources, and visualization. CY: data curation. SX: conceptualization, writing—review and editing, and supervision. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (Grants 31971836 and 30970247), the Hunan Province Natural Science Fund for Distinguished Young Scholars (Grant 11JJ1007), the Chongqing municipal education commission (Grant KJQN201900533), and the Chongqing Bureau of Human Resources and Social Security Post-doctoral Funding (0019).
We thank Yanan Liu for the critical reading of the manuscript. We thank all members of our laboratories for their helpful assistance during the research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.944763/full#supplementary-material
Supplementary Table 1. RLP gene information in B. napus, B. rapa and B. oleracea.
Supplementary Table 2. Ka/Ks in A.thaliana and B. napus, B. rapa and B. oleracea pairs.
Supplementary Table 3. Primers in this study.
Supplementary Figure 1. Gene and protein structures of RLP genes in B. napus. The green bar represents CDS, the black line represents intron, and the blue bar represents UTR. The pink bar stands for the LRR motif, the yellow bar for the transmembrane domain, and the red for the signal peptide.
Supplementary Figure 2. Motif and conserved domain of RLP in B. napus. Motif labeled as motif 1-motif 10. The green bar represents CDS, the blue bar represents UTR. The yellow bar stands for the conserved domain PLN03150 and the pink bar for PLN00113.
Supplementary Figure 3. Motifs in BnRLPs.
Supplementary Figure 4. Syntenic relationship of RLP-encoding genes between A. thaliana and Brassicaceae species.
Supplementary Figure 5. Cis-acting element in BnRLP promoter.
Supplementary Figure 6. Tissue specificity of upregulated BnRLPs under S. sclerotiorum infection. The box plot represents dispersion and expression degree. Gene expression was detected with at least three biological repeats and three experiment repeats, and statistical analysis showed that compared with controls, the expression of all genes reached significant levels when elicited by S. sclerotiorum (p-value < 0.05).
Supplementary Figure 7. Protein similarity and identity of RLP families. Data on the upper right present the protein sequence identity, and data on the bottom left present the protein sequence similarity.
Albert, I., Böhm, H., Albert, M., Feiler, C. E., Imkampe, J., Wallmeroth, N., et al. (2015). An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1, 15140. doi: 10.1038/nplants.2015.140
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Bayer, P. E., Golicz, A. A., Tirnaz, S., Chan, C.-K. K., Edwards, D., and Batley, J. (2019). Variation in abundance of predicted resistance genes in the Brassica oleracea pangenome. Plant Biotechnol. J. 17, 789–800. doi: 10.1111/pbi.13015
Becker, M. G., Zhang, X., Walker, P. L., Wan, J. C., Millar, J. L., Khan, D., et al. (2017). Transcriptome analysis of the Brassica napus-Leptosphaeria maculans pathosystem identifies receptor, signaling and structural genes underlying plant resistance. Plant J. 90, 573–586. doi: 10.1111/tpj.13514
Belfanti, E., Silfverberg-Dilworth, E., Tartarini, S., Patocchi, A., Barbieri, M., Zhu, J., et al. (2004). The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc. Natl. Acad. Sci. USA. 101, 886–890. doi: 10.1073/pnas.0304808101
Boutte, J., Maillet, L., Chaussepied, T., Letort, S., Aury, J.-M., Belser, C., et al. (2020). Genome size variation and comparative genomics reveal intraspecific diversity in Brassica rapa. Front. Plant Sci. 11, 577536. doi: 10.3389/fpls.2020.577536
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Catanzariti, A.-M., Do, H. T. T., Bru, P., de Sain, M., Thatcher, L. F., Rep, M., et al. (2017). The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 89, 1195–1209. doi: 10.1111/tpj.13458
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Wang, T., He, X., Cai, X., Lin, R., Liang, J., et al. (2022). BRAD V3.0: an upgraded Brassicaceae database. Nucleic Acids Res. 50, D1432–D1441. doi: 10.1093/nar/gkab1057
Chen, I.-H., Huang, Y.-P., Tseng, C.-H., Ni, J.-T., Tsai, C.-H., Hsu, Y.-H., et al. (2017). Nicotiana benthamiana elicitor-inducible leucine-rich repeat receptor-like protein assists bamboo mosaic virus cell-to-cell movement. Front. Plant Sci. 8, 1736. doi: 10.3389/fpls.2017.01736
Chen, J.-Y., Huang, J.-Q., Li, N.-Y., Ma, X.-F., Wang, J.-L., Liu, C., et al. (2015). Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biol. 15, 148. doi: 10.1186/s12870-015-0508-3
Chen, X., Truksa, M., Shah, S., and Weselake, R. J. (2010). A survey of quantitative real-time polymerase chain reaction internal reference genes for expression studies in Brassica napus. Anal. Biochem. 405, 138–140. doi: 10.1016/j.ab.2010.05.032
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Deng, W., Wang, Y., Liu, Z., Cheng, H., and Xue, Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS ONE 9, e111988. doi: 10.1371/journal.pone.0111988
Duke, K. A., Becker, M. G., Girard, I. J., Millar, J. L., Dilantha Fernando, W. G., Belmonte, M. F., et al. (2017). The biocontrol agent Pseudomonas chlororaphis PA23 primes Brassica napus defenses through distinct gene networks. BMC Genomics 18, 467. doi: 10.1186/s12864-017-3848-6
Fan, L., Fröhlich, K., Melzer, E., Pruitt, R. N., Albert, I., Zhang, L., et al. (2022). Genotyping-by-sequencing-based identification of Arabidopsis pattern recognition receptor RLP32 recognizing proteobacterial translation initiation factor IF1. Nat. Commun. 13, 1294. doi: 10.1038/s41467-022-28887-4
Fritz-Laylin, L. K., Krishnamurthy, N., Tör, M., Sjölander, K. V., and Jones, J. D. G. (2005). Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 138, 611–623. doi: 10.1104/pp.104.054452
Hu, B., Jin, J., Guo, A., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jehle, A. K., Fürst, U., Lipschis, M., Albert, M., and Felix, G. (2013a). Perception of the novel MAMP eMax from different Xanthomonas species requires the Arabidopsis receptor-like protein ReMAX and the receptor kinase SOBIR. Plant Signal. Behav. 8, e27408. doi: 10.4161/psb.27408
Jehle, A. K., Lipschis, M., Albert, M., Fallahzadeh-Mamaghani, V., Fürst, U., Mueller, K., et al. (2013b). The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 25, 2330–2340. doi: 10.1105/tpc.113.110833
Jeong, S., Trotochaud, A. E., and Clark, S. E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1934. doi: 10.1105/tpc.11.10.1925
Jones, D. A., Thomas, C. M., Hammond-Kosack, K. E., Balint-Kurti, P. J., and Jones, J. D. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. doi: 10.1126/science.7973631
Kang, W.-H., and Yeom, S.-I. (2018). Genome-wide identification, classification, and expression analysis of the receptor-like protein family in tomato. Plant Pathol. J. 34, 435–444. doi: 10.5423/PPJ.OA.02.2018.0032
Kawchuk, L. M., Hachey, J., Lynch, D. R., Kulcsar, F., van Rooijen, G., Waterer, D. R., et al. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. USA. 98, 6511–6515. doi: 10.1073/pnas.091114198
Kondrashov, F. A., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2002). Selection in the evolution of gene duplications. Genome Biol. 3, RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008
Kourelis, J., and van der Hoorn, R. A. L. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299. doi: 10.1105/tpc.17.00579
Larkan, N. J., Lydiate, D. J., Parkin, I., A., P., Nelson, M. N., Epp, D. J., et al. (2013). The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197, 595–605. doi: 10.1111/nph.12043
Larkan, N. J., Ma, L., and Borhan, M. H. (2015). The Brassica napus receptor-like protein RLM2 is encoded by a second allele of the LepR3/Rlm2 blackleg resistance locus. Plant Biotechnol. J. 13, 983–992. doi: 10.1111/pbi.12341
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, W., Lu, J., Lu, K., Yuan, J., Huang, J., Du, H., et al. (2016). Cloning and Phylogenetic analysis of Brassica napus L. caffeic acid o-methyltransferase 1 gene family and its expression pattern under drought stress. PLoS ONE 11, e0165975. doi: 10.1371/journal.pone.0165975
Liebrand, T. W. H., van den Berg, G. C. M., Zhang, Z., Smit, P., Cordewener, J. H. G., America, A. H. P., et al. (2013). Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA. 110, 10010–10015. doi: 10.1073/pnas.1220015110
Ma, L., and Borhan, M. H. (2015). The receptor-like kinase SOBIR1 interacts with Brassica napus LepR3 and is required for Leptosphaeria maculans AvrLm1-triggered immunity. Front. Plant Sci. 6, 933. doi: 10.3389/fpls.2015.00933
Malnoy, M., Xu, M., Borejsza-Wysocka, E., Korban, S. S., and Aldwinckle, H. S. (2008). Two receptor-like genes, Vfa1 and Vfa2, confer resistance to the fungal pathogen Venturia inaequalis inciting apple scab disease. Mol. Plant Microbe Interact. 21, 448–458. doi: 10.1094/MPMI-21-4-0448
Nadeau, J. A., and Sack, F. D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700. doi: 10.1126/science.1069596
Nagaharu, U. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japanese Journal of Botany 7, 389–452.
Ono, E., Mise, K., and Takano, Y. (2020). RLP23 is required for Arabidopsis immunity against the grey mould pathogen Botrytis cinerea. Sci. Rep. 10, 13798. doi: 10.1038/s41598-020-70485-1
Petre, B., Hacquard, S., Duplessis, S., and Rouhier, N. (2014). Genome analysis of poplar LRR-RLP gene clusters reveals RISP, a defense-related gene coding a candidate endogenous peptide elicitor. Front. Plant Sci. 5, 111. doi: 10.3389/fpls.2014.00111
Ramonell, K., Berrocal-Lobo, M., Koh, S., Wan, J., Edwards, H., Stacey, G., et al. (2005). Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol. 138, 1027–1036. doi: 10.1104/pp.105.060947
Ron, M., and Avni, A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16, 1604–1615. doi: 10.1105/tpc.022475
Schnable, J. C., Freeling, M., and Lyons, E. (2012). Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol. Evol. 4, 265–277. doi: 10.1093/gbe/evs009
Shen, Y., and Diener, A. C. (2013). Arabidopsis thaliana resistance to fusarium oxysporum 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genet. 9, e1003525. doi: 10.1371/journal.pgen.1003525
Steidele, C. E., and Stam, R. (2021). Multi-omics approach highlights differences between RLP classes in Arabidopsis thaliana. BMC Genomics 22, 557. doi: 10.1186/s12864-021-07855-0
Taguchi-Shiobara, F., Yuan, Z., Hake, S., and Jackson, D. (2001). The fasciated ear2 gene encodes a leucine-rich repeat receptor-like protein that regulates shoot meristem proliferation in maize. Genes Dev. 15, 2755–2766. doi: 10.1101/gad.208501
Tirnaz, S., Bayer, P. E., Inturrisi, F., Zhang, F., Yang, H., Dolatabadian, A., et al. (2020). Resistance gene analogs in the brassicaceae: identification, characterization, distribution, and evolution. Plant Physiol. 184, 909–922. doi: 10.1104/pp.20.00835
Wang, G., Ellendorff, U., Kemp, B., Mansfield, J. W., Forsyth, A., Mitchell, K., et al. (2008). A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147, 503–517. doi: 10.1104/pp.108.119487
Wang, G., Long, Y., Thomma, B. P. H. J., de Wit, P. J. G. M., Angenent, G. C., and Fiers, M. (2010). Functional analyses of the CLAVATA2-like proteins and their domains that contribute to CLAVATA2 specificity. Plant Physiol. 152, 320–331. doi: 10.1104/pp.109.148197
Wang, G., Zhang, Z., Angenent, G. C., and Fiers, M. (2011). New aspects of CLAVATA2, a versatile gene in the regulation of Arabidopsis development. J. Plant Physiol. 168, 403–407. doi: 10.1016/j.jplph.2010.08.015
Wang, H. Z., Liu, G. H., Zheng, Y. B., Wang, X. F., and Yang, Q. (2004). Breeding of the Brassica napus cultivar Zhongshuang 9 with high-resistance to Sclerotinia sclerotiorum and dynamics of its important defense enzyme activity. Sci. Agric. Sin. 37, 23–28.
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wolf, S., van der Does, D., Ladwig, F., Sticht, C., Kolbeck, A., Schürholz, A.-K., et al. (2014). A receptor-like protein mediates the response to pectin modification by activating brassinosteroid signaling. Proc. Natl. Acad. Sci. USA. 111, 15261–15266. doi: 10.1073/pnas.1322979111
Wu, J., Liu, Z., Zhang, Z., Lv, Y., Yang, N., Zhang, G., et al. (2016a). Transcriptional regulation of receptor-like protein genes by environmental stresses and hormones and their overexpression activities in Arabidopsis thaliana. J. Exp. Bot. 67, 3339–3351. doi: 10.1093/jxb/erw152
Wu, J., Zhao, Q., Yang, Q., Liu, H., Li, Q., Yi, X., et al. (2016b). Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci. Rep. 6, 19007. doi: 10.1038/srep19007
Yang, H., Bayer, P. E., Tirnaz, S., Edwards, D., and Batley, J. (2020). Genome-wide identification and evolution of receptor-like kinases (RLKs) and receptor like proteins (RLPs) in Brassica juncea. Biology 10, 17. doi: 10.3390/biology10010017
Yang, Y., Chen, T., Ling, X., and Ma, Z. (2017). Gbvdr6, a gene encoding a receptor-like protein of cotton (Gossypium barbadense), confers resistance to verticillium wilt in Arabidopsis and upland cotton. Front. Plant Sci. 8, 2272. doi: 10.3389/fpls.2017.02272
Zhang, B., Li, P., Su, T., Li, P., Xin, X., Wang, W., et al. (2018). BrRLP48, encoding a receptor-like protein, involved in downy mildew resistance in Brassica rapa. Front. Plant Sci. 9, 1708. doi: 10.3389/fpls.2018.01708
Zhang, L., Kars, I., Essenstam, B., Liebrand, T. W. H., Wagemakers, L., Elberse, J., et al. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164, 352–364. doi: 10.1104/pp.113.230698
Zhang, W., Fraiture, M., Kolb, D., Löffelhardt, B., Desaki, Y., Boutrot, F. F. G., et al. (2013). Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241. doi: 10.1105/tpc.113.117010
Keywords: Brassica napus, receptor-like protein, evolution, expression pattern, Sclerotinia sclerotiorum
Citation: Li W, Lu J, Yang C and Xia S (2022) Identification of receptor-like proteins induced by Sclerotinia sclerotiorum in Brassica napus. Front. Plant Sci. 13:944763. doi: 10.3389/fpls.2022.944763
Received: 15 May 2022; Accepted: 11 July 2022;
Published: 16 August 2022.
Edited by:
Ryo Fujimoto, Kobe University, JapanReviewed by:
Intikhab Alam, Fujian Agriculture and Forestry University, ChinaCopyright © 2022 Li, Lu, Yang and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shitou Xia, eHN0b25lMDUwNUBodW5hdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.