- School of Life Sciences, Anhui Agricultural University, Hefei, China
Dendrobium officinale Kimura et Migo is a famous Chinese herb. D. officinale grows on rocks where the available phosphorus is low. The SPX family plays a critical role in maintaining Pi homeostasis in plants. In this paper, 9 SPX family genes were identified in the genome of D. officinale. Bioinformatics and qRT-PCR analysis showed that DoSPXs were involved in response to −Pi stress and had different expression patterns. DoSPX4, which had a unique expression pattern, was clustered with AtSPX4 and OsSPX4. Under −Pi treatment, the expression level of DoSPX4 reached a peak on 5 d in roots, while showing a downward trend in the aboveground parts. DoSPX4 was located on the cell membrane. Overexpression DoSPX4 promoted Pi content in the stem and the expression level of NtPHT1/2 in Nicotiana tabacum. The results of Yeast two-hybrid showed that DoSPX4 could interact with Phosphate High-Affinity Response factor (DoPHR2). These results highlight the role of DoSPX4 in response to low phosphorus, which provides a theoretical basis for further study on the response mechanism of −Pi in D. officinale.
Introduction
Phosphorus (Pi) plays an essential role in plant growth (Chen and Liao, 2017). The low content of available Pi in the soil is difficult to meet the Pi requirements accompany with plant growth. Plants increase effective Pi utilization by sensing and absorbing Pi in the soil, then adapt to low Pi environment in molecular and physiological levels.
Dendrobium officinale is a perennial herb which has a variety of pharmacological effects. It can grow on tree trunks, rocky cliffs, or fern surfaces in natural conditions. It is subjected to various environmental stresses in the harsh growth environment, among which low Pi stress is the main factor affecting its growth and development (Liu N. et al., 2018). The previous research found that D. officinale could respond to −Pi and accumulate effective active components in low phosphorus. The expression of the key genes in the secondary metabolism were significantly correlated with Pi concentration, while the early response genes to −Pi in D. officinale have not been reported (Liu L. et al., 2021).
Proteins, containing the SPX domain, have been identified as early response factors which participate in Pi signal transduction in plants (Zhou et al., 2021). The N-terminal of plant SPX protein contains a highly conserved SPX domain, named by the first letter of SYG1, PHO81, and XPR1 genes (Secco et al., 2012). At present, SPX has been classified into four subfamilies by the C-terminal domains of the protein. The four subfamilies are SPX (containing only one SPX domain), SPX-EXS (containing SPX and one EXS domain), SPX-MFS (containing SPX and MFS domain), and SPX-RING (containing ring-type star finger domain) (Wang et al., 2004; Chen et al., 2009; Chiou and Lin, 2011; Lin et al., 2013; Su et al., 2015; Wang et al., 2015; Liu J. et al., 2016; Zhang et al., 2016; Yue et al., 2017; Yang et al., 2018). Among them, the SPX subfamily plays an important role in the early stage of Pi signal recognition.
Generally, SPX can respond to Pi signals and then interact with MYB to change the transcriptional activation of downstream PSI genes. At present, Phosphorus-related MYB includes PHR (MYB-CC) and several R2R3-MYB. In Arabidopsis, AtSPX1/2/3/4 proteins are upstream regulators of AtPHR1 (Duan et al., 2008). AtSPX1 and AtSPX2 had functional redundancy. Both of them could regulate AtPHR1, and the extent of this interaction was affected by AtSPX3 (Puga et al., 2014). In stem, AtSPX4 was a repressor of AtPHR1 (Osorio et al., 2019). In Oryza sativa, there were 6 SPXs (OsSPX1–OsSPX6). Under Pi deficiency, the expression of OsSPX4 was downregulated, while the other five OsSPX genes showed an upward trend (Secco et al., 2012; Lv et al., 2014). OsSPX1/2/4 could interact with OsPHR2 and affect the regulation of OsPHR2 on downstream PSI genes (Shi et al., 2014; Wang et al., 2014). OsSPX3, OsSPX5, and OsSPX6 were homologous genes, which were involved in functional redundancy in response to phosphorus. OsSPX3 and OsSPX5 could form homodimers and participate in complex regulation in O. sativa (Shi et al., 2014). OsSPX4 was rapidly degraded by the proteasome pathway under low phosphorus, which had unique subcellular localization (Lv et al., 2014). These studies showed that SPX played an important role in phosphate response in plants, and SPX4 (OsSPX4 in O. sativa and AtSPX4 in A. thaliana) may have a different regulatory pattern from other SPXs.
Among them, gene prediction based on similarity comparison through published genome sequence has become the main method to screen key genes and analyze molecular mechanisms (Bhatt et al., 2021). It has become the main method to screen key genes and analyze molecular mechanism that gene prediction based on similarity comparison. The previous studies showed that low phosphorus could promote the accumulation of effective active substances in D. officinale (Liu L. et al., 2021). However, the molecular mechanism of D. officinale response to −Pi has not been reported. In this study, the bioinformatics analysis of the early Pi response factor DoSPX was carried out. The expression pattern of DoSPX in D. officinale was analyzed by qRT-PCR. DoSPX4 had a unique expression pattern, which was subcellular localization was further investigated. The interaction between DoSPX4 and DoPHR2 was studied by Yeast two-hybrid. The function of DoSPX4 to plant low phosphorus was verified by heterologous overexpression of DoSPX4 in N. tabacum.
Materials and Methods
Treatment and Preservation of Plant Materials
The D. officinale tissue culture seedlings used in the experiment were from the Anhui Provincial Engineering Technology Research Center for Development and Utilization of Regional Characteristic Plants, School of Life Sciences, Anhui Agricultural University. The seedlings of D. officinale with uniform size, shape, and color were cultured in the plant tissue culture room under a constant temperature of 25°C at 8L:16D photoperiod. The tissue culture seedlings were cultured on Murashige and Skoog medium (MS) for 8 months, then put into MS medium with different Pi concentrations. The 5 levels of KH2PO4 are used to set different Pi concentrations on MS medium (2.5, 1.25, 0.625, 0.0625, and 0 mM), and K+ in different Pi concentration media were supplemented with different concentrations of KCl. Samples were taken on 0, 1, 5, 10, and 40 days after treatment and put into a 10-ml centrifuge tube and immediately froze in liquid nitrogen and store it in the refrigerator at −80°C for standby. The three biological replicates were set for each treatment.
Identification of SPX Gene Family in Dendrobium officinale
By reference SPX protein sequences in Oryza sativa (Supplementary Table 1) and Arabidopsis thaliana (Supplementary Table 1), SPX proteins in D. officinale genome (Supplementary Table 1) sequence were selected with a threshold of E-value < 1E–5. Then the obtained sequences were submitted to CD-HIT1 (Fu et al., 2012) to remove the redundant sequence. Finally, the candidate sequences are submitted to SMART2 (Letunic et al., 2021) and PFAM3 (Mistry et al., 2021) to identify the conserved motifs. The basic information of protein sequence was obtained online by the ExPasy website4 (Gasteiger et al., 2003).
Construction of DoSPXs Phylogenetic Tree
The 7 SPX sequences in D. officinale were compared with the SPX protein sequences of 27 A. thaliana, 12 O. sativa and 12 Phalaenopsis equestris by ClustalW (Thompson et al., 1994). A phylogenetic tree was constructed using the MEGA7.0 NJ method with the bootstrap = 1,500 and beautified with ITOLS online website5 (Letunic and Bork, 2021). The names of the gene name and accession numbers of SPX can be found in Supplementary Table 2.
Cis-Acting Element Analysis
The TBTOOLS (Chen et al., 2020) was used to extract the promoter region of the genome sequence by using the genome annotation file of D. officinale. The 1,500 bp upstream sequences of SPX CDS was submitted to PLANTCARE6 (Lescot et al., 2002) and NEWPLACE7 (Higo et al., 1999) online websites to analyze the possible cis-acting elements of the promoter. Use TBTOOLS for drawing.
Subcellular Localization
In order to understand the subcellular localization of DoSPX4, the recombinant plasmid pCAMBIA1305-DoSPX1-GFP was constructed by amplifying the fragment of DoSPX4 by sense and antisense primers (Supplementary Table 3). The recombinant plasmid was introduced into Agrobacterium tumefaciens EHA105. EHA105 Infect tobacco leaves and DoSPX4 is transiently expressed in Nicotiana benthamiana leaf epidermal cells. A confocal laser microscope was used to observe the GFP fluorescence signal.
Yeast Two-Hybrid
In order to verify the interaction of DoSPX4 with DoPHR2, the AD-DoSPX4 recombinant vector was constructed by amplifying DoSPX4 open reading frame into AD, and the DoPHR2-BD recombinant vector was constructed by amplifying the DoPHR2 fast-play reading frame into BD, the primers were shown in Supplementary Table 3. The constructed AD-DoSPX4 and DoPHR2-BD were used to transform AH109 jointly, and the successfully transformed clones were screened on SD-WL, and the clones were screened on SD-HAWL. The X-α-Gal is used to identify positive interactions (SD-WL indicates the SD medium without Leu and Trp, and SD-HAWL means the SD medium without Ade, His, Leu, and Trp).
Nicotiana tabacum Transformation
In order to understand the function of DoSPX4, the recombinant plasmid pCAMBIA1305-DoSPX4 was constructed by amplifying the fragment DoSPX4 by sense and antisense primer (Supplementary Table 3), pCAMBIA1305-DoSPX4 transformed into A. tumefaciens GV3101. The GV3101 was transformed into N. tabacum leaf discs via an A. tumefaciens-mediated leaf disc procedure (Topping, 1988). Regenerated plants are obtained by inducing callus, budding, rooting, and transplanting. The positive strain is screened and selected by using 50 mg/L Hygromycin B and 200 mg/L antibacterial Cefotaxime. The transgenic N. tabacum were cultured in MS medium with 1.25 mM Pi concentration (HP) and 0.0625 mM Pi concentration (LP) for 7 days.
Determination of Available Pi Content
The content of available Pi in plants was determined by the ammonium molybdate method (Nanamori et al., 2004). To determine the Pi concentration of the transgenic N. tabacum, the transgenic N. tabacum was ground with liquid nitrogen and 10% (w/v) perchloric acid (PCA). The supernatant was centrifuged after 10 times dilution with 5% (w/v) PCA. The working solution [sulfuric acid-ammonium molybdate (solution A) and ascorbic acid solution (solution B) were mixed in proportion (6:1)] extracts the available Pi from the supernatant. The absorbance was measured at 820 nm by a UV spectrophotometer.
Quantitative Real-Time PCR Analysis
The RNA was extracted from the Liquid nitrogen quick-frozen plant tissues using a Plant Total RNA Isolation Kit (Sangon Biotech, Shanghai, China). A One Step RT-qPCR Kit (BBI Life Science, Shanghai, China) was used to obtain cDNA. 2× TaqMan Fast qPCR Master Mix (BBI Life Science, China) was used to execute qRT-PCR. Reaction conditions were performed according to Liu’s method (Liu L. et al., 2021). The qRT-PCR primers were designed using NCBI PRIMER-BLAST8 (Supplementary Table 3). Each experiment was set up with three biological replicates, and the results were calculated using 2–ΔΔCT method.
The Temporal Expression Patterns Analysis of DoSPXs
The transcriptome data of 8 D. officinale tissues (root, stem, leaf, flower buds, column, lip, and sepal) were downloaded from the NCBI SRA database (PRJNA348403). Trimmomatic (Bolger et al., 2014) is used to filter and trim data. A retrieval file of D. officinale genome was established by HISAT2 (Pertea et al., 2016), and high-quality reads were compared to D. officinale genome. Samtools (Li et al., 2009) was used for sorting and format conversion to obtain BAN format files. Finally, StringTie (Pertea et al., 2016) was assembled for sequence, and transcript abundance was estimated. TBOOLS (Chen et al., 2020) is used for the visualization of results.
Results
Identification of SPX Family Genes in Dendrobium officinale
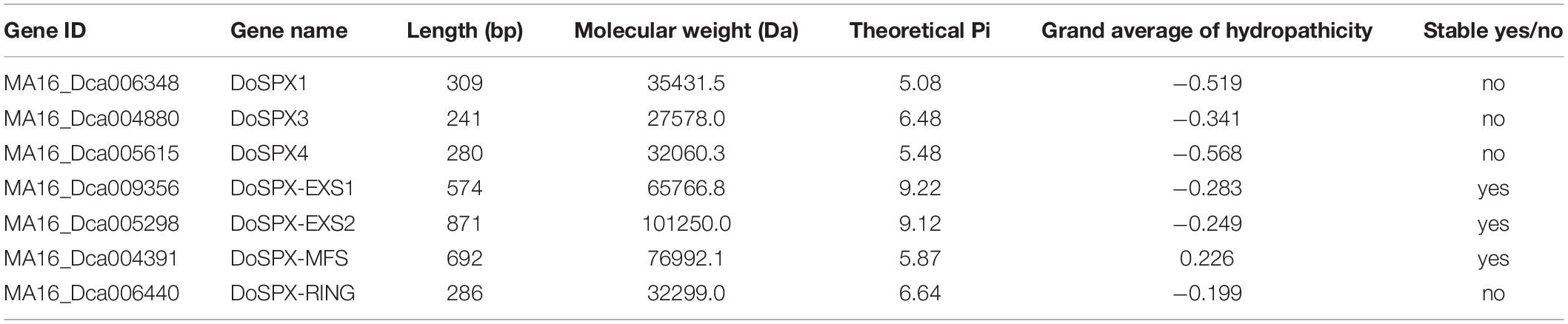
By blasting the published genome sequences of D. officinale using the conserved SPX sequences reported in O. sativa and A. thaliana, the possible DoSPX sequences were obtained, and the redundancy of the sequences with high similarity was removed, then verified these sequences with SMART and PFAM, the result was shown in Table 1. Seven possible DoSPX sequences were obtained, all of which contain SPX conserved domains. Combining with the naming method of A. thaliana, two of them contain EXS domains named DoSPX-EXS1 and DoSPX-EXS2. One sequence contains the MFS sequence, named DoSPX-MFS. A sequence containing the RING field, called DoSPX-RING. The three sequences containing only the SPX domain were named DoSPX1, DoSPX3, and DoSPX4. The amino acid sequence analysis showed that the size of all proteins was 2.75–10.12 kDa and the isoelectric point was 5.08–9.22. The average hydrophilic coefficient (GRAVY < 0) shows that the other six are hydrophilic proteins except for DoSPX-MFS.
System Evolution Analysis of DoSPXs
Through CLUSTERW alignment, the phylogenetic trees of D. officinale, P. equestris, O. sativa, and A. thaliana were established by the NJ method of MEGA7.0. The result was shown in Figure 1. It was found that all sequences were divided into four subclasses. The sequences of each subclass of SPX in D. officinale were well classified from those in O. sativa and A. thaliana. DoSPX1 is clustered with AtSPX1, 2, and OsSPX1; DoSPX3 were clustered with OsSPX3, 5, 6, and AtSPX3; while DoSPX4 is clustered with AtSPX4, OsSPX4. DoSPX-EXS1, DoSPX-EXS2, DoSPX-MFS, and DoSPX-RING are well clustered with the three subfamilies reported in O. sativa and A. thaliana.
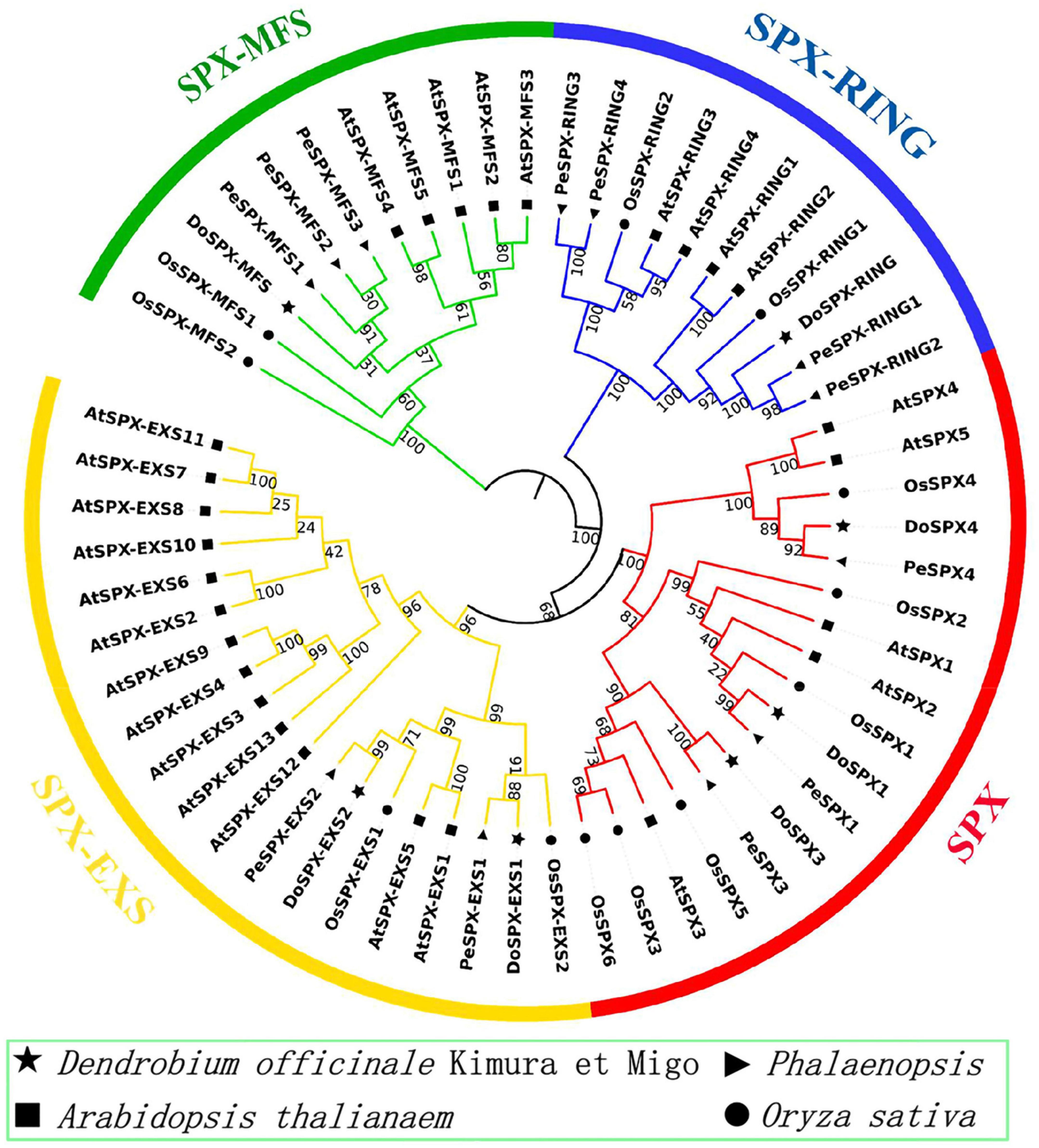
Figure 1. Phylogenetic analysis of SPX proteins in Dendrobium officinale, Phalaenopsis equestris, Oryza sativa, and Arabidopsis thaliana. The neighbor-joining (NJ) tree was created with 9 D. officinale, 12 P. equestris, 12 O. sativa, and 27 A. thaliana SPX protein sequences.
Analysis of DoSPXs Cis-Acting Elements
By analyzing the upstream promoter sequence of DoSPXs, many cis-acting elements necessary for plant growth, development, and stress response were excavated. The result was shown in Figure 2. In these elements, MSA like, CAT-box and BOX-4 related to plant growth and development; ERE and TGACG-motif were involved in response to hormones; TC-rich repeat were involved in response to abiotic stress. These results suggested that DoSPXs may participate in the growth, development, and stress response of D. officinale.

Figure 2. Analysis of the cis-acting elements in DoSPXs of D. officinale. (A) The number in each cell represents the number of cis-acting elements in different SPX genes. (B) Each bar diagram represents a different type of cis-acting element. (C) The pie chart represents the proportion of promoters in other groups.
Expression Pattern Analysis of DoSPXs
In order to understand the expression patterns of the DoSPXs in D. officinale, the expression level of the DoSPXs in different tissues was analyzed. The results were presented in heat map form in Figure 3. DoSPX1 and DoSPX4 showed similar expression patterns in various tissues and had high expression levels in D. officinale. DoSPX3 had high expression level in sepals, showed that DoSPX3 played an important role in the sepals. DoSPX-EXS1 had high expression level in the root tips and stem. DoSPX-EXS1, DoSPX-RING, and DoSPX-MFS had low expression levels in all eight tissues.
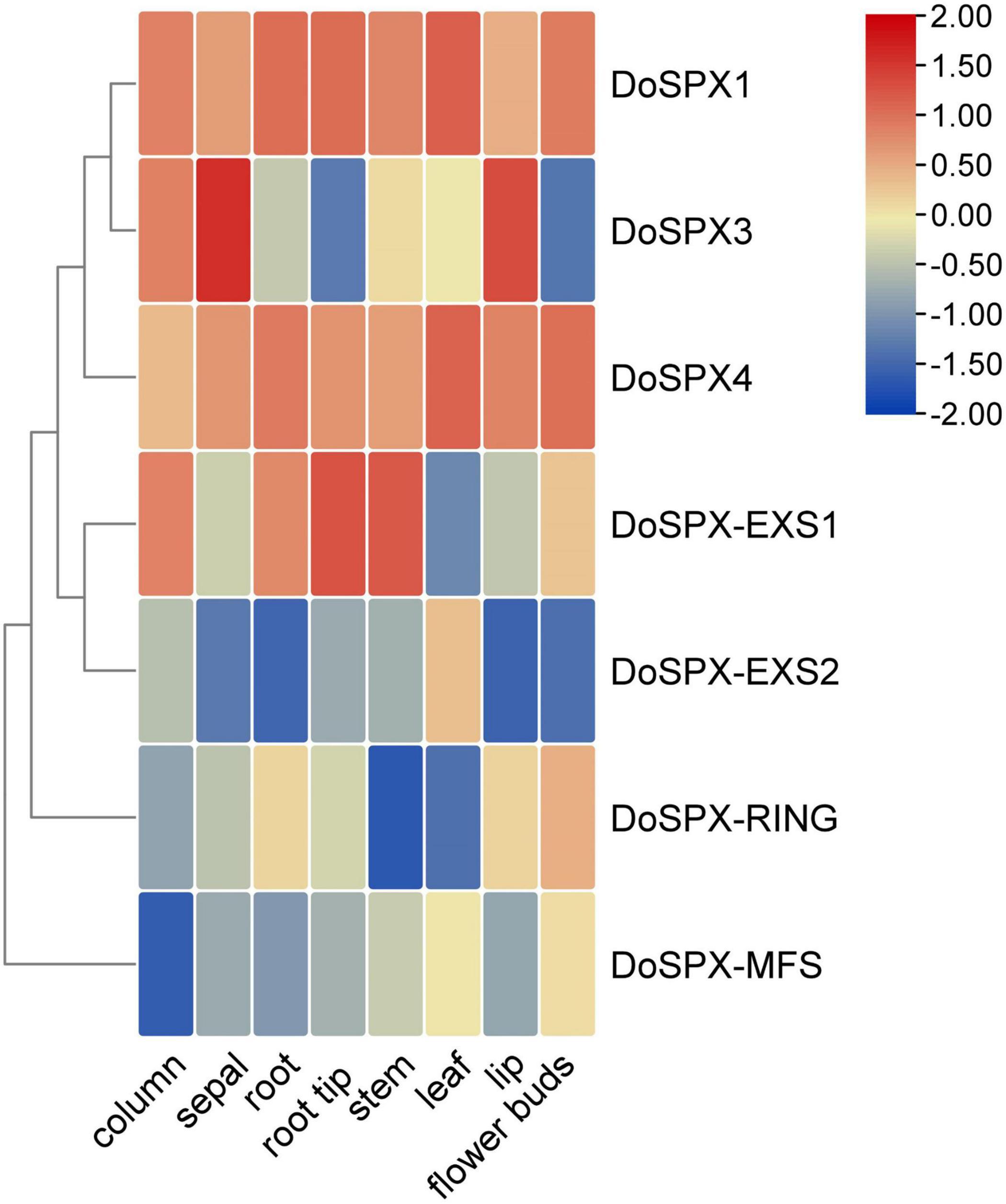
Figure 3. Expression profiles of DoSPX genes in 8 tissues of D. officinale. Blue to red indicates low to high expression levels of the gene.
In order to understand the response of DoSPXs to −Pi stress, D. officinale was taken at 1, 5, 10, and 40 days after the −Pi treatment and determined the expression level of DoSPXs. The results were shown in Figure 4. Under the −P, the transcript abundance of most PSI genes increased. However, each gene showed its unique expression pattern at 0 day of the −P treatment. In D. officinale buds, the expression levels of DoSPX-EXS1, DoSPX-MFS, and DoSPX-RING reached the highest at 1 day after the −P treatment, DoSPX1 and DoSPX3 reached the highest at 5 day after the −P treatment; DoSPX-EXS2 peaked at day 10. In roots, DoSPX1-DoSPX4, DoSPX-EXS1, DoSPX-EXS2, and DoSPX-MFS reached the highest expression level at day 5 after the −P treatment; their expression level increased with the increase of −Pi treatment time. These results suggested that DoSPXs mostly reached a high expression level in the early stage of −P stress, while DoSPX4 showed different changes from other genes in the buds under the −P stress. The expression of DoSPX4 reached its highest level at 0 day of −P treatment, and the expression down-regulated with −Pi treatment time.
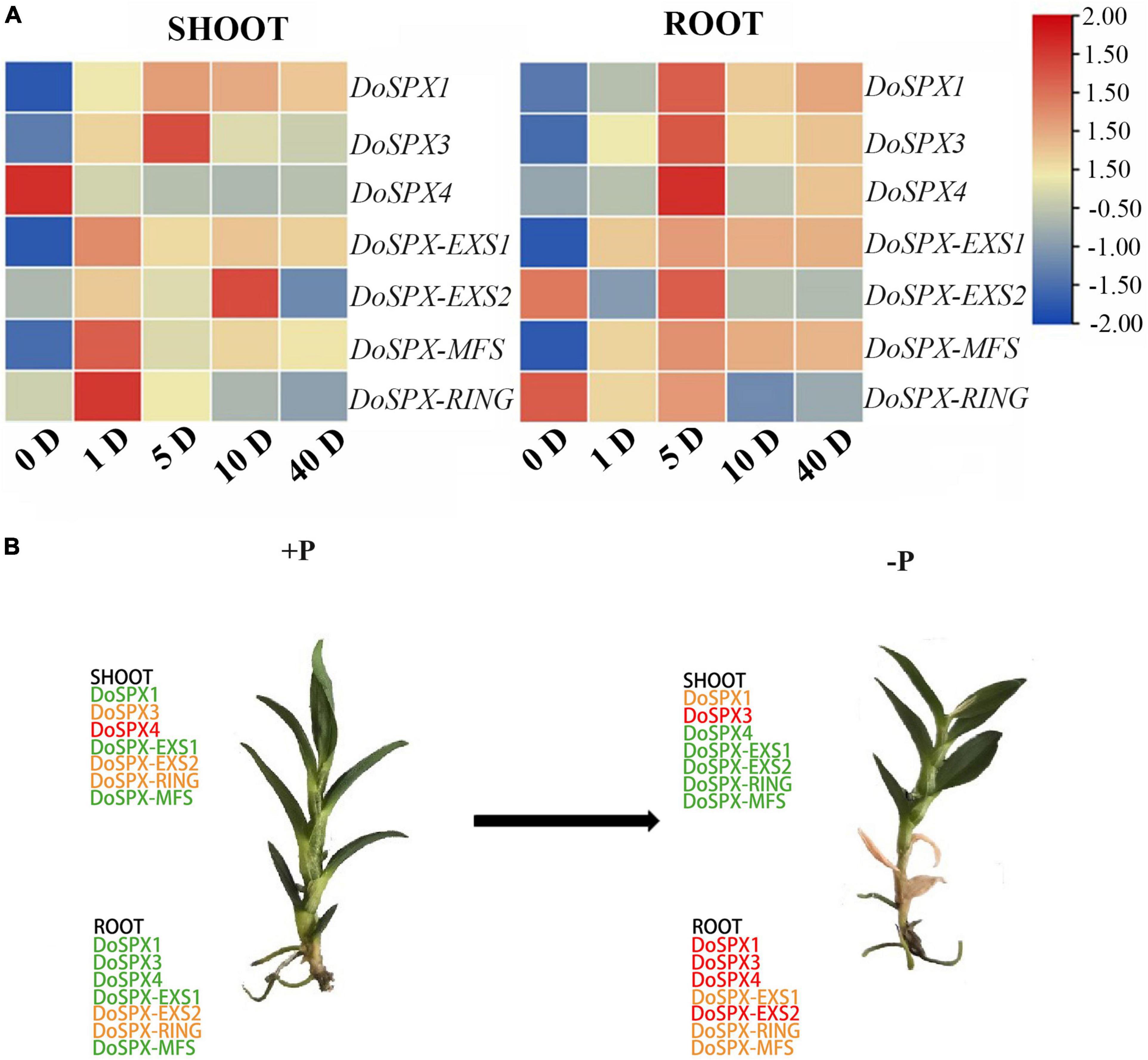
Figure 4. (A) Temporal and spatial expression patterns of PSI gene in shoot and roots under −P stress in D. officinale. The actin was used as an internal reference gene to regulate gene expression (B) SPX genes expression in shoot and root of D. officinale. Treated with −Pi for 5 day. Green, yellow, and red indicate low, medium, and high expression.
Subcellular Localization of DoSPX4
The CDS region without the stop codon of the DoSPX4 gene was cloned into the pCAMBIA1305.1-GFP vector, and the subcellular localization of DoSPX4 in epidermal cells of N. tabacum was observed by a laser confocal microscope. The results showed that DoSPX4-GFP recombinant protein had a fluorescence signal on the cell membrane, while GFP was distributed in the whole cell (Figure 5).
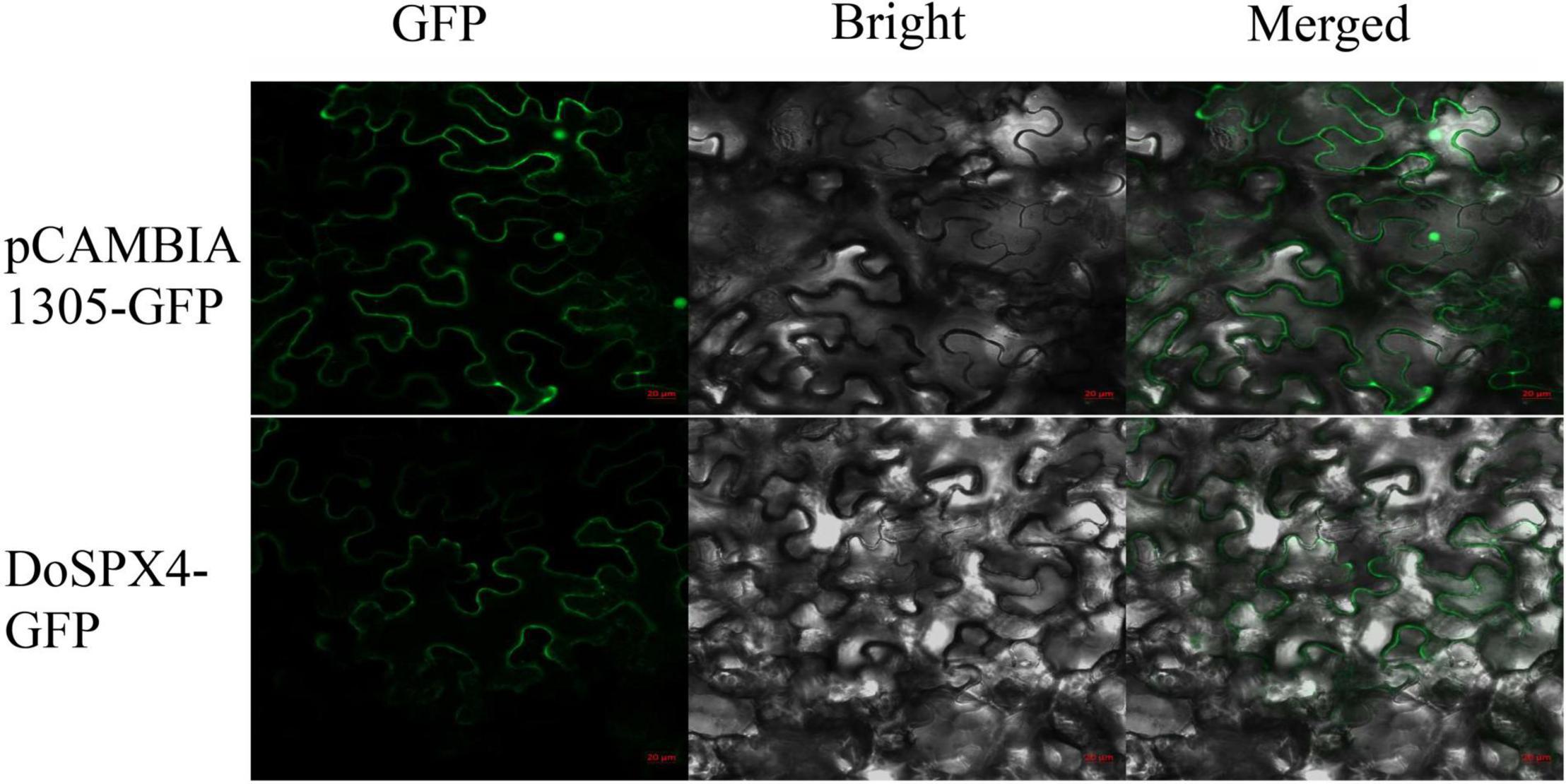
Figure 5. Subcellular localization of DoSPX4 protein in Nicotiana tabacum epidermal cells. The results were observed by confocal laser microscopy. The pictures show a green fluorescent field, a bright field and an overlay of two fields (merge). The numerical reading of the red ruler is 20 μm.
DoSPX4 Interact With DoPHR2 by Y2H
To identify the interaction between DoSPX4 and DoPHR2, the CDS of DoSPX4 was cloned into pGADT7, and the segment (C192aa-225aa) of DoPHR2 without transcriptional activation domain was cloned into the pGBKT7 vector. AD-DoSPX4 and BD-DoPHR2192aa–225aa were transformed into yeast strain AH109. The yeast strain could grow normally on SD/-Trp-Leu-His-Ade medium, indicating that DoSPX4 was tender enough to interact with DoPHR2192aa–225aa (Figure 6).
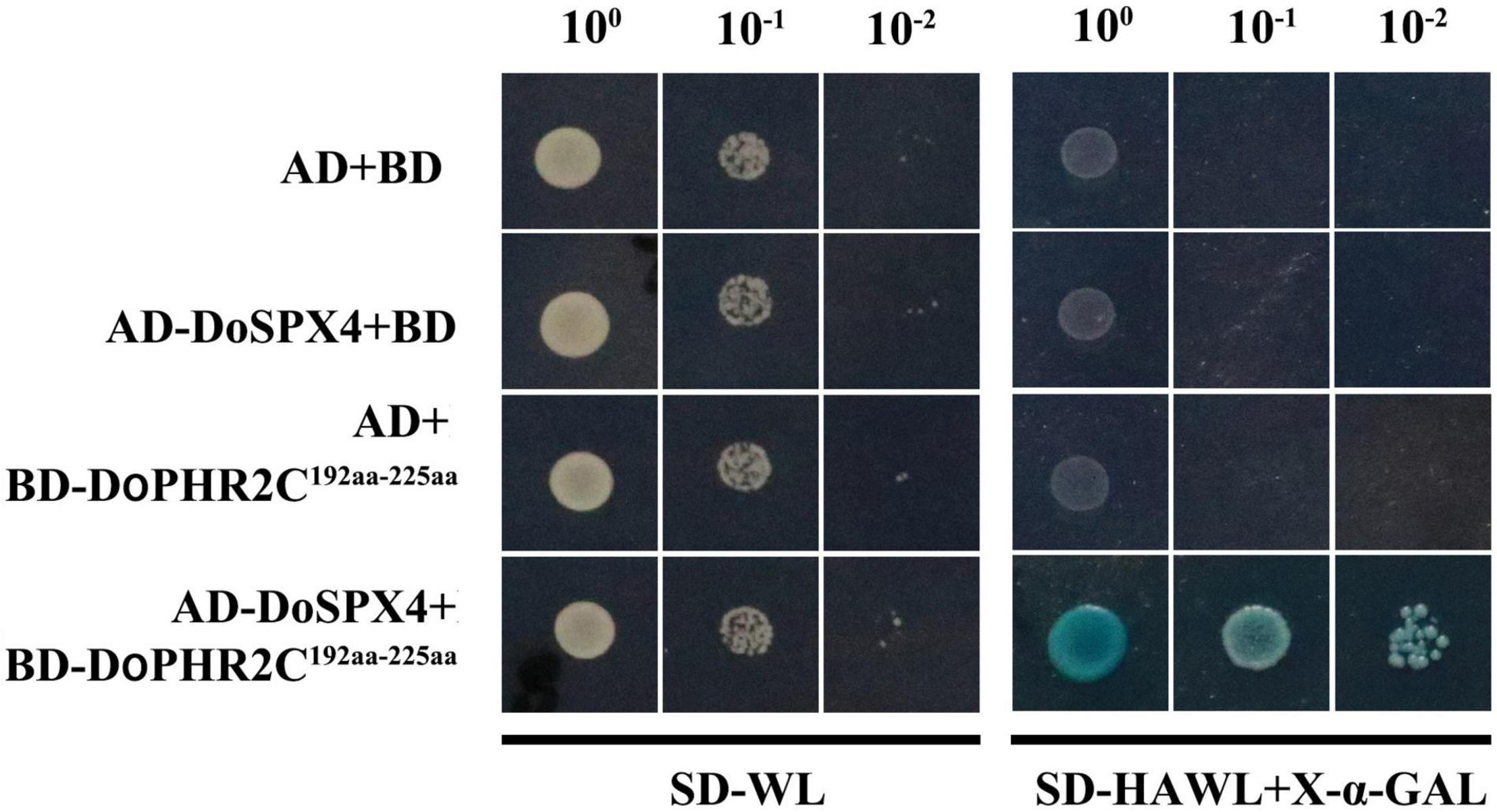
Figure 6. The DoSPX4 interacts with DoPHR2. The pGADT7 and pGBKT7 were used as the negative control. SD-WL indicates the SD medium without Leu and Trp, and SD-HAWL means the SD medium without Ade, His, Leu, and Trp.
Overexpression DoSPX4 in Nicotiana tabacum
To analyze the function of DoSPX4 in plants, we obtained transgenic N. tabacum overexpression DoSPX4. As shown in Figure 7, through −Pi treatment, the length of leaves and root in OE-DoSPX4 N. tabacum plants decreased significantly, and the root–shoot ratio was 1.97 times than that of the empty vector (EV) group.
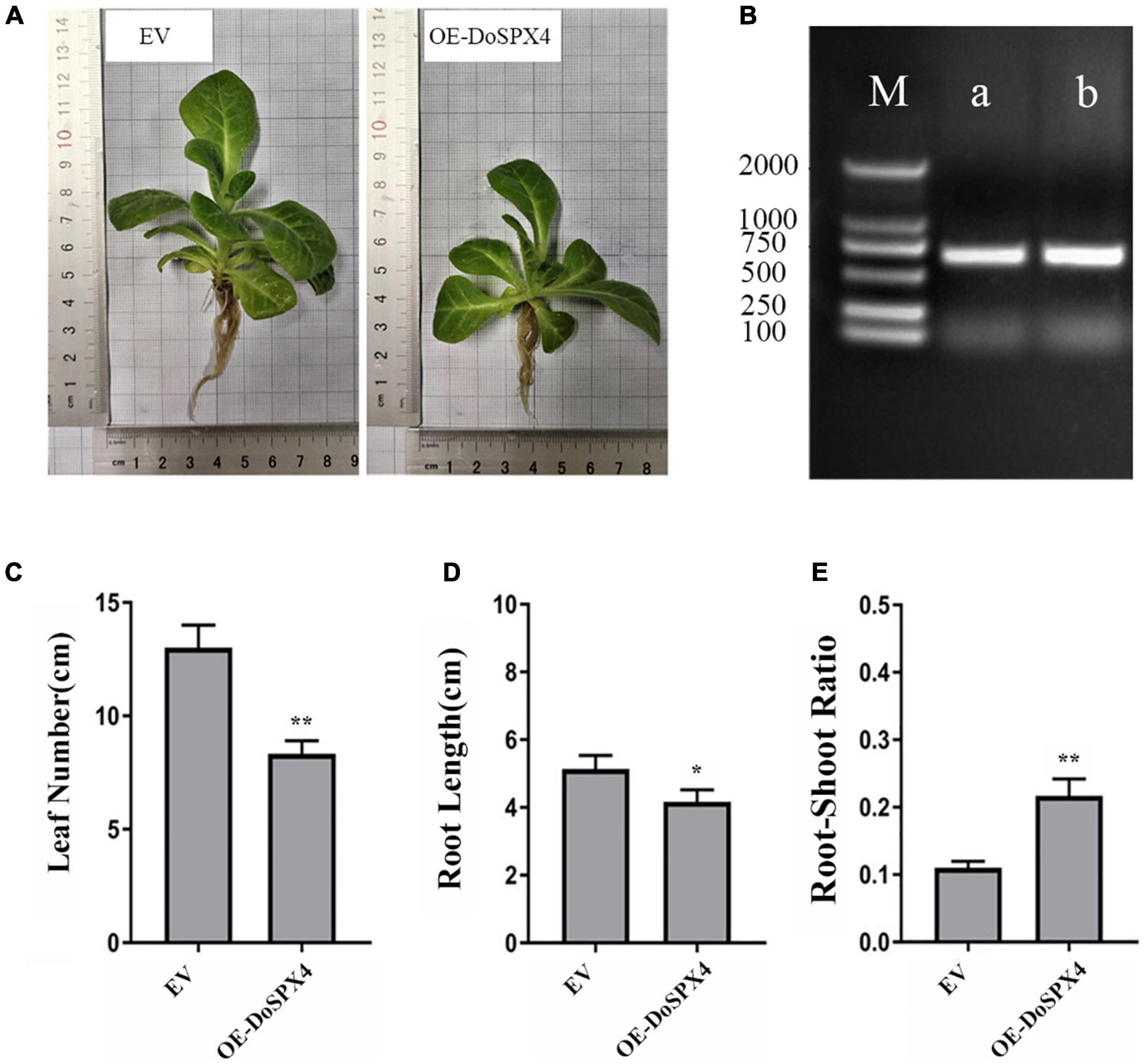
Figure 7. Transgene analysis. (A) Phenotypic difference of OE-DoSPX4 transgenic N. tabacum. (B) Identification of positive transgenic N. tabacum lines by PCR (M represent DL2000 DNA marker, a represent EMPTY VECTOR, b represent OE-DoSPX4). (C) The leaf number of OE-DoSPX4 transgenic N. tabacum. (D) The root length of OE-DoSPX4 transgenic N. tabacum. (E) The root–shoot ratio of OE-DoSPX4 transgenic N. tabacum. Each sample included at least three replicates, and the values are the means ± SDs. *Indicates that the difference is significant, **Indicates that the difference is very significant.
To understand the function of DoSPXs on Pi absorption and utilization, the available Pi contents of transgenic N. tabacum under + P and −P culture were measured (Figure 7). Under the −P stress, P content in the aboveground part of OE-DoSPX4 N. tabacum was significantly higher than that in the EV group. These results indicated that the DoSPX4 overexpression promoted the Pi accumulation in the shoot.
The expression levels of Pi response genes (NtPHRs and NtPTs) in OE-DoSPX4 N. tabacum were also detected (Figure 8). Under + P, the expression levels of NtPHR1 and NtPHR2 were significantly lower in OE-DoSPX4 N. tabacum than those in the control group. Under the −P condition, the expression levels of NtPHR1 and NtPHR2 were significantly up-regulated except the NtPHR2 in the aboveground of OE-DoSPX4 N. tabacum. NtPT1 and NtPT2 were basically induced in OE-DoSPX4 N. tabacum, especially under the −Pi stress. These results suggested that DoSPX4 may play a negative regulatory role in the expression of NtPHR1 and NtPHR2, and then affect the process of Pi transport from N. tabacum roots to shoot.
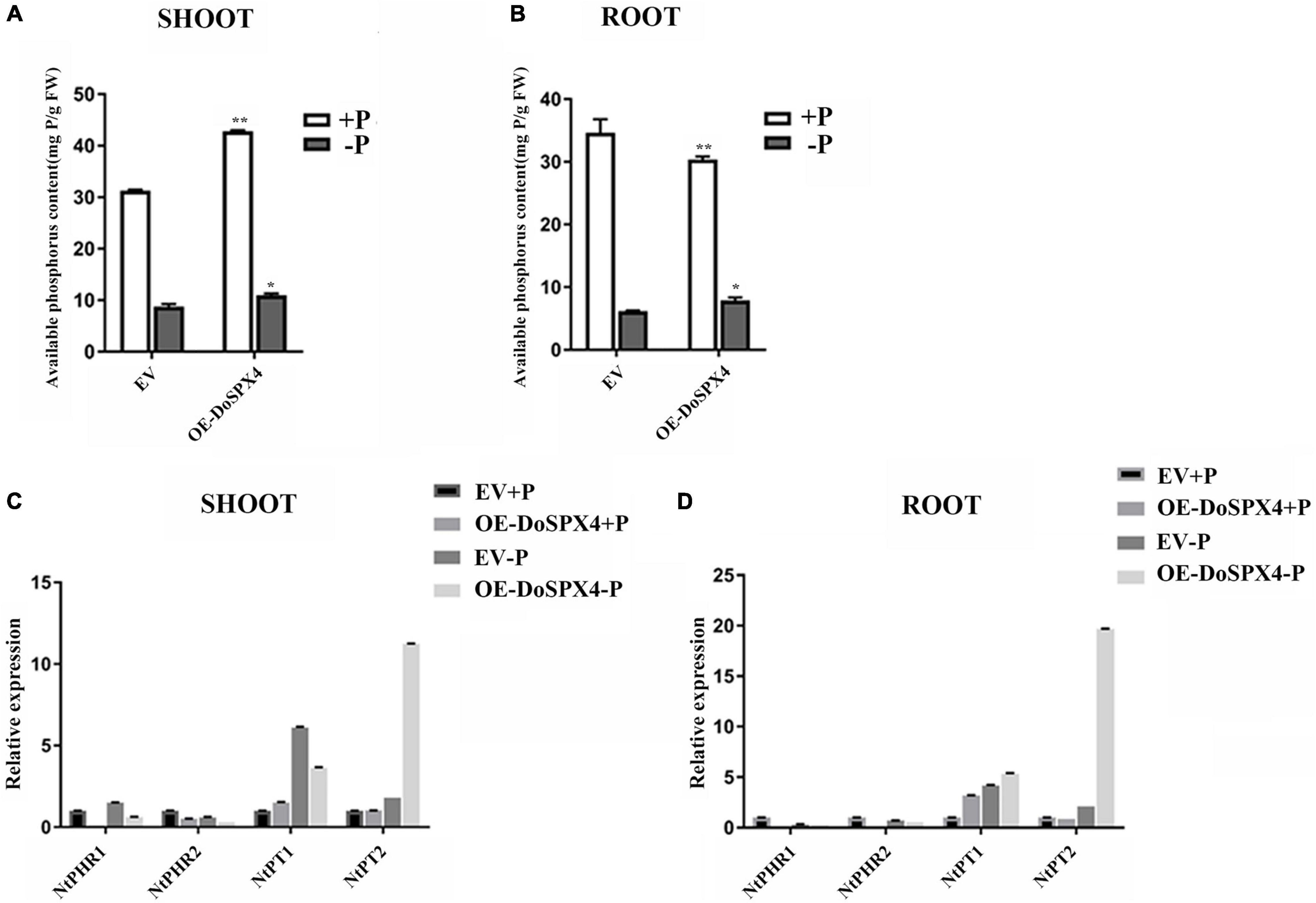
Figure 8. Determination of indices in transgenic N. tabacum. (A) The available Pi concentration in shoot in OE-DoSPX4 N. tabacum. (B) The available Pi concentration in root in OE-DoSPX4 N. tabacum. (C) The expression analysis of NtPHR1/2 and NtPT1/2 in shoot in OE-DoSPX4 N. tabacum. (D) The expression analysis of NtPHR1/2 and NtPT1/2 in root in OE-DoSPX4 N. tabacum. Each sample included at least three replicates, and the values are the means ± SDs. *Indicates that the difference is significant, **Indicates that the difference is very significant.
Discussion
The organic Pi that plants can absorb in the environment cannot satisfy the need of plants (Grennan, 2008). Medicinal plants have also evolved complex mechanisms to adapt to the −Pi responses (Liu L. et al., 2016, 2018; Wang et al., 2020). The proteins, containing the SPX domain, participate in the molecular regulatory network of plant response to Pi stress (Liu N. et al., 2018). Through the genome analysis of D. officinale, we obtained seven DoSPX proteins. DoSPX proteins have strong homology in P. equestris and D. officinale, suggesting that the regulatory network of plants responding to −Pi stress may be conserved. Through phylogenetic analysis and conservative motif analysis, we found that seven DoSPXs belong to 4 subfamilies (SPX, SPX-MFS, SPX-EXS, and SPX-RING) and have homology with O. sativa and A. thaliana.
The cis-acting elements of DoSPXs promoter region include MSA-like elements involved in cell cycle regulation and CAT-box elements involved in meristem development, which indicates that DoSPXs may be involved in the growth of D. officinale. In addition, the promoter region contains ERF, TGACG-motif, TC-rich, and ARE elements, which can respond to ethylene, plant hormones, plant defense and stress (Feng et al., 2020; Huo et al., 2021; Zhang et al., 2022). These results indicated that DoSPXs may involve in abiotic stress such as invasion and drought.
Tissue special expression showed that DoSPXs had different expression patterns, which indicates that DoSPXs may play different functions. The expression level of DoSPX1 and DoSPX3 was induced in the roots and shoot of D. officinale under −P and reached the peak at day 5, which indicated that DoSPX1 and DoSPX3 may participate in the early response of D. officinale to −P, this was similar to the expression pattern of AtSPX1/2/3 (Duan et al., 2008; Puga et al., 2014) and OsSPX1/2/3/5/6 (Wang et al., 2009; Shi et al., 2014).
Under the −P, DoSPX4 had a different expression pattern compared to DoSPX1 and DoSPX3. According to homology comparison analysis, DoSPX4 had high homology with OsSPX4 (Lv et al., 2014) and AtSPX4 (Duan et al., 2008). In A. thaliana, the localization of AtSPX4 is different from AtSPX1 and AtSPX3, which shows the uniqueness of the function of AtSPX4 in A. thaliana Pi response. The subcellular localization analysis indicates that DoSPX4 is located on the membrane, which is consistent with AtSPX4 and OsSPX4. These results imply that DoSPX4 may have similar functions to AtSPX4 and OsSPX4, but this still needs a lot of experimental to prove.
It has been reported that SPX can interact with MYB-CC transcription factor PHR and then affect the downstream PSI genes (Lv et al., 2014). InsPs can promote the interaction between SPX and the MYB-CC domain of PHR (Duan et al., 2008). The C-terminal of DoPHR2 contains the MYB-CC domain. At Yeast two-hybrid test showed that DoSPX4 interacted with the C-terminal of DoPHR2. The results indicated that DoSPX4 worked by DoPHR2 in −P. However, the recognition of SPX protein conserved domain by InsPs and the regulation of DoPHR2 at the protein level need an in-depth study.
The DoSPX4 was overexpressed in N. tabacum, which can observe the changes in the root system, Pi content, and Pi transporter expression levels. In OE-DoSPX4 N. tabacum, the root–shoot ratio increased, which was conducive to the absorption of Pi from the environment. the qRT-PCR analysis found that the expression of NtPHR1/2 and NtPT1/2 in OE-DoSPX4 transgenic N. tabacum shoot and root increased. The effective Pi content decreased in the root, while the effective Pi content increased in the aboveground part. It is speculated that DoSPX4 is involved in the induction of NtPT1/2 by NtPHR1/2. NtPT1/2 is a high-affinity phosphate transporter responsible for the long-distance transport of Pi in plants. The high expression of NtPT1/2 promotes the transport of Pi from root to aboveground part in vivo, which increases the Pi content of aboveground parts. The absorption and utilization efficiency of Pi in N. tabacum was improved (Figure 9).
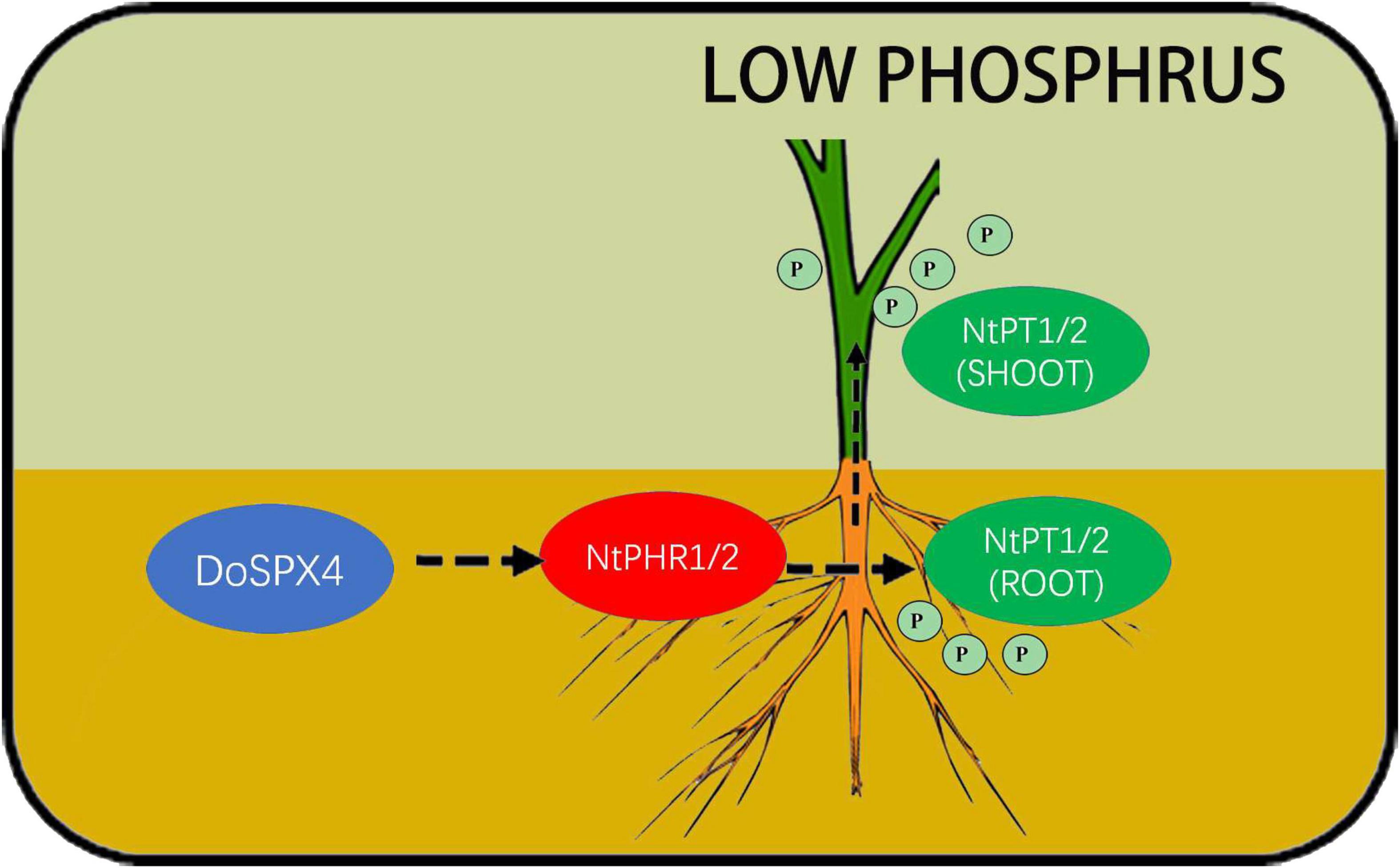
Figure 9. Possible DoSPX4 impact mechanism. DoSPX4 promoted the expression of NtPHR1/2 in roots, induced the expression of NtPT1/2 and high-affinity phosphate transporter in roots and shoot, and promoted the transportation of Pi from roots to stems in transgenic N. tabacum in low phosphorus.
Conclusion
In this study, seven SPX family proteins were identified from D. officinale. The qRT-PCR analysis showed the SPX family had different expression patterns. The DoSPX4 was located in the cell membrane and had the ability to interact with DoPHR2. The heterologous expression of N. tabacum showed that DoSPX4 could activate NtPHR1/2, increase the expression of NtPT1/2 in roots and stems, and promote the transport of Pi from roots to shoot. These results provide an experimental basis for further study on the adaptation mechanism of D. officinale to low phosphorus.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
HF organized and designed the experiment. LL and HX conducted the experiment and completed the manuscript writing. JS and HS analyzed the data and put forward valuable suggestions to XS and LT in the process of manuscript modification. All authors read and approved the manuscript.
Funding
This work was supported by the Anhui University Natural Science Foundation (KJ2019A0181 and K1942003), the Hefei Natural Science Foundation (2022036), and the Anhui Natural Science Foundation (1908085MC59).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thanks to the authors for their contributions to the experiment, and thanks to the foundation for supporting the project.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.943788/full#supplementary-material
Footnotes
- ^ http://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index.cgi
- ^ http://smart.embl-heidelberg.de/
- ^ http://pfam.xfam.org/
- ^ http://web.expasy.org/protparam/
- ^ https://itol.embl.de/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ https://www.dna.affrc.go.jp/PLACE/?action=newplace
- ^ https://www.ncbi.nlm.nih.gov/tools/primer-blast/
References
Bhatt, P., Rene, E. R., Huang, Y. H., Lin, Z. Q., Pang, S. M., Zhang, W. P., et al. (2021). Systems biology analysis of pyrethroid biodegradation in bacteria and its effect on the cellular environment of pests and humans. J. Environ. Chem. Eng. 9:106582. doi: 10.1016/j.jece.2021.106582
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., and Liao, H. (2017). Engineering crop nutrient efficiency for sustainable agriculture. J. Integr. Plant Biol. 59, 710–735. doi: 10.1111/jipb.12559
Chen, Y. F., Li, L. Q., Xu, Q., Kong, Y. H., Wang, H., and Wu, W. H. (2009). The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21, 3554–3566. doi: 10.1105/tpc.108.064980
Chiou, T. J., and Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Duan, K., Yi, K. K., Dang, L., Huang, H. J., Wu, W., and Wu, P. (2008). Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 54, 965–975. doi: 10.1111/j.1365-313X.2008.03460.x
Feng, K., Hou, X. L., Xing, G. M., Liu, J. X., Duan, A. Q., Xu, Z. S., et al. (2020). Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 40, 750–776. doi: 10.1080/07388551.2020.1768509
Fu, L. M., Niu, B. F., Zhu, Z. W., Wu, S. T., and Li, W. Z. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Grennan, A. K. (2008). Phosphate accumulation in plants: signaling. Plant Physiol. 148, 3–5. doi: 10.1104/pp.104.900269
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Huo, Y. B., Zhang, B., Chen, L., Zhu, C., Zhang, X., and Zhu, C. (2021). Isolation and functional characterization of the promoters of miltiradiene synthase genes, TwTPS27a and TwTPS27b, and interaction analysis with the transcription factor TwTGA1 from Tripterygium wilfordii. Plants (Basel) 10:418. doi: 10.3390/plants10020418
Lescot, M., Déhais, P., Thijs, G., Zhang, J., Zhang, X., and Zhu, C. S. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., and Homer, N. (2009). 1000 genome project data processing subgroup The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lin, W. Y., Huang, T. K., and Chiou, T. J. (2013). Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25, 4061–4074. doi: 10.1105/tpc.113.116012
Liu, J., Fu, S., Yang, L., Luan, M., Zhao, F., Luan, S., et al. (2016). Vacuolar SPX-MFS transporters are essential for phosphate adaptation in plants. Plant Signal. Behav. 11:e1213474. doi: 10.1038/ncomms11095
Liu, L., Xiang, H. X., Shen, H. M., Dong, Y. X., Sun, X., Cai, Y. P., et al. (2021). Effects of low phosphorus stress on the main active ingredients and antioxidant activities of Dendrobium officinale. Ind. Crop Prod. 173:114095. doi: 10.1016/j.indcrop.2021.114095
Liu, L., Yang, D. F., Liang, T. Y., Zhang, H. H., and Liang, Z. S. (2016). Phosphate starvation promoted the accumulation of phenolic acids by inducing the key enzyme genes in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 35, 1933–1942. doi: 10.1007/s00299-016-2007-x
Liu, L., Yang, D. F., Xing, B. C., Zhang, H., and Liang, Z. (2018). Salvia castanea hairy roots are more tolerant to phosphate deficiency than Salvia miltiorrhiza hairy roots based on the secondary metabolism and antioxidant defenses. Molecules 23:1132. doi: 10.3390/molecules23051132
Liu, N., Shang, W., Li, C., Jia, L. H., Wang, X., Xing, G., et al. (2018). Evolution of the SPX gene family in plants and its role in the response mechanism to phosphorus stress. Open Biol. 8:170231. doi: 10.1098/rsob.170231
Lv, Q., Zhong, Y., Wang, Y., Zhang, L., Shi, J., Wu, Z., et al. (2014). SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26, 1586–1597. doi: 10.1105/tpc.114.123208
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Nanamori, M., Shinano, T., Wasaki, J., Yamamura, T., Rao, I. M., and Osaki, M. (2004). Low phosphorus tolerance mechanisms: phosphorus recycling and photosynthate partitioning in the tropical forage grass, Brachiaria hybrid cultivar Mulato compared with rice. Plant Cell Physiol. 45, 460–469. doi: 10.1093/pcp/pch056
Osorio, M. B., Ng, S., Berkowitz, O., De Clercq, I., Mao, C. Z., and Shou, H. X. (2019). SPX4 acts on phr1-dependent and -independent regulation of shoot phosphorus status in arabidopsis. Plant Physiol. 1, 332–352. doi: 10.1104/pp.18.00594
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Puga, M. I., Mateos, I., Charukesi, R., Wang, Z., Franco-Zorrilla, J. M., de Lorenzo, L., et al. (2014). SPX1 is a phosphate-dependent inhibitor of phosphate starvation response 1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 14947–14952. doi: 10.1073/pnas.1404654111
Secco, D., Wang, C., Arpat, B. A., Wang, Z. Y., Poirier, Y., Tyerman, S. D., et al. (2012). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 4, 842–851. doi: 10.1111/j.1469-8137.2011.04002.x
Shi, J., Hu, H., Zhang, K., Zhang, W., Yu, Y. M., Wu, Z., et al. (2014). The paralogous SPX3 and SPX5 genes redundantly modulate Pi homeostasis in rice. J. Exp. Bot. 65, 859–870. doi: 10.1093/jxb/ert424
Su, T., Xu, Q., Zhang, F. C., Chen, Y., Li, L. Q., Wu, W. H., et al. (2015). WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 167, 1579–1591. doi: 10.1104/pp.114.253799
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Topping, J. F. (1988). Tobacco transformation. Methods Mol. Biol. 81, 362–372. doi: 10.1385/0-89603-385-6:365
Wang, B., Zhang, T. X., Du, H. W., Zhao, Q., and Meng, X. C. (2020). Effect of peg on secondary metabolites in suspension cells of scutellaria baicalensis georgi. Acta Med. Mediterr. 36, 2307–2312. doi: 10.19193/0393-6384_2020_4_359
Wang, C., Yue, W., Ying, Y., Wang, S., Secco, D., Liu, Y., et al. (2015). Rice SPX-major facility superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. Plant Physiol. 169, 2822–2831. doi: 10.1104/pp.15.01005
Wang, Y., Ribot, C., Rezzonico, E., and Poirier, Y. (2004). Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135, 400–411. doi: 10.1104/pp.103.037945
Wang, Z. Y., Hu, H., Huang, H. J., Duan, K., Wu, Z. C., and Wu, P. (2009). Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and pistarvation signaling in rice. J. Integr. Plant Biol. 51, 663–674. doi: 10.1111/j.1744-7909.2009.00834.x
Wang, Z. Y., Ruan, W. Y., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111, 14953–14958. doi: 10.1073/pnas.1404680111
Yang, J., Wang, L., Mao, C. Z., and Lin, H. H. (2018). Characterization of the rice NLA family reveals a key role for OsNLA1 in phosphate homeostasis. Rice 10:52. doi: 10.1186/s12284-017-0193-y
Yue, W. H., Ying, Y. H., Wang, C., Zhao, Y., Dong, C. H., Whelan, J., et al. (2017). OsNLA1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters. Plant J. 90, 1040–1051. doi: 10.1111/tpj.13516
Zhang, B., Song, Y. F., Zhang, X. D., Wang, Q. N., Li, X. Q., He, C. Z., et al. (2022). Identification and expression assay of calcium-dependent protein kinase family genes in Hevea brasiliensis and determination of HbCDPK5 functions in disease resistance. Tree Physiol. 42, 1070–1083. doi: 10.1093/treephys/tpab156
Zhang, J. Y., Zhou, X., Xu, Y., Yao, M. L., Xie, F. B., Gai, J. Y., et al. (2016). Soybean SPX1 is an important component of the response to phosphate deficiency for Pi homeostasis. Plant Sci. 248, 82–91. doi: 10.1016/j.plantsci.2016.04.010
Keywords: gene family, SPX, PHR, Dendrobium officinale, gene function
Citation: Liu L, Xiang H, Song J, Shen H, Sun X, Tian L and Fan H (2022) Genome-Wide Analysis of DoSPX Genes and the Function of DoSPX4 in Low Phosphorus Response in Dendrobium officinale. Front. Plant Sci. 13:943788. doi: 10.3389/fpls.2022.943788
Received: 14 May 2022; Accepted: 06 June 2022;
Published: 11 July 2022.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Pankaj Bhatt, Purdue University, United StatesPengguo Xia, Zhejiang Sci-Tech University, China
Nai-Dong Chen, West Anhui University, China
Copyright © 2022 Liu, Xiang, Song, Shen, Sun, Tian and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honghong Fan, aGhmYW4wNTUxQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
 Lin Liu†
Lin Liu† Haoxin Xiang
Haoxin Xiang Honghong Fan
Honghong Fan