
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 15 July 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.943669
This article is part of the Research TopicGenetic Improvement of Triticeae Crops Based on High-throughput Phenotyping: Molecular Design for Yield, Resistance and ToleranceView all 14 articles
Powdery mildew is one of the most devastating foliar diseases in wheat production. The wild relative Thinopyrum ponticum (2n = 10x = 70) has been widely used in wheat genetic improvement due to its superior resistance to both biotic and abiotic stresses. In the present study, two wheat-Th. ponticum introgression lines named SN0293-2 and SN0293-7 were developed from the progenies of a cross between the octoploid Trititrigia SNTE20 and common wheat, including the elite cultivar Jimai 22. They had a novel powdery mildew resistance gene (temporarily named PmSN0293) putatively from Th. ponticum pyramided with Pm2 and Pm52, exhibiting excellent Pm resistance at both the seedling and adult stages. Sequential GISH-FISH detected no signal of Th. ponticum in these two lines but a pair of T1BL·1RS in SN0293-2. Chromosomal structural variations were also observed obviously in SN0293-2 and SN0293-7. Through the Wheat 660K SNP array, 157 SNPs, 134 of which were on 6A, were found to be specific to Th. ponticum. Based on the data combined with DNA re-sequencing, seven specific markers, including one CAPS marker on 2B and six CAPS and Indel markers on 6A, were developed, confirming their wheat-Th. ponticum introgression nature. Furthermore, the two lines displayed positive plant height and produced more kernels and higher 1,000-grain weight. Excellent resistance with desirable agronomic traits makes them valuable in wheat breeding programs.
As one of the most important staple food crops, wheat provides more than 20% of the calories and the protein for the world's population (Braun et al., 2010). The need for the production of wheat is continuously growing because of the increasing population (Hawkesford et al., 2013). However, it is threatened by many diseases which reduce yield and decrease quality.
Powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is one of the most devastating wheat diseases across the world. It can affect wheat photosynthesis and consequently decrease plant growth and grain filling, resulting in yield reductions (Zhang et al., 2020). Although fungicides can effectively control this disease, they also cause environmental pollution and cost increases. Host resistance is considered to be the most economical and environment-friendly means to do so (Wang et al., 2005; Liu et al., 2017). Therefore, it is a key step to explore and utilize resistance genes in wheat breeding programs. To date, more than 100 designated powdery mildew (Pm) resistance genes/alleles in 63 loci (Pm1-Pm68) have been documented (He et al., 2021; Gao et al., 2022). Some of them have been cloned, such as the broad-spectrum resistance gene Pm21 from Dasypyrum villosum (2n= 2x = 14, VV) (He et al., 2018; Xing et al., 2018). However, with the rapid evolution of new Bgt isolates, the single resistance gene is easily overcome. For instance, the well-known Pm8 on rye (Secale cereale L., 2n = 2x = 14, RR) chromosome arm 1RS has lost its function against new Bgt isolates, such as E09 (Ren et al., 2017). Hence, it is of great necessity to explore new Pm genes and pyramid multiple ones to broaden the resistance spectrum.
Wild relatives of common wheat carry many valuable genes that can be used for wheat improvement. The tall wheatgrass Thinopyrum ponticum (Podp.) Barkworth & D. R. Dewey (2n =10x =70, StStStStEeEeEbEbExEx or JJJJJJJSJSJSJS) has long been known to have superior resistance to both biotic and abiotic stresses, including powdery mildew, stem rust, leaf rust, stripe rust, eyespot and Fusarium head blight (Li and Wang, 2009). Because of its ability to readily be crossed with wheat, many genes for disease resistance have been introduced into wheat. Among them, eleven were formally documented, including Lr19, Lr24, Lr29, Sr24, Sr25, S26, Sr43, Yr69, Cmc2, Fhb7, and Pm51 (Sarma and Knott, 1966; Hart et al., 1976; Whelan and Lukow, 1990; Procunier et al., 1995; Mago et al., 2005; Li and Wang, 2009; Niu et al., 2014; Zhan et al., 2014; Hou et al., 2016; Wang et al., 2020). Pm51 is the only Pm gene designated officially from Th. ponticum so far. Due to the allodecaploid nature, Th. ponticum has a large and complex genome, and resistance to a disease might be associated with its different chromosomes (Chen et al., 1998; Li and Wang, 2009). Therefore, there might be novel Pm gene(s) to be explored in Th. ponticum.
SNTE20, a wheat-Th. ponticum partial amphiploid with powdery mildew resistance, was previously developed in our lab (He et al., 2013). In the present study, crosses between SNTE20 and common wheat were carried out, and consequently two Trititrigia introgression lines were generated, SN0293-2 and SN0293-7, both of which were resistant to powdery mildew. The two introgression lines were then characterized by combined methods of morphology, disease evaluation, sequential genomic in situ hybridization-fluorescence in situ hybridization (GISH-FISH), and molecular marker analyses.
Materials used in this study included Th. ponticum, rye, SNTE20, Yannong15 (YN15), Shannongfu63 (SNF63), Jimai22 (JM22), Shannong224 (SN224), SN0293-2, SN0293-7, CH7086, and Huixianhong (HXH). Th. ponticum (accession No. R431) was provided by Prof. Zhensheng Li, the former Northwest Institute of Botany, the Chinese Academy of Sciences, Yangling, China. Wheat-Th. ponticum octoploid SNTE20 was developed from the multiple cross Th. ponticum/YN15//SNF63. YN15, SNF63, and JM22 are wheat cultivars. SN224 is a T1BL·1RS translocation line with dwarf stems. SN0293-2 and SN0293-7 were generated from a hybrid of the cross SNTE20/YN15//SN224/3/JM22 (Supplementary Figure 1). CH7086 carrying Pm51 was provided by Prof. Xiaojun Zhang, the former Crop Science Institute, Shanxi Academy of Agricultural Sciences, Taiyuan, China.
Fifteen plants were grown in each 1.5 m long row, with 25 cm spacing between the rows. Agronomic traits, including plant height, spike length, spikelets per spike, kernels per spike, and thousand kernels weight, were recorded at the Experimental Station of Shandong Agricultural University. Each trait was averaged on 10 plants.
Seedling resistance assessment was performed in a growth chamber using the Bgt isolate E09, following the method described by Zhao et al. (2013). Seedlings were grown in rectangular plastic trays (5 cm × 5 cm; 10 plants per tray) and inoculated with fresh Bgt conidiospores obtained from the susceptible cultivar HXH at the one-leaf stage. After approximately 2 weeks, when symptoms were severe on HXH, infection types (ITs) on the plants were described using a 0–4 infection scale: 0–2 scores indicating resistance and 3–4 susceptibility.
At the adult stage, resistance to powdery mildew was evaluated after natural infection in field-grown plants at the Experimental Station of Shandong Agricultural University over three growing seasons (2018–2020). The most severe reaction type in a given year was considered to be the final resistance result. HXH was planted perpendicular and adjacent to the test rows to serve as an inoculum spreader and a susceptible control. The disease symptoms were recorded three times at weekly intervals after flowering, and the most severe infection score was used as the final response. The ITs of powdery mildew at the adult stage were scored using a 0–9 scale: 0–4 scores indicating resistance and 5–9 susceptibility (Li et al., 2011).
The chromosomes were prepared following the method described by Kato et al. (2004). The purified total genomic DNA extracted from Th. ponticum or rye was labeled with Texas Red-5-dCTP probes, with the sheared genomic DNA from YN15 as a blocker. GISH analysis was performed as described by Fu et al. (2012). For FISH analysis, oligonucleotide probes, including TAMRA (6-carboxytetramethylrhodamine)-labeled oligonucleotides pAs1-1, pAs1-3, pAs1-4, pAs1-6, AFA-3, and AFA-4 and FAM (6-carboxyfluorescein)-labeled oligonucleotides pSc119.2-1 and (GAA)10, were used. All probes were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). FISH analysis was performed as described by Huang et al. (2018). The chromosomes were counterstained with 4, 6-diamidino-2-phenylindole (DAPI), and the images were captured with a fluorescence microscope (Olympus BX60) equipped with a CCD (charge-coupled device) camera.
The Axiom Wheat 660K Genotyping Array was used to genotype SN0293-2 and their parents YN15, SNF63, JM22, SN224, SNTE20 and Th. ponticum. The SNP typing data were processed using Microsoft Excel 2016 software, regarding SNPs present in SN0293-2, SNTE20 and Th. ponticum and absent in the comment wheat parents as the putatively specific ones of Th. ponticum. Library construction and high-throughput sequencing of SN0293-2, SNF63, and SNTE20 were performed by Berry Genomics Company (Beijing, China). The raw reads were subjected to a quality check and then filtered by fastp to remove adapter sequences and low-quality bases (Chen et al., 2018). High-quality reads were mapped to the Triticeae repeat database mipsREdat 9.3p (PGSB Repeat Database) using Burrows-Wheeler Aligner in order to filter out repeat noises, non-mapped reads (from the genome region of non-repeat sequences) were fished by homemade scripts and then used for variant calling (Li and Durbin, 2010). GATK 3.8 (https://www.software.broadinstitute.org/gatk) was used to call out all the variations, including SNPs and InDels. Markers of cleaved amplified polymorphic sequences (CAPS) were designed on the basis of SNPs, and the corresponding restriction endonucleases were used for restriction digestion. InDel markers were designed according to the results of resequencing. Briefly, the corresponding 500-bp both upstream and downstream sequences of the InDels larger than 10 bp were fished from the Chinese Spring reference on the website WheatOmics (Ma et al., 2021). About 1-kb fragments corresponding the InDels were used to perform local BLAST to identify genome specific fragments for marker development (identity > 95%, number of hits ≤ 3). The conserved sequences flanking the target region were then used to design primers.
DNA was isolated from young leaf tissues following a standard CTAB method. The PCR cycling condition was as follows: 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, appropriate anneal temperature (50–60°C) for 40 s, 72°C for 1 min, and a final extension at 72°C for 5 min, and the amplification products were kept at 10°C using a Bio-Rad 9600 Thermal Cycler (Hercules, USA). PCR products amplified by CAPS markers were separated through 1% agarose electrophoresis and PCR products amplified by Indel markers were separated on 8% non-denatured polyacrylamide gels (39 acrylamide: 1 bisacrylamide).
Restriction endonuclease (HaeIII, SacII, BstEII) were selected for the CAPS marker and the digestion reaction was carried out according to the manufacturer's instructions (New England Biolabs, England). The digestion was conducted in a 10 μL reaction volume, contained 50 ng PCR products and digested at 37°C or 65°C (depending on the enzyme). The digestion reaction products were separated by 2% agarose electrophoresis.
At the seedling stage, SN0293-2, SN0293-7, and its parents were inoculated with the Bgt isolate E09, and the disease reaction was assessed once the control HXH displayed thoroughly susceptible symptoms. Th. ponticum, SNTE20, JM22, SN0293-2, and SN0293-7 were all immune to E09 (IT = 0), while the common wheat parents YN15, SNF63, and SN224 appeared to be susceptible with an IT score of 4 (Figure 1A), suggesting that the resistance to powdery mildew at the seeding stage in SN0293-2 and SN0293-7 originated from either Th. ponticum or JM22. The seedling reactions of SN0293-2, SN0293-7, and 42 wheat genotypes carrying documented Pm genes and gene combinations to 25 Bgt isolates were simultaneously recorded (Supplementary Table 1). The susceptible cultivar Chancellor exhibited a high IT (4). SN0293-2 and SN0293-7 were resistant to 23 isolates, indicating their broad-spectrum resistance. Additionally, Pm51, the only documented powdery mildew resistance gene putatively derived from Th. ponticum, was nearly immune to the isolate E20 (Zhan et al., 2014), but SN0293-2 and SN0293-7 were susceptible to E20, indicating that the resistance genes in SN0293-2 and SN0293-7 were different from Pm51.
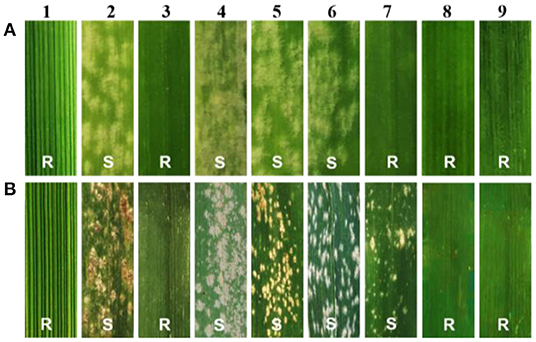
Figure 1. Evaluation of powdery mildew resistance. (A) The seedling stage. (B) The adult stage. 1–9 refer to Th. ponticum, HXH, SNTE20, SNF63, YN15, SN224, JM22, SN0293-2, and SN0293-7, respectively.
At the adult stage, resistance to powdery mildew was tested in field over three growing seasons (2018–2020), and the most severe reaction type observed in a given year was considered to be the final result. It was found that SN0293-2, SN0293-7, Th. ponticum, and SNTE20 were resistant to powdery mildew, whereas YN15, SNF63, SN224, JM22, and CH7086 showed susceptibility (Figure 1B and Supplementary Figure 2). Thus, the resistance at the adult stage in SN0293-2 and SN0293-7, temporarily designated PmSN0293, was putatively inherited from Th. ponticum and different from Pm51. Previous studies have found that JM22 contains powdery mildew resistance genes Pm2 and Pm52 (Cao et al., 2010; Qu et al., 2020). To determine whether SN0293-2 and SN0293-7 had these two genes or not, we amplified the genomes of them and their parents by using the markers Xcfd81 and Xicssl326, which were known to be linked with Pm2 and Pm52, respectively. It was showed that both SN0293-2 and SN0293-7 contained the two genes derived from JM22 (Figure 2). The results above indicated that SN0293-2 and SN0293-7 might carry a new powdery mildew resistance gene PmSN0293 from Th. ponticum pyramided with Pm2 and Pm52.
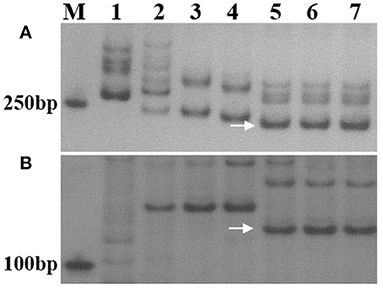
Figure 2. Amplified results of specific markers Xcfd-81 and Xicssl326. (A) Xcfd-81. (B) Xicssl326. Lanes: M, DL2000 marker; 1–7 refer to Th. ponticum, SNTE20, YN15, SN224, JM22, SN0293-2, and SN0293-7, respectively. Arrows indicate specific bands.
GISH, probed with the total genomic DNA of Th. ponticum and blocked with the genomic DNA of YN15, revealed that both SN0293-2 and SN0293-7 had 42 chromosomes and no alien signal was detected (Figures 3A,B). Because the parent SN224 is a T1BL·1RS translocation line, the rye genomic DNA was further used as a probe. Two chromosome arms with red coloration alongside the blue chromosome arms of wheat were found in SN0293-2 and no hybridization signal in SN0293-7, indicating the rye chromosome arm 1RS was present in SN0293-2 and absent in SN0293-7 (Figures 3C,D).
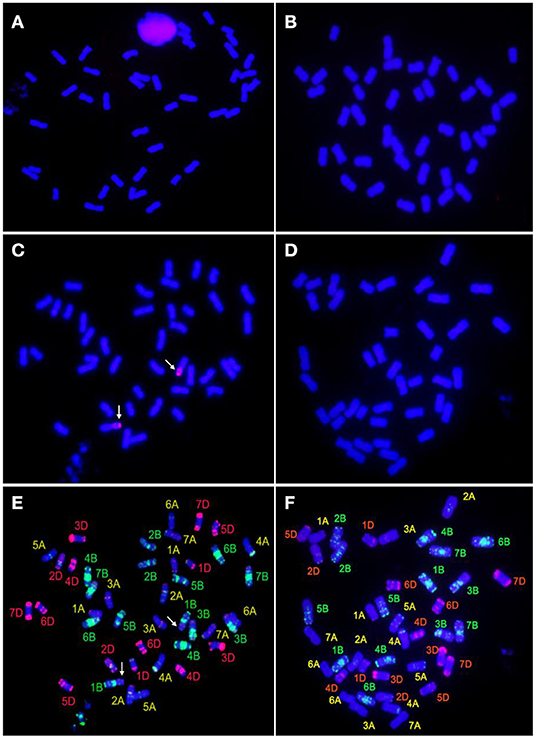
Figure 3. GISH-FISH analyses of SN0293-2 and SN0293-7. (A) GISH patterns of SN0293-2 probed with total genomic DNA of Thinopyrum ponticum. (B) GISH patterns of SN0293-7 probed with total genomic DNA of Th. ponticum. (C) GISH patterns of SN0293-2 probed with total genomic DNA of rye. (D) GISH patterns of SN0293-7 probed with total genomic DNA of rye. (E) FISH patterns of SN0293-2. (F) FISH patterns of SN0293-7. Arrows indicate the translocation T1BL·1RS.
After removing the GISH signals, the same slides were subjected to FISH analysis. The 1RSs in SN0293-2 were found in the form of T1BL·1RS, inheriting from the parent SN224 (Figure 3E). Further, FISH patterns of SN0293-2 and SN0293-7 in Figures 3E,F were compared with those of their parents (Figure 4). Differences between SN0293-2 and its parents were detected in the terminal region of 1DS, 2DS, 2DL, 6BL, and 7AL, and the middle of 3BL. As for SN0293-7, the terminal of 1DS, 2DL, 6BS, and 7AL as well as the middle of 1BL and 3BL appeared to be different from its parents. This suggested that chromosomes underwent structural variations with the formation of SN0293-2 and SN0293-7. In addition, differences between SN0293-2 and SN0293-7 were also present apart from the chromosome T1BL·1RS. For instance, red signals existed in the terminal regions of 1DS, 6BS, and 7AS in SN0293-2, but they disappeared in the corresponding regions of SN0293-7. The green signals in the telomere of SN0293-7 differed from those of SN0293-2.
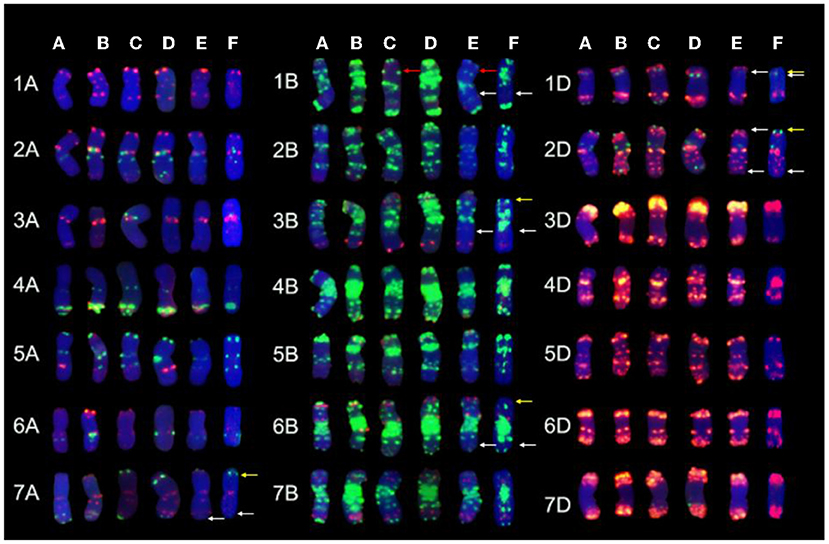
Figure 4. Chromosome comparison of FISH patterns among SN0293-2, SN0293-7, and their parents. (A) SNTE20. (B) YN15. (C) SN224. (D) JM22. (E) SN0293-2. (F) SN0293-7. The red, white, and yellow arrows indicate 1RS, FISH differences of SN0293-2 and SN0293-7, respectively.
A total of 157 specific SNP loci of Th. ponticum were obtained by using wheat 660K SNP array to analyze Th. ponticum and its parents, involving 9 chromosomes of three wheat subgenomes, of which 134 were located on chromosome 6A and mainly distributed in the physical interval of 60–110 Mb (Figure 5). DNA resequencing data showed that a total of 111 Indels larger than 10 bp were identified in the above 50 Mb region of chromosome 6A. These Indels were identical between the two lines SN0293 and SNTE20 containing Th. ponticum fragments, but different from the common wheat line SNF63. According to the results of Wheat 660K SNP array and resequencing data, three CAPS markers (CAPS421-HaeIII, CAPS761-SacI, CAPS564-BstEII) and four Indel markers (Indel192, Indel101, Indel752, Indel753) were obtained (Table 1). Th. ponticum specific bands were amplified in SN0293-2 and SN0293-7 (Figure 6), in which CAPS421-HaeIII was located on wheat chromosome 2B, CAPS761-SacI, CAPS564-Bst II, Indel192, Indel101, Indel752, and Indel753 were located on chromosome 6A. The results showed that SN0293-2 and SN0293-7 inherited genetic components from Th. ponticum, so they were wheat-Th. ponticum introgression lines.
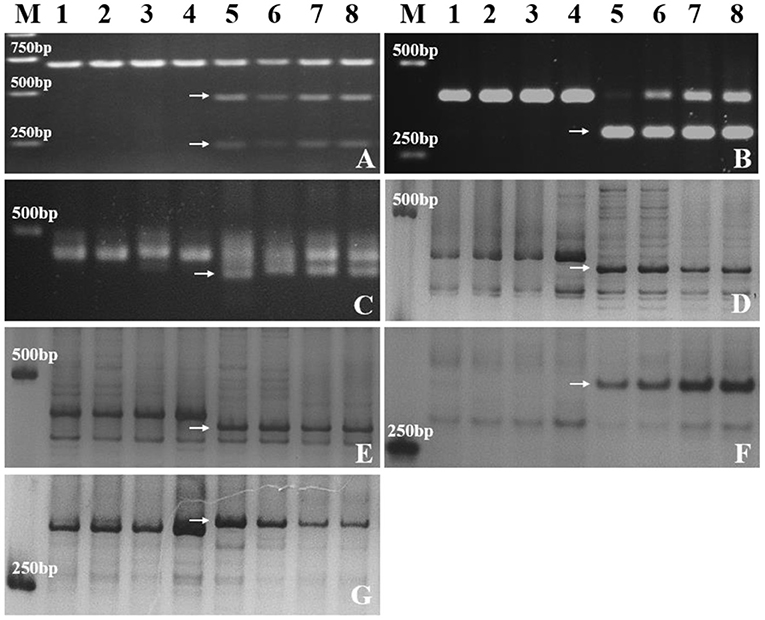
Figure 6. Molecular markers analysis of SN0293-2, SN0293-7 and their parents. (A) CAPS421-HaeIII. (B) CAPS761-SacI. (C) CAPS564-BstEII. (D) Indel192. (E) Indel101. (F) Indel752. (G) Indel753. Lanes: M, DL2000 marker; 1–8 refer to SNF63, YN15, SN224, JM22, Th. ponticum, SNTE20, SN0293-2, and SN0293-7, respectively. Arrows indicate specific bands of Th. ponticum.
The agronomic traits of SN0293-2 and SN0293-7 were compared with those of their parents (Figure 7). SN0293-2 and SN0293-7 displayed average plant heights of 65.4 ± 3.1 cm and 69.3 ± 4.2 cm, respectively, which were lower than the parents SNTE20 (105.2 ± 3.9 cm), YN15 (84.0 ± 2.4 cm) and JM22 (83.5 ± 2.7 cm) except SN224 (61.0 ± 3.6 cm). They shared about equal numbers of spike length and spikelets as the common wheat parents. However, SN0293-2 and SN0293-7 produced more kernels per spike (56 ± 5 and 67 ± 3, respectively) and higher 1,000-grain weight (53.8 ± 1.7 and 53.0 ± 1.3, respectively). These agronomic data indicated that the two introgression lines probably carried genes that were beneficial for wheat breeding.

Figure 7. Plants, spikes, and kernels of SN0293-2, SN0293-7, and their parents. (A) Plants. (B) Spikes. (C) Kernels. 1–6 refer to SNTE20, YN15, SN224, JM22, SN0293-2, and SN0293-7, respectively. Scale bar = 2 cm.
Wild relatives of wheat have been serving as valuable gene reservoirs due to their resistance to many biotic and abiotic stresses. In the past few decades, wide crosses have been employed to incorporate useful genes into common wheat. The tall grass Th. ponticum was extensively used in wheat genetic improvement. The elite cultivars, such as Xiaoyan 6, Gaoyou 503, Xiaoyan 60, and Xiaoyan 81, were widely planted in China and had played an important role in wheat production (Li et al., 2015; Luo et al., 2021; Yang et al., 2022). Meanwhile, numerous chromosome engineering materials were generated, including partial amphiploids (Zheng et al., 2015), additions (Li et al., 2016), substitutions (Wang et al., 2019; Li et al., 2021), translocations (Yang et al., 2022) and introgression lines (Zhan et al., 2014).
In the present study, Th. ponticum, SNTE20 and wheat-Th. ponticum introgression lines SN0293-2 and SN0293-7 displayed excellent resistance to powdery mildew. Up to now, 11 genes have been officially designated from Th. ponticum (Li and Wang, 2009; Liu et al., 2020). Among them, only one gene named Pm51 was responsible for resistance to powdery mildew. At the seedling stage, the two introgression lines were susceptible to the Bgt isolate E20 (Supplementary Table 1), while Pm51 was nearly immune (Zhan et al., 2014). As for the Bgt isolate E09, common wheat parents SNF63, YN15, and SN224 showed susceptibility but JM22 displayed immunity because JM22 carried two resistance genes Pm2 and Pm52 (Cao et al., 2010; Qu et al., 2020). Molecular marker analyses demonstrated that both SN0293-2 and SN0293-7 carried these two powdery mildew resistance genes as well. At the adult plant stage, JM22 and CH7086 exhibited susceptibility to powdery mildew similarly to the other common wheat parents (Figure 1 and Supplementary Figure 2), suggesting that Pm2, Pm51, and Pm52 lost resistance in Tai'an, Shandong Province, China. However, resistance of the two introgression lines remained as well as Th. ponticum and SNTE20. Seen from the results above, the seedling resistance of SN0293-2 and SN0293-7 originated from either Th. ponticum or JM22, and the adult plant resistance of them was putatively derived from Th. ponticum obviously different from Pm51. Segregating populations are being developed for mapping the adult plant resistance. In addition, 1RS, herein, had no resistance to powdery mildew since the T1BL·1RS translocation SN224 exhibited consistent susceptibility at both the seeding stage and adult plant stage.
GISH and FISH have been used as powerful tools to detect alien chromosomes or chromosome segments and chromosomal structural variations in distant hybridization. In this study, one pair of T1BL·1RS was confirmed by GISH-FISH in SN0293-2, but no cytological evidence for Th. ponticum was found in SN0293-2 and SN0293-7, even though some structural variations were demonstrated between these two lines and their parents. The alien segments must be too small to be detected for cytogenetic tools due to the accuracy limitation. Recently, the wheat SNP array has been employed to identify wild relatives and their derivatives with wheat. Zhou et al. (2018) constructed the genetic linkage map of Agropyron cristatum and characterized a number of wheat-A. cristatum chromosome lines. Several wheat-Th. ponticum translocation and introgression lines were also subjected to Wheat 660K SNP array and deletion events were detected (Li et al., 2019; Yang et al., 2021, 2022). In order to determine whether SN0293-2 and SN0293-7 inherited genetic components from Th. ponticum or not, the Axiom Wheat 660K Genotyping Array were also used in the present study. One hundred and fifty-seven SNPs were found to be specific to Th. ponticum. These SNPs referred to 9 chromosomes of wheat genomes, 134 of which were located on the wheat chromosome 6A (Figure 5). Then three CAPs markers were developed and showed specificity to Th. ponticum in SN0293-2 and SN0293-7. Similarly, four Indel markers belonging to homoeologous group 6 were obtained according to the re-sequence data (Table 1 and Figure 6). It was indicated that the wheat SNP array has higher accuracy than cytological methods and could effectively work in identification of wheat-Th. ponticum introgression lines.
Most of the alien chromosome lines experience more or less penalties of agronomic traits because of linkage drags of the whole alien chromosomes or large alien segments they carry. For instance, many addition lines generally have longer growth duration and smaller grains due to the introduced alien chromosomes. However, introgression lines have small alien segments that GISH cannot detect, generally resulting in no or less genetic drag. Therefore, germplasms of this type with target genes usually exhibit positive agronomic traits and are considered to be the most valuable materials for wheat breeding. In order to breed disease-resistant lines with favorable agronomic traits, SN224 and JM22 were used as male parents. The former is a breeding line with dwarf stems developed in our lab. The latter is an elite cultivar with the largest promotion area in the last decade in China (Jia et al., 2020). JM22 was also employed as the last parent to improve agronomic traits and to provide two Pm genes. Consequently, the wheat-Th. ponticum introgression lines SN0293-2 and SN0293-7 carried a new resistance gene putatively from Th. ponticum and pyramided Pm2 and Pm52 derived from JM22. They also showed positive agronomic traits, such as more kernels and higher 1,000-weight, enhancing the application potential in wheat breeding programs.
Two wheat-Th. ponticum introgression lines were developed and named SN0293-2 and SN0293-7, respectively. Besides Pm2 and Pm52, they possessed a new powdery mildew resistance gene PmSN0293 putatively from Th. ponticum obviously different from Pm51 reported previously. Seven markers were obtained and confirmed to be specific to Th. ponticum. Superior resistance to powdery mildew at both the seedling and adult plant stages and positive agronomic traits give the two introgression lines great potential to be used in wheat breeding programs.
The data presented in the study are deposited in the Genome Sequence Archive in BIG Data Center repository (https://bigd.big.ac.cn/), accession number PRJCA009783
YB designed the research. ML and YY performed the experiments. FN and XL analyzed the data and developed the specific markers. HW contributed to the development of the materials. ML and YB wrote the manuscript. All authors approved the final version of the manuscript.
This project was supported by the National Natural Science Foundation of China (No. 32071998), the National Key Research and Development Program of China (No. 2016YFD0102004-02) and Key R&D Program of Shandong Province (Major Science and Technology Innovation Project) (2021LZGC009).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We sincerely thank Prof. Yilin Zhou at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences, for providing powdery mildew assessment at the seedling stage.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.943669/full#supplementary-material
Braun, H. J., Atlin, G., and Payne, T. (2010). “Multi-location testing as a tool to identify plant response to global climate change,” in Climate Change and Crop Production, ed M. P. Reynolds (CABI Climate Change Series), Oxfordshire: CABI, 115–138. doi: 10.1079/9781845936334.0115
Cao, X., Zhou, Y., Duan, X., Song, Y., He, W., Ding, K., et al. (2010). Postulation of wheat powdery mildew resistance genes in 101 wheat cultivars (lines) from major wheat regions in China. J. Triticeae Crops 30, 948–953. doi: 10.7606/j.issn.1009-1041.2010.05.030
Chen, Q., Conner, R. L., Laroche, A., and Thomas, J. B. (1998). Genome analysis of Thinopyrum intermedium and Th. ponticum using genomic in situ hybridization. Genome 41, 580–586. doi: 10.1139/g98-055
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Fu, S., Lv, Z., Qi, B., Guo, X., Li, J., Liu, B., et al. (2012). Molecular cytogenetic characterization of wheat-Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium Head Blight. J. Genet. Genomics 39, 103–110. doi: 10.1016/j.jgg.2011.11.008
Gao, H., Xu, X., Ai, P., Luo, F., Guo, P., and Ma, P. (2022). Identification of the powdery mildew resistance in Chinese wheat cultivar Heng 4568 and its evaluation in marker-assisted selection. Front. Genet. 13, 819844. doi: 10.3389/fgene.2022.819844
Hart, G. E., Mcmillin, D. E., and Sears, E. R. (1976). Determination of the chromosomal location of a glutamate oxaloacetate transaminase structural gene using Triticum-Agropyron translocations. Genetics 83, 49–61. doi: 10.1093/genetics/83.1.49
Hawkesford, M. J., Araus, J. L., Park, R., Calderini, D., Miralles, D., Shen, T. M., et al. (2013). Prospects of doubling global wheat yields. Food Energy. Secur. 2, 34–48. doi: 10.1002/fes3.15
He, F., Xu, J., Qi, X., Bao, Y., Li, X., Zhao, F., et al. (2013). Molecular cytogenetic characterization of two partial wheat Elytrigia elongate amphilploids resistant to powdery mildew. Plant Breed 132, 553–557. doi: 10.1111/pbr.12104
He, H., Liu, R., Ma, P., Du, H., Zhang, H., Wu, Q., et al. (2021). Characterization of Pm68, a new powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
He, H., Zhu, S., Zhao, R., Jiang, Z., Ji, Y., Ji, J., et al. (2018). Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 11, 879–882. doi: 10.1016/j.molp.2018.03.004
Hou, L., Jia, J., Zhang, X., Li, X., Yang, Z., Ma, J., et al. (2016). Molecular mapping of the stripe rust resistance gene Yr69 on wheat chromosome 2AS. Plant Dis.100, 1717–1724. doi: 10.1094/PDIS-05-15-0555-RE
Huang, X., Zhu, M., Zhuang, L., Zhang, S., Wang, J., Chen, X., et al. (2018). Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor. Appl. Genet. 131, 1967–1986. doi: 10.1007/s00122-018-3126-2
Jia, M., Xu, H., Liu, C., Mao, R., Li, H., Liu, J., et al. (2020). Characterization of the powdery mildew resistance gene in the elite wheat cultivar Jimai 23 and its application in marker-assisted selection. Front. Genet. 11, 241. doi: 10.3389/fgene.2020.00241
Kato, A., Lamb, J. C., and Birchler, J. A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. U.S.A. 101, 13554–13559. doi: 10.1073/pnas.0403659101
Li, H., Boshoff, W., Pretorius, Z. A., Zheng, Q., Li, B., and Li, Z. (2019). Establishment of wheat-Thinopyrum ponticum translocation lines with resistance to Puccinia graminis f. sp. tritici Ug99. J. Genet. Genomics 46, 405-407. doi: 10.1016/j.jgg.2019.07.005
Li, H., and Durbin, R. (2010). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., and Wang, X. (2009). Thinopyrum ponticum and Th. intermedium: the promising source of resistance to fungal and viral diseases of wheat. J. Genet. Genomics 36, 557–565. doi: 10.1016/S1673-8527(08)60147-2
Li, H., Zheng, Q., Pretorius, Z. A., Li, B., Tang, D., and Li, Z. (2016). Establishment and characterization of new wheat-Thinopyrum ponticum addition and translocation lines with resistance to Ug99 races. J. Genet. Genomics 43, 573–575. doi: 10.1016/j.jgg.2016.07.004
Li, H. J., Wang, X. M., Song, F. J., Wu, C. P., Wu, X. F., Zhang, N., et al. (2011). Response to powdery mildew and detection of resistance genes in wheat cultivars from China. Acta Agron. Sin. 37, 943–954. doi: 10.3724/SP.J.1006.2011.00943
Li, M., Wang, Y., Liu, X., Li, X., Wang, H., and Bao, Y. (2021). Molecular cytogenetic identification of a novel wheat-Thinopyrum ponticum 1JS (1B) substitution line resistant to powdery mildew and leaf rust. Front. Plant. Sci. 12, 727734. doi: 10.3389/fpls.2021.727734
Li, Z. S., Li, B., Zheng, Q., and Li, H. (2015). “Review and new progress in wheat wide hybridization for improving the resistance to biotic and abiotic stresses,” in Advances in Wheat Genetics: From Genome to Field, eds Y. Ogihara, S. Takumi, H. Handa, et al. (Tokyo: Springer), 377–385. doi: 10.1007/978-4-431-55675-6_43
Liu, C., Han, R., Wang, X. L., Gong, W. P., Cheng, D. G., Cao, X. Y., et al. (2020). Research progress of wheat wild hybridization, disease resistance genes transfer and utilization. Sci. Agric. Sin. 53, 1287–1308. doi: 10.3864/j.issn.0578-1752.2020.07.001
Liu, W., Koo, D. H., Xia, Q., Li, C., Bai, F., Song, Y., et al. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat. Theor. Appl. Genet. 130, 841–848. doi: 10.1007/s00122-017-2855-y
Luo, Q., Zheng, Q., Hu, P., Yang, G., Li, H., Li, B., et al. (2021). Mapping QTL for agronomic traits under two levels of salt stress in a new constructed RIL wheat population. Theor. Appl. Genet. 134, 171–189. doi: 10.1007/s00122-020-03689-8
Ma, S., Wang, M., Wu, J., Guo, W., Chen, Y., Li, G., et al. (2021). WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14:1965–1968. doi: 10.1016/j.molp.2021.10.006
Mago, R., Bariana, H. S., Dundas, I. S., Spielmeyer, W., Lawrence, G. J., Pryor, A. J., et al. (2005). Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor. Appl. Genet. 111, 496–504. doi: 10.1007/s00122-005-2039-z
Niu, Z., Klindworth, D. L., Yu, G. L., Friesen, T., Chao, S., et al. (2014). Development and characterization of wheat lines carrying stem rust resistance gene Sr43 derived from Thinopyrum ponticum. Theor. Appl. Genet. 127, 969–980. doi: 10.1007/s00122-014-2272-4
Procunier, J. D., Townley-Smith, T. F., Fox, S., Prashar, S., Gray, M., Kim, W. K., et al. (1995). PCR-based RAPD/DGGE markers linked to leaf rust resistance genes Lr29 and Lr25 in wheat (Triticum aestivum L.). J. Genet. Breed. 49, 92–97.
Qu, Y., Wu, P., Hu, J., Chen, Y., Shi, Z., Qiu, D., et al. (2020). Molecular detection of the powdery mildew resistance genes in winter wheats DH51302 and Shimai 26. J. Integr. Agric 19, 931–940. doi: 10.1016/S2095-3119(19)62644-4
Ren, T., Tang, Z., Fu, S., Yan, B., Tan, F., Ren, Z., et al. (2017). Molecular cytogenetic characterization of novel wheat-rye T1RS∙1BL translocation lines with high resistance to diseases and great agronomic traits. Front. Plant. Sci. 8, 799. doi: 10.3389/fpls.2017.00799
Sarma, D., and Knott, D. R. (1966). The transfer of leaf-rust resistance from Agropyron to Triticum by irradiation. Genome 8, 137–143. doi: 10.1139/g66-018
Wang, H., Sun, S., Ge, W., Zhao, L., Hou, B., Wang, K., et al. (2020). Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368, a5435. doi: 10.1126/science.aba5435
Wang, S., Wang, C., Wang, Y., Wang, Y., Chen, C., and Ji, W. (2019). Molecular cytogenetic identification of two wheat-Thinopyrum ponticum substitution lines conferring stripe rust resistance. Mol. Breed. 39, 143. doi: 10.1007/s11032-019-1053-9
Wang, Z., Li, L., He, Z., Duan, X., Zhou, Y., Chen, X., et al. (2005). Seedling and adult plant resistance to powdery mildew in Chinese bread wheat cultivars and lines. Plant Dis. 89, 457–463. doi: 10.1094/PD-89-0457
Whelan, E. D. P., and Lukow, O. M. (1990). The genetics and gliadin protein characteristics of a wheat-alien translocation that confers resistance to colonization by the wheat curl mite. Genome 33, 400–404. doi: 10.1139/g90-061
Xing, L., Hu, P., Liu, J., Witek, K., Zhou, S., Xu, J., et al. (2018). Pm21 from Haynaldia villosa encodes a CC-NBS-LRR protein conferring powdery mildew resistance in wheat. Mol. Plant 11, 874–878. doi: 10.1016/j.molp.2018.02.013
Yang, G., Boshoff, W. H. P., Li, H., Pretorius, Z. A., Luo, Q., Li, B., et al. (2021). Chromosomal composition analysis and molecular marker development for the novel Ug99-resistant wheat-Thinopyrum ponticum translocation line WTT34. Theor. Appl. Genet. 134, 1587–1599. doi: 10.1007/s00122-021-03796-0
Yang, G., Tong, C., Li, H., Li, B., Li, Z., and Zheng, Q. (2022). Cytogenetic identification and molecular marker development of a novel wheat-Thinopyrum ponticum translocation line with powdery mildew resistance. Theor. Appl. Genet. doi: 10.1007/s00122-022-04092-1
Zhan, H., Li, G., Zhang, X., Li, X., Guo, H., Gong, W., et al. (2014). Chromosomal location and comparative genomics analysis of powdery mildew resistance gene Pm51 in a putative wheat-Thinopyrum ponticum introgression line. PLoS ONE 9, e113455. doi: 10.1371/journal.pone.0113455
Zhang, R., Xiong, C., Mu, H., Yao, R., Meng, X., Kong, L., et al. (2020). Pm67, a new powdery mildew resistance gene transferred from Dasypyrum villosum chromosome 1V to common wheat (Triticum aestivum L.). Crop J. 9, 882–888. doi: 10.1016/j.cj.2020.09.012
Zhao, Z., Sun, H., Song, W., Lu, M., Huang, J., Wu, L., et al. (2013). Genetic analysis and detection of the gene MlLX99 on chromosome 2BL conferring resistance to powdery mildew in the wheat cultivar Liangxing 99. Theor. Appl. Genet. 126, 3081–3089. doi: 10.1007/s00122-013-2194-6
Zheng, Q., Luo, Q., Niu, Z., Li, H., Li, B., Xu, S. S., et al. (2015). Variation in chromosome constitution of the Xiaoyan series partial amphiploids and its relationship to stripe rust and stem rust resistance. J. Genet. Genomics 42, 657–660. doi: 10.1016/j.jgg.2015.08.004
Keywords: wheat, powdery mildew, Thinopyrum ponticum, introgression, GISH, FISH
Citation: Li M, Yuan Y, Ni F, Li X, Wang H and Bao Y (2022) Characterization of Two Wheat-Thinopyrum ponticum Introgression Lines With Pyramiding Resistance to Powdery Mildew. Front. Plant Sci. 13:943669. doi: 10.3389/fpls.2022.943669
Received: 14 May 2022; Accepted: 02 June 2022;
Published: 15 July 2022.
Edited by:
Pengtao Ma, Yantai University, ChinaReviewed by:
Huagang He, Jiangsu University, ChinaCopyright © 2022 Li, Yuan, Ni, Li, Wang and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinguang Bao, YmFveWluZ3VhbmdAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.